Abstract
We have developed a novel class of antisense agents, RNA Lassos, which are capable of binding to and circularizing around complementary target RNAs. The RNA Lasso consists of a fixed sequence derived from the hairpin ribozyme and an antisense segment whose size and sequence can be varied to base pair with accessible sites in the target RNA. The ribozyme catalyzes self-processing of the 5′- and 3′-ends of a transcribed Lasso precursor and ligates the processed ends to produce a circular RNA. The circular and linear forms of the self-processed Lasso coexist in an equilibrium that is dependent on both the Lasso sequence and the solution conditions. Lassos form strong, noncovalent complexes with linear target RNAs and form true topological linkages with circular targets. Lasso complexes with linear RNA targets were detected by denaturing gel electrophoresis and were found to be more stable than ordinary RNA duplexes. We show that expression of a fusion mRNA consisting of a sequence from the murine tumor necrosis factor-α (TNF-α) gene linked to luciferase reporter can be specifically and efficiently blocked by an anti-TNF Lasso. We also show in cell culture experiments that Lassos directed against Fas pre-mRNA were able to induce a change in alternative splicing patterns.
INTRODUCTION
The specific inhibition of gene expression dependent on base pairing between an inhibitor and its DNA or RNA target has generated substantial interest, due to the potential ability to target any sequence by applying the simple rules of Watson–Crick recognition. Inhibitors of this type, broadly referred to as antisense molecules, have widely been used as tools for functional genomics and target validation, and to some extent as drugs. They can be classified into several groups according to their various mechanisms of action: (i) DNA-based oligonucleotides that recruit RNase H to degrade the target RNA following formation of the DNA–RNA duplex, including oligodeoxynucleotides (ODNs) that are either unmodified or contain certain modifications such as phosphorothioate and 2′-fluoro groups (1,2); (ii) steric blockers, which do not induce target cleavage but block expression of the target, including phosphorodiamidate morpholino oligomers (PMOs), N3′-P5′ phosphorodiamidates, 2′-O-methoxyethyl RNAs, locked nucleic acids (LNA), and peptide nucleic acids (PNA) (3–7); (iii) unmodified antisense RNAs that can be expressed intracellularly from appropriate vectors (8–13); (iv) catalytic nucleic acids (ribozymes and deoxyribozymes) that can hybridize to and cleave target RNAs (14,15); and (v) short interfering RNAs (siRNAs) and short hairpin RNAs (shRNAs), which cause cleavage of target RNAs via the naturally existing mechanism of RNA interference (RNAi) (16,17). Although the mechanisms of posttranscriptional gene silencing by si/shRNAs may involve more steps than those for ordinary antisense agents, all of these mechanisms have in common the hybridization of antisense guide sequences with the complementary site in target mRNA (18).
Here, we introduce a new type of antisense agent, the RNA Lasso, which contains both a ribozyme, whose sequence is constant (target-independent), and an antisense sequence, whose size and sequence can be varied to match accessible sites in the target RNA. This design allows the Lasso to pair with a target RNA, intertwining the two RNAs, and then to undergo self-ligation, resulting in circularization of the Lasso around the target. The ribozyme chosen to provide self-circularization is the hairpin ribozyme (HPR) because of its ability to catalyze both cleavage and ligation of substrate RNAs efficiently. Lassos can be conveniently synthesized by in vitro transcription, in which case cis-cleavage by the HPR allows excision of the Lasso from a longer transcript by trimming both the 5′- and 3′-ends of Lasso molecules. This reaction creates ends that are substrates for a second HPR reaction, ligation in cis, which results in circularization of the molecule. The circular and linear Lasso forms exist in equilibrium, the position of which depends on the Lasso sequence and solution conditions. These processing reactions can occur spontaneously when the Lasso is expressed in cells.
In experiments using fragments of murine tumor necrosis factor (TNF-α) mRNA as targets, we show that Lassos having the appropriate TNF-specific antisense sequences can efficiently bind to and circularize around the target without cleaving it. This sequence-specific binding leads to the formation of complexes that are more stable than ordinary RNA–RNA duplexes used as controls. We also demonstrate that these anti-TNF Lassos can specifically and efficiently block expression of a model TNF-Luciferase reporter gene in an in vitro translation assay. In addition, we show that Lassos targeting the intron–exon junction of Fas receptor pre-mRNA are able to alter the pattern of alternative splicing in cultured cells.
MATERIALS AND METHODS
Preparation of Lassos
Transcription templates for all Lassos were prepared in two steps as described below using synthetic oligodeoxynucleotides provided by IDT (Coralville, IA, USA). All sequences are shown in the 5′–3′ direction if not otherwise indicated.
ATR1
First, two synthetic oligodeoxynucleotides, partially complementary at their 3′-ends, TGACAGTCCTGTCCGTATGACAGAGAAGTCAACCAGAGAAACAC ACGTTGTGGTATATTACCTGGTCAAGAGAAG and AAACAGGACGGTCAGTTCTCTTCAAGGGACAAG GCTCGGGAAAAAAAGGAACAGGGAACTTCTCTTGACCAGG, were annealed and filled in by the Klenow fragment of DNA polymerase I (Promega, Madison, WI, USA) to create a double-stranded DNA fragment. Then two extended oligodeoxynucleotide primers, GGCTCGAATTCTAATACGACTCACTATAGGGTGACAGTCCTGTCCG and AAACAGGACGGTCAGTTC, were used to extend, add a T7 promoter sequence and amplify the DNA fragment from the previous step by PCR using Taq DNA polymerase (Promega).
ALR562-2
Two partially complementary oligodeoxynucleotides, CGTCCGTATGACGAGAGAAGCCTACCA GAGAAACACACGACGTAAGTCGTGGTACATTACCTGGTACAAGCCTT and GTTGTTGTTGTTGTTGT TGTTCTCTTCAAGGGACAAGGCTTGTACCAGGTAATGTACCACGACTTACGTC, were annealed and filled in using Klenow extension to create a double-stranded DNA fragment. Then the two extended oligodeoxynucleotide primers, TAATACGACTCACTATAGGGAGGCTGTCCTCGTCCGTATGACGAGAGAAGC and GACGAGGACAGCCTTGGTTGTTGTTGTTGTTGTTGTTGTTGTTCTCTTC, were used to extend, add a T7 promoter sequence and amplify the DNA fragment from the previous step by PCR.
FAS1, FAS2 and FAS3
In vitro transcription templates encoding the anti-Fas Lassos were prepared similarly to the anti-TNF Lassos. First, the pairs of partially overlapping oligodeoxynucleotides (FAS 1-1 and FAS 1–2 for Fas1; FAS 2–1 and FAS 2–2 for Fas2; and FAS 3–1 and FAS 3–2 for Fas 3) were annealed and filled in using Klenow extension to create a double-stranded DNA fragment. These fragments were simultaneously extended, T7 promoter sequence added and amplified by PCR using the following oligodeoxynucleotide primers: FAS 1–3 and FAS 1–4 for Fas1; FAS 2–3 and FAS 2–4 for Fas2; FAS 3–3 and FAS 3–4 for Fas3. The sequences of oligodeoxynucleotides described in this section are shown below:
CGTCCGTATGACGAGAGAAGCGAACCAGAGA AACACACGACGTAAGTCGTGGTACATTACCTGGTAACGC (FAS1–1); TGTTGTTGTTGTTGTTGTTCAACCTACAGGATCCAGATCGCGTTACCAGGTAATGTACCACG (FAS 1–2); TAATACGACTCACTATAGGGTCGCGGTCCTCGTCCGTATGACGAGAGAAG (FAS 1–3); GACGAGGACCGCGAGTTGTTGTTGTTGTTGTTGTTGTTGTTCAAC (FAS 1–4); CGTCCGTATGACGAGAGAAGTAGACCAGAGAAACACACGACGTAAGTCGTGGTACATTACCTGG (FAS 2–1); TGTTGTTGTTGTTGTTCAATGTTCCAACCTACAGGAGTTACCAGGTAATGTACCACGACTTACG (FAS 2–2); TAATACGACTCACTATAGGGCTACAGTCCTCGTCCGTATGACGAGAGAAG (FAS 2–3); GACGAGGACTGTAGTGTGTTGTTGTTGTTGTTGTTGTTCAATGTTCC (FAS 2–4); CGTCCGTATGACGAGAGAAGT AGACCAGAGAAACACACGACGTAAGTCGTGGTACATTACCTGGTAACTTAG (FAS 3–1); GTTGTTGTTGTTGTTGTTCCTACAGGATCCAGATCTAAGTTACCAGGTAATGTACCACGAC (FAS 3–2); TAATACGACTCACTATAGGGCTACAGTCCTCGTCCGTATGACGAGAGAAG (FAS 3–3); GACGAGGACTGTAGGTTGTTGTTGTTGTTGTTGTTGTTCCTAC (FAS 3–4).
Precursors to RNA Lassos were produced by in vitro transcription with T7 RNA polymerase (Ambion, Austin, TX, USA). The reaction was typically performed in 6 mM Mg2+, 2 mM spermidine–HCl, 40 mM Tris–HCl (pH 7.5) for 3 h at 37°C. Following transcription, the reaction products were treated with DNase I to degrade the transcription template and desalted using a G-50 microspin column pre-equilibrated with 1× TE buffer (Amersham/GE Healthcare, Picataway, NJ, USA). Internally 32P-labeled RNA Lassos were produced by addition of [α32P]CTP to the transcription reaction. The 5′-end labeling of ATR1 self-processing products was performed posttranscription with [γ32P]-ATP (Amersham) and T4 polynucleotide kinase (NEB, Ipswich, MA, USA) by incubation at 37°C for 60 min. The 3′-end labeling was performed using [32P]pCp (Amersham) and T4 RNA ligase (NEB).
Purification of individual linear and circular ATR1 forms
Following transcription of the ATR1 DNA template in the presence of [α32P]CTP, ATR1 was incubated for 30 min in 50 mM Tris–HCl, pH 7.5, 10 mM MgCl2 (Lasso processing buffer, TM) to allow complete Lasso self-processing. The Lasso species were separated by denaturing electrophoresis in polyacrylamide gels (d-PAGE) containing 6% polyacrylamide, 8 M urea and 0.5× TBE. Prior to loading onto the gel, an equal volume of denaturing loading buffer was added to the RNA (92% formamide, 12 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue). Internally 32P-labeled ATR1 species were visualized by autoradiography, and the bands corresponding to the fastest (LL) and the slowest migrating (CL) Lasso forms were excised from the gel. RNA was eluted from the gel slices by crushing and soaking in 1× TE buffer. It should be noted that use of standard RNA elution buffers may result in partial interconversion of the HPR circular and linear forms (19). The eluted RNA was purified using RNase-free Costar Spin-X columns (Corning, NY, USA) and analyzed by 6% d-PAGE to confirm the purity of the individual Lasso species. It is important to avoid ethanol precipitation of the individual Lasso species because this procedure can also promote HPR activity (19). To monitor conversion of linear to circular ATR1 and vice versa, each eluted RNA was incubated in TM buffer at 37°C for 20 min prior to d-PAGE analysis.
Preparation of RNA targets
TNF
The transcription template for the model TNF RNA target was prepared by PCR using the primers GAATTCGATTTAGGTGACACTATAGAAGAGCTCTTCTGTCTACTGAACTTCGG and GTTGGACTCTGAGCCATAATCC. The resulting DNA fragment, corresponding to nucleotides 280–965 of the murine TNF-α gene, was inserted into plasmid pGEM-4z (20), which allows transcription of the insert from a SP6 promoter. Nonradioactive TNF685 RNA (685 nt) was produced by in vitro transcription with SP6 RNA polymerase using the MegaScript kit (Ambion) according to the manufacturer's protocol. Internally, 32P-labeled TNF685 RNA was prepared by performing a small-scale transcription reaction in the presence of [α32P]-CTP. The eluted RNA was centrifuged through RNase-free Spin-X columns to remove gel fragments and analyzed by 6% d-PAGE to confirm the integrity and purity of the transcription products.
Circular RNA target, CMT
The transcription template for the linear model target (LMT), which contains the 22-nt TNF target site, was prepared by annealing the partially complementary DNA oligonucleotides, TAATACGACTCACTATAGGGTCAAAAGTTTCGACTGCAC CTGACGAACAACAACAACAACAACAACAACGTTCTCTTCAAGGGAC and TATGAGTTTCGACTGCACCTGACGGTTGTTGTTGTTGTTGTTGTTGTTAGCCTT GTCCCTTGAAGAGAACGTTGTTGTTGTTG, followed by extension of 3′-ends using the Klenow polymerase. The resulting double-stranded DNA containing a T7 promoter was transcribed using T7 RNA polymerase to produce LMT RNA. This RNA was treated with calf intestinal alkaline phosphatase (CIAP, NEB) to remove its 5′-triphospate followed by phenol–chloroform extraction to remove CIAP, and finally precipitated by ethanol. Then a single phosphate was added to the 5′-end of LMT by treating with [γ32P]-ATP and T4 polynucleotide kinase followed by phenol–chloroform extraction and ethanol precipitation. To convert to the circular form, LMT was treated with T4 RNA ligase (NEB) for 4 h at 16°C as described by Beaudry and Perreault (21). The ligation product, 32P-labeled CMT, had a slower electrophoretic mobility than LMT. CMT was purified using 9% d-PAGE, eluted from the gel by crushing and soaking in 1× TE buffer. The eluted RNA was centrifuged through RNase-free spin-X columns and precipitated by ethanol.
The circular nature of CMT was confirmed by its reduced gel mobility in comparison to the linear LMT (22) as well as by partial alkaline-induced cleavage and phosphatase treatment (23,24). Briefly, 5′-32P-labeled LMT and CMT labeled at its ligation site were first incubated with increasing amounts of NaOH (10–100 mM) for 5 min at 25°C. An analysis of the cleavage products using 9% d-PAGE revealed that LMT generated a normal ladder corresponding to cleavage products fragments differing by one nucleotide, while CMT generated a main band comigrating with LMT with no intermediate bands between these two forms. This pattern corresponds to circular RNA since each single cleavage produces linear RNA molecules with various ends but the same length. In the second assay, the 32P-labeled LMT and CMT were incubated with or without CIAP at 37°C for 10 min and analyzed by 9% d-PAGE. As expected, 5′-end labeled LMT was dephosphorylated whereas the CMT retained its 32P-label.
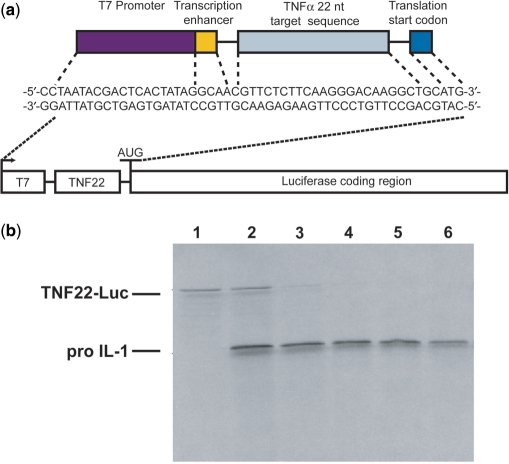
TNF22-Luc
The model target message TNF22-Luc was constructed by inserting upstream of the luciferase translation start codon a T7 promoter followed by the 22-nt TNF-α sequence that is complementary to the antisense sequence of ATR1. This was accomplished by hybridization of appropriate synthetic DNA to form the sequence shown in Figure 6a flanked by NcoI sites. This fragment was digested by NcoI (Invitrogen, Carlsbad, CA, USA) and inserted into the pGL3 Control Vector (Promega) at its NcoI site using T4 DNA ligase (Invitrogen). The resultant vector was cloned in DH5 bacterial cells and isolated using QIAprep columns (Qiagen). It was then linearized by BamH1 (NEB) at the 3′-end of the luciferase gene, treated by proteinase K and purified by phenol–chloroform extraction and ethanol precipitation. The TNF22-Luc mRNA (1923 nt) was synthesized by in vitro transcription of this linearized vector using the T7 Ribomax large-scale RNA kit (Promega).
Figure 6.
Inhibition of in vitro translation of a TNF-luciferase reporter mRNA by Lasso ATR1. (a) Schematic of TNF22-luc. (b) Products of translation of TNF22-Luc mRNA and pro-IL-1 mRNA (negative control) in rabbit reticulocyte lysate in the presence increasing amounts of Lasso ATR1 and 35S-labeled methionine. Lane 1 contains only the TNF22-Luc mRNA RNA, while lanes 2–6 contain both TNF22-Luc and pro-IL-1 mRNAs. Lanes 3–6 additionally contain 20-, 40-, 80- and 160-fold molar excess of ATR1 Lasso with respect to the targets. The translation products were analyzed by 12% SDS–PAGE.
Pro-IL1β
The plasmid constructs and in vitro transcription reaction used to generate proIL-1β mRNA for in vitro translation experiments were done as described by Jobling et al. (25).
Fas
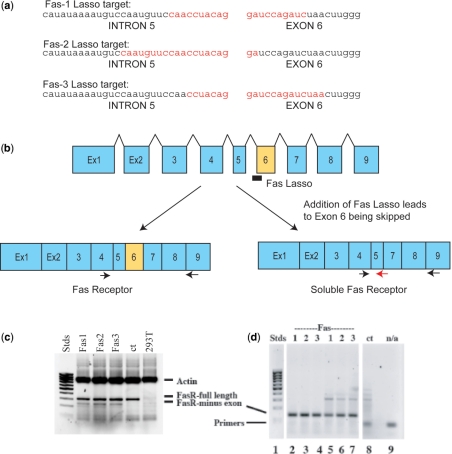
A DNA fragment encoding the Fas RNA target including the intron 5/exon 6 junction sequence (Figure 7a) was generated by PCR from genomic DNA of human Jurkat T lymphocytes (Promega) using a forward primer to Intron 5, CATGGAATCATCAAGGAATG (FasInt5-f), and a reverse primer to Exon 6, GGTAAGAATGAGGCAAATC (FasEx6-r). The PCR-amplified DNA fragment was cloned into the pCR2.1 vector downstream of the T7 promoter (to yield pCR2.1-Fas) using the TA-cloning kit (Invitrogen). Target Fas RNA (347 nt) was transcribed in large scale from the linearized pCR2.1-Fas vector using the T7 MEGAscript kit (Ambion). For in vitro binding studies, internally 32P-labeled Fas target RNA was prepared by performing a small-scale transcription in the presence of [α32P]-CTP.
Figure 7.
RNA Lassos can redirect splicing of Fas pre-mRNA. (a) Target sequences (red) of Lassos Fas1, Fas2 and Fas3 that span the intron 5/exon 6 splice junction. (b) Schematic of intron/exon structure of Fas receptor primary transcript (not to scale). The Lasso target site (intron 5/exon 6 junction) chosen to prevent inclusion of exon 6 in the final mRNA product is shown. The PCR primer site locations for analysis are shown as horizontal arrows. (c) RT–PCR amplification (using the exon 4 and exon 9 primers, black arrows) of products of Fas splicing in the presence of Lassos Fas1, Fas2 or Fas3. No shorter DNA fragment corresponding to Fas lacking exon 6 was seen in the absence of Fas Lassos (ct). β-actin serves as a loading control. Stds is a DNA ladder (100–1000 bp). (d) Confirmation that Fas Lassos lead to exon 6 skipping revealed by PCR analysis using a downstream primer spanning the exon 5 and exon 7 junction (red arrow) along with the exon 4 upstream primer, which are unable to amplify a product that includes exon 6. Lanes 2–4 and 5–7 correspond to independent experiments with anti-Fas Lassos 1, 2 and 3. Control lane 8 corresponds to the RT–PCR products obtained with cells that were not transfected by Lassos (ct), and lane 9 contains no PCR template (n/a).
In vitro binding assays
Typically, 5 µl reactions containing trace amounts of internally 32P-labeled Lasso RNAs were incubated with target RNAs (1.4 µM) at 37°C for 15–60 min (as indicated) in buffer containing 50 mM Tris–HCl, pH7.5 plus either 10 mM MgCl2 (TM buffer, +Mg condition) or 10 mM EDTA (−Mg condition). In addition, TM–formamide buffer containing 50 mM Tris–HCl, 10 mM MgCl2, 20% formamide has been used where indicated. For thermal dissociation experiments, the complexes were subsequently postincubated for 2 min at 50, 65, 80 or 95°C as indicated followed by immediate chilling on ice to prevent rehybridization. Reactions were terminated by addition of an equal volume of denaturing loading buffer containing 90% formamide, 12 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol. Complexes were loaded onto 6% polyacrylamide gels containing 8 M urea and 0.5× TBE, and were electrophoresed. The products were detected and quantified using phosphorimager Molecular Imager FX and Molecular Analyst 2.1 software for Macintosh (Bio-Rad, Hercules, CA, USA).
In vitro translation assay
A total of 0.8 µg TNF22-Luc target mRNA or pro-IL-1 mRNA (used as a nonspecific control) was preincubated with 20-, 40-, 80- and 160-fold molar excess of Lasso ATR1 over the target (yielding final Lasso concentrations of 1, 2, 4 and 8 μM, respectively) as indicated in Figure 6b for 1 h at 37°C in 10 mM MgCl2, 50 mM Tris–acetate (pH 7.5). Equal aliquots of these solutions were then mixed with FLEXI rabbit reticulocyte lysate translation system components (Promega). Following a 40 min incubation with rabbit reticulocyte lysate in the presence of 35S-labeled methionine (Amersham/GE Healthcare), the translation products were separated by nondenaturing 12% PAGE–SDS according to the manufacturer's protocol. Products were detected by radiography. The autoradiogram was scanned and translation levels of TNF-α relative to pro-IL-1 were quantified by the program UN-SCAN-IT Gel (Silk Scientific, Orem, UT, USA).
Splicing redirection experiments
Human Jurkat lymphocytes (E6-1 derivative, ATCC #TIB-152) were transfected with the anti-Fas Lassos using oligofectamine (Invitrogen). Total RNA from ∼1.2 × 106 treated Jurkat cells was isolated by Trizol (Invitrogen) according to the manufacturer's protocol 24–48 h after transfection. RT–PCR was performed on the extracted total cellular RNA with Superscript II reverse transcriptase (Invitrogen) using an anchored oligo(dT)20 primer (Invitrogen) followed by 40 cycles of PCR. A forward primer corresponding to nucleotides 410–430 (FasEx4-f, 5′-CTACTGTATGTGAACACTGTG-3′) and a reverse primer corresponding to nucleotides 721–702 (FasEx9-r, 5′-TCATGACTCCAGCAATAGTG-3′) were used in the PCR step. In a control PCR experiment, β-actin sense primer corresponding to nucleotides 578–609 (5′-ATCTGGCACCACACCTTCTACAATGAGCTGCG-3′) and antisense primer complementary to nucleotides 1415–1384 (5′-CGTCATACTCCTGCTTGCTGATCCACATCTGC-3′) were used as described in Shiraki et al. (26). The resulting PCR-amplified DNA fragments corresponding to full-length Fas (316 bp), Fas minus exon 6 (259 bp) and β-actin (837 bp) were separated by electrophoresis on 3% agarose gels and stained with ethidium bromide to visualize bands. PCR products were excised from the gel, cloned and sequenced (Retrogen, San Diego, CA, USA). An additional PCR reaction was performed with the same forward primer but the reverse primer spanned the Exon5–Exon7 junction (5′-CTTCCTTTCTCTTCACTTCC-3′). This set of primers will amplify a 110-bp product only if splicing is redirected to skip Exon6.
RESULTS
Design of a prototype RNA Lasso
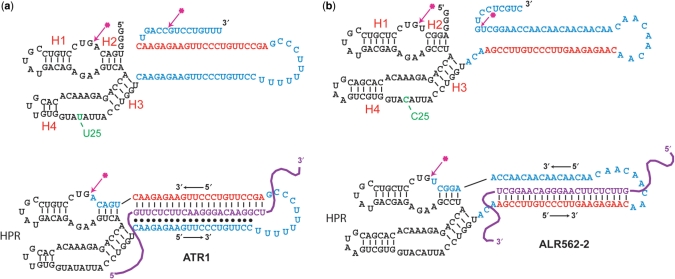
The sequence and secondary structure of a prototype Lasso, ATR1, is shown in Figure 1a. ATR1 contains three functional elements: an HPR sequence with its substrate sequences at both ends, an antisense sequence (22-nt) that can hybridize to the coding region of TNF-α to form two turns of an A-form helix (27) and a sequence that can bind to that duplex to form a triplex. The HPR sequence was adapted from a previously described minimal HPR construct, mini-M36 (19,28), by substituting the TNF-specific antisense and triplex forming sequences for the 36-nt sequence of loop 2. The triplex forming sequence was included with the expectation that triplex formation would help promote circularization of the Lasso around its target RNA by placing the 3′-end of the Lasso near its 5′-end (Figure 1a). The target sequence for ATR1 was chosen primarily because it contained an extensive polypyrimidine tract favorable for triplex formation. Although accessibility of this target site in the RNA targets used was not specifically investigated, our results suggest that ATR1 molecules bind efficiently with the corresponding target sequences both in vitro and in cell lysates (see below).
Figure 1.
Structures of the TNF-specific RNA Lassos ATR1 (a) and ALR 562-2 (b). The top structures correspond to unprocessed lasso transcripts (pre-Lasso), while the bottom structures depict fully processed lassos circularized around the murine TNF-α target RNA. Antisense sequences within the Lassos are shown in red, hairpin ribozyme sequences are in black, linker sequences are in blue, the U25→C25 substitution is in green, and target RNA is purple. The ribozyme cleavage/ligation sites are marked by arrows. Potential base pairing of a triplex strand is indicated by black dots.
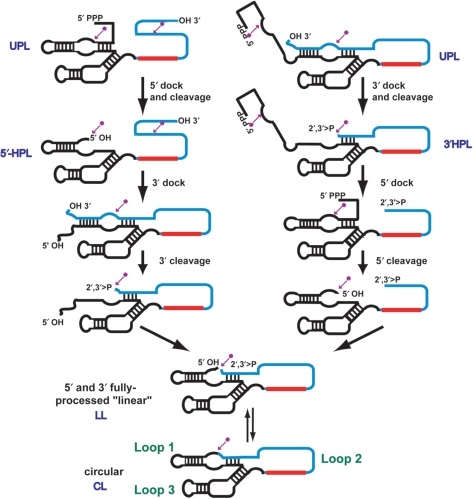
Self-processing of Lasso transcripts
Upon transcription in vitro, the unprocessed Lasso precursor (UPL) undergoes HPR-catalyzed sequential, intramolecular cleavage at its 5′- and 3′-ends, both of which contain ribozyme cleavage/ligation sites (Figure 2). The first cleavage results in either a 5′- or 3′-half-processed molecule (5′-HPL and 3′-HPL, respectively), and the second cleavage yields a fully processed linear Lasso (LL). LL can then undergo intramolecular ligation, also catalyzed by the HPR, to produce the circular Lasso form (CL). The last step is reversible, and LL and CL coexist in an equilibrium distribution. Similar self-processing schemes have been described previously for HPR derivatives (28–30).
Figure 2.
Schematic of self-processing (cleavage and ligation) of the RNA Lasso precursor. Self-processing of a primary Lasso transcript (pre-Lasso) having 5′-triphosphate (5′-PPP) and 3′-hydroxyl (3′-OH) groups can proceed by either the right or left pathway. Self-cleavage catalyzed by the HPR generates specific fragments having 2′,3′-cyclic phosphate (2′,3′ > P) and 5′-hydroxyl (5′-OH) termini. The fully 5′- and 3′-processed RNA Lasso exists as an equilibrium between circular and linear forms. The cleavage/ligation sites are marked by magenta arrows with asterisks. Loops 1, 2 or 3 indicated in the bottom structure can be used for insertion of an antisense sequence. Here, the antisense sequence (red) is inserted into loop 2 (blue). Species are identified as unprocessed Lasso (UPL), 5′-half-processed Lasso (5′-HPL), 3′-half-processed Lasso (3′-HPL), 5′- and 3′-processed ‘linear’ Lasso (LL) and circular Lasso (CL).
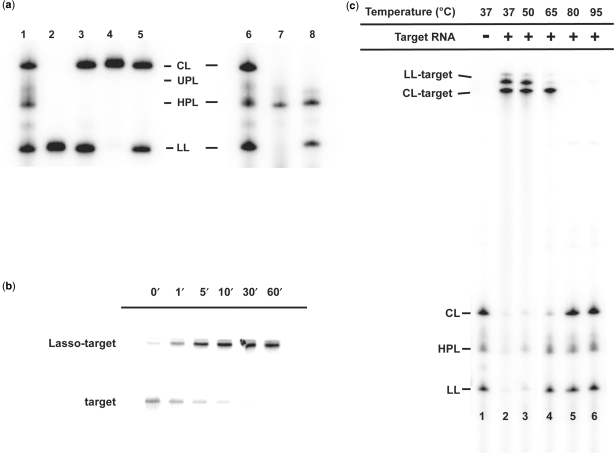
In the case of ATR1, self-processing is only partially completed upon its synthesis via in vitro transcription in the presence of 6 mM Mg2+ (see Materials and Methods section). Additional incubation in buffer containing 50 mM Tris–HCl (pH 7.5) and 10 mM MgCl2 (TM buffer) enables completion of self-processing (Figure 3a, lanes 1 and 6). By comparing the electrophoretic mobilities and analyzing the 5′- and 3′-ends of each Lasso species (see below), we assigned electrophoretic bands for the following processing products of [ATR 1]: UPL (136 nt), 5′-HPL (128 nt) and 3′-HPL (127 nt), LL and CL (both 117 nt).
Figure 3.
Transcription, self-processing, and target binding of Lasso ATR1. (a) Lane 1 shows internally 32P-labeled products of ATR1 transcription and spontaneous self-processing during the transcription reaction. Lane 2 shows gel-purified LL (linear Lasso), and lane 3 its partial conversion to CL (circular Lasso) through self-ligation after incubation in TM buffer (50 mM Tris–HCl, 10 mM MgCl2) for 1 h at 37°C. Lane 4 shows gel-purified CL, and lane 5 its partial conversion to LL through self-cleavage after the same incubation as for lanes 2 and 3. Lanes 6–8 compare internally 32P-labeled ATR1 Lasso species (lane 6, the same as lane 1) with 3′-end-labeled (lane 7) and 5′-end labeled ATR1 (lane 8). Because of efficient Lasso self-processing, the unprocessed Lasso (UPL) and the half-processed Lasso (comigrating 5′-HPL and 3′-HPL) forms are present in relatively small amounts (lanes 1 and 6). (b) Time-course of binding of internally 32P-labeled TNF685 RNA target with an excess of non-radioactive Lasso ATR1 transcription mixture in TM buffer at 37°C for the indicated time periods. (c) Internally 32P-labeled ATR1 incubated with or without an excess of nonradioactive TNF685 fragment (as indicated) in TM buffer for 2 h at 37°C. Samples in lanes 3–6 were additionally incubated for 2 min at elevated temperatures as indicated and transferred immediately to ice to prevent rehybridization of the complexes. The products were analyzed by electrophoresis on denaturing 6% polyacrylamide gels (d-PAGE).
Several lines of evidence support the identification of the slowest migrating band as the circular RNA species. First, the lower electrophoretic mobility of CL in comparison to LL is consistent with the properties of circular RNAs longer than 42 nt, which migrate more slowly than their linear counterparts (22,28). Second, incubation of gel-purified CL and LL bands in buffer that supports HPR catalytic activity showed that these bands interconvert only with each other (Figure 3a, lanes 2–5). The assignments of CL and LL were further supported by 32P-end-labeling analysis (29). CL could not be labeled by [32P]pCp and T4 RNA ligase at its 3′-end (Figure 3a, lane 7), or by [γ32P]ATP and T4 polynucleotide kinase at its 5′-OH end (Figure 3a, lane 8), consistent with CL being a circular molecule without free ends. LL could not be labeled at the 3′-end, consistent with its having a 2′,3′-cyclic phosphate, but it could be 5′-end kinased (see Figure 3a, lanes 7–8), consistent with its having a 5′-OH. The only bands that could be 3′-end-labeled were UPL and 5′-HPL (Figure 3a, lanes 7), confirming that they have free 3′-OH groups as expected. Specific 32P-labeling of 5′-HPL and LL at their 5′-ends (Figure 3a, lane 8) established that these are the only species containing a free 5′-OH. Additional confirmation of the circular nature of CL was provided by partial RNA cleavage of 32P-labeled CL under mild alkaline conditions (23). Under these conditions, the internally 32P-labeled CL produced a band comigrating with LL, with no intermediate bands appearing between the putative circular and linear RNA species (data not shown). This pattern is expected of a circular nucleic acid since a single alkali-induced cleavage will produce a linear RNA with the same length and electrophoretic mobility as LL.
Formation of strong and specific Lasso-target complexes
Binding of Lasso ATR1 with target TNF685, a 685-nt fragment of TNF-α mRNA containing the target of the ATR1 antisense sequence (nt 562–583 of the full-length mRNA) was performed in solutions supporting HPR catalysis. Complex formation was analyzed by gel mobility shift assays under both nondenaturing (data not shown) and denaturing conditions (Figure 3b and c). We found that Lasso-target complexes were stable under both conditions. d-PAGE was adopted as the standard method for analyzing these strong complexes. We confirmed that the mobility shifts of the Lasso-target complexes were the same whether the Lassos or the targets were radio-labeled (labeling was by transcription in the presence of [α-32P]-CTP). In time-course experiments, complex formation was 50% complete in 1 min (t1/2) and 95% complete within 10 min (Figure 3b).
Products of self-processing of the ATR1 form two differently shifted complexes with TNF685 (Figure 3c, lanes 1–2). To identify them and compare their relative thermostability, these complexes were additionally incubated (post-incubated) at increasing temperatures after complex formation at 37°C and analyzed by d-PAGE. After post-incubation at 50°C, virtually all ATR1 species were shifted into one or the other of the two complexes (Figure 3c, lanes 2–3). However, after post-incubation at 65°C, the lower shifted complex remained intact while the upper one dissociated, and bands that comigrated only with the linear Lasso species (LL and HPL) reappeared (Figure 3c, compare lanes 1 and 4). After post-incubation at 80°C, the lower shifted complex also dissociated, and a band comigrating with the free circular (CL) Lasso appeared (Figure 3c, lanes 5–6). We interpret from these results that the faster migrating complex band was CL bound to target RNA, whereas the slower migrating complex band corresponded to LL bound to target RNA. Therefore, the circular Lasso-target complex appeared to be more thermostable than the linear Lasso-target complexes.
We found that even linear Lassos can form complexes that are more stable than ordinary RNA–RNA duplexes. In chase experiments, we used 20-nt sense or antisense synthetic RNAs that correspond to the TNF-α target site. We observed that even a 7- to 14-fold excess of these short RNAs could not displace the TNF-specific Lassos from long TNF RNA targets when incubated with the preformed Lasso-target complexes (Supplementary Figure 1).
In negative control experiments, no complex formation was observed between TNF685 target and mini-M36 (19,28), which contains the HPR portion of the Lasso but lacks sequence complementary to the TNF target (data not shown). In addition, complexes between ATR1 and unrelated target RNAs that did not contain the ATR1 binding site were not detected even by nondenaturing PAGE (data not shown). As expected, cleavage of the RNA targets in trans by the Lasso-HPR domain were not observed.
Circular Lassos bind target RNAs via linear intermediates
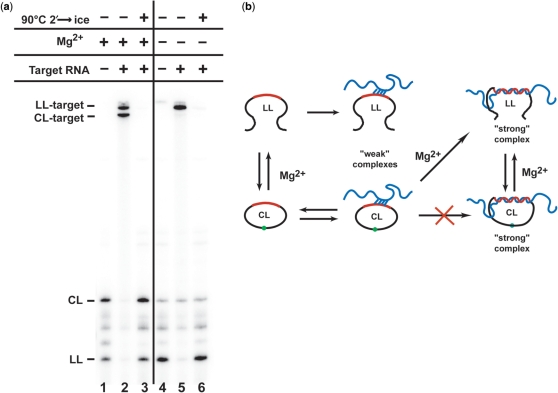
Sterically, it should not be a topological problem for linear Lasso species to hybridize with a target RNA and then circularize around it. But, how can a circular Lasso species bind to the linear RNA target and circularize around it? To address this question, we incubated 32P-labeled ATR1 transcripts with excess target TNF685 in two different solutions: (i) TM buffer containing 10 mM Mg2+ (+Mg), in which the CL and LL Lasso species are able to interconvert, or (ii) buffer containing 10 mM EDTA and no added Mg2+ (−Mg), in which LL cannot circularize and CL cannot interconvert into LL because the HPR is inactive under these conditions (Figure 4). Because the transcription reaction to synthesize ATR1 contains Mg2+ ions, initial self-processing of the Lasso RNA occurs during transcription. However, additional incubation in +Mg buffer allows equilibration between LL and CL. Under the +Mg conditions, both LL and CL species bound to the target, and two distinct complexes that correspond to the circular and linear Lasso-target complexes were detected by d-PAGE (Figure 4a, lanes 1–2). When these complexes were postincubated under highly denaturing conditions (90°C, 50% formamide, 10 mM EDTA) in which both CL and LL forms can be displaced from the complexes but cannot interconvert, both dissociated forms were detected (Figure 4a, lane 3). Under the −Mg conditions, the original ATR1 transcript did not undergo complete self-processing and the LL form predominated over CL (Figure 4a, lane 4). Under this condition, LL was completely shifted into the corresponding complex, which dissociated only under highly denaturing conditions (Figure 4a, lanes 5–6), whereas the CL species, here present at low levels, did not form a strong complex detectable by d-PAGE (Figure 4a, lane 5). This discrimination is more obvious when the gel is overexposed (data not shown). It is possible that less stable complexes between CL species and the target could form in the absence of Mg2+ but would then dissociate (and therefore not be detected) under d-PAGE conditions. These results suggest that, in the presence of Mg2+, circular Lasso species must convert to LL through self-cleavage either before, or as result of, target binding in order to form strong complexes. Once bound to the target RNA, the linear Lasso can religate, circularizing around the target as schematically depicted in Figure 4b. The similar CL/LL ratios before and after ATR1 binding to targets (Figure 4a, lanes 1 and 3) show that this Lasso can circularize around a target as efficiently as it circularizes in the absence of target.
Figure 4.
Binding of circular and linear ATR1 forms to TNF RNA target depends on presence of Mg2+. (a) Internally 32P-labeled ATR1 transcripts incubated at 37°C for 2 h in TM buffer containing 10 mM MgCl2 (lanes 1–3) or in buffer containing 10 mM EDTA (lanes 4–6) either without target (lanes 1 and 4) or in the presence of nonradioactive TNF685 target (lanes 2–3 and 5–6). Samples in lanes 3 and 6 were additionally postincubated at 90°C for 2 min to dissociate the Lasso-target complexes. The products were analyzed by 6% d-PAGE. (b) Schematic of the mechanism of formation of complexes between Lasso (with antisense region shown in red) and target RNA (blue) under −Mg (top) and +Mg (bottom) conditions.
The triplex-forming domain is not required for Lasso circularization around the target
To study if the triplex-forming domain present in ATR1 is required for circularization of the Lasso around the target, we prepared Lasso ALR-562-2, which lacked the triplex-forming sequence of ATR1 but had the same antisense sequence (compare Figure 1a and b). In ALR-562-2, the antisense sequence was moved further from the HPR cleavage/ligation site than in ATR1 and a linker was substituted for the triplex-forming sequence to keep the loop lengths similar in both Lassos. The (AAC)8 sequence of the linker was chosen to minimize formation of secondary structures that could affect HPR folding. The HPR domain in ALR562-2 was also modified to stabilize the catalytically active structure of HPR. These HPR modifications, which have been described previously (31), included a U→C substitution at position 25 in the ribozyme core, extension of the substrate stem H1 by one G–C base pair, and substitution of a GNRA-type tetraloop for the 3-nt H4 terminal loop.
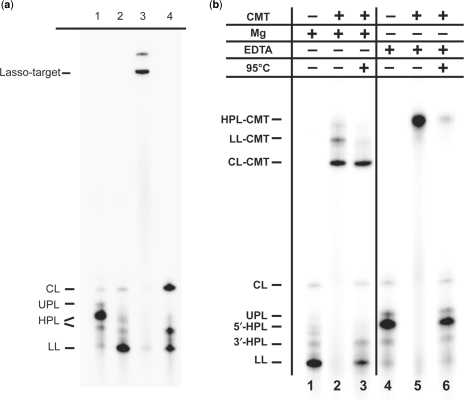
Products of ALR562-2 transcription and its self-processing were characterized by the same methods as with ATR1 (data not shown). The 5′-HPL species was identified as the major product of ALR562-2 formed during transcription in the presence of 6 mM Mg2+, whereas additional incubation in the TM buffer yielded mostly the fully processed LL species (Figure 5a, lanes 1–2). When internally labeled LL was incubated with an excess of target TNF685 in TM–formamide buffer, a high yield of a strong Lasso-target complex was detected by d-PAGE (Figure 5a, lane 3). However, the kinetics of ALR562-2 binding to TNF685 were found to be about 10-fold slower than with ATR1 under similar conditions (data not shown). When the ALR562-2 complex with TNF685 was postincubated under highly denaturing conditions (90°C, 50% formamide, 10 mM EDTA), the displaced Lassos were mostly the circular form (Figure 5a, lane 4), whereas ALR562-2 that had not been exposed to the target was predominantly in the linear form (Figure 5a, lane 2). Therefore, the triplex-forming domain present in ATR1 is not required for Lasso circularization on the target but may promote hybridization since ATR1 does binds to target with faster kinetics than the non-triplex containing ALR562-2.
Figure 5.
Lasso ALR562-2 binding to linear TNF685 and a circular target (CMT). (a) Products of self-processing of internally 32P-labeled ALR562-2 in transcription buffer containing 6 mM Mg2+ (lane 1) and in buffer containing 50 mM Tris–HCl, 10 mM MgCl2, 20% formamide (lane 2). Lane 3: formation of the strong complex between ALR562-2 (from lane 2) and nonradioactive TNF685 target provided in excess. Lane 4: products of dissociation of the complex from Lane 3 upon addition of 50% formamide and 10 mM EDTA and post-incubation for 2 min at 90°C. The products were analyzed by 6% d-PAGE and bands were assigned to Lasso-target complexes as well as unbound circular (CL), unprocessed (UPL), half-processed (HPL) and linear (LL) Lasso forms as indicated. (b) Demonstration of topological linkage between CMT and Lasso ALR562-2. Internally 32P-labeled Lasso ALR562-2 was incubated in TM–formamide buffer containing 50 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 20% formamide (lanes 1–3) or 50 mM Tris–HCl (pH 7.5), 10 mM EDTA, 20% formamide (lanes 4–6) for 2 h at 37°C either alone (lanes 1 and 4) or with the CMT target (lanes 2–3 and 5–6). Samples in lanes 3 and 6 were additionally postincubated at 95°C for 5 min followed by quenching on ice. The products were analyzed by denaturing 6% PAGE and assigned to complexes of CMT with various forms of the Lasso (HPL-CMT, LL-CMT and CL-CMT) as well as unbound circular (CL), unprocessed (UPL), half-processed (5′-HPL and 3′-HPL) and linear (LL) forms as indicated.
Theoretically, the triplex-forming strand of ATR1 could form five U•A-U and six C+•G-C Hoogsteen base-triplets, whose formation typically requires mildly acidic conditions to have a protonated cytosine (32). In addition, there are five potential A•U-A and four G•G-C reverse-Hoogsteen base-triplets, whose formation is pH-independent although sterically constrained because the major groove of a typical RNA duplex is not wide enough to accommodate the third strand (33,34). We found that formation of strong complexes between ATR1 and the TNF target increased when the pH was changed from 5.2 to 8.0, while the equilibrium between the circular and linear Lasso forms was not pH-dependent (see Supplementary Figure 2). Because ATR1 circularized around the target more efficiently at pH ⩾ 6.5 than at pH ⩽ 6, if an ATR1-target RNA triplex structure is formed, it appears to hinder rather than promote circularization of the Lasso bound to target RNA.
Lassos can form true topologically linked complexes
To verify that circular Lassos are truly intertwined with the target RNA in their strong complexes, we examined the binding of Lasso ALR562-2 with a CMT, which was prepared by circularization of a corresponding linear form (LMT) (see Supplementary Figure 3). These two targets CMT and LMT, each 120 nt in length, contain the 22-nt TNF sequence flanked by two (AAC)8 linkers and a hairpin-forming element that facilitates LMT circularization by RNA ligase (21).
The partially and fully self-processed forms of 32P-labeled ALR562-2, which were primarily the 5′-HPL and LL forms, were incubated with an excess of CMT, either (i) in buffer containing 10 mM Mg2+ (+Mg), in which case the 5′-HPL was completely converted into LL form while LL and CL forms were able to interconvert (Figure 5b, lanes 1–3), or (ii) in buffer containing 10 mM EDTA and no added Mg2+ (−Mg), in which case the ribozyme was inactive, 5′-HPL could not be converted into the LL form, and no CL could be formed from LL (Figure 5b, lanes 4–6). All Lasso species bound CMT and were retarded upon denaturing polyacrylamide gel electrophoresis. Under the +Mg conditions, three discrete bands corresponding to different forms of Lasso-target complexes were detected (Figure 5b, lane 2), whereas under the −Mg conditions, only one major complex band was observed (Figure 5b, lane 5). The identity of each complex was assigned by analyzing the Lasso dissociation products after post-incubation of these complexes under highly denaturing −Mg conditions (90°C, 50% formamide, 10 mM EDTA). We reasoned that under highly denaturing conditions all nontopologically linked complexes between Lasso and CMT should dissociate and free Lasso species should re-appear on d-PAGE. Indeed, we found that all Lasso-CMT complexes formed in the absence of free Mg2+ dissociated under highly denaturing conditions, and the major labeled species released was 5′-HPL, as expected (Figure 5b, lane 6). Thus, we assigned the major band (Figure 5b, lane 5) to the complex of CMT with 5′-HPL. This band also corresponds to the top-shifted, minor band in Figure 5b, lane 2. When Lasso–CMT complexes that formed in the presence of Mg2+ were postincubated at 90°C, the two uppermost gel-shifted bands disappeared, and the LL and 5′-HPL forms of the Lassos reappeared (compare Figure 5b, lanes 2 and 3). We assigned the middle shifted band (Figure 5b, lane 2) to the CMT complex with LL and the bottom-shifted band to the complex with CL, which survived the highly denaturing conditions. Since the CL species did not reappear upon incubation at 90°C (Figure 5b, lane 3) as was the case with the linear target, we concluded that the only surviving band represented a true topologically linked complex between circular Lasso and circular target RNA. In subsequent experiments, we confirmed that this complex did not dissociate even after incubation at 95°C for 10 min (data not shown). The minor shifted bands that appeared in lanes 3 and 6 (Figure 5b) presumably result from partial rehybridization, which may occur during the loading of samples on the gel. In control experiments with the linear target LMT, we did not observe any complexes that could survive the highly denaturing conditions (data not shown).
Lassos can block translation efficiently in vitro
Having demonstrated that Lassos are able to bind rapidly to and form strong complexes with model RNA targets, we tested whether the Lasso ATR1 would be able to block expression of a model mRNA via an in vitro translation assay. This model mRNA, TNF22-Luc, contained the 22-nt TNF sequence that was the target of ATR1 placed upstream of the luciferase gene (Figure 6a). ATR1 was added in increasing amounts to equimolar mixtures of TNF22-Luc and pro-IL-1β mRNAs (a nonspecific control RNA). Following a 40 min incubation with a rabbit reticulocyte lysate in the presence of 35S-labeled methionine, the translation products were separated by SDS–PAGE. At 20- and 40-fold excess of Lasso to target RNA (Figure 6b, lanes 3 and 4), in vitro translation was inhibited by 80 and 92%, respectively, with only a modest reduction in expression of pro-IL-1β (16 and 26%, respectively).
We also demonstrated the superiority of Lasso ATR1, which can circularize around the target, over an antisense RNA that cannot not circularize (AT, derived from ATR1 by deleting the HPR domain), in the inhibition of target RNA translation (see Supplementary Figure 4). In these experiments, we used the same TNF22-Luc mRNA target (Figure 6a) along with a negative control target, which was the same sequence as TNF22-Luc but without the TNF sequence targeted by ATR1 and AT. Transcription of these constructs using T7 RNA polymerase produced mRNAs that were then incubated with either ATR1 or AT RNA and then provided as a template for translation using the rabbit reticulocyte lysate system. Luciferase activity assays revealed that, for an optimal ATR1:target molar ratio of 30:1, Lasso ATR1 provided >95% knockdown of translation of TNF22-Luc mRNA, whereas antisense AT RNA reduced the efficacy of translation only by about 5%.
RNA Lassos can redirect splicing of Fas pre-mRNA
To test for biological activity of Lassos, we decided to evaluate their ability to alter splicing patterns by interfering with the splicing machinery at crucial mRNA sequences. Specifically, we tried to skew Fas pre-RNA splicing towards production of mRNA encoding soluble Fas. To this end, we designed three anti-Fas Lassos (Fas1, Fas2 and Fas3) targeting the Fas pre-mRNA intron 5/exon 6 boundary sequences as well upstream to the splice branch point sequence (Figure 7a and Supplementary Figure 5). We confirmed that the anti-Fas Lassos have the ability to access the target sites and form topologically linked complexes in vitro in tests similar to those described above. The efficiency (rate and yield) of binding of the Fas-specific Lassos to Fas were found to be moderate (data not shown) in comparison to those for the TNF-specific Lassos with their target RNA, but we decided to test the anti-Fas lassos in cells without further in vitro optimization. We reasoned that a cellular environment would provide different Lasso-target binding parameters that could not be predicted based only on the in vitro tests.
Human Jurkat lymphocytes were transfected with the anti-Fas Lassos as described in Materials and Methods section. Total RNA and protein were isolated from the cells in 24–48 h after the transfection. The total RNA was analyzed by RT-PCR using the specific primers (Figure 7b). The resulting PCR-amplified DNA fragments corresponding to full-length Fas (231 bp) and Fas minus exon 6 (169 bp) were analyzed on agarose gels to determine the extent of redirection of splicing (Figure 7c). To confirm that transfection of the Jurkat cells with anti-Fas Lassos results in redirection of Fas pre-mRNA splicing such that exon 6 is excluded, an additional PCR analysis was performed (Figure 7d). In two independent transfection experiments, the 112-bp product lacking exon 6 was only observed following transfection with Lassos directed against the intron 5/exon 6 splice junction. Finally, this fragment was excised from the gel, purified, cloned and sequenced. As expected, no exon 6 sequence was found.
DISCUSSION
In this study, we have introduced a novel class of antisense-based agents called RNA Lassos that contain both HPR and antisense domains. In contrast to the conventional use of ribozymes as sequence-specific nucleases (15,35), RNA Lassos do not cleave their RNA targets. Instead, RNA Lassos hybridize to and circularize around them forming intertwined Lasso-target complexes. Complexes with circular targets are topologically linked and thus cannot be dissociated even under highly denaturing conditions. Lasso complexes with linear RNA targets, however, can be dissociated under highly denaturing conditions although they were found to be more thermostable than if they were simple antisense RNA–RNA duplexes (Figure 3c). Higher thermostability has the potential for higher potency in gene inhibition, as has been demonstrated for chemically modified oligonucleotides such as 2′-OMe, N3′-P5′, morpholino phosphorodiamidates and LNA, which also bind to their targets with high affinity (3–7).
The HPR, a naturally existing ribozyme, was chosen for use in the Lasso design for several reasons, including its catalytic activity (both in vitro and in vivo) as well as the folded, stable structure of its catalytic domain (36,37). In the presence of Mg2+, the HPR catalyzes a reversible, site-specific RNA cleavage and ligation reaction, generating 2′,3′-cyclic phosphate and 5′-OH termini upon scission or reformation of a 3′–5′ phosphodiester bond upon ligation (36). Because HPR substrate sequences are incorporated into both the 5′- and 3′-ends of the primary Lasso transcripts, intramolecular cleavage reactions at each end followed by ligation in cis result in a circular RNA molecule. The size and sequence of the loops connecting the HPR substrate and enzymatic domains can be modified substantially without dramatic effects on HPR catalytic activity as long as the loop sequences do not prevent the formation of catalytically active HPR structures (28,38–40). Although the HPR is particularly well-suited for this application, other ribozymes that combine both cleavage and ligation activities could be evolved by in vitro selection (41) and used in place of the HPR in Lassos.
RNA Lassos bear some resemblance to DNA padlock probes (42,43), which also circularize around polynucleotide targets. However, the two differ in important respects. Padlock probes are 70–100 nt ODNs whose ends can bind to adjacent sites on a target nucleic acid in such a way that they can be ligated. Ligation is accomplished by addition of a DNA ligase and yields a circular padlock probe linked to the target. In contrast, Lassos are RNAs of 120–130 nt, are generally made by transcription in vitro, and use an internal HPR domain to self-process the ends of the primary Lasso transcripts. This reaction produces ends that, using the same HPR activity, can reversibly circularize in a nontarget-dependent manner with no need for an added ligase. Unlike with padlock probes, the ends of the Lasso are not bound to the polynucleotide target prior to ligation. The ability to be transcribed allows Lassos to be expressed from DNA vectors in vivo, whereupon they can spontaneously self-process and self-circularize within the cell. Padlock probes, on the other hand, must be prepared by chemical synthesis and require careful purification to isolate products that can be ligated in a target-dependent manner.
In our initial Lasso design, exemplified by ATR1, we included a triplex-forming sequence with the expectation that this feature would promote Lasso circularization around target RNA by guiding the 3′-end of the Lasso to fold back such that it would aid in bringing the ends into close proximity for efficient self-ligation (Figure 1a). We found that both ATR1 and a variant (ALR562) that lacked the triplex-forming element could circularize around the same target (Figures 3c and 5a). We therefore concluded that the triplex-forming domain was not required for the Lasso to circularize around the target RNA. Of the two Lasso designs, the simpler one lacking the triplex-forming sequence is more versatile because there is no requirement for the target region to have the homopurine character needed for stable triplex formation. Comparison of the binding and circularization properties of these two Lassos indicates that variations in the sequences of both the HPR domain and the linkers surrounding the antisense domain can affect the rate of target hybridization and the efficiency of self-ligation. Interestingly, circularization of Lasso ALR562-2 is promoted by binding to the target whereas circularization of ATR1 is not. Although unintended, this result indicates that target-dependent Lasso circularization can be achieved in ways other than through triplex formation. The study of such mechanisms will be published elsewhere (A. Dallas, B.H. Johnston, and S.A. Kazakov, manuscript in preparation).
Because of the modular nature of the RNA Lasso, it can be modified easily to swap in antisense sequences for any gene of interest. The sequence and length of the antisense sequence can be varied to optimize the specificity of target recognition as well as the rate of hybridization and yield of complex. Antisense sequence lengths in the range 18–25 nt appear to be optimal for specifying a unique target sequence within the genome while minimizing the formation of stable, partially mismatched duplexes at unintended target sites (i.e. off-target effects). However, if one takes into account variations in the number of genes expressed in specific tissues at a given time (44) as well as estimates of the amount of mRNA sequences that are thought to be inaccessible because of intramolecular structure and intermolecular macromolecular interactions (45), as little as 14 nt may be sufficient to confer specificity.
We have demonstrated that both linear and circular Lasso species can efficiently hybridize to and circularize around target RNAs (Figures 3 and 4). Under target-binding conditions, fully processed Lasso molecules exist as interconverting linear and circular forms like any HPR derivatives. The structures of the linear and circular forms are probably similarly folded except that in the linear form, the 5′- and 3′-termini are not covalently linked (see bottom structures in Figure 2). We speculate that this interaction provides Lassos with an inherent potential to be more sequence specific than ordinary antisense RNA based on two factors. One factor is their circularity: circular antisense DNAs are known to provide greater sequence selectivity (i.e. less tolerance for mismatches) than corresponding linear molecules (46–48). The other factor is the presence of a stringency element, a feature also shared with molecular beacons. Like a DNA molecular beacon (49), the Lasso cannot fully pair with a target without disrupting its own structure. It first interacts with a target through formation of a few base pairs, but to fully pair its antisense sequence, its termini must unfold, disrupting intramolecular pairing interactions, to allow extension of the target-antisense helix. It can then refold into the ‘circular’ conformation where the termini are adjacent to each other. Based on this model, we would expect the specificity of any given Lasso to be influenced by its GC-content as well as the accessibility of its target sequence since internal structure in targets as well as the antisense molecule can act as a stringency element. A study on how Lasso sequence manipulations affect their sequence specificity will be described elsewhere (A. Dallas, B.H. Johnston, and S.A. Kazakov, manuscript in preparation).
There is now clear evidence of the in vivo efficacy of RNA-based drugs, both synthetic and vector expressed (50). Lassos can be generated by either in vitro transcription or in vivo expression from vectors. Like other circular RNA molecules (23,51,52), Lassos should be more stable in biological fluids than linear RNAs since circularization prevents RNA digestion by exonucleases. Circularizable antisense molecules could also provide more potent knockdown of cellular targets than linear ones because of the enhanced thermostability of their complexes with targets (42,53).
In this study, we demonstrated that expression of a TNF22-Luc reporter construct could be blocked efficiently and sequence specifically by Lasso RNAs in a model in vitro translation assay. Similar luciferase fusion constructs have been previously used for screening of large numbers of antisense agents, both in cell lysates and in cells. The inhibitory profiles obtained for such gene surrogates were usually found to be similar to those observed in the corresponding endogenous genes assayed at the mRNA level (54). For our studies, we used murine TNF-α as a model target RNA, as the human form of this cytokine has been validated as a therapeutic target for treatment of chronic inflammation associated with many diseases, including rheumatoid arthritis, psoriasis and HCV-related liver necrosis (55). Since the target in the TNF22-Luc reporter is upstream of the AUG start codon, we speculate that the mechanism of translation inhibition is steric interference with the formation of the initiation complex or ribosome scanning.
Another possible application of the Lasso is to regulate alternative splicing. In this work we demonstrated the modest ability of several Lassos to redirect splicing of Fas-pre-mRNA. Many tumor cells express Fas ligand as a way to kill Fas-expressing tumor-infiltrating lymphocytes (TILs). The interaction between Fas and Fas ligand, which results in apoptosis of TILs, is an increasingly attractive cancer therapeutic target (26,56). The ability to redirect splicing could allow skipping of an exon of Fas that is responsible for membrane anchoring, resulting in a soluble receptor that cannot transduce signal. Thus, not only is the receptor downregulated, resulting in unresponsiveness of the affected cells, but secretion of the soluble receptor that is the new splicing product results in binding and inactivation of extracellular ligand with the result that neighboring cells are also protected.
The efficacy of anti-Fas Lassos described in this work was less than those provided by alternative antisense approaches using synthetic morpholino oligonucleotides (57–59). It remains to be seen to what extent this efficacy can be improved by selecting better target sequences and adjusting the design of the Lasso. It is well known that, on average, <5% of tested antisense agents yield high potency in cells (60–62). Randomization of certain sequences in Lassos together with in vitro selection methods can identify more potent Lasso molecules (unpublished results).
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
National Institutes of Health (1R43CA65179, 1R43GM54956, 2R44GM54956 to B.H.J., 1R43CA103317 to R.L.K., 1R43AI074255 to S.A.K.). The Open Access publication charges were partially waived by Oxford University Press.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Attila A. Seyhan for recommendations on the design of the HPR domain in ALR562-2, Babak N. Alizadeh for help in chemiluminescence-based in vitro translation assays and Michelle Stroud for assistance in the splicing redirection experiments. We also thank the Department of Chemistry and Biochemistry at Brigham Young University for providing the equipment to perform the analysis of in vitro translation assays.
REFERENCES
- 1.Crooke ST. Antisense strategies. Curr. Mol. Med. 2004;4:465–487. doi: 10.2174/1566524043360375. [DOI] [PubMed] [Google Scholar]
- 2.Dias N, Stein CA. Antisense oligonucleotides: basic concepts and mechanisms. Mol. Cancer Ther. 2002;1:347–355. [PubMed] [Google Scholar]
- 3.Braasch DA, Corey DR. Novel antisense and peptide nucleic acid strategies for controlling gene expression. Biochemistry. 2002;41:4503–4510. doi: 10.1021/bi0122112. [DOI] [PubMed] [Google Scholar]
- 4.Heasman J. Morpholino oligos: making sense of antisense? Dev. Biol. 2002;243:209–214. doi: 10.1006/dbio.2001.0565. [DOI] [PubMed] [Google Scholar]
- 5.Kurreck J. Antisense technologies. Improvement through novel chemical modifications. Eur. J. Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 6.Toulme JJ. New candidates for true antisense. Nat. Biotechnol. 2001;19:17–18. doi: 10.1038/83451. [DOI] [PubMed] [Google Scholar]
- 7.Vester B, Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43:13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- 8.Chadwick DR, Lever AM. Antisense RNA sequences targeting the 5′ leader packaging signal region of human immunodeficiency virus type-1 inhibits viral replication at post-transcriptional stages of the life cycle. Gene Ther. 2000;7:1362–1368. doi: 10.1038/sj.gt.3301254. [DOI] [PubMed] [Google Scholar]
- 9.De Backer MD, Raponi M, Arndt GM. RNA-mediated gene silencing in non-pathogenic and pathogenic fungi. Curr. Opin. Microbiol. 2002;5:323–329. doi: 10.1016/s1369-5274(02)00319-3. [DOI] [PubMed] [Google Scholar]
- 10.Ji Y, Yin D, Fox B, Holmes DJ, Payne D, Rosenberg M. Validation of antibacterial mechanism of action using regulated antisense RNA expression in Staphylococcus aureus. FEMS Microbiol. Lett. 2004;231:177–184. doi: 10.1016/S0378-1097(03)00931-5. [DOI] [PubMed] [Google Scholar]
- 11.Ji Y, Zhang B, Van SF, Horn, Warren P, Woodnutt G, Burnham MK, Rosenberg M. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science. 2001;293:2266–2269. doi: 10.1126/science.1063566. [DOI] [PubMed] [Google Scholar]
- 12.Varga LV, Toth S, Novak I, Falus A. Antisense strategies: functions and applications in immunology. Immunol. Lett. 1999;69:217–224. doi: 10.1016/s0165-2478(99)00082-6. [DOI] [PubMed] [Google Scholar]
- 13.Yin D, Ji Y. Genomic analysis using conditional phenotypes generated by antisense RNA. Curr. Opin. Microbiol. 2002;5:330–333. doi: 10.1016/s1369-5274(02)00315-6. [DOI] [PubMed] [Google Scholar]
- 14.Scherer LJ, Rossi JJ. Approaches for the sequence-specific knockdown of mRNA. Nat. Biotechnol. 2003;21:1457–1465. doi: 10.1038/nbt915. [DOI] [PubMed] [Google Scholar]
- 15.Schubert S, Kurreck J. Ribozyme- and deoxyribozyme-strategies for medical applications. Curr. Drug Targets. 2004;5:667–681. doi: 10.2174/1389450043345092. [DOI] [PubMed] [Google Scholar]
- 16.Li M, Rossi JJ. Lentiviral vector delivery of siRNA and shRNA encoding genes into cultured and primary hematopoietic cells. Methods Mol. Biol. 2005;309:261–272. doi: 10.1385/1-59259-935-4:261. [DOI] [PubMed] [Google Scholar]
- 17.Sano M, Kato Y, Akashi H, Miyagishi M, Taira K. Novel methods for expressing RNA interference in human cells. Methods Enzymol. 2005;392:97–112. doi: 10.1016/S0076-6879(04)92006-X. [DOI] [PubMed] [Google Scholar]
- 18.Zhang YC, Taylor MM, Samson WK, Phillips MI. Antisense inhibition: oligonucleotides, ribozymes, and siRNAs. Methods Mol. Med. 2005;106:11–34. [PubMed] [Google Scholar]
- 19.Kazakov SA, Balatskaya SV, Johnston BH. Ligation of the hairpin ribozyme in cis induced by freezing and dehydration. RNA. 2006;12:446–456. doi: 10.1261/rna.2123506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kisich KO, Freedland SJ, Erickson KL. Factors altering ribozyme-mediated cleavage of tumor necrosis factor-alpha mRNA in vitro. Biochem. Biophys. Res. Commun. 1997;236:205–211. doi: 10.1006/bbrc.1997.6934. [DOI] [PubMed] [Google Scholar]
- 21.Beaudry D, Perreault JP. An efficient strategy for the synthesis of circular RNA molecules. Nucleic Acids Res. 1995;23:3064–3066. doi: 10.1093/nar/23.15.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrison B, Zimmerman SB. Polymer-stimulated ligation: enhanced ligation of oligo- and polynucleotides by T4 RNA ligase in polymer solutions. Nucleic Acids Res. 1984;12:8235–8251. doi: 10.1093/nar/12.21.8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puttaraju M, Perrotta AT, Been MD. A circular trans-acting hepatitis delta virus ribozyme. Nucleic Acids Res. 1993;21:4253–4258. doi: 10.1093/nar/21.18.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saville BJ, Collins RA. RNA-mediated ligation of self-cleavage products of a Neurospora mitochondrial plasmid transcript. Proc. Natl Acad. Sci. USA. 1991;88:8826–8830. doi: 10.1073/pnas.88.19.8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jobling SA, Auron PE, Gurka G, Webb AC, McDonald B, Rosenwasser LJ, Gehrke L. Biological activity and receptor binding of human prointerleukin-1 beta and subpeptides. J. Biol. Chem. 1988;263:16372–16378. [PubMed] [Google Scholar]
- 26.Shiraki K, Tsuji N, Shioda T, Isselbacher KJ, Takahashi H. Expression of Fas ligand in liver metastases of human colonic adenocarcinomas. Proc. Natl Acad. Sci. USA. 1997;94:6420–6425. doi: 10.1073/pnas.94.12.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saenger W. Principles of Nucleic Acid Structure. Springer: New York; 1984. [Google Scholar]
- 28.Feldstein PA, Bruening G. Catalytically active geometry in the reversible circularization of ‘mini-monomer’ RNAs derived from the complementary strand of tobacco ringspot virus satellite RNA. Nucleic Acids Res. 1993;21:1991–1998. doi: 10.1093/nar/21.8.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kazakov SA, Balatskaya SV, Johnston BH. In: Structure, Motion, Interaction, and Expression of Biological Macromolecules. Sarma RH, Sarma MH, editors. Vol. 2. Adenine Press: Albany, New York; 1998. pp. 155–161. [Google Scholar]
- 30.Pieper S, Vauleon S, Muller S. RNA self-processing towards changed topology and sequence oligomerization. Biol. Chem. 2007;388:743–746. doi: 10.1515/BC.2007.067. [DOI] [PubMed] [Google Scholar]
- 31.Esteban JA, Banerjee AR, Burke JM. Kinetic mechanism of the hairpin ribozyme. Identification and characterization of two nonexchangeable conformations. J. Biol. Chem. 1997;272:13629–13639. doi: 10.1074/jbc.272.21.13629. [DOI] [PubMed] [Google Scholar]
- 32.Cheng YK, Pettitt BM. Stabilities of double- and triple-strand helical nucleic acids. Prog. Biophys. Mol. Biol. 1992;58:225–257. doi: 10.1016/0079-6107(92)90007-s. [DOI] [PubMed] [Google Scholar]
- 33.Roberts RW, Crothers DM. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science. 1992;258:1463–1466. doi: 10.1126/science.1279808. [DOI] [PubMed] [Google Scholar]
- 34.Semerad CL, Maher L.J., 3rd Exclusion of RNA strands from a purine motif triple helix. Nucleic Acids Res. 1994;22:5321–5325. doi: 10.1093/nar/22.24.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tafech A, Bassett T, Sparanese D, Lee CH. Destroying RNA as a therapeutic approach. Curr. Med. Chem. 2006;13:863–881. doi: 10.2174/092986706776361021. [DOI] [PubMed] [Google Scholar]
- 36.Fedor MJ. Structure and function of the hairpin ribozyme. J. Mol. Biol. 2000;297:269–291. doi: 10.1006/jmbi.2000.3560. [DOI] [PubMed] [Google Scholar]
- 37.Wilson TJ, Nahas M, Ha T, Lilley DM. Folding and catalysis of the hairpin ribozyme. Biochem. Soc. Trans. 2005;33:461–465. doi: 10.1042/BST0330461. [DOI] [PubMed] [Google Scholar]
- 38.Komatsu Y, Koizumi M, Sekiguchi A, Ohtsuka E. Cross-ligation and exchange reactions catalyzed by hairpin ribozymes. Nucleic Acids Res. 1993;21:185–190. doi: 10.1093/nar/21.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Komatsu Y, Kanzaki I, Koizumi M, Ohtsuka E. Construction of new hairpin ribozymes with replaced domains. Nucleic Acids Symp. Ser. 1995:223–224. [PubMed] [Google Scholar]
- 40.Berzal-Herranz A, Burke JM. Ligation of RNA molecules by the hairpin ribozyme. Methods Mol. Biol. 1997;74:349–355. doi: 10.1385/0-89603-389-9:349. [DOI] [PubMed] [Google Scholar]
- 41.Joyce GF. Directed evolution of nucleic acid enzymes. Annu. Rev. Biochem. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- 42.Nilsson M, Malmgren H, Samiotaki M, Kwiatkowski M, Chowdhary BP, Landegren U. Padlock probes: circularizing oligonucleotides for localized DNA detection. Science. 1994;265:2085–2088. doi: 10.1126/science.7522346. [DOI] [PubMed] [Google Scholar]
- 43.Nilsson M, Barbany G, Antson DO, Gertow K, Landegren U. Enhanced detection and distinction of RNA by enzymatic probe ligation. Nat. Biotechnol. 2000;18:791–793. doi: 10.1038/77367. [DOI] [PubMed] [Google Scholar]
- 44.Nielsen PE. Peptide nucleic acids: on the road to new gene therapeutic drugs. Pharmacol. Toxicol. 2000;86:3–7. doi: 10.1034/j.1600-0773.2000.pto860102.x. [DOI] [PubMed] [Google Scholar]
- 45.Bergeron LJ, Ouellet J, Perreault JP. Ribozyme-based gene-inactivation systems require a fine comprehension of their substrate specificities; the case of delta ribozyme. Curr. Med. Chem. 2003;10:2589–2597. doi: 10.2174/0929867033456486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kool ET. Molecular recognition by circular oligonucleotides: increasing the selectivity of DNA binding. J. Am. Chem. Soc. 1991;113:6265–6266. [Google Scholar]
- 47.Kool ET. Circular oligonucleotides: new concepts in oligonucleotide design. Annu. Rev. Biophys. Biomol. Struct. 1996;25:1–28. doi: 10.1146/annurev.bb.25.060196.000245. [DOI] [PubMed] [Google Scholar]
- 48.Wang S, Kool ET. Circular RNA oligonucleotides. Synthesis, nucleic acid binding properties, and a comparison with circular DNAs. Nucleic Acids Res. 1994;22:2326–2333. doi: 10.1093/nar/22.12.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 50.Eckstein F. The versatility of oligonucleotides as potential therapeutics. Expert Opin. Biol. Ther. 2007;7:1021–1034. doi: 10.1517/14712598.7.7.1021. [DOI] [PubMed] [Google Scholar]
- 51.Bohjanen PR, Colvin RA, Puttaraju M, Been MD, Garcia-Blanco MA. A small circular TAR RNA decoy specifically inhibits Tat-activated HIV-1 transcription. Nucleic Acids Res. 1996;24:3733–3738. doi: 10.1093/nar/24.19.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Puttaraju M, Been MD. Circular ribozymes generated in Escherichia coli using group I self-splicing permuted intron-exon sequences. J. Biol. Chem. 1996;271:26081–26087. doi: 10.1074/jbc.271.42.26081. [DOI] [PubMed] [Google Scholar]
- 53.Gryaznov S, Lloyd D. Oligonucleotide clamps having diagnostic and therapeutic applications. United States Patent No. 6048974. 2000 [Google Scholar]
- 54.Husken D, Asselbergs F, Kinzel B, Natt F, Weiler J, Martin P, Haner R, Hall J. mRNA fusion constructs serve in a general cell-based assay to profile oligonucleotide activity. Nucleic Acids Res. 2003;31:e102. doi: 10.1093/nar/gng103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor PC, Williams RO, Feldmann M. Tumour necrosis factor alpha as a therapeutic target for immune-mediated inflammatory diseases. Curr. Opin. Biotechnol. 2004;15:557–563. doi: 10.1016/j.copbio.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 56.Mercatante D, Kole R. Modification of alternative splicing pathways as a potential approach to chemotherapy. Pharmacol. Ther. 2000;85:237–243. doi: 10.1016/s0163-7258(99)00067-4. [DOI] [PubMed] [Google Scholar]
- 57.Sierakowska H, Agrawal S, Kole R. Antisense oligonucleotides as modulators of pre-mRNA splicing. Methods Mol. Biol. 2000;133:223–233. doi: 10.1385/1-59259-215-5:223. [DOI] [PubMed] [Google Scholar]
- 58.Skordis LA, Dunckley MG, Yue B, Eperon IC, Muntoni F. Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc. Natl Acad. Sci. USA. 2003;100:4114–4119. doi: 10.1073/pnas.0633863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vacek M, Sazani P, Kole R. Antisense-mediated redirection of mRNA splicing. Cell Mol. Life Sci. 2003;60:825–833. doi: 10.1007/s00018-003-3042-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grunweller A, Wyszko E, Bieber B, Jahnel R, Erdmann VA, Kurreck J. Comparison of different antisense strategies in mammalian cells using locked nucleic acids, 2′-O-methyl RNA, phosphorothioates and small interfering RNA. Nucleic Acids Res. 2003;31:3185–3193. doi: 10.1093/nar/gkg409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miyagishi M, Hayashi M, Taira K. Comparison of the suppressive effects of antisense oligonucleotides and siRNAs directed against the same targets in mammalian cells. Antisense Nucleic Acid Drug Dev. 2003;13:1–7. doi: 10.1089/108729003764097296. [DOI] [PubMed] [Google Scholar]
- 62.Sohail M, Southern EM. Selecting optimal antisense reagents. Adv. Drug Deliv. Rev. 2000;44:23–34. doi: 10.1016/s0169-409x(00)00081-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.