Abstract
Severe heat stress causes protein denaturation in various cellular compartments. If Saccharomyces cerevisiae cells grown at 24°C are preconditioned at 37°C, proteins denatured by subsequent exposure to 48–50°C can be renatured when the cells are allowed to recover at 24°C. Conformational repair of vital proteins is essential for survival, because gene expression is transiently blocked after the thermal insult. Refolding of cytoplasmic proteins requires the Hsp104 chaperone, and refolding of lumenal endoplasmic reticulum (ER) proteins requires the Hsp70 homologue Lhs1p. We show here that conformational repair of heat-damaged glycoproteins in the ER of living yeast cells required functional Hsp104. A heterologous enzyme and a number of natural yeast proteins, previously translocated and folded in the ER and thereafter denatured by severe heat stress, failed to be refolded to active and secretion-competent structures in the absence of Hsp104 or when an ATP-binding site of Hsp104 was mutated. During recovery at 24°C, the misfolded proteins persisted in the ER, although the secretory apparatus was fully functional. Hsp104 appears to control conformational repair of heat-damaged proteins even beyond the ER membrane.
INTRODUCTION
All organisms acquire tolerance toward otherwise lethal high temperatures if they are first preconditioned at a moderately high temperature at which their heat shock genes are activated (Parsell and Lindquist, 1993). In Saccharomyces cerevisiae, the heat shock protein Hsp104 is indispensable for acquisition of thermotolerance (Sanchez and Lindquist, 1990; Sanchez et al., 1992). Hsp104 belongs to the HSP100/Clp family of ATPases and has two essential nucleotide-binding sites (Parsell et al., 1991; Schirmer et al., 1996). It promotes survival of yeast cells exposed to 48–50°C after preconditioning at 37°C by disaggregating heat-denatured proteins in the cytosol (Parsell et al., 1994). Recently, Hsp104 was shown to be able to modulate the conformational status of the cytoplasmic yeast prion protein (Chernoff et al., 1995; Schirmer and Lindquist, 1997).
The structure of endoplasmic reticulum (ER)-located proteins distorted by severe heat stress can be repaired by an ATP-dependent mechanism (Jämsäet al., 1995). According to the yeast genome sequence, there is no Hsp104 homologue in the ER; thus, the repair process in the ER must depend on other chaperones. Indeed, we found that the Lhs1 protein is involved in conformational repair in the yeast ER (Saris et al., 1997). Lhs1p belongs to the Hsp110 subgroup of the Hsp70s and shares 24% identical amino acids with the ER-resident Hsp70 chaperone BiP/Kar2p (Rasmussen, 1994; Craven et al., 1997), which functions in ER translocation and folding of newly synthesized polypeptides (Vogel et al., 1990; Simons et al., 1995; Holkeri et al., 1998). Under physiological conditions, Lhs1p assists efficient ER translocation of a subset of proteins, especially at low temperatures (Baxter et al., 1996; Craven et al., 1996; Hamilton and Flynn, 1996). In the absence of Lhs1p, previously heat-denatured proteins were not refolded but degraded. In normal cells, Lhs1p was found in association with heat-affected but not native reporter proteins, according to coimmunoprecipitation experiments (Saris et al., 1997; Saris and Makarow, 1998). Mammalian cells harbor an Lhs1p homologue in the ER (Lin et al., 1993; Chen et al., 1996). In vitro experiments suggest that it is involved in translocation (Dierks et al., 1996), whereas no data are available regarding a role in conformational repair. Also, Lhs1p, like Hsp104, is dispensable at physiological temperature but is necessary for the acquisition of thermotolerance (Sanchez and Lindquist, 1990; Saris et al., 1997). Lhs1p enhances survival after severe heat stress by 10-fold, and Hsp104 enhances survival by as much as 1000-fold. Unlike other chaperones, they do not function in de novo protein folding or suppression of aggregation, but they seem to be specialized in repair functions after stress. Hsp104 directly disaggregates denatured proteins (Glover and Lindquist, 1998), whereas the nature of Lhs1p function is not known. Here we show that refolding of fully translocated and folded, and thereafter heat-denatured, artificial and natural secretory yeast proteins in the S. cerevisiae ER requires functional Hsp104.
MATERIALS AND METHODS
Strains and Media
The following yeast strains were used in this study: H1 (Mata ade2-101 ura3-52 leu2-3 leu2-112 suc2Δ9 gal2), H4 (Mata sec18-1 ura3-52 trp1-289 leu2-3,122), H393 (Matα sec18-1 ura3-52 trp1-289 leu2-3 112 URA3::HSP150Δ-β-lactamase) (Simonen et al., 1994), H453 (Mata ura3-1 his3-11,15 leu2-3,112 trp1-2 ade2-1 can1-100 hsp104::LEU2) (Sanchez and Lindquist, 1990), and H534 (Matα sec18-1 ade2-1 his3-11,15 trp1 hsp104::LEU2 URA3::HSP150Δ-β-lactamase) (Saris et al., 1997). Strains H826 (Mata ura3-1 his3-11,15 leu2-3,112 trp1-2 ade 2-1 can1-100 hsp104::LEU2 URA3::HSP104) and H836 (Mata sec18-1 ura3-52 trp1-289 leu2-3,112 hsp104::Kan-Cre-Lox URA3::HSP104) were created by integrating the HSP104 gene in plasmid pKTH4681 (see below) into the genome of strains H453 and H835, respectively. Strain H835 (Mata sec18-1 ura3-52 trp1-289 leu2-3,112 hsp104::Kan-Cre-Lox) was obtained by transformation of strain H4 with an hsp104 disruption cassette (see below). Strains H835 and H836 were transformed with integrative plasmid pKTH4660 containing the Hsp150Δ-β-lactamase gene (Paunola et al., 1998) to create strains H850 (Mata sec18-1 ura3-52 trp1-289 leu2-3,112 hsp104::Kan-Cre-Lox URA3::HSP104 LEU2::HSP150Δ-β-lactamase) and H851 (Mata sec18-1 ura3-52 trp1-289 leu2-3,112 hsp104::Kan-Cre-Lox LEU2::HSP150Δ-β-lactamase), respectively.
Strains H924 (Mata ura3-1 his3-11 leu2-3,112 trp1-2 ade2-1 can1-100 hsp104::LEU2 URA3::HSP104-K218T) and H941 (Mata sec18-1 ura3-52 trp1-289 leu2-3,112 hsp104::Kan-Cre-Lox LEU2::HSP150Δ-β-lactamase URA3::HSP104-K218T) were created by integrating plasmid pKTH4720 (see below) into yeast strains H453 and H851, respectively. Transformations were done with the lithium acetate method (Hill et al., 1991). Yeast strains were grown at 24°C in shake flasks overnight to early logarithmic phase in YPD medium or in synthetic complete (SC) medium lacking methionine and cysteine. Escherichia coli strain DH5α (Sambrook et al., 1989), used as the host for cloning of the HSP104 gene, was grown on Luria-Bertani medium supplemented with ampicillin (100 μg/ml).
Cloning, Deletion, and Rescue of HSP104
To clone the HSP104 gene, a 3327-base pair (bp) PCR fragment containing the whole coding region as well as 600 bp upstream from the start codon was amplified from cosmid 1F17 with primers 71526 (5′ATTGTCATCGATTCAAAGGCG3′) and 71527 (5′CATCAGACTAGTTAATCTAGGTCATCATC3′). The PCR product was digested with ClaI and SpeI (Promega, Madison, WI) and ligated to pKTH4685 cut with the same enzymes to produce plasmid pKTH4780. The cloning vector pKTH4685 was constructed by cloning the ADC1 transcription terminator from pAAH5 (Ammerer, 1983) as a 450-bp HindIII-BamHI fragment to the NotI site of pBluescript SK+ plasmid (Stratagene, La Jolla, CA). Standard cloning and transformation methods according to Sambrook et al. (1989) were used. Plasmid pKTH4681 was constructed by first cutting pKTH4680 with SacI and SalI, then isolating the 3.8-kilobase fragment containing the HSP104 gene and the ADC1 terminator, and finally ligating the fragment with pFL34 (Bonneaud et al., 1991) cut with the same enzymes. Before integration into yeast strain H453 to construct rescue strain H826, pKTH4681 was linearized with NcoI. Deletion of the HSP104 gene from the yeast genome was performed with the use of the loxP-kanMX-loxP disruption cassette method (Guldener et al., 1996). The disruption cassette was amplified from plasmid pUG6 (Guldener et al., 1996) with PCR primers 73148 (5′CAACTACACGTACCATAAAATATACAGAATATATGAACG-ACAGCTGAAGCTTCGTACGC3′) and 73149 (5′CTTGTTCGAAAG-TTTTTAAAAATCACACTATATTAAATTAGCATAGGCCACTA-GTGGATCTG3′) containing 40-bp homologous stretches (boldface nucleotides) upstream and downstream, respectively, of HSP104 in the yeast genome. Western analysis, performed as described previously (Saris et al., 1997) with the use of anti-Hsp104 antiserum (1:2000) (Stressgen, Victoria, British Columbia, Canada), confirmed the absence and presence of Hsp104.
Site-directed mutagenesis of Hsp104 was performed with the use of PCR. Primers 82007 (5′CCAGGTATCGGTACGACCGC3′) and 82008 (5′GCGGTCGTACCGATACCTGG3′), hybridizing to opposite DNA strands, were used to introduce mutation K218T into HSP104. The 5′ end of HSP104 was amplified with primers 71526 and 82007, and the 3′ terminus was amplified with primers 82008 and 71527 with pKTH4680 as a template. The full-length mutant gene was generated by amplifying the hybridized 5′ and 3′ terminal fragments with primers 71526 and 71527. The PCR fragment was cloned into pKTH4685 like the wild-type gene (see above). The mutation was verified from the resulting plasmid pKTH4719 by sequencing. The HSP104-K218T expression plasmid pKTH4720 was constructed and integrated into the yeast genome like plasmid pKTH4681.
Metabolic Labeling and Immunoprecipitation
For metabolic labeling with [35S]methionine/cysteine (1000 Ci/mmol; Amersham Pharmacia Biotech UK, Little Chalfont, England), the yeast cells were grown in SC medium lacking methionine and cysteine and labeled in the same medium (25 × 106 cells/0.5 ml). Immunoprecipitation of lysed cell samples or medium samples was performed with β-lactamase antiserum (1:100) or carboxypeptidase Y antiserum (1:100) and protein A–Sepharose for 2 h at 4°C, as described previously (Saris et al., 1997). Protein-bound glycans were labeled with d-[2-3H]mannose (10.3 Ci/mmol; Amersham Pharmacia Biotech UK) in YPD medium containing 0.1% glucose. Labeling was stopped by reconstituting the glucose concentration to 4%. The cell wall was removed by zymolyase digestion, and the spheroplasts were lysed in 0.1% SDS. To determine protein-bound 3H radioactivity, the radioactivity incorporated in the presence of 100 μg/ml cycloheximide was subtracted from the total incorporated radioactivity.
Other Materials and Methods
For thermotolerance assays, cells were preincubated in Wassermann tubes for 1 h at 37°C before shifting them to 50°C. Duplicate samples of 2 × 106 cells were removed after 20 min, diluted, and plated onto YPD plates. The colonies were counted after 4 d of incubation at 24°C (Sanchez and Lindquist, 1990; Saris et al., 1997). Determination of the β-lactamase activity was performed according to Simonen et al. (1994), and the Km values were determined according to Paunola et al. (1998). Glucose consumption by the yeast cells was followed by determining the glucose concentration in the medium of duplicate cell samples with the use of the Gluco-quant kit of Boehringer Mannheim (Indianapolis, IN).
RESULTS
Heat-denatured Hsp150Δ-β-Lactamase Fails to Be Refolded in the ER in the Absence of Hsp104
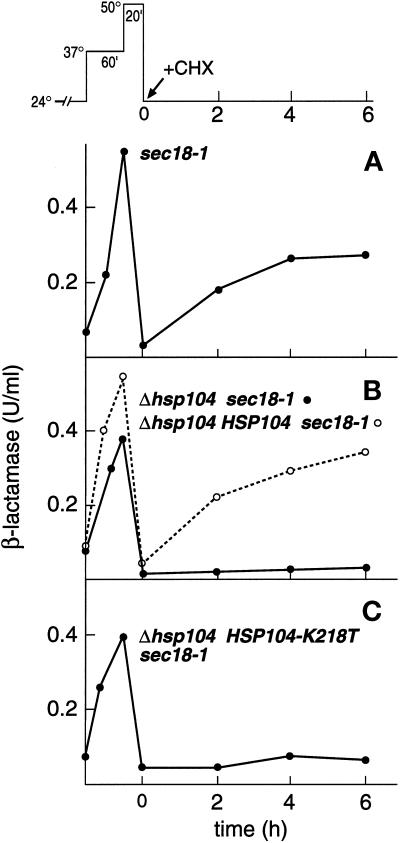
We have shown previously that severe heat shock at 48–50°C, after preconditioning at 37°C, resulted in aggregation, inactivation, and loss of secretion competence of an ER-confined reporter enzyme, Hsp150Δ-β-lactamase. Once the cells were returned to physiological temperature (24°C), the aggregates were solubilized and the enzymatic activity and secretion competence of the reporter enzyme were resumed (Jämsäet al., 1995; Saris et al., 1997; Saris and Makarow, 1998) (see top of Figure 1 for thermal treatments of cells). Here we show first for reference the reactivation of heat-inactivated Hsp150Δ-β-lactamase in sec18-1 cells (H393; see Table 1 for strains). The cells were preincubated at 37°C to condition them to survive a subsequent thermal insult and to accumulate β-lactamase activity in the ER (Figure 1A). The temperature-sensitive sec18-1 mutation inhibits transport of exocytic proteins from the ER to the Golgi at nonpermissive temperature (37°C) (Novick et al., 1981). A 20-min incubation at 50°C resulted in inactivation of the reporter enzyme. When the cells were returned to 24°C, 51% of the original β-lactamase activity was recovered in 4–6 h, even though protein synthesis was inhibited by cycloheximide (CHX) (Figure 1A).
Figure 1.
Dependence of reactivation of heat-inactivated Hsp150Δ-β-lactamase on Hsp104. (Top) Scheme of thermal treatments. After growth at 24°C, cells were incubated at 37°C (preconditioning), thereafter at 50°C (thermal insult), and then at 24°C (recovery). Strains H393 (A), H534 (B, ●), H850 (B, ○), and H941 (C) were preincubated for 10 min at 37°C, pelleted, resuspended in prewarmed medium, and incubated at 37°C for 1 h and then at 50°C for 20 min, after which CHX was added and the cells remained at 24°C. The β-lactamase activity of lysed cell samples was determined and plotted against incubation time.
Table 1.
Relevant features of yeast strains
| Designation | Relevant mutation | Artificial reporter | Source |
|---|---|---|---|
| H1 | None | ||
| H4 | sec18-1 | Novick et al., 1981 | |
| H393 | sec18-1 | Hsp150Δ-β-lactamase | Simonen et al., 1994 |
| H453 | Δhsp104a | Sanchez and Lindquist, 1990 | |
| H534 | Δhsp104asec18-1 | Hsp150Δ-β-lactamase | Saris and Makarow, 1998 |
| H826 | Δhsp104aHSP104b | This work | |
| H835 | Δhsp104asec18-1 | This work | |
| H836 | Δhsp104aHSP104bsec18-1 | This work | |
| H850 | Δhsp104aHSP104bsec18-1 | Hsp150Δ-β-lactamase | This work |
| H851 | Δhsp104asec18-1 | Hsp150Δ-β-lactamase | This work |
| H924 | Δhsp104 HSP104-K218T | This work | |
| H941 | Δhsp104 HSP104-K218T sec18-1 | Hsp150Δ-β-lactamase | This work |
See MATERIALS AND METHODS for complete genotypes.
Different disruption strategies were used; see MATERIALS AND METHODS.
HSP104 rescue strain.
To study the role of Hsp104 in the refolding events in the ER, the reactivation experiment was repeated with a strain from which the HSP104 gene had been deleted (Δhsp104 sec18-1). In the absence of Hsp104, β-lactamase activity accumulated in the ER at 37°C and was inactivated at 50°C, as described above (Figure 1B, ●). Pulse-chase experiments confirmed that Hsp150Δ-β-lactamase was translocated into the ER in Δhsp104 mutants as in normal cells (see below). However, after the cells were shifted to 24°C, very little of the catalytic activity was resumed (Figure 1B, ●). When the wild-type HSP104 gene was returned to the genome of Δhsp104 sec18-1 cells, reactivation of the heat-denatured reporter enzyme was rescued (Figure 1B, ○). Mutation of lysine 218 to threonine of one of the two nucleotide-binding sites of Hsp104 destroys its ability to confer thermotolerance to cells (Parsell et al., 1991). To determine whether nucleotide binding was required for the effect of Hsp104 on the refolding events inside of the ER, we replaced the wild-type HSP104 gene with an HSP104-K218T variant. Very little heat-denatured β-lactamase activity was recovered in this mutant (Figure 1C). In none of the strains was β-lactamase activity secreted to the medium during the 6-h recovery period (data not shown). We conclude that functional Hsp104 was required for reactivation of heat-inactivated Hsp150Δ-β-lactamase inside of the ER.
Metabolic Activity of Δhsp104 Cells after Thermal Insult
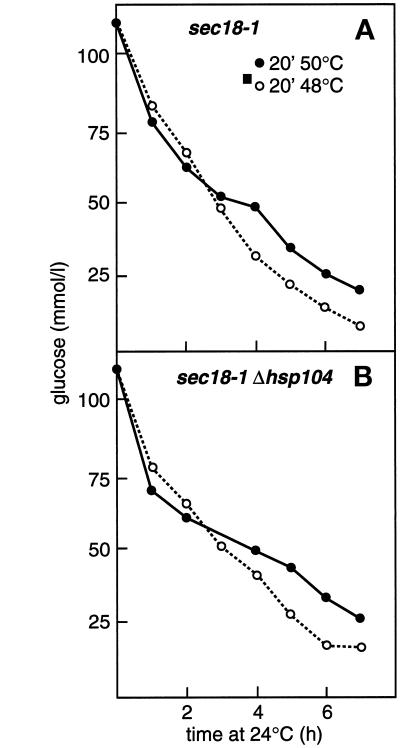
Most normal cells, when preincubated at 37°C for 30–60 min followed by 20–30 min at 48–50°C, form colonies in 3–4 d when plated at 24°C, whereas in the absence of Hsp104, only 0.1–1% survive (Sanchez and Lindquist, 1990). In our hands, 0.4% of Δhsp104 cells (H453), 0.2% of HSP104-K218T cells (H924), and 40% of the HSP104 rescue cells (H826) acquired thermotolerance. Thus, it was important to establish that the Δhsp104 deletion strains remained metabolically active during the experiments described above. This was studied by following their rate of glucose consumption. sec18-1 (Figure 2A) and Δhsp104 sec18-1 mutants (Figure 2B) consumed glucose from the medium equally vigorously after preconditioning at 37°C and subsequent thermal insult at 48°C (○) or 50°C (●). In this experiment, we used a fivefold higher cell density than normal to increase the sensitivity of the assay. Therefore, in none of the other experiments did the glucose concentration of the medium become limiting. We conclude that in the absence of Hsp104 metabolic activity was not compromised for at least 7 h after severe heat stress; thus, lack of reactivation of Hsp150Δ-β-lactamase in the experiment described above was not due to abrupt cell death.
Figure 2.
Glucose consumption of cells after thermal insult in the absence and presence of Hsp104. H393 (A) and H534 (B) cells were resuspended in YPD medium (25 × 107 cells/ml) and incubated for 1 h at 37°C and then for 20 min at 48°C (○) or 50°C (●). The cells were pelleted and resuspended in fresh YPD medium and shifted to 24°C. Duplicate samples were withdrawn at the indicated times, and the glucose concentration of the medium was determined and plotted against incubation time at 24°C.
Resumption of Secretion Competence of Heat-denatured Hsp150Δ-β-Lactamase
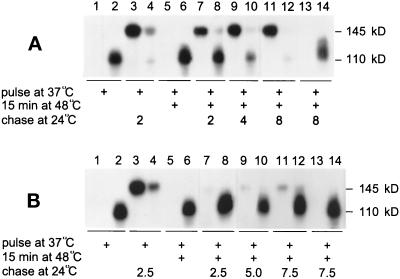
In sec18-1 cells, the heat-denatured β-lactamase fusion protein gradually becomes secretion competent during recovery of the cells at 24°C (Saris and Makarow, 1998). Next we studied whether the lack of Hsp104 affected resumption of the secretion competence of Hsp150Δ-β-lactamase. The experiment was first performed on sec18-1 cells for reference. To avoid excessively long recovery periods, a less severe thermal insult (15 min at 48°C) was applied. The cells were first labeled at 37°C with [35S]methionine/cysteine, after which the medium and lysed cells were subjected to immunoprecipitation. SDS-PAGE analysis revealed the ER form (110 kDa) of the reporter in the lysate (Figure 3A, lane 2) and no protein in the medium (lane 1). The ER form carries single mannose residues on serine and threonine residues, whereas the mature form has extended O-glycans (Paunola et al., 1998). When similarly labeled cells were shifted directly to 24°C for 2 h without any thermal insult, most of the protein was detected in the medium in fully O-glycosylated form (145 kDa; lane 3) and very little was found in the lysate (lane 4). When parallel cells labeled at 37°C underwent the 48°C treatment, the ER form still persisted in the cells (lane 6) and no protein was found in the medium (lane 5). After a shift of parallel cells from 48 to 24°C, less than half of the molecules were secreted after 2 h (lanes 7 and 8). After 4 h, most of the Hsp150Δ-β-lactamase was in the medium (lanes 9 and 10), and after 8 h, all of the Hsp150Δ-β-lactamase was in the medium (lanes 11 and 12). Sodium azide in the recovery mixture inhibited the appearance of the reporter in the medium (lanes 13 and 14). This experiment was repeated in Δhsp104 sec18-1 cells. This time, metabolically labeled Hsp150Δ-β-lactamase persisted in the ER form for at least 7.5 h (Figure 3B, lanes 8, 10, and 12) and very little was secreted (lanes 7, 9, and 11). The secretory apparatus of Δhsp104 cells was functional during the experiment (see below). Thus, Hsp104 was required not only for reactivation but also for resumption of the secretion competence of heat-denatured Hsp150Δ-β-lactamase.
Figure 3.
The role of Hsp104 in the resumption of the secretion competence of Hsp150Δ-β-lactamase. sec18-1 (A; H393) and Δhsp104 sec18-1 (B; H534) cells were preincubated for 10 min at 37°C, labeled with [35S]methionine/cysteine for 60 min at the same temperature, and incubated for 15 min at 48°C. The labeling medium was replaced by SC medium with excess unlabeled methionine and cysteine, and the cells were shifted to 24°C for the indicated times (lanes 1, 2, and 5–12). In lanes 3 and 4, the 48°C treatment was omitted. In lanes 13 and 14, sodium azide was included in the recovery mixture. The culture medium samples (lanes with uneven numbers) and lysed cell samples (lanes with even numbers) were immunoprecipitated with β-lactamase antiserum and analyzed by SDS-PAGE. Mature (145 kDa) and ER-specific (110 kDa) Hsp150Δ-β-lactamase are indicated.
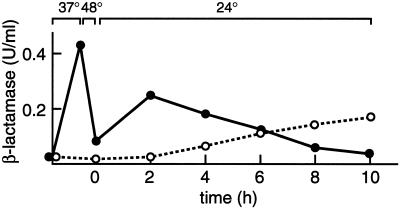
To establish a relationship between the folding state and the secretion competence of Hsp150Δ-β-lactamase, the protein was again accumulated in the ER in sec18-1 cells at 37°C and inactivated by a thermal insult of 20 min at 48°C. CHX was added, and the cells were shifted to 24°C. During the first hours of recovery, about half of the initial catalytic activity was resumed, but it remained intracellular (Figure 4, ●). Only after reactivation did the activity start to be slowly secreted to the medium (○). The Km value for nitrocefin of the reactivated and thereafter secreted Hsp150Δ-β-lactamase molecules was 44 μM, and that of molecules secreted normally at 24°C in the absence of any thermal treatments was 47 μM. The Km value of authentic E. coli β-lactamase is 49 μM (Paunola et al., 1998). Thus, the structural features essential for catalytic activity, which had been destroyed by the thermal insult, were reestablished during the recovery period. Disulfide bonds were not reduced by the thermal insult (data not shown). Resumption of the secretion competence of the β-lactamase activity shown in Figure 4 was slower than that shown in Figure 3A, apparently because of the more severe thermal insult. The very slow rate of secretion must have been due to slow refolding, because the secretion kinetics of copies synthesized during the recovery period were normal (see below).
Figure 4.
Resumption of the secretion competence of β-lactamase activity. sec18-1 cells (H393) were treated as described in the scheme at the top of Figure 1, except that the thermal insult was for 20 min at 48°C. The β-lactamase activity of cell lysates (●) and medium samples (○) was determined and plotted against time.
Resumption of Exocytosis after Severe Heat Stress in the Absence of Hsp104
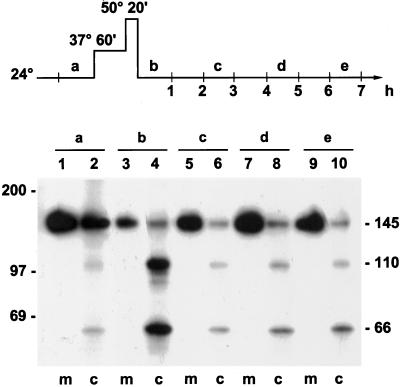
The experiment described above (Figure 3B) on the secretion competence of Hsp150Δ-β-lactamase in the absence of Hsp104 was performed in a sec18-1 mutant to accumulate the reporter enzyme in the ER before the thermal insult. Irreversible ER retention could thus have resulted in irreversible misfolding of the temperature-sensitive Sec18 protein, leading to cessation of membrane traffic. This is why we needed to confirm that exocytosis operated in heat-treated sec18-1 cells even in the absence of Hsp104. Parallel Δhsp104 sec18-1 cell samples were incubated at 24, 37, and 50°C, and then again at 24°C, and labeled during a 1-h period at 24°C before the shift to 37°C, as well as during the indicated 1-h periods during recovery at 24°C (scheme at top of Figure 5). The lysed cell samples were subjected to immunoprecipitation followed by SDS-PAGE analysis. At 24°C before heat treatment, most of the newly synthesized reporter protein molecules were detected in the medium in the fully glycosylated 145-kDa form (Figure 5, sample a, lane 1). Some molecules remained cell associated in the cytoplasmic (66 kDa; Paunola et al., 1998), the ER (110 kDa), and the mature forms (lane 2). When the labeling was performed after the thermal insult during the first hour of recovery, some of the cell-associated molecules were cytoplasmic, some represented the ER form, and some were mature (sample b, lane 4). A few molecules were detected in the medium (lane 3). This suggests that translocation and ER exit were retarded immediately after the thermal insult. However, during the third, fifth, and even seventh hours of recovery (samples c, d, and e, respectively), very little of the protein was detected in the cell lysates (lanes 6, 8, and 10), whereas most of it was detected in the medium (lanes 5, 7, and 9). The Hsp150Δ-β-lactamase molecules synthesized during recovery at 24°C must have been disulfide bonded, because in reduced form the protein fails to leave the ER (Simonen et al., 1994). Similar data were obtained for normal cells that lacked sec mutations and expressed normal Hsp104 (H335; data not shown). Expression of the Hsp150Δ-β-lactamase was driven by the HSP150 promoter, which confers expression at 24°C but is up-regulated at 37°C (Russo et al., 1993).
Figure 5.
Recovery of synthesis and modification and secretion of Hsp150Δ-β-lactamase in the absence of Hsp104. The experimental design is shown in the scheme at the top of the figure. Parallel samples of Δhsp104 sec18-1 cells (H534) were labeled with 35S during successive 1-h periods at 24°C (samples a–e), lysed, immunoprecipitated with β-lactamase antiserum, and analyzed by SDS-PAGE. The cytoplasmic (66 kDa), ER-specific (110 kDa), and mature (145 kDa) forms of Hsp150Δ-β-lactamase and the molecular mass markers are indicated. m, medium; c, cell lysate.
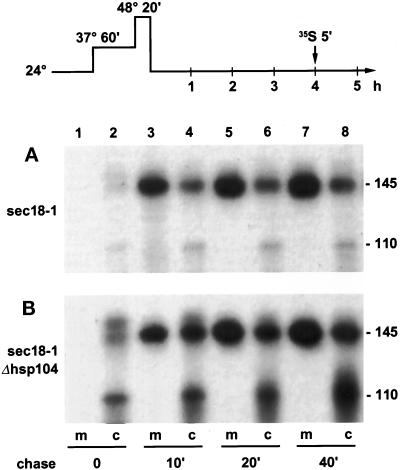
To examine more closely the secretion kinetics of newly synthesized Hsp150Δ-β-lactamase molecules, we incubated sec18-1 and Δhsp104 sec18-1 cells at 37 and 50°C and then for 4 h at 24°C. The cells were then labeled with 35S for 5 min and chased with CHX at 24°C (scheme at top of Figure 6). Immunoprecipitation with β-lactamase antiserum showed that most of the Hsp150Δ-β-lactamase was in the medium after a chase of 10 min in the case of sec18-1 cells (Figure 6A). In the case of Δhsp104 sec18-1 cells, most of the protein was in the medium after a chase of 20 min (Figure 6B, lanes 3 and 4), and even after 40 min of chase some of the reporter protein appeared to reside in the ER (lanes 7 and 8). In both strains, the intracellular forms of Hsp150Δ-β-lactamase were barely visible right after the pulse (Figure 6, A and B, lanes 2). This is probably due to the glycan heterogeneity of the intracellular forms. We conclude that synthesis, translocation, disulfide bonding, and O-glycosylation, as well as exocytosis of the reporter, functioned in most cells for at least 7 h of recovery, irrespective of the presence of Hsp104 and the sec18-1 mutation.
Figure 6.
Secretion kinetics of newly synthesized Hsp150Δ-β-lactamase. H393 (A) and H534 (B) cells were treated as indicated in the scheme at the top of the figure. After 4 h of recovery at 24°C, the cells were labeled with 35S for 5 min and then chased with CHX at 24°C for the indicated times. The medium (m) and cell lysate (c) samples were subjected to precipitation with β-lactamase antiserum and analyzed by SDS-PAGE. Mature (145 kDa) and ER-specific (110 kDa) Hsp150Δ-β-lactamase are indicated.
Heat-affected Carboxypeptidase Y Fails to Acquire Secretion Competence without Functional Hsp104
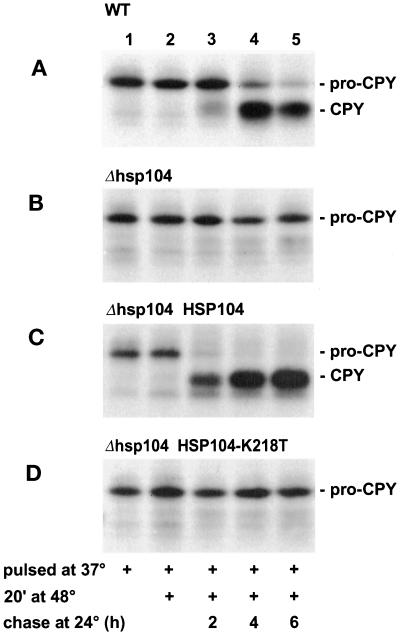
Next we expanded the experiments to a natural yeast glycoprotein, the vacuolar protease carboxypeptidase Y (CPY). The secretion competence of proCPY has been widely used as a measure of its folding state (Simons et al., 1995; Silberstein et al., 1998). Normally, preproCPY loses its signal peptide and acquires in the ER primary N-glycans, which are extended in the Golgi. The propeptide is removed in the vacuole, yielding mature CPY. Thus, the biosynthetic state of CPY reveals its intracellular location (Stevens et al., 1982). In the experiments described below, we used cells lacking sec mutations. Wild-type cells were first incubated at 37°C for 1 h and then labeled for 5 min at 37°C (Figure 7A) to serve as a control. Immunoprecipitation of the cell lysates and SDS-PAGE analysis revealed mostly ER-specific proCPY (67 kDa) (lane 1). Similarly labeled parallel cells were shifted for 20 min to 48°C and then chased at 24°C. After the thermal insult at 48°C, proCPY persisted (lane 2), and after 2 h at 24°C, a little proCPY had been processed to mature CPY (lane 3). After 4 h (lane 4) and 6 h (lane 5) at 24°C, most of the protein was in the mature form and thus apparently in the vacuole. When the thermal insult was omitted, proCPY was in the vacuole in <30 min (Saris and Makarow, 1998). In the Δhsp104 strain, proCPY was detected after the 37°C pulse as described above (Figure 7B, lane 1). However, after the thermal insult and 2–6 h at 24°C, proCPY persisted (lanes 2–5), showing that it had not acquired transport competence but remained in the ER. In the Δhsp104 HSP104 rescue strain, heat-affected proCPY reached the vacuole with kinetics similar to those in normal cells (Figure 7C). In cells expressing Hsp104-K218T instead of Hsp104, proCPY persisted in the ER form (Figure 7D). These data confirm that permanent ER retention in the absence of Hsp104 was not caused by irreversible conformational distortion of the mutant Sec18 protein, or of other components required for intracellular transport, but by failure of the reporter proteins to gain a secretion-competent conformation.
Figure 7.
The role of Hsp104 in the resumption of the secretion competence of proCPY. H1 (A), H453 (B), H826 (C), and H924 (D) cells were preincubated for 1 h and then pulse labeled for 5 min with [35S]methionine/cysteine at 37°C (lanes 1). Parallel cells remained at 48°C for 20 min, after which the labeling medium was changed to SC medium with excess unlabeled methionine and cysteine and the cells were shifted to 24°C for 2 h (lanes 2), 4 h (lanes 3), or 6 h (lanes 4). The cells were lysed and subjected to immunoprecipitation with CPY antiserum followed by SDS-PAGE analysis. ProCPY (67 kDa) and CPY(61 kDa) are indicated.
Hsp104 Is Required for Resumption of Secretion Competence of Bulk Cell Wall Proteins
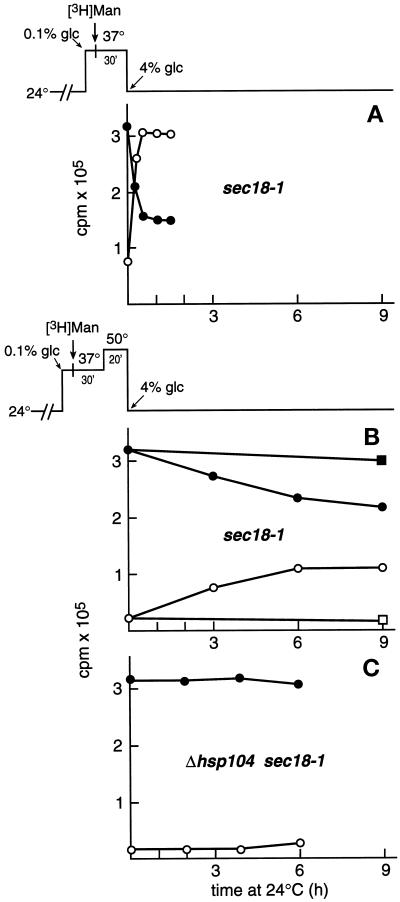
Finally, to examine the physiological relevance of Hsp104 in refolding events in the ER, we extended the studies to a number of authentic cell wall mannoproteins. The S. cerevisiae cell wall is composed of a glucan polymer and numerous extensively N- and O-glycosylated mannoproteins. The fate of heat-affected newly synthesized bulk mannoproteins was studied in the absence and presence of Hsp104. First we established the assay. sec18-1 cells were preincubated for 15 min at 37°C and labeled for 30 min at 37°C with [3H]mannose in low-glucose medium (0.1%) to achieve efficient incorporation of radioactivity (scheme at top of Figure 8A). The glucose concentration of the medium was then increased to 4% to stop labeling, and the cells were shifted to 24°C to reverse the sec18-1 block. Samples were withdrawn after different times, the cell walls were removed by zymolyase digestion, and the 3H radioactivity of the lysed spheroplasts (●) and the cell wall material (○) was counted. Most of the radioactivity had reached the cell wall <1 h after the shift of the cells to 24°C (Figure 8A), thus representing cell wall mannoproteins. The remaining spheroplast-associated radioactivity evidently represented resident glycoproteins of the secretory compartment. Negligible amounts of radioactivity were detected in the medium (data not shown). Next, similarly 3H-labeled sec18-1 cells were treated for 20 min at 50°C before being shifted to 24°C (scheme at top of Figure 8B). The spheroplast-associated radioactivity decreased very slowly (Figure 8B, ●), with a concomitant increase in cell wall–associated radioactivity (○). The addition of sodium azide to the recovery mixture inhibited transport of the radioactivity to the cell wall (▪ and □). When the experiment was repeated on Δhsp104 sec18-1 cells, the 3H radioactivity remained intracellular (Figure 8C). This finding demonstrates that loss of transport competence as a result of heat-inflicted damage and subsequent Hsp104-dependent refolding apparently affected a large number of different cell wall glycoproteins and was thus a physiologically relevant phenomenon.
Figure 8.
The role of Hsp104 in the fate of heat-affected cell wall mannoproteins. Strains H4 (A and B) and H534 (C) were preincubated in YPD medium containing 0.1% glucose for 15 min at 37°C and labeled with [3H]mannose for 30 min. (A) The cells were pelleted, washed, and resuspended in fresh YPD medium containing 4% glucose and returned to 24°C (see scheme at top). (B and C) The 3H-labeled cell samples were incubated for 20 min at 50°C before being shifted to 24°C (see scheme at top). Duplicate samples were withdrawn, washed, and subjected to cell wall removal. The spheroplast-associated (●) and cell wall–associated (○) 3H radioactivity was counted and plotted against incubation time at 24°C.
DISCUSSION
We show here that repair of heat-damaged glycoproteins inside of the yeast ER depends on the cytoplasmic chaperone Hsp104. Fully translocated and folded, and thereafter in vivo heat denatured, Hsp150Δ-β-lactamase failed to be refolded at physiological temperature to an active and secretion-competent form in the absence of Hsp104 and remained in the ER. Similar results were obtained when one of the two ATP-binding sites of Hsp104 was inactivated by site-directed mutagenesis. In control cells harboring an intact HSP104 gene, as well as in rescue strains in which the wild-type HSP104 gene was returned to a Δhsp104 mutant, Hsp150Δ-β-lactamase gained back its catalytic activity and was thereafter slowly secreted to the medium. The Km value for nitrocefin of the secreted molecules was the same as that of authentic E. coli β-lactamase. This result shows that the structural features essential for catalytic activity of the heat-inactivated molecules were slowly reestablished. The crystal structure of E. coli TEM1 β-lactamase is a tight globule with a disulfide bond close to the active site on the interior of the molecule (Jelsch et al., 1992). The role of Hsp104 in refolding events in the ER must be physiologically relevant, because it controlled the conformational repair of a number of authentic yeast cell wall mannoproteins as well as vacuolar CPY. The conformational status of the yeast glycoproteins was monitored by following their secretion competence, as in studies in which misfolding was brought about by drugs or mutant ER chaperones instead of heat (Simons et al., 1995; Silberstein et al., 1998).
Although the Δhsp104 deletion strains cannot form colonies after severe heat stress, they remained viable for a long time, enabling us to study their physiology after severe heat stress. After preconditioned Δhsp104 cells were shifted from 48–50°C back to physiological temperature (24°C), they were able to synthesize, translocate, disulfide bond, glycosylate, and secrete Hsp150Δ-β-lactamase for at least 7 h. This was true even for Δhsp104 cells harboring the sec18-1 mutation that were used to accumulate the reporters in the ER before thermal insult in some of the experiments. This finding shows that the temperature-sensitive secretion block caused by the mutant Sec18 protein was fully reversible after severe heat stress even in the absence of Hsp104. The death of heat-treated Δhsp104 cells, the direct cause of which is unknown (Parsell and Lindquist, 1993), thus must have been due to later events. We conclude that irreversible ER retention of heat-damaged secretory Hsp150Δ-β-lactamase, vacuolar CPY, and cell wall mannoproteins in the absence of Hsp104 was not due to cell death or cessation of membrane traffic but more likely was due to irreversible conformational damage of the reporter proteins.
Recently, Hsp104 was shown in vitro to act directly in the solubilization of previously denatured cytosolic proteins, together with the Hsp70 member Ssa1p and its cochaperone Ydj1p, an Hsp40 member (Glover and Lindquist, 1998). The three proteins were suggested to function as a chaperone machine in which Hsp104 first remodels aggregated proteins, making them accessible for Ssa1/Ydj1p, which then provide a primary refolding function. Genetic data support the interaction of Hsp104 and Hsp70 (Sanchez et al., 1993). Our present observations cannot be explained by current models. To affect the refolding events in the ER lumen, Hsp104 could transiently interact, directly or indirectly, with the cytosolic portion of a membrane protein, whose lumenal portion would be involved with the ER repair mechanism. For example, BiP/Kar2p exerts its effect from the ER lumen to the cytosolic aspect of the ER membrane via Sec63p, a membrane-spanning protein with cytosolic and lumenal portions, to promote protein translocation (Lyman and Schekman, 1995). Nuclear fusion, which follows mating of haploid S. cerevisiae cells, directly requires BiP/Kar2p and another lumen ER protein, Jem1p (Ng and Walter, 1996; Brizzio et al., 1999). Whether Lhs1p, which is required for conformational repair in the ER (Saris et al., 1997), interacts with Sec63p or other membrane-spanning proteins is not known. Transient interactions are likely to be difficult to preserve for detection. Indeed, the interaction of Hsp104 with Ssa1p and Ydj1p is labile as a result of its dynamic nature (Glover and Lindquist, 1998). In fact, Hsp104’s cooperation with Ssa1p and Ydj1p could provide a link to the ER membrane, because the latter are thought to function in posttranslational translocation of polypeptides into the ER (Deshaies et al., 1988; Caplan et al., 1992).
One explanation of how Hsp104 could affect the refolding mechanism in the ER is that it performs its normal cytoplasmic repair function. This scenario holds that the cytoplasmic portion of a transmembrane protein required for the lumenal repair functions would be heat damaged. Hsp104 would repair the damage, restoring the activity of the lumenal portion in the refolding of target proteins. However, the mechanisms responsible for translocation and modification of proteins, as well as for exocytosis, involving tens if not hundreds of cytosolic proteins and cytosol-facing domains of membrane proteins, did survive the thermal insult in the absence of Hsp104 or were rapidly refolded by other chaperones. It would be surprising if the chaperone mechanism involved in repair functions were more vulnerable to heat than these mechanisms. Another possibility is that the interaction of Hsp104 with the ER refolding mechanism is more specific. The unfolded protein response exemplifies the transfer of information from the ER lumen to the cytosol via the transmembrane protein kinase Ire1p/Ern1p (Cox et al., 1993; Mori et al., 1993). In any case, Hsp104’s function in the refolding events in the ER lumen, as in conferring thermotolerance, requires its ATPase activity. Hsp104 appears to have a pivotal role in the survival of heat-stressed cells, controlling the repair of damaged proteins not only in the cytosol but even beyond the ER membrane.
ACKNOWLEDGMENTS
We thank Anna Liisa Nyfors for technical assistance, Dr. Leevi Kääriäinen for valuable comments on the manuscript, Dr. Susan Lindquist for yeast strains, and Dr. R. Himmelreich for cosmid 1F17. This work was supported by the Academy of Finland (grants 38017 and 41409). M.M. is a Biocentrum Helsinki fellow.
REFERENCES
- Ammerer G. Expression of genes in yeast using the ADC1 promoter. Methods Enzymol. 1983;10:192–201. doi: 10.1016/0076-6879(83)01014-9. [DOI] [PubMed] [Google Scholar]
- Baxter BK, James P, Evans T, Craig EA. SSI1 encodes a novel Hsp70 of the Saccharomyces cerevisiae endoplasmic reticulum. Mol Cell Biol. 1996;16:6444–6456. doi: 10.1128/mcb.16.11.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneaud M, Ozier-Kalogeropoulos O, Li G, Labouesse M, Minvielle-Sebastia L, Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- Brizzio V, Khalfan W, Huddler D, Beh CT, Andersen SSL, Latterich M, Rose MD. Genetic interactions between KAR7/s71, KAR8/JEM1, KAR5, and KAR2 during nuclear fusion in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:609–626. doi: 10.1091/mbc.10.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan AJ, Cyr CM, Douglas MG. Ydj1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell. 1992;71:1143–1155. doi: 10.1016/s0092-8674(05)80063-7. [DOI] [PubMed] [Google Scholar]
- Chen X, Easton D, Oh H, Lee-Yoon D, Liu X, Subjek JR. The 170 kDa glucose regulated stress protein is a large HSP70-HSP110-like protein of the endoplasmic reticulum. FEBS Lett. 1996;380:68–72. doi: 10.1016/0014-5793(96)00011-7. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov G, Liebman SW. Role of chaperone Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;74:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Craven A, Tyson JR, Stirling CJ. A novel subfamily of the Hsp70s in the endoplasmic reticulum. Trends Cell Biol. 1997;7:277–282. doi: 10.1016/S0962-8924(97)01079-9. [DOI] [PubMed] [Google Scholar]
- Craven RA, Egerton M, Stirling CJ. A novel Hsp70 of the yeast ER lumen is required for the efficient translocation of a number of protein precursors. EMBO J. 1996;15:2640–2650. [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Sanders SL, Feldheim DA, Scheckman R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature. 1988;332:800–805. doi: 10.1038/332800a0. [DOI] [PubMed] [Google Scholar]
- Dierks T, Volkmer J, Schlenstedt G, Jung C, Sandholzer U, Zachmann K, Schlotterhose P, Neifer K, Schmidt B, Zimmermann R. A microsomal ATP-binding protein involved in efficient protein transport into the mammalian endoplasmic reticulum. EMBO J. 1996;15:6931–6942. [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Guldener U, Heck S, Fiedler T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;13:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton G, Flynn GC. Cer1p, a novel Hsp70-related protein required for posttranslational endoplasmic reticulum translocation in yeast. J Biol Chem. 1996;271:30610–30613. doi: 10.1074/jbc.271.48.30610. [DOI] [PubMed] [Google Scholar]
- Hill J, Donald KAIG, Griffiths DE. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 1991;19:5791. doi: 10.1093/nar/19.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holkeri H, Paunola E, Makarow M. Dissection of the translocation and chaperoning functions of yeast BiP/Kar2p in vivo. J Cell Sci. 1998;111:749–757. doi: 10.1242/jcs.111.6.749. [DOI] [PubMed] [Google Scholar]
- Jämsä E, Vakula N, Arffman A, Kilpeläinen I, Makarow M. In vivo reactivation of heat-denatured protein in the endoplasmic reticulum of yeast. EMBO J. 1995;14:6028–6033. doi: 10.1002/j.1460-2075.1995.tb00291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelsch C, Enfant F, Masson JM, Samama JP. β-Lactamase TEM1 of E. coli crystal structure determination at 2.5 Å resolution. FEBS Lett. 1992;299:723–733. doi: 10.1016/0014-5793(92)80232-6. [DOI] [PubMed] [Google Scholar]
- Lin H, Masso-Welch P, Cai J, Shen J, Subjek JR. The 170-kDa glucose-regulated stress protein is an endoplasmic reticulum protein that binds immunoglobulin. Mol Biol Cell. 1993;4:1109–1119. doi: 10.1091/mbc.4.11.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman SK, Schekman R. Interaction between BiP and Sec63p is required for the completion of protein translocation into the ER of Saccharomyces cerevisiae. J Cell Biol. 1995;131:1163–1171. doi: 10.1083/jcb.131.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Ma W, Gething M-J, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- Ng DTW, Walter P. ER membrane protein complex required for nuclear fusion. J Cell Biol. 1996;132:499–509. doi: 10.1083/jcb.132.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Ferro S, Scheckman R. Order of events in the yeast secretory pathway. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Lindquist S. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Sanchez Y, Stitzel JD, Lindquist S. Hsp104 is a highly conserved protein with two essential nucleotide-binding sites. Nature. 1991;53:270–273. doi: 10.1038/353270a0. [DOI] [PubMed] [Google Scholar]
- Paunola E, Suntio T, Jämsä E, Makarow M. Folding of active β-lactamase in the yeast cytoplasm prior to translocation into the endoplasmic reticulum. Mol Biol Cell. 1998;9:817–827. doi: 10.1091/mbc.9.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SW. Sequence of a 20.7 kb region of yeast chromosome XI includes the NUP100 gene, an open reading frame (ORF) possibly representing a nucleoside diphosphate kinase gene, tRNA for His, Val and Trp in addition to seven ORFs with weak or no significant similarity to known proteins. Yeast. 1994;10:S69–S74. doi: 10.1002/yea.320100009. [DOI] [PubMed] [Google Scholar]
- Russo P, Simonen M, Uimari A, Teesalu T, Makarow M. Dual regulation by heat and nutrient stress of the yeast HSP150 gene encoding a secretory glycoprotein. Mol Gen Genet. 1993;239:273–280. doi: 10.1007/BF00281628. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Sanchez Y, Lindquist S. Hsp104 is required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Parsell DA, Taulien J, Vogel JL, Craig EA, Lindquist S. Genetic evidence for a functional relationship between Hsp104 and Hsp70. J Bacteriol. 1993;175:6484–6491. doi: 10.1128/jb.175.20.6484-6491.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Taulien J, Borkovich KA, Lindquist S. Hsp104 is required for tolerance to many forms of stress. EMBO J. 1992;11:2357–2364. doi: 10.1002/j.1460-2075.1992.tb05295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saris N, Holkeri H, Craven RA, Stirling C, Makarow M. The Hsp70 homolog Lhs1p is involved in a novel function of the yeast endoplasmic reticulum, refolding and stabilization of heat-denatured proteins. J Cell Biol. 1997;137:813–824. doi: 10.1083/jcb.137.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saris N, Makarow M. Transient ER retention as a stress response: conformational repair of heat-damaged proteins to secretion-competent structures. J Cell Sci. 1998;111:1575–1582. doi: 10.1242/jcs.111.11.1575. [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Glover JR, Singer MA, Lindquist S. Hsp100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- Schirmer EC, Lindquist S. Interactions of the chaperone Hsp104 with yeast Sup35 and mammalian PrP. Proc Natl Acad Sci USA. 1997;94:13932–13937. doi: 10.1073/pnas.94.25.13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberstein S, Schlenstedt G, Silver P, Gilmore R. A role for the DnaJ homolog Scj1p in protein folding in the yeast endoplasmic reticulum. J Cell Biol. 1998;143:921–933. doi: 10.1083/jcb.143.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonen M, Jämsä E, Makarow M. The role of the carrier protein and disulfide formation in the folding of β-lactamase fusion proteins in the endoplasmic reticulum of yeast. J Biol Chem. 1994;269:13889–13892. [PubMed] [Google Scholar]
- Simons JF, Ferro-Novick S, Rose MD, Helenius A. BiP/Kar2p serves as a molecular chaperone during carboxypeptidase Y folding in yeast. J Cell Biol. 1995;130:41–49. doi: 10.1083/jcb.130.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens T, Esmon B, Scheckman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982;30:439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- Vogel JP, Misra MD, Rose MD. Loss of BiP/GRP78 function blocks translocation of secretory proteins in yeast. J Cell Biol. 1990;110:1885–1895. doi: 10.1083/jcb.110.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]