Abstract
The molecular mechanism of human mitochondrial translation has yet to be fully described. We are particularly interested in understanding the process of translational termination and ribosome recycling in the mitochondrion. Several candidates have been implicated, for which subcellular localization and characterization have not been reported. Here, we show that the putative mitochondrial recycling factor, mtRRF, is indeed a mitochondrial protein. Expression of human mtRRF in fission yeast devoid of endogenous mitochondrial recycling factor suppresses the respiratory phenotype. Further, human mtRRF is able to associate with Escherichia coli ribosomes in vitro and can associate with mitoribosomes in vivo. Depletion of mtRRF in human cell lines is lethal, initially causing profound mitochondrial dysmorphism, aggregation of mitoribosomes, elevated mitochondrial superoxide production and eventual loss of OXPHOS complexes. Finally, mtRRF was shown to co-immunoprecipitate a large number of mitoribosomal proteins attached to other mitochondrial proteins, including putative members of the mitochondrial nucleoid.
INTRODUCTION
Mitochondria play a vital role in a wide variety of cellular processes in eukaryotic cells. They possess their own genomic DNA (mtDNA) that encodes 13 proteins, along with 22 tRNA and 2 ribosomal RNA (1). All protein products of the mitochondrial genome are part of the multi-subunit complexes involved in oxidative phosphorylation. Synthesis of these proteins is carried out on a specialized translational apparatus located within the organelle. The components of the mitochondrial translational machinery—translational factors and mitoribosomes—are distinct from those in the cytosol and generally resemble bacterial counterparts (2).
Our understanding of the mechanisms of mitochondrial translation is far from complete. A limited number of factors involved in mitochondrial translation initiation, elongation and termination have been identified and characterized (3–7). The mechanism of the last step of protein synthesis, the disassembly of the post-termination complex, has not yet been explored in human mitochondria.
Due to the monophyletic α-proteobacterial origin of mitochondria, our knowledge of ribosome recycling in prokaryotes can serve as a rough model for mitochondrial processes, although it is highly likely that they will prove to be important variations. In the bacterial system, when a ribosome reaches a stop codon, the nascent polypeptide is released by the coordinated actions of a sequence-specific class I release factor (RF1 or RF2) and a sequence-independent class II release factor [RF3, (8)]. The resulting post-termination complex containing mRNA and deacylated tRNA in the P/E site (9), has to be disassembled and ribosomal subunits need to be recycled to initiate a new round of protein synthesis. This process is catalysed by the ribosome recycling factor (RRF). RRF is universally conserved in eubacteria and deletion of the gene-encoding RRF (frr) has been shown to be lethal for Escherichia coli (10). Mycoplasma genitalium, with one of the smallest genomes reported to date, retains RRF despite losing RF2 and RF3, further supporting a key role for this factor in prokaryotic translation (11).
The mechanism of bacterial ribosome recycling has been addressed by several independent laboratories [for example, (12–14)]. Conformational changes mediated by RRF bound to the 70S ribosome can disrupt the intersubunit bridges leading to separation of the monosome. This process requires GTP hydrolysis that is triggered by elongation factor G (EFG). Subsequently, initiation factor 3 (IF3) binds to the 30S subunit preventing re-association of the free subunits. More recently, emphasis has been placed on the exact role played by bacterial RRF in this process. Cryoelectron microscopy data on E. coli RRF binding to 70S post-termination complexes is consistent with a spontaneous movement of RRF from around the peptidyltransferase centre in the monosome to a site on the 50S subunit, resulting in the cleavage of intersubunit bridges and subunit dissociation (15). However, no such structural reorganization was found on X-ray crystallography of a similar but not identical Thermus thermophilus system (16).
To date, no human mitochondrial ribosome recycling factor has been characterized. A candidate, mtRRF, was proposed several years ago from bioinformatic analyses of several overlapping EST sequences (7) but with no supportive functional studies or investigation of subcellular localization. In this article, we report that this putative mitochondrial recycling factor is indeed a mitochondrial protein. Furthermore, we show a direct interaction of purified mtRRF with ribosomes of both bacterial and mammalian mitochondrial origin in vitro and in vivo. To assess the physiological function of mtRRF, we have examined the ability of human mtRRF to rescue yeast strains deleted of their endogenous factor and the effect of mtRRF depletion in cultured human cells. We observed aggregation of mitoribosomes, increased ROS and profound changes in mitochondrial morphology. Our results imply that analogous to the bacterial situation, this protein is essential for mitochondrial function and viability of human cells.
METHODS
Tissue culture manipulations
Human HeLa cells were cultured (37°C, humidified 5% CO2) in Eagle's modified essential medium (Sigma-Aldrich Co.Ltd, Dorset, UK) supplemented with 10% (v/v) fetal calf serum (FCS), 1 × non-essential amino acids and 2 mM l-glutamine. Flp-In™T-Rex™-293 cells (HEK293T, Invitrogen Ltd, Paisley, UK) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FCS, 50 µg/ml uridine and 1 × non-essential amino acids in the presence of 10 µg/ml Blasticidin S and 100 µg/ml Zeocin (Invitrogen). Post-transfection selection was effected with Hygromycin B (100 µg/ml). For growth on respiratory substrates, medium was glucose-free DMEM (Gibco, UK), 0.9 mg/ml galactose, 1 mM sodium pyruvate, 10% (v/v) FCS, non-essential amino acids and 2 mM l-glutamine. For growth-curve analyses, galactose medium included 50 µg/ml uridine.
Production of GFP-, FLAG- and GST-fusion constructs and cloning into yeast expression vectors
The original human mtRRF clone was obtained from MGC (MGC:17776; Acc No BC013049), but primer positions described below are relative to the reference sequence NM_138777.2.
To generate a fusion construct that would allow expression of mtRRF with GFP at the C-terminus, nucleotides 102–894 of the mtRRF cDNA were amplified using primers for 5′-CACACATGATCAGATTGTCTTCAGTCATGG-3′ and Rev 5′-CACACATGATCAAGTTCTTTGGTCTTCACTGC-3′ (BclI sites underlined). This was digested and cloned in-frame upstream of GFP in pGFP3 (pcDNA3.1 Invitrogen, containing the GFP open reading frame (ORF) and multiple cloning site kindly donated by Dr D Elliott, Newcastle University).
The glutathione-S-transferase (GST)-fusion constructs were made of the full-length (FL) or N-terminal deleted (NDel) mtRRF. These were generated by amplifying nucleotide 116 or nucleotides 324–940, respectively. The FL forward primer incorporated a BclI site, whilst the NDel included an EcoR1 site (FL for 5′-CACACCTGATCAATGGCCTTGGGATTAAAG-3′; NDel for 5′-CACACCGAATTCCCAGAGTGAATATTAATGCT-3′) and the same reverse primer was used for both amplification reactions (Rev 5′-AATTATGCGGCCGCACTGGGCTCTGGAGTATT-3′ containing a NotI site). Amplicons and vectors were digested to allow in-frame downstream fusion of mtRRF-GST in pGEX-6P-1 or pGEX-6P-2, respectively (GE Healthcare, Amersham, UK).
To generate a FLAG-tagged mtRRF construct, the following primers were used to amplify nucleotides 115–900. The region corresponding to the FLAG tag is italicized and EcoRV sites are underlined for 5′-CTTTCTTGATATCCCACCATGGCCTTGGGATTAAAGTGCTTCCGCATGGTCCA-3′ and Rev 5′-CTCTCCGATATCCTACTTATCGTCGTCATCCTTGTAATC TCCAAGGAGTTCTTTGG-3′.
A FL mtRRF PCR product (nucleotides 101–980) was generated using primers for 5′-CACACCTGATCAGGAUUGUCUUCAGUCAUG-3′ and Rev 5′-CACACCTGATCAGAGAGAAGTCCCAATGTGC-3′ (BclI site underlined). This product was cloned into pGEM-Teasy (Promega, Southampton,UK), excised by NotI digestion and subcloned into NotI-digested Saccharomyces cerevisiae URA3 expression vector pFL61 (17) or the Schizosaccharomyces pombe ura4 expression vector pTG1754 [a gift from Transgene, (18)], which both contain a strong constitutive promoter.
Transient transfection of HeLa cells, microscopy and image capture
HeLa cells were grown on coverslips to 50% confluency, transfected with GFP fusion vector (1 µg) using SuperFect (Qiagen, Sussex, UK) as recommended and cultured for a further 24 h prior to incubation with Mitotracker Red CM-H2XRos (1 µM final, Invitrogen). After brief fixation (4% paraformaldehyde/PBS, 15 min at room temperature), cells were mounted in Vectashield (H-1200 Vector Laboratories Ltd, Peterborough, UK) and visualized by fluorescence microscopy using a Leica (Nussloch, Germany) DMRA. Images were recorded as a Z-series (0.5 µm slices) using a cooled CCD camera and imaging system (Spot–II Diagnostics Instruments, Sterling Heights, MI, USA).
Immunocytochemistry
HeLa cells (3 × 105) plated on coverslips were incubated with Mitotracker Red CMH2XRos, as described above, fixed in 3% paraformaldehyde, incubated with anti-mtRRF polyclonal antibodies (1/300) for 4 h, washed, and then incubated with AlexaFluor-488 conjugated anti-rabbit antibodies (Molecular Probes, Invitrogen, Paisley, UK) for 1 h. Signals were visualized by microscopy as described above.
Mitochondrial import assays
Assays were performed and assessed using isolated rat liver mitochondria following the methods described in ref. (6) but with reticulocyte lysates programmed with the relevant RNA species.
Over-expression and purification of mtRRF–GST
Escherichia coli strain Rosetta(DE3)pLysS (Novagen, Merck Biosciences Ltd, Nottingham, UK) was transformed with constructs for the over-expression of the human mitochondrial RRF. Bacteria were induced at 0.5 A600 with 1 mM IPTG, overnight at 16°C. Pelleted cells were resuspended in 50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM lysozyme, 1 mM EDTA, 1 mM PMSF and sonicated on ice 10 × 10 s (Soniprep 150). Post-centrifugation (30 000g for 20 min, 4°C), the supernatant was filtered (0.45 µm Corning) and incubated for 3 h at 4°C with glutathione Sepharose 4B beads (GE Healthcare). Beads were extensively washed prior to elution in 50 mM Tris–HCl pH 7.8, 150 mM NaCl, 20 mM reduced glutathione, 1 mM PMSF.
For cleavage and removal of the GST on the Sepharose, beads were incubated at 4°C overnight in 50 mM Tris–HCl pH 7.8, 150 mM NaCl, 1 mM PMSF supplemented with 1 mM EDTA, 1 mM DTT and 24 µl/ml PreScission Protease (GE Healthcare). Eluted recombinant protein was stored at 4°C.
Production of anti-mtRRF antibodies
Recombinant human mtRRF purified as described above was used as an antigen to raise rabbit antisera. Antibody generation and affinity purification was performed by Eurogentec, Belgium.
In vitro ribosome binding assay
Escherichia coli ribosomes were prepared as described in ref. (6). Reactions (50 µl) combined purified recombinant mtRRFΔ69 and 70S ribosomes (1 µM) in 10 mM Tris–HCl pH 7.2, 10 mM Mg(OAc)2, 80 mM NH4Cl, 1 mM DTT at room temperature for 30 min. The mixture was centrifuged through 10% (v:v) sucrose (150 µl in binding buffer) for 1 h in Beckman–Coulter air-driven ultracentrifuge (30 p.s.i.). Fractions (4 × 50 µl) were collected and analysed by western blot.
Stable transfection of HEK293T cells with FLAG-tagged mitochondrially targeted proteins
Cells were transfected at ∼30% confluency using Superfect (Qiagen). The vectors, pOG44 expressing FRT recombinase, and pcDNA5/FRT/TO containing sequences of the genes to be expressed (FLAG-tagged mtRRF or mitochondrially targeted luciferase) were combined to give a total of 2 µg DNA in a 9:1 ratio, and mixed with Superfect prior to a 3-h incubation with the cells. Selection with Hygromycin B (100 µg/ml) commenced 2 days later. Independent colonies were isolated, propagated and analysed for induction (1 µg/ml tetracycline) by western analysis using anti-FLAG antibodies (Sigma).
Isokinetic sucrose-gradient analysis of mitochondrial ribosomes
Total cell lysates (0.5 mg) were loaded on a linear sucrose gradient [10–30% (v:v), 1 ml] in 10 mM Tris–HCl pH 7.2, 10 mM Mg(OAc)2, 80 mM NH4Cl, 1 mM PMSF and centrifuged for 2 h at 100 000g at 4°C. Fractions of 100 µl were collected and analysed by western blot.
Affinity purification and elution of FLAG peptides
Mitochondria were isolated from HEK293T cells over-expressing FLAG-mtRRF or -mtLUC essentially as described in ref. (6) and treated with proteinase K (5 µg/1 mg of mitochondria; 30 min 4°C) followed by PMSF (1 mM) inhibition. Pelleted mitochondria were resuspended in lysis buffer (supplemented with 10 mM Mg(OAC)2 as indicated, although no difference in the composition of protein precipitate could be measured without supplement). Immunoprecipitation was performed with a-FLAG-Gel following manufacturer's recommendations (Sigma Aldrich, St Louis, MO, USA). Elution was effected with FLAG peptide. RNase A (5 µg/ml) and EDTA (50 mM) were added as indicated.
Mass spectrometry analysis and protein identification
Mass spectrometric analysis of immunoprecipitated complexes was performed as described in Supplementary Material.
Yeast growth conditions, plasmid and general strain constructions and complementation assays
All yeast strains used in this study are detailed in Table S2. General yeast media and genetic techniques/transformation protocols were as described in ref. (18). In brief, fermentable media contained 2% glucose, non-fermentable conditions were 3% (v:v) glycerol/ethanol or 2% (v:v) glycerol with 0.1% glucose or for S. pombe only 2% (w:v) galactose/0.1% (w:v) glucose.
Production of S. pombe Δrrf1 strain
A single S. pombe protein, encoded by the gene SPBC1709.09, was found to have significant similarities to both S. cerevisiae Rrf1 (Figure S4) and E. coli RRF. The ORF was PCR amplified from genomic DNA and cloned into the NotI site of the pTG1754 and pFL61 expression plasmids for S. pombe and S. cerevisiae, respectively. An S. pombe Δrrf1::KanR strain (NB331) was constructed in the wild-type recipient strain NB205-6A by using a PCR disruption strategy with hybrid rrf1-KanR oligonucleotides as described in ref. (19), removing 629 bp encompassing the 735 bp rrf1Sp ORF (starting 8 bp before the ATG). Of 50, 22 clones showed delayed growth on glycerol medium and were proved to contain the correct insertion by PCR. Crossing to wild-type followed by sporulation and tetrad analysis showed a 2:2 cosegregation of the geneticin resistance and slow glycerol growth. Transformation with the Rrf1pSp-encoding plasmid restored growth on galactose, confirming that the mitochondrial DNA was intact in this strain.
Spectroscopic analyses
Cells from Δrrf1Sp transformants were dried, mixed with sodium dithionite to fully reduce the cytochromes and frozen in liquid nitrogen before recording the absorbance of the samples at wavelengths from 630 nm to 490 nm (spectrophotometer Cary 400Ò, Varian, San Fernando, CA). Peaks were the following: cytochrome c (548 nm), cytochrome c1 (554 nm), cytochrome b (560 nm) and cytochrome aa3 (603 nm).
siRNA constructs, transfection and RT–PCR analysis
The following sequences targeting mtRRF were tested for efficiency of mRNA and protein depletion. Nucleotide positions are relative to reference sequence NM_138777.2, siRNA ORF1 5′ GGACACCAUUAGGCUAAUA dTdT 3′ (nucleotides 211–229); siRNA ORF2 5′ GAGAAAUGCUGGUGAAACU dTdT 3′ (nucleotides 684–702); siRNA ORF3 5′ GACAGUGCAUGAAAGACAA dTdT 3′ (nucleotides 799–817). Experiments were performed with siRNA ORF2 unless otherwise specified. Treatment of stably transfected and induced HEK293T cells was performed with siRNA UTR1 5′ GUAUUCUUGUUGCACUUAA dTdT 3′ (nucleotides 1633–1651) targeting the 3′-UTR region that was absent in the inducibly expressed form of mtRRF. Transfections were performed on 20% confluent cells with Oligofectamine (Invitrogen) in Optimem-I medium (Gibco) with final concentrations of 0.2 µM siRNA. Control non-targetting siRNA and custom siRNA duplexes were purchased, purified and pre-annealed from Eurogentec. Quantification of mtRRF mRNA was performed with TaqMan Gene Expression Assay (Applied Biosystems, Warrington, UK) using ABI Prism sequence detector system. Data were normalized to GAPDH as an endogenous control. Probe kits were ordered from Applied Biosystems—HS01067555_g1 (mtRRF) and HS999999905_m1 (GAPDH).
Cell preparations, western analysis and Blue Native-PAGE
Schizosaccharomyces pombe mitochondria was purified from cells grown either in complete glucose medium or in minimal glucose medium lacking uracil as described in ref. (19) with Roche protease inhibitors present at all stages. Yeast proteins were separated by 12% SDS–PAGE and immobilized on nitrocellulose (Schleicher & Schuell, London, UK) by wet transfer in 25 mM Tris, 192 mM glycine, 0.02% (w/v) SDS and 20% (v/v) ethanol. Blots were hybridized with mouse (anti-human Hsp60, Sigma H-3525; anti-FLAG, Sigma H-1804) or rabbit antibodies [anti-mtRRFHs (this work); -Cox2Sp, (20) -Arg8Sc (21); -EFTuSp (18)].
Human cell lysates were prepared, separated by SDS–PAGE and immobilized by wet transfer as described (6). Proteins of interest were bound by overnight incubation (4°C) with antibodies [mtRRF, GDH (Lightowlers Group, Newcastle); Porin, COX I, COX II, SDH 70K and ND6 (MitoSciences, Eugene, OR); β-actin (Sigma); DAP3, MRLP3 (Abcam, Cambridge, UK)] followed by HRP-conjugated secondary antibodies (DAKO) and visualized by ECL-plus (GE Healthcare).
Blue native gel electrophoresis was performed as described in ref. (22) with 30 µg protein loaded per lane and wet transfer as above.
Free radicals and mitochondrial mass measurements
MitoSOX (Molecular Probes) prepared in DMSO, diluted to 5 µM in DMEM medium-lacking serum was added to cells for 10 min at 37°C. Cells were washed before re-suspension in 2 ml DMEM-lacking serum and passed through a FACScan flow cytometer (Becton Dickinson Ltd, Oxford, UK, equipped with 488 nm Argon laser). Data were collected for forward and side scatter together with autofluorescence of unstained cells, gating eliminated debris and apoptotic cells from the analyses. After acquisition, data were analysed by WinMDI software (version 2.8). Experiments were performed on four independent occasions with triplicate measurements based on 10 000 events. Dihydrorhodamine 123 (DHR, Molecular Probes) and nonyl acridine orange (NAO, Molecular Probes) were resuspended in DMSO, diluted to 30 mM (DHR) or 10 mM (NAO) in DMEM-lacking serum and incubated with cells at 37°C for 30 (DHR) min or 10 (NAO) min. Cells were then washed before resuspension in 2 ml DMEM-lacking serum and green fluorescence emission analysed as above.
Electron microscopy studies
Cells were grown as detailed above, pelleted and fixed in 2% glutaraldehyde in Sorensen's phosphate buffer. Samples were than post-fixed in 1% osmium tetroxide in the same buffer, dehydrated in alcohol and embedded in Epoxy resin (TAAB). Ultrathin sections were stained with 2% aqueous uranyl acetate and lead citrate and examined using a Philips CM100 transmission electron microscope (EM Research Services, Newcastle University).
In vivo mitochondrial protein synthesis
Protein translation in HeLa cells was performed essentially as described (23). Cells were incubated with radiolabelled methionine for 15 min.
Respirometry
High-resolution respirometry was performed at 37°C with an Oroboros Oxygraph-2K (Oroboros Instruments, Austria) essentially as detailed in ref. (6).
Statistical analysis
Students unpaired t-tests were used to determine the significance of values as indicated in the figure legends. Values of P < 0.05 were recorded as statistically significant.
RESULTS
MtRRF, the human homologue of bacterial RRF, is a mitochondrial protein
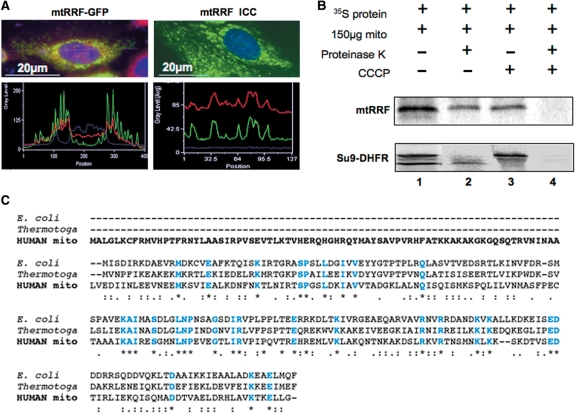
To confirm mitochondrial localization of the putative human mtRRF predicted by in silico studies (7), a mtRRF–GFP fusion construct was used to transfect HeLa cells. This chimera included all but the terminal 2 residues of mtRRF fused N-terminal to GFP. Cells were cultured for 24 h prior to staining with Mitotracker CM-H2XRos, to visualize mitochondria. Fluorescence microscopy images and line scan analysis (Figure 1A, left panels) of transfected cells demonstrated co-localization of mtRRF–GFP with Mitotracker Red. In contrast to the well-defined, discrete pattern produced by mtRRF–GFP, expression of the GFP protein alone produced a diffuse stain (data not shown).
Figure 1.
mtRRF is a mitochondrial protein with an extended N-terminal pre-sequence. (A) Human mtRRF is targeted to mitochondria. Left panels show HeLa cells transiently transfected (24 h) with a mtRRF–GFP fusion construct. Cells were stained to visualize nuclei (DAPI blue) and mitochondria (Mitotracker red). Fluorescence images and linescans confirmed mitochondrial localization of mtRRF–GFP by superimposition of green and red signals (lower left). The image reflects three-independent transfections. Endogenous mtRRF in HeLa cells was visualized by immunocytochemistry (upper right) using affinity purified anti-mtRRF and FITC secondary. DAPI-stained nuclei (blue) and Mitotracker the mitochondria (red). Mitochondrial localization of mtRRF was confirmed by superimposition of linescan green and red fluorescence (lower right). The image reflects three-independent transfections. (B) Human mtRRF is imported into mitochondria. FL 35S-radiolabelled mtRRF was in vitro synthesized and incubated with rat liver mitochondria (lane 1). Under import conditions a single product is visible (lane 2) that is protected from proteinase K (lane 3), but degraded by treatment with proteinase K and FCCP uncoupler (lane 4). Control import reactions contained DHFR with a mitochondrial pre-sequence (Su9) showing the FL pre-protein and the matured form under import conditions. (C) Sequence alignment (CLUSTALW) indicates an extensive N-terminal pre-sequence when compared with E. coli or Thermotoga maritima. Identity to these RRFs is indicated in blue and by *, high levels of similarity by a colon ‘:’ and lower levels by a fullstop ‘.’.
To verify, if localization of the mtRRF–GFP fusion protein reflects the behaviour of the endogenous protein, immunocytochemistry was performed. Polyclonal antisera against FL recombinant protein were raised and anti-mtRRF antibodies affinity were purified. Immunocytochemistry (Figure 1A, right panels) indicated similar mitochondrial localization of both the endogenous and mtRRF–GFP.
To confirm that mtRRF is indeed imported into the mitochondrial matrix and not merely localized to the organelle, import assays were performed with isolated mitochondria and in vitro translated 35S-labelled mtRRF (Figure 1B). Resistance of the radiolabelled protein to proteinase K (lane 2) confirmed successful mitochondrial uptake. Import resulted in cleavage of the mitochondrial targeting pre-sequence of Su9-DHFR but interestingly not of mtRRF. This is consistent with bioinformatic predictions (http://www.cbs.dtu.dk/services/TargetP) being unable to identify a clear cleavage signal in the mtRRF sequence (7). Alignment of the mtRRF sequence with bacterial counterparts indicates that optimal alignment begins at position 80 of the FL mtRRF (Figure 1C). Since our results show that this long N-terminal extension is not a cleavable import signal but an integral part of the mature protein, it suggests that it may be of functional importance specific to mitochondria.
mtRRF binds E. coli ribosomes in vitro and can associate with human mitoribosomes
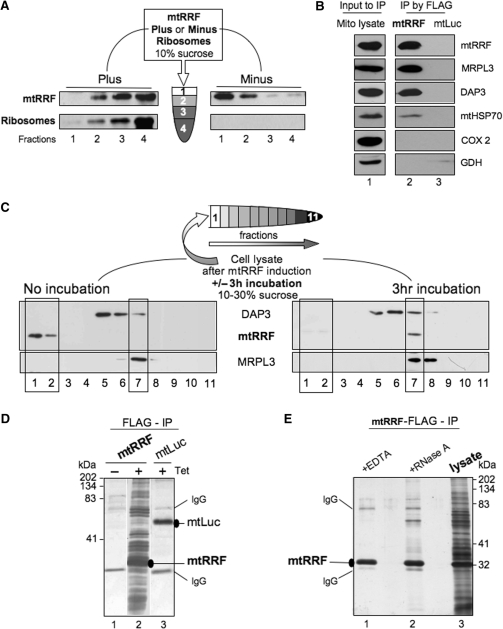
Direct interaction of prokaryotic RRF with ribosomes in the absence of other translation factors has been demonstrated (16,24). Binding experiments were therefore performed to determine if human mtRRF could play a similar role to its bacterial homologues. Isolation of FL over-expressed mtRRF showed that the protein tended to aggregate (Figure S1). Therefore, a short N-terminal truncation mtRRFΔ69 was used as this deletion significantly increased solubility of the recombinant protein. Comparative circular dichroism analysis of both the FL and truncated mtRRF confirmed that removal of the N-terminus did not cause any major structural changes (data not shown). In vitro binding assays showed that mtRRF localized to the rapidly sedimenting ribosomal fraction only in the presence of E. coli ribosomes (Figure 2A), indicating a direct interaction.
Figure 2.
Human mtRRF associates with bacterial ribosomes in vitro and mitoribosomes in vivo. (A) Human mtRRF binds E. coli ribosomes. Recombinant mtRRF (mtRRFΔ69) was incubated with or without 70S ribosomes prior to sedimentation through 10% (w:v) sucrose. Fractions were subjected to western analysis. (B) mtRRF associates with mitoribosomes. FLAG-mtRRF and mtLuc HEK293T cells were induced, mitochondria isolated. Lysate was prepared (lane 1) from which proteins were immunoprecipitated via the FLAG epitope (lanes 2–3). Western blot analysis used antibodies to mtRRF, large 39S and small 28S mitoribosomal subunits (MRPL3, DAP3), mitochondrial chaperone (mtHSP70), complex IV (COX 2) and a matrix protein (glutamate dehydrogenase -GDH). (C) Pre-incubation of mitochondria allows detection of mtRRF binding to mitoribosomes. Lysate was prepared from HEK293T cells after induction of FLAG-mtRRF and was either separated immediately or post 3 h incubation on a 10–30% (v:v) isokinetic sucrose gradient. Fractions were analysed by western blot with antibodies to mtRRF, MRPL3 (39S mitoribosomal subunit) or DAP3 (28S mitoribosomal subunit). A similar time-dependent mitoribosomal association in HeLa cell lysate was noted for the endogenous mtRRF (data not shown). (D) HEK293T cells were induced (±Tet) for FLAG-mtRRF (lanes 1–2) or control mtluc (lane 3) expression and mitochondria isolated. Co-immunoprecipitating proteins were separated through a 12% SDS–PAG and visualized by silver stain. mtRRF, mtluc and anti-FLAG IgG are indicated. (E) Preparations of the mtRRF IP were also treated with EDTA (lane 1) or RNase A (lane 2). Mitochondrial lysate is shown in lane 3.
To determine whether mtRRF could interact with mitoribosomes in isolated organelles, FLAG epitope-tagged mtRRF was inducibly expressed in human Flip-In T-REx HEK293 cells (HEK293T) as described in procedures. Subsequently, complexes containing the fusion protein were isolated from solubilized mitochondria by anti-FLAG affinity purification and FLAG peptide elution. To control for non-specific binding, FLAG-tagged mitochondrially targeted luciferase (mtluc) was expressed in cells and immunoprecipitated by the same procedure. Western analysis revealed a specific interaction of mtRRF and mitoribosomal proteins (Figure 2B). Mitoribosomes were very efficiently precipitated with FLAG-mtRRF. Taken together these data show that mtRRF can tightly associate with mitoribosomes.
Bacterial RRF is only transiently associated with ribosomes in vivo, partially occluding the P and A sites on binding to the post-termination complex (9,15) so it was surprising to find that the majority of mtRRF was bound to mitoribosomes. To investigate this further, HEK293T cells were induced as above and lysates were immediately subjected to isokinetic sucrose-gradient centrifugation. This mtRRF over-expression was not deleterious to cell growth (data not shown) but led to a slight decrease in mitoribosome formation (Figure 2C cf. Figure 5A). No association of mtRRF with mitoribosomes at steady state was detectable. However, binding became evident on incubation (3 h) of the lysate prior to centrifugation (mimicking the IP procedure) potentially due to dissociation or hydrolysis of charged tRNAs that would naturally occupy most A/P sites. This dependence on pre-incubation was not a consequence of mtRRF over-expression, as a similar effect was noted with HeLa cell lysate (Figure 5A, data not shown). Further, association was not due to non-specific interactions promoted by the pre-incubation step as interactions were lost either on RNase or EDTA treatment (Figure 2E, lanes 1 and 2).
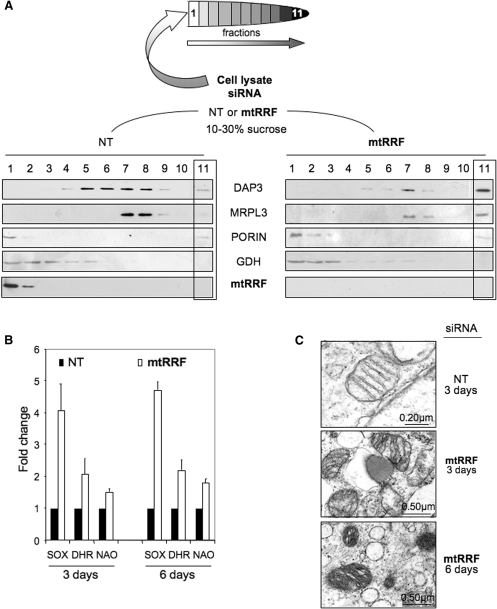
Figure 5.
Reduction of mtRRF results in redistribution of ribosomal proteins, increased ROS and mitochondrial dysmorphism. (A). HeLa lysates were prepared from cells treated with mtRRF or non-targeted (NT) siRNA (3 days). After separation through 10–30% sucrose gradients, fractions were analysed by western blot using antibodies against the small (DAP3) and large (MRPL3) mitochondrial ribosomal subunits, mtRRF, porin as a mitochondrial membrane marker and glutamate dehydrogenase (GDH, matrix). (B) Superoxide levels are increased after mtRRF depletion. HeLa cells were exposed to mtRRF (white) or non-targeted (NT, black) siRNA over 6 days and superoxide and peroxide levels were measured with mitoSOX (superoxide) or DHR (peroxide) and compared to untreated controls. The fold increase is shown as a mean ± SEM from minimally three repeat experiments. Mitochondrial mass per cell was also measured using the cardiolipin selective dye NAO. (C) HeLa cells depleted of mtRRF show altered morphology. EM micrographs show HeLa cells treated with mtRRF (3 and 6 days) or non-targeted (NT, 3 days) siRNA. The images reflect mitochondria from two preparations.
Mitoribosomes associated with numerous mitochondrial proteins are co-immunoprecipitated with mtRRF
Immunoprecipitation (IP) experiments using mtRRF as bait revealed an association with two mitoribosomal proteins, presumably indicating the presence of the large (39S) and small (28S) ribosomal subunits (MRPs) (Figure 2B). To confirm the presence of other MRPs and determine which if any additional proteins were present, IP experiments were performed on lysates from a protease/DNase-treated HEK293T mitoplast fraction. After elution of specifically bound proteins with FLAG peptide, the eluant was separated by gel electrophoresis and gel slices subjected to LC-MS/MS as detailed in Supplementary Material. The mitoplast enrichment procedure yielded a total of 167 identified mitochondrial proteins (Tables 1 and S1), 73 of which were indeed MRPs. Although MRPs constituted most of the purified immunoprecipitated proteins, detailed LC-MS/MS analysis revealed some remaining cytosolic contaminants (data available on request) but also allowed us to identify other mitochondrial proteins that co-immunoprecipitated with mtRRF (Table 1). The second largest group were proteins that had been previously reported as components of human mitochondrial nucleoids (25–27). Among other proteins interacting with mitoribosomes were factors involved in intra-organellar translation, nucleic acid binding proteins and chaperones/proteases, all proteins that might be expected to associate with ribosomal components or nascent peptides. Several polypeptides known to be involved in metabolism were also identified.
Table 1.
Predicted function of the mitochondrial proteins identified after anti-FLAG mtRRF affinity precipitation
| Protein typea | Immunoprecipitationhr |
||
|---|---|---|---|
| mtRRFb | mtRRF + EDTAc | mtRRF + RNased | |
| Mitoribosomal proteinse | 73 | 1 | 0 |
| Nucleoidf | |||
| Class I | 25 (31) | 2 | 0 |
| Class II | 6 (10) | 0 | 0 |
| Class III | 6 (16) | 0 | 0 |
| Nucleoidg | 5 (6) | 1 | 0 |
| Translation associated | 6 | 1h | 1h |
| Nucleic acid binding/modifying | 13 | 0 | 0 |
| Metabolic | 23 | 0 | 0 |
| Chaperone/protease | 5 | 0 | 0 |
| Transporter | 4 | 0 | 0 |
| Unknown function | 6 | 0 | 0 |
| TOTAL | 167j | 7i | 1 |
Known or predicted function of identified mitochondrial proteins.
Immunoprecipitation from mitoplasts after protease and DNase shaving.
EDTA-washed immunoprecipitant from crude mitochondria.
RNase-treated immunoprecipitant from crude mitochondria.
Putative nucleoid proteins as recently classified by Bogenhagen et al. (25) (of total proteins).
Putative nucleoid proteins as identified by He et al. (26) (of total proteins).
Mitochondrial ribosomal recycling factor, mtRRF.
Three cytosolic ribosomal proteins were identified in the precipitation following EDTA treatment.
Does not include any non-mitochondrial, hypothetical or predicted proteins that were found.
Human mtRRF improves the respiratory competence of S. pombe Δrrf1
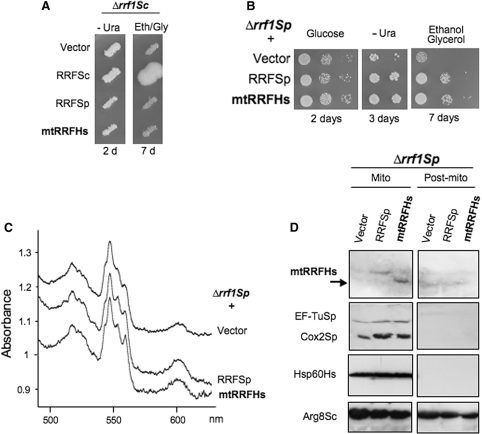
To gain insight into the in vivo function of human mtRRF, we first examined whether it could replace the endogenous mtRRF of the budding yeast S. cerevisiae. Respiratory deficiency due to loss of S. cerevisiae mtRRF (Δrrf1Sc) was reportedly associated with depletion of cytochrome c oxidase activity (28) or total loss of mtDNA (29). Thus, for complementation analysis we used Δrrf1Sc strains with different nuclear and mitochondrial backgrounds (Supplementary Material). In the W303 background, Δrrf1Sc cells showed a mild to pronounced respiratory deficiency depending on the mitochondrial intron content (Figure S2), but retained wild-type mtDNA. This allowed direct complementation tests by transformation with expression plasmids carrying either the natural S. cerevisiae gene, or human or S. pombe (fission yeast) homologues. However, neither the S. pombe nor the human homologue complemented the S. cerevisiae mutant (Figure 3A, data not shown).
Figure 3.
Human mtRRF suppresses a partial respiratory deficiency in Δrrf1 fission yeast. (A) Human mtRRF cannot rescue the Δrrf respiratory defect in budding yeast. Saccharomyces cerevisiae transformants of Δrrf1 strain MG38 carrying the control vector pFL44L or URA3 plasmids producing the S. cerevisiae, S. pombe or human mtRRF were replica-plated on non-fermentable media (eth/gly + glycerol 0.1% glucose) and incubated at 28°C. (B–D) Human mtRRF restores respiratory capacity in Δrrf1 fission yeast. Schizosaccharomyces pombe lacking endogenous RRF (Δrrf1Sp, NB331) was transformed with vector alone (vector) or expressing human (RRFHs) or S. pombe (RRFSp) mtRRF (B). Serial dilutions of three Δrrf1Sp transformants were spotted on complete glucose, uracil-free minimal or complete gly/eth media and grown at 28 °C for the times indicated. (C) Δrrf1Sp transformants were grown on 2% glycerol/0.1% glucose (vector) or ethanol/glycerol medium (RRFSp or RRFHs) before recording whole-cell cytochrome spectra. Cytochrome b + c1 and aa3 correspond to Complex III and Complex IV, respectively. (D) Mitochondria and post-mitochondrial supernatants from transformants grown in glucose medium minus uracil were separated by 12% PAGE and analysed by western blot with antibodies recognizing human mtRRF, S. pombe mtEF-Tu and mt-encoded Cox2, human mt-Hsp60 and S. cerevisiae mtArg8 (this also recognizes S. pombe Arg1 in mitochondria and an unknown protein in the supernatant).
As the fission yeast S. pombe is regarded as a closer evolutionary relative to human than S. cerevisiae, particularly in mitochondrial physiology (19), we inactivated the gene coding the S. pombe mtRRF homologue (rrf1Sp) to test if it could be replaced by human mtRRF. Deletion of rrf1Sp slightly but significantly impaired growth on non-fermentable carbon sources (Figure 3B), lowered overall content of mt-encoded cytochromes (Figure 3C) and decreased steady-state levels of mt-encoded Cox2 protein (Figure 3D), consistent with a role in mitochondrial translation. In addition, western analysis of S. pombe mtRRF protein confirmed mitochondrial location (Figure S3). Transfectants of this strain expressing FL human mtRRF were able to fully restore normal growth on non-fermentable medium, levels of mt-encoded cytochromes and Cox2 protein, correlating with weak but reproducibly detectable mtRRFHs in only the mitochondrial fraction (Figure 3B–D). Thus, human mtRRF clearly performs the same function as S. pombe mtRRF consistent with interaction with the S. pombe mitochondrial translation machinery.
Loss of mtRRF in human cells results in reduced growth rate and cell death
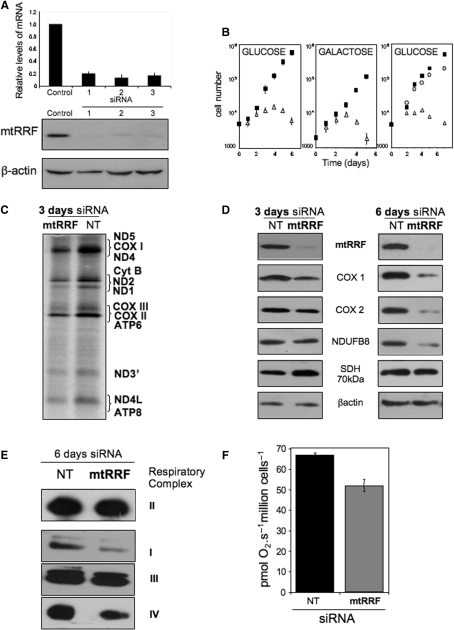
To determine the function of mtRRF in human mitochondria, three siRNA specific to the human mtRRF mRNA were designed and used to transfect HeLa and HEK293T cells. A non-targeting siRNA (NT) was used to confirm specificity. A reduction of 80–90% of mtRRF mRNA levels were observed 2 days post-transfection with a consequent dramatic reduction in mtRRF protein by day 3 (Figure 4A). To investigate the long-term effect of mtRRF depletion, HeLa cells were retransfected with siRNA on the third day of the experiment. Interestingly, the cells transfected with mtRRF siRNA(ORF2) showed a significant reduction in growth compared to NT controls (Figure 4B). After 3 days of mtRRF siRNA treatment, cells developed morphological changes and depletion for more than 6 days was lethal. If cells were cultured in glucose-free media supplemented with galactose to force dependence on oxidative phosphorylation, the effect of mtRRF depletion was even more profound (Figure 4B). These observations suggest that mtRRF is an essential protein and its loss affects mitochondrial functions.
Figure 4.
Depletion of human mtRRF severely affects cell viability and compromises mitochondrial translation. (A) siRNA-mediated depletion of human mtRRF. HeLa cells were exposed to three different siRNA molecules all in the mtRRF-coding sequence or an control siRNA for 6 days and RNA or cell lysates were prepared. Levels of mtRRF transcript compared to control siRNA transfected cells were quantified by real-time PCR (upper graph, mean ± SEM from four-independent transfections). Western blots (30 µg lysate/lane) in the lower panel were performed with antibodies against mtRRF to confirm depletion and β-actin as a loading. The blot accurately reflects three experiments. (B) Depletion of mtRRF causes a severe growth defect. Multiple aliquots of HeLa cells (left and centre panels) were exposed to targeted (mtRRF, open triangles) or non-targeted (NT, black squares) siRNA for 6 days in glucose (left) or galactose (centre) media, cells counted and presented in a semi-log plot. Counts were made of HEK293T transfectants (right panel) treated with non-targeted siRNA (NT, black squares) or siRNA directed against the endogenous mtRRF 3′-UTR with (open circles) or without (open triangles) concomitant inducible expression of FLAG-mtRRF. Numbers are a mean ± SEM of four independent wells. (C) In vivo mitochondrial proteins synthesis is partially affected by mtRRF depletion. HeLa cells were grown in the presence of control (NT) or targeted (mtRRF) siRNA. Cytosolic protein synthesis was inhibited with emetine prior to labelling mitochondrial proteins with 35S-methionine (15 min). Equal amounts of lysate (20 µg) were separated by 15% SDS–PAGE and gels analysed by PhosphorImager. Polypeptides designation is as described in ref. (23). (D) Steady-state levels of mtDNA-encoded proteins are affected by depletion of mtRRF. Western blots show analysis of cell lysates after 3 or 6 days siRNA treatment (mtRRF or NT) with antibodies against mitochondrial translation products or proteins sensitive to mitochondrial translation inhibition (cytochrome c oxidase subunits I and II—COX I and 2; complex I subunit NDUFB8) or nuclear-encoded complex II SDH 70 kDa protein and β-actin as a loading control. The blot accurately reflects three repeat experiments. (E) OXPHOS complexes are reduced at steady-state levels after mtRRF depletion. HeLa cells were exposed (6 days) to mtRRF- or NT-targeted siRNA prior to BN–PAGE. Complexes were visualized with antisera to complex I, anti-39 kDa; complex II, anti-SDH 70 kDa; complex III, anti-core 2, complex IV, anti-COII. The blot accurately reflects three experiments. SDH is a complex only comprising proteins encoded in the cytosol. (F) Respiratory coupling is modestly affected in cells depleted of human mtRRF. HeLa cells were exposed (3 days) to mtRRF-or NT-targeted siRNA prior to high resolution respirometry (as in Methods), three measures of respiratory control and capacity were made.
To further ensure that the growth phenotype was specific to mtRRF depletion, an siRNA designed against the 3′-UTR of mtRRF was introduced to HEK293T cells, concomitant with inducible expression of the exogenous FLAG-mtRRF lacking this 3′-UTR. Expression of mtRRF after transfection of the cells with 3′-UTR-specific mtRRF siRNA restored normal growth (Figure 4B) consistent with the growth phenotype being solely due to depletion of mtRRF.
Depletion of mtRRF partially affects mitochondrial translation and inhibits mitoribosomal disassembly
What is the mechanism causing the growth phenotype? It has been shown that in bacteria and yeast mitochondria, inactivation of RRF inhibits protein synthesis (12,29). To determine if the same is true for the mtRRF, we investigated the effect of protein depletion on an in vivo mitochondrial protein synthesis. In the presence of mtRRF siRNA, there was partial inhibition of mitochondrial protein translation (Figure 4C). Although the effect was not as dramatic as in S. cerevisiae Δrrf1 (29), it must be recognized that even partial translation defects result in instability and loss of mtDNA, which in turn produce a greater impairment of mitochondrial protein synthesis. The relative synthesis of most of the components was significantly altered (Figure 4C, data not shown). Consequently, the steady-state levels of mtDNA-encoded proteins were progressively affected as detected by western blot (Figure 4D). Although we did not observe significant reduction of the protein levels after short siRNA treatment (2–3 days), longer treatment resulted in decreased steady-state levels of mitochondrially encoded proteins of up to 40% when compared to non-targeting controls (day 6). These results were consistent with the decreased levels of fully assembled OXPHOS complexes after 6 days siRNA treatment, as revealed by Blue Native gel electrophoresis (Figure 4E). Moreover, polarographic measurements showed that respiration rates of the siRNA-treated cells decreased, consistent with affected function of OXPHOS complexes (Figure 4F).
To further decipher the exact mechanism of mtRRF function in mitochondrial translation, we looked at the sedimentation profile of HeLa cell mitoribosomes in isokinetic sucrose density gradients following mtRRF depletion. Western analysis of mitoribosomes from cells treated for 4 days with NT or mtRRF siRNA (Figure 5A) showed a striking contrast in distribution of DAP3 representing the small ribosomal subunit. mtRRF siRNA caused a shift from fractions 5–8 to mainly 7. This fraction has proteins from both mitochondrial ribosomal subunits and represents monosome accumulation. Moreover, only after mtRRF depletion where both ribosomal subunits detected in the heaviest fraction (F11), suggesting aggregation of the ribosomal proteins. We conclude that on depletion of mtRRF, monosomes are poorly dissociated into single subunits and non-functional monosomes accumulate, producing aggregates. These data are consistent with mtRRF functioning as a factor responsible for disassembly of ribosomes in human mitochondria.
Depletion of mtRRF causes increased mitochondrial ROS and changes in mitochondrial morphology
The observed aggregation of mitoribosomes may have a direct influence on mitochondrial function and may be a primary cause of cell death. Stress conditions in mitochondria are frequently associated with elevated levels of reactive oxygen species (ROS). To assess stress due to ROS accumulation in siRNA-treated cells, superoxide and peroxide/peroxynitrite levels were measured by mitoSOX and DHR, respectively. A statistically significant increase of ROS levels over the NT control was seen at day 3, a difference that was maintained through to day 6 (Figure 5B).
Increased ROS is often paralleled by augmented mitochondrial biogenesis and mtRRF depletion also resulted in a significant increase of mitochondrial mass as measured by NAO fluorescence, a dye that binds selectively to cardiolipin in the mitochondrial inner membrane (Figure 5B).
These abnormalities were reflected in the progressive morphological changes of mitochondria revealed by EM (Figure 5C 3 days and 6 days mtRRF siRNA) where the longer treatment resulted in a dramatic increase in mitochondrial density. Many more tightly packed membranous sheets were observed, which was consistent with increased mitochondrial biogenesis, whereas the numerous ‘black spots’ detected in the matrix compartment are consistent with mitoribosome aggregates.
DISCUSSION
Our studies presented here provide the first evidence that this candidate functions as the human mitochondrial ribosome recycling factor: (i) mtRRF is a mitochondrial protein, (ii) mtRRF is able to bind bacterial ribosomes in vitro and can tightly associate with human mitoribosomes in vivo, (iii) expression of human mtRRF suppresses the partial Δrrf1 respiratory deficiency in fission yeast in vivo and (iv) upon depletion of mtRRF, the relative level of mitochondrial monosomes increases whereas the free ribosomal subunits are reduced, indicating a defect in ribosome disassembly as has previously been reported on depletion of the bacterial counterpart (12).
Our results show that although mtRRF can associate tightly with mitoribosomes, the amount of interaction is enormously increased by pre-incubation. There are many possible hypotheses to explain this observation, but it is interesting to note that high resolution studies have shown that RRF binds bacterial ribosomes that have adopted a ratchet conformation at a site spanning both P and A site (15). Therefore, it may be speculated that tRNA bound to these sites may prevent interaction with mtRRF allowing occupancy only when these sites become vacant.
Why does expression of human mtRRF restore the respiratory function of an rrf1 mutant only in S. pombe and not in S. cerevisiae? Human mtRRF shows a stronger identity to S. pombe than the S. cerevisiae orthologue (EBI tools 21.1% versus 16.8%; Figure S4). There is an overall greater similarity between fission yeast and human mtRNAs (30) and these two species share a highly conserved set of general translation factors (18). In bacterial systems, functional studies with heterologous RRF and EFG proteins showed poor activity on the post-termination ribosomal complex compared to assays using RRF and EFG from the same species, underlining the importance of interplay between RRF and EFG [for example, (31,32)]. In light of these findings, it is also possible that human mtRRF was unable to rescue the growth phenotype of S. cerevisiae Δrrf1 due to a weak interaction with the budding yeast 70S mitoribosome or mtEFG that also has only limited similarity with the human mtEFG.
Depletion experiments revealed that mtRRF is essential for cell viability, with depletion causing gross mitochondrial dysmorphology and dysfunction. One of the primary effects of mtRRF depletion, which seemed to precede measurable translation abnormalities, was elevated ROS production (evident by day two post-transfection). The mechanisms that lead to mitochondrial ROS overproduction, although often associated with mitochondrial dysfunction are not always clear. Our recent work on depletion of the mitochondrial release factor mtRF1a also resulted in a profound increase in mitochondrial ROS without any measurable effect on mitochondrial translation (6). It is possible that increased ROS production could be a direct result of mitoribosome aggregation, but other subtle indirect effects cannot be ruled out. Our results are another example that even a small disruption of interplay between mitochondrial translation, respiration and ROS production may lead to profound cellular dysfunction.
In this study, lack of ribosome recycling led to a variety of deleterious downstream consequences. Shortage of available functionally active mitoribosomes eventually affected translation, and subsequently resulted in decreased steady-state levels of mitochondrially encoded proteins, hence reduced amounts of fully assembled complexes (6 days post-transfection). It was surprising, however, that a measurable decrease in mitochondrial protein synthesis took so long to manifest. It is possible that mitoribosomes are in excess in human organelles (33) with translation being initiated by mitoribosomes that had spontaneously disassembled. A similar process could explain the partial growth defect in yeasts lacking RRF. Another intriguing explanation could be that translation may be able to initiate from monosomes that have terminated translation, processed back along the mt-mRNA in a 3′ to 5′manner to reach the 5′-terminus and then scanned to locate the closest initiation codon, all without the monosome being dissociated. Although controversial, it must be considered that there is currently no model to explain how the mitoribosome naturally locates to the mt-mRNA initiation codon in the absence of a 5′-cap or Shine–Dalgarno sequence.
Finally, proteomic analysis of the factors that were eluted from immunoprecipitated mtRRF-FLAG, revealed 73 MRPs, several translation-associated factors and putative nucleoid proteins on the production and protease/DNase treatment of mitoplasts (Table 1). Nucleoids are dynamic structures containing mtDNA and proteins involved in maintenance, replication and transcription of the mitochondrial genome [reviewed in (34)]. These structures are well described in bacteria and close association between nucleoids and ribosomes has been previously measured in the Gram positive bacteria Bacillus subtilis, with this association being dependent on active transcription (35). Coupling of transcription and translation in human mitochondria has been frequently suggested (36,37), but there has been only one report showing interaction of mtRNA polymerase with a mitoribosomal protein, MRPL12, an interaction that was shown in vitro to stimulate transcription (38). In a recent report on proteins associated with nucleoids, 15 mitoribosomal proteins were identified, along with assorted factors that also function in protein synthesis (25). This observation is consistent with our data, but the authors were concerned that MRPs may have been present due to adventitious co-purification. Such a criticism could also be levelled at our observation, particularly as we were unable to completely remove cytosolic contamination, even in the mitoplasting experiments. Without further experimentation, this remains only tentative evidence of a direct physical link between mitoribosome and nucleoid. However, these proteomic studies also revealed several other candidate proteins that may be associated with the translational machinery and/or nucleoids. These findings open exciting avenues for further investigations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
AFM and ANR (grant JCJC06-0163 to I.K., M.G. and N.B.); MEST-CT-FP6-503684 (to J.R. and M.W.); EUMITOCOMBAT (LSHM-CT-2004-503116 to J.W., J.S. and R.N.L.). Funding for open access charge: Wellcome Trust Grant 074454/Z/04/Z.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank L Spremulli (North Carolina, USA) for the initial construct. TD Fox for anti-Arg8Sc antibody and M Yoshida for the S. pombe RRF-FLAG2His6 plasmid. R.N.L. and Z.M.A.C.-L. wish to thank the Wellcome Trust for continuing support.
REFERENCES
- 1.Anderson S, Bankier AT, Barrell BG, De Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 2.Pel HJ, Grivell LA. Protein synthesis in mitochondria. Mol. Biol. Rep. 1994;19:183–194. doi: 10.1007/BF00986960. [DOI] [PubMed] [Google Scholar]
- 3.Chung HK, Spremulli LL. Purification and characterization of elongation factor G from bovine liver mitochondria. J. Biol. Chem. 1990;265:21000–21004. [PubMed] [Google Scholar]
- 4.Koc EC, Spremulli LL. Identification of mammalian mitochondrial translational initiation factor 3 and examination of its role in initiation complex formation with natural mRNAs. J. Biol. Chem. 2002;277:35541–35549. doi: 10.1074/jbc.M202498200. [DOI] [PubMed] [Google Scholar]
- 5.Schwartzbach CJ, Spremulli LL. Bovine mitochondrial protein synthesis elongation factors. Identification and initial characterization of an elongation factor Tu-elongation factor Ts complex. J. Biol. Chem. 1989;264:19125–19131. [PubMed] [Google Scholar]
- 6.Soleimanpour-Lichaei HR, Kuhl I, Gaisne M, Passos JF, Wydro M, Rorbach J, Temperley R, Bonnefoy N, Tate W, Lightowlers R, et al. mtRF1a is a human mitochondrial translation release factor decoding the major termination codons UAA and UAG. Mol. Cell. 2007;27:745–757. doi: 10.1016/j.molcel.2007.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Spremulli LL. Identification and cloning of human mitochondrial translational release factor 1 and the ribosome recycling factor. Biochim. Biophys. Acta. 1998;1443:245–250. doi: 10.1016/s0167-4781(98)00223-1. [DOI] [PubMed] [Google Scholar]
- 8.Petry S, Weixlbaumer A, Ramakrishnan V. The termination of translation. Curr. Opin. Struct. Biol. 2008;18:70–77. doi: 10.1016/j.sbi.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Agrawal RK, Sharma MR, Kiel MC, Hirokawa G, Booth TM, Spahn CM, Grassucci RA, Kaji A, Frank J. Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: functional implications. Proc. Natl Acad. Sci. USA. 2004;101:8900–8905. doi: 10.1073/pnas.0401904101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janosi L, Shimizu I, Kaji A. Ribosome recycling factor (ribosome releasing factor) is essential for bacterial growth. Proc. Natl Acad. Sci. USA. 1994;91:4249–4253. doi: 10.1073/pnas.91.10.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janosi L, Ricker R, Kaji A. Dual functions of ribosome recycling factor in protein biosynthesis: disassembling the termination complex and preventing translational errors. Biochimie. 1996;78:959–969. doi: 10.1016/s0300-9084(97)86718-1. [DOI] [PubMed] [Google Scholar]
- 12.Hirokawa G, Demeshkina N, Iwakura N, Kaji H, Kaji A. The ribosome-recycling step: consensus or controversy? Trends Biochem. Sci. 2006;31:143–149. doi: 10.1016/j.tibs.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Peske F, Rodnina MV, Wintermeyer W. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol. Cell. 2005;18:403–412. doi: 10.1016/j.molcel.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 14.Zavialov AV, Hauryliuk VV, Ehrenberg M. Splitting of the posttermination ribosome into subunits by the concerted action of RRF and EF-G. Mol. Cell. 2005;18:675–686. doi: 10.1016/j.molcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 15.Barat C, Datta PP, Raj VS, Sharma MR, Kaji H, Kaji A, Agrawal RK. Progression of the ribosome recycling factor through the ribosome dissociates the two ribosomal subunits. Mol. Cell. 2007;27:250–261. doi: 10.1016/j.molcel.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Weixlbaumer A, Petry S, Dunham CM, Selmer M, Kelley AC, Ramakrishnan V. Crystal structure of the ribosome recycling factor bound to the ribosome. Nat. Struct. Mol. Biol. 2007;14:733–737. doi: 10.1038/nsmb1282. [DOI] [PubMed] [Google Scholar]
- 17.Minet M, Dufour ME, Lacroute F. Cloning and sequencing of a human cDNA coding for dihydroorotate dehydrogenase by complementation of the corresponding yeast mutant. Gene. 1992;121:393–396. doi: 10.1016/0378-1119(92)90150-n. [DOI] [PubMed] [Google Scholar]
- 18.Chiron S, Suleau A, Bonnefoy N. Mitochondrial translation: elongation factor tu is essential in fission yeast and depends on an exchange factor conserved in humans but not in budding yeast. Genetics. 2005;169:1891–1901. doi: 10.1534/genetics.104.037473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiron S, Gaisne M, Guillou E, Belenguer P, Clark-Walker GD, Bonnefoy N. Studying mitochondria in an attractive model: Schizosaccharomyces pombe. Methods Mol. Biol. 2007;372:91–105. doi: 10.1007/978-1-59745-365-3_7. [DOI] [PubMed] [Google Scholar]
- 20.Gaisne M, Bonnefoy N. The COX18 gene, involved in mitochondrial biogenesis, is functionally conserved and tightly regulated in humans and fission yeast. FEMS Yeast Res. 2006;6:869–882. doi: 10.1111/j.1567-1364.2006.00083.x. [DOI] [PubMed] [Google Scholar]
- 21.Steele DF, Butler CA, Fox TD. Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc. Natl Acad. Sci. USA. 1996;93:5253–5257. doi: 10.1073/pnas.93.11.5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nijtmans LG, Henderson NS, Holt IJ. Blue native electrophoresis to study mitochondrial and other protein complexes. Methods. 2002;26:327–334. doi: 10.1016/S1046-2023(02)00038-5. [DOI] [PubMed] [Google Scholar]
- 23.Chomyn A. In vivo labeling and analysis of human mitochondrial translation products. Methods Enzymol. 1996;264:197–211. doi: 10.1016/s0076-6879(96)64020-8. [DOI] [PubMed] [Google Scholar]
- 24.Ishino T, Atarashi K, Uchiyama S, Yamami T, Saihara Y, Yoshida T, Hara H, Yokose K, Kobayashi Y, Nakamura Y. Interaction of ribosome recycling factor and elongation factor EF-G with E. coli ribosomes studied by the surface plasmon resonance technique. Genes Cells. 2000;5:953–963. doi: 10.1046/j.1365-2443.2000.00382.x. [DOI] [PubMed] [Google Scholar]
- 25.Bogenhagen DF, Rousseau D, Burke S. The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 2008;283:3665–3675. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- 26.He J, Mao CC, Reyes A, Sembongi H, Di Re M, Granycome C, Clippingdale AB, Fearnley IM, Harbour M, Robinson AJ, et al. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J. Cell. Biol. 2007;176:141–146. doi: 10.1083/jcb.200609158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Bogenhagen DF. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 2006;281:25791–25802. doi: 10.1074/jbc.M604501200. [DOI] [PubMed] [Google Scholar]
- 28.Kanai T, Takeshita S, Atomi H, Umemura K, Ueda M, Tanaka A. A regulatory factor, Fil1p, involved in derepression of the isocitrate lyase gene in Saccharomyces cerevisiae–a possible mitochondrial protein necessary for protein synthesis in mitochondria. Eur. J. Biochem. 1998;256:212–220. doi: 10.1046/j.1432-1327.1998.2560212.x. [DOI] [PubMed] [Google Scholar]
- 29.Teyssier E, Hirokawa G, Tretiakova A, Jameson B, Kaji A, Kaji H. Temperature-sensitive mutation in yeast mitochondrial ribosome recycling factor (RRF) Nucleic Acids Res. 2003;31:4218–4226. doi: 10.1093/nar/gkg449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schafer B, Hansen M, Lang BF. Transcription and RNA-processing in fission yeast mitochondria. RNA. 2005;11:785–795. doi: 10.1261/rna.7252205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito K, Fujiwara T, Toyoda T, Nakamura Y. Elongation factor G participates in ribosome disassembly by interacting with ribosome recycling factor at their tRNA-mimicry domains. Mol. Cell. 2002;9:1263–1272. doi: 10.1016/s1097-2765(02)00547-6. [DOI] [PubMed] [Google Scholar]
- 32.Rao AR, Varshney U. Specific interaction between the ribosome recycling factor and the elongation factor G from Mycobacterium tuberculosis mediates peptidyl-tRNA release and ribosome recycling in Escherichia coli. EMBO J. 2001;20:2977–2986. doi: 10.1093/emboj/20.11.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King MP, Attardi G. Post-transcriptional regulation of the steady-state levels of mitochondrial tRNAs in HeLa cells. J. Biol. Chem. 1993;268:10228–10237. [PubMed] [Google Scholar]
- 34.Kucej M, Butow RA. Evolutionary tinkering with mitochondrial nucleoids. Trends Cell. Biol. 2007;17:586–592. doi: 10.1016/j.tcb.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 35.Mascarenhas J, Weber MH, Graumann PL. Specific polar localization of ribosomes in Bacillus subtilis depends on active transcription. EMBO Rep. 2001;2:685–689. doi: 10.1093/embo-reports/kve160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol. Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 37.Fontanesi F, Soto IC, Horn D, Barrientos A. Assembly of mitochondrial cytochrome c-oxidase, a complicated and highly regulated cellular process. Am. J. Physiol. Cell Physiol. 2006;291:C1129–C1147. doi: 10.1152/ajpcell.00233.2006. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z, Cotney J, Shadel GS. Human mitochondrial ribosomal protein MRPL12 interacts directly with mitochondrial RNA polymerase to modulate mitochondrial gene expression. J. Biol. Chem. 2007;282:12610–12618. doi: 10.1074/jbc.M700461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koc EC, Burkhart W, Blackburn K, Moyer MB, Schlatzer DM, Moseley A, Spremulli LL. The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J. Biol. Chem. 2001;23:43958–69. doi: 10.1074/jbc.M106510200. [DOI] [PubMed] [Google Scholar]
- 40.CadverKoc E, Burkhart W, Blackburn K, Moseley A, Spremulli LL. The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J. Biol. Chem. 2001;276:19363–19374. doi: 10.1074/jbc.M100727200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.