Abstract
In plants, fatty acids are de novo synthesized predominantly in plastids from acetyl-coenzyme A. Although fatty acid biosynthesis has been biochemically well studied, little is known about the regulatory mechanisms of the pathway. Here, we show that overexpression of the Arabidopsis (Arabidopsis thaliana) LEAFY COTYLEDON1 (LEC1) gene causes globally increased expression of fatty acid biosynthetic genes, which are involved in key reactions of condensation, chain elongation, and desaturation of fatty acid biosynthesis. In the plastidial fatty acid synthetic pathway, over 58% of known enzyme-coding genes are up-regulated in LEC1-overexpressing transgenic plants, including those encoding three subunits of acetyl-coenzyme A carboxylase, a key enzyme controlling the fatty acid biosynthesis flux. Moreover, genes involved in glycolysis and lipid accumulation are also up-regulated. Consistent with these results, levels of major fatty acid species and lipids were substantially increased in the transgenic plants. Genetic analysis indicates that the LEC1 function is partially dependent on ABSCISIC ACID INSENSITIVE3, FUSCA3, and WRINKLED1 in the regulation of fatty acid biosynthesis. Moreover, a similar phenotype was observed in transgenic Arabidopsis plants overexpressing two LEC1-like genes of Brassica napus. These results suggest that LEC1 and LEC1-like genes act as key regulators to coordinate the expression of fatty acid biosynthetic genes, thereby representing promising targets for genetic improvement of oil production plants.
Fatty acids and fatty acid-derived complex lipids are some of the most important macromolecules in all living organisms. Fatty acids and lipids are not only essential components of the cellular membranes and cellular signaling molecules, but also are major energy reserves in storage tissues such as seeds. In plant cells, fatty acids are de novo synthesized predominantly from acetyl-CoA in plastids. The reaction is initiated by the formation of malonyl-CoA from acetyl-CoA, catalyzed by acetyl-CoA carboxylase (ACCase). Subsequently, fatty acid synthase catalyzes the transfer of malonyl moiety to acyl carrier protein (ACP) by adding two carbons to the growing chain, leading to the formation of C16:0- and C18:0-ACP, which, upon chain elongation and desaturation reactions, can form a variety of fatty acid derivatives at the acyl chains. A major portion of acyl chains is then exported into the cytoplasm for the synthesis of complex lipids (Slabas and Fawcett, 1992; Ohlrogge and Browse, 1995; Harwood, 1996). Most enzymes of the fatty acid biosynthesis pathway have been biochemically characterized, and genes encoding most enzymes have been cloned from various species (Slabas and Fawcett, 1992; Ohlrogge and Browse, 1995; Harwood, 1996; Millar et al., 2000; Beisson et al., 2003). In Arabidopsis (Arabidopsis thaliana), the plastidial fatty acid synthetic pathway includes at least 24 enzymes or subunits, which are encoded by 46 nuclear genes and one plastidial gene, respectively (Beisson et al., 2003). Among these enzymes, ACCase is a major control point of the pathway (Slabas and Fawcett, 1992; Ohlrogge and Jaworski, 1997; Baud et al., 2003; Sasaki and Nagano, 2004).
In higher plants, the biosynthesis of most fatty acids is physiologically coupled with seed development. Several key regulators controlling seed maturation have been identified in Arabidopsis, including LEAFY COTYLEDON1 (LEC1), LEC2, ABSCISIC ACID INSENSITIVE3 (ABI3), and FUSCA3 (FUS3). Mutations in these genetic loci result in similar but distinctive phenotypes during seed development (Brocard-Gifford et al., 2003; To et al., 2006). LEC2 (Stone et al., 2001), ABI3 (Giraudat et al., 1992), and FUS3 (Luerssen et al., 1998) all belong to the plant-specific B3 transcription factor family, and LEC1 is an NFY-B-type or CCAAT-binding factor-type transcription factor (Lotan et al., 1998). Both LEC1 and LEC2 act as positive regulators upstream of ABI3 and FUS3, which, in turn, function partially redundantly to control the expression of seed storage protein (SSP) genes (Baud et al., 2002; Kroj et al., 2003; Kagaya et al., 2005b; To et al., 2006). Limited evidence suggests that these four genetic loci may also be involved in the regulation of fatty acid metabolism (Lotan et al., 1998; Ogas et al., 1999; Kroj et al., 2003; Kagaya et al., 2005b; To et al., 2006; Wang et al., 2007). However, direct evidence for this regulatory mechanism is not available.
Several other genes or genetic loci downstream from LEC1, LEC2, FUS3, and ABI3 have been identified to play an important role in the control of seed maturation. In particular, the wrinkled1 (wri1) mutant was identified in an elegantly designed genetic screen aimed at reducing seed oil levels by sorting incompletely filled and wrinkled-like seeds from normal seeds through centrifugation of mutagenized M2 seeds (Focks and Benning, 1998). The wri1 mutant seeds showed a decreased incorporation of Suc and Glc into triacylglycerols (TAGs), presumably caused by reduced activities of key glycolytic enzymes, including hexokinase and pyrophosphate-dependent phosphofructokinase (Focks and Benning, 1998). WRI1 was characterized as encoding a transcription factor containing an AP2/EREB domain, and overexpression of the transcription factor gene resulted in an increased TAG level in both seeds and leaves (Cernac and Benning, 2004). A more recent study revealed that LEC2 directly regulated WRI1, which, in turn, controlled the expression of a subset of genes involved in late glycolysis and fatty acid biosynthesis as well as the biosynthesis of biotin and lipoic acids (Baud et al., 2007a).
Despite these advances, the regulatory mechanism of fatty acid metabolism is not well understood. Here, we report a systematic analysis of the regulatory role of LEC1 in fatty acid biosynthesis. We found that inducible overexpression of LEC1 causes globally elevated expression of fatty acid synthetic genes and a substantially increased level of major fatty acid species. Overexpression of LEC1 ectopically activates FUS3, ABI3, and WRI1, but not LEC2. Consistent with these results, LEC1-regulated fatty acid biosynthesis is partially dependent on FUS3, ABI3, and WRI1. Our observations suggest that LEC1 acts as a key regulator to coordinate fatty acid biosynthesis in Arabidopsis.
RESULTS
Inducible Overexpression of LEC1 in Transgenic Arabidopsis Plants
Previous studies revealed that overexpression of LEC1 induced the expression of SSP and oleosin genes (Lotan et al., 1998). However, constitutive overexpression of LEC1 caused seedling lethality (Lotan et al., 1998), thus rendering it difficult to study LEC1 function. Therefore, we generated transgenic Arabidopsis plants carrying a pER8-LEC1 transgene, in which a LEC1 cDNA was placed under the control of an estradiol-inducible promoter (Zuo et al., 2000). For clarity, hereafter we refer to pER8-LEC1 transgenics treated with estradiol as LEC1-OX-inducible (LEC1-OXi) plants.
Except for the microarray experiments, all other experiments were repeated with at least three to five independent transgenic lines, and similar results were obtained. Results obtained from a representative line (line 23) are shown. Most experiments were carried out using both wild-type and transgenic plants treated with dimethyl sulfoxide (DMSO; a solvent for estradiol) as controls, which showed similar phenotypes. For conciseness, we only present data obtained from wild-type plants as controls.
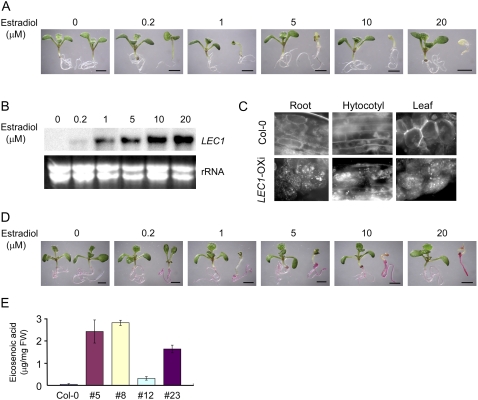
When germinated and grown in the presence of estradiol, LEC1-OXi seedlings showed a strong growth-inhibitory phenotype (Fig. 1A), characterized as yellowish cotyledons and rarely initiated true leaves. The transgenic phenotype was well correlated to the inducer concentrations (Fig. 1A) and the expression level of the LEC1 transgene (Fig. 1B). In rare cases, somatic embryos could be formed in LEC1-OXi plants. When stained with Nile Red, a large number of oil bodies were observed in vegetative tissues of LEC1-OXi transgenic plants but not in wild-type plants (Fig. 1C). Moreover, positive staining of Fat Red 7B was observed in LEC1-OXi plants but not in wild-type plants (Fig. 1D). Consistent with these observations, the accumulation of TAG, indicated by levels of eicosenoic acid (C20:1) in Arabidopsis, was increased in LEC1-OXi plants (Fig. 1E; Tables I–III). These results suggest that overexpression of LEC1 causes the accumulation of lipids and the formation of oil bodies in vegetative tissues, including roots, hypocotyls, cotyledons, and leaves.
Figure 1.
Characterization of pER8-LEC1 transgenic plants. A, Ten-day-old pER8-LEC1 plants germinated and grown on MS medium in the absence (left) or presence (right) of different concentrations of estradiol as indicated at top. Bars = 2 mm. B, Northern-blot analysis of LEC1 expression in transgenic plants. Total RNA was prepared from 10-d-old pER8-LEC1 transgenic plants treated with different concentrations of estradiol as indicated at top for 16 h. Each lane contained 20 μg of RNA. C, Oil bodies (white dots in the bottom panels; stained with Nile Red) in vegetative tissues of pER8-LEC1 transgenic seedlings germinated and grown in the presence of 10 μm estradiol for 5 d. D, Fat Red 7B staining of 10-d-old pER8-LEC1 transgenic plants germinated and grown on MS medium in the absence (left) or presence (right) of different concentrations of estradiol as indicated at top. Bars = 2 mm. E, The accumulation of eicosenoic acid (C20:1; a fatty acid maker for the formation of TAG) in pER8-LEC1 transgenic seedlings germinated and grown in the presence of 10 μm estradiol for 10 d. Four independent transgenic lines (line numbers are given below the graph) were analyzed. Data obtained from line 23 are presented hereafter. See Table I for more details. FW, Fresh weight.
Table I.
Fatty acid compositions of wild-type (Col-0) and LEC1-OXi seedlings at different times after germination
Wild-type and pER8-LEC1 seedlings were germinated and grown on MS medium with (LEC1-OXi) or without (Col-0) estradiol for varying times as indicated and then collected for fatty acid analysis as described in “Materials and Methods.” Data reported are mean values of three independent experiments (three biological repeats) ± se. All values are expressed as μg mg−1 fresh weight. ND, Not detectable.
| Fatty Acid | Day 5
|
Day 10
|
Day 15
|
|||
|---|---|---|---|---|---|---|
| Col-0 | LEC1-OXi | Col-0 | LEC1-OXi | Col-0 | LEC1-OXi | |
| 16:0 | 0.47 ± 0.08 | 0.62 ± 0.10 | 0.59 ± 0.09 | 0.70 ± 0.15 | 0.60 ± 0.12 | 1.12 ± 0.02 |
| 18:0 | 0.10 ± 0.00 | 0.15 ± 0.00 | 0.08 ± 0.01 | 0.21 ± 0.01 | 0.07 ± 0.01 | 0.22 ± 0.00 |
| 18:1 (Δ9) | 0.13 ± 0.07 | 0.34 ± 0.04 | 0.08 ± 0.03 | 0.82 ± 0.16 | 0.07 ± 0.01 | 1.10 ± 0.21 |
| 18:1 (Δ11) | 0.05 ± 0.02 | 0.10 ± 0.03 | 0.03 ± 0.01 | 0.36 ± 0.06 | ND | 0.38 ± 0.03 |
| 18:2 | 0.67 ± 0.15 | 1.11 ± 0.04 | 0.57 ± 0.01 | 1.78 ± 0.36 | 0.52 ± 0.08 | 2.75 ± 0.63 |
| 18:3 | 0.99 ± 0.14 | 2.18 ± 0.11 | 1.08 ± 0.20 | 2.68 ± 0.63 | 1.20 ± 0.09 | 3.53 ± 0.22 |
| 20:0 | 0.01 ± 0.05 | 0.05 ± 0.01 | ND | 0.12 ± 0.05 | ND | 0.15 ± 0.01 |
| 20:1 | 0.13 ± 0.04 | 0.94 ± 0.12 | 0.04 ± 0.04 | 1.64 ± 0.17 | ND | 1.58 ± 0.36 |
| 20:2 | ND | 0.13 ± 0.04 | ND | 0.22 ± 0.04 | ND | 0.16 ± 0.02 |
| 22:0 | ND | 0.04 ± 0.01 | ND | 0.03 ± 0.01 | ND | 0.05 ± 0.01 |
| 22:1 | 0.02 ± 0.00 | 0.13 ± 0.01 | ND | 0.22 ± 0.09 | ND | 0.26 ± 0.10 |
| Sum | 2.57 ± 0.55 | 5.97 ± 0.51 | 2.47 ± 0.39 | 8.78 ± 1.73 | 2.46 ± 0.31 | 11.3 ± 1.61 |
Table II.
Fatty acid contents in wild-type and LEC1-OXi seedlings treated with different concentrations of estradiol
Ten-day-old wild-type and pER8-LEC1 seedlings germinated and grown on MS medium containing different concentrations of estradiol as indicated were used for fatty acid analysis as described in “Materials and Methods.” Treatment with 25 μm estradiol did not significantly alter the fatty acid level compared with treatment with 10 μm estradiol (data not shown). Data reported are mean values of three independent experiments (three biological repeats) ± se. All values are expressed as μg mg−1 fresh weight. ND, Not detectable.
| Fatty Acid | Estradiol Level
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 0 μm
|
1 μm
|
5 μm
|
10 μm
|
|||||
| Col-0 | LEC1-OXi | Col-0 | LEC1-OXi | Col-0 | LEC1-OXi | Col-0 | LEC1-OXi | |
| 16:0 | 0.50 ± 0.04 | 0.55 ± 0.02 | 0.60 ± 0.00 | 0.72 ± 0.05 | 0.57 ± 0.08 | 0.77 ± 0.02 | 0.52 ± 0.02 | 0.70 ± 0.15 |
| 18:0 | 0.10 ± 0.03 | 0.12 ± 0.00 | 0.12 ± 0.03 | 0.19 ± 0.02 | 0.13 ± 0.06 | 0.24 ± 0.05 | 0.11 ± 0.03 | 0.21 ± 0.01 |
| 18:1 | 0.19 ± 0.01 | 0.16 ± 0.01 | 0.10 ± 0.04 | 0.45 ± 0.19 | 0.12 ± 0.01 | 0.64 ± 0.07 | 0.09 ± 0.06 | 1.17 ± 0.24 |
| 18:2 | 0.53 ± 0.06 | 0.61 ± 0.20 | 0.53 ± 0.01 | 1.53 ± 0.14 | 0.55 ± 0.00 | 1.71 ± 0.11 | 1.45 ± 0.10 | 3.16 ± 0.59 |
| 18:3 | 1.29 ± 0.13 | 1.56 ± 0.11 | 1.35 ± 0.12 | 2.04 ± 0.14 | 1.26 ± 0.17 | 2.15 ± 0.22 | 1.34 ± 0.09 | 3.48 ± 0.38 |
| 20:0 | ND | ND | ND | 0.09 ± 0.07 | ND | 0.11 ± 0.04 | ND | 0.12 ± 0.05 |
| 20:1 | ND | ND | ND | 1.14 ± 0.21 | ND | 1.40 ± 0.15 | ND | 1.51 ± 0.17 |
| 20:2 | ND | ND | ND | 0.13 ± 0.06 | ND | 0.17 ± 0.05 | ND | 0.22 ± 0.04 |
| 22:0 | ND | ND | ND | 0.01 ± 0.01 | ND | 0.03 ± 0.00 | ND | 0.03 ± 0.01 |
| 22:1 | ND | ND | ND | 0.12 ± 0.02 | ND | 0.16 ± 0.07 | ND | 0.22 ± 0.09 |
| Sum | 2.61 ± 0.27 | 3.00 ± 0.34 | 2.70 ± 0.20 | 6.42 ± 0.91 | 2.63 ± 0.32 | 7.38 ± 0.78 | 3.51 ± 0.30 | 10.82 ± 1.73 |
Table III.
Fatty acid compositions in individual seeds and seedlings of wild-type and LEC1-OXi plants
The same batches of wild-type (Col-0) and pER8-LEC1 seeds were used in the analysis. Ten-day-old seedlings germinated and grown in the presence (LEC1-OXi) or absence (Col-0) of estradiol were collected for fatty acid analysis as described in “Materials and Methods.” Randomly collected samples (100 seeds or seedlings per sample) were mixed and then used for extraction of total fatty acids and lipids. Mean values of fatty acid methyl esters in individual seeds or seedlings were calculated by dividing raw data by 100. Data presented are mean values of three independent experiments (three biological repeats) ± se. All values are expressed as μg seed−1 or μg seedling−1 as applicable. ND, Not detectable.
| Fatty Acid | Seed
|
Seedling
|
||
|---|---|---|---|---|
| Col-0 | pER8-LEC1 | Col-0 | LEC1-OXi | |
| C16:0 | 0.36 ± 0.07 | 0.39 ± 0.11 | 0.39 ± 0.03 | 1.16 ± 0.03 |
| C16:1 | 0.02 ± 0.01 | 0.02 ± 0.00 | 0.02 ± 0.01 | 0.01 ± 0.01 |
| C16:3 | 0.01 ± 0.00 | 0.01 ± 0.01 | 0.37 ± 0.08 | 0.05 ± 0.08 |
| C18:0 | 0.12 ± 0.01 | 0.11 ± 0.01 | 0.07 ± 0.01 | 0.30 ± 0.03 |
| C18:1 | 0.48 ± 0.06 | 0.51 ± 0.14 | 0.09 ± 0.10 | 1.38 ± 0.11 |
| C18:2 | 1.31 ± 0.12 | 1.28 ± 0.21 | 0.45 ± 0.04 | 2.55 ± 0.12 |
| C18:3 | 1.06 ± 0.23 | 1.15 ± 0.16 | 1.16 ± 0.12 | 3.78 ± 0.43 |
| C20:0 | 0.05 ± 0.01 | 0.04 ± 0.01 | ND | 0.18 ± 0.05 |
| C20:1 | 0.93 ± 0.11 | 0.91 ± 0.07 | 0.01 ± 0.00 | 2.09 ± 0.22 |
| C20:2 | 0.07 ± 0.02 | 0.05 ± 0.01 | ND | 0.23 ± 0.04 |
| C22:1 | 0.06 ± 0.02 | 0.04 ± 0.02 | ND | 0.28 ± 0.07 |
| Sum | 4.47 ± 0.66 | 4.51 ± 0.80 | 2.56 ± 0.39 | 12.01 ± 1.19 |
Increased Fatty Acid Accumulation in LEC1-OXi Plants
To determine the fatty acid levels, we analyzed major fatty acid species in LEC1-OXi plants by gas chromatography-mass spectrometry. In LEC1-OXi plants, most assayed fatty acid species showed an increased level at varying degrees, reaching a higher level around 10 to 15 d after germination (Table I). At day 10, the C16:0 and C18:0 levels in LEC1-OXi were increased approximately 18% and 160%, respectively, compared with the wild type. Several unsaturated species also displayed a substantially increased level. Compared with that in the wild type, C18:2 and C18:3 were increased 2- to 3-fold, whereas both C18:1 (Δ9) and C18:1 (Δ11) were increased more than 10-fold. Several species of very-long-chain unsaturated fatty acids showed a more substantially increased level in LEC1-OXi plants, owing to undetectable levels in the wild type (Table I). The increased fatty acid level in LEC1-OXi plants was strictly dependent on the inducer concentrations (Table II).
Because of the growth-inhibitory effect of overexpression of LEC1, the increased level of fatty acids observed in LEC1-OXi seedlings might result from incomplete degradation of lipids in seeds. If de novo synthesis of fatty acids did not occur in LEC1-OXi seedlings, one would expect a gradually decreased level or at least an unaltered level of fatty acids during the postgerminative growth. However, the longer the period of postgerminative growth, the higher the level of fatty acid observed in LEC1-OXi seedlings (Table I), suggesting that the increased level of fatty acids in LEC1-OXi seedlings was unlikely to be caused by impaired degradation of lipids in seeds. To further confirm this view, we measured and calculated mean levels of fatty acids in individual LEC1-OXi seeds and seedlings. In the absence of estradiol, pER8-LEC1 seeds (4.51 μg seed−1) accumulated fatty acids at a level comparable to that of wild-type (Columbia-0 [Col-0]) seeds (4.47 μg seed−1). When germinated and grown on medium containing estradiol, LEC1-OXi seedlings (12.01 μg seedling−1) accumulated a higher level of fatty acids than wild-type seedlings (2.56 μg seedling−1). More importantly, each LEC1-OXi seedling produced a higher level of fatty acids than each seed (12.01 μg seedling−1 versus 4.51 μg seed−1; Table III), demonstrating that de novo synthesis of fatty acids indeed occurred in LEC1-OXi seedlings during the postgerminative growth. Taken together, these results suggest that LEC1 positively regulates major steps of de novo fatty acid biosynthesis, including condensation, chain elongation, and desaturation.
Differentially Expressed Genes in LEC1-OXi Are Mainly Involved in Fatty Acid Metabolism and Embryogenesis
To better understand the molecular mechanism of the LEC1-regulated fatty acid biosynthesis, we attempted to identify differentially expressed genes in LEC1-OXi transgenic plants by gene profiling using the Affymetrix ATH1 microarray (Supplemental Fig. S1). In germinating LEC1-OXi seeds, the fatty acid level had no detectable alterations when treated with estradiol for 6 h and was marginally increased when treated for 1 d (data not shown), but it was substantially elevated at day 5 (Table I). Therefore, we used RNA samples prepared from wild-type and LEC1-OXi seedlings at day 4 after germination for the microarray experiment. Whereas most differentially expressed genes identified from this experiment might not be the primary responsive genes of LEC1, they should include those directly related to fatty acid metabolism. We identified 687 differentially expressed genes with log2 ratios greater than 1.00 or less than −1.00, among which 425 were up-regulated (Supplemental Table S1) and 262 were down-regulated (Supplemental Table S2).
A functional analysis of these 687 differentially expressed genes revealed that the expression of genes involved in lipid metabolism and embryogenesis was significantly altered in LEC1-OXi seedlings (Table IV; Supplemental Fig. S2). More than 13% of down-regulated genes were related to general protein synthesis machinery (Table IV; Supplemental Table S2). In particular, ribosomal protein genes showed a substantially reduced expression level. This result suggests that overexpression of LEC1 may slow the protein synthesis rate.
Table IV.
Functional classification of differentially expressed genes in LEC1-OXi transgenic plants
Functional classification of the differentially expressed genes was performed using the biological process category of Arabidopsis Gene Ontology (http://www.geneontology.com). Percentage (far right column) refers to the ratio of genes of each functional category relative to total up-regulated or down-regulated differentially expressed genes identified in the microarray experiment.
| Category | Up-Regulated Differentially Expressed Genes
|
|||||
|---|---|---|---|---|---|---|
| ≥10 | 6–10 | 3–6 | 1–3 | Total | Percentage | |
| log2 ratio | ||||||
| Metabolism | ||||||
| Lipid metabolism | 3 | 8 | 9 | 28 | 48 | 11.3 |
| Carbohydrate metabolism | 0 | 1 | 5 | 27 | 33 | 7.8 |
| Nucleic acid | 0 | 0 | 1 | 11 | 12 | 2.8 |
| Amino acid and protein | 0 | 4 | 2 | 14 | 20 | 4.7 |
| Growth and development | ||||||
| Storage protein | 11 | 1 | 1 | 1 | 14 | 3.3 |
| Embryo and seed development | 0 | 3 | 4 | 5 | 12 | 2.8 |
| Flower development | 1 | 0 | 4 | 2 | 7 | 1.6 |
| Cell wall | 0 | 3 | 1 | 2 | 6 | 1.4 |
| Cytoskeleton structure | 0 | 0 | 0 | 15 | 15 | 3.5 |
| Hormone | 0 | 1 | 1 | 3 | 5 | 1.2 |
| Stress/defense response | 1 | 3 | 2 | 9 | 15 | 3.5 |
| Cell regulation | ||||||
| Transcriptional regulation | 0 | 0 | 5 | 31 | 36 | 8.4 |
| Protein degradation | 0 | 1 | 2 | 13 | 16 | 3.8 |
| Signaling transduction | 0 | 0 | 7 | 31 | 38 | 8.9 |
| Electron transport | 0 | 0 | 3 | 8 | 11 | 2.6 |
| Transport facilitation | 0 | 3 | 2 | 23 | 28 | 6.6 |
| Unknown function | 1 | 3 | 14 | 60 | 78 | 18.4 |
| Other | 1 | 5 | 6 | 19 | 31 | 7.3 |
| Category | Down-Regulated Differentially Expressed Genes
|
|||
|---|---|---|---|---|
| ≤−2 | −2 to −1 | Total | Percentage | |
| log2 ratio | ||||
| Metabolism | ||||
| Lipid metabolism | 7 | 3 | 10 | 3.8 |
| Carbohydrate metabolism | 6 | 6 | 12 | 4.6 |
| Nucleic acid | 4 | 11 | 15 | 5.7 |
| Amino acid and protein | 6 | 29 | 35 | 13.4 |
| Growth and development | ||||
| Embryo and seed development | 3 | 6 | 9 | 3.4 |
| Other development | 2 | 3 | 5 | 1.9 |
| Hormone | 1 | 3 | 4 | 1.5 |
| Stress/defense response | 10 | 7 | 17 | 6.5 |
| Cell regulation | ||||
| Transcriptional regulation | 7 | 12 | 19 | 7.3 |
| Protein degradation | 1 | 7 | 8 | 3.1 |
| Signal transduction | 4 | 9 | 13 | 5.0 |
| Electron transport | 2 | 8 | 10 | 3.8 |
| Transport facilitation | 9 | 13 | 22 | 8.4 |
| Photosynthesis | 6 | 3 | 9 | 3.4 |
| Unknown function | 21 | 34 | 55 | 21 |
| Other | 5 | 14 | 19 | 7.3 |
In the up-regulated genes, the most apparent alteration was observed in genes related to fatty acid and lipid metabolism, including 48 genes or 11.3% of all up-regulated genes (Table IV; Supplemental Table S1). In addition, 33 genes (7.5%) were related to carbohydrate metabolism. Notably, the expression of several genes in the glycolysis pathway was significantly increased, including a Suc synthase gene (At5g49190), a putative Glc-6-P 1-dehydrogenase gene (G6PD; At1g09420), and a pyruvate kinase gene (At5g52920; Supplemental Table S1), implying an increased flux of Suc and hexose required for fatty acid de novo synthesis (Rawsthorne, 2002; Schwender et al., 2003). In the Arabidopsis genome, six G6PD genes were identified and biochemically characterized. Of these, At1g09420, annotated as G6PD4 localized in plastid, appeared to be a unique member that did not group with the other five genes. In contrast to other members, G6PD4 recombinant protein showed very low enzymatic activity (Wakao and Benning, 2005). Because all G6PD recombinant proteins rapidly lost their biochemical activities upon purification (Wakao and Benning, 2005), the putative G6PD4 protein might have a higher activity in planta or might have a higher specificity on unknown substrates. Plastidic pyruvate kinase is a heteromeric complex, of which At5g52920 encodes a β1-subunit (Andre et al., 2007). Loss-of-function mutations in At5g52920 (the pkp1 or pkp2 mutant) caused a wrinkled-like phenotype of seeds and a significant reduction of seed oil content compared with the wild type (Andre et al., 2007; Baud et al., 2007b). Moreover, the PKP1/PKP2 activity was also required for embryo development, seed germination, and seedling establishment, presumably by regulating sugar conversion and carbon flux (Andre and Benning, 2007; Andre et al., 2007; Baud et al., 2007b).
In addition, genes implicated in embryo development were also up-regulated in LEC1-OXi plants (Supplemental Table S1). In particular, expression of LEAFY COTYLEDON1-LIKE (L1L; At5g47670; Fig. 3A; Kwong et al., 2003), WRI1 (At3g54320; Cernac and Benning, 2004), and PICKLE (PKL; At2g25170; Ogas et al., 1999) was up-regulated. L1L functions redundantly with its close homolog LEC1 during embryogenesis (Kwong et al., 2003). WRI1, a target of LEC2, controls the expression of a subset of genes encoding enzymes of late glycolysis and fatty acid synthesis (Baud et al., 2007a). However, LEC2 expression was unaltered in LEC1-OXi plants (Fig. 3A). Because PKL can repress LEC1 expression (Ogas et al., 1997, 1999), LEC1 is likely to be involved in the control of PKL expression by a feedback mechanism. Lastly, consistent with previous studies on the regulatory role of LEC1 in the At2S3 gene (Kroj et al., 2003; Kagaya et al., 2005b; To et al., 2006), 14 SSP genes were up-regulated. Taken together, these four groups of genes accounted for 25.2% of all up-regulated genes, suggesting that LEC1 directly or indirectly controls the expression of a large repertoire of genes involved in embryo or seed maturation.
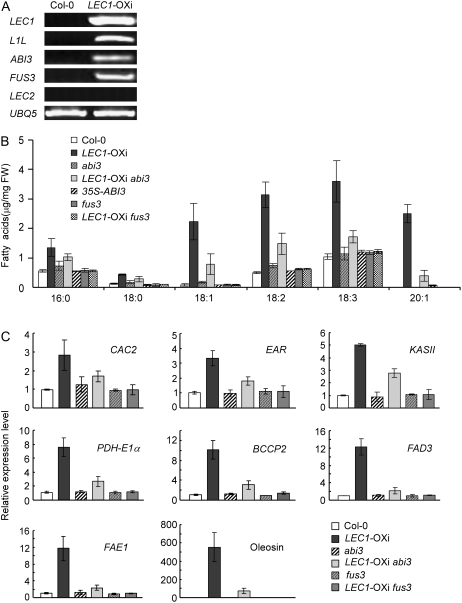
Figure 3.
ABI3 and FUS3 are required for LEC1 function in fatty acid biosynthesis. A, RT-PCR analysis of the expression of LEC1, L1L, ABI3, FUS3, and LEC2 in wild-type and LEC1-OXi plants germinated and grown in the presence of 10 μm estradiol for 4 d. See Figure 2C legend for other technical details. B, Fatty acid contents in 10-d-old seedlings germinated and grown in the presence of 10 μm estradiol. Data presented are mean values of three independent experiments. FW, Fresh weight. Error bars denote sd. C, Quantitative RT-PCR analysis of the expression of representative fatty acid synthetic genes in 4-d-old seedlings germinated and grown in the presence of 10 μm estradiol. See Figure 2B legend for other technical details.
LEC1 Positively Regulates Fatty Acid Biosynthetic Genes
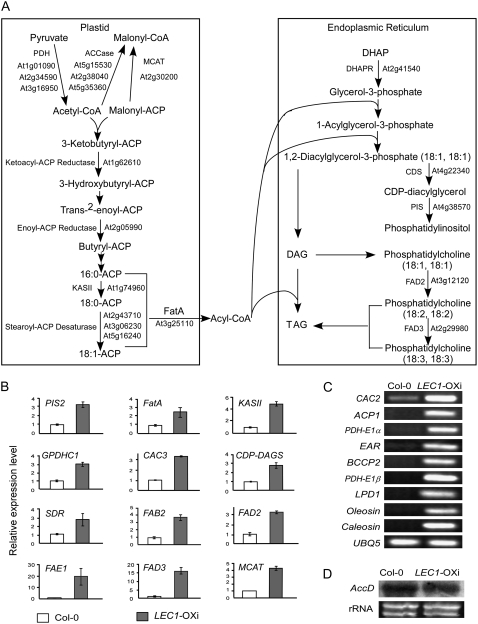
In Arabidopsis, 46 nuclear genes were annotated to encode 24 enzymes or subunits in the plastidial fatty acid synthetic pathway (Beisson et al., 2003). Two additional genes (At1g52670 and At5g08415) were recently annotated as components of this pathway (www.Arabidopsis.org), bringing the total number of the nuclear genes to 48. In the plastidial pathway, 18 genes were up-regulated in LEC1-OXi plants, which encoded 14 of 24 enzymes or enzymatic subunits of the pathway (58.3%; Fig. 2, A–C; Table IV; Supplemental Tables S1 and S3). Among these enzymes, ACCase, a key enzyme of the pathway, consists of four subunits: α-, β-, and biotin-subunits and a biotin carboxyl carrier protein. The β-subunit is encoded by the plastidial genome, whereas the other three subunits are encoded by the nuclear genome. In LEC1-OXi plants, expression of the α-subunit (At2g38040), biotin-subunit (At5g35360), and biotin carboxyl carrier protein genes (At5g15530 and At1g52670) was increased (Fig. 2, A–C; Supplemental Tables S1 and S3). However, expression of the plastidial gene β-subunit (AtCg00500) remained largely unaltered (Fig. 2D).
Figure 2.
Up-regulated differentially expressed genes in the fatty acid biosynthesis pathway in LEC1-OXi plants. A, Diagram of the fatty acid biosynthetic pathway with major steps in the plastid and the cytosol (endoplasmic reticulum). Up-regulated differentially expressed genes in LEC1-OXi plants are indicated by the Arabidopsis Genome Initiative codes. See Supplemental Table S3 for a complete list of the up-regulated differentially expressed genes of the pathway and their full names. B, Expression of representative fatty acid synthetic genes analyzed by quantitative real-time RT-PCR using total RNA prepared from wild-type and LEC1-OXi seedlings germinated and grown in the presence of 10 μm estradiol for 4 d. The relative expression level of each gene was normalized with that of ACTIN7. Error bars indicate sd values of three independent experiments. C, Expression of representative fatty acid synthetic genes analyzed by RT-PCR. The PCR was cycled 26 times for all samples. RNA was prepared from seedlings germinated and grown in the presence of 10 μm estradiol for 4 d. D, Expression of the β-subunit (AtCg00500) gene analyzed by northern blot. See Figure 1B for technical details.
Downstream from ACCase, a key reaction is to transfer a malonyl moiety from malonyl-CoA to ACP by malonyl-CoA:ACP transacylase (MCAT or FabD) to form malonyl-ACP. In LEC1-OXi plants, expression of a putative MCAT gene (At2g30200) was elevated. In addition, several genes encoding stearoyl-ACP desaturases (At2g43710 or FAB2/SSI2, At3g02630, and At5g16240; Kachroo et al., 2001) were also up-regulated (Fig. 2B; Supplemental Table S3). Stearoyl-ACP desaturase catalyzes a reaction by insertion of a cis double bond at position 9 of the C18:0-ACP. Thus, this result is consistent with the observation that C18:1 was one of the fatty acid species that showed the highest increased level in LEC1-OXi plants (Tables I and II). The expression of ACP1 (At5g27200; Bonaventure and Ohlrogge, 2002) was also significantly increased. Moreover, expression of several other genes was moderately increased, including a putative α-subunit of pyruvate dehydrogenase (At1g01090 or PDH-E1α) and an enoyl-ACP reductase (At2g05990 or EAR/MOD1; Mou et al., 2000).
In addition to genes in the plastidial pathway, other fatty acid synthetic genes also showed an elevated expression level in LEC1-OXi plants (Fig. 2; Supplemental Table S3), including two genes encoding ω-6 fatty acid desaturase (FAD2; At3g12120), ω-3 fatty acid desaturase (FAD3; At2g29980), and FATTY ACID ELONGASE1 (FAE1 or KCS1; At4g34520; James et al., 1995; Millar and Kunst, 1997). Lastly, consistent with the formation of oil bodies in the transgenic plants (Fig. 1C), expression of six oleosin genes was significantly increased.
Taken together, these results indicate that LEC1 positively coordinates the expression of a large number of fatty acid biosynthetic genes, thereby controlling fatty acid biosynthesis and the accumulation of lipids.
LEC1 Function Is Partially Dependent on ABI3, FUS3, and WRI1
In LEC1-OXi plants, both ABI3 and FUS3 showed an increased expression level (Fig. 3A), consistent with previous observations that LEC1 acts upstream of ABI3 and FUS3 (Kroj et al., 2003; Kagaya et al., 2005b; To et al., 2006). To explore possible interactions of these genetic loci in the regulation of fatty acid biosynthesis and lipid accumulation, we examined the LEC1-OXi phenotype in the abi3 or fus3 mutant background. Compared with that in LEC1-OXi, overexpression of LEC1 in the abi3 or fus3 mutant background showed a lower level of several fatty acid species (Fig. 3B). Remarkably, LEC1-OXi fus3 plants showed a similar phenotype as the wild type, suggesting that FUS3 is critical for fatty acid biosynthesis regulated by LEC1. Consistently, LEC1-up-regulated expression of several fatty acid synthetic genes was reduced in LEC1-OXi abi3 and was nearly unaltered in LEC1-OXi fus3 plants compared with LEC1-OXi plants (Fig. 3C).
Because overexpression of FUS3 caused increased expression of several fatty acid biosynthetic genes and higher fatty acid levels (Wang et al., 2007; H. Tan, J. Mu, and J. Zuo, unpublished data), we then asked if overexpression of ABI3 could have a similar phenotype. Overexpression of ABI3 resulted in an increased eicosenoic acid (20:1) level (Fig. 3B), consistent with the results obtained from a previous study, in which eicosenoic acid was found to be reduced in abi3 seeds (Finkelstein and Somerville, 1990). However, other examined fatty acid species did not show apparently altered levels in the abi3 mutant and 35S-ABI3 transgenic plants (Fig. 3B).
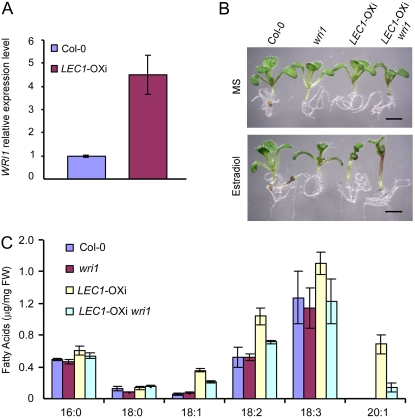
Previous studies showed that WRI1 acts downstream of LEC2 to regulate the biosynthesis of storage compounds, including fatty acids (Cernac and Benning, 2004; Baud et al., 2007a). In LEC1-OXi plants, WRI1 expression was increased (Fig. 4A), implying that WRI1 might also function downstream of LEC1. To test this possibility, we generated LEC1-OXi wri1 plants and analyzed their phenotype. When germinated and grown in the presence of 10 μm estradiol, a concentration used in most experiments in this study, less than 50% of LEC1-OXi wri1 seeds germinated, and most of the seedlings did not survive. This phenotype is somewhat expected, because wri1 is hypersensitive to ABA and sugar (Cernac et al., 2006) and overexpression of LEC1 activates ABI3 and FUS3 (Fig. 3). Therefore, we analyzed LEC1-OXi wri1 at lower concentrations of the chemical inducer estradiol. We found that LEC1-OXi wri1 plants germinated and grown in the presence of 0.1 μm estradiol had a germination rate of more than 90% and showed an apparent growth phenotype (Fig. 4B; data not shown). Moreover, the fatty acid level was lower in LEC1-OXi wri1 plants than in LEC1-OXi plants (Fig. 4C). In particular, the C20:1 level of LEC1-OXi wri1 plants was 5-fold lower that than of LEC1-OXi plants (Fig. 4C). These results suggest that LEC1 acts upstream of WRI1 to regulate fatty acid biosynthesis.
Figure 4.
LEC1-regulated fatty acid biosynthesis is partially dependent on WRI1. A, Quantitative RT-PCR analysis of WRI1 expression in wild-type (Col-0) and LEC1-OXi plants germinated and grown in the presence of 10 μm estradiol for 4 d. Results are mean values of three independent experiments. See Figure 2B legend for technical details. Error bars denote sd. B, Ten-day-old seedlings germinated and grown in the absence (MS) or presence of 0.1 μm estradiol. Bars = 2 mm. C, Fatty acid levels in Col-0, wri1, LEC1-OXi, and LEC1-OXi wri1 seedlings germinated and grown in the presence of 0.1 μm estradiol for 10 d. Results are mean values obtained from three independent experiments. FW, Fresh weight. Error bars indicate sd. [See online article for color version of this figure.]
Taken together, these results suggest that LEC1-regulated fatty acid biosynthesis is partially dependent on ABI3, FUS3, and WRI1.
BnLEC1 and BnL1L Function Similarly as Arabidopsis LEC1
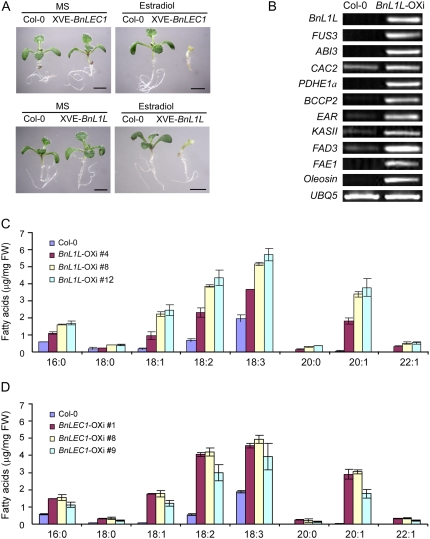
The finding that LEC1 is a key regulator for fatty acid biosynthesis prompted us to ask if the orthologous genes of LEC1 in other species have a similar function. This is particularly important for oil production crops such as oilseed rape (Brassica napus). To test this feasibility, we cloned BnLEC1 and BnL1L genes and analyzed their functions in transgenic Arabidopsis plants. BnLEC1 and BnL1L share significant homology with their Arabidopsis orthologs (Supplemental Fig. S3). Under the control of an estradiol-inducible promoter, BnLEC1 and BnL1L were stably transformed into wild-type Arabidopsis plants. Inducible overexpression of BnLEC1 and BnL1L showed a phenotype similar to that of LEC1-OXi plants, including growth inhibition (Fig. 5A), an increased fatty acid level (Fig. 5, C and D), and up-regulated expression of representative fatty acid synthetic genes (Fig. 5B; data not shown). Note that the C20:1 level was significantly increased in all tested transgenic lines of BnL1L and BnLEC1 (Fig. 5, C and D), indicating the increased accumulation of lipids in these transgenic plants.
Figure 5.
Functional characterization of BnLEC1 and BnL1L1. A, Ten-day-old seedlings germinated and grown in the absence (MS) or presence of 10 μm estradiol. Bars = 2 mm. B, RT-PCR analysis of the expression of representative fatty acid synthetic genes in BnL1L-OXi seedlings germinated and grown in the presence of 10 μm estradiol for 4 d (transgenic line 12). C, Fatty acid levels in pER10-BnL1L transgenic seedlings germinated and grown in the presence of 10 μm estradiol for 10 d. Three independent transgenic lines were tested. Results are mean values obtained from two independent experiments. Error bars indicate sd. Note that the levels of C20:0, C20:1, and C22:1 were under the detection limit in wild-type seedlings (Col-0; dark blue bars). FW, Fresh weight. D, Fatty acid levels in pER10-BnLEC1 transgenic seedlings analyzed as described for C. [See online article for color version of this figure.]
DISCUSSION
Fatty acids are a class of prominent metabolites that are essential for the growth and development of all living organisms. Although the biochemistry of fatty acid biosynthesis has been well studied, the regulatory mechanism of fatty acid metabolism is not well understood in plants, a major source of consumable lipids in the world. In this study, we found that overexpression of the Arabidopsis transcription factor gene LEC1 resulted in elevated expression of a large number of genes in the fatty acid biosynthesis pathway. Consistent with this observation, the accumulation of major fatty acid species and lipids was substantially increased in LEC1-OXi transgenic plants. Thus, LEC1 appears to act as a key regulator for fatty acid biosynthesis and lipid accumulation in Arabidopsis.
Regulation of fatty acid biosynthesis has been proposed to take place at multiple levels, of which transcriptional control has been considered a major means to regulate the pathway (Ohlrogge and Jaworski, 1997; Millar et al., 2000). Expression of fatty acid synthetic genes is expected to be coordinately regulated, and this regulatory mode appears to be partly pertinent to key fatty acid synthetic genes during seed maturation, including those encoding major subunits of ACCase (Ruuska et al., 2002; Baud et al., 2003). Coincident with these observations, more than half of plastidial fatty acid synthetic genes are up-regulated in LEC1-OXi plants. In particular, several nuclear genes encoding ACCase subunits were up-regulated in a proportional manner in LEC1-OXi transgenic plants. However, expression of the plastidial genome-encoded β-subunit remained nearly unaltered. This result is not fully in agreement with previous observations that the expression of all four subunit genes of ACCase is closely coordinated to maintain a constant molar stoichiometric ratio of the transcript concentrations (Ke et al., 2000; O'Hara et al., 2002; Sasaki and Nagano, 2004). We speculate that a functionally excessive amount of β-subunit transcript or protein is present in plastids, thereby sufficing for an increased ACCase activity caused by the elevated levels of the other three subunits. This view is consistent with the observation that many enzymes of fatty acid biosynthesis are indeed present in functional excess (Ohlrogge and Jaworski, 1997).
We noticed that the expression of ACCase and fatty acid synthase genes was only moderately increased, whereas the expression of several genes involved in acyl chain elongation and desaturation reactions showed a more remarkable increase in LEC1-OXi plants. Consequently, while levels of C16:0 and C18:0, two end products of the condensation reactions, were moderately elevated, unsaturated C18 and C20 species were increased more significantly. A lower inducible expression level of ACCase genes is therefore consistent with the model that ACCase is a key switch to control fatty acid flux, in which the switch must be highly dynamic in responding to variable signals and must be kept at a low threshold to allow tight control. Therefore, ACCase may act as a sensor or a gating system to monitor the overall flux of fatty acid biosynthesis.
In LEC1-OXi plants, the expression of several SSP genes was highly inducible, similar to that observed previously (Kagaya et al., 2005a, 2005b). Overexpression of LEC1 causes an elevated expression level of At2S3, a representative SSP gene, in an ABI3- and FUS3-dependent manner (Kagaya et al., 2005a, 2005b). This regulatory mechanism is similar to that of fatty acid synthesis as revealed in this study. However, regulation of fatty acid synthesis appears to employ a more sophisticated mechanism. Whereas LEC1-regulated fatty acid biosynthesis is partially dependent on ABI3, the fus3 mutation appears to completely suppress the LEC1-OXi phenotype. This result suggests that FUS3 is essential for the LEC1 function in the regulation of fatty acid biosynthesis and possibly other developmental programs as well. In agreement with this notion, ABI3 may play an indirect role, likely acting as a cofactor to regulate At2S3, whereas FUS3 and LEC2 are able to directly activate At2S3 expression (Kroj et al., 2003).
Does FUS3 act directly on fatty acid synthetic genes? Although FUS3 and LEC2 are able to directly activate At2S3 expression (Kroj et al., 2003), currently available evidence does not reveal a direct link between any B3 transcription factors and fatty acid synthetic genes, implying the presence of additional regulatory components between them. Indeed, LEC2 has been shown to directly target WRI1, which appears to regulate a subset of fatty acid synthetic genes (Baud et al., 2007a). We noticed that several putative target genes of WRI1 in the fatty acid synthesis pathway (At5g15530, At2g05990, At1g01090, and At4g34520; Baud et al., 2007a) are also substantially up-regulated in LEC1-OXi plants. On the other hand, because LEC1 and LEC2 do not appear to function in a linear pathway (To et al., 2006), it is not surprising that LEC2 did not show a differentially expressed pattern in LEC1-OXi plants. However, the expression of WRI1 was up-regulated in LEC1-OXi plants, suggesting that WRI1 might also be regulated by a LEC2-independent mechanism. In agreement with this notion, the wri1 mutation partially suppresses the LEC1-OXi phenotype, suggesting that WRI1 acts downstream of LEC1 and LEC2, possibly in two parallel or partially redundant pathways, as suggested previously in studies on the expression of SSP genes (Kroj et al., 2003; To et al., 2006).
Most, if not all, of the past efforts to increase oil production in seeds were made by manipulating key enzyme genes in the fatty acid biosynthesis pathway. In some cases, multiple genes were co-overexpressed in transgenic plants. However, many of these efforts have not been able to substantially increase oil production, owing to apparent bottlenecks in the fatty acid flux (Thelen and Ohlrogge, 2002; Jaworski and Cahoon, 2003; Cahoon et al., 2007). These results are somewhat expected, because fatty acid biosynthesis appears to be controlled by a coordinated regulatory mechanism. This coregulation mechanism is not only pertinent to major steps of the fatty acid and lipid metabolism pathways but also requires the coordination of key components in carbohydrate metabolism, in particular the regulation of Suc and hexose flux (Rawsthorne, 2002; Ruuska et al., 2002; Wakao and Benning, 2005; Andre et al., 2007; Baud et al., 2007b; Wakao et al., 2008). Thus, in addition to the manipulation of key regulatory reactions such as ACCase, coregulation of other key genes is essential for an increased flux of the entire pathway. Together with previous studies of LEC2, WRI1, and FUS3, the results presented in this study suggested that manipulation of this coregulation mode is feasible by overexpression of LEC1 or L1L from both Arabidopsis and Brassica. In this regard, this class of genes is a promising target for genetic manipulations of oil-producing crops and plants to improve both the quantity and quality of lipids.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Col-0 and Landsberg erecta (Ler) accessions of Arabidopsis (Arabidopsis thaliana) were used in this study. Plants were grown under continuous white light at 22°C in soil or on Murashige and Skoog (MS) agar (half-strength MS salts, 3% Suc, and 0.8% agar) as described previously (Sun et al., 2003). Seeds were sterilized, washed extensively with water, and then sown on MS agar plates with or without estradiol. The plate was kept at 4°C for 2 to 3 d and then transferred to a tissue culture room (22°C and continuous white light). Treatment by estradiol prior to or after stratification (directly sowing seeds on estradiol-containing medium or adding estradiol onto medium immediately after stratification) did not have any apparent effects on the LEC1-OXi phenotype described in this study.
The abi3-5 mutant was in the Ler background (Ooms et al., 1993), and 35S-ABI3 transgenic lines were in the Col-0 background (Zhang et al., 2005). A fus3 knockout mutant (SALK_039162) was obtained from the Arabidopsis Biological Resource Center (Alonso et al., 2003). A wri1-3 knockout mutant was a gift of Dr. Loïc Lepiniec. The pER8-LEC1 abi3, pER8-LEC1 fus3, and pER8-LEC1 wri1 mutant plants were obtained by crossing of the transgenic lines (two lines were tested) with abi3-5, fus3, and wri1-3, respectively. The pER8-LEC1 construct was also used to transform abi3-5 mutant plants. Mutant plants homozygous for both pER8-LEC1 and abi3 loci were identified from F3 or T2 generations and used for subsequent experiments. Similar results were obtained from these two sets of materials. In the analysis of LEC1-OXi abi3 plants, a randomly collected F2 population obtained from a cross between Col-0 and Ler or DMSO-treated LEC1-OXi abi3 seedlings were used as controls.
Plasmid Construction and Generation of Transgenic Plants
The estradiol-inducible vectors (XVE vectors pER8 and pER10) have been described previously (Zuo et al., 2000). The two vectors are identical except that pER8 carries a hygromycin selection marker and pER10 carries a kanamycin selection marker. A LEC1 cDNA clone was obtained by reverse transcription (RT)-PCR and then cloned into pER8. BnLEC1 and BnL1L genomic clones were amplified by PCR and then cloned into pER10. All constructs were verified by extensive restriction digestion and DNA sequencing analysis. These constructs were transformed into Agrobacterium tumefaciens strain GV3101, which was then used for the transformation of Arabidopsis by vacuum infiltration as described (Bechtold et al., 1993).
Analysis of Fatty Acids and Lipids
Extraction and analysis of fatty acids were carried out essentially as described (Poirier et al., 1999). After extraction with 1 mL of hexane and 2 mL of 0.9% (w/v) NaCl, the fatty acyl methyl esters (the organic phase) were used for analysis by gas chromatography-mass spectrometry (GC-MS) using methyl heptadecanoate as an internal standard. GC-MS analysis was performed on a Perkin-Elmer Turbomass gas chromatograph/mass spectrometer equipped with a BPX-70 (30 m × 0.25 mm) chromatography column. The initial column temperature was 120°C, increasing at 10°C min−1 to 150°C. The column temperature was then raised at 4°C min−1 to 230°C and held for 10 min at the final temperature. After the run, peaks corresponding to each fatty acid species were identified by their characteristic retention times. Concentrations of each sample were normalized against the internal control.
Staining of seedlings with Fat Red 7B (Sigma China) was carried out essentially as described (Ogas et al., 1997). To detect oil bodies, seedlings were infiltrated with an aqueous solution of Nile Red (Molecular Probes) for 30 min at room temperature as described (Greenspan et al., 1985). After brief rinsing with distilled water, samples were visualized with a microscope (Olympus Bx51/Bx52).
Microarray Analysis
The pER8-LEC1 transgenic seedlings were germinated and grown in the presence of 10 μm estradiol for 4 d. The control sample was germinated and grown under identical conditions without estradiol but containing 0.1% DMSO. Total RNA was prepared from fresh or frozen plant materials using the RNeasy Plant Mini Kit (Qiagen China). The first-strand cDNA was synthesized and then hybridized with the ATH1 oligonucleotide chips as described by the manufacturer (Affymetrix).
The microarray hybridization data were collected and analyzed using related R (http://www.r-project.org/) packages provided by Bioconductor (http://www.biocoductor.org/). In brief, genes differentially expressed between wild-type and mutant plants were selected by first removing “absent” genes, which were never detected to be expressed in the experiments, then a two-sided t test was applied to the remaining genes in order to test the expression difference between wide-type and mutant plants. To avoid multiple testing problems, raw P values were then adjusted into false discovery rate (FDR) using the Benjamini and Hochberg approach (Benjamini and Hochberg, 1995). Finally, differentially expressed genes were defined as those with FDR less than 0.2. Functional analysis of differentially expressed genes was carried out using the biological process category of Arabidopsis Gene Ontology. The hierarchical map of Gene Ontology annotation was constructed according to the ontology tree provided by the Gene Ontology Web site (http://www.geneontology.com), as described previously (Zheng and Wang, 2008). Ontology categories that are significantly enriched among differentially expressed genes (hypergeometric test and FDR less than 0.1) were displayed as boxes in the map.
Analysis of Gene Expression by Northern Blot, RT-PCR, and RT-Quantitative PCR
Total RNA was prepared by the Plant RNeasy Prep Kit (Qiagen China and Qiagen Hong Kong; microarray experiments) or the Trizol reagent (Invitrogen; other experiments) according to the manufacturers' instructions. RNA Northern blotting, RT-quantitative PCR (qPCR), and semiquantitative RT-PCR analyses were carried out as described previously (Sun et al., 2003, 2005; Feng et al., 2006). UBIQUITIN5 (At3g62250) and ACTIN7 (At5g09810) were used as internal controls in RT-PCR and RT-qPCR, respectively. All primer pairs used in the RT-PCR and RT-qPCR analyses are listed in Supplemental Table S4.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EU371726 (BnLEC1), EU371727 (BnL1L), and GSE12137 (microarray data).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Correlation analysis of differentially expressed genes in two biological repeats of the microarray experiment.
Supplemental Figure S2. Biological process ontology map of differentially expressed genes between wild-type and LEC1-OXi plants.
Supplemental Figure S3. Amino acid sequence alignment of AtLEC1, AtL1L, BnLEC1, and BnL1L proteins.
Supplemental Table S1. List of up-regulated genes in LEC1-OXi plants.
Supplemental Table S2. List of down-regulated genes in LEC1-OXi plants.
Supplemental Table S3. Up-regulated fatty acid synthetic genes in LEC1-OXi plants.
Supplemental Table S4. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Drs. Loïc Lepiniec, Nam-Hai Chua, Ligeng Ma, and Zhizhong Gong for mutant seeds. We are grateful to Ms. Shanting Hao for advice and help on GC-MS analysis. We thank Dr. Yongbiao Xue for critically reading the manuscript.
This work was supported by grants from the Ministry of Science and Technology of China (grant nos. 2006CB101601, 2007CB948203, and 2007AA021402), the National Natural Science Foundation of China (grant nos. 30670196, 30600047, 30125025, and 30221002), and the Chinese Academy of Sciences (grant no. KSCX2–YW–N–015).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jianru Zuo (jrzuo@genetics.ac.cn).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Andre C, Benning C (2007) Arabidopsis seedlings deficient in a plastidic pyruvate kinase are unable to utilize seed storage compounds for germination and establishment. Plant Physiol 145 1670–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre C, Froehlich JE, Moll MR, Benning C (2007) A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell 19 2006–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40 151–160 [Google Scholar]
- Baud S, Guyon V, Kronenberger J, Wuilleme S, Miquel M, Caboche M, Lepiniec L, Rochat C (2003) Multifunctional acetyl-CoA carboxylase 1 is essential for very long chain fatty acid elongation and embryo development in Arabidopsis. Plant J 33 75–86 [DOI] [PubMed] [Google Scholar]
- Baud S, Mendoza MS, To A, Harscoet E, Lepiniec L, Dubreucq B (2007. a) WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J 50 825–838 [DOI] [PubMed] [Google Scholar]
- Baud S, Wuilleme S, Dubreucq B, de Almeida A, Vuagnat C, Lepiniec L, Miquel M, Rochat C (2007. b) Function of plastidial pyruvate kinases in seeds of Arabidopsis thaliana. Plant J 52 405–419 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Ser III Sci Vie 316 1194–1199 [Google Scholar]
- Beisson F, Koo AJK, Ruuska S, Schwender J, Pollard M, Thelen JJ, Paddock T, Salas JJ, Savage L, Milcamps A, et al (2003) Arabidopsis genes involved in acyl lipid metabolism: a 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a Web-based database. Plant Physiol 132 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57 289–300 [Google Scholar]
- Bonaventure G, Ohlrogge JB (2002) Differential regulation of mRNA levels of acyl carrier protein isoforms in Arabidopsis. Plant Physiol 128 223–235 [PMC free article] [PubMed] [Google Scholar]
- Brocard-Gifford IM, Lynch TJ, Finkelstein RR (2003) Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiol 131 78–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon EB, Shockey JM, Dietrich CR, Gidda SK, Mullen RT, Dyer JM (2007) Engineering oilseeds for sustainable production of industrial and nutritional feedstocks: solving bottlenecks in fatty acid flux. Curr Opin Plant Biol 10 236–244 [DOI] [PubMed] [Google Scholar]
- Cernac A, Andre C, Hoffmann-Benning S, Benning C (2006) WRI1 is required for seed germination and seedling establishment. Plant Physiol 141 745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac A, Benning C (2004) WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J 40 575–585 [DOI] [PubMed] [Google Scholar]
- Feng H, An F, Zhang S, Ji Z, Ling HQ, Zuo J (2006) Light-regulated, tissue-specific, and cell differentiation-specific expression of the Arabidopsis Fe(III)-chelate reductase gene AtFRO6. Plant Physiol 140 1345–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Somerville CR (1990) Three classes of abscisic acid (ABA)-insensitive mutations of Arabidopsis define genes that control overlapping subsets of ABA responses. Plant Physiol 94 1172–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focks N, Benning C (1998) wrinkled1: a novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol 118 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan P, Mayer EP, Fowler SD (1985) Nile Red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol 100 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood JL (1996) Recent advances in the biosynthesis of plant fatty acids. Biochim Biophys Acta 1301 7–56 [DOI] [PubMed] [Google Scholar]
- James DW Jr, Lim E, Keller J, Plooy I, Ralston E, Dooner HK (1995) Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon activator. Plant Cell 7 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski J, Cahoon EB (2003) Industrial oils from transgenic plants. Curr Opin Plant Biol 6 178–184 [DOI] [PubMed] [Google Scholar]
- Kachroo P, Shanklin J, Shah J, Whittle EJ, Klessig DF (2001) A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc Natl Acad Sci USA 98 9448–9453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Okuda R, Ban A, Toyoshima R, Tsutsumida K, Usui H, Yamamoto A, Hattori T (2005. a) Indirect ABA-dependent regulation of seed storage protein genes by FUSCA3 transcription factor in Arabidopsis. Plant Cell Physiol 46 300–311 [DOI] [PubMed] [Google Scholar]
- Kagaya Y, Toyoshima R, Okuda R, Usui H, Yamamoto A, Hattori T (2005. b) LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol 46 399–406 [DOI] [PubMed] [Google Scholar]
- Ke J, Wen TN, Nikolau BJ, Wurtele ES (2000) Coordinate regulation of the nuclear and plastidic genes coding for the subunits of the heteromeric acetyl-coenzyme A carboxylase. Plant Physiol 122 1057–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroj T, Savino G, Valon C, Giraudat J, Parcy F (2003) Regulation of storage protein gene expression in Arabidopsis. Development 130 6065–6073 [DOI] [PubMed] [Google Scholar]
- Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ (2003) LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93 1195–1205 [DOI] [PubMed] [Google Scholar]
- Luerssen H, Kirik V, Herrmann P, Misera S (1998) FUSCA3 encodes a protein with a conserved VP1/ABI3-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J 15 755–764 [DOI] [PubMed] [Google Scholar]
- Millar AA, Kunst L (1997) Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J 12 121–131 [DOI] [PubMed] [Google Scholar]
- Millar AA, Smith MA, Kunst L (2000) All fatty acids are not equal: discrimination in plant membrane lipids. Trends Plant Sci 5 95–101 [DOI] [PubMed] [Google Scholar]
- Mou Z, He Y, Dai Y, Liu X, Li J (2000) Deficiency in fatty acid synthase leads to premature cell death and dramatic alterations in plant morphology. Plant Cell 12 405–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas J, Cheng JC, Sung ZR, Somerville C (1997) Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277 91–94 [DOI] [PubMed] [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C (1999) PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci USA 96 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara P, Slabas AR, Fawcett T (2002) Fatty acid and lipid biosynthetic genes are expressed at constant molar ratios but different absolute levels during embryogenesis. Plant Physiol 129 310–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J (1995) Lipid biosynthesis. Plant Cell 7 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge JB, Jaworski JG (1997) Regulation of fatty acid synthesis. Annu Rev Plant Physiol Plant Mol Biol 48 109–136 [DOI] [PubMed] [Google Scholar]
- Ooms JJJ, Leon-Kloosterziel KM, Bartels D, Koornneef M, Karssen CM (1993) Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana (a comparative study using abscisic acid-insensitive abi3 mutants). Plant Physiol 102 1185–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Ventre G, Caldelari D (1999) Increased flow of fatty acids toward β-oxidation in developing seeds of Arabidopsis deficient in diacylglycerol acyltransferase activity or synthesizing medium-chain-length fatty acids. Plant Physiol 121 1359–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawsthorne S (2002) Carbon flux and fatty acid synthesis in plants. Prog Lipid Res 41 182–196 [DOI] [PubMed] [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Nagano Y (2004) Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci Biotechnol Biochem 68 1175–1184 [DOI] [PubMed] [Google Scholar]
- Schwender J, Ohlrogge JB, Shachar-Hill Y (2003) A flux model of glycolysis and the oxidative pentosephosphate pathway in developing Brassica napus embryos. J Biol Chem 278 29442–29453 [DOI] [PubMed] [Google Scholar]
- Slabas AR, Fawcett T (1992) The biochemistry and molecular biology of plant lipid biosynthesis. Plant Mol Biol 19 169–191 [DOI] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer R, Goldberg RB, Harada JJ (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Hirose N, Wang X, Wen P, Xue L, Sakakibara H, Zuo J (2005) Arabidopsis SOI33/AtENT8 gene encodes a putative equilibrative nucleoside transporter that is involved in cytokinin transport in planta. J Integr Plant Biol 47 588–603 [Google Scholar]
- Sun J, Niu QW, Tarkowski P, Zheng B, Tarkowska D, Sandberg G, Chua NH, Zuo J (2003) The Arabidopsis AtIPT8/PGA22 gene encodes an isopentenyl transferase that is involved in de novo cytokinin biosynthesis. Plant Physiol 131 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen JJ, Ohlrogge JB (2002) Metabolic engineering of fatty acid biosynthesis in plants. Metab Eng 4 12–21 [DOI] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F (2006) A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakao S, Andre C, Benning C (2008) Functional analyses of cytosolic glucose-6-phosphate dehydrogenases and their contribution to seed oil accumulation in Arabidopsis. Plant Physiol 146 277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakao S, Benning C (2005) Genome-wide analysis of glucose-6-phosphate dehydrogenases in Arabidopsis. Plant J 41 243–256 [DOI] [PubMed] [Google Scholar]
- Wang H, Guo J, Lambert K, Lin Y (2007) Developmental control of Arabidopsis seed oil biosynthesis. Planta 226 773–783 [DOI] [PubMed] [Google Scholar]
- Zhang X, Garreton V, Chua NH (2005) The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev 19 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Wang XJ (2008) GOEAST: a Web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res 36 W358–W363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH (2000) An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.