Abstract
Resectable non-small-cell lung cancer (NSCLC) patients have poor prognosis, with 30–50% relapsing within 5 years. Current staging criteria do not fully capture the complexity of this disease. Survival could be improved by identification of those early-stage patients who are most likely to benefit from adjuvant therapy. Molecular classification by using mRNA expression profiles has led to multiple, poorly overlapping signatures. We hypothesized that differing statistical methodologies contribute to this lack of overlap. To test this hypothesis, we analyzed our previously published quantitative RT-PCR dataset with a semisupervised method. A 6-gene signature was identified and validated in 4 independent public microarray datasets that represent a range of tumor histologies and stages. This result demonstrated that at least 2 prognostic signatures can be derived from this single dataset. We next estimated the total number of prognostic signatures in this dataset with a 10-million-signature permutation study. Our 6-gene signature was among the top 0.02% of signatures with maximum verifiability, reaffirming its efficacy. Importantly, this analysis identified 1,789 unique signatures, implying that our dataset contains >500,000 verifiable prognostic signatures for NSCLC. This result appears to rationalize the observed lack of overlap among reported NSCLC prognostic signatures.
Keywords: biomarkers, systems biology, mRNA quantitation, substaging
Non-small-cell lung cancer (NSCLC) is the predominant histological type of lung cancer, accounting for up to 85% of cases (1). Tumor stage is the best established and validated predictor of patient survival (2). When identified at an early stage, NSCLC is primarily treated by surgical resection, which is potentially curative. However, 30–60% of patients with stage IB to IIIA NSCLC die within 5 years after surgery, primarily from tumor recurrence (3). These relapses have been postulated to arise from a reservoir of cells beyond the resection site, such as microscopic residual tumors at the resection margin, occult systemic metastases, or circulating tumor cells. Such a reservoir could potentially be eliminated with an adjuvant systemic therapy, such as chemotherapy. Indeed, this type of adjuvant therapy is routinely applied in the treatment of other solid tumors, including breast (4) and colorectal cancer (5, 6).
Randomized clinical trials have confirmed the benefit of adjuvant chemotherapy in stage II to IIIA NSCLC patients, but the benefit in stage I remains controversial (7–10). However, even in stage I the overall survival is only 70%, which suggests that there is a subpopulation of stage I patients who have more aggressive tumors. In theory, these patients might benefit from postoperative adjuvant chemotherapy. In contrast, there may be subpopulations of stage II or IIIA patients who have such good prognoses that they may neither need nor derive benefit from adjuvant therapy.
Several groups have attempted to identify these subpopulations by studying the mRNA expression profiles of surgically excised tumor samples by using high-density microarray platforms (11–17). Other groups, including our own, have reported smaller prognostic signatures assayed by quantitative reverse-transcriptase PCR (RT-PCR) (18). However, the specific signatures identified by these groups show minimal overlap (19), and it is unclear why this is so. Ein-Dor and coworkers (20) demonstrated that biological heterogeneity leads to thousands of samples being required to identify robust and reproducible subsets for most tumor types. These conclusions are supported by the finding that thousands of genes display intratumor heterogeneity, likely caused by the diversity of tumor microenvironments and cell populations (21, 22). We hypothesized that different statistical methods handle disease heterogeneity in different ways and thus play a major role in the lack of overlap among reported NSCLC prognostic signatures.
Results
Classifier Training.
To determine the impact of alternative statistical methods on prognostic marker identification, we considered our previously published 147-patient, 158-gene RT-PCR NSCLC dataset. This dataset had been analyzed by using high concordance-index as a criterion, which identified a 3-gene classifier capable of separating patients into groups with significantly different prognoses (19). The majority of signatures developed for NSCLC used linear or risk-score methods to classify patients (11, 13, 14, 16, 23), which are unable to capture nonlinear interactions among genes. For example, regulatory networks make substantial use of “or” logic: A cell may respond to hypoxic conditions by up-regulating HIF1A or down-regulating VHL. Such relationships cannot generally be captured by linear methods. We thus developed a nonlinear semisupervised method by coupling unsupervised pattern recognition to gradient descent optimization. We call this algorithm modified Steepest Descent, or mSD (supporting information (SI) Fig. S1).
Applying mSD to a training dataset of 147 NSCLC patients generated a prognostic signature comprising 6 genes: syntaxin 1A (STX1A), hypoxia inducible factor 1A (HIF1A), chaperonin containing TCP1 subunit 3 (CCT3), MHC Class II DP beta 1 (HLA-DPB1), v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (MAFK), and ring finger protein 5 (RNF5). Table S1 gives additional information on these genes.
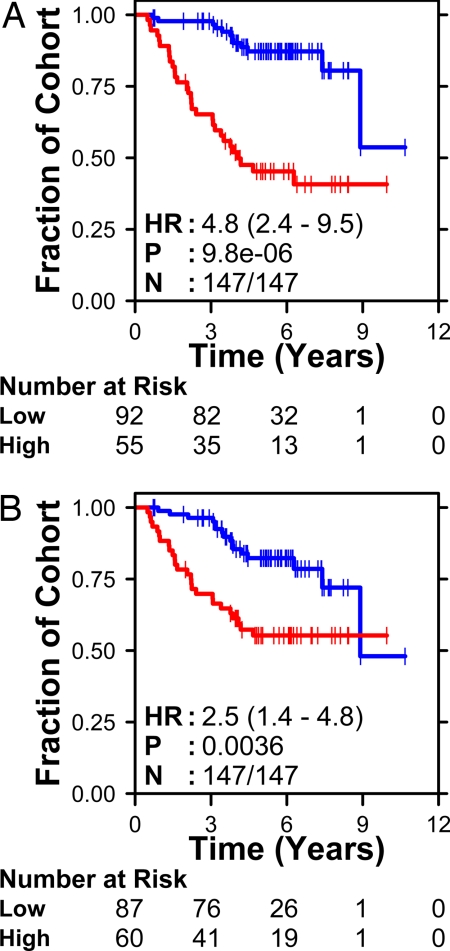
We visualized the mSD signature by using unsupervised pattern recognition and found that the 6 genes were largely uncorrelated (Fig. S2). The signature separated the 147 training patients into groups with significantly different survivals (P = 2.14 × 10−8; log-rank test) (Fig. 1A). Both patient prognosis and treatment are strongly affected by clinical stage, and our previous analysis showed it to be a significant covariate in the training dataset (19). Accordingly, we adjusted for the effects of stage by using Cox proportional-hazards modeling and showed that the mSD molecular signature was independent of clinical stage (HR 4.8, P < 0.001). We also performed a preliminary validation by using leave-one-out cross-validation (24). The 6-gene signature divided patients into 2 groups with significantly different outcome during cross-validation (Fig. 1B) (HR: 2.5, P = 0.0036). The six-gene signature leads to similar patient classifications in the training dataset as our earlier 3-gene signature (SI Text and Table S2).
Fig. 1.
Classifier development. The mSD algorithm was trained on an RT-PCR dataset of 158 genes in 147 NSCLC patients. The resulting 6-gene classifier separated patients into 2 groups with significantly different outcomes (A). Leave-one-out cross-validation again identified 2 groups with significantly different outcomes (B). The number of patients at risk at each time interval in the molecularly defined good- and poor-prognosis groups is listed below each survival curve. The stage-adjusted hazard ratio (HR), P value (Wald test), and number of patients classified (N) are given on each survival curve.
Classifier Validation.
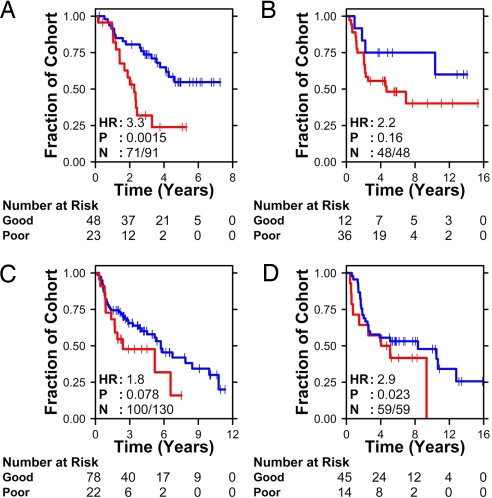
To validate our 6-gene signature, we tested its ability to stratify patients into groups with different prognosis by using 4 independent publicly available datasets from Duke University (25), the University of Michigan (16), and the Prince Charles Hospital (13, 14). These datasets represent 2 versions of Affymetrix arrays (U133Plus2.0, Duke; U133A, Michigan) and a custom cDNA array (Prince Charles). Two of these studies comprise exclusively squamous cell carcinomas (13, 16), one exclusively adenocarcinomas (14), and one both (25). Each dataset was analyzed separately, as outlined in SI Text. The molecular stratifications are plotted in Fig. 2. The 6-gene signature was prognostic in all 4 independent patient cohorts, with hazard ratios ranging from 1.4 (P = 0.08) to 3.3 (P = 0.002). The validation on the 2 datasets from Prince Charles is notable because 1 gene from our 6-gene signature (RNF5) and 2 of the 4 normalization genes were not present on the array platform. Despite this missing information, the mSD signature classified patients into groups with significantly different outcomes (Fig. 2 B and D). In the 2 Affymetrix datasets (Fig. 2 A and C), ≈10% of patients had expression profiles equidistant from the 2 training clusters. These patients were not classified; in practice these equivocal classifications would be assigned to standard clinical practice.
Fig. 2.
Classifier validation. To validate the 6-gene classifier, we classified patients from 4 independent datasets. (A) Mixed adenocarcinomas and squamous cell carcinomas profiled with Affymetrix HG-U133Plus2 arrays by Potti et al. (15). (B) Adenocarcinomas profiled on cDNA arrays by Larsen et al. (13). (C) Squamous cell carcinomas profiled on Affymetrix HG-U133A arrays by Raponi et al. (16). (D) Squamous cell carcinomas profiled on cDNA arrays by Larsen et al. (14). The number of patients at risk in each molecularly-defined group is indicated at several time points. The stage-adjusted hazard ratio (HR), P value (Wald test), and the number of patients successfully classified (N) are also shown.
Pooled Validation.
In addition to the 4 datasets analyzed in Fig. 1, a number of small or older NSCLC datasets exist. We combined the data from the 4 validation datasets with that from a previous study of adenocarcinomas on the older Hu6800 Affymetrix array (11), a study of adenocarcinomas on the relatively old U95Av2 Affymetrix array (12), and small adenocarcinoma and squamous cell carcinoma datasets on Affymetrix U133A arrays from a pooled study (23). This procedure generated a cohort of 589 patients taken from 8 datasets. This cohort was separated into 2 groups by using the 6-gene signature (Fig. S3A). The resulting groups showed significant stage-adjusted differences in survival with a hazard ratio of 1.6 (95% CI 1.2–2.2; P = 7.6 × 10−4). The 6-gene signature was also capable of separating Stage I patients from this cohort into 2 groups with different survival (Fig. S3B), with a hazard ratio of 1.5 (95% CI 1.1 to 2.2; P = 0.02). These results for Stage I patients were adjusted for clinical stage (IA vs. IB), demonstrating that our molecular classification improves upon existing staging criteria. The hazard ratios in this pooled analysis are somewhat compressed by the addition of older and less-sensitive microarray platforms, but nevertheless the results are statistically significant consistent in a very large patient cohort. The extensive validation of our 6-gene signature compares favorably to other published NSCLC signatures (Fig. S4). Table S3 summarizes all validation datasets.
Permutation Analysis.
This 6-gene classifier shows partial overlap with the 3-gene classifier identified previously from the same dataset by using risk-score methods. We questioned whether other small prognostic signatures could be identified from this 158-gene dataset. To test this question comprehensively, we mapped our 158 genes in 4 test datasets (11, 12, 16, 25). In total, 113 genes were common to these 4 datasets, and adding additional datasets greatly reduced this number. We restricted subsequent analyses to the 113 genes profiled in all 4 datasets. We then generated 10 million permutations of 6 genes and tested their prognostic capability in these 4 datasets. For each subset, we calculated its statistical significance by using the log-rank test, as before.
In the training set, the mSD signature was superior to 99.999% of the 10 million unique signatures tested, as measured by the statistical significance of the separation between the 2 patient groups. Although few signatures performed as well as the mSD signature, a large number showed statistical significance. In total, 16.4% of all 6-gene signatures were significant at P < 0.05. This proportion is 3.28-fold greater than the 5% expected by chance alone and reflects a statistically significant enrichment (P < 2.2 × 10−16; proportion test).
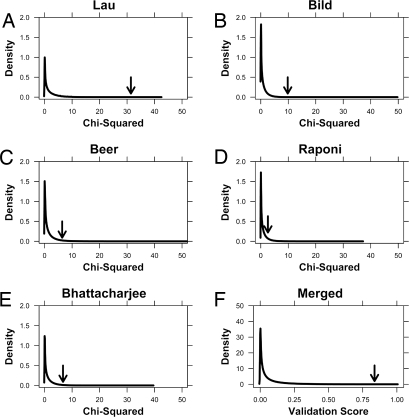
The distribution of all 10 million 6-gene signatures is shown in Fig. 3A as a kernel density estimate. Kernel density estimates are an established method of estimating the probability density function of a random variable. They can be thought of as smoothed histograms, where the y axis reflects the likelihood of observing the value specified by the x axis. In Fig. 3A, the x axis indicates the χ2 value from the log-rank analysis. The higher the χ2, the smaller (more significant) the P value for differential prognoses between the 2 predicted groups. Thus, more effective prognostic signatures lie to the right of the plot.
Fig. 3.
Permutation validation. Ten million 6-gene signatures were generated at random from our training dataset. The ability of each signature to separate the training dataset into 2 groups with significantly different prognoses was evaluated using the log-rank test. The kernel density of the χ2 values from this log-rank test was generated (A). The x axis indicates the χ2 values: Larger values indicate a lower P value and hence a more statistically significant separation of patient groups in the training dataset. The y axis gives the kernel density, which reflects the probability distribution of the dataset. Higher values indicate a larger fraction of the population, akin to a smoothed histogram. The performance of the mSD signature is marked with an arrow. These 10 million trained signatures were then tested in 4 independent datasets. Kernel density estimates, as above, are provided for each test dataset (B–E). Each test dataset is labeled with the first author of the study. The performance of the mSD signature is marked with an arrow. Finally, to demonstrate the significance of the mSD signature across all 4 test datasets we generated a validation score by multiplying the percentile rankings of each signature in each of the 4 test datasets. Higher values thus correspond to improved validation across all 4 datasets. The performance of the mSD signature is marked with an arrow.
We next compared the validation of the mSD signature with that of the 10 million random signatures. For each test dataset (11, 12, 16, 25), the distribution of validation rates was again plotted as kernel density estimates. For each kernel density estimate in the training dataset, we marked the performance of the 6-gene mSD signature in that dataset with an arrow (Fig. 3 B–E). The mSD signature performs well in each of the 4 datasets but with some variability. The lower bound was the squamous-cell-carcinoma dataset reported by Raponi et al. (16), where our classifier was among the top 10.4% of all signatures. The upper bound was the dataset reported by Potti and coworkers (15), where it was among the top 0.14% of all signatures. Summary data from all permutation analyses are presented in Table S4. The raw permutation data are also available (www.cs.utoronto.ca/∼juris/data/PNAS08/PNAS_permutation_data.zip).
These data demonstrate the efficacy of our 6-gene signature in 4 distinct testing datasets. Whereas our signature performed among the top 10% of all signatures in each test dataset, it was not the single best signature in any single dataset. Rather, its strength is its validation in 4 independent datasets. To compare the validation of our signature across all 4 test datasets, we calculated its percentile ranking in each dataset and took the product of these rankings. The resulting validation score provides a measure of the interdataset reproducibility of a signature. Only 1,789 of the 10 million signatures tested perform better than the mSD signature across all 4 validation datasets. Thus, the mSD signature was superior to 99.98% of signatures tested (Fig. 3F). The small difference in performance of the mSD signature in the training and testing datasets (99.999% vs. 99.982%) indicates minimal over-fitting on our training dataset.
Enrichment Analysis.
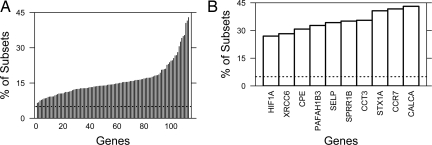
Having used our large permutation dataset to rank our 6-gene prognostic signature, we next tested whether specific genes were enriched in prognostic signatures. For each gene, we calculated the percentage of signatures containing each gene that were statistically significant (P < 0.05, log-rank test). At this threshold we expect 5% of signatures to be significant by chance alone. When we plotted the percentages for the 113 gene set (Fig. 4A), most genes were enriched over this baseline, with enrichment values ranging from 6.7%–43.1%. This elevation likely reflects the enrichment of our test dataset for putative prognostic genes (19).
Fig. 4.
Prognostic genes. For each gene, we calculated the fraction of 6-gene signatures containing each gene that are statistically significant at P < 0.05 (A). A zoom-in on the 10 most enriched genes is also shown (B). The horizontal line represents the 5% level expected by chance alone, the y axis gives the fraction of signatures containing that gene that are significant at P < 0.05 and individual genes are on the x axis.
To focus on specific genes, we considered the 10 most highly enriched genes (Fig. 4B). Both genes shared by our mSD and risk-score signatures are present on this list (STX1A, 3rd, and HIF1A, 10th), as are 1 additional gene from the mSD signature (CCT3, 4th) and 1 additional gene from the risk-score signature (CCR7, 4th). Genes on this list are highly effective in prognostic signatures, independent of the other genes they are combined with, and may therefore represent unique aspects of disease initiation or progression. Table S5 provides the enrichment values for all 113 genes.
Discussion
We hypothesized that the observed lack of overlap in reported prognostic signatures for NSCLC resulted from the use of different statistical techniques. To test this hypothesis, we trained 2 distinctive algorithms on a single dataset to determine if identical signatures would be found. For training, we selected a real-time PCR dataset of 158 genes assessed in 147 patients, which we had used previously to identify a 3-gene signature by using linear risk-score methods (19). To provide a counterpoint to this linear analysis, we then developed a semisupervised algorithm by coupling unsupervised pattern-recognition and gradient-descent algorithms. We call this new algorithm mSD.
The application of mSD to the same 147-patient training dataset identified a 6-gene signature. This signature stratified NSCLC patients into 2 groups with different outcomes in 4 independent public datasets (Fig. 2). These datasets included 3 different array platforms and both squamous cell carcinoma and adenocarcinoma patients. Beyond these validation datasets, a number of other smaller or older studies exist. We combined 4 such datasets with the 4 validation datasets to generate a cohort of 589 patients drawn from 8 published studies. The 6-gene signature performed well, both on the entire cohort (Fig. S3A) and when Stage I patients were considered separately (Fig. S3B). This validation suggests that our signature may identify a cohort of Stage I patients who have the potential to benefit from adjuvant therapy. Importantly, all validations include adjustments for clinical stage, indicating that our signature is independent of traditional staging criteria, which remain the standard method for determining treatment and predicting outcome, although other factors such as age and grade also play roles.
Clinical implementation of this 6-gene signature would be straightforward. For each patient, RT-PCR analysis would be performed for the 6 prognostic and 4 housekeeping genes. After normalization, Euclidean distances will determine if the patient's profile most resembles good or poor prognosis tumors—a similar approach to that of 2 major breast-cancer studies (26, 27). The 6-gene signature can be used even if some of the PCR reactions fail or data are otherwise unavailable, as shown by successful validation in 2 cDNA microarray datasets where 1 signature and 2 normalization genes were not present on the array platform (13, 14).
We have validated our 6-gene signature in 8 of 11 recent NSCLC microarray studies (Fig. S4). The 8 included studies are themselves quite heterogeneous, with differences in both clinical and technical covariates. Clinically, the studies had varying patient-inclusion criteria, with some studies including patients with only some stages (11, 23) or histologies (11–14). Technically, studies varied in the fraction of tumor sample included in each sample, the protocols used to extract RNA, and the microarray platforms used to assess mRNA levels. The ability of the 6-gene signature to handle these many confounding factors may reflect both our secondary validation design (19) and the nonlinear nature of the mSD algorithm. The 3 omitted studies include 1 where the raw array data has not yet been deposited in a public database (18) and 2 where identifiers to link the expression data to clinical covariates do not appear to have been provided (15). This extensive validation was only possible because of the public availability of a large number of previous studies, highlighting the benefit of earlier work in the field.
Two genes (STX1A and HIF1A) are common to both the 3- and 6-gene signatures (19). This partial overlap led us to hypothesize that additional small prognostic signatures could be identified from our training dataset. To test this, we trained 10 million sets of 6 genes in our PCR dataset and tested each in 4 independent validation datasets. In both the training and testing datasets, our 6-gene classifier is superior to 99.98% of prognostic signatures (Fig. 3F). This technique provides a universal method for evaluating both specific prognostic signatures and the algorithms used to generate them.
These results demonstrate that a very large number of potential prognostic signatures exists. Our permutation study focused on 113 genes that were profiled in 5 separate studies. This small dataset can generate ≈2.5 billion unique 6-gene signatures. If, as our results suggest, 0.02% of these can be verified in multiple independent validation cohorts, then a minimum of 500,000 verifiable 6-gene prognostic signatures exist. This large number may explain the poor genewise overlap observed in prognostic signatures from different groups (19). It will be critical to determine if this conclusion can be generalized to other datasets and sizes of prognostic signature.
A detailed comparison of verifiable prognostic signatures might reveal common features. Our initial univariate analysis shows that some specific genes were highly enriched in statistically significant prognostic signatures (Fig. 4B). In particular, signatures containing calcitonin-related polypeptide alpha were statistically significant 43% of the time, implicating it in disease etiology. Overall, 3 genes in the mSD signature were enriched in prognostic signatures. Additional study of verifiable prognostic signatures might reveal other such insights. For example, certain pathways might be captured by all signatures, but represented by a number different of genes. Gene–gene interactions could be determined from pairs of genes co-occurring at a high frequency.
Our approach may provide a template for future studies to develop reproducible, mRNA-based signatures for cancer and other complex diseases. We started by using a high-quality training dataset enriched for prognostic markers. By keeping this dataset small, we minimized the problems of over-fitting that arise from using thousands of genes. Next, we used a nonlinear algorithm that dynamically learned patient groupings (i.e., a semisupervised algorithm). Finally, we extensively validated our results, by using cross-validation, multiple external datasets, and permutation-type analyses. Application of this protocol to the development of other signatures may be fruitful.
In summary, we developed a semisupervised algorithm and used it to demonstrate that a single training dataset can yield multiple prognostic signatures. The 6-gene signature identified by this algorithm was validated in multiple testing datasets and with a permutation analysis. This permutation analysis suggests a rationale for the number and diversity of distinct NSCLC prognostic markers identified.
Materials and Methods
Prognostic Signature Identification by mSD.
To identify a subset of genes whose mRNA expression profile is predictive of patient prognosis, we combined feature selection by greedy forward selection with unsupervised pattern recognition. We term this procedure mSD, and it is described in detail in SI Text. Briefly, this iterative algorithm adds genes to an existing classifier based on their ability to maximize the significance of a log-rank test on patient groups identified by k-medians clustering.
Training Dataset.
A previously published RT-PCR dataset of 158 genes assessed in 147 NSCLC patients (19) was used for training. Data were normalized as described in ref. 28. Training used the original clinical annotation; subsequent survival analyses were performed by using updated annotations, which increased patient follow-up by an average of 5.2 months (Table S2).
Cross-Validation.
To estimate the generalization error of the mSD method, we performed leave-one-out cross-validation (29). Each of the 147 patients was classified by using clusters defined with the remaining 146 patients. Euclidean distances were used to classify patients, and significance was assessed with a stage-adjusted Cox proportional-hazards model.
Independent Validation Datasets.
Four independent public datasets were used for validation (13, 14, 16, 25): Details of the validation procedure are presented in the SI Text. Briefly, the normalized data were downloaded, and a unique probe for each of the 6 genes was identified in each dataset. Median-scaling and housekeeping gene normalization (to the geometric mean of ACTB, BAT1, B2M, and TBP levels) was performed (28). Euclidean distances to the training clusters were used to classify each patient. Survival differences were assessed by using stage-adjusted Cox proportional-hazards models.
Pooled Analysis.
We combined patients from the 4 validation datasets described above with 4 older or smaller NSCLC datasets (11, 12, 23). These 589 patients were classified as described above, with Cox modeling to identify survival differences. Details are given in SI Text.
Permutation Analysis.
To determine the number of 6-gene classifiers (signatures) that could be generated from our 158-gene training dataset, we performed a permutation analysis. We tested the prognostic capability of all 10 million combinations of the 6 genes. For each combination we divided the patients into 2 groups by using k-means clustering and calculated significance by using log-rank analysis. The distribution of subsets with prognostic significance (χ2 > 3.84 or P < 0.05) in the training dataset was visualized by using Gaussian density plots.
Supplementary Material
Acknowledgments.
We thank Melania Pintilie for outstanding statistical advice; Richard Lu for computer system support; Davina Lau for updated clinical follow-up data; Christian Cumbaa for advice on machine-learning; and members of the Tsao, Jurisica, and Penn labs for critical commentary. F.A.S is the Clive Taylor Chair in Lung Cancer Research; M.-S.T. is the M. Qasim Choksi Chair in Lung Cancer Translational Research; L.Z.P. is Canada Chair in Molecular Oncology; and I.J. is Canada Chair in Integrative Computational Biology. This work was supported by the National Cancer Institute of Canada (L.Z.P., I.J., M.S.T., S.D.D.); Natural Sciences and Engineering Research Council (I.J.); Princess Margaret Hospital Foundation (I.J.); Genome Canada through the Ontario Genome Institute (I.J., S.D.D.); IBM (I.J.); and fellowships from the PreCarn Foundation (P.C.B.), the Natural Sciences and Engineering Research Council (P.C.B.), and the Canadian Institutes of Health Research's Excellence in Radiation Research for the 21st Century Strategic Training Initiative in Health Research Program (P.C.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809444106/DCSupplemental.
References
- 1.Tsuboi M, et al. The present status of postoperative adjuvant chemotherapy for completely resected non-small cell lung cancer. Ann Thorac Cardiovasc Surg. 2007;13:73–77. [PubMed] [Google Scholar]
- 2.Mountain CF. Staging classification of lung cancer. A critical evaluation. Clin Chest Med. 2002;23:103–121. doi: 10.1016/s0272-5231(03)00063-7. [DOI] [PubMed] [Google Scholar]
- 3.Mountain CF. Revisions in the International System for Staging Lung Cancer. Chest. 1997;111:1710–1717. doi: 10.1378/chest.111.6.1710. [DOI] [PubMed] [Google Scholar]
- 4.Jones KL, Buzdar AU. A review of adjuvant hormonal therapy in breast cancer. Endocr Relat Cancer. 2004;11:391–406. doi: 10.1677/erc.1.00594. [DOI] [PubMed] [Google Scholar]
- 5.Zaniboni A, Labianca R. Adjuvant therapy for stage II colon cancer: An elephant in the living room? Ann Oncol. 2004;15:1310–1318. doi: 10.1093/annonc/mdh342. [DOI] [PubMed] [Google Scholar]
- 6.Gramont A. Adjuvant therapy of stage II and III colon cancer. Semin Oncol. 2005;32(6) Suppl 8:11–14. doi: 10.1053/j.seminoncol.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 7.NSCLC Group. Chemotherapy in non-small cell lung cancer: A meta-analysis using updated data on individual patients from 52 randomised clinical trials. BMJ. 1995;311:899–909. [PMC free article] [PubMed] [Google Scholar]
- 8.Winton T, et al. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med. 2005;352:2589–2597. doi: 10.1056/NEJMoa043623. [DOI] [PubMed] [Google Scholar]
- 9.Douillard JY, et al. Adjuvant vinorelbine plus cisplatin versus observation in patients with completely resected stage IB-IIIA non-small-cell lung cancer (Adjuvant Navelbine International Trialist Association [ANITA]): A randomised controlled trial. Lancet Oncol. 2006;7:719–727. doi: 10.1016/S1470-2045(06)70804-X. [DOI] [PubMed] [Google Scholar]
- 10.Kato H, et al. A randomized trial of adjuvant chemotherapy with uracil-tegafur for adenocarcinoma of the lung. N Engl J Med. 2004;350:1713–1721. doi: 10.1056/NEJMoa032792. [DOI] [PubMed] [Google Scholar]
- 11.Beer DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 12.Bhattacharjee A, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsen JE, et al. Expression profiling defines a recurrence signature in lung squamous cell carcinoma. Carcinogenesis. 2007;28:760–766. doi: 10.1093/carcin/bgl207. [DOI] [PubMed] [Google Scholar]
- 14.Larsen JE, et al. Gene expression signature predicts recurrence in lung adenocarcinoma. Clin Cancer Res. 2007;13:2946–2954. doi: 10.1158/1078-0432.CCR-06-2525. [DOI] [PubMed] [Google Scholar]
- 15.Potti A, et al. A genomic strategy to refine prognosis in early-stage non-small-cell lung cancer. N Engl J Med. 2006;355:570–580. doi: 10.1056/NEJMoa060467. [DOI] [PubMed] [Google Scholar]
- 16.Raponi M, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006;66:7466–7472. doi: 10.1158/0008-5472.CAN-06-1191. [DOI] [PubMed] [Google Scholar]
- 17.Sun Z, Wigle DA, Yang P. Non-overlapping and non-cell-type-specific gene expression signatures predict lung cancer survival. J Clin Oncol. 2008;26:877–883. doi: 10.1200/JCO.2007.13.1516. [DOI] [PubMed] [Google Scholar]
- 18.Chen HY, et al. A 5-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 19.Lau SK, et al. Three-gene prognostic classifier for early-stage non small-cell lung cancer. J Clin Oncol. 2007;25:5562–5569. doi: 10.1200/JCO.2007.12.0352. [DOI] [PubMed] [Google Scholar]
- 20.Ein-Dor L, Zuk O, Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc Natl Acad Sci USA. 2006;103:5923–5928. doi: 10.1073/pnas.0601231103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bachtiary B, et al. Gene expression profiling in cervical cancer: An exploration of intratumor heterogeneity. Clin Cancer Res. 2006;12:5632–5640. doi: 10.1158/1078-0432.CCR-06-0357. [DOI] [PubMed] [Google Scholar]
- 22.Blackhall FH, et al. Stability and heterogeneity of expression profiles in lung cancer specimens harvested following surgical resection. Neoplasia. 2004;6:761–767. doi: 10.1593/neo.04301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu Y, et al. A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med. 2006;3:e467. doi: 10.1371/journal.pmed.0030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simon R, Radmacher MD, Dobbin K, McShane LM. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J Natl Cancer Inst. 2003;95:14–18. doi: 10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
- 25.Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 26.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 27.van 't Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 28.Barsyte-Lovejoy D, et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66:5330–5337. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- 29.Duda RO, Hart PE, Stork DG. Pattern Classification. 2nd ed. New York: Wiley; 2001. p. 654. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.