Abstract
T cell receptor signaling processes are controlled by the integrated actions of families of protein tyrosine kinases (PTKs) and protein tyrosine phosphatases (PTPases). Several distinct cytosolic protein tyrosine phosphatases have been described that are able to negatively regulate TCR signaling pathways, including SHP-1, SHP-2, PTPH1, and PEP. Using PTPase substrate-trapping mutants and wild type enzymes, we determined that PTPN4/PTP-MEG1, a PTPH1-family member, could complex and dephosphorylate the ITAMs of the TCR ζ subunit. In addition, the substrate-trapping derivative augmented basal and TCR-induced activation of NF-κB in T cells. To characterize the contribution of this PTPase in T cells, we developed PTPN4-deficient mice. T cell development and TCR signaling events were comparable between wild type and PTPN4-deficient animals. The magnitude and duration of TCR-regulated ITAM phosphorylation, as well as overall protein phosphorylation, was unaltered in the absence of PTPN4. Finally, Th1- and Th2-derived cytokines and in vivo immune responses to Listeria monocytogeneswere equivalent between wild type and PTPN4-deficient mice. These findings suggest that additional PTPases are involved in controlling ITAM phosphorylations.
Keywords: T cells, signal transduction, protein kinases/phosphatases, transgenic/knockout, cell differentiation
1. Introduction
The T cell receptor (TCR) is a multimeric complex consisting of the ligand-binding chains (αβ or γδ TCR) non-covalently associated with the signal-transducing dimers, CD3 γε, δε, and TCR ζζ (Call et al., 2004). TCR recognition of cognate peptide/MHC complexes on APCs results in a cascade of intracellular signaling processes (reviewed in (Wange and Samelson, 1996)). These intracellular signals are critical for T cell development, cytokine production, and/or proliferation. Seconds after TCR cross-linking, the cytoplasmic tail of CD3 ε undergoes a conformation (Gil et al., 2002; Gil et al., 2005). This conformational change, along with simultaneous or subsequent activation of protein tyrosine kinases (PTKs), results in increased protein tyrosine phosphorylations (Gil et al., 2005; Minguet et al., 2007). The protein phosphorylation pathways commence once the Src-family PTKs, Lck and/or Fyn, are activated and re-localized to the CD3 subunits (Palacios and Weiss, 2004). There, the kinases phosphorylate the two tyrosine residues located within the ITAMs (reviewed in (Cambier, 1995; Reth, 1989; Weiss, 1993)). ITAMs are a conserved signaling motif and are present in one or more copies in the cytoplasmic tails of the CD3 γ, δ, ε and TCR ζ subunits. ITAMs comprise the sequence YxxLx(6–8)YxxL, with the phosphorylation of the tyrosine residues providing the critical molecular determinant for TCR-mediated signal transmission (Pitcher and van Oers, 2003; Weiss and Littman, 1994). ITAMs also form the basis of antigen receptor signal transduction for the B cell receptor, selected activating NK cell receptors, and particular Ig-receptors (Humphrey et al., 2005; Reth, 1992; Weiss and Littman, 1994).
In T cells, the bi-phosphorylated ITAMs are bound by the tandem SH2 domains of ZAP-70, a Syk-family PTK (Hatada et al., 1995). ZAP-70 is catalytically activated upon tyrosine phosphorylation by Src PTKs (Chan et al., 1995; Wange et al., 1995). The signaling cascade continues with activated ZAP-70 phosphorylating several adaptor proteins, kinases and effector proteins (Jordan et al., 2003; Samelson, 2002). These latter molecules participate in the activation of multiple transcription factors, including NFAT, AP-1 and NF-κB, which results in the production of different cytokines (Huang and Wange, 2004; Li and Verma, 2002; Rao et al., 1997; Waldmann, 2006).
TCR-induced increases in protein tyrosine phosphorylations are transient, with most phosphoproteins returning to baseline levels within minutes of stimulation. The transient nature of phosphotyrosine induction is partly a consequence of changes in the enzymatic activity and/or distribution of protein tyrosine phosphatases (PTPases). PTPases are a family of enzymes that dephosphorylate tyrosine residues on a large number of TCR-regulated proteins. It is estimated that T cells express approximately 60 of the 107 potential PTPases identified in the human genome (Alonso et al., 2004; Gjorloff-Wingren et al., 2000; Mustelin et al., 2005). The identity of the PTPases that dephosphorylate components of the TCR signaling pathway have been elucidated by the characterization of PTPase-deficient or -mutant mice (Mustelin and Tasken, 2003; Veillette et al., 2002). Two key PTPases characterized in this manner include SHP-1 and SHP-2, homologous enzymes containing tandem SH2 domains followed by a catalytic domain (Tonks and Neel, 2001). SHP-1 is expressed specifically in hematopoietic cells and is mutated in motheaten mice. Thymocytes from these mutant mice have elevated levels of phosphoproteins compared to normal mice (Lorenz et al., 1996; Pani et al., 1996; Tsui et al., 1993). These analyses and biochemical characterizations of SHP-1 suggest that this PTPase dephosphorylates positive regulatory tyrosine residues in the catalytic domains of Src- and ZAP-70-family PTKs (Brockdorff et al., 1999). SHP-2, in contrast to SHP-1, is ubiquitously expressed and functions as a positive regulator of the Ras/ERK pathway following TCR signaling (Frearson and Alexander, 1998; Nguyen et al., 2006). One report suggested that SHP-2 can dephosphorylate the phospho-ITAMs of TCR ζ (Lee et al., 1998). Yet, neither SHP-1 nor SHP-2 has been conclusively shown to dephosphorylate ITAMs. Another PTPase implicated in TCR signaling is PEP, or Pest-domain Enriched Tyrosine Phosphatase. Mechanistically, a proline-rich domain of PEP constitutively associates with the SH3 domain of the PTK Csk, enabling the PEP/Csk complex to localize near the PTKs involved in TCR signaling (Cloutier and Veillette, 1996; Gjorloff-Wingren et al., 1999; Veillette et al., 2002). The targeted disruption of PEP results in increased numbers of effector/memory T cell subsets compared to wild type mice (Hasegawa et al., 2004). Yet, naïve T cells from these mice have similar TCR-induced protein tyrosine phosphorylation pathways as wild type mice, suggesting that other PTPases dephosphorylate the ITAMs.
Alternative approaches to identify PTPases that regulate signaling pathways include the use of substrate-trapping derivatives of PTPases to entrap phosphoproteins (Flint et al., 1997; Walchli et al., 2000). Using such PTPase screens, we identified PTPH1 (PTPN3) as a PTPase that interacts with and dephosphorylates TCR ζ, suggesting that PTPH1 is an essential regulator of TCR signaling (Sozio et al., 2004). This is consistent with transcriptional reporter assays in T cells, wherein the over-expression of PTPH1 inhibited TCR-induced NFAT activation (Han et al., 2000). Although mice containing a targeted deletion of PTPH1 exhibit enhanced growth, normal T cell development, TCR signaling events, and T cell effector functions is maintained (Bauler et al., 2007; Pilecka et al., 2007). These results suggested that additional PTPases could target the TCR ITAMs and potentially negatively regulate TCR-mediated signaling. One candidate PTPase is PTPN4/PTP-MEG1, a PTPH1-family member that contains highly homologous N-terminal FERM-, central PDZ-, and C-terminal catalytic-domains (Gu et al., 1991; Park et al., 2000). Transcriptional reporter assays in T cells containing elevated levels of PTPN4 suppressed TCR-induced NFAT and AP-1 activation (Han et al., 2000).
To determine whether PTPN4 negatively regulates TCR signaling events, we performed biochemical assays to characterize the functional interaction between PTPN4 and phospho-ζ. Findings from these experiments indicated that PTPN4 could interact with and dephosphorylate TCR ζ. In addition, a substrate-trapping derivative of PTPN4 potentiated NF-κB activation, even prior to TCR engagement. To fully define the contribution of PTPN4 in lymphocytes, we generated PTPN4-deficient mice. We report here that PTPN4 gene disrupted mice had normal T cell development in the thymus and similar T cell subsets in the secondary lymphoid organs. T cells from these mice had normal TCR-induced signaling pathways. In addition, these mice exhibited a similar capacity to produce both Th1- and Th2-derived cytokines, such as IL-2, IL-4, IL-5, IL-13, and IFN-γ, following TCR/CD28 cross-linking. Finally, PTPN4-deficient mice had comparable abilities to clear primary Listeria monocytogenes infections as wild type littermates. These findings suggest that multiple PTPase-families are likely involved in the regulation of ITAM phosphorylations, providing for effective compensatory mechanisms in the absence of PTPN4.
2. Materials and Methods
2.1. Antibodies
The 145-2C11 hybridoma (anti-CD3 ε) was obtained from American Type Culture Collection (ATCC). The 35.71 (anti-CD28) hybridoma was kindly provided by Dr. James Allison (Memorial Sloan-Kettering Cancer Center). Antibodies were purified from hybridoma culture supernatants with PA or PG affinity chromatography procedures (Harlow and Lane, 1988). C305.2 (anti-TCRβ) and 1F6 (anti-Lck) were obtained from Dr. Arthur Weiss (University of California San Francisco). The following antibodies were used for Western blotting: anti-β-actin (4967; Cell Signaling Technologies), anti-FLAG (M2; Sigma Aldrich), anti-phosphotyrosine (4G10; Upstate Biotechnology), anti-IκBα (sc-371; Santa Cruz Biotechnology), anti-MAPK (Erk-1/2) and -phosphoMAPK (M8159; Sigma), anti-p42/44 (9102; Cell Signaling), anti-phospho-SAPK/JNK (9255; Cell Signaling), anti-SAPK/JNK (9252; Cell Signaling). The anti-TCR ζ (6B10.2) antibody has been previously described (van Oers et al., 1995). Anti-PTPN4 specific antibodies were generously provided by Dr. Philip Majerus (Washington University), or purchased from Orbigen (Orbigen, Inc.). Horseradish peroxidase (HRP)-conjugated goat anti-mouse Ig, goat anti-rabbit Ig (Bio-Rad Laboratories), or HRP-conjugated goat anti-mouse IgG2b (Invitrogen Corp.) were used as secondary antibodies. The following antibodies were utilized in multicolor flow cytometry (purchased from BD Pharmingen): APC-Cy7-B220, PerCP 5.5-CD4, PE-Cy7-CD8, FITC-CD25, PE-CD69, APC-Cy7-CD11b. Pacific Blue-CD3 was purchased from eBiosciences, and PE-Texas Red-CD69 and PE-Texas Red-CD62L were purchased from Invitrogen Corp. Cell populations were analyzed with either FACSCaliber or LSRII flow cytometers (Becton-Dickenson) using Cell Quest (BD) and/or FlowJo software (Treestar).
2.2. Cloning of PTPN4
Full-length PTPN4 was cloned from RNA isolated from the murine thymus, spleen, or testes. For thymic tissue, RNA was isolated from a single cell suspension of thymocytes using the Trizol extraction procedure according to the manufacturers’ instructions (Invitrogen Corp). One-three µg of total RNA was reverse transcribed with oligo-dT using the Thermoscript RT-PCR system from Invitrogen. The full-length cDNA for murine PTPN4 (mTEP) was amplified using either High Fidelity Pfu (Clontech) or LA-Taq (Takara Inc., distributed by Fisher Scientific) with 5’ sense (GTGTGGACAGTAATGACCGC) and 3’ anti-sense (CCCAGTACTTGTTCCAACC) oligonucleotide primers. The PCR reactions were performed for 32–35 cycles under the following conditions: 94°C for 30 sec, 56°C for 30 sec, and 68°C for 4 min. When the reactions were performed with Pfu, Taq was added during the last 5 cycles to provide for oligonucleotide overhangs. The PCR products were resolved on 1 % agarose gels, excised, and extracted with QIAquick Gel extraction columns (Qiagen Sciences). An aliquot was cloned by TOPO-TA cloning procedures into the pCR2.1-TOPO vector (Invitrogen). The complete cDNA sequence for PTPN4 was confirmed by automated dsDNA sequencing procedures. For generating a substrate-trapping derivative of PTPN4, an Asp to Ala point mutation was introduced in the PTPase domain using the Quick-change site-directed mutagenesis procedure according to the manufacturer’s instructions (Stratagene Inc.)(Flint et al., 1997). When used as substrate-traps in pull-down experiments, the catalytic domain of PTPN4 containing the Asp to Ala mutation was subcloned into the pGEX-2TK vector (GE-Biosciences).
2.3. Cell lines and transfection procedures
The Jurkat T cell line (E6.1) was generously provided by Dr. Virginia Shapiro (University of Pennsylvania). The COS-7 and HEK 293T/17 cell lines were purchased from ATCC. For transient transfections of COS-7 cells, plasmid DNA was transfected into the cells with the Fugene 6 transfection reagent according to the manufacturer’s instructions (Roche Molecular). Standard calcium phosphate transfection procedures were used for HEK 293T/17 cells. For transfections into T cells, 10 × 106 Jurkat T cells were washed once and resuspended in 0.4 ml of serum-free RPMI. Twenty µg of an AP-1 luciferase reporter or 20 µg of an NF-κB luciferase reporter were co-transfected with 2.5, 5 or 10 µg of either wild-type or DA PTPN4 (Finco and Baldwin, 1995; Shapiro et al., 1996). The total amount of DNA per transfection was standardized by the additional of pcDEF3 empty vector. Electroporation was performed using a Gene Pulser II (BioRad) at 250 volts, 950µF. After electroporation, cells were resuspended in 10 mls of RPMI with 5% FCS (Gibco BRL) (supplemented with Penicillin, Streptomycin, and L-glutamine). The following day, live cells were counted by trypan blue exclusion (Bio-Whittaker), and 1 × 10 5 cells were either left unstimulated or stimulated with antibodies to TCR (C305, 1:1000 final dilution) and CD28 (1 µg/ml, Caltag) for 7 hours. Stimulations were performed in triplicate per transfection, and the relative luciferase units were compared. Luciferase assays were performed as previously described (Shapiro et al., 1996).
2.4. Lysates, immunoprecipitations, GST-pull-downs, and immunoblotting
Cells were lysed in either a 1% or 0.2% Triton X-100 containing buffers (50 mM Tris-Cl, pH 7.6, 100 mM NaCl, 10% glycerol lysis buffer) supplemented with protease (10 µg/ml aprotinin, leupeptin, benzamidine, and 0.25 mM PMSF) and phosphatase inhibitors (1 mM sodium fluoride, sodium orthovanadate, sodium molybdate). In some experiments, the cells were sonicated in a 50 mM Tris-Cl, pH 8.0 buffer containing protease inhibitors. Proteins were immunoprecipitated with antibodies conjugated to either protein A- or protein G-Sepharose beads (GE Biosciences). GST-fusion proteins, pull-downs, and Western blotting procedures have been described elsewhere (Sozio et al., 2004).
2.5. Generation of PTPN4-deficient mice
Genomic DNA was obtained from 129/Sv mice. PCR reactions were used to amplify a region of PTPN4 containing exons 20–23 and another region spanning exon 24. A loxP site was inserted between exon 20 and 21 by PCR cloning. To generate the long targeting arm, the gene fragment, including exon 20-loxP-exons 21–23, was digested and subcloned into the NotI-BstZI sites of the pGK-neo plasmid (a generous gift from Dr. Toru Miyazaki, University of Tokyo, Japan). The PCR primers were: (intron 19–20) 5’ ATC GCG GCC GCC AGA CAT GTC CAG CCT GTG GCC; (exon 21) 3’ CCT TTT AAA AGG ACC CGT GTA GCA TC; (loxP) 5’ AGC TAT AAC TTC GTA TAG CAT ACA TTA TAC GAA GTT AT; (loxP) 3’ AGC TAT AAC TTC GTA TAA TGT ATG CTA TAC GAA GTT AT; (exon 20) 5’ ATC GCG GCC GCC AAA TGT TCT TTG 11 TAG ACT TGC; (exon 23) 3’ CTC GGC CGT CTA AAT TCT CAG AGC TGG GG. The second fragment containing exon 24 was cloned into BamH1-KpnI sites downstream of a floxed Neor cassette. The PCR primers were: (exon 24) 5’ CCA GCT CTG AGA ATT TAG ACA GAA GGA TCC CTG GG; (exon 24) 3’ GAT GGT ACC AGT ATT GAT GCT GGA GCA ACG. The pGK-neo-PTPN4 targeting fragment was electroporated into SM-1 embryonic stem cells by the University of Texas Southwestern Transgenic Technology Center. Following G418 and gancyclovir selection, 576 ES cell clones were screened by Southern blotting. For Southern blot analysis, 10 µg of purified genomic DNA was digested with the Xba I restriction enzyme at 37°C for 12–18 h. DNA fragments were separated by gel electrophoresis and transferred to a positively charged nylon membrane (Bio-Rad Laboratories). Hybridization with radiolabelled probe 7 was performed in a sodium phosphate buffer (0.5 M NaHPO4, pH 7.2, 7% SDS, 1 mM EDTA, 1% BSA) at 60°C for 4–12 h. Probe 7 recognizes a region spanning exon 19 external to the targeting construct. The probe was generated with the PCR primers: (intron 18–19) 5’ ACA GGA TCC CAA AGG AAT AGC AAC CCC C; (intron 19–20) 3’ AGT AAG CTT GCA CAG TAG ATT AGG CCC TAA AC. One ES cell clone (3F6), identified by Southern blotting, contained the targeted allele. This clone was expanded for injections into C57BL/6 blastocysts. The resulting chimeric male mice were mated to C57BL/6 female mice. Heterozygous mice containing the targeted allele were interbred to produce homozygous mice (PTPN4fl/fl). These mice were crossed with Lck-Cre, MORE-Cre and CAG-Cre transgenic lines (Jackson Laboratories) to obtain a targeted deletion of PTPN4 in the thymus or the entire mouse, respectively. The mice have been backcrossed onto C57BL/6 mice for over 5 generations. Mice were genotyped by PCR, with the targeted disruption confirmed by Southern blotting with Probe 7 (Fig. 2). Genomic DNA was isolated either from the tail or thymus for PCR reactions. The PCR primers were: (exon 20) 5’ CCT GGA ATG ACA ATG TCT TGT GCC; (intron 23–24) 3’ GTC CTC GAA CTC AGA AAT CCA CCT; (intron 21–22) 3’ GGT TGC AAT GTT ATT CAC CAT GGA GAG. The isolation of messenger RNA for PTPN4 was described in section 2.2. Controls for GAPDH were included: 5’ TGG CAA AGT GGA GAT TGT TGC C; 3’ AAG ATG GTG ATG GGC TTC CCG. The University of Texas Southwestern Institutional Animal Care and Use Committee approved all animal use and all protocols.
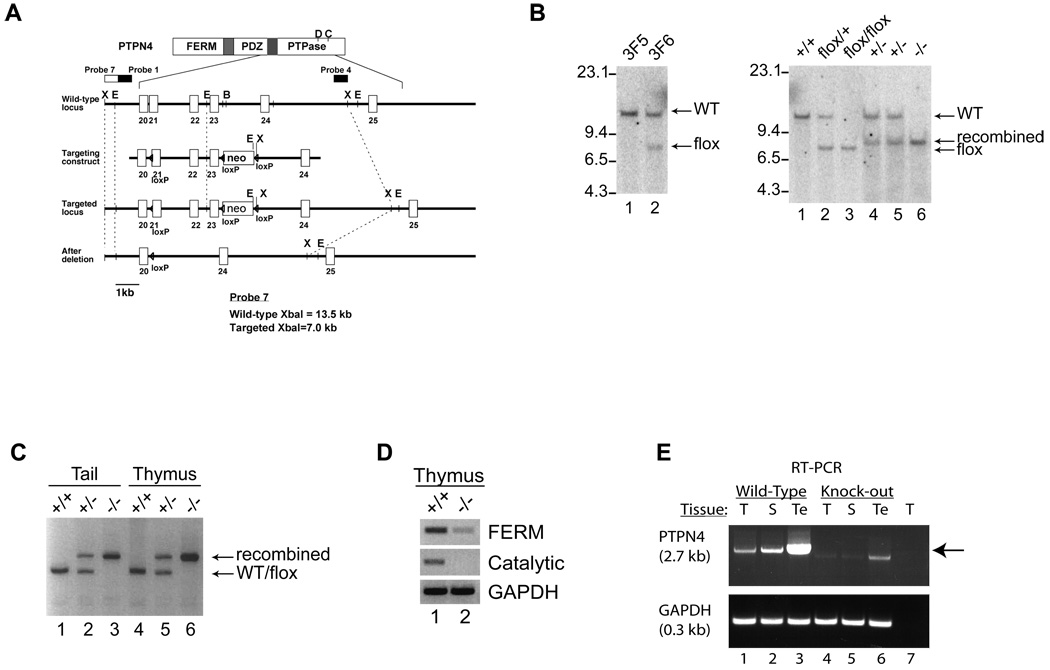
Fig. 2. Generation of PTPN4 knockout mice.
(A) A schematic diagram of the FERM, PDZ, and catalytic domain (PTPase) of PTPN4. The targeting construct was designed with a loxP site between exon 20 and 21 followed by loxP sites flanking the neomycin cassette. Probe 7 was used in Southern blot analysis to detect the targeted locus and the KO locus. B, Bam HI; E, Eco RI; X, Xba I; neo, neomycin cassette. (B) Two ES cell clones (left panel: control, 3F5; positive for the targeted allele, 3F6) and F1 progeny from B6 crosses (right panel: lanes 1–3) were analyzed by Southern blotting to confirm the presence of the flanking loxP (floxed) targeted allele. Progeny from MORE-Cre crosses were examined for recombination of the floxed allele (wild type mice, +/+; heterozygotes, +/−; knock-out mice, −/−). Genomic DNA was digested with XbaI and hybridized with radiolabelled probe 7. Wild type (WT), targeted (flox), and recombined alleles are indicated. (C) To detect recombination of the targeted allele in mice, DNA was purified from either the tail or thymus of WT and PTPN4-deficient mice and amplified with PCR primers. (D) PTPN4 mRNA from the thymus of WT and PTPN4 knockout mice was amplified using RT-PCR for the full-length and the catalytic regions of PTPN4. (E) Equivalent amounts of messenger RNA, isolated from the thymus (T), spleen (S), and testes (Te) of WT (lanes 1–3) and PTPN4 knockout mice (lanes 4–6) were reversed transcribed into cDNA using primers for PTPN4 (upper panel, 2.7 kb) or GAPDH (lower panel, 0.3 kb). No product was detected when the reverse transcriptase was excluded from the reaction (lane 7).
2.6. Functional assays of lymphocytes
Cells were isolated from the thymus, lymph nodes, or spleen and processed for flow cytometry and signaling studies using procedures outlined elsewhere (Becker et al., 2007; Pitcher et al., 2005). The cytokine production was measured by Cytometric Bead Array Flex Set assay (BD Pharmingen) and the upregulation of T cell activation markers was analyzed by cell surface staining and flow cytometry. For proliferation assays, cells were isolated from the lymph nodes and depleted of B220-expressing cells (B cells) using magnetic beads (Invitrogen Corp). Purified T cells were resuspended at 107 cells/ml in PBS and labeled with 1 µM CFSE for 10 min at 37°C. Labeled cells were stimulated for 72 h with 3 µg/ml plate-bound anti-CD3 ε (145-2C11) and 1 µg/ml anti-CD28 (37.51) or 50 ng/ml PMA and 1 µM ionomycin. Proliferation was assayed by flow cytometry. Listeria monocytogenes infections were undertaken with the assistance of Dr. James Forman (UT Southwestern Medical Center) using procedures outlined elsewhere (Berg et al., 2002; Berg et al., 2005). T cell polarization studies were done as described (Farrar et al., 2001; Persky et al., 2005).
3. Results
3.1. PTPN4 dephosphorylates tyrosine phosphorylated TCR ζ
We reported previously that TPH1 associates with and dephosphorylates tyrosine phosphorylated TCR ζ (phospho-ζ (Sozio et al., 2004). Since PTPN4 is a highly homologous family member, we examined whether PTPN4 could also regulate phospho-ζ. First, we examined the ability of a substrate-trapping derivative of PTPN4 to complex phospho-ζ. The catalytic domain of PTPN4 was engineered as a substrate-trapping mutant and expressed as a GST-fusion protein. This GST-fusion protein was used in pull-down assays with cell lysates containing phospho-ζ. The PTPN4 substrate-trapping mutant bound to phospho-ζ at comparable levels to the substrate trapping GST-PTPH1 (Fig. 1A, lanes 1–2). SHP-1 and SHP-2 substrate-trapping mutants failed to associate with phospho-ζ, consistent with our earlier findings (Fig. 1A, lanes 3–4)(Sozio et al., 2004). While both the phosphorylated and non-phosphorylated pools of TCR ζ were immunoprecipitated with ζ-specific mAbs, only phospho-ζ was complexed by the PTPN4 and PTPH1 substrate-trapping mutants (Fig. 1A, lane 5 versus 1–2). These results indicated that the catalytic domains of both PTPN4 and PTPH1 are capable of complexing phospho-ζ.
Fig. 1. PTPN4 dephosphorylates TCR ζ and negatively regulates TCR-mediated signaling.
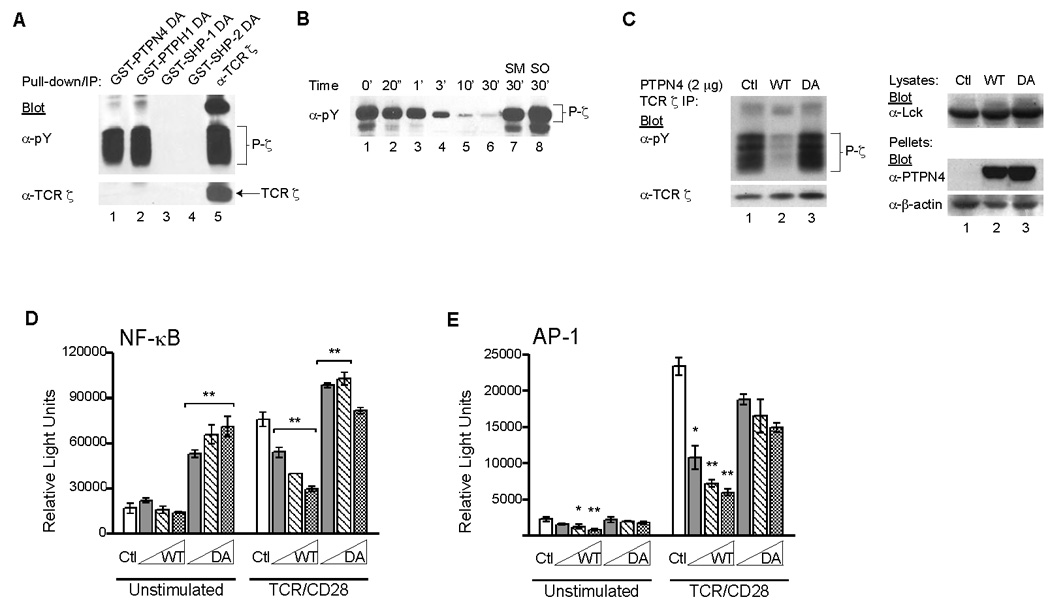
(A) COS7 cells were co-transfected with Lck and TCR ζ. Cell lysates were subsequently prepared and mixed with GST-fusion proteins containing the catalytic domains of the indicated PTPases that were engineered as substrate-traps. Alternatively, TCR ζ was directly immunoprecipitated. The pull-downs and immunoprecipitates were resolved by SDS-PAGE and Western blotted with anti-phosphotyrosine (pY) followed by anti-TCR ζ. Data are representative of three independent experiments. (B) Recombinant PTPN4, purified from Sf9 insect cells, was incubated with purified, in vitro-phosphorylated ζ for the indicated time points. Two samples were incubated in the presence of 0.5 mM sodium molybdate (SM, lane 7) or sodium orthovanadate (SO, lane 8) for 30 min. The proteins from each sample were immunoblotted with phosphotyrosine-specific mAb. Data are representative of three independent experiments. (C) Lck and TCR ζ were co-transfected with a control vector (ctl, lane 1), full-length wild type (WT, lane 2) PTPN4, or a substrate-trapping mutant (DA, lane 3) of full-length PTPN4 into HEK 293T cells. After 48 h, the cells were lysed, and TCR ζ was directly immunoprecipitated from the cell lysates. The precipitates were Western blotted with anti-pY and anti-TCR ζ mAbs. Total cell extracts were also prepared and immunoblotted with anti-Lck mAbs. The insoluble fraction was immunoblotted with anti-PTPN4 and anti-β-actin mAbs. Data are representative of five independent experiments. (D) Jurkat T cells were co-transfected with 20 µg of NF-κB reporter construct and 2.5, 5.0, and 10 µg of WT or DA PTPN4. Total DNA was equalized with empty vector. Luciferase activity was measured in unstimulated and TCR/CD28-stimulated cells. Data are shown as mean ± SD (n = 3; **p < 0.01). Results are representative of three independent experiments. (E) AP-1 transcriptional activity was measured as described in (D). Luciferase activity was measured in unstimulated and TCR-stimulated cells. Data are shown as mean ± SD (n = 3; *p < 0.05; **p < 0.01). Results are representative of two independent experiments.
We next examined whether full-length PTPN4 could dephosphorylate TCR ζ using two distinct assays. First, recombinant PTPN4, purified from insect cells, was incubated with phospho-ζ for different lengths of time. Reductions in phospho-ζ were evident within 1 min, and TCR ζ was completely dephosphorylated by 30 min (Fig. 1B, lanes 2–6 versus 1). The addition of sodium molybdate (SM) or sodium orthovanadate (SO), two PTPase-specific inhibitors, completely blocked the phosphatase activity of PTPN4 when added for the entire 30 min incubation (Fig. 1B, lanes 7–8). In a separate approach, wild type or substrate-trapping derivatives of PTPN4 were transfected into HEK 293T cells in combination with TCR ζ and the Lck PTK. Phospho-ζ was detected as a number of distinct phosphorylated intermediates in the absence of PTPN4 (Fig. 1C, lane 1). Over-expression of wild type (WT) PTPN4 reduced the amount of phospho-ζ 10–20 fold (Fig. 1C, lane 2 versus 1). The PTPN4 substrate-trapping mutant (DA) did not affect the levels of phospho-ζ (Fig. 1C, lane 3). Using similar co-transfection assays, PTPN4 also dephosphorylated DAP-12, an ITAM-containing protein expressed in NK cells (data not shown). However, unlike PTPH1, PTPN4 did not dephosphorylate Lck at tyrosine residue 394 (data not shown) (Sozio et al., 2004). Taken together, these findings demonstrated that PTPN4 dephosphorylates phospho-ITAMs.
Previous reports have shown that overexpression of PTPN4 modestly inhibited NFAT/AP-1-controlled luciferase expression following TCR stimulation in Jurkat cells (Han et al., 2000). To determine the role of PTPN4 in various TCR signaling pathways, we assessed the effects of PTPN4 overexpression on both NF-κB- and AP-1-driven luciferase reporter constructs. Jurkat T cells were co-transfected with either an NF-κB- or an AP-1- reporter construct and increasing amounts of WT or DA PTPN4. The cells were then left untreated or stimulated with anti-CD3 and anti-CD28 mAb, lysed, and luciferase activities were measured. WT PTPN4 significantly inhibited TCR-induced activation of NF-κB (Fig. 1D, p < 0.01). The over-expression of the substrate-trapping derivative (DA PTPN4) increased the levels of NF-κB activation both before and after stimulation (Fig. 1D, p < 0.01). This finding suggested that the substrate-trapping mutant potentiated the NF-κB signaling pathway, possibly by protecting a tyrosine-phosphorylated protein involved in TCR-mediated NF-κB activation. These findings were not seen in non-T cells (data not shown). We also examined the effects of PTPN4 on the transcriptional activity of AP-1. Increasing amounts of WT PTPN4 suppressed both basal and TCR-induced AP-1 activity (Fig. 1E, p < 0.05 or p < 0.01). The overexpression of DA PTPN4 had minimal effects on AP-1 activity either before or after TCR stimulation (Fig. 1E). Taken together, these results indicate that PTPN4 negatively regulates the TCR-mediated signaling pathway involved in both NF-κB and AP-1 activation.
3.2. Development of PTPN4-deficient mice
To investigate the role of PTPN4 in T cell functions, we generated a PTPN4 conditional knockout mouse. PTPN4 is predominantly expressed in testis as 3.2, 3.7, and 7.0 kb transcripts, with the 7.0 kb transcript also present in thymic tissue (data not shown)(Park et al., 2000). A targeting vector was designed with loxP sites preceding exon 21, and flanking a neomycin cassette following exon 23 (Fig. 2A). This construct was designed to disrupt the catalytic domain of PTPN4. One ES cell clone (3F6), identified by the presence of a floxed allele, was used to generate chimeric mice (Fig. 2B). Two independent chimeric lines were established. The wild type (WT, 13.5 kb) and targeted (flox, 7.0 kb) alleles were identified by Southern blotting (Fig. 2A–B). To delete PTPN4 in T cells, homozygous mice containing the targeted allele (PTPN4flox/flox) were backcrossed onto Lck-Cre transgenic mice. Despite multiple backcross attempts, the targeted allele was never deleted. Consequently, the PTPN4-targeted mice were backcrossed onto the MORE-Cre transgenic line. In this line, the Cre recombinase is controlled by the Mox2 promoter which is ubiquitously expressed as early as E5 of embryonic development (Tallquist and Soriano, 2000). Recombination of the PTPN4-targeted allele was confirmed by analyzing genomic DNA, isolated from both the tail and thymus, by Southern blotting and PCR techniques (Fig. 2B and C). PTPN4-deficient mice were born at normal Mendelian ratio with no apparent developmental or phenotypic defects. Male mice were fertile. The targeted disruption of PTPN4 was confirmed by RT-PCR and dsDNA sequencing (Fig. 2D). To analyze different tissues for PTPN4, PTPN4 was cloned by RT-PCR procedures using equivalent amounts of total RNA prepared from the thymus, spleen, and testes of wild type and PTPN4-deficient mice (Fig. 2E, lanes 1–6). Full-length cDNA for PTPN4 was generated from the thymus, spleen, and testes of wild type mice (lanes 1–3). Contrasting this, a truncated fragment of 2.2 kb was barely visible in tissues from the knockout mice (Fig. 2E, lanes 4–6). Similar levels of GAPDH were cloned from all tissues examined, except when the reverse transcriptase was excluded from the reaction (Fig. 2E, lanes 1–6 versus 7). The weak band seen in the KO tissue may have been background reactivity, or a truncated fragment of PTPN4. Northern blotting revealed a truncated, poorly expressed mRNA for PTPN4 in the testes of KO mice. Importantly, such a truncated product, if it were PTPN4, would not have any catalytic activity. PTPN4 protein expression is extremely difficult to detect. In wild type mice (WT), using thymus, lymph node, brain, and testes extracts, we could not detect protein expression (data not shown). This is due to several factors. First, most cell types cannot tolerate elevated levels of PTPN4 as they undergo cell cycle arrest (Gu and Majerus, 1996; Gu et al., 1996; Zhang et al., 1995; Zhang et al., 1999). Second, PTPN4 becomes trapped in the detergent insoluble pellet following most cell lysis procedures (data not shown) (Bretscher et al., 2002).
3.3. PTPN4 is not essential for normal T cell development
To determine whether PTPN4 was required for T cell development, thymocytes and peripheral T cell populations were compared between PTPN4 knockout mice and WT littermates. The cellularity in the thymus, lymph nodes, and spleen were similar between WT and PTPN4-deficient mice. Thymocytes from WT and PTPN4-deficient littermates were stained with a panel of antibodies to characterize the various thymic subsets. The percentages of the double negative (DN), double positive (DP), and CD8+ or CD4+ single positive (SP) populations in PTPN4-null mice were similar to those detected in WT littermates (Fig. 3A). The DN population is comprised of four developmental stages that are defined by CD44 and CD25 expression. Deletion of PTPN4 did not appear to impair the transition of thymocytes through the four DN stages (Fig. 3A). The intensities of CD3 and CD5 surface expression were also similar between WT and PTPN4-deficient DP and SP thymocyte subpopulations (Fig. 3B). In summary, we conclude that PTPN4 is not required for early T cell development in the thymus.
Fig. 3. T cell development and peripheral T cell populations are similar in PTPN4 knockout mice and wild type littermates.
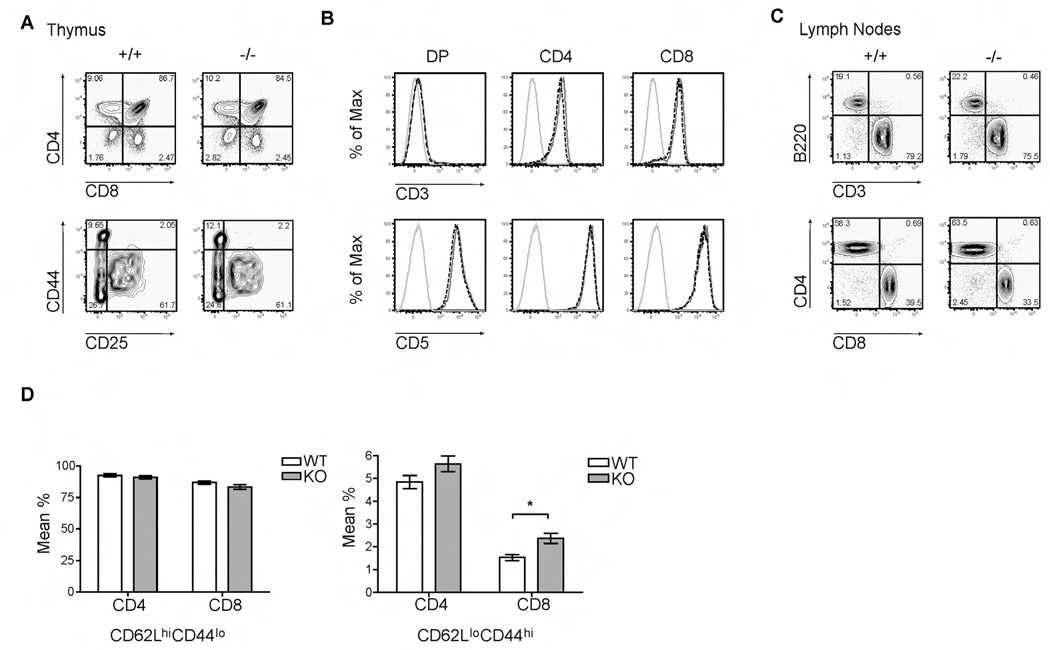
(A) Thymocytes isolated from WT (+/+) and KO (−/−) mice were stained with anti-CD4-PerCP 5.5 and anti-CD8-PE-Cy7 Abs. The double negative populations (CD4−CD8−B220−CD11b−) were further distinguished by staining cells with anti-CD44-APC and anti-CD25-FITC Ab. All cells were analyzed by flow cytometry. Data are representative of six experiments. (B) Gated double positive (DP) and CD4+ and CD8+ single positive populations from a representative WT mouse (gray solid line) or knockout mouse (black dashed line) were examined for CD3 and CD5 surface expression. Thymocytes were stained with anti-CD4-PerCP 5.5, anti-CD8-PE-Cy7, anti-CD3-Pacific Blue, and anti-CD5-PE Abs. An isotype control for CD3 or CD5 was used (light gray line). All cells were analyzed by flow cytometry. Data are represented as overlaid histograms and comparable to six independent experiments. (C) Lymph node cells of the indicated mice were isolated and stained with anti-CD3-Pacific Blue, anti-B220-APC-Cy7, anti-CD4-PerCP 5.5, and anti-CD8-PE-Cy7 Abs to examine peripheral lymphocytes populations by flow cytometry. Data are representative of six independent experiments. (D) Peripheral CD8+ and CD4+ T cells were stained with anti-CD62L-PE-Texas Red and anti-CD44-APC Abs to examine naïve and effector/memory subsets by flow cytometry. Data are shown as mean % ± SD (n = 4; *p = 0.003) and representative of three independent experiments.
Next, we characterized the peripheral lymphocyte populations from the lymph nodes and spleen in both WT and PTPN4-null mice. WT and PTPN4-deficient mice contained equivalent percentages of B and T cells in the lymph nodes (Fig. 3C). Moreover, the knockout mice had normal proportions of mature CD4+ and CD8+ T cells when compared to WT littermates (Fig. 3C). Similar findings were observed for the spleen (data not shown). In several experiments, an examination of the naïve and effector/memory cells in the peripheral CD4 and CD8 populations revealed an increase in CD62LloCD44hi effector/memory subset in PTPN4 KO mice when compared to WT littermates (Fig. 3D). These findings were consistent in 4 of 8 experiments. We do not know the reason for the variability.
3.4. Normal TCR signal transduction in the absence of PTPN4
The results from the substrate-trapping experiments suggested that the phosphorylated TCR ζ ITAMs were a likely PTPN4 substrate. To characterize the effects of PTPN4-deficiency on TCR signaling events, the phosphorylation state of various intracellular signaling proteins were analyzed prior to and following anti-CD3/CD28 stimulation. The 21-kDa phosphorylated form of TCR ζ (p21) is constitutively phosphorylated in thymocytes and peripheral lymphocytes, while the 23-kDa phosphorylated form of ζ (p23) is induced following TCR cross-linking (van Oers et al., 1994; van Oers et al., 1993; van Oers et al., 2000). The magnitude and duration of p21 and p23 phosphorylation were similar between WT and PTPN4-deficient cells (Fig. 4A, lanes 1–4 versus 6–8). These findings suggested that either the TCR ζ ITAMs are not the primary target of PTPN4, or that an effective compensatory mechanism exists with other PTPases (Fig. 4B and data not shown). Additionally, the phosphorylation of Lck, ZAP-70, Erk1/2, p38, and Jnk1/2 was comparable in intensity and duration in WT and PTPN4 knockout cells (Fig. 4A and B) (data not shown). Finally, whole cell lysates were immunoblotted for the presence of tyrosine-phosphorylated proteins. The absence of PTPN4 had no effect on overall protein tyrosine phosphorylation patterns in the first 90 minutes of T cell activation (Fig. 4A and B). These findings implied that PTPN4 does not regulate the tyrosine phosphorylation of TCR ζ or other common signaling molecules.
Fig. 4. TCR-mediated signals are unaltered in the absence of PTPN4.
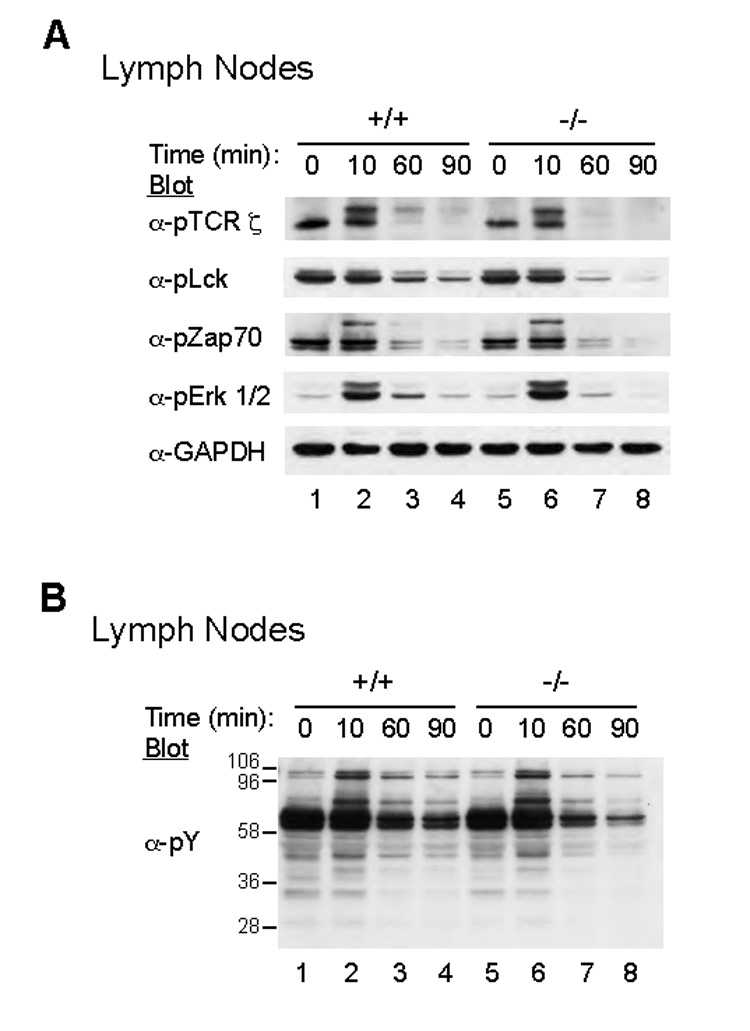
(A) PTPN4 WT (+/+) and knockout (−/−) T cells were left untreated or stimulated for 10, 60, and 90 minutes with anti-CD3 mAbs. The cells were lysed and the lysates were Western immunoblotted with antibodies specific for phospho-ζ, phospho-Lck, phospho-ZAP70, phospho-Erk1/2, and GAPDH. (B) Total protein tyrosine phosphorylation was detected from whole cell lysates by Western blotting with anti-phosphotyrosine mAb (pY). Results are representative of three independent experiments.
3.5. PTPN4-deficient T cells have normal cytokine production and T cell effector functions
Since a PTPN4-deficiency had no effect on T cell development, we wanted to determine whether the effector functions of these cells were altered. Peripheral T cells were isolated from PTPN4 knockout mice and WT littermates, stimulated with anti-CD3/CD28 mAb, and assayed for the upregulation of selected activation markers such as CD69. The induction of CD69 on CD4+ or CD8+ T cells appeared to be similar between WT and KO mice (Fig. 5A). To examine whether PTPN4 affected T cell proliferation, purified T cells from WT and knockout mice were labeled with CFSE and stimulated with plate-bound anti-CD3/anti-CD28 mAbs. Following three days of stimulation, the extent of cell division was compared by examining the degree of CFSE dilution. Cell division was similar in CD4+ or CD8+ T cells when comparing WT and PTPN4 knockout mice (Fig. 5B). Additionally, no differences in apoptosis were detected between WT and knockout cells following stimulation (data not shown).
Fig. 5. T cells from PTPN4-deficient mice have normal effector functions.
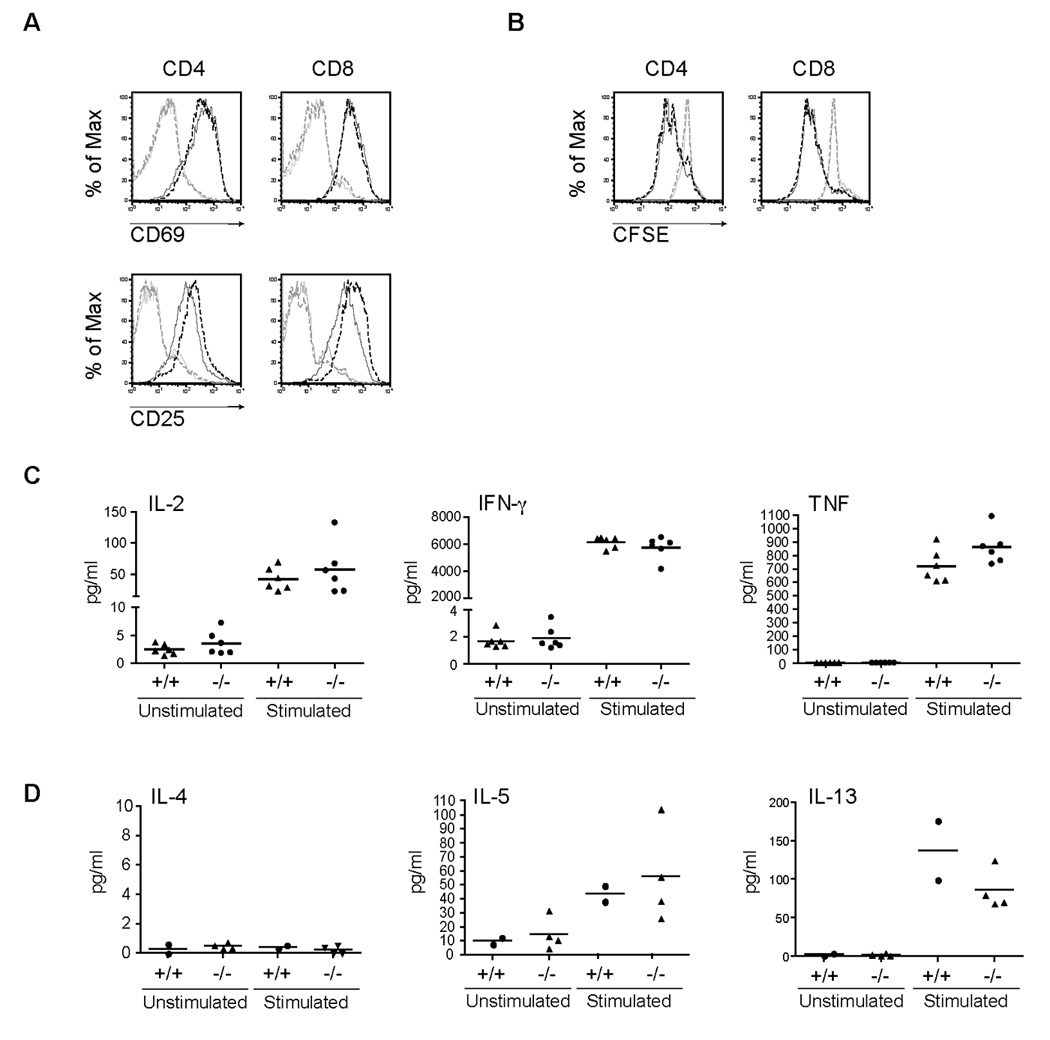
(A) Purified T cells from a representative WT mouse (solid lines) or PTPN4 knockout mouse (dashed lines) were stimulated with anti-CD3/CD28 antibodies. Cells were harvested on Day 3 and stained with anti-CD4-APC, anti-CD8-PE-Cy7, anti-CD69 PE, and anti-CD25-FITC Abs to analyze upregulation of activation markers. Data are represented as overlaid histograms (light gray lines, unstimulated cells; black lines, stimulated cells). Results are representative of three independent experiments. (B) Purified T cells from a representative WT (solid lines) or PTPN4 knockout mouse (dashed lines) were labeled with CFSE and stimulated. Cell division was assessed by CFSE dilution by flow cytometry. Data are shown as overlaid histograms (light gray lines, unstimulated cells; black lines, stimulated cells). Data are representative of three independent experiments. Culture supernatants were collected from unstimulated and anti-CD3/CD28 stimulated T cells from WT (+/+) or PTPN4 knockout (−/−) mice (n = 6). (C) IL-2, IFN-γ, and TNF and (D) IL-4, IL-5, and IL-13 were measured from culture supernatants using the CBA assay.
To determine whether the types and levels of distinct cytokines were regulated by PTPN4, different Th1- and Th2-specific cytokines were measured from the culture supernatant of CD3/CD28-stimulated T cells from WT and knockout mice. The production of Th1-controlled cytokines, including IL-2, TNF, and IFN-γ cytokines were similar between PTPN4 KO and WT cells (Fig. 5C). In addition, those cytokines that are characteristic of Th2-polarized cells (IL-4, IL-5, IL-13) were similar between PTPN4-deficient T cells and WT littermates (Fig. 5D). These results suggested that PTPN4 is not required for TCR-mediated cytokine production and/or T cell polarization. In order to determine whether PTPN4 knockout mice could mount an appropriate immune response during bacterial infections, PTPN4 knockout and WT littermate mice were infected with 50,000 CFU of Listeria monocytogenes that expressed the OVA protein (LM-OVA). We initially examined innate effector responses since a CD8+ T cell response is important for bacterial clearance and our findings suggested increased numbers of CD8 memory/effector populations (Fig. 3) (Berg et al., 2003). The liver and spleen were harvested three days post-infection and the CFUs/organ were determined. The ability of PTPN4 knockout mice to clear an LM-OVA infection was similar to wild type mice as the CFUs in the liver (118,000 and 145,000 CFU respectively) and spleen (9 × 106 and 6 × 106 CFU, respectively) were comparable (Fig. 6 A and B). In addition, the CD8+ naïve and memory compartments were comparable between wild type and PTPN4 knockout mice and following infections (data not shown). Preliminary findings also revealed similar adaptive immune responses to LM-OVA (data not shown). These data show that PTPN4 is not required for effective bacterial clearance.
Fig. 6. PTPN4 knockout mice and wild type littermates have similar responses to Listeria monocytogenes infections.
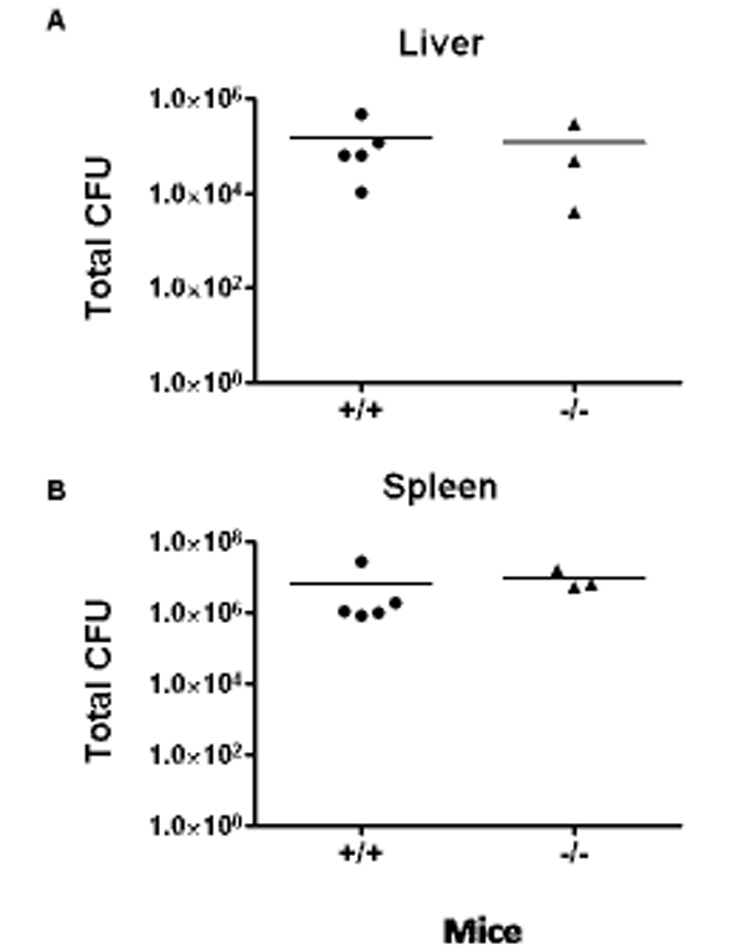
WT and PTPN4 knockout mice were infected with LM intravenously. Three days later, the spleens (A) and livers (B) were removed. Homogenates from these organs were prepared, diluted, and aliquots then plated on brain-heart infusion medium plates. After an overnight culture, the number of bacterial colonies was determined/organ. Data are presented from six mice/group. The data are similar between two independent experiments.
4. Discussion
In this report, we provide evidence that the protein tyrosine phosphatase PTPN4 interacts with and dephosphorylates the ITAMs of the TCR ζ subunit. Transcriptional reporter assays demonstrate that overexpression of catalytically active PTPN4 suppresses NF-κB and AP-1 activation, supporting the notion that PTPN4 is a regulator of TCR signaling. Further, the substrate-trapping form of PTPN4 potentiates NF-κB activity in T cells. To define the role of PTPN4 in vivo, PTPN4-deficient mice were developed. The targeted deletion of PTPN4 does not impair T cell development in the thymus or periphery. Finally, the PTPN4-deficiency does not impact T cell-dependent cytokine production or immune responses to bacterial infections. These experiments suggest that additional PTPases are likely involved in the in vivo regulation of TCR signaling pathways.
We characterized PTPN4 based on its ability to bind and dephosphorylate the ITAMs in TCR ζ, suggesting that PTPN4 has a critical functional contribution to TCR signaling. However, our analyses of PTPN4-deficient mice revealed normal T cell development and normal peripheral T cell functions. In fact, all of the tyrosine phosphorylated proteins induced following TCR signaling, including the ITAMs of TCR ζ, appeared unaffected by the absence of the intact PTPN4 catalytic domain. We considered several alternate explanations to account for the minimal effects of a PTPN4-deficiency on TCR signaling. First, considerable functional redundancy and/or compensatory mechanisms may exist between PTPN4 and other PTPases. One likely candidate is PTPH1, a homologous-PTPN4 family member that also complexes to and dephosphorylates TCR ζ. Interestingly, PTPH1-deficient mice have normal T cell development and TCR signaling functions (Bauler et al., 2007). Thus, PTPN4 and PTPH1 could coordinately regulate TCR signaling pathways. The combined disruption of PTPN4 and PTPH1 will be necessary to resolve this issue. Functional redundancy has been observed in other studies of PTPase-deficient mice. The absence of Hematopoietic Protein Tyrosine Phosphatase (HePTP) in thymocytes led to an increase in ERK activation following stimulation (Gronda et al., 2001). Yet, these mice maintain normal T cell development and lymphocyte activation, suggesting that a HePTP-deficiency is compensated (Gronda et al., 2001). Similarly, the targeted deletion of PEP resulted in the expansion of effector/memory T cell subsets, but had no affect on T cell development or naïve T cell functions (Hasegawa et al., 2004). These data indicate that functional redundancy and/or compensatory mechanisms are common among PTPases.
A second possible explanation is that the phosphorylated ITAMs of TCR ζ and the other TCR subunits are not the primary targets of PTPN4. PTPN4 contains both FERM and PDZ domains, with the FERM domain responsible for localizing PTPN4 to the detergent-insoluble cytoskeleton. That restricted localization may prevent PTPN4 from targeting the CD3 ITAMs. More recent reports have revealed the presence of ITAM-like molecules in signaling pathways mediated by adhesion molecules, lectin receptors, and chemokine receptors (Abram and Lowell, 2007). Ezrin and moesin are two such proteins that contain ITAM-like sequences and are found in the cytoskeleton in association with the adhesion molecule PSGL-1 (Alonso-Lebrero et al., 2000; Lankes and Furthmayr, 1991). Therefore, PTPN4, through its localization to the actin cytoskeleton, may participate in the regulation of multiple signaling pathways that utilize ITAM-like proteins (Abram and Lowell, 2007). Alternatively, PTPN4 might dephosphorylate completely unrelated proteins, such as valosin-containing protein (VCP), since PTPH1, the PTPN4 homolog, is know to regulate the phosphorylation state of VCP (Zhang et al., 1999). VCP is an AAA-ATPase that regulates ubiquitin-dependent protein degradation, in part through an interaction with Npl4/Ufd1 and IκBα (Dai et al., 1998; Meyer et al., 2000; Wojcik et al., 2006). Our finding that the overexpression of the substrate-trapping derivative of PTPN4 potentiates the NF-κB pathway provides a model where the tyrosine phosphorylation of VCP enhances ubiquitin-dependent IκBα degradation and the ensuing NF-κB activation. It is interesting to note that our initial studies of the PTPN4-deficient mice suggested that the Th2-regulated cytokines, IL-4 and IL-13 were elevated in response to TCR stimulation, when compared to WT littermates. As the PTPN4 KO mice were further backcrossed onto a C57Bl/6 line, this cytokine difference was no longer obvious between the KO and WT littermates. Further experiments are necessary to explore the possibility that PTPN4 regulates IL-4 pathways through NF-κB, an effect that might only be apparent in susceptible strains of mice.
It is also possible that PTPN4 may have a non-redundant role in activated rather than naïve T cells, much like PEP. This might be due to the possible upregulation of PTPN4 in activated T cells, much like with PTPH1, which is more highly expressed in peripheral lymphocytes following TCR/CD28 stimulation (Bauler et al., 2007). While we could not detect endogenous protein in T cells or testes, where PTPN4 is expressed, an increased expression in actively proliferating cells might be required for controlling the extent of ubiquitination-dependent protein degradation. If both PTPN4 and PTPH1 function in this capacity, only the combined disruption of both enzymes would reveal their contribution to TCR signaling. Another PTPN4 knockout mouse was recently developed. In this knockout mouse, there was a selective defect in cerebellar long-term depression (Kina et al., 2007). No studies were undertaken with lymphocytes from these mice. Many knockout mice have developmental problems that complicate phenotypic changes to a specific genetic modification. Alternate approaches have included the modulation of gene expression in temporal and tissue-specific manner. In our initial attempt to modulate PTPN4 expression in lymphocytes, we mated the PTPN4-targeted mice with a second set of mice expressing Lck-Cre in order to eliminate PTPN4 in thymocytes. As we could not detect recombination, we resorted to a complete knockout of PTPN4. We also attempted to knock-down PTPN4 in lymphocytes using siRNA procedures. Such approaches yielded knock-downs of only two-fold. Finally, tetracycline-regulated expression of wild type and substrate-trapping derivatives of PTPN4 were attempted without success.
5. Conclusion
In summary, PTPN4 regulates ITAM phosphorylation and NF-κB activation in transfected T cells. The development of PTPN4 knockout mice revealed that this PTPase is not critical for T cell functions, suggesting that compensatory mechanisms exist among many PTPase-families.
Acknowledgements
This work was supported in part by grants from the National Institutes of Health 5T32AI005284 (to JAY, AB, AW), AI42953 and AI69249 (to NvO). We thank Angela Mobley for assistance with flow cytometry and Sean Murray for technical advice. We thank Jonathan Huber and Hilario Ramos for technical advice on T cell polarizations and James Forman for helpful scientific discussions. We also thank Laura DeFord-Watts, Lisa Pitcher, and Srividya Subramanian for critical review of the manuscript.
Abbreviations
- PTK
Protein tyrosine kinase
- PTPase
Protein tyrosine phosphatase
- WT
Wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abram CL, Lowell CA. The expanding role for ITAM-based signaling pathways in immune cells. Sci STKE. 2007;2007:re2. doi: 10.1126/stke.3772007re2. [DOI] [PubMed] [Google Scholar]
- Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Alonso-Lebrero JL, Serrador JM, Dominguez-Jimenez C, Barreiro O, Luque A, del Pozo MA, Snapp K, Kansas G, Schwartz-Albiez R, Furthmayr H, Lozano F, Sanchez-Madrid F. Polarization and interaction of adhesion molecules P-selectin glycoprotein ligand 1 and intercellular adhesion molecule 3 with moesin and ezrin in myeloid cells. Blood. 2000;95:2413–2419. [PubMed] [Google Scholar]
- Bauler TJ, Hughes ED, Arimura Y, Mustelin T, Saunders TL, King PD. Normal TCR signal transduction in mice that lack catalytically active PTPN3 protein tyrosine phosphatase. J Immunol. 2007;178:3680–3687. doi: 10.4049/jimmunol.178.6.3680. [DOI] [PubMed] [Google Scholar]
- Becker AM, Deford-Watts LM, Wuelfing C, van Oers NS. The Constitutive Tyrosine Phosphorylation of CD3{zeta} Results from TCR-MHC Interactions That Are Independent of Thymic Selection. J Immunol. 2007;178:4120–4128. doi: 10.4049/jimmunol.178.7.4120. [DOI] [PubMed] [Google Scholar]
- Berg RE, Cordes CJ, Forman J. Contribution of CD8+ T cells to innate immunity: IFN-gamma secretion induced by IL-12 and IL-18. Eur J Immunol. 2002;32:2807–2816. doi: 10.1002/1521-4141(2002010)32:10<2807::AID-IMMU2807>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Berg RE, Crossley E, Murray S, Forman J. Memory CD8+ T cells provide innate immune protection against Listeria monocytogenes in the absence of cognate antigen. J Exp Med. 2003;198:1583–1593. doi: 10.1084/jem.20031051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg RE, Crossley E, Murray S, Forman J. Relative contributions of NK and CD8 T cells to IFN-gamma mediated innate immune protection against Listeria monocytogenes. J Immunol. 2005;175:1751–1757. doi: 10.4049/jimmunol.175.3.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- Brockdorff J, Williams S, Couture C, Mustelin T. Dephosphorylation of ZAP-70 and inhibition of T cell activation by activated SHP1. Eur J Immunol. 1999;29:2539–2550. doi: 10.1002/(SICI)1521-4141(199908)29:08<2539::AID-IMMU2539>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Call ME, Pyrdol J, Wucherpfennig KW. Stoichiometry of the T-cell receptor-CD3 complex and key intermediates assembled in the endoplasmic reticulum. Embo J. 2004;23:2348–2357. doi: 10.1038/sj.emboj.7600245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambier JC. New nomenclature for the Reth motif (or ARH1/TAM/ARAM/YXXL) Immunol. Today. 1995;16:110. doi: 10.1016/0167-5699(95)80105-7. [DOI] [PubMed] [Google Scholar]
- Chan AC, Dalton M, Johnson R, Kong G-H, Wang T, Thoma R, Kurosaki T. Activation of ZAP-70 kinase activity by phosphorylation of tyrosine 493 is required for lymphocyte antigen receptor function. EMBO J. 1995;14:2499–2508. doi: 10.1002/j.1460-2075.1995.tb07247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier J-F, Veillette A. Association of inhibitory tyrosine protein kinase p50csk with protein tyrosine phosphatase PEP in T cells and other hematopoeitic cells. EMBO J. 1996;15:4909–4918. [PMC free article] [PubMed] [Google Scholar]
- Dai RM, Chen E, Longo DL, Gorbea CM, Li CC. Involvement of valosin-containing protein, an ATPase Co-purified with IkappaBalpha and 26 S proteasome, in ubiquitin-proteasome-mediated degradation of IkappaBalpha. J Biol Chem. 1998;273:3562–3573. doi: 10.1074/jbc.273.6.3562. [DOI] [PubMed] [Google Scholar]
- Farrar JD, Ouyang W, Lohning M, Assenmacher M, Radbruch A, Kanagawa O, Murphy KM. An instructive component in T helper cell type 2 (Th2) development mediated by GATA-3. J Exp Med. 2001;193:643–650. doi: 10.1084/jem.193.5.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finco TS, Baldwin AS. Mechanistic aspects of NF-kappa B regulation: the emerging role of phosphorylation and proteolysis. Immunity. 1995;3:263–272. doi: 10.1016/1074-7613(95)90112-4. [DOI] [PubMed] [Google Scholar]
- Flint AJ, Tiganis T, Barford D, Tonks NK. Development of "substrate-trapping" mutants to identify physiological substrates of protein tyrosine phosphatases. Proc Natl Acad Sci U S A. 1997;94:1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frearson JA, Alexander DR. The phosphotyrosine phosphatase SHP-2 participates in a multimeric signaling complex and regulates T cell receptor (TCR) coupling to the ras/mitogen-activated protein kinase (MAPK) pathway in Jurkat T cells. J. Exp. Med. 1998;187:1417–1426. doi: 10.1084/jem.187.9.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil D, Schamel WW, Montoya M, Sanchez-Madrid F, Alarcon B. Recruitment of Nck by CD3 epsilon reveals a ligand-induced conformational change essential for T cell receptor signaling and synapse formation. Cell. 2002;109:901–912. doi: 10.1016/s0092-8674(02)00799-7. [DOI] [PubMed] [Google Scholar]
- Gil D, Schrum AG, Alarcon B, Palmer E. T cell receptor engagement by peptide-MHC ligands induces a conformational change in the CD3 complex of thymocytes. J Exp Med. 2005;201:517–522. doi: 10.1084/jem.20042036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjorloff-Wingren A, Saxena M, Han S, Wang X, Alonso A, Renedo M, Oh P, Williams S, Schnitzer J, Mustelin T. Subcellular localization of intracellular protein tyrosine phosphatases in T cells [In Process Citation] Eur J Immunol. 2000;30:2412–2421. doi: 10.1002/1521-4141(2000)30:8<2412::AID-IMMU2412>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Gjorloff-Wingren A, Saxena M, Williams S, Hammi D, Mustelin T. Characterization of TCR-induced receptor-proximal signaling events negatively regulated by the protein tyrosine phosphatase PEP. Eur J Immunol. 1999;29:3845–3854. doi: 10.1002/(SICI)1521-4141(199912)29:12<3845::AID-IMMU3845>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Gronda M, Arab S, Iafrate B, Suzuki H, Zanke BW. Hematopoietic protein tyrosine phosphatase suppresses extracellular stimulus-regulated kinase activation. Mol Cell Biol. 2001;21:6851–6858. doi: 10.1128/MCB.21.20.6851-6858.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Majerus PW. The properties of the protein tyrosine phosphatase PTPMEG. J Biol Chem. 1996;271:27751–27759. doi: 10.1074/jbc.271.44.27751. [DOI] [PubMed] [Google Scholar]
- Gu M, Meng K, Majerus PW. The effect of overexpression of the protein tyrosine phosphatase PTPMEG on cell growth and on colony formation in soft agar in COS-7 cells. Proc Natl Acad Sci U S A. 1996;93:12980–12985. doi: 10.1073/pnas.93.23.12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu MX, York JD, Warshawsky I, Majerus PW. Identification, cloning, and expression of a cytosolic megakaryocyte protein-tyrosine-phosphatase with sequence homology to cytoskeletal protein 4.1. Proc Natl Acad Sci U S A. 1991;88:5867–5871. doi: 10.1073/pnas.88.13.5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Williams S, Mustelin T. Cytoskeletal protein tyrosine phosphatase PTPH1 reduces T cell antigen receptor signaling. Eur J Immunol. 2000;30:1318–1325. doi: 10.1002/(SICI)1521-4141(200005)30:5<1318::AID-IMMU1318>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. New York: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hasegawa K, Martin F, Huang G, Tumas D, Diehl L, Chan AC. PEST domain-enriched tyrosine phosphatase (PEP) regulation of effector/memory T cells. Science. 2004;303:685–689. doi: 10.1126/science.1092138. [DOI] [PubMed] [Google Scholar]
- Hatada MH, Lu X, Laird ER, Green J, Morgenstern JP, Lou M, Marr CS, Phillips TB, Ram MK, Theriault K, Zoller MJ, Karas JL. Molecular basis for the interactions of the protein tyrosine kinase ZAP-70 with the T cell receptor. Nature. 1995;377:32–38. doi: 10.1038/377032a0. [DOI] [PubMed] [Google Scholar]
- Huang Y, Wange RL. T cell receptor signaling: beyond complex complexes. J Biol Chem. 2004;279:28827–28830. doi: 10.1074/jbc.R400012200. [DOI] [PubMed] [Google Scholar]
- Humphrey MB, Lanier LL, Nakamura MC. Role of ITAM-containing adapter proteins and their receptors in the immune system and bone. Immunol Rev. 2005;208:50–65. doi: 10.1111/j.0105-2896.2005.00325.x. [DOI] [PubMed] [Google Scholar]
- Jordan MS, Singer AL, Koretzky GA. Adaptors as central mediators of signal transduction in immune cells. Nat Immunol. 2003;4:110–116. doi: 10.1038/ni0203-110. [DOI] [PubMed] [Google Scholar]
- Kina S, Tezuka T, Kusakawa S, Kishimoto Y, Kakizawa S, Hashimoto K, Ohsugi M, Kiyama Y, Horai R, Sudo K, Kakuta S, Iwakura Y, Iino M, Kano M, Manabe T, Yamamoto T. Involvement of protein-tyrosine phosphatase PTPMEG in motor learning and cerebellar long-term depression. Eur J Neurosci. 2007;26:2269–2278. doi: 10.1111/j.1460-9568.2007.05829.x. [DOI] [PubMed] [Google Scholar]
- Lankes WT, Furthmayr H. Moesin: a member of the protein 4.1-talin-ezrin family of proteins. Proc Natl Acad Sci U S A. 1991;88:8297–8301. doi: 10.1073/pnas.88.19.8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K-M, Chuang E, Griffin M, Khattri R, Hong DK, Zhang W, Straus D, Samelson LE, Thompson CB, Bluestone JA. Molecular basis of T cell inactivation by CTLA-4. Science. 1998;282:2263–2266. doi: 10.1126/science.282.5397.2263. [DOI] [PubMed] [Google Scholar]
- Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- Lorenz U, Ravichandran KS, Burakoff SJ, Neel BG. Lack of SHPTP1 results in src-family kinase hyperactivation and thymocyte hyperresponsiveness. Proc. Natl. Acad. Sci. USA. 1996;93:9624–9629. doi: 10.1073/pnas.93.18.9624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. Embo J. 2000;19:2181–2192. doi: 10.1093/emboj/19.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minguet S, Swamy M, Alarcon B, Luescher IF, Schamel WW. Full activation of the T cell receptor requires both clustering and conformational changes at CD3. Immunity. 2007;26:43–54. doi: 10.1016/j.immuni.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Mustelin T, Tasken K. Positive and negative regulation of T-cell activation through kinases and phosphatases. Biochem J. 2003;371:15–27. doi: 10.1042/BJ20021637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustelin T, Vang T, Bottini N. Protein tyrosine phosphatases and the immune response. Nat Rev Immunol. 2005;5:43–57. doi: 10.1038/nri1530. [DOI] [PubMed] [Google Scholar]
- Nguyen TV, Ke Y, Zhang EE, Feng GS. Conditional deletion of Shp2 tyrosine phosphatase in thymocytes suppresses both pre-TCR and TCR signals. J Immunol. 2006;177:5990–5996. doi: 10.4049/jimmunol.177.9.5990. [DOI] [PubMed] [Google Scholar]
- Palacios EH, Weiss A. Function of the Src-family kinases, Lck and Fyn, in T-cell development and activation. Oncogene. 2004;23:7990–8000. doi: 10.1038/sj.onc.1208074. [DOI] [PubMed] [Google Scholar]
- Pani G, Fischer K-D, Mlinaric-Rascan I, Siminovitch KA. Signaling capacity of the T cell antigen receptor is negatively regulated by the PTP1C tyrosine phosphatase. J. Exp. Med. 1996;184:839–852. doi: 10.1084/jem.184.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KW, Lee EJ, Lee S, Lee JE, Choi E, Kim BJ, Hwang R, Park KA, Baik J. Molecular cloning and characterization of a protein tyrosine phosphatase enriched in testis, a putative murine homologue of human PTPMEG. Gene. 2000;257:45–55. doi: 10.1016/s0378-1119(00)00351-6. [DOI] [PubMed] [Google Scholar]
- Persky ME, Murphy KM, Farrar JD. IL-12, but not IFN-alpha, promotes STAT4 activation and Th1 development in murine CD4+ T cells expressing a chimeric murine/human Stat2 gene. J Immunol. 2005;174:294–301. doi: 10.4049/jimmunol.174.1.294. [DOI] [PubMed] [Google Scholar]
- Pilecka I, Patrignani C, Pescini R, Curchod ML, Perrin D, Xue Y, Yasenchak J, Clark A, Magnone MC, Zaratin P, Valenzuela D, Rommel C, van Huijsduijnen RH. Protein-tyrosine Phosphatase H1 Controls Growth Hormone Receptor Signaling and Systemic Growth. J Biol Chem. 2007;282:35405–35415. doi: 10.1074/jbc.M705814200. [DOI] [PubMed] [Google Scholar]
- Pitcher LA, Mathis MA, Young JA, Deford LM, Purtic B, Wulfing C, van Oers NS. The CD3 gammaepsilon/deltaepsilon signaling module provides normal T cell functions in the absence of the TCR zeta immunoreceptor tyrosine-based activation motifs. Eur J Immunol. 2005;35:3643–3654. doi: 10.1002/eji.200535136. [DOI] [PubMed] [Google Scholar]
- Pitcher LA, van Oers NSC. T cell receptor signal transmission:who gives an ITAM. Trends in Immunology. 2003;24:554–560. doi: 10.1016/j.it.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- Reth M. Antigen receptor tail clue. Nature. 1989;338:383–384. [PubMed] [Google Scholar]
- Reth M. Antigen receptors on B lymphocytes. Ann. Rev. Immunol. 1992;10:97–121. doi: 10.1146/annurev.iy.10.040192.000525. [DOI] [PubMed] [Google Scholar]
- Samelson LE. Signal transduction mediated by the T cell antigen receptor: the role of adapter proteins. Annu Rev Immunol. 2002;20:371–394. doi: 10.1146/annurev.immunol.20.092601.111357. [DOI] [PubMed] [Google Scholar]
- Shapiro VS, Mollenauer MN, Greene WC, Weiss A. c-rel regulation of IL-2 gene expression may be mediated through activation of AP-1. J Exp Med. 1996;184:1663–1669. doi: 10.1084/jem.184.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozio MS, Mathis MA, Young JA, Walchli S, Pitcher LA, Wrage PC, Bartok B, Campbell A, Watts JD, Aebersold R, Van Huijsduijnen RH, van Oers NS. PTPH1 is a predominant protein-tyrosine phosphatase capable of interacting with and dephosphorylating the T cell receptor zeta subunit. J Biol Chem. 2004;279:7760–7769. doi: 10.1074/jbc.M309994200. [DOI] [PubMed] [Google Scholar]
- Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000;26:113–115. doi: 10.1002/(sici)1526-968x(200002)26:2<113::aid-gene3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Tonks NK, Neel BG. Combinatorial control of the specificity of protein tyrosine phosphatases. Curr. Opin. Cell. Biol. 2001;13:182–195. doi: 10.1016/s0955-0674(00)00196-4. [DOI] [PubMed] [Google Scholar]
- Tsui HW, Siminovitch KA, deSouza L, Tsui FWL. Motheaten and motheaten viable mice have mutations in the haematopoietic cell phosphatase gene. Nature Genetics. 1993;4:124–129. doi: 10.1038/ng0693-124. [DOI] [PubMed] [Google Scholar]
- van Oers NSC, Killeen N, Weiss A. ZAP-70 is constitutively associated with tyrosine phosphorylated TCR z in murine thymocytes and lymph node T cells. Immunity. 1994;1:675–685. doi: 10.1016/1074-7613(94)90038-8. [DOI] [PubMed] [Google Scholar]
- van Oers NSC, Tao W, Watts JD, Johnson P, Aebersold R, Teh H-S. Constitutive tyrosine phosphorylation of the T cell receptor (TCR) ζ subunit: Regulation of TCR-associated protein kinase activity by TCR ζ. Mol. Cell. Bio. 1993;13:5771–5780. doi: 10.1128/mcb.13.9.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oers NSC, Tohlen B, Malissen B, Moomaw CR, Afendis S, Slaughter C. The 21- and 23- kDa forms of TCR ζ are generated by specific ITAM phosphorylations. Nature Immunology. 2000;1:322–328. doi: 10.1038/79774. [DOI] [PubMed] [Google Scholar]
- van Oers NSC, von Boehmer H, Weiss A. The pre-TCR complex is functionally coupled to the TCR ζ subunit. J. Exp. Med. 1995;182:1585–1590. doi: 10.1084/jem.182.5.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillette A, Latour S, Davidson D. Negative regulation of immunoreceptor signaling. Annu Rev Immunol. 2002;20:669–707. doi: 10.1146/annurev.immunol.20.081501.130710. [DOI] [PubMed] [Google Scholar]
- Walchli S, Curchod ML, Gobert RP, Arkinstall S, Hooft van Huijsduijnen R. Identification of tyrosine phosphatases that dephosphorylate the insulin receptor. A brute force approach based on "substrate-trapping" mutants. J Biol Chem. 2000;275:9792–9796. doi: 10.1074/jbc.275.13.9792. [DOI] [PubMed] [Google Scholar]
- Waldmann TA. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat Rev Immunol. 2006;6:595–601. doi: 10.1038/nri1901. [DOI] [PubMed] [Google Scholar]
- Wange RL, Guitian R, Isakov N, Watts JD, Aebersold R, Samelson LE. Activating and inhibitory mutations in adjacent tyrosines in the kinase domain of ZAP-70. J. Biol. Chem. 1995;270:18730–18733. doi: 10.1074/jbc.270.32.18730. [DOI] [PubMed] [Google Scholar]
- Wange RL, Samelson LE. Complex complexes: signaling at the TCR. Immunity. 1996;5:197–205. doi: 10.1016/s1074-7613(00)80315-5. [DOI] [PubMed] [Google Scholar]
- Weiss A. T cell antigen receptor signal transduction: a tale of tails and cytoplasmic protein-tyrosine kinases. Cell. 1993;73:209–212. doi: 10.1016/0092-8674(93)90221-b. [DOI] [PubMed] [Google Scholar]
- Weiss A, Littman DR. Signal transduction by lymphocyte antigen receptors. Cell. 1994;76:263–274. doi: 10.1016/0092-8674(94)90334-4. [DOI] [PubMed] [Google Scholar]
- Wojcik C, Rowicka M, Kudlicki A, Nowis D, McConnell E, Kujawa M, DeMartino GN. Valosin-containing protein (p97) is a regulator of endoplasmic reticulum stress and of the degradation of N-end rule and ubiquitin-fusion degradation pathway substrates in mammalian cells. Mol Biol Cell. 2006;17:4606–4618. doi: 10.1091/mbc.E06-05-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SH, Eckberg WR, Yang Q, Samatar AA, Tonks NK. Biochemical characterization of a human band 4.1-related protein-tyrosine phosphatase, PTPH1. J Biol Chem. 1995;270:20067–20072. doi: 10.1074/jbc.270.34.20067. [DOI] [PubMed] [Google Scholar]
- Zhang SH, Liu J, Kobayashi R, Tonks NK. Identification of the cell cycle regulator VCP (p97/CDC48) as a substrate of the band 4.1-related protein-tyrosine phosphatase PTPH1. J Biol Chem. 1999;274:17806–17812. doi: 10.1074/jbc.274.25.17806. [DOI] [PubMed] [Google Scholar]