Abstract
Trailer Hitch (Tral or LSm15) and enhancer of decapping-3 (EDC3 or LSm16) are conserved eukaryotic members of the (L)Sm (Sm and Like-Sm) protein family. They have a similar domain organization, characterized by an N-terminal LSm domain and a central FDF motif; however, in Tral, the FDF motif is flanked by regions rich in charged residues, whereas in EDC3 the FDF motif is followed by a YjeF_N domain. We show that in Drosophila cells, Tral and EDC3 specifically interact with the decapping activator DCP1 and the DEAD-box helicase Me31B. Nevertheless, only Tral associates with the translational repressor CUP, whereas EDC3 associates with the decapping enzyme DCP2. Like EDC3, Tral interacts with DCP1 and localizes to mRNA processing bodies (P bodies) via the LSm domain. This domain remains monomeric in solution and adopts a divergent Sm fold that lacks the characteristic N-terminal α-helix, as determined by nuclear magnetic resonance analyses. Mutational analysis revealed that the structural integrity of the LSm domain is required for Tral both to interact with DCP1 and CUP and to localize to P-bodies. Furthermore, both Tral and EDC3 interact with the C-terminal RecA-like domain of Me31B through their FDF motifs. Together with previous studies, our results show that Tral and EDC3 are structurally related and use a similar mode to associate with common partners in distinct protein complexes.
Proteins of the (L)Sm (Sm and Like-Sm) family are found in all domains of life and play critical roles in RNA metabolism (reviewed in references 26 and 54). These proteins have the Sm fold, which comprises an N-terminal α-helix stacked on top of a five-stranded β-barrel-like structure (10, 25, 38, 44, 48, 49). Sm domains often oligomerize to form hexameric or heptameric rings that stably or transiently associate with single-stranded RNA. Eubacterial and archeal genomes encode between 1 and 3 LSm paralogs, which form monohexameric or monoheptameric rings, while eukaryotes encode more than 18 (L)Sm paralogs that assemble into heteroheptameric rings of different composition and function (reviewed in references 26 and 54).
Although much is known about (L)Sm proteins consisting of a single Sm domain, proteins possessing an N-terminal Sm domain followed by C-terminal extensions with additional domains are less well characterized. These include LSm12 to LSm16 (1, 2). LSm12 is characterized by a C-terminal protein methyltransferase domain (1, 2). The LSm13-16 proteins share a divergent form of the Sm domain and a central FDF motif (1, 2). The FDF motifs of LSm13-15 are embedded in low-complexity regions rich in glycine and arginine (1, 2). In contrast, in LSm16 (known as enhancer of decapping-3 and referred to as EDC3 hereafter) the FDF motif is followed by a conserved C-terminal YjeF_N domain that adopts a divergent Rossman fold similar to one in the N-terminal domain of bacterial YjeF (1, 2, 32).
EDC3 (LSm16) is known to enhance bulk mRNA decapping in yeast and is required for the decapping-dependent regulation of RPS28B mRNA and YRA1 pre-mRNA (4, 16, 29). In agreement with this, EDC3 interacts with the decapping enzyme DCP2, the decapping activator DCP1, and other proteins that vary, depending on the species (13, 20, 50). For instance, in human cells EDC3 associates with the RNA helicase RCK/p54 and Ge-1 (also known as EDC4 or human enhancer of decapping large subunit Hedls) (20). In Drosophila melanogaster, EDC3 associates with DCP1, DCP2, and Me31B (the D. melanogaster ortholog of RCK/p54) (50). These interactions are mediated by specific EDC3 domains. Indeed, EDC3 interacts with DCP1 through its N-terminal LSm domain, with DCP2 through the linker region between the LSm domain and the FDF motif, and with Me31B through the FDF motif (50). In addition, EDC3 self-associates through its C-terminal YjeF_N domain (32, 50). Similarly, the FDF motif and the YjeF_N domain of the Saccharomyces cerevisiae Edc3 protein mediate the interaction with the yeast Me31B ortholog (Dhh1p) and self-association, respectively, demonstrating that these interactions are conserved (13, 21, 32, 33).
LSm13 to LSm15 represent a group of orthologous proteins in different species. These include S. cerevisiae Scd6p (LSm13), D. melanogaster Trailer Hitch (Tral or LSm15), Caenorhabditis elegans CAR-1, and vertebrate RAP55 (LSm14) (1, 2). Although the function of Scd6p remains unclear, available evidence on Tral, CAR-1, and RAP55 suggest that they fulfill closely related functions in the respective species. Like EDC3, these proteins are characterized by the presence of an N-terminal LSm domain and a central FDF motif.
In D. melanogaster oocytes, Tral associates with Me31B, the translational repressor CUP and the RNA-binding protein YPS (5, 6, 53). All of these proteins colocalize in cytoplasmic ribonucleoprotein (RNP) granules and play roles in the localization and translational regulation of maternal mRNAs during oogenesis and embryogenesis (53). CAR-1 and Xenopus laevis RAP55 (xRAP55) are also components of RNP granules in the germ line and associate with Me31B orthologues (CGH-1 and Xp54, respectively), as well as with YPS orthologues (CEY-2-4 and FRGY2) (3, 6, 12, 36, 47). Moreover, like EDC3, in somatic cells Tral and its orthologs localize to mRNA processing bodies or P bodies, where mRNAs destined for translational repression and/or degradation accumulate (17, 18, 47, 55).
The observation that both Tral and EDC3 share a similar domain organization, localize to P bodies in D. melanogaster cells, and associate with Me31B suggests that these proteins may have related functions. In the present study, we further characterized Tral and EDC3 in D. melanogaster S2 cells. We show that Tral and EDC3 form distinct protein complexes: Tral associates with DCP1, Me31B, and CUP, whereas, as shown before, EDC3 coimmunoprecipitates with DCP1, Me31B, and DCP2 (50). Similar to EDC3, Tral interacts with DCP1 via the LSm domain and interacts with Me31B via the FDF motif. Furthermore, the Tral LSm domain is sufficient for P-body targeting.
To begin to elucidate the molecular function of Tral, we determined the solution structure of its N-terminal LSm domain. The domain adopts a divergent Sm fold related to that of EDC3. We show that the structural integrity of this domain is required for Tral both to interact with DCP1 and CUP and to localize to P bodies. Together with our previous study on EDC3 (50), these results show that the divergent LSm domains of Tral and EDC3 lack the oligomerization and RNA-binding properties of canonical (L)Sm proteins but have acquired common novel functionalities, including DCP1 binding and P-body targeting.
MATERIALS AND METHODS
DNA constructs and transfection of S2 cells.
cDNAs encoding full-length Tral, DCP1, DCP2, EDC3, Me31B, HPat, CUP, LSm1, LSm4, and LSm7 proteins or protein domains were amplified with primers containing appropriate restriction sites, using a (dT)15-primed S2 cDNA library as a template. The amplified cDNAs were cloned into a vector allowing the expression of green fluorescent protein (GFP) or λN-hemagglutinin (HA)-peptide fusions (pAc5.1B-EGFP or pAc5.1B-λN-HA, respectively) as described previously (18, 19, 50). Mutants of the D. melanogaster Tral LSm domain were generated by site-directed mutagenesis using a Stratagene QuikChange mutagenesis kit and the oligonucleotide sequences in Table S1 in the supplemental material. All constructs were fully sequenced to confirm the presence of the mutations and the absence of additional mutations. Additional information on plasmids and oligonucleotides used in the present study is provided in Table S1 in the supplemental material.
Western blots and coimmunoprecipitations.
Antibodies to D. melanogaster Tral were raised in rats immunized with a Tral protein fragment encompassing residues 401 to 533 (FDF motif) expressed in E. coli as a glutathione S-transferase (GST) fusion. Antibodies to D. melanogaster EDC3 were raised in rats immunized with an EDC3 protein fragment encompassing residues 339 to 440 (FDF motif) expressed in E. coli as a GST fusion.
Transfections of S2 cells were performed in six-well dishes using Effectene transfection reagent (Qiagen). For coimmunoprecipitations cells were collected 3 days after transfection, washed with phosphate-buffered saline (PBS), and lysed for 15 min on ice in NET buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1 mM EDTA, and 0.1% Triton X-100) supplemented with protease inhibitors. Cells were spun at 16,000 × g for 15 min at 4°C. Anti-HA antibodies (Covance Research Products) were added to the supernatants (2.5 μl/2 × 106 cells). After 1 h at 4°C, 25 μl of protein G-agarose (Roche) was added, and the mixtures were rotated 1 h at 4°C. Beads were washed three times with NET buffer and once with NET buffer without Triton X-100. Bound proteins were eluted with sample buffer.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. Membranes were blocked in PBS containing 5% fat-free milk powder and 0.3% Tween 20. Western blotting was performed with polyclonal anti-HA antibodies (1:1,000; Sigma catalog number H6908), anti-Tral (1:2,000), anti-EDC3 (1:2,000), or anti-GFP antibodies (1:2,000) using a CDP-Star chemiluminescent immunoblot system (Western-Star kit; Tropix) as recommended by the manufacturer.
Fluorescence microscopy.
Three days after transfection, S2 cells were allowed 15 min to adhere to poly-d-lysine-coated coverslips, washed once in serum-free medium, and fixed with 4% paraformaldehyde in PBS for 15 min, followed by 5 min of incubation in methanol at −20°C. After fixation, cells were washed in PBS, permeabilized for 5 min with PBS containing 0.5% Triton X-100, and washed again with PBS. For the detection of HA fusions, cells were stained with monoclonal anti-HA antibody (Covance Research Products) diluted 1:1,000 in PBS containing 1% bovine serum albumin. Alexa Fluor 594-coupled goat secondary antibody (Molecular Probes) was used in a dilution of 1:1,000. Cells were mounted using Fluoromount-G (Southern Biotechnology Associates, Inc.). Images were acquired by using a Leica TCS SP2 confocal microscope.
Purification of the Tral LSm domain.
The LSm domain of D. melanogaster Tral (UniProtKB entry Q9VTZ0; M1 to P84) was amplified from a (dT)15-primed S2 cell cDNA library and cloned into the pETM60 vector (derived from pET24-d; Novagen). The protein was expressed in the E. coli strain BL21(DE3) Rosetta 2 at 20°C overnight. To uniformly label Tral LSm domain with 15N/13C or 15N, cells were grown in M9 minimal medium supplemented with 15NH4Cl with or without 13C6-labeled glucose. Cell lysates were purified by affinity chromatography using a nickel-nitrilotriacetic acid HiTrap chelating HP column (GE Healthcare), followed by cleavage of the tag with overnight exposure to TEV protease. The protein was purified to homogeneity by two subsequent gel filtrations using a HiLoad 26/60 Superdex 75 preparative-grade column (GE Healthcare). The purity of the resulting protein, consisting of the cloned sequence plus additional four residues at the N terminus, was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Samples for nuclear magnetic resonance (NMR) at 0.5 to 0.8 mM were prepared in PBS (pH 7.1) containing 0.02% sodium azide and 1 mM dithiothreitol.
Solution structure of the LSm domain of D. melanogaster Tral.
All spectra were recorded at 25°C on Bruker DMX600, DMX750, and AVANCE900 spectrometers. Backbone sequential assignments were completed by using standard triple-resonance experiments implemented using selective proton flipback techniques for fast pulsing (15). Aliphatic side chain assignments and aromatic assignments were completed as described before (9). Stereospecific assignments and the resulting χ1 rotamer assignments were determined for 27 of 46 prochiral CβH2 protons and for the CγH3 groups of two of four valine residues. Assignments of χ1 rotamers were also available for all nine isoleucine residues and four of five threonine residues. Assignments of χ2 rotamers were made for five of nine isoleucine and five of seven leucine residues.
Distance data were derived from a set of four 3D-NOESY spectra, including the heteronuclear edited NNH- and CNH-NOESY spectra (14), in addition to conventional 15N- and 13C-HSQC-NOESY spectra and a 2D-NOESY spectrum recorded on an unlabeled sample. Distance restraints, dihedral angle restraints (applied for the 75 high-confidence predictions found by the program TALOS) (11), 53 direct coupling constant restraints (included for the backbone φ angles) (52), and 29 hydrogen bond restraints were derived as detailed elsewhere (51).
MEXICO experiments using five mixing times (50, 100, 150, 200, and 250 ms) determined rates of exchange of HN protons with water (22, 28). A 15N{1H} heteronuclear NOE experiment was run at 900 MHz with a 3-s proton presaturation time.
Refinement was carried out by comparing experimental and back-calculated 15N-HSQC-NOESY, 13C-HSQC-NOESY, CNH-NOESY, and NNH-NOESY spectra (in-house software). This process adjusted side chain rotamers for several residues.
Structures were calculated with XPLOR (NIH version 2.9.4) using standard protocols with modifications as described previously (9). For the final set, 50 structures were calculated, and 22 were chosen on the basis of the lowest restraint violations. An average structure was calculated and regularized to give a structure representative of the ensemble (see Fig. 5, 6, and 8).
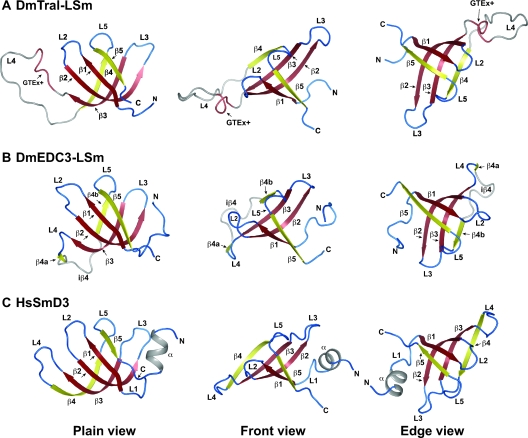
FIG. 5.
Structure of the LSm domain of D. melanogaster Tral (Tral-LSm). (A) NMR structure of Tral-LSm. (B) NMR structure of the D. melanogaster EDC3 LSm domain (DmEDC3-LSm; PDB-ID: 2rm4) (according to reference 50). (C) Crystal structure of human SmD3 (HsSmD3; PDB-ID: 1d3b-A) (according to reference 20). β-strands belonging to the Sm1 motif are colored in red, and β-strands belonging to the Sm2 motif are colored in yellow (according to references (20 and 50). Shown in gray are the N-terminal α-helix in HsSmd3 (that is absent in Tral and EDC3), the extended loop L4 from Tral, and the iβ4 insertion that is unique to EDC3. The GTEx+ motif of Tral is shown in salmon. On the left is a plain view of the open β-barrel with the open side on the top. The center shows the front view, and an edge view is shown on the right.
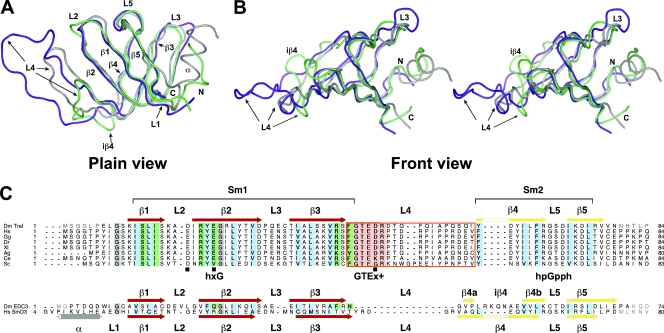
FIG. 6.
Structure-based alignment of the D. melanogaster Tral LSm domain. (A and B) Superposition of the structures of Tral-LSm (purple), DmEDC3-LSm (lime), and HsSmD3 (gray), represented as tubes. (A) Plain view. (B) Front view (stereo) illustrating the different orientations of loops L3 and L4. (C) Alignment of Tral and EDC3 LSm domains with human SmD3. Secondary structure elements are colored as in Fig. 5. The Sm1 and Sm2 motifs are indicated. Residues from the hydrophobic core that define the Sm1 and Sm2 motifs are shaded blue; conserved glycines are shaded gray. Gray letters indicate amino acids absent in the presented structures. Tral residues mutagenized in the present study, together with residues shown to mediate DCP1 binding in EDC3, are shaded in green. The loop L4 in Tral LSm is boxed in orange, and the GTEx+ motif is shaded in salmon. Black squares indicate acidic residues clustered around Tral residue R21. Abbreviations and accession numbers are as follows: Hs, Homo sapiens (gi:71648673 for RAP55 and gi:74007795 for SmD3); Dm, Drosophila melanogaster (gi:24665977); Gg, Gallus gallus (gi:61098250); Xl, Xenopus laevis (gi:117165637); Dr, Danio rerio (gi:42476222); Ag, Anopheles gambiae (gi:158286595); Ce, Caenorhabditis elegans (gi:17509741); and Sc, Saccharomyces cerevisiae (gi:1066486).
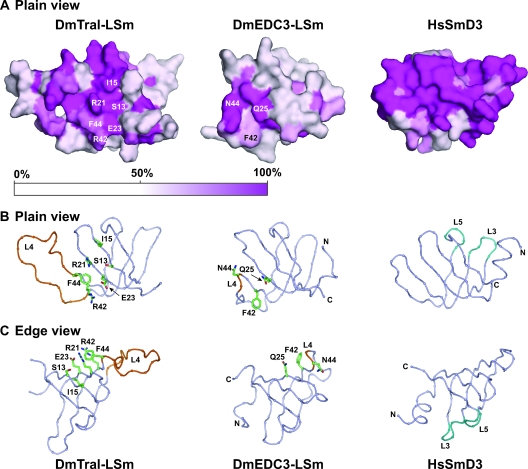
FIG. 8.
Probing functionally relevant residues in Tral-LSm. (A) Surface representation (plain view) of the structures colored by sequence conservation comparing the seven metazoan species shown in Fig. 6 and Caenorhabditis briggsae. For this figure the yeast orthologs (Scd6p and Edc3p) were excluded because S. cerevisiae Edc3p is highly divergent from metazoa EDC3 (for instance, the linker region between the LSm domain and FDF motif is absent in S. cerevisiae Edc3p, making the alignment with the metazoan proteins difficult). Color ramp by identity: fuchsia (100%) to white (50% or less). (B and C) Tube representation with Cα carbons as spheres in plain view (B) and edge view (C). Loop L4, which is structurally distinct in Tral-LSm and DmEDC3-LSm, is highlighted in orange. Surface residues mutagenized in this and previous studies (50) are drawn as sticks with carbons in green, oxygens in red, and nitrogens in blue. Residues mutagenized in Tral-LSm are located in topologically similar positions to residues in EDC3-LSm that are required for DCP1 binding (Q25, F42, and N44 in EDC3). These residues are on the opposite side from the RNA-binding residues in loops L3 and L5 in canonical (L)Sm proteins (colored cyan on HsSmD3).
Protein structure accession numbers.
The coordinates for the D. melanogaster Tral-LSm and zebrafish RAP55-LSm structure ensembles have been deposited in the Protein Data Bank (PDB) under access codes 2vxe and 2vxf, respectively.
RESULTS
Tral and EDC3 associate with DCP1 and Me31B to form distinct protein complexes.
Tral interacts with the translational regulators CUP and Me31B in D. melanogaster embryos (5, 6, 53). while in D. melanogaster Schneider cells (S2 cells) Me31B associates with EDC3 and the ortholog of Pat1 (HPat) (19, 50). EDC3 also interacts with DCP1 and DCP2 (13, 20, 50). These interactions suggest that Tral could be part of a protein network consisting of CUP, Me31B, DCP1, DCP2, EDC3, and HPat. To test this, we transiently expressed HA-tagged versions of these proteins in S2 cells and assayed whether endogenous Tral could be coimmunoprecipitated from transfected cell lysates by using anti-HA antibodies. In parallel, we analyzed the association of endogenous EDC3 with these proteins, using in this case HA-tagged Tral instead of HA-EDC3.
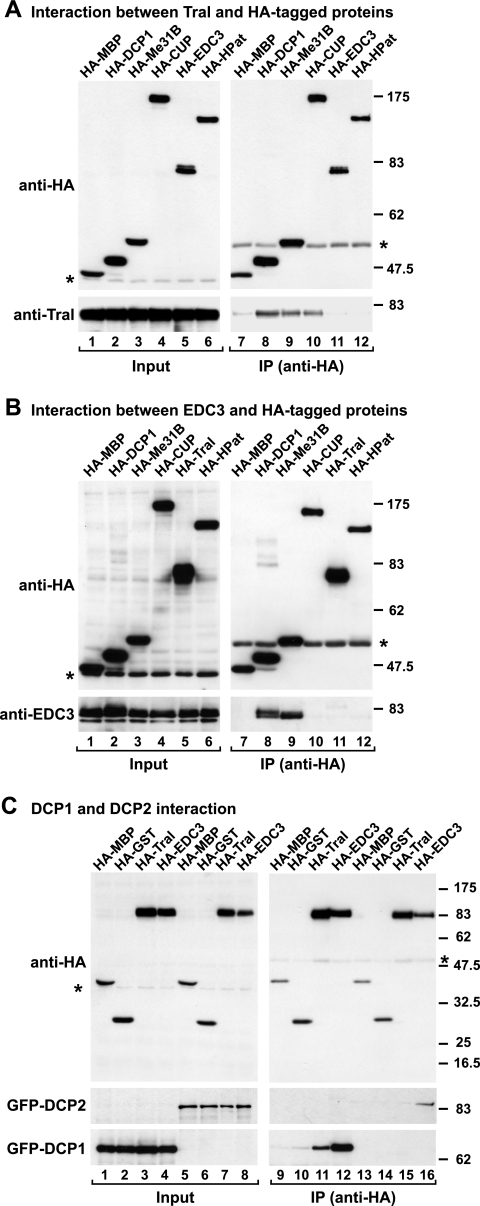
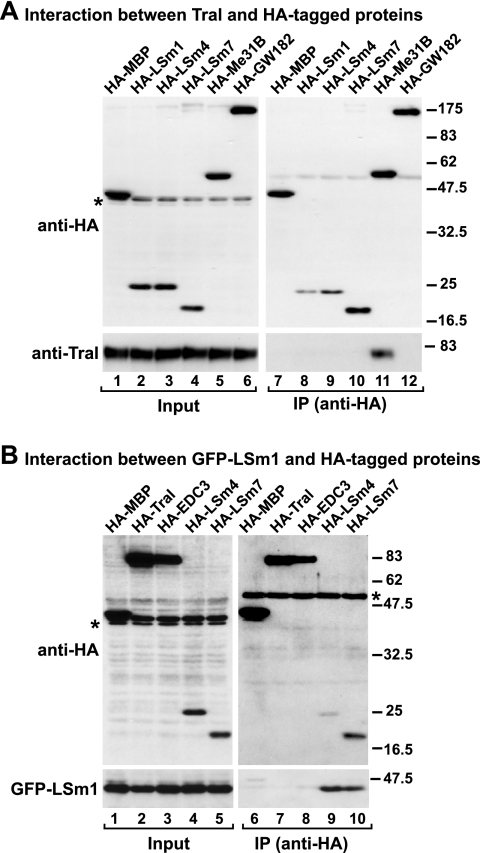
Our results show that endogenous Tral coimmunoprecipitates with HA-DCP1, HA-Me31B, and HA-CUP (Fig. 1A, lanes 8 to 10). HA-EDC3 and HA-HPat did not coimmuniprecipitate Tral above the background levels observed with the negative control, an HA-tagged version of maltose-binding protein (HA-MBP) (Fig. 1A, compare lanes 11 and 12 with lane 7). Under similar conditions, endogenous EDC3 coimmunoprecipitated with HA-DCP1 and HA-Me31B, as reported before (Fig. 1B, lanes 8 and 9) (50), but not with HA-CUP, HA-Tral, or HA-HPat (Fig. 1B, lanes 10 to 12). The lack of coimmunoprecipitation between Tral and EDC3 suggests that these proteins assemble into distinct complexes.
FIG. 1.
Tral interacts with DCP1, Me31B, and CUP. (A and B) Epitope HA-tagged versions of MBP, DCP1, Me31B, CUP, EDC3 (or Tral), and HA-HPat were transiently expressed in S2 cells, as indicated above the panels. Cell lysates were immunoprecipitated by using a monoclonal anti-HA antibody. Inputs (1.25%) and immunoprecipi- tates (30%) were analyzed by Western blotting using a polyclonal anti-HA antibody. The presence of endogenous Tral (A) or EDC3 (B) in the immunoprecipitates was tested by Western blotting with anti-Tral or anti-EDC3 antibodies. In panel A immunoprecipitations were performed in the presence of RNase A. Asterisks indicate cross-reactivity of the polyclonal anti-HA antibody with an endogenous protein (Input panels) or cross-reactivity with the immunoglobulin heavy chain by the secondary antibody (IP panels). (C) Epitope HA-tagged versions of MBP, GST, Tral, or EDC3 were transiently coexpressed in S2 cells with GFP-DCP1 or GFP-DCP2 as indicated. Cell lysates were immunoprecipitated using a monoclonal anti-HA antibody. Inputs (1.25%) and immunoprecipitates (30%) were analyzed by Western blotting with polyclonal anti-HA and anti-GFP antibodies.
The interaction between Tral and DCP1, Me31B, or CUP was observed in the presence of RNase A (Fig. 1A). Furthermore, the interaction between Tral and DCP1 or Me31B was observed with recombinant proteins coexpressed in E. coli, although binding was not stoichiometric (data not shown), suggesting that these interactions are direct. Consistent with our results, recombinant X. laevis RAP55 and Xp54 also associate in the presence of RNase A (47).
It is noteworthy that the association of Tral with Me31B and of CAR-1 with CGH-1 (in D. melanogaster and C. elegans embryos, respectively) was reported as either RNase sensitive (3, 6) or RNase resistant (53). One possible explanation for these differences is that the RNase treatment reduces the interaction between Tral and Me31B, allowing the proteins to remain bound or dissociate, depending on the experimental conditions and/or the relative concentration of other Me31B partners that may compete with Tral for binding to Me31B.
Because HA-DCP2 is expressed at very low levels in S2 cells, its association with endogenous Tral or EDC3 could not be tested. However, in independent experiments, we coexpressed GFP fusions of DCP1 or DCP2 with HA-Tral or HA-EDC3 and did observe an interaction between Tral and DCP1, but not DCP2 (Fig. 1C, lanes 11 and 15, respectively). In contrast, EDC3 coimmunoprecipitated both DCP1 and DCP2, as reported previously (Fig. 1C, lanes 12 and 16, respectively) (50).
Although we do not know whether all of these interactions are direct or take place simultaneously, we can rule out the possibility that under our experimental conditions we are coimmunoprecipitating large protein aggregates (e.g., submicroscopic P-bodies), because under these conditions we could not detect an association between Tral and additional P-body components, including EDC3, HPat, DCP2, LSm1, LSm4, LSm7, or GW182 (Fig. 1A and C and see Fig. 7 below).
FIG. 7.
The Tral LSm domain is not incorporated into the LSm1-7 ring. (A) HA-tagged versions of MBP, LSm1, LSm4, LSm7, Me31B, and GW182 were transiently expressed in S2 cells. Cell lysates were immunoprecipitated with a monoclonal anti-HA antibody. Inputs and immunoprecipitates were analyzed by Western blotting with a polyclonal anti-HA antibody. The presence of endogenous Tral in the immunoprecipitates was tested by Western blotting with anti-Tral antibodies. (B) HA-tagged versions of MBP, Tral, EDC3, LSm4, and LSm7 were transiently coexpressed in S2 cells with GFP-LSm1. Cell lysates were immunoprecipitated with a monoclonal anti-HA antibody. Inputs and immunoprecipitates were analyzed with polyclonal anti-HA and anti-GFP antibodies.
In summary, Tral and EDC3 have common interacting partners, but they do not interact with each other (Fig. 1A and B). Moreover, CUP coimmunoprecipitates with Tral but not with EDC3 (Fig. 1A and B), whereas DCP2 coimmunoprecipitates with EDC3 but not with Tral (Fig. 1C). Taken together, these results indicate that Tral and EDC3 associate with DCP1 and Me31B in distinct protein complexes, which might comprise other distinct proteins.
A modular domain organization enables Tral to interact with multiple partners.
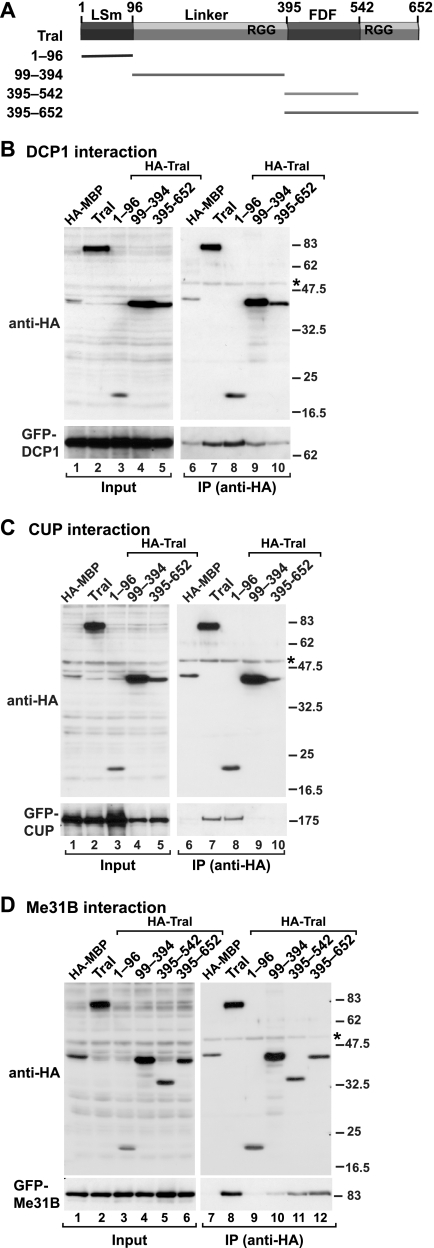
Similar to EDC3, Tral is characterized by an N-terminal domain with a predicted Sm fold and a central FDF motif (Fig. 2A) (1, 2). The FDF motif is embedded in segments rich in glycine and arginine residues (RGG-rich regions [1, 2]). To delineate the domains of Tral that mediate the interactions described above, we coexpressed HA-tagged Tral protein fragments with GFP-tagged DCP1, CUP, or Me31B. Tral fragment boundaries were selected based on sequence alignments of orthologous proteins from various species (Fig. 2A).
FIG. 2.
Tral interacts with DCP1, CUP, and Me31B through specific domains. (A) Domain architecture of Tral. Tral homologs contain an LSm domain and an FDF motif. The FDF motif is flanked by sequences rich in glycine and arginine residues (RGG-rich regions). The numbers above the protein outline represent amino acid positions at fragment boundaries for the D. melanogaster protein. The protein domains sufficient for the localization to P bodies and the interaction with DCP1, CUP (1-96), and Me31B (395-542) are indicated. (B to D) HA-tagged Tral or the indicated Tral protein fragments were cotransfected in S2 cells with GFP fusions of DCP1 (B), CUP (C), or Me31B (D) as indicated. Cell lysates were immunoprecipitated using a monoclonal anti-HA antibody. Inputs and immunoprecipitates were analyzed by Western blotting with polyclonal anti-HA and anti-GFP antibodies as described in Fig. 1C.
We found that both GFP-DCP1 and GFP-CUP coimmunoprecipitated with full-length HA-Tral, as well as with a protein fragment, including the LSm domain (Fig. 2B and C, lanes 7 and 8). In fact, the Tral LSm domain alone was sufficient for these interactions (Fig. 2B and C, lane 8). In contrast, GFP-Me31B coimmunoprecipitated with full-length Tral and with fragments containing the FDF motif (Fig. 2D, lanes 8, 11, and 12). The FDF-containing fragment alone (amino acids 395 to 542) interacted with Me31B, although less efficiently than the full-length protein (Fig. 2D, lane 11), suggesting additional sequences may contribute to this binding. Indeed, it has been shown that the RGG regions flanking the FDF motif enhance the interaction between human RAP55 and the human Me31B ortholog RCK/p54 (47).
Together, our results confirm that Tral has a modular domain organization with specific functionalities for the LSm domain and the FDF motif. Interestingly, these functionalities are conserved in the equivalent domains of EDC3, which like Tral, interacts with DCP1 and Me31B through its LSm domain and FDF motif, respectively (13, 50).
The FDF motifs of Tral and EDC3 interact with the C-terminal RecA-like domain of Me31B.
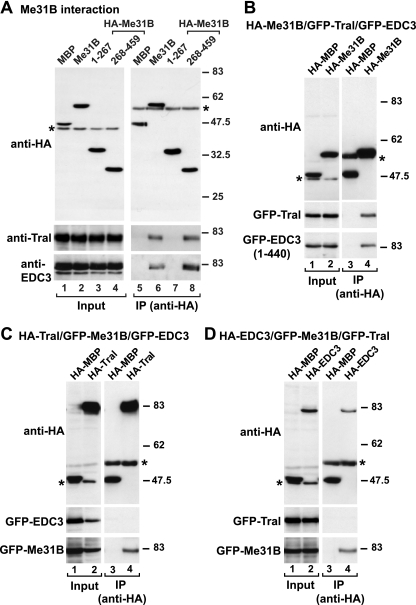
By comparing the Tral and EDC3 protein fragments that are sufficient to interact with Me31B, it is apparent that the highest sequence similarity between these fragments is confined to the FDF motif; this suggests a similar mode of interaction with Me31B. Me31B is a DEAD-box RNA helicase, which, like other members of this protein family, contains two RecA-like domains (8). In agreement with the idea that Tral and EDC3 use a similar mode to associate with Me31B, both endogenous Tral and EDC3 coimmunoprecipitated with the C-terminal RecA-like domain of Me31B (Fig. 3A, Me31B 268-459).
FIG. 3.
Tral and EDC3 interact with the C-terminal RecA-like domain of Me31B. (A) HA-tagged Me31B or the indicated Me31B protein fragments were cotransfected in S2 cells. Cell lysates were immunoprecipitated with a monoclonal anti-HA antibody. The presence of endogenous Tral or EDC3 in the immunoprecipitates was tested by Western blotting with anti-Tral or anti-EDC3 antibodies as described in Fig. 1A and B. (B to D) S2 cells were cotransfected with mixtures of three plasmids. In panel B the plasmids encoded HA-Me31B, GFP-Tral, and GFP-EDC3 (fragment 1-440). In panel C, the plasmids encoded HA-Tral, GFP-Me31B, and GFP-EDC3. In panel D, the mixture consisted of HA-EDC3, GFP-Me31B, and GFP-Tral. Cell lysates were immunoprecipitated with a monoclonal anti-HA antibody. In all panels, HA-MBP served as a negative control. Inputs and immunoprecipitates were analyzed by Western blotting with polyclonal anti-HA and anti-GFP antibodies as described in Fig. 1C.
Because Tral and EDC3 do not interact with each other, but both interact with the C-terminal RecA-like domain of Me31B via their FDF motifs, we hypothesized that their interactions with Me31B may be mutually exclusive. To investigate this possibility further, we cotransfected S2 cells with mixtures of plasmids expressing the three proteins. We used three different mixtures each containing a plasmid for expression of one of the proteins with an HA tag and the other two proteins with GFP tags. We assayed whether the proteins fused to GFP could be coimmunoprecipitated from transfected cell lysates using anti-HA antibodies. We observed that HA-Me31B coimmunoprecipitated both GFP-Tral and GFP-EDC3, showing that Me31B does indeed interact with both of these proteins (Fig. 3B). In contrast, HA-Tral coimmunoprecipitated GFP-Me31B, but not GFP-EDC3 (Fig. 3C), whereas HA-EDC3 coimmunoprecipitated GFP-Me31B, but not GFP-Tral (Fig. 3D). Thus, Tral and EDC3 use a similar mode to associate with Me31B to form distinct alternative protein complexes.
The LSm domain is sufficient to target Tral to P-bodies.
In previous studies in D. melanogaster S2 cells, we showed that Tral localizes to P bodies but is not required for P-body integrity (18). The domains of Tral required for P-body localization were previously defined for human RAP55 (LSm14), which is highly related to D. melanogaster Tral; however, different results were reported. Yang et al. (55) showed that a protein fragment comprising the two C-terminal RGG and the FDF motifs, but not the LSm domain, was necessary and sufficient to target the protein to P bodies in human cells. In contrast, Tanaka et al. (47) reported that the LSm domain of RAP55 was important for P-body localization.
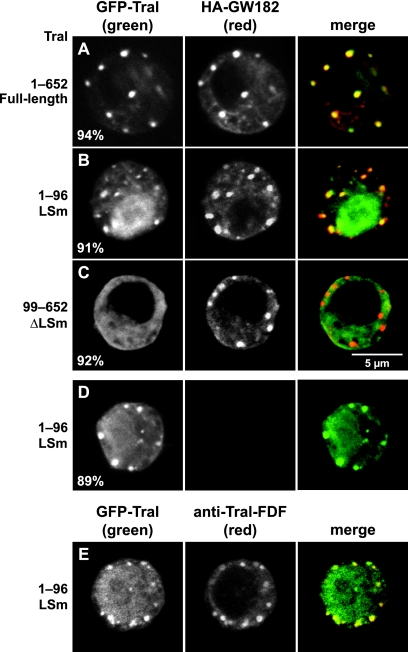
We therefore investigated the role of the different Tral domains in P-body localization in D. melanogaster (Fig. 4A to E). We cotransfected S2 cells with two expression vectors, one encoding GFP fused to Tral fragments and the other encoding HA-tagged GW182, a P-body marker in metazoa that does not coimmunoprecipitate with Tral (see below Fig. 7A) (17). We found a GFP fusion protein containing the Tral LSm domain accumulated in cytoplasmic foci, although to a lesser extent than the full-length protein (Fig. 4B versus Fig. 4A). Indeed, the LSm domain alone was also detected throughout the cytoplasm and within the nucleus (Fig. 4B). Nevertheless, the foci formed by the Tral LSm domain were similar to those observed with full-length Tral, as judged by the colocalization with HA-GW182 (Fig. 4A and 4B). The Tral LSm domain formed foci also when expressed alone, indicating that these foci are not induced by the overexpression of GW182 (Fig. 4D). Moreover, endogenous Tral was detected in Tral-LSm foci using an antibody raised against the Tral FDF motif, indicating that these foci correspond to endogenous P bodies (Fig. 4E).
FIG. 4.
The LSm domain of Tral is necessary and sufficient for P-body localization. (A to E) Confocal fluorescent micrographs of fixed S2 cells expressing GFP-tagged fusions of full-length Tral or the protein fragments indicated on the left. In panels A to C the cells were cotransfected with HA-GW182. In panel E the cells were stained with affinity-purified anti-Tral antibodies (FDF motif). The merged images show the GFP signal in green and the HA or anti-Tral signal in red. The fraction of cells exhibiting a staining identical to that shown in the representative panel was determined by scoring 100 cells in two independent transfections performed per protein. Scale bar, 5 μm.
In contrast to the Tral-LSm domain, a protein fragment lacking the LSm domain (residues 99 to 652) spread throughout the cell and did not accumulate in HA-GW182 foci (Fig. 4C). Because this protein fragment comprises the Me31B-interaction domain, we conclude that the interaction with Me31B is not sufficient for P-body targeting. Together, our results indicate that the Tral LSm domain is sufficient to direct the protein to P bodies, although the diffuse staining with this domain alone suggests that additional sequences in the full-length protein contribute to its accumulation in P bodies. These results are similar to those obtained for the LSm domain of D. melanogaster EDC3, which is also sufficient for P-body targeting (50).
In summary, the LSm domain of Tral mediates binding to CUP and DCP1 and accumulates in P-bodies. To delineate the molecular bases for these functions, we determined the solution structure of the LSm domain of D. melanogaster Tral by NMR.
Structure of the LSm domain of Tral.
The D. melanogaster Tral LSm domain (Tral-LSm, residues M1 to P84) comprises a structured region (residues 7 to 79) that adopts the predicted Sm-like fold (1, 2). The remaining terminal residues are unstructured and the loop (L4) between β-strands β3 and β4 of the Sm fold is less well defined than the rest of the structure (residues 44 to 61; Fig. 5A). The ensemble of 22 lowest-energy NMR structures (see Fig. S1 in the supplemental material) has a good root mean square deviation (RMSD; calculated over 52 structured residues; K11-F44, I61-V78) of 0.16 Å for backbone atoms and of 0.69 Å for all nonhydrogen atoms (Tables 1 and 2). The restraint violations are also very low, with the final set having on average four violations of distance restraints greater than 0.08 Å per structure and one dihedral restraint violation greater than 1°.
TABLE 1.
Structural statisticsa
| Parameterb | SA | 〈SA〉r |
|---|---|---|
| RMSD from distance restraints (Å) | ||
| All (n = 342) | 0.020 ± 0.001 | 0.019 |
| Intraresidue (n = 67) | 0.014 ± 0.001 | 0.013 |
| Interresidue sequential (n = 111) | 0.015 ± 0.001 | 0.015 |
| Medium range (n = 28) | 0.020 ± 0.003 | 0.018 |
| Long range (n = 107) | 0.028 ± 0.001 | 0.027 |
| H-bond (n = 29) | 0.011 ± 0.001 | 0.011 |
| RMSD from dihedral restraints (n = 248) | 0.15 ± 0.01 | 0.14 |
| RMSD from J-coupling restraints (Hz) (n = 51) | 0.83 ± 0.01 | 0.88 |
| H-bond restraints (avg Å/avg °)c (n = 26) | 2.21 ± 0.14/10.4 ± 4.8 | 2.20 ± 0.14/10.3 ± 4.9 |
| H-bond restraints, minimum-maximum (Å/°)c | 1.97-2.50/1.7-23.5 | 1.95-2.50/1.4-23.9 |
| Deviations from ideal covalent geometry | ||
| Bonds (Å, 10−3) | 4.91 ± 0.03 | 4.86 |
| Angles (°) | 0.616 ± 0.004 | 0.613 |
| Impropers (°) | 1.57 ± 0.06 | 1.55 |
| Structure quality indicatorsd | ||
| Ramachandran map regions (%) | 90.9/9.1/0.0/0.0 | 93.5/6.5/0.0/0.0 |
Values are given as means ± the standard deviation where applicable. Structures are labeled as follows: SA, the set of 22 final simulated annealing structures; 〈SA〉, the mean structure calculated by averaging the coordinates of SA structures after fitting over secondary structure elements; 〈SA〉r, the structure obtained by regularizing the mean structure under experimental restraints.
n, Number of restraints of each type.
Hydrogen bonds were restrained by treating them as pseudocovalent bonds (see Materials and Methods). The average and minimum/maximum for distances and acceptor antecedent angles are stated for restrained hydrogen bonds.
Determined using the program PROCHECK (29). Percentages are for residues in allowed/additionally allowed/generously allowed/disallowed regions of the Ramachandran map.
TABLE 2.
Atomic RMSDa
| Comparison | Category | Secondary structure (avg Å ± SD)b | 〈SA〉 vs 〈SA〉rc (Å) |
|---|---|---|---|
| SA vs 〈SA〉 | Backbone | 0.16 ± 0.05 | 0.15 |
| All | 0.69 ± 0.07 | 0.70 | |
| SA vs 〈SA〉r | Backbone | 0.21 ± 0.06 | |
| All | 0.87 ± 0.09 |
Based on heavy atom superimpositions. Values are given as means ± the standard deviation where applicable. Structures are labeled as follows: SA, the set of 22 final simulated annealing structures; 〈SA〉, the mean structure calculated by averaging the coordinates of SA structures after fitting over secondary structure elements; 〈SA〉r, the structure obtained by regularizing the mean structure under experimental restraints.
Defined as residues K11-F44, I61-V78.
RMSD for superimposition over-ordered residues.
The structured part of Tral-LSm corresponds to the core of canonical (L)Sm proteins represented by human SmD3 (HsSmD3; Fig. 5A to C) (25) and described in the SCOP database (http://scop.mrc-lmb.cam.ac.uk/scop/ [39]). The fold consists of a five-stranded open β-barrel (β1 to β5), where β-strands β2 and β3 are strongly bent by β-bulges. The edges of the barrel are formed by β-strands β4 and β5, which are connected by a conserved helical turn (loop L5) that completes the barrel on the open side (Fig. 5A to C). The N-terminal α-helix, a hallmark of the (L)Sm protein family, is absent as such from the structure (Fig. 5C versus 5A). The structure confirms and confines the Tral LSm domain as an independent folding unit and suggests that residues beyond N79 are part of the unstructured region of Tral that links the LSm domain to the FDF motif.
The LSm domain of Tral adopts a divergent Sm fold similar to that of EDC3.
A search of the PDB using the DALI server (24) indicates that one of the closest structural neighbors to Tral-LSm is the LSm domain of the human Sm protein SmD3 (DALI Z-score of 7.3 for PDB entry 1d3b-A). A superposition of Tral-LSm with HsSmD3 (18% identity over 50 alignable Cα atoms) yields a low RMSD value of 1.4 Å (Fig. 6A and B).
Tral-LSm is also similar to the LSm domains of human and D. melanogaster EDC3 (HsEDC3-LSm and DmEDC3-LSm, respectively). The identity between Tral-LSm and HsEDC3-LSm or DmEDC3-LSm is 30 or 14%, respectively, over 50 alignable Cα positions. This results in a high degree of structural similarity (Fig. 5 and 6), as reflected by an RMSD of 1.6 Å in both cases.
A structure-based alignment (Fig. 6C), which includes HsSmD3, DmEDC3-LSm, and Tral-LSm domains from various species shows that Tral contains the Sm1 (β1 to β3) and Sm2 (β4 and β5) motifs (23, 25, 45); these contribute a characteristic set of hydrophobic side chains that pack together to form the core of the protein (Fig. 6C, residues shadowed in blue). Thus, from a structural point of view, the N-terminal domain of Tral belongs to the Sm superfamily of proteins. However, both Tral and EDC3 LSm domains are divergent members of this superfamily, since both domains lack the characteristic N-terminal α-helix.
The main differences between Tral-LSm and EDC3-LSm are found in β-strand β4 and loop L4 (linking β-strands β3 and β4). Tral-LSm has a continuous β-strand β4, which in human and D. melanogaster EDC3-LSm are disrupted into strands β4a and β4b by insertions (iβ4) of four and seven residues, respectively (Fig. 5 and 6) (50). Moreover, in EDC3-LSm domains, loop L4 consists of a tight and highly conserved β-turn, whereas in Tral-LSm this loop is 16 residues longer and comprises a highly conserved GTEX+ motif (where “X” is any residue and “+” is a positively charged residue [2]) followed by 12 nonconserved residues (Fig. 5A and 6C). The GTEX+ motif is also present in S. cerevisiae Scd6p, C. elegans CAR-1, and vertebrate RAP55 (Fig. 6C) (2).
In the Tral-LSm structure, the GTEX+ motif shows increasing RMSD values in the C-terminal direction making this motif the starting point for a partially disordered region of L4. Except for NOE contacts between I55 and T46, no long-range structural information could be obtained for this region. Nevertheless, for the structure calculations, short-range information over the E47-Q54 and A56-I61 regions were used when available, resulting in a relatively well-defined loop in all calculated structures (see Fig. S1 in the supplemental material).
Surprisingly, the deposited solution structure of the LSm domain of the zebrafish RAP55 (Brachyodanio rerio PDB-ID 2FB7) appears only partially similar to that of Tral-LSm, despite a sequence identity of 64% over the structured region. In particular, the complete C-terminal half of strand β3 is absent from the Sm fold, resulting in an entirely different orientation of loop L4. Furthermore, loop L4 is much more open and unstructured than observed in Tral-LSm. However, the chemical shifts data submitted for RAP55-LSm in the context of a structural genomics project indicate no substantial deviation from the Tral-LSm structure. Careful reevaluation of the submitted data allows the calculation of an alternative structure for RAP55-LSm (see Fig. S2 in the supplemental material) that is basically identical to Tral-LSm, indicating that Drosophila Tral and vertebrate RAP55 are true orthologs.
The LSm domain of Tral does not form multimeric (L)Sm rings.
Canonical (L)Sm proteins multimerize in a head-to-tail fashion via an antiparallel arrangement of the β-strands β4 and β5. This leads to six- or seven-membered homo- or heteromeric rings with a continuous inner β-sheet (10, 25, 27, 38, 44, 48, 49). Similar multimerization properties were therefore predicted for the LSm domain of Tral and EDC3 (2). However, we previously showed that EDC3-LSm is monomeric in solution (50). Similarly, the NMR measurements with Tral-LSm show no significant multimerization at concentrations up to 0.8 mM, as reflected by a diffusion coefficient of 1.09 (± 0.1) × 10−10 m2/s at 25°C.
We therefore tested in vivo whether Tral forms heteromeric associations with members of the LSm1-7 complex (7). Endogenous Tral did not detectably coimmunoprecipitate with HA-tagged LSm1, LSm4, or LSm7 (Fig. 7A) but did associate with HA-Me31B (which served as a positive control). These results indicate that, in contrast to LSm1 or LSm8, Tral is unlikely to function as an alternative subunit in the assembly of a specialized LSm complex, as suggested previously (2, 55). The failure of Tral to interact with LSm1, LSm4, or LSm7 is probably not caused by the overexpression of these proteins, because overexpressed HA-LSm7 or HA-LSm4 coprecipitate GFP-LSm1 (Fig. 7B, lanes 9 and 10). LSm7 does not directly contact LSm1 within the LSm1-7 ring; therefore, the coimmunoprecipitation of GFP-LSm1 with HA-LSm7 indicates that these overexpressed proteins assemble into LSm1-7 rings together with additional endogenous proteins.
We also tested whether Tral could homo-oligomerize in vivo. However, in contrast to EDC3, which self-associates through the YjeF_N domain (13, 21, 32, 33, 50), no interaction was observed either between GFP-Tral and HA-Tral or between HA-Tral and the endogenous protein (data not shown), suggesting that Tral does not oligomerize.
Finally, Tral-LSm did not detectably interact with an RNA oligonucleotide consisting of eight uridines in an analytical gel filtration experiment (data not shown). Binding studies with poly(U)-Sepharose indicated that the LSm domain of CAR-1 was able to bind RNA (3); however, the protein fragment used in the present study included sequences beyond the Sm fold that may have contributed to this binding. Our results, together with previous studies on EDC3-LSm (50), indicate that two of the hallmarks of canonical (L)Sm proteins—ring formation and RNA-binding properties—are absent in the divergent LSm domains of Tral and EDC3.
Probing the role of surface residues in DCP1 and CUP interaction and P-body localization.
Another difference between the Tral and EDC3 LSm domains is reflected by the degree of conservation of individual surface residues (Fig. 8A); overall, Tral-LSm shows higher conservation than EDC3-LSm. Tral shows conservation of both the hxG signature in the Sm1 motif, and the Gpph signature in the Sm2 motif (x, any amino acid; p, polar; h, hydrophobic [2]), whereas in EDC3 only the hxG signature is present (Fig. 6C).
To locate functionally relevant regions on Tral-LSm, we performed mutational analyses and examined the effect of the mutations on DCP1 and CUP binding, as well as on P-body localization (Fig. 8B and C and Fig. 9). We focused on highly conserved residues exposed on the β-sheet surface, on the side of Tral-LSm opposite to loops L3 and L5 [these loops provide RNA-binding residues in canonical (L)Sm proteins; Fig. 8]. The mutated residues are topologically equivalent to EDC3-LSm domain residues involved in the interaction with DCP1 (Fig. 8) (50).
FIG. 9.
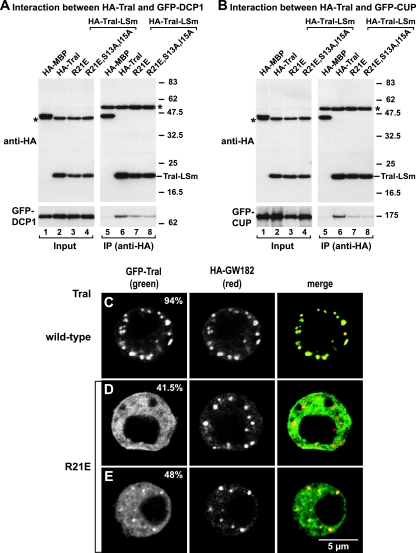
Effect of Tral-LSm mutations on DCP1 and CUP interaction, as well as on P-body localization. (A and B) HA-tagged MBP, Tral-LSm, or the indicated Tral-LSm mutants were cotransfected in S2 cells with GFP-DCP1 or GFP-CUP, as indicated. Cell lysates were immunoprecipitated with a monoclonal anti-HA antibody and analyzed by Western blotting as described in Fig. 2. Asterisks indicate cross-reactivity of the antibodies as described in Fig. 1. (C to E) Confocal fluorescent micrographs of fixed S2 cells expressing GFP-tagged fusions of full-length Tral wild type or the R21E mutant. Cells were cotransfected with HA-GW182. The merged images show the GFP signal in green and the HA signal in red. The fraction of cells exhibiting a staining identical to that shown in the representative panel was determined by scoring 100 cells in two to three independent transfections performed per protein. Scale bar, 5 μm.
In particular, the invariant residues S13 and I15 in strand β1 and the aromatic residue F44 in loop L4 were substituted with alanines (S13A, I15A, and F44A; Fig. 6C, 8B, and 8C). The charged residues R21 and E23 in strand β2, as well as R42 in strand β3, were substituted by residues having the opposite charge (R21E, E23K, and R42E). We also tested the effect of deleting the conserved GTEx+ motif in loop L4 (Fig. 6C, 8B, and 8C).
Substitutions of single conserved residues (i.e., E23K, R42E, and F44A) did not affect the interaction between Tral and DCP1 or CUP; one exception was the charge substitution R21E, which reduced binding to both proteins without affecting Tral expression levels (Fig. 9A and B). Deletion of the GTEx+ motif in loop L4, or substitutions of more than one conserved residue (R21E,E23K or R21E,S13A,I15A) had effects comparable to the single R21E substitution alone (Fig. 9A and B and data not shown).
In the context of the full-length protein, the R21E substitution also affected P-body localization. Indeed, cells expressing a GFP fusion of a protein carrying this substitution had fewer and smaller GFP-containing foci, although GW182 foci were not affected (Fig. 9C versus Fig. 9D and E). To quantify this effect, the number of cells displaying GFP foci was counted. Whereas 94% of cells expressing wild-type GFP-Tral displayed GFP foci (Fig. 9C), only 48% of cells expressing GFP-Tral-R21E displayed reduced numbers of foci, which were also smaller in size (Fig. 9E). Moreover, ca. 43% of cells expressing the mutant protein had no detectable GFP foci (Fig. 9D).
To verify whether the strong phenotype caused by the R21E mutation can exclusively be interpreted in terms of a direct interaction of R21 with partners such as DCP1 or CUP, we analyzed the structural stability of the mutated LSm domain by NMR. We found that the mutant domain is partially unfolded in vitro, possibly due to the repulsive charges of E23, D19, and D48 (Fig. 6, black squares) that are no longer compensated for by R21. Similar results were obtained when R21 was substituted with alanine (data not shown). Thus, we could not prevent the interaction of Tral with DCP1 and CUP and its accumulation in P-bodies by single point mutations and could observe an effect only with the partial destabilization of the entire LSm domain.
In summary, although the LSm domains of Tral and EDC3 both interact with DCP1, the detailed modes of interaction appears to be different. Indeed, in DmEDC3, residues Q25, F42, and N44 play a critical role in the interaction with DCP1 (50). In particular, a single substitution of Q25 with alanine abolished EDC3-DCP1 interaction (50). In contrast, substitution of residue E23, which is at the equivalent structural position in DmTral-LSm, did not affect the association with DCP1, instead, the structural integrity of the LSm domain is required for this function.
DISCUSSION
In this study we show that Tral and EDC3 are structurally related proteins that associate with common partners in distinct protein complexes. The N-terminal domains of Tral and EDC3 adopt a divergent Sm fold that mediates their interaction with the decapping activator DCP1 and is sufficient for P-body targeting. In Tral, this domain also mediates the interaction with the translational regulator CUP. Tral and EDC3 share an additional common partner, the RNA helicase Me31B. Both proteins interact with the C-terminal RecA-like domain of Me31B via their FDF motifs, suggesting that their binding is mutually exclusive. This agrees with the conclusion that Tral and EDC3 function in distinct protein complexes. EDC3-containing complexes are known to play a role in mRNA decapping (4, 13, 16, 19, 20, 29). The localization of Tral in P bodies, its association with decapping activators, and translational repressors suggest that this protein functions in translational repression and/or mRNA degradation.
Role of EDC3 and Tral in P-body assembly.
P bodies are cytoplasmic domains that accumulate a variety of proteins involved in mRNA degradation, translational repression, mRNA surveillance, and RNA-mediated gene silencing, together with their mRNA targets (17, 41). The mechanisms leading to P-body assembly are not fully understood, but several P-body components, including RNA, are required for P-body integrity (17, 41). In yeast cells, P-body assembly occurs through parallel redundant pathways, requiring either Edc3 or LSm4 (13). In multicellular organisms, several P-body components are required for P-body formation, since depleting them disperses the remaining P-body components throughout the cytoplasm (17, 41).
In vertebrates the Tral ortholog RAP55 is among the essential P-body components (47, 55). In contrast, in D. melanogaster S2 cells, depleting either Tral or EDC3 does not affect P bodies (18), suggesting that these proteins are not essential for P-body assembly or that multiple redundant pathways lead to P-body formation. Similarly, CAR-1 is not required for P-granule formation in C. elegans (3). Still, Tral and EDC3 likely contribute to the assembly of RNP particles. Both proteins have a modular domain organization that allows them to interact with additional P-body components and probably with multiple RNPs, bringing them in close proximity and thereby facilitating the nucleation of P bodies.
The LSm domains of Tral and EDC3 accumulate in P bodies.
Over the past few years, the number of proteins shown to localize to P bodies has increased dramatically (17, 41). A question that remains open is what causes these proteins to accumulate into P bodies. Some proteins may passively accumulate in P bodies as components of mRNP complexes. In this case, RNA-binding domains or protein-protein interaction domains likely mediate their localization to P bodies. However, the aggregation of individual mRNPs into large granules detectable by light microscopy requires either (i) multidomain proteins that can bridge more than one mRNP or (ii) specific proteins or protein domains that self-aggregate.
One such domain has been described in S. cerevisiae LSm4 (13). LSm4 consists of a canonical LSm domain followed by a C-terminal extension rich in glutamine and arginine residues (Q/N-rich domain), which is not part of the Sm fold. This extension is required to localize LSm4 to P bodies (13, 34). As mentioned above, LSm4 is partially redundant with Edc3 for P-body assembly in yeast cells, but in cells lacking Edc3, P-body assembly relies on the Q/N-rich extension of LSm4. Indeed, a truncated version of LSm4 lacking the Q/N-rich region cannot sustain P-body assembly in the absence of Edc3 (13). It was proposed that the Q/N-rich extension of LSm4 has prion-like properties and promotes P-body formation by aggregating with itself or with additional Q/N-rich domains (13), similar to the assembly mechanisms for Q/N-rich domains in prions (35).
In contrast to LSm4, the LSm domains of Tral and EDC3 are sufficient to localize them to P bodies. These domains lack Q/N prion-like features and remain monomeric in solution, indicating that they are not self-aggregating domains. Furthermore, these domains do not bind RNA, suggesting that they accumulate in P bodies through protein-protein interactions. Nevertheless, in EDC3, mutations that disrupt DCP1-binding do not affect P-body localization (50), suggesting that interactions with additional P-body component(s) drive this protein into P bodies. In Tral, we could not identify mutations that reduced its accumulation in P bodies without affecting the folding of the LSm domain.
Cellular role of Tral.
RAP55 is the vertebrate ortholog of Tral and was originally identified in the salamander Pleurodeles waltl as a component of cytoplasmic RNP particles containing translationally repressed maternal mRNAs (30). Orthologous proteins have been described in several eukaryotic species, including C. elegans (CAR-1), X. laevis (xRAP55), and mammals (RAP55) (3, 6, 30, 42, 46, 47). Like Tral, these proteins localize in diverse cytoplasmic RNP granules that share components with P-bodies and serve as storage sites for translationally inactive mRNAs in germ cells (3, 6, 36, 43, 46, 47). For instance, in young oocytes Tral, xRAP55 and murine RAP55 localize to the Balbiani body, a large organelle aggregate that includes mitochondria, endoplasmic reticulum, germinal granule proteins, and RNAs that become incorporated into germ cells in the developing embryos (36, 42, 43, 53).
RNP granules containing Tral, CAR-1, or xRAP55 comprise additional proteins with roles in translational repression and/or mRNA decapping. These include, Me31B and its orthologs (C. elegans CGH-1 and X. laevis Xp54), the Y-box domain-containing proteins (D. melanogaster YPS, C. elegans CEY-2-4, and X. laevis FRGY2), which are major components of maternal RNP granules, as well as the eIF4E-binding proteins CUP and 4E-T (3, 6, 36, 47, 53). Moreover, the D. melanogaster fragile-X mental retardation protein (dFMRP), which is involved in translational repression, colocalizes with Tral in RNP-containing granules in both embryos and neuronal cells (5, 37). Tral and CAR-1-containing granules in germ cells also include DCP1 and additional components of somatic P-bodies (5, 31, 46).
The localization of Tral and its orthologs in RNP granules in germ cells and young embryos and their association with proteins involved in translational repression, together with the conservation of these interactions in eukaryotes, suggest that these proteins play a fundamental role in regulating translation of maternal mRNAs during oogenesis and early embryogenesis (3, 6, 36, 46, 47, 53). In agreement with this, xRAP55 represses translation both in vivo and in vitro (47). Nevertheless, the precise molecular mechanism by which Tral orthologs exert their regulatory functions remains to be established.
Depleting or mutating Scd6p, CAR-1, or Tral alters endoplasmic reticulum morphology and causes diverse developmental phenotypes (3, 6, 40, 43, 46, 47, 53). These phenotypes are likely due to the misregulation of specific mRNAs. However, only few mRNA targets of these proteins are known (53). It is also not yet clear whether these targets are conserved and whether Tral orthologs recognize specific cis-acting sequence elements on regulated mRNAs.
CAR-1 and xRAP55 expression is confined to the germ line and early embryos (3, 6, 30, 47). In contrast, human RAP55 is ubiquitously expressed (55) and localizes to P bodies in somatic cells at rest and in stress granules in cells exposed to heat shock or oxidative stress (47, 55). Similarly, Tral expression is not confined to the germ line, the protein is detected in S2 cells and in neurons (5, 17, 18), where it is also a component of neuronal RNP granules and participates in neuronal translation regulation (5). The localization of human RAP55 and Tral in somatic P bodies and neuronal granules suggests that, in addition to their role in regulating maternal mRNAs, these proteins have acquired more general roles in mRNA metabolism.
Based on the structural similarity between Tral and EDC3 and their association with common partners, one might anticipate that these proteins represent alternative subunits in the assembly of DCP1- and Me31B-containing complexes. These complexes may perform dual and partially overlapping functions; they may repress translation (e.g., in oocytes) or enhance decapping (e.g., in somatic cells), depending on the additional partners with which they associate.
Supplementary Material
Acknowledgments
We are grateful to Horst Kessler, Michael Sattler, and the staff of the Bavarian NMR Centre at the Technical University, Munich, Germany, for access to spectrometers and technical support.
This study was supported by the Max Planck Society, by a grant from the Deutsche Forschungsgemeinschaft (FOR855), and by the Sixth Framework Programme of the European Commission through the SIROCCO Integrated Project LSHG-CT-2006-037900. A.E. is the recipient of a fellowship from the Portuguese Foundation for Science and Technology.
Footnotes
Published ahead of print on 2 September 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Albrecht, M., and T. Lengauer. 2004. Novel Sm-like proteins with long C-terminal tails and associated methyltransferases. FEBS Lett. 56918-26. [DOI] [PubMed] [Google Scholar]
- 2.Anantharaman, V., and L. Aravind. 2004. Novel conserved domains in proteins with predicted roles in eukaryotic cell-cycle regulation, decapping and RNA stability. BMC Genomics 545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Audhya, A., F. Hyndman, I. X. McLeod, A. S. Maddox, J. R. Yates III, A. Desai, and K. Oegema. 2005. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J. Cell Biol. 171267-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badis, G., C. Saveanu, M. Fromont-Racine, and A. Jacquier. 2004. Targeted mRNA degradation by deadenylation-independent decapping. Mol. Cell 155-15. [DOI] [PubMed] [Google Scholar]
- 5.Barbee, S. A., P. S. Estes, A. M. Cziko, J. Hillebrand, R. A. Luedeman, J. M. Coller, N. Johnson, I. C. Howlett, C. Geng, R. Ueda, A. H. Brand, S. F. Newbury, J. E. Wilhelm, R. B. Levine, A. Nakamura, R. Parker, and M. Ramaswami. 2006. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 52997-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boag, P. R., A. Nakamura, and T. K. Blackwell. 2005. A conserved RNA-protein complex component involved in physiological germline apoptosis regulation in Caenorhabditis elegans. Development 1324975-4986. [DOI] [PubMed] [Google Scholar]
- 7.Bouveret, E., G. Rigaut, A. Shevchenko, M. Wilm, and B. Seraphin. 2000. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 191661-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, Z., J. Coller, R. Parker, and H. Song. 2005. Crystal structure and functional analysis of DEAD-box protein Dhh1p. RNA. 111258-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coles, M., M. Hulko, S. Djuranovic, V. Truffault, K. Koretke, J. Martin, and A. N. Lupas. 2006. Common evolutionary origin of swapped-hairpin and double-psi beta barrels. Structure 141489-1498. [DOI] [PubMed] [Google Scholar]
- 10.Collins, B. M., L. Cubeddu, N. Naidoo, S. J. Harrop, G. D. Kornfeld, I. W. Dawes, P. M. Curmi, and B. C. Mabbutt. 2003. Homomeric ring assemblies of eukaryotic Sm proteins have affinity for both RNA and DNA. Crystal structure of an oligomeric complex of yeast SmF. J. Biol. Chem. 27817291-17298. [DOI] [PubMed] [Google Scholar]
- 11.Cornilescu, G., F. Delaglio, and A. Bax. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13289-302. [DOI] [PubMed] [Google Scholar]
- 12.Decker, C. J., and R. Parker. 2006. CAR-1 and trailer hitch: driving mRNP granule function at the ER? J. Cell Biol. 173159-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Decker, C. J., D. Teixeira, and R. Parker. 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179437-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diercks, T., M. Coles, and H. Kessler. 1999. An efficient strategy for assignment of cross-peaks in 3D heteronuclear NOESY experiments. J. Biomol. NMR 15177-180. [DOI] [PubMed] [Google Scholar]
- 15.Diercks, T., M. Daniels, and R. Kaptein. 2005. Extended flip-back schemes for sensitivity enhancement in multidimensional HSQC-type out-and-back experiments. J. Biomol. NMR 33243-259. [DOI] [PubMed] [Google Scholar]
- 16.Dong, S., C. Li, D. Zenklusen, R. H. Singer, A. Jacobson, and F. He. 2007. YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA. Mol. Cell 25559-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eulalio, A., I. Behm-Ansmant, and E. Izaurralde. 2007. P bodies: at the crossroads of posttranscriptional pathways. Nat. Rev. Mol. Cell. Biol. 89-22. [DOI] [PubMed] [Google Scholar]
- 18.Eulalio, A., I. Behm-Ansmant, D. Schweizer, and E. Izaurralde. 2007. P-body formation is a consequence, not the cause of RNA-mediated gene silencing. Mol. Cell. Biol. 273970-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eulalio, A., J. Rehwinkel, M. Stricker, E. Huntzinger, S.-F. Yang, T. Doerks, S. Dorner, P. Peer Bork, M. Boutros, and E. Izaurralde. 2007. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 212558-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fenger-Grøn, M., C. Fillman, B. Norrild, and J. Lykke-Andersen. 2005. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell 20905-915. [DOI] [PubMed] [Google Scholar]
- 21.Fromont-Racine, M., A. E. Mayes, A. Brunet-Simon, J. C. Rain, A. Colley, I. Dix, L. Decourty, N. Joly, F. Ricard, J. D. Beggs, and P. Legrain. 2000. Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins. Yeast 1795-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gemmecker, G., W. Jahnke, and H. Kessler. 1993. Measurement of fast proton exchange rates in isotopically labeled compounds. J. Am. Chem. Soc. 11511620-11621. [Google Scholar]
- 23.Hermann, H., P. Fabrizio, V. A. Raker, K. Foulaki, H. Hornig, H. Brahms, and R. Lührmann. 1995. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein-protein interactions. EMBO J. 142076-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holm, L., and C. Sander. 1995. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20478-480. [DOI] [PubMed] [Google Scholar]
- 25.Kambach, C., S. Walke, R. Young, J. M. Avis, E. de la Fortelle, V. A. Raker, R. Lührmann, J. Li, and K. Nagai. 1999. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 96375-387. [DOI] [PubMed] [Google Scholar]
- 26.Khusial, P., R. Plaag, and G. W. Zieve. 2005. LSm proteins form heptameric rings that bind to RNA via repeating motifs. Trends Biochem. Sci. 30522-528. [DOI] [PubMed] [Google Scholar]
- 27.Kilic, T., S. Sanglier, A. van Dorsselaer, and D. Suck. 2006. Oligomerization behavior of the archaeal Sm2-type protein from Archaeoglobus fulgidus. Protein Sci. 152310-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koide, S., W. Jahnke, and P. E. Wright. 1995. Measurement of intrinsic exchange rates of amide protons in a 15N-labeled peptide. J. Biomol. NMR 6306-312. [DOI] [PubMed] [Google Scholar]
- 29.Kshirsagar, M., and R. Parker. 2004. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics 166729-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereo chemical quality of protein structures. J. Appl. Cryst. 26283-291. [Google Scholar]
- 30.Lieb, B., M. Carl, R. Hock, D. Gebauer, and U. Scheer. 1998. Identification of a novel mRNA-associated protein in oocytes of Pleurodeles waltl and Xenopus laevis. Exp. Cell Res. 245272-281. [DOI] [PubMed] [Google Scholar]
- 31.Lin, M. D., S. J. Fan, W. S. Hsu, and T. B. Chou. 2006. Drosophila decapping protein 1, dDcp1, is a component of the oskar mRNP complex and directs its posterior localization in the oocyte. Dev. Cell 10601-613. [DOI] [PubMed] [Google Scholar]
- 32.Ling, S. H. M., C. J. Decker, M. A. Walsh, M. She, R. Parker, and H. Song. 2008. Crystal structure of human Edc3 and its functional implications. Mol. Cell. Biol. 285965-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mariño-Ramírez, L., and J. C. Hu. 2002. Isolation and mapping of self-assembling protein domains encoded by the Saccharomyces cerevisiae genome using lambda repressor fusions. Yeast 19641-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mazzoni, C., I. D'Addario, and C. Falcone. 2007. The C terminus of the yeast Lsm4p is required for the association to P-bodies. FEBS Lett. 5814836-4840. [DOI] [PubMed] [Google Scholar]
- 35.Michelitsch, M. D., and J. S. Weissman. 2000. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc. Natl. Acad. Sci. USA 9711910-11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minshall, N., M. H. Reiter, D. Weil, and N. Standart. 2007. CPEB interacts with an ovary-specific eIF4E and 4E-T in early Xenopus oocytes. J. Biol. Chem. 28237389-37401. [DOI] [PubMed] [Google Scholar]
- 37.Monzo, K., O. Papoulas, G. T. Cantin, Y. Wang, J. R. Yates, 3rd, and J. C. Sisson. 2006. Fragile X mental retardation protein controls trailer hitch expression and cleavage furrow formation in Drosophila embryos. Proc. Natl. Acad. Sci. USA 103:18160-18165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mura, C., D. Cascio, M. R. Sawaya, and D. S. Eisenberg. 2001. The crystal structure of a heptameric archaeal Sm protein: implications for the eukaryotic snRNP core. Proc. Natl. Acad. Sci. USA 985532-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murzin, A. G., S. E. Brenner, T. Hubbard, and C. Chothia. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247536-540. [DOI] [PubMed] [Google Scholar]
- 40.Nelson, K. K., and S. K. Lemmon. 1993. Suppressors of clathrin deficiency: overexpression of ubiquitin rescues lethal strains of clathrin-deficient Saccharomyces cerevisiae. Mol. Cell. Biol. 13521-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker, R., and U. Sheth. 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell 25635-646. [DOI] [PubMed] [Google Scholar]
- 42.Pepling, M. E., J. E. Wilhelm, A. L. O'Hara, G. W. Gephardt, and A. C. Spradling. 2007. Mouse oocytes within germ cell cysts and primordial follicles contain a Balbiani body. Proc. Natl. Acad. Sci. USA 104:187-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Röper, K. 2007. Rtnl1 is enriched in a specialized germline ER that associates with ribonucleoprotein granule components. J. Cell Sci. 1201081-1092. [DOI] [PubMed] [Google Scholar]
- 44.Schumacher, M. A., R. F. Pearson, T. Moller, P. Valentin-Hansen, and R. G. Brennan. 2002. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 213546-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Séraphin, B. 1995. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J. 142089-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Squirrell, J. M., Z. T. Eggers, N. Luedke, B. Saari, A. Grimson, G. E. Lyons, P. Anderson, and J. G. White. 2006. CAR-1, a protein that localizes with the mRNA decapping component DCAP-1, is required for cytokinesis and ER organization in Caenorhabditis elegans embryos. Mol. Biol. Cell 17336-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanaka, K. J., K. Ogawa, M. Takagi, N. Imamoto, K. Matsumoto, and M. Tsujimoto. 2006. RAP55, a cytoplasmic mRNP component, represses translation in Xenopus oocytes. J. Biol. Chem. 28140096-40106. [DOI] [PubMed] [Google Scholar]
- 48.Thore, S., C. Mayer, C. Sauter, S. Weeks, and D. Suck. 2003. Crystal structures of the Pyrococcus abyssi Sm core and its complex with RNA. Common features of RNA binding in archaea and eukarya. J. Biol. Chem. 2781239-1247. [DOI] [PubMed] [Google Scholar]
- 49.Törö, I., S. Thore, C. Mayer, J. Basquin, B. Seraphin, and D. Suck. 2001. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 202293-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tritschler, F., A. Eulalio, V. Truffault, M. D. Hartmann, S. Helms, S. Schmidt, M. Coles, E. Izaurralde, and O. Weichenrieder. 2007. A divergent Sm-fold in EDC3 proteins mediates DCP1-binding and P-body targeting. Mol. Cell. Biol. 27:8600-8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Truffault, V., M. Coles, T. Diercks, K. Abelmann, S. Eberhardt, H. Luttgen, A. Bacher, and H. Kessler. 2001. The solution structure of the N-terminal domain of riboflavin synthase. J. Mol. Biol. 309949-960. [DOI] [PubMed] [Google Scholar]
- 52.Vuister, G. W., and A. Bax. 1993. Quantitative, J. correlation: a new approach for measuring homonuclear three-bond J. (HNH.alpha.) coupling constants in 15N-enriched proteins. J. Am. Chem. Soc. 1157772-7777. [Google Scholar]
- 53.Wilhelm, J. E., M. Buszczak, and S. Sayles. 2005. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev. Cell 9675-685. [DOI] [PubMed] [Google Scholar]
- 54.Wilusz, C. J., and J. Wilusz. 2005. Eukaryotic Lsm proteins: lessons from bacteria. Nat. Struct. Mol. Biol. 121031-1036. [DOI] [PubMed] [Google Scholar]
- 55.Yang, W. H., J. H. Yu, T. Gulick, K. D. Bloch, and D. B. Bloch. 2006. RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA 12547-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.