SUMMARY
The spliceosome is both compositionally and conformationally dynamic. Each transition along the splicing pathway presents an opportunity for progression, pausing or discard, allowing splice site choice to be regulated throughout both the assembly and catalytic phases of the reaction.
INTRODUCTION
A number of excellent reviews have been published over the past decade (including Staley and Guthrie, 1998; Burge et al., 1999; Cramer et al., 2001; Hastings and Krainer, 2001; Brow, 2002; Jurica and Moore, 2003; Nilsen, 2003; Stark and Lührmann, 2006; Valadkhan, 2007; Hertel, 2008; House and Lynch, 2008), addressing numerous aspects of pre-mRNA splicing and its coordination with other nuclear events. This review will focus on selected themes, highlighting the dynamic nature of both the assembly and catalytic phases. In the resulting view of the splicing reaction, unidirectional linear pathways describing progression are often insufficient, if not misleading. We aim to emphasise that multiple transitions in spliceosome assembly and catalysis can be modulated by alterations in the identity or activity of spliceosomal components, or by modulation of the stability of interactions between the pre-mRNA substrate and the spliceosome. For a given intron and set of cellular conditions, one or more of these transitions will limit splicing and thus be available as a potential point of regulation. Changes in the efficiency of any transition in the entire splicing pathway can therefore result in regulated – and thus alternative – splicing.
THE DYNAMICS OF SPLICEOSOME ASSEMBLY
The classical, sequential view of spliceosome assembly (reviewed in Burge et al., 1999) holds that the 5’ splice site (5’SS) is first bound by U1 snRNP, then the branch site (BS) and 3’SS by U2 snRNP and associated protein factors to form a pre-spliceosome, also known as complex A. The [U4/U6.U5] tri-snRNP joins the complex to form complex B, and a series of conformational and compositional changes result, including the loss of U1 and U4 snRNPs to leave U2/5/6. Recruitment of the CDC5L complex (known as the NTC in S. cerevisiae) follows (Makarov et al., 2002), giving rise to an activated spliceosome. Assembly can be stimulated or repressed by the binding of general or specific splicing factors to snRNPs and pre-mRNA; snRNPs can also interact both with pre-mRNA and with each other. Spliceosome assembly is thus highly cooperative, and the fact that many interactions can occur independently of one another in fact results in an assembly cascade that does not follow a single obligatory trajectory, but instead can occur via multiple pathways.
An extreme example of cooperative assembly is the penta-snRNP, which contains all five snRNPs in the absence of pre-mRNA and can be isolated from S. cerevisiae. Although the penta-snRNP alone is not competent to catalyse splicing, addition of nuclease-treated cell extract restores its activity without requiring disassembly and reassembly (Stevens et al., 2002). Together with the observation of splicing in the absence of U1 recruitment in multiple systems (reviewed in Burge et al., 1999), this finding suggests that there is unlikely to be any essential compositionally defined assembly intermediate prior to the activated spliceosome. Rather, a number of potential assembly pathways can lead to this same point. For a given substrate, the stabilities of intermediates along a given pathway might be such that this mode of assembly is observed to the exclusion of all others. In such cases, stable intermediates may be isolated (for example, the purification of complex A by Behzadnia et al., 2006), but this does not mean that the assembly pathway in question is an obligate one for any substrate.
Spliceosome assembly appears to be driven by the stochastic association of snRNPs with pre-mRNA. Data from photobleaching experiments using various GFP-tagged spliceosomal components are consistent with free diffusion of snRNPs within the nucleus and their transient, random association with pre-mRNA (Rino et al., 2007 and references therein). Two major roles of factors stimulating or repressing assembly in such a system would be to increase or decrease the local concentration of snRNPs on or near a transcript, and to modulate the stability of snRNP-pre-mRNA and inter-snRNP interactions. Given that the CTD of RNA polymerase II is one of the multiple regulators that can interact with snRNPs to modulate assembly (reviewed in Cramer et al., 2001), future studies must address how assembly is modulated in response to transcriptional events and chromatin structure. In addition, it is possible that future work will identify more entry points into the spliceosome assembly pathway – for example, “re-initiation” by postcatalytic U2/5/6 complexes.
Another convergence point during spliceosome assembly exists due to the two possible orientations of interaction between the U1 and U2 snRNPs. U1-U2 interaction can occur across the intron to be removed by the spliceosome that will ultimately contain these two snRNPs (intron definition); in multi-intron genes, however, this interaction can also occur across an exon such that U1 bound at the 5’SS of the downstream intron interacts with U2 bound to the BS-3’SS of the upstream intron (exon definition). A reporter gene with long exons is spliced efficiently only if the flanking introns are short (Sterner et al., 1996), and increasing intron length in a model substrate favours exon definition (Fox-Walsh et al., 2005), consistent with the interpretation that simple binding kinetics determine the predominant assembly pathway for a given pre-mRNA substrate. It is not known whether multiple U1 snRNPs can interact with a single U2, or vice versa. If such polymeric interactions are not possible, inter-snRNP interactions promoting exon definition at a given splice site will preclude intron definition. An exon-defined assembly intermediate must therefore make the transition to an intron-defined state in order for functional splicing to proceed. Indeed, splicing can be inhibited by preventing the establishment of intron definition, for example by hnRNP L binding to an exonic splicing silencer and stabilising exon definition interactions between U1 and U2 (House and Lynch, 2006). hnRNP L has a stimulatory effect when bound within an intron, likely due to facilitation of a cross-intron U1–U2 interaction (Hui et al., 2005). Similarly, the binding of SR proteins within an intron can inhibit splicing (Ibrahim el et al., 2005), whereas SR binding within exons is generally stimulatory (reviewed in Hastings and Krainer, 2001). These observations are consistent with the existence of mutually exclusive interactions during exon- and intron-defined states.
Circular exons, the predicted products of splicing from an exon-defined state, have been detected in several systems (see for example Bailleul, 1996). The formation of such products, even at a slow rate, suggests that the maintenance of U1–U2 interaction across an exon (i.e. with the wrong polarity) is not sufficient to prevent the formation of a catalytically competent spliceosome. It is therefore likely that a polarity sensing mechanism normally exists to distinguish between exon- and intron-defined complexes. U1–U2 interactions, when intron-defined, may provide a binding surface for the [U4/U6.U5] tri-snRNP, with exon-defined complexes normally lacking such a surface. The recruitment of non-tri-snRNP proteins is also likely to play a role in this transition, with candidates for proteins involved in polarity sensing expected to bind only to intron-defined complexes. Although no mechanism or factors responsible for such polarity sensing are known, DEK, a chromatin-associated protein not required for early assembly but important for 3’SS definition and splicing catalysis (Soares et al., 2006), could represent one such factor. Proteomic analysis of human spliceosome assembly intermediates suggests that DEK joins the complex during or after the exon definition-intron definition transition (Sharma et al., 2008). In addition, there is no DEK homologue in S. cerevisiae, whose almost exclusively single-intron genes presumably lack an exon-defined stage.
THE ROLE OF RNA STRUCTURE
Consistent with many alternative splicing factors having a role in increasing or decreasing the local concentration of snRNPs on transcripts, a large number of sequence-specific RNA binding proteins have been shown to modulate spliceosome assembly (for example Jensen et al., 2000). Several protein motifs that bind single stranded RNA have been characterised, and these are commonly found in splicing factors. Consistent with their action in a single-stranded state, a set of splicing enhancers and silencers has been confirmed bioinformatically to be more single stranded than bulk sequence, and to function more effectively when placed in the loop than the stem of a hairpin structure (Hiller et al., 2007). The formation and stabilisation of secondary structure around such regulatory elements is therefore a potential mechanism to reduce their effects on splicing.
All evidence suggests that splice sites themselves must be single stranded in order to allow spliceosome assembly, with secondary structure inhibiting U1 and U2 snRNP binding. Inclusion of the 3’SS in a hairpin is inhibitory for splicing, although this can be overcome by the presence of a single stranded ‘helper’ downstream 3’SS, likely recognised during assembly (Liu et al., 1995). A particularly elegant example of alternative splicing regulation by direct modulation of secondary structure around splice sites is the control of alternative splicing by a thiamine pyrophosphate (TPP) riboswitch in Neurospora crassa (Cheah et al., 2007). When TPP concentration is low, the pre-mRNA adopts a structure such that an otherwise favoured downstream 5’SS is occluded, and the branch region is flexible. Splicing proceeds using a suboptimal upstream 5’SS to produce mRNA encoding a functional NMT1 protein. When TPP concentration is high, however, conformational changes in the riboswitch cause structure around the favoured downstream 5’SS to be disrupted, leading to the predominant production of a longer mRNA containing uORFs that prevent NMT1 translation; in addition, the branch region is partially occluded, yielding an overall decrease in splicing efficiency. It is likely that similar examples of splicing regulation, mediated by proteins or small molecules, will be discovered in other systems: how common such mechanisms of splicing regulation will prove to be remains an open question.
Secondary structure is not always inhibitory to splicing; for example, the S. cerevisiae RP51B intron contains complementary sequences close to the 5’SS and BS that bring the ends of the intron together and aid spliceosome assembly (Charpentier and Rosbash, 1996), and it is possible that this is a common way to increase the efficiency of U1–U2 binding and intron definition. The splicing of exon clusters 4 and 6 in the well-characterised Dscam gene in Drosophila provides two further examples of stimulatory secondary structures. Disruption of the iStem – a large hairpin loop downstream of exon 3 – interferes with the splicing of all twelve exon 4 variants (Kreahling and Graveley, 2005), although the mechanism by which this stem stimulates exon 4 splicing remains unclear. The basis of maintenance of mutually exclusive splicing in the exon 6 cluster, however, is better understood. Each exon 6 variant is preceded by a selector sequence complementary to a docking site downstream of exon 5. Interaction between a given selector sequence and the docking site leads to splicing of the following exon and, as the docking site is thus removed from the transcript, the inclusion of further exon 6 variants is suppressed under normal conditions (Graveley, 2005). Knockdown of the hnRNP protein hrp36 leads to the inclusion of multiple exon 6 variants, suggesting that this protein mediates the repression of splicing across the cluster (Olson et al., 2007).
hnRNP proteins normally act as general inhibitors of splicing: they are antagonised by the generally activating SR proteins (reviewed in Hastings and Krainer, 2001). There is increasing evidence that SR proteins exert at least some of their stimulatory effect via the stabilisation of RNA-RNA interactions during both spliceosome assembly and splicing catalysis. A pre-mRNA with a 5’ exon as short as one nucleotide can undergo SR protein-dependent splicing in HeLa extract, suggesting a post-assembly role for these proteins (Hertel and Maniatis, 1999). The arginine-serine rich (RS) domain of a natural SR protein, or an artificial domain comprising multiple RS repeats, when tethered to pre-mRNA, directly contacts the branch site and facilitates prespliceosome formation (Shen et al., 2004); it is thought that the BS is already base paired to U2 snRNA in such assembly intermediates (Xu and Query, 2007). Defects due to SR protein depletion can be suppressed by increasing the strength of the interaction between the 5’SS and U6 snRNA that is required for the first catalytic step (Kim and Abelson, 1996; Shen and Green, 2006). Subsequent work has demonstrated that an RS domain can be crosslinked to the intronic 5’SS region bound to U6 snRNA during the first step of splicing, and subsequently to the exonic 5’SS region bound to U5 snRNA during the second step (Shen and Green, 2007). This argues for the involvement of enhancer-recruited SR proteins not only in assembly but also during the catalytic phase of splicing.
Data concerning the role of SR proteins in alternative splicing have generally been interpreted with assembly in mind: such data might now need to be reconsidered, as the stabilisation of duplexes could produce diverse phenotypes during the dynamic and structurally complex catalytic phase. It is also possible that hnRNP proteins might exert some of their effect via the disruption of RNA-RNA interactions – most simply by sequence sequestration. The mechanistic basis for duplex stabilisation by SR proteins remains unclear, as does the issue of whether this stabilisation is general or protein-duplex specific, with defined SR proteins stabilising only certain duplexes.
THE TRANSITION BETWEEN THE TWO CHEMICAL STEPS
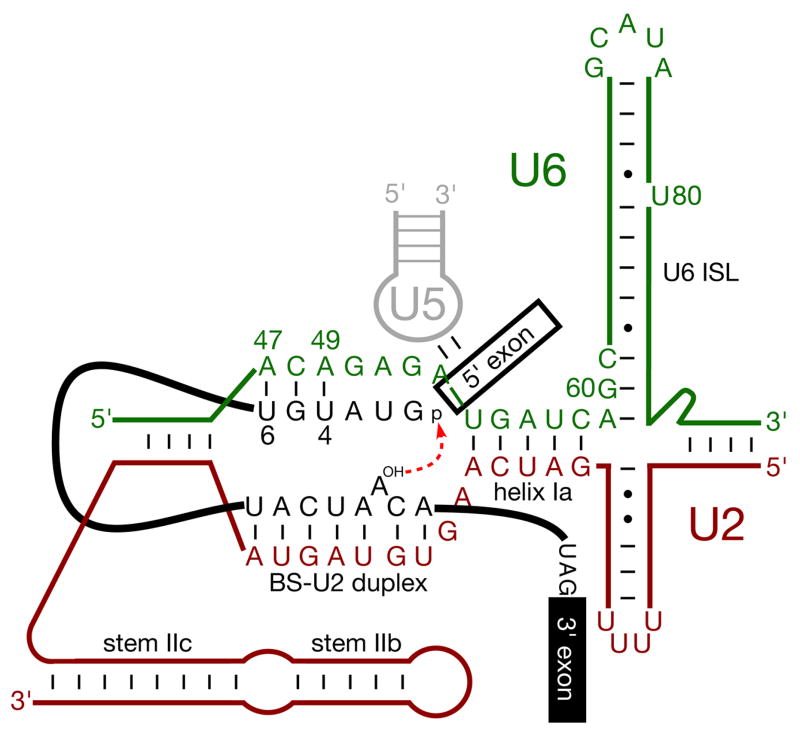
Splicing catalysis consists of two successive transesterification reactions: in the first step, the 2’ hydroxyl of the BS nucleotide nucleophilically attacks the 5’SS to yield a lariat intermediate and a free 5’ exon; in the second step, this free exon nucleophilically attacks the 3’SS, producing mRNA and an excised lariat intron (Fig. 1). The 3’SS remains sensitive to nuclease degradation until after the first step; this suggests that it enters the active site after first step catalysis (Schwer and Guthrie, 1992) – a repositioning event that would require removal of the newly-formed branch structure of the lariat intermediate from the catalytic centre. Indeed, multiple lines of evidence suggest that the 3’SS replaces the branch structure, with the 5’ exon remaining in a fixed position relative to loop 1 of U5 snRNA. Crosslinks between U5 loop 1 and the terminal nucleotide of the 5’ exon can be chased through both steps of splicing: those between loop 1 and position +2 of the intron, however, can be chased into a lariat intermediate but not a lariat product (Sontheimer and Steitz, 1993), suggesting disruption of U5-intron 5’SS interaction following the first step and consistent with genetic and biochemical interactions between loop 1 and the 3’SS during the second step (Crotti et al., 2007 and references therein). Further crosslinking studies show juxtaposition of the 5’SS and U6 snRNA positions U47-A51 during the first step (proposed RNA-RNA interactions in the first step catalytic centre are shown in Fig. 2), and positions G39-A44 when in the lariat intermediate branch structure (Sawa and Abelson, 1992). Genetic evidence also supports the disruption of the 5’SS-U6 pairing required for the first step of splicing: the 5’SS A3C mutation (/GUAUGU to /GUCUGU) hyperstabilises pairing between the 5’SS and U6; this mutation inhibits the second step, but can be suppressed by mutations that destabilise 5’SS-U6 pairing, presumably restoring the duplex to near wild-type stability (Konarska et al., 2006).
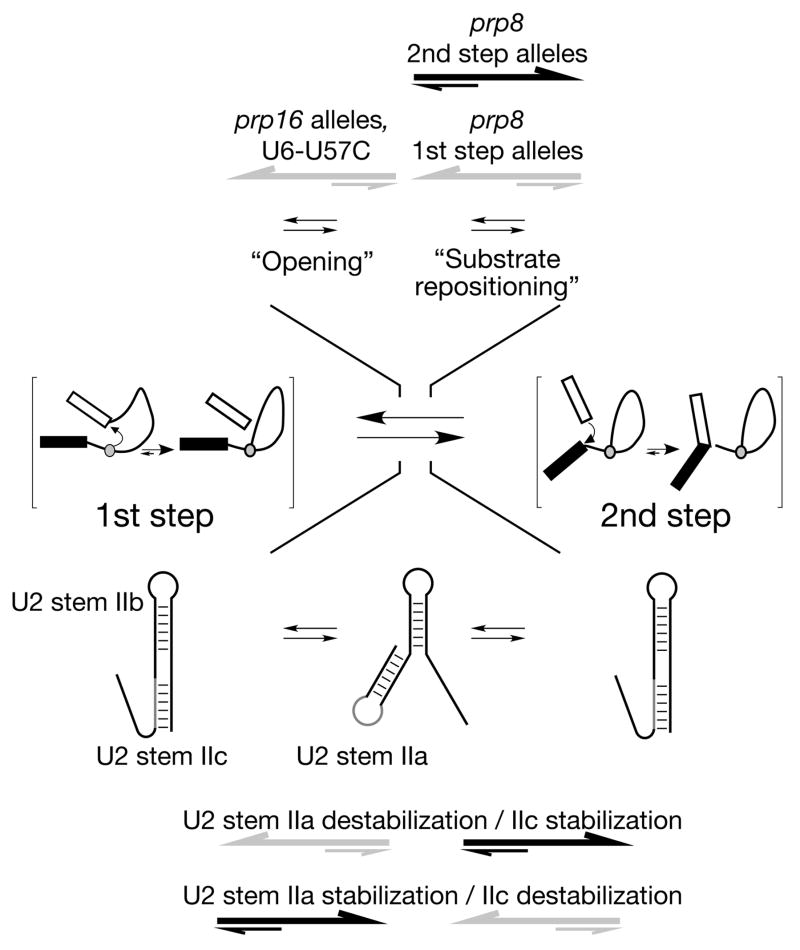
Figure 1. Two-state conformational model of the catalytic spliceosome.
Spliceosomal conformations competent to carry out the first and second steps are in competition with one another. In addition, two opposing classes of prp8 alleles modulate an event in the first-to-second step transition distinct from that modulated by alleles of prp16 and U6 snRNA (upper) (Liu et al., 2007); thus, two distinct events (‘opening’ and ‘substrate repositioning’) can be distinguished. Accompanying conformational changes in U2 snRNA stem II (lower) parallel these two events (Hilliker et al., 2007; Perriman and Ares, 2007).
Figure 2. Schematic of RNA:RNA interactions that contribute to the first step of splicing.
Pre-mRNA is shown in black, U2 snRNA in red, U5 snRNA in grey, and U6 snRNA in green; numbering corresponds to S. cerevisiae snRNAs and indicates U6 nucleotides discussed in this review. Nucleophilic attack of the 5′SS by BS (the first catalytic step of the splicing reaction) is shown.
The splicing defect due to a hyperstabilised 5’SS-U6 helix can be suppressed not only directly by duplex destabilisation, but also by the U6 U57A mutation, a variety of mutations in Prp8 (Konarska et al., 2006) or deletion of the NTC component Isy1 (Villa and Guthrie, 2005). Mutations that suppress the second step defect of an A3C intron also increase the efficiency of the second step for a variety of other intron mutations, including those at the branch site adenosine and 3’SS; commensurate with their stimulation of the second step, these suppressors inhibit the first step of splicing. There also exists the opposite class of spliceosomal mutants: those that increase the efficiency of the first step at the expense of the second (Query and Konarska, 2004). The existence of two opposing classes of suppressor allele, each capable of suppressing a wide range of intron mutations, suggests that suppression is not necessarily via direct contact between mutated bases/amino acids, but that the spliceosomal conformations competent to carry out the first and second steps are in competition with one another. Mutations that stabilise the first step conformation relative to the second will stimulate the first step and inhibit the second, while the opposite will be true for those that cause relative stabilisation of the second step conformation. As a mutation is more likely to disrupt an interaction than to form a new one, it is likely that relative stabilisation takes the form of destabilisation of the competing conformation, such that most first step suppressors would destabilise the second step conformation and vice versa. At present, however, the molecular basis of the action of these general suppressor mutations is unknown. This two-state model provides a mechanism by which modulation of the stabilities of conformational states of the catalytic spliceosome can modulate the efficiency of splicing of suboptimal substrates. Such tolerance of suboptimal splice sites is manifested in metazoa as alternative splicing, meaning that local or global modulation of conformational stability during catalysis could impact the splicing pattern of individual transcripts, or of classes thereof, respectively.
Recent detailed analysis of the interplay between global suppressor mutations has led to a refinement of the two-state model (Fig. 1). First and second step suppressor point mutations combined in the same molecule of Prp8, a large and exceptionally well-conserved U5 snRNP protein that makes contacts with the 5’SS, BS and 3’SS (reviewed in Grainger and Beggs, 2005), cancel one another’s effects and produce a phenotype resembling wild-type Prp8. A different effect is observed, however, when Prp8 second step suppressor mutations are combined with first step suppressors in U6 snRNA or Prp16, the ATPase that modulates the first-to-second step transition (Burgess and Guthrie, 1993); in this instance, the suppressors act in concert such that both the first and second steps are improved. Cancellation by the opposing classes of prp8 allele indicates that these prp8 alleles act at the same kinetic step as one another, whereas the additive nature of the Prp16/U6-Prp8 suppressor pairs requires that they be affecting different kinetic steps in the transition. These observations have led to a model in which the transition between the two steps has multiple phases, requiring an ‘opening’ step (affected by prp16 and U6 mutants) and a repositioning step (affected by prp8 mutants), followed by ‘closure’ into the second step conformation (Liu et al., 2007).
The suggestion that transitions traditionally considered as one step actually comprise multiple phases has important implications for future proteomic, biochemical and structural studies of the spliceosome. A complex containing a lariat intermediate and a free 5’ exon could plausibly be in one of at least four conformations that are currently compositionally and conformationally undefined: post-first step but pre-opening, open but un-repositioned, open and repositioned, or closed pre-second step. In the first or fourth case the purified complex would be competent to carry out first or second step catalysis, respectively, whereas the open complexes are presumably not catalytically competent. Accurately ascertaining the state of purified complexes is therefore essential in order to allow coherent and reliable insights into the mechanism of splicing. As will be discussed below, there is an emerging view that multiple transitions along the splicing pathway resemble one another, so it is possible that the transition between the catalytic steps is not the only one composed of several smaller remodelling events.
CONFORMATIONAL TOGGLING, ASYMMETRY, AND THE RE-USE OF MOTIFS
Alleles of prp22, the ATPase involved in transitions during and after the second step of splicing, produce a cold-sensitive phenotype due to an mRNA release defect; a screen for suppressors of this phenotype identified a prp8 allele (Schneider et al., 2004) subsequently shown also to act as a general suppressor of first step splicing defects (Liu et al., 2007). Indeed, all known first step suppressor alleles of prp8 suppress prp22 defects. This observation is consistent with the hypothesis that these alleles destabilise the second step conformation of the spliceosome, thereby stimulating the surrounding steps on the splicing pathway (first step catalysis and mRNA release), but could also suggest some degree of similarity between these flanking states. Consistent with the existence of such similarity, a growing number of interactions appear to be disrupted and re-form at defined stages of the splicing reaction – i.e. to toggle.
Many RNA-RNA interactions between snRNAs, as well as between snRNAs and pre-mRNA, have been identified, and take the form of generally short intra- and intermolecular helices (reviewed in Brow, 2002). Some of these interactions are mutually exclusive with others, which suggests that they might exist only transiently, or may toggle between competing conformations. As previously noted, during the first catalytic step the UGU trinucleotide at positions 4-6 of the 5’SS base pairs with a conserved ACA in U6 snRNA (positions 47–49) (Kim and Abelson, 1996). During the second step, when the 5’SS is in the branch of the lariat intermediate, it is in proximity to U6 positions 42–44 (Sawa and Abelson, 1992): interestingly, this region of U6 has also been shown to bind the 5’SS in early complexes (Johnson and Abelson, 2001; Chan et al., 2003). Thus, the same binding site may be used for the 5’SS before and after its involvement in first step catalysis. Indeed, spliceosomal states even further apart on the splicing pathway display surprising similarities: the ATPase Brr2 disrupts the interaction between U4 and U6 snRNAs during spliceosome assembly, allowing U6 to interact with U2 and the 5’SS (Raghunathan and Guthrie, 1998). It has recently been demonstrated that Brr2 is activated by the GTP-bound form of the U5 snRNP protein Snu114 and repressed by its GDP-bound form, and that Brr2 activation is required for spliceosome disassembly as well as U4/U6 unwinding (Small et al., 2006). GTP hydrolysis by Snu114 is not required for Brr2 activation, suggesting a mechanism of action resembling that of classical G proteins. Although the GAP and GEF acting on Snu114 have not been identified, it is tempting to speculate that the relevant conformational states of the spliceosome might perform these roles, akin to the action of the signal recognition particle itself as the GEF for SR-β (Helmers et al., 2003).
Recent work provides another example of conformational toggling (Hilliker et al., 2007; Perriman and Ares, 2007). The dynamic stem II region of U2 snRNA can form two mutually exclusive interactions, known as stem IIa and stem IIc. Stabilisation of stem IIa stimulates prespliceosome formation but inhibits the first catalytic step, which can be promoted by stem IIc stabilisation or IIa destabilisation. Disruption of stem IIc suppresses the splicing defect due to prp16 mutation, whereas disruption of IIa (and therefore relative IIc stabilisation) suppresses second step splicing defects. These data are consistent with a model whereby stems IIa and IIc toggle, coexisting with the previously discussed open and closed forms of the spliceosome, respectively, with stem IIc therefore present during catalysis and IIa during repositioning (Fig. 1).
Although more examples of conformational toggling will likely be identified, many spliceosomal interactions are unlikely to reoccur once disrupted – for example the Prp28-mediated replacement of U1 by U6 at the 5’SS (Staley and Guthrie, 1999). In fact, a general theme in intermolecular interactions involving U6 snRNA is that of asymmetry. U6 mutations that disrupt a structure often have a more severe phenotype than corresponding mutations in the interacting partner, and incomplete suppression by compensatory changes is common. For example, mutations on the U6 side of U2/U6 helix Ia are substantially more severe than those on the U2 side (Madhani and Guthrie, 1992); this suggests that helix Ia does not remain intact throughout the entire splicing reaction, and that the interactions of the U6 component when not engaged in this helix are more critical for splicing than those of the U2 component. Similar asymmetry is observed for the conserved AGC triad of U6, which can interact with sequences in U4 snRNA, with U2 snRNA (to form helix Ib), and within U6 to extend the intramolecular stem loop (ISL) (reviewed in Brow, 2002). Most mutations in AGC are viable if accompanied by a compensatory mutation in U2 that restores helix Ib, but some substitutions such as G60Y cannot be suppressed in this manner (Hilliker and Staley, 2004). An identical AGC motif in domain 5 of group II self-splicing introns acts as a metal binding site crucial for catalysis (reviewed in Pyle, 2008) and, as noted by Hilliker and Staley, spliceosomal AGC mutations that cannot be suppressed by U2 are expected to interfere with metal binding.
A second metal bound in domain 5 of group II introns is thought to mediate a docking interaction (reviewed in Pyle, 2008), and again an analogous metal binding motif exists in the ISL of U6. U80 (S. cerevisiae numbering) is bulged from the ISL and binds magnesium (Yean et al., 2000). The formation of this U80 bulge occurs after U4/U6 unpairing (reviewed in Brow, 2002), and suppression data indicate the delicate balance of relative stabilities required to allow both structures to form, and thus permit splicing (McManus et al., 2007). However, the importance of specifying not only the nature but also the precise timing of interactions within the spliceosome is illustrated by the complex behaviour of this nucleotide. When substituted by 4-thio-uridine, U80 forms a site-specific crosslink with a nucleotide well upstream of the branch site of actin pre-mRNA (Ryan et al., 2004); in addition, an Fe-BABE group tethered at the +10 position of the 5’SS stimulates cleavage at the human equivalent of U80 (U74) (Rhode et al., 2006). Although it is possible that these biochemical data correspond to off-pathway intermediates, it is also plausible that they indicate that at least some of the groups responsible for catalysis are sequestered by other interactions until the immediate pre-catalytic state of the spliceosome.
Spliceosome conformations can also be affected by transient protein modifications. For example, in addition to the known effects of the phosphorylation state of SR and other proteins on spliceosome assembly, Snu114 and the U2 snRNP protein SF3b155 appear to be dephosphorylated for the second step of splicing (Shi et al., 2006 and references therein). Similar effects are likely to be uncovered for many splicing factors, and indeed Prp8 ubiquitinylation has recently been shown to affect spliceosome assembly (Bellare et al. 2008)
ATPASES AND FIDELITY
Although the short duplexes involved in spliceosome assembly and catalysis may be able to unwind naturally to facilitate conformational changes, perhaps in concert with other remodelling events, DExD/H ATPases represent a major class of spliceosomal proteins (reviewed in Cordin et al., 2006). Many DExD/H ATPases have been shown to have RNA unwindase activity correlating with ATPase activity in vitro. Cyt19, a DExD/H protein that promotes group I intron splicing in vivo and in vitro, does so by acting as a non-specific chaperone, resolving kinetic traps along the folding pathway (Mohr et al., 2006); similarly, splicing of the ai5γ group II intron in S. cerevisiae is stimulated by Mss116, and splicing activity in vitro correlates with Mss116 ATPase and unwinding activity (Del Campo et al., 2007). However, the same study reported a residual, ATPase-independent unwinding activity for Mss116, and recent data suggest that Dbp5, which is involved in mRNA export, functions to remodel RNPs only in its ADP-bound form, with ATP hydrolysis thus acting as a conformational switch rather than a power stroke (Tran et al., 2007). It therefore remains unclear whether ATPase/unwindase activity is the mechanistic basis of all, or indeed any, spliceosomal activity of DExD/H ATPases.
Most spliceosomal ATPases are currently thought to facilitate a single transition along the splicing pathway, although the example of Brr2 demonstrates the possibility of a single ATPase acting multiple times (Fig. 3). The question of how ATPase activity is limited to the correct stage(s) remains an open one. Binding of one ATPase to the spliceosome is not necessarily mutually exclusive with the presence of others, as illustrated by the persistent presence of Prp43 from early complexes until disassembly (reviewed in Jurica and Moore, 2003) during which time many other ATPases act. It is, however, possible that the binding sites for some ATPases share common elements, such that mutually exclusive subsets exist. A requirement for cofactors to stimulate ATPase activity is one mechanism by which activity could be temporally regulated: the helicase activity of Prp43 is stimulated by Ntr1, and this stimulation is required for Prp43’s role in spliceosome disassembly (Tanaka et al., 2007). The recruitment of a cofactor or, in the event of an ATPase interacting with multiple spliceosomal components, the conformation of the spliceosome itself, could therefore activate an individual ATPase among several simultaneously bound, and repress others such that inappropriate conformational changes are not induced.
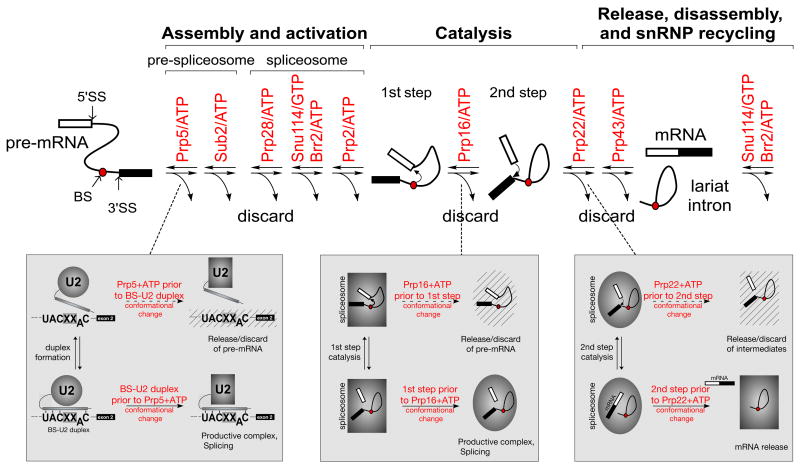
Figure 3. NTPase-associated steps during splicing offer opportunities for kinetic discrimination of suboptimal pre-mRNA substrates.
(upper) Schematic of transitions facilitated by DExD/H-box ATPases and the Snu114 GTPase during pre-mRNA splicing. SS, splice site; BS, branch site. (lower) Characterised examples of kinetic proofreading mediated by spliceosomal ATPases: (left) Altered competition between BS-U2 pairing and the conformational change mediated by the Prp5 ATPase changes the fidelity of BS selection. (center) Altered competition between the first catalytic step and Prp16 ATPase activity affects the fidelity of splice site usage in this step. (right) Altered competition between the second catalytic step and Prp22 ATPase affects second step splice site fidelity.
By stimulating conformational transitions within the spliceosome, DExD/H ATPases play an integral role in the maintenance of splicing fidelity. Prp16, which as previously noted facilitates opening following the first step, was isolated as a suppressor of a branch site mutation in S. cerevisiae, and the relaxation of the requirement for adenosine at the branch site was subsequently shown to be due to ATPase impairment, resulting in the kinetic proofreading model of splicing fidelity (Burgess and Guthrie, 1993). According to this model, functional progression into the second step occurs if catalysis precedes Prp16 ATP hydrolysis, whereas substrate discard results if catalysis has not occurred before ATP hydrolysis. ATPase-deficient prp16 alleles reduce fidelity and suppress splicing defects by allowing more time for catalysis to occur, resulting in the progression of suboptimal substrates, which would otherwise be discarded, through the first step.
An important prediction of the kinetic proofreading model is that each ATPase-mediated conformational change affords opportunity for such a progression/discard branch in the splicing pathway: recent work has indeed demonstrated analogous behaviour for two more spliceosomal ATPases, consistent with the generality of this mechanism. S. cerevisiae spliceosomes assembled on 3’SS mutant substrates and purified after the first step proceed through the second in the presence of mutant Prp22 protein deficient for ATPase and/or unwindase activity, or in the absence of ATP (Fig. 3, Mayas et al., 2006). Genetic work in S. cerevisiae has also shown kinetic proofreading of branch site-U2 snRNA interaction by Prp5 (Fig. 3). ATPase-deficient prp5 alleles suppress mutations flanking the BS that destabilise its pairing to U2, but suppression can be superseded by re-stabilising this interaction, either by compensatory mutations in U2 or intron mutations that generate extra upstream base pairs. The level of suppression by prp5 alleles correlates inversely with their ATPase activity. Prp5 proteins from organisms in which the branch site is less highly conserved than in S. cerevisiae have lower ATPase activity, thus providing a mechanism by which the fidelity of branch site selection is reduced in these organisms (Xu and Query, 2007). This work identifies the first structure to be in direct competition with the activity of a spliceosomal DExD/H ATPase, but the precise molecular consequences of ATPase activity remain to be elucidated for this and other ATPase-mediated transitions.
Important mechanistic questions regarding kinetic proofreading remain to be addressed: for example, the direct targets of spliceosomal ATPases are unknown. In addition, although each ATPase-mediated step is an opportunity for discard, the nature of this discard is enigmatic. For example, it is possible that the conformational change resulting from ATPase activity is not compatible with binding of the substrate for the previous step, such that it would cause the spliceosome to fall apart: alternatively, an active disassembly cascade could be triggered by such a conformational change. It is even possible that the spliceosome may need to undergo several conformational changes resembling functional progression along the splicing pathway. A role in discard for Prp43 and Ntr1, which cooperate in spliceosome disassembly (Pandit et al., 2006) might suggest that discard is mechanistically similar to progression. Some support for this model is provided by the observation that discarded intermediates are degraded in the cytoplasm (Hilleren and Parker, 2003); this finding implies that discard, like mRNA release, is coupled to nuclear export.
THE SECOND CATALYTIC STEP
The second step of splicing remains substantially less well-characterised than the first. In addition to early recognition at the stage of complex A formation, the 3’SS has been proposed to be selected after the first step via a simple scanning mechanism as the first AG dinucleotide, or the second if sufficiently close to the first AG, downstream of the branch site (yeast) or polypyrimidine tract (metazoa) (Smith et al., 1993; Anderson and Moore, 1997; Chua and Reed, 2001). Distance from the branch site is an important determinant of 3’SS strength: Prp22, Slu7 and Prp18 are all dispensable in vitro for introns with short BS-3’SS distances (Schwer and Gross, 1998 and references therein), and Prp22 mutants stimulate the use of non-AG splice sites closer to the branch (Mayas et al., 2006). In addition, the splicing of genes with short BS-3’SS spacing is unaffected by knockout of the second step factor Prp17 in S. cerevisiae (Sapra et al., 2004). Aside from proximity to the branch site or polypyrimidine tract, what constitutes a favoured 3’SS remains unclear. The YAG 3’SS consensus sequence has not been extended by bioinformatic investigation, although it seems possible that local RNA structure may play a role in determining the quality of a 3’SS for the second step; such bioinformatics, along with genetic screens to search for possible determinants of YAG strength, may reveal higher complexity.
Experimental investigation of the second step is hindered by several factors: the apparent ability of the spliceosome to assemble on one 3’SS and use another on the same pre-mRNA is one such hindrance. Previously noted experiments in plants demonstrating that a 3’SS sequestered in a hairpin could act as a splice acceptor only in the presence of a downstream 3’SS suggested the possibility of re-specifying the 3’SS after early assembly (Liu et al., 1995). Work on autoregulatory splicing by sex-lethal in Drosophila has provided evidence that a 3’SS can play a critical role in exon definition but not be preferred for catalysis (Penalva et al., 2001). It is therefore possible that 3’SS requirements for assembly and catalysis are, from an experimental point of view, at best not necessarily identical and at worst obligately distinct. An ‘ideal’ 3’SS could thus represent a balance between assembly-competent and catalytically favoured states; this, and potential effects due to the almost unavoidable presence of nearby AG dinucleotides in natural genes, must be taken into account in any systematic analysis of 3’SS quality.
A second obstacle to the investigation of the second step is that any active site component required for both steps will presumably exert its effects at the stage of first step catalysis, thus rendering the investigation of its role in the second step technically difficult. Although many spliceosomal components involved in the second step are dispensable for the first, such as the 3’SS itself (Chiara and Reed, 1995), Slu7 (Chua and Reed, 1999) and loop 1 of U5 snRNA (O'Keefe et al., 1996), several shared active site components might exist. Indirect information about the components of the second step active site could be derived from knowledge about the second step binding site of the lariat branch. The branch structure of the lariat intermediate must be repositioned and bound during the second step, consistent with the second step defect of 5’SS and BS mutants being suppressed by spliceosomal alleles that improve the second step (Query and Konarska, 2004; Konarska et al., 2006; Mayas et al., 2006). Although the 5’SS portion of the branch structure appears to reposition relative to U6 as previously discussed, the nature of this interaction as well as the interacting partner of the branch site region, which appears to unpair from U2 snRNA during or after first step catalysis (Smith et al., 2007), is unknown.
In fact, temporal epistasis – that is, the manifestation of mutations that cause multiple sequential defects in a pathway only at their earliest point of action – can also impede the study of first step catalysis due to the long preceding assembly phase. Systems in which assembly can be bypassed, at least in part (for example Konforti and Konarska, 1994; Anderson and Moore, 1997; Mayas et al., 2006; Valadkhan et al., 2007), represent possible ways to circumvent this problem.
Many proteins join the spliceosome between the two catalytic steps (reviewed in Jurica and Moore, 2003). In vitro depletion/reconstitution studies, together with genetic work in vivo, have provided clues as to the function of many second step factors. Loop 1 of U5 snRNA appears to act as a ‘platform’ on which the 5’ and 3’ exons are juxtaposed for ligation (Crotti et al., 2007), and interacts functionally with Prp18 – a protein that also binds Slu7 (Bacikova and Horowitz, 2002). Slu7 depletion leads to a loss of 3’SS fidelity, and also appears to destabilise free 5’ exon binding to the pre-second step spliceosome (Chua and Reed, 1999). Prp22 crosslinks directly to the 3’SS following the action of Prp16 (McPheeters et al., 2000) and is also involved in mRNA release (Schwer and Gross, 1998; Wagner et al., 1998). Interestingly, although all of these factors are essential in vivo, the requirement for each can be at least partially bypassed in vitro (Schwer and Gross, 1998; Chua and Reed, 1999; Segault et al., 1999; Crotti et al., 2007), suggesting both that the second step spliceosome is fairly robust, and that these factors do not form interactions strictly necessary for catalytic events. Instead, a likely possibility is that they all contribute to stabilisation of the second step conformation relative to the first. Elucidation of the interactions made by these factors will be necessary to understand how they can impact 3’SS use and therefore alternative splicing.
ALTERNATIVE SPLICING
In the absence of repression, strong splice sites give rise to constitutive splicing; alternative splicing therefore represents the suppression of optimal splice sites and/or the use of those that are suboptimal. Most alternatively spliced introns are thought to be controlled by multiple splicing enhancer and silencer elements whose activity depends on their location relative to splice sites; these regulatory elements are thought to affect splicing predominantly through corresponding RNA binding proteins that exert their effects by altering a specific step in the spliceosome assembly pathway (reviewed in Black, 2003). However, modulation of splicing efficiency is theoretically possible at any stage of the splicing reaction. Each splicing event, depending on the introns and exons involved, will be limited by one or more transitions, and will as such be sensitive to modulation of their efficiency while being insensitive to all but the most major changes in that of non-limiting steps. Given the diversity of pre-mRNA substrates, the multi-step nature of the spliceosome assembly and catalysis pathway, and the enormous number of factors involved in the splicing of every transcript, it is almost certain that examples exist in nature of splicing regulation at every possible stage. The existence of natural splicing events at least partially limited by post-assembly transitions has already been demonstrated: the splicing of overlapping but non-identical sets of endogenous introns is sensitive to the knockdown or mutation of core spliceosomal proteins important for the catalytic phase of the reaction (Park et al., 2004; Pleiss et al., 2007); this sensitivity affords opportunities for splicing regulation during catalysis. The ability to regulate the splicing of an individual intron by modulating the local activity or concentration of a given protein, or that of a class of introns via more global changes, together with the combinatorial effects of regulation of multiple transitions, can allow a robust and specific regulation of splicing events without a necessary requirement for large numbers of individual splicing factors to exert this control. Thus, in order to understand alternative splicing, specific factors acting on each transcript need not necessarily be sought, and the entire reaction through assembly, catalysis and disassembly must be considered.
The stimulation or repression of spliceosome assembly naturally represents a common mechanism of splicing regulation, and may indeed be the most prevalent in higher eukaryotes. Such assembly-based regulation might, however, be more subtle and complex than commonly thought. Virtually all metazoan transcripts contain multiple sequences capable of being recognised and used as splice sites, and much silencer-based repression may therefore require kinetic competition between these sites, serving predominantly to redirect rather than strictly to repress spliceosome assembly and/or catalysis.
Alternative splicing is not normally considered to occur in S. cerevisiae, but this organism’s ease of genetic manipulation and strong splice site consensus requirements allow many different events in splicing to artificially be made limiting for gene expression, facilitating detailed mechanistic analysis of suboptimal splice site use. Recent data showing that the splicing of meiosis-specific genes is repressed outside meiosis (Juneau et al., 2007) also potentially identify a system in which true alternative splicing can be studied in this organism.
CONCLUDING REMARKS
Enormous amounts of data exist regarding regulated and cell-type-specific splicing patterns (reviewed in Moore and Silver, 2007). Specific splicing factors that affect the splicing of small numbers of transcripts must exert their effects via spliceosomal transitions and through core components, both of which are finite in number. This imposes a limit on the number of possible unique mechanisms of splicing regulation. In addition to the discovery of more specific splicing factors, we anticipate that more widespread and varied regulatory roles will be discovered for core spliceosomal components themselves. A clear, mechanistic description of the splicing process is necessary to explain regulated splicing, and the detailed analysis of various alternative splicing systems is also likely to identify additional transitions in the splicing pathway. Much work in the mechanistic splicing field is justifiably focused on the generation of spliceosome preparations suitable for X-ray crystallography. The high-resolution structures that will hopefully result from this work should resolve many of the questions posed in this review, and provide invaluable information about the mechanistic details of the splicing reaction. The preparation of large quantities of sufficiently pure, conformationally homogeneous spliceosomes, however, remains a serious challenge. Further and more detailed knowledge of the dynamic behaviour of the spliceosome will aid such crystallographic efforts, and will also be necessary for the rationalisation and accurate interpretation of their results.
Acknowledgments
We offer our apologies to those whose work we could not cite directly due to space constraints. We thank Doug Black, Beth Moorefield, Erik Sontheimer, Jon Staley and Juan Valcarcel for comments on the manuscript, numerous other colleagues for sharing pre-publication data to which we cannot refer, and Allison Amend for introducing us to the poem from which the title is derived.
Footnotes
From Mutability (1816) by P.B. Shelley: full text available at http://www.litscape.com/author/Percy_Bysshe_Shelley/Mutability.html
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson K, Moore MJ. Bimolecular exon ligation by the human spliceosome. Science. 1997;276:1712–1716. doi: 10.1126/science.276.5319.1712. [DOI] [PubMed] [Google Scholar]
- Bacikova D, Horowitz DS. Mutational analysis identifies two separable roles of the Saccharomyces cerevisiae splicing factor Prp18. RNA. 2002;8:1280–1293. doi: 10.1017/s1355838202023099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailleul B. During in vivo maturation of eukaryotic nuclear mRNA, splicing yields excised exon circles. Nucleic Acids Res. 1996;24:1015–1019. doi: 10.1093/nar/24.6.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadnia N, Hartmuth K, Will CL, Lührmann R. Functional spliceosomal A complexes can be assembled in vitro in the absence of a penta-snRNP. RNA. 2006;12:1738–1746. doi: 10.1261/rna.120606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Brow DA. Allosteric cascade of spliceosome activation. Annu Rev Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- Burge CB, Tuschl TH, Sharp PA. Splicing of precursors to mRNAs by the spliceosomes. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 2. New York: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- Burgess SM, Guthrie C. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 1993;73:1377–1391. doi: 10.1016/0092-8674(93)90363-u. [DOI] [PubMed] [Google Scholar]
- Chan SP, Kao DI, Tsai WY, Cheng SC. The Prp19p-associated complex in spliceosome activation. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- Charpentier B, Rosbash M. Intramolecular structure in yeast introns aids the early steps of in vitro spliceosome assembly. RNA. 1996;2:509–522. [PMC free article] [PubMed] [Google Scholar]
- Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- Chiara MD, Reed R. A two-step mechanism for 5′ and 3′ splice-site pairing. Nature. 1995;375:510–513. doi: 10.1038/375510a0. [DOI] [PubMed] [Google Scholar]
- Chua K, Reed R. The RNA splicing factor hSlu7 is required for correct 3′ splice-site choice. Nature. 1999;402:207–210. doi: 10.1038/46086. [DOI] [PubMed] [Google Scholar]
- Chua K, Reed R. An upstream AG determines whether a downstream AG is selected during catalytic step II of splicing. Mol Cell Biol. 2001;21:1509–1514. doi: 10.1128/MCB.21.5.1509-1514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Cramer P, Srebrow A, Kadener S, Werbajh S, de la Mata M, Melen G, Nogues G, Kornblihtt AR. Coordination between transcription and pre-mRNA processing. FEBS Lett. 2001;498:179–182. doi: 10.1016/s0014-5793(01)02485-1. [DOI] [PubMed] [Google Scholar]
- Crotti LB, Bacikova D, Horowitz DS. The Prp18 protein stabilizes the interaction of both exons with the U5 snRNA during the second step of pre-mRNA splicing. Genes Dev. 2007;21:1204–1216. doi: 10.1101/gad.1538207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Campo M, Tijerina P, Bhaskaran H, Mohr S, Yang Q, Jankowsky E, Russell R, Lambowitz AM. Do DEAD-box proteins promote group II intron splicing without unwinding RNA? Mol Cell. 2007;28:159–166. doi: 10.1016/j.molcel.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox-Walsh KL, Dou Y, Lam BJ, Hung SP, Baldi PF, Hertel KJ. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc Natl Acad Sci U S A. 2005;102:16176–16181. doi: 10.1073/pnas.0508489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger RJ, Beggs JD. Prp8 protein: at the heart of the spliceosome. RNA. 2005;11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell. 2005;123:65–73. doi: 10.1016/j.cell.2005.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings ML, Krainer AR. Pre-mRNA splicing in the new millennium. Curr Opin Cell Biol. 2001;13:302–309. doi: 10.1016/s0955-0674(00)00212-x. [DOI] [PubMed] [Google Scholar]
- Helmers J, Schmidt D, Glavy JS, Blobel G, Schwartz T. The beta-subunit of the protein-conducting channel of the endoplasmic reticulum functions as the guanine nucleotide exchange factor for the beta-subunit of the signal recognition particle receptor. J Biol Chem. 2003;278:23686–23690. doi: 10.1074/jbc.C300180200. [DOI] [PubMed] [Google Scholar]
- Hertel KJ. Combinatorial control of exon recognition. J Biol Chem. 2008;283:1211–1215. doi: 10.1074/jbc.R700035200. [DOI] [PubMed] [Google Scholar]
- Hertel KJ, Maniatis T. Serine-arginine (SR)-rich splicing factors have an exon-independent function in pre-mRNA splicing. Proc Natl Acad Sci U S A. 1999;96:2651–2655. doi: 10.1073/pnas.96.6.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Zhang Z, Backofen R, Stamm S. Pre-mRNA Secondary Structures Influence Exon Recognition. PLoS Genet. 2007;3:e204. doi: 10.1371/journal.pgen.0030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren PJ, Parker R. Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. Mol Cell. 2003;12:1453–1465. doi: 10.1016/s1097-2765(03)00488-x. [DOI] [PubMed] [Google Scholar]
- Hilliker AK, Mefford MA, Staley JP. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes Dev. 2007;21:821–834. doi: 10.1101/gad.1536107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker AK, Staley JP. Multiple functions for the invariant AGC triad of U6 snRNA. RNA. 2004;10:921–928. doi: 10.1261/rna.7310704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House AE, Lynch KW. An exonic splicing silencer represses spliceosome assembly after ATP-dependent exon recognition. Nat Struct Mol Biol. 2006;13:937–944. doi: 10.1038/nsmb1149. [DOI] [PubMed] [Google Scholar]
- House AE, Lynch KW. Regulation of alternative splicing: more than just the ABCs. J Biol Chem. 2008;283:1217–1221. doi: 10.1074/jbc.R700031200. [DOI] [PubMed] [Google Scholar]
- Hui J, Hung LH, Heiner M, Schreiner S, Neumuller N, Reither G, Haas SA, Bindereif A. Intronic CA-repeat and CA-rich elements: a new class of regulators of mammalian alternative splicing. EMBO J. 2005;24:1988–1998. doi: 10.1038/sj.emboj.7600677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim el C, Schaal TD, Hertel KJ, Reed R, Maniatis T. Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc Natl Acad Sci U S A. 2005;102:5002–5007. doi: 10.1073/pnas.0500543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Musunuru K, Lewis HA, Burley SK, Darnell RB. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proc Natl Acad Sci U S A. 2000;97:5740–5745. doi: 10.1073/pnas.090553997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TL, Abelson J. Characterization of U4 and U6 interactions with the 5′ splice site using a S. cerevisiae in vitro trans-splicing system. Genes Dev. 2001;15:1957–1970. doi: 10.1101/gad.895601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneau K, Palm C, Miranda M, Davis RW. High-density yeast-tiling array reveals previously undiscovered introns and extensive regulation of meiotic splicing. Proc Natl Acad Sci U S A. 2007;104:1522–1527. doi: 10.1073/pnas.0610354104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Kim CH, Abelson J. Site-specific crosslinks of yeast U6 snRNA to the pre-mRNA near the 5′ splice site. RNA. 1996;2:995–1010. [PMC free article] [PubMed] [Google Scholar]
- Konarska MM, Vilardell J, Query CC. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol Cell. 2006;21:543–553. doi: 10.1016/j.molcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Konforti BB, Konarska MM. U4/U5/U6 snRNP recognizes the 5′ splice site in the absence of U2 snRNP. Genes Dev. 1994;8:1962–1973. doi: 10.1101/gad.8.16.1962. [DOI] [PubMed] [Google Scholar]
- Kreahling JM, Graveley BR. The iStem, a long-range RNA secondary structure element required for efficient exon inclusion in the Drosophila Dscam pre-mRNA. Mol Cell Biol. 2005;25:10251–10260. doi: 10.1128/MCB.25.23.10251-10260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HX, Goodall GJ, Kole R, Filipowicz W. Effects of secondary structure on pre-mRNA splicing: hairpins sequestering the 5′ but not the 3′ splice site inhibit intron processing in Nicotiana plumbaginifolia. EMBO J. 1995;14:377–388. doi: 10.1002/j.1460-2075.1995.tb07012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Query CC, Konarska MM. Opposing classes of prp8 alleles modulate the transition between the catalytic steps of pre-mRNA splicing. Nat Struct Mol Biol. 2007;14:519–526. doi: 10.1038/nsmb1240. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71:803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Lührmann R. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- Mayas RM, Maita H, Staley JP. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat Struct Mol Biol. 2006;13:482–490. doi: 10.1038/nsmb1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, Schwartz ML, Butcher SE, Brow DA. A dynamic bulge in the U6 RNA internal stem-loop functions in spliceosome assembly and activation. RNA. 2007;13:2252–2265. doi: 10.1261/rna.699907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters DS, Schwer B, Muhlenkamp P. Interaction of the yeast DExH-box RNA helicase prp22p with the 3′ splice site during the second step of nuclear pre-mRNA splicing. Nucleic Acids Res. 2000;28:1313–1321. doi: 10.1093/nar/28.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S, Matsuura M, Perlman PS, Lambowitz AM. A DEAD-box protein alone promotes group II intron splicing and reverse splicing by acting as an RNA chaperone. Proc Natl Acad Sci U S A. 2006;103:3569–3574. doi: 10.1073/pnas.0600332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Silver PA. Global analysis of mRNA splicing. RNA. 2007;14:197–203. doi: 10.1261/rna.868008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW. The spliceosome: the most complex macromolecular machine in the cell? Bioessays. 2003;25:1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- O'Keefe RT, Norman C, Newman AJ. The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell. 1996;86:679–689. doi: 10.1016/s0092-8674(00)80140-3. [DOI] [PubMed] [Google Scholar]
- Olson S, Blanchette M, Park J, Savva Y, Yeo GW, Yeakley JM, Rio DC, Graveley BR. A regulator of Dscam mutually exclusive splicing fidelity. Nat Struct Mol Biol. 2007;14:1134–1140. doi: 10.1038/nsmb1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S, Lynn B, Rymond BC. Inhibition of a spliceosome turnover pathway suppresses splicing defects. Proc Natl Acad Sci U S A. 2006;103:13700–13705. doi: 10.1073/pnas.0603188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci U S A. 2004;101:15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penalva LO, Lallena MJ, Valcarcel J. Switch in 3′ splice site recognition between exon definition and splicing catalysis is important for sex-lethal autoregulation. Mol Cell Biol. 2001;21:1986–1996. doi: 10.1128/MCB.21.6.1986-1996.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriman RJ, Ares M., Jr Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes Dev. 2007;21:811–820. doi: 10.1101/gad.1524307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 2007;5:e90. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle AM. Group II Introns: Catalysts for Splicing, Genomic Change and Evolution. 1. London: Royal Society of Chemistry; 2008. [Google Scholar]
- Query CC, Konarska MM. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol Cell. 2004;14:343–354. doi: 10.1016/s1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- Raghunathan PL, Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol. 1998;8:847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- Rhode BM, Hartmuth K, Westhof E, Lührmann R. Proximity of conserved U6 and U2 snRNA elements to the 5′ splice site region in activated spliceosomes. EMBO J. 2006;25:2475–2486. doi: 10.1038/sj.emboj.7601134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rino J, Carvalho T, Braga J, Desterro JM, Lührmann R, Carmo-Fonseca M. A stochastic view of spliceosome assembly and recycling in the nucleus. PLoS Comput Biol. 2007;3:2019–2031. doi: 10.1371/journal.pcbi.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan DE, Kim CH, Murray JB, Adams CJ, Stockley PG, Abelson J. New tertiary constraints between the RNA components of active yeast spliceosomes: a photo-crosslinking study. RNA. 2004;10:1251–1265. doi: 10.1261/rna.7060404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapra AK, Arava Y, Khandelia P, Vijayraghavan U. Genome-wide analysis of pre-mRNA splicing: intron features govern the requirement for the second-step factor, Prp17 in Saccharomyces cerevisiae and Schizosaccharomyces pombe. J Biol Chem. 2004;279:52437–52446. doi: 10.1074/jbc.M408815200. [DOI] [PubMed] [Google Scholar]
- Sawa H, Abelson J. Evidence for a base-pairing interaction between U6 small nuclear RNA and 5′ splice site during the splicing reaction in yeast. Proc Natl Acad Sci USA. 1992;89:11269–11273. doi: 10.1073/pnas.89.23.11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S, Campodonico E, Schwer B. Motifs IV and V in the DEAH box splicing factor Prp22 are important for RNA unwinding, and helicase-defective Prp22 mutants are suppressed by Prp8. J Biol Chem. 2004;279:8617–8626. doi: 10.1074/jbc.M312715200. [DOI] [PubMed] [Google Scholar]
- Schwer B, Gross CH. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 1998;17:2086–2094. doi: 10.1093/emboj/17.7.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Guthrie C. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J. 1992;11:5033–5039. doi: 10.1002/j.1460-2075.1992.tb05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segault V, Will CL, Polycarpou-Schwarz M, Mattaj IW, Branlant C, Lührmann R. Conserved loop I of U5 small nuclear RNA is dispensable for both catalytic steps of pre-mRNA splicing in HeLa nuclear extracts. Mol Cell Biol. 1999;19:2782–2790. doi: 10.1128/mcb.19.4.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Kohlstaedt LA, Damianov A, Rio DC, Black DL. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat Struct Mol Biol. 2008;15:183–191. doi: 10.1038/nsmb.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Green MR. RS domains contact splicing signals and promote splicing by a common mechanism in yeast through humans. Genes Dev. 2006;20:1755–1765. doi: 10.1101/gad.1422106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Green MR. RS domain-splicing signal interactions in splicing of U12-type and U2-type introns. Nat Struct Mol Biol. 2007;14:597–603. doi: 10.1038/nsmb1263. [DOI] [PubMed] [Google Scholar]
- Shen H, Kan JL, Green MR. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol Cell. 2004;13:367–376. doi: 10.1016/s1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- Shi Y, Reddy B, Manley JL. PP1/PP2A phosphatases are required for the second step of Pre-mRNA splicing and target specific snRNP proteins. Mol Cell. 2006;23:819–829. doi: 10.1016/j.molcel.2006.07.022. [DOI] [PubMed] [Google Scholar]
- Small EC, Leggett SR, Winans AA, Staley JP. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol Cell. 2006;23:389–399. doi: 10.1016/j.molcel.2006.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CW, Chu TT, Nadal-Ginard B. Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol Cell Biol. 1993;13:4939–4952. doi: 10.1128/mcb.13.8.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Query CC, Konarska MM. trans-splicing to spliceosomal U2 snRNA suggests disruption of branch site-U2 pairing during pre-mRNA splicing. Mol Cell. 2007;26:883–890. doi: 10.1016/j.molcel.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares LM, Zanier K, Mackereth C, Sattler M, Valcarcel J. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science. 2006;312:1961–1965. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- Sontheimer EJ, Steitz JA. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993;262:1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- Stark H, Lührmann R. Cryo-electron microscopy of spliceosomal components. Annu Rev Biophys Biomol Struct. 2006;35:435–457. doi: 10.1146/annurev.biophys.35.040405.101953. [DOI] [PubMed] [Google Scholar]
- Sterner DA, Carlo T, Berget SM. Architectural limits on split genes. Proc Natl Acad Sci U S A. 1996;93:15081–15085. doi: 10.1073/pnas.93.26.15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens SW, Ryan DE, Ge HY, Moore RE, Young MK, Lee TD, Abelson J. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol Cell. 2002;9:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Aronova A, Schwer B. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev. 2007;21:2312–2325. doi: 10.1101/gad.1580507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EJ, Zhou Y, Corbett AH, Wente SR. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol Cell. 2007;28:850–859. doi: 10.1016/j.molcel.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Valadkhan S. The spliceosome: caught in a web of shifting interactions. Curr Opin Struct Biol. 2007;17:310–315. doi: 10.1016/j.sbi.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Valadkhan S, Mohammadi A, Wachtel C, Manley JL. Protein-free spliceosomal snRNAs catalyze a reaction that resembles the first step of splicing. RNA. 2007;13:2300–2311. doi: 10.1261/rna.626207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa T, Guthrie C. The Isy1p component of the NineTeen Complex interacts with the ATPase Prp16p with to regulate the fidelity of pre-mRNA splicing. Genes Dev. 2005;19:1894–1904. doi: 10.1101/gad.1336305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JD, Jankowsky E, Company M, Pyle AM, Abelson JN. The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J. 1998;17:2926–2937. doi: 10.1093/emboj/17.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YZ, Query CC. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol Cell. 2007;28:838–849. doi: 10.1016/j.molcel.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yean SL, Wuenschell G, Termini J, Lin RJ. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature. 2000;408:881–884. doi: 10.1038/35048617. [DOI] [PMC free article] [PubMed] [Google Scholar]