Abstract
Ovarian serous carcinoma (OSC) is the most common and lethal histologic type of ovarian epithelial malignancy. Mutations of TP53 and dysfunction of the Brca1 and/or Brca2 tumor-suppressor proteins have been implicated in the molecular pathogenesis of a large fraction of OSCs, but frequent somatic mutations in other well-established tumor-suppressor genes have not been identified. Using a genome-wide screen of DNA copy number alterations in 36 primary OSCs, we identified two tumors with apparent homozygous deletions of the NF1 gene. Subsequently, 18 ovarian carcinoma-derived cell lines and 41 primary OSCs were evaluated for NF1 alterations. Markedly reduced or absent expression of Nf1 protein was observed in 6 of the 18 cell lines, and using the protein truncation test and sequencing of cDNA and genomic DNA, NF1 mutations resulting in deletion of exons and/or aberrant splicing of NF1 transcripts were detected in 5 of the 6 cell lines with loss of NF1 expression. Similarly, NF1 alterations including homozygous deletions and splicing mutations were identified in 9 (22%) of 41 primary OSCs. As expected, tumors and cell lines with NF1 defects lacked mutations in KRAS or BRAF but showed Ras pathway activation based on immunohistochemical detection of phosphorylated MAPK (primary tumors) or increased levels of GTP-bound Ras (cell lines). The TP53 tumor-suppressor gene was mutated in all OSCs with documented NF1 mutation, suggesting that the pathways regulated by these two tumor-suppressor proteins often cooperate in the development of ovarian carcinomas with serous differentiation.
Introduction
Ovarian epithelial cancer (OvCa) is the most lethal type of gynecologic cancer in much of the industrialized world. It is a morphologically and biologically heterogeneous disease. Morphological criteria define four major types of primary ovarian adenocarcinomas—serous, mucinous, endometrioid, and clear cell. Molecular studies have offered support for the notion that the different histologic types of OvCas likely represent distinct disease entities. Ovarian serous carcinoma (OSC) is the most common histologic type of epithelial OvCa, comprising roughly 70% of diagnoses and almost always presents at advanced stage [1]. Mutations of TP53 and dysfunction of the Brca1 and/or Brca2 tumor-suppressor proteins have been implicated in the molecular pathogenesis of a large fraction of OSCs [2–7].
However, relatively little is known about other somatic genetic alterations that are present in a substantial fraction of OSCs. Recent comprehensive sequence-based analyses of somatic mutations in potentially oncogenic kinases in OvCa suggest that a number of genes encoding kinases may each be mutated at low frequency in OSCs, such as the LRRK2, STK36, ALPK2, and AKT1 genes (see http://www.sanger.ac.uk/genetics/CGP/Studies/Kinases/) [8,9]. In addition, despite the fact that the TP53 gene is somatically mutated in 60% or more of OSCs and the BRCA1 and/or BRCA2 genes are inactivated by genetic and/or epigenetic mechanisms in more than of 80% of OSCs, prior studies have not offered supporting evidence for the notion that somatic mutations in other well-established tumor-suppressor genes, such as the retinoblastoma (RB1), neurofibromatosis type 1 or type 2 (NF1 and NF2, respectively), Wilms tumor 1 (WT1), phosphatase and tensin homolog (PTEN), or the adenomatous polyposis coli (APC) genes, are common in OSCs.
Our initial genome-wide analysis of DNA copy number alterations in 36 primary OSCs identified two tumors with apparent homozygous deletions of the NF1 gene and several additional tumors with suspected hemizygous loss of NF1. Prompted by this finding, we screened 18 OvCa cell lines and 41 primary OSCs for NF1 alterations. We report here that mutational defects leading to reduced or absent Nf1 expression are common in the OvCa primary tumor specimens and cell lines analyzed and lead to Ras pathway activation. In addition, we found that all tumors with documented NF1 alterations also harbored mutations of the TP53 tumor-suppressor gene, suggesting the pathways regulated by these two tumor-suppressor proteins often cooperate in OSC pathogenesis.
Materials and Methods
Tumor Samples
Forty-one OSC samples were analyzed in this study, including 18 tumors from the Cooperative Human Tissue Network/Gynecologic Oncology Group Tissue Bank (Columbus, OH), 2 tumors from the New York Presbyterian Hospital (Weill Medical College of Cornell University), 11 tumors from the University of Michigan Health System, and 10 tumors from the Kumamoto University Hospital (Kumamoto, Japan). Primary tumor tissues were manually dissected with microscope guidance to ensure that each tumor sample contained a minimum of 70% tumor cells. Analysis of tissues from human subjects was approved by the University of Michigan's Institutional Review Board (IRB-MED 2001-0568 and 1999-0428).
Cell Lines
The OSC-derived cell lines SKOV-3 and CAOV3 [10] and TOV-21G (derived from a clear cell carcinoma) and the colorectal carcinoma cell line HCT116 were obtained from the American Type Culture Collection (Manassas, VA). Ovarian serous carcinoma cell lines HOC-1, HOC-7, HOC-8, and HEY [11,12] were a gift from L. Dubeau (USC School of Medicine, Los Angeles, CA). Ovarian carcinoma cell lines (histologic type not specified) A1847, A2780, OVCAR-4, OVCAR-5, OVCAR-8, and OVCAR-10 and OSC-derived cell lines PEO1 and PEO4 [13] were a gift from T. Hamilton (Fox Chase Cancer Center, Philadelphia, PA). Ovarian serous carcinoma cell lines OVCA420, OVCA429, OVCA432, and DOV13 [10] were a gift from D. Fishman (Northwestern University, Chicago, IL). IOSE-80 ovarian surface epithelial cells immortalized with SV40 large T antigen were a gift from N. Auersperg (University of British Columbia, Vancouver, Canada). All cell lines were maintained in DMEM with 10% FBS.
DNA, RNA, and cDNA Preparation
DNA, RNA, and cDNA were prepared using standard techniques. Briefly, genomic DNA was isolated from cultured cells or frozen tissue sections using SDS/proteinase K digestion followed by phenol/chloroform extraction. Total RNA was extracted from cultured cells or frozen tissue sections with Trizol (Invitrogen, San Diego, CA) according to the manufacturer's protocol. First-strand cDNA was synthesized from DNaseI-treated mRNA samples using random hexamer primers (Amersham Biosciences, Piscataway, NJ) and Superscript II (Invitrogen).
Representational Oligonucleotide Microarray Analysis and Data Analysis
Microarrays bearing 42,000 oligonucleotides designed to hybridize Bgl-II restriction fragments of human genome were custom-synthesized by Nimblegen Systems (Madison, WI) [14]. Briefly, tumor and normal human/male genomic DNA (1 µg each) were digested by Bgl-II enzyme (New England Biolabs, Ipswich, MA), and purified by QIAquick polymerase chain reaction (PCR) purification kit (Qiagen, Valencia, CA), followed by overnight ligation with the adapters (Bgl-12: gatctgctgcgt, Bgl-24: tcagcatcgagactgaacgcagca) to provide an anchoring site for subsequent PCR amplification to generate genome representations. Subsequent steps of genome representation amplification, array hybridization, and data acquisition/analysis were performed essentially as previously described [15]. The hybridized microarrays were scanned with an Axon 4000B scanner (Molecular Devices, Sunnyvale, CA) with pixel size set to 5 µm.
Southern Blot Analysis
For Southern blot analysis, 10 µg of DNA was digested with EcoRI for 12 to 18 hours at 37°C. The digested DNA was separated in 0.8% agarose gels and transferred overnight to positively charged nylon membranes (Hybond; Amersham Biosciences). Hybridization was carried out in Rapid-hyb buffer (Amersham Biosciences) to [α-32P]dCTP-labeled probes using standard procedures as described previously [16]. Three probes located at different positions within the NF1 locus and a control probe on the same chromosomal arm (DHX40 at 17q23.1) were used. The oligonucleotide sequences used to generate each probe are provided in Table W1.
Northern Blot Analysis
Ten micrograms of total RNA was separated in 0.9% agarose gels containing formaldehyde and transferred to Hybond-N+ membranes (Amersham Biosciences). NF1 and GAPDH probes were generated by PCR and labeled with 32P-dCTP with a random primer kit (Invitrogen). The NF1-specific probe corresponding to nucleotides 5999 to 6658 (NM_001042492) of the human NF1 cDNA and the GAPDH control probe were amplified using the primer sequences indicated in Table W1. Northern blot hybridization to 32P-labeled probes was carried out by standard methods. Membranes were exposed to phosphor-imager screens and scanned by phosphorimager (Molecular Dynamics, Sunnyvale, CA). The arbitrary units for intensity of photon emissions represent the level of NF1 mRNA normalized to GAPDH.
Protein Truncation Test
The protein truncation test (PTT) assay was performed as described by Heim et al. [17]. Briefly, 2 to 5 µg of mRNA was reverse-transcribed, and the entire NF1 cDNA was analyzed using the TNT Quick Coupled Transcription/Translation system (Promega Corp., Madison, WI) in overlapping segments using five pairs of primers (Table W1). The RT-PCR products were transcribed in vitro, and then in vitro translation was performed in the presence of 35S-methionine. The PTT samples were separated on 4% to 20% SDS-PAGE gels, which were dried and subjected to autoradiography. For each sample with aberrant PTT product(s), the corresponding regions of cDNA and, in most cases, genomic DNA, were amplified and sequenced. The primers used for amplification and sequencing of individual exons were those described by Schirinzi et al. [18].
Western Blot Analysis
Cells were lysed in radioimmunoprecipitation assay lysis buffer [50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS] containing complete protease inhibitor cocktail (Roche, Indianapolis, IN). The protein concentration was analyzed by bicinchoninic acid assay (Pierce, Rockford, IL). Subsequently, 100 µg of each cell lysate was separated on a 4% to 20% SDS-PAGE gel and then transferred to Immobilon P membranes (Millipore, Bedford, MA) by semidry electroblotting. Western blot analysis was carried out using anti-Nf1 antibody at a 1:1000 dilution (sc-67; Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Expression of β-actin was used as a loading control and was detected with anti-actin polyclonal antibody (Sigma-Aldrich, St. Louis, MO) at a dilution of 1:1000. The horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin (Pierce) was used at a 1:10,000 dilution. Antigen-antibody complexes were detected by enhanced chemiluminescence (ECL; Amersham Biosciences) and exposure to X-Omat film (Kodak, Rochester, NY). The cell line HCT116 is known to have frameshift mutations in both NF1 alleles and hence serves as a control for extracts lacking Nf1 expression [19].
Ras Activation Assay
Levels of activated Ras in selected cell lines were detected with a Ras activation assay kit (Upstate USA, Inc., Charlottesville, VA) per the manufacturer's protocol. Briefly, cells were starved in serum-free DMEM for 3 hours at 37°C. Cells were lysed, and the protein concentration was determined by the bicinchoninic acid method. A total of 250 µg of cellular lysate was incubated with Raf-1 RBD agarose (10 µl) at 4°C for 45 minutes. The agarose pull-downs were washed three times with lysis buffer, boiled with 2x Laemmli sample buffer, and separated on SDS-PAGE gels, followed by Western blot analysis using a monoclonal anti-Ras antibody at a 1:3000 dilution (Upstate USA, Inc.). To determine the levels of total Ras protein, 15 µg of total cellular lysates was electrophoresed on polyacrylamide gels followed by immunoblot analysis with the monoclonal anti-Ras antibody. Subsequently, the blot was reprobed with anti-actin antibody (Sigma-Aldrich) at a dilution of 1:1000.
Immunohistochemical Analysis of p53 and pMAPK
Five-micrometer sections were cut from blocks of formalin-fixed paraffin-embedded tissue for routine staining with hematoxylin and eosin. Immunohistochemical staining was conducted using the avidinbiotin-peroxidase method. Sections were immunostained using rabbit polyclonal anti-active MAPK Pab at a 1:200 dilution (pTEpY; Promega Corp.) or with mouse monoclonal anti-TP53 antibody (1:100 dilution, clone D0-7; Invitrogen). Immunostaining for nuclear and/or cytoplasmic pMAPK was scored on a four-tiered scale for intensity (-, absent; +, weak; ++, moderate; and +++, strong). Immunostaining for nuclear TP53 was scored as negative (absent, weak, or focal) or positive (strong and diffuse).
Sequencing of TP53, KRAS, and BRAF
Mutational analyses of the TP53 (exons 3–10), KRAS (exon 2), and BRAF (exons 11 and 15) genes were performed in the OSC samples using published and custom primer sequences as indicated in Table W1. Pearson's product moment correlation analysis (using the S-Plus statistical package) was used to measure the correlation between TP53 and NF1 mutations in the primary OSCs, and the Fisher's exact test was used to determine the significance of the association between mutations of these genes.
Results
A Subset of Primary OSCs Harbor Homozygous Deletions of the NF1 Gene
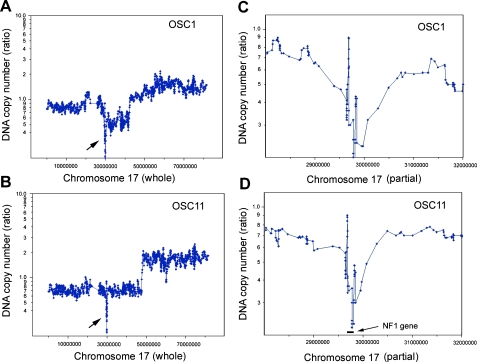
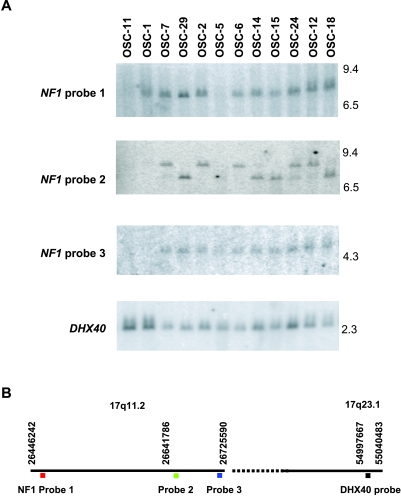
We pursued comprehensive analyses of DNA copy number alterations in 78 primary OvCas using a representational oligonucleotide microarray analysis (ROMA) approach. The full analysis of DNAcopy number changes in OvCas will be presented elsewhere. However, 2 of 36 OSCs studied (OSC-1 and OSC-11) were found to show copy number changes consistent with potential homozygous deletions involving the NF1 locus at chromosome band 17q11.2. Figure 1 shows moving average plots depicting the homozygous deletions identified by ROMAin these two tumors. To generate these plots, a moving window of five data points was used to average the raw DNAcopy number along the whole chromosome 17. On the basis of the ROMA data, four additional tumors (OSC-2, OSC-5, OSC-24, and OSC-25) showed evidence for loss of one NF1 allele. To confirm the presence of NF1 homozygous deletions, we performed Southern blot studies of selected primary OSCs, including the tumors with apparent homozygous or hemizygous deletions of the NF1 gene. The NF1 gene is very large, spanning nearly 300 kb, with 57 common exons and at least 3 alternatively spliced exons. Three DNA sequence probes derived from widely spaced regions of the NF1 locus were used for the Southern blot analysis, and the NF1-specific hybridization intensities were compared to those for a control probe (DHX40), located telomeric to NF1 on chromosome 17q. Genomic DNA from tumor OSC-11 did not show a detectable hybridization signal with any of the three NF1 probes (Figure 2), suggesting that both chromosome 17q deletions in this tumor affected the entire NF1 locus. OSC-1 showed no hybridization signal for probe 2 or 3 but showed hybridization to probe 1, whereas DNA from OSC-5 did not hybridize with probe 1 or 2 but retained NF1 sequences detected by probe 3. These results indicate that, in tumor OSC-1, there was a homozygous deletion involving a telomeric region of the NF1 gene (i.e., the 5′ end of the NF1 gene), and in tumor OSC-5, there was a homozygous deletion of a more centromeric (3′) portion of the NF1 gene. OSC-5 is one of the primary tumors suspected to have at least hemizygous deletion of NF1 based on ROMA.
Figure 1.
ROMA detects homozygous deletions including NF1 in primary OSCs. Using a moving average algorithm of the window of five data points, the raw DNA copy number ratio of tumor over normal reference was smoothened and plotted against the chromosomal position of all the oligonucleotide probes on chromosome 17 for tumor samples OSC-1 (A) and OSC-11 (B). The arrows point at the apparent homozygous deletions including the NF1 locus. To provide greater resolution of the deleted region, an area of 4 Mb across the NF1 deletion is shown for OSC-1 (C) and OSC-11 (D). Location of NF1 is as indicated in panel (D). Chromosomal positions of ROMA oligonucleotide probes were based on the NCBI build 34 (hg16 assembly).
Figure 2.
Southern blot analysis reveals homozygous deletions of NF1 in primary OSCs. (A) Southern blot analysis of EcoRI-digested OSC DNA samples using three probes at different locations within the NF1 gene. To control for loading, the blots were rehybridized with a probe for DHX40, which lies telomeric to NF1 on chromosome 17q. (B) The relative location of each probe on chromosome 17 is shown in the diagram.
Reduced or Absent NF1 Gene and Protein Expression in OvCa Cell Lines
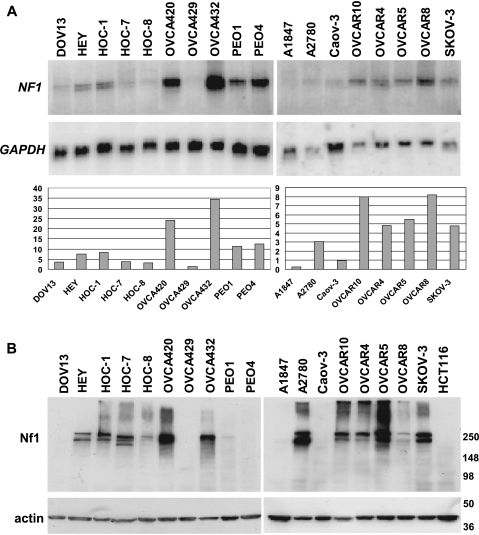
To explore the possibility that the NF1 gene might be inactivated in a significant fraction of OSCs, we pursued analysis of NF1 transcript and protein levels in a panel of 18 human OvCa cell lines. Twelve of the ovarian cancer cell lines are known to have been derived from OSCs [10–13]. The other six lines were derived from ovarian carcinomas of unspecified histologic subtype. However, given the frequency of OSC as a fraction of all OvCa diagnoses, many of the lines of unspecified type are likely to have been derived from serous tumors. NF1 transcripts range in size from 11 to 13 kb owing to alternative splicing and differences in the extent of 3′ untranslated sequences [20,21]. On the basis of Northern blot analysis, NF1 transcript levels varied significantly among the ovarian carcinoma cell lines, with high expression seen in OVCA420 and OVCA432 and minimal NF1 gene expression seen in several other cell lines including DOV13, HOC-7, HOC-8, OVCA429, A1847, A2780, and CAOV3 (Figure 3A). To assess expression of the Nf1 protein in the OvCa lines, Western blot analysis with an antibody directed against a carboxyl-terminal region of the Nf1 protein was pursued (Figure 3B). The HCT116 colon carcinoma cell line is known to have frameshift mutations in both NF1 alleles and hence serves as a negative control for Nf1 expression [19]. Of the 18 OvCa lines examined, five cell lines (DOV13, OVCA429, PEO4, A1847, and CAOV3) had essentially undetectable Nf1 protein expression. One additional cell line, PEO1, expressed only very minimal levels of Nf1. The findings suggest that perhaps more than one-third of OSCs might have mutational or epigenetic defects leading to NF1 inactivation.
Figure 3.
Expression of NF1 is reduced or absent in several ovarian carcinoma cell lines. (A) NF1 transcripts in the indicated cell lines were detected by Northern blot analysis, using expression of GAPDH as a loading control. Relative expression of NF1 in each cell line normalized to GAPDH (arbitrary units) is indicated. (B) Nf1 protein levels in the same cell lines were determined by Western blot analysis using an anti-Nf1 antibody. Detection of actin was used as a loading control.
NF1 Mutations Are Present in OvCa Lines with Reduced or Absent Nf1 Expression
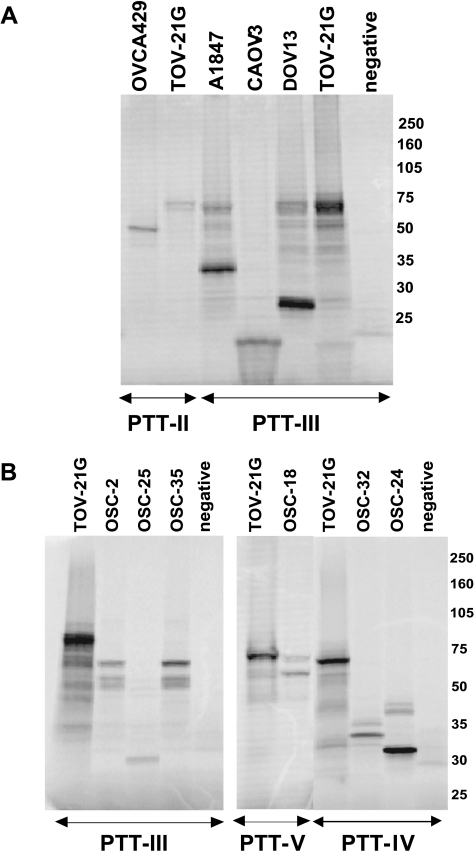
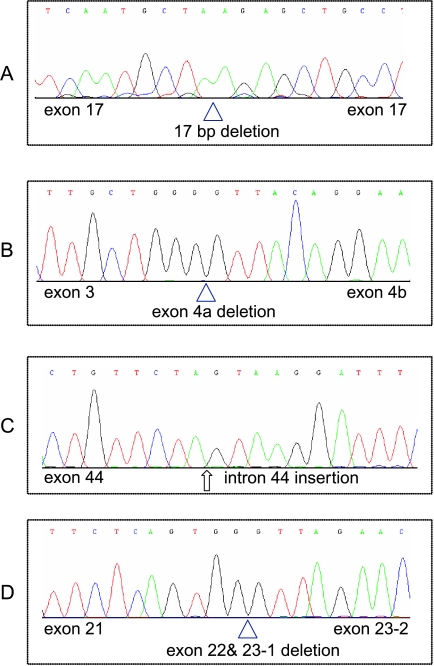
Protein truncation test assays were performed on the six OvCa lines (OVCA429, CAOV3, DOV13, PEO4, PEO1, and A1847) with low or absent Nf1 protein expression to ascertain a molecular basis for altered expression in the lines. cDNA was prepared from mRNA extracted after cells were cultured in the presence of puromycin, which inhibits nonsense-mediated decay of mRNA [22]. The entire NF1 coding region was examined by in vitro transcription and translation in five overlapping fragments (segments I–V). We were able to detect aberrant PTT products in five of the six cell lines, either as altered sized RT-PCR fragments (data not shown) or as truncated peptide fragments in segments I, II, or III in the PTT assay (Figure 4A). The corresponding cDNA and genomic DNA fragments from each cell line were cloned and sequenced (data summarized in Table 1). The cell line OVCA429 shows an aberrant PTT product in segment II, and the corresponding cDNA and genomic DNA both show a 17 base pair (bp) deletion in exon 17. The CAOV3 line has an aberrant PTT product in segment III, and sequencing of the corresponding cDNA revealed deletion of exons 22 and 23-1 and inclusion of 102 bp from the intron downstream of exon 23-2. We were unable to amplify these exons from genomic DNA by PCR, but amplification using adjacent intron primers revealed deletion of 1448 bp from this region. The A1847 line also showed an aberrant peptide in segment III, and the cDNA had an insertion of 101 bp from the intron downstream of exon 23-2. Sequencing of genomic DNA from A1847 revealed a single nucleotide change (C to G) within this cryptic exon, but the mechanism by which this change may lead to inclusion of intronic sequences in the NF1 transcript is not clear. DOV13 has a splicing mutation at the +1 position of exon 23-1, resulting in deletion of exon 23-1 from the cDNA. A germ line NF1 mutation identical to the presumed somatic mutation in DOV13 has been reported earlier [23]. In the cell line PEO4, PCR amplification of cDNA segment I yielded a RT-PCR product that was smaller than expected (data not shown). Our inability to detect a corresponding truncated protein fragment in the PTT assay is likely caused by the very small size of the predicted protein product (≈12 kDa). Sequencing of the cDNA revealed deletion of the entirety of exon 4a, and at the genomic level, mutation at the splice junction of exon 4a (-2, acceptor site, A to T), which leads to the skipping of this exon. Mutation at the same site in this splice junction (A to G) has also been reported previously [24]. The PTT assay failed to detect any mutations in PEO1 cells. This result is perhaps not unexpected, because a previous study that screened for NF1 germ line mutations using PTT only had a combined mutation detection rate of 56% [25]. Overall, our studies provide evidence for Nf1 inactivation in 6 (33%) of 18 OvCa lines evaluated, with definitive inactivating mutations identified in 5 of the 6 lines.
Figure 4.
The PTT assay shows truncated Nf1 peptides in ovarian carcinoma cell lines and primary tumors. (A) PTT was performed using five overlapping cDNA segments encompassing the entire NF1 coding region. Representative examples of truncated peptides in segments II and III from the indicated cell lines are shown. The TOV21G cell line expresses normal levels of full-length NF1, and it was used to determine the size of the expected (normal) PTT products in each segment. (B) Representative examples of truncated peptides in Segments III, IV and V from the indicated OSC samples are shown. The TOV21G cell line expresses normal levels of NF1, and it was used to determine the size of the expected (normal) PTT products in each segment.
Table 1.
NF1 Mutations Detected in OvCa Cell Lines and Primary Tumors by PTT.
| ID | cDNA Sequencing (NM_001042492, ATG as +1) | Genomic DNA Sequencing | Protein Change | Mutation Type |
| PEO4 | Del* c†291–481 (exon 4a) | A478T (first nucleotide of exon 4b, NM_001042492) | aa 97, PTC‡ at 109 | Splicing |
| CAOV3 | Del c3711-3977 (exons 22 and 23-1), c4110ins§102 bp | Del 1448 bps (exons 22 and 23-2 and partial introns 21 to 23-2) | aa1237, PTC 1245 | deletion/splicing |
| OVCA429 | Del c2943-2959 (exon 17) | Del 2943-2959 (NM_001042492) | aa 981, PTC 981 | deletion |
| A1847 | c4110 Ins101 bp from intron 23-2 | C-G intron 23a (at 304314067 NT_010799) | aa1371, PTC 1379 | Splicing |
| DOV13 | del c3873 -3977 (exon 23-1) | 3974 (exon 23-1) + 1 (G to T) (NM_001042492) | aa1291, PTC 1298 | splicing |
| OSC-2 | Duplication 662 bp (exons 24–27b) | No mutation detected | aa 1370, PTC 1379 | splicing |
| OSC-18 | Ins 178 bp (exons 44–45) | No mutation detected | aa 2602, PTC 2609 | splicing |
| OSC-24 | Del c5549-5751 (exon 30) | Del 34 nucleotides 5′ exon 30, Ins ATG | aa 1849, PTC 18451 | deletion/splicing |
| OSC-25 | Del c3711-3977 (exons 22 & 23-1) | No mutation detected | aa 1237, PTC 1245 | splicing |
| OSC-32 | Del exon 31 | No mutation detected | aa 1916, PTC 1921 | splicing |
| OSC-35 | Duplication 662 bp (exons 24–27b) | No mutation detected | aa 1370, PTC 1379 | splicing |
Del = deletion.
Sequence changes identified in cDNA are indicated by “c” before the number.
PTC = premature termination codon.
Ins = insertion.
NF1 Mutations Are Present in Primary OSCs
We screened a collection of 41 OSCs (including 29 of the 36 OSCs evaluated by ROMA) for NF1 mutations using the PTT assay. Aberrant PTT products affecting segment III, IV, or V were detected in six tumors (Figure 4B and Table 1). Sequencing of the cDNA corresponding to the aberrant PTT products identified aberrant transcripts in all six cases. cDNA from two of the tumors, OSC-2 and OSC-35, showed duplications of exons 24 to 27b (662 bp), whereas OSC-25 has deletion of exons 22 and 23-1. OSC-24 and OSC-32 showed deletions of exons 30 and 31, respectively. Finally, OSC-18 had an insertion of 178 bp corresponding to the entire intron between exons 44 and 45. Chromatograms of NFI cDNA sequence alterations in representative cell lines and primary tumors are shown in Figure 5. Genomic DNA isolated from OSC-24 revealed a deletion of 34 nucleotides encompassing the 5′ splice site of exon 30, with an insertion of ATG in its place. Genomic DNA sequencing of the corresponding NF1 exons and exon-intron boundaries in the other tumors failed to reveal mutations. This might reflect the likelihood that a significant fraction of mutations that alter appropriate splicing of NF1 transcripts are distant from the intron-exon boundaries; these mutations presumably create novel splice sites or activate cryptic splice sites within exonic or intronic sequences [26]. Alternatively, splicing errors have also been shown to occur in the absence of identifiable sequence alterations in the NF1 gene, with tumors showing nearly twice the amount of aberrant transcript as normal tissues [27]. Overall, we were able to document NF1 alterations in 9 (22%) of 41 primary OSCs, including three tumors with homozygous deletions of sizable portions of the NF1 gene and six tumors with localized mutations leading to premature truncation of the Nf1 protein. Three of these six tumors also showed evidence for hemizygous loss of NF1 based on ROMA.
Figure 5.
NF1 cDNA sequence alterations in representative cell lines and primary tumors. Chromatograms showing (A) absence of 17 bp of exon 17 from the cDNA of cell line OVCA429, (B) deletion of exon 4a in cDNA from PEO4, (C) portion of 178 bp insertion from intron 44 between exons 44 and 45 in tumor OSC-18, and (D) deletion of exons 22 and 23-1 in cDNA from tumor OSC-25.
The Ras Pathway Is Activated in Ovarian Carcinoma Cell Lines and Primary OSCs with NF1 Mutations
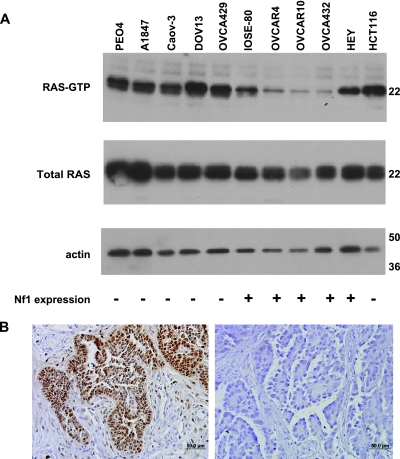
The Nf1 (neurofibromin) protein is a 2839 amino acid polypeptide, with a domain homologous to the catalytic domain of GTPase activating proteins (GAPs) [28]. This domain, called the GAP-related domain (GRD), regulates Ras activity by accelerating the conversion of GTP-bound “active” Ras to its inactive GDP-bound form [29]. Previous studies have shown that reduced or absent Nf1 protein expression in neurofibrosarcoma cell lines results in high levels of the active GTP-bound form of Ras (Ras-GTP) [30]. To determine whether NF1 mutations and/or concomitant reduction of Nf1 expression are associated with increased levels of active Ras-GTP in OSCs, an assay for activated Ras was performed in representative cell lines. Cell lines with markedly reduced or absent Nf1 expression (PEO4, A1847, CAOV3, DOV13, and OVCA429) had higher Ras-GTP levels compared to the cell lines with robust expression of Nf1 protein (Figure 6A). One OvCa line, HEY, which showed readily detectable levels of Nf1 protein (Figure 3B), also manifested high levels of active Ras-GTP.
Figure 6.
The Ras pathway is activated in ovarian carcinoma cell lines and primary OSCs. (A) Ras-GTP and total Ras levels in the ovarian carcinoma cell lines were detected using a Ras activation assay kit as described in the Materials and Methods section. Representative data are shown. The blot was reprobed with antiactin as a loading control. HCT116, which does not express NF1, and HEY, which expresses mutant KRas (G12D), were used as a positive controls for Ras-GTP levels. Expression of Nf1 in each cell line [low or absent (-) versus readily detectable (+)] is indicated below each lane. (B) Immunohistochemical analysis of pMAPK in primary OSCs; representative examples are shown. OSC-32 (left panel) has a NF1 mutation and shows strong nuclear staining for pMAPK in the tumor cells, with absence of staining in the non-neoplastic stromal cells. No NF1 alterations were detected in OSC-33, which is negative for pMAPK expression (right panel).
A major consequence of Ras pathway activation is the phosphorylation of MAPK [31]. Hence, we examined our collection of primary OSCs for the presence of active MAPK by immunohistochemistry using an antibody directed against the active phosphorylated form of p44/42 MAPK. As shown in Figure 6B and summarized in Table 2, eight of nine primary OSCs with documented NF1 alterations showed strong (++ or +++) pMAPK expression. Among the remaining OSCs studied, ∼41% (12/29) also stained positively for pMAPK. In these tumors, NF1 mutations may have been missed by our detection approach or MAPK may be activated by an alternative mechanism, such as activating mutations of upstream signaling components, including the transmembrane receptor tyrosine kinase epidermal growth factor receptor (EGFR), or HER-2/Neu.
Table 2.
Summary of the Mutational Analysis and Immunohistochemistry in OSC Tissues.
| Tumor ID | Clinical Data | ROMA-NF1 Deletions | NF1 Mutations Type | pMAPK (IHC) | p53 (IHC) | TP53Mutations (Exon 3–10) | KRAS Exon 2,3 | BRAFExon 11,15 | |||||
| Age | Stage | Grade | Exon | Nucleotide | Codon | Type | |||||||
| OSC-1 | 83 | 3C | 3 | homo | Deletion | +++ | + | 8 | G824A | C275Y | Missense | WT | WT |
| OSC-2 | 61 | 4 | 3 | hemi | Splicing | ++ | + | 5 | A395G | K132R | Missense | WT | WT |
| OSC-3 | 43 | 3C | 3 | WT | - | + | 7 | A701G | Y234C | Missense | WT | WT | |
| OSC-4 | 66 | 3C | 3 | WT | - | - | WT | WT | WT | ||||
| OSC-5 | 58 | 3B | 2 | hemi | Deletion | +++ | + | 5 | C380T | S127F | Missense | WT | WT |
| OSC-6 | 57 | 3C | 2 | WT | ++ | - | WT | WT | WT | ||||
| OSC-7 | 44 | 3C | 3 | WT | - | - | WT | WT | WT | ||||
| OSC-8 | 57 | 3C | 3 | WT | +++ | + | 5 | A491G | K164E | Missense | WT | WT | |
| OSC-9 | 67 | 1C | 3 | WT | +++ | + | 5 | T537G | H179Q | Missense | WT | WT | |
| OSC-10 | 53 | 2C | 2 | WT | ND | + | 5 | A395G | K132R | Missense | WT | WT | |
| OSC-11 | 60 | 4 | 2 | homo | Deletion | +++ | + | 4 | C215G | P72R | Missense | WT | WT |
| OSC-12 | 44 | 3D | 3 | WT | - | + | 9 | G976T | E326stop | Nonsense | WT | WT | |
| OSC-13 | 67 | 4 | 1 | WT | - | + | 5 | A395G | K132R | Missense | WT | WT | |
| OSC-14 | 40 | 3C | 2 | WT | ++ | - | 5 | C499T | Q167stop | Nonsense | WT | WT | |
| OSC-15 | 74 | 3C | 3 | WT | + | - | 5 | C497G | S166stop | Nonsense | WT | WT | |
| OSC-16 | 67 | 3C | 3 | WT | ++ | + | 6 | C569T | P190L | Missense | WT | WT | |
| OSC-17 | 62 | 3C | 3 | WT | + | - | WT | WT | WT | ||||
| OSC-18 | 59 | 1A | 2 | Splicing | +++ | - | 7 | 746delG | 344stop | Frameshift | WT | WT | |
| OSC-19 | 52 | 3 | 3 | WT | ND | ND | 7 | C742T | R248W | Missense | WT | WT | |
| OSC-20 | 65 | 2 | 3 | WT | +++ | + | 7 | C725T | C242F | Missense | WT | WT | |
| OSC-21 | 19 | 4 | 1 | WT | - | - | WT | WT | WT | ||||
| OSC-22 | 30 | 4 | 1 | WT | - | - | WT | WT | WT | ||||
| OSC-23 | 58 | 3C | 2 | WT | - | - | 8 | C916T | R306Stop | Nonsense | WT | WT | |
| OSC-24 | 53 | 3C | 2 | hemi | Deletion/splicing | - | + | 8 | C817G | R273G | Missense | WT | WT |
| OSC-25 | 58 | 3C | 3 | hemi | Splicing | +++ | + | 8 | C844T | R282W | Missense | WT | WT |
| OSC-26 | 73 | 3C | 2 | WT | - | + | 8 | C844T | R282W | Missense | WT | WT | |
| OSC-27 | 41 | 1C | 2 | WT | ND | - | 7 | 689delC | 246stop | Frameshift | WT | WT | |
| OSC-28 | 61 | 2C | 1 | WT | - | - | 6 | C637T | R213Stop | Nonsense | WT | WT | |
| OSC-29 | 49 | 3C | 1 | WT | ++ | - | WT | G12R | WT | ||||
| OSC-30 | 42 | 3C | 3 | WT | +++ | - | WT | WT | WT | ||||
| OSC-31 | 50 | 3C | 2 | WT | - | - | WT | WT | WT | ||||
| OSC-32 | 54 | 3C | 3 | Splicing | +++ | + | 5 | G517A | V173M | Missense | WT | WT | |
| OSC-33 | 60 | 3C | 3 | WT | - | - | 6 | C637T | R213Stop | Nonsense | WT | WT | |
| OSC-34 | 61 | 3 | 1 | WT | +++ | - | WT | WT | WT | ||||
| OSC-35 | 43 | 3C | 3 | Splicing | +++ | + | 6 | A659G | Y220C | Missense | WT | WT | |
| OSC-36 | 56 | 4 | 3 | WT | - | + | 6 | A659G | Y220C | Missense | WT | ND | |
| OSC-37 | 48 | 3C | 3 | WT | ++ | + | 6 | T581G | L194R | Missense | WT | WT | |
| OSC-38 | 68 | 1A | 3 | WT | - | - | 7 | 689delC | 246stop | Frameshift | WT | WT | |
| OSC-39 | 32 | 1B | 1 | WT | ++ | - | WT | WT | WT | ||||
| OSC-40 | 68 | 3C | 3 | WT | ++ | - | 5 | 455delC | 169stop | Frameshift | WT | WT | |
| OSC-41 | 73 | 3C | 3 | WT | - | + | 8 | G796A | G266R | Missense | WT | WT | |
OSCs with NF1 Defects Harbor Frequent Mutations in TP53 But Not KRAS or BRAF
Previous studies provide evidence for two major pathways in the pathogenesis of OSCs [32,33]. Low-grade OSCs are less common but have a high prevalence of activating mutations of the KRAS or BRAF proto-oncogenes and low prevalence of inactivating mutations of the TP53 tumor-suppressor gene. Indeed, nearly 70% of low-grade OSCs and their putative precursor lesions (serous borderline tumors) have either KRAS or BRAF mutations [34,35]. In contrast, high-grade OSCs comprise most OSCs, and in these tumors, KRAS and BRAF mutations are rare, whereas TP53 mutations are common [35,36]. We analyzed all 41 OSCs for mutations of TP53 (exons 3–10), KRAS (codons 12 and 13), and BRAF (exons 11 and 15). None of the OSCs had BRAF mutations, and only one tumor had a KRAS mutation (Table 2). Immunostaining for p53 protein was also performed in all but 1 of the 41 tumors (Table 2). Strong and diffuse nuclear p53 expression of the type seen in tumors with missense TP53 mutations was identified in 20 (50%) of 40 OSCs (data not shown). To confirm the presence of missense mutations and to detect mutations leading to loss or truncation of the p53 protein, TP53 exons 3–10 were sequenced. Overall, we identified TP53 mutations in 30 (73%) of 41 tumors. Of these, 21 were missense, 4 were frameshift, and 5 were nonsense mutations. Notably, all nine tumors with inactivated NF1 had documented TP53 mutations (Table 2). The association between NF1 and TP53 mutations was statistically significant (P = .041, 1-tail Fisher's exact test) and there was a positive correlation between mutations of NF1 and TP53 (r = 0.3211308, P = .0406; Pearson's product moment correlation).
Discussion
Although progress has been made in defining genetic alterations underlying the pathogenesis of OvCa, including the identification of specific mutations and gene expression patterns characteristic of the various morphological subtypes of OvCa, much work would seem to remain before we will have a full accounting of the key gene defects contributing to the development of OvCa. Because genomewide analysis of DNA copy number alterations in primary OvCas suggested the possibility of homozygous deletions of the NF1 gene in a subset of OSCs, we pursued in-depth molecular analyses to assess the frequency and mechanisms underlying NF1 inactivation in OSCs. We provide data here that mutational defects leading to reduced or absent Nf1 expression were found in 5 of 18 OvCa cell lines and 9 (22%) of 41 primary OSCs. In addition, given what is known about the role of the Nf1 protein in negatively regulating the activity of Ras proteins, we found, not unexpectedly, that OSCs with NF1 mutations lacked KRAS or BRAF mutations. All tumors with documented NF1 alterations were found to harbor mutations of the TP53 tumor-suppressor gene.
Neurofibromatosis type 1 (NF1) is an autosomal dominant genetic disorder affecting approximately 1 in 3000 individuals [37,38]. The most common manifestations are café-au-lait macules, neurofibromas, Lisch nodules, skin-fold freckling, bony dysplasia and learning disabilities [38]. In addition to neurofibromas, those with neurofibromatosis type 1 are at increased risk for development of other neoplasms, including malignant peripheral nerve sheath tumors, gliomas, and gastrointestinal stromal tumors. Interestingly, coinactivation of TP53 by deletion or by point mutation in conjunction with NF1 inactivation has been shown to be a negative prognostic marker in NF1 patients with malignant peripheral nerve sheath tumors [39,40]. A number of studies have characterized germ line NF1 mutations in individuals with neurofibromatosis type 1 and identified a diverse spectrum of mutations that includes small deletions and insertions, missense and nonsense point mutations, and mutations that affect splicing [17,23,24,41–44]. There are no clear mutational hotspots. Because more than 70% of NF1 mutations are predicted to result in truncation of the gene product [25], PTT has been widely used to screen for germ line (constitutional) NF1 mutations.
The Ras proteins have central roles in the regulation of cell proliferation and differentiation, and mutational activation of Ras signaling contributes to the development of many types of cancer. Ras proteins function as molecular switches in signaling pathways that transmit signals from the cell membrane to the nucleus [31,45,46]. Ras cycles between the inactive GDP-bound and active GTP-bound forms, signaling to downstream effectors that regulate basic cellular functions including cell proliferation, differentiation, and apoptosis. Nf1 acts as a Ras-GTPase activating protein (Ras-GAP), which catalyzes hydrolysis of Ras-GTP to Ras-GDP, with resultant down-regulation of downstream signaling through Raf, Ral/Cdc42, PLC, and PI3K. Loss of Nf1 function has been shown to deregulate Ras signaling in many types of cells, including Schwann cells, astrocytes, hematopoietic cells, mast cells, and melanocytes [47–49]. Moreover, NF1 mutations and/or loss of expression have been identified in several different types of tumors, including melanomas, colorectal carcinomas, small cell lung carcinomas, and transitional cell carcinomas [50–54]. Only a few studies in the published literature have addressed the role of NF1 in ovarian cancer pathogenesis, and to the best of our knowledge, no studies have described comprehensive analysis of NF1 mutations in ovarian cancers. Interestingly, Salud et al. [55] described a 29-year-old woman with neurofibromatosis type 1 who developed epithelial ovarian cancer. We believe ours is the first comprehensive study undertaken to determine the frequency of NF1 mutations in a sizable collection of OSC primary tumors and ovarian cancer cell lines.
We identified NF1 alterations in 5 (28%) of 18 ovarian carcinoma-derived cell lines and 9 (22%) of 41 primary OSCs. Evidence for biallelic inactivation of NF1 was obtained for six of the nine primary tumors (three with homozygous deletion and three with hemizygous deletion and mutation). The actual prevalence of inactivating NF1 mutations in OSCs is likely higher, because our mutation detection strategy was based entirely on the PTT assay, which fails to detect a third or more of NF1 mutations [42]. The high frequency of splicing defects identified in our analysis is in keeping with other studies in the published literature, which reported high rates of splicing mutations in NF1 [41,42]. Although absence of matched normal tissue precluded us from more definitively determining whether OSC tumors or cell lines with mutations of one NF1 allele had allelic deletions of the other copy, frequent allelic losses at the NF1 locus in ovarian cancers have been reported by others [56,57].
As mentioned previously, a major consequence of Ras pathway activation is the phosphorylation of MAPK. Notably, in their analysis of active MAPK in OSC, Hsu et al. [58] showed that 41% of high-grade OSCs expressed the active (phosphorylated) form of MAPK (pMAPK) by immunohistochemistry, although KRAS or BRAF mutations were not present in these tumors. Our findings confirm the paucity of KRAS and BRAF mutations and suggest that MAPK activation in OSCs may be largely attributable to Nf1 inactivation. All but one of our OSCs with documented NF1 alterations expressed pMAPK. Several additional tumors without demonstrable NF1 mutations also highly expressed pMAPK, suggesting that these tumors harbor NF1 mutations missed by PTT or mutations of other genes upstream of MAPK, such as HER-2/Neu or EGFR.
Most studies have shown that ∼50% to 80% of “typical” (i.e., high-grade, high-stage) OSCs have mutations in TP53 [36,59,60]. Mutations of KRAS in these tumors are much less common (0–12%), and BRAF mutations are extremely rare [33]. Our mutational data are in agreement with these published reports. TP53 mutations were detected in 30 (73%) of 41 primary OSCs. No BRAF mutations were found, and only one tumor had mutant KRAS. The co-occurrence of TP53 and NF1 mutations in our series of OSCs suggests the pathways regulated by these two tumor-suppressor proteins often cooperate in the development of ovarian carcinomas with serous differentiation. Additional studies, for example, in genetically engineered mice with conditionally mutant P53 and NF1 alleles, will be required to test this possibility.
Supplementary Material
Acknowledgments
The authors thank Lora Hedrick Ellenson for providing ovarian carcinoma samples for analysis.
Abbreviations
- OSC
ovarian serous carcinoma
- OvCa
ovarian epithelial cancer
- PTT
protein truncation test
Footnotes
This work was supported by the National Cancer Institute (2P30 CA046592 and 2RO1 CA94172) and University of Michigan Comprehensive Cancer Center's Tissue Procurement Core facility.
This article refers to a supplementary material, which is designated by Table W1 and is available online at www.neoplasia.com.
References
- 1.Seidman JD, Horkayne-Szakaly I, Haiba M, Boice CR, Kurman RJ, Ronnett BM. The histologic type and stage distribution of ovarian carcinomas of surface epithelial origin. Int J Gynecol Pathol. 2004;23:41–44. doi: 10.1097/01.pgp.0000101080.35393.16. [DOI] [PubMed] [Google Scholar]
- 2.Hilton JL, Geisler JP, Rathe JA, Hattermann-Zogg MA, DeYoung B, Buller RE. Inactivation of BRCA1 and BRCA2 in ovarian cancer. J Natl Cancer Inst. 2002;94:1396–1406. doi: 10.1093/jnci/94.18.1396. [DOI] [PubMed] [Google Scholar]
- 3.Wang C, Horiuchi A, Imai T, Ohira S, Itoh K, Nikaido T, Katsuyama Y, Konishi I. Expression of BRCA1 protein in benign, borderline, and malignant epithelial ovarian neoplasms and its relationship to methylation and allelic loss of the BRCA1 gene. J Pathol. 2004;202:215–223. doi: 10.1002/path.1507. [DOI] [PubMed] [Google Scholar]
- 4.Schuijer M, Berns EM. TP53 and ovarian cancer. Hum Mutat. 2003;21:285–291. doi: 10.1002/humu.10181. [DOI] [PubMed] [Google Scholar]
- 5.Pal T, Permuth-Wey J, Betts JA, Krischer JP, Fiorica J, Arango H, LaPolla J, Hoffman M, Martino MA, Wakeley K, et al. BRCA1 and BRCA2 mutations account for a large proportion of ovarian carcinoma cases. Cancer. 2005;104:2807–2816. doi: 10.1002/cncr.21536. [DOI] [PubMed] [Google Scholar]
- 6.Willner J, Wurz K, Allison KH, Galic V, Garcia RL, Goff BA, Swisher EM. Alternate molecular genetic pathways in ovarian carcinomas of common histological types. Hum Pathol. 2007;38:607–613. doi: 10.1016/j.humpath.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Salani R, Kurman RJ, Giuntoli R, II, Gardner G, Bristow R, Wang TL, Shih IM. Assessment of TP53 mutation using purified tissue samples of ovarian serous carcinomas reveals a higher mutation rate than previously reported and does not correlate with drug resistance. Int J Gynecol Cancer. 2008;18:487–491. doi: 10.1111/j.1525-1438.2007.01039.x. [DOI] [PubMed] [Google Scholar]
- 8.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage S, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 10.Kim JH, Skates SJ, Uede T, Wong KK, Schorge JO, Feltmate CM, Berkowitz RS, Cramer DW, Mok SC. Osteopontin as a potential diagnostic biomarker for ovarian cancer. JAMA. 2002;287:1671–1679. doi: 10.1001/jama.287.13.1671. [DOI] [PubMed] [Google Scholar]
- 11.Buick RN, Pullano R, Trent JM. Comparative properties of five human ovarian adenocarcinoma cell lines. Cancer Res. 1985;45:3668–3676. [PubMed] [Google Scholar]
- 12.Masazza G, Lucchini V, Tomasoni A, Peccatori F, Lampasona V, Giudici G, Mangioni C, Biondi A, Giavazzi R. Malignant behavior and resistance to cisplatin of human ovarian carcinoma xenografts established from the same patient at different stages of the disease. Cancer Res. 1991;51:6358–6362. [PubMed] [Google Scholar]
- 13.Langdon SP, Lawrie SS, Hay FG, Hawkes MM, McDonald A, Hayward IP, Schol DJ, Hilgers J, Leonard RC, Smyth JF. Characterization and properties of nine human ovarian adenocarcinoma cell lines. Cancer Res. 1988;48:6166–6172. [PubMed] [Google Scholar]
- 14.Lucito R, Healy J, Alexander J, Reiner A, Esposito D, Chi M, Rodgers L, Brady A, Sebat J, Troge J, et al. Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kendall J, Liu Q, Bakleh A, Krasnitz A, Nguyen KC, Lakshmi B, Gerald WL, Powers S, Mu D. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci USA. 2007;104:16663–16668. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu R, Lin L, Beer DG, Ellenson LH, Lamb BJ, Rouillard JM, Kuick R, Hanash S, Schwartz DR, Fearon ER, et al. Amplification and overexpression of the L-MYC proto-oncogene in ovarian carcinomas. Am J Pathol. 2003;162:1603–1610. doi: 10.1016/S0002-9440(10)64294-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heim RA, Kam-Morgan LN, Binnie CG, Corns DD, Cayouette MC, Farber RA, Aylsworth AS, Silverman LM, Luce MC. Distribution of 13 truncating mutations in the neurofibromatosis 1 gene. Hum Mol Genet. 1995;4:975–981. doi: 10.1093/hmg/4.6.975. [DOI] [PubMed] [Google Scholar]
- 18.Schirinzi A, Drmanac S, Dallapiccola B, Huang S, Scott K, De Luca A, Swanson D, Drmanac R, Surrey S, Fortina P. Combinatorial sequencing-byhybridization: analysis of the NF1 gene. Genet Test. 2006;10:8–17. doi: 10.1089/gte.2006.10.8. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, Montmain G, Ruano E, Upadhyaya M, Dudley S, Liskay RM, Thibodeau SN, Puisieux A. Neurofibromatosis type 1 gene as a mutational target in a mismatch repair-deficient cell type. Hum Genet. 2003;112:117–123. doi: 10.1007/s00439-002-0858-4. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, O'Connell P, Breidenbach HH, Cawthon R, Stevens J, Xu G, Neil S, Robertson M, White R, Viskochil D. Genomic organization of the neurofibromatosis 1 gene (NF1) Genomics. 1995;25:9–18. doi: 10.1016/0888-7543(95)80104-t. [DOI] [PubMed] [Google Scholar]
- 21.Vandenbroucke I, Vandesompele J, De Paepe A, Messiaen L. Quantification of NF1 transcripts reveals novel highly expressed splice variants. FEBS Lett. 2002;522:71–76. doi: 10.1016/s0014-5793(02)02887-9. [DOI] [PubMed] [Google Scholar]
- 22.Lim SK, Sigmund CD, Gross KW, Maquat LE. Nonsense codons in human beta-globin mRNA result in the production of mRNA degradation products. Mol Cell Biol. 1992;12:1149–1161. doi: 10.1128/mcb.12.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han SS, Cooper DN, Upadhyaya MN. Evaluation of denaturing high performance liquid chromatography (DHPLC) for the mutational analysis of the neurofibromatosis type 1 (NF1) gene. Hum Genet. 2001;109:487–497. doi: 10.1007/s004390100594. [DOI] [PubMed] [Google Scholar]
- 24.Kluwe L, Friedrich RE, Korf B, Fahsold R, Mautner VF. NF1 mutations in neurofibromatosis 1 patients with plexiform neurofibromas. Hum Mutat. 2002;19:309. doi: 10.1002/humu.9018. [DOI] [PubMed] [Google Scholar]
- 25.Osborn MJ, Upadhyaya M. Evaluation of the protein truncation test and mutation detection in the NF1 gene: mutational analysis of 15 known and 40 unknown mutations. Hum Genet. 1999;105:327–332. doi: 10.1007/s004399900135. [DOI] [PubMed] [Google Scholar]
- 26.Zatkova A, Messiaen L, Vandenbroucke I, Wieser R, Fonatsch C, Krainer AR, Wimmer K. Disruption of exonic splicing enhancer elements is the principal cause of exon skipping associated with seven nonsense or missense alleles of NF1. Hum Mutat. 2004;24:491–501. doi: 10.1002/humu.20103. [DOI] [PubMed] [Google Scholar]
- 27.Kaufmann D, Leistner W, Kruse P, Kenner O, Hoffmeyer S, Hein C, Vogel W, Messiaen L, Bartelt B. Aberrant splicing in several human tumors in the tumor suppressor genes neurofibromatosis type 1, neurofibromatosis type 2, and tuberous sclerosis 2. Cancer Res. 2002;62:1503–1509. [PubMed] [Google Scholar]
- 28.DeClue JE, Cohen BD, Lowy DR. Identification and characterization of the neurofibromatosis type 1 protein product. Proc Natl Acad Sci USA. 1991;88:9914–9918. doi: 10.1073/pnas.88.22.9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bollag G, McCormick F. Ras regulation. NF is enough of GAP. Nature. 1992;356:663–664. doi: 10.1038/356663a0. [DOI] [PubMed] [Google Scholar]
- 30.Basu T, Gutmann D, Fletcher JA, Glover TW, Collins FS, Downward J. Aberrant regulation of ras proteins in malignant tumour cells from type 1 neurofibromatosis patients. Nature. 1992;356:713–715. doi: 10.1038/356713a0. [DOI] [PubMed] [Google Scholar]
- 31.McKay MM, Morrison DK. Integrating signals from RTKs to ERK/MAPK. Oncogene. 2007;26:3113–3121. doi: 10.1038/sj.onc.1210394. [DOI] [PubMed] [Google Scholar]
- 32.Singer G, Kurman RJ, Chang HW, Cho SK, Shih I-M. Diverse tumorigenic pathways in ovarian serous carcinoma. Am J Pathol. 2002;160:1223–1228. doi: 10.1016/s0002-9440(10)62549-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shih I-M, Kurman RJ. Ovarian tumorigenesis: a proposed model based on morphological and molecular genetic analysis. Am J Pathol. 2004;164:1511–1518. doi: 10.1016/s0002-9440(10)63708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singer G, Shih I-M, Truskinovsky A, Umudum H, Kurman RJ. Mutational analysis of K-ras segregates ovarian serous carcinomas into two types: invasive MPSC (low-grade tumor) and conventional serous carcinoma (high-grade tumor) Int J Gynecol Pathol. 2003;22:37–41. doi: 10.1097/00004347-200301000-00009. [DOI] [PubMed] [Google Scholar]
- 35.Singer G, Oldt R, III, Cohen Y, Wang BG, Sidransky D, Kurman RJ, Shih I-M. Mutations in BRAF and KRAS characterize the development of low-grade ovarian serous carcinoma. J Natl Cancer Inst. 2003;95:484–486. doi: 10.1093/jnci/95.6.484. [DOI] [PubMed] [Google Scholar]
- 36.Singer G, Stohr R, Cope L, Dehari R, Hartmann A, Cao DF, Wang TL, Kurman RJ, Shih I-M. Patterns of p53 mutations separate ovarian serous borderline tumors and low- and high-grade carcinomas and provide support for a new model of ovarian carcinogenesis: a mutational analysis with immunohistochemical correlation. Am J Surg Pathol. 2005;29:218–224. doi: 10.1097/01.pas.0000146025.91953.8d. [DOI] [PubMed] [Google Scholar]
- 37.Rasmussen SA, Friedman JM. NF1 gene and neurofibromatosis 1. Am J Epidemiol. 2000;151:33–40. doi: 10.1093/oxfordjournals.aje.a010118. [DOI] [PubMed] [Google Scholar]
- 38.Ferner RE. Neurofibromatosis 1 and neurofibromatosis 2: a twenty first century perspective. Lancet Neurol. 2007;6:340–351. doi: 10.1016/S1474-4422(07)70075-3. [DOI] [PubMed] [Google Scholar]
- 39.Holtkamp N, Atallah I, Okuducu AF, Mucha J, Hartmann C, Mautner VF, Friedrich RE, Mawrin C, von Deimling A. MMP-13 and p53 in the progression of malignant peripheral nerve sheath tumors. Neoplasia. 2007;9:671–677. doi: 10.1593/neo.07304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Upadhyaya M, Kluwe L, Spurlock G, Monem B, Majounie E, Mantripragada K, Ruggieri M, Chuzhanova N, Evans DG, Ferner R, et al. Germline and somatic NF1 gene mutation spectrum in NF1-associated malignant peripheral nerve sheath tumors (MPNSTs) Hum Mutat. 2008;29:74–82. doi: 10.1002/humu.20601. [DOI] [PubMed] [Google Scholar]
- 41.Ars E, Serra E, Garcia J, Kruyer H, Gaona A, Lazaro C, Estivill X. Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum Mol Genet. 2000;9:237–247. doi: 10.1093/hmg/9.2.237. [DOI] [PubMed] [Google Scholar]
- 42.Messiaen LM, Callens T, Mortier G, Beysen D, Vandenbroucke I, Van Roy N, Speleman F, Paepe AD. Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum Mutat. 2000;15:541–555. doi: 10.1002/1098-1004(200006)15:6<541::AID-HUMU6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 43.Ars E, Kruyer H, Morell M, Pros E, Serra E, Ravella A, Estivill X, Lazaro C. Recurrent mutations in the NF1 gene are common among neurofibromatosis type 1 patients. J Med Genet. 2003;40:e82. doi: 10.1136/jmg.40.6.e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wimmer K, Roca X, Beiglbock H, Callens T, Etzler J, Rao AR, Krainer AR, Fonatsch C, Messiaen L. Extensive in silico analysis of NF1 splicing defects uncovers determinants for splicing outcome upon 5′ splice-site disruption. Hum Mutat. 2007;28:599–612. doi: 10.1002/humu.20493. [DOI] [PubMed] [Google Scholar]
- 45.Adjei AA. Blocking oncogenic Ras signaling for cancer therapy. J Natl Cancer Inst. 2001;93:1062–1074. doi: 10.1093/jnci/93.14.1062. [DOI] [PubMed] [Google Scholar]
- 46.Harrisingh MC, Lloyd AC. Ras/Raf/ERK signalling and NF1. Cell Cycle. 2004;3:1255–1258. doi: 10.4161/cc.3.10.1182. [DOI] [PubMed] [Google Scholar]
- 47.Bollag G, Clapp DW, Shih S, Adler F, Zhang YY, Thompson P, Lange BJ, Freedman MH, McCormick F, Jacks T, et al. Loss of NF1 results in activation of the Ras signaling pathway and leads to aberrant growth in haematopoietic cells. Nat Genet. 1996;12:144–148. doi: 10.1038/ng0296-144. [DOI] [PubMed] [Google Scholar]
- 48.Gutmann DH, Wu YL, Hedrick NM, Zhu Y, Guha A, Parada LF. Heterozygosity for the neurofibromatosis 1 (NF1) tumor suppressor results in abnormalities in cell attachment, spreading and motility in astrocytes. Hum Mol Genet. 2001;10:3009–3016. doi: 10.1093/hmg/10.26.3009. [DOI] [PubMed] [Google Scholar]
- 49.Ingram DA, Hiatt K, King AJ, Fisher L, Shivakumar R, Derstine C, Wenning MJ, Diaz B, Travers JB, Hood A, et al. Hyperactivation of p21(ras) and the hematopoietic-specific Rho GTPase, Rac2, cooperate to alter the proliferation of neurofibromin-deficient mast cells in vivo and in vitro. J Exp Med. 2001;194:57–69. doi: 10.1084/jem.194.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aaltonen V, Bostrom PJ, Soderstrom KO, Hirvonen O, Tuukkanen J, Nurmi M, Laato M, Peltonen J. Urinary bladder transitional cell carcinogenesis is associated with down-regulation of NF1 tumor suppressor gene in vivo and in vitro. Am J Pathol. 1999;154:755–765. doi: 10.1016/S0002-9440(10)65322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson MR, Look AT, DeClue JE, Valentine MB, Lowy DR. Inactivation of the NF1 gene in human melanoma and neuroblastoma cell lines without impaired regulation of GTP.Ras. Proc Natl Acad Sci USA. 1993;90:5539–5543. doi: 10.1073/pnas.90.12.5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cacev T, Radosevic S, Spaventi R, Pavelic K, Kapitanovic S. NF1 gene loss of heterozygosity and expression analysis in sporadic colon cancer. Gut. 2005;54:1129–1135. doi: 10.1136/gut.2004.053348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Furukawa K, Yanai N, Fujita M, Harada Y. Novel mutations of neurofibromatosis type 1 gene in small cell lung cancers. Surg Today. 2003;33:323–327. doi: 10.1007/s005950300074. [DOI] [PubMed] [Google Scholar]
- 54.Ahlquist T, Bottillo I, Danielsen SA, Meling GI, Rognum TO, Lind GE, Dallapiccola B, Lothe RA. RAS signaling in colorectal carcinomas through alteration of RAS, RAF, NF1, and/or RASSF1A. Neoplasia. 2008;10:680–686. doi: 10.1593/neo.08312. 2 p following 686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salud A, Porcel JM, Capdevila F, Felip E, Rovirosa MA, del Campo JM. Ovarian cancer in a female patient with von Recklinghausen's disease. Med Clin (Barc) 1991;96:138–140. [PubMed] [Google Scholar]
- 56.Schildkraut JM, Collins NK, Dent GA, Tucker JA, Barrett JC, Berchuck A, Boyd J. Loss of heterozygosity on chromosome 17q11-21 in cancers of women who have both breast and ovarian cancer. Am J Obstet Gynecol. 1995;172:908–913. doi: 10.1016/0002-9378(95)90020-9. [DOI] [PubMed] [Google Scholar]
- 57.Wertheim I, Tangir J, Muto MG, Welch WR, Berkowitz RS, Chen WY, Mok SCH. Loss of heterozygosity of chromosome 17 in human borderline and invasive epithelial ovarian tumors. Oncogene. 1996;12:2147–2153. [PubMed] [Google Scholar]
- 58.Hsu CY, Bristow R, Cha MS, Wang BG, Ho CL, Kurman RJ, Wang TL, Shih I-M. Characterization of active mitogen-activated protein kinase in ovarian serous carcinomas. Clin Cancer Res. 2004;10:6432–6436. doi: 10.1158/1078-0432.CCR-04-0893. [DOI] [PubMed] [Google Scholar]
- 59.Schuijer M, Berns EM. TP53 and ovarian cancer. Hum Mutat. 2003;21:285–291. doi: 10.1002/humu.10181. [DOI] [PubMed] [Google Scholar]
- 60.Kupryjanczyk J, Thor AD, Beauchamp R, Merritt V, Edgerton SM, Bell DA, Yandell DW. p53 gene mutations and protein accumulation in human ovarian cancer. Proc Natl Acad Sci USA. 1993;90:4961–4965. doi: 10.1073/pnas.90.11.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.