Abstract
Perforin-mediated lymphocyte cytotoxicity is critical for pathogen elimination and immune homeostasis. Perforin disruption of target cell membranes is hypothesized to require binding of a calcium-dependent, lipid-inserting, C2 domain. In a family affected by hemophagocytic lymphohistiocytosis, a severe inflammatory disorder caused by perforin deficiency, we identified 2 amino acid substitutions in the perforin C2 domain: T435M, a previously identified mutant with disputed pathogenicity, and Y438C, a novel substitution. Using biophysical modeling, we predicted that the T435M substitution, but not Y438C, would interfere with calcium binding and thus cytotoxic function. The capacity for cytotoxic function was tested after expression of the variant perforins in rat basophilic leukemia cells and murine cytotoxic T lymphocytes. As predicted, cells transduced with perforin-T435M lacked cytotoxicity, but those expressing perforin-Y438C displayed intact cytotoxic function. Using novel antibody-capture and liposome-binding assays, we found that both mutant perforins were secreted; however, only nonmutated and Y438C-substituted perforins were capable of calcium-dependent lipid binding. In addition, we found that perforin-Y438C was capable of mediating cytotoxicity without apparent proteolytic maturation. This study clearly demonstrates the pathogenicity of the T435M mutation and illustrates, for the first time, the critical role of the human perforin C2 domain for calcium-dependent, cytotoxic function.
Introduction
Perforin plays an essential role in lymphocyte-mediated cytotoxicity as it is required for the delivery of granzymes to the cytosol of target cells, leading to death by apoptosis of tumor cells or virally infected cells.1 Absence or severe alteration of perforin function in natural killer (NK) cells and cytotoxic T lymphocytes (CTLs) leads to familial hemophagocytic lymphohistiocytosis type 2 (FHLH2), a life-threatening immunologic disorder of infancy caused by mutations in the gene encoding for perforin (PRF1).2 Animal studies in perforin-deficient mice suggest that the hemophagocytosis and cytokine storm results from a sustained exposure of cytotoxic lymphocytes to infectious antigens.3
Early studies demonstrated that perforin-mediated cytotoxic function is calcium (Ca)–dependent.4,5 The cytotoxicity of purified perforin is abolished by Ca-chelating agents, such as ethylenediaminetetraacetic acid. The C2 domain of perforin is hypothesized to be the critical domain for conferring Ca-dependent binding of perforin to target cell membranes6 as it is homologous to conserved C2 domains from other Ca-dependent, lipid-binding proteins, such as synaptotagmin, rabphilin, phospholipase A2, and protein kinase C family members.7–9 Mutation of aspartate residues in the predicted Ca binding loops in murine perforin led to a loss of lytic activity.10 Uellner et al6 first noted the proteolytic maturation of human perforin and hypothesized that proteolytic removal of the carboxyl-terminus allowed a conformational change, exposing the C2 domain. Their studies suggested that the mature, but not precursor, perforin was capable of Ca-dependent cytotoxic function.
The finding of missense mutations in the perforin C2 domain in association with FHLH22,11–18 suggests that the C2 domain is physiologically relevant for cytotoxic function. It is assumed that all FHLH2-associated mutations in the C2 domain abolish cytotoxic function; however, this has not been rigorously demonstrated. Indeed, the pathogenicity of perforin C2 mutation, T435M, has been disputed. Although this variant has been clearly associated with patients with hemophagocytic lymphohistiocytosis,14,18 introduction of the equivalent amino acid substitution into murine perforin (T434M) had no impact on lytic function when tested in vitro in a model system.10
Consistent with this diagnostic dilemma, we identified healthy adults from a consanguineous family who were compound heterozygous for a combination of 3 naturally occurring perforin variants: A91V, a functional polymorphism,19–21 and 2 amino acid substitutions in the perforin C2 domain, the disputed T435M, and a novel variant, Y438C. As one of their children died of hemophagocytic lymphohistiocytosis 20 years earlier, perforin function and expression were assayed in all available family members and strongly suggested that the T435M mutation was pathogenic. Based on the structures of several crystallized C2 domains, we generated a revised structural model for the perforin C2 domain that predicted impairment of Ca binding with the T435M, but not Y438C, substitution. As we recently established a model system to assess the biochemical consequences of missense mutations on perforin folding and maturation,19 we extended these studies in the current report to test our hypothesis that the T435M, but not Y438C, substitution would abolish perforin cytotoxicity.
Methods
Clinical assessment of family members
Family members were tested for the presence of mutations in PRF1 as previously described13 with modifications listed in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Detection of perforin expression by flow cytometry in family members and healthy controls was performed using anti-T-cell receptor-αβ fluorescein isothiocyanate, anti-CD8 peridinin chlorophyll protein (PerCP), and anti-CD56 allophycocyanin antibodies (BD Biosciences, San Jose, CA) for surface staining and antiperforin phycoerythrin (PE; clone δG9) or isotype controls for intracellular staining. CD56+ T cells were excluded from the analysis of NK cells. NK function was tested by standard 51Cr release assay using freshly drawn peripheral blood mononuclear cells as effectors.22
Structural modeling
Structural homologues of the perforin C2 domain were identified using PSI-BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and the Conserved Domain Search website (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). Sequence alignments of perforin and homologues were done with T-Coffee (http://tcoffee.vital-it.ch) sequence alignment program (Table 1; Figures S1–S3). The alignment with type I C2 domains led to higher scores and fewer deletions/insertions compared with the type II sequences. Three modeling programs were used in tandem to model the perforin C2 domain according to both type I and type II topologies: (1) The Robetta server identified 2CHD, a type I C2 domain, as the top structural template used for modeling. (2) The (PS)2 server (http://ps2.life.nctu.edu.tw/) was used to generate both type I and type II C2 domain models for perforin. The program identified 1DQV, a Ca-bound type I C2 domain, as the best template. Using user-defined templates, all 7 type I C2 domains successfully generated C2-like structures. In contrast, of the 5 type II templates used, only 3 generated final models that showed appropriate C2 topology. 3) The Swiss-Model server (http://swissmodel.expasy.org/) was used to generate 6 type I C2 domain models (Ca-bound templates 1A25, 1DQV (both C2 domains), 2CM5, and 3RPB, and Ca-free model 2CHD). Potential side chain rotamer configurations for the T435M and Y438C mutations were compared using the Mutagenesis Wizard in Pymol.
Cell culture
EL4 cells (ATCC, Manassas, VA) were maintained in Dulbecco modified Eagle medium with 10% equine serum. For metabolic studies, 100 ng/mL concanamycin A (CMA) or 10 μg/mL E64 was added to cultures of cells overnight.
Retroviral transduction
Generation of retroviral constructs, retroviral supernatants from Phoenix Eco packaging cells, and transduction of RBL1 and RBL-2H3 cells were performed as previously described.19 The same retroviral supernatants were used to transduce murine splenocytes. Prf1−/− mice were crossed with P14 T-cell receptor transgenic mice (P14.Prf1−/−). Splenocytes from P14.Prf1−/− mice were loaded with cognate antigen (100 ng/mL gp33-41 peptide, abbreviated gp33 throughout text), synthesized by the Molecular Resource Center at National Jewish Medical Center (Denver, CO) for 1 hour at 37°C, washed with Hanks buffered saline solution, resuspended in growth media (Dulbecco modified Eagle medium with 10% fetal bovine serum, and incubated at 37°C). Retroviral transduction was performed 48 hours later via spinfection (2 hours at 500g) and 4 μg/mL polybrene (Sigma-Aldrich, St Louis, MO). After transduction, the cells were washed and expanded in media containing murine IL-2 (10 ng/mL). Detection of green fluorescent protein (GFP) and perforin in transduced rat basophilic leukemia (RBL), and CTL was performed by flow cytometry as previously described.19 Only RBL-2H3 cells required GFP sorting a result of lower transduction efficiency (< 30%). This use of mice in this study was regulated by the Biosafety Office at Cincinnati Children's Hospital.
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis/Western blot
Western blots were probed by P1-8 perforin antibody (Kamiya Biomedical, Seattle, WA) under either reducing or nonreducing conditions.19 Equal loading was monitored by the detection of actin with AC-15 clone (Sigma-Aldrich). Antiperforin antibody 2d4 was a gift from Dr Griffiths.23 Antiperforin antibody pf344 (Mabtech, Nacka, Sweden) was a gift from Dr Froelich.
RBC lytic assays
RBL-2H3 cells were precoated with anti-2,4-dinitrophenol IgE, coincubated with 2,4-dinitrophenol-labeled, 51Cr-loaded red blood cells (RBCs), and lysis measured as previously described.19
Degranulation with antibody capture assay (RBL-2H3)
Cells were stimulated with phorbol myristate acetate (PMA) at 50 ng/mL and ionomycin at 1 μg/mL for 1 hour at 37°C. RBL-2H3 cells were treated with PMA and ionomycin (P/I) in the presence of 2 μg an anti-perforin “capture” antibody, δG9, at 37°C in RPMI with 10% fetal bovine serum. The anti-perforin antibody was included to “capture” perforin in the supernatant as it was secreted, thus increasing the detection of the protein. After stimulation, the RBL-2H3 cells were pelleted and resuspended in lysis buffer. The antibody/perforin complex in the supernatant was precipitated by adding protein A agarose, 1 hour at 4°C, and the pellet washed in 2% NP-40 lysis buffer. The pellets were resuspended with Laemmli buffer (nonreducing) and heated to 85°C for 10 minutes. The samples were analyzed by Western blot as described in “Sodium dodecyl sulfate polyacrylamide gel electrophoresis/Western blot” with P1-8 antibody.
Target cell–mediated degranulation capture assay (murine CTL)
P14.Prf1−/− CTLs expressing human perforins were isolated by centrifugation through Ficolle-Hypaque 72 to 96 hours after retroviral transduction and coincubated with gp33-loaded EL4 target cells at a 1:1 ratio in growth medium for 2 hours at 37°C. The growth medium contained 2 μg capture antibody, δG9. After stimulation, the antibody/perforin complex was precipitated as described for the RBL2H3 capture assay.
Murine CTL cytotoxicity assay
EL4 cells (targets) were incubated with 51Cr and loaded with 100 ng/mL GP33 peptide for 1 hour at 37°C. Murine CTLs were Ficolled 48 to 72 hours after transduction. Viable cells were coincubated with target EL4 cells at various effector/target ratios, by quadruplicate in RPMI 10% fetal bovine serum. Chromium release was measured after 4 hours of coincubation as described previously.19
Generation of liposomes
Lipids at 2:1 molar ratio of cholesterol (5 mg) and dioleoyl phosphatidylcholine (10 mg; Avanti Polar Lipids, Alabaster, AL) were dissolved in chloroform, dried under vacuum, and rehydrated with 1 mL of phosphate-buffered saline. The resulting liposomes were then labeled with 1,1′-dioctadecyl-3,3,3′,3′-tetramethyl-indodicarbocyanine-5,5′-disulfonic acid (DiD) by adding an equal volume of a 25-μM solution to the liposomes. The liposomes were then serially extruded through 0.4-, 0.2-, and 0.1-μm pore membranes and stored under argon gas.
Ca-dependent lipid binding of secreted perforin
RBL-2H3 cells (2 × 106) expressing perforin were resuspended in 200 μL secretion buffer, Hanks buffered saline solution, pH 6.9, 1% fatty acid free bovine serum albumin, and 2 μL fluorescent liposomes (“Generation of liposomes”). After 20 minutes of stimulation with P/I at 37°C with agitation, 10 μL PE-conjugated δG9 antibody was added to the tube for the final 60 minutes at 37°C. After stimulation, the RBL-2H3 cells were removed by centrifugation, and the liposome-containing supernatant was analyzed by flow cytometry.
Results
Family study of C2 domain variant perforins
We studied perforin expression, function, and sequence in healthy family members of a child who died 20 years ago from hemophagocytic lymphohistiocytosis. The child presented at 8 months of age with a clinical picture that clearly fulfilled the current diagnostic criteria of hemophagocytic lymphohistiocytosis: fever, hepatosplenomegaly, cytopenias, hypofibrinogenemia, hypertriglyceridemia, and visible hemophagocytosis in liver and bone marrow.24 The unaffected family members presented for genetic evaluation of perforin deficiency as the child's DNA was not available for genotyping.
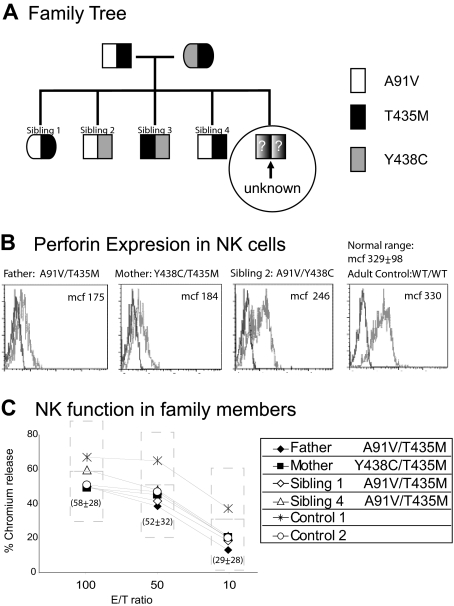
As shown in Figure 1A, each healthy family member was a compound heterozygous carrier of a combination of 3 amino acid substitutions. The father contributed a polymorphic allele leading to the A91V substitution, a functional perforin variant previously identified in healthy adult carriers and in patients with hemophagocytic syndromes.25–27 Both parents shared a second perforin amino acid substitution, T435M. The T435M substitution has been reported in only 2 patients presenting with hemophagocytic lymphohistiocytosis, and its pathogenicity has been disputed.10,14,18 The mother contributed a third variant allele, leading to the Y438C substitution, a novel perforin variant. In the healthy siblings and parents, all combinations of the 3 alleles were represented except T435M/T435M.
Figure 1.
Genotype, expression, and function of perforin in a family affected by FHLH2. (A) The family tree illustrates the parents and siblings of a child with unknown genotype who died of HLH as an infant. All family members are compound heterozygous for 3 different amino acid transitions in perforin. Compared with the parents, sibling 2 has a unique combination of 2 alleles but is healthy. The only remaining combination that is unique is homozygous T435M, the predicted genotype of the child. (B) Flow cytometry was used to detect perforin in NK cells from parents and siblings. Each combination leads to detectable perforin expression, represented by the light gray histogram. The dark gray histogram represents the isotype control. A representative adult control is shown for comparison. The MCF of perforin in the NK cells of the mother and father were less than 2 SDs below normal compared with healthy adults (n = 13 controls, average MCF = 329 ± 98 [2 SD]). (C) The NK function in the family members and 2 healthy adult controls was tested by chromium release assay, using K562 cells as targets and freshly isolated peripheral blood mononuclear cells as effectors. Percentage of chromium release was calculated as (cpm of sample − cpm of blank)/(cpm of maximum − cpm of blank). Shown is the percentage chromium release at 3 effector/target ratios (E/T). The normal range from 10 healthy adults is shown for comparison at each E/T ratio (dashed box; mean ± 2 SD). All persons were noted to have normal NK percentages at the time of the assay.
To determine whether perforin expression was influenced by one or both alleles, we assayed perforin expression by flow cytometry in NK cells from all available family members. Reduced perforin signal (mean channel fluorescence [MCF]) was noted in the NK cells of the parents, compared with healthy adult controls. A single population was noted (Figure 1B), although the MCF of each histogram was decreased, indicating that the perforin detection in all NK cells is decreased but not absent. Additional family members with the same allele combinations had equivalent staining (not shown).
To study the impact of compound heterozygous perforin mutations on lymphocyte-mediated cytotoxicity, NK function was tested in the parents and 2 of the siblings. Cytotoxic activity was measured as the percentage of chromium released from K562 target cells. No diminution of cytotoxic function was detected in the family members (Figure 1C), compared with 2 healthy controls of the day and compared with the normal range in the laboratory, indicating that at least one allele was present in each family member, which was responsible for cytotoxic function comparable with that of normals. Of the 4 combinations of perforin missense mutations possible in the family, only the biallelic T435M/T435M was unrepresented by healthy family members. Therefore, we hypothesized that T435M was a pathogenic mutation. As earlier studies had shown that the equivalent mutation in the mouse perforin, T434M, did not adversely impact lytic function,10 we sought to examine the impact of this mutation on human perforin structure and function while also testing the novel variant, PRF1-Y438C.
Sequence comparison across C2 domains
Both the T435M and Y438C mutations are located in the C2 domain of perforin, a region responsible for binding phospholipid membranes in other proteins.7–9 We first compared the T435 and Y438 residues in perforins across species (Figure S1) and determined that T435 is perfectly conserved across all mammalian species and is immediately adjacent to a conserved aspartate previously predicted to bind Ca.10 The Y438 is also highly conserved across mammalian species, replaced by a similar hydrophobic residue, phenylalanine, only in the platypus.
We then compared C2 domains from other Ca-dependent lipid-binding proteins. The perforin C2 domain has been modeled previously according to 2 different C2 classes: a type I topology18 or a type II topology.6,10,12 Both topologies comprise a beta sandwich with 2 4-stranded beta sheets, but the arrangement of beta strands within each sheet is different. As a result of the strand permutation, the Ca-binding loops are on opposite ends of the C2 domain in the 2 topologies and have different chemical environments. We identified by BLAST the C2 domains with the highest sequence identity to human perforin and found that the type I C2 sequences showed significantly better alignments to the sequence of the human perforin than type II sequences (Figures S2,S3). According to the type I alignment, the equivalent of T435 is usually threonine or a conservative substitution serine; whereas, according to the type II topology, the equivalent residue to T435 is serine, proline, or methionine. The Y438 is conserved as a tyrosine or phenylalanine in either alignment. Finally, all 4 aspartate residues predicted to bind Ca by molecular modeling and structure function studies of murine perforin10 are perfectly conserved in the type I alignment.
Modeling the effects of variations within the C2 domain
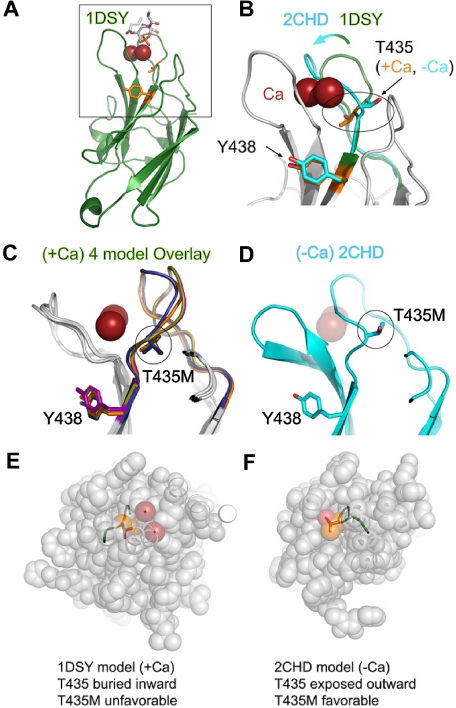
Using the type I topology alignment, we assessed the structural implications of the T435M and Y438C substitutions in perforin with regard to Ca binding or C2 domain stability. We observed that the T435 side chain adopted opposite orientations in the Ca-free versus Ca-bound models (Figure 2; Table S1). The substitution with methionine at amino acid 435 did not disrupt folding in the Ca-free model (2CHD), thus explaining the near-normal levels of protein found in the family members. However, the methionine substitution at amino acid 435 was not tolerated in the Ca-bound models where the side chain was oriented inward toward the protein core; the interior cavity containing the T435 side chain was too small to accommodate the long methionine side chain. Therefore, the T435M mutation would destabilize the Ca-bound conformation and, as a result, interfere with cytotoxic function. Because of its localization away from the Ca-binding site, the Y438C substitution was not predicted to disrupt Ca binding and did not exhibit any folding clashes, suggesting that this substitution may be well tolerated. To test our hypothesis that the T435M, but not the Y438C, mutation disrupted perforin function, we expressed both variant perforins in 2 independent cell lines containing secretory granules: RBL cells and CTLs from perforin-deficient mice.
Figure 2.
Structural modeling of perforin missense mutations in the C2 domain. (A) Homology model of the perforin C2 domain based on the C2 domain of 1DSY (protein kinase Cα) bound to Ca and phosphatidylserine. Res spheres represent Ca ions; phosphatidylserine are sticks colored according to atom type, and side chains of T435 and Y438 are orange sticks. The box highlights the zoomed view shown in panel B. (B) Magnified view of Ca-binding region. An overlay is shown of perforin C2 models based on 1DSY (green, as in panel A) and 2CHD, a Ca-free C2 domain from rabphilin-3A (cyan). The Ca-binding BC loop is highlighted to illustrate the structural differences. Note that T435 adopts opposite orientations in the Ca-bound versus Ca-free models. (C) Side view overlay of 4 representative Ca-bound type I perforin C2 domain models built by the PS2 server, based on 1A25, 1DQV, 1DSY, and 2CM6. Note the similar positions of T435 and Y438 side chains in all models. (D) Side view of the perforin C2 model based on the Ca-free 2CHD structure, with the same orientation as panel C. Note that the BC loop in the Ca-free form would clash with the Ca ions (transparent red spheres). (E,F) Top views of perforin C2 models based on Ca-bound 1DSY (E) or Ca-free 2CHD (F). In both panels, the C2 domain has been sliced identically by an approximately horizontal plane just above the Ca ions in the 1DSY model. This view reveals that, in the Ca-bound model, the T435 side chain is pointed inward and surrounded by atoms from other residues. In contrast, T435 is directed outward and is solvent-accessible in the Ca-free form. Thus, the T435M mutation would be tolerated only in the Ca-free conformation of the C2 domain.
Expression of mutant perforins in RBL cells
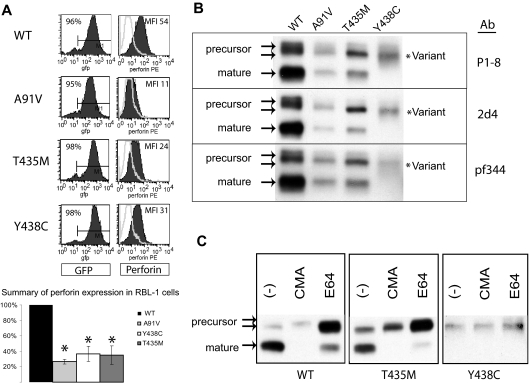
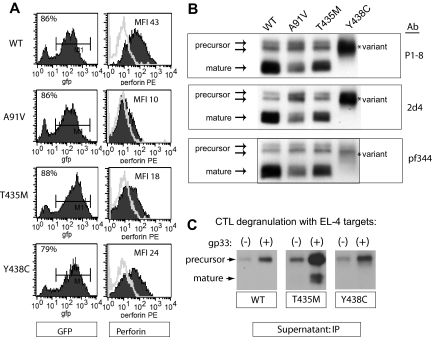
In RBL-1 cells, we previously demonstrated that human, wild-type perforin (PRF1-WT) was stably expressed and correctly processed after transduction with perforin-containing retroviral vectors.19 We used the same approach to analyze individually the effect of Y438C and T435M mutations on perforin expression and maturation. Mutant and WT perforin cDNAs were cloned into GFP-containing retroviral vectors and then introduced into RBL-1 cells by retroviral transduction. Flow cytometry histograms and results from 3 independent transductions are represented in Figure 3A. PRF1-A91V, -T435M, and -Y438C exhibited statistically decreased detection compared with PRF1-WT.
Figure 3.
Expression of human perforin in RBL cells. (A) Flow cytometry of RBL-1 cells expressing recombinant human perforin cDNA, wild-type (WT) or mutant, after retroviral transduction. The retroviral vector contains GFP in cis with the perforin cDNA. GFP expression is shown in the left column, perforin expression in the right column. The results of 3 independent transductions are summarized in the bar graph with the MFI of the WT protein set at 100% and the MFI of all mutant perforins compared with WT. *Statistical significance compared with WT (2-tailed Student t test, P < .05). (B) Western blot of WT and mutant perforins expressed in RBL-1 cells. Three different monoclonal antibodies were used (P1-8, 2d4, and Pf-344). The precursor and mature isoforms seen in the WT perforin are marked by arrows. The lysates were generated under nonreducing conditions. (C) Metabolic study: Western blot of lysates from perforin-expressing, RBL-1 cells treated overnight with an alkalinizing agent (CMA), a cysteine protease inhibitor (E64), or no treatment (control; −).
Proteolytic maturation of mutant perforins
We then compared the maturation patterns by Western blot of PRF1-T435M and -Y438C to WT perforin expressed in RBL-1 cells. As shown in Figure 3B, PRF1-WT, -A91V, and -T435M were processed to both precursor and mature forms, suggesting that these mutant perforins fold properly and undergo proteolytic maturation during transport to the secretory granule. In contrast, no mature perforin was detected for PRF1-Y438C; only a diffusely stained band was seen with a slightly increased electrophoretic mobility compared with the precursor WT perforin.
To discern whether the mature band of PRF1-Y438C was present in an altered conformation inaccessible to the P1-8 antibody, we used 2 additional antibodies, neither of which identified a mature isoform (pf344, 2d4). In addition, we looked for additional evidence of perforin proteolytic maturation by metabolic studies. Mature band formation can be inhibited by treating cells overnight with CMA, an agent that alkalinizes the secretory granule, or with E64, a cysteine protease inhibitor to interrupt processing. As shown in Figure 3C, treatment with either CMA or E64 prevented the maturation of PRF1-WT, -A91V, and -T435M. In contrast, the PRF1-Y438C band(s) were not affected, inconsistent with mature perforin. As the PRF1-Y438C perforin form identified by Western blot had a slightly increased mobility compared with PRF1-WT precursor, we refer to this as a novel, variant perforin.
Lytic function of PRF1-T435M and -Y438C expressed in RBL-2H3 cells
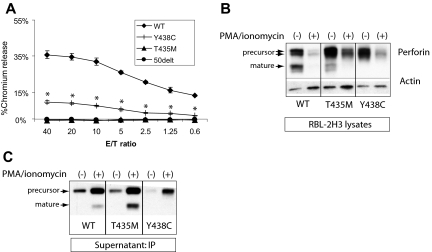
We tested the lytic function of PRF1-T435M and -Y438C by expressing the human perforin cDNA in RBL-2H3 cells (the RBL-1 do not degranulate effectively to IgE-triggered stimuli) as previously described for both PRF1-WT and PRF1-A91V.19 WT and mutant perforins were successfully introduced into RBL-2H3 cells and sorted for GFP expression. RBL-2H3 cells expressing WT perforin lysed RBC targets, as seen in a representative experiment in Figure 4A. Previously, we have shown that PRF1-A91V exhibited lytic function when expressed in RBL-2H3 cells, although reduced compared with WT.19,20 In the current study, we noted that PRF1-Y438C also displayed significantly reduced, but not absent, lytic function. Consistent with the prediction from molecular modeling, the human PRF1-T435M lacked lytic function, comparable with PRF1-50delT, a nonsense mutation associated with FHLH2.28 Our results are in direct contrast to the mouse equivalent mutation perforin-T434M, which showed intact rather than absent function.10
Figure 4.
Lytic function of perforin expressed in RBL-2H3 cells. (A) RBL-2H3 cells were transduced with perforin-expressing retrovirus, GFP sorted, and used in lytic assays against 2,4,6-trinitrobenzenesulphonic acid (TNBS)-treated RBC targets. To achieve cell conjugation, RBL-2H3 cells were coated with anti 2,4-dinitrophenol IgE and mixed with RBCs at various effector/target ratios (E/T). Percentage lysis was a measure of chromium release from targets. The negative control used RBL-2H3 cells expressing a severely truncated perforin cDNA, 50delT. Each sample is performed in quadruplicate to obtain the mean plus or minus SE. *Statistical significance of Y438C compared with WT or 50delT (2-tailed Student t test, P < .01). T435M was not different from 50delT at each E/T ratio. (B) Degranulation of perforin from transduced RBL-2H3 cells was achieved by treating with PMA/ionomycin for 2 hours. Shown are the Western blots from the cell lysates; perforin is detected by P1-8 and isoforms marked by arrows. Actin was probed to control for protein amount. WT indicates wild-type human perforin. (C) Detection of degranulated perforin from RBL-2H3 cells was achieved by PMA/ionomycin treatment for 1 hour in the presence of a capture antibody in the medium, δG9. The perforin/δG9 complex was precipitated with protein A agarose and then run on nondenaturing gels followed by Western blot with P1-8.
Secretion of WT and mutant perforins
Previous studies in NK cell and RBL cell lines expressing perforin suggested that mature perforin is localized to the dense secretory granule.6,19 The precursor form of WT perforin is assumed to be restricted to prelysosomal vesicles or confined to the trans Golgi network. Because human PRF1-T435M was processed to mature perforin, we suspected that it was appropriately packaged in the secretory granule and able to be degranulated. Therefore, we stimulated the perforin-expressing, RBL-2H3 cells with PMA and ionomycin (P/I) for 2 hours (Figure 4B) and analyzed the perforin remaining in the cells by Western blot compared with control cells treated with dimethyl sulfoxide. As expected, mature perforin disappeared with degranulation in RBL-2H3 cells expressing PRF1-WT and -T435M. Consistent with the lytic potential of PRF1-Y438C, the variant perforin also underwent significant degranulation, as did precursor perforin for PRF1-WT and -T435M. The degranulation of precursor forms was somewhat surprising, as previous studies in NK cell lines suggested that these forms may not be delivered to the dense secretory granule.6
Development of a novel capture assay to detect extracellular perforin isoforms
We suspected that the PRF1-Y438C might be cleaved to a mature form after secretion from the cell. Therefore, we developed a novel assay to capture perforin in the supernatant after degranulation. RBL-2H3 cells were stimulated with P/I in the presence of a capture antibody, δG9, in the medium. After P/I stimulation, the antibody/perforin complex in the supernatant was precipitated with protein A agarose and probed by Western blot with the P1-8, antiperforin antibody. The results are shown in Figure 4C.
In the absence of stimulation, precursor perforins from PRF1-WT, -T435M, and -Y438C were detected at low levels in the supernatant of RBL-2H3 cells, suggesting that low level degranulation may occur in the absence of trigger. Alternatively, precursor perforin may be redirected to a constitutive secretory pathway as previously described.23,29 No mature isoforms were detected under these conditions. On stimulation with P/I, mature isoforms of PRF1-WT and -T435M perforin were detectable in the supernatant, and the levels of precursor perforin in the supernatant increased as well, demonstrating the clear secretion of both mature and precursor isoforms. The intermediate form of PRF1-Y438C was detected as the only form after stimulation, arguing against extracellular conversion to a mature isoform as the mechanism for lytic function; however, additional forms of perforin may be present in the medium that are not recognized by δG9. Although 2 precursor forms are noted when perforin is expressed in RBL cells (Figure 3B), a single band was detected in the supernatant after P/I stimulation (Figure 4C), suggesting at least one form was retained whereas the other was secreted.
Expression of WT and mutant human perforins in murine CTL
The RBL cell system is ideal for studying the biochemical features of human perforin, but not optimal for measuring cytotoxic function of perforin mutations, because mast cells do not contain granzyme A or B.19,30–32 In contrast, perforin-deficient murine CTLs represent a physiologic cell model to assay perforin/granzyme-mediated cytotoxicity. A recent study demonstrated that human PRF1-WT and PRF1-A91V, expressed in CTLs from perforin knockout mice (Prf1−/−), restored cytotoxic function.20 Therefore, we introduced PRF1-WT, -T435M, and -Y438C into perforin-deficient murine CTLs by retroviral transduction. We crossed Prf1−/− mice with the T-cell receptor transgenic line, P14; the T-cell receptor expressed in these mice recognizes the gp33 peptide from lymphocytic choriomeningitis virus. Splenocytes from P14.Prf1−/− mice were stimulated with gp33 peptide 48 hours before retroviral transduction. Efficiency of transduction was high, with 70% to 80% of cells transduced, as assessed by GFP detection, in each of 3 independent transductions (Figure 5A). Perforin expression as measured by flow cytometry was comparable with that obtained in RBL-1 cells: PRF1-A91V, -T435M, and -Y438C were all detected at reduced levels.
Figure 5.
Expression of human perforin in murine CTLs. Murine splenocytes from T-cell receptor transgenic, perforin-deficient mice (P14.Prf1−/−) were transduced with bi-cistronic retroviruses containing GFP and perforin cDNA after stimulation with cognate antigen, as described in “Retroviral transduction.” (A) Flow cytometric detection of GFP and perforin after transduction. These cells were not sorted as transduction was uniform and efficient. (B) Western blot of WT and mutant perforins expressed in murine CTL. Three different monoclonal antibodies were used (P1-8, 2d4, and Pf-344). The precursor and mature isoforms seen in the WT perforin are marked by arrows. The lysates were generated under nonreducing conditions. (C) Detection of degranulated perforin from murine CTLs. Degranulation of splenocytes was achieved by coincubation of antigen-presenting, target cells (gp33-labeled EL4) with transduced splenocytes at an effector/target ratio of 5:1 for 2 hours. For a negative control, effectors were coincubated with EL4 target cells without gp33 peptide labeling. The degranulated perforin was “captured” and immunoprecipitated from the supernatant by δG9 antiperforin antibody as described in Figure 3.
Maturation of human perforin expressed in murine CTLs
Previous studies indicated that human perforin was functional when expressed in murine CTLs20 but did not address whether human perforin undergoes typical proteolytic maturation in murine CTLs. We analyzed human perforin expressed by murine CTLs by Western blots and confirmed that it is correctly processed in murine cells; precursor and mature isoforms are noted for WT perforin (Figure 5B). As previously seen in RBL cells, PRF1-A91V and -T435M also exhibit mature isoforms, whereas PRF1-Y438C was exceptional for a lack of mature isoforms and the presence of a novel, intermediate isoform. Consistent with studies in RBL cells, staining with 3 different antibodies showed the same pattern of perforin isoforms.
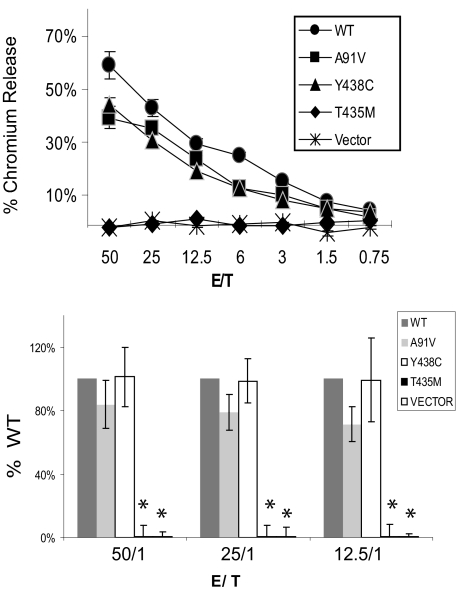
Cytotoxic function of mutant perforins expressed in murine CTL
The cytotoxic function of murine CTLs expressing human WT and mutant perforins was tested by killing of gp33-loaded, EL4 target cells. Three independent transductions and cytotoxic assays were performed. Because the retroviral transduction was highly efficient (80% GFP+ cells) and transduction was consistent between each vector, CTLs were not sorted before the assay. Results from 1 representative cytotoxic assay and the summary of 3 experiments are shown in Figure 6. PRF1-A91V had minimally decreased cytotoxic activity (not statistically significant) in our assays. Consistent with our predictions from molecular modeling and results in RBL-2H3 cells, PRF1-T435M had no cytotoxic capacity. Despite an absence of mature band in Western blots, PRF1-Y438C exhibited no impairment of cytotoxic function.
Figure 6.
Cytotoxic function of perforin expressed in murine P14/Prf1−/− splenocytes. Murine splenocytes expressing human perforins were coincubated with gp33-loaded EL4 target cells in a standard chromium release assay. Cytotoxicity was measured by the percentage of chromium released from targets. The negative control cells were transduced with vector alone (no perforin). WT indicates wild-type human perforin. The top panel is a single representative assay. Each E/T coincubation is performed in quadruplicate to generate a mean plus or minus SE. The bottom panel is the data summarized from 3 independent retroviral transductions followed by cytotoxicity assays, graphed as a percentage of WT function for each assay. Only the T435M and vector alone were different from WT. *Statistical significance (2-tailed Student t test, P < .01).
Capture of extracellular perforin secreted from murine CTLs
To determine whether WT and mutant perforins were effectively released from secretory granules of murine splenocytes during effector/target conjugate formation, we modified our “capture” assay to detect perforin in the medium during physiologic degranulation rather than P/I stimulation. Degranulation of perforin from murine CTL was achieved by a 2-hour coincubation with gp33-loaded EL4 targets in the presence of the δG9 antibody. Perforin was readily detected in the media from murine splenocytes secreting PRF1-WT, -T435M, and -Y438C (Figure 5C). As noted previously in RBL-2H3 cells, no mature isoforms of PRF1-Y438C were noted in the media after degranulation, demonstrating that the PRF1-Y438C variant was either not converted to mature isoforms on degranulation (Figure 5C) or the mature perforin was unstable in the media, preventing detection. Interestingly, WT precursor perforin was also readily detected in the supernatant after stimulation of murine CTLs (Figure 5C) or RBL-2H3 (Figure 4C). Experiments are ongoing to assess whether the precursor perforin secretion is via the secretory granule or via a secretory pathway separate from the secretory granule.23
Calcium-dependent binding of perforin to liposomes
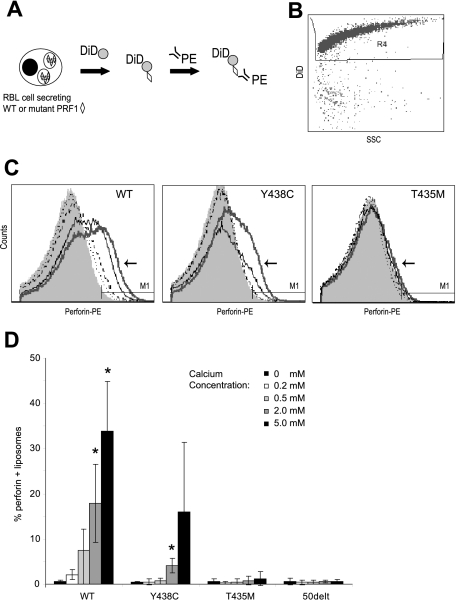
Highly purified perforin has been shown to bind to liposomes and/or target cells in a Ca-dependent manner.33,34 To test our hypothesis that the T435M mutation prevented Ca binding, we designed a novel “readout” of calcium-dependent lipid attachment, by measuring the binding of wild-type and mutant perforins to liposomes. We achieved this by stimulating perforin secretion from RBL-2H3 cells with P/I in medium containing liposomes and increasing Ca concentrations (0-5 mM CaCl2). After 20 minutes of P/I stimulation, PE-conjugated δG9 antiperforin antibody was added to the supernatant for 60 minutes at 37°C to bind the liposome/perforin complex. The RBL-2H3 cells were removed by centrifugation, and we analyzed the percentage of liposomes with detectable perforin bound by flow cytometry (Figure 7).
Figure 7.
Ca-dependent binding of perforin to liposomes. (A) Binding assay: RBL-2H3 cells expressing WT or mutant perforins (◇) were stimulated by P/I in the presence of DiD-labeled liposomes ( ) in a minimum buffer with defined Ca concentrations. Twenty minutes into the stimulation, PE-conjugated δG9 antiperforin antibody (linear depiction of antibody) was added to the medium. Sixty minutes later, the RBL-2H3 cells were pelleted and the liposome-containing supernatant analyzed by flow cytometry. (B) Scatter plots of dye-labeled (DiD) liposomes vs side scatter. To analyze perforin staining, we gated on these brightly labeled liposomes. (C) Histograms showing perforin staining in the presence of buffers of increasing Ca concentration. In each figure, the histogram for the 5 mM of Ca buffer is indicated by an arrow and thick gray line. The thin black line represents 2 mM of Ca; dashed black line, 0.5 mM of Ca; dotted black line, 0.2 mM of Ca. (D) Summary of 3 experiments: PRF1-WT and -Y438C, but not PRF1-T435M, show a dose-dependent signal with increasing Ca concentrations. *Statistical significance compared with 0 mM Ca (2-tailed Student t test, P < .01). PRF1-50delt (nonsense mutation) is shown as a negative control.
) in a minimum buffer with defined Ca concentrations. Twenty minutes into the stimulation, PE-conjugated δG9 antiperforin antibody (linear depiction of antibody) was added to the medium. Sixty minutes later, the RBL-2H3 cells were pelleted and the liposome-containing supernatant analyzed by flow cytometry. (B) Scatter plots of dye-labeled (DiD) liposomes vs side scatter. To analyze perforin staining, we gated on these brightly labeled liposomes. (C) Histograms showing perforin staining in the presence of buffers of increasing Ca concentration. In each figure, the histogram for the 5 mM of Ca buffer is indicated by an arrow and thick gray line. The thin black line represents 2 mM of Ca; dashed black line, 0.5 mM of Ca; dotted black line, 0.2 mM of Ca. (D) Summary of 3 experiments: PRF1-WT and -Y438C, but not PRF1-T435M, show a dose-dependent signal with increasing Ca concentrations. *Statistical significance compared with 0 mM Ca (2-tailed Student t test, P < .01). PRF1-50delt (nonsense mutation) is shown as a negative control.
Wild-type perforin secreted from RBL-2H3 cells was detected on liposomes in a Ca-dose–dependent manner (Figure 7B). The increase in perforin detection with a rise in Ca may reflect increased amounts of perforin binding to the liposome or an increased detection of perforin, a result of a conformational change as shown by Metkar et al.33 The PRF1-Y438C was also detectable on liposomes after secretion from RBL-2H3 cells, although the overall staining was lower than PRF1-WT, except at the highest concentration of Ca. This may be a result of reduced affinity for binding of either Ca or δG9 or reduced perforin secreted from the RBL cells. In contrast to the functional perforins, PRF1-T435M was not detected on liposomes at any Ca concentration. These studies show clearly the pathogenicity of the T435M mutation, predicted to interrupt Ca binding by molecular modeling of the C2 domain. In addition, we have demonstrated for the first time that the absence of a mature form of variant perforin does not preclude Ca binding to the C2 domain.
Discussion
In our previous studies of mutant perforins associated with FHLH2, we expressed 21 mutant perforins in RBL cells and categorized them by the presence of precursor and/or mature isoforms detectable by Western blot: class 1, with evidence of maturation; class 2, with absence of maturation; and class 3, in which no native forms of perforin were detected.19 The current work validates and extends our initial observations, demonstrating comparable perforin expression and maturation in murine CTLs. We have introduced a novel technique to assess the secretion of perforin from either RBL-2H3 or murine CTLs, and we were surprised to find that both precursor and mature perforins are “captured” in the medium from stimulated cells. In addition, we assessed the ability of mutant perforin secreted from RBL cells to bind to liposome targets, allowing us to test the Ca-dependent, lipid-binding capacity of secreted mutant perforins for the first time. Specifically, our studies illustrate the critical role of lipid binding by the Ca-bound C2 domain for maintaining cytotoxic function of human perforin in CTLs.
Perforin is the only member of pore-forming proteins to contain a Ca-binding C2 domain required for binding the lipid membrane,35 and its sequence is highly conserved across mammalian species. Based on alignments, we have determined that human perforin shares the highest sequence similarity with C2 domains that contain a type I fold. This model allowed us to correctly predict that folding of the T435M substitution would be tolerated only in the Ca-free configuration because of constraints of the Ca-binding pocket in the human perforin. The differences in functional outcomes between the human and murine equivalent of this mutation suggest that there may be subtle differences between the Ca-binding loops between species.10 Future structural studies are required to test the type I model and to explain the differences between the human and murine Ca-binding pocket.
We present clear evidence for the pathogenicity of the previously disputed T435M mutation in HLH. McCormick et al described one family with a child diagnosed with early-onset HLH, who carried both T435M on a single allele and a polymorphism in the CD45 gene of unclear clinical significance.18 A second patient who presented with lymphoma and hemophagocytic syndrome also carried the T435M mutation; however, this patient was heterozygous for another variant in C2, T450M, complicating the interpretation of the pathogenicity of each allele.14 In the current study, in vitro cytotoxicity and lipid binding experiments, together with predictions based on a revisited molecular model of C2 domain, strongly implicate T435M as pathogenic for FHLH2. Importantly, each healthy family member that carries this allele also expresses a potentially functional allele, Y438C or A91V.
Y438C is a newly described amino acid substitution with impaired maturation when expressed in RBL or mouse CTL cells, as only a variant, nonmature form is detected. Surprisingly, PRF1-Y438C displayed both lytic activity toward RBCs and cytotoxic activity toward nucleated target cells. Our clinical data support this finding: the NK activity of the mother, a carrier for both Y438C and T435M, was intact. Assuming T435M is nonfunctional, according to our results, Y438C may be responsible, even in mono-allelic state, for the NK activity seen. However, we cannot say with certainty that the Y438C variant offers no risk toward developing FHLH2. There may be subtle defects in function of this unusual variant, not revealed by the standard 4-hour chromium release assay.
The functionality of PRF1-Y438C despite impaired maturation suggests that the cytotoxic function of perforin may be associated with either precursor or mature form, contradicting the previously held view that only mature perforin is cytotoxic. The importance of precursor perforin function in target cell killing in healthy persons may be limited, as the degranulation of mature perforin and granzymes via the secretory granule pathway leads to rapid cell death of target cells. In our previous studies, we identified 7 FHLH2-associated mutations (in the MACPF domain) that led to absent proteolytic maturation when perforin was expressed in RBL cells (class 2).19 We hypothesize that cytotoxic function may be dependent on the secretion of precursor perforin from lymphocytes expressing these mutant perforins, and studies to define the secretory and cytotoxic capacity of these unique variants are ongoing in our laboratory.
In conclusion, analysis of naturally occurring perforin mutations provides a unique opportunity to understand the pathogenesis of FHLH2 and to gain understanding in the basic cellular mechanisms of perforin-mediated cytotoxicity.
Supplementary Material
Acknowledgments
The authors thank Dr Alexandra Filipovich and Dr Eduardo Fernandez Cruz who provided valuable mentorship and unique opportunities for translational research, Catherine Terrell for technical assistance, Dr Gillian Griffiths for the gift of the 2d4 antibody, and Dr Christopher Froelich for the gift of the pf344 antibody and insight into appropriate buffers to study perforin binding.
R.U.M. was supported by a clinical immunology fellowship from Fundación de Investigación Biomédica, Hospital General Universitario Gregorio Marañón, Universidad Complutense de Madrid. K.A.R. was supported by grants from the National Institutes of Health: K12 Child Health Research Career Development Award, American Academy of Allergy, Asthma and Immunology/Altana, and by the Doris Duke Charitable Foundation. A.B.H. was supported by funds from the State of Ohio Eminent Scholar Program. M.B.J. was supported by National Institutes of Health (grant RO1-HL091769) and a grant from United States Immunodeficiency Network.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: R.U.M. designed and executed immunologic, genetic, and experimental studies, reviewed the literature, and drafted/revised the manuscript; J.G. performed clinical and immunophenotyping of the family, coordinated all work performed in Spain, and revised the manuscript; E.C. performed clinical characterization of the patients and revised the manuscript; C.R.-S. performed PRF1 sequencing and revision of the manuscript; V.L. and B.O. designed and executed experimental studies and revised the manuscript; A.B.H. executed the computer modeling studies, reviewed the existing literature related to biophysical modeling, and drafted/revised the manuscript; J.S. helped design and construct experimental vectors and revised the manuscript; M.B.J. provided all murine splenocytes, designed murine splenocyte transduction and cytotoxic studies, produced liposomes, helped design liposome-based studies, and revised the manuscript; and K.A.R. designed and executed experimental studies, reviewed the existing literature, and drafted/revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kimberly A. Risma, Division of Allergy/Immunology, Cincinnati Children's Hospital Medical Center,3333 Burnet Avenue, Cincinnati, OH 45229-2000; e-mail: Kimberly.risma@cchmc.org.
References
- 1.Trambas CM, Griffiths GM. Delivering the kiss of death. Nat Immunol. 2003;4:399–403. doi: 10.1038/ni0503-399. [DOI] [PubMed] [Google Scholar]
- 2.Stepp SE, Dufourcq-Lagelouse R, Le Deist F, et al. Perforin gene defects in familial hemophagocytic lymphohistiocytosis. Science. 1999;286:1957–1959. [PubMed] [Google Scholar]
- 3.Jordan MB, Hildeman D, Kappler J, Marrack P. An animal model of hemophagocytic lymphohistiocytosis (HLH): CD8+ T cells and interferon gamma are essential for the disorder. Blood. 2004;104:735–743. doi: 10.1182/blood-2003-10-3413. [DOI] [PubMed] [Google Scholar]
- 4.Podack ER, Konigsberg PJ. Cytolytic T cell granules: isolation, structural, biochemical, and functional characterization. J Exp Med. 1984;160:695–710. doi: 10.1084/jem.160.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henkart PA, Millard PJ, Reynolds CW, Henkart MP. Cytolytic activity of purified cytoplasmic granules from cytotoxic rat large granular lymphocyte tumors. J Exp Med. 1984;160:75–93. doi: 10.1084/jem.160.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uellner R, Zvelebil MJ, Hopkins J, et al. Perforin is activated by a proteolytic cleavage during biosynthesis which reveals a phospholipid-binding C2 domain. EMBO J. 1997;16:7287–7296. doi: 10.1093/emboj/16.24.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rizo J, Sudhof TC. C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem. 1998;273:15879–15882. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- 8.Nalefski EA, Falke JJ. The C2 domain calcium-binding motif: structural and functional diversity. Protein Sci. 1996;5:2375–2390. doi: 10.1002/pro.5560051201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponting CP, Parker PJ. Extending the C2 domain family: C2s in PKCs delta, epsilon, eta, theta, phospholipases, GAPs, and perforin. Protein Sci. 1996;5:162–166. doi: 10.1002/pro.5560050120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voskoboinik I, Thia MC, Fletcher J, et al. Calcium-dependent plasma membrane binding and cell lysis by perforin are mediated through its C2 domain: a critical role for aspartate residues 429, 435, 483, and 485 but not 491. J Biol Chem. 2005;280:8426–8434. doi: 10.1074/jbc.M413303200. [DOI] [PubMed] [Google Scholar]
- 11.Ishii E, Ueda I, Shirakawa R, et al. Genetic subtypes of familial hemophagocytic lymphohistiocytosis: correlations with clinical features and cytotoxic T lymphocyte/natural killer cell functions. Blood. 2005;105:3442–3448. doi: 10.1182/blood-2004-08-3296. [DOI] [PubMed] [Google Scholar]
- 12.Feldmann J, Menasche G, Callebaut I, et al. Severe and progressive encephalitis as a presenting manifestation of a novel missense perforin mutation and impaired cytolytic activity. Blood. 2005;105:2658–2663. doi: 10.1182/blood-2004-09-3590. [DOI] [PubMed] [Google Scholar]
- 13.Molleran Lee S, Villanueva J, Sumegi J, et al. Characterisation of diverse PRF1 mutations leading to decreased natural killer cell activity in North American families with haemophagocytic lymphohistiocytosis. J Med Genet. 2004;41:137–144. doi: 10.1136/jmg.2003.011528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clementi R, Locatelli F, Dupre L, et al. A proportion of patients with lymphoma may harbor mutations of the perforin gene. Blood. 2005;105:4424–4428. doi: 10.1182/blood-2004-04-1477. [DOI] [PubMed] [Google Scholar]
- 15.Katano H, Ali MA, Patera AC, et al. Chronic active Epstein-Barr virus infection associated with mutations in perforin that impair its maturation. Blood. 2004;103:1244–1252. doi: 10.1182/blood-2003-06-2171. [DOI] [PubMed] [Google Scholar]
- 16.Ueda I, Kurokawa Y, Koike K, et al. Late-onset cases of familial hemophagocytic lymphohistiocytosis with missense perforin gene mutations. Am J Hematol. 2007;82:427–432. doi: 10.1002/ajh.20878. [DOI] [PubMed] [Google Scholar]
- 17.Feldmann J, Le Deist F, Ouachee-Chardin M, et al. Functional consequences of perforin gene mutations in 22 patients with familial haemophagocytic lymphohistiocytosis. Br J Haematol. 2002;117:965–972. doi: 10.1046/j.1365-2141.2002.03534.x. [DOI] [PubMed] [Google Scholar]
- 18.McCormick J, Flower DR, Strobel S, Wallace DL, Beverley PC, Tchilian EZ. Novel perforin mutation in a patient with hemophagocytic lymphohistiocytosis and CD45 abnormal splicing. Am J Med Genet A. 2003;117:255–260. doi: 10.1002/ajmg.a.10010. [DOI] [PubMed] [Google Scholar]
- 19.Risma KA, Frayer RW, Filipovich AH, Sumegi J. Aberrant maturation of mutant perforin underlies the clinical diversity of hemophagocytic lymphohistiocytosis. J Clin Invest. 2006;116:182–192. doi: 10.1172/JCI26217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voskoboinik I, Sutton VR, Ciccone A, et al. Perforin activity and immune homeostasis: the common A91V polymorphism in perforin results in both presynaptic and postsynaptic defects in function. Blood. 2007;110:1184–1190. doi: 10.1182/blood-2007-02-072850. [DOI] [PubMed] [Google Scholar]
- 21.Zur Stadt U, Beutel K, Weber B, Kabisch H, Schneppenheim R, Janka G. A91V is a polymorphism in the perforin gene not causative of an FHLH phenotype. Blood. 2004;104:1909. doi: 10.1182/blood-2004-02-0733. author reply 1910. [DOI] [PubMed] [Google Scholar]
- 22.Egeler RM, Shapiro R, Loechelt B, Filipovich A. Characteristic immune abnormalities in hemophagocytic lymphohistiocytosis. J Pediatr Hematol Oncol. 1996;18:340–345. doi: 10.1097/00043426-199611000-00002. [DOI] [PubMed] [Google Scholar]
- 23.Isaaz S, Baetz K, Olsen K, Podack E, Griffiths GM. Serial killing by cytotoxic T lymphocytes: T cell receptor triggers degranulation, re-filling of the lytic granules and secretion of lytic proteins via a non-granule pathway. Eur J Immunol. 1995;25:1071–1079. doi: 10.1002/eji.1830250432. [DOI] [PubMed] [Google Scholar]
- 24.Henter JI, Horne A, Arico M, et al. HLH-2004: Diagnostic and therapeutic guidelines for hemophagocytic lymphohistiocytosis. Pediatr Blood Cancer. 2007;48:124–131. doi: 10.1002/pbc.21039. [DOI] [PubMed] [Google Scholar]
- 25.Trizzino A, zur Stadt U, Ueda I, et al. Genotype-phenotype study of familial haemophagocytic lymphohistiocytosis due to perforin mutations. J Med Genet. 2008;45:15–21. doi: 10.1136/jmg.2007.052670. [DOI] [PubMed] [Google Scholar]
- 26.Santoro A, Cannella S, Trizzino A, Lo Nigro L, Corsello G, Arico M. A single amino acid change A91V in perforin: a novel, frequent predisposing factor to childhood acute lymphoblastic leukemia? Haematologica. 2005;90:697–698. [PubMed] [Google Scholar]
- 27.Zhang K, Johnson JA, Biroschak J, et al. Familial haemophagocytic lymphohistiocytosis in patients who are heterozygous for the A91V perforin variation is often associated with other genetic defects. Int J Immunogenet. 2007;34:231–233. doi: 10.1111/j.1744-313X.2007.00679.x. [DOI] [PubMed] [Google Scholar]
- 28.Lee SM, Sumegi J, Villanueva J, et al. Patients of African ancestry with hemophagocytic lymphohistiocytosis share a common haplotype of PRF1 with a 50delT mutation. J Pediatr. 2006;149:134–137. doi: 10.1016/j.jpeds.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Griffiths GM, Argon Y. Structure and biogenesis of lytic granules. Curr Top Microbiol Immunol. 1995;198:39–58. doi: 10.1007/978-3-642-79414-8_3. [DOI] [PubMed] [Google Scholar]
- 30.Shiver JW, Su L, Henkart PA. Cytotoxicity with target DNA breakdown by rat basophilic leukemia cells expressing both cytolysin and granzyme A. Cell. 1992;71:315–322. doi: 10.1016/0092-8674(92)90359-k. [DOI] [PubMed] [Google Scholar]
- 31.Shiver JW, Henkart PA. A noncytotoxic mast cell tumor line exhibits potent IgE-dependent cytotoxicity after transfection with the cytolysin/perforin gene. Cell. 1991;64:1175–1181. doi: 10.1016/0092-8674(91)90272-z. [DOI] [PubMed] [Google Scholar]
- 32.Voskoboinik I, Thia MC, De Bono A, et al. The functional basis for hemophagocytic lymphohistiocytosis in a patient with coinherited missense mutations in the perforin (PFN1) gene. J Exp Med. 2004;200:811–816. doi: 10.1084/jem.20040776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Metkar SS, Wang B, Froelich CJ. Detection of functional cell surface perforin by flow cytometry. J Immunol Methods. 2005;299:117–127. doi: 10.1016/j.jim.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Blumenthal R, Millard PJ, Henkart MP, Reynolds CW, Henkart PA. Liposomes as targets for granule cytolysin from cytotoxic large granular lymphocyte tumors. Proc Natl Acad Sci U S A. 1984;81:5551–5555. doi: 10.1073/pnas.81.17.5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosado CJ, Kondos S, Bull TE, et al. The MACPF/CDC family of pore-forming toxins. Cell Microbiol. 2008;10:1765–1774. doi: 10.1111/j.1462-5822.2008.01191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.