Abstract
Most of the activities of IFN-γ are the result of STAT1-mediated transcriptional responses. In this study, we show that the BRCA1 tumor suppressor acts in concert with STAT1 to differentially activate transcription of a subset of IFN-γ target genes and mediates growth inhibition by this cytokine. After IFN-γ treatment, induction of the cyclin-dependent kinase inhibitor, p21WAF1, was synergistically activated by BRCA1, whereas the IRF-1 gene was unaffected. Importantly, the differential induction of p21WAF1 was impaired in breast cancer cells homozygous for the mutant BRCA1 5382C allele. Biochemical analysis illustrated that the mechanism of this transcriptional synergy involves interaction between BRCA1 aa 502–802 and the C-terminal transcriptional activation domain of STAT1 including Ser-727 whose phosphorylation is crucial for transcriptional activation. Significantly, STAT1 proteins mutated at Ser-727 bind poorly to BRCA1, reinforcing the importance of Ser-727 in the recruitment of transcriptional coactivators by STAT proteins. These findings reveal a novel mechanism for BRCA1 function in the IFN-γ-dependent tumor surveillance system.
IFN-γ is a cytokine that plays an important role in both innate and adaptive immunity (1). At the cellular level, IFN-γ mediates activation of an antiviral state and causes cell growth arrest at the G1 phase of the cell cycle (2–6). The IFN-γ response has also been postulated to be part of an endogenous tumor surveillance system (7, 8). The biological effects of IFN-γ are mediated through a heterodimeric transmembrane receptor which is capable of activating the Janus kinase (JAK)-STAT pathway, leading to tyrosine phosphorylation of the STAT1α protein (9). Phosphorylation of STAT1α renders it competent for dimerization, nuclear translocation, and DNA binding. The C terminus of STAT1 contains the transcription activation domain (10, 11), which is modified by phosphorylation at Ser-727 in response to IFN-γ treatment (12, 13). The phosphorylation of STAT1 on Ser-727 enhances the recruitment of transcriptional coactivators such as the CBP/p300 histone acetyltranferases and the DNA replication factor, MCM5 (14–16).
BRCA1, first identified as a breast cancer susceptibility gene, encodes a 1,863-aa protein with a N-terminal Really Interesting New Gene (RING) finger domain and a C-terminal acidic domain termed the BRCA1 C terminus (BRCT) (17, 18). This protein may act at a number of points in nuclear function and growth control (19–23) and has primarily been implicated in the DNA repair process when cells are treated with DNA-damaging agents (24–26).
Recent findings have also implicated BRCA1 as a transcriptional regulator (27, 28). Ectopic expression of BRCA1 activates several promoter reporter genes, especially those regulated by p53 such as p21WAF1 and MDM2. This ability to regulate gene expression implies that BRCA1 functions as a transcriptional coactivator which can enhance p53-dependent gene regulation (29). BRCA1 peptides have also been copurified with the RNA polymerase II holoenzyme, and these sequences can act as transcriptional activation domains (30–32). Importantly, tumor-associated mutations in the BRCT domain abolish this transcriptional activation activity (27, 28). Recently, it has been reported that the p300/CBP coactivator interacts with BRCA1, supporting that BRCA1 plays a role in transcription regulation (33).
In this study, we demonstrate transcriptional enhancement between IFN-γ-activated STAT1α and BRCA1. The cooperation between these proteins results from direct binding of Ser-phosphorylated STAT1α to BRCA1. We further show that the BRCA1-STAT1-dependent transcriptional enhancement is promoter specific. The BRCA1 protein is required for IFN-γ stimulation of the p21WAF1 gene, but not for IFN-γ stimulation of the IRF-1 gene. This differential regulation of IFN-γ-responsive genes is impaired in BRCA1 mutant cells. Based on these results, we suggest that BRCA1 is a critical component of IFN-γ-regulated antitumor responses.
Materials and Methods
Cell Culture and Antibodies.
293, 293T, 2fTGH, and U3A cells were maintained in DMEM supplemented with 10% FBS (GIBCO/BRL). G8 cells were grown in the same medium containing G418 (100 mg/ml, GIBCO/BRL). HCC1937 cells were purchased from American Type Culture Collection and maintained in RPMI medium 1640 supplemented with 10% FBS and 1 mM sodium pyruvate. Sf9 cells were maintained in Sf-900 II SFM (GIBCO/BRL) supplemented with 10% FBS. IFN-γ was purchased from PeProTech (Rocky Hill, NJ). IFN-γ (10 ng/ml) was added 5 h before reverse transcription–PCR (RT-PCR), or 12 h before Western blot analysis. Anti-BRCA1 antibody (Ab-1) was purchased from Calbiochem, and anti-STAT1 (E-23) and glutathione S-transferase (GST) (Z-5) were from Santa Cruz Biotechnology.
RT-PCR.
RT-PCR reactions were performed as described (34). Primers used for this assay are: glyceraldehyde 3-phosphatase dehydrogenase (5′-GTGAAGGTCGGAGTCAAC-3′ and 5′-TGGAATTTGCCATGGGTG-3′), p21WAF1 (5′-GACACCACTGGAGGGTGACT-3′ and 5′-CAGGTCCACATGGTCTTCCT-3′), and IRF-1 (5′-ATGAGACCCTGGCTAGAG-3′ and 5′-AAGCATCCGGTACACTCG-3′). The relative expression levels of p21WAF1 and IRF-1 were normalized to the expression levels of glyceraldehyde 3-phosphatase dehydrogenase.
Plasmid Construction.
Expression vectors for wild-type or Ser-727A mutant human STAT1 cDNAs with the FLAG tag were described elsewhere (12, 35). BRCA1 cDNA with NotI linker was cloned at the NotI site of pEBG vector (36) to express GST-fused full-length BRCA1 in mammalian cells. BRCA1/pEFN for the luciferase assay was constructed by subcloning the BRCA1 cDNA into the NotI site of pEF-BOS (37). Plasmid pGAL4DBD was generated by transferring a HindIII–BamHI fragment containing the GAL4DBD in pSG424 (a gift from A. N. A. Monteiro, Cornell University, New York, NY) to pcDNA3 (Invitrogen). pGAL4AD was generated by transferring a HindIII–BamHI fragment containing GAL4AD in pACT2 (CLONTECH) to pcDNA3. To generate a series of STAT1 segments fused with GAL4DBD, BamHI–NotI fragments of STAT1(1–52, 107–279, 185–279, 289–374, 383–487, 712–750) cloned in pGEX5X vector (34) were transferred to pGAL4DBD. Amino acids 498–568 of STAT1 was PCR-amplified by a set of primers, 5′-GGGATCCTTTCAGAAGTGCTGAGTTGGCAGTTT-3′ and 5′-GGCGGCCGCGTGTTTTTTAATGAGTTCTAGGAT-3′, and aa 569–679 was PCR amplified by 5′-GGGATCCTGCTCCCTCTCTGGAATGATGGGTGC-3′ and 5′-GGCGGCCGCTCCAAAGGCATGGTCTTTGTCAAT-3′. These fragments were also cloned into the BamHI–NotI-digested pGAL4DBD. GST-BRCA1 segments were described (24). The BamHI–NotI BRCA1 (502–802) fragment was subcloned into pGAL4AD to generate pGAL4AD-BRCA1(502–802). Luciferase reporter plasmid, 3x Ly6e-LUC, has been described (12).
Immunoprecipitation and GST-Affinity Purification Assays.
Cell extracts were prepared in Extraction Buffer C [50 mM Tris (pH 8)/120 mM NaCl/0.5% Nonidet P-40], with the addition of 100 mM NaF/200 μM sodium orthovanadate/100 μg/ml of PMSF/20 μg/ml of aprotinin/10 μg/ml of leupeptin. Whole-cell extract (20 μg) was loaded per lane. For immunoprecipitation, 1.0 mg of whole-cell extract was used with 1–2 μg of antibodies. The secondary antibodies were peroxidase-conjugated goat anti-mouse or rabbit IgG from Jackson ImmunoResearch. Signals were developed by chemiluminescence (Renaissance, NEN). For GST-affinity purification assays, whole-cell extracts prepared from 293T cells treated with IFN-γ for 15 min were used. Briefly, 500 μg of lysates were incubated with about 2 μg of GST-fusion proteins for 1 h at 4°C. Then, glutathione beads were added to each sample, and samples were further rotated for 1 h at 4°C. After extensive washing with NET-N buffer [20 mM Tris (pH 8.0)/100 mM NaCl/1 mM EDTA/0.5% Nonidet P-40/100 mM NaF/200 μM sodium orthovanadate], samples were loaded on a 7.5% SDS/PAGE gel, and blotted with anti-STAT1 antibody.
Luciferase Assay.
Basic procedures have been described (29). For each cell culture, 1 μg of 3x Ly6e-LUC with or without 2 μg of BRCA1/pEFN were transfected. Cells were treated with IFN-γ (10 ng/ml) for 6 h before luciferase assay. β-Galactosidase activity was measured as an internal control by cotransfection of pcDNA3.1/His/LacZ (Invitrogen).
Generation of Adenovirus.
The protocol has been reported (38). Briefly, cDNAs of BRCA1 or LacZ was subcloned into pShuttle-cytomegalovirus, and recombination was performed in a BJ5180 bacterial cell line. Recombinant virus was isolated, amplified, and analyzed for protein expression. Cells were infected with LacZ (Ad-LacZ) or BRCA1 (Ad-BRCA1) viruses at a multiplicity of infection of 5 or 100, respectively.
Results
BRCA1 and IFN-γ Cooperation in Activation of STAT1 Responsive Promoter Elements.
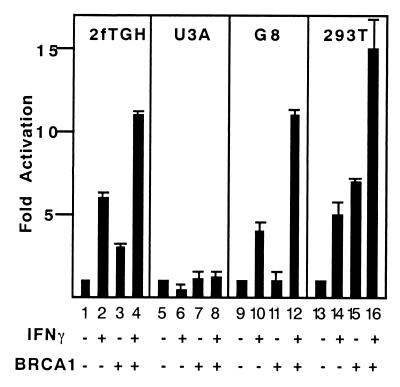
p21WAF1 promoter contains IFN-γ response elements to which activated STAT1 dimers bind, contributing to the inhibition of cell growth (39). Because IFN-γ activates the p21WAF1 promoter through these STAT1 elements, we asked if BRCA1 might also be involved in the IFN-γ pathway leading to p21WAF1 gene expression. The ability of BRCA1 to activate a STAT responsive reporter gene, 3x Ly6e-LUC, was tested in a cotransfection assay. A BRCA1 expression vector was cotransfected with 3x Ly6e-LUC into several cell lines. In the 2fTGH cell line, approximately sixfold induction of the reporter gene was observed with IFN-γ stimulation (Fig. 1, lane 2). Transfection with a BRCA1 expression vector modestly increased the basal transcription level in these cells. On IFN-γ treatment, the transcriptional activation in the BRCA1-expressing cells increased to 11-fold (lane 4). To clarify the role of STAT1 in this transcriptional enhancement with BRCA1, the same experiment was performed in the STAT1-deficient U3A cell line (40, 41). No activation of 3x Ly6e-LUC was observed in U3A cells regardless of BRCA1 expression or IFN-γ treatment (lanes 5–8).
Figure 1.
BRCA1 expression enhances IFN-γ-dependent transcription. 2fTGH, U3A, G8, and 293T cells were transfected with a STAT reporter gene (3x Ly6e-LUC) plus BRCA1 expression constructs or empty vectors for 40 h and followed by a 6-h IFN-γ treatment (10 ng/ml) as indicated. Extracts were prepared, and the luciferase assay was performed. The combination of transfected plasmids and IFN-γ treatment is shown at the bottom. Luciferase light units are plotted on the y axis as fold activation in response to IFN-γ. Relative luciferase activity was normalized to a cotransfected β-galactosidase internal control.
Complementation of the U3A cells by stable transfection with a STAT1α expression vector produced a line, G8, in which IFN-γ responses are fully restored (11, 12, 34, 35). Similar results were obtained in G8 cells after IFN-γ treatment with cotransfection of BRCA1 (Fig. 1, lanes 9–12). These results indicate that the additive enhancement between BRCA1 and IFN-γ requires, at least, the STAT1α protein. To demonstrate the generality of this effect in an efficiently transfectable cell line, a similar experiment was performed in 293T cells. IFN-γ stimulation of 3x Ly6e-LUC in 293T cells resulted in an approximately fivefold increase in luciferase activity (lane 14). In these cells, expression of BRCA1 increased the basal activity of the reporter from five- to sevenfold over background (lane 15), but again a strong enhancement was observed on IFN-γ stimulation, leading to a 15-fold increase in reporter gene activity (lane 16).
Differential Regulation of Endogenous IFN-γ Target Genes by BRCA1.
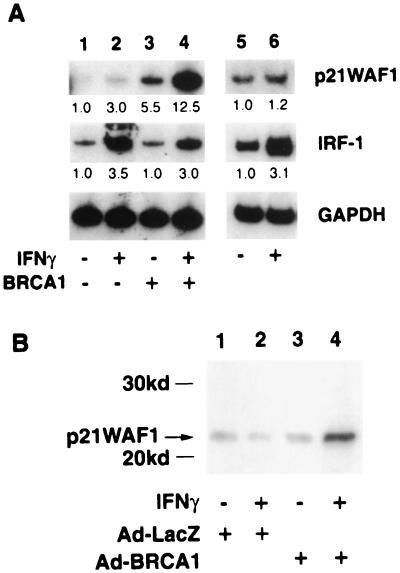
The response of two endogenous genes known to be transcriptionally induced by IFN-γ stimulation was next tested. Both p21WAF1 and IRF-1 are known to be induced by activated STAT1 in response to IFN-γ stimulation (39, 42); 293T cells were transfected with control vector or BRCA1 expression plasmid, and treated with IFN-γ for 4 h. Total RNA was extracted and subjected to RT-PCR analysis for detection of endogenous p21WAF1 or IRF-1 mRNA expression. For IRF-1, a low basal mRNA level was stimulated 3.5-fold in response to IFN-γ treatment (Fig. 2A Middle, lane 2). The basal mRNA level of p21WAF1 was extremely low in our assay, but IFN-γ treatment resulted in a threefold stimulation (Fig. 2A Upper, lane 2). Expression of BRCA1 had no apparent effect on the levels or inducibility of the IRF-1 mRNA (Fig. 2A Middle, lane 3), but had a dramatic effect on expression of p21WAF1 (Fig. 2A Upper, lane 3). The combination of IFN-γ treatment and BRCA1 transfection resulted in more than 12-fold induction compared with IFN-γ treatment alone or BRCA1 expression alone (Fig. 2A Upper, lane 4). In contrast, IRF-1 did not show any further activation by BRCA1 after IFN-γ treatment (Fig. 2 Upper, lane 4). We also tested other IFN-γ target genes such as SMAD7 and IP10, but the synergistic effect of BRCA1 on IFN-γ stimulation was not observed (data not shown). Consistent with our findings using a STAT responsive reporter gene, these results indicate that endogenous IFN-γ-responsive genes may be regulated by a BRCA1–STAT1 complex, and suggests that BRCA1 acts as a specific and not general coactivator of gene expression.
Figure 2.
IFN-γ responsive genes are differentially regulated by BRCA1. (A) Quantitative RT-PCR analysis of p21WAF1 and IRF-1. 293T cells were transiently transfected with vector (lanes 1 and 2) or BRCA1 expression plasmid for 48 h (lanes 3 and 4). Before total RNA extraction, 293T cells and HCC1937 breast cancer cells were treated with IFN-γ (10 ng/ml, 4 h) (lanes 2, 4, and 6). Fragments of p21WAF1, IRF-1, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were PCR amplified with [α-32P]dCTP by appropriate primers (see Materials and Methods). After 5% acrylamide gel electrophoresis, gels were dried and autoradiography performed. No amplification was observed without reverse transcription of the RNA (not shown). Relative induction from phosphorimaging of genes is indicated under the panels. (B) Expression level of p21WAF1 protein in G8 cells treated with recombinant adenovirus plus IFN-γ. Cells were infected with Ad-LacZ or Ad-BRCA1 virus for 2 days. Twelve hours before assay, cells were treated with IFN-γ, and immunoblot analysis was done to study the p21WAF1 protein level.
To further investigate the requirement for BRCA1 in this differential regulation of IFN-γ target genes, RT-PCR analysis was performed in the breast cancer cell line, HCC1937, homozygous for the BRCA1 5382C mutation (43). This insertion mutation causes premature termination of the BRCA1 protein resulting in the lack of a functional BRCT domain in the C terminus. Expression of IRF-1 mRNA was increased by IFN-γ stimulation of HCC1937 cells (Fig. 2A Middle, lanes 5 and 6), but IFN-γ-dependent induction of p21WAF1 was abolished in these BRCA1 mutant cells (Fig. 2A Upper, lanes 5 and 6).
Induction of p21WAF1 was also examined by Western blot analysis by using G8 cells; these cells were infected with recombinant adenovirus of LacZ or BRCA1, and cells were treated with IFN-γ for 5 h before assay. Like the induction of p21 mRNA shown by RT-PCR analysis of 293T cells, p21WAF1 protein was significantly increased by IFN-γ and BRCA1 (Fig. 2B, lane 4). Induction of p21WAF1 protein was not observed in STAT1-negative U3A cells (data not shown).
BRCA1 Interacts with the STAT1α Transcriptional Activation Domain.
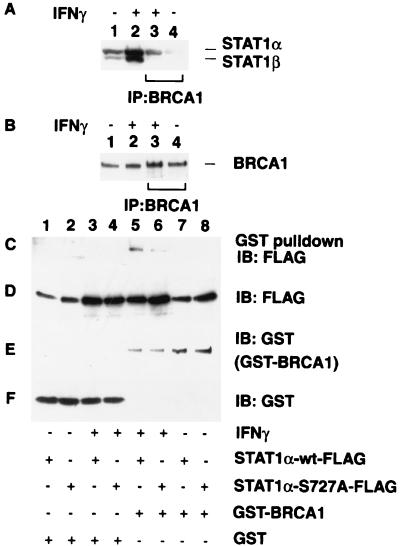
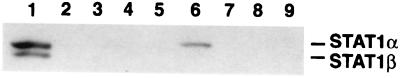
To determine if STAT1 and BRCA1 form a physical complex, the endogenous BRCA1 was immunoprecipitated with an anti-BRCA1 mAb from extracts of 293T cells which were unstimulated or stimulated with IFN-γ for 12 h. Because of the low affinity of the anti-BRCA1 antibody, large amounts of cell extract were required for these immunoprecipitation experiments. IFN-γ treatment increased the level of cellular STAT1α and STAT1β proteins two- to threefold, but the endogenous BRCA1 level was not altered (Fig. 3 A and B, lanes 1 and 2). STAT1 was specifically coimmunoprecipitated with BRCA1 after IFN-γ stimulation (Fig. 3A, lanes 3 and 4). The coprecipitated STAT1 is primarily the full-length STAT1α (Fig. 3A, lane 3), but a minor amount of STAT1β could also be detected in the immunoprecipitates of BRCA1, likely because of mixed heterodimers of STAT1α and STAT1β.
Figure 3.
STAT1α and BRCA1 form a complex both in vitro and in vivo. (A and B) Coimmunoprecipitation of endogenous BRCA1 and STAT1α from IFN-γ-treated 293T cells. BRCA1 was immunoprecipitated from 1.2 mg of IFN-γ-treated (10 ng/ml, 12 h) (lane 3) or untreated (lane 4) cells. Total cell lysates (20 μg) were also analyzed for the positive control of protein expression (lanes 1 and 2). Samples were separated by 6% SDS/PAGE, and immunoblotted with (A) anti-STAT1 antibody or (B) anti-BRCA1 antibody . (C) Copurification of STAT1 with BRCA1. GST-BRCA1 and FLAG-tagged STAT1α or STAT1αSer-727A were cotransfected into 293T cells. For controls, separate plates were transfected with GST vector alone or GST-BRCA1 expression vector with or without FLAG-tagged STAT1α expression vector in the combinations shown at the bottom of the panel. Cells were treated with IFN-γ as indicated (10 ng/ml, 12 h) (lanes 3–6), and GST or GST-BRCA1 was purified from extracts with glutathione beads. Samples were separated by 6% SDS/PAGE, and immunoblotted with anti-FLAG antibody. (D, E, and F) Detection of exogenously expressed proteins. Total cell lysates (20 μg) were separated by 6% (D and E) or 10% (F) SDS/PAGE, and the expression level of FLAG-tagged STAT1α (D), GST-BRCA1 (E), and GST (F) were confirmed by immunoblot analysis by using anti-FLAG or anti-GST antibody.
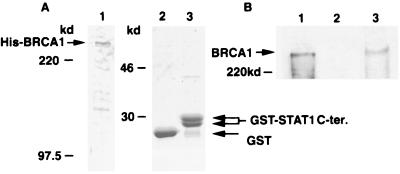
In the activation of STAT1, IFN-γ induces Ser-727 phosphorylation which is crucial for maximal transcriptional activation (12, 44). To test the importance of Ser-727, in vitro binding assays were performed by cotransfection of 293T cells with expression vectors encoding a GST-BRCA1 fusion protein and expression vectors encoding FLAG-tagged versions of either the wild-type or Ser-727A STAT1α. When cells were not treated with IFN-γ, no STAT1 copurification was observed (Fig. 3C, lanes 7 and 8), nor was any STAT1 protein recognized by the GST protein alone even after IFN-γ treatment (Fig. 3C, lanes 1–4). In contrast, after IFN-γ stimulation, GST-BRCA1 was capable of binding the wild-type STAT1α protein (Fig. 3C, lane 5). The Ser-727A mutant was also capable of binding BRCA1 after IFN-γ treatment, but the interaction was greatly reduced compared with that of wild-type STAT1α (Fig. 3C, lane 6). This suggests that Ser-727 plays an important role in BRCA1 binding, and is consistent with the reduced ability of this mutant to bind cofactors and regulate transcription (12, 15). Although the in vitro interaction between STAT3 and BRCA1 was also tested, we could not detect the binding (data not shown). To further confirm the binding of BRCA1 to the STAT1 C-terminal region, we performed the coprecipitation assay by using purified proteins. Full-length His-tagged BRCA1 was partially purified from baculovirus-infected Sf9 cells through Ni column (Fig. 4A, lane 1). BRCA1 protein (500 ng) was incubated with 1 μg of a GST fusion protein expressing the STAT1α C-terminal 38 aa or GST alone. After precipitation with glutathione beads, coprecipitated BRCA1 was detected by immunoblot analysis (Fig. 4B, lane 3). The STAT1 C terminus clearly bound to the BRCA1 protein whereas the GST carrier did not. This result suggests that the two proteins are able to bind directly.
Figure 4.
Purified BRCA1 binds to STAT1α C terminus 38 aa. (A) Purification of full-length BRCA1 and GST-STAT1α C terminus. Full-length BRCA1 was partially purified through a Ni column from Sf9 cells infected with recombinant baculovirus, BVC8B, expressing His-tagged BRCA1. GST-fused STAT1α C terminus was bacterially expressed, and purified through glutathione beads. Coomassie staining of BRCA1 (1 μg, lane 1), GST (5 μg, lane 2), and GST-STAT1C terminus (5 μg, lane 3) are shown. Arrows indicate the purified proteins. (B) Detection of binding of purified BRCA1 to STAT1α C terminus. Purified BRCA1 (500 ng) was incubated with 1 μg of GST (lane 2) or GST-STAT1α C terminus (lane 3). After precipitation with glutathione beads, samples were subjected to immunoblot analysis by using anti-BRCA1 antibody, Ab-1. Purified BRCA1 (200 ng) was loaded as a control (lane 1).
Collectively, these protein interaction results indicate that the C-terminal transcription activation domain of STAT1α can serve as a specific binding site for BRCA1 and that the phosphorylation acceptor, Ser-727, can enhance the recruitment BRCA1.
STAT1α Binds to the Central Region of BRCA1.
The region of BRCA1 that interacts with STAT1α was next determined. Lysate from IFN-γ-treated 293T cells were incubated with six different BRCA1-GST fusion proteins (24) bound to glutathione Sepharose beads, and bound proteins were released and immunoblotted to detect STAT1. The GST-BRCA1 segment containing aa 502–802 specifically bound to the STAT1α protein but not STAT1β (Fig. 5, lane 6), consistent with results indicating that BRCA1 preferentially binds to STAT1α (Fig. 3). These results lend further support to the notion that BRCA1 binds directly to the C-terminal 38-aa transcriptional activation domain of STAT1α that is absent in STAT1β.
Figure 5.
Identification of the STAT1-binding segment of BRCA1. Six GST-BRCA1 fusion proteins (aa 1–324; 260–553; 502–802; 758–1,064; 1,005–1,313; and 1,314–1,863) were generated in E. coli, and used for an in vitro binding assay (from lanes 4 to 9, respectively). GST protein alone was used for lane 3. Approximately equal amounts of each GST-fusion protein, bound to glutathione-Sepharose beads, were incubated with an extract of IFN-γ-treated (10 ng/ml, 15 min) 293T cells. Bound proteins were recovered and separated electrophoretically. Total lysate (20 μg) was loaded as a control (lane 1). Lane 2 contains no protein. The separated proteins were immunoblotted for STAT1. This STAT1 antiserum recognizes a 91-kDa (STAT1α), not 84-kDa (STAT1β), protein bound to GST-BRCA1 (aa 502–802) in lane 6, showing that STAT1α specifically binds to BRCA1.
IFN-γ Induces Binding of BRCA1 to the STAT1α Transcriptional Activation Domain.
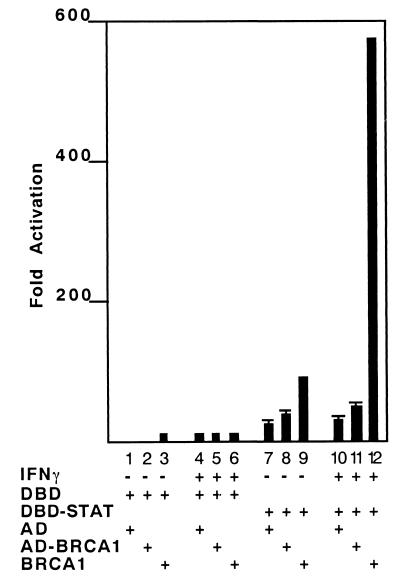
To more directly test the function of the interaction between STAT1 and BRCA1, a mammalian two-hybrid assay was performed. The BRCA1 segment containing aa 502–802 was fused with the heterologous GAL4 activation domain (GAL4AD). The STAT1 C-terminal segment containing transactivation and the BRCA1-binding domain (aa 712–750) was also expressed as a fusion protein with a GAL4 DNA-binding region. The hybrid proteins were coexpressed with or without IFN-γ stimulation, and activity of a GAL4 reporter was measured. As shown in Fig. 6, GAL4DBD-STAT1(712–750) stimulated the reporter gene activity up to 16-fold, and the reporter gene activity was only weakly increased by IFN-γ stimulation up to 20-fold (Fig. 6, lanes 7 and 10), consistent with other reports using artificial GAL4DBD-STAT1(712–750) proteins (15). An antibody specifically recognizing phosphorylated Ser-727 of STAT1α equally reacted with GAL4DBD-STAT1(712–750) with or without IFN-γ treatment (data not shown), suggesting that this hybrid protein is at least partially phosphorylated constitutively. Coexpression of GAL4AD-BRCA1(502–802) with GAL4DBD-STAT1(712–750) further activated the reporter gene by up to twofold (lanes 8 and 11). No other domain of STAT1 tested was capable of either activating transcription by itself or cooperating with BRCA1 (data not shown). These results confirm that the region of BRCA1 between aa 502–802 interacts with the C-terminal transcriptional activation domain of STAT1α.
Figure 6.
Mammalian two-hybrid analysis demonstrates functional interaction between BRCA1 and STAT1. Both BRCA1 (502–802) segment fused with GAL4AD (AD-BRCA1) and full-length BRCA1 (BRCA1) can hyperactivate a STAT1α C-terminal transcriptional activation domain fused with GAL4 DBD (DBD-STAT). The reporter plasmids contain four copies of GAL4-binding sites upstream of the luciferase gene. Each construct shown at the bottom of the panel was expressed in 293T cells transiently with (+) or without (−) IFN-γ treatment (10 ng/ml, 6 h). Relative activity was normalized with a β-galactosidase control. In all cases, luciferase activity was determined at 48 h posttransfection.
To evaluate the ability of the complete BRCA1 protein to cooperate with the STAT1α transcriptional activation domain, the activity of GAL4DBD-STAT1(712–750) was examined with and without full-length BRCA1 cotransfection. Although GAL4DBD-STAT1(712–750) activated the reporter gene when coexpressed with BRCA1 (Fig. 6, lane 9), IFN-γ stimulation enhanced activation more than 500-fold over basal activity (Fig. 6, lane 12). This result strengthens the conclusion that the BRCA1 protein interacts functionally with the STAT1α transcriptional activation domain in IFN-γ signaling.
Discussion
The present studies provide the first demonstration that BRCA1 is required for induction of a subset of IFN-γ target genes as a coactivator for the transcription factor, STAT1α. The mechanism of transcriptional cooperation between the two proteins involves interaction between the BRCA1 residues 502–802 and the STAT1 transcriptional activation domain.
Studies from several laboratories have implied that the BRCA1 protein might be able to function as a transcriptional activator. BRCA1 can stimulate transcription of several promoter reporter genes, including the cyclin-dependent kinase inhibitor, p21WAF1. Our results demonstrate a critical role for STAT1–BRCA1 interaction in activation of the p21WAF1 gene by IFN-γ. It is not known if STAT1 requires wild-type p53 for maximal activation of the p21WAF1 gene, but it is important to note that the 2fTGH, U3A, and G8 cell lines express wild-type p53 (G. R. Stark, personal communication).
The C-terminal 38 aa of STAT1α have an intrinsic transcriptional activation activity when fused with the GAL4DBD (ref. 15 and Fig. 6). Although phosphorylation of the Ser-727 in this region is required for maximal transcriptional activation, little is known concerning how Ser-727 phosphorylation regulates STAT1 activity. Recent work has illustrated that the C-terminal region of STAT1α has a capacity to bind to multiple cellular proteins including CBP/p300 and MCM5, and that this binding can be enhanced by Ser-727 phosphorylation after IFN-γ stimulation (15). We postulate that the recruitment of specific pol II coactivators by IFN-γ-activated STAT1 dimers relies in part on the coordination with other promoter-bound factors. It is conceivable that the choice of coactivators determines the differential gene regulation leading to subsets of biological outcomes. Our mammalian two-hybrid analysis showed that the full-length BRCA1 is a more potent activator of STAT1 than the isolated STAT1-binding region. It is possible that the full-length BRCA1 protein is itself posttranslationally modified by IFN-γ treatment, resulting in greater capacity for transcriptional activation.
Although many studies suggest a transcriptional activation function for BRCA1 based on reporter gene activation, it is unclear how mutations at the BRCA1 locus that are found in breast and ovarian cancer affect specific gene regulation. Our evidence showing that IFN-γ induction of p21WAF1, not IRF-1, is impaired in BRCA1 mutant cells, and that p21WAF1, not IRF-1, can be stimulated synergistically by IFN-γ and BRCA1, clearly demonstrates that STAT1 requires BRCA1 for specific gene induction events. Whereas the mutant form of BRCA1 expressed in HCC1937 cells still retains the STAT1-binding region, the protein lacks the BRCT domain. Because this BRCA1 variant is likely to retain the capacity to bind to STAT1, the impaired induction of p21WAF1 in HCC1937 cells after IFN-γ stimulation suggests that the mutant BRCA1 may serve as a dominant-negative protein which interferes with the link between activated STAT1 and the basal transcriptional machinery. In fact, we examined the reconstitution of BRCA1 in HCC1937 cells by Ad-BRCA1 infection, but p21WAF1 expression was not restored on IFN-γ stimulation (data not shown). Further research is required to determine if BRCA1 mutations exist which do not bind to STAT1, but retain the intact BRCT domain.
The differential regulation of the IFN-γ target genes by BRCA1/STAT1 coactivation raises the question of how BRCA1 achieves promoter specificity. There are currently no data in support of the BRCA1 protein directly contacting DNA, but this issue requires further investigation. Recent studies of the ability of the BRCT domain to initiate transcription in vitro determined that BRCA1 prefers a supercoiled template and was virtually nonfunctional from a linear template (45). The conclusion drawn from these studies is that the BRCT transcriptional activity is highly dependent on the negative superhelical topology of the template DNA. Because negatively supercoiled DNA contains a high percentage of single-stranded regions (46), this finding suggests that BRCA1 prefers partially single-stranded promoters. It is worth noting that the other transcription factor known to collaborate with BRCA1 on this promoter, p53, has a well-studied ability to bind single-stranded DNA (37, 47). We suggest that altered chromatin structure of the p21WAF1 promoter might be a major determinant of the differential regulation of the STAT1 target genes by BRCA1. In this respect, it is noteworthy that the three factors known to be recruited by the STAT1α transcriptional activation domain, CBP/p300, MCM5, and BRCA1 all have functions that have been linked to DNA topology.
A unique biological role of IFN-γ in antitumor immunity has been recently postulated (7, 8). Genetic abnormalities affecting proteins involved in IFN-γ signaling might impede the initial control of nascent tumor cell proliferation. Our findings provide evidence supporting this concept. The loss of BRCA1 function would decrease IFN-γ-dependent gene regulation events that limit cell growth. We demonstrated that the p21WAF1 gene was synergistically activated by BRCA1 and IFN-γ in intact cells, but this synergy is impaired in cells lacking a functional BRCA1 protein. It is possible that the disturbance of the p21WAF1 induction by IFN-γ provides an early growth advantage to nascent tumor cells, allowing them to bypass the initial antitumor actions of IFN-γ. Our demonstration that IFN-γ may control cell growth regulation through direct STAT1–BRCA1 interaction provides not only a new insight into the mechanisms behind IFN-γ-mediated growth inhibition, but also provides insight into the differential regulation of gene expression depending on recruitment of transcriptional coactivators. In addition, these findings present a new mechanism by which disturbance of STAT1/BRCA1-mediated gene expression might contribute to cancer development.
Acknowledgments
We thank many colleagues for their generous sharing of reagents and stimulating discussion. In particular, we thank Drs. George R. Stark for a gift of cell lines and Ralph Scully for BRCA1 GST constructs. We also thank Drs. H. Hanafusa and J. E. Darnell for continued encouragement and comments on this manuscript. This work was supported by National Institute of Health Grant 1P50CA68425 (SPORE) to S.A.A., RO1CA78356 to S.W.L., and R01CA79892 to T.O, who is also a recipient of an Incentive Award from Mount Sinai School of Medicine. During the course of these studies, C.M.H. was a Special Fellow of the Leukemia Society of America and is currently the Ira M. Jacobson, MD Liver Scholar of the American Liver Foundation, and recipient of the New York City Council Speaker's Fund for Biomedical Research.
Abbreviations
- STAT
signal transducers and activators of transcription
- GST
glutathione S-transferase
- RT-PCR
reverse transcription–PCR
- GAL4DBD
GAL4 DNA-binding domain
- BRCT
BRCA1 C terminus
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.080469697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.080469697
References
- 1.Boehm U, Klamp T, Groot M, Howard J C. Annu Rev Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 2.Balkwill F R, Oliver R T D. Int J Cancer. 1977;20:500–505. doi: 10.1002/ijc.2910200405. [DOI] [PubMed] [Google Scholar]
- 3.Balkwill F, Taylor-Papadimitriou J. Nature (London) 1978;274:798–801. doi: 10.1038/274798a0. [DOI] [PubMed] [Google Scholar]
- 4.Bromberg J, Horvath C M, Wen Z, Schreiber R D, Darnell J E., Jr Proc Natl Acad Sci USA. 1996;93:7673–7678. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horvath C M, Darnell J E., Jr J Virol. 1996;70:647–650. doi: 10.1128/jvi.70.1.647-650.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin S, Kikuchi T, Pledger W J, Tamm I. Science. 1986;233:356–359. doi: 10.1126/science.3726533. [DOI] [PubMed] [Google Scholar]
- 7.Coughlin C M, Salhany K E, Gee M S, LaTemple D C, Kotenko S, Ma X, Gri G, Wysocka M, Kim J E, Liu L, et al. Immunity. 1998;9:25–34. doi: 10.1016/s1074-7613(00)80585-3. [DOI] [PubMed] [Google Scholar]
- 8.Kaplan D H, Shankaran V, Dighe A S, Stockert E, Aguet M, Old L J, Schreiber R D. Proc Natl Acad Sci USA. 1998;95:7556–7561. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 10.Schindler C, Fu X-Y, Improta T, Aebersold R, Darnell J E., Jr Proc Natl Acad Sci USA. 1992;89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muller M, Laxton C, Briscoe J, Schindler C, Improta T, Darnell J E, Jr, Stark G R, Kerr I M. EMBO J. 1993;12:4221–4228. doi: 10.1002/j.1460-2075.1993.tb06106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wen Z, Zhong Z, Darnell J E., Jr Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 13.Wen Z, Darnell J E., Jr Nucleic Acids Res. 1997;25:2062–2067. doi: 10.1093/nar/25.11.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J J, Vinkemeier U, Gu W, Chakravarti D, Horvath C M, Darnell J E., Jr Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J J, Zhao Y, Chait B T, Lathem W W, Ritzi M, Knippers R, Darnell J E., Jr EMBO J. 1998;17:6963–6971. doi: 10.1093/emboj/17.23.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horvai A E, Xu L, Korzus E, Brard G, Kalafus D, Mullen T-M, Rose D W, Rosenfield M G, Glass C K. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miki Y, Swensen J, Shattuck-Eidens D, Futreal P A, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett L M, Ding W, et al. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 18.Koonin E V, Altschul S F, Bork P. Nat Genet. 1996;13:266–268. doi: 10.1038/ng0796-266. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Tombline G, Weber B L. Cell. 1998;92:433–436. doi: 10.1016/s0092-8674(00)80936-8. [DOI] [PubMed] [Google Scholar]
- 20.Holt J T, Thompson M E, Szabo C, Robinson-Benion C, Arteaga C L, King M C, Jensen R A. Nat Genet. 1996;12:298–302. doi: 10.1038/ng0396-298. [DOI] [PubMed] [Google Scholar]
- 21.Holt J T. Ann NY Acad Sci. 1997;833:34–41. doi: 10.1111/j.1749-6632.1997.tb48590.x. [DOI] [PubMed] [Google Scholar]
- 22.Somasundaram K, Zhang H, Zeng Y X, Houvras Y, Peng Y, Wu G S, Licht J D, Weber B L, El-Deiry W S. Nature (London) 1997;389:187–190. doi: 10.1038/38291. [DOI] [PubMed] [Google Scholar]
- 23.Tait D L, Obermiller P S, Redlin-Frazier S, Jensen R A, Welcsh P, Dann J, King M C, Johnson D H, Holt J T. Clin Cancer Res. 1997;3:1959–1968. [PubMed] [Google Scholar]
- 24.Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston D M. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 25.Scully R, Chen J, Ochs R L, Keegan K, Hoekstra M, Feunteun J, Livingston D M. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 26.Gowen L C, Avrutskaya A V, Latour A M, Koller B H, Leadon S A. Science. 1998;281:1009–1012. doi: 10.1126/science.281.5379.1009. [DOI] [PubMed] [Google Scholar]
- 27.Chapman M S, Verma I M. Nature (London) 1996;382:678–679. doi: 10.1038/382678a0. [DOI] [PubMed] [Google Scholar]
- 28.Monteiro A N, August A, Hanafusa H. Proc Natl Acad Sci USA. 1996;93:13595–13599. doi: 10.1073/pnas.93.24.13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ouchi T, Monteiro A N, August A, Aaronson S A, Hanafusa H. Proc Natl Acad Sci USA. 1998;95:2302–2306. doi: 10.1073/pnas.95.5.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scully R, Anderson S F, Chao D M, Wei W, Ye L, Young R A, Livingston D M, Parvin J D. Proc Natl Acad Sci USA. 1997;94:5605–5610. doi: 10.1073/pnas.94.11.5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neish A S, Anderson S F, Schlegel B P, Wei W, Parvin J D. Nucleic Acids Res. 1998;26:847–853. doi: 10.1093/nar/26.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson S F, Schlegel B P, Nakajima T, Wolpin E S, Parvin J D. Nat Genet. 1998;19:254–256. doi: 10.1038/930. [DOI] [PubMed] [Google Scholar]
- 33.Pao G M, Janknecht R, Ruffner H, Hunter T, Verma I M. Proc Natl Acad Sci USA. 2000;97:1020–1025. doi: 10.1073/pnas.97.3.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horvath C M, Stark G R, Kerr I M, Darnell J E., Jr Mol Cell Biol. 1996;16:6957–6964. doi: 10.1128/mcb.16.12.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horvath C M, Wen Z, Darnell J E., Jr Genes Dev. 1995;9:984–994. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
- 36.Mayer B J, Baltimore D. Mol Cell Biol. 1994;14:2883–2894. doi: 10.1128/mcb.14.5.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oberosler P, Hloch P, Ramsperger U, Stahl H. EMBO J. 1993;12:2389–2396. doi: 10.1002/j.1460-2075.1993.tb05893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He T C, Zhou S, da Costa L T, Yu J, Kinzler K W, Vogelstein B. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chin Y E, Kitagawa M, Su W C, You Z H, Iwamoto Y, Fu X Y. Science. 1996;272:719–722. doi: 10.1126/science.272.5262.719. [DOI] [PubMed] [Google Scholar]
- 40.Pellegrini S, John J, Shearer M, Kerr I M, Stark G R. Mol Cell Biol. 1989;9:4605–4612. doi: 10.1128/mcb.9.11.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKendry R, John J, Flavell D, Muller M, Kerr I M, Stark G R. Proc Natl Acad Sci USA. 1991;88:11455–11459. doi: 10.1073/pnas.88.24.11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, Furia A, Miyata T, Taniguchi T. Cell. 1989;58:729–739. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- 43.Tomlinson G E, Chen T T, Stastny V A, Virmani A K, Spillman M A, Tonk V, Blum J L, Schneider N R, Wistuba I I, Shay J W, et al. Cancer Res. 1998;58:3237–3242. [PubMed] [Google Scholar]
- 44.Zhu X, Wen Z, Xu L Z, Darnell J E., Jr Mol Cell Biol. 1997;17:6618–6623. doi: 10.1128/mcb.17.11.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haile D T, Parvin J D. J Biol Chem. 1999;274:2113–2117. doi: 10.1074/jbc.274.4.2113. [DOI] [PubMed] [Google Scholar]
- 46.Parvin J D, Sharp P A. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 47.Jayaraman J, Prives C. Cell. 1995;81:1021–1029. doi: 10.1016/s0092-8674(05)80007-8. [DOI] [PubMed] [Google Scholar]