Abstract
Fused Toes (FTS) is a member of a small group of inactive variant E2 ubiquitin-conjugating enzyme domain-containing proteins of unknown function. Through proteomic analysis of FTS complexes purified from human embryonic kidney 293T cells, we identified a new multiprotein complex, the FHF complex, containing FTS, members of the microtubule-binding Hook family of coiled-coil proteins (Hook1, Hook2, and Hook3), and a previously uncharacterized 107-kDa protein, FTS and Hook Interacting Protein (FHIP). FTS associated with a conserved C-terminal motif in Hook proteins in the yeast two-hybrid system and in tissue culture cells, and Hook proteins were found to form homo- and heterodimers. The ∼500-kDa FHF complex contained all three Hook proteins, and small interfering RNA depletion experiments suggest that Hook proteins can interact interchangeably within this complex. Hook proteins as well as FTS interact with members of both the class B and class C components of the homotypic vesicular protein sorting (HOPS) complex. Depletion of FTS by RNA interference affects both the trafficking of epidermal growth factor from early-to-late endosome/lysosomes and the efficiency by which overexpression of the HOPS component Vps18 promotes clustering of lysosomal-associated membrane protein 1-positive endosome/lysosomes. These data suggest that the FTS/Hook/FHIP complex functions to promote vesicle trafficking and/or fusion via the HOPS complex.
INTRODUCTION
E2 ubiquitin-conjugating (UBC) enzymes constitute a family of proteins containing a conserved protein fold and in humans, 40 such proteins have been identified (Pickart and Eddins, 2004; Winn et al., 2004). The largest class of UBC-domain–containing proteins are catalytically active and can be charged with a ubiquitin-like (Ubl) protein via an E1-activating enzyme in an ATP-dependent reaction. Charged E2s transfer their cognate Ubl to appropriate substrates via an associated E3 ubiquitin ligase (Pickart and Eddins, 2004; Dye and Schulman, 2007). In contrast, a subset of proteins with the UBC domain lack a critical active site cysteine residue and therefore do not participate directly in ubiquitin transfer. This subfamily of proteins is generally referred to as variant E2s. The two best understood variant E2s are the TSG101 protein and the UEV/Mms2 protein. TSG101, the mammalian orthologue of yeast Vps23, plays an important role in the sorting of ubiquitinated proteins at the multivesicular body (MVB). TSG101 is a component of ESCRT-I, which also contains Vps37, Vps28, and Mvb12 (Hurley and Emr, 2006; Kostelansky et al., 2006, 2007). TSG101 forms a complex with ubiquitin (Sundquist et al., 2004) and is thought to function in the recognition of ubiquitinated cargo on the endosomal surface and to facilitate transfer of this cargo from ESCRT-I to ESCRT-II during sorting (Katzmann et al., 2001; Kostelansky et al., 2006). hMms2/UBE2V2, in contrast, forms a heterodimer with the catalytically active Ubc13 ubiquitin-conjugating enzyme, and together these proteins promote lysine-63–linked ubiquitin transfer from Ubc13 to substrates (Chan and Hill, 2001). In this process, the hMms2/UBE2V3 protein functions to bind the donor ubiquitin in a manner that orients its lysine-63 side chain toward the acceptor ubiquitin thioester linked to the active site of Ubc13 (Eddins et al., 2006, 2007). Thus, both of the characterized variant E2 proteins function in ubiquitin binding. Several variant E2s in mammals have yet to be studied in detail.
In this study, we examined the molecular function of poorly understood variant E2 referred to as FTS. The FTS gene was initially identified as one of six genes deleted in a mouse mutant called Fused Toes, due to defects in limb development, and referred to as FT1/FTS (Lesche et al., 1997). To date, direct linkage of the FTS gene to the phenotype of this mouse has not emerged. Subsequently, FTS was identified in an interaction screen with AKT1 and proposed to control Akt phosphorylation by PDK1 (Remy and Michnick, 2004). However, this study relied on overexpression, and the endogenous function of FTS is yet to be determined.
Unlike active E2s that bind their E3s only transiently, variant E2s frequently form relatively tight complexes with other proteins. To begin to understand the function of FTS, we used a proteomic approach to identify human FTS-associated proteins. We found that FTS assembles into a tightly associated multiprotein complex containing three members of the Hook family of coiled-coil proteins as well as a previously uncharacterized protein (C11ORF56) that we refer to as p107FHIP (FTS/Hook Interacting Protein). For simplicity, we refer to this complex as the FHF complex, reflecting its major components FTS, Hook proteins, and FHIP. Hook proteins were first described in Drosophila, where mutations in hook (hk) lead to defects in endocytic trafficking of proteins in the sevenless tyrosine kinase signaling pathway (Kramer and Phistry, 1996). In particular, trafficking of the sevenless ligand Bride of Sevenless (Boss) through the endosomal pathway is defective in hk mutant animals, apparently reflecting defects in maturation of MVBs (Kramer and Phistry, 1996; Sunio et al., 1999).
Mammals express 3 hk-related proteins Hook1, Hook2, and Hook3 (Walenta et al., 2001). These proteins share a common central coiled-coil domain, which has been shown to promote dimerization of Hook proteins, as well as an N-terminal domain that is thought to facilitate interaction with microtubules, possibly indirectly (Walenta et al., 2001). Hook1 displays 58 and 48% identity with Hook3 and Hook2 throughout their length, but the C-terminal 100 residues are somewhat more divergent (41% identity for Hook1 and Hook3 and 42% identity for Hook1 and Hook2) (Supplemental Figure S1). Hook proteins are cytoplasmic, and in some cases they display enriched localization with cellular organelles (Walenta et al., 2001). For example, Hook3 is enriched in the cis-Golgi compartment, and this association involves sequences that are C-terminal to the central coiled-coil domain. Hook2 is also localized throughout the cytoplasm (Walenta et al., 2001), but it is enriched at centrosomes (Szebenyi et al., 2007).
Although Hook proteins have been linked to the endocytic pathway, their precise biological functions are poorly understood, as are the number and types of interactions that they participate in. Recent work has suggested a potential link between Hook1 and the homotypic vacuolar protein sorting (HOPS) complex (Richardson et al., 2004). HOPS is composed of subcomplexes containing the class C VPS proteins (Vps18, Vps16, and Vps11) and the class B VPS proteins (Vps39/Vam6 and Vps41/Vam2). Class B VPS proteins have been implicated in tethering vesicles to microtubules via soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins, whereas class C VPS proteins have been linked to tethering of vesicles to actin filaments (Richardson et al., 2004). Components of the HOPS complex interact with target membrane-SNAREs and have been implicated in homotypic fusion of both early and late endosome/lysosomes in mammalian cells. In previous coimmunoprecipitation studies in mammalian cells, Hook1 was found to interact with Vps18 (Richardson et al., 2004), whereas hk interacts with the Drosophila Vps18 orthologue Deep Orange (Dor) in the two-hybrid system (Lloyd et al., 1998; Sevrioukov et al., 1999). Mutants in Dor, as well as in light (Vps41) and carnation (Vps33), affect the accumulation of pigments granules in the Drosophila eye, organelles whose biogenesis resembles that of the lysosome (Lloyd et al., 1998). Interestingly, human Hook1 was demonstrated previously to coprecipitate with class B subunits Vps39 and Vps41 on microtubules and to coprecipitate with class C subunits Vps18 and Vps16 on actin filaments in vitro (Richardson et al., 2004). These interactions with cytoskeletal components are interesting because vesicle trafficking and fusion are coordinated spatially and rely on transport and tethering via cytoskeletal scaffolds.
Through a series of biochemical experiments, we found that all three Hook proteins interact with each other and with FTS and p107FHIP in an endogenous cytoplasmic complex that migrates at ∼500 kDa on gel filtration columns, the FHF complex. FTS uses its β-sheet surface (analogous to the surface in TSG101 used to interact with ubiquitin; Sundquist et al., 2004) to interact with a conserved helical motif in the extreme C terminus of the Hook proteins. Depletion of Hook1 leads to increased abundance of Hook3 and Hook3 in the FTS immunoprecipitates, suggesting that Hook2 or Hook3 can take the place of Hook1 in the FHF complex. In addition, we find that the FHF complex cannot only interact with Vps18 but also can associate with an additional class C VPS component Vps16 as well as the class B components Vps39 and Vps41 in mammalian cells. Loss-of-function studies using small interfering RNAs (siRNAs) targeting FHF complex components indicate that trafficking of epidermal growth factor (EGF) from early to late endosome/lysosomes is delayed when members of this complex are absent, suggesting that the FHF complex promotes endosomal trafficking. Moreover, previous studies have linked overexpression of HOPS complex components with the clustering and fusion of both early and late endocytic components (Mullock et al., 2000; Poupon et al., 2003; Richardson et al., 2004). We find that the ability of Vps18 to promote lysosomal clustering is compromised upon depletion of FTS, Hook proteins, and p107FHIP; moreover, overexpression of FTS or Hook proteins leads to lysosomal-associated membrane protein (LAMP) 1-positive endosomal clusters. These data suggest that the FHF complex may help to coordinate vesicle movement, tethering, or both via the HOPS complex.
MATERIALS AND METHODS
Plasmids and Antibodies
Open reading frames (ORFs) for FTS, Hook1, Hook2, Hook3, p107FHIP (C11ORF56), Vps16, Vps18, Vps39, and Vps41 were obtained by polymerase chain reaction (PCR) by using cDNAs as templates. ORFs were ligated into pENTR, sequence verified, and recombined into the appropriate vector using in vitro recombination (Invitrogen, Carlsbad, CA). For expression of glutathione transferase (GST)-fusion proteins in mammalian cells, pDEST27 (cytomegalovirus-GST) was used as a recipient for homologous recombination of the appropriate pDONR plasmid. Mutagenesis was performed using a QuikChange mutagenesis kit (Stratagene, La Jolla, CA). Plasmids containing cDNAs for Hook proteins as well as the corresponding antibodies were generously provided by Helmut Kramer (University of Texas Southwestern, Dallas, TX). Plasmids expressing Vps18 and Vps39 and the corresponding antibodies were generously provided by Robert Piper (University of Iowa, Iowa City, IA). Crude anti-FTS was from Stephen Michnick (University of Montreal, Montreal, QC, Canada), and for some experiments, the antibody was purified using immobilized GST-FTS from bacteria. Anti-actin was from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-early endosome antigen (EEA) 1, anti-CD63, and anti-LAMP1 antibodies were from BD Biosciences Transduction Laboratories (Lexington, KY), AbD Serotec (Raleigh, NC), and Development Studies Hybridoma Bank (University of Iowa), respectively.
Cell Culture and Transfections
To generate cells stably expressing FTS, pMSCV-FLAG-hemagglutinin (HA)-FTS DNA was transfected into Pheonix packaging cells and packaged virus used to infect human embryonic kidney (HEK)293T cells at a multiplicity of infection of 1. After 2 d, cells were selected in puromycin (1 μg/ml). Transient transfections were performed using FuGENE 6 (Roche Diagnostics, Indianapolis, IN) and the indicated plasmid DNAs. Cells were harvested at the indicated times before analysis.
siRNA transfections used 20 nM of the indicated siRNA (or pool of the indicated siRNAs) using oligofectamine (Invitrogen). In cases where both siRNA and expression plasmids were transfected, the siRNAs were first transfected followed 48 h later by plasmid transfection by using FuGENE 6. The sequences of all siRNAs used in this study were purchased from Invitrogen or Ambion (Austin, TX) and had the following sequences: siFTS-1, UAAUGCAGAGCGAUAAGAUGGCUGC; siFTS-2, UGCAAAUGCUCUCUUCACAUCCA GC; siHOOK1–1, UUAACAUGAAGAUCUUGAACCUGCC; siHOOK1–2, AUUUGAUGAAGAACUUGUGCCAUGG; siHOOK2–1, AUCUGGUUCAGCACAUAGGCUACGG; siHOOK2–2, AUGCUGAACCGAUUCUUCCAGCGUC; siHOOK3–1, UUAACUCUUCCUCUAGACUGACAGU; siHOOK3–2, UUAUCUGCUUUCUUUGAUUCUUCGG; siFHIP-1, GGCGGG CUGAGCAACUGAAACUAUU; and siFHIP-3, CACUCUUCAUGAGUUCCCUGGAGUU.
Protein Interactions and Mass Spectrometry
For tandem purification of FLAG-HA-FTS, lysates from cells grown on ten 15-cm dishes were incubated with anti-FLAG M2 antibodies (500 μl of beads at 1 mg/ml antibody; Sigma-Aldrich, St. Louis, MO) for 4 h at 4°C followed by elution with a 3X FLAG-peptide (250 μg/ml). The anti-FLAG eluate was then subjected to purification using anti-HA affinity resin (100 μl of 2 mg/ml) and subsequently eluted with HA peptide (250 μg/ml) (Jin et al., 2006). Where noted, the FLAG immunoprecipitation was omitted. Eluted proteins were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) on a 4–20% gradient gel. Bands were excised and subjected to mass spectrometry on an LTQ ion trap mass spectrometer (Thermo Fisher Scientific, Waltham, MA), or for eluted protein complexes, proteins were precipitated using trichloroacetic acid, digested, with trypsin, and analyzed by liquid chromatography/tandem mass spectrometry (LC/MS/MS).
For determining interactions in mammalian cells, cells were lysed in 50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, and 10 mM NaF with protease inhibitors (Roche Diagnostics), unless otherwise noted. Where noted, 0.5% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) was substituted for NP-40 as detergent. Cleared lysates were incubated with the indicated antibodies or affinity resin at 4°C for 1–2 h, and proteins were eluted from washed resins by using 2X SDS-PAGE sample buffer. Proteins were subjected to SDS-PAGE and immunoblotted using the indicated antibodies.
For two-hybrid measurements, the indicated ORF was recombined into either a GAL4 activation domain or GAL4 DNA binding domain fusion expression plasmid and transformed into PJ69-4A cells with selection for Trp and Leu auxotrophy, respectively, as described previously (Xu et al., 2003). Cells were then plated on media lacking Trp, Leu, Ade, and His and strain growth monitored. Two-hybrid library screens using a HeLa cell activation domain library were performed as described previously (Xu et al., 2003).
EGF Translocation and Endosomal Clustering Assays
For EGF translocation assays, HeLa cells were serum starved for 2 h and treated with 500 ng/ml Texas Red-conjugated EGF (EGF-TR) on ice. Cells were left on ice for 15 min and then transferred to 19.5°C to initiate the internalization and incubated for 1 h before washing the cells with phosphate-buffered saline. Cells were then transferred to 37°C to initiate EGF transit through the endocytic system. At the indicated times, cells were fixed with 4% paraformaldehyde. Cells were then incubated in 3% bovine serum albumin (BSA) in PBS followed by anti-EEA1, anti-CD63, or anti-LAMP1 antibodies at 1:200 dilution. Secondary antibodies were coupled to Alexa488 (Invitrogen). Images were captured using a FluoView confocal laser-scanning microscope (Olympus, Tokyo, Japan).
Quantification was performed using the program MetaMorph (Molecular Devices, Sunnyvale, CA). A median filter of 32 × 32 was applied to reduce the background for each image. Threshold was set for images from both fluorescein isothiocyanate (FITC) and cyanine (Cy) 3 filter channels and kept constant throughout the analysis. Colocalization was analyzed in 30–45 cells among three separate slides, representing three independent experiments. The marker area was defined as the sum of all the contiguous pixels of green signals that met the color threshold. The EGF area was defined as the sum contiguous pixels of red signals that met the color threshold. The percentage of areas of EGF that colocalize with endosome markers were recorded. Statistical significance was determined using a t test (mean ± SEM) is shown.
For Vps18-dependent endosomal clustering, HeLa cells were transfected with 20 nM of the indicated siRNA (or pool of the indicated siRNAs) by using Oligofectamine. After 2 d, cells were again transiently transfected with 1 μg GFP-Vps18 per well for 12-well plates by using FuGENE 6. Cells were fixed in 4% paraformaldehyde 60 h post-GFP-Vps18 transfection. Saponin extraction before fixation was performed as described previously (Richardson et al., 2004). Cells were then incubated in 3% BSA in PBS and then with anti-LAMP1 antibody at 1:200 dilution. Secondary antibodies were coupled to Alexa598 (Invitrogen). Images were captured using an Olympus FluoView confocal laser-scanning microscope. Parallel transfections were performed for immunoblot analysis to demonstrate depletion of the indicated proteins. To detect p107FHIP, cells were cotransfected with a vector expressing GST-p107FHIP, and the level of expression determined by blotting with anti-GST antibodies.
Slides were scanned for GFP–Vps18 clusters at high exposure to detect both dispersed Vps18 staining and clustered staining. Individual cluster-positive cells were then imaged through both FITC and Cy3 filter channels at lower exposure to ensure the maximum pixel intensity within the linear range of the charge-coupled device camera. Lamp1 signals at low exposures that are above a threshold intensity were selected in the Cy3 channel. The area and intensity of the selected pixels are integrated to obtain the integrated intensity and plotted for individual cells. More than 30 cells from three different slides representing three independent experiments were analyzed for each condition by determining the integrated pixel intensity above the threshold for Cy3. Statistical analysis on three independent experiments was performed using either Student's t test or Fisher (F)-test.
Gel Filtration
HeLa cells transfected with indicated siRNAs were lysed in 50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, and 10 mM NaF with protease inhibitors (Roche Diagnostics). Lysate (100 μl) was then loaded onto Superdex 200 column (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). The column was washed with 50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% NP-40, and 10 mM NaF. Each fraction (0.5 ml/fraction) was collected, precipitated with trichloroacetic acid (TCA), and analyzed by Western blot. In some experiments as detailed in figure legends, HEK293T cell lysates were used. To analyze FLAG—HA–FTS complexes by gel filtration, the complex was isolated from four 15-cm plates of HEK293T/FLAG-HA-FTS cells, the protein eluted with anti-HA antibodies and subjected to gel filtration. Column fractions were either subjected to immunoblotting with the indicated antibodies or precipitated with TCA, trypsinized, and subjected to LC/MS/MS.
RESULTS
FTS Associates with Mammalian Hook Proteins
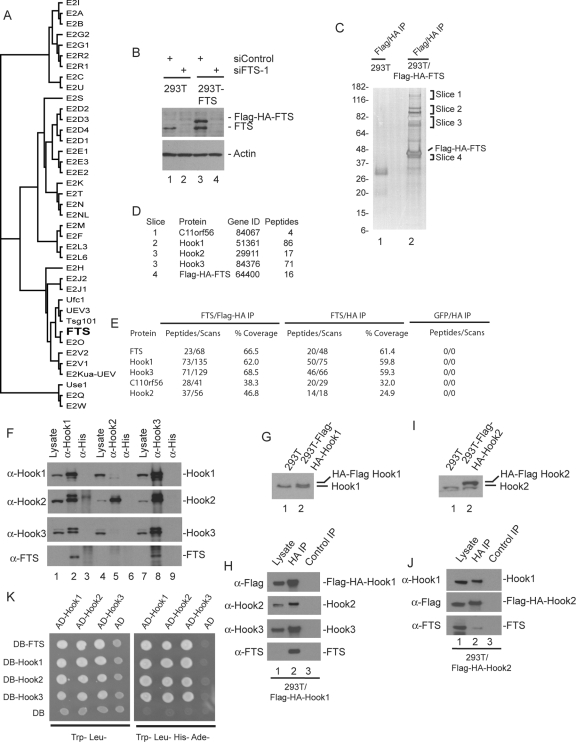
FTS is a member of a subfamily of ubiquitin-conjugating enzymes that lack a catalytic cysteine residue (Figure 1A). The physiological function of FTS is unknown. To begin to understand FTS function, we used a proteomic approach. We created HEK293T cells that stably express FLAG-HA-FTS under control of the long terminal repeat (LTR) promoter via a murine stem cell virus (MSCV)-based retrovirus at near endogenous levels (Figure 1B). Sequential FLAG and HA or single HA immune complexes were prepared from extracts of these cells as well as control cells, and proteins were analyzed by SDS-PAGE coupled with mass spectrometry (Figure 1, C and D; see Materials and Methods) or by direct LC/MS/MS analysis of proteins eluted from the anti-HA column using an HA peptide (Figure 1E and Supplemental Table 1). Several proteins were found in both the tandem and HA immune complexes that were absent in immune complexes from control cells expressing either empty vector or FLAG-HA-GFP. The most abundant specific proteins identified were the mammalian orthologues of the Drosophila hk protein (Hook1, Hook2, and Hook3) as well as the previously uncharacterized protein C11ORF56 (Figure 1, D and E). Although FTS was previously found to interact with Akt and PDK1 when overexpressed (Remy and Michnick, 2004), neither of these protein kinases were detected in FTS complexes.
Figure 1.
Proteomic analysis of FTS complexes identifies a new complex containing Hook proteins and C11ORF56/p107FHIP. (A) Phylogenetic tree of the human E2 conjugating family, generated using ClustalW. The variant E2 protein FTS lacking the active site cysteine residue found in catalytically active E2 enzymes is shown in bold. (B) Immunoblot analysis of extracts from HEK293T cells or HEK293T cells stably expressing FLAG-HA-FTS from an MSCV-based retrovirus by using anti-FTS antibodies. To demonstrate the specificity of the antibodies, cells were previously transfected with control siRNA or an siRNA targeting FTS. Blots were stripped and reprobed with actin as a loading control. (C and D) SDS-PAGE and mass spectral analysis of FLAG-HA-FTS complexes. Tandem anti-FLAG/anti-HA immune complexes from HEK293T cells or HEK293T/FLAG-HA-FTS cells separated on a 4–20% gradient SDS-PAGE gel and stained with silver (C). The indicated gel slices were analyzed by mass spectrometry (D). The number of independent peptides identified for each protein is shown. These proteins were not found in the negative control (lane 1). (E) LC/MS/MS analysis of tandem and single HA purified FLAG–HA–FTS complexes. Tandem (FLAG-HA) or single anti-HA purified complexes derived from four 15-cm dishes of cells were eluted with HA peptide, precipitated with trichloroacetic acid to remove the eluting peptide, trypsinized, and subjected to LC/MS/MS. Cells expressing FLAG-HA-GFP were used in parallel as a control. The number of independent peptides as well as the total number of scans for these peptides is indicated, as well as the coverage of each protein. (F) Extracts from HEK293T cells were subjected to immunoprecipitation using anti-Hook1, anti-Hook2, or anti-Hook3 antibodies (or anti-His tag antibodies as a negative control), and complexes were subjected to immunoblotting by using the indicated antibodies. Lysates (5% of input) were included in the blot. Note that under these conditions, the crude FTS antisera used in this experiment does not readily detect FTS in crude cell extracts, and FTS is highly enriched in the anti-Hook1 and Hook3 immune complexes. (G) HEK293T cells stably expressing FLAG-HA-Hook1 were generated and extracts examined by immunoblotting using antibodies against Hook1. (H) Anti-HA or control immune complexes from HEK293T/FLAG-HA-Hook1 cells were immunoblotted with the indicated antibodies, with lysate (5%) as a positive control. Note that under these conditions, the crude FTS antisera used in this experiment does not readily detect FTS in crude cell extracts, and FTS is highly enriched in the anti-Hook1 and Hook3 immune complexes. (I) HEK293T cells stably expressing FLAG-HA-Hook2 were generated and extracts examined by immunoblotting using antibodies against Hook2. (J) Anti-HA or control immune complexes from HEK293T/FLAG-HA-Hook2 cells were immunoblotted with the indicated antibodies, with lysate (5%) as a positive control. In this experiment, affinity-purified anti-FTS was used, which was capable of detecting FTS present in cell extracts (lane 1). (K) Two hybrid analysis of FTS and Hook proteins. The indicated open reading frames were cloned into either pDB or pAD vectors by using empty vectors as negative controls (see Materials and Methods) and transformed into PJ69-4A cells. Cells were plated on either Trp−, Leu− media to select for plasmids or on Trp−, Leu−, His−, Ade− to demonstrate the two hybrid interaction.
Using available anti-Hook antibodies (Walenta et al., 2001), we found that anti-FLAG immune complexes from cells stably expressing FLAG-HA-FTS contained Hook1, Hook2, and Hook3 (Supplemental Figure S2A), confirming the proteomic results. To validate the interaction between endogenous Hook proteins and FTS, Hook proteins were individually immunoprecipitated from HEK293T cells, and immunoblots were probed for endogenous FTS and Hook proteins. Hook1 and Hook3 immune complexes are highly enriched in both FTS and Hook2 (Figure 1F, lanes 2 and 8). Hook1 and Hook3 also were found to reciprocally associate with each other (Figure 1F, lanes 2 and 8). As an independent approach, we generated cells stably expressing FLAG-HA-Hook1 at endogenous levels (Figure 1G) and examined anti-HA immune complexes for associated proteins by using immunoblotting. Hook1 associated with Hook2, Hook3, and FTS (Figure 1H, lane 2). We found that immune complexes prepared from available Hook2 antibodies contained Hook2 but lacked FTS and displayed very low levels of Hook1 and Hook3 (Figure 1F, lanes 5). The simplest interpretation of this result is that the available Hook2 antibodies preferentially precipitate free Hook2 protein. To examine this question further, we generated cells expressing FLAG-HA-Hook2 at approximately threefold higher than endogenous levels (Figure 1I). Anti-HA immune complexes from these cells revealed the association of endogenous Hook1 and FTS with FLAG-HA-Hook2 (Figure 1J, lane 2). Finally, although available FTS antibodies perform poorly for immunoprecipitation, we were able to observe endogenous Hook1 and Hook3 in anti-FTS immune complexes (Supplemental Figure S2B). These data indicate that FTS is capable of interacting with one or more Hook proteins and that endogenous Hook proteins form complexes with each other.
Anatomy of the Hook–FTS Complex
The finding that Hook proteins coimmunoprecipitate each other led us to examine their ability to form homo- and heterotypic interactions by using the two-hybrid system. Each Hook protein was found to interact with itself and with each of the other Hook proteins (Figure 1K). Previous studies suggest that Drosophila hk can homodimerize via its coiled-coil domain (Kramer and Phistry, 1996). In addition, each Hook protein was found to interact with FTS in the two-hybrid system, suggesting that Hook proteins share a binding site for FTS (Figure 1K).
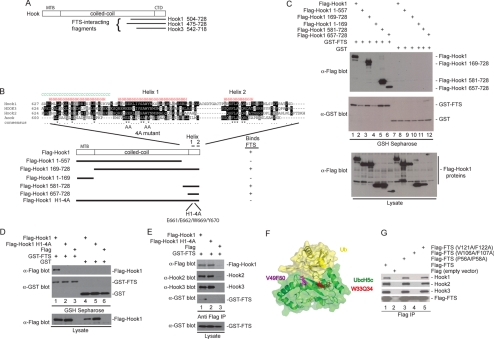
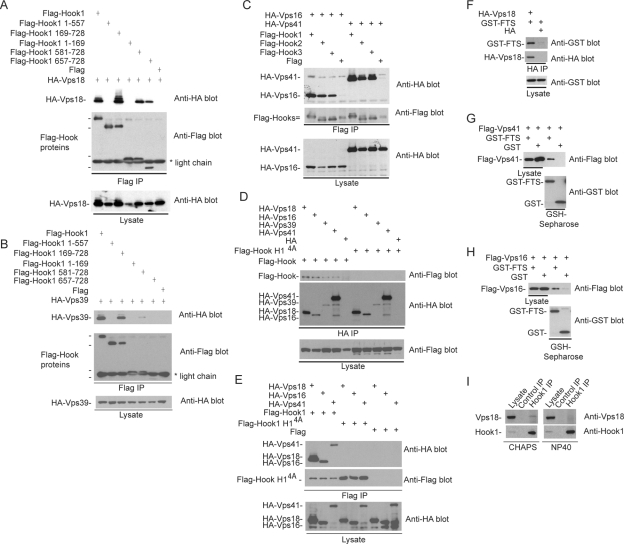
We next sought to determine the structural requirements for the Hook–FTS interaction. A parallel two-hybrid screen using a HeLa cell library and FTS as bait identified two Hook1-interacting clones as well as a single Hook3 interacting clone. Each of these clones initiated in the C-terminal half of the coiled-coil domain and extended to the C terminus of the protein (Figure 2A), placing the FTS interacting domain in the C terminus of Hook proteins. A series of mutants in Hook1 were generated, including Hook1 lacking the C terminus (residues 1–557) and two Hook1 fragments containing only the C-terminal region (residues 581–728 and 657–728) (Figure 2B). In addition, fragments either lacking the N terminus (residues 169–728) or containing only the N terminus (1–169) were generated (Figure 2B). Plasmids expressing these constructs as FLAG-tagged fusions were transfected with plasmids expressing either GST-FTS or GST alone. After 48 h, proteins associated with glutathione-Sepharose were examined by Western blotting. Hook11-557 and Hook11-169 failed to interact with GST-FTS (Figure 2C, lanes 2 and 4). In contrast, all three constructs containing the extreme C terminus of Hook1 associated with GST-FTS (Figure 2C, lanes 3, 5, and 6). Similar results were seen with Hook2 and Hook3 (Supplemental Figure S2D; data not shown). Interestingly, although Hook1657-728 was present in extracts from cells coexpressing GST-FTS (Figure 3C, lane 6), it was absent in extracts from cells coexpressing GST (lane 12). One explanation for this is that Hook1657-728 is unstable in the absence of an FTS binding partner. Indeed, addition of the proteasome inhibitor MG132 to cells before harvesting led to the accumulation of Hook1657-728 in the absence of coexpressed GST-FTS (Supplemental Figure S2C).
Figure 2.
Anatomy of the FHF complex. (A) A two-hybrid screen for proteins that interact with FTS by using a HeLa cells activation domain library was performed in PJ69-4A cells. Two Hook1 clones and one Hook3 clone were recovered. The location of the FTS-interacting clones on the Hook domain structure is shown. (B) Schematic of Hook1 fragments generated along with the sequence of the C-terminal FTS interacting region of Hook proteins and the results of binding studies. The positions of 2 helices in the C terminus of Hook1 (as determined using the JPRED secondary structure prediction tool; http://www.compbio.dundee.ac.uk/∼www-jpred/) are shown. The positions of mutations in helix 1 within the Hook14A mutant are indicated by “A” for alanine. (C) A C-terminal fragment of Hook1 is necessary and sufficient for interaction with FTS. HEK293T cells were transfected with vectors expressing the indicated proteins and after 48 h, cell extracts were generated and incubated with GSH-Sepharose. Bound proteins and control lysates were subjected to immunoblotting with the indicated antibodies. (D) The Hook14A mutant cannot bind FTS. Experiments were performed as described in C. (E) The Hook14A mutant maintains interaction with Hook2 and Hook3. HEK293T cells were transfected with vectors expressing the indicated proteins and after 48 h, cell extracts were generated and incubated with anti-FLAG (M2) resin. Bound proteins and control lysates were subjected to immunoblotting with the indicated antibodies. (F) A structure of UbcH5 bound to ubiquitin (PDB code: 2fuh), showing the residues in UbcH5 that correspond structurally to the W106/F107 and V121/F122 residues mutated in FTS, based on sequence alignments using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html). (G) FTSW106AF107A fails to interact with Hook proteins. Plasmids expressing the indicated FLAG-tagged wild-type or mutant FTS proteins were transfected into HEK293T cells, and after 48 h, cell extracts were generated and incubated with anti-FLAG (M2) resin. Bound proteins and control lysates were subjected to immunoblotting with the indicated antibodies.
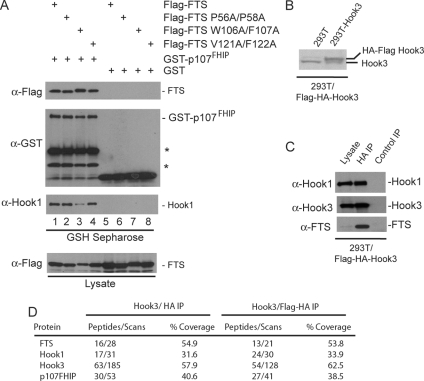
Figure 3.
p107FHIP interacts with FTS and Hook proteins to form the FHF complex. (A) GST-p107FHIP interacts with FTS and Hook1. Plasmids expressing the indicated FLAG-tagged wild-type or mutant FTS proteins were transfected into HEK293T cells together with plasmids expressing either GST-p107FHIP or GST and after 48 h, cell extracts were generated and incubated with GSH-Sepharose. Bound proteins and control lysates were subjected to immunoblotting with the indicated antibodies. (B–D) HEK293T cells stably expressing FLAG-HA-Hook3 were generated, and extracts were examined by immunoblotting using antibodies against Hook3 (B). Anti-HA or control immune complexes from HEK293T/FLAG-HA-Hook3 cells were immunoblotted with the indicated antibodies (C) or eluted with HA peptide before trypsinization and LC/MS/MS analysis (D). The number of independent peptides as well as the total number of scans for these peptides is indicated, as well as the coverage of each protein.
Secondary structure analysis of Hook1 revealed the presence of two putative helices at the extreme C terminus (Figure 2B). These two helices were well conserved with Drosophila hk. We found that a quadruple mutant Hook1 protein in which C-terminal helix 1 residues E661, E662, W669, and Y670 were mutated to alanine (the Hook14A mutant) was unable to bind GST-FTS (Figure 2D, lane 2). In contrast, this mutant protein efficiently associated with endogenous Hook2 and Hook3 proteins (Figure 2E, lane 2), indicating that neither the C-terminal helix 1 nor the interaction with FTS is required for Hook–Hook interaction. Together, these data indicate that FTS interacts with Hook proteins via a conserved helix near the C terminus of Hook proteins.
FTS Mutants Defective in Hook Binding
Structural studies of the catalytically active ubiquitin conjugating enzyme UbcH5 have demonstrated that it can bind ubiquitin through the β-sheet region that constitutes the core of the UBC fold (Brzovic et al., 2006), and it is distinct from the surface that makes up the active site. TSG101 is known to interact with ubiquitin through the same side of the UBC fold but makes the most productive interactions with an extended loop (referred to as the “β-tongue”), which is absent in UbcH5 (Sundquist et al., 2004). Given the ability of this β-sheet surface to function in protein binding, we wanted to test the role of this and other regions of FTS in association with Hook proteins. Structural modeling revealed W33Q34 and V49F50 in UbcH5 corresponds to W106F107 and V121F122 in FTS (Figure 2F; data not shown). Mutation of V121 and F122 to alanine had no effect on the ability of FLAG-FTS to associate with endogenous Hook1, Hook2, or Hook3 in transfected cells (Figure 2G, lane 5). In contrast, FTSW106AF107A failed to associate with Hook proteins in this assay (Figure 2G, lane 4). We also found that mutation of P56 and P58 located outside the UBC domain had no effect on association with Hook proteins (Figure 2G, lane 3). Thus, FTS seems to require its central β-sheet region to interact with C-terminal helix 1 in Hook proteins.
The Hook–FTS Complex Assembles with a Novel Protein, p107FHIP
As noted, FLAG–HA–FTS complexes contained a previously uncharacterized 107-kDa protein encoded by C11ORF56, as determined by mass spectrometry (Figure 1, D and E). Apparent orthologues were found in mouse, zebrafish, Drosophila, Caenorhabditis elegans, and a weakly related protein was identified in Saccharomyces cerevisiae (Supplemental Figure S3A; data not shown). C11ORF56 displays weak similarity to two other previously uncharacterized proteins, NP_001103447 and a retinoic acid inducible transcript 16 (RAI16) (Supplemental Figure S3A). The C-terminal portion of C11ORF56 contains a region that displays similarity to domain in S. cerevisiae Rud3/YOR216C referred to as the GRAB domain, which is thought to facilitate localization with Golgi structures (Gillingham et al., 2004). Two distinct isoforms of C11ORF56 have been identified (isoform 1, NP_115503.2 and isoform 2, NP_001092264.1), which differ by 14 amino acids in the central region of the protein. Positive identification of the longer isoform 1 was seen by mass spectrometry of C11ORF56 in the FLAG–HA–Hook1 complex via identification of a tryptic peptide overlapping the region deleted in isoform 2 (R.HHAPSPPRPEHASWARGGPSR.E; Charge: +3; XCorr 1.6947; ΔCn 0.0611). Using multiple approaches, we have found that this protein is a component of the FTS–Hook complex; therefore, we refer to this protein as p107FHIP (FTS-Hook Interacting Protein).
We first examined the association of Hook and FTS with p107FHIP by using transient transfection. Vectors expressing GST-p107FHIP (or GST as a negative control) were cotransfected together with vectors expressing FLAG-FTS or FTS mutant proteins. GST-p107FHIP was purified using GSH-Sepharose and associated proteins examined by immunoblotting. GST-p107FHIP was found to associate efficiently with FTS and all three FTS mutants (Figure 3A, lanes 1–4), whereas GST alone did not (lanes 5–8). In addition, GST-p107FHIP was found to associate with endogenous Hook1 (Figure 3A, lanes 1–4). Interestingly, the association of Hook1 with GST-p107FHIP was significantly reduced in the context of expression of FTSW106AF107A, which interacts poorly with Hook1. This result suggests that p107FHIP interacts with Hook1 primarily via association with FTS, although we cannot exclude direct contact with Hook proteins.
To verify the interaction of p107FHIP with the Hook complex, we used cells that stably express FLAG–HA–Hook3 complex at near endogenous levels (Figure 3B). As expected, HA immune complexes from these cells contain endogenous Hook1 and FTS (Figure 3C). These complexes were subjected to direct LC/MS/MS analysis after either dual FLAG/HA purification or HA only purification, revealing the presence of abundant peptides for p107FHIP (Figure 3D). These data indicate that p107FHIP is a component of a complex containing Hook proteins and FTS. We did not observe peptides containing the unique tryptic products of p107FHIP isoform 2, but we cannot exclude that this isoform also interacts with the Hook–FTS complex.
FTS, Hook Proteins and p107FHIP Assemble into a Single Major Complex of ∼500 kDa
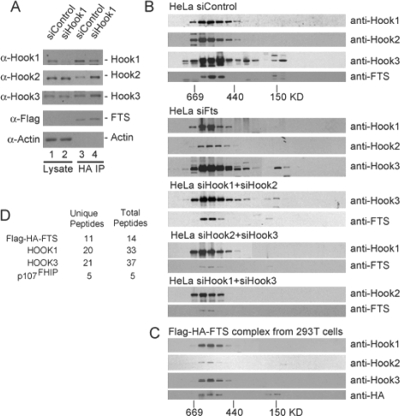
Although the results presented thus far indicate that FTS can interact with Hook proteins individually and Hook proteins can interact with each other, the oligomeric state of the complex was unknown. In principle, FTS and p107FHIP could form independent complexes with individual Hook proteins, with each Hook protein forming hetero- and homodimers. Alternatively, FTS and p107FHIP could associate with a single complex containing all three Hook proteins. Indeed, coiled-coil proteins have been shown to form stable dimers or trimers (e.g., GCN4) (Harbury et al., 1993, 1994). Initially, we found that upon depletion of Hook1 from HEK293T/FLAG-HA-FTS cells, the levels of both Hook2 and Hook3 increased proportionally within the FTS immune complex (Figure 4A, lanes 3 and 4) but were unchanged in lysates (Figure 4, lanes 1 and 2). This result suggests that Hook2 or Hook3 may assemble interchangeably into the Hook/FTS complex and may substitute for Hook1. To examine this question further, we used gel filtration of both purified FLAG–HA–FTS/Hook/p107FHIP complexes and cell extracts followed by immunoblotting or mass spectrometry analysis. Analysis of extracts indicated that FTS and Hook proteins migrated as a single major complex at ∼500 kDa (Figure 4B). Depletion of FTS had no detectable effect on the migration properties of Hook proteins, consistent with the idea that Hook proteins interact with each other independently of FTS (Figure 4B and Supplemental Figure S2E). Moreover, depletion of Hook1 and Hook2 did not alter the migration properties of Hook3 or FTS (Figure 4B and Supplemental Figure S2E). Likewise, depleting Hook2 and Hook3 or Hook1 and Hook3 does not affect the migration of the remaining Hook protein (Figure 4B and Supplemental Figure S2E). If Hook proteins do not interact interchangeably within the complex, the expectation would be that depletion of any particular family member would lead to a reduction in the apparent mass of the complex by ∼80 kDa. The absence of such a change in mass upon deletion of any single Hook protein or multiple Hook proteins suggests that the complex is an obligate multimer, with Hooks assembling interchangeably to form the complex. One caveat to this conclusion, however, is that the migration properties of complexes containing coiled-coil proteins are not always sensitive to loss of subunits, because the elongated structure dominates the migration properties.
Figure 4.
FTS, Hook proteins, and p107FHIP form a single major complex as assessed by gel filtration. (A) Effect of Hook1 depletion on the abundance of Hook2 and Hook3 proteins in complexes with FTS. Extracts were made from HEK293T/FLAG-HA-FTS cells transfected with the indicated siRNAs and subjected to immunoprecipitation with anti-HA antibodies. Blots were probed with the indicated antibodies. (B) Gel filtration analysis of FTS/Hook complexes in wild-type cells and in cells depleted of either FTS or Hook proteins. HeLa cells were transfected with the indicated siRNAs for 48 h before lysis (see Supplemental Figure S2 for depletion). Extracts were subjected to gel filtration by using a Superdex 200 column as described in Materials and Methods (B). Aliquots of fractions were analyzed by immunoblotting using the indicated antibodies. Anti-FTS antibodies were affinity purified. (C) Gel filtration analysis of FLAG–HA–FTS complexes purified from HEK293T cells. Complexes purified as described in Figure 1 were subjected to gel filtration using a Superdex 200 column, and fractions were analyzed by immunoblotting. (D) LC/MS/MS analysis of purified FLAG–HA–FTS complexes after gel filtration. FLAG–HA–FTS complexes purified as described in Figure 1 were subjected to gel filtration as described in B. Fractions corresponding to the peak of Hook/FTS proteins by immunoblotting (C) were precipitated with trichloroacetic acid, trypsinized, and subjected to LC/MS/MS analysis to identify p107FHIP. The number of unique peptides and total scans for the relevant proteins are indicated.
To confirm the presence of p107FHIP in the 500-kDa complex, we examined the components of FLAG-HA-FTS/Hook complexes purified from HEK293T/FLAG-HA-FTS cells using mass spectrometry. The purified complex migrated at a position indistinguishable from the complex observed in crude extracts (Figure 4C). Fractions corresponding to the 500-kDa size range contained peptides derived from p107FHIP, FTS, as well as Hook proteins (Figure 4D). The size of the complex is larger than anticipated based on the presence of three Hook proteins, one FTS molecule and one p107FHIP molecule. This could reflect the likely elongated structure of coiled-coil protein containing complexes, which increases the apparent size of proteins in gel filtration analysis. Alternatively, multiple molecules of p107FHIP may be present in the complex. Together, these studies indicate that FTS, Hook proteins, and FHIP interact in an ∼500-kDa complex, the FHF complex.
The FHF Complex Associates with the HOPS Complex
The experiments described thus far indicate that Hook, FTS, and p107FHIP form a stable soluble complex. However, previous work suggests that endogenous Hook1 can also interact with endogenous Vps18, a component of the class C HOPS complex (Richardson et al., 2004). Likewise, Drosophila hk interacts with Drosophila Vps18/Dor in the two-hybrid system (Lloyd et al., 1998). Using transient transfection, we found that FLAG-tagged Hook1 interacts with HA-tagged Vps16, Vps18, Vps39, and Vps41 and that Vps16 and Vps41 also interacted with Hook2 and Hook3 (Figure 5, A–C). The pattern of interactions of Vps18 with a panel of Hook1 deletions paralleled that found with FTS interaction, indicating that Vps18 proteins interact with Hook1 via its C-terminal domain (Figure 5A).Vps39 displayed weaker interaction with the C-terminal Hook fragments than did Vps18 (Figure 5B). As with FTS, FLAG-Hook14A containing mutations in helix 1 of the C terminus of Hook1 failed to interact with Vps18, Vps16, Vps39, or Vps41 in reciprocal immunoprecipitation experiments (Figure 5, D and E). In parallel, we found that HA-tagged Vps18, Vps41, and Vps16 can interact with GST-FTS in transfected cells (Figure 5, F–H). Given the interaction seen between transfected FTS/Hook proteins and VPS proteins in transient expression experiments, we revisited the question of why VPS proteins were not observed in the initial FTS complex mass spectrometry analysis. We reasoned that the more stringent detergent used for the proteomic experiments may reduce interactions between the FHF and HOPS complexes. We found that endogenous Vps18 was readily seen in anti-Hook1 immune complexes from HeLa cells by using CHAPS as detergent, whereas Vps18 was undetectable in Hook1 immune complexes from cells lysed with buffer containing NP-40 (Figure 5I). Presumably, transient expression of FTS, Hook, or VPS components allows visualization of the interaction even in the presence of NP-40. Together with the finding that endogenous Hook1 can interact with endogenous Vps18 (Richardson et al., 2004), we conclude that the FHF complex can assemble with the HOPS complex.
Figure 5.
FTS and Hook proteins associate with class B and class C components of the HOPS complex. (A–E) Plasmids expressing the indicated HA-tagged Vps or FLAG-tagged Hook constructs were transfected into HEK293T cells and after 48 h, protein complexes were purified using the indicated anti-HA or anti-FLAG (M2) antibodies. Washed complexes and lysates as loading controls were subjected to SDS-PAGE and immunoblotting. (F–H) Plasmids expressing the indicated HA-tagged Vps or GST-tagged FTS constructs were transfected into HEK293T cells and after 48 h, protein complexes were purified using GSH-Sepharose. Washed complexes and lysates as loading controls were subjected to SDS-PAGE and immunoblotting. (I) Association of endogenous Hook1 with endogenous Vps18. HeLa cells were lysed in either buffer containing NP-40 or buffer containing CHAPS (see Materials and Methods), and cleared extracts were subjected to immunoprecipitation by using anti-Hook1 antibodies. Immune complexes were immunoblotted with either anti-Vps18 or anti-Hook1 antibodies.
Components of the FHF Complex Promote Lysosomal Clustering upon Overexpression of Vps18
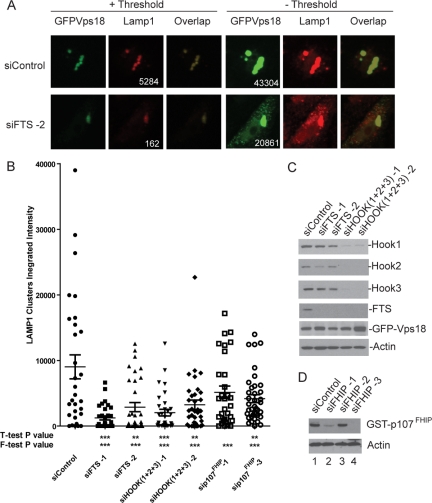
The ability of HOPS components to promote clustering of early (EEA1) and late (LAMP1) endosome/lysosomes has frequently been used to assay the homotypic fusion activity of this complex (Mullock et al., 2000; Caplan et al., 2001; Kim et al., 2001; Poupon et al., 2003; Richardson et al., 2004). To examine the potential functional interplay between the FHF complex and the HOPS complex, we asked whether depletion of components of the FHF complex affected the ability of overexpressed Vps18 to promote late endosomal/lysosomal clustering in HeLa cells. As shown previously (Poupon et al., 2003; Richardson et al., 2004), transfection of green fluorescent protein (GFP)-Vps18 led to the clustering of both GFP-Vps18 and LAMP1-positive endosome/lysosomes in a juxtanuclear pattern in ∼40% of GFP–Vps18-positive cells (Figure 6A and Supplemental Figure S4), a phenotype that was not observed by expression of GFP alone (data not shown). Quantitative imaging of the integrated intensity of GFP-Vps18 and LAMP1 staining in >35 individual cells revealed a range LAMP1 staining intensity, with a mean near 9000 pixels. In contrast depletion of FTS by using two independent siRNAs led to a substantial reduction in the integrated intensity of LAMP1 associated with GFP–Vps18 clusters; 1000 for siFTS-1 and 3000 for siFTS-2 (Figure 6B), despite the presence of similar overall levels of GFP-Vps18 (Figure 6C). In the absence of thresholding (used for quantification; see Materials and Methods), dispersed LAMP1 and GFP–Vps18-positive endosomes were frequently seen throughout the cytoplasm in cells depleted of FTS but were rare in cells transfected with control siRNA (Figure 6A). The cells shown in Figure 6A represent an example of cells displaying the mean integrated intensity for each condition. Similarly, simultaneous depletion of all three Hook proteins or p107FHIP led to a reduction in LAMP1-positive staining in GFP–Vps18 clusters (Figure 6B and Supplemental Figure S4), again with similar total levels of GFP-Vps18 (Figure 6C). In this context, the depletion of FHIP had a somewhat weaker effect than that seen with depletion of either FTS or Hook proteins. In all cases, the effects observed reached statistical significance using the F-test (***p < 0.001). The frequency of cells displaying LAMP1 staining intensity >10,000 pixels within the GFP–Vps18 cluster is reduced upon depletion of FTS, Hook, and p107FHIP proteins (Supplemental Figure S5). These data indicate that FTS functions to promote vesicle clustering and/or fusion promoted by Vps18.
Figure 6.
Depletion of FHF complex components reduces the ability of Vps18 to promote late endosome/lysosome clustering. (A) HeLa cells were transfected with control siRNA or siRNA targeting FTS. After 48 h, cells were transfected with a plasmid expressing GFP-Vps18, and 60 h later, late endosomal/lysosomal clusters were examined by immunofluorescence using anti-LAMP1 antibodies in conjunction with detection with Alexa598-conjugated secondary antibodies (red). GFP-Vps18 was identified by GFP fluorescence (green). To determine the integrated intensity for LAMP1 within clusters, a threshold (+ Threshold) was applied such that the maximal pixel signal was in the linear range. In the absence of threshold (− Threshold), individual vesicles not present within clusters can be seen in cells wherein FTS was depleted. Integrated intensities of Vps18 before thresholding (including both clustered Vps18 and dispersed Vps18), as well as LAMP1 aggregates after thresholding, are presented. The integrated intensities for LAMP1 and GFP-Vps18 are shown. The integrated intensities for GFP-Vps18 ranged from ∼20,000 to ∼40,000 over all the cells analyzed. (B) Quantification of the effects of FTS depletion on Vps18-mediated endosomal clustering. The integrated intensity of LAMP1 within GFP–Vps18-positive clusters was determined using MetaMorph software as described in Materials and Methods for 10–15 cells in each of three independent experiments (30–45 cells total). The mean ± SEM is indicated. Depletion of the indicated proteins displayed statistical significance using the F-test (***p < 0.001). (C–D) Immunoblotting of extracts from cells transfected with the indicated siRNAs as described in A. Extracts were separated by SDS-PAGE and blots probed with the indicated antibodies. The anti-FTS antibodies used here were affinity purified. To demonstrate depletion of p107FHIP, cells were cotransfected with a vector expressing GST-p107FHIP and extracts blotted with anti-GST antibodies.
Analysis of FTS levels in cells depleted of all three Hook proteins (Figure 6C) revealed a striking dependence of Hook proteins for accumulation of FTS. With two independent sets of siRNAs simultaneously targeting Hook1, Hook2, and Hook3, FTS levels were undetectable, whereas the levels of GFP-Vps18 or actin as a control were unchanged (Figure 6C, lanes 4 and 5). These data are consistent with the idea that FTS assembles with Hook proteins in vivo and that FTS becomes unstable or unable to accumulate in the absence of its interaction with the Hook complex.
FTS Is Required for Timely Transit of EGF from Early-to-Late Endocytic Organelles
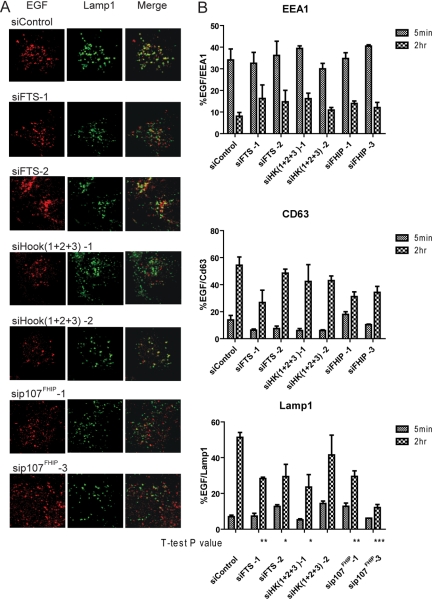
To determine further whether the FHF complex contributes to endocytic trafficking, we examined transit of EGF-TR from EEA1-positive early endosomes to LAMP1-positive late endosome/lysosomes by using immunofluorescence coupled with quantitative microscopic analysis (see Materials and Methods) with and without depletion of FHF complex components by RNAi. We also examined the late endosome/lysosome marker CD63. Entry of EGF into EEA1-positive endosomes at 5 min was largely unaffected by depletion of FHF components with two independent siRNAs for each protein, indicating that the FHF complex is not required for early steps in the process (Figure 7, A and B). After 2 h, when control cells had very low levels of EGF-positive EEA1-positive endosomes (8%), cells depleted of FTS displayed higher levels of EGF-positive EEA1 positive endosomes (∼15%) (Figure 7, A and B). At 2 h, ∼55% of EGF-positive endosomes were LAMP1 positive, whereas cells lacking FTS displayed only ∼30% EGF/LAMP1-positive endosomes (Figure 7C; p < 0.01 for siFTS-1 and p < 0.05 for siFTS-2). Depletion of FHIP or simultaneous depletion of all three Hook proteins with 2 independent siRNAs also led to a reduction in EGF-positive/LAMP1-positive endosomes (Figure 7, A and B). The late endosome marker CD63 displayed an intermediate pattern: at 2 h, 55% of EGF-positive endosomes were CD63 positive in control cells, whereas cells depleted of FTS displayed 25–50% colocalization (Figure 7B), although this difference did not reach statistical significance. These data suggest a delay in transit of EGF from early-to-late endosome/lysosomes.
Figure 7.
The FHF complex promotes timely trafficking of EGF through the endosomal pathway. (A) Fluorescently labeled EGF (EGF-TR, Texas Red) was used in endosomal trafficking assays as described in Materials and Methods. At either 5 min or 2 h after initiation of EGF trafficking, cells were fixed and stained with antibodies against EEA1, CD63, or LAMP1 by using anti-FITC detection (green). Cells were imaged by confocal microscopy. (B) The extent of overlap between the indicated endosomal markers and EGF-TR was determined using MetaMorph software as described in Materials and Methods. For each condition, 30–50 cells were imaged from three independent experiments. The percentage of EGF-TR that colocalizes with marked endosomes.
As a further test of the role of FHF complex proteins in EGF trafficking, we examined the loss of EGF from cells over time by using a previously published assay (Bishop et al., 2002) that monitors decay of fluorescent EGF from cells (Supplemental Figure S7). Cells were initially transfected with control siRNA or siRNA targeting FTS or Hook1/Hook2/Hook3. After 48 h, the kinetics of decay of the internalized TR-EGF was monitored by microscopy and image analysis (Supplemental Figure S7). In siControl-treated cells, TR-EGF decayed with a half-time of ∼100 min (Supplemental Figure S7, A and E). Depletion of FTS or TSG101 extended the decay half-time to ∼150 and ∼160 min, respectively (Supplemental Figure S7, A and E; data not shown). Expressing an RNAi-resistant FTS construct reversed the delay seen with siFTS alone (Supplemental Figure S7B). In contrast, expression of a mutant FTS protein (FTSW106AF107A) failed to reverse the extension in TR-EGF decay when expressed at the same level as the wild-type protein (Supplemental Figure S7, C and F), suggesting that the interaction between Hook proteins and FTS is important for TR-EGF decay. Consistent with a role for Hook proteins, codepletion of all three Hook proteins led to a ∼30-min delay in TR-EGF decay, whereas depletion of individual Hook proteins had little effect (Supplemental Figure S7D).
DISCUSSION
FTS is a poorly understood member of the variant E2 ubiquitin-conjugating enzyme family. In this study, we set out to identify FTS-associated proteins with the hope of elucidating its cellular function. FTS was found to interact with all three human Hook proteins (Hook1, Hook2, and Hook3) based on mass spectrometry, immunological analysis, and the two-hybrid system. This interaction involves a conserved helix in the C terminus of Hook1 and the β-sheet surface of the UBC domain of FTS. We also identified a previously uncharacterized open reading frame C11ORF56 encoding a 107-kDa protein (p107FHIP) as an FTS-Hook Interacting Protein. This protein was found in FLAG–HA–FTS, FLAG–HA–Hook1, and FLAG–HA–Hook3 complexes and comigrated with the major 500-kDa Hook–FTS complex on gel filtration. p107FHIP lacks known protein interaction domains but contains four short regions that may form coiled-coils (Supplemental Figure S3B). Due to the absence of appropriate immunological reagents, it is unclear at present whether p107FHIP is exclusively associated with the Hook complex or whether it may interact with other proteins. Attempts to stably express tagged versions of p107FHIP for mass spectrometry have thus far been unsuccessful. Previous overexpression studies have suggested that FTS interacts with the Akt1 protein kinase (Remy and Michnick, 2004). We failed to identify Akt1 in association with FTS under the conditions used here.
The biological functions of mammalian Hook proteins are poorly defined. Mutants in Drosophila hk lead to defects in the accumulation of the transmembrane ligand boss in multivesicular bodies during sevenless tyrosine kinase signaling, suggesting a role for hk in the endocytic process (Kramer and Phistry, 1996; Kramer and Phistry, 1999). Mammalian Hook proteins localize generally throughout the cytoplasm, but they also can be closely associated with cytoplasmic organelles, the centrosome in Hook2 and the Golgi in Hook3 (Walenta et al., 2001; Szebenyi et al., 2007). Overexpression of deletion mutants of Hook3 lacking the C-terminal domain required for tethering to Golgi affects Golgi morphology (Walenta et al., 2001) A portion of overexpressed Hook 2 has been reported to interact with overexpressed centriolin via sequences contained in its C terminus and overexpression of C-terminal fragments of Hook2 affect the localization of centrosomal markers (Szebenyi et al., 2007). The closest Hook ortholog in C. elegans is Zyg-12, which has been implicated in the attachment of the centrosome to the nuclear membrane (Malone et al., 2003). Zyg-12 contains a conserved coiled-coil sequence, but unlike Drosophila hk, it lacks sequence conservation in the C-terminal region we implicate in the interaction with FTS and p107FHIP. Our attempts to examine the localization of endogenous FTS have been adversely affected by the lack of suitable antibodies for immunofluorescence.
There is also uncertainty as to the oligomeric state of Hook proteins. Drosophila express a single Hook protein, hk, and hk homodimerizes through its central coiled-coil domain (Kramer and Phistry, 1996). In contrast, other studies based on immunoprecipitation of mammalian Hook proteins indicated that they do not homo- or heterodimerize (Walenta et al., 2001). Our results strongly indicate that mammalian Hook proteins can interact through homotypic and heterotypic interactions. Hook1 and Hook3 immune complexes contain Hook1, Hook2, and Hook3 proteins in addition to FTS, whereas FTS complexes contain Hook1, Hook2, and Hook3 in a 500-kDa complex, as determined by gel filtration. Interestingly, the majority of soluble Hook1, Hook2, and Hook3 protein, as well as the bulk of FTS, are present in this 500-kDa complex, as determined by immunoblotting of gel filtration fractions. Given the association seen with different affinity reagents, we conclude that human Hook proteins do physically interact with each other via their coiled-coils. The nominal molecular mass of a complex containing one molecule of each Hook protein (83–84 kDa), one molecule of p107FHIP (107 kDa), and one molecule of FTS (33 kDa)–390 kDa–is smaller than the apparent size based on gel filtration. However, given the coiled-coil nature of Hook proteins, they likely form an elongated structure, which would migrate at a larger size than expected. The stoichiometry of proteins within the complex is unclear at present. Because each Hook protein can, in principle, interact with FTS, there may be multiple FTS molecules present in the complex. We note, however, that Hook2 levels were lower than that of Hook1 and Hook3 in FTS immune complexes, as determined by mass spectral scan number (Figure 1E), suggesting that its abundance in the complex could be lower than that of Hook1 and Hook3. We note that the Hook2 antibodies used in this study failed to efficiently immunoprecipitate Hook1, Hook3, or FTS, although they did precipitate Hook2 itself. In contrast, stably expressed epitope-tagged Hook2 associated with Hook proteins as well as FTS.
Given previous work indicating that Hook1 can interact with the Vps18 subunit of the HOPS complex in mammalian cells (Richardson et al., 2004) and Drosophila hk can interact with the Vps18 orthologue Dor in the two-hybrid system (Lloyd et al., 1998), we examined the ability of Hook proteins and FTS to assemble with components of the HOPS complex. Hook proteins, as well as FTS, associated with Vps18, Vps16, Vps39, and Vps41 in transfected cells. This interaction required the C-terminal domain of Hook1, based on deletion mapping. Moreover, the quadruple point mutant in a C-terminal helix of Hook1 (Hook14A), which cannot bind FTS, failed to interact with Vps16, Vps18, Vps39, or Vps41. We also found that endogenous Hook1 associates with endogenous Vps18 more efficiently in CHAPS buffer than in NP-40 buffer, providing an explanation for why the HOPS complex was not seen in FTS complexes by mass spectrometry. We speculate that the affinity of the HOPS complex for the FTS/Hook complex is weaker than the interaction of the individual complexes. The interactions we have defined in this study are summarized in Figure 8.
Figure 8.
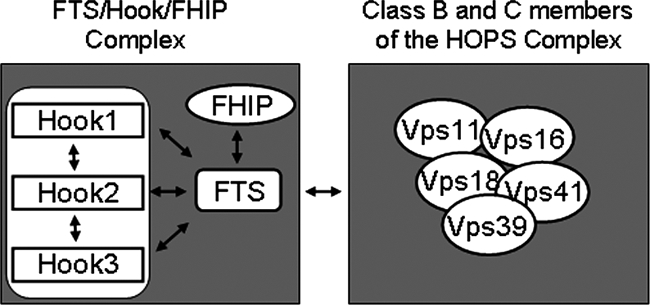
Interactions observed within the FHF complex and between the FHF complex and the HOPS complex. See text for details.
Components of the HOPS complex are thought to promote tethering of vesicles for homotypic fusion via SNARE proteins. This complex associates either directly or indirectly with SNARE proteins, Rab proteins, and Sec1/Munc-like (S/M-like) proteins (Waters and Pfeffer, 1999; Whyte and Munro, 2002). Components of HOPS complexes colocalize with EEA1 and LAMP1-positive endosomes, suggesting roles in both early and late endosomal events. Overexpression of Vps18 promotes clustering of both early and late endosome/lysosomes, whereas Vps39 overexpression promotes primarily late endosomal clustering, although some of the differences seen may reflect differences in the cell types used (Caplan et al., 2001; Poupon et al., 2003; Richardson et al., 2004). These clustering events are thought to reflect the tethering of homotypic vesicles by the HOPS complex on route to fusion. Given the interaction between the FHF complex and HOPS components, we examined whether components of the FHF complex are involved in Vps18-dependent clustering. We found that depletion of FTS by RNAi resulted in a reduction in the extent of LAMP1-positive endosomal clustering, as measured by the integrated intensity of LAMP1 fluorescence in GFP–Vps18 clusters. Similar results were found upon depletion of all three Hook proteins as well as p107FHIP. The reduced size GFP–Vps18, LAMP1-positive endosomal clusters upon depletion of components of the complex suggests that the FHF complex promotes enlargement or maintenance of clusters but may not be required for initiation of clustering. Interestingly, we also found that overexpression of FTS or Hook proteins could promote clustering of LAMP1-positive structures (Supplemental Figure S6). These results further link the FHF complex, together with the HOPS complex, to late lysosomal/endosomal tethering.
Vesicle transport and fusion events are coordinated by the cytoskeleton (Qualmann et al., 2000). Previous studies have indicated that the class C Vps components in the HOPS complex can bind to actin filaments, whereas class B components precipitate with microtubules (Richardson et al., 2004). Interestingly, Hook1 was found to precipitate with both microtubules and actin filaments (Richardson et al., 2004). The N terminus of Hook proteins share a domain that is capable of associating with microtubules, although it is not clear whether this interaction is direct (Walenta et al., 2001). We have found that cells depleted of FTS display a delay in the transit of EGF from early-to-late endosomes/lysosomes. Given the interaction of the FHF complex with the HOPS complex coupled with the effects of FTS depletion on endosomal clustering and EGF trafficking to late endosomae/lysosomes, we speculate that the FHF complex may function at the interface between vesicle tethering and the cytoskeleton. Further studies are required to elucidate the temporal and spatial relationships that govern FHF complex function and its interaction with the HOPS complex.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Piper, H. Kramer, and S. Michnick for reagents; R. Piper for helpful suggestions and discussions; and M. Kuroda for access to MetaMorph software. M.E.S. was supported by a postdoctoral fellowship from the American Cancer Society. This work was supported by National Institutes of Health (NIH) grants GM-70565 and AG-11085 (to J.W.H.) and NIH grant GM-067945 (to S.P.G.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-05-0473) on September 17, 2008.
REFERENCES
- Bishop N., Horman A., Woodman P. Mammalian class E vps proteins recognize ubiquitin and act in the removal of endosomal protein-ubiquitin conjugates. J. Cell Biol. 2002;157:91–101. doi: 10.1083/jcb.200112080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzovic P. S., Lissounov A., Christensen D. E., Hoyt D. W., Klevit R. E. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol. Cell. 2006;21:873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Caplan S., Hartnell L. M., Aguilar R. C., Naslavsky N., Bonifacino J. S. Human Vam6p promotes lysosome clustering and fusion in vivo. J. Cell Biol. 2001;154:109–122. doi: 10.1083/jcb.200102142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan N. L., Hill C. P. Defining polyubiquitin chain topology. Nat. Struct. Biol. 2001;8:650–652. doi: 10.1038/90337. [DOI] [PubMed] [Google Scholar]
- Dye B. T., Schulman B. A. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu. Rev. Biophys. Biomol. Struct. 2007;36:131–150. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- Eddins M. J., Carlile C. M., Gomez K. M., Pickart C. M., Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat. Struct. Mol. Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- Eddins M. J., Varadan R., Fushman D., Pickart C. M., Wolberger C. Crystal structure and solution NMR studies of Lys48-linked tetraubiquitin at neutral pH. J. Mol. Biol. 2007;367:204–211. doi: 10.1016/j.jmb.2006.12.065. [DOI] [PubMed] [Google Scholar]
- Gillingham A. K., Tong A. H., Boone C., Munro S. The GTPase Arf1p and the ER to Golgi cargo receptor Erv14p cooperate to recruit the golgin Rud3p to the cis-Golgi. J. Cell Biol. 2004;167:281–292. doi: 10.1083/jcb.200407088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbury P. B., Kim P. S., Alber T. Crystal structure of an isoleucine-zipper trimer. Nature. 1994;371:80–83. doi: 10.1038/371080a0. [DOI] [PubMed] [Google Scholar]
- Harbury P. B., Zhang T., Kim P. S., Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- Hurley J. H., Emr S. D. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Arias E. E., Chen J., Harper J. W., Walter J. C. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Katzmann D. J., Babst M., Emr S. D. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- Kim B. Y., Kramer H., Yamamoto A., Kominami E., Kohsaka S., Akazawa C. Molecular characterization of mammalian homologues of class C Vps proteins that interact with syntaxin-7. J. Biol. Chem. 2001;276:29393–29402. doi: 10.1074/jbc.M101778200. [DOI] [PubMed] [Google Scholar]
- Kostelansky M. S., Schluter C., Tam Y. Y., Lee S., Ghirlando R., Beach B., Conibear E., Hurley J. H. Molecular architecture and functional model of the complete yeast ESCRT-I heterotetramer. Cell. 2007;129:485–498. doi: 10.1016/j.cell.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostelansky M. S., Sun J., Lee S., Kim J., Ghirlando R., Hierro A., Emr S. D., Hurley J. H. Structural and functional organization of the ESCRT-I trafficking complex. Cell. 2006;125:113–126. doi: 10.1016/j.cell.2006.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer H., Phistry M. Mutations in the Drosophila hook gene inhibit endocytosis of the boss transmembrane ligand into multivesicular bodies. J. Cell Biol. 1996;133:1205–1215. doi: 10.1083/jcb.133.6.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer H., Phistry M. Genetic analysis of hook, a gene required for endocytic trafficking in Drosophila. Genetics. 1999;151:675–684. doi: 10.1093/genetics/151.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesche R., Peetz A., van der Hoeven F., Ruther U. Ft1, a novel gene related to ubiquitin-conjugating enzymes, is deleted in the Fused toes mouse mutation. Mamm. Genome. 1997;8:879–883. doi: 10.1007/s003359900604. [DOI] [PubMed] [Google Scholar]
- Lloyd V., Ramaswami M., Kramer H. Not just pretty eyes: Drosophila eye-colour mutations and lysosomal delivery. Trends Cell Biol. 1998;8:257–259. doi: 10.1016/s0962-8924(98)01270-7. [DOI] [PubMed] [Google Scholar]
- Malone C. J., Misner L., Le Bot N., Tsai M. C., Campbell J. M., Ahringer J., White J. G. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- Mullock B. M., et al. Syntaxin 7 is localized to late endosome compartments, associates with Vamp 8, and is required for late endosome-lysosome fusion. Mol. Biol. Cell. 2000;11:3137–3153. doi: 10.1091/mbc.11.9.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C. M., Eddins M. J. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Poupon V., Stewart A., Gray S. R., Piper R. C., Luzio J. P. The role of mVps18p in clustering, fusion, and intracellular localization of late endocytic organelles. Mol. Biol. Cell. 2003;14:4015–4027. doi: 10.1091/mbc.E03-01-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B., Kessels M. M., Kelly R. B. Molecular links between endocytosis and the actin cytoskeleton. J. Cell Biol. 2000;150:F111–F116. doi: 10.1083/jcb.150.5.f111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy I., Michnick S. W. Regulation of apoptosis by the Ft1 protein, a new modulator of protein kinase B/Akt. Mol. Cell. Biol. 2004;24:1493–1504. doi: 10.1128/MCB.24.4.1493-1504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S. C., Winistorfer S. C., Poupon V., Luzio J. P., Piper R. C. Mammalian late vacuole protein sorting orthologues participate in early endosomal fusion and interact with the cytoskeleton. Mol. Biol. Cell. 2004;15:1197–1210. doi: 10.1091/mbc.E03-06-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrioukov E. A., He J. P., Moghrabi N., Sunio A., Kramer H. A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila. Mol. Cell. 1999;4:479–486. doi: 10.1016/s1097-2765(00)80199-9. [DOI] [PubMed] [Google Scholar]
- Sundquist W. I., Schubert H. L., Kelly B. N., Hill G. C., Holton J. M., Hill C. P. Ubiquitin recognition by the human TSG101 protein. Mol. Cell. 2004;13:783–789. doi: 10.1016/s1097-2765(04)00129-7. [DOI] [PubMed] [Google Scholar]
- Sunio A., Metcalf A. B., Kramer H. Genetic dissection of endocytic trafficking in Drosophila using a horseradish peroxidase-bride of sevenless chimera: hook is required for normal maturation of multivesicular endosomes. Mol. Biol. Cell. 1999;10:847–859. doi: 10.1091/mbc.10.4.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szebenyi G., Hall B., Yu R., Hashim A. I., Kramer H. Hook2 localizes to the centrosome, binds directly to centriolin/CEP110 and contributes to centrosomal function. Traffic. 2007;8:32–46. doi: 10.1111/j.1600-0854.2006.00511.x. [DOI] [PubMed] [Google Scholar]
- Walenta J. H., Didier A. J., Liu X., Kramer H. The Golgi-associated hook3 protein is a member of a novel family of microtubule-binding proteins. J. Cell Biol. 2001;152:923–934. doi: 10.1083/jcb.152.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M. G., Pfeffer S. R. Membrane tethering in intracellular transport. Curr. Opin. Cell Biol. 1999;11:453–459. doi: 10.1016/s0955-0674(99)80065-9. [DOI] [PubMed] [Google Scholar]
- Whyte J. R., Munro S. Vesicle tethering complexes in membrane traffic. J. Cell Sci. 2002;115:2627–2637. doi: 10.1242/jcs.115.13.2627. [DOI] [PubMed] [Google Scholar]
- Winn P. J., Religa T. L., Battey J. N., Banerjee A., Wade R. C. Determinants of functionality in the ubiquitin conjugating enzyme family. Structure. 2004;12:1563–1574. doi: 10.1016/j.str.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Xu L., Wei Y., Reboul J., Vaglio P., Shin T. H., Vidal M., Elledge S. J., Harper J. W. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425:316–321. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.