Abstract
DNA structure is a critical element in determining its function. The DNA molecule is capable of adopting a variety of non-canonical structures, including three-stranded (i.e. triplex) structures, which will be the focus of this review. The ability to selectively modulate the activity of genes is a long-standing goal in molecular medicine. DNA triplex structures, either intermolecular triplexes formed by binding of an exogenously applied oligonucleotide to a target duplex sequence, or naturally occurring intramolecular triplexes (H-DNA) formed at endogenous mirror repeat sequences, present exploitable features that permit site-specific alteration of the genome. These structures can induce transcriptional repression and site-specific mutagenesis or recombination. Triplex-forming oligonucleotides (TFOs) can bind to duplex DNA in a sequence specific fashion with high affinity, and can be used to direct DNA-modifying agents to selected sequences. H-DNA plays important roles in vivo and is inherently mutagenic and recombinogenic, such that elements of the H-DNA structure may be pharmacologically exploitable. In this review we discuss the biological consequences and therapeutic potential of triple helical DNA structures. We anticipate that the information provided will stimulate further investigations aimed toward improving DNA triplex-related gene targeting strategies for biotechnological and potential clinical applications.
Keywords: triplex DNA, DNA triple helices, triplex-forming oligonucleotides, H-DNA, unusual DNA structures, genetic instability
1. Introduction
The DNA of a single cell contains all of the genetic information necessary for life’s processes. Friedrich Miescher discovered DNA in 1868, yet it took more than 70 years to demonstrate that it is the molecule that carries genetic information [1]. Once this was realized, tremendous effort has been made to better understand both the structure and function of DNA. Not only does the DNA primary nucleic acid sequence define the genetic code, its secondary structure plays important roles in regulating gene expression such that the formation of multi-stranded DNA structures at specific sites in the genome can influence many cellular functions. DNA can form multi-stranded helices through either folding of one of the two strands or association of two, three, or four strands of DNA. A well-established multi-stranded DNA structure, triple helical DNA (triplex DNA), both naturally occurring intramolecular H-DNA structures, and triplex-forming oligonucleotide (TFO)-targeted intermolecular triplexes will be the focus of this review.
Triple-helical nucleic acids were first described in 1957 by Felsenfeld and Rich [2], who demonstrated that polyuridylic acid and polyadenylic acids strands in a 2:1 ratio were capable of forming a stable complex. In 1986, it was demonstrated that a short (15-mer) mixed-sequence triplex-forming oligonucleotide (TFO) formed a stable specific triple helical DNA complex [3]. The third strand of DNA in the triplex structure (i.e. the TFO) follows a path through the major groove of the duplex DNA. The specificity and stability of the triplex structure is afforded via Hoogsteen hydrogen bonds [4], which are different from those formed in classical Watson-Crick base pairing in duplex DNA. Because purines contain potential hydrogen bonds with incoming third strand bases, the binding of the third strand is to the purine-rich strand of the DNA duplex [5,6].
2. Classification of DNA triple helices
Since the original discovery of triple helical nucleic acids, a number of triplex DNA structures that form under various conditions in vitro and/or in vivo have been identified (reviewed in [7–9]). These include intermolecular triplexes (with a pyrimidine third strand “Y:RY”, a purine strand or mixed pyrimidine/purine third strand “R:RY”), and intramolecular triplexes (H-DNA).
2.1. Intermolecular triplexes
Intermolecular triplexes are formed when the triplex-forming strand originates from a second DNA molecule (Figure 1). Intermolecular triplexes have attracted much attention because of their potential therapeutic application in inhibiting the expression of genes involved in cancer and other human diseases, for targeting disease genes for inactivation, for stimulating DNA repair and/or homologous recombination pathways, for inducing site-specific mutations, and for interfering with DNA replication. An example of triplex formation with a polypurine TFO sequence specific for the human c-MYC P2 promoter is shown in Figure 2.
Figure 1. Schematic representation of intermolecular DNA triplex formation.
In the target duplex, the purine and pyrimidine strands are shown in blue and yellow, respectively. The TFO, which binds to the purine-rich strand of the target duplex through the major groove, is indicated in red.
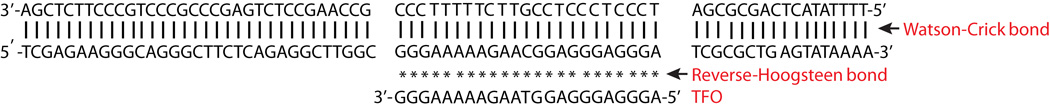
Figure 2. Triplex-forming sequences in the human c-MYC gene.
The TFO is placed in an antiparallel orientation relative to the target duplex from the human c-MYC P2 promoter. Vertical lines indicate Watson-Crick hydrogen bonds and stars indicate reverse Hoogsteen hydrogen bonding.
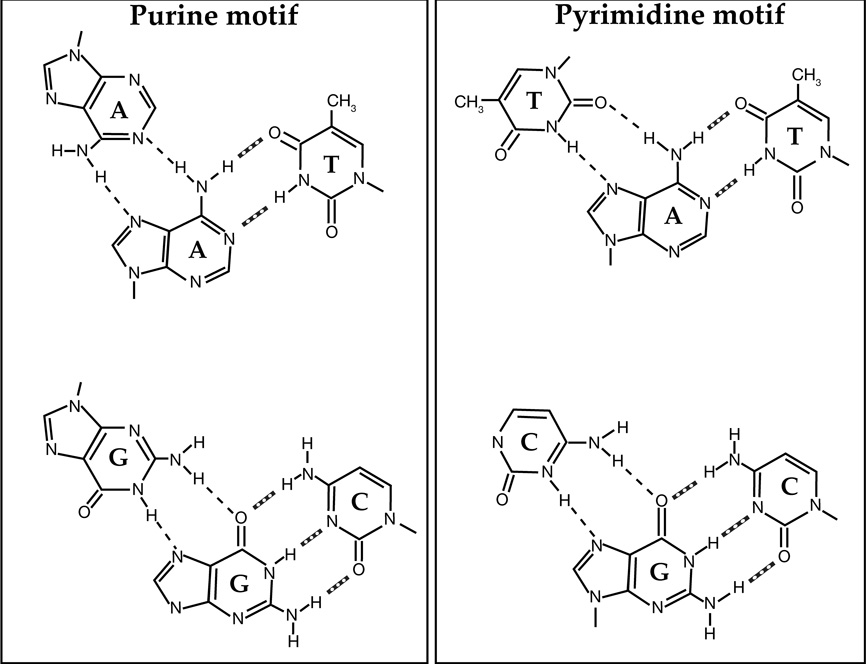
Triplex formation occurs in two motifs, distinguished by the orientation of the third strand with respect to the purine-rich strand of the target duplex. Typically, polypyrimidine third strands (Y) bind to the polypurine strand of the duplex DNA via Hoogsteen hydrogen bonding in a parallel fashion (i.e. in the same 5’ to 3’, orientation as the purine-rich strand of the duplex), whereas the polypurine third strand (R) binds in an antiparallel fashion to the purine strand of the duplex via reverse-Hoogsteen hydrogen bonds [6,10,11]. In the antiparallel, purine motif, the triplets are G:G-C, A:A-T, and T:A-T; whereas in the parallel, pyrimidine motif, the canonical triples are C+:G-C and T:A-T triplets (where C+ represents a protonated cytosine on the N3 position) The hydrogen bonding schemes found in purine and pyrimidine motif triplexes are depicted in Figure 3. Antiparallel GA and GT TFOs form stable triplexes at neutral pH, while parallel CT TFOs bind well only at acidic pH so that N3 on cytosine in the TFO is protonated [12], substitution of C with 5-methyl-C permits binding of CT TFOs at physiological pH [13,14] as 5-methyl-C has a higher pK than does cytosine. For both motifs, contiguous homopurine-homopyrimidine runs of at least 10 base pairs are required for TFO binding, since shorter triplexes are not substantially stable under physiological conditions, and interruptions in optimum sequence can greatly destabilize the triplex structure [15–20]. If purine bases are randomly distributed between two duplex strands, the consecutive third-strand bases should switch from one strand of the duplex to the other, resulting in a structural distortion of the sugar-phosphate backbone and lack of stacking interactions. This is energetically unfavorable, and therefore the most appropriate duplex target for triplex formation contains consecutive purine bases in one strand. Thus, an ideal target for triplex formation is the presence of a homopurine sequence in one strand of duplex and a homopyrimidine sequence in the complementary strand.
Figure 3. Schematic representation of canonical base triplets formed in purine and pyrimidine triplex motifs.
Watson-Crick base pairing is illustrated by dotted lines, and Hoogsteen base pairing by broken lines.
Triplex formation is kinetically slow compared to duplex annealing [21–25]. However, once formed, triplexes are very stable, exhibiting half-lives on the order of days [21,23]. Formation of both intramolecular and intermolecular triplexes depends on several factors including length, base composition, divalent cations, and temperature [7]. The affinity and specificity of TFO binding are critical features to their success of a gene targeting molecules. The dissociation constants (Kds) of TFOs for their target duplexes typically range from 10−7 to 10−10 M, making them feasible gene targeting agents.
2.2. Intramolecular triplexes (H-DNA)
In an intramolecular triplex or H-DNA structure, the third strand is provided by one of the strands of the same duplex DNA molecule at a mirror repeat sequence (Figure 4). Four isomers of intramolecular triplexes can exist depending on the strand that serves as the third strand. Intramolecular triplexes are also known as H-DNA or *H-DNA, depending on whether the third strand of the triplex is Py- or Pu-rich, respectively. These structures will be discussed at length in Section 5–Section 9.
Figure 4. H-DNA (intramolecular triplex DNA).
In the polypurine-polypyrimidine tract with mirror repeat symmetry, one of the single strands (shown in blue) folds back and forms triplex structure and the other strand (shown in yellow) is left unpaired.
3. Potential applications of DNA triplex formation in therapeutics
3.1. Targeting genes as an approach to molecular-targeted therapeutics
The ability to target specific genes to modulate their structure and/or function in the genome has far-reaching implications in biology, biotechnology, and medicine. TFOs represent near-ideal molecules for this purpose because of their ability to bind duplex DNA with high affinity and specificity. Facile chemistries for TFO modification are also available, allowing the covalent attachment of DNA damaging agents, for example, to target damage to specific sites in a genome. Oligonucleotides have been designed to target nucleic acids as well as proteins in a variety of applications. A potential advantage of targeting DNA rather than RNA or protein is the limited number of copies to be targeted. In addition, TFOs unlike antisense oligonucleotides or siRNAs can be used to target damage specifically to mutate or inactivate a gene. Thus, in a biological context, antigene approaches may provide some advantages over antisense strategies. Other general positive features of oligonucleotide-based approaches are: facile synthesis of the reagents; the availability of a variety of chemical modifications (to the bases, sugar-phosphate backbone, and the 5’ and 3’ ends to improve cellular uptake, target binding, specificity, and stability; and availability of efficient delivery systems.
3.2. Abundance of TFO binding sites in mammalian genomes
The gene products involved in many important biological processes such as cell signaling, proliferation, and carcinogenesis have now been identified, providing plausible targets for gene modulation. Triplex technology represents an approach to regulate these processes by manipulating the structure and/or function of these critical genes. However, in order to target any gene of interest, there must be unique TFO binding sites in those genes. To determine the number of potential TFO binding sites available in mammalian genomes, we designed an algorithm to search the entire human and mouse genomes for such sites, and were surprised to find ~2 million in each of these mammalian genomes [26]. We found that most annotated genes in both the mouse and human genomes contain at least one unique TFO binding site, and these sites are enriched in the promoter and /or transcribed gene regions.
3.3. Modulating gene expression via triplex formation
The first demonstration of TFO-directed transcription inhibition was reported by the Hogan group two decades ago [5]. Since that time, many groups have published reports demonstrating the ability of TFOs to inhibit the expression of a number of genes in a variety of systems. Mammalian genes that have been targeted by TFOs are summarized by Wu et al. (2007) [27]. However, obstacles to this approach have limited the potential of triplex technology as a consistent and reproducible method of transcriptional regulation. Several limitations to this technology include TFO delivery and uptake into cells, TFO stability once in the cells, lack of optimal target site binding affinity and specificity due to intracellular salt concentrations and pH, displacement by DNA metabolic activities (e.g. transcription, replication, and repair), and chromatin structure which may present a barrier to target site accessibility. As examples, physiologic concentrations of potassium can facilitate the formation of G-quadruplex structures on G-rich purine TFOs, thereby preventing triplex formation [15,21,28], and in the case of pyrimidine TFOs, binding can be inhibited by physiologic pH due to the requirement for cytosine protonation at N3 [14]. It is also possible that alternative activities of the TFO in the cell preclude its intended purpose in binding its duplex target to inhibit gene expression. TFOs have been demonstrated to act as “decoy” oligonucleotides, which bind transcription factors, such that they are not available to bind their duplex consensus sequences for transcription activation [29]. Aptamer effects of TFOs have also been observed in which the oligonucleotides can bind proteins and inhibit their activity. Efforts have been made to overcome these limitations, largely through chemical modification of the TFO, which we discuss in detail in Section 4.
3.4. Directing site-specific DNA damage
Another potential strategy for the development of TFOs as “therapeutic agents” is their utility as targeted DNA damaging agents. This approach differs from TFO-directed transcription inhibition in that TFO-directed DNA damage has been shown to stimulate mutation, recombination, and DNA repair at the targeted sites. Thus, this application has the potential to directly inactivate genes, rather than transiently regulating gene expression. As an example, triplex formation can be used to direct site-specific DNA damage and thereby induce DNA repair synthesis locally, independent of replication synthesis [30–32]. We have used TFOs targeted to the c-MYC gene to stimulate repair synthesis in the presence of an antitumor antimetabolite, gemcitabine, to increase its incorporation into DNA. We found that when used in combination, c-MYC-specific TFOs significantly increased the effectiveness of gemcitabine in inhibiting the growth of human breast tumor cells [33].
Physical studies of the triplex structure reveal that binding of the TFO induces structural distortions in the underlying duplex even though the Watson-Crick hydrogen bonding is preserved [34,35]. These features, along with facile chemistries to covalently couple DNA damaging agents to the TFOs, make triplexes attractive probes for directing site-specific DNA damage [36–38]. Targeting DNA damage to a specific site via triplex formation can be used to induce mutations [32,39,40] and/or recombination in vitro and in vivo [41–43], presumably through recognition of the triplex structures as damage by the repair machinery of the cell [31,32,44–46]. Thus, triplex technology provides a mechanism to modify gene structure and function in living organisms [39,47]. Conjugation of TFOs with photoactivatable chemical groups such as psoralen (which requires UVA irradiation for activation) allows one to control the timing of damage (i.e. after the TFO has bound its target) to reduce non-specific, collateral damage to the rest of the genome [37,38,48,49]. These features make TFOs powerful reagents for controlled gene manipulation in mammalian systems.
4. Approaches to Improve the Efficacy of TFOs in Biological Systems
As discussed above (Section 3.3), there are many factors that can limit the efficacy of triplex technology in cellular systems. Improvements in TFO chemistries are under active investigation, as these modifications could considerably increase the efficacy of antigene oligonucleotide therapeutics.
4.1 Chemical modifications of TFOs
To improve the binding affinity, selectivity and stability of oligonucleotides inside the cell, a number of modifications have been made to the bases, the backbone, the 5’ or/or 3’ ends, and/or the sugar moiety of oligonucleotides. Some examples of base modifications to TFOs designed to bind in the antiparallel purine triplex motif include the substitution of guanine by 6-thioguanine [28,50] or the substitution of adenine by 7-dezaxanthine [51]. These modifications have assisted in preventing the formation of unwanted intramolecular secondary structures within the TFOs. In the parallel pyrimidine triplex motif, the substitution of cytosine by 5-methylcytosine, N6-methyl-8-oxo-2-deoxyadenosine [52,53] or pseudoisocytodines [54] has been used to reduce the pH dependence of triplex formation. At neutral pH, the substitution of thymine by 5-propynyluracil stabilizes triplex formation [55].
Modifications of the natural phosphodiester backbone have been designed to improve TFO binding affinity and stability. Among these are chemical modifications that result in neutral or cationic backbones to reduce the electrostatic repulsion between the negatively charged phosphodiester backbone of the TFO with that of the target DNA duplex. Examples of such modifications include thioate linkages [56,57], N3’ − P5’ phosphoramidates [58,59], and morpholino phosphoramidate linkages [60]. The cationic phosphoramidate linkages, N,N-diethylethylenediamine and N,N-dimethylaminopropylamine, confer increases in TFO binding affinity and intracellular activity [61]. Other promising oligonucleotide backbone modifications that allow for improvements in triplex formation include peptide nucleic acids (PNAs) and locked nucleic acids (LNAs). PNAs are non-ionic nucleic acid analogs in which the sugar-phosphate backbone is replaced by an N-aminoethyl-glycine-based polyamide structure. PNAs bind to single–stranded DNA via Watson-Crick base pairing and can also form triple helices through Hoogsteen base pairing with the DNA/PNA duplex [62,63]. The neutral polyamide backbone was designed to minimize non-specific electrostatic effects that often are observed with DNA oligonucleotides. PNAs, under certain conditions, can bind to any DNA duplex sequence by a strand-invasion mechanism. Based on this property, PNAs have great potential as DNA targeting “drugs” [64]. Moreover, PNAs are resistant to both nucleases and proteases, and their neutral backbone increases their hybridization affinities to complementary RNA and DNA strands [65]. PNAs have been used successfully to target chromosomal DNA at transcription start sites to inhibit gene expression in cells [66–68]. In LNA molecules, the deoxyribose moiety is modified by introducing a methylene bridge between the 2’-O, 4’-O, and 4’-C. This bridge results in a locked 3’-endo conformation, which reduces the flexibility of the ribose, and allows for stable triplex formation [69–72]. LNA modifications within a TFO can increase the binding affinity to the target duplex DNA, and can increase resistance to digestion by nucleases [73]. Beane et al., have demonstrated that LNAs can recognize chromosomal target sequences and efficiently block endogenous expression of the progesterone and androgen receptors [74]. An LNA analogue ENA, containing a 2’-O, 4’-Cethylene bridge has also been reported to form stable triplex at physiological pH [75].
Because RNA:DNA duplexes are more stable than DNA:DNA duplexes, many researchers are interested in modifying the ribose moiety in oligonucleotides to test the effect of binding and stability in triplex formation. Inoue et al., have reported that 2’-O-methyloligoribonucleotide derivatives are not subject to cleavage by RNase H, and that a 2’-O-methylribonucleotide:RNA duplex is thermally more stable than an RNA:RNA duplex [76]. 2’-O-methylribose (2’-OMe) and 2’-O-aminoethylribose (2’-AE) modified oligonucleotides have been synthesized and successfully used to induce mutations in cells via the formation of DNA:DNA:RNA triple helices [77,78]. Interestingly, extensive modification of TFOs with 2’-OMe and 2’-AE residues leads to increased binding affinity in vitro, but reduced activity in vivo [79]. Bridged nucleic acids (BNAs) have been used to overcome the requirement for long purine runs for efficient triplex formation [80]. For example, a 2’-O,4’-C-methyleneribonucleic acid containing 2-pyridone-modified bases can recognize target duplexes containing a CG inversion with high affinity, without compromising selectivity, thereby increasing the TFO binding code TFO binding code [81].
The vast majority of 5’ or 3’ end modifications include covalent attachment of DNA damaging agents to direct site-specific DNA damage via triplex formation. Examples of reactive molecules that have been conjugated to TFOs include 2-amino-6-vinylpurine, haloacetyl amide, aryl nitrogen mustard, N4,N4-etheno-5-methyldeoxycytidine, and various psoralen derivatives. All of these induce site-specific cross-linking and/or alkylation in the target duplex DNA molecule [37,38,82–86]. Moreover, end modifications have also been used to prevent cellular exonucleases from degrading the TFO once inside cells. A successful example of such a 3’-end modification includes a 3’-phosphopropyl amine, which prevents TFO degradation following intravenous or intraperitoneal injection into mice [87].
4.2. Delivery of TFOs to nuclear target sites
A variety of delivery reagents are available to facilitate the cellular uptake of oligonucleotides in tissue culture and in vivo studies. Cationic lipids, cationic polymers, and cell penetrating peptides represent a few examples. While these strategies have been widely and successfully applied to deliver oligonucleotides into cells, uptake still remains a limitation and improvements to these techniques are being developed. A number of chemical modifications have been incorporated into TFOs to facilitate their uptake. These include, but are not limited to: polyamine analogs [88] cholesterol derivatives [89] nuclear targeting peptide conjugates [90]; 6-phosphate-bovine serum albumin [91]; and polypropylenimine dendrimers [92].
In eukaryotic cells DNA is packaged into chromatin, such that this packaging of the DNA into nucleosomes may present a limitation to TFO accessibility to their duplex targets in vivo. Chemical modifications to TFOs (discussed above) may allow TFOs to compete with the chromatin structure, thereby enhancing the efficacy of triplex technology in biological systems. Interestingly, a tetrapeptide within the DNA binding domain of human high mobility group B1 (HMGB1) protein has been shown to stabilize triplex formation in the purine motif [93]. HMGB1 has also been demonstrated to specifically bind to triplex structures with high affinity [94]. HMGB1 is a very abundant non-histone chromatin-associated protein, thus it is possible that this protein may play a role in triplex formation and stabilization in chromatin.
A better understanding of the molecular mechanisms of action and the requirements for optimization of cellular stability, nuclear delivery, and DNA binding of these oligonucleotide therapeutics will certainly enhance their clinical potential.
5. H-DNA conformation and its occurrence in genomic DNA
Eukaryotic genomes contain many S1 nuclease sensitive sites with a common feature being runs of polypurine-polypyrimidine sequences. These types of sequences are capable of adopting non-canonical DNA structures. For example, H-DNA, or intramolecular triplex DNA is a structure in which half of the pyrimidine tract swivels its backbone parallel to the purine strand in the underlying duplex, or the purine strand (in *H-DNA) binds to the purine strand of the underlying duplex in an antiparallel orientation, to form a triple helical DNA structure [95]. The complimentary strand remains single stranded [96], and is therefore sensitive to S1 nuclease activity. In an H-DNA structure, the conformation is maintained by T-A*T or C-G*C+ Hoogsteen hydrogen bonding in the major groove of the DNA. Similar to intermolecular triplexes, formation of a stable C-G*C+ hydrogen bond requires protonation of cytosine at N3, which explains the pH dependency of this type of H-DNA structure. In fact, this structure was called H-DNA based on the requirement of H+ to protonate the cytosines in the swiveled third strand. The other form of intramolecular triplex, *H-DNA, which is maintained by T-A*A or C-G*G base triplets, can be formed at neutral pH and requires bivalent cations such as Mg2+ for stability. Hence, a polypurine-polypyrimidine stretch with mirror repeat symmetry can form H-DNA readily. Interestingly, protonated base triads such as C-G* A+ can be incorporated into an intermolecular pyrimidine-purine*purine triplex conformation. For example, this triad can be formed between plasmids containing a d(C)n(G)n duplex sequence and a d(AG)n oligonucleotide as the third strand, or intramolecularly on a G10TTAA(AG)5 sequence in a supercoiled plasmid, forming a triplex structure containing C-G*G and C-G*A+ base triads under acid conditions, but without a requirement for bivalent cations [97]. Therefore, the formation of an *H-DNA structure is more versatile and does not require sequences containing perfect mirror symmetry. For both types of H-DNA (H-DNA and *H-DNA, both will be referred to as H-DNA in the following text), two isoforms are possible depending on whether the 3’ half or the 5’ half of the third strand is involved in the triplex structure formation. However, the isoform in which the 3’ half of polypyrimidine strand is included in the triplex structure, or when the 5’ half of the polypurine strand is part of the triplex is favored [98,99].
5.1. Abundance of genomic H-DNA-forming sequences
Computer generated sequence analysis of genomic DNA from various species resulted in several interesting observations related to DNA structural elements. Regions of 15–30 contiguous purine or pyrimidine tracts are greatly overrepresented in all eukaryotic species examined, ranging from yeast to human [100,101]. In the N. tobacum chloroplast genome, the most abundant regions of contiguous purine or pyrimidine tracts are found in the following order: intergenic regions; 3’-downstream and 5’-upstream (promoter) regions; 5’ and 3’ untranslated regions; introns; and coding regions [101]. However, only very few of these tracts were found in prokaryotic genomes (and were most often located in intergenic regions [101]). Naturally occurring sequences capable of adopting H-DNA structures are very abundant in mammalian cells (~1 in every 50,000 bp in humans [102]). Genome-wide scanning of the human genome for long (>100 bp) polypurine-polypyrimidine sequences suggested that most polypurine.polypyrimidine sequences were located in introns, followed by promoters and 5’ or 3’ untranslated regions, and were enriched in genes involved in cell signaling and cell communication [103]. In addition, the genes containing long polypurine-polypyrimidine sequences tended to undergo alternative splicing, were more susceptible to chromosomal translocations, and were expressed at lower levels compared to other genes [103]. Interestingly, there seems to be a different distribution of potential H-DNA-forming sequences in S. cerevisiae [104]; the occurrence of polypurine-polypyrimidine sequences with the length of mirror repeat >10 bp was highest in the promoter region, followed by exons, while very few were found in intron and intergenic regions, which is very different from the other reports of such sequences in eukaryotic genomes. It is not yet clear if this discrepancy reflects differences between the yeast and human genomes, or if this difference is due to a bias in the searching programs used.
5.2. Detecting H-DNA in vitro and in vivo
There are a variety of methods to test the presence of an H-DNA conformation in vitro or on constructs containing the sequences of interest in E. coli using chemicals such as diethyl pyrocarbonate, chloroacetaldehyde, osmium tetroxide (OsO4), dimethyl sulfate, or psoralen to modify nucleotides specifically in single-stranded DNA or double-stranded DNA [105,106]. Raghavan et al. (2006) recently summarized chemical probing procedures to detect non-B DNA structures in mammalian genomic DNA using either bisulfite or KMnO4/OsO4 [107]. Triplex-specific monoclonal antibodies have been developed and used to probe the conformation of H-DNA in vivo [108,109]. The triplex-specific antibodies showed significant binding to the nuclei and this binding could be inhibited by competing triplex DNA [110]. Another approach to identify H-DNA conformations in vivo takes advantage of the single-stranded region exposed in an H-DNA structure, which is capable of hybridization with complementary fluorescence modified single-stranded DNA by “in situ non-denaturing" hybridization. Results from both hybridization probing signals and H-DNA antibody immunostaining were found to be consistent [111]. These studies provide evidence for the existence of H-DNA structures in vivo. However, these experiments were not performed under physiological conditions. It is very difficult to detect H-DNA in vivo in living cells under native conditions due to the transient and dynamic nature of the H-DNA conformation, the complexity of genomic DNA, the presence of DNA binding proteins, and the lack of specific probes that are highly selective to H-DNA.
6. H-DNA induces genetic instability
Bacolla et al. (2006) found that genes carrying long polypurine-polypyrimidine sequences are more susceptible to chromosomal translocations [103]. Certain “fragile site” or “hotspot” regions of the genome are mapped in or near sequences that have the potential to adopt non-B DNA conformations. For example, a segment in the promoter of the human c-MYC gene capable of adopting H-DNA [112], overlaps with the one of major breakage hotspots found in c-MYC-induced lymphomas and leukemias [113–117]. An H-DNA structure is also found in the BCL-2 gene major breakpoint region in follicular lymphomas, and disruption of the H-DNA conformation markedly reduces the frequency of translocation events, supporting a role for the H-DNA structure in the formation of the oncogenic translocations [118]. Autosomal dominant polycystic kidney disease (ADPKD) is one of the most frequent inherited single gene disorders, and mutations in the PKD1 gene account for up to 85% of ADPKD cases [119]. The 21st intron of the human PKD1 gene was found to contribute to the high mutation rate of the gene in both the germ line and somatic cells [120]. This unstable region contains a 2.5 kb polypyrimidine tract composed of 97% C+T sequences, and 23 H-DNA-forming mirror repeats [121] which forms H-DNA when tested in vitro [122]. In addition to the truncation and missense mutations, further analysis revealed complex germ line rearrangements in a 5.8 kb fragment harboring the polypyrimidine tracts of introns 21 and 22 in patients [123]. We have demonstrated that H-DNA structures are intrinsically mutagenic in mammalian cells [124]. Either the endogenous H-DNA-forming sequence from the human c-MYC promoter where a breakpoint hotspot is found in diseases such as Burkitt’s lymphoma [125–127], or model H-DNA-forming sequences, induced mutation frequencies ~20-fold above background levels in a supF reporter gene in COS-7 cells. Approximately 80% of the mutations were large-scale deletions and/or rearrangements. The structures of the junctions at deletion breakpoints suggested that the H-DNA-forming plasmids had undergone DNA double-strand breaks (DSBs) that were subsequently processed via a non-homologous end-joining pathway. In Figure 5 we outline potential H-DNA-induced deletion or translocation pathways. Further, DSBs were found near the H-DNA locus on the plasmids recovered from mammalian cells [124]. Similarly, Bacolla et al (2004) found that the 2.5-kbp polypyrimidine sequence in the human PKD1 gene (as discussed above), induced DSBs at the regions that form H-DNA, and resulted in large-scale deletions in E. coli [128]. The mechanisms by which H-DNA induces DSBs and genetic instability are still under investigation, though there is evidence that the DNA replication, transcription, and repair proteins may be involved.
Figure 5. H-DNA structure-induced genetic instability in mammalian cells.
DSBs (chromosomal breakage) surrounding the H-DNA are generated by as yet undefined enzymes. Non-homologous end-joining repair at DSBs in mammalian cells can result in large-scale deletions, translocations and rearrangements. Adapted from [162].
6.1. Influences of H-DNA on DNA replication and repair
The existence of H-DNA structures may effectively impede the activity and accuracy of DNA polymerases in replication. In vitro, H-DNA structures impose a very strong barrier for Taq DNA polymerase [129]. In a primer extension assay, purified DNA polymerase β showed a 4-fold decrease of polymerase accuracy at a template AG11 versus its complementary CT11 sequence, and was suggested to be initiated by polymerase dissociation and re-association events due to the H-DNA structure formed at the AG11 repeats during synthesis [130]. Purified calf thymus polymerase á was arrested at microsatellite sequences capable of forming H-DNA structures, and also exhibited higher error frequencies [131]. Long GAA repeats (> 40 repeats), which form H-DNA, stalled replication fork progression on plasmids in S. cerevisiae [132]. An H-DNA-forming sequence GA(20) in the simian virus 40 (SV40) genome slowed growth in monkey CV1 cells, resulting in lower titers and smaller plaques [133]. GA repeats reduced the rate of nucleotide incorporation into the SV40 genome, resulting in stalled replication forks [133,134]. E. coli strains transformed with plasmids containing the 2.5-kbp polypyrimidine sequence from the human PKD1 gene grew slower than those had shorter inserts, and this effect correlated with the level of negative supercoiling of the plasmid DNA in vivo, suggesting that the H-DNA structures formed were responsible for the cell growth retardation [135]. Although direct evidence is still not available, it is possible that H-DNA structures formed in mammalian genomes could modulate DNA replication and result in DNA breakage and genetic instability.
Interestingly, our unpublished data suggest that H-DNA is able to induce DNA breakage and mutagenesis in HeLa cell extracts in the absence of replication (Wang and Vasquez, unpublished results), indicating that other factors, such as the DNA transcription, or repair machinery might be involved in the mutagenesis. In E. coli, the H-DNA-forming sequence from the human PKD1 gene activated an SOS response and induced significant NER-dependent cell lysis [135]. The same H-DNA-forming sequence also induced deletions in the adjacent GFP reporter gene in E. coli, but the mutation frequency was greatly reduced in mismatch repair (MMR) MutS and MutL deficient strains, implicating the MMR pathway in H-DNA structure formation, recognition, and/or processing in bacteria [128].
H-DNA structures have been implicated in stimulating homologous recombination. The single-stranded region in the H-DNA structure could potentially invade a duplex containing a homologous sequence, thereby forming a D-loop structure, which could induce homologous recombination. Alternatively, the single-stranded regions from two H-DNA structures could form Watson-Crick base pairs, and subsequently convert into a Holliday junction, an intermediate that leads to homologous recombination (Figure 6) [105,136]. Furthermore, H-DNA-induced DSBs in mammalian cells are also sources of recombination. In fact, H-DNA-forming sequences are often hotspots of recombination, for example, H-DNA sequences were found at sites of unequal sister chromatid exchange in the Cγ2a and Cγ2b heavy chain genes in the MPC-11 mouse myeloma cell line [137]. H-DNA-forming sequences cloned in plasmids near a neomycin resistance gene stimulated homologous recombination between plasmids in the EJ human bladder cancer cell line, and the level of recombination stimulation was proportional to the ability of the sequences to adopt an H-DNA conformation [138].
Figure 6. A model for H-DNA induced recombination.
(A) The single strand region of the H-DNA structure may invade and pair with a complementary strand of an homologous duplex. (B) The third strand in the H-DNA structure could form Watson-Crick base pairs with the released single strand from the homologous duplex to form a double four-way Holliday junction. (C) and (D) The junction can be rotated and resolved to non-crossover and crossover products. Adapted from [136].
7. H-DNA is implicated in transcription regulation
Sequences that are capable of forming H-DNA are found in promoter regions of genes more frequently than expected by random distribution of bases in eukaryotic genomes, suggesting that they may be involved in the regulation of gene expression [139]. There are many published reports that H-DNA can either up-regulate or down-regulate gene expression, depending on a number of factors, including the location of H-DNA in a gene, and the adjacent sequences and elements. In bacteria, when an H-DNA-forming sequence was inserted in a β-lactamase promoter, lacZ gene expression was increased significantly [140]; when an H-DNA structure was located at the coding region or between the promoter and coding sequence, a strong down-regulation of gene transcription was observed [141–143].
While a considerable body of evidence exists to support the notion that H-DNA within or near genes can affect gene expression, the mechanisms that are involved are complex and largely undefined. Brahmachari et al. (1997) reported that insertion of H-DNA-forming sequences within a lacZ reporter gene did not significantly inhibit gene expression in mammalian COS cells [142]. However, the presence of similar sequences upstream of a lacZ reporter gene led to a several-fold reduction of gene expression in mammalian cells, which was in contrast to the results from similar studies in E. coli [130]. Several studies performed either in vitro or in vivo in eukaryotic systems resulted in different conclusions, demonstrating either up-regulation [144,145], down-regulation [142,146–149] or no effect of H-DNA on gene transcription [150,151]. One obvious explanation for these differences is that there may be very different mechanisms involved under each particular condition and thus, should be studied case by case.
The transcriptional activity of minimal mouse albumin promoters in HeLa cells containing various mutant H-DNA-forming sequences derived from the human c-MYC promoter can be predicted by the ability of the particular sequences to form H-DNA and not by repeat number, position, or the number of mutant base pairs [144]. The H-DNA-forming sequence from the c-MYC promoter serves as a cis-acting element interacting with ribonucleoprotein and other transcription factors in mammalian cells and cell nuclear extracts [152]. When an H-DNA-forming sequence is located in close proximity to a TATA box, it results in an unwound region adjacent to the TATA box, destabilizing the T-A hydrogen bonds. This structural alteration in the TATA region may inhibit the initial recognition process by transcription factors at the transcription initiation site which may require double-stranded DNA [98]. H-DNA is a relatively rigid structure; once formed a 130° bend is induced in the DNA strand, which may bring distant cis-elements in close proximity to promoters to regulate transcription initiation [149,153]. Alternatively, an H-DNA structure formed between two adjacent cis-elements may disrupt the cooperation between proximal elements [149]. For example, an H-DNA-forming sequence located ~1.8 kbp upstream of the transcription start of the rat hsp70.1 stress gene, increased transcription of a reporter gene by abolishing the effect of an adjacent putative silencing element in rat hepatoma cells [154].
Although transcribed regions are not the most H-DNA-enriched regions in mammalian genomes, the ability of H-DNA-forming sequences to adopt H-DNA conformations may be enhanced in these regions. H-DNA formation can be induced by the negative supercoiling generated by the transcription machinery, and in some cases, further stabilized by the binding of nascent purine-rich RNA to the single-stranded DNA in H-DNA structure. Once formed, these structures may inhibit or block the transcription machinery [147]. We have recently reported that an S1-sensitive element from the c-MYC promoter, which has the potential to form either a H-DNA or a quadruplex structure, blocks T7 RNA polymerase in an in vitro assay, resulting in partial transcription product arrest. Further, various nucleotide substitutions were introduced to specifically destabilize either the triplex structure or the quadruplex structure, and the result suggested that the triplex structure, but not quadruplex, was responsible for the transcription arrest [155].
8. Modulating H-DNA structure as a potential gene targeting strategy
Anticancer agents that target DNA are among the most effective agents in cancer therapeutics, but are often extremely toxic due to lack of specificity for the tumor cells. Although the mechanisms by which H-DNA influences DNA metabolism are not well understood, it is clear that it plays important roles in a variety of DNA processes, and the unique structure of H-DNA provides a potential target for a the development of a new class of more selective DNA-based therapeutics.
8.1. Targeting the H-DNA structure
A potential promising therapeutic target is the single-stranded DNA region exposed in an H-DNA structure. When a plasmid containing H-DNA and a polypurine oligonucleotide complementary to the single-stranded polypyrimidine region of the H-DNA structure were transfected into HeLa cells, up to 90% of the oligonucleotides could be recovered after 24 hours, while the same oligonucleotides were predominantly eliminated when co-transfected with a control plasmid unable to form H-DNA [156]. Ohno et al. (2002) used complementary fluorescence-modified single-stranded DNA to detect H-DNA conformation in vivo [111]. Although the biological function of this oligonucleotide binding was not explored in these particular studies (e.g. whether or not complementary oligonucleotide binding can stabilize the H-DNA structure, the effect on gene expression, DNA replication and genetic instability), the high specificity and affinity of binding and the stability of the oligonucleotide once bound to H-DNA makes this a reasonable approach to develop potential gene targeting therapies. Expanded GAA.TTC trinucleotide repeats in intron 1 of the frataxin gene are involved in Friedreich’s ataxia (FRDA) disease etiology, and it is known that these repeats can reduce gene expression by forming H-DNA structures [157]. The use of an oligonucleotide that binds the single-stranded GAA sequence during transcription resulted in inhibition of H-DNA formation in the frataxin gene, and to a subsequent frataxin gene specific increase in full-length transcript, providing a therapeutic strategy to prevent or treat FRDA [158]. In addition to the effect of destabilizing H-DNA formation to regulate gene expression, H-DNA complementary oligonucleotides may also be covalently attached to DNA damaging agents to direct DNA damage to the corresponding H-DNA structure, similar to the sequence-specific delivery of DNA-damaging agents by TFOs (see Section 3.4 above).
8.2. Stabilizing H-DNA
Triplex-specific monoclonal antibodies have been developed to detect H-DNA structure in vivo, and to regulate H-DNA-related effects on DNA metabolic processes, via either structure stabilization, destabilization, and/or interference with the related trans-acting proteins. Introduction of triplex-specific monoclonal antibodies into permeabilized mouse myeloma cells inhibited DNA replication and total transcription in the nuclei by ~20%, resulting in a significant decrease in cell growth without an increase in cell death [110].
In addition to interacting with the H-DNA structure directly, molecules that interfere with H-DNA-binding proteins may also be useful in modulating H-DNA-related metabolism. Cellular nucleic acid binding protein (CNBP) binds to the polypurine single-stranded region of H-DNA and increases H-DNA-induced transcription in vivo. Purine-rich oligonucleotides that effectively competed with the binding of CNBP to the H-DNA-forming sequence in the human c-MYC promoter eliminated ~90% of H-DNA-mediated transcription in an in vitro transcription reaction [159].
Positively charged Lys- and Arg-rich oligopeptides, and spermine have been shown to stabilize the H-DNA conformation by interacting with the unpaired single stranded region and by neutralization of electrostatic repulsion of negatively charged phosphates in triplex molecule [160]. Small chemical compounds such as these have the advantage of efficient cellular uptake. A promising class of H-DNA-modulating agents includes DNA minor groove binders or intercalators which can bind to triplex structures. While minor groove binders typically destabilize triplexes, intercalators often stabilize this conformation by providing an aromatic surface area for stacking with triplex bases in an intercalation complex [161]. Most of these compounds were evaluated on intermolecular triplex structures, however, such molecules have the potential to modulate H-DNA as well, and it will be of interest to evaluate them on H-DNA structures. The antitumor agent, benzo[e]pyridoindole (BePI), has a higher affinity for triplex than for duplex DNA, and stabilizes H-DNA structures formed on plasmids inserted between the promoter and the coding sequence of the β-lactamase gene. The presence of BePI enhanced the H-DNA-induced transcription inhibition through the β-lactamase gene in E. coli cells, demonstrating the potential of small molecules as H-DNA modulators in cells [143].
9. Concluding Remarks
The formation of triplex DNA, either in an intramolecular fashion from the same DNA molecule, or in an intermolecular fashion by delivery of a TFO into cells, has very attractive application potential. Naturally occurring intramolecular triplexes play important roles in regulating DNA metabolism and gene function, and are inherently mutagenic and recombinogenic. Regulating H-DNA conformation or specifically interfering with H-DNA-related interactions using small molecules or oligonucleotides presents a promising gene targeting strategy. In the past decades, numerous published findings support the utility of TFOs as sequence-specific gene targeting agents for modulating gene function and/or modifying DNA sequence, and great efforts have been made to increase the efficiency and specificity of this gene targeting approach. However, the mechanisms by which (inter- and intramolecular) triplex structures regulate DNA metabolism, such as replication, transcription, recombination, and mutation, are still under investigation. An important topic of future efforts toward this goal includes identifying the proteins that recognize and bind to triplex DNA, stabilize or unwind triplex DNA, and that cleave or repair the triplex structure. Indeed, rather remarkable first steps have already been made toward characterizing the structure and function of triplex DNA. These efforts have been valuable in designing more specific and efficient triplex-related gene targeting agents.
ACKNOWLEDGEMENTS
We thank Dr. Rick A. Finch for critical reading of the manuscript. Support was provided by an NIH/NCI grant to K.M.V. (CA93729).
Abbreviations
- 2’-OMe
2’-O-methylribose
- 2’-AE
2’-O-aminoethylribose
- ADPKD
autosomal dominant polycystic kidney disease
- BePI
benzo[e]pyridoindole
- BNAs
bridged nucleic acids
- CNBP
cellular nucleic acid binding protein
- DSBs
double-strand breaks
- FRDA
Friedreich’s ataxia
- HMGB1
high mobility group B1
- LNAs
locked nucleic acids
- MMR
mismatch repair
- NER
nucleotide excision repair
- OsO4
osmium tetroxide
- PNAs
peptide nucleic acids
- TFOs
triplex-forming oligonucleotides
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Avery OT, Macleod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type iii. J Exp Med. 1944;79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felsenfeld G, Rich A. Studies on the formation of two- and three-stranded polyribonucleotides. Biochim Biophys Acta. 1957;26:457–468. doi: 10.1016/0006-3002(57)90091-4. [DOI] [PubMed] [Google Scholar]
- 3.Dervan PB. Design of sequence-specific DNA-binding molecules. Science. 1986;232:464–471. doi: 10.1126/science.2421408. [DOI] [PubMed] [Google Scholar]
- 4.Hoogsteen K. The structure of crystals containing a hydrogen-bonded complex of 1-methylthymine and 9-methyladenine. Acta Cryst. 1959;12:822–823. [Google Scholar]
- 5.Cooney M, Czernuszewicz G, Postel EH, Flint SJ, Hogan ME. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988;241:456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- 6.Beal PA, Dervan PB. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991;251:1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- 7.Frank-Kamenetskii MD, Mirkin SM. Triplex DNA structures. Annu Rev Biochem. 1995;64:65–95. doi: 10.1146/annurev.bi.64.070195.000433. [DOI] [PubMed] [Google Scholar]
- 8.Vasquez KM, Glazer PM. Triplex-forming oligonucleotides: principles and applications. Q Rev Biophys. 2002;35:89–107. doi: 10.1017/s0033583502003773. [DOI] [PubMed] [Google Scholar]
- 9.Bissler JJ. Triplex DNA and human disease. Front Biosci. 2007;12:4536–4546. doi: 10.2741/2408. [DOI] [PubMed] [Google Scholar]
- 10.Letai AG, Palladino MA, Fromm E, Rizzo V, Fresco JR. Specificity in formation of triple-stranded nucleic acid helical complexes: studies with agarose-linked polyribonucleotide affinity columns. Biochemistry. 1988;27:9108–9112. doi: 10.1021/bi00426a007. [DOI] [PubMed] [Google Scholar]
- 11.Durland RH, Kessler DJ, Gunnell S, Duvic M, Pettitt BM, Hogan ME. Binding of triple helix forming oligonucleotides to sites in gene promoters. Biochemistry. 1991;30:9246–9255. doi: 10.1021/bi00102a017. [DOI] [PubMed] [Google Scholar]
- 12.Lee JS, Johnson DA, Morgan AR. Complexes formed by (pyrimidine)n. (purine)n DNAs on lowering the pH are three-stranded. Nucleic Acids Res. 1979;6:3073–3091. doi: 10.1093/nar/6.9.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin SB, Kao CF, Lee SC, Kan LS. DNA triplex formed by d-A-(G-A)7-G and d-mC-(T-mC)7-T in aqueous solution at neutral pH. Anticancer Drug Des. 1994;9:1–8. [PubMed] [Google Scholar]
- 14.Singleton SF, Dervan PB. Influence of pH on the equilibrium association constants for oligodeoxyribonucleotide-directed triple helix formation at single DNA sites. Biochemistry. 1992;31:10995–11003. doi: 10.1021/bi00160a008. [DOI] [PubMed] [Google Scholar]
- 15.Cheng AJ, Van Dyke MW. Monovalent cation effects on intermolecular purine-purine-pyrimidine triple-helix formation. Nucleic Acids Res. 1993;21:5630–5635. doi: 10.1093/nar/21.24.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orson FM, Klysik J, Bergstrom DE, Ward B, Glass GA, Hua P, Kinsey BM. Triple helix formation: binding avidity of acridine-conjugated AG motif third strands containing natural, modified and surrogate bases opposed to pyrimidine interruptions in a polypurine target. Nucleic Acids Res. 1999;27:810–816. doi: 10.1093/nar/27.3.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng YK, Pettitt BM. Stabilities of double- and triple-strand helical nucleic acids. Prog Biophys Mol Biol. 1992;58:225–257. doi: 10.1016/0079-6107(92)90007-s. [DOI] [PubMed] [Google Scholar]
- 18.Cheng AJ, Van Dyke MW. Oligodeoxyribonucleotide length and sequence effects on intermolecular purine-purine-pyrimidine triple-helix formation. Nucleic Acids Res. 1994;22:4742–4747. doi: 10.1093/nar/22.22.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aich P, Ritchie S, Bonham K, Lee JS. Thermodynamic and kinetic studies of the formation of triple helices between purine-rich deoxyribo-oligonucleotides and the promoter region of the human c-src proto-oncogene. Nucleic Acids Res. 1998;26:4173–4177. doi: 10.1093/nar/26.18.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gowers DM, Fox KR. DNA triple helix formation at oligopurine sites containing multiple contiguous pyrimidines. Nucleic Acids Res. 1997;25:3787–3794. doi: 10.1093/nar/25.19.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vasquez KM, Wensel TG, Hogan ME, Wilson JH. High-affinity triple helix formation by synthetic oligonucleotides at a site within a selectable mammalian gene. Biochemistry. 1995;34:7243–7251. doi: 10.1021/bi00021a040. [DOI] [PubMed] [Google Scholar]
- 22.Alberti P, Arimondo PB, Mergny JL, Garestier T, Helene C, Sun JS. A directional nucleation-zipping mechanism for triple helix formation. Nucleic Acids Res. 2002;30:5407–5415. doi: 10.1093/nar/gkf675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.James PL, Brown T, Fox KR. Thermodynamic and kinetic stability of intermolecular triple helices containing different proportions of C+*GC and T*AT triplets. Nucleic Acids Res. 2003;31:5598–5606. doi: 10.1093/nar/gkg782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rusling DA, Broughton-Head VJ, Tuck A, Khairallah H, Osborne SD, Brown T, Fox KR. Kinetic studies on the formation of DNA triplexes containing the nucleoside analogue 2′-O-(2-aminoethyl)-5-(3-amino-1-propynyl)uridine. Org Biomol Chem. 2008;6:122–129. doi: 10.1039/b713088k. [DOI] [PubMed] [Google Scholar]
- 25.Paes HM, Fox KR. Kinetic studies on the formation of intermolecular triple helices. Nucleic Acids Res. 1997;25:3269–3274. doi: 10.1093/nar/25.16.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaddis SS, Wu Q, Thames HD, DiGiovanni J, Walborg EF, MacLeod MC, Vasquez KM. A web-based search engine for triplex-forming oligonucleotide target sequences. Oligonucleotides. 2006;16:196–201. doi: 10.1089/oli.2006.16.196. [DOI] [PubMed] [Google Scholar]
- 27.Wu Q, Gaddis SS, MacLeod MC, Walborg EF, Thames HD, DiGiovanni J, Vasquez KM. High-affinity triplex-forming oligonucleotide target sequences in mammalian genomes. Mol Carcinog. 2007;46:15–23. doi: 10.1002/mc.20261. [DOI] [PubMed] [Google Scholar]
- 28.Olivas WM, Maher LJ., 3rd Overcoming potassium-mediated triplex inhibition. Nucleic Acids Res. 1995;23:1936–1941. doi: 10.1093/nar/23.11.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borgatti M, Lampronti I, Romanelli A, Pedone C, Saviano M, Bianchi N, Mischiati C, Gambari R. Transcription factor decoy molecules based on a peptide nucleic acid (PNA)-DNA chimera mimicking Sp1 binding sites. J Biol Chem. 2003;278:7500–7509. doi: 10.1074/jbc.M206780200. [DOI] [PubMed] [Google Scholar]
- 30.Wu Q, Christensen LA, Legerski RJ, Vasquez KM. Mismatch repair participates in error-free processing of DNA interstrand crosslinks in human cells. EMBO Rep. 2005;6:551–557. doi: 10.1038/sj.embor.7400418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vasquez KM, Christensen J, Li L, Finch RA, Glazer PM. Human XPA and RPA DNA repair proteins participate in specific recognition of triplex-induced helical distortions. Proc Natl Acad Sci U S A. 2002;99:5848–5853. doi: 10.1073/pnas.082193799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang G, Seidman MM, Glazer PM. Mutagenesis in mammalian cells induced by triple helix formation and transcription-coupled repair. Science. 1996;271:802–805. doi: 10.1126/science.271.5250.802. [DOI] [PubMed] [Google Scholar]
- 33.Christensen LA, Finch RA, Booker AJ, Vasquez KM. Targeting oncogenes to improve breast cancer chemotherapy. Cancer Res. 2006;66:4089–4094. doi: 10.1158/0008-5472.CAN-05-4288. [DOI] [PubMed] [Google Scholar]
- 34.Kan LS, Callahan DE, Trapane TL, Miller PS, Ts'o PO, Huang DH. Proton NMR and optical spectroscopic studies on the DNA triplex formed by d-A-(G-A)7-G and d-C-(T-C)7-T. J Biomol Struct Dyn. 1991;8:911–933. doi: 10.1080/07391102.1991.10507857. [DOI] [PubMed] [Google Scholar]
- 35.Johnson KH, Durland RH, Hogan ME. The vacuum UV CD spectra of G.G.C triplexes. Nucleic Acids Res. 1992;20:3859–3864. doi: 10.1093/nar/20.15.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moser HE, Dervan PB. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 37.Vasquez KM, Wensel TG, Hogan ME, Wilson JH. High-efficiency triple-helix-mediated photo-cross-linking at a targeted site within a selectable mammalian gene. Biochemistry. 1996;35:10712–10719. doi: 10.1021/bi960881f. [DOI] [PubMed] [Google Scholar]
- 38.Perkins BD, Wilson JH, Wensel TG, Vasquez KM. Triplex targets in the human rhodopsin gene. Biochemistry. 1998;37:11315–11322. doi: 10.1021/bi980525s. [DOI] [PubMed] [Google Scholar]
- 39.Vasquez KM, Narayanan L, Glazer PM. Specific mutations induced by triplex-forming oligonucleotides in mice. Science. 2000;290:530–533. doi: 10.1126/science.290.5491.530. [DOI] [PubMed] [Google Scholar]
- 40.Christensen LA, Conti CJ, Fischer SM, Vasquez KM. Mutation frequencies in murine keratinocytes as a function of carcinogenic status. Mol Carcinog. 2004;40:122–133. doi: 10.1002/mc.20026. [DOI] [PubMed] [Google Scholar]
- 41.Faruqi AF, Datta HJ, Carroll D, Seidman MM, Glazer PM. Triple-helix formation induces recombination in mammalian cells via a nucleotide excision repair-dependent pathway. Mol Cell Biol. 2000;20:990–1000. doi: 10.1128/mcb.20.3.990-1000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasquez KM, Marburger K, Intody Z, Wilson JH. Manipulating the mammalian genome by homologous recombination. Proc Natl Acad Sci U S A. 2001;98:8403–8410. doi: 10.1073/pnas.111009698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Datta HJ, Chan PP, Vasquez KM, Gupta RC, Glazer PM. Triplex-induced recombination in human cell-free extracts. Dependence on XPA and HsRad51. J Biol Chem. 2001;276:18018–18023. doi: 10.1074/jbc.M011646200. [DOI] [PubMed] [Google Scholar]
- 44.Barre FX, Giovannangeli C, Helene C, Harel-Bellan A. Covalent crosslinks introduced via a triple helix-forming oligonucleotide coupled to psoralen are inefficiently repaired. Nucleic Acids Res. 1999;27:743–749. doi: 10.1093/nar/27.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guieysse AL, Praseuth D, Giovannangeli C, Asseline U, Helene C. Psoralen adducts induced by triplex-forming oligonucleotides are refractory to repair in HeLa cells. J Mol Biol. 2000;296:373–383. doi: 10.1006/jmbi.1999.3466. [DOI] [PubMed] [Google Scholar]
- 46.Rogers FA, Vasquez KM, Egholm M, Glazer PM. Site-directed recombination via bifunctional PNA-DNA conjugates. Proc Natl Acad Sci U S A. 2002;99:16695–16700. doi: 10.1073/pnas.262556899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vasquez KM, Wilson JH. Triplex-directed modification of genes and gene activity. Trends Biochem Sci. 1998;23:4–9. doi: 10.1016/s0968-0004(97)01158-4. [DOI] [PubMed] [Google Scholar]
- 48.Havre PA, Gunther EJ, Gasparro FP, Glazer PM. Targeted mutagenesis of DNA using triple helix-forming oligonucleotides linked to psoralen. Proc Natl Acad Sci U S A. 1993;90:7879–7883. doi: 10.1073/pnas.90.16.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Majumdar A, Puri N, McCollum N, Richards S, Cuenoud B, Miller P, Seidman MM. Gene targeting by triple helix-forming oligonucleotides. Ann N Y Acad Sci. 2003;1002:141–153. doi: 10.1196/annals.1281.013. [DOI] [PubMed] [Google Scholar]
- 50.Rao TS, Durland RH, Seth DM, Myrick MA, Bodepudi V, Revankar GR. Incorporation of 2′-deoxy-6-thioguanosine into G-rich oligodeoxyribonucleotides inhibits G-tetrad formation and facilitates triplex formation. Biochemistry. 1995;34:765–772. doi: 10.1021/bi00003a009. [DOI] [PubMed] [Google Scholar]
- 51.Milligan JF, Krawczyk SH, Wadwani S, Matteucci MD. An anti-parallel triple helix motif with oligodeoxynucleotides containing 2′-deoxyguanosine and 7-deaza-2′-deoxyxanthosine. Nucleic Acids Res. 1993;21:327–333. doi: 10.1093/nar/21.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krawczyk SH, Milligan JF, Wadwani S, Moulds C, Froehler BC, Matteucci MD. Oligonucleotide-mediated triple helix formation using an N3-protonated deoxycytidine analog exhibiting pH-independent binding within the physiological range. Proc Natl Acad Sci U S A. 1992;89:3761–3764. doi: 10.1073/pnas.89.9.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xodo LE, Manzini G, Quadrifoglio F, van der Marel GA, van Boom JH. Effect of 5-methylcytosine on the stability of triple-stranded DNA--a thermodynamic study. Nucleic Acids Res. 1991;19:5625–5631. doi: 10.1093/nar/19.20.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ono A, Chen CN, Kan LS. DNA triplex formation of oligonucleotide analogues consisting of linker groups and octamer segments that have opposite sugar-phosphate backbone polarities. Biochemistry. 1991;30:9914–9912. doi: 10.1021/bi00105a015. [DOI] [PubMed] [Google Scholar]
- 55.Lacroix L, Mergny JL. Chemical modification of pyrimidine TFOs: effect on i-motif and triple helix formation. Arch Biochem Biophys. 2000;381:153–163. doi: 10.1006/abbi.2000.1934. [DOI] [PubMed] [Google Scholar]
- 56.Lacoste J, Francois JC, Helene C. Triple helix formation with purine-rich phosphorothioate-containing oligonucleotides covalently linked to an acridine derivative. Nucleic Acids Res. 1997;25:1991–1998. doi: 10.1093/nar/25.10.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carbone GM, McGuffie EM, Collier A, Catapano CV. Selective inhibition of transcription of the Ets2 gene in prostate cancer cells by a triplex-forming oligonucleotide. Nucleic Acids Res. 2003;31:833–843. doi: 10.1093/nar/gkg198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Escude C, Giovannangeli C, Sun JS, Lloyd DH, Chen JK, Gryaznov SM, Garestier T, Helene C. Stable triple helices formed by oligonucleotide N3′-->P5′ phosphoramidates inhibit transcription elongation. Proc Natl Acad Sci U S A. 1996;93:4365–4369. doi: 10.1073/pnas.93.9.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giovannangeli C, Diviacco S, Labrousse V, Gryaznov S, Charneau P, Helene C. Accessibility of nuclear DNA to triplex-forming oligonucleotides: the integrated HIV-1 provirus as a target. Proc Natl Acad Sci U S A. 1997;94:79–84. doi: 10.1073/pnas.94.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Basye J, Trent JO, Gao D, Ebbinghaus SW. Triplex formation by morpholino oligodeoxyribonucleotides in the HER-2/neu promoter requires the pyrimidine motif. Nucleic Acids Res. 2001;29:4873–4880. doi: 10.1093/nar/29.23.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vasquez KM, Dagle JM, Weeks DL, Glazer PM. Chromosome targeting at short polypurine sites by cationic triplex-forming oligonucleotides. J Biol Chem. 2001;276:38536–38541. doi: 10.1074/jbc.M101797200. [DOI] [PubMed] [Google Scholar]
- 62.Cherny DI, Malkov VA, Volodin AA, Frank-Kamenetskii MD. Electron microscopy visualization of oligonucleotide binding to duplex DNA via triplex formation. J Mol Biol. 1993;230:379–383. doi: 10.1006/jmbi.1993.1154. [DOI] [PubMed] [Google Scholar]
- 63.Nielsen PE, Egholm M. Strand displacement recognition of mixed adenine-cytosine sequences in double stranded DNA by thymine-guanine PNA (peptide nucleic acid) Bioorg Med Chem. 2001;9:2429–2434. doi: 10.1016/s0968-0896(01)00244-9. [DOI] [PubMed] [Google Scholar]
- 64.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 65.Hanvey JC, Peffer NJ, Bisi JE, Thomson SA, Cadilla R, Josey JA, Ricca DJ, Hassman CF, Bonham MA, Au KG, et al. Antisense and antigene properties of peptide nucleic acids. Science. 1992;258:1481–1485. doi: 10.1126/science.1279811. [DOI] [PubMed] [Google Scholar]
- 66.Hu J, Corey DR. Inhibiting gene expression with peptide nucleic acid (PNA)--peptide conjugates that target chromosomal DNA. Biochemistry. 2007;46:7581–7589. doi: 10.1021/bi700230a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaihatsu K, Huffman KE, Corey DR. Intracellular uptake and inhibition of gene expression by PNAs and PNA-peptide conjugates. Biochemistry. 2004;43:14340–14347. doi: 10.1021/bi048519l. [DOI] [PubMed] [Google Scholar]
- 68.Zhilina ZV, Ziemba AJ, Nielsen PE, Ebbinghaus SW. PNA-nitrogen mustard conjugates are effective suppressors of HER-2/neu and biological tools for recognition of PNA/DNA interactions. Bioconjug Chem. 2006;17:214–222. doi: 10.1021/bc0502964. [DOI] [PubMed] [Google Scholar]
- 69.Sorensen MD, Meldgaard M, Raunkjaer M, Rajwanshi VK, Wengel J. Branched oligonucleotides containing bicyclic nucleotides as branching points and DNA or LNA as triplex forming branch. Bioorg Med Chem Lett. 2000;10:1853–1856. doi: 10.1016/s0960-894x(00)00362-0. [DOI] [PubMed] [Google Scholar]
- 70.Sun BW, Babu BR, Sorensen MD, Zakrzewska K, Wengel J, Sun JS. Sequence and pH effects of LNA-containing triple helix-forming oligonucleotides: physical chemistry, biochemistry, and modeling studies. Biochemistry. 2004;43:4160–4169. doi: 10.1021/bi036064e. [DOI] [PubMed] [Google Scholar]
- 71.Brunet E, Alberti P, Perrouault L, Babu R, Wengel J, Giovannangeli C. Exploring cellular activity of locked nucleic acid-modified triplex-forming oligonucleotides and defining its molecular basis. J Biol Chem. 2005;280:20076–20085. doi: 10.1074/jbc.M500021200. [DOI] [PubMed] [Google Scholar]
- 72.Hojland T, Kumar S, Babu BR, Umemoto T, Albaek N, Sharma PK, Nielsen P, Wengel J. LNA (locked nucleic acid) and analogs as triplex-forming oligonucleotides. Org Biomol Chem. 2007;5:2375–2379. doi: 10.1039/b706101c. [DOI] [PubMed] [Google Scholar]
- 73.Frieden M, Hansen HF, Koch T. Nuclease stability of LNA oligonucleotides and LNA-DNA chimeras. Nucleosides Nucleotides Nucleic Acids. 2003;22:1041–1043. doi: 10.1081/NCN-120022731. [DOI] [PubMed] [Google Scholar]
- 74.Beane RL, Ram R, Gabillet S, Arar K, Monia BP, Corey DR. Inhibiting gene expression with locked nucleic acids (LNAs) that target chromosomal DNA. Biochemistry. 2007;46:7572–7580. doi: 10.1021/bi700227g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koizumi M, Morita K, Daigo M, Tsutsumi S, Abe K, Obika S, Imanishi T. Triplex formation with 2′-O,4′-C-ethylene-bridged nucleic acids (ENA) having C3′-endo conformation at physiological pH. Nucleic Acids Res. 2003;31:3267–3273. doi: 10.1093/nar/gkg416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Inoue H, Hayase Y, Imura A, Iwai S, Miura K, Ohtsuka E. Synthesis and hybridization studies on two complementary nona(2′-O-methyl)ribonucleotides. Nucleic Acids Res. 1987;15:6131–6148. doi: 10.1093/nar/15.15.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Majumdar A, Khorlin A, Dyatkina N, Lin FL, Powell J, Liu J, Fei Z, Khripine Y, Watanabe KA, George J, Glazer PM, Seidman MM. Targeted gene knockout mediated by triple helix forming oligonucleotides. Nat Genet. 1998;20:212–214. doi: 10.1038/2530. [DOI] [PubMed] [Google Scholar]
- 78.Puri N, Majumdar A, Cuenoud B, Natt F, Martin P, Boyd A, Miller PS, Seidman MM. Targeted gene knockout by 2′-O-aminoethyl modified triplex forming oligonucleotides. J Biol Chem. 2001;276:28991–28998. doi: 10.1074/jbc.M103409200. [DOI] [PubMed] [Google Scholar]
- 79.Alam MR, Majumdar A, Thazhathveetil AK, Liu ST, Liu JL, Puri N, Cuenoud B, Sasaki S, Miller PS, Seidman MM. Extensive sugar modification improves triple helix forming oligonucleotide activity in vitro but reduces activity in vivo. Biochemistry. 2007;46:10222–10233. doi: 10.1021/bi7003153. [DOI] [PubMed] [Google Scholar]
- 80.Imanishi T, Obika S. BNAs: novel nucleic acid analogs with a bridged sugar moiety. Chem Commun (Camb) 2002:1653–1659. doi: 10.1039/b201557a. [DOI] [PubMed] [Google Scholar]
- 81.Obika S, Hari Y, Sekiguchi M, Imanishi T. Stable oligonucleotide-directed triplex formation at target sites with CG interruptions: strong sequence-specific recognition by 2′,4′-bridged nucleic-acid-containing 2-pyridones under physiological conditions. Chemistry. 2002;8:4796–4802. doi: 10.1002/1521-3765(20021018)8:20<4796::AID-CHEM4796>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 82.Nagatsugi F, Kawasaki T, Tokuda N, Maeda M, Sasaki S. Site-directed alkylation to cytidine within duplex by the oligonucleotides containing functional nucleobases. Nucleosides Nucleotides Nucleic Acids. 2001;20:915–919. doi: 10.1081/NCN-100002458. [DOI] [PubMed] [Google Scholar]
- 83.Povsic T. Sequence-Specific Alkylation of Double-helical DNA by Oligonucleotide-directed Triple Helix formation. J Am Chem Soc. 1990;112:9428–9430. [Google Scholar]
- 84.Reed MW, Lukhtanov EA, Gorn V, Kutyavin I, Gall A, Wald A, Meyer RB. Synthesis and reactivity of aryl nitrogen mustard-oligodeoxyribonucleotide conjugates. Bioconjug Chem. 1998;9:64–71. doi: 10.1021/bc970134a. [DOI] [PubMed] [Google Scholar]
- 85.Shaw J-P, Milligan JF, Krawczyk SH, Matteucci M. Specific, High-Efficiency, Triplex-Mediated Cross-Linking to duplex DNA. J Am Chem Soc. 1991;113:7765–7766. [Google Scholar]
- 86.Takasugi M, Guendouz A, Chassignol M, Decout JL, Lhomme J, Thuong NT, Helene C. Sequence-specific photo-induced cross-linking of the two strands of double-helical DNA by a psoralen covalently linked to a triple helix-forming oligonucleotide. Proc Natl Acad Sci U S A. 1991;88:5602–5606. doi: 10.1073/pnas.88.13.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zendegui JG, Vasquez KM, Tinsley JH, Kessler DJ, Hogan ME. In vivo stability and kinetics of absorption and disposition of 3′ phosphopropyl amine oligonucleotides. Nucleic Acids Res. 1992;20:307–314. doi: 10.1093/nar/20.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thomas RM, Thomas T, Wada M, Sigal LH, Shirahata A, Thomas TJ. Facilitation of the cellular uptake of a triplex-forming oligonucleotide by novel polyamine analogues: structure-activity relationships. Biochemistry. 1999;38:13328–13337. doi: 10.1021/bi991004n. [DOI] [PubMed] [Google Scholar]
- 89.Cheng K, Ye Z, Guntaka RV, Mahato RI. Enhanced hepatic uptake and bioactivity of type alpha1(I) collagen gene promoter-specific triplex-forming oligonucleotides after conjugation with cholesterol. J Pharmacol Exp Ther. 2006;317:797–805. doi: 10.1124/jpet.105.100347. [DOI] [PubMed] [Google Scholar]
- 90.Rogers FA, Manoharan M, Rabinovitch P, Ward DC, Glazer PM. Peptide conjugates for chromosomal gene targeting by triplex-forming oligonucleotides. Nucleic Acids Res. 2004;32:6595–6604. doi: 10.1093/nar/gkh998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ye Z, Cheng K, Guntaka RV, Mahato RI. Receptor-mediated hepatic uptake of M6P-BSA-conjugated triplex-forming oligonucleotides in rats. Bioconjug Chem. 2006;17:823–830. doi: 10.1021/bc060006z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Santhakumaran LM, Thomas T, Thomas TJ. Enhanced cellular uptake of a triplex-forming oligonucleotide by nanoparticle formation in the presence of polypropylenimine dendrimers. Nucleic Acids Res. 2004;32:2102–2112. doi: 10.1093/nar/gkh526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jain A, Akanchha S, Rajeswari MR. Stabilization of purine motif DNA triplex by a tetrapeptide from the binding domain of HMGBI protein. Biochimie. 2005;87:781–790. doi: 10.1016/j.biochi.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 94.Reddy MC, Christensen J, Vasquez KM. Interplay between human high mobility group protein 1 and replication protein A on psoralen-cross-linked DNA. Biochemistry. 2005;44:4188–4195. doi: 10.1021/bi047902n. [DOI] [PubMed] [Google Scholar]
- 95.Lyamichev VI, Mirkin SM, Frank-Kamenetskii MD. Structures of homopurine-homopyrimidine tract in superhelical DNA. J Biomol Struct Dyn. 1986;3:667–669. doi: 10.1080/07391102.1986.10508454. [DOI] [PubMed] [Google Scholar]
- 96.Htun H, Dahlberg JE. Single strands, triple strands, and kinks in H-DNA. Science. 1988;241:1791–1796. doi: 10.1126/science.3175620. [DOI] [PubMed] [Google Scholar]
- 97.Malkov VA, Voloshin ON, Veselkov AG, Rostapshov VM, Jansen I, Soyfer VN, Frank-Kamenetskii MD. Protonated pyrimidine-purine-purine triplex. Nucleic Acids Res. 1993;21:105–111. doi: 10.1093/nar/21.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Potaman VN, Ussery DW, Sinden RR. Formation of a combined H-DNA/open TATA box structure in the promoter sequence of the human Na,K-ATPase alpha2 gene. J Biol Chem. 1996;271:13441–13447. doi: 10.1074/jbc.271.23.13441. [DOI] [PubMed] [Google Scholar]
- 99.Htun H, Dahlberg JE. Topology and formation of triple-stranded H-DNA. Science. 1989;243:1571–1576. doi: 10.1126/science.2648571. [DOI] [PubMed] [Google Scholar]
- 100.Behe MJ. An overabundance of long oligopurine tracts occurs in the genome of simple and complex eukaryotes. Nucleic Acids Res. 1995;23:689–695. doi: 10.1093/nar/23.4.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bucher P, Yagil G. Occurrence of oligopurine.oligopyrimidine tracts in eukaryotic and prokaryotic genes. DNA Seq. 1991;1:157–172. doi: 10.3109/10425179109020767. [DOI] [PubMed] [Google Scholar]
- 102.Schroth GP, Ho PS. Occurrence of potential cruciform and H-DNA forming sequences in genomic DNA. Nucleic Acids Res. 1995;23:1977–1983. doi: 10.1093/nar/23.11.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bacolla A, Collins JR, Gold B, Chuzhanova N, Yi M, Stephens RM, Stefanov S, Olsh A, Jakupciak JP, Dean M, Lempicki RA, Cooper DN, Wells RD. Long homopurine*homopyrimidine sequences are characteristic of genes expressed in brain and the pseudoautosomal region. Nucleic Acids Res. 2006;34:2663–2675. doi: 10.1093/nar/gkl354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zain R, Sun JS. Do natural DNA triple-helical structures occur and function in vivo? Cell Mol Life Sci. 2003;60:862–870. doi: 10.1007/s00018-003-3046-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sinden RR. DNA structure and function. San Diego: Academic Press; 1994. [Google Scholar]
- 106.Mirkin SM, Frank-Kamenetskii MD. H-DNA and related structures. Annu Rev Biophys Biomol Struct. 1994;23:541–576. doi: 10.1146/annurev.bb.23.060194.002545. [DOI] [PubMed] [Google Scholar]
- 107.Raghavan SC, Tsai A, Hsieh CL, Lieber MR. Analysis of non-B DNA structure at chromosomal sites in the mammalian genome. Methods Enzymol. 2006;409:301–316. doi: 10.1016/S0076-6879(05)09017-8. [DOI] [PubMed] [Google Scholar]
- 108.Lee JS, Burkholder GD, Latimer LJ, Haug BL, Braun RP. A monoclonal antibody to triplex DNA binds to eucaryotic chromosomes. Nucleic Acids Res. 1987;15:1047–1061. doi: 10.1093/nar/15.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Agazie YM, Lee JS, Burkholder GD. Characterization of a new monoclonal antibody to triplex DNA and immunofluorescent staining of mammalian chromosomes. J Biol Chem. 1994;269:7019–7023. [PubMed] [Google Scholar]
- 110.Agazie YM, Burkholder GD, Lee JS. Triplex DNA in the nucleus: direct binding of triplex-specific antibodies and their effect on transcription, replication and cell growth. Biochem J. 1996;316(Pt 2):461–466. doi: 10.1042/bj3160461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ohno M, Fukagawa T, Lee JS, Ikemura T. Triplex-forming DNAs in the human interphase nucleus visualized in situ by polypurine/polypyrimidine DNA probes and antitriplex antibodies. Chromosoma. 2002;111:201–213. doi: 10.1007/s00412-002-0198-0. [DOI] [PubMed] [Google Scholar]
- 112.Kinniburgh AJ. A cis-acting transcription element of the c-myc gene can assume an H-DNA conformation. Nucleic Acids Res. 1989;17:7771–7778. doi: 10.1093/nar/17.19.7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Joos S, Haluska FG, Falk MH, Henglein B, Hameister H, Croce CM, Bornkamm GW. Mapping chromosomal breakpoints of Burkitt's t(8;14) translocations far upstream of c-myc. Cancer Res. 1992;52:6547–6552. [PubMed] [Google Scholar]
- 114.Haluska FG, Tsujimoto Y, Croce CM. The t(8;14) breakpoint of the EW 36 undifferentiated lymphoma cell line lies 5′ of MYC in a region prone to involvement in endemic Burkitt's lymphomas. Nucleic Acids Res. 1988;16:2077–2085. doi: 10.1093/nar/16.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Saglio G, Grazia Borrello M, Guerrasio A, Sozzi G, Serra A, di Celle PF, Foa R, Ferrarini M, Roncella S, Borgna Pignatti C, et al. Preferential clustering of chromosomal breakpoints in Burkitt's lymphomas and L3 type acute lymphoblastic leukemias with a t(8;14) translocation. Genes Chromosomes Cancer. 1993;8:1–7. doi: 10.1002/gcc.2870080102. [DOI] [PubMed] [Google Scholar]
- 116.Care A, Cianetti L, Giampaolo A, Sposi NM, Zappavigna V, Mavilio F, Alimena G, Amadori S, Mandelli F, Peschle C. Translocation of c-myc into the immunoglobulin heavy-chain locus in human acute B-cell leukemia. A molecular analysis. Embo J. 1986;5:905–911. doi: 10.1002/j.1460-2075.1986.tb04302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wilda M, Busch K, Klose I, Keller T, Woessmann W, Kreuder J, Harbott J, Borkhardt A. Level of MYC overexpression in pediatric Burkitt's lymphoma is strongly dependent on genomic breakpoint location within the MYC locus. Genes Chromosomes Cancer. 2004;41:178–182. doi: 10.1002/gcc.20063. [DOI] [PubMed] [Google Scholar]
- 118.Raghavan SC, Chastain P, Lee JS, Hegde BG, Houston S, Langen R, Hsieh CL, Haworth IS, Lieber MR. Evidence for a triplex DNA conformation at the bcl-2 major breakpoint region of the t(14;18) translocation. J Biol Chem. 2005;280:22749–22760. doi: 10.1074/jbc.M502952200. [DOI] [PubMed] [Google Scholar]
- 119.Peters DJ, Spruit L, Saris JJ, Ravine D, Sandkuijl LA, Fossdal R, Boersma J, van Eijk R, Norby S, Constantinou-Deltas CD, et al. Chromosome 4 localization of a second gene for autosomal dominant polycystic kidney disease. Nat Genet. 1993;5:359–362. doi: 10.1038/ng1293-359. [DOI] [PubMed] [Google Scholar]
- 120.Watnick TJ, Piontek KB, Cordal TM, Weber H, Gandolph MA, Qian F, Lens XM, Neumann HP, Germino GG. An unusual pattern of mutation in the duplicated portion of PKD1 is revealed by use of a novel strategy for mutation detection. Hum Mol Genet. 1997;6:1473–1481. doi: 10.1093/hmg/6.9.1473. [DOI] [PubMed] [Google Scholar]
- 121.Van Raay TJ, Burn TC, Connors TD, Petry LR, Germino GG, Klinger KW, Landes GM. A 2.5 kb polypyrimidine tract in the PKD1 gene contains at least 23 H-DNA-forming sequences. Microb Comp Genomics. 1996;1:317–327. doi: 10.1089/mcg.1996.1.317. [DOI] [PubMed] [Google Scholar]
- 122.Blaszak RT, Potaman V, Sinden RR, Bissler JJ. DNA structural transitions within the PKD1 gene. Nucleic Acids Res. 1999;27:2610–2617. doi: 10.1093/nar/27.13.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ariyurek Y, Lantinga-van Leeuwen I, Spruit L, Ravine D, Breuning MH, Peters DJ. Large deletions in the polycystic kidney disease 1 (PKD1) gene. Hum Mutat. 2004;23:99. doi: 10.1002/humu.9208. [DOI] [PubMed] [Google Scholar]
- 124.Wang G, Vasquez KM. Naturally occurring H-DNA-forming sequences are mutagenic in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:13448–13453. doi: 10.1073/pnas.0405116101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wiener F, Ohno S, Babonits M, Sumegi J, Wirschubsky Z, Klein G, Mushinski JF, Potter M. Hemizygous interstitial deletion of chromosome 15 (band D) in three translocation-negative murine plasmacytomas. Proc Natl Acad Sci U S A. 1984;81:1159–1163. doi: 10.1073/pnas.81.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Akasaka T, Akasaka H, Ueda C, Yonetani N, Maesako Y, Shimizu A, Yamabe H, Fukuhara S, Uchiyama T, Ohno H. Molecular and clinical features of non-Burkitt's, diffuse large-cell lymphoma of B-cell type associated with the c-MYC/immunoglobulin heavy-chain fusion gene. J Clin Oncol. 2000;18:510–518. doi: 10.1200/JCO.2000.18.3.510. [DOI] [PubMed] [Google Scholar]
- 127.Kovalchuk AL, Muller JR, Janz S. Deletional remodeling of c-myc-deregulating chromosomal translocations. Oncogene. 1997;15:2369–2377. doi: 10.1038/sj.onc.1201409. [DOI] [PubMed] [Google Scholar]
- 128.Bacolla A, Jaworski A, Larson JE, Jakupciak JP, Chuzhanova N, Abeysinghe SS, O'Connell CD, Cooper DN, Wells RD. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc Natl Acad Sci U S A. 2004;101:14162–14167. doi: 10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hoyne PR, Maher LJ., 3rd Functional studies of potential intrastrand triplex elements in the Escherichia coli genome. J Mol Biol. 2002;318:373–386. doi: 10.1016/S0022-2836(02)00041-4. [DOI] [PubMed] [Google Scholar]
- 130.Eckert KA, Mowery A, Hile SE. Misalignment-mediated DNA polymerase beta mutations: comparison of microsatellite and frame-shift error rates using a forward mutation assay. Biochemistry. 2002;41:10490–10498. doi: 10.1021/bi025918c. [DOI] [PubMed] [Google Scholar]
- 131.Hile SE, Eckert KA. Positive correlation between DNA polymerase alpha-primase pausing and mutagenesis within polypyrimidine/polypurine microsatellite sequences. J Mol Biol. 2004;335:745–759. doi: 10.1016/j.jmb.2003.10.075. [DOI] [PubMed] [Google Scholar]
- 132.Krasilnikova MM, Mirkin SM. Replication stalling at Friedreich's ataxia (GAA)n repeats in vivo. Mol Cell Biol. 2004;24:2286–2295. doi: 10.1128/MCB.24.6.2286-2295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rao BS, Manor H, Martin RG. Pausing in simian virus 40 DNA replication by a sequence containing (dG-dA)27.(dT-dC)27. Nucleic Acids Res. 1988;16:8077–8094. doi: 10.1093/nar/16.16.8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rao BS. Pausing of simian virus 40 DNA replication fork movement in vivo by (dG-dA) n.(dT-dC)n tracts. Gene. 1994;140:233–237. doi: 10.1016/0378-1119(94)90549-5. [DOI] [PubMed] [Google Scholar]
- 135.Bacolla A, Jaworski A, Connors TD, Wells RD. Pkd1 unusual DNA conformations are recognized by nucleotide excision repair. J Biol Chem. 2001;276:18597–18604. doi: 10.1074/jbc.M100845200. [DOI] [PubMed] [Google Scholar]
- 136.Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability. Mutat Res. 2006 doi: 10.1016/j.mrfmmm.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 137.Weinreb A, Collier DA, Birshtein BK, Wells RD. Left-handed Z-DNA and intramolecular triplex formation at the site of an unequal sister chromatid exchange. J Biol Chem. 1990;265:1352–1359. [PubMed] [Google Scholar]
- 138.Rooney SM, Moore PD. Antiparallel, intramolecular triplex DNA stimulates homologous recombination in human cells. Proc Natl Acad Sci U S A. 1995;92:2141–2144. doi: 10.1073/pnas.92.6.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Praseuth D, Guieysse AL, Helene C. Triple helix formation and the antigene strategy for sequence-specific control of gene expression. Biochim Biophys Acta. 1999;1489:181–206. doi: 10.1016/s0167-4781(99)00149-9. [DOI] [PubMed] [Google Scholar]
- 140.Kato M, Shimizu N. Effect of the potential triplex DNA region on the in vitro expression of bacterial beta-lactamase gene in superhelical recombinant plasmids. J Biochem (Tokyo) 1992;112:492–494. doi: 10.1093/oxfordjournals.jbchem.a123927. [DOI] [PubMed] [Google Scholar]
- 141.Sarkar PS, Brahmachari SK. Intramolecular triplex potential sequence within a gene down regulates its expression in vivo. Nucleic Acids Res. 1992;20:5713–5718. doi: 10.1093/nar/20.21.5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Brahmachari SK, Sarkar PS, Raghavan S, Narayan M, Maiti AK. Polypurine/polypyrimidine sequences as cis-acting transcriptional regulators. Gene. 1997;190:17–26. doi: 10.1016/s0378-1119(97)00034-6. [DOI] [PubMed] [Google Scholar]
- 143.Duval-Valentin G, de Bizemont T, Takasugi M, Mergny JL, Bisagni E, Helene C. Triple-helix specific ligands stabilize H-DNA conformation. J Mol Biol. 1995;247:847–858. doi: 10.1006/jmbi.1995.0185. [DOI] [PubMed] [Google Scholar]
- 144.Firulli AB, Maibenco DC, Kinniburgh AJ. Triplex forming ability of a c-myc promoter element predicts promoter strength. Arch Biochem Biophys. 1994;310:236–242. doi: 10.1006/abbi.1994.1162. [DOI] [PubMed] [Google Scholar]
- 145.Rustighi A, Tessari MA, Vascotto F, Sgarra R, Giancotti V, Manfioletti G. A polypyrimidine/polypurine tract within the Hmga2 minimal promoter: a common feature of many growth-related genes. Biochemistry. 2002;41:1229–1240. doi: 10.1021/bi011666o. [DOI] [PubMed] [Google Scholar]
- 146.Xu G, Goodridge AG. Characterization of a polypyrimidine/polypurine tract in the promoter of the gene for chicken malic enzyme. J Biol Chem. 1996;271:16008–16019. doi: 10.1074/jbc.271.27.16008. [DOI] [PubMed] [Google Scholar]
- 147.Grabczyk E, Fishman MC. A long purine-pyrimidine homopolymer acts as a transcriptional diode. J Biol Chem. 1995;270:1791–1797. doi: 10.1074/jbc.270.4.1791. [DOI] [PubMed] [Google Scholar]
- 148.Maiti AK, Brahmachari SK. Poly purine.pyrimidine sequences upstream of the beta-galactosidase gene affect gene expression in Saccharomyces cerevisiae. BMC Mol Biol. 2001;2:11. doi: 10.1186/1471-2199-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Michel D, Chatelain G, Herault Y, Harper F, Brun G. H-DNA can act as a transcriptional insulator. Cell Mol Biol Res. 1993;39:131–140. [PubMed] [Google Scholar]
- 150.Lu Q, Teare JM, Granok H, Swede MJ, Xu J, Elgin SC. The capacity to form H-DNA cannot substitute for GAGA factor binding to a (CT)n*(GA)n regulatory site. Nucleic Acids Res. 2003;31:2483–2494. doi: 10.1093/nar/gkg369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pahwa GS, Maher LJ, 3rd, Hollingsworth MA. A potential H-DNA element in the MUC1 promoter does not influence transcription. J Biol Chem. 1996;271:26543–26546. doi: 10.1074/jbc.271.43.26543. [DOI] [PubMed] [Google Scholar]
- 152.Davis TL, Firulli AB, Kinniburgh AJ. Ribonucleoprotein and protein factors bind to an H-DNA-forming c-myc DNA element: possible regulators of the c-myc gene. Proc Natl Acad Sci U S A. 1989;86:9682–9686. doi: 10.1073/pnas.86.24.9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tiner WJ, Sr, Potaman VN, Sinden RR, Lyubchenko YL. The structure of intramolecular triplex DNA: atomic force microscopy study. J Mol Biol. 2001;314:353–357. doi: 10.1006/jmbi.2001.5174. [DOI] [PubMed] [Google Scholar]
- 154.Fiszer-Kierzkowska A, Wysocka A, Jarzab M, Lisowska K, Krawczyk Z. Structure of gene flanking regions and functional analysis of sequences upstream of the rat hsp70.1 stress gene. Biochim Biophys Acta. 2003;1625:77–87. doi: 10.1016/s0167-4781(02)00592-4. [DOI] [PubMed] [Google Scholar]
- 155.Belotserkovskii BP, di Silva E, Tornaletti S, Wang G, Vasquez KM, Hanawalt PC. A triplex-forming sequence from the human c-Myc promoter interferes with DNA transcription. J Biol Chem. 2007 doi: 10.1074/jbc.M704618200. [DOI] [PubMed] [Google Scholar]
- 156.Michel D, Chatelain G, Herault Y, Brun G. The long repetitive polypurine/polypyrimidine sequence (TTCCC)48 forms DNA triplex with PU-PU-PY base triplets in vivo. Nucleic Acids Res. 1992;20:439–443. doi: 10.1093/nar/20.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Potaman VN, Oussatcheva EA, Lyubchenko YL, Shlyakhtenko LS, Bidichandani SI, Ashizawa T, Sinden RR. Length-dependent structure formation in Friedreich ataxia (GAA)n*(TTC)n repeats at neutral pH. Nucleic Acids Res. 2004;32:1224–1231. doi: 10.1093/nar/gkh274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Grabczyk E, Usdin K. Alleviating transcript insufficiency caused by Friedreich's ataxia triplet repeats. Nucleic Acids Res. 2000;28:4930–4937. doi: 10.1093/nar/28.24.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Michelotti EF, Tomonaga T, Krutzsch H, Levens D. Cellular nucleic acid binding protein regulates the CT element of the human c-myc protooncogene. J Biol Chem. 1995;270:9494–9499. doi: 10.1074/jbc.270.16.9494. [DOI] [PubMed] [Google Scholar]
- 160.Potaman VN, Sinden RR. Stabilization of intramolecular triple/single-strand structure by cationic peptides. Biochemistry. 1998;37:12952–12961. doi: 10.1021/bi972510k. [DOI] [PubMed] [Google Scholar]
- 161.Escude C, Nguyen CH, Kukreti S, Janin Y, Sun JS, Bisagni E, Garestier T, Helene C. Rational design of a triple helix-specific intercalating ligand. Proc Natl Acad Sci U S A. 1998;95:3591–3596. doi: 10.1073/pnas.95.7.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wang G, Vasquez KM. Z-DNA, an active element in the genome. Front Biosci. 2007;12:4424–4438. doi: 10.2741/2399. [DOI] [PubMed] [Google Scholar]