Abstract
Little is known about the protein complexes required for microRNA formation and function. Here we used native gel electrophoresis to identify miRNA ribonucleoprotein complexes (miRNPs) in Caenorhabditis elegans. Our data reveal multiple distinct miRNPs that assemble on the let-7 miRNA in vitro. The formation of these complexes is affected but not abolished by alg-1 or alg-2 null mutations. The largest complex (M*) with an estimated molecular mass of >669 kDa cofractionates with the known RISC factors ALG-1, VIG-1, and TSN-1. The M* complex and two complexes, M3 and M4, with similar molecular weights of ∼500 kDa, also assemble on all other miRNAs used in our experiments. Two smaller complexes, M1 (∼160 kDa) and M2 (∼250 kDa), assemble on the members of the let-7 miRNAs family but not lin-4 or mir-234, and their formation is highly dependent on specific sequences in the 5′ seed region of let-7. Moreover, an unidentified protein, p40, which only appears in the M1 and M2 complexes, was detected by UV triggered cross-linking to let-7 but not to lin-4. The cross-linking of p40 to let-7 is also dependent on the let-7 sequence. Another unidentified protein, p13, is detected in all let-7 binding complexes and lin-4 cross-linked products. Our data suggest that besides being present in certain large miRNPs with sizes similar to reported RISC, the let-7 miRNA also assembles with specific binding proteins and forms distinct small complexes.
Keywords: miRNA, siRNA, let-7, RISC
INTRODUCTION
MicroRNAs (miRNAs) belong to a class of noncoding small RNAs and have been shown to regulate gene expression through repressing translation and/or cleaving mRNAs (Bartel 2004; Valencia-Sanchez et al. 2006). let-7 is one of the founding members of the miRNA family and was identified through genetic mutation as a temporal patterning regulator in Caenorhabditis elegans (Reinhart et al. 2000; Ambros 2001; Banerjee and Slack 2002; Grosshans and Slack 2002; Pasquinelli and Ruvkun 2002). Previous studies indicated that in C. elegans, let-7 regulates lin-41 and let-60 by binding to sequences in the 3′-untranslated regions (3′-UTRs) of these mRNAs, which are partially complementary to let-7 (Lee et al. 1993; Wightman et al. 1993; Slack et al. 2000; Vella et al. 2004). However, the mechanism of action of miRNAs, including let-7, in regulating gene expression is still not clear. let-7 is a useful model for understanding miRNA-mediated regulation because it has been extensively studied in C. elegans development and is conserved in many organisms. Moreover, because human let-7 regulates the let-60 homolog, the RAS oncogene in lung tissue (Calin et al. 2004a,b; Takamizawa et al. 2004; Johnson et al. 2005), studies on let-7 are also important for human cancer research.
In C. elegans, systematic genetic analysis has indicated that a number of proteins function in the miRNA pathway (Ketting et al. 1999; Tabara et al. 1999; Grishok et al. 2001). For example, DCR-1, an RNase III-like enzyme, is involved in the processing of hairpin-like pre-miRNAs to generate mature miRNAs (Bernstein et al. 2001; Billy et al. 2001; Grishok et al. 2001; Hutvágner et al. 2001; Ketting et al. 2001). ALG-1 and ALG-2, which belong to the Argonaute (Ago) family, play crucial roles in let-7 and lin-4 miRNA-mediated regulation (Grishok et al. 2001) and associate with let-7 miRNP complexes (Hutvágner et al. 2004). TSN-1 (Tudor staphylococcal nuclease homolog-1) and VIG-1 (vasa intronic gene homolog-1), which were found associated with RNA interference activity in Drosophila, have been shown to also be required for proper let-7-mediated regulation in C. elegans (Caudy et al. 2003). Two let-7-containing ribonucleoprotein complexes with sizes of 500 kDa and 250 kDa, similar to the sizes of siRNA-induced silencing complexes (siRISC) found in Drosophila (Pham et al. 2004), have been identified by gel filtration of C. elegans extracts (Caudy et al. 2003). Among them, TSN-1 and VIG-1 comigrate with let-7 in the 250-kDa protein complex (Caudy et al. 2003). Further studies on miRNA-containing ribonucleoprotein complexes and discoveries of additional unknown cofactors in these complexes would be helpful to understand miRNA related mechanisms of action.
In this study, we used native gel electrophoresis to characterize let-7 miRNP complexes in C. elegans. Our data indicate the presence of multiple and distinct ribonucleoprotein complexes assembling with let-7 in vitro. These complexes are affected by alg-1 and alg-2 null mutations, suggesting that they are relevant to miRNA function. In addition to large complexes with estimated molecular mass ranges from ∼500 kDa to >669 kDa, which also assemble with other miRNAs, two small complexes of ∼250 kDa and ∼160 kDa seem specific to the let-7 miRNA family, and their formation depends on the sequence in the 5′ seed region. By UV cross-linking, we detected two unidentified proteins, p40 and p13, which may directly associate with let-7. p13 appears in all let-7 binding complexes and is also cross-linked to lin-4. p40 appears only in the small complexes, and its appearance is dependent on the let-7 sequence.
RESULTS
The let-7 miRNA forms multiple ribonucleoprotein complexes in C. elegans extracts
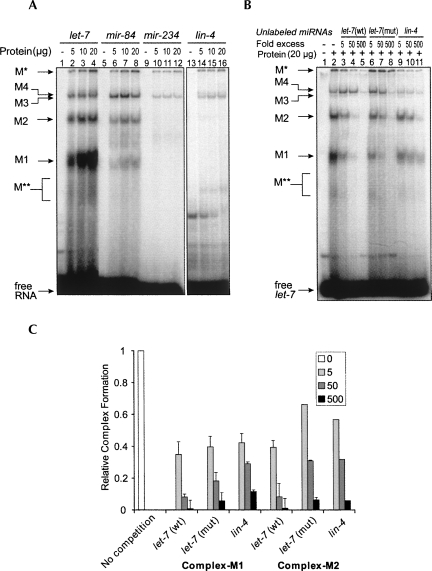
To elucidate the molecular mechanism of the let-7 miRNA in its function, we have identified and characterized multiple let-7 miRNA ribonucleoprotein (miRNP) complexes in C. elegans. We first prepared cytosolic extracts from mixed-stage C. elegans animals and examined whether these extracts supported the formation of miRNPs. Results from gel retardation experiments using end-labeled mature let-7 miRNA indicated that let-7 could form at least four distinct miRNP complexes. In this study, we refer to these complexes as M1, M2, M3, and M4 (Fig. 1A). In addition to the M1–M4 complexes, we found another large complex (M*), which was not capable of entering the gel, and a small and less distinct complex (M**). These complexes were sensitive to SDS and proteinase K (data not shown), suggesting that they contain protein components. The reaction mixture for gel retardation contained a 250-fold excess of tRNA over labeled let-7 to reduce the background caused by nonspecific RNA binding proteins.
FIGURE 1.
Multiple ribonucleoprotein complexes assemble with let-7 in vitro. (A) Radiolabeled let-7, mir-84, mir-234, or lin-4 was incubated with different amounts of C. elegans cell extracts and analyzed by native gel electrophoresis. The ribonucleoprotein complexes named in this study are indicated on the left. (B) Up to 500-fold excess of unlabeled let-7(wt), let-7(mut), and lin-4 RNAs were used to compete for the formation of let-7 binding complexes. (C) Quantification of fold decrease in the complexes M1 and M2 in B. The reaction without any unlabeled RNA served as control. The bars represent the error from two independent experiments.
We tested for complex formation with lin-4, mir-234, and a member of the let-7 miRNA family, mir-84 (Lim et al. 2003; Abbott et al. 2005). mir-84 formed all the same complexes as let-7 did albeit with reduced efficacy for M1 and M2 (Fig. 1A). Interestingly, lin-4 and mir-234 formed low amounts of M* and M3/M4 complexes but no M1 and M2 complexes. This indicated that M1 and M2 complexes preferentially assemble with let-7 or members of the let-7 miRNA family. In contrast, the M* and M3/M4 complexes seem to lack this selectivity. We also compared the ability of unlabeled lin-4, wild-type let-7, or mutant let-7(n2853) RNA (which contains a G-to-A base substitution at position 5 of let-7) (Reinhart et al. 2000) to compete for complex formation. We found that lin-4 did not compete for formation of the M1 and M2 complexes as efficiently as let-7 did, which is consistent with the inability of lin-4 to form these complexes (Fig. 1B,C). The competition by mutant let-7 was also weaker than that by wild-type let-7, although the difference was less pronounced. All of the above observations point to the possibility that formation of the M1 and M2 complexes is specific to let-7 and is dependent on the let-7 sequence (see below for further studies).
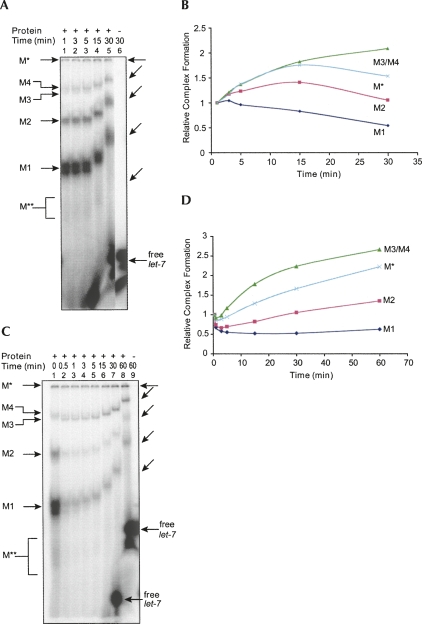
We performed time-course experiments to observe the formation of these complexes. Since it was not possible to arrest the formation of these complexes, we followed the protocols used in previous studies on RISC formation in Drosophila and loaded the binding reaction mixture directly into a running native gel at each time point (Pham et al. 2004). In the first set of experiments, we performed the binding reaction without a competitor miRNA. We found that the M3/M4 complexes increased in abundance over the time course of the experiment. The M* complex at first accumulated at a rate similar to that of M3/4, then the abundance slightly decreased (Fig. 2A,B). The M1 complex formed quickly and abundantly, then decreased over the time course of the experiment. The abundance of M2 increased for a short period and then decreased to its original amount. In the second set of experiments, we started the binding reaction with radiolabeled let-7 miRNA, and after 1 min of incubation, complex formation was competed by a 20-fold excess of unlabeled let-7 miRNA. The M* and M3/M4 complexes were almost not affected in formation and still increased their binding to labeled let-7 over time (Fig. 2C,D). The smaller complexes, especially M1, were very susceptible to the competition by unlabeled let-7, resulting in low amounts of the complexes carrying labeled let-7. After that, the formation of M1 complex was barely changed over time, while the M2 complex slightly increased its binding to labeled let-7 (Fig. 2C,D). A possible explanation is that the formation of smaller complexes M1 and M2 rapidly reaches the equilibrium state and these complexes could become less stable, and/or bound RNAs in the M1 and M2 complexes are quickly able to exchange and so was rapidly replaced by excess unlabeled let-7 (Fig. 2C). The larger complexes M* and M3/M4 may form more slowly and could be more stable, or the incorporated let-7 is less exchangeable. This may also explain why certain concentrations of unlabeled let-7 did not compete for the formation of M3/M4 complexes (Fig. 1B). Studies with Drosophila extracts indicate that siRNAs enter into multiple complexes and that the larger complexes arise from smaller ones (Pham et al. 2004; Tomari et al. 2004). It is also possible that the accumulation of the larger complexes M3/M4 is due to the transition from smaller complexes M1 or M2, and also a consequence of higher stability.
FIGURE 2.
let-7 miRNPs form in different ways. (A) Time-course analysis of miRNP assembly. Gel retardation samples were withdrawn at different durations of time and loaded into a running native gel. The differential mobility of the complexes is due to time differences between each loading. Lane 6 corresponds to the free label loaded at the same time as lane 5. The shifted positions of complexes are also shown by slanted arrows on the right. In lanes 1–3, the free probe ran off the gel because of earlier loading. (B) Amounts of complexes in A at different time intervals compared to the beginning of the reaction (1 min). (C) A gel retardation reaction was chased with an excess (20-fold) of unlabeled let-7 miRNA after the first minute of incubation. At indicated time intervals, the samples were withdrawn and analyzed as described in A. (D) Amounts of complexes in C at different time intervals compared to the beginning of the pulse chase (0.5 min after addition of unlabeled let-7 miRNA).
Fractionation studies of let-7 binding complexes
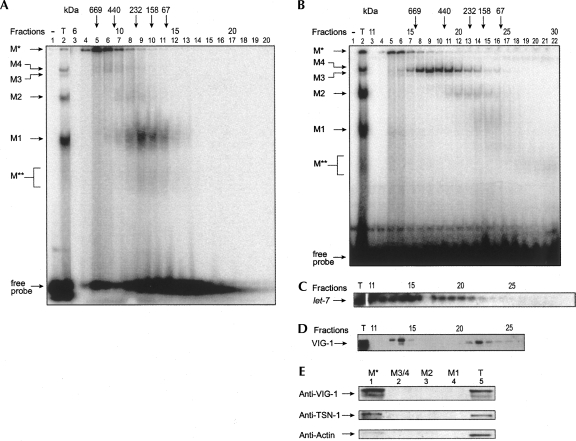
Gel filtration experiments with C. elegans extracts have shown that let-7 miRNA cofractionates in protein complexes of ∼500 and ∼250 kDa in size (Caudy et al. 2003). We performed similar experiments and fractionated radiolabeled let-7 miRNP complexes on a superdex-200 gel exclusion gravity column. We found radiolabeled M*, M3/M4, and M2 complexes migrating at >669 kDa, ∼500 kDa, and ∼250 kDa, respectively. M1 and M** complexes migrated at ∼160 kDa (Fig. 3A). Interestingly, we found slightly different results when we first fractionated the native extracts and then assayed for let-7 miRNP formation via gel shift (Fig. 3B). While the sizes of the complexes remained approximately the same, the relative abundance of the label in each complex was different. M1 and M2 complexes, the abundant forms in the nonfractionated extract (Fig. 3B, lane 2), formed poorly upon addition of radiolabeled let-7 miRNA in fractionated extracts (Fig. 3B, lanes 10–18). One explanation is that fractionation may segregate some necessary associating factors for formation of the M1 and M2 complexes and causes a reduction in new let-7 incorporation or the instability of these complexes. Pre-formed complexes containing let-7 miRNA (Fig. 3A) may be less affected or more stable in the fractionation process.
FIGURE 3.
Fractionation assay of let-7 miRNPs. (A) Fractionation of pre-formed complexes with radiolabeled let-7 miRNA by gel filtration. The positions of eluted molecular weight markers are represented by arrows. (B) C. elegans cytosolic extracts were fractionated, and then each fraction was assayed for miRNP formation with radiolabeled let-7 miRNA. (T) Complex formed with 20 μg of protein of total extracts. (C) RNAs in fractionated samples from B were analyzed by Northern blot for the presence of endogenous let-7 miRNA. (T) RNA isolated from total extract. (D) Indicated fractions were assayed for the presence of VIG-1 protein by Western blot. (E) Proteins isolated from indicated let-7 binding complexes were separated by SDS-PAGE and analyzed for the presence of VIG-1, TSN-1, or actin proteins by Western blot. (T) Total extract.
The complex M*, which was unable to enter the 5% native gel, was also observed in fractions that correspond to the void volume, which contain very-high-molecular-weight protein complexes of >669 kDa. Previous fractionation studies by Caudy et al. (2003) showed that endogenous let-7 comigrates with complexes of ∼500 kDa and ∼250 kDa. Since we have identified a >669-kDa complex (M*), in addition to the ∼500-kDa and ∼250-kDa complexes, assembling with exogenous let-7, we investigated how endogenous let-7 miRNA fractionates in our extracts. We isolated RNA from each fraction from the experiment shown in Figure 3B and probed for the presence of endogenous let-7 miRNA by Northern blot. We found that the majority of the let-7 miRNA fractionated in the void volume corresponding to M* and significant amounts of let-7 also cofractionated with the ∼500-kDa (M3/M4) complexes (Fig. 3C). Less let-7 miRNA cofractionated with the ∼250-kDa (M2) complex, and negligible amounts of let-7 miRNA corresponded to the ∼160-kDa (M1) complex. Considering the rapid formation and the abundance of M1 and M2 complexes when incubated with exogenous let-7, lower amounts of endogenous let-7 cofractionated with the smaller complexes M1 and M2 also point to their higher susceptibility or exchange rate for the substrate. This observation reveals the existence of a major let-7-containing complex of >669 kDa in addition to the previously found ∼500-kDa and ∼250-kDa complexes (Caudy et al. 2003). The difference between these two sets of results could be due to the extracts used for fractionation. Caudy and colleagues used ribosome-associated extracts for gel filtration, whereas we have used a 16,100g supernatant, which corresponds to the total cytosolic extract.
VIG-1 and TSN-1 have previously been shown to cofractionate with an ∼250-kDa complex, but not with an ∼500-kDa miRNP complex (Caudy et al. 2003). We examined our fractions for the presence of VIG-1 protein. We found VIG-1 cofractionating with a complex that peaks at ∼200 kDa overlapping partially with M1 and M2 (∼160 kDa and ∼250 kDa) complexes but not with M3/M4 (∼500 kDa) complexes (Fig. 3D). To our surprise, we found VIG-1 also cofractionated with the larger complex M* (>669 kDa). We isolated the proteins from these complexes from native gels and analyzed them by Western blot for the presence of VIG-1 and TSN-1. We found VIG-1 and TSN-1 proteins only in the M* complex but not in the M1, M2, or M3/M4 complexes (Fig. 3E). A control protein, actin, was barely present in the proteins isolated from the M* complex compared to the total proteins, indicating preferential enrichment of VIG-1 and TSN-1 proteins in M*, but not in other complexes. Although earlier fractionation studies (Caudy et al. 2003) suggested the presence of VIG-1 in an ∼250-kDa complex, Western blot analysis of proteins from these complexes suggests that VIG-1 is not present in our M1 (∼160 kDa) or M2 (∼250 kDa) complexes, but instead is present only in the M* complex. Therefore, in addition to the ∼250-kDa complex identified in the earlier study from C. elegans (Caudy et al. 2003), we have found that a larger let-7-containing complex M* (>669 kDa) also contains VIG-1 and TSN-1. However, since our Western blot analysis did not reveal that VIG-1 or TSN-1 associates with M1 (∼160 kDa) or M2 (∼250 kDa) complexes (Fig. 3E), cofractionation of VIG-1 with M1 and M2 complexes could be coincidental.
Formation of let-7 miRNP complexes is affected by alg-1 or alg-2 mutations
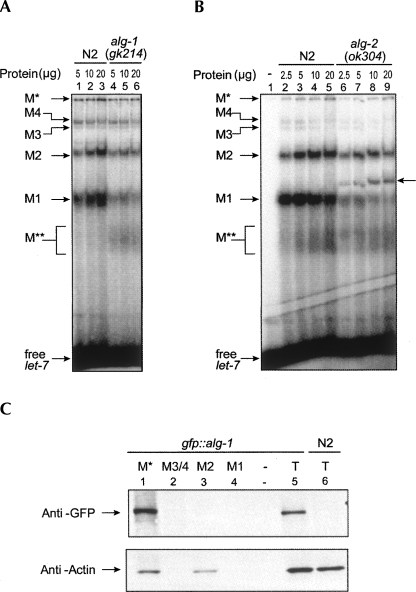
In C. elegans, genetic studies have shown that two closely related Argonaute proteins, alg-1 and alg-2 (referred to here as alg-1/2), are necessary for maturation and/or activity of lin-4 and let-7 miRNAs (Grishok et al. 2001). Coimmunoprecipitation experiments from C. elegans extracts have shown that ALG1/2 proteins interact with the let-7 miRNA (Hutvágner et al. 2004). ALG1/2 are homologous to RDE-1, which is essential for RNAi in C. elegans (Tabara et al. 1999), and to the Argonaute-like protein Ago2, which is known to be present in siRISC of human and Drosophila extracts (Hammond et al. 2001; Hutvágner and Zamore 2002; Martinez et al. 2002; Mourelatos et al. 2002) and responsible for the cleavage activity (Liu et al. 2004; Meister et al. 2004). Therefore, we asked whether Argonaute-like genes alg-1 and alg-2 are necessary for the formation of these complexes. We prepared extracts from alg-1(gk214) and alg-2(ok304) knockout mutants and assayed for the formation of let-7 binding complexes. We observed a substantial decrease in the amount of M1 and M2 complexes and a slight decrease in the amount of other complexes in both alg-1 and alg-2 deletion mutants (Fig. 4A,B). Interestingly, the absence of ALG-2 also leads to the formation of a distinct complex intermediate in size between complexes M1 and M2 (Fig. 4B). Since alg-1 and alg-2 have been shown necessary for maturation and activity of the let-7 miRNA (Grishok et al. 2001), and hence to mediate proper developmental timing, the susceptibility to alg-1 or alg-2 mutations of the let-7 miRNP complexes discovered here implies the relevance of these complexes to let-7 function. Extracts prepared from other developmental timing (heterochronic) defective mutants lin-28(n719) and lin-41(ma104) did not affect formation of let-7 miRNP complexes (data not shown), suggesting that these effects are specific to alg-1 and alg-2 and not associated with the heterochronic pathway per se. Since extracts prepared from the alg-1(gk214) and alg-2(ok304) null mutants (Grishok et al. 2001) did not completely abolish the formation of complexes, we speculate that ALG-1 or ALG-2 may be partially substituting for one another. Such redundancy between alg-1 and alg-2 has already been observed for their functions in temporal patterning, miRNA biogenesis, and miRNA function (Grishok et al. 2001). However, we are unable to directly test this hypothesis because the alg-1; alg-2 double mutant is lethal.
FIGURE 4.
Formation of the let-7 complexes is affected by alg-1 or alg-2 mutations. (A) Gel retardation of labeled let-7 miRNA by extracts prepared from wild-type (N2) or alg-1(gk214) animals. (B) Gel retardation of labeled let-7 miRNA by extracts prepared from wild-type (N2) or alg-2(ok304) animals. Arrow corresponding to lanes 5–9 indicate the appearance of an extra complex in alg-2(ok304) mutants. (C) Proteins isolated from indicated let-7 binding complexes formed in the cell extract prepared from a transgenic line of C. elegans that expresses GFP-tagged ALG-1 were separated by SDS-PAGE. The presence of GFP-tagged ALG-1 or actin proteins was analyzed by Western blot. (T) Total extract.
We have assayed the presence of ALG-1 protein in different let-7 binding complexes in cell extract prepared from a transgenic line of C. elegans that expresses GFP-tagged ALG-1. Similar to VIG-1 and TSN-1, ALG-1 was detected only in the M* complex (Fig. 4C). This is consistent with previous results showing that ALG-1 cofractionated only with a single complex larger than 650 kD in size (Tops et al. 2006) and suggests that only the M* complex is a candidate for functional miRISC. The reason that the smaller complexes M1 and M2, which do not contain ALG-1 protein, are more susceptible to alg-1 or alg-2 mutations is not clear. One speculation is that these complexes may play roles after the formation or functioning of Ago-containing let-7 miRISC or prior to this in a feedback loop, and their formation is affected by the absence of ALG-1 or ALG-2 proteins. Another explanation is that expression of some factors required for these complexes is regulated by alg-1 and alg-2.
Sequence requirements for let-7 miRNP formation
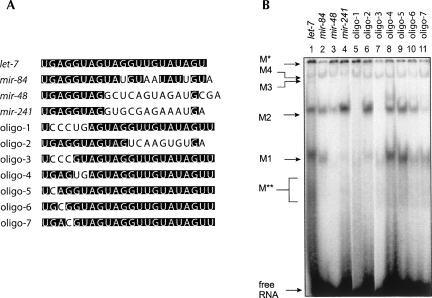
The M1 and M2 complexes assemble with let-7 and mir-84, a member of the let-7 family, but not lin-4 or mir-234 (Fig. 1). In contrast, the M* and M3/4 complexes are able to assemble with all miRNAs we tested so far, although in varying amounts. Thus, the M1 and M2 complexes appear specific to the let-7 family, while the others are not. To test this hypothesis, we applied the same assay to two other members of the let-7 family, mir-48 and mir-241, and also to artificial chimeric miRNAs designed by mixing the sequences of let-7 and lin-4. All short RNAs were heated before incubation with cell extracts to reduce the possibility of secondary structure formation. mir-48 and mir-241 are identical to let-7 in the 5′ seed region but less conserved in the sequences 3′ to the seed. mir-48 and mir-241 formed the M2 complex but only very small amounts of M1 complexes (Fig. 5B, lanes 3,4). Oligo-1, which contains the lin-4 seed and the let-7 sequence outside the seed, formed a very small amount of M1 complex but did not form M2 (Fig. 5B, lane 5). Oligo-2, which contains nucleotides 1 to 11 of let-7, covering the seed region, and nucleotides 12 to 21 of lin-4, formed a significant amount of M2 complex in addition to the small amount of M1 complex (Fig. 5B, lane 6). This suggests that the formation of the M2 complex is responding to the sequence of the let-7 seed region. On the other hand, all the four RNAs formed much smaller amounts of the M1 complex, compared to let-7 and mir-84, implying that intense formation of M1 complex may require the entire sequence to be more like let-7.
FIGURE 5.
The let-7 sequence is required for formation of the M1 and M2 complexes. (A) Sequence alignment of RNA oligonucleotides used in the gel retardation analysis in C. elegans extracts. (Solid boxes) Nucleotides identical to the let-7 sequence at the same positions. (B) RNA oligonucleotides listed in A were analyzed for ribonucleoprotein complex formation by native gel electrophoresis.
We further investigated the requirement of the let-7 seed sequence for complex formation. We modified the let-7 miRNA by replacing nucleotides in the seed region with the corresponding lin-4 sequences (Fig. 5A, oligo-3 to oligo-7). Interestingly, replacing the nucleotides 2∼4 completely abolished the M2 complex formation and also reduced the amount of M1 complex significantly (Fig. 5B, oligo-3, lane 7), whereas replacing nucleotides 5 and 6 did not show obvious negative effects (Fig. 5B, oligo-4, lane 8). This suggests that the nucleotides 2∼4 in the let-7 seed are required for M2 complex formation and are also important for M1. Then we modified each single nucleotide from the nucleotides 2 to 4 and found that single nucleotide substitutions at nucleotides 3 or 4 showed negative effects on formation of both the M1 and M2 complexes (Fig. 5B, lanes 9–11). In contrast, the existence of M3/M4 complexes was not affected by these sequence modifications. These results argue again that the M1 and M2 complexes are specific to let-7 and also reveal a putative cis-element on let-7 required for the formation of these complexes.
Sequence-dependent binding of proteins to let-7
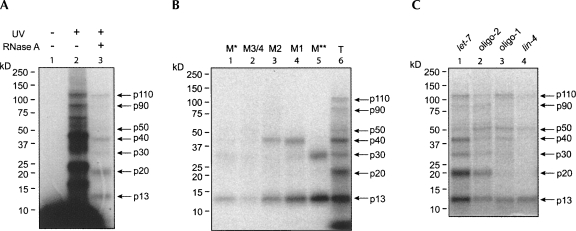
Although we did not find miRISC components like ALG-1, TSN-1, or VIG-1 in the M1 and M2 complexes, their susceptibility to alg-1 and alg-2 mutations implied a relevance to miRNA function. In addition, their formation depends on the let-7 sequence, suggesting that they may play roles in let-7-related functions. We sought to identify proteins present in these let-7 miRNP complexes and to determine whether those associating proteins are specific to let-7. To detect let-7-associating proteins, we used ultraviolet light (254 nm wavelength) to cross-link RNA to protein in a reaction mixture containing C. elegans extract and 5′-radiolabeled let-7 miRNA. Proteins cross-linked to let-7 were assayed by SDS-PAGE with or without RNase A treatment. After RNase A digestion, only proteins that were cross-linked to the 5′-end of let-7 could be detected and would be predicted to migrate close to positions corresponding to their molecular weights. At least seven cross-linked proteins, in sizes of ∼110, ∼90, ∼50, ∼40, ∼30, ∼20, and ∼13 kDa, were detected with medium to strong signals (Fig. 6A), indicating their close spatial relationships to let-7 or even a direct association with let-7.
FIGURE 6.
let-7 specific binding proteins were detected by UV cross-linking. (A) Radiolabeled let-7 was incubated with cell extract, and proteins were cross-linked to the probe with UV light. After RNase A treatment, cross-linked proteins were analyzed by SDS-PAGE and autoradiography. (B) Radiolabeled let-7 was incubated with cell extract, and let-7 binding complexes were separated on a native gel. Gel pieces containing each complex were sliced out and exposed to UV light. Cross-linked proteins were eluted and treated by RNase A and then analyzed by SDS-PAGE. (C) Radiolabeled let-7, lin-4, oligo-1, and oligo-2 RNAs were incubated with cell extract and exposed to UV light. Cross-linked proteins were treated by RNase A and T1 and analyzed by SDS-PAGE.
We performed an “in-gel” UV cross-linking to see whether these proteins are present in the let-7 binding complexes we discovered. After the let-7 binding complexes were separated on a native gel, we sliced the gel into pieces containing each let-7 binding complex and exposed them to UV light. Proteins eluted from each gel piece were assayed by SDS-PAGE. Only p13, p30, and p40 were observed after electrophoresis, indicating that other proteins may lose their associations with let-7 during electrophoresis. p13 was present in all complexes, suggesting its relevance in protein components among the let-7 binding complexes. Interestingly, p40 appeared to be present only in M1 and M2 complexes, and p30 was only present in the less distinct M** complex (Fig. 6B). These results also reveal that these let-7 binding complexes may share common components but could still contain specific factors.
To determine the requirements of these let-7 binding proteins for target sequences, we performed UV cross-linking with reaction mixtures in which other miRNAs, including lin-4 and artificial ones containing parts of let-7 sequences, were radiolabeled at the 5′-end and incubated with cell extract, respectively. Oligo-2, which contains the seed region of let-7 and is capable of forming abundant M2 complex in addition to a low amount of M1 complex (Fig. 5B), was cross-linked to all proteins that were cross-linked to let-7, but the signals seemed to be weaken (except for p50) (Fig. 6C, lane 2). Oligo-1, which is modified in the seed region and only formed a low amount of M1 complex and no M2 complex (Fig. 5B), was not cross-linked to p20 and showed less association with p30 and p40 (Fig. 6C, lane 3). Only p110, p50, and p13 were detected in the cross-linking reaction using lin-4, in which only the M* and M3/M4 complexes were detected. The presence of proteins specifically cross-linked to the let-7 miRNA seems to correspond to formation of the M1 and M2 complexes, which was highly dependent on the let-7 sequence (Fig. 5). These results suggest sequence-specific association of proteins to let-7, like the binding of p40, and indicate that let-7 associates with a set of specific proteins to assemble distinct complexes. However, some associating proteins discovered here, for example, p13, are not specific to let-7 since they were also cross-linked to lin-4 and may represent common factors in miRNPs.
DISCUSSION
We have observed the formation of multiple let-7 miRNP complexes in C. elegans, ranging in size from 160 to >669 kDa, similar to several RISC complexes described in previous studies (Hammond et al. 2000; Nykanen et al. 2001; Martinez et al. 2002; Pham et al. 2004). Our results also indicate that these let-7 miRNP complexes form in different ways and with different stabilities. A trend we observed is that the larger complexes of >669 kDa and ∼500 kDa have lower exchange rates for the substrate and higher stabilities. This is supported by observations that these larger complexes are more resistant to competitive unlabeled let-7 and to the fractionating process. Although the smaller M1 and M2 complexes form rapidly when exogenous let-7 is added into a binding reaction, the bound substrates seem to exchange actively. Moreover, we found that the major fractions of endogenous let-7 were cofractionated with the larger complexes but not the M1 and M2 complexes; hence, it is possible that the M1 and M2 complexes are intermediate complexes with less stability and the stable large complexes M* and M3/M4 are more likely the major complexes that maintain bound let-7 in vivo. The M* complex might represent a functional miRISC since it was cofractionated with abundant endogenous let-7 and contains ALG-1 and two known factors, VIG-1 and TSN-1, which have been shown to associate with RISC and let-7 (Caudy et al. 2002, 2003). It is worth noting that in Drosophila extracts, RISC activity has been observed in the R* complex, similar to M*, which does not enter the gel and fractionates in the void volume, as well as in an 80S complex that is likely to be a part of the R* complex (Pham et al. 2004).
The dynamic nature of RISC complexes described in results from Pham et al. (2004) and Tomari et al. (2004) suggests that smaller siRNA-associated complexes convert into larger, active complexes. It is possible that miRNA-associated complexes also perform a similar assembly pathway. let-7 could be transferred from the M1 and M2 complexes to the larger complexes, or a portion of the smaller complexes with incorporated substrate further assembles to stable larger complexes. However, our results show that free let-7 is able to enter into larger complexes in fractionated extracts in which M1 and M2 are absent. Thus the formation of larger complexes M* and M3/M4 can be separated from the formation of M1 and M2. In addition, some other miRNAs we have tested, including lin-4, mir-234, and several artificially designed short RNAs containing parts of let-7 or lin-4 sequences, are all able to form the larger complexes M* and M3/M4 but not the M1 and M2 complexes. It is possible that most miRNAs can enter large miRNP complexes of ∼500 kDa or >669 kDa to accomplish their function, but some miRNAs, including let-7, also enter multiple complexes with smaller sizes. These smaller miRNP complexes may play an aforementioned role in dynamic formation of the larger and active complexes or have unknown functions. For now in our experiments, we found that formation of the smaller M1 and M2 complexes is susceptible to alg-1 or alg-2 null mutations although ALG-1 is not detected in these complexes. We speculate that the M1 and M2 complexes may function at the steps prior to or following the formation of Argonaute-containing complexes.
While there seems to be no specificity for the formation of the M* and M3/M4 complexes when different miRNAs were used, interestingly, the M1 and M2 complexes seem specific to the let-7 miRNA family. We investigated the sequence specificity for complex formation by using a few artificially designed miRNAs containing parts of let-7 or lin-4 sequences. Changing or replacing different parts of the let-7 sequence can dramatically affect the formation of M1 and M2 complexes. We found that nucleotides 2 to 4 of let-7 are required for the formation of M2 and are also important for M1. Here we cannot eliminate the possibility that changes in the sequence alter secondary structure and hence affect complex formation. However, the most plausible short intramolecular base pairs predicted by mfold (Zuker 2003) in the sequences of the let-7 family members, which all form some amount of the M2 complex, are different from each other (data not shown), suggesting that a special secondary structure is not required for complex formation. Also, there is no clear correlation between the predicted structures of the short RNAs tested in this study and their abilities to form the M1 and M2 complexes. In addition, the free energy changes (ΔG) estimated by mfold for all chimeric short RNAs to form secondary structure are similar to those for let-7 and lin-4 (data not shown), which are presumed to associate with RISC component proteins in physiological conditions and to serve as single-stranded RNA fragments for base-pairing with their targets. Thus we propose that our results are more likely due to the changes in the primary sequences.
By UV cross-linking, we detected a number of proteins associating with let-7 in a binding reaction in which labeled let-7 was incubated with cell extract. Among them, p40 was specifically detected in the M1 and M2 complexes, and its cross-linking to the miRNA is highly sensitive to changes in the let-7 sequence. p40 is hence a strong candidate for a let-7-specific associating factor. p13 was detected in all let-7 binding complexes, including M* and M3/M4 that were also formed with other miRNAs we tested. p13 was also cross-linked to lin-4. One speculation is that p13 is not specific to let-7 and could generally exist in many miRNPs formed with different miRNAs. However, in our experiments, more abundant p13 was cross-linked to let-7, perhaps due to the formation of M1 and/or M2 complexes. This implies that the formation of smaller let-7 complexes, which may contain some let-7-specific binding proteins, also contribute to recruitment of other binding proteins, which may be miRISC components and lack a preference of selectivity to miRNAs. Recent studies have shown that mouse Lin28 is able to bind pri-let-7 and pre-let-7 miRNAs and specifically inhibits the pri-let-7 processing in embryonic cells (Viswanathan et al. 2008). This has revealed a possibility that some RNA-binding proteins selectively associate with the pri-miRNAs or pre-miRNAs and manipulate their biogenesis or function. Our results indicate that the mature let-7 miRNA could associate with specific proteins and be incorporated into distinct complexes as well. Since our complexes formed in the lin-28-null mutant, we have likely identified novel let-7 binding proteins in addition to LIN-28. It remains to be seen what the functions of these proteins are.
MATERIALS AND METHODS
Preparation of C. elegans extracts
Wild-type (N2) animals were grown at 20°C in 50 mL of S medium (Stiernagle 1999), supplemented with Escherichia coli HB101. After 48–60 h of growth, the mixed-staged animals were collected by sedimentation on ice. The animals were washed with 0.1 M NaCl three times and frozen at −80°C. Packed animals (1 mL) were thawed on ice and washed twice with 4 volumes of wash buffer (50 mM Tris-HCl at pH 7.5 and 10 mM potassium acetate) followed by centrifugation at 3000 rpm for 2 min. The animals were resuspended in 4.0 mL of homogenization buffer (50 mM Tris-HCl at pH 7.5, 10 mM potassium acetate, 5 mM DTT, 10 U/mL SUPERase-In [Ambion], and 1× Complete protease inhibitor [Roche]) and incubated for 20 min on ice with intermittent agitation. The animals were transferred to a 7-mL glass dounce homogenizer for 30 strokes on ice using pestle B. The homogenate was transferred to a 15-mL tube, 2 mM magnesium acetate was added, and the concentration of potassium acetate was adjusted to 100 mM. The homogenate was incubated for 20 min on ice with intermittent agitation and then centrifuged at 16,100g for 30 min at 4°C. The supernatant was stored at −80°C and used as cell extracts in this study.
In the experiments using alg-1 and alg-2 mutants (Fig. 4), wild-type (N2) or mutant animals were grown on NGM plates with E. coli OP50 as bacterial food. Mixed-staged animals were collected from 10 plates (∼100 μL of packed animals) and washed three times by M9 buffer (22 mM KH2PO4, 42 mM Na2HPO4, 85 mM NaCl, and 1 mM MgSO4). These animals were frozen at −80°C until use. The animals were washed and homogenized using a pellet pestle (Kimble-Knotes) and the lysis buffers described above.
RNA oligonucleotides
Synthetic RNAs were obtained from Dharmacon Research Inc. or Integrated DNA Technologies, Inc. as follows:
let-7(wt) (5′-UGAGGUAGUAGGUUGUAUAGU-3′);
let-7(mut) (5′-UGAGAUAGUAGGUUGUAUAGU-3′);
mir-84 (5′-UGAGGUAGUAUGUAAUAUUGUA-3′);
mir-48 (5′-UGAGGUAGGCUCAGUAGAUGCGA-3′);
mir-241 (5′-UGAGGUAGGUGCGAGAAAUGA-3′);
mir-234 (5′-UUAUUGCUCGAGAAUACCCUU-3′);
lin-4 (5′-UCCCUGAGACCUCAAGUGUGA-3′);
oligo-1 (5′-UCCCUGAGUAGGUUGUAUAGUU-3′);
oligo-2 (5′-UGAGGUAGUAGUCAAGUGUGA-3′);
oligo-3 (5′-UCCCGUAGUAGGUUGUAUAGUU-3′);
oligo-4 (5′-UGAGUGAGUAGGUUGUAUAGUU-3′);
oligo-5 (5′-UCAGGUAGUAGGUUGUAU AGUU-3′);
oligo-6 (5′-UGCGGUAGUAGGUUGUAUAGUU-3′); and
oligo-7 (5′-UGACGUAGUAGGUUGUAUAGUU-3′).
To monitor ribonucleoprotein complex formation in vitro, oligonucleotides were labeled at the 5′-end using T4 polynucleotide kinase and [γ-32P]ATP. Reaction mixtures (25 μL) contained 1× kinase buffer (NEB, 50 mM Tris at pH 7.5, 5 mM MgCl2), 2.0 μM RNA, 50 μCi of [γ-32P]ATP (3000 Ci/mmol, NEN), and 25 units of T4 polynucleotide kinase. Reactions were performed for 30 min at 37°C. Free [γ-32P]ATP nucleotides were removed by passing the mixture through a Spandex G-25 column (Roche). The specific activities of the RNAs were typically 1 × 106 c.p.m./pmol.
Native gel electrophoresis
RNA binding reaction mixtures (10 μL) containing 20 mM Tris (pH 7.5), 1 mM magnesium acetate, 1 mM calcium chloride, 0.01% NP-40, 2 mM ATP, 250 ng/μL E. coli tRNA, 4 × 105 c.p.m. end-labeled RNA (∼40 nM), and different amounts of extracts were incubated for 60 min at room temperature and then loaded onto a 5% native polyacrylamide gel. The native gel was subjected to electrophoresis in 1× TBE at 100 V for 120 min, dried on 3 MM Whatman paper, and exposed to a PhosphorImager plate. The PhosphorImager signals were quantified using ImageQuant 5.1 software (Molecular Dynamics).
Gel filtration
For fractionation of pre-formed let-7 binding complexes, a gravity column was prepared using an Econo-Column (Bio-Rad) column and 2 mL of packed Superdex-200 beads (Amersham-Pharmacia), which were equilibrated with gel filtration buffer (20 mM Tris-HCl at pH 7.5, 50 mM potassium acetate, 1 mM magnesium acetate, 1 mM calcium chloride, and 5% glycerol). One hundred microliters of RNA-binding reaction containing 200 μg of protein was carried out for 60 min and separated on the column by gravity. Around 30 fractions (∼100 μL each) were collected assayed by native gel electrophoresis as described above.
For gel filtration of crude C. elegans extracts, 400-μL extracts with 6 mg/mL total protein were loaded onto a Superdex-200 HR 10/30 column that was equilibrated with gel filtration buffer. The proteins were separated at 4°C at the rate of 0.35 mL/min using AKTA FPLC (Pharmacia), and 60 fractions (0.5 mL each) were collected. To monitor the ability to form let-7 binding complexes, samples of each fraction were mixed with radiolabeled let-7 and assayed for miRNP formation by native gel electrophoresis as described above.
To detect endogenous let-7, indicated fractions (300 μL) were mixed with 100 μL of 4× proteinase K buffer (400 mM Tris-HCl at pH 7.5 and 4% SDS). Proteinase K was added to a final concentration of 450 μg/mL, and the reaction was carried out for 20 min at 65°C. Then the samples were extracted by phenol/chloroform/isoamyl alcohol (25:24:1), and the RNAs were precipitated in the presence of 20 μg of glycogen. The RNAs were separated on a 10% TBE urea gel and transferred to a nylon membrane. The let-7 miRNA was probed by end-labeled DNA oligonucleotide complementary to let-7 (Johnson et al. 2003).
UV cross-linking
RNA binding reaction mixtures (20 μL) containing 1 mg/mL protein from cell extract and 32P-labeled miRNA were placed onto a pre-chilled 96-well plate on ice and exposed to 254-nm light for 15 min in a Stratalinker 2400 (Stratagene). The reactions were treated by 5 U/mL RNase A and/or 50 U/mL RNase T1 for 30 min at room temperature and then analyzed by denaturing gel electrophoresis using a Criterion XT Bis-Tris 4%–12% gradient gel (Bio-Rad) and autoradiography.
For the “in-gel” UV cross-linking experiment, a 150-μL RNA-binding reaction containing 1 mg/mL protein from cell extract and 32P-labeled miRNA was subjected to electrophoresis on a 5% native preparative gel. Without drying, the gel was exposed to a BioMax MS film (Kodak) for 1 h. The film was aligned with the gel, and the gel slices containing each complex were individually excised. For samples subjected to UV cross-linking, gel slices were placed on a clean glass plate on ice and exposed to 254-nm light for 15 min. The gel slices were minced into small pieces. For the RNase A treatment, the gel pieces were mixed with 150 μL of diluted RNase A (250 U/mL in 50 mM Tris-HCl at pH 7.5 buffer) and incubated for 30 min at room temperature. Then 50 μL of 4× sample buffer (Bio-Rad) were added, and the samples were agitated vigorously on a vortex mixer for 3 h. To extract protein efficiently, the gel slices were frozen and thawed twice intermittently. Eluted proteins were analyzed by denaturing gel electrophoresis as described above.
Immunoblotting
Proteins in gel filtration fractions or eluted from native gel slices were separated on SDS denaturing gels and transferred to nitrocellulose membranes (Schleicher and Schuell). Polyclonal anti-VIG-1, anti-TSN-1, and anti-GFP (A11122; Molecular Probes) antibodies, and monoclonal anti-Actin antibody (clone C4; ICN Pharmaceuticals) were used as primary antibodies. Western blots were developed by ECL (Amersham Biosciences).
ACKNOWLEDGMENTS
We thank G. Hannon and R. Ketting for providing anti-VIG-1, anti-TSN-1 antibodies. We thank C. Mello for providing the gfp∷alg-1 construct. We thank C. Crews for advice during the initial stages of this work. We thank H. Grosshans, A. Kerscher, D. Banerjee, R. Niwa, and S. Roush for critical reading of this manuscript and all the Slack lab members for their support during the investigation. This work was supported by the Culpepper Fund (Rockefeller Fund), the Robert Leet and Clara Guthrie Patterson Trust, an NIH grant to F.J.S. (GM62594), and the Anna Fuller Fund Fellowship and the Anna D. Barker Fellowship in Basic Cancer Research (AACR) to S.-P.C.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.551208.
REFERENCES
- Abbott, A.L., Alvarez-Saavedra, E., Miska, E.A., Lau, N.C., Bartel, D.P., Horvitz, H.R., Ambros, V. The let-7 microRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans . Dev. Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros, V. microRNAs: Tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- Banerjee, D., Slack, F. Control of developmental timing by small temporal RNAs: A paradigm for RNA-mediated regulation of gene expression. Bioessays. 2002;24:119–129. doi: 10.1002/bies.10046. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., Caudy, A.A., Hammond, S.M., Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Billy, E., Brondani, V., Zhang, H., Muller, U., Filipowicz, W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl. Acad. Sci. 2001;98:14428–14433. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin, G.A., Liu, C.G., Sevignani, C., Ferracin, M., Felli, N., Dumitru, C.D., Shimizu, M., Cimmino, A., Zupo, S., Dono, M., et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl. Acad. Sci. 2004a;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin, G.A., Sevignani, C., Dumitru, C.D., Hyslop, T., Noch, E., Yendamuri, S., Shimizu, M., Rattan, S., Bullrich, F., Negrini, M., et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. 2004b;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudy, A.A., Myers, M., Hannon, G.J., Hammond, S.M. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes & Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudy, A.A., Ketting, R.F., Hammond, S.M., Denli, A.M., Bathoorn, A.M., Tops, B.B., Silva, J.M., Myers, M.M., Hannon, G.J., Plasterk, R.H. A micrococcal nuclease homologue in RNAi effector complexes. Nature. 2003;425:411–414. doi: 10.1038/nature01956. [DOI] [PubMed] [Google Scholar]
- Grishok, A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., Mello, C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Grosshans, H., Slack, F.J. Micro-RNAs: Small is plentiful. J. Cell Biol. 2002;156:17–21. doi: 10.1083/jcb.200111033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, S.M., Bernstein, E., Beach, D., Hannon, G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Boettcher, S., Caudy, A.A., Kobayashi, R., Hannon, G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Hutvágner, G., Zamore, P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Hutvágner, G., McLachlan, J., Pasquinelli, A.E., Balint, E., Tuschl, T., Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Hutvágner, G., Simard, M.J., Mello, C.C., Zamore, P.D. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:e98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, S.M., Lin, S.Y., Slack, F.J. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev. Biol. 2003;259:364–379. doi: 10.1016/s0012-1606(03)00202-1. [DOI] [PubMed] [Google Scholar]
- Johnson, S.M., Grosshans, H., Shingara, J., Byrom, M., Jarvis, R., Cheng, A., Labourier, E., Reinert, K.L., Brown, D., Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Haverkamp, T.H., van Luenen, H.G., Plasterk, R.H. mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., Plasterk, R.H. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans . Genes & Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, R.C., Feinbaum, R.L., Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14 . Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Lau, N.C., Weinstein, E.G., Abdelhakim, A., Yekta, S., Rhoades, M.W., Burge, C.B., Bartel, D.P. The microRNAs of Caenorhabditis elegans . Genes & Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Carmell, M.A., Rivas, F.V., Marsden, C.G., Thomson, J.M., Song, J.J., Hammond, S.M., Joshua-Tor, L., Hannon, G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Martinez, J., Patkaniowska, A., Urlaub, H., Luhrmann, R., Tuschl, T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- Meister, G., Landthaler, M., Patkaniowska, A., Dorsett, Y., Teng, G., Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Mourelatos, Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M., Dreyfuss, G. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes & Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykanen, A., Haley, B., Zamore, P.D. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- Pasquinelli, A.E., Ruvkun, G. Control of developmental timing by microRNAs and their targets. Annu. Rev. Cell Dev. Biol. 2002;18:495–513. doi: 10.1146/annurev.cellbio.18.012502.105832. [DOI] [PubMed] [Google Scholar]
- Pham, J.W., Pellino, J.L., Lee, Y.S., Carthew, R.W., Sontheimer, E.J. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila . Cell. 2004;117:83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans . Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Slack, F.J., Basson, M., Liu, Z., Ambros, V., Horvitz, H.R., Ruvkun, G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- Stiernagle, T. Maintenance of C. elegans. C. elegans: A practical approach . In: Hope I.A., editor. Oxford University Press; New York: 1999. pp. 51–67. [Google Scholar]
- Tabara, H., Sarkissian, M., Kelly, W.G., Fleenor, J., Grishok, A., Timmons, L., Fire, A., Mello, C.C. The rde-1 gene, RNA interference, and transposon silencing in C. elegans . Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Takamizawa, J., Konishi, H., Yanagisawa, K., Tomida, S., Osada, H., Endoh, H., Harano, T., Yatabe, Y., Nagino, M., Nimura, Y., et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Tomari, Y., Du, T., Haley, B., Schwarz, D.S., Bennett, R., Cook, H.A., Koppetsch, B.S., Theurkauf, W.E., Zamore, P.D. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- Tops, B.B., Plasterk, R.H., Ketting, R.F. The Caenorhabditis elegans Argonautes ALG-1 and ALG-2: Almost identical yet different. Cold Spring Harb. Symp. Quant. Biol. 2006;71:189–194. doi: 10.1101/sqb.2006.71.035. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez, M.A., Liu, J., Hannon, G.J., Parker, R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes & Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Vella, M.C., Choi, E.Y., Lin, S.Y., Reinert, K., Slack, F.J. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes & Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan, S.R., Daley, G.Q., Gregory, R.I. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman, B., Ha, I., Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans . Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]