Abstract
We compare three Arabidopsis (Arabidopsis thaliana) complex glycan1 (cgl1) alleles and report on genetic interaction with staurosporin and temperature sensitive3a (stt3a). STT3a encodes a subunit of oligosaccharyltransferase that affects efficiency of N-glycan transfer to nascent secretory proteins in the endoplasmic reticulum; cgl1 mutants lack N-acetyl-glucosaminyltransferase I activity and are unable to form complex N-glycans in the Golgi apparatus. By studying CGL1-green fluorescent protein fusions in transient assays, we show that the extra N-glycosylation site created by a point mutation in cgl1 C5 is used in planta and interferes with folding of full-length membrane-anchored polypeptides in the endoplasmic reticulum. Tunicamycin treatment or expression in the stt3a-2 mutant relieved the folding block, and migration to Golgi stacks resumed. Complementation tests with C5-green fluorescent protein and other N-glycosylation variants of CGL1 demonstrated that suppression of aberrant N-glycosylation restores activity. Interestingly, CGL1 seems to be functional also as nonglycosylated enzyme. Two other cgl1 alleles showed splicing defects of their transcripts. In cgl1 C6, a point mutation affects the 3′ splice site of intron 14, resulting in frame shifts; in cgl1-T, intron 11 fails to splice due to insertion of a T-DNA copy. Introgression of stt3a-2 did not restore complex glycan formation in cgl1 C6 or cgl1-T but suppressed the N-acetyl-glucosaminyltransferase I defect in cgl1 C5. Root growth assays revealed synergistic effects in double mutants cgl1 C6 stt3a-2 and cgl1-T stt3a-2 only. Besides demonstrating the conditional nature of cgl1 C5 in planta, our observations with loss-of-function alleles cgl1 C6 and cgl1-T in the stt3a-2 underglycosylation background prove that correct N-glycosylation is important for normal root growth and morphology in Arabidopsis.
N-Glycosylation of secreted proteins is a vital function in all eukaryotic cells. N-Glycans attached to glycoproteins support proper folding in the endoplasmic reticulum (ER) lumen (a prerequisite for vesicle-mediated protein export to the cis-Golgi), increase hydration at the protein surface, and confer enhanced stability against proteolytic degradation. During cotranslational import into the ER, most secretory proteins are glycosylated on Asn residues within the conserved motif [Asn-X(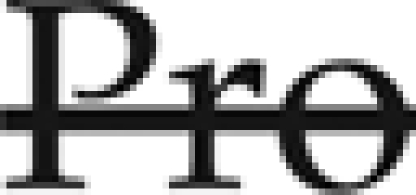 )-Ser/Thr] by the function of oligosaccharyltransferase (OST). OST catalyzes the en bloc transfer of Glc3Man9GlcNac2 core N-glycans from dolichol-pyrophosphate lipid anchors onto nascent polypeptides in the ER lumen. During folding, glycoproteins undergo a sequence of interactions with ER-resident carbohydrate-binding (i.e. lectin) chaperone complexes. Folding and oligomerization are monitored and reported by transient, cyclic deglucosylation/reglucosylation of the N-glycan moieties, which regulates binding to the ER chaperones calnexin and calreticulin (for a recent review, see Hebert et al., 2005). Inhibition of core N-glycosylation by the nucleoside antibioticum tunicamycin (for review, see Elbein, 1987) or mutations abrogating OST activity in the ER lumen activates the unfolded protein response (Koizumi et al., 1999; Martinez and Chrispeels, 2003; for review, see Patil and Walter, 2001; Lai et al., 2007) and ER-associated degradation (Bonifacino and Weissman, 1998; Plemper and Wolf, 1999; Parodi, 2000). While prolonged activation of the unfolded protein response results in lethality (Silberstein et al., 1995; Koizumi et al., 1999; Iwata and Koizumi, 2005; Hauptmann et al., 2006, and refs. therein), unfolded protein response and ER-associated degradation operate together as part of the quality-control surveillance in the secretory system of eukaryotes (Ellgaard et al., 1999; Hebert et al., 2005; Banerjee et al., 2007). Only when folding of single polypeptide chains and assembly of multisubunit protein complexes are completed are glycoproteins released and transported from the ER to the cis-Golgi via SNARE-mediated vesicle-budding and fusion events (Chatre et al., 2005; Moreau et al., 2007).
)-Ser/Thr] by the function of oligosaccharyltransferase (OST). OST catalyzes the en bloc transfer of Glc3Man9GlcNac2 core N-glycans from dolichol-pyrophosphate lipid anchors onto nascent polypeptides in the ER lumen. During folding, glycoproteins undergo a sequence of interactions with ER-resident carbohydrate-binding (i.e. lectin) chaperone complexes. Folding and oligomerization are monitored and reported by transient, cyclic deglucosylation/reglucosylation of the N-glycan moieties, which regulates binding to the ER chaperones calnexin and calreticulin (for a recent review, see Hebert et al., 2005). Inhibition of core N-glycosylation by the nucleoside antibioticum tunicamycin (for review, see Elbein, 1987) or mutations abrogating OST activity in the ER lumen activates the unfolded protein response (Koizumi et al., 1999; Martinez and Chrispeels, 2003; for review, see Patil and Walter, 2001; Lai et al., 2007) and ER-associated degradation (Bonifacino and Weissman, 1998; Plemper and Wolf, 1999; Parodi, 2000). While prolonged activation of the unfolded protein response results in lethality (Silberstein et al., 1995; Koizumi et al., 1999; Iwata and Koizumi, 2005; Hauptmann et al., 2006, and refs. therein), unfolded protein response and ER-associated degradation operate together as part of the quality-control surveillance in the secretory system of eukaryotes (Ellgaard et al., 1999; Hebert et al., 2005; Banerjee et al., 2007). Only when folding of single polypeptide chains and assembly of multisubunit protein complexes are completed are glycoproteins released and transported from the ER to the cis-Golgi via SNARE-mediated vesicle-budding and fusion events (Chatre et al., 2005; Moreau et al., 2007).
During passage through the various Golgi cisternae, surface-accessible N-glycans of plant glycoproteins (Faye et al., 1986) are further modified by eight enzymatic steps, involving mannosidases and glycosyltransferases, en route to their final destination: the plasma membrane, endosomes, or vacuoles (Fitchette-Laine et al., 1997; Lerouge et al., 1998). The second and committing step for complex N-glycan formation in the Golgi apparatus is catalyzed by N-acetyl-glucosaminyltransferase I (GnTI). After removal of terminal Mans, GnTI transfers GlcNAc to Man5GlcNac2 attached to polypeptides. This modification is a prerequisite for complex glycan formation (von Schaewen et al., 1993), characterized by the attachment of core α1,3-Fuc and β1,2-Xyl residues. GnTI shows class II membrane-protein topology, consisting of the N-terminal cytosolic domain followed by a single-span transmembrane domain (or membrane anchor), hinge region, and a large C-terminal luminal domain for enzymatic activity. In accordance with their proposed location, GnTI-GFP fusion proteins were shown to label Golgi stacks in plant cells (Essl et al., 1999; Dixit and Cyr, 2002; Grebe et al., 2003). Interestingly, Arabidopsis (Arabidopsis thaliana) ethyl methanesulfonate (EMS) mutants complex glycan1 (cgl1) C5 and C6, which are defective in GnTI activity and lack complex modified N-glycans, display no obvious aberrant phenotype under standard growth conditions (von Schaewen et al., 1993), albeit occasional enhanced susceptibility to stray pathogen infection (A. von Schaewen, unpublished data) and a slightly extended flowering period (Boyes et al., 2001) have been reported.
For cgl1 C5, Strasser et al. (2005) reported previously that a second N-glycosylation site was created by a point mutation in the Arabidopsis GnTI sequence (Wenderoth and von Schaewen, 2000). In transfected insect cells, they observed that, when synthesized with a cleavable signal peptide, soluble wild-type but not C5 protein was secreted into the culture medium. Since cell-retained C5 polypeptides carried two high-Man N-glycans, the authors concluded that neoglycosylation might interfere with protein folding in the ER (Strasser et al., 2005). However, the effects of the D→144N change on the maturation and function of CGL1 were not tested directly. Our report aims (1) to elucidate the molecular nature of cgl1 alleles, (2) to determine whether CGL1 C5 polypeptides accumulate in the ER of plant cells, (3) to monitor effects of the D→144N change on complex glycosylation in combination with epistatic mutant stt3a-2, which affects number but not quality of the N-glycan decoration, and (4) to check whether combination of two basic N-glycosylation defects might provoke phenotypic deviations in Arabidopsis.
RESULTS AND DISCUSSION
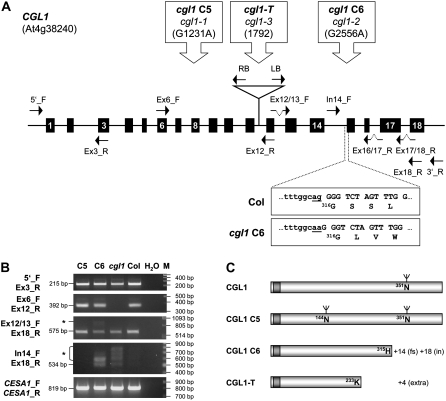
Novel cgl1 C6 and cgl1-T Mutants Produce CGL1 Transcripts with Splicing Defects
We addressed the molecular basis for missing complex glycan formation in three available Arabidopsis cgl1 alleles. Mutants cgl1 C5 and C6 were originally identified in a screen of EMS-mutagenized M2 seedlings (von Schaewen et al., 1993) and shown to contain mRNA levels comparable to those of the wild type (Wenderoth and von Schaewen, 2000). Sequence analyses of PCR-amplified genomic DNA fragments proved that both result from single G-to-A base changes. For cgl1 C5, our data confirmed the change at position 1,231 reported by Strasser et al. (2005). Besides altering a conserved sequence motif, the mutation creates a new N-glycosylation site (Strasser et al., 2005) in the context of Arabidopsis GnTI (Wenderoth and von Schaewen, 2000).
For cgl1 C6, we established that the G-to-A change at position 2,556 alters the 3′ splice site of intron 14 (Fig. 1A). Insertion mutant cgl1-T was identified in the SIGnAL-SALK database. Genomic PCR analyses confirmed that a single T-DNA resides within intron 11 (Supplemental Fig. S1A). Sequence analyses of both border fragments located the T-DNA insertion at position 1,792 without any base deletion.
Figure 1.
Molecular defects in Arabidopsis cgl1 alleles. A, Schematic depiction of the Arabidopsis CGL1 locus (At4g38240). Exons are shown as black boxes. Orientation and position of PCR primers are indicated by arrows, and the T-DNA insertion is indicated by the triangle. Details are given above the sequence (boxed). Below are magnifications of the borders between intron 14 and exon 15 in Col and cgl1 C6. Col, Columbia wild type; cgl1 C5 and C6, EMS mutation lines; cgl1-T, T-DNA insertion line. B, RT-PCR analyses confirm the presence of extra sequences in C6 (cgl1 C6) and cgl1 (cgl1-T) compared with wild-type (Col) and C5 (cgl1 C5) mRNA. CGL1 primers binding upstream (5′_F + Ex3_R), flanking (Ex6_F + Ex12_R), or downstream (Ex12/13_F + Ex18_R) of the T-DNA insertion show that unit length mRNA is missing from cgl1 (cgl1-T). PCRs with intron 14 sense and exon 18 antisense (In14_F + Ex18_R) primers indicate that a subset of C6 (cgl1 C6) and cgl1 (cgl1-T) mRNA molecules contain nonspliced downstream introns (asterisks). Control PCR with CELLULOSE SYNTHASE A1 (CESA1) primers verified similar quality of all reverse-transcribed mRNA samples. C, Primary structure of CGL1 wild type compared with mutant CGL1 C5, CGL1 C6, and CGL1-T polypeptides. The N-terminal membrane anchor is shaded darker, and the positions of N-glycosylation sites are indicated. Full-length CGL1 and CGL1 C5 polypeptides terminate with amino acid 443. Truncated CGL1 versions of cgl1 C6 and cgl1-T mutants are shown to scale. fs, Frame shift; in, intron.
In order to confirm the predicted molecular defects in cgl1 C6 and cgl1-T at the transcript level, we characterized the structures of mutant CGL1 transcripts by reverse transcription (RT)-PCR and sequence analyses. For cgl1 C6, sequencing the products amplified with Ex12/13_F and Ex18_R (Fig. 1B) revealed that intron 14 is aberrantly spliced by shifting the 3′ splice site to the next G, introducing a translational frame shift (Fig. 1A). Larger PCR products found in cgl1 C6 were aberrant transcripts that retain intron 15, 16, and 17 (Fig. 1B; Supplemental Fig. S1C). For cgl1-T, transcripts were characterized using primers binding upstream or flanking the T-DNA insertion. RT-PCR with upstream primer pairs 5′_F and Ex3_R detected wild-type transcript levels; however, PCR using primer pairs Ex6_F and Ex12_R flanking the T-DNA insertion yielded no product for cgl1-T (Fig. 1B). This demonstrated that cgl1-T does not produce intact CGL1 transcripts. Since we could detect chimeric CGL1:T-DNA transcripts (Supplemental Fig. S1B), it appears that mature CGL1-T transcripts contain unspliced intron 11 with an additional 4.5 kb of T-DNA sequence. In both cgl1 C6 and cgl1-T, the mutations create premature stop codons a few codons downstream of the mutated sites (Fig. 1C).
The stt3a-2 Mutation Rescues cgl1 C5 by Suppressing Neoglycosylation
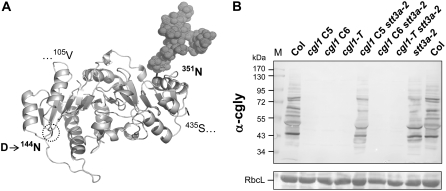
Due to the D→144N change, CGL1 C5 polypeptides most likely acquire a second N-glycan (Strasser et al., 2005) upon cotranslational insertion into the ER membrane. Based on a three-dimensional model of the catalytic domain, the D→144N change lies in a buried region of the protein, whereas native glycan 351N is exposed at the surface (Fig. 2A). Strasser et al. (2005) compared the secretion of soluble versions of rabbit and Arabidopsis GnTI proteins in insect cells. They deduced that neoglycosylated C5 polypeptides are inactive due to aberrant folding and do not leave the ER.
Figure 2.
Effects of mutated CGL1 proteins on complex glycan formation in Arabidopsis single and double N-glycosylation mutants. A, Three-dimensional model of the catalytic domain of Arabidopsis GnTI (CGL1 105V to 435S) with glycan 351N exposed at the protein surface. The D144N change in cgl1 C5 creates a new N-glycosylation site in a region buried inside the native CGL1 protein (darkened in the circled region) that interfered with the secretion of soluble C5 polypeptides from insect cells (Strasser et al., 2005). B, Immunoblot analysis with complex glycan antiserum (α-cgly; colorimetric detection). CGL1 C5 protein regains activity in the stt3a-2 mutant background. Note that the complex glycosylation pattern of double mutant cgl1 C5 stt3a-2 matches exactly to that of the stt3a-2 parent. The loading reference RbcL (for Rubisco large subunit) of the Ponceau S-stained blot is shown at bottom. M, Molecular mass standards (in kD).
In order to determine whether reduced frequency of N-glycosylation can specifically restore GnTI function of CGL1 C5 in planta, the viable stt3a-2 allele was introgressed into all three Arabidopsis cgl1 lines. STT3a encodes an isoform of OST subunit STT3, and the stt3a-2 mutation was previously shown to cause underglycosylation of glycoproteins (Koiwa et al., 2003). Both double mutants cgl1 C6 stt3a-2 and cgl1-T stt3a-2 failed to react with complex glycan antiserum on immunoblots (“nonstainers”; Fig. 2B). This, however, was not the case for the cgl1 C5 stt3a-2 double mutant lines. Compared with the wild type and the cgl1 single mutants, cgl1 C5 stt3a-2 displayed intermediate complex glycosylation patterns on immunoblots (Supplemental Fig. S2). It is noteworthy that the complex glycosylation pattern and intensity in cgl1 C5 stt3a-2 match exactly those of the stt3a parent (Fig. 2B). Thus, despite the D→144N change in a highly conserved GnTI motif (Wenderoth and von Schaewen, 2000; Strasser et al., 2005), CGL1 C5 polypeptides appear to retain sufficient activity to restore complex glycosylation in the stt3a mutant.
CGL1 C5 Is Aberrantly Glycosylated in Planta
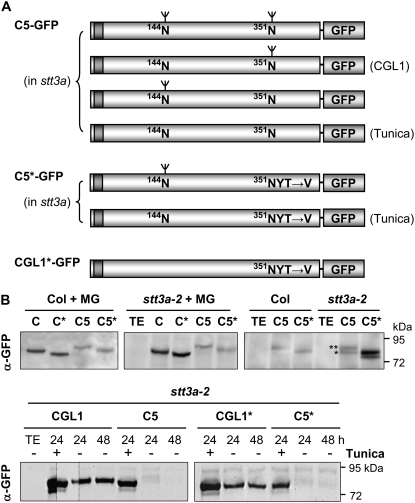
In order to test the N-glycosylation status of CGL1 variants, cDNA fragments encoding wild-type and mutated CGL1 variants were fused to GFP (Fig. 3A) and expressed in Arabidopsis wild-type and stt3a-2 mutant protoplasts. The variants of CGL1 tested include C5, CGL1* (lacking native glycosylation site 351N), and C5* (lacking native glycosylation site 351N but containing aberrant glycosylation site 144N). Experiments were conducted in both the presence and absence of proteasome inhibitor MG-132 to prevent potential degradation of aberrant fusion proteins (Lee and Goldberg, 1998), which gave similar results (representative data shown in Fig. 3B). Consistent with the expected N-glycosylation states, CGL1-GFP variants displayed different mobilities in anti-GFP immunoblot analyses of transformed protoplasts. In both wild-type and stt3a mutant protoplasts, CGL1 variants showed similar electrophoretic mobility (Fig. 3B, top left and center). Furthermore, protein levels of C5-GFP and C5*-GFP were lower than those of wild-type CGL1-GFP and unglycosylated CGL1*-GFP in both genotypes. In stt3a-2 protoplasts, we occasionally observed additional bands for C5-GFP and C5*-GFP (Fig. 3B, top right), whose mobility is consistent with underglycosylated polypeptide forms. Interestingly, treatment with the N-glycosylation inhibitor tunicamycin promoted the accumulation of C5-GFP and C5*-GFP in both wild-type and stt3a genotypes (Fig. 3B, bottom). This indicated that aberrant N-glycosylation interferes with the stability of CGL1 C5 polypeptides in plant cells. Nevertheless, since complex glycosylation is restored in the cgl1 C5 stt3a-2 double mutant, it appears that small amounts of underglycosylated CGL1 C5 polypeptides regain catalytic activity.
Figure 3.
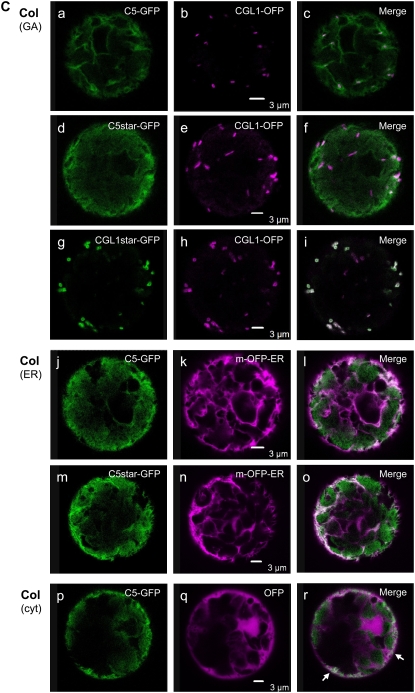
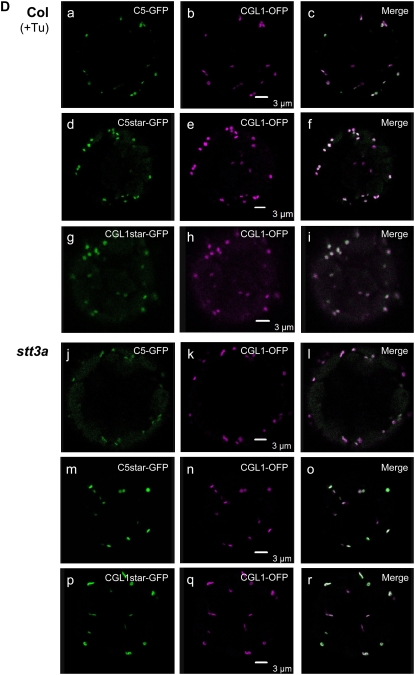
Analysis of CGL1-GFP N-glycosylation variants in Arabidopsis protoplasts. A, Cartoon of recombinant GFP fusions designed for subcellular localization and complementation tests. N-Glycosylation variants theoretically produced in stt3a-2 mutant cells are encompassed by braces. C5-, C5*-, and CGL1*-GFP variants were created by site-directed mutagenesis of the CGL1 cDNA sequence. Protein versions corresponding to CGL1-GFP in the wild type or with tunicamycin treatment are indicated at right. B, Immunoblot analyses of plasmid-transfected Arabidopsis protoplasts (routinely harvested between 24 and 48 h) using GFP antiserum. Top, Aberrantly glycosylated C5- and C5*-GFP polypeptides are at the detection limit in both Col wild-type and stt3a-2 mutant protoplasts, independent of proteasome inhibitor MG-132 (+MG; 20 μm f.c.). Col, Columbia wild type; C, CGL1-GFP; C*, CGL1*-GFP; N-glycan number is marked by asterisks. Bottom, Tunicamycin treatment (+Tunica) greatly improved immunodetection of nonglycosylated C5- and C5*-GFP polypeptides (here shown for stt3a-2 protoplasts). Note that natively glycosylated CGL1-GFP also dominates in stt3a-2 (compare + and − Tunica lanes; the area within the dotted lines shows a shorter blot exposure). TE, mock-transfected buffer controls. Molecular mass standards (in kD) are indicated. C, Confocal laser-scanning microscopy analyses of Arabidopsis wild-type protoplasts coexpressing CGL1-GFP variants in combination with the Golgi control CGL1-OFP [Col (GA)]. For better visibility, C5star and CGLstar replace C5* and CGL1* labeling in confocal laser-scanning microscopy images. In Col wild-type protoplast, C5- and C5star-GFP are visible in the ER [Col (ER)] and in parts of the cytosol [Col (cyt); the cytosolic control of C5star-GFP is not shown]. Arrows point to overlapping signals (which appear white in merged images). D, C5- and C5star-GFP label Golgi stacks in Col wild-type protoplasts treated with tunicamycin [Col (+Tu); a–i] and without the drug in stt3a-2 mutant protoplasts (j–r). Green, GFP (test constructs); magenta, OFP (control constructs); white, colocalization. Images were recorded 24 h after transfection. Bars = 3 μm.
Aberrant Glycosylation of CGL1 C5 Polypeptides Prevents Their Migration to Golgi Stacks
Since aberrant glycosylation of CGL1 C5 polypeptides may also affect Golgi targeting, we determined the subcellular location of fluorescent protein fusions using confocal laser-scanning microscopy. In transfected Arabidopsis protoplasts, CGL1-GFP and CGL1-OFP (for orange-shifted monomeric red fluorescent protein [mRFP]) both labeled Golgi stacks, as indicated by shape, size, and characteristic stop-and-go movements (Nebenführ et al., 1999; Supplemental Videos S1 and S2). Furthermore, fluorescent signals were redistributed into ER/Golgi-hybrid compartments upon incubation with brefeldin A (data not shown). When mutated C5-GFP and C5*-GFP constructs were analyzed in ecotype Columbia (Col) wild-type protoplasts, green fluorescence was not detected in Golgi stacks labeled by CGL1-OFP but faint GFP signals could be seen in the ER and in certain areas of the cytosol (Fig. 3C, arrows). This indicates that C5-GFP and C5*-GFP are not delivered to the Golgi apparatus but are likely degraded in the ER.
In the presence of tunicamycin, Col wild-type protoplasts transformed with either C5-GFP or C5*-GFP constructs developed a typical Golgi pattern within 24 h after transfection that colocalized with CGL1-OFP used as an internal control (Fig. 3D, a–c and d–f). This showed that blocking N-glycosylation can restore protein folding of CGL1 C5, independent of the altered amino acid motif, so that migration to Golgi stacks resumes (Supplemental Video S1A). Expression of C5-GFP or C5*-GFP constructs in protoplasts of the stt3a-2 mutant resulted in Golgi labeling within 24 h (Fig. 3D, j–l and m–o), similar to tunicamycin-treated wild-type protoplasts. In the stt3a-2 mutant background, however, signal intensities of Golgi stacks were much lower than in tunicamycin-treated wild-type protoplasts, perhaps because only a small proportion of C5-GFP and C5*-GFP polypeptides are underglycosylated and allowed to exit the ER (Fig. 3D; Supplemental Fig. S3).
N-Glycan Profiling by Peptide:N-Glycosidase F Supports the Notion That CGL1-GFP Arrives in Golgi Stacks and CGL1 C5-GFP Polypeptides Are Retained in the ER
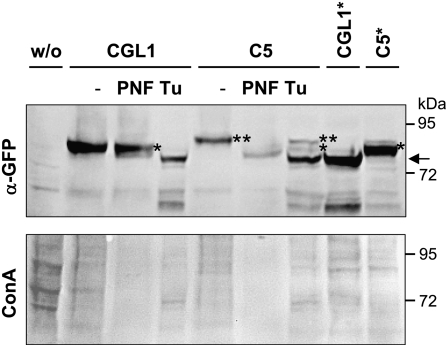
In order to biochemically confirm the Golgi localization of CGL1 variants, structures of N-glycans attached to CGL1 polypeptides were analyzed using peptide:N-glycosidase F (PNGase F). PNGase F cleaves N-glycans that do not contain core Fuc modification. Since addition of core Fucs occurs in the Golgi apparatus, PNGase F-resistant N-glycans can be regarded as a hallmark for successful delivery to Golgi stacks. In order to obtain CGL1 variants in sufficient quantity, all constructs were transiently expressed in Nicotiana benthamiana leaves. Subcellular localization and immunoblot profiles of CGL1-GFP variants expressed in agroinfiltrated N. benthamiana leaf tissue were consistent with those expressed in Arabidopsis protoplasts (Supplemental Fig. S4; Supplemental Video S2). We found that C5-GFP displayed a downward mobility shift after PNGase F treatment. The lower Mr form produced after PNGase F digestion migrated similar to nonglycosylated CGL1*-GFP or C5-GFP produced in the presence of tunicamycin. In contrast, CGL1-GFP hardly shifted at all (Fig. 4). These findings establish that CGL1-GFP arrives in Golgi stacks, where core fucosylation of its N-glycan occurs, but C5-GFP decorated with two high-Man N-glycans remains in the ER. Taken together, our in planta approaches unequivocally show that CGL1 C5 is likely retained in the ER because of aberrant N-glycosylation. Prevention of aberrant N-glycosylation of C5-GFP and C5*-GFP polypeptides alleviated misfolding in the ER and enabled membrane-bound delivery to Golgi stacks. Next, we set out to determine whether this can explain the restoration of GnTI function in the cgl1 C5 stt3a-2 double mutant.
Figure 4.
Comparison of untreated (−), PNGase F-treated (PNF), and tunicamycin-infiltrated (Tu) samples by GFP immunoblot analyses (top). Protein extracts were prepared from agroinfiltrated N. benthamiana leaves and treated without or with PNGase F. This glycosidase can only release ER-type N-glycans lacking core Fucs. Success of the treatment is indicated by reduced concanavalin A binding to the stripped GFP blot (shown below). CGL1 (−GFP) is largely PNGase F resistant (core Fucs attached), whereas C5 (−GFP) is PNGase F sensitive (no core Fucs) and migrates similar to the tunicamycin-infiltrated control (arrow). Asterisks indicate number of N-glycans. ConA, Concanavalin A (jack bean lectin, binds preferentially to Man-terminated glycans); w/o, without binary construct (Agrobacterium strain 19K only). Molecular mass standards (in kD) are indicated.
CGL1 C5-GFP Restores Complex Glycan Maturation in cgl1-T stt3a-2 But Not in cgl1-T
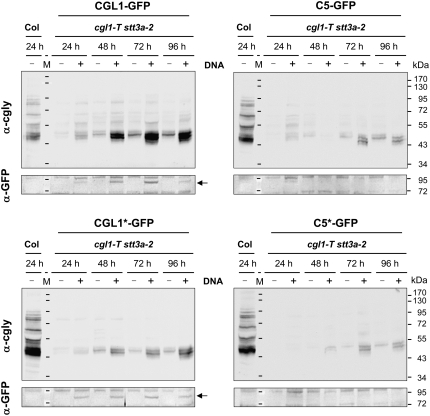
The ability of the N-glycosylation variants of CGL1 to restore complex N-glycan production in planta was analyzed by transient expression of CGL1-GFP constructs in cgl1-T mutant protoplasts. In addition, the impact of CGL1 underglycosylation was assessed by expressing the same variants in cgl1-T stt3a-2 protoplasts. In principle, all CGL1-GFP variants expressed in cgl1-T stt3a-2 restored complex glycan formation within 96 h on a fraction of proteins (traveling along the secretory route) compared with mock-transfected controls (minus DNA; Fig. 5). The extent of recovery varied among constructs and correlated with expression level of the GFP fusions. Best recovery was observed with CGL1-GFP (wild-type situation) followed by nonglycosylated CGL1*-GFP. These two protein fusions were clearly detectable with GFP antiserum (Fig. 5, arrows in bottom left sections). C5-GFP and C5*-GFP showed delayed and less efficient recovery of complex glycosylation in the cgl1-T stt3a-2 background, consistent with low abundance of the GFP fusions. In cgl1-T mutant protoplasts, only CGL1-GFP and CGL1*-GFP displayed Golgi labeling and produced complex modified glycoproteins (data not shown).
Figure 5.
Complementation test of CGL1-GFP variants in protoplasts of the Arabidopsis cgl1-T stt3a-2 double mutant. Protoplasts were transfected with the fusion constructs indicated and harvested at 24 to 96 h after transfection. Cells were extracted in buffer with 0.5% SDS and subjected to immunoblot analyses, first with GFP antiserum (bottom sections), followed by complex glycan antiserum after stripping the GFP blot (top sections). +, DNA-transfected samples; −, mock-transfected buffer controls. Arrows point to visible GFP signals. Col, Columbia wild-type protoplasts; M, molecular mass standards (in kD).
Since all CGL1-GFP variants were localized to the Golgi apparatus in cgl1-T stt3a-2 protoplasts (data not shown), we conclude that complementation depends upon delivery of correctly folded CGL1 variants to Golgi stacks. As already observed for Col wild-type and stt3a-2 mutant protoplasts (Fig. 3B), C5-GFP and C5*-GFP signals were hardly detectable in cgl1-T stt3a-2 protoplasts (Fig. 5, bottom right sections). This might be due to a generally reduced stability of aberrantly glycosylated polypeptides in protoplasts. Nevertheless, this demonstrates that minimal amounts of CGL1 C5 protein are functional and can account for the recovery of complex glycosylation in cgl1 C5 stt3a-2 double mutant lines (Fig. 2B; Supplemental Fig. S2). Probably, this can also explain why transgenic tobacco (Nicotiana tabacum) plants with less than 3% GnTI activity produce normal levels of complex glycans (Strasser et al., 2004) and indicates that “low-stainer” plants obtained by an antisense approach in tobacco and potato (Solanum tuberosum; Wenderoth and von Schaewen, 2000) are essentially devoid of GnTI.
Deviant Root Growth of Arabidopsis Mutants with Combined N-Glycosylation Defects
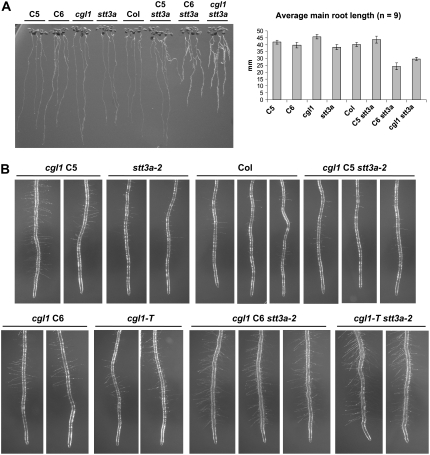
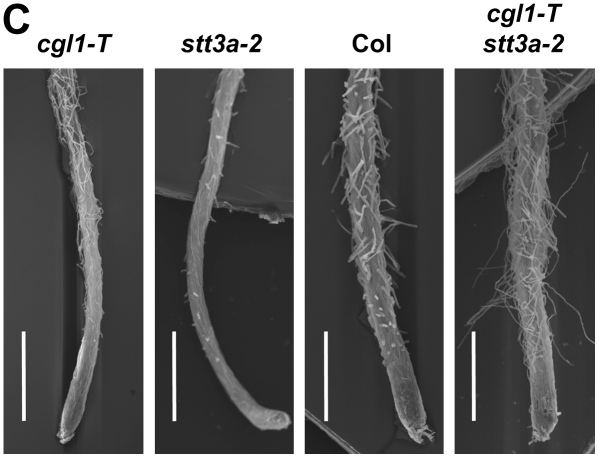
None of the tested single or double mutants with N-glycosylation defects displayed obvious growth phenotypes in soil (data not shown). However, differences in root growth were detected on vertical agar plates (Fig. 6). The most striking morphological deviations (i.e. shorter, more branched, and hairy roots) were displayed by cgl1 C6 stt3a-2 and cgl1-T stt3a-2 double mutants (Fig. 6A). Root growth of cgl1 C5 stt3a-2 seedlings resembled the wild-type and single mutant parents, which can be explained by suppression of the cgl1 C5 defect in the stt3a-2 mutant background (described above). Close-up images of root tips show that all cgl1 alleles produce longer root hairs compared with Col wild type or stt3a-2 (Fig. 6B). In cgl1 C6 stt3a-2 and cgl1-T stt3a-2 double mutants, stunted growth of the root hair zone probably accounts for the bushy appearance (Fig. 6A). Scanning electron microscopy images of root tips show that root hairs of the cgl1-T stt3a-2 double mutant are much thinner and longer than those of the Col wild type (Fig. 6C). The latter seems to be a feature of true loss-of-function cgl1 alleles, since roots of stt3a-2 single and cgl1 C5 stt3a-2 double mutants grown in parallel have shorter hairs (Fig. 6B). Together, these findings point to problems with expansion and probably also division of cells in the root elongation zone as a consequence of combining two basic N-glycosylation defects (i.e. less frequent core glycosylation in the ER combined with aberrant N-glycan modification in the Golgi). An obvious glycoprotein candidate known to affect root tip morphology when mutated in Arabidopsis is KORRIGAN1 (KOR1/RSW2), a class II membrane protein with eight predicted N-glycosylation sites (Nicol et al., 1998). The β1,4-d-glucanase activity of KOR1 seems to play crucial roles for cellulose biosynthesis at the plasma membrane (Peng et al., 2002) and during cytokinesis (Zuo et al., 2000; Lane et al., 2001). In accordance, we observed altered SDS gel migration of KOR1 protein in cgl1-T, stt3a-2, and mutants with other N-glycosylation defects (Kang et al., 2008). In the past, root hair phenotypes have been associated with defects in various secreted enzymes. Among them is KOJAK, a cellulose synthase-like protein found in Arabidopsis and rice (Oryza sativa; Favery et al., 2001; Kim et al., 2007), glycoproteins affecting the synthesis of extracellular matrix components in Arabidopsis (e.g. LRX1; Baumberger et al., 2001; Diet et al., 2006), and COBRA (Roudier et al., 2005; Ko et al., 2006). In this context, it is noteworthy that the latter seems to be indispensable for oriented root cell expansion.
Figure 6.
Root phenotypes of Arabidopsis single and double N-glycosylation mutants. A, Arabidopsis seedlings after 7 d of growth at 21°C on vertical hard agar plates (1.5% agar, 3% Suc, and MS medium, pH 5.7) with light source from the top, and bar diagram of primary root lengths (se indicated by error bars; n = 9). Col, Columbia wild type; C5, cgl1 C5; C6, cgl1 C6; cgl1, cgl1-T; stt3a, stt3a-2. B, Close-up images of the main roots show that all cgl1 single mutants produce long root hairs. Bushy root tips of the double mutants cgl1 C6 stt3a-2 and cgl1-T stt3a-2 are indicative of a stunted root hair zone when stt3a-2 is combined with true loss-of-function cgl1 alleles. Note that root tips of the cgl1 C5 stt3a-2 double mutant resemble those of the Col wild type and the stt3a-2 parent (compare also with the complex glycan patterns in Fig. 2B). C, Scanning electron micrographs of root tips of 3-d-old seedlings. Bars = 0.5 mm.
CONCLUSION
Recovery of complex glycan patterns in the cgl1 C5 stt3a-2 double mutant initially indicated that underglycosylated CGL1 C5 enzyme can rescue the cgl1 defect in Arabidopsis. By studying CGL1-GFP glycosylation variants in Arabidopsis wild type and selected N-glycosylation mutants, we demonstrate that aberrant neoglycosylation also obstructs ER folding of membrane-anchored CGL1 C5 polypeptides in planta. Recovery of complex glycan patterns in the stt3a-2 mutant background showed that the D→144N replacement in CGL1 C5 probably has a less severe effect compared with an equivalent change in rabbit GnTI, which did not create a glycosylation site but substantially affected catalytic activity (Strasser et al., 2005). Analyses of altered N-glycan decoration of CGL1-GFP variants in Arabidopsis protoplasts proved that either no or native glycosylation is compatible with folding of CGL1 polypeptides in the ER and delivery to Golgi stacks. The fact that all CGL1-GFP variants principally complemented the cgl1 defect over time (in protoplasts of the cgl1-T stt3a-2 double mutant) supports the notion that low amounts of functional GnTI suffice to guarantee complex glycosylation in plants. Since no deviant phenotype was detected in aerial parts of true cgl1 stt3a double mutants, we reasoned that combination of two basic N-glycosylation defects might influence only a subset of all glycoproteins synthesized during certain stages of Arabidopsis development. Indeed, stunted root growth and altered root hair morphology indicate that among the glycoproteins synthesized during root cell elongation are prime targets affected.
Outlook
STT3 is a proposed catalytic subunit of OST and encoded by paralogous genes in Arabidopsis. Although our recent study indicates that STT3a plays a predominant role in N-glycosylation of KOR1 (Kang et al., 2008), it remains to be investigated whether the same applies to CGL1 C5, using stt3b mutants. Since MG-132 did not improve the detection of aberrantly folded CGL1 polypeptides, it will be informative to study possible mechanisms by which these misfolded membrane proteins are cleared from the ER of plant cells. As interesting question remains, whether changes analogous to CGL1 C5 introduced into potato and tobacco GnTI (146D→N), which both create extra N-glycosylation sites in addition to single native sites at position 203N (compared with 351N in Arabidopsis; Wenderoth and von Schaewen, 2000), are used in planta and also lead to ER retention of the mutated proteins in wild-type cells.
According to the analyses presented here, cgl1 C5 defines a conditional Arabidopsis allele and only cgl1 C6 and cgl1-T can be considered true loss-of-function mutants. It is conceivable that the leakiness of the cgl1 C5 mutation becomes relevant under stressful conditions (Kang et al., 2008) and at certain developmental stages (e.g. rapid root growth), when massive production of secreted proteins ensues. Clearly, more detailed root growth analyses are required to identify potential downstream targets of altered N-glycosylation in Arabidopsis. Since knockout mutants are indispensable for unbiased further studies involving cgl1 alleles in existing (Kang et al., 2008) and new combinations, seeds of all mutants will be deposited with the Arabidopsis stock centers using synonymous new allele numbering: cgl1-1 (for cgl1 C5), cgl1-2 (for cgl1 C6), and cgl1-3 (for cgl1-T; Fig. 1A).
Finally, our data demonstrate that small amounts of functional GnTI protein are sufficient to produce nearly wild-type levels of complex glycans in plants. Considering that conventional gene silencing often does not yield complete loss of expression, more robust strategies (e.g. double-stranded RNA interference) are needed to study the role of reduced complex glycosylation in crop plants.
MATERIALS AND METHODS
Plant Growth
Arabidopsis (Arabidopsis thaliana Col) plants were grown in soil under a long-day regime (16 h of light, 21°C). Backcrossed lines (more than seven rounds) of cgl1 C5#5 and cgl1 C6#22 EMS mutants (von Schaewen et al., 1993; Wenderoth and von Schaewen, 2000) were used for all analyses. Information on T-DNA insertion lines was retrieved from the SIGnAL database at http://signal.salk.edu (Alonso et al., 2003). Intron line cgl1-T (SALK_073650) was provided by the Nottingham Arabidopsis Stock Centre, and stt3a-2 (SALK_058814) was provided by the Arabidopsis Biological Resource Center (Koiwa et al., 2003). T3 individuals were analyzed for the presence of the T-DNA insertion by genomic PCR (Supplemental Fig. S1; data not shown). To generate double mutants, anthers of a previously established stt3a-2 BiP-GUS line (H. Koiwa, unpublished data) were used for pollination of emasculated cgl1 mutant flowers. F1 individuals that showed GUS staining in roots were selected (successful mates), checked for binding of complex glycan antiserum on dot-blotted leaf extracts (“stainers”; von Schaewen et al., 1993), and allowed to self. Among the segregating F2 progeny, double mutant candidates were checked for homozygosity of the T-DNA insertion at the stt3a-2 locus (see below).
For protoplast isolation, Arabidopsis seeds were surface sterilized with 12% hypochloride, placed on agar medium (0.8% agar, 1% Suc, and 0.5 Murashige and Skoog [MS] salts plus vitamins, pH 5.8), and kept at 4°C for 3 d. Plates were transferred to a growth room with artificial illumination (Phillips TLD, 36W/827 and 36W/840 twin bulb sets). One-week-old seedlings were transferred to Magenta boxes (Sigma) and grown in sterile culture for 4 to 5 weeks under a long-day regime (16 h of light/21°C, 8 h of dark/18°C) prior to harvesting sterile leaf material.
For root growth analyses, Arabidopsis seeds were surface sterilized, placed on hard agar plates (1.5% agar, 3% Suc, and MS salts plus vitamins, pH 5.8), and stratified for 3 d (Koiwa et al., 2003). Seedlings of similar growth stages were transferred to a new plate and kept in a vertical position in a growth chamber (Percival E36L, equipped with Phillips F17T8/ TL741 17W neon light bulbs) under a short-day regime (8 h of light/20°C, 16 h of dark/18°C) for 5 to 7 d.
Nicotiana benthamiana plants were grown in soil in the greenhouse with permanent fertilization (2‰ NPK 14:6:24) and additional illumination (12 h of light/22°C; Phillips 400W/645 Master HPI-T Plus light bulbs, 250 μE at plant height). Three-week-old plants were used for Agro-leaf infiltrations.
PCR Analyses of Genomic DNA Fragments
Genomic DNA was isolated from small amounts of leaf tissue (Dilworth and Frey, 2000). PCR fragments were amplified with Phusion High-Fidelity DNA Polymerase (Finnzymes) and either subcloned into EcoRV-digested pBSK (Stratagene) or sequenced directly to identify the point mutation in C5 (1,235G→A) and in C6 (2,556G→A). To determine the exact position of the T-DNA insertion in cgl1-T, T-DNA border fragments (Phusion amplified with primer pair 5′_F/RBa1 or LBa1/3′_R) were subcloned into pJET (Fermentas) and sequenced using primers LBb1 and RBa1. Homozygosity at the C5 locus was determined either by 3-(cyclohexylamino)propanesulfonic acid (CAPS) analyses (Neff et al., 1998) using C5-CAPS primers and FokI digests or by direct sequencing of genomic PCR products using CGL1_336F or CGL1_503R (see Supplemental Table S1 for primers used in this study).
RT-PCR Analyses
Total RNA from leaf tissue was isolated with the guanidinium HCl procedure (Logemann et al., 1987). Five-microgram aliquots were treated with RNase-free DNase I (Fermentas), primed with 30 ng of oligo(dT) (30-mers), and used for reverse transcription of first-strand cDNA with RevertAid H Minus M-MuLV reverse transcriptase (Fermentas) in 20-μL reactions. PCR amplification was conducted with various primer combinations (10 μm each) using 0.1-μL reverse transcriptase aliquots, 1 mm dNTP mix, and BioTherm Taq-DNA Polymerase (Genecraft) in the buffer provided.
Bioinformatics
To obtain a three-dimensional structure model, the amino acid sequence of Arabidopsis CGL1 was analyzed with Swiss-model, an automated comparative protein-modeling server at http://swissmodel.expasy.org/workspace. A glycosylated protein model of CGL1 was created with GlyProt for in silico glycosylation of proteins (Glycosciences.de of the Deutsches Krebsforschungszentrum Heidelberg) at http://www.glycosciences.de/modeling/glyprot/php/main.php.
Cloning of Reporter Constructs and Site-Directed Mutagenesis
For OFP fusion constructs, we started from pOFP-ΔNco (T. Meyer, personal communication), a derivative of plant expression vector pSY526 with cauliflower mosaic virus 35S-mRFP cassette. The second NcoI site in the mRFP coding region was eliminated by silent mutagenesis using the Quick Change PCR mutagenesis protocol (Stratagene) and Phusion High-Fidelity DNA Polymerase (Finnzymes). The OFP-ER control was obtained by multistep cloning. First, a 950-bp HindIII-EcoRI fragment, comprising the 35S promoter and signal sequence of pBIN m-gfp5-ER (Haseloff et al., 1997), was subcloned in pBSK (Stratagene). From pOFP-ΔNco, a 680-bp fragment was amplified with primer pair OFP-ER_F/R and inserted 3′ of the signal sequence into the pBSK subclone via EcoRI-SacI. The entire open reading frame (signal sequence-OFP-HDEL) was excised and inserted into plant expression vector pGFP2 (Kost et al., 1998) via XbaI-SacI (replacing GFP) and termed pOFP2-ER. Short and full-length CGL1-OFP constructs were assembled in pOFP-ΔNco (via XhoI-NcoI) after reverse transcription of leaf mRNA (Col wild type) and PCR amplification of cDNA fragments with primer combination CGL1-OFP_F and CGL-s-OFP_R or CGL-OFP_R, respectively. GFP fusion constructs were obtained by transfer of short and full-length CGL1-cDNA fragments (via XhoI-NcoI) into pGFP2 (Kost et al., 1998) after mutagenesis of the NcoI site in the cauliflower mosaic virus 35S promoter. To generate C5-GFP, the G-to-A point mutation (CGL1 position 1,235) was introduced into CGL1 by site-directed mutagenesis with SDM primer pair C5mut_F/R. Alteration of the native N-glycosylation context in CGL1 was done after consulting an amino acid alignment of plant GnTI sequences (Wenderoth and von Schaewen, 2000). CGL1*-GFP and C5*-GFP constructs were created with SDM primer pairs C5*mut_F/R and CGL1*mut_F/R, respectively. All changes were confirmed by sequence analyses. Constructs were first tested by transient expression in protoplasts. Binary constructs were obtained by inserting expression cassettes into pGPT.VII (Walter et al., 2004) via XhoI-SacI and introduced into Agrobacterium tumefaciens strain GV2260 (Deblaere et al., 1985) by direct transformation (Höfgen and Willmitzer, 1988).
Transient Expression in Arabidopsis Protoplasts
Protoplasts were prepared from leaves of 4- to 5-week-old Arabidopsis plants in sterile culture and transfected with the polyethylene glycol method as described by Damm et al. (1989). Routinely, 5 × 105 protoplasts were incubated with 30 μg of pure plasmid DNA (NucleoBond AX kits; Macherey-Nagel) premixed in Tris-EDTA buffer up to a total volume of 50 μL. As a precaution, cefotaxime (0.1 mg mL−1 final concentration [f.c.]; Duchefa) was added to the final protoplast cultivation medium. Where indicated, proteasome inhibitor MG-132 (20 μm f.c.; Calbiochem) or glycosylation inhibitor tunicamycin (5 μm f.c.; Serva) was either added already to protoplasts on ice (i.e. before transfection) or solely included during protoplast cultivation, which gave similar results. Tunicamycin-treated protoplasts were vital for about 24 h, but most cells died within 48 h of incubation, similar to a study with BY2 cells (Iwata and Koizumi, 2005). In contrast, protoplasts of the stt3a-2 mutant were vital for more than 3 d.
Infiltration of Nicotiana benthamiana Leaves
Agro-leaf infiltration of soil-grown N. benthamiana plants was essentially done as described by Schöb et al. (1997). Fresh agrobacteria cultures (grown under selection in liquid YT medium at 28°C) were diluted (1:50) in YT medium, 10 mm MES, pH 5.6, supplemented with 100 μm acetosyringone (in dimethyl sulfoxide; Fluka), and incubated at 28°C overnight in a shaking-water bath. Cells were harvested by centrifugation (2,500g, 4°C for 15 min), adjusted to 1 OD600 with infiltration buffer (10 mm MES, pH 5.6, 10 mm MgCl2, and 100 μm acetosyringone), and incubated for 2 to 3 h at room temperature in the dark. To suppress gene silencing, Agrobacterium strain 19K (C58C1:p19, expressing a viral anti-silencing protein; Voinnet et al., 2000) was grown in parallel, adjusted to 2 OD600, incubated as above, and mixed (1:1) with the recombinant test strains prior to infiltration. Leaves were infiltrated from the lower epidermis using a rubber clamp injection device holding a syringe (custom made by the shop at Westfälische Wilhelms-Universität). Tunicamycin (5–10 μm f.c.) was either added to the Agrobacterium suspension or infiltrated the following day. The latter improved detection of the GFP fusions.
Fluorescence Microscopy
Light microscopy was performed with a confocal laser-scanning microscope (Leica TCS SP2/AOBS) set up for simultaneous three-channel color detection. Fluorescing cells were spotted under an inverse microscope (Leica DM IRE2 UV) equipped with filter sets for detecting GFP (L5; Leica) or OFP (Phycoerythrin; AF Analysentechnik). One of two water-dipping objectives was used: 63× (0.9 W) for submerged leaf tissue on Petriperm plates (flexible bottom; Greiner), or 63× (1.2 W corrected) for protoplasts in a Bachhofer chamber (coverslip bottom). Confocal laser-scanning microscopy images were recorded digitally using the Leica confocal software. Excitation/emission wavelengths were 488/490 to 510 nm for GFP and 543/580 to 600 nm for OFP (orange-shifted mRFP or DsRed). As a reference, chloroplast fluorescence was recorded above 700 nm. During coexpression analyses, care was taken that real-time recorded signals did not bleed through into other detection channels.
Protein Extraction and PNGase F Treatment
Equal amounts of cultivated protoplasts were cooled on ice and harvested by centrifugation (5 min at 1,000g, 4°C). Supernatants were discarded and cell pellets were frozen in liquid nitrogen. Leaf discs were excised with a cork borer and immediately frozen in liquid nitrogen. Concomitant with thawing in protein extraction buffer (50 mm HEPES-NaOH, pH 7.5, 250 mm NaCl, 1 mm EDTA, 1% β-mercaptoethanol, 0.6% SDS, and 1 mm Pefabloc SC; Serva), a tip of spatula polyvinylpolypyrrolidone (Sigma) was added to prevent protein oxidation. Samples were ground with a potter, and cleared supernatants were used for further analyses. Prior to PNGase F treatment, cleared leaf extracts were boiled for 3 to 5 min. Heated samples were diluted with extraction buffer to reduce SDS to 0.2% (f.c.) and supplemented with Triton X-100 (1% f.c.) and EDTA (10 mm f.c.). One unit of PNGase F (Roche) was added to each sample, mixed, and kept at 37°C for 18 h or longer prior to SDS-PAGE. Mock-incubated samples were treated in the same way, except for omitting the enzyme. Successful PNGase F digests reduce concanavalin A binding to blotted glycoproteins in the presence of Ca2+ and Mn2+ ions, as revealed by peroxidase-coupled affinoblot detection (Faye and Chrispeels, 1985).
Immunoblot Analyses
Protein extracts were separated by SDS-PAGE followed by western-blot transfer using the MiniProtean-III system (Bio-Rad). In brief, SDS gels were rinsed with transfer buffer (25 mm Tris, 192 mm Gly, and 5% methanol) and electrotransferred to a nitrocellulose membrane (PROTRAN; Schleicher & Schüll). Blotted proteins were stained with Ponceau S (0.3% [w/v] in 3% TCA; Serva) on the blot, documented with a scanner, and destained with TBST (20 mm Tris, pH 7.4, 150 mm NaCl, and 0.1% Tween 20) prior to 1-h incubation of the membrane in blocking buffer (2% nonfat milk powder in TBST). Incubation in primary antibodies diluted in blocking buffer (1:300–1:50,000 depending on the antiserum and detection system used) was either at room temperature for 2 h or at 4°C overnight. Blots were washed with TBS (20 mm Tris-HCl, pH 7.4, 500 mm NaCl), TBST, and TBS (10 min each) and incubated for 1 h with goat anti-rabbit horseradish peroxidase conjugate (Bio-Rad), diluted 1:3,000 in blocking buffer for colorimetric detection, where indicated (von Schaewen et al., 1993; Wenderoth and von Schaewen, 2000), and otherwise 1:20,000 for chemiluminescence detection with the ECL Advance Western Blotting Detection Kit (Amersham/GE Healthcare) and washed again with TBS, TBST, and TBS (as above) prior to development. For additional probing, blots were incubated in blot-strip buffer (2% SDS, 0.1 m β-mercaptoethanol in 62.5 mm Tris, pH 6.7; ECL detection kit manual) at 50°C for 30 min followed by extensive washing with TBST. Anti-GFP serum (rabbit) was purchased from Molecular Probes/Invitrogen. A polyclonal GnTI antiserum directed against the catalytic domain of potato (Solanum tuberosum) GnTI (Wenderoth and von Schaewen, 2000) was purified using His-GnTI antigen immobilized on blot-strips. Those were incubated with crude antiserum at room temperature for 3 h, followed by two 5-min washes with TBS and two 5-min washes with phosphate-buffered saline. Bound antibodies were eluted by two 10-min incubations in 0.1 m Gly buffer, pH 2.5, followed by immediate adjustment to pH 7.4 using 1 M Tris-HCl, pH 9.
Root Growth Analyses
Root growth assays were conducted on hard agar plates as described previously (Koiwa et al., 2003). Photographs were taken at day 5 with a digital camera (Nikon D100, AF NIKKOR lens) prior to measuring root lengths. Close-up images were recorded with a light-sensitive digital camera (Leica DC 300F) attached to a stereomicroscope (Olympus SZ61).
Scanning Electron Microscopy
Seedlings were removed from hard agar plates and taken through a series of dehydration steps with increasing ethanol concentrations. Seedlings were kept in absolute ethanol, then dried with a critical point drier (Emitech K850), gold coated with a vacuum-sputter device (Emitech K550x), and examined by scanning electron microscopy (Hitachi S-3000N) at 15 kV under high vacuum. Micrographs were recorded digitally.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. A simple T-DNA insertion characterizes the cgl1-T allele (SALK_073650).
Supplemental Figure S2. Complex glycosylation recovers in C5 stt3a-2 double mutants.
Supplemental Figure S3. Signal intensities of labeled Golgi stacks differ in Col wild-type and stt3a-2 mutant protoplasts.
Supplemental Figure S4. Analysis of CGL1-GFP and C5-GFP variants in Agro-infiltrated N. benthamiana leaves, coexpressing the m-OFP-ER reference.
Supplemental Table S1. PCR primers used in this study.
Supplemental Video S1. Plasmid DNA-transfected Arabidopsis protoplasts (after 24-h cultivation), coexpressing C5-GFP and Golgi control CGL1-OFP in the presence of tunicamycin.
Supplemental Video S2. Agro-infiltrated N. benthamiana leaves (2 dpi), and protoplasts released thereof coexpressing C5-GFP or C5*-GFP plus the m-OFP-ER control.
Supplementary Material
Acknowledgments
We are grateful to Klaus Tenberge (Institute for Botany, Westfälische Wilhelms-Universität Münster) for basic instructions on scanning electron microscopy. Several colleagues shared their research material with us. We thank Benedikt Kost (University of Warwick, United Kingdom) for plasmid vector pGFP2, Shaoul Yalovsky (Tel Aviv University, Israel) for plasmid vector pSY526, and Sarah Hodge (Medical Research Council, Cambridge, United Kingdom) for pBIN m-gfp5-ER. KOR1-specific antibodies were provided by the group of Herman Höfte (INRA, Versailles, France), and Agrobacterium strain 19K was provided by the group of Jörg Kudla (Institute for Botany, Westfälische Wilhelms-Universität Münster). This study received excellent technical support from Kerstin Fischer, Olessja Becker, Meenu Vikram, and Jae Sook Kang.
This work was supported by the Deutsch Forschungsgemeinschaft (grant nos. SCHA 541/7 and HBFG 112–480 to A.v.S.), by the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service Designing Food for Health Program (grant no. 2008–34402–19195 to H.K.), and by the Texas AgriLife Research Federal Initiative (grant no. 2007–118409 to H.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Antje von Schaewen (schaewen@uni-muenster.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Banerjee S, Vishwanath P, Cui J, Kelleher DJ, Gilmore R, Robbins PW, Samuelson J (2007) The evolution of N-glycan-dependent endoplasmic reticulum quality control factors for glycoprotein folding and degradation. Proc Natl Acad Sci USA 104 11676–11681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N, Ringli C, Keller B (2001) The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev 15 1128–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Weissman AM (1998) Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu Rev Cell Dev Biol 14 19–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Gorlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatre L, Brandizzi F, Hocquellet A, Hawes C, Moreau P (2005) Sec22 and Memb11 are v-SNAREs of the anterograde endoplasmic reticulum-Golgi pathway in tobacco leaf epidermal cells. Plant Physiol 139 1244–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm B, Schmidt R, Willmitzer L (1989) Efficient transformation of Arabidopsis thaliana using direct gene transfer to protoplasts. Mol Gen Genet 217 6–12 [DOI] [PubMed] [Google Scholar]
- Deblaere R, Bytebier B, De Greve H, Debroeck F, Schell J, van Montagu M, Leemans J (1985) Efficient octopine Ti plasmid-derived vectors of Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res 13 4777–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diet A, Link B, Seifert GJ, Schellenberg B, Wagner U, Pauly M, Reiter W-D, Ringli C (2006) The Arabidopsis root hair cell wall formation mutant lrx1 is suppressed by mutations in the RHM1 gene encoding a UDP-l-rhamnose synthase. Plant Cell 18 1630–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth E, Frey J (2000) A rapid method for high throughput DNA extraction from plant material for PCR amplification. Plant Mol Biol Rep 18 61–64 [Google Scholar]
- Dixit R, Cyr R (2002) Golgi secretion is not required for marking the preprophase band site in cultured tobacco cells. Plant J 29 99–108 [DOI] [PubMed] [Google Scholar]
- Elbein AD (1987) Inhibitors of the biosynthesis and processing of N-linked oligosaccharide chains. Annu Rev Biochem 56 497–534 [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Molinari M, Helenius A (1999) Setting the standards: quality control in the secretory system. Science 286 1882–1888 [DOI] [PubMed] [Google Scholar]
- Essl D, Dirnberger D, Gomord V, Strasser R, Faye L, Glossl J, Steinkellner H (1999) The N-terminal 77 amino acids from tobacco N-acetylglucosaminyltransferase I are sufficient to retain a reporter protein in the Golgi apparatus of Nicotiana benthamiana cells. FEBS Lett 453 169–173 [DOI] [PubMed] [Google Scholar]
- Favery B, Ryan E, Foreman J, Linstead P, Boudonck K, Steer M, Shaw P, Dolan L (2001) KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev 15 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye L, Chrispeels MJ (1985) Characterization of N-linked oligosaccharides by affinoblotting with concanavalin A-peroxidase and treatment of the blots with glycosidases. Anal Biochem 149 218–244 [DOI] [PubMed] [Google Scholar]
- Faye L, Johnson KD, Chrispeels MJ (1986) Oligosaccharide side chains of glycoproteins that remain in the high-mannose form are not accessible to glycosidases. Plant Physiol 81 206–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitchette-Laine AC, Gomord V, Cabanes M, Michalski JC, Saint Macary M, Foucher B, Cavelier B, Hawes C, Lerouge P, Faye L (1997) N-Glycans harboring the Lewis a epitope are expressed at the surface of plant cells. Plant J 12 1411–1417 [DOI] [PubMed] [Google Scholar]
- Grebe M, Xu J, Möbius W, Ueda T, Nakano A, Geuze HJ, Rook MB, Scheres B (2003) Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr Biol 13 1378–1387 [DOI] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauptmann P, Riel C, Kunz-Schughart LA, Frohlich KU, Madeo F, Lehle L (2006) Defects in N-glycosylation induce apoptosis in yeast. Mol Microbiol 59 765–778 [DOI] [PubMed] [Google Scholar]
- Hebert DN, Garman SC, Molinari M (2005) The glycan code of the endoplasmic reticulum: asparagine-linked carbohydrates as protein maturation and quality-control tags. Trends Cell Biol 15 364–370 [DOI] [PubMed] [Google Scholar]
- Höfgen R, Willmitzer L (1988) Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res 16 9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Koizumi N (2005) Unfolded protein response followed by induction of cell death in cultured tobacco cells treated with tunicamycin. Planta 220 804–807 [DOI] [PubMed] [Google Scholar]
- Kang JS, Frank J, Kang CH, Kajiura H, Vikram M, Ueda A, Kim S, Bahk JD, Triplett B, Fujiyama K, et al (2008) Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc Natl Acad Sci USA 105 5933–5938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CM, Park SH, Je BI, Park SH, Park SJ, Piao HL, Eun MY, Dolan L, Han CD (2007) OsCSLD1, a cellulose synthase-like D1 gene, is required for root hair morphogenesis in rice. Plant Physiol 143 1220–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Kim JH, Jayanty SS, Howe GA, Han KH (2006) Loss of function of COBRA, a determinant of oriented cell expansion, invokes cellular defence responses in Arabidopsis thaliana. J Exp Bot 57 2923–2936 [DOI] [PubMed] [Google Scholar]
- Koiwa H, Li F, McCully MG, Mendoza I, Koizumi N, Manabe Y, Nakagawa Y, Zhu J-H, Rus A, Pardo JM, et al (2003) The STT3a subunit isoform of the Arabidopsis thaliana oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell 15 2273–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi N, Ujino T, Sano H, Chrispeels MJ (1999) Overexpression of a gene that encodes the first enzyme in the biosynthesis of asparagine-linked glycans makes plants resistant to tunicamycin and obviates the tunicamycin-induced unfolded protein response. Plant Physiol 121 353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua NH (1998) A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J 16 393–401 [DOI] [PubMed] [Google Scholar]
- Lai E, Teodoro T, Volchuk A (2007) Endoplasmic reticulum stress: signaling the unfolded protein response. Physiology (Bethesda) 22 193–201 [DOI] [PubMed] [Google Scholar]
- Lane DR, Wiedemeier A, Peng L, Höfte H, Vernhettes S, Desprez T, Hocart CH, Birch RJ, Baskin TI, Burn JE, et al (2001) Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-beta-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol 126 278–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Goldberg AL (1998) Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol 8 397–403 [DOI] [PubMed] [Google Scholar]
- Lerouge P, Cabanes-Macheteau M, Rayon C, Fischette-Laine AC, Gomord V, Faye L (1998) N-Glycosylation biosynthesis in plants: recent developments and future trends. Plant Mol Biol 38 31–48 [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L (1987) Improved method for the isolation of RNA from plant tissues. Anal Biochem 163 16–20 [DOI] [PubMed] [Google Scholar]
- Martinez IM, Chrispeels MJ (2003) Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15 561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau P, Brandizzi F, Hanton S, Chatre L, Melser S, Hawes C, Satiat-Jeunemaitre B (2007) The plant ER-Golgi interface: a highly structured and dynamic membrane complex. J Exp Bot 58 49–64 [DOI] [PubMed] [Google Scholar]
- Nebenführ A, Gallagher LA, Dunahay TG, Frohlick JA, Mazurkiewicz AM, Meehl JB, Staehelin LA (1999) Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol 121 1127–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Neff JD, Chory J, Pepper AE (1998) dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J 14 387–392 [DOI] [PubMed] [Google Scholar]
- Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Höfte H (1998) A plasma membrane-bound putative endo-1,4-beta-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J 17 5563–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parodi AJ (2000) Role of N-oligosaccharide endoplasmic reticulum processing reactions in protein folding and degradation. Biochem J 348 1–13 [PMC free article] [PubMed] [Google Scholar]
- Patil C, Walter P (2001) Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr Opin Cell Biol 13 349–355 [DOI] [PubMed] [Google Scholar]
- Peng L, Kawagoe Y, Hogan P, Delmer D (2002) Sitosterol-beta-glucoside as primer for cellulose synthesis in plants. Science 295 147–150 [DOI] [PubMed] [Google Scholar]
- Plemper RK, Wolf DH (1999) Retrograde protein translocation: eradication of secretory proteins in health and disease. Trends Biochem Sci 24 266–270 [DOI] [PubMed] [Google Scholar]
- Roudier F, Fernandez AG, Fujita M, Himmelspach R, Borner GH, Schindelman G, Song S, Baskin TI, Dupree P, Wasteneys GO, et al (2005) COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 17 1749–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöb H, Kunz C, Meins F Jr (1997) Silencing of transgenes introduced into leaves by agro-infiltration: a simple, rapid method for investigation of sequence requirements for gene silencing. Mol Gen Genet 256 581–585 [DOI] [PubMed] [Google Scholar]
- Silberstein S, Collins PG, Kelleher DJ, Gilmore R (1995) The essential OST2 gene encodes the 16-kD subunit of the yeast oligosaccharyltransferase, a highly conserved protein expressed in diverse eukaryotic organisms. J Cell Biol 131 371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R, Altmann F, Glössl J, Steinkellner H (2004) Unaltered complex N-glycan profiles in Nicotiana benthamiana despite drastic reduction of beta1,2-N-acetylglucosaminyltransferase I activity. Glycoconj J 21 275–282 [DOI] [PubMed] [Google Scholar]
- Strasser R, Stadlmann J, Svoboda B, Altmann F, Glossl J, Mach L (2005) Molecular basis of N-acetylglucosaminyltransferase I deficiency in Arabidopsis thaliana plants lacking complex N-glycans. Biochem J 387 385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Lederer C, Baulcombe DC (2000) A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103 157–167 [DOI] [PubMed] [Google Scholar]
- von Schaewen A, Sturm A, O'Neill J, Chrispeels MJ (1993) Isolation of a mutant Arabidopsis plant that lacks N-acetyl glucosaminyl transferase I and is unable to synthesize Golgi-modified complex N-linked glycans. Plant Physiol 102 1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40 428–438 [DOI] [PubMed] [Google Scholar]
- Wenderoth I, von Schaewen A (2000) Isolation and characterization of plant N-acetylglucosaminyltransferase I (GntI) cDNA sequences: functional analyses in the Arabidopsis cgl mutant and in antisense plants. Plant Physiol 123 1097–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Nishizawa N, Wu Y, Kost B, Chua NH (2000) KORRIGAN, an Arabidopsis endo-1,4-beta-glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. Plant Cell 12 1137–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.