Abstract
The Smad2 and Smad3 (Smad2/3) proteins are principally involved in the transmission of transforming growth factor β (TGF-β) signaling from the plasma membrane to the nucleus. Many transcription factors have been shown to cooperate with the Smad2/3 proteins in regulating the transcription of target genes, enabling appropriate gene expression by cells. Here we identified 1,787 Smad2/3 binding sites in the promoter regions of over 25,500 genes by chromatin immunoprecipitation on microarray in HaCaT keratinocytes. Binding elements for the v-ets erythroblastosis virus E26 oncogene homolog (ETS) and transcription factor AP-2 (TFAP2) were significantly enriched in Smad2/3 binding sites, and knockdown of either ETS1 or TFAP2A resulted in overall alteration of TGF-β-induced transcription, suggesting general roles for ETS1 and TFAP2A in the transcription induced by TGF-β-Smad pathways. We identified novel Smad binding sites in the CDKN1A gene where Smad2/3 binding was regulated by ETS1 and TFAP2A. Moreover, we showed that small interfering RNAs for ETS1 and TFAP2A affected TGF-β-induced cytostasis. We also analyzed Smad2- or Smad3-specific target genes regulated by TGF-β and found that their specificity did not appear to be solely determined by the amounts of the Smad2/3 proteins bound to the promoters. These findings reveal novel regulatory mechanisms of Smad2/3-induced transcription and provide an essential resource for understanding their roles.
Members of the transforming growth factor β (TGF-β) family are multifunctional proteins that regulate various biological processes, including cell growth, differentiation, apoptosis, motility, and extracellular matrix production, and thus play essential roles in embryonic development and the pathogenesis of various diseases (47). TGF-β transduces signals through heteromeric complexes of type I (TβR-I) and type II (TβR-II) serine/threonine kinase receptors and intracellular Smad proteins (18). After TGF-β binding, TβR-II phosphorylates TβR-I, which then phosphorylates Smad2 and Smad3 at the C-terminal SSXS motif. The phosphorylated Smad2 and Smad3 (Smad2/3) proteins then form oligomers with or without Smad4 that translocate to the nucleus, where they regulate the transcription of target genes. Activated Smad oligomers have been reported to bind to sequences, termed the Smad binding elements (SBEs), containing the (C)AGAC element (13, 26, 52, 54). In addition, Smad3 and Smad4 directly bind to AP-1 sites (TGA[G/C]TCA) with or without JUN (57). A GC-rich sequence was also identified as a Smad binding site (19, 28, 30, 33).
Regulation of TGF-β-induced gene expression is frequently modulated by other transcription factors and cofactors, which are induced by various stimuli and are often expressed in a cell- and tissue-specific context (18, 38). These factors provide target specificity to Smad complexes, since Smad3 and Smad4 alone have relatively low binding affinity for the SBEs. As a result, restricted types of receptor family and Smad family members can induce appropriate sets of gene expression to execute the broad range of biological responses to TGF-β stimuli.
Chromatin immunoprecipitation (ChIP) combined with oligonucleotide tiling microarray technologies (ChIP-chip) is an emerging method for the identification of transcription factor binding sites (3, 22, 27). In the present study, we performed ChIP-chip analysis of Smad2/3 binding sites of promoter regions of known human genes. We found many previously unidentified regions with novel target genes in proximity. Even for the reported target gene for cyclin-dependent kinase inhibitor 1A (CDKN1A, which encodes the p21 protein), novel Smad binding regions were detected with greater significance than previously identified binding positions. Motif analyses revealed that canonical SBE sites were enriched in Smad2/3 binding regions and related to a greater change in expression with TGF-β stimulation. In addition, v-ets erythroblastosis virus E26 oncogene homolog (ETS) and transcription factor AP-2 (TFAP2) (also termed AP-2) binding sites were identified as significantly enriched motifs in Smad binding sites. In knockdown experiments, ETS1 and TFAP2A (also known as AP-2α) appeared to strengthen the binding of Smad2/3 to target promoters and affect transcriptional responses. We also obtained gene expression profiles by knockdown of either Smad2 or Smad3 and found that Smad2 or Smad3 dependency of gene expression is not solely determined by the total amounts of the Smad2/3 proteins bound to the promoters. These findings are essential to determining where Smad2/3 bind and how Smad2- or Smad3-dependent gene expression is determined. These results also suggest, for the first time, that ETS1 and TFAP2A might frequently participate in TGF-β-induced transcription.
MATERIALS AND METHODS
Cell culture.
HaCaT and 293T cells were maintained in Dulbecco's modified Eagle's medium (catalog no. 11965; GIBCO/Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum, 100 U/ml penicillin G, and 100 μg/ml streptomycin. Cells were grown in a humidified atmosphere with 5% CO2 at 37°C.
Promoter-reporter constructs, cDNA constructs, and chemicals.
Human SERPINE1 and CDKN1A promoter reporters (−2300 to +8) were as described previously (29, 41). Human CDKN1A SBR1 (+1135 to +2234) and SBR2 (+3543 to +5387) and human CDC6 (−4378 to +67) promoter-reporters were constructed by a PCR-based approach. Human constitutively active type I TGF-β receptor cDNA and ETS1 cDNA were described previously (31, 46). Human TFAP2A and JUN cDNAs were constructed by a PCR-based approach. All of the DNAs constructed were verified by sequencing. TGF-β3 was from Novartis (Basel, Switzerland). TGF-β type I receptor kinase inhibitor A-44-03 was as described previously (17).
Antibodies.
We used commercially available antibodies as follows: mouse anti-Smad2/3 (BD Biosciences, Franklin Lakes, NJ), anti-α-tubulin (DM1A; Sigma, St. Louis, MO), anti-lamin A/C (BD Biosciences), anti-CDKN1A (EMD Chemicals, NJ), rabbit anti-ETS1 (C-20; Santa Cruz, CA), anti-TFAP2A (C-18; Santa Cruz), and anti-JUN (H79; Santa Cruz). Mouse immunoglobulin G1 (IgG1) (MB002; R&D Systems, Minneapolis, MN) and rabbit IgG (SouthernBiotech, Birmingham, AL) were used as controls.
RNA interference and oligonucleotides.
Stealth small interfering RNAs (siRNAs) were purchased from Invitrogen as follows: human ETS1 (sense, 5′-GGAGAUGGCUGGGAAUUCAAACUUU-3′), TFAP2A (sense, 5′-CCGUCUCCGCCAUCCCUAUUAACAA-3′), JUN (sense, 5′-UCCUGAAACAGAGCAUGACCCUGAA-3′), CDKN1A, (sense, 5′-GAACUUCGACUUUGUCACCGAGACA-3′), Smad2 (sense, 5′-AAUGGAGUGAGUAUAGUCAUCCAGA-3′), Smad3 (sense, 5′-AGAUCUUCAGGUUGCAUCCUGGUGG-3′), and control oligonucleotides (SKU no. 12935-200; sequence not available). siRNAs were introduced into HaCaT cells with the Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer's instructions.
ChIP.
Cells were cultured in 15-cm plates to approximately 80% confluence, and one plate was used per immunoprecipitation. Cells were fixed with 1% formaldehyde for 10 min at room temperature with swirling. Glycine was added to a final concentration of 0.125 M, and the incubation was continued for an additional 5 min. Cells were washed twice with ice-cold phosphate-buffered saline, harvested by scraping, pelleted, and resuspended in 1 ml of sodium dodecyl sulfate (SDS) lysis buffer (50 mM Tris-HCl [pH 8.1], 1% SDS, 10 mM EDTA, protease inhibitors [P8340]). Samples were sonicated four times for 15 s each time at intervals of 30 s with a UH-50 sonicator (SMT, Japan). Alternatively, 0.2 ml of lysis buffer per 10-cm cell culture plate and a Bioruptor UCW-201 (output, H; 15 cycles of 30 s of sonication with 30-s intervals; Cosmobio, Japan) were used for samples for some of the conventional ChIP-quantitative PCR (qPCR) analyses. Samples were centrifuged at 14,000 rpm at 8°C for 10 min. After removal of a control aliquot (whole-cell extract), supernatants were diluted 10-fold in ChIP dilution buffer (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, Complete EDTA-free protease inhibitors [Roche Diagnostics, Rotkreuz, Switzerland]). Samples were incubated at 4°C overnight in 2-methacryloyloxyethyl phosphorylcholine polymer-treated 15-ml polypropylene tubes (Assist, Japan) with protein A or anti-mouse IgG-Dynabeads that had been preincubated with 5 to 10 μg of antibodies in phosphate-buffered saline-0.5% bovine serum albumin. The beads were then moved to 1.7-ml siliconized tubes (catalog no. 3207; Corning, Corning, NY) and washed five times with ChIP wash buffer (50 mM HEPES-KOH [pH 7.0], 0.5 M LiCl, 1 mM EDTA, 0.7% deoxycholate, 1% Igepal CA630) and once with TE buffer (pH 8.0). Immunoprecipitated samples were eluted and reverse cross-linked by incubation overnight at 65°C in elution buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 1% SDS). Genomic DNA was then extracted with a PCR purification kit (Qiagen).
Sample preparation for tiling array.
ChIP and control input DNA samples were amplified by two cycles of in vitro transcription and hybridized on separate Affymetrix human promoter 1.0 oligonucleotide tiling arrays as described previously (27, 49). Two biologically independent ChIP and control samples were amplified and hybridized individually.
Analysis of tiling array data.
Enrichment values (ChIP/control input DNA) were calculated with the MAT algorithm as described previously (25, 49).
Sequence analysis.
Interspecies sequence conservation analysis data were obtained from the CEAS website (23). CEAS calculates an average phastCons score for each single base pair position and generates an average conservation plot. The extent of the conservation of the SBRs can be estimated by comparison of the middle of the plot to both ends of the plot. The phastCons score was obtained from the University of California Santa Cruz genome server, which uses human, chimpanzee, mouse, rat, rabbit, macaque, dog, cow, armadillo, elephant, tenrec, opossum, chicken, frog, zebra fish, tetraodon, and fugu genome sequences. The conservation scores can be interpreted as probabilities that each base is in a conserved element, as described previously (48). Data on enriched binding motifs were also obtained from the CEAS website, where the position-weighted matrix method was used for identification. We summarized all of the enriched matrices for the same transcription factor and used the results for analyses. TRANSFAC also identified enriched AP-1 sites and, to a lesser extent, AP-2 and ETS sites. We also used a pattern-based approach to determine the frequency of each motif in SBRs compared to random sequences, where the following motifs were used for identification: AP-1, TGASTCA; AP-2, GCCNNNRGS; ETS, SMGGAWR. Pairwise analysis with each enriched motif was performed with Fisher's exact probability test.
Immunoblotting.
Subcellular fractionation of HaCaT cells was performed with NE-PER nuclear and cytoplasmic extraction reagent (Thermo Fisher Scientific, IL) according to the manufacturer's protocol. After centrifugation, protein concentrations of cell lysates were quantified with a DC protein assay kit (Bio-Rad, Hercules, CA) where indicated. SDS-gel electrophoresis and immunoblotting were performed as described previously (16), with an LAS-3000 mini lumino-image analyzer (Fuji Film, Tokyo, Japan).
Luciferase assay.
Cells in 24-well plates were transfected with combinations of promoter-reporter constructs and expression plasmids by Lipofectamine LTX (Invitrogen). The total amount of transfected DNA was adjusted to the same quantity with empty vector. Twenty-four hours later, cells were treated with TGF-β for an additional 24 to 48 h and lysed. Luciferase activities in the lysates were measured with the Dual-Luciferase Reporter System (Promega, Madison, WI) as described previously (31). For normalization, pGL4.75-SV40-hRluc was cotransfected. Where indicated, siRNAs were transfected 24 h before reporter transfection. All of the samples were prepared in triplicate, and results were averaged.
Reverse transcription (RT)-qPCR.
Total RNAs were extracted with TRIZOL (Invitrogen). First-strand cDNAs were synthesized with PrimeScript reverse transcriptase (Takara Bio Inc., Shiga, Japan) and oligo(dT)13-18 primers (Invitrogen).
qPCR analysis.
Real-time qPCR analysis was performed with Platinum Sybr green qPCR SuperMix-UDG, ROX (Invitrogen), and the ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Foster City, CA) (31). Amplification data were quantified by the standard curve method. Detected signals were confirmed to be specific by a dissociation protocol. All of the samples were run in duplicate or triplicate, and the results were averaged.
Thymidine incorporation assay.
Cells were seeded at a density of 1.0 × 104/well in 24-well plates and cultured overnight. At 24 h posttransfection of siRNA, cells were stimulated with TGF-β. Twenty-four hours later, cells were labeled with [3H]thymidine for 2 h. Thymidine incorporation into the trichloroacetic acid-insoluble fraction was analyzed as described previously (16).
RNA extraction and microarray expression analysis.
Total RNAs were extracted with TRIZOL (Invitrogen). The experimental procedures for GeneChip (Affymetrix, Santa Clara, CA) were performed according to the Affymetrix GeneChip expression analysis technical manual. Briefly, total RNA was used to synthesize biotin-labeled cRNA, which was then hybridized to a GeneChip Human U133 plus 2.0 oligonucleotide array (Affymetrix). After being washed, the arrays were stained with streptavidin-phycoerythrin and analyzed on an Affymetrix scanner to collect the image data. Microarray Suite software 5.0 (Affymetrix) was used to calculate the average difference (AD) for each gene probe set, shown as the gene expression intensity value. The AD values were normalized for each array so that the average of all AD values was 100. One array datum was obtained for each sample. Obtained data were verified by qPCRs for various transcripts, and we had no conflicting results between the array data and qPCR data. Affymetrix probe IDs were converted to gene symbols by use of DAVID (12). To determine the significance of enrichment of SBRs in either upregulated or downregulated genes by TGF-β, pairwise analysis of the target genes with SBRs and without SBRs was performed with Fisher's exact probability tests.
For information on primer sequences and for bed format files of the SBRs, see the supplemental material.
The ChIP-chip and expression microarray data have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO series accession number GSE11710 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE11710).
RESULTS
Identification of Smad2/3 binding sites in promoters of human genes with HaCaT cells.
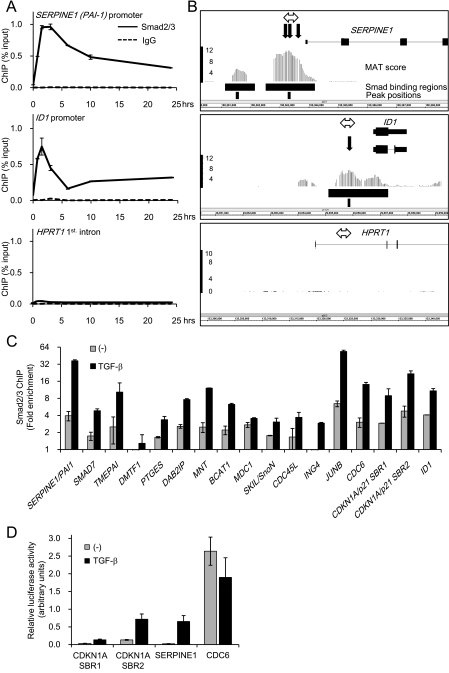
To identify Smad2/3 binding sites in promoters of known human genes, we performed ChIP-chip analysis with human keratinocyte HaCaT cells, in which responses to TGF-β have been well defined (2, 20). With a Smad2/3-specific antibody (see Fig. S1 in the supplemental material), binding of Smad2/3 to known targets of TGF-β, e.g., the plasminogen activator inhibitor type 1 (PAI-1/SERPINE1) promoter (13) and the inhibitor of differentiation 1 (ID1) promoter (28), was detected by ChIP-qPCR (Fig. 1A). ChIP-chip analysis with this antibody identified 1,787 SBRs by a MAT-scoring method (25) (P < 1.0 × 10−5; false discovery rate, <5%; see Table S1 in the supplemental material). As shown in Fig. 1B, Smad2/3 binding sites were detected in the promoter regions of SERPINE1 and ID1 with peak positions near the known SBEs. We then verified 17 regions among the identified SBRs by ChIP-qPCR and observed significant TGF-β-induced Smad2/3 binding to all of the regions (Fig. 1C). We also performed promoter-reporter assays of SERPINE1, CDKN1A, and the cell division cycle 6 homolog (CDC6), which contained the identified SBRs. We found two SBRs, termed SBR1 and SBR2, in CDKN1A (see Fig. 5). As shown in Fig. 1D, we found that TGF-β induced SERPINE1 and CDKN1A transcriptional activity but reduced CDC6 transcriptional activity.
FIG. 1.
Identification of Smad2/3 binding sites in promoters of human genes with HaCaT cells. (A) Time course of Smad2/3 binding to known target promoters. HaCaT cells were treated with TGF-β, formalin fixed at the indicated times, and harvested. Chromatin precipitated by anti-Smad2/3 or control IgG was reverse-cross-linked, and the obtained genomic fragments were quantified by real-time PCR. Data were normalized by input DNA. IgG, mouse control IgG1. Error bars represent standard deviations. (B) Comparison of obtained ChIP-chip signals with published Smad binding positions at promoters of the known target genes of TGF-β. Obtained probe signals were transformed to MAT scores for each position, and peak signal positions were determined from the significant Smad binding regions in the SERPINE1 or ID1 promoter. The HPRT1 intronic region is shown as a control. The upper black bars represent significant Smad binding regions, and the lower bars indicate peak positions. The bold black arrows are reported Smad binding positions. The double-headed arrows are positions of amplicons analyzed in panel A. (C) Validation of Smad2/3 binding by ChIP-qPCR. HaCaT cells were treated as for panel A, and Smad2/3-bound DNA at 1.5 h after TGF-β treatment was quantified by real-time PCR. Values are presented as n-fold enrichment over the mean value of control Smad2/3 ChIPs (HPRT1, HOXA13, and KIF5B promoters). SBR1/2, Smad binding region 1/2 (see Fig. 5A). Error bars represent standard errors. (D) Validation of the TGF-β-induced transcriptional activity of identified Smad2/3 binding regions. HaCaT cells were transfected with the luciferase reporter constructs as indicated and stimulated with TGF-β. Error bars represent the standard errors.
FIG. 5.
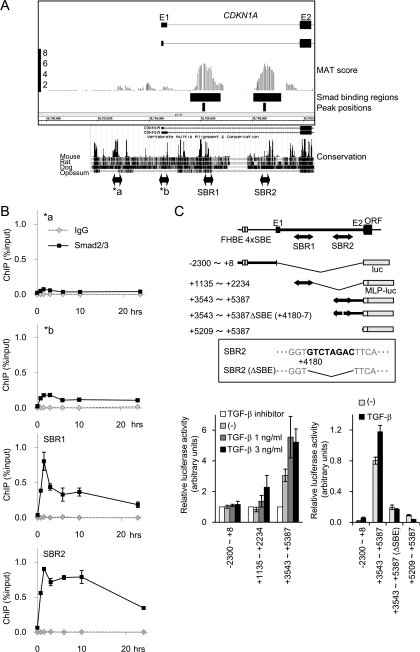
Novel Smad binding regions in the first intron of the CDKN1A gene. (A) Schematic representation of novel Smad binding regions in the CDKN1A gene. MAT scores, significant Smad2/3 binding regions, and peak positions are shown as in Fig. 1B. Interspecies conservation of genomic sequences was obtained from the University of California Santa Cruz genome browser and is shown in the lower panel. E1/E2, exons 1 and 2; a/b, positions of amplicon analyzed in panel B. (B) Validation of Smad2/3 binding to the CDKN1A intronic regions by ChIP-qPCR. HaCaT cells were treated as for Fig. 1A, and Smad2/3 ChIP samples were quantified by ChIP-qPCR, with each primer amplifying the position indicated in panel A. (C) Confirmation of TGF-β-induced transcriptional response in CDKN1A SBR1/2 by luciferase reporter assays. The upper panel shows a schematic representation of the reporter constructs used. The inset shows the identified SBE within SBR2 and its mutant used in the following experiment. (Lower left) HaCaT cells were transfected with the reporters indicated and treated with TGF-β or TGF-β type I receptor kinase inhibitor A44-03 (represented as TGF-β inhibitor, 1 μM), and luciferase activities were determined. (Lower right) HaCaT cells were transfected with the reporters indicated and treated with TGF-β, and luciferase activities were determined. MLP, minimal luciferase promoter; FHBE 4×SBE, reported forkhead transcription factor binding element and four tandem SBEs (45). Error bars represent the standard deviations.
SBRs are enriched near transcription start sites and frequently found in upregulated genes.
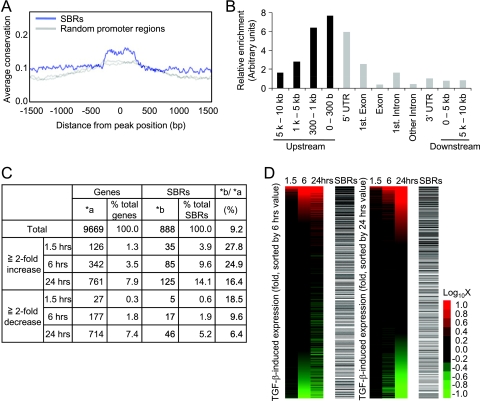
We then examined the overall characteristics of SBRs and compared them to TGF-β-induced gene expression. The sequences of the identified SBRs were relatively conserved in the genomes of vertebrates, suggesting the importance of these regions during evolution (Fig. 2A). We next determined the relative positions of the SBRs from the transcription start sites of RefSeq genes. Although the SBRs were widely distributed from distant upstream regions to intronic regions, they were enriched near the transcription start sites (Fig. 2B). As a result, we identified 888 genes that had SBRs between 5 kb upstream from the transcription start sites and the first intron of their promoters (see Table S2 in the supplemental material). We therefore compared the binding of Smad2/3 to the expression of nearby genes. Results of expression arrays were in agreement with previous analyses of upregulation or downregulation of known target genes, with 1.3% of the genes more than twofold upregulated at 1.5 h by TGF-β stimulation and more genes (7.9%) affected after 24 h of stimulation (Fig. 2C) (35, 53). SBRs were significantly enriched in the early upregulated genes (P < 0.0001) and present in 27.8% of them. Enrichment of SBRs was also observed for the genes upregulated at 6 and 24 h after stimulation (P < 0.0001, Fig. 2D). In contrast, enrichment of Smad2/3 binding was not significant for the downregulated genes (Fig. 2C and D), though there were several novel downregulated genes, e.g., CDC6 and prostaglandin E synthase (PTGES) (Fig. 1C and D; see Fig. S2 in the supplemental material; data not shown).
FIG. 2.
The SBRs are enriched near transcription start sites and frequently found in upregulated genes. (A) Smad binding regions are evolutionarily conserved. The average conservation of Smad binding regions was determined by CEAS analysis tools (23) by using phastCons score (48) information for analysis. As a control, two sets of randomly selected promoter regions were analyzed and plotted as gray lines. (B) Smad binding regions are enriched near transcription start sites. The distribution of the peak positions of Smad binding regions relative to the nearby RefSeq gene was determined, and results were normalized by the distribution of probes designed for the array used. UTR, untranslated region. (C) Summary of expression array results compared to ChIP-chip analysis. A total of 9,669 genes that had values of more than 100 at at least one time point for one of their probes (n = 14,754) were used for the following analysis. Upregulated or downregulated genes were determined compared to 0 h values. The positions of peak signals of SBRs relative to the nearby RefSeq genes were first determined, and regions more than 5 kb upstream from the transcription start site and downstream from the first intron were filtered out. (D) Smad binding regions were enriched at upregulated genes. Probe signals were sorted by the change in expression at 6 (left) or 24 (right) h with TGF-β stimulation and compared to the presence of SBRs (black bars). Changes in expression were visualized by heat mapping.
Colocalization of ETS and TFAP2 sites with canonical SBEs and AP-1 sites.
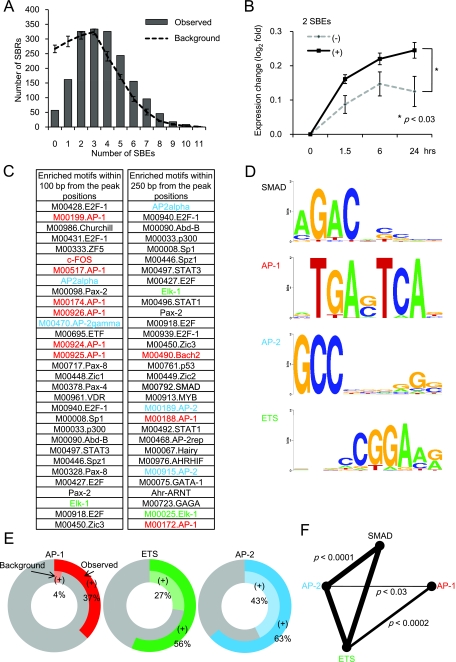
To characterize the DNA sequences in SBRs, we first counted the SBEs (AGAC or GTCT) within 250 bp of the peak signal positions. The numbers of SBEs in SBRs were significantly higher than those of background sequences, with only a few regions with no SBEs or only one SBE (Fig. 3A). Of note, target genes with SBRs containing multiple SBEs were significantly induced by TGF-β compared to those with one SBE or no SBEs (Fig. 3B). On the other hand, inverted tandem repeats of SBEs, GTCTAGAC (54), were rarely observed and found in only 14 SBRs (0.8%).
FIG. 3.
Enrichment of ETS and TFAP2 sites in SBRs. (A) Canonical Smad binding elements are enriched in SBRs. GTCT or AGAC SBEs were counted from repetitive-sequence-masked DNA sequences of 250 bp from the peak position of each SBR (Observed) and compared to the average result of five shuffled sequences (Background). Error bars represent the standard deviations. (B) Number of SBEs correlated with the magnitude of the TGF-β response. Changes in expression, compared to 0 h, of all of the target genes with SBRs were log2 transformed and subdivided by the number of SBEs (zero to one versus two or more). Mean values are shown. Error bars represent the standard deviations. Significance of difference by SBE number was determined by repeated-measures analysis of variance. (C) Identification of enriched transcription factor binding motifs in SBRs. DNA sequences of 100 or 250 bp from the peak position of each SBR were analyzed by CEAS, and the top 31 significantly enriched motifs are shown. There were many matrices defined for the same transcription factors, and AP-1, ETS, and AP-2 sites are categorized and colored red, green, and blue, respectively. The Smad binding element (M00792.SMAD) is shown in bold characters. As a control, we analyzed randomly selected promoter regions by CEAS and confirmed that enrichment of AP-1, ETS, and AP-2 sites was less significant than in Smad2/3 ChIP-chip results (data not shown). (D) Schematic representation of some of the enriched transcription factor binding motifs. Representative matrix data for each enriched transcription factor binding site are shown by Weblogo (8) and were obtained from the CEAS web tool. Sites in panel C: SMAD, M00792.SMAD; AP-1, M00199; AP-2, AP-2alpha; ETS, Elk-1. (E) Frequencies of AP-1, AP-2, and ETS sites in Smad2/3 binding regions. Frequencies of AP-1, AP-2, and ETS sites 250 bp from the peak position of SBRs were determined by a pattern-based identification approach (outer circles) and compared to their shuffled background sequences (inner circles). (F) Smad, ETS, and AP-2 sites appeared to be found in the same Smad binding regions. Pairwise analysis of each enriched motif was performed with Fisher's exact probability test, with significance presented as lines. Broader lines indicate greater significance.
We further analyzed the SBRs by CEAS (23) and TRANSFAC (39) and observed many significantly enriched transcription factor binding motifs near the peak signal positions determined by the position-weighted matrix method. Indeed, SBE was found to be significantly enriched in the SBRs by this analysis; however, its significance was weaker than those of other binding motifs (Fig. 3C). One of the enriched motifs observed within 100 bp from the peak signals was several types of AP-1 site, where Smad complexes have been reported to directly bind, instead of canonical SBE (57). E2F sites were also significantly enriched in the SBRs (Fig. 3C). They are known to serve as GC-rich Smad binding sequences, where Smad proteins cooperate with E2F4/5 and p107 to repress gene expression (4, 19, 36). There were other transcription factor binding sites, including Sp1 and p53 sites, where cooperation with Smad signaling has been reported (7, 41, 43). In addition, we found unexpectedly significant enrichment of AP-2 sites (present in 63% of all SBRs) to which a family of TFAP2 molecules bind (Fig. 3C to E). ETS binding sites, including Elk-1 binding sites (Fig. 3C), and other ETS family binding motifs (data not shown) were also enriched in many of the SBRs (56% of all SBRs). JUN is one of the transcription factors that bind to AP-1 sites. We verified the binding of JUN, ETS1, and TFAP2A to SBRs with or without TGF-β treatment by ChIP-qPCR (see Fig. S3 in the supplemental material). We found increased binding of these factors to SBRs upon TGF-β stimulation, although some induced binding was observed in SBRs that did not have these motifs.
We next determined whether the presence of each motif correlates with that of others. There were significantly positive correlations between the SBEs, ETS-binding sites, and AP-2-binding sites (P < 0.0001, Fig. 3F). Positive correlations were also found between the SBEs and other transcription factor binding motifs, including those of Sp1, p53, E2F, thyroid transcription factor 1 (TTF-1), VDR, Myc (P < 0.0001), Myb (P < 0.0003), and TCF3 (P < 0.03, data not shown). On the other hand, AP-1 sites occurred independently of SBEs, suggesting that AP-1 sites, which were found in about 37% of the SBRs (Fig. 3E), might serve as Smad binding elements instead of canonical SBEs.
Knockdown of ETS1, TFAP2A, or JUN alters TGF-β signaling.
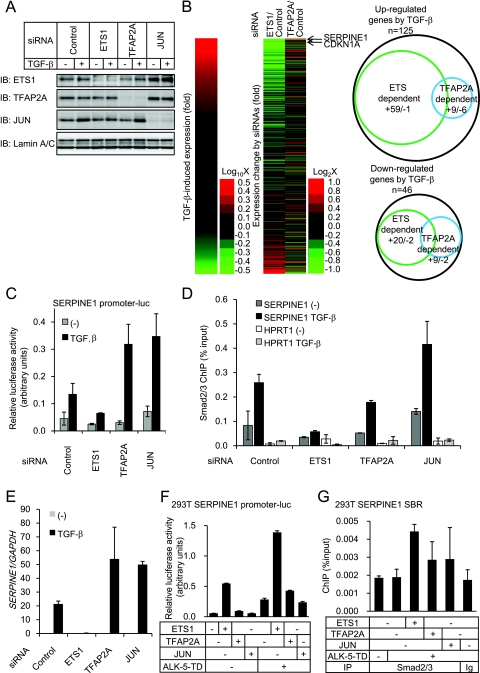
Of the ETS family genes, ETS1 was expressed in HaCaT cells (data not shown), and cooperation of ETS1 with TGF-β signaling for expression of parathyroid hormone-like hormone (PTHLH/PTHrP) in breast cancer cells has been reported (37). TFAP2A was also constitutively expressed in HaCaT cells (data not shown). We found that Smad2/3 bound to the promoters of ETS1 and TFAP2A in the presence of TGF-β (see Fig. S3 in the supplemental material). Although expression of ETS1 mRNA was upregulated by TGF-β and that of TFAP2A mRNA was downregulated by TGF-β (data not shown), their protein levels early after stimulation appeared to be unaffected (Fig. 4A). Expression of the JUN protein was induced by TGF-β (Fig. 4A), as previously reported (50). To address the roles of ETS1 and TFAP2A in TGF-β-induced transcription, ETS1 and TFAP2A were knocked down with siRNAs (Fig. 4A). The cells were then treated with TGF-β, and expression array analyses were performed. Significant impairment of the TGF-β-induced change in the expression of target genes by ETS1 siRNA was observed for both upregulated and downregulated genes at 1.5 and 24 h (P < 0.0001, Fig. 4B and data not shown). Upregulation of 59 genes by TGF-β at 24 h (47.2% of the upregulated genes with SBRs) was reduced more than twofold by ETS1 siRNA (Fig. 4B, right panel). On the other hand, downregulation of 20 genes by TGF-β at 24 h (43.5% of the downregulated genes with SBRs) was attenuated more than twofold by ETS1 siRNA. Essentially the same impairment of the TGF-β effect was obtained with the other siRNA targeting ETS1, as determined by RT-qPCR (data not shown). Silencing the expression of TFAP2A either enhanced or decreased the TGF-β-induced changes in gene expression, but the effect of knockdown of TFAP2A appeared to be less significant than that of ETS1 siRNA (Fig. 4B). As ETS1 siRNA remarkably affected the expression of target genes of TGF-β, we calculated the minimal spacing length between the SBE and the ETS binding site for each SBR. Of the 1,075 SBRs that contained both ETS binding sites and SBEs, 668 had a 50-bp or smaller spacing length but there was no constant distance between them (data not shown).
FIG. 4.
Knockdown of ETS1 and TFAP2A affects target gene expression. (A) Knockdown of ETS1, TFAP2A, or JUN protein in HaCaT cells. Cells were transfected with siRNAs as indicated, 48 h later treated with 3 ng/ml TGF-β for 3 h, and harvested. A 30-μg portion of the nuclear fraction of the cell lysate was applied. The upper three panels show ETS1, TFAP2A, and JUN protein levels, while the lower panel shows lamin A/C as a loading control. IB, immunoblot. (B) Summary of the effects of ETS1 and TFAP2A siRNAs on TGF-β-induced gene expression at 24 h after stimulation. HaCaT cells were transfected with siRNAs as indicated and 48 h later treated with 1 ng/ml TGF-β for 0 or 24 h, and then total RNAs were extracted. For the identified Smad2/3 target genes near the SBRs (Fig. 2C), probe signals were sorted by their TGF-β-induced change in expression in the cells transfected with control siRNA (24 h/0 h, left). The effects of the ETS1 and TFAP2A siRNAs on target gene expression at 24 h after TGF-β stimulation were determined, compared to a control siRNA, and are shown in the same order (right). Arrows indicate the positions where CDKN1A and SERPINE1 reside. For genes either upregulated or downregulated twofold by TGF-β, the effects of the ETS1 and TFAP2A siRNAs are shown as Venn diagrams. A plus sign indicates the number of genes whose TGF-β-induced expression change was inhibited twofold or more by the siRNAs. A minus sign indicates the number of genes whose TGF-β-induced expression change was enhanced twofold or more by the siRNAs. (C) Luciferase reporter activity induced by TGF-β in siRNA-transfected HaCaT cells. Cells were serially transfected with siRNAs and reporter constructs and left unstimulated or stimulated with TGF-β, and reporter activities were determined. Error bars represent the standard errors. (D) Effects of JUN, ETS1, and TFAP2A siRNAs on Smad2/3 ChIP. HaCaT cells were transfected with siRNAs as indicated, left unstimulated or treated 72 h later with 1 ng/ml TGF-β for 1.5 h, and harvested. Smad2/3 binding to the SERPINE1 promoter region or HPRT1 intron as a control was determined by ChIP-qPCR. Error bars represent the standard deviations. (E) Effects of JUN, ETS1, and TFAP2A siRNAs on SERPINE1 mRNA expression. HaCaT cells were transfected with the indicated siRNAs, left unstimulated or stimulated 30 h later with 1 ng/ml of TGF-β, and incubated for an additional 24 h. SERPINE1 expression was determined by RT-qPCR and normalized by GAPDH expression. Error bars represent the standard deviations. (F) Effects of forced expression of ETS1, TFAP2A, or JUN on SERPINE1 promoter-reporter activity in 293T cells. Cells were transfected with the indicated plasmids, and luciferase activities were determined 24 h later. ALK-5-TD: constitutively active type I TGF-β receptor. Error bars indicate the standard deviations. (G) Exogenous ETS1 enhances Smad2/3 binding to the SERPINE1 promoter in 293T cells. Cells were transfected with the indicated plasmid and 24 h later fixed and harvested. ChIP-qPCR was performed as described for panel D. Error bars represent the standard errors. Ig, control IgG.
We then examined the effects of these transcription factors on the activation of the SERPINE1/PAI1 promoter. In addition to ETS and AP-2 sites, there was an AP-1 site in the SBR of SERPINE1. We therefore also knocked down the expression of JUN (Fig. 4A). TGF-β induced transcriptional activity of the promoter, which was inhibited by ETS1 siRNA and enhanced by either TFAP2A or JUN siRNA (Fig. 4C). Moreover, when the effects of these siRNAs on Smad2/3 binding to the SBR were determined by ChIP, ETS1 siRNA and, to a lesser extent, TFAP2A siRNA repressed Smad2/3 binding, whereas JUN siRNA enhanced it (Fig. 4D). We then examined the effect of these siRNAs on the expression of SERPINE1 mRNA (Fig. 4E) and obtained effects similar to those observed in the reporter assay (Fig. 4C). We also examined the effect of exogenous ETS1 in the 293T cells which lack the expression of ETS1 (data not shown). Forced expression of ETS1 resulted in synergistic upregulation of the SERPINE1 promoter activity with the constitutively active type I TGF-β receptor ALK-5-TD (Fig. 4F) and enhancement of Smad2/3 binding to the SERPINE1 promoter (Fig. 4G). In contrast, effects of exogenous TFAP2A and JUN on SERPINE1 transcription and Smad2/3 binding to its promoter were not clear (Fig. 4F and G), which may in part be due to high expression levels of these genes in 293T cells (data not shown). Therefore, in the SERPINE1 promoter, ETS1 enhances both Smad2/3 binding and transcription. On the other hand, TFAP2A and JUN repress transcription, although the mechanisms of repression appear to differ between TFAP2A and JUN.
Novel cis-regulatory regions in the CDKN1A intron for TGF-β-induced transcription.
CDKN1A/p21 is one of the important direct targets of TGF-β signaling for cytostasis, and previous analysis has revealed several transcription factors that participate in its regulation (10, 21, 41, 43, 45). By ChIP-chip analysis, we identified two novel SBRs in the first intronic region of the CDKN1A gene, designated Smad binding region 1 (SBR1) and SBR2 (Fig. 5A). Consistent with these findings, Smad2/3 bound to SBR1 and SBR2 much more strongly than to the known Smad2/3 binding regions (41, 45) by ChIP-qPCR (Fig. 5B). Luciferase reporter assays confirmed the induction of transcriptional activity of SBR1 and SBR2 by TGF-β (Fig. 1D and 5C). SBR2 contained inverted tandem SBEs (GTCTAGAC) that are rarely found but likely to be essential for TGF-β-induced transcriptional activity in this region (Fig. 5C).
ETS1, TFAP2A, and JUN affect CDKN1A expression and cytostasis.
Through transcription factor binding motif analyses (Fig. 3C to E), we identified ETS, AP1, and AP2 sites in both CDKN1A SBR1 and SBR2. We then examined the effects of knockdown of ETS1, TFAP2A, and JUN on the TGF-β-induced transcription of CDKN1A through SBR1 or SBR2. The transcriptional activity of the SBR1 reporter appeared to be predominantly regulated by JUN, since knockdown of JUN greatly enhanced its activity (Fig. 6A). Knockdown of either ETS1 or TFAP2A reduced TGF-β-induced transcriptional activity, but the effects of knockdown of these genes were relatively small compared to that of JUN. In the SBR2 reporter, TGF-β-induced transcriptional activity was strongly inhibited by both ETS1 and TFAP2A siRNAs. We further examined, by ChIP-qPCR, whether these transcription factors affect the recruitment of Smad2/3 to the promoter regions. Knockdown of ETS1 resulted in reduced Smad2/3 binding to both of these regions upon TGF-β stimulation. TFAP2A siRNA also appeared to inhibit Smad2/3 binding to SBR1, though only a weak effect was observed for Smad2/3 binding to SBR2. On the other hand, JUN knockdown enhanced Smad2/3 binding to SBR1 in the presence of TGF-β stimulation, although binding to SBR2 was not significantly affected by JUN knockdown (Fig. 6B). We further analyzed the importance of the ETS binding site in CDKN1A SBR2 and observed severe impairment of transcriptional activity upon ETS site deletion, which was similar to the effect of SBE deletion (Fig. 6C).
FIG. 6.
ETS1, TFAP2A, and JUN affect TGF-β-induced CDKN1A expression and cytostasis. (A) Luciferase reporter activity induced by TGF-β in siRNA-transfected HaCaT cells. Cells were serially transfected with siRNA and reporter constructs and left unstimulated or stimulated with TGF-β, and reporter activities were determined. Error bars represent the standard errors. (B) Effects of JUN, ETS1, and TFAP2A siRNAs on Smad2/3 ChIP. HaCaT cells were transfected with siRNAs as indicated, left unstimulated or treated 72 h later with 1 ng/ml TGF-β for 1.5 h, and harvested. Smad2/3 binding to the indicated promoter regions was determined by ChIP-qPCR. Error bars represent the standard deviations. (C) Deletion of an ETS site in CDKN1A SBR2 results in loss of TGF-β-induced transcription. (Upper panel) Partial DNA sequence of CDKN1A SBR2 with identified SBE and ETS binding site. Numbers indicate relative positions from the transcription start site. (Lower panel) HaCaT cells were transfected with luciferase reporter constructs as indicated and stimulated with TGF-β. Transcriptional activity was then determined. ΔSBE, CDKN1A +3543 to +5387 reporter construct with deletion of SBE (+4180 to +4187); ΔETS, deletion construct of ETS binding site (+4211 to +4217). (D) Effects of JUN, ETS1, and TFAP2A siRNAs on CDKN1A mRNA expression. HaCaT cells were treated as for Fig. 4E, and CDKN1A expression was determined by RT-qPCR. Error bars represent the standard deviations. (E) Immunoblotting (IB) of CDKN1A protein in siRNA-transfected cells. HaCaT cells were transfected with the siRNAs indicated, treated with 3 ng/ml TGF-β for 3 h, and lysed. The top panel shows CDKN1A expression, and the bottom panel shows α-tubulin as a loading control. (F) Thymidine incorporation assay of siRNA-transfected HaCaT cells. Cells were transfected with siRNAs and 24 h later treated with TGF-β as indicated, and [3H]thymidine incorporation was determined. Values are n-fold changes relative to no TGF-β treatment. Error bars represent the standard deviations. (G) Knockdown of CDKN1A by siRNA. HaCaT cells were transfected with CDKN1A siRNA as for panel D, and CDKN1A expression was determined by RT-qPCR at the indicated time points after treatment with TGF-β (1 ng/ml). Error bars represent the standard deviations. (H) Thymidine incorporation assay of HaCaT cells transfected with the CDKN1A siRNA. Cells were treated as for panel F, and the effect of CDKN1A siRNA was determined. Error bars represent the standard deviations.
Consistent with these findings, ETS1 siRNA inhibited the induction of both CDKN1A mRNA and protein by TGF-β (Fig. 6D and E). Conversely, JUN knockdown enhanced CDKN1A induction by TGF-β. In contrast to the results of the reporter assay, TFAP2A siRNA also enhanced the expression of CDKN1A, suggesting that TFAP2A has additional effects on the expression of CDKN1A through other mechanisms. When we determined the effects of these factors on the TGF-β-induced cytostatic response, we found that cytostasis was inhibited by ETS1 siRNA but was enhanced by TFAP2A or JUN siRNA (Fig. 6F). Accordingly, we confirmed that TGF-β-induced cytostasis was impaired when CDKN1A was knocked down by an siRNA (Fig. 6G and H). Although TGF-β induces cell growth arrest through various molecules, including c-Myc and CDKN2B/p15 (47), these findings suggest that regulation of CDKN1A expression appears to play a central role in TGF-β-induced cell growth arrest.
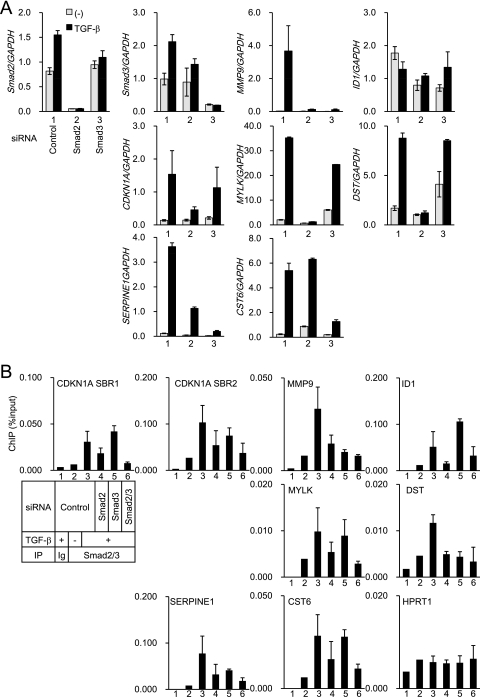
Comparison of the Smad2- or Smad3-specific gene expression profiles and Smad2/3 binding to the promoters.
Several reports have revealed that Smad2 and Smad3 have distinct roles in the expression of some target genes and exhibit certain different biological functions (32). To address this issue, we selectively knocked down Smad2 or Smad3 and obtained gene expression profiles by microarrays. The efficiency and specificity of the siRNAs were confirmed by RT-qPCR (Fig. 7A), expression microarray, and immunoblot analyses (data not shown). The efficiency of Smad3 silencing was relatively weak, as determined by RT-qPCR, but almost no Smad3 band was detected by immunoblotting (data not shown). As a result, we identified several Smad2- or Smad3-dependent target genes. TGF-β-induced myosin light chain kinase (MYLK) and dystonin (DST) expression was strongly inhibited only by the Smad2 siRNA, and cystatin E/M (CST6) expression was selectively inhibited by the Smad3 siRNA. TGF-β-induced changes in expression of CDKN1A, SERPINE1, matrix metallopeptidase 9 (MMP9), and ID1 was repressed by both Smad2 and Smad3 siRNAs. Of these, SERPINE1 expression was more dependent on Smad3 than on Smad2, while CDKN1A expression was more dependent on Smad2 than on Smad3. Smad2 or Smad3 dependency was confirmed by RT-qPCR with other siRNAs (data not shown).
FIG. 7.
Effects of Smad2 and Smad3 siRNAs on TGF-β-induced gene expression and Smad2/3 binding to the promoters. (A) HaCaT cells were transfected with Smad2 or Smad3 siRNAs and 48 h later stimulated with 1 ng/ml TGF-β for 24 h. Levels of gene expression were determined by RT-qPCR and normalized by GAPDH. Each lane represents cells treated with control siRNA (lane 1), Smad2 siRNA (lane 2), or Smad3 siRNA (lane 3). Error bars represent the standard deviations. (B) HaCaT cells were transfected with siRNAs, 48 h later treated with TGF-β for 1.5 h, and fixed. ChIP-qPCR was performed as for Fig. 4D and normalized by input DNA. For double knockdown of Smad2 and Smad3, the total amounts of siRNA were adjusted to be the same as in the other samples. Each lane represents cells treated with control siRNA (lanes 1 to 3), Smad2 siRNA (lane 4), Smad3 siRNA (lane 5), or Smad2 and Smad3 siRNAs (lane 6). Samples were immunoprecipitated (IP) with control IgG (lane 1) or Smad2/3 antibody (lanes 2 to 6). Cells were not treated with TGF-β in lane 2. Error bars represent the standard deviations.
We then knocked down the expression of Smad2, Smad3, or both and determined Smad2/3 binding to the promoters of these genes by ChIP-qPCR. Using the input cell lysate used for the ChIP experiment, we confirmed that the Smad2 and/or Smad3 proteins were efficiently knocked down (data not shown). Unexpectedly, we found that Smad2/3 binding to these promoters was only partially affected by knockdown of either Smad2 or Smad3 (Fig. 7B). An efficient decrease in Smad2/3 binding to these promoters was achieved only when both Smad2 and Smad3 were knocked down. Thus, when the expression of either Smad2 or Smad3 is silenced, another one may bind to the promoters and the total amounts of Smad2/3 bound to the promoters are only weakly affected. These findings suggest that specific effects of Smad2 or Smad3 on target gene expression are determined not only by the total amounts of Smad2/3 bound to the promoters but by other Smad2- or Smad3-specific functions on the promoters. We finally obtained the SBR sequences of either Smad2-specific or Smad3-specific target genes and compared the enriched motifs. ETS, AP-1, and AP-2 sites were significantly enriched in both SBRs of Smad2- and Smad3-specific target genes. Thus, other cofactors may determine Smad2- or Smad3-specific gene expression, which need to be determined in the future.
DISCUSSION
Following the discovery of the Smad family, regulation of gene expression by TGF-β signaling has been extensively studied. Several different motifs have been reported as TGF-β-responsive elements based on the analysis of different target genes. The wide variety of motifs essential to the regulation of gene expression by TGF-β is due, at least in part, to the presence of many Smad binding partners. In this regard, recent studies have focused on the analysis of cell- and tissue-specific and context-dependent regulation of gene expression by TGF-β (38). However, this has made it difficult to predict the regulatory factors and Smad binding positions of specific target promoters. The landscape of Smad2/3 binding regions presented here thus reveals more general aspects of transcriptional regulation by Smad2/3.
ETS1 and TFAP2A as major transcription factors that regulate the function of Smad2/3.
ETS1 is a member of the ETS domain family of genes. ETS1 expression is induced by several growth factor signals, and roles for ETS1 in hematopoietic development, angiogenesis, and anti- or proapoptosis have been reported (15). On the other hand, overexpression of ETS1 is observed in many types of cancers and is related to a poor prognosis, an invasive phenotype, or metastasis of cancer (15). ETS1 has also been reported to participate in the regulation of some specific target genes of TGF-β. Lindemann et al. (37) showed that ETS1 binds both SBE and ETS binding motifs in cooperation with Smad proteins at the PTHrP/PTHLH promoter and activates its transcription in MDA-MB231 breast cancer cells. Indeed, we identified the same SBR and ETS binding site in it by ChIP-chip (see Fig. S4A and B in the supplemental material). Moreover, we found that ETS1 siRNA impaired PTHLH expression in HaCaT cells (see Fig. S4C in the supplemental material), suggesting that some of the SBRs, and even the regulation by ETS1, are commonly observed in different types of cells. Of note, TFAP2A siRNA enhanced PTHLH expression, further suggesting its engagement in TGF-β signaling. Shirakihara et al. (46) reported the functional engagement of ETS1 in the TGF-β-induced epithelial-to-mesenchymal transition in NMuMG normal murine mammary gland cells via induction of the δEF1 gene. We found TGF-β-induced δEF1 expression in HaCaT cells, but in contrast to the effect on PTHLH, ETS1 siRNA did not affect its expression (data not shown). Thus, some effects of ETS1 on TGF-β signaling may be induced in a cell type-specific fashion. The present study also suggests, for the first time, that the effects of ETS1 in TGF-β-induced transcription are generally observed due to frequent copresentation of its binding sites with SBE. It is difficult, however, to determine whether ETS binding sites in all of the identified SBRs are required for exhibition of the regulatory effects of ETS1. We also found that some TGF-β-target genes that were affected by ETS1 siRNA lacked ETS sites in the analyzed SBR sequences (data not shown). This was probably due to the presence of ETS sites outside the sequence analyzed or to effects of ETS1 on TGF-β signaling other than promotion of DNA binding of Smad2/3. We found a physical interaction of ETS1 or TFAP2A with Smad2/3 (see Fig. S5A and B in the supplemental material), which may account for such additional effects of ETS1 on TGF-β signaling. This physical interaction may also adversely affect TGF-β signaling when ETS1 overexpression occurs in cancer, since there is an antagonistic effect of exogenously overexpressed ETS1 on TGF-β signaling in fibroblasts (9). In contrast, our finding that loss of ETS1 inhibited CDKN1A induction by TGF-β may also be important for some pathological condition to evade the cytostatic program. Taken together, these findings suggest that the resultant effects of ETS1 on TGF-β signaling may be important for many of the Smad2/3-binding target genes.
To our knowledge, the most enriched motif, AP-2, has not been reported to be a regulator of TGF-β signaling. The family of TFAP2 genes plays important roles in ectodermal development, especially in craniofacial development (56). TFAP2A was also reported to inhibit cancer cell proliferation via enhancement of phorbol ester-induced CDKN1A expression (55). On the other hand, several studies have demonstrated upregulation of TFAP2A in cancers (6, 40), suggesting complex roles for TFAP2A in carcinogenesis. Our findings suggested that TFAP2A is involved in the modulation of TGF-β-induced transcriptional activity in both positive and negative fashions, and unlike the finding of a previous study (55), TFAP2A appeared to inhibit CDKN1A expression upon TGF-β stimulation in HaCaT cells. Although we could not determine the critical region in the CDKN1A promoter, TFAP2A may function as a CDKN1A gene repressor, as reported for the regulation of C/EBPα and K3 keratin expression (5, 24). In addition, our findings regarding the effects of TFAP2A on the SERPINE1 promoter in Smad-mediated transcription (Fig. 4C and D) suggest that TFAP2A may form an inhibitory complex including Smad2/3 and enhance their binding to the SERPINE1 gene promoter.
Possible role of the AP-1 site as another Smad2/3 binding element.
Unlike the ETS and AP-2 sites, AP-1 sites did not coexist with SBEs but were remarkably enriched near the peak Smad binding positions. One of the transcription factors, JUN, which binds AP-1 sites, appeared to repress Smad2/3 binding and their transcription of some of the targets with SBRs. These findings, together with those of previous studies (57), suggest that AP-1 sites may function as Smad binding elements in some SBRs, without the involvement of SBEs. However, we were not able to determine which of the motifs within the SBRs are actually bound by Smad2/3. Improvement of the resolution of SBRs may solve this question in the future.
Possible roles of other regulatory factors in TGF-β-induced transcription.
There are also several transcription factor binding sites other than AP-1, AP-2, and ETS enriched in SBRs. Although we have not determined their roles in the present study, these enriched motifs contained binding sites for known cofactors of TGF-β signaling and may also function as modulators of subsets of target genes. Some of these motifs were bound by tissue-specific factors, e.g., TTF-1, which is expressed in the thyroid and lung (34). Thus, analysis of the effects of these tissue-specific factors on TGF-β signaling may reveal novel tissue-specific regulation of TGF-β signaling in the future.
The previous definition of “direct” target genes of TGF-β signaling depended principally on their early responses. However, we found that Smad binding early after TGF-β stimulation was also significantly correlated with later changes in the expression of upregulated genes. This may reflect a requirement for additional molecules, e.g., ETS1 and TFAP2A, for transcription or a modification of chromatin status by histone modification or chromatin remodeling (44, 51). As a result, previous studies may have overlooked a substantial number of the direct target genes of Smad signaling because of their later expression change by TGF-β stimulation.
In the case of known target genes of TGF-β, we also found novel SBRs and novel regulatory mechanisms. Although previous studies examined the regulation of CDKN1A by its upstream promoter sequence, the present study revealed that the first intronic region may also serve as an important cis-regulatory region. Since JUN, TFAP2A, and ETS1 are upregulated by many stimuli, the first intronic region may function as a regulatory center for various signals. Likewise, there may be previously overlooked intronic regions in other genes which are important for regulation by TGF-β, since first introns are the most enriched downstream regions where SBRs are located. Moreover, in addition to CDKN1A, we also identified Smad2/3 binding sites in the SMAD7 and JUNB promoters, which differed from those reported in previous studies (data not shown; 11, 26, 42). Thus, transcriptional regulation of such target genes by the identified transcription factors in the SBRs should be reevaluated in the future.
What determines Smad2- and Smad3-specific gene expression?
The experiments with Smad2 and Smad3 siRNAs revealed that differences in the affinity of the Smad complexes for the promoters modulated by Smad2 or Smad3 are not sufficient to explain their functional differences. Some Smad2- or Smad3-specific cofactors may determine their transcriptional activity on different promoters. Our attempt to determine such factors based on the SBR sequences was, however, not successful in revealing the difference between Smad2 and Smad3. Improvement of the resolution of SBRs may help in the identification of specific motifs in the future.
Some targets of the Smad pathway remain to be identified.
There are some problems associated with the ChIP-based analyses performed in the present study. One is that the number of SBRs may differ, depending on the threshold (see Fig. S6A in the supplemental material). Non-Smad pathways of TGF-β signaling that are transmitted by signaling molecules other than members of the Smad family have been reported (14). However, we could not determine whether “expression-defined” target genes without significant Smad binding in ChIP-chip were actual targets of non-Smad pathways. These target genes may have SBRs when the cutoff threshold is changed. It is also possible that these target genes have Smad binding sites distantly located from the promoter regions. These target genes without SBRs may also be induced by de novo transcription factors regulated by SBRs. We analyzed published cycloheximide-treated expression microarray data (1) and identified some cycloheximide-sensitive genes without SBRs, e.g., that for fibronectin 1 (see Fig. S6B in the supplemental material; data not shown). Another problem is that, as transcription factors form huge complexes with various cofactors, specific epitopes recognized by antibodies may be masked by components in the Smad complexes. This may explain why some reported Smad binding regions exhibited weak signals in the present study.
Nonetheless, the findings presented here solved some of the remaining problems related to Smad signaling, i.e., where Smad proteins bind and which transcription factors are associated with Smad proteins at the SBRs in general, by a high-throughput method. The present findings are thus of great importance in understanding the mechanisms of TGF-β-induced transcriptional regulation.
Supplementary Material
Acknowledgments
We thank Takuya Shirakihara and Masao Saitoh for ETS1 cDNA. We also thank Noriko Kaneniwa, Hiroko Meguro, Aki Hanyu, Yasushi Yuki, and Etsuko Kobayashi for technical assistance.
This study was mainly supported by a grant from the Genome Network Project from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan, and partly supported by KAKENHI (Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan).
Footnotes
Published ahead of print on 27 October 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Akiyoshi, S., M. Ishii, N. Nemoto, M. Kawabata, H. Aburatani, and K. Miyazono. 2001. Targets of transcriptional regulation by transforming growth factor beta: expression profile analysis using oligonucleotide arrays. Jpn. J. Cancer Res. 92257-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boukamp, P., R. T. Petrussevska, D. Breitkreutz, J. Hornung, A. Markham, and N. E. Fusenig. 1988. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 106761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll, J. S., C. A. Meyer, J. Song, W. Li, T. R. Geistlinger, J. Eeckhoute, A. S. Brodsky, E. K. Keeton, K. C. Fertuck, G. F. Hall, Q. Wang, S. Bekiranov, V. Sementchenko, E. A. Fox, P. A. Silver, T. R. Gingeras, X. S. Liu, and M. Brown. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 381289-1297. [DOI] [PubMed] [Google Scholar]
- 4.Chen, C. R., Y. Kang, P. M. Siegel, and J. Massague. 2002. E2F4/5 and p107 as Smad cofactors linking the TGF-β receptor to c-myc repression. Cell 11019-32. [DOI] [PubMed] [Google Scholar]
- 5.Chen, T. T., R. L. Wu, F. Castro-Munozledo, and T. T. Sun. 1997. Regulation of K3 keratin gene transcription by Sp1 and AP-2 in differentiating rabbit corneal epithelial cells. Mol. Cell. Biol. 173056-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiefari, E., A. Brunetti, F. Arturi, J. M. Bidart, D. Russo, M. Schlumberger, and S. Filetti. 2002. Increased expression of AP2 and Sp1 transcription factors in human thyroid tumors: a role in NIS expression regulation? BMC Cancer 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cordenonsi, M., S. Dupont, S. Maretto, A. Insinga, C. Imbriano, and S. Piccolo. 2003. Links between tumor suppressors: p53 is required for TGF-β gene responses by cooperating with Smads. Cell 113301-314. [DOI] [PubMed] [Google Scholar]
- 8.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 141188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Czuwara-Ladykowska, J., V. I. Sementchenko, D. K. Watson, and M. Trojanowska. 2002. Ets1 is an effector of the transforming growth factor β (TGF-β) signaling pathway and an antagonist of the profibrotic effects of TGF-β. J. Biol. Chem. 27720399-20408. [DOI] [PubMed] [Google Scholar]
- 10.Datto, M. B., Y. Yu, and X. F. Wang. 1995. Functional analysis of the transforming growth factor β responsive elements in the WAF1/Cip1/p21 promoter. J. Biol. Chem. 27028623-28628. [DOI] [PubMed] [Google Scholar]
- 11.Denissova, N. G., C. Pouponnot, J. Long, D. He, and F. Liu. 2000. Transforming growth factor β-inducible independent binding of SMAD to the Smad7 promoter. Proc. Natl. Acad. Sci. USA 976397-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis, G., Jr., B. T. Sherman, D. A. Hosack, J. Yang, W. Gao, H. C. Lane, and R. A. Lempicki. 2003. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4P3. [PubMed] [Google Scholar]
- 13.Dennler, S., S. Itoh, D. Vivien, P. ten Dijke, S. Huet, and J.-M. Gauthier. 1998. Direct binding of Smad3 and Smad4 to critical TGFβ-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 173091-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derynck, R., and Y. E. Zhang. 2003. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature 425577-584. [DOI] [PubMed] [Google Scholar]
- 15.Dittmer, J. 2003. The biology of the Ets1 proto-oncogene. Mol. Cancer 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ebisawa, T., K. Tada, I. Kitajima, K. Tojo, T. K. Sampath, M. Kawabata, K. Miyazono, and T. Imamura. 1999. Characterization of bone morphogenetic protein-6 signaling pathways in osteoblast differentiation. J. Cell Sci. 1123519-3527. [DOI] [PubMed] [Google Scholar]
- 17.Ehata, S., A. Hanyu, M. Hayashi, H. Aburatani, Y. Kato, M. Fujime, M. Saitoh, K. Miyazawa, T. Imamura, and K. Miyazono. 2007. Transforming growth factor-β promotes survival of mammary carcinoma cells through induction of antiapoptotic transcription factor DEC1. Cancer Res. 679694-9703. [DOI] [PubMed] [Google Scholar]
- 18.Feng, X. H., and R. Derynck. 2005. Specificity and versatility in TGF-β signaling through Smads. Annu. Rev. Cell Dev. Biol. 21659-693. [DOI] [PubMed] [Google Scholar]
- 19.Frederick, J. P., N. T. Liberati, D. S. Waddell, Y. Shi, and X. F. Wang. 2004. Transforming growth factor β-mediated transcriptional repression of c-myc is dependent on direct binding of Smad3 to a novel repressive Smad binding element. Mol. Cell. Biol. 242546-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geng, Y., and R. A. Weinberg. 1993. Transforming growth factor β effects on expression of G1 cyclins and cyclin-dependent protein kinases. Proc. Natl. Acad. Sci. USA 9010315-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomis, R. R., C. Alarcon, W. He, Q. Wang, J. Seoane, A. Lash, and J. Massague. 2006. A FoxO-Smad synexpression group in human keratinocytes. Proc. Natl. Acad. Sci. USA 10312747-12752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horak, C. E., M. C. Mahajan, N. M. Luscombe, M. Gerstein, S. M. Weissman, and M. Snyder. 2002. GATA-1 binding sites mapped in the β-globin locus by using mammalian chIp-chip analysis. Proc. Natl. Acad. Sci. USA 992924-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji, X., W. Li, J. Song, L. Wei, and X. S. Liu. 2006. CEAS: cis-regulatory element annotation system. Nucleic Acids Res. 34W551-W554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang, M. S., Q. Q. Tang, J. McLenithan, D. Geiman, W. Shillinglaw, W. J. Henzel, and M. D. Lane. 1998. Derepression of the C/EBPα gene during adipogenesis: identification of AP-2α as a repressor. Proc. Natl. Acad. Sci. USA 953467-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson, W. E., W. Li, C. A. Meyer, R. Gottardo, J. S. Carroll, M. Brown, and X. S. Liu. 2006. Model-based analysis of tiling-arrays for ChIP-chip. Proc. Natl. Acad. Sci. USA 10312457-12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jonk, L. J., S. Itoh, C. H. Heldin, P. ten Dijke, and W. Kruijer. 1998. Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-β, activin, and bone morphogenetic protein-inducible enhancer. J. Biol. Chem. 27321145-21152. [DOI] [PubMed] [Google Scholar]
- 27.Kaneshiro, K., S. Tsutsumi, S. Tsuji, K. Shirahige, and H. Aburatani. 2007. An integrated map of p53-binding sites and histone modification in the human ENCODE regions. Genomics 89178-188. [DOI] [PubMed] [Google Scholar]
- 28.Kang, Y., C. R. Chen, and J. Massague. 2003. A self-enabling TGF-β response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol. Cell 11915-926. [DOI] [PubMed] [Google Scholar]
- 29.Keeton, M. R., S. A. Curriden, A. J. van Zonneveld, and D. J. Loskutoff. 1991. Identification of regulatory sequences in the type 1 plasminogen activator inhibitor gene responsive to transforming growth factor β. J. Biol. Chem. 26623048-23052. [PubMed] [Google Scholar]
- 30.Kim, J., K. Johnson, H. J. Chen, S. Carroll, and A. Laughon. 1997. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature 388304-308. [DOI] [PubMed] [Google Scholar]
- 31.Koinuma, D., M. Shinozaki, A. Komuro, K. Goto, M. Saitoh, A. Hanyu, M. Ebina, T. Nukiwa, K. Miyazawa, T. Imamura, and K. Miyazono. 2003. Arkadia amplifies TGF-β superfamily signalling through degradation of Smad7. EMBO J. 226458-6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kretschmer, A., K. Moepert, S. Dames, M. Sternberger, J. Kaufmann, and A. Klippel. 2003. Differential regulation of TGF-β signaling through Smad2, Smad3 and Smad4. Oncogene 226748-6763. [DOI] [PubMed] [Google Scholar]
- 33.Labbé, E., C. Silvestri, P. A. Hoodless, J. L. Wrana, and L. Attisano. 1998. Smad2 and Smad3 positively and negatively regulate TGF-β-dependent transcription through the forkhead DNA-binding protein FAST2. Mol. Cell 2109-120. [DOI] [PubMed] [Google Scholar]
- 34.Lazzaro, D., M. Price, M. de Felice, and R. Di Lauro. 1991. The transcription factor TTF-1 is expressed at the onset of thyroid and lung morphogenesis and in restricted regions of the foetal brain. Development 1131093-1104. [DOI] [PubMed] [Google Scholar]
- 35.Levy, L., and C. S. Hill. 2005. Smad4 dependency defines two classes of transforming growth factor β (TGF-β) target genes and distinguishes TGF-β-induced epithelial-mesenchymal transition from its antiproliferative and migratory responses. Mol. Cell. Biol. 258108-8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, J. M., P. P. Hu, X. Shen, Y. Yu, and X. F. Wang. 1997. E2F4-RB and E2F4-p107 complexes suppress gene expression by transforming growth factor β through E2F binding sites. Proc. Natl. Acad. Sci. USA 944948-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindemann, R. K., P. Ballschmieter, A. Nordheim, and J. Dittmer. 2001. Transforming growth factor β regulates parathyroid hormone-related protein expression in MDA-MB-231 breast cancer cells through a novel Smad/Ets synergism. J. Biol. Chem. 27646661-46670. [DOI] [PubMed] [Google Scholar]
- 38.Massagué, J., J. Seoane, and D. Wotton. 2005. Smad transcription factors. Genes Dev. 192783-2810. [DOI] [PubMed] [Google Scholar]
- 39.Matys, V., E. Fricke, R. Geffers, E. Gossling, M. Haubrock, R. Hehl, K. Hornischer, D. Karas, A. E. Kel, O. V. Kel-Margoulis, D. U. Kloos, S. Land, B. Lewicki-Potapov, H. Michael, R. Munch, I. Reuter, S. Rotert, H. Saxel, M. Scheer, S. Thiele, and E. Wingender. 2003. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31374-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McPherson, L. A., G. W. Woodfield, and R. J. Weigel. 2007. AP2 transcription factors regulate expression of CRABPII in hormone responsive breast carcinoma. J. Surg. Res. 13871-78. [DOI] [PubMed] [Google Scholar]
- 41.Moustakas, A., and D. Kardassis. 1998. Regulation of the human p21/WAF1/Cip1 promoter in hepatic cells by functional interactions between Sp1 and Smad family members. Proc. Natl. Acad. Sci. USA 956733-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagarajan, R. P., J. Zhang, W. Li, and Y. Chen. 1999. Regulation of Smad7 promoter by direct association with Smad3 and Smad4. J. Biol. Chem. 27433412-33418. [DOI] [PubMed] [Google Scholar]
- 43.Pardali, K., A. Kurisaki, A. Moren, P. ten Dijke, D. Kardassis, and A. Moustakas. 2000. Role of Smad proteins and transcription factor Sp1 in p21(Waf1/Cip1) regulation by transforming growth factor-β. J. Biol. Chem. 27529244-29256. [DOI] [PubMed] [Google Scholar]
- 44.Ross, S., E. Cheung, T. G. Petrakis, M. Howell, W. L. Kraus, and C. S. Hill. 2006. Smads orchestrate specific histone modifications and chromatin remodeling to activate transcription. EMBO J. 254490-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seoane, J., H. V. Le, L. Shen, S. A. Anderson, and J. Massague. 2004. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117211-223. [DOI] [PubMed] [Google Scholar]
- 46.Shirakihara, T., M. Saitoh, and K. Miyazono. 2007. Differential regulation of epithelial and mesenchymal markers by δEF1 proteins in epithelial mesenchymal transition induced by TGF-β. Mol. Biol. Cell 183533-3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siegel, P. M., and J. Massague. 2003. Cytostatic and apoptotic actions of TGF-β in homeostasis and cancer. Nat. Rev. Cancer 3807-821. [DOI] [PubMed] [Google Scholar]
- 48.Siepel, A., G. Bejerano, J. S. Pedersen, A. S. Hinrichs, M. Hou, K. Rosenbloom, H. Clawson, J. Spieth, L. W. Hillier, S. Richards, G. M. Weinstock, R. K. Wilson, R. A. Gibbs, W. J. Kent, W. Miller, and D. Haussler. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 151034-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wendt, K. S., K. Yoshida, T. Itoh, M. Bando, B. Koch, E. Schirghuber, S. Tsutsumi, G. Nagae, K. Ishihara, T. Mishiro, K. Yahata, F. Imamoto, H. Aburatani, M. Nakao, N. Imamoto, K. Maeshima, K. Shirahige, and J. M. Peters. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451796-801. [DOI] [PubMed] [Google Scholar]
- 50.Wong, C., E. M. Rougier-Chapman, J. P. Frederick, M. B. Datto, N. T. Liberati, J. M. Li, and X. F. Wang. 1999. Smad3-Smad4 and AP-1 complexes synergize in transcriptional activation of the c-Jun promoter by transforming growth factor β. Mol. Cell. Biol. 191821-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xi, Q., W. He, X. H. Zhang, H. V. Le, and J. Massague. 2008. Genome-wide impact of the BRG1 SWI/SNF chromatin remodeler on the transforming growth factor β transcriptional program. J. Biol. Chem. 2831146-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yingling, J. M., M. B. Datto, C. Wong, J. P. Frederick, N. T. Liberati, and X. F. Wang. 1997. Tumor suppressor Smad4 is a transforming growth factor β-inducible DNA binding protein. Mol. Cell. Biol. 177019-7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zavadil, J., M. Bitzer, D. Liang, Y. C. Yang, A. Massimi, S. Kneitz, E. Piek, and E. P. Bottinger. 2001. Genetic programs of epithelial cell plasticity directed by transforming growth factor-β. Proc. Natl. Acad. Sci. USA 986686-6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zawel, L., J. L. Dai, P. Buckhaults, S. Zhou, K. W. Kinzler, B. Vogelstein, and S. E. Kern. 1998. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol. Cell 1611-617. [DOI] [PubMed] [Google Scholar]
- 55.Zeng, Y. X., K. Somasundaram, and W. S. el-Deiry. 1997. AP2 inhibits cancer cell growth and activates p21WAF1/CIP1 expression. Nat. Genet. 1578-82. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, J., S. Hagopian-Donaldson, G. Serbedzija, J. Elsemore, D. Plehn-Dujowich, A. P. McMahon, R. A. Flavell, and T. Williams. 1996. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature 381238-241. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, Y., X. H. Feng, and R. Derynck. 1998. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription. Nature 394909-913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.