Abstract
HIT (histidine triad)1 proteins, named for a motif related to the sequence HφHφHφφ, (φ a hydrophobic amino acid) are a superfamily of nucleotide hydrolases and transferases, which act on the α-phosphate of ribonucleotides, and contain a ∼30 kDa domain that is typically either a homodimer of ∼15 kDa polypeptides with two active-sites or an internally, imperfectly repeated polypeptide that retains a single HIT active site. On the basis of sequence, substrate specificity, structure, evolution and mechanism, HIT proteins can be classified into the Hint branch, which consists of adenosine 5′-monophosphoramide hydrolases, the Fhit branch, which consists of diadenosine polyphosphate hydrolases, and the GalT branch, which consists of specific nucleoside monophosphate transferases including galactose-1-phosphate uridylyltransferase, diadenosine tetraphosphate phosphorylase, and adenylylsulfate:phosphate adenylytransferase. At least one human representative of each branch is lost in human diseases. Aprataxin, a Hint branch hydrolase, is mutated in ataxia-oculomotor apraxia syndrome. Fhit is lost early in development of many epithelially derived tumors. GalT is deficient in galactosemia. Additionally, ASW is an avian Hint family member that has evolved to have unusual gene expression properties and the complete loss of its nucleotide binding-site. The potential roles of ASW and Hint in avian sexual development are discussed in an accompanying manuscript. Here we review what is known about biological activities of HIT proteins, the structural and biochemical bases for their functions, and propose a new enzyme mechanism for Hint and Fhit that may account for the differences between HIT hydrolases and transferases.
Mammalian Hint and GalT Structurally Define the HIT Superfamily
Galactose-1-phosphate uridylyltransferase (GalT), the second enzyme in the Leloir pathway of galactose utilization (1), was crystallized and its structure determined to understand the mechanistic basis for transferring UMP between glucose-1-phosphate and galactose-1-phosphate (2-4). Histidine triad nucleotide-binding protein (Hint), purified as a purine nucleoside/nucleotide-binding protein from rabbit heart cytosol (5) and shown to have a widely conserved sequence (6), was crystallized and its structure determined to establish whether the conserved sequence formed the basis for a new mode of nucleotide-binding (7). GalT (2) and Hint are both dimers (8) but the GalT monomer is more than twice the size of the Hint dimer. Though GalT (9) and Hint (6) homologs could be cloned by similarity and the crystal structures (2, 8) co-existed in protein databases, mutual similarity was not noted until inspection of the rabbit Hint crystal structure (7) and computational analysis (10) indicated that a GalT monomer and a Hint dimer can be superimposed and that their nucleotide-binding sites retain some of the same residues for binding a nucleoside monophosphate (7) (Figure 1). The level of sequence similarity of Hint and GalT is difficult to distinguish from noise in a standard BLAST search but can be picked up by PSI-BLAST (11). Nonetheless, the structures of Hint and GalT and modes of nucleotide-binding are convincingly similar and the mechanisms are related as discussed below.
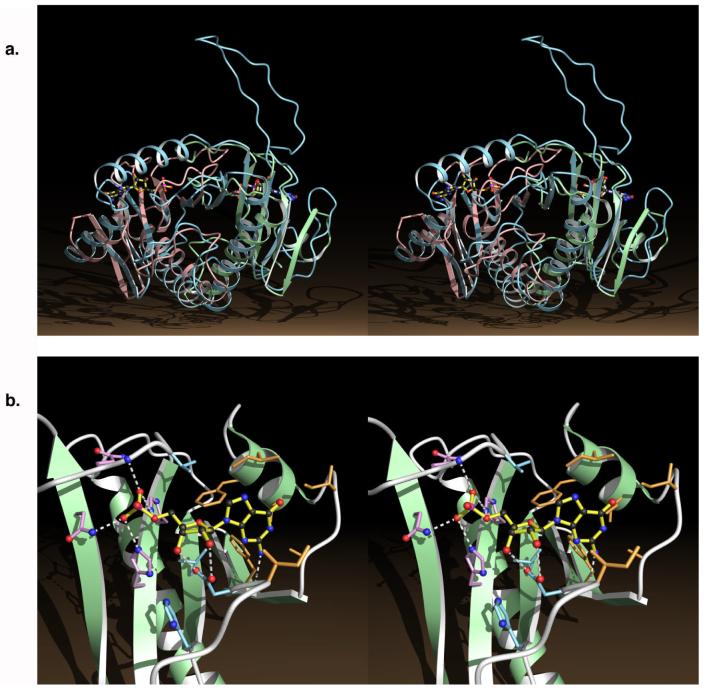
Figure 1.
Structural definition of HIT proteins. A) Stereo view of the GalT half-barrel (blue) superimposed on the Hint dimer (green and pink). The half-barrel consists of nine (GalT) or ten (Hint) anti-parallel β strands that enclose a pair of anti-parallel α helices. GalT possesses a distinct loop structure and a helix that connects the N-terminal portion (right) of the half-barrel with the C-terminal portion (left) of the half-barrel. The GalT monomer has a single Hint-homologous active-site in its N-terminal portion below the large loop where UMP is bound. UMP bound to GalT superimposes with GMP bound in the right hand Hint monomer. B) Highly conserved residues in Hint define a new mode of nucleotide-binding on top of the β-sheet. One of the two identical active sites is shown bound to GMP (yellow). The figure is based on reported X-ray structures (2, 7) and analysis (7).
Hint and Hnt1 are Enzymes with Adenosine 5′-Monophosphoramidase Activity and Hnt1 Enzyme Activity Positively Regulates Yeast Kin28
Because Hint genes are found in virtually every sequenced genome and appear to be the common link to fragile histidine triad (Fhit) branch and GalT branch sequences, it is considered that Hint is the ancestral HIT protein (7) and Hint homologs constitute Branch I of the HIT superfamily. Branch I includes at least four human, three chicken and two yeast proteins (Table 1). Unfortunately, certain sequence annotations of HIT proteins persistently refer to the HIT motif as a zinc binding site or to Hint homologs as protein kinase C interactors or inhibitors. These annotations are based on data that has been publicly withdrawn in the case of protein kinase C (12) and on a paper whose data prove that zinc does not bind (8), as we have reviewed earlier (13). Crystallographic analysis demonstrated that fourteen of the most highly conserved residues in the HIT protein superfamily mediate interactions with the purine base, the ribose and the 5′ monophosphate (7). However, while this analysis indicated that Hint homologs are conserved as nucleotide-binding proteins, the enzymatic and physiological functions of Hint homologs were not immediately clear.
Table 1.
A Genomic View of HIT Proteins: Three Branches in Three Genomes
| Branch |
H. sapiens Gene |
(a.a.)map | locusID |
G. gallus Gene(a.a.) |
map |
S. cerevisiae Gene (a.a.) |
map |
|---|---|---|---|---|---|---|---|
| I) Hint | HINT1 | (126) 5q31.2 | 3094 | HINT (126) | Z | HNT1 (158) | YDL125C |
| HINT2 | (163) 9p11.2 | 84681 | ? | --- | |||
| HINT3 | (182) 6q22.33 | 135114 | ? | --- | |||
| HINTP1 | (126) 7q35 | 168334 | ? | --- | |||
| APTX | (342)9p13.3 | 54840 | APTX (?) | ? | HNT3 (217) | YOR258W | |
| --- | ASW (130) | W | --- | ||||
| II) Fhit | FHIT | (147) 3p14.2 | 2272 | FHIT (147) | ? | HNT2 (217) | YDR305C |
| III) GalT | GALT | (379) 9p13 | 2592 | GALT (?) | ? | GAL7 (366) | YBR018C |
| ? | ? | APA1 (321) | YCL050C | ||||
| ? | ? | APA2 (325) | YDR530C |
Gene names are official except chicken APTX, FHIT and GALT, which have been observed as mRNAs but not mapped. Human map positions and locusIDs are from LocusLink (http://www.ncbi.nlm.nih.gov:80/LocusLink/index.html) (14). Chicken and Saccharomyces genomic databases are at http://www.thearkdb.org/browser?species=chicken (15) and http://genomewww. stanford.edu/Saccharomyces (16), respectively. Sizes of primary translation products are for monomers of the largest splice forms. The size of chicken Fhit is inferred from an expressed sequence tag (EST).
Specific substrates for rabbit Hint and its yeast ortholog, Hnt1, emerged alongside physiological assays for the yeast HNT1 gene (17). While deletion of hnt1 was tolerated by haploid yeast cells on glucose media at all temperatures and deletion of hnt1 did not lead to a galactose- phenotype at 30 °C, hnt1 deletion produced cells that are galactose- at 38-39 °C. Rabbit Hint and yeast Hnt1 were both purified from E. coli and treated with a panel of nucleotides. As Fhit and GalT both cleave substrates that can be described as NMP-X (Figure 2), a large panel of dinucleoside polyphosphates (ApnA), nucleoside di- and triphosphates, nucleotide sugars, and other compounds were tested as rabbit Hint and yeast Hnt1 substrates. The only compounds hydrolyzed at greater than ∼1 nmol min-1 μg-1 were the adenosine 5′-monophosphoramide linked compounds AMP morpholidate, AMP-N-alanine methyl ester, AMP-N-ε-(N-α-acetyl lysine methyl ester), and AMP-NH2. AMP-NH2 was hydrolyzed at greater than 1,000,000 s-1 M-1 by both rabbit and yeast enzymes. This activity was at least 200,000-fold dependent upon the His116 nucleophile of Hnt1, which corresponds to His112 of mammalian Hint. Just as yeast Hnt1 enzyme activity was destroyed by the H116A mutation, so the hnt1-H116A allele was without function in supporting growth of yeast cells on galactose at 39 °C. Because the Hnt1-H116A protein is not unstable, it was concluded that hydrolysis of an Hnt1 substrate is required for growth of yeast cells at 39 °C (17).
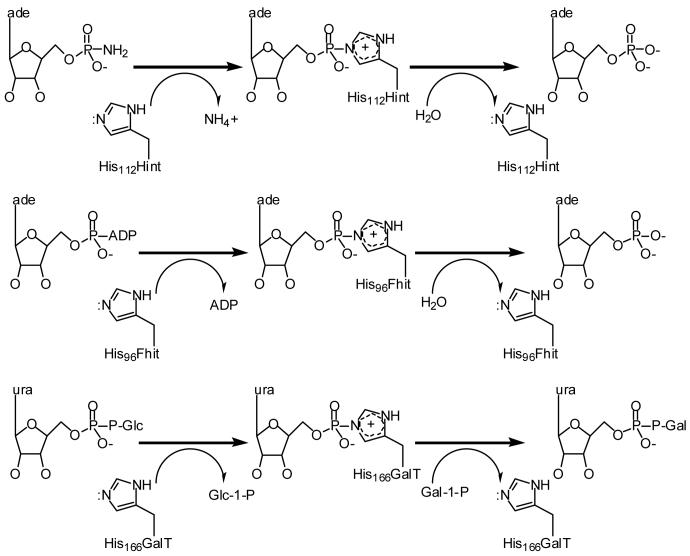
Figure 2.
Substrates and forward reaction schemes for prototypical members of the three branches of the HIT superfamily. Note that Hint and Fhit hydrolyze nucleotidylated enzyme intermediates while the GalT nucleotidylated intermediate, stable to water, transfers to a second substrate, namely a phosphorylated sugar.
A major clue to Hint function was provided by identification of a two-hybrid interaction between human Cdk7 and human Hint1 (18). Cdk7 is the kinase catalytic subunit of CAK, the Cdc2 activating kinase complex, consisting of Cdk7, cyclin H and MAT-1 (19-21). Additionally, this heterotrimer is part of a larger complex termed general transcription factor TFIIH that contains the major RNA polymerase II C-terminal domain kinase activity (22, 23). Adding to the significance of the findings, the yeast Cdk7 homolog, Kin28, had a weak two-hybrid interaction with yeast Hnt1 and it was demonstrated that the growth of a temperature sensitive mutant in kin28 was impaired by hnt1 deletion on galactose media (18). Though the two-hybrid isolation of Hint as a Cdk7 interactor initially suggested a physical basis for regulation (18), expression of hnt1-H116A in kin28-ts backgrounds was as deleterious as the hnt1 knockout, indicating that Hnt1 enzyme activity is a positive regulator of Kin28 function on galactose media (17).
Though fungal and plant orthologs of mammalian enzymes are typically 50% identical at the amino acid sequence level, the S. cerevisiae Hint1 ortholog Hnt1 is only 22% identical, maintaining little more than the residues that form the nucleotide-binding site and the dimer interface (7). To address the possibility that Hnt1 enzymatic activity is not only necessary but sufficient for Kin28 regulation, rabbit Hint was expressed in place of yeast Hnt1 and showed full suppression of every hnt1 Δ phenotype (17).
One obvious mechanism by which loss of Hnt1 enzyme activity might result in inhibition of a protein kinase was that the as yet unidentified in vivo substrate of Hint, provisionally termed AMP-X, which might be AMP-NH2 or a chemically related molecule, could be an ATP-competitive protein kinase inhibitor. Further, because Cdk7 is both CAK and the kinase component of TFIIH, it was considered that Hnt1 enzyme activity might be a positive regulator of either Kin28 or Cak1. Consistent with the possibility that Cak1 might be the target of Hnt1 regulation, cak1-ts mutants are synthetically less viable with hnt1 mutations. However, when hnt1 mutations were put into a strain that contains CDC28 alterations that bypass the essential function of Cak1, poor growth was observed, indicating that Hnt1 does not act as a positive regulator of Cak1 (17). In fact, in the cak1 Δ hnt1 Δ background, poor growth was observed with glucose and galactose as carbon sources. This experiment excluded Cak1 as the target of Hnt1 regulation and also excluded the possibility that the galactose-dependent nature of hnt1 phenotypes might have been due to AMP-X being produced solely as a byproduct of the Leloir pathway of galactose utilization.
With genetic experiments having shown that Hnt1 functions downstream of Cak1, and that hnt1 mutations synergize with nonphosphorylatable Kin28, overexpressed Kin28, and temperature-sensitive mutations in kin28, ccl1 and tfb3 (17), it remains to be determined precisely what the nature of Hnt1 regulation is and why the defect is easier to discern on galactose media than on glucose. Four models that account for all available data are shown in Figure 3. In each model, the glucose-essential function of Kin28 is depicted as residing in the TFIIH complex. Kin28 is also shown as existing in a CAK-like trimer consisting of Kin28, Ccl1 and Tfb3, which has recently been shown to exist in vivo (24), that we refer to as TFIIK. We further illustrate the TFIIK trimer as reversibly dissociable to its constituent polypeptides, with the equilibrium favoring monomers at higher temperature and as a function of temperature-sensitive alleles of Kin28, Ccl1 and Tfb3. It is known that the combination of temperature-sensitive Tfb3 and nonphosphorylatable Kin28 leads to Kin28 proteolysis (25). On the contrary, hnt1 deletion with nonphosphorylatable Kin28 is synthetically lethal on galactose media but hnt1 deletion does not destabilize the Kin28 polypeptide (17). For these reasons, each model hypothesizes that the Kin28 monomer, released from TFIIK at elevated temperature and with mutations in Cak1, Kin28, Ccl1 or Tfb3, is susceptible to inhibition in the absence of Hnt1 enzyme activity, and that the hnt1-inhibited form of Kin28 is disadvantaged in complex formation. Additionally, each model suggests that the hnt1-inhibited form of Kin28 is stable and inhibits growth on galactose. Further, each model hypothesizes that with mutations in Cak1, Kin28, Ccl1 or Tfb3 combined with loss of Hnt1, sufficient Kin28 is drawn down to the poorly complexing form to deplete Kin28 from TFIIH and thus reduce viability on glucose.
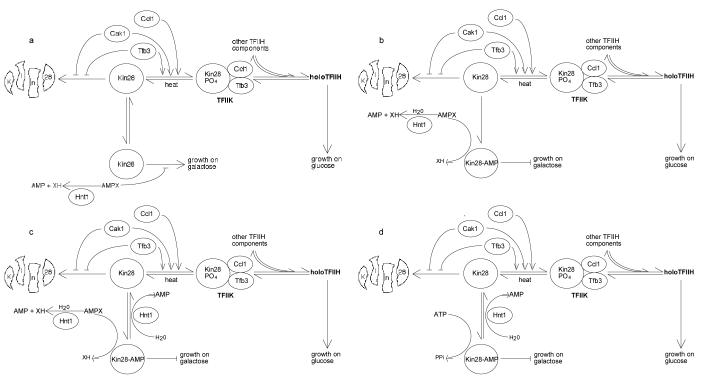
Figure 3.
Models for regulation of Kin28 function by Hnt1 enzyme activity. See text for details.
The models consider alternate mechanisms by which Hnt1 enzyme activity may regulate Kin28 function. In model A, the small molecule Hnt1 substrate provisionally termed AMP-X is a noncovalent inhibitor of Kin28 and the function of Hnt1 is to reduce the concentration of this molecule in the nucleocytoplasm. In model B, AMP-X is an Hnt1 substrate that covalently modifies Kin28. In model C, AMP-X is an Hnt1 substrate that adenylylates Kin28 as in model B and, additionally, Kin28-AMP is an Hnt1 substrate. Finally, in model D, Kin28 becomes adenylylated by virtue of reaction with ATP and Kin28-AMP is the only Hnt1 substrate. In models A, B and C, small molecules are proposed to be important Hnt1 substrates. In models C and D, adenylylated Kin28 protein is proposed to be an important Hnt1 substrate. Experiments are in progress to test these models.
Aprataxin: A Zn-finger-containing Hint Family Member That is Mutated in Ataxia-Oculomotor Apraxia
Ataxia-oculomotor apraxia syndrome (AOA) is an early onset, progressive neurological disease characterized by balance and facial motor problems, cerebellar atrophy and hypoalbuminemia, whose neurological symptoms are difficult to distinguish from ataxia-telangiectasia (26). At the halfway point of a survey intended to catalog every case of hereditary ataxia in Portugal, AOA is second in occurrence only to Friedreich’s among the autosomal recessive ataxias (27). Despite literature that is sparse outside of Portugal (27) and Japan (28), AOA has been found in many ethnic groups and does not appear to be a Portuguese or Japanese disease but rather a disease that is rare but ubiquitous (26). Two groups succeeded in positional cloning of an AOA disease locus to the APTX gene encoding Aprataxin on 9p13 (29, 30).
Human Aprataxin is expressed in two splice forms (30). The minor splice form encodes a predicted polypeptide of 168 amino acids consisting of a Hint domain and an apparent C(2)H(2) zinc finger C-terminal to the Hint domain (29, 30). The Hint domain of Aprataxin shows 31% amino acid identity with rabbit and human Hint in an 86 amino acid stretch that begins with the first beta strand and ends with the HIT motif in the fifth and last strand (7). Though this segment does not include helix 1 and conserved sequences C-terminal to strand 5, it appears to be a minimum Hint domain we predict to dimerize and possess adenosine 5′-monophosphoramidase activity. C-terminal to the Hint domain, Aprataxin has the sequence CX(2)CX(12)HX(3)H that may be a nucleic acid-binding motif consisting of a single zinc finger (31, 32).
The major splice form of Aprataxin is extended 174 amino acids at the N-terminus with respect to the minor splice form. Residues 6 through 102 of the N-terminus of Aprataxin are 41% identical with the N-terminus of human polynucleotide kinase (PNK), an enzyme with 5′-DNA kinase, 3′-phosphatase activity that is located C-terminal to this domain (33, 34). The observation that PNK and Aprataxin share an N-terminal domain suggested that Aprataxin may have a role in single strand break repair (30). Indeed the PNK and Aprataxin N-terminal domain, termed PANT (30), was recognized as a forkhead-associated (FHA) domain by Keith Caldecott (personal communication). FHA domains are phosphothreonine-binding protein association domains that have been found in a variety of proteins involved in responses to DNA damage (35-38).
Many eukaryotes appear to have multiple Hint family members, one or more of which are Hint homologs, and one which is an Aprataxin homolog (Table 1). These sequences can be distinguished by degree of similarity to Hint and Aprataxin. Though Aprataxin homologs only align with Hint sequences inclusive of the β sheet, they are typically larger than Hint homologs because they are extended at the N and C-termini and sometimes by insertions between helix 2 and strand 4. To date, we have only observed PANT/FHA domains in Aprataxin-homologous proteins from vertebrates. Invertebrate Aprataxin homologs, such as S. cerevisiae Hnt3 (17), possess a domain structure similar to the minor splice form of human Aprataxin (30). The molecular targets and pathways involving Aprataxin and its homologs remain to be discovered. With identification of such targets, it is hoped that new light can be shed on brain development and motor coordination.
Evolutionary of HIT Proteins: Bacteria to Birds
The observation that Hint homologs are nearly ubiquitous and that they are encoded in the smallest observed bacterial genomes suggested that Hint was present at the cellular root of the tree of life (7). Given the ability of Hint homologs to hydrolyze AMP-NH2 and to hydrolyze AMP linked to a lysine sidechain (17), we speculate that Hint may have evolved as an enzyme to cleanse spontaneously adenylylated proteins, which are potentially a consequence of life at high temperature with high levels of ATP. During evolution, certain proteins may have developed the ability to react in a suicide manner with adenosine nucleotides leading to specific sites of high-occupancy adenylylation. In addition, specific proteins may have become sensitive to noncovalent effects of accumulated Hint substrates. Developments such as these could be responsible for specificity in the consequences of loss of Hint homologs, such as has been observed in yeast. Specificity in function of Hint homologs clearly exists because hnt1 phenotypes are easy to observe in cells that are HNT3+ (17) and because the genetics of AOA indicate that affected individuals have loss of function APTX mutations (29, 30) but ought to contain normal HINT1-3 genes.
As will be discussed below, it appears that early to midway through eukaryotic evolution, a HIT protein underwent mutation and selection to become a Fhit-homologous Branch II enzyme. As illustrated in Figure 2, Fhit substrate specificity differs from that of Hint with Fhit homologs preferring to hydrolyze substrates such as AppppA (39), ApppA (40), γ-(m-Nitrobenzyl) adenosine 5′-O-triphosphate (41), and ApppBODIPY (42). This amounts to an altered specificity for the adenylylation step of the reaction because adenylylation of each of these substrates produces an identical Fhit-AMP intermediate that, like Hint-AMP, is enzymatically hydrolyzed. Thus, if the original enzyme was a Branch I enzyme, the new enzyme was altered to change its adenylylation specificity from attack of AMP-NXH to attack of ApppX.
We have previously observed that a HIT protein may have undergone a tandem duplication with addition of a connecting helix and loss of one active site to produce a GalT-like nucleotidyl transferase (7). As shown in Figure 2, GalT and other Branch III enzymes have substrates that conform to the structure NMP-X, and become nucleotidylated with release of the leaving group. Unlike Hint and Fhit intermediates, nucleotidylated GalT intermediates are stable in 55 M H2O (3, 43). Not being hydrolases, Branch III enzymes enforce dependence of the intermediate reacting with a second substrate, typically phosphate or a phosphorylated hexose, to produce product. A possible basis for the differences between HIT hydrolases and transferases is provided in the mechanistic section below.
Apart from Aprataxin (29, 30), which is a divergent member of the Hint branch, and the branch II and branch III enzymes Fhit (44) and GalT (45), the human genome appears to encode at least two additional Hint homologs (46). As shown in Table 1, HINT2, HINT3 and the putative pseudogene HINTP1 appear to be encoded at 9p11.2, 6q22.33 and 7q35, respectively. Expressed sequence tags (ESTs) and/or cDNAs corresponding to HINT2 and HINT3 have been deposited, indicating more restricted expression patterns than the prototypical HINT (now HINT1) gene at 5q31.2 (47). HINT2, expressed in liver, is predicted to encode a Hint homolog with a 35 amino acid N-terminal extension with respect to the canonical 126 amino acid rabbit and human Hint. HINT3 is predicted to encode a primary translation product of 182 amino acids, extended 31 and 25 amino acids at the N and C-termini, respectively. An ortholog of Hint3 including the C-terminal extension can be found in several other metazoans and these genes are clearly expressed at the mRNA level. The HINTP1 locus is apparently intronless and its expression is supported by no publicly available EST. Conceptual translation of HINTP1 produces a primary translation product of 126 amino acids with the peculiar substitution of two conserved histidine residues, H51R and H110Q. In the absence of evidence for expression of HINTP1, this locus has been annotated as a pseudogene. The physiological basis for the complexity of Hint homologs remains to be determined.
Perhaps the most remarkable sequence related to Hint occurs in birds on the W chromosome. ASW, which has every chemically important residue and nearly all nucleoside-binding residues mutated with respect to Hint, is an apparently catalytically inactive Hint family member encoded in an array of approximately 40 tandem repeats on avian W chromosomes (48, 49) whose mRNA may be stabilized by a unique mechanism (50). A canonical Hint homolog is encoded on avian Z chromosomes (49). Female birds are the heterogametic (WZ) sex while males are the homogametic (ZZ) sex. As the genetic basis for sex determination is not known in birds (51) and ASW is expressed in a temporal and special manner that could relate to feminization (48, 49), it will be important to investigate the function of ASW and avian Hint genetically, biochemically and structurally. Heterodimerization as a mode of ASW inhibition of Hint was suggested in the absence of knowledge that Hint function depends on enzyme activity (48-50). However, because Hint is a dimeric enzyme (7) that shows no cooperativity with respect to substrate (17), it is not obvious how heterodimerization of one inactive subunit with one active subunit would reduce specific activity by more than 50%. In an accompanying paper we suggest three mechanisms by which ASW may plausibly function as a dominant negative form of Hint to promote feminization in avian development (52). The most interesting single feature in the ASW sequence that could make this molecule not only negative but dominant is Gln127, which is substituted for Trp123 in Hint, a residue that is buried across the Hint dimer interface in the interior of the other monomer (7). Our molecular model of the proposed Hint-ASW heterodimer predicts that ASW Gln127 may pull Hint His114 out of its active conformation, accounting for dominant negativity (52).
Fhit Homologs: Tumor Suppressors and ApnA Hydrolases
Branch II of the HIT superfamily consists of orthologs of human Fhit. The first member of this branch to be characterized biochemically (53), cloned (39) and characterized physiologically (54) was aph1, a diadenosine tetraphosphate (AppppA) hydrolase from S. pombe. As aph1 possesses the canonical HφHφHφφ motif, this enzyme was recognized as a member of the HIT superfamily (39), then populated by Branch I sequences, which were not yet known to encode nucleotide-binding proteins or enzymes. In addition, careful sequence inspection allowed these investigators to recognize that aph1 is distantly related to the HXHXQφφ-containing AppppA phosphorylases (39), which we now recognized as members of Branch III of the HIT superfamily. Thus, despite absence of statistically convincing sequence similarity and presence of mechanistic difference between hydrolases and phosphorylases, prescient observations made upon cloning aph1 (39) and later expanded in the review literature (55) form the basis for our current classification of three branches of HIT proteins.
Study of aph1-related molecules took on a sense of urgency with the discovery that the gene at the human chromosome 3 fragile site, which is disrupted in a familial chromosomal translocation associated with early-onset renal carcinoma and is lost in many cancers of the digestive tract, is the human aph1 homolog (44). Spanning the chromosome 3 fragile site FRA3B, which is the most fragile site in the human genome, and possessing a HIT motif, the gene was named FHIT for fragile histidine triad gene (44). It is now clear that FHIT deletions are among the earliest and most frequent genetic alterations in development of tumors in environmentally-exposed tissues such as the lung (56, 57). Because the chromosomal location of FHIT is fragile and susceptible to deletions, the null hypothesis stated that losses at the FHIT locus are not contributing causes of cancer but rather consequences of the genome instability of p53- tumors (58). Though the kinetics of FHIT inactivation in the lung are inconsistent with this hypothesis (loss of Fhit protein occurs earlier and more frequently than TP53 mutation (56)), it was critically important to determine whether re-expression of the FHIT gene in fhit- cancer cell lines suppressed tumorigenicity. This has been demonstrated convincingly in several experimental systems (59-66) and has been recently reviewed (67).
The murine fhit gene is encoded on a syntenic portion of chromosome 14 and also spans a fragile site (68, 69). Murine fhit is not an essential gene and fhit-/- mice survive and develop a wide range of tumors including lymphomas and sebaceous tumors at higher incidence than wild-type mice (70). Taking advantage of the fhit gene’s susceptibility to mutational inactivation, it was discovered that treatment of fhit+/- mice with low doses of the carcinogen N-nitrosomethylbenzylamine (NMBA) lead rapidly to development of Fhit- stomach and sebaceous tumors (71). Interestingly, stomach and sebaceous tumors are observed in a subset of hereditary nonpolyposis colon cancer (HNPCC) termed Muir-Torre syndrome that is thought to be due to loss of mismatch repair genes, potentially indicating that loss of FHIT is an important target in HNPCC (71).
Because loss of FHIT in development of lung cancer occurs in preneoplastic lesions (56) and FHIT reexpression in fhit-/- cells suppresses tumor formation by induction of apoptosis (60, 61, 63, 64, 66), introduction of FHIT-expressing viruses has been explored as a preventative strategy in the NMBA-tumor induction model. Remarkably, administration of FHIT via viral vectors shortly after NMBA administration prevents tumor development (65), presumably by killing the cells that suffer their second hit at fhit.
Though FHIT re-expression in fhit- human or mouse cells induces death by apoptosis (60, 61, 63, 64, 66), relatively little can be said about the cellular pathway through which Fhit acts. There are three clues, however. First, the tumor suppressing function of Fhit does not depend on hydrolysis of ApnA (59, 62) but may depend on forming a complex with ApnA (72). Second, there is a Rosetta Stone relationship (73, 74) between Fhit and Nit, a novel member of the nitrilase superfamily (75, 76). Third, Fhit homologs are found through a sufficient swath of eukaryotic evolution to predate many players in developmentally regulated apoptotic pathways and are likely to have a more fundamental cellular function, whose alteration triggers apoptotic consequences in animals.
Fhit homologs from different organisms vary in their substrate specificity for AppppA and diadenosine triphosphate (ApppA) but the enzymes that have been characterized kinetically bind both nucleotides very specifically (42, 76). Whereas aph1 from S. pombe prefers AppppA (53), human Fhit (40) and S. cerevisiae Hnt2 (77, 78) prefer ApppA. Fhit homologs from human and C. elegans do not discriminate significantly between ApppA and AppppA in terms of Km, both enzymes being saturated by both substrates in the low micromolar range (42, 76), and thus Fhit homologous enzymes are thought to discriminate between substrates in the kcat term (42, 76). Moreover, as the deadenylylation step for ApppA and AppppA is not different, these enzymes must discriminate on the basis of the rate of adenylylation of substrates that are bound with nearly identical Km values (42).
ApnA are made in side reactions of certain tRNA synthetases (79) and are the subject of a comprehensive recent review (80). Because the functional consequences of elevated ApnA are not known, we considered three possible connections between the tumor suppressing function of Fhit and ApnA (72). First, elevated ApnA may have a proliferative or anti-apoptotic function that is potentiated by loss of Fhit. Second, ApnA may signal through Fhit in an antiproliferative or pro-apoptotic manner that is lost with loss of Fhit. Third, the tumor suppressing function may be ApnA-independent (72). Because the H96N mutant of Fhit had been shown to be extremely deficient in enzymatic activity (40) and yet functional in tumor suppression (59), it was important to characterize the nature of the biochemical defect. The mutant turned out to be more than 4,000,000-fold reduced in kcat and less than four-fold increased in the Km term with respect to the wild-type enzyme in hydrolysis of ApppA (72). This was clearly inconsistent with the idea that Fhit must reduce the concentration of ApnA to function in tumor suppression. Because the H96N mutant retained a low micromolar Km for ApppA and was found crystallographically to bind nonhydrolyzable ApppA in a manner nearly identical to that of wild-type Fhit, it was argued that the tumor suppressing function of Fhit may depend on forming an ApnA complex (72). Furthermore, the crystallographically defined Fhit dimer surface that becomes filled with two ApnA molecules was proposed as a possible site of effector interactions (72). The critical test of this model depends either on discovery of an effector that binds Fhit-ApnA, potentially in a manner that retards hydrolysis, or showing that Fhit mutants that are defective in substrate-binding are poor tumor suppressors (72). These are both active research areas.
In the course of cloning FHIT-homologous genes from invertebrates, it was discovered that flies and worms encode Fhit in a fusion protein containing a novel ∼300 a.a. polypeptide at its N-terminus that is a member of the nitrilase superfamily (75). Analysis of domain fusion events in sequenced genomes suggested that such fusions might be “Rosetta Stones” that decode a previously hidden cellular or biochemical interaction between proteins unrelated by sequence (73, 74). It was argued that fusions between domains that share a common phylogenetic profile (both genes are either represented in or both genes are absent from a genome) and which are coordinately expressed in organisms in which they are not fused are most likely to be indicative of a common cellular pathway (81).
Despite the fact that the “Nit” domain of fly and worm NitFhit had not been previously observed in any mammal, NIT genes were discovered in human and mouse as separate genes from FHIT, and murine nit1 and murine fhit were shown to have nearly identical patterns of tissue-specific mRNA accumulution (75). Furthermore, discovery of homologs of the Nit domain of NitFhit in frogs, fission yeast and budding yeast supported the co-evolution of these polypeptides in the organisms that have Fhit homologs (76). The crystal structure of worm NitFhit showed that Nit is a tetramer that binds two Fhit dimers and that the C-terminus of Nit is conserved to interact with Fhit (76). The Nit-interacting surface of Fhit is not the Fhit-ApnA surface (76) but the architecture of the 200 kDa NitFhit complex is such that it could potentially bind two Fhit-ApnA interacting molecules at opposite poles of the 140 Å tetramer.
The active site of all members of the nitrilase superfamily was inferred from the NitFhit structure to be a Glu-Lys-Cys catalytic triad (76). The nitrilase superfamily consists of at least 13 branches of enzymes, members of only one of which have been shown to possess nitrilase activity (82). Eight branches of the nitrilase superfamily have either demonstrated or predicted amide hydrolase or amide condensation activities for a variety of substrates and domain fusion events have been common in the nitrilase superfamily (82). Biochemical studies have yet to identify a substrate for the Nit active site of NitFhit or a Nit ortholog.
In addition to fungi (53, 77, 78), mammals (44, 68, 69), and invertebrates (75), Fhit homologs can be found in the genomes of microsporidia such as E. cuniculi (83), mycetozoa such as D. discoidium (unpublished), and green plants such as A. thaliana (84). According to the tree of eukaryotic life constructed from protein data (85), unless Fhit is found in plants due to a lateral transfer, the earliest rift that would have preceded appearance of Fhit was the separation of plants from animals, fungi, microsporidia and mycetozoa. It will therefore be important to look for Fhit homologs in organisms such as Tetrahymena, Plasmodium, Trypanosomes and Euglena as they are thought to be on the plant side of the plant/animal rift (85) that potentially preceded the appearance of Fhit. Because several apoptotic players including p53 and Bcl-2 family proteins are in animal-specific pathways, it is hard to reckon the primary function of Fhit being in direct management of apoptosis even though the biological effect of FHIT reexpression in fhit- cells has been apoptotic (60, 61, 63, 64, 66). A role more fundamental to eukaryotic cell biology, such as regulation of protein localization, which could have consequences that result in apoptosis in animal cells, seems more likely.
GalT-related Enzymes are Specific Nucleoside Monophosphate Transferases that Contain a HXHXQφφ Motif
Branch III of the HIT superfamily consists of GalT homologs and other related nucleotide transferases. The second step in the Leloir pathway for galactose utilization requires galactose-1-phosphate to be transferred to UMP to form UDP-galactose (1). As shown in Figure 2, the reaction is catalyzed by GalT, which consumes UDP-glucose to form a covalent UMP intermediate with His166 (43, 86) releasing glucose-1-phosphate, and then reacts with galactose-1-phosphate to form UDP-galactose. Humans with mutations in the GALT gene are the largest class of galactosemics and are initially diagnosed with failure to thrive in the first days of life. Despite being put on a galactose-free diet, the long-term outcomes for GALT galactosemics include problems with cataracts and intelligence (87).
S. cerevisiae (88) and E. coli (89) GalT sequences were similar enough to guide cloning of human GalT (9). GalT sequences co-existed with Hint sequences (6, 90) in the databases for several years without notice. In fact, the first Hint structure (8) and the first GalT structure (2) were initially considered to be unrelated to any other solved structures before Hint-nucleotide structures were solved (7). Independently, computational approaches based on structural coordinates (10) and on sequence alone (11) were able to detect the similarity.
While the Leloir pathway and thus GalT homologs may be extant in all organisms that utilize galactose as a carbon source (1), at least two other enzymes exist in subsets of organisms that are recognizable by sequence and mechanism to be members of Branch III of the HIT superfamily. S. cerevisiae and some other fungi and bacteria possess AppppA phosphorylases (91-93) that produce ATP and ADP (not AMP) with overall retention of configuration (94). Like GalT, the reaction proceeds through a covalent adenylylated intermediate (95) with release of ATP and uses inorganic phosphate rather than water to phosphorylyze the adenylylated intermediate to produce the ADP product. The three S. cerevisiae Branch III enzymes, Gal7, Apa1 and Apa2, are included in Table 1. Finally, Thiobacillus denitrificans possesses a GalT-related enzyme that is now termed adenylylsulfate:phosphate adenylytransferase whose NMP-X substrate is AMP-SO4 (96). After producing a covalent adenylylated intermediate that was detected by mass spectroscopy, this enzyme reacts with inorganic phosphate to produce ADP (96). Branch III enzymes are recognizable by related sequences that include a HXHXQφφ motif and mechanisms that depend on transferring the covalently bound nucleoside monophosphate to phosphate or a phosphorylated second substrate, rather than hydrolyzing it with water (Figure 2).
Rotaviruses Utilize an Enzyme Resembling Hint That May Have Arisen Via Convergent Evolution
Rotaviruses, which are major causes of diarrhea in humans and other animals, consist of six structural and six nonstructural proteins encoded by eleven segments of double stranded RNA. Nonstructural protein two (NSP2) is a conserved basic protein that has been shown genetically to be required for RNA packaging (97, 98), which possesses intrinsic single stranded RNA-binding activity and magnesium-dependent nucleoside triphosphatase activity for GTP, ATP, UTP and CTP to the corresponding nucleoside diphosphates (99). The crystal structure of the NSP2 octomer revealed that an ∼80 residue segment within the C-terminal domain of the 317 amino acid NSP2 monomer adopts a Hint-like three dimensional fold that is superimposible with beta strands 1 through 3, helix 2, and strands 4 through 5 of Hint (100). Though NSP2 is an octomer, NSP2 has no dimer interface that aligns with Hint’s dimer interface at helix 2 and strand 4, nor is there sequence conservation with Hint that would have indicated similarity by descent (100). While His221 of NSP2 can be aligned with His112 of Hint, no other residues important for nucleotide-binding or hydrolysis in Hint are found at corresponding positions in NSP2. The fact that NSP2 hydrolyzes any nucleoside triphosphate in a magnesium-dependent manner at the γ-phosphate while HIT proteins are specific for adenylylation or uridylylation in a cation-independent manner at the α phosphate suggest that NSP2 is not a HIT protein. However, we wish to revisit our tentative exclusion of NSP2 from the HIT superfamily in light of a nucleotide-bound NSP2 crystal structure. If NSP2 has a mode of nucleotide recognition that is nearly isosteric with that of Hint, one might be able to consider NSP2 a distant relative that has substituted a metal and new sequences for conserved residues, lost the characteristic dimer, and altered the site of nucleotide chemistry. For now, we agree that the Hint-NSP2 resemblance is striking but convergent (100), and not mechanistically related.
Mechanistic Insights in the HIT Superfamily: Hydrolases Versus Transferases and a Potential Role for Acid-Base Catalysis
Truly related sequences not only fold similarly but function similarly so the extensive mechanistic dissection of GalT serves as a model for the entire HIT superfamily. In this regard, it is clear that both the nucleotidylation and the nucleotidyl transferase steps depicted in Figure 2 proceed with inversion of configuration at the α phosphate, leading to overall retention of configuration (101, 102). The GalT mechanism is thus extremely symmetrical: a UDP-hexose brings UMP in for covalent nucleotidylation of His168 producing a hexose-1-phosphate leaving group, and a hexose-1-phosphate retraces those steps to reclaim the nucleotide as part of a UDP-hexose product. The residues involved in recognizing the hexose-1-phosphate leaving/attacking groups have been defined crystallographically and, indeed, glucose and galactose are bound at the same site (4). The key difference between the HIT hydrolases in the Hint and Fhit branches and the transferases in the GalT branch is that hydrolases do not await a second substrate to accept the histidine-bound nucleotide but instead are competent to transfer the nucleotide to water. Might the nearly isosteric nature of the GalT leaving and attacking groups be the basis for GalT-UMP intermediates being resistant to hydrolysis? The existence of AppppA phosphorylases and adenylylsulfate:phosphate adenylytransferase makes us argue against this idea. Both of these enzymes are GalT-related transferases (96) that go through covalent intermediates (95, 96) with known stereochemistry (overall retention) in the case of AppppA phosphorylase (94). Because the AppppA leaving group, ATP, is not isosteric with the second substrate, phosphate, it is difficult to argue that substrate-binding sites remote from the nucleotidylated histidine enforce transfer rather than hydrolysis. We argue instead that Branch III enzymes lack the final histidine residue in the HIT motif, which is necessary to activate water for hydrolysis.
Kinetic studies with O18H2O and with stereospecific α-thio labeled γ-(m-Nitrobenzyl) adenosine 5′-O-triphosphate demonstrated that Fhit hydrolyzes substrates with overall retention of configuration and addition of water to the AMP product (41). Given physical evidence (103) for a covalently adenylylated intermediate and the stereochemical evidence (41) for retention of configuration, it is reasonable to assume that the hydrolytic water comes “in-line” from the position formerly occupied by the α-β bridging oxygen of a Fhit substrate. The fact that Branch I and II enzymes and the non-GalT Branch III enzymes are less symmetrical than GalT (i.e., water or inorganic phosphate as the nucleotide acceptor as opposed to another hexose-1-phosphate) does not make us think that the geometry of hydrolysis or phosphorylysis is any different than the geometry of nucleotide transfer in GalT (4).
Taken this way, some of the catalytic efficiency of Fhit may derive from the enzyme providing the same entry route to a polarized water as it provided an exit route for ADP. In the case of Hint, whose substrates include phosphoramides such as AMP-NH2 and AMP linked to amino acids (17), the hydrolytic water’s final approach to attack the enzyme adenylylate should retrace the initial escape path of the amine leaving group. In both cases, the enzyme has a role in getting the leaving group to leave the confines of the active site, which is the area in which a leaving group can reattack. Altering the ionization state and/or solvating the leaving group is thus important because, by jettisoning the leaving group into bulk solvent, what was microscopically reversible becomes effectively irreversible.
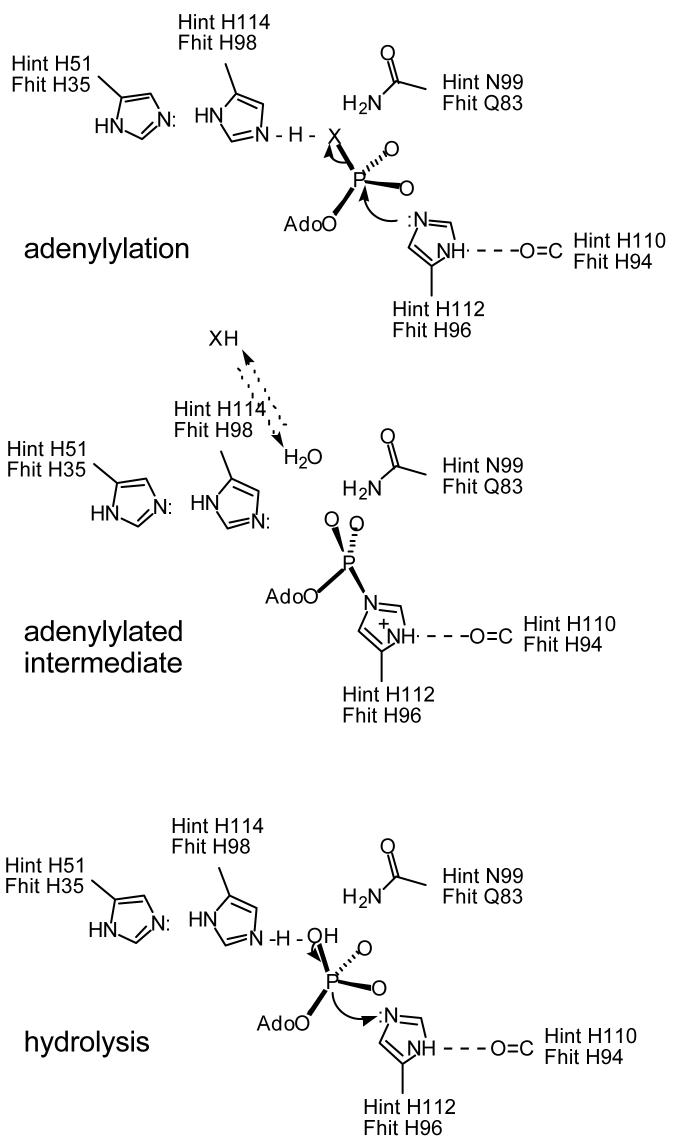
We make these points to disagree with one aspect of the Fhit mechanism proposed as a result of stereochemical analysis (41). According to that proposed mechanism, His98, the final His of the HIT motif, which aligns with Gln168 of GalT, makes a hydrogen bond with an α phosphate oxygen of an ApppA substrate. In our structures of wild-type and mutant Fhit bound to ApppA analog, IB2, which has a phosphorothio substitution on one α phosphate and substitution of a methylene group for the other α-β bridging oxygen (104), we observed His98 to be positioned to interact with the α-β bridging oxygen (72) and not an α phosphate oxygen like Gln168 of GalT (3). The distance between the His98 εN and the α-β bridging oxygen in the co-crystal structure of Fhit H96N with nonhydrolyzable ApppA was 2.5 Å (72), which is in the range of low barrier hydrogen bonds that are frequently found at enzyme active sites (105). As shown in Figure 4, we propose that the final His in HIT hydrolases is positioned to interact with the amine or ADP leaving group and the attacking water as a general acid-base catalyst. The fourth invariant His residue in HIT hydrolases, Hint His51/Fhit His35, is positioned to stabilize the proposed positive charge of Hint His114/Fhit His98 in the ground state. As we interpret our X-ray structures (7, 72), the exit path for the leaving group and the entry path for the hydrolytic water is between Hint His114/Fhit His98 and Hint Asn99/Fhit Gln83. Referring back to Figure 1B, which is a product complex between Hint and GMP (7), the α phosphate oxygen interpreted to result from water addition is the lower left oxygen shown between His114 and Asn99. In the case of Fhit, this mechanism also accounts for the catalytic roles for His35 and His98 in addition to the role of the nucleophilic His96, as defined by mutagenesis (40). Further, in an accompanying paper (52), we describe how alteration of the position of the proposed general acid base catalyst, His114 in chicken Hint, in the proposed Hint-ASW heterodimer (48-50) may reduce the activity of the heterodimer substantially more than two-fold.
Figure 4.
Proposed catalytic mechanism for HIT hydrolases. Hint and Fhit substrates are depicted as AMP-X and their leaving groups as XH. Upon adenylylation of the middle His of the HIT motif, the atom at position X (a phosphoramide N for Hint substrates and the α-β bridging O for Fhit substrates) receives a low barrier hydrogen bond from the C-terminal His of the HIT motif. Transfer of this proton is postulated to favor leaving over readenylylation. The C-terminal His is then proposed to accept a low barrier hydrogen bond from the incoming water molecule that hydrolyzes the adenylylated intermediate.
Though we have structural but not kinetic evidence for Hint His114/Fhit His98 as a general acid-base, this mechanism is compelling for Hint because the amine product must be protonated to leave, and the incoming water would gain nucleophilicity while restoring His114 to an acid. In the case of Fhit, we hesitated to suggest that His98 protonates an ADP or ATP leaving group because the terminal phosphate has two negative charges in bulk solvent at pH 7. We now consider, however, that donation of a proton from His98 would render the terminal oxygen a poor nucleophile to reattack the α phosphate. In fact, a low barrier hydrogen bond may be so favorable to form that the rate-limiting step in adenylylation may be getting the leaving group to take the proton and go. We have previously observed that substrates with fundamentally unaltered chemical lability such as ApppA, AppppA, ApppBODIPY and GpppBODIPY are cleaved with increasingly poor rates suggesting that slow substrates may prefer to reattack than to exit (42). Ultimately the hydrogen bond network between His35 and His98 is postulated to promote transfer of the His98 Nε proton to the leaving group, rendering the adenylylation reaction not immediately reversible and His98 basic to activate ahydrolytic water.
The most common galactosemic mutation in human GALT, Q188R, corresponds to E. coli mutant Q168R that has been characterized and found to be reduced a million-fold in rates of uridylylation and deuridylylation (106). It will be interesting to characterize a site-directed Q168H mutant in vitro to see whether alteration of the HXHXQφφ motif to a hydrolase-like HXHXHφφ motif is sufficient to produce some hydrolytic activity. Though deletions and mutations in GALT are not usually lethal once galactose is eliminated from the diet (87), it is possible that mutations in GALT which produce UDP-hexose hydrolase activity have not been observed because of the potentially lethal consequences of consuming UDP-glucose.
ACKNOWLEDGMENT
In preparing this review, it would be difficult to overstate the value of the collected works of Dr. Perry A. Frey and co-workers on GalT structure and function, and the contributions of Drs. Preston Garrison and Larry D. Barnes on identifying enzymatic activities and the sequence and structural basis for function in the HIT superfamily. Additionally, I thank Drs. Dagmar Ringe, Charles P. Scott, Kay Huebner, Helen C. Pace, Keith W. Caldecott and G. Michael Blackburn for helpful discussions.
This work was supported by grants from the NIH (CA75954 and CA77738).
Footnotes
- HIT
- histidine triad
- φ
- hydrophobic amino acid
- GalT
- galactose-1-phosphate uridylyltranferase
- Hint
- histidine triad nucleotide-binding protein
- Fhit
- fragile histidine triad
- APTX
- Aprataxin gene
- ASW
- avian sex-specific W-linked gene
- EST
- expressed sequence tag
- ApnA
- diadenosine polyphosphate
- CAK
- Cdc2 activating kinase
- TFIIH
- general transcription factor TFIIH
- TFIIK
- mammalian Cdk7-cyclin H-MAT1 or yeast Kin28-Ccl1-Tfb3 trimer
- AOA
- ataxia-oculomotor apraxia syndrome
- PNK
- polynucleotide kinase
- PANT
- polynucleotide kinase and Aprataxin N-terminus
- FHA
- forkhead associated
- NMBA
- N-nitrosomethylbenzylamine
- AppppA
- diadenosine tetraphosphate
- ApppA
- diadenosine triphosphate
- HNPCC
- hereditary nonpolyposis colon cancer
- NSP2
- rotavirus nonstructural protein two
REFERENCES
- 1.Frey PA. FASEB J. 1996;10:461–70. [PubMed] [Google Scholar]
- 2.Wedekind JE, Frey PA, Rayment I. Biochemistry. 1995;34:11049–11061. doi: 10.1021/bi00035a010. [DOI] [PubMed] [Google Scholar]
- 3.Wedekind JE, Frey PA, Rayment I. Biochemistry. 1996;35:11560–11569. doi: 10.1021/bi9612677. [DOI] [PubMed] [Google Scholar]
- 4.Thoden JB, Ruzicka FJ, Frey PA, Rayment I, Holden HM. Biochemistry. 1997;36:1212–22. doi: 10.1021/bi9626517. [DOI] [PubMed] [Google Scholar]
- 5.Gilmour J, Liang N, Lowenstein JM. Biochem. J. 1997;326:471–7. doi: 10.1042/bj3260471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seraphin B. DNA Seq. 1992;3:177–179. doi: 10.3109/10425179209034013. [DOI] [PubMed] [Google Scholar]
- 7.Brenner C, Garrison P, Gilmour J, Peisach D, Ringe D, Petsko GA, Lowenstein JM. Nat. Struct. Biol. 1997;4:231–238. doi: 10.1038/nsb0397-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lima C, Klein MG, Weinstein IB, Hendrickson WA. Proc. Natl. Acad. Sci. U.S.A. 1996;93:5357–5362. doi: 10.1073/pnas.93.11.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reichardt JK, Berg P. Mol. Biol. Med. 1988;5:107–22. [PubMed] [Google Scholar]
- 10.Holm L, Sander C. Structure. 1997;5:165–71. doi: 10.1016/s0969-2126(97)00176-7. [DOI] [PubMed] [Google Scholar]
- 11.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraser ED, Walsh MP. FEBS Lett. 1991;294:285–9. doi: 10.1016/0014-5793(91)81450-m. [DOI] [PubMed] [Google Scholar]
- 13.Brenner C, Bieganowski P, Pace HC, Huebner K. J. Cell. Physiol. 1999;181:179–187. doi: 10.1002/(SICI)1097-4652(199911)181:2<179::AID-JCP1>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pruitt KD, Maglott DR. Nucleic Acids Res. 2001;29:137–40. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu J, Mungall C, Law A, Papworth R, Nelson JP, Brown A, Simpson I, Leckie S, Burt DW, Hillyard AL, Archibald AL. Nucleic Acids Res. 2001;29:106–10. doi: 10.1093/nar/29.1.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ball CA, Dolinski K, Dwight SS, Harris MA, Issel-Tarver L, Kasarskis A, Scafe CR, Sherlock G, Binkley G, Jin H, Kaloper M, Orr SD, Schroeder M, Weng S, Zhu Y, Botstein D, Cherry JM. Nucleic Acids Res. 2000;28:77–80. doi: 10.1093/nar/28.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bieganowski P, Garrison PN, Hodawadekar SC, Faye G, Barnes LD, Brenner C. J. Biol. Chem. 2002;277:10852–10860. doi: 10.1074/jbc.M111480200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korsisaari N, Makela TP. J. Biol. Chem. 2000;275:34837–34840. doi: 10.1074/jbc.C000505200. [DOI] [PubMed] [Google Scholar]
- 19.Fisher RP, Morgan DO. Cell. 1994;78:713–24. doi: 10.1016/0092-8674(94)90535-5. [DOI] [PubMed] [Google Scholar]
- 20.Fisher RP, Jin P, Chamberlin HM, Morgan DO. Cell. 1995;83:47–57. doi: 10.1016/0092-8674(95)90233-3. [DOI] [PubMed] [Google Scholar]
- 21.Devault A, Martinez AM, Fesquet D, Labbe JC, Morin N, Tassan JP, Nigg EA, Cavadore JC, Doree M. EMBO J. 1995;14:5027–36. doi: 10.1002/j.1460-2075.1995.tb00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serizawa H, Makela TP, Conaway JW, Conaway RC, Weinberg RA, Young RA. Nature. 1995;374:280–2. doi: 10.1038/374280a0. [DOI] [PubMed] [Google Scholar]
- 23.Shiekhattar R, Mermelstein F, Fisher RP, Drapkin R, Dynlacht B, Wessling HC, Morgan DO, Reinberg D. Nature. 1995;374:283–7. doi: 10.1038/374283a0. [DOI] [PubMed] [Google Scholar]
- 24.Keogh MC, Cho EJ, Podolny V, Buratowski S. Mol. Cell. Biol. 2002;22:1288–97. doi: 10.1128/mcb.22.5.1288-1297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimmelman J, Kaldis P, Hengartner CJ, Laff GM, Koh SS, Young RA, Solomon MJ. Mol. Cell. Biol. 1999;19:4774–87. doi: 10.1128/mcb.19.7.4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aicardi J, Barbosa C, Andermann E, Andermann F, Morcos R, Ghanem Q, Fukuyama Y, Awaya Y, Moe P. Ann. Neurol. 1988;24:497–502. doi: 10.1002/ana.410240404. [DOI] [PubMed] [Google Scholar]
- 27.Barbot C, Coutinho P, Chorao R, Ferreira C, Barros J, Fineza I, Dias K, Monteiro J, Guimaraes A, Mendonca P, do Ceu Moreira M, Sequeiros J. Arch. Neurol. 2001;58:201–5. doi: 10.1001/archneur.58.2.201. [DOI] [PubMed] [Google Scholar]
- 28.Koike R, Tanaka H, Tsuji S. Neurol. Med. 1998;48:237–242. [Google Scholar]
- 29.Date H, Onodera O, Tanaka H, Iwabuchi K, Uekawa K, Igarashi S, Koike R, Hiroi T, Yuasa T, Awaya Y, Sakai T, Takahashi T, Nagatomo H, Sekijima Y, Kawachi I, Takiyama Y, Nishizawa M, Fukuhara N, Saito K, Sugano S, Tsuji S. Nat. Genet. 2001;29:184–8. doi: 10.1038/ng1001-184. [DOI] [PubMed] [Google Scholar]
- 30.Moreira MC, Barbot C, Tachi N, Kozuka N, Uchida E, Gibson T, Mendonca P, Costa M, Barros J, Yanagisawa T, Watanabe M, Ikeda Y, Aoki M, Nagata T, Coutinho P, Sequeiros J, Koenig M. Nat. Genet. 2001;29:189–93. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- 31.Pedone PV, Ghirlando R, Clore GM, Gronenborn AM, Felsenfeld G, Omichinski JG. Proc. Natl. Acad. Sci. U.S.A. 1996;93:2822–6. doi: 10.1073/pnas.93.7.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Omichinski JG, Pedone PV, Felsenfeld G, Gronenborn AM, Clore GM. Nat. Struct. Biol. 1997;4:122–32. doi: 10.1038/nsb0297-122. [DOI] [PubMed] [Google Scholar]
- 33.Jilani A, Ramotar D, Slack C, Ong C, Yang XM, Scherer SW, Lasko DD. J. Biol. Chem. 1999;274:24176–86. doi: 10.1074/jbc.274.34.24176. [DOI] [PubMed] [Google Scholar]
- 34.Karimi-Busheri F, Daly G, Robins P, Canas B, Pappin DJ, Sgouros J, Miller GG, Fakhrai H, Davis EM, Le_Beau MM, Weinfeld M. J. Biol. Chem. 1999;274:24187–94. doi: 10.1074/jbc.274.34.24187. [DOI] [PubMed] [Google Scholar]
- 35.Durocher D, Henckel J, Fersht AR, Jackson SP. Mol. Cell. 1999;4:387–94. doi: 10.1016/s1097-2765(00)80340-8. [DOI] [PubMed] [Google Scholar]
- 36.Li J, Smith GP, Walker JC. Proc. Natl. Acad. Sci. U.S.A. 1999;96:7821–6. doi: 10.1073/pnas.96.14.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yaffe MB, Cantley LC. Nature. 1999;402:30–1. doi: 10.1038/46925. [DOI] [PubMed] [Google Scholar]
- 38.Durocher D, Taylor IA, Sarbassova D, Haire LF, Westcott SL, Jackson SP, Smerdon SJ, Yaffe MB. Mol. Cell. 2000;6:1169–82. doi: 10.1016/s1097-2765(00)00114-3. [DOI] [PubMed] [Google Scholar]
- 39.Huang Y, Garrison PN, Barnes LD. Biochem. J. 1995;312:925–932. doi: 10.1042/bj3120925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barnes LD, Garrison PN, Siprashvili Z, Guranowski A, Robinson AK, Ingram SW, Croce CM, Ohta M, Huebner K. Biochemistry. 1996;35:11529–11535. doi: 10.1021/bi961415t. [DOI] [PubMed] [Google Scholar]
- 41.Abend A, Garrison PN, Barnes LD, Frey PA. Biochemistry. 1999;38:3668–3676. doi: 10.1021/bi981895j. [DOI] [PubMed] [Google Scholar]
- 42.Draganescu A, Hodawadekar SC, Gee KR, Brenner C. J. Biol. Chem. 2000;275:4555–4560. doi: 10.1074/jbc.275.7.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong LJ, Frey PA. J. Biol. Chem. 1974;249:2322–4. [PubMed] [Google Scholar]
- 44.Ohta M, Inoue H, Cotticelli MG, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T, Croce CM, Huebner K. Cell. 1996;84:587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- 45.Shih LY, Suslak L, Rosin I, Searle BM, Desposito F. Am. J. Med. Genet. 1984;19:539–43. doi: 10.1002/ajmg.1320190316. [DOI] [PubMed] [Google Scholar]
- 46.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 47.Brzoska PM, Chen HY, Levin NA, Kuo WL, Collins C, Fu KK, Gray JW, Christman MF. Genomics. 1996;36:151–156. doi: 10.1006/geno.1996.0435. [DOI] [PubMed] [Google Scholar]
- 48.O’Neill M, Binder M, Smith C, Andrews J, Reed K, Smith M, Miller C, Lambert D, Sinclair A. Dev. Genes Evol. 2000;210:243–249. doi: 10.1007/s004270050310. [DOI] [PubMed] [Google Scholar]
- 49.Hori T, Asakawa S, Itoh Y, Shimizu N, Mizuno S. Mol. Biol. Cell. 2000;11:3645–60. doi: 10.1091/mbc.11.10.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lemaire R, Prasad J, Kashima T, Gustafson J, Manley JL, Lafyatis R. Genes Dev. 2002;16:594–607. doi: 10.1101/gad.939502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ellegren H. EMBO Rep. 2001;2:192–6. doi: 10.1093/embo-reports/kve050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pace HC, Brenner C. Biochemistry submitted. 2002 [Google Scholar]
- 53.Robinson AK, de la Pena CE, Barnes LD. Biochim. Biophys. Acta. 1993;1161:139–148. doi: 10.1016/0167-4838(93)90207-8. [DOI] [PubMed] [Google Scholar]
- 54.Ingram SW, Barnes LD. Biochem. J. 2000;350:663–9. [PMC free article] [PubMed] [Google Scholar]
- 55.Huebner K, Garrison PN, Barnes LD, Croce CM. Annu. Rev. Genet. 1998;32:7–31. doi: 10.1146/annurev.genet.32.1.7. [DOI] [PubMed] [Google Scholar]
- 56.Sozzi G, Pastorino U, Moiraghi L, Tagliabue E, Pezzella F, Ghirelli C, Tornielli S, Sard L, Huebner K, Pierotti MA, Croce CM, Pilotti S. Cancer Res. 1998;58:5032–5037. [PubMed] [Google Scholar]
- 57.Huebner K, Sozzi G, Brenner C, Pierotti MA, Croce CM. Adv. Oncology. 1999;15:3–10. [Google Scholar]
- 58.Boldog F, Gemmill RM, West J, Robinson M, Robinson L, Li EF, Roche J, Todd S, Waggoner B, Lundstrom R, Jacobson J, Mullokandov MR, Klinger H, Drabkin HA. Hum. Mol. Genet. 1997;6:193–203. doi: 10.1093/hmg/6.2.193. [DOI] [PubMed] [Google Scholar]
- 59.Siprashvili Z, Sozzi G, Barnes LD, McCue P, Robinson AK, Eryomin V, Sard L, Tagliabue E, Greco A, Fusetti L, Schwartz G, Pierotti MA, Croce CM, Huebner K. Proc. Natl. Acad. Sci. U.S.A. 1997;94:13771–13776. doi: 10.1073/pnas.94.25.13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ji L, Fang B, Yeh N, Fong K, Minna JD, Roth JA. Cancer Res. 1999;59:3333–3339. [PubMed] [Google Scholar]
- 61.Sard L, Accornero P, Tornielli S, Delia D, Bunone G, Campiglio M, Colombo MP, Gramegna M, Croce CM, Pierotti MA, Sozzi G. Proc. Natl. Acad. Sci. U.S.A. 1999;96:8489–8492. doi: 10.1073/pnas.96.15.8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Werner NS, Siprashvili Z, Fong LYY, Marquitan G, Schroder JK, Bardenheuer W, Seeber S, Huebner K, Schutte J, Opalka B. Cancer Res. 2000;60:2780–2785. [PubMed] [Google Scholar]
- 63.Dumon KR, Ishii H, Vecchione A, Trapasso F, Baldassarre G, Chakrani F, Druck T, Rosato EF, Williams NN, Baffa R, During MJ, Huebner K, Croce CM. Cancer Res. 2001;61:4827–4836. [PubMed] [Google Scholar]
- 64.Ishii H, Dumon KR, Vecchione A, Trapasso F, Mimori K, Alder H, Mori M, Sozzi G, Baffa R, Huebner K, Croce CM. Cancer Res. 2001;61:1578–1584. [PubMed] [Google Scholar]
- 65.Dumon KR, Ishii H, Fong LYY, Zanesi N, Fidanza V, Mancini R, Vecchione A, Baffa R, Trapasso F, During MJ, Huebner K, Croce CM. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3346–3351. doi: 10.1073/pnas.061020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roz L, Gramegna M, Ishii H, Croce CM, Sozzi G. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3615–20. doi: 10.1073/pnas.062030799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ishii H, Dumon KR, Vecchione A, Fong LY, Baffa R, Huebner K, Croce CM. J. Am. Med. Assoc. 2001;286:2441–9. doi: 10.1001/jama.286.19.2441. [DOI] [PubMed] [Google Scholar]
- 68.Pekarsky Y, Druck T, Cotticelli MG, Ohta M, Shou J, Mendrola J, Montgomery JC, Buchberg AM, Siracusa LD, Manenti G, Fong LY, Dragani TA, Croce CM, Huebner K. Cancer Res. 1998;58:3401–8. [PubMed] [Google Scholar]
- 69.Glover TW, Hoge AW, Miller DE, Ascara-Wilke JE, Adam AN, Dagenais SL, Wilke CM, Dierick HA, Beer DG. Cancer Res. 1998;58:3409–14. [PubMed] [Google Scholar]
- 70.Zanesi N, Fidanza V, Fong LY, Mancini R, Druck T, Valtieri M, Rudiger T, McCue PA, Croce CM, Huebner K. Proc. Natl. Acad. Sci. U.S.A. 2001:10250–10255. doi: 10.1073/pnas.191345898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fong LYY, Fidanza V, Zanesi N, Lock LF, Siracusa LD, Mancini R, Siprashvili Z, Ottey M, Martin SE, Dolsky R, Druck T, McCue PA, Croce CM, Huebner K. Proc. Natl. Acad. Sci. U.S.A. 2000;97:4742–4747. doi: 10.1073/pnas.080063497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pace HC, Garrison PN, Robinson AK, Barnes LD, Draganescu A, Rosler A, Blackburn GM, Siprashvili Z, Croce CM, Huebner K, Brenner C. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5484–9. doi: 10.1073/pnas.95.10.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. Science. 1999;285:751–3. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 74.Enright A, Iliopoulos I, Kyrpides N, Ouzounis C. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 75.Pekarsky Y, Campiglio M, Siprashvili Z, Druck T, Sedkov Y, Tillib S, Draganescu A, Wermuth P, Rothman JH, Huebner K, Buchberg AM, Mazo A, Brenner C, Croce CM. Proc. Natl. Acad. Sci. U.S.A. 1998;95:8744–8749. doi: 10.1073/pnas.95.15.8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pace HC, Hodawadekar SC, Draganescu A, Huang J, Bieganowski P, Pekarsky Y, Croce CM, Brenner C. Curr. Biol. 2000;10:907–17. doi: 10.1016/s0960-9822(00)00621-7. [DOI] [PubMed] [Google Scholar]
- 77.Chen J, Brevet A, Blanquet S, Plateau P. J. Bacteriol. 1998;180:2345–9. doi: 10.1128/jb.180.9.2345-2349.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rubio-Texeira M, Varnum JM, Bieganowski P, Brenner C.BMC Mol. Biol 20023, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zamecnik PC, Stephenson ML, Janeway CM, Randerath K. Biochem. Biophys. Res. Commun. 1966;24:91–7. doi: 10.1016/0006-291x(66)90415-3. [DOI] [PubMed] [Google Scholar]
- 80.McLennan AG. Pharmacol. Ther. 2000;87:73–89. doi: 10.1016/s0163-7258(00)00041-3. [DOI] [PubMed] [Google Scholar]
- 81.Marcotte E, Pellegrini M, Thompson M, Yeates T, Eisenberg D. Nature. 1999;402:83–86. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- 82.Pace HC, Brenner C. Genome Biol. 2001;2:reviews0001.1–reviews0001.9. doi: 10.1186/gb-2001-2-1-reviews0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, Delbac F, El Alaoui H, Peyret P, Saurin W, Gouy M, Weissenbach J, Vivares CP. Nature. 2001;414:450–3. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 84.Sato S, Nakamura Y, Kaneko T, Katoh T, Asamizu E, Kotani H, Tabata S. DNA Res. 2000;7:31–63. doi: 10.1093/dnares/7.1.31. [DOI] [PubMed] [Google Scholar]
- 85.Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. Science. 2000;290:972–7. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- 86.Kim J, Ruzicka F, Frey PA. Biochemistry. 1990;29:10590–3. doi: 10.1021/bi00499a003. [DOI] [PubMed] [Google Scholar]
- 87.Elsas LJ, 2nd, Lai K. Genet. Med. 1998;1:40–8. doi: 10.1097/00125817-199811000-00009. [DOI] [PubMed] [Google Scholar]
- 88.Tajima M, Nogi Y, Fukasawa T. Yeast. 1985;1:67–77. doi: 10.1002/yea.320010108. [DOI] [PubMed] [Google Scholar]
- 89.Lemaire HG, Muller-Hill B. Nucleic Acids Res. 1986;14:7705–11. doi: 10.1093/nar/14.19.7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pearson JD, DeWald DB, Mathews WR, Mozier NM, Zurcher-Neely HA, Heinrikson RL, Morris MA, McCubbin WD, McDonald JR, Fraser ED, Vogel HJ, Kay CM, Walsh MP. J. Biol. Chem. 1990;242:4583–4591. [PubMed] [Google Scholar]
- 91.Plateau P, Fromant M, Schmitter JM, Buhler JM, Blanquet S. J. Bacteriol. 1989;171:6437–45. doi: 10.1128/jb.171.12.6437-6445.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Plateau P, Fromant M, Schmitter JM, Blanquet S. J. Bacteriol. 1990;172:6892–9. doi: 10.1128/jb.172.12.6892-6899.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Avila DM, Robinson AK, Kaushal V, Barnes LD. J. Bacteriol. 1991;173:7875–80. doi: 10.1128/jb.173.24.7875-7880.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lazewska D, Guranowski A. Nucleic Acids Res. 1990;18:6083–8. doi: 10.1093/nar/18.20.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Booth JW, Guidotti G. J. Biol. Chem. 1995;270:19377–19382. doi: 10.1074/jbc.270.33.19377. [DOI] [PubMed] [Google Scholar]
- 96.Bruser T, Selmer T, Dahl C. J. Biol. Chem. 2000;275:1691–8. doi: 10.1074/jbc.275.3.1691. [DOI] [PubMed] [Google Scholar]
- 97.Ramig RF, Petrie BL. J. Virol. 1984;49:665–73. doi: 10.1128/jvi.49.3.665-673.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen D, Gombold JL, Ramig RF. Virology. 1990;178:143–51. doi: 10.1016/0042-6822(90)90387-7. [DOI] [PubMed] [Google Scholar]
- 99.Taraporewala Z, Chen D, Patton JT. J. Virol. 1999;73:9934–43. doi: 10.1128/jvi.73.12.9934-9943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jayaram H, Taraporewala Z, Patton JT, Prasad BVV. Nature. 2002;417:311–315. doi: 10.1038/417311a. [DOI] [PubMed] [Google Scholar]
- 101.Frey PA, Wong LJ, Sheu KF, Yang SL. Methods Enzymol. 1982;87:20–36. doi: 10.1016/s0076-6879(82)87004-3. [DOI] [PubMed] [Google Scholar]
- 102.Arabshahi A, Brody RS, Smallwood A, Tsai TC, Frey PA. Biochemistry. 1986;25:5583–9. doi: 10.1021/bi00367a036. [DOI] [PubMed] [Google Scholar]
- 103.Lima CD, Klein MG, Hendrickson WA. Science. 1997;278:286–90. doi: 10.1126/science.278.5336.286. [DOI] [PubMed] [Google Scholar]
- 104.Blackburn GM, Liu XH, Rosler A, Brenner C. Nucleosides Nucleotides. 1998;17:301–308. doi: 10.1080/07328319808005178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cleland WW, Frey PA, Gerlt JA. J. Biol. Chem. 1998;273:25529–32. doi: 10.1074/jbc.273.40.25529. [DOI] [PubMed] [Google Scholar]
- 106.Geeganage S, Frey PA. Biochemistry. 1998;37:14500–14507. doi: 10.1021/bi9815546. [DOI] [PubMed] [Google Scholar]