Abstract
Polymorphonuclear leukocyte (PMN) recruitment to sites of inflammation is initiated by selectin-mediated PMN tethering and rolling on activated endothelium under flow. Cell rolling is modulated by bulk cell deformation (mesoscale), microvillus deformability (microscale), and receptor-ligand binding kinetics (nanoscale). Selectin-ligand bonds exhibit a catch-slip bond behavior, and their dissociation is governed not only by the force but also by the force history. Whereas previous theoretical models have studied the significance of these three “length scales” in isolation, how their interplay affects cell rolling has yet to be resolved. We therefore developed a three-dimensional computational model that integrates the aforementioned length scales to delineate their relative contributions to PMN rolling. Our simulations predict that the catch-slip bond behavior and to a lesser extent bulk cell deformation are responsible for the shear threshold phenomenon. Cells bearing deformable rather than rigid microvilli roll slower only at high P-selectin site densities and elevated levels of shear (≥400 s−1). The more compliant cells (membrance stiffness = 1.2 dyn/cm) rolled slower than cells with a membrane stiffness of 3.0 dyn/cm at shear rates >50 s−1. In summary, our model demonstrates that cell rolling over a ligand-coated surface is a highly coordinated process characterized by a complex interplay between forces acting on three distinct length scales.
Keywords: immersed boundary method, Monte Carlo simulation, cell adhesion, cell deformation, viscoelastic microvillus, fluid shear, P-selectin
the recruitment of free flowing polymorphonuclear leukocytes (PMNs) to sites of inflammation or infection is a key step in the body's innate immune response. Targeting of PMNs to these sites is viewed as a multistep process with the sequential involvement of distinct adhesion molecules on PMN and endothelial cell (EC) surfaces (26). This process is initiated by selectin-mediated PMN tethering and rolling along the EC surface, followed by integrin-dependent firm adhesion, before PMN extravasation into the tissue space. This cascade of highly regulated molecular events is dictated by local circulatory hemodynamics and the mechanical and kinetic properties of participating adhesion molecules (26, 42).
The ability of selectins (P-, L-, and E-selectin) to mediate leukocyte tethering and rolling in shear flow is attributed to the fast association (kf) and dissociation (kr) rates of selectin-ligand pairs as well as the high tensile strength of the bonds under rapid loading (2, 16, 20). Although the dynamics of adhesion is primarily controlled by the physical chemistry of adhesion molecules (7), cellular material properties such as cell deformation and microvillus viscoelasticity may critically affect selectin-mediated PMN recruitment to ECs. Cell deformation is predicted to increase the contact area between the cell and substrate, leading to improved adhesion. The importance of cell deformability in the PMN adhesion process is supported by both theoretical (22) and experimental (35, 47) studies showing that PMNs roll significantly more slowly and relatively more smoothly than do solid microspheres coated with a matched density of P-selectin glycoprotein ligand (PSGL)-1 molecules. When deformation is reduced, for example, by fixing PMNs with formaldehyde, faster rolling is observed on P-selectin-coated surfaces in shear flow compared with untreated PMNs (30, 35, 37). Further support of the importance of cell deformation is provided by experiments in which PMN deformation is increased, as when PMNs are treated with actin cytoskeleton-disrupting cytochalasins. In this case, PMNs roll more slowly than untreated PMNs (30, 40). Cell rolling may also be impacted by the viscoelasticity of the microvilli (39), which can stretch to form tethers rather than allow adhesive interactions to break (38). Finally, the type and spatial distribution of the receptors, which determine the avidity of the PMN-substrate interaction, also play a key role in cell rolling. In summary, there are multiple mechanisms ranging from the molecular to cellular level that may affect PMN tethering and rolling on ECs. However, their relative roles have yet to be deconvoluted from a computational viewpoint.
Both in vivo and in vitro studies have documented the shear threshold phenomenon for all three selectins (17, 28), in which the number of rolling PMNs first increases and then decreases while monotonically increasing wall shear stress. How cell-substrate adhesive contacts can be amplified by the application of progressively stronger dispersive hydrodynamic forces is counterintuitive. Force is traditionally thought to shorten the lifetimes of receptor-ligand bonds, referred to as slip bonds, as it lowers the energy barrier between bound and free states (12). However, recent experimental observations have suggested that selectins exhibit a catch-slip bond transition where increasing tensile forces initially prolong and subsequently diminish bond lifetimes (29, 48). Indeed, a recent computational study (6) has suggested that the catch-slip behavior of selectin bonds is primarily responsible for the shear threshold effect. Since shear threshold phenomena have been observed at low levels of shear (50–100 s−1) (28), it has been hypothesized but yet to be tested that cellular features do not significantly contribute to it (6).
Receptor-mediated cell rolling has been systematically investigated in computational studies by adhesive dynamics, in which the cell is modeled as a nondeformable sphere in shear flow near a plane with a Monte Carlo method for simulating receptor-ligand interactions (7). The aforementioned computational model was recently extended to account for the microvillus deformability (5) and the two-pathway kinetic model of selectin-ligand binding (16) in an effort to explain the shear threshold phenomenon (6). However, the adhesive dynamics algorithm does not account for cell deformation. Other computational models such as the elastic ring (13, 14) and compound drop (34) models, designed to account for cell deformability during rolling in shear flow, have been limited to a two-dimensional (2-D) representation of cell rolling. Recently, a three-dimensional (3-D) viscous drop model was proposed that simulated deformation of a cell bound to a substrate in a channel (25). However, since the simulations in this study were conducted for small intervals of time (4 ms), it was unclear whether the cell shapes had reached steady-state deformation (25). Balazs and colleagues (1, 44) developed a 3-D computational model that integrates the lattice Boltzmann model for hydrodynamics and lattice spring model for micromechanics of elastic solids to investigate how deformability of elastic capsules affects their motion on chemically and mechanically patterned planar surfaces in shear flow. These simulations predict that the trajectory of a capsule on a patterned substrate is dictated by its compliance (1, 44). However, in all the aforementioned studies simulating deformable cells, the kinetics of cell-substrate binding were represented by deterministic relationships, preventing these models from capturing the intermittent, jerky, “stop-and-go” cell rolling pattern.
We (22, 23) have recently developed a 3-D model simulating receptor-mediated rolling of a deformable cell on a selectin-coated surface in a linear shear field. The model parameters were chosen to represent PSGL-1-mediated PMN rolling on a P-selectin-decorated planar surface. To this end, the immersed boundary method (IBM) (15) was used to simulate the motion of an elastic capsule near a plane in a linear shear field. The IBM was coupled to the Hookean spring model to simulate the force-dependent kinetics of receptor-ligand interactions (12), whereas the stochastic behavior of P-selectin-PSGL-1 bond formation and breakage was simulated by the Monte Carlo method. Theoretical predictions of the kinetics of receptor-ligand interactions under external force have been predominantly carried out using the Hookean spring (Dembo) (12) and Bell (3) models. More recently, a two-pathway kinetic model (16) was proposed to account for the “catch-to-slip” bond transition under force (29). Briefly, bond dissociation can occur via either of two pathways: fast or slow, depending on the force applied to the bond (16). At low forces, state 1 (fast dissociation) is more populated, so bond dissociation is fast. As the force increases, state 2 (slow dissociation) becomes more populated, so bond dissociation is slow, leading to a catch bond behavior.
In this article, we extended our recently developed 3-D computational model of cell rolling to incorporate the three different kinetic models of receptor-ligand binding and to account for microvillus deformability. Our primary objective was to compare and contrast the relative contributions of the three distinct length scales involved in cell rolling to shear threshold phenomenon, namely, 1) bulk cell deformation (mesoscale), 2) microvillus deformability (microscale), and 3) receptor-ligand binding kinetics (nanoscale). We also wanted to evaluate which of the two most widely used kinetic models (Dembo vs. Bell) gives predictions that are in close agreement with the recently developed two-pathway model used to capture catch-slip bond behavior (16). Given the variability that is observed in any stochastic process, we also incorporated a deterministic representation of receptor-ligand binding into our code to compare the differences in cell rolling that were observed between the deformable and rigid microvilli models.
COMPUTATIONAL MODEL
The rolling of PSGL-1-bearing modeled leukocytes, decorated with either rigid or deformable microvilli, on a P-selectin-coated plane surface in a linear shear field was simulated by combining 1) IBM to solve the Navier-Stokes equation for motion of an elastic capsule near a plane in a linear shear field (15, 21, 22, 36); 2) the finite-element method to solve the constitutive equation for the neo-Hookean membrane of the spherical capsule (15, 21, 22); and 3) the Monte Carlo method for simulating the formation and breakage of receptor-ligand bonds with kinetic rate constants based on either the Hookean spring model (12), Bell model (3), or the two-pathway model (16). Bond formation and rupture are considered to be stochastic events, and the results obtained are validated using a deterministic model for receptor-ligand binding.
IBM
The cell is modeled as a 3-D elastic capsule containing a Newtonian liquid, and the motion of the fluid inside and outside of the elastic capsule is governed by the following continuity (Eq. 1) and Navier-Stokes (Eq. 2) equations:
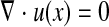 |
(1) |
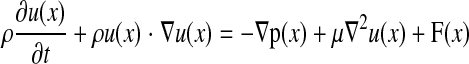 |
(2) |
which are discretized by finite differencing on a uniform Cartesian grid. ρ and μ are the density and viscosity of the fluid, respectively; u(x) and p are the velocity and pressure, respectively, at fluid grid nodes x(x,y,z); and t is time. F(x) is the total force acting on each of the fluid grid nodes comprising the force exerted by the plane, elastic capsule, and receptor-ligand bonds. The inertial term in Eq. 2 is neglected because the Reynolds number for flow of microscopic particles under physiological flow conditions is very small. The stationary plane over which fluid flow occurs is modeled as a network of nodes interconnected by stiff springs similar to a previous IBM implementation simulating flow over a solid surface (27). A linear shear field is generated by applying a constant force along a plane parallel to and far from the stationary surface.
In the IBM, the computational domain is composed of an Eulerian Cartesian fluid grid x(x,y,z) and a Langragian triangular finite-element grid X(X,Y,Z) that tracks cell motion and deformation (21, 22). At the beginning of each time step t, restoring forces due to the displacement of nodes in the elastic membrane and the stationary plane as well as those due to the stretching of bonds are calculated. These forces, denoted by F(X) and located at immersed boundary nodes X, are distributed to the nearby fluid grid nodes x, using the appropriately chosen function Dh(X − x) as follows:
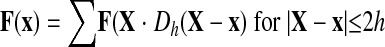 |
(3) |
where h is the uniform grid spacing and Dh is the 3-D discrete δ-function given by the following:
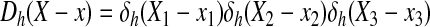 |
(4) |
and
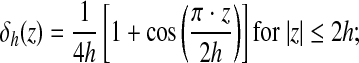 |
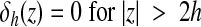 |
(5) |
Similarly, the velocity at membrane node X is calculated from the velocity at fluid grid node x using the δ-function as follows:
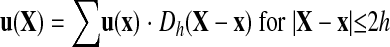 |
(6) |
This also implies a velocity continuity condition on the membrane surface since its velocity matches that of the fluid. Periodic boundary conditions on the velocity and pressure are imposed, and the fast Fourier transform method is used to solve the flow equations (21, 22, 36).
Membrane Constitutive Equation
The elastic membrane is assumed to have an initial spherical stress-free shape. Cell membrane mechanics are described by the neo-Hookean membrane model, which assumes that the membrane material is incompressible and isotropic. The strain energy density (W) of a neo-Hookean membrane is given by the following:
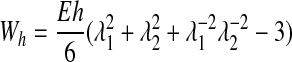 |
(7) |
where λ1 and λ2 are the principal stretch ratios, E is Young's modulus, and h is membrane thickness.
The finite-element implementation of the force [F(x)] calculation of the membrane was based on the models developed by Charrier JM et al. (8) and Eggleton CD and Popel AS (15). The membrane is discretized into triangular finite elements to obtain the forces acting at the discrete nodes of the membrane surface, which are then distributed onto the fluid grid as described above. Given the displacement of the three nodes of an element, its state of strain (λ1 and λ2) is obtained. The material properties of the element determine the forces that are required to maintain the element in a given state of strain/stress. The principle of virtual work is used to calculate the forces at the three nodes of an element. The resultant force [F(X)] on membrane node X is simply the sum of the forces exerted by the triangular elements attached to that node. An equal and opposite force acts on the fluid in the manner described by the IBM. Further details of the numerical implementation and validation of the model can be found in Refs. 8 and 15.
The uniformly discretized fluid grid used in the simulations has 64 × 64 × 128 nodes, (with the longest dimension aligned with the flow), and the finite-element grid of each cell has 10,240 triangular elements. The spatial order accuracy of the IBM is second order globally except near the interface, where it is first order. The numerical method is semi-implicit and first order in time.
A time step of 10−6 s was used to ensure numerical stability and to better resolve bond dynamics. This time step is smaller than the characteristic time for the elastic response of the membrane, which is defined as Eh/μR (where R is the cell radius) and is ∼10−4. The computations require 2 GB of memory and run times of 4–6 wk depending on the CPUs employed and other hardware characteristics. Our simulations were conducted on an IBM P-series (2528 Power 4+ CPUs) and a SUN Fire Server. Since the objective of this work was to investigate the dynamics of PMN rolling and to reduce computational cost, we neglected PMN sedimentation and initialized our simulations with the cell placed 75 nm above the planar wall. Thus, PMNs were modeled as neutrally buoyant in light of their relative density to that of suspending medium. The distance between the PMN membrane and substrate evolves under the governing equations used in our simulations (22).
Microvillus Deformability
In this model, bonds between the cell and surface form and break based on bond kinetics, causing the cell to advance forward in the direction of flow. Under the application of a pulling force exerted by the bond, the microvillus stretches as described by Shao and coworkers (39). Briefly, the viscoelastic nature of the microvillus is captured by modeling it as a Hookean spring if the force is <45 pN (extension regime) or as a long thin membrane cylinder for forces >45 pN (tethering regime). In the extension regime, the force exerted by the bond on the microvillus (Fm) is given by:
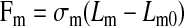 |
(8) |
whereas in the high force regime, Fm is:
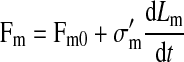 |
(9) |
where Fm0 is the threshold force between the two regimes, σm and σm′ are the microvillus spring constants, Lm is the microvillus length at time t, and Lm0 is the resting microvillus length.
Assuming that all bonds have the same direction vector as that of the bound microvillus, the system can be likened to a parallel set of Hookean springs (receptor-ligand bonds) in series with either another Hookean spring or a viscoelastic membrane (microvillus). Fm can then be expressed as a function of the resting bond length (Lb0) as follows:
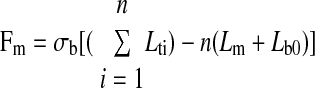 |
(10) |
where σb is the spring constant for the bond, Lb is the bond length at time t, n is the number of bonds, and Lti is the distance between the base of the microvillus on the cell surface and the base of the bond on the selectin coated plane (Lti = Lm + Lb). Based on the magnitude of the force acting on the microvillus, its length can then be obtained using Eqs. 11 or 12. In the extension regime,
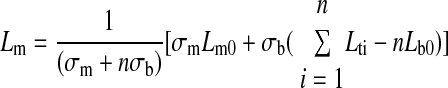 |
(11) |
whereas in the tethering regime,
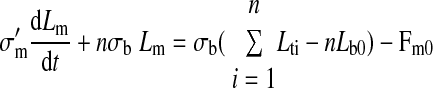 |
(12) |
Receptor-Ligand Binding Kinetics
Dembo model.
According to the Hookean spring model, the forward and reverse rate constants for receptor-ligand interactions under external force are given by Ref. 12 as follows:
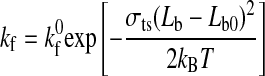 |
(13) |
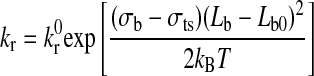 |
(14) |
where kf0 and kr0 are the forward and reverse rate constants at equilibrium distance Lb0; kf and kr are the forward and reverse rate constants at distance (Lb − Lb0) from equilibrium; kB is the Boltzmann constant; T is the absolute temperature; and σb and σts represent the spring constants in the bound and transition state, respectively.
Bell model.
The Bell model off rate is given by Ref. 3 as follows:
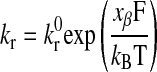 |
(15) |
where xβ is the reactive compliance, kBT is the thermal energy, and F is the force acting on the bond. The reactive compliance is a parameter with units of length that describes the degree to which force facilitates bond breakage; a smaller value of reactive compliance means that the bond is less susceptible to breakage by force. To satisfy the Boltzmann distribution at equilibrium, the forward rate (kf) must depend on separation distance according to Ref. 5 as follows:
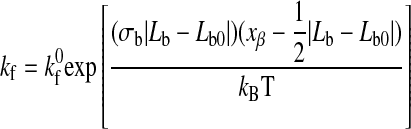 |
(16) |
where Lb is the length of the possible new bond and kf0 is the unstressed association constant.
Two-pathway model.
Evans and coworkers (16) recently proposed a two-state model for bond dissociation to incorporate the catch-slip bond transition. It was proposed that there are two pathways for dissociation: pathway 1 is very fast and dominates at low forces, whereas pathway 2 is slower and dominates in the high shear regime. The pathways originate from two possible bound configurations. Dissociation via the two pathways occurs with rates of k1rup and k2rup for pathway 1 and 2, respectively. It was assumed that the off rate for the fast pathway is constant at k1rup. For the slow pathway, on the other hand, dissociation was proposed to follow the Bell model for an exponential increase in off rate with force, so k2rup = k20exp(f/fβ) (16), where fβ = kBT/xβ.
The dominant dissociation pathway is determined by the occupancy ratio of the two states (16). The states were assumed to be in equilibrium at all times (fast equilibration) with a small difference in energy between state 2 and state 1 (ΔE21). According to the Boltzmann distribution, this energy difference sets the equilibrium occupancy ratio (f0) of state 1 to state 2 at zero force. However, force applied to the bond causes a shift in the energy of each state, resulting in a change in the energy difference between the states. The occupancy ratio of the two states, then, changes exponentially with applied force with a scale of f12 = kBT/Δx12. So, while pathway 1 may dominate at low forces where equilibrium favors the occupancy of state 1, at higher forces, pathway 2 dominates as the equilibrium shifts to favor the occupancy of state 2. For bonds exhibiting this type of catch-slip behavior, the off rate is given by the following:
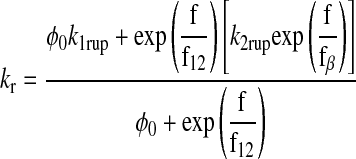 |
(17) |
where φ0 = exp(ΔE21/kBT) and is the equilibrium constant between the two states at zero force. The values of the different parameters used in our simulations are shown in Table 1.
Table 1.
Model parameters
| Parameter | Definition | Value | Reference |
|---|---|---|---|
| R | Leukocyte radius, μm | 3.75 | 43 |
| nL | No. of P-selectin glycoprotein ligand-1 molecules per cell | 15,000 | 33 |
| nR | P-selectin (receptor) site density, receptors/μm2 | 15 or 150 | 47 |
| Lb0 | Equilibrium bond length, μm | 0.1 | 18 |
| kf0 | Unstressed on rate, s−1 | 1 | 31 |
| kf0 | Unstressed off rate, s−1 | 1 | 31 |
| σb | Bond spring constant, dyn/cm | 1 | 12 |
| σts | Transition state spring constant, dyn/cm | 0.99 | 12 |
| σm | Microvillus spring constant, pN/μm | 43 | 39 |
| σm′ | Effective viscosity of tether, pN·s·μm−1 | 11 | 39 |
| Eh | Membrane stiffness, dyn/cm | 1.2 or 3.0 | |
| γ | Shear rate, s−1 | 10–800 | |
| μ | Fluid viscosity, cP | 0.8 | |
| ρ | Fluid density, g/ml | 1 | |
| T | Temperature, K | 310 | |
| xβ | Reactive compliance, Å | 0.3 | 5 |
| Lm0 | Equilibrium length of microvillus, μm | 0.35 | 39 |
| nm | No. of microvilli per cell | 252 | 9 |
| Fm0 | Threshold microvillus force, pN | 45 | 5 |
| k1rup | Failure rate along the low impedance pathway, s−1 | 12 | 16 |
| k2rup | Failure rate along the high impedance pathway, s−1 | 1 | 6 |
| ΔE21 | Difference in energy between the two bound states, kBT | 5 kBT | 16 |
| f12 | Span of force for which the switch between the two states occurs, pN | 6 | 16 |
kB, Boltzmann constant.
Deterministic model.
To calibrate the results obtained using our stochastic model for receptor-ligand binding, we used the deterministic model recently proposed by Truskey and colleagues (24). If the separation distance between a microvillus and the selectin-coated plane becomes equal to or less than the unstressed bond length (Lb0), the microvillus forms a link with the plane. The link is characterized by the time-dependent number of bonds, nb = nb(t). It is considered to be broken if the number of bonds becomes zero. The velocity field and shape of the leukocyte are found using IBM as described above.
The bond surface density nb = nb(t) is found by integrating the kinetic equation as follows:
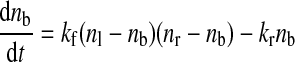 |
(18) |
The deterministic model of bond formation calculates the time-averaged number of bonds present between the cell surface and substrate for any given condition. However, the deterministic model does not simulate the formation and rupture of individual bonds, and, as a result, bond lifetimes as well as individual bond lengths and forces cannot be calculated. Using the deterministic model, it is possible to determine a steady-state value of the translational velocity, which can be directly compared with the time-averaged rolling velocity calculated from the stochastic model. A detailed study on the comparison of deterministic versus stochastic model simulations was conducted by Lauffenburger and colleagues (10), where they showed that these models agreed very well in a relatively narrow window of parameters (a category in which our simulations fall), whereas significant differences have been observed outside this range.
Bond formation and rupture.
The force acting on the bond is given by the following:
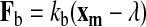 |
(19) |
The stochastic nature of receptor-ligand interactions are included using the Monte Carlo simulation. In time interval Δt, the probability that a receptor will bind (Pb) is as follows (19):
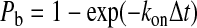 |
(20) |
where the kon = kfAL(nL − nb). AL is the surface area on ligand-coated plane accessible to each receptor, whereas (nL − nb) is the density of the unbound ligand. Similarly, the probability for rupture of a bond (Pr) is as follows (19):
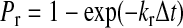 |
(21) |
At each time step, the probabilities of bond formation and breakage are compared with random numbers (Pran1 and Pran2) between 0 and 1. Pb > Pran1 indicates bond formation, whereas Pr > Pran2 indicates bond rupture. PSGL-1 molecules are assumed to be concentrated on the tips of PMN microvilli (32). The time of receptor-ligand bond formation and breakage as well as the number of bonds per cell and per microvillus were recorded for every time step. A time step of 10−6 s was used to simulate cell rolling for a period of 1 s for both the stochastic and deterministic models.
All parameters used in the simulations reported herein were selected to match the best-available measurements in the literature for the rolling of deformable PMNs on P-selectin substrates (Table 1).
RESULTS
Effects of P-selectin Site Density and Bulk Cell Deformation on PMN Rolling in Shear Flow
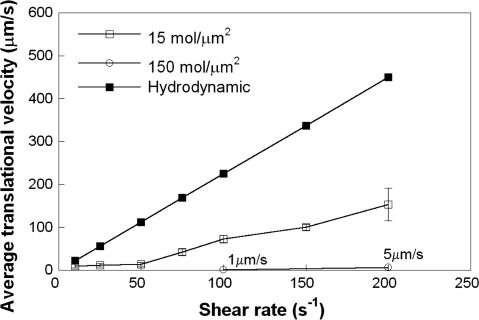
In this work, we extended our previously described 3-D computational model (22) to account for nanoscale (receptor-ligand binding kinetics), microscale (microvillus deformability), and mesoscale (bulk cell deformation) parameters to delineate their relative contributions to PSGL-1-dependent leukocyte rolling on a P-selectin-coated substrate in shear flow. As a first step, we examined the effect of selectin site density on cell rolling over a wide range of physiological shear rates. The cell was modeled as a deformable, spherical capsule of radius 3.75 μm decorated with PSGL-1 molecules localized on the tips of rigid microvilli. The membrane stiffness value of 3 dyn/cm was chosen to approximate the rolling behavior of a sphere with relatively moderate deformation, as used in previous 2-D viscous drop models of cell rolling (34). The Dembo model (12) was used to simulate receptor-ligand binding kinetics using the parameter values shown in Table 1. As shown in Fig. 1, for a P-selectin site density of 15 molecules/μm2, the average translational velocity varied little with shear for shear rates up to 50 s−1. Above this shear threshold level, cells rolled progressively faster with increasing shear rates (Fig. 1). Moreover, increasing the P-selectin site density from 15 to 150 molecules/μm2 resulted in a pronounced decrease in the average translational velocity of PSGL-1-decorated, modeled cells (Fig. 1). This differential rolling behavior is attributed to the fact that the number of cell-substrate bonds, which counteract the dispersive hydrodynamic forces, increased as the selectin site density was augmented (from 15 ± 1 to 150 ± 8 bonds for a selectin site density of 15 and 150 molecules/μm2, respectively, at a shear rate of 100 s−1).
Fig. 1.
Effect of P-selectin site density and shear rate on cell rolling. Hydrodynamic velocities and average translational velocities of modeled polymorphonuclear leukocytes (PMNs) rolling over P-selectin-coated surfaces with site densities of 15 and 150 molecules/μm2 were calculated as a function of shear rate (10–200 s−1). The membrane stiffness of the modeled cells was 3 dyn/cm. Receptor-ligand binding kinetics was governed by the Dembo model. The microvilli were assumed to be rigid cylindrical rods. Data represent means ± SE from 3 independent simulations.
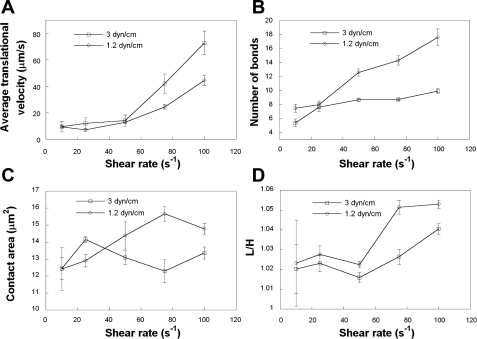
Several in vitro experiments have illustrated that cell deformation influences cell rolling behavior. To investigate the effect of this mesoscale parameter on cell rolling under the action of hydrodynamic shear, we recorded the average translational velocities of modeled cells with membrane stiffness values of 3.0 or 1.2 dyn/cm. The latter value was chosen to closely match the extent of leukocyte deformation in vivo (11, 41) for relatively low levels of shear stress typical of the venous circulation. Although the average translational velocities for the two modeled cells were comparable in the low shear regime, the more compliant cells (Eh = 1.2 dyn/cm) rolled slower than cells with a membrane stiffness of 3.0 dyn/cm at elevated levels of shear (>50 s−1; Fig. 2A). This is ascribed to the higher number of cell-substrate bonds associated with the more compliant cell at >50 s−1 (Fig. 2B). This is a direct consequence of the increased cell-substrate contact area (Fig. 2C) due to the greater cell deformation (L/H) index (11, 22, 41) of the more compliant cell (Fig. 2D) noted at elevated levels of shear.
Fig. 2.
Effect of membrane stiffness on cell rolling behavior. Average translational velocities (A), average numbers of bonds (B), cell-substrate contact areas (C), and cell deformation (L/H ratio) indexes (D) of modeled, deformable PMNs rolling over P-selectin-coated surfaces at prescribed shear rates were computed for membrane stiffness values of 3 and 1.2 dyn/cm. The P-selectin site density was 15 molecules/μm2. Receptor-ligand binding kinetics was governed by the Dembo model. The microvilli were assumed to be rigid cylindrical rods. Data represent means ± SE from 3 independent simulations.
Effects of Microvillus Deformability on PMN Rolling in Shear Flow
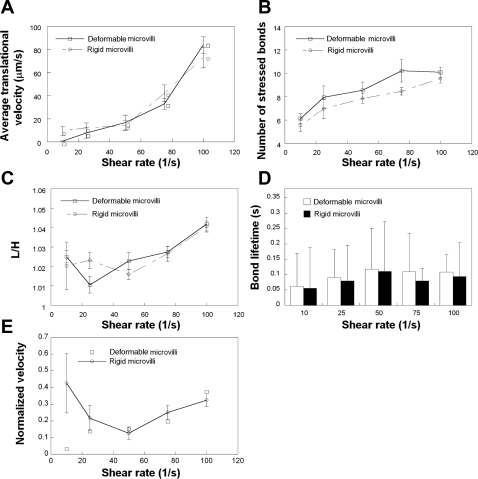
Micropipette assays have disclosed that under the action of a pulling force (Fm), a microvillus can be extended like an elastic spring if Fm < 45 pN or a tether can be formed if Fm > 45 pN (39). To quantify the effects of microvillus deformability on cell rolling, we simulated the rolling of cells bearing either rigid or viscoelastic microvilli over a plane surface coated with P-selectin molecules at a site density of 15 molecules/μm2 for shear rates varying from 10 to 100 s−1. Both modeled cells exhibited similar average translational velocities over the entire range of prescribed shear rates except for 10 s−1 (Fig. 3A). Along these lines, the average number of stressed bonds (defined as those whose length was greater than the equilibrium bond length) as well as the L/H index and average bond lifetime were not significantly altered by the viscoelastic nature of the microvillus at any given shear rate (Figs. 3, B–D). The difference in the velocity observed between modeled cells bearing rigid (9.6 ± 4.0 μm/s) versus viscoelastic (0.7 ± 0.1 μm/s) microvilli at 10 s−1 is presumably due to the fact that the force absorbed by the engagement of a small number of extensible microvilli is comparable with the magnitude of the dispersive hydrodynamic force prevalent at this low shear.
Fig. 3.
Effect of microvillus deformability on cell rolling at low P-selectin site densities. Average translational velocities (A), numbers of stressed bonds (B), L/H indexes (C), average bond lifetimes (D), and normalized velocities (E) of modeled PMNs, decorated with either deformable or rigid microvilli, rolling over P-selectin-coated surfaces at prescribed shear rates were computed for a membrane stiffness value of 3 dyn/cm. The P-selectin site density was 15 molecules/μm2. Receptor-ligand binding kinetics was governed by the Dembo model. Data represent means ± SE except for bond lifetimes, which are expressed as means ± SD, from 3 independent simulations.
It is noteworthy that the average bond lifetime increased with increasing shear rate and reached a maximum at 50 s−1 (Fig. 3D). This increase in bond lifetime with shear can be explained by the higher number of receptor-ligand bonds that are progressively formed as shear increases from 10 to 50 s−1, which results in a diminished load on any individual bond. These observations are in accord with the postulate of Chen and Springer (9), by which an increase in bond number with shear rate acts as an automatic braking system and could lead to the shear threshold effect. Related to the increase in bond number, our simulations predict that velocity normalized by hydrodynamic velocity shows a minimum at 50 s−1 for modeled cells bearing rigid microvilli (Fig. 3E), which is indicative of the shear threshold phenomenon.
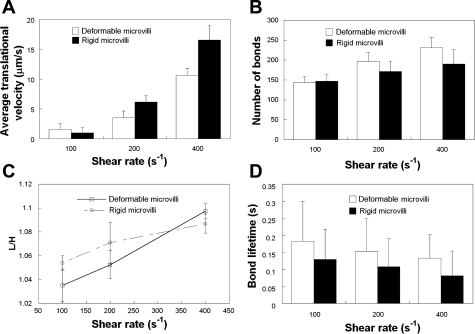
Since the viscoelastic microvillus is in series with the receptor-ligand bond(s), it can potentially act as a “shock/force absorber” over a wide range of shear rates (39). In such a case, the average lifetime of a bond attached to a deformable microvillus would be expected to be higher than that of a bond linked to a rigid microvillus. However, this is in clear contrast to the data obtained at the low selectin site density of 15 molecules/μm2 (Fig. 3D). We speculate that this may be due to the fact that only one bond per microvillus is involved in the cell-substrate contact (data not shown) under these prescribed simulation conditions, and, as a result, the force absorbed by the microvillus is not sufficient to significantly deform it for a prolonged period of time. To this end, we wanted to subject the deformable microvilli to sustained high-magnitude forces by increasing the average number of bonds associated per microvillus during a cell-substrate contact event. To accomplish this, we allowed cells bearing either rigid or deformable microvilli to roll over surfaces coated with P-selectin at a site density of 150 molecules/μm2 for shear rates ranging from 100 to 400 s−1. Cells bearing rigid microvilli rolled faster than those coated with deformable microvilli at elevated levels of shear; this difference became more prominent as the shear rate increased from 200 to 400 s−1 (Fig. 4A). Along these lines, the average number of bonds between the cell and selectin-coated substrate (Fig. 4B) and average bond lifetime (Fig. 4C) were lower for the rigid microvilli model. Interestingly, the L/H index was similar for both modeled cells (Fig. 4D), suggesting that the difference in cell rolling is based solely on microvillus deformability and is not a consequence of bulk cell deformation.
Fig. 4.
Effect of microvillus deformability on cell rolling at high P-selectin site densities. Average translational velocities (A), numbers of stressed bonds (B), L/H indexes (C), and average bond lifetimes (D) of modeled PMNs, decorated with either deformable or rigid microvilli, rolling over P-selectin-coated surfaces at prescribed shear rates were computed for a membrane stiffness value of 3 dyn/cm. The P-selectin site density was 150 molecules/μm2. Receptor-ligand binding kinetics was governed by the Dembo model. Data represent means ± SE except for bond lifetimes, which are expressed as means ± SD, from 3 independent simulations.
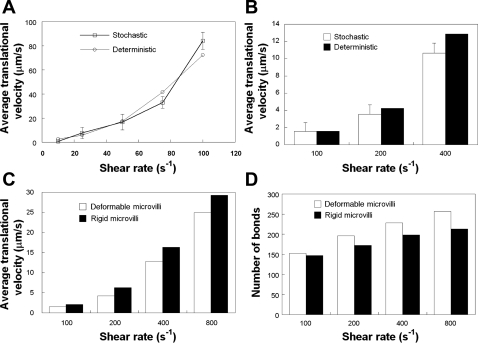
We next compared the results predicted by the probabilistic model for receptor-ligand binding with those acquired using a deterministic model of bond formation and rupture (24). Our analysis revealed that the cell translational velocities obtained using the stochastic model were in excellent agreement with those of the deterministic model at both low (Fig. 5A) and high (Fig. 5B) selectin site densities on the substrate. Using the deterministic model, we also confirmed that cells bearing deformable microvilli roll slower than those decorated with rigid microvilli (Fig. 5C), presumably because of the lower force exerted on the bonds attached to deformable microvilli. This is a consequence of the fact that the average number of bonds associated with the deformable microvilli is greater than that associated with the rigid microvilli at elevated levels of hydrodynamic shear (Fig. 5D). This is in direct agreement with the concept that the deformable microvillus acts as a “force absorber,” thereby increasing the lifetimes of the bonds attached to it.
Fig. 5.
Comparison of the stochastic and deterministic models of cell rolling at low (A) and high (B–D) P-selectin site densities. Average translational velocities of modeled PMNs, decorated with deformable microvilli, rolling over surfaces coated with P-selectin at a site density of 15 (A) or 150 (B) molecules/μm2 were computed for a membrane stiffness value of 3 dyn/cm. Average translational velocities (C) and numbers of bonds (D) for cells decorated with either rigid or deformable microvilli rolling over P-selectin-coated substrates (150 molecules/μm2) were computed using the deterministic model for a membrane stiffness value of 3 dyn/cm. In all these calculations, receptor-ligand kinetics was governed by the Dembo model. Data from the stochastic model represent means ± SE from 3 independent simulations.
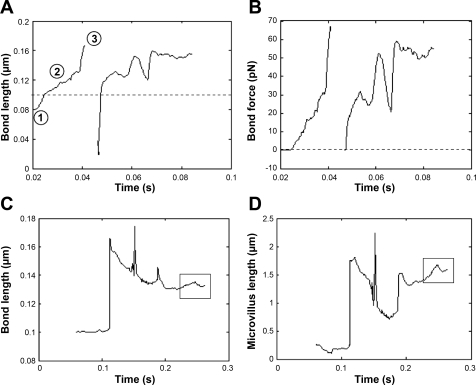
Figure 6A shows a sample trace for bond length for the rigid microvillus model. Bond formation occurred when the distance between the tip of the microvillus and selectin-coated plane surface was ∼0.08 μm, which was slightly less than the resting bond length (0.1 μm) indicated by the horizontal dotted line in Fig. 6A. As the cell moved in the direction of fluid flow, the bond length increased beyond its resting length, resulting in bond stretching. The corresponding force trace for the bond is shown in Fig. 6B. Since the receptor-ligand bonds were modeled as Hookean springs, bond force increased linearly upon bond stretching. Note that in our model, unlike a true Hookean spring, the bond force is assumed to be zero when the bond length is less than the equilibrium bond length. This bond ruptures when the force reaches ∼70 pN. Due to the probabilistic nature of receptor-ligand interactions, the same PSGL-1 molecule is engaged in a binding interaction with a P-selectin molecule as the simulation proceeds in time and the cell moves forward. Also interesting to note is the “valley” observed in both traces. This is indicative of the fact that receptor-ligand bond forces may occasionally pull the cell in a direction opposite to fluid flow, thereby reducing the lengths of some bonds.
Fig. 6.
Sample length and force traces for individual bonds and microvilli of modeled PMNs rolling over P-selectin substrates. Bond lengths (A) and bond forces (B) were tracked for an individual bond attached to a rigid microvillus as a function of time. Bond lengths (C) and microvillus lengths (D) were also recorded for the case of a deformable microvillus. Whereas the microvillus length followed the same pattern as the bond initially, it is noteworthy that microvillus length increased, whereas the bond length remained nearly constant, later in the simulation (indicated by boxes in C and D). The P-selectin site density was 15 molecules/μm2, and the cell was subjected to a uniform shear rate of 10 s−1. The membrane stiffness was 3 dyn/cm.
We also recorded the variation of bond (Fig. 6C) and microvillus (Fig. 6D) lengths at low selectin site densities for cells bearing deformable microvilli. Whereas the microvillus length followed the same pattern as the bond initially, it is noteworthy that microvillus length increased, whereas the bond length remained nearly constant, later in the simulation (indicated by the boxes in Fig. 6, C and D), presumably because the microvillus no longer acts like a Hookean spring but as a viscous tether.
Effects of Receptor-Ligand Binding Kinetics on Cell Rolling
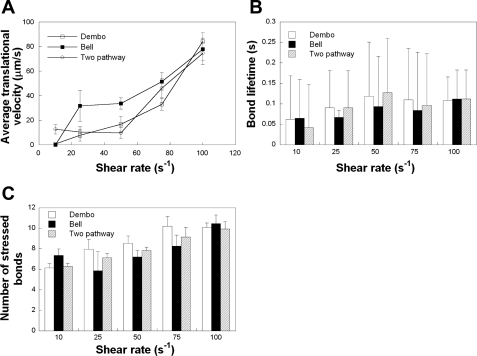
To investigate the effect of catch-slip bond behavior on the rolling of modeled, deformable cells, we implemented the two-pathway kinetic model of bond dissociation into our code and compared it with the results obtained with the Bell and Dembo models over a wide range of shear rates (10–100 s−1). In these simulations, cells bearing deformable microvilli with PSGL-1 molecules localized at the tips were allowed to roll over P-selectin-coated surfaces. A P-selectin site density of 15 molecules/μm2 was used to minimize the number of receptor-ligand bonds formed during the rolling process. As a first step, we recorded the average translational velocities of modeled cells as a function of shear rate. The velocity profile predicted by the two-pathway model exhibited the closest resemblance to the shear threshold effect, wherein the average translational velocity of the cell first decreased with increasing shear, reached a minimum at 50 s−1, and then increased markedly at higher shear rates (Fig. 7A). The translational velocity predicted by the Bell model was higher than that predicted by the two-pathway model at low shear rates (25–50 s−1), but collapsed into the two-pathway model in the high shear regime. This is presumably because when the cell is subjected to high hydrodynamic forces, the force-dependent Bell model term in Eq. 17 becomes more significant. On the other hand, the Dembo model showed good agreement with the two-pathway model over the entire range of shear rates (except for 10 s−1) used in our simulations.
Fig. 7.
Comparison among Dembo, Bell, and two-pathway models of receptor-ligand binding. Average translational velocities (A), average bond lifetimes (B), and numbers of stressed bonds (C) of modeled PMNs subjected to uniform levels of shear (10–100 s−1) were calculated using the three different kinetic models. In these simulations, the P-selectin site density on the substrate was 15 molecules/μm2, and the membrane stiffness of modeled cells bearing deformable microvilli was 3 dyn/cm. Data represent means ± SE except for bond lifetimes, which are expressed as means ± SD, from 3 independent simulations.
Whereas the average translational velocity is a parameter that quantifies the rolling behavior of the cell as a whole and includes contributions from all the receptor-ligand bonds formed during the simulation, bond lifetimes disclose the effect of receptor-ligand binding kinetics on individual bonds. The bond lifetimes calculated using the two-pathway model displayed a maximum at 50 s−1 (Fig. 7B). Interestingly, the bond lifetimes computed using the Dembo model were in good agreement with those calculated using the two-pathway model, whereas the Bell model predicted similar values only in the high shear regime. Along these lines, the average number of stressed bonds showed a steady increase with increasing shear rate for both the two-pathway and Dembo models (Fig. 7C). The Bell model predicted similar values at high shear rates only (Fig. 7C).
DISCUSSION
To our knowledge, this is the first 3-D computational model that integrates three distinct length scales involved in cell rolling on a ligand-coated surface: bulk cell deformation (mesoscale), microvillus deformability (microscale), and receptor-ligand binding kinetics (nanoscale). The major findings of this work are as follows: 1) the catch-slip bond transition governed by nanoscale receptor-ligand binding kinetics is a major factor that influences the shear threshold effect; 2) bulk cell deformation can partially account for the existence of the shear threshold phenomenon in PMN tethering and rolling on sparsely coated P-selectin surfaces; and 3) cells decorated with deformable microvilli roll slower compared with those associated with rigid microvilli only at high P-selectin site densities and elevated levels of shear.
Both in vivo and in vitro studies have documented the shear threshold phenomenon, in which adhesion goes through a maximum with shear rate (17, 28). Whether this phenomenon is solely the result of the selectin-ligand binding kinetics (6) or the mesoscale/microscale physical characteristics of the cell (whole cell and/or microvillus deformation) contribute to it have not been previously resolved. Our simulations using deformable, modeled cells bearing rigid microvilli disclose that the normalized translational velocities, computed using the Dembo model, exhibit a distinct minimum at 50 s−1, akin to the shear threshold effect. Interestingly, the average bond lifetimes displayed a maximum at this shear threshold level, which is in accord with the experimentally reported level of shear threshold for P-selectin-dependent rolling (28). Using modeled cells decorated with deformable microvilli, both the Bell and Dembo models were able to predict a local minimum in normalized velocity, although this was not as pronounced as in the two-pathway kinetic model. Taken together, these data clearly demonstrate that the catch-slip bond transition plays a pivotal role in regulating the shear threshold phenomenon. However, the detection of a minimum in normalized velocity of deformable cells using the Dembo model (Fig. 3E), which for the parameters chosen herein predicts slip bond behavior, suggests that whole cell deformation, even if limited (Fig. 2D), can play an auxiliary role in this process, presumably by further augmenting the encounter rate of receptor-ligand pairs under shear.
Theoretical studies by Weinbaum and colleagues have predicted the presence of sizeable compressive forces on the tips of microvilli, especially on the small population (5–10%) of long microvilli (>0.5 μm), due to gravitational-hydrodynamic interactions (49). Thus, another plausible explanation for the threshold phenomenon could be that these compressive forces increase with shear rate and that a minimum compressive force on microvilli tips is required to initiate receptor-ligand binding (45, 49).
The effect of bulk cell deformation on rolling becomes more evident at elevated levels of shear rate. Progressively increasing hydrodynamic forces cause larger deformations, especially in cells with relatively more compliant membranes. This deformation reduces the hydrodynamic force experienced by the cell and increases the cell-substrate contact area (Fig. 2, C and D). Consequently, a higher number of receptor-ligand bonds with prolonged lifetimes are formed, which are responsible for the slower rolling of more compliant cells. It is worth noting that the effect of cell deformation on rolling is dependent not only on the magnitude of the imposed hydrodynamic shear but also on the selectin site density on the substrate. More specifically, at low selectin densities, cells with the stiffer membrane roll faster at ≥75 s−1 (Fig. 2A), whereas at higher selectin densities, this differential rolling behavior becomes apparent at shear rates ≥200 s−1 (22). This may be explained by the fact that at low selectin densities, smaller differences in cell deformation have a higher impact on the accessibility of selectins to ligands on the cell surface and thus on rolling velocity.
In accord with experimental data (35), our simulations also predict that microvillus deformability stabilizes cell rolling under shear by augmenting the lifetime of receptor-ligand bonds (Fig. 4D). However, the effect of microvillus deformability on rolling becomes evident at elevated levels of shear and high selectin densities, which correspond to the engagement of a large number (∼8) of receptor-ligand bonds per microvillus (22). In contrast, the engagement of a single bond per microvillus is not sufficient to alter the magnitude of the cell translational velocity over the entire range of shear rates examined herein, except for 10 s−1. Cumulatively, these observations suggest that significant and prolonged microvillus deformation is necessary for the stabilization of cell rolling under shear. A microvillus exploration parameter study (5) revealed that stiffer microvilli (high σm values) result in faster rolling, whereas a minimum in rolling velocity occurs at an intermediate value of microvillus membrane viscosity (σ′m), which is close to the reported physiological value (39) used in these simulations.
A critical question is whether the microvilli of a free-flowing leukocyte will be able to penetrate the endothelial glycocalyx layer (EGL) and initiate tethering and rolling in vivo, in view of the fact that EGL, which covers the entire vasculature including venular microvessels, has a 0.5 μm thickness (45, 46). Although the average length of microvilli is 0.35 μm, a detailed ultrastructural study by Bruehl et al. (4) has shown that the actual length varies from 0.15 to 0.7 μm. The impact of a small percentage (5–10%) of longer microvilli (>0.5 μm) dramatically changes leukocyte behavior. The long microvilli on a leukocyte experiencing rolling in vivo are subjected to high compressive forces due to the gravitational-hydrodynamic interaction and are thus predicted to easily penetrate the EGL thickness and form new tethering attachments (45).
It is of note that at 10 s−1, the translational velocity of modeled cells bearing deformable microvilli is dramatically lower than that of cells with rigid microvilli when the Dembo model was employed to capture receptor-ligand binding kinetics. Most importantly, only at this low shear rate (10 s−1) are the velocities predicted by either the Dembo or Bell model strikingly similar. We speculate that the force absorbed by the engagement of a small number of extensible microvilli is sufficient to effectively counteract the low hydrodynamic force prevalent at 10 s−1 and slow the cell rolling velocity. In contrast, the two-pathway model yielded a significantly higher average translational velocity relative to the Bell or Dembo model at 10 s−1. This can be explained by the fact that at this low shear rate, and correspondingly low hydrodynamic force, as the cell marches forward, bonds are likely to be subjected to low loading rates, which can also be facilitated by microvillus elongation and hence behave like catch bonds and rupture via the slow dissociation pathway.
In conclusion, we used a 3-D computational model to demonstrate that cell rolling over ligand-coated surfaces is influenced by three distinct length scales; whereas at the nanoscale level, receptor-ligand binding kinetics is a major contributing factor in defining the shear threshold phenomenon, at the mesoscale level, bulk cell deformation plays a supporting role in this process. Furthermore, the deformability of microvilli is responsible for stabilizing cell rolling in shear flow at physiological levels of shear and high selectin density on the substrate. Although experimental evidence has suggested that receptor-ligand binding is a stochastic process, the results predicted by our mathematically deduced deterministic model are in excellent agreement with those obtained using the probabilistic depiction. Future work will focus on extending the model to include the internal structure of the cell (e.g., viscoelastic properties of the nucleus and cytoplasm) and the nonuniform length of microvilli as well as the use of other constitutive models to describe microvillus viscoelasticity (e.g., the Kelvin-Voigt model).
GRANTS
This work was supported by National Institute of Allergy and Infectious Diseases Grant AI-063366 and a Mid-Atlantic American Heart Association grant-in-aid.
Acknowledgments
The authors thank Dr. Kit Yan Chan for helpful discussions.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Alexeev A, Verberg R, Balazs AC. Patterned surfaces segregate compliant microcapsules. Langmuir 23: 983–987, 2007. [DOI] [PubMed] [Google Scholar]
- 2.Alon R, Hammer DA, Springer TA. Lifetime of the P-selectin-carbohydrate bond and its response to tensile force in hydrodynamic flow. Nature 374: 539–542, 1995. [DOI] [PubMed] [Google Scholar]
- 3.Bell GI Models for the specific adhesion of cells to cells. Science 200: 618–627, 1978. [DOI] [PubMed] [Google Scholar]
- 4.Bruehl RE, Springer TA, Bainton DF. Quantitation of L-selectin distribution on human leukocyte microvilli by immunogold labeling and electron microscopy. J Histochem Cytochem 44: 835–844, 1996. [DOI] [PubMed] [Google Scholar]
- 5.Caputo KE, Hammer DA. Effect of microvillus deformability on leukocyte adhesion explored using adhesive dynamics simulations. Biophys J 89: 187–200, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caputo KE, Lee D, King MR, Hammer DA. Adhesive dynamics simulations of the shear threshold effect for leukocytes. Biophys J 92: 787–797, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang KC, Tees DF, Hammer DA. The state diagram for cell adhesion under flow: leukocyte rolling and firm adhesion. Proc Natl Acad Sci USA 97: 11262–11267, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charrier JM, Shrivastava S, Wu R. Free and constrained inflation of elastic membranes in relation to thermoforming–non-axisymmetric problems. J Strain Anal Eng Design 24: 55–74, 1989. [Google Scholar]
- 9.Chen S, Springer TA. An automatic braking system that stabilizes leukocyte rolling by an increase in selectin bond number with shear. J Cell Biol 144: 185–200, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cozens-Roberts C, Lauffenburger D, Quinn J. Receptor-mediated cell attachment and detachment kinetics. I. Probabilistic model and analysis. Biophys J 58: 841–856, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Damiano ER, Westheider J, Tozeren A, Ley K. Variation in the velocity, deformation, and adhesion energy density of leukocytes rolling within venules. Circ Res 79: 1122–1130, 1996. [DOI] [PubMed] [Google Scholar]
- 12.Dembo M On peeling an adherent cell from a surface. In: Lectures in Mathematics in Life Sciences, Some Mathematical Problems in Biology. Providence, RI: American Mathematical Society, 1994, p. 51–77.
- 13.Dong C, Cao J, Struble EJ, Lipowsky HH. Mechanics of leukocyte deformation and adhesion to endothelium in shear flow. Ann Biomed Eng 27: 298–312, 1999. [DOI] [PubMed] [Google Scholar]
- 14.Dong C, Lei XX. Biomechanics of cell rolling: shear flow, cell-surface adhesion, and cell deformability. J Biomech 33: 35–43, 2000. [DOI] [PubMed] [Google Scholar]
- 15.Eggleton CD, Popel AS. Large deformation of red blood cell ghosts in a simple shear flow. Phys Fluids 10: 1834–1845, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans E, Leung A, Heinrich V, Zhu C. Mechanical switching and coupling between two dissociation pathways in a P-selectin adhesion bond. Proc Natl Acad Sci USA 101: 11281–11286, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finger EB, Puri KD, Alon R, Lawrence MB, von Andrian UH, Springer TA. Adhesion through L-selectin requires a threshold hydrodynamic shear. Nature 379: 266–269, 1996. [DOI] [PubMed] [Google Scholar]
- 18.Fritz J, Katopodis AG, Kolbinger F, Anselmetti D. Force-mediated kinetics of single P-selectin/ligand complexes observed by atomic force microscopy. Proc Natl Acad Sci USA 95: 12283–12288, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammer DA, Apte SM. Simulation of cell rolling and adhesion on surfaces in shear flow: general results and analysis of selectin-mediated neutrophil adhesion. Biophys J 63: 35–57, 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanley WD, Wirtz D, Konstantopoulos K. Distinct kinetic and mechanical properties govern selectin-leukocyte interactions. J Cell Sci 117: 2503–2511, 2004. [DOI] [PubMed] [Google Scholar]
- 21.Jadhav S, Chan KY, Konstantopoulos K, Eggleton CD. Shear modulation of intercellular contact area between two deformable cells colliding under flow. J Biomech 40: 2891–2897, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jadhav S, Eggleton CD, Konstantopoulos K. A 3-D computational model predicts that cell deformation affects selectin-mediated leukocyte rolling. Biophys J 88: 96–104, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jadhav S, Eggleton CD, Konstantopoulos K. Mathematical modeling of cell adhesion in shear flow: application to targeted drug delivery in inflammation and cancer metastasis. Curr Pharm Des 13: 1511–1526, 2007. [DOI] [PubMed] [Google Scholar]
- 24.Khismatullin DB, Truskey GA. 3D numerical simulation of receptor-mediated leukocyte adhesion to surfaces; effects of cell deformabilty and viscoelasticity. Phys Fluids 17: 031505, 2005.
- 25.Khismatullin DB, Truskey GA. A 3D numerical study of the effect of channel height on leukocyte deformation and adhesion in parallel-plate flow chambers. Microvasc Res 68: 188–202, 2004. [DOI] [PubMed] [Google Scholar]
- 26.Konstantopoulos K, Kukreti S, McIntire LV. Biomechanics of cell interactions in shear fields. Adv Drug Delivery Res 33: 141–164, 1998. [DOI] [PubMed] [Google Scholar]
- 27.Lai MC, Peskin CS. An immersed boundary method with formal second-order accuracy and reduced numerical viscosity. J Comp Physiol 160: 705–719, 2000. [Google Scholar]
- 28.Lawrence MB, Kansas GS, Kunkel EJ, Ley K. Threshold levels of fluid shear promote leukocyte adhesion through selectins (CD62L,P,E). J Cell Biol 136: 717–727, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marshall BT, Long M, Piper JW, Yago T, McEver RP, Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature 423: 190–193, 2003. [DOI] [PubMed] [Google Scholar]
- 30.McCarty OJ, Tien N, Bochner BS, Konstantopoulos K. Exogenous eosinophil activation converts PSGL-1-dependent binding to CD18-dependent stable adhesion to platelets in shear flow. Am J Physiol Cell Physiol 284: C1223–C1234, 2003. [DOI] [PubMed] [Google Scholar]
- 31.Mehta P, Cummings RD, McEver RP. Affinity and kinetic analysis of P-selectin binding to P-selectin glycoprotein ligand-1. J Biol Chem 273: 32506–32513, 1998. [DOI] [PubMed] [Google Scholar]
- 32.Moore KL, Patel KD, Bruehl RE, Li F, Johnson DA, Lichenstein HS, Cummings RD, Bainton DF, McEver RP. P-selectin glycoprotein ligand-1 mediates rolling of human neutrophils on P-selectin. J Cell Biol 128: 661–671, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore KL, Varki A, McEver RP. GMP-140 binds to a glycoprotein receptor on human neutrophils: evidence for a lectin-like interaction. J Cell Biol 112: 491–499, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.N'Dri NA, Shyy W, Tran-Son-Tay R. Computational modeling of cell adhesion and movement using a continuum-kinetics approach. Biophys J 85: 2273–2286, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park EY, Smith MJ, Stropp ES, Snapp KR, DiVietro JA, Walker WF, Schmidtke DW, Diamond SL, Lawrence MB. Comparison of PSGL-1 microbead and neutrophil rolling: microvillus elongation stabilizes P-selectin bond clusters. Biophys J 82: 1835–1847, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peskin CS, Mcqueen DM. A 3-dimensional computational method for blood-flow in the heart. 1. Immersed elastic fibers in a viscous incompressible fluid. J Comp Physiol 81: 372–405, 1989. [Google Scholar]
- 37.Ramachandran V, Williams M, Yago T, Schmidtke DW, McEver RP. Dynamic alterations of membrane tethers stabilize leukocyte rolling on P-selectin. Proc Natl Acad Sci USA 101: 13519–13524, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidtke DW, Diamond SL. Direct observation of membrane tethers formed during neutrophil attachment to platelets or P-selectin under physiological flow. J Cell Biol 149: 719–730, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shao JY, Ting-Beall HP, Hochmuth RM. Static and dynamic lengths of neutrophil microvilli. Proc Natl Acad Sci USA 95: 6797–6802, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheikh S, Nash GB. Treatment of neutrophils with cytochalasins converts rolling to stationary adhesion on P-selectin. J Cell Physiol 174: 206–216, 1998. [DOI] [PubMed] [Google Scholar]
- 41.Smith ML, Smith MJ, Lawrence MB, Ley K. Viscosity-independent velocity of neutrophils rolling on p-selectin in vitro or in vivo. Microcirculation 9: 523–536, 2002. [DOI] [PubMed] [Google Scholar]
- 42.Springer TA Traffic signals on endothelium for lymphocyte recirculation and leukocyte emigration. Annu Rev Physiol 57: 827–872, 1995. [DOI] [PubMed] [Google Scholar]
- 43.Tandon P, Diamond SL. Kinetics of beta2-integrin and L-selectin bonding during neutrophil aggregation in shear flow. Biophys J 75: 3163–3178, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Usta OB, Alexeev A, Balazs AC. Fork in the road: patterned surfaces direct microcapsules to make a decision. Langmuir 23: 10887–10890, 2007. [DOI] [PubMed] [Google Scholar]
- 45.Weinbaum S, Tarbell JM, Damiano ER. The structure and function of the endothelial glycocalyx layer. Annu Rev Biomed Eng 9: 121–167, 2007. [DOI] [PubMed] [Google Scholar]
- 46.Weinbaum S, Zhang X, Han Y, Vink H, Cowin SC. Mechanotransduction and flow across the endothelial glycocalyx. Proc Natl Acad Sci USA 100: 7988–7995, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yago T, Leppanen A, Qiu H, Marcus WD, Nollert MU, Zhu C, Cummings RD, McEver RP. Distinct molecular and cellular contributions to stabilizing selectin-mediated rolling under flow. J Cell Biol 158: 787–799, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yago T, Wu J, Wey CD, Klopocki AG, Zhu C, McEver RP. Catch bonds govern adhesion through L-selectin at threshold shear. J Cell Biol 166: 913–923, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao Y, Chien S, Weinbaum S. Dynamic contact forces on leukocyte microvilli and their penetration of the endothelial glycocalyx. Biophys J 80: 1124–1140, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]