Abstract
Saccharomyces cerevisiae cells are capable of responding to mating pheromone only prior to their exit from the G1 phase of the cell cycle. Ste5 scaffold protein is essential for pheromone response because it couples pheromone receptor stimulation to activation of the appropriate mitogen-activated protein kinase (MAPK) cascade. In naïve cells, Ste5 resides primarily in the nucleus. Upon pheromone treatment, Ste5 is rapidly exported from the nucleus and accumulates at the tip of the mating projection via its association with multiple plasma membrane-localized molecules. We found that concomitant with its nuclear export, the rate of Ste5 turnover is markedly reduced. Preventing nuclear export destabilized Ste5, whereas preventing nuclear entry stabilized Ste5, indicating that Ste5 degradation occurs mainly in the nucleus. This degradation is dependent on ubiquitin and the proteasome. We show that Ste5 ubiquitinylation is mediated by the SCFCdc4 ubiquitin ligase and requires phosphorylation by the G1 cyclin-dependent protein kinase (cdk1). The inability to efficiently degrade Ste5 resulted in pathway activation and cell cycle arrest in the absence of pheromone. These findings reveal that maintenance of this MAPK scaffold at an appropriately low level depends on its compartment-specific and cell cycle-dependent degradation. Overall, this mechanism provides a novel means for helping to prevent inadvertent stimulus-independent activation of a response and for restricting and maximizing the signaling competence of the cell to a specific cell cycle stage, which likely works hand in hand with the demonstrated role that G1 Cdk1-dependent phosphorylation of Ste5 has in preventing its association with the plasma membrane.
Scaffold proteins play a pivotal role in spatial and temporal regulation of multitiered mitogen-activated protein kinase (MAPK) cascades (8, 30, 107). Scaffold protein function can be controlled at several different levels, including phosphorylation, oligomerization, and subcellular localization, which can dramatically influence signaling (5, 21, 61).
A well-characterized scaffold-dependent MAPK pathway drives the mating pheromone response in budding yeast Saccharomyces cerevisiae (15). The occupancy of a heterotrimeric G-protein-coupled receptor by pheromone results in release of its associated membrane-tethered Gβγ (Ste4-Ste18) complex. Ste5 scaffold protein (917 residues) is recruited to the plasma membrane via its association with this freed Gβγ (106) and by additional multivalent contacts with membrane phospholipids mediated by an N-terminal amphipathic α-helix (PM motif) (111) and an internal PH domain (34). Because Ste5 is also able to bind a MAPK kinase kinase (Ste11), a MAPK kinase (Ste7), and two MAPKs (Fus3 and Kss1) (102), membrane recruitment of Ste5 delivers these components to the plasma membrane. Membrane localization of Ste5 juxtaposes its passenger kinases to Ste20, a p21-activated protein kinase that also interacts with membrane phospholipids (94) and requires plasma membrane-tethered and GTP-loaded Cdc42 for its activation (56, 58, 60). GTP-bound Cdc42 is generated in this vicinity via other Gβγ-recruited effectors, especially Far1, which binds the Cdc42 guanine nucleotide exchange factor, Cdc24 (14, 98). Once activated, Ste20 directly phosphorylates and activates the Ste11 MAPK kinase kinase, triggering the MAPK cascade (24, 114).
In naïve haploid cells, Ste5 undergoes continuous nucleocytoplasmic shuttling but is located predominantly in the nucleus (53, 66). In response to pheromone, this flux is dramatically shifted in favor of export, elevating the cytosolic pool of Ste5, thereby raising the number of molecules available for membrane recruitment (66, 79). Pheromone-induced nuclear export of Ste5 requires the exportin, Msn5/Ste21 (66).
Little is known about why Ste5 is located in the nucleus in unstimulated cells. It has been suggested that passage of Ste5 through the nucleus modifies it in an as yet undefined manner to make it “competent” to subsequently promote signaling at the membrane (66, 103). However, other evidence indicates that nuclear shuttling of Ste5 is not necessary for its translocation to the plasma membrane or its function (34, 79, 111) and that reimport into the nucleus contributes to pathway downregulation following initial stimulation (53). It has remained obscure, mechanistically speaking, how nuclear localization of Ste5 contributes to the regulation of pathway activation and signal flux.
Given that Ste5 is the least abundant component of this entire signaling system (≤500 molecules per haploid cell) (38), we suspected that dynamic regulation of the location and level of this scaffold protein provides a critically important control point for influencing the timing, potency, duration, and specificity of signaling in this pathway. Indeed, as described here, we found that the subcellular localization of Ste5 and cell cycle progression have dramatic effects in controlling the stability of Ste5. Our findings provide new insights about the physiological importance of Ste5 nuclear localization and G1 cyclin-dependent protein kinase 1 (CDK1) action in establishment and maintenance of the conditions that preserve signaling fidelity in this system.
MATERIALS AND METHODS
Yeast strains and media.
Strains (Table 1) were cultivated at the indicated temperature (23, 30, or 37°C) in standard rich (yeast extract-peptone [YP]) or defined minimal (synthetic complete [SC]) medium (91) containing either 2% glucose (Glc), 2% raffinose and 0.2% sucrose (Raf-Suc), or 2% galactose (Gal), supplemented, where necessary, with appropriate nutrients to maintain selection for plasmids. For expression of genes under the control of the GAL1 promoter, cells were pregrown to mid-exponential phase in SC containing Raf-Suc and then Gal was added (final concentration, 2%) for various times, as indicated. To retard pheromone proteolysis, culture medium was preadjusted to pH 3.5 with Na succinate (final concentration, 50 mM) (17). Standard techniques for propagation and genetic manipulation of yeast were used (91).
TABLE 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| W303-1A | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | 91 |
| BYB69 | W303-1A lys2Δ::hisG ste5Δ::LYS2 | 50 |
| YAB5a | W303-1A lys2Δ::hisG bar1::hisG ste5Δ::LYS2 | This study |
| YAB8b | W303-1A lys2Δ::hisG bar1::hisG ste5Δ::LYS2 msn5Δ::TRP1 | This study |
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | 10 |
| RCY9321 | BY4741 fus3Δ::KlURA3 kss1Δ::CgHIS3c | R. E. Chen (this laboratory) |
| MHY753 | MATahis3-Δ200 leu2Δ1 ura3-52 lys2-801 trp1Δ63 ade2-101 | 36 |
| MHY754 | MHY753 cim3-1 | 36 |
| BKY48-5C | MATα leu2-3 ura3-52 erg6Δ::LEU2 | 33 |
| BYB84 | MATagal2 leu2 prb1-1122 pep4-3 prc1-407 trp1 ura3-52 ste5Δ | 50 |
| YLG18d | BYB84 ste11Δ::hisG | This study |
| HMK30 | W303-1A msn5Δ::TRP1 | 53 |
| YMK101e | W303-1A msn5Δ::TRP1 yrb1-51ts | This study |
| JTY4427 | MATamsn5Δ::TRP1 yrb1-52ts | 53 |
| JTY4299 | W303-1A msn5Δ::TRP1 apc10-22ts | This study |
| JTY4300 | W303-1A msn5Δ::TRP1 cdc34-2ts | This study |
| JTY4301 | W303-1A msn5Δ::TRP1 cdc4-1ts | This study |
| JTY4327 | W303-1A apc10-22ts | This study |
| JTY4329 | W303-1A cdc34-2ts | This study |
| JTY4330 | W303-1A cdc4-1ts | This study |
| BF264-15D | MATα ade1 his2 leu2-3,112 trp1-1 | 65 |
| YLG90f | BF264-15D msn5Δ::hphNT1 | This study |
| YLG91f | BF264-15D cdc28-13ts msn5Δ::hphNT1 | This study |
| JTY2142 | BF264-15D ura3Δns cln1Δ cln2Δ cln3Δ PGAL1-CLN2 | C. Wittenberg |
| YLG92f | BF264-15D ura3Δns cln1Δ cln2Δ cln3Δ PGAL1-CLN2 msn5Δ::hphNT1 | This study |
Strain YAB5 was a spore from a cross between strains BYB69 and W303-1B bar1::hisG (gift of K. Nasmyth, Institute of Molecular Pathology, Vienna, Austria).
MSN5 was deleted from the genome by a one-step PCR-based gene replacement using appropriate synthetic oligonucleotide primers and vector pRS314 (92) as the template DNA. Authentic transplacement of the MSN5 locus was verified by genomic PCR using primers within the TRP1 gene and in the msn5 flanking region.
KlURA3 indicates that the URA3 gene was from Kluyveromyces lactis, and CgHIS3 indicates that the HIS3 gene was from Candida glabrata.
STE11 was deleted from the genome by standard one-step PCR-mediated gene replacement (64) with the hisG::URA3::hisG cassette contained on vector pNKY51 (1). Gene replacements were verified by PCR, and Ura− transformants were then selected on medium containing 5-fluoroorotic acid.
Strain YMK101 was a spore from a cross between W303-1A yrb1-51 (YMK94) and W303-1B msn5::TRP1 (YMK68), a MATα derivative of HMK30.
MSN5 was deleted from the genome by a one-step PCR-based gene replacement using appropriate synthetic oligonucleotide primers and pRS41H (96) as the template DNA. Hygromycin B-resistant colonies were then selected, and gene replacements were confirmed by PCR and reduced mating efficiency.
Plasmids and recombinant DNA methods.
Standard procedures for construction and propagation of plasmid DNA in Escherichia coli strain DH5α were used (40, 84). Plasmids pPP1968 (pSTE5-STE5-3xGFP, CEN) (111), pCJ6 (pGAL1-HIS6-MYC-STE5, 2μm), pCJ117 (pGAL1-HIS6-MYC-STE5, CEN), pCJ80 (pGAL1-HIS6-MYC-STE5-GFP, CEN) (34, 49), and pUB223 (pCUP1-UBI K48R G76A) (57) have been described. To construct C-terminally FLAG3-tagged Ste5, the FLAG3 epitope was amplified from p3FLAG-KanMX (35) by PCR using a synthetic oligonucleotide that included the sequence corresponding to an endogenous XhoI site near the 3′ end of the STE5 coding sequence as the upstream primer and an oligonucleotide that included a stop codon and an XbaI site as the downstream primer. The resulting PCR product was subcloned as an XhoI-XbaI fragment into pCJ98 (which encodes an untagged version of Ste5 behind the GAL1 promoter; C. Inouye and J. Thorner, unpublished results), generating pLG92. In-frame attachment of the FLAG3 epitope tag was confirmed by direct DNA sequence analysis. Low-copy-number and multicopy plasmids expressing a green fluorescent protein (GFP)-Ste5 fusion under the control of a constitutive promoter (NOP1) were constructed in two steps. First, the STE5 coding region was amplified as an NcoI-BamHI fragment using primers OSTE5-1 (5′-GGG GGC CAT GGG TAT GGA AAC TCC TAC AGA C-3′) and OSTE5-2 (5′-GGG GGG ATC CCT ATA TAT AAT CCA TAT GG-3′) from YEp13-STE5 (gift of V. L. MacKay, University of Washington, Seattle) and inserted into the corresponding sites of pUN100-NOP1prom-ProtA-TEV-ADH1term (45). The STE5-containing insert was then released as a PstI fragment and inserted into the PstI sites of CEN vector pRS315 and 2μm DNA vector pRS425 (92) into which had been inserted before (as an ApaI-SpeI fragment) an ADE2-NOP1prom-GFP(S65T)-TEV-GAL4term cassette (B. Senger and E. Hurt, unpublished results), yielding pNOP(G1AL)-GFP-Ste5 and pNOP(G2AL)-GFP-Ste5, respectively. YEplac195-NOP1prom-myc-STE5 (plasmid 1729 in Hurt lab collection) was constructed by ligating a SacI-PstI NOP1prom-3Xmyc fragment from pRS315-NOP1prom-myc-NUP116-C (3) into the corresponding sites of YEplac195, yielding YEplac195-NOP1prom-myc (plasmid 1728 in Hurt lab collection), and then inserting therein the STE5-containing PstI fragment described immediately above. Unless otherwise indicated, all tagged Ste5 derivatives were fully functional, as judged by ability to rescue the sterility of ste5Δ cells. To generate an untagged version of a nuclear localization signal mutant [Ste5(NLSm)] under the control of its native promoter, pPP2105 (111) was digested with EagI (to excise a 13Xmyc tag) and religated, yielding pLG120. DNA sequence analysis confirmed that the in-frame stop codon of the STE5 open reading frame was still intact.
Measurement of reporter gene expression.
To measure FUS1-lacZ reporter gene expression, exponentially growing cells carrying a high-copy FUS1-lacZ reporter plasmid (YEpU-FUS1Z) (4) were incubated with and without 1 μM α-factor for 60 min, and the level of β-galactosidase activity present was measured using a colorimetric substrate as described previously in detail (4).
Protein extraction.
In most experiments, protein was extracted from yeast cells by alkaline lysis and trichloroacetic acid (TCA) precipitation by the method of Yaffe and Schatz (116). Briefly, cells were harvested by centrifugation, and the resulting pellets were flash-frozen in liquid N2. For analysis, the pellets were resuspended by vortex mixing in 150 μl of 1.85 M NaOH and 7.4% β-mercaptoethanol and incubated for 10 min on ice, and then ice-cold TCA (final concentration, 25%) was added. After 10 min on ice, the precipitated protein was collected by centrifugation at maximum speed in a microcentrifuge at 4°C, washed twice with ice-cold acetone to remove residual TCA, and resuspended in an appropriate volume (32 μl per A600 unit) of 5% sodium dodecyl sulfate (SDS) in 0.1 M untitered Tris base. For SDS-polyacrylamide gel electrophoresis (PAGE) of such samples, 8 μl (per 1.0 A600 unit) of 5× concentrated SDS-PAGE sample buffer was added to each sample, and after the samples were boiled for 2 min and clarified by centrifugation in a microcentrifuge, a portion (typically ∼10 μl, the equivalent of ∼0.25 A600 unit) of the resulting supernatant solution was subjected to SDS-PAGE on an 8% slab gel, transferred to a nitrocellulose membrane, and analyzed by immunoblotting.
Where noted, protein was extracted from yeast cells by SDS solubilization and glass bead lysis. Briefly, samples (∼10 A600 units) of exponentially growing cells were harvested, washed in ice-cold sterile phosphate-buffered saline (PBS), and resuspended in 200 μl of 2× SDS-PAGE sample buffer to which was added ∼100 μl of acid- and ether-washed glass beads (400- to 500-nm diameter), followed by boiling for 3 min and three 10-s bursts of vigorous vortex mixing. After clarification by centrifugation in a microcentrifuge, appropriate portions (to achieve equivalent loading based on the initial A600 and extract volume) were analyzed by SDS-PAGE and immunoblotting.
Antibodies and immunoblotting.
Proteins resolved in slab gels were transferred to nitrocellulose filter paper, incubated with the appropriate primary antibodies and then incubated with appropriate secondary antibodies. Proteins were then visualized and quantified using standard chemiluminescence detection for horseradish peroxidase-conjugated secondary antibodies or using an infrared imaging system (Odyssey v2.1 software; Li-Cor Biosciences, Inc., Lincoln, NE) for infrared dye-conjugated secondary antibodies.
To detect Ste5 in experiments in which it was immunoprecipitated and/or overexpressed, affinity-purified polyclonal rabbit anti-Ste5 immunoglobulin G (IgG) (44) was used. To detect Ste5 expressed at its endogenous (or near endogenous) level, a different polyclonal rabbit anti-Ste5 antiserum (gift of K. R. Benjamin, Molecular Sciences Institute, Berkeley, CA) was used. The other primary antibodies used in this study were mouse anti-c-myc monoclonal antibody (MAb) 9E10 (28), mouse antiubiquitin MAb (Santa Cruz Biotechnology, Inc.), rabbit polyclonal anti-Pgk1 antiserum (6), rabbit polyclonal anti-Cdc12 antiserum (101), and rabbit polyclonal anti-Yrb1 antiserum (53).
Measurement of Ste5 half-life.
The stability of Ste5 in vivo was measured using either a protein synthesis shutoff technique or a GAL1 promoter-based pulse-chase method. Similar results were obtained by using either approach. For analysis by protein synthesis shutoff, cells expressing Ste5 at its endogenous level were grown to mid-exponential phase and then subjected to the test condition (e.g., temperature shift, etc.) immediately after the addition of cycloheximide (CHX) (final concentration of 10 μg/ml), and samples were taken at the indicated times. In experiments involving pheromone, cells were pregrown to mid-exponential phase in YPD (yeast extract-peptone-dextrose) buffered with 50 mM Na succinate (pH 3.5) and then treated with α-factor (final concentration of 3 μM) for 30 min followed by addition of the CHX. For analysis by Gal-induced pulse-chase, cells expressing STE5 from the GAL1 promoter were grown to mid-exponential phase in medium containing Raf-Suc, and then STE5 expression was induced by the addition of Gal (final concentration, 2%). After a 2-h period of synthesis, the cells were collected, washed with fresh medium containing Glc (2%) and resuspended in the same medium, and samples were taken at the indicated times.
Effect of proteasome inhibitor MG132.
To allow for efficient entry of MG132, an erg6Δ mutant was used (59). The erg6Δ cells carrying a URA3-marked 2μm DNA plasmid expressing His6-myc-Ste5 from the GAL1 promoter were grown to mid-exponential phase at 30° in SC medium lacking Ura but containing Raf-Suc, synthesis of Ste5 was induced by the addition of 2% Gal for 2 h, and the cells were then collected and washed with and resuspended in medium containing 2% Glc as described above. The resulting cell suspension was split into two equal portions, which were then incubated in the absence (solvent dimethyl sulfoxide [DMSO] alone) or presence of MG132 (final concentration of 50 μM; added from a concentrated stock in DMSO), and samples were taken at the indicated times. The cells in each sample were harvested by centrifugation and snap-frozen in liquid N2, and the resulting pellets were lysed by vigorous vortex mixing with glass beads in ice-cold lysis buffer (0.1% Tween 20, 125 mM potassium acetate, 4 mM MgCl2, 0.5 mM EDTA, 5 mM sodium bisulfite, 20 mM Tris-HCl [pH 7.2]) containing 12.5% glycerol, 1 mM dithiothreitol, and a commercial protease-inhibitor mix (Complete EDTA-free; Roche Diagnostics, Inc.). From the resulting lysates (1 mg total protein), myc-tagged Ste5 was immunoprecipitated using mouse ascitic fluid containing anti-c-myc MAb 9E10, as described previously (49). Immune complexes were solubilized in SDS-PAGE sample buffer, and after boiling and clarification, they were analyzed by SDS-PAGE and immunoblotting.
Special growth conditions in the presence of a trace of detergent also make S. cerevisiae cells more permeable to MG132 (63), which permitted analysis of endogenous Ste5. Briefly, strain W303-1A carrying an empty URA3 vector was pregrown to mid-exponential phase in a synthetic medium (0.17% yeast nitrogen base without ammonium sulfate or amino acids [Difco]) supplemented with 0.1% proline and the other necessary supplements for growth of these cells and 2% Glc. The cells were then reinoculated at an A600 of 0.5 into a fresh sample of the same medium also containing 0.003% SDS and grown for another 3 h, whereupon the culture was divided into two equal portions and then incubated in the absence (solvent DMSO alone) or presence of MG132 (final concentration of 75 μM from a concentrated stock in DMSO). After 30 min, CHX (final concentration of 50 μg/ml) was added to stop protein synthesis, and commencing 10 min later, samples were taken at the indicated times.
Fluorescence microscopy.
Live cells expressing GFP fusions to Ste5 were grown to mid-exponential phase (and in some experiments, then treated with 5 μM α-factor), concentrated by brief centrifugation, spotted onto an agarose pad, and examined immediately thereafter under an epifluorescence microscope (model BH-2; Olympus) using a 100× objective equipped with a GFP band-pass filter (Chroma Technology Corp.). Images were collected using a charged-coupled-device camera (Olympus), recorded with Magnafire SP imaging software (Optronics), and digitally processed using Photoshop (Adobe).
In vivo ubiquitinylation assay.
Strain BYB84 (ste5Δ) was cotransformed with a plasmid (pLG92) expressing Ste5-FLAG3 from the GAL1 promoter and a plasmid (pUB233) expressing a His6-myc-tagged mutant ubiquitin, Ubi(K48R G76A), from the Cu2+-inducible CUP1 promoter. As controls, the same strain was transformed with each of these plasmids alone, along with the corresponding other empty vector. Transformants were grown to mid-exponential phase in selective medium with Raf-Suc, and then His6-myc-Ubi(K48R G76A) expression was induced by the addition of 250 μM CuSO4 for 4 h, followed by induction of Ste5-FLAG3 expression by the addition of 2% Gal for 2 h. Cells were harvested by centrifugation, washed once with ice-cold sterile PBS containing 5 mM N-ethylmaleimide (NEM), and frozen in liquid N2. The resulting cell pellets (equivalent to ∼40 A600 units) were lysed by the alkaline lysis and TCA precipitation method described above, and the precipitated protein was collected by centrifugation at 30,000 × g for 30 min at 4°C in a refrigerated preparative centrifuge (Sorvall RC-5B). The protein pellet was solubilized in 6 M guanidinium hydrochloride containing 5 mM NEM and buffered with 100 mM sodium phosphate and 10 mM Tris-HCl (both pH 8.0) (buffer A). After the protein was completely redissolved, the solution was clarified by centrifugation at 16,000 × g for 30 min at 4°C. His6-myc-Ubi(K48R G76A)-tagged proteins in the resulting supernatant fraction were recovered by incubation at 4°C for 4 h with 100 μl of a 50:50 slurry of Ni2+-charged nitrilotriacetate-derivatized agarose beads (HIS-Select HC nickel affinity gel; Sigma Chemical Co.). The beads were washed successively once with 1 ml of buffer A, twice with 8 M urea containing 5 mM NEM and buffered with 100 mM sodium phosphate and 10 mM Tris-HCl (both pH 8.0) (buffer B), and finally twice with buffer B that was adjusted to pH 6.3. Bound proteins were then eluted with 200 μl of 2× SDS-PAGE sample buffer and analyzed by SDS-PAGE and immunoblotting.
RESULTS
Pheromone stimulation stabilizes Ste5.
Molecular modeling and simulations predict that the level of a scaffold protein can profoundly influence the output of a signaling pathway (13, 30, 90). Measurement of the cellular content of each component of the yeast mating pheromone response pathway indicates that Ste5 is the most limiting factor required for signal propagation (38; T. M. Thomson, K. R. Benjamin, A. Bush, T. Love, R. C. Yu, A. Gordon, A. Colman-Lerner, D. Endy, and R. Brent, submitted for publication). Correspondingly, we have found that Ste5 overexpression dramatically potentiates signaling in response to pheromone by nearly an order of magnitude and, importantly, can elicit robust pathway output even in the absence of pheromone when present at a high enough level (Fig. 1A). This response occurs even though active G1 Cdk1 (Cln-Cdc28) is present in these cells and has been demonstrated to block Ste5 binding to the plasma membrane (93). Therefore, we reasoned that, normally, there may be additional mechanisms that set the appropriate level of Ste5 to maximize its ability to mount a stimulus-evoked signal while minimizing its capacity to inadvertently cause stimulus-independent signaling. Hence, we explored whether any aspect of the spatial and temporal regulation of Ste5 function and localization during the cell cycle and in response to pheromone might contribute to controlling its cellular level.
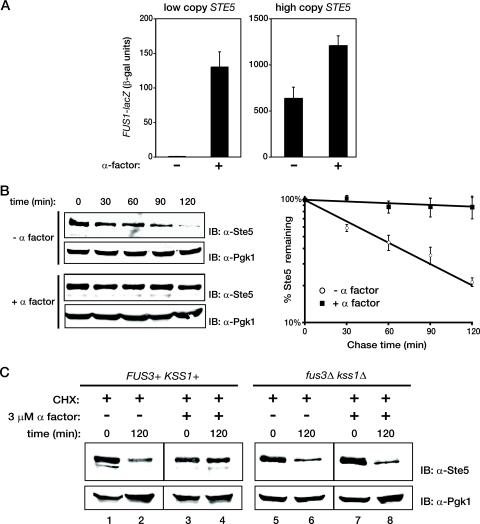
FIG. 1.
Effect of Ste5 overexpression on signaling output and of pheromone treatment on Ste5 stability. (A) Exponentially growing cultures of ste5Δ cells (YAB5) carrying a high-copy-number plasmid expressing a FUS1-lacZ reporter gene (YEpU-FUS1Z) and expressing GFP-STE5 from the NOP1 promoter on either a low-copy-number plasmid (left) or a high-copy-number plasmid (right) were treated in the absence (−) or presence (+) of 1 μM α-factor for 60 min, and then the specific activity of β-galactosidase (black columns) was measured {values in Miller units [β-galactosidase (β-gal)] represent the means of three independent trials, and the error bars represent the standard deviations of those means}. The cultures overexpressing GFP-Ste5 grew more slowly and displayed a high proportion of shmoo-shaped cells. (B) (Left) Wild-type (W303) cells were grown to mid-exponential phase and either treated with 3 μM α-factor for 30 min (+ α factor) or not treated with α-factor (− α factor), followed by the addition of CHX (final concentration, 10 μg/ml) to stop any further protein synthesis. Samples were then taken every 30 min as indicated above the gels, and the level of Ste5 was assessed by SDS-PAGE and immunoblotting with rabbit polyclonal anti-Ste5 antiserum (IB: α-Ste5). For a control for equivalent loading in the lanes, the same samples were also immunoblotted with rabbit polyclonal anti-Pgk1 antiserum (IB: α-Pgk1). The results of a representative experiment are shown. (Right) To determine the time dependence of Ste5 degradation, the amount of Ste5 in each sample was quantified using an infrared imaging system (Li-Cor Odyssey version 2.1 software) and normalized to the corresponding content of Pgk1, and the logs of the means of the values so obtained for three independent experiments (performed as described for the left panels) were plotted against time after CHX was added. Error bars represent the standard errors of the means. (C) Wild-type cells (BY4741) or an otherwise isogenic fus3Δ kss1Δ derivative (RCY9321) were grown to mid-exponential phase and either treated (+) or not treated (−) with 3 μM α-factor for 30 min followed by the addition (+) of CHX (10 μg/ml), and 2 h later, a sample was taken and the amounts of Ste5 and Pgk1 were determined as described above for panel B.
To begin to address this issue, we monitored the endogenous level of Ste5 over time in the absence or presence of pheromone (after blocking any further new synthesis by treatment of the cells with CHX) by immunoblotting (Fig. 1B). We also measured Ste5 stability via an independent method for pulse-chase analysis, which utilized Glc-imposed repression after a burst of Gal-induced GAL1 promoter-dependent expression (see later figures). Very similar results were obtained by either method. During vegetative growth, Ste5 appears to be a moderately unstable protein (Fig, 1B, left top) with a half-life (t1/2) of ∼60 min (Fig. 1B, right), in agreement with previous observations (31). However, we found that after cells were exposed to pheromone (Fig. 1B, left bottom), Ste5 remained almost completely stable throughout the time course analyzed (t1/2 ≫ 2 h) (Fig. 1B, right).
Ste5 has been shown to be directly phosphorylated by Fus3 in response to pheromone in vivo (43), in yeast cell extracts (31, 52), and in vitro with purified proteins (7, 11). Indeed, we did not observe pheromone-induced stabilization of Ste5 in a fus3Δ kss1Δ double mutant (Fig. 1C, compare lane 8 with lane 4), indicating that pheromone-stimulated inhibition of Ste5 turnover requires MAPK action. However, the MAPK-mediated phosphorylation of Ste5 (or another cellular component) could reduce the rate of Ste5 degradation directly or could do so indirectly because MAPK function is required to stimulate relocalization of Ste5 to the cytosol, where it may be more stable.
Preventing nuclear export increases the rate of Ste5 degradation.
Nuclear export and plasma membrane recruitment of Ste5 are rapidly stimulated in response to pheromone (53, 66, 79, 89) and occur concomitantly with the pheromone-induced stabilization of Ste5 we observed, as monitored by examination of GFP-tagged Ste5 expressed at a near endogenous level (Fig. 2A). This correlation led us to consider the possibility that Ste5 is susceptible to degradation in the nucleus, but not in the cytosol. Thus, the observed stabilization caused by pheromone stimulation might be primarily a consequence of the increase in the cytoplasmic pool of Ste5 after its pheromone-induced nuclear export. If this view is correct, then the steady-state level of Ste5 should be decreased and its rate of turnover increased if Ste5 is prevented from escaping the nucleus. Therefore, we examined the stability of Ste5 in cells lacking exportin Msn5, which clearly trapped more of the GFP-tagged Ste5 population in the nucleus (Fig. 2B). Indeed, for cells expressing myc-tagged Ste5 at a near endogenous level from the low-level NOP1 promoter, the steady-state level of Ste5 was substantially reduced in msn5Δ cells compared to otherwise isogenic MSN5+ cells, as assessed by immunoblotting of equivalent amounts of cell extract (Fig. 2C; see also Fig. 3A). Correspondingly, we found that the rate of turnover of Ste5 (as assessed by examining endogenous Ste5 using the protein synthesis shutoff method) (Fig. 2D, left) is more rapid in msn5Δ cells (t1/2 of ∼30 min) than it is in otherwise isogenic MSN5+ cells (Fig. 2D, right). These results indicate that, when trapped in the nucleus, Ste5 is more rapidly degraded, suggesting that the nucleus is the cellular compartment where Ste5 degradation occurs.
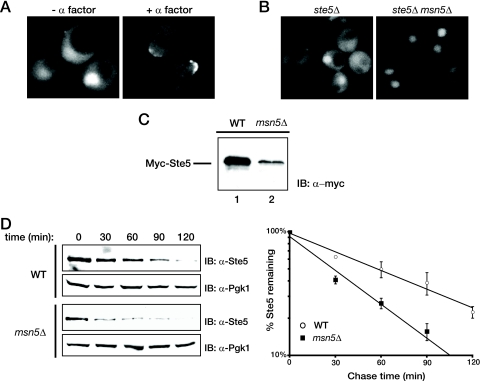
FIG. 2.
Stabilization of Ste5 is concomitant with its exit from the nucleus. (A) An exponentially growing culture of an ste5Δ mutant (BYB69) expressing Ste5-GFP3 from the STE5 promoter on a CEN plasmid was not treated (− α factor) or treated with 3 μM α-factor (+ α factor). After 45 min, samples of the cells were visualized by standard epifluorescence microscopy as described in Materials and Methods. (B) Exponentially growing cultures of ste5Δ cells (YAB5) or otherwise isogenic ste5Δ msnΔ cells (YAB8 expressing Ste5-GFP3 as in panel A) were visualized by standard fluorescence microscopy. (C) Wild-type (WT) cells (W303) or an otherwise isogenic msn5Δ derivative (HMK30) were transformed with a plasmid expressing Myc-tagged Ste5 from the NOP1 promoter and grown to mid-exponential phase. Extracts were prepared from samples of cell paste (samples with the same weight [wet weight]) by vigorous vortex mixing with glass beads, and the steady-state level of Myc-Ste5 present was analyzed by SDS-PAGE and immunoblotting with an anti-c-Myc monoclonal antibody (IB: α-myc) (MAb 9E10), as described in Materials and Methods. (D) Rate of Ste5 degradation in wild-type cells (W303) or in an otherwise isogenic msn5Δ derivative (HMK30) was monitored after the addition of CHX and plotted as described in the legend to Fig. 1B.
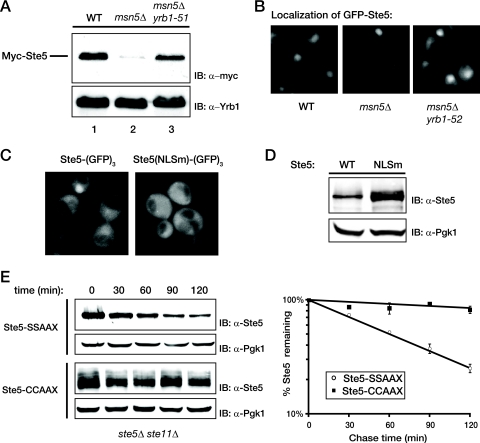
FIG. 3.
Retention of Ste5 in the cytosol prevents degradation. (A) Wild-type (WT) cells and otherwise isogenic msn5Δ and msn5Δ yrb1-51 derivatives were transformed with a plasmid expressing Myc-tagged Ste5 from the NOP1 promoter and grown at 23°C until mid-exponential phase, and cell extracts were prepared by vigorous vortex mixing with glass beads. The amount of Ste5 present was assessed by SDS-PAGE and immunoblotting with an anti-c-Myc MAb 9E10 (IB: α-myc). For a control for equivalent loading in the lanes, the same samples were also immunoblotted with rabbit polyclonal anti-Yrb1 antiserum (IB: α-Yrb1). (B) The same cells shown in panel A were transformed with a plasmid expressing GFP-tagged Ste5 from the NOP1 promoter, grown at 23°C until mid-exponential phase, and then visualized by fluorescence microscopy as described in Materials and Methods. Representative fields are shown. (C) Exponentially growing cultures of ste5Δ cells (YAB5) carrying a CEN vector expressing either Ste5-GFP3 or Ste5(NLSm)-GFP3 from the STE5 promoter were visualized by standard fluorescence microscopy. Representative fields are shown. (D) An ste5Δ mutant (BYB69) transformed with a CEN vector expressing either untagged wild-type Ste5 or untagged Ste5(NLSm), each from the STE5 promoter, was grown to mid-exponential phase, harvested, and lysed by the TCA precipitation method as described in Materials and Methods, and the resulting extracts were analyzed by SDS-PAGE and immunoblotting with rabbit polyclonal anti-Ste5 antiserum. For a control for equivalent loading in the lanes, the same samples were also immunoblotted with rabbit polyclonal anti-Pgk1 antiserum. (E) A ste5Δ ste11Δ strain (YLG18) expressing from the GAL1 promoter on a CEN plasmid either Ste5-CCAAX or, as a control, Ste5-SSAAX were pregrown in raffinose-containing medium until early exponential phase, and expression was induced by the addition of galactose. After 2 h, the cells were washed and resuspended in glucose-containing medium to shut off further expression, and samples were taken every 30 min. (Left) The level of Ste5 was determined by immunoblotting with polyclonal anti-Ste5 antiserum, and (right) the results of three independent experiments were plotted as described in the legend to Fig. 1B. For Ste5-CCAAX, the multiple bands represent various states of modification (farnesylation and/or palmitoylation, with or without proteolytic AAX removal accompanied [or not] thereafter by carboxymethylation).
Preventing nuclear entry stabilizes Ste5.
If Ste5 destruction occurs exclusively in the nucleus, we reasoned that preventing its nuclear import should stabilize Ste5, mimicking the stabilization observed upon pheromone-induced nuclear export. Therefore, we examined the effect of preventing Ste5 import into the nucleus in three different ways. First, we determined the steady-state level of Ste5 in otherwise isogenic MSN5+ cells, msn5Δ cells, and msn5Δ cells also carrying temperature-sensitive mutations in the yeast RanBP1 ortholog (yrb1-51ts or yrb1-52ts). Yrb1 is a key coregulator of the Ran GTPase cycle that controls nucleocytoplasmic trafficking of proteins in yeast (72, 88). Loss-of-function yrb1 mutations prevent efficient hydrolysis of Ran-bound GTP in the cytosol. The resulting high cytosolic level of Ran-GTP impedes nuclear import because Ran-GTP binding to importins prevents cargo binding (39). We have demonstrated before that the yrb1-51 and yrb1-52 alleles impede overall protein import into the nucleus, including, specifically, import of a GFP-Ste5 fusion (53). Strikingly, introduction of either of these yrb1ts mutations into msn5Δ cells increased the steady-state level of myc-tagged Ste5 in cell extracts compared to that in msn5Δ cells alone (Fig. 3A) and increased the visible cytoplasmic pool of GFP-tagged Ste5 (Fig. 3B), consistent with the conclusion that the sole reason that the absence of Msn5 destabilizes Ste5 is because more Ste5 is sequestered in the nucleus where it is inherently less stable than when it is in the cytosol.
As a second approach to address this issue, we examined the level of Ste5(NLSm), a variant in which multiple substitution mutations (E53A, K54A, R57A, F58V, Q59A, R60A, S61A, and S62A) have been introduced into a primary nuclear localization signal (66, 79). In our hands, these mutations significantly reduce (but do not eliminate) nuclear localization of Ste5 (Fig. 3C), as we have observed previously for a deletion (deletion of residues 26 to 99) that removes this entire sequence (C. Inouye, N. Dhillon, and J. Thorner, unpublished results). Nonetheless, consistent with the conclusion that efficient Ste5 degradation requires its presence in the nucleus, we found that the steady-state level of Ste5(NLSm) was reproducibly higher than that of wild-type Ste5 expressed in an identical manner (Fig. 3D), providing further evidence that preventing nuclear import impedes Ste5 degradation.
As a third independent means to address whether preventing Ste5 from entering the nucleus stabilizes this protein, we measured the half-life of otherwise normal Ste5 that we artificially tethered to the plasma membrane by attaching in frame to its C-terminal end the S-palmitoylated and S-farnesylated C-terminal CCAAX box of yeast Ras2 (62, 86). Both Ste5-CCAAX and, as a control, Ste5-SSAAX (an otherwise identical derivative that lacks the two Cys residues, so it cannot be modified by either lipophilic substituent) were expressed from the GAL1 promoter, at which time synthesis was turned off by the addition of Glc. Because this targeting of Ste5 to the membrane causes pathway activation (79, 111), we used a ste5Δ ste11Δ strain for these experiments to block any downstream signaling. We found that the Ste5-SSAAX derivative, which presumably enters the nucleus freely, was degraded with a half-life indistinguishable from that of normal Ste5 (t1/2 of ∼60 min) (Fig. 3E). In contrast, all of the Ste5-CCAAX species were markedly stabilized (Ste5-CCAAX migrated as several bands representing the S-farnesylated, the S-palmitoylated, and the doubly modified derivatives, with and without proteolytic -AAX removal and subsequent carboxymethylation, as observed for other CCAAX box-containing molecules, such as the Gγ subunit, Ste18 [48]) (Fig. 3E). Most strikingly, the degree of stabilization achieved by tethering Ste5 to the plasma membrane in the absence of pheromone (t1/2 ≫ 2 h) was equivalent to the stabilization of Ste5 evoked in response to pheromone (compare Fig. 1B and Fig. 3E), suggesting that the nuclear export and membrane localization of Ste5 triggered by pheromone are sufficient to explain its pheromone-induced stabilization. Indeed, only the change in subcellular localization can account for how Ste5 degradation is prevented in response to pheromone (as opposed to Fus3-dependent modification of Ste5 or any other downstream signaling events) since no signaling occurs in the ste11Δ cells we used for these particular experiments.
Thus, impeding efficient exit of Ste5 from the nucleus increased its rate of breakdown, whereas blocking efficient access of Ste5 to the nucleus markedly stabilized the protein. Taken together, these findings argued strongly that degradation of Ste5 takes place primarily, if not exclusively, in the nucleus.
Ste5 is degraded in a ubiquitin- and proteasome-dependent manner.
In budding yeast, protein degradation can occur via at least three distinct mechanisms: (i) inherent sensitivity to a compartment-specific protease (113), (ii) targeting by the ubiquitin-proteasome system (100), and (iii) reclamation via autophagy and digestion by vacuolar proteases (67). The fact that Ste5 degradation occurs in the nucleus made the third possibility unlikely. To distinguish between the first two possibilities, we examined the effect of blocking the activity of the 26S proteasome on the rate of Ste5 turnover. We utilized a strain carrying a temperature-sensitive mutation in CIM3, which encodes one of the six AAA+-ATPases of the 19S cap of the 26S proteasome that is required for degradation of ubiquitinylated substrates and localized mainly to the nucleus throughout the cell cycle (82). After the shift to a restrictive temperature, Ste5 was markedly stabilized in a cim3-1ts mutant (36) compared to the otherwise isogenic CIM3+ control cells (Fig. 4A), suggesting that Ste5 degradation occurs in a proteasome-dependent manner.
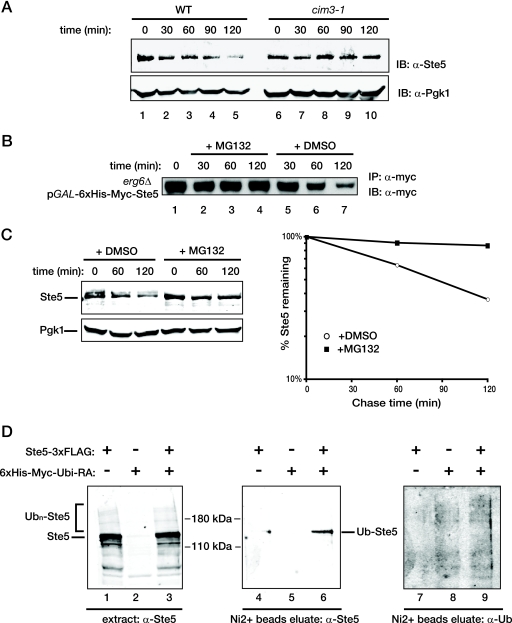
FIG. 4.
Ste5 is degraded in a ubiquitin- and proteasome-dependent manner. (A) A wild-type (WT) strain (MHY753) and an otherwise isogenic strain carrying a temperature-sensitive mutation, cim3-1, in the 19S cap of the proteasome (MHY754) were grown to mid-exponential phase at 26°C, shifted to 37°C for 30 min, and treated with CHX, and samples were taken every 30 min, in which the levels of Ste5 and Pgk1 were assessed, as described in the legend to Fig. 1B. (B) BKY48-5C, an erg6Δ mutant permeable to proteasome inhibitor MG132, expressing His6-myc-Ste5 under the control of the GAL1 promoter was grown to mid-exponential phase in raffinose-containing medium, and then Ste5 expression was induced by the addition of galactose. After 2 h, the culture was washed and resuspended in medium containing 2% glucose to shut off any further Ste5 expression (time zero), and half of the culture was incubated with MG132 (final concentration of 50 μM, added from a concentrated stock in DMSO) (+ MG132) or with the same volume of solvent alone (+ DMSO). At the times indicated above the gel, samples were taken, the cells were lysed, and the resulting extracts were subjected to immunoprecipitation using anti-c-Myc MAb 9E10 (IP: α-myc) as described in Materials and Methods. The amount of Myc-tagged Ste5 in the resulting immune complexes was analyzed by SDS-PAGE and immunoblotting with the same antibody (MAb 9E10) (IB: α-myc). (C) To permit entry of MG132 into a wild-type strain (W303), cells pregrown in standard minimal medium supplemented with the appropriate nutrients were incubated for 3 h in a synthetic medium supplemented with 0.1% proline as the nitrogen source and containing 0.003% SDS. One half of the culture received either solvent alone (DMSO) or MG132 (final concentration, 75 μM) in the same solvent as indicated above the gel. After 30 min, CHX (final concentration, 50 μg/ml) was added, and 10 min later, samples were taken at the time points indicated above the gel. The levels of Ste5 and Pgk1 were analyzed (left) and plotted (right) as described in the legend to Fig. 1B. (D) To determine whether Ste5 is a ubiquitinylated protein in vivo during vegetative growth, a ste5Δ mutant (BYB84) was cotransformed with a CEN vector expressing Ste5-FLAG3 from the GAL1 promoter and another CEN vector expressing His6-Myc-Ubi(K48R G76A) from the CUP1 promoter or with each plasmid alone along with the corresponding empty vector as controls. After induction of expression, cells were lysed under denaturing conditions, and a sample of each lysate was analyzed by SDS-PAGE and immunoblotting with polyclonal anti-Ste5 antiserum (lanes 1 to 3). The remaining portion of each lysate was incubated with over Ni2+-loaded NTA beads to enrich for proteins covalently tagged by the His6-Myc-Ubi(K48R G76A). Ubiquitinylated Ste5 (Ub-Ste5) was detected by immunoblotting the eluate of the bead-bound proteins with rabbit polyclonal anti-Ste5 antiserum (lanes 4 to 6), and total ubiquitinylated proteins were detected with a monoclonal antiubiquitin antibody (α-Ub) (lanes 7 to 9).
To confirm this conclusion in an independent manner, we examined the effect on Ste5 degradation of treating cells with MG132, a well-characterized peptide aldehyde inhibitor of the proteasome (59). Using either the GAL1 promoter-based pulse-chase method to study the turnover of epitope-tagged Ste5 in drug-permeable erg6Δ cells (Fig. 4B) or the CHX-dependent protein synthesis shutoff method to analyze endogenous Ste5 under growth conditions that enhance the permeability of wild-type cells to MG132 (Fig. 4C), we found that treatment with MG132, but not with the solvent alone (DMSO), clearly stabilized Ste5. These results indicate that the turnover of Ste5 observed in naïve cells is mediated by the proteasome.
It is well established that degradation by the 26S proteasome requires addition of a chain of at least four K48-linked ubiquitin molecules to the target protein (97, 100). Therefore, we next examined whether Ste5 is ubiquitinylated in vivo. Because the level of endogenous Ste5 is quite low, we used several tactics to permit detection of its ubiquitinylated species. We overexpressed an epitope-tagged derivative, Ste5-FLAG3, in ste5Δ cells in which a His6- and myc-tagged mutant ubiquitin, Ubi(K48R G76A), was also overexpressed from the CUP1 promoter. Overexpression of the mutant ubiquitin allowed for successful competition with endogenous ubiquitin for attachment to targets, but the K48R substitution prevents polymerization of K48-linked chains, making the target a poor substrate for recognition by the proteasome. Moreover, once attached, the G76A substitution inhibits hydrolytic removal of protein-linked ubiquitin by deubiquitinating enzymes, whose activity is often unleashed upon cell lysis (57). To further inhibit promiscuous deubiquitinating activity and help preserve ubiquitin modification, cell extracts were prepared under denaturing conditions and a sulfhydryl alkylating agent, NEM, was also included in every buffer because all known deubiquitinating enzymes have an active site Cys that is essential for activity (20).
Under these conditions, GAL1 promoter-driven expression of Ste5-FLAG3 clearly elevated the amount of Ste5 present, as assessed by immunoblotting with polyclonal anti-Ste5 antiserum (Fig. 4D, left), and overexpression of His6-myc-Ubi(K48R G76A) clearly increased the level and number of ubiquitinylated species that could be captured by binding to a Ni2+-nitrilotriacetic acid (Ni2+-NTA) affinity resin, as judged by immunoblotting elutes of the bead-bound proteins with an antiubiquitin MAb (Fig. 4D, right). We found that, when coexpressed with His6-Myc-Ubi(K48R G76A), a readily detectable amount of Ste5-FLAG3 was retained on the Ni2+-NTA affinity resin, whereas when either was expressed alone, there was little or no Ste5-FLAG3 bound to the beads (Fig. 4D, middle). Based on the degree of retardation of its mobility, the bound Ste5-FLAG3 species appeared to be monoubiquitinylated, as expected, due to the abundant overexpression of the nonpolymerizing Ubi(K48R G76A) mutant. These data demonstrate that Ste5 can undergo ubiquitinylation in vivo, consistent with the observed proteasome-dependent turnover of endogenous Ste5 in vegetatively growing cells.
Ste5 RING-H2 domain has no role in Ste5 degradation.
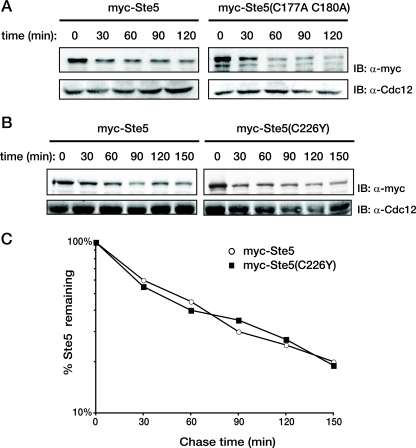
Substrate-specific ubiquitinylation requires a ubiquitin ligase (E3) that acts as a matchmaker by recruiting both the target and an appropriate ubiquitin-charged ubiquitin-conjugating enzyme (E2). Two major classes of E3 include those with a HECT domain and those with a Zn2+-binding RING (or RING-H2) domain (78). Ste5 contains a RING-H2 domain in its N terminus (residues 177 to 229) (50). Substitution mutations in certain residues whose side chains serve as Zn2+ ligands, such as Ste5(C177A C180A), are loss-of-function alleles and cripple positive functions of Ste5 in signaling, including its ability to bind Gβγ and to self-associate (29, 50, 103, 115). However, hyperactive alleles have also been identified in the Ste5 RING-H2 domain, such as Ste5(C226Y) (89). One way to explain the phenotype of the latter mutation would be if the RING-H2 domain also has a negative function, like autoubiquitinylation for self-degradation. However, using the GAL shutoff method, we found that neither the loss-of-function allele, Ste5(C177A C180A), nor the hyperactive allele, Ste5(C226Y), conferred any significant stabilization (Fig. 5), as judged by their initial steady-state level or the rate of degradation in comparison to wild-type Ste5. Therefore, we sought to identify the specific enzymes that act in trans to ubiquitinylate Ste5 for its nucleus-specific and proteasome-mediated degradation.
FIG. 5.
RING-H2 domain mutations in Ste5 do not affect its stability. (A) A ste5Δ strain (BYB84) expressing genes under control of the GAL1 promoter on a CEN vector. A strain expressing His6-Myc-tagged versions of either wild-type Ste5 or Ste5(C177A C180A) as indicated above the gels were pregrown in raffinose-containing medium until early exponential phase, and expression was induced by the addition of galactose for 2 h. Cells were then washed and resuspended in glucose-containing medium to shut off further expression, and portions of the culture were taken every 30 min thereafter. These cell samples were lysed by vortex mixing with glass beads and resolved by SDS-PAGE, and the level of Ste5 was determined by immunoblotting with anti-c-myc MAb 9E10 (IB: α-myc). For a control for equivalent loading in the lanes, the same samples were also immunoblotted with rabbit polyclonal anti-Cdc12 antiserum (IB: α-Cdc12). (B) The half-lives of wild-type His6-Myc-tagged versions of wild-type Ste5 or Ste5(C226Y) expressed from the GAL1 promoter on a 2μm DNA plasmid were determined in ste5Δ cells (BYB84) using the GAL shutoff protocol as described above for panel A. (C) The results of the experiment in panel B were quantified, normalized to the level of Cdc12 in the same samples, and plotted on a log scale against time after expression was shut off.
SCFCdc4 is responsible for Ste5 degradation.
Mating pheromone action arrests cells in G1 and stabilizes Ste5. Therefore, it seemed reasonable that the ubiquitinylation machinery that promotes the G1-to-S transition might be involved in Ste5 degradation. One factor critical for the G1-to-S transition is the nuclear E3 comprising the Skp1-Cullin/Cdc53-F box protein (SCF) complex that contains a RING protein (Hrt1/Rbx1), its associated E2 (Cdc34), and the F-box-containing substrate recognition factor, Cdc4 (76, 110). Cdc4 is localized exclusively to the nucleus, whereas the core subunits of the SCF complex and Cdc34 are found in both the nucleus and the cytoplasm (9, 16, 37).
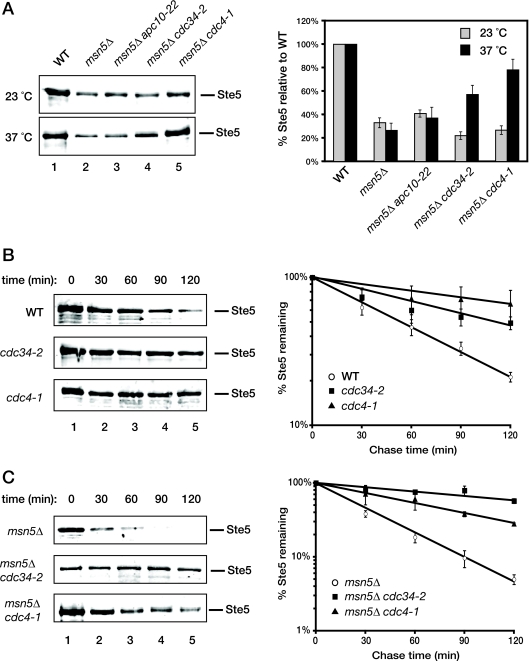
To examine whether the SCFCdc4 complex is the major E3 that mediates degradation of Ste5 in the nucleus, we analyzed the effects of temperature-sensitive mutations (cdc34-2 and cdc4-1) in two essential components of SCFCdc4 on the steady-state level of endogenous Ste5. For a control, we also examined a temperature-sensitive mutation (apc10-22) that ablates the function of another nuclear E3, the anaphase-promoting complex, which regulates cell cycle progression at other stages (74). To enhance our ability to detect any stabilizing effect of these mutations, we used msn5Δ cells, in which Ste5 is trapped in the nucleus and displays an accelerated rate of degradation, as we have demonstrated already (Fig. 1 and 2). Indeed, we found that, at the restrictive temperature (but not at the permissive temperature), the steady-state level of Ste5 was reproducibly increased in msn5Δ cells carrying the cdc34-2 or cdc4-1 mutation (Fig. 6A). In contrast, in msn5Δ cells carrying the apc10-22 mutation, there was no significant change in Ste5 level at either temperature, compared to that observed in msn5Δ cells alone (Fig. 6A). The increase in level conferred by the absence of functional Cdc4 or Cdc34 is not an indirect effect of arresting cell cycle progression in G1 because the cim3-1ts proteasome mutation conferred a similar degree of Ste5 stabilization (Fig. 4) but causes arrest of cell cycle progression at the G2-to-M transition (36).
FIG. 6.
Ste5 turnover requires the SCFCdc4 ubiquitin ligase. (A) (Left) Exponentially growing cultures of wild-type (WT), msn5Δ, msn5Δ apc10-22, msn5Δ cdc34-2, and msn5Δ cdc4-1 cells were each divided into two equal portions, which were then incubated for 2 h at either a permissive temperature (23°C) or a restrictive temperature (37°C). The cells were then harvested, and protein was extracted by the TCA method. The resulting extracts were resolved by SDS-PAGE and immunoblotted with anti-Ste5 antiserum. (Right) The level of Ste5 was quantified, normalized to the level of Pgk1 in the same samples (not shown), and plotted. Values represent the averages from two independent experiments, and the error bars represent the standard error of those means. (B) (Left) Exponentially growing cultures of wild-type, cdc34-2, or cdc4-1 cells were shifted to 37°C for 45 min followed by the addition of cycloheximide (10 μg/ml) for 10 min. Samples were then taken at the time points indicated above the gels, and the level of endogenous Ste5 was analyzed by SDS-PAGE and immunoblotting with polyclonal anti-Ste5 antiserum. (Right) The level of endogenous Ste5 in wild-type, cdc34-2, or cdc4-1 cells was normalized to the level of Pgk1 in the same samples (not shown) and plotted on a log scale against time after the addition of CHX. Values represent the averages of three independent experiments conducted as shown in the left panels, and the error bars represent the standard errors of those means. (C) Exponentially growing cultures of msn5Δ, msn5Δ cdc34-2, or msn5Δ cdc4-1 cells were shifted to 37°C for 45 min followed by the addition of CHX (10 μg/ml) for 10 min, and then analyzed as explained above for panel B.
To confirm these conclusions, we examined the effects of the cdc4-1 and cdc34-2 mutations on the rate of Ste5 degradation in both wild-type and msn5Δ cells at a restrictive temperature using the CHX-dependent protein synthesis shutoff method. Consistent with their observed effects on the steady-state level of Ste5, both the cdc34ts and the cdc4ts mutations dramatically reduced the rate of Ste5 turnover, both in wild-type cells where Ste5 is able to shuttle between the nucleus and cytoplasm (Fig. 6B) and even more dramatically when Ste5 was trapped in the nucleus by the lack of Msn5 (Fig. 6C). We found in these experiments and most others in which cdc4 and cdc34 mutants were compared (see below) that inactivation of Cdc4 had a stronger effect in preventing Ste5 degradation than inactivation of Cdc34, suggesting that perhaps other E2 enzymes can also associate and operate with SCFCdc4, albeit less efficiently than Cdc34. These data indicate that the SCFCdc4 complex makes a major contribution to marking Ste5 for its ubiquitin- and proteasome-mediated destruction in the nucleus.
Regulation of Ste5 degradation by G1 CDK phosphorylation.
Mechanistic and structural studies have revealed that substrate phosphorylation precedes substrate recognition by SCFCdc4 and subsequent ubiquitin addition to accessible Lys residues in the target (77, 95). The MAPK-mediated phosphorylation of Ste5 induced in response to pheromone (31, 43, 52) cannot be responsible for marking Ste5 for destruction because, as we have shown here, MAPK action is required to stabilize Ste5 by promoting its nuclear export, allowing it to escape the ubiquitin- and proteasome-mediated degradation that occurs in the nucleus (Fig. 1, 2, and 3). Therefore, phosphorylation by some other kinase is presumably responsible for marking Ste5 for destruction in the nucleus. Indeed, it has been observed that, during vegetative growth, Ste5 is phosphorylated by the major G1 CDK, Cln2-Cdc28 (31, 93). Moreover, it is well established that, as the level of Cln2-Cdc28 activity rises and cells commit to entry into a new cycle, signaling through the mating pheromone response pathway is inhibited (18, 69). However, the molecular basis for this effect has been controversial. Some suggest that G1 CDK-mediated phosphorylation blocks signaling in this pathway by interfering with Ste11 function (105), whereas others claim Ste20 function is impeded (70). Yet others have provided evidence that Cln2-Cdc28-dependent phosphorylation prevents stable association of Ste5 with the plasma membrane (93). However, in light of the cumulative results we have presented here and the clear-cut role that G1 CDK phosphorylation has in marking other SCFCdc4 substrates for destruction (71), we reasoned that a contributing mechanism by which phosphorylation of Ste5 by Cln2-Cdc28 helps prevent signal propagation as cells commit to START and exit G1 is via lowering the overall level of Ste5 by marking it for SCFCdc4-dependent ubiquitinylation and subsequent degradation by nuclear proteasomes (82).
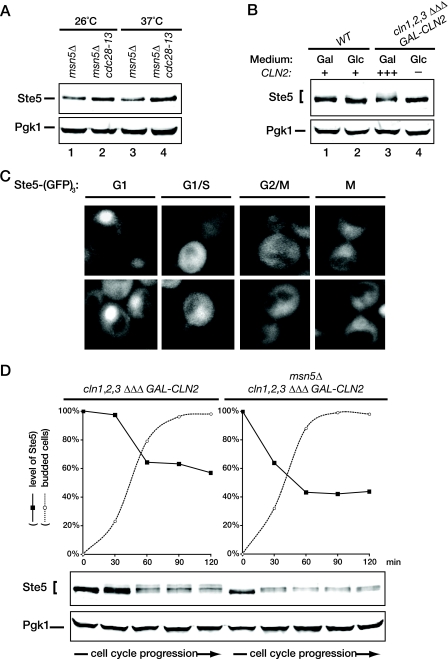
To confirm this idea, we first examined the steady-state level of endogenous Ste5 in asynchronous cultures of msn5Δ cells and in otherwise isogenic msn5Δ cells carrying the temperature-sensitive cdc28-13 allele (19). We found that the nuclear pool of Ste5 was measurably elevated at the restrictive temperature in the msn5Δ cdc28-13 cells compared to the msn5Δ control (Fig. 7A), and a modest effect of the cdc28-13 mutation was even observed at the permissive temperature. These particular experiments were performed with asynchronous cells shifted to 37°C, and it has been reported before that the cdc28-13ts mutant arrests with a significant fraction of the population in cell cycle stages other than G1 when shifted to 37 or 38°C (81, 112). Once Cln-bound Cdc28 has initiated Ste5 breakdown at the G1/S boundary, any cell that has passed that point in the cell cycle will have a much lower content of Ste5 than that of a G1 cell. Hence, the observed increase in Ste5 level in the cdc28-13 mutant underestimates the effect of the loss of G1 Cdk1 phosphorylation on stabilizing Ste5 compared to cells uniformly arrested in G1.
FIG. 7.
High G1 CDK activity promotes Ste5 degradation. (A) Exponentially growing cultures of an msn5Δ derivative (YLG90) of an wild-type strain (BF264-15D) and an msn5Δ derivative (YLG91) of an otherwise isogenic cdc28-13 strain were divided into two equal portions, which were incubated for 3 h at either the permissive temperature (23°C) or restrictive temperature (37°C). The cells were then harvested, and protein was extracted by the TCA method. The resulting extracts were resolved by SDS-PAGE and immunoblotted with anti-Ste5 antiserum. For a control for equivalent loading in the lanes, the same samples were also immunoblotted with rabbit polyclonal anti-Pgk1 antiserum. (B) Wild-type (WT) cells (BF264-15D) and otherwise isogenic cln1Δ cln2Δ cln3Δ GAL1-CLN2 cells (JTY2142) were pregrown in YP medium with 2% Gal to early exponential phase, at which point each culture was divided into two equal portions. One portion was washed and resuspended in YP medium with 2% Glc to repress Cln2 expression (lanes 2 and 4), and the other portion was maintained in YP medium with 2% Gal (lanes 1 and 3). After 2.5 h, the cells were harvested, and the content of Ste5 and Pgk1 (as a loading control) in each culture was analyzed as described above for panel A. (C) To determine whether the level of Ste5 in the nucleus correlates with cell cycle position, exponentially growing cultures of ste5Δ cells (YAB5) expressing Ste5-GFP3 from the STE5 promoter on a CEN vector were visualized by fluorescence microscopy (∼80% of the cells displayed detectable fluorescence). The pattern of Ste5 localization in representative cells from each phase of the cell cycle, as determined by their budding pattern, is shown. (D) To examine the cell cycle dependence of Ste5 degradation, cells of a cln1Δ cln2Δ cln3Δ GAL1-CLN2 strain or an otherwise isogenic msn5Δ derivative of the same strain were arrested by depletion of the G1 cyclin Cln2 by growth in medium containing 5% Glc for 3 h. Then, the cells were released from G1 arrest by resuspension in medium containing 1% Gal-1% Raf-0.2% Suc. Thereafter, samples were collected at 30 min intervals over the next 2 h, and the content of Ste5 and Pgk1 (as a loading control) in each sample was analyzed as described above for panel A. Measurement of the budding index by examination under the microscope was used to confirm the resumption of cell cycling (n = 200 cells at each time point).
Therefore, as an independent approach to specifically address whether Cln-activated Cdc28 phosphorylation affects Ste5 stability, we analyzed the effects of Cln2 depletion and Cln2 overexpression on the steady-state level of endogenous Ste5. For this purpose, we examined cultures of a strain (cln1Δ cln2Δ cln3Δ) that requires expression of CLN2 from the GAL1 promoter to grow. For wild-type cells of the same lineage, a shift of carbon source (from Gal to Glc) had no effect on the steady-state level of Ste5 (Fig. 7B, lanes 1 and 2). By contrast, when the cln1Δ cln2Δ cln3Δ (pGAL-CLN2) cells were grown on Gal, where Cln2 is overproduced, the level of Ste5 was lower than in wild-type cells and present as multiple isoforms of lower mobility, diagnostic of phosphorylation (Fig. 7B, lane 3), in agreement with the findings of Strickfaden et al. (93). After shift of the culture of cln1Δ cln2Δ cln3Δ (pGAL-CLN2) cells to Glc for 150 min, the level of Ste5 increased significantly and the slower-migrating isoforms disappeared (Fig. 7B, lane 4). These results are consistent with the hypothesis that Cln2-Cdc28-mediated phosphorylation is a prelude to the SCFCdc4-dependent destruction of Ste5 in the nucleus.
The above findings predict that the level of nuclear Ste5 should be highest in unbudded cells and should fall rather precipitously as cells commit to the cell cycle and exit G1. Close inspection of random fields of cells in exponentially growing cultures of cells expressing Ste5-GFP3 at a near endogenous level (from the STE5 promoter on a CEN plasmid in ste5Δ cells) showed exactly this expected pattern (Fig. 7C). Unbudded and newborn cells have the brightest nuclear signal. The Ste5 signal is distinctly fainter in cells with very small buds, indicative of cells that have initiated S phase. Cells with large buds that have not undergone mitosis have the faintest Ste5 signal, and in cells undergoing mitosis (and prior to cytokinesis), the level of Ste5 starts to rise again. Thus, the level of Ste5 does indeed correlate with cell cycle phase.
Despite the cumulative evidence that only haploid cells in G1 are susceptible to stimulation of the pheromone response pathway (12, 18, 85), it has been observed that cells synchronized in M phase by arrest of a cdc15ts mutation at the nonpermissive temperature are capable of responding to pheromone, as judged by induction of the transcript of a pheromone-induced gene (FUS1) (69). Our finding that the level of Ste5-GFP3 starts to rise during M phase could reconcile these results, if the amount of Ste5 continues to increase in cells held at the cdc15 block and reaches a level above the threshold required for signaling competence.
Apparent cell cycle-dependent heterogeneity in the level of Ste5 was noted in a prior study (66). However, in that work, a Ste5-myc9 derivative overexpressed from the CUP1 promoter on a high-copy-number (2μm DNA) plasmid was examined by indirect immunofluorescence, and therefore, it could not be ruled out that nonuniform segregation of the 2μm DNA plasmid and/or nonuniform cell permeabilization prior to antibody decoration was responsible for the reported differences in staining at different stages of the cell cycle.
As an additional independent method to demonstrate the cell cycle- and Cln2-dependent change in the level of Ste5, we synchronized both cln1Δ cln2Δ cln3Δ (pGAL-CLN2) cells and cln1Δ cln2Δ cln3Δ (pGAL-CLN2) cells also carrying an msn5Δ mutation in G1 by shifting the cells from Gal-containing medium to Glc-containing medium and then released them from the block by returning them to Gal-containing medium and monitored the level of Ste5. Strikingly, in agreement with the need for active Cln2-Cdc28 to promote both entry into the cell cycle and Ste5 degradation, the level of Ste5 was highest in unbudded cells and subsequently decreased substantially as the cells progressed through the cell cycle, concomitant with the onset of budding (Fig. 7D, left). As expected, the nuclear pool of Ste5 was subject to even more rapid degradation once the cells entered the cell cycle (Fig. 7D, right). We noted that in the cln1Δ cln2Δ cln3Δ (pGAL-CLN2) cells, Ste5 became hyperphosphorylated, in agreement with the findings of Strickfaden et al. (93). Moreover, Ste5 phosphorylation was clearly accompanied by a concomitant decrease in its level (Fig. 7D, lower panels). In marked contrast, Strickfaden et al. (93) did not observe any change in the level of Ste5 during the cell cycle, but in their work, they used a Ste5(ΔNLS) allele that is largely restricted to the cytoplasm. Based on the findings we have presented here, the behavior observed for Ste5(ΔNLS) is exactly what would be predicted because it escapes SCFCdc4-dependent ubiquitinylation and proteasome-mediated destruction in the nucleus. In this regard, we noted that, in msn5Δ cells, where Ste5 is more restricted to the nucleus, the phosphorylated species with the lowest mobility are absent (Fig. 7D, right), most likely because they are the most susceptible to SCFCdc4-dependent ubiquitinylation and therefore undergo the most rapid degradation, in agreement with the known preference of SCFCdc4 for substrates that have been phosphorylated at multiple sites (68, 71, 95). Taken together, these data argue that the level of Ste5 is regulated in a cell cycle-dependent manner because phosphorylation by G1 cyclin-activated Cdk1 during passage through “START” targets Ste5 for ubiquitinylation by the SCFCdc4 complex. Thereby, this mechanism would help ensure that, once cells have exited G1, the level of Ste5 is below the critical threshold necessary to mount a pheromone response.
SCFCdc4-controlled degradation of Ste5 prevents spurious MAPK pathway activation.
As mentioned earlier, it has long been recognized that only haploid cells in G1 are susceptible to stimulation of the pheromone response pathway (12, 18, 85). Our findings suggest that the cells are competent to respond only in this phase of the cell cycle at least in part because the level of Ste5 is at its maximum in early G1. In other words, our results indicate that nucleus- and G1-to-S-specific degradation of Ste5 may be an important mechanism to restrict the capacity of cells to activate this MAPK pathway to G1. If so, inefficient nuclear degradation of Ste5 might raise its level above the threshold necessary to trigger signaling and thus elicit pathway activation even in the absence of any pheromone stimulus. Activation of this pathway elicits several well-characterized responses, including growth inhibition, arrest in the G1 (unbudded) phase of the cell cycle, and induction of the shmoo morphology (51, 102).
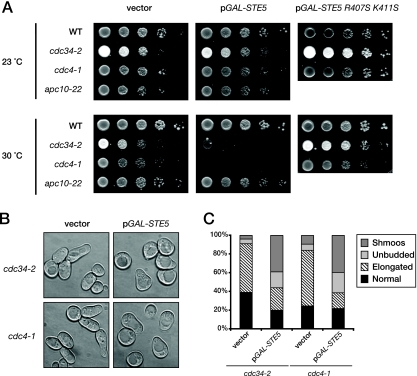
Consistent with our hypothesis, we found that modest overexpression of Ste5 was well-tolerated by wild-type cells but was growth inhibitory to cells carrying mutations in components of SCFCdc4 (cdc34-2ts or cdc4-1ts) when they were incubated at a semipermissive temperature at which they are still able to grow quite well (Fig. 8A). In other words, when cells are capable (at 23°C) of degrading Ste5 via G1 Cdk1-initiated and SCFCdc4-dependent ubiquitinylation in the nucleus, they can continue to grow even when Ste5 is overexpressed from the GAL promoter presumably because G1 Cdk1-dependent destruction of Ste5 and/or G1 Cdk1-dependent displacement of Ste5 from the plasma membrane (93) can keep pace with the elevated STE5 expression and prevent any signaling in cycling cells. However, under conditions (at 30°C) where SCFCdc4-mediated degradation is only slightly compromised, three readouts of the induction of pheromone response all occurred—growth inhibition (Fig. 8A), G1-specific arrest (Fig. 8B and C), and shmoo formation (Fig. 8B and C)—even though active G1 Cdk1 is present in these cells. Thus, G1 Cdk1 phosphorylation of Ste5 is not sufficient to prevent signaling in the absence of efficient Ste5 degradation. These findings indicate that, to prevent Ste5-dependent signaling, G1 Cdk1-dependent turnover and G1 Cdk1-dependent blockade of its membrane recruitment are both necessary.
FIG. 8.
Ubiquitin-dependent proteolysis of Ste5 prevents inadvertent MAPK signaling. (A) To examine the effect of elevated Ste5 expression when its nuclear degradation is compromised, serial dilutions of exponentially growing wild-type (WT) (W303) and otherwise isogenic cdc34-2, cdc4-1, and apc10-22 mutants carrying either an empty CEN vector or the same vector expressing from the GAL1 promoter either wild-type Ste5 or the signaling-defective mutant Ste5(R407S K411S), were spotted onto plates containing SC medium supplemented with Gal and Ura and grown for 3 days at the indicated temperatures (23 and 30°C). (B) To examine the basis of the growth arrest observed, cultures of cdc34-2 and cdc4-1 cells carrying either an empty vector control (left) of the same plasmid expressing Ste5 from the GAL1 promoter (right) were grown in raffinose-containing at 30°C until early exponential phase, at which time galactose was added (final concentration, 2%). After 5 h, cells were visualized by light microscopy. Each panel depicts a collage of representative cells. (C) The percentages of normal cells, cells with elongated buds, large unbudded (G1-arrested) cells, and overt shmoos in the cell populations shown in panel B were determined by microscopic examination of these cultures (n = 200 cells for each culture).
In fact, these results provide an independent confirmation that Ste5 is an authentic SCFCdc4 target because it has been observed before that overexpression of demonstrated substrates of SCFCdc4 inhibits the growth of cells carrying temperature-sensitive mutations in components of the SCFCdc4 complex at a semipermissive temperature (2, 23, 109). Elevated levels of expression of Ste5 were somewhat more deleterious to the cdc4ts mutant than to the cdc34ts mutant (Fig. 8A), in agreement with the greater Ste5 stabilization conferred, in most of our experiments, by the loss of Cdc4 function (Fig. 6). Moreover, this behavior was specific to cells with compromised SCFCdc4 function because cells harboring a mutation (apc10-22) in another nuclear E3, the anaphase-promoting complex, grew just as well at 30°C as the wild-type control when Ste5 was overexpressed, in agreement with the lack of effect of this mutation on Ste5 degradation (Fig. 6).
The observed growth inhibition caused by Ste5 overexpression when SCFCdc4 function is compromised could be due to competition for the more limited pool of SCFCdc4 available and the resulting inability to efficiently degrade other substrates that need to be eliminated to permit optimal cell cycle progression (e.g., Sic1). Alternately, because the degradation of Ste5 itself is impeded, the observed growth inhibition could arise instead from elevated MAPK pathway activation and ensuing cell cycle arrest due to the increased level of signaling. As one means to address this issue, we used, as a negative control, a signaling-defective Ste5 mutant, Ste5(R407S K411S), which we have demonstrated before is expressed at the same level as wild-type Ste5 (34). Hence, as a Cdc34- and SCFCdc4 substrate, it should impede Cdc34- and SCFCdc4-mediated degradation of Sic1 and other substrates to the exact same extent as overexpressed wild-type Ste5 does. We found that when the signaling-defective mutant of Ste5 was overexpressed, no growth inhibition was observed (Fig. 8A, right). Likewise, we found that overexpression of wild-type Ste5 had no effect on the growth of cdc34ts and cdc4ts cells at 30°C if they also lacked Ste11, the MAPK kinase kinase of the signaling cascade (data not shown). These two controls support the conclusion that the growth arrest arises from enhanced Ste5-mediated signaling and is not due to cdc4ts or cdc34ts mutations sensitizing the cells to elevation of the level of any SCFCdc4 substrate.
In this same regard, the growth arrest caused by Ste5 overexpression cannot be attributed to an inability to degrade another SCFCdc4 substrate, the CDK inhibitor Far1 (9, 46). Like Sic1, Far1 should be stabilized by the cdc4ts and cdc34ts mutations in the vector-only control cells (Fig. 8B and C), and perhaps even more so in the cells also overexpressing the signaling-defective Ste5 mutant (data not shown). However, in both cases, the population consisted mainly of large budded cells with markedly elongated buds, the known phenotype for cells with the cdc34ts and cdc4ts mutations, which delay the final stages of the G1/S transition (42). In marked contrast, the majority of the population (>60%) in the culture overexpressing wild-type Ste5 were present as large unbudded cells and shmoos (Fig. 8B and C). Our results are consistent with a previous finding that, even in the absence of pheromone-induced signaling, overexpression of Far1 in cdc34ts cells under semipermissive conditions caused G1 arrest, but not shmoo formation (46). Hence, shmoo formation evoked under the same conditions by Ste5 overexpression provides a second criterion by which we conclude that the inability to efficiently degrade Ste5 leads to elevated signaling.
Finally, when Ste5 expressed at its endogenous level was spared from efficient SCFCdc4-dependent degradation by depletion of G1 cyclins (Fig. 7D), in excess of 30% of the unbudded cells that accumulated displayed a clear-cut shmoo-like morphology (data not shown). Taken together, all of the above findings argue strongly that the nucleus-specific, ubiquitin-dependent, cell cycle-regulated destruction of Ste5 is required to prevent inadvertent pathway activation in the absence of pheromone and is an important mechanism for setting the proper threshold for the optimal spatial and temporal control of signaling flux through the pheromone response pathway.
DISCUSSION
The scaffold protein Ste5 is essential for signal propagation in the mating pheromone-initiated MAPK signaling cascade. Despite its crucial importance, little was known about how Ste5 itself is regulated. Here we have shown that the low steady-state level of Ste5 in vegetatively growing cells is maintained via its ubiquitin- and proteasome-mediated degradation specifically in the nucleus and that this degradation is promoted by G1 cyclin- and CDK1-dependent phosphorylation of Ste5 during exit from G1, thereby marking Ste5 as a substrate for the SCFCdc4 ubiquitin ligase. Conversely, we also show here that pheromone action stabilizes Ste5 by stimulating its Msn5-dependent export from the nucleus and that, once in the cytosol or at the plasma membrane, the half-life of Ste5 is markedly prolonged. Given these new insights, restriction of Ste5 ubiquitinylation and degradation to the nucleus make physiological sense because, once exported to the cytoplasm and thus spared from destruction, Ste5 is free to deliver its bound MAPK cascade components to the cell membrane, thereby potentiating and sustaining signaling. In addition, we provide compelling evidence that the nucleus-restricted, SCFCdc4- and cell cycle-dependent degradation of Ste5 is an important mechanism that contributes to keeping the level of Ste5 below the threshold that could cause spurious stimulus-independent pathway activation. Finally, this mechanism provides an additional satisfying explanation for why only yeast cells that have not yet exited G1 have the capacity to respond efficiaciously to a pheromone signal.
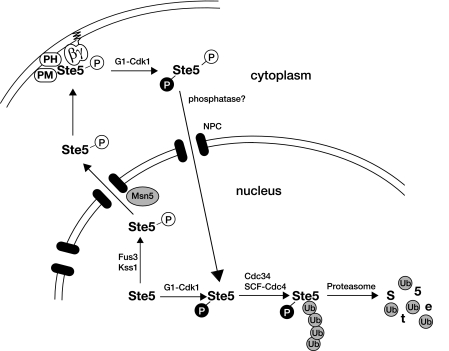
Our findings are summarized schematically in Fig. 9. In essence, the cell has two mechanisms for turning Ste5 off that likely operate hand in hand—G1 Cdk1-initiated displacement of Ste5 from the membrane (93) may allow it to shuttle into the nucleus where it can undergo ubiquitin- and proteasome-mediated destruction, as we have shown, and conversely, lowering the level of Ste5 via its G1 Cdk1-initiated degradation in the nucleus as we have described reduces the cytoplasmic pool of Ste5 that would need to be excluded from the membrane. Thus, both mechanisms ensure that the maximum amount of membrane-binding-competent Ste5 is available in a G1 cell and act together to drop the level of signaling-competent Ste5 below the threshold needed for pheromone-evoked signaling as cells passage out of G1.
FIG. 9.
Control of Ste5 degradation by compartmentalization and protein kinase action. In naïve cells, Ste5 undergoes nucleocytoplasmic shuttling, with the highest concentration residing in the nucleus. In early G1, prior to the buildup of G1 cyclins (like Cln2) and Cln-dependent activation of Cdk1/Cdc28, the nuclear pool of Ste5 is at a maximum, which can be rapidly drawn upon if the cells encounter mating pheromone. Pheromone-evoked MAPK (Fus3 and Kss1)-dependent phosphorylation of Ste5 stimulates its Msn5-dependent export from the nucleus, and once Ste5 is in the cytosol where it is stable, the amount of Ste5 that can be recruited to the plasma membrane to potentiate signaling becomes correspondingly elevated. This mechanism provides a self-reinforcing feed-forward loop that strengthens and sustains signaling. However, once the levels of the G1 cyclins (especially Cln2) have risen sufficiently, Cdk1(Cdc28) becomes activated. Active Cln-bound Cdk1 promotes the processes required for exit from G1 and entry into S phase (e.g., phosphorylation of the CDK inhibitor Sic1, thereby marking it for its SCFCdc4-dependent and proteasome-mediated destruction). Likewise, active Cln-bound Cdk1 also phosphorylates Ste5; in the cytosol, the G1 Cdk1-dependent modifications block membrane binding, and in the nucleus, G1 Cdk1 phosphorylation initiates SCFCdc4-dependent and proteasome-mediated destruction. Removal of this scaffold protein and its displacement from the membrane obviate the ability of the cells to mount a productive pheromone response at any subsequent stage of the cell division cycle. When the cells return to G1, when Ste5 is stable, an adequate amount of Ste5 builds up and is again available to promote an efficacious response to pheromone, if the cells encounter this signal. See text for further discussion. Abbreviations: PH, pleckstrin homology domain; PM, plasma membrane-binding motif; P, phosphate; NPC, nuclear pore complex: Ub, ubiquitin.
As we demonstrated here, Ste5 appears to be an only moderately unstable protein in naïve cells (t1/2 of ∼60 min). However, this overall rate of turnover is misleading because Ste5 undergoes continuous nucleocytoplasmic shuttling and, as we showed, the molecules that are in the cytosol are spared from degradation. Indeed, when exit of Ste5 from the nucleus is impeded by lack of the exportin Msn5, Ste5 turnover was commensurately more rapid (t1/2 of ∼30 min). We have evidence that another karyopherin, the exportin Crm1/Xpo1, also contributes to the basal (as opposed to the pheromone-stimulated) exit of Ste5 from the nucleus (I. Macara, C. Sette, and J. Thorner, unpublished results). If Ste5 were confined exclusively to the nucleus, its intrinsic rate of breakdown in that compartment might approach that observed for bona fide cell cycle regulators, such as the G1 cyclin Cln2 (57, 83) and the S-phase CDK inhibitor Sic1 (68) (t1/2 of ∼5 to 10 min). Indeed, the rather precipitous drop in the cellular content of Ste5 we observed in synchronized msn5Δ cells as they exit G1 indicates that the degradation of Ste5 in the nucleus may be rapid enough to provide the kind of irreversible switch-like transition necessary to execute a dramatic reduction in the capacity of the cell to respond to pheromone once cells have committed to entering the cell cycle.
Our evidence argues that regulation by the ubiquitin-proteasome system contributes in a significant way to setting the critical threshold for Ste5 function. We found that wild-type cells tolerate an increase in the level of Ste5, whereas cells in which G1-specific degradation of Ste5 in the nucleus is compromised displayed response pathway activation in the absence of pheromone, as judged by growth inhibition, G1-specific arrest, and the formation of characteristic shmoos. Normally, elevation of stable cytosolic Ste5 is achieved following initial exposure to pheromone via its self-reinforcing export from the nucleus. Thus, overall, the mechanism we have uncovered helps maximize the ability of the cell to mount an efficacious response if the cell encounters pheromone, provided it is in the G1 phase of the cell cycle, while minimizing the likelihood that spurious activation of a response will occur in the absence of an authentic upstream signal, even if the cell is in G1. Akin to Ste5, reducing the rate of turnover of KSR (kinase suppressor of Ras), a pseudokinase that acts as a scaffold in the mammalian Ras-Raf-MEK-ERK (extracellular signal-regulated kinase) pathway, appears to be important for achieving sustained activation of a MAPK-dependent signaling response after receptor tyrosine kinase stimulation (80).
Catalysis of Ste5 ubiquitinylation by SCFCdc4 is presumably the rate-limiting step for Ste5 degradation because, once the first ubiquitin is attached, this E3 is known to be a highly processive enzyme (77). On the other hand, studies of other targets of SCFCdc4 indicate that the specific position in the substrate to which the ubiquitin chain is attached can profoundly influence the rate of subsequent proteasomal proteolysis (75). It might be interesting to explore whether the exact site(s) of Ste5 ubiquitin modification influences the kinetics, amplitude, duration, and cell cycle dependence of signaling. The fact that SCFCdc4 can bind only to substrates that have been prephosphorylated by G1 cyclin-activated Cdk1 (68) provides a built-in mechanism, first, to ensure that even nuclear Ste5 is spared from degradation in G1 when the level of Cln-bound Cdc28 is low and, second, to initiate rapid and efficient degradation of Ste5 once a sufficient level of Cln-bound Cdk1 is present to trigger exit from G1. This role for Cln-bound Cdk1-mediated phosphorylation in dramatically lowering the cellular content of Ste5 provides a distinctly different mechanism for impeding Ste5-dependent signaling than that proposed by Strickfaden et al. (93). They provided evidence that Ste5 phosphorylated at multiple sites in its N-terminal region (including T4, S11, T29, S43, S69, S71, S81, and T102) by G1 cyclin-bound Cdk1 has a much reduced affinity for the plasma membrane, which should inhibit signal propagation and contribute to restricting pathway activation to the G1 phase of the cell cycle. We showed here that depletion of Cln2 results in a marked increase in the level of Ste5; conversely, overexpression of Cln2 results in a reduction in the steady-state level of Ste5. Correspondingly, we also observed that the highest accumulation of GFP-tagged Ste5 occurs in unbudded (G1-phase) cells and that inactivation of Cdk1 resulted in accumulation of an even higher level of Ste5. Although G1 Cdk1-dependent reduction in the level of Ste5 and G1 Cdk1-dependent inhibition of membrane association are distinct processes, both likely contribute to preventing Ste5 from acting at any stage of the cell cycle other than G1, as already discussed.
Given this dual mechanism for blocking Ste5-dependent signaling, it is not hard to see how, as reported by Strickfaden et al. (93), abnormally high Cdk1 activity (due to overexpression of CLN2 from the GAL promoter) could still block signaling by a mutant, Ste5(NLSm), that is impaired in its nuclear import or by an overexpressed hyperactive allele, Ste5-Q59L. We showed that overexpression of CLN2 from the GAL promoter not only results in elevation of Cdk1-mediated phosphorylation of Ste5 (Fig. 7B) but also drives Ste5 degradation (Fig. 7D). Thus, Ste5-Q59L and even Ste5(NLSm), which, in our hands, is still able to enter the nucleus (Fig. 3C), will still be subject to Cdk1- and SCFCdc4-mediated degradation. Indeed, we have shown that as long as the G1 Cdk1-dependent and SCFCdc4-mediated destruction of Ste5 is not compromised, the cell can tolerate GAL-driven overexpression of wild-type Ste5 even when CLN2 is expressed at its endogenous level, and the cell continues to divide (Fig. 8A). Thus, again, it is easy to envision that an elevated rate of phosphorylation (due to GAL-driven CLN2 expression) can—via the combined mechanisms of promoting Ste5 degradation and inhibiting Ste5 membrane association—readily block signaling even by Ste5(NLSm) or the overexpressed hyperactive Ste5 allele.
Versions of Ste5 tethered to the plasma membrane artificially never enter the nucleus, yet they are fully capable of supporting a robust mating pheromone response (34, 79). By contrast, an msn5Δ xpo1-1ts double mutant exhibits completely defective mating, even at a permissive temperature, whereas both an msn5Δ single mutant and an xpo1-1ts single mutant display reduced but readily detectable mating (C. Sette and J. Thorner, unpublished results). Thus, normally, initiation of pheromone response requires that at least some cytosolic pool of Ste5 be present via its exit from the nucleus (Fig. 9). If pheromone is present to generate free Gβγ to recruit that initial pool of Ste5 to the plasma membrane, activation of some Fus3 ensues. The activated MAPK then translocates to the nucleus (99), where it phosphorylates Ste5 (31), thereby greatly stimulating its Msn5-dependent nuclear export (66), which, as we have shown here, allows the scaffold to escape SCFCdc4-mediated destruction and permits the now stable protein to accumulate at the tip of the mating projection. Thus, the nuclear pool of Ste5 serves as a storehouse and reservoir to supply the full complement of Ste5 that is needed to mount an effective and sustained signal, and continuous low-level shuttling of some of the scaffold into the cytosol from that depot gives the cell the capacity to constantly be on guard for the presence of a pheromone signal. The basal level of Fus3 activity may be important for promoting the Msn5-dependent shuttling of these pioneer Ste5 molecules into the cytosol because, as would be predicted from our findings, it has recently been reported that fus3Δ cells display a lower steady-state level of Ste5 (117), just as we observed for cells lacking Msn5.
Prior work suggested that reimport of Ste5 into the nucleus contributes to attenuation of pheromone-evoked signaling (53) and reduces inadvertent signaling in naïve cells (55, 111). Our discovery that Ste5 degradation can occur only in the nucleus now provides a physiologically meaningful rationale for these findings. In this regard, an observation made by Bhattacharyya et al. (7) is of some interest. They found that binding of a fragment of Ste5 to Fus3 stimulates the activity of this MAPK in vitro, which results in phosphorylation of Ste5 on T287, and that, as judged by site-directed mutagenesis at least, this residue in Ste5 appears to be important for downregulation of signaling in vivo. In light of our findings, it will be interesting to determine whether phosphorylation at this site occurs in vivo and, if so, whether modification at this site increases nuclear import or prevents nuclear export of Ste5 (or both), thus contributing to signal attenuation by promoting degradation of Ste5 in the nuclear compartment.
Initially, it was an attractive hypothesis that the RING-H2 domain (residues 177 to 229) in Ste5 might contribute to its own destruction via autoubiquitinylation. The observation that two-hybrid analysis of an N-terminal fragment of Ste5 identified the ubiquitin-conjugating enzyme (E2) Ubc4 as an interacting partner and that this interaction required the RING-H2 domain (P. Pryciak, University of Massachusetts Medical Center, personal communication) provided some support for this hypothesis. However, as we showed here, several mutations in the RING-H2 motif that have clear phenotypic effects on Ste5 function did not detectably alter its rate of turnover in vivo. Furthermore, when incubated individually with ATP, commercially obtained E1, and purified recombinant versions of each yeast E2, bacterially expressed and purified fragments of Ste5 containing its RING-H2 domain or full-length Ste5 did not show any detectable autoubiquitinylation activity under conditions where another recombinant yeast RING-containing protein, Apc11, displayed copious autoubiquitinylation (L. Garrenton, A. Saviñon-Tejeda, and J. Thorner, unpublished results). Therefore, even if the RING-H2 domain in Ste5 can promote ubiquitinylation in vivo (when Ste5 is appropriately modified and/or associated with the right factors), we suspect that Ste5 will not act as an E3 on itself, but on some other component of the pheromone response pathway.
Evidence from a variety of organisms implicates ubiquitinylation in modulating MAPK signaling through effects exerted at a variety of levels (assembly of protein kinase-containing complexes, subcellular localization of MAPKs and/or their substrates, or degradation of MAPKs and/or their substrates) (reviewed in reference 54). In yeast, Ste5 joins a growing list of components of the mating pheromone response pathway reportedly regulated by ubiquitin- and proteasome-mediated destruction. However, for many of these proteins, including Ste2 (47), Gpa1 (87), Ste7 (104), Ste11 (26), and Ste12 (27), pheromone treatment apparently stimulates the ubiquitin- and proteasome-mediated degradation of these molecules as a means to downregulate and/or terminate signaling. In contrast, as we have shown here, pheromone treatment rescues Ste5 from the ubiquitin- and proteasome-mediated degradation it would otherwise undergo in the nucleus. In this respect, the behavior of Ste5 resembles that of Far1, which serves as a scaffold protein for the Cdc42 guanine nucleotide exchange factor, Cdc24, in polarized morphogenesis of the shmoo (108) and which also serves as an inhibitor of the G1 CDK (73). As for Ste5, pheromone treatment promotes nuclear export of Far1, and because Far1 is another target of the SCFCdc4, its exit from the nucleus results in stabilization of the protein (9, 46). It makes sound physiological sense that the cell would employ one common mechanism to make certain that the two scaffold proteins pivotal to the execution of the two major physiological branches of the mating response are simultaneously stabilized and thus available in the cytosol to associate with the plasma membrane where they need to act. Other SCFCdc4 targets (Sic1, Cdc6, and Gcn4) are also stabilized when sequestered away from nuclear Cdc4 (22, 25). Likewise, in mammalian cells, changes in subcellular localization control the rate of ubiquitinylation and subsequent degradation of key regulatory proteins, including the p53 tumor suppressor (41) and the CDK inhibitor p27Kip1(32). Our work shows that similar mechanisms can be extended to the scaffold proteins that control MAPK signaling.
Acknowledgments
We thank P. Pryciak, C. Wittenberg, L. Hicke, and K. Benjamin for the generous gift of reagents, communication of unpublished results, and/or useful advice. We thank A. Connery, A. Saviñon-Tejeda, R. Chen, and other members of the Thorner laboratory for technical assistance and/or helpful discussions.
This work was supported by NIH Predoctoral Traineeship GM07232 and NCI Predoctoral Traineeship CA09041 (to L.S.G), by Ku-1235 of the Deutsche Forschungsgemeinschaft and Long-Term Fellowship of the Human Frontier Science Program Organization (to M.K.), by NIH research grant GM21841 (to J.T.), and by facilities provided by the Cancer Research Laboratory of the University of California, Berkeley.
Footnotes
Published ahead of print on 10 November 2008.
REFERENCES
- 1.Alani, E., L. Cao, and N. Kleckner. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai, C., P. Sen, K. Hofmann, L. Ma, M. Goebl, J. W. Harper, and S. J. Elledge. 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86263-274. [DOI] [PubMed] [Google Scholar]
- 3.Bailer, S. M., C. Balduf, J. Katahira, A. Podtelejnikov, C. Rollenhagen, M. Mann, N. Pante, and E. Hurt. 2000. Nup116p associates with the Nup82p-Nsp1p-Nup159p nucleoporin complex. J. Biol. Chem. 27523540-23548. [DOI] [PubMed] [Google Scholar]
- 4.Bardwell, L., J. G. Cook, J. X. Zhu-Shimoni, D. Voora, and J. Thorner. 1998. Differential regulation of transcription: repression by unactivated mitogen-activated protein kinase Kss1 requires the Dig1 and Dig2 proteins. Proc. Natl. Acad. Sci. USA 9515400-15405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bashor, C. J., N. C. Helman, S. Yan, and W. A. Lim. 2008. Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics. Science 3191539-1543. [DOI] [PubMed] [Google Scholar]
- 6.Baum, P., J. Thorner, and L. Honig. 1978. Identification of tubulin from the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 754962-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharyya, R. P., A. Remenyi, M. C. Good, C. J. Bashor, A. M. Falick, and W. A. Lim. 2006. The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science 311822-826. [DOI] [PubMed] [Google Scholar]
- 8.Bhattacharyya, R. P., A. Remenyi, B. J. Yeh, and W. A. Lim. 2006. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu. Rev. Biochem. 75655-680. [DOI] [PubMed] [Google Scholar]
- 9.Blondel, M., J. M. Galan, Y. Chi, C. Lafourcade, C. Longaretti, R. J. Deshaies, and M. Peter. 2000. Nuclear-specific degradation of Far1 is controlled by the localization of the F-box protein Cdc4. EMBO J. 196085-6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14115-132. [DOI] [PubMed] [Google Scholar]
- 11.Breitkreutz, A., L. Boucher, and M. Tyers. 2001. MAPK specificity in the yeast pheromone response independent of transcriptional activation. Curr. Biol. 111266-1271. [DOI] [PubMed] [Google Scholar]
- 12.Bücking-Throm, E., W. Duntze, L. H. Hartwell, and T. R. Manney. 1973. Reversible arrest of haploid yeast cells in the initiation of DNA synthesis by a diffusible sex factor. Exp. Cell Res. 7699-110. [DOI] [PubMed] [Google Scholar]
- 13.Burack, W. R., and A. S. Shaw. 2000. Signal transduction: hanging on a scaffold. Curr. Opin. Cell Biol. 12211-216. [DOI] [PubMed] [Google Scholar]
- 14.Butty, A. C., P. M. Pryciak, L. S. Huang, I. Herskowitz, and M. Peter. 1998. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science 2821511-1516. [DOI] [PubMed] [Google Scholar]
- 15.Chen, R. E., and J. Thorner. 2007. Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 17731311-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi, W. J., M. W. Clark, J. X. Chen, and A. Y. Jong. 1990. The CDC4 gene product is associated with the yeast nuclear skeleton. Biochem. Biophys. Res. Commun. 1721324-1330. [DOI] [PubMed] [Google Scholar]
- 17.Ciejek, E., and J. Thorner. 1979. Recovery of S. cerevisiae a cells from G1 arrest by alpha factor pheromone requires endopeptidase action. Cell 18623-635. [DOI] [PubMed] [Google Scholar]
- 18.Colman-Lerner, A., A. Gordon, E. Serra, T. Chin, O. Resnekov, D. Endy, C. G. Pesce, and R. Brent. 2005. Regulated cell-to-cell variation in a cell-fate decision system. Nature 437699-706. [DOI] [PubMed] [Google Scholar]
- 19.Cross, F. R., and C. M. Blake. 1993. The yeast Cln3 protein is an unstable activator of Cdc28. Mol. Cell. Biol. 133266-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D'Andrea, A., and D. Pellman. 1998. Deubiquitinating enzymes: a new class of biological regulators. Crit. Rev. Biochem. Mol. Biol. 33337-352. [DOI] [PubMed] [Google Scholar]
- 21.Dard, N., and M. Peter. 2006. Scaffold proteins in MAP kinase signaling: more than simple passive activating platforms. Bioessays 28146-156. [DOI] [PubMed] [Google Scholar]
- 22.Deshaies, R. J. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15435-467. [DOI] [PubMed] [Google Scholar]
- 23.Deshaies, R. J., V. Chau, and M. Kirschner. 1995. Ubiquitination of the G1 cyclin Cln2p by a Cdc34p-dependent pathway. EMBO J. 14303-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drogen, F., S. M. O'Rourke, V. M. Stucke, M. Jaquenoud, A. M. Neiman, and M. Peter. 2000. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 10630-639. [DOI] [PubMed] [Google Scholar]
- 25.Elsasser, S., Y. Chi, P. Yang, and J. L. Campbell. 1999. Phosphorylation controls timing of Cdc6p destruction: a biochemical analysis. Mol. Biol. Cell 103263-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esch, R. K., and B. Errede. 2002. Pheromone induction promotes Ste11 degradation through a MAPK feedback and ubiquitin-dependent mechanism. Proc. Natl. Acad. Sci. USA 999160-9165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esch, R. K., Y. Wang, and B. Errede. 2006. Pheromone-induced degradation of Ste12 contributes to signal attenuation and the specificity of developmental fate. Eukaryot. Cell 52147-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evan, G. I., G. K. Lewis, G. Ramsay, and J. M. Bishop. 1985. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 53610-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng, Y., L. Y. Song, E. Kincaid, S. K. Mahanty, and E. A. Elion. 1998. Functional binding between Gbeta and the LIM domain of Ste5 is required to activate the MEKK Ste11. Curr. Biol. 8267-278. [DOI] [PubMed] [Google Scholar]
- 30.Ferrell, J. E., Jr. 2000. What do scaffold proteins really do? Sci. STKE 2000PE1. [DOI] [PubMed] [Google Scholar]
- 31.Flotho, A., D. M. Simpson, M. Qi, and E. A. Elion. 2004. Localized feedback phosphorylation of Ste5p scaffold by associated MAPK cascade. J. Biol. Chem. 27947391-47401. [DOI] [PubMed] [Google Scholar]
- 32.Frescas, D., and M. Pagano. 2008. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat. Rev. Cancer 8438-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaber, R. F., D. M. Copple, B. K. Kennedy, M. Vidal, and M. Bard. 1989. The yeast gene ERG6 is required for normal membrane function but is not essential for biosynthesis of the cell-cycle-sparking sterol. Mol. Cell. Biol. 93447-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garrenton, L. S., S. L. Young, and J. Thorner. 2006. Function of the MAPK scaffold protein, Ste5, requires a cryptic PH domain. Genes Dev. 201946-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gelbart, M. E., T. Rechsteiner, T. J. Richmond, and T. Tsukiyama. 2001. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol. Cell. Biol. 212098-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghislain, M., A. Udvardy, and C. Mann. 1993. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature 366358-362. [DOI] [PubMed] [Google Scholar]
- 37.Goebl, M. G., L. Goetsch, and B. Byers. 1994. The Ubc3 (Cdc34) ubiquitin-conjugating enzyme is ubiquitinated and phosphorylated in vivo. Mol. Cell. Biol. 143022-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gordon, A., A. Colman-Lerner, T. E. Chin, K. R. Benjamin, R. C. Yu, and R. Brent. 2007. Single-cell quantification of molecules and rates using open-source microscope-based cytometry. Nat. Methods 4175-181. [DOI] [PubMed] [Google Scholar]
- 39.Görlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15607-660. [DOI] [PubMed] [Google Scholar]
- 40.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 41.Harris, S. L., and A. J. Levine. 2005. The p53 pathway: positive and negative feedback loops. Oncogene 242899-2908. [DOI] [PubMed] [Google Scholar]
- 42.Hartwell, L. H., J. Culotti, J. R. Pringle, and B. J. Reid. 1974. Genetic control of the cell division cycle in yeast. Science 18346-51. [DOI] [PubMed] [Google Scholar]
- 43.Hasson, M. S. 1992. Analysis of Saccharomyces cerevisiae pheromone response: biochemical and genetic characterization of the Ste5 protein. Ph.D. thesis. University of California, Berkeley, Berkeley.
- 44.Hasson, M. S., D. Blinder, J. Thorner, and D. D. Jenness. 1994. Mutational activation of the STE5 gene product bypasses the requirement for G protein β and γ subunits in the yeast pheromone response pathway. Mol. Cell. Biol. 141054-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hellmuth, K., D. M. Lau, F. R. Bischoff, M. Kunzler, E. Hurt, and G. Simos. 1998. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol. Cell. Biol. 186374-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henchoz, S., Y. Chi, B. Catarin, I. Herskowitz, R. J. Deshaies, and M. Peter. 1997. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 113046-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hicke, L., B. Zanolari, and H. Riezman. 1998. Cytoplasmic tail phosphorylation of the alpha-factor receptor is required for its ubiquitination and internalization. J. Cell Biol. 141349-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirschman, J. E., and D. D. Jenness. 1999. Dual lipid modification of the yeast Gγ subunit Ste18p determines membrane localization of Gβγ. Mol. Cell. Biol. 197705-7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inouye, C., N. Dhillon, T. Durfee, P. C. Zambryski, and J. Thorner. 1997. Mutational analysis of STE5 in the yeast Saccharomyces cerevisiae: application of a differential interaction trap assay for examining protein-protein interactions. Genetics 147479-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inouye, C., N. Dhillon, and J. Thorner. 1997. Ste5 RING-H2 domain: role in Ste4-promoted oligomerization for yeast pheromone signaling. Science 278103-106. [DOI] [PubMed] [Google Scholar]
- 51.Knaus, M., P. Wiget, Y. Shimada, and M. Peter. 2005. Control of cell polarity in response to intra- and extracellular signals in budding yeast. Novartis Found. Symp. 26947-58. [PubMed] [Google Scholar]
- 52.Kranz, J. E., B. Satterberg, and E. A. Elion. 1994. The MAP kinase Fus3 associates with and phosphorylates the upstream signaling component Ste5. Genes Dev. 8313-327. [DOI] [PubMed] [Google Scholar]
- 53.Künzler, M., J. Trueheart, C. Sette, E. Hurt, and J. Thorner. 2001. Mutations in the YRB1 gene encoding yeast ran-binding-protein-1 that impair nucleocytoplasmic transport and suppress yeast mating defects. Genetics 1571089-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laine, A., and Z. Ronai. 2005. Ubiquitin chains in the ladder of MAPK signaling. Sci. STKE 2005re5. [DOI] [PubMed] [Google Scholar]
- 55.Lamson, R. E., S. Takahashi, M. J. Winters, and P. M. Pryciak. 2006. Dual role for membrane localization in yeast MAP kinase cascade activation and its contribution to signaling fidelity. Curr. Biol. 16618-623. [DOI] [PubMed] [Google Scholar]
- 56.Lamson, R. E., M. J. Winters, and P. M. Pryciak. 2002. Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20. Mol. Cell. Biol. 222939-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lanker, S., M. H. Valdivieso, and C. Wittenberg. 1996. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science 2711597-1601. [DOI] [PubMed] [Google Scholar]
- 58.Leberer, E., D. Dignard, D. Harcus, D. Y. Thomas, and M. Whiteway. 1992. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein beta gamma subunits to downstream signalling components. EMBO J. 114815-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee, D. H., and A. L. Goldberg. 1996. Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J. Biol. Chem. 27127280-27284. [DOI] [PubMed] [Google Scholar]
- 60.Leeuw, T., C. Wu, J. D. Schrag, M. Whiteway, D. Y. Thomas, and E. Leberer. 1998. Interaction of a G-protein beta-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature 391191-195. [DOI] [PubMed] [Google Scholar]
- 61.Levchenko, A., J. Bruck, and P. W. Sternberg. 2000. Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc. Natl. Acad. Sci. USA 975818-5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Linder, M. E., and R. J. Deschenes. 2007. Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 874-84. [DOI] [PubMed] [Google Scholar]
- 63.Liu, C., J. Apodaca, L. E. Davis, and H. Rao. 2007. Proteasome inhibition in wild-type yeast Saccharomyces cerevisiae cells. BioTechniques 42158-162. [DOI] [PubMed] [Google Scholar]
- 64.Lorenz, M. C., R. S. Muir, E. Lim, J. McElver, S. C. Weber, and J. Heitman. 1995. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene 158113-117. [DOI] [PubMed] [Google Scholar]
- 65.Lörincz, A. T., and S. I. Reed. 1986. Sequence analysis of temperature-sensitive mutations in the Saccharomyces cerevisiae gene CDC28. Mol. Cell. Biol. 64099-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mahanty, S. K., Y. Wang, F. W. Farley, and E. A. Elion. 1999. Nuclear shuttling of yeast scaffold Ste5 is required for its recruitment to the plasma membrane and activation of the mating MAPK cascade. Cell 98501-512. [DOI] [PubMed] [Google Scholar]
- 67.Nair, U., and D. J. Klionsky. 2005. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J. Biol. Chem. 28041785-41788. [DOI] [PubMed] [Google Scholar]
- 68.Nash, P., X. Tang, S. Orlicky, Q. Chen, F. B. Gertler, M. D. Mendenhall, F. Sicheri, T. Pawson, and M. Tyers. 2001. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414514-521. [DOI] [PubMed] [Google Scholar]
- 69.Oehlen, L. J., and F. R. Cross. 1994. G1 cyclins CLN1 and CLN2 repress the mating factor response pathway at Start in the yeast cell cycle. Genes Dev. 81058-1070. [DOI] [PubMed] [Google Scholar]
- 70.Oehlen, L. J., and F. R. Cross. 1998. Potential regulation of Ste20 function by the Cln1-Cdc28 and Cln2-Cdc28 cyclin-dependent protein kinases. J. Biol. Chem. 27325089-25097. [DOI] [PubMed] [Google Scholar]
- 71.Orlicky, S., X. Tang, A. Willems, M. Tyers, and F. Sicheri. 2003. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112243-256. [DOI] [PubMed] [Google Scholar]
- 72.Ouspenski, I. I., U. W. Mueller, A. Matynia, S. Sazer, S. J. Elledge, and B. R. Brinkley. 1995. Ran-binding protein-1 is an essential component of the Ran/RCC1 molecular switch system in budding yeast. J. Biol. Chem. 2701975-1978. [DOI] [PubMed] [Google Scholar]
- 73.Peter, M. 1997. The regulation of cyclin-dependent kinase inhibitors (CKIs). Prog. Cell Cycle Res. 399-108. [DOI] [PubMed] [Google Scholar]
- 74.Peters, J. M. 2002. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell 9931-943. [DOI] [PubMed] [Google Scholar]
- 75.Petroski, M. D., and R. J. Deshaies. 2003. Context of multiubiquitin chain attachment influences the rate of Sic1 degradation. Mol. Cell 111435-1444. [DOI] [PubMed] [Google Scholar]
- 76.Petroski, M. D., and R. J. Deshaies. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 69-20. [DOI] [PubMed] [Google Scholar]
- 77.Petroski, M. D., and R. J. Deshaies. 2005. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell 1231107-1120. [DOI] [PubMed] [Google Scholar]
- 78.Pickart, C. M. 2004. Back to the future with ubiquitin. Cell 116181-190. [DOI] [PubMed] [Google Scholar]
- 79.Pryciak, P. M., and F. A. Huntress. 1998. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gbetagamma complex underlies activation of the yeast pheromone response pathway. Genes Dev. 122684-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Razidlo, G. L., R. L. Kortum, J. L. Haferbier, and R. E. Lewis. 2004. Phosphorylation regulates KSR1 stability, ERK activation, and cell proliferation. J. Biol. Chem. 27947808-47814. [DOI] [PubMed] [Google Scholar]
- 81.Reed, S. I., and C. Wittenberg. 1990. Mitotic role for the Cdc28 protein kinase of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 875697-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Russell, S. J., K. A. Steger, and S. A. Johnston. 1999. Subcellular localization, stoichiometry, and protein level of 26S proteasome subunits in yeast. J. Biol. Chem. 27421943-21952. [DOI] [PubMed] [Google Scholar]
- 83.Salama, S. R., K. B. Hendricks, and J. Thorner. 1994. G1 cyclin degradation: the PEST motif of yeast Cln2 is necessary, but not sufficient, for rapid protein turnover. Mol. Cell. Biol. 147953-7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 85.Samokhin, G. P., L. V. Lizlova, J. D. Bespalova, M. Titov, and V. N. Smirnov. 1981. The effect of alpha-factor on the rate of cell-cycle initiation in Saccharomyces cerevisiae: alpha-factor modulates transition probability in yeast. Exp. Cell Res. 131267-275. [DOI] [PubMed] [Google Scholar]
- 86.Schafer, W. R., and J. Rine. 1992. Protein prenylation: genes, enzymes, targets, and functions. Annu. Rev. Genet. 26209-237. [DOI] [PubMed] [Google Scholar]
- 87.Schauber, C., L. Chen, P. Tongaonkar, I. Vega, and K. Madura. 1998. Sequence elements that contribute to the degradation of yeast G alpha. Genes Cells 3307-319. [DOI] [PubMed] [Google Scholar]
- 88.Schlenstedt, G., D. H. Wong, D. M. Koepp, and P. A. Silver. 1995. Mutants in a yeast Ran binding protein are defective in nuclear transport. EMBO J. 145367-5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sette, C., C. J. Inouye, S. L. Stroschein, P. J. Iaquinta, and J. Thorner. 2000. Mutational analysis suggests that activation of the yeast pheromone response mitogen-activated protein kinase pathway involves conformational changes in the Ste5 scaffold protein. Mol. Biol. Cell 114033-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shao, D., W. Zheng, W. Qiu, Q. Ouyang, and C. Tang. 2006. Dynamic studies of scaffold-dependent mating pathway in yeast. Biophys. J. 913986-4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 92.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 12219-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Strickfaden, S. C., M. J. Winters, G. Ben-Ari, R. E. Lamson, M. Tyers, and P. M. Pryciak. 2007. A mechanism for cell-cycle regulation of MAP kinase signaling in a yeast differentiation pathway. Cell 128519-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Takahashi, S., and P. M. Pryciak. 2007. Identification of novel membrane-binding domains in multiple yeast Cdc42 effectors. Mol. Biol. Cell 184945-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tang, X., S. Orlicky, Z. Lin, A. Willems, D. Neculai, D. Ceccarelli, F. Mercurio, B. H. Shilton, F. Sicheri, and M. Tyers. 2007. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell 1291165-1176. [DOI] [PubMed] [Google Scholar]
- 96.Taxis, C., and M. Knop. 2006. System of centromeric, episomal, and integrative vectors based on drug resistance markers for Saccharomyces cerevisiae. BioTechniques 4073-78. [DOI] [PubMed] [Google Scholar]
- 97.Thrower, J. S., L. Hoffman, M. Rechsteiner, and C. M. Pickart. 2000. Recognition of the polyubiquitin proteolytic signal. EMBO J. 1994-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Toenjes, K. A., D. Simpson, and D. I. Johnson. 2004. Separate membrane targeting and anchoring domains function in the localization of the S. cerevisiae Cdc24p guanine nucleotide exchange factor. Curr. Genet. 45257-264. [DOI] [PubMed] [Google Scholar]
- 99.van Drogen, F., V. M. Stucke, G. Jorritsma, and M. Peter. 2001. MAP kinase dynamics in response to pheromones in budding yeast. Nat. Cell Biol. 31051-1059. [DOI] [PubMed] [Google Scholar]
- 100.Varshavsky, A. 2005. Regulated protein degradation. Trends Biochem. Sci. 30283-286. [DOI] [PubMed] [Google Scholar]
- 101.Versele, M., and J. Thorner. 2004. Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4. J. Cell Biol. 164701-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang, Y., and H. G. Dohlman. 2004. Pheromone signaling mechanisms in yeast: a prototypical sex machine. Science 3061508-1509. [DOI] [PubMed] [Google Scholar]
- 103.Wang, Y., and E. A. Elion. 2003. Nuclear export and plasma membrane recruitment of the Ste5 scaffold are coordinated with oligomerization and association with signal transduction components. Mol. Biol. Cell 142543-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang, Y., Q. Ge, D. Houston, J. Thorner, B. Errede, and H. G. Dohlman. 2003. Regulation of Ste7 ubiquitination by Ste11 phosphorylation and the Skp1-Cullin-F-box complex. J. Biol. Chem. 27822284-22289. [DOI] [PubMed] [Google Scholar]
- 105.Wassmann, K., and G. Ammerer. 1997. Overexpression of the G1-cyclin gene CLN2 represses the mating pathway in Saccharomyces cerevisiae at the level of the MEKK Ste11. J. Biol. Chem. 27213180-13188. [DOI] [PubMed] [Google Scholar]
- 106.Whiteway, M. S., C. Wu, T. Leeuw, K. Clark, A. Fourest-Lieuvin, D. Y. Thomas, and E. Leberer. 1995. Association of the yeast pheromone response G protein beta gamma subunits with the MAP kinase scaffold Ste5p. Science 2691572-1575. [DOI] [PubMed] [Google Scholar]
- 107.Whitmarsh, A. J., and R. J. Davis. 1998. Structural organization of MAP-kinase signaling modules by scaffold proteins in yeast and mammals. Trends Biochem. Sci. 23481-485. [DOI] [PubMed] [Google Scholar]
- 108.Wiget, P., Y. Shimada, A. C. Butty, E. Bi, and M. Peter. 2004. Site-specific regulation of the GEF Cdc24p by the scaffold protein Far1p during yeast mating. EMBO J. 231063-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Willems, A. R., S. Lanker, E. E. Patton, K. L. Craig, T. F. Nason, N. Mathias, R. Kobayashi, C. Wittenberg, and M. Tyers. 1996. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell 86453-463. [DOI] [PubMed] [Google Scholar]
- 110.Willems, A. R., M. Schwab, and M. Tyers. 2004. A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta 1695133-170. [DOI] [PubMed] [Google Scholar]
- 111.Winters, M. J., R. E. Lamson, H. Nakanishi, A. M. Neiman, and P. M. Pryciak. 2005. A membrane binding domain in the Ste5 scaffold synergizes with Gbeta-gamma binding to control localization and signaling in pheromone response. Mol. Cell 2021-32. [DOI] [PubMed] [Google Scholar]
- 112.Wittenberg, C., and S. I. Reed. 2005. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene 242746-2755. [DOI] [PubMed] [Google Scholar]
- 113.Wolf, D. H. 1992. Proteases as biological regulators. Introductory remarks. Experientia 48117-118. [DOI] [PubMed] [Google Scholar]
- 114.Wu, C., M. Whiteway, D. Y. Thomas, and E. Leberer. 1995. Molecular characterization of Ste20p, a potential mitogen-activated protein or extracellular signal-regulated kinase kinase (MEK) kinase kinase from Saccharomyces cerevisiae. J. Biol. Chem. 27015984-15992. [DOI] [PubMed] [Google Scholar]
- 115.Yablonski, D., I. Marbach, and A. Levitzki. 1996. Dimerization of Ste5, a mitogen-activated protein kinase cascade scaffold protein, is required for signal transduction. Proc. Natl. Acad. Sci. USA 9313864-13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yaffe, M. P., and G. Schatz. 1984. Two nuclear mutations that block mitochondrial protein import in yeast. Proc. Natl. Acad. Sci. USA 814819-4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yu, L., M. Qi, M. A. Sheff, and E. A. Elion. 2008. Counteractive control of polarized morphogenesis during mating by mitogen-activated protein kinase Fus3 and G1 cyclin-dependent kinase. Mol. Biol. Cell 191739-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]