Abstract
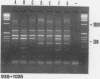
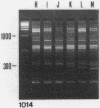
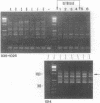
A typing procedure for methicillin-resistant Staphylococcus aureus (MRSA) based on the polymerase chain reaction (PCR) amplification of both mecA sequences and variable DNA sequences as present in the prokaryotic genome has been developed. Two primers based on the sequences of DNA repeats as discovered in gram-negative members of the family Enterobacteriaceae allow detection of variable regions in the genome of a gram-positive bacterium such as S. aureus, as does a newly described arbitrary primer. This procedure, enabling the detection of 23 different genotypes in a collection of 48 MRSA isolates, was validated by comparisons with phage typing studies. It appeared that within the same group of isolates only 13 different phagovars could be identified. Combination of the results from both phage typing and genotyping allowed the discrimination of 34 of 48 isolates. However, depending on the primer-variable complexity of the PCR fingerprints, which could also be modulated by combination of PCR primers, clear homologies between the groups defined by either phage typing or fingerprinting were observed. An analysis of an MRSA outbreak in a geriatric institution showed a collection of genetically homogeneous isolates. In agreement with phage typing, PCR fingerprinting revealed the identical natures of the MRSA strains isolated from all patients.
Full text
PDF


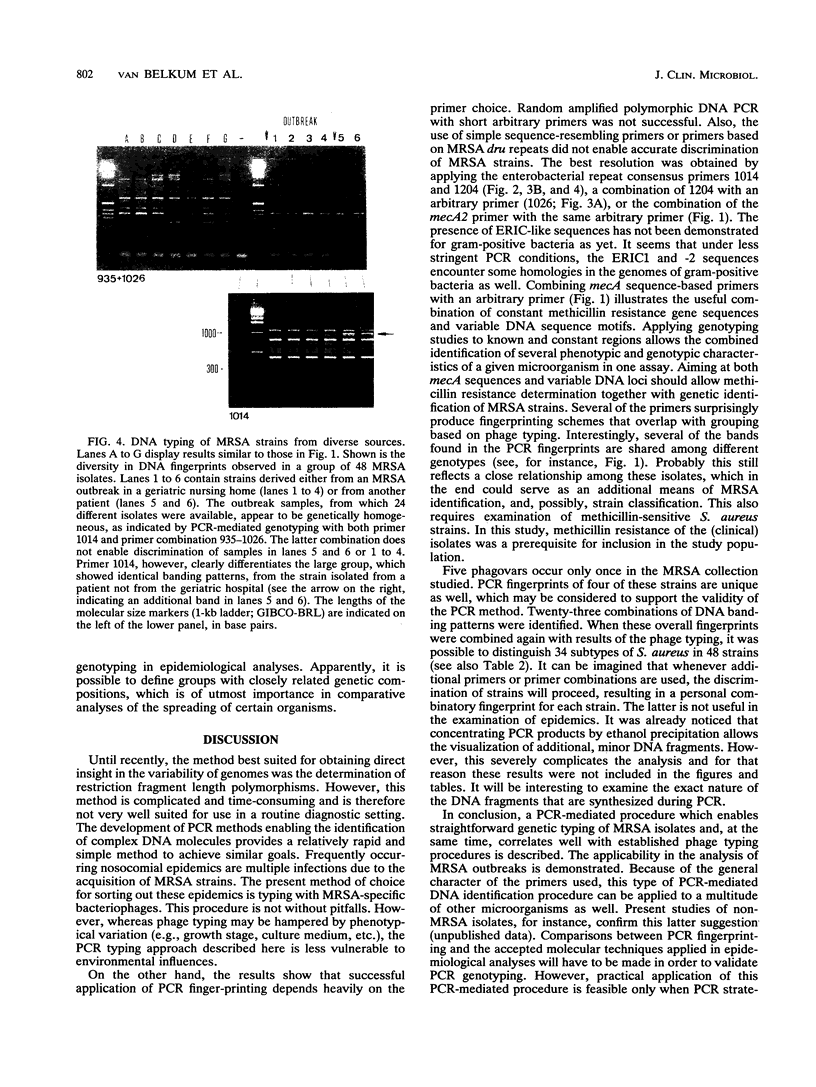

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Archer G. L., Pennell E. Detection of methicillin resistance in staphylococci by using a DNA probe. Antimicrob Agents Chemother. 1990 Sep;34(9):1720–1724. doi: 10.1128/aac.34.9.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg H. M., Rimland D., Kiehlbauch J. A., Terry P. M., Wachsmuth I. K. Epidemiologic typing of Staphylococcus aureus by DNA restriction fragment length polymorphisms of rRNA genes: elucidation of the clonal nature of a group of bacteriophage-nontypeable, ciprofloxacin-resistant, methicillin-susceptible S. aureus isolates. J Clin Microbiol. 1992 Feb;30(2):362–369. doi: 10.1128/jcm.30.2.362-369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boom R., Sol C. J., Salimans M. M., Jansen C. L., Wertheim-van Dillen P. M., van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990 Mar;28(3):495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano-Anollés G., Bassam B. J., Gresshoff P. M. DNA amplification fingerprinting using very short arbitrary oligonucleotide primers. Biotechnology (N Y) 1991 Jun;9(6):553–557. doi: 10.1038/nbt0691-553. [DOI] [PubMed] [Google Scholar]
- Costas M., Cookson B. D., Talsania H. G., Owen R. J. Numerical analysis of electrophoretic protein patterns of methicillin-resistant strains of Staphylococcus aureus. J Clin Microbiol. 1989 Nov;27(11):2574–2581. doi: 10.1128/jcm.27.11.2574-2581.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh S. H., Byrne S. K., Zhang J. L., Chow A. W. Molecular typing of Staphylococcus aureus on the basis of coagulase gene polymorphisms. J Clin Microbiol. 1992 Jul;30(7):1642–1645. doi: 10.1128/jcm.30.7.1642-1645.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackbarth C. J., Chambers H. F. Methicillin-resistant staphylococci: genetics and mechanisms of resistance. Antimicrob Agents Chemother. 1989 Jul;33(7):991–994. doi: 10.1128/aac.33.7.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans P. W., Schuitema A. R., Van Soolingen D., Verstynen C. P., Bik E. M., Thole J. E., Kolk A. H., van Embden J. D. Specific detection of Mycobacterium tuberculosis complex strains by polymerase chain reaction. J Clin Microbiol. 1990 Jun;28(6):1204–1213. doi: 10.1128/jcm.28.6.1204-1213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiwa N. M., Van Gemert G. W., Raap A. K., Van de Rijke F. M., Mulder A., Lens P. F., Salimans M. M., Zwaan F. E., Van Dorp W., Van der Ploeg M. Rapid detection of human cytomegalovirus DNA in peripheral blood leukocytes of viremic transplant recipients by the polymerase chain reaction. Transplantation. 1989 Jul;48(1):72–76. doi: 10.1097/00007890-198907000-00017. [DOI] [PubMed] [Google Scholar]
- Jordens J. Z., Hall L. M. Characterisation of methicillin-resistant Staphylococcus aureus isolates by restriction endonuclease digestion of chromosomal DNA. J Med Microbiol. 1988 Oct;27(2):117–123. doi: 10.1099/00222615-27-2-117. [DOI] [PubMed] [Google Scholar]
- Melchers W., van den Brule A., Walboomers J., de Bruin M., Burger M., Herbrink P., Meijer C., Lindeman J., Quint W. Increased detection rate of human papillomavirus in cervical scrapes by the polymerase chain reaction as compared to modified FISH and southern-blot analysis. J Med Virol. 1989 Apr;27(4):329–335. doi: 10.1002/jmv.1890270413. [DOI] [PubMed] [Google Scholar]
- Murakami K., Minamide W., Wada K., Nakamura E., Teraoka H., Watanabe S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991 Oct;29(10):2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OEDING P. Serological typing of Staphylococci. Acta Pathol Microbiol Scand Suppl. 1952 Jun;93:356–365. [PubMed] [Google Scholar]
- Prevost G., Jaulhac B., Piemont Y. DNA fingerprinting by pulsed-field gel electrophoresis is more effective than ribotyping in distinguishing among methicillin-resistant Staphylococcus aureus isolates. J Clin Microbiol. 1992 Apr;30(4):967–973. doi: 10.1128/jcm.30.4.967-973.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryffel C., Bucher R., Kayser F. H., Berger-Bächi B. The Staphylococcus aureus mec determinant comprises an unusual cluster of direct repeats and codes for a gene product similar to the Escherichia coli sn-glycerophosphoryl diester phosphodiesterase. J Bacteriol. 1991 Dec;173(23):7416–7422. doi: 10.1128/jb.173.23.7416-7422.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Sompolinsky D., Samra Z., Karakawa W. W., Vann W. F., Schneerson R., Malik Z. Encapsulation and capsular types in isolates of Staphylococcus aureus from different sources and relationship to phage types. J Clin Microbiol. 1985 Nov;22(5):828–834. doi: 10.1128/jcm.22.5.828-834.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubukata K., Nakagami S., Nitta A., Yamane A., Kawakami S., Sugiura M., Konno M. Rapid detection of the mecA gene in methicillin-resistant staphylococci by enzymatic detection of polymerase chain reaction products. J Clin Microbiol. 1992 Jul;30(7):1728–1733. doi: 10.1128/jcm.30.7.1728-1733.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal S., Hoskins J., Flokowitsch J. E., Wu C. Y., Preston D. A., Skatrud P. L. Detection of methicillin-resistant staphylococci by using the polymerase chain reaction. J Clin Microbiol. 1992 Jul;30(7):1685–1691. doi: 10.1128/jcm.30.7.1685-1691.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versalovic J., Koeuth T., Lupski J. R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991 Dec 25;19(24):6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Genomic fingerprinting using arbitrarily primed PCR and a matrix of pairwise combinations of primers. Nucleic Acids Res. 1991 Oct 11;19(19):5275–5279. doi: 10.1093/nar/19.19.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. G., Kubelik A. R., Livak K. J., Rafalski J. A., Tingey S. V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990 Nov 25;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A., De Jonckheere J., Quint W. G. Genotyping Naegleria spp. and Naegleria fowleri isolates by interrepeat polymerase chain reaction. J Clin Microbiol. 1992 Oct;30(10):2595–2598. doi: 10.1128/jcm.30.10.2595-2598.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]