Abstract
Previous studies have shown that loss of the type B histone acetyltransferase Hat1p leads to defects in telomeric silencing in Saccharomyces cerevisiae. We used this phenotype to explore a number of functional characteristics of this enzyme. To determine whether the enzymatic activity of Hat1p is necessary for its role in telomeric silencing, a structurally conserved glutamic acid residue (Glu-255) that has been proposed to be the enzymes catalytic base was mutated. Surprisingly neither this residue nor any other acidic residues near the enzymes active site were essential for enzymatic activity. This suggests that Hat1p differs from most histone acetyltransferases in that it does not use an acidic amino acid as a catalytic base. The effects of these Hat1p mutants on enzymatic activity correlated with their effects on telomeric silencing indicating that the ability of Hat1p to acetylate substrates is important for its in vivo function. Despite its presumed role in the acetylation of newly synthesized histones in the cytoplasm, Hat1p was found to be a predominantly nuclear protein. This subcellular localization of Hat1p is important for its in vivo function because a construct that prevents its accumulation in the nucleus caused defects in telomeric silencing similar to those seen with a deletion mutant. Therefore, the presence of catalytically active Hat1p in the cytoplasm is not sufficient to support normal telomeric silencing. Hence both enzymatic activity and nuclear localization are necessary characteristics of Hat1p function in telomeric silencing.
Hat1p is a member of the GNAT2 family of histone acetyltransferases (1). This enzyme was originally identified in Saccharomyces cerevisiae but is broadly conserved throughout eukaryotes (2–8). Hat1p serves as a paradigm for type B histone acetyltransferases that were originally distinguished from type A histone acetyltransferases on the basis of a number of criteria (9). First, type B histone acetyltransferases have the ability to acetylate free histones but are inactive on nucleosomal substrates. Second, type B histone acetyltransferases are thought to be involved in the acetylation of newly synthesized histones that correlates with the process of chromatin assembly and hence are likely to function in the cytoplasm.
Consistent with its designation as a type B histone acetyltransferase, Hat1p was originally isolated from yeast cytoplasmic extracts (3). In addition, Hat1p can readily acetylate free histones but has no activity with nucleosomal histones as substrate. The histone specificity of Hat1p is also consistent with its identification as a type B histone acetyltransferase as the enzyme is specific for histone H4 lysines 5 and 12 (for recombinant yeast Hat1p), which matches the evolutionarily conserved pattern of acetylation found on newly synthesized histone H4 (2, 3, 10, 11).
Relative to most other histone acetyltransferases, which exist in large, multisubunit complexes, Hat1p is found in comparatively simple complexes in yeast cells (12). When isolated from the cytoplasm, yeast Hat1p is found associated with Hat2p (3). Hat2p is a WD repeat protein that is a homolog of the Rbap46/48 proteins that are components of a variety of chromatin-modifying complexes. The association of Hat1p with Hat2p is a conserved interaction as similar complexes have also been isolated from a number of eukaryotes (4–6, 13).
Although Hat1p was originally isolated from cytoplasmic extracts, subsequent studies demonstrate that this enzyme is predominantly nuclear in most organisms examined (4, 5, 13, 14). In S. cerevisiae, when in the nucleus, Hat1p exists in a distinct complex (referred to as the NuB4 complex) that contains Hif1p in addition to Hat2p (15–17). Hif1p is an H3/H4-specific histone chaperone that has chromatin assembly activity in vitro (15). It is not known whether a complex comparable to the yeast NuB4 complex also exists in other eukaryotes.
Surprisingly, evidence suggests that both the cytoplasmic and nuclear Hat1p-containing complexes are stably associated with histones H3 and H4. For example, Hat1p, Hat2p, and Kap123p are the primary proteins that co-purify with cytoplasmic histone H3/H4 (18). In addition, the purified NuB4 complex has also been co-purified with histones H3 and H4 (15). Further the association of histone H4 with Hif1p in the nucleus is dependent on the presence of Hat1p and Hat2p (15, 16). Together these results suggest that Hat1p may not be solely functioning in a catalytic capacity and that the association of Hat1p complexes with histones H3 and H4 may be involved in the import of these histones into the nucleus and their delivery to chromatin assembly factors.
Genetic analyses of HAT1 are consistent with a role for this enzyme in chromatin assembly. Deletion of the HAT1 gene by itself does not result in any observable phenotype (2, 3). The first phenotype associated with the loss of Hat1p was uncovered when a deletion of the HAT1 gene was combined with mutations in specific lysine residues in the histone H3 NH2-terminal tail. These mutants displayed a defect in telomeric silencing (19). Telomeric silencing is a phenomenon that results from the transcriptional repression that occurs when genes are in proximity to telomeric heterochromatin structure (20). Placing reporter genes such as URA3 or ADE2 near telomeres allows telomeric silencing to be used as a sensitive in vivo assay to monitor telomeric chromatin structure. Telomeric silencing can be affected by mutations in a number of chromatin modifying activities including several chromatin assembly factors (21–26). Subsequent studies also demonstrated that hat1Δ mutants (usually in combination with H3 NH2-terminal tail mutations) are also sensitive to DNA-damaging agents (27–29).
All of the previous in vivo characterizations of S. cerevisiae Hat1p have used gene deletions. In the present study, we used defined mutations to determine whether specific properties of the enzyme are necessary for the role of Hat1p in telomeric chromatin structure in vivo. These studies demonstrated that the catalytic activity of Hat1p is necessary for the in vivo function of the enzyme in telomeric silencing. However, Hat1p does not appear to use an acidic residue as the catalytic base in the enzyme reaction as predicted from structural studies of Hat1p and other histone acetyltransferases. In addition, the catalytic activity of Hat1p is not sufficient for its cellular function as cells expressing a catalytically active form of Hat1p that is excluded from the nucleus showed defective telomeric silencing. These results indicate that both its acetyltransferase activity and its nuclear localization are important for the in vivo function of Hat1p in telomeric silencing.
MATERIALS AND METHODS
Plasmids—The COOH-terminal portion of the S. cerevisiae HAT1 gene harboring a TAP tag was PCR-amplified from the genome of XAY1 (15) and cloned into the pCR 2.1 TOPO vector to generate pEM 6 according to manufacturer's instructions (Invitrogen). Glutamate to glutamine mutations at positions 162 (E162Q) and 255 (E255Q) and aspartate to asparagine substitution at position 256 (D256N) or the combination of E255Q and D256N were generated by site-directed mutagenesis of pEM 6 (QuikChange site-directed mutagenesis kit, Stratagene) resulting in vectors pEM 23, 9, 10, and 11. Mutant alleles were confirmed by DNA sequencing.
Strains—HAT1 was Myc-tagged at the COOH terminus in UCC1111 with and without a nuclear export signal (NES) derived from protein kinase inhibitor (PKI) to generate EMY31 and EMY35, respectively (19, 30) (Table 1). The presence of the epitope tag was confirmed by both PCR and Western blot using antibodies against c-Myc. Mutant versions of HAT1 harboring the TAP tag were amplified by PCR from pEM 23, 9, 10, and 11 and incorporated into the genome of UCC1111 (wild type) by standard LiAc transformation methods (31). Correct incorporation and expression of the mutant alleles were verified by PCR, sequencing, and Western blotting with peroxidase-anti-peroxidase complex (Sigma).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Ref. or source |
|---|---|---|
| UCC1111 | MAT adh4::URA3-TEL (VII-L) hhf1-hht1::LEU2 hhf2-hht2::MET15/pRS412 (ADE2 CEN ARS)-HHF2-HHT2 | Kelly et al. (19) |
| ASY50 | MAT adh4::URA3-TEL (VII-L) hhf1-hht1::LEU2 hhf2-hht2::MET15 hat1::LYS2/pRS412 (ADE2 CEN ARS)-HHF2-HHT2 | Kelly et al. (19) |
| XAY1 | MATα ade2–::hisG his3–200 leu2–0 lys2–0 met15–0 trp1–63 ura3–0 adh4::URA3-TEL (VII-L) hhf2-hht2::MET15 hhf1-hht1::LEU2, pRS412 (ADE2 CEN ARS)-HHF2-HHT2 HAT1-TAP Tag (TRP1) | This study |
| EMY31 | MATα ade2–::hisG his3–200 leu2–0 lys2–0 met15–0 trp1–63 ura3–0 adh4::URA3-TEL (VII-L) hhf2-hht2::MET15 hhf1-hht1::LEU2, pRS412 (ADE2 CEN ARS)-HHF2-HHT2 HAT1-Myc Tag (TRP1) | This study |
| EMY35 | MATα ade2–::hisG his3–200 leu2–0 lys2–0 met15–0 trp1–63 ura3–0 adh4::URA3-TEL (VII-L) hhf2-hht2::MET15 hhf1-hht1::LEU2, pRS412 (ADE2 CEN ARS)-HHF2-HHT2 HAT1-NES-Myc Tag (TRP1) | This study |
| EMY15 | MATα ade2–::hisG his3–200 leu2–0 lys2–0 met15–0 trp1–63 ura3–0 adh4::URA3-TEL (VII-L) hhf2-hht2::MET15 hhf1-hht1::LEU2, pRS412 (ADE2 CEN ARS)-HHF2-HHT2 HAT1E255Q-TAP Tag (TRP1) | This study |
| EMY26 | MATα ade2–::hisG his3–200 leu2–0 lys2–0 met15–0 trp1–63 ura3–0 adh4::URA3-TEL (VII-L) hhf2-hht2::MET15 hhf1-hht1::LEU2, pRS412 (ADE2 CEN ARS)-HHF2-HHT2 HAT1D256N-TAP Tag (TRP1) | This study |
| EMY28 | MATα ade2–::hisG his3–200 leu2–0 lys2–0 met15–0 trp1–63 ura3–0 adh4::URA3-TEL (VII-L) hhf2-hht2::MET15 hhf1-hht1::LEU2, pRS412 (ADE2 CEN ARS)-HHF2-HHT2 HAT1E255Q, D256N-TAP Tag (TRP1) | This study |
| EMY40 | MATα ade2–::hisG his3–200 leu2–0 lys2–0 met15–0 trp1–63 ura3–0 adh4::URA3-TEL (VII-L) hhf2-hht2::MET15 hhf1-hht1::LEU2, pRS412 (ADE2 CEN ARS)-HHF2-HHT2 HAT1E162Q-TAP Tag (TRP1) | This study |
Whole Cell Extracts—Yeast whole cell extracts from wild type, hat1 point mutant, and hat1Δ strains were prepared as described previously (32) with the following modification: extracts were dialyzed against DN(50) buffer (DN buffers contain 25 mm Tris, 0.1 mm EDTA, 10% glycerol, and the concentration (in mm) of NaCl given in parentheses) before use.
Column Chromatography—Dialyzed yeast whole cell extracts were centrifuged at 10,000 × g for 10 min. Resulting clarified extracts were applied to a HiTrap DEAE fast flow column (Amersham Biosciences) equilibrated with DN(50) buffer. The column was washed with 10 column volumes, and proteins were eluted with a 20-column volume gradient from DN(50) to DN(1000).
Western Blotting—Western blots were performed and visualized using an ECL Plus chemiluminescence detection kit according to the manufacturer's instructions (Amersham Biosciences). The signal was detected by scanning on a Storm PhosphorImager, and the intensity of specific protein bands was obtained with ImageQuant 5 software.
Histone Acetyltransferase Activity Assays—Liquid histone acetyltransferase assays were performed using free chicken histones or a synthetic peptide encoding the first 28 amino acids of the histone H4 NH2-terminal tail as the substrate (3). Reactions were performed in a final volume of 50 μl in buffer DN(50) containing 0.1 μm [3H]acetyl coenzyme A (6.1 Ci/mmol, ICN Biomedicals Inc.) and 1 mg/ml chicken erythrocyte core histones or 5 μg of H4 peptide. Chicken histones were purified as described previously (33). Reactions were incubated at 37 °C for either 30 or 60 min and then transferred to P-81 filters (Whatman). The filters were washed three times with 250 ml of 50 mm NaHCO3 (pH 9.0) for 10 min followed by a quick acetone rinse and then allowed to air dry. The amount of 3H bound to the filters was quantified by liquid scintillation counting. A portion of the assay mixture was also resolved by 18% SDS-PAGE, prepared for fluorography, and exposed to x-ray film.
Telomeric Silencing Assays—Telomeric silencing assays were performed as described previously (19). Isolated colonies of the indicated strains were used to inoculate overnight cultures in rich medium. Cultures were normalized by A600, collected by centrifugation, and resuspended 200 μl of water. 10-fold serial dilutions of the cell suspensions were made, and 10 μl of each dilution was spotted onto synthetic complete plates and synthetic complete plates containing 0.1% 5-fluoroorotic acid (5-FOA). The plates were then incubated for 3 days at 30 °C. At least three individual isolates of each strain were tested.
Immunofluorescence Microscopy—Immunofluorescence localization was performed as described previously with the following modification (15). Hat1p-Myc and Hat1p-NES-Myc were visualized with Cy3-conjugated secondary antibodies. A hat1Δ strain (ASY50) was used as a negative control.
RESULTS
Hat1p was originally isolated based on its ability to acetylate histones. However, an important outstanding question is what characteristics of this protein are important for its function in the cell. The goal of this study was to probe whether the catalytic activity and subcellular localization of Hat1p are necessary for the in vivo role of Hat1p in telomeric silencing.
Mutational Analysis of the Hat1p Active Site—To determine whether the histone acetyltransferase activity of Hat1p is essential for its function in telomeric silencing, we sought to generate point mutants that would inactivate the enzymatic activity of Hat1p. The mutational analysis of Hat1p is greatly facilitated by the presence of a crystal structure for this enzyme (34). This crystal structure was obtained in the presence of acetyl coenzyme A (Ac-CoA), which provides important information on the location of the active site of the enzyme.
The crystal structures of several other histone acetyltransferases have also been determined. These include other GNAT family enzymes such as P/CAF and Gcn5p as well as the MYST family histone acetyltransferase Esa1p (35–41). Surprisingly despite the fact that Hat1p displays almost no primary sequence similarity with these histone acetyltransferases, as noted by Marmorstein and co-workers (35, 36, 39), their active sites display a remarkable level of structural similarity. Of particular interest is the presence of a structurally conserved glutamic acid residue (Glu-255 in Hat1p, Glu-173 in yGcn5p, Glu-570 in P/CAF, and Glu-338 in Esa1p). For yGcn5p, P/CAF, and Esa1p, this acidic amino acid has been shown to function as an essential catalytic base (Fig. 1A) (36, 39, 42–44). This residue acts by extracting a proton from the Nε-lysine of the histone substrate. The deprotonated lysine can then participate in a nucleophilic attack on the Ac-CoA that leads to the transfer of the acetyl group to the histone. The importance of acidic amino acids serving as catalytic bases is underscored by the observation that the recently identified histone acetyltransferase Rtt109p also requires a specific aspartic acid residue for catalytic activity (45). The importance of Hat1p Glu-255 is also suggested by a phylogenetic analysis. Search of the NCBI data base with the Hat1p sequence identifies homologs in 84 eukaryotic organisms. Glu-255 is strictly conserved in every one of these Hat1p homologs (data not shown).
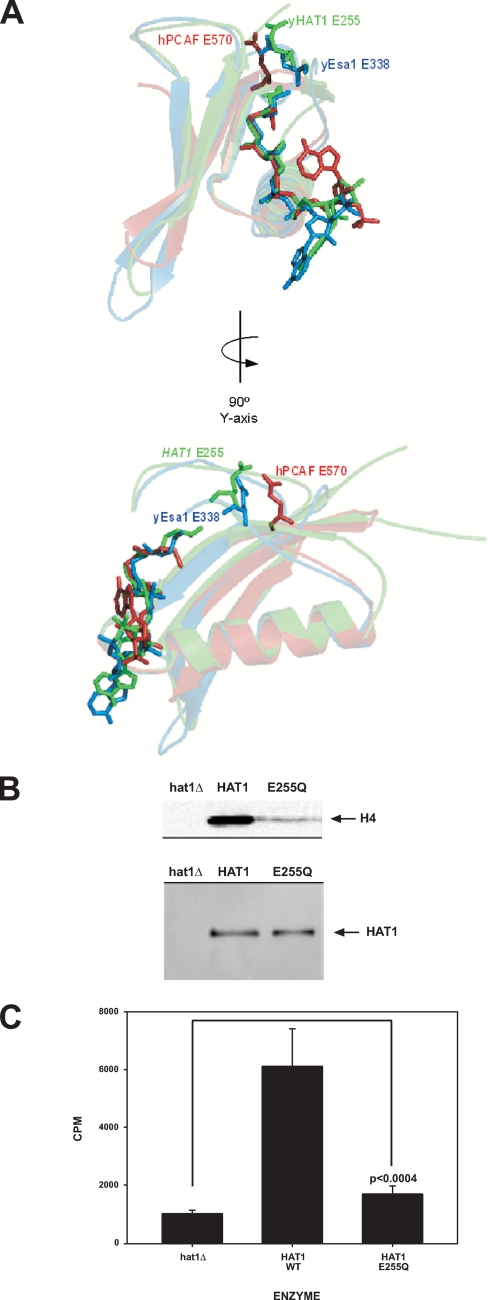
FIGURE 1.
Glu-255 is not an essential catalytic base for Hat1p. A, superimposition of the active sites of Hat1p (green), Esa1p (blue), and P/CAF (red). Acetyl-CoA and the conserved glutamic acid residues are shown in stick figure. Protein models were created using Pymol software (DeLano Scientific). B, extracts from hat1Δ, HAT1, wild type (WT), and HAT1 E255Q strains (as indicated) were fractionated by DEAE chromatography, and peak fractions were assayed for histone acetyltransferase activity using chicken erythrocyte histones and [3H]Ac-CoA as substrates. Reactions were resolved by SDS-PAGE, and the gels were processed for fluorography and exposed to film (top panel). Migration of histone H4 is indicated. The levels of Hat1p in the peak DEAE fractions were determined by Western blot analysis using peroxidase-anti-peroxidase complex to visualize Hat1p-TAP fusion (bottom panel). The absence of Hat1p in the hat1Δ extract was confirmed by Western blots probed with α-Hat1p antibodies. C, fractions used in B were also assayed with a peptide composed of the first 28 amino acids of yeast histone H4, and histone acetyltransferase activity was monitored by scintillation counting. Results are the average of six independent assays with the error bars representing the S.D. of the mean. p value was determined by unpaired t test of the E255Q mutant relative to the hat1Δ mutant as indicated.
These observations focused attention on Glu-255 of Hat1p as an obvious target for the generation of a mutant that would completely eliminate the catalytic activity of this enzyme. We chose to test the effect of Hat1p mutations on protein expressed in yeast and present in its native complex(es). Therefore, a site-directed mutant that changed HAT1 Glu-255 to glutamine was constructed on a DNA fragment that contained the HAT1 open reading frame with a COOH-terminal TAP tag and a TRP1 selectable marker. The fragment was then transformed into yeast and integrated into the genome at the normal HAT1 locus. Integration of the HAT1 mutation into the genome also helped ensure that the mutant was expressed at normal levels in the cell (see Fig. 3). Whole cell extracts were then prepared from wild type and the HAT1 E255Q strains, and the extracts were fractionated by DEAE chromatography. The bulk of cellular Hat1p eluted as a well defined peak from the DEAE column, and assays of this partially purified protein give a reliable measure of Hat1p activity because contaminants that inhibit the histone acetyltransferase assays in the crude extract are removed at this step (3). Also as expected, Hat1p-associated factors Hat2p and Hif1p co-eluted with Hat1p from the DEAE column.
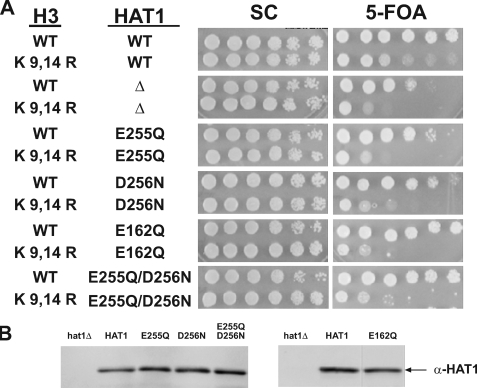
FIGURE 3.
HAT1 active site mutants are defective in telomeric silencing. A, point mutants of hat1 harboring the indicated histone H3 alleles were assayed for telomeric silencing. Telomeric silencing was measured by spotting 10-fold serial dilutions of cells on synthetic complete plates (SC) and synthetic complete plates containing 5-FOA. Plates were photographed after 3 days of growth at 30 °C. The relevant genotypes of the strains are indicated on the left. Assays were performed on at least three independent isolates from each strain. B, Western blot analyses were performed on normalized total protein samples to verify equivalent protein expression utilizing peroxidase-anti-peroxidase antibody. WT, wild type.
The peak Hat1p-containing fractions from the wild type and E255Q extracts were then assayed for histone acetyltransferase activity. In addition, an extract from a hat1Δ strain was fractionated in an identical fashion, and the equivalent DEAE fraction, based on conductivity, was assayed as a negative control. As seen in Fig. 1B, in assays using purified histones as substrates, the E255Q mutant clearly retains a significant level of histone acetyltransferase activity that is well above background. Assays were also performed with a peptide substrate that represents the first 28 amino acids of yeast histone H4. Again the HAT1 E255Q mutant had a reduced but detectable level of activity. Based on the peptide substrate assays, the E255Q mutation caused a ∼10-fold reduction in the enzymatic activity of Hat1p. This is in contrast to comparable mutations in Gcn5p and Esa1p that reduced catalytic activity to background levels (42, 44, 46, 47). These results show that, although this residue is important for the full activity of the enzyme, Glu-255 may not be an essential catalytic base for Hat1p despite its structural and phylogenetic conservation.
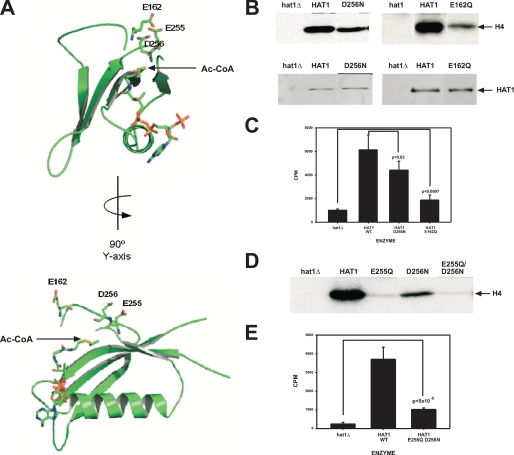
As all other GNAT and MYST histone acetyltransferases that have been examined use a glutamic acid residue as a catalytic base, we explored the crystal structure of Hat1p to identify other acidic amino acids that are in proximity to the active site and might serve as a catalytic base (34). This analysis identified two additional acidic residues, Asp-256 and Glu-162. The location of these amino acids in the active site of Hat1p is shown in Fig. 2A. Both of these residues lie near the acetyl group of the Ac-CoA moiety and hence could function in catalysis. These residues are also highly conserved evolutionarily. Of the 84 identified Hat1p homologs, Glu-162 is conserved in 70 (83%). Among the remainder, 13 have an aspartic acid residue at this position. In only one organism, rice, has an acidic amino acid not been retained at this position. For Asp-256, this residue is conserved in 75 of the 84 homologs (89%). Of the remainder, only five of the homologs do not have an acidic residue at this site.
FIGURE 2.
Mutational analysis of acidic amino acids in the Hat1p active site. A, a representation of the active site of Hat1p showing acetyl-CoA and acidic residues near the active site in stick figure. Protein models were created using Pymol software (DeLano Scientific). B–E, extracts from strains containing the indicated form of Hat1p were assayed for histone acetyltransferase activity using full-length histones (B and D) or a histone H4 NH2-terminal tail peptide (C and E). Samples were processed and analyzed as described in the legend to Fig. 1. WT, wild type. Error bars represent the S. D. p values were determined by unpaired t test of the indicated samples.
As above, partially purified Hat1p, containing D256N or E162Q mutations, was assayed using full-length histones or an H4 NH2-terminal tail peptide (Fig. 2, B and C). Mutation of Asp-256 had a moderate effect on Hat1p activity, whereas mutation of Glu-162 resulted in defects that were of a magnitude similar to those seen with the E255Q mutation. These results indicate that these residues, although also likely contributing to catalysis, are not essential catalytic bases for Hat1p.
The presence of adjacent acidic residues in the active site of Hat1p (Glu-255 and Asp-256) that are not observed in other GNAT or MYST enzymes raises the possibility that these residues might have redundant functions in Hat1p catalysis. To test this possibility, we expressed in yeast a form of Hat1p in which both residues are mutated (E255Q/D256N). As seen in Fig. 2D, the activity of this enzyme was significantly above background and is very similar to that seen for the E255Q single mutant when assayed with either full-length histones or with a histone H4 NH2-terminal tail peptide. This suggests that these residues do not play a redundant role in Hat1p catalysis. Hence our mutational analysis of acidic residues in the active site of Hat1p suggests that this enzyme may not function through the same mechanism seen with most histone acetyltransferases.
Impact of Hat1p Active Site Mutations on Telomeric Silencing—Monitoring the transcriptional repression of telomere proximal reporter genes (telomeric silencing) has proven to be a sensitive in vivo assay for many factors that modulate chromatin structure. To assess the role of the histone acetyltransferase activity of Hat1p in telomeric silencing, we determined the effect of the HAT1 E255Q, D256N, E162Q, and E255Q/D256N mutations on the expression of a URA3 reporter positioned near the telomere of the left arm of chromosome VII. This was assayed by spotting 10-fold serial dilutions of strains on plates in the presence and absence of 5-FOA. 5-FOA is toxic to cells expressing the URA3 gene (48). Hence cells with a telomereproximal URA3 gene can grow in the presence of 5-FOA because the gene is silenced by telomeric chromatin structure. However, mutations that disrupt this repressive chromatin environment lead to expression of URA3 and loss of viability in the presence of 5-FOA.
The role of Hat1p in telomeric silencing is at least partially redundant with specific lysine residues in the histone H3 NH2-terminal tail. Hence as demonstrated previously (and seen in Fig. 3A), combining a deletion of HAT1 with a mutation in histone H3 resulted in a marked decrease in viability on 5-FOA plates in strains containing a telomeric URA3 gene (15, 16, 19). To detect the effect of the HAT1 active site mutations on telomeric silencing, these mutations were also combined with the histone H3 mutations. Although, as described above, these residues are not essential for Hat1p catalysis, these mutations cause a range of defects in enzymatic activity. As seen in Fig. 3A, these mutations in HAT1 also resulted in significantly decreased viability in 5-FOA medium. The effect of these mutations was slightly less severe than that seen with the complete deletion. In addition, viability was slightly higher in the HAT1 D256N mutant relative to the E255Q, E162Q, and E255Q/D256N mutants. Importantly as shown in Fig. 3B, Hat1p levels were equal in all the HAT1 mutants. Hence the impact of the HAT1 active site mutations on telomeric silencing roughly correlates with their effect on the catalytic activity of the enzyme. Thus, the function of Hat1p in telomeric silencing appeared to be critically dependent on its catalytic activity because even the moderate loss of enzymatic activity observed with the D256N mutant resulted in a significant defect in telomeric silencing.
Hat1p Nuclear Localization Is Important for Telomeric Silencing—Traditionally the primary function of type B histone acetyltransferases has been viewed as the acetylation of newly synthesized histones in the cytoplasm (9). An expanded role for Hat1p in the metabolism of histones was suggested by the observation that, in many species, this enzyme is primarily localized to the nucleus (4, 5, 13, 14). Further, in yeast, Hat1p is found in a distinct complex in the nucleus (known as the NuB4 complex) that, in addition to Hat2p, contains the histone chaperone Hif1p (15, 16). Interestingly the NuB4 complex was also found to be stably associated with histones H3 and H4 (15). Coupled with the observation that the cytoplasmic Hat1p-Hat2p complex is stably associated with histone H4, these results suggest that the type B histone acetyltransferase complex may not be acting in a strictly catalytic capacity but also playing a role in the escort of histones from the cytoplasm into the nucleus and to sites of chromatin assembly (18).
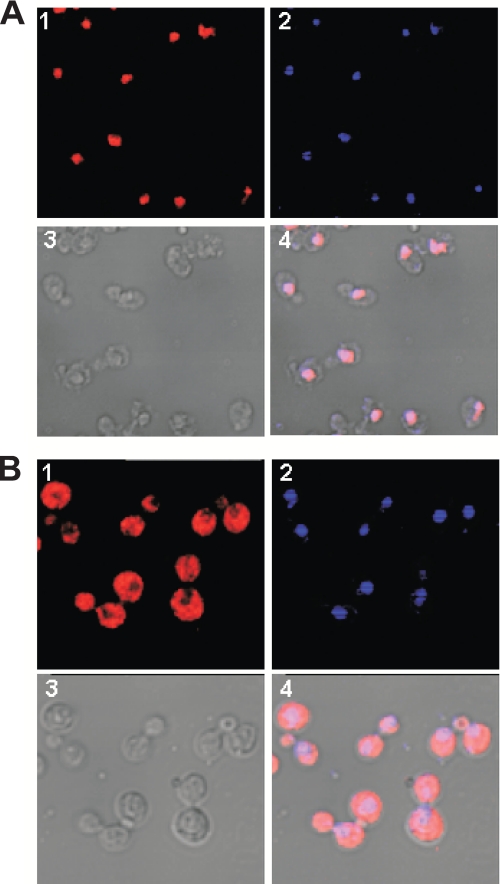
To test this model, we sought to determine whether the presence of a catalytically active form of Hat1p in the cytoplasm was sufficient for its in vivo function. To prevent the accumulation of Hat1p in the nucleus, we fused the NES of mammalian PKI as well as a Myc epitope to the COOH terminus of Hat1p. The PKI NES has been shown to be effective in relocalizing yeast nuclear proteins to the cytoplasm (30, 49). The effect of the PKI NES on Hat1p localization was then determined by immunofluorescence microscopy. Fig. 4A shows the localization of Myc-tagged wild type Hat1p. In agreement with previous reports, Hat1p was found to be a predominantly nuclear protein (15, 16). The PKI NES had a dramatic effect on the subcellular localization of Hat1p. As seen in Fig. 4B, there was a significant increase in the amount of Hat1p in the cytoplasm. In addition, Hat1p appeared to be largely excluded from the nucleus because there was a marked absence of staining in the region of the cell that corresponded to the nucleus.
FIGURE 4.
The NES of PKI effectively excludes Hat1p from the nucleus. Subcellular localization of Hat1p was determined by indirect immunofluorescence and confocal microscopy. Cells from strain EMY31 (HAT1-Myc; A) or EMY35 (HAT1-Myc-NES; B) were probed with anti-Myc primary antibody. Hat1p was visualized with a Cy3-conjugated secondary antibody. The images shown are Cy3 (1), 4′,6-diamidino-2-phenylindole (2), differential interference contrast (3), and merge (4).
Although the PKI NES was effective at relocalizing Hat1p to the cytoplasm, it was important to confirm that the presence of the NES did not decrease the catalytic activity of the enzyme. Hat1p was partially purified from the HAT1-Myc and HAT1-Myc-NES strains as described above. Histone acetyltransferase assays performed with these Hat1p preparations indicated that the NES did not affect the catalytic activity of Hat1p. It has been demonstrated previously that the absence of Hat2p causes a 10-fold decrease in the catalytic activity of Hat1p (3, 4). Hence this result also indicated that the cytoplasmically localized Hat1p had retained its association with Hat2p.
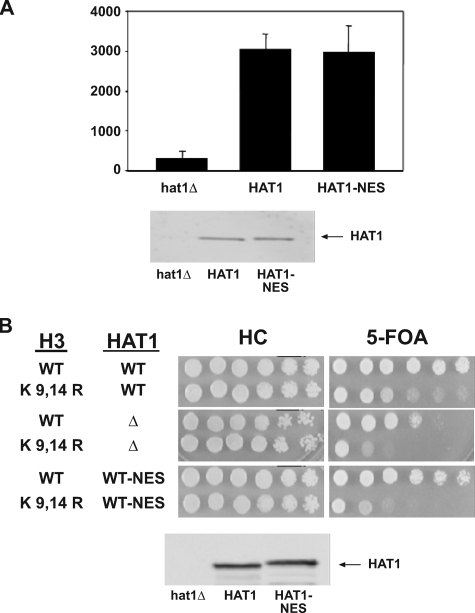
To determine whether nuclear localization is necessary for the in vivo function of the Hat1p complex, telomeric silencing assays (using a telomeric URA3 reporter) were performed in strains expressing either wild type HAT1 or the HAT1-NES fusion. As shown in Fig. 5B, fusion of the NES to Hat1p resulted in a decrease in viability in the presence of 5-FOA similar to that seen with hat1Δ. The defect in telomeric silencing was not because of an overall decrease in the amount of Hat1p protein because Western blot analysis demonstrated that the Hat1p-NES fusion was present at normal levels (Fig. 5B). Hence the presence of catalytically active Hat1p in the cytoplasm was not sufficient to support telomeric silencing. This suggested that the ability to localize to the nucleus is necessary for the in vivo function of Hat1p or its associated factors.
FIGURE 5.
Hat1p nuclear localization is important for telomeric silencing. A, partially purified whole cell extracts from strains EMY31 (HAT1-Myc), ASY50 (hat1Δ), and EMY35 (HAT1-NES-Myc) were quantitated by Western blot (bottom panel), and normalized protein samples from each were subjected to liquid histone acetyltransferase assays using free histones and [3H]Ac-CoA as substrates (top panel). ASY50 was normalized to Hat2p levels (data not shown). Assays were performed in triplicate, and results are shown as the average of these three independent samples. B, strains EMY31 (HAT1-Myc), ASY50 (hat1Δ), and EMY35 (HAT1-NES-Myc) harboring the indicated histone H3 alleles were assayed for telomeric silencing as described in the legend to Fig. 3 (top panel). Western blot analysis of equal amounts of total protein using α-Myc antibodies indicated equal levels of Hat1p expression (bottom panel). WT, wild type; SC, synthetic complete plates.
DISCUSSION
The results presented here extend our understanding of the type B histone acetyltransferase Hat1p in a number of ways. First mutational analysis of the active site of the enzyme indicated that Glu-255 of Hat1p did not function as a catalytic base as predicted from structural comparisons with other histone acetyltransferases. Further Hat1p did not appear to use other acidic amino acids that are located near the active site as an essential base. In addition, in vivo analysis of these mutants suggested that the catalytic activity of the enzyme was essential for its role in telomeric silencing. Finally the ability of Hat1p to localize to the nucleus was essential for it to function in telomeric silencing because a catalytically active form of the enzyme that was restricted to the cytoplasm did not support normal telomeric gene silencing.
Catalytic Mechanism of Hat1p—Enzymes that transfer an acetyl group from Ac-CoA to a substrate typically use one of two general mechanisms. One mechanism involves the initial transfer of the acetyl group to a nucleophile on the enzyme followed by subsequent transfer to the substrate. The second mechanism involves the formation of a ternary complex where direct transfer of the acetyl group from Ac-CoA to the substrate is achieved (50). For most histone acetyltransferases, regardless of the mechanism used, the reaction involves a general base, which is almost always a glutamic acid residue, that deprotonates the histone lysine residue to facilitate its nucleophilic attack on the acetyl group. Structural studies from a number of GNAT and MYST histone acetyltransferases have shown that there is a structurally conserved glutamic acid residue that performs this function (35, 36, 39, 42–44).
For Hat1p, Glu-255 has been proposed to serve as the general catalytic base for this enzyme. Hence this residue was an obvious candidate for creating a Hat1p point mutant that lacks catalytic activity and that could be used to determine whether the ability of Hat1p to acetylate histones was essential for its function in telomeric silencing. Although mutation of Glu-255 to glutamine, which cannot function as a base, had a significant effect on catalytic activity (∼10-fold), this level of reduction was significantly lower than would be expected if this residue were playing an essential catalytic role. Mutation of other acidic amino acids in the active site of Hat1p, including the adjacent aspartic acid residue (Asp-256), also failed to identify a single residue essential for enzymatic activity. These results suggest that, although Hat1p has significant structural similarity to other histone acetyltransferases such as Gcn5p, P/CAF, and Esa1p, it may function through a distinct mechanism. Perhaps as originally proposed by Dutnall et al. (34) the chemical environment of the Hat1p active site may promote or stabilize the deprotonated state of substrate lysine residues thus obviating the need for a specific acidic amino acid to serve as an active site base.
The possibility that Hat1p may not require an acidic amino acid to function as an essential catalytic base is reminiscent of the mechanism recently proposed for the histone acetyltransferase p300 (51). The crystal structure of p300 indicates that it is distantly related to the GNAT and MYST enzymes. p300 appears to use a “hit and run” mechanism that uses a tryptophan residue to stabilize the neutral form of the histone lysine residue rather than a catalytic base.
Interestingly the yeast histone acetyltransferase Rtt109p, which has been shown to possess structural similarity with p300, shares with Hat1p the presence of adjacent acidic residues (Asp-287 and Asp-288) near its active site (52–54). However, unlike Hat1p, mutation of both aspartic acid residues of Rtt109p (to asparagines) results in a much greater loss of catalytic activity than altering either residue individually. Curiously mutation of Rtt109p Asp-287 and Asp-288 to alanine causes a significantly greater loss of catalytic activity than the corresponding asparagine mutants (45). As the asparagine residues are unlikely to function as catalytic bases, this may reflect the fact that post-translational conversion of asparagine to aspartic acid can occur in some proteins (55–58).
In Vivo Function of Type B Histone Acetyltransferases—It is clear that more detailed kinetic studies, using purified recombinant enzyme, will be required to unravel the precise mechanism used by Hat1p in the acetylation of histones. Regardless of the mechanisms involved, the in vivo analysis of the Hat1p active site mutants suggests that the catalytic activity of Hat1p is necessary for its role in telomeric silencing. Although the defect in telomeric silencing observed with each mutant roughly correlated with the level of enzymatic activity, it was somewhat surprising that mutants that displayed only a moderate decrease in Hat1p enzymatic activity still caused a significant decrease in telomeric silencing. As it has previously been demonstrated that a histone H4 K12R allele (but not an H4 K5R allele) phenocopies a hat1Δ, this result suggests that telomeric silencing is highly sensitive to the acetylation status of histone H4 lysine 12 in a manner that is functionally redundant with the acetylation of histone H3 (19).
Although the enzymatic activity of Hat1p is necessary for its function in vivo, it is not sufficient. This was demonstrated by the observation that a catalytically active form of Hat1p that is restricted to the cytoplasm does not support normal telomeric silencing. This result is not consistent with the original models of type B histone acetyltransferase function that suggested that these enzymes simply modified newly synthesized histones in the cytoplasm and then released them to proceed with subsequent steps in chromatin assembly such as association with histone chaperones and nuclear import (9). Rather, this result suggests that activities of Hat1p that occur subsequent to the cytoplasmic acetylation of newly synthesized histone H4 are important for its in vivo function. These activities could potentially include a role for Hat1p in facilitating the nuclear import of histone H4 or a more direct role for Hat1p in the assembly of histones into chromatin. An alternative possibility is that, in the context of the cell, Hat1p is only catalytically active when it is in the nucleus. Although this model is unlikely, it is difficult to definitively rule out. As the Hat1p-mediated acetylation of histone H4 is a transient modification, the loss of Hat1p activity in yeast does not measurably impact the steady state level of histone H4 acetylation under normal conditions (16, 59). Hence it is difficult to determine whether restricting Hat1p to the cytoplasm has an impact on the acetylation state of histone H4 in the cell.
Although the precise role that Hat1p plays in telomeric silencing is not known, it requires both the catalytic activity and nuclear localization of the enzyme. It is likely that Hat1p influences telomeric chromatin structure through its involvement in chromatin assembly. An important question is whether this involvement is direct or indirect. For example, Hat1p might increase the overall rate of chromatin assembly by modifying and facilitating the nuclearimport of histone H3-H4 complexes that are then used by other chromatin assembly factors, such as CAF-1, Asf1p, or the Hir complex. In this model, Hat1p would function to increase the efficiency of chromatin assembly near telomeres and hence help promote formation of silent chromatin structure. A second possibility is that the assembly of histone H4 that has been acetylated at lysine 12 by Hat1p (along with the functionally redundant modification of histone H3) may be specifically required near telomeres for a subsequent step in the assembly or maintenance of silent chromatin structure. Alternatively, Hat1p, in association with Hif1p, may help promote formation of silent chromatin by directly participating in the assembly of chromatin near telomeres. Finally a recent report links Hat1p to the origin recognition complex, and, therefore, loss of nuclear Hat1p could indirectly influence telomeric chromatin structure through an impact on DNA replication (60). Importantly these models are not mutually exclusive, and Hat1p may influence telomeric chromatin structure by multiple mechanisms.
Acknowledgments
We thank Dr. Russ Hille for many helpful discussions and Craig Hemann for generous assistance with molecular models.
This work was supported, in whole or in part, by National Institutes of Health Grant GM062970. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GNAT, GCN5-related histone N-acetyltransferase; TAP, tandem affinity purification; NES, nuclear export signal; PKI, protein kinase inhibitor; 5-FOA, 5-fluoroorotic acid; Ac-CoA, acetyl coenzyme A; P/CAF, p300/CBP-associating factor (CBP, cAMP-response element-binding protein (CREB)-binding protein).
References
- 1.Neuwald, A. F., and Landsman, D. (1997) Trends Biochem. Sci. 22 154-155 [DOI] [PubMed] [Google Scholar]
- 2.Kleff, S., Andrulis, E. D., Anderson, C. W., and Sternglanz, R. (1995) J. Biol. Chem. 270 24674-24677 [DOI] [PubMed] [Google Scholar]
- 3.Parthun, M. R., Widom, J., and Gottschling, D. E. (1996) Cell 87 85-94 [DOI] [PubMed] [Google Scholar]
- 4.Verreault, A., Kaufman, P. D., Kobayashi, R., and Stillman, B. (1998) Curr. Biol. 8 96-108 [DOI] [PubMed] [Google Scholar]
- 5.Imhof, A., and Wolffe, A. P. (1999) Biochemistry 38 13085-13093 [DOI] [PubMed] [Google Scholar]
- 6.Eberharter, A., Lechner, T., Goralik-Schramel, M., and Loidl, P. (1996) FEBS Lett. 386 75-81 [DOI] [PubMed] [Google Scholar]
- 7.Chang, L., Loranger, S. S., Mizzen, C., Ernst, S. G., Allis, C. D., and Annunziato, A. T. (1997) Biochemistry 36 469-480 [DOI] [PubMed] [Google Scholar]
- 8.Lusser, A., Brosch, G., Lopez-Rodas, G., and Loidl, P. (1997) Eur. J. Cell Biol. 74 102-110 [PubMed] [Google Scholar]
- 9.Brownell, J. E., and Allis, C. D. (1996) Curr. Opin. Genet. Dev. 6 176-184 [DOI] [PubMed] [Google Scholar]
- 10.Sobel, R. E., Cook, R. G., Perry, C. A., Annunziato, A. T., and Allis, C. D. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 1237-1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chicoine, L. G., Schulman, I. G., Richman, R., Cook, R. G., and Allis, C. D. (1986) J. Biol. Chem. 261 1071-1076 [PubMed] [Google Scholar]
- 12.Lee, K. K., and Workman, J. L. (2007) Nat. Rev. Mol. Cell Biol. 8 284-295 [DOI] [PubMed] [Google Scholar]
- 13.Lusser, A., Eberharter, A., Loidl, A., Goralik-Schramel, M., Horngacher, M., Haas, H., and Loidl, P. (1999) Nucleic Acids Res. 27 4427-4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmad, A., Nagamatsu, N., Kouriki, H., Takami, Y., and Nakayama, T. (2001) Nucleic Acids Res. 29 629-637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ai, X., and Parthun, M. R. (2004) Mol. Cell 14 195-205 [DOI] [PubMed] [Google Scholar]
- 16.Poveda, A., Pamblanco, M., Tafrov, S., Tordera, V., Sternglanz, R., and Sendra, R. (2004) J. Biol. Chem. 279 16033-16043 [DOI] [PubMed] [Google Scholar]
- 17.Parthun, M. R. (2007) Oncogene 26 5319-5328 [DOI] [PubMed] [Google Scholar]
- 18.Mosammaparast, N., Guo, Y., Shabanowitz, J., Hunt, D. F., and Pemberton, L. F. (2002) J. Biol. Chem. 277 862-868 [DOI] [PubMed] [Google Scholar]
- 19.Kelly, T. J., Qin, S., Gottschling, D. E., and Parthun, M. R. (2000) Mol. Cell. Biol. 20 7051-7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottschling, D. E., Aparicio, O. M., Billington, B. L., and Zakian, V. A. (1990) Cell 63 751-762 [DOI] [PubMed] [Google Scholar]
- 21.Kaufman, P. D., Kobayashi, R., and Stillman, B. (1997) Genes Dev. 11 345-357 [DOI] [PubMed] [Google Scholar]
- 22.Kaufman, P. D., Cohen, J. L., and Osley, M. A. (1998) Mol. Cell. Biol. 18 4793-4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enomoto, S., and Berman, J. (1998) Genes Dev. 12 219-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Enomoto, S., McCune-Zierath, P. D., Gerami-Nejad, M., Sanders, M. A., and Berman, J. (1997) Genes Dev. 11 358-370 [DOI] [PubMed] [Google Scholar]
- 25.Sharp, J. A., Fouts, E. T., Krawitz, D. C., and Kaufman, P. D. (2001) Curr. Biol. 11 463-473 [DOI] [PubMed] [Google Scholar]
- 26.Krawitz, D. C., Kama, T., and Kaufman, P. D. (2002) Mol. Cell. Biol. 22 614-625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benson, L. J., Phillips, J. A., Gu, Y., Parthun, M. R., Hoffman, C. S., and Annunziato, A. T. (2007) J. Biol. Chem. 282 836-842 [DOI] [PubMed] [Google Scholar]
- 28.Barman, H. K., Takami, Y., Ono, T., Nishijima, H., Sanematsu, F., Shibahara, K., and Nakayama, T. (2006) Biochem. Biophys. Res. Commun. 345 1547-1557 [DOI] [PubMed] [Google Scholar]
- 29.Qin, S., and Parthun, M. R. (2002) Mol. Cell. Biol. 22 8353-8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henderson, B. R., and Eleftheriou, A. (2000) Exp. Cell Res. 256 213-224 [DOI] [PubMed] [Google Scholar]
- 31.Adams, A., Gottschling, D. E., Kaiser, C. A., and Stearns, T. (1997) Methods in Yeast Genetics, pp. 99-102, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 32.Schultz, M. C., Choe, S. Y., and Reeder, R. H. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 1004-1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng, H. P., Scherl, D. S., and Widom, J. (1993) Biochemistry 32 7824-7831 [DOI] [PubMed] [Google Scholar]
- 34.Dutnall, R. N., Tafrov, S. T., Sternglanz, R., and Ramakrishnan, V. (1998) Cell 94 427-438 [DOI] [PubMed] [Google Scholar]
- 35.Yan, Y., Barlev, N. A., Haley, R. H., Berger, S. L., and Marmorstein, R. (2000) Mol. Cell 6 1195-1205 [DOI] [PubMed] [Google Scholar]
- 36.Clements, A., Rojas, J. R., Trievel, R. C., Wang, L., Berger, S. L., and Marmorstein, R. (1999) EMBO J. 18 3521-3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin, Y., Fletcher, C. M., Zhou, J., Allis, C. D., and Wagner, G. (1999) Nature 400 86-89 [DOI] [PubMed] [Google Scholar]
- 38.Rojas, J. R., Trievel, R. C., Zhou, J., Mo, Y., Li, X., Berger, S. L., Allis, C. D., and Marmorstein, R. (1999) Nature 401 93-98 [DOI] [PubMed] [Google Scholar]
- 39.Trievel, R. C., Rojas, J. R., Sterner, D. E., Venkataramani, R. N., Wang, L., Zhou, J., Allis, C. D., Berger, S. L., and Marmorstein, R. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 8931-8936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poux, A. N., Cebrat, M., Kim, C. M., Cole, P. A., and Marmorstein, R. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 14065-14070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poux, A. N., and Marmorstein, R. (2003) Biochemistry 42 14366-14374 [DOI] [PubMed] [Google Scholar]
- 42.Tanner, K. G., Trievel, R. C., Kuo, M. H., Howard, R. M., Berger, S. L., Allis, C. D., Marmorstein, R., and Denu, J. M. (1999) J. Biol. Chem. 274 18157-18160 [DOI] [PubMed] [Google Scholar]
- 43.Berndsen, C. E., Albaugh, B. N., Tan, S., and Denu, J. M. (2007) Biochemistry 46 623-629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, L., Liu, L., and Berger, S. L. (1998) Genes Dev. 12 640-653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han, J., Zhou, H., Horazdovsky, B., Zhang, K., Xu, R. M., and Zhang, Z. (2007) Science 315 653-655 [DOI] [PubMed] [Google Scholar]
- 46.Yan, Y., Harper, S., Speicher, D. W., and Marmorstein, R. (2002) Nat. Struct. Biol. 9 862-869 [DOI] [PubMed] [Google Scholar]
- 47.Langer, M. R., Tanner, K. G., and Denu, J. M. (2001) J. Biol. Chem. 276 31321-31331 [DOI] [PubMed] [Google Scholar]
- 48.Boeke, J. D., Trueheart, J., Natsoulis, G., and Fink, G. R. (1987) Methods Enzymol. 154 164-175 [DOI] [PubMed] [Google Scholar]
- 49.Stade, K., Ford, C. S., Guthrie, C., and Weis, K. (1997) Cell 90 1041-1050 [DOI] [PubMed] [Google Scholar]
- 50.Hodawadekar, S. C., and Marmorstein, R. (2007) Oncogene 26 5528-5540 [DOI] [PubMed] [Google Scholar]
- 51.Liu, X., Wang, L., Zhao, K., Thompson, P. R., Hwang, Y., Marmorstein, R., and Cole, P. A. (2008) Nature 451 846-850 [DOI] [PubMed] [Google Scholar]
- 52.Lin, C., and Yuan, Y. A. (2008) Structure (Lond.), in press
- 53.Stavropoulos, P., Nagy, V., Blobel, G., and Hoelz, A. (2008) Proc. Natl. Acad. Sci. U. S. A. [DOI] [PMC free article] [PubMed]
- 54.Tang, Y., Holbert, M. A., Wurtele, H., Meeth, K., Rocha, W., Gharib, M., Jiang, E., Thibault, P., Verrault, A., Cole, P. A., and Marmorstein, R. (2008) Nat. Struct. Mol. Biol. 15 738-745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ichiyama, S., Kurihara, T., Li, Y. F., Kogure, Y., Tsunasawa, S., and Esaki, N. (2000) J. Biol. Chem. 275 40804-40809 [DOI] [PubMed] [Google Scholar]
- 56.Ichiyama, S., Kurihara, T., Kogure, Y., Tsunasawa, S., Kawasaki, H., and Esaki, N. (2004) Biochim. Biophys. Acta 1698 27-36 [DOI] [PubMed] [Google Scholar]
- 57.Stephenson, R. C., and Clarke, S. (1989) J. Biol. Chem. 264 6164-6170 [PubMed] [Google Scholar]
- 58.Pries, F., Kingma, J., and Janssen, D. B. (1995) FEBS Lett. 358 171-174 [DOI] [PubMed] [Google Scholar]
- 59.Poveda, A., and Sendra, R. (2008) FEBS J. 275 2122-2136 [DOI] [PubMed] [Google Scholar]
- 60.Suter, B., Pogoutse, O., Guo, X., Krogan, N., Lewis, P., Greenblatt, J. F., Rine, J., and Emili, A. (2007) BMC Biol. 5 38. [DOI] [PMC free article] [PubMed] [Google Scholar]