Abstract
Indigenous chickens (IC) in developing countries provide a useful resource to detect novel genes in mitochondrial and nuclear genomes. Here, we investigated the nature and level of genetic diversity in IC from five distinct regions of Sri Lanka using a PCR-based resequencing method. Additionally, we investigated the relatedness of IC to different species of junglefowls including Ceylon (CJF; Gallus lafayetti), a subspecies that is endemic to Sri Lanka, green (Gallus varius), grey (Gallus sonneratii), and red (Gallus gallus) junglefowl. A total of 140 birds including eight CJF were used to screen 613 bp of IC and 675 bp of CJF control region of the mitochondrial DNA sequence for single nucleotide polymorphisms (SNPs) and other variants. We detected and validated 44 SNPs, which formed 42 haplotypes and six haplogroups in IC. The SNPs observed in the CJF were distinct and the D-loop appeared to be missing a 62 bp found in IC and the red junglefowl (RJF). Among the six haplogroups of IC, only one was region-specific. Estimates of haplotype and nucleotide diversities ranged from 0.901 to 0.965 and from 0.011 to 0.013, respectively. Estimates of genetic divergence were inconsistent but generally low in all the regions. Further, variation among individuals within regions accounted for 92% of the total molecular variation among birds. The Sri Lankan indigenous chickens were more closely related to red and grey junglefowls than to CJF, suggesting a multiple origin. The molecular information on genetic diversity revealed in our study may be useful in developing genetic improvement and conservation strategies to better utilize indigenous Sri Lankan chicken resources.
Keywords: Indigenous chicken, Genetic diversity, Sri Lanka
Introduction
Diversity among farm animals within and among countries is of major interest to the scientific community as it is a significant resource for livestock development and for responding to changing needs and production requirements. With increasing world population, there is concern that the growing demands for animal products are eroding these genetic resources especially in developing countries, where most of the diversity is found. In recognition of this concern, many efforts have begun to characterize animals in developing countries to provide a foundation for developing sustainable genetic improvement approaches. Chief among these efforts is the program by the Food and Agricultural Organization (FAO) of the United Nations to develop a Global Strategy for the Management of Farm Animal Genetic Resources or FAnGR (Gibson et al., 2005; http://www.fao.org/ag/cgrfa/AnGR.htm).
Some efforts to understand diversity in native or indigenous chickens (IC) in developing countries have been described. Using microsatellites and DNA pools, Hillel et al. (2003) evaluated variation in 52 chicken populations that included selected and unselected chicken stocks. Though estimates varied, it was observed that unselected populations were more diverse. In another global diversity assessment in IC and commercial chickens, Granevitze et al. (2007) reported that populations from Asian countries that included China and Vietnam had high microsatellite-based heterozygosities that reflect their management histories. Mwacharo et al. (2007) also recently used microsatellites to assess genetic diversity among African IC from Ethiopia, Kenya, Sudan and Uganda. The populations appeared to cluster by region as could be expected from geographic (or reproductive) isolation. A similar analysis of relatedness within and among Chinese IC using microsatellites was reported by Qu et al. (2006). The indigenous breeds were also highly diverse and clustered by geographic regions.
Efforts to understand genetic diversity in commercial and non-commercial chickens have also involved characterization of variation in the mitochondrial genome. Though relatively few, the mitochondrial DNA (mtDNA) based studies have also provided insight into the maternal origins of chickens (Akishinonomiya et al., 1994; Liu et al. 2006; Niu et al., 2002). In a diversity study involving the D-loop of the mitochondria, Oka et al. (2007) evaluated genetic variation within and among Japanese IC and also assessed their routes of introduction into Japan. Using haplogroups, they showed that indigenous chickens in Japan have multiple origins that include both game and non-game chickens. Given the consensus on the Asian origins of domestic chickens (Akishinonomiya et al., 1994; 1996), characterization of IC in Asian countries thus continues to be of interest.
Sri Lankan IC are geographically isolated from the rest of the Indian sub-continent. This isolation may be responsible for the Ceylon Junglefowl (CJF, Gallus lafayetti) being endemic to Sri Lanka (Ceylon). Like with other Gallus species, the relationship between CJF and the domestic chicken as well as with other Galliformes remains of interest to scientists. There is a lack of consensus, for example, on whether the domestic chicken has a mono- or polyphyletic origin that could include the CJF. Recently, Krieg et al. (2007) used annotated retroposed element activity to find the evolutionary evidence of Galliformes. Using the fixation pattern of the transposed elements in different galliformes, they revealed that CJF and Red Junglefowl hold very close taxonomic positions in the phylogenetic tree of Galliformes. However, contribution of CJF in evolutionary process of domestic chicken is not yet been fully revealed. As a crossroads of ancient sea trade routes that connect Asia and the western world, Sri Lanka has been enriched with a variety of animal germplasms including chicken, which eventually developed into a distinct indigenous population. Diversity and relatedness among Sri Lankan IC have been little examined. Using randomly amplified polymorphic DNA analysis, Silva and Rajapaksha (2005) reported that the Sri Lankan IC, when considered as a relatively homogenous group, were more closely related to commercial chicken, the Rhode Island Red, than to CJF. To determine if chickens within Sri Lanka are a homogenous group and to further evaluate the relatedness of the IC with CJF, chickens from five different geographical locations were evaluated for genetic diversity in the mitochondrial D-loop. The data may provide some useful information in the ongoing debate about whether chickens have a single (Fumihito et al., 1996; Hillel et al., 2003) or multiple origins (Moiseyeva et al., 1998).
Materials and Methods
Samples
Blood samples were collected on FTA cards (Whatman, Inc.) from a total of 132 IC in five different geographical regions including north-central (NCP), north-western (NWP), southern (SP), uva (UP), and western (WP) provinces (Figure 1). Within each region, samples were collected from several birds from multiple households in different villages. To minimize the chances that the birds used from each village were related, a single bird was used from each household. The households within each village from which each bird was used were approximately 0.5 to one mile apart. Blood was also collected on FTA cards from 9 birds identified as CJF at the national Zoo (3 birds) and from the wild in central Sri Lanka (6 birds).
Figure 1.
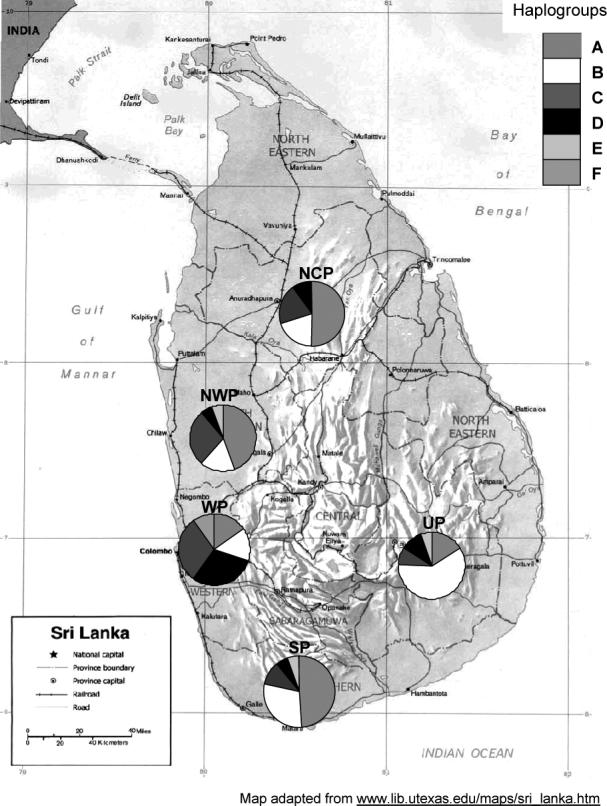
Sampling sites in Sri Lanka. NCP, NWP, SP, UP, and WP represent north central province, north western province, western province, uva province and southern province, respectively. Regional frequency distribution of the haplogroups is shown by each pie chart. The number of birds sampled from each region is presented in Table 2.
Molecular analysis
Extraction of DNA from the FTA cards was according to the recommended protocol of the manufacturer (Whatman, Inc.). The genomic DNA was used for PCR as previously described by Guan et al. (2007) with a minor modification. Though the forward primer, as described by Guan et al. (2007) was 5’ AGGACTACGGCTTGAAAAGC 3’, the reverse primer, 5’ GCGATCACGGACTAAAGAGG 3’, was developed for the present work using GenBank sequence with accession number NC_001323.. The amplicons, of expected size of 613 bp, were processed, sequenced, and the sequences analyzed using the approach of Guan et al. (2007).
Population genetic analyses
The sequences were initially analyzed using CLUSTALW (Higgins et al., 1994) as previously described (Guan et al., 2007). Using the DnaSP software (version 4.10.9, Rozas et al., 2003), the following statistics were estimated from the sequence comparisons: haplotype diversity (h), nucleotide diversity (π), genetic differentiation (FST) and mismatch distribution based on pairwise differences among all haplotypes (Rogers and Harpending, 1992). Parsimony network analysis of IC was done using TCS software version 1.21 (Clement et al., 2000). To further evaluate the partitioning of sequence variation in the five regions, analysis of molecular variance or AMOVA among the IC from the five regions was evaluated using ARLEQUIN (version 3.01, Excoffier, 2006). The D statistics (Tajima, 1989) were used to estimate whether the D-loop data in the IC, though relatively small in size, was consistent with the expectation of neutrality.
Phylogenetic analyses
Genetic relatedness among birds from the five regions as well as between the IC and CJF, green (Gallus varius), grey (Gallus sonneratii), and red (Gallus gallus) junglefowls were assessed using PAUP* version 4.0 (Swofford, 2002). Publicly available sequences in Genbank for the junglefowls, other than CJF were used in the analyses. Phylogenetic trees were constructed using the neighbor-joining method. One thousand bootstrap replicates were used to assess confidence in the grouping (Felsenstein & Kishino,1993).
Results and Discussion
A total of 613 and 675 bp of the mitochondrial D-loop sequence from IC and CJF, respectively, were involved in the analyses. The difference in sequence length, considering that the amplicons sequenced were from the same primer pair, is due to an insertion of 62 bp in the CJF (Figure S1) at position 356 of the Genbank reference Gallus gallus mtDNA sequence (Accession number NC_001323). This insertion in the CJF has previously been reported, based on sequence from a single bird, to be about 61 bp and to also occur in the grey junglefowl (GrJF, Nishibori et al., 2005). The sequences flanking this insertion showed on average, the percentage similarity of 81.5% with the IC D-loop sequence. One of the CJF samples obtained from the Zoo, CJF141, lacked this insertion. Additionally, the sequence of the CJF141 also had a 99.5% sequence identity with the IC D-loop sequence as shown in Figure S1. Therefore, CJF141 was removed from further analyses as we believe it is most likely an IC.
Within the IC, a total of 42 haplotypes were detected from 44 polymorphic sites (Table 1). The sequences of all the haplotypes have been submitted to Genbank and assigned accession numbers. The haplotypes ranged in frequency from less than 1 to 12%. Only three haplotypes, SLvtHap1, 26 and 32, were observed in all five regions of Sri Lanka and 31 were detected in only one region. Sixty one percent of the haplotypes were unique to the SP region (Figure 1). Six haplogroups (A-F) based on shared SNPs as shown in Table S1 (Supplement) were identified from the 42 haplotypes. All but two, haplogroups E and F, were found in all the regions sampled (Figure 1). The haplogroups ranged in frequency from 0.02 to 0.33. In the CJF 21 SNPs formed six haplogroups (Table S2).
Table 1.
Mitochondrial D-loop haplotype frequencies and distribution in Sri Lankan indigenous chicken.
| Haplotype | Accession number | Number1 |
Frequency2 | ||||
|---|---|---|---|---|---|---|---|
| NCP | NWP | WP | UP | SP | |||
| SLvtHap1 | EU199906 | 5 | 2 | 1 | 4 | 2 | 0.1061 |
| SLvtHap2 | EU199907 | 1 | - | - | - | - | 0.0076 |
| SLvtHap3 | EU199908 | 1 | - | - | - | - | 0.0076 |
| SLvtHap4 | EU199909 | - | - | - | - | 2 | 0.0152 |
| SLvtHap5 | EU199910 | - | 1 | - | - | - | 0.0076 |
| SLvtHap6 | EU199911 | - | - | - | - | 1 | 0.0076 |
| SLvtHap7 | EU199912 | - | - | - | - | 1 | 0.0076 |
| SLvtHap8 | EU199913 | 1 | - | - | 1 | - | 0.0152 |
| SLvtHap9 | EU199914 | - | - | - | - | 1 | 0.0076 |
| SLvtHap10 | EU199915 | - | 1 | - | - | - | 0.0076 |
| SLvtHap11 | EU199916 | - | 2 | - | - | - | 0.0152 |
| SLvtHap12 | EU199917 | - | - | - | - | 1 | 0.0076 |
| SLvtHap13 | EU199918 | 1 | 2 | 1 | - | 1 | 0.0379 |
| SLvtHap14 | EU199919 | 1 | - | - | - | - | 0.0076 |
| SLvtHap15 | EU199920 | - | - | - | - | 1 | 0.0076 |
| SLvtHap16 | EU199921 | - | - | 1 | - | 2 | 0.0227 |
| SLvtHap17 | EU199922 | - | - | - | - | 1 | 0.0076 |
| SLvtHap18 | EU199923 | - | - | - | - | 2 | 0.0152 |
| SLvtHap19 | EU199924 | - | - | - | 1 | - | 0.0076 |
| SLvtHap20 | EU199925 | - | - | - | - | 2 | 0.0152 |
| SLvtHap21 | EU199926 | - | - | - | - | 1 | 0.0076 |
| SLvtHap22 | EU199927 | - | - | - | 1 | - | 0.0076 |
| SLvtHap23 | EU199928 | - | - | - | 1 | - | 0.0076 |
| SLvtHap24 | EU199929 | - | 1 | 1 | 10 | 5 | 0.1288 |
| SLvtHap25 | EU199930 | - | - | - | - | 1 | 0.0076 |
| SLvtHap26 | EU199931 | 1 | 2 | 1 | 3 | 1 | 0.0606 |
| SLvtHap27 | EU199932 | - | - | - | 1 | - | 0.0076 |
| SLvtHap28 | EU199933 | 1 | - | - | - | - | 0.0076 |
| SLvtHap29 | EU199934 | 1 | - | - | - | - | 0.0076 |
| SLvtHap30 | EU199935 | 1 | - | 1 | 4 | 4 | 0.0758 |
| SLvtHap31 | EU199936 | - | - | - | 2 | - | 0.0152 |
| SLvtHap32 | EU199937 | 3 | 4 | 5 | 3 | 2 | 0.1288 |
| SLvtHap33 | EU199938 | 1 | 1 | 1 | - | 1 | 0.0303 |
| SLvtHap34 | EU199939 | - | - | - | - | 1 | 0.0076 |
| SLvtHap35 | EU199940 | 1 | - | 2 | 2 | 2 | 0.0530 |
| SLvtHap36 | EU199941 | 1 | 1 | 4 | 2 | - | 0.0606 |
| SLvtHap37 | EU199942 | - | 1 | - | - | - | 0.0076 |
| SLvtHap38 | EU199943 | - | - | - | - | 1 | 0.0076 |
| SLvtHap39 | EU199944 | - | - | - | - | 1 | 0.0076 |
| SLvtHap40 | EU199945 | - | - | - | 1 | - | 0.0076 |
| SLvtHap41 | EU199946 | - | - | - | 1 | - | 0.0076 |
| SLvtHap42 | EU199947 | - | - | 2 | - | - | 0.0152 |
Where NCP, NWP, WP, UP and SP represent north central province, north western province, western province, uva province and southern province, respectively.
The cumulative frequency of the indigenous chicken population.
The diversity indices for IC ranged from 0.901 to 0.965 and from 0.011 to 0.13 for h and π, respectively (Table 2). Pairwise genetic (Fst) and nucleotide divergence (dxy) estimates were significant for most of the comparisons (Table 3). Nucleotide divergence between CJF and each of the IC populations were also significant with Fst ranging from 0.921 to 0.932 (P<0.05). Both estimates of Fst and dxy between the CJF and IC populations were several-fold higher than those between IC populations. The negative Fst values indicate negligible variation between the regions compared. Within the IC, birds from SP and WP were most divergent according to the estimates of inter-population nucleotide divergence. Further, the analysis of molecular variation revealed significantly high (92%) within region variation (P<0.05).
Table 2.
Sampling sites, sample size (n), haplotype distribution (f), and haplotype (h) and nucleotide diversities (π), with standard deviations in parentheses, in the indigenous chickens (IC) of Sri Lanka and Ceylon Junglefowl (CJF) based on mitochondrial D-loop sequence comparisons.
| Sampling sites | n | f | h | π |
|---|---|---|---|---|
| NCP | 20 | 14 | 0.932 (0.044) | 0.013 (0.001) |
| NWP | 18 | 11 | 0.935 (0.038) | 0.013 (0.001) |
| WP | 20 | 11 | 0.905 (0.044) | 0.013 (0.001) |
| UP | 37 | 15 | 0.901 (0.033) | 0.011 (0.001) |
| SP | 37 | 23 | 0.965 (0.015) | 0.012 (0.001) |
| CJF |
8 |
6 |
0.929 (0.084) |
0.012 (0.002) |
| Total (IC & CJF) | 140 | 48 | 0.947 (0.008) | 0.017 (0.001) |
| Total (only IC) | 132 | 42 | 0.947 (0.009) | 0.013 (0.000) |
Table 3.
Inter-region haplotype (above diagonal) and nucleotide divergence (below diagonal)
| NCP | SP | UP | NWP | WP | CJF | |
|---|---|---|---|---|---|---|
| NCP | −0.007 | 0.107* | −0.025 | 0.070 | 0.923 | |
| SP | 0.012 | 0.066* | 0.014 | 0.130* | 0.926 | |
| UP | 0.013 | 0.012 | 0.123* | 0.183* | 0.932 | |
| NWP | 0.013 | 0.013 | 0.013 | 0.049* | 0.921 | |
| WP | 0.014 | 0.014 | 0.014 | 0.014 | 0.923 | |
| CJF | 0.062 | 0.061 | 0.060 | 0.061 | 0.063 |
Significantly different at P<0.05
The consensus of an unrooted neighbor-joining (NJ) tree shows three distinct clusters for RJF, GrJF and IC, the green junglefowl, and the CJF (Figure 2). Within the RJF, GrJF and IC cluster, the RJF appears to be most closely related to haplogroup E and the GrJF to haplogroup A.
Figure 2.
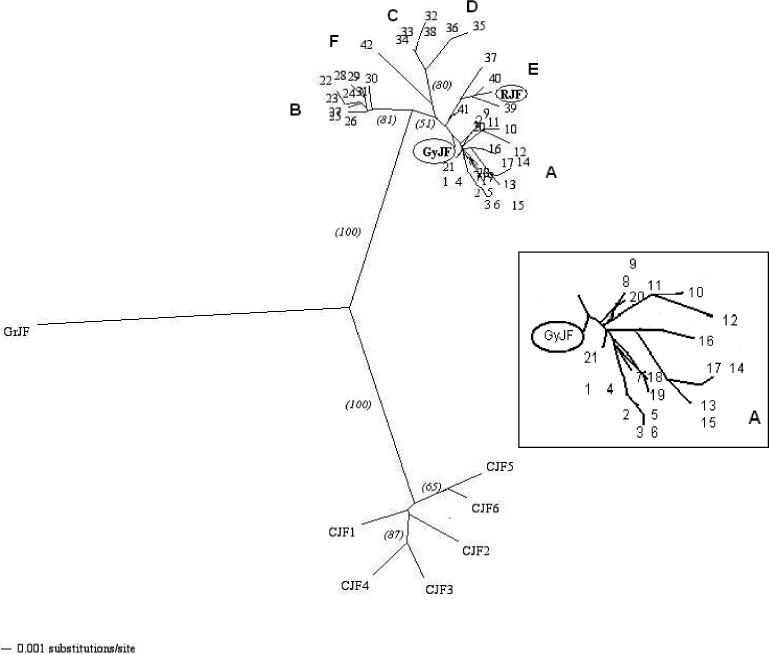
An unrooted neighbor-joining tree relating the mitochondrial D-loop haplotypes observed in the indigenous chickens of Sri Lanka (haplotypes 1−42) and the Ceylon (haplotypes CJF1-CJF6), Red (RJF, Accession number 71658078), Grey (GyJF, Accession number 71040179) and Green junglefowls (GrJF, Accession number 71040235). A - E refer to haplogroups, with D being that of the Genbank reference sequence, accession number NC_001323. Numbers in parentheses represent boostrap values from 1,000 replicates. Inset gives the enlargement of Haplgroup A
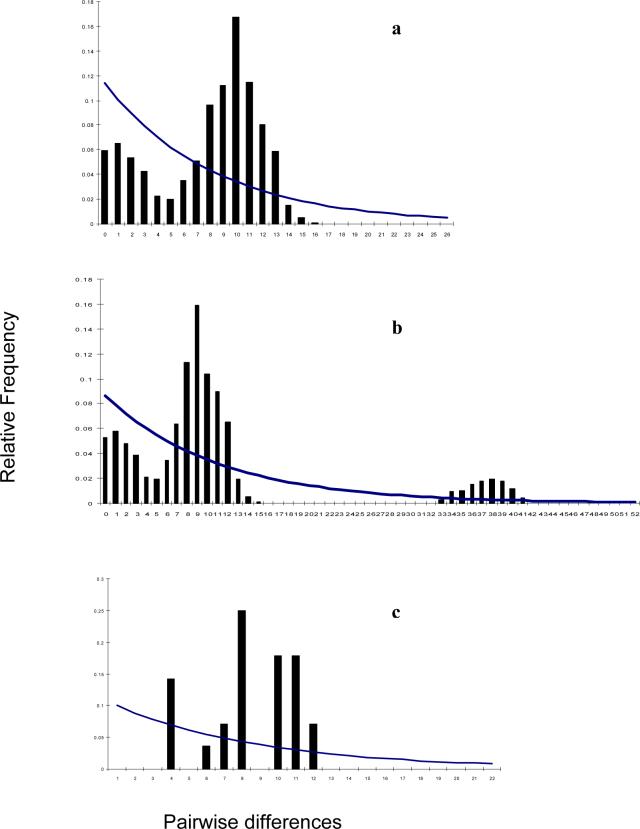
In both CJF and IC, Tajima's D statistics for neutrality test (data not presented) was not significantly different from zero (P>0.10). The average pairwise nucleotide differences were 10.63, 7.80 and 7.90 between IC and CJF, among IC, and among CJF, respectively. The distribution of observed mismatches of pairwise differences for IC and CJF populations are given in Figure 3. The IC population deviated from expected values and demonstrated a bimodal pattern of distribution (raggedness r=0.0129, calculated with parameters θfinal = ∞, θinitial = 2.878, τ = 4.920). The combined mismatch distribution analysis of IC+CJF (based on the parameters θfinal = ∞, θinitial = 9.432, τ = 1.204) showed two major peaks (raggedness r=0.0126) at around 1 and 9 differences and a smaller peak around 38. The mismatch analysis among birds within each of the five geographic regions sampled also showed a bimodal pattern (data not shown).
Figure 3.
Frequency distribution of the number of sequence differences observed in pairwise comparisons of 613 bp of mitochondrial D-loop of indigenous chickens (IC) of Sri Lanka and Ceylon Junglefowl (CJF). a. comparisons within the IC. b. Comparisons among IC and the CJF. c comparisons within the CJF. The within-indigenous chickens comparisons lags behind both the intra CJF and inter-CJF and indigenous chickens derivations.
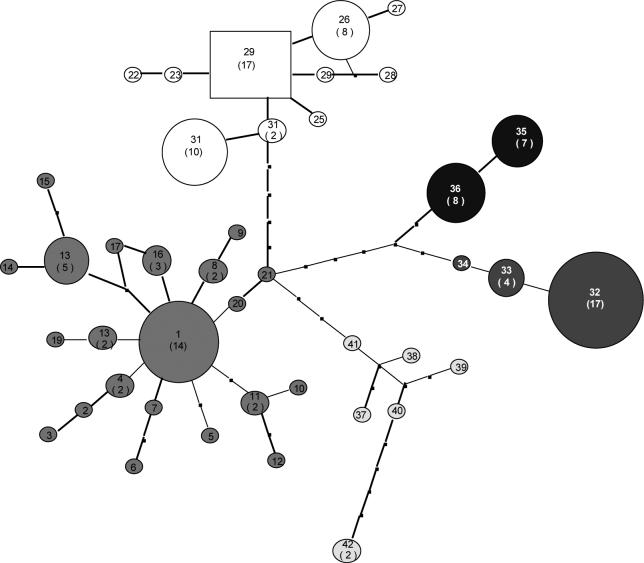
The parsimony network analysis of IC haplotypes revealed five distinct groups of haplotypes with on average 7−8 bp difference (Figure 4). The five groups correspond to five of the six haplogroups identified in this study, and described in Table S1 and Figure 1. The root sequence (95% probability) is one of the most frequent haplotypes and it was included in haplogroup B. Haplogroup A was genetically diverse compared to other groups. The data further suggest that Haplogroups C, D, and E have diverged from haplogroup B and that haplogroup F, which is region specific, diverged from E. Haplogroups D and C appear to be more closely related. The network analysis also appears to support the wide geographic distribution in Sri Lanka of haplogroups A-D that was observed from the mismatch data.
Figure 4.
Parsimony network (minimum spanning tree) of 613-bp partial D-loop sequences from indigenous chickens in Sri Lanka. Circles represent individual haplotypes as described in Table 1. The line connecting two circles, independent of length, indicates a single base pair difference between the two haplotypes. The rectangle indicates the root haplotype based on 95% probability. Filled dots on the line represent intermediate haplotypes (theoretical) not found in the present analysis. The size of each circle is proportional to the frequency of the haplotype. The number in the circle indicate the haplotype and those in parenthesis correspond to the number of individuals with that haplotype (when number >1). Shading indicates haplogroups as described in Figure 1.
The diversity estimates observed in our study were higher than the diversity indices reported for Japanese IC of 0.0016 (Oka et al., 2007), for Chinese IC of 0.045 (Niu et al., 2002), and for Indian IC of 0.66 (Pirany et al., 2007). The high genetic diversities revealed among Sri Lankan IC could be attributed to the evolutionary history of the population. The country's geographic location on the ancient trade routes, which connected East Asia to Europe and Middle East enabled the exchange of agriculturally important genetic materials including chicken. (Chandrasiri, 2002, McClellan & Dorn, 2006).
Since the CJF is endemic to Sri Lanka, the current comparison with the IC provides the first direct evidence that it is not the progenitor of the native domesticated chickens in the country. Our data shows that the IC are closely related to both RJF and GrJF, which appears to support reports of multiple origins of domestic chickens (Nishibori et al., 2005; Liu et al., 2006; and Oka et al, 2007). This, however, is inconsistent with earlier reports of a monophyletic origin of domestic chickens (Fumihito et al., 1994; 1996).
Given the free movement of birds and farming communities across Sri Lanka, it is not surprising that the current work detected no genetic subdivision of IC. However, the relatively high number of unique haplotypes in different geographical regions may be due to the breeding pattern practiced in rural areas where the birds are raised in free range and there is limited or no exchange of breeding birds among farms (Gunaratne et al., 1993). This tends to minimize inbreeding, and thus causing the relatively high level of haplotype diversity observed.
In summary, the current analyses suggest that in Sri Lanka, the IC have a relatively high level of genetic diversity that is consistent across a wide geographic area. The random geographic distribution of haplogroups indicates an extensive resource both for conservation and/or genetic improvement by breeding. This could be especially useful if divergent haplotypes and/or haplogroups are crossed to take advantage of heterosis. Our data, though based on only a few junglefowls and thus requiring further studies, appears to suggest that the Sri Lankan IC may have originated from either red or GrJF and not from the CJF that is endemic to the country.
Supplementary Material
Figure S1 The alignment of mitochondrial D-loop sequences of Sri Lankan Indigenous Chicken (IC), nine Ceylon Junglefowl and GenBank sequence of domestic chicken (accession number NC_001323). Note that Ceylon Junglefowl has 62 bp in two insertions at positions 356 (51 bp) and 360 (11bp) (position number is according to NC_001323) as indicated by arrows. Also note that the there is a 3 bp difference in numbering of GenBank sequence and IC sequence. This difference is due to three additional repeats of C found in all IC sequences at position 49 (not shown in the figure).
Table S1a. Haplotypes and substitution sites in the D-loop region (between 1−613 bp) of mtDNA of indigenous Chicken. Horizontal dotted lines separate haplogroups A-F, respectively.
Table S1b. Haplotypes and substitution sites in the D-loop region (between 1−675 bp) of mtDNA of Ceylon Junglefowl
Acknowledgements
We are grateful to the following organizations and individuals for support given during the study: the Fulbright mutual educational exchange program for supporting the visiting scholar, National Institute of Health and Virginia Tech for financial support, Ranga Kumara and Priyantha Perera (University of Peradeniya, Sri Lanka) for sample collection, and all the farmers in sampling locations.
Footnotes
Supplementary Material
The following supplementary material is available for the article online at .....
Table S1a Haplotypes and substitution sites of indigenous Chicken (between segment of 1−613 bp)
Table S1b Haplotypes and substitution sites of Ceylon Junglefowl (between segment of 1−675 bp)
Figure S1 The alignment of mitochondrial D-loop sequences of Sri Lankan Indigenous Chicken (IC), nine Ceylon Junglefowl, GenBank sequence of domestic chicken
References
- Akishinonomiya F, Miyake T, Sumi S, Takada M, Ohno S, Kondo N. One subspecies of the red jungle fowl (Gallus gallus gallus) suffices as the matriarchic ancestor of all domestic breeds. Proceedings of National Academy of Sciences of the United States of America. 1994;91:12505–12509. doi: 10.1073/pnas.91.26.12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akishinonomiya F, Miyake T, Takada M, Shingu R, Endo T, Gojobori T, Kondo N, Ohno S. Monophyletic origin and unique dispersal patterns of domestic fowls. Proceedings of National Academy of Sciences of the United States of America. 1996;93:6792–6795. doi: 10.1073/pnas.93.13.6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasiri ADN. Country Report on the State of Animal Genetic Resources (Sri Lanka) The Food and Agriculture organization of the United Nations; Rome: 2002. [Google Scholar]
- Clement M, Pasoda D, Crandall K. TCS: a computer program to estimate gene genealogies. Molecular Ecology. 2000;9:1657–1660. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L. Arlequin ver. 3.01. Computational and Molecular Population Genetics Lab. Zoological Institute; University of Berne: 2006. [Google Scholar]
- Felsenstein J, Kishino H. Is there something wrong with the bootstrap on phylogenies? A reply to Hillis and Bull. Systematic Biology. 1993;42:193–200. [Google Scholar]
- Gibson J, Gamage S, Hanotte O, Iniguez L, Maillard JC, Rischkowsky B, Semambo D, Toll J. Option and strategies for the conservation of farm animal genetic resources: Report of an International Workshop (7−10 November 2005, France) CGIAR System-wide Genetic Resources Programme (SGRO)/Biodiversity International; Rome, Italy: 2006. [Google Scholar]
- Granevitze Z, Hillel J, Chen GH, Cuc NTK, Feldman M, Eding H, Weigend S. Genetic diversity within chicken populations from different continents and management histories. Animal Genetics. 2007;38:576–583. doi: 10.1111/j.1365-2052.2007.01650.x. [DOI] [PubMed] [Google Scholar]
- Guan X, Geng T, Silva P, Smith EJ. Mitochondrial DNA sequence and haplotype variation analysis in the chicken (Gallus gallus). Journal of Heredity. 2007;98:723–726. doi: 10.1093/jhered/esm094. [DOI] [PubMed] [Google Scholar]
- Gunaratne SP, Chandrasiri ADN, Hemaltha WAPM, Roberts JA. The feed resource base for scavenging village chicken in Sri Lanka. Tropical Animal Health and Production. 1993;26:249–257. doi: 10.1007/BF02250880. [DOI] [PubMed] [Google Scholar]
- Higgins D, Thompson J, Gibson JD, Higgins DG, Gobson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillel J, Groenen MA, Tixier-Boichard M, Korol AB, David L, Kirshner VM, Burke T, Barre-Dirie A, Crooijmans RP, Elo K, Feldman MV, Freidlin PJ, Maki-Tanila A, Oortwijn M, Thomson P, Vignal A, Wimmers K, Weigend S. Biodiversity of 52 chicken populations assessed by microsatellite typing of DNA pools. Genetics Selection and Evolution. 2003;35:533–557. doi: 10.1186/1297-9686-35-6-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegs JO, Matzke A, Churakov G, Kuritzin A, Mayr G, Brosius J, Schmitz J. Waves of genomic hitchhikers shed light on the evolution of gamebirds (Aves: Galliformes). BMC Evolutionary Biology. 2007;7:190. doi: 10.1186/1471-2148-7-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wu G, Yao Y, Miao Y, Luikart G, Baig M, Beja-Pereira A, Ding Z, Palanichamy MG, Zhang Y. Multiple maternal origins of chickens: Out of the Asian Jungles. Molecular Phylogenetics and Evolution. 2006;38:12–19. doi: 10.1016/j.ympev.2005.09.014. [DOI] [PubMed] [Google Scholar]
- McClellan JE, Dorn H. Science and technology in world history: An introduction. 2nd Edition JHU Press; Baltimore, USA: 2006. [Google Scholar]
- Moiseyeva IG. Ancient evidence for the origin and distribution of domestic fowl.. Proceedings of the 10th European Conference “The Poultry Industry Towards the 21st Century”; Jerusalem. 21−28 June, 1998.1998. pp. 244–245. [Google Scholar]
- Mwacharo JM, Nomra K, Hanada H, Jianlin H, Hanotte O, Amano T. Genetic relationships among Kenyan and other East African indigenous chickens. Animal Genetics. 2007;38:485–490. doi: 10.1111/j.1365-2052.2007.01641.x. [DOI] [PubMed] [Google Scholar]
- Nishibori M, Shimogiri T, Hayashi T, Yasue H. Molecular evidence for hubridization of species in the genus Gallus except for Gallus varius. Animal Genetics. 2005;36:367–375. doi: 10.1111/j.1365-2052.2005.01318.x. [DOI] [PubMed] [Google Scholar]
- Niu D, Fu Y, Luo J, Ruan H, Yu X, Chen G, Zhang Y. The origin and genetic diversity of Chinese indigenous chicken breeds. Biochemical Genetics. 2002;40:163–174. doi: 10.1023/a:1015832108669. [DOI] [PubMed] [Google Scholar]
- Oka T, Ino Y, Nomura K, Kawashima S, Kuwayama T, Hanada H, Amano T, Takada M, Takahata N, Hayashi Y, Akishinonomiya F. Analysis of mtDNA sequences shows Japanese native chicken have multiple origins. Animal Genetics. 2007;38:287–293. doi: 10.1111/j.1365-2052.2007.01604.x. [DOI] [PubMed] [Google Scholar]
- Pirany N, Romanov MN, Ganpule SP, Dewagowda G, Prasad DT. Microsatellite analysis of genetic diversity in Indian chicken populations. Journal of Poultry Science. 2007;44:19–28. [Google Scholar]
- Qu L, Li X, Chen K, Yang H, Zhang L, Wu G, Hou Z, Xu G, Yang N. Evaluation of genetic diversity in Chinese indigenous chicken breeds using microsatellite markers. Science in China Series C: Life Sciences. 2006;49:332–341. doi: 10.1007/s11427-006-2001-6. [DOI] [PubMed] [Google Scholar]
- Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sánchez-Delbarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics methods and protocols: methods in molecular biology. Humana Press; Totowa, NJ: 2000. pp. 365–386. Available from http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi. [DOI] [PubMed] [Google Scholar]
- Silva LP, Rajapaksha WRKJS. Preliminary investigation of genetic characterization of native and endemic fowl types of Sri Lanka. In: Makkar HPS, Viljoen GJ, editors. Application of gene-based Technologies for Improving Animal Production and Health in Developing Countries. Springer Publishers, P.O. Box 17, 3300 AA Dordrecht; The Netherlands: 2005. pp. 593–604. [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods) 4.0. Sinauer Associates; Sunderland, Massachusetts: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The alignment of mitochondrial D-loop sequences of Sri Lankan Indigenous Chicken (IC), nine Ceylon Junglefowl and GenBank sequence of domestic chicken (accession number NC_001323). Note that Ceylon Junglefowl has 62 bp in two insertions at positions 356 (51 bp) and 360 (11bp) (position number is according to NC_001323) as indicated by arrows. Also note that the there is a 3 bp difference in numbering of GenBank sequence and IC sequence. This difference is due to three additional repeats of C found in all IC sequences at position 49 (not shown in the figure).
Table S1a. Haplotypes and substitution sites in the D-loop region (between 1−613 bp) of mtDNA of indigenous Chicken. Horizontal dotted lines separate haplogroups A-F, respectively.
Table S1b. Haplotypes and substitution sites in the D-loop region (between 1−675 bp) of mtDNA of Ceylon Junglefowl