Abstract
Ataxia telangiectasia–mutated gene (ATM) is a 350-kDa protein whose function is defective in the autosomal recessive disorder ataxia telangiectasia (AT). Affinity-purified polyclonal antibodies were used to characterize ATM. Steady-state levels of ATM protein varied from undetectable in most AT cell lines to highly expressed in HeLa, U2OS, and normal human fibroblasts. Subcellular fractionation showed that ATM is predominantly a nuclear protein associated with the chromatin and nuclear matrix. ATM protein levels remained constant throughout the cell cycle and did not change in response to serum stimulation. Ionizing radiation had no significant effect on either the expression or distribution of ATM. ATM immunoprecipitates from HeLa cells and the human DNA-dependent protein kinase null cell line MO59J, but not from AT cells, phosphorylated the 34-kDa subunit of replication protein A (RPA) complex in a single-stranded and linear double-stranded DNA–dependent manner. Phosphorylation of p34 RPA occurred on threonine and serine residues. Phosphopeptide analysis demonstrates that the ATM-associated protein kinase phosphorylates p34 RPA on similar residues observed in vivo. The DNA-dependent protein kinase activity observed for ATM immunocomplexes, along with the association of ATM with chromatin, suggests that DNA damage can induce ATM or a stably associated protein kinase to phosphorylate proteins in the DNA damage response pathway.
INTRODUCTION
Cells respond to DNA damage by activating checkpoint pathways that delay progression through the cell cycle. This cell cycle delay provides the necessary time for the cell to assess and repair the damage before reentering the cell cycle. If the damage is determined to be beyond repair, the cell may undergo apoptosis to prevent mutations from being propagated. When mammalian cells are exposed to ionizing radiation (IR) or radiomimetic drugs, a signal transduction pathway is activated that arrests cells in G1, S, and/or G2 phases of the cell cycle. The G1 arrest is the best characterized and is dependent on a functional p53 response that leads to transcriptional activation of the G1-specific cyclin-dependent kinase inhibitor p21/WAF1/CIP (Kastan et al., 1991). The S and G2 checkpoints seem to be p53 independent because p53-defective cells retain these checkpoints (El-Deiry et al., 1993). Although the existence of DNA damage–dependent checkpoint pathways has been known for some time, the molecular mechanism(s) by which the cell senses the DNA double-strand breaks and converts this information into a growth arrest signal remains unclear.
Ataxia telangiectasia (AT)1 is an autosomal recessive disorder characterized by cerebellar ataxia, dilated blood vessels in the eyes and skin (oculocutaneous telangiectasias), immunodeficiency, hypersensitivity to IR, and a 100-fold increase in the risk of some types of cancer (for example, lymphoma) (Taylor et al., 1994). AT is estimated to effect between 1 in 40,000 and 1 in 100,000 live births. Significantly, the 1% of the general population that are estimated to be AT heterozygotes have a three- to fivefold greater risk of cancer (Swift et al., 1991). Many of these clinical symptoms may be attributed to the cellular defects observed in cell lines derived from AT patients. These include chromosomal instability, defects in the cytoskeleton (Taylor et al., 1994), decreased telomere length, failure to activate cell cycle arrest checkpoints in response to IR (Houldsworth and Lavin, 1980; Beamish and Lavin, 1994; Paules et al., 1995), and the inability to activate damage response pathways (p53 [Kastan et al., 1992], stress-activated protein kinase [Shafman et al., 1995], and replication protein A [RPA] [Liu and Weaver, 1993]). Because the AT-defective cell lines fail to activate the G1, S, and G2 cell cycle checkpoints, the defect is likely to occur early along the DNA damage response pathway that may be common among the three checkpoint pathways. Because AT cells are also extremely sensitive to IR and undergo apoptosis at very low doses, the AT gene product may also be involved in programmed cell death. How, or whether, this response is linked to the checkpoint arrest is unclear.
Although genetic analysis suggested that AT could be separated into five complementation groups, a single gene that is mutated in all complementation groups has been identified. Ataxia telangiectasia–mutated gene (ATM) was identified by positional cloning and localized to chromosome 11q22–23 (Savitsky et al., 1995a,b; Gilad et al., 1996). Northern blot analysis revealed that ATM is ubiquitously expressed in all human tissues that were examined. The ATM cDNA encodes a 3056-amino acid protein with a calculated molecular weight of 350 kDa. The carboxyl terminus of ATM shares significant homology with the kinase domain of phosphatidylinositol 3 (PI3)-kinase. ATM also shares sequence homology with portions of the yeast RAD3 gene. Although the functional significance of these homologies remains speculative, it is clear that ATM is a member of an emerging family of exceptionally large (250- to 460-kDa) proteins that are involved in cell cycle regulation or DNA damage recognition and repair (reviewed in Hunter, 1995; Lavin et al., 1995; Meyn, 1995; Zakian, 1995; Jackson, 1996). The family members share 30% identity and 60% similarity in the PI3-like kinase domain and 20% identity and 50% similarity in the RAD3 domain. This family includes the Saccharomyces cerevisiae proteins TEL1 and MEC1 (Greenwell et al., 1995; Morrow et al., 1995), Schizosaccharomyces pombe RAD3 (Bentley et al., 1995), Drosophila MEI-41 (Hari et al., 1995), ATM-related protein kinase (ATR) (Cimprich et al., 1996) and human DNA-PKcs (Hartley et al., 1995). These genes may be functionally related because mutations in these genes exhibit many of the phenotypes observed in AT-defective cells.
Although ATM is a member of a family of proteins that share a PI3-like kinase domain, ATM does not seem to function as a lipid kinase but as a protein kinase. It has recently been reported that ATM immunoprecipitates can phosphorylate GST–IκB fusion protein in vitro (Jung et al., 1997). However, the specificity of the protein kinase is uncertain given that protein kinase activity was also observed in immunoprecipitates from AT cells. When the kinase domain of ATM was overexpressed and immunoprecipitated from atm knock-out mouse cells, the immunocomplex was found to phosphorylate c-abl (Baskaran et al., 1997). Although ATM and c-abl coimmunoprecipitated from lymphoid cell lines (Shafman et al., 1997), the c-abl interaction domain of ATM was not included in the construct used to overexpress the kinase domain. Overexpression of the ATM kinase domain was also found to complement partially the radiosensitivity and the IR-induced S-phase arrest of AT-cells (Morgan et al., 1997). For all of these studies, it is unknown whether the correction of the radiosensitivity or the phosphorylation of c-abl was simply caused by an overly active protein kinase domain resulting from the loss of important regulatory domains that lie outside the kinase domain. Support for the importance of the nonkinase domain in ATM function has come from both genetic analysis of ATM mutants as well as the finding that the putative leucine zipper region of ATM (that lies N-terminal to the kinase domain) behaved as a transdominant-negative mutant by rendering an AT-like phenotype to transfected RKO cells (Morgan et al., 1997).
In an effort to examine and understand the molecular mechanism of ATM in the DNA damage response, we produced affinity-purified antibodies to ATM and used them to characterize the properties of ATM with regard to cell cycle expression, subcellular localization, and response to IR. Furthermore, ATM immunoprecipitates obtained from HeLa, MO59K, and the DNA-PK null MO59J cells phosphorylated the p34 subunit of the RPA complex in a single-stranded and linear double-stranded DNA–dependent manner. This protein kinase activity was not detected in cells from the ATM null AT2SF cell line. Phosphoamino acid analysis showed that p34 RPA is phosphorylated on serine and threonine residues by the ATM-associated protein kinase. Moreover, we demonstrate that some of the residues phosphorylated by the ATM-associated protein kinase in vitro are the same as those observed when RPA is phosphorylated in vivo in response to IR. The ATM-associated DNA-dependent protein kinase activity suggests a mechanism by which ATM or an associated subunit can detect DNA damage and activate an associated protein kinase that phosphorylates substrates that lead to cell cycle arrest.
MATERIALS AND METHODS
Cell Culture
Cells were grown in a humidified incubator at 37°C and 5% CO2. HeLa cells were grown as monolayers in RPMI medium supplemented with 10% fetal bovine serum (FBS). K562 and Jurkat cells were grown as suspension cultures in RPMI supplemented with 10% FBS. Primary human and AT-defective fibroblast cell lines were obtained from the Coriell cell repository (Camden, NJ). GM08333A (primary human fibroblasts), GM03487C (AT3Be), GM09607A (AT2SF), and GM05823B (AT5BI) were grown as monolayers in MEM supplemented with 20% FBS and 2× concentrations of amino acids and vitamins. Normal human, AT3ABR, AT5ABR, and AT3Be lymphocytes, a generous gift of Dr. Tim Shafman (Dana-Farber Cancer Center, Cambridge, MA), were maintained in RPMI supplemented with 15% FBS. MO59K and MO59J cells, generously donated by Dr. Joan Turner (Cross Cancer Institute, Edmonton, Alberta, Canada), were maintained in a 1:1 mixture of DMEM and Ham’s F-12 media supplemented with 10% FBS and essential amino acids.
Cell Synchronization, Elutriation, and Irradiation
HeLa cells were split 1:4 and allowed to attach to the plate for 8 h. Cells were synchronized at the G1–S boundary by double thymidine block. Normal human fibroblasts (NHF) were serum starved for 24 h in 0.5% FBS in MEM with 2× concentrations of amino acids and vitamins. Cells were stimulated with 20% FBS and harvested at various times after stimulation. K562 cells were synchronized by centrifugational elutriation, as described by Kauffman et al. (1990), with a JE5.0 rotor with a 40 ml chamber in a Beckman J6-MC centrifuge (Beckman Instruments, Fullerton, CA). Cells were treated with IR by a Shephard 81–14R cesium-137 beam irradiator (J.L. Shepard, San Fernando, CA) at a dose rate of 1.32 Gy/min.
Bacterial Expression of ATM Fragments and Generation of ATM Antibodies
Three nonoverlapping ATM cDNA fragments derived from K562 cells were cloned into either pMAL or pET28 expression vectors to facilitate expression and purification of recombinant ATM fusion proteins. A 2.8-kb cDNA that begins at an internal EcoRI site at nucleotide (nt) 6604 and extends to an EcoRI linker at the 3′-untranslated region of ATM was subcloned into the EcoRI site of pBluescript II SK. The resultant plasmid was digested with PstI and SalI that flank the cDNA to release an ∼1.8-kb Pst fragment (from nt 6604 to 8404 of the ATM cDNA) and an ∼1.4-kb PstI/SalI fragment that encodes from nt 8404 in ATM to the 3′ end. These cDNAs were subcloned into the corresponding sites in the expression vectors to create pET28-ATMPst1.8 and pET28-ATMPst/SalI.4. In addition, an 821-bp EcoRI fragment that is derived from nt 932 to 1753 of the ATM cDNA was subcloned into pMAL to create pMAL-ATMRI821.
High-level protein expression from pET28-ATMPst1.8, pET28-ATMPst/SalI.4, and pMAL-ATMRI821 was achieved in Escherichia coli BL21 DE3 by inducing cultures that reached an OD600 of 0.6 with 2 mM isopropyl β-d-thiogalactopyranoside. Cultures were incubated for 3–4 h at 30°C, harvested, resuspended in 1/10 volume of PBS and protease inhibitors, and lysed by sonication. The His-tagged ATMPst1.8 and ATMPst/SalI.4 fusion proteins were found to be insoluble, and their purification over a Ni-chelate column was facilitated after dissolving in 6 M GuHCl according to the manufacturer’s instructions. The maltose-binding protein (MBP)–ATMRI821 was soluble and was affinity purified over an amylose column.
Purified recombinant proteins were used to immunize rabbits. Sera were collected after two boosts and tested for their ability to identify the corresponding recombinant proteins by Western blots. To distinguish antibody titers that were specifically directed to the ATM portion of the MBP–ATM fusion protein, the MBP fusion protein was digested with Factor Xa and the digestion products used for Western blots.
For affinity purification of the ATM antibodies, purified fusion proteins at concentrations of 5–10 mg/ml were covalently coupled to 1.5 ml of Affi-gel10 (Bio-Rad, Richmond, CA). Sera were incubated overnight with the beads at 4°C, the bound beads were washed with 25 column volumes of Tris-buffered saline (20 mM Tris-HCl, pH 7.5, 0.5 M NaCl) and were eluted with 150 mM NaCl and 0.5% acetic acid, and the fractions were neutralized with 1 M Tris-HCl, pH 9.0. Fractions were monitored at OD280, and the peak fractions were combined, desalted, concentrated, and stored in PBS and 50% glycerol at −80°C. In the case of the MBP–ATM samples, the sera were incubated successively over two columns that were covalently coupled with MBP to reduce the titer of MBP antibodies before passing over the MBP–ATM column. The affinity-purified antibodies Ab1, Ab2, and Ab3 were directed against epitopes encoded by PstI.8, PstI.4, and RI821, respectively.
Immunodetection
For Western blots, cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% Nonidet P-40 [NP-40], 0.5% deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0) with protease inhibitors (10 μg/ml AEBSF, 10 μg/ml leupeptin, 5 μg/ml pepstatin, 5 μg/ml chymotrypsin, 10 μg/ml aprotinin), and the insoluble fraction was pelleted by centrifugation at 160,000 × g for 10 min at 4°C. Total cellular proteins were separated on a 4–12% gradient or on 8.5% low cross-linking (121:1, acrylamide:bis-acrylamide) SDS-PAGE gels (recipe provided by D. Chan and Dr. S. P. Lees-Miller, University of Calgary, Alberta, Canada). Proteins were transferred to Immobilin-P membranes (Millipore, Bedford, MA). The membranes were incubated with ATM antibodies at a 1:1500 dilution (final concentration between 0.3 and 0.4 μg/ml) in I-block (Tropix, Bedford, MA). After extensive washing with I-block, the membranes were incubated with a 1:30,000 dilution of alkaline phosphatase–conjugated goat anti-rabbit (Sigma, St. Louis, MO). The membranes were washed three times in I-block and three times in assay buffer (100 mM diethanolamine, pH 10, 1 mM MgCl2) and were incubated with CPD-Star chemiluminescent detection substrate (Tropix), according to the manufacturer’s protocol. ATM was visualized by exposing membranes to x-ray film. Antibodies to DNA-PK and Ku proteins were generously provided by Dr. S. P. Lees-Miller (University of Calgary). Antibodies to the ATM-related protein kinase ATR were a generous gift of Dr. M. Hoekstra (Icos, Bothell, WA).
For immunofluorescence experiments, cells were grown on glass coverslips, fixed with 3.7% paraformaldehyde in PBS for 5 min, and permeabilized with 0.1% Triton X-100 in PBS for 2 min. Coverslips were incubated with 1 μg/ml affinity-purified ATM IgG diluted in KB buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.1% BSA, 0.2% Triton X-100) for 1 h in a humidified 37°C incubator. The coverslips were washed three times for 5 min each in PBS and incubated with biotinylated-goat anti-rabbit IgG (Jackson ImmunoResearch Labs, West Grove, PA) diluted in KB buffer for 1 h at 37°C. The coverslips were washed in PBS and then incubated with UltraAvidin Texas-Red (Leinco Technologies, Baldwin, MO) in KB for 30 min at 37°C. The coverslips were mounted onto glass slides with VectaShield antifade (Vector Labs, Burlingame, CA), and the slides were visualized on a Nikon Microphot-SA with the use of a 100× Neofluor objective. Images were captured with a charge-coupled device MTI CCD72S camera (Dage MTI, Michigan City, IN) that was controlled by IP LabSpectrum software (Signal Analytics, Vienna, VA).
Subcellular Fractionation
Normal human fibroblasts and HeLa cells were fractionated as described (Frangioni et al., 1992). Asynchronously growing cells were swollen in hypotonic buffer (10 mM Tris, pH 7.4, 0.2 mM MgCl2, 5 mM KCl), dounce homogenized, and returned to isotonic conditions by the addition of sucrose and EDTA, and the nuclei were pelleted at 1000 × g for 5 min at 4°C. The supernatant was then centrifuged at 100,000 × g for 1 h at 4°C. The S100 fraction was removed and diluted in an equal volume of 2× RIPA (300 mM NaCl, 2% NP-40, 1% deoxycholate, 0.2% SDS, 100 mM Tris, pH 8.0). The membrane fraction (100,000 × g pellet) was resuspended in 1× RIPA buffer. Each fraction was brought to a final volume of 750 μl with 1× RIPA. Equal volumes (hence, proteins from equal cell numbers) of each fraction (whole cell, cytoplasm, nucleus, and membranes) were separated on 4–12% SDS-PAGE gels, and ATM was detected by Western blotting with Ab1 and was confirmed with Ab3.
Nuclear fractionation was performed as described previously with minor modifications (Staufenbiel and Deppert, 1984; Liao et al., 1995). Cells grown to 80% confluence were washed three times with PBS and lysed in KM buffer (10 mM N-morpholinoethanesulfonic acid, pH 6.2, 10 mM NaCl, 1.5 mM MgCl2, 10% glycerol) containing 1% digitonin, 1 mM EGTA, and 5 mM DTT. Nuclei were pelleted by centrifugation at 1000 × g, and the supernatant was saved. The nuclei were washed three times with KM buffer and then extracted with KM buffer containing 100 μg/ml DNase I. The nuclei were pelleted, and the supernatant was saved. The nuclei were then extracted with 2 M NaCl, 1 mM EGTA, and 5 mM DTT in KM buffer. The nuclei were again pelleted, and the supernatant was saved. Finally the nuclei were lysed in RIPA buffer, and the insoluble fraction was pelleted by centrifugation at 160,000 × g for 10 min at 4°C. The soluble fraction of the RIPA lysate and all the previously saved supernatants were diluted to 1 ml with a final concentration of 400 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, and 50 mM Tris, pH 8.0. ATM was immunoprecipitated with a 1:200 dilution (final concentration of ∼3 μg/ml) of Ab1, Ab3, or nonimmune rabbit IgG for 3 h at 4°C followed by incubation with 30 μl of a 50% slurry of Protein A sepharose (Sigma). The sepharose beads were pelleted and washed three times with RIPA buffer. Proteins were removed from the beads by adding 1× SDS sample buffer and boiling for 5 min. The proteins were separated on 8.5% low cross-linking gels as described above.
Protein Kinase Assays
Cells were lysed in PBS containing 10% glycerol and 1% NP-40 with protease and phosphatase inhibitors (10 mM NaF, 1 mM Na3VO4, 60 mM β-glycerophosphate, 100 nM microcystin), and the insoluble fraction was pelleted by centrifugation at 160,000 × g for 10 min at 4°C. Lysates were incubated with a 1:200 dilution (final concentration of ∼3 μg/ml) of nonimmune rabbit IgG, Ab1, Ab3, or peptide-blocked Ab3 for 3 h at 4°C followed by incubation with 30 μl of a 50% slurry of Protein A sepharose (Sigma). For peptide-blocking controls, Ab3 was incubated with a 10-fold molar excess of the R1821 fusion protein for 3 h before addition to the cell lysates. The immunocomplexes were pelleted and washed five times with lysis buffer and three times with protein kinase buffer (20 mM HEPES, pH 7.4, 10 mM MgCl2, 10 mM MnCl2, protease and phosphatase inhibitors). Kinase reactions were performed at 37°C by incubating the ATM-bound beads in 25 μl of protein kinase buffer supplemented with 50 μM ATP, 2 mM DTT, 1 μM protein kinase inhibitor (Sigma), 0.2 μg of single-stranded M13 DNA (ssDNA), 1 μg of HindIII-digested λ DNA (dsDNA), 50 μCi [γ-32P]ATP, and 0.5 μg of RPA (generous gifts of Dr. Marc Wold, University of Iowa, Ames, IA, and Dr. George Iliakis, Thomas Jefferson University, Philadelphia, PA). After 15 min, the ATP concentration was increased to 500 μM to drive the reaction to completion, and the reaction was stopped by the addition of SDS-PAGE–loading buffer. The proteins were separated on 12% SDS-PAGE gels, fixed, dried, and exposed to x-ray film.
Phosphoamino Acid and Phosphopeptide Analysis
Phosphorylated p34 RPA was excised from dried gels, minced with a razor blade, rehydrated in 50 mM NH4CO3, pH 8.05, with 10 μg of trypsin (Sigma), and digested 16 h at 37°C. To ensure complete cleavage, we added another 10 μg of trypsin and continued the incubation for another 3 h. Peptides were lyophilized and resuspended in 100 μl 6N HCl and boiled at 100°C for 1 h. The sample was frozen, lyophilized, resuspended in 50 μl of water that contained unlabeled phosphoamino acid standards, and then relyophilized. The sample was resuspended in 3 μl of water, spotted onto cellulose TLC (0.1-mm-thick) plates (VWR, Philadelphia, PA), and electrophoresed at 1300 V for 20 min in each direction in pH 3.5 buffer (5% glacial acetic acid, 0.5% pyridine) (van der Geer et al., 1993). After electrophoresis, the plates were sprayed with ninhydrin to stain phosphoamino acid standards. Radiolabeled phosphoamino acids were visualized after exposure to x-ray film.
Phosphorylated p34 RPA was obtained either by the in vitro protein kinase reactions described above or by immunoprecipitation of in vivo phosphate-labeled RPA. HeLa cells were grown to 80% confluence and starved in phosphate-free media for 30 min. The cells were then labeled with 1.0 mCi/ml [32P]orthophosphate for 2 h, irradiated with 10 Gy of IR, and lysed in RIPA buffer 1 h after irradiation. RPA was immunoprecipitated with a 1:100 dilution of mouse monoclonal anti-RPA antibody (a generous gift of Dr. Marc Wold, University of Iowa) followed by incubation with 30 μl of a 50% slurry of Protein G sepharose (Sigma). Proteins were separated on a 12% PAGE gel, dried, and exposed to x-ray film as above. Phosphopeptide mapping was performed by standard methods (van der Geer et al., 1993). The p34 RPA phosphorylated bands were cut out of the dried gel, removed from the drying paper, and minced. The minced gel slices were incubated for 24 h at 37°C with 10 μg of trypsin (Sigma) in 80 μl of 50 mM NH4HCO3, pH 8.0. Another 10 μg of trypsin was added, and the peptides were incubated for 3 h at 37°C. The gel fragments were pelleted and washed three times with water and once with methanol. The washes and original supernatant containing the phosphorylated peptides were lyophilized, resuspended in 15 μl of 25 mM Tris-HCl, pH 8.5, and 1 mM EDTA containing 0.02 μg/ml endoproteinase lys-C (sequencing grade) (Boehringer Mannheim, Indianapolis, IN), and incubated for 24 h at 37°C. Another 0.015 μg of endoproteinase lys-C was added to the peptides and incubated another 3 h at 37°C. The peptides were again lyophilized, resuspended in 3 μl of water, and loaded onto TLC (0.1-mm-thick) plates (VWR). The peptides were separated in the first dimension by electrophoresis for 1.5 h at 1000 V toward the cathode in pH 3.5 buffer The peptides were separated in the second dimension by ascending chromatography in 62.55% isobutyric acid, 4.8% pyridine, 2.9% acetic acid, 1.9% butanol, 27.85% water. The plates were dried, and the peptides were visualized by the use of a Fujix BAS 1000 PhosphorImager (Fuji Medical Systems, Stamford, CT).
RESULTS
Characterization of Antibodies Raised against ATM
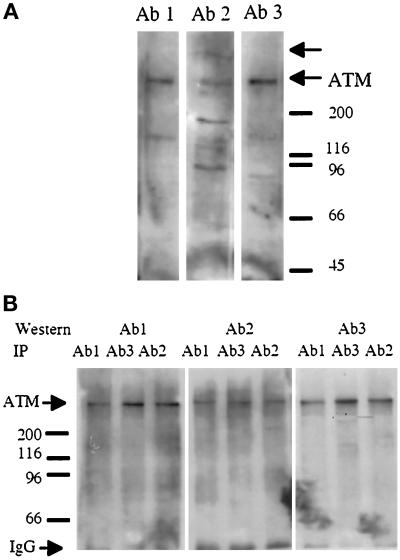
We raised three sets of rabbit polyclonal antibodies against nonoverlapping domains of ATM that were expressed as bacterial fusion proteins. All antibodies were affinity purified on columns that were coupled with the corresponding ATM fusion protein. Ab1 was raised against the RAD3 domain (aa 2138–2739), Ab2 was raised against the kinase domain (aa 2740–3056), and Ab3 was raised against a domain of unknown function near the N terminus (aa 247–521). In Western blots of HeLa whole-cell lysates that were separated on 4–12% gradient gels, all three antibodies recognized a protein of ∼350 kDa, the calculated size of ATM (Figure 1A). Additional bands identified by Ab2 are likely to be other proteins that contain a PI3-like kinase domain, such as the 450-kDa DNA-PK (Figure 1A, top arrow). The 350-kDa band was only detected by ATM antibodies because no signal was detected with preimmune IgG (our unpublished results). A 350-kDa protein was also detected in NHF, but this band was not detected in several fibroblast lines that were derived from AT-deficient individuals (see Figure 2B). To confirm further the specificity of the antibodies, we found each antibody to immunoprecipitate a ∼350-kDa band from HeLa extracts that was recognized by the other two antibodies on Western blots (Figure 1B). We also found that the immunoprecipitated 350-kDa protein was not DNA-PK or ATR because these two proteins were not detectable when the ATM immunoprecipitates were probed with DNA-PK and ATR antibodies (our unpublished results). The combined data demonstrate that the ∼350-kDa protein recognized by the three antibodies is ATM.
Figure 1.
Characterization of antibodies raised against ATM. (A) Three antibodies raised against ATM recognize a 350-kDa band in Western blots of HeLa whole-cell lysates (20 μg) that were separated on 4–12% gradient SDS-PAGE gels. (B) Each of the three ATM antibodies immunoprecipitated a 350-kDa band that is recognized by the other two antibodies on Western blots.
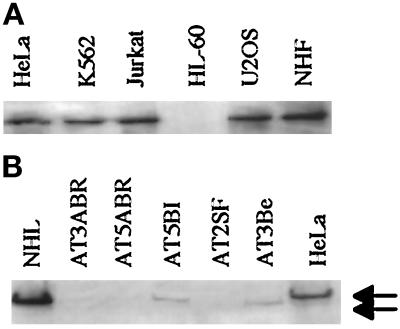
Figure 2.
Comparison of ATM protein levels in different cell lines. (A) Whole-cell lysates of 106 NHF and HeLa and U2OS cells and of 107 K562, Jurkat, and HL-60 cells probed with Ab3. (B) Whole-cell lysates (20 μg) of normal human lymphocytes (NHL) and of AT3ABR, AT5ABR, AT5BI, AT2SF, AT3Be, and HeLa cells separated and probed with Ab3. Top arrow, full-length ATM; bottom arrow, truncated ATM in AT3Be.
ATM Protein Varies among Different Cell Lines
We compared the ATM protein levels in various normal and transformed cell lines and in cell lines derived from AT patients. ATM was most abundant in the NHF and in transformed HeLa and U2OS cell lines. ATM protein levels in the K562 and Jurkat cell lines were 10–15% of that observed for an equivalent number of adherent cells that were examined (Figure 2A). Unexpectedly, ATM was not detectable in the highly radiosensitive HL-60 cells (Chapman, Fox Chase Cancer Center, personal communication). The molecular basis for the ATM deficiency in HL-60 cells is unclear. Of the five AT-defective cell lines examined in this study, the mutations for three of the lines are known. AT3ABR and AT5ABR have mutations that cause premature termination at amino acids 2755 and 2137, respectively (Shafman et al., 1997). AT5BI is a compound heterozygote in which each allele has a two-amino acid deletion (amino acids 1079–1080 and 2426–2427) (Savitsky et al., 1995a; Gilad et al., 1996). The mutations in AT3Be and AT2SF are unknown. AT3ABR, AT5ABR, and AT2SF cells did not express detectable amounts of ATM, whereas low levels of ATM were detected in the AT5BI mutant (Figure 2B, top arrow). Interestingly, the AT3Be mutant expressed a truncated peptide of ∼300 kDa (Figure 2B, bottom arrow) that was detected by Ab1 that was generated against amino acids 2138–2739 that span the RAD3-homology domain of ATM.
Subcellular Localization of ATM
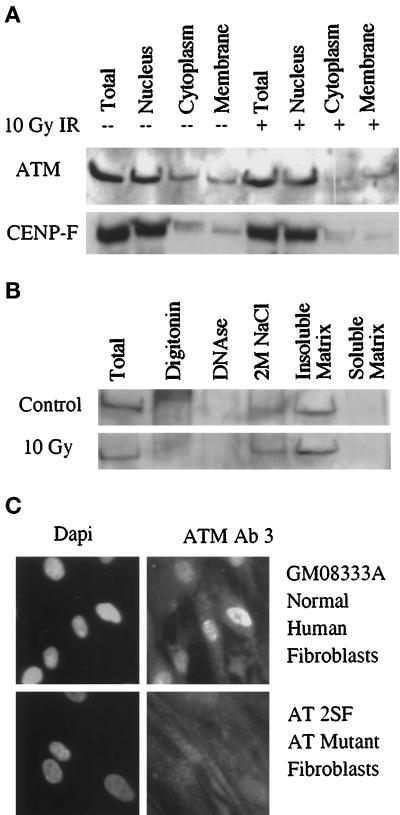
Because AT cell lines are defective in their response to DNA damage, we wanted to determine the subcellular localization of ATM to obtain clues as to its function. We initially undertook a biochemical approach by fractionating cells to examine the distribution of ATM in various subcellular compartments. When NHF were fractionated, ATM was found to be highly enriched in nuclei (Figure 3A, top). The purity of the subcellular fractions was determined by probing the Western blots with an antibody raised against the 367-kDa nuclear matrix protein CENP-F (Figure 3A, bottom) (Liao et al., 1995). Because CENP-F is predominantly a nuclear protein, these results indicate that there is slight but detectable contamination of the cytoplasmic and membrane fractions from the nuclear fraction (see below). IR had no significant effect on the distribution pattern of either protein.
Figure 3.
Subcellular localization of ATM. (A) Irradiated and unirradiated normal human fibroblasts fractionated as described in MATERIALS AND METHODS. Protein from unfractionated cells and from the nucleus, cytoplasm, and membrane fractions of an equal number of cells was probed with Ab3. Purity of the nuclear fraction was assessed by probing the same filter for the 367-kDa nuclear matrix protein CENP-F. (B) Subnuclear localization of ATM. NHF were sequentially extracted with 1% digitonin, 100 μg/ml DNase I, 2 M NaCl, and RIPA buffer (soluble matrix). ATM was immunoprecipitated with Ab1 from each of the soluble fractions that were adjusted to identical composition to minimize differences in immunoprecipitation efficiency. The insoluble pellet was directly dissolved in SDS sample buffer and loaded as the insoluble matrix. ATM was also immunoprecipitated from an equal number of NHF to serve as a control for the efficiency of ATM recovery (Total). The filter was probed with Ab3. (C) Immunofluorescence staining with Ab3 (right) of NHF (top) and AT2SF cell lines (bottom). Nuclei were visualized by DAPI staining (left).
To probe further the biochemical interactions of ATM in the nucleus, nuclei were separated into chromatin and nuclear matrix fractions (Staufenbiel and Deppert, 1984; Liao et al., 1995) and then probed for ATM (Figure 3B, top). Cytoplasmic proteins were released by extracting the cells with digitonin, which binds to cholesterol and disrupts the plasma membrane but leaves the nuclear membrane intact. This procedure was used to minimize the possibility that some ATM might leak out of the nucleus and contaminate the cytoplasmic fractions. ATM was not detected in the cytoplasmic fraction obtained by digitonin lysis (Figure 3B). Thus the cytoplasmic signal observed when cells were disrupted by dounce homogenization (Figure 3A) is likely caused by leakage of ATM from the nucleus. When nuclei were digested with DNase I, no detectable amount of ATM was released. A large fraction of ATM was solubilized when the nuclease-treated nuclei were extracted with 2 M NaCl. However, a substantial fraction of the total nuclear pool of ATM remained in the insoluble fraction that consists of the nuclear matrix (Figure 3B). Similar results were obtained with HeLa cells (our unpublished results). This extraction profile indicates that ATM is associated with the chromatin and the nuclear matrix. Because ATM is involved in the cellular response to DNA damage, we investigated whether the chromatin-associated fraction of ATM was altered after IR-induced damage. No quantitative changes in the subnuclear distribution of ATM were observed for normal human fibroblasts (Figure 3B, bottom) or HeLa cells (our unpublished results) 1 h after being exposed to 10 Gy of IR.
The subcellular localization of ATM was also examined by indirect immunofluorescence staining. Staining with antibodies raised against ATM confirmed the fractionation data because ATM was localized exclusively in the nucleus of NHFs (Figure 3C, top) and other human cell lines such as HeLa and U2OS (our unpublished results). This pattern was absent in the AT cell line AT2SF (Figure 3C, bottom), confirming the specificity of the antibodies. As above, the nuclear localization of ATM in NHF was not noticeably changed 1 h after exposure to 10 Gy of IR (our unpublished results). Because there is a weakly cross-reacting band at 120 kDa that was identified by Ab1 (Figure 1), these results were confirmed with Ab3 that had been preabsorbed with proteins from HeLa whole-cell lysates of <200 kDa.
ATM Protein Expression during the Cell Cycle and after Exposure to γ-Radiation
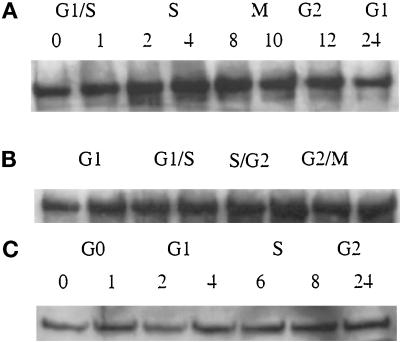
We next examined the expression pattern of ATM during the cell cycle in four different cell lines by the use of different synchronization methods. HeLa cells were synchronized at the G1–S boundary by a double thymidine block. At regular intervals after release from the G1–S block, equal cell numbers were analyzed for ATM content by Western blot. ATM levels did not fluctuate significantly during the cell cycle in HeLa cells (Figure 4A). To ensure that our finding was not an artifact produced by the method of synchronization or because of some unknown anomaly of HeLa cells, we repeated the experiment with Jurkat and K562 cells. These cells were enriched for discrete populations in different parts of the cell cycle by centrifugal elutriation. Consistent with the expression pattern in HeLa cells, ATM protein levels did not vary significantly during the cell cycle in elutriated K562 (Figure 4B) or Jurkat cells (our unpublished results). The modest increase in ATM levels in cells at late stages of the cell cycle is consistent with the increase in cell size. If the signal was corrected for equal protein, the level of ATM remains constant throughout the cell cycle. Because all of these cell lines are transformed, we investigated whether ATM protein levels might change after stimulation of serum-starved primary human fibroblasts. ATM protein levels were found to be insensitive to serum withdrawal or stimulation (Figure 4C). Using four different cell lines and three methods of synchronization, we determined that ATM is expressed throughout the cell cycle and its levels do not vary significantly at any point in the cell cycle.
Figure 4.
Comparison of the steady-state levels of ATM protein across the cell cycle. (A) Ab1 immunoblot of HeLa whole-cell lysates synchronized by double thymidine block and harvested at 1–24 h after release. (B) Ab1 immunoblot of K562 whole-cell lysates after separation by centrifugal elutriation. (C) Ab1 immunoblot of NHF serum starved for 24 h in 0.5% FBS and harvested at 1–24 h after stimulation with 20% FBS.
We examined the effects of γ-irradiation on the expression of ATM. HeLa cells were exposed to 2 Gy (IC50) or 10 Gy (IC99) of IR, and the steady-state levels of ATM were probed at various times after irradiation. Exposure to 2 or 10 Gy did not significantly alter the levels of ATM measured 1–24 h after irradiation (Figure 5, A and B). These experiments were repeated with both NHF (Figure 5C) and K562 cells (our unpublished results). ATM protein levels were unchanged 1–24 h after 2 or 10 Gy of IR in both of these cell lines.
Figure 5.
The effect of IR on expression of ATM protein. (A) Ab1 immunoblot of HeLa cells treated with 2 Gy of IR and harvested 1–24 h after treatment. (B) Ab1 immunoblot of HeLa cells treated with 10 Gy of IR and harvested 1–24 h after treatment. (C) Ab1 immunoblot of NHF treated with 10 Gy of IR and harvested 1–24 h after treatment.
ATM-associated Protein Kinase Activity
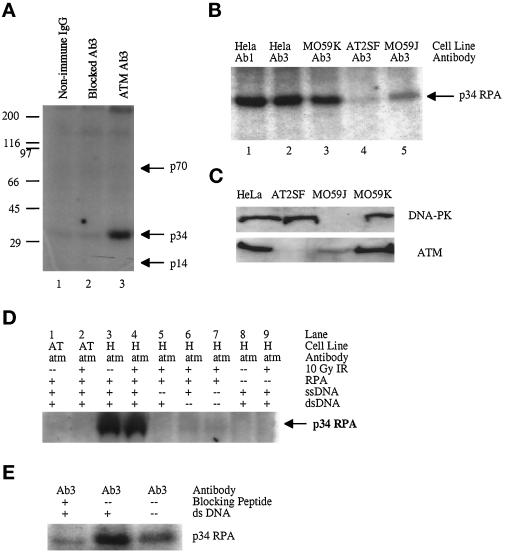
AT cells exhibit a decreased and delayed ability to phosphorylate the p34 subunit of the RPA complex (RPA p34) in response to IR (Liu and Weaver, 1993). To investigate whether p34 RPA might be an in vitro substrate for ATM, we immunoprecipitated ATM from HeLa cells that were exposed to 10 Gy of IR and tested the immunocomplex for its ability to phosphorylate p34 RPA. Because phosphorylation of p34 RPA is known to be stimulated when it is bound to ssDNA (Wold, 1997), circular M13 ssDNA was included in the kinase reaction. If ATM is important for recognizing dsDNA breaks that are induced by radiation, its protein kinase activity may only be stimulated under conditions that mimic the radiation-induced DNA damage response. We therefore supplemented the kinase reaction mix with HindIII-digested λ DNA that consisted of a mixture of different-sized linear dsDNAs (500 bp to >20 kb). In the presence of ssDNA and linear dsDNA, the ATM immunoprecipitate specifically phosphorylated the p34 subunit of RPA, whereas the p14 and p70 subunits were not phosphorylated (Figure 6A, lane 3). The RPA protein kinase activity was specific for the ATM immunocomplex because RPA remained unphosphorylated when the kinase reaction was performed with immunoprecipitates obtained with nonimmune rabbit IgG or peptide-blocked Ab3 (Figure 6A, lanes 1 and 2). This protein kinase activity was observed when ATM was immunoprecipitated with both Ab1 and Ab3 (Figure 6B, lanes 1 and 2). Moreover, ATM is required for the protein kinase activity because immunoprecipitates from AT2SF cells that lacked detectable levels of ATM also failed to phosphorylate RPA (Figure 6B, lane 4). Because p34 RPA is known to be phosphorylated by DNA-PK/Ku and cdc2 (Niu et al., 1997), ATM immunoprecipitates from HeLa cells were probed with antibodies to these protein kinases. We were unable to detect coimmunoprecipitating DNA-PK/Ku, cdc2, or cdk2 (our unpublished results). To eliminate the possibility that the observed DNA-dependent protein kinase activity is caused by contaminating DNA-PK activity, we performed these kinase assays in human MO59J glioblastoma cells that have been shown to contain no DNA-PK (Allalunis-Turner et al., 1995). As shown in Figure 6C, HeLa, MO59K, and AT2SF all contain approximately equal levels of DNA-PK, whereas MO59J cells contain no detectable DNA-PK. Interestingly, as reported previously, the MO59J cells also contain decreased levels of ATM (Chan et al., 1998). Although HeLa and MO59K cells immunoprecipitated high levels of protein kinase activity (Figure 6B, lanes 1–3), consistent with the lower steady-state level of ATM in the MO59J cells, the ATM-associated protein kinase activity was significantly lower relative to that in cell lines that contain higher steady-state levels of ATM (Figure 6B, lane 5).
Figure 6.
Phosphorylation of p34 RPA by ATM-associated protein kinase. All kinase reactions were performed with immunoprecipitates from equal amounts of cellular protein. (A) ATM immunoprecipitated from HeLa cells with nonimmune rabbit IgG (Nonimmune IgG, lane 1), peptide-blocked Ab3 (Blocked, lane 2), or Ab3 (ATM Ab3, lane 3) and incubated with [γ-32P]ATP, ssDNA, dsDNA, and purified RPA. (B) ATM protein kinase activity immunoprecipitated from HeLa (lanes 1 and 2), MO59K (lane 3), AT2SF (lane 4), and MO59J (lane 5) cells. (C) Lysate (50 μg) from HeLa (lane 1), AT2SF (lane 2), MO59J (lane 3), and MO59K (lane 4) cells probed with antibodies to DNA-PK (top) or ATM Ab3 (bottom). (D) ATM-associated protein kinase activity in AT2SF cells (AT) (lanes 1 and 2) or in HeLa cells (H) (lanes 3–9) with purified RPA used as a substrate (lanes 1–7). (E) Kinase reactions performed on immunoprecipitates from MO59J cells with blocked Ab3 (lane 1) and Ab3 (lanes 2 and 3) in the presence (lanes 1 and 2) or absence (lane 3) of dsDNA.
Although the initial kinase assays were performed with extracts prepared from irradiated cells, we found that ATM immunoprecipitates obtained from unirradiated cells phosphorylated p34 RPA as efficiently as those seen for irradiated samples as long as linear dsDNA was included in the kinase reaction (Figure 6D, compare lanes 3 and 4). Consistent with the finding that p34 RPA phosphorylation is stimulated by ssDNA, no phosphorylation of p34 RPA by the ATM immunocomplex was detected when ssDNA was omitted from the reaction mix (Figure 6D, lane 5). Interestingly, only background levels of p34 RPA phosphorylation were observed when linear dsDNA was omitted from the reaction (Figure 6D, lane 7). Consistent with the results obtained from HeLa cells, the protein kinase activity immunoprecipitated from MO59J cells was also stimulated by dsDNA (Figure 6E, compare lanes 2 and 3).
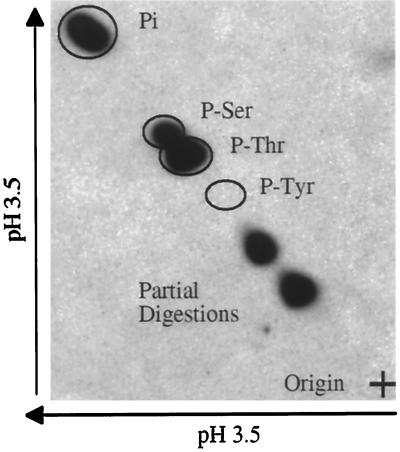
Phosphoamino acid analysis of the phosphorylated p34 RPA revealed that the ATM-associated protein kinase phosphorylated serine and threonine residues, but no detectable phosphotyrosine was observed (Figure 7). The combined data show that p34 RPA phosphorylation was mediated by a DNA-dependent serine and threonine protein kinase that is present specifically in the ATM immunocomplex.
Figure 7.
ATM-associated protein kinase phosphorylation of p34 RPA on serine and threonine residues. Two-dimensional phosphoamino acid analysis of phosphorylated p34 RPA is shown.
Phosphopeptide Mapping of p34 RPA Phosphorylated by the ATM-associated Protein Kinase Activity
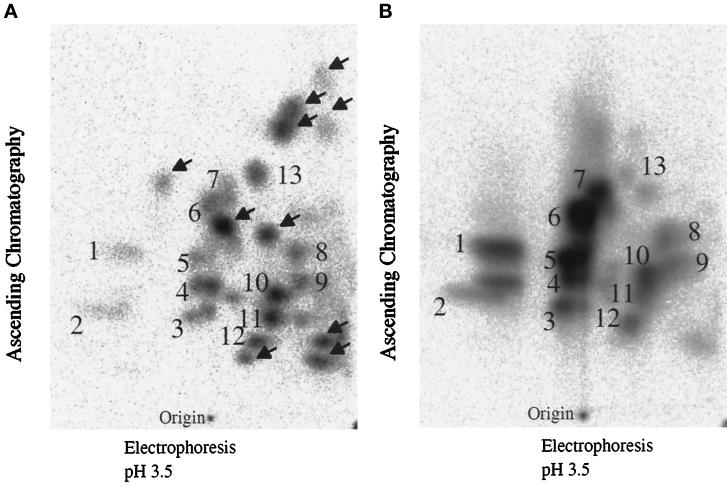
To determine whether the phosphorylation of p34 RPA by the ATM-associated protein kinase occurs at physiologically relevant sites, we compared the phosphopeptide maps of p34 RPA phosphorylated in vitro by the ATM-associated protein kinase and in vivo after exposure to IR. The majority of the phosphopeptides observed in the in vivo sample were also observed when p34 RPA was phosphorylated by the ATM-associated protein kinase activity (Figure 8B, numbered areas). The additional spots present in the in vivo phosphopeptide map (Figure 8A, arrows) may be caused by other protein kinases that also phosphorylate p34 RPA or by minor contaminating phosphoproteins that coimmunoprecipitated and comigrated with p34 RPA.
Figure 8.
Phosphopeptide maps of p34 RPA phosphorylated in vitro by ATM-associated protein kinase and in vivo in response to IR. In vivo–phosphorylated p34 RPA (A) and in vitro–phosphorylated p34 RPA (B) were digested with trypsin and endoproteinase lys-C and separated by electrophoresis in pH 3.5 buffer in the first dimension and ascending chromatography in the second dimension. Numbered areas correspond to phosphopeptides that are common in the two maps; arrows in A identify peptides that are specific to the in vivo sample.
DISCUSSION
We raised and affinity purified three sets of rabbit polyclonal antibodies against ATM that specifically recognize ATM by Western blotting, immunoprecipitation, and immunofluorescence. These antibodies specifically identified an ∼350-kDa protein, the predicted size of ATM, on Western blots. Each antibody immunoprecipitated a 350-kDa protein that is recognized by the other two antibodies on Western blots. Because this protein contains epitopes that are recognized by three antibodies that were raised against nonoverlapping regions of ATM, this protein must be ATM or a closely related protein. Because these antibodies failed to detect a 350-kDa protein in the majority of AT cell lines examined, the protein identified in the non-AT cell lines must be ATM. These findings are consistent with those of other groups that have raised antibodies against ATM (Chen and Lee, 1996; Lakin et al., 1996; Brown et al., 1997). Finally, the protein that was immunoprecipitated by these ATM antibodies was not recognized by antibodies directed against DNA-PK or to ATR, a ATM-related protein kinase (Cimprich et al., 1996; Keegan et al., 1996).
ATM protein levels were found to vary considerably among different cell lines, ranging from undetectable in many AT cell lines and HL-60 cells to high levels in HeLa, U2OS, and normal human fibroblasts. Although cells with no detectable ATM (AT cell lines and HL-60) are extremely sensitive to IR, the steady-state protein levels of ATM (measured per cell or per milligram of total protein) did not correlate with radioresistance as measured by the surviving fraction at 2 Gy (Chan et al., 1998). Similar findings have been reported for DNA-PK (Allalunis-Turner et al., 1995), suggesting that the amount of these proteins may not be rate limiting in the cellular DNA damage response and that other factors may contribute toward the overall radiosensitivity of a cell.
ATM steady-state protein levels did not vary significantly during the cell cycle as measured in four different cell lines synchronized by three different procedures. These quantitative data extend the findings of Brown et al. (1997) who used immunofluorescence staining to demonstrate that ATM is present throughout the cell cycle. Consistent with data reported previously by others (Lakin et al., 1996; Brown et al., 1997), ATM protein levels were unchanged from 1 to 24 h after exposure to IR. This lack of observable changes in ATM in response to DNA damage is in marked contrast to the changes in p53 and other proteins that are induced upon DNA damage. The constitutive expression of ATM during the cell cycle is consistent with its role as a sensor of DNA damage.
Others have demonstrated that ATM is a nuclear protein (Chen and Lee, 1996; Lakin et al., 1996; Brown et al., 1997) and that ATM colocalizes with chromatin-associated proteins on meiotic chromosomes (Keegan et al., 1996; Plug et al., 1997). However, our data provide the first biochemical evidence that ATM is associated with chromatin in somatic cells. Subcellular fractionation of NHF and HeLa cells showed that ATM is a nuclear protein that was partially solubilized after nuclei were digested with DNase I and extracted with high salt. This property suggests that ATM is associated with chromatin where it may monitor DNA damage and activate the DNA damage response pathway.
In three of the five AT cell lines examined, ATM was not detected even though the antibodies recognized epitopes that lie upstream of the mutations that prematurely truncated the protein. Although the low sample number prevents any firm conclusion, it is interesting to speculate that elements near the carboxyl terminus of ATM are important for its stability. If these elements are deleted, the protein becomes unstable and is degraded. This may explain the high incidence of null mutations observed in samples from AT patients, as even short carboxyl-terminal truncations would be predicted to destabilize ATM and thus produce a null phenotype (Gilad et al., 1996).
Two of the five AT cell lines examined expressed detectable amounts of ATM protein. The molecular nature of the AT3Be mutant has not been identified. However, our antibodies identified an ∼300-kDa protein that we believe is the mutant AT3Be protein. On the basis of our hypothesis that mutations near the carboxyl terminus destabilize ATM, we predict that AT3Be has an in-frame deletion that does not disrupt the putative stabilization domain. Because AT3Be was recognized by antibodies raised against amino acids 2138–2739 within the RAD3-like domain, the deletion is predicted to lie upstream of this domain.
The compound heterozygote AT5BI has a different two-amino acid deletion in each of its ATM alleles (amino acids 1079–1080 and 2426–2427) (Savitsky et al., 1995a; Gilad et al., 1996). Even though AT5BI cells accumulated detectable levels of ATM protein, they nevertheless exhibit an AT phenotype that consists of radiation sensitivity and the failure to activate cell cycle checkpoints. Lakin et al. (1996) have also found AT patients in which small deletions lead to detectable protein levels. Thus, these mutant alleles must define a critical functional domain within a region of ATM that shares no significant similarities with other known proteins.
We demonstrate that ATM immunocomplexes from HeLa cells phosphorylated p34 RPA. Because neither the p14 nor the p70 subunits of RPA were phosphorylated in these assays, the ATM-associated protein kinase activity exhibits substrate specificity. The ATM-associated protein kinase activity could not be attributed to some contaminating protein kinase that nonspecifically coprecipitated with ATM because no protein kinase activity was observed in ATM immunoprecipitates derived from AT cells or in immunoprecipitates obtained from HeLa lysates with nonimmune IgG or when the ATM antibodies were blocked with excess peptide. Although DNA-PK and cdc2 kinase have been shown to phosphorylate p34 RPA in vitro (Fried et al., 1996), we are confident that this activity is not caused by contaminating DNA-PK or cdc2 because neither DNA-PK, the Ku proteins, cdc2, nor cdk2 coimmunoprecipitated with ATM. Moreover, in MO59J cells that lack the catalytic subunit of DNA-PK, ATM-associated protein kinase was still detected, even though this cell line expressed lower levels of ATM per cell than did HeLa or MO59K cells. The combined data suggest that either ATM or a tightly associated protein kinase is responsible for the phosphorylation of p34 RPA in vitro.
AT cells are defective in their ability to phosphorylate the p34 subunit of RPA in response to IR (Liu and Weaver, 1993). Moreover, this defect is also observed in yeast with mutations in MEC1, a yeast homologue of ATM (Brush et al., 1996). We demonstrate that the ATM-associated protein kinase phosphorylates p34 RPA on peptides that are phosphorylated in vivo in response to IR. This indicates that the ATM-associated protein kinase may be responsible for the phosphorylation of p34 RPA in response to IR. Interestingly, phosphorylation of RPA in vitro by the ATM-associated protein kinase activity induces a shift of mobility on SDS-PAGE gels similar to that observed after IR-treatment in vivo (our unpublished results). Although the use of different proteases precludes direct comparison of the phosphopeptide data, the number of residues in p34 RPA that were phosphorylated in response to UV irradiation is similar to our in vitro and in vivo findings (Zernik-Kobak et al., 1997). Ongoing work will determine whether the peptides phosphorylated by the ATM-associated protein kinase are the same as those phosphorylated in response to UV irradiation.
Our data support the hypothesis that p34 RPA is an in vivo substrate for the ATM-associated protein kinase and are consistent with the recent findings of Plug et al. (1997) who demonstrated that ATM and RPA colocalize on synapsed meiotic chromosomes. However, our data seem to be inconsistent with the finding that overexpression of the ATM kinase domain in AT cells corrected the radiosensitivity of these cells and yet failed to hyperphosphorylate the p34 subunit of RPA (Morgan and Kastan, 1997). The discrepancy can be accounted for in several ways; as suggested by the authors, the overexpressed kinase domain may not correct all of the phenotypes associated with defective ATM (Morgan and Kastan, 1997; Morgan et al., 1997). Furthermore, they suggested that the phosphorylation of p34 RPA can be dissociated from radiation sensitivity. Perhaps the overexpression of an unregulated kinase can restore radioresistance through alternative pathways that do not lead to the hyperphosphorylation of p34 RPA. This possibility is supported by the recent finding that overexpression of the ATR kinase domain can also correct radiosensitivity in AT cells (Cliby et al., 1998). However, the effects of overexpression of the ATR kinase domain on the phosphorylation of p34 RPA were not investigated in this report.
In contrast to the ATM kinase overexpression studies, when the leucine zipper region of ATM was overexpressed in RKO cells, it increased radiosensitivity in RKO cells even though p34 RPA was phosphorylated in response to IR (Morgan and Kastan, 1997). Although these experiments were designed so that the transfected leucine zipper region would interfere with endogenous ATM function, whether the effects obtained with this mutant were mediated via ATM is unknown. Even if this was the case, it is possible that ATM functions that are important for radioresistance were disrupted, and yet the ability to phosphorylate p34 RPA was not effected. A second possibility is that although the predicted mobility shift of p34 was observed in the RKO cells containing the leucine zipper construct, the phosphorylated sites may be different from those normally seen when ATM function is intact.
The ATM-associated protein kinase appears to be in a protein complex. In HeLa cells labeled with [32P]orthophosphate or [35S]methionine, we have observed six coimmunoprecipitating bands that are not observed in AT2SF cells (our unpublished data). Moreover, gel filtration data show that ATM exists in a protein complex of over two million daltons (our unpublished data). Similar unpublished findings have been reported by Chen and Lee (1996). Therefore, we cannot eliminate the possibility that a protein that coimmunoprecipitates with ATM, and not ATM itself, is the protein kinase responsible for the phosphorylation of p34 RPA. Even if ATM was the protein kinase that directly phosphorylates p34 RPA, we do not know whether the protein kinase activity depends on other associated subunits. Along this line, the ATM-associated protein kinase activity is eliminated when the immunoprecipitates were washed in either RIPA buffer or in kinase buffer supplemented with 1 M urea (our unpublished results).
The ATM-associated protein kinase was found to phosphorylate p34 RPA only when both ssDNA and linear dsDNA were present. Although we suspect that the requirement for ssDNA is specific to the p34 RPA substrate (Wold, 1997), we speculate that the requirement for linear dsDNA may mimic the radiation-induced DNA damage response by ATM. This possibility is supported by the finding that the addition of dsDNA to ATM immunocomplexes obtained from nonirradiated HeLa cells was sufficient to induce phosphorylation of p34 RPA to the same extent as that seen with parallel irradiated samples. Whether ATM directly binds to DNA or DNA binding is mediated via an associated subunit in a manner analogous to that for Ku and DNA-PK remains to be clarified. It is interesting that the ATM kinase domain alone does not exhibit a requirement for DNA when it was used to phosphorylate c-Abl in vitro (Baskaran et al., 1997). These data suggest that regions outside of the ATM kinase domain are responsible for the DNA dependence of ATM-associated protein kinase activity. Although Keegan et al. (1996) have observed that ATM is autophosphorylated in the absence of ssDNA and dsDNA, further work will be needed to determine whether autokinase activity is affected by the addition of exogenous DNA.
The finding that ATM immunoprecipitates obtained from irradiated cells exhibited no protein kinase activity until exogenous dsDNA was added to the reaction suggests that irradiation per se does not induce some stable modification that activates and maintains the ATM protein kinase activity during preparation of the cell extract and immunoprecipitation. These data suggest that the ATM protein kinase activity might have been inactivated when it was no longer exposed to damaged DNA that was present in the irradiated cell. If ATM protein kinase activity is critical for checkpoint-mediated cell cycle arrest, a mechanism must exist to turn off the protein kinase once the damage DNA is repaired so that the cells can resume their cell cycle. Direct coupling of the ATM protein kinase activity to damaged DNA would provide a sensitive method to inactivate the protein kinase and release arrested cells after their DNA is repaired.
ACKNOWLEDGMENTS
The authors thank Alec Belman for cloning the ATM cDNA, Dr. Sandra Jablonski for critical reading of the manuscript, Dr. Tim Shafman for cell lines, Dr. Susan Lees-Miller for DNA-PK and Ku antibodies, Dr. M. Hoekstra for antibodies to ATR, Dr. Marc Wold for purified human RPA and antibodies to RPA, Dr. Joan Turner for MO59 cells, and Dr. George Iliakis for MO59 cells and purified human RPA. D.P.G. is supported by grant CA-09035–21 from the National Institutes of Health. T.J.Y. is a scholar of the Leukemia Society of America and is supported by core grant CA06927 from the National Cancer Institute and by an appropriation from the Commonwealth of Pennsylvania.
Abbreviations used:
- AT
ataxia telangiectasia
- ATM
ataxia telangiectasia–mutated gene
- ATR
ATM-related protein kinase
- BSA
bovine serum albumin
- cdk2
cyclin-dependent kinase 2
- dsDNA
double-stranded DNA
- DTT
dithiothreitol
- FBS
fetal bovine serum
- GST
glutathione S-transferase
- IgG
immunoglubulin G
- IR
ionizing radiation
- MBP
maltose-binding protein
- NHF
normal human fibroblasts
- NP-40
Nonidet P-40
- nt
nucleotide
- PBS
phosphate-buffered saline
- PI3
phosphatidylinositol 3
- RIPA
radioimmunoprecipitation assay
- RPA
replication protein A
- ssDNA
single-stranded DNA
REFERENCES
- Allalunis-Turner MJ, Lintott LG, Barron GM, Day RS, Lees-Miller SP. Lack of correlation between DNA-dependent kinase activity and tumor cell radiosensitivity. Cancer Res. 1995;55:5200–5202. [PubMed] [Google Scholar]
- Baskaran R, et al. Ataxia telangiectasia mutant protein activates c-Abl tyrosine kinase in response to ionizing radiation. Nature. 1997;387:516–519. doi: 10.1038/387516a0. [DOI] [PubMed] [Google Scholar]
- Beamish H, Lavin MF. Radiosensitivity in ataxia-telangiectasia: anomalies in radiation-induced cell cycle delay. Int J Radiat Biol. 1994;65:175–184. doi: 10.1080/09553009414550211. [DOI] [PubMed] [Google Scholar]
- Bentley NJ, Holtzman DA, Flaggs GA, Keegan KS, DeMaggio A, Ford JC, Hoekstra M, Carr AM. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- Brown KD, Ziv Y, Sadanandan SN, Chessa L, Collins FS, Shiloh Y, Tagle DA. The ataxia-telangiectasia gene product, a constitutively expressed nuclear protein that is not up-regulated following genome damage. Proc Natl Acad Sci USA. 1997;94:1840–1845. doi: 10.1073/pnas.94.5.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush GS, Morrow DM, Hieter P, Kelly TJ. The ATM homologue MEC1 is required for phosphorylation of replication protein A in yeast. Proc Natl Acad Sci USA. 1996;93:15075–15080. doi: 10.1073/pnas.93.26.15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, D.W., Gately, D.P., Urban, S., Galloway, A.M., Lees-Miller, S.P., Yen, T.J., and Allalunis-Turner, J. (1998). Lack of correlation between ATM protein expression and tumor cell radiosensitivity. Int. J. Radiat. Biol. (in press). [DOI] [PubMed]
- Chen G, Lee EY-HP. The product of the ATM gene is a 370 kDa nuclear phosphoprotein. J Biol Chem. 1996;271:33693–33697. doi: 10.1074/jbc.271.52.33693. [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Shin TB, Leith CT, Schreiber SL. cDNA cloning and gene mapping of a candidate human cell cycle checkpoint protein. Proc Natl Acad Sci USA. 1996;93:2850–2855. doi: 10.1073/pnas.93.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliby WA, Roberts CJ, Cimprich KA, Stringer CM, Lamb JR, Scheiber SL, Friend SH. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzer KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- Frangioni JV, Beahm PH, Shifrin V, Jost CA, Neel BG. The nontransmembrane tyrosine phosphotase PTP-1B localizes to the endoplasmic reticulum via its 35 amino acid C-terminal sequence. Cell. 1992;68:545–560. doi: 10.1016/0092-8674(92)90190-n. [DOI] [PubMed] [Google Scholar]
- Fried LM, et al. The DNA damage response in DNA-dependent protein kinase-deficient SCID mouse cells: replication protein A hyperphosphorylation and p53 induction. Proc Natl Acad Sci USA. 1996;93:13825–13830. doi: 10.1073/pnas.93.24.13825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad S, et al. Predominance of null mutations in ataxia-telangiectasia. Hum Mol Genet. 1996;5:433–439. doi: 10.1093/hmg/5.4.433. [DOI] [PubMed] [Google Scholar]
- Greenwell PW, Kronmal SL, Porter SE, Gassenhuber J, Obermaier B, Petes TD. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell. 1995;82:823–829. doi: 10.1016/0092-8674(95)90479-4. [DOI] [PubMed] [Google Scholar]
- Hari KL, Santerre A, Sekelsky JJ, McKim KS, Boyd JB, Hawley RS. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell. 1995;82:815–821. doi: 10.1016/0092-8674(95)90478-6. [DOI] [PubMed] [Google Scholar]
- Hartley KO, Gell D, Smith GC, Zhang H, Divecha N, Connelly MA, Admon A, Lees-Miller SP, Anderson CW, Jackson SP. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell. 1995;8:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- Houldsworth J, Lavin MF. Effects of ionizing radiation on DNA synthesis in ataxia telangiectasia cells. Nucleic Acids Res. 1980;8:3709–3720. doi: 10.1093/nar/8.16.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. When is a lipid kinase not a lipid kinase? When it is a protein kinase. Cell. 1995;83:1–4. doi: 10.1016/0092-8674(95)90225-2. [DOI] [PubMed] [Google Scholar]
- Jackson SP. The recognition of DNA damage. Curr Opin Genet Dev. 1996;1:19–25. doi: 10.1016/s0959-437x(96)90005-2. [DOI] [PubMed] [Google Scholar]
- Jung M, Kondratyev A, Lee SA, Dimtchev A, Dritschilo A. ATM gene product phosphorylates IkB-a. Cancer Res. 1997;57:24–27. [PubMed] [Google Scholar]
- Kastan MB, Onyekwere O, Sidransky D, Vogelstein B, Craig RW. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- Kastan MB, Zhan Q, El-Deiry WS, Carrier F, Jacks T, Walsh WV, Plankett BS, Vogelstein B, Fornace AJ. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- Kauffman MG, Noga SJ, Kelly TJ, Donnenberg AD. Isolation of cell cycle fractions by counterflow centrifugal elutriation. Anal Biochem. 1990;191:41–46. doi: 10.1016/0003-2697(90)90384-l. [DOI] [PubMed] [Google Scholar]
- Keegan KS, et al. The Atr and Atm protein kinases associate with different sites along meiotically pairing chromosomes. Genes Dev. 1996;10:2423–2437. doi: 10.1101/gad.10.19.2423. [DOI] [PubMed] [Google Scholar]
- Lakin ND, Weber P, Stankovic T, Rottinghaus ST, Taylor AMR, Jackson SP. Analysis of the ATM protein in wild-type and ataxia telangiectasia cells. Oncogene. 1996;13:2707–2716. [PubMed] [Google Scholar]
- Lavin MF, Khanna KK, Beamish H, Sping K, Watters D. Relationship of the ataxia-telangiectasia protein ATM to phosphoinositide 3-kinase. Trends Biochem Sci. 1995;20:382–383. doi: 10.1016/s0968-0004(00)89083-0. [DOI] [PubMed] [Google Scholar]
- Liao H, Winkfein RJ, Mack G, Rattner JB, Yen TJ. CENP-F is a protein of the nuclear matrix that assembles onto kineticores at late G2 and is rapidly degraded after mitosis. J Cell Biol. 1995;130:507–518. doi: 10.1083/jcb.130.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu VF, Weaver DT. The ionizing radiation-induced replication protein A phosphorylation response differs between ataxia telangiectasia and normal human cells. Mol Cell Biol. 1993;13:7222–7231. doi: 10.1128/mcb.13.12.7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyn MS. Ataxia-telangiectasia and cellular responses to DNA damage. Cancer Res. 1995;55:5991–6001. [PubMed] [Google Scholar]
- Morgan SE, Kastan MB. Dissociation of radiation-induced phosphorylation of replication protein A from S-phase checkpoint. Cancer Res. 1997;57:3386–3389. [PubMed] [Google Scholar]
- Morgan SE, Lovly C, Pandita TJ, Shiloh Y, Kastan MB. Fragments of ATM which have dominant-negative or complimenting activity. Mol Cell Biol. 1997;17:2020–2029. doi: 10.1128/mcb.17.4.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow DM, Tagle DA, Shiloh Y, Collins FS, Hieter P. Tel1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint control gene MEC1. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- Niu H, Erdjument-Bromage H, Pan Z, Lee S, Tempst P, Hurwitz J. Mapping of amino acid residues in the p34 subunit of human single-stranded DNA-binding protein phosphorylated by DNA-dependent protein kinase and Cdc2 kinase in vitro. J Biol Chem. 1997;272:12634–12641. doi: 10.1074/jbc.272.19.12634. [DOI] [PubMed] [Google Scholar]
- Paules RS, Levecakou EN, Wilson SJ, Innes CL, Rhodes N, Tlsty TD, Galloway DA, Donehower LA, Tainsky MA, Kaufman WK. Defective G2 checkpoint function from individuals with familial cancer syndromes. Cancer Res. 1995;55:1763–1773. [PubMed] [Google Scholar]
- Plug AW, Peters AHFM, Xu Y, Keegan KS, Moekstra MF, Baltimore D, de Boer P, Ashley T. ATM and RPA in meiotic chromosome synapsis and recombination. Nat Genet. 1997;17:457–461. doi: 10.1038/ng1297-457. [DOI] [PubMed] [Google Scholar]
- Savitsky K, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995a;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- Savitsky K, Sfez S, Tagle DA, Ziv Y, Sartiel A, Collins FS, Shiloh Y, Rotman G. The complete sequence of the coding region of the ATM gene reveals similarity to cell cycle regulators in different species. Hum Mol Genet. 1995b;4:2025–2032. doi: 10.1093/hmg/4.11.2025. [DOI] [PubMed] [Google Scholar]
- Shafman T, et al. Interaction between ATM protein and c-Abl in response to DNA damage. Nature. 1997;287:520–523. doi: 10.1038/387520a0. [DOI] [PubMed] [Google Scholar]
- Shafman TD, Saleem A, Kryiakis J, Weichselbaum R, Kharbanda S, Kufe DW. Defective induction of stress-activated protein kinase activity in ataxia-telangiectasia cells exposed to ionizing radiation. Cancer Res. 1995;55:3242–3245. [PubMed] [Google Scholar]
- Staufenbiel M, Deppert W. Preparation of nuclear matrices from cultured cells: subfractionation of nuclei in situ. J Cell Biol. 1984;98:1886–1894. doi: 10.1083/jcb.98.5.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift M, Morrell D, Massey RB, Chase CL. Incidence of cancer in 161 families affected by ataxia-telangiectasia. N Engl J Med. 1991;325:1831–1836. doi: 10.1056/NEJM199112263252602. [DOI] [PubMed] [Google Scholar]
- Taylor AMR, Byrd PJ, McConville CM, Thacker S. Genetic and cellular features of ataxia telangiectasia. Int J Radiat Biol. 1994;65:65–70. doi: 10.1080/09553009414550091. [DOI] [PubMed] [Google Scholar]
- van der Geer P, Luo K, Sefton BM, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis on cellulose thin-layer plates. In: Hardie DG, editor. Protein Phosphorylation. Oxford: Oxford University Press; 1993. pp. 31–59. [Google Scholar]
- Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. In: Richardson CC, editor. Annual Review of Biochemistry. Palo Alto, CA: Annual Reviews; 1997. pp. 61–92. [DOI] [PubMed] [Google Scholar]
- Zakian VA. ATM-related genes: what do they tell us about the functions of the human gene? Cell. 1995;82:685–687. doi: 10.1016/0092-8674(95)90463-8. [DOI] [PubMed] [Google Scholar]
- Zernik-Kobak M, Vasunia K, Connelly M, Anderson CW, Dixon K. Sites of UV-induced phosphorylation of the p34 subunit of replication protein A from HeLa cells. J Biol Chem. 1997;272:23896–23904. doi: 10.1074/jbc.272.38.23896. [DOI] [PubMed] [Google Scholar]