Abstract
Type II topoisomerases resolve problematic DNA topologies such as knots, catenanes, and supercoils that arise as a consequence of DNA replication and recombination. Failure to remove problematic DNA topologies prohibits cell division and can result in cell death or genetic mutation. Such catastrophic consequences make topoisomerases an effective target for antibiotics and anticancer agents. Despite their biological and clinical importance, little is understood about how a topoisomerase differentiates DNA topologies in a molecule that is significantly larger than the topoisomerase itself. It has been proposed that type II topoisomerases recognize angle and curvature between two DNA helices characteristic of knotted and catenated DNA to account for the enzyme’s preference to unlink instead of link DNA. Here we consider the electrostatic potential of DNA juxtapositions to determine the possibility of juxtapositions occurring through Brownian diffusion. We found that despite the large negative electrostatic potential formed between two juxtaposed DNA helices, a bulk counterion concentration as small as 50 mM provides sufficient electrostatic screening to prohibit significant interaction beyond an interhelical separation of 3 nm in both hooked and free juxtapositions. This suggests that instead of electrostatics, mechanical forces such as those occurring in anaphase, knots, catenanes, or the writhe of supercoiled DNA may be responsible for the formation of DNA juxtapositions.
1. Introduction
Type II topoisomerases untie knots, unlink catenanes, and modulate DNA linking number. These potentially problematic topologies arise naturally as a consequence of DNA metabolism. Requiring ATP and magnesium, type II topoisomerases function by breaking both strands of one DNA double-helix (the gate, or G segment), passing an intact DNA helix (the transfer, or T segment) through the breaks while remaining covalently bound to the 5′ ends of the cleaved helix, and then religating the strands. Failure to remove problematic DNA topologies prohibits cell division and can result in cell death or genetic mutation. Such catastrophic consequences make topoisomerases effective targets for chemotherapeutics. Despite their biological and clinical importance, little is understood about how a type II topoisomerase differentiates DNA topologies in a molecule that is significantly larger than the topoisomerase itself.
1.1. Models of topoisomerase function
It is clear that type II topoisomerases recognize and bind DNA–DNA juxtapositions, or crossovers (Zechiedrich and Osheroff 1990, Howard et al 1991, Roca and Wang 1992, Corbett et al 1992, Timsit et al 1998, Strick et al 2000). How do type II enzymes distinguish problematic from benign DNA topology in binding to a DNA crossover? Type II topoisomerases utilize ATP and reduce concentrations of knots and catenanes by a factor of 16 to 90 below their equilibrium levels (Rybenkov et al 1997). Equilibrium concentrations of knots and catenanes are strongly correlated with DNA length. Rybenkov et al proposed a three-binding-sites model whereby topoisomerase shifts the equilibrium concentrations of knots and catenanes by shortening the effective DNA length. The enzyme binds two T segments and then tracks along the DNA, entrapping the knot or catenane link, to bind the G segment. The magnitude of the observed equilibrium shift, however, cannot be explained by this model, so the authors hypothesized that some additional active mechanism would be required.
Because the probability that type II topoisomerase would bind three strands concurrently is very low, Yan et al (1999, 2001) adapted the so-called kinetic proofreading model (Hopfield 1974, Ninio 1975) to explain type II topoisomerase accuracy. They proposed that a G segment-bound topoisomerase makes two consecutive bindings to a T segment. The first binding activates the topoisomerase, enabling it to perform DNA strand passage the second time it binds to the T segment. The model assumes that the constraints of knotted, catenated, and supercoiled topoisomers promote locality of DNA segments, and, therefore, make DNA segment collision more probable. It also assumes that the dissociation rate of the topoisomerase from the G segment is independent of the DNA topology. Consequently, if the rate of the second T segment collision and strand passage is greater than the dissociation rate, then the steady-state fractions of DNA topoisomers will shift away from equilibrium. However, computer simulations reveal that the shift in the steady-state fraction of knots as a result of this assumption is small compared to the experimentally observed steady state (Vologodskii et al 2001).
Vologodskii et al (2001) suggested that type II topoisomerase bends a segment of DNA into a hairpin with the entrance to the enzyme inside the hairpin. The idea is that the hairpin mechanism is a one-way gate—the T segment enters the open end of the hairpin and exits the closed end. Monte Carlo simulations showed that this mechanism can produce a decrease in steady-state levels of knots and catenanes.
What the above models have in common is that they assume type II topoisomerases cannot discern global DNA topology from preexisting local information. However, there are a number of local properties of DNA–DNA juxtapositions that might give the topoisomerase clues about the global topology of the DNA. For instance, whether or not the juxtaposition is left-handed or right-handed yields two possible geometric solutions to how a type II topoisomerase might bind to the juxtaposition (Timsit et al 1998). It is unknown whether or not one of these geometries is energetically favorable. Other clues may include preferred DNA binding or cleavage sequences, or the orientation of the major and minor grooves in the interhelical region.
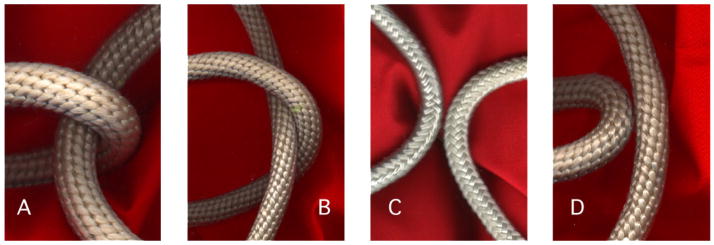
It has been proposed that type II topoisomerases recognize curvature and angle characteristic of juxtapositions found in knotted and catenated DNA (Buck and Zechiedrich 2004). A DNA juxtaposition is either ‘hooked’ or ‘free’ depending upon the curvature of the helices. In a hooked juxtaposition, each of the helices is curved towards the other, or one of the helices encloses the other (figures 1(A) and (B)). Similarly, a juxtaposition of two DNA helices where each of the helices is curved away from the other, or where the inner helix has greater curvature than the outer, is said to be free (figures 1(C) and (D)). Hooked juxtapositions are more likely to be found in knots, catenanes, and supercoils than free juxtapositions. However, type II topoisomerases show a preference for unknotting and unlinking over relaxing supercoils, so it is also necessary to consider the angle of the juxtaposition between the helices. The angle of minimum energy in a hooked juxtaposition is 90°, but writhed helices in supercoiled DNA tend to be juxtaposed at an angle of 60° (Bates and Maxwell 2005). Therefore, the Buck–Zechiedrich (BZ) model proposes that type II topoisomerases reduce concentrations of knotted and catenated DNA topologies by preferentially acting upon hooked juxtapositions at approximately right angles. New data support this model.
Figure 1.
Hooked and free juxtapositions. In hooked juxtapositions, (A) the helices are curved towards each other, or (B) one helix encloses the other. In free juxtapositions, (C) the helices are curved away from each other, or (D) one curvature of the outer helix is less than that of the inner helix.
Corbett et al published a 3.0 Å x-ray crystal structure of a ParC dimer of the Escherichia coli topoisomerase IV enzyme (2005). Although the activity of the dimer used was approximately one tenth that of the holoenzyme, the authors noted that the crystal structure of this dimer was capable of recognizing or promoting hooked juxtapositions of the G and T segments. In addition, Germe and Hyrien (2005) found that nucleosome deposition occurs on DNA plasmid catenanes: an event that would require sharp bending of the DNA in order to form a catenane node while simultaneously interacting with a nucleosome. They speculated that during the S phase, nucleosome deposition may force catenane nodes into hooked juxtapositions, promoting unlinking by type II topoisomerases.
1.2. Drivers of DNA–DNA interactions
The juxtaposition of DNA segments that compose the substrate of type II topoisomerases must form and persist on timescales that permit binding with the topoisomerase. Possible drivers of this persistence include mechanical forces such as those occurring in anaphase, and the stresses occurring in knots, catenanes, or the writhe of supercoiled DNA. In any case, the system would have to recognize two helices in contact and then pass one double strand through the other. Thus, in addition to the ATP-driven work done by a type II topoisomerase, bringing the two helices into contact also contributes to the equilibrium free energy.
One possible driving force for the formation and/or persistence of hooked or free juxtapositions is the electrostatic potential energy of DNA–DNA interactions. Hooked and free juxtapositions could have different free energies, which may affect the persistence of the juxtaposition. Thus a type II topoisomerase might be more likely to encounter the persistent juxtaposition and act.
Although DNA is a very negatively charged molecule, two interacting DNA segments are not always repelled (Gronbech-Jensen et al 1997). DNA condensation, a phenomenon that allows DNA to be packaged into very small volumes, enables segments of DNA to be separated by only one or two layers of saline solution. DNA condensation is promoted by the presence of low concentrations of condensing agents, such as multivalent cations, that screen the repulsive forces between DNA segments or decrease the favourability of DNA–solvent interactions. DNA condensation must, therefore, be a cooperative process because the free energy of condensation is just 10−2–10−1 kBT per base pair (Bloomfield 1997). An electrostatic analysis of a simple approximation of hooked juxtapositions showed that the interaction energy in vacuo was minimized when the angle of juxtaposition was 90°(Buck and Zechiedrich 2004). This tantalizing finding also showed that for some hooked geometries, two strands of like charge could actually be attracting at some (undetermined) close distance. Thus, it was possible that, given the close quarters in which DNA exists in the cell, hooked juxtapositions could become locally attracting. Clearly, before we can understand how DNA helices juxtapose and, thus, how topoisomerases act, we must better understand the role of electrostatics and electrostatic screening in DNA juxtapositions. Here we have performed a computational study of the electrostatics of hooked and free juxtapositions in vacuum and in electrolyte solution in order to determine the possibility of juxtapositions forming and persisting through Brownian diffusion.
2. Methods
2.1. DNA models
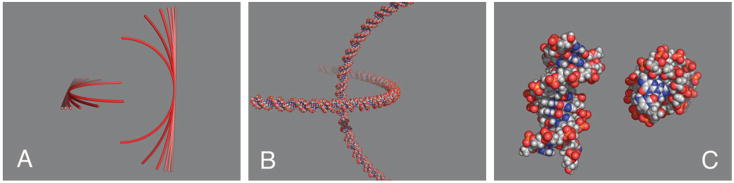
We used three different levels of models of DNA in the calculation of duplex–duplex interactions. We started with a simple charged-line approximation (figure 2(A)) for each duplex consisting of 368 negative point charges to model a 184 base pair (bp) segment (Manning 1969). To observe the effect of curvature on the interactions, the point charges were arranged on an arc in the x y-plane with a radius of curvature that varied (20, 40, 80, 160, 320 nm, and ∞). Given that the persistence length of relaxed DNA is approximately 150 bp in high concentrations of NaCl (Shore et al 1981), a 20 nm radius of curvature represents a case of relatively severe bending (coincidentally, nearer to the curvature reported by Cloutier and Widom (2004, 2005)). The average spacing between point charges, 0.17 nm, matched the 0.34 nm spacing between pairs of charged phosphate groups in the B form of DNA.
Figure 2.
Models of DNA juxtapositions. (A) 184 bp charged-line models with various curvatures, (B) 184 bp all-atom model, and (C) 12 bp all-atom model.
The results of the charged-line approximation were compared with two atomic models of DNA. In one atomic model, a 184 bp B-DNA double helix with random nucleotide sequence (figure 2(B)) was generated by the software package NAB (Macke and Case 1998). The helix was positioned on a 20 nm radius semicircle corresponding to the most pronounced curvature in the charged-line approximations. The partial charges and atomic radii were assigned to each atom according to the CHARMM27 nucleic acids force field (Foloppe and MacKerell 2000). This model is appropriate for implicit solvent models. For higher level calculations a 184 bp model is computationally prohibitive, so a 12 bp atomic model of B-DNA (figure 2(C)) with sequence d(CGTGTCCCTCTC) was used for molecular dynamics simulations and for graphical analysis of the counterion concentrations predicted by the Poisson–Boltzmann equation (Wong and Pettitt 2001).
In each of the models, a hooked juxtaposition of two identical helices was formed by rotating one helix 90° around the x-axis and 180° around the y-axis resulting in a 40 nm interaxial separation between the mid-points of the two helices. For all calculations, the interaxial separation is recorded as the distance along the x-axis between the helical axes of each molecule. The minimum atomic distance is the shortest distance between two atoms on different helices. The rigid translation along the x-axis of one helix through the other yields a free juxtaposition where the two helices are curved away from each other. The two helices are said to intersect when their interaxial separation is 0 nm. Note that infinite energies at the intersection were avoided by using an even number of charges so that no charge was placed directly on the x-axis.
2.2. Coulomb and Debye–Hückel energy calculations
We compare the bare Coulomb potential with increasingly sophisticated approximations to the screening in solution. The electrostatic potential of DNA juxtapositions (φ) was calculated in mean-field solvent and in vacuum using software written by GLR. Coulomb’s law
was used for in vacuo calculations, where n is the number of charges per strand, ri j is the distance between charges qi and qj in the individual strands, and ε is the dielectric constant of the solvent (ε = 1 in a vacuum). The Debye–Hückel (DH) equation
was next used to account for the screening effects of solvent and counterions present (Tombolato and Ferrarini 2005). A solvent dielectric (ε) of 78.54 and a Debye screening length (λD) corresponding to a 150 mM concentration of 1:1 electrolyte solution were assumed. The internal (self) energies of each helix were ignored. Interaction energies at interaxial separations from −40 nm to +40 nm were calculated by rigidly translating one of the helices along the x-axis towards, through, and away from the other helix.
2.3. Poisson–Boltzmann solutions
The next level of approximation takes into account the finite extent of the duplex excluded volume. The adaptive Poisson–Boltzmann Solver (APBS) software package (Baker et al 2001) was used to solve the nonlinear Poisson–Boltzmann (PB) equation
for atomic models of DNA juxtapositions where the electric field given by the term on the left-hand side is the superposition of the solute’s charge density ρ (r) and the Boltzmann-weighted ionic strength of the electrolytes, and the dielectric ε is now a function of the position vector r. The electrostatic interaction energy εI was calculated as the difference between the total electrostatic energy of the juxtaposition εJ and the internal electrostatic energies of the individual helices ε1, ε2, i.e., εI = εJ − ε1 − ε2.
Solutions to the PB equation for the 184 bp juxtapositions were completed under similar conditions as those used in the DH level calculations, namely the solvent dielectric constant was 78.54, the ionic strength was 150 mM, and the temperature was 300 K. The PB equation was solved at interhelical spacings of 2.0, 3.0, 4.0, and 5.0 nm, and with different duplex dielectrics ranging from that of a vacuum (1.0) to that of water (78.54) on a 513 × 513 × 481 grid (0.8 Å spacing) distributed over 64 processors with three focusing operations. Results (not shown) exhibited little dependence on the grid spacing or the processor overlap factor. Charges were discretized on the grid by a cubic B-spline. The electrostatic potential at the grid boundary was determined by the DH equation. The accessible surface of the helices determined by a solvent probe radius of 1.4 Å was smoothed by harmonic averaging to reduce grid effects.
The size of the grid required to solve the PB equation for 184 bp juxtapositions makes visual comparisons cumbersome. For this reason, the PB equation was also solved for the 12 bp models of juxtapositions using a much smaller 161 × 129 × 129 (0.5 Å spacing) grid with two focusing operations. A range of counterion concentrations (0.1, 1, 10, 25, 50, 100, 150, 200, 300 mM, …, 1000 mM) was examined. The PB equation was also solved for 12 bp juxtapositions using various internal dielectrics in order to observe any differences in interaction with the larger juxtapositions. Counterion concentrations were visualized with the software program PyMOL (DeLano 2002).
2.4. Molecular dynamics
A system of two identical 12 bp B-DNA helices juxtaposed in explicit solvent was constructed using a solvated DNA helix from a previous 10 ns simulation (Wong and Pettitt 2001) in order to reduce the amount of minimization time required to equilibrate the system. A recent study (Ponomarev et al 2004) has shown that counterion convergence in molecular dynamics simulations of DNA may require more than the typical 5–10 ns of equilibration. So to construct the system, a 50 × 30 × 30 Å3 box containing the DNA strand and two solvation shells of TIP3P (Jorgensen et al 1983, Foloppe and MacKerell 2000), water molecules and NaCl ions (1 343 TIP3P, 30 Na+, 15 Cl−) was excised from the previous simulation volume. The resulting box was duplicated and the two boxes were oriented at right angles. The minimum atomic distance between the helices was 0.98 nm. The remaining space of the new simulation volume (73 Å × 64 Å × 64 Å) was filled with randomly positioned TIP3P water molecules, sodium ions, and chlorine ions. The final system contained 30 055 atoms consisting of 9398 water molecules, 194 Na+ ions, and 150 Cl− ions ([NaCl] = 1.2 M). The 44 excess positive charges balance the negative charges on the DNA helices resulting in zero net charge on the system. We should note that although a bulk concentration of 150 mM NaCl is much more biologically relevant than the 1.2 M concentration we used in our simulation, it is difficult to accurately sample a system with such a low ionic strength because so few sodium and chlorine atoms would be included in the simulation volume.
The energy of the system was minimized by steepest descent for 200 steps followed by 290 ps of solvent equilibration with periodic velocity reassignment. The entire system was then equilibrated for 650 ps until the pressure and temperature stabilized. During the course of the simulation, the volume was resized to 72.5 Å × 63.5 Å × 63.5 Å to maintain pressure.
Production simulation occurred in the microcanonical ensemble (Allen and Tildesley 1987) at 300 K for 10 ns using ESP, a molecular dynamics software package developed in the Pettitt laboratory (Smith et al 1996). We used the velocity Verlet algorithm (Swope et al 1982) to integrate the equations of motion with a timestep of 2 fs and a short-range (Lennard-Jones) cutoff radius of 2.0 nm. Long-range electrostatic interactions were computed using a linked-list Ewald sum (de Leeuw et al 1980) using a real-space cutoff radius of 2.0 nm and a k-space cutoff radius of 8.7266 nm−1. Bond lengths and angles were constrained by the RATTLE algorithm (Andersen 1983).
3. Results
3.1. Coulombic interactions
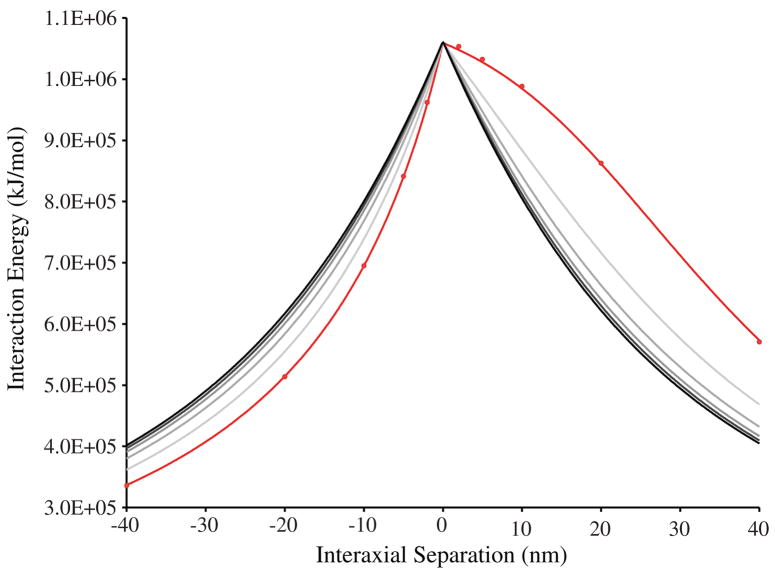
We began by considering the electrostatic interaction energies of two charged-line approximations of DNA in vacuo using Coulomb’s equation. Figure 3 plots the interaction energy as a function of interaxial separation for increasing helix curvatures. Positive values of interaxial separation represent hooked juxtapositions; negative values represent free juxtapositions. An asymmetry in the potential energy curves is immediately apparent. At an initial separation of 40 nm, the potential energy of a hooked juxtaposition of 20 nm radius helices (figure 3, red line) is 5.73 × 105 kJ mol−1 and increases monotonically with decreasing separation. As the helices approach intersection, the rate of the increasing potential energy decreases, eventually reaching a maximum potential energy of 1.06 × 106 kJ mol−1 at the intersection. In contrast, the potential energy of free juxtapositions falls off precipitously from the maximum as the separation increases. The predictions of the charged-line model with a 20 nm radius differ from those of the atomic model (figure 3, red circles) by less than 0.3% indicating good agreement between the high and low resolution models at these interhelical distances. The grey lines in the figure show the effect of curvature on the interaction energy. As the curvature of the two helices decreases, the asymmetry of the potential energy curves becomes less pronounced. At the limit, where the radius of curvature is infinite, the juxtaposition is neither hooked nor free so the interaction energy is symmetric, as expected.
Figure 3.
Interaction energies for DNA juxtapositions as a function of interhelical separation as predicted by Coulomb’s Law. Positive values of interaxial separation represent hooked juxtapositions; negative values represent free juxtapositions. The red line represents the greatest curvature (20 nm radius). Increasing radii of curvature (40, 80, 160, and 320 nm) are represented by progressively darker grey lines. The black line represents the limiting case where the radius of curvature is infinite. Red circles represent the solutions of an atomic DNA model with 20 nm radius.
3.2. Effects of counterion screening
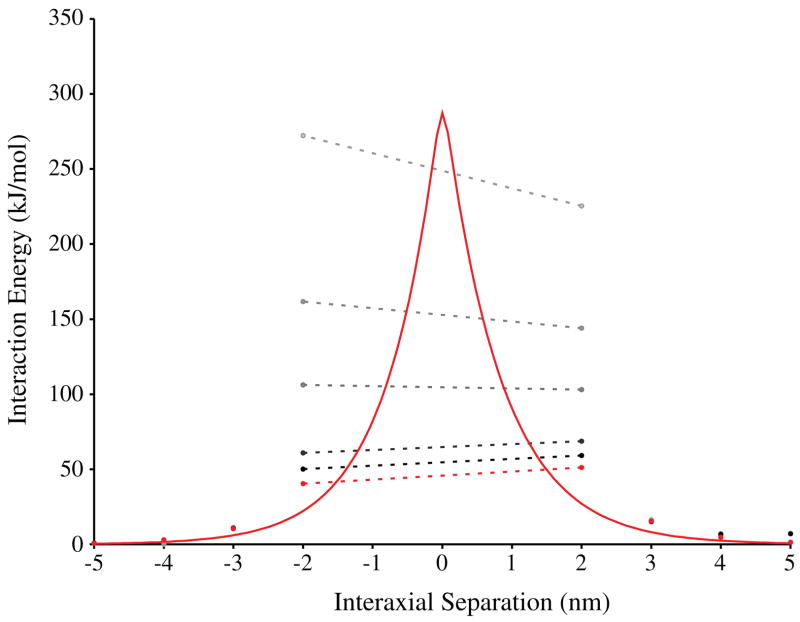
Because the environment of the cell nucleus is hardly a vacuum, much less salt free, we used the DH equation to incorporate roughly the screening effects of electrolytes and the solvent dielectric into our calculations. The results of the calculations on charged-line models of DNA juxtapositions are shown in figure 4 as a red line. At an interaxial separation of 5 nm, the interaction energy increases gradually from 0.7 kJ mol−1 to a maximum repulsion of 287 kJ mol−1 at 0 nm then decays rapidly to 0.4 kJ mol−1 at −5 nm. In contrast to the results of the calculations in figure 3, the potential predicted by the DH equation is imperceptibly asymmetric about the point of intersection, thus any difference in the interactions of hooked and free juxtapositions is nullified by electrolyte screening. The root-mean-square deviation (RMSD) of symmetric points in the charged-line model is only 1.7 kJ mol−1. The potential is largely independent of the radius of curvature exhibiting an RMSD of 0.83 kJ mol−1 between the 20 nm radius and linear configurations (not shown).
Figure 4.
Interaction energies of DNA juxtapositions at 300 K as predicted by the equations of Debye–Hückel and Poisson–Boltzmann. Positive values of interaxial separation represent hooked juxtapositions; negative values represent free juxtapositions. The red line represents the DH result of the charged-line model. Red circles are the DH result for the all atom model. Progressively darker grey circles are solutions of the PB equation for 184 bp helices in increasing internal dielectric constant of DNA (1.0, 2.0, 4.0, 20.0, and 78.54). The dotted lines connecting the circles demonstrate the asymmetry of the PB solutions for various internal dielectric constants.
At interhelical separations less than 3 nm, the DH predictions for the atomic model (red circles) diverge from those of the charged-line approximation, RMSD = 15.8 kJ mol−1. This deviation arises because the screening term, exp(−ri j/λD), in the DH equation makes the finite extent of the charge distribution significant at this resolution. In particular, as the interatomic distances ri j approach 0, the charged-line model lacks sufficient atomic detail and fails to include the excluded volume.
In order to better understand the electrostatic interactions occurring at interaxial separations less than 5 nm, we then calculated the interaction energy for a juxtaposition of two atomic-scale 184 bp helices by solving the PB equation, which accounts for variable charge distributions, dielectrics, and solvent accessibility. These solutions (figure 4, in grey) were strongly influenced by the chosen internal dielectric of DNA. The agreement between the PB and DH solutions improved as the choice of internal dielectric increased. An asymmetry in the PB-predicted interaction energies, depending on the choice of DNA dielectric, emerged as the interaxial separation approached 2 nm. When the internal dielectric constant of DNA is chosen to be less than 4, free juxtapositions appear to be more repulsive than hooked juxtapositions. The relationship reverses as the internal dielectric increases and is equivalent when the internal dielectric is chosen to be between 5 and 6. That the maximum interaction can switch from free juxtapositions to hooked juxtapositions by varying the internal dielectric of the solute is a curious result from a theoretical perspective. However, estimates of the dielectric constant for the interior of DNA are closer to 15 (Yang et al 1995), so we believe that the shifting asymmetry in the potential results from the model. The accuracy of PB solutions when two helices are in such close proximity is constrained by limitations in the PB equation, as will be discussed below.
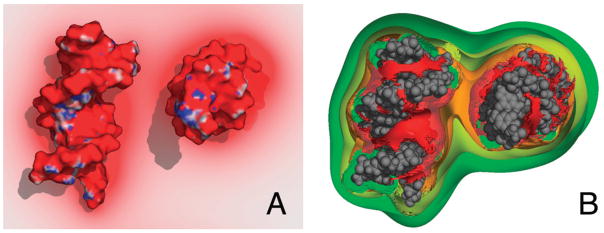
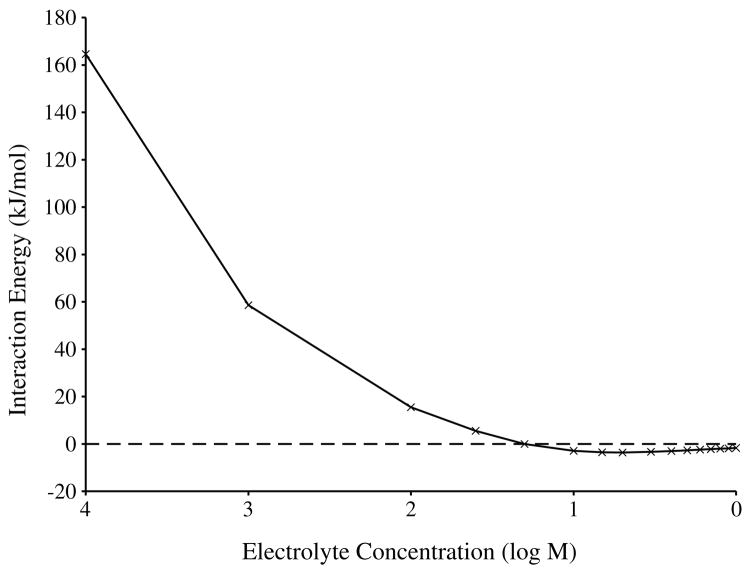
To analyse bulk concentrations of counterions and the screening of electrostatic potential in a DNA juxtaposition, we also solved the PB equation for a system of 12 bp helices at a separation of 3 nm. In figure 5(A), the screening effect of increasing counterion concentration on the interaction potential is qualitatively apparent. We observe a ‘necking’ of the negative electric field (minimum −2 kBT, red) into the interhelical region at low counterion concentrations (1:1, 150 mM), but at higher concentrations electrostatic screening is total, i.e., the electrostatic potential of the interhelical region is 0 kBT. This is reflected in the bulk counterion concentrations surrounding the helices (figure 5(B)). Large concentrations of counterions are recruited to the interhelical region to offset the negative potential. Further calculations (figure 6) showed that the interaction potential decreases rapidly as the counterion concentration increases to 50 mM, and then remains constant at about −3 kJ mol−1 for concentrations up to 1 M. Unlike in the PB calculations for the 184 bp models, the internal dielectric of these shorter helices had a negligible effect in these calculations.
Figure 5.
Poisson–Boltzmann predictions of (A) the electrostatic field from a minimum of −2 kB T (red) to a maximum of 2 kB T (blue), and (B) counterion concentrations (2 M, red; 1 M, orange; 0.5 M, yellow; 0.25 M, green) about a 12 bp DNA juxtaposition.
Figure 6.
Dependence of electrostatic interaction energy on the logarithm of bulk counterion concentration.
3.3. Simulation of juxtaposed helices
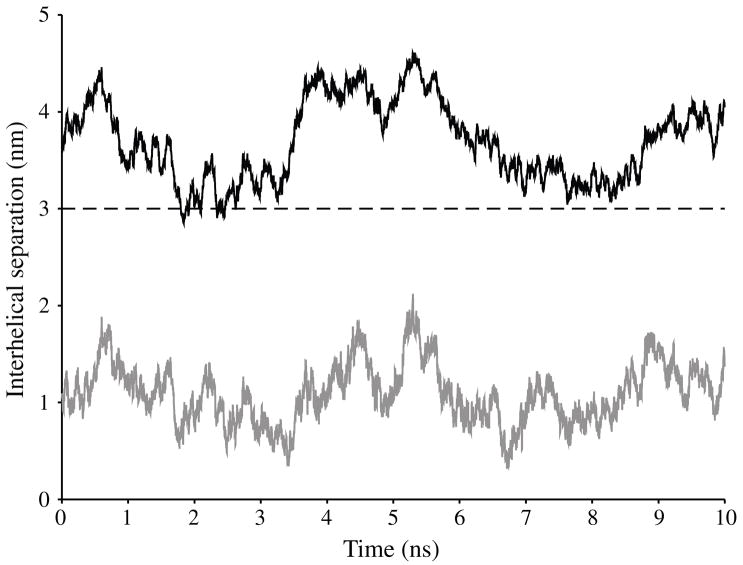
The PB results, along with those from the DH model, show that curvature and longer-range interactions (>3 nm) have minimal effect on the interaction energy between helices. Therefore, the interhelical potential energy is dominated by local interactions—beyond 3 nm the curvature and length of the strand do not have a significant effect on the interactions in saline solution. So simulation of relatively short, 12 bp, helices is reasonable given the practical limitations of computing resources. We performed a 10 ns molecular dynamics simulation of a juxtaposition of 12 bp helices in order to study the interactions of the helices with each other and the solution. During the course of the simulation, we observed no apparent net attraction between these DNA helices (figure 7). The mean interaxial separation during the simulation was 3.7 ± 0.4 nm. A minimum interhelical separation around 3 nm was reached on two separate occasions. The first minimum interhelical separation, 2.85 nm, was reached 1.83 ns into the simulation. At this minimum, the helices were aligned in parallel with phosphate groups on the minor grooves facing each other about 0.59 nm apart. The second minimum interhelical separation, 3.05 nm, occurred 7.63 ns into the simulation when the orientation of the helices was closer to perpendicular.
Figure 7.
Interhelical separations during a 10 ns course of molecular dynamics. The minimum separation between the helical axes of each DNA molecule is in black. The minimum separation between atoms of each DNA molecule is in grey.
4. Discussion
The results in this study greatly expand upon those previously reported (Buck and Zechiedrich 2004). In the previous results, it was found that hooked juxtapositions in a vacuum, without screening, could be locally attracting if two DNA helices were brought sufficiently close together. This attraction results from both the electrostatic approximation and the geometries considered. The previous model approximated the curvature of DNA by an angle between two straight segments of charged lines. Here, we apply Coulomb’s Law to hooked and free juxtapositions modelled by sections of circles and observe an asymmetric electrostatic potential curve. However, we found that this effect is caused by the interactions of distal sections of the DNA segments. These interactions appear to be negated by dielectric and counterion screening, leaving only the effects of local interactions.
Our mean-field solvent studies showed that hooked and free juxtapositions have nearly identical interaction energies in electrolytic solution, and that at separations greater than 3 nm these energies are strongly screened in bulk counterion concentrations greater than or equal to 50 mM. This number is significantly lower than the physiological concentration of salts, even when accounting for the routine 15% underestimation of counterion concentrations by PB (Pack et al 1990). Because of the efficacy of the counterion screening, DNA–DNA interactions are localized and the length of the model DNA molecule becomes irrelevant beyond two helical turns.
Further study of the electric field in the interhelical region for separations less than 3 nm is precluded by inherent limitations in the PB equation. First, the PB equation assumes that ionic species in the solvent are point-charges (zero radius). This hinders the accurate prediction of counterion concentrations in the interhelical region. Second, the PB equation assumes that dielectric screening of the solvent is constant. The dielectric constant is a bulk property of water and is inadequate to describe solvent screening effects in regions, such as in a solvation shell of DNA, where the orientations of the solvent molecules are strongly correlated. It has been previously shown that within 5 Å of a DNA helix, the dielectric constant of DNA in water is just half the bulk value (Young et al 1998). Clearly, these two assumptions will adversely affect the accuracy of PB solutions when only two or three layers of water separate the helices (<3 nm).
The importance of accurately modelling the solvent details in the interhelical region was demonstrated by our molecular dynamics simulation. The PB and DH equations predicted an electrostatic repulsion of 15.4 kJ mol−1 when the helices came within approximately 3 nm of each other. In our simulation, this situation occurred on two separate occasions and in both cases the helices apparently were repelled. The minimum distance between phosphate groups on the first of these encounters was 0.59 nm. This narrow region permitted just two or three layers of water molecules to fit between the helices and nominally screen their interaction. Substantially more energy than 15.4 kJ mol−1 would be required to repel a DNA helix in solution. In this case, we see how significant the repulsive force between the two helices is compared to what is predicted by the PB equation.
The results of our mean-field solvent studies and molecular dynamics simulation emphasize the point that solvent effects are important in determining the strength of molecular interactions when the distances approach that necessary for complexation with topoisomerase. More sophisticated and computationally expensive techniques, such as calculation of the potential of mean force along the interhelical separation coordinate, will be required to better understand the extent of DNA–DNA interactions. This will be the focus of future work.
Electron microscopy and atomic force microscopy reveal that the number of crossovers in supercoiled circular DNA appears to be highly dependent on the bulk concentration of Mg2+ in the solution (Bednar et al 1994, Lyubchenko and Shlyakhtenko 1997, Cherny and Jovin 2001). Given the importance of Mg2+ for type II topoisomerases, it would be beneficial to simulate multiple DNA segments with Mg2+ in order to see the coordination between the DNA, water, and the ions. Unfortunately, simulating multivalent ions is complicated. The large charge density of multivalent ions attracts many counterions from throughout the simulation volume, creating a concentrated solution of ions (Manning 1969). Additionally, these ions perturb the charge distribution and even the bond angle of nearby water molecules, significantly impacting the effective dielectric constant of the solvent (Lamm and Pack 1997). Accounting for these kinds of fluctuations in charge and bond angles is, at this time, computationally prohibitive. Larger multivalent cationic molecules, such as or the polyamines spermidine3+ and spermine4+, might provide an alternative means of simulating DNA condensation because their charge density is appreciatively smaller than divalent metal ions.
5. Conclusion
We have found that dielectric and counterion screening negate global effects on the electrostatic potential of DNA juxtapositions. The random association of type II topoisomerase and two DNA helices through Brownian diffusion is, therefore, not likely to be encouraged by electrostatically persistent hooked juxtapositions. We must conclude that, instead of electrostatics, mechanical forces occurring in anaphase, knots, catenanes, the writhe of supercoiled DNA, or forces generated by polymerases may be responsible for the formation of hooked DNA juxtapositions.
Acknowledgments
GLR is supported by a training fellowship from the Keck Center for Interdisciplinary Bioscience Training of the Gulf Coast Consortia (NLM Grant No 5T15LM07093). The Robert A Welch Foundation, and the National Institutes of Health (NIH) support BMP. The National Science Foundation (NSF) DMS0107747 supports GRB. Support for ELZ is provided by NSF (MCB-0090880), the Burroughs Wellcome Fund (BWF), and NIH (RO1 AI054830). We are indebted to the BWF Interfaces Program and the Program in Mathematics and Molecular Biology. This research was performed in part using the Molecular Science Computing Facility in the William R Wiley Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the US Department of Energy’s Office of Biological and Environmental Research and located at the Pacific Northwest National Laboratory. Pacific Northwest is operated for the Department of Energy by Battelle.
References
- Allen M, Tildesley D. Computer Simulation of Liquids. New York: Oxford University; 1987. [Google Scholar]
- Andersen H. J Comput Phys. 1983;52:24–34. [Google Scholar]
- Baker N, Sept D, Joseph S, Holst M, McCammon J. Proc Natl Acad Sci USA. 2001;98:10037–41. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates AD, Maxwell A. DNA Topology. Oxford: Oxford University Press; 2005. [Google Scholar]
- Bednar J, Furrer P, Stasiak A, Dubochet J, Egelman E, Bates A. J Mol Biol. 1994;235:825–47. doi: 10.1006/jmbi.1994.1042. [DOI] [PubMed] [Google Scholar]
- Bloomfield V. Biopolymers. 1997;44:269–82. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Buck G, Zechiedrich E. J Mol Biol. 2004;340:933–9. doi: 10.1016/j.jmb.2004.05.034. [DOI] [PubMed] [Google Scholar]
- Cherny D, Jovin T. J Mol Biol. 2001;313:295–307. doi: 10.1006/jmbi.2001.5031. [DOI] [PubMed] [Google Scholar]
- Cloutier T, Widom J. Mol Cell. 2004;14:355–62. doi: 10.1016/s1097-2765(04)00210-2. [DOI] [PubMed] [Google Scholar]
- Cloutier T, Widom J. Proc Natl Acad Sci USA. 2005;102:3645–50. doi: 10.1073/pnas.0409059102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett A, Zechiedrich E, Osheroff N. J Biol Chem. 1992;267:683–6. [PubMed] [Google Scholar]
- Corbett K, Schoeffler A, Thomsen N, Berger J. J Mol Biol. 2005;351:545–61. doi: 10.1016/j.jmb.2005.06.029. [DOI] [PubMed] [Google Scholar]
- DeLano W. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- de Leeuw S, Perram J, Smith E. Proc R Soc. 1980;A 373:27–56. [Google Scholar]
- Foloppe N, MacKerell A. J Comput Chem. 2000;21:86–104. [Google Scholar]
- Germe T, Hyrien O. EMBO Rep. 2005;6:729–35. doi: 10.1038/sj.embor.7400465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronbech-Jensen N, Mashl R, Bruinsma R, Gelbart W. Phys Rev Lett. 1997;78:2477–80. [Google Scholar]
- Hopfield J. Proc Natl Acad Sci USA. 1974;71:4135–9. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard M, Lee M, Hsieh T, Griffith J. J Mol Biol. 1991;217:53–62. doi: 10.1016/0022-2836(91)90610-i. [DOI] [PubMed] [Google Scholar]
- Jorgensen W, Chandrasekhar J, Madura J. J Chem Phys. 1983;79:926–35. [Google Scholar]
- Lamm G, Pack G. J Phys Chem. 1997;B 101:959–65. [Google Scholar]
- Lyubchenko Y, Shlyakhtenko L. Proc Natl Acad Sci USA. 1997;94:496–501. doi: 10.1073/pnas.94.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macke T, Case D. In: Molecular Modeling of Nucleic Acids. Leontis N, SantaLucia J, editors. Washington, DC: American Chemical Society; 1998. pp. 379–93. [Google Scholar]
- Manning G. J Chem Phys. 1969;51:924–33. [Google Scholar]
- Ninio J. Biochimie. 1975;57:587–95. doi: 10.1016/s0300-9084(75)80139-8. [DOI] [PubMed] [Google Scholar]
- Pack G, Lamm G, Wong L, Clifton D. In: Theoretical Biochemistry and Molecular Biophysics. Beveridge D, Lavery R, editors. New York: Adenine Press; 1990. pp. 237–46. [Google Scholar]
- Ponomarev S, Thayer K, Beveridge D. Proc Natl Acad Sci USA. 2004;101:14771–5. doi: 10.1073/pnas.0406435101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca J, Wang J. Cell. 1992;71:833–40. doi: 10.1016/0092-8674(92)90558-t. [DOI] [PubMed] [Google Scholar]
- Rybenkov V, Ullsperger C, Vologodskii A, Cozzarelli N. Science. 1997;277:690–3. doi: 10.1126/science.277.5326.690. [DOI] [PubMed] [Google Scholar]
- Shore D, Langowski J, Baldwin R. Proc Natl Acad Sci USA. 1981;78:4833–7. doi: 10.1073/pnas.78.8.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P, Holder M, Dang L, Feig M, Pettitt B. ESP. Houston, TX: University of Houston; 1996. [Google Scholar]
- Strick T, Croquette V, Bensimon D. Nature. 2000;404:901–4. doi: 10.1038/35009144. [DOI] [PubMed] [Google Scholar]
- Swope W, Andersen H, Berens P, Wilson K. J Chem Phys. 1982;76:637–49. [Google Scholar]
- Timsit Y, Duplantier B, Jannink G, Sikorav J. J Mol Biol. 1998;284:1289–99. doi: 10.1006/jmbi.1998.2281. [DOI] [PubMed] [Google Scholar]
- Tombolato F, Ferrarini A. J Chem Phys. 2005;122:54908. doi: 10.1063/1.1839859. [DOI] [PubMed] [Google Scholar]
- Vologodskii A, Zhang W, Rybenkov V, Podtelezhnikov A, Subramanian D, Griffith J, Cozzarelli N. Proc Natl Acad Sci USA. 2001;98:3045–9. doi: 10.1073/pnas.061029098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K, Pettitt B. Theor Chem Acc. 2001;106:233–5. [Google Scholar]
- Yan J, Magnasco M, Marko J. Nature. 1999;401:932–5. doi: 10.1038/44872. [DOI] [PubMed] [Google Scholar]
- Yan J, Magnasco M, Marko J. Phys Rev. 2001;E 63:031909. doi: 10.1103/PhysRevE.63.031909. [DOI] [PubMed] [Google Scholar]
- Yang L, Weerasinghe S, Smith P, Pettitt B. Biophys J. 1995;69:1519–27. doi: 10.1016/S0006-3495(95)80022-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M, Jayaram B, Beveridge D. J Phys Chem. 1998;B 102:7666–9. [Google Scholar]
- Zechiedrich E, Osheroff N. EMBO J. 1990;9:4555–62. doi: 10.1002/j.1460-2075.1990.tb07908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]