Abstract
Recent results indicate that nontranslating mRNAs in eukaryotic cells exist in distinct biochemical states that accumulate in P bodies and stress granules, although the nature of interactions between these particles is unknown. We demonstrate in Saccharomyces cerevisiae that RNA granules with similar protein composition and assembly mechanisms as mammalian stress granules form during glucose deprivation. Stress granule assembly is dependent on P-body formation, whereas P-body assembly is independent of stress granule formation. This suggests that stress granules primarily form from mRNPs in preexisting P bodies, which is also supported by the kinetics of P-body and stress granule formation both in yeast and mammalian cells. These observations argue that P bodies are important sites for decisions of mRNA fate and that stress granules, at least in yeast, primarily represent pools of mRNAs stalled in the process of reentry into translation from P bodies.
Introduction
The proper control of translation, mRNA degradation, and the subcellular localization of mRNAs is a key aspect of gene expression regulation in eukaryotic cells. Over the past few years, it has emerged that cytosolic mRNAs are in a dynamic equilibrium between different functional and subcellular locations. Translating mRNAs can be found in polysomes, whereas nontranslating mRNAs often accumulate in either stress granules or P bodies (Anderson and Kedersha, 2006; Eulalio et al., 2007; Parker and Sheth, 2007).
Stress granules have been primarily studied in mammalian cells and are dynamic aggregates of untranslating mRNAs in conjunction with a subset of translation initiation factors (eIF4E, eIF4G, eIF4A, eIF3, and eIF2), the 40S ribosomal subunit, and the poly(A) binding protein (Pabp; Anderson and Kedersha, 2006). Notable RNA binding proteins in stress granules include TIA-R, TIA-1, and G3BP, with the latter two possessing self-interaction domains that aid stress granule formation (Tourriere et al., 2003; Gilks et al., 2004). Stress granules are generally not present in normal cells and form in response to defects in translation initiation, including decreased function of eIF2 or eIF4A (Kedersha et al., 2002; Dang et al., 2006; Mazroui et al., 2006). Because stress responses often involve a transient inhibition of translation initiation, stress granules accumulate during many stress responses.
P bodies are RNA protein granules containing untranslating mRNAs complexed with a set of translation repressors, the mRNA decapping machinery and the 5′–3′ exonuclease Xrn1. P bodies are conserved structures present in yeast, plants, nematodes, flies, and mammalian cells (Anderson and Kedersha, 2006; Eulalio et al., 2007; Parker and Sheth, 2007). P bodies have been functionally implicated in mRNA decapping (Sheth and Parker, 2003; Cougot et al., 2004), nonsense-mediated decay (Unterholzner and Izaurralde, 2004; Sheth and Parker, 2006), mRNA storage (Brengues et al., 2005; Bhattacharyya et al., 2006), general translation repression (Holmes et al., 2004; Coller and Parker, 2005), microRNA-mediated repression (Jakymiw et al., 2005; Liu et al., 2005; Pillai et al., 2005) and viral life-cycles (Beliakova-Bethell et al., 2006; Beckham et al., 2007).
P bodies and stress granules in mammalian cells dock and undock from one another in a dynamic manner (Kedersha et al., 2005), suggesting that mRNPs are exchanged between P bodies and stress granules. One possibility is that mRNAs exiting translation accumulate in stress granules, where they are sorted either to return to translation, or are sent to P bodies for mRNA decay (Kedersha et al., 2005; Anderson and Kedersha, 2008). Alternatively, mRNAs exiting translation may first enter P bodies, and undergo a similar sorting process resulting in either mRNA decay, storage in a translationally silenced state, or a return to translation via a stress granule mRNP state. Given the existence of P bodies and stress granules as discrete, interacting cytoplasmic compartments containing nontranslating mRNA, determining the mechanisms by which these particles assemble and interact is important for understanding how cells regulate cytosolic mRNAs.
An ideal system for studying the relationship between stress granules and P bodies is yeast, as P-body assembly can be prevented or modified in various mutant strains (Coller and Parker, 2005; Decker et al., 2007; Teixeira and Parker, 2007). Recently, RNP granules containing eIF4E, eIF4G, and Pab1p, which are components of mammalian stress granules, have been described to form in yeast during glucose deprivation and to both overlap or be distinct from P bodies (Brengues and Parker, 2007; Hoyle et al., 2007). Moreover, these granules, referred to as EGP bodies, contain mRNA (Hoyle et al., 2007). In this work, we first demonstrate that by composition and assembly mechanisms, these eIF4E-, eIF4G-, and Pab1-containing granules are analogous to mammalian stress granules. More importantly, genetic and kinetic data indicate that stress granule assembly depends upon P-body formation, whereas P bodies form independently of stress granules. Our data suggest that formation of a P-body mRNP state is the key first step in deciding the fate of mRNAs after exit from polysomes during stress, and imply that stress granules represent sites of translation reinitiation.
Results
Yeast contain a distinct RNA granule with composition similar to mammalian stress granules
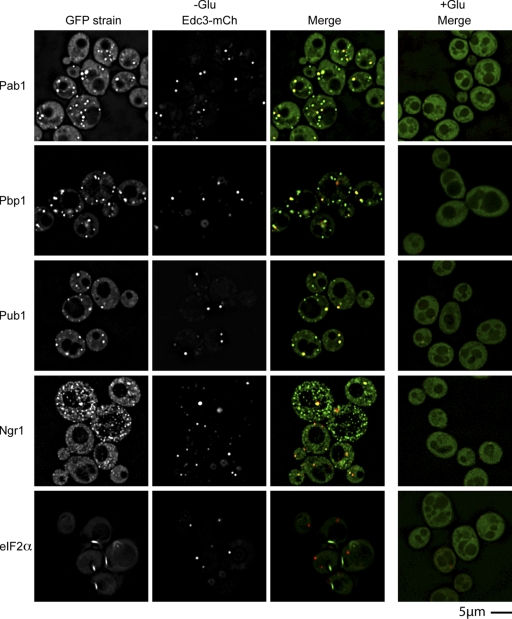
An unresolved issue is whether the eIF4E-, eIF4G-, and Pab1p-containing granules observed in Saccharomyces cerevisiae are analogous to mammalian stress. To determine this, we examined if they contained orthologues of additional mammalian proteins known to accumulate in stress granules using strains bearing a chromosomal C-terminal GFP fusion at the ORF of interest (Huh et al., 2003). Besides accepted yeast orthologues of mammalian stress granule proteins, other proteins were examined based on BLAST homology to other mammalian stress granule proteins (e.g., HuR and G3BP), or protein/genetic interaction data, where interactions with eIF4F factors, Pub1, Pbp1, and Ngr1 were given particular consideration. Each protein that formed foci was also examined for its localization to P bodies when coexpressing an Edc3-mCh or Dcp2-RFP fusion protein (unpublished data).
These experiments revealed that during glucose deprivation the yeast orthologues of many proteins seen in mammalian stress granules accumulate in cytoplasmic foci, which can be distinct from P bodies (Fig. 1). These included the translation initiation factors eIF4GI, eIF4GII, eIF4E, and Pab1, Pub1, and Ngr1, which are the yeast orthologues of mammalian TIA-1 and TIA-R, and Pbp1, which is the orthologue of Ataxin-2, a component of mammalian stress granules required for their assembly (Nonhoff et al., 2007; Fig. 1). Several other proteins also accumulate in these foci (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200807043/DC1), including Hrp1 and Gbp2, which are nuclear-cytoplasmic shuttling proteins; Nrp1 and Ygr250c, predicted RNA binding proteins; and Eap1, an eIF4E binding protein (Cosentino et al., 2000).
Figure 1.
Candidate yeast stress granule proteins form P body–distinct cytoplasmic foci during glucose deprivation. Log-phase wild-type cells expressing chromosomal GFP-tagged proteins and Edc3-mCh (pRP1574) were glucose deprived and examined. No GFP foci, and only faint P bodies were observed in +glucose control conditions, hence display of merge only.
To determine if all these proteins were components of a single granule type, we examined their colocalization with a Pub1-mCh fusion protein. We observed that Pub1-mCh colocalized almost completely with GFP fusions of Pab1, eIF4GI, eIF4GII, eIF4E, Pbp1, Ngr1, Eap1, Hrp1, Ygr250c, Gbp2, and Nrp1 (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200807043/DC1). This colocalization indicates that all these proteins are components of a single granule with a similar composition to mammalian stress granules. We refer to these granules as yeast stress granules and note that they are almost certainly the same RNP granules identified earlier as EGP bodies (Brengues and Parker, 2007; Hoyle et al., 2007).
In contrast to results with mammalian cells (Anderson and Kedersha, 2006), yeast stress granules did not contain the Prt1 subunit of eIF3 (unpublished data) or the α-subunit of eIF2, the latter of which forms distinct rod-shaped foci during glucose deprivation (Fig. 1; Campbell et al., 2005). This suggests that the multiple different types of mRNPs can accumulate to form stress granule–like RNPs under different conditions and in different organisms. Because stress granules are dynamic assemblies of mRNPs, this implies that the specific composition observed in the granule will depend on which step in a given mRNP transition process is rate limiting, which may change under differing cellular conditions (see Discussion). Additionally, mammalian P bodies often appear docked on the periphery of stress granules, whereas in yeast, when colocalization between P-body and stress granule components occurred, the overlap was more complete.
We also noted that the translation termination factors eRF1 and eRF3 accumulated in P bodies in a subset of cells, although the majority of these proteins remain cytoplasmic (Fig. S1). This is in agreement with the finding that eRF3 localizes in P bodies during stationary phase (Dori and Choder, 2007), and suggests that translation termination may be coupled to assembly of P-body mRNPs.
Properties of yeast stress granules
Additional experiments revealed several properties of yeast stress granules. First, as in mammalian cells, the assembly of yeast stress granules is blocked by trapping mRNAs in polysomes with cycloheximide (Fig. 2 A), arguing that yeast stress granules require nontranslating mRNA for assembly, which is also supported by the fact that these granules contain mRNAs (Hoyle et al., 2007). Second, unlike mammalian cells, we observed that several other stresses including oxidative stress (3 mM H2O2 for 15 min), hypo-osmotic stress (incubation in H2O + dextrose), and hyper-osmotic stress (incubation in 1 M KCL + dextrose) had little or no effect on the induction of yeast stress granules, although all of these stresses increased P bodies to varying degrees (Fig. 2 B). This suggests that most stresses in yeast that decrease translation generally lead to the accumulation of mRNAs in P bodies.
Figure 2.
Yeast stress granules are sensitive to cycloheximide, and stimulated by a constitutively active allele of the GCN2 kinase. (A) Log-phase yRP840 cells transformed with pRP1659 were glucose deprived and examined. Cycloheximide-treated cells (100 μg/ml) were preincubated with the drug for 1 min before harvest. (B) Cells identical to those in A were subject to the following stresses. Oxidative stress: shift to SC media containing 3 mM H2O2 or a mock treatment (H2O) for 15 min. Hyperosmotic stress: shift to SC media containing 1 M KCL for 15 min. Hypotonic stress: shift to H2O containing 2% dextrose for 15 min. SC media was used as a mock osmotic control. (C) Cells similar to those in A, but additionally transformed with either pRP1663 (Gcn2c) or pRS416 (empty vector control) were glucose deprived and examined for altered stress granule/P-body assembly.
We also examined the effect upon stress granule formation in yeast of expressing a constitutively active allele of the Gcn2 kinase (Gcn2c; Ramirez et al., 1992), which phosphorylates and decreases eIF2α function. The logic of this experiment was that in mammalian cells, stress granule assembly is often stimulated by phosphorylation of eIF2α at position Ser51 by stress-inducible kinases such as Gcn2 (Kedersha et al., 1999, 2002). This phosphorylation leads to decreased translation initiation rates, causing accumulation of nontranslating mRNAs in stress granules. We observed that Gcn2c expression in mid-log cultures slightly increased P-body size and brightness over the vector control, and occasional stress granules were visible, though these almost always localized with P bodies (Fig. 2 C; Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200807043/DC1). More strikingly, in the presence of Gcn2c, both stress granules and P bodies were strongly enhanced during glucose deprivation (Fig. 2 C), with stress granules being brighter, larger, and more frequent in cells compared with those expressing an empty vector control. This suggests that, as in mammalian cells, accumulation of mRNAs stalled in translation initiation due to decreased eIF2 function leads to increased formation of stress granules.
Specific proteins affect yeast stress granule assembly
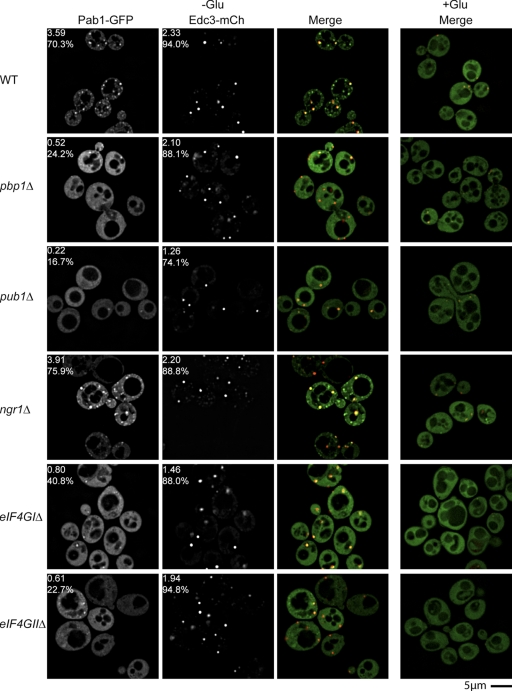
To determine how yeast stress granules assemble, we examined how the absence of specific proteins affected the assembly of either stress granules during glucose deprivation (monitored using Pab1-GFP or Pub1-mCh) or P bodies (monitored with Edc3-mCh). An initial set of null strains were chosen based on homology to factors reported to affect mammalian stress granule assembly (Kedersha et al., 1999; Nonhoff et al., 2007). We then quantified the effects of specific mutations on stress granules by blind scoring of the percentage of cells with stress granules, the average number of granules per cell, and the average size of stress granule foci (see Materials and methods, and Table S1). In these experiments, the absence of stress granules was not due to large changes in expression of the marker protein as verified by Western blot analysis or overall fluorescence levels (Fig. S3 [available at http://www.jcb.org/cgi/content/full/jcb.200807043/DC1] and unpublished data). These experiments revealed the following important points.
First, we observed that pbp1Δ strains showed a strong reduction in stress granule formation as judged by both the number of cells exhibiting Pab1 and Pub1 foci, and the average number of foci observed, although P bodies still formed at wild-type levels (Fig. 3; and see Fig. 6). This is consistent with studies of the mammalian orthologue Ataxin-2, where siRNA depletion of the protein leads to reduction in stress granules, but has no effect on P-body formation (Nonhoff et al., 2007).
Figure 3.
Mutations that inhibit stress granule assembly do not significantly affect P-body assembly. Log-phase BY4741 wild-type and isogenic knockout strains were transformed with pRP1657, glucose deprived, and examined for altered stress granule/P-body assembly. Numbers indicate average foci number per cell, and percentage of cells with foci.
Figure 6.
Formation of Pub1-mCh foci predominantly mirrors null strain sensitivity trends exhibited by Pab1-GFP. Various yeast deletion strains and wild-type isogenic controls were transformed with pRP1661 or pRP1662, according to strain auxotrophies. Log-phase strains were glucose deprived and examined for altered stress granule assembly. Images are collapsed Z-stacks.
Second, we observed that pub1Δ strains also displayed a strong decrease in the average number and percentage of cells with stress granules as judged by Pab1 localization, but with little effect on P bodies. Pub1 is the yeast homologue of TIA-1, which is thought to self-aggregate and promote stress granule assembly by virtue of a glutamine-rich prion domain (Kedersha et al., 1999), which is conserved in Pub1 (Michelitsch and Weissman, 2000). In contrast, strains lacking the yeast orthologue of TIA-R, Ngr1, did not show a defect in stress granule formation (Fig. 3), consistent with prior observations in mammalian cells (Gilks et al., 2004). The similar role in stress granule assembly for Pbp1, Pub1, and their orthologues in mammalian cells strongly argues that yeast and mammalian stress granules are related structures.
Third, we observed that strains with decreased levels of eIF4G showed decreases in the amount of stress granules formed, again with no clear defects in P-body formation. Yeast has two genes for eIF4G. Strains lacking eIF4GII showed a strong decrease in stress granules as judged by both Pab1p and Pub1, although the degree of inhibition was not as severe as that of either pbp1Δ or pub1Δ strains (Fig. 3, and see Fig. 6). In contrast, strains lacking eIF4GI showed a strong reduction in stress granules as judged by Pab1p (Fig. 3), but had no effect on Pub1 foci (see Fig. 6). These results indicate that both eIF4G1 and eIF4GII can affect the assembly of stress granules and that the recruitment of Pub1p into stress granules is differentially affected by the different eIF4G paralogues. Interestingly, although P bodies increase with defects in certain initiation factor mutants (ts alleles of eIF4E, eIF3, and pab1Δ; Teixeira et al., 2005; Brengues and Parker 2007), we did not observe an increase in P bodies with eIF4GIΔ or eIF4GIIΔ strains, which may reflect redundancy between these paralogues, or as-yet unappreciated roles for specific initiation factors in mRNP transitions between P bodies and stress granules.
These results identify Pub1, Pbp1, and eIF4G proteins as playing important roles in the assembly of yeast stress granules, while also critically demonstrating that yeast P bodies can form independently of stress granules.
Stress granule formation is not required for translation repression or mRNA stabilization
Stress granule formation occurs during stresses that lead to a reduction in translation initiation rates, and it has been proposed that the formation of the stress granules might play a role in such translation repression (Kedersha and Anderson, 2002; Anderson and Kedersha, 2008). In addition, many stresses globally stabilize mRNAs (Gowrishankar et al., 2006; Hilgers et al., 2006), and it has been inferred that such stabilization might be due to stress granule formation (Kedersha et al., 2005; Stöhr et al., 2006). Using the mutants that prevented the formation of stress granules, we examined whether stress granule formation affected translation repression and/or mRNA stabilization during glucose deprivation in yeast.
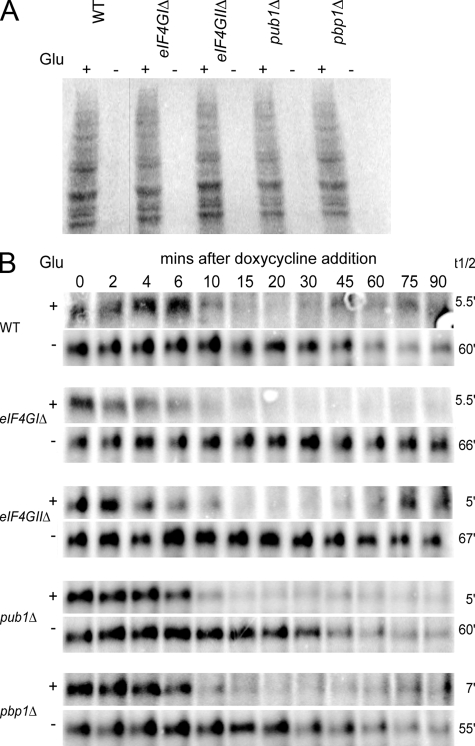
We observed that the pub1Δ, pbp1Δ, eIF4GIΔ, and eIF4GIIΔ strains, all of which are defective in stress granule formation, were able to repress translation similar to wild-type strains in response to glucose deprivation as judged by the incorporation of S35 into new proteins (Fig. 4 A; unpublished data). This observation indicates that stress granule formation is not required for global translation repression during glucose deprivation.
Figure 4.
Stress granule assembly mutants are not deficient in their ability to both translationally repress and stabilize mRNA during stress. (A) Log-phase BY4741 wild-type and isogenic knockout strains were washed and incubated in media +/− glucose at 30°C for 10 min, followed by 35S-met/cys labeling for 5 min. Lysates were prepared and separated by SDS-PAGE for PhosphorImager analysis. (B) Log-phase BY4741 WT and isogenic knockout strains transformed with pRP1192 were resuspended in media +/− glucose, followed by doxycycline addition to transcriptionally repress the MFA2-pG mRNA reporter. Thus, only decay of existing MFA2-pG mRNA was examined. Time points were taken and analyzed via Northern blot. mRNA half-lives (right) are indicated.
We also observed that the MFA2pG reporter mRNA, which is stabilized in response to glucose deprivation in yeast (Hilgers et al., 2006), was also clearly stabilized in wild-type, pub1Δ, pbp1Δ, eIF4GIIΔ, and eIF4GIIΔ strains (Fig. 4 B). This indicates that stress granule formation is not required for the major stabilization of mRNAs that occurs during glucose deprivation. Additionally, no differences in MFA2pG decay in the absence of stress were observed in any of the stress granule assembly mutants (Fig. 4 B). Thus, stress granule assembly is unlikely to function in protecting mRNAs from premature or aberrant decay.
Yeast stress granule formation is dependent on P-body assembly
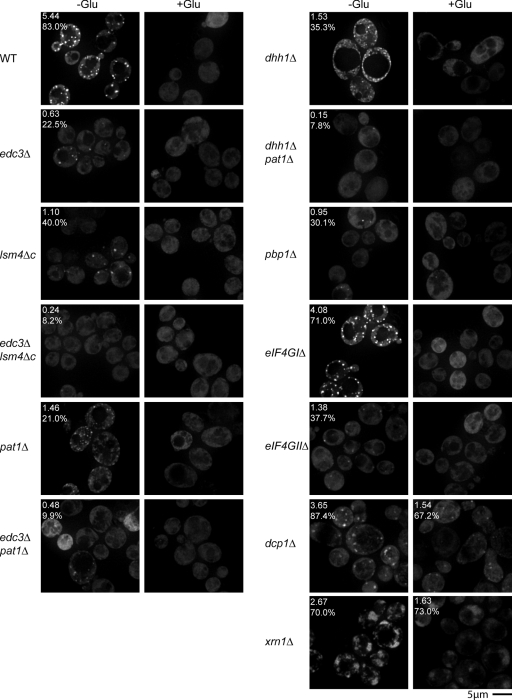
An unresolved issue is the nature of interactions between stress granules and P bodies. During translation repression, mRNAs may be initially routed to stress granules, in which mRNAs are either retained in storage before reentry into translation, or are instead sent to P bodies for decay (Anderson and Kedersha 2006, 2008). An alternative possibility is that mRNAs exiting translation first assemble into an mRNP that can accumulate in P bodies, followed either by degradation, retention for storage, or reentry into translation, which might lead to the formation of stress granules when steps in translation initiation are inhibited. As discussed above, the observation that P bodies can form in the absence of stress granules argues that both P bodies and stress granules form independently, or that P bodies are precursors to stress granules. To distinguish these two possibilities, we examined how defects in P-body assembly affected stress granule formation by taking advantage of mutant strains that inhibit P-body formation by limiting aggregation of mRNPs into larger P bodies (Coller and Parker, 2005; Decker et al., 2007). All images were quantified in a blind manner for the effects of each mutation on the percentage of cells with stress granules or P bodies, as well as average foci size and the average number of each foci per cell (see Materials and methods and Table S1).
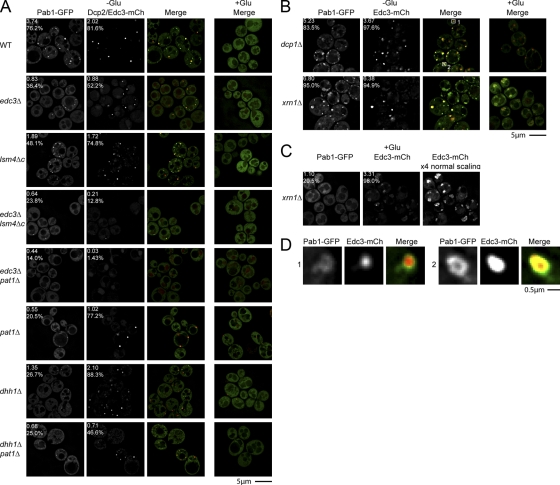
To examine possible effects of P-body formation of stress granules, we first examined the effects of deletions of Edc3, which affects P-body aggregation, or the C-terminal tail of Lsm4, which has a prion-type domain that contributes to aggregation of P bodies (Decker et al., 2007). In addition, we examined the effects of an edc3Δ lsm4Δc double mutant where P bodies are dramatically reduced. An important result was that edc3Δ lsm4Δc yeast, which are greatly reduced in P-body formation, were similarly strongly inhibited for formation of stress granules as judged by Pab1-GFP (Fig. 5 A) or Pub1-mCh (see Fig. 6). Occasional Pab1 or Pub1 foci were seen in a small fraction of cells, but they were fainter, smaller (Table S1), and at least in the case of Pab1, typically colocalized with what faint P bodies remained visible (Fig. 5 A). The edc3Δ and lsm4Δc strains also showed reductions in stress granule formation, but similar to their partial effects on P-body formation, stress granules were reduced but could still form in edc3Δ and lsm4Δc strains (Fig. 5 A). We interpreted these observations to suggest that the formation of stress granules was enhanced by P bodies.
Figure 5.
Mutations that inhibit P-body formation also inhibit stress granule assembly. (A) Various yeast deletion strains and wild-type isogenic controls were transformed with pRP1657, pRP1658, or pRP1659, according to auxotrophies and genetic properties of the strains; bottom three panels feature Edc3-mCh as a P-body marker, others feature Dcp2-mCh. Glucose deprivation and quantitation as shown in Fig. 3. (B) Decapping (dcp1Δ) and 5′–3′decay (xrn1Δ) mutant strains were transformed with pRP1659. (C) Individual Pab1-GFP and Edc3-mCh images from +glucose xrn1Δ control cells, highlighting large and diffuse nature of constitutive Edc3-mCh foci (more visible at 4x scaling intensity) and occasional large and diffuse Pab1-GFP foci. (D) Zoom panels from dcp1Δ images in B (white boxes), showing examples of Pab1-GFP forming donut-like foci, with Edc3 foci overlapping the Pab1-GFP holes.
To extend this analysis, we wished to examine additional mutants with defects in P-body assembly or mutants lacking individual P-body protein components. Because edc3Δ lsm4Δc strains can still form a few P bodies, we desired to create a strain with an even stronger defect in P-body assembly. We hypothesized that an edc3Δ pat1Δ strain might be extremely defective in P-body formation because it would (1) lack Edc3; (2) lack Pat1, which can contribute to the formation of P bodies (Teixeira and Parker, 2007); and (3) would be unable to use the Lsm4 C-terminal domain to assemble P bodies because the recruitment of the Lsm1-7p complex to P bodies is dependent on Pat1p (Teixeira and Parker, 2007). Indeed, examination of an edc3Δ pat1Δ strain showed an even stronger block to P-body formation than the edc3Δ lsm4Δc strain, although a few cells could still form small and faint P bodies (Fig. 5 A). Moreover, the edc3Δ pat1Δ showed an even stronger defect in stress granule formation (Fig. 5 A). This result identifies the edc3Δ pat1Δ strain as the most defective strain for P-body formation, and provides additional evidence that P bodies are required for stress granule formation.
Further experiments revealed that strains lacking either Pat1 or Dhh1 proteins also showed defects in stress granule formation. In pat1Δ strains, P-body formation was reduced and stress granules showed a corresponding reduction (Fig. 5 A and Fig. 6). Interestingly, in dhh1Δ strains, stress granules were reduced compared with wild-type strains, yet P-body formation was normal (Fig. 5 A). This suggests that Dhh1 might have a specific role in stress granule assembly. Finally, examination of a dhh1Δ pat1Δ strain, known to be deficient in both P-body formation and the ability to translationally repress (Coller and Parker, 2005), revealed a strong block to stress granule formation (Fig. 5 A and Fig. 6). P bodies were significantly decreased in number, though not as severely as in edc3Δ lsm4Δc or edc3Δ pat1Δ strains, arguing that in this case, a combination of impaired P-body assembly, fewer mRNAs exiting translation, and/or an inability to transition from P bodies to stress granules may account for this defect.
These results indicate that formation of stress granules in yeast is enhanced by existing P bodies. Moreover, in all these cases, the reduction or absence of stress granules is not due to large changes in Pab1p-GFP expression, as verified by Western blot (Fig. S3). One simple interpretation of these observations is that mRNAs move from P bodies to stress granules during transitions between different mRNP complexes, and that the formation of stress granules is facilitated by a preexisting pool of untranslating mRNPs in P bodies (see Discussion).
A prediction of stress granules forming from mRNAs in P bodies is that strains with enlarged P bodies should show enhanced assembly of stress granules. To test this prediction, we examined stress granule formation in strains defective in mRNA decapping (dcp1Δ) and 5′ to 3′ degradation of mRNAs (xrn1Δ), where P bodies are increased (Sheth and Parker, 2003; Teixeira and Parker, 2007). Strikingly, we observed that during glucose deprivation, stress granules were larger and more numerous in xrn1Δ and dcp1Δ strains than in wild-type cells (Fig. 5 B). Interestingly, in dcp1Δ strains, Pab1 often formed donut-like structures with Edc3 foci visible at the center (Fig. 5 D). These results are consistent with the mRNAs in stress granules predominantly being derived from P bodies. We also observed that both stress granules (as judged by Pab1 and Pub1) and P bodies (as judged by Edc3) were more diffuse in xrn1Δ and dcp1Δ strains than in wild-type cells, suggesting some alteration in their underlying morphology (Fig. 5, B and C). Interestingly, both dcp1Δ and xrn1Δ strains showed significantly more Pub1 foci than Pab1 foci in the absence of stress (Fig. 6, Fig. 5, and Table S1), suggesting that the kinetics of mRNA binding and dissociation differs between Pab1 and Pub1. One explanation for this may be that under nonstress conditions, Pub1 is removed from mRNAs in P bodies as the mRNA is degraded.
Temporal analysis indicates stress granules first form in conjunction with P bodies
The dependence of stress granule assembly on P-body formation suggests that stress granules may form by mRNPs in P bodies exchanging proteins to form an mRNP poised to reenter translation. This model makes two predictions. First, it predicts that stress granules would form after P bodies during stress responses. Second, it predicts that the components of stress granules would first associate with mRNPs in P bodies, followed by maturation of such a complex into a stress granule, as the translation repression/decapping complex was exchanged for translation factors. To test these predictions, we followed the formation of yeast stress granules and P bodies during glucose deprivation by time-lapse microscopy using Pab1-GFP as a marker of stress granules and Dcp2-mCh or Edc3-mCh as a marker of P bodies.
We observed that from 0–7 min after the onset of glucose deprivation stress, P bodies were strongly induced whereas stress granules, as judged by Pab1-GFP, were predominantly absent. However, wherever faint foci were present, they were colocalized with P bodies (Fig. 7 A; Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200807043/DC1). As time progressed, Pab1-GFP foci became brighter within P bodies and then started to form additional cytoplasmic foci which were separate from bright preexisting P bodies. Interestingly, a very faint signal of Dcp2-mCh or Edc3-mCh was often colocalized with these newly “P body–distinct” stress granule foci (Fig. 7 B; unpublished data). Additionally, P bodies that become enriched with Pab1 often showed a partially decreased signal for Dcp2-mCh or Edc3-mCh over time, whereas the Pab1 signal maintains or intensifies, suggesting some type of maturation process is indeed occurring. These observations indicate that yeast stress granules form after P bodies, primarily in conjunction with preexisting P bodies.
Figure 7.
Yeast temporal analysis reveals accumulation of Pab1 in P bodies before formation of P body–distinct stress granules. (A) Log-phase yRP840 strain, transformed with pRP1660, was glucose deprived for 5 min and followed over time. For technical reasons, identical cells at the 0-min time point are not shown; instead a typical image of this time point is shown. Orange arrowheads indicate initial accumulation of Pab1-GFP in preexisting P bodies. Purple arrowheads indicate appearance of P body–distinct stress granules. Images are typical of multiple independent experiments. Images were taken from Video 1 (available at http://www.jcb.org/cgi/content/full/jcb.200807043/DC1). (B) Zoom panels from the 19-min time point (white boxes). Arrowheads indicate common occurrence of faint P-body foci colocalizing with stress granules. Purple arrowhead indicates same foci in A, turquoise arrowheads indicate location of additional faint P bodies. Right: Dcp2-mCh image at 10x normal scaling intensity, indicating relative difference in Dcp2-mCh abundance between P bodies and stress granules. Asterisk indicates auto-fluorescent vacuolar signal.
Induction of mammalian P bodies and stress granules share kinetic properties with yeast
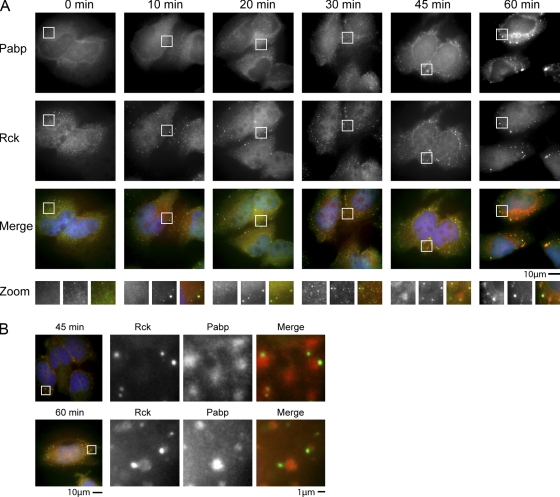
The above results demonstrate that stress granules in yeast form after P bodies during stress and require existing P bodies for efficient accumulation. To examine the relationship between stress granule formation and P bodies in mammalian cells, we examined a time course of stress granule and P-body formation in HeLa cells after arsenite stress using Pabp and Rck (Dhh1 homologue) as markers of stress granules and P bodies, respectively. Similar to yeast cells, we observed that after arsenite stress in HeLa cells, P bodies first increase in number, size, and intensity within 10 min, with no initial change in Pabp distribution (Fig. 8 A). However, in contrast to the strong colocalization of Pab1-GFP and P-body foci when stress granules are first forming in yeast, stress granules in mammalian cells primarily first appear as small foci distinct from P bodies, although occasional colocalization of Pabp and Rck was seen. At later time points, “classical” morphology stress granules appear that are usually docked to P bodies (Fig. 8 A, “45 min”). These results suggest that stress granules in mammalian cells may form independently of P bodies, or that the biochemical activity of P bodies that enhances stress granule formation may be present in multiple small P bodies throughout the cytosol in mammalian cells (see Discussion).
Figure 8.
Temporal analysis of arsenite-stressed HeLa cells reveals P bodies increase in size and number before formation of stress granules. (A) HeLa cells were fixed at several time points after arsenite stress, from 0 (no arsenite added) to 60 min. Endogenous Pabp (red) and Rck (green) were used as stress granule and P-body markers, respectively. DAPI staining (blue) reveals the nucleus. Images are typical of three independent experiments. (B) Representative images of HeLa cells after 45 and 60 min of arsenite stress, demonstrating a greater accumulation of Rck in stress granules at 60 min.
We also observed that Rck, the mammalian orthologue of yeast Dhh1, accumulated in P bodies for the first 45 min of arsenite stress in HeLa cells, but then at 1 h began to accumulate in large stress granules also (Fig. 8 B). This accumulation of Rck in stress granules at late time points during stress was also recently observed in another study (Wilczynska et al., 2005; Mollet et al., 2008). Although speculative, one possibility is that Rck may associate with mRNAs in P bodies and then play a role in those mRNAs transitioning into stress granules, perhaps as an intermediate in an mRNP remodeling process. Together, these data suggest that like yeast, at least some mammalian mRNAs may transition through P bodies before accumulating in stress granules.
Discussion
Stress granules form in S. cerevisiae
Several observations argue that an RNA protein granule equivalent to mammalian stress granules forms in S. cerevisiae during glucose deprivation. This was first suggested by observations that Pab1p, eIF4E, and eIF4G aggregate during glucose deprivation into foci that can overlap with, or be distinct from P bodies, and which contain mRNA (Brengues and Parker, 2007; Hoyle et al., 2007). Moreover, these granules contain many proteins analogous to those seen in mammalian stress granules including Pub1, Ngr1, and Pbp1, orthologues of the mammalian stress granule proteins TIA-1, TIA-R, and Ataxin-2 (Fig. 1). Like mammalian stress granules, yeast stress granule assembly is blocked by cycloheximide (Fig. 2 A), and enhanced by phosphorylation of eIF2α (Fig. 2 C). Moreover, Pub1 and Pbp1, whose orthologues affect stress granule assembly in mammalian cells (Kedersha et al., 1999; Nonhoff et al., 2007), are required for yeast stress granule assembly (Fig. 3). By these criteria of similar composition, similar assembly mechanisms, and forming when translation initiation is compromised, we consider these foci as yeast stress granules.
P bodies enhance the formation of yeast stress granules
Several observations indicate that P bodies enhance the assembly of yeast stress granules, whereas P bodies form independently of stress granules. The key observation is that strains strongly inhibited for P-body formation (edc3Δ lsm4Δc, edc3Δ pat1Δ, dhh1Δ, and pat1Δ) also cause a strong inhibition of stress granule formation (Fig. 5, Fig. 6), whereas strains defective in stress granule formation (pub1Δ, pbp1Δ, and eIF4GIIΔ) form P bodies normally (Fig. 3). Formally, the individual proteins within P bodies that affect stress granule formation may have separate roles in stress granule formation, although this is unlikely as many of these P-body proteins (e.g., Edc3, Lsm4, and Pat1) are not observed in stress granules. Furthermore, whenever occasional faint stress granules were detected in P body–deficient strains, they usually colocalized with the residual faint P bodies, reinforcing the dependency of stress granule formation on P bodies (Fig. 5). In addition, during inhibition of translation by glucose deprivation, P bodies form first, followed by accumulation of Pab1 in association with P bodies, with independent stress granules appearing last (Fig. 7). This order of events was also observed in a parallel study, where eIF4E rather than Pab1 was used as a marker of EGP bodies, which are equivalent to yeast stress granules (Hoyle et al., 2007). We interpret these observations to indicate that preexisting P bodies enhance the formation of yeast stress granules.
mRNPs in stress granules may primarily arrive via P bodies
A simple explanation for the role of P bodies in promoting stress granule formation is that mRNAs exiting translation first enter P bodies, and then subsequently transition to a stress granule state before reentering translation (Fig. 9). This model explains both why stress granules form in proximity to P bodies, and why P bodies would enhance the formation of stress granules. A predominant directionality of mRNAs moving from P bodies into stress granules and then onto translation is also consistent with observations that PGK1 mRNAs are detected in P bodies after 10 min of glucose deprivation (Teixeira et al., 2005), and in EGP bodies (stress granules) after 30 min of glucose deprivation (Hoyle et al., 2007). Additionally, mRNAs in yeast P bodies can return to translation (Brengues et al., 2005), and components of P bodies function as translation repressors, and can be required for translation repression during glucose deprivation (Holmes et al., 2004; Coller and Parker, 2005; Pilkington and Parker, 2008). Moreover, we observe that translation termination factors can associate with P bodies and not stress granules (Fig. 1), which is consistent with the mRNPs in P bodies representing those that have just exited translation and those in stress granules being those preparing to reenter translation. We interpret these observations to argue that most yeast mRNAs move from P bodies to stress granules, although we cannot rule out that some mRNAs, depending on cellular context or transcript-specific factors, may move from translation directly to stress granules, or from stress granules to P bodies.
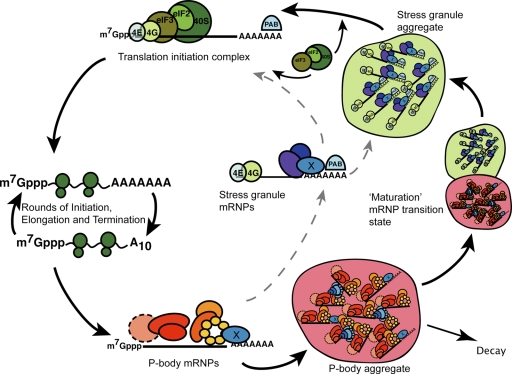
Figure 9.
Model for predominant cytoplasmic flow of mRNAs through P-body and stress granule mRNP states. After exit from polysomes, mRNAs are bound by P-body components, forming a P-body mRNP state that could either target the mRNA for decay, or for a return to translation, initially via transition into a stress granule mRNP state. Factors affecting this decision process may include specific mRNA binding proteins (factor “X”) or the presence of a poly(A) tail. Transition into a stress granule aggregate is favored by initial accumulation and mRNP remodeling in a P-body aggregate, though direct mRNP remodeling not involving visible cytoplasmic aggregates may also occur (dashed arrow). Having achieved a stress granule mRNP state, mRNAs would acquire additional translational components (eIF2, eIF3, and 40S subunits) before reentering translation.
We envision two possible and overlapping mechanisms by which P bodies could enhance the rate of forming a stress granule. First, a P-body mRNP might contain specific proteins that interact with stress granule components and promote the transition from a P-body mRNP to a stress granule state. A second model is that P bodies enhance the formation of stress granules by providing a specific location for the assembly of stress granule mRNPs, which then allows them to aggregate efficiently due to their high local concentration. In this latter model, P bodies are not required for the formation of the individual mRNPs that make up stress granules, but function to facilitate their aggregation. In either case, it is important to note that stress granules represent pools of mRNAs primarily poised to reenter translation, which is also supported by the observation that stress granules disassemble faster than P bodies when yeast are triggered to reenter growth from stationary phase (Brengues and Parker, 2007).
Several points argue that at least some mRNAs will move from P bodies to stress granules in mammalian cells. First, the increase in P bodies before stress granules in HeLa cells during stress (Fig. 8 A) is consistent with mRNAs exiting translation first entering a P-body state. Second, mammalian mRNAs within P bodies can return to translation, suggesting they can transition from P bodies to a state associated with translation factors (Bhattacharyya et al., 2006). Third, mammalian P bodies can form independently of stress granules (Kedersha et al., 2005; Kwon et al., 2007), although whether stress granules are dependent on preexisting P bodies has not been tested because it has not yet been possible to abolish formation of P bodies in mammalian cells during stress. Fourth, the initial concentration of the Rck protein in P bodies followed by its accumulation in stress granules is consistent with mRNPs transitioning from P bodies to stress granules (Fig. 8 B). Fifth, induction of stress granules by arsenite treatment in HeLa cells leads to a “burst” of stress granules forming in association with P bodies (Mollet et al., 2008), which is consistent with stress granules forming from mRNAs exiting P bodies. However, in contrast to yeast, mammalian stress granules, as assessed by Pabp, primarily appear first as distinct units without interaction with P bodies (Fig. 8), suggesting mammalian stress granules might be able to form independently of P bodies, or there might be dispersed small P bodies, below microscopic detection, under these nascent stress granules.
Implications for the nature and function of stress granules
Our results argue that stress granules do not function in translation repression or broadly preventing mRNA degradation during stress. Evidence that stress granules are not required for translation repression is that yeast strains defective in stress granule formation are still capable of translation repression during glucose deprivation (Fig. 4 A), and mammalian cells unable to form stress granules can still repress translation in response to inhibition of HDAC6 function (Kwon et al., 2007). Similarly, the enhanced stability of mRNAs under stress is not strongly reduced in strains lacking stress granules (Fig. 4 B). Finally, FRAP evidence indicating the rapid movement of both RNA binding proteins and mRNA in and out of stress granules (Kedersha et al., 2005; Mollet et al., 2008) argues against a role for stress granules in prolonged mRNA storage.
An alternative possibility is that stress granules may function to increase the translation of mRNAs, particularly during stress where initiation is limiting. Note that this might be particularly important when the rates of translation initiation are compromised, under which conditions the aggregation of stalled initiation complexes together might serve to enhance translation initiation by increasing the local concentration of translation initiation factors, and/or preventing alternative fates of the mRNA such as degradation and/or disassembly of the translation initiation complex. Strikingly, all of the factors required for yeast stress granule assembly can positively influence mRNA function: (1) eIF4G is a translation initiation factor; (2) Pub1 can stabilize and enhance the translation of some mRNAs (Ruiz-Echevarria and Peltz, 2000; Duttagupta et al., 2005); and (3) Pbp1 can enhance the translation of the HO mRNA (Tadauchi et al., 2004). However, it is worth noting that certain components of yeast stress granules have also been implicated in negatively regulating mRNA translation and stability (Buu et al., 2004; Vasudevan et al., 2005). Important areas of future work will be to determine what role stress granule proteins play in affecting mRNP transitions between polysomes and nontranslating states, as well as the consequences to the cell when stress granules cannot form.
A working model for the cytosolic mRNA cycle
The observations discussed above suggest a working model for the dynamics of mRNAs in the yeast cytosol with the following properties (Fig. 9). First, mRNAs that are engaged in translation are present in polysomes and distributed throughout the cytosol. Second, when translation initiation is compromised, either due to stochastic events on individual mRNAs or in response to stress inhibition of initiation, mRNAs cease translating and assemble with translation repressors and components of P bodies forming a translationally quiescent “P-body mRNP”, which can aggregate into P bodies. Third, whether assembled into a larger aggregate or not, the P-body mRNP can either be retained in this state, be decapped and degraded, or undergo protein exchange reactions that remove the P-body components and load translation initiation factors, thereby transitioning the mRNA into the type of mRNP seen in stress granules. When translation initiation is not limiting, these mRNAs would form polysomes and fail to form stress granules. In contrast, when specific steps in translation initiation are rate limiting, the accumulation of mRNPs at this stage would lead to the formation of stress granules. An important implication of this model is that transitions in mRNP composition within P bodies will be critical in determining the fate of mRNAs.
This model for cytosolic mRNA dynamics provides explanations for many observations about cytosolic mRNP granules. First, it suggests that different mRNP granules represent a continuum of mRNP states, and that different granules may be apparent if subsets of mRNPs aggregate via different mechanisms. Second, it suggests that the composition of a given granule may vary depending on the rate-limiting step in its formation and/or resolution, which may depend on the type of cellular stress encountered. For example, stress granules in mammalian cells during arsenite stress contain eIF2 and eIF3 consistent with a late block in translation initiation (Anderson and Kedersha, 2006). In contrast, stress granules in yeast induced by glucose deprivation do not contain eIF2 and eIF3, suggesting the process of translation initiation is inhibited before the joining of eIF3 and eIF2 in these conditions. Third, delays in transitions between mRNP states may cause fused or hybrid granules. Together, it suggests that it will be important in future work to consider cytosolic RNP granules as representing a continuum of mRNP states and not discreet and unique granule types.
Materials and methods
Yeast strains and growth conditions
The genotypes of strains used in this study are listed in Table S2 (available at http://www.jcb.org/cgi/content/full/jcb.200807043/DC1). Strains expressing proteins C-terminally tagged with GFP were obtained either from genomic libraries or constructed following the polymerase chain reaction (PCR)–based gene modification method described previously (Longtine et al., 1998). Strains were grown on synthetic complete (SC) medium supplemented with appropriate amino acids and 2% glucose (Glu) as a carbon source. Strains were grown at 30°C unless otherwise stated. Yeast strains were transformed by standard techniques.
HeLa cells grown to 50–70% confluency were subject to a 1 μM arsenite stress and paraformaldehyde fixed at different time points from 0 (no arsenite added) to 60 min. Staining of endogenous Pabp (red) and Rck (green) were used as markers of stress granules and P bodies, respectively, with DAPI staining (blue) to reveal the nucleus. Images are representative of three independent experiments.
HeLa cell growth conditions and immunofluorescence
HeLa cells were grown and maintained in DMEM media supplemented with 10% fetal bovine serum, l-glutamine (2 mM; Sigma-Aldrich), and Pen-Strep (100 U/ml penicillin, 100 μg/ml streptomycin; Cellgro). Flasks/slides were incubated in a 37°C incubator with a 5% CO2 concentration. Protocols for arsenite stress and immunofluorescent staining were derived from Kedersha and Anderson, (2007). In brief, cells were passaged onto 8-well chamber slides (Nalge Nunc Intl.), and allowed to grow for 1.5 to 2 d to reach 50–70% confluency. Cells were then subjected to a 1-μM arsenite stress and paraformaldehyde fixed. Slides were mounted using Vectashield with DAPI (Vector Laboratories). αPabp mouse monoclonal IgG (1:200 dilution; Santa Cruz Biotechnology, Inc.) and goat αmouse Texas red (1:200; Santa Cruz Biotechnology, Inc.) were used for Pabp staining. αRCK rabbit polyclonal IgG (1:200; MBL International) and donkey αrabbit Alexa 488 (1:200; Invitrogen) were used for RCK staining.
Plasmids
Plasmids pRP1657–1660 were generated by PstI digestion of pRP1362 and pRP1363 (Brengues and Parker, 2007), and repaired via homologous recombination with a PCR product bearing promoter sequence and the complete ORF sequence of either EDC3 (amplified from plasmids pRP1432 using oRP1407 and oRP1408) or DCP2 (amplified from pRP1205 using oRP1409 and oRP1410). A restriction site unique to the vectors was introduced at the 3′ end of the Edc3 ORF (XhoI) and the Dcp2 ORF (BsiWI). Next, these sites were used to gap the vectors again, followed by homologous recombination repair with a PCR product containing a seven amino acid linker sequence, the mCherry ORF, and PGK1 3′ UTR sequence (amplified from pRP1400 plasmid using oRP1411 [Edc3-specific]/oRP1412 [Dcp2 specific] and oRP1413). To check expression of Pab1-GFP in various mutant strains via Western analysis, αGFP mouse monoclonal (1:1,000; Covance) and goat αmouse-HRP conjugate (1:1,000; Thermo Fisher Scientific) were used.
To generate pRP1574 and pRP1575, pRP1657 and pRP1659 were digested with Acc65I to excise PAB1-GFP, the ADH1 3′ UTR sequence and part of the PAB1 promoter sequence, followed by re-ligation of the vector backbones. To generate pRP1661 and pRP1662, pRP1574 and pRP1575 were XhoI digested and gap repaired by homologous recombination using a PCR product bearing the PUB1 promoter and complete ORF sequence, which was amplified from yeast genomic DNA using oRP1414 and oRP1415. Oligo sequences for all cloning and additional plasmid properties are listed in Table S2.
Microscopy
For glucose depletion and control experiments, yeast cultures were grown to OD600 of 0.3–0.4 in the appropriate SC media. Next, cells were collected by brief centrifugation, washed in fresh SC medium +/− 2% glucose, resuspended in fresh SC medium, and incubated in a flask in a shaking 30°C water bath for 10 min. Cells were harvested and washed once more as described above, before spotting on slides and immediate microscopic examination at room temperature.
All images were acquired using a Deltavision RT microscope system running softWoRx 3.5.1 software (Applied Precision, LLC), using an Olympus 100x, oil-immersion 1.4 NA objective. They were collected as 512 × 512 pixel files with a CoolSnapHQ camera (Photometrics) using 1 × 1 binning for yeast and 2 × 2 binning for mammalian cells. All yeast images were deconvoled using standard softWoRx deconvolution algorithms (enhanced ratio, low noise filtering). ImageJ (Abramoff et al., 2004) was used to adjust all images to equal contrast ranges according to the experiment conducted or protein examined, unless explicitly stated otherwise. To optimize yeast colocalization accuracy, single plane images were used, with mCherry imaging immediately after GFP imaging (gap of ∼1 s). Images of Pub1-mCh in the null series of strains were Z-series compilations of 6–10 images per stack.
Image quantitation
Quantified datasets involving Pab1-GFP and P-body markers represent the analysis of at least three independent experiments; Pub1-mCh null datasets involved the analysis of at least two independent experiments. All scoring analyses were done in a blind manner, with a minimum of 50 cells scored for P-body data, and 100 cells scored for Pab1-GFP or Pub1-mCH data. To count and measure foci size, ImageJ smoothing, thresholding, and analyze particle functions were used—this was done manually to avoid errors due to differential strengths of cytoplasmic signals between cells arising from stochastic variation and/or potentially different copy numbers of plasmids between yeast cells.
35S-Met incorporation assays
Cells from wild-type and isogenic deletion strains were grown in YEPD to mid-log phase (OD600 0.3–0.35), and resuspended to equal optical density units. Samples were washed twice in YEP +/− glucose, followed by resuspension in YEP +/− glucose, and incubation in a flask placed in a 30°C water bath shaker for 10 min. Samples were concentrated, washed as before, resuspended in one-tenth the original volume, and labeled with 35S met/cys for 5 min. Cells were harvested by a quick spin and frozen immediately in dry ice. Proteins were extracted by boiling cell pellets for 10 min in 50 μl lysis buffer (2% SDS, 90 mM Hepes, pH 7.5, and 30 mM DTT), spinning for 2 min in a microfuge, and transferring the supernatant to a new tube. An aliquot of this was combined with denaturing protein loading buffer and run on a 10% SDS-PAGE gel, dried down and imaged on a Typhoon PhosphorImager.
Northern blot analysis/transcriptional shut-off
Northern blot analysis was conducted as previously described (Caponigro et al., 1993). Deletion strains were transformed with pRP1192, which bears a MFA2-polyG mRNA reporter under control of a tetracycline-sensitive promoter. oRP121 was used to probe levels of the MFA2 message. Yeast cultures were grown to mid log (OD600 0.3–0.35) in glucose containing SC media, concentrated and resuspended in fresh media, to which doxycycline was added at a final concentration of 2 μg/ml. Time points were taken thereafter by quick spinning and freezing cell pellets on dry ice. Alternatively, after concentration, cultures were resuspended in media lacking glucose for 10 min, after which doxycycline was added and subsequent time points taken.
Online supplemental material
Table S1 lists P bodies and stress granule quantitation in experiments involving null strains and Gcn2c. Table S2 details properties of yeast strains, plasmids, and oligos used in this work. Fig. S1 shows additional proteins which form P body–distinct foci during glucose deprivation. Fig. S2 shows that such proteins essentially completely colocalize with Pub1-mCh, suggesting a common granule state. Fig. S3 shows that Pab1-GFP levels are similar in glucose deprived P-body or stress granule assembly mutant strains. Video 1 shows formation of P bodies and stress granules over time during glucose deprivation. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200807043/DC1.
Supplementary Material
Acknowledgments
We thank Dr A. Hinnebusch for providing a plasmid that expresses a constitutively active allele of the Gcn2 kinase and C. Decker for unpublished reagents. Additional thanks to A. Webb for assistance with blind scoring of microscopic images and C. Boswell for assistance with microscopy. Final thanks to all Parker laboratory members, E. Forbes and family members for useful discussions and support.
Funding was provided by the Howard Hughes Medical Institute.
Abbreviations used in this paper: mRNP, messenger ribonucleoprotein; Pabp, poly(A) binding protein; YEPD, yeast extract peptone dextrose.
References
- Abramoff, M.D., P.J. Magelhaes, and S.J. Ram. 2004. Image processing with ImageJ. Biophotonics Intl. 11:36–42. [Google Scholar]
- Anderson, P., and N. Kedersha. 2006. RNA granules. J. Cell Biol. 172:803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, P., and N. Kedersha. 2008. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33:141–150. [DOI] [PubMed] [Google Scholar]
- Beckham, C.J., H.R. Light, T.A. Nissan, P. Ahlquist, R. Parker, and A. Noueiry. 2007. Interactions between brome mosaic virus RNAs and cytoplasmic processing bodies. J. Virol. 81:9759–9768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliakova-Bethell, N., C. Beckham, T.H. Jr. Giddings, M. Winey, R. Parker, and S. Sandmeyer. 2006. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA. 12:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya, S.N., R. Habermacher, U. Martine, E.I. Closs, and W. Filipowicz. 2006. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb. Symp. Quant. Biol. 71:513–521. [DOI] [PubMed] [Google Scholar]
- Brengues, M., and R. Parker. 2007. Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol. Biol. Cell. 18:2592–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues, M., D. Teixeira, and R. Parker. 2005. Movement of eukaryotic mRNA between polysomes and cytoplasmic processing bodies. Science. 310:486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buu, L.M., L.T. Jang, and F.J. Lee. 2004. The yeast RNA-binding protein Rbp1p modifies the stability of mitochondrial porin mRNA. J. Biol. Chem. 279:453–462. [DOI] [PubMed] [Google Scholar]
- Campbell, S.G., N.P. Hoyle, and M.P. Ashe. 2005. Dynamic cycling of eIF2 through a large eIF2B-containing cytoplasmic body: implications for translational control. J. Cell Biol. 170:925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro, G., D. Muhlrad, and R. Parker. 1993. A small segment of the MAT alpha 1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol. Cell. Biol. 13:5141–5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller, J., and R. Parker. 2005. General translational repression by activators of mRNA decapping. Cell. 122:875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosentino, G.P., T. Schmelzle, A. Haghighat, S.B. Helliwell, M.N. Hall, and N. Sonenberg. 2000. Eap1p, a novel eukaryotic translation initiation factor 4E-associated protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:4604–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot, N., S. Babajko, and B. Séraphin. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165:31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang, Y., N. Kedersha, W.K. Low, D. Romo, M. Gorospe, R. Kaufman, P. Anderson, and J.O. Liu. 2006. Eukaryotic initiation factor 2 alpha-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 281:32870–32878. [DOI] [PubMed] [Google Scholar]
- Decker, C.J., D. Teixeira, and R. Parker. 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179:437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dori, D., and M. Choder. 2007. Conceptual modeling in systems biology fosters empirical findings: the mRNA lifecycle. PLoS ONE. 2:e872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duttagupta, R., B. Tian, C.J. Wilusz, D.T. Khounh, P. Soteropoulos, M. Ouyang, J.P. Dougherty, and S.W. Peltz. 2005. Global analysis of Pub1p targets reveals a coordinate control of gene expression through modulation of binding and stability. Mol. Cell. Biol. 25:5499–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio, A., I. Behm-Ansmant, and E. Izaurralde. 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 8:9–22. [DOI] [PubMed] [Google Scholar]
- Gilks, N., N. Kedersha, M. Ayodele, L. Shen, G. Stoecklin, L.M. Dember, and P. Anderson. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 15:5383–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar, G., R. Winzen, O. Dittrich-Breiholz, N. Redich, M. Kracht, and H. Holtmann. 2006. Inhibition of mRNA deadenylation and degradation by different types of cell stress. Biol. Chem. 387:323–327. [DOI] [PubMed] [Google Scholar]
- Hilgers, V., D. Teixeira, and R. Parker. 2006. Translation-independent inhibition of mRNA deadenylation during stress in Saccharomyces cerevisiae. RNA. 12:1835–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, L.E., S.G. Campbell, S.K. De Long, A.B. Sachs, and M.P. Ashe. 2004. Loss of translational control in yeast compromised for the major mRNA decay pathway. Mol. Cell. Biol. 24:2998–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle, N.P., L.M. Castelli, S.G. Campbell, L.E. Holmes, and M.P. Ashe. 2007. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell Biol. 179:65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W.K., J.V. Falvo, L.C. Gerke, A.S. Carroll, R.W. Howson, A. Belle, N. Dephoure, E.K. O'Shea, and J.S. Weissman. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691. [DOI] [PubMed] [Google Scholar]
- Jakymiw, A., S. Lian, T. Eystathioy, S. Li, M. Satoh, J.C. Hamel, M.J. Fritzler, and E.K. Chan. 2005. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 7:1267–1274. [DOI] [PubMed] [Google Scholar]
- Kedersha, N., and P. Anderson. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30:963–969. [DOI] [PubMed] [Google Scholar]
- Kedersha, N., and P. Anderson. 2007. Mammalian stress granules and processing bodies. Methods Enzymol. 431:61–81. [DOI] [PubMed] [Google Scholar]
- Kedersha, N.L., M. Gupta, W. Li, I. Miller, and P. Anderson. 1999. RNA-binding proteins TIA-1 and TIA-R link the phosphorylation of eIF2-a to the assembly of mammalian stress granules. J. Cell Biol. 147:1431–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N., S. Chen, N. Gilks, W. Li, I.J. Miller, J. Stahl, and P. Anderson. 2002. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 13:195–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., G. Stoecklin, M. Ayodele, P. Yacono, J. Lykke-Andersen, M.J. Fritzler, D. Scheuner, R.J. Kaufman, D.E. Golan, and P. Anderson. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, S., Y. Zhang, and P. Matthias. 2007. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 21:3381–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., M.A. Valencia-Sanchez, G.J. Hannon, and R. Parker. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7:719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., A. McKenzie 3rd, D.J. Demarini, N.G. Shah, A. Wach, A. Brachat, P. Philippsen, and J.R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Mazroui R., R. Sukarieh, M.E. Bordeleau, R.J. Kaufman, P. Northcote, J.Tanaka, I. Gallouzi, and J. Pelletier. 2006. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol. Biol. Cell. 17:4212–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelitsch, M.D., and J.S. Weissman. 2000. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc. Natl. Acad. Sci. USA. 97:11910–11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet, S., N. Cougot, A. Wilczynska, F. Dautry, M. Kress, E. Bertrand, and D. Weil. 2008. Translationally repressed mRNA transiently cycles through stress granules during stress. Mol. Biol. Cell. 19:4469–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonhoff, U., M. Ralser, F. Welzel, I. Piccini, D. Balzereit, M.L. Yaspo, H. Lehrach, and S. Krobitsch. 2007. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol. Biol. Cell. 18:1385–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, R., and U. Sheth. 2007. P-bodies and the control of mRNA translation and degradation. Mol. Cell. 25:635–646. [DOI] [PubMed] [Google Scholar]
- Pilkington, G.R., and R. Parker. 2008. Pat1 contains distinct functional domains that promote P-body assembly and activation of decapping. Mol. Cell. Biol. 28:1298–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai, R.S., S.N. Bhattacharyya, C.G. Artus, T. Zoller, N. Cougot, E. Basyuk, E. Bertrand, and W. Filipowicz. 2005. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 309:1573–1576. [DOI] [PubMed] [Google Scholar]
- Ramirez, M., R.C. Wek, C.R. Vazquez de Aldana, B.M. Jackson, B. Freeman, and A.G. Hinnebusch. 1992. Mutations activating the yeast eIF-2 alpha kinase GCN2: isolation of alleles altering the domain related to histidyl-tRNA synthetases. Mol. Cell. Biol. 12:5801–5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Echevarria, M.J., and S.W. Peltz. 2000. The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell. 101:741–751. [DOI] [PubMed] [Google Scholar]
- Sheth, U., and R. Parker. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 300:805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth, U., and R. Parker. 2006. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 125:1095–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöhr, N., M. Lederer, C. Reinke, S. Meyer, M. Hatzfeld, R. Singer, and S. Hüttelmaier. 2006. ZBP1 regulates mRNA stability during cellular stress. J. Cell Biol. 175:527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadauchi, T., T. Inada, K. Matsumoto, and K. Irie. 2004. Posttranscriptional regulation of HO expression by the Mkt1-Pbp1 complex. Mol. Cell. Biol. 24:3670–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira, D., and R. Parker. 2007. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 18:2274–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira, D., U. Sheth, M.A. Valencia-Sanchez, M. Brengues, and R. Parker. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 11:371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourriere, H., K. Chebli, L. Zekri, B. Courselaud, J.M. Blanchard, E. Bertrand, and J. Tazi. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160:823–831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Unterholzner, L., and E. Izaurralde. 2004. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell. 16:587–596. [DOI] [PubMed] [Google Scholar]
- Vasudevan, S., N. Garneau, D. Tu Khounh, and S.W. Peltz. 2005. p38 mitogen-activated protein kinase/Hog1p regulates translation of the AU-rich-element-bearing MFA2 transcript. Mol. Cell. Biol. 25:9753–9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynska, A., C. Aigueperse, M. Kress, F. Dautry, and D. Weil. 2005. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J. Cell Sci. 118:981–992. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.