Abstract
Haemophilus parasuis is the agent responsible for causing Glässer's disease, but little is known about the pathogenic determinants of this major pig disease. Here we describe, for the pathogenic strain Nagasaki, the molecular characterization of 13 trimeric autotransporters as assessed by the presence of YadA C-terminal translocator domains which were classified into three groups. All passenger domains possess motifs and repeats characteristic of adhesins, hemagglutinins, and invasins with various centrally located copies of collagen-like repeats. This domain architecture is shared with two trimeric autotransporter proteins of H. somnus 129Pt. Genomic comparison by microarray hybridization demonstrated homologies among H. parasuis virulent strains and high divergence with respect to nonvirulent strains. Therefore, these genes were named vtaA (virulence-associated trimeric autotransporters). The sequencing of 17 homologous vtaA genes of different invasive strains highlighted an extensive mosaic structure. Based also on the presence of DNA uptake signal sequences within the vtaA genes, we propose a mechanism of evolution by which gene duplication and the accumulation of mutations and recombinations, plus the lateral gene transfer of the passenger domain, led to the diversity of this multigene family. This study provides insights to help understand the tissue colonization and invasiveness characteristic of H. parasuis pathogenic strains.
Adhesion to host tissues is an important step for bacterial colonization and survival (23, 39). Structures present at the surface of bacteria called adhesins mediate interactions with receptors of host cells. Adhesins can be very different in nature; among them a family of trimeric autotransporters, also termed AT-2 (21), are present in gram-negative bacteria (12). AT-2 adhesins have the capacity to bind eukaryotic cells (10, 28) as well as extracellular matrix proteins (45). These proteins are made up of an N-terminal signal peptide, a passenger domain, and a C-terminal translocator domain responsible for the pore-forming capacity in the outer membrane (12). The passenger domain often contains multiple repeats (Hep_Hag) and motifs (HIM) characteristic of adhesins and hemagglutinins (4). Although there is considerable amino acid diversity between the translocator domains of different bacterial species, they form a β-barrel through which the passenger domain transits (22, 55). Their immunogenicity makes them good candidates for vaccine development (9, 11). Trimeric autotransporters have been fully or partially characterized in terms of structure, function, and immunological properties for Haemophilus influenzae, Moraxella catarrhalis, Haemophilus ducreyi, Neisseria meningitidis, or Yersinia spp. However, not all studies of pathogenic bacteria benefit from the wealth of information generated for important human pathogens. This is the case for Haemophilus parasuis, a gram-negative bacillus classified in the Pasteurellaceae family, where the molecules which mediate adhesion to host tissues or virulence factors are largely unknown.
H. parasuis is commonly found in the upper respiratory tract of healthy conventional pigs. Some strains can migrate into the lungs, causing pneumonia (34), and disseminate to produce a severe systemic disease, characterized by fibrinous polyserositis, arthritis, and meningitis, known as Glässer's disease (52). Fifteen serovars have been described so far, but many strains cannot be typed with the current sera (25). Although there is not a strict correlation between the expression of a given serovar and the degree of pathogenicity of H. parasuis strains, it is commonly stated that bacteria exhibiting serovar 5 are highly virulent, while strains of serovar 3 are not virulent (41). Another striking feature of this bacterium is its genetic variability. When a multilocus typing method was applied to 120 field and 11 reference strains, 109 sequence types were found. Interestingly, two divergent branches were observed, one of them including most of the virulent strains isolated from systemic sites of diseased animals (37). Although molecules which mediate adhesion to host tissues or virulence factors are largely unknown in H. parasuis, a recent study has shown that H. parasuis strains of serotypes 5 and 4 (often associated with virulence) preferentially bind and invade porcine brain endothelial cells in vitro. Since the invasion was not abolished by proteinase K treatment, it was concluded that the putative invasin was not likely to be a protein (54). However, many of the described bacterial adhesins are proteins (39).
In this report, we describe 13 paralog genes of the highly pathogenic strain H. parasuis Nagasaki and 17 homologues from different invasive strains coding for VtaA proteins, which, unusually for AT-2, contained collagen triple helix repeats. The passenger domains were relatively conserved with vtaA homologues from pathogenic strains but highly divergent with those of nonpathogenic strains. Furthermore, this multigene family has likely evolved by the duplication and lateral gene transfer of at least modules of the passenger domain.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Haemophilus parasuis strains (Table 1) were cultured on chocolate agar plates (BioMérieux, Inc.) at 37°C in a 5% CO2 atmosphere until they reached confluence. HP and F9 strains were isolated from necropsied pigs in the diagnostic service of HIPRA S.A. Laboratories (Spain) and the veterinary school of Autonomous University of Barcelona, Spain, respectively.
TABLE 1.
Haemophilus parasuis strains used for the genomic comparison of the AT-2 genes
| Strain identification | Site of isolation | Pathology | Glässer | Serovar | Source or reference |
|---|---|---|---|---|---|
| Nagasaki | Brain | Septicemia | + | 5 | Reference strain (33) |
| SW114 | Nose | Without lesions | NDa | 3 | Reference strain (33) |
| F9 | Nose | Without clinical signs | ND | NTb | Veterinary School, Autonomous University of Barcelona |
| D74 | ND | Without lesions | ND | 9 | Reference strain (25) |
| HP-3123 | Nose | Enteritis by Escherichia coli | − | NT | HIPRA S.A. Laboratories |
| HP-1205 | Pericardium | Pericarditis and pleuritis | + | 11 | HIPRA S.A. Laboratories |
| HP-1302 | Brain | Polyserositis | + | 1 | HIPRA S.A. Laboratories |
| HP-1319 | Brain | Polyserositis | + | NT | HIPRA S.A. Laboratories |
| HP-2163 | Joint | Pneumonia and arthritis | + | NT | HIPRA S.A. Laboratories |
| HP-2269 | Joint | Pneumonia and arthritis | + | NT | HIPRA S.A. Laboratories |
| HP-33 | Lung | Pneumonia | ND | 1 | HIPRA S.A. Laboratories |
ND, not documented.
NT, not typeable; +, positive; −, negative.
Genomic DNA purification, fragmentation, and construction of genomic libraries.
Bacteria were resuspended in phosphate-buffered saline (pH 7.3), recovered by centrifugation, and washed twice in phosphate-buffered saline. Genomic DNA from H. parasuis Nagasaki was extracted using commercial kits (Qiagen) and partially digested with Sau3AI and RsaI (New England Biolabs) or sonicated. Afterwards, the sonicated DNA fragments were end repaired with T4 DNA polymerase (New England Biolabs). DNA fragments ranging from 450 to 1,500 bp were separated in agarose gels and purified. The genomic libraries were constructed following standard methods (47). DNA fragments were inserted into the BamHI or SmaI cloning sites of the dephosphorylated vector pUC19 using the Quick ligation kit (New England Biolabs), and competent Escherichia coli cells (Invitrogen) were transformed. The isolation of recombinant plasmid DNA was carried out using the NucleoSpin 96 flash kit (Macherey-Nagel).
Sequencing, sequence assembly, and gene annotation.
Plasmid inserts were sequenced using BigDye Terminator v3.1 chemistry (Applied Biosystems) with the M13 universal primers. Sequence assembly into contigs was accomplished with the programs PHRED (15, 16), PHRAP, and Consed (19). Homology search and open reading frame determinations were performed using the programs BlastX, BlastN, and BlastP (1) in the GenBank nonredundant database (http://www.ncbi.nlm.nih.gov/BLAST/) and GeneMark (http://opal.biology.gatech.edu/GeneMark/heuristic_hmm2.cgi), respectively, using a heuristic approach (6). The domain architecture of the translated genes was studied by searching the Pfam database (4) at the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/Software/Pfam/).
DNA microarray construction, hybridization, and analysis.
Eighty-five recombinant plasmids, containing nonoverlapping or partially overlapping coding sequences of the vtaA genes, were selected from the assembled contigs. A fragment of the 16S rRNA gene, which was 98% identical to all H. parasuis 16S rRNA genes, was used as a positive control as were 60 DNA fragments corresponding to single-copy genes well conserved in bacteria (ribosomal proteins, RNA and DNA polymerases, metabolical enzymes, etc.). A 750-bp polynucleotide coding for a mammal prion protein was chosen as the negative control. Inserts of the selected clones were amplified by PCR, using universal M13 sequencing primers, purified, and spotted in triplicate onto UltraGAPS slides (Corning).
One to 2 μg of genomic DNA was randomly labeled (Bioprime DNA labeling system; Invitrogen) using a modified procedure (30). DNA from the Nagasaki strain was labeled with dCTP-Cy3 for use as the hybridization control strain, and DNAs from the other strains, Nagasaki included, were labeled with dCTP-Cy5 (Amersham). Equal amounts of labeled genomic DNA from the reference strain (Nagasaki) and one of the test strains were then pooled and placed onto prehybridized slides for hybridization performed in the ArrayBooster AB410 (Advalitix). After washing, the slides were scanned with a 4000 microarray scanner (Perkin Elmer). The fluorescence intensity (FI) signals from Cy5 and Cy3 were analyzed with the program QuantArray (GSI Lumonics). After subtracting the local background, channel intensity normalization was performed with signals obtained from the 16S rRNA gene fragment. The signal intensities for triplicate spots with threefold increases compared to the negative controls (prion gene) were considered positives. Ratios (Cy5 test strain/Cy3 reference strain or Cy5 reference strain/Cy3 reference strain) were calculated for each spot and averaged. Ratios below 0.3 or ranging between 0.3 and 0.8 indicated high divergence or the absence of or moderate variability of the corresponding DNA fragments in the test strain, respectively, while ratios above 1.5 represented genomic regions duplicated in the test strain. Identity between the strains was indicated by ratios ranging between 0.8 and 1.5 (42, 46). Gene divergence was also estimated using the following formula: (ΣFI single-copy gene fragments/number of single-copy gene fragments)/(ΣFI vtaA passenger domain/number of passenger domain fragments).
DNA amplification of translocator YadA domains.
For PCRs, 15 ng of genomic DNA and 20 pmol of each forward and reverse primer (see Fig. S1B in the supplemental material) were used. Amplification cycles were preceded by 3 min at 94°C and consisted of 20 cycles of 45 s of denaturing at 94°C, 45 s of annealing at 68°C, and 1 min and 45 s of extension at 72°C, with a final 5-min extension at 72°C. The results were analyzed by electrophoresis on a 2% agarose gel and Sybr gold staining.
DNA amplification of vtaA genes.
Amplifications were carried out using AccuPrime Taq DNA polymerase high fidelity (Invitrogen), with 150 ng of genomic DNA, 40 pmol of the forward primer and 20 pmol of the corresponding reverse primer (see Fig. S1A and B in the supplemental material) with a first denaturation step of 2 min at 94°C, followed by 30 cycles at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 68°C for 15 min. All amplifications were completed with a further extension period of 15 min at 68°C, followed by an overnight incubation at 37°C. The PCR products were analyzed by electrophoresis on a 0.8% agarose gel and stained with Sybr gold (Invitrogen).
Cloning of the vtaA genes.
Long PCR products were purified from gels using the Nucleospin Extract II system (Macherey Nagel) and cloned into the pCR-Blunt II-TOPO or pCR-XL-TOPO cloning vector (Invitrogen). After transformation into E. coli, the positive recombinant plasmids were sequenced with a combination of specific primers designed every 400 to 500 bp in the different genes.
Sequence analysis, phylogeny, and predictions.
Nucleotide and amino acid sequences were aligned using ClustalW (51). Comparisons of motifs, repeats, and domains were done using local BLAST (1). SignalP (5) and PSORTb (18) were used to search for signal peptides. Structure homology modeling was performed with SWISS-MODEL (3). A selection pressure analysis was done using the SNAP (27) and Data Monkey (40) Web servers. Pasteurellaceae uptake signal sequence (USS) (37, 43) detection was done using BioEdit (20), as were Kyte and Doolittle scale mean hydrophobicity profiles. All phylogenetic analyses were bootstrap consensus trees constructed by neighbor joining (Kimura 2 parameter distance); maximum parsimony and minimum evolution (maximum composite likelihood distance) were performed using MEGA4.0 (50). Finally, recombination was detected using the RDP, GENECONV, Chimaera, MaxChi, BootScan, SiScan, and 3Seq algorithms included in the RDP2 package (31).
Accession numbers.
Sequences reported in this work have been registered in the GenBank database under the accession numbers EU678322 to EU678351.
RESULTS AND DISCUSSION
Identification and structural characterization of trimeric autotransporters of H. parasuis.
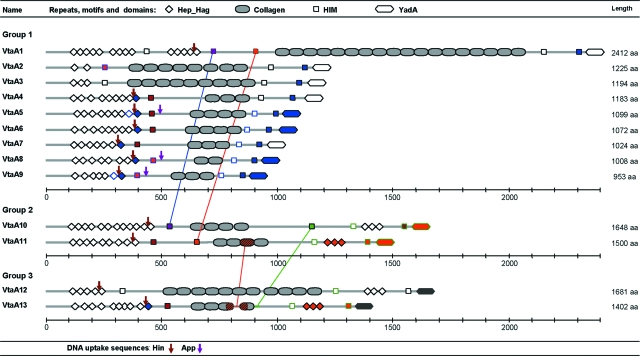
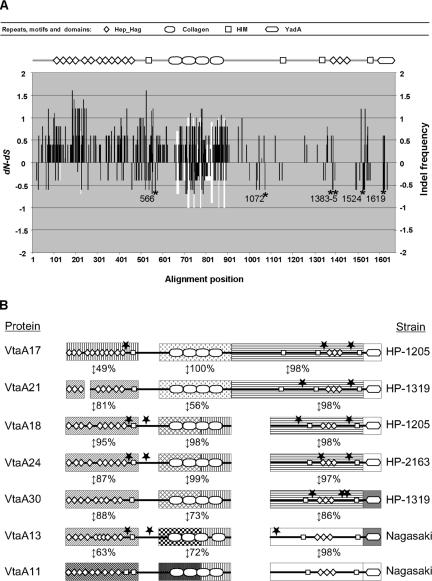
H. parasuis strain Nagasaki is the reference strain of the highly virulent serovar 5 (41). Experimental reproduction of the disease has been successfully accomplished with this strain in several laboratories (2, 35). For these reasons, this strain was selected for genome sequencing. The genome of H. parasuis (Nagasaki) was estimated to have a length of 2.4 Mbp by pulsed-field gel electrophoresis (data not shown). Ninety-nine percent of the genome was sequenced using a shotgun strategy, where 33,935 reads were assembled into 240 contigs. Annotation was performed, and 13 in silico-translated genes had significant homology (an E value of <e−05) with bacterial proteins belonging to the functional family of autotransporters, adhesins, and hemagglutinins now named trimeric autotransporters or the AT-2 family (12, 21). The architecture of these proteins was determined by comparing them to the Pfam database. All of them were made up of multiple Hep_Hag repeats (accession number PF05658), HIM motifs (PF05662), and a unique C-terminal YadA translocator domain (PF03895). These repeats, motifs, and domains are characteristic of adhesins and hemagglutinins that can be found in many bacterial pathogens. All 13 proteins had, in their central part, a variable number (ranging from 2 to 18) of collagen triple helix repeats (PF01391) inserted between the HIM motifs (Fig. 1). From these results, it was clear that the genes and gene products described above formed a family which we called vtaA (virulence-associated trimeric autotransporters) or VtaA, respectively, and individual genes were termed vtaA1 to vtaA13. This multigene family was not clustered but rather spread throughout the genome since none of the contigs contained more than one vtaA gene (data not shown). Multiple alignments of the 5′ end of all the genes (see Fig. S1 in the supplemental material) showed a good conservation of the first 207 nucleotides, suggesting a common function for this region, but no signal peptide was found using bioinformatics. However, a gene fusion constructed with a genomic fragment of 497 bp, which included 116 nucleotides upstream of the vtaA4 initiation codon, followed by 378 bp of coding nucleotides and a promoterless and leaderless phoA (44), demonstrated that an active promoter was present and that a signal peptide was encoded. Indeed, after the transformation of the plasmid, alkaline phosphatase activity was restored, indicating the presence of the fusion protein in the periplasmic space of E. coli DH5α. Furthermore, a closer analysis of the first VtaA amino acids indicated conservation with an extended signal peptide characteristic of many gamma- and betaproteobacterium autotransporters. Indeed, the motif MNKIF/Y was fully conserved, and the first 72 amino acids of VtaA contained alternating charged small hydrophilic regions with more extended hydrophobic regions (13).
FIG. 1.
Protein architecture of in silico-translated vtaA genes as determined by a Pfam database search. Only the 13 paralog molecules of the Nagasaki strain are represented. They display the same basic structure. Division into three groups is based on the sequence homologies of the YadA translocator domains. Repeats, motifs, and domains of the same color are 100% identical, and their distribution along VtaA passenger domains of different groups supports intrachromosomal recombination events. DNA USS identical to those found in H. influenzae (ACCGCACTT) and A. pleuropneumoniae (ACAAGCGGT) are represented by arrows. They never occur in sequences coding for repeats or motifs but rather in connecting regions. This type of sequence is necessary for natural transformation in bacteria and provides support for lateral gene transfer events.
Trimeric autotransporter genes and their translated proteins could be divided into three groups based on the sequence comparison of the YadA domain (Fig. S1B). It is not known how these amino acid variations affect the C-terminal region in terms of function. Structure homology modeling was performed with SWISS-MODEL using the crystal structure of Hia (Protein Data Bank 2gr7A) as the best matching template. All VtaA proteins were of very similar structures, with a propensity to form four β-strands at the C terminus (see Fig. S2 in the supplemental material), as found for Yersinia, Moraxella, and Haemophilus influenzae homologues (22, 32). Therefore, all three C-terminal YadA domains can most likely function as outer membrane anchors, with the characteristic β-barrel structure of autotransporters fulfilling all requisites for the AT-2 protein family assignment.
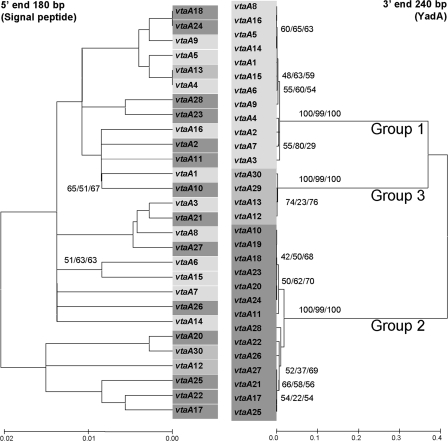
When phylogenetic reconstructions using the conserved 5′ (coding for the signal peptide) and 3′ (coding for the YadA domain) ends were compared, they revealed completely different topologies (Fig. 2). For the 3′ end, three branches with good bootstrap values give further support to the existence of the three groups deduced from the sequence comparison of the YadA domains. When the 5′ end was studied, no branch was supported by high bootstrap values, although genetic distances were much shorter. This suggests that the two extremes of the genes evolve following different patterns. Alternatively, the existence of recombination events involving parts, if not all, of the passenger domain could not be excluded.
FIG. 2.
Phylogenetic analysis and comparison of the 5′ extended signal peptide (without the first 27 nucleotides corresponding to the primer used for gene amplification) and 3′ YadA ends (without the terminal nucleotides corresponding to the amplification primers) of all vtaA genes sequenced in this study. Bootstrap consensus trees were constructed by neighbor joining (Kimura 2 parameter distance) and maximum parsimony and minimum evolution (maximum composite likelihood distance) using 1,000 bootstraps. Only neighbor-joining trees are shown, although bootstrap values above 50 are indicated for the tree methodologies used. The sequences of vtaA19 and vtaA29 could not be used in phylogenetic reconstructions using the 5′ end due to a deletion in this area and a lack of amplification with the 5′-end primer for each gene, respectively. The grouping observed for the 5′ end is different from that of the 3′ end, indicating different rates of evolution in the extremities of each gene. Bootstrap values at the 3′ end support the subdivision in three groups of the translocator domain.
Functional mapping of the YadA mature protein of Yersinia enterocolitica revealed that amino acids 21 to 81 and 83 to 103 are involved in binding to neutrophils and the collagen triple helix, respectively (45). These amino acid positions correspond to the first Hep_Hag and HIM repeats of the YadA protein. In H. parasuis, Hep_Hag and HIM variable multiple repeats are found at the N-terminal part of all VtaA proteins, suggesting similar functionalities. In groups 2 and 3, additional Hep_Hag repeats are inserted in between two HIM repeats upstream from the C-terminal YadA domain (Fig. 1). The sequence variabilities of the Hep_Hag repeats and HIM motifs are rather high (from 20 to 100% identities), without a strict homology between paralogs at the same position (Fig. 1), suggesting small intragenomic recombination events in the vtaA genes. However, the N-terminal regions (from the end of the signal peptide up to the last HIM motif situated before the first collagen repeat) of group 1 passenger domains seem to be more closely related between them than with their paralogs of groups 2 and 3. This was determined by pairwise comparisons of the N-terminal regions previously described (see Table S1 in the supplemental material).
Of the 376 bacterial proteins possessing collagen triple helix repeats described in the Pfam database, only two are associated with YadA C-terminal domains. These (GenBank accession numbers YP_719837 and YP_718689) belong to a member of the family Pasteurellaceae, Haemophilus somnus (strain 129Pt), whose genome has been sequenced (8). The function of surface bacterial proteins bearing collagen repeats has been best described for Firmicutes, where these proteins have been shown to be involved in adhesion to human lung fibroblasts via the α2β1 integrin inducing intracellular signaling and cell spreading (24). Therefore, the presence of collagen domains in AT-2 molecules is rather exceptional and might confer unique biological properties to H. somnus and H. parasuis. Moreover, although H. somnus 129Pt is considered a nonpathogenic strain, it colonizes genital mucosal surfaces, and of the 10 AT-2 genes found in the genome, two display collagen repeats (8). Thus, the presence of these collagen repeats in VtaA proteins suggests again that this protein family is involved in adhesion. The insertion of multiple collagen repeats in between Hep_Hag and HIM repeats and motifs that are potential collagen-binding repeats raises several questions about the structure and function of this multigene family. As for Hep_Hag or HIM repeats, the sequences of collagen repeats are highly variable within and between VtaA proteins.
Given that some H. parasuis strains, including the Nagasaki strain, can be isolated from multiple systemic sites, together with the implication of AT-2 in virulence in other bacteria, we suspected that VtaA proteins could contribute to the adhesive/invasive capacity of this bacterium. To address this question, a panel of invasive and noninvasive strains was compared at the genomic level (Tables 1 and 2).
TABLE 2.
Results of the comparative genomic microarray for one representative of each H. parasuis Nagasaki AT-2 group
| Group and gene size | Position covereda | Domain coveredb | Relative FI ratiosc
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Noninvasive tested strains
|
Invasive tested strains
|
||||||||||||
| SW114 | F9 | D74 | HP-3123 | HP-1205 | HP-1302 | HP-1319 | HP-2163 | HP-2269 | HP-33 | Nagasaki | |||
| Group 1, vtaA9 (2,859 bp) | 26 to 713 | SP + Hep_Hag 1 to 5 | 0.07 | 0.06 | 0.07 | 0.03 | 0.70 | 0.50 | 0.45 | 0.70 | 0.18 | 0.65 | 1 |
| 415 to 996 | Hep_Hag 2 to 7 | 0.10 | 0.09 | 0.06 | 0.05 | 0.70 | 0.60 | 0.40 | 0.86 | 0.17 | 0.57 | 0.95 | |
| 1567 to 2097 | Col 1 to 3 | 0.08 | 0.11 | 0.07 | 0.04 | 1.10 | 0.77 | 0.58 | 1.35 | 0.42 | 1 | 1.15 | |
| 1901 to 2568 | Col 2 to 3 + HIM 2 to 3 | 0.16 | 0.10 | 0.12 | 0.05 | 1 | 0.83 | 0.55 | 1.25 | 0.42 | 1 | 1.14 | |
| 2437 to 2773 | HIM 3 + YadA | 0.24 | 0.28 | 0.19 | 0.17 | 1.20 | 1.10 | 0.75 | 1.22 | 0.77 | 1.22 | 0.96 | |
| Group 2, vtaA11 (4,500 bp) | 143 to 506 | SP + Hep_Hag 1 to 3 | 0.13 | 0.13 | 0.12 | 0.11 | 0.38 | 0.38 | 0.12 | 0.12 | 0.14 | 0.25 | 0.99 |
| 713 to 1435 | Hep_Hag 5 to 9 + HIM 1 | 0.13 | 0.16 | 0.10 | 0.02 | 0.70 | 0.70 | 0.42 | 0.70 | 0.32 | 0.65 | 1.03 | |
| 901 to 1617 | Hep_Hag 7 to 9 + HIM 1 | 0.20 | 0.26 | 0.23 | 0.13 | 0.70 | 0.80 | 0.45 | 0.70 | 0.56 | 0.70 | 1.06 | |
| 2100 to 2532 | Col 1 to 2 | 0.23 | 0.42 | 0.30 | 0.20 | 0.60 | 0.40 | 0.30 | 0.35 | 0.50 | 0.61 | 1.07 | |
| 2586 to 3301 | Col 3 to 4 | 0.24 | 0.24 | 0.20 | 0.14 | 0.80 | 0.80 | 0.70 | 1.00 | 0.75 | 0.88 | 1.07 | |
| 2819 to 3570 | Col 4 + HIM 3 | 0.25 | 0.19 | 0.16 | 0.24 | 0.90 | 1.00 | 0.80 | 0.95 | 0.90 | 0.83 | 0.88 | |
| 4125 to 4500 | HIM 4 + YadA | 0.41 | 0.65 | 0.27 | 0.33 | 1.10 | 1.40 | 0.90 | 1.00 | 1.20 | 1.00 | 0.97 | |
| Group 3, vtaA12 (5,043 bp) | 1 to 585 | SP + Hep_Hag 1 to 3 | 0.30 | 0.42 | 0.37 | 0.29 | 0.90 | 0.75 | 0.65 | 0.60 | 1.26 | 0.90 | 1.02 |
| 1484 to 1873 | Col 1 to 2 | 0.19 | 0.20 | 0.14 | 0.15 | 1.80 | 1.90 | 0.50 | 0.50 | 1.60 | 1.70 | 1.03 | |
| 1949 to 2400 | Col 3 to 5 | 0.07 | 0.05 | 0.05 | 0.05 | 1.80 | 2.10 | 0.55 | 0.55 | 2 | 1.85 | 1.04 | |
| 2483 to 3091 | Col 6 to 9 | 0.12 | 0.12 | 0.09 | 0.06 | 2.25 | 1.70 | 0.55 | 0.54 | 2 | 2.35 | 1.20 | |
| 2654 to 2991 | Col 7 to 8 | 0.09 | 0.08 | 0.04 | 0.06 | 2 | 2.50 | 0.40 | 0.39 | 1.80 | 2.70 | 1.16 | |
| 2936 to 3291 | Col 8 to 10 | 0.08 | 0.08 | 0.07 | 0.04 | 2.50 | 2.40 | 0.40 | 0.70 | 1.70 | 1.60 | 1.18 | |
| 3175 to 3795 | Col 10 to 11 + HIM 2 | 0.26 | 0.29 | 0.26 | 0.20 | 1.65 | 1.90 | 0.85 | 1 | 1.90 | 1.90 | 1.02 | |
| 3996 to 4751 | Hep_Hag 6 to 8 + HIM 3 | 1.10 | 1.47 | 0.95 | 0.96 | 1.30 | 1.46 | 0.08 | 1.20 | 1.80 | 1.28 | 1.01 | |
| 4554 to 5030 | HIM 3 + YadA | 0.90 | 0.90 | 0.75 | 0.65 | 0.80 | 1.10 | 0.32 | 1.10 | 1.20 | 0.90 | 0.99 | |
Refers to the nucleotide positions probed in relation to each vtaA gene.
The numbers indicate repeats. SP, signal peptide; col, collagen.
Values indicate Cγ5/Cγ3 ratios; bold values indicate regions of high divergence or absence in relation to H. parasuis Nagasaki vtaA genes.
Genomic comparison of vtaA genes using a microarray: the passenger domain discriminates invasive and noninvasive strains.
A microarray containing 85 DNA fragments representative of all 13 vtaA genes was used to compare 11 H. parasuis strains. The results for one member of each vtaA group are shown in Table 2; similar hybridization patterns were found in all other genes (see Table S2 in the supplemental material). Cy5 Nagasaki strain/Cy3 Nagasaki strain ratios varied between 0.88 and 1.2, indicating that genomic DNA was evenly labeled regardless of the dyes used. With the exception of a few DNA regions, mostly found at the 5′ terminus, all vtaA genes in the invasive strains were conserved or moderately divergent (Cy5/Cy3 ratios above 0.3) compared to those of the Nagasaki reference strain. Collagen repeats found in the vtaA12 gene were duplicated (Cy5/Cy3 ratios above 1.5) in the DNA of invasive strains HP-1205, HP-1302, HP-2163, and HP-33. Very high divergence was the main feature observed for noninvasive strains (Cy5/Cy3 ratios below 0.3). The existence of homologous vtaA genes of groups 2 and 3 in nonpathogenic strains was suggested because the 3′ ends of genes vtaA11 and vtaA12 and the two first collagen repeats of vtaA11 were moderately divergent or conserved. However, some of the results observed, especially for the genes of groups 1 and 2, could be due to high-copy-number differences between invasive and noninvasive strains. For this reason, the fluorescence signals of 60 H. parasuis (Nagasaki) single-copy gene fragments that were highly homologous among bacteria were analyzed. First, the Cy5/Cy3 ratios were between 0.75 and 1.2, independent of the pathogenic status of the strain, ruling out the possibility that noninvasive strains belonged to a lineage very different than that of the Nagasaki strain. Second, the existence of vtaA homologues coding for passenger domains in noninvasive strains was supported by the fact that Cy5 fluorescence intensities were in many instances largely superior to values obtained with negative controls. Furthermore, the average Cy5 fluorescence intensities in conserved single-copy genes were three times higher than those obtained from the vtaA passenger domains. Therefore, AT-2 molecules exist in noninvasive strains, but their passenger domains are drastically divergent compared to those in invasive strains. These conclusions could be reached because the average length of conserved single-gene-copy fragments deposited in the microarray was nearly equal to that of gene fragments representing the passenger domains of vtaA genes. Overall, most of the DNA diversity occurs in the 5′ Hep_Hag, HIM, and collagen triple helix repeats.
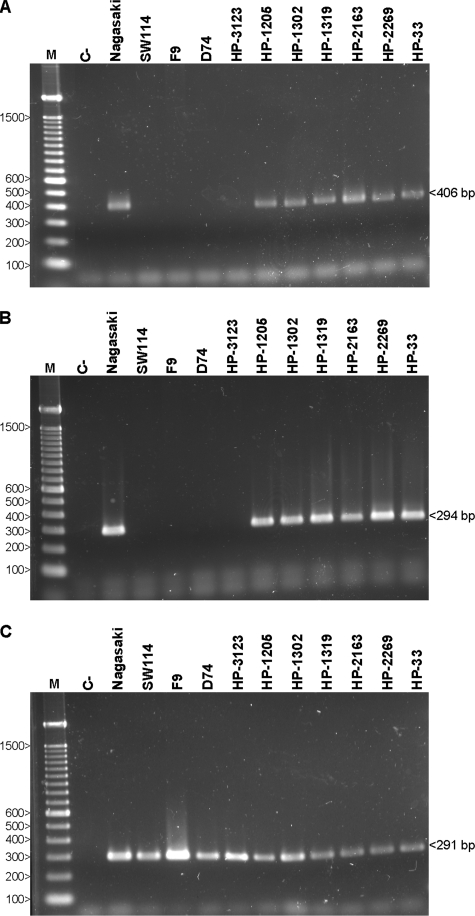
Moreover, PCR amplifications of the YadA translocator coding domain were also performed (Fig. 3). The presence of the YadA domains of groups 1 and 2 was confirmed in virulent strains but not in nonvirulent strains. The YadA domains of the group 3 vtaA genes were detected in all the strains. This validated the microarray results where 3′ termini of noninvasive AT-2 molecules were highly divergent, divergent, and conserved for groups 1, 2, and 3, respectively.
FIG. 3.
DNA amplification of vtaA translocator domains. The combinations of primers YADAF1/PADHR1 (A), YADAF2/PADHR2 (B), and YADAF3/PADHR3B (C) were used to specifically amplify vtaA YadA coding domains of groups 1, 2, and 3, respectively. PCR products were run in 2% agarose gels. The molecular sizes, in bp, of the standard (M) are shown on the left. C−, negative control.
These results demonstrate a striking polymorphism among vtaA genes in H. parasuis strains. There is a clear dichotomy between pathogenic and nonpathogenic strains, the latter being very divergent from the vtaA genes of the Nagasaki strain. These pathogenic strains adhere to lung epithelial cells and brain endothelial cells; they also grow on serosa and can cross the blood-brain barrier. These biological properties are not shared with noninvasive strains, which colonize only the upper respiratory tract (36, 53, 54).
Therefore, the observation that vtaA genes are relatively conserved among virulent isolates but highly divergent in noninvasive strains is of significant importance and strengthens the hypothesis that these molecules play a role in virulence. However, it is not known if the AT-2 molecules of noninvasive strains are functional or pseudogenes. Since the presence of AT-2 molecules has been reported in all gammaproteobacteria, it is very unlikely that noninvasive strains will not possess a full set of these proteins. Due to high sequence variability, it was not possible in this study to better characterize AT-2 passenger domains from nasal H. parasuis isolates. Assessing the repertoire of AT-2 of noninvasive strains will certainly clarify some of the molecular mechanisms related to the colonization and invasion of host tissues.
Sequencing of vtaA genes of H. parasuis invasive strains: assessing the diversity of the AT-2 repertoire.
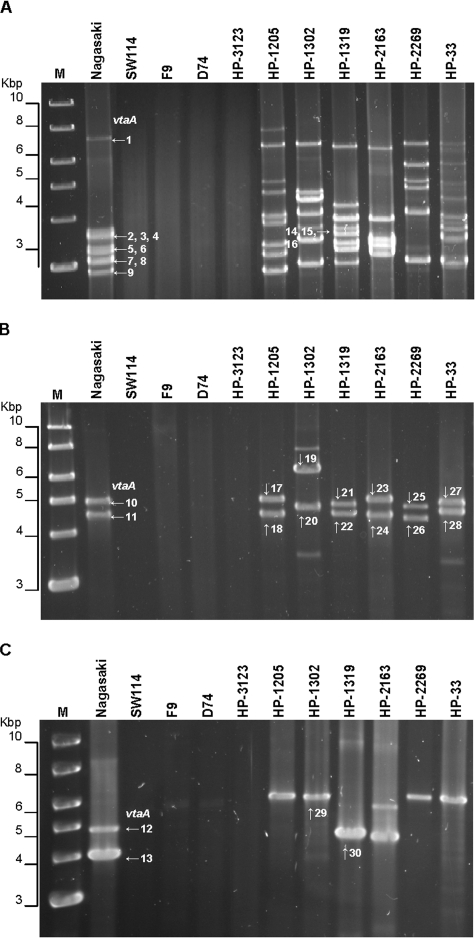
To gain further insights into the diversity of the vtaA genes, long PCRs were performed using a 5′-end primer (PADHF) common to all Nagasaki vtaA genes and one of the 3′-end primers (see Fig. S1 in the supplemental material) specific for each vtaA group (PADHR1, PADHR2, and PADHR3B, respectively). The DNA of nonvirulent strains was not amplified. The PCR products obtained from the Nagasaki strain were of the expected size, and with the exception of the genes of group 3, vtaA orthologs were found in all virulent strains (Fig. 4). The failure to detect the PCR products of group 3 in all but two strains (HP-1302 and HP-1319) was most likely due to small sequence variations at the 5′ ends of orthologs and poor PCR efficiency. When a primer (C5F) 151 bp downstream of the start codon was used in combination with the group 3-specific primer (PADHR3B), the PCR products appeared in all tracks corresponding to virulent strains. No amplification compatible with the expected size for the vtaA molecules was observed in the nonvirulent strains (Fig. 3C). Size polymorphisms were detected affecting mostly the genes of group 1. The various numbers of bands (ranging from 5 to 9 for group 1, 2 to 4 for group 2, and 1 to 2 for group 3) seen for different isolates showed that each invasive strain could have a different number of vtaA genes. All PCR products of the Nagasaki strain were cloned and sequenced. The sequences of the Nagasaki clones were in perfect agreement with those of the contigs, demonstrating correct assembly. A sample of invasive strain homologues was cloned and sequenced as indicated in Fig. 4, and the full-length sequence for vtaA30 (from strain HP-1319) was achieved by directly sequencing the 5′ end of the PCR product obtained with the primer combination PADHF and PADHR3B. All homologous VtaA proteins displayed the same basic structure as described previously, with variable numbers of Hep_Hag and HIM repeats and motifs at the 5′ end and multiple collagen repeats located centrally. The peculiar structure observed in groups 2 and 3 between the last collagen repeat and the YadA domain—three Hep_Hag repeats inserted in the middle of two HIM motifs—was also conserved between orthologs (see Fig. S3 in the supplemental material).
FIG. 4.
DNA amplification of vtaA genes. The combinations of primers PADHF/PADHR1 (A), PADHF/PADHR2 (B), and C5F/PADHR3B (C) were used to specifically amplify vtaA genes of groups 1, 2, and 3, respectively. PCR products were run in 0.8% agarose gels. PCR products, vtaA1 to vtaA30, selected for sequencing are indicated by arrows. The molecular sizes, in Kbp, of the standard (M) are shown on the left.
Nucleotide and amino acid global multiple alignments were difficult to perform with all molecules. For example, a strict homologue of vtaA11 (group 2, smallest PCR product) could not be found. In the case of vtaA19, a deletion event was observed at the 5′ end and will be discussed later. However, global multiple alignments could best be achieved with orthologs of vtaA10 (group 2, largest PCR product) from strains HP-1205 (vtaA17), HP-33 (vtaA27), HP-2163 (vtaA23) and HP-2269 (vtaA25). Overall identities at the nucleotide level ranged from 96 to 88% and at the protein level from 92 to 86%. Most of the variability was clustered in the regions coding for collagen, Hep_Hag and HIM repeats and motifs at the 5′ termini of the molecules. Selective pressure on these molecules was examined using this alignment (Fig. 5A). Globally, the microevolution of vtaA10 and its orthologs seems to be driven by positive selection (mean difference between nonsynonymous and synonymous changes [dN − dS] of 0.599163), but the dN − dS values varied between different parts of the molecule. Thus, positive selection is concentrated in the Hep_Hag repeats and HIM motifs at the 5′ parts of the molecules. Indeed, from the end of the signal peptide up to the collagen repeats, the substitutions were mainly nonsynonymous. The collagen repeats showed both synonymous and nonsynonymous substitutions together with a higher frequency of insertion-deletion (indel) positions. This particular evolutionary pattern is probably caused by the structural requirements of collagen repeats (GXY amino acid repeats) that conserve the glycine position and allow others to change, although there is a higher frequency of prolines in the second position. Moreover, this part of the molecule seems to be an indel hot spot, as shown by the variable number of collagen repeats. In the region between the collagen repeats and the YadA domain, there were fewer substitutions, indicating that the C-terminal part of the molecule is more conserved; five out of six positions that were statistically significant under purifying selection (dN < dS) were concentrated in this area. There is an apparent contradiction between the function devoted to the N-terminal end (adhesion to host molecules), which should require a certain degree of conservation, and its high variability. However, as it is exposed to the external media, the N-terminal part could be subjected to the pressure of the host immune response, requiring a faster evolution as described for other bacterial surface molecules (17).
FIG. 5.
Selective pressure of vtaA10 orthologs and mosaic structure of VtaA molecules. (A) A multiple alignment of vtaA17, vtaA27, vtaA23, and vtaA25 was performed for selective pressure determination. The dN − dS for each codon position of the vtaA10 strict homologs is indicated by the black bars and the frequency of indels by the white bars. The alignment position axis is given in amino acid numbers. For the whole molecule, the mean dN − dS was 0.599163 with a log likelihood of −10,670.5. Six positions (indicated by asterisks) were detected as negatively selected by the SLAC codon-based maximum likelihood method (P < 0.05). The protein architecture of VtaA10 is represented at the top of the figure; a positive selective pressure applies to the part of the gene coding for the N-terminal Hep_Hag and HIM repeats and motifs, respectively. (B) To illustrate the mosaic structure of a sample of VtaA molecules, modules of the passenger domain were selected and compared using BlastP. Pairwise amino acid identities are indicated as percentages. Potential recombination breakpoints detected by more than five of the seven algorithms included in the RDP2 package with statistical significance (P < 0.05) are indicated by stars. Modules and connective regions are not to scale, and blanks indicate indels.
For strains HP-1319 and HP-1302, the sequences of group 2 VtaA21 and VtaA19 (larger PCR products in Fig. 4) showed good conservation with VtaA10 in the region spanning from the YadA domain up to all, or part, of the collagen repeats. From this point, divergence was such that these molecules could not be considered strict homologues of VtaA10 (high amino acid substitutions with numerous gaps). The first 421 amino acids of VtaA21 displayed between 81 and 75% identities with the N termini of VtaA13, VtaA30 (group 3), VtaA18, and VtaA24 (HP-1205 and HP-2163 group 2 lower PCR product in Fig. 4), respectively, with an indel of 42 amino acids. These four N termini (VtaA13, -30, -18, and -24) shared between 87 and 95% identities between them (see Table S1 in the supplemental material). VtaA18 and VtaA24 displayed a nearly perfect alignment between them (96% identity at the protein level) for the whole molecule and shared nearly the same collagen repeats and connective region up to the beginning of the translocator domain of VtaA30. However, this connective region of VtaA13 was more similar to VtaA11 (98% identity at the protein level) than any other gene, including its group 3 paralog VtaA12 and ortholog VtaA30. Only the C-terminal part of the VtaA13 collagen repeats was similar to those of VtaA18, VtaA24, and VtaA30. Figure 5B illustrates the mosaic structure of the VtaA molecules, highlighting an extreme evolutionary complexity. Duplication of an ancestral vtaA group 2 or 3 gene followed by indel events does not entirely explain the diversity observed at the N terminus and the collagen repeats. Indeed, the comparison of the signal peptide and the translocator domain of these orthologs using phylogenetic reconstructions yielded incongruent trees not only between paralogs but also between orthologs (Fig. 2). Further support for the existence of recombination events was provided by searching for recombination signals for the vtaA genes included in Fig. 5B. Nineteen potential breakpoints were detected by more than five of the seven methods included in the RDP2 software package with statistical significance (P < 0.05). Interestingly, all of these breakpoints were usually located out of the predicted repeats and motifs (Fig. 5B), before and after the first HIM motif, after the collagen repeats, or surrounding the three Hep_Hag repeats characteristic of group 2 and 3 YadA domains. Altogether, this provided evidence for extensive recombination affecting part if not all of the passenger domain between different strains, evoking frequent lateral gene transfer as already suggested by the phylogenetic analysis of AT-2 proteins of gram-negative bacteria (26).
Although several plasmids have been described in H. parasuis (29, 48) and the genome of the Nagasaki strain has numerous bacteriophages inserted into it (result not shown), so far no vtaA genes have been found adjacent to or embedded in these mobile elements. Alternatively, natural transformation (7), without the need of a vector, could permit lateral transfer (43). In that respect, multiple DNA USS of different types have been detected in the passenger domain of many vtaA genes (Fig. 1; see also Fig. S3 in the supplemental material). Moreover, it is well known that several strains can coexist in the same farm, even in the same animal (38), increasing the probability of lateral gene transfer between them.
Genomic rearrangements in the vicinity of vtaA genes.
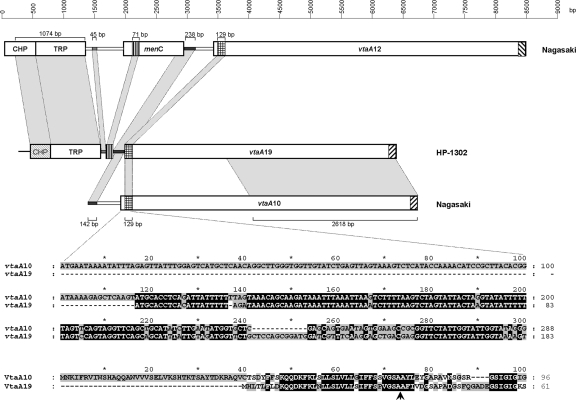
Genomic DNA amplification of strain HP-1302 with the group 2-specific primers yielded an unusually large 6,235-bp product (Fig. 4B). The sequence of this amplicon revealed a 25-bp repeat in both extremities and in opposite directions corresponding to the translocator group 2 domain primer which begins with the last codon of this vtaA gene (see Fig. S1B in the supplemental material). As shown in Fig. 6, although truncated at the signal peptide, a vtaA gene (vtaA19) was detected in one extremity with no interruptions (stop codons or frameshifts). The other extremity was characterized by a pseudogene (a conserved hypothetical protein) followed by a cation transporter, but no vtaA gene fragment was detected. Primers were designed along the 6,235-bp amplicon to amplify genomic DNA to rule out the possibility that we were working with an artifact. A comparison with the genome of Nagasaki revealed that the pseudogene and the transporter were highly conserved in strain HP-1302. However, in the Nagasaki strain, the conserved hypothetical protein was in frame with a methionine start codon. The menC gene (O-succinylbenzoate synthase) was absent in the HP-1302 amplicon, with only 71 nucleotides remaining, indicating a deletion event rather than an insertion in the Nagasaki genome in which the next gene was vtaA12 (group 3). The deletion affected one part of the signal peptide and one part of its adjacent intergenic space. Interestingly, the first 34 amino acids of VtaA19 were predicted as a signal peptide with a probability of 0.993. However, the expression of this gene is questionable since the promoter has likely been deleted. It is also unknown if the duplication of the last 25 nucleotides of vtaA19 was concomitant with the observed deletion. A careful examination of the Nagasaki genome did not reveal orphan DNA fragments outside their respective vtaA genes. Nonetheless, this result provides further support for the extreme plasticity of vtaA genes and their surroundings.
FIG. 6.
Comparison of the genomic regions upstream of genes vtaA19, vtaA10, and vtaA12. Boxes represent genes. Abbreviations: CHP, conserved hypothetical protein; TRP, divalent cation transporter; menC, gene encoding O-succinylbenzoate synthase. Light gray shaded areas correspond to high-homology regions (identities of 90% or more) shared between the different genes or intergenic spaces whose length in bp are indicated by values below or above the brackets. With the exception of 71 nucleotides, the entire menC gene has been deleted in HP-1302, and the CHP gene in this strain is a pseudogene with numerous frameshifts. The signal peptide region is magnified to show the exact extent of the deletion in vtaA19. The arrow indicates the most probable cleavage site of the signal peptide.
Proposed evolutionary model of the vtaA multigene family.
This report describes for the first time an appreciable sampling of AT-2 gene sequences within a single bacterial species, allowing us to explore some new aspects of the evolution of these molecules. First, compared to the Pasteurellaceae genomes sequenced to date, a main feature, only shared with H. somnus, is the high copy number of AT-2 genes. Strikingly, no vtaA pseudogenes have been found in the genome of the Nagasaki strain or vtaA orthologs examined, although vtaA19 seems to have no functional promoter. Also, from the fact that vtaA genes are very divergent between invasive and noninvasive strains and that a high positive selective pressure is observed at the most external part of the mature proteins, a major function for these molecules in relation to the adaptation of the bacterium to its host can be deduced. However, the repertoire of passenger domains shows a high variability with a mosaic structure not compatible with a single mechanism of punctual mutations accompanied by small indel events. Only lateral gene transfer of the whole or part of the passenger domain together with intrachromosomal rearrangements can account for the mosaic structure observed. Three translocator domains (groups 1 to 3) have been characterized. Apparently only group 1 is associated with a particular passenger domain repertoire, but since a limited number of orthologs have been sequenced in this group, recombination with other translocators cannot be excluded. In that respect, the nearest homologs of the N-terminal Hep_Hag and HIM repeats of VtaA11 (group 2) are VtaA5 and VtaA16 (group 1), having a 71% identity with both. The observation that the N-terminal domain architectures of VtaA1 and VtaA29 (group 3) are very similar (Fig. 1; see also Fig. S3 in the supplemental material), with a 71% identity, provides further support for the latter hypothesis. The fact that vtaA genes of group 1 had a significantly higher copy number in all invasive strains indicates evolutionary fates different from those for the genes of groups 2 and 3. The mechanism underlying this multiplication is unclear, but it results in the spreading of vtaA genes all over the genome; this has also been noticed for H. somnus (8). However, the observation that small terminal DNA fragments of the group 2 translocator can be found inverted in strain HP-1302 and the incomplete deletion upstream of vtaA19 is reminiscent of transposition-like activity (14, 49).
The phylogenetic analysis of YadA domains was difficult to perform due to substitution saturation (56). Phylogenetic reconstructions were not robust, and bootstrap values were usually low (see Fig. S4 in the supplemental material). Since the reliability of the internal branches was low, it was hard to conclude whether the three YadA domains of H. parasuis were derived from a common ancestor or if they were acquired independently. The three groups were not clustered together, but they were branched with other Pasteurellaceae YadA domains. Furthermore, H. parasuis was the only member of the family Pasteurellaceae to show three different groups of YadA domains, although some members (H. somnus, Pasteurella multocida, Actinobacillus succinogenes, and Actinobacillus pleuropneumoniae) showed two unrelated branches of YadA domains. Curiously, group 3, which is the most conserved among H. parasuis strains, had only one ortholog in A. succinogenes. This topology suggests that the YadA domains of H. parasuis and other Pasteurellaceae could have several origins rather than a common origin. However, since each group is found in more than one member of the family Pasteurellaceae, they may have been acquired before its divergence. Alternatively, YadA domains could have been expanded by intensive lateral gene transfer among Pasteurellaceae after being acquired by one of its members. Interestingly, group 1 Pasteurellaceae are clustered with one Burkholderia ambifaria YadA domain (with a bootstrap value of 30), suggesting the existence of lateral gene transfer.
The most intriguing feature is the presence of collagen repeats in all VtaA molecules of invasive strains. No collagen repeats are found in the genome of H. parasuis (Nagasaki) outside the vtaA genes. They display some common evolutionary features such as the upstream Hep_Hag and HIM repeats: variable number of repeats, accumulation of mutations and insertions/deletions, and in many instances better homology between orthologs than paralogs. However, their presence in only two gram-negative bacterial species (H. parasuis and H. somnus) suggests that they have been acquired. The presence of collagen repeats in all vtaA genes sequenced from invasive strains could be explained by an insertion event into a single vtaA ancestor, followed by the serial duplication of this gene. Alternatively, those repeats could have been interchanged by the recombination of parts of the passenger domain with other vtaA genes of different groups without collagen repeats. Both mechanisms would lead to homogeneous vtaA gene pools. The fact that many caudoviruses possess collagen repeats in some of their proteins is of interest (see the Pfam database for PF01391). In that respect, multiple bacteriophages of the same species as those harboring collagen repeats are inserted in the genome of H. parasuis. Because Pasteurellaceae and Streptococcaceae share the same host ecological niche (the respiratory tract), it is tempting to hypothesize that the donor DNA was from Firmicutes.
Supplementary Material
Acknowledgments
This work was supported by grant AGL2002-01760 from the Ministry of Science and Technology (Spain) and HIPRA Laboratories, S.A.
We thank Laia Viladevall for assistance and Roger Lahoz from the DNA Microchips and Sequencing Service of the Autonomous University of Barcelona. We also thank Olga Francino from the Unitat de Genètica i Millora Animal of the Autonomous University of Barcelona for useful help with sequencing and Raquel Rivas and Marta Perez from CReSA for technical assistance. Reference strains were kindly donated by P. Blackall from the Animal Research Institute (Queensland, Australia).
Footnotes
Published ahead of print on 14 November 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amano, H., M. Shibata, N. Kajio, and T. Morozumi. 1994. Pathologic observation of pigs intranasally inoculated with serovar 1, 4, and 5 of Haemophilus parasuis using immunoperoxidase method. J. Vet. Med. Sci. 56639-644. [DOI] [PubMed] [Google Scholar]
- 3.Arnold, K., L. Bordoli, J. Kopp, and T. Schwede. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22195-201. [DOI] [PubMed] [Google Scholar]
- 4.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved predictions of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. [DOI] [PubMed] [Google Scholar]
- 6.Besemer, J., and M. Borodovsky. 1999. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 273911-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bigas, A., M. E. Garrido, A. M. Rozas, I. Badiola, J. Barbé, and M. Llagostera. 2005. Development of a genetic manipulation system for Haemophilus parasuis. Vet. Microbiol. 105223-228. [DOI] [PubMed] [Google Scholar]
- 8.Challacombe, J. F., A. J. Duncan, T. S. Brettin, D. Bruce, O. Chertkov, J. C. Detter, C. S. Han, M. Misra, P. Richardson, R. Tapia, N. Thayer, G. Xie, and T. J. Inzana. 2007. Complete genome sequence of Haemophilus somnus (Histophilus somni) strain 129Pt and comparison to Haemophilus ducreyi 35000HP and Haemophilus influenzae Rd. J. Bacteriol. 1891890-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, D., V. Barniak, K. R. VanDerMeid, and J. C. McMichael. 1999. The levels and bactericidal capacity of antibodies directed against the UspA1 and UspA2 outer membrane proteins of Moraxella (Branhamella) catarrhalis in adults and children. Infect. Immun. 671310-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole, L. E., T. H. Kawula, K. L. Toffer, and C. Elkins. 2002. The Haemophilus ducreyi serum resistance antigen DsrA confers attachment to human keratinocytes. Infect. Immun. 706158-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comanducci, M., S. Bambini, B. Brunelli, J. Adu-Bobie, B. Arico, B. Capecchi, M. M. Giulani, V. Masignani, L. Santini, S. Savino, D. M. Granoff, D. A. Caugant, M. Pizza, M. Rappuoli, and M. Mora. 2002. NadA, a novel vaccine candidate of Neisseria meningitidis. J. Exp. Med. 1951445-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotter, S. E., N. K. Surana, and J. W. St. Geme III. 2005. Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 13199-205. [DOI] [PubMed] [Google Scholar]
- 13.Desvaux, M., L. M. Cooper, N. A. Filenko, A. Scott-Tucker, S. M. Turner, J. A. Cole, and I. R. Henderson. 2006. The unusual extended signal peptide of the type V secretion system is phylogenetically restricted. FEMS Microbiol. Lett. 26422-30. [DOI] [PubMed] [Google Scholar]
- 14.Doublet, B., P. Butaye, H. Imberechts, D. Boyd, M. R. Mulvey, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Salmonella genomic island 1 multidrug resistance gene clusters in Salmonella enterica serovar Agona isolated in Belgium in 1992 to 2002. Antimicrob. Agents Chemother. 482510-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8186-194. [PubMed] [Google Scholar]
- 16.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8175-185. [DOI] [PubMed] [Google Scholar]
- 17.Finlay, B. B., and G. McFadden. 2006. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124767-782. [DOI] [PubMed] [Google Scholar]
- 18.Gardy, J. L., M. R. Laird, F. Chen, S. Rey, C. J. Walsh, M. Ester, and F. S. L. Brinkman. 2005. PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21617-623. [DOI] [PubMed] [Google Scholar]
- 19.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8195-202. [DOI] [PubMed] [Google Scholar]
- 20.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 4195-98. [Google Scholar]
- 21.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoiczyk, E., A. Roggenkamp, M. Reichenbecher, A. Lupas, and J. Heesemann. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 195989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hultgren, S. J., S. Abraham, M. Caparon, P. Falk, J. W. St. Geme III, and S. Normark. 1993. Pilus and nonpilus bacterial adhesins: assembly and function in cell recognition. Cell 73887-901. [DOI] [PubMed] [Google Scholar]
- 24.Humtsoe, J. O., J. K. Kim, Y. Xu, D. R. Keene, M. Hook, S. Lukomski, and K. K. Wary. 2005. A streptococcal collagen-like protein interacts with the alpha2beta1 integrin and induces intracellular signaling. J. Biol. Chem. 28013848-13857. [DOI] [PubMed] [Google Scholar]
- 25.Kielstein, P., and V. J. Rapp-Gabrielson. 1992. Designation of 15 serovars of Haemophilus parasuis on the basis of immunodiffusion using heat-stable antigen extracts. J. Clin. Microbiol. 30862-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, D. S. H., Y. Chao, and M. H. Saier, Jr. 2006. Protein-translocating trimeric autotransporters of gram-negative bacteria. J. Bacteriol. 1885655-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korber, B. 2000. HIV signature and sequence variation analysis, p. 55-72. In A. G. Rodrigo, H. Gerald, and G. H. Learn (ed.), Computational analysis of HIV molecular sequences. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 28.Laarmann, S., D. Cutter, T. Juehne, S. J. Barenkamp, and J. W. St. Geme. 2002. The Haemophilus influenzae Hia autotransporter harbours two adhesive pockets that reside in the passenger domain and recognize the same host cell receptor. Mol. Microbiol. 46731-743. [DOI] [PubMed] [Google Scholar]
- 29.Lancashire, J. F., T. D. Terry, P. J. Blackall, and M. P. Jennings. 2005. Plasmid-encoded Tet B tetracycline resistance in Haemophilus parasuis. Antimicrob. Agents Chemother. 491927-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, J. Y., J. R. Pollack, F. L. Chou, C. A. Rees, A. T. Christian, J. S. Bedford, P. O. Brown, and M. H. Ginsberg. 2002. Physical mapping of genes in somatic cell radiation hybrids by comparative genomic hybridization to cDNA microarrays. Genome Biol. 3research0026.1-0026.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin, D. P., C. Williamson, and D. Posada. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21260-262. [DOI] [PubMed] [Google Scholar]
- 32.Meng, G., N. K. Surana, J. W. St. Geme III, and G. Waksman. 2006. Structure of the outer membrane translocator of Haemophilus influenzae Hia trimeric autotransporter. EMBO J. 252297-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morozumi, T., and J. Nicolet. 1986. Morphological variations of Haemophilus parasuis strains. J. Clin. Microbiol. 23138-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Müller, G., H. Köhler, R. Diller, A. Rassbach, A. Berndt, and D. Schimmel. 2003. Influences of naturally occurring and experimentally induced porcine pneumonia on blood parameters. Res. Vet. Sci. 7423-30. [DOI] [PubMed] [Google Scholar]
- 35.Nielsen, R. 1993. Pathogenicity and immunity studies of Haemophilus parasuis serotypes. Acta Vet. Scand. 34193-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oliveira, S., and C. Pijoan. 2004. Computer-based analysis of Haemophilus parasuis protein fingerprints. Can. J. Vet. Res. 6871-75. [PMC free article] [PubMed] [Google Scholar]
- 37.Olvera, A., M. Cerda-Cuellar, and V. Aragon. 2006. Study of the population structure of Haemophilus parasuis by multilocus sequence typing. Microbiology 1523683-3690. [DOI] [PubMed] [Google Scholar]
- 38.Olvera, A., M. Cerdà-Cuéllar, M. Nofrarías, E. Revilla, J. Segalés, and V. Aragon. 2007. Dynamics of Haemophilus parasuis genotypes in a farm recovered from an outbreak of Glässer's disease. Vet. Microbiol. 123230-237. [DOI] [PubMed] [Google Scholar]
- 39.Pizarro-Cerda, J., and P. Cossart. 2006. Bacterial adhesion and entry into host cells. Cell 124715-727. [DOI] [PubMed] [Google Scholar]
- 40.Pond, S. L., and S. D. Frost. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 212531-2533. [DOI] [PubMed] [Google Scholar]
- 41.Rapp-Gabrielson, V. J., S. R. Oliveira, and C. Pijoan. 2006. Haemophilus parasuis, p. 681-690. In B. E. Straw, J. J. Zimmerman, S. D'Allaire, and D. J. Taylor (ed.), Diseases of swine, 9th ed. Blackwell Publishing, Oxford, United Kingdom.
- 42.Read, T., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 42381-86. [DOI] [PubMed] [Google Scholar]
- 43.Redfield, R. J., A. W. Findlay, J. Bossé, S. J. Kroll, A. D. S. Cameron, and J. H. E. Nash. 2006. Evolution of competence and DNA uptake specificity in the Pasteurellaceae. BMC Evol. Biol. 682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez-Quiñones, F., S. Hernandez-Alles, S. Alberti, P. V. Escriba, and V. Benedi. 1994. A novel plasmid series for in vitro production of phoA translational fusions and its use in the construction of Escherichia coli PhoE:PhoA hybrid proteins. Gene 151125-130. [DOI] [PubMed] [Google Scholar]
- 45.Roggenkamp, A., H. R. Neuberger, A. Flugel, T. Schmoll, and J. Heesemann. 1995. Substitution of two histidine residues in YadA protein of Yersinia enterocolitica abrogates collagen binding, cell adherence and mouse virulence. Mol. Microbiol. 161207-1219. [DOI] [PubMed] [Google Scholar]
- 46.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 9714668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook, J., and R. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 48.San Millan, A., J. A. Escudero, A. Catalan, S. Nieto, F. Farelo, M. Gubert, M. A. Moreno, L. Domínguez, and B. Gonzalez-Zorn. 2007. β-Lactam resistance in Haemophilus parasuis is mediated by plasmid pB1000 bearing blaROB-1. Antimicrob. Agents Chemother. 512260-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siguier, P., J. Filée, and M. Chandler. 2006. Insertion sequences in prokaryotic genomes. Curr. Opin. Microbiol. 9526-531. [DOI] [PubMed] [Google Scholar]
- 50.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 51.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 244876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vahle, J. L., J. S. Haynes, and J. J. Andrews. 1995. Experimental reproduction of Haemophilus parasuis infection in swine: clinical, bacteriological, and morphologic findings. J. Vet. Diagn. Investig. 7476-480. [DOI] [PubMed] [Google Scholar]
- 53.Vahle, J. L., J. S. Haynes, and J. J. Andrews. 1997. Interaction of Haemophilus parasuis with nasal and tracheal mucosa following intranasal inoculation of cesarean derived colostrum deprived (CDCD) swine. Can. J. Vet. Res. 61200-206. [PMC free article] [PubMed] [Google Scholar]
- 54.Vanier, G., A. Szczotka, P. Friedl, S. Lacouture, M. Jacques, and M. Gottschalk. 2006. Haemophilus parasuis invades porcine brain microvascular endothelial cells. Microbiology 152135-142. [DOI] [PubMed] [Google Scholar]
- 55.Wollmann, P., K. Zeth, A. N. Lupas, and D. Linke. 2006. Purification of the YadA membrane anchor for secondary structure analysis and crystallization. Int. J. Biol. Macromol. 393-9. [DOI] [PubMed] [Google Scholar]
- 56.Xia, X., Z. Xie, M. Salemi, L. Chen, and Y. Wang. 2003. An index of substitution saturation and its application. Mol. Phylogenet. Evol. 261-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.