Abstract
Both protein and solid-state nanopores are under intense investigation for the analysis of nucleic acids. A crucial advantage of protein nanopores is that site-directed mutagenesis permits precise tuning of their properties. Here, by augmenting the internal positive charge within the α-hemolysin pore and varying its distribution, we increase the frequency of translocation of a 92-nt single-stranded DNA through the pore at +120 mV by ≈10-fold over the wild-type protein and dramatically lower the voltage threshold at which translocation occurs, e.g., by 50 mV for 1 event·s−1·μM−1. Further, events in which DNA enters the pore, but is not immediately translocated, are almost eliminated. These experiments provide a basis for improved nucleic acid analysis with protein nanopores, which might be translated to solid-state nanopores by using chemical surface modification.
Keywords: DNA sequencing, electroosmosis, nanopore, protein engineering, single-molecule detection
Pores with diameters of a few to hundreds of nanometers, “nanopores,” are being developed for a wide variety of analytical applications (1–5). Nanopores can be fabricated by using particle beams and etchants to treat various substrates, including silicon nitride (3, 6) and plastics, for instance poly(ethylene terephthalate) (4). Alternatively, protein pores such as α-hemolysin (αHL) can be used (1, 5). In both cases, to be detected, an analyte must travel into the pore by electrodiffusion, but few systematic attempts have been made to optimize this process. In the present work, we show how the manipulation of charge on the internal surface of the αHL pore can be used to improve the rate of capture of an important analyte, DNA.
The αHL protein nanopore is advantageous in sensing applications because it can be engineered with subnanometer precision with reference to the high resolution crystal structure (7). Further, the αHL pore is far more stable than commonly held, operating as normal at temperatures approaching 100 °C (8). In stochastic sensing, a binding site for a family of analytes is formed within the pore by site-directed mutagenesis or targeted chemical modification (1). The concentration of an analyte is estimated from the number of binding events per unit time to a single pore. An analyte is identified through the signature provided by the amplitude and mean duration of the individual binding events. In this way, a great variety of analytes have been examined including: cations, anions, organic molecules, and various polymers (1). For example, all 4 DNA bases can be detected as nucleoside monophosphates by an αHL pore equipped with an aminocyclodextrin adapter (9). In addition, individual covalent chemical reactions occurring within the lumen of the pore can be observed (5), offering a basis for the detection of reactive molecules (10).
Polymers can also be analyzed from the characteristics of transit events through the αHL pore (11–15). Studies of nucleic acids have been especially intensive, following the demonstration of the transit of single strands in 1996 (16). The examination of mean transit times and current amplitudes during transit have allowed the analysis of single-strand nucleic acid length (2), base composition (17), phosphorylation state (18) and secondary structure (19). The formation (20, 21) and dissociation of nucleic acid double strands (22–24) and hairpins (25, 26) have also been examined. In addition, the interactions of individual proteins with nucleic acids have been observed (27–30). Similarly, protein-ligand interactions can be examined indirectly, when the ligand is incorporated into nucleic acid strands (31).
Nucleic acid analysis with nanopores may also permit DNA sequencing (16, 32, 33) at a low cost commensurate with the $1,000 genome (http://grants.nih.gov/grants/guide/rfa-files/RFA-HG-04-003.html) (34, 35). Possible means of nanopore sequencing include the direct reading of bases in single strands as they pass through the pore (16), the reading of strands moved through the pore with the aid of an enzyme (30) and the detection or stripping of oligonucleotide probes on single strands as they are translocated (36, 37). All of these approaches require the efficient capture of DNA strands from solution.
DNA capture by nanopores occurs above a threshold potential (38) and even then it is inefficient. Rough estimates suggest that, at voltages close to the threshold, ≈1 DNA molecule in every 1,000 that collides with the pore is translocated (39). By contrast, at the highest accessible applied potentials (approximately +300 mV), ≈20% of the collisions result in capture (and most likely translocation) (40). In the present work, we lined the lumen of the αHL pore with positively charged groups by using site directed mutagenesis (Fig. 1). Through this manipulation, we both increased the rate of DNA translocation and lowered the voltage threshold for translocation.
Fig. 1.
Sections through the αHL nanopore (PDB: 7AHL). (A) Charge distribution in the WT αHL nanopore. Positively charged amino acids of αHL are colored in blue and negatively charged amino acids in red. (B) Sites in the αHL nanopore modified in this work. Lys-8 is in purple. The constriction formed by the ion pair Glu-111/Lys-147 is colored in green. Met-113, Thr-115, Thr-117, Gly-119, Asn-121, Asn-123 and Thr-125 are in orange.
Results
Electrical Recording of the Interactions of DNA with the WT αHL Nanopore.
Single-channel electrical recordings in planar lipid bilayers were carried out in 1 M KCl, 25 mM Tris·HCl (pH 8.0), containing 100 μM EDTA. Under these conditions, the WT αHL pore had a unitary conductance of 1.04 ± 0.01 nS (+120 mV, n = 23). After the addition of a 92-nucleotide synthetic ssDNA to the cis compartment (0.5 to 2.5 μM), the current through a single pore was interrupted by short-lived blockades (16). At +120 mV, the interactions of the DNA molecules with the WT nanopore produced 5 different current patterns (types A–E) that are combinations of three conductance levels: the open state, a midamplitude state and a low-amplitude state (Fig. 2A) (41). Events shorter than 10 μs were judged to represent transient collisions of DNA with the mouth of the pore and were ignored in our analysis. As substantiated below, in all 5 types of event, DNA is captured by the pore, but only in type A and C events is the DNA translocated through the pore, from the cis to the trans side.
Fig. 2.
Interactions of DNA with the αHL pore at +120 mV. (A) Type A event: A simple low-amplitude event (interpreted as the direct translocation of DNA through the pore). Type B event: A simple midamplitude event (DNA is captured in the vestibule and exits the pore from the side of entry, the cis side). Type C event: A midamplitude signal, followed by a low-amplitude signal within the same event (DNA is captured in the vestibule before it translocates through the β barrel to exit at the trans side). Type D event: A low-amplitude signal, followed by a midamplitude signal within the same event (DNA enters the β barrel directly and then retracts into the vestibule to exit the pore from the cis side). Type E event: A midamplitude signal, followed by a low-amplitude signal, followed by a second midamplitude signal within the same event (DNA explores the vestibule, threads into the β barrel, but retracts back into the vestibule and exits the pore from cis side). (B) Events histogram showing mean residual currents for 2,500 events expressed as fractions of the open pore current through the WT αHL pore. The peaks representing type A and type B events are labeled. The type C, D, and E events, characterized by their mean event currents, appear between the two major peaks. (C) Histogram of the DNA translocation times for the type A events. DNA (0.95 μM) was presented from the cis chamber in 1 M KCl, 100 μM EDTA, 25 mM Tris·HCl (pH 8.0).
In a histogram displaying the mean current associated with each event (Fig. 2B), the current levels are separated into 2 peaks centered at 0.11 times (low amplitude) and 0.58 times (mid amplitude) the open-pore current. The type A events characterized by a fractional current of 0.11 (Fig. 2B) had a most likely duration of 0.14 ± 0.01 ms (n = 7) as determined from a Gaussian fit to a histogram of the event durations (Fig. 2C) (17). The peak in the events histogram at a fractional current of 0.58 comprises type B current blockades, which show a midstate current level only (Fig. 2B). Type C, D, and E events contain both midamplitude and low-amplitude current levels. Type C events corresponded to 10% of the total number of events, type D events 2% of the total and type E events only 0.6%. The type C, D, and E events are scattered between the two main peaks in the histogram, reflecting their wide distribution of mean currents.
DNA Interactions with Engineered Nanopores.
We made a variety of homoheptameric αHL pores in which the charge distribution within the lumen was altered. The pores were homoheptamers, so the mutations appear in all 7 subunits, e.g., M113R has a ring of 7 positively charged side chains at position 113. The mutants were made by using the WT or RL2 genes as templates. The RL2 gene encodes 4 amino acid replacements in the barrel domain (Val-124 → Leu, Gly-130 → Ser, Asn-139 → Gln, Ile-142 → Leu) and one at the cis entrance of the pore (Lys-8 → Ala) (42). The properties of the mutants differed significantly from the WT pore with respect to the open-pore current and the frequency of occurrence of DNA translocation events (Table 1). The distribution of the 5 types of interaction of DNA with the nanopore (Fig. 2A) also depended on the mutant [supporting information (SI) Fig. S1 and Table 2]. For pores other than WT, RL2, A8R-RL2, and A8K-RL2, the numbers of captures without translocation were negligible and therefore the term “translocation” is used henceforth. Nevertheless, in all cases, the number of type A and type C events was used to determine the frequency of DNA translocation.
Table 1.
DNA translocation through the αHL nanopore at +120 mV
| αHL pore | τ, ms | f, s−1·μM−1 | IB | I+120mV, pA |
|---|---|---|---|---|
| WT | 0.141 ± 0.004 (n = 6) | 3.0 ± 0.2 (n = 12) | 0.11 (n = 7) | 125 ± 1 (n = 23) |
| RL2 | 0.147 ± 0.003 (n = 4) | 0.28 ± 0.04 (n = 5) | 0.10 (n = 4) | 124 ± 2 (n = 16) |
| A8R-RL2 | 0.15 ± 0.01 (n = 7) | 7.6 ± 0.7 (n = 5) | 0.10 (n = 7) | 125 ± 3 (n = 10) |
| A8K-RL2 | 0.16 ± 0.03 (n = 3) | 3.0 ± 0.3 (n = 3) | 0.11 (n = 3) | 123 ± 3 (n = 7) |
| M113R-WT | 0.138 ± 0.003 (n = 5) | 23 ± 2 (n = 9) | 0.04 (n = 5) | 140 ± 2 (n = 13) |
| E111N-WT | 0.28 ± 0.04 (n = 10) | 18 ± 1 (n = 9) | 0.11 (n = 10) | 137 ± 3 (n = 15) |
| M113K-RL2 | 0.092 ± 0.002 (n = 3) | 4.0 ± 0.3 (n = 3) | 0.05 (n = 3) | 129 ± 2 (n = 10) |
| M113R-RL2 | 0.141 ± 0.004 (n = 4) | 4.4 ± 0.6 (n = 7) | 0.03 (n = 5) | 131 ± 2 (n = 18) |
| T115R-RL2 | 0.23 ± 0.02 (n = 3) | 2.2 ± 0.1 (n = 7) | 0.02 (n = 3) | 122 ± 2 (n = 16) |
| T117R-RL2 | 0.35 ± 0.04 (n = 8) | 1.6 ± 0.1 (n = 7) | 0.03 (n = 4) | 113 ± 6 (n = 9) |
| G119R-RL2 | 0.292 ± 0.002 (n = 4) | 1.3 ± 0.1 (n = 7) | 0.02 (n = 4) | 111 ± 1 (n = 8) |
| N121R-RL2 | 0.30 ± 0.01 (n = 9) | 0.43 ± 0.03 (n = 10) | 0.03 (n = 6) | 96 ± 2 (n = 13) |
| N123R-RL2 | 0.22 ± 0.02 (n = 5) | 0.43 ± 0.04 (n = 8) | 0.03 (n = 4) | 77 ± 2 (n = 10) |
| M113D-RL2 | Not observed (n = 3) | Not observed (n = 3) | Not observed (n = 3) | 112 ± 3 (n = 3) |
τ is the most probable translocation time; f is the frequency of translocation (type A and C events) normalized to 1 μM DNA; IB is the fractional residual current during a type A event and I+120mV is the open-pore current at +120mV. The errors are expressed as the standard error of the mean and the number of experiments is in parentheses. For IB, the errors were all within ±0.01.
Table 2.
Distribution of the types of DNA event as a percentage of the total
| αHL pore | Type A | Type B | Type C | Type D | Type E |
|---|---|---|---|---|---|
| WT | 67.6 ± 2.9 | 19.6 ± 2.7 | 10.2 ± 1.2 | 2.0 ± 0.5 | 0.6 ± 0.3 |
| M113R-WT | 94.1 ± 1.7 | 1.2 ± 0.3 | 3.3 ± 1.2 | 1.4 ± 0.2 | 0 |
| E111N-WT | 97.5 ± 0.5 | 0.6 ± 0.1 | 1.1 ± 0.4 | 0.8 ± 0.2 | 0 |
| RL2 | 84.0 ± 3.2 | 7.3 ± 2.3 | 4.8 ± 0.7 | 3.5 ± 0.7 | 0.4 ± 0.2 |
| M113R-RL2 | 91.4 ± 1.3 | 1.7 ± 0.5 | 4.1 ± 0.8 | 2.6 ± 0.2 | 0.2 ± 0.1 |
The error is the standard error of the mean for four (WT, E111N-WT and M113R-WT) or five (M113R-RL2 and RL2) experiments.
Effects of Point Mutations on DNA Translocation.
The effects of charged amino acid side chains on the passage of DNA through the αHL pore were examined at +120 mV (Fig. 3). Lys-8 is the charged residue in the closest proximity to entering DNA. The substitution of Lys-8 with Ala (in the mutant RL2) decreased the number of translocation events per unit time by almost an order of magnitude, whereas substitution with Arg (in A8R-RL2) produced a 3-fold increase in the frequency of translocation (Table 1). The additional mutations in RL2 (with respect to WT) did not affect DNA translocation, as confirmed by reintroducing Lys-8 into the RL2 background (A8K-RL2, Table 1 and Fig. S1).
Fig. 3.
The effects of charged side-chains on the frequency of DNA translocation at +120 mV. (A) M113R-WT (0.46 μM DNA, cis). (B) E111N-WT (0.47 μM). (C) WT (0.53 μM). (D) M113R-RL2 (0.74 μM). (E) RL2 (0.60 μM). (F) M113D-RL2 (2.0 μM). M113D-RL2 showed no current blockades at up to +300 mV. Additional conditions are described in the legend to Fig. 2.
Removal of the positive charge from the cis entrance of the pore, combined with the introduction of a positive charge just below the constriction, restored (M113K-RL2) or even slightly enhanced (M113R-RL2) the frequency of translocation (Fig. 3 and Table 1). The effect of charge is cumulative as showed by the 10-fold increase in translocation frequency of M113R-WT compared with WT and the increase of 2 orders of magnitude when M113R-WT is compared with RL2 (Fig. 3 and Table 1). By contrast with the effects of positive charge, the placement of negative charge just below the constriction (M113D-RL2) eliminated DNA capture at all potentials tested (+100 to +300 mV). In accord with these findings, removal of the negative charge at the constriction (E111N-WT) increased the frequency of translocation 6-fold.
Amino acid substitutions within the β barrel of αHL also affected the frequency of DNA translocation. Individual replacements of the amino acids at positions 111, 113, 115, 117, 119, 121 and 123 with Arg (RL2 background) produced αHL pores that showed a progressive decrease in the translocation frequency as the Arg residue became located further from the cis entrance (Fig. 4 and Table 1). By contrast, when DNA was added to the trans side of the protein, the effect was reversed; the translocation of DNA molecules through the pore was enhanced by a ring of arginines close to the trans entrance when compared with one near the constriction site (Fig. S2).
Fig. 4.
Effects of arginine substitution in the β barrel of αHL on the frequency of DNA translocation through the pore at +120 mV. The mutants in the RL2 background are M113R, T113R, T113R, G113R, N113R, and N113R. The line is a exponential fit. Additional conditions are described in the legend to Fig. 2.
Voltage Dependence of DNA Translocation and Reduction of the Voltage Threshold for Translocation.
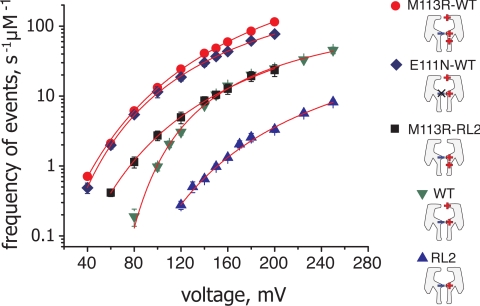
We found that the frequency of DNA translocation f(V) was described well by
where fmax is the rate of DNA translocation at a hypothetical infinite applied potential, V0 is the threshold for DNA translocation, and a larger value of b gives a faster approach to fmax (Fig. 5). More positive charge in the lumen produced pores with a greatly reduced threshold and a steeper approach to fmax (Table S1). Further, pores with no additional charge at the constriction had lower values of fmax (300 ± 60 s−1·μM−1 and 300 ± 150 s−1·μM−1 for WT and RL2, respectively), whereas additional positive charge at both the constriction and the cis entrance gave high values of fmax (7,300 ± 2,200 s−1·μM−1 and 1,600 ± 200 s−1·μM−1 for M113R-WT and E111N-WT, respectively). A pore with a ring of additional charge at the constriction, but no charge at the cis entrance (M113R-RL2), displayed an intermediate value of fmax (990 ± 300 s−1·μM−1).
Fig. 5.
Voltage dependence of the frequency of DNA translocation plotted on a logarithmic scale. Only type A and C events are included in the analysis. Red circles, M113R-WT; blue diamonds, E111N-WT; black squares, M113R-RL2; green inverted triangles, WT; blue triangles, RL2. The lines show fits of the data with Eq. 1.
Discussion
We have enhanced the frequency of translocation of DNA molecules by the αHL pore by using site-directed mutagenesis. The introduction or removal of charged groups within the lumen had a profound effect on DNA translocation. Increasing the positive charge near the internal constriction increased the DNA translocation frequency at +120 mV by almost an order of magnitude (Fig. 3), and greatly decreased the threshold voltage for DNA translocation (Fig. 5). By contrast, eliminating the positive charge at the cis entrance decreased the frequency of DNA translocation by 10-fold. The effect of charge at the constriction is surprising, because the cis entrance of the pore is 5 nm away and under the experimental conditions of 1 M KCl the Debye length is 0.3 nm. Therefore, charged residues at the constriction are not expected to affect the capture of DNA through electrostatic interactions that extend into the bulk solution. Rather some form of “action at a distance” must be proposed, the most likely being rapid, reversible sampling of the pore lumen by DNA from the cis compartment and electroosmosis, both of which are discussed here.
Scheme for the Interaction of DNA with the αHL Pore.
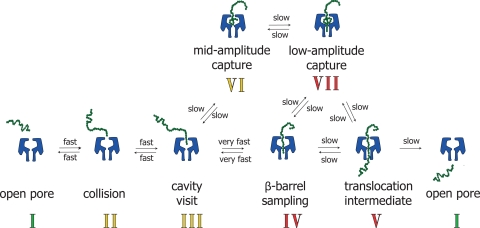
A scheme for DNA capture and translocation that incorporates reversible sampling of the lumen is depicted (Fig. 6). Before a typical translocation event (via intermediate V, Fig. 6), a DNA molecule collides with the pore (II). Upon visiting the vestibule (III), the DNA is transported by electrophoresis into the narrow β barrel (IV). Because the pores used here are anion selective, electroosmotic solvent flow occurs in the same direction (cis to trans) and assists DNA transport (43). At this point, the effect of the applied field on the DNA is strongest (44) and it is pulled through the pore into the trans compartment (V → I), the only irreversible step in the scheme. The translocation intermediate (V) was observed experimentally by the abrupt reduction of the current to 90% of the open pore value (during type A and C events), an assignment that is supported by several observations in the literature (16, 44–46) (SI Text). For example, Kasianowicz and coworkers showed that the cis addition of ssDNA produces short-lived current blockades of 85–100% of the open pore current. PCR analysis of the trans chamber revealed that DNA had passed through the αHL pore (16).
Fig. 6.
Model for the interaction of DNA with the αHL pore. To pass through the pore (V), DNA must first collide with the cis entrance (II), be transported through the vestibule (III) and enter the β barrel (IV). The DNA can prolong its visit within the vestibule and produce midamplitude and low-amplitude events corresponding to VI and VII. All steps are reversible with exception of translocation. In type A events, DNA collides with the pore, enters the vestibule and then directly translocates through the β barrel to exit on the trans side. Sampling of the barrel (III → IV) during this process is too fast to observe. In type B events, DNA is captured by the pore but instead of penetrating the β barrel it has a prolonged interaction with the vestibule to produce the midamplitude event (I → II → III → VI). The DNA exits the pore from the cis entrance from which it entered (VI → III → II → I). Like type B events, type C events first show a midamplitude current (I → II → III → VI), but then one end of the DNA enters the β barrel (VII) and translocation occurs (VII → V → I). In type D events, the DNA enters the β barrel (I → II → III → IV) but then stops translocating (VII), retracts back into the vestibule (VI) and exits the pore from the cis side (VI → III → II → I). Finally, in type E events, the DNA begins to translocate as in type C events (I → II → III → VI → VII → V), but then retracts into the vestibule (V → VII → VI) and exits the pore from the cis side as in type D events (VI → III → II → I). Intermediates expected to give midamplitude events are numbered in yellow, although II and III would be too short for observation. Intermediates expected to give low-amplitude events are numbered in red, although IV would be too short for observation.
In the case of the WT-αHL pore, not all of the current blockades were due to straightforward DNA translocation. In one-third of the events, a midamplitude level was observed (Fig. 2A). This current signature represents DNA molecules captured within the αHL vestibule (VI, Fig. 6), as supported by the fact that occupancy of the vestibule is associated with a lowered conductance, but not one reduced to the extent caused by occupancy of the barrel (44, 45, 47, 48) (SI Text). For example, when DNA duplexes are tethered within the vestibule, a 38% reduction of the pore current is observed (44). Capture within the vestibule is similar to the phenomenon of entropic trapping described by Han and Craighead (49).
DNA molecules captured in the midamplitude configuration (VI, Fig. 6) can either exit the pore from the cis compartment (type B event, Fig. 2A), or proceed into the β barrel producing a low-amplitude signal (intermediate VII). From this configuration, the DNA can either complete translocation through the pore and exit on the trans side (type C event, Fig. 2A, via intermediate V, Fig. 6) or retract back into the vestibule and exit on the cis side (type E event, Fig. 2A). Finally, DNA can visit the vestibule, move at once into the β barrel to produce a low-amplitude signal, but instead of proceeding through the pore (type A event) reverse into the vestibule to produce a midamplitude signal, before exiting on the cis side (type D, Fig. 2A). Butler and colleagues recently observed similar phenomena with 6 homopolymeric deoxyribo- and ribonucleotide 50-mers and presented a detailed analysis of the origin of the current levels (41). In further support of our scheme, we have also observed a few events with multiple mid-to-low (VI → VII) steps (Fig. S3). In addition, we have observed type A–E events with additional DNAs, both with and without weak secondary structure (D. Japrung, unpublished data). The 92-mer used in the present work is unlikely to possess stem-loop structures, but the existence of helical structure within single-stranded nucleic acids remains a contentious area (50).
Effects of Charge in the Lumen of the Pore.
A ring of 7 positive charges introduced by site-directed mutagenesis near the internal constriction increased the frequency of translocation of DNA molecules (Fig. 3), and reduced the fraction of captures with midamplitude components (Fig. 2 A and B). However, according to our proposed scheme, the DNA is within a Debye length of the residues forming the constriction only as intermediate IV, V or VII (Fig. 6) and cannot sense these charged groups from bulk solution. Therefore, to explain the effect of charge at the constriction, we propose that a DNA molecule enters and quickly exits the top end of the β barrel (sampling, III → IV, Fig. 6) many times before either retracting back into the cis compartment or proceeding to intermediate V, VI or VII. An increase in positive charge at the constriction (Glu-111 → Asn or Met-113 → Arg) increases the affinity of DNA for the pore at this location. Therefore, in these mutants the number of molecules that return to the vestibule after sampling the β barrel is reduced and the rate of occurrence of direct DNA translocation events (type A) is increased. In addition, because the fraction of time spent as intermediate III is decreased, the probability of a midamplitude event (VI) is reduced and fewer type B to E events occur. The sampling of the β barrel by ssDNA was observed at +100 mV for short DNA overhangs protruding from a duplex DNA tethered near the cis entrance (44). For untethered DNA molecules, we propose that the sampling of the barrel is much quicker and not recorded because of bandwidth limitations.
The elimination of a charge at the cis entrance of the pore in the RL2 mutant (Lys-8 → Ala) reduced the fraction of type B and C events (Table 2), and decreased the frequency of DNA translocation by an order of magnitude (Fig. 3). We attribute these effects to a reduced affinity of DNA for the pore. A weaker interaction with the pore during collisions with the entrance and visits to the vestibule (intermediates II and III) reduces the overall number of observable captures.
Importantly, we have observed an increased number of capture events with αHL pores with increased internal positive charge with several additional DNAs and RNAs, as described here for the 92-mer. Therefore, the phenomena we describe are not peculiar to a specific DNA sequence.
Role of the β Barrel.
The depth to which DNA samples the β barrel (step IV) was investigated by introducing a ring of arginines at various positions. When DNA was added from the cis side, we found that the DNA translocation frequency decreased approximately exponentially as the ring was moved from the constriction (M113R-RL2) toward the trans entrance (N123R-RL2) of the pore (Fig. 4), suggesting that DNA molecules can sample the β barrel deeply. When presented from the cis side, the DNA must occasionally come close to residue 119, which is 2.3 nm away from the constriction. Accordingly, when the DNA was added on the trans side of the pore, more translocation events were observed (at −120 mV) when the ring of arginines was closer to the trans entrance (Fig. S2). As the ring of charges is moved toward the trans entrance, the unitary conductance of the pore is decreased by approximately 2-fold (Table 1), but this is unlikely to be the sole cause of a change in DNA translocation frequency of more than 10-fold. Further, in other cases, the trend is not seen. For example, RL2 has a similar conductance to the WT pore, but a far lower translocation frequency.
Electroosmotic Effect.
In the preceding discussion, we have ignored the differences in electroosmotic solvent flow through the various αHL pores. For example, the WT αHL pore is slightly anion selective (P+/P− = 0.78, 1 M NaCl cis and 0.2 M NaCl trans) and, therefore, there will be a net flow of water from cis to trans under a positive potential (43). The M113R-WT pore is more anion selective (P+/P− = 0.38, 1 M NaCl cis and 0.2 M NaCl trans) (43) than the WT pore and the increased frequency of DNA translocation with this mutant could be due, at least in part, to enhanced electroosmotic flow. A simple calculation, based on equation 2 in ref. 43, assuming that 5 molecules of water are carried per ion, suggests that with a weakly anion selective pore (P+/P− = 0.9) only two additional molecules of DNA are transported to the mouth of the pore per second in a 1 μM DNA solution. With a moderately selective pore (P+/P− = 0.5), 23 additional molecules of DNA arrive per second and with a more strongly selective pore (P+/P− = 0.1) an additional 53. These numbers are of a similar order of magnitude to the effects seen here.
However, several lines of evidence suggest that electroosmosis does not have a dominant effect on DNA translocation through the αHL pore at +120 mV. For example, E111N-WT shows a similar translocation frequency to M113R-WT, but it is less anion selective (P+/P− = 0.71, 1 M NaCl cis and 0.2 M NaCl trans) (43). The mutant RL2 (P+/P− = 0.68, 0.9 M KCl cis and 0.3 M KCl trans) (R. Madathil, unpublished data) has a similar ion selectivity to WT αHL, but the DNA translocation frequency with RL2 is 10-fold lower (Table 1). RL2 also shows the same DNA translocation frequency as N123R-RL2 (Fig. 5), which has a higher anion selectivity (P+/P− = 0.24, 0.3 M NaCl cis and 1.0 M NaCl trans) (R. Madathil, unpublished data). Finally, if the trend in DNA translocation frequencies observed when a ring of arginines is placed at different positions within the β barrel were due to electroosmosis, we would not expect the trend to reverse when DNA is threaded from the trans rather than the cis side. Previous workers have recognized the possibility that electroosmosis might affect the rate of DNA translocation through nanopores (51, 52), whereas others have ignored the effect (39, 40) or considered it an unlikely contributor under the prevailing conditions (53). It is also possible that electroosmosis has a role in decreasing the type B–E events by promoting laminar flow through the vestibule.
Movileanu and colleagues (13, 15) have explored the passage of positively charged peptides through αHL pores with additional negative charge in the lumen. Both the rate of capture and the translocation time were increased, which they explained in terms of reduced barriers to transit. The differences between nucleic acids, for which the translocation time is not greatly altered (τ, Table 1), and peptides will require further experimentation to resolve, but one obvious distinction is the higher charge density on nucleic acids.
Voltage Dependence of DNA Translocation.
The frequency of DNA translocation events through αHL pores has a strong nonlinear dependence on the applied potential (Fig. 5A). At the low applied potentials of our experiments, the oligonucleotides that visit the vestibule (III, Fig. 6) do not sense a strong potential drop [<10 mV at an applied potential of +100 mV (44)]. Further, the contributions of local potentials from fixed charges in the protein are expected to be small, given the high ionic strength. Therefore, only intermediates IV and V (Fig. 6) in which the DNA is inside the β barrel and subjected to a strong electric field are expected to be significantly influenced by the applied potential. The existence of a voltage threshold for DNA translocation and the steep dependence of the frequency of translocation on potential at <150 mV (Fig. 5) suggests that the energy barrier for DNA translocation is large and only a fraction of the DNA molecules that reach the β barrel (intermediate IV) pass through the entire pore. Increasing the charge near the constriction lowers the overall barrier and more DNA molecules can pass with a lower voltage threshold (Table S1). Other groups working with WT αHL pores have observed the threshold and the steep voltage dependence and fitted their observations to exponential functions (38–40).
At higher potentials (V >150 mV), the voltage dependence of DNA translocation is less steep (Fig. 5). Groups, working with the WT αHL pore, have described a linear dependence on voltage at elevated potentials (39, 40), but in our case the dependence is stronger. Interestingly, the maximum frequency of DNA translocation (fmax) for WT αHL and the mutant pores varies considerably (Fig. 5), the hallmark of electroosmosis (52), which may play a greater role when the applied potential is high.
Conclusions
We have used site-directed mutagenesis to increase the frequency of ssDNA translocation through αHL pores, and at the same time the voltage threshold for DNA translocation has been lowered. The most likely mechanisms for the increased frequency of translocation are rapid sampling of the interior of the pore by the DNA (Fig. 6), increased electroosmotic flow (51, 52) or a combination of the two that depends on the applied potential. The newly engineered pores or derivatives of them will be useful for enhancing the sensitivity of αHL as a biosensor of nucleic acids (2, 17–25). An additional potential application is in nanopore sequencing, where the bases in a single DNA strand are read off one by one during translocation (16, 30, 32–37). In this area, progress has been made on base identification (9, 54) and in controlling the rate at which DNA moves through the pore (29, 30). An increase in the frequency of ssDNA translocation through αHL pores, as demonstrated here, would reduce the dead-time between reads. By analogy, engineered pores might be used to increase the capture efficiency of individual bases during exonuclease sequencing (9), which would be vital for accurate reads. Finally, these results with protein nanopores might be translated to solid-state nanopores by using chemical surface modification (55).
Methods
Full details of the experimental procedures can be found in SI Text.
Protein Preparation.
αHL protein was produced by expression in an E. coli in vitro transcription and translation (IVTT) system and assembled into heptamers on rabbit red blood cell membranes. Mutants αHL genes were prepared by either QuikChange or cassette mutagenesis.
Planar Bilayer Recordings.
Lipid bilayers were formed from 1,2-diphytanoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids). Both compartments of the recording chamber contained 0.4 mL of 1 M KCl, 25 mM Tris·HCl, pH 8, with 100 μM EDTA. The αHL pores and the DNA were added to the cis compartment, which was connected to ground. The 92-nt DNA was 5′-AAAAAAAAAAAAAAAAAAAAATTCCCCCCCCCCCCCCCCCCCCCTTAAAAAAAAAATTCCCCCCCCCCTTAAAA A AAAAATTCCCCCCCCCC-3′.
Supplementary Material
Acknowledgments.
This work was supported by grants from the National Institutes of Health and the Medical Research Council. H.B. is the holder of a Royal Society Wolfson Research Merit Award.
Footnotes
Conflict of interest statement: Hagan Bayley is the Founder, a Director, and a share-holder of Oxford Nanopore Technologies, a company engaged in the development of nanopore sequencing technology. This article was not supported by Oxford Nanopore Technologies.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808296105/DCSupplemental.
References
- 1.Bayley H, Cremer PS. Stochastic sensors inspired by biology. Nature. 2001;413:226–230. doi: 10.1038/35093038. [DOI] [PubMed] [Google Scholar]
- 2.Deamer DW, Branton D. Characterization of nucleic acids by nanopore analysis. AccChemRes. 2002;35:817–825. doi: 10.1021/ar000138m. [DOI] [PubMed] [Google Scholar]
- 3.Dekker C. Solid-state nanopores. Nat Nanotechnol. 2007;2:209–215. doi: 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- 4.Sexton LT, Horne LP, Martin CR. Developing synthetic conical nanopores for biosensing applications. Mol Biosyst. 2007;3:667–685. doi: 10.1039/b708725j. [DOI] [PubMed] [Google Scholar]
- 5.Bayley H, Luchian T, Shin S-H, Steffensen MB. In: Single Molecules and Nanotechnology. Rigler R, Vogel H, editors. Heidelberg: Springer; 2008. pp. 251–277. [Google Scholar]
- 6.Li J, et al. Ion-beam sculpting at nanometre length scales. Nature. 2001;412:166–169. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- 7.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1865. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 8.Kang X, Gu L-Q, Cheley S, Bayley H. Single protein pores containing molecular adapters at high temperatures. Angew Chem Int Ed. 2005;44:1495–1499. doi: 10.1002/anie.200461885. [DOI] [PubMed] [Google Scholar]
- 9.Astier Y, Braha O, Bayley H. Toward single molecule DNA sequencing: Direct identification of ribonucleoside and deoxyribonucleoside 5′-monophosphates by using an engineered protein nanopore equipped with a molecular adapter. J Am Chem Soc. 2006;128:1705–1710. doi: 10.1021/ja057123+. [DOI] [PubMed] [Google Scholar]
- 10.Wu HC, Bayley H. Single-molecule detection of nitrogen mustards by covalent reaction within a protein nanopore. J Am Chem Soc. 2008;130:6813–6819. doi: 10.1021/ja8004607. [DOI] [PubMed] [Google Scholar]
- 11.Movileanu L, Cheley S, Bayley H. Partitioning of individual flexible polymers into a nanoscopic protein pore. Biophys J. 2003;85:897–910. doi: 10.1016/S0006-3495(03)74529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robertson JW, et al. Single-molecule mass spectrometry in solution using a solitary nanopore. Proc Natl Acad Sci USA. 2007;104:8207–8211. doi: 10.1073/pnas.0611085104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolfe AJ, Mohammad MM, Cheley S, Bayley H, Movileanu L. Catalyzing the translocation of polypeptides through attractive interactions. J Am Chem Soc. 2007;129:14034–14041. doi: 10.1021/ja0749340. [DOI] [PubMed] [Google Scholar]
- 14.Mohammad MM, Prakash S, Matouschek A, Movileanu L. Controlling a single protein in a nanopore through electrostatic traps. J Am Chem Soc. 2008;130:4081–4088. doi: 10.1021/ja710787a. [DOI] [PubMed] [Google Scholar]
- 15.Mohammad MM, Movileanu L. Excursion of a single polypeptide into a protein pore: Simple physics, but complicated biology. Eur Biophys J. 2008;37:913–925. doi: 10.1007/s00249-008-0309-9. [DOI] [PubMed] [Google Scholar]
- 16.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci USA. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Rapid nanopore discrimination between single polynucleotide molecules. Proc Natl Acad Sci USA. 2000;97:1079–1084. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, Dunning JE, Huang APH, Nyamwanda JA, Branton D. DNA heterogeneity and phosphorylation unveiled by single-molecule electrophoresis. Proc Natl Acad Sci USA. 2004;101:13472–13477. doi: 10.1073/pnas.0405568101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid and polyuridylic acid as homopolymers or as segments within single RNA molecules. BiophysJ. 1999;77:3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howorka S, Movileanu L, Braha O, Bayley H. Kinetics of duplex formation for individual DNA strands within a single protein nanopore. Proc Natl Acad Sci USA. 2001;98:12996–13001. doi: 10.1073/pnas.231434698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howorka S, Cheley S, Bayley H. Sequence-specific detection of individual DNA strands using engineered nanopores. Nat Biotechnol. 2001;19:636–639. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- 22.Sauer-Budge AF, Nyamwanda JA, Lubensky DK, Branton D. Unzipping kinetics of double-stranded DNA in nanopores. Phys Rev Lett. 2003;90:238101–238104. doi: 10.1103/PhysRevLett.90.238101. [DOI] [PubMed] [Google Scholar]
- 23.Nakane J, Wiggin M, Marziali A. A nanosensor for transmembrane capture and identification of single nucleic acid molecules. Biophys J. 2004;87:615–621. doi: 10.1529/biophysj.104.040212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tropini C, Marziali A. Multi-nanopore force spectroscopy for DNA analysis. Biophys J. 2007;92:1632–1637. doi: 10.1529/biophysj.106.094060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vercoutere W, et al. Rapid discrimination among individual DNA hairpin molecules at single-nucleotide resolution using an ion channel. Nat Biotechnol. 2001;19:248–252. doi: 10.1038/85696. [DOI] [PubMed] [Google Scholar]
- 26.Winters-Hilt S, et al. Highly accurate classification of Watson–Crick basepairs on termini of single DNA molecules. Biophys J. 2003;84:967–976. doi: 10.1016/S0006-3495(03)74913-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Astier Y, Kainov DE, Bayley H, Tuma R, Howorka S. Stochastic detection of motor protein-RNA complexes by single-channel current recording. Chem Phys Chem. 2007;8:2189–2194. doi: 10.1002/cphc.200700179. [DOI] [PubMed] [Google Scholar]
- 28.Hornblower B, et al. Single-molecule analysis of DNA-protein complexes using nanopores. Nat Methods. 2007;4:315–317. doi: 10.1038/nmeth1021. [DOI] [PubMed] [Google Scholar]
- 29.Benner S, et al. Sequence-specific detection of individual DNA polymerase complexes in real time using a nanopore. Nature Nanotechnol. 2007;2:718–724. doi: 10.1038/nnano.2007.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cockroft SL, Chu J, Amorin M, Ghadiri MR. A single-molecule nanopore device detects DNA polymerase activity with single-nucleotide resolution. J Am Chem Soc. 2008;130:818–820. doi: 10.1021/ja077082c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kasianowicz JJ, Henrickson SE, Weetall HH, Robertson B. Simultaneous multianalyte detection with a nanometer-scale pore. Analyt Chem. 2001;73:2268–2272. doi: 10.1021/ac000958c. [DOI] [PubMed] [Google Scholar]
- 32.Deamer DW, Akeson M. Nanopores and nucleic acids: Prospects for ultrarapid sequencing. Trends Biotechnol. 2000;18:147–151. doi: 10.1016/s0167-7799(00)01426-8. [DOI] [PubMed] [Google Scholar]
- 33.Branton D, et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bayley H. Sequencing single molecules of DNA. Curr Opin Chem Biol. 2006;10:628–637. doi: 10.1016/j.cbpa.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 35.Schloss JA. How to get genomes at one ten-thousandth the cost. Nat Biotechnol. 2008;26:1113–1115. doi: 10.1038/nbt1008-1113. [DOI] [PubMed] [Google Scholar]
- 36.Ling XS, Bready B, Pertsinidis A. 2007 US patent application no. 20070190542. [Google Scholar]
- 37.Soni GV, Meller A. Progress toward ultrafast DNA sequencing using solid-state nanopores. Clin Chem. 2007;53:1996–2001. doi: 10.1373/clinchem.2007.091231. [DOI] [PubMed] [Google Scholar]
- 38.Henrickson SE, Misakian M, Robertson B, Kasianowicz JJ. Driven DNA transport into an asymmetric nanometer-scale pore. PhysRevLett. 2000;85:3057–3060. doi: 10.1103/PhysRevLett.85.3057. [DOI] [PubMed] [Google Scholar]
- 39.Meller A. Dynamics of polynucleotide transport through nanometre-scale pores. J Phys Condens Matt. 2003;15:R581–R607. [Google Scholar]
- 40.Nakane J, Akeson M, Marziali A. Evaluation of nanopores as candidates for electronic analyte detection. Electrophoresis. 2002;23:2592–2601. doi: 10.1002/1522-2683(200208)23:16<2592::AID-ELPS2592>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 41.Butler TZ, Gundlach JH, Troll MA. Determination of RNA orientation during translocation through a biological nanopore. Biophys J. 2006;90:190–199. doi: 10.1529/biophysj.105.068957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheley S, Gu L-Q, Bayley H. Stochastic sensing of nanomolar inositol 1,4,5-trisphosphate with an engineered pore. ChemBiol. 2002;9:829–838. doi: 10.1016/s1074-5521(02)00172-2. [DOI] [PubMed] [Google Scholar]
- 43.Gu L-Q, Cheley S, Bayley H. Electroosmotic enhancement of the binding of a neutral molecule to a transmembrane pore. Proc Natl Acad Sci USA. 2003;100:15498–15503. doi: 10.1073/pnas.2531778100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howorka S, Bayley H. Probing distance and electrical potential within a protein pore with tethered DNA. Biophys J. 2002;83:3202–3210. doi: 10.1016/S0006-3495(02)75322-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Movileanu L, Cheley S, Howorka S, Braha O, Bayley H. Location of a constriction in the lumen of a transmembrane pore by targeted covalent attachment of polymer molecules. J GenPhysiol. 2001;117:239–251. doi: 10.1085/jgp.117.3.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sánchez-Quesada J, Saghatelian A, Cheley S, Bayley H, Ghadiri MR. Single molecule DNA rotaxanes of a transmembrane pore protein. Angew Chem Int Ed. 2004;43:3063–3067. doi: 10.1002/anie.200453907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin H, et al. Nanoscale protein pores modified with PAMAM dendrimers. J Am Chem Soc. 2007;129:9640–9649. doi: 10.1021/ja0689029. [DOI] [PubMed] [Google Scholar]
- 48.Jung Y, Cheley S, Braha O, Bayley H. The internal cavity of the staphylococcal α-hemolysin pore accommodates ≈175 exogenous amino acid residues. Biochemistry. 2005;44:8919–8929. doi: 10.1021/bi0473713. [DOI] [PubMed] [Google Scholar]
- 49.Han J, Craighead HG. Characterization and optimization of an entropic trap for DNA separation. Anal Chem. 2002;74:394–401. doi: 10.1021/ac0107002. [DOI] [PubMed] [Google Scholar]
- 50.Seol Y, Skinner GM, Visscher K, Buhot A, Halperin A. Stretching of homopolymeric RNA reveals single-stranded helices and base-stacking. Phys Rev Lett. 2007;98:158103. doi: 10.1103/PhysRevLett.98.158103. [DOI] [PubMed] [Google Scholar]
- 51.Chen P, et al. Atomic layer deposition to fine-tune the surface properties and diameters of fabricated nanopores. Nano Lett. 2004;4:1333–1337. doi: 10.1021/nl0494001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong CT, Muthukumar M. Polymer capture by electro-osmotic flow of oppositely charged nanopores. J Chem Phys. 2007;126:164903. doi: 10.1063/1.2723088. [DOI] [PubMed] [Google Scholar]
- 53.Gershow M, Golovchenko JA. Recapturing and trapping single molecules with a solid-state nanopore. Nat Nanotechnol. 2007;2:775–779. doi: 10.1038/nnano.2007.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ashkenasy N, Sánchez-Quesada J, Bayley H, Ghadiri MR. Recognizing a single base in an individual DNA strand: A step toward nanopore DNA sequencing. Angew Chem Int Ed. 2005;44:1401–1404. doi: 10.1002/anie.200462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wanunu M, Meller A. Chemically modified solid-state nanopores. Nano Lett. 2007;7:1580–1585. doi: 10.1021/nl070462b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.