Abstract
Severe acute respiratory syndrome (SARS) is an infectious disease caused by the human coronavirus, SARS-CoV. The main viral protease, SARS 3CLpro is a validated target for the development of antiviral therapies. Since the enzyme is a homodimer and the individual monomers are inactive, two approaches are being used to develop inhibitors: enzyme activity inhibitors that target the active site and dimerization inhibitors. Dimerization inhibitors are usually targeted to the dimerization interface and need to compete with the attractive forces between subunits in order to be effective. In this paper, we show that the dimerization of SARS 3CLpro is also under allosteric control and that additional and energetically more favorable target sites away from the dimerization interface may also lead to subunit dissociation. We previously identified a cluster of conserved serine residues (Ser139, Ser144 and Ser147) located adjacent to the active site of 3CLpro that could effectively be targeted to inactivate the protease. Mutation of any of these serine residues to alanine had a debilitating effect on the catalytic activity of 3CLpro. In particular, the mutation of Ser147, which does not make any contact with the opposing subunit and is located approximately 9Å away from the dimer interface, totally inhibited dimerization and resulted in a complete loss of enzymatic activity. The finding that residues away from the dimer interface are able to control dimerization, defines alternative targets for the design of dimerization inhibitors.
Keywords: SARS, SARS 3CLpro, protease, SARS-CoVMpro, Dimerization, Analytical Ultracentrifugation, Calorimetry, Thermodynamics
Severe acute respiratory syndrome (SARS) is an upper respiratory tract disease caused by a novel human coronavirus, SARS-CoV (1-3). Since the emergence of the virus in late 2002, over 8,000 people have been infected, resulting in a mortality exceeding 50% for patients over age 65 with an estimated 10% mortality overall. Currently there are no effective treatments available against SARS. The recent identification of a SARS-like coronavirus (SL-CoV) circulating in the Chinese horseshoe bat population emphasizes the possibility of a reemergence of infection, calling for the development of antiviral therapies targeting SARS-CoV (4). The SARS-CoV main protease, 3CLpro (also called Mpro) has been identified as a key target for drug development. SARS 3CLpro plays a pivotal role in the life cycle of the coronavirus by the extensive proteolytic processing of two highly conserved polyproteins: pp1a (486 kDa) and pp1ab (790 kDa), which encode for essential viral replicase proteins. The functional importance of 3CLpro in the coronavirus life cycle makes it a highly attractive target for drug design. Several types of small molecule inhibitors targeting 3CLpro have been identified either by high-throughput screening, computational docking or structure-based drug design, some of which include bifunctional aryl boronic acid compounds, substrate analogue inhibitors, HIV protease inhibitors and dimerization inhibitors based on the N-terminal residues of 3CLpro (5-18).
In all available crystal structures SARS 3CLpro is a 68 kDa homodimer, with the two protomers of the dimer oriented almost perpendicular to one another, as shown in Figure 1A (18-21). Each monomer consists of three domains and contains a catalytic dyad of His41 and Cys145 located in a cleft between the first two domains. A water molecule is hydrogen bonded to His41; a position usually occupied by the third member of the catalytic triad in other cysteine proteases (18, 20, 21). The catalytic residues lie within a chymotrypsin-like double β-barrel fold comprised of the first two domains of the protein (residues 1-184). The catalytic domains are connected by a long loop region (residues 185-200) to a third globular domain composed of five anti-parallel α-helices (residues 201-306). A surface area of ~2300 Å (~1250Å apolar and 950Å polar) is buried at the dimer interface. The N-terminus, or “N-finger”, comprised of residues 1-8 makes extensive interactions with the second domain of the parent protomer and the third domain of the opposing monomer near the active site, thereby preserving the shape of the S1 pocket through hydrogen bond interactions with the oxyanion loop (18, 19). The positioning of these residues prompted speculation that the N-terminus is likely to be important in mediating dimerization and enzymatic activity of 3CLpro (18-21). The third alpha helical domain has also been proposed to mediate dimerization and substrate recognition either through positioning of the N-terminus or by holding domain two and the long loop connected to the third domain in a catalytically competent orientation (18, 20-22).
Figure 1.
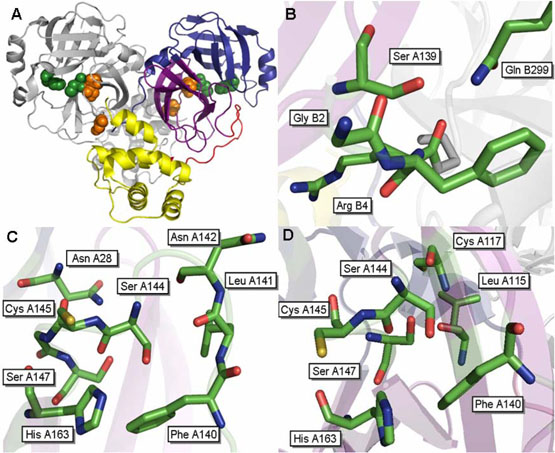
Structure of the SARS 3CLpro dimer at pH 7.6 [pdb file 1UK3 (18)]. (A) Wild type SARS 3CLpro displayed in ribbon representation. Domain 1 (residues 1-100) is shown in blue, domain 2 (residues 101-183) are colored purple, the loop connecting these two domains to domain 3 is colored red and domain 3 (residues 201-306) is shown in yellow. The catalytic residues, His41 and Cys145, are located in the cleft between the first two domains and are displayed as green spheres. The cluster of serines, Ser139, Ser144 and Ser147 are displayed as orange spheres. Important residues surrounding (B) S139A, (C) S144A and (D) S147A are shown.
Kinetic studies have demonstrated that the specific activity of SARS 3CLpro increases with protein concentration, indicating that the dimer is the functional form of the protein [(23, 24), this work]. The enzyme exists as a dimer in solution over a wide range of concentrations (12, 24-27). Reported dimerization dissociation constants (Kd) for SARS 3CLpro range from low nanomolar affinities (~0.35 nM) to high micromolar affinities (~230 μM). Differences in the values depend on the technique used to measure the Kd as well as the presence of N- or C-terminal extensions or affinity tags. In addition, handling of the protease during purification and storage appears to significantly affect the overall stability of the protease, thereby affecting activity and dimerization measurements.
Residues at the dimer interface that are essential for SARS 3CLpro dimerization and activity have been identified by several groups. The N- and C-termini of the protease are important for maintaining the oligomeric state of 3CLpro, with the majority of interactions being either ionic or hydrophobic. Chou et al (26) observed that both enzyme dimerization and activity decrease at higher salt concentrations and at lower pH, and that a salt bridge between Arg4 and Glu290 was important for these interactions. Mutation of Glu290 to Ala resulted in a complete loss of dimerization and activity while a mutation in Arg4 resulted in an approximately five-fold loss in dimerization and only a moderate loss of activity. Another study showed that deletion of the first three residues of the N-terminus resulted in only a moderate loss in activity and dimerization, while deletion of the first four residues of the N-terminus resulted in a dramatic loss in both areas, supporting the importance of Arg4 (28). Furthermore, it has been observed that deletion of the first seven residues at the N-terminus of 3CLpro interfered with protease activity and dimerization (16, 28), though one group reported diminished activity but no change in oligomeric state when the same mutation was made (29). Mutation of Met6 to Ala completely interferes with both dimerization and enzymatic activity (16). Met6 is proposed to form hydrophobic interactions with Tyr126 and Phe140 of the opposing chain. In addition, two peptides derived from the N-terminus of the protein have been shown to inhibit the activity of 3CLpro, one of which has been shown to prevent dimerization, supporting the idea that the N-terminus plays a role in maintaining the oligomeric state of 3CLpro (8, 16). Deletion of the third alpha helical domain also interferes with dimerization and activity [(27-29)and unpublished data from this laboratory]. The third domain alone has been observed to dimerize on its own, leading to the hypothesis that the role of this domain may be the regulation of enzymatic activity through dimerization (27).
All of the mutations affecting the dimerization of SARS 3CLpro reported so far involve residues which are located directly at the dimer interface. Here we provide data which demonstrates that dimerization is also controlled by long range cooperative interactions in 3CLpro. Mutation of conserved Ser147 to Ala, located approximately 9 Å away from the dimer interface, inhibited dimerization and resulted in an inactive enzyme. The location of Ser147 suggests alternative sites for SARS 3CLpro dimerization inhibitors.
EXPERIMENTAL PROCEDURES
Cloning
cDNA encoding full-length SARS 3CLpro (Tor2 strain, GenBank entry AY274119) was previously cloned into a pET 100 vector (Champion pET Directional TOPO Expression and Cloning Kit, Invitrogen) which carries an N-terminal polyhistidine tag, an enterokinase cleavage site, and ampicillin resistance (5). Mutations at selected positions (S139A, S144A and S147A) were introduced using an in vitro site directed mutagenesis kit (QuickChange, Stratagene) with the pET-SARS 3CLpro vector as the template. The sequences of the primers used to generate the mutants were as follows: S139A: 5′ CCA TTA AAG GTG CTT TCC TTA ATG G 3′; S144A: TTC CTT AAT GGA GCA TGT GGT AGT G 3′; and S147A: GGA TCA TGT GGT GCT GTT GGT TTT AAC ATT G 3′. Following the PCR reaction carried out with Pfu turbo DNA polymerase (Stratagene), the parent template was degraded with a DpnI (Stratagene) digestion reaction for 1 hour at 37 °C. One shot TOPO10 competent cells (Invitrogen) were transformed with the PCR product for plasmid amplification. The plasmid was then purified and mutations confirmed by DNA sequencing. This procedure was used to generate single, double and triple mutations of the serines to alanine using plasmids and primers corresponding to the residue change.
Protein Expression and Purification
Recombinant SARS 3CLpro was expressed as a soluble fraction in BL21 Star DE3 E. coli competent cells (Invitrogen). The construct begins with residue Ser1, and therefore does not contain the full N-terminal auto-cleavage site of the protein. Cells were grown in LB supplemented with ampicillin (50 μg/mL) at 37 °C, induced with 1 mM IPTG when the optical density (as determined by absorbance at 600 nm) was 0.8 or greater, and harvested after 4 hours. Cells were resuspended in lysis buffer (50 mM potassium phosphate (pH 7.8), 400 mM sodium chloride, 100 mM potassium chloride, 10% glycerol, 0.5% Triton-X, and 10 mM imidazole). The cells were broken by sonicating on ice for short pulses of one second followed by three seconds off for a total of 16 minutes. Cell debris was collected by centrifugation (20,000g at 4 °C for 45 min). The supernatant was filtered using a 0.45 μm pore size filter (Millipore) and applied directly to a nickel affinity column (HiTrap Chelating HP, Amersham Biosciences) that had been pre-equilibrated with binding buffer (50 mM sodium phosphate, 0.3 M sodium chloride, 10 mM imidazole, pH 8.0). The protease was eluted with a linear gradient of 50 mM sodium phosphate, 0.3 M sodium chloride, 250 mM imidazole, pH 8.0. After elution, the protein was buffer exchanged into 10 mM Tris-HCl pH 7.5, and loaded onto a Q-sepharose anion exchange column (Amersham Biosciences). The protease was eluted with a gradient of 10 mM Tris-HCl, 1 M NaCl, pH 7.5. The pooled fractions containing 3CLpro were buffer exchanged into storage buffer (10 mM sodium phosphate, 10 mM sodium chloride, 1 mM TCEP, 1 mM EDTA, pH 7.4) and digested for 24 h at 4 °C with enterokinase (Invitrogen, 0.1 units per 112 μg of protease) to remove the N-terminal polyhistidine tag. The enterokinase was removed by incubation with EK-away resin (Invitrogen). The reaction mixture was passed through a nickel affinity column to remove undigested protease. The protease was buffer exchanged into storage buffer, concentrated to 10 mg/mL and used immediately for experiments. The sample was more than 95% pure, as assessed by SDS-PAGE.
Kinetics
The catalytic activities of wild type and mutant SARS 3CLpro were determined using a fluorescence-based peptide cleavage assay with a commercially available fluorogenic substrate, Dabcyl-KTSAVLQSGFRKME-Edans (Genesis Biotech, Taiwan), which corresponds to the N-terminal auto-cleavage site of the protease (24). The change in fluorescence intensity was monitored in a Cary Eclipse fluorescence spectrophotometer (Varian) with 355 nm and 538 nm excitation and emission wavelengths respectively. The experiments were performed in 10 mM sodium phosphate, 10 mM sodium chloride, 1 mM EDTA, 1 mM TCEP, pH 7.4. Kinetic parameters such as Km and kcat were determined by initial rate measurement of substrate cleavage at 25 °C. The reaction was initiated by the addition of substrate such that final concentration varied from 1-60 μM.
Analytical Ultracentrifugation
Sedimentation velocity and equilibrium experiments were conducted using a Beckman-Coulter XL-I analytical ultracentrifuge. Wild type and mutant samples were prepared by exhaustive dialysis into 10 mM Tris-HCl, 0.1 M NaCl, 1 mM TCEP, 1 mM EDTA, pH 7.5 and then concentrated to 1 mg/mL. For sedimentation velocity experiments, reference (420 μL) and sample (400 μL) solutions were loaded into double-sector centerpieces and mounted in a Beckman An60Ti rotor. Protein concentrations for these experiments were between 0.25mg/ml (7.4 μM) and 1 mg/ml (29.5 μM). Experiments were performed at 20 °C with a rotor speed of 50,000 rpm. Sample absorbance at 280 nm was monitored in a continuous mode with no delay and a step size of 0.003 cm without averaging. Multiple scans at different time points were fit to a continuous size distribution using SEDFIT version 9.3b (30). The c(s) distribution in SEDFIT was exported to SEDPHAT where experimental values were corrected to S20, w (31). A partial specific volume of 0.7331 cm3/g for wild type 3CLpro and 0.7334 cm3/g for the S147A mutant were used in all calculations based on the amino acid sequence of the protein. The solvent density and viscosity were calculated using SEDNTERP (Philo, J. website http://www.jphilo.mailway.com/default.htm). Sedimentation equilibrium experiments were performed in the same buffer as sedimentation velocity experiments at concentrations between 0.1 and 2 mg/ml (between 2.9 μM and 59.1 μM). Reference (120 μl) and sample (110 μl) solutions were loaded into six sector centerpieces and mounted in a Beckman An60Ti rotor. Samples were equilibrated at 20 °C and three speeds: 15,000 rpm, 20,000 rpm and 25,000 rpm. Sample absorbance at 280 or 250 nm was collected in step mode with a step size of 0.001 cm and 10 replicates at each radial position. The program WinMatch (Biotechnology-Bioservices Center, University of Connecticut) was used to determine the point at which samples reached equilibrium. Equilibrium was typically attained for all samples between 24 and 30 hours. All samples were visually inspected for clarity to ensure no precipitation had occurred during the runs. Dissociation constants were determined by global fitting of multiple concentrations and rotor speeds in SEDPHAT using the monomer-dimer equilibrium model (32).
Differential Scanning Calorimetry
The heat capacities of wild type 3CLpro and the serine to alanine mutants were measured as a function of temperature with a high-precision differential scanning VP-DSC microcalorimeter (Microcal Inc., Northampton, MA). Protein samples and reference solutions were properly degassed and carefully loaded into the cells to avoid bubble formation. Thermal denaturation scans were performed with freshly prepared buffer-exchanged protease solutions using a temperature scan rate of 1°C/min. Data was analyzed by software developed in this laboratory.
RESULTS AND DISCUSSION
Enzymatic Activity
We previously identified a highly conserved cluster of serine residues: Ser139, Ser144 and Ser147, which lie adjacent to the active site of SARS 3CLpro (5). The location of this serine cluster in the structure of 3CLpro is shown in Figure 1A. Serines 139 and 147 are absolutely conserved in all known coronavirus 3CL proteases, while Ser144 is conserved in 55% of the reported sequences.
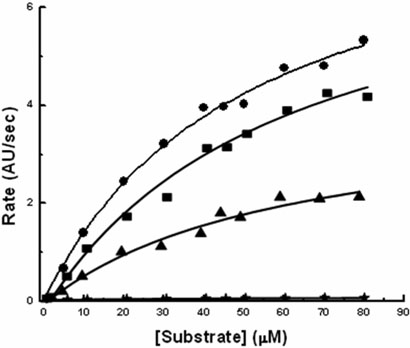
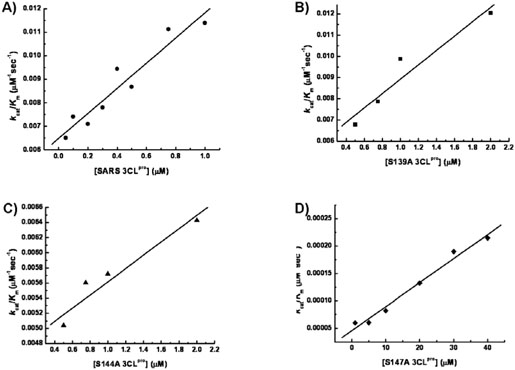
The role played by the three serine residues in the catalytic activity of SARS 3CLpro was evaluated by generating serine to alanine mutants at the three positions. The catalytic activities of wild type 3CLpro and the mutants S139A, S144A and S147A were measured at 1 μM enzyme concentration using a fluorescence-based cleavage assay in order to obtain the catalytic rate constants (kcat) and the Michaelis constant (Km). The results are shown in Figure 2. The wild type 3CLpro has a Km of 56.42 ± 9.18 μM and a kcat of 0.643 ± 0.051 sec−1. The resulting Km and kcat are of the same magnitude as those previously reported with the same substrate(24). Table 1 shows the kinetic parameters obtained for wild type, S139A, S144A and S147A. All of the proteases exhibited very similar Km's, suggesting that their ability to bind the substrate with similar affinities was not impaired. S139A had a catalytic constant, kcat, similar to the wild type while the S144A value was about half. The most significant effect was observed for S147A which had a 160-fold drop in kcat with respect to the wild type enzyme. Consistent with previously published observations(23, 33), the activity of the wild type SARS 3CLpro and the three mutants showed a linear correlation with increasing enzyme concentration as shown in Figure 3.
Figure 2.
The enzymatic activities of wild type 3CLpro (●), S139A (■), S144A (▲), and S147A (♦) were determined at 25°C with an enzyme concentration of 1 μM in 10mM NaCl, 10mM NaPi, 1mM EDTA, 1mM TCEP (pH 7.4). The initial velocity, which was measured as arbitrary fluorescence units per second, is plotted as a function of substrate concentration.
Table 1.
Summary of Enzyme Kinetics, Structural Stability and Dimerization Data
| Protein | Km (μM)a | kcat (sec−1) a |
kcat/ Kma (mM−1 ·sec−1) |
Tm (°C) b | ΔTm b (°C) |
Kd (μM) c |
|---|---|---|---|---|---|---|
| WT | 56.42 ± 9.18 | 0.643 ± 0.051 | 11.4 | 56.50 ± 0.03 | 0 | 1.3 ± 1.8 |
| S139A | 61.35 ± 9.38 | 0.615 ± 0.102 | 10.02 | 55.58 ± 0.02 | − 0.92 | 12.9 ± 1.7 |
| S144A | 60.21 ± 10.64 | 0.344 ± 0.046 | 5.71 | 58.40 ± 0.01 | + 1.90 | 3.3 ± 1.4 |
| S147A | 52.87 ± 13.24 | 0.004 ± 0.001 | .075 | 54.78 ± 0.02 | − 1.72 | --- |
Kinetic values were obtained by global non-linear least squares fit of initial rate measurements at increasing substrate concentrations at 1 μM enzyme concentration.
Structural stability values were obtained by fitting excess heat capacity versus temperature data using software from this laboratory.
Dimerization Kd values were calculated from the global analysis of sedimentation equilibrium data at concentrations of 0.1 mg/ml (2.9 μM), 0.25 mg/ml (7.4 μM), 0.5 mg/ml (14.8 μM), 0.75 mg/ml (22.1 μM), 1 mg/ml (29.5 μM) and 2 mg/ml (59.1 μM) for each protein using the monomer-dimer self-association model using the program SEDPHAT(32).
Figure 3.
The catalytic efficiencies (kcat/Km) of (A) wild type 3CLpro, (B) S139A, (C) S144A and (D) S147A are plotted as a function of increasing protease concentration. The kcat and km values were obtained by global non-linear least square fit of the initial rate measurements at increasing substrate concentrations. The experiments were performed with in experimental conditions similar to that shown in Fig.2.
Structural Stability of Wild Type 3CLpro and Serine Mutants
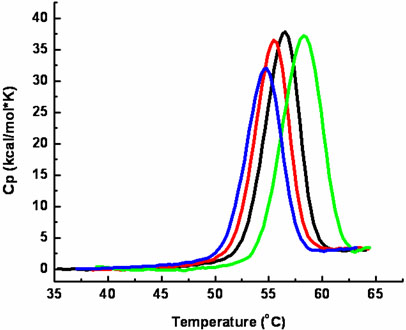
To verify that the mutants were properly folded and to assess the effect of the serine to alanine mutations, the structural stability of the wild type and mutant proteases was measured by differential scanning calorimetry. Figure 4 shows the temperature dependence of the heat capacity function of wild type 3CLpro, S139A, S144A and S147A at pH 7.4 and 10 mM NaCl. The denaturation transition for wild type 3CLpro is centered at 56.50 ± 0.1°C at a protein concentration of 0.2 mg/mL (5.9 μM) and temperature scanning rate (1°C/min) used in these experiments. This value is approximately 4°C higher than our previously reported value (5) which was measured using a construct containing five additional amino acids at the N-terminus of the protein. The transition temperature is similar to the value obtained by circular dichroism (CD)(27), but lower than others reported values (~60°C) obtained at faster scanning rates (29), as expected for irreversible denaturation. The S139A mutant had a slight effect on the stability on the protease with a downward shift in Tm of 0.9°C. The S144A mutation resulted in a 1.9°C upward shift in the transition temperature. The S147A mutation resulted in a downward shift in the transition temperature of approximately 1.7°C. Besides the changes in denaturation temperature, these experiments confirmed that all four proteins were properly folded and indicate that the serine to alanine mutations did not significantly affect the overall stability of the recombinant proteins. Furthermore, similar results were obtained throughout the length of time required for other experiments (e.g. sedimentation equilibrium measurements) confirming the structural integrity of the proteins in all experiments presented here. The areas under the curves, ΔH, were also similar for all proteins, indicating the absence of any significant structural changes. A full thermodynamic analysis could not be performed with any of the proteins due to precipitation once the transitions were complete which lead to a lack of reversibility in the microcalorimetric scans. This irreversibility was observed regardless of buffer conditions used (pH values between 3 and 9; NaCl concentrations between 0 and 3M).
Figure 4.
Thermal denaturation of WT (black), S139A (red), S144A (green) and S147A (blue) 3CLpro as determined by differential scanning calorimetry. Excess heat capacity is plotted as a function of temperature. Calorimetric scans for both proteins were performed at identical concentrations (0.2 mg/mL) at a scanning rate of 1°C per minute.
Quaternary Structure of Wild Type 3CLpro and Serine Mutants
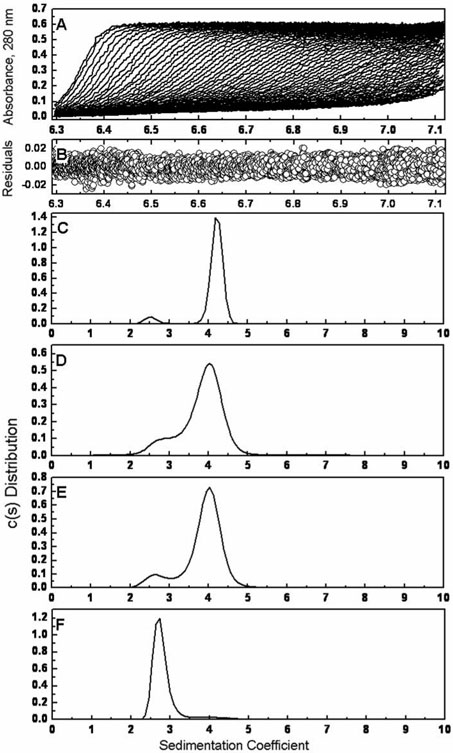
To investigate the effects of the serine to alanine mutations on the quaternary structure of SARS 3CLpro, sedimentation velocity ultracentrifugation was performed at 20°C on all constructs at concentrations between 0.25 mg/ml (7.4 μM) and 1 mg/ml (29.5 μM). Figure 5 shows typical sedimentation velocity profiles at 0.5mg/ml (14.8 μM) obtained using direct boundary modeling with the program SEDFIT (30). The experimental data is well accounted for by the continuous sedimentation coefficient model, c(s), as indicated by the plot of the residuals in Figure 5B. The sedimentation data for wild type 3CLpro in Figure 5C shows the presence of two peaks, at S1 = 2.5 and S2 = 4.0. These results are consistent with previous reported values in the literature (16, 26, 28). There did not appear to be a concentration dependence of the peak positions of c(s) in the concentration range tested. Similar results were obtained for the S139A and S144A proteins, shown in figures 5D and 5E. Despite being located directly at the dimer interface, Serine 139 does not play a dominating role in enzyme dimerization. Similarly, even though kinetic measurements show that S144A is less active than the wild type, the sedimentation profile was similar, albeit with a slight decrease in dimer population, suggesting that this mutation directly affects catalysis. Like the wild type, neither of these mutants showed a concentration dependence on the peak position of c(s).
Figure 5.
Sedimentation velocity ultracentrifugation of wild type 3CLpro and the serine mutants. The sedimentation of the four recombinant proteins was carried out with a Beckman Coulter XL-I analytical ultracentrifuge at 20°C and 50,000 rpm at concentrations between 0.25 mg/ml and 1 mg/ml. (A) Sedimentation velocity absorbance trace of WT 3CLpro at 280 nm. (B) Residuals of the experimental fit of WT 3CLpro at 0.5 mg/ml (14.8 μM). Continuous sedimentation coefficient distributions at 0.5 mg/ml of (C) WT 3CLpro, (D) S139A, (E) S144A and (F) S147A are shown.
Analysis of the S147A sedimentation velocity data unexpectedly produced a primary peak at 2.7S, as shown in Figure 5F. An additional small, broad peak can be seen at ~4.0S, corresponding to the formation of a minimal amounts of dimer. This peak can be seen to increase slightly with increasing protein concentration. The peak positions of this mutant also remained constant within the range of the protein concentrations tested. The presence of the S147A dimer is short lived however, as ultracentrifugation experiments performed later than twenty four hours after purification yielded solely a monomeric species. This correlates well with the lack of enzymatic activity observed in the kinetics measurements.
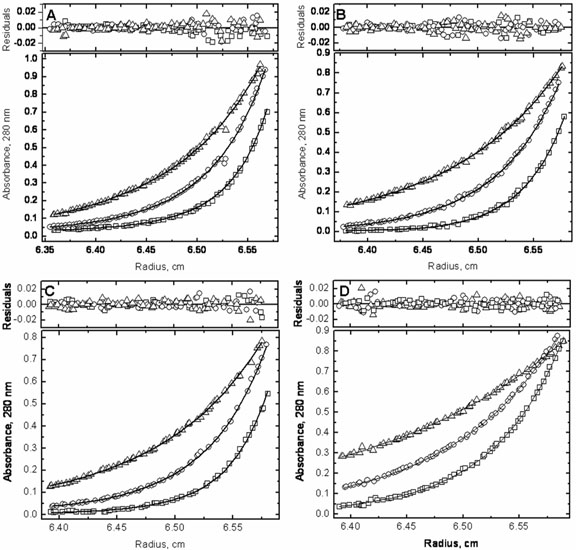
To obtain dimer dissociation constants for the four constructs, sedimentation equilibrium experiments were performed with wild type 3CLpro, S139A, S144A and S147A. Global analysis for each of the proteins at multiple concentrations between 0.1 mg/ml (2.9 μM) and 2 mg/ml (59.1 μM), and at multiple rotor speeds (15,000 rpm, 20,000 rpm, and 25,000 rpm) was carried out with the program SEDPHAT using the monomer-dimer self association model (32). Figure 6 shows the results of the global fitting of each set of data at 0.5 mg/ml (14.8 μM). All sets of experimental data fit well to the monomer-dimer model; the molecular weight obtained for the wild type monomeric species was 33,889 ± 441 daltons, which correlates exceedingly well with the expected monomeric weight of 33,846 kDa as estimated by the protein sequence. The dissociation constant for the wild type 3CLpro dimer was determined to be 1.3 ± 1.8 μM, which is slightly higher than previously reported values of 0.19 μM (26) and 0.28 μM (28) obtained using sedimentation velocity, and slightly lower than a recently reported Kd value of 14 μM obtained also by sedimentation equilibrium ultracentrifugation (16). The slight discrepancy between the two sedimentation equilibrium measurements may be due to the extra amino acid on the N-terminus of the protein used in the previously published work. It has been observed that additional amino acids on the N-terminus of the protease can increase the measured Kd value or interfere with dimerization completely [(34) and unpublished data from this laboratory]. If additional amino acids corresponding to a 3CLpro cleavage site are added to the N-terminus of the protein, the protein is able to cleave them off through trans-processing, albeit with a lower dimerization Kd. If the added residues do not correspond to a cleavage site, as with the previously published sedimentation equilibrium experiments, then the extra residue cannot be cleaved off and may have an effect on the dimerization Kd.
Figure 6.
Sedimentation equilibrium ultracentrifugation was carried out and globally analyzed with wild type 3CLpro and the three serine mutants at concentrations between 0.1 and 2 mg/ml (between 2.9 and 59.1 μM). The data sets at 0.5 mg/ml (14.8 μM) are shown for (A) wild type 3CLpro, (B) S139A, (C) S144A and (D) S147A. The lower graphs in each panel show the raw data at 15,000 rpm (Δ), 20,000 rpm (○), and 25,000 rpm (□) and the corresponding global fits displayed in a continuous line. The upper graphs in each panel show the residuals for the given fits.
Dissociation constants for the three mutants were also obtained, the results of which are shown in Figure 6 and Table 1. S139A showed a slight decrease in its ability to dimerize with a Kd of 12.9 ± 1.7 μM, and a molecular weight of the monomeric species calculated as 32,737 ± 342 daltons. The decrease in catalytic efficiency is less than the one expected from the weaker dimerization constant, suggesting a better intrinsic catalytic activity of the mutant over the wild type. On the other hand, the decrease in catalytic activity observed with S144A was not mirrored by a large decrease in dimerization, confirming the sedimentation velocity results. The dimerization Kd obtained for the mutant was 3.3 ± 1.4 μM and the molecular weight obtained for the monomer was 33,466 ± 1121 daltons. The reduction in S144A activity is apparently not due to a weaker dimerization, but to a direct effect on the catalytic activity of the enzyme. It must be noted that this enzyme is structurally more stable than the wild type (Table 1) and that this added stability might reflect itself in a lower catalytic efficiency.
The sedimentation equilibrium profile of S147A fit extremely well to a monomeric distribution, as evidenced by the well dispersed residuals seen in Figure 6D. The molecular weight obtained for the monomer was 32,641 ± 210 daltons, a value consistent with that obtained from the sedimentation velocity experiments. If we assume that only the dimer is active and that the S147A mutant dimer has an intrinsic kcat similar to that of the wild type, the Kd for S147A could be expected to be around 300 μM; i.e. more than two orders of magnitude weaker than the wild type.
In summary, we have carried out mutational analysis on a cluster of conserved serine residues (Ser139, Ser144 and Ser147) found adjacent to the active site of SARS 3CLpro. We examined the effects of separately mutating each of these residues to alanine on the activity, stability and oligomeric state of the protease using a fluorescence-based cleavage assay, differential scanning calorimetry, sedimentation velocity ultracentrifugation and sedimentation equilibrium sedimentation. Ser139, which makes interactions at the dimer interface, does not appear to play a significant role in maintaining the dimeric state of the protease, showing only slight decrease in activity, thermal stability and dimerization as compared to wild type 3CLpro when mutated to alanine. Ser144, located closer to the active site cavity, showed a two-fold decrease in catalytic efficiency when mutated to alanine compared to that of the wild type, but maintained a similar dimeric state and showed an increase in thermal stability by 1.9°C. Mutation of Ser147, located approximately 9 Å away from the dimer interface, to alanine resulted in an approximately 150-fold loss in catalytic efficiency in comparison to the wild type, and an inability of the protease to dimerize. This demonstrates that dimerization in SARS 3CLpro can be controlled by long range cooperative interactions, suggesting that targeting cavities nearby Ser147 with small molecule inhibitors may provide an alternative way of inactivating the protease.
Structural Analysis
According to the existing crystallographic structures, serine 139 is the only residue of the three considered in this paper directly located at the dimerization interface. Serines 144 and 147 are closer to the active site cavity and by visual inspection do not appear to play a key role in the dimerization of the enzyme. The interactions made by Ser139 are not bidirectional. Figure 1B shows the residues surrounding Ser139 of chain A at pH 7.5 [pdb structure 1UK3 (18)]. In this structure, Ser139 of chain A makes hydrogen bonds to the backbone of Gly2 from chain B. Conversely, no side chain interactions are made by Ser139 of chain B with residues in chain A. Mutation of Ser139 to Ala disrupts the dimerization interface and weakens the dimerization constant by about 10-fold but does not interfere directly in the catalytic activity of the enzyme.
Serine 144 is located closer to the active site in comparison to Ser139. Figure 1C shows the residues surrounding Ser144. The side chain hydroxyl of Ser144 forms hydrogen bonds with the side chain and backbone of Asn142. The backbone carbonyl of Ser144 interacts with the backbone amide of Ser147, and the side chain of Asn28. Mutation of Ser 144 to Ala results in a more structurally stable enzyme as reflected in the calorimetric data. The Km and the dimerization constant remain essentially the same while the catalytic constant, kcat, is reduced by half, suggesting that the structural rearrangement induced by the mutant reduces the catalytic efficiency of the enzyme.
The Ser147 to Ala mutation induces the most dramatic activity and structural effects even though this residue is 9Å away from the dimer interface. Ser147 makes several backbone and side chain interactions with residues which are involved in an extensive hydrogen bonding network within domains I and II in the protein. The backbone of Ser147 interacts with the backbones of residues Ser144 and His163. Incidentally, His163 is important for catalytic activity, since it comprises the S1 subsite that contributes to the specificity of substrate binding (18). The side chain of Ser147 makes two interactions with surrounding residues. The side chain hydroxyl of Ser147 hydrogen bonds to the backbones of Cys117 and Ser144. Cys117 forms two additional hydrogen bonds with Asn28 and Leu115. Asn28 may be important in maintaining the structural integrity in and around the active site region of the protease. This residue makes several key interactions, including two interactions with the backbone carbonyl of the catalytic residue Cys145. The hydrogen bonds made by Asn28 to the backbones of Cys117 and Gly120 may be critical in maintaining the position of the beta strand on which they reside, which is connected to and runs anti-parallel to the beta strand containing Tyr126. In turn, Tyr126 forms a hydrophobic pocket with Phe140 of the same chain and Met6 of the opposing chain. Met6 is the closest residue of the opposing monomer to Ser147, which is positioned approximately 9Å away. As previously mentioned, there is evidence suggesting that Met6 plays a role in maintaining dimerization, potentially through this hydrophobic interaction with Tyr126 and Phe140 (16). The loss of Asn28 interactions with Cys117 and Gly120 may result in the repositioning of this beta sheet and the disruption of this hydrophobic interaction. Additionally, structural rearrangements due to the loss of these interactions may also interfere with the hydrogen bond that Asn28 makes with the backbone carbonyl Gly143. Gly143 lies at the top of the loop which contains Phe140 which is part of the hydrophobic pocket. Furthermore, the elimination of the hydroxyl group in the S147A mutation not only eliminates the hydrogen bonds made by this group but also allows some rearrangements due to the smaller van der Waals volume of the Ala mutation.
Conclusions
These studies reveal the possibility of new strategies for engineering inhibitors of SARS 3CLpro by targeting sites that are away from the dimer interface but involved in the allosteric control of dimerization. Since Ser147 is conserved in all known coronavirus 3CL proteases, this observation opens the intriguing possibility that small molecules targeting this site may have a broad antiviral spectrum against infections caused by different coronaviruses.
Acknowledgements
We kindly thank Cynthia Wolberger for the use of her Beckman XL-I analytical ultracentrifuge.
Supported by the National Institutes of Health grant GM57144
ABBREVIATIONS
- SARS
Severe Acute Respiratory Syndrome
- SARS Co-V
Severe Acute Respiratory Syndrome Coronavirus
- 3CLpro
Chymotrypsin-Like Protease
- DMSO
Dimethyl sulfoxide
- SDS-PAGE
Sodium Dodecyl Sulfate Polyacrylamide Electrophoresis
- DSC
Differential Scanning Calorimetry
- CD
Circular Dichroism
- IPTG
Isopropyl-beta-D-thiogalactopyranoside
- TCEP
Tris(2-carboxyethyl)phosphine hydrochloride
- EDTA
Ethylenediaminetetraacetic acid
- EK
Enterokinase
REFERENCES
- 1.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling A-E, Humphrey CD, Shieh W-J, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang J-Y, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ, SARS Working Group A Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 2.Peiris J, Lai S, Poon L, Guan Y, Yam L, Lim W, Nicholls J, Yee W, Yan W, Cheung M. Coronavirus as a possible cause of severe acute respiratory syndrome. The Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peiris J, Chu C, Cheng V, Chan K, Hung I, Poon L, Law K, Tang B, Hon T, Chan C. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. The Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lau SKP, Woo PCY, Li KSM, Huang Y, Tsoi H-W, Wong BHL, Wong SSY, Leung S-Y, Chan K-H, Yuen K-Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. PNAS. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bacha U, Barrila J, Velazquez-Campoy A, Leavitt SA, Freire E. Identification of Novel Inhibitors of the SARS Coronavirus Main Protease 3CLpro. Biochemistry. 2004;43:4906–4912. doi: 10.1021/bi0361766. [DOI] [PubMed] [Google Scholar]
- 6.Blanchard JE, Elowe NH, Huitema C, Fortin PD, Cechetto JD, Eltis LD, Brown ED. High-Throughput Screening Identifies Inhibitors of the SARS Coronavirus Main Proteinase. Chemistry & Biology. 2004;11:1445–1453. doi: 10.1016/j.chembiol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu CM, Cheng VCC, Hung IFN, Wong MML, Chan KH, Chan KS, Kao RYT, Poon LLM, Wong CLP, Guan Y, Peiris JSM, Yuen KY. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59:252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding L, Zhang X-X, Wei P, Fan K, Lai L. The interaction between severe acute respiratory syndrome coronavirus 3C-like proteinase and a dimeric inhibitor by capillary electrophoresis. Analytical Biochemistry. 2005;343:159–165. doi: 10.1016/j.ab.2005.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh AK, Xi K, Ratia K, Santarsiero BD, Fu W, Harcourt BH, Rota PA, Baker SC, Johnson ME, Mesecar AD. Design and Synthesis of Peptidomimetic Severe Acute Respiratory Syndrome Chymotrypsinlike Protease Inhibitors. J. Med. Chem. 2005;48:6767–6771. doi: 10.1021/jm050548m. [DOI] [PubMed] [Google Scholar]
- 10.Jain RP, Pettersson HI, Zhang J, Aull KD, Fortin PD, Huitema C, Eltis LD, Parrish JC, James MNG, Wishart DS, Vederas JC. Synthesis and Evaluation of Keto-Glutamine Analogues as Potent Inhibitors of Severe Acute Respiratory Syndrome 3CLpro. J. Med. Chem. 2004;47:6113–6116. doi: 10.1021/jm0494873. [DOI] [PubMed] [Google Scholar]
- 11.Kao RY, Tsui WHW, Lee TSW, Tanner JA, Watt RM, Huang J-D, Hu L, Chen G, Chen Z, Zhang L. Identification of Novel Small-Molecule Inhibitors of Severe Acute Respiratory Syndrome-Associated Coronavirus by Chemical Genetics. Chemistry & Biology. 2004;11:1293–1299. doi: 10.1016/j.chembiol.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee T-W, Cherney MM, Huitema C, Liu J, James KE, Powers JC, Eltis LD, James MNG. Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide. Journal of Molecular Biology. 2005;353:1137–1151. doi: 10.1016/j.jmb.2005.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y-C, Huang V, Chao T-C, Hsiao C-D, Lin A, Chang M-F, Chow L-P. Screening of drugs by FRET analysis identifies inhibitors of SARS-CoV 3CL protease. Biochemical and Biophysical Research Communications. 2005;333:194–199. doi: 10.1016/j.bbrc.2005.05.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shie J-J, Fang J-M, Kuo T-H, Kuo C-J, Liang P-H, Huang H-J, Wu Y-T, Jan J-T, Cheng Y-SE, Wong C-H. Inhibition of the severe acute respiratory syndrome 3CL protease by peptidomimetic [alpha],[beta]- unsaturated esters. Bioorganic & Medicinal Chemistry. 2005;13:5240–5252. doi: 10.1016/j.bmc.2005.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shie J-J, Fang J-M, Kuo C-J, Kuo T-H, Liang P-H, Huang H-J, Yang W-B, Lin C-H, Chen J-L, Wu Y-T, Wong C-H. Discovery of Potent Anilide Inhibitors against the Severe Acute Respiratory Syndrome 3CL Protease. J. Med. Chem. 2005;48:4469–4473. doi: 10.1021/jm050184y. [DOI] [PubMed] [Google Scholar]
- 16.Wei P, Fan K, Chen H, Ma L, Huang C, Tan L, Xi D, Li C, Liu Y, Cao A, Lai L. The N-terminal octapeptide acts as a dimerization inhibitor of SARS coronavirus 3C-like proteinase. Biochemical and Biophysical Research Communications. 2006;339:865–872. doi: 10.1016/j.bbrc.2005.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu C-Y, Jan J-T, Ma S-H, Kuo C-J, Juan H-F, Cheng Y-SE, Hsu H-H, Huang H-C, Wu D, Brik A, Liang F-S, Liu R-S, Fang J-M, Chen S-T, Liang P-H, Wong C-H. Small molecules targeting severe acute respiratory syndrome human coronavirus. PNAS. 2004;101:10012–10017. doi: 10.1073/pnas.0403596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang H, Yang M, Ding Y, Liu Y, Lou Z, Zhou Z, Sun L, Mo L, Ye S, Pang H, Gao GF, Anand K, Bartlam M, Hilgenfeld R, Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. PNAS. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan J, Verschueren KHG, Anand K, Shen J, Yang M, Xu Y, Rao Z, Bigalke J, Heisen B, Mesters JR. pH-dependent Conformational Flexibility of the SARS-CoV Main Proteinase (Mpro) Dimer: Molecular Dynamics Simulations and Multiple X-ray Structure Analyses. Journal of Molecular Biology. 2005;354:25–40. doi: 10.1016/j.jmb.2005.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R. Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of Anti-SARS Drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 21.Anand K, Palm GJ, Mesters JR, Siddell SG, Ziebuhr J, Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha helical domain. The EMBO Journal. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ziebuhr J, Snijder EJ, Gorbalenya AE. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 23.Fan K, Wei P, Feng Q, Chen S, Huang C, Ma L, Lai B, Pei J, Liu Y, Chen J, Lai L. Biosynthesis, Purification, and Substrate Specificity of Severe Acute Respiratory Syndrome Coronavirus 3C-like Proteinase. J. Biol. Chem. 2004;279:1637–1642. doi: 10.1074/jbc.M310875200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo C-J, Chi Y-H, Hsu JT-A, Liang P-H. Characterization of SARS main protease and inhibitor assay using a fluorogenic substrate. Biochemical and Biophysical Research Communications. 2004;318:862–867. doi: 10.1016/j.bbrc.2004.04.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S, Chen L.-l., Luo H.-b., Sun T, Chen J, Ye F, Cai J.-h., Shen J.-k., Shen X, Jiang H.-l. Enzymatic activity characterization of SARS coronavirus 3C-like protease by fluorescence resonance energy transfer technique. Acta Pharmacologica Sinica. 2005;26:99–106. doi: 10.1111/j.1745-7254.2005.00010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chou C-Y, Chang H-C, Hsu W-C, Lin T-Z, Lin C-H, Chang G-G. Quaternary Structure of the Severe Acute Respiratory Syndrome (SARS) Coronavirus Main Protease. Biochemistry. 2004;43:14958–14970. doi: 10.1021/bi0490237. [DOI] [PubMed] [Google Scholar]
- 27.Shi J, Wei Z, Song J. Dissection Study on the Severe Acute Respiratory Syndrome 3C-like Protease Reveals the Critical Role of the Extra Domain in Dimerization of the Enzyme: DEFINING THE EXTRA DOMAIN AS A NEW TARGET FOR DESIGN OF HIGHLY SPECIFIC PROTEASE INHIBITORS. J. Biol. Chem. 2004;279:24765–24773. doi: 10.1074/jbc.M311744200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu W-C, Chang H-C, Chou C-Y, Tsai P-J, Lin P-I, Chang G-G. Critical Assessment of Important Regions in the Subunit Association and Catalytic Action of the Severe Acute Respiratory Syndrome Coronavirus Main Protease. J. Biol. Chem. 2005;280:22741–22748. doi: 10.1074/jbc.M502556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S, Chen L, Tan J, Chen J, Du L, Sun T, Shen J, Chen K, Jiang H, Shen X. Severe Acute Respiratory Syndrome Coronavirus 3C-like Proteinase N Terminus Is Indispensable for Proteolytic Activity but Not for Enzyme Dimerization: BIOCHEMICAL AND THERMODYNAMIC INVESTIGATION IN CONJUNCTION WITH MOLECULAR DYNAMICS SIMULATIONS. J. Biol. Chem. 2005;280:164–173. doi: 10.1074/jbc.M408211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuck P. Size-Distribution Analysis of Macromolecules by Sedimentation Velocity Ultracentrifugation and Lamm Equation Modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Analytical Biochemistry. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- 32.Vistica J, Dam J, Balbo A, Yikilmaz E, Mariuzza RA, Rouault TA, Schuck P. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Analytical Biochemistry. 2004;326:234–256. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 33.Shan Y-F, Li S-F, Xu G-J. A novel auto-cleavage assay for studying mutational effects on the active site of severe acute respiratory syndrome coronavirus 3C-like protease. Biochemical and Biophysical Research Communications. 2004;324:579–583. doi: 10.1016/j.bbrc.2004.09.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu M-F, Kuo C-J, Chang K-T, Chang H-C, Chou C-C, Ko T-P, Shr H-L, Chang G-G, Wang AH-J, Liang P-H. Mechanism of the Maturation Process of SARS-CoV 3CL Protease. J. Biol. Chem. 2005;280:31257–31266. doi: 10.1074/jbc.M502577200. [DOI] [PMC free article] [PubMed] [Google Scholar]