Abstract
The cystic fibrosis transmembrane conductance regulator (CFTR)1 is an epithelial chloride channel mutated in patients with cystic fibrosis. Its expression and functional interactions in the apical membrane are regulated by several PDZ (PSD-95, discs large, zonula occludens-1) proteins, which mediate protein-protein interactions, typically by binding C-terminal recognition motifs. In particular, the CFTR-associated ligand (CAL) limits cell-surface levels of the most common disease-associated mutant ΔF508-CFTR. CAL also mediates degradation of wild-type CFTR, targeting it to lysosomes following endocytosis. Nevertheless, wild-type CFTR survives numerous cycles of uptake and recycling. In doing so, how does it repeatedly avoid CAL-mediated degradation? One mechanism may involve competition between CAL and other PDZ proteins including Na+/H+ Exchanger-3 Regulatory Factors 1 and 2 (NHERF1 and NHERF2), which functionally stabilize cell-surface CFTR. Thus, to understand the biochemical basis of WT-CFTR persistence, we need to know the relative affinities of these partners. However, no quantitative binding data are available for CAL or the individual NHERF2 PDZ domains, and published estimates for the NHERF1 PDZ domains conflict. Here we demonstrate that the affinity of the CAL PDZ domain for the CFTR C-terminus is much weaker than those of NHERF1 and NHERF2 domains, enabling wild-type CFTR to avoid premature entrapment in the lysosomal pathway. At the same time, CAL’s affinity is evidently sufficient to capture and degrade more rapidly cycling mutants, such as ΔF508-CFTR. The relatively weak affinity of the CAL:CFTR interaction may provide a pharmacological window for stabilizing rescued ΔF508-CFTR in patients with cystic fibrosis.
CFTR is a cAMP-activated, ATP-gated chloride channel. It plays a central role in maintaining fluid and ion homeostasis in epithelial tissues and is mutated in patients with cystic fibrosis (CF) (1). Although CFTR is subject to rapid endocytosis (2), this appears to be coupled with a highly efficient constitutive recycling mechanism (e.g refs. 3, 4). As a result, mature CFTR exhibits a long functional half-life (5, 6), requiring individual molecules to cycle through the endocytic pathway dozens or even hundreds of times.
The endocytic uptake and recycling of CFTR are controlled by a wide variety of proteins whose interactions are only incompletely understood (1, 7). However, three proteins have been shown to be closely implicated in the regulation of CFTR trafficking and localization: NHERF1 (8–10) and NHERF2 (11), which each contain two PDZ domains, and CAL (12), which contains one. PDZ domains are modular protein-protein interaction domains whose binding affinity is typically dominated by interactions with the C-terminus of the target protein. In addition to the terminal carboxylate group, such PDZ domains specifically bind a few side chains, and are generally classified according to the resulting sequence motifs (13). Consistent with this pattern, NHERF1, NHERF2, and CAL all bind the cytoplasmic C-terminus of CFTR.
Overexpression of CAL dramatically shortens the half-life of mature CFTR (12) by promoting its lysosomal degradation following endocytosis (14). Given that CAL and CFTR can interact during endocytic trafficking pathways, and that their interaction favors CFTR degradation, how does wild-type CFTR achieve such high levels of recycling efficiency?
CAL’s effect can be blocked by concomitant expression of NHERF1 (12), a protein that is known to promote post-endocytic recycling as opposed to degradation for a number of receptors (15, 16). Both NHERF1 and NHERF2 exert favorable influences on Cl− currents mediated by CFTR, stabilizing its apical membrane expression levels and mediating interactions with a network of regulatory proteins (1, 7). Consistent with this observation, cell-surface levels of the most common disease-associated CFTR mutation (ΔF508-CFTR) can be enhanced in airway epithelial cells either by overexpression of NHERF1 (17) or by suppression of endogenous CAL (18).
As a result, the recycling efficiency of CFTR is significantly affected by its relative affinities for these functionally antagonistic partners: CAL on the one hand, and NHERF1 and NHERF2 on the other. However, at present, no information is available on the affinity of CAL binding either to CFTR or to any of its other known binding partners, with the exception of the β1 adrenergic receptor (β1-AR) (19). Similarly, for NHERF2, no information is available on the binding affinities of the individual PDZ domains for CFTR. For NHERF1, data are available, but published studies present conflicting estimates ranging from 14 to 300 nM for the PDZ1 domain and from 16 to 4800 nM for the PDZ2 domain (7, 8, 20, 21).
This highlights a common problem in understanding protein-protein interactions: while it is comparatively easy to identify interactions qualitatively, it is much more difficult to quantitate their affinities and even harder to do so in a way that permits meaningful comparison. In the case of PDZ proteins, completely different techniques have often been used to study the binding of different partners. In particular, solution- and surface-based methods have tended to yield significantly different estimates, often due to avidity effects in surface-based approaches. Side-by-side comparisons of multiple interactions with a single method have only recently become available (e.g. ref. 22), and cross-methodological comparisons of individual, biochemically well-defined PDZ interactions remain scarce.
Here we report a systematic fluorescence polarization (FP) analysis of CFTR-PDZ interactions. Parallel measurements of the binding of the CFTR C-terminus to the NHERF1 PDZ1 domain using FP, isothermal titration calorimetry (ITC) and surface-plasmon resonance (SPR) provide a complete thermodynamic profile of the interaction and resolve conflicting published data on the affinity of the interaction. Our FP assay provides the first data on the CFTR-binding affinities of the PDZ domains of CAL and NHERF2, and directly comparable binding data for the NHERF1 interaction. Finally, we have used FP shifts to determine the relative affinities of CAL for known binding partners other than CFTR. Taken together, our studies reveal that CAL and CFTR bind each other with very low affinity compared to their other interactions. As a result, the CAL:CFTR complex may be susceptible to selective pharmacological targeting as a means of retaining rescued ΔF508-CFTR at the apical membrane. Furthermore, the weak affinity of CFTR for CAL compared to NHERF1 provides a biochemical explanation for the efficient post-endocytic recycling of WT CFTR.
EXPERIMENTAL PROCEDURES
Recombinant Protein Expression Vectors
For a list of protein abbreviations, see Table 1. NHERF1 (SwissProt accession #O41745) and NHERF2 (SwissProt accession #Q15599) constructs were subcloned into the pET16b expression vector (Novagen) on an NdeI/BamHI fragment generated by PCR. In each case, the coding sequence was introduced in frame with the start codon and N-terminal decahistidine purification tag of the vector, followed by an SSGHIEGRH linker. The NHERF1 PDZ1 domain (N1P1; residues 1-139) and the NHERF2 PDZ1 (N2P1; residues 1-151) and PDZ2 (N2P2; residues 142-280) domains were subcloned from cDNA constructs provided by the Dartmouth CF Core Facility (originally generous gifts of Dr. K. Foskett, University of Pennsylvania and Dr. M. Donowitz, Johns Hopkins University, respectively). The NHERF1 PDZ2 domain (N1P2; residues 132-299) was subcloned from a full-length protein isolated from a human adult kidney cDNA library (BioChain Institute). GYGH binding-site mutations were introduced into the NHERF1 PDZ domains (N1P1: F26H, N1P2: F166H) using the QuickChange II Site Directed Mutagenesis Kit (Stratagene).
Table 1.
Molecular weights and experimental and calculated molar extinction coefficients for the protein constructs used in binding studies.
| Protein | Abbn. | Mol. Wt. | E280 nm (cm−1·M−1) | |
|---|---|---|---|---|
| Experimental | Calculated | |||
| NHERF1 PDZ1 | N1P1 | 17555 | 2464 | 2980 |
| NHERF1 PDZ2 | N1P2 | 21128 | 1599 | 1490 |
| NHERF2 PDZ1 | N2P1 | 19592 | 12615 | 12490 |
| NHERF2 PDZ2 | N2P2 | 17672 | 3105 | 2980 |
| CAL PDZ | CALP | 12689 | 3034 | 2980 |
| Full-length CAL | CAL | 49875 | 29583 | 24410 |
Molecular weights and molar extinction coefficients (E280nm) were calculated for each of the constructs used in binding studies. Experimental E280nm values were determined by measurement of absorbance values for a stock solution that was also subjected to quantitative amino-acid analysis.
The pHCAL1 vector encoding the decahistidine-tagged full-length CAL (SwissProt accession #Q9HD26-2) construct used for cross-linking studies (H10-CAL) has been described previously (18). In addition, for binding studies, we developed a vector (pHCAL3) for a full-length CAL construct that is based on pHCAL1, but contains a 3C-protease recognition sequence (LEVLFQ*G) between the polyhistidine tag and the CAL protein sequence (H10-3C-CAL).
The vectors encoding the wild-type CAL PDZ domain (CALP; residues 278-362) and the binding-site mutant (CALP-D; S294D/T296E/K340D/K342E) have also been described previously (18). The S-tagged CAL PDZ construct (CALP-S) used to validate peptide-array data was constructed in a modified pET16b vector. The coding sequence is identical to CALP, except that the stop codon is replaced by a C-terminal dipeptide linker, followed by the S-tag sequence (DPKETAAAKFERQHMDS). All constructs were verified by DNA sequencing at the Dartmouth Molecular Biology Core Facility.
Protein Expression and Purification
Full-length CAL and all PDZ constructs were expressed, harvested, and lysed, and the lysates clarified as previously described (18), except that NHERF1 and NHERF2 PDZ domains were expressed in Rosetta 2 (DE3) cells (Novagen). Immobilized metal-affinity and size-exclusion chromatographic (SEC) purification of the PDZ domains of NHERF1, NHERF2 and CAL was performed essentially as previously described for CALP (18), with two exceptions. First, the elution profile was modified for the CALP protein to provide a sharper elution. Second, the buffer used for SEC of CALP was supplemented with 1.5 mM TCEP (Tris(2-carboxyethyl)phosphine hydrochloride), 1 mM ATP, 2 mM MgCl2, 1 mM L-glutamate, and 5% glycerol. Full-length CAL proteins (H10-CAL and H10-3C-CAL) were purified as described (18). Lysis and chromatography buffers contained variable concentrations (0, 0.1, and 1 mM) of ATP.
Following SEC, H10-3C-CAL was further treated by digestion with polyhistidine-tagged human rhinovirus 3C protease (Novagen) at a 1:5 mass ratio at 4°C for 20 hours to remove the polyhistidine purification tag and most of the cleavage site. The resulting protein (described as “CAL” below) contains the full-length wild-type CAL sequence, together with additional N-terminal Gly-Pro linker. Imidazole was added to the protein to a final concentration of 40 mM before application to a 1 ml HisTrap™ HP column (GE Healthcare), which had been pre- Tris, pH 8.5, 150 mM NaCl, 1 mM DTT, 0.1 mM equilibrated with 5 CV of TBS-CAL (50 mM ATP), containing 40 mM imidazole. The cleaved CAL protein was collected in the flow-through fraction, and then dialyzed in TBS-CAL containing 0.02% NaN3 and 25 mM Tris.
Proteins were concentrated using Amicon Ultra-15 5000 MWCO (NHERF1 and NHERF2 PDZ domains) or Amicon Centriprep YM-10 (H10-CAL and CAL) or YM-3 concentrators (CAL PDZ domain constructs) (Millipore). Following concentration, proteins were dialyzed into storage buffer: 25–50 mM NaH2PO4, pH 7.4 (N1P1, N1P2, and N2P1), 25 mM Tris, pH 7.5 (N2P2) or 25 mM Tris, pH 8.5 (CAL PDZ and full-length proteins); 150 mM NaCl; 0.02% NaN3. Buffers included 0.1 mM TCEP or 1 mM DTT, and for H10-CAL and CAL proteins, 0.1 mM ATP. Following concentration, for each protein, oligomeric homogeneity was assessed by analytical SEC, purity was assessed by SDS-PAGE, and the relative molar mass was verified using matrix-assisted laser desorption/ionization time-of-flight spectrometry on an Applied Biosystems Voyager DE Pro System. Protein stock solutions were supplemented with 5–10% (v/v) glycerol and flash frozen for storage.
Protein Concentration Determination
The absorbance of a protein stock solution at 280 nm ( ) was assessed for each protein. The amino-acid content of the same stock was determined by quantitative amino-acid analysis (Keck Biotechnology Resource Laboratory, Yale University), providing an experimental estimation of the extinction coefficient for each protein (Table 1). Bradford reagent (Biorad) was used for routine detection during purification and concentration.
Circular Dichroism
Protein constructs were diluted to ~0.5 mg/mL in either 25 mM NaH2PO4, pH 7.4, 150 mM NaCl (NHERF1 and NHERF2 PDZ domains) or 25 mM Tris-HCl pH 8.5, 150 mM NaCl (CAL PDZ and full-length CAL). Proteins were equilibrated at 25 °C in a Jasco J-715 spectropolarimeter, and circular dichroism spectra measured from 210 to 240 nm. After incubation at 25 °C for ≥2 h, spectra were acquired again and compared to assess structural integrity. In addition, protein samples were heated stepwise to ≥55 °C and spectra determined from 210–240 nm to assess structural integrity as a function of temperature.
Peptide Ligands
Peptides were synthesized by the Yale Keck Biotechnology Resource, Genscript, or Tufts University Core Facility. As standard nomenclature, native peptide sequences are indicated by an abbreviation of the target protein name and a subscript indicating the number of C-terminal residues included. N-terminal modifications are indicated by a prefix. In some cases, a fluorescein moiety was attached synthetically to the peptide N-terminus, either directly (F*-) or via a diglycine linker (F*GG-). In other cases, an N-terminal cysteine residue (C-) or cysteine-diglycine linker (CGG-) was added to the sequence. Cysteine-tagged peptides were used without further modification as inhibitors, or were labeled for use as reporters (F*C-) by adding fluorescein-5-maleimide (Molecular Probes) at a ~50% molar excess in 100 mM NaH2PO4, pH 7.4, 5 mM EDTA and incubating for 1 hour on ice in the dark. Coupled peptides were purified by SEC, using a 10 mL Sephadex G-15 (Pharmacia) column equilibrated in 25 mM NaH2PO4, pH 7.4, 150 mM NaCl, 0.02% NaN3. Purity was assessed by matrix-assisted laser desorption/ionization time-of-flight spectrometry on an Applied Biosystems Voyager DE Pro System, following desalting using Omix C18 pipette tips (Varian) and target preparation using 2,5-dihydroxybenzoic acid matrix (Aldrich). Maleimide-coupled fluorescent peptide concentrations were determined using the molar extinction coefficient for fluorescein (Molecular Probes). A control peptide sequence (“C-SCR6”; C-QVTLDR) was also synthesized, corresponding to a scrambled version of the C-CFTR6 sequence.
Fluorescence Polarization
Fluorescence polarization data were measured on a Spectramax M5 microplate reader (Molecular Devices) at 24°C. For Kd measurements with a given fluorescently labeled peptide, a stock solution of protein was incubated for 10–30 minutes at room temperature in FP buffer (storage buffer, supplemented to a final concentration of 0.1 mg/mL bovine IgG (Sigma) and 0.5 mM Thesit (Fluka)) containing 30 nM fluorescent peptide. The pre-equilibrated protein:reporter peptide mixture was then serially diluted into FP buffer containing 30 nM fluorescent peptide and allowed to incubate for 10 minutes. Following centrifugation for 4 minutes at 1200 × g to remove air bubbles, 40 μL aliquots were transferred to HE low-volume, black 96-well plates (Molecular Devices). The plates were again centrifuged for 4 minutes at 1200 × g, placed in the microplate reader, and allowed to equilibrate at 24 °C for an additional 15 minutes prior to measurement.
Competition binding experiments were performed similarly. A single stock solution was prepared in FP buffer containing fixed concentrations of both fluorescently labeled reporter peptide and protein. This mixture was allowed to equilibrate for 20–60 minutes at RT. Unlabeled competitor peptide was dissolved and serially diluted in DMSO (Fluka). Each serial dilution was aliquoted at 1/20th final volume, to which was added 19/20th volume of the protein:reporter mixture. The final reporter peptide concentration was 30 nM, and the final protein concentration was 0.25 – 3.0 × Kd, depending on the measurement. Plates were mixed by vibration, centrifuged, and allowed to incubate for an additional 15 minutes at 24 °C in the microplate reader before measurement.
Fluorescence polarization was determined at an excitation wavelength of 485 nm and an emission wavelength of 525 nm as
| (1) |
where Ivv and Ivh represent the vertically and horizontally polarized emission signals obtained with vertically polarized excitation, and g represents the assay-specific polarization bias, which was separately determined in each buffer. The time of equilibration and salt concentration, and the requirement for detergent or carrier protein, were determined to minimize nonspecific peptide binding. Fluorescence intensities were monitored to ensure no change in reporter fluorescence quantum yield upon binding, and also to exclude any light-scattering contribution to the measured polarization. For analysis, data were converted to anisotropy values.
Direct binding data were fit to a model for a single-site
| (2) |
binding equilibrium. A non-linear least-squares algorithm (Kaleidagraph) was used to fit the experimental anisotropy (rexp) to the anisotropy calculated by the following equation:
| (3) |
where [L]tot = total reporter peptide concentration and rL and rPL = the fluorescence anisotropies of the free and bound ligands, respectively, for the case in which fluorescence lifetime and quantum yield are unaffected by protein binding. The concentration of protein:ligand complex [PL] was determined as:
| (4) |
where [P]tot = total protein concentration and Kd = equilibrium dissociation constant of complex formation.
Competition isotherms were fit using a non-linear least-squares fitting algorithm implemented in Excel using the SOLVER function. rexp was fit to
| (5) |
where the free protein concentration [P] in the presence of both reporter and competitive inhibitor was calculated as a function of the total protein and ligand concentrations and the equilibrium dissociation constants Kd (known) and Ki (fit), respectively, by exact analytical solution of the resulting cubic equation (see eqn. 13, ref. 23). For illustrative purposes, four-parameter logistic curve fits are shown in figures to illustrate relative IC50 values.
Peptide-Array Binding Studies
As reported previously, a library of 6223 C-termini (11-mers) of human proteins was prepared, using an optimized method for generating inverted peptides (24), and incubated with the CAL PDZ domain. The peptide sequences were generated by a MultiPep SPOT-robot (INTAVIS Bioanalytical Instruments AG, Cologne, Germany; Software LISA, in-house software).
The peptide array was pre-washed with EtOH (1 × 10 min), then washed with Tris-buffered saline (TBS), pH 8.0 (3 × 10 min), and then blocked for 4 h with blocking buffer (blocking reagent [Sigma-Genosys] in TBS pH 8.0, containing 5% sucrose). The membranes were incubated with polyhistidine-tagged CALP or with S-tagged CALP-S (20 μg/ml) in blocking buffer overnight at 4°C, and then washed with TBS pH 8.0 (3 × 10 min). CALP was detected using a mouse anti-polyHis antibody (Sigma; 1:2600 in blocking buffer, 2 h at RT) followed by horseradish-peroxidase conjugated anti-mouse antibody (Calbiochem; 1:2000 in blocking buffer, 1 h at RT). The CALP-S domain was detected using a horseradish-peroxidase conjugated S-protein (Novagen; 1:5000 in blocking buffer, 2 h at RT). In either case, detection was carried out with Uptilight HRP blot chemiluminescent substrate (Uptima) with an exposure time of 1 min. The signal intensities were recorded as Boehringer Light Units (BLU) using the LumiAnalyst™ software. Background-subtracted intensities were used to identify tight-binding peptide sequences.
Isothermal Titration Calorimetry
For calorimetry experiments, NHERF1 PDZ1 was dialyzed versus 25 mM sodium phosphate or 25 mM tricine, pH 7.4, 150 mM NaCl, 1 mM TCEP, 0.02% NaN3. Dialysis buffers were degassed and aliquots of a peptide representing the C-terminal 10 residues of CFTR (CFTR10; 1471TEEEVQDTRL1480) were solubilized in the respective outer dialysis buffers. Experiments were conducted at 15, 25, or 35°C on a MicroCal VP-ITC calorimeter. A 2 μL injection of peptide over 3.4 seconds preceded 57-5μL injections over 10.3 seconds each. The system was allowed to equilibrate between injections for 450 seconds while stirring at a rate of 300 rpm. Thermograms were manually adjusted to account for the heat of dilution by the peptide. Titrations in the presence of buffers (tricine, phosphate) of known, but distinct, heats of ionization, allowed us to estimate the level of proton release during binding (NH+). In phosphate buffer, the associated heat of protonation represented <2% of the heat of reaction, and was therefore ignored during fitting. Thermograms obtained in tricine buffer at 15 and 35 °C were of higher quality than those in phosphate buffer, so that tricine ΔH data were fit to determine ΔCp.
Surface Plasmon Resonance
Sensorgrams were measured at 25 °C using a BIAcore X system (GE Healthcare). CFTR C-terminal decamer peptides containing an N-terminal cysteine (C-CFTR10) were coupled at low density to a CM4 chip. The chip was activated with a 1:1 mixture of N-hydroxysuccinimide:1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (GE Healthcare), followed by N-[β-maleimidopropionic acid] hydrazide, trifluoroacetic acid salt (Pierce) in 10 mM sodium borate and 1 M NaCl. After blocking excess amines with ethanolamine hydrochloride (pH 7.0) (Fluka), the peptide was bound to the chip in HBS (10 mM HEPES, pH 7.0, 150 mM NaCl, and 0.005% (v/v) Tween-20 (Calbiochem)). Excess maleimide reactive groups were then blocked with 50 mM cysteine in 0.1M sodium acetate, pH 4.0 and 1 M NaCl. The C-CFTR10 peptide was captured at 23 response units. N1P1 was injected over the C-CFTR10 chip in HBS at a flow rate of 20 μl/min for 3 minutes, followed by a 3 minute wash-out. The sensor chip was regenerated between successive injections with 10 mM glycine-HCl, pH 1.5 (GE Healthcare). The control surface response was subtracted from the peptide-coupled surface response to create a response curve. Data were analyzed using the BIAevaluation software package (GE Healthcare). Curves were corrected by zeroing at actual injection time and subtracting the response generated by a buffer blank injection. They were globally fitted to a kinetic model of a simple 1:1 bimolecular binding (A + B = AB). Results obtained at multiple flow rates confirmed that mass transport was not rate limiting.
Analytical ultracentrifugation
Immediately following preparative SEC of H10-CAL, a protein sample from the trailing edge of the peak was concentrated in a Centricon concentrator (50000 MWCO; Millipore) to yield an . Sedimentation velocity ultracentrifugation was carried out at 35,000 rpm at 20 °C in a Beckman ProteomeLab XL-A centrifuge equipped with absorbance optics and an AN-60 rotor. Absorbance scans were performed at 70 sec intervals at a wavelength of 280 nm using a 0.003 cm scan step. The partial specific volume of the protein (ν̄), buffer density (ρ) and viscosity (η) were estimated from the protein sequence and buffer composition using the program SEDNTERP (J. Philo, D. Hayes, T. Laue, unpublished). Alternate scans from 1 to 349 were analyzed using the program SEDFIT87 (25) to determine the distribution of protein concentration as a function of sedimentation coefficient, c(s). Fitting of multiple-component equilibria was performed using the program SEDANAL v3.61 (26).
Cross-Linking
Purified H10-CAL protein was incubated at a final concentration of 0.2 mg/ml with bis[sulfosuccinimidyl] suberate (BS3; Pierce) at a concentration of 0.28 mM in 25 mM sodium phosphate buffer, pH 8.5, 150 mM NaCl, 1 mM DTT, 0.1 mM ATP, 0.02% NaN3, representing a 2-fold stoichiometric excess of cross-linker to primary amine reactive groups in the H10-CAL protein. Samples were incubated at RT for 5, 15, 30, or 60 minutes prior to addition of ethanolamine (GE Healthcare) to a final concentration of 100 mM. The 0 minute sample had ethanolamine added prior to cross-linker. Following ethanolamine quenching, samples were incubated on ice for an additional 5 min prior to addition of ½ volume of 3× reducing sample buffer. Samples were resolved under reducing conditions by SDS-PAGE on precast 4–15% Tris-HCl gradient gels (Biorad) and visualized using Bio-safe Coomassie G-250 stain (Biorad).
RESULTS
Protein Purification and Characterization
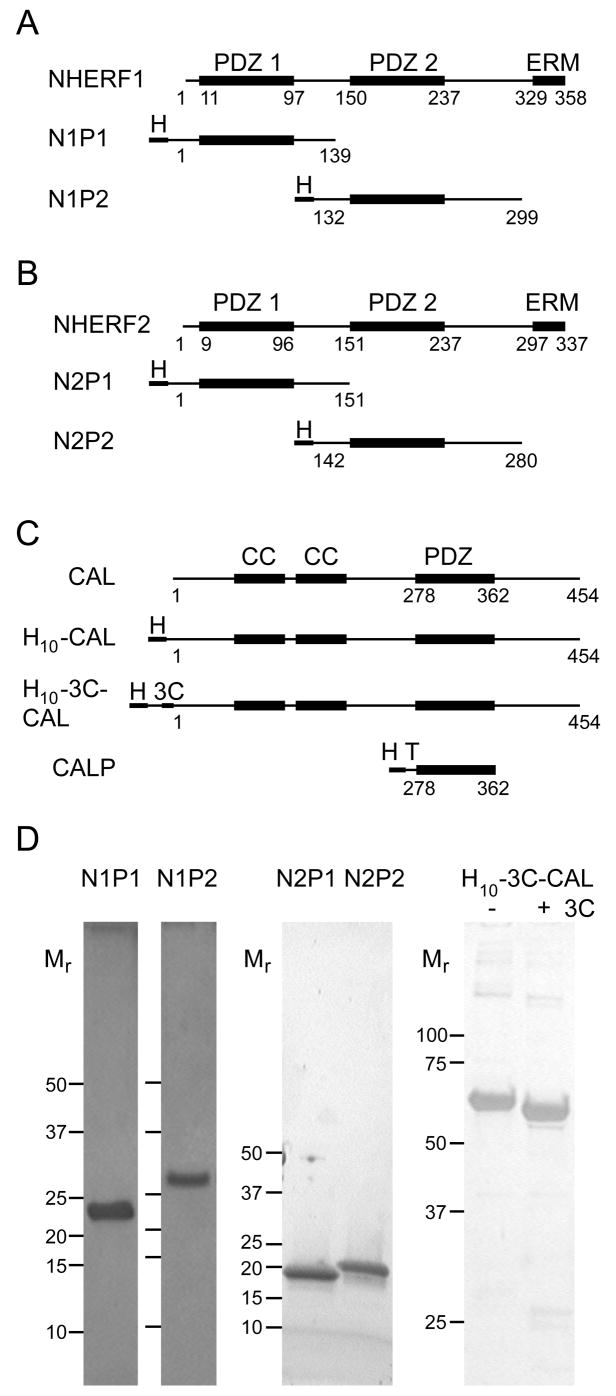
In order to determine the relative binding affinities of the NHERF1, NHERF2, and CAL PDZ domains for the CFTR C-terminus, we developed a system to supply milligram quantities of purified recombinant proteins. An appropriate purification scheme had already been established for the CAL PDZ domain (CALP) (18, 27). Published reports also had described purification of both NHERF1 PDZ domains for biochemical studies (20). We subcloned corresponding constructs for the NHERF1 and NHERF2 PDZ1 and PDZ2 domains (N1P1, N1P2, N2P1, and N2P2) and purified them to homogeneity (Figs. 1A and 1B). We also subcloned a novel full-length CAL construct (H10-3C-CAL) with a cleavable polyhistidine tag, for use in binding studies (Fig. 1C). All proteins were obtained with excellent purity and yield (Fig. 1D).
Figure 1. Purified proteins used in binding studies.
Schematic representations are shown for the domain structures (top line) of (A) NHERF1, (B) NHERF2, and (C) CAL, together with boundaries of the recombinant constructs used in this study (lower lines). PDZ, coiled coil (CC), and ezrin-radixin-moesin binding (ERM) domains are indicated above the native sequences (top line in A–C). The position of included domains (thick lines), decahistidine purification tags (H), TEV protease cleavage sites (T) and 3C-protease cleavage sites (3C) are indicated schematically for each construct. (D) Silver- (N1P1, N1P2, H10-3C-CAL) or Coomassie- (N2P1 and N2P2) stained SDS-PAGE gels are shown for previously unpublished protein constructs used in these studies. Mr standards are marked to the left of each lane. For the PDZ domains, Mr standards are shown at 50, 37, 25, 20, 15, 10 kD. For H10-3C-CAL, they are shown at 100, 75, 50, 37 and 25 kD. The dye front was run off the gel for H10-3C-CAL, to resolve more clearly the molar-mass shift before (−) and after (+) 3C-protease treatment. Protein Mr were separately validated by mass spectrometry for each construct.
Since affinity measurements require a rigorous knowledge of protein concentration, we calibrated the molar extinction coefficients of all purified constructs used in binding studies by performing quantitative amino-acid analysis. The experimental values agree well with those calculated from the protein sequences (Table 1). Finally, in order to ensure protein stability at the temperatures required for the binding assays described below, we performed circular dichroism studies for each construct as a function of temperature. All constructs were stable above the maximum temperature used in our binding assays. Furthermore, circular dichroism spectra before and after prolonged incubation at 25°C indicated no significant change over 2–4 hours, corresponding to the time-scales of our experiments (data not shown).
Solution-Binding Analysis of NHERF1 PDZ:CFTR Interactions
Published data report generally nanomolar affinities for binding of the CFTR C-terminal peptide sequence to the NHERF1 PDZ domains (7, 8, 20, 21). However, the different estimates conflict, ranging over several orders of magnitude. Furthermore, all available data are based on solid-state techniques that can overestimate protein affinity due to avidity effects. Preliminary experiments suggested that the CAL-CFTR interaction exhibits relatively low affinity and would not be amenable to solid-state studies. As a result, in order to obtain directly comparable affinites for all five PDZ domains, and if possible, to resolve the conflicting estimates for NHERF1-CFTR affinity, we decided to establish a fluorescence polarization (FP) assay. FP is a sensitive, solution-state technique capable of analyzing peptide-protein interactions across a wide range of affinities.
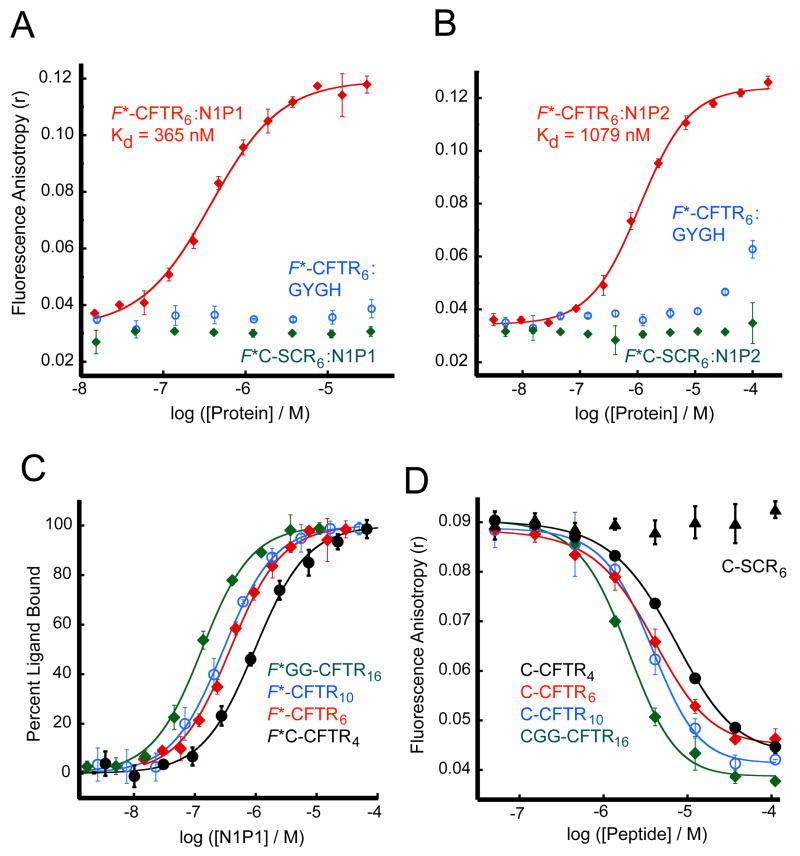
Given the strong reported affinity of the NHERF1 PDZ domains for CFTR, we focused first on obtaining solution-state binding data for their interactions. A reporter peptide was synthesized corresponding to the last six residues of the CFTR sequence (1475VQDTRL1480), with a fluorescein moiety covalently attached to the N-terminus (F*-CFTR6). In the absence of binding proteins, the free peptide exhibited low levels of fluorescence polarization, as expected (Fig. 2A and 2B). Furthermore, when incubated with increasing concentrations of the NHERF1 PDZ1 (N1P1) and PDZ2 (N1P2) domains, a saturable, dose-dependent shift was observed in the fluorescence polarization of the samples, consistent with the formation of a peptide:protein complex with increased hydrodynamic radius. When converted to the related quantity of fluorescence anisotropy and fit to an equation for single-site binding, the observed isotherms yielded affinities of 365 ± 35 nM for N1P1 and 1079 ± 79 nM for N1P2 (Figs. 2A and 2B, red curves).
Figure 2. Solution-state binding assay reveals high affinity of the NHERF1 PDZ domains for the CFTR C-terminus.
(A and B) Fluorescence anisotropy data were obtained in the presence of increasing concentrations of NHERF1 PDZ1 (N1P1) (A) and PDZ2 (N1P2) (B) in the presence of reporter peptides corresponding to the C-terminal six residues of CFTR (F*-CFTR6, red) or a scrambled control sequence (F*C-SCR6, green), each covalently coupled to fluorescein. Curve fits (solid lines) yielded estimates of Kd of 365 ± 35 nM and 1079 ± 79 nM, respectively. Parallel titrations were performed with the GYGH mutants (blue) of the N1P1 (A) and N1P2 (B) domains. (C) Fluorescein-labeled peptides corresponding to the C-terminal four (F*C-CFTR4, black, filled circles), six (F*-CFTR6, red, filled diamonds), ten (F*-CFTR10, blue, open circles), or sixteen (F*GG-CFTR16, green, open diamonds) residues of CFTR were incubated at 30 nM each with increasing concentrations of the N1P1 domain, and the fluorescence anistropy determined. Curves were fit (solid lines) as in (A–B), yielding estimates of Kd shown in Table 2. (D) Residual fluorescence anistropy was determined for a mixture of F*-CFTR6 (30 nM) and N1P1 (1 μM ≈ 3 × Kd) incubated with increasing concentrations of unlabeled C-CFTR4 (black, filled circles), C-CFTR6 (red, filled diamonds), C-CFTR10 (blue, open circles), CGG-CFTR16 (green, open diamonds) or a scrambled control peptide (C-SCR6) (black, triangles), shown with logistic curve fits (solid lines). Values shown are mean ± SD, n=3.
In order to assess the peptide-sequence specificity of the observed FP shift, we tested the ability of a fluorescent reporter peptide with a scrambled sequence (F*C-SCR6) to interact with both domains: neither domain caused an FP shift of the scrambled reporter (Figs. 2A and 2B, green curves). Furthermore, to determine whether the detected interaction involved the targeted PDZ site, we mutated the phenylalanine residue in the canonical “GYGF” carboxylate-binding motif of the N1P1 (F26) and N1P2 (F166) PDZ binding pockets (ref. 28, and PDB entry 2OZF, unpublished) to histidine. In both cases, titration with the “GYGH” mutant domain caused either no FP shift or only a weak shift at significantly higher protein concentrations (Figs. 2A and 2B, blue curves), indicative of a severely attenuated interaction affinity.
Length Dependence of CFTR Affinity for NHERF1
While the four C-terminal residues encode much of the affinity of typical PDZ binding interactions, studies have shown that upstream residues may contribute as well (29–31). In order to characterize this effect for the NHERF1 PDZ domains and to establish a baseline length for cross-domain binding comparisons, we performed both direct binding (Fig. 2C) and competition experiments (Fig. 2D) in parallel with peptides containing the C-terminal 4, 6, 10, and 16 residues of CFTR, in order to determine the effect of peptide length on affinity. As shown in Table 2, the tetrapeptide encodes much of the affinity of the interaction. The affinity of the peptides longer than six amino acids increased only modestly, consistent with observations in other PDZ domains (31). However, because other reports have observed an influence of residues between P-5 and P-9 (29–31), we used decamer peptides for further experiments. At each length, the presence of an N-terminal fluorescein moiety in direct (Kd) titrations caused a small (< 2.2-fold), but consistent increase in affinity compared to competition (Ki) studies performed using an unlabeled peptide.
Table 2.
NHERF1 PDZ1 binding affinity depends on peptide length.
| Peptidea | Sequenceb | Kd (nM) | Ki (nM) |
|---|---|---|---|
| F*C-CFTR4 | F*CDTRL | 949 ± 89 | |
| C-CFTR4 | CDTRL | 1520 ± 78 | |
| F*-CFTR6 | F*VQDTRL | 365 ± 35 | |
| C-CFTR6 | CVQDTRL | 670 ± 97 | |
| F*-CFTR10 | F*TEEEVQDTRL | 279 ± 21 | |
| C-CFTR10 | CTEEEVQDTRL | 597 ± 77 | |
| F*GG-CFTR16 | F*GGAALKEETEEEVQDTRL | 116 ± 10 | |
| CGG-CFTR16 | CGGAALKEETEEEVQDTRL | 209 ± 64 |
Kd values were determined by fluorescence anisotropy measurements following titration of the indicated fluorescently labeled sequences with increasing concentrations of N1P1. Ki values were determined by titration of the indicated peptide sequences in the presence of 30 nM F*-CFTR6 and 1 μM N1P1 and determining the fluorescence anisotropy of the reporter ligand. Data were fit using equations 3 and 4 (Kd) or equation 5 (Ki).
F*- and F*GG represent fluorescein groups coupled covalently to the peptide amino-terminus. F*C- represents a fluorescein group maleimide-coupled to an N-terminal cysteine side chain. C- and CGG- represent unmodified N-terminal linkers. Subscript indicates length of CFTR C-terminal sequence included.
CFTR sequences are underlined.
Thermodynamic Analysis of N1P1-CFTR Binding
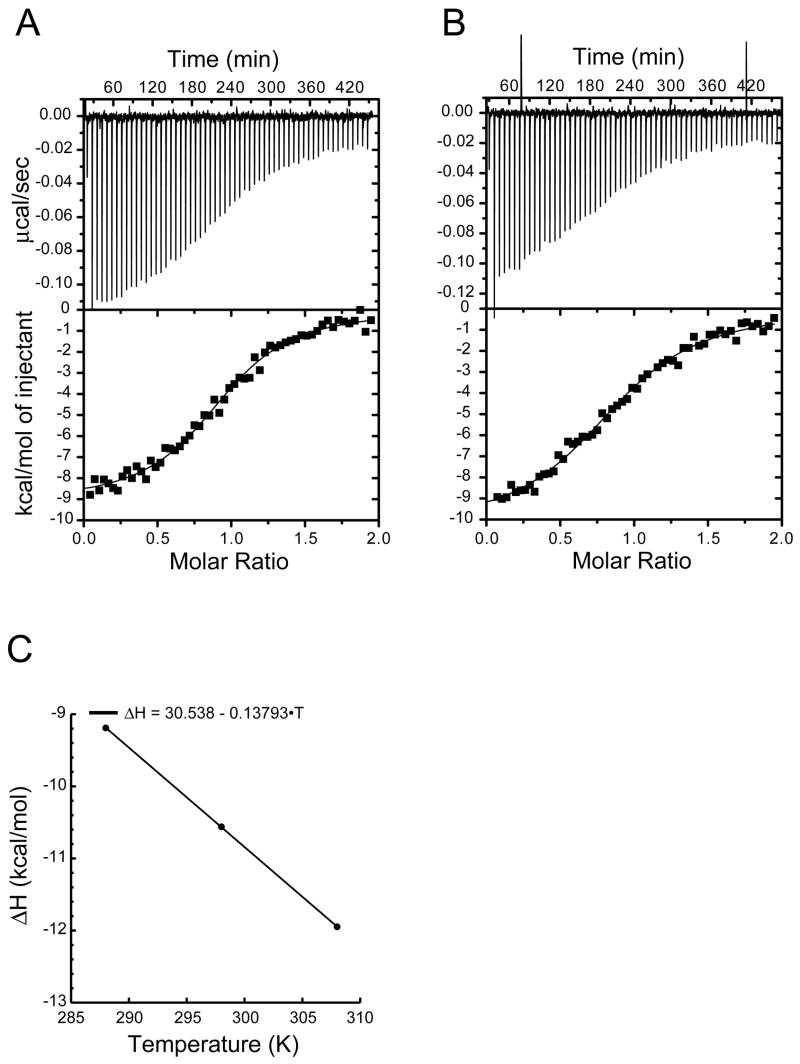
A detailed analysis of the enthalpy and entropy of binding processes can provide important insights into the stereochemistry of the interaction, but such information is available only for a limited subset of PDZ domains (e.g. refs. 31, 32). Here, we performed ITC measurements on the N1P1-CFTR binding interaction using the unmodified decamer peptide CFTR10 (1471TEEEVQDTRL1480). Representative thermograms are shown in tricine (Fig. 3A) and phosphate (Fig. 3B) buffers. The different heats of protonation of the buffers allowed us to determine the level of proton release upon binding (NH+) as 0.19 per mole, consistent with a negligible (<2%) heat of protonation in phosphate buffer. Fitting of the phosphate thermograms yielded an estimate for Kd of 787 ± 55 nM, which is closely comparable to the submicromolar value obtained using FP (Ki = 597 ± 77 nM for a non-fluorescent, cysteine-coupled decamer; Table 2).
Figure 3. The N1P1:CFTR binding interaction is enthalpically driven.
(A) Thermogram of CFTR10 injected into NHERF1 PDZ1 solution in tricine (top) with a binding isotherm fit using a single-site model (bottom), yielding an estimate for ΔH= −9.11 kcal/mol. (B) The equivalent experiment was performed in sodium phosphate buffer, yielding an estimate for ΔH= −10.37 kcal/mol and permitting determination of NH+ Fitting of the titration to a single-site model (solid line) yielded a Kd value of 787 ± 55 nM. (C) Equivalent thermograms were obtained in tricine buffer at 15, 25, and 35 °C and fit to a single-site model. The tricine ΔH values were plotted as a function of temperature and fitted with a linear function to permit estimation of ΔCp (−138 cal/mol/K).
Estimates of ΔH and ΔS are −10.37 ± 0.19 kcal/mol and −6.84 cal/mol/K, respectively. Comparison of the ΔH values obtained at 15, 25, and 35°C established that ΔCp = −138 cal/mol/K (Fig. 3C), consistent with values observed for other PDZ:peptide interactions (e.g refs. 31, 32). Since ΔCp is typically constant for protein-ligand interactions (33), linear extrapolation to 37 °C, gives a value of -12.2 kcal/mol for ΔH. The equilibrium dissociation constant can also be extrapolated to physiological temperature (ref. 34, equation 2), providing an estimate of 1.6 μM. Based on these values, at 37 °C ΔG is −8.2 kcal/mol, and TΔS is −4.0 kcal/mol, indicating that binding is enthalpically driven at physiological temperature.
Furthermore, the enthalpy and heat capacity estimates allow us to calculate the net solvent-accessible surface area sequestered by the interaction as 1365 Å2, with polar and apolar contributions of 671 Å2 and 694 Å2, respectively (35).
Comparison of Solid-State and Solution-State Binding Measurements: N1P1-CFTR Affinity by SPR
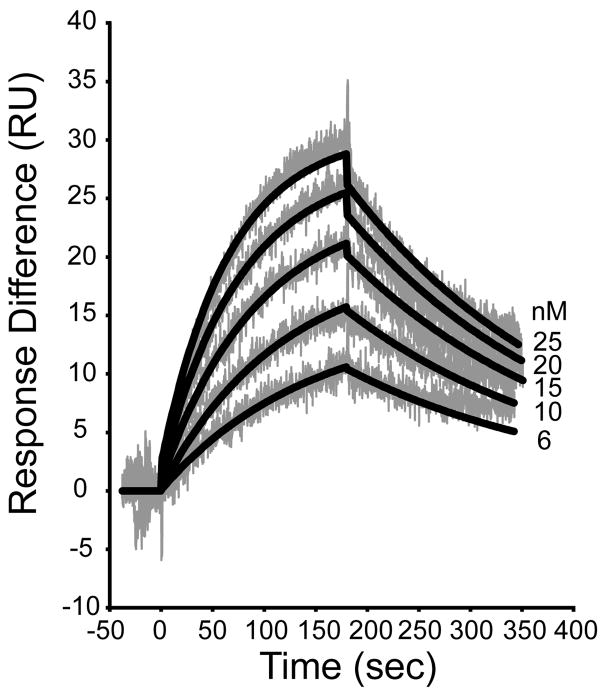
Because our solution-based affinity values for the N1P1-CFTR interaction differ significantly from most (8, 20, 21) but not all (7) data obtained using SPR techniques, we also performed SPR measurements with our protein in order to assess the comparability of our protein construct to those used in other work. Following surface immobilization of a peptide containing a cysteine residue followed by the C-terminal 10 residues of CFTR (C-CFTR10), we obtained a dose-dependent binding signal (Fig. 4). Global fits of the data over a restricted range of analyte concentrations (6 to 25 nM) yielded estimates of ka = 4.3 × 105 M−1s−1, kd = 4.4 × 10−3 s−1, and Kd = 10.3 nM (Fig. 4), in good agreement with previous values (8, 20, 21). However, attempts to fit globally a broader range of protein concentrations were less successful, generating large, systematically varying residuals (not shown). As a result, although we believe these data demonstrate the comparability of our protein to previously studied constructs, we were not able to determine the affinity of this interaction using SPR with immobilized peptide ligands and protein analytes.
Figure 4. Solid-state binding assay provides a higher estimate of affinity for the N1P1:CFTR C-terminal interaction over a limited concentration range.
An undecameric peptide containing an N-terminal cysteine and the C-terminal 10 residues of CFTR (C-CFTR10) was immobilized by maleimide coupling to an activated CM4 chip in a Biacore X System. The indicated concentrations of purified N1P1 were allowed to interact with the surface for 180 sec (association phase) and then washed out for a further 180 sec (dissociation phase), each at 20 μL/min. The response difference between the peptide labeled and control surfaces (grey) was measured in response units, and the resulting data were fit globally using the BIAevaluation software (black curves), yielding an estimate for Kd = 10.3 nM.
NHERF2 Affinity for CFTR
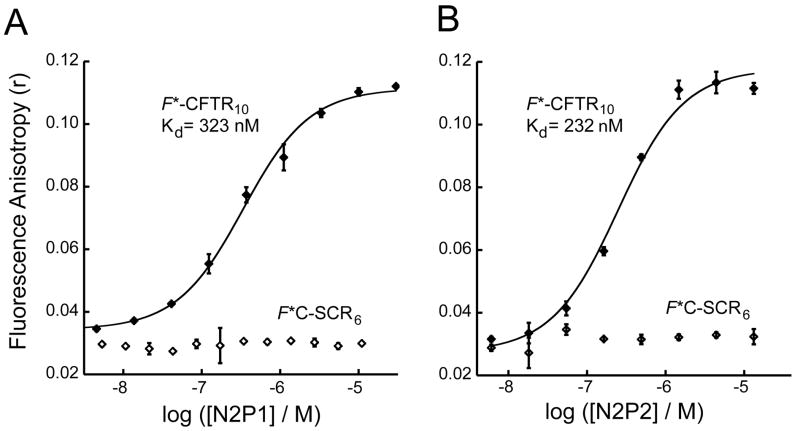
Since NHERF2, like NHERF1, functionally stabilizes CFTR activity at the apical membrane, we also expressed and purified the NHERF2 PDZ1 (N2P1) and PDZ2 (N2P2) domains (Figs. 1B and 1D) for binding analysis. Titration of fluorescent CFTR decamer reporter peptide with increasing concentrations of both NHERF2 domains led to a dose-dependent and saturable increase in fluorescence polarization similar to that seen with the NHERF1 domains (Fig. 5). Fitting of the data to a single-site binding model yielded estimates for Kd of 323 ± 32 nM and 232 ± 46 nM for the N2P1 and N2P2 domains, respectively. In each case, the control titration with a scrambled peptide showed no change in FP as a function of protein concentration (Fig. 5). The higher affinity of the N2P2 domain compared to N2P1 is consistent with previously published qualitative results (11).
Figure 5. NHERF2 PDZ domains bind the CFTR C-terminus with high affinity.
(A and B) Fluorescence anisotropy data were obtained for 30 nM F*-CFTR10 (filled diamonds) or a scrambled control peptide F*C-SCR6 (open diamonds) in the presence of increasing concentrations of N2P1 (A) or N2P2 (B). Single binding-site fits (solid lines) are shown for N2P1 (A; Kd = 323 ± 32 nM) and N2P2 (B; Kd = 232 ± 46 nM). Values shown are mean ± SD, n=3.
Identifying Novel Higher Affinity CAL Reporter Ligands: Peptide Arrays
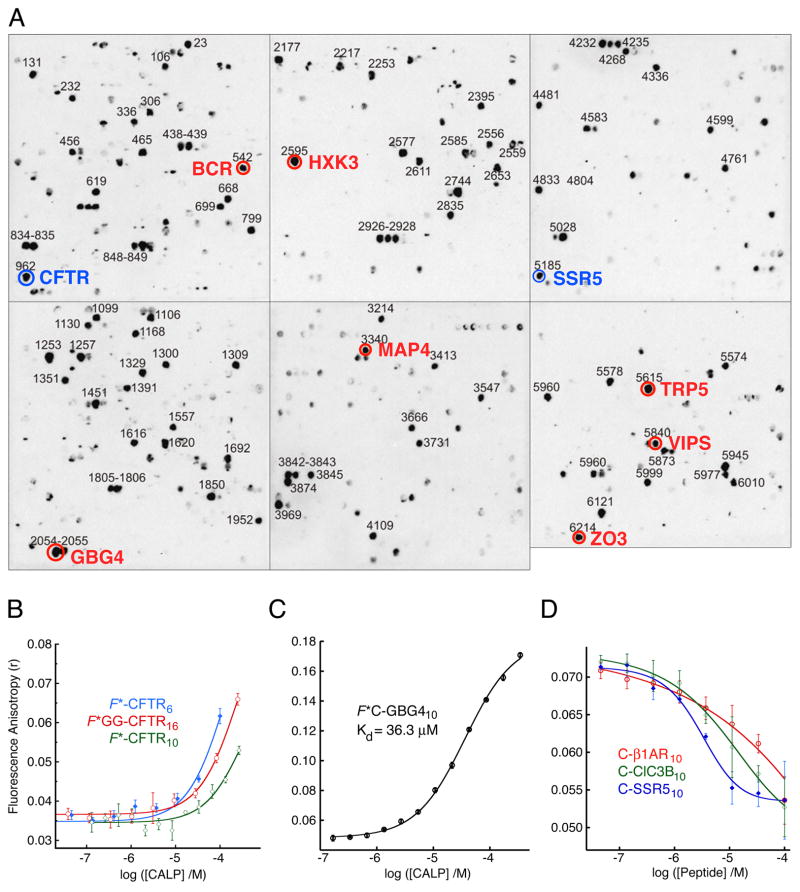
In comparison to its relatively strong interactions with the NHERF1 and NHERF2 PDZ domains, the fluorescein-labeled CFTR reporter peptides containing 6, 10, or 16 amino acids exhibited increased FP only at the highest tested concentrations of the CALP protein (Fig. 6B). Furthermore, none of the FP binding isotherms exhibited saturation.
Figure 6. Identification of higher-affinity CAL reporter ligands.
(A) A peptide array containing 6223 N-terminally immobilized human protein C-termini is shown following incubation with polyhistidine-tagged CALP protein and chemiluminescent immunodetection of bound protein. Blue circles highlight known CAL binders CFTR (12, 18) and SSR5 (37). Red circles highlight novel candidate sequences tested for CAL binding as decameric peptides with an N-terminal cysteine for coupling to fluorescein-5-maleimide. (B) Fluorescence anisotropy was determined for F*-CFTR6 (blue, filled diamonds), F*-CFTR10 (green, open diamonds), or F*GG-CFTR16 (red, open circles) peptides in the presence of increasing concentrations of the CALP protein and fit (solid lines) to a single-site model. (C) The fluorescence anisotropy of the GBG4 reporter (F*C-GBG410) increases with increasing concentration of CAL PDZ domain. When fit to a single-site binding model (solid line), an estimate of 36.3 ± 1.7 μM is obtained for Kd. (D) Logistic fits (solid lines) are shown for incubations of F*C-GBG410 reporter (30 nM) with 10 μM CALP (~0.25 × Kd) in the presence of increasing concentrations of peptides corresponding to the validated CAL ligands β1-AR (C-β1AR10 red, open circles, Ki = 410 ± 190 μM), ClC-3B (C-ClC3B10, green, open diamonds, Ki = 110 ± 60 μM), and SSR5 (C-SSR510, blue, filled diamonds, Ki = 22.6 ± 7.1 μM). Values shown are mean ± SD, n=3.
Since it was impossible to achieve saturation with direct titration of CALP, we sought a higher affinity reporter peptide, both to characterize better the affinity of the CAL PDZ domain for the CFTR C-terminus by competition binding, and also to ensure that our purified CALP construct was biochemically active. In order to survey a large population of candidate reporter peptides efficiently, we utilized the HUMLIB array of 6223 human C-terminal undecamer peptides (36).
Purified CALP was incubated with the HUMLIB array, and protein binding was quantified via chemiluminescent detection of the polyhistidine purification tag (Fig. 6A) or of a construct modified to incorporate an S-tag (not shown). Sequence analysis of the peptides showing the highest specific binding signals yielded the binding motif -S/T-X-L-COOH. Validated CAL binding epitopes were identified among the peptide binders (CFTR, ref. 18, and SSR5, ref. 37, blue circles in Fig. 6A), confirming the ability of the array to detect physiologically relevant interactions.
Seven novel candidate higher-affinity peptides were identified based on strong binding signals in both assays (red circles in Fig. 6A). Corresponding reporter peptides were synthesized with an N-terminal cysteine residue for fluorescein-5-maleimide coupling. Labeled peptides were incubated with increasing concentrations of CALP, and the level of fluorescence polarization detected. The highest affinity interaction was observed with the fluorescein-labeled guanine nucleotide-binding protein gamma-4 subunit precursor (F*C-GBG410), which exhibited a Kd value of 36.3 ± 1.7 μM (Fig. 6C).
In search of even higher affinity reporter ligands, we synthesized three additional peptides, with an N-terminal cysteine residue followed by the C-terminal 10 residues of three validated CAL binding partners: β1-adrenergic receptor (β1-AR, ref. 19), the chloride channel ClC-3B (38), and somatostatin receptor subtype 5 (SSR5, ref. 37). Each was tested for its ability to inhibit the interaction of 30 nM F*C-GBG410 peptide with CALP at low protein concentrations (10 μM, i.e. ~0.25 × Kd), to facilitate inhibition even by weakly binding peptides (Fig. 6D). The C-SSR510 sequence was the most potent inhibitor tested, with a Ki value vs. the F*C-GBG410 reporter of 22.6 ± 7.1 μM, compared to values of 110 ± 60 μM and 410 ± 190 μM, respectively, for C-ClC3B10 and C-β1AR10. For comparison, the Ki value of the C-GBG410 sequence vs. F*C-GBG410 reporter was 109 ± 12 μM, suggesting that the SSR5 sequence has significantly higher affinity.
SSR5 As a CAL PDZ Reporter Ligand
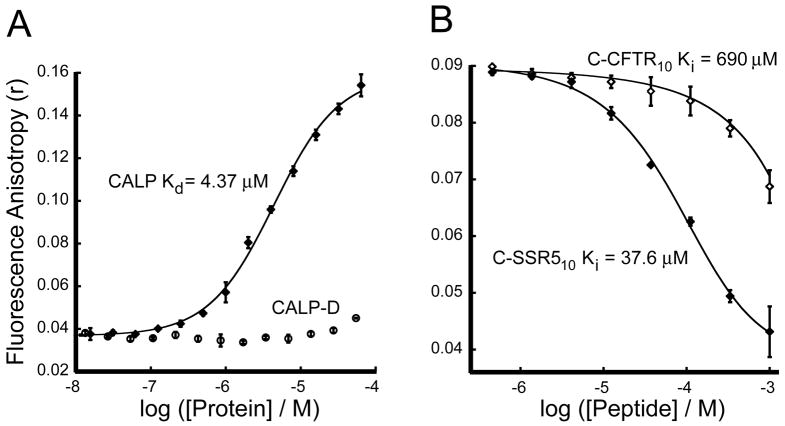
When the SSR5 C-terminal decamer was covalently labeled with fluorescein (F*C-SSR510), addition of increasing concentrations of purified CALP revealed a clear increase in peptide fluorescence polarization (Fig. 7A), analogous to that seen for the CFTR:NHERF1 PDZ interactions (Figs. 2A and 2B). The fitted Kd value was 4.37 ± 0.31 μM. This value is significantly lower than the Ki value determined vs. F*C-GBG410 reporter. To confirm this difference and to obtain a more accurate estimate for the SSR5-CALP Ki, we performed an additional competition binding assay, using C-SSR510 peptide to compete with F*C-SSR510 reporter peptide incubated with CALP at a higher relative concentration (5 μM, ~1 × Kd for F*C-SSR510). The Ki value of 37.6 ± 4.0 μM (Fig. 7B) is almost 9-fold weaker than the Kd value of the corresponding fluorescein-labeled F*C-SSR510 peptide. Compared with the smaller (<2.2-fold) affinity enhancements seen with the fluorescein moiety in N1P1 binding peptides, this suggests that fluorescein interacts more favorably with the CAL PDZ domain, providing a greater enhancement of reporter sensitivity.
Figure 7. CALP binds the C-terminus of CFTR with very low affinity.
(A) Direct binding analysis of a high affinity reporter: Fluorescence anisotropy was determined following titration of 30 nM F*C-SSR510 reporter peptide with increasing concentrations of CALP (diamonds) or CALP-D mutant (circles). Curve fitting (solid line) yields an estimate for Kd = 4.37 ± 0.31 μM. (B) Competition binding analysis: Fluorescence anisotropy data were obtained for 30 nM F*C-SSR510 and 5 μM CALP (~ 1 × Kd) incubated with increasing concentrations of either unlabeled C-SSR510 (filled diamonds, Ki = 37.6 ± 4.0 μM) or unlabeled C-CFTR10 (open diamonds, Ki = 690 ± 120 μM). Fits are shown as solid lines. Values shown are mean ± SD, n=3.
To determine whether the observed interaction was indeed mediated by the CAL PDZ binding pocket, we attempted to purify a “GLGH” mutant of CALP, targeting the carboxylate-binding motif. Although analogous mutant constructs successfully disrupted the CFTR:NHERF1 PDZ interactions (Figs. 2A and 2B), the CALP-GLGH mutant domain proved biochemically unstable. However, we had previously shown that the CALP-D mutant (a) involves residues located in the binding pocket, (b) preserves the structure of the CAL PDZ domain, and (c) disrupts the CALP:CFTR interaction in vitro (18). Titration of the F*C-SSR510 reporter peptide with increasing concentrations of CALP-D exhibited only a very weak interaction (Fig. 7A).
The CAL PDZ Domain Binds CFTR Weakly
Using the F*C-SSR510 reporter peptide, we attempted to determine more accurately the affinity of the CFTR C-terminus for the CAL PDZ domain, using C-CFTR10 peptide to displace the CALP binding of F*C-SSR510 reporter (Fig. 7B). Fitting of the displacement yielded a Ki value for the CALP:CFTR affinity of 690 ± 120 μM, although this is only a rough estimate, because inhibition was incomplete even at a CFTR peptide concentration as high as 1mM.
Oligomerization Stoichiometry of Full-length CAL
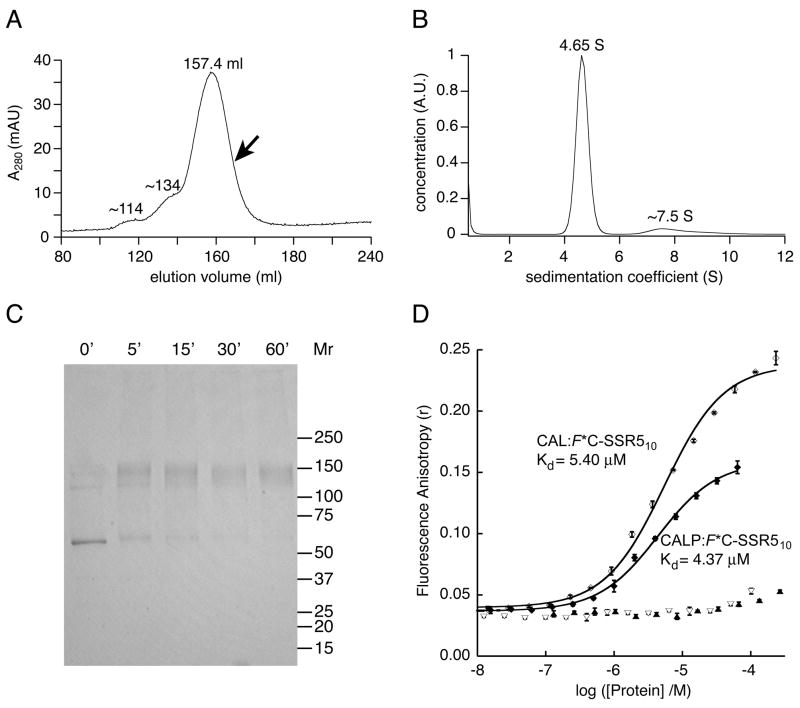
To determine whether the CFTR-binding interaction of the CAL PDZ domain depended on its molecular context, we first purified and characterized full-length CAL (18). It has been shown that CAL forms homomultimers within the cellular environment (12, 39), but the stoichiometry of oligomerization has never been determined. To assure the correct assembly of full-length CAL in vitro and to assess its oligomeric state quantitatively, size-exclusion chromatography (SEC) and analytical ultracentrifugation experiments were performed, providing a shape-independent estimate of molecular weight. During SEC, H10-CAL protein eluted in a single, dominant peak, with two small, higher molecular-weight shoulders (Fig. 8A). Calibration of the relative elution volume of the main CAL peak (157.4 ml) against the inverse molecular diffusion constants (1/D20,w) of standard proteins yielded an estimate for D20,w of 3.88 × 10−7 cm2/sec. A fraction of the SEC-purified protein (arrow in Fig. 8A) was subjected to velocity sedimentation analysis. As for the SEC chromatogram, analysis of the distribution of sedimentation coefficients (c(s)) revealed a predominant peak, with a sedimentation coefficient of 4.65 S (Fig. 8B). Using the Svedberg equation, these two parameters provide a shape-independent relative molar mass (Mr) estimate of 109 kD for CAL. The predicted Mr of the CAL monomer is 52.5 kD, suggesting a dimeric association. A dimeric association is also supported by chemical cross-linking (Fig. 8C) and by sedimentation equilibrium data (not shown).
Figure 8. The CAL PDZ domain recapitulates the affinity of full-length dimeric CAL for CFTR.
(A) Size-exclusion chromatography (SEC) of H10-CAL reveals a dominant peak at Ve=157.4 ml. Two additional minor peaks are seen as well, with smaller elution volumes. The arrow indicates the position of the fraction collected for velocity sedimentation experiments. Standard proteins were also subjected to SEC and used to calibrate the diffusion coefficient (D20,w) of H10-CAL. (B) The sedimentation coefficient concentration distribution c(s) (solid line) was determined for SEC-purified CAL using velocity sedimentation analysis. The major peak is shown at 4.65 S, together with minor faster-sedimenting components exhibiting a mode of ~7.5 S and a mean of ~9.1 S. (C) A Coomassie-stained SDS-PAGE gel is shown of H10-CAL samples cross-linked with BS3. The reaction was quenched with ethanolamine at the time indicated, revealing the time-dependent formation of a covalent dimer that was essentially complete at 60 min. (D) Fluorescence anisotropy data were obtained in the presence of increasing concentrations of CAL for 30 nM F*C-SSR510 (open diamonds) or 30 nM F*-CFTR10 (open triangles). The solid line represents a fit of the F*C-SSR510 data to a single-site binding model (Kd = 5.40 ± 0.47 μM). The interaction with F*-CFTR10 was too weak to be fit. For comparison, equivalent titrations are shown for the CALP protein (closed diamonds and closed triangles, respectively). Values shown are mean ± SD, n=3.
Full-length Dimeric CAL Binds CFTR With Low Affinity
In titrations with F*C-SSR510 reporter peptide, full-length CAL exhibits an affinity (Kd = 5.40 ± 0.47 μM) very similar to that obtained in parallel titrations with the CALP construct (Fig. 8D). Furthermore, even though only minimal FP shifts could be achieved for the titrations with the CFTR C-terminal reporter peptide, it is clear that the full-length CAL protein does not have a significantly higher affinity for this sequence than the isolated CAL PDZ domain. Thus, the binding characteristics of the isolated CAL PDZ domain appear to reflect those of the same domain embedded in the full-length protein. The larger fluorescence polarization values obtained at saturating concentrations of the full-length protein as opposed to the isolated CAL PDZ domain correspond to the larger hydrodynamic radius of the dimeric full-length protein.
The Relative Affinity of CAL Binding Partners
Using the F*C-SSR510 reporter sequence that we identified for the CAL PDZ domain, we also performed a systematic analysis of all ligands reported to bind either to CAL or to its neuronal splice variant (40). Unlabeled peptides corresponding to the C-termini of a total of 18 interactors (including GBG4) were tested in an inhibition binding assay with the CAL PDZ domain, which is common to both CAL isoforms (Table 3). With the exception of neuroglycan C (NGC), all peptides were observed to inhibit the CALP:reporter interaction, with Ki values ranging from 37.6 ± 4.0 μM (for C-SSR510) to 1940 ± 920 μM (for NMDA receptor subunit sequence C-NR2A10). With the exception of β-catenin, C-CFTR10 was the weakest of the non-neuronal CAL binders.
Table 3.
Ki values for CAL binding peptides.
| Binding partnera | Sequence | Ki (μM) |
|---|---|---|
| C-SSR510 | CANGLMQTSKL | 37.6 ± 4.0 |
| C-AIC3A10 | CHRTCYLVTQL | 42.2 ± 4.2 |
| C-FRZ810 | CYPKQMPLSQV | 47 ± 10 |
| C-GBG410 | CREKKFFSTILb | 63 ± 15 |
| C-CL110 | CDGQMQLVTSL | 88.3 ± 6.3 |
| C-ClC3B10 | CEVYLLNSTTL | 93.0 ± 8.5 |
| C-NLG210 | CLPHPHSTTRV | 106.7 ± 1.7 |
| C-SDK210 | CAPIAGFSSFV | 150 ± 20 |
| C-NLG10 | CHPHSHSTTRV | 158 ± 19 |
| C-RTKN10 | CQPRTWLQSPV | 229 ± 16 |
| C-FRZ510 | CYHKQVSLSHV | 451 ± 42 |
| C-β1AR10 | CRPGFASESKV | 510 ± 110c |
| C-GluRδ210 | CGNDPDRGTSI | 560 ± 130 |
| C-CFTR10 | CTEEEVQDTRL | 690 ± 120 |
| C-GluRδ110 | CALDTSHGTSI | 875 ± 56 |
| C-βCAT10 | CNQLAWFDTDL | 1110 ± 480d |
| C-NR2A10 | CKKMPSIESDV | 1940 ± 920d |
| C-NGC10 | CDVNSLQNNLT | n.b.d.e |
Ki values were determined by titration of the indicated peptide sequences in the presence of 30 nM F*C-SSR510 and 5 μM CALP and determining the fluorescence anisotropy of the reporter ligand. Data were fit using equation 5. Bold entries reflect binding partners identified only on the basis of yeast two-hybrid data.
Full names, accession numbers, and references are presented in Supplementary Table S1.
The native sequence contains a Cys at position 8, instead of a Ser.
Previously reported value: 0.035 μM (19).
Large uncertainty reflects substantially incomplete displacement at highest concentration tested.
n.b.d.: no binding detected.
DISCUSSION
In order to understand how CAL, NHERF1, and NHERF2 compete to regulate CFTR endocytic processing, it is essential to move beyond the enumeration of binding interactions and instead to compare their affinities quantitatively. This requires the application of binding assays that can cover a wide spectrum of affinities, ranging in strength from nanomolar to submillimolar. For CAL and NHERF2, no data has been available on the affinity of the individual PDZ domains. In the case of NHERF1, estimates have been available, but conflict. As a result, a major goal of our studies has been to establish a firm basis for quantitative comparison, and thus for understanding the observation that CFTR efficiently escapes CAL-mediated degradation through repeated rounds of uptake and recycling.
High Affinity of NHERF1 and NHERF2 for CFTR
Here, we have shown that the N1P1 domain binds with CFTR C-terminal sequences with Ki values for fluorescein-free peptides ranging from 949 nM for the tetrapeptide to 209 nM for the hexadecapeptide (Table 2). These values compare well with the 298 nM affinity detected for binding to the C-terminal 70 residues of CFTR (41), indicating that most of the affinity is captured by the interaction with the C-terminal 6 residues, and virtually all of it by the final 16 residues.
Some previously published data had supported even higher affinity estimates (8, 20, 21). However, these earlier studies used SPR to detect the binding of analytes expressed as GST fusion proteins, an approach that can lead to avidity effects due to GST dimerization (42, 43).
The N1P2 domain also exhibits strong binding to the CFTR C-terminus, with a Kd value of 1079 nM. Our construct includes flanking sequences that have been shown to be important in determining the affinity of the isolated N1P2 domain (20): weak interactions are reported for the core 90-amino acid PDZ domain (9, 41), whereas robust binding is reported for constructs that include the flanking residues 132-143 and 248-299 (refs. 20, 21 and Fig. 2).
Although N1P2 can be blocked by an intramolecular interaction with the NHERF1 C-terminus in the full-length protein (41), this interaction is released upon binding of the NHERF1 C-terminus to the ezrin FERM domain. As a result, in vivo the N1P2 domain may also participate in CFTR binding at the apical membrane, where NHERF1 can deploy both PDZ domains. Each of these component interactions exhibits an affinity for the CFTR C-terminus that places it among the tightest physiological PDZ binders (44).
The NHERF2 PDZ domains, whose interactions facilitate CFTR functional associations with effector proteins such as protein kinase A (11), also exhibit high affinity for the CFTR C-terminus (Fig. 5). As a result, strong binding to CFTR may be a common feature of the NHERF family proteins, which appear to be important both in the stabilization of CFTR at the apical membrane and in coupling it to both effectors and regulatory targets (1). In the case of the N1P1 domain, our thermodynamic analysis suggests that this affinity is based on a large binding interface and a strongly favorable enthalpy of interaction (ΔH ~ −12.2 kcal/mol at 37° C), compared to other PDZ domains (e.g. refs. 31, 32).
Low Affinity of CAL for CFTR
In contrast, CAL’s weak affinity for CFTR places it firmly on the other end of the affinity spectrum, with an estimated Ki value of >600 μM (Figs. 6B and 7B), the weakest of any PDZ domain for a confirmed physiological target. Overall, the CAL PDZ domain exhibits weak interactions with all known binding partners: even the strongest has a Ki value of only 37.6 μM (Table 3). The full-length CAL protein exhibits affinities similar to those of the isolated PDZ domain both for CFTR and for SSR5 (Fig. 8D), demonstrating that the observed weak binding is physiologically relevant, and not simply due to a loss of flanking sequences in the PDZ-only construct. Furthermore, the accessibility of the PDZ domain in the full-length protein confirms that it is not subject to intramolecular block.
Since CAL mediates post-endocytic degradation of CFTR (12, 14), the low underlying affinity of the interaction may be essential for CFTR to avoid a potentially fatal interaction through many rounds of endocytic recycling. At the same time, based on our functional studies (18), CAL appears to have retained just enough “stickiness” for CFTR to capture molecules engaged in accelerated recycling due to functional or folding defects, without degrading significant quantities of intact WT protein.
In part, the weak inherent affinity of CAL may be compensated for by local concentration effects, due to co-localization with CFTR during intracellular trafficking. It is also possible that the CAL:CFTR interaction may be stabilized by dimerization. Here, we have shown that full-length CAL assembles as a dimer (Fig. 8), and although the oligomeric state of CFTR is not well-understood, it also appears able to form dimers (1). Finally, it is possible that CAL binding may be strengthened by interactions with CFTR sequence elements upstream of the C-terminal hexadecamer, either by direct binding or via effects on the conformation of the C-terminal epitope itself. In fact, there is evidence that distal residues modulate CFTR trafficking, although it is not known whether these effects are mediated via specific PDZ interactions (45, 46).
In spite of these considerations, it is likely that the affinity of the CAL:CFTR interaction remains modest in vivo. In general, PDZ interaction affinities are dominated by the contributions of the C-terminal residues (31). Furthermore, since PDZ binding buries a significant fraction of the accessible surface of a target peptide, the conformational interactions of the C-terminus with other regions of the intact protein are as likely to have an unfavorable effect on PDZ binding (e.g. via steric hindrance) as to have a favorable effect (e.g. via entropic stabilization). Experimental data using longer C-terminal constructs are also consistent with a weak overall affinity. We have previously expressed a GST-fusion construct corresponding to the C-terminal 104 residues (1377-1480, ref. 18). The fusion protein contains all residues C-terminal to the second nucleotide-binding domain. It also oligomerizes and should therefore benefit from the chelate effect when interacting with dimeric full-length CAL. Nevertheless, when immobilized on glutathione beads, the ability of the fusion protein to pull-down either full-length CAL or its PDZ domain required high protein concentrations and very gentle washing (18). Thus, although an exact understanding of CFTR:PDZ interactions will ultimately require studies with intact CFTR, the values reported here are likely to reflect relative physiological affinities. They also provide a baseline for quantitating potential modulatory effects of upstream binding or conformational determinants.
Quantitating Low-Affinity PDZ Binding Interactions
The affinities of physiological PDZ:peptide interactions are typically 0.5 – 20 μM (13, 22, 44). However, PDZ domains can bind peptides with affinities that span a range of more than four orders of magnitude, from the 10–20 nM interactions observed in mutagenetically optimized binding interactions (e.g. ref. 47) to >100 μM interactions (e.g. refs. 22, 48).
Although they pose technical challenges to quantitation, the low-affinity PDZ interactions can be physiologically significant. The PSD-95/SAP90 PDZ1 domain binds the kainate receptor subunit GluR6 with an affinity of only 160 μM (48). Nevertheless, inhibitors of this interaction have been shown to disrupt kainate receptor clustering in cultured neurons (49).
In general, surprisingly modest affinity appears to be sufficient for the physiological activity of a number of PDZ domains, and may even be required to permit the transient, rapidly reversible interactions required to hand off binding partners within a network of interactions. A recent survey of mouse PDZ interactions found that 40% of observed “hits” exhibited an affinity between 20 and 100 μM (22), and even the latter value represented an arbitrary cut-off that may miss physiologically relevant interactions. In the proteomic survey, CAL failed to bind a single peptide in the training set with sufficient affinity to be further analyzed (22), even though it has been clearly shown to modulate CFTR surface expression in vivo (12, 18, 19).
Such weak, but physiologically relevant interactions may be representative of a significant fraction of the PDZ population. The approach developed here, coupling peptide-array technology with an FP binding assay to identify and then exploit relatively high affinity reporter ligands, should prove broadly useful in quantifying them.
CAL: A Potential Pharmacological Target
Even though CAL exhibits a Ki value in excess of 600 μM for the CFTR C-terminus, our previous work has shown that it plays a physiologically important role in CFTR trafficking. In cultured CF airway epithelial cells, endogenous CAL strongly reduces the cell-surface levels of functional, temperature-rescued ΔF508-CFTR (18). Nearly 90% of CF patients are homozygous or heterozygous for the ΔF508-CFTR allele (50). The resulting mutant protein, which lacks the single amino acid Phe508, matures inefficiently, exhibits decreased channel activity, and is degraded more rapidly than wild-type protein (1). As a result, net chloride efflux is impaired, leading to a breakdown of airway mucociliary clearance and ultimately to the development of chronic lung infections that are currently the primary cause of CF mortality (51).
Because mature ΔF508-CFTR protein retains some functional activity (52–54), the identification of chemical chaperones (“correctors”) and compounds that stimulate ΔF508-CFTR channel activity (“potentiators”) remains a major therapeutic goal (55). Yet even when rescued, ΔF508-CFTR is subject to accelerated endocytosis that leads to a dramatically reduced half-life in both heterologous (5) and airway epithelial (6) cell lines. Given its short half-life, stabilization of ΔF508-CFTR could offer an additional, complementary approach for increasing the net amount of apical chloride channel activity. We have shown that a structurally conservative mutation of the CAL binding pocket abrogates CAL-mediated degradation of CFTR (18). As a result, pharmacological targeting of the pocket could mimic the effects of CAL knock-down in stabilizing ΔF508-CFTR cell-surface expression.
However, the ultimate feasibility of targeting a specific protein:PDZ interaction will depend on the ability to circumvent the bi-directional promiscuity of these molecular relationships. A given target protein generally binds to a number of different PDZ proteins (see e.g. refs. 22, 44). At the same time, PDZ proteins typically interact with multiple target proteins. As a result, selective inhibition requires not only pharmacological specificity for a given PDZ domain, but also that the targeted partner of that domain be displaced more easily than the other partners.
In order to assess the underlying feasibility of targeting the CAL:CFTR interaction, we have therefore analyzed the binding of multiple partners to both proteins using our FP assay, which permits direct comparison across the entire physiologically relevant range of affinities. Although the CAL PDZ domain has been reported to bind a number of receptors, only a single affinity estimate has previously been available, for β1-AR (19). Here, we have systematically determined solution-state affinities for all reported CAL partners (Table 3), revealing that CAL binds CFTR very weakly compared to other CAL interactors, with the exception of a few, generally neuron-specific partners. Thus, CFTR should be more susceptible to competitive displacement than other CAL partners.
At the same time, CFTR is known to interact with a number of PDZ proteins. Here, we have also shown that the affinity of CAL for the CFTR C-terminus is much lower than those of two members of the NHERF-family of proteins, both of which contribute to its apical membrane retention and activity (1). This affinity profile should facilitate the identification of inhibitors that can displace CFTR from CAL but not from the NHERF proteins. Overall, our studies show that the CAL:CFTR C-terminal interaction is at the weak end of the spectrum for both partners. This should favor our efforts to identify selective pharmacological inhibitors, as a basis for stabilizing rescued mutant CFTR.
Supplementary Material
Supplementary Table S1 describes the full name, abbreviation, SwissProt/TrEMBL accession number, and source reference for CAL binding partners analyzed in this work. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
The authors gratefully acknowledge the collaborative support of Drs. B. Stanton, J. Cheng (Johns Hopkins Univ.), W. Guggino (Johns Hopkins Univ.) and R. Volkmer (Charité Universitätsmedizin). We thank Dr. H. Higgs for assistance with FP assay development; Drs. N. Grossoehme and D. Wilcox (Dartmouth College) for support with isothermal titration calorimetry measurements; Dr. J. Gosse for advice on microplate FP assays; Dr. J. Bodwell for providing access to the Spectramax microplate FP reader; K. Karlson and the Dartmouth CF Core Facility for providing cDNA constructs; Dr. D. Mierke (Dartmouth College) for suggestions regarding PDZ binding-site mutants; J. Piro and J. van Dieck for assistance with PDZ cloning and expression; and M. Schowalter for excellent technical assistance.
Footnotes
This work was supported in part by grants from the Cystic Fibrosis Foundation (MADDEN06P0 and STANTO97R0) and the NIH (grants P20-RR018787 from the Institutional Development Award (IDeA) Program of the NCRR and R01-DK075309 from NIDDK). P.B. was supported by the Deutsche Forschungsgemeinschaft (DFG grant VO 885/3-1).
The abbreviations used are: CFTR, cystic fibrosis transmembrane conductance regulator; PDZ, PSD-95, discs large, zonula occludens-1; CAL, CFTR-Associated Ligand; NHERF1, Na+/H+ Exchanger-3 Regulatory Factor-1; NHERF2, Na+/H+ Exchanger-3 Regulatory Factor-2; CF, cystic fibrosis; DTT, dithiothreitol; TCEP, Tris(2-carboxyethyl)phosphine hydrochloride; SPR, surface-plasmon resonance; ITC, isothermal titration calorimetry; FP, fluorescence polarization; SEC, size-exclusion chromatography; BS3, bis[sulfosuccinimidyl] suberate.
References
- 1.Guggino WB, Stanton BA. Molecular switches regulating CFTR trafficking and activity. Nat Rev Mol Cell Biol. 2006;7:426–436. doi: 10.1038/nrm1949. [DOI] [PubMed] [Google Scholar]
- 2.Prince LS, Workman RB, Jr, Marchase RB. Rapid endocytosis of the cystic fibrosis transmembrane conductance regulator chloride channel. Proc Natl Acad Sci USA. 1994;91:5192–5196. doi: 10.1073/pnas.91.11.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swiatecka-Urban A, Duhaime M, Coutermarsh B, Karlson KH, Collawn J, Milewski M, Cutting GR, Guggino WB, Langford G, Stanton BA. PDZ domain interaction controls the endocytic recycling of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 2002;277:40099–40105. doi: 10.1074/jbc.M206964200. [DOI] [PubMed] [Google Scholar]
- 4.Sharma M, Pampinella F, Nemes C, Benharouga M, So J, Du K, Bache KG, Papsin B, Zerangue N, Stenmark H, Lukacs GL. Misfolding diverts CFTR from recycling to degradation: quality control at early endosomes. J Cell Biol. 2004;164:923–933. doi: 10.1083/jcb.200312018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lukacs GL, Chang XB, Bear C, Kartner N, Mohamed A, Riordan JR, Grinstein S. The ΔF508 mutation decreases the stability of cystic fibrosis transmembrane conductance regulator in the plasma membrane. Determination of functional half-lives on transfected cells. J Biol Chem. 1993;268:21592–21598. [PubMed] [Google Scholar]
- 6.Swiatecka-Urban A, Brown A, Moreau-Marquis S, Renuka J, Coutermarsh B, Barnaby R, Karlson KH, Flotte TR, Fukuda M, Langford GM, Stanton BA. The short apical membrane half-life of rescued ΔF508-CFTR results from accelerated endocytosis ΔF508-CFTR in polarized human airway epithelial cells. J Biol Chem. 2005;280:36762–36772. doi: 10.1074/jbc.M508944200. [DOI] [PubMed] [Google Scholar]
- 7.Li C, Naren AP. Macromolecular complexes of cystic fibrosis transmembrane conductance regulator and its interacting partners. Pharmacol Ther. 2005;108:208–223. doi: 10.1016/j.pharmthera.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Short DB, Trotter KW, Reczek D, Kreda SM, Bretscher A, Boucher RC, Stutts MJ, Milgram SL. An apical PDZ protein anchors the cystic fibrosis transmembrane conductance regulator to the cytoskeleton. J Biol Chem. 1998;273:19797–19801. doi: 10.1074/jbc.273.31.19797. [DOI] [PubMed] [Google Scholar]
- 9.Wang SS, Raab RW, Schatz PJ, Guggino WB, Li M. Peptide binding consensus of the NHE-RF-PDZ1 domain matches the C-terminal sequence of cystic fibrosis transmembrane conductance regulator (CFTR) FEBS Lett. 1998;427:103–108. doi: 10.1016/s0014-5793(98)00402-5. [DOI] [PubMed] [Google Scholar]
- 10.Hall RA, Ostedgaard LS, Premont RT, Blitzer JT, Rahman N, Welsh MJ, Lefkowitz RJ. A C-terminal motif found in the β2-adrenergic receptor, P2Y1 receptor and cystic fibrosis transmembrane conductance regulator determines binding to the Na+/H+ exchanger regulatory factor family of PDZ proteins. Proc Natl Acad Sci USA. 1998;95:8496–8501. doi: 10.1073/pnas.95.15.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun F, Hug MJ, Lewarchik CM, Yun CHC, Bradbury NA, Frizzell RA. E3KARP mediates the association of ezrin and protein kinase A with the cystic fibrosis transmembrane conductance regulator in airway cells. J Biol Chem. 2000;275:29539–29546. doi: 10.1074/jbc.M004961200. [DOI] [PubMed] [Google Scholar]
- 12.Cheng J, Moyer BD, Milewski M, Loffing J, Ikeda M, Mickle JE, Cutting GR, Li M, Stanton BA, Guggino WB. A Golgi-associated PDZ domain protein modulates cystic fibrosis transmembrane regulator plasma membrane expression. J Biol Chem. 2002;277:3520–3529. doi: 10.1074/jbc.M110177200. [DOI] [PubMed] [Google Scholar]
- 13.Nourry C, Grant SG, Borg JP. PDZ domain proteins: plug and play! Sci STKE 2003. 2003:RE7. doi: 10.1126/stke.2003.179.re7. [DOI] [PubMed] [Google Scholar]
- 14.Cheng J, Wang H, Guggino WB. Modulation of mature cystic fibrosis transmembrane regulator protein by the PDZ domain protein CAL. J Biol Chem. 2004;279:1892–1898. doi: 10.1074/jbc.M308640200. [DOI] [PubMed] [Google Scholar]
- 15.Li JG, Chen C, Liu-Chen LY. Ezrin-radixin-moesin-binding phosphoprotein-50/Na+/H+ exchanger regulatory factor (EBP50/NHERF) blocks U50,488H-induced down-regulation of the human kappa opioid receptor by enhancing its recycling rate. J Biol Chem. 2002;277:27545–27552. doi: 10.1074/jbc.M200058200. [DOI] [PubMed] [Google Scholar]
- 16.Cao TT, Deacon HW, Reczek D, Bretscher A, von Zastrow M. A kinase-regulated PDZ-domain interaction controls endocytic sorting of the beta2-adrenergic receptor. Nature. 1999;401:286–290. doi: 10.1038/45816. [DOI] [PubMed] [Google Scholar]
- 17.Guerra L, Fanelli T, Favia M, Riccardi SM, Busco G, Cardone RA, Carrabino S, Weinman EJ, Reshkin SJ, Conese M, Casavola V. Na+/H+ exchanger regulatory factor isoform 1 overexpression modulates cystic fibrosis transmembrane conductance regulator (CFTR) expression and activity in human airway 16HBE14o- cells and rescues ΔF508 CFTR functional expression in cystic fibrosis cells. J Biol Chem. 2005;280:40925–40933. doi: 10.1074/jbc.M505103200. [DOI] [PubMed] [Google Scholar]
- 18.Wolde M, Fellows A, Cheng J, Kivenson A, Coutermarsh B, Talebian L, Karlson K, Piserchio A, Mierke DF, Stanton BA, Guggino WB, Madden DR. Targeting CAL as a negative regulator of ΔF508-CFTR cell-surface expression: an RNA interference and structure-based mutagenetic approach. J Biol Chem. 2007;282:8099–8109. doi: 10.1074/jbc.M611049200. [DOI] [PubMed] [Google Scholar]
- 19.He J, Bellini M, Xu J, Castleberry AM, Hall RA. Interaction with cystic fibrosis transmembrane conductance regulator-associated ligand (CAL) inhibits β-adrenergic receptor surface expression. J Biol Chem. 2004;279:50190–50196. doi: 10.1074/jbc.M404876200. [DOI] [PubMed] [Google Scholar]
- 20.Raghuram V, Mak DOD, Foskett JK. Regulation of cystic fibrosis transmembrane conductance regulator single-channel gating by bivalent PDZ-domain-mediated interaction. Proc Natl Acad Sci USA. 2001;98:1300–1305. doi: 10.1073/pnas.031538898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JH, Richter W, Namkung W, Kim KH, Kim E, Conti M, Lee MG. Dynamic regulation of cystic fibrosis transmembrane conductance regulator by competitive interactions of molecular adaptors. J Biol Chem. 2007;282:10414–10422. doi: 10.1074/jbc.M610857200. [DOI] [PubMed] [Google Scholar]
- 22.Stiffler MA, Chen JR, Grantcharova VP, Lei Y, Fuchs D, Allen JE, Zaslavskaia LA, MacBeath G. PDZ domain binding selectivity is optimized across the mouse proteome. Science. 2007;317:364–369. doi: 10.1126/science.1144592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang ZX. An exact mathematical expression for describing competitive binding of two different ligands to a protein molecule. FEBS Lett. 1995;360:111–114. doi: 10.1016/0014-5793(95)00062-e. [DOI] [PubMed] [Google Scholar]
- 24.Boisguérin P, Ay B, Radziwill G, Fritz RD, Moelling K, Volkmer R. Characterization of a putative phosphorylation switch: adaptation of SPOT synthesis to analyze PDZ domain regulation mechanisms. Chembiochem. 2007;8:2302–2307. doi: 10.1002/cbic.200700518. [DOI] [PubMed] [Google Scholar]
- 25.Dam J, Schuck P. Calculating sedimentation coefficient distributions by direct modeling of sedimentation velocity concentration profiles. Methods Enzymol. 2004;384:185–212. doi: 10.1016/S0076-6879(04)84012-6. [DOI] [PubMed] [Google Scholar]
- 26.Stafford WF, Sherwood PJ. Analysis of heterologous interacting systems by sedimentation velocity: curve fitting algorithms for estimation of sedimentation coefficients, equilibrium and kinetic constants. Biophys Chem. 2004;108:231–243. doi: 10.1016/j.bpc.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 27.Piserchio A, Fellows A, Madden DR, Mierke DF. Association of the Cystic Fibrosis Transmembrane Regulator with CAL: Structural Features and Molecular Dynamics. Biochemistry. 2005;44:16158–16166. doi: 10.1021/bi0516475. [DOI] [PubMed] [Google Scholar]
- 28.Karthikeyan S, Leung T, Ladias JAA. Structural basis of the Na+/H+ exchanger regulatory factor PDZ1 interaction with the carboxyl-terminal region of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 2001;276:19683–19686. doi: 10.1074/jbc.C100154200. [DOI] [PubMed] [Google Scholar]
- 29.Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, Chan AC, Anderson JM, Cantley LC. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 30.Lim IA, Hall DD, Hell JW. Selectivity and promiscuity of the first and second PDZ domains of PSD-95 and synapse-associated protein 102. J Biol Chem. 2002;277:21697–21711. doi: 10.1074/jbc.M112339200. [DOI] [PubMed] [Google Scholar]
- 31.Saro D, Li T, Rupasinghe C, Paredes A, Caspers N, Spaller MR. A thermodynamic ligand binding study of the third PDZ domain (PDZ3) from the mammalian neuronal protein PSD-95. Biochemistry. 2007;46:6340–6352. doi: 10.1021/bi062088k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma SC, Rupasinghe CN, Parisien RB, Spaller MR. Design, synthesis, and evaluation of linear and cyclic peptide ligands for PDZ10 of the multi-PDZ domain protein MUPP1. Biochemistry. 2007;46:12709–12720. doi: 10.1021/bi7008135. [DOI] [PubMed] [Google Scholar]
- 33.O’Brien R, Chowdry BZ, Ladbury JE. Isothermal titration calorimetry of biomolecules. In: Harding SE, Chowdry BZ, editors. Protein-Ligand Interactions: Hydrodynamics and Calorimetry. Oxford University Press; 2001. pp. 263–286. [Google Scholar]
- 34.Fukada H, Sturtevant JM, Quiocho FA. Thermodynamics of the binding of L-arabinose and of D-galactose to the L-arabinose-binding protein of Escherichia coli. J Biol Chem. 1983;258:13193–13198. [PubMed] [Google Scholar]
- 35.Xie D, Freire E. Molecular basis of cooperativity in protein folding. V Thermodynamic and structural conditions for the stabilization of compact denatured states. Proteins. 1994;19:291–301. doi: 10.1002/prot.340190404. [DOI] [PubMed] [Google Scholar]
- 36.Boisguérin P, Leben R, Ay B, Radziwill G, Moelling K, Dong L, Volkmer-Engert R. An improved method for the synthesis of cellulose membrane-bound peptides with free C termini is useful for PDZ domain binding studies. Chem Biol. 2004;11:449–459. doi: 10.1016/j.chembiol.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 37.Wente W, Stroh T, Beaudet A, Richter D, Kreienkamp HJ. Interactions with PDZ domain proteins PIST/GOPC and PDZK1 regulate intracellular sorting of the somatostatin receptor subtype 5. J Biol Chem. 2005;280:32419–32425. doi: 10.1074/jbc.M507198200. [DOI] [PubMed] [Google Scholar]
- 38.Gentzsch M, Cui L, Mengos A, Chang XB, Chen JH, Riordan JR. The PDZ-binding chloride channel ClC-3B localizes to the Golgi and associates with cystic fibrosis transmembrane conductance regulator-interacting PDZ proteins. J Biol Chem. 2003;278:6440–6449. doi: 10.1074/jbc.M211050200. [DOI] [PubMed] [Google Scholar]
- 39.Neudauer CL, Joberty G, Macara IG. PIST: a novel PDZ/coiled-coil domain binding partner for the rho-family GTPase TC10. Biochem Biophys Res Commun. 2001;280:541–547. doi: 10.1006/bbrc.2000.4160. [DOI] [PubMed] [Google Scholar]
- 40.Yue Z, Horton A, Bravin M, DeJager PL, Selimi F, Heintz N. A novel protein complex linking the δ2 glutamate receptor and autophagy: implications for neurodegeneration in lurcher mice. Neuron. 2002;35:921–933. doi: 10.1016/s0896-6273(02)00861-9. [DOI] [PubMed] [Google Scholar]
- 41.Li J, Dai Z, Jana D, Callaway DJ, Bu Z. Ezrin controls the macromolecular complexes formed between an adapter protein Na+/H+ exchanger regulatory factor and the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 2005 doi: 10.1074/jbc.M502305200. [DOI] [PubMed] [Google Scholar]
- 42.Ladbury JE, Lemmon MA, Zhou M, Green J, Botfield MC, Schlessinger J. Measurement of the binding of tyrosyl phosphopeptides to SH2 domains: a reappraisal. Proc Natl Acad Sci U S A. 1995;92:3199–3203. doi: 10.1073/pnas.92.8.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuck P. Reliable determination of binding affinity and kinetics using surface plasmon resonance biosensors. Curr Opin Biotechnol. 1997;8:498–502. doi: 10.1016/s0958-1669(97)80074-2. [DOI] [PubMed] [Google Scholar]
- 44.Stiffler MA, Grantcharova VP, Sevecka M, MacBeath G. Uncovering quantitative protein interaction networks for mouse PDZ domains using protein microarrays. J Am Chem Soc. 2006;128:5913–5922. doi: 10.1021/ja060943h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Milewski MI, Mickle JE, Forrest JK, Stafford DM, Moyer BD, Cheng J, Guggino WB, Stanton BA, Cutting GR. A PDZ-binding motif is essential but not sufficient to localize the C terminus of CFTR to the apical membrane. J Cell Sci. 2001;114:719–726. doi: 10.1242/jcs.114.4.719. [DOI] [PubMed] [Google Scholar]
- 46.Milewski MI, Lopez A, Jurkowska M, Larusch J, Cutting GR. PDZ-binding motifs are unable to ensure correct polarized protein distribution in the absence of additional localization signals. FEBS Lett. 2005;579:483–487. doi: 10.1016/j.febslet.2004.11.106. [DOI] [PubMed] [Google Scholar]
- 47.Reina J, Lacroix E, Hobson SD, Fernandez-Ballester G, Rybin V, Schwab MS, Serrano L, Gonzalez C. Computer-aided design of a PDZ domain to recognize new target sequences. Nat Struct Biol. 2002;9:621–627. doi: 10.1038/nsb815. [DOI] [PubMed] [Google Scholar]
- 48.Piserchio A, Pellegrini M, Mehta S, Blackman SM, Garcia EP, Marshall J, Mierke DF. The PDZ1 domain of SAP90. Characterization of structure and binding. J Biol Chem. 2002;277:6967–6973. doi: 10.1074/jbc.M109453200. [DOI] [PubMed] [Google Scholar]
- 49.Piserchio A, Salinas GD, Li T, Marshall J, Spaller MR, Mierke DF. Targeting specific PDZ domains of PSD-95; structural basis for enhanced affinity and enzymatic stability of a cyclic peptide. Chem Biol. 2004;11:469–473. doi: 10.1016/j.chembiol.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 50.Cheng SH, Gregory RJ, Marshall J, Paul S, Souza DW, White GA, O’Riordan CR, Smith AE. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- 51.Boucher RC. Airway surface dehydration in cystic fibrosis: pathogenesis and therapy. Annu Rev Med. 2007;58:157–170. doi: 10.1146/annurev.med.58.071905.105316. [DOI] [PubMed] [Google Scholar]
- 52.Dalemans W, Barbry P, Champigny G, Jallat S, Dott K, Dreyer D, Crystal RG, Pavirani A, Lecocq JP, Lazdunski M. Altered chloride ion channel kinetics associated with the ΔF508 cystic fibrosis mutation. Nature. 1991;354:526–528. doi: 10.1038/354526a0. [DOI] [PubMed] [Google Scholar]
- 53.Drumm ML, Wilkinson DJ, Smit LS, Worrell RT, Strong TV, Frizzell RA, Dawson DC, Collins FS. Chloride conductance expressed by ΔF508 and other mutant CFTRs in Xenopus oocytes. Science. 1991;254:1797–1799. doi: 10.1126/science.1722350. [DOI] [PubMed] [Google Scholar]
- 54.Denning GM, Anderson MP, Amara JF, Marshall J, Smith AE, Welsh MJ. Processing of mutant cystic fibrosis transmembrane conductance regulator is temperature-sensitive. Nature. 1992;358:761–764. doi: 10.1038/358761a0. [DOI] [PubMed] [Google Scholar]
- 55.Roomans GM. Pharmacological approaches to correcting the ion transport defect in cystic fibrosis. Am J Respir Med. 2003;2:413–431. doi: 10.1007/BF03256668. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1 describes the full name, abbreviation, SwissProt/TrEMBL accession number, and source reference for CAL binding partners analyzed in this work. This material is available free of charge via the Internet at http://pubs.acs.org.