Abstract
There are three iron superoxide dismutases in Arabidopsis thaliana: FE SUPEROXIDE DISMUTASE1 (FSD1), FSD2, and FSD3. Their biological roles in chloroplast development are unknown. Here, we show that FSD2 and FSD3 play essential roles in early chloroplast development, whereas FSD1, which is found in the cytoplasm, does not. An fsd2-1 fsd3-1 double mutant had a severe albino phenotype on agar plates, whereas fsd2 and fsd3 single knockout mutants had pale green phenotypes. Chloroplast development was arrested in young seedlings of the double mutant. The mutant plants were highly sensitive to oxidative stress and developed increased levels of reactive oxygen species (ROS) during extended darkness. The FSD2 and FSD3 proteins formed a heteromeric protein complex in the chloroplast nucleoids. Furthermore, transgenic Arabidopsis plants overexpressing both the FSD2 and FSD3 genes showed greater tolerance to oxidative stress induced by methyl viologen than did the wild type or single FSD2- or FSD3-overexpressing lines. We propose that heteromeric FSD2 and FSD3 act as ROS scavengers in the maintenance of early chloroplast development by protecting the chloroplast nucleoids from ROS.
INTRODUCTION
Under normal growth conditions, the production of reactive oxygen species (ROS) in cells is low, but under biotic and abiotic stress conditions, production is enhanced; this affects many cellular functions by damaging nucleic acids, oxidizing proteins, and causing lipid peroxidation (Polle, 2001). Plants have well-developed defense systems against ROS, involving both enzymatic and nonenzymatic mechanisms (Alscher et al., 2002). Superoxide dismutases (SODs) constitute the first line of cellular defense against ROS; they enzymatically and rapidly convert superoxide (O2−) and water (H2O) to hydrogen peroxide (H2O2) and molecular oxygen (O2) (for review, see Bowler et al., 1992). Most plants contain a number of SOD isozymes. SODs are classified by their metal cofactors into three known types: iron SOD (FeSOD), manganese SOD (MnSOD), and copper–zinc SOD (Cu/ZnSOD). Phospholipid membranes are impermeable to O2−, and multiple SODs are present for the removal of O2− in various compartments of plant cells where O2− radicals are formed (Takahashi and Asada, 1983). Chloroplasts bear a particular risk of oxygen toxicity, because molecular O2 can be photoreduced to O2− by electrons from photosystem I (PSI) (Mehler, 1951). Therefore, chloroplast SODs play a very important role in preventing the oxidative damage associated with photosynthesis.
FeSOD is found in both prokaryotes and plants but not in animals (Alscher et al., 2002). Several conserved regions of FeSOD sequences are present in plants and cyanobacteria but absent in nonphotosynthetic bacteria (Bowler et al., 1994), suggesting that the FeSOD gene originated in the plastid genome and moved to the nuclear genome after endosymbiosis during evolution. Three FeSOD genes (FE SUPEROXIDE DISMUTASE1 [FSD1], FSD2, and FSD3), three Cu/ZnSOD genes (COPPER/ZINC SUPEROXIDE DISMUTASE1 [CSD1], CSD2, and CSD3), and one MnSOD gene (MANGANESE SUPEROXIDE DISMUTASE1 [MSD1]) are encoded in the Arabidopsis thaliana genome (Kliebenstein et al., 1998). Subcellular localization studies have suggested that the FSD proteins and CSD2 are localized in chloroplasts and that CSD1, CSD3, and MSD1 are localized in the cytoplasm, peroxisome, and mitochondria, respectively. To date, several proteomic studies of different plant cell organelles have been reported (for reviews, see Peck, 2005; Baginsky and Gruissem, 2006). From the results of these studies, FSD1 protein has been identified among chloroplast proteins (Kleffmann et al., 2004), in the peripheral thylakoid (Peltier et al., 2002), stroma (Peltier et al., 2006), and envelope (Ferro et al., 2003) of purified Arabidopsis chloroplasts, as well as in the plasma membrane (Marmagne et al., 2004) and mitochondrial membrane (Brugière et al., 2004) in Arabidopsis cell suspensions. FSD2 and CSD2 proteins have been identified only in chloroplasts (Kleffmann et al., 2004). However, this approach has limitations, because chloroplasts contain a large amount of ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) and a few other highly abundant photosynthetic proteins, which may prevent the detection of proteins that are present at low levels (Baginsky et al., 2005). Therefore, the precise subcellular localization of the three FeSOD isozymes has not yet been determined.
There have been many reports of the production of oxidative stress–tolerant transgenic plants that produce SODs. Transgenic tobacco (Nicotiana tabacum) plants that overproduce chloroplast Cu/ZnSOD from pea (Pisum sativum) show increased resistance to methyl viologen (MV)–induced membrane damage (Gupta et al., 1993). Overexpression of an FeSOD gene in tobacco chloroplasts was more effective in protecting against MV-induced damage than overexpression of an MnSOD gene, but it did not confer tolerance to H2O2, singlet oxygen (1O2), or abiotic stresses such as chilling and salinity (Van Camp et al., 1996). These results suggest that localization of SOD in the intrachloroplast regions where superoxide is generated is important for the acquisition of oxidative stress tolerance and that overproduction of one enzyme is not enough to have a substantial impact on ROS-scavenging capacity.
Prompt scavenging of ROS by many participating enzymes is necessary for normal plant or cell growth (Pnueli et al., 2003; Rizhsky et al., 2003; Miller et al., 2007), and a detailed analysis of mutants is important if we are to understand the functions and interactions of such enzymes. Knockdown Arabidopsis plants containing a T-DNA insert in the promoter of CSD2 (knockdown SOD [KD-SOD]) showed growth retardation and abnormal chloroplasts (Rizhsky et al., 2003). The KD-SOD plants had reduced numbers of the granal thylakoids and reduced photosynthetic activity, but because of the induction of several antioxidant genes, including an FeSOD, they were more tolerant to oxidative stress. These results suggest that CSD2 is essential for protecting the chloroplasts in higher plants from photooxidative damage. In the unicellular cyanobacterium Synechococcus, which possesses two SODs, MnSOD in the thylakoids and FeSOD in the cytosol, a mutant strain lacking FeSOD is sensitive to photooxidative stress, and its PSI complex is inactivated (Herbert et al., 1992; Samson et al., 1994; Thomas et al., 1998). However, the role of FeSOD isozymes in maintaining chloroplast function has not yet been documented.
To determine the function of chloroplast FeSOD, we collected Arabidopsis tagged mutant lines with mutations in the three FeSOD genes (FSD1, FSD2, and FSD3) and analyzed the phenotypes of these mutants. The phenotypes produced by the three mutant fsd1 alleles were normal, whereas the two mutant alleles of fsd2 and the two of fsd3 resulted in pale green phenotypes. The fsd2 fsd3 double mutant had a severe albino phenotype. High levels of superoxide were found in the leaves of fsd2 and fsd3 mutants grown in continuous darkness, suggesting that FSD2 and FSD3 can detoxify the superoxide radicals produced by photosynthesis. The FSD2 and FSD3 proteins were localized to the chloroplasts in transgenic tobacco plants, and the FSD1 protein was localized to the cytosol. We also show by both in vitro and in vivo analysis that FSD2 and FSD3 proteins form a heterocomplex. We discuss the essential roles of chloroplast FSD2 and FSD3, which function as a heteroduplex in ROS scavenging at the plastid nucleoid during early chloroplast development.
RESULTS
Phenotypes of fsd1, fsd2, fsd3, and fsd2-1 fsd3-1 Mutants
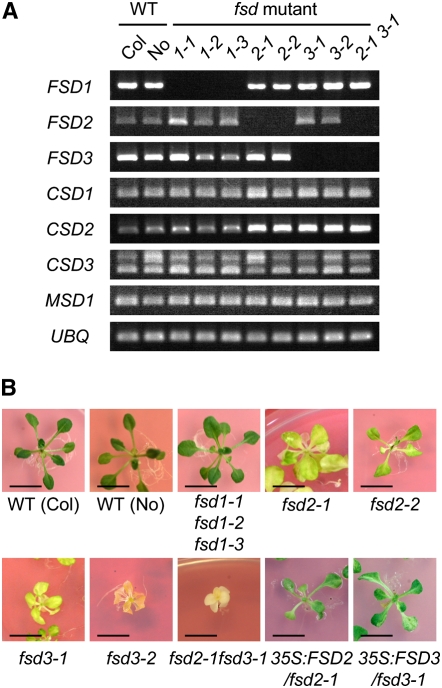
A pale green mutant, named apg8 (for albino or pale green8), was identified in a screen of 702 RIKEN Dissociation (Ds)-tagged lines (Kuromori et al., 2004; Ito et al., 2005) that had disrupted nucleus-encoded chloroplast proteins. The Ds transposon was inserted in a gene encoding FSD2 in Arabidopsis. Therefore, we renamed the apg8 mutant fsd2-1. Three FeSOD group proteins, FSD1, FSD2, and FSD3, are encoded in the Arabidopsis genome. To investigate the functions of these Arabidopsis FeSODs, we searched for lines with Ds or T-DNA insertions within the FSD1, FSD2, and FSD3 genes. From the SALK (Alonso et al., 2003), SAIL (Sessions et al., 2002), and GABI-Kat (Rosso et al., 2003) collections, seven independent lines were identified and named fsd1-1 (SALK_029455), fsd1-2 (GABI_740E11), fsd1-3 (GABI_341D04), fsd 2-1 (11-6562-1), fsd2-2 (SALK_080457), fsd3-1 (SALK_103228), and fsd3-2 (SAIL_224_E05) (see Supplemental Figure 1 online). In these mutants, the Ds or T-DNA segregated as a single locus. Homozygous plants were obtained by self-pollination. RT-PCR analysis showed that the transcripts of the corresponding genes were absent in the mutant plants, whereas the transcripts of CSD2 were upregulated in both fsd2 and fsd3 mutants (Figure 1A). The two mutant fsd2 lines and the two mutant fsd3 lines had pale green leaves; the leaves of the fsd3 lines were paler than those of the fsd2 lines (Figure 1B). Both fsd2 and fsd3 mutants were retarded in growth but could flower and produce seeds. By contrast, the fsd1 mutants grew normally and did not have any obviously abnormal phenotypes under normal growth conditions (Figure 1B). Overexpression of FSD2 and FSD3 cDNAs driven by the constitutive cauliflower mosaic virus 35S promoter functionally complemented the fsd2-1 and fsd3-1 mutants, respectively (Figure 1B). These results suggest that both FSD2 and FSD3 are required for normal plant growth.
Figure 1.
Visible Phenotypes of Mutants with Knockout of FSD Genes.
(A) RT-PCR analysis of expression of the Arabidopsis SOD gene family in wild type and Ds/T-DNA–tagged mutant plants. RT-PCR demonstrated that the transcripts of the disrupted FSD genes were absent from whole plants of the fsd1, fsd2, and fsd3 mutants. Transcript levels of CSD2 specifically accumulated in fsd2-1, fsd2-2, fsd3-1, fsd3-2, and fsd2-1 fsd3-1 mutants. The polymorphism of RT-PCR products of CSD3 was due to the presence or absence of a 95-bp first intron. The RT-PCR band of the UBIQUITIN1 gene was used as a loading control. Two replicates were performed.
(B) fsd1, fsd2, and fsd3 single mutants, fsd2-1 fsd3-1 double mutants and their wild types (Columbia [Col]; Nossen [No]), and the fsd2-1 mutant complemented with the 35S:FSD2 construct and the fsd3-1 mutant complemented with the 35S:FSD3 construct. All strains except fsd2-1 are in the Columbia background; fsd2-1 is Nossen background. Three-week-old plants (wild type, fsd1, 35S:FSD2/fsd2-1, and 35S:FSD3/fsd3-1) and 5-week-old plants (fsd2, fsd3, and fsd2-1 fsd3-1) were grown on GM agar plates, and their visible phenotypes were observed. Bar = 1 cm.
We then crossed fsd2-1 with fsd3-1 to obtain a double mutant, named fsd2-1 fsd3-1. RT-PCR analysis confirmed that both the FSD2 and FSD3 transcripts were absent in the double mutant plants (Figure 1A). In contrast with the single mutant plants, the double mutant plants had a more severe albino phenotype when grown on agar plates (Figure 1B). A detailed study revealed that the leaf color of the double mutant was dramatically reduced compared with those of the single mutants, whereas the roots grew as well as the wild-type roots. The aerial organs (leaves and rosettes) developed normally in double mutant plants grown on agar medium containing 1% sucrose, but when they were transferred to soil, all of the plants withered and died and failed to produce progeny. We measured the chlorophyll fluorescence of the mutants by pulse amplitude–modulated fluorometry to investigate their photosynthetic activity (see Supplemental Table 1A online). After 1 week under normal light conditions, the primary maximum photochemical efficiency of photosystem II (PSII; Fv/Fm) and nonphotochemical quenching were significantly decreased in both fsd2-1 and fsd3-1 plants compared with wild-type and fsd1-1 plants (P < 0.001, with Bonferroni correction), whereas no significant effects on the fraction of open PSII centers were observed (P > 0.05). This indicates that PSII itself, and not the downstream photosynthetic apparatus of PSII, such as cytochrome b/f complexes or PSI, was impaired in the fsd2-1 and fsd3-1 plants.
fsd2 and fsd3 Are Sensitive to Oxidative Stress
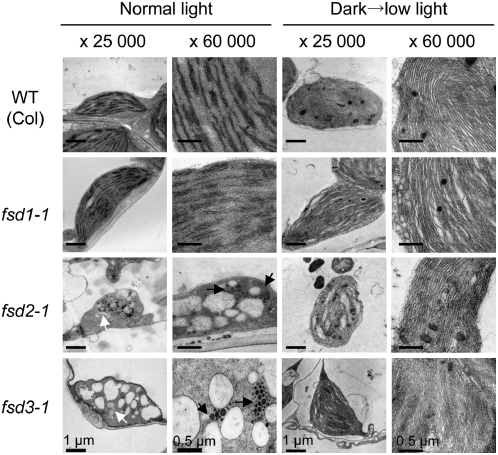
We used transmission electron microscopy to investigate morphological changes in the plastids of fsd2 and fsd3 pale green leaves (Figure 2). We found abnormal plastids in the severely albino tissues of fsd2-1 and fsd3-1 mutants. The plastids were highly vacuolated and lacked internal membrane structures such as single stromal thylakoids or stacked grana thylakoids, and they contained abnormal suborganelle structures. In addition, there were a considerable number of densely stained globular structures, probably plastoglobuli. However, when the mutants were grown in darkness for 10 d and then exposed to low light (7 μmol·m−2·s−1) for 3 h, the plastids of fsd2-1 and fsd3-1 cotyledons had phenotypes similar to those of the wild type and the fsd1-1 mutant grown under the same conditions. They formed stromal-thylakoid–like membranes instead of stacked grana thylakoids. These results suggest that the greening processes of fsd2-1 and fsd3-1 mutants are normal (i.e., like those of wild-type and fsd1-1 plants) under low-light conditions and that both FSD2 and FSD3 proteins play important roles in chloroplast development, particularly in the maintenance of thylakoid membranes.
Figure 2.
Transmission Electron Micrographs of Plastids in fsd Mutants.
Leaves of wild-type (Columbia [Col] ecotype) and three FSD knockout mutant (fsd1-1, fsd2-1, and fsd3-1) plants were grown under a 16-h-day/8-h-night cycle for 10 d (left) or exposed to low light (photon flux density of 7 μmol·m−2·s−1) for 3 h after being grown in darkness for 10 d (right). Images were observed at low (×25,000) and high (×60,000) magnifications. White and black arrows indicate the abnormal suborganelle and the densely stained globular structures, respectively.
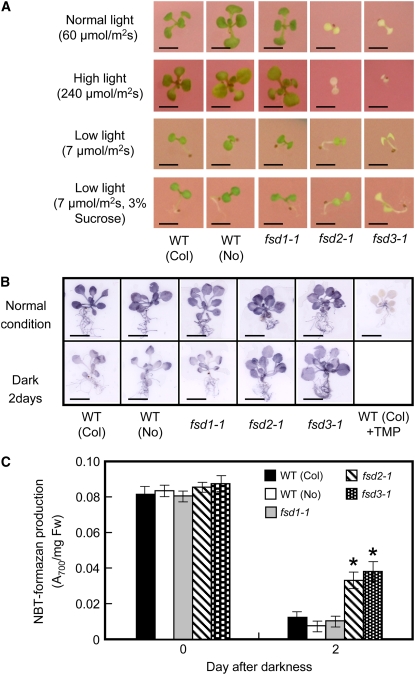
Ten-day-old seedlings of both fsd2-1 and fsd3-1 mutants did not show a wild-type response when subjected to changes in light intensity (Figure 3A). Severe growth retardation and the albino phenotype were observed in fsd2-1 and fsd3-1 plants grown under strong light (240 μmol·m−2·s−1). By contrast, accumulation of anthocyanin was observed in normally grown seedlings of wild-type and fsd1-1 plants under strong-light conditions. Under low-light conditions (7 μmol·m−2·s−1), the stems of seedlings of both wild-type and fsd1-1 plants grew fast, the cotyledons remained folded, and the photosynthetic machinery was immature. By contrast, the fsd2-1 and fsd3-1 mutant seedlings were pale green with expanded cotyledons, but the green was darker than that seen in these plants under normal light conditions. Moreover, low light caused a slight increase in chlorophyll accumulation in fsd2-1 and fsd3-1 leaves but a markedly reduced accumulation of chlorophyll in wild-type and fsd1-1 leaves during greening (see Supplemental Table 1B online). When fsd2-1 and fsd3-1 plants were transferred to a medium supplemented with 3.0% (w/v) sucrose, their phenotypes improved, even when grown under low-light conditions, compared with those on a medium with 1.0% (w/v) sucrose under low-light conditions (Figure 3A; see Supplemental Table 1C online).
Figure 3.
O2−-Mediated Growth Inhibition of fsd1-1, fsd2-1, fsd3-1, and Wild-Type Plants under Different Light Conditions.
(A) Visible phenotypes of wild-type (Columbia [Col] and Nossen [No] ecotypes) and mutant plants at 10 d after germination and growth under various light conditions on agar plates supplemented with 1% or 3% sucrose, as indicated at left. Bar = 2 mm.
(B) Accumulation of O2− in 3-week-old plants grown under normal light conditions (top) and then transferred to the dark for 2 d (bottom). Before staining with NBT, wild-type plants were incubated with 1 mM tetramethyl piperidinooxy (TMP) as a negative control (far right column). Bar = 1 cm.
(C) Colorimetric quantification of NBT-formazan production in plant extracts. Three-week-old plants were grown under normal light conditions (0 d after darkness) and then transferred to the dark for 2 d (2 d after darkness). Formazan precipitates were eluted completely by 2 M KOH/DMSO, and the amount of NBT-formazan was determined as A700. The results shown are means ± sd of four samples. The asterisks indicate that the numbers are significantly higher than those of the wild-type plants (t test, P < 0.01). Fw, fresh weight.
To investigate the removal of the ROS produced by photosynthesis in plants, we detected superoxide production by the reduction of nitroblue tetrazolium (NBT). Formazan, produced upon the reduction of NBT by superoxide, appears strongly in the leaves of wild-type and mutant plants, and the reduction of NBT is suppressed by the application of a specific ROS scavenger of O2−, tetramethyl piperidinooxy, which was used as a negative control (Rodríguez-Serrano et al., 2006). When plants were incubated for 2 d at 22°C in darkness, formazan production was suppressed in both wild-type and fsd1-1 plants. By contrast, its formation was not reduced in the pale green seedlings of the fsd2-1 and fsd3-1 mutants (Figure 3B). Quantification of the formazan spots demonstrated that wild-type and fsd1-1 mutant plants produced smaller amounts of formazan precipitates than the fsd2-1 and fsd3-1 mutant plants during a prolonged dark period (Figure 3C). These results suggest that the fsd2-1 and fsd3-1 mutations have a negative effect on the capacity to scavenge superoxide radicals.
Subcellular Localization
Multiple alignments of plant and bacterial FeSODs revealed that the N-terminal sequences were not conserved and that long N-terminal sequences were present in the FSD2 and FSD3 proteins (see Supplemental Figure 2 online). FSD2 and FSD3 contained a putative N-terminal chloroplast-targeting sequence predicted by the TargetP (Emanuelsson et al., 2000), Predotar (Small et al., 2004), WoLF PSORT (Horton et al., 2007), and PCLR (Schein et al., 2001) programs. However, TargetP, Predotar, and PCLR failed to detect the N-terminal targeting sequence in FSD1 (see Supplemental Table 2 online).
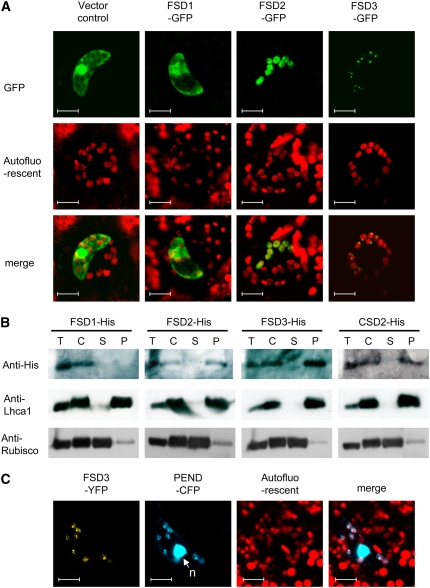
To confirm the chloroplast localization of these proteins in plant cells, we generated constructs containing full-length cDNAs of FSD1, FSD2, and FSD3 fused to a synthetic green fluorescent protein (sGFP) at their C termini (FSD1-GFP, FSD2-GFP, and FSD3-GFP, respectively). Transient expression of FSD-GFP fusion proteins in the epidermal cells of tobacco leaves transformed by means of particle bombardment resulted in GFP fluorescence in various subcellular compartments (Figure 4A). The FSD2-GFP and FSD3-GFP fusion proteins colocalized with the chloroplasts, whereas FSD1-GFP proteins were detected in both the cytoplasm and the nucleus. FSD2-GFP signals were detected in the entire chloroplast, but FSD3-GFP signals were detected only in discrete regions inside the chloroplasts. To confirm the localization of FSD2 and FSD3 proteins in chloroplasts, we prepared chloroplast fractions from transgenic lines producing His-tagged fusion proteins of FSD1, FSD2, FSD3, and CSD2, with the tags fused to the C termini. Immunoblot analysis using anti-His tag antibody revealed that FSD2 and FSD3 localized to the thylakoids but not to the stroma and FSD1 localized to the cytosol (Figure 4B). Application of antibodies against the thylakoid lumenal protein light-harvesting chlorophyll protein complex and against the stromal Rubisco complex confirmed that the thylakoid fraction was reasonably pure. These results suggest that FSD2 and FSD3 are tightly attached to the stromal side of the thylakoid membranes, because they do not have transmembrane domains. The membrane association of FSD2 and FSD3 is similar to that of CSD2 in spinach (Spinacia oleracea) (Ogawa et al., 1995).
Figure 4.
Subcellular Localization of FSD Proteins.
(A) Intracellular accumulation of FSD1-GFP, FSD2-GFP, FSD3-GFP, or GFP alone (vector control) in tobacco leaves transiently transformed by particle bombardment. The green fluorescence of GFP fusion proteins (top) and the red autofluorescence of chlorophyll (middle) were monitored separately using a confocal laser scanning microscope, and the two fluorescence images were merged (bottom). Bars = 20 μm.
(B) Immunoblot analysis of protein extracts prepared from the indicated Arabidopsis plants transformed with His tag fusions driven by the cauliflower mosaic virus 35S promoter. Each protein fraction was prepared from 3-week-old plants. Total protein (T) is the entire protein content of the cell. Intact chloroplasts (C) were lysed, and then soluble (supernatant [S]) and insoluble (pellet [P]) proteins (10 μg/lane) were separated on a 12.5% SDS-PAGE gel and probed with anti-His antibody. The blot was also probed with anti-RBC-L (for large subunit of Rubisco; stromal protein) and anti-Lhca1 (for subunit of the light-harvesting chlorophyll–protein complex I; thylakoid protein) antibodies as controls for each fraction.
(C) Colocalization of FSD3-YFP (yellow) and PEND-CFP (blue) fluorescence with chloroplast nucleoids. The arrow indicates the large fluorescent spot that represents the nucleus (n) of a guard cell. Bar = 20 μm.
In the mature chloroplast, plastid nucleoids are located within the stroma as small particles mostly associated with thylakoids (Kuroiwa, 1991). We analyzed whether FSD3 was colocalized with PEND (for plastid envelope DNA binding), a well-characterized DNA binding protein in the inner envelope membrane of the developing chloroplast (Sato et al., 1993), by transiently coexpressing FSD3–yellow fluorescent protein (YFP) and a modified PEND–cyan fluorescent protein (CFP) fusion protein consisting of the N-terminal region of the PEND homolog of Arabidopsis (Terasawa and Sato, 2005). Fluorescence signals of PEND-CFP were observed in the cell nucleus and chloroplast nucleoids (Figure 4C); this finding differs from that of a previous report of stable PEND-CFP transformants of Arabidopsis (Terasawa and Sato, 2005), in which only plastid nucleoids were observed without interference by cell nuclei. This difference may be because we transiently expressed Arabidopsis PEND-CFP fusion proteins in tobacco. Figure 4C shows that the FSD3-YFP signals colocalized with the PEND-CFP signals in the chloroplasts, indicating that FSD3 colocalized with the nucleoids in mature leaf chloroplasts.
Expression Analysis of Plastid-Encoded Genes and FeSOD
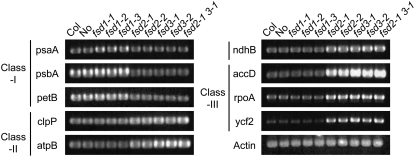
We then examined the expression of nucleus-encoded plastid RNA polymerase (NEP) and/or plastid-encoded plastid RNA polymerase (PEP) genes in both wild-type and fsd mutant plants (Figure 5). The psaA, psbA, and petB genes were selected as PEP-dependent genes (class I); accD, rpoA, and ycf2 were chosen as NEP-dependent genes (class III); and atpB, clpP, and ndhB were used as both PEP- and NEP-dependent genes (class II). RT-PCR analysis using primer sets (Nagashima et al., 2004) specific for each gene showed that pale green and albino mutants (i.e., fsd2, fsd3, and fsd2 fsds3 mutants) accumulated decreased amounts of mRNA for class I genes and increased amounts of mRNA for class III genes in the total mRNA pool. In the case of the class II genes—especially ndhB—accumulation of the mRNA in the fsd2 and fsd3 mutants was higher than in the wild-type and fsd1 plants. These results suggest that the chloroplast expression system that utilizes PEP is deficient in the fsd2 and fsd3 mutants.
Figure 5.
Expression Analysis of NEP-Dependent and PEP-Dependent Genes.
RT-PCR analysis for plastid-encoding genes. Total RNA fractions were isolated from wild-type (Columbia [Col] and Nossen [No] ecotypes) and fsd mutant plants. psaA, psbA, and petB were selected as NEP-dependent genes (class I); clpP, atpB, and ndhB were selected as both PEP- and NEP-dependent genes (class II); and accD, rpoA, and ycf2 were selected as PEP-dependent genes (class III). The ACTIN gene was used as a control. Two replicates were performed.
FSD2 and FSD3 transcripts could not be detected in RNA gel blots of wild-type plants, but quantitative RT-PCR showed their accumulation in all organs tested except roots, with maximum amounts in immature siliques, whereas transcripts of FSD1 were detected strongly in all organs except flowers and mature seeds (see Supplemental Figure 3A online). FSD2 and FSD3 mRNA rapidly accumulated in response to oxidative stresses such as high light, cold, and MV treatment (see Supplemental Figure 3B online). In corroboration of the results of our RT-PCR analysis, examination of the Genevestigator database (http://www.genevestigator.ethz.ch) revealed that the expression profiles of FSD2 and FSD3 by growth stage and stress response differ from those of FSD1 but are similar to each other. We analyzed the expression of FeSODs throughout the development of Arabidopsis using tissue-specific staining patterns in transgenic plants expressing β-glucuronidase (GUS) driven by a specific FeSOD promoter (FSDpro:GUS). GUS staining of FSD1pro:GUS transgenic Arabidopsis plants was strong in all organs of seedlings and young plants and weak in mature siliques (see Supplemental Figure 4 online). In the FSD3pro:GUS plants, GUS staining was observed in the cotyledons, leaf primordia, and young leaves but not in mature leaves, stems, flowers, or mature siliques. GUS staining was not observed in FSD2pro:GUS plants. The GUS staining pattern suggests that FSD3 functions in the early stage of chloroplast development.
FSD2 and FSD3 Form Heterocomplexes
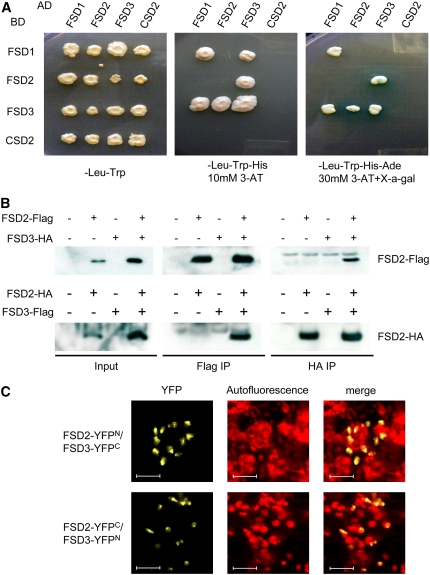
There have been several reports that FeSODs in plants form homodimers (Salin and Bridges, 1980, 1982; Duke and Salin, 1985; Kwiatowski et al., 1985; Almansa et al., 1994; Gómez et al., 2004). To examine whether the FSD2 and FSD3 proteins form homodimers and heterodimers with each other, we examined the interactions among the three FSDs and CSD2 by yeast two-hybrid assays. However, yeast cells producing full-length proteins of FSD2, FSD3, or CSD2 were not viable, even on plates containing a low-stringency medium such as SD/–Leu/–Trp, probably because they have functional chloroplast transit peptides and are thus likely misdirected to intracellular compartments in the yeast. Therefore, we transformed yeast cells with FSD2, FSD3, or CSD2 sequences that lacked transit peptide regions. Coproduction of FSD2 with FSD3 resulted in positive β-galactosidase activity and growth on SD/–Leu/–Trp/–His/–adenine (Ade)/30 mM 3-amino-1,2,4-triazole (3-AT) plates (Figure 6A). Similar interactions between FSD1 and FSD1, FSD1 and FSD3, and FSD3 and FSD3 were observed, with faint β-galactosidase activity. Among these, complexes between FSD1 and FSD3 are not formed in plants because of the different subcellular localizations of these proteins (Figures 4A and 4B). Taken together, these results led us to speculate that FSD1 and FSD3 exist as homodimers and that FSD2 and FSD3 form a heterodimer in planta. To confirm the FSD2 and FSD3 interaction in vivo, we performed coimmunoprecipitation assays using leaf cell extracts (Figure 6B). FSD2 with a C-terminal Flag epitope tag (FSD2-Flag) and FSD3 with a C-terminal hemagglutinin (HA) epitope tag (FSD3-HA) were coproduced in leaves of Nicotiana benthamiana following agroinfiltration. Leaf extracts were subsequently immunoprecipitated with the anti-HA antibody. FSD2-Flag was coimmunoprecipitated with FSD3-HA using the anti-HA antibody. Similar results were obtained by immunoprecipitating FSD2-HA with the anti-Flag antibody when leaf extracts expressing both FSD2 with a C-terminal HA epitope tag (FSD2-HA) and FSD3 with a C-terminal Flag epitope tag (FSD3-Flag) were used. Neither FSD2 nor FSD3 could interact with itself, because immunoprecipitation using anti-Flag or anti-HA antibody was not observed when FSD2-Flag and FSD2-HA, or FSD3-Flag and FSD3-HA, were cotransformed in leaf cells.
Figure 6.
Detection of FSD2 and FSD3 Heteromeric Complexes in Plastids.
(A) Interaction between FSD1, FSD2, FSD3, and CSD2 proteins in yeast two-hybrid assays. Four clones of yeast containing each combination of bait (BD) and prey (AD) vectors were grown on selection medium with drop out of several amino acids. Media used to assess the strength of interaction between bait and prey were as follows: SD-Leu-Trp, SD-Leu-Trp-His + 10 mM 3-AT, and SD-Leu-Trp-His-Ade + 30 mM 3-AT + X-α-Gal.
(B) Coimmunoprecipitation assay revealing in vivo interactions between FSD2 and FSD3. The combinations of constructs indicated above each blot were transiently expressed in N. benthamiana. Proteins detected by immunoblot are indicated at right. FSD2-Flag coimmunoprecipitated with FSD3-HA using the anti-HA antibody (HA IP), and immunoblotting was performed with the anti-Flag antibody. Similarly, FSD2-HA coimmunoprecipitated with FSD3-Flag when using the anti-Flag antibody (Flag IP), followed by detection with the anti-HA antibody.
(C) Visualization of protein interaction in plastids by the BiFC assay. YFP epifluorescence microscopy images show tobacco epidermal cells in leaves transiently expressing constructs encoding the fusion proteins indicated. Merge indicates an overlay of the YFP and autofluorescence chlorophyll images. Each image is representative of at least two experiments. Bars = 20 μm.
We then tried to visualize the protein interaction between FSD2 and FSD3 in chloroplasts using bimolecular fluorescence complementation (BiFC) in transiently transformed tobacco leaves. BiFC involves the reconstitution of YFP fluorescence upon association of nonfluorescent, N-terminal (NY) and C-terminal (CY) fragments of YFP, brought together as a consequence of their fusion with interacting proteins (Walter et al., 2004). GFP fluorescence in tobacco cells producing full-length FSD2 and FSD3 fused with sGFP (FSD2-GFP and FSD3-GFP) was detected in the chloroplast (Figure 4A). This observation suggested that BiFC fluorescence associated with the FSD2–FSD3 interaction would be observed as if the two fluorescence images of FSD2-GFP and FSD3-GFP in Figure 4A were combined. Indeed, when we analyzed tobacco leaves coproducing fusions of FSD2-NY and FSD3-CY, a strong YFP fluorescence signal was detected in specific regions of the chloroplast nucleoids and weakly in the entire chloroplast (Figure 6C). By contrast, no FSD2 and FSD3 homodimerization-induced YFP fluorescence was observed in epidermal cells that were transformed with FSD2-NY and FSD2-CY, and FSD3-NY and FSD3-CY, respectively. Taken together, these results strongly support the hypothesis that FSD2 and FSD3 form a heterodimer that functions in chloroplast nucleoids.
Both Recombinant FSD2 and FSD3 Retain SOD Activity
To obtain a large amount of soluble and active FSD2 and FSD3 proteins, we induced the expression of N-terminally His-tagged or glutathione S-transferase (GST)–tagged cDNA fragments of FSD2 and FSD3 in transgenic Escherichia coli by adding isopropylthio-β-galactoside. However, His-FSD3 was exclusively recovered in inclusion bodies and was absent from the soluble fraction when subjected to SDS-PAGE, followed by Coomassie blue staining, and was not detected in the protein fraction purified on a nickel column (see Supplemental Figure 5 online). The enzyme activities of His-FSD2 and GST-FSD3 were then further analyzed. SOD activity signals on the native PAGE gels were detected with both of the recombinant proteins, indicating that both FSD2 and FSD3 proteins have specific enzymatic activity (see Supplemental Figure 6A online). GST-FSD3 had SOD activity that was at least 10 times that of His-FSD2 in several active bands (monomer, dimer, trimer, etc.). We investigated whether the formation of FSD2 and FSD3 heterodimers caused an elevation in enzyme activity levels. Addition of His-FSD2 and GST-FSD3 led to increased SOD activity. However, no significant difference in the SOD activity was observed between a mixture of His-FSD2 and GST-FSD3 and the sum of the individual His-FSD2 and GST-FSD3 activities (see Supplemental Figure 6A online). To further analyze the interaction between His-FSD2 and GST-FSD3 in vitro, we performed pull-down assays. For this purpose, GST and GST-FSD3 were immobilized on glutathione–Sepharose columns and assayed for their ability to pull down recombinant His-FSD2 by immunoblotting with anti-His antibodies. GST-FSD3 weakly interacted with His-FSD2 but not with GST alone (see Supplemental Figure 6B online). These data indicated that FSD2 weakly interacts with FSD3 directly in vitro; under these conditions, SOD activity due to the formation of a His-FSD2/GST-FSD3 heterodimer was not significantly higher than that produced by each protein individually.
Transgenic Arabidopsis Plants Overexpressing Both FSD2 and FSD3 Are Tolerant to Oxidative Stress
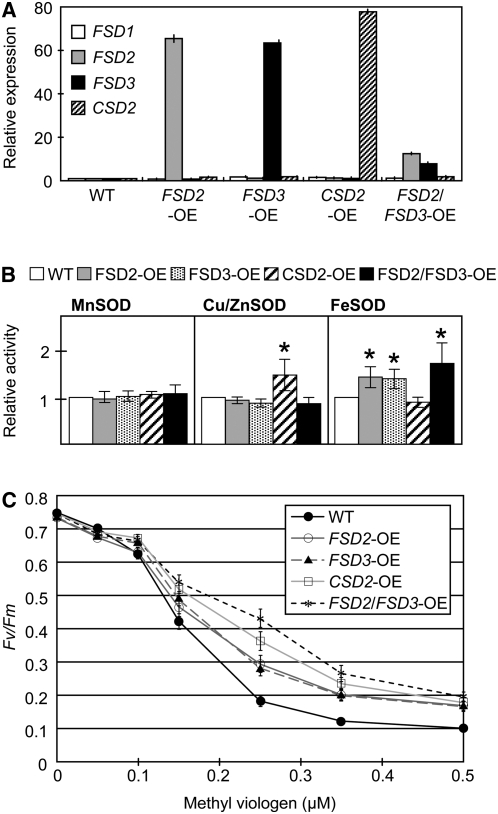
To further analyze the functions of FSD2 and FSD3, we generated transgenic lines overexpressing FSD2, FSD3, and CSD2 individually and FSD2 and FSD3 together. There were no obvious morphological or developmental differences between the transgenic and wild-type plants. However, seeds of transgenic plants overexpressing FSD2 and FSD3 together had a 50% reduction in germination compared with those of the wild-type plants, after treatment at 4°C for 3 d in darkness. Furthermore, after germination, no morphological differences were observed in transgenic plants overexpressing FSD2 and FSD3 compared with wild-type plants. Quantitative RT-PCR analysis revealed strong overexpression of the transcripts in T3 plants of homozygous transgenic lines (Figure 7A). Transgenic lines expressing FSD2, FSD3, and CSD2 had high levels of the corresponding mRNAs, whereas FSD2 and FSD3 expression in plants overexpressing both of these genes was only slightly increased compared with those of the wild type. Overexpression of FSD2 was suppressed by the induction of FSD3 expression in 35S:FSD2 plants retransformed with 35S:FSD3, indicating the existence of a reciprocal regulation between FSD2 and FSD3 expression. Relatively high levels of FeSOD and Cu/ZnSOD activity were observed in transgenic plants compared with those of the wild type, while the activity of MnSOD was almost similar to each other (Figure 7B). MV causes an increase in the production of superoxide radicals in chloroplasts. We investigated the effect of MV on the transgenic plants by measuring the MV-dependent decrease in activity of the PSII reaction center under normal light conditions at 30 μmol·m−2·s−1. The leaves of transgenic plants expressing FSD2 and FSD3 individually and combined showed less inactivation of PSII, as indicated by Fv/Fm, than did wild-type plants (Figure 7C). These results indicate that transgenic plants were more tolerant to MV than were wild-type plants. The differences between transgenic and control plants were significant in the presence of 0.25 μM MV (wild type versus FSD2-OE/FSD3-OE, P < 0.05; wild type versus CSD2-OE/double-OE, P < 0.001, with Bonferroni correction; see Supplemental Table 3 online). Under this level of exposure, transgenic plants expressing both FSD2 and FSD3 transgenes showed less inactivation of PSII than did wild-type plants and transgenic plants expressing a single transgene (FSD2-OE/FSD3-OE versus double-OE, P < 0.01). Taken together, these data indicate that overexpression of both FSD2 and FSD3 together improves tolerance to MV and protects chloroplasts against light-dependent oxidative stress; this also supports the hypothesis that FSD2 and FSD3 act as a heterodimer.
Figure 7.
MV-Induced Oxidative Stress Tolerance of FSD Transgenic Lines.
(A) Relative expression levels of transgenes in transgenic lines overexpressing FSD2 (FSD2-OE), FSD3 (FSD3-OE), CSD2 (CSD2-OE), and FSD2/FSD3 (FSD2/FSD3-OE) cDNAs, as determined by quantitative RT-PCR using 3-week-old seedlings grown on agar plates. The levels were normalized to the wild type. Three biological replicates were performed for each experiment. Ubiquitin was used as an internal control. Error bars indicate sd (n = 4).
(B) SOD activity of the transgenic lines. Total proteins extracted from 3-week-old plants were subjected to 10% SDS-PAGE, and their SOD activity was visualized as negatively staining bands. Histograms of relative areas of peak intensities of negatively staining bands of MnSOD (left), Cu/ZnSOD (middle), and FeSOD (right) were generated. Peak areas were measured using the ImageJ program after eliminating background signals. Relative measured peak areas of transgenic lines were normalized to that of the wild type. Three biological replicates were performed for each experiment. Error bars indicate sd (n = 5 to 7). The asterisks indicate numbers significantly higher than those of the wild-type plants (t test, P < 0.05).
(C) Comparison of MV-dependent decreases in Fv/Fm in wild-type plants and four transgenic lines exposed to the free-radical–generating herbicide, MV. Values are means ± se of at least 60 leaf disc measurements.
DISCUSSION
We investigated the roles of two Arabidopsis FeSODs, FSD2 and FSD3, in chloroplasts and showed that loss of function of each protein caused marked damage to proper development of the plastid (Figure 2). Ds- and T-DNA–tagged knockout mutants of FSD2 and FSD3 had a pale green phenotype, whereas three lines of FSD1 mutants with different T-DNA–tagged alleles grew as well as wild-type plants (Figure 1B). Since CSD2 has existed as a FSD1-related functional homolog in the cytosol (Kliebenstein et al., 1998), these results suggest that cytoplasmic FSD1 does not play a crucial role in determining plant development under normal growth conditions. However, there is a possibility that fsd1 mutants may respond to severe oxidative stress in soil with low copper content, because FSD1 transcripts were detected only under this condition (Abdel-Ghany et al., 2005). Chloroplast FSD2 and FSD3 play indispensable roles during the early stages of plastid development, but partial functional differences in FSD2 and FSD3 exist, because FSD2 and FSD3 double mutants had severely albino phenotypes, unlike the pale green phenotypes of the single mutants (Figure 1). However, the phenotypes of fsd2-1 and fsd3-1, which could be partly reversed by growth under low-light conditions (Figure 3), could not be reversed by induction of CSD2, the Cu/ZnSOD in chloroplasts (Figure 1A). We tried to obtain a homozygous csd2-1 mutant using PCR analysis of 20 independent progeny of the heterozygous csd2-1/+ mutant, but we were not successful, as described previously (Rizhsky et al., 2003), possibly because loss of function of CSD2 caused developmental inhibition during seed development and resulted in an embryo-lethal phenotype. The chloroplasts of knockdown CSD2 (KD-SOD) plants are smaller than those of wild-type plants and, like those of mutants deficient in FSD2 and FSD3 (Figure 2), contain fewer stacks of grana thylakoids (Rizhsky et al. 2003). Taken together, our findings reveal that functional specialization of FSD2, FSD3, and CSD2 is achieved via differences in both production and localization within the chloroplast. Transcript levels of FSD2 accumulated to a lesser degree than did those of FSD3 in young seedlings (Figure 1A); this corresponded to the different color-loss phenotypes seen in fsd2-1 and fsd3-1 (Figure 1B). Kliebenstein et al. (1998) reported the induction of FSD2 mRNA expression in response to UV irradiation and high light levels. Our expression analysis of SOD genes revealed that expression of the transcripts of all three chloroplast SOD genes increased in response to high light, cold, and MV treatment, but expression of FSD1 that is localized to the cytosol (Figures 4A and 4B) did not (see Supplemental Figure 3 online). We speculate that when ROS production increases under oxidative stress conditions, the three SOD enzymes in the chloroplast act cooperatively to remove superoxide radicals.
The functional differences among FSD2, FSD3, and CSD2 may be related to their different localizations in the chloroplast. FSD3-GFP production was localized to specific regions of the chloroplast, and its location correlated with the position of PEND-CFP in plastid nucleoids, whereas FSD2-GFP was uniformly dispersed in chloroplasts. CSD2 is present in the chloroplast stroma (Kliebenstein et al., 1998), and a chloroplast CSD isoform from spinach leaves is localized mainly at the stromal face of the thylakoid membranes (Ogawa et al., 1995). A homolog of Arabidopsis FSD3 was recently found in a highly purified soluble RNA polymerase (sRNAP) preparation from mustard (Sinapis alba) (Pfannschmidt et al., 2000), and both FSD2 and FSD3 were also identified by electrospray ionization ion-trap tandem mass spectrometry analysis as components of transcriptionally active chromosomes (TACs) from Arabidopsis and mustard (Pfalz et al., 2006). sRNAP and TACs include plastid protein preparations in which subunits of the PEP core are present. Interestingly, the T-DNA–tagged mutants of three TAC components (ptac2, ptac6, and ptac12) had albino or pale green phenotypes on agar plates (Pfalz et al., 2006). The expression patterns of plastid-encoded genes in all of the ptac mutants resemble those of the fsd2 and fsd3 mutants, which showed defects in PEP-dependent transcription: the expression of class I plastid genes with PEP promoters was downregulated relative to the wild type, and the expression of class III genes with NEP promoters was upregulated (Figure 5A). The phenotypic similarity of the fsd2/3 and ptac mutant plants suggests that FSD2 and FSD3 are functional components of the PEP complex and function in plastid gene expression in plastid nucleoids. Our microscopic observations also showed that FSD3 is specifically located in the chloroplast nucleoids and that FSD2 and FSD3 form a heterocomplex in this region (Figures 4C and 6C). The existence of FSD proteins in the core complex of PEP shows the importance of FSD2 and FSD3 in thylakoid-associated chloroplast transcription. One implication of this physical proximity is that newly synthesized transcript must be protected from damage by superoxide radicals generated as a photosynthetic by-product. Furthermore, chloroplast transcription is modulated by the redox-reactive reagents. H2O2, which is rapidly produced from O2− by the action of SOD enzymes, might act as a transcriptional signal in the chloroplast, as has been shown to be the case in oxyR-dependent transcription in bacterial systems (Åslund et al., 1999). Studies of H2O2-mediated signaling have demonstrated that the induction of chloroplast ndh genes under photooxidative stress is mediated by H2O2 (Casano et al., 2001). As described in a previous report, the transcription kinase cpCK2—one of the best characterized components of sRNAP—plays a role in the regulation of plastid gene expression via phosphorylation and redox signaling (Baginsky et al., 1997, 1999; Ogrzewalla et al., 2002). Sulfite reductase also participates in organelle nucleoid organization (Cannon et al., 1999; Chi-Ham et al., 2002; Sekine et al., 2002). Thus, in summary, H2O2 or some other redox signal, which is generated mainly in the chloroplast, induces the expression of plastid genes involved in photosynthesis. Both FSD2 and FSD3, therefore, might be functional components of the PEP complex and might function in gene expression in plastid nucleoids.
We showed that two recombinant FSD2 and FSD3 proteins with SOD activity can form a heterocomplex (see Supplemental Figure 6 online) and that transgenic plants overproducing both FSD2 and FSD3 proteins accumulate more heterocomplex than those overproducing single FSDs (Figures 6C and 7A). Transgenic overproducers of both FSD2 and FSD3 showed higher tolerance to MV-induced oxidative stress than did single overproducers (Figure 7C). We also showed that disruption of the FSD2–FSD3 heterocomplex caused the accumulation of reactive oxygen in prolonged darkness (Figures 3B and 3C). During formation of the heteroduplex, FSD2 may bind to high-density FSD3 proteins located at thylakoid membrane–associated nucleoids in plastids. This physical interaction between the two SOD proteins in the nucleoid may raise their level of activity in scavenging superoxide radicals in the plastid transcriptional complex. We also speculate that the activated SOD heterodimer functions as a signal transducer that coordinates transcriptional and translational events in plastids.
In conclusion, FSD2 and FSD3 can form a heterodimer with strong ROS scavenging activity in plastid nucleoids; this FSD2–FSD3 heterodimer may protect TACs in early chloroplast development from ROS produced in the thylakoid during photosynthesis. Because overproduction of FSD2 and FSD3 improved tolerance to ROS stress, this complex may be a novel target for improving the tolerance of plants to stress from ROS damage caused by severe environmental conditions such as strong light and drought.
METHODS
Plant Growth Conditions and Genetic Analysis
Arabidopsis thaliana seeds were sown on GM agar plates, and seedlings were grown in CF-405 chambers (TOMY-Seiko) at 22°C with a 16-h photoperiod (Myouga et al., 2006). Single-knockout FeSOD mutants were obtained from the ABRC (Ohio State University) and GABI-Kat (Bielefeld University). The fsd2-1 fsd3-1 double mutant was obtained by crossing flowers of fsd2-1 plants with pollen from fsd3-1. All genotypes were initially screened in the F2 generation and then confirmed in the F3 and F4 generations. The genotypes at various loci were determined by PCR analysis. Standard cloning techniques were used to make the plant transformation constructs and to generate transgenic Arabidopsis (Columbia ecotype). The multiple independent transgenic lines producing each fusion protein reported here were T-DNA homozygous plants in the F3 generation that were selected and tested by PCR and RNA gel blot analyses for high expression of the transgenes.
Fusion Gene Constructs
Putative full-length cDNA clones of Arabidopsis FeSODs were obtained from the RIKEN Bioresource Center (a gift from Masatomo Kobayashi). Clones used were FSD1 (RAFL02-01-K01), FSD2 (RAFL15-24-J05), FSD3 (RAFL06-83-B22), and CSD2 (RAFL09-24-G05). To prepare FSD-GFP/YFP, FSD-His, FSD-Flag, and FSD-HA epitope tag fusion proteins, the open reading frames (ORFs) of the FeSODs were fused to the N termini of GFP/YFP, His, Flag, and HA, respectively. To prepare promoter-GUS fusion constructs, the FeSOD promoter sequences were fused to GUS. Primer sequences are listed in Supplemental Table 4 online. PCR was performed using PrimeSTAR HS DNA polymerase (Takara) in accordance with the manufacturer's recommended conditions. Using Gateway cloning technology (Ohara and Temple, 2001), FeSOD ORFs were cloned into the pGWB2 (35S promoter, no tag), pGWB5 (35S promoter, C-terminal [C]-sGFP), pGWB8 (35S promoter, C-6xHis), pGWB11 (35S promoter, C-Flag), pGWB14 (35S promoter, C-3xHA) (Nakagawa et al., 2007), and pH35GY (35S promoter, C-YFP) (Kubo et al., 2005) vectors. Promoter sequences of FeSODs were cloned into the pGWB3 (no promoter, C-GUS) vector. For PEND-CFP constructs, the DNA fragments encoding the N-terminal region of PEND (88 amino acids) amplified from genomic DNA were inserted into the pH35GC (35S promoter, C-CFP) vector. After transfection in DH5α, stably transformed lines were cultured with appropriate antibiotics.
RNA Extraction and Expression Analyses
Total RNA was isolated from 3-week-old seedlings grown on GM agar plates using an RNeasy plant mini kit (Qiagen) according to the manufacturer's protocol and then treated with RNase-free DNase I (Promega) to degrade any remaining DNA. First-strand cDNA synthesis was performed from 2 mg of total RNA using a random hexamer with the SuperScript II reverse transcriptase kit (Invitrogen). For RT-PCR, cDNAs were amplified using ExTaq DNA polymerase (Takara) and gene-specific primers (see Supplemental Table 4 online). The number of amplification cycles was reduced to 25 from 35 to evaluate and quantify any differences among transcript levels before the levels reached saturation. PCR fragments were separated on a 1% (w/v) agarose gel containing a 1:10,000 dilution of Sybr safe (Invitrogen) dye. DNA was visualized with a UV light transilluminator. For quantitative RT-PCR, the cDNAs were amplified using TaKaRa SYBR Premix ExTaq in an ABI PRISM 7500 real-time PCR instrument (Applied Biosystems). PCR was optimized, and reactions were performed in triplicate. The transcript level was standardized based on cDNA amplification of reference genes such as ACTIN2 and UBIQUITIN1. Relative gene expression data were generated using the wild type as the reference (for basal expression comparisons). Primers used for real-time PCR amplifications are described in Supplemental Table 4 online. Primers specific for plastid-encoded genes used for RT-PCR analysis have been described (Nagashima et al., 2004).
Measurement of Photosynthetic Parameters and Chlorophyll Content
Photosynthetic parameters were calculated from chlorophyll fluorescence data collected with a portable pulse amplitude modulator fluorometer (MINI-PAM; Walz). The maximum efficiency of PSII photochemistry was determined as the ratio of variable to maximum chlorophyll fluorescence (Fv/Fm), and the chlorophyll fluorescence parameter fractions of open PSII centers and nonphotochemical quenching (Fm/Fm′ − 1) were determined as described (Krause and Weis, 1991; Kramer et al., 2004). Before measurement, fully expanded leaves taken from plants grown under normal conditions for 3 weeks were dark-adapted for 20 min. Chlorophyll content was assayed as described (Myouga et al., 2006).
Measurement of ROS in Leaves
ROS were quantified by precipitation in leaves by a method used to monitor O2− radicals, as described previously (Hoffmann et al., 2005). To quantify formazan generation, leaves were ground in liquid nitrogen and solubilized in a mixture of 2 M KOH and DMSO at a ratio of 1:1.167 (v/v). The amount of formazan was determined as A700. As negative controls, seedlings were incubated with 1 mM tetramethyl piperidinooxy, an O2− scavenger.
Microscopy Analysis
GUS-stained Arabidopsis leaves were observed with an MZ APO (Leica) or a BX60 (Olympus) light microscope and recorded with a VB-7000 digital camera (Keyence). Subcellular localization of several FSD fusion proteins was analyzed using GFP, YFP, and CFP fluorescence. GFP, YFP, and CFP fluorescence of transiently transformed tobacco (Nicotiana tabacum cv SR1) leaves was observed by confocal laser scanning microscopy (LSM510 Meta; Zeiss). For transmission electron microscopy, images were obtained as described (Motohashi et al., 2001).
Purification and Fractionation of Chloroplast Proteins
Fresh leaves from transgenic and control Arabidopsis plants were homogenized in extraction buffer (50 mM HEPES-KOH, pH 7.5, 330 mM sorbitol, 1 mM MgCl2, 1 mM MnCl2, 2 mM EDTA, pH 8.0, 0.2% BSA, and 2 mM sodium ascorbate). Intact chloroplasts were isolated by centrifugation of cell lysates through a Percoll density gradient (40 to 80%), essentially as described previously (Bartlett et al., 1982). The integrity of the chloroplasts was assessed by phase-contrast light microscopy (Walker et al., 1987), and >90% of chloroplasts were found to be intact. To prepare the chloroplast fractions, isolated chloroplasts were osmotically lysed in a hypotonic buffered solution (5 mM Tricine-NaOH [pH 7.8], 5 mM NaCl, and 1 mM MgCl2) and further ruptured by two freeze–thaw cycles. The suspension of broken chloroplasts was then centrifuged for 1 h at 100,000g. The soluble fraction containing the stroma and envelopes was removed and used directly for further analyses. The pellet, containing crude thylakoid membranes, was resuspended in the hypotonic buffered solution described above.
Immunoblot Analysis
Protein extraction, SDS-PAGE, and immunoblot analysis were performed essentially as described (Motohashi et al., 2001). Leaf tissue for protein analysis was collected from 3-week-old plants grown under standard conditions. The antibodies used in this report are monoclonal anti-His antibody (GE Healthcare), monoclonal anti-Flag antibody (F1804; Sigma-Aldrich), monoclonal anti-HA antibody (H9658; Sigma-Aldrich), and horseradish peroxidase–conjugated antibody against mouse IgG (GE Healthcare) for detection with the ECL detection system (GE Healthcare).
Yeast Two-Hybrid Assay
A commercial yeast two-hybrid system (Clontech) was used to test interactions among FSD1, FSD2, FSD3, and CSD2. The plasmids used in yeast two-hybrid assays were generated by PCR amplification of ORFs of FSDs and CSD2 that removed the predicted chloroplast transit peptides, followed by insertion of the resulting amplimers between the EcoRI and BamHI sites of both plasmid pGBKT7 DNA-BD/bait and pGADT7 AD/library (Clontech), respectively. Saccharomyces cerevisiae strain AH109 was cotransfected by the lithium acetate method (Schiestl and Gietz, 1989) with both bait and library plasmids. His prototrophs were selected on medium-stringency plates containing triple-dropout medium (lacking Leu, Trp, and His) supplemented with 10 mM 3-AT to suppress background colony growth. His and Ade prototrophs were selected on high-stringency plates containing quadruple-dropout medium (lacking Leu, Trp, His, and Ade) supplemented with both 30 mM 3-AT and 40 mg/L X-α-Gal to detect reporter gene expression.
BiFC Assay
The BiFC method was used to visualize FSD2–FSD3 interactions in living plant cells (Walter et al., 2004). The cDNA regions encoding FSD2 and FSD3 were amplified from plasmid templates containing the corresponding cDNA by PCR using gene-specific primers (see Supplemental Table 4 online) and cloned into each of the BiFC vectors, pUC-SPYNE and pUC-SPYCE. The constructs were delivered into leaf cells of tobacco by particle bombardment using a Bio-Rad PDS-1000 He particle delivery system according to the manufacturer's instructions. The BiFC-induced fluorescence was detected by confocal laser scanning microscopy after 24 h of incubation at 22°C in the dark.
Coimmunoprecipitation Assay
FSD with a C-terminal Flag epitope tag (FSD-Flag) and FSD with a C-terminal HA epitope tag (FSD-HA) were coproduced in leaves of Nicotiana benthamiana by agroinfiltration using Agrobacterium tumefaciens strain GV3101. Agrobacterium culture and infiltration were performed as described (Llave et al., 2000). Protein extraction and immune complex formation were performed as described (Kurihara et al., 2006), with minor modifications. The extraction protein (1 mL) was incubated at 4°C for 2 h with either anti-Flag–agarose (A2220; Sigma-Aldrich) or anti-HA–agarose (A2095; Sigma-Aldrich) in a 40-μL bed volume. The immune complexes were centrifuged for 30 s at 700g and washed three times in 500 μL of 50 mM Tris-HCl (pH 7.4) and 150 mM NaCl. Finally, 50 μL of 2× SDS-PAGE sample buffer was added to each sample, and the samples were boiled for 3 min. Undissolved agarose was removed by centrifugation at 700g for 5 s, and the supernatant was used for further analysis. The protein samples were separated by SDS-PAGE and immunoblotted using specific antibodies against the tagged proteins.
Purification of Recombinant Proteins and GST Pull-Down Assay
For SOD activity analysis in vitro, recombinant proteins of His-FSD2 (residues 47 to 305) and GST-FSD3 (residues 42 to 263) from which the predicted chloroplast transit peptides had been removed were cloned into the pQE30 (Qiagen) and pGEX-5X-1 (GE Healthcare) vectors, respectively. His-FSD2 was produced in Escherichia coli strain M15[pREP4] and purified with nickel-nitrilotriacetic acid agarose (Qiagen). GST-FSD3 was produced in E. coli strain BL21 and purified with glutathione–Sepharose 4B (GE Healthcare) in accordance with the manufacturer's instructions. For the GST pull-down assay, GST and GST-FSD3 fusion proteins were incubated on a rotating wheel for 2 h at 4°C with glutathione–Sepharose 4B beads (GE Healthcare) in a total volume of 500 μL of homogenization buffer (HB; 25 mM Tris-HCl [pH 7.5], 100 mM NaCl, 2 mM EDTA, and 0.5% Triton X-100). After the fusion-protein–coated beads had been washed three times with HB, a total volume of 500 μL of homogenization buffer containing His-FSD2 fusion proteins was incubated with the beads for 2 h at 4°C. After the incubation, the beads were washed three times with HB and resuspended with 50 μL of 2× sample buffer. The mixture was boiled for 2 min to dissociate the protein complex from the beads and then centrifuged for 30 s at 700g. The supernatant protein fractions were separated by 12.5% SDS-PAGE for immunoblot analysis with an anti-His antibody (GE Healthcare).
Measurement of SOD Activity
SOD was assayed by the method of Beauchamp and Fridovich (1971) by visualizing its ability to inhibit the photochemical reduction of NBT. For SOD visualization, protein samples were separated by 10% nondenaturing PAGE in Tris-Gly buffer (pH 8.3). The gel was then soaked in 0.1% (w/v) NBT solution for 15 min, rinsed with distilled water, and held for another 15 min in 100 mM potassium phosphate buffer (pH 7.0) containing 0.028 mM riboflavin and 28 mM N,N,N′,N′-tetramethyl-ethylenediamine. After being washed with distilled water, the gel was illuminated for 15 min on a light box under a light intensity of 30 μmol·m−2·s−1 to initiate the photochemical reaction. The intensities of the protein bands were quantified using ImageJ imaging software (National Institutes of Health; http://rsb.info.nih.gov/ij). SOD activity was verified by the application of KCN or H2O2. KCN is an inhibitor of Cu/ZnSOD, whereas H2O2 inhibits both Cu/ZnSOD and FeSOD. The MnSOD activity is not inhibited by either chemical.
Analysis of Oxidative Stress Tolerance
MV damage of plants was analyzed as described by Slooten et al. (1995), with some modifications. Leaf discs of ∼0.3 cm2 from wild-type and transgenic plants were preincubated in the dark at 22°C overnight in water or in an aqueous solution of MV (0.05, 0.10, 0.15, 0.25, 0.35, and 0.50 μM) to generate superoxide radicals. They were then illuminated for 2.5 h at a photon flux density of 30 μmol·m−2·s−1 and subsequently incubated at 22°C for 2 h in the dark. MV-dependent oxygen radical damage was measured as Fv/Fm, a measure of the activity of the PSII reaction centers (Krause and Weis, 1991), with a pulse amplitude modulator fluorometer (Walz). The average values of at least 60 leaf discs at seven different MV concentrations were used to calculate the MV-induced decrease in Fv/Fm.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: FSD1 (At4g25100, NP_19420), FSD2 (At5g51100, NP_19923), FSD3 (At5g23310, NP_197722), CSD1 (At1g08830), CSD2 (At2g28190), CSD3 (At5g18100), MSD1 (At3g10920), UBIQUITIN1 (At3g52590), ACTIN2 (At3g18780), PEND (At3g15170), Ec FSD (BAA15422), Ss FSD (BAA18027), and Os FSD (BAA37131).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Structures of FSD1, FSD2, and FSD3 Genes and Their Ds/T-DNA Insertion Sites.
Supplemental Figure 2. SOD Gene Family from Arabidopsis and Other Species.
Supplemental Figure 3. RT-PCR Analysis of Transcripts of FSD1, FSD-2, FSD-3, and CSD2 in Wild-Type Plants.
Supplemental Figure 4. Activity of the FSD1 and FSD3 Promoters Is Tissue-Specific.
Supplemental Figure 5. Total Soluble Protein Profiles of Isopropylthio-β-Galactoside–Induced E. coli Cells Containing the Tagged Constructs Indicated.
Supplemental Figure 6. SOD Activity of Recombinant FSD2 and FSD3 Proteins.
Supplemental Table 1. Restoration of Greening in Pale Green Mutants Grown under Low-Light Conditions.
Supplemental Table 2. Prediction of Targeting Sequences and Subcellular Localization of SOD in Arabidopsis by Different Software Tools.
Supplemental Table 3. Bonferroni Multiple Comparison Test.
Supplemental Table 4. Nucleotide Sequences of Gene-Specific Primers.
Supplementary Material
Acknowledgments
We thank Masae Kouno (RIKEN) and Noriko Nagai (RIKEN) for their technical support in managing the plant growth facility; Yukio Kurihara (RIKEN) for his help with the coimmunoprecipitation study; Mitsuoka Araki (RIKEN) and Sachiko Oyama (RIKEN) for their contribution to DNA sequencing; Bernd Weisshaar (Max Planck Institute for Plant Breeding Research) for providing T-DNA mutants that were generated in the context of the GABI-Kat program; Tsuyoshi Nakagawa (Shimane University) for providing the binary vector pGWB series; Taku Demura (RIKEN) for providing the pH35GY and pH35GC vectors; and Jörg Kudla (Münster University) for providing BiFC vectors. We also thank Masatomo Kobayashi of the RIKEN Bioresource Center for providing RAFL cDNA clones. This work was supported by the RIKEN Plant Science Center, Japan.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Kazuo Shinozaki (sinozaki@rtc.riken.jp).
Online version contains Web-only data.
References
- Abdel-Ghany, S.E., Müller-Moulé, P., Niyogi, K.K., Pilon, M., and Shikanai, T. (2005). Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 17 1233–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almansa, M.S., del Río, L.A., and Sevilla, F. (1994). Characterization of an iron-containing superoxide dismutase from a higher plant, Citrus limonum. Physiol. Plant. 90 339–347. [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Alscher, R.G., Erturk, N., and Heath, L.S. (2002). Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 53 1331–1341. [PubMed] [Google Scholar]
- Åslund, F., Zheng, M., Beckwith, J., and Storz, G. (1999). Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc. Natl. Acad. Sci. USA 96 6161–6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginsky, S., and Gruissem, W. (2006). Arabidopsis thaliana proteomics: From proteome to genome. J. Exp. Bot. 57 1485–1491. [DOI] [PubMed] [Google Scholar]
- Baginsky, S., Kleffmann, T., von Zychlinski, A., and Gruissem, W. (2005). Analysis of shotgun proteomics and RNA profiling data from Arabidopsis thaliana chloroplasts. J. Proteome Res. 4 637–640. [DOI] [PubMed] [Google Scholar]
- Baginsky, S., Tiller, K., and Link, G. (1997). Transcription factor phosphorylation by a protein kinase associated with chloroplast RNA polymerase from mustard (Sinapis alba). Plant Mol. Biol. 34 181–189. [DOI] [PubMed] [Google Scholar]
- Baginsky, S., Tiller, K., Pfannschmidt, T., and Link, G. (1999). PTK, the chloroplast RNA polymerase-associated protein kinase from mustard (Sinapis alba L.), mediates redox control of plastid in vitro transcription. Plant Mol. Biol. 39 1013–1023. [DOI] [PubMed] [Google Scholar]
- Bartlett, S.G., Grossman, A.R., and Chua, N.-H. (1982). In vitro synthesis and uptake of cytoplasmically-synthesized chloroplast proteins. In Methods in Chloroplast Molecular Biology, M. Edelman, R.B. Hallick, and N.-H. Chua, eds (Amsterdam: Elsevier Biomedical), pp. 1081–1091.
- Beauchamp, C., and Fridovich, I. (1971). Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44 276–287. [DOI] [PubMed] [Google Scholar]
- Bowler, C., van Camp, W., van Montagu, M., and Inzé, D. (1994). Superoxide dismutases in plants. Crit. Rev. Plant Sci. 13 199–218. [Google Scholar]
- Bowler, C., van Montagu, M., and Inzé, D. (1992). Superoxide dismutase and stress tolerance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43 83–116. [Google Scholar]
- Brugière, S., Kowalski, S., Ferro, M., Seigneurin-Berny, D., Miras, S., Salvi, D., Ravanel, S., d'Hérin, P., Garin, J., Bourguignon, J., Joyard, J., and Rolland, N. (2004). The hydrophobic proteome of mitochondrial membranes from Arabidopsis cell suspensions. Phytochemistry 65 1693–1707. [DOI] [PubMed] [Google Scholar]
- Cannon, G.C., Ward, L.N., Case, C.I., and Heinhorst, S. (1999). The 68 kDa DNA compacting nucleoid protein from soybean chloroplasts inhibits DNA synthesis in vitro. Plant Mol. Biol. 39 835–845. [DOI] [PubMed] [Google Scholar]
- Casano, L.M., Martín, M., and Sabater, B. (2001). Hydrogen peroxide mediates the induction of chloroplastic Ndh complex under photooxidative stress in barley. Plant Physiol. 125 1450–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi-Ham, C.L., Keaton, M.A., Cannon, G.C., and Heinhorst, S. (2002). The DNA-compacting protein DCP68 from soybean chloroplasts is ferredoxin:sulfite reductase and co-localizes with the organellar nucleoid. Plant Mol. Biol. 49 621–631. [DOI] [PubMed] [Google Scholar]
- Duke, M.V., and Salin, M.L. (1985). Purification and characterization of an iron-containing SOD from a eukaryote, Ginkgo biloba. Arch. Biochem. Biophys. 243 305–314. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300 1005–1016. [DOI] [PubMed] [Google Scholar]
- Ferro, M., Salvi, D., Brugiere, S., Miras, S., Kowalski, S., Louwagie, M., Garin, J., Joyard, J., and Rolland, N. (2003). Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol. Cell. Proteomics 2 325–345. [DOI] [PubMed] [Google Scholar]
- Gómez, J.M., Jiménez, A., Olmos, E., and Sevilla, F. (2004). Location and effects of long-term NaCl stress on superoxide dismutase and ascorbate peroxidase isoenzymes of pea (Pisum sativum cv. Puget) chloroplasts. J. Exp. Bot. 55 119–130. [DOI] [PubMed] [Google Scholar]
- Gupta, A.S., Heinen, J.L., Holaday, A.S., Burke, J.J., and Allen, R.D. (1993). Increased resistance to oxidative stress in transgenic plants that overexpress chloroplastic Cu/Zn superoxide dismutase. Proc. Natl. Acad. Sci. USA 90 1629–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert, S.K., Samson, G., Fork, D.C., and Laudenbach, D.E. (1992). Characterization of damage to photosystems I and II in a cyanobacterium lacking detectable iron superoxide dismutase activity. Proc. Natl. Acad. Sci. USA 89 8716–8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, A., Hammes, E., Plieth, C., Desel, C., Sattelmacher, B., and Hansen, U.P. (2005). Effect of CO2 supply on formation of reactive oxygen species in Arabidopsis thaliana. Protoplasma 227 3–9. [DOI] [PubMed] [Google Scholar]
- Horton, P., Park, K.J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C.J., and Nakai, K. (2007). WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 35 W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., Motohashi, R., Kuromori, T., Yoshiteru, N., Seki, M., Kamiya, A., Mizukado, S., Sakurai, T., and Shinozaki, K. (2005). A resource of 5814 Dissociation transposon-tagged and sequence-indexed lines of Arabidopsis transposed from start loci on chromosome 5. Plant Cell Physiol. 46 1149–1153. [DOI] [PubMed] [Google Scholar]
- Kleffmann, T., Russenberger, D., von Zychlinski, A., Christopher, W., Sjolander, K., Gruissem, W., and Baginsky, S. (2004). The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 14 354–362. [DOI] [PubMed] [Google Scholar]
- Kliebenstein, D.J., Monde, R.A., and Last, R.L. (1998). Superoxide dismutase in Arabidopsis: An eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 118 637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, D.M., Johnson, G., Kiirats, O., and Edwards, G.E. (2004). New fluorescence parameters for the determination of QA redox state and excitation energy fluxes. Photosynth. Res. 79 209–218. [DOI] [PubMed] [Google Scholar]
- Krause, G.H., and Weis, E. (1991). Chlorophyll fluorescence and photosynthesis: The basics. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42 313–349. [Google Scholar]
- Kubo, M., Udagawa, M., Nishikubo, N., Horiguchi, G., Yamaguchi, M., Ito, J., Mimura, T., Fukuda, H., and Demura, T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara, Y., Takashi, Y., and Watanabe, Y. (2006). The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12 206–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa, T. (1991). The replication, differentiation, and inheritance of plastids with emphasis on the concept of organelle nuclei. Int. Rev. Cytol. 128 1–62. [Google Scholar]
- Kuromori, T., Hirayama, T., Kiyosue, Y., Takabe, H., Mizukado, S., Sakurai, T., Akiyama, K., Kamiya, A., Ito, T., and Shinozaki, K. (2004). A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J. 37 897–905. [DOI] [PubMed] [Google Scholar]
- Kwiatowski, J., Safianowska, A., and Kaniuga, Z. (1985). Isolation and characterization of an iron-containing superoxide dismutase from tomato leaves, Lycopersicon esculentum. Eur. J. Biochem. 146 459–466. [DOI] [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., and Carrington, J.C. (2000). Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc. Natl. Acad. Sci. USA 97 13401–13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmagne, A., Rouet, M.A., Ferro, M., Rolland, N., Alcon, C., Joyard, J., Garin, J., Barbier-Brygoo, H., and Ephritikhine, G. (2004). Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Mol. Cell. Proteomics 3 675–691. [DOI] [PubMed] [Google Scholar]
- Mehler, A.H. (1951). Studies on reactions of illuminated chloroplasts. II. Stimulation and inhibition of the reaction with molecular oxygen. Arch. Biochem. Biophys. 33 339–351. [DOI] [PubMed] [Google Scholar]
- Miller, G., Suzuki, N., Rizhsky, L., Hegie, A., Koussevitzky, S., and Mittler, R. (2007). Double mutants deficient in cytosolic and thylakoid ascorbate peroxidase reveal a complex mode of interaction between reactive oxygen species, plant development and response to abiotic stresses. Plant Physiol. 144 1777–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi, R., Nagata, N., Ito, T., Takahashi, S., Hobo, T., Yoshida, S., and Shinozaki, K. (2001). An essential role of a TatC homologue of a Delta pH-dependent protein transporter in thylakoid membrane formation during chloroplast development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98 10499–10504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myouga, F., Motohashi, R., Kuromori, T., Nagata, N., and Shinozaki, K. (2006). An Arabidopsis chloroplast-targeted Hsp101 homologue, APG6, has an essential role in chloroplast development as well as heat-stress response. Plant J. 48 249–260. [DOI] [PubMed] [Google Scholar]
- Nagashima, A., Hanaoka, M., Motohashi, R., Seki, M., Shinozaki, K., Kanamaru, K., Takahashi, H., and Tanaka, K. (2004). DNA microarray analysis of plastid gene expression in a mutant deficient in a plastid transcription factor sigma, SIG2. Biosci. Biotechnol. Biochem. 68 694–704. [DOI] [PubMed] [Google Scholar]
- Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., Toyooka, K., Matsuoka, K., Jinbo, T., and Kimura, T. (2007). Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104 34–41. [DOI] [PubMed] [Google Scholar]
- Ogawa, K., Kanematsu, S., Takabe, K., and Asada, K. (1995). Attachment of CuZn-superoxide dismutase to thylakoid membranes at the site of superoxide generation (PSI) in spinach chloroplasts: Detection by immuno-gold labeling after rapid freezing and substitution method. Plant Cell Physiol. 36 565–573. [Google Scholar]
- Ogrzewalla, K., Piotrowski, M., Reinbothe, S., and Link, G. (2002). The plastid transcription kinase from mustard (Sinapis alba L.). A nuclear-encoded CK2-type chloroplast enzyme with redox-sensitive function. Eur. J. Biochem. 269 3329–3337. [PubMed] [Google Scholar]
- Ohara, O., and Temple, G. (2001). Directional cDNA library construction assisted by the in vitro recombination reaction. Nucleic Acids Res. 29 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck, S.C. (2005). Update on proteomics in Arabidopsis. Where do we go from here? Plant Physiol. 138 591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier, J.B., Cai, Y., Sun, Q., Zabrouskov, V., Giacomelli, L., Rudella, A., Ytterberg, A.J., Rutschow, H., and van Wijk, K.J. (2006). The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Mol. Cell. Proteomics 5 114–133. [DOI] [PubMed] [Google Scholar]
- Peltier, J.B., Emanuelsson, O., Kalume, D.E., Ytterberg, J., Friso, G., Rudella, A., Liberles, D.A., Soderberg, L., Roepstorff, P., von Heijne, G., and van Wijk, K.J. (2002). Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14 211–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz, J., Liere, K., Kandlbinder, A., Dietz, K.J., and Oelmüller, R. (2006). pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18 176–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt, T., Ogrzewalla, K., Baginsky, S., Sickmann, A., Meyer, H.E., and Link, G. (2000). The multisubunit chloroplast RNA polymerase A from mustard (Sinapis alba L.). Integration of a prokaryotic core into a larger complex with organelle-specific functions. Eur. J. Biochem. 267 253–261. [DOI] [PubMed] [Google Scholar]
- Pnueli, L., Liang, H., and Mittler, R. (2003). Growth suppression, abnormal guard cell response, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (Apx1)-deficient Arabidopsis plants. Plant J. 34 187–203. [DOI] [PubMed] [Google Scholar]
- Polle, A. (2001). Dissecting the superoxide dismutase-ascorbate-glutathione pathway in chloroplasts by metabolic modeling. Computer simulations as a step towards flux analysis. Plant Physiol. 126 445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky, L., Liang, H., and Mittler, R. (2003). The water-water cycle is essential for chloroplast protection in the absence of stress. J. Biol. Chem. 278 38921–38925. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Serrano, M., Romero-Puertas, M.C., Zabalza, A., Corpas, F.J., Gómez, M., Del Río, L.A., and Sandalio, L.M. (2006). Cadmium effect on oxidative metabolism of pea (Pisum sativum L.) roots. Imaging of reactive oxygen species and nitric oxide accumulation in vivo. Plant Cell Environ. 29 1532–1544. [DOI] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Salin, M.L., and Bridges, S.M. (1980). Isolation and characterization of an iron-containing superoxide dismutase from a eucaryote, Brassica campestris. Arch. Biochem. Biophys. 201 369–374. [DOI] [PubMed] [Google Scholar]
- Salin, M.L., and Bridges, S.M. (1982). Isolation and characterization of an iron-containing superoxide dismutase from water lily, Nuphar luteum. Plant Physiol. 69 161–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson, G., Herbert, S.K., Fork, D.C., and Laudenbach, D.E. (1994). Acclimation of the photosynthetic apparatus to growth irradiance in a mutant strain of Synechococcus lacking iron superoxide dismutase. Plant Physiol. 105 287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, N., Albrieux, C., Joyard, J., Douce, R., and Kuroiwa, T. (1993). Detection and characterization of a plastid envelope DNA binding protein which may anchor plastid nucleoids. EMBO J. 12 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schein, A.I., Kissinger, J.C., and Ungar, L.H. (2001). Chloroplast transit peptide prediction: A peek inside the black box. Nucleic Acids Res. 29 e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl, R.H., and Gietz, R.D. (1989). High efficiency transformation of intact cells using single stranded nucleic acids as a carrier. Curr. Genet. 16 339–346. [DOI] [PubMed] [Google Scholar]
- Sekine, K., Hase, T., and Sato, N. (2002). Reversible DNA compaction by sulfite reductase regulates transcriptional activity of chloroplast nucleoids. J. Biol. Chem. 277 24399–24404. [DOI] [PubMed] [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slooten, L., Capiau, K., Van Camp, W., Van Montagu, M., Sybesma, C., and Inzé, D. (1995). Factors affecting the enhancement of oxidative stress tolerance in transgenic tobacco overexpressing manganese superoxide dismutase in the chloroplast. Plant Physiol. 107 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, I., Peeters, N., Legeai, F., and Lurin, C. (2004). Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4 1581–1590. [DOI] [PubMed] [Google Scholar]
- Takahashi, M.A., and Asada, K. (1983). Superoxide anion permeability of phospholipid membranes and chloroplast thylakoids. Arch. Biochem. Biophys. 226 558–566. [DOI] [PubMed] [Google Scholar]
- Terasawa, K., and Sato, N. (2005). Visualization of plastid nucleoids in situ using the PEND-GFP fusion protein. Plant Cell Physiol. 46 649–660. [DOI] [PubMed] [Google Scholar]
- Thomas, D.J., Avenson, T.J., Thomas, J.B., and Herbert, S.K. (1998). A cyanobacterium lacking iron superoxide dismutase is sensitized to oxidative stress induced with methyl viologen but is not sensitized to oxidative stress induced with norflurazon. Plant Physiol. 116 1593–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Camp, W., Capiau, K., Van Montagu, M., Inzé, D., and Slooten, L. (1996). Enhancement of oxidative stress tolerance in transgenic tobacco plants overproducing Fe-superoxide dismutase in chloroplasts. Plant Physiol. 112 1703–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, D., Zoran, G., Cerovic, Z., and Robinson, S. (1987). Isolation of intact chloroplasts: General principles and criteria of integrity. Methods Enzymol. 148 145–157. [Google Scholar]
- Walter, M., Chaban, C., Schutze, K., Batistic, O., Weckermann, K., Nake, C., Blazevic, D., Grefen, C., Schumacher, K., Oecking, C., Harter, K., and Kudla, J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40 428–438. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.