Abstract
Exposure to ultraviolet light induces a number of forms of damage in DNA, of which (6–4) photoproducts present the most formidable challenge to DNA replication. No single DNA polymerase has been shown to bypass these lesions efficiently in vitro suggesting that the coordinate use of a number of different enzymes is required in vivo. To further understand the mechanisms and control of lesion bypass in vivo, we have devised a plasmid-based system to study the replication of site-specific T–T(6–4) photoproducts in chicken DT40 cells. We show that DNA polymerase ζ is absolutely required for translesion synthesis (TLS) of this lesion, while loss of DNA polymerase η has no detectable effect. We also show that either the polymerase-binding domain of REV1 or ubiquitinated PCNA is required for the recruitment of Polζ as the catalytic TLS polymerase. Finally, we demonstrate a previously unappreciated role for REV1 in ensuring bypass synthesis remains in frame with the template. Our data therefore suggest that REV1 not only helps to coordinate the delivery of DNA polymerase ζ to a stalled primer terminus but also restrains its activity to ensure that nucleotides are incorporated in register with the template strand.
INTRODUCTION
DNA damage-induced mutations arise in consequence of the use of specialized DNA polymerases to synthesize across base lesions that have arrested DNA replication, a process known as translesion synthesis (TLS). These polymerases, which are found in all organisms, possess active sites that are more tolerant of damage to, or distortions of, the DNA template that would stall the replicative polymerases (1,2). Although deployment of TLS allows replication to be completed, it does so at the cost of an increased error rate. This is not only because most DNA lesions are non- or mis-instructional, but also because the TLS polymerases themselves are error-prone. Although it is likely that the use of TLS is frequently anti-mutagenic at a given DNA lesion, the high intrinsic error rate of many of these enzymes means that the access of the translesion polymerases to nascent 3′-termini must be tightly regulated.
Originally identified in the budding yeast Saccharomyces cerevisiae, the ubiquitination of the DNA sliding clamp PCNA (POL30) at lysine 164 by RAD6/RAD18 appears to be a conserved mechanism in eukaryotes for recruitment of translesion polymerases (3,4). The modification is proposed to increase the affinity of the clamp for the Y-family translesion polymerases (Pols η, ι, κ and REV1) through an interaction with ubiquitin-binding domains located within the polymerases themselves (5). A B-family polymerase, DNA polymerase ζ, is also required for effective translesion synthesis. Polζ comprises two subunits, REV3, which includes the catalytic domain and REV7, whose functions are less well understood (6). REV7 also interacts with the extreme C-terminus of REV1 (7), a domain that additionally interacts with each of the Y family polymerases (8). Polζ is conserved from yeast to mammals, and although vertebrate REV3 is much larger than its yeast counterpart, the function of the additional sequence is unclear.
Monoubiquitination of PCNA is also needed for a more poorly understood error-free recombinational mode of bypass in S. cerevisiae (9), thought to be a form of template switching (10). In this context, the initial ubiquitin conjugated to PCNA provides the seed for subsequent lysine 63-linked polyubiquitination by RAD5/MMS2/UBC13 (3). Although the function of the polyubiquitin chain is currently unclear, RAD5 has also been shown to be a helicase capable of catalysing fork regression in vitro (11). There is evidence that many aspects of this mechanism are conserved in higher eukaryotes although to date there is only relatively limited genetic data to support its importance (12–14).
In the study presented here, we employ the genetically tractable chicken cell line DT40, which currently provides the most versatile genetic system for studying vertebrate DNA damage tolerance mechanisms (15). Homologues of the S. cerevisiae RAD6 epistasis group, which comprises genes involved in DNA damage tolerance, are present in DT40 and are well conserved between chicken and human (16). Ubiquitination of PCNA plays an important role in DNA damage tolerance in DT40 (17,18), although the role of this modification is not as central as it is in yeast (17–20) and RAD18 does not appear to be the sole ubiquitin ligase responsible (18). Further, the C-terminal region of the Y-family polymerase REV1 has acquired greater prominence in the control of lesion bypass in DT40 compared with yeast and is able to act independently of PCNA ubiquitination in coordinating translesion synthesis (19).
A particularly powerful method for studying DNA damage tolerance is to monitor the outcome of the replication of a defined lesion at a known site in shuttle plasmids. Of the many approaches taken to this problem, we were drawn to an experimental arrangement devised by Lawrence and colleagues that seemed well suited for adaptation to use in a vertebrate cell system. The system comprises a plasmid capable of episomal replication in which a thymine–thymine pyrimidine (6–4) pyrimidone photoproduct is incorporated into each strand, staggered 28-bp apart. Each photoproduct is placed opposite a C–C mismatch (21.22). This arrangement allows the unbiased detection, in recovered replicated copies, of TLS on either strand or error-free bypass.
Although produced in UV-irradiated DNA at only about a third of the frequency of the cyclobutane pyrimidine dimer, the (6–4) photoproduct presents a much more potent block to replication (23) by introducing very significant distortion into the DNA backbone with the 3′ base orientated almost perpendicular to the 5′ base (24,25). Indeed, in vitro, no single polymerase is able to efficiently bypass this lesion leading to the proposal that, in vivo, bypass is a two-step process with one polymerase incorporating opposite the 3′T and a second performing the second incorporation and extension (26).
To allow direct comparison with the work of Zhang and Lawrence, we have devised a plasmid capable of replicating in DT40 into which we incorporate a pyrimidine–pyrimidone T–T(6–4) photoproduct as a model DNA replication-stalling lesion. Using this system, we find that TLS is used some ten times more frequently in DT40 than has been reported for S. cerevisiae (22). Interestingly, loss of PCNA ubiquitination does not affect the ability of the cells to perform error-free bypass, but does play a role in recruitment of DNA polymerase ζ, which is essential for TLS of this lesion, despite no known direct interaction between ubiquitin and Polζ. Further, we show that REV1 not only acts in parallel with PCNA ubiquitination to facilitate Polζ-dependent bypass, but that it also modifies the catalytic behaviour of Polζ, restraining its synthetic activity to ensure the frame of bypass is maintained.
MATERIALS AND METHODS
Plasmid construction
The pQ1 plasmid was constructed from two halves. First, pentameric Gal4 sites provided by the oligonucleotides 5′-CGCGA(CGGAGGACAGTACTCCGCT)5A and 5′-CGCGT(AGCGGAGTACTGTCCTCCG)5T were ligated into an MluI site created in pIRES2-EGFP (Clontech, St. Germain-en-Laye, France) by site-directed mutagenesis using the primers 5′-CAATGTATCTTAACGCGTAAATTGTAAG and 5′-CTTACAATTTACGCGTTAAGATACATTG. This ligation preserved an MluI site downstream of the insertion. After filling in the only MfeI site and religation, the KanR region was removed from the plasmid using MluI and NsiI. The replacement AmpR gene was provided by a pBluescript plasmid (Stratagene, Amsterdam, The Netherlands) which had a polylinker inserted into the blunt PsiI site using the oligonucleotides 5′-GAATTCGGTACCCATATGCTGCAG and 5′-CTGCAGCATATGGGTACCGAATTC to allow for the cloning of lesion-containing oligonucleotides into the finished pQ1. A region of the modified pBluescript was amplified using the primers 5′-GGTACGCGTCGCGCCCTGTAGCGGCGC (containing an MluI site) and 5′-GCACCACTGCAGTGGGAACATGTGAGCAAAAGGCC (containing a PstI site flanked by two halves of a BstXI site) and cut with MluI and BstXI (which mimics a PstI cut in the primer). The cut PCR product was ligated into the MluI and NsiI cut modified pIRES2-EGFP, giving pQ1ΔCDC6. One EcoRI site and two PstI sites were removed silently from human CDC6 by site-directed mutagenesis, and the product was amplified using primers 5′-GAGTCGACCATGCCTCAAACCCGATCCCAG and 5′-GAGGATCCTTAAGGCAATCCAGTAGCTAAG containing SalI and BamHI sites. The DNA-binding domain of GAL4 was cut out of pDBLeu (Stratagene) using HindIII and SalI. The GAL4DBD-hCDC6 fusion was assembled in pBluescript (Stratagene) containing a modified polylinker (NheI–HindIII–SalI–BamHI), and subcloned into pQ1ΔCDC6 using NheI and BamHI.
The kanamycin-resistant pQ2 control plasmid was based on pQ1. A SacI site was created in pQ1 between the AmpR cassette and the pUC origin by site-directed mutagenesis using the primers 5′-CCCTTAACGTGAGAGCTCGTTCCACTGAGCG and 5′-CGCTCAGTGGAACGAGCTCTCACGTTAAGGG. The AmpR cassette was removed using PstI and SacI, and replaced with the KanR cassette amplified from pCR2.1-TOPO using the primers 5′-CACTGCAGGGCGCAAGGGCTGCTAAAGG and 5′-CAGAGCTCAGAAGAACTCGTCAAGAAGG containing sites for PstI and SacI, respectively.
Oligonucleotides containing a T–T(6–4) photoproduct were synthesized using a previously described building block (27). The following oligonucleotides were used: TTF1 (AATTGTCCACCTC-T(6–4)T-CCTGTATTCTTAGTACCTACTGACGCTAGCTCGATCCATGCA), TTR1 (TGGATCGA-T(6–4)T-TAGCGTCAGTAGGTACTAAGAATACAGGGCGAGGTGGAC), TTR2 (TGGATCGAGCTAGCGTCAGTAGGTACTAAGAATACAGG-T(6–4)T-GAGGTGGAC). In the lesion-free control oligonucleotides, TTFC and TTRC, the photoproducts were replaced by the dinucleotide GC. The inserts for the lesion-containing constructs were annealed pairs of oligonucleotides as follows: QTs (TTF1 and TTR1), QTo (TTF1 and TTR2), QTc (TTFC and TTRC). The annealed inserts were ligated into pQ1 in two steps as described (22). The final ligation mix was found to contain over 75% circular product, and was used for transfection without a gel purification step.
Cell culture and transfection
DT40 cells were grown at 37°C in RPMI medium supplemented with 7% fetal bovine serum and 3% chicken serum. The DT40 lines null for polh, rev3, rad18 and rev1 and the pcnaK164R and rev1 pcnaK164R mutants have been described previously (17,28–31). Selection cassettes were excised from the rev1 and pcnaK164 lines, together with the AID transgene these lines contain, by the addition of 50 nM 5-hydroxytamoxifen to the media for Cre induction. A puromycin resistant XPA knockout construct (32) was used to disrupt the single xpa allele and recreate the xpa cell line as well as generate double mutants. The hREV1 reconstitutions have been described (19,20). Colony survival assay on methylcellulose-containing medium was performed as previously described (29). All transient transfections were performed using an Amaxa nucleofector device (Amaxa, Cologne, Germany) with reagent T according to the manufacturer's instructions. Three million DT40 cells were transfected and immediately recovered into 5 ml warm medium. The amount of plasmid transfected was optimized so as to produce <10% EGFP-positive cells to limit the number of cells expected to take up more than one copy of the plasmid. For transfections with ligated shuttle plasmid, the ligation product of 1 µg pQ1 was used (containing ∼0.1 µg ligated circular plasmid), supplemented with 0.4 µg pQ2 in experiments used for the measurement of plasmid replication efficiency. EGFP expression from the plasmid was analysed at various time points after transfection using a Becton–Dickinson FacsCalibur flow cytometer.
Plasmid extraction and bacterial transformation
Cells transfected with lesion-containing plasmids were cultured for 48 h before plasmids were extracted using a simplified Hirt protocol. PBS washed cells were resuspended in buffer P1 from a Qiagen plasmid miniprep kit (Qiagen, Crawley, UK), followed by a 5-min lysis in buffer P2 and neutralization in N3. Plasmid DNA was recovered from the supernatant of a high-speed spin by the addition of glycogen and isopropanol precipitation. Dried pellets were dissolved in 10 µl enzyme digest mix containing 10 U DpnI and incubated for 30 min. DNA was once more recovered by isopropanol/ethanol precipitation, dissolved in water and used to transform DH10B electrocompetent cells (Invitrogen, Paisley, UK) using a Bio-Rad Gene Pulser (Bio-Rad, Hemel Hempstead, UK) at 200 Ω, 0.25 μF and 1.8 kV. In experiments assaying plasmid replication efficiency, 80% of transfections were plated on ampicillin plates and 20% on kanamycin-containing medium.
RESULTS
A replicating plasmid in DT40 cells
To exploit the powerful genetics afforded by DT40 for monitoring lesion bypass in replicating plasmids, we first set out to identify a plasmid that replicated reliably in this cell line. We initially explored the use of pEPI-1 (33), a gift of Professor Hans Lipps. This plasmid expresses GFP from a CMV promoter and contains a human matrix attachment region. In human cells, this leads to the chromatinization of the plasmid and its replication and stable maintenance during prolonged culture. In DT40, however, pEPI was subject to aggressive degradation following transfection and was poorly maintained (data not shown). Copies of the plasmid that have been replicated in DT40 become resistant to cleavage by the restriction enzyme DpnI due to loss of Dam methylation. Although some replication of pEPI did take place, as assessed by the recovery of DpnI-resistant copies of the plasmid from Hirt supernatants, the frequency with which such copies were recovered was very low.
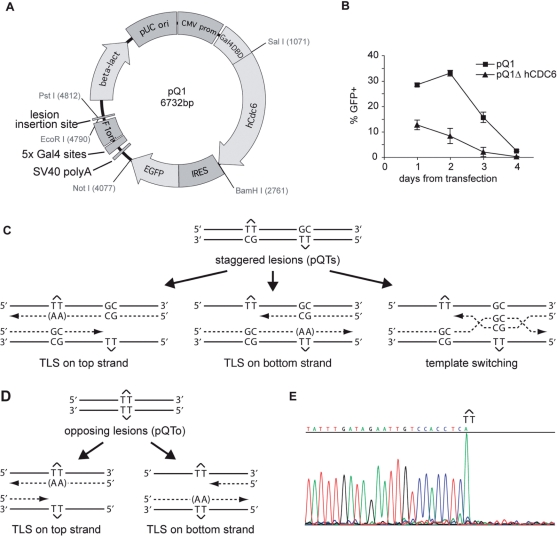
We therefore constructed a plasmid, pQ1, based on the observation that tethering CDC6 to DNA is sufficient to create an origin of replication in human cells (34). pQ1 consists of a CMV promoter-driven human CDC6 fused with an N-terminal GAL4 DNA-binding domain. An internal ribosomal entry signal drives expression of enhanced green fluorescent protein. Five repeats of the GAL4 recognition sequence are placed in an A:T base pair rich region of the plasmid 200-bp upstream of a cloning site into which the lesion-containing oligonucleotide is ligated (Figure 1A).
Figure 1.
A system for monitoring lesion bypass in a replicating plasmid. (A) Layout of the pQ1 shuttle plasmid. pUC ori, pUC bacterial replication origin; CMV prom, cytomegalovirus promoter; Gal4-DBD, GAL4 DNA-binding domain; hCdc6, human CDC6 coding sequence; IRES, internal ribosome entry sequence; EGFP, enhanced green fluorescent protein; 5 × Gal4 sites, pentameric Gal4 binding sites. (B) A daily time course of the percentage of wild-type DT40 cells expressing GFP after transfection with 5 µg of pQ1 or a variant lacking the human CDC6 reading frame (pQ1ΔCDC6). Error bars represent 1 SD. (C) A schematic of the staggered arrangement of T–T(6–4) photoproducts in the construct pQTs, with the dinucleotide GC placed opposite each lesion, and 28 bp between the lesion with the possible outcomes of DNA replication over the area. TLS may occur on either the top or the bottom strand, with the most common base insertion shown as (AA). Alternatively, the nascent strand of the sister chromatid may be used as an alternative undamaged template; one possible layout for such a template switching mechanism is illustrated. (D) A schematic representation of the opposing arrangement of T–T(6–4) photoproducts in the construct pQTo and the possible outcomes of DNA replication, only by TLS, over the lesion. (E) Part of a DNA sequencing reaction of the pQTo construct is shown to illustrate the purity of the preparation. The sequencing polymerase stalls at the lesion, and inserts an adenosine opposite the 3′T.
To assess the replication and maintenance of this plasmid following transient transfection, wild-type DT40 cells were transfected with 5 µg pQ1 or pQ1ΔCDC6, a variant of pQ1 lacking the HsCDC6 open reading frame. The percentage of GFP positive cells was monitored by flow cytometry at 24 h (the earliest point at which strong GFP expression could be detected) and then at 24 h intervals thereafter. In the absence of CDC6, the percentage of GFP positive cells declined steadily over four days after transfection. However, with GAL4-HsCDC6 present, the percentage of GFP positive cells increased for 2 days before the plasmid began to be lost from the population (Figure 1B). The decay in GFP expression in both cases was accompanied by extensive degradation of the plasmid as assessed by Southern blot (data not shown). Although replication of pQ1 is tempered by competing nucleolytic degradation, DpnI-resistant plasmid was readily recovered 48 h following transfection providing further evidence that pQ1 is replicated.
Incorporation of T–T(6–4) photoproducts into pQ1 to assess translesion synthesis and error-free bypass
T–T(6–4) photoproduct-containing oligonucleotides were synthesized as previously described (27) and were ligated into pQ1 using the two-step technique described by the Lawrence laboratory (21,22) so as to maximize the amount of covalently closed plasmid (see also Materials and methods section). A separate photoproduct placed on each strand of DNA makes damage bypass necessary for any replicated plasmid product. The photoproducts were arranged in one of two ways. In the staggered conformation (pQTs), the lesions are separated by 28 intervening base pairs and placed opposite a GpC mismatch. Replicated copies can thus result from TLS on the top strand or bottom strand. Error-free bypass or excision repair of the lesions is reported by the presence of GpC at the site of the dimer (Figure 1C). Although GpC could be inserted by TLS, evidence available to date suggests that this combination of base insertions opposite a T–T(6–4) photoproducts would be unusual (35). We also created an unphysiological substrate in which the lesions are placed opposite each other (pQTo). This arrangement can only lead to replicated copies by TLS, or by deletion (Figure 1D). The effectiveness of the block to DNA synthesis posed by the photoproducts in these plasmids and the purity of the preparations is illustrated by DNA sequencing (Figure 1E). Note that the sequencing polymerase inserts an A opposite the 3′T of the photoproduct before further extension is blocked.
Efficiency of replication of pQ1 harbouring T–T(6–4) photoproducts in wild-type and nucleotide excision repair-defective DT40 cells
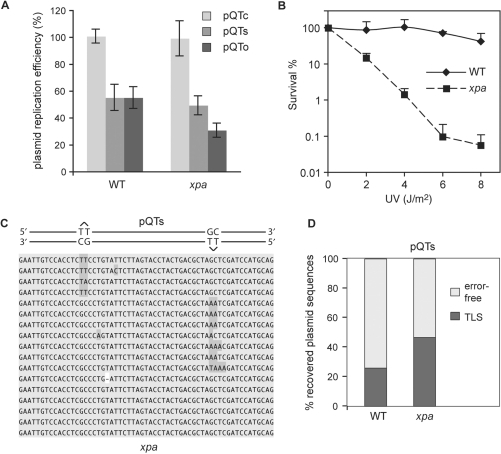
We next assessed the efficiency with which DT40 is able to replicate pQTs and pQTo. To do this, we cotransfected fixed amounts of a kanamycin-resistant version of pQ1 (called pQ2) with the lesion-containing pQ1-derived ampicillin-resistant constructs. Following plating of the recovered plasmid separately on both ampicillin and kanamycin-containing plates we normalized the number of AmpR colonies to the internal KanR control. In comparison to a plasmid containing a ligated lesion-free oligo (pQTc), wild-type cells were able to replicate both pQTs and pQTo with 50% efficiency (Figure 2A). As T–T(6–4) photoproducts are excellent substrates for nucleotide excision repair, the efficiency of replication might be expected to be significantly enhanced by repair of the lesion before replication. We therefore performed the same experiments in cells in which the XPA locus was disrupted (32). Disruption of XPA renders DT40 significantly sensitive to UV light (Figure 2B) but surprisingly had little effect on the replication efficiency of pQTs (Figure 2A) suggesting that the plasmid can replicate efficiently with the lesion in place. There was a greater impact of XPA disruption on the efficiency of replication of pQTo consistent with this being a more difficult replication substrate for which there is no option to perform error-free bypass.
Figure 2.
Lesion bypass in wild-type and xpa cells. (A) Efficiency of replication of shuttle plasmids transfected into WT and xpa cells, and recovered 48 h later. A ligated lesion-free control preparation pQTc as well as two different lesion-containing constructs, pQTs and pQTo were used. These AmpR constructs were co-transfected with the KanR pQT2 plasmid, allowing for the normalization of the number of recovered replicated pQ1-derived constructs against the internal pQ2 control. The average and SEM of 3–5 experiments is shown. (B) Colony survival assay measures the UV light sensitivity of wild-type (WT) and xpa mutant cell lines. Diamonds, WT; squares, xpa. Error bars represent 1 SD. For clarity, only positive error is shown. (C) Example sequences of replicated pQTs plasmids recovered from xpa cells are shown, aligned with a schematic drawing of pQTs. The sequences are sorted, demonstrating TLS on the top strand (inserting mostly AA in the reverse direction, top set), TLS on the bottom strand (middle set), and error-free bypass (bottom set). The proportions are not representative. (D) The proportion of TLS versus error-free bypass in pQTs sequences recovered from wt or xpa cells, shown as percentage of the total. A total of over 80 sequences are shown as a sum of three or more independent experiments.
DT40 cells make extensive use of translesion synthesis in bypassing T–T(6–4) photoproducts, which in the majority of cases is accurate
To assess the mode of bypass used in producing replicated copies of pQTs we sequenced the plasmids recovered from wild-type and xpa DT40 and subjected to DpnI digestion. As expected, we were able to identify examples of TLS on the top strand and bottom strand. We also found in both wild-type and xpa cells abundant error-free processing, as evidenced by GpC bases at the site of the dimer (Figure 2C). Since these products are found in copies of the plasmid that had replicated in DT40 cells in the absence of nucleotide excision repair we believe that they reflect ‘error-free’ recombinational bypass of the photoproduct.
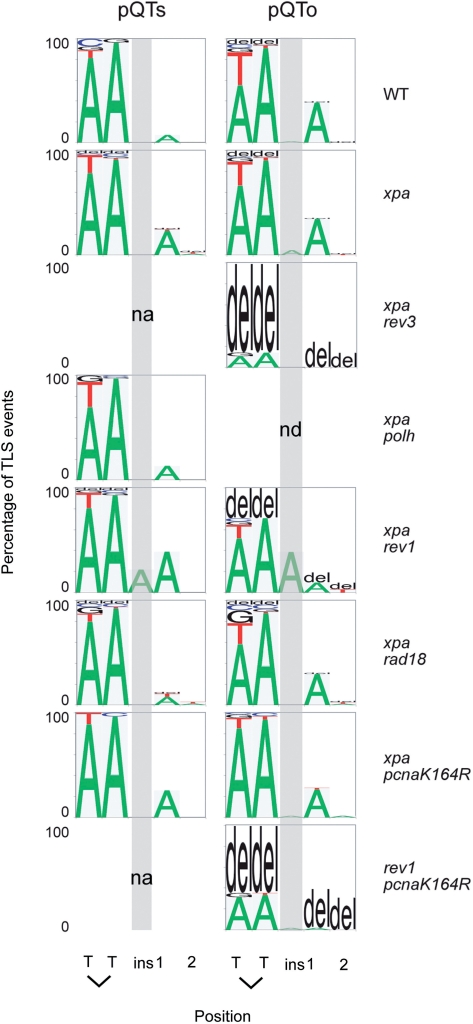
In wild-type cells, TLS was used in 27% of cases. This increased to 47% in xpa cells (Figure 2D and Table 1). By comparison, a similar construct was replicated using TLS in only 4% of cases when introduced into excision repair-defective S. cerevisiae (22). The pattern of TLS was very similar in WT and xpa cells (Figures 2C and 5; see also Supplementary Table S1 for raw figures for all TLS spectrum data). In over 50% of cases, TLS was accurate with ApA being inserted opposite the T–T photoproduct. Two common misinsertions could be observed: a T instead of A opposite the 3′ T of the lesion, and an A instead of the correct base at the 5′+1 position, i.e. at the first inserted base after the lesion is bypassed. We also observed infrequent insertion of other bases or deletion of two or more bases at the site of the lesion (Figures 2C and 5). Although our staggered lesion reporter system cannot distinguish TLS incorporating GpC from error-free bypass, the more commonly expected GpA is not frequently seen. We also classified the outcomes of bypass in sequences obtained from the QTo construct, in which bypass can be effected only by TLS or deletion. Because two lesions are opposite each other, there is no a priori information on the direction of TLS in replicated sequences. However, after the analysis of TLS outcomes from pQTs, we found that we can predict the direction of TLS in pQTo from the resulting sequence in 98% of cases (for example, the insertion of AA is very much more common than TT). This allowed us to draw up the spectrum of base insertions in the pQTo construct as well. It is very similar to that seen for pQTs (Figure 5) and confirms that GpC is only extremely rarely seen in bypass of T–T(6–4) photoproducts.
Table 1.
Type of bypass in recovered pQTs plasmids
| TLS | TLS (%) | Error-free | Error-free (%) | Total | No. expts. | |
|---|---|---|---|---|---|---|
| WT | 27 | 27.0 | 73 | 73.0 | 100 | 3 |
| xpa | 70 | 46.7 | 80 | 53.3 | 150 | 3 |
| xpa rev3 | 1 | 0.8 | 132 | 99.2 | 133 | 3 |
| xpa polh | 64 | 54.2 | 54 | 45.8 | 118 | 5 |
| xpa rev1 | 42 | 27.5 | 111 | 72.5 | 153 | 5 |
| xpa rad18 | 65 | 53.7 | 56 | 46.3 | 121 | 3 |
| xpa pcnaK164R | 27 | 31.4 | 59 | 68.6 | 86 | 5 |
| rev1 pcnaK164R | 3 | 3.3 | 88 | 96.7 | 91 | 3 |
| rev1 : hREV1 | 14 | 36.8 | 24 | 63.2 | 38 | 4 |
| rev1 : hREV1cat | 31 | 44.3 | 39 | 55.7 | 70 | 3 |
| rev1 : hREV1ΔC | 7 | 6.7 | 97 | 93.3 | 104 | 3 |
| Escherichia coli | 2 | 5.0 | 38 | 95.0 | 40 | 2 |
The number of sequences generated by TLS and by error-free bypass is shown, followed by the percentage in each group. The total number of sequences analysed is presented in the column ‘Total’ and the number of independent experiments performed to generate each database shown in ‘No. expts.’. The spectrum of bypass obtained following introduction of the plasmid directly into E. coli is also shown.hREV1cat = hREV1[D570A/E571A]; hREV1ΔC = hREV1[1–1137].
Figure 5.
The pattern of nucleotide incorporation opposite the T–T(6–4) photoproduct. Data for pQTs and pQTo are shown individually. Error-free pQTs sequences are excluded from the analysis. The percentage of each nucleotide incorporated at each position is indicated by the size of the letter of the nucleotide in the column; del, deletion. The incorporation positions indicated are at the 3′T and 5′T of the lesion followed by the next two bases in the template, indicated 1 and 2. Insertions before the + 1 position are indicated in the column ‘ins’, which is also indicated by a shaded grey box. The pQTs plasmids recovered from the xpa rev3 and rev1 pcnaK164R lines had replicated almost exclusively by an error-free mechanism and generated to few mutant sequences to be plotted, indicated by ‘na’ (not applicable); nd, not done.
In addition, occasional base substitutions were seen in other regions of the oligonucleotide insert but these were at low frequency, exhibited no clear genetic dependence and will not be considered further here.
DNA polymerase ζ is absolutely required for bypass of a 6–4PP by translesion synthesis
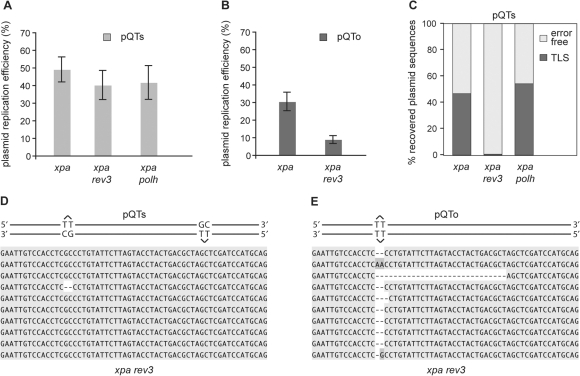
We next examined the requirement for DNA polymerases ζ and η for TLS of the T–T(6–4) photoproducts. Both polymerases have been implicated in a two-step model for bypass of this lesion (36). Again to eliminate interference from nucleotide excision repair, we disrupted the XPA locus in both rev3 and polh DT40 lines (28,30). Disruption of REV3, the catalytic subunit of Polζ, did not result in a significant decrease in overall replication efficiency of pQTs (Figure 3A). However, analysis of the pattern of bypass revealed an almost complete absence of TLS in xpa rev3 cells, with only one out of 133 sequences (0.8%) showing evidence of error-prone bypass, and this was a two base deletion (Figure 3C and D, Table 1). All other sequences were the result of error-free bypass. In agreement with this, the replication of pQTo, in which no donor exists for error-free bypass, is severely compromised in xpa rev3 cells (Figure 3B). Sequence from the recovered replicated plasmids revealed an almost complete absence of TLS (Figure 3C). Instead, we observed the deletion of two or more bases covering the site of the lesion, suggesting inefficient lesion bypass by template misalignment in the absence of REV3 (Figure 3E). Xpa polh cells exhibited unexplained but reproducibly poorer survival following transfection than the other lines tested here and sufficient replication efficiency data were only obtained with the pQTs plasmid. These data showed that xpa polh cells are able to replicate pQTs with similar efficiency to wild type (Figure 3A) and that the spectrum of TLS in this line is essentially identical to that in WT and xpa cells (Figure 5). In particular, there was no change in the low frequency with which G was incorporated opposite the 3′T of the lesion. Together, these data do not support Polη playing a significant role in the bypass of (6–4) photoproducts in this system. However, they demonstrate the critical importance of Polζ.
Figure 3.
REV3 is required for TLS over the T–T(6–4) photoproduct. (A) Replication efficiency of the pQTs shuttle plasmid in xpa, xpa rev3 and xpa polh cells, normalized against an internal pQ2 control as in Figure 2. The average and SEM of 3–5 experiments is shown. (B) Replication efficiency of the pQTo shuttle plasmid in xpa and xpa rev3 cells. (C) The proportion of TLS versus error-free bypass in pQTs sequences recovered from xpa, xpa rev3 and xpa polh cells, shown as a percentage of the total. (D and E) Aligned example sequences from replicated pQTs (D) and pQTo (E) constructs recovered from xpa rev3 cells.
PCNA ubiquitination and the C-terminal region of REV1 play complementary roles in coordinating [6–4] photoproduct bypass
Although the ubiquitination of PCNA is absolutely central to DNA damage bypass in yeast, recent work has demonstrated that, in DT40, the C-terminus of REV1 can also coordinate lesion bypass independently of PCNA ubiquitination (19,20). We have proposed that these two pathways of TLS coordination define temporally separate modes of bypass, with REV1 acting at stalled replication forks and PCNA ubiquitination at post-replicative gaps (19). The experimental protocol described here allowed us to directly examine the contribution made by REV1 and ubiquitination of PCNA to TLS at a defined DNA lesion.
Disruption of PCNA ubiquitination either by mutation of RAD18 or the target lysine at position 164 in PCNA in xpa cells did not affect the overall efficiency of replication of pQTs or of pQTo (Figure 4A). Examination of the pattern of bypass, however, revealed a modest decrease in the frequency with which TLS was used in the pcnaK164R mutant cells, but not in the rad18 cells (Figure 4C). This difference is likely to be explained by the existence of RAD18-independent ubiquitination of PCNA in DT40 cells (18). Perhaps more surprising is the robust error-free bypass in cells lacking PCNA ubiquitination. This suggests that loss of post-replicative TLS can be readily compensated for by error-free modes of gap filling. Further, it suggests that either error-free bypass by template switching can take place efficiently without PCNA ubiquitination or that classical homologous recombination can very efficiently substitute for the loss of PCNA ubiquitination-dependent error-free bypass.
Figure 4.
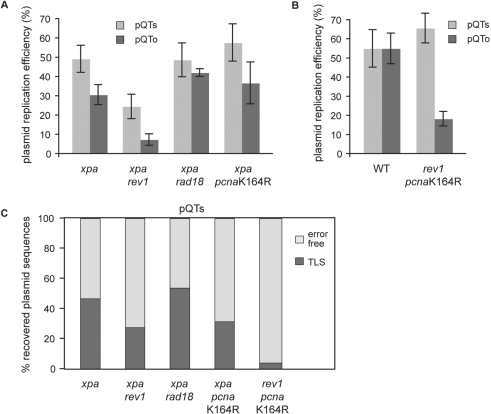
The dual control of TLS by REV1 and PCNA ubiquitination. (A and B) Replication efficiency of the pQTs and pQTo shuttle plasmids in xpa, xpa rev1, xpa rad18 and xpa pcnaK164R cells (A), and in wt and rev1 pcnaK164R cells (B), normalized against an internal pQ2 control as in Figure 2. The average and SEM of 3–5 experiments is shown. (C) The proportion of TLS versus error-free bypass in pQTs sequences recovered from the indicated cell lines, shown as a percentage of the total.
The xpa rev1 cells, in contrast, do show a decrease in the efficiency with which they can replicate both pQTs and pQTo, when compared to xpa cells [P = 0.039 and 0.009, respectively (unpaired t-test); Figure 4A]. We also observed a decrease in the use of TLS within replicated copies of pQTs from 47% to 27% following disruption of REV1 (Figure 4C, Table 1). Further, the proportion of accurate TLS (i.e. incorporation of ApA) decreased from 54% in xpa cells to 30% in xpa rev1 cells. This suggests that a proportion of REV1-dependent bypass events cannot be substituted by error-free bypass, but that as previously proposed, REV1-dependent TLS plays a substantially non-overlapping role to TLS dependent on PCNA ubiquitination in DT40 (19,20). Importantly, neither loss of REV1 nor PCNA-ubiquitination significantly alter the spectrum of nucleotides inserted during bypass in the remaining plasmids replicated by TLS (Figure 5), suggesting that in this context both mechanisms are controlling the same polymerase, Polζ.
To test this, we examined bypass in rev1 pcnaK164R cells. Despite repeated attempts, we were unable to disrupt the XPA locus in this line, probably reflecting the anticipated exquisite sensitivity such a line would exhibit to many forms of DNA damage, and so we compared its response with that of wild-type cells. While we did not observe a reduction of pQTs replication in rev1 pcnaK164R cells, the efficiency of replicating the pQTo construct was significantly reduced (P = 0.006, unpaired t-test) (Figure 4B). The analysis of the bypass pattern is even more revealing, as very few sequences (3/91) showed use of TLS (Figure 4C, Table 1). Thus, the inactivation of both REV1 and PCNA ubiquitination resulted in an 8.2-fold reduction of the use of TLS compared to the wild-type. Although a proportion of the ‘error-free’ bypass seen in the rev1 pcnaK164R cells may be due to excision repair, the reduction in the use of TLS far exceeds the difference seen between xpa and wild-type cells (Figure 2D) and the less than 2-fold reduction seen when each factor is disrupted on its own in the xpa background (Figure 4C).
These results argue that DNA polymerase ζ can be recruited to PCNA by distinct ubiquitin- and REV1-dependent mechanisms and that in the absence of both methods of recruitment TLS is almost completely disabled.
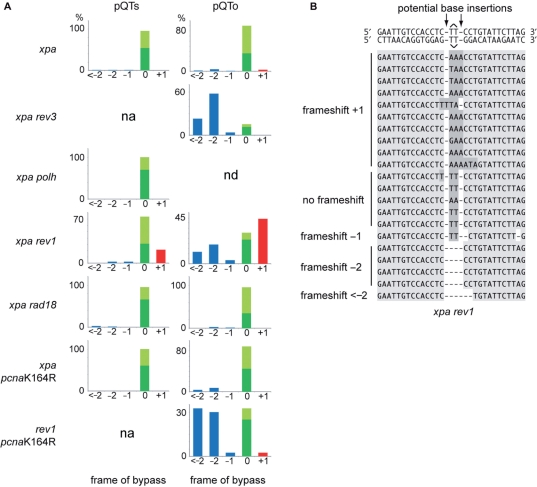
REV1 is required to ensure the maintenance of frame during TLS of a T–T(6–4) photoproduct
While we find that PCNA ubiquitination and REV1 have separate and likely non-overlapping roles in REV3 recruitment, we also considered whether either mechanism affects the accuracy of TLS. An analysis of TLS events in both pQTs and pQTo constructs concentrating on the frame of bypass (i.e. the number of bases inserted opposite the lesion) reveals a striking defect in xpa rev1 cells (Figure 6A). The loss of REV1 is prominently associated with the incorporation of a third base opposite the T–T(6–4) photoproduct, always an A, resulting in a +1 frameshift (22% in pQTs, 39% in pQTo), although there is also an increase in deletions (33% in pQTo) (Figure 6A and B). This was observed in both of two independently derived rev1 mutants (17,29). In contrast, in almost all cases (over 95%), TLS in wild-type and xpa cells is accomplished without any alteration in frame, in other words gain or loss of bases relative to the template. This is the case even when TLS involves a run of three or more bases (Figures 2C and 6A). PCNA ubiquitination-defective and Polη-deficient cells both exhibited comparable patterns to wild-type and xpa cells (Figure 6A).
Figure 6.
REV1 is required for avoiding frameshifts during TLS. (A) The outcome of TLS in the cell lines indicated. Sequences are classified according to frameshifts (incorrect number of bases inserted opposite the lesion), with deletions shown as <−2, −2 or −1 (blue), the correct two-base insertion as 0 (green), and an extra base as +1 (red). More than one extra base was never observed. The non-frameshifted sequences are further classified, shown as a stacked column, as correct bypass (AA inserted, dark green) or incorrect bypass (light green). In case of pQTs (on the left), the error-free sequences were removed from the analysis; n/a, not applicable. The pQTs samples from the xpa rev3 and rev1 pcnaK164R cell lines gave almost exclusively error-free results, and therefore could not be plotted, indicated as ‘na’ (not applicable); nd, not done. (B) Aligned example sequences from replicated pQTo constructs recovered from xpa rev1 cells. The different types of frameshift are annotated on the left.
Interestingly, loss of PCNA ubiquitination appears to suppress the +1 frameshifts seen in the rev1 mutant (Figure 6A). While this suggests that this (i.e. the frame-maintenance) function of REV1 predominantly operates in UbPCNA-dependent post-replicative bypass, the absolute numbers of TLS events in rev1 pcnaK164R cells is rather low, especially for pQTs (Supplementary Table 1). This conclusion can therefore only be drawn tentatively.
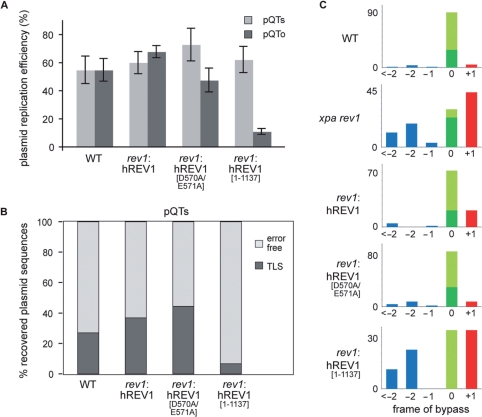
As discussed earlier, the function of REV1 in bypass of lesions that are not its catalytic substrates depends on the capacity of its C-terminus to bind the other TLS polymerases, including Polζ (7,8). To ascertain whether the maintenance of frame during bypass depends on this C-terminal polymerase-binding domain, we examined T–T photoproduct bypass in pQTo in rev1 cells complemented with human REV1 (hREV1), an hREV1 catalytic mutant (hREV1[D570A/E571A]) and an hREV1 lacking the C-terminal polymerase-binding domain (hREV1[1–1137]) (19,20). Although these experiments were performed without the disruption of the XPA locus, they revealed that rev1 cells expressing hREV1[1–1137], exhibit a defect in the efficiency with which pQTo was replicated (Figure 7A) and less frequent use of TLS in pQTs (Figure 7B and Table 1) compared with wild-type, hREV1 and hREV1[D570AE571A] complemented cells. Absence of the C-terminus of REV1, but not disruption of the catalytic domain of REV1, also revealed a substantial elevation in +1 frameshifts associated with bypass of the T–T photoproduct (Figure 7C). Together, these data suggest that the C-terminal polymerase-binding activity of REV1 is required to ensure accurate maintenance of frame during bypass of a (6–4) photoproduct by DNA polymerase ζ.
Figure 7.
The C-terminal domain of REV1 is required for its role in restraining Polζ. (A) Replication efficiency of the pQTs and pQTo shuttle plasmids assayed in wt cells and in rev1 mutant cells complemented with full length human REV1 (hREV1), a catalytically inactive point mutant (hREV1[D570AE571A]) and a C-terminal truncation starting at residue 1138 (hREV1[1–1137]). The average and SEM of 3–5 experiments is shown. (B) The proportion of TLS versus error-free bypass in pQTs sequences recovered from the indicated cell lines, shown as a percentage of the total. (C) The outcome of TLS in pQTo in the cell lines indicated, classified according to frameshifts as in Figure 6.
DISCUSSION
The nature of the error-free bypass in pQ-derived plasmids
These experiments reveal clear evidence for the existence of an error-free mode of DNA damage bypass in a vertebrate cell. The T–T(6–4) photoproduct is a very potent replicative block. Placing two of these lesions on opposite strands, staggered by 28 bp and opposite a GC mismatch, allows us to monitor both translesion replication and recombinational damage avoidance. However, the T–T(6–4) photoproduct is also an excellent target for nucleotide excision repair on account of the significant distortion it introduces into the DNA duplex. Our experimental configuration is unable to distinguish between error-free bypass and excision repair as both will result in plasmid in which the T–T at the photoproduct appears to have been replicated as GpC. However, the approximate 2-fold increase in the number of sequences generated by TLS following disruption of XPA (Figure 2D), a key factor involved in both transcription coupled and global genome repair (37), suggests that no more than about 50% of the (6–4) photoproducts in the pQ plasmids are subject to nucleotide excision repair before replication in wild-type cells.
In terms of pathways other than nucleotide excision repair that could trigger the excision of damage and lead to an ‘error-free’ outcome, there is no evidence that base excision repair can act at (6–4) photoproducts. Although the human mismatch recognition heterodimer MSH2/6 (hMutSα) can bind (6–4)T–T photoproduct-containing mismatches (38), more recent evidence suggests that this binding does not stimulate excision in vitro (39). Further, an msh2 mutant DT40 behaved no differently from wild-type in the proportion of pQTs replicated by TLS (data not shown).
Various assays have consistently shown that budding yeast relies much more heavily on error-free recombinational modes of bypass of DNA lesions than on TLS. Zhang and Lawrence (22) noted that TLS was used only 4% of the time, the remainder being error-free and the result of either RAD5-dependent template switching or RAD52-dependent homologous recombination. Similar figures have been arrived at by the Fuchs and Prakash (40,41) labs, who reported TLS being used in 8% of bypass events at an N-2-acetylaminofluorene (AAF) adduct and 6% at an abasic site, respectively, the remainder being the result of other error-free processes. In the experiments we present here, the fraction of lesion containing plasmid replicated by nucleotide excision repair defective cells was similar to that found in yeast by Zhang and Lawrence (49% versus 54.7%) (22). However, the fraction generated by translesion replication was very different (47% versus 4% of recovered plasmids) suggesting that DT40 relies much more heavily on TLS than do yeast cells. This may reflect a greater pressure to avoid potentially destabilizing recombination when replicating a larger and more complex genome.
A further surprising observation in our DT40 experiments is the lack of impact the loss of PCNA ubiquitination has on error-free bypass. In yeast, mutation of RAD18, which in this organism abrogates PCNA ubiquitination (3), results in complete loss of translesion replication and a very substantial decrease in error-free bypass (22). In contrast, neither loss of RAD18 nor mutation of K164 of PCNA has any impact on the overall efficiency of replication. Although pcnaK164R cells exhibit a decrease in use of TLS, loss of PCNA ubiquitination does not significantly impact on the efficiency with which the lesion-containing plasmids are replicated, nor does it reduce the frequency of error-free outcomes. This suggests that other pathways readily compensate for any role played by this modification in activating error-free bypass in DT40. Further work is needed to investigate the genetic requirements of error-free bypass in this system.
Which polymerases are catalytically active in the bypass of (6–4) photoproducts?
Our data suggest that Polζ plays an absolutely central role in TLS across the (6–4) photoproduct in vivo, even though in vitro studies suggest that no one polymerase is able to efficiently bypass this lesion on its own (2). Two possible scenarios can be advanced to explain this paradox. In the first, the bypass of (6–4) photoproducts is a two-step process with one polymerase inserting a base opposite the 3′T and a second polymerase, Polζ, extending from the resulting mismatch (36,42). In the second, Polζ is capable of the complete bypass reaction in vivo, aided by the presence of accessory factors absent from the in vitro assays (35).
In support of the two polymerase model, in vitro both Polη and Polι (the product of the RAD30B locus) are capable of incorporating the first base opposite the 3′T of a T–T(6–4) photoproduct, Polη preferring G and Polι G or A (36,42). Although neither can extend, this is efficiently performed by Polζ, which inserts the correct A opposite the 5′T. Consistent with these observations, insertion of G opposite the 3′T is observed in vivo in yeast and in one reporter system was shown to be dependent on Polη (43). However, in a different sequence context the Lawrence group reported much less frequent dGMP insertion at the 3′T in their wild-type strain, which although reduced by disruption of Polη was not eliminated (35). The work presented here reveals that, in DT40, incorporation of G at the 3′T is infrequent, the most common base used being A. Further, disruption of Polη (the sole RAD30 homologue in chickens, which appear to lack Polι) does not lead to any diminution in efficiency of bypass in our assay nor to any significant change in the pattern of incorporation opposite either base of the photoproduct. This suggests that Polη does not play a significant role in bypass of a T–T(6–4) photoproduct in DT40.
Which other polymerases could catalyse incorporation at the 3′T? It could be achieved by the one of the replicative polymerases, as seen in the bacterial polymerase reaction shown in Figure 1E. Although, genetic evidence has implicated the Pol32 subunit of DNA polymerase δ in TLS in yeast (44) and Polδ has been shown to insert A opposite a thymine glycol in vitro (45), firm evidence for the replicative polymerases playing a role in catalysis at a (6–4) photoproduct in vivo is lacking and indeed is intrinsically difficult to obtain. Even if Polδ did incorporate A at opposite the 3′T, it is not unlikely that, in vivo, this would trigger the proofreading activity of the enzyme. Polκ, the vertebrate homologue of Escherichia coli DinB (POLIV), is another member of the Y family of translesion polymerases, which we have not studied in this article as it has limited in vitro ability to incorporate or extend opposite a (6–4) photoproduct (46–49).
So could Polζ in DT40 be catalysing the complete bypass reaction in vivo as has been previously suggested for yeast (35)? There is yet to be a formal biochemical analysis of the polymerase activity of a vertebrate Polζ and somewhat conflicting in vitro results have been reported for the ability of the yeast enzyme (yPolζ) to bypass a (6–4) photoproduct. One group reported no detectable bypass by yPolζ (36,42), showing strong inhibition at the 3′T, while another reported inefficient bypass when yPolζ was used in a molar excess to the template (50). Drosophila Polζ, which resembles the yeast enzyme, has also been found to be incapable of nucleotide incorporation opposite a (6–4) photoproduct (51). Despite its reluctance to incorporate bases opposite lesions in vitro, Polζ has a remarkable ability to extend from a primer terminus mismatched with a lesion-containing base, which it extends almost as efficiently as on an undamaged template (42,52). It is this latter property that has contributed significantly to the notion of bypass being a two-step process. However, these in vitro studies do not place Polζ in its physiological context with other potentially crucial factors such as PCNA and RPA and the possibility remains that in vivo Polζ is capable of insertion at both the 3′ and 5′T of the photoproduct, and of extension from these inserted bases. We have shown here that loss of two factors, REV1 and ubiquitinated PCNA, result in decreased efficiency and accuracy of Polζ-dependent TLS and that, further, the phenotype of the rev1 pcnaK164R doubly mutant cells is very similar to that of the rev3 mutant in terms of bypass, suggesting that either factor can act independently through Polζ. In the case of Polζ, this raises an interesting paradox since the only demonstrated ubiquitin-binding activity associated with Polζ is indirectly via the UBM domains of REV1. Thus, the precise mechanism by which PCNA ubiquitination promotes Polζ-dependent translesion synthesis remains unclear.
Finally, it is noteworthy that nucleotide incorporation preferences exhibited by Polζ in the in vitro (6–4) photoproduct bypass experiments of Guo et al. (50), discussed above, closely matches what we report here for in vivo bypass. Thus, a number of rather indirect lines of evidence point to Polζ being the sole catalytic activity responsible for bypass of this lesion in vivo. However, further work is needed to formally distinguish whether bypass requires one or two translesion polymerases.
The roles played by REV1 during lesion bypass
REV1 possesses very limited catalytic activity against a small number of lesions, such as abasic sites, and exhibits no ability to bypass a photoproduct (53,54). Indeed, the role of REV1 in bypass of UV damage tolerance is independent of its catalytic function (19). The heart of this non-catalytic function of REV1 resides in its C-terminus, which likely acts as an adaptor between the replisome and the incoming catalytic TLS polymerase (20,55). In the case of the (6–4)T–T photoproduct, we show here that the relevant polymerase coordinated by REV1 is Polζ, which interacts with REV1 via its non-catalytic REV7 subunit (7).
The dependence of Polζ activity on REV1 in vivo in budding yeast is well established. Early genetic data demonstrated that REV3/7 were important for all induced mutagenesis while the requirement for REV1 was somewhat dependent on the context, appearing important only at base substitution but not frameshift alleles (56,57). A potential explanation for this was provided by a study from the Fuchs group that showed that REV1 was required for bypass of a AAF adduct when the primer terminus was at the lesion, but not when the template slipped effectively bypassing the lesion by deletion (58), suggesting that REV1 was required for Polζ to efficiently extend from a mismatched terminus in vivo. The ability of Polζ to generate complex + 1 frameshifts in yeast is also dependent on REV1 (59). However, since this function of Polζ was entirely dependent on REV1, it was not possible to distinguish between a potential role for REV1 in maintaining accurate contact between Polζ and the template from simple failure to recruit Polζ at all.
Since the recruitment of Polζ in DT40 is only partially dependent on REV1, we have been able to show the presence of REV1 also modifies the catalytic behaviour of Polζ ensuring that it incorporates nucleotides in frame with the damaged template. Two mechanisms could be envisaged to account for this observation. The first is that REV1 is required to recruit an exonuclease that trims excess nucleotides synthesized by Polζ during bypass. The second, which we favour, is that REV1 structurally restrains Polζ, forcing it to ‘count’ the number of nucleotides it incorporates relative to the template. The elucidation of the exact mechanism by which it does this is unclear and will likely require formal structural studies of the complexes formed by REV1 and Polζ at the stalled replisome. Interestingly, in this context, a recent study has implicated REV1 in maintaining the stability of trinucleotide repeats in S. cerevisiae that is independent of Polζ (60) suggesting that REV1 may play a role in avoidance of aberrant secondary structure formation during general replication. It will thus be interesting to determine the extent to which these functions of REV1 are related and how they apply to other lesions, DNA structures and polymerases in both general and translesion replication.
Supplementary Data
Supplementary Data are available at NAR Online.
FUNDING
Medical Research Council; Leukaemia Research Fund grant number 04046. Funding for open access charge: Medical Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Shunichi Takeda and Jean-Marie Buerstedde for generously sharing constructs and cell lines. We would like to thank members of the Sale group and Sue Cotterill for discussions and critical comments on the article.
REFERENCES
- 1.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 3.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 4.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 5.Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 6.Gan GN, Wittschieben JP, Wittschieben BO, Wood RD. DNA polymerase ζ (pol zeta) in higher eukaryotes. Cell Res. 2008;18:174–183. doi: 10.1038/cr.2007.117. [DOI] [PubMed] [Google Scholar]
- 7.Murakumo Y, Ogura Y, Ishii H, Numata S, Ichihara M, Croce CM, Fishel R, Takahashi M. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J. Biol. Chem. 2001;276:35644–35651. doi: 10.1074/jbc.M102051200. [DOI] [PubMed] [Google Scholar]
- 8.Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 2003;22:6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broomfield S, Hryciw T, Xiao W. DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat Res. 2001;486:167–184. doi: 10.1016/s0921-8777(01)00091-x. [DOI] [PubMed] [Google Scholar]
- 10.Higgins NP, Kato K, Strauss B. A model for replication repair in mammalian cells. J. Mol. Biol. 1976;101:417–425. doi: 10.1016/0022-2836(76)90156-x. [DOI] [PubMed] [Google Scholar]
- 11.Blastyak A, Pinter L, Unk I, Prakash L, Prakash S, Haracska L. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu RK, Brun J, Ramaekers C, Theys J, Weng L, Lambin P, Gray DA, Wouters BG. Lysine 63-polyubiquitination guards against translesion synthesis-induced mutations. PLoS Genet. 2006;2:e116. doi: 10.1371/journal.pgen.0020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Motegi A, Sood R, Moinova H, Markowitz SD, Liu PP, Myung K. Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. J. Cell Biol. 2006;175:703–708. doi: 10.1083/jcb.200606145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Unk I, Hajdu I, Fatyol K, Hurwitz J, Yoon JH, Prakash L, Prakash S, Haracska L. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc. Natl Acad. Sci. USA. 2008;105:3768–3773. doi: 10.1073/pnas.0800563105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sale JE. Immunoglobulin diversification in DT40: a model for vertebrate DNA damage tolerance. DNA Repair. 2004;3:693–702. doi: 10.1016/j.dnarep.2004.03.042. [DOI] [PubMed] [Google Scholar]
- 16.Sale JE, Ross AL, Simpson LJ. Analysis of DNA replication damage bypass and its role in immunoglobulin repertoire development. Sub-cellular Biochem. 2006;40:271–294. doi: 10.1007/978-1-4020-4896-8_16. [DOI] [PubMed] [Google Scholar]
- 17.Arakawa H, Moldovan GL, Saribasak H, Saribasak NN, Jentsch S, Buerstedde JM. A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biol. 2006;4:e366. doi: 10.1371/journal.pbio.0040366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simpson LJ, Ross AL, Szuts D, Alviani CA, Oestergaard VH, Patel KJ, Sale JE. RAD18-independent ubiquitination of proliferating-cell nuclear antigen in the avian cell line DT40. EMBO Rep. 2006;7:927–932. doi: 10.1038/sj.embor.7400777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edmunds CE, Simpson LJ, Sale JE. PCNA Ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol. Cell. 2008;30:519–529. doi: 10.1016/j.molcel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 20.Ross AL, Simpson LJ, Sale JE. Vertebrate DNA damage tolerance requires the C-terminus but not BRCT or transferase domains of REV1. Nucleic Acids Res. 2005;33:1280–1289. doi: 10.1093/nar/gki279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozgenc AI, Szekeres ES, Lawrence CW. In vivo evidence for a recA-independent recombination process in Escherichia coli that permits completion of replication of DNA containing UV damage in both strands. J. Bacteriol. 2005;187:1974–1984. doi: 10.1128/JB.187.6.1974-1984.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Lawrence CW. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc. Natl Acad. Sci. USA. 2005;102:15954–15959. doi: 10.1073/pnas.0504586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd. ASM Press: Washington, DC; 2006. [Google Scholar]
- 24.Kim JK, Choi BS. The solution structure of DNA duplex-decamer containing the (6-4) photoproduct of thymidylyl(3′–>5′)thymidine by NMR and relaxation matrix refinement. Eur. J. Biochem. 1995;228:849–854. doi: 10.1111/j.1432-1033.1995.tb20331.x. [DOI] [PubMed] [Google Scholar]
- 25.Kim JK, Patel D, Choi BS. Contrasting structural impacts induced by cis-syn cyclobutane dimer and (6-4) adduct in DNA duplex decamers: implication in mutagenesis and repair activity. Photochem. Photobiol. 1995;62:44–50. doi: 10.1111/j.1751-1097.1995.tb05236.x. [DOI] [PubMed] [Google Scholar]
- 26.Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 27.Iwai S, Shimizu M, Kamiya H, Ohtsuka E. Synthesis of a phosphoramidite coupling unit of the pyrimidine (6-4) pyrimidone photoproduct and its incorporation into oligodeoxynucleotides. J. Am. Chem. Soc. 1996;118:7642–7643. [Google Scholar]
- 28.Kawamoto T, Araki K, Sonoda E, Yamashita YM, Harada K, Kikuchi K, Masutani C, Hanaoka F, Nozaki K, Hashimoto N, et al. Dual roles for DNA polymerase η in homologous DNA recombination and translesion DNA synthesis. Mol. Cell. 2005;20:793–799. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 29.Simpson LJ, Sale JE. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J. 2003;22:1654–1664. doi: 10.1093/emboj/cdg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sonoda E, Okada T, Zhao GY, Tateishi S, Araki K, Yamaizumi M, Yagi T, Verkaik NS, van Gent DC, Takata M, et al. Multiple roles of Rev3, the catalytic subunit of polζ in maintaining genome stability in vertebrates. EMBO J. 2003;22:3188–3197. doi: 10.1093/emboj/cdg308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamashita YM, Okada T, Matsusaka T, Sonoda E, Zhao GY, Araki K, Tateishi S, Yamaizumi M, Takeda S. RAD18 and RAD54 cooperatively contribute to maintenance of genomic stability in vertebrate cells. EMBO J. 2002;21:5558–5566. doi: 10.1093/emboj/cdf534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okada T, Sonoda E, Yamashita YM, Koyoshi S, Tateishi S, Yamaizumi M, Takata M, Ogawa O, Takeda S. Involvement of vertebrate polκ in Rad18-independent postreplication repair of UV damage. J. Biol. Chem. 2002;277:48690–48695. doi: 10.1074/jbc.M207957200. [DOI] [PubMed] [Google Scholar]
- 33.Piechaczek C, Fetzer C, Baiker A, Bode J, Lipps HJ. A vector based on the SV40 origin of replication and chromosomal S/MARs replicates episomally in CHO cells. Nucleic Acids Res. 1999;27:426–428. doi: 10.1093/nar/27.2.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeda DY, Shibata Y, Parvin JD, Dutta A. Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells. Genes Dev. 2005;19:2827–2836. doi: 10.1101/gad.1369805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibbs PE, McDonald J, Woodgate R, Lawrence CW. The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics. 2005;169:575–582. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson RE, Haracska L, Prakash S, Prakash L. Role of DNA polymerase eta in the bypass of a (6-4) TT photoproduct. Mol. Cell Biol. 2001;21:3558–3563. doi: 10.1128/MCB.21.10.3558-3563.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, Lawrence CW, Li GM, Hays JB. Specific binding of human MSH2.MSH6 mismatch-repair protein heterodimers to DNA incorporating thymine- or uracil-containing UV light photoproducts opposite mismatched bases. J. Biol. Chem. 1999;274:16894–16900. doi: 10.1074/jbc.274.24.16894. [DOI] [PubMed] [Google Scholar]
- 39.Wang H, Hoffman PD, Lawrence C, Hays JB. Testing excision models for responses of mismatch-repair systems to UV photoproducts in DNA. Environ. Mol. Mutagen. 2006;47:296–306. doi: 10.1002/em.20206. [DOI] [PubMed] [Google Scholar]
- 40.Baynton K, Bresson-Roy A, Fuchs RP. Analysis of damage tolerance pathways in Saccharomyces cerevisiae: a requirement for Rev3 DNA polymerase in translesion synthesis. Mol. Cell Biol. 1998;18:960–966. doi: 10.1128/mcb.18.2.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pages V, Johnson RE, Prakash L, Prakash S. Mutational specificity and genetic control of replicative bypass of an abasic site in yeast. Proc. Natl Acad. Sci. USA. 2008;105:1170–1175. doi: 10.1073/pnas.0711227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 43.Bresson A, Fuchs RP. Lesion bypass in yeast cells: Pol η participates in a multi-DNA polymerase process. EMBO J. 2002;21:3881–3887. doi: 10.1093/emboj/cdf363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haracska L, Unk I, Johnson RE, Johansson E, Burgers PM, Prakash S, Prakash L. Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson RE, Yu SL, Prakash S, Prakash L. Yeast DNA polymerase ζ (zeta) is essential for error-free replication past thymine glycol. Genes Dev. 2003;17:77–87. doi: 10.1101/gad.1048303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson RE, Prakash S, Prakash L. The human DINB1 gene encodes the DNA polymerase Polθ. Proc. Natl Acad. Sci. USA. 2000;97:3838–3843. doi: 10.1073/pnas.97.8.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohashi E, Bebenek K, Matsuda T, Feaver WJ, Gerlach VL, Friedberg EC, Ohmori H, Kunkel TA. Fidelity and processivity of DNA synthesis by DNA polymerase κ, the product of the human DINB1 gene. J. Biol. Chem. 2000;275:39678–39684. doi: 10.1074/jbc.M005309200. [DOI] [PubMed] [Google Scholar]
- 48.Washington MT, Johnson RE, Prakash L, Prakash S. Human DINB1-encoded DNA polymerase κ is a promiscuous extender of mispaired primer termini. Proc. Natl Acad. Sci. USA. 2002;99:1910–1914. doi: 10.1073/pnas.032594399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Yuan F, Wu X, Wang M, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Error-free and error-prone lesion bypass by human DNA polymerase κ in vitro. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo D, Wu X, Rajpal DK, Taylor JS, Wang Z. Translesion synthesis by yeast DNA polymerase ζ from templates containing lesions of ultraviolet radiation and acetylaminofluorene. Nucleic Acids Res. 2001;29:2875–2883. doi: 10.1093/nar/29.13.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takeuchi R, Oshige M, Uchida M, Ishikawa G, Takata K, Shimanouchi K, Kanai Y, Ruike T, Morioka H, Sakaguchi K. Purification of Drosophila DNA polymerase ζ by REV1 protein-affinity chromatography. Biochem. J. 2004;382:535–543. doi: 10.1042/BJ20031833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lawrence CW, Hinkle DC. DNA polymerase ζ and the control of DNA damage induced mutagenesis in eukaryotes. Cancer Surv. 1996;28:21–31. [PubMed] [Google Scholar]
- 53.Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Wu X, Rechkoblit O, Geacintov NE, Taylor JS, Wang Z. Response of human REV1 to different DNA damage: preferential dCMP insertion opposite the lesion. Nucleic Acids Res. 2002;30:1630–1638. doi: 10.1093/nar/30.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo C, Tang TS, Bienko M, Parker JL, Bielen AB, Sonoda E, Takeda S, Ulrich HD, Dikic I, Friedberg EC. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol. Cell Biol. 2006;26:8892–8900. doi: 10.1128/MCB.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lawrence CW, Christensen RB. Ultraviolet-induced reversion of cyc1 alleles in radiation-sensitive strains of yeast. I. rev1 Mutant strains. J. Mol. Biol. 1978;122:1–21. doi: 10.1016/0022-2836(78)90104-3. [DOI] [PubMed] [Google Scholar]
- 57.Lawrence CW, O’Brien T, Bond J. UV-induced reversion of his4 frameshift mutations in rad6, rev1, and rev3 mutants of yeast. Mol. Gen. Genet. 1984;195:487–490. doi: 10.1007/BF00341451. [DOI] [PubMed] [Google Scholar]
- 58.Baynton K, Bresson-Roy A, Fuchs RP. Distinct roles for Rev1p and Rev7p during translesion synthesis in Saccharomyces cerevisiae. Mol. Microbiol. 1999;34:124–133. doi: 10.1046/j.1365-2958.1999.01583.x. [DOI] [PubMed] [Google Scholar]
- 59.Harfe BD, Jinks-Robertson S. DNA polymerase ζ introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol. Cell. 2000;6:1491–1499. doi: 10.1016/s1097-2765(00)00145-3. [DOI] [PubMed] [Google Scholar]
- 60.Collins NS, Bhattacharyya S, Lahue RS. Rev1 enhances CAG.CTG repeat stability in Saccharomyces cerevisiae. DNA Repair) 2007;6:38–44. doi: 10.1016/j.dnarep.2006.08.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.