Abstract
End joining of double-strand breaks (DSBs) requires Ku proteins and frequently involves base pairing between complementary terminal sequences. To define the role of terminal base pairing in end joining, two oppositely oriented HO endonuclease cleavage sites separated by 2.0 kb were integrated into yeast chromosome III, where constitutive expression of HO endonuclease creates two simultaneous DSBs with no complementary end sequence. Lack of complementary sequence in their 3′ single-strand overhangs facilitates efficient repair events distinctly different from when the 3′ ends have a 4-bp sequence base paired in various ways to create 2- to 3-bp insertions. Repair of noncomplementary ends results in a set of nonrandom deletions of up to 302 bp, annealed by imperfect microhomology of about 8 to 10 bp at the junctions. This microhomology-mediated end joining (MMEJ) is Ku independent, but strongly dependent on Mre11, Rad50, and Rad1 proteins and partially dependent on Dnl4 protein. The MMEJ also occurs when Rad52 is absent, but the extent of deletions becomes more limited. The increased gamma ray sensitivity of rad1Δ rad52Δ yku70Δ strains compared to rad52Δ yku70Δ strains suggests that MMEJ also contributes to the repair of DSBs induced by ionizing radiation.
Ionizing radiation and radiomimetic chemicals induce a variety of DNA lesions, the most lethal of which is the DNA double-strand break (DSB). Eukaryotes have thus evolved a number of different mechanisms to repair DSBs (19). Two types of mechanisms for repairing DSBs are homologous recombination (56), wherein the break is repaired by recombination between homologous sequences; and nonhomologous end joining (NHEJ), which involves the rejoining of DNA ends by ligation (31). In Saccharomyces cerevisiae, three different mechanisms of homologous recombination have been described: gene conversion, break-induced replication (BIR), and single-strand annealing (SSA), all of which are dependent on a set of genes known as the RAD52 epistasis group (43). Homologous recombination initiates with the resection of DSB ends by a 5′-to-3′ exonuclease to produce long 3′-ended single-stranded DNA (ssDNA), which in gene conversion or BIR will invade a homologous donor sequence and act as a primer of new DNA synthesis (43). In SSA, complementary strands of homologous regions flanking a DSB can anneal, producing an intermediate that has two nonhomologous 3′-end tails that must be removed before new DNA synthesis and ligation can occur (54).
The second pathway is NHEJ, also called illegitimate recombination, in which little or no homology is required to join the ends of the DNA (35). At least two types of end-joining reactions, both of which are independent of RAD52 but dependent on the Ku proteins, have been described. One pathway is the precise joining of short overhanging, complementary ends, such as those produced by restriction endonucleases (1, 29). This is a highly efficient process, whereby most ends are successfully rejoined without any alteration of the DNA information (1, 29). However, when the ends are not complementary or when the continued presence of an endonuclease precludes precise religation, the break can be joined by an alternative, imprecise NHEJ pathway (38). The imprecise NHEJ mechanism involves alignment of overhanging ends by pairing of as few as 1 bp, followed by gap filling by a DNA polymerase or trimming of a few bases, resulting in the ligation of ends with either the insertion or the deletion of a few base pairs (38). In budding yeast, imprecise NHEJ is relatively inefficient, allowing only about 1 in 1,000 cells to survive (38).
NHEJ in yeast and mammals requires gene products that are distinct from those needed for homologous recombination. In all organisms studied, NHEJ requires the heterodimeric DNA binding proteins Ku70 and Ku80 (Yku70p and Yku80p in budding yeast), and the DNA ligase IV and the associated factor XRCC4 (Dnl4p and Lif1p, respectively, in budding yeast) (6). In S. cerevisiae, the Rad50p-Mrellp-Xrs2p complex also plays an essential role in promoting intermolecular DNA joining by Dnl4p-Lif1p (4). The product of the NEJ1 gene, whose transcription is strongly regulated by yeast mating type, facilitates the nuclear localization of Lif1p (13, 25, 41, 67, 70). Pol4p and Fen1p/Rad27p are required for DNA polymerization or removal of 5′-flap intermediates, respectively, during the end-processing steps of NHEJ when the terminal bases are damaged or not fully compatible for simple ligation (65, 72, 73).
In addition, substantial evidence exists for an alternative end-joining mechanism that is Ku and Rad52 independent (11, 26, 34, 74). Although very little is known about the factors required for the Ku- and Rad52-independent DSB repair process, the repair events that occur by this process appear dependent on DNA microhomologies and are frequently associated with large deletions (11, 34, 74).
Many studies on DSB repair rely on the ability to create site-specific strand breaks on a plasmid or chromosomes either by transformation of restriction endonuclease-cleaved plasmids or by the induction of HO or I-SceI endonuclease in vivo (1, 29, 32, 34, 70). The DSBs caused by these agents have the terminal ends with either 5′ or 3′ 4-bp overhangs that can reanneal by their complementary sequences. These studies uncovered the use of complementary base pairing (even as few as 1 bp) of the ends for alignment prior to ligation as the distinct feature of Ku-dependent NHEJ (38, 72). In vivo, however, cells may frequently encounter breaks whose ends are incompatible for alignment by any base pairing. Many DSB causative agents such as ionizing radiation or oxygen free radicals, as well as various forms of endogenous damage, are in fact believed to generate DSBs with a diverse set of biochemical end configurations (35). The key question then is whether the repair of breaks with no complementary end sequences occurs by the same mechanisms formulated from studies using the ends with the complementary sequences.
To address this question, we have developed a system that can induce unique DSBs in vivo with no complementary end sequences. Surprisingly, the repair of DSBs without complementary end sequence occurs by a mechanism distinctly different from that which operates for the repair of complementary overhanging ends. The repair of such breaks involves joining by base pairing between opposite single strands, yielding deletions with short stretches of homology (microhomology). We discovered that the microhomology-mediated end joining (MMEJ) is independent of Ku or Rad52 proteins, but dependent on Mre11, Rad50, and Rad1 proteins.
MATERIALS AND METHODS
Strains and plasmids.
All strains are derivatives of JKM179, which has the genotype hoΔ MATα hmlΔ::ADE1 hmrΔ::ADE1 ade1-100 leu2-3,112 lys5 trp1::hisG ura3-52 ade3::GAL::HO (29). SLY18 and SLY19 were constructed by one-step gene replacement method, replacing MATα with an EcoRI- and HindIII-cleaved MATα::URA3::HO cut site fragment of pSL18 and pSL19, respectively. The YKU70 deletion derivatives of SLY18 (EMK10) and SLY19 (EMK11) were constructed by using a PCR-derived KANMX module flanked by short terminal sequences homologous to the ends of the YKU70 open reading frame (68). The rad14::KANr strain was constructed by the same PCR-derived KANMX module. Construction of the rad52::TRP1 (EMK12) or rad1::LEU2 (EMK13) derivatives of SLY19 was done by using plasmid pJH182 (38) or pRHB113 (44), respectively.
The MRE11 (JLM30) or RAD50 (JLM33) deletion derivatives of SLY19 were made by crosses between SLY19 and SLY46 (MATa mre11::hisG) or JKM125 (MATa rad50::hisG), respectively. The DNL4 deletion derivative of SLY19 was constructed by the one-step gene deletion method by using a HindIII and SacI fragment of pJJ252 (67). The rad1 yku70 double-gene deletion derivative of SLY19 (JLM31) was constructed by a cross between EMK13 and JKM173 (MATa yku70::KANr). Other double or triple mutants were generated by crosses between each single- or double-mutant strain and then selected for the appropriate marked segregant after tetrad dissection.
Both pSL18 and pSL19 were generated from pJH220 that has had URA3 inserted at the promoter region of MATα1 and -α2 by cloning an SmaI and HincII fragment of pJH245 that contains a 117-bp MATa HO cut site into the SmaI site of URA3. The orientations of a cloned SmaI and HincII fragment in pSL18 and pSL19 are such that pSL18 has the two HO recognition sites that are in a direct orientation, whereas the two HO cut sites in pSL19 are in the opposite orientation.
HO endonuclease induction.
Cells were grown in preinduction medium (YEP-glycerol) at 30°C and then spread onto plates containing either galactose (YEPGAL) or glucose (YEPD). The frequency of survival after a HO-induced DSB was determined by dividing the number of colonies growing on YEPGAL by the number of colonies growing on YEPD (27, 29).
Determination of gamma ray sensitivity.
Cells grown in YEPD media that reach logarithmic culture (optical density at 600 nm [OD600] of ∼0.5) were plated onto YEPD plates and then irradiated with the indicated dose of gamma ray (137Cs source at a dose rate of 0.808 krad/min) and examined along with the unirradiated control plates after 3 or 4 days of growth at 30°C.
Analysis of repair junctions.
Initial analyses of repair events were carried out by checking the uracil auxotrophy and the mating phenotype of the colonies growing on YEPGAL plates by replica plating onto plates containing SC medium lacking uracil and the complementation mating phenotype test (53).
To analyze sequence of the repair junction, PCR was performed directly on yeast cells by using the following sets of primers: pX (5′-GTAAACGGTGTCCTCTGTAAGG-3′) and p2 (5′-TCGAAAGATAAACAACCTCC-3′) for amplifying Ura− survivors, pM (5′-ATGTCTAGTATGCTGGATTTAAAC-3′) and pURA3-2 (5′-GAACCGTGGATGATGTGGTCTCTA-3′) for amplifying repair junctions of the MATa cut site from Ura+ survivors, and p12(5′-CTAGCTGAGCATGTGAGGCC-3′) and pL (5′-ACATTGGGAACAAGAGCAAGACG-3) for the repair junctions of the MATα cut site from Ura+ survivors (38). All DNA sequencing was performed with an Applied Biosystems 377 DNA sequencer and dye terminator chemistry according to the manufacturer's instructions.
RESULTS
Repair of DSBs without complementary ends results in deletions.
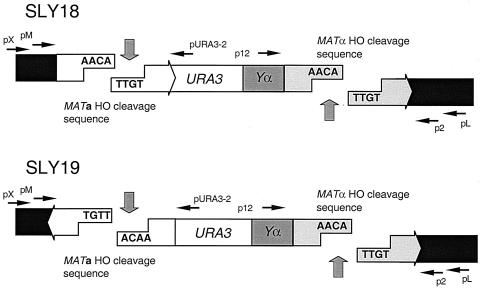
We have created yeast strains SLY18 and SLY19, in which the MAT locus of chromosome III contains two HO cleavage sites, a 117-bp segment from MATa and the normal MATα sequence, separated by ∼2.0 kb of DNA containing the 1.2-kb URA3 gene and 0.8 kb of the Yα region (Fig. 1). These strains contain a galactose-inducible HO endonuclease gene integrated at the ADE3 locus in the genome to efficiently induce two site-specific breaks simultaneously (27). Deletion of the two silent donor sequences, HML and HMR, prevents homologous recombination and restricts repair of the DSBs by DNA end-joining mechanisms (27, 29). SLY18 and SLY19 are identical except for the orientation of HO cleavage sites, such that SLY18 has two HO cleavage sites in a direct orientation, but SLY19 has the oppositely oriented HO cleavage sites (Fig. 1). Upon induction of HO endonuclease, both sites are cut simultaneously, liberating a DNA sequence containing the URA3 fragment and the Yα region, leaving complementary 4-base 3′-overhanging ends in SLY18 or 3′-overhanging ends with no complementary sequences in SLY19.
FIG. 1.
HO cleavage sites in the MAT locus of chromosome III in SLY18 and SLY19. After HO endonuclease induction, two complementary breaks are generated in SLY18, whereas in SLY19, noncomplementary breaks will be formed. The locations of primers that were used for PCR amplification and sequence analysis of the repair junctions from survivors are indicated by arrows.
We measured the survival rate of SLY18 and SLY19 strains upon induction of persistent HO endonuclease-generated DSBs by plating cells onto galactose-containing media. To survive from these breaks, cells must become HO endonuclease resistant by end-joining reactions that alter the HO recognition sequence to prevent recutting by HO endonuclease (29). As noted above, homologous recombination does not occur in these strains because of the absence of a homologous template.
SLY18 cells survived the two HO endonuclease-induced DSBs at a frequency of 1.2 × 10−4, which is approximately 10 times lower than that in an isogenic strain with a single HO-induced DSB (Table 1) (27). About 35% of survivors were Ura+; these could arise by several different mechanisms, including the imprecise NHEJ of both HO cleavage sites (described below) or, on rare occasions, situations in which DSBs were not made by HO. All of these repair events retained the sequences between the two cleavage sites, including URA3, and depended on Ku-dependent NHEJ, because the deletion of the YKU70 gene dramatically impaired cell survival in SLY18 (Table 1). Sequence analysis of 20 PCR-amplified HO cleavage sites of the Ura+ survivors indicated that all of these repair events involve either 2- or 3-bp insertions or 3-bp deletions at the HO recognition sequences, as was seen previously from Ku-dependent NHEJ of a single DSB with the same 4-bp 3′-overhanging ends (38; data not shown). Approximately 65% of the survivors became Ura− and had repaired the DSBs by joining the ends that were initially 2 kb apart.
TABLE 1.
Survival frequency after induction of persistent HO breaks
| Strain | Genotype | Frequency of survival (fold reduction)a | Frequency of survivor type
|
|
|---|---|---|---|---|
| Ura+ | Ura− | |||
| SLY18 | Wild type | (1.22 ± 0.6) × 10−4 (1) | 4.33 × 10−5 | 7.87 × 10−5 |
| EMK10 | yku70Δ | (4.87 ± 2.7) × 10−6 (25) | 4.16 × 10−6 | 7.08 × 10−7 |
| SLY19 | Wild type | (6.00 ± 1.7) × 10−4 (1) | 9.60 × 10−5 | 5.04 × 10−4 |
| EMK11 | yku70Δ | (4.43 ± 1.6) × 10−4 (1.35) | 0 | 4.43 × 10−4 |
| EMK12 | rad52Δ | (7.57 ± 2.2) × 10−4 (0.79) | 5.25 × 10−5 | 7.05 × 10−4 |
| JLM30 | mre11Δ | (4.00 ± 2.4) × 10−5 (15) | 6.40 × 10−6 | 3.36 × 10−5 |
| JLM33 | rad50Δ | (1.33 ± 0.4) × 10−4 (4.5) | 0 | 1.33 × 10−4 |
| EMK13 | rad1Δ | (1.30 ± 0.3) × 10−4 (4.6) | 2.89 × 10−5 | 1.01 × 10−4 |
| JLM31 | rad1Δ yku70Δ | (9.93 ± 4.5) × 10−6 (60.4) | 6.45 × 10−6 | 3.48 × 10−6 |
| JLM32 | dnl4Δ | (1.34 ± 0.8) × 10−4 (4.47) | 1.09 × 10−5 | 1.23 × 10−4 |
Reduction was calculated by dividing the survival frequency of each mutant strain by that of either parental strain SLY18 or SLY19. Each value represents the average from at least three independent experiments ± standard deviation.
Unexpectedly, the survival frequency of SLY19 cells was 5 times higher than that of SLY18 (6.0 × 10−4, Table 1). The majority of SLY19 survivors arising in the continuous presence of HO were Ura−, accounting for 84% of survivors. The observed difference in the survival frequency between SLY18 and SLY19 is mainly due to a 6-fold increase in the repair events that give Ura− cells (7.7 × 10−5 versus 5.0 × 10−4, respectively), although there is also a twofold increase in Ura+ events (Table 1).
To gain insight into the repair events that generate Ura− survivors in SLY18 and SLY19, we amplified the junction sequences by using a set of oligonucleotides (p2 and pX) that anneal to the regions 5′ to the 117-bp MATa HO recognition site and 3′ to the MATα HO cleavage site (Fig. 1). The PCR amplification of Ura− survivors in SLY18 indicated that this class of survivors indeed arises by repair events joining the two distant ends through imprecise NHEJ with the loss of the URA3 gene and Yα sequence. Sequence analysis of these PCR products confirmed that most of these repair events occur by imprecise NHEJ using at least a 1-bp overlap upon ligation, very similar to those seen previously for a single HO cleavage site (38; data not shown). In contrast, the PCR products from the Ura− colonies in SLY19 are frequently smaller and more heterogeneous, suggesting that they arise from a repair process different from that in SLY18. Sequence analysis of 46 PCR products from the Ura− survivors in SLY19 demonstrated that many of them are associated with the deletions of up to 302 bp. Twenty-nine of 46 of the repair events were a 62-bp deletion lacking 2 bp at the centromere proximal end and 60 bp at the other, annealed at a 9-bp sequence of imperfect homology (Table 2).
TABLE 2.
Junctional sequences observed in survivors of SLY19 and in the rad52 and yku70 gene deletion derivativesa
| Strain | Junctional sequence | Modifications | No. of overlapping bases | No. of events | |
|---|---|---|---|---|---|
| Parental | TTTATAAAATTATACTGTT | GTATAATTTTATAAACCCTGGTTTTGGTTTTGTAGA | |||
| AAATATTTTAATATG | TTGTCATATTAAAATATTTGGGACCAAAACCAAAACATCT | ||||
| SLY19 | TTTATAAAATTATA(CTGTT) | AACAGTATAATTTTATAAACCCTGGTTTTGGTTTTGTAGAGG | −5/0 | 1 | 1 |
| TTTATAAAATTATAC(TGTT) | (A) ACAGTATAATTTTATAAACCCTGGTTTTGGTTTTGTAGAG | −4/−1 | 2 | 4 | |
| TTTATAAAATTATACTGT(T) | (AACA) GTATAATTTTATAAACCCTGGTTTTGGTTTTGTAGAG | −1/−4 | 2 | 1 | |
| TTTATAAAATTATACTG(TT) | (AACAGTATAATTTTATAAACC) CTGGTTTTGGTTTTGTAGAG | −2/−21 | 3 | 1 | |
| TTTATAAAATTATA(CTGTT) | (AACAG) TATAATTTTATAAACCCTGGTTTTGGTTTTGTAGAG | −5/−5 | 4 | 2 | |
| TTTATAAAATTATA(CTGTT) | (AACAGTATAATT) TTATAAACCCTGGTTTTGGTTTTGTAGAG | −5/−12 | 10 (12) | 2 | |
| TTTATAAAATTATACTG(TT) | (AACAGTATAA---GAGTGGTTGACGAATAATTATG) CTGAAG | −2/−60 | 9 (10) | 29 | |
| TTTATAAAATTAT(ACTGTT) | (AACAGTATAA---GAGTGGTTGACGAAT) AATTATGCTGAAG | −6/−53 | 9 (10) | 5 | |
| TTTATAAAAT(TATACTGTT) | (AACAGTATAA--------GTCTATGTATTTG) TATAAAATAT | −9/−293 | 8 | 1 | |
| EMK11 (yku70Δ) | TTTATAAAATTATACTG(TT) | (AACAGTATAA---GAGTGGTTGACGAATAATTATG) CTGAAG | −2/−60 | 9 (10) | 29 |
| TTTATAAAATTAT(ACTGTT) | (AACAGTATAA---GAGTGGTTGACGAAT) AATTATGCTGAAG | −6/−53 | 9 (10) | 1 | |
| EMK13 (rad52Δ) | TTTATAAAATTATACTGT(T) | (AACA) GTATAATTTTATAAACCCTGGTTTTGGTTTTGTAGA | −1/−4 | 2 | 1 |
| TTTATAAAATTATA(CTGTT) | (AACAG) TATAATTTTATAAACCCTGGTTTTGGTTTTGTAGAG | −5/−5 | 4 | 3 | |
| TTTATAAAATTATA(CTGTT) | (AACAGTATAATT) TTATAAACCCTGGTTTTGGTTTTGTAGAG | −5/−12 | 10 (12) | 4 | |
| TTTATAA(AATTATACTGTT) | (AACAG) TATAATTTTATAAACCCTGGTTTTGGTTTTGTAGAG | −12/−5 | 10 (12) | 2 | |
| GTTTATAAA(ATTATACTGTT) | (AACAGTATAA) TTTTATAAACCCTGGTTTTGGTTTTGTAGAG | −11/−11 | 10 (12) | 8 | |
| AAAACCAGGGTTT(AT--TGTT) | (AACAGTATAATTTTATAAACCCT) GGTTTTGGTTTTGTAGAG | −16/−23 | 10 (12) | 2 | |
Shown are the nucleotide sequences at the sites of joining between two HO breaks in Ura− survivors of SLY19 and their mutant derivatives. The original sequences that constitute noncomplementary single-stranded ends are underlined. Microhomologies with overlapping sequences are shown in boldface. The deletions are shown in parentheses. Negative numbers in the “Modifications” column indicate the number of nucleotides deleted in either side of the DSBs. In cases of imperfect overlap, mismatches with several matching bases on either side were only considered as microhomologies. The number of overlapping bases is shown to indicate the total number of overlapping sequences (in parentheses) and the identical sequences among them.
Microhomology-mediated deletional end joining does not require Ku and Rad52 proteins.
To determine whether the deletion-associated repair events seen in SLY19 require Ku, we created yku70Δ derivatives of SLY18 and SLY19—EMK10 and EMK11, respectively—and examined the survival frequency upon induction of persistent HO breaks. Although the deletion of the YKU70 gene in SLY18 dramatically impaired cell survival, deletion of YKU70 in SLY19 only caused a mild reduction (1.35-fold) in cell survival (Table 1). Interestingly, all of the survivors from EMK11 became Ura−. PCR amplification and sequence analysis of the repair junctions of these survivors demonstrated that the majority of the repair events involve the same 62-bp deletion that we saw in SLY19, being mediated by 9 bp of imperfect microhomology (Table 2). Thus, this type of repair, which comprises at least 60% of all survivors in SLY19, is independent of Ku.
We then asked if the MMEJ repair events in SLY19 require Rad52-mediated SSA (54). EMK12, a rad52Δ derivative of SLY19, survived continuous HO cleavage with the same frequency as SLY19 (Table 1). Moreover, the frequency of Ura− colonies among survivors in SLY19-rad52Δ is indistinguishable from that of SLY19, indicating that RAD52 does not contribute to cell survival in this setting. Interestingly, deletion of RAD52 affects the extent of deletion, so that most of the repair events involved deletions ranging from 5 to 39 bp (Table 2). Nonetheless, the great majority of the repair events involve deletions and occur via 10 to 12 bp of imperfect microhomology.
From these data, we concluded that chromosome breaks with no complementary ends are repaired predominantly through the MMEJ pathway that is independent of Ku and Rad52.
MMEJ requires Mre11 protein.
Mre11 is a multifunctional protein that plays roles in homologous recombination, NHEJ, telomere length maintenance, and cell cycle checkpoint regulation (16). Mre11 forms a heterotrimeric complex with Rad50 and Xrs2 and exhibits double-strand 3′-to-5′ exonuclease activity as well as single-strand endonuclease activity (46, 63, 64). The Mre11 complex has also been reported to mediate end-to-end DNA association and annealing of complementary ssDNA (4, 8, 9).
To ask whether Mre11 is required for MMEJ in SLY19, we created an mre11Δ derivative of SLY19 (JLM30) and examined its survival upon induction of HO-induced DSBs. The deletion of MRE11 reduced cell survival by 15-fold (Table 1). Most mre11Δ survivors were Ura−; rare Ura+ survivors maintained the original HO recognition sequences, suggesting that they arose from inefficient HO expression. The reduction in cell survival in JLM30 cannot be solely attributed to the lack of Ku-dependent NHEJ, since deletion of YKU70 only caused a 1.3-fold reduction in survival. The results therefore indicate that Ku-independent MMEJ requires Mre11.
We characterized the repair events of the individual survivors in the absence of MRE11 (JLM30) by PCR amplification and analysis of the junctional sequences (Table 3). The most frequent type of repair in mre11Δ survivors is still the deletion of 62 bp, which was the most frequent class in SLY19. Other repair events also involve microhomologies, but with smaller deletions. Smaller deletions could be expected because of inefficient 5′-to-3′ degradation of breaks in mre11Δ cells (28).
TABLE 3.
Junctional sequences observed in survivors of the mre11, rad1, rad1 yku70, and dnl4 gene deletion derivatives of SLY19a
| Strain | Junctional sequence | Modifi- cations | No. of overlapping bases | No. of events | |
|---|---|---|---|---|---|
| Parental | TTTATAAAATTATACTGTT | GTATAATTTTATAAACCCTGGTTTTGGTTTTGTAGA | |||
| AAATATTTTAATATG | TTGTCATATTAAAATATTTGGGACCAAAACCAAAACATCT | ||||
| JLM30 (mre11Δ) | TTTATAAAATTATA(CTGTT) | (AACAG)TATAATTTTATAAACCCTGGTTTTGGTTTTGTAG | −5/−5 | 4 | 1 |
| TTTATAAAATTATA(CTGTT) | (AACAGTATAATT)TTATAAACCCTGGTTTTGGTTTTGTAG | −5/−12 | 10 (12) | 2 | |
| TTTATAA(AATTATACTGTT) | (AACAG)TATAATTTTATAAACCCTGGTTTTGGTTTTGTAG | −12/−5 | 10 (12) | 2 | |
| AAACC(AGGGTTTA--TGTT) | (AACAGTATAATTTTAT)AAACCCTGGTTTTGGTTTTGTAG | −23/−16 | 10 (12) | 4 | |
| TTTATAAAATTATACTG(TT) | (AACAGTATAA---GAGTGGTTGACGAATAATTATG)CTGA | −2/−60 | 9 (10) | 9 | |
| TTTATAAAATTAT(ACTGTT) | (AACAGTATAA---GAGTGGTTGACGAAT)AATTATGCTGA | −6/−53 | 9 (10) | 1 | |
| ACCAAAAACC(AAA--TGTT) | (AACAGTATAATTTTAT)AAACCCTGGTTTTGGTTTTGTAG | −29/−16 | 5 | 1 | |
| EMK13 (rad1Δ) | TTTATAAAATTATACTGTT | (AA)CCAGTATAATTTTATAAACCCTGGTTTTGGTTTTGTA | 0/−2,+1 | 0 | 1 |
| TTTATAAAATTATAC(TGTT) | (A)ACAGTATAATTTTATAAACCCTGGTTTTGGTTTTGTAG | −4/−1 | 2 | 7 | |
| TTTATAAAATTATACTGT(T) | (AACA)GTATAATTTTATAAACCCTGGTTTTGGTTTTGTAG | −1/−4 | 2 | 2 | |
| TTTATAAAATTATAC(TGTT) | (AACA)GTATAATTTTATAAACCCTGGTTTTGGTTTTGTAG | −4/−4 | 0 | 5 | |
| TTTATAAAATTA(TACTGTT) | AACAGTATAATTTTATAAACCCTGGTTTTGGTTTTGTAGAG | −7/0 | 1 | 1 | |
| TTTATAAAATTATA(CTGTT) | (AACAG)TATAATTTTATAAACCCTGGTTTTGGTTTTGTAG | −5/−5 | 4 | 2 | |
| AACCAG(GGTTT---CTGTT) | (AA)CAGTATAATTTTATAAACCCTGGTTTTGGTTTTGTAG | −21/−2 | 10 (12) | 1 | |
| TTTATAAA(ATTATACTGTT) | (AACAGTATAA)TTTTATAAACCCTGGTTTTGGTTTTGTAG | −11/−11 | 10 (12) | 1 | |
| JLM31 (rad1Δ yku70Δ) | TTTATAAAATTATA(CTGTT) | (AACAGTATAATT)TTATAAACCCTGGTTTTGGTTTTGTAG | −5/−12 | 10 (12) | 1 |
| TTTATAAAATTATACTG(TT) | (AACAGTATAATTTTATAAACC)CTGGTTTTGGTTTTGTAG | −2/−21 | 3 | 1 | |
| AAAACCAGGGTTT(AT--TGTT) | (AACAGTATAATTTTATAAACCCT)GGTTTTGGTTTTGTAG | −16/−23 | 10 (12) | 2 | |
| TTTATAAAATTATACTG(TT) | (AACAGTATAA---GAGTGGTTGACGAATAATTATG)CTGA | −2/−60 | 9 (10) | 3 | |
| JLM32 (dnl4Δ) | TTTATAAAATTATA(CTGTT) | (AACAGTATAATT)TTATAAACCCTGGTTTTGGTTTTGTAG | −5/−12 | 10 (12) | 1 |
| TTTATAAAATTATACTG(TT) | (AACAGTATAA---GAGTGGTTGACGAATAATTATG)CTGA | −2/−60 | 9 (10) | 17 | |
Top strand sequences at the sites of joining between two HO breaks in Ura− survivors of the mre11, rad1, rad1 yku70, and dnl4 gene deletion derivatives of SLY19 are shown, as described in Table 2. The insertions are shown in italic.
The Mre11, Rad50, and Xrs2 proteins form a stable complex, and their strong association is substantiated by the identical phenotypes of their deletion mutations (2, 3, 23). To determine whether Rad50 is needed for MMEJ, we deleted RAD50 in SLY19 (JLM33) and measured the survival frequency after induction of noncomplementary HO breaks. Deletion of the RAD50 gene caused a 4.5-fold reduction in survival, and all rad50Δ survivors were Ura−. The repair events in rad50Δ survivors are indistinguishable from those in mre11Δ (data not shown), supporting the idea that the entire Mre11-Rad50 complex is needed for MMEJ.
Roles of Rad1 and Dnl4/Lig4 in MMEJ.
Yeast Rad1 and Rad10 proteins form a stable endonuclease complex that is required for several different DNA repair pathways, including nucleotide excision repair (NER) and recombination repair (7, 55, 61). The RAD1 and RAD10 genes are also required for SSA involving repeated sequences flanking a DSB (12, 18, 59). These genetic observations suggested to us that Rad1 and Rad10 may be needed for MMEJ by removing the 3′-flap DNA.
To test this idea, we created a rad1Δ derivative of SLY19 (EMK13) and examined its survival frequency after induction of HO breaks. In the absence of RAD1, there was a 4.6-fold decrease in survival compared to SLY19 (Table 1). Although the majority of survivors were Ura−, there were many Ura+ survivors that could arise from imprecise NHEJ. To further determine the basis of survival in the absence of Rad1p, we PCR amplified and analyzed Ura− rad1Δ survivors. In the absence of the Rad1p, MMEJ disappeared; instead, cells survived the HO-induced DSBs primarily by end joining, similar to Ku-dependent NHEJ (Table 3; described below). The presence of rare survivors of the RAD1 gene deletion strain that appeared to repair DSBs by MMEJ suggests the existence of a Rad1-independent 3′-flap removal mechanism, as with SSA (5). Nevertheless, our results strongly implicate the Rad1 protein in MMEJ.
To ascertain the role of Rad1 in MMEJ, we deleted both RAD1 and YKU70 from SLY19 and induced HO endonuclease-created DSBs. We reasoned that if Ku-independent MMEJ requires Rad1 protein, the absence of both Rad1 and Yku70 should eliminate Ku-dependent and -independent end joining to cause a severe reduction in cell survival. Indeed, deletion of both RAD1 and YKU70 genes led to a synergistic 60-fold reduction in survival (Table 1). Characterization of rare survivors revealed that all have repaired the HO-induced DSB by MMEJ. These data confirm the requirement for Rad1 in MMEJ and further support the existence of a Rad1-independent, inefficient mechanism of removing 3′ flaps.
In S. cerevisiae, there are two distinct DNA ligases (50, 60). Cdc9, the essential yeast homolog of DNA ligase I is required for the joining of Okazaki fragments at the replication fork and is also involved in Rad52-dependent homologous recombination and single-strand break repair (17, 22, 37, 62). Importantly, there is in vitro evidence that ligase I may be involved in using minihomologies to join noncomplementary DNA ends (21, 45, 51). The other yeast ligase, Dnl4p, is a homolog of mammalian DNA ligase IV and functions in the NHEJ pathway of DSB repair (52, 58, 71). To identify the DNA ligase that is involved in Ku- and Rad52-independent MMEJ, we deleted the DNL4 gene in SLY19 (JLM32) and determined the survival frequency after persistent HO endonuclease-generated DSBs. The absence of Dnl4 reduced survival about 4.5-fold (Table 1). Characterization of individual survivors of the dnl4Δ version of SLY19 after persistent DSBs showed that all the Ura+ survivors were derived from inefficient HO induction, and all of the Ura− colonies were the result of the MMEJ (Table 3). The lack of Ku-dependent imprecise NHEJ events among dnl4Δ survivors is consistent with its known role in this process (52, 58, 71). In addition, the lower rate of survival of dnl4Δ cells compared to yku70Δ cells after the persistent HO breaks implicates Dnl4 in MMEJ. Nevertheless, there was much less inhibition of DSB end repair than was seen, for example, in rad1Δ yku70Δ or mre11Δ cells. The moderate MMEJ impairment in the dnl4Δ mutant indicated that another ligase (very likely Cdc9) can also function in MMEJ.
Repair of radiation damage by the Ku- and Rad52-independent mechanism requires Rad1.
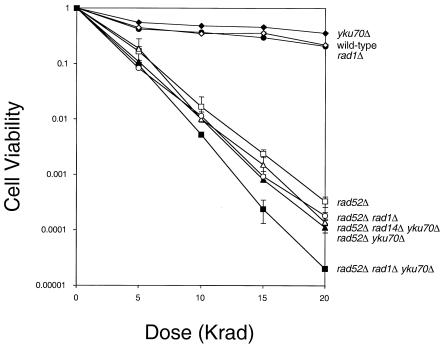
Ionizing radiation such as gamma rays can cause DSBs; hence, cell survival depends on successful DSB repair (30). Although the majority of DSBs induced by ionizing radiation are repaired through Rad52-dependent homologous recombination and Ku-dependent NHEJ, in the absence of functional Rad52 and Ku proteins, cell survival after gamma irradiation should rely on MMEJ. We thus created a rad1Δ yku70Δ rad52Δ triple-mutant strain and examined cell survival after gamma irradiation. Our results show that the triple-mutant strain is significantly more sensitive to gamma irradiation than the yku70Δ rad52Δ double-mutant strain (Fig. 2). This epistatic relationship further confirms the existence of the Ku- and Rad52-dependent DNA repair process that requires Rad1 protein. The effect of the RAD1 gene deletion on gamma-ray hypersensitivity in the absence of Rad52 and Yku70 is not due to defective NER, since the deletion of another essential component of NER, RAD14, did not affect the radiation sensitivity of yku70Δ rad52Δ cells (49) (Fig. 2).
FIG. 2.
Epistasis analyses of the effects of RAD1, RAD52, and YKU70 on gamma-ray hypersensitivity. Logarithmic cultures of yeast cells were harvested and resuspended in phosphate-buffered saline, plated onto YEP-dextrose media, and irradiated with the indicated dose of gamma rays. After 3 to 4 days of growth, surviving colonies were counted, and the survival rate was calculated by comparison to the number of colonies from the mock-treated cells. The percentages of survival of RAD+ (solid circles), rad1Δ (open diamonds), yku70Δ (solid diamonds), rad52Δ (open squares), rad52Δ yku70Δ (solid triangles), rad1Δ rad52Δ (open circles), rad14Δ rad52Δ yku70Δ (open triangles), and rad1Δ rad52Δ yku70Δ (solid squares) cells are shown. Each experimental point represents the average of three independent experiments.
DISCUSSION
We developed a genetic assay to create site-specific DSBs with noncomplementary 4-bp, 3′-overhanging end sequences in vivo and discovered that the repair of these DSBs is primarily mediated by MMEJ, a highly error-prone mechanism independent of Ku or Rad52 proteins, but dependent on Rad1, Mre11, and Rad50 proteins and partially dependent on Dnl4 proteins. Moreover, this pathway appears to contribute to the repair of ionizing radiation damage in the absence of Ku and Rad52 proteins.
Much of what we know about DNA DSB repair and recombination both in yeast and mammals has come from studies that induce chromosomal DSBs in vivo (20, 43). Two classes of causative agents have been used to create DSBs: either the random, unspecified number of breaks with the complex biochemical end configurations (such as ionizing radiation) or a site-specific endonuclease-generated DSB whose ends contain either 3′ or 5′ overhangs with complementary sequences (such as HO endonuclease or EcoRI) (32, 43). The site-specific endonucleases such as HO and I-SceI have been particularly useful not only to generate a site-specific DSB synchronously from a large proportion of cells, but also because the repair of DNA damage in real time can be followed by monitoring the repair products at specific sites in a quantitative manner (20, 43). These studies demonstrated that the end joining is mediated by base pairing of overlapping sequences of DNA ends and requires a set of proteins including Ku proteins (43). However, many chromosome breaks that occur during normal cell growth may not possess such overlapping complementary end sequences, including blunt ends and resected but noncomplementary sequences (35). We thus developed an assay to create site-specific DSBs with noncomplementary end sequences in vivo and explored whether the repair of such ends occurs by the same principles as those deduced from the repair of DSBs with the complementary end sequences. Surprisingly, we discovered that the lack of complementary sequences at the ends prompted a cell to rely on repair primarily by a Ku-independent MMEJ mechanism. This repair pathway is quite efficient and highly competitive against the Ku-dependent imprecise end joining in our assay condition. In addition, all of the joints mediated by the Ku-independent end joining resulted in deletions that apparently involve annealing of as few as 8 bp of almost identical sequences located at some distance from each side of the HO break. Many of these overlapping sequences are AT rich and are frequently interrupted by a few mismatches. However, the fact that other sequences in the same interval with similar microhomologies are not utilized for MMEJ evades definition of the effective microhomology for the Ku-independent end joining.
The requirement of short stretches of homology for Ku- and Rad52-independent end repair is consistent with a number of previous studies (74). In yeast, Ku- and Rad52-deficient cells can still repair DSBs, albeit less efficiently (48). These repair events were almost always associated with long one-sided deletions and extensive imperfect overlapping sequences at the junctions, as shown here (74). Moreover, plasmid molecules cleaved with an EcoRI restriction enzyme and transformed into the Ku-deficient strains were rejoined with the deletions ranging from 6 to 811 bp in length, with junctional overlapping sequences of 3 to 15 bp (1).
Evidence for Ku- and Rad52-independent DSB repair is not limited to yeast cells. In Ku-deficient mice, some lymphocyte development was observed, suggesting that there is a Ku-independent pathway capable of resolving the DSBs created during V(D)J recombination (15). A few of these repair events were characterized and were revealed to identify extensive deletions in their repair junctions. Moreover, a Ku86-deficient cell line showed increased frequencies of imprecise end joining, which can be attributed to Ku- and Rad52-independent DSB repair (24, 33). In the chicken B-cell line DT-40, the ability to repair DSBs was partially retained in the absence of Rad54 and Ku proteins (57). Finally, several groups have reported the partial purification of a Ku- and Rad52-independent end-joining activity by using cell extracts from Xenopus eggs, calf thymus, and human lymphoblast cells (14, 26, 36). Therefore, the available evidence strongly supports the existence of a highly mutagenic DSB repair mechanism that is independent of Ku and Rad52 proteins, and the use of the microhomology seems universal for this repair mechanism.
Our study has provided further evidence of such Rad52- and Ku-independent end-joining events: in this case, for a well-defined DSB generated in chromosomal DNA that has its normal chromatin composition as opposed to naked, restriction endonuclease-digested DNA transformed into yeast. Moreover, we show that this Ku-independent pathway creates nonrandom deletions and is surprisingly efficient—more efficient in fact than the apparently less complex process of misaligning 4-bp 3′-overhanging complementary sequences together to form small insertions and deletions. In addition, we show that this process is strongly dependent on Mre11, Rad50, and Rad1 (and presumably Rad10) but only partially dependent on DNA ligase IV.
In many ways, the MMEJ seems mechanistically similar to the Rad52-dependent SSA. However, we suggest that they represent two distinct repair mechanisms based on their different genetic requirements. Unlike most SSA that depends on Rad52 protein, the efficiency of MMEJ is not affected by the absence of Rad52, although it affects the extent of deletions. Notably, rare SSA has been reported to occur in the absence of Rad52 proteins when it involves very long (several kilobase) lengths of homology that can compensate for the lack of Rad52 activity (42). In contrast, the MMEJ involves short stretches of imperfect sequences of only about 10 bp, supporting the idea that it should rely on a distinct mechanism that is different from SSA.
Furthermore, whereas the Mre11 complex is largely dispensable for SSA (70), it plays a critical role in MMEJ. Biochemically, the Mre11 complex from both yeast and humans exhibits 3′-to-5′ resection activity on double-stranded DNA as well as single-stranded endo- and exonuclease activities (39, 45, 47, 63, 66). More importantly, the purified human Mre11 protein mediates the annealing of complementary single-stranded molecules in vitro (8). Although the lack of efficient ssDNA formation in mre11 strains may affect the MMEJ, we favor the model that the ssDNA annealing activity implicated in human Mre11 may underlie the critical involvement of Mre11 in this process. Such a model is consistent with the minor role of Mre11 in SSA, which should also require the degradation of 5′ ends to uncover the flanking homologous sequences.
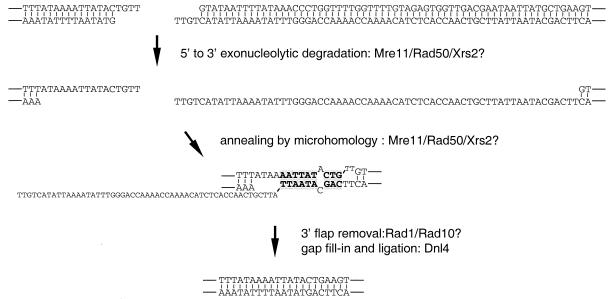
Taken together, we propose the model for MMEJ illustrated in Fig. 3. The model assumes that a microhomology uncovered by the Mre11-Rad50-dependent exonucleolytic processing of DSBs could mediate annealing of broken ends, and then unpaired 3′ flaps would be removed by the Rad1-Rad10 endonuclease complex to prime for gap synthesis and ligation (Fig. 3).
FIG. 3.
Model for MMEJ. Shown is the joining event that is the most frequent in SLY19. Putative end processing of 4-bp 3′ noncomplementary overhangs created by the cleavage of two HO endonuclease recognition sequences will expose the microhomology that mediates annealing. Subsequent 3′-flap removal that is mostly dependent on Rad1-Rad10 will prime the fill-in synthesis and ligation by Dnl4. Microhomologies are shown in boldface. Unpaired 3′ flaps are shown as small letters. The gene products that may function at each step are listed to the right of the arrows.
One of the key questions that emanate from our study is why the breaks with no complementary end sequences are the preferred substrates for the Ku-independent end joining, but not for Ku-dependent NHEJ. Although it is yet unclear, we imagine that some factor or factors may preferentially bind to and stabilize the base pairing between complementary sequences at the overhanging ends to inhibit the processing of DNA ends. Consequently, the microhomology that is needed for Ku-independent MMEJ may be prevented from being exposed by exonucleolytic resection of the DSB ends. In fact, Paull and Gellert had previously reported that Mre11 exonuclease activity is sensitive to the structure and the sequence of the ends (46). Addition of mismatched DNA ends promotes degradation of DNA by Mre11, whereas cohesive ends strongly inhibit it (46). In this context, the base pairing of ends may inhibit the search for microhomology that will only be exposed by 5′-to-3′-end degradation.
The crystal structure of Ku70/Ku80 (69), combined with data suggesting that Ku binds at the border between ssDNA and double-stranded DNA (10), implies that Ku will be an effective mediator of religation and end joining when the overhanging tails are short, but Ku may not be able to effect end joining when the base pairing occurs away from the DSB ends. Consequently, it may be that the Mre11-Rad50-Xrs2 complex, with its annealing and bridging activities (4, 8, 9), takes over these functions in order to promote deletion formation. The binding of Ku to unresected ends may also make intrinsically inefficient the misaligned, Ku-dependent end-joining events, so that Ku-independent, Mre11-Rad50-Xrs2-dependent MMEJ actually becomes more efficient.
Ku- and Rad52-independent DSB repair is an error-prone process, because many of the repair events are associated with large deletions (1, 33). In most cases, the ends are rejoined by base pairing between opposite single strands, yielding junctions with short stretches of homology (microhomology) (1, 14, 24, 33). Such a mechanism will contribute significantly to the genetic instability observed in many cancer cells. In fact, it was reported that the absence of the tumor suppressor BRCA1 frequently causes the cells to repair DSBs using the repair events reminiscent of MMEJ (75). In addition, high levels of chromosomal aberrations were reported in cells deficient in homologous recombination and Ku-dependent NHEJ after exposure to gamma irradiation (57). In yeast, Myung et al. (40) identified spontaneous translocations and deletions involving long imperfect homologous sequences at the junctions that are independent of Rad52 and Ku proteins, but dependent on Mre11 and highly similar to those observed from cancer cells.
In summary, we have developed an assay to assess the repair of chromosomal breaks with no complementary end sequences in vivo. This assay enabled us to discover that these breaks are repaired primarily and efficiently by a Ku-independent but Mre11-, Rad50-, and Rad1-dependent MMEJ mechanism. This assay should be highly useful to dissect the mechanistic and genetic requirements of this highly mutagenic repair process that may be of great relevance to genetic instability.
Acknowledgments
We thank Patrick Sung, Alan Tomkinson, Grzegorz Ira, and members of the Lee and Haber laboratory for helpful comments on the manuscript.
This work was supported in part by a Leukemia and Lymphoma Society Special fellowship, a Howard Hughes Medical Institute New Faculty Start Up award, An American Cancer Society Institutional Research grant, the Sydney Kimmel Foundation for Cancer Research (S.E.L.), and DOE grant ER01-63229 (J.E.H.).
REFERENCES
- 1.Boulton, S. J., and S. P. Jackson. 1996. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 24:4639-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bressan, D. A., H. A. Olivares, B. E. Nelms, and J. H. Petrini. 1998. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics 150:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chamankhah, M., and W. Xiao. 1999. Formation of the yeast Mre11-Rad50-Xrs2 complex is correlated with DNA repair and telomere maintenance. Nucleic Acids Res. 27:2072-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, L., K. Trujillo, W. Ramos, P. Sung, and A. E. Tomkinson. 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8:1105-1115. [DOI] [PubMed] [Google Scholar]
- 5.Colaiacovo, M. P., F. Paques, and J. E. Haber. 1999. Removal of one nonhomologous DNA end during gene conversion by a RAD1- and MSH2-independent pathway. Genetics 151:1409-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Critchlow, S. E., and S. P. Jackson. 1998. DNA end-joining: from yeast to man. Trends Biochem. Sci. 23:394-398. [DOI] [PubMed] [Google Scholar]
- 7.Davies, A. A., E. C. Friedberg, A. E. Tomkinson, R. D. Wood, and S. C. West. 1995. Role of the Rad1 and Rad10 proteins in nucleotide excision repair and recombination. J. Biol. Chem. 270:24638-24641. [DOI] [PubMed] [Google Scholar]
- 8.de Jager, M., M. L. Dronkert, M. Modesti, C. E. Beerens, R. Kanaar, and D. C. van Gent. 2001. DNA-binding and strand-annealing activities of human Mre11: implications for its roles in DNA double-strand break repair pathways. Nucleic Acids Res. 29:1317-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Jager, M., J. van Noort, D. C. van Gent, C. Dekker, R. Kanaar, and C. Wyman. 2001. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell 8:1129-1135. [DOI] [PubMed] [Google Scholar]
- 10.Falzon, M., J. W. Fewell, and E. L. Kuff. 1993. EBP-80, a transcription factor closely resembling the human autoantigen Ku, recognizes single- to double-strand transitions in DNA. J. Biol. Chem. 268:10546-10552. [PubMed] [Google Scholar]
- 11.Feldmann, E., V. Schmiemann, W. Goedecke, S. Reichenberger, and P. Pfeiffer. 2000. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 28:2585-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fishman-Lobell, J., and J. E. Haber. 1992. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science 258:480-484. [DOI] [PubMed] [Google Scholar]
- 13.Frank-Vaillant, M., and S. Marcand. 2001. NHEJ regulation by mating type is exercised through a novel protein, Lif2p, essential to the ligase IV pathway. Genes Dev. 15:3005-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottlich, B., S. Reichenberger, E. Feldmann, and P. Pfeiffer. 1998. Rejoining of DNA double-strand breaks in vitro by single-strand annealing. Eur. J. Biochem. 258:387-395. [DOI] [PubMed] [Google Scholar]
- 15.Gu, Y., S. Jin, Y. Gao, D. T. Weaver, and F. W. Alt. 1997. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J. recombination. Proc. Natl. Acad. Sci. USA 94:8076-8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haber, J. E. 1998. The many interfaces of Mre11. Cell 95:583-586. [DOI] [PubMed] [Google Scholar]
- 17.Henricksen, L. A., J. Veeraraghavan, D. R. Chafin, and R. A. Bambara. 2002. DNA ligase I competes with FEN1 to expand repetitive DNA sequences in vitro. J. Biol. Chem. 277:22361-22369. [DOI] [PubMed] [Google Scholar]
- 18.Ivanov, E. L., and J. E. Haber. 1995. RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:2245-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson, S. P. 2002. Sensing and repairing DNA double-strand breaks. Carcinogenesis 23:687-696. [DOI] [PubMed] [Google Scholar]
- 20.Jasin, M. 1996. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet. 12:224-228. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, A. P., and M. P. Fairman. 1996. The identification and characterization of mammalian proteins involved in the rejoining of DNA double-strand breaks in vitro. Mutat. Res. 364:103-116. [DOI] [PubMed] [Google Scholar]
- 22.Johnston, L. H., and K. A. Nasmyth. 1978. Saccharomyces cerevisiae cell cycle mutant cdc9 is defective in DNA ligase. Nature 274:891-893. [DOI] [PubMed] [Google Scholar]
- 23.Johzuka, K., and H. Ogawa. 1995. Interaction of Mre11 and Rad50: two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics 139:1521-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabotyanski, E. B., L. Gomelsky, J. O. Han, T. D. Stamato, and D. B. Roth. 1998. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 26:5333-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kegel, A., J. O. Sjostrand, and S. U. Astrom. 2001. Nej1p, a cell type-specific regulator of nonhomologous end joining in yeast. Curr. Biol. 11:1611-1617. [DOI] [PubMed] [Google Scholar]
- 26.Labhart, P. 1999. Ku-dependent nonhomologous DNA end joining in Xenopus egg extracts. Mol. Cell. Biol. 19:2585-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, S. E., D. A. Bressan, J. H. J. Petrini, and J. E. Haber. 2002. Complementation between N-terminal Saccharomyces cerevisiae mre11 alleles in DNA repair and telomere length maintenance. DNA Repair 1:27-40. [DOI] [PubMed] [Google Scholar]
- 28.Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. D. Kolodner, and J. E. Haber. 1998. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94:399-409. [DOI] [PubMed] [Google Scholar]
- 29.Lee, S. E., F. Paques, J. Sylvan, and J. E. Haber. 1999. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr. Biol. 9:767-770. [DOI] [PubMed] [Google Scholar]
- 30.Leskov, K. S., T. Criswell, S. Antonio, J. Li, C. R. Yang, T. J. Kinsella, and D. A. Boothman. 2001. When X-ray-inducible proteins meet DNA double strand break repair. Semin. Radiat. Oncol. 11:352-372. [DOI] [PubMed] [Google Scholar]
- 31.Lewis, L. K., and M. A. Resnick. 2000. Tying up loose ends: nonhomologous end-joining in Saccharomyces cerevisiae. Mutat. Res. 451:71-89. [DOI] [PubMed] [Google Scholar]
- 32.Lewis, L. K., J. W. Westmoreland, and M. A. Resnick. 1999. Repair of endonuclease-induced double-strand breaks in Saccharomyces cerevisiae: essential role for genes associated with nonhomologous end-joining. Genetics 152:1513-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang, F., and M. Jasin. 1996. Ku80-deficient cells exhibit excess degradation of extrachromosomal DNA. J. Biol. Chem. 271:14405-14411. [DOI] [PubMed] [Google Scholar]
- 34.Liang, F., P. J. Romanienko, D. T. Weaver, P. A. Jeggo, and M. Jasin. 1996. Chromosomal double-strand break repair in Ku80-deficient cells. Proc. Natl. Acad. Sci. USA 93:8929-8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lieber, M. R. 1999. The biochemistry and biological significance of nonhomologous DNA end joining: an essential repair process in multicellular eukaryotes. Genes Cells 4:77-85. [DOI] [PubMed] [Google Scholar]
- 36.Mason, R. M., J. Thacker, and M. P. Fairman. 1996. The joining of non-complementary DNA double-strand breaks by mammalian extracts. Nucleic Acids Res. 24:4946-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore, C. W. 1982. Ligase-deficient yeast cells exhibit defective DNA rejoining and enhanced gamma ray sensitivity. J. Bacteriol. 150:1227-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore, J. K., and J. E. Haber. 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2164-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreau, S., J. R. Ferguson, and L. S. Symington. 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19:556-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myung, K., C. Chen, and R. D. Kolodner. 2001. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411:1073-1076. [DOI] [PubMed] [Google Scholar]
- 41.Ooi, S. L., D. D. Shoemaker, and J. D. Boeke. 2001. A DNA microarray-based genetic screen for nonhomologous end-joining mutants in Saccharomyces cerevisiae. Science 294:2552-2556. [DOI] [PubMed] [Google Scholar]
- 42.Ozenberger, B. A., and G. S. Roeder. 1991. A unique pathway of double-strand break repair operates in tandemly repeated genes. Mol. Cell. Biol. 11:1222-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.PÂques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.PÂques, F., and J. E. Haber. 1997. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6765-6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paull, T. T., and M. Gellert. 1998. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell 1:969-979. [DOI] [PubMed] [Google Scholar]
- 46.Paull, T. T., and M. Gellert. 2000. A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc. Natl. Acad. Sci. USA 97:6409-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paull, T. T., and M. Gellert. 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 13:1276-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pfeiffer, P., W. Goedecke, and G. Obe. 2000. Mechanisms of DNA double-strand break repair and their potential to induce chromosomal aberrations. Mutagenesis 15:289-302. [DOI] [PubMed] [Google Scholar]
- 49.Prakash, S., and L. Prakash. 2000. Nucleotide excision repair in yeast. Mutat. Res. 451:13-24. [DOI] [PubMed] [Google Scholar]
- 50.Ramos, W., N. Tappe, J. Talamantez, E. C. Friedberg, and A. E. Tomkinson. 1997. Two distinct DNA ligase activities in mitotic extracts of the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 25:1485-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramsden, D. A., T. T. Paull, and M. Gellert. 1997. Cell-free V(D)J recombination. Nature 388:488-491. [DOI] [PubMed] [Google Scholar]
- 52.Schar, P., G. Herrmann, G. Daly, and T. Lindahl. 1997. A newly identified DNA ligase of Saccharomyces cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes Dev. 11:1912-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 54.Sugawara, N., G. Ira, and J. E. Haber. 2000. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 20:5300-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sung, P., P. Reynolds, L. Prakash, and S. Prakash. 1993. Purification and characterization of the Saccharomyces cerevisiae RAD1/RAD10 endonuclease. J. Biol. Chem. 268:26391-26399. [PubMed] [Google Scholar]
- 56.Symington, L. S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66:630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takata, M., M. S. Sasaki, E. Sonoda, C. Morrison, M. Hashimoto, H. Utsumi, Y. Yamaguchi-Iwai, A. Shinohara, and S. Takeda. 1998. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17:5497-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Teo, S. H., and S. P. Jackson. 1997. Identification of Saccharomyces cerevisiae DNA ligase IV: involvement in DNA double-strand break repair. EMBO J. 16:4788-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomas, B. J., and R. Rothstein. 1989. The genetic control of direct-repeat recombination in Saccharomyces: the effect of rad52 and rad1 on mitotic recombination at GAL10, a transcriptionally regulated gene. Genetics 123:725-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Timson, D. J., M. R. Singleton, and D. B. Wigley. 2000. DNA ligases in the repair and replication of DNA. Mutat. Res. 460:301-318. [DOI] [PubMed] [Google Scholar]
- 61.Tomkinson, A. E., A. J. Bardwell, L. Bardwell, N. J. Tappe, and E. C. Friedberg. 1993. Yeast DNA repair and recombination proteins Rad1 and Rad10 constitute a single-stranded-DNA endonuclease. Nature 362:860-862. [DOI] [PubMed] [Google Scholar]
- 62.Tomkinson, A. E., N. J. Tappe, and E. C. Friedberg. 1992. DNA ligase I from Saccharomyces cerevisiae: physical and biochemical characterization of the CDC9 gene product. Biochemistry 31:11762-11771. [DOI] [PubMed] [Google Scholar]
- 63.Trujillo, K. M., and P. Sung. 2001. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J. Biol. Chem. 276:35458-35464. [DOI] [PubMed] [Google Scholar]
- 64.Trujillo, K. M., S. S. Yuan, E. Y. Lee, and P. Sung. 1998. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 273:21447-21450. [DOI] [PubMed] [Google Scholar]
- 65.Tseng, H. M., and A. E. Tomkinson. 2002. A physical and functional interaction between yeast Pol4 and Dnl4-Lif1 links DNA synthesis and ligation in nonhomologous end joining. J. Biol. Chem. 277:45630-45637. [DOI] [PubMed] [Google Scholar]
- 66.Usui, T., T. Ohta, H. Oshiumi, J. Tomizawa, H. Ogawa, and T. Ogawa. 1998. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95:705-716. [DOI] [PubMed] [Google Scholar]
- 67.Valencia, M., M. Bentele, M. B. Vaze, G. Herrmann, E. Kraus, S. E. Lee, P. Schar, and J. E. Haber. 2001. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature 414:666-669. [DOI] [PubMed] [Google Scholar]
- 68.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 69.Walker, J. R., R. A. Corpina, and J. Goldberg. 2001. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412:607-614. [DOI] [PubMed] [Google Scholar]
- 70.Wilson, T. E. 2002. A genomics-based screen for yeast mutants with an altered recombination/end-joining repair ratio. Genetics 162:677-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilson, T. E., U. Grawunder, and M. R. Lieber. 1997. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature 388:495-498. [DOI] [PubMed] [Google Scholar]
- 72.Wilson, T. E., and M. R. Lieber. 1999. Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase beta (Pol4)-dependent pathway. J. Biol. Chem. 274:23599-23609. [DOI] [PubMed] [Google Scholar]
- 73.Wu, X., T. E. Wilson, and M. R. Lieber. 1999. A role for FEN-1 in nonhomologous DNA end joining: the order of strand annealing and nucleolytic processing events. Proc. Natl. Acad. Sci. USA 96:1303-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu, X., and A. Gabriel. 2003. Ku-dependent and Ku-independent end-joining pathways lead to chromosomal rearrangements during double-strand break repair in Saccharomyces cerevisiae. Genetics 163:843-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhong, Q., C. F. Chen, P. L. Chen, and W. H. Lee. 2002. BRCA1 facilitates microhomology-mediated end joining of DNA double strand breaks. J. Biol. Chem. 277:28641-28647. [DOI] [PubMed] [Google Scholar]