Abstract
The transcriptional coactivator paralogs p300 and CBP contain acetyltransferase domains (HAT) and catalyze the lysine acetylation of histones and other proteins as an important aspect of their functions. Prior studies revealed that the basic leucine zipper domain (b-ZIP) of transcription factor ATF-2 (also called CRE-BP1) can interact with the CBP HAT domain. In this study, we have examined the ATF-2 b-ZIP interaction with the p300 HAT domain and shown that p300 HAT autoacetylation can enhance the binding affinity. Pull-down assays revealed that hyperacetylated p300 HAT is more efficiently retained by immobilized ATF-2 b-ZIP than hypoacetylated p300 HAT. Loop deleted p300 HAT lacking autoacetylation was retained about as well as hyperacetylated p300 HAT, suggesting that the loop and ATF-2 compete for p300 HAT binding. While, ATF-2 b-ZIP is a weak inhibitor of hypoacetylated p300 HAT acetylation of a histone H4 peptide, hyperacetylated p300 HAT is much more potently inhibited by ATF-2 b-ZIP. Moreover, we showed that ATF-2 b-ZIP could serve as an acetyltransferase substrate for p300 HAT. Using mass spectrometry, two p300 HAT lysine acetylation sites were mapped in ATF-2 b-ZIP. Immunoprecipitation-western blot analysis with anti-acetyl-lysine antibody revealed that ATF-2 can undergo reversible acetylation in vivo. Mutational analysis of the two ATF-2 b-ZIP acetylation sites revealed their potential contributions to ATF-2-mediated transcriptional activation. Taken together, these studies suggest multiple roles for protein acetylation in the regulation of transcription by p300/CBP and ATF-2.
Keywords: HAT, histone acetylatransferase; ATF-2, activating transcription factor-2; b-ZIP, basic leucine zipper domain; HDACs, histone deacetylases; TSA, trichostatin; PMA, Phorbol myristate; LPS, lipopolysacharrides; PCAF, p300/CBP associated factor; CRE, cyclic AMP response element; TNF, tumor necrosis factor; CBP, CREB binding protein; CREB, cyclic AMP binding protein; VMA intein, comes from sacchromyces cerevisiae nuclear gene called SceVMA); MESNA, Sodium 2-mercapto ethanesulfonate, KK/RR, Double arginine mutant at K357, K374; KK/AA, Double alanine mutant at K357, K374; KK/QQ, Double glutamine mutant at K357, K374; AEBSF, aminoethyl-benzene sulfonyl fluoride
Protein acetylation by histone acetyltransferases (HATs) is critical to the regulation of gene expression and cell signaling (1-3). Among the characterized HATs, the transcriptional coactivator paralogs p300 and CBP have received special attention for their broad and important contributions to cell growth and differentiation (4-9). The congenital Rubenstein-Taybi syndrome resulting from heterozygous loss of CBP results in a variety of developmental abnormalities including predisposition to cancer (9). Both CBP and p300 show distinct and overlapping functions in gene regulation and a wide variety of proteins are known to interact with these proteins (4-9). In particular, the KIX and C/H3 domains of p300/CBP have been found to recruit a range of transcription factors and other proteins including p53 (10), p73 (11), STAT1 (12), NFκB (13,14), MyoD (15,16), BRCA1 (17), c-Fos, CREB (18), c-Jun, and c-Myb (19,20) (Figure 1B). In general, these transcription factor interactions are thought to help localize p300/CBP to the requisite promoters on chromatin where p300/CBP is thought to catalyze targeted histone acetylation (4-9, 21,22). However, for several of these and other proteins, p300/CBP is thought to acetylate the transcription factors themselves on one or more lysines, modulating their functions (10-14). The p300 HAT domain has been targeted by small molecule inhibitors for potential therapeutic applications (23,24).
Fig 1.
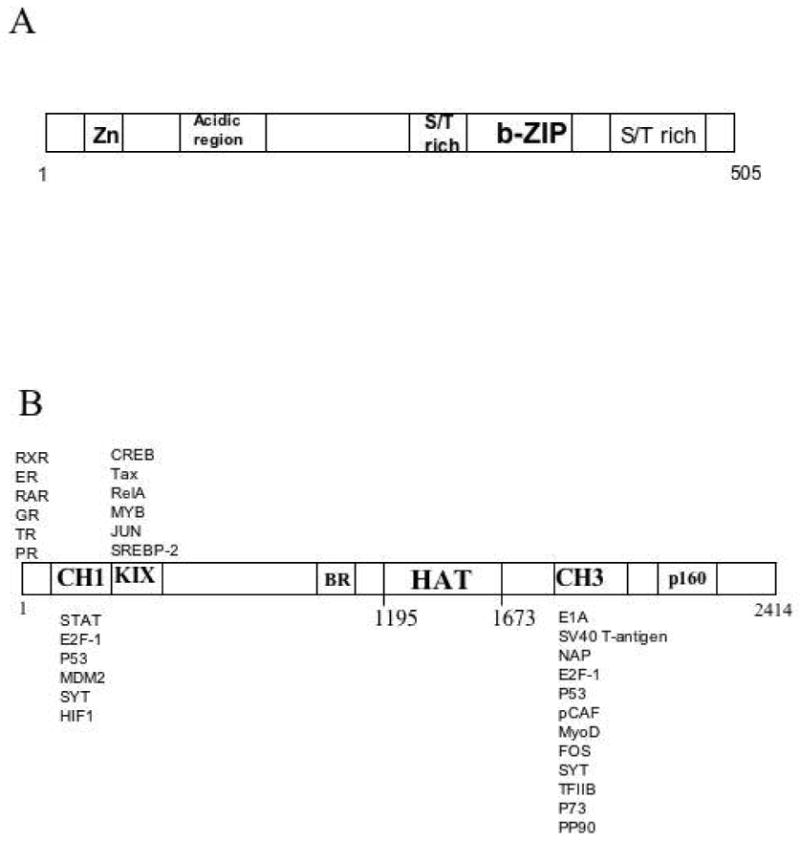
ATF-2 and p300 architecture. A) Domain structure of ATF-2. B) Schematic representation of p300 domain structure with selected interacting proteins.
The transcription factor ATF-2 has been shown previously to bind to CBP in vivo and in vitro (4,25). ATF-2 is a member of the ATF/CREB family and contains a C-terminal b-ZIP DNA binding domain and an N-terminal activation domain regulated by stress activated protein kinases (Figure 1) (26,27,28). ATF-2 can form homodimers and heterodimers with c-Jun and bind to the cAMP response element (CRE) in DNA (29-31). Like CBP, ATF-2 is a key developmental gene as knockout mice die shortly after birth due to neonatal respiratory distress (32). Domain mapping studies showed that ATF-2 associates with CBP via the CBP HAT domain and the ATF-2 b-ZIP domain (4). However, detailed biochemical analysis of this interaction with purified p300/CBP and ATF-2 protein domains has not yet been carried out.
It has been shown previously that the p300 HAT domain can be extensively autoacetylated, with a high proportion of acetylation sites located in a proteolytically sensitive loop (33, 34). Loop autoacetylation of p300 is associated with catalytic activation, somewhat analogous to the regulation of many protein kinases by autophosphorylation (33). Since generation of p300 HAT by standard recombinant expression produces protein containing ∼10 acetylated Lys residues, an alternative method, expressed protein ligation (35, 36), was employed (33) which affords p300 HAT in hypoacetylated form (<3 acetylation events). In this approach, p300 HAT domain is produced by semisynthesis in which catalytically impaired recombinant p300 C-terminal thioester fragment aa 1284-1652 is ligated to N-Cys containing synthetic peptide (aa 1653-1666) (33). Hyperacetylated p300 HAT can be generated in vitro simply by incubating with acetyl-CoA.
In this study, we describe a biochemical analysis of the binding of p300 HAT domain with the ATF-2 b-ZIP domain. We show that this interaction can be modulated by p300 autoacetylation. Moreover, p300 HAT can catalyze the acetylation of ATF-2 b-ZIP and two Lys acetylation sites have been mapped. ATF-2 is shown to be acetylated in vivo and mutation of the b-ZIP acetylation sites can modulate the transcriptional activation properties of ATF-2. Taken together, these experiments reveal multiple roles for protein acetylation in the p300/CBP-ATF-2 gene regulatory pathway.
Experimental Procedures
General Reagents
Ampicillin, BSA, acetylated bovine serum albumin, dithiothreitol, EDTA, glutathione, Protein A and Protein G sepharose, HEPES, glutathione agarose were purchased from Sigma. N-α-Fmoc-amino acids were purchased from Novabiochem (San Diego); unlabeled acetyl-CoA, 14C-labelled acetyl-CoA, and 14C-labeled acetylated bovine serum albumin were purchased from Amersham Biosciences. Oligonucleotide primers were obtained from Integrated DNA Technologies (Coralville, Iowa). Phenol, trifluoroacetic acid, and IPTG were obtained from Fisher Scientific. Quickchange mutagenesis and the sequences were confirmed by DNA sequencing. Anti-ATF-2 and anti-acetyl-lysine antibodies were purchased from Santa Cruz Biotechnology, Upstate Biotechnology and Cell signaling Technologies. Dual Glow luciferase kit was obtained from Promega Corporation. Glutathione S-transferase was expressed and purified from a standard GST expression plasmid using glutathione agarose.
Peptide synthesis
p300 C-terminal peptide CMLVELHTQSQDRF for expressed protein ligation and H4-15 peptide Ac-GRGKGGKGLGKGGAK for acetyltransferase assays were prepared using the Fmoc strategy. Peptides were purified (>95% homogeneity) by reversed phase (C-18) high performance liquid chromatography as described previously using a gradient of water-acetontitrile (0.05% trifluoroacetic acid). Electrospray mass spectrometry of peptide confirmed the correct structures.
Preparation of Wt p300 HAT and p300 HAT-ΔLoop
These semisynthetic proteins were prepared and purified following previously described procedures (33). Briefly, pTYB2 expression plasmids encoding p300 HAT protein (aa 1287-1652) or (aa 1287-1652 with an internal deletion of aa 1523-1554) containing an M1652G mutation fused to VMA intein-chitin binding domain was grown in E. coli BL21(DE3)-RIL cells to A600 of 0.45 at which point the incubator temperature was reduced to 16°C and media allowed to cool. After 15 min, protein expression was induced by addition of IPTG to a final concentration of 0.5 mM. Cells (1 L) were then grown for 16 h at 16°C, harvested by centrifugation, resuspended in intein lysis buffer (25 mM HEPES (pH 7.9), 500 mM NaCl, 10% glycerol, 1 mM MgSO4, and 2 mM PMSF) and lysed by two passages through a French press cell. The lysate was cleared by centrifugation and applied to a 12 ml chitin column after extensive washing. Excess buffer was drained and this immobilized fusion protein was treated with 200 mM MESNA to generate the thioester and ligated to 10 mg synthetic peptide aa 1653-1666 (CMLVELHTQSQDRF) over 16 h at room temperature. Fractions containing semisynthetic p300 HAT were pooled and concentrated before being applied to a Mono-S HR5/5 (Amersham Biosciences) strong cation exchange column for further purification. Fractions containing purified protein (>90%), as determined by SDS-PAGE analysis, were pooled and concentrated to ∼5 mg/ml as measured by Bradford assay. Following concentration, 5% glycerol was added before flash freezing in liquid N2 and samples were stored at -80°C. Semisynthetic proteins showed the correct molecular weights as determined by MALDI (matrix-assisted laser ddesorption/ionization) TOF (time-of-flight) mass spectrometry.
Purification of GST-ATF-2-b-ZIP
pGEX-4T-3 plasmid encoding basic leucine zipper domain (aa 349-415) was grown in E.coli BL21 (DE3)-RIL cells to A600 of 0.45 at which point the incubator temperature was reduced to 16°C and media allowed to cool. After 15 min, protein expression was induced by addition of IPTG to a final concentration of 1.0 mM. Cells (1 L) were then grown for 16 h at 16°C, harvested by centrifugation, re-suspended in lysis buffer (20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1.0% NP-40, 10% glycerol, 5 mM EDTA, 5 mM DTT and 2 mM PMSF) and lysed by two passages through a French press cell. The lysate was cleared by centrifugation and applied to a 10 ml glutathione agarose column. The GST beads were eluted extensively (>5 column volumes) with wash buffer (20 mM Tris-HCl (pH 8.0), 300 mM NaCl, 1.0% NP-40, 10% glycerol, 5 mM EDTA, 5 mM DTT and 2 mM PMSF). The protein was eluted with elution buffer (20 mM Tris-HCl (pH 8.0), 10 mM reduced glutathione, 5 mM DTT and 2 mM PMSF), fractions were analyzed by 10% (w/v) SDS-PAGE, and fractions containing recombinant GST-ATF-2-b-ZIP (>90% purified) were pooled and dialyzed to remove glutathione and concentrated to 2 mg/mL. GST-ATF-2-b-ZIP was stored in 10% glycerol, 20 mM Tris, pH 7.4, and 1 mM DTT at -80°C.
Preparation of hyperacetylated p300
Semisynthetic hypoacetylated p300 HAT domain (10 μM) was incubated with acetyl-CoA (125 μM) in reaction buffer (50 mM HEPES pH 7.9, 0.1 mM EDTA, 1 mM DTT and 50 μg/ml bovine serum albumin) for 1 h at 30°C (33, 34). For comparative analysis of binding studies and acetyltransferase assays (see below), hypoacetylated p300-ΔLoop (10 μM) and p300 HAT (10 μM) were incubated with desulfo-CoA (125μM) in reaction buffer above in the absence of acetyl-CoA.
GST-ATF-2-b-ZIP pull down assays
GST-ATF-2-b-ZIP (1 mg/ml) immobilized on glutathione agarose resin in 16 μl incubation buffer (20 mM HEPES pH 7.9, 5 mM DTT, 1 mM EDTA) was incubated with wt hyper- or hypoacetylated p300 HAT domain or hypoacetylated p300 HAT-ΔLoop in binding buffer (50 mM HEPES pH 7.9, 0.1 mM EDTA, 1 mM DTT and 50 μg/ml bovine serum albumin, 30 μL volumes) at 16°C for 20 min. The resultant samples were centrifuged at 10,000g for 1 min and supernatants were collected. The pellets were washed twice with 0.1 ml of wash buffer (20 mM HEPES pH 7.9, 5 mM DTT, 1 mM EDTA, 50 mM NaCl, 50 μM CoASH). Following washing, the samples were treated separately with 5×SDS gel loading buffer and analyzed using 10% SDSPAGE. The dried gels were scanned and analyzed by Scion Image analysis (Scion Corporation).
Acetyltransferase assays
For the peptide H4-15 substrate, H4-15 (100 μM) was incubated with or without GST or GST-ATF-2-b-ZIP, in the presence of p300 HAT domain (40 nM) in hyperacetylated or hypoacetylated state in assay buffer (50 mM HEPES, pH 7.9, 0.1 mM EDTA, 1 mM DTT and 50 μg/ml bovine serum albumin, 30 μL) for 10 min at 30°C prior to initiation of reaction by the addition of 14C-acetyl-CoA (20 μM unless otherwise specified) for 0-1 min whereupon the reaction was quenched with 6 μL of 5X SDS gel loading buffer and analyzed using 16% Tris-Tricine SDSPAGE (34, 35). For acetylation of GST-ATF-2 b-ZIP, up to 5 μM protein substrate was incubated with 40 nM of p300 HAT domain in presence of 14C-acetyl-CoA (20 μM) for 0-4 min with and the mixture was quenched with 6X-SDS loading solution. After running out on 10% SDS-PAGE, the gels were dried and the radioactivity quantified by phosphorimage analysis (Molecular Dynamics) relative to a 14C-bovine serum albumin (Amersham) standard. Assays were performed in duplicates and these generally agreed within 20%.
ATF-2-b-ZIP acetylation for mass spectrometry analysis
GST-ATF-2-b-ZIP (5 μM) was incubated with or without wt p300 HAT domain (250 nM) in presence of acetyl-CoA (2 mM) in assay buffer (50 mM HEPES pH 7.9, 0.1 mM EDTA, 1 mM DTT and 50 μg/ml bovine serum albumin) for 1 h. The proteins were digested with trypsin (20:1 w/w) in 25 mM ammonia bicarbonate at 37°C for 18 h. Digested peptide mixtures were separated by reversed phase HPLC on a C18 column with an acetonitrile/water/0.1% trifluoroacetic acid solvent system. The collected fractions were dried completely and resuspended in 3 μl of water. 0.5 μL of the sample was spotted on a MALDI target followed by the addition of 0.5 μL of α-cyano-4-hydroxycinnamic acid matrix and was allowed to dry at room temperature. The MS and MS/MS spectra were acquired in the positive ion mode using a Kratos Analytical (Manchester, UK) AXIMA-CFR MALDI-TOF mass spectrometer equipped with a pulsed extraction source, a 337-nm pulsed nitrogen laser and a curved-field reflectron. The acceleration voltage was 20 kV. For the MS/MS spectra metastable fragmentation was enhance by increasing the MALDI laser power. No collision gas was used.
In vivo acetylation of ATF-2, immunoprecipitation and western blotting
293 cells and Cos-7 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Life technologies, Gaithersburg, MD, USA) and 1% antibiotics. Cells were treated with LPS, cycloheximide and phorbol myristate (PMA) with or without HDAC inhibitors (5 mM sodium butyrate, 10 mM nicotinamide and 100 ng/ml TSA) for 8 h. Cells were then harvested and lysed at 4°C by gentle scrapping in lysis buffer (50 mM Tris-HCl (pH 7.5), 250 mM NaCl, 5 mM EDTA, 1 mM DTT, 0.1% NP-40 (w/v), 0.5 mM AEBSF, 1 mg/ml aprotinin/leupeptin, 2 mM NaF and 0.5 mM Na-vanadate). Lysates were further clarified by centrifugation at 10,000g for 20 min at 4°C. The protein concentrations of lysates were estimated by Bradford reagent. Lysates were pre-cleared for 5 min with 10 μl protein G sepharose and immunoprecipitated at 4°C for overnight with mouse monoclonal anti-ATF-2 (sc-242, Santa Cruz Biotechnology) followed by precipitation with 30 μl protein G for 2 h at 4°C. After two washes with PBS buffer (140 mM NaCl, 2.7 mM KCl,10 mM Na2HPO4, 1.8 mM KH2PO4, 2 mM EDTA, 0.1% (w/v) NP-40, Roche protease inhibitor cocktail “COMPLETE”), the immunoprecipitates were run out on 8% SDS-PAGE, transferred to nitrocellulose membrane, immunoblotted with rabbit monoclonal anti-ATF-2 (9226, Cell Signaling Technology), stripped with Pierce stripping buffer and re-probed with rabbit polyclonal anti-acetyl lysine antibody (Upstate 06-933) and detected using ECL chemiluminescence as per manufacturer instructions (Super-signal, Pierce).
Mutagenesis of full-length human ATF-2 human plasmid
pACT-ATF-2 (36) was subjected to site-directed mutations at K357 and K374 which were created by the QuikChange mutagenesis procedure (Stratagene). Primers containing ∼18 nucleotides on either side of the mutation were made. Following PCR assays according to the manufacturer's protocol, the DNA was digested with DpnI and transformed into E. coli. Positive clones were isolated, and the introduction of the mutation was confirmed by sequence analysis. Using the above methodology KK/RR, KK/AA, KK/QQ, K357R, K357A, K357Q, K374R, K374A, K374Q mutants were generated.
Luciferase Assays
Cos-7 cells were plated in 6-well flat bottomed plates on the day prior to transfection at a density of 2 X 104 cells/well in DMEM. Transfections were performed using Lipofectamine-2000 (Invitrogen). The reporter CRE-Luc containing sequence placed at 5′ to the gene of luciferase was employed (37). Cells were co-transfected with 1μg pCRE-Luc (stratagene), 30 ng of pRL-TK-Renilla (pRL-TK from Promega), and equal amounts of pACT plasmids expressing wt-ATF-2, KK/RR, KK/AA, KK/QQ, K357R, K357A, K357Q, K374R, K374A, K374Q for 36 h and treated with HDAC inhibitors (5 mM sodium butyrate, 10 mM nicotinamide and 100 ng/ml TSA) for 8 h. Cells were lysed and firefly luciferase and Renilla luciferase activities were measured with luciferase assay reagent (Dual-Glow luciferase Reporter Assay system, Promega) using a luminometer. Three replicates were performed and the standard errors are shown in Figure 8. The lysates were further analyzed for amount of ATF-2 expressed in different transfection reactions by immunoprecipitation and immunoblotting with anti-ATF-2 antibody (mouse monoclonal anti-ATF-2 (sc-242, Santa Cruz Biotechnology) followed by precipitation with 30 μl protein G for 2 h at 4°C. After two washes with PBS buffer (140 mM NaCl, 2.7 mM KCl,10 mM Na2HPO4, 1.8 mM KH2PO4, 2 mM EDTA, 0.1% (w/v) NP-40, Roche protease inhibitor cocktail “COMPLETE”), the immunoprecipitates were run out on 8% SDS-PAGE, transferred to nitrocellulose membrane, immunoblotted with rabbit monoclonal anti-ATF-2 (9226, Cell Signaling Technology)).
Fig 8.
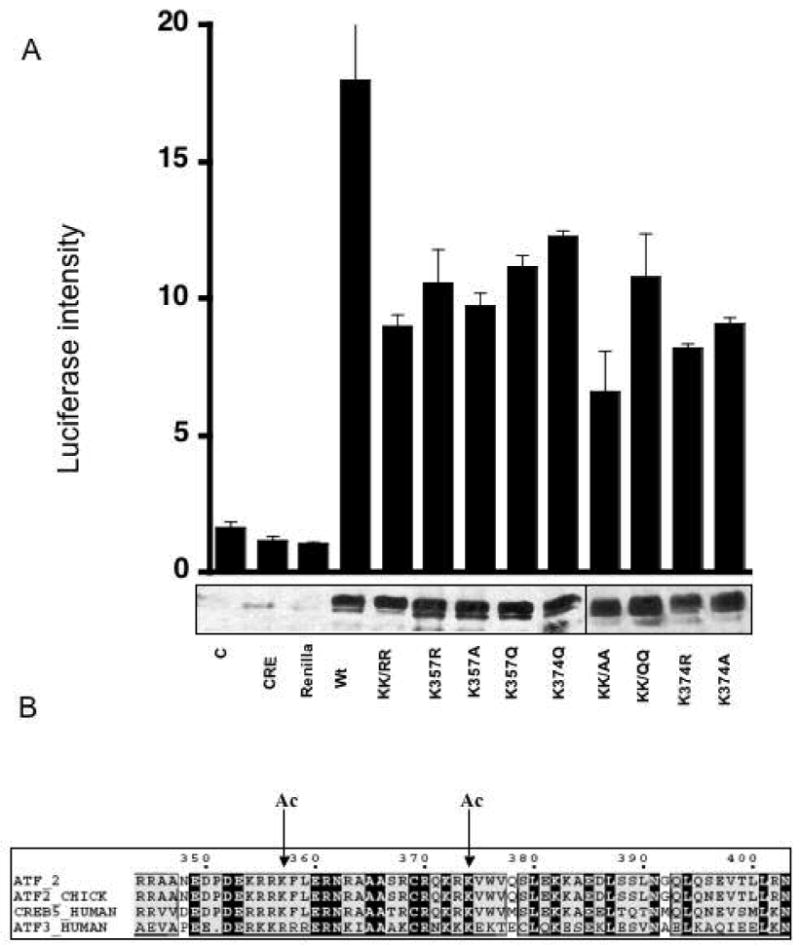
Mutational Analysis of Lys-357 and Lys-374 in ATF-2 on transcriptional regulation using a CREB luciferase reporter plasmid. A) Cell lysates were analyzed for luciferase activity. The bars from left to right are as follows: C-control; CRE–pCRE-luc plasmid alone; Renilla- Renilla plasmid alone; Wt-ATF-2, pCRE-luc, Renilla together (as for all of the ATF-2 mutants to the right of Wt); KKRR-double lysine to arginine mutant; K357R; K357A; K357Q; K374Q; KKAA-double lysine to alanine mutant; KKQQ-double lysine to glutamine mutant; K374R; K374A. Anti-ATF-2 immunoblots show comparable transfection efficiency and ATF-2 stability in overexpressing conditions. B) Sequence alignment of acetylated lysines-357 and 374 in human ATF-2 b-ZIP domain show partial or full conservation in related transcription factors human CREB5, human ATF3, and chicken ATF-2.
Results
ATF-2-b-ZIP Interaction with p300 HAT
Recombinant GST-ATF-2-b-ZIP protein was immobilized on glutathione resin and incubated with semisynthetic p300 HAT protein. It was found that GST-ATF-2-b-ZIP could induce selective retention of p300 HAT compared to resin or GST alone, consistent with prior cellular studies (Figure 2A and B). However, the hyperacetylated p300 HAT (lane 3) was more efficiently pulled down by GST-ATF-2-b-ZIP compared with hypoacetylated p300 HAT (lane 5). Lanes 2, 4, and 6 show that the p300 HAT was evenly loaded in these binding studies since the supernatant levels remain consistent. Since many of the acetylation sites in hyperacetylated p300 HAT reside in a flexible loop region (33, 34), we investigated pull-down of the loop deleted form of p300 HAT (p300 HAT-Δloop). Strikingly, p300 HAT-Δloop, not subject to autoacetylation, showed retention on immobilized GST-ATF-2-b-ZIP (lane 1) that was similar to that of hyperacetylated p300 HAT (lane 3). These studies suggest that the p300 autoacetylated loop is not necessary for p300 HAT-ATF-2 interaction. However, the unacetylated loop may negatively influence the interaction of p300 HAT with ATF-2-b-ZIP as loop autoacetylation confers enhanced affinity of p300 HAT binding to ATF-2-b-ZIP.
Fig 2.
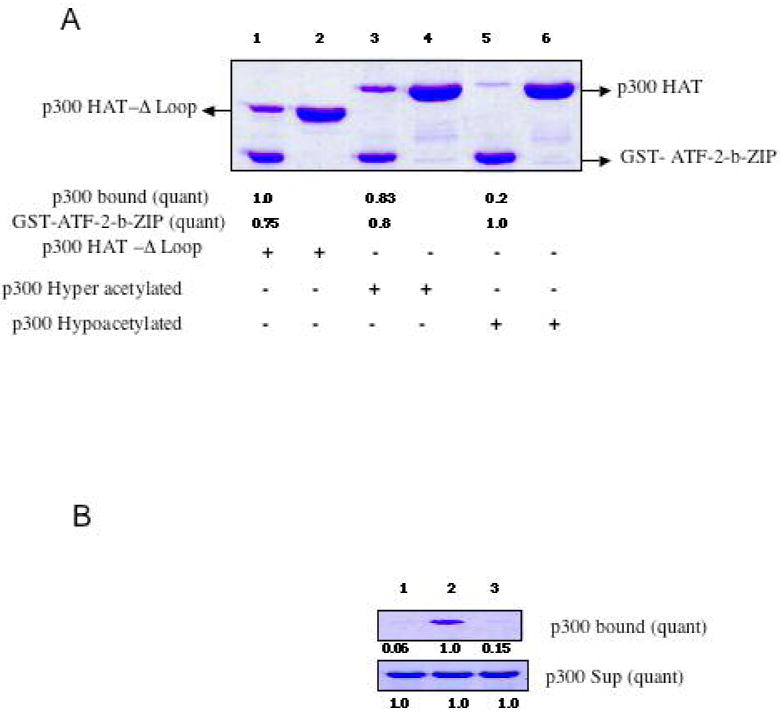
Affinity binding assays with immobilized GST-ATF-2-b-ZIP. A) Comparative pull-downs of wt hyperacetylated p300 HAT, wt hypoacetylated p300 HAT, and hypoacetylated p300 HAT-Δloop. SDS-PAGE are stained with Coomassie and intensity values derived from duplicate measurements. B) Control experiments with lanes 1 and 3 employing resin only and GST bound to resin and lane 2 containing GST-ATF-2-b-ZIP each treated with hyperacetylated p300 HAT protein as in A.
We next investigated the p300 HAT/GST-ATF-2-b-ZIP interaction by analyzing GST-ATF-2-b-ZIP's effects on p300-mediated acetylation of a histone-derived peptide substrate H4-15. It was found that GST-ATF-2-b-ZIP but not GST alone could inhibit the acetylation of H4-15 peptide substrate by hyperacetylated p300 in a concentration-dependent manner of the inhibitor (Figure 3). Inhibition of hypoacetylated p300 by GST-ATF-2-b-ZIP was much less (Figure 3), which was especially noteworthy because of the lower baseline rate of hypoacetylated p300 compared with hyperacetylated p300. The potency and selectivity of GST-ATF-2-b-ZIP inhibition was most striking with the hyperacetylated form of p300 HAT at the short time points. As shown in Figure 3B and 3C, at 30 s reaction time, 1 μM acetylation of GST-ATF-2-b-ZIP resulted in 50% inhibition of p300 HAT activity whereas 5 μM of GST-ATF-2-b-ZIP led to less than 20% hypoacetylated p300 HAT at this time point. GST alone did not significantly inhibit p300 HAT whether hypo- or hyperacetylated at these time points (Figure 3B and 3C). These results suggest that ATF-2-b-ZIP binds more tightly to hyperacetylated p300 HAT, consistent with the pull-down experiments (Figure 2). Taken together, these data suggest a structural model in which loop autoacetylation exposes a binding site for ATF-2-b-ZIP, blocking p300 HAT-mediated histone acetylation (Figure 4).
Fig 3.
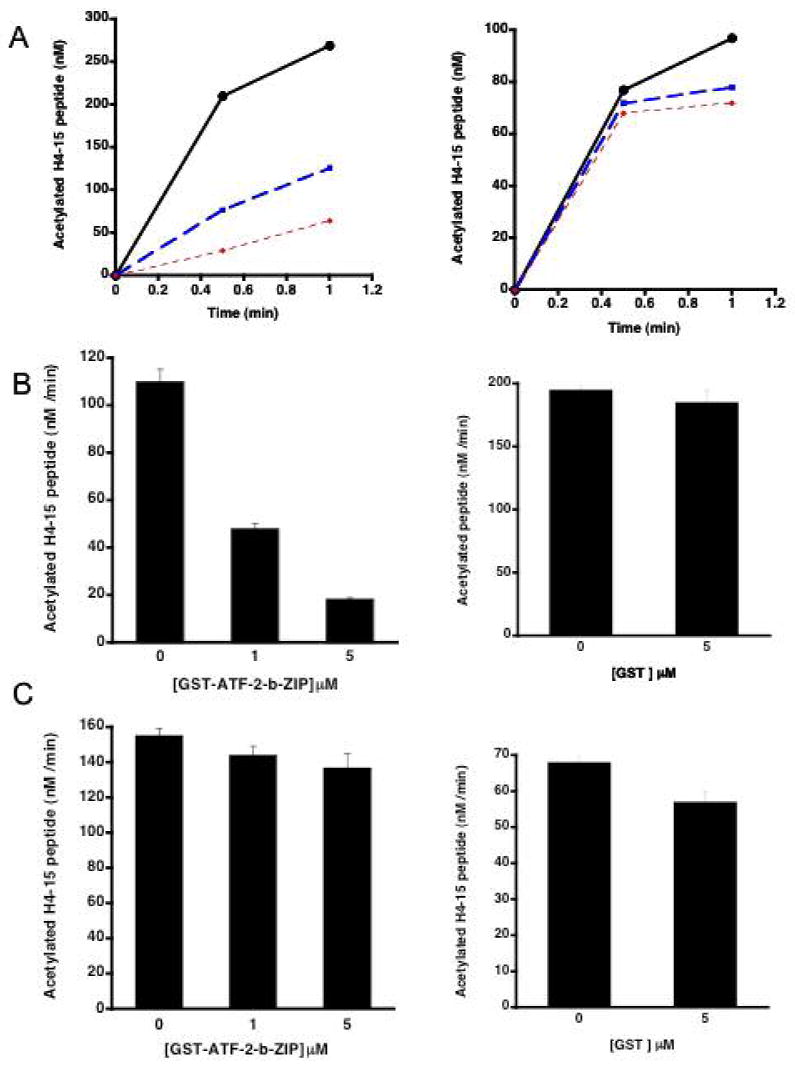
ATF-2-b-ZIP domain inhibits hyperacetylated p300 activity A) Time course of hyperacetylated (left) or hypoacetylated p300 HAT (right) (40 nM) used to acetylate peptide H4-15 peptide (100 μM) in the presence of GST-ATF-2-b-ZIP domain (0, 1, 5 μM; black, blue, red, respectively). B) Replot of 30 s time point with hyperacetylated p300 HAT data in A (left) and control using 5 μM GST in place of GST-ATF-2-b-ZIP (right). C) Replot of 30 s time point with hypoacetylated p300 HAT data in A (left) and control using 5 μM GST in place of GST-ATF-2-b-ZIP (right).
Fig 4.
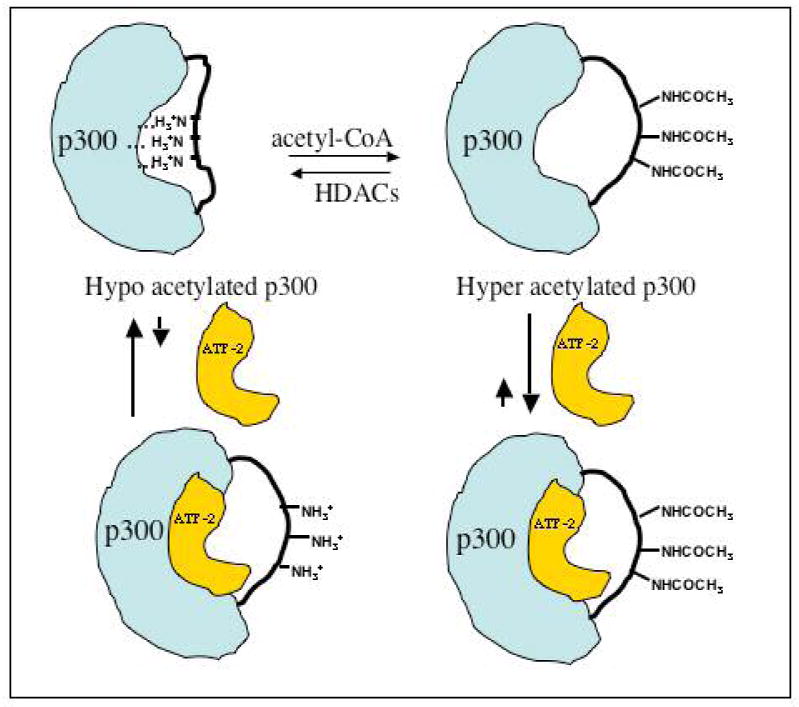
Proposed model to explain enhanced affinity of p300 HAT for ATF-2-b-ZIP for hyperacetylated p300 HAT.
ATF-2-b-ZIP as a p300 substrate
We also investigated the effect of ATF-2 on p300 HAT autoacetylation. Analogous to p300 HAT inhibition of histone peptide acetylation, GST-ATF-2-b-ZIP could block p300 HAT autoacetylation (data not shown). Accompanying this inhibition of autoacetylation was the rather interesting appearance of radiolabeling of GST-ATF-2-b-ZIP, consistent with p300-mediated acetylation of GST-ATF-2-b-ZIP (Figure 5). In comparison to GST-ATF-2-b-ZIP, GST alone was minimally acetylated by p300 HAT (Figure 5B and C) suggesting that the targeted lysines were within the ATF-2 moiety. Kinetic analysis of p300-mediated GST-ATF-2-b-ZIP acetylation showed a time-dependent increase in GST-ATF-2-b-ZIP acetylation by p300 HAT over 4 min (Figure 5A). There was little evidence of saturation of GST-ATF-2-b-ZIP up to 5 μM of protein, suggesting that the apparent Km for GST-ATF-2-b-ZIP was greater than 5 μM. Interestingly, hyperacetylated p300 HAT showed about 1.8-fold greater efficiency than hypoacetylated p300 HAT in catalyzing acetyl transfer to GST-ATF-2-b-ZIP. The increased rate with hyperacetylated p300 may reflect tighter interaction with ATF-2-b-ZIP or a relief of autoinhibition by loop displacement.
Fig 5.
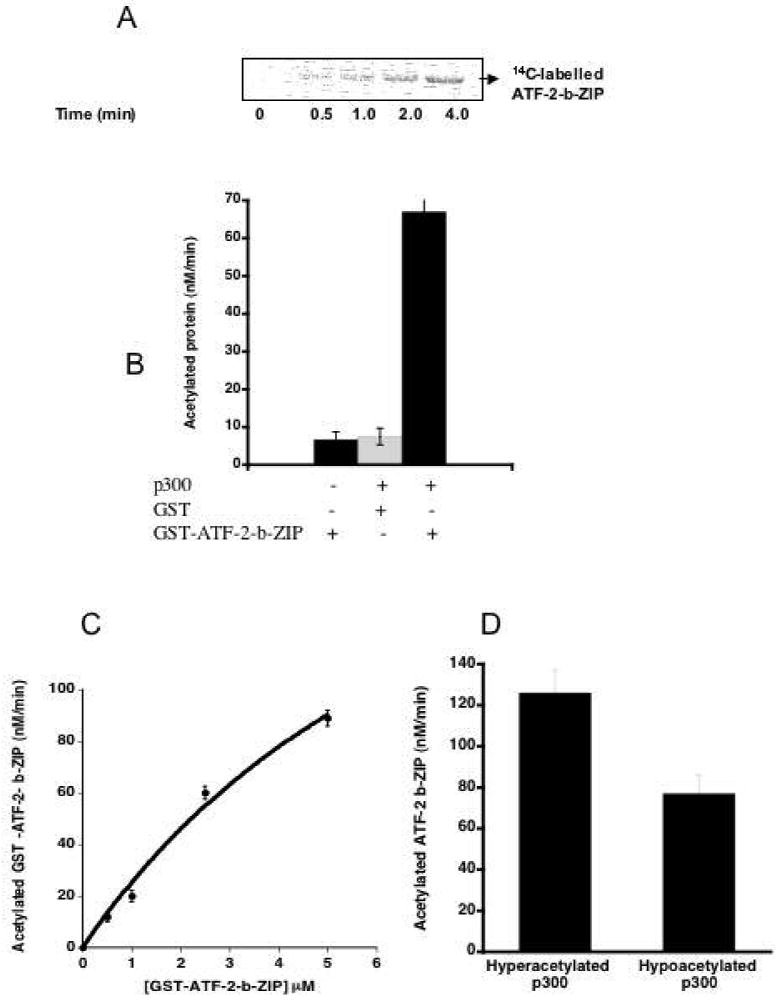
ATF-2-b-ZIP is acetylated by p300 HAT. A) hypoacetylated p300 HAT-mediated acetylation of GST-ATF-2-b-ZIP (2.5 μM) vs. time with 40 nM p300 HAT (relative acetylation values at 0.5 min, 38%; 1 min, 50%, 2 min, 68%, and 4 min, 100%). B) Acetylation of GST-ATF-2-b-ZIP (5 μM) vs. GST (5 μM) by hypoacetylated p300 HAT (0 or 40 nM). C) Concentration dependence of acetylation of GST ATF-2-b-ZIP by hypoacetylated p300 HAT. D) Comparison of hyperacetylated p300 HAT and hypoacetylated p300 HAT catalyzed acetylation of GST-ATF-2-b-ZIP. GST-ATF-2-b-ZIP (5 μM) incubated with hyper/hypoacetylated p300 HAT domain (40 nM) for 4 min.
To further probe the ATF-2-b-ZIP modification, we undertook a mass spectrometric analysis of b-ZIP modification. Using a combination of tryptic digestion and MALDI mass spectrometry, we obtained unequivocal evidence for two acetylation sites in ATF-2-b-ZIP at Lys-357 and Lys-374 (Fig. 6). Tryptic digestion at lysine and argine residues (but not acetylated lysine residues) resulted in two peptides: KAcFLER and KAcVWVQSLEK, whose MS/MS spectra revealed the sequence-specific fragmentation shown in Figures 6a and 6b, respectively, consistent with acetylation at these sites. The sequences of the two acetylation site-containing peptides were unambiguously determined by the series of b- and y- types fragment ions (38) presented in the MALDI-MSMS spectra. The b2 fragments in both spectra contain acetylated lysine residues. In addition, modified lysine residues were further confirmed by the two marker ions at m/z 143 (acetylated lysine immonium ion) and m/z 126 (loss of NH3 from the acetylated lysine immonium ion). It should be noted that each of these sites contains several proximal basic residues, with two Arg and one Lys on the N-terminal side of Lys-357 and one Lys and one Arg on the N-terminal side of Lys-374. The preference for nearby positively charged residues in p300 substrate acetylation sites has been noted and discussed previously (35).
Fig 6.
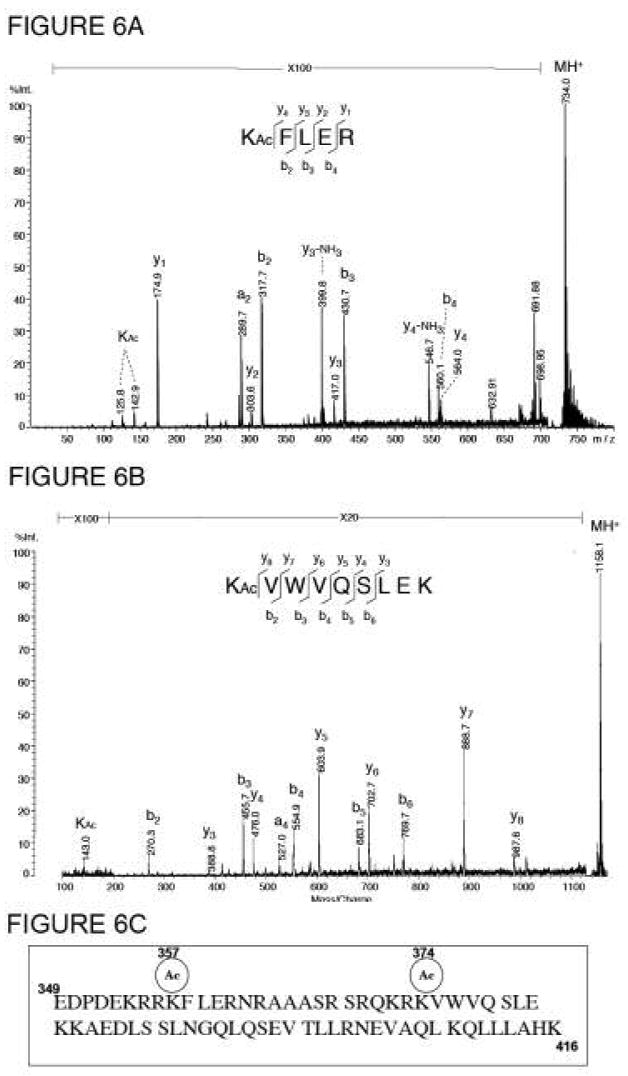
MALDI MS/MS spectra of two acetylated peptides obtained from the tryptic digest of the GST-ATF-2 b-ZIP domain: A) KAcFLER and B) KAcVWVQSLEK; the peaks labeled as KAc represents the characteristic ions of acetylated lysine residue. The y-series ions labeled on the spectra refer to cleavages at the peptide amide bonds with positive charge retention on the C-terminal fragment, according to the nomenclature by Biemann (46). C) Sequence of ATF-2 b-ZIP indicating acetylation sites at K357 and K374.
ATF-2 acetylation in vivo
To investigate the possibility that endogenous ATF-2 is subject to acetylation in vivo, we carried out immunoprecipitation-western blot analysis with and without anti-acetyl-lysine antibodies (Fig. 7). We found that ATF-2 showed significant staining with anti-acetyl-lysine antibodies in both 293 and Cos 7 cell lines. This staining was greatly enhanced by the addition of HDAC inhibitors, suggesting that ATF-2 acetylation is governed by the equlibrium resulting from the opposing actions of acetyltransferases and deacetylases. It is known that ATF-2 transcriptional activation can be stimulated by stress conditions such as inflammatory mediators or protein synthesis inhibition (26-31). However, unlike responses to changes in phosphorylation (26-31), enhanced acetylation of ATF-2 was not observed by pre-treatment with stress agents TNF-α, PMA, or cycloheximide, suggesting that these phosphorylations by MAP kinases are not correlated with lysine acetylation.
Fig 7.
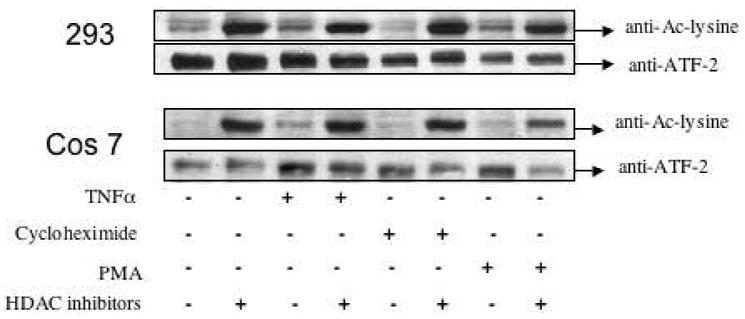
ATF-2 acetylation in cell culture. 293 and Cos 7 cells were treated with TNFα, cycloheximide, PMA, or HDAC inhibitors as indicated and ATF-2 was immunoprecipitated and blotted with anti-ATF-2 and anti-acetyl-Lys as indicated.
Role of ATF-2 Acetylation in Transcription
We employed a transcriptional reporter assay containing a CRE promoter driving the production of luciferase in Cos 7 cells (37). As shown in Figure 8, the endogenous expression of ATF-2 in these cells was relatively low, thus allowing a simple comparison of transfected wt and mutant ATF-2 induced transcription. As can be seen, wt ATF-2 transfected cells led to a 10-fold enhanced luciferase signal compared to controls (Figure 8). However, the non-acetylatable ATF-2 double mutant KK357/374RR (KK/RR) showed 2-fold less transcriptional activation compared to wt ATF-2 in these experiments (Figure 8). A series of other single and double Lys mutant constructs displayed similar results to KK//4RR suggesting that each of these Lys residues is required for maximal ATF-2 transcriptional enhancement (Figure 8). It is noteworthy that Lys replacements with Arg (charge conservation with Lys), Gln (potential mimic of acetyl-Lys) or Ala all showed similar reductions indicating a specific need for the physiologic side chain and/or acetylated lysine.
Discussion
Since the molecular identification of HATs and histone deacetylases in the mid-1990's, reversible protein acetylation has become an increasingly commonly observed mode of gene regulation (5). In this study, we have explored two potential new roles for protein acetylation. On the one hand, we have found that reversible autoacetylation of p300 HAT regulates its binding interaction with the ATF-2-b-ZIP domain. On the other hand, we have shown that p300 HAT can catalyze the acetylation of the ATF-2-b-ZIP domain on at least two Lys sites. Given the importance of ATF-2 and p300/CBP in gene regulation, these findings may have significance for the control of a broad array of transcriptional events.
Mechanistically, the enhanced association of ATF-2 and p300 HAT offers an unusual example of acetylation modulating protein-protein interaction. It is now well-accepted that bromo-domains can bind to acetyl-lysine sequences in proteins (39). Moreover, there are examples cases where lysine acetylation can directly modulate protein-DNA interactions (10, 40-41). However, there is limited information regarding situations where protein acetylation enhances a protein-protein interaction without bromo-domain involvement (12, 20). In the ATF-2-b-ZIP case, it is proposed that the role of p300 autoacetylation on ATF-2 interaction is indirect since hypoacetylated p300 HAT lacking its regulatory loop was able to bind to ATF-2-b-ZIP about as well as hyperacetylated p300 HAT containing its regulatory loop. These findings should stimulate further efforts to search for a broader range of mechanisms of how lysine acetylation can affect protein-protein interactions.
The enhanced affinity of autoacetylated p300 for ATF-2 may serve two important functions. First, it may strengthen the targeting of p300 to ATF-2 regulated genes (26-31). Second, since ATF-2 can block p300's catalytic action on histone peptide substrate, it may suppress histone acetylation at a given promoter (22). The relative significance of these two functions should be a consideration in future transcriptional studies. In this light, the apparent ability of p300 conformational changes to modulate other protein-protein interactions in transcriptional regulation should be considered (42, 43). It has been shown that binding of p300 to the pre-initiation complex can be modulated by autoacetylation of p300 (42).
An increasing number of acetyltransferase protein substrates of p300/CBP have been uncovered and in this report we add ATF-2 (10-20). It has been reported several years ago that ATF-2 may have intrinsic acetyltransferase activity (44). While this has not yet been further confirmed, the acetylation of ATF-2-b-ZIP is clearly p300 HAT dependent in vitro. We demonstrate that ATF-2 can be acetylated on Lys-357 and Lys-374 and that these sites are mutationally sensitive for transcriptional regulation from the CRE reporter. These lysine modifications may exert at least two influences on ATF-2. Since these lysines are within the DNA binding domain (45), acetylation may change the conformation or affinity of the ATF-2-DNA complex. Alternatively, Lys modifications could affect the intramolecular inhibitory interaction between the N- and C-terminal domains of ATF-2 (4). Distinguishing among these possibilities will require careful biochemical characterization of full-length ATF-2. It is also noteworthy that one of these acetylation sites (Lys-374) is conserved in the transcription factors CREB5 and ATF3 indicating potential regulatory significance in these pathways.
Acknowledgments
We thank Dr. Narendrakumar Ramanan, for assistance with transcriptional analysis, Prof. P. Fafournoux INRA, France and for construct pACT-ATF-2. We thank the NIH for support (U54RR020839 to R.J.C. and D.W.; GM062437 to P.A.C.; GM060293 to R.M.)
References
- 1.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 2.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 3.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 4.Sano Y, Tokitou F, Dai P, Maekawa T, Yamamoto T, Ishii S. CBP alleviates the intramolecular inhibition of ATF-2 function. J Biol Chem. 1998;273:29098–29105. doi: 10.1074/jbc.273.44.29098. [DOI] [PubMed] [Google Scholar]
- 5.Kimura A, Matsubara K, Horikoshi M. A decade of histone acetylation: marking eukaryotic chromosomes with specific codes. J Biochem (Tokyo) 2005;138:647–662. doi: 10.1093/jb/mvi184. [DOI] [PubMed] [Google Scholar]
- 6.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Kalkhoven E. CBP and p300: HATs for different occasions. Biochem Pharmacol. 2004;68:1145–1155. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 8.Marmorstein R. Structural and chemical basis of histone acetylation. Novartis Found Symp. 2004;259:78–98. [PubMed] [Google Scholar]
- 9.Gayther SA, Batley SJ, Linger L, Bannister A, Thorpe K, Chin SF, Daigo Y, Russell P, Wilson A, Sowter HM, Delhanty JD, Ponder BA, Kouzarides T, Caldas C. Mutations truncating the EP300 acetylase in human cancers. Nat Genet. 2000;24:300–303. doi: 10.1038/73536. [DOI] [PubMed] [Google Scholar]
- 10.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 11.Costanzo A, Merlo P, Pediconi N, Fulco M, Sartorelli V, Cole PA, Fontemaggi G, Fanciulli M, Schiltz L, Blandino G, Balsano C, Levrero M. DNA damage-dependent acetylation of p73 dictates the selective activation of apoptotic target genes. Mol Cell. 2002;9:175–186. doi: 10.1016/s1097-2765(02)00431-8. [DOI] [PubMed] [Google Scholar]
- 12.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 13.Chen LF, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 14.Greene WC, Chen LF. Regulation of NF-kappaB action by reversible acetylation. Novartis Found Symp. 2004;259:208–217. [PubMed] [Google Scholar]
- 15.Yuan W, Condorelli G, Caruso M, Felsani A, Giordano A. Human p300 protein is a coactivator for the transcription factor MyoD. J Biol Chem. 1996;271:9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]
- 16.Puri PL, Avantaggiati ML, Balsano C, Sang N, Graessmann A, Giordano A, Levrero M. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 1997;16:369–383. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pao GM, Janknecht R, Ruffner H, Hunter T, Verma IM. CBP/p300 interact with and function as transcriptional coactivators of BRCA1. Proc Natl Acad Sci U S A. 2000;97:1020–1025. doi: 10.1073/pnas.97.3.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 19.Tomita A, Towatari M, Tsuzuki S, Hayakawa F, Kosugi H, Tamai K, Miyazaki T, Kinoshita T, Saito H. c-Myb acetylation at the carboxyl-terminal conserved domain by transcriptional co-activator p300. Oncogene. 2000;19:444–451. doi: 10.1038/sj.onc.1203329. [DOI] [PubMed] [Google Scholar]
- 20.Sano Y, Ishii S. Increased affinity of c-Myb for CREB-binding protein (CBP) after CBP-induced acetylation. J Biol Chem. 2001;276:3674–3682. doi: 10.1074/jbc.M006896200. [DOI] [PubMed] [Google Scholar]
- 21.Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 22.Kundu TK, Palhan VB, Wang Z, An W, Cole PA, Roeder RG. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol Cell. 2000;3:551–61. doi: 10.1016/s1097-2765(00)00054-x. [DOI] [PubMed] [Google Scholar]
- 23.Lau OD, Kundu TK, Soccio RE, Ait-Si-Ali S, Khalil EM, Vassilev A, Wolffe AP, Nakatani Y, Roeder RG, Cole PA. HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol Cell. 2000;5:589–595. doi: 10.1016/s1097-2765(00)80452-9. [DOI] [PubMed] [Google Scholar]
- 24.Bandyopadhyay D, Okan NA, Bales E, Nascimento L, Cole PA, Medrano EE. Down-regulation of p300/CBP histone acetyltransferase activates a senescence checkpoint in human melanocytes. Cancer Res. 2002;62:6231–6239. [PubMed] [Google Scholar]
- 25.Kawasaki H, Song J, Eckner R, Ugai H, Chiu R, Taira K, Shi Y, Jones N, Yokoyama KK. p300 and ATF-2 are components of the DRF complex, which regulates retinoic acid- and E1A-mediated transcription of the c-jun gene in F9 cells. Genes Dev. 1998;12:233–245. doi: 10.1101/gad.12.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta S, Campbell D, Dérijard B, Davis RJ. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 27.Livingstone C, Patel G, Jones N. ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J. 1995;14:1785–1797. doi: 10.1002/j.1460-2075.1995.tb07167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Dam H, Wilhelm D, Herr I, Steffen A, Herrlich P, Angel P. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J. 1995;14:1798–1811. doi: 10.1002/j.1460-2075.1995.tb07168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hai T, Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci U S A. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nomura N, Zu YL, Maekawa T, Tabata S, Akiyama T, Ishii S. Isolation and characterization of a novel member of the gene family encoding the cAMP response element-binding protein CRE-BP1. J Biol Chem. 1993;268:4259–4266. [PubMed] [Google Scholar]
- 31.Matsuda S, Maekawa T, Ishii S. Identification of the functional domains of the transcriptional regulator CRE-BP1. J Biol Chem. 1991;266:18188–18193. [PubMed] [Google Scholar]
- 32.Reimond AM, Grusby MJ, Kosaras B, Fries JWU, Mori R, Maniwa S, Clauss IM, Collins T, Sidman RL, Glimcher MJ, Glimcher LH. Chondrodysplasia and neurological abnormalities in ATF-2-deficient mice. Nature. 1996;379:262–265. doi: 10.1038/379262a0. [DOI] [PubMed] [Google Scholar]
- 33.Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J, Levrero M, Sartorelli V, Cotter RJ, Cole PA. Regulation of the p300 HAT domain via a novel activation loop. Nat Struct, Mol, Biol. 2004;11:308–315. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- 34.Karanam B, Jiang L, Wang L, Kelleher NL, Cole PA. Kinetic and mass spectrometric analysis of p300 HAT autoacetylation. J Biol Chem. 2007 doi: 10.1074/jbc.M608813200. in press (epub: http://www.jbc.org/cgi/content/full/281/52/40292) [DOI] [PubMed]
- 35.Thompson PR, Kurooka H, Nakatani Y, Cole PA. Transcriptional coactivator protein p300. Kinetic characterization of its histone acetyltransferase activity. J Biol Chem. 2001;276:33721–33729. doi: 10.1074/jbc.M104736200. [DOI] [PubMed] [Google Scholar]
- 36.Monzen K, Hiroi Y, Kudoh S, Akazawa H, Okaa T, Takimoto E, Hayashi D, Hosoda T, Kawabata M, Miyazono K, Ishii S, Yazaki Y, Nagaia R, Komuro I. Smads, TAK1, and their common target ATF-2 play a critical role in cardiomyocyte differentiation. J Cell Biol. 2001;153:687–698. doi: 10.1083/jcb.153.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salas TR, Reddy SA, Clifford JL, Davis RJ, ikuchi A, Lippman SM, Menter DG. Alleviating the suppression of glycogen synthase kinase-3 by Akt Leads to the phosphorylation of cAMP-response element-binding protein and its transactivation in intact cell nuclei. J Biol Chem 278. 2003;42:41338–41346. doi: 10.1074/jbc.M302972200. [DOI] [PubMed] [Google Scholar]
- 38.Roepstorff P, Fohlmann J. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed Mass Spectrom. 1984;11:601. doi: 10.1002/bms.1200111109. [DOI] [PubMed] [Google Scholar]
- 39.Zheng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 40.Cleary J, Sitwala KV, Khodadoust MS, Kwok RP, Mor-Vaknin M, Cole PA, Markovitz DM. p300/CBP-associated factor drives DEK protein into interchromatin granule clusters. J Biol Chem. 2005;280:31760–31767. doi: 10.1074/jbc.M500884200. [DOI] [PubMed] [Google Scholar]
- 41.Guidez F, Howell L, Isalan M, Cebrat M, Alani RM, Ivins S, Hormaeche I, McConnell MJ, Pierce S, Cole PA, Licht J, Zelent A. Histone acetyltransferase activity of p300 is required for transcriptional repression by the promyelocytic leukemia zince finger protein. Mol Cell Biol. 2005;25:5552–5566. doi: 10.1128/MCB.25.13.5552-5566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turnell AS, Stewart GS, Grand RJ, Rookes SM, Martin A, Yamano H, Elledge SJ, Gallimore PH. The APC/C and CBP/p300 cooperate to regulate transcription and cell cycle progression. Nature. 2005;438:690–695. doi: 10.1038/nature04151. [DOI] [PubMed] [Google Scholar]
- 43.Black JC, Choi JE, Lombardo SR, Carey M. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell. 2006;23:809–818. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 44.Kawasaki H, Schiltz L, Chiu R, Itakura K, Taira K, Nakatani Y, Yokoyama KK. ATF-2 has intrinsic acetyltransferase activity which is modulated by phosphorylation. Nature. 2000;405:195–200. doi: 10.1038/35012097. [DOI] [PubMed] [Google Scholar]
- 45.Panne D, Maniatis T, Harrison SC. Crystal structure of ATF-2/c-Jun and IRF-3 bound to the interferon-beta enhancer. EMBO J. 2004;23:4384–4393. doi: 10.1038/sj.emboj.7600453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biemann K. Appendix 5. Nomenclature for peptide fragment ions (positive ions) Meth Enz. 1990;193:886–887. doi: 10.1016/0076-6879(90)93460-3. [DOI] [PubMed] [Google Scholar]


