Abstract
Summary: ATP synthase, a double-motor enzyme, plays various roles in the cell, participating not only in ATP synthesis but in ATP hydrolysis-dependent processes and in the regulation of a proton gradient across some membrane-dependent systems. Recent studies of ATP synthase as a potential molecular target for the treatment of some human diseases have displayed promising results, and this enzyme is now emerging as an attractive molecular target for the development of new therapies for a variety of diseases. Significantly, ATP synthase, because of its complex structure, is inhibited by a number of different inhibitors and provides diverse possibilities in the development of new ATP synthase-directed agents. In this review, we classify over 250 natural and synthetic inhibitors of ATP synthase reported to date and present their inhibitory sites and their known or proposed modes of action. The rich source of ATP synthase inhibitors and their known or purported sites of action presented in this review should provide valuable insights into their applications as potential scaffolds for new therapeutics for human and animal diseases as well as for the discovery of new pesticides and herbicides to help protect the world's food supply. Finally, as ATP synthase is now known to consist of two unique nanomotors involved in making ATP from ADP and Pi, the information provided in this review may greatly assist those investigators entering the emerging field of nanotechnology.
INTRODUCTION
ATP synthase (F0F1) is a multisubunit, membrane-associated protein complex that catalyzes the phosphorylation of ADP to ATP at the expense of a proton motive force generated by an electron transport chain in energy-transducing membranes (303, 387). In some organisms, it also works in the reverse direction by hydrolyzing ATP and generating an electrochemical proton gradient across a membrane to support locomotion or nutrient uptake. ATP synthase is present in all living organisms and is located in the membranes of mitochondria, bacteria, and chloroplast thylakoids as well as on the surfaces of various cell types, including endothelial cells (269, 270), keratinocytes (58), and adipocytes (206).
ATP synthase is an exceptionally complicated protein complex. It is divided into two sectors, a soluble globular F1 catalytic sector and a membrane-bound F0 proton-translocating sector (Fig. 1) (304, 305). Even the simplest form of ATP synthase, found in nonphotosynthetic eubacteria, contains eight different subunit types, while the chloroplast and photosynthetic bacterial ATP synthase each consists of nine different subunit types (42, 331). The ATP synthase from mitochondria is much more complicated and, excluding regulators, is reported to date to consist of 15 and 17 different subunit types in animals and yeasts (or fungi), respectively (305, 413).
FIG. 1.
Current view of the structure of mitochondrial ATP synthase from metazoans. F1 is composed of α, β, γ, δ, and ɛ subunits, and F0 consists of a, b, c, d, e, f, g, A6L, and OSCP. IF1 is a regulatory protein. The coordinates of the subunits used in the structural model are 1E79 for the α, β, γ, δ, and ɛ subunits; 1ABV for the N-terminal domain of OSCP; 2CLY for F6, d, and the hydrophilic part of the b subunit; 1GMJ for IF1; and 1B9U for the transmembrane part of the b subunit. The ac10 subcomplex was modeled using the coordinates of the a and c subunits from 1C17, and the other subunits in the model were constructed manually using Quanta. No positions are assigned to the factor B and the e subunit. Here and where indicated in the other figure legends, the coordinates of protein structures were obtained from the PDB.
ATP synthase is associated directly or indirectly with various human diseases. One form of Leigh syndrome, a neurodegenerative disease which causes a neuromuscular disorder with a 50% survival rate to 3 years of age, is the consequence of a severe impairment of ATP synthesis. This is due to a mutation in subunit a of ATP synthase (99). The neuropathy, ataxia, retinitis pigmentosa syndrome and the familial bilateral striatal necrosis are also caused by the dysfunction of ATP synthase due to mutations within the same subunit (93, 396). In Batten's disease, a lysosomal storage disease also known as neuronal ceroid lipofuscinoses or Kufs' disease, the subunit c of ATP synthase has been found as a predominant storage protein (298, 299). In addition, in Alzheimer's disease or presenile dementia, which is a progressive and degenerative disease that attacks the brain, a deficiency of ATP synthase has been observed in mitochondria (357). A low expression of the ATP synthase β subunit and the cytosolic accumulation of the α subunit are detected in Alzheimer's disease, and the intraneuronal cytosolic accumulation of the α subunit is implicated in the neurodegenerative process (73, 208, 367). Moreover, the ATP synthase on the cell surface of endothelial cells has been reported to have an important role in the angiogenesis process required for tumor growth (269-271, 422). Additionally, the ATP synthase F6 subunit circulating in the blood has been recognized to be involved in the increase of blood pressure (293, 294). Finally, the β subunit of ATP synthase has been identified as a target protein for innate antitumor cytotoxicity mediated by natural killer and interleukin 2-activated killer cells (91).
ATP synthase has also been demonstrated and suggested as a good molecular target for drugs in the treatment of various diseases and the regulation of energy metabolism (16, 38, 72, 193, 202, 367). One of the drugs developed for the treatment of tuberculosis, R207910, was shown to be active against a number of drug-resistant strains of Mycobacterium tuberculosis and to eradicate M. tuberculosis infection rapidly and effectively (15, 313, 340). The drug has been revealed to block the synthesis of ATP by targeting subunit c of ATP synthase. Another drug, Bz-423, which was developed for therapy of the autoimmune disorder systemic lupus erythematosus, kills pathogenic lymphocytes selectively by inducing apoptosis in lymphoid cells (41). Significantly, Bz-423 has been found to inhibit the mitochondrial ATP synthase by binding to the subunit known as oligomycin sensitivity-conferring protein (OSCP) (193). In addition, the inhibition of nonmitochondrial ATP synthase resulted in the inhibition of cytosolic lipid droplet accumulation, suggesting ATP synthase as a molecular target for antiobesity drugs (16). Finally, the inhibition of ATP synthase has been suggested for an antiangiogenic therapeutic strategy to block tumor angiogenesis (17, 59, 269-271, 422). Here, the reaction of ATP synthase inhibitors with the nonmitochondrial ATP synthase of endothelial cells has been shown to inhibit markedly the migration and proliferation of endothelial cells with little effect on intracellular ATP (17).
The aim of this review is to provide insight and encouragement into the development of new ATP synthase-directed agents. We have meticulously categorized most of the natural and synthetic inhibitors of ATP synthase reported to date in accordance with physical/chemical characteristics of the inhibitors and have summarized the current knowledge of the modes of action of these inhibitors. The information provided in this review should prove to be an invaluable resource, not only for obtaining information about the interactions of known effectors, primarily inhibitors of ATP synthase, but for generating new ideas for the development of numerous additional ATP synthase-directed agents that can be used (i) in the treatment of human and animal diseases, (ii) in agriculture as pesticides or herbicides, and (iii) in the developing field of nanotechnology to understand the mechanics of nanomotor function.
PEPTIDE INHIBITORS
α-Helical Basic Peptide Inhibitors
The α-helical basic peptide inhibitors bind to F1 and inhibit ATPase activity (Table 1). Inhibitors in this group include α-helical structures containing basic residues, which appear to be crucial for their inhibitory activities. The α-helical basic peptide inhibitors include the bacterial/chloroplast ɛ subunit, melittin, the presequence of yeast cytochrome oxidase subunit IV (WT and its synthetic derivatives), and possibly the inhibitor protein (IF1) (Fig. 2A).
TABLE 1.
α-Helical basic peptide inhibitors
| Name | Amino acid sequence (species)a | Source | Inhibitory potency (reference) |
|---|---|---|---|
| Bacterial/chloroplast ɛ subunit | MTLNLCVLTPNRSIWNSEVKEIILSTNSGQIGVLPNHAPTATAVDIGILRIRLNDQWLTLALMGGFARIGNNEITILVNDAERGSDIDPQEAQQTLEIAEANLRKAEGKRQKIEANLALRRARTRVEASNTISS (spinach) | Natural regulatory peptide | 1-3 ɛ mol/molc CF1(-ɛ)b (spinach Ca2+-ATPase) (332); ∼0.73 μg/μgc (spinach CF1-Ca2+-ATPase) (284); ∼15 nMc (EF1-ATPase) (372); 100 nMc (EF1-ATPase, rotation rate of 60-nm beads) (282); 10 nMd (EF1-ATPase) (386); 2.1 nMe (Thermosynecoccus ascicula F1, αβγ complex) (212); 94% inhibition at 10 ɛ mol/mol CF1(-ɛ) (spinach Ca2+-ATPase) (289) |
| IF1 | MAVTALAARTWLGVWGVRTMQARGFGSDQSENVDRGAGSIREAGGAFGKREQAEEERYFRAQSREQLAALKKHHEEEIVHHKKEIERLQKEIERHKQKIKMLKHDD (human) | Natural regulatory peptide | 0.25 μMc (bovine heart MF1-ATPase) (143); 1.2 μMc at 21°C and 0.84 μM at 37°C (bovine heart MF1-ATPase) (446); 300 μg/mg proteinc (T. pyriformis SMP-ATPase) (404); 34 μg/mg proteinc (C. asciculate SMP-ATPase) (439); 0.24 μMd (rat liver MF1-ATPase) (229) |
| Melittin | GIGAVLKVLTTGLPALISWIKRKRQQ-NH2 | Apis mellifera (honey bee) | 5 μMc (bovine heart MF1-ATPase) (52); 12 μMc (bovine heart MF1-ATPase) (143) |
| WTf | MLSLRQSIRFFKPATRTLCSSRYLL-NH2 | Subunit IV of yeast cytochrome c oxidase | 16 μMc (bovine heart MF1-ATPase) (52) |
| Δ11,12 | MLSLRQSIRFPATRTLCSSRYLL-NH2 | Synthetic | 29 μMc (bovine heart MF1-ATPase) (52) |
| Syn-A2 | MLSRLSLRLLSRLSLRLLSRYLL-NH2 | Synthetic | 42 nMc (bovine heart MF1-ATPase) (52); 290 nMc (bovine heart MF1-ATPase) (143); 1.7 μMc (Bacillus PS3 F1-ATPase) (143) |
| Syn-C | MLSSLLRLRSLSLLRLRLSRYLL-NH2 | Synthetic | 58 nMc (bovine heart MF1-ATPase) (52); 160 nM (bovine heart MF1-ATPase) (143); 1.6 μMc (Bacillus PS3 F1-ATPase) (143) |
Where a species is indicated, sequences vary with species.
CF1 without ɛ subunit.
I50.
Ki.
Kd.
Leader sequence of subunit IV of yeast cytochrome c oxidase.
FIG. 2.
Structures of peptide inhibitors. (A) α-Helical basic peptide inhibitors. The coordinates of the inhibitors are 1BSN for the bacterial/chloroplast ɛ subunit, 1GMJ for IF1, and 2MLT for melittin. (B) Angiostatin and enterostatin. The coordinate for the structure is 1KI0. (C) Tentoxin and tentoxin analogs. (D) Leucinostatins and efrapeptins.
The bacterial/chloroplast ɛ subunit, composed of ∼120 to 140 amino acid residues, is an endogenous inhibitory subunit in F1, and inhibits ATPase activities of isolated and membrane-bound bacterial F1 (BF1) and chloroplast F1 (CF1) (198, 284, 332, 372, 386). The inhibition is reversible and noncompetitive with substrates (372, 386). It has no inhibitory effect on ATP synthesis and is required in the chloroplast ATP synthase for ATP synthesis in the light (289, 389, 402). The inhibition of F1-ATPase by the ɛ subunit is controlled by the electrochemical gradient and ADP/ATP balance (389), and the C-terminal α-helical domain is responsible for its inhibitory activity (168, 212, 289). At high proton motive forces and low ATP concentrations, the C-terminal α-helical domain of the ɛ subunit performs large conformational changes from the hairpin conformation to a “lifted-up” extended conformation, shifting its position ∼70 Å to interact with the α3β3 hexagon ring (389, 402). In the “lifted-up” extended conformation, the C-terminal helix lies close to the β-DELSEED motif of the β subunit, and the direct electrostatic interaction between the β-DELSEED motif and the basic residues in the C-terminal domain of the ɛ subunit leads to the inhibition of ATP hydrolysis (168).
IF1 is a natural regulatory peptide of 56 to 87 residues found in mitochondria (Fig. 2A). It binds to F1 with a 1:1 stoichiometric ratio and inhibits the ATP hydrolysis of mitochondrial ATP synthase without affecting ATP synthesis. The inhibition is reversible and noncompetitive, and the binding of IF1 to F1 requires the presence of ATP (178, 228, 229, 409). IF1 is more potent against the whole membrane-bound ATP synthase (F0F1-ATPase) complex than isolated F1 (144, 409, 411). IF1 inhibits the ATPase activity of mitochondrial ATP synthase and has no ATPase inhibitory effect against BF1 (143). The yeast IF1 can cross-react with animal F1, whereas the potato IF1 shows no inhibitory effect against animal F1 (60, 319). IF1 proteins from animals are considerably (18 to 31 residues) longer than those from plants and fungi (176). In a study of truncated bovine IF1 for inhibitory activity, the minimal inhibitory sequence was shown to localize within residues 14 to 47 (411). The adjoining residues 10 to 13 and 48 to 56 are considered to play a stabilizing role. In the crystal structure of F1 with IF1, the N-terminal domain of IF1 is bound at the interface between αDP and βDP subunits and also has contacts with βTP386, αE355, and the γ subunit (61). It has been suggested that the inhibitory mode of action of IF1 could be similar to that of the bacterial ɛ subunit (260, 402). IF1 is considered to play its inhibitory role by impeding the closure of the αDP-βDP catalytic interface to prevent the hydrolysis of bound ATP (61, 141). Cross-linking and intrinsic phosphorescence decay studies implicate IF1 as being functionally associated with the mitochondrial ɛ subunit (260, 373). Both proteins are in close proximity in the crystal structure of the F1-IF1 complex (141).
Melittin, which is a 26-residue peptide known as the principal active component of bee venom and which has a powerful anti-inflammatory effect, inhibits the ATPase activity of F1 (52, 143). The 25-residue presequence of yeast cytochrome oxidase subunit IV (WT) and its synthetic derivatives, Syn-A2, Syn-C, and Δ11,12, also inhibit ATP hydrolysis by F1 (52, 143). Melittin, WT, Syn-A2, and Syn-C (and possibly Δ11,12) form basic and amphiphilic α-helical structures (191, 337, 338, 393). Melittin, Syn-A2, and Syn-C have been suggested to bind to F1 at the same site as IF1 (143), and WT and Δ11,12, which are derivatives of Syn-A2 and Syn-C, are considered to also play similar inhibitory roles. Syn-A2 and Syn-C are very effective inhibitors among amphiphilic peptide inhibitors, showing 50% inhibitory (I50) values of about 40 to 50 nM for inhibition of bovine F1-ATPase activity (52). Syn-A2 inhibits the ATPase activity of bovine F1 noncompetitively in a parabolic manner, whereas Syn-C exhibits mixed inhibition and melittin shows noncompetitive hyperbolic inhibition (52).
Angiostatin and Enterostatin
Angiostatin is a 57-kDa N-terminal fragment of a larger protein, plasmin, which is also a fragment of plasminogen. Angiostatin has a triangular structure with three to five contiguous kringle domains, and it acts as a natural angiogenesis inhibitor (Fig. 2B) (1). It binds to the α and β subunits of ATP synthase and inhibits its ATP hydrolysis (269, 270). In an experiment with bovine F1 and human angiostatin, the angiostatin bound strongly to F1 and completely inhibited ATPase activity (269). Angiostatin was also found to inhibit ATP generation by the nonmitochondrial ATP synthase located on endothelial cells that comprise the human umbilical vein, with 1 μM angiostatin inhibiting about 81% of the ATP synthesis activity (270). However, no ATP synthesis by plasma membrane ATP synthase was reported in human vascular endothelial cells (325), and the inhibition of ATP synthesis of nonmitochondrial ATP synthase by ATP synthase-specific inhibitors is still controversial.
Enterostatin is a pentapeptide released from procolipase during dietary fat digestion (Fig. 2B). Enterostatin binds to the ATP synthase β subunit and inhibits ATP synthesis (38, 39, 301). Binding of enterostatin to the mitochondrial ATP synthase in insulinoma cells leads to an ∼31% decrease of ATP production accompanied by an increase in thermogenesis and oxygen consumption (38). The binding of enterostatin to F1 is inhibited by β-casomorphin, a peptide derived from the digestion of β-casein in milk (38, 39, 301).
Tentoxin and Its Derivatives
The properties and inhibitory potencies of tentoxin and its analogs are summarized in Table 2. Tentoxin is a natural cyclic tetrapeptide produced by phytopathogenic fungi, Alternaria species (19, 257, 342). In aqueous solution, tentoxin exists as four interconverting conformations in different proportions (51, 37, 8, and 4%) resulting from a “conformational peptide flip” (318). At low concentrations, tentoxin acts as an uncompetitive inhibitor of the ATPase activity of CF1 derived from certain sensitive plant species but not of homologous CF1s from chloroplasts of some other plant species. Also, tentoxin does not inhibit the ATPase activity of F1s derived from bacteria or mitochondria (19, 378, 380). Tentoxin also inhibits ATP synthesis in chloroplasts from the sensitive species. In contrast to the above, tentoxin at high concentrations strongly stimulates ATPase activity of CF1 (379) and partially reactivates the proton transport-coupled activity of the membrane-bound CF0F1 (369). Based on labeling studies, tentoxin-susceptible CF1 is considered to contain a high-affinity inhibitory binding site and one or two low-affinity stimulatory binding sites (69, 265, 317, 350). The binding of tentoxin to a low-affinity binding site releases the inhibitory effect caused by binding of tentoxin to the high-affinity binding site and reactivates the enzyme. The binding of a tentoxin molecule to the third site with very low affinity results in overactivation (265). In the crystal structure of the CF1-tentoxin complex, a tentoxin molecule is bound at the high-affinity binding site located in a cleft at an αβ subunit interface. Here, it blocks the contact between αArg-297 and βAsp-83 (153, 155), restrains the movements of these residues, and also restrains conformational changes at the catalytic interface. This may arrest the catalytic αβ interface in the closed conformation and thereby hinder its transformation into the open conformation (153, 155).
TABLE 2.
Tentoxin and tentoxin analogs
| Name or abbreviation | Sequence | Molecular formula | Inhibitory potency (reference) |
|---|---|---|---|
| Tentoxin | Cyclo-(l-N-methyl-Ala1-l-Leu2-N-methyl-ΔZPhe3-Gly4) | C22H30N4O4 | ∼0.6 mol/mola (spinach CF1-ATPase) (179); 50 nMa (spinach CF1(-ɛ)-ATPase) (69); 0.4-0.6 μMa (lettuce chloroplasts, photophosphorylation) (380); 10 nMb (spinach CF1(-ɛ)-ATPase) (350); 30-60 μMb (60°C, TF1-ATPase) (351); 8-10 nMc (spinach CF1(-ɛ)-ATPase) (350, 351) |
| MeSer1-TTX | Cyclo-(l-N-methyl-Ser1-l-Leu2-N-methyl-ΔZPhe3-Gly4) | C22H30N4O5 | 50 nMa (spinach CF1(-ɛ)-ATPase) (69); 0.5 μMa with 2 min incubation and 0.1 μMa with 30 min incubation in the dark (spinach thylakoids, ATP synthesis) (316); 15 nMc (spinach CF1(-ɛ)-ATPase) (351) |
| Ala1-TTX | Cyclo-(l-Ala1-l-Leu2-N-methyl-ΔZPhe3-Gly4) | C21H28N4O4 | 34 nMc (spinach CF1(-ɛ)-ATPase) (351) |
| Sar1-TTX | Cyclo-(l-N-methyl-Gly1-l-Leu2-N-methyl-ΔZPhe3-Gly4) | C21H28N4O4 | 45 nMc (spinach CF1(-ɛ)-ATPase) (351) |
| Gly1-TTX | Cyclo-(l-Gly1-l-Leu2-N-methyl-ΔZPhe3-Gly4) | C20H26N4O4 | 34 nMc (spinach CF1(-ɛ)-ATPase) (351) |
| MeSer(Bn)1-TTX | Cyclo-(l-N-methyl-Ser(Bn)1-l-Leu2-N-methyl-ΔZPhe3-Gly4) | C29H36N4O5 | 0.5 μMa (spinach CF1(-ɛ)-ATPase) (69); 0.5 μMc (spinach CF1(-ɛ)-ATPase) (351) |
| MeGlu1-TTX | Cyclo-(l-N-methyl-Glu1-l-Leu2-N-methyl-ΔZPhe3-Gly4) | C24H32N4O6 | 5 μMa (spinach CF1(-ɛ)-ATPase) (69) |
| MeGlu(tBu)1-TTX | Cyclo-(l-N-methyl-Glu(tBu)1-l-Leu2-N-methyl-ΔZPhe3-Gly4) | C28H41N4O6 | 2 μMa (spinach CF1(-ɛ)-ATPase) (69); 1.5 μMc (spinach CF1(-ɛ)-ATPase) (351) |
| Lys2-TTX | Cyclo-(l-N-methyl-Ala1-l-Lys2-N-methyl-ΔZPhe3-Gly4) | C22H31N5O4 | 3 μMa (spinach CF1(-ɛ)-ATPase); 2 μMc (spinach CF1(-ɛ)-ATPase) (351) |
| Lys(Z)2-TTX | Cyclo-(l-N-methyl-Ala1-l-Lys(Z)2-N-methyl-ΔZPhe3-Gly4) | C30H37N5O6 | 1 μMa (spinach CF1(-ɛ)-ATPase) (69); 0.75 μMc (spinach CF1(-ɛ)-ATPase) (351) |
| MeΔTyr3-TTX | Cyclo-(l-N-methyl-Ala1-l-Leu2-N-methyl-ΔZTyr3-Gly4) | C22H30N4O5 | 0.05 μMa (spinach CF1(-ɛ)-ATPase) (69); 12 nMc (spinach CF1(-ɛ)-ATPase) (351) |
| Tyr(Me)3-TTX | Cyclo-(l-N-methyl-Ala1-l-Leu2-N-methyl-ΔZTyr(Me)3-Gly4) | C23H32N4O5 | 0.05 μMa (spinach CF1(-ɛ)-ATPase) (69); 10 nMc (spinach CF1(-ɛ)-ATPase) (351) |
| ΔPhe3-TTX | Cyclo-(l-N-methyl-Ala1-l-Leu2-ΔZPhe3-Gly4) | C21H28N4O4 | 0.8 μMc (spinach CF1(-ɛ)-ATPase) (351) |
| Dihydro-TTX | Cyclo-(l-N-methyl-Ala1-l-Leu2-N-methyl-Phe3-Gly4) | C22H32N4O4 | 0.5 μMc (spinach CF1(-ɛ)-ATPase) (351) |
| Iso3-TTX | Cyclo-(l-N-methyl-Ala1-l-Leu2-N-methyl-ΔEPhe3-Gly4) | C22H30N4O4 | 8.7 μMc (spinach CF1(-ɛ)-ATPase) (351) |
I50.
Ki.
Kd.
MeSer1-TTX, Ala1-TTX, Sar1-TTX, Gly1-TTX, MeSer(Bn)1-TTX, MeGlu1-TTX, MeGlu(tBu)1-TTX, Lys2-TTX, Lys(Z)2-TTX, MeΔTyr3-TTX, MeΔTyr(Me)3-TTX, ΔPhe3-TTX, dihydro-TTX, and Iso3-TTX are synthetic analogs of tentoxin in which an amino acid residue is mutated at the residue number indicated (316, 351) (Fig. 2C). MeSer1-TTX appears to inhibit isolated CF1 and the membrane-bound enzyme (CF0CF1) in thylakoids and proteoliposomes the same way and with the same efficiency as tentoxin. However, MeSer1-TTX exhibits much weaker reactivation of CF1 than tentoxin at high concentrations (69). On the other hand, MeΔTyr(Me)3-TTX shows similar activities as tentoxin in both inhibitory and stimulatory potencies (69). MeSer(Bn)1-TTX, MeGlu1-TTX, Glu(tBu)1-TTX, Lys2-TTX, and MeSer1-TTX analogs exhibit inhibitory activities with lower affinities but show no stimulatory effects (69).
Leucinostatins and Efrapeptins
The leucinostatins (A to D, H, and K) are nonapeptide antibiotics produced by Paecilomyces (Fig. 2D and Table 3). Leucinostatin A is produced by Paecilomyces lilacinus, P. marquandii, and P. abruptus (434), leucinostatin B by P. lilacinus, and P. marquandii (266), leucinostatin C by P. lilacinus (259), leucinostatin D by P. lilacinus and P. marquandii (259, 339), and leucinostatin H and K by P. marquandii (259, 339). Leucinostatins adopt an α-helical conformation, and contains three Aib residues and some uncommon amino acid residues (71). Different types of leucinostatin differ in the kinds of amino acid at position 2 (Dec or Leu) and in the substitution pattern at the terminal nitrogen atom [-N(CH3)2, -NHCH3, -NH2, or -NO(CH3)2]. Leucinostatins bind to the F0 part of ATP synthases (127, 404, 439) and inhibit oxidative phosphorylation in mitochondria and photophosphorylation in chloroplasts (224, 242, 328). Leucinostatins have no inhibitory activity on isolated F1-ATPase (127, 439).
TABLE 3.
Leucinostatins and efrapeptins
| Name | Molecular formula | Source | Synonyms | Inhibitory potency (reference) |
|---|---|---|---|---|
| Leucinostatin | A, C62H111N11O13; B, C61H109N11O13; C, C60H107N11O13; D, C56H101N11O11; H, C57H103N11O12; K, C62H111N11O14 | A, P. lilacinus, P. marquandii, and P. abruptus; B, P. lilacinus and P. marquandii; C, P. lilacinus; D, P. lilacinus and P. marquandii; H and K, P. marquandii | A, A20668, paecilotoxin A, CC-1014; B, paecilotoxin B; C, paecilotoxin C; D, paecilotoxin D; H, paecilotoxin H; K, paecilotoxin K | 11 μg/mg proteina (Crithidia asciculate SMP-ATPase) (439); 2 μg inhibitor/mla (spinach chloroplast, photophosphorylation) (242); 0.1-0.4 μg/mg protein (rat liver mitochondria, ATPase) (328) |
| Efrapeptin | C, C80H137N18O16+; D, C81H139N18O16+; E, C82H141N18O16+; F, C82H141N18O16+; G, C83H143N18O16+ | Tolypocladium species | Efrastatin, A23871 | 0.56 mol/mol F1a (bovine heart MF1-ATPase) (83); 70 ng/mla (C. asciculate MF1-ATPase) (173); 0.3 μMa (human umbilical vein endothelial cell, nonmitochondrial ATP synthase, ATP synthesis) (17); 0.5 μg/mla (R. rubrum chromatophores, photophosphorylation) (241); 0.05-0.5 μg of inhibitor/mg proteina (T. pyriformis SMP-ATPase) (404); 21.5 μMb (EF1-ATPase) (436); 10 nMc (bovine heart MF1-ATPase) (83); complete inhibition at 2.4 mol inhibitor/mol enzyme (bovine heart SMP-ATPase and ATP synthesis) (83) |
I50.
Ki.
Kd.
Efrapeptins are a group of lipophilic peptide antibiotics (efrapeptins C to G) produced by Tolypocladium species (Fig. 2D and Table 3). Efrapeptin inhibits both ATP hydrolysis and ATP synthesis reactions of the ATP synthase from mitochondria, chloroplasts, and photosynthetic bacteria by binding at the F1 catalytic domain (2, 164, 173, 224, 232, 241, 242). Efrapeptin inhibits the ATP synthase from some, but not all, nonphotosynthetic bacteria, including thermophilic Bacillus strain PS3 (343, 436). The mode of inhibition by efrapeptin during ATP synthesis is competitive with ADP and phosphate (83). Efrapeptin also binds to the nonmitochondrial ATP synthase of endothelial cells and inhibits extracellular ATP synthesis (17). In the crystal structure of the F1-ATPase-efrapeptin complex, a single efrapeptin molecule is bound in the large central cavity of F1 lined with βE, αE, αTP, and the α-helical structure of the γ subunit. The binding of efrapeptin is stabilized predominantly by hydrophobic interactions between efrapeptin and the residues in the cavity and also by two potential intermolecular hydrogen bonds (2). Efrapeptin is believed to inhibit the ATP synthase by preventing the βE subunit from converting into a nucleotide binding conformation.
POLYPHENOLIC PHYTOCHEMICALS, ESTROGENS, AND STRUCTURALLY RELATED COMPOUNDS
Phytochemicals are naturally occurring bioactive nonnutrient compounds derived from plants. They possess chemopreventive or chemotherapeutic effects associated with reduced risk of various diseases, including cancer, and they bind to multiple molecular targets in the body (30, 286, 395). Phytochemicals are categorized into various groups, and among these are the polyphenolic phytochemicals. Some of the polyphenolic phytochemicals, many of which are phytoestrogens, bind to the ATP synthase and inhibit its ATPase activity. (Fig. 3) (143, 448, 449). The effects of polyphenolic phytochemicals on the ATPase activity of ATP synthase are additive, and the phenolic structures that comprise the polyphenolic phytochemicals play an important role in their inhibitory potencies (448). Two or more phenolic structures appear to be required, and the position of hydroxy groups seems to affect significantly the inhibitory effectiveness of polyphenolic phytochemicals on the ATP synthase (448).
FIG. 3.
Structures of polyphenolic phytochemicals, estrogens, and structurally related compounds. (A) Stilbenes. SITS, 4-Acetamido-4′-isothiocyanostilbene 2,2′-disulfonate; DIDS, 4,4′-diisothiocyanatostilbene-2,2′-disulfonic acid. (B) Flavones and isoflavones. (C) Other polyphenolic phytochemicals. ECG, epicatechin gallate; EGCG, epigallocatechin gallate. (D) Steroidal estradiols and estrogen metabolites.
Some endogenous and synthetic estrogens also target ATP synthase. Endogenous steroidal estradiols and estrogen metabolites and synthetic nonsteroidal stilbene estrogens bind to mitochondrial ATP synthase and inhibit its ATPase activity (450, 451).
Stilbenes
Stilbenes consist of two phenolic rings linked by a spacer containing a double bond (Fig. 3A). Stilbene phytoalexins, resveratrol, and piceatannol are natural phytochemicals found in grapevine organs such as berries, leaves, canes, and roots. They inhibit the ATPase activity of mitochondrial ATP synthase by targeting the F1 catalytic headpiece (Table 4) (325, 448, 449). The mode of inhibition by resveratrol is mixed (448). In contrast to the above, resveratrol and piceatannol show no inhibition of ATPase activity of F1 from thermophilic Bacillus strain PS3 (TF1) (143). Resveratrol and piceatannol bind to a hydrophobic pocket between the hydrophobic tip in the C-terminal region of the γ subunit and the hydrophobic inside of an annulus provided by the βTP subunit (142). The binding of these inhibitors, stabilized by hydrophobic interactions and hydrogen bonds, is believed to block the rotation of the γ subunit, inhibiting both the hydrolysis and synthesis of ATP. Resveratrol and piceatannol are bound to a single binding site in F1, and there are no equivalent sites between the γ subunit and either the βDP or βE subunit.
TABLE 4.
Stilbenes
| Name or abbreviation | Molecular formula | Source | Other names | Inhibitory potency, I50 (reference) |
|---|---|---|---|---|
| Resveratrol | C14H12O3 | Grapes and red wine | 3,4′,5-Stilbenetriol; 3,4′,5-trihydroxystilbene | 27.7 μM (rat brain SMP, ATP synthesis) (448); 14 μM (rat liver MF1-ATPase) (449); 19 μM (rat brain M F0F1-ATPase) (448); 6.4 μM (bovine heart MF1-ATPase) (143); 2 μM (human umbilical vein endothelial cell, nonmitochondrial ATP synthase, ATP synthesis) (17) |
| Piceatannol | C14H12O4 | Seeds of Euphorbia lagascae | 3,5,3′,4′-Tetrahydroxystilbene; 3-hydroxyresveratol | 8-9 μM (rat brain MF0F1 ATPase) (448, 449); 4 μM (rat liver MF1-ATPase) (449); 6.1 μM (bovine heart MF1-ATPase) (143); 1.5 μM (human umbilical vein endothelial cell, nonmitochondrial ATP synthase, ATP synthesis) (143); ∼70% inhibition at 10 μM (bovine heart MF1-ATPase) (325) |
| DES | C18H20O2 | Synthetic | Diethylstilbestrol; (E)-4,4′-(1,2-diethyl-1,2-ethenediyl)bisphenol; 4,4′-dihydroxydiethylstilbene; (E)-3,4-bis(4-hydroxyphenyl)-3-ascic; Acnestrol; Antigestil; Comestrol; Cyren; Desma; Dibestrol; Distilbene; Estrobene; Pabestrol; Stilbetin; Vagestrol | 10 μM (rat liver MF0F1-ATPase) (252); 10-25 μM (rat brain MF0F1-ATPase) (451) |
| SITS | C17H14N2O7S3 | Synthetic | 4-Acetamido-4′-isothiocyanostilbene 2,2′-disulfonate | ∼1.3 μM (V. parahaemolyticus F0F1-ATPase) (290); 95% inhibition at 25 μM (V. parahaemolyticus F1-ATPase) (344) |
| DIDS | C16H10N2O6S4 | Synthetic | 4, 4′-d-Isothiocyanatostilbene-2,2′-disulfonic acid; diisothiocyanatostilbene-2,2-disulfonic acid | 20.9 μM (rat liver MF1ATPase) (40) |
Diethylstilbestrol (DES) is a synthetic nonsteroidal estrogen. DES targets F0 and inhibits both ATPase and ATP-dependent proton translocation activities of both membrane-bound and isolated F0F1 from mitochondria (252, 451). DES inhibits membrane-bound F0F1 with half-maximal and maximal inhibitory effects at about 10 and 60 μM, respectively (252). For the isolated F0F1, the concentration for 50% inhibition is 10 μM, and maximal inhibition of ATPase activity is about 90%. In contrast, DES has little effect on the ATPase activity of the F1 moiety, exhibiting only ∼20% inhibition at 60 μM. The binding site of DES is considered to be structurally distinct from other types of F0 inhibitors, as DES provides no protection against the inhibition of the F0F1 complex by N,N′-dicyclohexylcarbodiimide (DCCD), which is protected by oligomycin, venturicidin, and tricyclohexyltin. The combination of DES and DCCD produces a synergic inhibitory effect at low concentrations (<20 μM).
4-Acetamido-4′-isothiocyanostilbene 2,2′-disulfonate and 4,4′-di-isothiocyanatostilbene-2,2′-disulfonic acid are structurally very analogous and have been known as anion exchanger inhibitors. They also bind to ATP synthase and inhibit its catalytic activity. 4-Acetamido-4′-isothiocyanostilbene 2,2′-disulfonate strongly inhibits the ATPase activity of both F1 and F0F1 from Vibrio parahaemolyticus (290, 344). 4,4′-Di-isothiocyanatostilbene-2,2′-disulfonic acid also inhibits both the hydrolysis and synthesis of ATP in submitochondrial particles (SMP) and also ATP hydrolysis of isolated F1 from rat liver mitochondria (40).
Flavones and Isoflavones
Flavones and isoflavones are flavonoid-related polyphenolic compounds. Flavones and isoflavones differ in the position of a phenyl group on the 4H-1-benzopyr-4-one skeleton. Flavones are produced in various plants, whereas isoflavones are produced almost exclusively by beans. The flavones, quercetin, kaempferol, morin, and apigenin inhibit ATP hydrolysis (Fig. 3B). Specifically, quercetin inhibits the ATPase activities of mitochondrial F1 (MF1) and F0F1 (223, 448, 449) and also these activities in spinach chloroplasts (96), Escherichia coli (130), and Clostridium thermoaceticum (190). However, quercetin inhibits neither the ATPase activity of TF1 (343), a thermophilic bacterial ATP synthase, nor the ATP synthetic activity of mitochondrial ATP synthase (F0F1) (223). In contrast, quercetin has a stimulatory effect on photophosphorylation (218). Kaempferol and morin have inhibitory potencies similar to that of quercetin on the ATPase activity of mitochondrial F0F1, while apigenin, in which the 3-hydroxyl group in the chromone moiety is absent, shows about half the inhibitory potency (Table 5) (448).
TABLE 5.
Flavones and isoflavones
| Name | Molecular formula | Source | Other names | Inhibitory potency (reference) |
|---|---|---|---|---|
| Quercetin | C15H10O7 | Various plants | 3,3′,4′,5,7-Pentahydroxyflavone; natural yellow 10; meletin; flavin meletin; quercetol; Xanthaurine | 5 kmol/mola (232), 85 μMa (343) (bovine heart MF1-ATPase); 180 μMa (bovine heart SMP-ATPase) (343); 50 μMa (rat brain F0F1-ATPase) (448); 3 μMa (rat liver F1-ATPase) (449); 2 kmol/mola (spinach CF1-ATPase) (232); 2.6 μg/mg proteina (C. asciculate SMP-ATPase) (439); 0.2 mMb (pig heart MF1-ATPase) (100); 27 μMc (bovine heart MF1-ATPase) (232); 46% inhibition at 5 μM (C. thermoaceticum membrane-bound F0F1-ATPase) (190) |
| Kaempferol | C15H10O6 | Delphinium, witch-hazel, grapefruit, and other plant sources | Kempferol; campherol; indigo yellow; nimbecetin; pelargidenolon; populnetin; rhamnolutein; 3,4′,5,7-tetrahydroxyflavone; trifolitin | 55 μMa (rat brain MF0F1-ATPase) (448) |
| Morin | C15H10O7 | Various plants | 2′,3,4′,5,7-Pentahydroxyflavone; 2′,4′,5,7-tetrahydroxyflavan-3-ol; 3,5,7,2′,4′-pentahydroxyflavonol; al-morin; aurantica; calico yellow; osage orange | 60 μMa (rat brain MF0F1-ATPase) (448) |
| Apigenin | C15H10O5 | Parsley, artichoke, basil, celery and other plants | 4′,5,7-Trihydroxyflavaone; 2-(p-hydroxyphenyl)-5,7-dihydroxychromone; apigenol; chamomile; spigenin | 105 μMa (rat brain MF0F1-ATPase (448) |
| Genistein | C15H10O5 | Soybean | 4′,5,7-Trihydroxyisoflavone; genisteol; genisterin; prunetol; sophoricol; differenol A | 55 μMa (rat brain MF0F1-ATPase) (448); 10% inhibition at 50 μM (rat liver F1-ATPase) (449) |
| Biochanin A | C16H12O5 | Soybean | Biochanin; 4′-methylgenistein; 5,7-dihydroxy-4′-methoxyisoflavone; CCRIS 5449; 5,7-dihydroxy-4′-methoxyisoflavone | 65 μMa (rat brain MF0F1-ATPase) (448) |
| Daidzein | C15H10O4 | Soybean | 4′,7-Dihydroxyisoflavone; daidzeol; 7-hydroxy-3-(4-hydroxyphenyl)-4-benzopyrone | 127 μMa (rat brain MF0F1-ATPase) (448) |
I50.
Ki.
Kd.
Genistein, biochanin A, and daidzein are isoflavone phytoalexins found in soybeans. Genistein inhibits noncompetitively both the ATP hydrolysis and ATP synthesis activities of mitochondrial ATP synthase, most likely by targeting F0 (448, 449). Biochanin A inhibits the ATPase activity of mitochondrial F0F1 with an inhibitory potency similar to that of genistein. Compared to genistein and biochanin, daidzein contains only one hydroxyl group in the 4-chromone moiety and shows about half the inhibitory potency (448).
Other Polyphenolic Phytochemicals
Catechins are flavonoid compounds called flavan 3-ols. They are abundant in green tea, which includes four main catechins, epicatechin, epicatechin gallate, epigallocatechin, and epigallocatechin gallate. Among the catechins, epicatechin gallate and epigallocatechin gallate are inhibitors of the ATP hydrolysis activity of ATP synthase (Fig. 3C) (448). Epigallocatechin gallate, in which one more hydroxyl group is attached in the catechol moiety of epicatechin gallate, shows about three times higher potency than epicatechin gallate in the inhibition of ATPase activity of mitochondrial F0F1.
Grape seed proanthocyanidin extract, curcumin, an active ingredient of the Indian curry spice, and phloretin from apples inhibit the ATPase activity of mitochondrial F0F1. Theaflavin, a phytochemical from tea, and tannic acid, anionic polymers from the bark of trees, also exhibit inhibitory effects on the ATPase activity of mitochondrial F0F1 (Table 6) (448).
TABLE 6.
Other polyphenolic phytochemicals
| Name or abbreviation | Molecular formula | Source | Other names | Inhibitory potency, I50 (reference) |
|---|---|---|---|---|
| ECG | C22H18O10 | Green tea | (−)Epicatechin gallate; epicatechin-3-gallate; epicatechin-3-galloyl ester | 45 μM (rat brain MF0F1-ATPase) (448) |
| EGCG | C22H18O11 | Green tea | (−)-Epigallocatechin gallate; (−)-epigallocatechin gallate; (−)-epigallocatechin-3-O-gallate; CCRIS 3729; tea catechin | 17 μM (rat brain MF0F1-ATPase) (448) |
| GSPE | C31H28O12 | Grape seed | Grape seed proanthocyanidin extract; polyhydroxyflavan-3-ol | 30 μg of inhibitor/ml (rat brain F0F1-ATPase) (448) |
| Curcumin | C21H20O6 | Curcuma longa | Natural yellow 3; 1,7-bis(4-ascicul-3-methoxyphenyl)-1,6-heptadiene-3,5-dione | 40 μM (rat brain MF0F1 ATPase) (448) |
| Phloretin | C15H14O5 | Mainly from apples | Phloretol; 2′,4′,6′-trihydroxy-3-(p-hydroxyphenyl)propiophenone; dihydronaringenin; β-(p-hydroxyphenyl)-2,4,6-trihydroxypropiophenone | 40% inhibition at 70 μM (rat brain MF0F1-ATPase) (448) |
| Theaflavin | C29H24O12 | Tea | 1,8-Bis((2R,3R)-3,5,7-trihydroxy-2H-1-benzopyran-2-yl)-3,4,6-trihydroxy-5H-benzocyclohepten-5-one | 20 μg of inhibitor/ml (rat brain F0F1-ATPase) (448) |
| Tannic acid | A mixture of related compounds (mainly glucose esters of gallic acid) | Bark of trees | Gallotannic acid; gallotannin; glycerite; tannin | 5 μg of inhibitor/ml (rat brain F0F1-ATPase) (448) |
Steroidal Estradiols and Estrogen Metabolites
Endogenous steroidal estradiols and estrogen metabolites have inhibitory effects on mitochondrial ATP synthase (Fig. 3D and Table 7) (451). Two catecholestrogens, 4-hydroxyestradiol and 2-hydroxyestradiol, inhibit the ATPase activity of the mitochondrial ATP synthase, and the 4-hydroxyestradiol is about twofold more effective than the 2-hydroxyestradiol. 17β-Estradiol and 17α-estradiol inhibit the ATPase activity of solubilized brain mitochondrial fractions by 7 and 25% at 14 and 42 μM, respectively. Two micoestrogens, α-zearalenol and β-zearalenol, also inhibit mitochondrial F0F1-ATPase activity. The I50 value of α-zearalenol is about 50 μM, and the inhibitory potency of α-zearalenol is about three- to fourfold stronger than that of β-zearalenol. The mechanism of inhibition by the steroidal estradiols and estrogen metabolites is not defined clearly, but the ATP synthase OSCP subunit has been identified as an estradiol binding protein, and it has been suggested that the inhibition is mediated by the binding of estrogens to OSCP (450).
TABLE 7.
Steroidal estradiols and estrogen metabolites
| Name | Molecular formula | Source | Other names | Inhibitory potency, I50 (reference) |
|---|---|---|---|---|
| 4-Hydroxyestradiol | C18H24O3 | Natural estrogen | 4-Hydroxyestradiol-17β; 4-hydroxy-17-β−estradiol; estra-1,3,5(10)-triene-3,4,17-β-triol | 55 μM (rat brain MF0F1-ATPase) (451) |
| 2-Hydroxyestradiol | C18H24O3 | Natural estrogen | (17β)-Estra-1,3,5(10)-triene-2,3,17-triol; estra-1,3,5(10)-triene-2,3,17-β-triol | 110 μM (rat brain MF0F1-ATPase) (451) |
| 17-α-Estradiol | C18H24O2 | Natural estrogen | 1,3,5-Estratriene-3,17-α-diol; 3,17-dihydroxyestratriene; 3,17-α-dihydroxyoestra-1,3,5(10)-triene; epiestradial; epiestradiol; estra-1,3,5(10)-triene-3,17α-diol; oestra-1,3,5(10)-triene-3,17α-diol; estradiol-17-α; α-estradiol | 25% inhibition at 42 μM (rat brain MF0F1-ATPase) (451) |
| 17-β-Estradiol | C18H24O2 | Natural estrogen | 1,3,5-Estratriene-3,17-β-diol; 17-β-estra-1,3,5(10)-triene-3,17-diol; 17-β-OH-estradiol; 17-β-OH-estradiol; 17-β-oestra-1,3,5(10)-triene-3,17-diol; 17β-oestra-1,3,5(10)-triene-3,17-diol; 3,17-epidihydroxyestratriene; 3,17-epidihydroxyoestratriene; 3,17-β-dihydroxy-1,3,5(10)-oestratriene; 3,17-β-estradiol; 3,17-β−estradiol; Aerodiol; Aquadiol | 7% inhibition at 14 μM (rat brain MF0F1-ATPase) (451) |
| α-Zearalenol | C18H24O5 | Natural mycoestrogen | (4S,8R,12E)-8,16,18-Trihydroxy-4-methyl-3-oxabicyclo[12.4.0]octadeca-12,15,17,19-tetraen-2-one; trans-zearalenol | 50 μM (rat brain MF0F1-ATPase) (451) |
| β-Zearalanol | C18H24O5 | Natural mycoestrogen | (8S,12E)-8,16,18-Trihydroxy-4-methyl-3-oxabicyclo[12.4.0]octadeca-12,15,17,19-tetraen-2-one | 150-200 μM (rat brain MF0F1-ATPase) (451) |
POLYKETIDE INHIBITORS
Polyketides are polymers of two-carbon ketide units synthesized by polyketide synthases. Macrolides belong to the polyketide class and contain a macrolide ring, a large lactone ring to which one or more deoxy sugars, usually cladinose and desosamine, are attached (Fig. 4). Some natural macrolides, apoptolidin, cytovaricin, oligomycin, ossamycin, and venturicidin are elaborated by Nocardiopsis spp. and various strains of Streptomyces and are known as potent inhibitors of ATP synthase (Table 8) (205, 207, 225, 330, 358, 359). The binding sites of the macrolide inhibitors are located within the F0 part of the complex.
FIG. 4.
Structures of polyketide inhibitors.
TABLE 8.
Polyketide inhibitors
| Name | Molecular formula | Source | Other names | Inhibitory potency (reference) |
|---|---|---|---|---|
| Oligomycin | A, C45H74O11; B, C45H72O12; C, C45H74O10; D, C44H72O11; E, C45H72O13; F, C46H76O11 | A, B, and C, Streptomyces diastratochroogenes; D, Streptomyces griseus, Streptomyces aureofaciens, Streptomyces rutgersensis | D, Rutamycin, 26-demethyl-oligomycin A, A272 | 152 μg inhibitor/mg proteina (E. coli membrane vesicle, pH gradient formation) (311); 7.1 μg inhibitor/mg proteina (C. asciculate SMP-ATPase) (439); 2.0-3.0 μg inhibitor/mg proteina (S. cerevisiae SMP-ATPase) (150, 151); A, 0.3 μMa (human NCI-60 cell lines, F0F1-ATPase) (348); 15 ng inhibitor/mg proteinb (N. crassa SMP-ATPase) (112); 0.21 μMb (bovine heart MF0F1-ATPase) (85); 95% inhibition at 0.4 μg inhibitor/mg protein (bovine heart SMP-ATPase) (140); D, 75% inhibition at 0.5 μg/ml (rat liver SMP-ATPase) (423) |
| Peliomycin | C46H76O14 | Various strains of Streptomyces | 4.5 μg inhibitor/mg proteina (S. cerevisiae SMP-ATPase) (150) | |
| Venturicidin | A, C41H57NO11; B, C40H64NO10; X, C34H54O7 | Streptomyces aureofaciens, Streptomyces griseolus, Streptomyces halstedii, Streptomyces xanthophaeus, Streptomyces hygroscopicus | X, botrycidin | 9 μg inhibitor/mg proteina (E. coli pH gradient formation by membrane vesicle) (311); 11 μg inhibitor/mg proteina (E. coli membrane-bound ATPase) (311); 0.13 μg inhibitor/mg proteina (150); 0.06-0.18a (A and B) and 11.0a (X) μg inhibitor/mg protein (S. cerevisiae SMP-ATPase) (151); 5-11 μg inhibitor/mg proteina (T. pyriformis) (404); 3.0 μg/mg proteina (C. asciculate SMP-ATPase) (439); 0.5 μMa (spinach thylakoids, photophosphorylation) (447); 0.5 μMa (spinach thylakoids, ATPase) (447)a |
| Ossamycin | C50H87NO14 | S. hygroscopicus subsp. ossamyceticus | 1.3 μg of inhibitor/mg proteina (S. cerevisiae SMP-ATPase) (150); 46 μg of inhibitor/mg proteina (E. coli pH gradient formation by membrane vesicle) (311); 8 μMa (human NCI-60 cell lines, F0F1-ATPase) (348) | |
| Apoptolidin | C58H96O21 | Nocardiopsis sp. | 4-5 μMb (S. cerevisiae membrane-bound F0F1-ATPase) (349); 18 μMa (human NCI-60 cell lines, F0F1-ATPase) (348) | |
| Cytovaricin | C48H82O15 | Streptomyces sp. strain H-230 | H-230 | 1 μMa (human NCI-60 cell lines, F0F1-ATPase) (348); 0.4 μMb (S. cerevisiae membrane-bound F0F1-ATPase) (349) |
I50.
Ki.
Oligomycins are a closely related group of 26-membered macrolides with both lactone moieties and double bonds. Oligomycins are produced in various strains of Streptomyces. They include six different types, A, B, C, D, E, and F, based on the R groups attached to the macrolide ring and sugar. Oligomycin D is also named rutamycin. Other specific oligomycins include peliomycin and botrycidin; the latter is known also as venturicidin X. Oligomycin inhibits ATP synthases from mitochondria and the chromatophores of photosynthetic bacteria (85, 150, 151, 253, 311, 347, 360). However, it has no or only a weak effect on photophosphorylation activity in chloroplasts and on membrane-bound ATPase activity of nonphotosynthetic bacteria (22, 36, 118, 285, 311, 376). Mutagenesis studies that cause resistance to oligomycin in yeast implicate a target site residing at the interface of subunits a and c, with an involvement of both Gly23 and Glu59 of the N- and C-terminal transmembrane helices of subunit c, respectively (97, 192, 280). Yeast Glu59 of subunit c is equivalent to E. coli Asp61, located in the middle of the membrane, and is believed to be involved in proton translocation that drives ATP synthesis.
Peliomycin, produced from various strains of Streptomyces (323, 358), is cytotoxic to mammalian cells, with limited antimicrobial and antifungal activities. The inhibitory properties of peliomycin on ATP synthesis by oxidative phosphorylation in mitochondria mimic those of rutamycin (423).
Venturicidin consists of three different types, A, B, and X, where venturicidin X is an aglycone of venturicidin A or B (401). It binds to subunit c of the ATP synthase and inhibits both proton translocation and membrane-bound ATPase activities from bacteria, chloroplasts, and mitochondria (62, 251, 311, 423, 447). The region conferring venturicidin resistance or hypersensitivity in ATP synthase is located in the middle of the membrane, and most of this region overlaps with that for oligomycin resistance (123, 131, 280).
Ossamycin is a 24-membered macrolide produced in Streptomyces hygroscopicus subsp. ossamyceticus (209, 359). Ossamycin inhibits both the ATPase and oxidative phosphorylation activities of mitochondrial ATP synthase (150, 423). It has no direct effect on E. coli F1 (EF1) or F0, but it does inhibit ATP-driven proton transport by uncoupling ATP hydrolysis from proton transport (311). The binding site of ossamycin in mitochondrial ATP synthase lies close to the boundaries of regions that cause oligomycin and venturicidin resistance in subunit c. This site contains residues Leu53 to Leu57 (yeast sequence) in the C-terminal transmembrane helix (131).
Apoptolidin and cytovaricin are 20- and 26-membered macrolides found in Nocardiopsis spp. and Streptomyces sp. strain H-230, respectively. Both apoptolidin and cytovaricin inhibit membrane-bound mitochondrial ATP synthase. The precise binding sites of apoptolidin and cytovaricin are not yet defined. However, they are believed to be located at regions where oligomycin and ossamycin bind, as the chemical backbones of these inhibitors are structurally similar to those of oligomycin and ossamycin (349).
ORGANOTIN COMPOUNDS AND STRUCTURAL RELATIVES
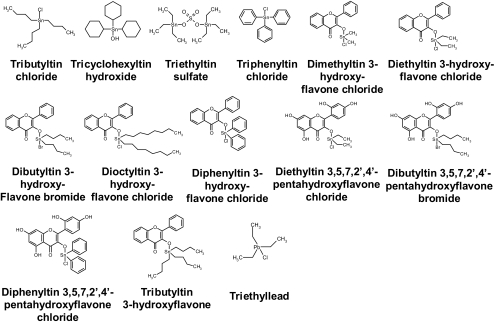
Organotin compounds are organic compounds that contain tin. They are classified as R4Sn, R3SnX, R2SnX2, and RSnX3. Among these, R3SnX organotin compounds have been used as biocides and pesticides and are known to inhibit ATP synthase (Fig. 5) (148-150, 190, 252, 403-405, 418, 437). Some R4Sn organotin compounds, such as tributyltin 3-hydroxyflavone, also inhibit ATP synthase (405). The organotin compounds inhibit both ATP hydrolysis and ATP synthesis catalyzed by the membrane-bound and isolated F0F1 complex. However, they have no effect on the ATPase activity of isolated F1 (Table 9). Organotin compounds react noncovalently with the ATP synthase, and the inhibitory effect of the compounds is reversed by mono- and dithiols such as dithiothreitol and mercaptoethanol (437). The sites of action of organotin compounds are located in the ion channel within subunit a. Here, they are believed to inhibit ATP synthase by competing with Na+ or H+ for the same binding site (418). Diorganotin-3-hydroxyflavone complexes such as dibutyltin 3-hydroxyflavone bromide and diphenyltin 3-hydroxyflavone chloride show a marked fluorescence enhancement on binding to mitochondrial ATP synthase (405).
FIG. 5.
Structures of organotin compounds and structural relatives.
TABLE 9.
Organotin compounds and structural relatives
| Name | Molecular formula | Other names | Inhibitory potency (reference) |
|---|---|---|---|
| Tributyltin chloride | C12H27ClSn | TBT-Cl; tributylchlorostannane; chlorotributyltin; tri-n-butyltin chloride; monochlorotributyltin; tri-n-butylchlorotin; tributylstannyl chloride | 200 nMb (E. coli and I. tartaricus F0F1-ATPase) (418); 47% inhibition at 1 μM and 87% inhibition at 5 μM (C. thermoaceticum membrane-bound F0F1-ATPase) (190); 80% inhibition at 1 μM (TF0F1-ATPase) (403) |
| Tricyclohexyltin hydroxide | C18H34OSn | Cyhexatin; tricyclohexylhydroxytin; hydroxytricyclohexylstannane; tricyclohexylhydroxystannane; tricyclohexylstannanol; Plictran; tricyclohexylstannium hydroxide | 92.9% inhibition at 37 μM (rat liver MF0F1-ATPase) (252) |
| Triethyltin sulfate | C12H30O4SSn2 | Triethylstannium hydrogen sulfate; bis(triethyltin) sulfate; triethylhydroxytin sulfate | 0.13 μg of inhibitor/mg proteina (S. cerevisiae SMP-ATPase) (150, 151); 3-7 μg of inhibitor/mg proteina (T. pyriformis SMP-ATPase) (404); 1.2 μg/mg proteina (C. asciculate SMP-ATPase) (439) |
| Triphenyltin chloride | C18H15ClSn | Chlorotriphenylstannane; chlorotriphenyltin; triphenylchlorotin | <10 μMa (bovine heart SMP-ATPase) (437) |
| Dimethyltin 3-hydroxyflavone chloride | C17H15ClO3Sn | 12-13 nmol inhibitor/mg proteina (rat liver SMP-ATPase) (405) | |
| Diethyltin 3-hydroxyflavone chloride | C19H19ClO3Sn | 1.5 nmol inhibitor/mg proteina (rat liver SMP-ATPase) (405) | |
| Dibutyltin 3-hydroxyflavone bromide | C23H27BrO3Sn | 0.7-0.9 nmol inhibitor/mg proteina (rat liver SMP-ATPase) (405) | |
| Dioctyltin 3-hydroxyflavone chloride | C31H43ClO3Sn | 12-13 nmol inhibitor/mg proteina (rat liver SMP-ATPase) (405) | |
| Diphenyltin 3-hydroxyflavone chloride | C27H19ClO3Sn | 1.5 nmol inhibitor/mg proteina (rat liver SMP-ATPase) (405) | |
| Diethyltin 3,5,7,2′,4′-pentahydroxy flavone chloride | C19H19ClO7Sn | 5-6 nmol inhibitor/mg proteina (rat liver SMP-ATPase) (405) | |
| Dibutyltin 3,5,7,2′,4′-pentahydroxy flavone bromide | C23H27BrO7Sn | 0.6-0.8 nmol inhibitor/mg proteina (rat liver SMP-ATPase) (405) | |
| Diphenyltin 3,5,7,2′,4′-pentahydroxy flavone chloride | C27H19ClO7Sn | 3.5-4 nmol inhibitor/mg proteina (rat liver SMP-ATPase) (405) | |
| Tributyltin 3-hydroxyflavone | C27H36O3Sn | 1.5-2 nmol inhibitor/mg proteina (rat liver SMP-ATPase) (405) | |
| Triethyllead | C6H15ClPb | Triethylplumbane | 16-17 μMa (rat liver SMP-ATPase) (275) |
I50.
Ki.
POLYENIC α-PYRONE DERIVATIVES
α-Pyrone (or 2-pyrone) is a six-membered cyclic unsaturated ester. Its derivatives are widely distributed in nature, and some α-pyrone-containing mycotoxins, such as aurovertin, citreoviridin, and asteltoxin, inhibit ATP synthase by targeting F1 (Fig. 6).
FIG. 6.
Structures of polyenic α-pyrone derivatives.
Aurovertin is an antibiotic from Calcarisporium arbuscula. Five different types of aurovertins (A to E) have been reported (Table 10). Aurovertin inhibits the ATPase activity of F1 from mitochondria and mesophilic bacteria (108, 189), whereas it has no inhibitory effect on thermophilic TF1 (196, 343). It binds to the ATP synthase β subunit and inhibits its ATPase activity uncompetitively (108, 189). There are two or three binding sites for aurovertin in F1 in the presence of ADP: one high-affinity site (Kd [dissociation constant] of 0.2 to 1 μM) and the others (one or two) of lower affinity (Kd of 3 to 6 μM) (188, 416). In contrast, two high-affinity sites are observed in the presence of ATP (188). In the crystal structure of one F1-aurovertin complex (410), two aurovertin B molecules are bound at two equivalent sites within the βTP and βE subunits. These sites are located in a cleft between the nucleotide binding and C-terminal domains of the subunits and do not overlap with the nucleotide binding sites. In βTP, the pyrone ring of aurovertin interacts with α-Glu399 of αTP. However, in βE the pyrone ring has no equivalent interaction with αE, as the aurovertin bound in βE is too far from αE. The interactions between aurovertin and amino acids are mainly hydrophobic. In βDP, the interface between αDP and βDP is tightly packed, making the aurovertin binding pocket inaccessible (410). In the binding of aurovertin to F1, β-Arg398 (E. coli sequence) appears to play an important role, as mutations in this residue confer aurovertin resistance (230, 231, 424). In bacteria that are naturally resistant to aurovertin, the β-Arg398 residue is replaced with other amino acid residues (172, 343). Aurovertin is believed to inhibit F1 by preventing catalytic interface closure involved in the cyclic interconversion of catalytic sites (410, 430). In addition, aurovertin increases the affinity of F1 for phosphate (307). Aurovertin fluoresces weakly at 470 nm, and this is enhanced by 50- to 60-fold when aurovertin binds to F1 (74, 136, 232). The fluorescence increase is considered to be due to the limited mobility of aurovertin at its binding site and has been used to monitor inhibition of F1-ATPase activity (74, 136).
TABLE 10.
Polyenic α-pyrone derivatives
| Name | Molecular formula | Source | Inhibitory potency (reference) |
|---|---|---|---|
| Aurovertin | A, C27H34O9; B, C25H32O8; C, C24H30O8; D, C25H32O9; E, C23H30O7 | C. arbuscula | 9.2 μmol/mg proteina and 25 μMc (aurovertin A, bovine heart MF1-ATPase) (232); 2 μMa (aurovertin B, EF1-ATPase) (353); 17-30 nmol/mg proteina and 0.1 μMc (aurovertin B, bovine heart MF1-ATPase) (232); 2 nmol/mg proteina and 0.6 μMc (aurovertin C, bovine heart SMP) (232); 0.9 μMa (aurovertin D, EF1-ATPase) (353); 1 μMa (aurovertin D, EF1-ATPase) (436); 9-20 nmol/mg proteina and 60 nMc (aurovertin D, bovine heart MF1-ATPase) (232); 1.6 μmol/mg proteina and 22 μMc (aurovertin E, bovine heart SMP) (232); 80 nMa (rat liver MF1-ATPase) (108); 66% inhibition at 10 μM (bovine heart MF1-ATPase) (325) |
| Citreoviridin | A, C23H30O6; B, unknown; C, C23H30O6; D, C24H32O6 | A, Penicillium citreoviride, Penicillium toxicarium, Penicillium ochrosalmoneum, Aspergillus terreus; B, A. terreus; C, A. terreus; D, A. terreus | 60 μMa (EF1-ATPase) (353); 1.11 μmol/mg proteina (bovine heart MF1-ATPase) (232); 2 μMb (S. cerevisiae MF1-ATPase) (136); 4.23 μMb (354) (bovine heart MF1-ATPase); 2.82 μMb (354), 6.1 μMb (354) (bovine heart SMP-ATPase); 3.1 μMc (232), 4.1 μMc (354) (bovine heart MF1-ATPase); 60 μMc (EF1-ATPase) (353) |
| Asteltoxin | C23H30O7 | A. stellatus Curzi, E. variecolor | 10 μMa (EF1-ATPase) (352); ∼450 nMa (state 3 respiration of rat liver mitochondria) (200); 8 μMc (EF1-ATPase) (352) |
I50.
Ki.
Kd.
Aurovertin B has been tested for the treatment of breast cancer cells as an anticancer agent and has shown strong inhibition of the proliferation of breast cancer cell lines, whereas it showed little influence on normal cells (180). Aurovertin B induced apoptosis of cancer cells and arrested their cell cycles in G0/G1 phase.
Citreoviridin, produced by some molds of the genera Penicillium and Aspergillus, inhibits the ATPase activities of F1 from bacteria and mitochondria by binding to the ATP synthase β subunit (136, 353) (Table 10). However, ATP synthases from some species are resistant (404, 439). In sensitive species, citreoviridin acts as an uncompetitive inhibitor of ATP hydrolysis by soluble and membrane-bound ATP synthase and as a noncompetitive inhibitor of ATP synthesis by the membrane-bound ATP synthase enzyme (354). The binding of citreoviridin to F1 or its isolated β subunit is noncompetitive with respect to aurovertin (136). Although the binding site of citreoviridin within the β subunit is not clarified, it has been suggested that citreoviridin and aurovertin interact at separate sites (136). Citreoviridin fluoresces weakly at 530 nm when irradiated at 380 nm. However, unlike aurovertin, enhancement is not observed when bound to F1 (233). Light converts citreoviridin to its stereoisomer, isocitreoviridin, which has no effect on either ATP hydrolysis or ATP synthesis catalyzed by ATP synthase (354).
Asteltoxin is made in Aspergillus stellatus Curzi and Emericella variecolor. It contains a unique 2,8-dioxabicyclooctane ring and inhibits both BF1 and MF1 with a stoichiometry of 1:1 in the presence of ADP (Table 10) (200, 352). As asteltoxin fails to inhibit aurovertin-resistant mutants, it is believed to bind to the same site as aurovertin (352). Asteltoxin binding to F1 shows an enhancement of fluorescence (emission maximum, 470 nm; excitation maximum, 385 nm). The ADP-stimulatory effect and the Mg2+-quenching effect on the fluorescence enhancement of asteltoxin binding are similar to those observed for aurovertin. However, the stimulatory effect on phosphate binding to F1 observed with aurovertin is not observed with asteltoxin (352).
CATIONIC INHIBITORS
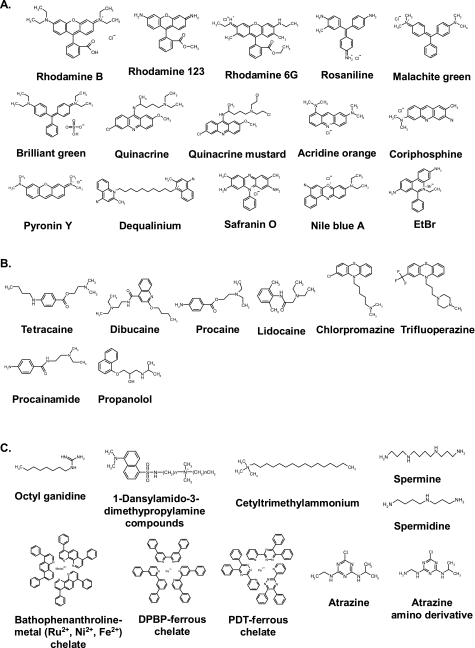
Amphiphilic Cationic Dyes
Amphiphilic cationic dyes containing a basic amine group and a lipophilic portion (Fig. 7A) inhibit the ATPase activities of both F1 and F0F1. Most exhibit a stronger inhibitory effect on the ATPase activity of F0F1 than on that of F1 (Table 11).
FIG. 7.
Structures of cationic inhibitors. (A) Amphiphilic cationic dyes. EtBr, ethidium bromide. (B) TALAs and related compounds. (C) Other organic cations. DPBP, 4,4-diphenyl-2,2-bipyridine; PDT, 3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine.
TABLE 11.
Amphiphilic cationic dyes
| Name or abbreviation | Molecular formula | Other names | Inhibitory potency (reference) |
|---|---|---|---|
| Rhodamine B | C28H31ClN2O3 | N-(9-(2-Carboxyphenyl)-6-(diethylamino)-3H-xanthen-3-ylidene)-N-ethylethanaminium chloride; rheonine B; rhodamine O; rhodamine S | 475 μMa (bovine heart MF1-ATPase) (52); 125 μMa (bovine heart MF0F1-ATPase) (52) |
| Rhodamine 123 | C21H17ClN2O3 | 3,6-Diamino-9-(2-(methoxycarbonyl)phenyl)xanthylium chloride; RH 123 | 270 μMa (bovine heart MF1-ATPase) (52); 141 μMa (bovine heart MF0F1-ATPase) (52); 580 μMa (EF0F1-ATPase) (268); 177 μMb (rat liver MF1-ATPase) (113) |
| Rhodamine 6G | C28H31ClN2O3 | Basic rhodamine yellow; rhodamine J | 10 μMa (bovine heart MF1-ATPase) (52); 27 μMa (bovine heart MF1-ATPase) (143); 2 μMa (bovine heart MF0F1-ATPase) (52); 34 μMa (EF0F1-ATPase) (268); 2.4 μMb (S. cerevisiae MF1-ATPase) (433); 1.95 μMb (S. cerevisiae MF0F1-ATPase) (433); 1.91 μMb (S. cerevisiae SMP-ATPase) (433) |
| Rosaniline | C20H20ClN3 | Magenta base; 4-((4-aminophenyl)(4-imino-2,5-cyclohexadien-1-ylidene)methyl)-2-methylbenzenamine | 15 μMa (bovine heart MF1-ATPase) (52); 16 μMa (bovine heart MF0F1-ATPase) (52) |
| Malachite green | C23H25N2Cl | Aniline green; benzal green; Victoria green; (4-(4-dimethylaminobenzhydriylidene)cyclo-hexa-2,5-dienylidene)dimethylammonium chloride | 14 μMa (bovine heart MF1-ATPase) (52); 7 μMa (bovine heart MF0F1-ATPase) (52) |
| Brilliant green | C27H33N2.HO4S | Basic green 1; (4-(4-(diethylamino)benzhydrylene)cyclohexa-2,5-dien-1-ylidene)diethylammonium hydrogen sulfate | 27 μMa (EF0F1-ATPase) (268) |
| Quinacrine | C23H30ClN3O | 2-Methoxy-6-chloro-9-diethylaminopentylaminoacridine; 3-chloro-7-methoxy-9-(1-methyl-4-diethylaminobutylamino)acridine; mepacrine | 580 μMa (EF0F1-ATPase) (268); 580 μMa (bovine heart MF1-ATPase) (220); 440 μMb (bovine heart MF1-ATPase) (220) |
| Quinacrine mustard | C23H28Cl3N3O | Quinacrine mustard dihydrochloride; 2-methoxy-6-chloro-9-(3-(ethyl-2-chloroethyl)aminopropylamino)acridine dihydrochloride; 9-[4-(bis(2-chloroethyl)amino)-1-methylbutylamino]-6-chloro-2-methoxyacridine dihydrochloride | 5.3 μMa (EF0F1-ATPase) (268); 27 μMc (bovine heart MF1-ATPase) (53) |
| Acridine orange | C17H19N3Cl | 3,6-Acridinediamine, N,N,N′,N′-tetramethyl-, monohydrochloride; 3,6-bis(dimethylamino)acridine hydrochloride; rhoduline orange | 180 μMa (bovine heart MF1-ATPase) (52); 1 μMa (bovine heart MF0F1-ATPase) (52); 68 μMa (EF0F1-ATPase) (268) |
| Coriphosphine | C16H17N3.HCl | Coriphosphine O; coriphosphine OX; 3-amino-6-(dimethylamino)-2-methylacridine monohydrochloride | 480 μMa (bovine heart MF1-ATPase) (52); 16 μMa (bovine heart MF0F1-ATPase) (52) |
| Pyronin Y | C17H19ClN2O | Pyronine; pyronin G | 1.65 mMa (bovine heart MF1-ATPase) (52); 10 μMa (bovine heart MF0F1-ATPase) (52); 70 μMa (EF0F1-ATPase) (268) |
| Dequalinium | C30H40N4 | 1,1′-(1,10-Decanediyl)bis(4-amino-2-methyl-quinolinium | 8 μMa (52), 12 μMa (452), 46 μMa (143) (bovine heart MF1-ATPase); 24 μMa (EF0F1-ATPase) (268); 50 μMa (TF1-ATPase, photoinactivation) (296);19 mM (Bacillus PS3 ATPase, αβγ complex) (143); 4 μMb (spinach CF1, Ca2+-ATPase) (329); 12.5 μMc (TF1-ATPase) (296); 12.5 μMc (bovine heart MF1-ATPase) (452) |
| Safranin O | C20H19ClN4 | Basic red 2; 3,7-diamino-2,8-dimethyl-5-phenylphenazinium chloride; safranine T | 1.14 mMa (bovine heart MF1-ATPase) (52); 175 μMa (bovine heart MF0F1-ATPase) (52); 330 μMa (EF0F1-ATPase) (268) |
| Nile blue A | C20H20N3OCl | Nile blue; Nile blue AX; 5-amino-9-(diethylamino)benzo(a)phenoxazine-7-ium chloride | >2,000 μMa (bovine heart MF1-ATPase) (52); 16 μMa (bovine heart MF0F1) (52) |
| EtBr | C21H20BrN3 | Ethidium bromide; homidium bromide; AI3-62997; 2,7-diamino-10-ethyl-9-phenylphenanthridinium bromide | 220 μMa (S. cerevisiae MF1-ATPase) (82); 250 μMa (Trypanosoma cruzi F0F1-ATPase) (66); 279 μMb (S. cerevisiae MF1-ATPase) (433); 256 μMb (S. cerevisiae MF0F1-ATPase) (433); 263.6b μM (S. cerevisiae SMP-ATPase) (433) |
I50.
Ki.
Kd.
Rhodamines are a group of fluorone dyes made by fusing an amino derivative of phenol with phthalic anhydride, and they include rhodamine B, rhodamine 123, and rhodamine 6G. Rhodamine B and rhodamine 123 inhibit the ATPase activity of MF1 from bovine heart in a parabolic, noncompetitive manner, whereas inhibition by rhodamine 6G is mixed (433). In contrast, rhodamine 6G acts as an uncompetitive inhibitor of MF1 and as a noncompetitive inhibitor for isolated and membrane-bound ATP synthase F0F1 from yeast (433). Rhodamine B and rhodamine 123 are considered to bind F1 at more than one binding sites, while rhodamine 6G at high concentrations is believed to bind at least two binding sites (52). The precise location of rhodamine 6G binding sites in the three-dimensional structure of F1 has yet to be identified (143).
Rosaniline, malachite green, and brilliant green are closely related in structure. Rosaniline and malachite green inhibit MF1 in a parabolic mixed fashion, indicating at least two binding sites at high concentrations (52).
Quinacrine inhibits reversibly the ATPase activities of EF1 and bovine MF1 with a similar inhibitory potency (220, 268). This agent inhibits the ATP hydrolysis activity of F1 competitively when Mg2+ is at a constant concentration and ATP at a variable concentrations (220, 268). Quinacrine mustard is a quinacrine derivative in which a diethyl group attached to the tertiary amino group is replaced by a bischloroethyl groups. The quinacrine mustard binds to F1 and alkylates β subunits. The inhibition of the ATPase activity of F1 by quinacrine mustard is irreversible (220) and is due, at least in part, to modification of one or more of the carboxylic acid side chains in the β subunit DELSEED region and possibly also to modification of unspecified amino acid side chains between residues β302 and 356 in the bovine sequence (53). The rate of inactivation of MF1 and TF1 by quinacrine mustard is inhibited by ATP, whereas the rate of inactivation of EF1 is stimulated by ATP (54).
Acridine orange and coriphosphine are acridine derivatives that inhibit the ATPase activity of MF1 in a mixed fashion (52). Pyronin Y, a xanthene derivative, inhibits the ATPase activities of F0F1 from mitochondria and E. coli (52, 268). Here, the inhibitory effect on the mitochondrial ATPase is more potent for F0F1 (>100-fold) than for F1 (52).
Dequalinium is a quinoline derivative that inhibits the ATPase activities of F1 from both mitochondria and bacteria (52, 268, 296, 329, 452). Dequalinium inhibits chloroplast Ca2+-ATPase, whereas it stimulates chloroplast Mg2+-ATPase (329). The inhibition of ATPase activity by dequalinium is reversible, hyperbolic, and noncompetitive for MF1 and TF1 in the dark (52, 268, 296, 329, 452). A long lag is observed in the inhibition of TF1 by dequalinium that is not observed for the inhibition of MF1 (296). Dequalinium, upon illumination at 350 nm, inactivates F1-ATPase with pseudo-first-order kinetics (296, 329, 452, 454). This is accompanied by derivatization of βPhe420 in TF1 (296), βMet183 in CF1 (329), and αPhe403, αPhe406, and a side chain within residues 440 to 459 of the β subunit in bovine heart MF1 (454).
Safranin O inhibits the ATPase activities of membrane-bound F0F1 from both bovine heart mitochondria and E. coli (52, 268). Safranin O also inhibits soluble MF1 with weaker inhibitory potency (52). Nile blue A inhibits the ATPase activity of membrane-bound F0F1 from mitochondria, whereas it has no inhibitory effect on isolated F1 (52). Ethidium bromide inhibits noncompetitively ATP hydrolysis by both MF1 and F0F1 from Saccharomyces cerevisiae (82, 433), with similar inhibitory potencies (66, 82).
TALAs and Related Compounds
Tertiary amine local anesthetics (TALAs) are composed of an aromatic portion, an intermediate chain, and a terminal amine group (Fig. 7B) (370). The intermediate chain contains either an ester (tetracaine and procaine) or an amide (dibucaine and lidocaine) group. In procainamide, the ester group in procaine is replaced with an amide. Chlorpromazine and trifluoroperazine are cationic phenothiazine derivatives. The TALAs are known to inhibit primarily sodium influx through sodium-specific ion channels in the neuronal cell membrane. However, they can also bind to ATP synthases from mitochondria and some bacteria and can inhibit ATP hydrolysis activity (Table 12) (76, 406).
TABLE 12.
Tertiary amine local anesthetics and related compounds
| Name | Molecular formula | Other names | Inhibitory potency, I50 (reference) |
|---|---|---|---|
| Tetracaine | C15H24N2O2 | Dicaine; 2-(dimethylamino)ethyl p-(butylamino)benzoate; dimethylaminoethyl p-butyl-aminobenzoate; p-butylaminobenzoyl-2-dimethylaminoethanol | 0.7-0.83 mM (76), 1.1 mM (406), 1.95 mM (343) (bovine heart MF1-ATPase); 1.4 mM (76), 1.79 mM (343) (bovine heart SMP-ATPase) |
| Dibucaine | C20H29N3O2 | 2-Butoxy-N-(2-(diethylamino)ethyl) cinchoninamide; 2-butoxy-N-(2-DEAE) quinoline-4-carboxamide; cincainum; cinchocaine; Dermacaine; dibucainum; Nupercaine; Percamine; Sovcaine; α-butyloxycinchonic acid-γ-diethylethylenediamine | 0.19-0.5 mM (bovine heart MF1-ATPase) (76); 0.26 mM (bovine heart SMP-ATPase) (76); 29% inhibition at 1 mM (M. phlei F1-ATPase) (4); 55.7% inhibition at 1 mM (M. phlei membrane-bound ATPase) (4) |
| Procaine | C13H20N2O2 | 2-DEAE-4-aminobenzoate; DEAE p-aminobenzoate; p-aminobenzoyldiethylaminoethanol; procain; Spinocaine | 1.8 mM (343), 15-17 mM (76) (bovine heart MF1-ATPase); 8.4 mM (343), 9.5 mM (76) (bovine heart SMP-ATPase) |
| Lidocaine | C14H22N2O | 2-(Diethylamino)-N-(2,6-imethylphenyl) acetamide; cappicaine; Duncaine; Esracaine; Isicaine; Lidocaine; Maricaine; xycaine; Xylocaine | 12-16 mM (76), 18.2 mM (343) (bovine heart MF1-ATPase); 10 mM (76), 22 mM (343) (bovine heart SMP-ATPase) |
| Chlorpromazine | C17H19N2SCl | 2-Chloro-10-(3-(dimethylamino)propyl) phenothiazine; Aminazin; Aminazine; Chlor-Promanyl; Chlorderazin; Chlorpromados; Contomin; Elmarin; Esmind; Fenactil; Largactil; Megaphen; Novomazina; Proma; Phenactyl; Promactil; Propaphenin; Prozil; Psychozine; Sanopron; Thorazine; Torazina; Wintermin | 50 μM(54), 60 μM (343), 50-150 μM (221) (bovine heart MF1-ATPase); 26 μM (76), 450 μM (343) (bovine heart SMP-ATPase); 150 μM (EF1-ATPase) (54); 30.8-56.0 μM (bovine heart MF0F1-ATPase) (87); 6.5-12 μM (bovine heart MF0F1, photoinactivation) (87) |
| Trifluoperazine | C21H24F3N3S | 10-(3-(4-Methyl-1-piperazinyl)propyl)-2-(trifluoromethyl)phenothiazine; trifluoromethylperazine | 17.2-30.5 μM (bovine heart MF0F1-ATPase) (87); 3.0-5.5 μM (bovine heart MF0F1, photoinactivation) (87) |
| Procainamide | C13H21N3O | 4-Amino-N-(2-(diethylamino)ethyl)benzamide | 17-35 mM (76), 33 mM (343) (bovine heart MF1-ATPase); 31 mM (bovine heart SMP-ATPase) (76) |
| Propranolol | C16H21NO2 | 1-((1-Methylethyl)amino)-3-(1-naphthalenyloxy)-2-propanol | 210 μM (343), 0.87-1.4 mM (76) (bovine heart MF1-ATPase); 310 μM at 37°C and 880 μM at 60°C (TF1-ATPase) (343); 660 μM (343), 840 μM (76) (bovine heart SMP-ATPase) |
TALAs inhibit both membrane-bound and soluble MF1. Inhibition of MF1 is reversible, and the concentration ranges for inhibition are near those for blocking nerve conduction (76). The hydrophobicity of TALAs seems to determine their relative affinities for F1, as the inhibitory potencies are directly correlated with the octanol/water partition coefficient (76). Among the TALAs, procainamide shows activation of the ATPase activity of F1 at low concentrations prior to its inhibition of F1 at high concentrations. This is not observed with other TALAs (76). The mechanism of the inhibitory action of TALAs on MF1 is still controversial, with one view implicating the induction of the structural dissociation of the multisubunit structure of F1 (76) and a second view the interaction with the catalytic sites of F1 (221).
In contrast to the case for the mitochondrial ATP synthase, the TALAs inhibit bacterial ATP synthases selectively. For example, they exhibit no inhibition of F1 from the thermophilic bacterium PS3 under the conditions tested (343). However, tetracaine and dibucaine do inhibit the ATPase activity of the membrane-bound ATP synthase from the bacterium Mycobacterium phlei (4), whereas procaine and lidocaine show no inhibitory effects. In addition, tetracaine and dibucaine show no or partial inhibition of the ATPase activity of soluble F1, in contrast to full inhibition of the ATPase activity of the membrane-bound ATP synthase. Upon inhibition (uncompetitive) of the membrane-bound ATP synthase from M. phlei by tetracaine and dibucaine, proton conductivity is markedly inhibited. Tetracaine and DCCD are not mutually exclusive in binding to the ATP synthase from M. phlei, and they appear to bind to separate binding sites within the proton-translocating “F0” region (4).
Chlorpromazine and trifluoroperazine interact with various subunit types of F1 and F0. Both bind to membrane-bound subunits more readily than to soluble subunits, with trifluoroperazine binding to hydrophobic subunits more extensively than chlorpromazine (88). The binding sites of chlorpromazine and trifluoroperazine are not identical and mutually nonexclusive (87, 88). Upon photoactivation with UV light, the phenothiazine moiety of chlorpromazine and trifluoroperazine forms covalent bonds with the ATP synthase, leading to its irreversible inhibition. In other studies, chlorpromazine has been shown to protect MF1 and EF1 against both cold-induced dissociation and inactivation by DCCD (54). This agent is believed to cause inhibition by interacting with the catalytic site at position βGlu188 (bovine sequence). However, in other studies, chlorpromazine has been shown to stimulate the ATPase activity of TF1 both at 37°C and at low concentrations (below 0.6 mM) at 23°C. It shows no inhibition up to 1.2 mM at 37°C or 60°C (54).
Propranolol is a nonselective beta blocker for the treatment of hypertension. It is not a TALA and has no ester or amide group in the intermediate chain. However, it is structurally analogous to TALAs. The main action of propanolol is to block the action of epinephrine on both β1- and β2-adrenergic receptors, but it also targets ATP synthase. Propranolol inhibits the mitochondrial ATPase activities of both membrane-bound ATP synthase and isolated F1 (76, 343). It also inhibits TF1 at both 37°C and 60°C with nearly the same effective concentrations as that for inhibition of membrane-bound mitochondrial ATP synthase (76, 343).
Other Organic Cations
Alkylguanidines (Fig. 7C) that possess an alkyl chain of more than six carbons inhibit the ATPase activities of both membrane-bound and isolated MF1 (92, 300). The inhibition by octylguanidine, an alkylguanidine, is fully reversible, and the octylguanidine prevents cold-induced dissociation of F1 (92).
1-Dansylamido-3-dimethypropylamine compounds are dansylated organic cationic inhibitors (Fig. 7C). They inhibit both ATP hydrolysis and ATP synthesis at similar concentrations (116). The 1-dansylamido-3-dimethypropylamine compounds inhibit the ATPase activities of both isolated and membrane-bound F1 and exhibit more potent inhibitory effect on the membrane-bound F1 than the isolated enzyme. The 1-dansylamido-3-dimethypropylamine compounds with longer alkyl groups (decyl and hexadecyl) have stronger inhibitory activity than those with short groups (propyl and hexyl) (Table 13). The binding site(s) of these compounds is not clarified but is considered to be located on the β subunit (116).
TABLE 13.
Other organic cations
| Name or abbreviation | Molecular formula | Other names | Inhibitory potency (reference) |
|---|---|---|---|
| Octyl guanidine | C9H21N3 | 300 μMa (bovine heart SMP- and MF1-ATPase) (92); 330 μMa (rat liver SMP-ATPase) (300) | |
| 1-Dansyl amido-3-dimethypropylamine compounds | C20H32N3O2S (n = 2) C23H38N3O2S (n = 5) C27H46N3O2S (n = 9) C33H58N3O2S (n = 15) | 1.4 mMa (n = 2), 0.4 mMa (n = 5), 7.9 μMa and 4.4 μMb (n = 9), and 3.4 μMa (n = 15) (bovine heart SMP-ATPase) (116) | |
| Cetyltrimethylammonium | C19H42N | Cetrimonium; cetrimonum; cetyltrimethylammonium; hexadecyltrimethylammonium; trimethylhexadecylammonium | 80 μMb (bovine heart MF1-ATPase) (31) |
| Spermine | C7H19N3 | 4-Azaoctamethylenediamine | Inhibitory effect at 1-2 mM range (185); ∼55% inhibition at 2 mM with 2 mM Mg2+ (rat liver MF1-ATPase) (185) |
| Spermidine | C10H26N4 | 1,4-Bis(aminopropyl) butanediamine; diaminopropyltetramethylenediamine | Inhibitory effect at 2.5-5 mM range (rat liver MF1-ATPase) (185) |
| Bathophenan throline-metal (Ru2+, Ni2+, Fe2+) chelate | C24H16N2, 3C24H16N2·Ru, 3C24H16N2·Ni, 3C24H16N2·Fe | 1,10-Bathophenanthroline; 4,7-diphenyl-1,10-phenanthroline; bathophenanthroline ruthenium(II); Ru-Tdpa; tris(4,7-diphenyl-1,10-phenanthroline)ruthenium (II); 4,7-diphenyl-1,10-phenanthroline-ferrous chelate; BPh3Fe2+ | For BPh, almost complete inhibition at 5 μM (bovine heart MF1) (315); for BPh3·Fe2+, 30 nmol/mg proteinb (N. crassa SMP-ATPase) (112); 100% inhibition at 0.67 μM (bovine heart MF1-ATPase) (63) |
| DPBP-ferrous chelate | 3C22H16N2·Fe | 4,4-Diphenyl-2,2-bipyridine | 85% inhibition at 0.67 μM and 99% inhibition at 3.33 μM (bovine heart MF1-ATPase) (63) |
| PDT-ferrous chelate | 3C20H14N4·Fe | 3-(2-Pyridyl)-5,6-diphenyl-1,2,4-triazine | 73% inhibition at 0.67 μM and 95% inhibition at 3.33 μM (bovine heart MF1-ATPase) (63) |
| Atrazine | C8H14ClN5 | 6-Chloro-N-ethyl-N′-(propan-2-yl)-1,3,5-triazine-2,4-diamine | |
| Atrazine amino derivative | C7H13ClN6 | N-(Aminomethyl)-6-chloro-N′-(propan-2-yl)-1,3,5-triazine-2,4-diamine |
I50.
Ki.
Cetyltrimethylammonium inhibits the ATPase activities of soluble and membrane-bound F1 in a noncompetitive manner (31). The inhibition is reversible and can be reversed by dilution. The inhibition of membrane-bound F1 shows a more complex pattern than that of isolated F1 with a sigmoidal dependence on the concentration of cetyltrimethylammonium. Also, cetyltrimethylammonium potentiates inhibition of membrane-bound ATP synthase by oligomycin, and vice versa. It lowers the Ki of the ATP synthase for oligomycin by about 1 order of magnitude. The inhibitory effect by cetyltrimethylammonium is believed to be due to an interaction of negatively charged residues buried in a hydrophobic environment of F1.
Spermine and spermidine are polyamines distributed widely in nature. Both activate the ATPase activity of membrane-bound ATP synthase at low physiological concentrations (312, 374) and inhibit it at high concentrations (185). Spermine and spermidine also inhibit the ATPase activity of isolated F1. Inhibition by spermine (1 to 2 mM range) is much greater than that by spermidine (2.5 to 5 mM range) and is uncompetitive with variable concentrations of ATP in the presence of Mg2+ but competitive when both ATP and Mg2+ concentrations are variable. Spermine and spermidine bind to ATP, an event that is inhibited by Mg2+. In fact, the inhibition of the ATPase activities of membrane-bound and isolated F1 by polyamines is considered to be due to their direct binding to ATP. In contrast to their ATPase-inhibitory actions, spermine and spermidine stimulate catalysis in SMP of both succinate-dependent ATP synthesis and Pi-ATP exchange (185).
Octahedral bathophenanthroline (BPh3)-metal chelates inhibit MF1 in an uncoupler-reversible fashion (63-65, 315). They bind to the ATP synthase β subunit and form a complex with a stoichiometic ratio of 3 mol BPh3-Me2+/mol F1. Full inhibition is observed with 0.67 μM of BPh3-Fe2+ for MF1 from bovine heart (63). BPh3-Fe2+ competes with aurovertin for binding to the β subunit. The inhibition is relieved by addition of uncouplers of oxidative phosphorylation via a process that involves direct interaction of the uncouplers with the inhibitory chelates. In fact, inhibitor-uncoupler adducts are believed to be formed (63). BPh3-Ni2+ and BPh3-Ru2+ are equally efficient inhibitors in the uncoupler-reversible inhibition of MF1 (63, 65). Moreover, BPh3-Fe2+ protects F1 from cold-induced dissociation and light-induced inactivation by Rose bengal in an uncoupler-reversible manner (64). The related chelates 4,4-diphenyl-2,2-bipyridine and 3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine with Fe2+ also inhibit MF1, but with weaker inhibitory potencies than BPh3-metal chelates (63).
Atrazine is a globally used triazine herbicide that inhibits photosynthetic electron transport by binding the plastoquinone binding protein in photosystem II (382). Atrazine also targets ATP synthase from sperm and mitochondria, inhibiting the ATP synthesis activity of ATP synthase (170). The amino derivative of atrazine in which a terminal methyl group is replaced with an amino group is more potent in inhibition of ATP synthesis.
SUBSTRATES AND SUBSTRATE ANALOGS
Phosphate Analogs
Arsenate mimics the γ-phosphate of ATP. It inhibits ATP synthesis at the active site of ATP synthase by competing with phosphate (Fig. 8 and Table 14) (81, 264, 307). Arsenate blocks the Pi↔ H2O exchange and also the ATP↔ Pi exchange catalyzed by the ATP synthase (201) and is a more effective inhibitor when the concentration of phosphate is low (307). Thus, at 40 μM phosphate, 4.6 mM arsenate inhibits phosphate binding to bovine heart MF1 by 84%.
FIG. 8.
Structures of phosphate analogs.
TABLE 14.
Phosphate analogs
| Name or abbreviation | Molecular formula | Inhibitory potency (reference) |
|---|---|---|
| Arsenate | AsO4 | 84% inhibition at 4.6 mM at low conc of phosphate (40 μM) (bovine heart MF1-ATPase) (307) |
| Aluminum fluoride | AlF3 and AlF4− | 10 μMa of AlCl3 in the presence of 5 mM NaF and 100 μM ADP (bovine heart MF1-ATPase) (243) |
| Beryllium fluoride | BeF+, BeF2, and BeF3− | 10 μMa of BeCl2 in the presence of 5 mM NaF and 100 μM ADP (bovine heart MF1-ATPase) (243); 20 μMa of BeCl2 in the presence of 2.5 mM NaF with 80 μM ADP with 50 mM Cl− (45 min incubation), 20 mM SO42− (14 min incubation), or 20 mM SO3− (2 min incubation) (bovine heart MF1-ATPase) (187) |
| Scandium fluoride | ScFx | 60 μMa and 95% inhibition at 0.3 mM in the presence of 2.5 mM MgSO4, 1 mM ADP, and 10 mM NaF (279) |
| Vanadate | VO43− and VO3− | VO43−, 50% inhibition in ∼45 min and ∼80% inhibition in ∼2 h at 200 μM in the presence of 200 μM each of MgCl2 and ADP (rat liver MF1-ATPase) (210); VO3−, 30% inhibition at 300 μM (V. parahaemolyticus F1-ATPase) (344) |
| Magnesium fluoride | MgFx | 50% inhibition at 1mM NaADP, 1 mM NaF, and 11-12 mM MgCl2 with 5-12 h preincubation (EF1-ATPase) (5) |
| Sulfite | SO32− | 3.5 mMa and maximal 70% at 10 mM (P. denitrificans F0F1, ATP synthesis) (295) |
| Thiophosphate | SPO33− | Km, 1.5 μM in the presence of 1 mM from 4.5 μM in the absence (pea SMP-ATP synthesis) (254) |
| Azide | N3− | ∼10−5 Mb (bovine heart MF1-ATPase) (412); ∼25 μMb (EF1-ATPase) (287); 71% inhibition at 1 mM (V. parahaemolyticus F1-ATPase) (344); >90% inhibition at 0.5 mM (EF1-ATPase) (287); 55% inhibition at 500 μM (C. thermoaceticum F1-ATPase) (190) |
| ANPP | C6H4N4O6P | 25 μMa (spinach CF1-ATPase, photoinactivation) (321); 60 μMb in the dark (bovine heart MF1-ATPase) (227) |
I50.
Ki.
The phosphate analogs aluminum fluoride and beryllium fluoride also bind to the catalytic sites of ATP synthase by mimicking the γ-phosphate of ATP (48, 107, 195, 243, 256). The inhibition by these fluorides of aluminum and beryllium involves ADP, Mg2+, and the fluoride ion (F−). In fact, no inhibition occurs without fluoride. Inhibition also occurs when IDP, GDP, or CDP replaces ADP (187, 243). Aluminum fluoride and beryllium fluoride inhibit F1 to the same extent via a “quasi-irreversible” process (243). The inhibitory species recognized by F1 are AlF3 and AlF4− for aluminum fluoride (48, 256) and BeF+, BeF2, and BeF3− for beryllium fluoride (187, 195). In crystals of F1 grown with ADP and one of the inhibitors (AlF4− or BeF3−), two catalytic sites are occupied, one in the βTP subunit and the other in the βDP subunit (195, 256). Only one catalytic site, βDP, is occupied with aluminum fluoride (AlF3) in the crystal grown in the presence of ADP, adenylyl imidodiphosphate (AMP-PNP), and the inhibitor. No bound aluminum fluoride or beryllium fluoride is found in the α and βE subunits. Three basic residues located in the vicinity of the γ-phosphate site, βLys162, βArg189, and αArg373, are involved in coordination of the inhibitors and are considered to provide charge stabilization (256).
Scandium fluoride (ScFx) binds to F1 of ATP synthase and inhibits its ATPase activity (279). ScFx forms a tight-binding inhibitory ternary complex with MgADP at the catalytic sites, and the MgADP·ScFx complex acts as a transition state analog. The inhibition by ScFx is Mg2+ dependent, and ADP is also required for strong inhibition. The inhibition is reversible, and the ATPase activity is slowly regained in a single exponential reactivation process.
Two vanadate species, VO43− and VO3−, inhibit F1-ATPase (77, 210, 211, 344). Orthovanadate (VO43−) binds to the catalytic sites and forms a transition-like state MgADP·Vi-F1 complex in the presence of ADP and Mg2+. The inhibition of rat liver MF1 by orthovanadate is reversible, with a restoration of original activity to a level close to 90% (210, 211), whereas EF1 is resistant to orthovanadate (6). In the presence of UV and O2, the cleavage of the β subunit from rat liver MF1 occurs at position Ala158 in the P-loop (210, 211). In the crystal structure of F1 with vanadate from the same source, one vanadate ion is found in each catalytic site of the β subunit (77). The vanadate in this transition-like state is located in a charged pocket surrounded by βLys162, βGlu188, βArg189, and βArg260 and is complexed with ADP and Mg2+. Moreover, the vanadate is positioned closer to P-loop βAla158 than is phosphate in the F1-ADP,Pi ground state structure. It has been proposed that the positioning of βAla158 closer to the γ-phosphate of ATP in the transition state may help facilitate the dehydration of ADP and Pi (to give water) and therefore facilitate ATP synthesis (77).
Magnesium fluoride inhibits F1 by acting also as an apparent transition state analog in combination with MgADP (5). Like vanadate, it mimics the γ-phosphate of ATP in the transition state. The inhibition is slow and reversible and requires ADP.
Sulfite is known as an effective activator of F1-ATPase. However, it can also play a role as an inhibitor of the reversal of ATP synthase as a mixed-type inhibitor in the presence of ADP and phosphate (Fig. 8 and Table 14). The sulfite diminishes the rate of ATP synthesis of Paracoccus denitrificans with an I50 of 3.5 mM (295). The mechanism of sulfite inhibition is uncertain, but it has been suggested that the action of inhibitory ADP is involved in the binding of nucleotides to noncatalytic sites (249), and the binding of sulfite to the noncatalytic sites increases the Ki for inhibitory ADP (295, 327).
Thiophosphate is a group of compounds in which a phosphorus atom is bonded to one or more sulfur and zero or more oxygen atoms, and it is found in a number of insecticides. A thiophosphate, SPO33−, has been shown to inhibit ATP synthesis in mitochondria (254, 363). It inhibits the Pi↔ ATP exchange in SMP from bovine heart mitochondria competitively and also inhibits ATP synthesis noncompetitively with respect to ADP without a change in Km for ADP (363). In contrast, in pea SMP, thiophosphate decreases the Km of the enzyme for ADP (254).
Azide inhibits the ATPase activity of F1 from mitochondria, bacteria, and chloroplasts (25, 46, 126, 274, 278, 287, 391, 412). Azide has no inhibitory effect on ATP synthesis (25). The inhibition by azide is noncompetitive (287, 391) and occurs only in the presence of ADP and ATP (274). The binding of inhibitory azide requires prior binding of both ADP and Mg2+ (160, 278). Azide binds to the catalytic site in βDP of F1 and resides adjacent to the β-phosphate of ADP, mimicking the nonbridging oxygen atom of the γ-phosphate (46). The binding of azide in the βDP catalytic site is very tight, and the azide is closely associated via hydrogen bonds with βLys162 in the P-loop and αArg373 (46). The inhibition is dependent on ATP concentration (274) and is reversed by addition of phosphate, possibly by competing for the azide binding site (262, 274).
Azido-2-nitrophenyl phosphate (ANPP) is a photoaffinity phosphate analog in which the 4-azido-2-nitrophenyl group is attached to phosphate (Fig. 8 and Table 14). ANPP inhibits F1 as a competitive inhibitor in the dark by specifically targeting γ-phosphate binding sites within the nucleotide binding pockets on the β subunit of isolated F1 or on both α and β subunits of membrane-bound F0F1 (154, 227). However, upon photoirradiation with visible light, ANPP inactivates the enzyme by binding covalently to these subunits. This occurs most frequently on βTyr 311, together with βIle304 and βGln308 in MF1, and on the analogous βTyr 328, together with βVal329 and βPro330 in CF1 (133, 258). Phosphate added before photoirradiation protects the photoinactivation by ANPP. The stoichiometry for full photoinactivation of F1 is approximately 1 mol of ANPP/mol of CF1 (321).
Divalent Metal Ions
Divalent metal ions are usually activators of F1, but in their free form, they can also function as inhibitors at high concentrations (47, 98, 174, 278, 291, 365). Free Mg2+ acts as a linear competitive inhibitor (98, 365). The inhibition of CF1 by free Mg2+ requires the presence of a tightly bound ADP at the catalytic site (160, 278). The Ki values are variable, and CF1 and BF1 are about 2 orders of magnitude more sensitive to the inhibition by free Mg2+ than is MF1. Free Mn2+ and Ca2+ ions also inhibit F1-ATPase in a competitive manner and are more effective than free Mg2+ in inhibition of CF1 (Table 15) (174).
TABLE 15.
Divalent metal ions
| Name | Inhibitory potency (reference) |
|---|---|
| Inhibitory free Mg2+ | 2.8 mMa (P. blakesleeanus MF1-ATPase) (98); 3 mMa (ox heart MF1-ATPase) (365); 20 μMa (lettuce CF1-ATPase) (174); 7 μMa (R. rubrum F1-ATPase) (291); 10-15 μMb and 4 μMb (spinach CF1-ATPase) (278) |
| Inhibitory free Mn2+ | 5 μMa (lettuce CF1-ATPase) (174) |
| Inhibitory free Ca2+ | 5-7 μMa (lettuce CF1-ATPase) (174) |
Ki.
Kd.
Purine Nucleotides and Nucleotide Analogs
Excess free ATP is also an inhibitor of ATP synthase (Tables 16 and 17 and Fig. 9A) (98, 291, 365). Inhibition of ATPase activity of F1 by free ATP can be competitive (in the photosynthetic bacterium Rhodospirillum rubrum [291], biphasic (in Phycomyces blakesleeanus [98], or second order/parabolic (in ox heart mitochondria (365).
TABLE 16.
Properties of purine nucleotides and nucleotide analogs
| Name or abbreviation | Molecular formula | Source | Other names |
|---|---|---|---|
| Excess free ATP | C10H16N5O13P3 | Natural | Adenosine triphosphate; adenosine 5′-triphosphate |
| ADP | C10H15N5O10P2 | Natural | Adenosine diphosphate; adenosine 5′-diphosphate |
| GTP | C10H16N5O14P3 | Natural | Guanosine triphosphate; guanosine 5′-triphosphate |
| FTP | C10H16N5O13P3 | Synthetic | Formycin triphosphate; formycin 5′-triphosphate; formycin A 5′-triphosphate |
| TNP-ATP | C16H17N8O19P3 | Synthetic; fluorescent | 2′,3′-O-(2,4,6-Trinitrophenyl) adenosine 5′-triphosphate |
| TNP-ADP | C16H15N8O16P2 | Synthetic; fluorescent | 2′,3′-O-(2,4,6-Trinitrophenyl) adenosine 5′-triphosphate |
| TNP-Ado | C16H13N8O10 | Synthetic; fluorescent | (2′,3′)-O-(2,4,6-Trinitrocyclohexadienylidine)adenosine |
| Lin-benzo-ADP | C14H17N5O10P2 | Synthetic; fluorescent | Linear-benzoadenosine diphosphate |
| AP4A | C20H28N10O19P4 | Natural extracellular mediator | Diadenosine tetraphosphate; 5′,5′′′-diadenosine tetraphosphate |
| AP5A | C20H14N10O22P5 | Natural extracellular mediator | Diadenosine pentaphosphate |
| AP6A | C20H30N10O25P6 | Natural extracellular mediator | Diadenosine hexaphosphate; diadenosine 5′,5′′′′-P1,P6-hexaphosphate |
| AMP-PNP | C10H17N6O12P3 | Synthetic | Adenylyl imidodiphosphate; p[NH]ppA; γ-imino-ATP |
| GMP-PNP | C10H17N6O13P3 | Synthetic | 5′-Guanylyl imidodiphosphate; p[NH]ppG |
| IMP-PNP | C10H16N5O13P3 | Synthetic | Inosine-5′-[(β,γ)-imido]triphosphate |
| AMP(CH2)P | C11H17N5O9P2 | Synthetic | Adenosine 5′-methylenediphosphate; adenosine-5′-(α,β-methylene)-diphosphate; α,β-methylene ADP |
| RhATP | C10H23N5O16P3Rh (tridentate RhATP) | Synthetic | Bidentate RhATP, bidentate tetraaquarhodium-adenosine 5′-triphosphate [Rh(H2O)4ATP]; tridentate RhATP, tridentate triaquarhodium-adenosine 5′-triphosphate [Rh(H2O)3ATP] |
| CrATP or Cr(NH3)4ATP | C10H26CrN5O17P3 (bidentate CrATP) | Synthetic | Monodentate CrATP, monodentate pentaaquachromium-adenosine 5′-triphosphate [Cr(H2O)5ATP]; monodentate Cr(NH3)4ATP, Monodentate tetraaminemonoaquachromium-adenosine 5′-triphosphate [Cr(NH3)4(H2O)ATP]; bidentate CrATP, bidentate tetraaquachromium-adenosine 5′-triphosphate [Cr(H2O)4ATP]; bidentate Cr(NH3)4ATP, bidentate tetraaminechromium-adenosine 5′-triphosphate; bidentate Cr(NH3)2ATP, bidentate biaminebiaquachromium-adenosine 5′-triphosphate [Cr(NH3)2(H2O)2ATP] |
| Co(NH3)4ATP | C10H34CoN9O13P3 | Synthetic | Bidentate tetraaminecobalt-adenosine 5′-triphosphate; bidentate cobalt(III)tetraamine-adenosine 5′-triphosphate |
| 3′-O-Acetyl-ATP | C16H17N8O19P3 | Synthetic | Acetyl adenosine triphosphate |
| 3′-O-Acetyl-ADP | C16H16N8O16P2 | Synthetic | Acetyl adenosine diphosphate |
| 3′-O-Caproyl-ADP | C16H25N5O11P2 | Synthetic | Caproyl adenosine diphosphate |
| 3′-O-Enanthyl-ADP | C17H27N5O11P2 | Synthetic | Enanthyl adenosine diphosphate |
| 3′-O-Caprylyl-ADP | C18H29N5O11P2 | Synthetic | Caprylyl adenosine diphosphate |
| F-ADP/DMAN-ADP | C23H23N6O11P2 | Synthetic; fluorescent | 3′-O-[1-(5-Dimethylamino)-naphthoyl]adenosine diphosphate; 3′-O-(5-dimethylaminonaphthoyl-1)-adenosine diphosphate |
| F-ATP | C23H25N6O14P3 | Synthetic; fluorescent | 3′-O-[1-(5-Dimethylamino)-naphthoyl]adenosine triphosphate; 3′-O-(5-dimethylaminonaphthoyl-1)-adenosine triphosphate |
| 3′-O-(1-Naphthoyl)-ADP/N-ADP | C21H21N5O11P2 | Synthetic; fluorescent | 3′-O-(Naphthoyl-1)adenosine diphosphate; 3-NP-ADP |
| 3′-O-(1-Naphthoyl)-ATP | C21H22N5O14P3 | Synthetic; fluorescent | 3′-O-(Naphthoyl-1)adenosine triphosphate |
| 3′-O-(2-Naphthoyl)-ADP | C21H22N5O14P3 | Synthetic | 3′-O-(2-Naphthoyl)-adenosine 5′-diphosphate |
| BzATP | C24H24N5O15P3 | Synthetic; photoreactive | 3′-O-(4-Benzoyl) benzoyl ATP; 3′-O-(4-benzoyl)benzoyladenosine 5′-triphosphate |
| BzADP | C24H23N5O12P2 | Synthetic; photoreactive | 3′-O-(4-Benzoyl) benzoyl ADP; 3′-O-(4-benzoyl)benzoyladenosine 5′-diphosphate |
| t-Butylacetyl-ADP | C16H25N5O11P2 | Synthetic | tert-Butylacetyl-adenosine 5′-diphosphate |
| 3′-O-Phenylacetyl-ADP | C18H21N5O11P2 | Synthetic | 3′-O-Phenylacetyl-adenosine 5′-diphosphate |
| 3′-O-Phenylbutyryl-ADP | C20H25N5O11P2 | Synthetic | 3′-O-Phenylbutyryl-adenosine 5′-diphosphate |
| 3′-O-Benzoyl-ADP | C17H19N5O11P2 | Synthetic | 3′-O-Benzoyl-ADP |
| 3′-O-[N-(2-Nitrophenyl)-γ-aminobutyryl]-ADP | C20H25N7O13P2 | Synthetic | 3′-O-[N-2-Nitrophenyl-γ-aminobutyryl]-adenosine 5′-diphosphate |
| 3′-O-[N-(4-Nitrophenyl)-γ−aminobutyryl]-ADP | C20H25N7O13P2 | Synthetic | 3′-O-[N-(4-Nitrophenyl)-γ-aminobutyryl]-adenosine 5′-diphosphate |
| 3′-O-(1-Naphthylacetyl)-ADP | C22H23N5O11P2 | Synthetic | 3′-O-(1-Naphthylacetyl)-adenosine 5′-diphosphate |
| 3′-O-(2-Naphthyl acetyl)-ADP | C22H23N5O11P2 | Synthetic | 3′-O-(2-Naphthylacetyl)-adenosine 5′-diphosphate |
| 3′-O-(1-Anthranoyl)-ADP | C25H23N5O11P2 | Synthetic | 3′-O-(1-Anthranoyl)-adenosine 5′-diphosphate |
| 3′-O-(9-Anthranoyl)-ADP | C25H23N5O11P2 | Synthetic | 3′-O-(9-Anthranoyl)-adenosine 5′-diphosphate |
| FSBI | C17H15FN4O8S | Synthetic | 5′-p-Fluorosulfonylbenzoylinosine; 5′-4-Fsbi |
| FSBA | C17H16FN5O7S | Synthetic | 5′-p-Fluorosulfonylbenzoyladenosine; 5′-(4-(fluorosulfonyl)benzoyl)adenosine; 5-Fsba |
| FSBɛA | C19H16FN5O7S | Synthetic | 5′-p-Fluorosulfonylbenzoylethenoadenosine; 5′-(4-fluorosulfonylbenzoyl)-1,N(6)-ethenoadenosine; FSB epsilon A; Fsbn-ethenoadenosine |
| AP2-PL | C18H22N6O12P2 | Synthetic | Adenosine diphosphopyridoxal; PLP-AMP; ADP-pyridoxal; pyridoxal 5′-diphospho-5′-adenosine; 5′-adenosine-5′-diphosphopyridoxal |
| AP3-PL | C18H23N6O15P3 | Synthetic | Adenosine triphosphopyridoxal; adenosine 5′-(tetrahydrogen triphosphate), mono((4-formyl-5-hydroxy-6-methyl-3-pyridinyl)methyl) ester |
| AP4-PL | C18H24N6O18P4 | Synthetic | Adenosine tetraphosphopyridoxal; adenosine tetraphosphate pyridoxal |
| oATP | C10H14N5O13P3 | Synthetic | 2′,3′-Dialdehyde of ATP; dial-ATP |
| oADP | C10H13N5O10P2 | Synthetic | 2′,3′-Dialdehyde of ADP |
| oAMP | C10H12N5O7P | Synthetic | 2′,3′-Dialdehyde of AMP |
| Cibacron blue | C29H17N7O11S3Cl | Synthetic; protein synthesis inhibitor | |
| BzAF | C34H21NO7 | Synthetic; photoreactive | 4-Benzoyl(benzoyl)-1-amidofluorescein |
TABLE 17.
Inhibitory potencies of purine nucleotides and nucleotide analogs
| Name or abbreviation | Inhibitory potency (reference) |
|---|---|
| Excess free ATP | 1 Mg2+/8-10 ATPa (R. rubrum F1-ATPase) (291); 1 mMb (R. rubrum F1-ATPase) (291); 5% inhibition at 12 mM (P. blakesleeanus F1-ATPase) (98) |
| ADP | 15-17 μMb (bovine heart MF1-ATPase) (272, 362); 8.6-9 μMb (rat liver SMP-ATPase) (263, 272) |
| GTP | 5-10 μMa for 15 min (spinach CF1-ATPase) (159) |
| FTP | 5-10 μMa for 15 min (spinach CF1-ATPase) (159) |
| TNP-ATP | 100 nMa (bovine heart MF1-ATPase) (277); 5.5 nMb (157), 25 nMb (214) (bovine heart MF1-ATPase); 21 nMb (bovine heart SMP-ATPase) (157); noncatalytic sites 0.2 μMc and catalytic sites < 0.001, 0.023, 1.39 μMc (EF1-ATPase) (429) |
| TNP-ADP | 15-20 nMa (bovine heart MF1-ATPase) (277); 8 nMb (214), 10 nMb (157) (bovine heart MF1-ATPase); 1.3 μMb (bovine heart SMP-ATP synthesis) (157); noncatalytic sites 6.5 μMc and catalytic sites 0.008, 1.3, 1.3 μMc (EF1-ATPase) (429) |
| TNP-Ado | 33 μMb (bovine heart MF1-ATPase) (214) |
| Lin-benzo-ADP | 16 μMb (bovine heart MF1-ATPase) (199); 0.2 μMc (EF1-ATPase) (424); <10 nMc and 1-2 μMc (bovine heart MF1-ATPase) (428) |
| AP4A | 18 μMa (bovine heart MF1-ATPase) (417) |
| AP5A | ∼60% inhibition at 520 μM in 10 min (bovine heart MF1-ATPase) (417); no inhibition up to 100 μM (bovine heart MF1-ATPase) (325) |
| AP6A | ∼80% inhibition at 520 μM in 80 min (bovine heart MF1-ATPase) (417) |
| AMP-PNP | 0.5 μMb (37), 0.33 μMb (306), 0.32 μMb (361), 14 nMb (84) (bovine heart MF1-ATPase); 0.16 μMb (bovine heart SMP-ATPase) (306); 0.92 μMb (255), 0.3 μMb (361) (rat liver MF1-ATPase); 1.3 μMb (rat liver SMP-ATPase) (255); 0.6b (EF1-ATPase) (436) |
| GMP-PNP | 12.3 μMb (bovine heart MF1-ATPase) (37); 300 μMb (rat liver MF1-ATPase) (361) |
| IMP-PNP | 105 μMb with bicarbonate (bovine heart MF1-ATPase) (362) |
| AMP(CH2)P | Km, 2.8 μM in the presence of 1 mM inhibitor from 0.85 μM in its absence (pea SMP-ATP synthesis) (254) |
| RhATP | Bi- and tridentate RhATP, 300 μMb (bovine heart MF1-ATPase) (383) |
| CrATP or Cr(NH3)4ATP | Monodentate CrATP, 78 μMb (bovine heart MF1-ATPase) (383); monodentate Cr(NH3)4ATP, 500 μMb (bovine heart MF1-ATPase) (383); bidentate CrATP, 1 mMb (bovine heart MF1-ATPase) (383) and 170 μMb (S. cerevisiae MF1-ATPase) (432); bidentate Cr(NH3)4ATP, 100 μMb (bovine heart MF1-ATPase) (383); tridentate CrATP, 150 μMb (S. cerevisiae MF1-ATPase) (432) |
| Co(NH3)4ATP | Bidentate Co(NH3)4ATP, 400 μMb (bovine heart MF1-ATPase) (384) |
| 3′-O-Acetyl-ATP | 400 nMb (bovine heart MF1-ATPase) (394) |
| 3′-O-Acetyl-ADP | 55.3-85 μMa (bovine heart SMP, oxidative phosphorylation) (355, 356) |
| 3′-O-Caproyl-ADP | 1.7 μMa (bovine heart SMP-oxidative phosphorylation) (355) |
| 3′-O-Enanthyl-ADP | 2.7 μMa (bovine heart SMP-oxidative phosphorylation) (355) |
| 3′-O-Caprylyl-ADP | 1.7 μMa (bovine heart SMP-oxidative phosphorylation) (355) |
| DMAN-ADP/F-ADP | 0.25 μMa (bovine heart SMP-oxidative phosphorylation) (356); 40 nMb (bovine heart SMP-oxidative phosphorylation) (356); 9.8 μMb (bovine heart SMP-uncoupled ATPase) (356); 50 nMc (bovine heart MF1-ATPase) (397) |
| F-ATP | 2.1 μMa (bovine heart SMP-oxidative phosphorylation) (356); 0.3 μMb (bovine heart SMP, oxidative phosphorylation) (356); 12-27 μMb (bovine heart SMP-uncoupled ATPase) (356) |
| 3′-O-(1-Naphthoyl)-ADP/N-ADP | 300-350 nMa (bovine heart SMP-oxidative phosphorylation) (355, 356); 4.6 μMb (bovine MF1-ATPase) (240); 9 μMb (bovine heart SMP-ATPase) (240); 48 nMb (bovine heart SMP-oxidative phosphorylation) (355); 20-50 nMc (bovine MF1-ATPase) (397) |
| 3′-O-(1-Naphthoyl)-ATP | 2.0 μMa (bovine heart SMP-oxidative phosphorylation) (356) |
| 3′-O-(2-Naphthoyl)-ADP | 5.0 μMa (bovine heart SMP-oxidative phosphorylation) (356) |
| BzATP | 0.85 μMb (bovine heart MF1-ATPase) (3); ∼6 μMb (TF1-ATPase) (8); 1.6 μMc (bovine heart MF1-ATPase) (3) |
| BzADP | 0.72 μMb (bovine heart MF1-ATPase) (3) |
| t-butylacetyl-ADP | 1.5 μMa (bovine heart SMP-oxidative phosphorylation) (355) |
| 3′-O-Phenylacetyl-ADP | 3.2-3.6 μMa (bovine heart SMP-oxidative phosphorylation) (355, 356) |
| 3′-O-Phenylbutyryl-ADP | 1.3-4.6 μMa (bovine heart SMP-oxidative phosphorylation) (355, 356); 0.2 μMb (bovine heart SMP-oxidative phosphorylation) (355) |
| 3′-O-Benzoyl-ADP | 6.0 μMa (bovine heart SMP-oxidative phosphorylation) (355, 356) |
| 3′-O-[N-(2-Nitrophenyl)-γ− aminobutyryl]-ADP | 0.55 μMa (bovine heart SMP-oxidative phosphorylation) (356) |
| 3′-O-[N-(4-Nitrophenyl)-γ− aminobutyryl]-ADP | 0.76 μMa (bovine heart SMP-oxidative phosphorylation) (356) |
| 3′-O-(1-Naphthylacetyl)-ADP | 0.8 μMa (bovine heart SMP-oxidative phosphorylation) (356) |
| 3′-O-(2-Naphthylacetyl)-ADP | 0.8 μMa (bovine heart SMP-oxidative phosphorylation) (356) |
| 3′-O-(1-Anthranoyl)-ADP | 0.56 μMa (bovine heart SMP-oxidative phosphorylation) (356) |
| 3′-O-(9-Anthranoyl)-ADP | 5.9 μMa (bovine heart SMP-oxidative phosphorylation) (355, 356, 397); 1.1 μMb (bovine heart SMP-oxidative phosphorylation) (355) |
| FSBI | 0.5 mMc for reversible binding (bovine heart MF1-ATPase) (49) |
| FSBA | 45% (1 h) and 65% (2 h) inhibition at 0.8 mM (bovine heart SMP-ATPase) (51); 0.23 mMc for reversible binding (pig heart MF1-ATPase) (101) |
| FSBεA | 250 μMc (bovine heart MF1-ATPase) (414) |
| AP2-PL | ∼150 μMc (α subunit of EF1-ATPase) (326); 30% inhibition at 50 μM (EF1-ATPase) (288) |
| AP3-PL | 18 μMa (EF1-ATPase) (288); 2.5 μMa with Mg2+ and 10 μMa without Mg2+ (EF1-ATPase) (184) |
| AP4-PL | 18 μMa (EF1-ATPase) (288) |
| oATP | 10 mMb (M. phlei F1-ATPase) (219); 1.05 mol/mol of ATPasea (ox heart MF1-ATPase) (239); 40% inhibition at 1 mol/mol ATPase without ADP and 60 min incubation (ox heart MF1-ATPase) (239) |
| oADP | 80 μMc (bovine heart MF1-ATPase) (217) |
| oAMP | 90% inhibition at 3 mM (bovine heart MF1-ATPase) (95) |
| Cibacron blue | 600 μMa (rat liver MF1-ATPase) (28) |
| BzAF | 50 μMb in the dark (bovine heart MF1-ATPase) (297); 58 μMc (bovine heart MF1-ATPase) (297) |
I50.
Ki.
Kd.
FIG. 9.
Structures of purine nucleotides and nucleotide analogs. (A) Nucleotides and nucleotide analogs. (B) Azidonucleotides.
ADP is a substrate for F1, but preincubation of F1 with ADP and Mg2+ induces hysteretic inhibition (32, 102, 261). The inhibition arises when medium Mg2+ combines with F1 to which ADP is bound to only a single catalytic site in the absence of bound Pi. The onset of the inhibition is rather slow (seconds to minutes). The Mg2+ADP-induced inhibition can be slowly and partially reversed by addition of ATP in the absence of Mg2+ (272), and the recovery of ATPase activity requires the binding of ATP at a noncatalytic site. The recovery is promoted by anions such as bicarbonate and sulfite (272, 412). The inhibition can arise from the medium ADP, but ADP produced at the catalytic site by ATP hydrolysis can also start Mg2+ADP-induced inhibition. F1 from chloroplasts is more readily inhibited than F1 from mitochondria, whereas EF1 is not susceptible to Mg2+ADP-induced inhibition under conditions where Mg2+ is not in huge excess (6, 106). The Mg2+ADP-induced inhibition of F1 also occurs in the intact ATP synthase with no or low proton motive force. However, sufficient proton motive force can drive the ATP synthase to remove the inhibitory Mg2+ADP without altering net ATP synthesis (47).
GTP and formycin 5′-triphosphate (FTP) bind to empty noncatalytic sites on CF1 in the presence of Mg2+ and inhibit its ATPase activity (159). Binding of GTP or FTP to two sites causes more inhibition than binding to one site, and the GTP has stronger inhibitory potency than FTP. With GTP or FTP bound at two noncatalytic sites, the GTP inhibits the ATPase activity about 90%, and the FTP about 80%. After a 15-min incubation period, about 50% maximal inhibition is achieved with 5 to 10 μM GTP or FTP for spinach CF1-ATPase.
2′,3′-O-(2,4,6-trinitrophenyl) ATP (TNP-ATP) and TNP-ADP are ribose-modified chromophoric and fluorescent analogs of ATP and ADP in which a trinitrophenyl group is attached to the 2′ and 3′ hydroxyls of ribose (Fig. 9A). These compounds have been used widely for various assays of ATP binding to proteins. Both compounds are potent inhibitors of F1 with high affinity, and the TNP-ATP is hydrolyzable by F1 from mitochondria, chloroplasts, and bacteria (157, 219, 273, 368, 429). The inhibition of ATP hydrolysis by TNP-ATP or TNP-ADP has been reported to be competitive (157) or biphasic (277). These nucleotide analogs bind to both catalytic and noncatalytic sites of F1. Their binding is noncooperative at the three noncatalytic sites and cooperative at the three catalytic sites (429).
2-Azido-TNP-ATP, a 2-azido derivative of TNP-ATP, inhibits F1 catalyzed ATP hydrolysis biphasically (Fig. 9B and Table 18) (276). Bicarbonate decreases the degree of inhibition by 2-azido-TNP-ATP. The Km and Vmax for 2-azido-TNP-ATP hydrolysis are similar to those for TNP-ATP hydrolysis. Upon UV illumination of the F1-ATPase complex with the bound 2-azido-TNP-ATP, it is incorporated into the complex covalently and inactivates the F1-ATPase irreversibly.
TABLE 18.
Azidonucleotides
| Name | Molecular formula | Other names | Inhibitory potency (reference) |
|---|---|---|---|
| 8-Azido-ATP | C10H15N8O13P3 | 8-Azidoadenosine 5′-triphosphate | 1 mMb (bovine heart SMP, phosphorylation) (371); 1 mMb (bovine heart SMP-ATPase) (371); 88% inhibition at 1.7 mM (bovine heart MF1-ATPase) (419); complete inhibition at 2 inhibitor bound mol/mol F1 (bovine heart MF1-ATPase) (110, 420) |
| 8-Azido-ADP | C10H14N8O10P2 | 8-Azidoadenosine 5′-diphosphate | 86% inhibition at 1.7 mM (bovine heart MF1-ATPase) (419); full inhibition at 1.9-2 mol bound inhibitor/mol F1 (134, 419) |
| 2-Azido-ATP | C10H15N8O13P3 | 2-Azidoadenosine 5′-triphosphate | 52% inhibition at 1.8 (0.8 covalent) inhibitor mol/mol F1 (EF1-ATPase) (425); complete inhibition at 0.92 inhibitor bound mol/mol F1 (bovine heart MF1-ATPase) (408) |
| 2-Azido-ADP | C10H14N8O10P2 | 2-Azidoadenosine 5′-diphosphate; 1-azidoadenosine-3′,5′-bisphosphate | 5 μMc in the dark (bovine heart MF1-ATPase) (45); full inhibition at 1.9-2 mol bound inhibitor/mol F1 (45, 134) |
| 2-Azido-TNP-ATP | C17H20N11O18P3 | 2-N3-TNP-ATP | |
| ANA-ADP | C21H17N8O11P2 | 3′-O-[5-Azidonaphthoyl]-ADP | 11 μMb in the dark (bovine heart MF1-ATPase) (240); total inactivation at 2 mol/mol F1 (bovine heart MF1-ATPase) (240) |
| 8-Azido-FSBA | C17H15FN8O7S | 5′-p′-Fluorosulfonylbenzoyl-8-azidoadenosine; 8-N3-FSBA | 0.47 mMc in the dark (bovine heart MF1-ATPase) (453) |
| 2-Azido-AMP-PNP | C10H15N9O12P3 | 2-Azidoadenyl-5′-yl imidodiphosphate | 4 μMb (bovine heart MF1-ATPase) (109) |
| 8-Azido-AMP-PNP | C10H15N9O12P3 | 8-Azidoadenyl-5′-yl imidodiphosphate | 460 μMb (bovine heart MF1-ATPase) (109) |
| NAP4-ADP | C20H24N10O13P2 | N-4-Azido-2-nitrophenyl-γ−aminobutyryl-ADP | 2.0 μMa (bovine heart SMP-oxidative phosphorylation) (355); 0.6 mMb in the dark (bovine heart MF1-ATPase) (244); 0.5 μMb (bovine heart SMP-oxidative phosphorylation) (355) |
| NAP4-AMP-PNP | C20H23N11O15P3 | Nap4-PPNHP; NAP4-AdoPP[NH]P; N-4-azido-2-nitrophenyl γ−aminobutyryl-5-adenylyl imidodiphosphate; N-4-Azido-2-nitrophenyl-γ-aminobutyryl-AdoPP[NH]P | 3 μMc in the dark (bovine heart MF1-ATPase) (247) |
| NAP3-ATP | C19H23N10O16P3 | 3′-O-{3-[N-(4-Azido-2-nitrophenyl) amino]propionyl}adenosine 5′-triphosphate; arylazido aminopropionyl ATP | 43% maximum inhibition at 36 μM with 15 min photoreaction (bovine heart MF1-ATPase) (341) |
| NAP3-ADP | C19H22N10O13P2 | 3′-O-[3-[N-(Azido-2-nitrophenyl) amino]propionyl]adenosine 5′-diphosphate; arylazido-β-alanyl-ADP; arylazido aminopropionyl ADP | 80% inhibition at 50 μM in the dark (pig heart MF1-ATPase) (117) |
| NAB-ATP | C17H18N9O16P3 | 3′(2′)-O-(2-Nitro-4-azidobenzoyl)adenosine 5′-triphosphate | Km of 0.85 mM, maximal 40-45% modification (bovine heart MF1-ATPase) (216) |
| NAB-GTP | C17H18N9O17P3 | 3′(2′)-O-(2-Nitro-4-azidobenzoyl)guanosine 5′-triphosphate | Maximal 40-45% modification (bovine heart MF1-ATPase) (216) |
| ANP-ADP | C19H27N9O13P2 | 3′-O-[3-(4-Azido-2-nitrophenyl)propionyl]-ADP | 1.3 μMa (bovine heart SMP-oxidative phosphorylation) (355); 50 μMb (bovine heart MF1-ATPase) (426); 0.2 μMb (bovine heart SMP-oxidative phosphorylation) (355, 426); full inhibition at 3 mol/mol F1 (bovine heart MF1-ATPase) (426) |
I50.
Ki.
Kd.
Linear-benzoadenosine diphosphate (lin-benzo-ADP) is a fluorescent adenine-modified ADP analog in which the adenine ring is laterally extended by the insertion of a benzene ring between the pyrimidine and imidazole ring (Fig. 9A) (199, 428). Lin-benzo-ADP binds to all six nucleotide binding sites. The affinities for lin-benzo-ADP to three α subunits and one β subunit of MF1 from bovine heart are low (Kd = 1 to 2 μM), whereas the affinities for the other two β subunits are very high (Kd <10 nM) (428). Inhibition by lin-benzo-ADP is competitive and has complex kinetics of inhibition. Lin-benzo-ADP is fluorescent, and its fluorescence spectrum is extensively quenched by adding F1. As expected, this fluorescence quenching is reversed by adding ADP (199).
5′,5′-Diadenosine oligophosphates (APxA) are compounds which have a chain of phosphoryl groups linking two adenosine moieties. The APxA that have a long chain of phosphoryl groups (AP4A, AP5A, and AP6A) has been shown to inhibit the ATP hydrolysis activity of MF1, whereas compounds that have a shorter chain (AP2A and AP3A) showed stimulatory effects (417). The inhibition by AP4A, AP5A, and AP6A required the presence of at least one vacant noncatalytic site, and the maximal level of inhibition was 80%. AP4A was the most potent, and its stoichiometry for maximal inhibition was near 1 mol/mol of F1. In contrast, a contradictory result has also been reported in the inhibition of the same enzyme by AP5A, and no inhibition was observed up to 100 μM (325).
AMP-PNP is a nonhydrolyzable ATP analog in which the terminal bridge oxygen of the triphosphate moiety is replaced by an NH group (444). AMP-PNP has been used widely in kinetic studies of F1 and has been found to be a potent competitive inhibitor in ATPase assays of either the soluble or membrane-bound enzyme from bovine heart (37, 147, 306, 361) However, AMP-PNP is reported to be noncompetitive in ATPase assays with membrane-bound rat liver F1 (361). The Ki values reported are variable (14 nM to 0.5 μM) (37, 84, 255, 306, 361). AMP-PNP has no effect on the ATP synthesis activity of ATP synthase, although it is a potent inhibitor of F1-catalyzed ATP hydrolysis (302, 306). It binds to both catalytic and noncatalytic sites, and when it is bound to the latter sites, it induces hysteretic inhibition to the same extent as ADP (34, 37).
Guanylyl imidodiphosphate (GMP-PNP) and inosine-5′-[(β,γ)-imido]triphosphate (IMP-PNP) are analogs of GTP and ITP, respectively, in which the bridge oxygen atom between the β and γ phosphorus atoms is replaced by an NH group. The inhibition by GMP-PNP versus GTP and ITP is competitive (361), whereas inhibition versus ATP is competitive (37) or mixed (361). Unlike AMP-PNP, GMP-PNP shows no induction of hysteretic inhibition (34). IMP-PNP inhibits ITP hydrolysis potently, whereas it inhibits ATP hydrolysis only at low concentrations of ATP below 100 μM (362). At high concentrations of ATP, IMP-PNP stimulates the rate of ATP hydrolysis. In contrast, the stimulation of ATP hydrolysis by IMP-PNP is not seen in the presence of bicarbonate, and IMP-PNP inhibits ATP hydrolysis competitively.
Adenosine 5′-methylenediphosphate is an analog of ADP in which the bridge oxygen atom between the α and β phosphorus atoms is replaced by a CH2 group. Adenosine 5′-methylenediphosphate inhibits ATP synthesis competitively with respect to ADP (254, 363) and inhibits Pi↔ ATP exchange uncompetitively (363).
Exchange-inert metal-nucleotide complexes are stable, inert octahedral complexes of Cr(III), Co(III), or Rh(III) with ATP and ADP (383). The exchange-inert metal-nucleotide complexes inhibit ATP synthase by binding to F1 (44, 158, 383, 384, 432). Chromium complexes of ATP and ADP, i.e., α,β-CrADP, β,γ-CrATP, and α,β,γ-CrATP, are competitive inhibitors of MF1 with respect to MgATP (383, 432). β,γ-CrATP and α,β,γ-CrATP inhibit F1 by binding at the catalytic site and α,β-CrATP by binding at a regulatory site (432). The binding sites show no significant selectivity for the steric arrangement of the chromium complexes. β,γ-CrATP and α,β,γ-CrATP bind to the catalytic site with the same affinity, although they have different steric arrangements of the chromium (β,γ-CrATP with monocyclic coordination at the metal ion and α,β,γ-CrATP with bicyclic coordination). Two diastereomers of α,β-CrADP (Λ and Δ isomers) also exert similar inhibitory effects (432). Monodentate Cr(NH3)4ATP, bidentate/tridentate RhATP, bidentate Cr(NH3)4ATP, and bidentate Co(NH3)4ATP are mixed noncompetitive inhibitors of F1 (44, 158, 383, 384). All the amine and aqua exchange-inert metal-nucleotide complexes are mutually exclusive during ATP hydrolysis and appear to bind the same site(s) (383).
3′-acetyl ATP and 3′-acetyl ADP are monoacetylated adenine nucleotides in which an acetyl group is attached to the 3′ hydroxyl group of ribose. 3′-Acetyl ATP and 3′-acetyl ADP inhibit the ATPase activity of MF1 in a competitive fashion with ATP and ADP, respectively (355, 394). They bind to catalytic sites, but no reactions occur; i.e., the 3′-acetyl ADP is not phosphorylated, and the 3′-acetyl ATP is not hydrolyzed (355).
3′-O-[1-(5-dimethylamino)-naphthoyl]ADP (F-ADP or DMAN-ADP) and 3′-O-(1-naphthoyl)ADP (N-ADP) are fluorescent analogs of ADP in which 5-dimethyl amino-naphthoyl and naphthoyl groups are attached to the 3′ hydroxyls of ribose, respectively (356, 397, 427). Both inhibitors are potent competitive inhibitors of both ATP hydrolysis and ATP synthesis and exhibit a much stronger inhibition of ATP synthesis than of ATP hydrolysis (356, 397). F-ADP binds to three sites in bovine heart MF1 with Kd values of 50 nM for all sites, whereas the N-ADP binds to two sites with Kd values of 20 to 50 nM (397). F-ADP binds approximately 10 times more strongly than F-ATP (3′-O-[1-(5-dimethylamino)-naphthoyl]ATP), whereas F-AMP (3′-O-[1-(5-dimethylamino)-naphthoyl]AMP) is not inhibitory (356). ANA-ADP (3′-O-[5-azidonaphthoyl]-ADP) is a photoreactive analog of N-ADP (Fig. 9B and Table 18). It binds to the same site as N-ADP but with a lower affinity, i.e., about 2.5 times lower than the Ki of N-ADP for bovine heart MF1. Upon illumination, ANA-ADP rapidly photoinactivates F1 (240).
3′-O-(4-Benzoyl)benzoyladenosine 5′-triphosphate (BzATP) and BzADP are ribose-modified photoactivatable analogs of ATP and ADP in which a photoreactive (4-benzoyl)benzoyl group is attached to the 3′ hydroxyls of ribose (Fig. 9A) (435). BzATP binds to the ATP synthase β subunits both isolated and complexed but binds only to isolated α subunits (33). BzATP and BzADP bind to the catalytic site as competitive and reversible inhibitors in the absence of illumination. However, under actinic illumination, BzATP and BzADP inactivate F1 irreversibly by covalently modifying the catalytic site (3, 8, 435).
Other 3′-O-substituted adenine nucleotides include 3′-O-phenylacetyl-ADP, 3′-O-phenylbutyryl-ADP, 3′-O-benzoyl-ADP, 3′-O-[N-(2-nitrophenyl)-γ-aminobutyryl]-ADP, 3′-O-[N-(4-nitrophenyl)-γ-aminobutyryl]-ADP, 3′-O-naphthoyl-(1)-ADP, 3′-O-naphthoyl-(1)-ATP, 3′-O-naphthoyl-(2)-ADP, 3′-O-naphthyl-(1)-acetyl-ADP, 3′-O-naphthyl-(2)-acetyl-ADP, 3′-O-5-dimethylaminonaphthoyl-(1)-ADP, 3′-O-5-dimethylaminonaphthoyl-(1)-ATP, 3′-O-anthranoyl-(1)-ADP, and 3′-O-anthranoyl-(9)-ADP (356). These inhibitors inhibit oxidative phosphorylation in bovine heart SMP with Ki values in the range of 0.3 to 5.9 μM (Table 17).
The fluorosulfonylbenzoyl nucleotides 5′-p-fluorosulfonylbenzoylinosine (FSBI), 5′-p-fluorosulfonylbenzoyladenosine (FSBA), and 5′-p-fluorosulfonylbenzoylethenoadenosine (FSBɛA) bind to F1 and inactivate the enzyme by modifying amino acid side chains of α and/or β subunits. FSBI binds to the β subunit reversibly and reacts covalently with a Tyr residue. The inactivation follows pseudo-first-order kinetics, and the residues modified are βTyr345 in bovine heart MF1 (49, 57) and βTyr364 in F1 from thermophilic bacterium PS3 (50). The modification of a Tyr residue in a single β subunit is sufficient to inactivate F1 completely (49).
FSBA binds reversibly to a single binding site on the β subunit of MF1 (101). This inactivates F1 irreversibly by forming a covalent bond via a process that follows pseudo-first-order kinetics (51, 101). The modified residues are αTyr244, αTyr300, and either βTyr368 or βHis427 (51, 56, 114, 407). The complete inactivation of F1-ATPase by FSBA requires the modification of all three copies of the β subunits, in contrast to that by FSBI (49). 8-azido-FSBA (5′-p-fluorosulfonylbenzoyl-8-azidoadenosine) binds to MF1 in the absence of light and inhibits ATPase activity. Upon illumination of the dark-inactivated F1, 8-azido-FSBA induces in high yield cross-linking between βHis427 and βTyr345 within the same β subunit (453).
FSBɛA binds to αTyr244 of MF1, inactivating ATPase activity with pseudo-first-order kinetics (152, 414). Maximal inactivation is achieved when FSBɛA modifies αTyr244 in one or two copies of the subunit. Inactivation of F1 by both FSBA and FSBɛA is stimulated by high concentrations of phosphate, whereas inactivation by FSBI is not greatly affected. Prior modification of F1 with FSBA completely prevents modification of αTyr244 by FSBɛA, while prior inactivation with FSBI allows considerable modification.
Adenosine oligophospho-pyridoxal compounds (APxPL) contain a chain of phosphoryl groups linking adenosine and pyridoxal moieties. Adenosine triphospho-pyridoxal (AP3-PL) binds to the catalytic sites of EF1 and inhibits hydrolytic activity by modifying α and β subunits. The stoichiometric ratio of binding of AP3-PL for complete inactivation of F1 is about 1 mol of AP3-PL per 1 mol F1 (288). Addition of Mg2+ increases the inhibitory potencies of AP3-PL and also causes a change in the ratio of modification of α and β subunits by AP3-PL from 4:1 in the absence of Mg2+ to 1:3 in its presence (184). The residues modified by AP3-PL are αLys201, βLys155, and βLys201 (184, 281, 390). Adenosine tetraphospho-pyridoxal (AP4-PL) binds to EF1 with the same concentration for half-maximal inactivation as AP3-PL and shows essentially the same absorption spectrum and binding kinetics (288). Adenosine diphospho-pyridoxal (AP2-PL or PLP-AMP) is a weak inhibitor compared to AP3-PL (288). It binds to αLys201 in the isolated α subunit from E. coli with a maximal stoichiometry of approximately 1 mol/mol (Kd of ∼150 μM). It also impairs the reconstitution of α subunits with β and γ subunits.
The 2′,3′-dialdehydes of ATP, ADP, and AMP (oATP, oADP, and oAMP) are periodate-oxidized derivatives of ATP, ADP, and AMP in which the ribose ring is opened (Fig. 9A). In the presence of Mg2+, oATP is a substrate and acts as a competitive inhibitor of ATP hydrolysis. Prolonged incubation of the enzyme with oATP inactivates F1-ATPase activity irreversibly with pseudo-first-order kinetics by modifying both α and β subunits (95, 219, 239). Similar inactivation kinetics are also observed with oADP, but the kinetics of inactivation are the same whether Mg2+ is present or absent (95). The type of subunits and stoichiometry for the binding of oADP to F1 are somewhat controversial; the binding of oADP to both α and β subunits with a stoichiometry of 2 to 3 mol oADP/mol F1 (95, 239) and the binding of oADP only to α subunits with a stoichiometry of 0.9 to 1 mol oADP/mol F1 (217) both have been proposed. oAMP also inactivates F1, while AMP is not a substrate for F1. Finally, both oADP and oAMP inactivate F1 more efficiently than does oATP (Table 17).
Cibacron blue and 4-benzoyl(benzoyl)-1-amidofluorescein (BzAF) are structural analogs of purine nucleotides. They bind to MF1 and inhibit ATPase activity (28, 297). BzAF contains a benzophenone moiety on one side of the molecule that is excitable by irradiation at ∼340 to 366 nm, and the irradiation of BzaF leads to the covalent insertion of BzAF into F1. BzAF also contains a fluorescein moiety on the other side of the molecule that fluoresces at >515 nm upon excitation at ∼460 to 490 nm. BzAF inhibits mitochondrial ATP synthase as a catalytic site-specific covalent modifying agent (297). Like BzATP, BzAF binds to F1 competitively with respect to ATP in the absence of illumination and forms a covalent bond with F1 upon actinic irradiation. The photoinactivation of F1 by BzAF follows pseudo-first-order kinetics.
8-Azido-ATP and 8-azido-ADP are adenine-modified analogs of ATP and ADP in which an azido group is attached to the carbon 8 of adenine (Fig. 9B). 8-Azido-ATP is a substrate of F1 and is hydrolyzed slowly by F1 in the dark (420). The Km for 8-azido-ATP is similar to that for ATP, but the Vmax of hydrolysis with 8-azido-ATP is only 6% of that observed with ATP (bovine heart MF1) (371). On irradiation at 350 to 360 nm, the 8-azido-ATP inactivates F1-ATPase by binding covalently to F1, where both α and β subunits are modified. About 2.5 to 3 times more 8-azido-ATP is bound to β than to α subunits in MF1 (175, 371), whereas almost equal amounts are bound at these two subunits in CF1 (421). The modified residues in the β subunit of bovine heart F1 are Lys301, Ile304, and Tyr311 (175). F1-ATPase activity is completely inhibited when 2 mol 8-azido-ATP binds per mol F1. Moreover, Mg2+ is not required for the binding (420). Interestingly, 8-azido-ADP is phosphorylated by ATP synthase in SMP at a very low rate in the dark. The Ki for 8-azido-ADP is about 1 mM for mitochondrial F0F1 from bovine heart, whereas the Ki for ADP is ∼20 nM for MF1 from the same source (371). Photolysis at 350 nm leads to the inactivation of ATP synthase, as the 8-azido-ADP preferentially binds to β subunits (133, 371). The ATPase activity of F1 is completely inhibited at 2 mol of 8-azido-ADP bound per 1 mol F1 (419). In the presence of fluoroaluminate, 8-azido-ADP modifies βTyr-345 (133).
2-Azido-ATP and -ADP are also adenine-modified analogs of ATP and ADP in which an azido group is attached to carbon 2 of adenine. 2-Azido-ADP photolabels β subunits exclusively upon photoirradiation, in contrast to 8-azido-ADP or -ATP, which modify both α and β subunits (86, 89, 419, 421). 2-Azido-ADP binds to F1 with an affinity similar to the affinity of ADP (45), and upon irradiation it modifies βLeu342, βIle344, βTyr345, βPro346, or βTyr368 (bovine heart MF1) (111, 132).
2- and 8-Azidoadenyl-5′-imidodiphosphate (2-azido-AMP-PNP and 8-azido-AMP-PNP) are derivatives of AMP-PNP. They bind to F1 at what appear to be both catalytic and noncatalytic sites (109). Under nonphotolytic conditions, 2-azido-AMP-PNP has a much higher inhibitory potency (Ki = 4 μM) than 8-azido-AMP-PNP (Ki = 460 μM).
3′-Arylazido butyryl ADP (NAP4-ADP) is a photoreactive derivative of ADP in which a photosensitive N-4-azido-2-nitrophenylaminobutyryl group is attached to the adenine ring of ADP (244). NAP4-ADP is a competitive inhibitor with respect to ATP, with a Ki value of 0.6 mM (bovine heart MF1). NAP4-ADP is a moderate inhibitor in the dark. However, upon photoirradiation with visible light, it inactivates F1 by binding covalently to both α and β subunits. NAP4-AMP-PNP (or NAP4-AdoPP[NH]P) is an analog of NAP4-ATP containing an NH group that replaces oxygen at the position of the terminal bridge oxygen of the triphosphate chain. NAP4-AMP-PNP binds to F1 with high affinity, and upon illumination, it inactivates F1 by covalently modifying α and β subunits (247). NAP4-AMP-PNP preferentially modifies the α subunit(s) at low concentrations, whereas it modifies α and β subunits equally at high concentrations.
3′-O-[3-[N-(Azido-2-nitrophenyl)amino]propionyl]ATP (NAP3-ATP) and NAP3-ADP are analogs of ATP and ADP in which a photoreactive N-4-azido-2-nitrophenylaminopropionyl group is attached to the adenine ring. NAP3-ATP acts as a substrate in the dark and shows photodependent inhibition associated with covalent modification of F1 upon illumination (117, 341). In contrast, NAP3-ADP, just like ADP, induces hysteretic inhibition of soluble F1 and membrane-bound F1, with the latter being more sensitive (117). The kinetics of inhibition is biphasic. Preincubation of MF1 from pig heart with NAP3-ADP in the dark inhibits ATPase activity about 80%, a value that is increased to 87% upon photoirradiation (117).
3′(2′)-O-(2-Nitro-4-azidobenzoyl)ATP (NAB-ATP) and NAB-GTP are 3′(2′)-O-(2-nitro-4-azidobenzoyl)-derivatives of ATP and GTP in which a 2-nitro-4-azidobenzoyl group is attached to the 2′ hydroxyls of ribose. NAB-ATP binds to the catalytic site of F1 and is hydrolyzed to NAB-ADP and inorganic phosphate (216). After hydrolysis, NAB-ADP remains bound to F1, whereas phosphate is dissociated. The F1·NAD-ADP complex is inactive, but in the presence of ATP, the bound NAB-ADP is released, resulting in the reactivation of ATPase activity. Illumination (300 to 380 nm) of F1 inhibited with NAB-ADP leads to its covalent binding to the enzyme. NAB-GTP has an inhibitory activity similar to that of NAB-ATP.
3′-O-[3-(4-Azido-2-nitrophenyl)propionyl]-ADP (ANP-ADP) is a photoreactive analog of ADP in which a 4-azido-2-nitrophenyl propionyl group is attached to the 3′ hydroxyls of ribose (Fig. 9B). ANP-ADP binds to nucleotide binding sites on F1, inhibiting both ATP hydrolysis and ATP synthesis (355, 426). Inhibition of F1 by ANP-ADP is competitive with ADP in the dark, but upon illumination, ANP-ADP inactivates F1 by covalently modifying α and β subunits. The stoichiometry for complete photoinactivation of F1 is 3 mol of ANP-ADP/mol of F1. The inhibition of F1 by the photolabeling is reversed by mild alkaline treatment due to the hydrolysis of the 3′-ester bond and release of the ADP moiety of the inhibitor (426).
AMINO ACID MODIFIERS
Amino Group Modifiers
Phenylglyoxal and butanedione are dicarbonylic Arg residue modifiers. They inactivate both membrane-bound and isolated F1 (Fig. 10A and Table 19) (43, 128, 129, 162, 248, 375, 381, 385). Inactivation by these agents follows pseudo-first-order kinetics (67, 128, 129, 248). Although the rate of inactivation is decreased in the presence of ADP and ATP (67, 128, 398), it is not significantly influenced by the presence of phosphate (398). Phenylglyoxal and butanedione also inhibit ATP↔ Pi exchange activity (43, 128, 162, 248, 385). Only one molecule of reagent per F1 active site is required for inactivation, with the binding site(s) believed to be located at or near this active site (128, 248).
FIG. 10.
Structures of amino acid residue modifiers. (A) Amino group modifiers. FDNB, 1-fluoro-2,4-dinitrobenzene. (B) Carboxyl group modifiers. CMCD, 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide metho-p-toluenesulfonate. (C) Cys/Tyr residue modifiers. DTNB, 5,5′-dithiobis(2-nitrobenzoic acid). (D) His residue modifiers.
TABLE 19.
Amino group modifiers
| Name or abbreviation | Molecular formula | Other names | Inhibitory potency, I50 (reference) |
|---|---|---|---|
| Phenylglyoxal | C8H6O2 | Benzoylcarboxaldehyde; phenylglyoxal; benzoylformaldehyde; phenylethanedione; α-oxobenzeneacetaldehyde | 25% inhibition at 2.7 μmol/mg protein (bovine heart SMP-ATPase) (162); 47.5% inhibition at 3 mM (E. coli F0F1-ATPase after F0 modification) (381); 33.5% inhibition at 20 mM (E. coli F0-liposome proton uptake) (381) |
| Butanedione | C4H6O2 | Diacetyl; dimethyl glyoxal; 2,3-butanedione; dimethyl diketone; butadione | 0.63 μmol/mg protein and ∼100% inhibition at 1.7 μmol/mg protein (bovine heart SMP-ATPase) (162) |
| FDNB | C6H3FN2O4 | 1-Fluoro-2,4-dinitrobenzene; dinitrofluorobenzene; 2,4-DNFB; 2,4-dinitro-1-fluorobenzene; 2,4-dinitrofluorobenzene; fluoro-1,3-dinitrobenzene; Sanger's reagent | 96% inhibition at about four 2,4-dinitrophenyl labels (bovine heart MF1-ATPase) (194) |
| Dansyl chloride | C12H12ClNO2S | 5-(Dimethylamino)-naphthalene-1-sulfonyl chloride; 1-chlorosulfonyl-5-dimethylaminonaphthalene; 1-dimethylaminonaphthalene-5-sulfonyl chloride; dansyl; DNS chloride |
1-Fluoro-2,4-dinitrobenzene is a Lys residue modifier that inhibits the hydrolytic activity of MF1 (11, 194, 250, 399). It modifies Lys162 (bovine sequence) in the P loop, the same residue to which the nitrobenzene (NBD) group migrates at pH 9 (194). Inhibition of ATPase activity follows first-order kinetics (399), with about four 2,4-dinitrophenyl labels required for 96% inhibition (194). Inhibition is reversed nearly 50% by dithiothreitol (11) and is protected effectively by ATP or Pi and slightly by ADP (399).
Dansyl chloride is an acyl chloride of 5-dimethylamino-1-naphthalenesulfonic acid. It modifies reactive amino groups of proteins. Dansyl chloride binds to MF1 and inhibits both ATP synthesis and membrane-bound ATPase activity to approximately the same extent (250).
Carboxyl Group Modifiers
Carbodiimides are compounds containing a N=C=C functional group. Some inhibit ATP synthase by modifying carboxyl residues residing within F1, F0, or both (Fig. 10B). DCCD and N-(2,2,6,6-tetramethylpeperidyl-1-oxyl)-N-(cyclohexyl)carbo-diimide (NCCD) are lipid-soluble carbodiimides. DCCD binds to both F1 and F0 of ATP synthases from mitochondria and some bacteria (137, 204, 400, 441) (Table 20). F1 from some bacteria, such as Helicobacter pylori, are insensitive to DCCD (36). DCCD reacts covalently with DCCD-sensitive F1 via a Glu residue in the β subunit. In F1 from E. coli, βGlu192 binds DCCD, while in bovine MF1, βGlu199, corresponding to E. coli βGlu192, is modified. In F1 from thermophilic Bacillus, βGlu181 (E. coli sequence) rather than βGlu192 is modified (137, 400, 441). Incorporation of 1 mol of DCCD into 1 mol of F1 results in 95% inhibition of the ATPase activity of EF1, and 2 mol of DCCD/mol F1 leads to complete inhibition (400). In the crystal structure of the F1-DCCD complex from bovine heart mitochondria, one molecule of DCCD is bound per F1 (137). In this structure, the βGlu199 of βDP located at the interface between βDP and αDP is modified. The covalently modified DCCD (dicyclohexyl-N-acylurea) is bound in a hydrophobic cleft with one face exposed to the solvent. Residues βVal164, βMet167, βVal420 and βPhe424 contribute to the binding of DCCD, and the steric hindrance involved is believed to inhibit F1 by blocking a conformational change from βDP to βE.
TABLE 20.
Carboxyl group modifiers
| Name or abbreviaion | Molecular formula | Other names | Inhibitory potency (reference) |
|---|---|---|---|
| DCCD | C13H22N2 | 1,3-Dicyclohexylcarbodiimide; N,N′-dicyclohexylcarbodiimide; bis(cyclohexyl)carbodiimide; carbodicyclohexylimide; N,N′-methanetetraylbiscyclo-hexaamine | 1.2 μg of inhibitor/mg proteina (S. cerevisiae SMP-ATPase) (150); 1-5 μg of inhibitor/mg proteina (T. pyriformis SMP-ATPase) (404); 200 μMa in less than 5 min and at ∼40 μMa in 30 min (R. rubrum F1-ATPase) (204); 1.9 μg/mg proteina (C. fasciculata SMP-ATPase) (439); 95% inhibition with 1 mol DCCD/mol F1 (EF1-ATPase) (400); maximal 70-80% inhibition at 30 μM (membrane-bound EF0F1-ATPase) (171); 47% inhibition at 5 μM (C. thermoaceticum membrane-bound F0F1-ATPase) (190); 97% inhibition with 2 mol inhibitor bound/mol F1 (bovine heart MF1-ATPase) (250); maximal inhibition at 1 mol inhibitor/mol F0 (bovine heart SMP-ATPase) (140); maximal inhibition at 2 mol inhibitor/mol F0 (bovine heart H+-translocation) (140); maximal inhibition at 1 mol inhibitor/mol F0 (E. coli membrane H+-translocation) (171) |
| NCCD | C16H28N3O | N-(2,2,6,6-Tetramethylpeperidyl-1-oxyl)-N(cyclohexyl)carbodiimide; N-(2,2,6,6-tetramethyl-1-oxypiperid-4-yl)-N′-cyclohexylcarbodiimide | 0.65 nmol/mg proteina (bovine heart SMP-ATPase) (24); 85% inhibition at 1 nmol NCCD/mg protein (bovine heart SMP-ATPase) (23) |
| CMCD | C14H28N3O·C7H8O3S | 1-Cyclohexyl-3-(2-morpholinoethyl)carbodiimidemetho-p-toluenesulfonate; N-cyclohexyl-N′-2-morpholinoethyl-carbodiimide-methyl-4-toluolsulfonate | 200 μMb (bovine heart MF1-ATPase) (186) |
| EDC | C8H17N3 | Ethyldimethylaminopropyl carbodiimide; 1-ethyl-3-[3-(dimethylamino)propyl] carbodiimide; (3-(dimethylamino) propyl)ethylcarbodiimide | 95% inhibition at 13 mol of EDC/mol F1 (EF1-ATPase) (236) |
| EEDQ | C14H17NO3 | N-Ethoxycarboxyl-2-ethoxy-1,2-dihydroquinoline | 200 μMa in less than 5 min and at ∼40 μMa in 30 min (R. rubrum F1-ATPase) (204); 70% inhibition at 400-600 μM (bovine heart MF1-ATPase) (250, 320); 75% inhibition at 400 μM (E. coli F1-ATPase) (320) |
| Woodward's reagent K | C11H11NO4S | 2-Ethyl-5-phenylisoxazolium-3′-sulfonate; N-ethyl-5-phenylisoxazolium-3′-sulfonate | 88% inhibition at 15 mM (E. coli F0-liposome proton uptake) (162) |
I50.
Kd.
DCCD, by binding F0 (35), also inhibits F0-mediated proton translocation and the ATPase activity of the coupled F0F1 complex. Here, DCCD is bound covalently to an essential carboxyl residue of subunit c at position 61 (E. coli sequence) (68, 122, 364). The stoichiometries for the maximal inhibition of function are 1 mol of DCCD/mol of F0, i.e., modification of 1 subunit c/F0 for inhibiting ATPase activity of ATP synthase and 2 mol of DCCD/mol F0 for inhibiting proton translocation (140, 171, 213).
NCCD is a lipid-soluble spin-labeled inhibitor of ATP synthase that targets the F0 of ATP synthase (23, 24). The binding site for NCCD is believed to be the same as that for DCCD, i.e., Asp61 of subunit c, as NCCD's binding to the ATP synthase is prevented by DCCD (24). Moreover, the mutant of Ala25 in subunit c, which is near Asp61, shows a greatly reduced inhibitory activity with NCCD (138).
1-Cyclohexyl-3-(2-morpholinoethyl)carbodiimide metho-p-toluenesulfonate and ethyldimethylaminopropyl carbodiimide (EDC) are water-soluble carbodiimides that modify a carboxyl group(s) in F1. 1-Cyclohexyl-3-(2-morpholinoethyl)carbodiimide metho-p-toluenesulfonate binds to F1 reversibly and likely modifies carboxyl groups near the catalytic sites (186). EDC inhibits F1 after modifying several carboxyl groups in β subunits. The inhibition by EDC is greatly reduced by Mg2+ (236). Incorporation of about 13 mol of EDC/mol F1 (E. coli) leads to 95% inhibition of ATPase activity. Here, two-thirds of the bound EDC is bound to β subunits, where it modifies multiple sites in a short segment (residues 162 to 194) (E. coli sequence) (236). EDC also promotes formation of intersubunit cross-links between subunits β and ɛ. The residues involved are βGlu381 and likely ɛSer108 (90).
N-Ethoxycarboxyl-2-ethoxy-1,2-dihydroquinoline (EEDQ) inhibits both MF1 and BF1 (Fig. 10B and Table 20) (204, 222, 250, 320, 322, 399). The inactivation by EEDQ is both pH and temperature dependent and also time and concentration dependent (204, 322). One mole of EEDQ binds to one mole of F1. The inactivation follows pseudo-first-order kinetics until 90 to 95% inactivation occurs (322). Inhibitions by EEDQ and DCCD are additive, suggesting that the binding sites of EEDQ and DCCD are either the same (204, 222) or located close to each other (320).
Woodward's reagent K inhibits both F1 and F0 (204, 381). The chemical modification of the β subunit of F1 from Rhodospirillum rubrum with this reagent results in loss of both phosphate and ATP binding capacities (203). However, ADP binding sites remain active. Chemical modification of F0 from E. coli by Woodward's reagent K inhibits both proton translocation and total ATPase activity (381).
Cys and Tyr Residue Modifiers
4-Chloro-7-nitrobenzofurazan (NBD-Cl) is a fluorescent adenine analog that labels Tyr or Cys residues (Fig. 10C and Table 21). It inhibits both the synthetic and hydrolytic activities of ATP synthases from bacteria, chloroplasts, and mitochondria by modifying an essential residue (βTyr311, bovine sequence) at the catalytic site(s) of F1 (12, 70, 119, 120, 245, 388, 415). Depending on the experimental conditions, other subunits, particularly the α subunit, are also modified by NBD-Cl (96, 121, 146, 283). In F1 modified by NBD-Cl, the Tyr-O-NBD linkage is unstable at alkaline pH. The NBD group from βTyr311 migrates to βLys162 in the P-loop at pH 9 as a consequence of O-to-N migration (13, 14, 121). The resulting NBD-N-Lys derivative of F1 is also catalytically inactive (14, 121). In a crystal structure of bovine MF1 covalently modified by NBD-Cl, the NBD-Cl is found in only one of three β subunits, βE (292). The βTyr311 residues in the βTP and βDP subunits are buried at the α-β subunit interfaces and are inaccessible to NBD-Cl. The NBD binding pocket is positioned in the central nucleotide binding domain with no hydrogen bonds between the NBD ring and the protein. NBD-Cl appears to inhibit F1 by preventing βE from undergoing a conformational change (292).
TABLE 21.
Cys/Tyr residue modifiers
| Name or abbreviation | Molecular formula | Other names | Inhibitory potency, I50 (reference) |
|---|---|---|---|
| NBD-Cl | C6H2ClN3O3 | NBF-Cl; 7-chloro-4-nitrobenzofurazan; 4-chloro-7-nitrobenzofurazan; 7-chloro-4-nitrobenzofurazan; 4-chloro-7-nitro-2,1,3-benzoxadiazole; 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole; 4-chloro-7-nitrobenzo-2-oxa-1,3-diazole | 4.5 μg of inhibitor/mg protein (T. pyriformis SMP-ATPase) (404); 68% inhibition at 50 μM (C. thermoaceticum F1-ATPase) (190); complete inhibition at 1 mol inhibitor bound/mol F1 (bovine heart F1-ATPase) (246); >90% inhibition at 1.4 mol inhibitor bound/mol F1 (TF1-ATPase) (415) |
| Tetranitromethane | CN4O8 | Tetan | 130 nmol/mg protein and ∼100% inhibition at 210 nmol/mg protein (bovine heart SMP-ATPase) (162); 2.5 mM (TF0 vesicle, proton conduction) (375); almost complete inhibition at 8 mM (TF0 vesicle, proton conduction) (375) |
| DFDNB | C6H2F2N2O4 | 1,5-Difluoro-2,4-dinitrobenzene; 4,6-difluoro-1,3-dinitrobenzene | Complete inhibition at 3 mol inhibitor/mol F1 (bovine heart MF1-ATPase) (7) |
| NEM | C6H7N1O2 | N-Ethylmaleimide; maleic acid N-ethylimide | ∼0.6 mM (S. pombe MF1-ATPase) (115); 74% inhibition at 1 mM (V. parahaemolyticus F1-ATPase) (344) |
| Bismuth subcitrate | C6H8O7Bi | CBS; colloidal bismuth subcitrate; tripotassium dicitratobismuthate | 73 μM (H. pylori F1-ATPase) (36) |
| Omeprazolea | C17H19N3O3S | 5-Methoxy-2-(((4-methoxy-3,5-dimethyl-2-pyridyl)methyl)sulfinyl)benzimidazole; Audazol; Omepral | 90 μM (without acid activation) and 43 μM (with acid activation) (H. pylori F1-ATPase) (36) |
| DTNB | C14H8N2O8S2 | 5,5′-Dithiobis(2-nitrobenzoic acid); dithionitrobenzoic acid; 2,2′-dinitro-5,5′-dithiodibenzoic acid; 3,3′-dithiobis(6-nitrobenzoic acid); dithiobisnitrobenzoic acid; Ellman's Reagent | 39% inhibition at 0.4 mM and 46% inhibition at 1.3 mM (bovine heart MF1-ATPase) (392) |
| PCMB | C7H5ClHgO2 | p-Chloromercuribenzoic acid; 4-carboxyphenylmercuric chloride; 4-chloromercuribenzoic acid | ∼90% inhibition at 4.5 mM (bovine heart SMP-ATPase) (438) |
| PCMS | C6H5ClHgO3S | p-Chloromercuribenzene sulfonate; 4-chloromercuribenzenesulfonate; PCMBS | 6 mM (bovine heart SMP-ATPase) (438) |
| Mersalyl | C13H16HgNO6.Na | O-((3-Hydroxymercuri-2-methoxypropyl)carbamoyl)phenoxy-acetic acid; (3-((2-(carboxymethoxy)benzoyl)amino)-2-methoxypropyl)hydroxymercury; mercuramide; mercusal; mersalyl acid | 70% inhibition at 130 μM (bovine heart MF0, proton conductivity) (445) |
| 2,2′-Dithiobispyridine | C10H8N2S2 | 2,2′-Dithiodipyridine; 2,2′-dipyridyl disulfide; 2PDS; bis(2-pyridinyl) disulfide | 55% inhibition at 1 mM (bovine heart MF0, proton conductivity) (445) |
| N-(7-Dimethylamino-4-methyl-coumarinyl)-maleimide | C16H14N2O4 | N-(4-Methyl-7-dimethylamino-3-coumarinyl)maleimide | 60% inhibition at 400 μM (bovine heart MF0, proton conductivity) (445) |
Omeprazole is converted to a cyclic sulfenamide with acid-activation.
Tetranitromethane and 1,5-difluoro-2,4-dinitrobenzene (DFDNB) modify Tyr residues. Tetranitromethane nitrates the Tyr residue of ATP synthase subunit c of the thermophilic bacterium PS3 and inhibits the proton conduction of TF0 (375). In contrast, tetranitromethane inhibits neither proton translocation nor ATPase activity of E. coli ATP synthase (381). However, DFDNB does inhibit the ATPase activity of MF1 (7, 55), with a molar ratio of 3 for complete inhibition. Here, inhibition is reversed by dithiothreitol. (7). Inactivation of F1 by DFDNB is believed to be due to modification of either βTyr311 (55) or another Tyr residue (7).
Thiol group reagents, N-ethylmaleimide (NEM), bismuth subcitrate, omeprazole, 5,5′-dithiobis(2-nitrobenzoic acid), p-chloromercuribenzoate (PCMB), p-chloromercuribenzene sulfonate (PCMS), mersalyl, 2,2′-dithiobispyridine, and N-(7-dimethylamino-4-methyl-coumarinyl)-maleimide inhibit ATP synthase by modifying Cys residues. Specifically, NEM inhibits the ATPase activity of F1s from fungi, some bacteria such as Vibrio parahaemolyticus, and some mitochondria, i.e., those from S. cerevisiae and Schizosaccharomyces pombe (115, 145, 344). The inactivation of F1 by NEM in sensitive cases is irreversible and protected by nucleotides (115). In contrast, the F1s from E. coli and bovine heart mitochondria are resistant to NEM (344, 366). NEM also binds various F0 polypeptides, inhibiting proton conduction (445). For example, NEM inhibits mitochondrial F0 from bovine heart while labeling 25-, 11-, and 9-kDa polypeptides (445).
Bismuth subcitrate and omeprazole are antiulcer drugs. They bind to sulfhydryl groups of F1 and form stable complexes (36). They inhibit the ATPase activity of F1 from Helicobacter pylori via a reaction that can be prevented and also reversed by mercaptan glutathione. At low pH, omeprazole is converted into a cyclic sulfonamide, and this form inhibits the ATPase activity of H. pylori F1 more potently than the form without acid activation (I50 = 43 μM when acid activated, compared to 90 μM without acid activation).
Regarding other sulfhydryl reactive agents, 5,5′-dithiobis(2-nitrobenzoic acid) inhibits the ATPase activity of nucleotide-depleted F1 (392). In contrast, it is inhibitory neither to native F1 nor to nucleotide-depleted F1 in the presence of either ADP or ATP.
PCMB, PCMS, and mersalyl are polar organic mercurials that target F0 of mitochondrial ATP synthase. Both PCMB and PCMS inhibit the ATP synthesis and ATPase activities of bovine heart ATP synthase. Thiols modified by the mercurials are different from those modified by NEM (438). In contrast to the case for NEM, inhibition by mercurials is reversed almost completely (PCMB) or partially (PCMS) by addition of dithiothreitol. Moreover, the binding of mercurials protects the ATP synthase from irreversible inhibition by DCCD. Mersalyl also inhibits proton conductivity by F0 from bovine heart mitochondria. Here, the inhibition is much more potent than that observed with PCMB and PCMS (445). Although mersalyl has no inhibitory effect at concentration of up to ≤50 μM, it inhibits proton conduction at higher concentrations (∼70% inhibition at 130 μM).
The sulfhydryl-reactive agents 2,2′-dithiobispyridine and N-(7-dimethylamino-4-methyl-coumarinyl)-maleimide also inhibit proton conductivity by F0 from bovine heart mitochondria (445). N-(7-Dimethylamino-4-methyl-coumarinyl)-maleimide has stronger inhibitory potencies than 2,2′-dithiobispyridine and NEM. N-(7-dimethylamino-4-methyl-coumarinyl)-maleimide shows no inhibition up to a concentration of 200 μM and inhibits proton conduction by 60% at 400 μM.
His Residue Modifiers
Diethyl pyrocarbonate and Rose bengal are His residue-modifying reagents (Fig. 10D and Table 22). Diethyl pyrocar-bonate modifies the ATP synthase β subunit, completely preventing the binding of phosphate. It also blocks the binding of ATP to a Mg2+-dependent low-affinity site (203, 381, 445). In contrast, the ADP binding capacity of the β subunit is not affected by modification with diethyl pyrocarbonate (203). Diethyl pyrocarbonate also modifies F0 from E. coli, inducing inhibition of proton uptake (381).
TABLE 22.
His and other amino acid residue modifiers
| Name | Molecular formula | Other names | Inhibitory potency, I50 (reference) |
|---|---|---|---|
| Diethyl pyrocarbonate | C6H10O5 | Baycovin; diethyl dicarbonate; diethyl oxydiformate; pyrocarbonic acid diethyl ester | >50% inhibition at 3 mM (E. coli F0, liposome proton uptake) (381) |
| Rose bengal | C20H2Cl4I4Na2O5 | Bengal rose | 75-85% inhibition at 0.2 μM (bovine heart F1-ATPase) (139) |
| Iodine | I2 | 40 μM (rat liver MF1-ATPase) (314) |
Rose bengal photooxidizes His residues of β subunits, causing conformational instability in F1 (139). About 60% of the His residues are photooxidized, causing 50% inactivation. This photochemical damage is prevented by various phenanthroline compounds.
Others
Iodine is an electron-dense heavy atom that reacts with and inactivates F1 (314). It behaves like a typical covalent inhibitor in its modification of amino acid residues. MgATP, MgADP, and phosphate fail to protect F1 from inhibition by iodine. Iodine preferentially labels the ATP synthase β subunit, although it also labels α and γ subunits to some extent. About 10 atoms of iodine are incorporated per F1 (rat liver mitochondria) under conditions where the labeling proceeds in a linear fashion. About two atoms of iodine are incorporated per β subunit.
PHYSICAL INHIBITORY FACTORS
High Hydrostatic Pressure
High hydrostatic pressure of above 60 to 80 MPa inactivates both F1 and the complete ATP synthase (F0F1) (Table 23) (105, 310, 377). At below 60 to 80 MPa, the hydrostatic pressure shows stimulatory effects on ATPase activity. However, both membrane-bound and isolated F0F1 from mitochondria are inhibited reversibly at high hydrostatic pressure, while soluble F1-ATPase is inactivated irreversibly due to reassociation with an altered hydrodynamic radius after decompression (105). In contrast to the case for the isolated mitochondrial ATP synthase, the inhibition of the isolated ATP synthase from chloroplasts is irreversible, showing no restoration after decompression (377). The inactivation is dependent on protein concentration (377). Inhibition by high hydrostatic pressure is believed to be associated with dissociation that impairs contacts essential for transmission of conformational information between those subunits needed for rotational catalysis (105, 377).
TABLE 23.
Physical inhibitory factors
| Factor | Inhibitory potency (reference) |
|---|---|
| Hydrostatic pressure | 850 barsa (bovine heart SMP-ATPase, 0.02 mg/ml) (105) |
| Far-UV irradiation | ∼80% inhibition within 15 min at 254 nm (bovine heart SMP-ATPase) (75) |
| Cold temp | 15-60 minb at 4°C (324), 4-40 mina at 0°C with different prepn (bovine heart MF1-ATPase) (308, 309) |
I50.
Half-life.
UV Irradiation
Mitochondrial ATPase activity is inhibited also by far-UV irradiation. UV light at 254 nm results in a time-dependent inhibition of both membrane-bound and soluble F1. Inhibition reaches its maximum level within 15 min after exposure of SMP to UV (75). This also induces the release of tightly bound adenine nucleotides from F1. Succinate, a substrate for the electron transport chain, partially protects against the detrimental effects of UV. Inhibition by UV is due to the photochemical modification of the essential Tyr residue located at the active site of F1 that induces subsequent structural changes in F1.
Low Temperature
The F1“catalytic” moiety of the ATP synthase (F0F1) is cold labile (308, 309, 324). Its ATPase activity decrease rapidly upon incubation at low temperature. The rate of inactivation is first order, and the half-life varies between 15 and 60 min with different preparations (324). The inactivation is not protected by ATP, ADP, or Mg2+ and is reversed by rewarming the enzyme solution under appropriate conditions (309). The inactivation by cold temperature is associated with the dissociation of the enzyme complex into subunits (309).
MISCELLANEOUS INHIBITORS
Polyborates are boron cluster compounds with a unique molecular structure and unusual chemical properties. Among the polyborates, dodecaborates ([B12H12]2−) and dicarbononaborates ([C2B9H11]−) inhibit ATPase activity of MF1, and dicarbononaborates have much stronger inhibitory potencies than dodecaborates (Fig. 11) (104). One of the dicarbononaborates, dichlorodicarbononaborate ([Cl2C2B9H10]−), that contains two chlorides inhibits competitively with respect to ATP the ATPase activities of both membrane-bound and soluble F1. The inhibition is due to a direct interaction of the reagent with the catalytic F1 moiety (104).
FIG. 11.
Miscellaneous inhibitors. TCS, tetrachlorosalicylanilide.
Almitrine is a piperazine-like agent that is known to be a respiratory stimulant that enhances respiration by acting as an agonist of peripheral chemoreceptors located on the carotid bodies. This agent inhibits mitochondrial ATP synthase in an uncompetitive manner (336). Also, it does not destroy the electrochemical proton gradient across the mitochondrial membrane that normally drives ATP synthesis (333-335). Thus, mitochondria treated with this agent remain intact despite the fact that this agent has debilitated their ATP synthase.
5-Hydroxynaphthalenedicarboxylic anhydride (HNA) inhibits the mitochondrial ATPase activity induced by 2,4-dinitrophenol and the ATPase activity of SMP induced by Mg2+ (165). HNA also inhibits the ATP-energized mitochondrial volume change. The inhibitory effects of HNA are similar to those of rutamycin.
R207910 is a diarylquinoline drug that has antimycobacterial activity. It inhibits mycobacterial ATP synthase and targets subunit c in F0 (15, 215, 313). The site of action of R207910 seems to be located close to an essential carboxyl residue, Asp61 of subunit c (E. coli sequence), as the mutations Asp32Val (Mycobacterium smegmatis) and Ala63Pro (M. tuberculosis) confer resistance to the drug. Also, the mycobacterial species naturally resistant to R207910 contains Met at position 63 in place of a conserved Ala in all sensitive mycobacteria (15, 181, 313). R207910 is an enantiomeric compound with two chiral centers. It adopts the lowest-energy conformation with the carbon alpha relative to the quinoline moiety R and the carbon beta S (135). The binding of the inhibitor to the binding site in ATP synthase is stereoselective, and its (S,R) stereoisomer is 2 orders of magnitude less inhibitory than R207910 (215). R207910 appears to act specifically on mycobacteria, and the range of MICs of R207910 is 0.03 to 0.12 μg/ml for 99% inhibition of the growth of M. tuberculosis strains (15). The killing effect of M. tuberculosis by R207910 is time dependent rather than concentration dependent (15), and R207910 acts synergically when combined with other tuberculous drugs (183, 237, 238).
Spegazzinine is a dihydroindole alkaloid from Aspidosperma chakensis Spegazzini (103). It inhibits uncompetitively the ATPase activities of both membrane-bound and isolated CF1 from spinach (10). Spegazzinine inhibits both cyclic and noncyclic photophosphorylation of isolated spinach chloroplasts. It also inhibits the mitochondrial ATPase activity of S. pombe (234) and slightly inhibits the mitochondrial ATPase activity of Tetrahymena pyriformis ST (404). In contrast, spegazzinine has no inhibitory effects on the ATPase activities of ATP synthases from Clostridium pasteurianum (78), Tritrichomonas foetus (235), and mitochondria of Crithidia fasciculata (439).
n-Butanol inhibits the ATPase activities of both membrane-bound and soluble MF1 (406). It inhibits the isolated F1 at the same or lower concentrations as it inhibits membrane-bound F1. Inhibition is temperature dependent. N-Butanol also shows partial inhibition of ATP synthesis.
Tetrachlorosalicylanilide is a lipophilic weak acid known as an H+ conductor. It inhibits the ATPase activities of both isolated F1 and F0F1 from Vibrio parahaemolyticus (290, 344). The concentration of tetrachlorosalicylanilide for 50% inhibition of F0F1-ATPase activity from V. parahaemolyticus is about 9 to 10 μM (290).
Dihydrostreptomycin is a polycationic aminoglycoside antibiotic drug produced from Streptomyces humidus. It significantly stimulates the ATPase activity of membrane-bound ATP synthase from bovine heart mitochondria in the concentration range of 1 to 5 mM. The stimulation is followed by inhibition at higher concentrations (161). Dihydrostreptomycin also inhibits the ATPase activity of isolated F1, but the stimulation of the ATPase activity observed in the inhibition of membrane-bound F1 at low concentrations of dihydrostreptomycin is not observed in the inhibition of isolated F1. The inhibition of ATPase activity of F1 by dihydrostreptomycin is noncompetitive. Dihydrostreptomycin also exhibits partial inhibition of proton conductivity of F0 in the ATP synthase devoid of its catalytic F1 moiety.
Suramin, a synthetic antiparasitic drug, is an inhibitor of various proteins in different cell types and also inhibits the binding of some growth factors to their receptors. Suramin also binds to ATP synthase and inhibits both F1-ATPase and membrane-bound F0F1-ATPase from mitochondria (28, 173). Suramin acts as a noncompetitive inhibitor of the membrane-bound ATPase and as a strictly competitive inhibitor of purified F1-ATPase (173). Half-maximal inhibition of rat liver F1-ATPase occurs at 40 μM suramin.
Bz-423 is an 1,4-benzodiazepine derivative known as a cytotoxic immunomodulatory drug that suppresses disease in lupus-prone mice by inducing apoptosis in autoreactive B and T lymphocytes (193). Bz-423 binds to the OSCP subunit of ATP synthase and inhibits both synthetic and hydrolytic activities of the enzyme. The inhibition of the ATPase activity of ATP synthase by Bz-423 leads to rapid generation of superoxide (O2−) from the respiratory chain within mitochondria and the initiation of apoptosis by the reactive oxygen species. Bz-423 affects both the Vmax and Km of the ATPase activity of ATP synthase and inhibits ATP synthesis in a concentration-dependent fashion.
Dimethyl sulfoxide (DMSO) inhibits the hydrolytic activities of BF1 and MF1 strongly at concentrations of above 30 to 40% (9, 345, 440). Inhibition by DMSO is reversible, affecting Vmax without a significant change in the Km (9, 440). In contrast, the synthesis of ATP by soluble F1 is promoted in the presence of DMSO (94, 197, 346). The effect of DMSO on the promotion of ATP synthesis by isolated F1 is considered to be due to an increase in affinity of F1 for phosphate at the catalytic site (197, 345).
Hypochlorous acid (HOCl) is a strong oxidant that is produced as a microbicide in activated neutrophils and monocytes by myeloperoxidase-catalyzed peroxidation of chloride ion (182). HOCl inhibits the ATPase activity of F1 in a biphasic fashion. The ATPase activity falls rapidly to 20 to 30% at low concentrations of HOCl and then slowly to zero at high concentrations (29). The biphasic mode of inhibition is attributed to two different inhibitory activities of HOCl: oxidative modification of intact F1 and subunit dissociation of F1 due to more extensive oxidation (29, 167). The target sites for HOCl are believed to be amino acid residues within nucleophilic side chains (167).
4,4′-Dichlorodiphenyltrichloroethane (DDT) is a synthetic organic insecticide and affects sodium ion channels in the neurons of DDT-sensitive insects, causing repetitive discharge by the increase and prolongation of membrane's negative after-potential, leading to spasms and eventual death. DDT binds to an unidentified 23-kDa protein in the F0 of mitochondrial ATP synthase and inhibits the ATPase activity of the enzyme (442, 443). The 23-kDa protein is present in DDT-sensitive insects but not in DDT-tolerant insects and mammals, and the prepared DDT-sensitive ATP synthase devoid of the 23-kDa protein is not inhibited by DDT (442, 443).
Diazoxide, a mitochondrial potassium channel activator, is a cardioprotective drug for short-term treatment of malignant hypertension. Diazoxide also binds to MF1 and potentiates the binding of IF1 to F1, inhibiting the ATPase activity of ATP synthase (79, 80). The inhibition by diazoxide is reversible, and the binding of one equivalent of diazoxide to F1 is sufficient to inhibit the F1-ATPase activity. The inhibitory effect of diazoxide is ATP dependent, and no inhibition is observed without Mg2+-ATP. The binding site of diazoxide is believed to be located within the nucleotide binding domain of the β subunit.
2-Hydroxy-5-nitrobenzyl bromide (HNB) stimulates the hydrolytic activity of F1 from bovine heart mitochondria at below 0.5 mM but exhibits a concentration-dependent inhibition of F1 from the same source at above 0.5 mM (26, 27). HNB is a Trp-modifying reagent. Its capacity to activate catalytic activity at below 0.5 mM is attributed to its covalent interaction with a single Trp residue in the ɛ subunit of F1 (26). In contrast, HNB's inhibitory effect at above 0.5 mM appears to be due to noncovalent, reversible, aspecific binding to F1. About 50% of the hydrolytic activity is inhibited at 2.5 mM.
A series of derivatives of benzodiazepine, 4-(N-arylimidazole)-substituted benzopyran, and N-[1-aryl-2-(1-imidazolo)ethyl]-guanidine have been synthesized and tested for the treatment of ischemic heart disease as cardioprotective agents (Table 24) (20, 21, 166). During ischemia, ATP is hydrolyzed by mitochondrial ATP synthase, leading to depletion of ATP. To prevent the ATP wastage in ischemia, the ATPase activity of ATP synthase should be inhibited selectively without affecting the ATP synthesis activity of the enzyme. Several inhibitors were proposed as potential compounds for drug design for ischemia.
TABLE 24.
Miscellaneous inhibitors
| Name or abbreviation | Molecular formula | Other names | Inhibitory potency (reference) |
|---|---|---|---|
| Dicarbopolyborate | C2B9H11 (Dicarbononaborate) | Mercapto and chloro derivatives of dicarbononaborates, ∼95% inhibition at 500-800 μM (rat liver MF1-ATPase) (104); dichlorodicarbononaborate, 170 μMb (rat liver MF1-ATPase) (104) | |
| Almitrine | C26H29F2N7 | 6-(4-(Bis(4-fluorphenyl)methyl)-1-piperazinyl)-N,N′-di-2-propenyl-1,3,5-triazin-2,4-diamin; 2,4-bis(allylamino)-6-(4-(bis(p-fluorophenyl)methyl)-1-piperazinyl)-s-triazine | 30 μMa (S. cerevisiae mitochondria, ATPase) (336) |
| 5-Hydroxy-1,2-naphthalene dicarboxylic anhydride | C12H6O4 | 6-Hydroxynaphtho(1,2-c)furan-1,3-dione; 5-hydroxynaphthalenedicarboxylic anhydride | Complete inhibition of ATPase induced by gramicidin at 30 μM (rat liver SMP-ATPase) (165) |
| R207910 | C32H31BrN2O2 | 1-(6-Bromo-2-methoxy-quinolin-3-yl)-4-dimethylamino-2-naphthalen-1-yl-1-phenyl-butan-2-ol; TMC207; compound J | 2.5 nMa (M. smegmatis membrane vesicles, ATP synthesis) (215); 99% inhibition in the range of 0.03-0.12 μg/ml (M. tuberculosis, growth) (15) |
| Spegazzinine | C21H28N2O3 | 18.5-24 μg inhibitor/mg proteina (S. pombe ATPase activity of cell extracts) (234); 100 μMa (spinach CF1-ATPase) (10); 80 μMa (spinach chloroplasts, photophosphorylation) (10) | |
| n-Butanol | C4H10O | 1-Butanol; propyl carbinol; n-butyl alcohol; 1-hydroxybutane; butyl hydroxide; Hemostyp; methylolpropane; propylcarbinol; propylmethanol | 160 mMa (bovine heart MF1-ATPase) (406) |
| TCS | C13H7Cl4NO2 | TCSA; tetrachlorosalicylanilide; 3,3′,4′,5-tetrachlorosalicylanilide; 3,5-dichlorosalicyl 3,4-dichloroanilide; 3,5-dichloro-N-(3,4-dichlorophenyl)-2-hydroxy-benzamide | 9-10 μMa (F0F1-ATPase from V. parahaemolyticus) (290); 71% inhibition at 25 μM (V. parahaemolyticus F1-ATPase) (344) |
| Dihydrostreptomycin | C21H41N7O12 | Abiocine; Vibriomycin | 38 mMb (bovine heart SMP- and isolated MF1-ATPase) (161) |
| Suramin | C51H40N6O23S6 | Belganyl; Naganol | 40 μMa (rat liver MF1-ATPase) (28); 0.7 μg/mla (C. fasciculata MF1-ATPase) (173) |
| Bz-423 | C27H21ClN2O2 | Bz-48 | 5 μMa (Ramos cells, ATP synthesis) (193) |
| DMSO | C2H6OS | Dimethyl sulfoxide | > 95% inhibition at 40% DMSO (vol/vol) (EF1-ATPase) (9); ∼60% inhibition at 50% DMSO (TF1-ATPase) (440) |
| Hypochlorous acid | HOCl | 75% inhibition at 125 μM HOCl/g cells (EF1-ATPase) (167); 50 μmol inhibitor/g cellsa (EF1-ATPase) (29) | |
| DDT | C14H9Cl5 | 4, 4′-Dichlorodiphenyltri-chloroethane; 4, 4′-DDT; p,p′-DDT; 1,1′-(2,2,2-trichloroethylidene)bis (4-chlorobenzene); Agritan; Chlorophenothan; 1,1,1-trichloro-2,2-bis(4,4′-dichlorodiphenyl) ethane; Detoxan | 50% lethal dose of 11 μg/mg (A. melllifera) |
| Diazoxide | C8H7ClN2O2S | 7-Chloro-3-methyl-2H-1,2,4-benzothiadiazine 1,1-dioxide; Eudemine; Hyperstat; Hypertonalum | Kd of IF1 to F1, 250 nM with 1 diazoxide equivalent/F1 from 760 nM without diazoxide (bovine MF1-ATPase) (80) |
| HNB | C7H6BrNO3 | 2-Hydroxy-5-nitrobenzyl bromide; Koshland's reagent I; 2-bromomethyl-4-nitrophenol; α-bromo-4-nitro-o-cresol | 2.5 mMa (bovine heart MF1-ATPase) (27) |
| N-Sulfonyl or N-alkyl-substituted tetrahydrobenzodiazepine derivatives | C28H30N4O3S | 1-(1H-Imidazol-4-ylmethyl)-4-[(4-methoxyphenyl)sulfonyl]-2-(2-phenylethyl)-2,3,4,5-tetrahydro-1H-1,4-benzodiazepine | 77 nMa (bovine heart MF0F1-ATPase) (166) |
| C22H23F3N4O2S | 1-(1H-Imidazol-4-ylmethyl)-2-(2-phenylethyl)-4-[(trifluoromethyl)sulfonyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepine | 77 nMa (bovine heart MF0F1-ATPase) (166) | |
| C31H36N4O2S | 4-[(4-tert-Butylphenyl)sulfonyl]-1-(1H-imidazol-4-ylmethyl)-2-(2-phenylethyl)-2,3,4,5-tetrahydro-1H-1,4-benzodiazepine | 8 nMa (bovine heart MF0F1-ATPase) (166) | |
| C32H38N4O2S | 4-[(4-tert-Butylphenyl)sulfonyl]-1-[(5-methyl-1H-imidazol-4-yl)methyl]-2-(2-phenylethyl)-2,3,4,5-tetrahydro-1H-1,4-benzodiazepine | 77 nMa (bovine heart MF0F1-ATPase) (166) | |
| C28H28Cl2N4O2S | 4-[(3,4-Dichlorophenyl)sulfonyl]-1-[(5-methyl-1H-imidazol-4-yl)methyl]-2-(2-phenylethyl)-2,3,4,5-tetrahydro-1H-1,4-benzodiazepine | 22 nMa (bovine heart MF0F1-ATPase) (166) | |
| 4-(N-Arylimidazole)-substituted benzopyran derivatives | C22H21ClN4O2 | 4-[(4-Chlorophenyl)(1H-imidazol-2-ylmethyl)amino]-3-hydroxy-2,2-dimethyl-3,4-dihydro-2H-chromene-6-carbonitrile | 3R, 4S enantiomer, 0.48 μMa (rat heart MF0F1-ATPase) (21) and 4 μMa (rat heart SMP-ATP synthesis) (21); 3S, 4R enantiomer, 0.24 μMa (rat heart SMP-ATPase) (21) and 3.8 μMa (rat heart SMP-ATP synthesis) (21); |
| C26H31ClN4O4S | 4-[(4-Chlorophenyl)(1H-imidazol-2-ylmethyl)amino]-2,2-dimethyl-6-(piperidin-1-ylsulfonyl)-3,4-dihydro-2H-chromen-3-ol | 3R, 4S enantiomer (BMS-199264), 0.48 μMa (rat heart SMP-ATPase) (21), 18 μMa (rat heart SMP-ATP synthesis) (21); ∼42% inhibition at 3 μM (ischemic rat heart SMP-ATPase) (156) | |
| N-[1-Aryl-2-(1-imidazolo)ethyl]-cyanoguanidine | C19H14Cl4N6 | 2-Cyano-1-(2,4-dichlorophenyl)-3-[1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl]guanidine | 0.6 μMa (bovine MF0F1-ATPase) (20) |
| derivatives | C21H14Cl2F6N6 | 1-{1-[2,5-Bis(trifluoromethyl)phenyl]-2-(1H-imidazol-1-yl)ethyl}-2-cyano-3-(2,4-dichlorophenyl)guanidine | 0.71 μMa (bovine MF0F1-ATPase) (20) |
| N-[1-Aryl-2-(1-imidazolo)ethyl]-acylguanidine derivatives | C26H19Cl3N6O | N-[(Z)-[(4-Chlorophenyl)amino]{[1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl]amino}methylidene]-3-cyanobenzamide | Racemic mixture, 33 nMa (bovine MF0F1-ATPase) (20); one enantiomer. 18 nMa (bovine MF0F1-ATPase) (20); the other enantiomer, >100 nMa (bovine MF0F1-ATPase) (20) |
| C25H19Cl4N5O | 4-Chloro-N-[(Z)-[(4-chlorophenyl)amino]{[1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl]amino}methylidene]benzamide | Racemic mixture, 82 nMa (bovine MF0F1-ATPase) (20) | |
| O-[1-Aryl-2-(1-imidazolo)ethyl]-thiourethane derivatives | C18H14Cl3N3OS | O-[1-(2,4-Dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl] (4-chlorophenyl)carbamothioate | 0.43 μMa (bovine heart MF0F1-ATPase) (20); >300 μMa (bovine heart MF0F1, ATP synthesis) (20) |
| C18H13Cl4N3OS | O-[1-(2,4-Dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl] (2,4-dichlorophenyl)carbamothioate | 30 nMa (bovine heart MF0F1-ATPase) (20) | |
| Dio-9 complex | Unknown (a mixture of at least 9 compounds) | 0.7 μg inhibitor/mg proteina (T. pyriformis SMP-ATPase) (404); ∼500 μg inhibitor/mg proteina (S. faecalis F1-ATPase) (169); 6.6 μg/mg proteina (C. fasciculata SMP-ATPase) (439) | |
| Ethanol | C2H5OH | Ethyl alcohol | 60% inhibition at about 7 μM (V. parahaemolyticus F0F1-ATPase) (290) |
| Zinc | Zn2+ | ∼100 μMa (V. parahaemolyticus F0F1-ATPase) (290) |
I50.
Ki.
N-Sulfonyl- or N-alkyl-substituted tetrahydrobenzodiazepine derivatives inhibit the mitochondrial ATPase activity of ATP synthase (166). The inhibition of ATP synthesis by these derivatives is much less potent than their inhibition of ATP hydrolysis. The derivatives with an N-sulfonyl moiety seem to have stronger inhibitory potencies than those with an N-alkyl moiety.
4-(N-Arylimidazole)-substituted benzopyran derivatives are inhibitors of ATP hydrolysis of mitochondrial ATP synthase (21, 156). The inhibition of ATP synthesis by these derivatives is about an order of magnitude less potent than that of ATP hydrolysis (21). Both the N-arylimidazole ring and benzopyran seem to be required for inhibition, since the removal of either from the structure causes a dramatic loss of inhibitory potency. BMS-199264 has been tested as a cardioprotective agent in ischemic rat hearts and showed selective inhibition of ATP hydrolase activity with no effect on ATP synthesis (156). It conserved ATP during ischemia, while it had no influence on preischemic ATP concentrations and cardiac function.
Cyano- and acylguanidine derivatives containing imidazoloethyl and aryl groups also inhibit the hydrolytic activity of mitochondrial ATP synthase (20). Inhibition by derivatives of N-[1-aryl-2-(1-imidazolo)ethyl]-cyanoguanidine and N-[1-aryl-2-(1-imidazolo)ethyl]-acylguanidine is selective for ATPase activity. No inhibition of ATP synthesis is observed up to 100 μM. In cyanoguanidine derivatives, the number and position of the chloride in aryl groups are believed to be important for their inhibitory activities. For example, the 2,4-dichloro analog is more potent than 2,3-dichloro and monochloro analogs in inhibiting the ATPase activity of F1. Two symmetrical enantiomers with an identical chemical composition also have different inhibitory potencies. For instance, one entiomer of N-[(Z)-[(4-chlorophenyl)amino]{[1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl]amino}methylidene]-3-cyanobenzamide inhibits the ATPase activity of bovine mitochondrial ATP synthase (F0F1) with an I50 of 18 nM, whereas the other entiomer has no inhibitory activity on the ATPase activity of the same enzyme (20).
O-[1-Aryl-2-(1-imidazolo)ethyl]-thiourethane derivatives also inhibit the ATPase activity of mitochondrial ATP synthase. Similar to the derivatives of N-[1-aryl-2-(1-imidazolo)ethyl]-cyanoguanidine and N-[1-aryl-2-(1-imidazolo)ethyl]-acylguanidine, the O-[1-aryl-2-(1-imidazolo)ethyl]-thiourethane derivatives also maintain selectivity for inhibition of ATPase activity of ATP synthase over ATP synthesis. For example, substitutions in the 1-aryl-2-imidazoloethyl and aniline moieties affect the inhibitory potencies of the derivatives, and halogen substitution in these moieties also seems to be favorable for promoting inhibition.
Dio-9 is a mixture of at least nine compounds, two of which have antibiotic properties (232). Dio-9 inhibits both ATPase and ATP synthase activities of mitochondria, chloroplasts, and bacteria (124, 125, 163, 169, 431). There is still much to be learned about the structures and chemical actions of the class of compounds comprising Dio-9.
Ethanol inhibits the ATPase activity of F0F1 from V. parahaemolyticus at concentrations of above 4% (290). In contrast, ethanol exhibits stimulatory effects on the ATPase activity of F1.
Zinc strongly inhibits the ATPase activities of both purified and membrane-bound F0F1 from V. parahaemolyticus (267, 290). The site of action of the zinc ion is considered to be located within F0 (290).
CONCLUSIONS
ATP synthase was previously considered to be located only in the mitochondrial inner membrane, the bacterial plasma membrane, and the chloroplast thylakoid membrane. It was also considered to be involved only in the synthesis of ATP or in the generation of a proton gradient. Now, however, significant evidence has accumulated that the ATP synthase is also present on the surfaces of multiple animal cell types and serves as a receptor for various ligands, participating in a number of cellular processes, including angiogenesis, lipid metabolism, the regulation of intercellular pH, and the cytolytic pathway of tumor cells (17, 38, 39, 72, 91, 202, 269). As the multiple roles of the cell surface ATP synthase are now beginning to be understood, this pivotal enzyme complex both at this location and its mitochondrial location is emerging as a molecular target for the treatment of various diseases.
The use of ATP synthase as a molecular target has multiple advantages. First, as it is indispensable for energy metabolism, if selectively targeted, it may be possible to eradicate some types of cancer. It may also provide an ideal target for controlling a number of other diseases because of its complex subunit composition. For example, it has been demonstrated already that a lupus drug, Bz-423, targets the OSCP of F0, whereas an antimycobacterial drug, R207910, binds to subunit c of F0 (15, 193, 313). In addition, it has been shown that resveratrol and piceatannol, potential antiangiogenesis agents, block tumor growth by binding to the β subunit of F1 (143, 449). Lastly, the high inhibitory specificity of ATP synthase inhibitors also suggests that this complex is an excellent target for the development of new insecticidal or herbicidal agents. For example, tentoxin is a strong inhibitor of CF1-ATPase from certain sensitive species such as spinach, potato, and lettuce, but it has little or no inhibitory effect on the same enzyme from insensitive species such as corn, tobacco, and radish, even though they exhibit high sequence and structural similarity (380). In addition, slight structural modifications of tentoxin can cause dramatic effects on the properties and inhibitory potencies of the inhibitor (316, 351). Finally, the drug R207910, developed for the treatment of tuberculosis, also shows a narrow selectivity in its inhibition of the ATP synthase in mycobacterial species (15).
The mitochondrial ATP synthase contains a number of supernumerary subunits that are absent in bacterial or chloroplast counterparts. The plasma membrane ATP synthase found in various types of animal cells also includes more subunit types than the bacterial and chloroplast ATP synthases. The roles of the supernumerary subunits are currently unknown or poorly defined, but evidence is accumulating that these “extra” subunits are also involved in cellular processes other than ATP synthesis. Thus, subunit F6 has been reported to be associated with regulating blood pressure. Additionally, subunit e has been reported to be involved in the regulation of the expression of the gene for subunit g of the ATP synthase (18) and also for that of the c-myc proto-oncogene (177, 226). The expression level of subunit e has also been shown to be highly sensitive to diverse physiologic changes and stresses. Although the detailed regulatory roles of subunits F6 and e and the roles of other supernumerary subunits require further investigation, it seems likely that they will be implicated in a multitude of cellular processes that will result in future use of the ATP synthase as a drug target.
In this review, we have provided detailed information about most natural and synthetic inhibitors of ATP synthases reported to date. Figure 12 summarizes the known or proposed sites of these ATP synthase inhibitors. About 270 inhibitors are described here and need further investigations to identify clearly or confirm their sites of actions and inhibitory mechanisms. When this mammoth task is accomplished, it will further heighten consideration of ATP synthase as a major target for new therapies for human and animal diseases and likely contribute also to the discovery of novel agents that may prove valuable in agriculture and other areas. In addition, the rich source of structures and other knowledge about ATP synthase inhibitors already provided in this review will likely prove invaluable as scaffolds for new drug discoveries in the near future.
FIG. 12.
Inhibitory sites of ATP synthase. The inhibitor binding sites in the ATP synthase as revealed by biochemical/structural studies are indicated by red circles, and the binding subunits in which the binding sites have not been completely clarified are indicated by green circles. The coordinates of each subunit in the structural model are the same as in Fig. 1.
Acknowledgments
P.L.P. is supported for work on ATP synthase by National Institutes of Health grants 5R01 CA10951 and 5P01 HL081427.
REFERENCES
- 1.Abad, M. C., R. K. Arni, D. K. Grella, F. J. Castellino, A. Tulinsky, and J. H. Geiger. 2002. The X-ray crystallographic structure of the angiogenesis inhibitor angiostatin. J. Mol. Biol. 3181009-1017. [DOI] [PubMed] [Google Scholar]
- 2.Abrahams, J. P., S. K. Buchanan, M. J. Van Raaij, I. M. Fearnley, A. G. Leslie, and J. E. Walker. 1996. The structure of bovine F1-ATPase complexed with the peptide antibiotic efrapeptin. Proc. Natl. Acad. Sci. USA 939420-9424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ackerman, S. H., C. Grubmeyer, and P. S. Coleman. 1987. Evidence for catalytic cooperativity during ATP hydrolysis by beef heart F1-ATPase. Kinetics and binding studies with the photoaffinity label BzATP. J. Biol. Chem. 26213765-13772. [PubMed] [Google Scholar]
- 4.Agarwal, N., and V. K. Kalra. 1984. Studies on the mechanism of action of local anesthetics on proton translocating ATPase from Mycobacterium phlei. Biochim. Biophys. Acta 764316-323. [DOI] [PubMed] [Google Scholar]
- 5.Ahmad, Z., and A. E. Senior. 2006. Inhibition of the ATPase activity of Escherichia coli ATP synthase by magnesium fluoride. FEBS Lett. 580517-520. [DOI] [PubMed] [Google Scholar]
- 6.Ahmad, Z., and A. E. Senior. 2004. Mutagenesis of residue βArg-246 in the phosphate-binding subdomain of catalytic sites of Escherichia coli F1-ATPase. J. Biol. Chem. 27931505-31513. [DOI] [PubMed] [Google Scholar]
- 7.Akhrem, A. A., M. A. Kisel, I. A. Kozlov, I. S. Tsybovsky, and E. N. Vulfson. 1985. The inhibition of mitochondrial F1-ATPase by 1,5-difluoro-2,4-dinitrobenzene. FEBS Lett. 187249-252. [DOI] [PubMed] [Google Scholar]
- 8.Aloise, P., Y. Kagawa, and P. S. Coleman. 1991. Comparative Mg2+-dependent sequential covalent binding stoichiometries of 3′-O-(4-benzoyl)benzoyl adenosine 5′-diphosphate of MF1, TF1, and the α3β3 core complex of TF1. The binding change motif is independent of the F1 γδɛ subunits. J. Biol. Chem. 26610368-10376. [PubMed] [Google Scholar]
- 9.Al-Shawi, M. K., and A. E. Senior. 1992. Effects of dimethyl sulfoxide on catalysis in Escherichia coli F1-ATPase. Biochemistry 31886-891. [DOI] [PubMed] [Google Scholar]
- 10.Andreo, C. S. 1978. Inhibition of energy-transducing functions of chloroplasts by spegazzinine. Arch. Biochem. Biophys. 186416-421. [DOI] [PubMed] [Google Scholar]
- 11.Andrews, W. W., and W. S. Allison. 1981. 1-Fluoro-2,4-dinitrobenzene modifies a tyrosine residue when it inactivates the bovine mitochondrial F1-ATPase. Biochem. Biophys. Res. Commun. 99813-819. [DOI] [PubMed] [Google Scholar]
- 12.Andrews, W. W., F. C. Hill, and W. S. Allison. 1984. Identification of the essential tyrosine residue in the β subunit of bovine heart mitochondrial F1-ATPase that is modified by 7-chloro-4-nitro[14C]benzofurazan. J. Biol. Chem. 2598219-8225. [PubMed] [Google Scholar]
- 13.Andrews, W. W., F. C. Hill, and W. S. Allison. 1984. Identification of the lysine residue to which the 4-nitrobenzofurazan group migrates after the bovine mitochondrial F1-ATPase is inactivated with 7-chloro-4-nitro[14C]benzofurazan. J. Biol. Chem. 25914378-14382. [PubMed] [Google Scholar]
- 14.Andrews, W. W., M. Yoshida, F. C. Hill, and W. S. Allison. 1984. Identification of an essential lysine residue in the β subunit of the F1-ATPase from the thermophilic bacterium, PS3, using 7-chloro-4-nitro[14C]benzofurazan. Biochem. Biophys. Res. Commun. 1231040-1046. [DOI] [PubMed] [Google Scholar]
- 15.Andries, K., P. Verhasselt, J. Guillemont, H. W. Gohlmann, J. M. Neefs, H. Winkler, J. Van Gestel, P. Timmerman, M. Zhu, E. Lee, P. Williams, D. de Chaffoy, E. Huitric, S. Hoffner, E. Cambau, C. Truffot-Pernot, N. Lounis, and V. Jarlier. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307223-227. [DOI] [PubMed] [Google Scholar]
- 16.Arakaki, N., T. Kita, H. Shibata, and T. Higuti. 2007. Cell-surface H+-ATP synthase as a potential molecular target for anti-obesity drugs. FEBS Lett. 5813405-3409. [DOI] [PubMed] [Google Scholar]
- 17.Arakaki, N., T. Nagao, R. Niki, A. Toyofuku, H. Tanaka, Y. Kuramoto, Y. Emoto, H. Shibata, K. Magota, and T. Higuti. 2003. Possible role of cell surface H+-ATP synthase in the extracellular ATP synthesis and proliferation of human umbilical vein endothelial cells. Mol. Cancer Res. 1931-939. [PubMed] [Google Scholar]
- 18.Arnold, I., K. Pfeiffer, W. Neupert, R. A. Stuart, and H. Schagger. 1998. Yeast mitochondrial F1F0-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J. 177170-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arntzen, C. J. 1972. Inhibition of photophosphorylation by tentoxin, a cyclic tetrapeptide. Biochim. Biophys. Acta 283539-542. [DOI] [PubMed] [Google Scholar]
- 20.Atwal, K. S., S. Ahmad, C. Z. Ding, P. D. Stein, J. Lloyd, L. G. Hamann, D. W. Green, F. N. Ferrara, P. Wang, W. L. Rogers, L. M. Doweyko, A. V. Miller, S. N. Bisaha, J. B. Schmidt, L. Li, K. J. Yost, H. J. Lan, and C. S. Madsen. 2004. N-[1-Aryl-2-(1-imidazolo)ethyl]-guanidine derivatives as potent inhibitors of the bovine mitochondrial F1F0 ATP hydrolase. Bioorg. Med. Chem. Lett. 141027-1030. [DOI] [PubMed] [Google Scholar]
- 21.Atwal, K. S., P. Wang, W. L. Rogers, P. Sleph, H. Monshizadegan, F. N. Ferrara, S. Traeger, D. W. Green, and G. J. Grover. 2004. Small molecule mitochondrial F1F0 ATPase hydrolase inhibitors as cardioprotective agents. Identification of 4-(N-arylimidazole)-substituted benzopyran derivatives as selective hydrolase inhibitors. J. Med. Chem. 471081-1084. [DOI] [PubMed] [Google Scholar]
- 22.Avron, M., and N. Shavit. 1965. Inhibitors and uncouplers of photophosphorylation. Biochim. Biophys. Acta 109317-331. [DOI] [PubMed] [Google Scholar]
- 23.Azzi, A., M. A. Bragadin, G. Neri, G. Farnia, and A. M. Tamburro. 1973. A spin-label carbodiimide as a probe for mitochondrial ATPase. FEBS Lett. 30249-252. [DOI] [PubMed] [Google Scholar]
- 24.Azzi, A., M. A. Bragadin, A. M. Tamburro, and M. Santato. 1973. Site-directed spin labeling of the mitochondrial membrane. Synthesis and utilization of the adenosine triphosphatase inhibitor (N-(2,2,6,6-tetramethyl-piperidyl-1-oxyl)-N′-(cyclohexyl)-carbodiimide). J. Biol. Chem. 2485520-5526. [PubMed] [Google Scholar]
- 25.Bald, D., T. Amano, E. Muneyuki, B. Pitard, J. L. Rigaud, J. Kruip, T. Hisabori, M. Yoshida, and M. Shibata. 1998. ATP synthesis by F0F1-ATP synthase independent of noncatalytic nucleotide binding sites and insensitive to azide inhibition. J. Biol. Chem. 273865-870. [DOI] [PubMed] [Google Scholar]
- 26.Baracca, A., S. Barogi, G. Lenaz, and G. Solaini. 1993. Interactions and effects of 2-hydroxy-5-nitrobenzyl bromide on the bovine heart mitochondrial F1-ATPase. Int. J. Biochem. 251269-1275. [DOI] [PubMed] [Google Scholar]
- 27.Baracca, A., D. Menegatti, G. Parenti Castelli, C. A. Rossi, and G. Solaini. 1990. Does 2-hydroxy-5-nitrobenzyl bromide react with the ɛ-subunit of the mitochondrial F1-ATPase? Biochem. Int. 211135-1142. [PubMed] [Google Scholar]
- 28.Barret, J. M., A. P. Ernould, M. H. Rouillon, G. Ferry, A. Genton, and J. A. Boutin. 1993. Studies of the potency of protein kinase inhibitors on ATPase activities. Chem. Biol. Interact. 8617-27. [DOI] [PubMed] [Google Scholar]
- 29.Barrette, W. C., Jr., D. M. Hannum, W. D. Wheeler, and J. K. Hurst. 1989. General mechanism for the bacterial toxicity of hypochlorous acid: abolition of ATP production. Biochemistry 289172-9178. [DOI] [PubMed] [Google Scholar]
- 30.Barta, I., P. Smerak, Z. Polivkova, H. Sestakova, M. Langova, B. Turek, and J. Bartova. 2006. Current trends and perspectives in nutrition and cancer prevention. Neoplasma 5319-25. [PubMed] [Google Scholar]
- 31.Barzu, O., F. Guerrieri, R. Scarfo, G. Capozza, and S. Papa. 1989. Effect of cetyltrimethylammonium on ATP hydrolysis and proton translocation in the F0-F1 H+-ATP synthase of mitochondria. J. Bioenerg. Biomembr. 21403-414. [DOI] [PubMed] [Google Scholar]
- 32.Bar-Zvi, D., and N. Shavit. 1982. Modulation of the chloroplast ATPase by tight ADP binding. Effect of uncouplers and ATP. J. Bioenerg. Biomembr. 14467-478. [DOI] [PubMed] [Google Scholar]
- 33.Bar-Zvi, D., M. Yoshida, and N. Shavit. 1996. Modification of domains of α and β subunits of F1-ATPase from the thermophylic bacterium PS3, in their isolated and associated forms, by 3′-O-(4-benzoyl)benzoyl adenosine 5′-triphosphate (BzATP). J. Bioenerg. Biomembr. 28471-481. [DOI] [PubMed] [Google Scholar]
- 34.Baubichon, H., C. Godinot, A. Di Pietro, and D. C. Gautheron. 1981. Competition between ADP and nucleotide analogues to occupy regulatory sites(s) related to hysteretic inhibition of mitochondrial F1-ATPase. Biochem. Biophys. Res. Commun. 1001032-1038. [DOI] [PubMed] [Google Scholar]
- 35.Beechey, R. B., A. M. Roberton, C. T. Holloway, and I. G. Knight. 1967. The properties of dicyclohexylcarbodiimide as an inhibitor of oxidative phosphorylation. Biochemistry 63867-3879. [DOI] [PubMed] [Google Scholar]
- 36.Beil, W., C. Birkholz, S. Wagner, and K. F. Sewing. 1995. Bismuth subcitrate and omeprazole inhibit Helicobacter pylori F1-ATPase. Pharmacology 50333-337. [DOI] [PubMed] [Google Scholar]
- 37.Belda, F. J., F. G. Carmona, F. G. Canovas, J. C. Gomez-Fernandez, and J. A. Lozano. 1983. A kinetic study of the interaction between mitochondrial F1 adenosine triphosphatase and adenylyl imidodiphosphate and guanylyl imidodiphosphate. Biochem. J. 210727-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berger, K., U. Sivars, M. S. Winzell, P. Johansson, U. Hellman, C. Rippe, and C. Erlanson-Albertsson. 2002. Mitochondrial ATP synthase—a possible target protein in the regulation of energy metabolism in vitro and in vivo. Nutr. Neurosci. 5201-210. [DOI] [PubMed] [Google Scholar]
- 39.Berger, K., M. S. Winzell, J. Mei, and C. Erlanson-Albertsson. 2004. Enterostatin and its target mechanisms during regulation of fat intake. Physiol. Behav. 83623-630. [DOI] [PubMed] [Google Scholar]
- 40.Bernardes, C. F., J. R. Meyer-Fernandes, O. B. Martins, and A. E. Vercesi. 1997. Inhibition of succinic dehydrogenase and F0F1-ATP synthase by 4,4′-diisothiocyanatostilbene-2,2′-disulfonic acid (DIDS). Z. Naturforsch. C 52799-806. [DOI] [PubMed] [Google Scholar]
- 41.Blatt, N. B., J. J. Bednarski, R. E. Warner, F. Leonetti, K. M. Johnson, A. Boitano, R. Yung, B. C. Richardson, K. J. Johnson, J. A. Ellman, A. W. Opipari, Jr., and G. D. Glick. 2002. Benzodiazepine-induced superoxide signals B cell apoptosis: mechanistic insight and potential therapeutic utility. J. Clin. Investig. 1101123-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borghese, R., P. Turina, L. Lambertini, and B. A. Melandri. 1998. The atpIBEXF operon coding for the F0 sector of the ATP synthase from the purple nonsulfur photosynthetic bacterium Rhodobacter capsulatus. Arch. Microbiol. 170385-388. [DOI] [PubMed] [Google Scholar]
- 43.Bossard, M. J., and S. M. Schuster. 1981. Catalysis of partial reactions of ATP synthesis by beef heart mitochondrial adenosine triphosphatase. J. Biol. Chem. 2561518-1521. [PubMed] [Google Scholar]
- 44.Bossard, M. J., and S. M. Schuster. 1981. Structural preferences for the binding of chromium nucleotides by beef heart mitochondrial ATPase. J. Biol. Chem. 2566617-6622. [PubMed] [Google Scholar]
- 45.Boulay, F., P. Dalbon, and P. V. Vignais. 1985. Photoaffinity labeling of mitochondrial adenosinetriphosphatase by 2-azidoadenosine 5′-[α-32P]diphosphate. Biochemistry 247372-7379. [DOI] [PubMed] [Google Scholar]
- 46.Bowler, M. W., M. G. Montgomery, A. G. Leslie, and J. E. Walker. 2006. How azide inhibits ATP hydrolysis by the F-ATPases. Proc. Natl. Acad. Sci. USA 1038646-8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boyer, P. D. 2000. Catalytic site forms and controls in ATP synthase catalysis. Biochim. Biophys. Acta 1458252-262. [DOI] [PubMed] [Google Scholar]
- 48.Braig, K., R. I. Menz, M. G. Montgomery, A. G. Leslie, and J. E. Walker. 2000. Structure of bovine mitochondrial F1-ATPase inhibited by Mg2+ ADP and aluminium fluoride. Structure Fold Des. 8567-573. [DOI] [PubMed] [Google Scholar]
- 49.Bullough, D. A., and W. S. Allison. 1986. Inactivation of the bovine heart mitochondrial F1-ATPase by 5′-p-fluorosulfonylbenzoyl[3H]inosine is accompanied by modification of tyrosine 345 in a single β subunit. J. Biol. Chem. 26114171-14177. [PubMed] [Google Scholar]
- 50.Bullough, D. A., and W. S. Allison. 1988. Inactivation of the F1-ATPase from the thermophilic bacterium PS3 by 5′-p-fluorosulfonylbenzoylinosine at 65 °C is accompanied by modification of β-tyrosine-364. Biochim. Biophys. Acta 934397-400. [DOI] [PubMed] [Google Scholar]
- 51.Bullough, D. A., and W. S. Allison. 1986. Three copies of the β subunit must be modified to achieve complete inactivation of the bovine mitochondrial F1-ATPase by 5′-p-fluorosulfonylbenzoyladenosine. J. Biol. Chem. 2615722-5730. [PubMed] [Google Scholar]
- 52.Bullough, D. A., E. A. Ceccarelli, D. Roise, and W. S. Allison. 1989. Inhibition of the bovine-heart mitochondrial F1-ATPase by cationic dyes and amphipathic peptides. Biochim. Biophys. Acta 975377-383. [DOI] [PubMed] [Google Scholar]
- 53.Bullough, D. A., E. A. Ceccarelli, J. G. Verburg, and W. S. Allison. 1989. Localization of sites modified during inactivation of the bovine heart mitochondrial F1-ATPase by quinacrine mustard using [3H]aniline as a probe. J. Biol. Chem. 2649155-9163. [PubMed] [Google Scholar]
- 54.Bullough, D. A., M. Kwan, P. K. Laikind, M. Yoshida, and W. S. Allison. 1985. The varied responses of different F1-ATPases to chlorpromazine. Arch. Biochem. Biophys. 236567-575. [DOI] [PubMed] [Google Scholar]
- 55.Bullough, D. A., J. G. Verburg, M. Yoshida, and W. S. Allison. 1987. Evidence for functional heterogeneity among the catalytic sites of the bovine heart mitochondrial F1-ATPase. J. Biol. Chem. 26211675-11683. [PubMed] [Google Scholar]
- 56.Bullough, D. A., M. Yoshida, and W. S. Allison. 1986. Sequence of the radioactive tryptic peptide obtained after inactivating the F1-ATPase of the thermophilic bacterium PS3 with 5′-p-fluorosulfonylbenzoyl[3H]adenosine at 65 °C. Arch. Biochem. Biophys. 244865-871. [DOI] [PubMed] [Google Scholar]
- 57.Bullough, D. A., S. Q. Zhuo, and W. S. Allison. 1991. Separate β subunits are derivatized with 14C and 3H when the bovine heart mitochondrial F1-ATPase is doubly labeled with 7-chloro-4-nitro[14C]benzofurazan and 5′-p-fluorosulfonylbenzoyl[3H]inosine. Biochim. Biophys. Acta 1057208-214. [DOI] [PubMed] [Google Scholar]
- 58.Burrell, H. E., B. Wlodarski, B. J. Foster, K. A. Buckley, G. R. Sharpe, J. M. Quayle, A. W. Simpson, and J. A. Gallagher. 2005. Human keratinocytes release ATP and utilize three mechanisms for nucleotide interconversion at the cell surface. J. Biol. Chem. 28029667-29676. [DOI] [PubMed] [Google Scholar]
- 59.Burwick, N. R., M. L. Wahl, J. Fang, Z. Zhong, T. L. Moser, B. Li, R. A. Capaldi, D. J. Kenan, and S. V. Pizzo. 2005. An inhibitor of the F1 subunit of ATP synthase (IF1) modulates the activity of angiostatin on the endothelial cell surface. J. Biol. Chem. 2801740-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cabezon, E., P. J. Butler, M. J. Runswick, R. J. Carbajo, and J. E. Walker. 2002. Homologous and heterologous inhibitory effects of ATPase inhibitor proteins on F-ATPases. J. Biol. Chem. 27741334-41341. [DOI] [PubMed] [Google Scholar]
- 61.Cabezon, E., M. G. Montgomery, A. G. Leslie, and J. E. Walker. 2003. The structure of bovine F1-ATPase in complex with its regulatory protein IF1. Nat. Struct. Biol. 10744-750. [DOI] [PubMed] [Google Scholar]
- 62.Capozza, G., O. Dmitriev, I. A. Krasnoselskaya, S. Papa, and V. P. Skulachev. 1991. The effect of F0 inhibitors on the Vibrio alginolyticus membrane ATPase. FEBS Lett. 280274-276. [DOI] [PubMed] [Google Scholar]
- 63.Carlsson, C., and L. Ernster. 1981. Uncoupler-reversible inhibition of mitochondrial ATPase by metal chelates of bathophenanthroline. I. General features. Biochim. Biophys. Acta 638345-357. [DOI] [PubMed] [Google Scholar]
- 64.Carlsson, C., and L. Ernster. 1981. Uncoupler-reversible inhibition of mitochondrial ATPase by metal chelates of bathophenanthroline. II. Comparison with other inhibitors. Biochim. Biophys. Acta 638358-364. [DOI] [PubMed] [Google Scholar]
- 65.Carlsson-Skwirut, C., and L. Ernster. 1982. Bathophenanthroline-ruthenium chelate, a fluorescent inhibitor of F1-ATPase. FEBS Lett. 14577-81. [DOI] [PubMed] [Google Scholar]
- 66.Cataldi de Flombaum, M. A., and A. O. Stoppani. 1986. Effects of ethidium bromide on the mitochondrial adenosine triphosphatase from Trypanosoma cruzi. Biochem. Int. 12513-519. [PubMed] [Google Scholar]
- 67.Cataldi de Flombaum, M. A., and A. O. Stoppani. 1982. Phenylglyoxal inactivation of the mitochondrial adenosine triphosphatase from Trypanosoma cruzi. Mol. Biochem. Parasitol. 5371-379. [DOI] [PubMed] [Google Scholar]
- 68.Cattell, K. J., C. R. Lindop, I. G. Knight, and R. B. Beechey. 1971. The identification of the site of action of NN′-dicyclohexylcarbodi-imide as a proteolipid in mitochondrial membranes. Biochem. J. 125169-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cavelier, F., C. Enjalbal, J. Santolini, F. Haraux, C. Sigalat, J. Verducci, and F. Andre. 1997. Analogs of tentoxin: tools for mechanistic investigations. Lett. Peptide Sci. 4283-288. [Google Scholar]
- 70.Ceccarelli, E. A., J. G. Verburg, S. Q. Zhuo, and W. S. Allison. 1989. Selectivity of modification when latent and activated forms of the chloroplast F1-ATPase are inactivated by 7-chloro-4-nitrobenzofurazan. Arch. Biochem. Biophys. 272400-411. [DOI] [PubMed] [Google Scholar]
- 71.Cerrini, S., D. Lamba, A. Scatturin, and G. Ughetto. 1989. The crystal and molecular structure of the α-helical nonapeptide antibiotic leucinostatin A. Biopolymers 28409-420. [DOI] [PubMed] [Google Scholar]
- 72.Champagne, E., L. O. Martinez, X. Collet, and R. Barbaras. 2006. Ecto-F1FO ATP synthase/F1 ATPase: metabolic and immunological functions. Curr. Opin. Lipidol. 17279-284. [DOI] [PubMed] [Google Scholar]
- 73.Chandrasekaran, K., K. Hatanpaa, S. I. Rapoport, and D. R. Brady. 1997. Decreased expression of nuclear and mitochondrial DNA-encoded genes of oxidative phosphorylation in association neocortex in Alzheimer disease. Brain Res. Mol. Brain Res. 4499-104. [DOI] [PubMed] [Google Scholar]
- 74.Chang, T., and H. S. Penefsky. 1973. Aurovertin, a fluorescent probe of conformational change in beef heart mitochondrial adenosine triphosphatase. J. Biol. Chem. 2482746-2754. [PubMed] [Google Scholar]
- 75.Chavez, E., and A. Cuellar. 1984. Inactivation of mitochondrial ATPase by ultraviolet light. Arch. Biochem. Biophys. 230511-516. [DOI] [PubMed] [Google Scholar]
- 76.Chazotte, B., G. Vanderkooi, and D. Chignell. 1982. Further studies on F1-ATPase inhibition by local anesthetics. Biochim. Biophys. Acta 680310-316. [DOI] [PubMed] [Google Scholar]
- 77.Chen, C., A. K. Saxena, W. N. Simcoke, D. N. Garboczi, P. L. Pedersen, and Y. H. Ko. 2006. Mitochondrial ATP synthase. Crystal structure of the catalytic F1 unit in a vanadate-induced transition-like state and implications for mechanism. J. Biol. Chem. 28113777-13783. [DOI] [PubMed] [Google Scholar]
- 78.Clarke, D. J., F. M. Fuller, and J. G. Morris. 1979. The proton-translocating adenosine triphosphatase of the obligately anaerobic bacterium Clostridium pasteurianum. 1. ATP phosphohydrolase activity. Eur. J. Biochem. 98597-612. [DOI] [PubMed] [Google Scholar]
- 79.Comelli, M., G. Metelli, and I. Mavelli. 2007. Downmodulation of mitochondrial F0F1 ATP synthase by diazoxide in cardiac myoblasts: a dual effect of the drug. Am. J. Physiol. Heart Circ. Physiol. 292H820-H829. [DOI] [PubMed] [Google Scholar]
- 80.Contessi, S., G. Metelli, I. Mavelli, and G. Lippe. 2004. Diazoxide affects the IF1 inhibitor protein binding to F1 sector of beef heart F0F1ATPsynthase. Biochem. Pharmacol. 671843-1851. [DOI] [PubMed] [Google Scholar]
- 81.Crane, R. K., and F. Lipmann. 1953. The effect of arsenate on aerobic phosphorylation. J. Biol. Chem. 201235-243. [PubMed] [Google Scholar]
- 82.Crosby, B., M. Boutry, and A. Goffeau. 1979. Inhibition of soluble yeast mitochondria ATPase by ethidium-bromide. Biochem. Biophys. Res. Commun. 88448-455. [DOI] [PubMed] [Google Scholar]
- 83.Cross, R. L., and W. E. Kohlbrenner. 1978. The mode of inhibition of oxidative phosphorylation by efrapeptin (A23871). Evidence for an alternating site mechanism for ATP synthesis. J. Biol. Chem. 2534865-4873. [PubMed] [Google Scholar]
- 84.Cross, R. L., and C. M. Nalin. 1982. Adenine nucleotide binding sites on beef heart F1-ATPase. Evidence for three exchangeable sites that are distinct from three noncatalytic sites. J. Biol. Chem. 2572874-2881. [PubMed] [Google Scholar]
- 85.Cunningham, C. C., and D. T. George. 1975. The relationship between the bovine heart mitochondrial adenosine triphosphatase, lipophilic compounds, and oligomycin. J. Biol. Chem. 2502036-2044. [PubMed] [Google Scholar]
- 86.Czarnecki, J. J., M. S. Abbott, and B. R. Selman. 1982. Photoaffinity labeling with 2-azidoadenosine diphosphate of a tight nucleotide binding site on chloroplast coupling factor 1. Proc. Natl. Acad. Sci. USA 797744-7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dabbeni-Sala, F., and P. Palatini. 1990. Mechanism of local anesthetic effect. Involvement of F0 in the inhibition of mitochondrial ATP synthase by phenothiazines. Biochim. Biophys. Acta 1015248-252. [DOI] [PubMed] [Google Scholar]
- 88.Dabbeni-Sala, F., G. Schiavo, and P. Palatini. 1990. Mechanism of local anesthetic effect on mitochondrial ATP synthase as deduced from photolabelling and inhibition studies with phenothiazine derivatives. Biochim. Biophys. Acta 1026117-125. [DOI] [PubMed] [Google Scholar]
- 89.Dalbon, P., F. Boulay, and P. V. Vignais. 1985. Exploration of nucleotide binding sites in the mitochondrial membrane by 2-azido-[α-32P]ADP. FEBS Lett. 180212-218. [DOI] [PubMed] [Google Scholar]
- 90.Dallmann, H. G., T. G. Flynn, and S. D. Dunn. 1992. Determination of the 1-ethyl-3-[(3-dimethylamino)propyl]-carbodiimide-induced cross-link between the β and ɛ subunits of Escherichia coli F1-ATPase. J. Biol. Chem. 26718953-18960. [PubMed] [Google Scholar]
- 91.Das, B., M. O. Mondragon, M. Sadeghian, V. B. Hatcher, and A. J. Norin. 1994. A novel ligand in lymphocyte-mediated cytotoxicity: expression of the β subunit of H+ transporting ATP synthase on the surface of tumor cell lines. J. Exp. Med. 180273-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.De Gomez-Puyou, M. T., A. Gomez-Puyou, and M. Beigel. 1976. On the mechanism of action of alkylguanidines in oxidative phosphorylation: their action on soluble F1. Arch. Biochem. Biophys. 173326-331. [DOI] [PubMed] [Google Scholar]
- 93.De Meirleir, L., S. Seneca, W. Lissens, E. Schoentjes, and B. Desprechins. 1995. Bilateral striatal necrosis with a novel point mutation in the mitochondrial ATPase 6 gene. Pediatr. Neurol. 13242-246. [DOI] [PubMed] [Google Scholar]
- 94.de Meis, L. 1989. Role of water in the energy of hydrolysis of phosphate compounds—energy transduction in biological membranes. Biochim. Biophys. Acta 973333-349. [DOI] [PubMed] [Google Scholar]
- 95.de Melo, D. F., M. Satre, and P. V. Vignais. 1984. Inactivation of beef heart mitochondrial F1-ATPase by the 2′,3′-dialdehyde derivatives of adenine nucleotides. FEBS Lett. 169101-106. [DOI] [PubMed] [Google Scholar]
- 96.Deters, D. W., E. Racker, N. Nelson, and H. Nelson. 1975. Partial resolution of the enzymes catalyzing photophosphorylation. XV. Approaches to the active site of coupling factor I. J. Biol. Chem. 2501041-1047. [PubMed] [Google Scholar]
- 97.Devenish, R. J., M. Prescott, G. M. Boyle, and P. Nagley. 2000. The oligomycin axis of mitochondrial ATP synthase: OSCP and the proton channel. J. Bioenerg. Biomembr. 32507-515. [DOI] [PubMed] [Google Scholar]
- 98.de Vicente, J. I., P. del Valle, F. Busto, D. de Arriaga, and J. Soler. 1991. Inhibition by excess of free ATP, and free Mg2+ ions of the mitochondrial F1-ATPase moiety from Phycomyces blakesleeanus. Biochem. Int. 24339-347. [PubMed] [Google Scholar]
- 99.de Vries, D. D., B. G. van Engelen, F. J. Gabreels, W. Ruitenbeek, and B. A. van Oost. 1993. A second missense mutation in the mitochondrial ATPase 6 gene in Leigh's syndrome. Ann. Neurol. 34410-412. [DOI] [PubMed] [Google Scholar]
- 100.Di Pietro, A., C. Godinot, M.-L. Bouillant, and D. C. Gautheron. 1975. Pig heart mitochondrial ATPase: properties of purified and membrane-bound enzyme. Effects of flavonoids. Biochemie 57959-967. [DOI] [PubMed] [Google Scholar]
- 101.Di Pietro, A., C. Godinot, J.-C. Martin, and D. C. Gautheron. 1979. Affinity labeling of catalytic and regulatory sites of pig heart mitochondrial F1-ATPase by 5′-p-fluorosulfonylbenzoyladenosine. Biochemistry 181738-1745. [DOI] [PubMed] [Google Scholar]
- 102.Di Pietro, A., F. Penin, C. Godinot, and D. C. Gautheron. 1980. “Hysteretic” behavior and nucleotide binding sites of pig heart mitochondrial F1 adenosine 5′-triphosphatase. Biochemistry 195671-5678. [DOI] [PubMed] [Google Scholar]
- 103.Djerassi, C., H. W. Brewer, H. Budzikiewicz, O. O. Orazi, and R. A. Corral. 1962. The structure of the aspidosperma alkaloids spegazzinine and spegazzinidine. J. Am. Chem. Soc. 843480-3485. [Google Scholar]
- 104.Drahota, Z., V. Mares, H. Rauchova, P. Saf, and M. Kalous. 1994. Inhibition of mitochondrial ATPase by dicarbopolyborate, a new enzyme inhibitor. J. Bioenerg. Biomembr. 26583-586. [DOI] [PubMed] [Google Scholar]
- 105.Dreyfus, G., H. Guimaraes-Motta, and J. L. Silva. 1988. Effect of hydrostatic pressure on the mitochondrial ATP synthase. Biochemistry 276704-6710. [DOI] [PubMed] [Google Scholar]
- 106.Du, Z. Y., and P. D. Boyer. 1990. On the mechanism of sulfite activation of chloroplast thylakoid ATPase and the relation of ADP tightly bound at a catalytic site to the binding change mechanism. Biochemistry 29402-407. [DOI] [PubMed] [Google Scholar]
- 107.Dupuis, A., J. P. Issartel, and P. Vignais. 1989. Direct identification of the fluoroaluminate and fluoroberyllate species responsible for inhibition of the mitochondrial F1-ATPase. FEBS Lett. 25547-52. [Google Scholar]
- 108.Ebel, R. E., and H. A. Lardy. 1975. Influence of aurovertin on mitochondrial ATPase activity. J. Biol. Chem. 2504992-4995. [PubMed] [Google Scholar]
- 109.Eckhardt, U., and W. G. Hanstein. 1993. Beef heart mitochondrial F1-ATPase: inhibition by azidoadenyl-5′-yl imidodiphosphates and cooperative binding of substrate. Biochim. Biophys. Acta 1144419-425. [DOI] [PubMed] [Google Scholar]
- 110.Edel, C. M., A. F. Hartog, and J. A. Berden. 1993. Identification of an exchangeable non-catalytic site on mitochondrial F1-ATPase which is involved in the negative cooperativity of ATP hydrolysis. Biochim. Biophys. Acta 1142327-335. [DOI] [PubMed] [Google Scholar]
- 111.Edel, C. M., A. F. Hartog, and J. A. Berden. 1992. Inhibition of mitochondrial F1-ATPase activity by binding of (2-azido-) ADP to a slowly exchangeable non-catalytic nucleotide binding site. Biochim. Biophys. Acta 1101329-338. [DOI] [PubMed] [Google Scholar]
- 112.Edwards, D. L., and B. W. Unger. 1978. Nuclear mutations conferring oligomycin resistance in Neurospora crassa. J. Biol. Chem. 2534254-4258. [PubMed] [Google Scholar]
- 113.Emaus, R. K., R. Grunwald, and J. J. Lemasters. 1986. Rhodamine 123 as a probe of transmembrane potential in isolated rat-liver mitochondria: spectral and metabolic properties. Biochim. Biophys. Acta 850436-448. [DOI] [PubMed] [Google Scholar]
- 114.Esch, F. S., and W. S. Allison. 1978. Identification of a tyrosine residue at a nucleotide binding site in the β subunit of the mitochondrial ATPase with p-fluorosulfonyl[14C]-benzoyl-5′-adenosine. J. Biol. Chem. 2536100-6106. [PubMed] [Google Scholar]
- 115.Falson, P., A. Di Pietro, and D. C. Gautheron. 1986. Chemical modification of thiol groups of mitochondrial F1-ATPase from the yeast Schizosaccharomyces pombe. Involvement of α- and γ-subunits in the enzyme activity. J. Biol. Chem. 2617151-7159. [PubMed] [Google Scholar]
- 116.Feldman, R. I., and D. S. Sigman. 1985. Membrane-specific inhibitors of the bovine heart mitochondrial ATPase. Biochim. Biophys. Acta 806277-282. [DOI] [PubMed] [Google Scholar]
- 117.Fellous, G., C. Godinot, H. Baubichon, A. Di Pietro, and D. C. Gautheron. 1984. Photolabeling on β subunit of the nucleotide site related to hysteretic inhibition of mitochondrial F1-ATPase. Biochemistry 235294-5299. [Google Scholar]
- 118.Ferguson, S. J., and P. John. 1977. The inhibitor-sensitivity of the plasma-membrane adenosine triphosphatase of Paracoccus denitrificans: comparison with the mitochondrial adenosine triphosphatase. Biochem. Soc. Trans. 51525-1527. [DOI] [PubMed] [Google Scholar]
- 119.Ferguson, S. J., P. John, W. J. Lloyd, G. K. Radda, and F. R. Whatley. 1974. Selective and reversible inhibition of the ATPase of Micrococcus denitrificans by 7-chloro-4-nitrobenzo-2-oxa-1,3 diazole. Biochim. Biophys. Acta 357457-461. [DOI] [PubMed] [Google Scholar]
- 120.Ferguson, S. J., W. J. Lloyd, M. H. Lyons, and G. K. Radda. 1975. The mitochondrial ATPase. Evidence for a single essential tyrosine residue. Eur. J. Biochem. 54117-126. [DOI] [PubMed] [Google Scholar]
- 121.Ferguson, S. J., W. J. Lloyd, and G. K. Radda. 1975. The mitochondrial ATPase. Selective modification of a nitrogen residue in the β subunit. Eur. J. Biochem. 54127-133. [DOI] [PubMed] [Google Scholar]
- 122.Fillingame, R. H. 1975. Identification of the dicyclohexylcarbodiimide-reactive protein component of the adenosine 5′-triphosphate energy-transducing system of Escherichia coli. J. Bacteriol. 124870-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fillingame, R. H., M. Oldenburg, and D. Fraga. 1991. Mutation of alanine 24 to serine in subunit c of the Escherichia coli F1F0-ATP synthase reduces reactivity of aspartyl 61 with dicyclohexylcarbodiimide. J. Biol. Chem. 26620934-20939. [PubMed] [Google Scholar]
- 124.Fisher, R. R., and R. J. Guillory. 1968. The action of Dio-9: energy-linked swelling of rat-liver mitochondria. Biochim. Biophys. Acta 162182-194. [DOI] [PubMed] [Google Scholar]
- 125.Fisher, R. R., and R. J. Guillory. 1967. Inhibition of the energy conservation reactions of Rhodospirillum rubrum by Dio-9. Biochim. Biophys. Acta 143654-656. [DOI] [PubMed] [Google Scholar]
- 126.Fitin, A. F., E. A. Vasilyeva, and A. D. Vinogradov. 1979. An inhibitory high affinity binding site for ADP in the oligomycin-sensitive ATPase of beef heart submitochondrial particles. Biochem. Biophys. Res. Commun. 86434-439. [DOI] [PubMed] [Google Scholar]
- 127.Frasch, A. C., J. J. Cazzulo, and A. O. Stoppani. 1978. Solubilization and some properties of the Mg2+-activated adenosine triphosphatase from Trypanosoma cruzi. Comp. Biochem. Physiol. B 61207-212. [DOI] [PubMed] [Google Scholar]
- 128.Frigeri, L., Y. M. Galante, W. G. Hanstein, and Y. Hatefi. 1977. Effects of arginine binding reagents on ATPase and ATP-Pi exchange activities of mitochondrial ATP synthetase complex (complex V). J. Biol. Chem. 2523147-3152. [PubMed] [Google Scholar]
- 129.Frigeri, L., Y. M. Galante, and Y. Hatefi. 1978. Interaction of complex V and F1-ATPase with [14C]phenylglyoxal. J. Biol. Chem. 2538935-8940. [PubMed] [Google Scholar]
- 130.Futai, M., P. C. Sternweis, and L. A. Heppel. 1974. Purification and properties of reconstitutively active and inactive adenosinetriphosphatase from Escherichia coli. Proc. Natl. Acad. Sci. USA 712725-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Galanis, M., J. R. Mattoon, and P. Nagley. 1989. Amino acid substitutions in mitochondrial ATP synthase subunit 9 of Saccharomyces cerevisiae leading to venturicidin or ossamycin resistance. FEBS Lett. 249333-336. [DOI] [PubMed] [Google Scholar]
- 132.Garin, J., F. Boulay, J. P. Issartel, J. Lunardi, and P. V. Vignais. 1986. Identification of amino acid residues photolabeled with 2-azido[α-32P]adenosine diphosphate in the β subunit of beef heart mitochondrial F1-ATPase. Biochemistry 254431-4437. [DOI] [PubMed] [Google Scholar]
- 133.Garin, J., L. Michel, A. Dupuis, J. P. Issartel, J. Lunardi, J. Hoppe, and P. Vignais. 1989. Photolabeling of the phosphate binding site of mitochondrial F1-ATPase by [32P]azidonitrophenyl phosphate. Identification of the photolabeled amino acid residues. Biochemistry 281442-1448. [DOI] [PubMed] [Google Scholar]
- 134.Garin, J., M. Vincon, J. Gagnon, and P. Vignais. 1994. Photolabeling of mitochondrial F1-H+ATPase by 2-azido[3H]ADP and 8-azido[3H]ADP entrapped as fluorometal complexes into the catalytic sites of the enzyme. Biochemistry 333772-3777. [DOI] [PubMed] [Google Scholar]
- 135.Gaurrand, S., S. Desjardins, C. Meyer, P. Bonnet, J. M. Argoullon, H. Oulyadi, and J. Guillemont. 2006. Conformational analysis of R207910, a new drug candidate for the treatment of tuberculosis, by a combined NMR and molecular modeling approach. Chem. Biol. Drug Des. 6877-84. [DOI] [PubMed] [Google Scholar]
- 136.Gause, E. M., M. A. Buck, and M. G. Douglas. 1981. Binding of citreoviridin to the β subunit of the yeast F1-ATPase. J. Biol. Chem. 256557-559. [PubMed] [Google Scholar]
- 137.Gibbons, C., M. G. Montgomery, A. G. Leslie, and J. E. Walker. 2000. The structure of the central stalk in bovine F1-ATPase at 2.4 A resolution. Nat. Struct. Biol. 71055-1061. [DOI] [PubMed] [Google Scholar]
- 138.Girvin, M. E., and R. H. Fillingame. 1994. Hairpin folding of subunit c of F1FO ATP synthase: 1H distance measurements to nitroxide-derivatized aspartyl-61. Biochemistry 33665-674. [DOI] [PubMed] [Google Scholar]
- 139.Glaser, E., E. Cadenas, S. Andell, and L. Ernster. 1988. Inhibition of the mitochondrial F1-ATPase by Rose Bengal mediated photooxidation. Interaction of the Fe2+ chelate of bathophenanthroline with the sensitizer. Acta Chem. Scand. B 42175-182. [DOI] [PubMed] [Google Scholar]
- 140.Glaser, E., B. Norling, J. Kopecky, and L. Ernster. 1982. Comparison of the effects of oligomycin and dicyclohexylcarbodiimide on mitochondrial ATPase and related reactions. Eur. J. Biochem. 121525-531. [DOI] [PubMed] [Google Scholar]
- 141.Gledhill, J. R., M. G. Montgomery, A. G. Leslie, and J. E. Walker. 2007. How the regulatory protein, IF1, inhibits F1-ATPase from bovine mitochondria. Proc. Natl. Acad. Sci. USA 10415671-15676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gledhill, J. R., M. G. Montgomery, A. G. Leslie, and J. E. Walker. 2007. Mechanism of inhibition of bovine F1-ATPase by resveratrol and related polyphenols. Proc. Natl. Acad. Sci. USA 10413632-13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gledhill, J. R., and J. E. Walker. 2005. Inhibition sites in F1-ATPase from bovine heart mitochondria. Biochem. J. 386591-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gomez-Fernandez, J. C., and D. A. Harris. 1978. A thermodynamic analysis of the interaction between the mitochondrial coupling adenosine triphosphatase and its naturally occurring inhibitor protein. Biochem. J. 176967-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gregory, R., and B. Hess. 1981. The sulphydryl content of yeast mitochondrial F1-ATPase and the stoichiometry of subunits. FEBS Lett. 129210-214. [DOI] [PubMed] [Google Scholar]
- 146.Gregory, R., D. Recktenwald, and B. Hess. 1981. The interaction of nucleotides with F1-ATPase inactivated with 4-chloro-7-nitrobenzofurazan. Biochim. Biophys. Acta 635284-294. [DOI] [PubMed] [Google Scholar]
- 147.Gresser, M. J., S. Beharry, and D. M. Moennich. 1984. Inhibition of mitochondrial F1-ATPase by adenylyl imidodiphosphate. Curr. Top. Cell. Regul. 24365-378. [DOI] [PubMed] [Google Scholar]
- 148.Griffiths, D. E. 1997. Cation adducts of mitochondrial ATPase subunit c: is subunit c a proton/cation exchanger? Biochem. Soc. Trans. 25388S. [DOI] [PubMed] [Google Scholar]
- 149.Griffiths, D. E. 1994. Dibutyltin-3-hydroxyflavone titrates a dissociable component (cofactor) of ATP synthase: an energy transfer component linked to the ubiquinone pool. Biochem. Soc. Trans. 22225S. [DOI] [PubMed] [Google Scholar]
- 150.Griffiths, D. E., and R. L. Houghton. 1974. Studies on energy-linked reactions: modified mitochondrial ATPase of oligomycin-resistant mutants of Saccharomyces cerevisiae. Eur. J. Biochem. 46157-167. [DOI] [PubMed] [Google Scholar]
- 151.Griffiths, D. E., R. L. Houghton, W. E. Lancashire, and P. A. Meadows. 1975. Studies on energy-linked reactions: isolation and properties of mitochondrial venturicidin-resistant mutants of Saccharomyces cerevisiae. Eur. J. Biochem. 51393-402. [DOI] [PubMed] [Google Scholar]
- 152.Grodsky, N. B., C. Dou, and W. S. Allison. 1998. Mutations in the nucleotide binding domain of the α subunits of the F1-ATPase from thermophilic Bacillus PS3 that affect cross-talk between nucleotide binding sites. Biochemistry 371007-1014. [DOI] [PubMed] [Google Scholar]
- 153.Groth, G. 2002. Structure of spinach chloroplast F1-ATPase complexed with the phytopathogenic inhibitor tentoxin. Proc. Natl. Acad. Sci. USA 993464-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Groth, G., D. A. Mills, E. Christiansen, M. L. Richter, and B. Huchzermeyer. 2000. Characterization of a phosphate binding domain on the α-subunit of chloroplast ATP synthase using the photoaffinity phosphate analogue 4-azido-2-nitrophenyl phosphate. Biochemistry 3913781-13787. [DOI] [PubMed] [Google Scholar]
- 155.Groth, G., and E. Pohl. 2001. The structure of the chloroplast F1-ATPase at 3.2 A resolution. J. Biol. Chem. 2761345-1352. [DOI] [PubMed] [Google Scholar]
- 156.Grover, G. J., K. S. Atwal, P. G. Sleph, F. L. Wang, H. Monshizadegan, T. Monticello, and D. W. Green. 2004. Excessive ATP hydrolysis in ischemic myocardium by mitochondrial F1F0-ATPase: effect of selective pharmacological inhibition of mitochondrial ATPase hydrolase activity. Am. J. Physiol. Heart Circ. Physiol. 287H1747-H1755. [DOI] [PubMed] [Google Scholar]
- 157.Grubmeyer, C., and H. S. Penefsky. 1981. The presence of two hydrolytic sites on beef heart mitochondrial adenosine triphosphatase. J. Biol. Chem. 2563718-3727. [PubMed] [Google Scholar]
- 158.Gruys, K. J., J. L. Urbauer, and S. M. Schuster. 1985. Metal-nucleotide structural characteristics during catalysis by beef heart mitochondrial F1. J. Biol. Chem. 2606533-6540. [PubMed] [Google Scholar]
- 159.Guerrero, K. J., L. L. Ehler, and P. D. Boyer. 1990. Guanosine and formycin triphosphates bind at non-catalytic nucleotide binding sites of CF1 ATPase and inhibit ATP hydrolysis. FEBS Lett. 270187-190. [DOI] [PubMed] [Google Scholar]
- 160.Guerrero, K. J., Z. X. Xue, and P. D. Boyer. 1990. Active/inactive state transitions of the chloroplast F1 ATPase are induced by a slow binding and release of Mg2+. Relationship to catalysis and control of F1 ATPases. J. Biol. Chem. 26516280-16287. [PubMed] [Google Scholar]
- 161.Guerrieri, F., S. Micelli, C. Massagli, E. Gallucci, and S. Papa. 1984. Interaction of the aminoglycoside antibiotic dihydrostreptomycin with the H+-ATPase of mitochondria. Biochem. Pharmacol. 332505-2510. [DOI] [PubMed] [Google Scholar]
- 162.Guerrieri, F., and S. Papa. 1981. Effect of chemical modifiers of amino acid residues on proton conduction by the H+-ATPase of mitochondria. J. Bioenerg. Biomembr. 13393-409. [DOI] [PubMed] [Google Scholar]
- 163.Guillory, R. J. 1964. The action of Dio-9: an inhibitor and an uncoupler of oxidative phosphorylation. Biochim. Biophys. Acta 89197-207. [DOI] [PubMed] [Google Scholar]
- 164.Gupta, S., S. B. Krasnoff, D. W. Roberts, and J. A. A. A. Renwick. 1992. Structure of efrapeptins from the fungus Tolypocladium niveum: peptide inhibitors of mitochondrial ATPase. J. Org. Chem. 572306-2313. [Google Scholar]
- 165.Hadler, H. I., and J. M. Demetriou. 1975. A new inhibitor of coupled oxidative phosphorylation, 5-hydroxynaphthalenedicarboxylic anhydride, a derivative of a carcinogenic polynuclear hydrocarbon. Biochemistry 145374-5378. [DOI] [PubMed] [Google Scholar]
- 166.Hamann, L. G., C. Z. Ding, A. V. Miller, C. S. Madsen, P. Wang, P. D. Stein, A. T. Pudzianowski, D. W. Green, H. Monshizadegan, and K. S. Atwal. 2004. Benzodiazepine-based selective inhibitors of mitochondrial F1F0 ATP hydrolase. Bioorg. Med. Chem. Lett. 141031-1034. [DOI] [PubMed] [Google Scholar]
- 167.Hannum, D. M., W. C. Barrette, Jr., and J. K. Hurst. 1995. Subunit sites of oxidative inactivation of Escherichia coli F1-ATPase by HOCl. Biochem. Biophys. Res. Commun. 212868-874. [DOI] [PubMed] [Google Scholar]
- 168.Hara, K. Y., Y. Kato-Yamada, Y. Kikuchi, T. Hisabori, and M. Yoshida. 2001. The role of the βDELSEED motif of F1-ATPase: propagation of the inhibitory effect of the ɛ subunit. J. Biol. Chem. 27623969-23973. [DOI] [PubMed] [Google Scholar]
- 169.Harold, F. M., J. R. Baarda, C. Baron, and A. Abrams. 1969. Dio 9 and chlorhexidine: inhibitors of membrane-bound ATPase and of cation transport in Streptococcus faecalis. Biochim. Biophys. Acta 183129-136. [DOI] [PubMed] [Google Scholar]
- 170.Hase, Y., M. Tatsuno, T. Nishi, K. Kataoka, Y. Kabe, Y. Yamaguchi, N. Ozawa, M. Natori, H. Handa, and H. Watanabe. 2008. Atrazine binds to F1F0-ATP synthase and inhibits mitochondrial function in sperm. Biochem. Biophys. Res. Commun. 36666-72. [DOI] [PubMed] [Google Scholar]
- 171.Hermolin, J., and R. H. Fillingame. 1989. H+-ATPase activity of Escherichia coli F1F0 is blocked after reaction of dicyclohexylcarbodiimide with a single proteolipid (subunit c) of the F0 complex. J. Biol. Chem. 2643896-3903. [PubMed] [Google Scholar]
- 172.Hicks, D. B., and T. A. Krulwich. 1990. Purification and reconstitution of the F1F0-ATP synthase from alkaliphilic Bacillus firmus OF4. Evidence that the enzyme translocates H+ but not Na+. J. Biol. Chem. 26520547-20554. [PubMed] [Google Scholar]
- 173.Higa, A. I., and J. J. Cazzulo. 1981. Mg2+-activated adenosine triphosphatase from Crithidia fasciculata: purification and inhibition by suramin and efrapeptin. Mol. Biochem. Parasitol. 3357-367. [DOI] [PubMed] [Google Scholar]
- 174.Hochman, Y., and C. Carmeli. 1981. Correlation between the kinetics of activation and inhibition of adenosinetriphosphatase activity by divalent metal ions and the binding of manganese to chloroplast coupling factor 1. Biochemistry 206287-6292. [DOI] [PubMed] [Google Scholar]
- 175.Hollemans, M., M. J. Runswick, I. M. Fearnley, and J. E. Walker. 1983. The sites of labeling of the β-subunit of bovine mitochondrial F1-ATPase with 8-azido-ATP. J. Biol. Chem. 2589307-9313. [PubMed] [Google Scholar]
- 176.Hong, S., and P. L. Pedersen. 2002. ATP synthase of yeast: structural insight into the different inhibitory potencies of two regulatory peptides and identification of a new potential regulator. Arch. Biochem. Biophys. 40538-43. [DOI] [PubMed] [Google Scholar]
- 177.Hong, S., and P. L. Pedersen. 2003. Subunit E of mitochondrial ATP synthase: a bioinformatic analysis reveals a phosphopeptide binding motif supporting a multifunctional regulatory role and identifies a related human brain protein with the same motif. Proteins 51155-161. [DOI] [PubMed] [Google Scholar]
- 178.Horstman, L. L., and E. Racker. 1970. Partial resolution of the enzyme catalyzing oxidative phosphorylation. XXII. Interaction between mitochondrial adenosine triphosphatase inhibitor and mitochondrial adenosine triphosphatase. J. Biol. Chem. 2451336-1344. [PubMed] [Google Scholar]
- 179.Hu, N., D. A. Mills, B. Huchzermeyer, and M. L. Richter. 1993. Inhibition by tentoxin of cooperativity among nucleotide binding sites on chloroplast coupling factor 1. J. Biol. Chem. 2688536-8540. [PubMed] [Google Scholar]
- 180.Huang, T. C., H. Y. Chang, C. H. Hsu, W. H. Kuo, K. J. Chang, and H. F. Juan. 2008. Targeting therapy for breast carcinoma by ATP synthase inhibitor aurovertin B. J. Proteome Res. 71433-1444. [DOI] [PubMed] [Google Scholar]
- 181.Huitric, E., P. Verhasselt, K. Andries, and S. E. Hoffner. 2007. In vitro antimycobacterial spectrum of a diarylquinoline ATP synthase inhibitor. Antimicrob. Agents Chemother. 514202-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hurst, J. K., and W. C. Barrette, Jr. 1989. Leukocytic oxygen activation and microbicidal oxidative toxins. Crit. Rev. Biochem. Mol. Biol. 24271-328. [DOI] [PubMed] [Google Scholar]
- 183.Ibrahim, M., K. Andries, N. Lounis, A. Chauffour, C. Truffot-Pernot, V. Jarlier, and N. Veziris. 2007. Synergistic activity of R207910 combined with pyrazinamide against murine tuberculosis. Antimicrob. Agents Chemother. 511011-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ida, K., T. Noumi, M. Maeda, T. Fukui, and M. Futai. 1991. Catalytic site of F1-ATPase of Escherichia coli. Lys-155 and Lys-201 of the β subunit are located near the γ-phosphate group of ATP in the presence of Mg2+. J. Biol. Chem. 2665424-5429. [PubMed] [Google Scholar]
- 185.Igarashi, K., K. Kashiwagi, H. Kobayashi, R. Ohnishi, T. Kakegawa, A. Nagasu, and S. Hirose. 1989. Effect of polyamines on mitochondrial F1-ATPase catalyzed reactions. J. Biochem. (Tokyo) 106294-298. [DOI] [PubMed] [Google Scholar]
- 186.Imedidze, E. A., I. A. Kozlov, V. A. Metel'skaia, and M. Mil'grom Ia. 1978. Inhibition of mitochondrial ATPase by water-soluble carbodiimide. Biokhimiia 431404-1413. [PubMed] [Google Scholar]
- 187.Issartel, J. P., A. Dupuis, J. Lunardi, and P. V. Vignais. 1991. Fluoroaluminum and fluoroberyllium nucleoside diphosphate complexes as probes of the enzymatic mechanism of the mitochondrial F1-ATPase. Biochemistry 304726-4733. [DOI] [PubMed] [Google Scholar]
- 188.Issartel, J. P., G. Klein, M. Satre, and P. V. Vignais. 1983. Aurovertin binding sites on beef heart mitochondrial F1-ATPase. Study with [14C]aurovertin D of the binding stoichiometry and of the interaction between aurovertin and the natural ATPase inhibitor for binding to F1. Biochemistry 223492-3497. [DOI] [PubMed] [Google Scholar]
- 189.Issartel, J. P., and P. V. Vignais. 1984. Evidence for a nucleotide binding site on the isolated β subunit from Escherichia coli F1-ATPase. Interaction between nucleotide and aurovertin D binding sites. Biochemistry 236591-6595. [DOI] [PubMed] [Google Scholar]
- 190.Ivey, D. M., and L. G. Ljungdahl. 1986. Purification and characterization of the F1-ATPase from Clostridium thermoaceticum. J. Bacteriol. 165252-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Iwadate, M., T. Asakura, and M. P. Williamson. 1998. The structure of the melittin tetramer at different temperatures. An NOE-based calculation with chemical shift refinement. Eur. J. Biochem. 257479-487. [DOI] [PubMed] [Google Scholar]
- 192.John, U. P., and P. Nagley. 1986. Amino acid substitutions in mitochondrial ATPase subunit 6 of Saccharomyces cerevisiae leading to oligomycin resistance. FEBS Lett. 20779-83. [DOI] [PubMed] [Google Scholar]
- 193.Johnson, K. M., X. Chen, A. Boitano, L. Swenson, A. W. Opipari, Jr., and G. D. Glick. 2005. Identification and validation of the mitochondrial F1F0-ATPase as the molecular target of the immunomodulatory benzodiazepine Bz-423. Chem. Biol. 12485-496. [DOI] [PubMed] [Google Scholar]
- 194.Joshi, V. K., and J. H. Wang. 1987. Cross-linking study of the quaternary fine structure of mitochondrial F1-ATPase. J. Biol. Chem. 26215721-15725. [PubMed] [Google Scholar]
- 195.Kagawa, R., M. G. Montgomery, K. Braig, A. G. Leslie, and J. E. Walker. 2004. The structure of bovine F1-ATPase inhibited by ADP and beryllium fluoride. EMBO J. 232734-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kagawa, Y., and N. Nukiwa. 1981. Conversion of stable ATPase to labile ATPase by acetylation, and the αβ and αγ subunit complexes during its reconstitution. Biochem. Biophys. Res. Commun. 1001370-1376. [DOI] [PubMed] [Google Scholar]
- 197.Kandpal, R. P., K. E. Stempel, and P. D. Boyer. 1987. Characteristics of the formation of enzyme-bound ATP from medium inorganic phosphate by mitochondrial F1 adenosinetriphosphatase in the presence of dimethyl sulfoxide. Biochemistry 261512-1517. [DOI] [PubMed] [Google Scholar]
- 198.Kato-Yamada, Y., D. Bald, M. Koike, K. Motohashi, T. Hisabori, and M. Yoshida. 1999. ɛ subunit, an endogenous inhibitor of bacterial F1-ATPase, also inhibits F0F1-ATPase. J. Biol. Chem. 27433991-33994. [DOI] [PubMed] [Google Scholar]
- 199.Kauffman, R. F., H. A. Lardy, J. R. Barrio, M. C. Bario, and N. J. Leonard. 1978. Dimensional probes of the enzyme binding sites of adenine nucleotides. Interaction of lin-benzoadenosine 5′-di- and triphosphate with mitochondrial ATP synthetase, purified ATPase, and the adenine nucleotide carrier. Biochemistry 173686-3692. [DOI] [PubMed] [Google Scholar]
- 200.Kawai, K., H. Fukushima, and Y. Nozawa. 1985. Inhibition of mitochondrial respiration by asteltoxin, a respiratory toxin from Emericella variecolor. Toxicol. Lett. 2873-77. [DOI] [PubMed] [Google Scholar]
- 201.Kayalar, C., J. Rosing, and P. D. Boyer. 1977. An alternating site sequence for oxidative phosphorylation suggested by measurement of substrate binding patterns and exchange reaction inhibitions. J. Biol. Chem. 2522486-2491. [PubMed] [Google Scholar]
- 202.Kenan, D. J., and M. L. Wahl. 2005. Ectopic localization of mitochondrial ATP synthase: a target for anti-angiogenesis intervention? J. Bioenerg. Biomembr. 37461-465. [DOI] [PubMed] [Google Scholar]
- 203.Khananshvili, D., and Z. Gromet-Elhanan. 1985. Evidence that the Mg-dependent low-affinity binding site for ATP and Pi demonstrated on the isolated β subunit of the F0·F1 ATP synthase is a catalytic site. Proc. Natl. Acad. Sci. USA 821886-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Khananshvili, D., and Z. Gromet-Elhanan. 1983. The interaction of carboxyl group reagents with the Rhodospirillum rubrum F1-ATPase and its isolated β-subunit. J. Biol. Chem. 2583720-3725. [PubMed] [Google Scholar]
- 205.Kihara, T., H. Kusakabe, G. Nakamura, T. Sakurai, and K. Isono. 1981. Cytovaricin, a novel antibiotic. J. Antibiot. (Tokyo) 341073-1074. [DOI] [PubMed] [Google Scholar]
- 206.Kim, B. W., H. J. Choo, J. W. Lee, J. H. Kim, and Y. G. Ko. 2004. Extracellular ATP is generated by ATP synthase complex in adipocyte lipid rafts. Exp. Mol. Med. 36476-485. [DOI] [PubMed] [Google Scholar]
- 207.Kim, J. W., H. Adachi, K. Shin-ya, Y. Hayakawa, and H. Seto. 1997. Apoptolidin, a new apoptosis inducer in transformed cells from Nocardiopsis sp. J. Antibiot. (Tokyo) 50628-630. [DOI] [PubMed] [Google Scholar]
- 208.Kim, S. H., R. Vlkolinsky, N. Cairns, and G. Lubec. 2000. Decreased levels of complex III core protein 1 and complex V β chain in brains from patients with Alzheimer's disease and Down syndrome. Cell. Mol. Life Sci. 571810-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Kirst, H. A., J. S. Mynderse, J. W. Martin, P. J. Baker, J. W. Paschal, J. L. Rios Steiner, E. Lobkovsky, and J. Clardy. 1996. Structure of the spiroketal-macrolide ossamycin. J. Antibiot. (Tokyo) 49162-167. [DOI] [PubMed] [Google Scholar]
- 210.Ko, Y. H., M. Bianchet, L. M. Amzel, and P. L. Pedersen. 1997. Novel insights into the chemical mechanism of ATP synthase. Evidence that in the transition state the γ-phosphate of ATP is near the conserved alanine within the P-loop of the β-subunit. J. Biol. Chem. 27218875-18881. [DOI] [PubMed] [Google Scholar]
- 211.Ko, Y. H., S. Hong, and P. L. Pedersen. 1999. Chemical mechanism of ATP synthase. Magnesium plays a pivotal role in formation of the transition state where ATP is synthesized from ADP and inorganic phosphate. J. Biol. Chem. 27428853-28856. [DOI] [PubMed] [Google Scholar]
- 212.Konno, H., T. Murakami-Fuse, F. Fujii, F. Koyama, H. Ueoka-Nakanishi, C. G. Pack, M. Kinjo, and T. Hisabori. 2006. The regulator of the F1 motor: inhibition of rotation of cyanobacterial F1-ATPase by the ɛ subunit. EMBO J. 254596-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kopecky, J., E. Glaser, B. Norling, and L. Ernster. 1981. Relationship between the binding of dicyclohexylcarbodiimide and the inhibition of H+-translocation in submitochondrial particles. FEBS Lett. 131208-212. [DOI] [PubMed] [Google Scholar]
- 214.Kormer, Z. S., I. A. Kozlov, Y. M. Milgrom, and I. Novikova. 1982. Using 2′(3′)-O-trinitrophenyl derivatives of adenine nucleotides to study the structure and mechanism of functioning of soluble mitochondrial ATPase. Eur. J. Biochem. 121451-455. [DOI] [PubMed] [Google Scholar]
- 215.Koul, A., N. Dendouga, K. Vergauwen, B. Molenberghs, L. Vranckx, R. Willebrords, Z. Ristic, H. Lill, I. Dorange, J. Guillemont, D. Bald, and K. Andries. 2007. Diarylquinolines target subunit c of mycobacterial ATP synthase. Nat. Chem. Biol. 3323-324. [DOI] [PubMed] [Google Scholar]
- 216.Kozlov, I. A., M. Milgrom Ya, M. B. Murataliev, and E. N. Vulfson. 1985. The nucleotide binding site of F1-ATPase which carries out uni-site catalysis is one of the alternating active sites of the enzyme. FEBS Lett. 189286-290. [DOI] [PubMed] [Google Scholar]
- 217.Kozlov, I. A., and Y. M. Milgrom. 1980. The non-catalytic nucleotide-binding site of mitochondrial ATPase is localised on the α-subunit of factor F1. Eur. J. Biochem. 106457-462. [DOI] [PubMed] [Google Scholar]
- 218.Krogmann, D. W., and M. L. Stiller. 1962. A naturally occurring cofactor for photosynthetic phosphorylation. Biochem. Biophys. Res. Commun. 746-49. [DOI] [PubMed] [Google Scholar]
- 219.Kumar, G., V. K. Kalra, and A. F. Brodie. 1979. Affinity labeling of coupling factor-latent ATPase from Mycobacterium phlei with 2′,3′-dialdehyde derivatives of adenosine 5′-triphosphate and adenosine 5′-diphosphate. J. Biol. Chem. 2541964-1971. [PubMed] [Google Scholar]
- 220.Laikind, P. K., and W. S. Allison. 1983. Quinacrine mustard inactivates the bovine heart mitochondrial F1-ATPase with the modification of the β subunit. J. Biol. Chem. 25811700-11704. [PubMed] [Google Scholar]
- 221.Laikind, P. K., T. M. Goldenberg, and W. S. Allison. 1982. On the mechanism of inhibition of the bovine heart F1-ATPase by local anesthetics. Biochem. Biophys. Res. Commun. 109423-427. [DOI] [PubMed] [Google Scholar]
- 222.Laikind, P. K., F. C. Hill, and W. S. Allison. 1985. The use of [3H]aniline to identify the essential carboxyl group in the bovine mitochondrial F1-ATPase that reacts with 1-(ethoxycarbonyl)-2-ethoxy-1,2-dihydroquinoline. Arch. Biochem. Biophys. 240904-920. [DOI] [PubMed] [Google Scholar]
- 223.Lang, D. R., and E. Racker. 1974. Effect of quercetin and F1 inhibitor on mitochondrial ATPase and energy-linked reactions in submitochondrial particles. Biochim. Biophys. Acta 333180-186. [DOI] [PubMed] [Google Scholar]
- 224.Lardy, H., P. Reed, and C. H. Lin. 1975. Antibiotic inhibitors of mitochondrial ATP synthesis. Fed. Proc. 341707-1710. [PubMed] [Google Scholar]
- 225.Lardy, H. A., D. Johnson, and M. W. Mc. 1958. Antibiotics as tools for metabolic studies. I. A survey of toxic antibiotics in respiratory, phosphorylative and glycolytic systems. Arch. Biochem. Biophys. 78587-597. [DOI] [PubMed] [Google Scholar]
- 226.Lauber, A. H., T. J. Barrett, M. Subramaniam, M. Schuchard, and T. C. Spelsberg. 1997. A DNA-binding element for a steroid receptor-binding factor is flanked by dual nuclear matrix DNA attachment sites in the c-myc gene promoter. J. Biol. Chem. 27224657-24665. [DOI] [PubMed] [Google Scholar]
- 227.Lauquin, G., R. Pougeois, and P. V. Vignais. 1980. 4-Azido-2-nitrophenyl phosphate, a new photoaffinity derivative of inorganic phosphate. Study of its interaction with the inorganic phosphate binding site of beef heart mitochondrial adenosine triphosphatase. Biochemistry 194620-4626. [DOI] [PubMed] [Google Scholar]
- 228.Lebowitz, M. S., and P. L. Pedersen. 1996. Protein inhibitor of mitochondrial ATP synthase: relationship of inhibitor structure to pH-dependent regulation. Arch. Biochem. Biophys. 330342-354. [DOI] [PubMed] [Google Scholar]
- 229.Lebowitz, M. S., and P. L. Pedersen. 1993. Regulation of the mitochondrial ATP synthase/ATPase complex: cDNA cloning, sequence, overexpression, and secondary structural characterization of a functional protein inhibitor. Arch. Biochem. Biophys. 30164-70. [DOI] [PubMed] [Google Scholar]
- 230.Lee, R. S., J. Pagan, M. Satre, P. V. Vignais, and A. E. Senior. 1989. Identification of a mutation in Escherichia coli F1-ATPase β-subunit conferring resistance to aurovertin. FEBS Lett. 253269-272. [DOI] [PubMed] [Google Scholar]
- 231.Lee, R. S., J. Pagan, S. Wilke-Mounts, and A. E. Senior. 1991. Characterization of Escherichia coli ATP synthase β-subunit mutations using a chromosomal deletion strain. Biochemistry 306842-6847. [DOI] [PubMed] [Google Scholar]
- 232.Linnett, P. E., and R. B. Beechey. 1979. Inhibitors of the ATP synthase system. Methods Enzymol. 55472-518. [DOI] [PubMed] [Google Scholar]
- 233.Linnett, P. E., A. D. Mitchell, M. D. Osselton, L. J. Mulheirn, and R. B. Beechey. 1978. Citreoviridin, a specific inhibitor of the mitochondiral adenosine triphosphatase. Biochem. J. 170503-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Lloyd, D., and S. W. Edwards. 1976. Mitochondrial adenosine triphosphatase of the fission yeast, Schizosaccharomyces pombe 972H−. Changes in activity and inhibitor-sensitivity in response to catabolite repression. Biochem. J. 160335-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Lloyd, D., D. G. Lindmark, and M. Muller. 1979. Adenosine triphosphatase activity of Tritrichomonas foetus. J. Gen. Microbiol. 115301-307. [DOI] [PubMed] [Google Scholar]
- 236.Lotscher, H. R., C. deJong, and R. A. Capaldi. 1984. Inhibition of the adenosinetriphosphatase activity of Escherichia coli F1 by the water-soluble carbodiimide 1-ethyl-3-[3-(dimethylamino)propyl]carbodiimide is due to modification of several carboxyls in the β subunit. Biochemistry 234134-4140. [DOI] [PubMed] [Google Scholar]
- 237.Lounis, N., T. Gevers, J. Van Den Berg, and K. Andries. 2008. Impact of the interaction of R207910 with rifampin on the treatment of tuberculosis studied in the mouse model. Antimicrob. Agents Chemother. 523568-3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Lounis, N., N. Veziris, A. Chauffour, C. Truffot-Pernot, K. Andries, and V. Jarlier. 2006. Combinations of R207910 with drugs used to treat multidrug-resistant tuberculosis have the potential to shorten treatment duration. Antimicrob. Agents Chemother. 503543-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Lowe, P. N., and R. B. Beechey. 1982. Interactions between the mitochondrial adenosinetriphosphatase and periodate-oxidized adenosine 5′-triphosphate, an affinity label for adenosine 5′-triphosphate binding sites. Biochemistry 214073-4082. [DOI] [PubMed] [Google Scholar]
- 240.Lubben, M., U. Lucken, J. Weber, and G. Schafer. 1984. Azidonaphthoyl-ADP: a specific photolabel for the high-affinity nucleotide-binding sites of F1-ATPase. Eur. J. Biochem. 143483-490. [DOI] [PubMed] [Google Scholar]
- 241.Lucero, H., W. I. Lescano, and R. H. Vallejos. 1978. Inhibition of energy conservation reactions in chromatophores of Rhodospirillum rubrum by antibiotics. Arch. Biochem. Biophys. 1869-14. [DOI] [PubMed] [Google Scholar]
- 242.Lucero, H. A., R. A. Ravizzine, and R. H. Vallejos. 1976. Inhibition of spinach chloroplasts photophosphorylation by the antibiotics leucinostatin and efrapeptin. FEBS Lett. 68141-144. [DOI] [PubMed] [Google Scholar]
- 243.Lunardi, J., A. Dupuis, J. Garin, J. P. Issartel, L. Michel, M. Chabre, and P. V. Vignais. 1988. Inhibition of H+-transporting ATPase by formation of a tight nucleoside diphosphate-fluoroaluminate complex at the catalytic site. Proc. Natl. Acad. Sci. USA 858958-8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Lunardi, J., G. J. Lauquin, and P. V. Vignais. 1977. Interaction of azidonitrophenylaminobutyryl-ADP, a photoaffinity ADP analog, with mitochondrial adenosine triphosphatase. Identification of the labeled subunits. FEBS Lett. 80317-323. [DOI] [PubMed] [Google Scholar]
- 245.Lunardi, J., M. Satre, M. Bof, and P. V. Vignais. 1979. Reactivity of the β subunit of Escherichia coli adenosine triphosphatase with 4-chloro-7-nitrobenzofurazan. Biochemistry 185310-5316. [DOI] [PubMed] [Google Scholar]
- 246.Lunardi, J., and P. V. Vignais. 1979. Adenine nucleotide binding sites in chemically modified F1-ATPase: inhibitory effect of 4-chloro-7-nitrobenzofurazan on photolabeling by arylazido nucleotides. FEBS Lett. 10223-28. [DOI] [PubMed] [Google Scholar]
- 247.Lunardi, J., and P. V. Vignais. 1982. Studies of the nucleotide-binding sites on the mitochondrial F1-ATPase through the use of a photoactivable derivative of adenylyl imidodiphosphate. Biochim. Biophys. Acta 682124-134. [DOI] [PubMed] [Google Scholar]
- 248.Marcus, F., S. M. Schuster, and H. A. Lardy. 1976. Essential arginyl residues in mitochondrial adenosine triphosphatase. J. Biol. Chem. 2511775-1780. [PubMed] [Google Scholar]
- 249.Matsui, T., E. Muneyuki, M. Honda, W. S. Allison, C. Dou, and M. Yoshida. 1997. Catalytic activity of the α3β3γ complex of F1-ATPase without noncatalytic nucleotide binding site. J. Biol. Chem. 2728215-8221. [DOI] [PubMed] [Google Scholar]
- 250.Matsuno-Yagi, A., and Y. Hatefi. 1984. Inhibitory chemical modifications of F1-ATPase: effects on the kinetics of adenosine 5′-triphosphate synthesis and hydrolysis in reconstituted systems. Biochemistry 233508-3514. [DOI] [PubMed] [Google Scholar]
- 251.Matsuno-Yagi, A., T. Yagi, and Y. Hatefi. 1985. Studies on the mechanism of oxidative phosphorylation: effects of specific F0 modifiers on ligand-induced conformation changes of F1. Proc. Natl. Acad. Sci. USA 827550-7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.McEnery, M. W., and P. L. Pedersen. 1986. Diethylstilbestrol. A novel F0-directed probe of the mitochondrial proton ATPase. J. Biol. Chem. 2611745-1752. [PubMed] [Google Scholar]
- 253.Melandri, A. B., E. Fabbri, and B. A. Melandri. 1975. Energy transduction in photosynthetic bacteria. VIII. Activation of the energy-transducing ATPase by inorganic phosphate. Biochim. Biophys. Acta 37682-88. [DOI] [PubMed] [Google Scholar]
- 254.Melanson, D. L., and M. S. Spencer. 1981. Kinetics of ATP synthesis in pea cotyledon submitochondrial particles. Plant Physiol. 68648-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Melnick, R. L., J. T. De Sousa, J. Magiure, and L. Packer. 1975. Action of the adenosine triphosphate analog, adenylyl imidodiphosphate in mitochondria. Arch. Biochem. Biophys. 166139-144. [DOI] [PubMed] [Google Scholar]
- 256.Menz, R. I., J. E. Walker, and A. G. Leslie. 2001. Structure of bovine mitochondrial F1-ATPase with nucleotide bound to all three catalytic sites: implications for the mechanism of rotary catalysis. Cell 106331-341. [DOI] [PubMed] [Google Scholar]
- 257.Meyer, W. L., L. F. Kuyper, R. B. Lewis, G. E. Templeton, and S. H. Woodhead. 1974. The amino acid sequence and configuration of tentoxin. Biochem. Biophys. Res. Commun. 56234-240. [DOI] [PubMed] [Google Scholar]
- 258.Michel, L., J. Garin, G. Girault, and P. V. Vignais. 1992. Photolabeling of the phosphate binding site of chloroplast coupling factor 1 with [32P]azidonitrophenyl phosphate. FEBS Lett. 31390-93. [DOI] [PubMed] [Google Scholar]
- 259.Mikami, Y., K. Yazawa, K. Fukushima, T. Arai, S. Udagawa, and R. A. Samson. 1989. Paecilotoxin production in clinical or terrestrial isolates of Paecilomyces lilacinus strains. Mycopathologia 108195-199. [DOI] [PubMed] [Google Scholar]
- 260.Minauro-Sanmiguel, F., C. Bravo, and J. J. Garcia. 2002. Cross-linking of the endogenous inhibitor protein (IF1) with rotor (γ, ɛ) and stator (α) subunits of the mitochondrial ATP synthase. J. Bioenerg. Biomembr. 34433-443. [DOI] [PubMed] [Google Scholar]
- 261.Minkov, I. B., A. F. Fitin, E. A. Vasilyeva, and A. D. Vinogradov. 1979. Mg2+-induced ADP-dependent inhibition of the ATPase activity of beef heart mitochondrial coupling factor F1. Biochem. Biophys. Res. Commun. 891300-1306. [DOI] [PubMed] [Google Scholar]
- 262.Minkov, I. B., and H. Strotmann. 1989. The effect of azide on regulation of the chloroplast H+-ATPase by ADP and phosphate. Biochim. Biophys. Acta 9737-12. [Google Scholar]
- 263.Mitchell, P., and J. Moyle. 1970. Influence of aurovertin on affinity of mitochondrial adenosine triphosphatase for ATP and ADP. FEBS Lett. 6309-311. [DOI] [PubMed] [Google Scholar]
- 264.Mitchell, R. A., B. F. Chang, C. H. Huang, and E. G. DeMaster. 1971. Inhibition of mitochondrial energy-linked functions by arsenate. Evidence for a nonhydrolytic mode of inhibitor action. Biochemistry 102049-2054. [DOI] [PubMed] [Google Scholar]
- 265.Mochimaru, M., and H. Sakurai. 1997. Three kinds of binding site for tentoxin on isolated chloroplast coupling factor 1. FEBS Lett. 41923-26. [DOI] [PubMed] [Google Scholar]
- 266.Mori, Y., M. Suzuki, K. Fukushima, and T. Arai. 1983. Structure of leucinostatin B, an uncoupler on mitochondria. J. Antibiot. (Tokyo) 361084-1086. [DOI] [PubMed] [Google Scholar]
- 267.Moritani, C., Y. Sakai, M. Tsuda, H. Kanazawa, and T. Tsuchiya. 1990. Characteristics of the H+-translocating adenosine triphosphatase of Vibrio parahaemolyticus. Chem. Pharm. Bull. (Tokyo) 38164-167. [DOI] [PubMed] [Google Scholar]
- 268.Moriyama, Y., V. Patel, and M. Futai. 1995. Quinacrine mustard and lipophilic cations inhibitory to both vacuolar H+-ATPase and F0F1-ATP synthase. FEBS Lett. 35969-72. [DOI] [PubMed] [Google Scholar]
- 269.Moser, T. L., D. J. Kenan, T. A. Ashley, J. A. Roy, M. D. Goodman, U. K. Misra, D. J. Cheek, and S. V. Pizzo. 2001. Endothelial cell surface F1-F0 ATP synthase is active in ATP synthesis and is inhibited by angiostatin. Proc. Natl. Acad. Sci. USA 986656-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Moser, T. L., M. S. Stack, I. Asplin, J. J. Enghild, P. Hojrup, L. Everitt, S. Hubchak, H. W. Schnaper, and S. V. Pizzo. 1999. Angiostatin binds ATP synthase on the surface of human endothelial cells. Proc. Natl. Acad. Sci. USA 962811-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Moser, T. L., M. S. Stack, M. L. Wahl, and S. V. Pizzo. 2002. The mechanism of action of angiostatin: can you teach an old dog new tricks? Thromb. Haemost. 87394-401. [PubMed] [Google Scholar]
- 272.Moyle, J., and P. Mitchell. 1975. Active/inactive state transitions of mitochondrial ATPase molecules influenced by Mg2+, anions and aurovertin. FEBS Lett. 5655-61. [DOI] [PubMed] [Google Scholar]
- 273.Muneyuki, E., T. Hisabori, T. Sasayama, K. Mochizuki, and M. Yoshida. 1996. The heterogeneous interaction of substoichiometric TNP-ATP and F1-ATPase from Escherichia coli. J. Biochem. (Tokyo) 120940-945. [DOI] [PubMed] [Google Scholar]
- 274.Muneyuki, E., M. Makino, H. Kamata, Y. Kagawa, M. Yoshida, and H. Hirata. 1993. Inhibitory effect of NaN3 on the F0F1 ATPase of submitochondrial particles as related to nucleotide binding. Biochim. Biophys. Acta 114462-68. [DOI] [PubMed] [Google Scholar]
- 275.Munter, K., M. Athanasiou, and C. Stournaras. 1989. Inhibition of cellular activities by triethyllead. Role of glutathione and accumulation of triethyllead in vitro. Biochem. Pharmacol. 383941-3945. [DOI] [PubMed] [Google Scholar]
- 276.Murataliev, M. B. 1995. Interaction of mitochondrial F1-ATPase with trinitrophenyl derivatives of ATP. Photoaffinity labeling of binding sites with 2-azido-2′,3′-O-(4,6-trinitrophenyl)adenosine 5′-triphosphate. Eur. J. Biochem. 232578-585. [DOI] [PubMed] [Google Scholar]
- 277.Murataliev, M. B., and P. D. Boyer. 1994. Interaction of mitochondrial F1-ATPase with trinitrophenyl derivatives of ATP and ADP. Participation of third catalytic site and role of Mg2+ in enzyme inactivation. J. Biol. Chem. 26915431-15439. [PubMed] [Google Scholar]
- 278.Murataliev, M. B., Y. M. Milgrom, and P. D. Boyer. 1991. Characteristics of the combination of inhibitory Mg2+ and azide with the F1 ATPase from chloroplasts. Biochemistry 308305-8310. [DOI] [PubMed] [Google Scholar]
- 279.Nadanaciva, S., J. Weber, and A. E. Senior. 2000. New probes of the F1-ATPase catalytic transition state reveal that two of the three catalytic sites can assume a transition state conformation simultaneously. Biochemistry 399583-9590. [DOI] [PubMed] [Google Scholar]
- 280.Nagley, P., R. M. Hall, and B. G. Ooi. 1986. Amino acid substitutions in mitochondrial ATPase subunit 9 of Saccharomyces cerevisiae leading to oligomycin or venturicidin resistance. FEBS Lett. 195159-163. [DOI] [PubMed] [Google Scholar]
- 281.Nakamoto, R. K., K. Shin, A. Iwamoto, H. Omote, M. Maeda, and M. Futai. 1992. Escherichia coli F0F1-ATPase. Residues involved in catalysis and coupling. Ann. N. Y. Acad. Sci. 671335-344. [DOI] [PubMed] [Google Scholar]
- 282.Nakanishi-Matsui, M., S. Kashiwagi, H. Hosokawa, D. J. Cipriano, S. D. Dunn, Y. Wada, and M. Futai. 2006. Stochastic high-speed rotation of Escherichia coli ATP synthase F1 sector: the ɛ subunit-sensitive rotation. J. Biol. Chem. 2814126-4131. [DOI] [PubMed] [Google Scholar]
- 283.Nelson, N., B. I. Kanner, and D. L. Gutnick. 1974. Purification and properties of Mg2+-Ca2+ adenosinetriphosphatase from Escherichia coli. Proc. Natl. Acad. Sci. USA 712720-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Nelson, N., H. Nelson, and E. Racker. 1972. Partial resolution of the enzymes catalyzing photophosphorylation. XII. Purification and properties of an inhibitor isolated from chloroplast coupling factor 1. J. Biol. Chem. 2477657-7662. [PubMed] [Google Scholar]
- 285.Nieuwenhuis, F. J., B. I. Kanner, D. L. Gutnick, P. W. Postma, and K. van Dam. 1973. Energy conservation in membranes of mutants of Escherichia coli defective in oxidative phosphorylation. Biochim. Biophys. Acta 32562-71. [DOI] [PubMed] [Google Scholar]
- 286.Nishino, H., M. Murakoshi, X. Y. Mou, S. Wada, M. Masuda, Y. Ohsaka, Y. Satomi, and K. Jinno. 2005. Cancer prevention by phytochemicals. Oncology 69(Suppl. 1)38-40. [DOI] [PubMed] [Google Scholar]
- 287.Noumi, T., M. Maeda, and M. Futai. 1987. Mode of inhibition of sodium azide on H+-ATPase of Escherichia coli. FEBS Lett. 213381-384. [DOI] [PubMed] [Google Scholar]
- 288.Noumi, T., M. Tagaya, K. Miki-Takeda, M. Maeda, T. Fukui, and M. Futai. 1987. Loss of unisite and multisite catalyses by Escherichia coli F1 through modification with adenosine tri- or tetraphosphopyridoxal. J. Biol. Chem. 2627686-7692. [PubMed] [Google Scholar]
- 289.Nowak, K. F., V. Tabidze, and R. E. McCarty. 2002. The C-terminal domain of the ɛ subunit of the chloroplast ATP synthase is not required for ATP synthesis. Biochemistry 4115130-15134. [DOI] [PubMed] [Google Scholar]
- 290.Ogawa, W., S. Izawa, Y. Sakai-Tomita, C. Moritani, M. Tsuda, K. Kinomura, S. Kitazawa, and T. Tsuchiya. 1994. F0F1-ATPase of Vibrio parahaemolyticus: purification using new detergents and characterization. Biochim. Biophys. Acta 118869-74. [DOI] [PubMed] [Google Scholar]
- 291.Oren, R., and Z. Gromet-Elhanan. 1979. Coupling factor ATPase complex of Rhodospirillum rubrum. Purification and characterization of an oligomycin and N,N′-dicyclohexylcarbodiimide-sensitive (Ca2+ + Mg2+)-ATPase. Biochim. Biophys. Acta 548106-118. [DOI] [PubMed] [Google Scholar]
- 292.Orriss, G. L., A. G. Leslie, K. Braig, and J. E. Walker. 1998. Bovine F1-ATPase covalently inhibited with 4-chloro-7-nitrobenzofurazan: the structure provides further support for a rotary catalytic mechanism. Structure 6831-837. [DOI] [PubMed] [Google Scholar]
- 293.Osanai, T., K. Magota, M. Tanaka, M. Shimada, R. Murakami, S. Sasaki, H. Tomita, N. Maeda, and K. Okumura. 2005. Intracellular signaling for vasoconstrictor coupling factor 6: novel function of β-subunit of ATP synthase as receptor. Hypertension 461140-1146. [DOI] [PubMed] [Google Scholar]
- 294.Osanai, T., S. Okada, K. Sirato, T. Nakano, M. Saitoh, K. Magota, and K. Okumura. 2001. Mitochondrial coupling factor 6 is present on the surface of human vascular endothelial cells and is released by shear stress. Circulation 1043132-3136. [DOI] [PubMed] [Google Scholar]
- 295.Pacheco-Moises, F., F. Minauro-Sanmiguel, C. Bravo, and J. J. Garcia. 2002. Sulfite inhibits the F1F0-ATP synthase and activates the F1F0-ATPase of Paracoccus denitrificans. J. Bioenerg. Biomembr. 34269-278. [DOI] [PubMed] [Google Scholar]
- 296.Paik, S. R., J. M. Jault, and W. S. Allison. 1994. Inhibition and inactivation of the F1 adenosinetriphosphatase from Bacillus PS3 by dequalinium and activation of the enzyme by lauryl dimethylamine oxide. Biochemistry 33126-133. [DOI] [PubMed] [Google Scholar]
- 297.Pal, P. K., and P. S. Coleman. 1990. Detecting precatalytic conformational changes in F1-ATPase with 4-benzoyl(benzoyl)-1-amidofluorescein, a novel fluorescent nucleotide site-specific photoaffinity label. J. Biol. Chem. 26514996-15002. [PubMed] [Google Scholar]
- 298.Palmer, D. N., G. Barns, D. R. Husbands, and R. D. Jolly. 1986. Ceroid lipofuscinosis in sheep. II. The major component of the lipopigment in liver, kidney, pancreas, and brain is low molecular weight protein. J. Biol. Chem. 2611773-1777. [PubMed] [Google Scholar]
- 299.Palmer, D. N., I. M. Fearnley, J. E. Walker, N. A. Hall, B. D. Lake, L. S. Wolfe, M. Haltia, R. D. Martinus, and R. D. Jolly. 1992. Mitochondrial ATP synthase subunit c storage in the ceroid-lipofuscinoses (Batten disease). Am. J. Med. Genet. 42561-567. [DOI] [PubMed] [Google Scholar]
- 300.Papa, S., M. Tuena de Gomez-Puyou, and A. Gomez-Puyou. 1975. On the mechanism of action of alkylguanidines on oxidative phosphorylation in mitochondria. Eur. J. Biochem. 551-8. [DOI] [PubMed] [Google Scholar]
- 301.Park, M., L. Lin, S. Thomas, H. D. Braymer, P. M. Smith, D. H. Harrison, and D. A. York. 2004. The F1-ATPase β-subunit is the putative enterostatin receptor. Peptides 252127-2133. [DOI] [PubMed] [Google Scholar]
- 302.Pedersen, P. L. 1975. Adenosine triphosphatase from rat liver mitochondria: separate sites involved in ATP hydrolysis and in the reversible, high affinity binding of ADP. Biochem. Biophys. Res. Commun. 64610-616. [DOI] [PubMed] [Google Scholar]
- 303.Pedersen, P. L. 2007. Transport ATPases into the year 2008: a brief overview related to types, structures, functions and roles in health and disease. J. Bioenerg. Biomembr. 39349-355. [DOI] [PubMed] [Google Scholar]
- 304.Pedersen, P. L., and L. M. Amzel. 1993. ATP synthases. Structure, reaction center, mechanism, and regulation of one of nature's most unique machines. J. Biol. Chem. 2689937-9940. [PubMed] [Google Scholar]
- 305.Pedersen, P. L., Y. H. Ko, and S. Hong. 2000. ATP synthases in the year 2000: evolving views about the structures of these remarkable enzyme complexes. J. Bioenerg. Biomembr. 32325-332. [DOI] [PubMed] [Google Scholar]
- 306.Penefsky, H. S. 1974. Differential effects of adenylyl imidodiphosphate on adenosine triphosphate synthesis and the partial reactions of oxidative phosphorylation. J. Biol. Chem. 2493579-3585. [PubMed] [Google Scholar]
- 307.Penefsky, H. S. 1977. Reversible binding of Pi by beef heart mitochondrial adenosine triphosphatase. J. Biol. Chem. 2522891-2899. [PubMed] [Google Scholar]
- 308.Penefsky, H. S., M. E. Pullman, A. Datta, and E. Racker. 1960. Partial resolution of the enzymes catalyzing oxidative phosphorylation. II. Participation of a soluble adenosine tolphosphatase in oxidative phosphorylation. J. Biol. Chem. 2353330-3336. [PubMed] [Google Scholar]
- 309.Penefsky, H. S., and R. C. Warner. 1965. Partial resolution of the enzymes catalyzing oxidative phosphorylation. VI. Studies on the mechanism of cold inactivation of mitochondrial adenosine triphosphatase. J. Biol. Chem. 2404694-4702. [PubMed] [Google Scholar]
- 310.Penniston, J. T. 1971. High hydrostatic pressure and enzymic activity: inhibition of multimeric enzymes by dissociation. Arch. Biochem. Biophys. 142322-332. [DOI] [PubMed] [Google Scholar]
- 311.Perlin, D. S., L. R. Latchney, and A. E. Senior. 1985. Inhibition of Escherichia coli H+-ATPase by venturicidin, oligomycin and ossamycin. Biochim. Biophys. Acta 807238-244. [DOI] [PubMed] [Google Scholar]
- 312.Peter, H. W., M. R. Pinheiro, and M. Silva Lima. 1981. Regulation of the F-ATPase from mitochondria of Vigna sinensis (L.) Savi cv. Pitiuba by spermine, spermidine, putrescine, Mg2+, Na+, and K+. Can. J. Biochem. 5960-66. [DOI] [PubMed] [Google Scholar]
- 313.Petrella, S., E. Cambau, A. Chauffour, K. Andries, V. Jarlier, and W. Sougakoff. 2006. Genetic basis for natural and acquired resistance to the diarylquinoline R207910 in mycobacteria. Antimicrob. Agents Chemother. 502853-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Petrone, G., D. N. Garboczi, and P. L. Pedersen. 1987. Mitochondrial ATP synthase complex: interaction of its F1 adenosinetriphosphatase moiety with the heavy atom iodine. Biochemistry 264016-4021. [DOI] [PubMed] [Google Scholar]
- 315.Phelps, D. C., K. Nordenbrand, B. D. Nelson, and L. Ernster. 1975. Inhibition of purified mitochondiral ATPase (F1) by bathophenanthroline and relief of the inhibition by uncouplers. Biochem. Biophys. Res. Commun. 631005-1012. [DOI] [PubMed] [Google Scholar]
- 316.Pinet, E., F. Cavelier, J. Verducci, G. Girault, L. Dubart, F. Haraux, C. Sigalat, and F. Andre. 1996. Synthesis, structure, and properties of MeSer1-tentoxin, a new cyclic tetrapeptide which interacts specifically with chloroplast F1 H+-ATPase differentiation of inhibitory and stimulating effects. Biochemistry 3512804-12811. [DOI] [PubMed] [Google Scholar]
- 317.Pinet, E., J. M. Gomis, G. Girault, F. Cavelier, J. Verducci, J. P. Noel, and F. Andre. 1996. Tentoxin has at least two binding sites on CF1 and ɛ-depleted CF1 ATPases isolated from spinach chloroplast. FEBS Lett. 395217-220. [DOI] [PubMed] [Google Scholar]
- 318.Pinet, E., J. M. Neumann, I. Dahse, G. Girault, and F. Andre. 1995. Multiple interconverting conformers of the cyclic tetrapeptide tentoxin, [cyclo-(L-MeAla1-L-Leu2-MePhe[(Z)Δ]3-Gly4)], as seen by two-dimensional 1H-nmr spectroscopy. Biopolymers 36135-152. [DOI] [PubMed] [Google Scholar]
- 319.Polgreen, K. E., J. Featherstone, A. C. Willis, and D. A. Harris. 1995. Primary structure and properties of the inhibitory protein of the mitochondrial ATPase (H+-ATP synthase) from potato. Biochim. Biophys. Acta 1229175-180. [DOI] [PubMed] [Google Scholar]
- 320.Pougeois, R. 1983. EEDQ probably reacts with the Mg2+-ATP catalytic sites of mitochondrial and bacterial F1-ATPases. FEBS Lett. 15447-50. [DOI] [PubMed] [Google Scholar]
- 321.Pougeois, R., G. J. Lauquin, and P. V. Vignais. 1983. Interaction of 4-azido-2-nitrophenyl phosphate, an inorganic phosphate photoreactive analogue, with chloroplast coupling factor 1. Biochemistry 221241-1245. [DOI] [PubMed] [Google Scholar]
- 322.Pougeois, R., M. Satre, and P. Vignais. 1978. N-Ethoxycarbonyl-2-ethoxy-1,2-dihydroquinoline, a new inhibitor of the mitochondrial F1-ATPase. Biochemistry 173018-3023. [DOI] [PubMed] [Google Scholar]
- 323.Price, K. E., A. Schlein, W. T. Bradner, and J. Lein. 1963. Peliomycin, a new cytotoxic agent. II. Biological properties. Antimicrob. Agents Chemother. 16195-99. [PubMed] [Google Scholar]
- 324.Pullman, M. E., H. S. Penefsky, A. Datta, and E. Racker. 1960. Partial resolution of the enzymes catalyzing oxidative phosphorylation. I. Purification and properties of soluble dinitrophenol-stimulated adenosine triphosphatase. J. Biol. Chem. 2353322-3329. [PubMed] [Google Scholar]
- 325.Quillen, E. E., G. C. Haslam, H. S. Samra, D. Amani-Taleshi, J. A. Knight, D. E. Wyatt, S. C. Bishop, K. K. Colvert, M. L. Richter, and P. A. Kitos. 2006. Ectoadenylate kinase and plasma membrane ATP synthase activities of human vascular endothelial cells. J. Biol. Chem. 28120728-20737. [DOI] [PubMed] [Google Scholar]
- 326.Rao, R., D. Cunningham, R. L. Cross, and A. E. Senior. 1988. Pyridoxal 5′-diphospho-5′-adenosine binds at a single site on isolated α-subunit from Escherichia coli F1-ATPase and specifically reacts with lysine 201. J. Biol. Chem. 2635640-5645. [PubMed] [Google Scholar]
- 327.Recktenwald, E., and B. Hess. 1977. Allosteric influence of anions on mitochondrial ATPase of yeast. FEBS Lett. 7625-28. [DOI] [PubMed] [Google Scholar]
- 328.Reed, P. W., and H. A. Lardy. 1975. Uncoupling and specific inhibition of phosphoryl transfer reactions in mitochondria by antibiotic A20668. J. Biol. Chem. 2503704-3708. [PubMed] [Google Scholar]
- 329.Ren, H. M., and W. S. Allison. 1997. Photoinactivation of the F1-ATPase from spinach chloroplasts by dequalinium is accompanied by derivatization of methionine β183. J. Biol. Chem. 27232294-32300. [DOI] [PubMed] [Google Scholar]
- 330.Rhodes, A., K. H. Fantes, B. Boothroyd, M. P. McGonagle, and R. Crosse. 1961. Venturicidin: a new antifungal antibiotic of potential use in agriculture. Nature 192952-954. [DOI] [PubMed] [Google Scholar]
- 331.Richter, M. L., R. Hein, and B. Huchzermeyer. 2000. Important subunit interactions in the chloroplast ATP synthase. Biochim. Biophys. Acta 1458326-342. [DOI] [PubMed] [Google Scholar]
- 332.Richter, M. L., W. J. Patrie, and R. E. McCarty. 1984. Preparation of the ɛ subunit and ɛ subunit-deficient chloroplast coupling factor 1 in reconstitutively active forms. J. Biol. Chem. 2597371-7373. [PubMed] [Google Scholar]
- 333.Rigoulet, M. 1990. Control processes in oxidative phosphorylation: kinetic constraints and stoichiometry. Biochim. Biophys. Acta 1018185-189. [DOI] [PubMed] [Google Scholar]
- 334.Rigoulet, M., L. Fraisse, R. Ouhabi, B. Guerin, E. Fontaine, and X. Leverve. 1990. Flux-dependent increase in the stoichiometry of charge translocation by mitochondrial ATPase/ATP synthase induced by almitrine. Biochim. Biophys. Acta 101891-97. [DOI] [PubMed] [Google Scholar]
- 335.Rigoulet, M., X. Leverve, E. Fontaine, R. Ouhabi, and B. Guerin. 1998. Quantitative analysis of some mechanisms affecting the yield of oxidative phosphorylation: dependence upon both fluxes and forces. Mol. Cell. Biochem. 18435-52. [PubMed] [Google Scholar]
- 336.Rigoulet, M., R. Ouhabi, X. Leverve, F. Putod-Paramelle, and B. Guerin. 1989. Almitrine, a new kind of energy-transduction inhibitor acting on mitochondrial ATP synthase. Biochim. Biophys. Acta 975325-329. [DOI] [PubMed] [Google Scholar]
- 337.Roise, D., S. J. Horvath, J. M. Tomich, J. H. Richards, and G. Schatz. 1986. A chemically synthesized pre-sequence of an imported mitochondrial protein can form an amphiphilic helix and perturb natural and artificial phospholipid bilayers. EMBO J. 51327-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Roise, D., F. Theiler, S. J. Horvath, J. M. Tomich, J. H. Richards, D. S. Allison, and G. Schatz. 1988. Amphiphilicity is essential for mitochondrial presequence function. EMBO J. 7649-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Rossi, C., L. Tuttobello, M. Ricci, C. G. Casinovi, and L. Radics. 1987. Leucinostatin D, a novel peptide antibiotic from Paecilomyces marquandii. J. Antibiot. 40130-133. [DOI] [PubMed] [Google Scholar]
- 340.Rubin, E. J. 2005. Toward a new therapy for tuberculosis. N. Engl. J. Med. 352933-934. [DOI] [PubMed] [Google Scholar]
- 341.Russell, J., S. J. Jeng, and R. J. Guillory. 1976. Arylazido aminopropionyl ATP, and active site directed photoaffinity reagent for mitochondrial adenosine triphosphatase. Biochem. Biophys. Res. Commun. 701225-1234. [DOI] [PubMed] [Google Scholar]
- 342.Saad, S. M., J. M. Halloin, and D. J. Hagedorn. 1970. Production, purification, and bioassay of tentoxin. Phytopathology 60415-418. [DOI] [PubMed] [Google Scholar]
- 343.Saishu, T., Y. Kagawa, and R. Shimizu. 1983. Resistance of thermophilic ATPase (TF1) to specific F1-atpase inhibitors including local anesthetics. Biochem. Biophys. Res. Commun. 112822-826. [DOI] [PubMed] [Google Scholar]
- 344.Sakai, Y., H. Kanazawa, M. Tsuda, and T. Tsuchiya. 1990. Rapid purification and characterization of F1-ATPase of Vibrio parahaemolyticus. Biochim. Biophys. Acta 101818-22. [DOI] [PubMed] [Google Scholar]
- 345.Sakamoto, J. 1984. Effect of dimethylsulfoxide on ATP synthesis by mitochondrial soluble F1-ATPase. J. Biochem. (Tokyo) 96483-487. [DOI] [PubMed] [Google Scholar]
- 346.Sakamoto, J., and Y. Tonomura. 1983. Synthesis of enzyme-bound ATP by mitochondrial soluble F1-ATPase in the presence of dimethylsulfoxide. J. Biochem. (Tokyo) 931601-1614. [DOI] [PubMed] [Google Scholar]
- 347.Salomon, A. R., D. W. Voehringer, L. A. Herzenberg, and C. Khosla. 2001. Apoptolidin, a selective cytotoxic agent, is an inhibitor of F0F1-ATPase. Chem. Biol. 871-80. [DOI] [PubMed] [Google Scholar]
- 348.Salomon, A. R., D. W. Voehringer, L. A. Herzenberg, and C. Khosla. 2000. Understanding and exploiting the mechanistic basis for selectivity of polyketide inhibitors of F0F1-ATPase. Proc. Natl. Acad. Sci. USA 9714766-14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Salomon, A. R., Y. Zhang, H. Seto, and C. Khosla. 2001. Structure-activity relationships within a family of selectively cytotoxic macrolide natural products. Org. Lett. 357-59. [DOI] [PubMed] [Google Scholar]
- 350.Santolini, J., F. Haraux, C. Sigalat, G. Moal, and F. Andre. 1999. Kinetic analysis of tentoxin binding to chloroplast F1-ATPase. A model for the overactivation process. J. Biol. Chem. 274849-858. [DOI] [PubMed] [Google Scholar]
- 351.Santolini, J., C. Minoletti, J. M. Gomis, C. Sigalat, F. Andre, and F. Haraux. 2002. An insight into the mechanism of inhibition and reactivation of the F1-ATPases by tentoxin. Biochemistry 416008-6018. [DOI] [PubMed] [Google Scholar]
- 352.Satre, M. 1981. The effect of asteltoxin and citreomontanine, two polyenic α-pyrone mycotoxins, on Escherichia coli adenosine triphosphate. Biochem. Biophys. Res. Commun. 100267-274. [DOI] [PubMed] [Google Scholar]
- 353.Satre, M., M. Bof, and P. V. Vignais. 1980. Interaction of Escherichia coli adenosine triphosphatase with aurovertin and citreoviridin: inhibition and fluorescence studies. J. Bacteriol. 142768-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Sayood, S. F., H. Suh, C. S. Wilcox, and S. M. Schuster. 1989. Effect of citreoviridin and isocitreoviridin on beef heart mitochondrial ATPase. Arch. Biochem. Biophys. 270714-721. [DOI] [PubMed] [Google Scholar]
- 355.Schafer, G., and G. Onur. 1979. 3′ Esters of ADP as energy-transfer inhibitors and probes of the catalytic site of oxidative phosphorylation. Eur. J. Biochem. 97415-424. [DOI] [PubMed] [Google Scholar]
- 356.Schafer, G., G. Onur, and M. Schlegel. 1980. Use of modified adenine nucleotides in mechanistic studies on oxidative phosphorylation: structure and space at the catalytic site. J. Bioenerg. Biomembr. 12213-232. [DOI] [PubMed] [Google Scholar]
- 357.Schagger, H., and T. G. Ohm. 1995. Human diseases with defects in oxidative phosphorylation. 2. F1F0 ATP-synthase defects in Alzheimer disease revealed by blue native polyacrylamide gel electrophoresis. Eur. J. Biochem. 227916-921. [DOI] [PubMed] [Google Scholar]
- 358.Schmitz, H., S. B. Deak, K. E. Crook, Jr., and I. R. Hooper. 1963. Peliomycin, a new cytotoxic agent. I. Production, isolation, and characterization. Antimicrob. Agents Chemother. 16189-94. [PubMed] [Google Scholar]
- 359.Schmitz, H., S. D. Jubinski, I. R. Hooper, K. E. Crook, Jr., K. E. Price, and J. Lein. 1965. Ossamycin, a new cytotoxic agent. J. Antibiot. 1882-88. [PubMed] [Google Scholar]
- 360.Scholes, P., P. Mitchell, and J. Moyle. 1969. The polarity of proton translocation in some photosynthetic microorganisms. Eur. J. Biochem. 8450-454. [DOI] [PubMed] [Google Scholar]
- 361.Schuster, S. M., R. E. Ebel, and H. A. Lardy. 1975. Kinetic studies on rat liver and beef heart mitochondrial ATPase. Evidence for nucleotide binding at separate regulatory and catalytic sites. J. Biol. Chem. 2507848-7853. [PubMed] [Google Scholar]
- 362.Schuster, S. M., R. J. Gertschen, and H. A. Lardy. 1976. Effect of inosine 5′-(β,γ-imido) triphosphate and other nucleotides on beef heart mitochondrial ATPase. J. Biol. Chem. 2516705-6710. [PubMed] [Google Scholar]
- 363.Schuster, S. M., G. D. Reinhart, and H. A. Lardy. 1977. Studies on the kinetic mechanism of oxidative phosphorylation. J. Biol. Chem. 252427-432. [PubMed] [Google Scholar]
- 364.Sebald, W., W. Machleidt, and E. Wachter. 1980. N,N′-dicyclohexylcarbodiimide binds specifically to a single glutamyl residue of the proteolipid subunit of the mitochondrial adenosinetriphosphatases from Neurospora crassa and Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 77785-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Selwyn, M. J. 1967. Preparation and general properties of a soluble adenosine triphosphatase from mitochondria. Biochem. J. 105279-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Senior, A. E. 1973. Relationship of cysteine and tyrosine residues to adenosine triphosphate hydrolysis by mitochondrial adenine triphosphatase. Biochemistry 123622-3627. [DOI] [PubMed] [Google Scholar]
- 367.Sergeant, N., A. Wattez, M. Galvan-valencia, A. Ghestem, J. P. David, J. Lemoine, P. E. Sautiere, J. Dachary, J. P. Mazat, J. C. Michalski, J. Velours, R. Mena-Lopez, and A. Delacourte. 2003. Association of ATP synthase α-chain with neurofibrillary degeneration in Alzheimer's disease. Neuroscience 117293-303. [DOI] [PubMed] [Google Scholar]
- 368.Shapiro, A. B., K. D. Gibson, H. A. Scheraga, and R. E. McCarty. 1991. Fluorescence resonance energy transfer mapping of the fourth of six nucleotide-binding sites of chloroplast coupling factor 1. J. Biol. Chem. 26617276-17285. [PubMed] [Google Scholar]
- 369.Sigalat, C., B. Pitard, and F. Haraux. 1995. Proton coupling is preserved in membrane-bound chloroplast ATPase activated by high concentrations of tentoxin. FEBS Lett. 368253-256. [DOI] [PubMed] [Google Scholar]
- 370.Skidmore, R. A., J. D. Patterson, and R. S. Tomsick. 1996. Local anesthetics. Dermatol. Surg. 22511-524. [DOI] [PubMed] [Google Scholar]
- 371.Sloothaak, J. B., J. A. Berden, M. A. Herweijer, and A. Kemp. 1985. The use of 8-azido-ATP and 8-azido-ADP as photoaffinity labels of the ATP synthase in submitochondrial particles: evidence for a mechanism of ATP hydrolysis involving two independent catalytic sites? Biochim. Biophys. Acta 80927-38. [DOI] [PubMed] [Google Scholar]
- 372.Smith, J. B., and P. C. Sternweis. 1977. Purification of membrane attachment and inhibitory subunits of the proton translocating adenosine triphosphatase from Escherichia coli. Biochemistry 16306-311. [DOI] [PubMed] [Google Scholar]
- 373.Solaini, G., A. Baracca, E. Gabellieri, and G. Lenaz. 1997. Modification of the mitochondrial F1-ATPase ɛ subunit, enhancement of the ATPase activity of the IF1-F1 complex and IF1-binding dependence of the conformation of the ɛ subunit. Biochem. J. 327443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Solaini, G., and B. Tadolini. 1984. Spermine binding to submitochondrial particles and activation of adenosine triphosphatase. Biochem. J. 218495-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Sone, N., K. Ikeba, and Y. Kagawa. 1979. Inhibition of proton conduction by chemical modification of the membrane moiety of proton translocating ATPase. FEBS Lett. 9761-64. [DOI] [PubMed] [Google Scholar]
- 376.Sone, N., M. Yoshida, H. Hirata, and Y. Kagawa. 1975. Purification and properties of a dicyclohexylcarbodiimide-sensitive adenosine triphosphatase from a thermophilic bacterium. J. Biol. Chem. 2507917-7923. [PubMed] [Google Scholar]
- 377.Souza, M. O., T. B. Creczynski-Pasa, H. M. Scofano, P. Graber, and J. A. Mignaco. 2004. High hydrostatic pressure perturbs the interactions between CF0F1 subunits and induces a dual effect on activity. Int. J. Biochem. Cell. Biol. 36920-930. [DOI] [PubMed] [Google Scholar]
- 378.Steele, J. A., R. D. Durbin, T. F. Uchytil, and D. H. Rich. 1978. Tentoxin. An uncompetitive inhibitor of lettuce chloroplast coupling factor 1. Biochim. Biophys. Acta 50172-82. [DOI] [PubMed] [Google Scholar]
- 379.Steele, J. A., T. F. Uchytil, and R. D. Durbin. 1978. The stimulation of coupling factor 1 ATPase by tentoxin. Biochim. Biophys. Acta 504136-141. [DOI] [PubMed] [Google Scholar]
- 380.Steele, J. A., T. F. Uchytil, R. D. Durbin, P. Bhatnagar, and D. H. Rich. 1976. Chloroplast coupling factor 1: a species-specific receptor for tentoxin. Proc. Natl. Acad. Sci. USA 732245-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Steffens, K., E. Schneider, B. Herkenhoff, R. Schmid, and K. Altendorf. 1984. Chemical modification of the F0 part of the ATP synthase (F1F0) from Escherichia coli. Effects on proton conduction and F1 binding. Eur. J. Biochem. 138617-622. [DOI] [PubMed] [Google Scholar]
- 382.Steinback, K. E., L. McIntosh, L. Bogorad, and C. J. Arntzen. 1981. Identification of the triazine receptor protein as a chloroplast gene product. Proc. Natl. Acad. Sci. USA 787463-7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Steinke, L., R. Bacon, and S. M. Schuster. 1987. The effects of exchange-inert metal-nucleotide complexes on the kinetics of beef heart mitochondrial F1-ATPase. Arch. Biochem. Biophys. 258482-490. [DOI] [PubMed] [Google Scholar]
- 384.Steinke, L., and S. M. Schuster. 1985. The effect of Co(III)(NH3)4ATP on the kinetics of beef heart mitochondrial ATPase. Arch. Biochem. Biophys. 238629-635. [DOI] [PubMed] [Google Scholar]
- 385.Steinmeier, R. C., and J. H. Wang. 1979. Reconstitution of oxidative phosphorylation by chemically modified coupling factor F1: differential inhibition of reactions catalyzed by F1 labeled with 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole or 2,3-butanedione. Biochemistry 1811-18. [DOI] [PubMed] [Google Scholar]
- 386.Sternweis, P. C., and J. B. Smith. 1980. Characterization of the inhibitory (ɛ) subunit of the proton-translocating adenosine triphosphatase from Escherichia coli. Biochemistry 19526-531. [DOI] [PubMed] [Google Scholar]
- 387.Stock, D., C. Gibbons, I. Arechaga, A. G. Leslie, and J. E. Walker. 2000. The rotary mechanism of ATP synthase. Curr. Opin. Struct. Biol. 10672-679. [DOI] [PubMed] [Google Scholar]
- 388.Sutton, R., and S. J. Ferguson. 1985. Tyrosine-311 of a β chain is the essential residue specifically modified by 4-chloro-7-nitrobenzofurazan in bovine heart mitochondrial ATPase. Eur. J. Biochem. 148551-554. [DOI] [PubMed] [Google Scholar]
- 389.Suzuki, T., T. Murakami, R. Iino, J. Suzuki, S. Ono, Y. Shirakihara, and M. Yoshida. 2003. F0F1-ATPase/synthase is geared to the synthesis mode by conformational rearrangement of ɛ subunit in response to proton motive force and ADP/ATP balance. J. Biol. Chem. 27846840-46846. [DOI] [PubMed] [Google Scholar]
- 390.Tagaya, M., T. Noumi, K. Nakano, M. Futai, and T. Fukui. 1988. Identification of α-subunit Lys201 and β-subunit Lys155 at the ATP-binding sites in Escherichia coli F1-ATPase. FEBS Lett. 233347-351. [DOI] [PubMed] [Google Scholar]
- 391.Takeda, K., J. Miki, H. Kanazawa, T. Tsuchiya, and M. Futai. 1985. Change of inhibitor sensitivities of Escherichia coli F1-ATPase due to a mutational substitution of Phe for Ser at residue 174 of the β subunit. J. Biochem. (Tokyo) 971401-1407. [DOI] [PubMed] [Google Scholar]
- 392.Tamura, J. K., and J. H. Wang. 1983. Changes in chemical properties of mitochondrial adenosinetriphosphatase upon removal of tightly bound nucleotides. Biochemistry 221947-1954. [DOI] [PubMed] [Google Scholar]
- 393.Terwilliger, T. C., and D. Eisenberg. 1982. The structure of melittin. I. Structure determination and partial refinement. J. Biol. Chem. 2576010-6015. [DOI] [PubMed] [Google Scholar]
- 394.Thomassen, J., and L. Klungsoyr. 1983. ATPase of bovine heart mitochondria. Modulation of ITPase activity by ATP, ADP, acetyl ATP and acetyl AMP. Biochim. Biophys. Acta 723114-122. [DOI] [PubMed] [Google Scholar]
- 395.Thomasset, S. C., D. P. Berry, G. Garcea, T. Marczylo, W. P. Steward, and A. J. Gescher. 2007. Dietary polyphenolic phytochemicals—promising cancer chemopreventive agents in humans? A review of their clinical properties. Int. J. Cancer. 120451-458. [DOI] [PubMed] [Google Scholar]
- 396.Thyagarajan, D., S. Shanske, M. Vazquez-Memije, D. De Vivo, and S. DiMauro. 1995. A novel mitochondrial ATPase 6 point mutation in familial bilateral striatal necrosis. Ann. Neurol. 38468-472. [DOI] [PubMed] [Google Scholar]
- 397.Tiedge, H., U. Lucken, J. Weber, and G. Schafer. 1982. High-affinity binding of ADP and of ADP analogues to mitochondrial F1-ATPase. Eur. J. Biochem. 127291-299. [DOI] [PubMed] [Google Scholar]
- 398.Ting, L. P., and J. H. Wang. 1980. Effect of phosphate and adenine nucleotides on the rate of labeling of functional groups at the catalytic sites of F1-ATPase. J. Bioenerg. Biomembr. 1279-93. [DOI] [PubMed] [Google Scholar]
- 399.Ting, L. P., and J. H. Wang. 1980. Functional groups at the catalytic site of F1 adenosine triphosphatase. Biochemistry 195665-5670. [DOI] [PubMed] [Google Scholar]
- 400.Tommasino, M., and R. A. Capaldi. 1985. Effect of dicyclohexylcarbodiimide on unisite and multisite catalytic activities of the adenosinetriphosphatase of Escherichia coli. Biochemistry 243972-3976. [DOI] [PubMed] [Google Scholar]
- 401.Tsunashima, K., M. Ide, H. Kadoi, A. Hirayama, and M. Nakata. 2001. Synthesis of the C15-C27 portion of venturicidins: a formal total synthesis of venturicidin X. Tetrahedron Lett. 423607-3611. [Google Scholar]
- 402.Tsunoda, S. P., A. J. Rodgers, R. Aggeler, M. C. Wilce, M. Yoshida, and R. A. Capaldi. 2001. Large conformational changes of the ɛ subunit in the bacterial F1F0 ATP synthase provide a ratchet action to regulate this rotary motor enzyme. Proc. Natl. Acad. Sci. USA 986560-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Ueno, H., T. Suzuki, K. Kinosita, Jr., and M. Yoshida. 2005. ATP-driven stepwise rotation of FoF1-ATP synthase. Proc. Natl. Acad. Sci. USA 1021333-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Unitt, M. D., and D. Lloyd. 1981. Effects of inhibitors on mitochondrial adenosine triphosphatase of Tetrahymena pyriformis ST. J. Gen. Microbiol. 126261-266. [DOI] [PubMed] [Google Scholar]
- 405.Usta, J., and D. E. Griffiths. 1992. Organotin-flavone complexes: a new class of fluorescent probes for F1F0ATPase. Biochem. Biophys. Res. Commun. 188365-371. [DOI] [PubMed] [Google Scholar]
- 406.Vanderkooi, G., J. Shaw, C. Storms, R. Vennerstrom, and D. Chignell. 1981. On the mechanism of action of anesthetics. Direct inhibition of mitochondrial F1-ATPase by n-butanol and tetracaine. Biochim. Biophys. Acta 635200-203. [DOI] [PubMed] [Google Scholar]
- 407.van der Zwet-de Graaff, I., A. F. Hartog, and J. A. Berden. 1997. Modification of membrane-bound F1 by p-fluorosulfonylbenzoyl-5′-adenosine: sites of binding and effect on activity. Biochim. Biophys. Acta 1318123-132. [DOI] [PubMed] [Google Scholar]
- 408.van Dongen, M. B., J. P. de Geus, T. Korver, A. F. Hartog, and J. A. Berden. 1986. Binding and hydrolysis of 2-azido-ATP and 8-azido-ATP by isolated mitochondrial F1: characterisation of high-affinity binding sites. Biochim. Biophys. Acta 850359-368. [DOI] [PubMed] [Google Scholar]
- 409.Van Heeke, G., L. Deforce, R. A. Schnizer, R. Shaw, J. M. Couton, G. Shaw, P. S. Song, and S. M. Schuster. 1993. Recombinant bovine heart mitochondrial F1-ATPase inhibitor protein: overproduction in Escherichia coli, purification, and structural studies. Biochemistry 3210140-10149. [DOI] [PubMed] [Google Scholar]
- 410.van Raaij, M. J., J. P. Abrahams, A. G. Leslie, and J. E. Walker. 1996. The structure of bovine F1-ATPase complexed with the antibiotic inhibitor aurovertin B. Proc. Natl. Acad. Sci. USA 936913-6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.van Raaij, M. J., G. L. Orriss, M. G. Montgomery, M. J. Runswick, I. M. Fearnley, J. M. Skehel, and J. E. Walker. 1996. The ATPase inhibitor protein from bovine heart mitochondria: the minimal inhibitory sequence. Biochemistry 3515618-15625. [DOI] [PubMed] [Google Scholar]
- 412.Vasilyeva, E. A., I. B. Minkov, A. F. Fitin, and A. D. Vinogradov. 1982. Kinetic mechanism of mitochondrial adenosine triphosphatase. Inhibition by azide and activation by sulphite. Biochem. J. 20215-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Velours, J., and G. Arselin. 2000. The Saccharomyces cerevisiae ATP synthase. J. Bioenerg. Biomembr. 32383-390. [DOI] [PubMed] [Google Scholar]
- 414.Verburg, J. G., and W. S. Allison. 1990. Tyrosine α244 is derivatized when the bovine heart mitochondrial F1-ATPase is inactivated with 5′-p-fluorosulfonylbenzoylethenoadenosine. J. Biol. Chem. 2658065-8074. [PubMed] [Google Scholar]
- 415.Verburg, J. G., M. Yoshida, and W. S. Allison. 1986. The use of dithionite reduction to identify the essential tyrosine residue in the F1-ATPase from the thermophilic bacterium, PS3, that reacts with 7-chloro-4-nitrobenzofurazan. Arch. Biochem. Biophys. 2458-13. [DOI] [PubMed] [Google Scholar]
- 416.Verschoor, G. J., P. R. van der Sluis, and E. C. Slater. 1977. The binding of aurovertin to isolated β subunit of F1 (mitochondrial ATPase). Stoicheiometry of β subunit in F1. Biochim. Biophys. Acta 462438-449. [DOI] [PubMed] [Google Scholar]
- 417.Vogel, P. D., and R. L. Cross. 1991. Adenine nucleotide-binding sites on mitochondrial F1-ATPase. Evidence for an adenylate kinase-like orientation of catalytic and noncatalytic sites. J. Biol. Chem. 2666101-6105. [PubMed] [Google Scholar]
- 418.von Ballmoos, C., J. Brunner, and P. Dimroth. 2004. The ion channel of F-ATP synthase is the target of toxic organotin compounds. Proc. Natl. Acad. Sci. USA 10111239-11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Wagenvoord, R. J., I. van der Kraan, and A. Kemp. 1979. Localisation of adenine nucleotide-binding sites on beef-heart mitochondrial ATPase by photolabelling with 8-azido-ADP and 8-azido-ATP. Biochim. Biophys. Acta 54885-95. [DOI] [PubMed] [Google Scholar]
- 420.Wagenvoord, R. J., I. Van der Kraan, and A. Kemp. 1977. Specific photolabelling of beef-heart mitochondrial ATPase by 8-azido-ATP. Biochim. Biophys. Acta 46017-24. [DOI] [PubMed] [Google Scholar]
- 421.Wagenvoord, R. J., G. J. Verschoor, and A. Kemp. 1981. Photolabelling with 8-azido-adenine nucleotides of adenine nucleotide-binding sites in isolated spinach chloroplast ATPase (CF1). Biochim. Biophys. Acta 634229-236. [DOI] [PubMed] [Google Scholar]
- 422.Wahl, M. L., D. J. Kenan, M. Gonzalez-Gronow, and S. V. Pizzo. 2005. Angiostatin's molecular mechanism: aspects of specificity and regulation elucidated. J. Cell. Biochem. 96242-261. [DOI] [PubMed] [Google Scholar]
- 423.Walter, P., H. A. Lardy, and D. Johnson. 1967. Antibiotics as tools for metabolic studies. X. Inhibition of phosphoryl transfer reactions in mitochondria by peliomycin, ossamycin, and venturicidin. J. Biol. Chem. 2425014-5018. [PubMed] [Google Scholar]
- 424.Weber, J., R. S. Lee, E. Grell, and A. E. Senior. 1992. Investigation of the aurovertin binding site of Escherichia coli F1-ATPase by fluorescence spectroscopy and site-directed mutagenesis. Biochemistry 315112-5116. [DOI] [PubMed] [Google Scholar]
- 425.Weber, J., R. S. Lee, S. Wilke-Mounts, E. Grell, and A. E. Senior. 1993. Combined application of site-directed mutagenesis, 2-azido-ATP labeling, and lin-benzo-ATP binding to study the noncatalytic sites of Escherichia coli F1-ATPase. J. Biol. Chem. 2686241-6247. [PubMed] [Google Scholar]
- 426.Weber, J., U. Lucken, and G. Schafer. 1985. Total number and differentiation of nucleotide binding sites on mitochondrial F1-ATPase. An approach by photolabeling and equilibrium binding studies. Eur. J. Biochem. 14841-47. [DOI] [PubMed] [Google Scholar]
- 427.Weber, J., M. Rogner, and G. Schafer. 1987. Novel approaches towards characterization of the high-affinity nucleotide binding sites on mitochondrial F1-ATPase by the fluorescence probes 3′-O-(1-naphthoyl)adenosine di- and triphosphate. Biochim. Biophys. Acta 89230-41. [DOI] [PubMed] [Google Scholar]
- 428.Weber, J., S. Schmitt, E. Grell, and G. Schafer. 1990. Differentiation of the nucleotide-binding sites on nucleotide-depleted mitochondrial F1-ATPase by means of a fluorescent ADP analogue. J. Biol. Chem. 26510884-10892. [PubMed] [Google Scholar]
- 429.Weber, J., and A. E. Senior. 1997. Binding of TNP-ATP and TNP-ADP to the non-catalytic sites of Escherichia coli F1-ATPase. FEBS Lett. 412169-172. [DOI] [PubMed] [Google Scholar]
- 430.Weber, J., and A. E. Senior. 1998. Effects of the inhibitors azide, dicyclohexylcarbodiimide, and aurovertin on nucleotide binding to the three F1-ATPase catalytic sites measured using specific tryptophan probes. J. Biol. Chem. 27333210-33215. [DOI] [PubMed] [Google Scholar]
- 431.West, K. R., and J. T. Wiskich. 1969. The action of Dio-9 on photophosphorylation. FEBS Lett. 3247-249. [DOI] [PubMed] [Google Scholar]
- 432.Wieker, H. J., and B. Hess. 1985. α,β-Bidentate CrADP abolishes the negative cooperativity of yeast mitochondrial F1-ATPase. Biochim. Biophys. Acta 80635-41. [DOI] [PubMed] [Google Scholar]
- 433.Wieker, H. J., D. Kuschmitz, and B. Hess. 1987. Inhibition of yeast mitochondrial F1-ATPase, F0F1-ATPase and submitochondrial particles by rhodamines and ethidium bromide. Biochim. Biophys. Acta 892108-117. [DOI] [PubMed] [Google Scholar]
- 434.Wiley, P. F., J. M. Koert, and L. J. Hanka. 1982. Two new polypeptide antibiotics, CC-1014 and CC-1014B. J. Antibiot. 351231-1233. [DOI] [PubMed] [Google Scholar]
- 435.Williams, N., and P. S. Coleman. 1982. Exploring the adenine nucleotide binding sites on mitochondrial F1-ATPase with a new photoaffinity probe, 3′-O-(4-benzoyl)benzoyl adenosine 5′-triphosphate. J. Biol. Chem. 2572834-2841. [PubMed] [Google Scholar]
- 436.Wise, J. G., T. M. Duncan, L. R. Latchney, D. N. Cox, and A. E. Senior. 1983. Properties of F1-ATPase from the uncD412 mutant of Escherichia coli. Biochem. J. 215343-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Yagi, T., and Y. Hatefi. 1984. Thiols in oxidative phosphorylation: inhibition and energy-potentiated uncoupling by monothiol and dithiol modifiers. Biochemistry 232449-2455. [DOI] [PubMed] [Google Scholar]
- 438.Yagi, T., and Y. Hatefi. 1987. Thiols in oxidative phosphorylation: thiols in the F0 of ATP synthase essential for ATPase activity. Arch. Biochem. Biophys. 254102-109. [DOI] [PubMed] [Google Scholar]
- 439.Yarlett, N., and D. Lloyd. 1981. Effects of inhibitors on mitochondrial adenosine triphosphatase of Crithidia fasciculata: an unusual pattern of specificities. Mol. Biochem. Parasitol. 313-17. [DOI] [PubMed] [Google Scholar]
- 440.Yoshida, M. 1983. The synthesis of enzyme-bound ATP by the F1-ATPase from the thermophilic bacterium PS3 in 50% dimethylsulfoxide. Biochem. Biophys. Res. Commun. 114907-912. [DOI] [PubMed] [Google Scholar]
- 441.Yoshida, M., W. S. Allison, F. S. Esch, and M. Futai. 1982. The specificity of carboxyl group modification during the inactivation of the Escherichia coli F1-ATPase with dicyclohexyl[14C]carbodiimide. J. Biol. Chem. 25710033-10037. [PubMed] [Google Scholar]
- 442.Younis, H. M., M. M. Abo-El-Saad, R. K. Abdel-Razik, and S. A. Abo-Seda. 2002. Resolving the DDT target protein in insects as a subunit of the ATP synthase. Biotechnol. Appl. Biochem. 359-17. [PubMed] [Google Scholar]
- 443.Younis, H. M., J. N. Telford, and R. B. Koch. 1978. Adenosine triphosphatase from cockroach coxal muscle mitochondria. Isolation, properties, and response to DDT. Pestic. Biochem. Physiol. 8271-277. [Google Scholar]
- 444.Yount, R. G., D. Babcock, W. Ballantyne, and D. Ojala. 1971. Adenylyl imidodiphosphate, an adenosine triphosphate analog containing a P-N-P linkage. Biochemistry 102484-2489. [DOI] [PubMed] [Google Scholar]
- 445.Zanotti, F., F. Guerrieri, Y. W. Che, R. Scarfo, and S. Papa. 1987. Proton translocation by the H+-ATPase of mitochondria. Effect of modification by monofunctional reagents of thiol residues in F0 polypeptides. Eur. J. Biochem. 164517-523. [DOI] [PubMed] [Google Scholar]
- 446.Zanotti, F., G. Raho, A. Gaballo, and S. Papa. 2004. Inhibitory and anchoring domains in the ATPase inhibitor protein IF1 of bovine heart mitochondrial ATP synthase. J. Bioenerg. Biomembr. 36447-457. [DOI] [PubMed] [Google Scholar]
- 447.Zhang, S., D. D. Letham, and A. T. Jagendorf. 1993. Inhibition of thylakoid ATPase by venturicidin as an indicator of CF1-CF0 interaction. Plant Physiol. 101127-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Zheng, J., and V. D. Ramirez. 2000. Inhibition of mitochondrial proton F0F1-ATPase/ATP synthase by polyphenolic phytochemicals. Br. J. Pharmacol. 1301115-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Zheng, J., and V. D. Ramirez. 1999. Piceatannol, a stilbene phytochemical, inhibits mitochondrial F0F1-ATPase activity by targeting the F1 complex. Biochem. Biophys. Res. Commun. 261499-503. [DOI] [PubMed] [Google Scholar]
- 450.Zheng, J., and V. D. Ramirez. 1999. Purification and identification of an estrogen binding protein from rat brain: oligomycin sensitivity-conferring protein (OSCP), a subunit of mitochondrial F0F1-ATP synthase/ATPase. J. Steroid. Biochem. Mol. Biol. 6865-75. [DOI] [PubMed] [Google Scholar]
- 451.Zheng, J., and V. D. Ramirez. 1999. Rapid inhibition of rat brain mitochondrial proton F0F1-ATPase activity by estrogens: comparison with Na+, K+-ATPase of porcine cortex. Eur. J. Pharmacol. 36895-102. [DOI] [PubMed] [Google Scholar]
- 452.Zhuo, S., and W. S. Allison. 1988. Inhibition and photoinactivation of the bovine heart mitochondrial F1-ATPase by the cytotoxic agent, dequalinium. Biochem. Biophys. Res. Commun. 152968-972. [DOI] [PubMed] [Google Scholar]
- 453.Zhuo, S., S. Garrod, P. Miller, and W. S. Allison. 1992. Irradiation of the bovine mitochondrial F1-ATPase previously inactivated with 5′-p-fluorosulfonylbenzoyl-8-azido-[3H]adenosine cross-links His-β427 to Tyr-β345 within the same β subunit. J. Biol. Chem. 26712916-12927. [PubMed] [Google Scholar]
- 454.Zhuo, S., S. R. Paik, J. A. Register, and W. S. Allison. 1993. Photoinactivation of the bovine heart mitochondrial F1-ATPase by [14C]dequalinium cross-links phenylalanine-403 or phenylalanine-406 of an α subunit to a site or sites contained within residues 440-459 of a β subunit. Biochemistry 322219-2227. [DOI] [PubMed] [Google Scholar]