Abstract
In order to gain a better understanding of gene expression during early malaria infection, we conducted microarray analysis of early blood responses in mice infected with erythrocytic-stage Plasmodium chabaudi. Immediately following infection, we observed coordinated and sequential waves of immune responses, with interferon-associated gene transcripts dominating by 16 h postinfection, followed by strong increases in natural killer (NK) cell-associated and major histocompatibility complex class I-related transcripts by 32 h postinfection. We showed by flow cytometry that the observed elevation in NK cell-associated transcripts was the result of a dramatic increase in the proportion of NK cells in the blood during infection. Subsequent microarray analysis of NK cells isolated from the peripheral blood of infected mice revealed a cell proliferation expression signature consistent with the observation that NK cells replicate in response to infection. Early proliferation of NK cells was directly observed in studies with adoptively transferred cells in infected mice. These data indicate that the early response to P. chabaudi infection of the blood is marked by a primary wave of interferon with a subsequent response by NK cells.
Plasmodium species are responsible for an estimated 330 to 660 million cases of malaria per year, creating a major health and economic burden in regions of endemicity (41, 43, 47). Infections by Plasmodium falciparum, the most lethal human malaria parasite, result in an estimated 1 to 2 million deaths per year (41, 55). Despite the fact much malaria goes untreated in regions of endemicity due to limited health care infrastructure or is inadequately treated due to antimalarial drug resistance and the high cost of effective therapeutics, the majority of malaria infections, especially those in adults, are uncomplicated. Thus, natural immunity to malarial infection is reasonably effective.
Rodent malaria species have been invaluable for our ability to examine immune responses to malaria infection (23, 53). Although no single model can recapitulate all forms of human disease, different parasite-host strain combinations allow the study of different manifestations of malaria infection. Infection of C57BL/6 mice with P. chabaudi AS is an extensively used model for studying immune responses to malaria infection, as parasite replication is controlled by the mice and eventually cleared from the bloodstream. In this regard, this model is the most similar to uncomplicated malaria in which the infection resolves naturally.
A key feature of the initial response to malaria infection is the early production of gamma interferon (IFN-γ). Plasma IFN-γ has been reported to be elevated by approximately 24 to 36 h following the infection of human peripheral blood mononuclear cells (PBMCs) in vitro and of blood in infected mice, and it is one of the first detectable cytokine responses (1, 10, 13, 25, 34). However, the cellular sources of IFN-γ induced by malarial infections have not been definitively identified, with natural killer (NK) cells, CD4+ T cells, NKT cells, γδ-T cells, and dendritic cells all proposed as potential sources (1, 10, 16, 17, 31, 33, 34, 38, 39, 48). In addition, other cytokines such as tumor necrosis factor (TNF) (7, 13, 33), interleukin-12, and interleukin-15 (reviewed in reference 53) are also generated during the very early response to Plasmodium infection, but the relative contributions of these cytokines to the overall immune response are unknown.
Early in vivo cellular responses to malarial infection have been less well characterized than cytokine responses have been. Early studies comparing inbred mouse strains identified NK cells as a mediator of differential susceptibility to infection with P. chabaudi (19), but in genetic studies susceptibility did not correlate with NK cell activity (46, 49). More-recent studies using antibody depletion have suggested that NK cells are essential for the full control of P. chabaudi and P. yoelii infection (7, 27, 33). NK cells can mediate antimicrobial effects either through cytokine production or through direct cytotoxicity; in the cases of P. chabaudi in the mouse and P. falciparum in vitro, cytokine production appears to account for the protective function of NK cells (4, 33). However, the significance of NK cell generation of IFN-γ during parasite infection has recently been questioned, with a report that γδ-T cells are the primary producers of IFN-γ during in vitro exposure of human PBMCs to parasitized erythrocytes (16).
In order to better characterize early responses to malaria infection, we studied the whole blood gene expression response to the erythrocytic infection of mice with P. chabaudi. We report that P. chabaudi infection stimulates coordinated, sequential waves of responses corresponding to an early interferon release followed by a dramatic expansion of NK cells in the blood.
MATERIALS AND METHODS
Mice.
C57BL/6 8- to 10-week-old female mice (Jackson Laboratories) were maintained according to UCSF IACUC guidelines. Lights were maintained on a 12-hour cycle (on from 0600 to 1800).
Parasites and infections.
P. chabaudi Landau ASS was obtained from MR4 (MRA-429) and maintained in C57BL/6 mice. Blood was harvested by cardiac puncture from an infected mouse near the time of peak parasitemia (typically day 8 to 10). Infected blood was diluted in Alsever's solution to 106 parasitized erythrocytes per 100 μl. Infection was initiated by the intraperitoneal injection of 106 parasitized erythrocytes or the mock injection of uninfected erythrocytes. At specific time points postinoculation, blood was harvested by cardiac puncture, and spleens were harvested for analysis (time zero was also collected postinoculation).
RNA isolation and amplification.
Samples for RNA preparation were directly immersed in RNAlater (AM7020; Ambion) upon harvest and stored at −80°C until RNA isolation. RNA from blood was isolated by using the mouse RiboPure-blood kit (AM1951; Ambion). RNA from blood was amplified in a single round using the amino allyl MessageAmp II aRNA amplification kit (AM1753; Ambion).
For cell populations purified by flow cytometry (see below), RNA was isolated with the RNAqueous-Micro kit (AM1931; Ambion). These samples required two rounds of amplification with amino allyl incorporation in the second round in order to generate enough sample for hybridization.
Microarray hybridization.
All samples were hybridized against a reference of pooled samples supplemented with amplified universal mouse reference RNA (740100; Stratagene). In all experiments, Cy5 was used to label the experimental sample and Cy3 was used to label the reference. Sample and reference amplified RNAs were combined and mixed with hybridization solution [0.25 mg/ml poly(A), 0.25 mg/ml yeast tRNA, 25 mM HEPES, pH 7.4, 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.225% sodium dodecyl sulfate], boiled, and hybridized to mouse whole-genome microarrays for 48 h at 65°C. The MEEBO microarray platform was utilized due to its high genome coverage and constitutive exonic design (54) (MEEBOChip and HEEBOChip Information Page; A. Alizadeh [http://alizadehlab.stanford.edu/]).
Microarray analysis.
Image data were extracted in Genepix 6 (Molecular Devices) and normalized and filtered in Acuity 4 (Molecular Devices). Data were ratio normalized, and control and poor-quality features (as determined by both visual examination and application of quantitative filters for saturation, feature diameter, and variance) were removed. Data were further filtered for spots that did not exhibit signal in any of the sample channels (based on the percentage of pixels in the feature above background and feature intensity) and for excessive missing data. The ratio of medians of the remaining data was transformed to log2 space, median centered by array, and median centered by gene.
The normalized and filtered time course data set consisted of 14,843 features. Methods and software used for analysis included hierarchical clustering using Xcluster (20), partitioning by self-organizing maps (SOMs) in Cluster 3.0 (12), and statistical analysis using significance analysis of microarrays (SAM) (52). All SAM analyses were conducted as two-class unpaired comparisons using a t statistic and a K-nearest-neighbor imputation of missing values.
Visual inspection of the original cluster revealed multiple classes of gene expression, with some genes induced early and others late. Two SAM were performed to capture all classes: one using the 56- to 72-h-infected time points versus uninfected samples, and another using 16- to 32-h-infected time points versus uninfected samples. Significance thresholds were applied using a twofold change and a 1% false-discovery rate. These late and early significant genes were combined into a single data set for further analysis.
The combined data set was subjected to a 1-by-3 SOM weighted on the infected samples in order to group them into three temporal classes, and genes in each node were ordered by a decreasing Pearson correlation with the average vector of that node.
Microarray data were visualized in Java Treeview (44). Significant gene sets were subjected to functional analysis using lexical analysis as implemented in LACK (26), gene ontology (GO) analysis with EASE (24), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis with the Pathway-Express program in the Onto-Tools suite (18).
Flow cytometry of leukocyte subsets.
Whole blood was harvested into Alsever's solution and spleens were harvested and homogenized in phosphate-buffered saline. Erythrocytes were lysed in 1× red blood cell lysis solution (eBioscience). Fc receptors on the leukocytes were blocked with anti-CD16/anti-CD32 antibody 2.4G2 (Becton Dickinson), stained with specific antibodies, and analyzed on an LSR II flow cytometer (Becton Dickinson). Antibodies used for leukocyte subset identification included fluorescein isothiocyanate-conjugated anti-Siglec-H (eBioscience), phycoerythrin-conjugated anti-F4/80 (eBioscience), peridinin chlorophyll protein-Cy5-conjugated anti-NK1.1 (BD), allophycocyanin (APC)-conjugated anti-CD11c-APC (BD), APC-Cy7-conjugated anti-CD3 (BD), phycoerythrin-Cy7-conjugted anti-Gr1 (eBioscience), and Pacific Blue-conjugated anti-B220 (BD). Leukocyte subsets were defined as B cells (B220+ Siglec-H−), T cells (CD3+ NK1.1−), NK cells (CD3− NK1.1+), NKT cells (CD3+ NK1.1+), monocytes (F4/80+ Gr1−), granulocytes (high-side light scatter SSChi Gr1+), myeloid dendritic cells (CD11chi Siglec-H−), and plasmacytoid dendritic cells (CD11cint Siglec-H+) (specific gating is depicted in Fig. S1 in the supplemental material). For expression profiling of subsets, cells were sorted on a FACSAria directly into 100 μl of lysis buffer provided with the RNAqueous-Micro kit (AM1931; Ambion) and amplified as described above.
CFSE labeling.
Carboxyfluorescein diacetate succinimidyl ester (CFSE) (V12883; Invitrogen) was prepared and used to stain splenocytes as per the manufacturer's protocol. Donor splenocytes harvested from C57BL/6J mice were stained for 10 min in 10 μM CFSE, and 4 × 107 cells were injected intravenously (tail vein) into recipient C57BL/6J mice. Mice were infected or mock infected shortly thereafter, and blood and spleens were harvested for flow cytometry on day 3 postinfection. Lymphocyte subsets were gated as described above, and CFSE-positive (CFSE+) cells of each lymphocyte subset were analyzed for dye dilution. Peak modeling was performed in FlowJo (Tree Star, Inc.).
Microarray data accession numbers.
All microarray data are available through the Gene Expression Omnibus under accession numbers GSE12249 and GSE12727.
RESULTS
Expression profiling of early immune responses to P. chabaudi infection in blood.
In order to gain a better understanding of innate immune responses during early erythrocytic infection, we used microarrays to profile gene expression in whole blood of P. chabaudi-infected mice. Blood was harvested at 8-h intervals for the first 72 h postinfection from two mock-infected and two P. chabaudi-infected mice and processed for microarray hybridization. As is typical for P. chabaudi, infections peaked roughly 7 days after infection, followed by a reduction in parasitemia by day 14 and clearance by day 21. Biological replicates of the early time points were averaged and genes were subjected to hierarchical clustering and significance analysis.
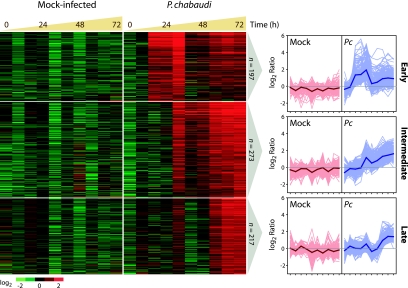
Hierarchical clustering revealed three dominant patterns of gene expression. Statistical analysis using SAM identified 687 genes as significantly belonging to one of these classes, and these genes were partitioned into three groups by use of SOMs (Fig. 1). Two aspects of the response were particularly striking. First, the response to infection was rapid, with significant changes in transcript abundance exhibited between 8 and 16 h postinfection. Second, the response occurred in coordinated waves of expression, suggesting that the nodes of the SOM may represent discrete functionalities. We refer to the three groups as early-, intermediate-, and late-response groups. The early pattern was characterized by a strong increase in relative transcript abundance by 16 h. The intermediate pattern was characterized by increased relative abundance at 32 h, which persisted through the last time point (72 h). The late pattern was characterized by increased relative abundance at 56 h, which persisted through 72 h.
FIG. 1.
Expression responses to P. chabaudi occur in coordinated waves. Genes that exhibited significant increases in relative abundance in response to infection from that seen for mock infection were identified by SAM and organized using SOMs. log2-transformed ratios of signal to reference are represented in a heat map using red to represent features with increased abundance and green to indicate decreased abundance. Each node was also represented as a series of line plots representing all data within the node, along with the average expression profile of the node (shown in bold). Pc, P. chabaudi. A list of gene names for each node can be found in Table S1 in the supplemental material.
Functional analysis of expression profiles.
Each of the primary expression profiles was independently analyzed for functional significance in order to assess whether the waves of increased gene expression represented discrete functional aspects of the immune response (see Fig. S2 in the supplemental material).
Early expression.
In many infections, antimicrobial defense is initiated by an early interferon response. In our analysis of gene expression, the early transient response to P. chabaudi included a large proportion of known interferon-responsive genes. Genes encoding STAT1 and STAT2, which transmit signals from interferon receptors and are upregulated by interferon (30), increased in relative mRNA abundance in response to P. chabaudi infection. STAT1 and STAT2 can form a complex which acts as a transcription factor to stimulate the expression of other interferon-regulated genes. Consistent with STAT1 and STAT2 activation, we observed an increased abundance of genes encoding downstream targets of interferon signaling in response to infection with P. chabaudi. The interferon regulatory factor 1 gene, which is also known to be upregulated in response to viral infection and interferon (30, 56), encodes a transcription factor that drives the expression of additional interferon-regulated genes. Downstream genes such as Cxcl9 and Cxcl10 encode chemokines which attract CXCR3-expressing lymphocytes to the site of infection; Il15 and Il18, which exhibit pleiotropic proliferative and activating effects on multiple leukocyte subsets; members of the p47 family of GTPase genes, such as Igtp, Ifi47 (IRG-47), Irgm (LRG-47), and Tgtp, which are required for full resistance to certain pathogens through an unknown mechanism (6, 9, 32, 51); and the antiviral oligoadenylate synthase genes (Oas1c, Oas1e, Oas1g, Oas2, Oas3, Oasl1, and Oasl2). Additionally, other genes of less-well-characterized function are present in the early transient response, including Ifi203 and Ifi205, which are members of the Ifi200 cluster (Ifi202-Ifi205 and human homologues Ifi16, Mnda, Aim2, and Ifix) and are believed to play a role in regulating cell proliferation; genes encoding proteins with tetratricopeptide repeats (Ifit1, Ifit2, and Ifit3); several genes encoding members of the p65 guanylate binding protein family (Gbp2, Gbp3, Gbp4, and Gbp5), which are of unknown function but associate with the parasitophorous vacuoles of intracellular Toxoplasma gondii (11); and other uncharacterized members of the p47 family of GTPase genes (Iigp1, Iigp2), some members of which are essential for the control of infection (see above). Most importantly, all of these genes are well-known indicators of the presence of interferon-induced transcription.
We observed that interferon-stimulated genes and other related inflammatory responses dominated the early response. These observations were confirmed in an unbiased fashion by using statistical tools to identify significant functions in the early transcriptional response. Significant GO categories included general processes referred to as “immune response,” “defense response,” and “cytokine activity.” Analysis of KEGG pathways corroborated the GO analysis, with the identification of the “cytokine and cytokine receptor interaction” pathway being significant. This analysis further identified the Jak-Stat signaling pathway, which is involved in interferon signaling, as significantly enriched. The results of the lexical analysis indicated that the early response was dominated by interferon-induced transcripts. In summary, these data suggest that the early response to P. chabaudi erythrocytic infection is a proinflammatory response predominantly mediated by interferon.
Intermediate expression.
The intermediate response was characterized by a dramatic change in function. Two major processes exhibited significantly enriched transcripts in the intermediate response: antigen presentation through major histocompatibility complex (MHC) class I and NK cell function.
MHC class I was specifically enriched in the intermediate response, including receptor genes belonging to both classical (H2-D and H2-K) and nonclassical (H2-Q7, H2-T9, H2-T10/22, and H2-T23) presentation and the MHC class I receptor-associated β2 microglobulin. MHC class I receptor transcript enrichment was detected only with intermediate responses, and not with early or late responses, indicating the specificity of the timing of this response. The overrepresentation of MHC class I during this response was statistically significant, as confirmed by GO analysis (“antigen presentation, endogenous antigen via MHC class I” and “antigen presentation, endogenous antigen”), pathway analysis (“antigen processing and presentation”), and lexical analysis (“histocompatibility” and “antigen”).
The intermediate response also included a dramatic increase in the representation of NK cell-associated genes. A majority of the lectin-like killer cell receptor (Klr) family (22 of 28 independent Klrs represented in the data set), which initiates both NK cell activating and inhibitory signals, exhibited significantly increased relative abundance by 32 h in response to infection with P. chabaudi. The key signaling adapter DAP12 (Tyrobp), which associates with activating NK cell receptors, was also significantly increased in abundance. NK cells are known to function primarily through cytokine generation and direct cytotoxicity against infected or tumor cells. Transcripts for molecules involved in cytotoxic mechanisms were also significantly increased in abundance (Prf1, GzmA, and FasL). The overrepresentation of NK cell-associated transcripts during the intermediate response was supported by the significance of GO categories and lexical terms reflecting the lectin-like nature of Klrs. In addition, the KEGG pathway “NK cell-mediated cytotoxicity,” which represents the apoptosis-inducing function of NK cells, was significantly enriched in the intermediate response.
Late expression.
The response initiated at 56 h postinfection was clearly inflammatory in nature but was less well functionally defined than that observed at the earlier time points. Of particular note, two TNF superfamily ligand genes, namely, Tnfsf13 and Tnfsf14, and a number of genes related to proteolysis were represented. Four members of the stefin genes (StfA1, StfA2, StfA3, and StfA2L1), which encode cysteine-protease inhibitors, were significantly increased in abundance, as confirmed by lexical analysis (“stefin”). Interestingly, transcripts for the cysteine protease cathepsins C, H, and S were also significantly increased at this time. Stefin A is a known inhibitor of cathepsin H, suggesting that the coexpression of this pair may be important for protection against self-inflicted damage. Additionally, transcripts for the serine protease inhibitor genes SerpinB2 and SerpinB10 were enriched in the late response, suggesting that a general protective response is coordinated with the upregulation of proteolytic activities.
Dynamics of cell composition of blood upon P. chabaudi infection.
Expression profiling of the innate immune response to P. chabaudi infection clearly suggested cell-type-specific signatures. To examine the contribution of infection-mediated blood leukocyte population changes to the whole-blood transcriptome, we used flow cytometry to analyze the major leukocyte components of the blood and spleen at daily time points for the first 3 days of infection (Fig. 2).
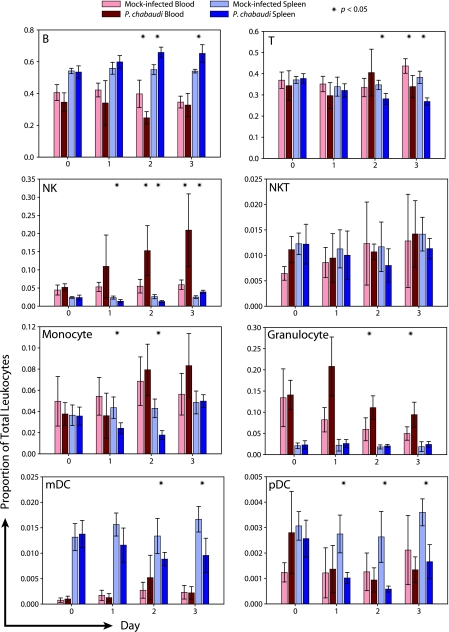
FIG. 2.
Frequencies of leukocyte subsets in the blood and spleen undergo reorganization after infection. Flow cytometry was used to identify the major leukocyte subsets in blood and spleens of mock- and P. chabaudi-infected mice during the first 3 days of infection. Data are reported as proportions of total leukocytes by organ and day. The data are presented as means with standard deviations from three to seven mice per group pooled from four independent experiments. Statistical comparisons were between uninfected and infected samples of the same time point and organ and utilized a two-tailed t test assuming unequal variances.
For uninfected blood, we observed that B lymphocytes (∼40% total) and T lymphocytes (35 to 40%) were the most abundant leukocytes, with a lower abundance of granulocytes (∼5 to 10%), monocytes (∼5%), and NK cells (∼5%). NKT (∼1%) and dendritic (<0.5%) cells made up a very minor fraction of blood leukocytes. These numbers are in line with previous reports on neutrophils and lymphocytes (14), as well as with the normal ranges for mouse hematology profiles (45). The numbers were similar for the spleen, except that in this organ there were larger proportions of B lymphocytes (∼55%) and dendritic cells (∼1%) but smaller proportions of NK cells (2 to 3%) and granulocytes (∼2%).
In response to P. chabaudi infection, there were only minor changes in leukocyte populations within 24 h. Granulocyte frequency increased in response to infection, although this was not statistically significant. By days 2 and 3, several significant changes occurred in the leukocyte populations of blood and spleen. On day 2, the frequency of B cells in the blood decreased to approximately half of the value for naïve or mock-infected mice. At the same time, NK cells exhibited a dramatic increase in blood frequency (∼5- to 10-fold). Intriguingly, there was a concomitant increase in B-cell frequency and a decrease in NK cell frequency in the spleen, suggesting that cells may be migrating between the spleen and peripheral blood. This conclusion was supported by absolute leukocyte counts, which demonstrated reciprocal responses in the blood and spleen that were consistent with a migration of B cells to the spleen and of NK cells to the blood (see Fig. S3 to S5 in the supplemental material). These changes generally persist through day 3, although NK frequency in the spleen increased by day 3.
Granulocyte frequencies in the blood of infected mice began to decline after a peak on day 1, although they remained significantly elevated, compared to what was seen for uninfected animals, through day 3. Additionally, a number of other populations (monocytes, dendritic cells, and T cells) decreased in frequency in the spleen, which could suggest migration out of the spleen, cell death, or a simple decrease in the overall proportion due to the proliferation and/or migration of other leukocyte subsets into the spleen.
NK cells do not upregulate Klr transcription in response to P. chabaudi infection.
We were particularly interested in understanding the dramatic increase in NK cell-associated transcripts in the blood in response to P. chabaudi infection. The increase in NK cell receptor transcripts in whole blood by day 3 might be explained by the 5- to 10-fold increase in NK cell frequency in the blood. Additionally, increased transcript abundance per cell might also contribute to the observed relative increase in whole blood. To explore the extent to which these possibilities contribute to the whole-blood transcriptome, we compared expression profiles of NK cells isolated from blood of mock-infected versus that of P. chabaudi-infected mice at 72 h postinfection by flow cytometry (Fig. 3). Interestingly, after P. chabaudi infection, Klrs were highly represented as enriched in whole blood (29 Klrs; median change, 3.25-fold; interquartile range, 2.36- to 4.68-fold) but not in isolated NK cells (median change, 0.87-fold; interquartile range, 0.80- to 0.98-fold). These data indicate that the increased abundance of Klr transcript observed in whole blood at 32 h was due primarily to the increase in NK cell frequency rather than to a transcriptional response to infection.
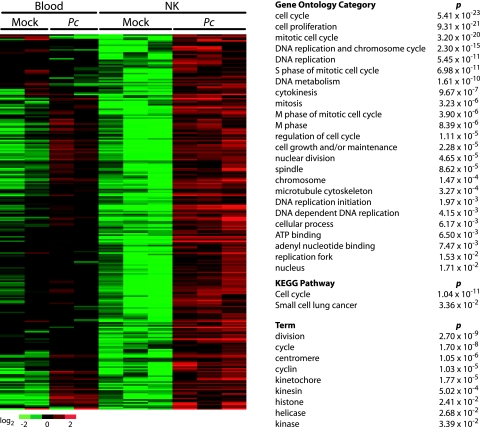
FIG. 3.
The transcriptional signature of NK cells suggests proliferation in response to infection. NK cells were isolated by flow cytometry from blood of three mock-infected and three P. chabaudi-infected mice at 72 h postinfection. Whole-blood samples from two mock-infected and two infected mice were also simultaneously profiled for expression. SAM was used to identify significantly overrepresented genes; these genes were organized by clustering and significant functional processes identified using GO, pathway analysis, and lexical analysis. The functional analyses (right) are for the set of genes which increased in abundance in NK cells upon P. chabaudi infection. Pc, P. chabaudi. A list of gene names can be found in Table S2 in the supplemental material.
Somewhat surprisingly, NK cells exhibited little transcriptional activity in pathways that would indicate a state of activation, such as those related to interferon or cytokine signaling. Only two genes associated with inflammatory responses were noteworthy: Ifi27, an IFN-α-inducible protein gene, and the gene for granzyme K, which is known to be produced by NK cells and to exhibit increased expression in patients with viral infections (3, 5).
NK cells undergo proliferation upon P. chabaudi infection.
While we observed only a few signatures that suggested that the NK cells might be activated, many key indicators of a proliferative response were upregulated by NK cells in response to infection with P. chabaudi (Fig. 3). These included genes with roles in the regulation of cell cycle, such as cyclin genes (Ccna2, Ccnb1, Ccnb2, Ccne1, and Ccne2), cell division cycle protein genes (Cdc2a, Cdc6, Cdc20, Cdc45l, Cdc73, Cdca2, Cdca3, Cdca5, and Cdca8), and cyclin-dependent kinase genes (Cdkl2, Cdkn3). Of particular note is that two probes representing Mki67, a marker indicative of cell proliferation, were significantly increased in abundance in NK cells after infection. In order to confirm these observations, these genes were subject to pathway and GO analyses in order to assess function. Two KEGG pathways associated with proliferative responses were identified as being significantly overrepresented in the expression programs of NK cells from infected mice: “cell cycle” (P = 1.04 × 10−11) and “small-cell lung cancer” (P = 3.36 × 10−2). GO was in accord with the pathway analysis; all of the 24 significantly overrepresented GO categories are related to cell cycle functions. Taken together with our observations that NK frequency in the blood increased (Fig. 2) and that absolute NK cell numbers increased in both blood and spleen by 72 h (see Fig. S4 and S5 in the supplemental material), these data suggest that NK cells may be undergoing proliferation within 2 or 3 days after P. chabaudi infection.
In order to determine if NK cells may undergo replication after infection as suggested by the expression profiles, we labeled naïve lymphocytes with CFSE and adoptively transferred 40 × 106 labeled cells into recipient mice. One half of the mice were mock infected and the other half were infected with P. chabaudi immediately after the adoptive transfer. Three days postinfection, we analyzed B, T, and NK cells in the blood and spleen by flow cytometry. These subsets were independently analyzed for the median fluorescence intensity of CFSE staining.
We first analyzed the proportion of each lymphocyte subset that was CFSE+ in blood and spleen after infection in order to assess migration. The proportion of B cells that were CFSE+ decreased in blood (mock, 3.1%; infected, 2.0%; P = 0.012) and increased in the spleen (mock, 2.6%; infected, 3.3%; P = 0.046) after infection, suggesting a redistribution of B cells from the blood to the spleen upon infection. This conclusion is consistent with our observation that absolute B-cell counts decreased in blood while increasing in the spleen (see Fig. S4 and S5 in the supplemental material). Changes in circulating CFSE+ NK cells did not reach statistical significance, but CFSE+ NK cells in the spleen decreased upon infection (mock, 5.9%; infected, 3.8%; P = 0.048), suggesting either a migration of NK cells from the spleen or the inability of adoptively transferred NK cells to reach the spleens of infected mice.
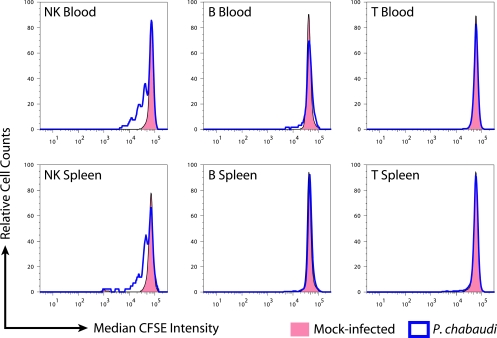
We next analyzed the degree of replication exhibited by each lymphocyte subset as measured by CFSE dilution. B and T cells in blood and spleen did not undergo marked replication within 3 days of the initiation of P. chabaudi infection (Fig. 4). By contrast, NK cells in both blood and spleen exhibited multiple peaks of CFSE dilution. Some cells appeared to have undergone as many as four or five rounds of replication by day 3, indicative of a rapid proliferative response. Peak modeling indicated that 44.0% (standard deviation = 7.6%) of NK cells in the blood and 56.4% (standard deviation = 8.2%) of NK cells in the spleen had undergone at least one round of replication. Our results indicate that in accord with the observed proliferation expression signature, NK cells, but not B or T cells, undergo early expansion in response to P. chabaudi infection.
FIG. 4.
NK cells in blood and spleen undergo division in response to infection. CFSE-labeled splenocytes were transferred to recipient mice, followed by either mock infection or P. chabaudi infection. After 72 h, blood and spleen were analyzed by flow cytometry for CFSE dilution by lymphocyte subset. The experiment was performed in triplicate; CFSE+ cells from the B, T, and NK subsets are represented in the histograms as relative cell counts.
DISCUSSION
We have utilized high-temporal-resolution gene expression profiling to characterize early immune responses to P. chabaudi infection in mice, which follows a nonlethal course resembling that of uncomplicated malaria in humans. We focused our attention on the blood, which has been largely neglected in expression analysis in rodent malaria models, presumably due to the technical challenges inherent in obtaining and working with small amounts of material. We also studied responses in the blood and spleen by flow cytometry. Our results demonstrate that extensive immune responses occur in both compartments in response to P. chabaudi infection. Specifically, an early interferon response is followed by a dramatic increase in circulating NK cells, which is at least partially explained by the observation that these cells rapidly proliferate.
The whole-blood expression response to P. chabaudi infection occurred in discrete waves of expression. The earliest responses to infection were initiated 8 to 16 h postinfection and were dominated by interferon-induced genes. The observation of interferon-associated responses at this early time point was not remarkable; rather, the significant finding was that interferon-associated responses were by far the predominant responses at the earliest time points. Given that TNF is thought to play a key role in the response to Plasmodium infection (29), it is possible that this cytokine may be an upstream mediator of the interferon responses, as TNF is reported to regulate the expression of type I and type II interferons (22, 56). Although we did not observe an increase in transcript abundance for some TNF-regulated genes, such as Ptgs2 and Il12, it remains possible that the subsets of leukocytes that respond to TNF are not as abundant as those that respond to interferons, resulting in our inability to detect the response. Regardless, interferon responses dominated the early response, consistent with a model in which IFN-γ is a central cytokine of the early innate immune response in P. chabaudi infection (50). We note, however, that little work has been conducted on the contribution of type I interferons to the early innate immune response to malaria infection. IFN-α is a typical product of the Tlr9-MyD88 signaling pathway, which has been proposed to detect Plasmodium infection. However, while IFN-α is produced in response to schizonts (40), hemazoin, the proposed malaria-specific Tlr9 ligand, did not induce detectable IFN-α (8, 37). Interestingly, one study using human PBMCs has shown that a blocking antibody to the type I interferon receptor abrogates the production of IFN-γ by NK cells (34), which is consistent with a human study reporting that type I interferon activity correlated with NK cell activity (36). Taken together, these studies indicate that both type I and type II interferons may be produced during the innate response to malaria (34) and that further studies exploring the interaction of these cytokines during the antimalarial response are warranted. Additionally, the earliest cellular source of IFN-γ remains controversial and will require clarification (2, 16).
A second wave in the expression response was initiated 24 to 32 h postinfection and was strongly dominated by transcripts associated with NK cells, especially the Klr family of NK cell receptors. Interestingly, this response could be attributed directly to changes in the NK cell frequency in the blood and not to differential expression within the NK cell population. Another immune-related response was initiated 48 to 56 h after infection. The changes in expression observed at this time point did not obviously correlate to any changes in cell frequency, so further work will be required to delineate the nature of this response. Overall, our data suggest that the immune response to P. chabaudi infection occurs in a series of discrete steps.
Analysis of the major blood leukocyte populations by flow cytometry and expression analysis revealed that changes in both leukocyte frequency and transcriptional activity contribute to the expression profiles observed for whole blood after P. chabaudi infection. We observed a sharp decrease in B-cell frequency in the blood and a concomitant increase in that in the spleen on day 2 postinfection. Results from the adoptive transfer of labeled cells and absolute leukocyte counts suggest that B cells migrate from the blood to the spleen after infection. NK cells exhibited the reciprocal response, with an increase in the blood and a decrease in the spleen on day 2 postinfection. Interestingly, by day 3, NK cells had increased in abundance in the spleen. However, previously labeled NK cells in the spleen had decreased by this time, suggesting either a migration of NK cells out of the spleen in response to infection or an inability of transferred NK cells to reach the spleen. Because we did not observe a preferential proliferation of a given Klr subset, as is observed a week after murine cytomegalovirus (MCMV) infection (15), and because we would expect cell death to impact both CFSE-labeled and unlabeled cells equally, we favor a model in which the initial decrease in splenic NK cells and the reciprocal increase in the blood after P. chabaudi infection are due to early migration followed by increases in both compartments due to proliferation.
There are two aspects of the NK cell expansion that distinguish it from the expansion of NK cells observed for other infections, such as that caused by MCMV. First, the expansion occurs in the blood, which is unlike the decrease in the NK cell frequency in the blood during MCMV infection (not shown). Second is the magnitude of the response; the expansion of NK cells in the blood triggered by P. chabaudi infection is considerably larger than that triggered by MCMV. These two aspects of P. chabaudi infection have not yet been reported for any other pathogen but could, at least in part, simply reflect the localization of the infectious agent.
We did not observe any enrichment for a particular Klr subset, in accord with previous observations showing that MCMV and vaccinia virus infections led to a generalized expansion of all mouse NK cell subsets at day 2 (15), although later after MCMV infection (day 4) subset selection became detectable. Interestingly, it has been reported that human NK cell subsets expressing high amounts of the CD94:NKG2A (Klrd1:Klrc1) receptor complex are preferentially activated by P. falciparum (1), suggesting the possibility that subsets expressing this receptor could also be preferentially expanded. However, our preliminary observations indicate that neither of these receptor subsets is preferentially expanded within 7 days after the initiation of mouse erythrocytic infection (not shown).
The contribution of NK cells to innate immune responses against malaria remains unclear. Early genetic experiments using P. chabaudi suggested that NK cell activity segregated independently of resistance to malaria (19, 46, 49), but more-recent depletion experiments with P. chabaudi and P. yoelii suggest a role in the direct control of parasite load (7, 27, 33). In pilot experiments, the depletion of NK cells during the early phase of infection did not impact the day or the magnitude of peak parasitemia (not shown). However, it is increasingly accepted that NK cells can produce cytokines that augment proinflammatory responses during P. chabaudi and P. falciparum infection (4, 33), and direct recognition of P. falciparum-infected erythrocytes by human NK cells has been described by use of in vitro systems (1, 2, 28, 34, 42). Further studies will be required to determine the impact of early stimulation of NK cells by malarial infection on the nature of the subsequent immune response.
Beyond our biological findings, our work has important general implications for the in vivo expression profiling of heterogeneous cell populations. First, our observations with Klr expression in NK cells demonstrate that some changes in transcript abundance are primarily a result of leukocyte frequency changes rather than transcriptional changes. Consideration of cell frequencies is thus an important aspect of the interpretation of whole-blood expression profiling (21, 35). Future studies should address overcoming the practical difficulties of obtaining such data in the field, as the collection of information on leukocyte frequencies greatly enhanced our interpretation of gene expression data. Second we have observed that some transcriptional profiles that are observed in individual cell populations are not visible in the context of whole organs (Fig. 3, top section of the heat map), which is an important limitation of the expression profiling of complex mixtures of cells. Although these observations are not surprising, our data provide formal proof that changes in cellular composition contribute directly to polyclonal expression profiles and, in some cases, can wholly explain observed changes in transcript abundance.
In summary, we have conducted a systematic exploration of early host responses to P. chabaudi infection. Our results indicate that the earliest immune response is mediated by interferon, followed by an expansion of NK cells. Although the relevance of these responses to the generation of protective immunity must still be addressed, the phenotypic observations that we have made will serve as an entry point to understanding the mechanistic basis of how immune responses to Plasmodium infection are initiated in vivo. We anticipate that gaining a comprehensive understanding of these processes will ultimately provide key insights into practical molecular correlates of protection.
Supplementary Material
Acknowledgments
We acknowledge the following for funding: NIH U01 AI53826 (J.L.D.), AI35800 (P.J.R.), AI35707 (P.J.R.), AI068129 (L.L.L.), and K23 AI060681 (S.P.), along with the David and Lucille Packard Foundation (J.L.D.), Howard Hughes Medical Institute (J.L.D.), the A. P. Giannini Foundation (C.C.K.), the Irvington Institute for Immunological Research (J.C.S.), and NIH Research Supplement for Underrepresented Minorities U01 AI152142-02S1 (A.M.). L.L.L. is an American Cancer Society Research Professor and P.J.R. is a Distinguished Clinical Scientist of the Doris Duke Charitable Foundation.
We thank Paige Nittler, Katherine Hermens, Sajeev Batra, and Jiri Gut for technical assistance.
Editor: J. F. Urban, Jr.
Footnotes
Published ahead of print on 29 September 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Artavanis-Tsakonas, K., K. Eleme, K. L. McQueen, N. W. Cheng, P. Parham, D. M. Davis, and E. M. Riley. 2003. Activation of a subset of human NK cells upon contact with Plasmodium falciparum-infected erythrocytes. J. Immunol. 1715396-5405. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas, K., and E. M. Riley. 2002. Innate immune response to malaria: rapid induction of IFN-gamma from human NK cells by live Plasmodium falciparum-infected erythrocytes. J. Immunol. 1692956-2963. [DOI] [PubMed] [Google Scholar]
- 3.Bade, B., H. E. Boettcher, J. Lohrmann, C. Hink-Schauer, K. Bratke, D. E. Jenne, J. C. Virchow, Jr., and W. Luttmann. 2005. Differential expression of the granzymes A, K and M and perforin in human peripheral blood lymphocytes. Int. Immunol. 171419-1428. [DOI] [PubMed] [Google Scholar]
- 4.Baratin, M., S. Roetynck, C. Lepolard, C. Falk, S. Sawadogo, S. Uematsu, S. Akira, B. Ryffel, J. G. Tiraby, L. Alexopoulou, C. J. Kirschning, J. Gysin, E. Vivier, and S. Ugolini. 2005. Natural killer cell and macrophage cooperation in MyD88-dependent innate responses to Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 10214747-14752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bratke, K., M. Kuepper, B. Bade, J. C. Virchow, Jr., and W. Luttmann. 2005. Differential expression of human granzymes A, B, and K in natural killer cells and during CD8+ T cell differentiation in peripheral blood. Eur. J. Immunol. 352608-2616. [DOI] [PubMed] [Google Scholar]
- 6.Carlow, D. A., S. J. Teh, and H. S. Teh. 1998. Specific antiviral activity demonstrated by TGTP, a member of a new family of interferon-induced GTPases. J. Immunol. 1612348-2355. [PubMed] [Google Scholar]
- 7.Choudhury, H. R., N. A. Sheikh, G. J. Bancroft, D. R. Katz, and J. B. De Souza. 2000. Early nonspecific immune responses and immunity to blood-stage nonlethal Plasmodium yoelii malaria. Infect. Immun. 686127-6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coban, C., K. J. Ishii, T. Kawai, H. Hemmi, S. Sato, S. Uematsu, M. Yamamoto, O. Takeuchi, S. Itagaki, N. Kumar, T. Horii, and S. Akira. 2005. Toll-like receptor 9 mediates innate immune activation by the malaria pigment hemozoin. J. Exp. Med. 20119-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collazo, C. M., G. S. Yap, G. D. Sempowski, K. C. Lusby, L. Tessarollo, G. F. Woude, A. Sher, and G. A. Taylor. 2001. Inactivation of LRG-47 and IRG-47 reveals a family of interferon gamma-inducible genes with essential, pathogen-specific roles in resistance to infection. J. Exp. Med. 194181-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couper, K. N., D. G. Blount, J. C. Hafalla, N. van Rooijen, J. B. de Souza, and E. M. Riley. 2007. Macrophage-mediated but gamma interferon-independent innate immune responses control the primary wave of Plasmodium yoelii parasitemia. Infect. Immun. 755806-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Degrandi, D., C. Konermann, C. Beuter-Gunia, A. Kresse, J. Wurthner, S. Kurig, S. Beer, and K. Pfeffer. 2007. Extensive characterization of IFN-induced GTPases mGBP1 to mGBP10 involved in host defense. J. Immunol. 1797729-7740. [DOI] [PubMed] [Google Scholar]
- 12.de Hoon, M. J., S. Imoto, J. Nolan, and S. Miyano. 2004. Open source clustering software. Bioinformatics 201453-1454. [DOI] [PubMed] [Google Scholar]
- 13.De Souza, J. B., K. H. Williamson, T. Otani, and J. H. Playfair. 1997. Early gamma interferon responses in lethal and nonlethal murine blood-stage malaria. Infect. Immun. 651593-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doeing, D. C., J. L. Borowicz, and E. T. Crockett. 2003. Gender dimorphism in differential peripheral blood leukocyte counts in mice using cardiac, tail, foot, and saphenous vein puncture methods. BMC Clin. Pathol. 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dokun, A. O., S. Kim, H. R. Smith, H. S. Kang, D. T. Chu, and W. M. Yokoyama. 2001. Specific and nonspecific NK cell activation during virus infection. Nat. Immunol. 2951-956. [DOI] [PubMed] [Google Scholar]
- 16.D'Ombrain, M. C., D. S. Hansen, K. M. Simpson, and L. Schofield. 2007. Gammadelta-T cells expressing NK receptors predominate over NK cells and conventional T cells in the innate IFN-gamma response to Plasmodium falciparum malaria. Eur. J. Immunol. 371864-1873. [DOI] [PubMed] [Google Scholar]
- 17.D'Ombrain, M. C., T. S. Voss, A. G. Maier, J. A. Pearce, D. S. Hansen, A. F. Cowman, and L. Schofield. 2007. Plasmodium falciparum erythrocyte membrane protein-1 specifically suppresses early production of host interferon-gamma. Cell Host Microbe 2130-138. [DOI] [PubMed] [Google Scholar]
- 18.Draghici, S., P. Khatri, A. L. Tarca, K. Amin, A. Done, C. Voichita, C. Georgescu, and R. Romero. 2007. A systems biology approach for pathway level analysis. Genome Res. 171537-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eugui, E. M., and A. C. Allison. 1980. Differences in susceptibility of various mouse strains to haemoprotozoan infections: possible correlation with natural killer activity. Parasite Immunol. 2277-292. [DOI] [PubMed] [Google Scholar]
- 20.Gollub, J., and G. Sherlock. 2006. Clustering microarray data. Methods Enzymol. 411194-213. [DOI] [PubMed] [Google Scholar]
- 21.Griffiths, M. J., M. J. Shafi, S. J. Popper, C. A. Hemingway, M. M. Kortok, A. Wathen, K. A. Rockett, R. Mott, M. Levin, C. R. Newton, K. Marsh, D. A. Relman, and D. P. Kwiatkowski. 2005. Genomewide analysis of the host response to malaria in Kenyan children. J. Infect. Dis. 1911599-1611. [DOI] [PubMed] [Google Scholar]
- 22.Haider, A. S., J. Cohen, J. Fei, L. C. Zaba, I. Cardinale, K. Toyoko, J. Ott, and J. G. Krueger. 2008. Insights into gene modulation by therapeutic TNF and IFNgamma antibodies: TNF regulates IFNgamma production by T cells and TNF-regulated genes linked to psoriasis transcriptome. J. Investig. Dermatol. 128655-666. [DOI] [PubMed] [Google Scholar]
- 23.Hernandez-Valladares, M., J. Naessens, and F. A. Iraqi. 2007. Gene-knockout mice in malaria research: useful or misleading? Trends Parasitol. 23522-526. [DOI] [PubMed] [Google Scholar]
- 24.Hosack, D. A., G. Dennis, Jr., B. T. Sherman, H. C. Lane, and R. A. Lempicki. 2003. Identifying biological themes within lists of genes with EASE. Genome Biol. 4R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, K. Y., W. W. Schultz, and F. B. Gordon. 1968. Interferon induced by Plasmodium berghei. Science 162123-124. [DOI] [PubMed] [Google Scholar]
- 26.Kim, C. C., and S. Falkow. 2003. Significance analysis of lexical bias in microarray data. BMC Bioinformatics 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitaguchi, T., M. Nagoya, T. Amano, M. Suzuki, and M. Minami. 1996. Analysis of roles of natural killer cells in defense against Plasmodium chabaudi in mice. Parasitol. Res. 82352-357. [DOI] [PubMed] [Google Scholar]
- 28.Korbel, D. S., K. C. Newman, C. R. Almeida, D. M. Davis, and E. M. Riley. 2005. Heterogeneous human NK cell responses to Plasmodium falciparum-infected erythrocytes. J. Immunol. 1757466-7473. [DOI] [PubMed] [Google Scholar]
- 29.Langhorne, J., F. R. Albano, M. Hensmann, L. Sanni, E. Cadman, C. Voisine, and A. M. Sponaas. 2004. Dendritic cells, pro-inflammatory responses, and antigen presentation in a rodent malaria infection. Immunol. Rev. 20135-47. [DOI] [PubMed] [Google Scholar]
- 30.Lehtonen, A., S. Matikainen, and I. Julkunen. 1997. Interferons up-regulate STAT1, STAT2, and IRF family transcription factor gene expression in human peripheral blood mononuclear cells and macrophages. J. Immunol. 159794-803. [PubMed] [Google Scholar]
- 31.Leisewitz, A. L., K. A. Rockett, B. Gumede, M. Jones, B. Urban, and D. P. Kwiatkowski. 2004. Response of the splenic dendritic cell population to malaria infection. Infect. Immun. 724233-4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacMicking, J. D., G. A. Taylor, and J. D. McKinney. 2003. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science 302654-659. [DOI] [PubMed] [Google Scholar]
- 33.Mohan, K., P. Moulin, and M. M. Stevenson. 1997. Natural killer cell cytokine production, not cytotoxicity, contributes to resistance against blood-stage Plasmodium chabaudi AS infection. J. Immunol. 1594990-4998. [PubMed] [Google Scholar]
- 34.Newman, K. C., D. S. Korbel, J. C. Hafalla, and E. M. Riley. 2006. Cross-talk with myeloid accessory cells regulates human natural killer cell interferon-gamma responses to malaria. PLoS Pathog. 2e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ockenhouse, C. F., W. C. Hu, K. E. Kester, J. F. Cummings, A. Stewart, D. G. Heppner, A. E. Jedlicka, A. L. Scott, N. D. Wolfe, M. Vahey, and D. S. Burke. 2006. Common and divergent immune response signaling pathways discovered in peripheral blood mononuclear cell gene expression patterns in presymptomatic and clinically apparent malaria. Infect. Immun. 745561-5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ojo-Amaize, E. A., L. S. Salimonu, A. I. Williams, O. A. Akinwolere, R. Shabo, G. V. Alm, and H. Wigzell. 1981. Positive correlation between degree of parasitemia, interferon titers, and natural killer cell activity in Plasmodium falciparum-infected children. J. Immunol. 1272296-2300. [PubMed] [Google Scholar]
- 37.Parroche, P., F. N. Lauw, N. Goutagny, E. Latz, B. G. Monks, A. Visintin, K. A. Halmen, M. Lamphier, M. Olivier, D. C. Bartholomeu, R. T. Gazzinelli, and D. T. Golenbock. 2007. Malaria hemozoin is immunologically inert but radically enhances innate responses by presenting malaria DNA to Toll-like receptor 9. Proc. Natl. Acad. Sci. USA 1041919-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perry, J. A., A. Rush, R. J. Wilson, C. S. Olver, and A. C. Avery. 2004. Dendritic cells from malaria-infected mice are fully functional APC. J. Immunol. 172475-482. [DOI] [PubMed] [Google Scholar]
- 39.Pichyangkul, S., P. Saengkrai, K. Yongvanitchit, A. Stewart, and D. G. Heppner. 1997. Activation of gammadelta T cells in malaria: interaction of cytokines and a schizont-associated Plasmodium falciparum antigen. J. Infect. Dis. 176233-241. [DOI] [PubMed] [Google Scholar]
- 40.Pichyangkul, S., K. Yongvanitchit, U. Kum-arb, H. Hemmi, S. Akira, A. M. Krieg, D. G. Heppner, V. A. Stewart, H. Hasegawa, S. Looareesuwan, G. D. Shanks, and R. S. Miller. 2004. Malaria blood stage parasites activate human plasmacytoid dendritic cells and murine dendritic cells through a Toll-like receptor 9-dependent pathway. J. Immunol. 1724926-4933. [DOI] [PubMed] [Google Scholar]
- 41.Roll Back Malaria, World Health Organization, and UNICEF. 2005. The world malaria report 2005. World Health Organization, Geneva, Switzerland.
- 42.Ronnblom, L., E. A. Ojo-Amaize, L. Franzen, H. Wigzell, and G. V. Alm. 1983. Plasmodium falciparum parasites induce interferon production in human peripheral blood ‘null’ cells in vitro. Parasite Immunol. 5165-172. [DOI] [PubMed] [Google Scholar]
- 43.Sachs, J., and P. Malaney. 2002. The economic and social burden of malaria. Nature 415680-685. [DOI] [PubMed] [Google Scholar]
- 44.Saldanha, A. J. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 203246-3248. [DOI] [PubMed] [Google Scholar]
- 45.Schalm, O. W., N. C. Jain, and E. J. Carroll. 1975. Veterinary hematology, 3rd ed. Lea & Febiger, Philadelphia, PA.
- 46.Skamene, E., M. M. Stevenson, and S. Lemieux. 1983. Murine malaria: dissociation of natural killer (NK) cell activity and resistance to Plasmodium chabaudi. Parasite Immunol. 5557-565. [DOI] [PubMed] [Google Scholar]
- 47.Snow, R. W., C. A. Guerra, A. M. Noor, H. Y. Myint, and S. I. Hay. 2005. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature 434214-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soulard, V., J. Roland, C. Sellier, A. C. Gruner, M. Leite-de-Moraes, J. F. Franetich, L. Renia, P. A. Cazenave, and S. Pied. 2007. Primary infection of C57BL/6 mice with Plasmodium yoelii induces a heterogeneous response of NKT cells. Infect. Immun. 752511-2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stevenson, M., S. Lemieux, and E. Skamene. 1984. Genetic control of resistance to murine malaria. J. Cell. Biochem. 2491-102. [DOI] [PubMed] [Google Scholar]
- 50.Su, Z., and M. M. Stevenson. 2000. Central role of endogenous gamma interferon in protective immunity against blood-stage Plasmodium chabaudi AS infection. Infect. Immun. 684399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor, G. A., C. M. Collazo, G. S. Yap, K. Nguyen, T. A. Gregorio, L. S. Taylor, B. Eagleson, L. Secrest, E. A. Southon, S. W. Reid, L. Tessarollo, M. Bray, D. W. McVicar, K. L. Komschlies, H. A. Young, C. A. Biron, A. Sher, and G. F. Vande Woude. 2000. Pathogen-specific loss of host resistance in mice lacking the IFN-gamma-inducible gene IGTP. Proc. Natl. Acad. Sci. USA 97751-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 985116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Urban, B. C., R. Ing, and M. M. Stevenson. 2005. Early interactions between blood-stage plasmodium parasites and the immune system. Curr. Top. Microbiol. Immunol. 29725-70. [DOI] [PubMed] [Google Scholar]
- 54.Verdugo, R. A., and J. F. Medrano. 2006. Comparison of gene coverage of mouse oligonucleotide microarray platforms. BMC Genomics 758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.World Health Organization. 2004. The world health report 2004. World Health Organization, Geneva, Switzerland.
- 56.Yarilina, A., K. H. Park-Min, T. Antoniv, X. Hu, and L. B. Ivashkiv. 2008. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat. Immunol. 9378-387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.