Abstract
The intronic enhancer (Eμ) of the immunoglobulin heavy chain (IgH) locus is critical for V region gene assembly. To determine Eμ's subsequent functions, we created an Igh allele with assembled VH gene but with Eμ removed. In mice homozygous for this Eμ-deficient allele, B cell development was normal and indistinguishable from that of mice with the same VH knockin and Eμ intact. In mice heterozygous for the Eμ-deficient allele, however, allelic exclusion was severely compromised. Surprisingly, this was not a result of reduced suppression of V-DJ assembly on the second allele. Rather, the striking breakdown in allelic exclusion took place at the pre-B to immature B cell transition. These findings reveal both an important role for Eμ in influencing the fate of newly arising B cells and a second checkpoint for allelic exclusion.
Ig heavy chain (IgH) genes are assembled through an ordered process of joining V, D, and J gene segments into a functional V region–coding sequence (VDJ recombination). A regulatory element, Eμ, lying just downstream of the J segments, was initially discovered as a transcriptional enhancer (1–3), but Eμ deletion studies subsequently demonstrated that it was essential for promoting efficient VDJ recombination as well (4–8). A model emerged in which Eμ promoted heavy chain variable region (VH) gene assembly and then served to enhance transcription of the newly formed IgH gene.
Although recombination can theoretically occur on either or both Igh alleles, only one allele is expressed in individual B cells, a phenomenon which is termed “allelic exclusion” (9). Allelic exclusion insures that individual B lymphocytes express antigen receptors (Ig) of a single antigen specificity, which is a fundamental requirement of the clonal selection theory put forth by MacFarlane Burnet (10) over 50 yr ago.
Several models have been proposed to explain allelic exclusion. In all models, it is assumed that VDJ recombination occurs at a low frequency and that two-thirds of such rearrangements result in a nontranslatable reading frame (nonfunctional rearrangement). Although these assumptions predict that cells with functional rearrangements on both alleles will be rare, they do not explain their almost complete absence from the normal B lymphocyte repertoire. Models to explain the absence of such cells fall into two general groups: those that invoke selective processes acting at the level of the cell and those that invoke feedback mechanisms affecting VDJ recombination itself. An early model in the former group proposed that expression of μ chains derived from both Igh loci would lead to toxic heavy chain levels resulting in cell death (11). Gene targeting experiments in which both Igh loci were modified to carry preassembled VH genes contradicted this model; in almost all B cells of these animals, both Igh alleles were expressed (12).
Studies of transformed precursor B cells and of Igμ transgenic mice supported an alternate regulated model of V-DJ recombination (13, 14). In this regulated model, it was proposed that μ chain expression from a productive Igh allele prevents V-DJ recombination on the other, precluding development of allelically included B lineage cells (9, 15). In Igμ transgenic mice, DJ assembly is common on the endogenous Igh loci of mature B cells, but full VDJ assembly is rare, suggesting that early expression of the Igμ transgene suppresses the second step (V-DJ recombination) in VH gene assembly within the endogenous loci. Although allelic exclusion is rarely perfect in transgenic mice (16), it has been thought that this results from the variable levels and/or timing of transgene expression as compared with IgH genes lying in their natural position within the Igh locus. Consistent with the “level” hypothesis, doubling a multicopy Ig transgene by mating led to both higher transgene expression levels and greater allelic exclusion than when the same transgene multimer was left hemizygous (17). Allelic exclusion appeared virtually complete when a preassembled variable region gene was inserted into one allele of the natural Igh locus by gene targeting (12).
The μ heavy chain encoded by a newly formed Igμ gene assembles with surrogate light chain (composed of VpreB1/2 and λ5) and with the signaling molecules Igα and Igβ to form a pre-B cell receptor (BCR) (18; for review see reference 19). It is through components of the preBCR that developing B cells sense successful assembly of an Igμ gene on one allele and signal arrest of any further assembly on the other allele (for review see references 19, 20). Even within the context of this feedback regulation, however, there would be opportunity for cells to arise with two functional Igμ alleles, if gene assembly and Igμ-mediated signaling were sufficiently separated in time.
In the present study, we provide evidence that a breach of allelic exclusion occurs quite regularly at the level of VH gene assembly in developing B cells but, under normal circumstances, this results in few, if any, detectable double producers. When the first allele to assemble a VH gene lacks Eμ, however, there is a dramatic increase in double producers both within the BM and among peripheral B cells. This striking effect is not the result of a failure to inhibit VH assembly on the second chromosome; rather, the change takes place at the pre-B to immature B cell transition. One of Eμ's functions, subsequent to VDJ recombination, is to facilitate the survival of newly generated B cells expressing a single Igh allele. In its absence, a second checkpoint for allelic exclusion is breached.
RESULTS
Generating a mouse strain that carries an Eμ-deficient VH-assembled Igha allele
Because VDJ recombination is rare on an allele lacking Eμ, B cell development is dramatically impaired in mice homozygous for this deletion (7, 8). This has made it difficult to assess Eμ's functions subsequent to VH gene assembly, particularly because it is possible that special circumstances (e.g., compensatory mutations) are required to generate the assembled IgH genes found in such animals. To circumvent this problem and also to study the behavior of an Eμ-deficient allele in competition with a WT one, we generated mice bearing an Igh allele that lacked Eμ but included a fully assembled VH upstream of the constant region (CH) gene cluster. Both core Eμ and flanking matrix attachment regions were deleted, each of which has been shown necessary for the expression of Igμ transgenes in developing B cells (21, 22).
A 3.7-kb region (including the most proximal DH gene segment, DQ52, all the JH gene segments, and Eμ) was replaced with a loxP-flanked neomycin resistance gene (neoR) and a fully assembled VH gene (neoRVHΔa; Fig. 1, gene targeting strategy). The VH gene and ∼2 kb of 5′ flanking sequence were isolated from the B1–8 hybridoma, which produces a 4-hydroxy-3-nitrophenyl acetyl-binding antibody (23, 24). Embryonic stem (ES) cells (E14.1 line) were transfected with the targeting vector, and clones undergoing the desired gene replacement were recovered by drug selection.
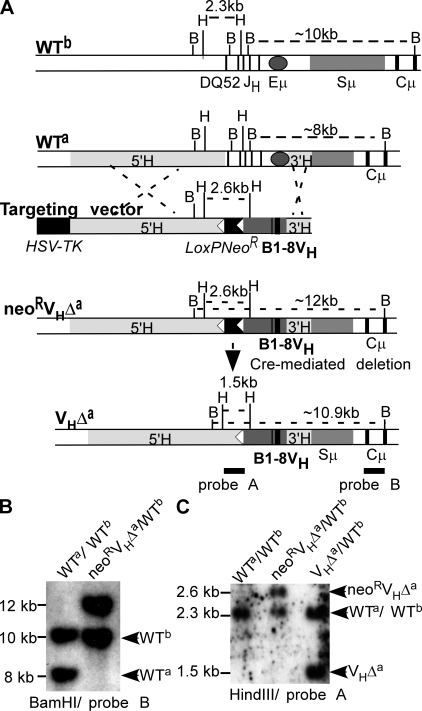
Figure 1.
Eμ deletion and VH gene insertion in an Igha locus. (A) Targeting strategy. WTb, WT Ighb locus. Thick vertical bars show exons for DQ52, JH1-4, and the first two exons of Cμ. Sμ, μ switch region; shaded oval, Eμ; B, BamHI; H, HindIII; WTa, unmodified Igha locus. Shaded boxes (5′H and 3′H) show regions of homology with targeting vector. Targeting vector: LoxPNeoR, loxP-flanked neomycin resistance gene (white arrowheads indicate loxP sites), B1-8VH promoter region (shaded), and coding sequences (thick vertical lines). neoRVHΔa: Igha allele after homologous recombination. VHΔa, modified Igha allele after neoR gene deletion. (B) Southern blot of liver DNA from WT (WTa/WTb) and mouse heterozygous for modified Igha allele before neoR deletion (neoRVHΔa/WTb). (C) Southern blot of liver DNA from WTa/WTb, neoR VHΔa/WTb, and heterozygous mutant mice after neoR deletion (VHΔa/WTb).
Igh locus structure in the ES clones and resulting mouse line was confirmed by genomic Southern blot (Fig. 1, A and B). An 8-kb BamHI fragment spanning Eμ and the first two exons of Cμ in the germline Igha locus (WTa) was replaced with a 12-kb BamHI fragment in the targeted locus of neoRVHΔa/WTb heterozygous mice. The 10-kb BamHI fragment derives from the WT Ighb allele (WTb). neoRVHΔa/WTb mice were mated to the EIIa-cre transgenic mouse line (25) to remove loxP-flanked neoR. A DNA probe (probe A) upstream of loxP-flanked neoR detected a 2.6-kb HindIII fragment from the neoRVHΔa locus that was reduced to 1.5 kb upon neoR deletion (VHΔa allele; Fig. 1, A and C). neoR deletion was also confirmed by PCR (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20081202/DC1; and see Materials and methods).
IgM from an Eμ-deficient allele can drive B cell development
It has been assumed that one of Eμ's important roles within the Igh locus is to activate transcription of newly assembled IgH genes. Other enhancers that can drive IgH gene transcription lie at the far 3′ end of the locus, but it has not been established whether or not this regulatory region, the 3′ regulatory region (3′ RR), is functional immediately after IgH gene formation in pro-B cells.
If transcription of newly formed IgH genes were Eμ dependent, expression of the VHΔa allele would be delayed until other transcription control elements (e.g., the 3′ RR) became functional. This would arrest B cell development at the pro-B to pre-B transition in homozygotes but not in mice heterozygous for the VHΔa allele because, in heterozygotes, VDJ recombination and IgH gene expression could take place on the other WT Igh allele. If transcription of newly formed IgH genes were Eμ-independent, B cell development would be normal both in homozygotes and heterozygotes and, because of feedback inhibition of V-DJ recombination, the B cells of VHΔa/WTb animals would express only the VHΔa allele.
B lineage cells were first examined in VHΔa/VHΔa animals and directly compared with VHEμa/VHEμa animals, a mouse strain which carries the same VH gene knockin but with Eμ left intact (12). As shown in Fig. 2 A, the proportion of B cells in the splenic lymphocytes of VHΔa/VHΔa mice was normal and did not differ from that in VHEμa/VHEμa mice (comparisons of multiple mice with these genotypes are summarized in Table S1, available at http://www.jem.org/cgi/content/full/jem.20081202/DC1). IgMa levels on the surface of resting splenic B lymphocytes was also the same, whether the B cells' Igh loci contained or lacked Eμ (Fig. 2 B). Consistent with surface IgM levels, quantitative measurements of Igμ messenger RNA (mRNA) produced by splenic B cells showed that the Eμ-deficient genes produced as much (or more) Igμ mRNA as Eμ-containing genes (Fig. 2 B).
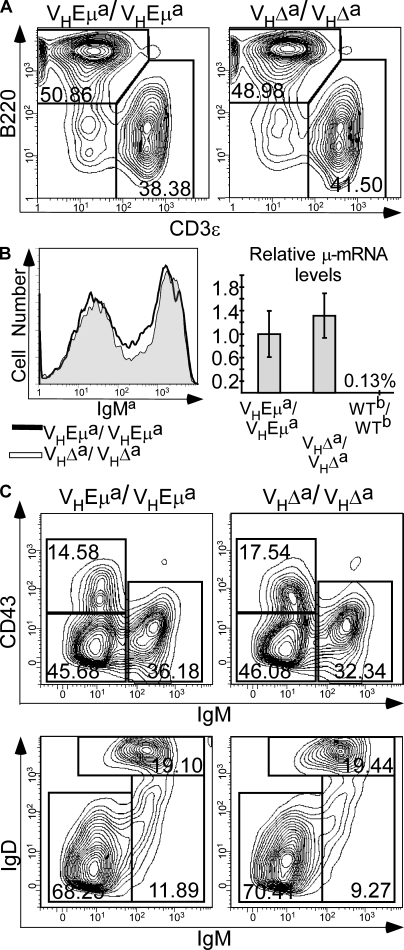
Figure 2.
Spleen and BM B cell profiles in homozygous VHEμa and VHΔa mice. (A) Flow cytometry profiles of splenic lymphocytes from adult mice stained for B220 (B cell marker) and CD3-ε (T cell marker). (B) Surface IgMa and relative Igμ transcript levels in splenic B cells. Left, histogram of spleen cells stained with antibody to IgMa. Right, quantitative RT-PCR results, using primers for B1-8i μ mRNA and normalized to hgprt1 mRNA (see Materials and methods). μ mRNA from VHEμa B cells are set as 1.0 and WTb/WTb is the negative control. (C) BM B cell subsets. Top, BM cells gated for B220+ cells and analyzed for CD43 and IgM expression. Bottom, BM B220+ cells analyzed for IgD and IgM. Data shown are representative of three animals of each genotype.
To eliminate the possibility that peripheral expansion of a small precursor population was masking a central defect, BM cells were also analyzed in these animals. As shown in Fig. 2 C, the pro-B (B220+IgM−CD43+), preB (B220+IgM−CD43−), and immature/mature B cell (B220+IgM+CD43−) subsets were found in similar proportions in VHEμa/VHEμa and VHΔa/VHΔa animals (Fig. 2 C, top; summary of data from multiple animals is in Table S1). When antibodies to IgD and IgM were used to distinguish immature (IgM+IgD−) from mature B cells (IgM+IgD+), these two subpopulations were also found at comparable levels (Fig. 2 C, bottom). The total number of B lineage cells in the BM of these homozygous mutant mouse lines was reduced with respect to WT (≥50% WT; Table S1) but was the same for the two mutant lines, indicating that the difference from WT was a function of the VH gene knockin by itself and was not affected by the presence/absence of Eμ. We conclude that an assembled IgH gene can be expressed in an Eμ-independent manner early in B cell development, allowing for the normal developmental progression of B lineage cells.
Allelic exclusion is dramatically compromised in the absence of Eμ
It was shown previously that B cells in mice heterozygous for the VHEμa (knockin) allele expressed only that allele, displaying what appeared to be perfect allelic exclusion (12). Because Igμ from the Eμ-deficient VHΔa allele could drive normal B cell development in homozygotes, we expected that B cells from mice heterozygous for this locus (VHΔa/WTb mice) would similarly express only the VHΔa allele and not the alternate WTb allele.
As shown in Fig. 3 A, IgM-positive BM cells from WTa/WTb mice expressed either IgMa or IgMb but not both. VHEμa/WTb BM B cells expressed only the VHEμa allele, which is consistent with earlier reports (12). Quite clearly, however, BM B cells from VHΔa/WTb mice included a subpopulation (∼10% of IgM+ cells) that broke the rules of allelic exclusion, displaying both alleles on the cell surface. This was even more striking in splenic B cells (∼20% of IgM+ cells) and in B cells of the peritoneal cavity (∼50% of IgM+ cells; Fig. 3 A). In stark contrast, VHEμa/WTb mice had no distinct population of double producers in the spleen, and such cells, although present in the peritoneal cavity, were much lower in number than in the VHΔa/WTb mice (∼7% vs. ∼50%; Fig. 3 A).
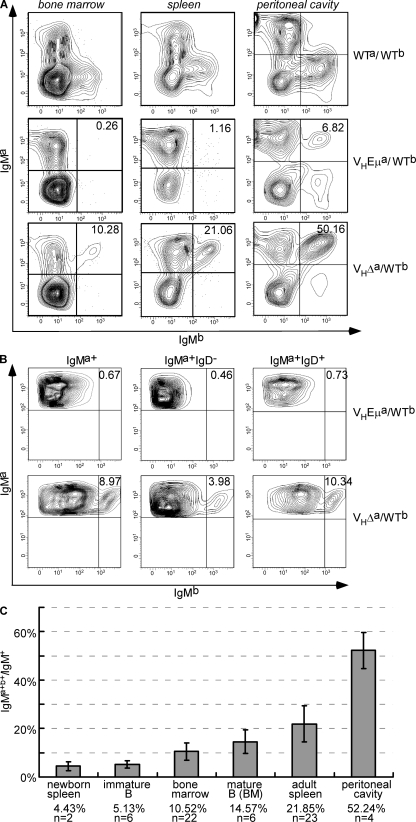
Figure 3.
IgM allotype expression in VHEμa/WTb and VHΔa/WTb mice. (A) Flow cytometry profiles of lymphocytes in BM, spleen, and the peritoneal cavity, stained with antibodies to IgMa and IgMb. (B) IgM allotype expression on immature and mature B cells of BM (enriched for B220+ cells). Plots are of cells gated for IgMa alone (left), cells that were IgMa+ and IgD− (middle; immature B cells), or IgMa+ and IgD+ cells (right; mature B cells) and then analyzed for IgMa and IgMb. In both A and B, the number in top right quadrant = (percentage of double-positive cells)/(total IgM+ cells). (C) Percentage of IgMa+b+/IgM+ cells at different stages of development and in different tissues. n = number of mice analyzed. Error bars show SD.
Further analyses of the IgMa+b+ peripheral B cells in VHΔa/WTb mice revealed that many were of the marginal zone phenotype in spleen and the B1-B cell phenotype in the peritoneal cavity (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20081202/DC1). In fact, among CD5+ cells in the peritoneal cavity, almost all proved to be double producers. Analyses of fetal liver and newborn spleen, however, provided no evidence that double producers were arising in greater numbers among B1 B cell progenitors (Fig. 3 C and Fig. S3).
To determine the frequency with which double producers were arising in the BM of adult mice, we used IgD/IgM expression to distinguish newly arising immature B cells (IgM+IgD−) from recirculating mature B cells (IgM+IgD+). Double producers made up a mean of 5% of the immature B cells (Fig. 3, B and C). These expanded to a mean of 15% of mature BM B cells and underwent further expansion in the spleen (Fig. 3, B and C). In summary, analyses of the lymphoid tissues of VHΔa/WTb mice revealed a profound defect in allelic exclusion, distinguishing these from VHEμa/WTb mice which carried an identical VH insertion but with Eμ intact.
To confirm at the molecular level that cells staining with both antiallotype reagents in VHΔa/WTb animals carried a successfully assembled Igμ gene on the Ighb allele, splenic IgMa+b+ cells were isolated and their genes examined by PCR. A primer specific for the WT Igh allele (JH4 primer; Fig. 4 D) and primers specific for three VH families were used to amplify and clone assembled VH genes on the WTb allele. 14 independent clones were sequenced, and all revealed a successfully assembled (productive) VH gene on the WTb allele (unpublished data).
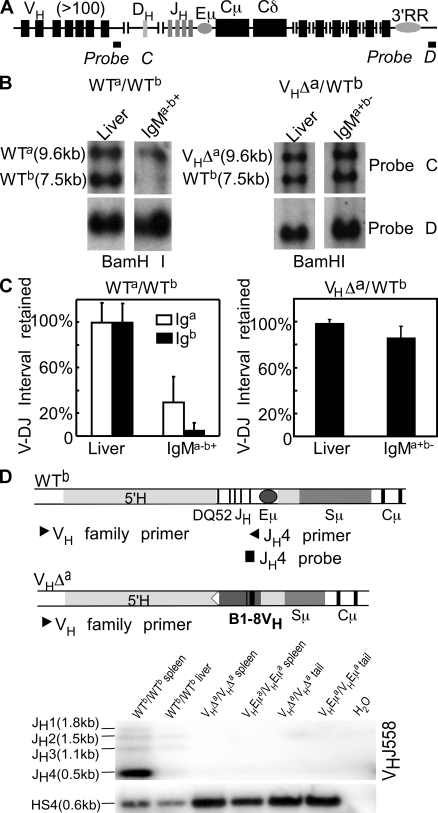
Figure 4.
V-DJ rearrangements on the WT Ighb allele of VHΔa/WTb mice. (A) Igh locus. Probe C detects a sequence deleted upon V-DJ joining. Probe D detects a sequence retained on both alleles in all cells. (B) Assay for V-DJ rearrangement on the Igha and Ighb alleles in WTa/WTb and VHΔa/WTb mice. Genomic DNA extracted from liver and from sorted IgMa−b+ spleen cells (WTa/WTb; left) and IgMa+b− spleen cells (VHΔa/WTb; right). DNAs were digested with BamHI and hybridized with probes C (top) and D (bottom). (C) Quantitative analysis (ImageQuant) of blot shown in B. A repeat experiment was done using liver and spleen cells from VHΔa/WTb animals. Band intensities in liver were set to 100%. Normalization was to probe D for WTa/WTb mice; normalization was to the Igha fragment in VHΔa/WTb mice. Error bars show SD. (D) Igh locus maps indicating PCR primers and probes for detecting V-DJ rearrangements on Ighb allele. 5′ primers specific for individual VH gene families (VH family primer) were used in combination with a 3′ primer (JH4 primer), which was present on the Ighb allele but missing on both the VHEμa and VHΔa alleles. The representative blot demonstrates allele-specific amplification. H2O, no DNA template control.
A productive IgH allele with or without Eμ significantly inhibits, but does not prohibit, DNA rearrangement on the alternate allele
Theoretically, only one-third of V-DJ rearrangements should result in a functional gene. The ∼5% double producers among immature BM B cells of VHΔa/WTb mice should correspond, therefore, to only 1/3 of the total attempts at VH gene assembly on the WTb allele. The vestiges of aberrant rearrangements should be detectable on the WTb alleles of IgMa+b− single producers in these mice.
To estimate the frequency of such rearrangements, we took advantage of a BamHI restriction fragment length polymorphism within the V-D interval. A probe lying within this interval (Fig. 4 A, probe C) detects a 9.6-kb BamHI fragment in liver DNA from 129/Ola (Igha) mice and a 7.5-kb BamHI fragment from C57BL/6 (Ighb) mice. Because of their location, these BamHI fragments would be deleted upon fusion of any VH with any DH gene segment on the Igha and Ighb alleles, respectively.
Splenic B cells were sorted from WTa/WTb F1 animals and from VHΔa/WTb mice. IgMa−b+ cells from WTa/WTb mice almost entirely lacked the BamHI fragment derived from the WTb allele (residual fragment results from contaminating cells in sorted populations) and retained only ∼30% germline (liver) levels of the larger BamHI fragment from the WTa allele (Figs. 4. B and C). A BamHI fragment mapping 3′ of the Igh loci (Fig. 4 A, probe D) was used to normalize DNA loading in these experiments. These results are consistent with earlier studies and demonstrate the inefficiency of the VH assembly process such that many B cells (in this case, ∼70%) have undergone unsuccessful VDJ gene assembly on the unexpressed Igha allele before they attempt and succeed at assembly on the Ighb allele (13, 26).
In B1–8 VH knockin mice, the B1–8 VH gene was inserted upstream of Cμ on the Igha allele and all germline JH genes were simultaneously removed. As noted earlier, the inserted B1–8 VH coding sequences were modified to inhibit VH gene replacement events, so the 9.6-kb BamHI fragment lying between the VH and DH genes on this Igha chromosome should not be lost in any B cells from these mice (early studies reported no evidence for V-D rearrangements [reference 13]). As shown in Fig. 4 B, this fragment did, in fact, remain in DNA isolated from IgMa+b− B cells from VHΔa/WTb mice. The smaller BamHI fragment from the WTb allele was also present in these cells and was present at 85–95% germline levels (Fig. 4 C). Quantification of the data showed no statistically significant difference from levels of this fragment in liver DNA (Fig. 4 C). We conclude that most cells expressing only the VHΔa allele have not undergone V-DJ rearrangement on the WTb allele.
To address this issue further, we turned to a more sensitive measure of V-DJ rearrangement. Semiquantitative PCR was used to quantify VDJ rearrangements on the WTb allele of IgMa+b− splenic B cells (single producers) using VH family primers and the WT Igh-specific primer described earlier. As shown in Fig. 5 A, V-DJ rearrangements on the WTb allele were readily detectable in IgMa+b+ double producers from the VHΔa/WTb mice (similar in abundance to Igh rearrangements in the IgMa+b− cells of WT mice). PCR products were also generated with each of the VH family primers when DNA was isolated from the single producers (IgMa+b− B cells) from VHΔa/WTb spleen, but these were somewhere between 1/5 and 1/25 as abundant as those seen in double producers or in the IgMa+b− B cells from WT mice. Importantly, this small but detectable level of V-DJ rearrangement on the WTb allele was also seen in the IgMa+b− spleen cells of VHEμa/WTb mice (Fig. 5 B). Consistent with the surface phenotype, these rearrangements, cloned from both animals, were nonproductive (18/18 from VHEμa/WTb mice and 18/19 from VHΔa/WTb mice; unpublished data).
Figure 5.
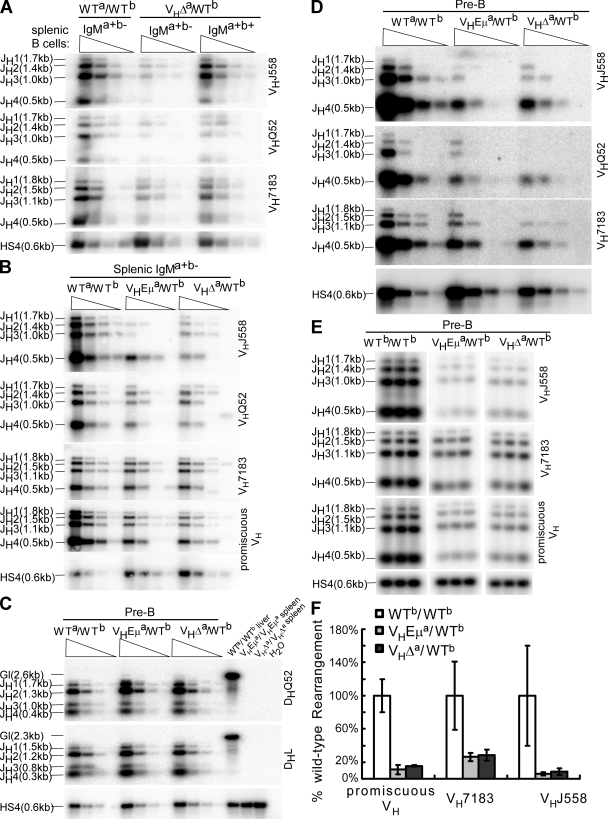
V-DJ rearrangements on the WT Ighb allele of VHEμa/WTb and VHΔa/WTb mice. (A) Splenic B cells from WTa/WTb and VHΔa/WTb, sorted on the basis of phenotype (IgMa+b− or IgMa+b+). PCRs were performed on fivefold serial dilutions of DNA templates (four lanes/cell type). PCR strategy detects V-DJ rearrangements on only the Ighb allele in VHΔa/WTb mice (both alleles in WTa/WTb mice). VDJ rearrangements involving each JH gene segment are indicated (JH1, JH2, etc.). VH family primers are indicated to the right of blots (VHJ558, etc.). PCR of HS4 (a 3′ RR element) was normalized for DNA input. (B) Like A, but a primer designed to anneal to all VH families (promiscuous VH 46) was included. (C) D-J rearrangements on the Ighb allele of pre-B cells (B220+CD43−IgM−) sorted from six mice of each genotype. The DHL primer anneals to most DH genes (47); DQ52 anneals only to DHQ52. Allele-specific 3′ primer (JH4) was used as in A. Liver DNA from WTa/WTb mice and spleen cell DNA from homozygous VHEμa and VHΔa mice were included as controls. H2O, no template control. (D) Pre-B cells were isolated as in C and PCR reactions using the VH gene family primers were performed as described in A. (E) Pre-B cells were isolated as in C. Sample dilutions determined to be in the linear range (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20081202/DC1) were prepared in triplicate for PCR. Representative data is shown. (F) Quantitative analysis of VDJ rearrangement blots. For each PCR reaction with a given VH primer, signals for JH1, JH2, JH3, and JH4 rearrangements were quantified and summed by ImageQuant. Values obtained for pre-B cells from VHEμa/WTb and VHΔa/WTb mice were normalized to the corresponding values for pre-B cells of WTb/WTb mice. Data were obtained from two pools of two mice each for WTb/WTb and three pools involving a total of seven mice for each of the VHEμa/WTb and VHΔa/WTb mouse lines. Error bars show SD.
The same PCR approach was used to estimate the frequency of WTb rearrangements in pre-B cells of both VHΔa/WTb and VHEμa/WTb mice. Pre-B cells comprise the pool that has just ceased IgH rearrangement and is beginning the process of Ig light chain gene assembly. Semiquantitative PCR assays of DNA from these cells showed that this pool had undergone D-J joining on the WTb allele at the same frequency as pre-B cells from normal WTa/WTb mice (Fig. 5 C). V-DJ joins were also evident on the WTb alleles of pre-B cells from both VHΔa/WTb and VHEμa/WTb mice. These, however, were both present at ∼1/5–1/25 the frequency seen in normal pre-B cells (Fig. 5 D).
To increase the resolution of these analyses, we used twofold dilutions of template, confirmed linearity (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20081202/DC1), and performed triplicate PCRs of diluted template samples. Pre-B cells from seven mice of the VHEμa/WTb genotype and seven of the VHΔa/WTb genotype were examined in three independent pools. Assembly of the WTb allele in these pre-B cells was compared with that of the two WT (WTa and WTb) alleles in WT mice (two pools of two mice each). Representative data are shown in Fig. 5 E and a quantitative analysis of all data is provided in Fig. 5 F.
V-region gene assembly on the WTb allele in the pre-B cells of both VHEμa/WTb and VHΔa/WTb mice was reduced relative to the WTa and WTb alleles in pre-B cells of WT mice (Fig. 5 F). The reduction, relative to WT mice, was greater for the more distal VHJ558 family than for the more proximal VH7183 family. We conclude that the VHΔa and VHEμa alleles were inhibiting, but not entirely prohibiting, V-DJ recombination on the allelic chromosome, and the feedback inhibition was largely, if not entirely, Eμ independent. Most importantly, however, the data showed that there was no significant difference in the amount of V-region assembly taking place on the WTb alleles of VHEμa/WTb and VHΔa/WTb pre-B cells. Assuming that some of these rearrangements were productive (confirmed by cloning as described in the next paragraph), both populations of pre-B cells included precursors to double-producing cells.
An Eμ-dependent checkpoint for allelic exclusion at the pre-B to immature B cell transition
Because the phenotypic outcome of gene assembly on the WTb allele was so dramatically different in the VHΔa/WTb and VHEμa/WTb mice (double producers evident in the immature and mature B cells of the former but not the latter mouse strain), we asked whether the VH gene rearrangements in the pre-B cells of these two mouse lines differed in some fundamental way. Pre-B cells were harvested from mice of both genotypes, their DNA was isolated, and assembled VH genes from the WTb allele were cloned and sequenced. 15/36 unique VH7183-DJ rearrangements cloned from pre-B cells of the VHEμa/WTb genotype were both in frame and lacked a stop codon (productive rearrangements), as were 12/36 unique VH7183-DJ rearrangements cloned from VHΔa/WTb pre-B cells. Closer examination of these productive rearrangements showed no fundamental differences among them in the DH segments used, the 7183 VH family members used, or the overall size of the junctions (Table S2, available at http://www.jem.org/cgi/content/full/jem.20081202/DC1). N nucleotides were found in the V-D junctions in >70% of the sequences, and 40% of these had three or more N nucleotides at this junction (mean length, 3.5 nt in VHEμa/WTb mice and 4.3 nt in VHΔa/WTb mice). Notably, this contrasts with light chain genes where N nucleotides are much more rare (∼10% of cells) and are found in much lower numbers/junction (generally 1 or 2 nt/junction) (27). We conclude that the precursors to double producers can be found in BM from both VHΔa/WTb and VHEμa/WTb mice and that VH assembly on the WTb allele has likely occurred before the pre-B cell stage (see Discussion).
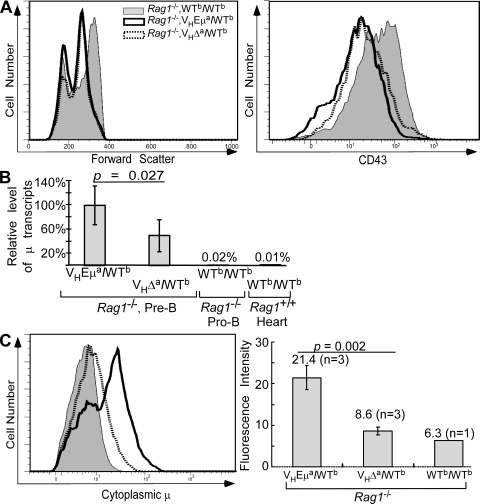
In an attempt to understand the mechanism through which Eμ might influence the development and selection of precursor B cells, we compared levels of μ mRNA generated from the VHEμa and VHΔa alleles in pre-B cells. To enrich for pre-B cells, VHEμa and VHΔa mice were backcrossed to Rag1−/− mice to generate VHEμa/WTb and VHΔa/WTb mice that lacked Rag-1 activity. In both of the resulting strains, B cell development was arrested at the pre-B cell stage because Ig light chain gene assembly was blocked. Age-matched mice of each genotype were killed and B220+ BM cells isolated (see Materials and methods). As expected, no IgM+ cells were present in these BM cells, and the bulk of cells were smaller and expressed lower levels of CD43 than comparable cells from WTb/WTb Rag1−/− littermates, which is consistent with their having progressed to the pre-B cell stage (Fig. 6 A). Total RNA was isolated from the B220+ BM cells of Rag1-deficient VHEμa/WTb and VHΔa/WTb mice (WTb/WTb; Rag1−/− littermates were controls) and Igμ mRNA was quantified by real-time RT-PCR (5′ primer for unique VDJ junction of VHB1-8 and 3′ primer for CH1 exon of Cμ; see Materials and methods).
Figure 6.
Igμ transcription and cytoplasmic Igμ protein levels in pre-B cells of mutant mice. (A) B220+ lymphocytes from BM of Rag1-deficient WTb/WTb, VHEμa/WTb, and VHΔa/WTb mice were analyzed for size (forward scatter) and for CD43 expression by FACS. Data shown are representative of three individual mice of each genotype analyzed. (B) Igμ mRNA levels in cells shown in A. Data were generated by quantitative RT-PCR, normalized to hgprt1 mRNA, and included two experiments, analyzing a total of five individual animals of each genotype. Negative controls were mRNA isolated from pro-B cells of a WTb/WTb Rag1−/− littermate and mRNA from C57BL/6 heart tissue (WTb/WTb, Rag1+/+). Statistical significance (P = 0.027) was obtained by a two-tailed Student's t test. Error bars show SD. (C) Cytoplasmic Igμ levels in B220+ BM cells of Rag1−/− mice. Left, histograms of cytoplasmic Igμ. Right, mean Igμ fluorescence in pre-B cells from multiple mice (n = number of mice analyzed). Error bars show SD.
Steady-state μ mRNA levels were roughly twice as high in cells expressing the VHEμa allele than in cells expressing the VHΔa allele (Fig. 6 B). This was in contrast to the finding in splenic B cells where the Eμ-deficient allele produced as much or more Igμ mRNA than its Eμ-containing counterpart (Fig. 2 C). Protein levels mirrored the mRNA levels; pre-B cells expressing only the VHΔa allele produced ∼1/2 the amount of Igμ protein produced by cells expressing the VHEμa allele IgH (Fig. 6 C). At this developmental stage, when pre-BCR and BCR-mediated signals are dictating B cell fate, μ heavy chain levels were measurably influenced by the presence or absence of Eμ.
Evidence for greater selective pressure on the developing B cells of VHΔa/WTb mice
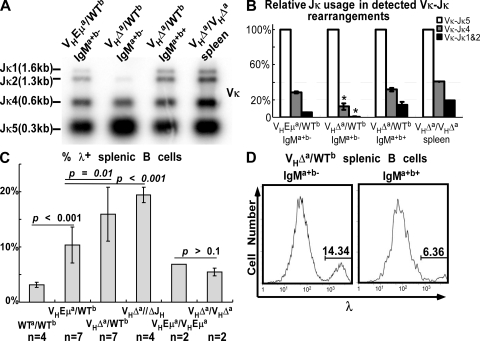
The reduced Igμ in VHΔa/WTb pre-B cells might be suboptimal for preBCR-mediated clonal expansion and/or BCR-mediated selection into the immature B cell pool. Survival of precursors expressing both alleles (double producers) in VHΔa/WTb mice might reflect a need for the increased Igμ levels achieved by biallelic expression. If this were true, IgMa+b− cells might circumvent this problem through alternate means, perhaps by expressing light chains that yielded BCRs with superior signaling properties. This would likely require light chain receptor editing. Receptor editing can be achieved through successive Vκ-Jκ rearrangements in which Vκ-Jκ joins involving the more upstream Jκ gene segments (Jκ1 and Jκ2) are replaced by joins to the more downstream Jκ segments (Jκ4 and Jκ5). A degenerate Vκ primer (VκD) (28) was used in combination with a primer downstream of Jκ5 (Jκ5e) to examine Jκ gene usage in splenic B cells of VHEμa/WTb and VHΔa/WTb animals. As shown in Fig. 7 (A and B; and Fig. S5, available at http://www.jem.org/cgi/content/full/jem.20081202/DC1), the IgMa+b− single producers of VHΔa/WTb mice uniquely showed a decrease in Vκ-Jκ joins involving Jκ1 and Jκ2 gene segments and a concomitant increase in joins involving Jκ5. The IgMa+b+ double producers of VHΔa/WTb mice were not similarly affected; these cells arose from precursors that would have been expressing both the VHΔa allele and the WTb allele where Eμ remains intact.
Figure 7.
Light chain editing in IgMa+b− cells of VHΔa/WTb mice. (A) Vκ-Jκ rearrangement analyses. PCR products detected with Jκ5 probe (see Materials and methods). (B) Quantitative analysis (ImageQuant) of blot shown in A and a repeat experiment. Detected Vκ-Jκ4 and Vκ-Jκ1 and 2 products were normalized to those for Vκ-Jκ5. *, P < 0.05 by a two-tailed Student's t test. Pie charts in Fig. S3 (available at http://www.jem.org/cgi/content/full/jem.20081202/DC1) show the relative increase in Vκ-Jκ5 rearrangements in IgMa+b− B cells from VHΔa/WTb animals. Error bars show SD. (C) λ+ splenic B cells in WT and mutant mice. Spleen cells were stained for both Igκ and Igλ and the percentage of λ+ cells was calculated as λ/(κ+λ). Genotypes are provided below bars. Data were pooled from four experiments. n = number of mice analyzed. P-values were calculated by a two-tailed Student's t test. Error bars show SD. (D) Histogram of cells stained for μa, μb, and λ. Left, single producers (IgMa+b−) from VHΔa/WTb mice; right, double producers (IgMa+b+) from VHΔa/WTb mice.
Another indicator of light chain receptor editing is an increase in the numbers of Igλ+ cells. In normal mice, Igλ+ cells make up only ∼5% of splenic B cells, and analyses of Igκ loci in Igλ-producing cells have suggested that Igλ genes are usually assembled only after the opportunity to express one of the two available Igκ alleles has been exhausted (29; for review see references 30, 31). We looked for Igλ-expressing splenic B cells in WTa/WTb, VHΔa/WTb, VHEμa/WTb, VHΔa/VHΔa, and VHEμa/VHEμa mice. We also examined hemizygous VHΔa/ΔJH mice (the ΔJH allele lacks all JH gene segments, prohibiting VH assembly on this allele) (32). The hypothesis was that both the VHΔa/WTb and VHΔa/ΔJH mice would have increased numbers of Igλ-expressing B cells because most of the B cells of the former and all B cells of the latter would have arisen from precursors expressing only the VHΔa allele. Consistent with this hypothesis, the frequency of Igλ producers among VHΔa/WTb splenic B cells was significantly higher than that in VHEμa/WTb mice (mean of 16 vs. 10%; P = 0.01; Igλ expressers were higher in both knockin strains relative to WT, presumably because of constraints imposed by predominant expression of a single structure heavy chain; Fig. 7 C). It was the single producers of the VHΔa/WTb mice, and not the double producers, that explained the increase in Igλ–expressing B cells relative to VHEμa/WTb mice (Fig. 7 D). Finally, consistent with the stated hypothesis, no such difference was observed when mice homozygous for the knockin alleles were compared (Fig. 7 C). In VHΔa/VHΔa mice, biallelic expression of the VHΔa allele would be expected to equal expression of one allele with Eμ.
DISCUSSION
The present study has shown that the intronic enhancer Eμ serves a critical function even after VH gene (VDJ) assembly is complete. In mice heterozygous for an Eμ-deficient but productively rearranged IgH gene (VHΔa/WTb), large numbers of IgH double-producing cells arose in peripheral tissues, breaking the rules of allelic exclusion. In contrast, a matched mouse line with the same VH knockin but with Eμ present (VHEμa/WTb) lacked these double-producing cells. In both animals, there was evidence that the Igμ product of the assembled IgH gene was signaling a shutdown of VH assembly on the Ighb allele, which is the primary mechanism postulated to ensure IgH allelic exclusion (Figs. 4 and 5). There was no measurable difference in the efficiency of this feedback inhibition whether Eμ was present or absent.
This latter finding was unexpected. Eμ was initially discovered as a transcriptional enhancer and believed to serve as such in newly formed IgH genes (1–3). In that capacity, Eμ could dictate preBCR levels and, thereby, signaling strength in a developing B lymphocyte. Even in Eμ's absence, however, pre-B cells were dominated by those that had undergone D-J, but not V-DJ, rearrangement on their Ighb alleles, demonstrating that feedback signals inhibiting this last step in recombination were no less efficient in the absence of Eμ.
Both VHΔa/WTb and VHEμa/WTb mice contained rare pre-B cells with V-DJ rearrangement on their Ighb alleles. Sequence analyses of these rearrangements revealed abundant N nucleotides in the V-D junction (Table S2), suggesting that they occurred in pro-B cells, the only developmental stage at which terminal transferase (TdT) is expressed (27, 33). Chromatin remodeling studies have documented that pro-B cells are the only developing B cells with contracted Igh alleles facilitating V-DJ rearrangements, especially those involving distal VH gene segments (e.g., VHJ558) (34). VH J558-DJ rearrangements on the Ighb allele were easily detected in pre-B cells of both VHΔa/WTb and VHEμa/WTb mice. These results suggest that subsequent to VH gene assembly, there is sufficient delay in the assembly of, or signaling through, the preBCR to allow RAG-mediated rearrangements on the second allele in a minority of cells before transition to the pre-B cell stage.
Signaling through the preBCR is required not only to down-regulate the RAG genes (terminating V-DJ recombination) but also to promote cell proliferation and differentiation to the pre-B cell stage (for review see reference 31). If the reduced Igμ chain level in VHΔa/WTb pro-B cells resulted in preBCR signals that were suboptimal for promoting this transition, we might expect that precursors to double producers (expressing Igμ from two alleles, one with Eμ present) would be enriched in the pre-B cell population of VHΔa/WTb mice. Similarly, if expression of the VHΔa were delayed relative to expression of the VHEμa allele, a higher proportion of pre-B cells in VHΔa/WTb mice would carry a functionally assembled VH gene on the WTb allele, as this would be required in the absence of VHΔa allele expression for transit to the pre-B cell stage. This was not the case, however; these precursors were present at the same or close to equal numbers in VHΔa/WTb and VHEμa/WTb mice (Fig. 5 and Fig. S4). Quantitative comparisons of V-DJ assembly on the WTb allele showed no significant difference between VHΔa/WTb and VHEμa/WTb pre-B cells, and cloned VH gene sequences from the WTb allele had the same ratio of productive to nonproductive rearrangements in both pre-B cell populations. In RAG-1−/− mice, the bulk of B-lineage cells in both mice assumed a pre-B cell phenotype, further supporting the idea that signaling through the Igμ chain (preBCR) to promote the pro- to pre-B cell transition was largely unaffected by loss of Eμ and lower Igμ levels (Fig. 6).
The transition from pre-B to immature B cell requires successful assembly of a functional Ig light chain gene, resulting in replacement of the preBCR with a BCR. The BCR on immature B cells serves two opposing purposes. On the one hand, it is required for survival (BCR ablation at any B cell stage tested leads to cell death), and it has been suggested that this BCR-transmitted tonic signaling is antigen independent (positive selection) (35). On the other hand, if the BCR signal is too strong (e.g., autoreactive BCR), the cell is induced to modify the antigen specificity of the BCR through receptor editing and, failing that, is induced to die (negative selection) (for review see references 30, 31). The nature and strength of the BCR signal in immature B cells, therefore, profoundly affects cell fate. We propose that reduced Igμ levels in emerging immature BM B cells of VHΔa/WTb mice result in BCR signaling that is generally below the threshold for positive selection. In these animals, the rare cells that express both Igh alleles (double producers) would have a selective advantage because the increased tonic signaling through the BCR could then reach the required threshold for survival.
If, in the absence of Eμ, BCR density is generally not optimal for effective tonic signaling, then how do the single producers in VHΔa/WTb animals reach the immature B cell stage? Both density and signaling strength can be affected by light chain through its effects on BCR stability (the strength of heavy/light chain association) and Ag specificity. B cells starting with a disability (low μ-chain levels) would have more stringent light chain requirements than normal, needing either light chains that form a stronger and more stable association with the VHB1-8-μ chain or those that form an innocuous antigen specificity with stronger basal signaling properties. Analyses of these cells were consistent with that prediction. The IgMa+b− cells in VHΔa/WTb and VHΔa/ΔJH mice showed greater use of downstream Jκ gene segments (evidence of successive V-J rearrangements on a single Igκ allele) and included a larger subpopulation of Igλ-producing cells as compared with IgMa+b− cells from VHEμa/WTb mice (Fig. 7). Also consistent with this model was the finding that light chain editing was increased neither in the IgMa+b+ cells from VHΔa/WTb animals (μ-chain emanates from both a WT allele and the VHΔa allele) nor in the B cells of VHΔa/VHΔa mice (μ-chain emanates from two copies of the VHΔa allele).
As described in the previous paragraph, we predicted that inferior BCR signals caused by reduced Igμ in IgMa+b− cells from both VHΔa/WTb and VHΔa/ΔJH mice would lead to light chain editing. Unable to sense that a light chain gene has been successfully assembled, the cell would continue to accumulate V-J rearrangements. A correlation between underexpressed BCRs and light chain editing has been described previously (36, 37). Reciprocally, strong autoreactive BCR signals can also lead to light chain editing. In this case, the cell uses light chain editing to replace the light chain component of an undesirable BCR.
A corollary to this model is that in VHEμa/WTb mice, neither single producers nor double producers have a below-threshold signaling problem. Rather, in these mice, threshold signaling is achieved whether expression is monoallelic (single producers) or biallelic (double producers). Cells producing two different heavy chains at equally high levels, however, would be at greater risk of creating a receptor with unacceptable autoreactive properties, placing double-producers in VHEμa/WTb mice at a selective disadvantage (i.e., subject to clonal deletion) relative to single producers. It has been estimated that ∼75% of newly arising immature B cells harbor an autoreactive BCR (38–40). A cell producing two different heavy chains, each combining with a single unique light chain would have, on average, only a 6% chance (as compared with the usual 25%) of avoiding expression of an autoreactive BCR. Moreover, autoantibody silencing through formation of an alternate light chain (light chain editing) would be compromised in such a cell because a new light chain might solve the problem with one of the heavy chains and yet create a new autoreactive receptor with the other.
In summary, feedback inhibition of V-DJ rearrangement on the WTb allele is evident in the pre-B cells of both VHΔa/WTb and VHEμa/WTb mice. Despite this inhibition, VHΔa/WTb and VHEμa/WTb mice have equal or close to equal numbers of pre-B cells that harbor two functionally assembled Igh alleles. Potential antigen specificity for both single and double producers is exactly the same in both animals after assembly with randomly generated Ig light chains. Nevertheless, the newly arising B cells differ significantly between these two animals, with allelic exclusion compromised only in the VHΔa/WTb mice. The ultimate effect on the mature B cell pool is profound. As described and shown in Fig. 3 (and Fig. S2), the ∼5% double producers among immature B cells in VHΔa/WTb mice expanded to comprise ∼20% of splenic B cells and ∼50% of B cells in the peritoneal cavity. We suggest that the difference in outcome in the VHΔa/WTb and VHEμa/WTb mice (the immature B cells arising/surviving from the precursors) is a result of selection pressures, both positive and negative, that favor double producers only in VHΔa/WTb mice. Allelic exclusion with respect to the Igh locus, therefore, can be viewed as, at minimum, a two-step process involving both a largely Eμ-independent (feedback inhibition of V-DJ recombination) and an Eμ-dependent (pre-B to immature B selection) phase. More generally, these results suggest that one of Eμ's important functions is to ensure that monoallelic expression of most newly assembled IgH genes is sufficient to support both B cell development and entry into the mature B cell pool. Anything that reduces that expression (variants in promoter sequences, enhancer sequences, or signaling components) puts the system at risk for promoting the development of allelically included cells with potentially autoreactive properties.
Unlike cells expressing two different light chains in which one is generally expressed on the surface to the exclusion of the other (41, 42), the mature B cells expressing two different heavy chains in VHΔa/WTb mice can be expected to express both types of receptor on the cell surface at comparable levels. It is likely of significance to issues of autoimmunity that expression of the VHΔa allele is low at the time of selection but high once cells reach the mature B cell compartment. Notably, many of the IgMa+b+ double producers in these animals occupy the marginal zone of spleen and constitute most, if not all, CD5+ B1 B cells in the peritoneal cavity. It is not clear at this point how precursors of IgMa+b+ double producers are selected into these mature B cell compartments. One possibility is that expressing the WTb allele fulfills the antigen-specificity requirements associated with B1 B cell development (43). As CD5+ B1cells have been shown to require unusually strong and often autoreactive BCR signals for survival (43), it is also reasonable to speculate that monoallelic expression of the VHΔa allele, even with the assistance of light chain editing, is insufficient to signal development and/or maintenance of this B cell subset, leading to selective expansion of the double producers into this B cell pool.
MATERIALS AND METHODS
Generating VHΔ mice
To generate the B1-8VDJΔEμ targeting vector, K. Rajewsky (Harvard Medical School, Boston, MA) and W. Muller (University of Manchester, Manchester, England, UK) provided us with a vector (B1-8iVDJ) (12) that we modified to replace the 3′ homology arm with a 1.4-kb sequence between Eμ and Sμ (nt 3880–5295, GenBank accession no. J00440). Both the B1-8VDJΔEμ and B1-8iVDJ targeting vectors included ∼2-kb natural 5′ flanking DNA to the B1-8VH gene, and the 5′ homology arm was comprised of 9 kb of DNA lying immediately upstream of DHQ52 in the murine Igh locus. B1-8VH coding sequences carried a TGT to TGC silent mutation at codon 92 to prevent VH replacement events (12). The ClaI–NotI fragment, serving as 3′homology arm in the B1-8iVDJΔEμ targeting vector, was generated by PCR, using DNA from the 129P2/OlaHsd-derived ES (E14.1) cell line as template. Primers used were the following: 5′-CTCATCGATTCGGTTGAACATGCTGGTTG-3′ (ClaI site underlined) and 5′-CTCGCGGCCGCAGTGTAGGCAGTAGAGTTTA-3′ (NotI site underlined).
The B1-8VDJΔEμ targeting vector was used to transfect ES cell line E14.1, and three appropriately targeted clones were used to generate chimeric mice (Gene Targeting and Transgenic Service Laboratories, Rockefeller University). Successful germline transmission was achieved from one of the lines. Positive offspring that were heterozygous for the neoRVHΔ allele were mated to EIIa-cre mice (C57BL/6J background; provided by H. Westphal, National Institutes of Health, Bethesda, MD) (25) to induce deletion of the neomycin resistance gene. The resulting mouse line was designated VHΔ.
VHEμ mice were previously described as B1-8i mice (12) (supplied by K. Rajewsky, CBR Institute for Biomedical Research, Boston, MA). Mice carrying Igh alleles that lacked the JH gene segments were obtained from The Jackson Laboratory (B6.129P2-Igh-Jtm1Cgn/J; stock #002438). The mutant allele lacking the JH gene segments is called ΔJH in the present studies.
The VH knockin for VHEμ was also accomplished in ES 14.1 cells. Both VHEμ and VHΔ mice were mated to C57BL/6J mice for allelic exclusion analyses. Progeny of intercrosses between VHEμa/WTb and VHΔa/WTb mice were also analyzed, and double producers were strictly correlated with the VHΔ allele. Mice were bred and maintained in animal facilities at Hunter College, City University of New York, and all mouse experiments were approved by the Hunter College Institutional Animal Care and Use Committee.
Southern blots
Genomic Southern blot analyses were performed as previously described (44). ∼20 μg of restriction enzyme–digested genomic DNAs were size fractionated on 0.8% agarose gels and the DNA was transferred to nylon membrane (GE Healthcare). Blots were hybridized with 32P-labeled probes generated by the random priming method (MegaPrime; GE Healthcare). Probe A (Fig. 1) is a 1.2-kb XhoI–HindIII fragment cloned from the B1-8iVDJ targeting vector (nt 25622029–25623228; GenBank accession no. NT_166318). Probe B (Fig. 1) is a 724-bp sequence that begins upstream of Cμ and covers CH1 and ∼1/2 of CH2 (nt 1214283–1215005; GenBank accession no. NT_114985). Probe B was generated by PCR, using E14.1 ES cell DNA as template. Primers used were the following: CμU1, 5′-CAAGGAAATAGCAGG GTGTAG-3′; and CμD1, 5′-CTTTGTTCTCGATGGTCACC-3′. Probe C (V-D interval; Fig. 4) is a 1,056-bp sequence ∼73 kb upstream of the most CH-distal D gene fragment DFL16.1 and ∼25 kb downstream of the most CH-proximal V gene fragment VH81X (nt 7471–8526; GenBank accession no. AC073553). Probe C was generated by PCR from C57BL/6 mouse genomic DNA using the following primers: E1f, 5′-CATCCAGATACAGCACTCCCTTGTGTC-3′; and E1r, 5′-GAAGGCCAGGACCAAGGATTGAATAC-3′. Probe D (Fig. 4) is a 1,469-bp sequence derived from a region ∼70 kb 3′ of the hs4 element within the Igh 3′ RR (nt 25343884–25345352; GenBank accession no. NT_166318). Probe D was generated by PCR from C57BL/6J mouse genomic DNA with the following primers: Df, 5′-CTGAAGTTGGATGTAGGCCTGAAACTG-3′; and Dr, 5′-CCTCCCAATGCTAAGTAGAAACAGACG-3′. The JH4 probe is a 156-bp probe for JH4 (nt 134365–134520; GenBank accession no. AC073553) that is used to detect DJ and VDJ rearrangements on the Ighb allele. The JH4 probe was generated by PCR from C57BL/6 mouse germline DNA with the following primers: JH4f, 5′-CTATGGACTACTGGGGTCAAGGAAC-3′; and JH4r, 5′-CAACTTCTCTCAGCCGGCTC-3′. The hs4 probe is a 601-bp probe for the hs4 element of the Igh 3′ RR (nt 25418984–25419584; GenBank accession no. NT_166318).
Flow cytometry
Generally, 106 cells were incubated at 4°C for 15 min in staining buffer (PBS, 5.6 mM glucose, 0.1% BSA, and 0.1% NaN3) with monoclonal antibodies (BD or SouthernBiotech). Single-cell suspensions prepared from BM and spleen were treated with ACK buffer (0.15 M NH4Cl, 10 mM KHCO3, and 0.1 mM EDTA, pH 7.3) to lyse erythrocytes. Cells from the peritoneal cavity were recovered by flushing the cavity with cold RPMI-1640 media (Mediatech, Inc.), containing 5% bovine calf serum (Thermo Fisher Scientific). In most cases, propidium iodide was added before analyses to exclude dead cells.
Side and forward scatter were used to gate on lymphocytes. FACS instruments were used for flow cytometry and sorting (FACScan, FACSCalibur, and FACSVantage; BD). In some cases, BM B lineage cells were first enriched by positive selection (MACS B220 MicroBeads kit; Miltenyi Biotec). Data were acquired with CellQuest or Pro CellQuest (FACS instruments) and then further analyzed with FlowJo software (Tree Star, Inc.).
Monoclonal antibodies were obtained from BD, except where indicated, and conjugated to fluorescein-isothiocyanate (anti–mouse CD3-ε, anti-IgM, anti–mouse IgMb, anti-IgD, anti–mouse CD21, and anti–mouse Igκ), phycoerythrin (anti–mouse IgMa, anti–mouse CD23, anti–mouse CD43, and anti–mouse Igλ [SouthernBiotech]), allophycocyanin (anti–mouse B220 and anti–mouse CD5), or biotin (anti–mouse IgMb, anti–mouse IgMa, and anti–mouse IgD [Southern Biotech]). Cells were then washed in washing buffer (PBS, 5.6 mM glucose, and 0.1% NaN3) and biotin-conjugated monoclonal antibodies were revealed with streptavidin-allophycocyanin (BD) or streptavidin-phycoerythrin (BD).
Cytoplasmic Igμ staining
BM cells were incubated with antibodies to surface antigens, washed, and then fixed and stained with rat anti–mouse IgM (eBioscience) using Cytofix/Cytoperm (BD).
PCR
Genotyping mice.
All genotyping PCR reactions were performed for 40 cycles with an annealing temperature of 60°C. PCR kits with HotStar Taq polymerase were used (QIAGEN), and the manufacturer's protocol was adhered to. Primers used were the following: number 2, 5′-CAGAGGGAGTTCACACAGAGCATG-3′ (within the 3′ homology region of B1-8iVDJΔEμ; nt 136031–136054; GenBank accession no. AC073553); number 4, 5′-TCTTTACAGTTACTGAGCACACAGGAC-3′ (immediately 5′ of the leader exon of B1-8VH; nt. 312629–312655; GenBank accession no. BN000872); and number 8 (Eμ), 5′-CTTCCCTCTGATTATTGGTCTCCATTC-3′ (nt 135733–135759; GenBank accession no. AC073553). These three primers were used together. Numbers 2 and 4 generate an 848-bp product from the targeted Igha locus, and numbers 2 and 8 generate a 322-bp product from the WT Ighb locus. The PCR products generated by these three primers (numbers 4, 8, and 2) overlap the region of VDJ insertion.
Confirming neoR deletion.
Primers used were the following: number 9, 5′-CCCACCATCACAGACCTTTCTCCATAG-3′ (within the 5′ homology region of the targeting vectors; nt 132108–132134; GenBank accession no. AC073553); and number 10, 5′-CTGAGGGCAGCAGTACAATGATGAGTC-3′ (within the 5′ flank of B1-8VH and ∼280 bp 3′ of loxP-flanked neoR; primer differs by two nucleotides [underlined] from nt 311488–311514; GenBank BN000872). Primers number 9 and 10 can generate a product only from the targeted Igha allele. The PCR product from the neoRVHΔa allele was ∼1,500 bp before neoR deletion and, from the VHΔa allele after neoR deletion, was 400 bp.
Detecting D-J and V-DJ rearrangements in B-lineage cells.
To analyze V-DJ rearrangements in isolated B-lineage cells, a previously described protocol (7) was followed with slight modification. Genomic DNA template was isolated from sorted cells, either by conventional procedures or by the following method: cells were lysed in 10 mM Tris-HCl, pH 8.0, and 0.1 mM EDTA (50 μl/105 cells), proteinase K was added to a concentration of 0.5 mg/ml, and the mixture was incubated for 2.5 h at 50 and 95°C for 10 min. Serial dilutions of the purified DNA (or cell lysates) were used as a template to specifically amplify D-J and V-DJ rearrangements derived only from WT Igh alleles. Allele specificity was achieved by using a 3′ primer that derived from sequences 3′ of JH4 and that were missing on the targeted Igha allele of both VHEμ and VHΔ mice. The 5′ primers were “degenerate” primers designed to anneal to the DH genes, one of three VH gene families (J558, Q52, or 7183 family), or all VH genes. Primers for DH families were the following: The primer for DHL, 5′-GGAATTCGMTTTTTGTSAAGGGATCTACTACTGTG-3′, is a degenerate primer that anneals to most murine DH sequences (45); and the primer for DHQ52, 5′-CCACAGGCTCGAGAACTTTAGCG-3′, anneals to the most JH-proximal murine DH sequence, DQ52 (7). Primers for VH families were the following: VHJ558 family primer, 5′-GCGAAGCTTARGCCTGGGRCTTCAGTGAAG-3′ (7); VHQ52 family primer, 5′-GCCAAGCTTCTCACAGAGCCTGTCCATCAC-3′ (7); VH7183 family primer, 5′-GCGAAGCTTGTGGAGTCTGGGGGAGGCTTA-3′ (7); and promiscuous VH primer, 5′-GGGAATTCGAGGTGCAGCTGCAGGAGTCTGG-3′ (46). Each of these primers was used in conjunction with the following primer, which lies 3′ of JH4 and cannot anneal to the targeted Igha alleles: 5′-AGGCTCTGAGATCCCTAGACAG-3′ (nt 134530–134551; GenBank accession no. AC073553). PCR products were size fractionated on 1.5% agarose gels, blotted, and probed with a 32P-labeled JH4 probe (described in Southern blots). DNA amounts were normalized to a PCR product derived from the Igh locus 3′ RR element hs4 present as one copy/haploid genome in all cells (as in reference 7). HS4 primers used were the following: 5′-CCAAAAATGGCCAGGCCTAGG-3′ (nt 25419564–25419584; GenBank accession no. NT_166318); and HS4-3′, 5′-AGGTCTACACAGGGGCTCTG-3′ (nt 25418984–25419003; GenBank accession no. NT_166318). PCR conditions for detecting V-DJ and D-J rearrangements were 30 cycles at 94°C for 1 min, 60°C for 1 min, and 72°C for 3 min. The exceptions were for data shown in Fig. 5 (E and F) and Fig. S2. In these instances, HS4 was amplified for 20 cycles instead of 30.
Vκ-Jκ rearrangements.
The analysis of Vκ-Jκ rearrangements was the same as for IgH VDJ rearrangements except that different primers and probes were used. A degenerate Vκ primer (VκD) (28) was used to detect all Vκ genes, and a primer downstream of Jκ5 (Jκ5e) was used to detect rearrangements, regardless of Jκ fragment used. Primers used were the following: VκD, 5′-GGCTGCAGSTTCAGTGGCAGTGGRTCWGGRAC-3′ (a degenerate primer that anneals to most Vκ gene sequences) (28); Jκ5e, 5′-CTGACACTGTATGCCACGTCAACTG-3′ (downstream of Jκ5; nt 3168393–3168418; Gene ID: 243469; derived from GenBank NC_000072). The Jκ5 probe used for detecting PCR products was generated by PCR, using the following primer pair: Jκ5f, 5′-GCTCACGTTCGGTGCTGGGAC-3′ (nt 3168251–3168271; Gene ID: 243469; derived from GenBank NC_000072); and Jκ5r, 5′-ATAATGAGCCCTCTCCATTTTCTCAAG-3′ (nt 3168367–3168393; Gene ID: 243469; derived from GenBank NC_000072).
Real-time RT-PCR
For analyses of Igμ transcripts in splenic B cells, B cells were enriched by negative selection (B cell isolation kit; Miltenyi Biotech). For analyses of Igμ transcripts in pre-B cells, enrichment was by positive selection for B220+ cells (B220 MicroBeads kit; Miltenyi Biotech). Total RNA was isolated with the RNeasy mini-prep kit (QIAGEN). Real-time RT-PCR analyses of Igμ mRNA transcribed from the VHΔ and VHEμa alleles were performed with the QuantiTect SYBR Green RT-PCR kit (QIAGEN). Igμ transcripts were amplified with a 5′ primer that annealed to the unique VDJ junction sequence of VHB1-8 and a 3′ primer that annealed to a sequence within the CH1 exon of Cμ. The VDJ junction primer used was 5′-CGCAAGATACGATTACTACGG-3′ and the Cμ primer used was 5′-GAAGACATTTGGGAAGGACTG-3′ (nt 140198–140218; GenBank accession no. AC073553). Samples were normalized using primers and probes for mRNA from the housekeeping gene hgprt1 (hypoxanthine/guanine phosphoribosyl transferase; Applied Biosystems; TaqMan gene expression assay ID: Mm03024075_m1) and the TaqMan one-step RT-PCR kit (Applied Biosystems). PCR reactions were performed on the 7500 Real-Time PCR System (Applied Biosystems) and analyzed with a program supplied by Applied Biosystems (User bulletin #2; http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf).
Online supplemental material.
Additional results show PCR-cloning results demonstrating that surface Ig phenotype correlated with cloned IgH genes from sorted spleen cells. Fig. S1 shows evidence of CRE-mediated neoR deletion. Fig. S2 shows that double producers were enriched among marginal zone B cells in spleen and B1 B cells in peritoneal cavity. Fig. S3 shows analyses of newborn spleen and fetal liver cells for double producers. Fig. S4 shows establishment of quantification in VH gene assembly PCR experiments. Fig. S5 shows pie charts quantifying relative use of Jκ segments in Vκ-Jκ rearrangements in WT versus mutant mice. Table S1 shows absolute numbers of B cell subsets in BM and B and T cells of spleen in mutant and WT mice. Table S2 shows DNA sequences of assembled VH genes cloned from WT and mutant mice. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20081202/DC1.
Supplementary Material
Acknowledgments
We are grateful to Dr. Klaus Rajewsky and Dr. Werner Muller for providing the VHEμ mice and B1-8i targeting vector and to Dr. Heiner Westphal for providing the EIIa-CRE mice. We gratefully acknowledge Dr. Kyoko Hayakawa, Dr. Christopher Roman, and Dr. Harinder Singh for helpful discussions and critical readings of the manuscript. We thank Mr. Joon Kim of the Hunter College Flow Cytometry Facility for expert help with cell sorting.
This work was supported by National Institutes of Health grant AI30653 to L.A. Eckhardt. The infrastructure and instrumentation at Hunter College are supported in part by a Research Centers in Minority Institutions award from the National Institutes of Health (RR-0307).
The authors have no conflicting financial interests.
Abbreviations used: 3′ RR, 3′ regulatory region; BCR, B cell receptor; ES, embryonic stem; IgH, Ig heavy chain; mRNA, messenger RNA.
References
- 1.Banerji, J., L. Olson, and W. Schaffner. 1983. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy-chain genes. Cell. 33:729–740. [DOI] [PubMed] [Google Scholar]
- 2.GillIes, S.D., and S. Tonegawa. 1983. Expression of cloned immunoglobulin genes introduced into mouse L cells. Nucleic Acids Res. 11:7981–7997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neuberger, M.S. 1983. Expression and regulation of immunoglobulin heavy chain genes transfected into lymphoid cells. EMBO J. 2:1373–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, J., F. Young, A. Bottaro, V. Stewart, R.K. Smith, and F.W. Alt. 1993. Mutations of the intronic IgH enhancer and its flanking sequences differentially affect accessibility of the JH locus. EMBO J. 12:4635–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serwe, M., and F. Sablitzky. 1993. V(D)J recombination in B cells is impaired but not blocked by targeted deletion of the immunoglobulin heavy chain intron enhancer. EMBO J. 12:2321–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakai, E., A. Bottaro, L. Davidson, B.P. Sleckman, and F.W. Alt. 1999. Recombination and transcription of the endogenous Ig heavy chain locus is effected by the Ig heavy chain intronic enhancer core region in the absence of the matrix attachment regions. Proc. Natl. Acad. Sci. USA. 96:1526–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perlot, T., F.W. Alt, C.H. Bassing, H. Suh, and E. Pinaud. 2005. Elucidation of IgH intronic enhancer functions via germ-line deletion. Proc. Natl. Acad. Sci. USA. 102:14362–14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Afshar, R., S. Pierce, D. Bolland, A. Corcoran, and E.M. Oltz. 2006. Regulation of IgH gene assembly: role of the intronic enhancer and 5′DQ52 region in targeting DHJH recombination. J. Immunol. 176:2439–2447. [DOI] [PubMed] [Google Scholar]
- 9.Jung, D., C. Giallourakis, R. Mostoslavsky, and F.W. Alt. 2006. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 24:541–570. [DOI] [PubMed] [Google Scholar]
- 10.Burnet, F.M. 1976. A modification of Jerne's theory of antibody production using the concept of clonal selection. CA Cancer J. Clin. 26:119–121. [DOI] [PubMed] [Google Scholar]
- 11.Wabl, M., and C. Steinberg. 1982. A theory of allelic and isotypic exclusion for immunoglobulin genes. Proc. Natl. Acad. Sci. USA. 79:6976–6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sonoda, E., Y. Pewzner-Jung, S. Schwers, S. Taki, S. Jung, D. Eilat, and K. Rajewsky. 1997. B cell development under the condition of allelic inclusion. Immunity. 6:225–233. [DOI] [PubMed] [Google Scholar]
- 13.Alt, F.W., G.D. Yancopoulos, T.K. Blackwell, C. Wood, E. Thomas, M. Boss, R. Coffman, N. Rosenberg, S. Tonegawa, and D. Baltimore. 1984. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 3:1209–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Storb, U. 1987. Transgenic mice with immunoglobulin genes. Annu. Rev. Immunol. 5:151–174. [DOI] [PubMed] [Google Scholar]
- 15.Mostoslavsky, R., F.W. Alt, and K. Rajewsky. 2004. The lingering enigma of the allelic exclusion mechanism. Cell. 118:539–544. [DOI] [PubMed] [Google Scholar]
- 16.Stall, A.M., F.G. Kroese, F.T. Gadus, D.G. Sieckmann, L.A. Herzenberg, and L.A. Herzenberg. 1988. Rearrangement and expression of endogenous immunoglobulin genes occur in many murine B cells expressing transgenic membrane IgM. Proc. Natl. Acad. Sci. USA. 85:3546–3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe, N., S. Nisitani, K. Ikuta, M. Suzuki, T. Chiba, and T. Honjo. 1999. Expression levels of B cell surface immunoglobulin regulate efficiency of allelic exclusion and size of autoreactive B-1 cell compartment. J. Exp. Med. 190:461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark, M.R., A.B. Cooper, L.D. Wang, and I. Aifantis. 2005. The pre-B cell receptor in B cell development: recent advances, persistent questions and conserved mechanisms. Curr. Top. Microbiol. Immunol. 290:87–103. [DOI] [PubMed] [Google Scholar]
- 19.Martensson, I.L., R.A. Keenan, and S. Licence. 2007. The pre-B-cell receptor. Curr. Opin. Immunol. 19:137–142. [DOI] [PubMed] [Google Scholar]
- 20.Papavasiliou, F., M. Jankovic, S. Gong, and M.C. Nussenzweig. 1997. Control of immunoglobulin gene rearrangements in developing B cells. Curr. Opin. Immunol. 9:233–238. [DOI] [PubMed] [Google Scholar]
- 21.Cockerill, P.N., M.H. Yuen, and W.T. Garrard. 1987. The enhancer of the immunoglobulin heavy chain locus is flanked by presumptive chromosomal loop anchorage element. J. Biol. Chem. 262:5394–5397. [PubMed] [Google Scholar]
- 22.Forrester, W.C., C. van Genderen, T. Jenuwein, and R. Grosschedl. 1994. Dependence of enhancer-mediated transcription of the immunoglobulin mu gene on nuclear matrix attachment regions. Science. 265:1221–1225. [DOI] [PubMed] [Google Scholar]
- 23.Reth, M., G.J. Hammerling, and K. Rajewsky. 1978. Analysis of the repertoire of anti-NP antibodies in C57BL/6 mice by cell fusion. I. Characterization of antibody families in the primary and hyperimmune response. Eur. J. Immunol. 8:393–400. [DOI] [PubMed] [Google Scholar]
- 24.Bothwell, A.L., M. Paskind, M. Reth, T. Imanishi-Kari, K. Rajewsky, and D. Baltimore. 1981. Heavy chain variable region contribution to the NPb family of antibodies: somatic mutation evident in a gamma 2a variable region. Cell. 24:625–637. [DOI] [PubMed] [Google Scholar]
- 25.Lakso, M., J.G. Pichel, J.R. Gorman, B. Sauer, Y. Okamoto, E. Lee, F.W. Alt, and H. Westphal. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA. 93:5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.ten Boekel, E., F. Melchers, and A. Rolink. 1995. The status of Ig loci rearrangements in single cells from different stages of B cell development. Int. Immunol. 7:1013–1019. [DOI] [PubMed] [Google Scholar]
- 27.Victor, K.D., K. Vu, and A.J. Feeney. 1994. Limited junctional diversity in kappa light chains. Junctional sequences from CD43+B220+ early B cell progenitors resemble those from peripheral B cells. J. Immunol. 152:3467–3475. [PubMed] [Google Scholar]
- 28.Schlissel, M.S., and D. Baltimore. 1989. Activation of immunoglobulin kappa gene rearrangement correlates with induction of germline kappa gene transcription. Cell. 58:1001–1007. [DOI] [PubMed] [Google Scholar]
- 29.Casellas, R., Q. Zhang, N.Y. Zheng, M.D. Mathias, K. Smith, and P.C. Wilson. 2007. Igκ allelic inclusion is a consequence of receptor editing. J. Exp. Med. 204:153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nemazee, D. 2006. Receptor editing in lymphocyte development and central tolerance. Nat. Rev. Immunol. 6:728–740. [DOI] [PubMed] [Google Scholar]
- 31.Monroe, J.G., and K. Dorshkind. 2007. Fate decisions regulating bone marrow and peripheral B lymphocyte development. Adv. Immunol. 95:1–50. [DOI] [PubMed] [Google Scholar]
- 32.Gu, H., Y.R. Zou, and K. Rajewsky. 1993. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell. 73:1155–1164. [DOI] [PubMed] [Google Scholar]
- 33.Li, Y.S., K. Hayakawa, and R.R. Hardy. 1993. The regulated expression of B lineage associated genes during B cell differentiation in bone marrow and fetal liver. J. Exp. Med. 178:951–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roldan, E., M. Fuxa, W. Chong, D. Martinez, M. Novatchkova, M. Busslinger, and J.A. Skok. 2005. Locus ‘decontraction’ and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat. Immunol. 6:31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bannish, G., E.M. Fuentes-Panana, J.C. Cambier, W.S. Pear, and J.G. Monroe. 2001. Ligand-independent signaling functions for the B lymphocyte antigen receptor and their role in positive selection during B lymphopoiesis. J. Exp. Med. 194:1583–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braun, U., K. Rajewsky, and R. Pelanda. 2000. Different sensitivity to receptor editing of B cells from mice hemizygous or homozygous for targeted Ig transgenes. Proc. Natl. Acad. Sci. USA. 97:7429–7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kouskoff, V., G. Lacaud, K. Pape, M. Retter, and D. Nemazee. 2000. B cell receptor expression level determines the fate of developing B lymphocytes: receptor editing versus selection. Proc. Natl. Acad. Sci. USA. 97:7435–7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wardemann, H., S. Yurasov, A. Schaefer, J.W. Young, E. Meffre, and M.C. Nussenzweig. 2003. Predominant autoantibody production by early human B cell precursors. Science. 301:1374–1377. [DOI] [PubMed] [Google Scholar]
- 39.Wardemann, H., J. Hammersen, and M.C. Nussenzweig. 2004. Human autoantibody silencing by immunoglobulin light chains. J. Exp. Med. 200:191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wardemann, H., and M.C. Nussenzweig. 2007. B-cell self-tolerance in humans. Adv. Immunol. 95:83–110. [DOI] [PubMed] [Google Scholar]
- 41.ten Boekel, E., F. Melchers, and A.G. Rolink. 1998. Precursor B cells showing H chain allelic inclusion display allelic exclusion at the level of pre-B cell receptor surface expression. Immunity. 8:199–207. [DOI] [PubMed] [Google Scholar]
- 42.Liu, S., M.G. Velez, J. Humann, S. Rowland, F.J. Conrad, R. Halverson, R.M. Torres, and R. Pelanda. 2005. Receptor editing can lead to allelic inclusion and development of B cells that retain antibodies reacting with high avidity autoantigens. J. Immunol. 175:5067–5076. [DOI] [PubMed] [Google Scholar]
- 43.Hardy, R.R., and K. Hayakawa. 2005. Development of B cells producing natural autoantibodies to thymocytes and senescent erythrocytes. Springer Semin. Immunopathol. 26:363–375. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, B., A. Alaie-Petrillo, M. Kon, F. Li, and L.A. Eckhardt. 2007. Transcription of a productively rearranged Ig VDJCalpha does not require the presence of HS4 in the Igh 3′ regulatory region. J. Immunol. 178:6297–6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlissel, M.S., L.M. Corcoran, and D. Baltimore. 1991. Virus-transformed pre-B cells show ordered activation but not inactivation of immunoglobulin gene rearrangement and transcription. J. Exp. Med. 173:711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kantor, A.B., C.E. Merrill, L.A. Herzenberg, and J.L. Hillson. 1997. An unbiased analysis of V(H)-D-J(H) sequences from B-1a, B-1b, and conventional B cells. J. Immunol. 158:1175–1186. [PubMed] [Google Scholar]
- 47.Schlissel, M., A. Voronova, and D. Baltimore. 1991. Helix-loop-helix transcription factor E47 activates germ-line immunoglobulin heavy chain gene transcription and rearrangement in a pre-T cell line. Genes Dev. 5:1367–1376. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.