Abstract
MNL1, the Candida albicans homologue of an orphan Msn2-like gene (YER130c in Saccharomyces cerevisiae) has no known function. Here we report that MNL1 regulates weak acid stress responses. Deletion of MNL1 prevents the long-term adaptation of C. albicans cells to weak acid stresses and compromises their global transcriptional response under these conditions. The promoters of Mnl1-dependent genes contain a novel STRE-like element (SLE) that imposes Mnl1-dependent, weak acid stress–induced transcription upon a lacZ reporter in C. albicans. The SLE (HHYYCCCCTTYTY) is related to the Nrg1 response element (NRE) element recognized by the transcriptional repressor Nrg1. Deletion of NRG1 partially restores the ability of C. albicans mnl1 cells to adapt to weak acid stress, indicating that Mnl1 and Nrg1 act antagonistically to regulate this response. Molecular, microarray, and proteomic analyses revealed that Mnl1-dependent adaptation does not occur in cells exposed to proapoptotic or pronecrotic doses of weak acid, suggesting that Ras-pathway activation might suppress the Mnl1-dependent weak acid response in dying cells. Our work defines a role for this YER130c orthologue in stress adaptation and cell death.
INTRODUCTION
All organisms must respond and adapt to environmental stresses if they are to survive adverse conditions. Microbes elicit a combination of specific and general stress responses that repair the damage generated by environmental stresses and restore cellular and metabolic homeostasis under the hostile conditions. These responses are particularly important in pathogenic microbes which have evolved molecular mechanisms to counteract the defenses of their host.
In the benign model yeast Saccharomyces cerevisiae, the so-called general stress response or environmental stress response, confers resistance to heat shock, pro-oxidants, osmotic shock, nutrient deprivation, alcohol, and weak acids (Gasch et al., 2000; Causton et al., 2001). This general stress response is largely coordinated by the transcription factors Msn2 and Msn4 (Estruch and Carlson 1993, Marchler et al., 1993). Under stress conditions Msn2 and Msn4 accumulate in the nucleus (Gorner et al., 1998; Jacquet et al., 2003) where they activate the transcription of stress genes containing STRE elements (CCCCT) in their promoters (Martinez-Pastor et al., 1996). This response is down-regulated by the Ras-cAMP-PKA pathway (Garreau et al., 2000). Protein kinase A (PKA)-mediated phosphorylation of Msn2 and Msn4 results in their cytoplasmic accumulation, thereby decreasing the expression of their target stress genes (Gorner et al., 1998, 2002).
The pathogenic yeast Candida albicans causes frequent infections of the oral and vaginal mucosa and potentially lethal systemic infections in severely immunocompromised individuals, including patients receiving transplants or chemotherapy (Odds, 1988). C. albicans occupies a variety of niches within the human body, encountering a range of stressful conditions as it interacts with its host and counteracts the immune system (Lorenz et al., 2004; Fradin et al., 2005). The inactivation of stress signaling pathways or stress genes increases the sensitivity of C. albicans to these environmental stresses and attenuates its virulence (Wysong et al., 1998; Alonso-Monge et al., 1999; Hwang et al., 2002; Martchenko et al., 2004; Fradin et al., 2005). A better understanding of how C. albicans responds to environmental stresses and how these responses are be linked to its cellular fate is important because this may facilitate the design of antifungal therapies that manipulate the endogenous stress and death responses of fungal pathogens (Ramsdale, 2005).
The regulation of stress responses in C. albicans appears to have diverged from those in benign model yeasts. Core transcriptional responses to stress differ significantly in C. albicans, S. cerevisiae, and Schizosaccharomyces pombe (Enjalbert et al., 2003, 2006), as do the roles of the stress activated protein kinases in these yeasts (Hog1/Sty1: Smith et al., 2004; Enjalbert et al., 2006). Furthermore, we show here that Msn2/4-like proteins have evolved different functions in C. albicans compared with S. cerevisiae.
In S. cerevisiae, the closely related Msn2/4 family of (C2H2)2 zinc finger transcription factors contains a third member; Yer130c. The partially redundant MSN2 and MSN4 genes represent paralogues that were generated by the whole genome duplication event that occurred during yeast evolution (Byrne and Wolfe, 2005). YER130C is derived from a separate locus in the ancestral yeast genome that existed before whole genome duplication. C. albicans and S. cerevisiae diverged before the whole genome duplication event (Montcalm and Wolfe, 2006). Hence C. albicans has two MSN2/4-like loci: MSN4 and MNL1 (Nicholls et al., 2004). C. albicans MSN4 is most closely related to, and therefore most likely to be orthologous to the S. cerevisiae paralogues MSN2 and MSN4. C. albicans MNL1 is most closely related to YER130c, and synteny comparisons in the CTG clade (i.e., those species that decode CTG as serine) indicate that these genes are orthologues.
Although Msn2 and Msn4 play key roles in stress adaptation in S. cerevisiae, the role of the putative transcription factor Yer130c still remains obscure. Yer130c plays no obvious role in stress adaptation as S. cerevisiae yer130c mutants display no obvious phenotypes. In C. albicans, the roles of Msn4 and Mnl1 are not known. They do not contribute to responses to mild heat shock, hydrogen peroxide, salt stress, starvation, or ethanol (Nicholls et al., 2004). Furthermore, microarray studies have confirmed that Msn4 and Mnl1 do not contribute significantly to the global transcriptional response of this pathogen to heat shock, hydrogen peroxide, or osmotic stresses (Nicholls et al., 2004). These findings are consistent with the limited cross-protection observed when C. albicans cells are exposed to sequential stresses (Enjalbert et al., 2003; Smith et al., 2004). Recently however, activation of the Ras-cAMP pathway has been shown to accelerate programmed cell death (reminiscent of apoptosis) in C. albicans after exposure to high doses of acetic acid (Phillips et al., 2006). This link between death-associated Ras signaling and the weak acid stress response prompted us to test whether the MSN4 and MNL1 genes contribute to the weak acid stress response in C. albicans.
At low ambient pHs, weak organic acids such as acetic acid enter cells by passive diffusion. When they encounter the high intracellular pH, weak acids dissociate to produce acid anions and protons. The antimicrobial effects of weak acids are thought to arise from a combination of factors that include intracellular acidification, which inhibits essential cellular functions such as glycolysis (Krebs et al., 1983), futile proton pumping that depletes ATP stores (Lambert and Stratford, 1999), and alterations in membrane permeability that occur when lipophilic anions integrate into the cell membrane (Holyoake et al., 1999). Adaptation to weak acid stress in S. cerevisiae involves extensive remodeling of the transcriptome and proteome (Gorner et al., 1998; de Nobel et al., 2001; Schüller et al., 2004) with many of these changes overlapping with those of the general stress response. The transcriptional changes induced by the weak acid, sorbate, are dependent on Msn2/Msn4, War1, and a fourth as yet unidentified transcription factor (Schüller et al., 2004). Weak acid tolerance, however, is not directly dependent on Msn2/Msn4 activity because the sensitivity of war1 cells to weak acid stress can be attributed almost entirely to a defect in the expression of Pdr12, an ATP-binding cassette (ABC) efflux pump (Piper et al., 1998).
In this article we explore the roles of C. albicans Msn2/4-like proteins in weak acid stress and death responses. We show that Mnl1 (IPF9113; orf19.6121) is required for the weak acid response in C. albicans, but that Msn4 is not essential for this response. We define the Mnl1 regulon in C. albicans and show that it is driven by a novel STRE-like promoter element. In addition, we reveal significant overlap between this Mnl1 regulon and that of the transcriptional repressor Nrg1. Finally we show that down-regulation of the Mnl1-dependent acetic acid stress response correlates with the commitment of C. albicans cells to programmed cell death under proapoptotic conditions.
MATERIALS AND METHODS
Strains and Culture Conditions
Strains are listed in Table 1. All C. albicans strains were transformed with CIp10 or CIp10 derivative plasmids to restore their URA3 status (Brand et al., 2004). All mnl1 and msn4 strains are null mutants (Nicholls et al., 2004). Cells were grown at 30°C in SC-pH 3.0 (2% glucose; 0.675% yeast nitrogen base containing (NH4)2SO4, 0.2% amino acids, pH 3.0). For disk-diffusion assays, 1 × 107 cells were spread on the surface of plates, 6-mm-diameter Whatman 3MM discs (Clifton, NJ) were placed on the lawn of cells, 10 μl of acetic acid (500 mM–7.5 M) was applied to each disk, and growth was examined after 12- and 24-h incubation. Growth inhibition in liquid culture was monitored at A620 nm, and viability by clonogenic survival and propidium iodide (PI)-exclusion assays (Phillips et al., 2003). For single-cell analyses, 1 × 107 cells were spread onto plates and examined at 400× magnification. The growth status of a minimum of 200 cells was assessed at regular intervals.
Table 1.
C. albicans strains used in this study
| Strains | Relevant genotype | Source |
|---|---|---|
| SC5314 | Wild type | Gillum et al. (1984) |
| CAF2–1 | URA3/ura3::λimm434 | Fonzi and Irwin (1993) |
| CA18 | ura3::λimm434/ura3::λimm434 ade2::hisG/ade2::hisG | Fonzi and Irwin (1993) |
| MMC4 | ura3::λimm434/ura3::λimm434 nrg1::hisG/nrg1::hisG | Murad et al. (2001) |
| MSC4 | ura3::λimm43434/ura3::λimm434 ade2::hisG/ade2::hisG | |
| mnl1::hisG/mnl1::hisG | Nicholls et al. (2004) | |
| MSC8 | ura3::λimm434/ura3::λimm434 ade2::hisG/ade2::hisG | |
| msn4::hisG/msn4::hisG | Nicholls et al. (2004) | |
| MSC12 | ura3::λimm434/ura3::λimm434 ade2::hisG/ade2::hisG | |
| mnl1::hisG/mnl1::hisG msn4::hisG/msn4::hisG | Nicholls et al. (2004) | |
| MSC16 | ura3::λimm434/ura3::λimm434 ade2::hisG/ade2::hisG | |
| pCRW3 (ADE2) pACT1 (URA3) | Nicholls et al. (2004) | |
| MSC17 | ura3::λimm434/ura3::λimm434ade2::hisG/ade2::hisG | |
| pCRW3 (ADE2), pACT1-MNL1 (URA3) | Nicholls et al. (2004) | |
| MSC18 | ura3::λimm434/ura3::λimm434 ade2::hisG/ade2::hisG | |
| pCRW3 (ADE2), pACT1-MSN4 (URA3) | Nicholls et al. (2004) | |
| SNC10 | ura3::λimm434/ura3::λimm434 ade2::hisG/ade2::hisG | |
| mnl1::hisG/mnl1::hisG msn4::hisG/msn4::hisG nrg1::hisG/nrg1::hisG | Nicholls et al. (2004) | |
| SNC101 | CAI8 CIp10 (URA3) | This study |
| SNC102 | MSC4 CIp10 (URA3) | This study |
| SNC103 | MSC8 CIp10 (URA3) | This study |
| SNC104 | MSC12 CIp10 (URA3) | This study |
| SNC105 | SNC10 CIp10 (URA3) | This study |
| SNC106 | MMC4 CIp10 (URA3) | This study |
| MRC1 | MSC4 pSLEA-LacZ (URA3) | This study |
| MRC2 | MSC4 pSLEB-LacZ (URA3) | This study |
| MRC3 | MSC4 pSLEC-LacZ (URA3) | This study |
| MRC4 | MSC4 pSLED-LacZ (URA3) | This study |
| MRC5 | MSC4 plac-basal (URA3) | This study |
| MRC6 | MSC8 pSLEA-LacZ (URA3) | This study |
| MRC7 | MSC8 pSLEB-LacZ (URA3) | This study |
| MRC8 | MSC8 pSLEC-LacZ (URA3) | This study |
| MRC9 | MSC8 pSLED-LacZ (URA3) | This study |
| MRC10 | MSC8 plac-basal (URA3) | This study |
| MRC11 | MSC12 pSLEA-LacZ (URA3) | This study |
| MRC12 | MSC12 pSLEB-LacZ (URA3) | This study |
| MRC13 | MSC12 pSLEC-LacZ (URA3) | This study |
| MRC14 | MSC12 pSLED-LacZ (URA3) | This study |
| MRC15 | MSC12 plac-basal (URA3) | This study |
| MRC16 | SNC10 pSLEA-LacZ (URA3) | This study |
| MRC17 | SNC10 pSLEB-LacZ (URA3) | This study |
| MRC18 | SNC10 pSLEC-LacZ (URA3) | This study |
| MRC19 | SNC10 pSLED-LacZ (URA3) | This study |
| MRC20 | SNC10 plac-basal (URA3) | This study |
| MRC21 | MMC4 pSLEA-LacZ (URA3) | This study |
| MRC22 | MMC4 pSLEB-LacZ (URA3) | This study |
| MRC23 | MMC4 pSLEC-LacZ (URA3) | This study |
| MRC24 | MMC4 pSLED-LacZ (URA3) | This study |
| MRC25 | MMC4 plac-basal (URA3) | This study |
Transcript Profiling
Wild-type (CAI8 containing CIp10) and mnl1Δ (MSC4 containing CIp10) strains were grown in triplicate to 1 × 107 cells ml−1 in 50 ml SC-pH 3.0 broth and then treated with 0, 20, 120, or 300 mM acetic acid. At various times thereafter, cells were frozen in liquid N2, and RNA was extracted as described by Enjalbert et al. (2003). A control RNA sample was made by pooling cells from 12 replicate exponential cultures grown in SC-pH 3.0. Dye labeling and hybridizations were performed as described previously (Enjalbert et al., 2003) using C. albicans microarrays (Eurogentec, Seraing, Belgium). Microarrays were read at 10-μm resolution using a Scan Array Lite scanner (Perkin Elmer-Cetus Life Sciences, Beaconsfield, United Kingdom). Signal intensities were quantified using QuantArray version 2.0 software (Packard Biosciences, Beaconsfield, United Kingdom). Data normalization was performed using GeneSpring software (Silicon Genetics, Redwood City, CA) by applying intensity-dependent normalization, where the expression ratios were reduced to the residual of the Lowess fit of the intensity versus ratio curve. To account for biological variability of individual genes, log(2) ratios for each gene in each sample were divided by the average of the logs of the ratios from the control hybridizations as described by Enjalbert et al. (2003). Genes with significant changes in transcript abundance relative to the control (false discovery rate set to 10%) were identified using the multiclass SAM v.1 “Significance Analysis of Microarrays” algorithm (Tusher et al., 2001). Promoter analyses were performed using GeneSpring with the find other potential regulatory sequences algorithm. All datasets are available from ArrayExpress at EBI (Accession numbers E-MEXP-1633, E-MEXP-1641, and E-MEXP-1645) and are also provided in the Supplementary Material (Sup1.xls, Sup2.xls, and Sup3.xls).
Proteomics
C. albicans CAF2–1 cells subjected to proteomic analysis were grown in the same way as for transcript profiling. Total protein extracts were prepared in ice-cold protein lysis solution containing 11 M urea, 3.7 M thiourea, 1.8 mM EDTA, 86 mM CHAPS, 13.3% glycerol, 6.6% carrier ampholyte, 100 mM DTT, 42 mM Tris (pH 10.8), and 2.95 μg ml−1 pepstatin (Yin et al., 2004) and stored at −80°C. Protein yields were assessed using standard Bradford assays and 1D gel electrophoresis before running 20 × 24-cm format 2D gels in the range pH 4–7 as described previously (Yin et al., 2004). Gels were run in quadruplicate, using separate biological replicates for each. Gels were stained with colloidal Coomassie blue and scanned at a resolution of 300 dpi (16-bit) using a Hewlett Packard Scanjet 5370C (Palo Alto, CA). Images of protein gels were analyzed using Phoretix 2D software (Nonlinear Dynamics, Newcastle upon Tyne, United Kingdom). Spot volumes were normalized against total spot volume and total spot area. Spots were chosen for further examination if, after log2 transformation of the fold change data, they displayed statistically significant increases or decreases in abundance according to multiclass SAM v.1 analysis (http://www-stat.stanford.edu/∼tibs/SAM/). Spots were then cut from gels and the proteins were identified by peptide mass fingerprinting as described previously (Yin et al., 2004). Database searches were performed with a Protein 1 System (Applied Biosystems, Foster City, CA) using AutoMS-Fit, or manually using MS-Fit (for local database searches) or MASCOT (Matrix Science, London, United Kingdom) software (http://www.matrixscience.com/) to interrogate the annotated Candida genome sequence available at http://www.pasteur.fr/recherche/unites/Galar_Fungail. All experimental data have been submitted to the Aberdeen proteomics facility (http://www.cogeme.abdn.ac.uk/ap-proteome-data.hti).
LacZ Reporter Constructs
LacZ reporter constructs were made using pLac-basal (García-Sánchez et al., 2005). Oligonucleotides containing different types of STRE-like elements (SLEs) or STRE elements (Figure 4A and Table 2) were cloned between the PstI and SalI sites of pLac-basal, upstream of the basal ADH1 promoter and Streptococcus thermophilus lacZ reporter. These SLE reporter plasmids and the empty control pLac-basal were linearized with StuI and transformed into C. albicans. Uri+ transformants containing a single copy of the plasmid integrated at the RPS1 locus were selected for analysis (Murad et al., 2000). β-Galactosidase activity was visualized by chloroform permeabilization of cells and X-Gal overlays and quantified in broth cultures using the Miller ONPG assay adapted for use in microtiter plates (Guarente, 1983; Ausubel et al., 1992; García-Sánchez et al., 2005).
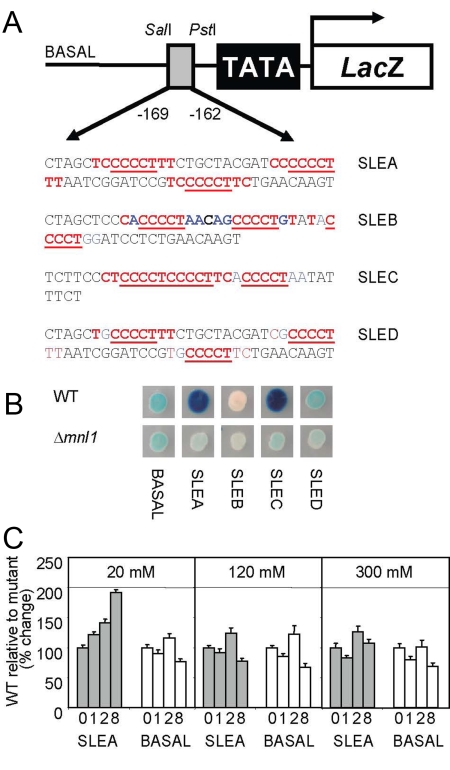
Figure 4.
SLE-mediated Mnl1-dependent gene expression in C. albicans in response to weak acid stress. (A) Diagram of the pLac-basal reporter showing the cloning of the regulatory elements upstream of the basal promoter (TATA) and the S. thermophilus lacZ gene: SLEA, three copies of an artificial consensus SLE element; SLEB, three copies of STRE; SLEC, a short sequence from the native orf19.5711 promoter containing three SLE motifs; and SLED, three copies of a mutant SLE with G next to the core CCCCT element. Red text indicates sequence that fits the consensus; blue text indicates nonconsensus sequence and the core CCCCT element is underlined in each construct. (B) Expression of the lacZ reporters in wild-type (CAI8) and mnl1 (MSC4) cells on solid SC-pH 3.0 medium containing 20 mM acetic acid after 24 h at 30°C. (C) Quantitative temporal activation of the SLEA-lacZ reporter in wild-type cells relative to mnl1 cells after exposure to 20, 120, or 300 mM acetic acid in SC-pH 3.0 for 0–8 h.
Table 2.
Oligomer sequences used in this study
| Oligo | 5′ to 3′ oligo sequence |
|---|---|
| SLEA-TOP | CTGCTGCAGGTCGACGGATCCGCTAGCTCCCCCTTTCTGCTACGATCCCCCCTTTAATCGGATCCGTCCCCCTTCTGAACAAGTCTGCAGAACCAATGCA |
| SLEA-BOT | TGCATTGGTTCTGCAGACTTGTTCAGAAGGGGGACGGATCCGATTAAAGGGGGGATCGTAGCAGAAAGGGGGAGCTAGCGGATCCGTCGACCTGCAGCAG |
| SLEB-TOP | CTGCTGCAGGTCGACGGATCCGCTAGCTCCCACCCCTAACAGCCCCTGTATACCCCTGGATCCTCTGAACAAGTCTGCAGAACCAATGCA |
| SLEB-BOT | TGCATTGGTTCTGCAGACTTGTTCAGAGGATCCAGGGGTATACAGGGGCTGTTAGGGGTGGGAGCTAGCGGATCCGTCGACCTGCAGCAG |
| SLEC-TOP | CTGCTGCAGGTCGACGGATCCGTCTTCCCTCCCCTCCCCTTCACCCCTAATATTTCTCTGCAGAACCAATGCA |
| SLEC-BOT | TGCATTGGTTCTGCAGAGAAATATTAGGGGTGAAGGGGAGGGGAGGGAAGACGGATCCGTCGACCTGCAGCAG |
| SLED-TOP | CTGCTGCAGGTCGACGGATCCGCTAGCTGCCCCTTTCTGCTACGATCGCCCCTTTAATCGGATCCGTGCCCCTTCTGAACAAGTCTGCAGAACCAATGCA |
| SLED-BOT | TGCATTGGTTCTGCAGACTTGTTCAGAAGGGGCACGGATCCGATTAAAGGGGCGATCGTAGCAGAAAGGGGCAGCTAGCGGATCCGTCGACCTGCAGCAG |
RESULTS
Mnl1, But Not Msn4, Is Required for the Weak Acid Stress Response in C. albicans
Ras-cAMP-PKA signaling down-regulates Msn2/4-mediated stress responses in S. cerevisiae (Garreau et al., 2000). Hence we reasoned that the acceleration of programmed cell death in C. albicans by Ras-cAMP-PKA hyperactivation (Phillips et al., 2006) might be mediated through the inactivation of Msn2/4-like proteins. Previous studies indicated that these two transcription factors, Msn4 and Mnl1, are not required for responses to a range of stresses in C. albicans (Nicholls et al., 2004). However, their contribution to the weak acid stress response had not been tested. Therefore we tested the sensitivity of C. albicans mnl1 and msn4 null mutants to acetic acid (Figure 1A). The mnl1 single and mnl1 msn4 double mutants were hypersensitive to this weak acid relative to wild-type and msn4 cells on plates and in liquid culture (MIC80 wild-type and msn4 22 mM, MIC80 mnl1 and mnl1 msn4 14 mM; see Supplementary Material, Sup3.xls). The hypersensitivity of mnl1 cells was suppressed by transformation with the plasmid pACT1-MNL1, confirming that this phenotype is attributable to Mnl1. Therefore, Mnl1 is required for the acetic acid stress response in C. albicans. Msn4 is not required for this response. This is in contrast to S. cerevisiae, where inactivation of the Mnl1 homologue (Yer130c) does not affect acetic acid resistance, whereas msn2 msn4 cells display acetic acid sensitivity (not shown).
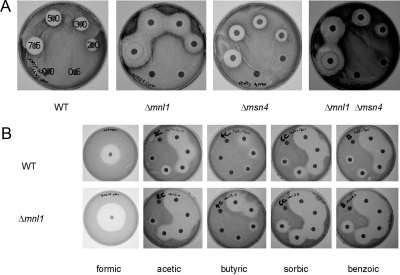
Figure 1.
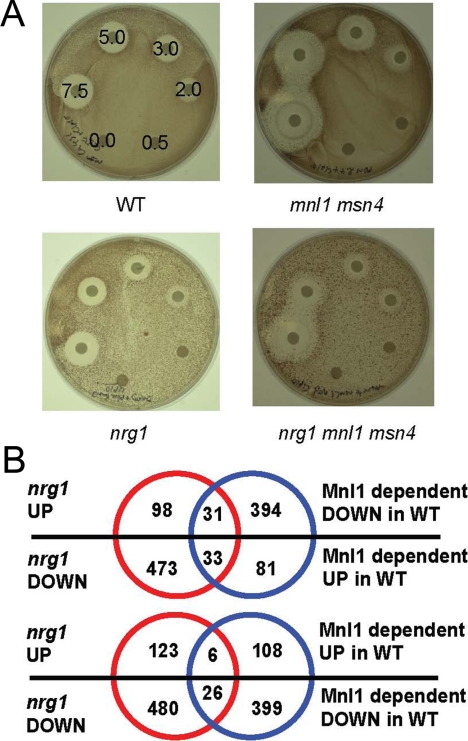
Inactivation of MNL1 causes a defect in weak acid stress responses. C. albicans strains were grown on solid SC-pH 3.0 medium containing weak acids applied on filter discs. All strains were transformed with the URA3-containing plasmid, CIp10. (A) Growth of C. albicans strains on medium containing varying concentrations of acetic acid (mM): wild type, CAI8; mnl1, MSC4; msn4, MSC8; mnl1 msn4, MSC12 (Table 1). (B) Effects of other weak acids on wild-type (CAI8) and mnl1 strains (MSC4): formic (7.5 M), and acetic; butyric; sorbic and benzoic acids (0.5–7.5 M).
To test whether the mnl1 phenotype reflects a defect in the weak acid stress response or susceptibility to the acetate anion, the assays were repeated at pH 8 when acetate is in its dissociated form. No difference was observed between wild-type and mnl1 strains at mild alkaline pH, indicating that the sensitivity of mnl1 cells is attributable to a defect in the weak acid stress response. This was confirmed by testing the sensitivity of the strains to other weak acids (Figure 1B). Mnl1 cells were most sensitive to one- and two-carbon weak acids (formic and acetic acid), but also were sensitive to four- and six-carbon chain weak acids (butyric and sorbic acid) and to an aromatic weak acid (benzoic acid). Msn4 cells were as resistant as wild-type cells to these weak acids.
It was possible that mnl1 mutants are sensitive to general stresses during growth at a low pH or to low pH itself. To test this we compared the sensitivities of wild-type, mnl1, msn4 and mnl1 msn4 mutants to oxidative, reductive, osmotic, heavy metal, and cell wall stresses at pH 3. The conditions, including 0.05–100 mM hydrogen peroxide, tert-butyl hydroperoxide, menadione or tunicamycin, 0.01–25 mM diamide, 0.0015–3 mg/ml calcofluor white, 0.009–18.83 mM caffeine, 0.195–4 mM cadmium sulfate or lead nitrate, 0.03–64 mM copper sulfate, 0.195–400 mM potassium chloride, 0.39–800 mM manganese sulfate, sodium chloride, calcium chloride, or magnesium chloride, 0.78–1.6 mM lithium chloride, 0.078–160 mM sodium nitrite or DTT, and 0.0015–3.2% SDS, were tested in broth microdilution assays. Sensitivities to these agents were also tested at other pHs, but no differences were observed between the strains (not shown). This suggests that Mnl1 contributes specifically to the weak acid stress response in C. albicans.
Mnl1 Is Required for the Adaptation of C. albicans to Acetic Acid Stress
Mnl1 might contribute to acetic acid stress resistance by regulating the acute, short-term response of C. albicans cells to acetic acid and/or by controlling the longer-term adaptive response to this type of stress. To address, this we examined the effects of acetic acid on wild-type and mutant strains in liquid culture at pH 3 (Figure 2A). Under these conditions wild-type cells exhibited a period of growth stasis (5–10 h) before recovering and growing at a normal rate. In contrast, mnl1 cells were unable to undergo this adaptation (up to 48 h). During the period of growth stasis there was no detectable killing of either wild-type or mutant cells, as revealed by PI staining and colony forming units (not shown).
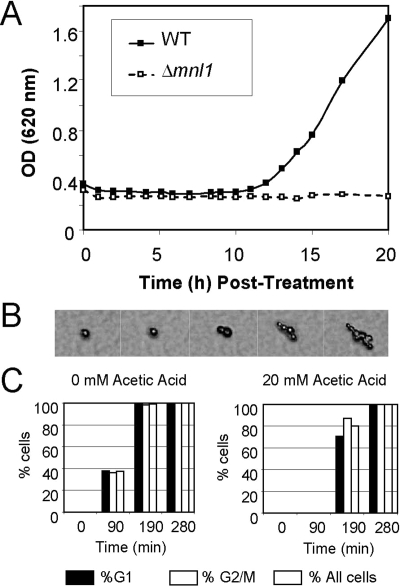
Figure 2.
Mnl1 is required for physiological adaptation to weak acid stress. (A) Growth of wild-type (CAI8) and mnl1 cells (MSC4) in SC-pH 3.0 containing 20 mM acetic acid. (B) Time-series micrographs of the recovery growth of a microcolony on SC-pH 3.0 medium containing 20 mM acetic acid. (C) The proportion of wild-type C. albicans cells (CAI8) in G1 or G2/M phase of the cell cycle at the onset of the experiment that resume growth on solid SC-pH3.0 medium in the presence or absence of 20 mM acetic acid: black bars, G1 unbudded cells (n = 495); white bars, G2/M cells with large buds (n = 487); hatched bars, all cells (n = 982). The y-axis shows the percentage of C. albicans cells that resume growth as a function of their cell cycle status at the start of an experiment (e.g., the percentage of all G1 cells that resume growth 90 min after plating).
In principle, growth recovery could have been due to a temporary physiological adaptation to acetic acid or to the selection of mutants with intrinsic resistance to acetic acid stress. To test this, cells that had “adapted” to acetic acid were isolated, regrown in the absence of the acetic acid, and then re-exposed to this weak acid. Once again these cells displayed growth stasis before resuming normal growth. Moreover the examination of single cells on solid agar containing 20 mM acetic acid at pH 3 revealed that the majority of the cells adapted and resumed growth at similar rates (Figure 2, B and C). Indeed cells at different phases of the cell cycle (according to their budding status) recovered at similar rates. We conclude that C. albicans undergoes a physiological adaptation to acetic acid stress before resuming growth and that Mnl1 is required for this adaptation.
Definition of the Mnl1 Regulon in C. albicans
We performed genome-wide expression profiling to define the C. albicans genes that are regulated by Mnl1 under weak acid stress conditions (20 mM acetic acid). RNA was harvested from wild-type and mnl1 cells during the static phase (0, 10, 30, 60, and 300 min) and just before wild-type cells resumed growth (600 min; Figure 2A). Transcript profiling revealed that during adaptation to acetic acid, a subset of 318 C. albicans genes was up-regulated more than 1.5-fold in wild-type cells relative to mnl1 cells. These changes were reproducible and statistically significant (SAM, 10% FDR: Figure 3A). A further 849 genes were up-regulated in both wild-type and mnl1 cells after exposure to the weak acid for 300 min during the late phase of adaptation. The validity of transcript profiling data were confirmed by Northern analysis of 17 mRNAs under the four experimental conditions (0, 20, 120, and 300 mM acetic acid; see Supplementary Data, Sup3.xls). The data indicate that extensive remodeling of C. albicans gene expression takes place during adaptation to this stress and that much of this remodeling is Mnl1-independent. However, 318 genes (∼5% of the C. albicans genome) are regulated in an Mnl1-dependent manner under these growth conditions. GO-Slim terms associated with this gene subset, loosely referred to as the Mnl1 regulon, are highly enriched for genes with stress related functions, including more granular GO-terms such as Response to Stress, Response to Oxidative Stress, cAMP Signals, and Glycogen Metabolism (Figure 3B and Supplementary Material, Sup3.xls).
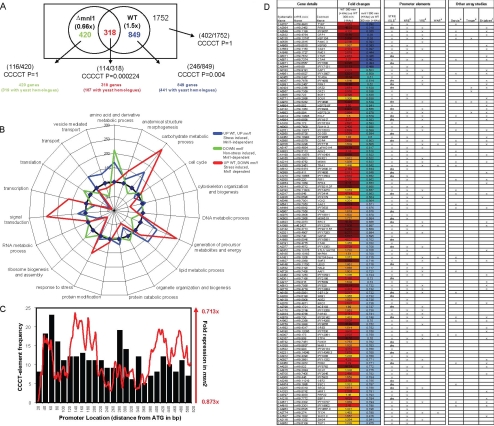
Figure 3.
Global analysis of C. albicans gene expression during exposure to weak acid stress. (A) Venn diagram showing the overlap between gene sets that are up-regulated after 300-min exposure to 20 mM acetic acid in wild-type CAI8 cells (relative to untreated cells) and those that are down-regulated in acetic acid–treated mnl1 cells (relative to acetic acid–treated wild-type cells). Numbers of genes containing STRE-like elements (CCCCT) are shown (numbers in brackets show CCCCT-containing genes/total number of genes in subset). p values indicate the chance, according to the hypergeometric probability distribution, of each sequence element occurring in each gene list compared with its overall frequency in noncoding regions of the genome. The 114 genes in the overlapping set that have a STRE-like element in the first 500 base pairs of their promoter are classed as Mnl1-dependent weak acid–responsive genes. (B) Radial plot showing frequency of GO-terms relative to all GO-terms associated with the arrayed genes with S. cerevisiae homologues. The categories shown cover the top 75% of all biological processes present in any one of the treatments. For example, genes involved with “Signal Transduction” are significantly overrepresented in the subset of genes that are induced in response to weak acid in an Mnl1-dependent manner. (C) Graph showing distribution of SLEs among promoters of Mnl1-regulated genes (■) and the mean fold difference in expression for these genes in mnl1 compared with wild-type cells (solid line indicating running average of expression for 20-base pair windows). (D) Mnl1-regulated genes showing those that contain STRE/SLE (1), NRE (2), YRE (3), or WAR elements (4). Also shown are genes (5) found to be regulated by Nrg1 in the study of García-Sánchez et al. (2005) (6), Treger's list of S. cerevisiae Msn2-regulated STRE containing genes (Treger et al., 1998), and (7) genes significantly altered under oxidative, heat shock, heavy metal, or osmotic stress; see Supplementary Material for a full list of genes.
Next we performed in silico analyses of Mnl1-dependent promoters to identify regulatory elements that might drive the expression of this regulon in C. albicans. A STRE-like element (CCCCT) was the most highly enriched element in these promoters (p = 0.000224) and is best characterized by the sequence HHYYCCCCTTYTY (SLE). One hundred fourteen of the 318 Mnl1-dependent genes that were up-regulated in wild-type cells under acetic acid stress conditions contain one or more SLEs in the first 500 base pairs of their promoters (Figure 3D and Supplementary Material, Sup3.xls). There was no significant enrichment of the SLE sequence in the 1752 remaining genes that passed the quality control criteria in all experiments. Closer examination of Mnl1-dependent genes revealed that those with three SLEs in the first 500 base pairs of their promoter were induced the most strongly and that the highest levels of induction were observed when SLEs were clustered between 120 and 200 base pairs or 380–500 base pairs upstream of the start codon (Figure 3C).
SLE-mediated, Mnl1-dependent Gene Induction during Acetic Acid Stress in C. albicans
To test whether Mnl1 can activate transcription via the SLE element during acetic acid stress in C. albicans, we constructed SLE-containing reporters using the pLac-basal plasmid. This plasmid contains the basal promoter region of the C. albicans ADH1 gene (Tripathi et al., 2002) cloned upstream of the Streptococcus thermophilus lacZ reporter (Uhl and Johnson, 2001) in the vector, CIp10 (Murad et al., 2000). Oligonucleotides containing different sequence elements were cloned upstream of the basal promoter to create the pLac-SLE plasmids (Figure 4A). pLac-SLEA contains synthetic repeats of the SLE consensus sequence (HHYYCCCCTTYTY), whereas pLac-SLEB carries classical STRE elements (CCCCT). pLac-SLEC contains natural SLEs from a 35 base-pair section of the IPF6629/AHP1 (orf19.2762) promoter, while pLac-SLED contains mutated SLE elements that have a nonstandard G residue immediately preceding the CCCCT core.
A single copy of each reporter plasmid was integrated into the genomes of wild-type and mutant C. albicans cells, and their expression was compared under weak acid stress conditions (20 mM acetic acid: Figure 4B). The control, basal reporter was expressed at similar levels in wild-type and mnl1 cells. In contrast, the reporters containing synthetic or natural SLEs were strongly induced in wild-type cells under these conditions (SLEA and SLEC). This induction was not observed in mnl1 cells. Furthermore, the reporter carrying mutated SLEs was not induced in wild-type cells (SLED). The temporal regulation of the SLEA reporter was analyzed, revealing that it was induced hours after the initial exposure to 20 mM acetic acid (Figure 4C). This was consistent with the role of Mnl1 during stress adaptation. We conclude that SLE elements mediate transcriptional activation by Mnl1 during the adaptive response to acetic acid stress.
STRE-containing reporters that lacked full SLE elements were not induced in response to acetic acid stress (SLEB and SLED: Figure 4B). This indicates that Mnl1 does not activate transcription via the classical STRE element under these conditions. This is consistent with our previous work, which showed that STRE-containing reporters are not induced in C. albicans under a range of stress conditions (Nicholls et al., 2004). These data reinforce the idea that there are significant differences between the Msn2/4-STRE regulon in S. cerevisiae and the Mnl1-SLE regulon C. albicans.
The Mnl1 and Nrg1 Regulons Overlap
Our analyses of SLE-containing promoters in C. albicans revealed that overall 60% of SLEs (and 70% of Mnl1-dependent SLEs; Figure 3D) match the consensus Nrg1 response element (NRE) sequence (MVCCCT: Murad et al., 2001b), which is recognized by the transcriptional repressor Nrg1. Furthermore, in 90% of promoters containing multiple SLEs, at least one of these elements matches the NRE motif. Nrg1 has been proposed to influence the general stress response in S. cerevisiae via similar regulatory elements (Vyas et al., 2005). Therefore, we examined whether Nrg1 modulates the acetic acid response in C. albicans.
First we compared the acetic acid sensitivity of nrg1 and wild-type cells. No significant difference in sensitivity was observed (Figure 5A). Next we deleted the NRG1 locus in mnl1 msn4 cells to generate a triple mutant. This mutant displayed a morphological phenotype similar to single nrg1 mutants (Braun et al., 2001; Murad et al., 2001a): mnl1 msn4 nrg1 cells were constitutively filamentous and invasive (not shown). Interestingly, the mnl1 msn4 nrg1 triple mutant was less sensitive to acetic acid stress (MIC80 20 mM) than the corresponding mnl1 or mnl1 msn4 mutants (MIC80 14 mM; Figure 5A). This phenotype was difficult to quantify because of the filamentous nature of nrg1 cells. Nevertheless, this partial rescue of the mnl1 acetic acid sensitivity was reproducible, and it suggests that Nrg1 plays a role that is antagonistic to Mnl1 under these conditions. This is consistent with the idea that the transcriptional activator Mnl1, and the transcriptional repressor Nrg1, regulate a common set of genes involved with adaptation to acetic acid stress. The validity of this idea was reinforced by comparing our mnl1 transcript profiling data with those for the nrg1 mutant (García-Sánchez et al., 2005). Many genes that are activated in an Mnl1-dependent manner are also repressed by Nrg1 (Figure 5B), and the overlap between these regulons is highly significant. This overlap includes MSN4 and PDE2. Pde2 is a phosphodiesterase that down-regulates Ras-cAMP signaling in C. albicans (Bahn et al., 2003; Jung and Stateva, 2003). Finally, loss of nrg1 de-represses the expression of lacZ under the control of SLEA (Supplementary Data, Sup3.xls).
Figure 5.
Functional overlap of the Mnl1 and Nrg1 regulons in C. albicans. (A) Growth of wild-type (CAI8), mnl1 (MSC4), mnl1 msn4 (MSC12), nrg1 (MMC4), and nrg1 mnl1 msn4 (SNC10) strains on solid SC-pH 3.0 medium after exposure to varying concentrations of acetic acid (mM). (B) Venn diagram showing the overlap between Mnl1 and Nrg1 regulons in C. albicans. The number of genes that are both up-regulated in nrg1 cells and down-regulated in mnl1 cells is highly significant (cumulative p values, P31 = 5.75 × 10−5, modal overlap expected = 9). Similarly, the number of C. albicans genes that are both down-regulated in nrg1 cells and up-regulated in mnl1 cells is highly significant (P33 = 3.85 × 10−3, modal overlap expected = 9). In contrast, the number of genes that are up- or down-regulated in both mutants are less significant: P6 = 0.909, modal overlap expected = 2; and P26 = 3.22 × 10−2, modal overlap expected = 36, respectively. Lists of genes in each overlapping category and their associated GO-terms are given in the Supplementary Material.
Mnl1-dependent Gene Expression Occurs in Stressed But Not Dying Cells
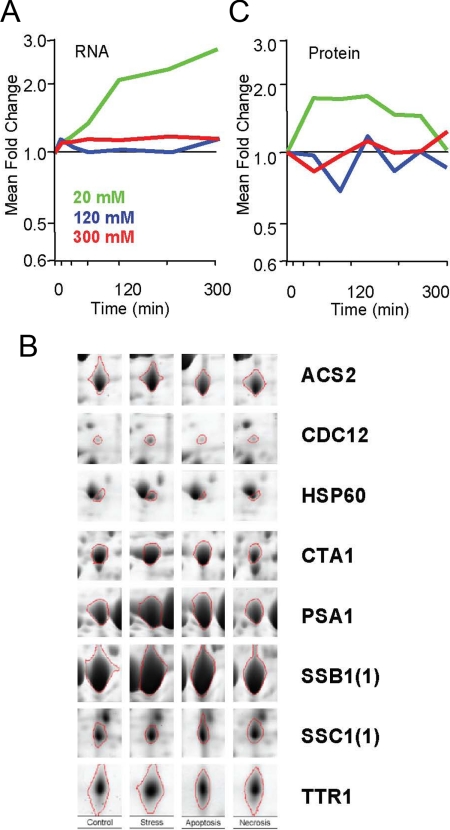
Elevated doses of acetic acid induce programmed cell death in C. albicans (Phillips et al., 2003). Given the contrasting outcomes of acetic acid stress adaptation and death responses, we compared the expression of the 114 Mnl1-dependent, SLE-containing genes in C. albicans cells exposed to different concentrations of acetic acid. Wild-type cells were treated with 20 mM (which induces weak acid stress), 120 mM (which promotes apoptosis), or 300 mM acetic acid (which activates necrosis in C. albicans), harvested at different time points, and subjected to transcript profiling. After exposure to 20 mM acetic acid, the mean induction level for these 114 genes increased over time (Figure 6A). In contrast, their mean induction levels remained unchanged under death inducing conditions. Similarly, the SLE-containing reporter, SLEA, was induced during stress adaptation, but was not induced in response to proapoptotic or pronecrotic doses of acetic acid (Figure 4C). These data indicate that Mnl1-dependent, SLE-containing genes are not induced in dying C. albicans cells.
Figure 6.
Mnl1-dependent gene activation occurs in stressed but not dying C. albicans cells. (A) Mean fold change in transcript levels for the subset of 114 Mnl1-dependent genes in wild-type C. albicans after exposure to 20, 120, or 300 mM acetic acid in SC-pH 3.0 for 0–300 min. (B) We have identified proteins corresponding to 13 Mnl1-dependent genes on 2D gels. The mean fold change in the levels of these proteins in wild-type C. albicans cells after exposure to 20, 120, or 300 mM acetic acid in SC-pH 3.0 for 0–300 min are presented. (C) Changes in levels of Acs1, Cta1, Hsp60, Psa1, Ttr1, Cdc12, Ssb1, and Ssc1 proteins are apparent in weak acid stress–treated (20 mM acetic acid) cells, but not in dying cells exposed to 120 or 300 mM acetic acid for 1 h. A full list of proteome changes associated with these treatment conditions is provided in the Supplementary Material (Sup3.xls) along with a reference 2D gel map.
To test this further we examined the expression of Mnl1-dependent, SLE-containing genes at the level of the proteome. We have identified proteins corresponding to 13 of these genes directly by 2D gel electrophoresis (Figures 3D and 6B). The regulation of these proteins accurately reflects that of Mnl1-dependent, SLE-containing genes. This class of proteins was induced under weak acid stress conditions, but remained relatively constant under death inducing conditions (Figure 6C).
DISCUSSION
The Adaptation of C. albicans to Weak Acids Depends on Mnl1
Several observations indicate that Mnl1 plays a key role in the regulation of the weak acid stress response of C. albicans. The inactivation of Mnl1 increases the sensitivity of C. albicans to a range of weak acids (Figure 1). This is due to the inability of mnl1 cells to undergo an adaptive response to acetic acid (Figure 2). This defect is only apparent at low pH, and the defect is not attributable to low pH per se, because the sensitivity of mnl1 strains to other stresses, at low pH, is the same as that of wild-type cells. Hence Mnl1 is required specifically for the weak acid response.
A second Msn2-like protein exists in C. albicans: Msn4 (Nicholls et al., 2004). Our data indicate that there are clear differences between the roles of Mnl1 and Msn4 in this yeast. Unlike Mnl1, Msn4 is not required for the response of C. albicans to weak acids (Figure 1). This contrasts with the situation in S. cerevisiae where Yer130c plays no obvious role in stress adaptation, whereas Msn2 and Msn4 are central to the core stress response and are required for the transcriptional and cellular responses to weak acids (Estruch and Carlson, 1993; Martinez-Pastor et al., 1996; Causton et al., 2001; Schüller et al., 2004).
In S. cerevisiae a third transcriptional regulator, War1, contributes to activate the regulation of weak acid stress responses (Schüller et al., 2004). Msn2, Msn4, and War1 appear to contribute to differing extents depending on the nature of the weak acid stress. War1 appears to be more important for responses to longer chain weak acids and is responsible for the activation of the transporter gene, PDR12 (Kren et al., 2003). However, PDR12 is not induced in response to acetic acid (Hatzixanthis et al., 2003), and Msn2 and Msn4 may be more important under these stress conditions. A WAR1 homologue has been identified in C. albicans (orf19.1035: Lebel et al., 2006). These authors report that C. albicans war1 cells are only sensitive to acetic acid in a trp1 background, suggesting that War1 is not essential for the response to acetic acid. WAR1 is however in the subset of 114 acetic acid–induced Mnl1-dependent genes we identified by transcript profiling (Figure 3D), but only five of these 114 genes contain a putative War1 recognition sequence similar to that described for WAR1 in S. cerevisiae (Kren et al., 2003). These observations suggest that Mnl1 is critical for the response of C. albicans to acetic acid, but they do not exclude a role for War1.
The Shape and Function of the Mnl1 Regulon
Our analyses of mnl1 mutant phenotypes indicated that Mnl1 contributes to the adaptation of C. albicans cells to weak acids. This suggests that Mnl1 might regulate late adaptation genes and possibly immediate early expression of stress genes. This was confirmed by our temporal analyses of the global transcriptional response of C. albicans to acetic acid. There were significant changes in the C. albicans transcriptome during the early and late phases of this response, and Mnl1 contributed to each phase. This is consistent with the available transcript profiling data for the immediate early (Schüller et al., 2004) and adaptative (de Nobel et al., 2001) responses to weak acid stress in S. cerevisiae, which share relatively few genes in common.
Our global analyses identified an acetic acid-induced, Mnl1-dependent regulon of 114 genes in C. albicans, many of which have well-described stress-related functions (Figure 3). It is highly significant that this regulon contains 21 transcriptional regulators. These include CRZ1 and CRZ2 (regulators of calcineurin function required for cell morphogenesis, azole tolerance, membrane stress responses, survival in serum, and virulence in mice: Santos and de Larrinoa, 2005), CAP1 (involved in multidrug resistance and oxidative stress responses: Alarco and Raymond 1999), BCR1 (implicated in biofilm formation: Nobile et al., 2005), CTA4 (induced by nitric oxide: Hromatka et al., 2005), CTA8 (response to stress), RIM101 (alkaline pH response and morphogenesis: Davis et al., 2000), RPN4 (a putative regulator of proteasomal functions and a component of the core stress response: Enjalbert et al., 2006), and SKO1 (induced by osmotic stress: Enjalbert et al., 2006). Mnl1 also controls the gene encoding the transcriptional regulator Tup1, which has a strong link to the repressor functions of Nrg1 and Rfg1 (Kadosh and Johnson, 2001; Murad et al., 2001a).
Additional stress response genes were up-regulated in our dataset, including GRE3, DDR48, and the so-called heat-shock proteins HSP104, HSP90, SSB1, and SSC1. Interestingly, both ERC3 (an ethionine resistance protein) and SEO2 (a suppressor of sulfoxide ethionine resistance) were also regulated by Mnl1. Calcium (PMC1) and copper (CRP1) transporters were up-regulated along with SNG4 (implicated as a drug transporter) and the putative multidrug resistance proteins CDR4, orf19.4779, and orf19.4551 (Sanglard et al., 1995).
Interestingly, several genes linked to Ras-cAMP signaling were up-regulated by Mnl1. These included PDE2 (a cAMP phosphodiesterase: Jung and Stateva, 2003) and NCE103 (a carbonic anhydrase gene implicated in the regulation of adenylate cyclase activity: Klengel et al., 2005). MSN4 also appeared in the list of MNL1 regulated genes.
Overall, there is considerable overlap between the acetic acid stress–induced genes identified here and C. albicans stress responsive genes described in other studies. Of 306 heat shock–inducible genes (Enjalbert et al., 2003), 97 show significant changes in our dataset. Furthermore, 50 of the 114 acetic acid stress–induced, Mnl1-dependent genes we have identified are also up-regulated in response to oxidative, osmotic, or heavy metal stresses (Enjalbert et al., 2006). Most of these shared genes are specifically responsive to oxidative stress, reinforcing the perceived links between weak acid stress and the production of reactive oxygen species (Piper, 1999; Phillips et al., 2003, Giannattasio et al., 2005).
Control of Adaptation by Mnl1
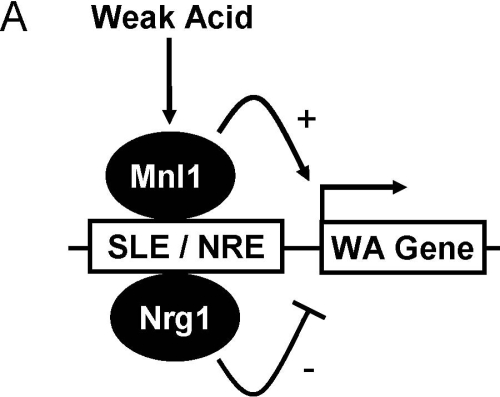
Our in silico analyses revealed a sequence element that is highly enriched in the promoters of acetic acid–induced genes. Interestingly, this promoter element (SLE: HHYYCCCCTTYTY) is distinct from the classical STRE element (CCCCT) that drives Msn2/4 activation in S. cerevisiae (Figure 3). The SLE imposes acetic acid–induced, Mnl1-dependent transcription upon a reporter gene in C. albicans (Figure 4). This expression pattern is blocked by a single base pair mutation close to the core of the of SLE sequence and is not observed for the classical STRE element in C. albicans (Figure 4). These observations suggest that Mnl1 regulates the transcription of many acetic acid–induced genes directly via the SLE (Figure 7).
Figure 7.
Model describing the potential roles of Mnl1 and Nrg1 in the adaptation of C. albicans to weak acid stress. As described in the text, a subset of C. albicans genes is induced in response to weak acid stress (20 mM acetic acid). The induction of many of these genes is dependent on Mnl1, which activates transcription in C. albicans through the promoter element, SLE. Adaptation to weak acid stress is dependent on Mnl1 and hence presumably on the activation of (some) Mnl1-dependent weak acid genes. The repressor Nrg1 may act antagonistically with Mnl1, probably via those SLE elements that overlap with the NRE consensus, though not all SLEs are NREs.
Of the 308 genes that were induced in response to acetic acid stress and down-regulated in mnl1 cells (Figure 3), 204 did not contain an SLE element in the 500-base pair region upstream of their open reading frame. How might their regulation by Mnl1 be explained? Clearly some of these genes might carry SLEs further upstream. In other cases, their regulation by Mnl1 might be indirect. As described above, the set of acetic acid–induced, Mnl1-dependent genes contains a large proportion of genes encoding transcriptional regulators. This suggests that Mnl1-mediated adaptation to acetic acid stress might involve the hierarchical control of numerous regulons in C. albicans. This is consistent with the idea that Mnl1 regulates some genes indirectly and might account for the absence of SLE elements in the promoters of some genes that were down-regulated in the mnl1 mutant.
There is overlap between the consensus sequences for the SLE (HHYYCCCCTTYTY) and NRE elements (MVCCCT: Murad et al., 2001b). On this basis we reasoned that there might be overlap between the Mnl1 and Nrg1 regulons in C. albicans. Our data are consistent with this idea. First, the inactivation of Nrg1 influences the weak acid sensitivity of mnl1 msn4 cells. Second, there is clear overlap between the gene sets that are regulated by Mnl1 and Nrg1 in C. albicans (Figure 5). Therefore, Nrg1 modulates the activity of the Mnl1 regulon in this pathogen (Figure 7).
The C. albicans genes that appear to be coregulated by Mnl1 and Nrg1 include AAF1, CDR4, CTA4, ERO1, GCF1, HSP104, NUP6, PDE2, SMF2, SSC1, TRA1, and TYE7, with MNL1 itself appearing as an Nrg1-regulated gene. In S. cerevisiae, 78 of 150 Nrg1 repressed genes that are induced by acid pH, appear to be regulated by Msn2/4 (Vyas et al., 2005). Moreover 106 of these 150 Nrg1-repressed genes are induced by overexpression of Msn2 and/or Msn4. We suggest that Mnl1/Yer130c (and Msn2/4) might overcome the transcriptional repression of some stress responsive genes by Nrg1 in both S. cerevisiae and C. albicans. In C. albicans, Mnl1 function primarily relates to a specific response to weak acid stress (Figure 7), rather than a general stress response.
Expression of the Mnl1 Regulon during Acetic Acid–induced Death in C. albicans
Ras-cAMP signaling down-regulates the Msn2/4-mediated stress response in S. cerevisiae (Gorner et al., 1998, 2002; Garreau et al., 2000). On the other hand, Ras-cAMP signaling accelerates acetic acid–induced cell death in C. albicans (Phillips et al., 2006). Therefore we compared the expression of the Mnl1 regulon during acetic acid stress and death responses in C. albicans (Figure 6). As described above, this regulon was activated during the weak acid stress response (induced by 20 mM acetic acid; Figure 3). Interestingly, the Mnl1 regulon was not activated in response to lethal doses of acetic acid that induce programmed cell death (120 mM) or necrotic death (300 mM; Figure 6). This contrasts with many other genes that are induced or repressed in dying cells (Contreras et al., 2002; Fernández-Arenas et al., 2007; see also Supplementary Array data files Sup1.xls and Sup2.xls). Furthermore, this expression pattern was reflected at the level of the C. albicans proteome (Figure 6). The fact that the Mnl1-mediated adaptive response to acetic acid is not activated when C. albicans cells are exposed to relatively high concentrations of weak acid is highly significant. First, an inability to respond appropriately to weak acid might contribute to cell death. Second, this observation is consistent with the idea that Ras-cAMP signaling might down-regulate the Mnl1-mediated adaptation of C. albicans cells, whereas accelerating programmed cell death. This link between Mnl1 regulation and cell death is reinforced by the observation that the deletion of MSN2, MSN4, and RIM15 decreases chronological lifespan in S. cerevisiae (Fabrizio et al., 2004).
In conclusion, our analyses have defined a key role for the Yer130c orthologue, Mnl1, during adaptation to weak acid stress and suggest a specific molecular link between the regulation of weak acid stress and death responses in C. albicans. Furthermore, our observations highlight the divergent regulation of stress responses between this pathogen and S. cerevisiae, which presumably reflect the contrasting niches they occupy.
Supplementary Material
ACKNOWLEDGMENTS
We thank Professor Wojtek Krzanowski (University of Exeter) for advice on the calculation of probabilities for overlapping gene sets and Geraldine Butler and Ken Wolfe for advice on yeast gene synteny. This work was supported by a United Kingdom Biotechnology and Biological Sciences Research Council (BBSRC) Grant BB/C501176/1. The BBSRC provided additional support for A.B., L.S., J.W., D.S., Z.Y., and Grants BBS/B/06679, BBS/B/13764, BB/C510391/1, and BB/D009308/1 for S.N.
Abbreviations used:
- NRE
Nrg1 response element
- SLE
STRE-like element.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-09-0946) on July 23, 2008.
REFERENCES
- Alarco A. M., Raymond M. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J. Bacteriol. 1999;181:700–708. doi: 10.1128/jb.181.3.700-708.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Monge R., Navarro-Garcia F., Molero G., Diez-Orejas R., Gustin M., Pla J., Sanchez M., Nombela C. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J. Bacteriol. 1999;181:3058–3068. doi: 10.1128/jb.181.10.3058-3068.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. Current Protocols in Molecular Biology. New York: Greene Publishing Associates and Wiley-Interscience; 1992. [Google Scholar]
- Bahn Y. S., Staab J., Sundstrom P. Increased high-affinity phosphodiesterase PDE2 gene expression in germ tubes counteracts CAP1-dependent synthesis of cyclic AMP, limits hypha induction and promotes virulence of Candida albicans. Mol. Microbiol. 2003;50:391–409. doi: 10.1046/j.1365-2958.2003.03692.x. [DOI] [PubMed] [Google Scholar]
- Brand A., MacCallum D. M., Brown A.J.P., Gow N.A.R., Odds F. C. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaroyt. Cell. 2004;3:900–909. doi: 10.1128/EC.3.4.900-909.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun B. R., Kadosh D., Johnson A. D. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 2001;20:4753–4761. doi: 10.1093/emboj/20.17.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne K. P., Wolfe K. H. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causton H. C., Ren B., Koh S. S., Harbison C. T., Kanin E., Jennings E. G., Lee T. I., True H. L., Lander E. S., Young R. A. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras R. H., Eberhardt I., Luyten W.H.M.L., Reekman R. J. Bax-responsive genes for drug target identification in yeast and fungi. 2002 Patent International Publication Number WO 02/064766 A2. [Google Scholar]
- Davis D., Wilson R. B., Mitchell A. P. RIM101-dependent and -independent pathways govern pH responses in Candida albicans. Mol. Cell Biol. 2000;20:971–978. doi: 10.1128/mcb.20.3.971-978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nobel H., Lawrie L., Brul S., Klis F., Davis M., Alloush H., Coote P. Parallel and comparative analysis of the proteome and transcriptome of sorbic acid-stressed Saccharomyces cerevisiae. Yeast. 2001;18:1413–1428. doi: 10.1002/yea.793. [DOI] [PubMed] [Google Scholar]
- Enjalbert B., Nantel A., Whiteway M. Stress induced gene expression in Candida albicans: absence of a general stress response. Mol. Biol. Cell. 2003;14:1460–1467. doi: 10.1091/mbc.E02-08-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert B., Smith D. A., Cornell M. J., Alam I., Nicholls S., Brown A.J.P., Quinn J. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol. Biol. Cell. 2006;17:1018–1032. doi: 10.1091/mbc.E05-06-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch F., Carlson M. Two homologous zinc finger genes identified by multicopy suppression in a SNF1 protein kinase mutant of Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:3872–3881. doi: 10.1128/mcb.13.7.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P., Pletcher S. D., Minois N., Vaupel J. W., Longo V. D. Chronological aging-independent replicative life span regulation by Msn2/Msn4 and Sod2 in Saccharomyces cerevisiae. FEBS Lett. 2004;557:136–142. doi: 10.1016/s0014-5793(03)01462-5. [DOI] [PubMed] [Google Scholar]
- Fernández-Arenas E., Cabezón V., Bermejo C., Arroyo J., Nombela C., Diez-Orejas R., Gil C. Integrated proteomic and genomic strategies bring new insight into Candida albicans response upon macrophage interaction. Mol. Cell Proteomics. 2007;6:460–478. doi: 10.1074/mcp.M600210-MCP200. [DOI] [PubMed] [Google Scholar]
- Fonzi W. A., Irwin M. Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin C., De Groot P., MacCallum D., Schaller M., Klis F., Odds F. C., Hube B. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol. Microbiol. 2005;56:397–415. doi: 10.1111/j.1365-2958.2005.04557.x. [DOI] [PubMed] [Google Scholar]
- García-Sánchez S., Mavor A. L., Russell C. L., Argimon S., Dennison P., Enjalbert B., Brown A.J.P. Global roles of Ssn6 in Tup1- and Nrg1-dependent gene regulation in the fungal pathogen, Candida albicans. Mol. Biol. Cell. 2005;16:2913–2925. doi: 10.1091/mbc.E05-01-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garreau H., Hasa R. N., Renault G., Estruch F., Boy-Marcotte E., Jacquet M. Hyperphosphorylation of Msn2 and Msn4 in response to heat shock and the diauxic shift is inhibited by cAMP in Saccharomyces cerevisiae. Microbiology. 2000;146:2113–2120. doi: 10.1099/00221287-146-9-2113. [DOI] [PubMed] [Google Scholar]
- Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., Botstein D., Brown P. O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio S., Guaragnella N., Corte-Real M., Passarella S., Marra E. Acid stress adaptation protects Saccharomyces cerevisiae from acetic acid-induced programmed cell death. Gene. 2005;354:93–98. doi: 10.1016/j.gene.2005.03.030. [DOI] [PubMed] [Google Scholar]
- Gillum A. M., Tsay E. Y., Kirsch D. R. Isolation of the Candida albicans gene for orotidine-5_-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- Gorner W., Durchschlag E., Martinez-Pastor M. T., Estruch F., Ammerer G., Hamilton B., Ruis H., Schüller C. Nuclear localisation of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorner W., Durchschlag E., Wolf J., Brown E. L., Ammerer G., Hamilton B., Ruis H., Schüller C. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 2002;21:135–144. doi: 10.1093/emboj/21.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Hatzixanthis K., Mollapour M., Seymour I., Bauer B. E., Krapf G., Schüller C., Kuchler K., Piper P. W. Moderately lipophilic carboxylate compounds are the selective inducers of the Saccharomyces cerevisiae Pdr12p ATP-binding cassette transporter. Yeast. 2003;20:575–585. doi: 10.1002/yea.981. [DOI] [PubMed] [Google Scholar]
- Holyoake C. D., Bracey D., Piper P. W., Kuchler K., Coote P. J. The Saccharomyces cerevisiae weak-acid-inducible ABC transporter Pdr12 transports fluorescein and preservative anions from the cytosol by an energy dependent mechanism. J. Bacteriol. 1999;181:4644–4652. doi: 10.1128/jb.181.15.4644-4652.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hromatka B. S., Noble S. M., Johnson A. D. Transcriptional response of Candida albicans to nitric oxide and the role of the YHB1 gene in nitrosative stress and virulence. Mol. Biol. Cell. 2005;16:4814–4826. doi: 10.1091/mbc.E05-05-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang C. S., Rhie G. E., Oh J. H., Huh W. K., Yim H. S., Kang S. O. Copper- and zinc-containing superoxide dismutase (Cu/ZnSOD) is required for the protection of Candida albicans against oxidative stresses and the expression of its full virulence. Microbiology. 2002;148:3705–3713. doi: 10.1099/00221287-148-11-3705. [DOI] [PubMed] [Google Scholar]
- Jacquet M., Renault G., Lallet S., De Mey J., Goldbeter A. Oscillatory nucleo-cytoplasmic shuttling of the general stress response transcriptional activators Msn2 and Msn4 in Saccharomyces cerevisiae. J. Cell Biol. 2003;161:497–505. doi: 10.1083/jcb.200303030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W. H., Stateva L. I. The cAMP phosphodiesterase encoded by CaPDE2 is required for hyphal development in Candida albicans. Microbiology. 2003;149:2961–2976. doi: 10.1099/mic.0.26517-0. [DOI] [PubMed] [Google Scholar]
- Kadosh D., Johnson A. D. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol. Cell. Biol. 2001;21:2496–2505. doi: 10.1128/MCB.21.7.2496-2505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T., et al. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr. Biol. 2005;15:2021–2026. doi: 10.1016/j.cub.2005.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs H. A., Wiggins D., Stubbs M., Sols A., Bedoya F. Studies on the mechanism of the antifungal action of benzoate. Biochem. J. 1983;214:657–663. doi: 10.1042/bj2140657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kren A., Magnum Y. M., Bauer B. E., Schüller C., Wolfger H., Hatzixanthis K., Mollapour M., Gregori C., Piper P., Kuchler K. War1p, a novel transcription factor controlling weak acid stress response in yeast. Mol. Cell. Biol. 2003;23:1775–1785. doi: 10.1128/MCB.23.5.1775-1785.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert R. J., Stratford M. Weak acid preservatives: modelling microbial inhibition and response. J. Appl. Microbiol. 1999;86:157–164. doi: 10.1046/j.1365-2672.1999.00646.x. [DOI] [PubMed] [Google Scholar]
- Lebel K., MacPherson S., Turcotte B. New tools for phenotypic analysis in Candida albicans: the WAR1 gene confers resistance to sorbate. Yeast. 2006;23:249–259. doi: 10.1002/yea.1346. [DOI] [PubMed] [Google Scholar]
- Lorenz M. C., Bender J. A., Fink G. R. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaroyt. Cell. 2004;3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler G., Schüller C., Adam G., Ruis H. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 1993;12:1997–2003. doi: 10.1002/j.1460-2075.1993.tb05849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martchenko M., Alarco A. M., Harcus D., Whiteway M. Superoxide dismutases in Candida albicans: transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Mol. Biol. Cell. 2004;15:456–467. doi: 10.1091/mbc.E03-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Pastor M. T., Marchler G., Schüller C., Marchler-Bauer A., Ruis H., Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress-response element. EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- Montcalm L. J., Wolfe K. H. Genome evolution in hemiascomycete yeasts. Mycota. 2006;13:19–34. [Google Scholar]
- Murad A. M., Lee P. R., Broadbent I. D., Barelle C. J., Brown A.J.P. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast. 2000;16:325–327. doi: 10.1002/1097-0061(20000315)16:4<325::AID-YEA538>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Murad A. M., d'Enfert C., Gaillardin C., Tournu H., Tekaia F., Talibi D., Marechal D., Marchais V., Cottin J., Brown A.J.P. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol. Microbiol. 2001a;42:981–993. doi: 10.1046/j.1365-2958.2001.02713.x. [DOI] [PubMed] [Google Scholar]
- Murad A. M., et al. NRG1 represses yeast hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 2001b;20:4742–4752. doi: 10.1093/emboj/20.17.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls S., Straffon M., Enjalbert B., Nantel A., Macaskill S., Whiteway M., Brown A.J.P. Msn2- and Msn4-like transcription factors play no obvious roles in the stress responses of the fungal pathogen Candida albicans. Eukaroyt. Cell. 2004;3:1111–1124. doi: 10.1128/EC.3.5.1111-1123.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C. J., Mitchell A. P. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr. Biol. 2005;15:1150–1155. doi: 10.1016/j.cub.2005.05.047. [DOI] [PubMed] [Google Scholar]
- Odds F. C. Candida and Candidosis. 2nd ed. London: Baillière Tindall; 1988. [Google Scholar]
- Phillips A. J., Sudbery I., Ramsdale M. Apoptosis induced by environmental stresses and amphotericin B in Candida albicans. Proc. Natl. Acad. Sci. USA. 2003;100:14327–14332. doi: 10.1073/pnas.2332326100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips A. J., Crowe J. D., Ramsdale M. Ras pathway signalling accelerates programmed cell death in the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. USA. 2006;103:726–731. doi: 10.1073/pnas.0506405103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper P., Mahe Y., Thompson S., Pandjaitan R., Holyoak C., Egner R., Muhlbauer M., Coote P., Kuchler K. The Pdr12 ABC transporter is required for the development of weak organic acid resistance in yeast. EMBO J. 1998;17:4257–4265. doi: 10.1093/emboj/17.15.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper P. W. Yeast superoxide dismutase mutants reveal a pro-oxidant action of weak organic acid food preservatives. Free Radic. Biol. Med. 1999;27:1219–1227. doi: 10.1016/s0891-5849(99)00147-1. [DOI] [PubMed] [Google Scholar]
- Ramsdale M. Programmed cell death and apoptosis in fungi. Mycota. 2005;13:113–146. [Google Scholar]
- Sanglard D., Kuchler K., Ischer F., Pagani J. L., Monod M., Bille J. Mechanisms of resistance to azole antifungal agents in C. albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos M., de Larrinoa I. F. Functional characterization of the Candida albicans CRZ1 gene encoding a calcineurin-regulated transcription factor. Curr. Genet. 2005;15:1–13. doi: 10.1007/s00294-005-0003-8. [DOI] [PubMed] [Google Scholar]
- Schüller C., Mamnun Y. M., Mollapour M., Krapf G., Schuster M., Bauer B. E., Piper P. W., Kuchler K. Global phenotypic analysis and transcriptional profiling defines the weak acid stress response regulon in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:706–720. doi: 10.1091/mbc.E03-05-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. A., Nicholls S., Morgan B. A., Brown A.J.P., Quinn J. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol. Biol. Cell. 2004;15:4179–4190. doi: 10.1091/mbc.E04-03-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treger J. M., Schmitt A. P., Simon J. R., McEntee K. Transcriptional factor mutations reveal regulatory complexities of heat shock and newly identified stress genes in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:26875–26879. doi: 10.1074/jbc.273.41.26875. [DOI] [PubMed] [Google Scholar]
- Tripathi G., Wiltshire C., Macaskill S., Tournu H., Budge S., Brown A.J.P. CaGcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J. 2002;21:5448–5456. doi: 10.1093/emboj/cdf507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher V., Tibshirani R., Chu C. Significance analysis of microarrays applied to ionizing radiation response. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl M. A., Johnson A. D. Development of Streptococcus thermophilus lacZ as a reporter gene for Candida albicans. Microbiology. 2001;147:1189–1195. doi: 10.1099/00221287-147-5-1189. [DOI] [PubMed] [Google Scholar]
- Vyas V. K., Berkey C. D., Miyao T., Carlson M. Repressors Nrg1 and Nrg2 regulate a set of stress-responsive genes in Saccharomyces cerevisiae. Eukaroyt. Cell. 2005;4:1882–1891. doi: 10.1128/EC.4.11.1882-1891.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysong D. R., Christin L., Sugar A. M., Robbins P. W., Diamond R. D. Cloning and sequencing of a Candida albicans catalase gene and effects of disruption of this gene. Infect. Immun. 1998;66:1953–1961. doi: 10.1128/iai.66.5.1953-1961.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z., Stead D., Selway L., Walker J., Riba-Garcia I., McLnerney T., Gaskell S., Oliver S. G., Cash P., Brown A.J.P. Proteomic response to amino acid starvation in Candida albicans and Saccharomyces cerevisiae. Proteomics. 2004;4:2425–2436. doi: 10.1002/pmic.200300760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.