Abstract
Ribosome biogenesis is an evolutionarily conserved pathway that requires ribosomal and nonribosomal proteins. Here, we investigated the role of the ribosomal protein S2 (Rps2) in fission yeast ribosome synthesis. As for many budding yeast ribosomal proteins, Rps2 was essential for cell viability in fission yeast and the genetic depletion of Rps2 caused a complete inhibition of 40S ribosomal subunit production. The pattern of pre-rRNA processing upon depletion of Rps2 revealed a reduction of 27SA2 pre-rRNAs and the concomitant production of 21S rRNA precursors, consistent with a role for Rps2 in efficient cleavage at site A2 within the 32S pre-rRNA. Importantly, kinetics of pre-rRNA accumulation as determined by rRNA pulse-chases assays indicated that a small fraction of 35S precursors matured into 20S-containing particles, suggesting that most 40S precursors were rapidly degraded in the absence of Rps2. Analysis of steady-state RNA levels revealed that some pre-40S particles were produced in Rps2-depleted cells, but that these precursors were retained in the nucleolus. Our findings suggest a role for Rps2 in a mechanism that monitors pre-40S export competence.
INTRODUCTION
Ribosomes are the molecular machine responsible for protein synthesis in all living cells. In eukaryotes, the 80S ribosome is composed of two unequal subunits with sedimentation coefficient of 40S and 60S. Both the small (40S) and large (60S) ribosomal subunits are ribonucleoprotein (RNP) complexes: the 40S subunit is composed of the 18S ribosomal RNA (rRNA) and roughly 30 ribosomal proteins, whereas the 60S subunit contains three rRNAs (25S, 5.8S and 5S) and approximately 50 ribosomal proteins. The synthesis and processing of rRNA, as well as the assembly between rRNAs and ribosomal proteins, occur in a nuclear compartment called the nucleolus. Although ribosome biogenesis is evolutionarily conserved, it has been most extensively studied in the yeast Saccharomyces cerevisiae. In the nucleolus of this organism, a large RNP complex is cotranscriptionally recruited to the nascent rRNA precursor that is synthesized by RNA polymerase I (1–3). This nascent pre-rRNA is chemically modified and rapidly processed into a (i) 20S precursor of the mature 18S rRNAs and a (ii) 27S precursor of the 5.8S and 25S rRNAs. RNA polymerase III is responsible for the synthesis of the fourth pre-RNA, the precursor of the mature 5S rRNA (4).
Advances in proteomic approaches in the past few years have led to a significant enhancement in the identification of factors associated with ribosome biogenesis in yeast (1,3,5–8). Specifically, affinity purification of distinct pre-ribosome intermediates coupled with mass spectrometry identified over 200 proteins with various predicted biochemical activities, including kinases, helicases, NTPases and nucleases. An important conclusion from these studies is that the machinery involved in the assembly of the small and large ribosomal subunits appears to be mostly independent of each other. Indeed, purification of 35S rRNA-associated pre-ribosomes (∼ 90S) and subsequent protein identification by mass spectrometry indicates that it mostly includes 40S subunit processing factors (1,3). Accordingly, RNP complexes enriched by the purification of proteins that were previously characterized as 60S assembly factors mainly identified 5.8S and 25S rRNA precursors, and were devoid of 90S-associated proteins and 18S pre-rRNAs (6,7). Interestingly, the identification of ribosomal proteins in distinct preribosome intermediates suggest that ribosomal proteins are not only structural components of ribosomes, but are actively involved in the processing and assembly of ribosomal subunits. Recent studies substantiate this scheme by demonstrating direct roles for ribosomal proteins in various steps of ribosome biogenesis, including pre-rRNA processing and/or nuclear export (9–15). As yet, however, the mechanisms by which ribosomal proteins are involved in ribosome biogenesis remain poorly understood.
Ribosomal subunits have to be exported from the nucleus to the cytoplasm to become actively involved in translation. Although knowledge about the molecular details of preribosome export is limited, genetic screening in budding yeast has been able to identify genes whose mutations result in nuclear accumulation of preribosomal subunits. Whereas specific nucleoporins and the major nuclear export receptor, Crm1, are necessary for export of both pre-40S and pre-60S particles (16–20), most of the identified factors required for nuclear export of preribosomes are dedicated for the small or the large subunit. This latter observation is consistent with the dichotomy between the machineries responsible for small and large ribosomal subunit assembly.
In budding yeast, pre-40S ribosomal subunits are exported from the nucleus as 20S pre-rRNA containing particles that are processed in the cytoplasm, generating 18S rRNAs and mature 40S subunits (21). Final processing of 40S precursors also occurs in the cytoplasm of mammalian cells (22), consistent with a cytosolic maturation step that is conserved throughout eukaryotes. Using localization of the 20S pre-rRNA or green fluorescent protein-tagged small subunit ribosomal proteins, mutations in ribosomal and nonribosomal protein-coding genes that result in nuclear accumulation of pre-40S have been identified in yeast (8,9,13,18,23,24). Using such strategies, the small subunit proteins Rps0, Rps2, Rps3, Rps5, Rps15, Rps18, Rps19 and Rps26 appear to be required for pre-40S nuclear export (9,13). For some of these aforementioned ribosomal proteins, however, it remains uncertain whether they are directly involved in the nuclear export of pre-40S (transit through the nuclear pore complex) or if they function in export competence (quality control events that allow small subunit assembly/export to proceed).
The 40S ribosomal protein S2 (Rps2) is present on the solvent side of the ribosome small subunit (25–27), and is related to the Escherichia coli S5 ribosomal protein (28). We and others have demonstrated that the protein arginine methyltransferase 3 (PRMT3 in mammals and Rmt3 in Schizosaccharomyces pombe) directly interacts and methylates Rps2 (29,30). Notably, deletion of rmt3 in fission yeast leads to hypomethylated Rps2 and causes a 40S ribosomal subunit deficit (31), while prmt3-deficient mice exhibit minute-like characteristics during embryogenesis (32). The biological role of Rps2 methylation remains to be determined, however. Herein, we report the functional analysis of fission yeast Rps2 in ribosome biogenesis. Our data indicate that Rps2 is required for efficient processing of the 32S pre-rRNA at site A2. Consequently, endonucleolytic cleavage at site A3 is kinetically favored relative to A2, resulting in the production of 21S pre-rRNAs. Importantly, our results revealed that most pre-40S subunits were rapidly degraded in the absence of Rps2. Yet, some 20S pre-rRNA containing particles were eventually produced in Rps2-depleted cells, but were not converted to mature small subunits because they were retained in the nucleolus. Our data suggest that Rps2 participates in pre-40S export competence rather than directly in the transport of 40S precursors from the nucleus to the cytoplasm.
MATERIALS AND METHODS
Strains, growth media and genetic methods
Cells were grown at 30°C in yeast extract medium with amino acid supplements (YES) and Edinburgh minimum medium (EMM) containing appropriate amino acid supplements. Schizosaccharomyces pombe cells were transformed with plasmids and PCR products by the lithium acetate method. Disruption of rps2 in a diploid strain was performed by PCR-mediated gene targeting using 100-nt oligonucleotides with 80-nt from the appropriate regions of the rps2 genomic sequence (33). The oligonucleotide sequences used for the construction of this strain are available upon request. Meiosis and sporulation were induced in selected heterozygote diploids by plating on malt extract agar and tetrads were dissected with a micromanipulator (MSM 200, Singer Instruments, UK). Strains FBY136 [ade6? leu1-32 ura4-D1 his3-D1 Δrps2::kanMX6 + pFB101] and FBY137 [ade6? leu1-32 ura4-D1 his3-D1 rps2+ + pFB101] were constructed as follows. Subsequent to the selection of rps2 heterozygote diploids by colony PCR, cells were transformed with plasmid pFB101 (see below), and the resulting asci were subjected to random spore analysis (34). Colonies were then screened for either wild-type (FBY137) or disrupted (FBY136) rps2 alleles by colony PCR. The nmt1+-dependent gene expression was repressed by the addition of 60 μM thiamine to the growth medium.
Plasmid constructs
The DNA sequence encoding S. pombe rps2 was amplified by PCR using genomic DNA extracted from fission yeast. The amplification was performed using the forward primer 5′-CCGCTCGAGCATGGCAGAAAGCGCACCCAG-3′ and the reverser primer 5′-CGCGGATCCTTAGTACTTCTTCTCAGTTTGC-3′. Following digestion of the 5′- and 3′-ends of the PCR product with XhoI and BamHI, respectively, the DNA was cloned into XhoI/BamHI-digested pREP3X (35), generating plasmid pFB101. The cDNA carrying the rps7 gene plus 750-nt of upstream regulatory sequences was amplified by PCR from fission yeast genomic DNA, using the following primer sets: the 5′ primer 5′-AAACTGCAGGAAAGATGCACTATGTGATGCCT-3′ and the 3′ primer 5′-GGAAGATCTCCCAAGCCCTCGCCGGTAGCAAC-3′. The PstI–BglII-digested PCR product was ligated to pSGP574 (a generous gift from Susan Forsburg) vector backbones previously digested with PstI and BamHI. This cloning strategy yielded a plasmid that removes the nmt1 promoter from the pSGP574 vector and expresses an Rps7-EGFP fusion under the control of the endogenous rps7 promoter.
Antibodies and protein analysis
Rabbit polyclonal antibodies specific to fission yeast Rps2 were raised at Covance Research Products (Denver, PA, USA) against a GST-Rps2 fusion protein purified from E. coli. Fission yeast total cell extracts were prepared by resuspending cells in ice-cold PBS-MT (1 × PBS supplemented with 2 mM MgCl2 and 1% Triton-X-100) containing a cocktail of protease inhibitors (Roche, Mississauga, ON, Canada) in a Fastprep (MP Biomedicals, Solon, OH, USA) using 0.5 mm glass beads. Clarified lysates were normalized for total protein concentration by using the Bradford protein assay (Bio-Rad, Inc.). Proteins were separated by SDS–PAGE (12%) and transferred to nitrocellulose membranes (Whatman, Springfield Mill, UK). Membranes were probed with rabbit polyclonal antibodies specific to Rmt1 (36) and Rps2 of S. pombe, as well as a mouse polyclonal antibody raised against human 60S ribosomal protein L7 (GeneTex).
Sucrose gradient fractionation of ribosomal subunits
The relative concentration of 40S and 60S ribosomal subunits was determined using low Mg2+ conditions as previously described (29) using extracts from log-phase fission yeast (OD600, 0.3–0.5). Briefly, cells were washed in ice-cold lysis buffer [20 mM Tris–HCl (pH 7.5), 50 mM KCl, 40 mM EDTA, 1 mM DTT and 0.2 mg/ml heparin] and resuspended in 300 μl of lysis buffer supplemented with protease (Roche) and RNase inhibitors (Promega, Madison, WI, USA). Cell suspensions were subjected to glass beads-mediated lysis using a Fastprep instrument (MP Biomedicals). After sedimentation of glass beads and cell debris by centrifugation, extracts were transferred to prechilled microcentrifuge tubes and centrifuged for an additional 15 min at 14 000 r.p.m. at 4°C. A total of eight A260 units of extracts were layered onto 5–45% sucrose gradients prepared in lysis buffer and centrifuged for 4 h at 39 000 r.p.m. and 4°C in a Beckman SW41 rotor. The gradients were then fractionated by upward displacement with 55% (w/v) sucrose using a gradient fractionator (Brandel, Gaithersburg, MD, USA) connected to a UV monitor (Teledyne Isco) for continuous measurement of the absorbance at 254 nm.
Pulse-chase analysis of pre-rRNA processing
The labeling of cells with [methyl-3H]-methionine was performed as described previously (29). Briefly, 50 ml cultures were grown at 25°C in EMM leu- to an OD600 of 0.2, at which point cultures were treated, or not treated, with 60 μm of thiamine. Following 6 h incubation with or without thiamine at 25°C, cells were harvested by centrifugation, and resuspended in 950 μl of EMM leu-containing 200 μCi of [methyl-3H]-methionine (GE Healthare, Piscataway, NJ, USA). Following a 8-min pulse, the labeled cells were diluted into 7 ml of EMM supplemented with 1 mg/ml l-methionine. At various time points, cell samples were taken and rapidly frozen in liquid nitrogen. RNAs were prepared using acidic hot phenol and 10 000 c.p.m. of radioactivity were resolved on 1.25% agarose-formaldehyde gels. RNAs were then transferred to Hybond-N+ (GE Healthcare) membranes, UV cross-linked and sprayed with Enhance (Perkin Elmer, Wlatham, MA, USA). The membranes were exposed to film for 2–4 weeks at −80°C.
Northern analysis
Steady-state levels of pre-rRNAs were assessed by northern analysis using oligonucleotides (numbered from 1–4, and 8 according to the scheme in Figure 5A) 1: (5′-GGT CTC TCT GCT GCC GG-3′), 2: (5′-CAT GGC TTA ATC TTT GAG AC-3′), 3: (5′-CGG TTT TAA TTG TCC TA-3′), 4: (5′-TGT TAC CTC TGG GCC C-3′) and 8: (5′-AAT TTC CAG TTA CGA AAA TTC TTG-3′) that were end-labeled with 50 μCi of [γ-32P]-ATP (Perkin Elmer, Wlatham, MA, USA; specific activity, 5000 Ci/mmol) by using T4 polynucleotide kinase (NEB). Total RNA was extracted as above, and 5 μg was loaded and resolved on 1.25% agarose–formaldehyde gels and transferred to and immobilized on nylon membranes. Prehybridization and hybridization were done in Church buffer (37). Washes were done in 2 × SSC/0.1% SDS and 0.1 × SSC/0.1% SDS, and the membranes were exposed to phosphor storage screens and analyzed using a Storm 860.
Figure 5.
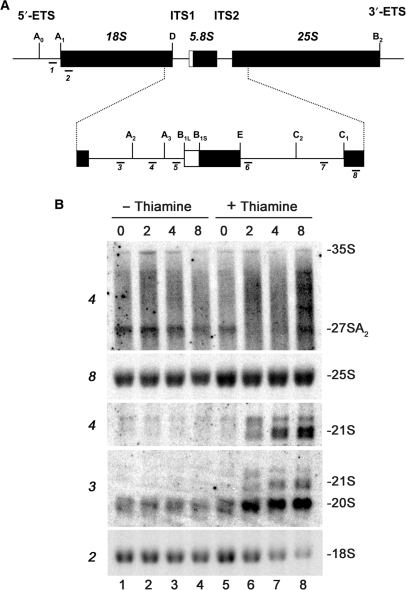
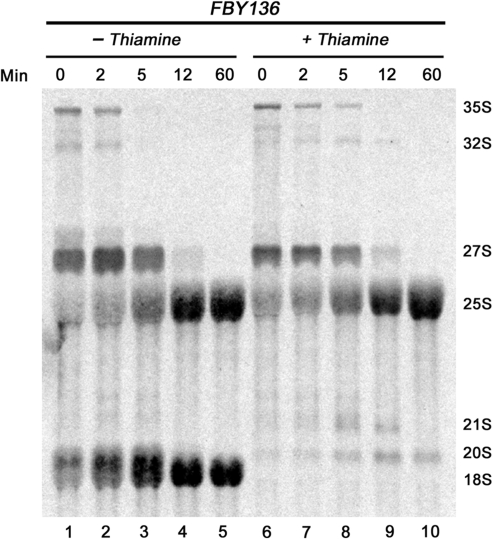
Depletion of Rps2 delays pre-rRNA processing at site A2 and leads to the accumulation of 20S pre-rRNA. (A) Structure of the ribosomal DNA locus. The ribosomal DNA encodes for the mature 18S, 5.8S and 25S rRNAs. The 18S–5.8S and 5.8S–25S rRNAs are interspaced with internal transcribed spacers 1 and 2 (ITS1 and ITS2), respectively. The 18S–5.8S–25S rRNAs are embedded into noncoding 5′- and 3′-external transcribed spacers (5′-ETS and 3′-ETS). Pre-rRNA processing sites are indicated as uppercase letters (A0–E) and oligonucleotide probes used for northern blotting are indicated as 1–8. (B) Strain FBY136 was grown to mid-log phase before being treated (lanes 5–8) or not (lanes 1–4) with thiamine. Equal amounts of RNA that were extracted from cell samples collected at the indicated time points (in hours) were separated on agarose–formaldehyde gels and transferred to nylon membranes for northern hybridization. The membranes were hybridized with specific oligonucleotides probes shown in (A) that are indicated to the left. rRNA species are indicated to the right.
Fluorescent in situ hybridization and fluorescence in live cells
Schizosaccharomyces pombe 5′ ITS1 rRNA was localized by fluorescence in situ hybridization as described (18). A DNA oligonucleotide complementary to the first 50 bases of ribosomal DNA internal transcribed spacer-1 (ITS-1), 5′-TTCCCAAAAAGTTAAAAGATGGAAATTTTTTAAAACCTTTTCATATAACTTTTC-3′, was synthesized with the Cy3 fluorophore at its 5′-end (Integrated DNA Technologies, Coralville, USA). Yeast strains were grown to a density of 1 × 107–4 × 107 and fixed with 2.4% formaldehyde for 90 min. Fixed samples were hybridized with the Cy3-labeled oligonucleotide at a concentration of 50 nM.
RESULTS
The 40S ribosomal protein S2 is essential for viability of fission yeast
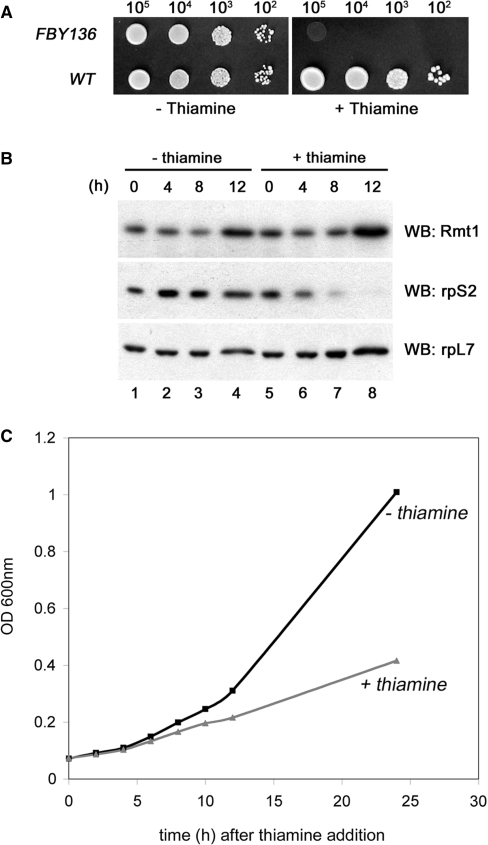
The gene encoding the 40S ribosomal protein S2 (Rps2) is essential in Saccharomyces cerevisiae (38). We therefore constructed a heterozygote diploid strain in which one of the two alleles of rps2 is disrupted to address whether rps2 is an essential gene in S. pombe. Spore germination after meiosis resulted in the growth of only geneticin-sensitive cells (data not shown), indicating that rps2 is essential for cell viability in S. pombe. To study the functional role of S. pombe Rps2 in ribosome biogenesis, we thus constructed a conditional strain (FBY136) in which the genomic copy of rps2 is deleted and plasmid-borne rps2 is expressed from the inducible/repressible nmt1+ promoter. Expression from the nmt1+ promoter is strongly repressed following thiamine addition (39). Whereas FBY136 grew as well as the control strain on thiamine-free medium, only the control strain was able to grow when thiamine was supplemented (Figure 1A). Accordingly, protein levels of Rps2 were markedly reduced when thiamine was added to the growth medium (Figure 1B). Western blot analyses demonstrated that levels of Rps2 were reduced by 50% and 90% by 4 h and 8 h, respectively, after the addition of thiamine (Figure 1B; lanes 5–7); Rps2 levels were unaffected in the absence of thiamine (Figure 1B; lanes 1–4). As controls, the expression level of the nonribosomal and ribosomal proteins Rmt1 and Rpl7, respectively, did not change in the presence or absence of thiamine (Figure 1B). Consistent with the direct role of depleting Rps2 in the thiamine-dependent growth arrest, growth rate began to slow down following ∼50–60% loss of Rps2 protein levels ∼5 h after thiamine addition (Fig. 1C). We conclude that the conditional S. pombe FBY136 strain specifically depletes cells of Rps2 in response to thiamine, and that Rps2 expression is essential for viability in fission yeast.
Figure 1.
Establishment of a conditional strain of S. pombe for the essential rps2 gene. (A) Tenfold serial dilutions of wild-type (rps2+) and rps2-null (rps2Δ; FBY136) cells that express plasmid-borne Rps2 from the nmt1 promoter were spotted onto EMM plates with (right) or without (left) thiamine. (B) Rps2-null cells that express plasmid-borne Rps2 under the control of the nmt1 promoter were grown to early log-phase, and shifted to medium containing (lanes 5–8) or not containing (lanes 1–4) thiamine for 0–12 h. Total cell extracts were prepared and analyzed by immunoblotting using antibodies specific to Rmt1 (upper panel), the 40S ribosomal protein S2 (Rps2; middle panel) and the 60S ribosomal protein L7 (rpL7; lower panel). (C) OD plot showing growth inhibition of FBY136 following addition of thiamine (0 h) to the culture medium.
Depletion of Rps2 results in a 40S ribosomal subunit deficit
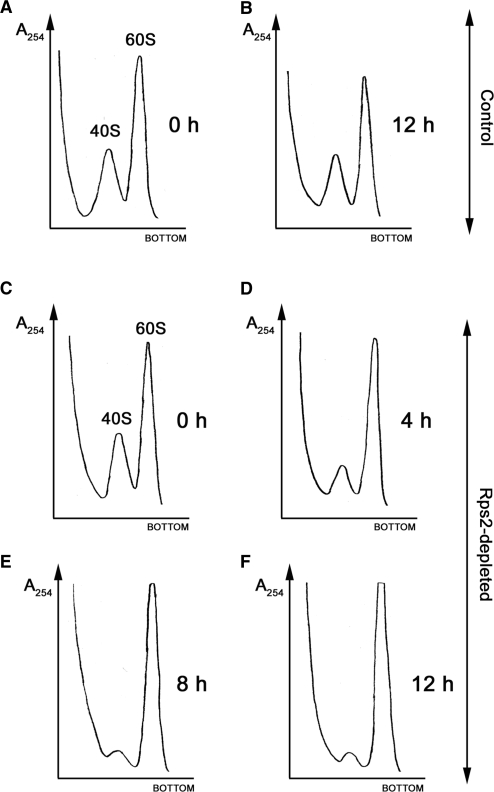
To begin to address the mechanism by which Rps2 depletion causes growth arrest, we examined ribosome profiles from extracts of cells that were previously grown in the presence or absence of thiamine. Depletion of Rps2 resulted in the reduction of free 40S subunits and polysomes as well as the accumulation of free 60S subunits (data not shown). To provide a quantitative analysis of the ratio between the small and large ribosomal subunits, we prepared cell extracts under low Mg2+ ion concentration that causes all cellular ribosomes to dissociate into separate subunits. After the centrifugation of extracts from Rps2-depleted cells on sucrose gradient, we detected a temporal decrease in 40S ribosomal subunit levels (Figure 2C–F). This deficit in small ribosomal subunit resulted in a gradual increase in the 60S:40S subunit ratio: 2.4, 5.4, 10.6 and 13.8 at 0, 4, 8 and 12 h, respectively, after the addition of thiamine (Figure 2C, D, E and F, respectively). The steady increase in 60S:40S ribosomal subunit ratio coincided with the time-dependent depletion of Rps2 (Figure 1B). As a control, cell extracts from strain FBY136 cultured in thiamine-free media showed similar 60S : 40S ratios at 0 h and 12 h (Figure 2A and B). These results indicate that Rps2 is required for the production of small ribosomal subunits and are similar to data reported for depletion of genes encoding other small subunit ribosomal proteins (9,11,12,15,40)
Figure 2.
Depletion of fission yeast Rps2 results in a deficit in 40S ribosomal subunits. Strain FBY136 was grown to early log-phase before being treated (C–F) or not treated (A and B) by the addition of thiamine to the culture media for 0 h (A and C), 4 h (D), 8 h (E) and 12 h (B and F). Cell extracts were prepared under low-Mg2+ ion concentration (40 mM EDTA) that causes all cellular ribosomes to dissociate into separate subunits. Eight A254 units of each extract were resolved on 5–45% sucrose gradients and the absorbance (A254) was continuously measured. The peaks corresponding to the 40S and 60S ribosomal subunits are indicated in (A and C).
Rapid turnover of nascent 40S precursors in the absence of Rps2
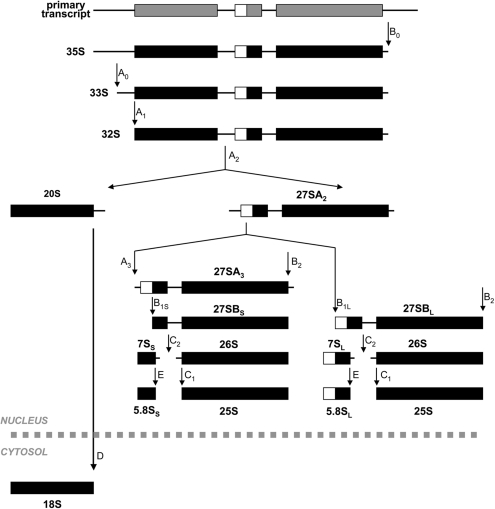
The aforementioned results indicating a 40S ribosomal subunit deficit upon Rps2 depletion could be the consequence of a block in small subunit synthesis. The synthesis and processing of ribosomal RNA precursors has been extensively studied in yeast (41,42). An outline of yeast pre-rRNA processing is shown in Figure 3. In yeast, rDNA repeats are transcribed by RNA polymerase I to generate a 35S rRNA precursor that is rapidly converted into 27S and 20S pre-rRNAs. The 27S pre-rRNA is processed into the mature 25S and 5.8S rRNAs found in the 60S ribosomal subunit, whereas the 20S pre-rRNA is cleaved in the cytosol to yield the mature 18S rRNA associated with the 40S subunit.
Figure 3.
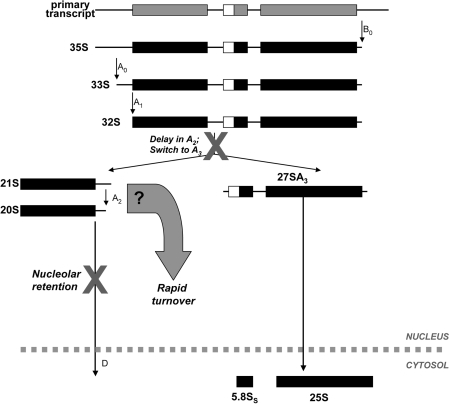
Pre-rRNA processing in yeast. The RNA polymerase I-dependent 35S precursor is first cleaved at site A0 to produce the 33S precursor, which is rapidly processed at site A1 to generate the 32S precursor. The 32S transcript is endonucleolytically cleaved at site A2 to produce the 20S and 27SA2 pre-rRNAs. The 20S RNA is exported to the cytoplasm for final processing at site D to generate the mature 18S rRNA. The 27SA2 precursor can be processed via two alternate pathways as indicated to generate the mature 5.8S and 25S rRNAs.
Because many of the nucleotides in the 35S rRNA precursor are subjected to posttranscriptional methylation, pre-rRNA processing is readily followed by metabolically labeling cells with [methyl-3H]methionine. We thus performed rRNA pulse-chase assays to examine the kinetics of pre-rRNA accumulation in the absence of Rps2. Strain FBY136 was grown in thiamine-containing medium for 6 h to deplete Rps2, and total RNA was then pulse labeled with [methyl-3H]methionine. Pre-rRNA processing was followed after chasing for 2, 5, 12 and 60 min with an excess of unlabeled methionine. As can be seen in Figure 4, the FBY136 strain cultured in the absence of thiamine (permissive condition) rapidly processed the 35S and 32S pre-rRNAs to 27S and 20S that were then matured to 25S and 18S rRNAs, respectively. FBY136 cells grown in nonpermissive conditions (with thiamine) showed only modest delays in the maturation of the 35S and 32S pre-rRNAs (compare lanes 6–8 to 1–3). Synthesis of mature 18S rRNA was severely compromised in Rps2-depleted conditions: 18S rRNA was not detected even after 60 min of chase as processing stopped after the production of 20S rRNA precursors (Figure 5). Importantly, the rRNA pulse-chases assays indicated that only a small fraction of 35S precursors matured into 20S-containing particles in Rps2-depleted cells (Figure 4, lanes 6–10), consistent with the active turnover of aberrant 40S precursors upon Rps2 depletion. The kinetics of 27S pre-rRNA processing into mature 25S rRNA were similar for cells cultured in permissive and nonpermissive conditions (Figure 4). These results indicate that Rps2 specifically functions in the assembly of the 40S ribosomal subunit and does not affect biogenesis of the large subunit, consistent with our ribosome profile data (Figure 2). The kinetics of pre-rRNA accumulation as determined by these experiments suggests that most nascent 40S precursors are rapidly degraded in the absence of Rps2.
Figure 4.
Inhibition in 18S rRNA synthesis in Rps2-depleted cells. Strain FBY136 was grown at 25°C in EMM medium to early log-phase and then treated (lanes 6–10) or not (lanes 1–5) with thiamine for 6 h. The cells were then pulse-labeled with [3H]-methionine for 8 min and chased with an excess of unlabeled methionine. Total RNA was extracted from cells samples harvested at the indicated time points and resolved on a 1.25% agarose–formaldehyde gel. The position of the rRNA species is indicated on the right.
Inhibition of 18S rRNA synthesis in Rps2-depleted cells is caused by a block in 20S pre-rRNA processing
We next analyzed steady-state RNA levels by northern hybridization to precisely determine the processing defects associated with Rps2 depletion. Total RNA prepared from the Rps2 conditional strain (FBY136) that was previously grown in permissive and nonpermissive conditions was hybridized to specific oligonucleotides probes (Figure 5A). As can be seen in Figure 5B, hybridization using an oligonucleotide that is complementary to segment D-A2 (probe 3; Figure 5A) of the internal transcribed spacer-1 (ITS1) demonstrated a time-dependent increase of 20S pre-rRNAs (lanes 5–8) in nonpermissive growth conditions, consistent with defective processing of the 20S rRNA precursor. The apparent discrepancy between the pulse-chase and northern analyses with respect to the levels of the 20S rRNA precursor is likely explained by the fact that the pulse-chase assay analyzes the kinetics of rRNA accumulation versus the steady-state RNA levels determined by northern hybridization (see Discussion section). As expected, the block in 20S pre-rRNA processing (Figure 4) resulted in a concomitant decrease in mature 18S rRNA (Figure 5B, lanes 5–8; probe 2). Northern analysis of total RNA from Rps2-depleted cells also detected the 21S pre-rRNA and lower levels of 27SA2 rRNA precursors (Figure 5B, lanes 5–8), as determined using a probe complementary to region A2–A3 of ITS1 (probe 4; Figure 5A). Hybridizations using a probe complementary to region A0–A1 (probe 1; Figure 5A) of the 5′-external transcribed region showed no accumulation of 22S and 23S pre-rRNAs (data not shown), indicating that processing at sites A0 and A1 is not significantly affected on Rps2 depletion. In summary, rRNA pulse-chase assays and northern blot experiments indicate that most 40S precursors undergo rapid turnover in the absence of Rps2; yet, the 40S precursors that are produced are completely inhibited in processing beyond the 20S pre-rRNA.
Retention of pre-40S ribosomal subunits in the nucleolus of Rps2-depleted cells
The 20S rRNA precursor is generated during pre-rRNA processing in the cell nucleus and matured by a cytoplasmic cleavage at site D to give the mature 18S rRNA [Figure 3; (21)]. Accordingly, the detection of 20S pre-rRNA in Rps2-depleted cells could be the consequence of the retention of pre-40S in the nucleus or the inhibition of cleavage at site D in the cytosol. We first used fluorescent in situ hybridization (FISH) to determine the subcellular localization of 20S rRNA precursors in strain FBY136 grown in nonpermissive conditions. As the 20S precursor is the main pre-rRNA detected by northern hybridization of total RNA using a probe complementary to region D-A2 of ITS1 (Figure 5 and data not shown), a fluorescently coupled oligonucleotide specific to the same region is expected to primarily show the steady-state localization of the 20S pre-rRNA. In permissive growth conditions, the 20S rRNA precursor was found concentrated in a nuclear region immediately adjacent to the DAPI-stained nucleoplasm, distinctive of nucleolar localization (Figure 6A, panels a–c); some staining was also detected in the cytoplasm. Such cytosolic and nucleolar staining using an ITS1-specific probe is consistent with localization studies of the 20S pre-rRNA in S. cerevisiae (13,18). Depletion of Rps2 resulted in the noticeable retention of the 20S pre-rRNA in a nuclear region that is distinct from the DAPI staining in >60% of the cells (Figure 6A, panels d–i). These results indicate that processing of the 20S pre-rRNA is compromised because the 40S precursors that are produced in the absence of Rps2 are retained in the nucleolus.
Figure 6.
Pre-40S ribosomal subunits accumulate in the nucleolus of Rps2-depleted cells. (A) FISH of pre-rRNA in strain FBY136 that was treated (d–i) or not treated (a–c) with thiamine for 6 h. Pre-rRNA was visualized with a probe complementary to the D-A2 segment of the ITS1 (panels a, d and g). DNA was labeled with DAPI (panels b, e and h) to visualize the nucleoplasm. (B) The GFP-tagged 40S ribosomal protein S7 (Rps7) was expressed in wild-type (panels c and d) and FBY136 (panels a and b) cells that were previously treated (panels b and d) or not treated (panels a and c) with thiamine for 6h before being visualized for GFP localization by live microscopy. White and red arrowheads indicate the nucleolus and nucleoplasm, respectively.
Nuclear export of the 20S rRNA precursor in yeast is mediated as part of a pre-40S ribosomal subunit (43). Therefore, nucleolar accumulation of 20S pre-rRNAs in Rps2-depleted cells (Figure 6A) is also expected for ribosomal proteins of the small subunit. To establish whether the localization of a small subunit ribosomal protein is perturbed upon Rps2 depletion, we generated a construct that expresses Rps7 from its endogenous promoter as N-terminal fusion to the green fluorescent protein (GFP). Sucrose gradient experiments indicated that the Rps7-GFP protein is incorporated into ribosomes (Supplementary Figure S1), suggesting that the plasmid-expressed ribosomal protein is functional. The Rps7-GFP fusion showed cytosolic and nucleolar localization in FBY136 and wild-type strains grown without thiamine (Figure 6B, panels a and c). In the presence of thiamine, however, the nucleolar levels of Rps7-GFP were increased in FBY136 cells as compared to the control strain (Figure 6B, compare panels b and d). Rps7-GFP was also detected in the nucleoplasm of Rps2-depleted cells, but not in control cells (compare panel b to panels a and d). We believe that the nucleoplasmic Rps7-GFP signal seen in nonpermissive conditions (Figure 6B) likely corresponds to the free ribosomal protein. Accordingly, sucrose gradient fractionation experiments demonstrated an increase in free Rps7-GFP protein after depletion of Rps2 (Supplementary Figure S1). Notably, this nuclear and nucleolar accumulation in nonpermissive conditions resulted in a corresponding decrease in the cytosolic localization of Rps7-GFP. Our FISH and GFP fluorescence results indicate that the 20S-containing particles produced in Rps2-depleted cells are retained in the nucleolus.
DISCUSSION
The advancement in proteomics and affinity purification tools has significantly contributed to the identification of trans-acting factors involved in various steps of ribosome biogenesis. In the case of ribosomal proteins, however, it has been difficult to ascertain whether their association with specific ribosome intermediates represents functional or structural roles. However, studies in multiple organisms point to important functions for ribosomal proteins in ribosome assembly. Accordingly, our findings support a critical role for the ribosomal protein Rps2 in pre-rRNA processing and pre-40S export competence.
Role of fission yeast Rps2 in the cleavage of the 32S pre-rRNA at site A2
Depletion of Rps2 resulted in the reduction of 27SA2 pre-rRNAs and a concomitant increase of 21S rRNA precursors (Figure 5). As indicated in Figure 7, such a phenotype of rRNA processing is indicative of delays in A2 cleavage within ITS1. The detection of 21S rRNA precursors (Figures 4 and 5) suggests that the rate of cleavage at site A3 exceeds cleavage at site A2 (Figure 7), thereby allowing the release of 27S rRNA precursors for the synthesis of 60S subunits. It should be noted that whereas the RNase MRP is responsible for A3 cleavage (44,45), the endonuclease responsible for A2 cleavage remains to be identified, although the putative nuclease Utp24 was recently proposed (46). Because accumulation of 22S and 23S pre-rRNAs were not detected in our northern analyses from Rps2-depleted cells, the results also suggest that Rps2 does not significantly contribute to cleavage at A0 and A1 in fission yeast.
Figure 7.
Function of Rps2 in fission yeast pre-rRNA processing. Our results indicate that Rps2 is required for efficient processing at site A2. In conditions of Rps2 insufficiency, the 32S precursor is cleaved at site A3, as suggested by the detection of 21S rRNA precursors. The kinetics of pre-rRNA accumulation as determined by pulse-chase assays suggest that most 40S precursors are rapidly degraded in the absence of Rps2. Whether 21S- or 20S-containing particles, or both, are subject to this rapid turnover remains to be determined. The remaining 20S-containing small subunit precursors are not processed into mature 18S rRNA in the cytoplasm because they are retained in the nucleolus.
Delays in A2 cleavage with little or no perturbation in A0–A1 processing have previously been reported in S. cerevisiae bearing mutations in the rrp8 (47), rrp5 (48) and yar1 (24) genes, as well as upon genetic depletion of the ribosomal proteins Rps0, Rps18, Rps19 and Rps21 (12,13,15). However, the molecular mechanism by which the product of these genes perturb 32S pre-rRNA processing at site A2 is still poorly understood. The small subunit (SSU) processome, a complex consisting of the U3 small nucleolar RNA and ∼40 proteins, is required for early pre-rRNA processing, including cleavage at A2 (49–51). Whereas Rps2 and the above-mentioned ribosomal and nonribosomal proteins have not been identified as components of the S. cerevisiae SSU processome, visualization of the processome structure by electron microscopy indicates that nonprocessome components can impact efficient formation of this RNP complex (50). It is therefore possible that processome-independent association of Rps2 with the nascent pre-rRNA causes structural rearrangements required for optimal cleavage at A2.
A role for Rps2 in pre-40S export competence
In yeast and mammalian cells, 40S precursors are generated in the nucleolus, but must be exported to the cytoplasm for final processing into mature small subunits (21,22). Cells that were depleted of Rps2 were completely inhibited in 18S rRNA synthesis and 40S subunit production, which is likely the cause of lethality in these cells. Importantly, the kinetics of pre-rRNA accumulation as determined by our rRNA pulse-chase assays (Figure 4) indicated that a small fraction of 35S rRNA precursors matured into 20S-containing particles in Rps2-depleted cells. As indicated in Figure 7, our results are consistent with the active turnover of aberrant 40S precursors in the absence of Rps2. Importantly, analyses of de novo-synthesized rRNA species in yeast strains depleted for small subunit ribosomal proteins indicate that 40S precursors stalled in maturation are not necessarily targeted for rapid degradation (9,11), suggesting that the rapid turnover of pre-40S observed upon Rps2 depletion is not a general consequence of the absence of a ribosomal protein. Two models, which are not mutually exclusive, could explain the low levels of 40S precursors detected in Rps2-depleted cells: (i) the efficient turnover of 21S-containing particles before reaching maturation into 20S pre-rRNAs, and (ii) slow processing of 21S into 20S pre-rRNA followed by rapid turnover of 20S-containing particles. Because we did not observe high levels of 21S pre-rRNA (Figure 5B) upon Rps2 depletion, which would be expected from the second model, we favor a mechanism, whereby 21S-containing particles are efficiently degraded. Interestingly, it does not appear that 21S-containing particles are intrinsically unstable precursors, since they can be processed directly into mature 18S rRNA in the cytoplasm (52). The stable incorporation of Rps2 into pre-40S particles could therefore directly control the inactivation of a surveillance mechanism. Alternatively, Rps2 could act indirectly via the structural rearrangement of a folding intermediate that triggers quality control pathways.
Although most 40S precursors were rapidly degraded upon depletion of Rps2, analysis of steady-state RNA levels by northern hybridization nevertheless detected 20S pre-rRNA in Rps2-depleted cells (Figure 5). These 40S precursors were retained in the nucleolus, however, as determined by the visualization of pre-rRNAs and ribosomal proteins in individual cells by FISH and GFP fluorescence, respectively. We thus conclude that Rps2-depleted cells did not produce mature 40S subunits because the remaining 20S pre-rRNA-containing particles did not reach the cytoplasm (Figure 7) for final processing by Fap7 and/or Nob1 (53,54). Furthermore, the absence of detectable levels of 20S pre-rRNA in the nucleoplasm of Rps2-depleted cells contrasts to the noticeable detection of this rRNA precursor in the nucleoplasm of Rps15-depleted cells (13). Accordingly, Rps15 appears to play a direct role in small subunit nuclear export. Taken together, our data suggest a role for S. pombe Rps2 in making 40S precursors export competent rather than being directly involved in the transport of pre-40S subunits from the nucleus to the cytoplasm. We thus propose that pre-40S particles produced in the absence of Rps2 are recognized as aberrant and retained in the nucleolus for degradation. Nucleolar retention of preribosomes has been previously observed in S. cerevisiae: in nop9-null cells (55) as well as in cells that express a temperature-sensitive allele of the HEAT repeat-containing protein, Sda1 (56). In fact, genetic evidence from the study of Dez et al. (56) indicates that the rapid turnover of pre-60S particles generated in the conditional sda1-2 strain requires the Trf4-Air1/2-Mtr4 polyadenylation (TRAMP) complex and the nuclear exosome. Whether TRAMP-dependent polyadenylation and the exosome are involved in the turnover of 40S precursors in Rps2-depleted cells remains to be determined.
Systematic approaches were used in S. cerevisiae to address the role of individual small subunit ribosomal proteins in ribosome biogenesis (9,57). These studies reveal that most ribosomal proteins of the small subunit are required for specific steps of 40S assembly. Consistent with our results in fission yeast, processing of the 20S pre-rRNA into 18S rRNA is inhibited upon depletion of budding yeast Rps2. In contrast to our FISH results, however, the cytoplasmic and nucleolar levels of the 20S pre-rRNA detected upon depletion of S. cerevisiae Rps2 were similar to a control strain (9). Yet, the same study shows that Rps2 is required for nuclear export of pre-40S particles based on biochemical data that used a dynamic maturation assay in which nuclear and cytoplasmic RNAs are fractionated after pulse-labeling (9). However, it is difficult to distinguish between nucleoplasmic and nucleolar sources of pre-rRNAs using such a pulse-labeling assay (9). Our study thus goes beyond these previous findings and provides evidence for the nucleolar retention of 20S-containing particles in the absence of Rps2.
The discrepancy between the ITS1 FISH results in Rps2-depleted budding (9) and fission (our study) yeasts is unclear; however, it could due to technical differences as the 2-h period allowed for depletion of Rps2 for the FISH analysis in budding yeast (9) was not sufficient to detect nucleolar accumulation of 20S rRNA precursors in our conditional strain of S. pombe (data not shown). It is also possible that Rps2 performs slightly different roles in small subunit assembly and export between both yeasts. Given that it is estimated that S. pombe diverged from S. cerevisiae roughly 1100 million years ago (58), functional differences for specific ribosomal proteins between fission and budding yeasts may not be that surprising. In fact, the biological function of S. pombe and mammalian Rps2 appears to be regulated by the protein arginine methyltransferase, PRMT3 (29,30), an enzyme encoded by a gene that is absent from the S. cerevisiae genome (59).
Our results have identified a role for the fission yeast Rps2 in 32S pre-rRNA processing at site A2. Importantly, we have established that Rps2 primarily functions in making pre-40S ribosomal subunits export competent. A detailed understanding of the different interactions occurring between Rps2, the pre-40S, and ribosome-associated proteins is an essential step toward the elucidation of the mechanisms that monitor preribosome quality control.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
Canadian Institutes for Health Research (to F.B.). F.B. is the recipient of a New Investigator Award from the CIHR. Funding for open access charge: CIHR Grant MOP-171704.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Pierre-Emmanuel Gleizes and François Dragon for critical reading of the article.
REFERENCES
- 1.Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gallagher JE, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 2004;18:2506–2517. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schafer T, Kuster B, Tschochner H, Tollervey D, et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 4.Weinmann R, Roeder RG. Role of DNA-dependent RNA polymerase 3 in the transcription of the tRNA and 5S RNA genes. Proc. Natl Acad. Sci. USA. 1974;71:1790–1794. doi: 10.1073/pnas.71.5.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassler J, Grandi P, Gadal O, Lessmann T, Petfalski E, Tollervey D, Lechner J, Hurt E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001;8:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- 6.Harnpicharnchai P, Jakovljevic J, Horsey E, Miles T, Roman J, Rout M, Meagher D, Imai B, Guo Y, Brame CJ, et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- 7.Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schafer T, Strauss D, Petfalski E, Tollervey D, Hurt E. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 2003;22:1370–1380. doi: 10.1093/emboj/cdg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferreira-Cerca S, Poll G, Gleizes PE, Tschochner H, Milkereit P. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell. 2005;20:263–275. doi: 10.1016/j.molcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Hofer A, Bussiere C, Johnson AW. Mutational analysis of the ribosomal protein Rpl10 from yeast. J. Biol. Chem. 2007;282:32630–32639. doi: 10.1074/jbc.M705057200. [DOI] [PubMed] [Google Scholar]
- 11.Jakovljevic J, de Mayolo PA, Miles TD, Nguyen TM, Leger-Silvestre I, Gas N, Woolford J.L., Jr The carboxy-terminal extension of yeast ribosomal protein S14 is necessary for maturation of 43S preribosomes. Mol. Cell. 2004;14:331–342. doi: 10.1016/s1097-2765(04)00215-1. [DOI] [PubMed] [Google Scholar]
- 12.Leger-Silvestre I, Caffrey JM, Dawaliby R, Alvarez-Arias DA, Gas N, Bertolone SJ, Gleizes PE, Ellis SR. Specific role for yeast homologs of the diamond blackfan anemia-associated Rps19 protein in ribosome synthesis. J. Biol. Chem. 2005;280:38177–38185. doi: 10.1074/jbc.M506916200. [DOI] [PubMed] [Google Scholar]
- 13.Leger-Silvestre I, Milkereit P, Ferreira-Cerca S, Saveanu C, Rousselle JC, Choesmel V, Guinefoleau C, Gas N, Gleizes PE. The ribosomal protein Rps15p is required for nuclear exit of the 40S subunit precursors in yeast. EMBO J. 2004;23:2336–2347. doi: 10.1038/sj.emboj.7600252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosado IV, Kressler D, de la Cruz J. Functional analysis of Saccharomyces cerevisiae ribosomal protein Rpl3p in ribosome synthesis. Nucleic Acids Res. 2007;35:4203–4213. doi: 10.1093/nar/gkm388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tabb-Massey A, Caffrey JM, Logsden P, Taylor S, Trent JO, Ellis SR. Ribosomal proteins Rps0 and Rps21 of Saccharomyces cerevisiae have overlapping functions in the maturation of the 3′ end of 18S rRNA. Nucleic Acids Res. 2003;31:6798–6805. doi: 10.1093/nar/gkg899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gleizes PE, Noaillac-Depeyre J, Leger-Silvestre I, Teulieres F, Dauxois JY, Pommet D, Azum-Gelade MC, Gas N. Ultrastructural localization of rRNA shows defective nuclear export of preribosomes in mutants of the Nup82p complex. J. Cell Biol. 2001;155:923–936. doi: 10.1083/jcb.200108142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho JH, Kallstrom G, Johnson AW. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 2000;151:1057–1066. doi: 10.1083/jcb.151.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moy TI, Silver PA. Nuclear export of the small ribosomal subunit requires the ran-GTPase cycle and certain nucleoporins. Genes Dev. 1999;13:2118–2133. doi: 10.1101/gad.13.16.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stage-Zimmermann T, Schmidt U, Silver PA. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell. 2000;11:3777–3789. doi: 10.1091/mbc.11.11.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas F, Kutay U. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J. Cell Sci. 2003;116:2409–2419. doi: 10.1242/jcs.00464. [DOI] [PubMed] [Google Scholar]
- 21.Udem SA, Warner JR. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J. Biol. Chem. 1973;248:1412–1416. [PubMed] [Google Scholar]
- 22.Rouquette J, Choesmel V, Gleizes PE. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 2005;24:2862–2872. doi: 10.1038/sj.emboj.7600752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milkereit P, Strauss D, Bassler J, Gadal O, Kuhn H, Schutz S, Gas N, Lechner J, Hurt E, Tschochner H. A Noc complex specifically involved in the formation and nuclear export of ribosomal 40 S subunits. J. Biol. Chem. 2003;278:4072–4081. doi: 10.1074/jbc.M208898200. [DOI] [PubMed] [Google Scholar]
- 24.Seiser RM, Sundberg AE, Wollam BJ, Zobel-Thropp P, Baldwin K, Spector MD, Lycan DE. Ltv1 is required for efficient nuclear export of the ribosomal small subunit in Saccharomyces cerevisiae. Genetics. 2006;174:679–691. doi: 10.1534/genetics.106.062117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marion MJ, Marion C. Ribosomal proteins S2, S6, S10, S14, S15 and S25 are localized on the surface of mammalian 40 S subunits and stabilize their conformation. A study with immobilized trypsin. FEBS Lett. 1988;232:281–285. doi: 10.1016/0014-5793(88)80753-1. [DOI] [PubMed] [Google Scholar]
- 26.Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 27.Wimberly BT, Brodersen DE, Clemons W.M., Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 28.All-Robyn JA, Brown N, Otaka E, Liebman SW. Sequence and functional similarity between a yeast ribosomal protein and the Escherichia coli S5 ram protein. Mol. Cell Biol. 1990;10:6544–6553. doi: 10.1128/mcb.10.12.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bachand F, Silver PA. PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. EMBO J. 2004;23:2641–2650. doi: 10.1038/sj.emboj.7600265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swiercz R, Person MD, Bedford MT. Ribosomal protein S2 is a substrate for mammalian PRMT3 (protein arginine methyltransferase 3) Biochem. J. 2005;386:85–91. doi: 10.1042/BJ20041466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bachand F, Lackner DH, Bahler J, Silver PA. Autoregulation of ribosome biosynthesis by a translational response in fission yeast. Mol. Cell Biol. 2006;26:1731–1742. doi: 10.1128/MCB.26.5.1731-1742.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swiercz R, Cheng D, Kim D, Bedford MT. Ribosomal protein rpS2 is hypomethylated in PRMT3-deficient mice. J. Biol. Chem. 2007;282:16917–16923. doi: 10.1074/jbc.M609778200. [DOI] [PubMed] [Google Scholar]
- 33.Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A., III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 34.Forsburg SL, Rhind N. Basic methods for fission yeast. Yeast. 2006;23:173–183. doi: 10.1002/yea.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 36.Perreault A, Lemieux C, Bachand F. Regulation of the nuclear poly(A)-binding protein by arginine methylation in fission yeast. J. Biol. Chem. 2007;282:7552–7562. doi: 10.1074/jbc.M610512200. [DOI] [PubMed] [Google Scholar]
- 37.Church GM, Gilbert W. Genomic sequencing. Proc. Natl Acad. Sci. USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 39.Maundrell K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- 40.Moritz M, Paulovich AG, Tsay YF, Woolford J.L., Jr Depletion of yeast ribosomal proteins L16 or rp59 disrupts ribosome assembly. J. Cell Biol. 1990;111:2261–2274. doi: 10.1083/jcb.111.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 42.Granneman S, Baserga SJ. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 2004;296:43–50. doi: 10.1016/j.yexcr.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 43.Zemp I, Kutay U. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 2007;581:2783–2793. doi: 10.1016/j.febslet.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 44.Lygerou Z, Allmang C, Tollervey D, Seraphin B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- 45.Lygerou Z, Mitchell P, Petfalski E, Seraphin B, Tollervey D. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev. 1994;8:1423–1433. doi: 10.1101/gad.8.12.1423. [DOI] [PubMed] [Google Scholar]
- 46.Bleichert F, Granneman S, Osheim YN, Beyer AL, Baserga SJ. The PINc domain protein Utp24, a putative nuclease, is required for the early cleavage steps in 18S rRNA maturation. Proc. Natl Acad. Sci. USA. 2006;103:9464–9469. doi: 10.1073/pnas.0603673103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bousquet-Antonelli C, Vanrobays E, Gelugne JP, Caizergues-Ferrer M, Henry Y. Rrp8p is a yeast nucleolar protein functionally linked to Gar1p and involved in pre-rRNA cleavage at site A2. RNA. 2000;6:826–843. doi: 10.1017/s1355838200992288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torchet C, Hermann-Le Denmat S. Bypassing the rRNA processing endonucleolytic cleavage at site A2 in Saccharomyces cerevisiae. RNA. 2000;6:1498–1508. doi: 10.1017/s1355838200000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bernstein KA, Baserga SJ. The small subunit processome is required for cell cycle progression at G1. Mol. Biol. Cell. 2004;15:5038–5046. doi: 10.1091/mbc.E04-06-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell. 2004;16:943–954. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 51.Wehner KA, Gallagher JE, Baserga SJ. Components of an interdependent unit within the SSU processome regulate and mediate its activity. Mol. Cell Biol. 2002;22:7258–7267. doi: 10.1128/MCB.22.20.7258-7267.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vos HR, Faber AW, de Gier MD, Vos JC, Raue HA. Deletion of the three distal S1 motifs of Saccharomyces cerevisiae Rrp5p abolishes pre-rRNA processing at site A(2) without reducing the production of functional 40S subunits. Eukaryot. Cell. 2004;3:1504–1512. doi: 10.1128/EC.3.6.1504-1512.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fatica A, Oeffinger M, Dlakic M, Tollervey D. Nob1p is required for cleavage of the 3′ end of 18S rRNA. Mol. Cell Biol. 2003;23:1798–1807. doi: 10.1128/MCB.23.5.1798-1807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Granneman S, Nandineni MR, Baserga SJ. The putative NTPase Fap7 mediates cytoplasmic 20S pre-rRNA processing through a direct interaction with Rps14. Mol. Cell Biol. 2005;25:10352–10364. doi: 10.1128/MCB.25.23.10352-10364.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomson E, Rappsilber J, Tollervey D. Nop9 is an RNA binding protein present in pre-40S ribosomes and required for 18S rRNA synthesis in yeast. RNA. 2007;13:2165–2174. doi: 10.1261/rna.747607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dez C, Houseley J, Tollervey D. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J. 2006;25:1534–1546. doi: 10.1038/sj.emboj.7601035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferreira-Cerca S, Poll G, Kuhn H, Neueder A, Jakob S, Tschochner H, Milkereit P. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol. Cell. 2007;28:446–457. doi: 10.1016/j.molcel.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 58.Heckman DS, Geiser DM, Eidell BR, Stauffer RL, Kardos NL, Hedges SB. Molecular evidence for the early colonization of land by fungi and plants. Science. 2001;293:1129–1133. doi: 10.1126/science.1061457. [DOI] [PubMed] [Google Scholar]
- 59.Bachand F. Protein arginine methyltransferases: from unicellular eukaryotes to humans. Eukaryot. Cell. 2007;6:889–898. doi: 10.1128/EC.00099-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.