Abstract
Changes in gene expression play an important role in species' evolution. Earlier studies uncovered evidence that the effect of mutations on expression levels within the primate order is skewed, with many small downregulations balanced by fewer but larger upregulations. In addition, brain-expressed genes appeared to show an increased rate of evolution on the branch leading to human. However, the lack of a mathematical model adequately describing the evolution of gene expression precluded the rigorous establishment of these observations. Here, we develop mathematical tools that allow us to revisit these earlier observations in a model-testing and inference framework. We introduce a model for skewed gene-expression evolution within a phylogenetic tree and use a separate model to account for biological or experimental outliers. A Bayesian Markov chain Monte Carlo inference procedure allows us to infer the phylogeny and other evolutionary parameters, while quantifying the confidence in these inferences. Our results support previous observations; in particular, we find strong evidence for a sustained positive skew in the distribution of gene-expression changes in primate evolution. We propose a “corrective sweep” scenario to explain this phenomenon.
THE genetic mechanisms underlying the phenotypic evolution of species are still poorly understood. More than 30 years ago, it was proposed that regulatory changes may have played a major role in the evolution of species and in particular in the rapid emergence of human-specific traits (King and Wilson 1975). It appears likely that in general, gene-expression levels are more closely related to the phenotypes upon which selection acts than the DNA sequence itself, motivating the study of their evolution. With the advent of microarray technology, the measurement of transcript levels on a genomewide scale and across species and individuals is now economical, opening the way for a systematic study of gene-expression evolution.
Quantitative traits such as transcript expression levels pose specific challenges. In contrast to sequence data, the variance of quantitative traits includes components of experimental error, and environmental and genetic variation, besides the evolutionary component of interest here. Separating these components is problematic, making it difficult to establish in particular cases whether or not the expression level of a gene has undergone a mutation. This difficulty may have contributed to the fact that previous studies have arrived at different conclusions as to the major modes of evolution of gene expression, ranging from neutral evolution (Khaitovich et al. 2004; De Meaux et al. 2005; Keightley et al. 2005), to stabilizing selection (Denver et al. 2005; Gilad et al. 2005; Lemos et al. 2005), to directional selection (Gilad et al. 2006).
Most recent studies of expression-level evolution have compared variances within and between species and classified genes as either differentially expressed or unchanged on the basis of thresholds of P-values obtained from multifactorial linear modeling of fluorescent probe log-intensity readings (Hsieh et al. 2003; Rifkin et al. 2003; Nuzhdin et al. 2004; Denver et al. 2005; Gilad et al. 2006; Oshlack et al. 2007). The loss of information inherent in such a dichotomous classification reduces the power of this approach. In addition, the environmental and genetic within-population components of the variance of these log-intensity readings may differ between species or experiments. These variance components can be difficult to measure (Gilad et al. 2006), but affect the power of the statistical tests and therefore weaken the conclusions reached by these studies.
A more principled approach to study quantitative traits is to explicitly model their evolution. In primates and flies, it was observed that the squared deviation of expression phenotypes increases linearly with divergence time (Rifkin et al. 2003; Khaitovich et al. 2004, 2005b). This observation is compatible with neutral diffusion-type models for quantitative trait evolution (Edwards and Cavalli-Sforza 1964; Felsenstein 1973; Lande 1976; Lynch and Hill 1986; Turelli et al. 1988; Lemos et al. 2005) as well as with directional and stabilizing selection over sufficiently short timescales (Felsenstein 2004; Khaitovich et al. 2004; Lemos et al. 2005). However, two aspects of gene-expression evolution are not very well captured by these diffusion-type models. First, while the traits themselves are continuous, their heritable component is encoded in DNA, and mutations may therefore be supposed to occur as discrete events rather than as a continuous diffusion. Although a continuous approximation is justifiable over long times, for evolution over short time intervals the granularity of the process might conceivably have an impact on observables. Second, the spectrum of expression-level changes exhibits a skew, so that while expression levels remain constant in expectation, this appears to be brought about by many small downregulations combined with fewer upregulations of a larger average magnitude (Khaitovich et al. 2005b), a feature not accounted for in existing diffusion-type models.
Here we introduce a new probabilistic model of gene-expression evolution that incorporates these characteristics. While probabilistic approaches have been used extensively to study nucleotide and amino acid sequences in an evolutionary perspective (methods reviewed in Durbin et al. 1998; Felsenstein 2004), relatively few authors have considered analogous methods, and in particular likelihood models, to investigate the evolution of expression data, the characteristics of which render the standard discrete-state models for nucleotide evolution inadequate (Felsenstein 1973; Oakley et al. 2005). Advantages of a probabilistic approach include the ability to do parameter inference with confidence intervals, to test the goodness-of-fit of alternative models, and to test hypotheses such as the existence of a phylogenetic signal.
We are particularly interested in investigating whether in recent evolution, more expression-level mutations have occurred in the human or in the chimpanzee branch. An analysis of gene-expression levels from human, chimpanzee, orangutan, and rhesus macaque samples previously suggested that more changes have occurred on the lineage leading up to humans (Enard et al. 2002b; Khaitovich et al. 2005b, 2006; Lemos et al. 2005). Here, we revisit these original observations, both the skew in the expression-mutation process and the excess of mutations in the human branch.
Our model has a number of features that distinguish it from previous approaches. One often-made assumption is that mutations cause changes in the relative transcript abundance, independent of the absolute level of expression. This would imply that the spectrum of expression changes on a logarithmic scale is independent of the absolute expression; however, we do not observe such independence. Here, instead of making this assumption, we replace the log transformation with a variance-stabilizing transformation that explicitly accounts for any level dependence of the interspecific variance, extending an approach introduced by Huber et al. (2002). In addition, we account for intraspecific variance and measurement errors by modeling the observed expression by a Gaussian distribution.
A second feature of our model is that we explicitly model outliers. Since the evolutionary model is relatively constrained, this outlier model ensures that nucleotide mutations resulting in mismatching probes, annotation errors, or indeed genes that have undergone strong directional selection do not dominate the final likelihood and thereby unduly influence parameter estimates. The proportion of genes that are deemed to be outliers is estimated alongside the other model parameters and provides an indication of the model fit and data quality. We chose the infinite-variance Cauchy distribution on a star-tree topology to model outliers, as this heavy-tailed distribution allows wide outliers to have relatively little effect on the likelihood.
To model the evolution along branches of the phylogenetic tree, we use the compound Poisson model introduced by Khaitovich et al. (2005b). In this model, mutations are modeled as discrete events that occur at a constant rate, and each mutation changes the intensity by a random amount that is drawn from a specified “jump-size” distribution. This distribution, which has mean 0, has two parameters determining its variance and skewness. A nonzero skewness confers a direction to the evolutionary process, and this time irreversibility allows us to infer rooted phylogenies without reference to an outgroup, even when expression profiles for only two species are available. We compute the likelihood of the data given the model and its parameters using an extension of Felsenstein's pruning algorithm (Felsenstein 1981). The fast Fourier transform algorithm allows an implementation that is efficient enough to use the Bayesian Markov chain Monte Carlo approach to infer parameters and credible intervals.
The expression level of a single gene, measured across a number of species, does not contain sufficient information to infer all model parameters. We therefore combine data across many genes by making two additional assumptions: that expression levels evolve independently for each gene, and that the evolutionary model is the same for all genes. While independence of expression will not hold in general, we show by simulation studies that the inference procedure is robust against quite substantial departures from independence. The second assumption, that genes in different categories (for example, those expressed in the brain vs. the liver) evolve according to the same rule, is not satisfied (see, e.g., Khaitovich et al. 2005a; Voolstra et al. 2007). Here, we have chosen not to complicate our analysis by differentiating between classes of genes, but rather to provide an initial, broad view of gene-expression evolution. Nevertheless, the variation of mutations rates and other evolutionary parameters across gene type is a topic that clearly warrants further investigation.
Here, we test the ability of our method to estimate branch lengths of two-species and three-species trees in a simulation study. We find that our model is indeed able to infer the correct phylogeny and branch lengths within their confidence intervals. We then apply our method to published sets of expression-profile data on the brain of humans and related primates, to infer the characteristics of the evolutionary process and the branch lengths of the phylogenetic tree relating these primates.
MATERIALS AND METHODS
Interspecies variance-stabilizing transformation:
The model defined below describes the evolution of a gene's “transformed expression,” E, rather than of the observed normalized intensity I. This transformed expression E is related to the observed intensity I through a transformation (unique up to a linear change of scale) that renders the interspecific variance vE independent of the expression level. If the transformation from normalized intensity is E = E(I), then 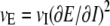 , so that vE is constant if
, so that vE is constant if 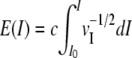 , with c and I0 arbitrary constants. For convenience, we first applied a log transformation to reduce the range of v and fitted a piecewise analytic function to the interspecific variance in log-transformed coordinates to compute E(I); this is where we depart from Huber et al. (2002), who use a two-parameter family for fitting. To simplify comparisons, we chose c so that the range of transformed expression levels roughly coincided with the range of log-transformed raw intensities (∼1–45). The same transformation was used for all species, and we ensured that no systematic across-species deviations remained by equalizing the within-species medians.
, with c and I0 arbitrary constants. For convenience, we first applied a log transformation to reduce the range of v and fitted a piecewise analytic function to the interspecific variance in log-transformed coordinates to compute E(I); this is where we depart from Huber et al. (2002), who use a two-parameter family for fitting. To simplify comparisons, we chose c so that the range of transformed expression levels roughly coincided with the range of log-transformed raw intensities (∼1–45). The same transformation was used for all species, and we ensured that no systematic across-species deviations remained by equalizing the within-species medians.
Expression evolution along a branch:
The evolution of expression levels along a branch of the phylogenetic tree is described by a compound Poisson process, with the rate parameter fixed to 1 so that branch lengths are measured in units of expected number of mutation events. This model was proposed by Khaitovich et al. (2005b); however, rather than using an extreme-value distribution to describe the changes of expression due to a single mutation (the jump-size distribution), we here use a two-parameter distribution consisting of an exponential distribution with density pE(x) = (1/a)e−(x+a)/a (x > −a if a > 0; x < −a if a < 0) convoluted with a Gauss kernel with density 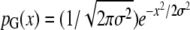 . The resulting distribution pD = pE * pG, which has mean 0, variance σ2 + a2, and skewness 2a3(a2 + σ2 )−3/2, has properties similar to the extreme value distribution (in particular, it has a one-sided heavy tail) but allows a better control of the skewness and simplifies the application of the Fourier transform.
. The resulting distribution pD = pE * pG, which has mean 0, variance σ2 + a2, and skewness 2a3(a2 + σ2 )−3/2, has properties similar to the extreme value distribution (in particular, it has a one-sided heavy tail) but allows a better control of the skewness and simplifies the application of the Fourier transform.
Starting from an initial expression of 0, the distribution of expression levels pY,t after the compound Poisson process Y has been allowed to run for a time t is calculated as pY,t = F−1{exp[t(F(pD) −1)]}, where F and F−1 are the Fourier and inverse-Fourier operators defined by 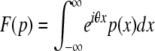 and
and 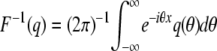 . To derive this, note that the probability that n mutations occur in the time interval [0, t] is e−ttn/n!, and the sum of n independent expression changes drawn from D is distributed as pD* … *pD (n times), where * is the convolution operator. Using F(f*g) = F(f)F(g), the distribution pY,t may be written as
. To derive this, note that the probability that n mutations occur in the time interval [0, t] is e−ttn/n!, and the sum of n independent expression changes drawn from D is distributed as pD* … *pD (n times), where * is the convolution operator. Using F(f*g) = F(f)F(g), the distribution pY,t may be written as 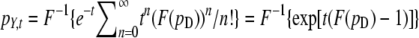 . Evaluating the Fourier transform, we obtain
. Evaluating the Fourier transform, we obtain
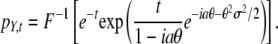 |
(1) |
Expression evolution in a phylogeny:
To calculate the likelihood of a configuration of expression levels on a binary phylogenetic tree, we use a reverse traversal algorithm analogous to Felsenstein's peeling algorithm. The algorithm computes partial-likelihood densities La(xa) representing the likelihood density of the observed transformed expressions at the collection of node a's descendant leaf nodes, conditional on its expression level xa. To compute these, we denote the immediate descendants of a by b and c. Let 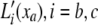 , be the “pulled-back” partial-likelihood densities of the expression at i (or its descendants if i is not a leaf node) conditional on the expression at a being xa. Integrating out all possible mutations yields
, be the “pulled-back” partial-likelihood densities of the expression at i (or its descendants if i is not a leaf node) conditional on the expression at a being xa. Integrating out all possible mutations yields 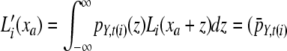 *
* 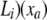 , where z denotes the increase of expression due to mutations along the branch from a to i, t(i) is the length of the branch connecting nodes i and a, and
, where z denotes the increase of expression due to mutations along the branch from a to i, t(i) is the length of the branch connecting nodes i and a, and 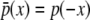 . In terms of these pulled-back likelihoods, the partial-likelihood density at a is
. In terms of these pulled-back likelihoods, the partial-likelihood density at a is 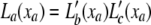 (note that this density lives on a space with as many dimensions as a has descendant leaves). This computation is potentially slow, since to compute this integral numerically, the x and z variables need to be discretized, and a naive implementation of the convolution is quadratic in the number of discretization bins. However, it can be computed in log-linear time by the fast Fourier transform algorithm, using the relation f*g = F−1{F(f)F(g)}. A further simplification is obtained by a direct computation of the kernel,
(note that this density lives on a space with as many dimensions as a has descendant leaves). This computation is potentially slow, since to compute this integral numerically, the x and z variables need to be discretized, and a naive implementation of the convolution is quadratic in the number of discretization bins. However, it can be computed in log-linear time by the fast Fourier transform algorithm, using the relation f*g = F−1{F(f)F(g)}. A further simplification is obtained by a direct computation of the kernel,
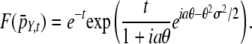 |
(2) |
The recursion ends with the computation of the likelihood density at the root. The full likelihood under the evolutionary model is obtained by integrating out the initial expression level with a suitable prior distribution Pr(x), which we chose to be uniform,
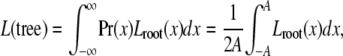 |
(3) |
where A is a suitably large bound.
Intraspecific variance, measurement error, and random physiological fluctuations were modeled for each gene in each species by initializing the partial-likelihood densities at the leaf nodes by a Gaussian distribution, with mean equal to the observed transformed expression, and variance equal to the observed variance.
Outlier model:
The outlier model, which allows for broad changes of expression level, makes the full model less susceptible to evolutionary and experimental outliers. The likelihood of the outlier model is independent of the phylogeny and is given by a product of Cauchy distributions,
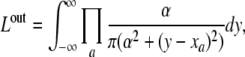 |
(4) |
where a runs over all external nodes and a is a scaling factor, which we set to α = 1. The final likelihood is obtained by considering the model where expression levels follow the outlier model with prior probability p and otherwise follow the evolutionary model, to obtain
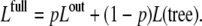 |
(5) |
Gene-expression levels are assumed to evolve independently, so that the compound likelihood for a set of genes is the product of the per-gene likelihoods. Finally, the full model P (ϕ, x), where ϕ represents parameters and x transformed expression levels, is obtained by multiplying the likelihood with a prior P(ϕ), which we chose to be uniform in all parameters. The algorithm to compute the likelihood under this model was implemented as an R module (available on request), using the FFTW package for computing fast Fourier transforms (Frigo and Johnson 2005). Discretization bins of size 0.1 were used across the range 1–45.
MCMC inference:
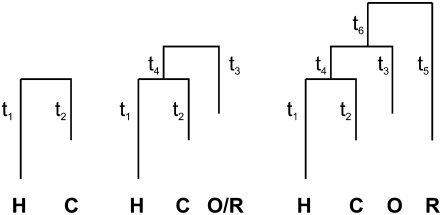
We implemented a Markov chain Monte Carlo (MCMC) scheme to sample the model parameters a, σ, and p, as well as the branch lengths (denoted by, e.g., t1, t2, t3, t4 for the three-species tree in Figure 1) from the posterior distribution defined by the model and the observed expression levels. Proposals consisted of the following types: (1) a normally distributed additive change to one of the branch lengths or one of the parameters a, σ, and p; (2) a translation of one of the internal nodes (including the root); (3) a change of sign of a; (4) swapping two neighboring branch lengths combined with a change of sign of a; (5) a rescaling of the branch lengths a and σ, leaving the total variance in the tree unchanged, but scaling the expected number of mutations; and (6) a rotation in a–σ space, changing the shape of the jump-size distribution, but leaving the total variance unchanged. These proposals were assigned different probabilities to optimize mixing, and each proposal (apart from types 3 and 4) had a scale parameter that was adjusted, during burn-in, to bring the acceptance ratio within the range 10–25%.
Figure 1.—
Branch-length parameters for the two-, three-, and four-species phylogenies considered. H, human; C, chimpanzee; R, rhesus macaque; O, orangutan.
Bayes factors were computed using models, by computing the ratio of posteriors P(x), themselves computed as 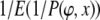 , where the expectation was taken over the posterior, using the samples from P(ϕ | x) generated in the MCMC run. Finally, we used our procedure to do approximate maximum-likelihood-ratio tests, by taking the maximum of the likelihood density across samples from runs on two nested models, accounting for the prior, and performing a standard likelihood-ratio test on the resulting two maxima.
, where the expectation was taken over the posterior, using the samples from P(ϕ | x) generated in the MCMC run. Finally, we used our procedure to do approximate maximum-likelihood-ratio tests, by taking the maximum of the likelihood density across samples from runs on two nested models, accounting for the prior, and performing a standard likelihood-ratio test on the resulting two maxima.
Simulation study:
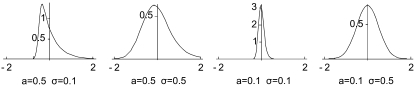
We generated expression data for samples of size 100, 1000, and 10,000 for (A) three different species under a given phylogeny (t1 = 2, t2 = 0.5, t3 = 1, t4 = 0.5; see Figure 1) and for (B) two different species under the phylogeny t1 = 2, t2 = 0.5. The intraspecific variance μ was fixed to 0.4. Samples were simulated for different values of parameters a (−0.1 and −0.5), σ (0.1 and 0.5), and p (0 and 0.01), corresponding in total to eight variations of the evolutionary model (see Figure 2 for an illustration of these variations). The expression level for the proportion p of outliers was drawn from a Gaussian distribution with standard deviation 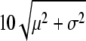 .
.
Figure 2.—
Examples of four distributions of change magnitudes used to simulate expression data, corresponding to different values of the shape parameters a and σ.
To test the sensitivity of the method to a lack of independence within the set of expression profiles, we generated additional sets of interdependent expression profiles for two species, taking a = −0.5, σ = 0.1, and p = 0 or 0.01. We first simulated expression profiles for 100, 1000, and 10,000 transcripts using the same protocol as above. We then replicated each transcript x times, where x was drawn from a geometric distribution with mean 6, to simulate sets of probes that refer to the same transcript or several coregulated genes. Finally, within each such set of probes, we added an error term drawn from a Gaussian distribution of mean 0 and standard deviation 1.8, similar to the distribution of residuals observed in real data, to simulate technical effects such as variation of hybridization efficiency as well as differences in expression between coregulated genes and alternative transcripts.
We generated 20 replicates for each combination of parameters (54 combinations, 1080 data sets). Each data set was analyzed using a 200,000-sample MCMC chain, including a burn-in period of 10,000 samples. Parameters were logged every 25 samples. After conservatively discarding the samples corresponding to the first half of each chain to allow for convergence, we estimated, for each parameter, the median of its posterior and its 95% credible interval, as well as the median and quartiles over 20 runs.
Normalization of primate expression array data:
We used our method to analyze a data set of expression profiles in brain for seven human (H), six chimpanzee (C), three orangutan (O), and six rhesus macaque (R) samples produced using Affymetrix U133plus2 arrays. On these arrays, 25mer oligonucleotide probes were designed to hybridize to specific transcript targets. Typically, 11 probes were designed per human target transcript, allowing both a reduction of noise and a removal of probe sequence-specific effects (Wu et al. 2004). We used the latest available human gene annotation (Ensembl r42, December 2006) to assign probes to target genes as described elsewhere (Dai et al. 2005). To minimize artifacts that result from hybridizing chimpanzee, orangutan, and macaque samples to arrays designed to assay human transcripts, prior to the analysis, we masked all oligonucleotide array probes that did not match perfectly the human (build March 2006), the chimpanzee (build March 2006), and the macaque genomes (build January 2006) as described elsewhere (Khaitovich et al. 2004). The resulting consensus set of probes contained on average 6 probes per target transcript (probe-set size distributions are shown in supplemental Figure 1). It must be noted that sequence differences specific to the orangutan genome were not masked by this procedure due to lack of the orangutan genome sequence information.
For all data sets, probe level data were normalized, adjusting for different backgrounds and overall hybridization intensities of individual arrays. This conservatively normalized data set with no secondary normalization (“nn”) was used in the subsequent analysis. In addition, to assess whether results were sensitive to further potential systematic differences between measurements that were not explicitly included in our model of expression evolution, probe-level data were quantile normalized across all samples, resulting in the quantile-normalized (“QQ”) data set. Transcript expression estimates for both data sets were obtained by robust fits of linear multichip probe-level intensity models (Bolstad 2004). See supplemental information for low-level analysis Q/C plots.
Our evolution model was then either (1) directly applied to probe-expression levels (86,818 probes) or (2) used for an analysis of gene-expression estimates from probe sets (11,309 probe sets). We performed analyses using no outgroup information (HC data sets), using information from one outgroup (HCR and HCO data sets), or using two outgroups (HCRO data sets). In total 16 data sets were analyzed, each labeled by the normalizing procedure and the included species, plus the letter “p” when probe rather than probe-set intensities were considered; for example, QQ-HCRp refers to the QQ data set of probe (p) intensities in H, C, and R.
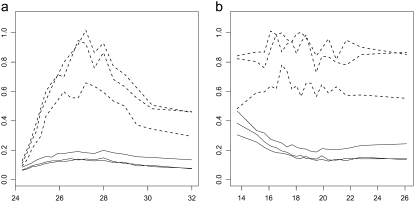
Major assumptions of the model were tested by exploring intra- and interspecific variances as well as the distribution skewness of the expression differences, in relation to transcript expression level. After QQ normalization, we variance stabilized each data set to decouple the interspecies variance from the mean intensity, according to the method described above. Figure 3 shows the relationship of these variances with mean intensity before and after this transformation for the QQ-HCR data set (plots based on other data sets were very similar). We checked that the skewness coefficient was constant across the variance-stabilized expression scale (supplemental Figure 2), as assumed by our evolutionary model. For low levels of expression, the intraspecies variance reaches levels comparable to the interspecies variance, most probably because of an increased influence of experimental noise (see Figure 4). In this range, the signal contains virtually no information, and to reduce computation time we removed these data before applying the model (overall, 44.9% of probes and 16.4% of probe sets were removed). Finally, we summarized the observed expression levels for each transcript and species by the median of the expression levels across independent samples (and within the probe set, when appropriate) and by the standard deviation of the median of these expression levels as estimated from a bootstrap procedure.
Figure 3.—
Relationship between the intraspecific (solid lines) and interspecific (dashed lines) standard deviation (y-axes) and the mean expression level (x-axes) before (a) and after (b) the interspecific variance-stabilizing transformation, in the QQ-HCR data set. The intraspecific variance is not stabilized and is accounted for explicitly on a probe-by-probe or a probe-set-by-probe-set basis in the model.
Figure 4.—
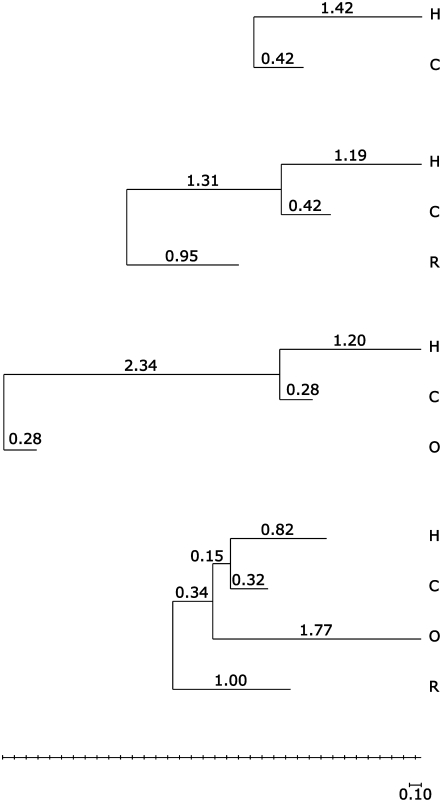
Phylogenetic trees inferred for QQ data sets. The labels H, C, O, and R refer to human, chimpanzee, orangutan, and rhesus macaque, respectively.
Supplemental information:
Supplemental information is available on the Genetics website. Data sets and low level analysis are provided at http://bioinf.boku.ac.at/pub/Chaix2008/.
RESULTS
Simulation study:
We evaluated the performance of our inference procedure on sets of simulated data, using parameters comparable to those estimated from our primate data set. Our primary interest is in the inference of the distribution of expression changes parameterized by a and σ (see Figure 2) and the ratio of the branch lengths of human to chimpanzee log2(t1/t2). Consequently, we focused mainly on their inference. Table 1 provides a representative sample of the simulation results; the full set of parameter inferences is given in supplemental Tables 1–3.
TABLE 1.
A representative sample of simulation results
| Outgroup | Probe sets | t1 | t2 | log2(t1/t2) | a | σ | p |
|---|---|---|---|---|---|---|---|
| No | No | 1.91 (1.72–2.08) | 0.36 (0.27–0.52) | 2.45 (1.83–2.75) | 0.50 (0.48–0.54) | 0.18 (0.12–0.18) | 0.00 (0.00–0.01) |
| Yes | No | 2.02 (1.86–2.16) | 0.49 (0.43–0.56) | 2.00 (1.83–2.24) | 0.50 (0.47–0.53) | 0.12 (0.10–0.14) | 0.01 (0.01–0.01) |
| No | Yes | 2.01 (1.39–2.40) | 0.50 (0.37–0.64) | 2.00 (1.49–2.51) | 0.43 (0.33–0.49) | 0.30 (0.22–0.35) | 0.00 (0.00–0.00) |
| True parameters | 2.00 | 0.50 | 2.00 | 0.50 | 0.10 | 0.01 | |
Shown are medians and 25 and 75% quartiles for estimates of branch lengths t1, t2, and log2(t1/t2) and shape parameters a, σ, and outlier proportion p over 20 simulated data sets with parameters close to values inferred from actual data (see Table 2). Inferences were made from data without (two-species tree) or with a single outgroup (three-species tree) and using either 1000 probes or 1000 probe sets, each consisting of a geometrically distributed number (mean 6) of identical probes. Full simulation results are presented in supplemental Tables 1–3.
Using a two-species tree with t1 = 2 and t2 = 0.5, we performed simulations for various sets of evolutionary parameters and gene counts. The branch-length ratio log2(t1/t2) and the parameters of the evolutionary model a and σ can be estimated accurately for a sufficient number of transcripts (typically >100) and moderate to large skewness (|a| ≥ σ), making the model sufficiently time irreversible. With insufficient skewness, the model becomes nearly reversible, resulting in conservative (near-zero) estimates of the log branch-length ratio. As expected, confidence intervals decrease with increasing number of genes and are reasonably small for n ≥ 1000 genes; for instance, for a = 0.5, σ = 0.1, p = 1% outliers, and n = 1000 genes, we estimate in the first of our 20 replicates a = 0.48 (95% C.I. 0.40–0.56) and σ = 0.13 (0.05–0.26). Replicating the simulation 20 times for 36 sets of parameters, we found that true parameters were outside estimated 95% C.I.'s in 5% of the cases for log2(t1/t2), 8% for a, and 6% for σ, suggesting no systematic biases in the parameter or confidence estimates (supplemental Table 1).
As for the three-species data sets, evolutionary parameters could be inferred with good accuracy even for as little as n = 100 genes when the model exhibits moderate to strong skew (|a| ≥ σ) (supplemental Table 2). When the skewness is small (|a| < σ), the number of mutation events (i.e., the total branch length) and the per-mutation variance a2 + σ2 become confounded: for instance, for a = 0.1, σ = 0.5, n = 1000 transcripts, and 1% of outliers, the total number of events inferred on the tree is overestimated (4.38 instead of 4) but the per-mutation variance is underestimated (0.20 instead of 0.26). However, the total variance along branches (number of events times per-mutation variance), and consequently the branch length ratio, can still be accurately determined, showing that the presence of outgroup data makes the procedure less reliant on the time irreversibility of the model. Replicating the simulation 20 times, we found that true parameters were outside estimated 95% C.I.'s in 9% of the cases for log2(t1/t2), in 12% of the cases for a, and in 8% of the cases for σ. We conclude that (i) when skewness is moderate to large (|a| ≥ σ), parameters and branch lengths can be consistently estimated for three-species and two-species data sets, and (ii) for small skewness, the model shows no evidence for nonzero skew and tends to equilibrate the branch lengths in the absence of an outgroup.
Finally, we evaluated the sensitivity of the method to the occurrence of interdependent expression profiles. This is obviously relevant when probe rather than probe-set intensities are analyzed, in which case each transcript is represented several times in the data set. Moreover, both technical limitations of the arrays (like cross-hybridization) and biologically related coregulated genes are likely to contribute to correlation in expression data. We simulated data sets in which each independent expression measure is represented x times, where x followed a geometric distribution with mean 6. We assumed that the within-probe-set measurement error followed a Gaussian distribution of mean 0 and standard deviation 1.8, corresponding to our observations in the data sets. For a substantially skewed jump-size distribution (a = 0.5 and σ = 0.1), we found that the method was able to accurately estimate log2(t1/t2) while the estimate for a was fairly accurate; the parameter σ was, however, somewhat overestimated (supplemental Table 3). For example, for a simulation of 6000 probes with a = 0.5, σ = 0.1, t1 = 2, t2 = 0.5, and 1% outliers, log2(t1/t2) was 2.00 (1.49–2.51), a = 0.43 (0.33–0.49), and σ = 0.30 (0.22–0.35). Replicating the simulation 20 times for six sets of parameters, we found that true parameters were outside estimated 95% C.I.'s in 2.5% of cases for log2(t1/t2), 22% for a, and 20% for σ.
Application to primate expression profiles:
We applied our method to sets of expression intensities in the brain for H, C, O, and R samples, removing probes that showed mismatches in either the chimpanzee or the rhesus macaque genomes (in comparison to the human genome) from all four data sets, and considered all possible combinations of data sets that included human and chimpanzee. Because the lack of genome sequence meant that probes that did not match the orangutan genome could not be masked (leading to notable differences in the distribution of intensities of orangutan in comparison to other species; see supplemental Figures 3 and 4), we first focus our presentation of the results on the HC and HCR sets and discuss the HCO and HCRO data sets later.
We first tested whether the data contained a significant phylogenetic signal, by running the MCMC procedure on all possible rooted phylogenies of human, chimpanzee, and macaque and comparing the statistical support for the true phylogeny to that for the alternative phylogenies. Indeed, we found strong support for the true phylogeny [Bayes factor 73 bits between true and next-best phylogeny, that is, chimpanzee closer to macaque than to human, “strong evidence” (Jeffreys et al. 2005); approximate maximum-likelihood test, 2Δ log(L) = 96, P < 10−5].
We next considered whether our data set exhibited a significant skew in the expression-mutation distribution or could be sufficiently well described with a zero-skew model (a = 0), which closely approximates a diffusion model. We found that the skewed compound Poisson model fit the data significantly better [Bayes factor 262 bits, “decisive evidence”; approximate maximum-likelihood test, 2Δ log(L) = 342, P < 10−5; both using the HCR set]. To illustrate the data, Figure 5 shows the inferred jump-size distribution from the HC set and histograms of simulated and observed expression differences.
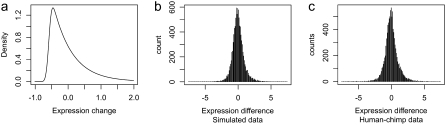
Figure 5.—
(a) Jump-size distribution for parameters inferred for the HC data set. (b) Histogram of expression differences simulated under the model, without outliers. Because expression-level mutations occur on either branch, the histogram is markedly less skewed than the jump-size distribution. (c) Observed expression differences in the HC data set, after normalization. The histogram is similar to the simulated data, although in the real data a larger number of outliers can be observed.
To investigate whether the expression-mutation distribution shows a skew of the type observed by Khaitovich et al. (2005a), namely, many small downregulations combined with relatively fewer but larger upregulations (positive skew; a > 0), we estimated parameters for all four species sets considered. For the two-species phylogeny, the model has a mathematical symmetry involving swapping the branches and changing the sign of a, and we consequently fixed the sign of a for the HC set, but not for the other sets. The estimates for a on the HC and HCR sets were 0.58 (95% C.I. 0.53–0.62) and 0.44 (0.41–0.46), respectively (see Table 2). We also applied our inference procedure directly on the probes (86,818 probes from 11,309 probe sets) to test the robustness of our results against errors in gene or chip annotations, resulting in similar estimates [HC, a = 0.53 (0.50–0.55); HCR, a = 0.31 (0.29–0.33); Table 3] and confirming the skew previously reported. The σ-parameter was estimated to be comparatively low at 0.07 (0.05–0.12) for HC and 0.08 (0.05–0.11) for HCR, and similar estimates were obtained on raw probe data [HC, 0.06 (0.05–0.1); HCR, 0.24 (0.22–0.26)]. To test whether different normalization procedures influenced these results, we also applied the inference procedure on the data set without interspecies QQ normalization (nn). Inferences on both the HC and the HCR sets were similar to those obtained on the QQ set (supplemental Tables 4 and 5).
TABLE 2.
Inferred human–chimp expression evolution parameters from probe-set data
| Data set | HC | HCR | HCO | HCRO |
|---|---|---|---|---|
| t1a | 1.42 (1.25–1.65) | 1.19 (1.10–1.31) | 1.20 (1.11–1.33) | 0.82 (0.77–0.88) |
| t2 | 0.42 (0.33–0.52) | 0.42 (0.36–0.48) | 0.28 (0.23–0.33) | 0.32 (0.29–0.35) |
| t3 | 0.95 (0.81–1.08) | 0.28 (0.16–0.41) | 1.77 (1.66–1.88) | |
| t4 | 1.31 (1.15–1.48) | 2.34 (2.09–2.62) | 0.15 (0.10–0.19) | |
| t5 | 1.0 (0.9–1.1) | |||
| t6 | 0.34 (0.26–0.41) | |||
| σ | 0.07 (0.05–0.12) | 0.08 (0.05–0.11) | 0.08 (0.05–0.13) | 0.27 (0.25–0.30) |
| a | 0.58 (0.53–0.62) | 0.44 (0.41–0.46) | 0.45 (0.42–0.47) | −0.43 (−0.45–0.41) |
| p | 0.01 (0–0.01) | 0.01 (0.01–0.01) | 0.01 (0.00–0.01) | 0.49 (0.03–0.97) |
| var | 0.34 (0.29–0.40) | 0.20 (0.18–0.22) | 0.21 (0.19–0.23) | 0.26 (0.25–0.28) |
| div × var | 0.63 (0.59–0.67) | 0.32 (0.30–0.34) | 0.31 (0.29–0.33) | 0.30 (0.29–0.31) |
| log2(t1/t2) | 1.8 (1.4–2.2) | 1.5 (1.3–1.8) | 2.1 (1.8–2.4) | 1.4 (1.2–1.6) |
| t1/t2 | 3.4 (2.7–4.4) | 2.9 (2.4–3.4) | 4.3 (3.5–5.4) | 2.6 (2.3–3.0) |
| n | 9,806 | 8,303 | 9,273 | 10,427 |
| ESS | 951 | 536 | 334 | 354 |
Estimates (posterior medians and 95% credible intervals) of branch lengths and model parameters from probe-set level data, using various choices of outgroups: none (HC, human–chimp), one outgroup (rhesus macaque, HCR; or orangutan, HCO), or two outgroup species (HCRO). For inferences from data without quantile–quantile normalization see supplemental Table 4.
t1, length of human branch; t2, length of chimpanzee branch; a and σ, shape parameters of the distribution of change magnitudes; p, proportion of outliers; var, event variance (a2 + σ2); div, expected number of events between human and chimpanzee (t1 + t2); n, number of probe sets in the data set; ESS, effective sample size for t1.
TABLE 3.
Inferred human–chimp expression evolution parameters from probe data
| Data set | HC | HCR | HCO | HCRO |
|---|---|---|---|---|
| t1a | 1.85 (1.72–2.01) | 1.06 (1.01–1.12) | 1.67 (1.59–1.78) | 0.81 (0.80–0.83) |
| t2 | 0.60 (0.53–0.67) | 0.50 (0.47–0.53) | 0.36 (0.32–0.40) | 0.41 (0.40–0.43) |
| t3 | 0.87 (0.76–0.98) | 0.07 (0.00–0.14) | 2.64 (2.59–2.68) | |
| t4 | 1.13 (1.02–1.23) | 4.51 (4.20–4.86) | 0.08 (0.07–0.09) | |
| t5 | 0.68 (0.66–0.69) | |||
| t6 | 0.57 (0.56–0.58) | |||
| σ | 0.06 (0.05–0.10) | 0.24 (0.22–0.26) | 0.06 (0.05–0.08) | 0.06 (0.05–0.06) |
| a | 0.53 (0.50–0.55) | 0.31 (0.29–0.33) | 0.35 (0.34–0.37) | −0.49 (−0.49–0.48) |
| p | 0.01 (0.00–0.01) | 0.01 (0.01–0.01) | 0.01 (0.01–0.01) | 0.21 (0.02–0.70) |
| var | 0.28 (0.26–0.31) | 0.16 (0.15–0.16) | 0.13 (0.12–0.14) | 0.24 (0.23–0.24) |
| div × var | 0.70 (0.68–0.72) | 0.25 (0.24–0.25) | 0.26 (0.26–0.27) | 0.29 (0.29–0.30) |
| log2(t1/t2) | 1.62 (1.46–1.81) | 1.10 (0.99–1.21) | 2.21 (2.06–2.40) | 0.97 (0.92–1.05) |
| t1/t2 | 3.07 (2.76–3.50) | 2.14 (1.99–2.32) | 4.63 (4.17–5.3) | 1.96 (1.89–2.08) |
| n | 50,759 | 44,668 | 40,160 | 55,802 |
| ESS | 191 | 180 | 171 | 20 |
Estimates (posterior medians and 95% credible intervals) of branch lengths and model parameters from probe level data are shown, using various choices of outgroups. For inferences on data without quantile–quantile normalization see supplemental Table 5.
Parameters are as in Table 2.
Estimates of the branch lengths from human (t1) and chimpanzee (t2) to their most recent common ancestor of human and chimp were consistent between the data sets, both for t1 [HC, 1.42 (1.25–1.65); HCR, 1.19 (1.1–1.31)] and for t2 [HC, 0.42 (0.33–0.52); HCR, 0.42 (0.36–0.48)], as expected since our simulations showed that in these parameter ranges (|a| > σ), the evolutionary parameters and branch lengths can be accurately estimated even for just two species. The ratio t1/t2 was estimated separately at 3.38 (2.66–4.43) for HC and 2.87 (2.43–3.42) for HCR. Similar estimates were obtained for raw probe data [HC, 3.07 (2.76–3.5); HCR, 2.14 (1.99–2.32)], suggesting that more gene-expression changes have occurred on the human than on the chimpanzee branch since the split with their most recent common ancestor.
Finally, we analyzed the data sets that include orangutan. On the HCO set, estimates for a and σ were 0.45 (0.42–0.47) and 0.08 (0.05–0.13), respectively, consistent with those on HCR and HC. The branch length estimates t1 and t2 are also consistent (Tables 2 and 3); however, the estimated branch length from the root to the human/chimpanzee ancestor is very large [t4 = 2.34 (2.09–2.62)], while the orangutan branch is very short at t3 = 0.28 (0.16–0.41). Analyzing all four species together (HCOR), we find that the inferred value of a changed sign [−0.43 (−0.45– −0.41)], and the branch to orangutan is now the longest in the tree [t3 = 1.77 (1.66–1.88)] (Figure 4). In addition, the estimate for the outlier model parameter p on this data set is 0.49 with a wide confidence interval (0.03–0.97); in contrast, in all other cases the upper 95% confidence limit for p did not exceed 0.02. We propose an interpretation of these results in the discussion.
DISCUSSION
We have introduced a model and an algorithm that allowed us to calculate the likelihood of gene-expression data evolving along a phylogenetic tree. The model, which is a departure from the diffusion or Brownian motion models often used for describing the evolution of quantitative traits, was specifically adapted for the study of gene-expression levels. Under the model, expression levels evolve along the branches of a phylogenetic tree by discrete mutation events, which occur stochastically at a certain rate. Each event modifies the expression intensity on an additive scale, by an amount drawn from a jump-size distribution that may exhibit a nonzero skewness. Using this model, we implemented a Bayesian MCMC procedure for the inference of parameters and branch lengths and for hypothesis testing.
The existence of a positively skewed mutation spectrum for expression levels was suggested previously by Khaitovich et al. (2005b), on the basis of the observation that the distributions of expression differences between human and chimpanzee were skewed. Assuming this positively skewed mutation spectrum, Khaitovich et al. (2005b) inferred that genes expressed in the brain have undergone more expression mutations in the human lineage than in the chimpanzee lineage. However, this conclusion depends crucially on the premise of a positively (rather than negatively) skewed mutation spectrum. Since the expression data in these studies were obtained using expression arrays designed for human samples, and mismatching probes for orangutan samples could not be reliably identified because of the lack of genome sequence, some doubt remained about these conclusions.
Here we revisit these observations. We developed an algorithm to calculate the likelihood of expression data under the skewed mutation model, enabling the inference of the phylogeny, evolutionary parameters, and confidence intervals using MCMC. We adjoined an outlier model allowing a proportion of data points to evolve essentially without constraint, to ensure that a small proportion of spurious data would not unduly bias the inferred parameters of the main model. Another innovation is the use of a variance-stabilizing transformation, which decouples the interspecies variance from the intensity, allowing the same evolutionary model to be applied to all transcripts whatever their intensity of expression. Finally, we accounted explicitly for intraspecific variation due to experimental error, as well as environmental and genetic variances. We applied the model to various combinations of H, C, O, and R data, each obtained using human microarrays. Probes that showed mismatches in homologous position in chimp or macaque were removed from the analysis. Because the same could not be done for orangutan, we separately analyzed data sets that included this primate.
We found clear evidence for the existence of a positive skew in the mutation spectrum of gene-expression levels in our analyses of the HC and HCR data sets, confirming the original observation by Khaitovich et al. (2005b). The inferred parameters remain essentially the same when all probes are used individually, rather than grouped per gene, and were also robust against changes to the normalization procedure. The simulation study showed that inferences in these parameter ranges are reliable, for both the two-species and the three-species case.
What could have caused the observed skew? A nonzero skewness in the mutation spectrum confers a direction to the evolutionary “arrow of time.” This does not necessarily imply that the underlying process involves selection (as opposed to purely mutational forces); for example, the high mammalian mutation rate of CpG dinucleotides results in a mutation bias directed away from CpGs, while an equivalent reverse bias does not exist, conferring a directionality to this process without involving selection (Lunter and Hein 2004). Nevertheless, it seems difficult to construct a plausible, purely mutational mechanism that would lead to a skewed spectrum of expression-level changes. Alternatively, we might consider a scenario involving selection to maintain expression levels at roughly constant values. Since transcription initiation involves many genomic sites, nucleotide mutations that influence its efficiency are likely to be relatively frequent. For example, our results suggest that between human and chimpanzee there have been on average ∼1.5 mutations per gene that affected expression levels. According to Kimura's neutral theory, negative (purifying) selection and drift will remove the majority of mutations, and those that become fixed and are observed as substitutions in interspecies comparisons will either be neutral or slightly deleterious (Ohta 1973). This process has a hidden directionality: random mutations increase entropy; in particular, they tend to decrease, rather than increase, the affinity of transcription factor binding sites. Thus, a succession of random (unselected) mutations may be expected to lead to a diminished expression of the transcript, building up an associated “fitness deficit.” This creates an opportunity for less frequent, “corrective” mutations that increase the efficiency of transcription by a relatively large amount to sweep to fixation, after which the process starts anew. Although the last step will of course have all the population-genetic characteristics of a selective sweep, it would be quite false to think of it as “positive selection” as it is generally understood, since no adaptation to a favored phenotype is involved. Rather, the conservation of a fixed desired phenotype (a particular expression level) is achieved as a dynamic equilibrium involving a combination of continuous deterioration by slightly deleterious mutations, counterbalanced by occasional corrective sweeps. The whole cycle would result in precisely the observed positive skew of the mutational spectrum and indeed appears a plausible model for the observed high turnover of transcription factor binding sites in mammals (Dermitzakis and Clark 2002) and flies (Moses et al. 2006).
It should be stressed that not all expression-level changing mutations that are fixed by drift are expected to cause a decrease in the level of expression. While our model predicts that about a third of mutations do increase expression levels, the proportion of mutations that are driven to fixation by selection is likely to be smaller. Although the long tail of the jump-size distribution (see Figure 5) might still suggest this proportion to be sizeable, the shape of the distribution should not be overinterpreted, as it is largely a modeling choice, and the asymptotics of the actual jump-size distribution might be different.
True positive selection acting upon expression-level mutations is also likely to have contributed to some extent and may also have influenced the observed spectrum. However, it appears implausible that adaptive changes would follow any systematic pattern (i.e., many small downregulations, fewer large upregulations), which would be necessary for it to leave its fingerprint on a genomewide analysis. We therefore propose the dynamic equilibrium of slightly deleterious mutations and occasional corrective sweeps, together acting to conserve the expression phenotype, as an explanation for the skewed mutation spectrum of gene-expression levels.
The pronounced skewness allowed us to infer the branch lengths of the human (t1) and chimp (t2) lineages separately, both with and without macaque as an outgroup. The inferred ratio t1/t2 ranges from 2.0 to 4.4, depending on whether macaque was included and whether probes or probe sets were used. This ratio is in the same order of magnitude as previous estimates: analyzing a different data set with a moment approach, Khaitovich et al. (2005a) found that for brain-expressed genes, the human branch was 3.3 times longer than the chimpanzee branch. Similarly, analyzing brain-expressed genes and restricting the analysis to those 5% exhibiting the highest divergence between human and chimpanzee, Enard et al. (2002a) inferred a phylogeny in which the human branch was 3.8-fold longer than the chimpanzee branch. Accepting dynamic stabilizing selection as the dominant mode of expression evolution, these observations could be explained by more efficient selection against slightly deleterious mutations affecting transcription factor binding sites in chimpanzees than in humans, as a result of a larger effective ancestral population size of chimpanzees (Harding and McVean 2004). Interestingly, such strong acceleration of expression changes on the human lineage is not observed in liver, heart, or kidney (Enard et al. 2002b; Khaitovich et al. 2005b, 2006; Lemos et al. 2005). At the same time, the skewed distribution of expression changes on the human and on the chimpanzee lineage is observed in other tissues, such as liver (Khaitovich et al. 2005b). Thus, the skew appears to be a general feature of expression evolutionary dynamics in primates. In contrast, the accelerated expression evolution on the human lineage may represent a special case, possibly reflecting an excess of positive selection on the gene-expression changes in the brain along the human lineage. Still, given the imperfections of microarray expression data and the paucity of human and chimpanzee samples analyzed thus far, more effort will be required to uncover the cause of this phenomenon with certainty.
The data sets that included orangutan (HCO and HCOR) gave various discordant results. For the HCO set, we found a highly skewed tree, with a very short branch leading to orangutan, while for HCOR, we inferred negative skewness, as well as a very large contribution of the outlier model (49%), while all other data sets support a very small outlier contribution (<2%). The orangutan data are the only set for which we were unable to remove mismatching probes because of the current lack of a genome sequence. The deviations of the overall distribution of orangutan expression levels compared to human, chimp, or macaque (supplemental Figure 3) indeed suggest that this data set is tainted by a small admixture of such mismatching probes, and the discordant inferences are all consistent with this explanation. Indeed, mismatching probes would cause an admixture of genes with apparently much reduced orangutan expression levels, favoring a model that allows occasional very large downregulations. This explains the aberrant tree inferred for the HCO set, since the positioning of the root near the orangutan effectively changes the direction of time along much of the branch toward orangutan. Adding the macaque to the phylogeny restricts the root position and forces the large downregulations to be modeled by changing the direction of the mutational skew.
It is satisfying that these data problems, which are nonobvious particularly after QQ normalization, were correctly diagnosed by the outlier model. It also serves to reemphasize the need for aggressive cleanup of expression data obtained using expression arrays, especially for cross-species experiments. In this study, the loss of affinity due to random mutations on probe targets within the orangutan genome appears sufficient, in number or effect, to overwhelm the phylogenetic patterns we observe in the other data sets.
The proposed evolutionary model is an approximation of the actual process of gene-expression evolution. Perhaps the most questionable assumption we make is that one set of parameters describes the evolution of expression patterns across all genes. We, however, expect that our main conclusions (fast evolution on the human branch and a distinct skew of the mutation spectrum) are robust against even quite strong deviations from this assumption, because the expected number of mutations on the phylogeny is modest (approximately four), assigning reasonable probability to instances with no mutations, while very fast-evolving genes are absorbed into the outlier model.
The Bayesian method introduced here was designed to estimate the phylogenetic tree and various parameters of the evolutionary model that relate several species on the basis of expression-profiles data. The method is based on a time-irreversible model of evolution, enabling the inference of branch lengths and the position of the root without recourse to an outgroup. We recover the known phylogeny and find evidence for a skewed spectrum of expression mutations, which we suggest is compatible with the action of stabilizing selection on gene-expression levels, leading to a dynamic equilibrium of slightly deleterious mutations and occasional corrective sweeps. We also find evidence for a larger number of expression-level changes in the human lineage compared to chimp, which taken by itself could point to a more efficient action of selection in the ancestral chimpanzee population owing to its larger ancestral population size, but is also compatible with extensive positive selection on brain-expressed genes in the human lineage. Gene-expression levels have long been suggested to underlie differences between species, and we hope that the method proposed here will help to better understand the role of gene regulatory changes in the evolution of our and other species.
Acknowledgments
We thank the two anonymous reviewers for their helpful comments. R.C. was supported by the European Molecular Biology Organization and the Fyssen foundation; G.L. by the Medical Research Council and the Engineering and Physical Sciences Research Council (code HAMKA); P.K. and M.S. by the Chinese Academy of Sciences and the Max Planck Society; and D.P.K. by the Vienna Science and Technology Fund, the Austrian Centre of Biopharmaceutical Technology, Austrian Research Centres Seibersdorf, and Baxter AG.
References
- Bolstad, B. M., 2004. Low level analysis of high-density oligonucleotide array data: background, normalization and summarization. Ph.D. Thesis, University of California, Berkeley, CA.
- Dai, M., P. Wang, A. D. Boyd, G. Kostov, B. Athey et al., 2005. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 33 e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Meaux, J., U. Goebel, A. Pop and T. Mitchell-Olds, 2005. Allele-specific assay reveals functional variation in the chalcone synthase promoter of Arabidopsis thaliana that is compatible with neutral evolution. Plant Cell 17 676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver, D. R., K. Morris, J. T. Streelman, S. K. Kim, M. Lynch et al., 2005. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat. Genet. 37 544–548. [DOI] [PubMed] [Google Scholar]
- Dermitzakis, E. T., and A. G. Clark, 2002. Evolution of transcription factor binding sites in mammalian gene regulatory regions: conservation and turnover. Mol. Biol. Evol. 19 1114–1121. [DOI] [PubMed] [Google Scholar]
- Durbin, R., S. Eddy, A. Krogh and G. Mitchison, 1998. Biological Sequence Analysis. Cambridge University Press, Cambridge, UK/London/New York.
- Edwards, A. F. W., and L. L. Cavalli-Sforza, 1964. Reconstruction of evolutionary trees, pp. 67–76 in Phenetic and Phylogenetic Classification, edited by W. H. Heywood and J. McNeill. Systematics Association, London.
- Enard, W., P. Khaitovich, J. Klose, S. Zollner, F. Heissig et al., 2002. a Intra- and interspecific variation in primate gene expression patterns. Science 296 340–343. [DOI] [PubMed] [Google Scholar]
- Enard, W., M. Przeworski, S. E. Fisher, C. S. Lai, V. Wiebe et al., 2002. b Molecular evolution of FOXP2, a gene involved in speech and language. Nature 418 869–872. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J., 1973. Maximum-likelihood estimation of evolutionary trees from continuous characters. Am. J. Hum. Genet. 25 471–492. [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J., 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17 368–376. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J., 2004. Inferring Phylogenies. Sinauer Associates, Sunderland, MA.
- Frigo, M., and S. G. Johnson, 2005. The design and implementation of FFTW3. Proc. IEEE 93 216–231. [Google Scholar]
- Gilad, Y., S. A. Rifkin, P. Bertone, M. Gerstein and K. P. White, 2005. Multi-species microarrays reveal the effect of sequence divergence on gene expression profiles. Genome Res. 15 674–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad, Y., A. Oshlack, G. K. Smyth, T. P. Speed and K. P. White, 2006. Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature 440 242–245. [DOI] [PubMed] [Google Scholar]
- Harding, R. M., and G. McVean, 2004. A structured ancestral population for the evolution of modern humans. Curr. Opin. Genet. Dev. 2004 667–674. [DOI] [PubMed] [Google Scholar]
- Hsieh, W. P., T. M. Chu, R. D. Wolfinger and G. Gibson, 2003. Mixed-model reanalysis of primate data suggests tissue and species biases in oligonucleotide-based gene expression profiles. Genetics 165 747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, W., A. von Heydebreck, H. Sultmann, A. Poustka and M. Vingron, 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18(Suppl. 1): S96–S104. [DOI] [PubMed] [Google Scholar]
- Jeffreys, A. J., R. Neumann, M. Panayi, S. Myers and P. Donnelly, 2005. Human recombination hot spots hidden in regions of strong marker association. Nat. Genet. 37 601–606. [DOI] [PubMed] [Google Scholar]
- Keightley, P. D., M. J. Lercher and A. Eyre-Walker, 2005. Evidence for widespread degradation of gene control regions in hominid genomes. PLoS Biol. 3 e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich, P., G. Weiss, M. Lachmann, I. Hellmann, W. Enard et al., 2004. A neutral model of transcriptome evolution. PLoS Biol. 2 E132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich, P., I. Hellmann, W. Enard, K. Nowick, M. Leinweber et al., 2005. a Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science 309 1850–1854. [DOI] [PubMed] [Google Scholar]
- Khaitovich, P., S. Paabo and G. Weiss, 2005. b Toward a neutral evolutionary model of gene expression. Genetics 170 929–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich, P., K. Tang, H. Franz, J. Kelso, I. Hellmann et al., 2006. Positive selection on gene expression in the human brain. Curr. Biol. 16 R356–R358. [DOI] [PubMed] [Google Scholar]
- King, M. C., and A. C. Wilson, 1975. Evolution at two levels in humans and chimpanzees. Science 188 107–116. [DOI] [PubMed] [Google Scholar]
- Lande, R., 1976. Natural selection and random genetic drift in phenotypic evolution. Evolution 30 314–334. [DOI] [PubMed] [Google Scholar]
- Lemos, B., C. D. Meiklejohn, M. Caceres and D. L. Hartl, 2005. Rates of divergence in gene expression profiles of primates, mice, and flies: stabilizing selection and variability among functional categories. Evol. Int. J. Org. Evol. 59 126–137. [PubMed] [Google Scholar]
- Lunter, G., and J. Hein, 2004. A nucleotide substitution model with nearest-neighbour interactions. Bioinformatics 20(Suppl. 1): I216–I223. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and W. G. Hill, 1986. Phenotypic evolution by neutral mutation. Evolution 40 915–935. [DOI] [PubMed] [Google Scholar]
- Moses, A. M., D. A. Pollard, D. A. Nix, V. N. Iyer, X. Y. Li et al., 2006. Large-scale turnover of functional transcription factor binding sites in Drosophila. PLoS Comput. Biol. 2 e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzhdin, S. V., M. L. Wayne, K. L. Harmon and L. M. McIntyre, 2004. Common pattern of evolution of gene expression level and protein sequence in Drosophila. Mol. Biol. Evol. 21 1308–1317. [DOI] [PubMed] [Google Scholar]
- Oakley, T. H., Z. Gu, E. Abouheif, N. H. Patel and W. H. Li, 2005. Comparative methods for the analysis of gene-expression evolution: an example using yeast functional genomic data. Mol. Biol. Evol. 22 40–50. [DOI] [PubMed] [Google Scholar]
- Ohta, T., 1973. Slightly deleterious mutant substitutions in evolution. Nature 246 96–98. [DOI] [PubMed] [Google Scholar]
- Oshlack, A., A. E. Chabot, G. K. Smyth and Y. Gilad, 2007. Using DNA microarrays to study gene expression in closely related species. Bioinformatics 23 1235–1242. [DOI] [PubMed] [Google Scholar]
- Rifkin, S. A., J. Kim and K. P. White, 2003. Evolution of gene expression in the Drosophila melanogaster subgroup. Nat. Genet. 33 138–144. [DOI] [PubMed] [Google Scholar]
- Turelli, M., J. H. Gillespie and R. Lande, 1988. Rate tests for selection on quantitative characters during macroevolution and microevolution. Evolution 42 1085–1089. [DOI] [PubMed] [Google Scholar]
- Voolstra, C., D. Tautz, P. Farbrother, L. Eichinger and B. Harr, 2007. Contrasting evolution of expression differences in the testis between species and subspecies of the house mouse. Genome Res. 17 42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Z., R. A. Irizarry, R. Gentleman, F. Martinez Murillo and F. Spencer, 2004. A model based background adjustment for oligonucleotide expression arrays. Department of Biostatistics Working Papers, Working Paper 1. Johns Hopkins University, Baltimore. http://www.bepress.com/jhubiostat/paper1/.