Summary
We present a case study illustrating the challenges of analyzing accelerometer data taken from a sample of children participating in an intervention study designed to increase physical activity. An accelerometer is a small device worn on the hip that records the minute-by-minute activity levels of the child throughout the day for each day it is worn. The resulting data are irregular functions characterized by many peaks representing short bursts of intense activity. We model these data using the wavelet-based functional mixed model. This approach incorporates multiple fixed effect and random effect functions of arbitrary form, the estimates of which are adaptively regularized using wavelet shrinkage. The method yields posterior samples for all functional quantities of the model, which can be used to perform various types of Bayesian inference and prediction. In our case study, a high proportion of the daily activity profiles are incomplete, i.e. have some portion of the profile missing, so cannot be directly modeled using the previously described method. We present a new method for stochastically imputing the missing data that allows us to incorporate these incomplete profiles in our analysis. Our approach borrows strength from both the observed measurements within the incomplete profiles and from other profiles, from the same child as well as other children with similar covariate levels, while appropriately propagating the uncertainty of the imputation throughout all subsequent inference. We apply this method to our case study, revealing some interesting insights into children's activity patterns. We point out some strengths and limitations of using this approach to analyze accelerometer data.
Keywords: Accelerometer, Bayesian inference, functional data analysis, missing data, mixed models, wavelets, wavelet regression
1 Introduction
Physical activity during childhood and adolescence has an impact on a number of different aspects of subsequent health and well-being. Studies have suggested that increased physical activity during the childhood and adolescent years can (1) help build and maintain healthy bones, muscles and joints; (2) help control weight, build lean muscles and reduce fat; (3) prevent or delay the development of hypertension, and help reduce blood pressure in some adolescents with hypertension; (4) reduce feelings of depression and anxiety; (5) positively influence mental health and feelings of independence; and (6) positively influence the development of self-esteem (CDC, 2000; CDC, 2001; Dietz 1998; Pearce, 1999; Strauss et al., 2001).
The systematic study of physical activity is complicated by the technical challenges of measurement. One common approach is to use self-report questionnaires, but these are known to correlate poorly with actual activity levels, often overestimating them (Matthews and Freedson 1995, Epstein et al. 1996, Coleman et al. 1997). Recently, more objective approaches have been used, including computerized accelerometers: small motion sensors that can digitally record minute-by-minute activity levels. Their small size and automatic measurement capabilities make them well-suited for objectively quantifying activity levels in large populations over long periods of time (Westerterp, 1999). They are being used increasingly to monitor activity levels in large surveillance studies and intervention trials, in children as well as adults (Rowlands, Eston, and Ingledew 1999; Gortmaker 2002; Going et al. 2003; Talbot et al. 2003; Algase et al. 2003; Abbott and Davies 2004). In this paper, we consider accelerometer data obtained from Planet Health (Gortmaker et al. 1999), a Boston-area study of a school-based intervention designed to reduce obesity in middle school youth by changing key physical activity and dietary risk factors.
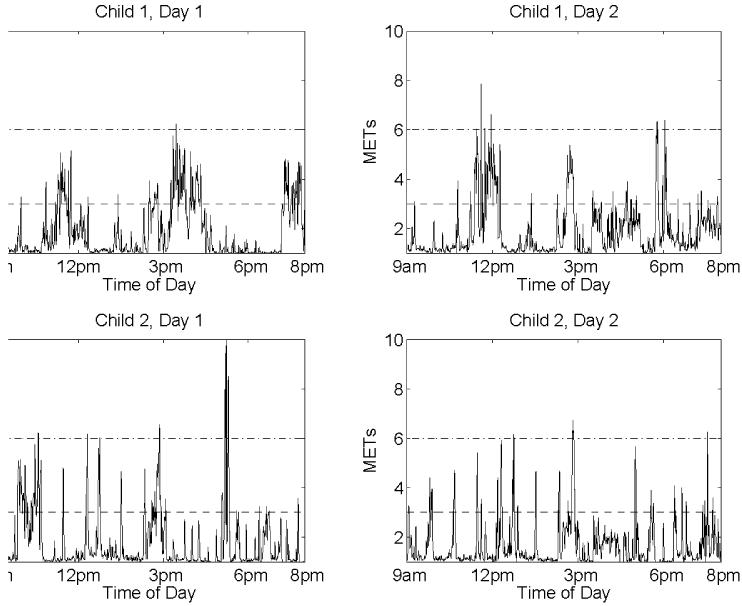
The Planet Health study used the TriTrac-R3D activity monitor (Hemokentics, Inc., Madison, WI), which is worn in a small pouch on the hip and provides minute-by-minute acceleration counts computed from motion sensors in three-dimensional space. The resulting daily profiles are irregular functional data characterized by many peaks representing short bursts of intense activity. Figure 1 contains two daily profiles from 9am to 8pm from two different children in the Planet Health study. Other accelerometers yield similar data. In studies involving accelerometers, it is standard practice to obtain 4 to 8 daily profiles for each subject in the study. These profiles can be analyzed directly, or used to calibrate and validate questionnaire-based measures of physical activity. In practice, many of the profiles are incomplete, having periods of time for which the activity levels are missing, for example, if the accelerometer is removed.
Figure 1. Sample profiles from 2 days for 2 different children, from 9am to 8pm.
Activity levels above 6 MET are considered vigorous, while activity levels between 3 and 6 MET are moderate.
Approaches currently used to analyze these data are based on simple summaries, such as the average daily activity level (Talbot et al. 2003), 30-minute averages (Going et al. 2003; Cradock et al. 2004), or the proportion of time spent above specific cutoff levels that correspond to sedentary, moderately vigorous, and vigorous activity (Rowlands, Eston and Ingledew 1999; Goldfield et al. 2000; Westerterp 2001; Abbott and Davies 2004). While these summaries are a reasonable starting point, they have serious limitations, mostly because they do not make full use of the rich information contained in the functional data. The proportions and daily averages do not account for time of the day variability, and the proportions do not even use the actual activity levels. The arbitrary binning inherent to taking 30-minute averages results in attenuation of the signal, and conclusions may be sensitive to the choice of endpoints for the bins. A second important limitation of these approaches is that they may not effectively handle the missing data in the incomplete profiles, which is a serious concern considering the large degree of missingness that can occur in these data.
Methods that model the functional profiles in their entirety have the potential to extract more information from the data compared with methods based on arbitrarily chosen summary measures. For example, these methods are able to identify at what time of day children are most active, detect covariate effects that vary throughout different times of the day, and allow the variability in activity levels to differ across time of day. Functional data analysis (FDA, Ramsay and Silverman 1997) is a general name for approaches that consider the functional profiles as single entities rather than simply a collection of individual data points. A key challenge with these approaches is that they must deal both with regularization, or borrowing strength across measurements within a profile, and replication, or combining information across multiple profiles to make inferences on the population from which they came.
The irregularity of accelerometer profiles makes them especially challenging to model in this manner. They clearly cannot be represented by simple parametric structures, and the most commonly-used nonparametric methods involve smoothing by kernels or splines, which are not well-suited for modeling functional data with many local features like peaks. Since there are typically multiple daily profiles per child, one must account for the possibility of correlation between profiles. Also, there are many factors potentially affecting the profiles, including the day of the week, time of year, school, and child-level characteristics like gender, age, and obesity status. The effects of these factors may vary by time of day. A suitable modeling approach should be able to simultaneously account for functional effects of multiple covariates like these, some of which are of interest to investigators, and others of which are potential confounders.
Morris and Carroll (2006) introduced a new method for analyzing functional data that accommodates these concerns. It is based on the functional mixed model, a generalization of the linear mixed model (Laird and Ware 1982) to functional data. This method is appropriate for modeling irregular profiles with many peaks, since regularization is accomplished using adaptive wavelet shrinkage. The functional mixed model allows multiple fixed effect functions of arbitrary form, which can simultaneously represent time-varying effects for multiple covariates, discrete or continuous in nature. It also allows multiple random effect functions of arbitrary form, which can model the correlation between profiles from the same child. The reasonably flexible assumptions made on the covariance matrices for the random effects and residual errors accommodate a broad range of nonstationary covariance structures. The output of the method consists of posterior samples for all quantities in the model, which can be used to perform Bayesian inference.
While the method of Morris and Carroll (2006) cannot handle incomplete profiles, it is based on a unified modeling approach, making it possible to develop rigorous methods for imputing the missing data. In this paper, we introduce new missing data methods for the wavelet-based functional mixed model. Briefly, our approach is to stochastically sample the missing regions of the incomplete profiles from their approximate posterior predictive distributions, as estimated from a preliminary model fit using only the complete daily profiles. This approach borrows strength both from the observed measurements within the incomplete profile and from other profiles, from the same child as well as other children with similar covariate levels, and appropriately propagates the uncertainty due to the imputation throughout any subsequent inference.
We apply these methods to analyze the TriTrac-R3D data from the Planet Health study in order to explore patterns in the children's activity level profiles, to identify factors related to activity levels, and to characterize the relative contributions of the day-to-day and child-to-child sources of variability in order to aid the design of future studies. We also assess the model fit, and discuss the strengths and limitations of this approach for analyzing accelerometer data.
The remainder of the paper is organized as follows. In Section 2, we describe the Planet Health study and explore the characteristics of the TriTrac-R3D accelerometer data from that study. In Section 3, we briefly overview wavelets and the wavelet-based functional mixed model, and in Section 4, we introduce new methods for imputing missing data for incomplete profiles in the wavelet-based functional mixed models setting. In Section 5, we present the results of our case study analysis, and in Section 6, discuss these results and assess the strengths and limitations of this approach for analyzing accelerometer data. A technical appendix contains the derivation of the posterior predictive distributions used to impute the missing data.
2 Accelerometer Data from the Planet Health Study
Childhood obesity is a major health problem in the United States. Planet Health was a school-based intervention designed to reduce obesity in middle school youth by changing key physical activity and dietary risk factors. The Planet Health study (Gortmaker et al. 1999) involved 10 Boston-area middle schools, which were paired up and randomized to either receive the intervention or serve as control schools. For each of the 1295 children involved in the study, various nutritional, behavioral, and health-related outcomes were measured at baseline (fall 1995) and follow-up (spring 1997). A subsample of 256 children was randomly selected to participate in a substudy focusing on the technical issue of objectively measuring physical activity using accelerometers. The activity levels of each child in the substudy were monitored using the TriTrac-R3D activity monitor for one or two four-day sessions between February and May 1997.
The TriTrac-R3D activity monitor is a pocket-sized motion sensor that, when worn on the hip in a nylon pouch, measures motion in three planes (horizontal, vertical, and mediolateral). The TriTrac stores data over several days, after which the data can be downloaded to a computer to provide a minute-by-minute record of movement in each dimension. The counts in the three planes may be combined into a single activity level for each minute by computing the vector magnitude. The vector magnitudes may then be converted into estimated energy expenditure per minute due to activity (EE). The TriTrac-R3D uses a proprietary formula involving the subjects weight to compute EE; Gortmaker (2002) calculated this formula, and then converted these into METS (metabolic units) using MET=(EE+BMR)/BMR, where BMR is the child's basal metabolic rate, estimated from their gender, age, and weight using the World Health Organization (WHO) equations (Recommended Daily Allowances 2000). The minimum activity level is 1 MET, which indicates the child is stationary, while activities yielding levels between 3 and 6 MET (e.g., walking) are considered moderately intense, and activities yielding levels of more than 6 MET (e.g., running) are considered vigorous (Rowlands et al. 2004).
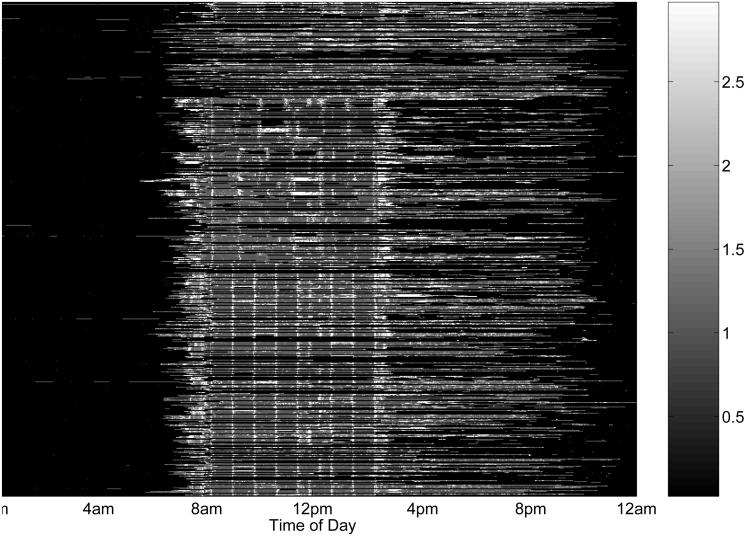
We focused on the weekday accelerometer profiles from children in the 5 control schools that did not receive the intervention. This data set consisted of 550 daily profiles from 112 children. Each daily profile contained 1440 measurements, the minute-by-minute activity levels for a single day, measured in METs. If no movement was recorded for 30 or more consecutive minutes, we assumed that the child removed the monitor, and the corresponding measurements were considered missing. Figure 2 contains a heatmap of the 550 daily profiles. A heatmap is a graphical device that is useful for representing high-dimensional data sets. Each row corresponds to a single profile and each column is one minute in the day. The color of the pixel (i, j) indicates the activity level of profile i at time tj, with higher activity levels represented by lighter colors. For this graph, we coded missing measurements as 0 (black), and censored moderate and vigorous activity levels (> 3.0 MET, white) to improve the contrast of the figure. The white dashed horizontal lines delineate the schools, which we will refer to as schools A, B, C, D, and E, respectively. Note the imbalance in the number of profiles obtained from each school.
Figure 2. Heat map of all weekday accelerometer profiles.
The rows are the daily profiles, ordered by school and child within school. The columns are the minutes of the day, from 12:01am through 12:00 midnight. The black regions of the plot indicate regions of the profiles where the activity level is missing. Since the minimum activity level is 1 MET, all non-missing activity levels are some shade of gray, not black. The white regions indicate bouts, or regions where the activity profile is > 3.0 MET. The dotted white lines mark the divisions between profiles from different schools.
There were a large number of missing measurements in the data. Supplementary material available on the first author's web site (http://biostatistics.mdanderson.org/Morris/papers_files/TriTrac_supplement.html) contains a plot of the proportion of daily profiles with nonmissing measurements as a function of the time of the day, and a histogram of the proportion of minutes missing from 9am-8pm for the 550 profiles. We focused our analysis on the data from 9am to 8pm, since most of the profiles were missing outside this region. Only 95 of the 550 profiles were complete from 9am to 8pm, and 187 profiles had more than 80% of this region missing. We focused on the 292 profiles that were at least 50% complete from 9am to 8pm, which were from 106 children. All of these profiles were obtained between February 10, 1997 and May 28, 1997. We also considered the criteria of 80% complete or 20% complete, and found similar results.
For each child, we had a number of covariate measurements, including school, race/ethnicity, gender, age, weight, height, body mass index (BMI), triceps skinfold (measured by skinfold calipers applied to the triceps), and average number of hours spent watching television per day. We also had a record of the day-of-week and the calendar date on which each profile was obtained.
We were interested in assessing how activity levels tended to vary throughout the day, across schools, across different days of the week, over time from early to late spring, and across various child-level covariates. We also were interested in assessing the relative variability from day-to-day and child-to-child, in order to provide design recommendations for future studies. We accomplished these goals by applying the wavelet-based functional mixed model to these data, after adapting the procedure to accomodate incomplete profiles. We assessed how well this model fit the data, and specifically checked how closely it predicted the frequency of bouts of moderate and vigorous activity for groups of children as a function of the time of day.
3 Wavelets and Functional Mixed Models
3.1 Brief Revew of Wavelets
Wavelets are basis functions that can be used to represent other functions, often very parsimoniously. A basic introduction to wavelets can be found in Vidakovic (1999). A wavelet series approximation to a continuous function y(t) is given by
| (1) |
where J is the number of scales, and k ranges from 1 to Kj, the number of coefficients at scale j. The functions ϕJ,k(t) and ψj,k(t) are wavelet basis functions that provide a location-scale decomposition of the observed function. They are dilations and translations of a father and mother wavelet function, ϕ(t) and ψ(t), respectively, with ϕj,k(t) = 2-j/2ϕ(2-j t - k) and ψj,k(t) = 2-j/2ψ(2-j t - k). The coefficients cJ,k, dJ,k, …, d1,k are the wavelet coefficients. The cJ,k are called the smooth coefficients, and represent smooth behavior of the function at coarse scale J, and the dj,k are called the detail coefficients, and represent deviations of the function at scale j, where smaller j correspond to finer scales. The wavelet coefficients at scale j essentially correspond to differences of averages of 2j-1 time units, spaced 2j units apart. In addition, by examining the phase properties of the wavelet bases, we can associate each wavelet coefficient on each scale with a specific set of time points.
Theoretically, each coefficient may be obtained by taking the inner product of the function and the corresponding wavelet basis function, although in practice more efficient approaches are used. If the function is sampled on an equally spaced grid of length T , then the coefficients may be computed using a pyramid-based algorithm called the discrete wavelet transform (DWT) in just O(T) operations. Applying the DWT to a row vector of observations y produces a row vector of wavelet coefficients d = (cJ,1, …, cJ,KJ, dJ,1, …, d1,K1). This transformation is a linear projection, so it may also be represented by matrix multiplication, d = yW′, with W′ being the DWT projection matrix. Similarly, the inverse discrete wavelet transform (IDWT) may be used to project wavelet coefficients back into the data space, and can also be represented by matrix multiplication by the IDWT projection matrix W , the transpose of the DWT projection matrix. We use the method implemented in the Matlab Wavelet Toolbox (Misiti, et al. 2000) for computing the DWT, although other implementations could just as well have been be used. Supplementary material (http://biostatistics.mdanderson.org/Morris/papers_files/TriTrac_supplement.html) describes how this implemenation deals with data for which T is not a power of 2 and discusses some of its properties.
Wavelets can be used to perform nonparametric regression using the following three-step procedure. First, noisy data y are projected into the wavelet domain using the DWT, yielding empirical wavelet coefficients d. The coefficients are then thresholded by setting to zero any coefficients smaller in magnitude than a specified threshold, and/or nonlinearly shrunken towards zero using one of a number of possible frequentist or Bayesian approaches. These result in estimates of the true wavelet coefficients, which would be the wavelet coefficients for the true function if there was no noise. Finally, these estimates are projected back to the original data domain using the IDWT, yielding a denoised nonparametric estimate of the true function. Because most signals may be represented by a small number of wavelet coefficients, yet white noise is distributed equally among all wavelet coefficients, this procedure yields denoised function estimates that tend to retain dominant local features of the function. In this paper, we refer to this property as adaptive regularization, since the function is regularized (i.e., denoised or smoothed) in a way that adapts to the characteristics of the function. This property makes the procedure useful for modeling functions with many local features like peaks. References on wavelet regression can be found in Chapters 6 and 8 of Vidakovic (1999), and in Donoho and Johnstone (1995), Chipman, Kolaczyk, and McCulloch (1997), Vidakovic (1998), Abramovich, Sapatinas, and Silverman (1998), Clyde, Parmigiani, and Vidakovic (1999), and Clyde and George (2000).
Most work in wavelet regression to date has been limited to the single-function case, with a few exceptions. For example, Brown, Fearn and Vannucci (2001) performed Bayesian variable selection on wavelet coefficients of functions serving as predictors for a scalar response. Chang and Vidakovic (2002) proposed a method to obtain a regularized estimate for the mean function over a sample of curves. Morris, et al. (2003) developed a wavelet-based method for analyzing hierarchical functional data, in which the functions are sampled from a nested hierarchical design. This method produces adaptively regularized estimates and inference for the overall mean function and individual profiles. Morris and Carroll (2006) generalized this work to the functional mixed models setting.
3.2 Functional Mixed Models
Suppose we observe n functional profiles Yi(t), i = 1, … ,n, all defined on the compact set . A functional mixed model for these profiles is given by
| (2) |
where Xij are covariates, Bj(t) are functional (time-varying) fixed effects, Zik are elements of the design matrix for functional random effects Uk(t), and Ei(t) are residual error processes. Here, we assume that Uk(t) are independent and identically distributed (iid) mean-zero Gaussian processes with covariance surface Q(t1, t2), and Ei(t) are iid mean-zero Gaussian processes with covariance surface S(t1, t2), with Uk(t) and Ei(t) being independent. Note that Q indicates the covariance across the random effect functions k = 1, … , m, and S indicates the covariance across the residual error processes for the n curves, after conditioning on the fixed and random effects. Note that the iid assumptions on the individual random effect functions and residual error processes makes Equation (2) a special case of the more general functional mixed model presented in Morris and Carroll (2006). Since the response, fixed effects, random effects, and residual error processes are all functional, heuristically this model can be viewed as fitting separate mixed models at each time point, but with an extra layer added for regularization, i.e. to borrow strength across observations within a function.
Suppose all observed profiles are sampled on the same equally spaced grid t of length T . Let Y be the n × T matrix containing the observed profiles on the grid, with each row containing one observed profile on the grid t. A discrete, matrix-based version of this mixed model can be written
| (3) |
The matrix X is an n × p design matrix containing (nonfunctional) covariates; B is a p × T matrix whose rows contain the corresponding fixed effect functions on the grid t. The quantity Bij denotes the effect of the covariate in column i of X on the response at time tj. The matrix U is an m × T matrix whose rows contain random effect functions on the grid t, and Z is the corresponding n × m design matrix. Each row of the n × T matrix E contains the residual error process for the corresponding observed profile. We assume that the rows of U are iid MVN(0, Q) and the rows of E are iid MVN(0, S), independent of U, with Q and S being T × T covariance matrices that are discrete approximations to the covariances surfaces in (2) on the grid.
This model is very flexible and can be used to represent a wide range of functional data. The fixed effect functions may be group mean functions, interaction functions, or functional linear effects for continuous covariates, depending on the structure of the design matrix. The random effect functions provide a convenient mechanism for modeling between-function correlation, for example when multiple profiles are obtained from the same individual. The model places no restrictions on the form of the fixed or random effect functions. Since the forms of the covariance matrices Q and S are also left unspecified, it is typically not feasible to fit this model without first making further assumptions.
3.3 Wavelet-Based Functional Mixed Models (WFMM)
Morris and Carroll (2006) did not fit model (4) directly, but instead projected the observed profiles into the wavelet space, then worked with the wavelet-space version of the model. This allowed the modeling to be done in a more parsimonious and computationally effcient manner, and enabled a convenient mechanism for adaptively regularizing the fixed effect functions.
The projection is accomplished by applying the discrete wavelet transform (DWT) to each row of Y, yielding a matrix of wavelet coeffcients D = YW′, where W′ is the DWT projection matrix. Row i of D contains the wavelet coeffcients for profile i, with the columns corresponding to individual wavelet coeffcients and double-indexed by scale j and location k. It is easy to show that the wavelet-space version of model (3) is
| (4) |
where each row of B* = BW′ contains the wavelet coeffcients corresponding to one of the fixed effect functions, each row of U* = UW′ contains the wavelet coeffcients for a random effect function, and E* = EW′ contains the wavelet-space residuals. The rows of U* and E* remain independent mean-zero Gaussians, but with covariance matrices Q* = WQW′ and S* = WSW′.
Motivated by the whitening property of the wavelet transform, many wavelet regression methods in the single-function setting assume that the wavelet coefficients for a given function are mutually independent. Morris and Carroll (2006) effectively make this assumption in the wavelet-based functional mixed model by constraining Q* and S* to be diagonal. Allowing the variance components to differ across both wavelet scale j and location k yields Q* = diag(qjk) and S* = diag(sjk). This assumption reduces the dimensionality of Q and S from T (T +1)/2 to T, while still accommodating a reasonably wide range of nonstationary within-profile covariance structures for both the random effects and residual error processes. For example, it allows heteroscedasticity and differing degrees of smoothness at different regions of the curves. Figure 1 of Morris and Carroll (2006) illustrates this point. By using transforms from wavelet packet tables (see Percival and Walden 2000, chapter 6), it may be possible to accommodate an even broader class of covariance matrices.
Morris and Carroll (2006) used a Markov Chain Monte Carlo scheme to generate posterior samples for quantities of model (4). Vague proper priors were used for the variance components, and independent mixture priors were used for the elements of B*. Specifically, the prior for Bijk*, the wavelet coeffcient at scale j and location k for fixed effect function i, was with γijk ~ Bernoulli (πij) and δ0 being a point mass at zero. This prior is commonly used in Bayesian implementations of wavelet regression, for example see Clyde, Parmigiani and Vidakovic (1998) and Abramovich, Sapatinas, and Silverman (1998). Use of this mixture prior causes the posterior mean estimates of the to be nonlinearly shrunken towards zero, which results in adaptively regularized estimates of the fixed effect functions. The parameters τij and πij are regularization parameters that determine the relative trade-off of variance and bias in the nonparametric estimation. They may either be prespecified or estimated from the data using an empirical Bayes method; see Morris and Carroll (2006) for details.
Posterior samples for each fixed effect function, , on the grid t are obtained by applying the IDWT to the posterior samples of the corresponding complete set of wavelet coeffcients , and similarly for the random effect functions Ui. If desired, posterior samples for the covariance matrices Q and S may also be computed using matrix multiplication Q(g) = WQ*(g)W′ and S(g) = WS*(g)W′, respectively. Since Q*(g) are S*(g) are diagonal, this may be accomplished in an equivalent but more effcient manner by applying the 2-dimensional version of the IDWT (2d-IDWT) to Q*(g) and S*(g) (Vannucci and Corradi, 1999). These posterior samples of the quantities in model (3) may subsequently be used to perform any desired Bayesian inference.
4 Handling Incomplete Profiles in WFMM
The method described above cannot be used to analyze data for which some profiles are incomplete, i.e., not fully observed on the grid t, because the first step of the method, the DWT, cannot be applied to incomplete profiles. Under these circumstances, one alternative is to simply perform a “complete case” analysis that uses only the complete profiles. However, this is ineffcient since it ignores information contained in the incomplete profiles. This problem is especially severe when a high proportion of observed profiles are incomplete, which is true for the TriTrac accelerometer data from the Planet Health study.
In this section, we introduce two missing data approaches that can be used to incorporate information from the incomplete profiles in the wavelet-based functional mixed model when there are at least some complete profiles available. Both methods involve imputation of “missing wavelet coeffcients” from predictive distributions at each iteration of the MCMC. Missing wavelet coeffcients are wavelet coeffcients for an individual profile, for which the support of the corresponding basis function intersect missing regions of the profile. In the first method, the imputation distributions are defined based on the posterior means of model parameters obtained from an MCMC applied to the complete case data, while in the second, posterior predictive means and variances are used. These approaches are appropriate to use when the data is missing completely at random (MCAR, Little and Rubin, 2002). Cradock, et al. (2004) described a validation study in which they essentially concluded that the missingness in the accelerometer data from the Planet Health study was MCAR.
Suppose we have an incomplete profile, Yi(t), that is observed on the set of times tO, but missing on the set of times tM, with tO ∪ tM = t, an equally spaced grid on the region T . From (3), the model for Yi, a row vector containing the full profile on the grid t, is given by Yi = XiB+ZiU+Ei, where Xi is 1×p, B is p×T, is Zi is 1×m, U is m×T, and Ei is 1×T. The rows of U are independently distributed as Normal{0,Q}, and Ei ~ Normal{0, S}. Let Ω be the vector containing the covariance parameters determining the matrices Q and S. The joint distribution of the profile on the grid, conditional on the fixed effects, covariance parameters, and random effects, is {Yi|B, Ω, U} ~ Normal(μi, Σi), with
| (5) |
| (6) |
Partition , with and being vectors of length and containing the observed and missing regions of the profile, respectively. Similarly partition and . By standard conditional normal calculations, we find that
| (7) |
with and . We refer to this conditional distribution as the imputation distribution. This distribution represents a linear regression of the missing data on the observed parts of the profile.
In the Bayesian paradigm, a natural way to deal with the missing data is to treat it as an unknown parameter vector to be updated at each iteration of the MCMC from its complete conditional distribution, which is the imputation distribution (7). While conceptually straightforward, this fully Bayesian approach is computationally infeasible here. It requires numerous applications of the DWT and IDWT and extra matrix inversions and multiplications at each iteration of the MCMC, since the missingness is in the data space and the MCMC is applied to the wavelet space version of the model.
A less computationally intensive alternative is to first perform a “complete case analysis” to obtain posterior samples of the model parameters, then use these to characterize the imputation distribution (7). This distribution may then be projected into the wavelet space to yield a distribution from which random draws of the “missing wavelet coeffcients” can be taken at each iteration of the MCMC. This approach does not require any DWTs, IDWTs, or extra matrix inversions or multiplications to be done inside the MCMC.
Following are the specific steps necessary to implement this approach, using the posterior mean estimates from the complete case run to characterize the mean and variance in (7).
Fit the wavelet-based functional mixed model to the set of complete profiles YC. Compute , , and , the posterior mean estimates of the fixed effect functions, random effect functions, and residual covariance matrix using the procedure described in Section 3.3.
-
For each incomplete profile Yi(t),
Compute and in (7), using the posterior mean estimates of the model quantities in place of μi and Σi.
Form the mean imputed profile Mi(t) on the grid t, Mi, by setting Mi(t) = Yi(t) for t ∈ tO and for t ∈ tM.
Form the covariance matrix for the imputed profile Vi(t1, t2) on t × t, by setting and Vi(t1, t2) = 0 if t1 ∈ tO or t2 ∈ tO.
Apply the DWT to the mean imputed profile, yielding the corresponding vector of wavelet coeffcients, , double-indexed by wavelet scale j and location k.
Apply the 2-d DWT to the covariance matrix for the imputed profile to compute the corresponding covariance matrix of the imputed wavelet coeffcients, . Because of our independence assumption between the wavelet coeffcients within a given function, we restrict attention to the diagonal elements of this matrix, . Each element of the vector , contains the imputation variance for a single wavelet coeffcient, double-indexed by scale j and location k.
Fit the wavelet-based functional mixed model to the full data set, including the complete and incomplete profiles. At each iteration of the MCMC, add a step whereby for each incomplete profile indexed by i, we draw each wavelet coeffcient dijk (j = 1, … , J, k = 1, … , Kj) from its wavelet-space imputation distribution, which is . For any “observed” wavelet coeffcients for which the corresponding wavelet basis function, ψjk(t), is completely contained within tO, the imputation variance will be 0 and the imputation mean will be dijk, the wavelet coeffcient we would have obtained by applying the DWT to the profile Yi if it were completely observed.
This procedure assumes that for each incomplete profile Yi, we have at least one complete profile from that child, since it conditions on estimates of the corresponding child-level random effect function in U from the complete case analysis. The procedure is easily adapted for profiles for which this is not true by substituting μi = XiB and Σi = Q + S for (5) and (6), respectively. Recall that in our example Q represents the child-to-child covariance and S the day-to-day covariance. Inclusion of the child-to-child variability tends to inflate the imputation variances.
This approach has several desirable qualities. It is easy to implement and makes full use of the functional data, borrowing strength both within and between profiles while performing the imputation. The borrowing of strength from within the profile is accomplished through the conditional distribution in (7), and the borrowing of strength between profiles occurs from the use of parameter estimates from the complete case analysis to characterize the distributions in (5) and (6). Because multiple draws are taken from the imputation distribution, the estimated variability of the imputation is propagated throughout any subsequent inference.
However, one weakness of this approach is that by conditioning on the parameter estimates from the complete profile model fit, it fails to propagate the uncertainty of the parameter estimation into the imputation variances. This problem can be fixed by using posterior predictive distributions instead, which integrate over this uncertainty; that is, replace the conditional mean and covariance in (5) and (6) in the imputation distribution in (7) by the posterior predictive mean and covariance, and , respectively. Given a set of G MCMC samples from the complete profile run, these may be estimated by the following equations:
| (8) |
| (9) |
with , and B(g) and U(g) being the posterior samples of B and U from iteration g of the complete case MCMC, and the posterior mean estimator of the within-function residual covariance matrix S. Details of the derivations of (8) and (9) are in the appendix. These distributions appropriately propagate the uncertainty in parameter estimation throughout the imputation distributions. They are not fully efficient, however, since they integrate over the posterior distribution of the parameters conditional on only the “complete case” data, while the fully Bayesian approach would integrate over the posterior conditional on all available data.
Note that because of the mixture prior placed on B*, the predictive distribution f(Yi|YC) is not multivariate normal. Thus, by representing this distribution by its first two moments and using the normal distribution to perform our imputation, we are using an approximation. This keeps the procedure computationally feasible, since sampling from the actual posterior predictive distribution would require multiple DWT and IDWT calculations within the MCMC, as was the case for the fully Bayesian approach. Our use of the posterior predictive means and variances improves upon the conditional approach because it propagates the uncertainty due to parameter estimation in the correct manner, as indicated by the second term in (9).
5 Analysis of TriTrac Data from Planet Health
5.1 Complete Case Analysis
We first modeled the set of complete profiles in order to obtain posterior predictive distributions to characterize the imputation distributions for the missing parts of the incomplete profiles. There were a total of 95 complete daily profiles from 61 children, i.e., had no missing observations between 9am and 8pm. Let YC be the 95 × 660 response matrix, each row of which contained the log accelerometer profile (in log METs) for one day from one child, sampled on the time grid t = {tm, m = 1, … ,660}, consisting of every minute from 9am to 8pm. We log transformed the data to stabilize the variance and make the data more symmetric to accommodate the Gaussian error assumptions underlying the functional mixed model.
We modeled these data using the functional mixed model (3), YC = XB + ZU + E. The matrix X was a 95×13 design matrix containing child and day-level covariates which correspond to the 13 × 660 matrix B, whose rows were the fixed effect functions on the grid t. Entry Bij described the effect of covariate i on the activity levels at time tj. The matrix Z was a 95 × 61 design matrix, with corresponding 61 × 660 matrix U, for which each row contained the random effect function for a single child on the grid t. The 95 × 660 matrix E contained the residual errors on the same grid. We assumed that the rows of U and E were mutually independent mean zero Gaussians with 660 × 660 covariance matrices Q and S, respectively.
The X matrix we used included columns corresponding to an overall functional “intercept”, a gender effect (=1 if boy, -1 if girl), an effect for higher tricep skinfolds (=1 if > 20, =-1 if ≤ 20), a body mass index (BMI) effect, an effect for the average number of hours per day watching television, day-of-week effects, a seasonal effect (=1 if after the beginning of daylight savings time [DST], =0 if before DST), and school effects. All effects were functional, meaning that they were allowed to vary as a function of time of day. Both the BMI and TV hours covariates were treated as continuous, with the BMI covariate mean-centered and the TV hours covariate standardized. School effects were only included for schools B, D, and E, since there were only 1 and 4 complete profiles from schools A and C, respectively out of the total of 11 and 15 that were at least 50% complete from these two schools. We determined this was not enough replication to estimate the corresponding school fixed effects.
Written out in scalar form, the model for profile i at time , was , where Uk(tj) were mean-zero Gaussians with if k = k′, 0 otherwise; Ei(tj) were mean-zero Gaussians with cov{Ei(tj),Ei′(tj′)} = Sjj′ if i = i′, 0 otherwise; and cov{Uk(tj),Ei(tj′)} = 0 for all i, j, j′, k.
We fit the wavelet-based functional mixed model described in Section 3.3 and obtained posterior samples for all parameters in this model. We chose a Daubechies wavelet with 4 vanishing moments (Daubechies 1992), and performed the wavelet decomposition to J = 8 levels. Maximum likelihood estimates were used as starting values for the MCMC for all model parameters. The regularization parameters were estimated using the empirical Bayes approach described in Morris and Carroll (2006), with πij ≈ 1 and τij chosen very large for the smooth coeffcients and the 2 coarsest levels of wavelet coeffcients to make the degree of shrinkage at these levels negligible. Vague proper priors were used for the variance components. After a burn-in of 1000, we obtained 20,000 MCMC samples of the model parameters, keeping every 10th. This took roughly 7 hours to run on our Xeon 3.2 GHz Windows 2000 machine with 2GB RAM, and the posterior samples occupied roughly 300MB. Trace plots and Metropolis-Hastings acceptance probabilities indicated that the MCMC had good convergence properties.
5.2 Imputation Distributions for Incomplete Profiles
We used these posterior samples to estimate the posterior predictive means and variances for each incomplete profile, and in (8) and (9). As described in Section 4, we used these estimates as the basis for the imputation distributions for the missing data.
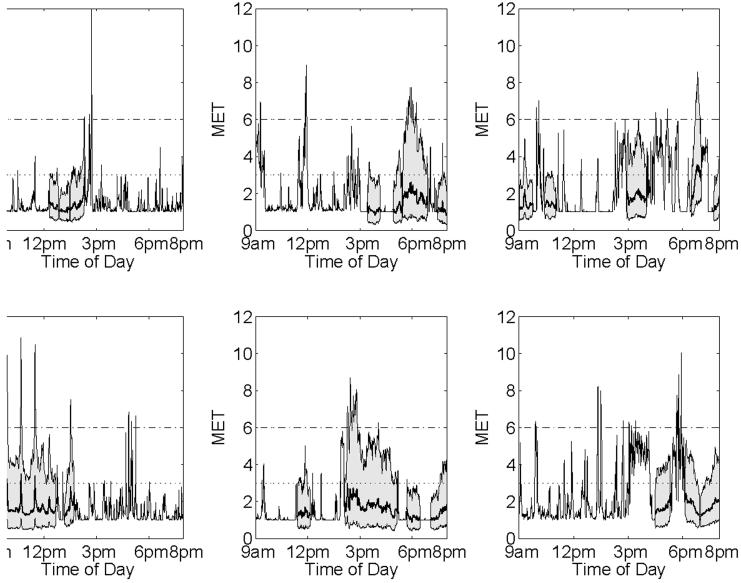
Figure 3 contains six incomplete profiles, with their missing regions replaced by the mean and 95% pointwise bounds of their imputation distributions. Note how the imputation distributions borrow strength from nearby observations and tend to “connect” the imputed data with nearby observed data. They also allow the magnitude of the nearby observed data to play a role in the imputation. Also, the imputation variances tend to be smaller for the values immediately adjacent to observed values, and in regions of the curves with less variability in the population.
Figure 3. Imputation distributions.
Some incomplete profiles, with mean and 95% pointwise bounds from the joint imputation distribution substituted for the missing regions, as indicated by the shaded regions. These are based on the normal approximation to the posterior predictive distribution, estimated from posterior samples obtained from a Markov Chain Monte Carlo scheme applied to the complete case data.
Using the DWT, we projected these imputation distributions into the wavelet space. At each iteration of the MCMC applied to the full data set, we took random draws from this distribution to fill in the “missing wavelet coeffcients.”
5.3 Model for Analysis of Full Data Set
We used the same underlying model for the full data analysis as for the complete case analysis, except we removed the functional intercept term and allowed each school to have its own fixed effect function, since the full data set contained enough profiles to reliably estimate them all.
We obtained posterior samples for all parameters in this model using an MCMC, with the missing wavelet coeffcients sampled from their imputation distributions at each iteration, as described in Section 4. We used the same method settings as used in the complete case analysis. We ran four parallel chains, each with a burn-in of 1000 and consisting of 2500 MCMC samples, keeping every 5. This took roughly 40 hours to run, and the posterior samples occupied roughly 300MB. For 87% of the variance components, the Metropolis-Hastings acceptance probabilities were between 0.25 and 0.50. Trace plots indicated good convergence properties.
5.4 Results
Analysis of Fixed Effect Functions
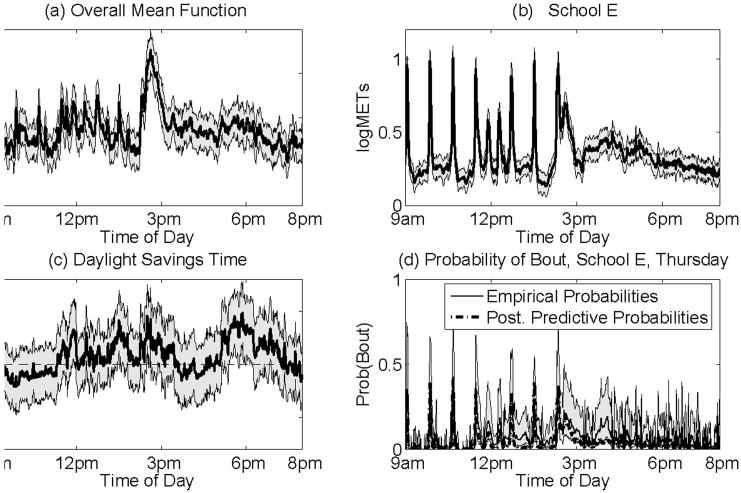
To consider the marginal time of the day effect on activity levels, we computed the overall mean curve by equally weighting each of the schools' fixed effects functions and conditioning on the mean values of all other covariates. Figure 4(a) contains the posterior mean and 90% posterior pointwise credible bounds for the overall mean curve in the logMETs scale. Throughout the school day, the curve is characterized by numerous spikes that indicate periods of time of coordinated activity, e.g., class switches and lunch periods for the different schools. There is a large spike between 2:15pm and 3:00pm corresponding to the end of the school day. The mean activity profiles are less spiky after school, indicating less coordinated activity across days and children. The mean activity levels remained relatively constant from 3:00pm to 6:30pm, then decreased later in the evening.
Figure 4. Results.
Posterior mean curves and 90% pointwise posterior credible intervals for the fixed effects curves for (a) overall mean curve, (b) school E, and (c) daylight savings time, plus (d) posterior predictive probabilities of bouts for a child from school E on Thursdays. The curve in (a) is obtained by averaging equally over the schools and conditioning on the mean value in the data set for all other covariates. The curve in (b) can be interpreted as the mean log profile on a Monday before daylight savings time for a child from school E with average BMI who is average with respect to the average number of hours of TV they watch per day. The curve in (c) is added to the mean log profile for any day after daylight savings time (April 6). The solid line in curve (d) indicates the wavelet-denoised empirical probabilities of bouts for each time point averaging over the 42 accelerometer profiles obtained on Thursdays from children enrolled at school E, along with 95% pointwise confidence bands computed using the method described in Bruce and Gao (1996). The dashed-dotted line indicates the posterior predictive probability of bouts from the functional mixed model for a child enrolled at school E on Thursday, for whom the levels of all other covariates are at their mean levels.
For each fixed effect function, we plotted the posterior mean curve and computed pointwise 90% posterior bounds. Two of these are plotted in Figure 4, panels (b) and (c), and other curves are available by request from the first author. Recall that the effects are additive on the log scale, so they are multiplicative on the MET scale. Exponentiating the values in these plots yields the multiplicative effects. These multiplicative effects can subsequently be converted into percent increases or decreases in the MET scale.
To further summarize the results, we computed the average percent increases/decreases corresponding to each fixed effect function, aggregating within the following time intervals, chosen based on the structure evident in the overall mean curve: (1) all day, 9am-8pm; (2) morning, 9am-11:30am; (3) lunch, 11:30am-12:30pm; (4) afternoon, 12:30pm-2:15pm; (5) going home, 2:15pm-3:00pm; (6) after school, 3:00pm-5:30pm; (7) early evening, 5:30pm-7:00pm; and (8) late evening, 7:00pm-8:00pm. The 90% credible intervals for these quantities are reported in square brackets.
The fixed effect curves for the different schools were characterized by numerous peaks present throughout the school day. These were most evident for school E; see Figure 4(b). This school provided the most daily profiles in this data set (146 out of 292), and it appeared to have the most regular schedule of the schools in the study; see Figure 2. The regular pattern of peaks reveals the school's class schedule. The large peaks in the morning and afternoon are exactly 48 minutes apart, and indicate class switches. The three peaks around noon are 24 minutes apart, and likely indicate different lunch periods. From the large extended spike in the afternoon, we surmise that school let out around 2:15pm. Although regular peaks were evident in the school's main effect curve, they were not obvious in any individual curve because of the high levels of day-to-day and child-to-child variability. This can be seen in the bottom two panels of Figure 1, which contain profiles from a child at this school.
We found that children tended to be less active on Tuesdays (3.3% decrease [-6.2%, -0.2%]), Thursdays (2.9% decrease [-5.5%, -0.2%]), and Fridays (3.6% decrease [-6.8%, -0.4%]), compared to Mondays, aggregating over the entire day. These effects were especially strong during the after-school period (3:00-5:30, Tuesday -7.7% [-12.6%, -2.2%], Thursday -7.8% [-12.5%, -3.1%], Friday -5.8% [-11.4%, -0.1%]) A similar but less pronounced trend was present on Wednesdays (overall 1.9% decrease [-5.0%, 1.4%], after school 5.1% decrease [-10.5%, 0.9%]).
In 1997, daylight savings time (DST) in Boston started on April 6th. We found that activity levels were generally higher after DST than before DST (overall 8.2% increase [-0.5%, 18.1%]). See Figure 4(c). This effect was especially strong as school was letting out (2:15-3:00, 25% increase [4.2%, 53.4%]) and in the early evening (5:30-7:00, 29.5% increase, [5.6%, 59.2%] ), a time period during which the sun was still out after DST, but had already set before DST.
Boys tended to be slightly more active than girls, on average, most notably over lunch (4.8% increase [0.5%, 9.1%]).
Variance Component Analysis
We summarized the relative day-to-day variability by ρ = Tr(S)/{T(Q + Tr(S)}, where Tr(Q) and Tr(S) were the traces of the child-to-child and day-to-day covariance matrices Q and S, respectively. The posterior mean of ρ was 90.9%, indicating 90.9% of the variability in the log accelerometer profiles was day-to-day and 9.1% child-to-child. The 90% posterior credible interval for ρ was [88.8%, 92.8%].
To evaluate how these sources of variability varied over t, we considered the child-to-child and day-to-day variances as a function of t, Vq(t) = Q(t, t) and Vs(t) = S(t, t), respectively (plots available in supplementary material available at (http://biostatistics.mdanderson.org/Morris/papers_files/TriTrac_supplement.html) describes how this implementation deals with data for. We found the child-to-child variability was greater during the school day than after school, while the day-to-day variability was greater after school than during the school day. The relative variability from day-to-day as a function of time of the day, ρ(t) = Vs(t)/{Vq(t) + Vs(t)}, revealed that the percent of variability from day-to-day increased from 85-90% during the school day up to 95-99% after 3pm.
Assessing Bouts
In the literature on accelerometer data, there has been a great deal of interest in studying “bouts,” or short bursts of intense activity. Here, we considered bouts to be periods of time during which the child was engaging in moderate or vigorous activity, which corresponded to activity levels of greater than 3.0 MET (Rowlands et al. 2004). In the existing literature, the data analysis performed frequently includes estimating the probabilities of bouts for different groups of individuals. In our wavelet-based functional mixed model approach, we did not directly model these probabilities, but the unified Bayesian framework underlying the method made it easy to compute these probabilities as a function of the time of day for any group of individuals using the posterior samples output from the MCMC.
Given a child with covariate levels X*, the posterior predictive probability that they experience a bout at time t in a hypothetical future daily activity profile Y*(t) is given by ϕWFMM(t = Pr{Y*(t) > 3.0|Y,X*} = ∫ I{Y*(t) > 3.0}f{Y*(t)|θ,X*)}f(θ|Y)dθ, where θ = {B, Q, S} are the fixed effect and covariance parameters in the model, and Y are the profiles we used to fit the model. This expression can be estimated from the G posterior samples θ(g) = {B(g), Q(g), S(g)}, g = 1, … , G, obtained from the MCMC, by , where , and Φ(x) is the standard normal cdf evaluated at x.
For example, we computed the probability of bouts as a function of the time of the day for Thursdays before DST for children from school E. This curve is given by the dashed line in Figure 4(d). To check how well our model captured these bouts, we also computed an empirical estimate of the probability of bouts for each time point averaging over the n = 42 accelerometer profiles obtained on Thursdays before DST from children enrolled at school E. The raw empirical estimator was . The solid line in Figure 4(d) is a wavelet denoised version of this curve, denoised using Sureshrink (Donoho and Johnstone, 1995) with soft thresholding. Although we did not directly model the probability of bouts, our wavelet-based functional mixed model appeared to do a reasonable job of capturing these features of the data.
6 Discussion
We first discuss the specific results of our analysis, and then assess the fitness of the wavelet-based functional mixed model for analyzing accelerometer data.
We were not surprised by the large daylight savings time (DST) effect we observed in the data. Considering the weather in Boston, naturally children tended to be more active after April 6th than before. In 1997, the sun set in Boston at times ranging from 5:10pm to 6:15pm from February 10th through April 5th, then from 7:16pm through 8:11pm from April 6th through May 28th. This may explain the very large spike in the DST effect between 5pm and 7pm.
In our relative variability analysis, we found that after adjusting for the covariates, the day-to-day dominated the child-to-child variability, especially during the after-school hours. In other words, the difference between the average log profiles of a very active and very inactive child with the same covariate levels is small compared to the variability in activity for a given child from day-to-day, especially after school. This suggests that it is important to get many days per child in order to accurately quantify each child's typical level of activity. While it is already typical in these studies to monitor each child for 4 to 8 days, it may be a better design to obtain even more days per child, especially if this does not result in much increase in cost for the study.
While not directly modeling the bouts, the wavelet-based functional mixed model yields estimates of the probability of bouts at different times of the day for children with different levels of the covariates. It appears that this model does a reasonable job of capturing these bouts, indicating that it is at least somewhat effective in modeling some of the tail behavior in the distributions. However, there is some evidence of attenuation in our model-based estimates of these probabilities, especially during the after-school period in which there is less coordinated activity and more day-to-day variability. These may be related to our underlying assumptions of normality and equal covariances across different groups of children. It is possible in our framework to allow different covariance parameters across different groups of children, and to relax the normality assumptions using scale mixtures of normals. These adaptations would accommodate heavier tails and may do an even better job of capturing these bouts.
The wavelet-based functional mixed model is a powerful tool for analyzing accelerometer data. It allows one to consider the joint functional effects of multiple covariates, and has the ability to model correlations between profiles obtained from the same individual using random effect functions. The functions are allowed to be of arbitrary form, and the within-function covariances are allowed to be nonstationary. The fixed effect functions are adaptively regularized using the principles of wavelet shrinkage. Less adaptive methods using kernels or splines would result in more attenuation of dominant local features in the fixed effects curves, possibly resulting in reduced power for inference.
It is also possible to introduce other random effect functions to account for covariance due to other clustering factors, such as school or neighborhood. In this analysis, we chose to model the schools using fixed effects because there were so few of them, but given more schools, we could use random effects. It would be a good idea to use more schools in these studies, since the school-to-school variability appears to be large, and it would be interesting and important to study a wider range of schools with different socioeconomic makeups.
Our model is linear in that we assume the effects are linear in the covariates. It would be interesting to consider generalizing or testing this assumption. However, this framework is still very flexible since the linear coefficients are allowed to be time-varying, and their functional form over time is left arbitrary. A much less flexible model, for example, would be to allow the overall mean function to be arbitrary, but to force the fixed effects to be constant over time. It would be interesting to develop formal testing procedures for testing whether it is necessary to allow the coefficient to be time-varying for a given covariate; this is a topic for future investigation.
Also, we note that this model is very flexible in its representation of the covariances of the profiles from day-to-day, S, and the covariances of the random effect profiles from child-to-child, Q. Our assumption of independence in the wavelet space reduces the dimensionality of these covariances from T(T +1)/2 to T , yet accommodates a reasonably wide range of nonstationary covariance structures. Different variance components are estimated for each wavelet coeffcient, which allows the day-to-day and child-to-child variability to differ across both scale and location. This accommodates various types of nonstationarity, allowing the day-to-day and child-to-child variances across profiles to vary over time, and also allowing the level of smoothness in the random effect functions and residual error processes to vary over time.
The missing data methods introduced in this paper allow the wavelet-based functional mixed model to be applied to data with incomplete profiles, which are commonly encountered in accelerometer data. In our case study, these methods allowed us to increase the number of profiles in the analysis by a factor of three. The resulting gain in precision is evident if one compares the posterior bounds from the full data and complete case analyses (not shown).
The wavelet-based functional mixed model method possesses some weaknesses and limitations. It is based on linear models, and so assumes additive errors (in our case study, additive on the log scale). Other types of variability may not be adequately picked up by these models. The normality assumption is somewhat restrictive, although as previously mentioned this could be relaxed using scale mixtures. As is true with nonfunctional mixed models, some models can be unstable to fit, especially when there is near-collinearity in the design matrices, or small effective sample sizes for some of the variance components. These problems are exacerbated in the functional context, since we are effectively fitting T simultaneous scalar mixed models. Also, it would be useful to have a global functional test for more formally assessing the significance of the fixed effect functions; this is an area we will investigate in the near future. The method is computationally and memory intensive; the analyses performed for this paper took a total of about 50 computer hours. However, this is not unreasonable considering the time it takes to collect the data. The method is also highly parallelizable, so it could be done in much less time whenever parallel computing resources are available.
While computationally intensive, the method is straightforward to implement. The minimal information one must specify to use our scripts are the Y , X, and Z matrices, the wavelet basis, the number of levels of decomposition, the number of MCMC samples, and the burn-in. By default, maximum likelihood starting values, empirical Bayes regularization parameters, Fisher's information-based proposal variances for the Metropolis-Hastings, and vague priors on the variance components are automatically computed.
Although straightforward to implement, this method is considerably complex, combining wavelets with mixed models and wrapping it all up inside a Markov Chain Monte Carlo. This begs the question of what all this complexity buys, of what this method can do that simpler approaches cannot. This functional data modeling approach can be used to perform the same types of standard analyses found in the existing literature, including analyses of average daily activity levels, 30-minute averages, and probabilities of bouts for different groups of individuals, but these can be done more effciently because we can effectively incorporate incomplete profiles into the analysis. More importantly, the functional approach opens new possibilities in terms of analyses that can be done and types of information that can be extracted from these rich data, for example, allowing us to perform inference on time-varying effects. Given the posterior samples from our model fit, we can perform nearly any type of inference we desire, functional or pointwise, on fixed effect functions, random effect functions, or covariance functions. The Bayesian approach propagates the uncertainty from the different sources of variability in the model and the different parameters estimated throughout any subsequent inference.
The wavelet-based functional mixed model, supplemented with the missing data methods introduced in this paper, comprises a promising tool for extracting information from accelerometer data in activity level studies.
Supplemental Material
Acknowledgements
Jeffrey Morris was supported by the National Cancer Institute(CA-107304), Louise Ryan was supported by CA-48061, and Brent Coull was supported by ES012044. Steven Gortmaker was supported by: National Institutes of Child Health and Human Development (HD-30780), and the Centers for Disease Control and Prevention (Prevention Research Centers Grants U48/CCU115807 and U48/DP00064-00S1). We thank the associate editor and two reviewers whose comments have led to a substantially improved paper.
Appendix A: Derivation of Posterior Predictive Means and Covariances
Suppose YC is an NC × T matrix whose rows contain NC complete profiles, observed on an equally spaced grid t of length T. Assume this matrix is modelled using the functional mixed model (3) with parameters Θ = {B, U, Q, S}. Let Yi be another profile following the same model that is independent of each profile in YC conditional on Θ. Here we show that, given a set of G posterior samples of (Θ|YC), we can estimate the posterior predictive mean and covariance matrix by the expressions in (8) and (9).
We start by showing (8). It is easy to show using alternating conditional expectation arguments that if X and Y are independent, conditional on Z, then E(X|Y) = EZ|Y{E(X|Z)}. Since Yi and YC are independent conditional on Θ, this means that . Estimating this expectation using the MCMC samples of Θ, we get .
To show (9), we first note that alternating conditional expectation arguments can be used to show that if X and Y are independent, conditional on Z, then var(X|Y) = EZ|Y {V AR(X|Z)}+ varZ|Y {E(X|Z)}. This means that we can write .
Since var(Yi|Θ) = S, the first term can be estimated by , the posterior mean estimator of S computed from the MCMC samples, which is the first term in (9). Next, we can write . This expression can be estimated using the MCMC samples by , which is the second part of (9).
References
- Abbott AP, Davies PS. Habitual Physical Activity and Physical Activity Intensity: Their Relation to Body Composition in 5.0-10.5-y-old Children. European Journal of Clinical Nutrition. 2004;58(2):285–291. doi: 10.1038/sj.ejcn.1601780. [DOI] [PubMed] [Google Scholar]
- Abramovich F, Sapatinas T, Silverman BW. Wavelet Thresholding via a Bayesian Approach. Journal of the Royal Statistical Society, Series B. 1998;60:725–749. [Google Scholar]
- Algase DL, Beattie E, Leitsch SA, Beel-Bates CA. Biomechanical Activity Devices to Index Wandering Behavior in Dementia. American Journal of Alzheimer's Disease and other Dementias. 2003;18(2):85–92. doi: 10.1177/153331750301800202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJ, Fearn T, Vannucci M. Bayesian Wavelet Regression on Curves with Application to a Spectroscopic Calibration Problem. Journal of the American Statistical Association. 2001;96:398–408. [Google Scholar]
- Bruce AG, Gao HY. Understanding WaveShrink: Variance and Bias Estimation. Biometrika. 1996;83:727–745. [Google Scholar]
- CDC . Technical Report, Centers for Disease Control and Prevention. 2000. Promoting Better Health for Young People Through Physical Activity and Sports: A Report to the President from the Secretary of Health and Human Services and the Secretary of Education. [Google Scholar]
- CDC NCCDPHP . Technical Report, Centers for Disease Control and Prevention. 2001. Physical Activity and Good Nutrition, Essential Elements to Prevent Chronic Disease and Obesity, at a Glance 2001. [PubMed] [Google Scholar]
- Chang W, Vidakovic B. Wavelet Estimation of a Base-Line Signal from Repeated Noisy Measurements by Vertical Block Shrinkage. Computational Statistics and Data Analysis. 2002;40:317–328. [Google Scholar]
- Chipman HA, Kolaczyk ED, McCulloch RE. Adaptive Bayesian Wavelet Shrinkage. Journal of the American Statistical Association. 1997;92:1413–1421. [Google Scholar]
- Clyde M, George EI. Flexible Empirical Bayes Estimation for Wavelets. Journal of the Royal Statistical Society, Series B. 2000;60:681–698. [Google Scholar]
- Clyde M, Parmigiani G, Vidakovic B. Multiple Shrinkage and Subset Selection in Wavelets. Biometrika. 1998;85:391–401. [Google Scholar]
- Coleman KJ, Saelens BE, Wiedrich-Smith MD, Finn JD, Epstein LH. Relationships Between TriTrac-R3D Vectors, Heart Rate, and Self-Report in Obese Children. Medicine and Science in Sports and Exercise. 1997;29(11):1535–1542. doi: 10.1097/00005768-199711000-00022. [DOI] [PubMed] [Google Scholar]
- Cradock AL, Weicha JL, Peterson KE, Sobol AM, Colditz GA, Gortmaker SL. Youth Recall and TriTrac Accelerometer Estimates of Physical Activity Levels. Medicine and Science in Sports and Exercise. 2004;36(3):525–532. doi: 10.1249/01.mss.0000117112.76067.d3. [DOI] [PubMed] [Google Scholar]
- Daubechies I. Ten Lectures on Wavelets. Society for Industrial and Applied Mathematics; Philadelphia, PA: 1992. [Google Scholar]
- Dietz WH. Health Consequences of Obesity in Youth: Childhood Predictors of Adult Disease. Pediatrics. 1998;101:518–558. [PubMed] [Google Scholar]
- Donoho DL, Johnstone IM. Adapting to Unknown Smoothness via Wavelet Shrinkage. Journal of the American Statistical Association. 1995;90:1200–1224. [Google Scholar]
- Epstein LH, Paluch RA, Coleman KJ, Vito D, Anderson K. Determinants of Physical Activity in Obese Children Assessed by Accelerometer and Self-Report. Medicine and Science in Sports and Exercise. 1996;28(9):1157–1164. doi: 10.1097/00005768-199609000-00012. [DOI] [PubMed] [Google Scholar]
- Going S, Thompson J, Cano S, Stewart D, Stone E, Harnack L, Hastings C, Norman J, Corbin C. The Effects of the Pathways Obesity Prevention Program on Physical Activity in American Indian Children. Preventive Medicine. 2003;37(6):S62–S69. doi: 10.1016/j.ypmed.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Goldfield GS, Kalakanis LE, Ernst MM, Epstein LH. Open-loop Feedback to Increase Physical Activity in Obese Children. International Journal of Obesity and Related Metabolic Disorders. 2000;24(7):888–892. doi: 10.1038/sj.ijo.0801248. [DOI] [PubMed] [Google Scholar]
- Gortmaker SL. The Role of Physical Activity Environment in Obesity Among Children and Adolescents in the Industrialized World. In: Chen C, Dietz WH, editors. Obesity in Childhood and Adolescents. Vol. 49. Vevey/Lippencott Wiliams & Wilkins; Nestec Ltd., Philadelphia: 2002. Nestle Nutrition Worshop Series, Pediatric Program. [Google Scholar]
- Gortmaker SL, Peterson K, Wiecha J, Sobol A, Dixit S, Fox M, Laird N. Reducing Obesity via a School-Based Interdisciplinary Intervention among Youth: Planet Health. Archives of Pediatric and Adolescent Medicine. 1999;153:409–418. doi: 10.1001/archpedi.153.4.409. [DOI] [PubMed] [Google Scholar]
- Johnstone IM, Silverman BW. Wavelet Threshold Estimators for Data with Correlated Noise. Journal of the Royal Statistical Society, Series B. 1997;59:319–351. [Google Scholar]
- Laird N, Ware JH. Random-Effects Models for Longitudinal Data. Biometrics. 1982;38:963–974. [PubMed] [Google Scholar]
- Little RJA, Rubin DB. Statistical Analysis with Missing Data. Wiley; Hoboken, NJ: 2002. [Google Scholar]
- Matthews CE, Freedson PS. Field Trial of a Three-Dimensional Activity Monitor: Comparison with Self-Report. Medicine and Science in Sports and Exercise. 1995;27(7):1071–1078. doi: 10.1249/00005768-199507000-00017. [DOI] [PubMed] [Google Scholar]
- Misiti M, Misiti Y, Oppenheim G, Poggi JM. Wavelet Toolbox For Use with Matlab: User's Guide. Mathworks, Inc.; Natick, MA: 2000. [Google Scholar]
- Morris JS, Carroll RJ. Wavelet-Based Functional Mixed Models. Journal of the Royal Statistical Society, Series B. 2006 doi: 10.1111/j.1467-9868.2006.00539.x. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JS, Vannucci M, Brown PJ, Carroll RJ. Wavelet-Based Non-parametric Modeling of Hierarchical Functions in Colon Carcinogenesis. Journal of the American Statistical Association. 2003;98:573–597. [Google Scholar]
- Pearce KD. Race, Ethnicity, and Physical Activity. Leisure Today JOPERD. 1999;70(1):25–28. [Google Scholar]
- Percival DB, Walden AT. Wavelet Methods for Time Series Analysis; New York: Cambridge: 2000. [Google Scholar]
- Ramsay JO, Silverman . Functional Data Analysis. Springer; New York: 1997. [Google Scholar]
- Recommended Dietary Allowances. 10th ed. National Academy Press; Washington, DC: 2000. [Google Scholar]
- Rowlands AV, Eston RG, Ingledew DK. Relationship between Activity Levels, Aerobic Fitness, and Body Fat in 8- to 10-yr-old Children. Journal of Applied Physiology. 1999;86(4):1428–1435. doi: 10.1152/jappl.1999.86.4.1428. [DOI] [PubMed] [Google Scholar]
- Rowlands AV, Thomas PW, Eston RG, Topping R. Validation of the RT3 Triaxial Accelerometer for the Assessment of Physical Activity. Medicine and Science in Sports and Exercise. 2004;36(3):518–524. doi: 10.1249/01.mss.0000117158.14542.e7. [DOI] [PubMed] [Google Scholar]
- Strauss RS, Rodzilshy D, Burack G, Colin M. Psychosocial Correlates of Physical Activity in Healthy Children. Archives of Pediatrics and Adolescent Medicine. 2001;155(8):897–902. doi: 10.1001/archpedi.155.8.897. [DOI] [PubMed] [Google Scholar]
- Talbot LA, Gaines JM, Huynh TN, Metter EJ. A Home-Based Pedometer-Driven Walking Program to Increase Physical Activity in Older Adults with Osteoarthritis of the Knee: A Preliminary Study. Journal of the American Geriatric Society. 2003;51(3):387–392. doi: 10.1046/j.1532-5415.2003.51113.x. [DOI] [PubMed] [Google Scholar]
- Vannucci M, Corradi F. Covariance Structure of Wavelet Coeffcients: Theory and Models in a Bayesian Perspective. Journal of the Royal Statistical Society, Series B. 1999;61:971–986. [Google Scholar]
- Vidakovic B. Statistical Modeling by Wavelets. Wiley; New York: 1999. [Google Scholar]
- Westerterp KR. Physical Activity Assessment with Accelerometers. International Journal of Obesity and Related Metabolic Disorders. 1999;23(Suppl 3):S45–S49. doi: 10.1038/sj.ijo.0800883. [DOI] [PubMed] [Google Scholar]
- Westerterp KR. Pattern and Intensity of Physical Activity. Nature. 2002;410:539. doi: 10.1038/35069142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.