Abstract
In our analysis of 5′ and 3′ end formation in plant mitochondria, we compared the major transcript ends of all mitochondrial protein-coding genes between the three Arabidopsis (Arabidopsis thaliana) accessions Columbia (Col), C24, and Landsberg erecta (Ler). Differences between transcript patterns were found for seven genes. For atp6-2, no transcripts at all were detected in Ler. This and further analyses suggest that the atp6-2 gene arrangement is absent from the mitochondrial DNA of this accession. All other transcript polymorphisms are attributed to variations at the 5′ termini and were consistently observed in all tissues investigated. mRNA phenotyping of reciprocal Col/Ler, Col/C24, and Ler/C24 F1 hybrids revealed the differing transcript patterns of ccmC to be inherited maternally, suggesting these to arise from differences in the mitochondrial DNA. Biparental inheritance was observed for the polymorphic transcripts of nad4, nad9, ccmB, and rpl5, indicating these differences to be caused by nuclear-encoded trans-factors. Deviant transcript patterns were tested in further accessions and were found in at least three additional accessions. Detailed examination of the nad4 and the nad9 transcripts demonstrates that the respective polymorphisms affect the major mRNAs of these genes. This study shows that natural genetic variation in Arabidopsis can also affect mitochondrial mRNA end processing. These variations can now be used to identify the nuclear genes responsible, as well as the mitochondrial cis-elements required, for 5′ end generation of mitochondrial transcripts.
Mitochondrial genomes in higher plants are extraordinarily large (up to 2.4 Mb), but usually encode less than 60 genes and therefore have large noncoding regions (Unseld et al., 1997; Kubo et al., 2000; Notsu et al., 2002; Handa, 2003; Clifton et al., 2004; Satoh et al., 2004; Ogihara et al., 2005; Sugiyama et al., 2005). This is accompanied by high genome complexity provoked by frequent DNA recombinations and rearrangements (Mackenzie, 2007; Kubo and Newton, 2008). Furthermore, complex transcript patterns are typical for plant mitochondria. These are partially due to the presence of multiple promoters per transcription unit, but also arise from a variety of posttranscriptional RNA-processing events. These processes include, most notably, cis- and trans-splicing, RNA editing, and formation of secondary 5′ and 3′ termini (Gagliardi and Binder, 2007). Detailed investigations of these posttranscriptional processes are still at the beginning and up to now have been predominantly descriptive. Although functional studies using in vitro or in organello systems have allowed some insight into the mechanisms of RNA editing, splicing, or 3′ end formation (Farré and Araya, 2001, 2002; Staudinger and Kempken, 2003; Takenaka and Brennicke, 2003; Perrin et al., 2004a, 2004b), still very little is known about the trans-factors active in these processes. One of the least explored maturation steps is the generation of mature 5′ ends of mitochondrial mRNAs. No in vitro or in organello systems are presently available to investigate this process and both cis-elements as well as trans-factors are mostly unknown (Forner et al., 2007; Gagliardi and Binder, 2007).
Because the purification of proteins required for mitochondrial RNA (mtRNA) processing has proven to be very difficult, so far mainly genetic approaches have been used to study such factors. For instance, two 3′ to 5′ exoribonucleases involved in the generation of 3′ ends (RNR1 and PNPase) have been characterized by reverse-genetics approaches and, very recently, a similar approach identified a protein required for trans-splicing of nad1 transcripts in Arabidopsis (Arabidopsis thaliana) mitochondria (Perrin et al., 2004a, 2004b; Falcon de Longevialle et al., 2007). Forward-genetics approaches have also been applied to identify genes for nuclear-encoded restorers of fertility in several cytoplasmic male sterility (CMS) systems. The polypeptides encoded by these genes are almost exclusively pentatricopeptide repeat proteins. They are involved in the modulation of the steady-state levels of respective CMS-specific transcripts either by influencing RNA stability or by endonucleolytic cleavage as, for instance, in the Boro CMS system in rice (Oryza sativa; Bentolila et al., 2002; Brown et al., 2003; Desloire et al., 2003; Kazama and Toriyama, 2003; Koizuka et al., 2003; Wang et al., 2006). However, these systems allow only the study of proteins involved in the modulation of these CMS-specific aberrant transcripts, whereas the normal functions of these genes remain unclear.
To identify additional genes encoding components of the plant mitochondrial processing machinery, other approaches are required. One such alternative line of investigation is map-based cloning exploiting the natural genetic variation in Arabidopsis. This method has been successfully applied to identify genes involved in a number of physiological processes (Shindo et al., 2007) and should also allow the identification of nuclear-encoded genes whose products are involved in different processes of plant mitochondrial mRNA maturation (Koornneef et al., 2004; Alonso-Blanco et al., 2005). In this respect, so far only the RNA editing status of mitochondrial transcripts have been analyzed in Columbia (Col) and Landsberg erecta (Ler) revealing differences in the editing extent at some sites (Bentolila et al., 2005, 2008).
To investigate whether the natural genetic variation in different accessions of Arabidopsis also affects 5′ and 3′ end formation of mitochondrial steady-state mRNAs, we analyzed transcripts of all mitochondrial protein-coding genes in the accessions Col, C24, and Ler in this respect. We describe here several mitochondrial mRNA-length polymorphisms, which are consistently observed in all tissues investigated. This strongly suggests that they are independent from tissue-specific or developmental effects. The analysis of reciprocal F1 hybrids attributes the polymorphisms either to nuclear-encoded loci or to differences in mitochondrial DNAs (mtDNAs). This analysis is a very promising prerequisite for the identification of mitochondrial cis-elements and nuclear-encoded trans-factors involved in 5′ end formation or mRNA stability.
RESULTS
Comparative Analysis of Mitochondrial Transcripts in Arabidopsis Accessions Col, Ler, and C24
As a prerequisite for the identification of mitochondrial cis-elements or nuclear-encoded trans-factors involved in mitochondrial mRNA 5′ and 3′ end formation, we systematically analyzed transcript termini of all mitochondrial protein-coding genes in Arabidopsis accessions Col, Ler, and C24. To this end, total RNA was prepared from above-ground parts of 14- to 17-d-old seedlings and the 5′ and 3′ termini of mitochondrial mRNAs were investigated by circularized RNA-reverse transcription-PCR (CR-RT-PCR) analyses with a set of primers successfully used for the mapping of mitochondrial mRNA termini in an Arabidopsis Col cell suspension culture (Forner et al., 2007). To verify the genetic status of the seed material, the genotype of these accessions was confirmed on the basis of nuclear, as well as mitochondrial, genetic markers (Supplemental Fig. S1; Jander et al., 2002; Törjek et al., 2003; Forner et al., 2005).
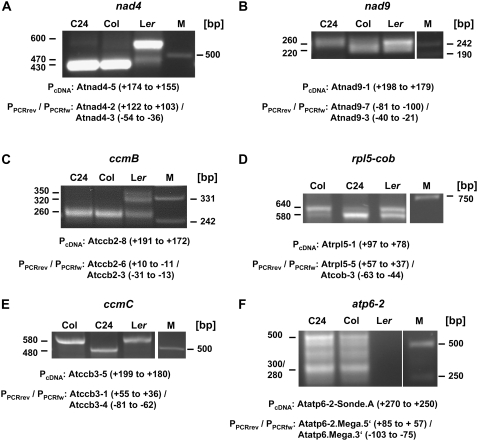
CR-RT-PCR product patterns differing between the accessions were found for transcripts from nad4, nad9, ccmB, rpl5, ccmC, atp6-2, and cox3 (Table I; Fig. 1). The latter has been described previously and is thus not included in this investigation (Forner et al., 2005). For nad4 and ccmC, alternative products were obtained in the different accessions: In C24 and Col, nad4 mRNA-derived products of about 430 bp consistent with previous results are observed (Forner et al., 2007), whereas in Ler a cDNA fragment of approximately 600 bp indicates the presence of an alternative nad4 mRNA end. In addition, a weak product of 470 bp is seen in Ler and, to an even lesser extent, in Col and C24 (Fig. 1A).
Table I.
Polymorphic mitochondrial transcript 5′ ends detected in Arabidopsis ecotypes Col, C24, and Ler
The transcript termini identified in the CR-RT-PCR products shown in Figure 1 are listed. The most prominent transcript 5′ ends are given in bold numbers. Products indicating polymorphic ends have been cloned and sequenced (**), directly sequenced (*), or both (†). –, End not detectable.
| Gene | Col
|
C24
|
Ler
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| PCR Product | 5′ End | 3′ End | PCR Product | 5′ End | 3′ End | PCR Product | 5′ End | 3′ End | |
| nad4 | 600 bp | −390** | +30 | 600 bp | (−396**) | +30 | 600 bp | −390** | +30 |
| 430 bp | −228 | +30 | 430 bp | −228† | +30 | – | – | – | |
| nad9 | 260 bp | −243 | +55 | 260 bp | −243* | +55 | 260 bp | −243 | +55 |
| 220 bp | −202 | +55 | – | – | – | 220 bp | −202 | +55 | |
| ccmB (ccb2) | – | – | – | – | – | – | 350 bp | −231* | +80 |
| – | – | – | – | – | – | 320 bp | −200* | +80 | |
| 260 bp | −140 | +80 | 260 bp | −140 | +80 | 260 bp | −140 | +80 | |
| rpl5-cob | 640 bp | −459 | +58 | 640 bp | −459 | +58 | 640 bp | −459 | +58 |
| 580 bp | −406 | +58 | 580 bp | −406 | +58 | 580 bp | −406 | +58 | |
| ccmC (ccb3) | 580 bp | −484/2 | −46 | – | – | – | 580 bp | −484/2 | −46 |
| − | − | − | 480 bp | 391/0* | −46 | – | – | – | |
| atp6-2 | 500 bp | −268 | +45 | 500 bp | −268 | +45 | – | – | – |
| 300/280 bp | −63/−44 | +45 | 300/280 bp | −63/−44 | +45 | – | – | – | |
Figure 1.
CR-RT-PCR analysis of mitochondrial mRNAs in different Arabidopsis accessions. The transcripts of all mitochondrial protein-coding genes were analyzed by CR-RT-PCR in the Arabidopsis accessions Col, C24, and Ler. PCR products separated on agarose gels are presented for those genes (designations given above the gel images) exhibiting transcript polymorphisms (A–F), except for cox3, which was investigated previously (Forner et al., 2005). Primers (P) used for cDNA first-strand synthesis (PcDNA) and PCR amplification (PPCRrev + PPCRfw), respectively, and their locations are given below the images. The primer positions refer to either the start (NATG, n=−1; PcDNA and PPCRrev) or the stop codon (TAAN, n=+1; PPCRfw). PCR product lengths are given to the left of the gel images. M, Size marker. Lengths of the marker fragments are given (in bp) on the right-hand side.
Likewise, products of identical sizes of about 580 bp were amplified from ccmC mRNAs for Col and Ler, whereas in C24 a 100-bp shorter product was obtained (Fig. 1E). For ccmB, two novel cDNA products were found in Ler (320 and 350 bp, respectively) in addition to the 260-bp product amplified from RNA of all three accessions (Fig. 1C).
The investigation of nad9 mRNAs yielded a complex picture. A CR-RT-PCR analysis with primers used in the examination of total RNA from the Col cell suspension culture repeatedly generated a weaker, but more focused product, within the expected size range in C24 in comparison to Col and Ler (data not shown). Investigation of these transcripts with a different 5′ primer (PPCRrev:Atnad9-7), designed to generate smaller cDNA fragments, and an inspection of the products on high-resolution agarose gels, revealed a complex pattern. In Col and Ler, products with a predominant size of about 220 bp are seen (Fig. 1B). This CR-RT-PCR product corresponds to the respective product obtained with RNA from the Col tissue culture (Forner et al., 2007). In C24 green tissue, this cDNA product is undetectable. In addition, high-resolution agarose gels now revealed a slightly longer cDNA product with a size around 260 bp in all three accessions (Fig. 1B), which indicates alternative 5′ or 3′ ends not resolved in the previous mapping analysis (Forner et al., 2007).
A particular polymorphism in mRNA termini is observed for rpl5. Here, in all accessions investigated, the same PCR products of 640 and 580 bp, respectively, are observed (Fig. 1D). However, there are consistently reproducible differences in the abundances of these two cDNA products: Both are equally abundant in Ler, but the longer cDNA is predominant in Col, whereas the shorter amplification product is found to be dominant in C24.
From transcripts of the atp6-2 gene, no PCR products were obtained in Ler, indicating a complete absence of this gene in this accession at least in the genomic arrangement anticipated from the sequence of this gene in C24 (Unseld et al., 1997). The atp6-2 pattern observed in the Col cell suspension culture is identical in green plants of both Col and C24 (Fig. 1F; Forner et al., 2007).
For all other mitochondrial genes, no differences in the CR-RT-PCR product patterns were observed between the three accessions (data not shown). In addition, no differences were found between the results obtained from RNA isolated from green plants and the CR-RT-PCR-based mapping data with RNA from Col cell suspension culture, except for the above-described nad9 (Forner et al., 2007). In a few instances, additional PCR products were amplified from RNA from green plants from all accessions. However, in all cases investigated, these turned out to be false positive products from other RNAs.
Natural Genetic Variation Affects mRNA 5′ Ends
To obtain detailed information about the observed polymorphisms, the PCR products with sizes deviating from the Col tissue culture studies were directly sequenced and/or sequenced after cloning (Forner et al., 2007).
In Col and C24 plants, sequencing of the 430-bp PCR products identifies main 5′ ends of the nad4 mRNAs at position −228, whereas in Ler the major 5′ terminus of this transcript maps to position −390 in the 600-bp cDNA product (Table I; Supplemental Fig. S2). This latter end is also present in Col and C24, but represents an RNA of only minor abundance in these accessions. Particularly in C24, the −390 end is only observed in a CR-RT-PCR approach with nested primers located upstream of the −228 end (Supplemental Fig. S3). In contrast, the 5′ mRNA end at position −228 is undetectable in Ler, even in a CR-RT-PCR approach specifically designed to detect this end (data not shown). This indicates a defect in the formation or stabilization of the nad4 transcript with the −228 5′ end in Ler. Sequencing of the 470-bp product present in all three accessions revealed a 5′ end at position −269 (data not shown).
Alternative ends are found for ccmC. In Col and Ler, the previously observed product of about 600 bp indicates the predominant mRNA to start 484/482 nucleotides upstream of the ATG codon (Forner et al., 2007). In C24, sequencing of the specific 480-bp product mapped the position of the 5′ ends 390 and 391 nucleotides upstream of the ATG (Supplemental Fig. S2).
Sequence analysis of nad9 cDNA confirmed the complex pattern indicated by the 260- and 220-bp PCR products. A major 5′ end is found at position −243 and a cluster of 5′ ends is present around position −202, the latter being in accordance with previous mapping results (Forner et al., 2007). In Col and Ler, both 5′ ends are found, whereas in C24 the 5′ termini around position −202 are not detectable. Thus, C24 contains only a single nad9 mRNA species with a 5′ end at position −243. For transcripts of this gene, again the genetic information to generate or stabilize certain transcripts seems to be missing in one accession (C24).
For the ccmB mRNA, a common 5′ end at −140 is present in the 260-bp cDNA fragment amplified in all accessions investigated. Additional 5′ termini at −200 and −231 were identified in the 320- and 350-bp products only in accession Ler (Supplemental Fig. S2).
The sequences of the CR-RT-PCR products revealed no alternative 3′ termini in any of these CR-RT-PCR polymorphisms. Thus, all transcript-size polymorphisms arise from alternative 5′ ends. The results of these analyses are summarized in Table I.
Alternative 5′ Ends Are Found at Major Transcripts
To analyze the polymorphic ends by an independent experimental approach and to see whether they are found at major abundant transcripts, the nad4 and nad9 mRNAs were investigated by northern-blot and primer extension analyses.
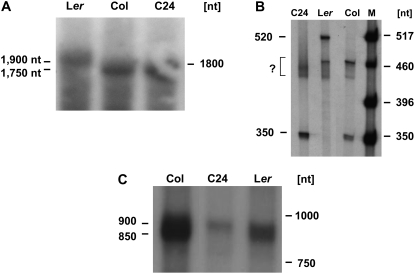
In total RNA preparations from Col and C24 seedlings, a nad4 probe detects a transcript of about 1,750 nucleotides as estimated in comparison to the cytoplasmic 18S rRNA (about 1,800 nucleotides) in the ethidium bromide-stained gel (Fig. 2A; data not shown). A larger nad4 mRNA of approximately 1,900 nucleotides is detected in Ler total RNA. The sizes of the nad4 mRNA observed in the northern analysis are consistent with the data obtained by the CR-RT-PCR analyses (1,746 and 1,908 nucleotides).
Figure 2.
Analyses of the nad4 and nad9 mRNA polymorphisms by independent experimental approaches. Primer extension and northern-blot analyses were carried out to investigate whether the polymorphisms observed by CR-RT-PCR affect the major transcripts. A, Northern-blot analysis of total RNA from seedlings with an nad4-specific probe covering positions +469 to +976 (approximately exon b) within the reading frame (positions are given with respect to the ATG, A=+1). The sizes of the major mRNAs are indicated in the left margin. B, Primer extension analysis of nad4 mRNAs carried out on total RNA from seedlings with oligonucleotide Atnad4-2 (+122–+103). Sizes of the extension products are indicated in the left margin. M, Size marker; lengths of marker fragments are given in the right margin (in nucleotides [nt]). Sizes of the products are given in the left margin. The products indicated by a question mark are of unknown origin because no corresponding CR-RT-PCR products were obtained. C, Northern-blot analysis of total RNA from cell suspension cultures (5 μg each) with a probe comprising the complete nad9 reading frame. The signals correspond to two mRNAs of approximately 900 and 850 nucleotides, respectively, in Col and Ler. In C24, only the larger nad9 mRNA is found. Size markers are given (in nt). The approximate sizes of the mRNA species are also given (in nt) in the left margin.
The different nad4 mRNA 5′ ends were also observed in a primer extension analysis of total RNA from seedlings (Fig. 2B). In Col and C24, extension of the primer Atnad4-2 yields a major cDNA of 350 nucleotides corresponding to the 5′ terminus 228 nucleotides upstream of the translation start codon. In Ler, the main extension product is 520 nucleotides and detects the 5′ mRNA end at −390. Longer expositions of the primer extension products also revealed the presence of the minor end at the −390 5′ end in Col, whereas no signal corresponding to the −228 nad4 mRNA 5′ end is visible in Ler. Faint signals are observed for the 5′ terminus at position −269, confirming the CR-RT-PCR results (data not shown). In addition, a number of cDNAs with sizes around 460 nucleotides were seen in all accessions. Because we never observed any corresponding CR-RT-PCR products, these were considered to be false positive signals as frequently obtained in primer extension analyses by spurious priming events.
In summary, the results of the northern, as well as of the primer extension analyses, confirm the nad4 mRNA transcript polymorphism between accessions Col/C24 and Ler with the absence of the −228 end in Ler and the concomitant appearance of the 5′ terminus at position −390. In addition, the transcript 5′ end polymorphism affects the abundant main steady-state mRNA species of nad4 in the different accessions (Figs. 1A and 2, A and B).
For the nad9 transcripts, northern-blot analysis carried out with total RNA preparations from cell suspension cultures of Col, C24, and Ler and an nad9-specific probe spanning the complete reading frame revealed two about equally abundant major transcript species to be present in Col and Ler (Fig. 2C). The approximate sizes of these mRNA species (900 and 850 nucleotides) are in agreement with those calculated from the above-described mapping data (871 and 830 nucleotides). As expected from the CR-RT-PCR results, only the longer nad9 transcripts are seen in the northern analysis of C24 RNA. Thus, these two experimental approaches consistently indicate that the three accessions generate the main nad9 mRNAs with a 5′ terminus at position −243, whereas mRNAs with equally prominent 5′ ends around −202 are present only in Col and Ler and absent from C24. In addition, these experiments show that also in the Col (and Ler) cell suspension cultures the major nad9 mRNAs with 5′ ends at −243 are present.
Taken together, these results demonstrate that the 5′ end polymorphisms observed in the CR-RT-PCR analysis affect the major nad4 and nad9 transcripts.
Transcript Analysis in Different Tissues and Developmental Stages
The differing transcript patterns in seedlings certainly reflect genetic differences between the three accessions. These might be attributed to accession-specific distinct expression patterns (of individual alleles, for instance) in certain tissues or developmental stages.
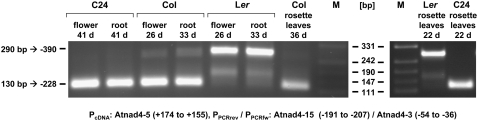
To evaluate this possibility, we investigated the polymorphic transcript patterns in roots, flowers, and rosette leaves from about 3-week-old plants from Col, C24, and Ler, respectively. For all six mitochondrial genes, the CR-RT-PCR product patterns obtained in the RNA preparations from the various tissues indicate that the transcript ends are identical between these tissues and developmental stages (Figs. 1 and 3; Supplemental Fig. S4). Thus, the observed mtRNA polymorphisms between the accessions are not restricted to seedlings, but are also observed in other tissues. These results imply that they can be found throughout the entire plant and in all developmental stages.
Figure 3.
Transcript ends in different tissues. CR-RT-PCR analyses of nad4 transcripts were carried out with total RNA from roots, flowers, and rosette leaves, respectively. Ages at harvest times are given in days (d). Primer locations are given as in Figure 1. Sizes of the PCR products (bp) and the position of corresponding 5′ transcript ends are indicated in the left margin. The lengths of the size marker fragments (lanes M) are given between the gel images.
Inheritance of the Transcript Polymorphisms in Reciprocal Crosses
The polymorphic transcript patterns could be caused by differing mitochondrial sequences or by differences in nuclear genes. In dicot plants, mtDNA is generally transmitted maternally, whereas the nuclear genome is inherited biparentally, with only rare exceptions from this rule having been reported recently for tobacco (Nicotiana tabacum; Svab and Maliga, 2007). Thus, the inheritance patterns of respective mRNAs in F1 offspring obtained after reciprocal crosses of the analyzed accessions should allow discrimination, whether a polymorphism is due to differences in the mitochondrial or nuclear DNA. For this analysis, the genotypes of the individual F1 plants were inspected by the analysis of appropriate nuclear as well as mitochondrial genetic markers (Supplemental Fig. S1; Jander et al., 2002; Törjek et al., 2003; Forner et al., 2005).
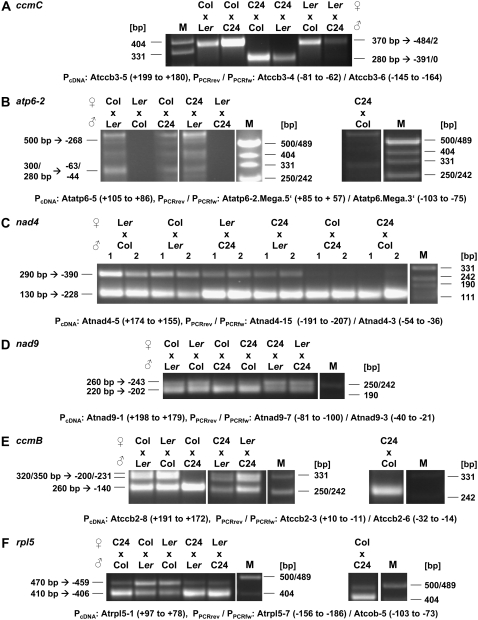
For ccmC and atp6-2, the PCR product patterns and thus the transcripts differ between the reciprocal hybrids. The C24-specific short cDNA fragment (280 bp) indicative of the ccmC transcript with a 5′ end at −390/−391 is only found in plants derived from crossings with C24 as female parent. Vice versa, the ccmC mRNAs with the further upstream-located 5′ terminus at −482/−484 indicated by the presence of an approximately 370-bp PCR product are seen only in F1 plants with a Col or Ler female parent. This points to a clear maternal inheritance of the length of the ccmC transcript, as to be expected from a difference in the mitochondrial genome (Fig. 4A). Likewise, the lack of atp6-2 mRNA-derived PCR products is restricted to F1 plants originating from a Ler female parent, whereas in F1 offspring derived from crossings with C24 and Col as maternal parent, the expected products are seen (Fig. 4B). Thus, the ccmC and atp6-2 mRNA polymorphisms are maternally inherited attributing them to differing cis-elements in the mtDNA (ccmC; see “Discussion”) or to the absence of the respective locus (atp6-2). To test the presence of the atp6-2 gene, a Southern-blot analysis of total DNA from the three accessions was performed. A probe representing atp6-2-specific sequences hybridizes to a 1.6-kb fragment in Col and in C24, which is expected from the mtDNA sequence from C24 and the mtDNA copy sequence in chromosome 2 of Col. No hybridization is seen in Ler. Consistent with this result, a PCR analysis with oligonucleotides annealing to the 5′ and 3′ extremities of the atp6-2-specific part revealed the expected product of 250 bp only in C24 and Col, but not in Ler (Supplemental Fig. S5). These data strongly suggest that the atp6-2 gene in the version seen in C24 is not present in Ler.
Figure 4.
mRNA 5′ end polymorphism analysis in the offspring of reciprocal crosses. The mode of inheritance of the transcript 5′ termini polymorphisms was investigated in F1 plants derived from reciprocal crosses between Col and C24, Col and Ler, as well as C24 and Ler. Total RNA from rosette leaves (C) or from flowers (A, B, D–F) was used in the CR-RT-PCR analyses. For nad4 (C), two individual plants (one and two) per crossing were examined. Details of the oligonucleotides used are given in legends of Figures 1 and 3. M, Marker fragment sizes are given on the right-hand side. Sizes of the PCR products (bp) and the position of corresponding 5′ transcript ends are indicated in the left margin.
Different modes of inheritance are observed for nad4, nad9, ccmB, and rpl5. For these genes, the CR-RT-PCR product patterns are identical between the respective reciprocal F1 hybrid plants. This strongly suggests that the transcript polymorphisms of these four mitochondrial genes are biparentally inherited, which is typical for nuclear genes. For the nad4 (−228), nad9 (−202), and ccmB (−200/−231) transcript 5′ ends, a dominant-recessive mode of inheritance of the presence or absence of these termini is observed (Fig. 4, C–E). These ends are present in all F1 plants derived from crossings in which at least one of the parents shows these termini independent of the gender. For rpl5, the relative abundances of the two transcript species are distinctively shifted toward the −406 end dominating in all hybrids with a C24 parent and toward the −459 terminus in the Col/Ler crosses (Fig. 4F). Thus, the C24 allele seems to be dominant over the Col and Ler alleles, whereas the Col allele seems to be dominant over the respective gene version in Ler.
Transcript End Analysis of Additional Accessions
To obtain first estimations as to whether these four distinct nuclear-encoded transcript polymorphisms might be assigned to one nuclear locus or to different loci, the nad4, nad9, ccmB, and rpl5 transcript ends were investigated in additional accessions (Table II).
Table II.
Distribution of nad4 (−228), nad9 (−202), ccmB (−200/−231), and rpl5 (−459:−406) 5′ ends in different Arabidopsis accessions
nad4 −228: −, Absence of the −228 5′ end and the presence of a major end at −390; +, these accessions have a major nad4 mRNA 5′ end at position −228. However, in these accessions the nad4 mRNAs with a −390 5′ end are also present in minor amounts. nad9 −202: −, Absence of the −5′ ends around position −202 and presence of the −243 5′ terminus; +, presence of both nad9 5′ ends. ccmB: + and − refer to the presence or absence of 5′ ends at positions −200 and −231. All other 5′ ends are found to be identical in all accessions investigated. rpl5: Preferential or equal abundance of rpl5 mRNA 5′ ends is given (−459 or −406, 1:1 for equal abundance). nd, Not determined; *, an extra PCR product has been obtained, but not yet further investigated.
| Accession | nad4, −228 | nad9, −202 | ccmB, −200/−231 | rpl5, −459:−406 |
|---|---|---|---|---|
| C24 | + | − | − | −406 |
| Col | + | + | − | −459 |
| Ler | − | + | + | 1:1 |
| Bay-0 | + | − | − | 1:1 |
| Bl-1 | + | − | − | −459 |
| Bla-10 | + | − | − | 1:1 |
| Calver | − | − | − | −406 |
| Can-0 | + | + | − | −459 |
| Co | nd | − | − | nd |
| Cvi | + | + | − | 1:1 |
| Dijon-1 | + | − | + | 1:1 |
| Dijon G | − | + | − | −406 |
| Ei | + | + | − | 1:1 |
| East Malling | − | − | − | −406 |
| Enkheim 1 | + | − | + | −459 |
| Est | + | + | − | 1:1 |
| Kas-1 | + | + | − | 1:1 |
| Kondara | + | + | − | 1:1 |
| La-1 | + | − | + | nd |
| Mt-0 | + | − | − | 1:1 |
| Nd | + | + | − | 1:1 |
| No-0 | + | − | − | nd |
| Oy-1 | + | − | − | nd |
| Pitztal-2 | + | − | + | 1:1 |
| Sha | + | + | − | 1:1 |
| Sorbo | + | + | − | 1:1 |
| Su-0 | nd | nd | − | nd |
| Tsu-1 | + | + | − | −459 |
| Van-0 | + | − | −* | 1:1 |
| Ws | + | − | − | −459 |
| Yo-0 | + | − | − | 1:1 |
In the additional accessions investigated, the nad4 −228 5′ end is not detectable in Calver, Dijon G, and East Malling, whereas the ccmB 5′ termini at −200/−231 are only observed in Dijon-1, Enkheim 1, and Pitztal-2. In contrast, a more balanced distribution is seen for the nad9 transcripts. The 5′ termini at −202 are found in 11 accessions, but are not detectable in 16 others. For rpl5, the ratio of the −459 and the −406 5′ mRNA ends is approximately 1:1 in more than one-half of the 23 accessions investigated for this trait. In five accessions, the −459 end dominates, and in three the −406 terminus is more abundant. Interestingly, the latter accessions are those that lack the nad4 −228 5′ end. Because no correlation between the preference of the rpl5 −406 and the absence of the nad4 −228 ends is seen in C24 (nad4, −228; rpl5, −406 predominant) and Ler (nad4, −390; rpl5, −406/−459=1:1), the nuclear factors required for the accumulation of nad4 mRNAs with the −228 5′ end and the preferential accumulation of an rpl5 transcript with the −406 5′ terminus, respectively, are probably not identical.
In summary, the uncorrelated distribution of the individual polymorphic transcript patterns in the various accessions suggests that the nuclear-encoded polymorphisms of the different mitochondrial transcripts can presumably be attributed to different loci rather than to different alleles of a single gene or locus.
DISCUSSION
Transcript End Polymorphisms in Different Arabidopsis Accessions
The systematic analysis of mitochondrial mRNAs in three Arabidopsis accessions revealed transcript polymorphisms for seven genes. The distinct transcripts found for cox3 had been analyzed in a separate report and were found to correlate with differences in the mtDNAs of the investigated accessions (Forner et al., 2005). The analysis of the atp6-2 and ccmC polymorphisms in F1 plants from respective crossings now also strongly suggests these polymorphisms to be caused by variations in the mtDNA (Fig. 4, A and B). The observed CR-RT-PCR product pattern obtained from atp6-2 transcripts as well as the inspection of the mtDNA by southern hybridization and PCR consistently suggests that the atp6-2 gene is completely absent from Ler or differs substantially from its form in C24 (Supplemental Fig. S5).
The generation of the alternative ccmC 5′ ends indicates that the 5′ end region of this gene varies in the investigated accessions. A comparison of the mtDNA sequence from C24 and the mtDNA copy in chromosome 2 of Col revealed that the sequences immediately surrounding the −484/2 and 391/0 ccmC 5′ ends are identical. However, a further upstream sequence stretch from positions −509 to −595 shows substantial differences between C24 and Col most likely also reflecting differences between the mtDNA of both accessions as seen for cox3 (Forner et al., 2005). The influence of these upstream sequences on the occurrence of the different ends remains as yet unknown, especially because the novel C24-specific ccmC 5′ end (−390/−391) is located directly downstream of a sequence that shows similarity to previously characterized promoter elements (Hoffmann and Binder, 2002; Kühn et al., 2005). Alternatively, this end might be generated posttranscriptionally.
The other 5′ end polymorphisms identified in this study all originate from differences between the respective nuclear genomes. Sequences of mtDNA (obtained from amplification of the respective regions) and cDNA surrounding the polymorphic ends of nad4 in Ler and nad9 in Col did not reveal any differences, whereas a single nucleotide exchange is observed 497 nucleotides upstream of the ATG of the ccmB gene in Ler (data not shown). In addition, the comparison of the mtDNA sequence from C24 and the mtDNA copy in chromosome 2 of Col at the rpl5 5′ ends revealed only one additional nucleotide in Col 486 nucleotides upstream of the 5′ end located at position −459. For the mRNAs of these mitochondrial genes, most likely nuclear-encoded trans-factors are involved, the specific roles of the proteins responsible for the nad4, nad9, ccmB, and rpl5 mRNA polymorphisms presently remaining open. In theory, there are three potential functions. First, transcription initiation might be affected by the malfunction of a specific transcription factor. Second, 5′ end processing may be impaired, for instance, by the restricted function of a processing factor. Third, the stability of certain mRNA species might be influenced. In the latter case, either increased or decreased stability could be the dominant trait. For nad9, any of three scenarios can explain the lack of the short mRNA in C24. However, because there is no promoter motif present at the 5′ terminus of this mRNA, it seems rather unlikely that the absence of the short nad9 mRNA is due to a failure of transcription initiation.
For nad4 mRNA 5′ ends, a complex picture is observed. It seems clear that essential genetic information for the occurrence of a stable nad4 mRNA with the −228 5′ terminus is missing in Ler. In turn, a slightly longer mRNA species with a further upstream-located end (−390) accumulates to substantial levels in this accession. Minor amounts of this latter RNA are also detectable in Col and C24. In addition, a minor 5′ end at −269 detectable in all three accessions is increased in Ler. Although we cannot definitely exclude disturbed transcription initiation, this seems to be unlikely for two reasons: There is no promoter motif present at the −228 5′ end of the short nad4 mRNA and abolished transcription initiation at −228 cannot reasonably explain the concomitant increase of the long nad4 transcript. Likewise, altered stability seems unlikely because this would require two parallel processes, the destabilization of the short RNA and an enhanced stability of the long nad4 mRNA with the −390 5′ end. The variable amounts of the large nad4 mRNA with the −390 5′ end rather suggest that this RNA species is an intermediate RNA, which is processed toward the (final) nad4 mRNA with the −228 5′ end. In Col, a large portion of this intermediate RNA species with the −390 5′ end is converted to RNA molecules with the −228 5′ end. In C24, this processing is almost complete and the intermediate RNA with the −390 5′ end is only detectable in a nested CR-RT-PCR approach. These quantitative differences are confirmed by the results of the primer extension analysis (Fig. 2B; data not shown). Thus, there are a defect allele in Ler and alleles of different strength for the generation of the short nad4 mRNA in Col and C24. This observation is in line with an impaired 5′ end processing at position −228 in Ler.
Alleles of different strength might also explain the polymorphism of rpl5 mRNAs. In all Arabidopsis accessions, RNAs with two different 5′ ends (−459 and −406) are present, but in different relative quantities (Fig. 1D). In Col, the gene encoding a factor required for the accumulation of the short RNA seems to be encoded by a weak allele, so only a small amount of this mRNA is present. In contrast, in C24, the short mRNA dominates, suggesting that there is a strong allele present in this accession. The strength of the allele encoding the respective protein in Ler seems to be intermediate. Interestingly, the amount of the long rpl5 mRNA with the −459 end is inverse proportional to the accumulation of the short end. This suggests that the short mRNA is processed from the long RNA or that both mRNAs derive from the same precursor RNA.
With the ccmB 5′ ends, several explanations are possible for the observed pattern. It is possible that Ler contains a weak allele encoding a protein required for the generation of the ccmB RNA with the −140 5′ terminus. This low-level processing leads indirectly to the accumulation of the longer mRNAs with ends at −200 and −231, respectively, although there seems to be no clear concomitant decrease of the ccmB mRNA with the −140 5′ end (Fig. 1C). In addition, one would expect a recessive inheritance of the appearance of the Ler-specific transcripts. However, the analysis of the F1 hybrids suggests a dominant inheritance of the presence of these transcripts, which is consistent with the assumption that there is additional genetic information for the ccmB mRNA with ends at positions −200 and −231 present in Ler (Fig. 4E). This, in turn, might be explained by defect alleles present in Col and C24 or by a gain-of-function mutation in Ler. This gain of function may involve a de novo mutation of a transcription factor, a processing factor, or a stability factor with a new function probably without disabling the old intrinsic function.
Natural Genetic Variation: A Promising Tool to Investigate Mitochondrial mRNA Processing
Despite intensive research over several decades, most of the cis- and trans-components required for the generation of mature 5′ and 3′ ends and/or the stability of mitochondrial mRNAs of higher plants are still unknown. Many mRNA termini have been mapped (Kühn et al., 2005; Forner et al., 2007), but, apart from a few exceptions, neither the cis-acting sequence elements nor the trans-factors required for the generation of these ends have as yet been identified (Hoffmann et al., 2001; Gagliardi and Binder, 2007). Because biochemical approaches are very difficult or even impossible, mainly genetic approaches have been applied to study components of RNA maturation. Map-based cloning using natural genetic variation might be an alternative approach to identify nuclear-encoded genes required for the maturation of normal mRNAs. The potential of this technique has been amply demonstrated for other traits (Koornneef et al., 2004; Alonso-Blanco et al., 2005). Now the characterization of mitochondrial mRNA phenotype variation reported here provides the basis to apply this method to identify the nuclear genes required for the accession-specific accumulation or absence of at least certain nad4, nad9, rpl5, and ccmB RNA species. As indicated by the distinct occurrence of the mRNA polymorphisms in more than 20 accessions (Table II), it is very likely that this approach may lead to four different nuclear genes encoding proteins operating in mtRNA metabolism.
Maturation of 5′ Ends of Mitochondrial mRNAs
Apart from atp6-2, all polymorphisms observed affect mitochondrial transcripts with different 5′ termini, these polymorphisms being due to differences in mitochondrial or nuclear DNA. No alternative 3′ end has been found so far. The reason for this bias is presently unknown, but there are several explanations. The number of accessions investigated might be too small. However, because it is broad enough to uncover several polymorphic 5′ ends, 3′ end polymorphisms must be at least comparatively rare. Another explanation could be that there are relatively few factors involved in the generation of many or all mature 3′ ends. Consequently, lack or constricted function of one of these few factors might have fatal consequences for the generation of mature RNAs of several mitochondrial genes and thus for the plant. This would prevent an easy manifestation of respective variants in the gene pool.
In turn, there might be many specific factors for the accumulation of transcripts with different 5′ termini, most of which may be needed only for the mRNA maturation of a single gene. The accumulation of a major transcript with a variant 5′ end seems to have no consequences for the fitness of a plant, indicating that the mitochondrial translation machinery exhibits enough flexibility to translate such transcripts. Alternatively, plants are able to tolerate reduced accumulation of one such protein or even the complete loss of this protein (Gutierres et al., 1997; Perales et al., 2005). This line of argument, however, then raises the question of why there is 5′ processing at all in plant mitochondria. Because correct 5′ processing (of at least some mRNAs) might not be a prerequisite for translation, this process probably does not have an essential regulatory function. The 5′ end processing systems in the long term more likely ensure that translatable mRNAs are created from large untranslatable precursor RNAs. This seems to be particularly important when considering the structural flexibility of the plant mitochondrial genome, which generates new DNA configurations by recombination (Mackenzie, 2007; Kubo and Newton, 2008). These may lead to the transcription of novel longer and untranslatable primary RNAs, which then need to be converted to mRNA molecules accessible for mitochondrial ribosomes. The data reported here provide a promising platform to systematically investigate 5′ processing, to identify the cis- and trans-factors involved, and to elucidate its biological significance.
MATERIALS AND METHODS
Plant Material, Plant Cultivation, and RNA Isolation
Arabidopsis (Arabidopsis thaliana) plants from different accessions were raised in a growth chamber at 21°C, 50% relative humidity, and a light flux of 80 to 160 μmol m−2 s−1 under a 16-h-light/8-h-dark regime. Plants were grown on soil composed of 80% (v/v) Fruhstorfer Erde SoMi 537 Traysubstrat (Hawita Gruppe), 20% (v/v) vermiculite grain size 2 to 3 mm (Isola-Mineralwolle-Werke), and 1.5 g/L Osmocote Exact Mini fertilizer (Scotts Deutschland). The green parts of seedlings were harvested after 14 to 17 d. Flowers and roots were taken at various time points, but not before 21 d after sowing. Leaves were taken from 21-d-old plants, unless stated otherwise. Leaves from F1 plants were harvested from 3- to 4-week-old plants.
The Arabidopsis cell suspension cultures were grown in the dark at 23°C on a shaker (120 rpm). The C24 and the Col cell suspension cultures have been described previously (Forner et al., 2005), whereas the Ler culture was freshly established. Cells were harvested 24 h after transfer to fresh medium.
Plant material was immediately shock frozen in liquid nitrogen after harvesting. Subsequently, approximately 100 mg were ground by mortar and pestle and used for isolation of total RNA with the RNeasy plant mini kit following the manufacturer's instructions (Qiagen).
CR-RT-PCR Analysis
CR-RT-PCR analysis was carried out as described previously with the established primer pairs unless stated otherwise (Forner et al., 2007).
Genotyping
PCR to verify the hybrid status of the F1 plants was carried out with GoTaq Flexi DNA polymerase under conditions recommended by the manufacturer (Promega). Templates were either residual DNA in total RNA preparations or total DNA prepared from the same plant material with the DNeasy plant mini kit (Qiagen) according to the instructions given in the manual. Approximately 1/30 of the RNA and 1/200 to 1/10 of the DNA preparations, respectively, was used per amplification reaction under standard conditions (Sambrook and Russel, 2001). The primer pairs used are detailed in the figure legends. Each PCR reaction—including the CR-RT-PCRs—consisted of 35 cycles.
Miscellaneous Methods
Sequencing of PCR products was done commercially (4base Lab). The resulting sequences were analyzed using the BLAST tools at the National Center for Biotechnology Information server (McGinnis and Madden, 2004).
Southern-blot and primer extension analyses, as well as basic methods of molecular biology, were carried out according to standard protocols (Sambrook and Russel, 2001).
Northern-blot experiments were carried out using RNA denatured by glyoxal separated on agarose gels and blotted onto Duralon-UV or Hybond-XL membranes (Stratagene). Probes were radioactively labeled with the Rediprime II random-prime DNA labeling system (GE Healthcare). Signals were visualized by exposition to x-ray films.
Agarose gels were photographed with a GeneFlash gel documentation system (Syngene).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Genotypes of accessions and respective F1 plants.
Supplemental Figure S2. Sequence analysis of polymorphic CR-RT-PCR products.
Supplemental Figure S3. CR-RT-PCR analysis of the nad4-390 5′ end.
Supplemental Figure S4. CR-RT-PCR analysis of RNA from different tissues.
Supplemental Figure S5. Analysis of the atp6-2 gene.
Supplementary Material
Acknowledgments
We are very grateful to Cornelia Guha for excellent technical assistance and thank all coworkers and students who directly or indirectly contributed to this project. F1 seeds were generated in the context of the GABI program, Creation of novel genetic variants of Arabidopsis.
This work was supported by the Deutsche Forschungsgemeinschaft (grant nos. Bi 590/6–1, 6–2, and 10–1) and by a fellowship of the Studienstiftung des deutsche Volkes (to J.F.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Stefan Binder (stefan.binder@uni-ulm.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso-Blanco C, Mendez-Vigo B, Koornneef M (2005) From phenotypic to molecular polymorphisms involved in naturally occurring variation of plant development. Int J Dev Biol 49 717–732 [DOI] [PubMed] [Google Scholar]
- Bentolila S, Alfonso AA, Hanson MR (2002) A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc Natl Acad Sci USA 99 10887–10892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila S, Chateigner-Boutin AL, Hanson MR (2005) Ecotype allelic variation in C-to-U editing extent of a mitochondrial transcript identifies RNA-editing quantitative trait loci in Arabidopsis. Plant Physiol 139 2006–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila S, Elliott LE, Hanson MR (2008) Genetic architecture of mitochondrial editing in Arabidopsis thaliana. Genetics 178 1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GG, Formanova N, Jin H, Wargachuk R, Dendy C, Patil P, Laforest M, Zhang J, Cheung WY, Landry BS (2003) The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J 35 262–272 [DOI] [PubMed] [Google Scholar]
- Clifton SW, Minx P, Fauron CM, Gibson M, Allen JO, Sun H, Thompson M, Barbazuk WB, Kanuganti S, Tayloe C, et al (2004) Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol 136 3486–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desloire S, Gherbi H, Laloui W, Marhadour S, Clouet V, Cattolico L, Falentin C, Giancola S, Renard M, Budar F, et al (2003) Identification of the fertility restoration locus, Rfo, in radish, as a member of the pentatricopeptide-repeat protein family. EMBO Rep 4 588–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon de Longevialle A, Meyer EH, Andres C, Taylor NL, Lurin C, Millar AH, Small ID (2007) The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 intron 1 in Arabidopsis thaliana. Plant Cell 19 3256–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré JC, Araya A (2001) Gene expression in isolated plant mitochondria: high fidelity of transcription, splicing, and editing of a transgene product in electroporated organelles. Nucleic Acids Res 29 2484–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré JC, Araya A (2002) RNA splicing in higher plant mitochondria: determination of functional elements in group II intron from a chimeric cox II gene in electroporated wheat mitochondria. Plant J 29 203–213 [DOI] [PubMed] [Google Scholar]
- Forner J, Weber B, Thuss S, Wildum S, Binder S (2007) Mapping of mitochondrial mRNA termini in Arabidopsis thaliana: t-elements contribute to 5′ and 3′ end formation. Nucleic Acids Res 35 3676–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner J, Weber B, Wietholter C, Meyer RC, Binder S (2005) Distant sequences determine 5′ end formation of cox3 transcripts in Arabidopsis thaliana ecotype C24. Nucleic Acids Res 33 4673–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi D, Binder S (2007) Expression of the plant mitochondrial genome. In D Logan, ed, Plant Mitochondria. Blackwell Publishing, Ames, IA, pp 50–95
- Gutierres S, Sabar M, Lelandais C, Chetrit P, Diolez P, Degand H, Boutry M, Vedel F, de Kouchkovsky Y, De Paepe R (1997) Lack of mitochondrial and nuclear-encoded subunits of complex I and alteration of the respiratory chain in Nicotiana sylvestris mitochondrial deletion mutants. Proc Natl Acad Sci USA 94 3436–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa H (2003) The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res 31 5907–5916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Binder S (2002) Functional importance of nucleotide identities within the pea atp9 mitochondrial promoter sequence. J Mol Biol 320 943–950 [DOI] [PubMed] [Google Scholar]
- Hoffmann M, Kuhn J, Däschner K, Binder S (2001) The RNA world of plant mitochondria. Prog Nucleic Acid Res Mol Biol 70 119–154 [DOI] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama T, Toriyama K (2003) A pentatricopeptide repeat-containing gene that promotes the processing of aberrant atp6 RNA of cytoplasmic male-sterile rice. FEBS Lett 544 99–102 [DOI] [PubMed] [Google Scholar]
- Koizuka N, Imai R, Fujimoto H, Hayakawa T, Kimura Y, Kohno-Murase J, Sakai T, Kawasaki S, Imamura J (2003) Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. Plant J 34 407–415 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D (2004) Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol 55 141–172 [DOI] [PubMed] [Google Scholar]
- Kubo T, Newton KJ (2008) Angiosperm mitochondrial genomes and mutations. Mitochondrion 8 5–14 [DOI] [PubMed] [Google Scholar]
- Kubo T, Nishizawa S, Sugawara A, Itchoda N, Estiati A, Mikami T (2000) The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNA(Cys)(GCA). Nucleic Acids Res 28 2571–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn K, Weihe A, Börner T (2005) Multiple promoters are a common feature of mitochondrial genes in Arabidopsis. Nucleic Acids Res 33 337–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie S (2007) The unique biology of mitochondrial genome instability. In D Logan, ed, Plant Mitochondria. Blackwell Publishing, Ames, IA, pp 36–49
- McGinnis S, Madden TL (2004) BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 32 W20–W25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notsu Y, Masood S, Nishikawa T, Kubo N, Akiduki G, Nakazono M, Hirai A, Kadowaki K (2002) The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol Genet Genomics 268 434–445 [DOI] [PubMed] [Google Scholar]
- Ogihara Y, Yamazaki Y, Murai K, Kanno A, Terachi T, Shiina T, Miyashita N, Nasuda S, Nakamura C, Mori N, et al (2005) Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Res 33 6235–6250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales M, Eubel H, Heinemeyer J, Colaneri A, Zabaleta E, Braun HP (2005) Disruption of a nuclear gene encoding a mitochondrial gamma carbonic anhydrase reduces complex I and supercomplex I + III2 levels and alters mitochondrial physiology in Arabidopsis. J Mol Biol 350 263–277 [DOI] [PubMed] [Google Scholar]
- Perrin R, Lange H, Grienenberger JM, Gagliardi D (2004. a) AtmtPNPase is required for multiple aspects of the 18S rRNA metabolism in Arabidopsis thaliana mitochondria. Nucleic Acids Res 32 5174–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin R, Meyer EH, Zaepfel M, Kim YJ, Mache R, Grienenberger JM, Gualberto JM, Gagliardi D (2004. b) Two exoribonucleases act sequentially to process mature 3′-ends of atp9 mRNAs in Arabidopsis mitochondria. J Biol Chem 279 25440–25446 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russel DW (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Satoh M, Kubo T, Nishizawa S, Estiati A, Itchoda N, Mikami T (2004) The cytoplasmic male-sterile type and normal type mitochondrial genomes of sugar beet share the same complement of genes of known function but differ in the content of expressed ORFs. Mol Genet Genomics 272 247–256 [DOI] [PubMed] [Google Scholar]
- Shindo C, Bernasconi G, Hardtke CS (2007) Natural genetic variation in Arabidopsis: tools, traits and prospects for evolutionary ecology. Ann Bot (Lond) 99 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger M, Kempken F (2003) Electroporation of isolated higher-plant mitochondria: transcripts of an introduced cox2 gene, but not an atp6 gene, are edited in organello. Mol Genet Genomics 269 553–561 [DOI] [PubMed] [Google Scholar]
- Sugiyama Y, Watase Y, Nagase M, Makita N, Yagura S, Hirai A, Sugiura M (2005) The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: comparative analysis of mitochondrial genomes in higher plants. Mol Genet Genomics 272 603–615 [DOI] [PubMed] [Google Scholar]
- Svab Z, Maliga P (2007) Exceptional transmission of plastids and mitochondria from the transplastomic pollen parent and its impact on transgene containment. Proc Natl Acad Sci USA 104 7003–7008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M, Brennicke A (2003) In vitro RNA editing in pea mitochondria requires NTP or dNTP, suggesting involvement of an RNA helicase. J Biol Chem 278 47526–47533 [DOI] [PubMed] [Google Scholar]
- Törjek O, Berger D, Meyer RC, Müssig C, Schmid KJ, Rosleff Sörensen T, Weisshaar B, Mitchell-Olds T, Altmann T (2003) Establishment of a high-efficiency SNP-based framework marker set for Arabidopsis. Plant J 36 122–140 [DOI] [PubMed] [Google Scholar]
- Unseld M, Marienfeld JR, Brandt P, Brennicke A (1997) The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat Genet 15 57–61 [DOI] [PubMed] [Google Scholar]
- Wang Z, Zou Y, Li X, Zhang Q, Chen L, Wu H, Su D, Chen Y, Guo J, Luo D, et al (2006) Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 18 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.