Abstract
Control of replication, transcription, recombination and repair requires proteins capable of finding particular DNA sequences in a background of a large excess of nonspecific sequences. Such recognition can involve direct readout, with direct contacts to the bases of DNA, or in some cases, through the less well characterized indirect readout mechanisms. In order to measure the relative contributions of direct and indirect readout by a sequence specific endonuclease, HincII, a mutant enzyme deficient in a direct contact was characterized, and surprisingly showed no loss of sequence specificity. The three dimensional crystal structure shows the loss of most of the direct readout contacts to the DNA, possibly capturing an early stage in target site recognition using predominately indirect readout to prescreen sites before full sequence interrogation.
Introduction
The problem of target site location for proteins that bind specific DNA sequences is a question of both specificity and speed, and affects such fundamental biological processes as DNA replication, transcription, recombination and repair (von Hippel, 2007). Proteins that bind specific DNA sequences must find and recognize their target sites in a vast excess of non-specific DNA. The importance of specificity is exemplified by the balance between affinity and sequence discrimination necessary for optimal regulation of the E.coli lac operon by the lac repressor (von Hippel and Berg, 1986). In addition, proteins must find their target sequences on a biologically relevant time scale. Early binding studies of lac repressor to DNA reveal an association rate constant that is faster than that predicted for three-dimensional diffusion, and such fast target site location has been termed “facilitated diffusion” (Riggs et al., 1970). Models of facilitated diffusion include one-dimensional sliding along the DNA (also known as scanning), transfer from one segment of DNA to another (possible if two DNA binding sites exist on the same protein), or dissociation/re-association near the site of DNA (also known as hopping)(Richter and Eigen, 1974; von Hippel and Berg, 1989). More recently, the fast diffusion of DNA binding proteins, including lac represssor, has been observed directly using single molecule methods (Blainey et al., 2006; Elf et al., 2007).
Investigations of DNA sequence discrimination using biochemical and structural studies suggest mechanisms for both specificity and rapid target site location. The structures of DNA binding proteins with their target sequences often show distortions of the bound DNA, with direct discriminatory contacts from the protein to the bases in the major groove (direct readout)(Garvie and Wolberger, 2001). However, in some structures specificity cannot be explained by direct readout as no direct contacts are observed to base edges, indicating that specificity occurs through indirect means (indirect readout)(Otwinowski et al., 1988). Indirect readout is more difficult to characterize but appears to involve such phenomena as water mediated hydrogen bonds to the DNA base edges, as well as the induction of DNA distortion to distinguish sequences energetically.
Structures of proteins bound to non-target DNA are thought to serve as models for the scanning mode of facilitated diffusion, showing limited contacts by the protein to the DNA bases in both the major and minor grooves, with relatively undistorted DNA (Kalodimos et al., 2004; Viadiu and Aggarwal, 2000; Winkler et al., 1993). However, these non-specific structures do little to explain target site recognition. For those proteins that distort DNA and make direct contacts in the major groove, such contacts would be impossible to make with undistorted DNA. Recognition of DNA sequences through direct contacts during target site location must then involve the interrogation of DNA sequences via similar DNA distortions. Observations of nonspecific DNA distortion by sequence specific DNA binding proteins has been observed by atomic force microscopy (Erie et al., 1994). However, DNA sequence interrogation using primarily direct readout with large DNA distortions could be costly in terms of the speed of finding a target site. Alternatively, some other property could pre-select only certain sequences for further testing via direct readout. Such pre-selection would expedite the target site location if rapid and selective. The pathway for target site location would then require the existence of an intermediate with more limited contacts and less DNA distortion.
To test the influences of direct and indirect readout on DNA target site location and specificity, we turned to a model system found in bacteria (Pingoud and Jeltsch, 2001). Bacteria protect themselves from invading phage DNA with sequence specific DNA cleaving enzymes known as restriction endonucleases (Jeltsch et al., 1996). The bacterial genome is protected from the endonuclease activity by a DNA methlytransferase that methylates the endonuclease recognition sequences, thus inhibiting cleavage at these sites. An invading phage does not have specifically methylated DNA, leaving the phage DNA susceptible to cleavage by the host endonuclease once the phage DNA is injected into the cell. These endonucleases must be sequence specific since nonspecific endonuclease activity could result in genomic DNA damage, and even death to the host bacterium. This bacterial survival system has evolved DNA endonucleases under the pressures of both time and specificity. The phage DNA must be recognized and cleaved fast enough to render it harmless to the bacterium, and the endonuclease must specifically recognize the phage DNA and not the host genome to ensure cell viability.
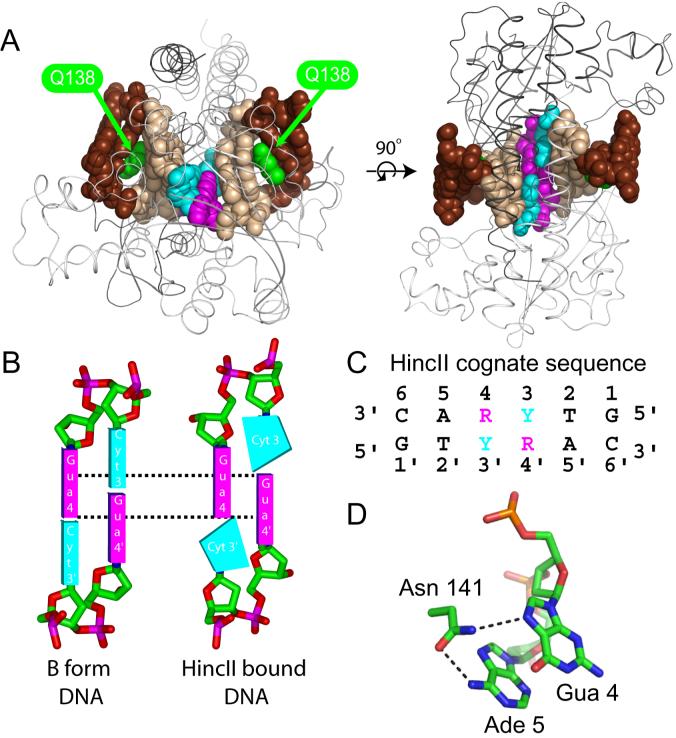
DNA sequence recognition has been investigated in the restriction endonuclease HincII from H. influenzae, which binds as a dimer and cleaves the DNA sequence GTYRAC (Y=C or T, R=A or G)(Kelly and Smith, 1970). The structure of wild type HincII bound to GTCGAC suggests that direct readout occurs at all bases except at the Y (C in the structure) (Fig.1A)(Horton et al., 2002). In addition, wild type HincII distorts the DNA through both intercalation and bending. The center YR step of the recognition sequence shows an unusual distortion as a result of the bending and the shifting of the base planes into the minor groove. The distortion is characterized by the unstacking of the pyrimidine at position 3 from the purine at position 4, and an increase in stacking of the purines at position 4 of each strand with each other (Fig.1B, numbering of the HincII recognition sequence given in Fig. 1C). This type of stacking has been seen in A form DNA, and is known as cross-strand purine stacking (CSPS)(Heinemann et al., 1987). The direct readout of the YR nucleotides occurs by a hydrogen bond from the side chain of HincII residue N141 to the N7 of the purine base at Pur 4 (Gua 4 in Fig. 1D). The side chain of N141 also hydrogen bonds to the N6 of Ade 5, a contact which both constitutes the direct readout at this nucleotide, but also orients the side chain for the contact to the N7 of Gua 4. Since no direct contacts are made by HincII to the nucleotide at position 3, the contact between N141 and Gua 4 is the sole direct readout of the center YR step of the recognition sequence. It is possible that the YR center step could be recognized solely through this direct readout, however the distortion at YR could also contribute to specificity via indirect readout. We therefore sought to measure the role of the DNA distortion at the center YR step in the specificity of HincII. Removal of the side chain N141 by mutation to alanine removes the direct readout of the center step purine, leaving any residual specificity for the center YR step to be performed by indirect readout. Unexpectedly, the N141A HincII mutant was found to cleave DNA only at the HincII cognate sequences GTYRAC, with no apparent cleavage at any other sequence. The DNA cleavage rate and binding affinity were both found to be greatly diminished by the mutation, but the persistent YR specificity in the absence of direct readout indicates that HincII uses indirect readout at these nucleotides. A crystal structure of N141A HincII bound to cognate DNA shows a surprising absence of direct readout at other bases in addition to those lost by the N141A mutation, as well as the loss of DNA intercalation, and may exhibit a conformation used by wild type HincII for sequence interrogation during target site location.
Figure 1. DNA binding and distortion by wild type HincII.
A. Two views of the structure of wild type HincII (two subunits shown as black or white ribbons) bound to cognate DNA (shown in space filling with the recognition site DNA in light brown and the center two base pairs of the YR step in cyan and magenta, respectively, with flanking DNA in dark brown). B. Cartoon depiction of B form DNA (left) with that of the center YR step of the HincII DNA recognition sequence bound to wild type HincII (right), in which the DNA bases of the same strand are unstacked and the purine bases from opposing strands exhibit greater stacking surface area (dashed lines) forming the cross-strand purine stack (CSPS). C. Numbering of the HincII recognition sequence used throughout the text. D. Hydrogen bonds between the side chain of N141 in wild type HincII and bound cognate DNA.
Results
Activity assay on plasmid DNA
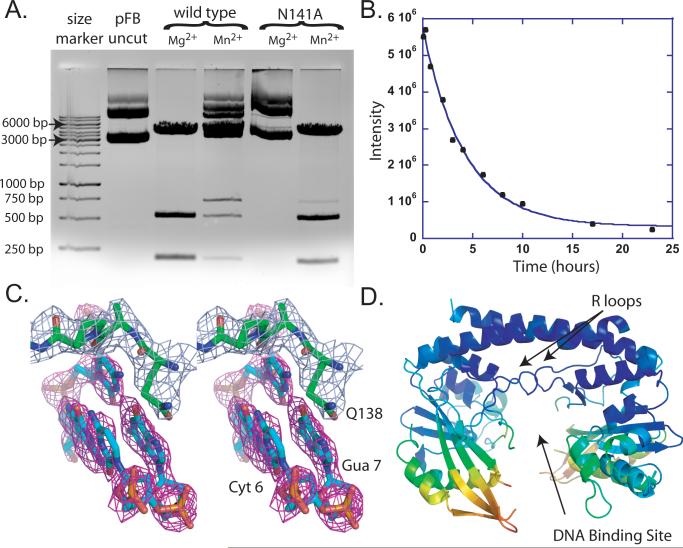
The DNA cleavage activity of N141A HincII was tested using a digestion assay with a plasmid bearing one each of the three HincII recognition sequences, and an additional 11 sites bearing the sequence GTNNAC, which are similar to HincII recognition sites but without YR at the center step. The wild type or mutant protein was incubated with the plasmid at 37°C overnight with 20 mM MgCl2 or 2 mM MnCl2. Wild type HincII cleaves the plasmid DNA completely in the presence of MgCl2 resulting in the three predicted fragments, and also cleaves the plasmid partially in the presence of MnCl2 (Fig. 2A). The band migrating third from the bottom of the gel is composed of the two faster migrating species prior to cleavage at the GTCGAC HincII site. In contrast, N141A HincII shows only slow cleavage of the plasmid in the presence of MgCl2, but cleaves almost completely with MnCl2. Significantly, the mutant HincII cleaves preferably at the HincII sites, as no other fragments except for the predicted three HincII cleavage products are apparent (Fig. 2A).
Figure 2. Functional and Structural analysis of N141A HincII.
A. Ethidium bromide stained agarose gel after electrophoresis of plasmid DNA incubated with wild type or N141A HincII. B. Single turnover cleavage assay of N141A HincII (5.0×10-6 M) and cognate DNA (1.0×10-7M). The integrated intensity of the uncleaved DNA is plotted versus time of incubation. The data are fit to a single exponential function (solid line). C. Stereo diagram of simulated annealing omit electron density (magenta, contoured at 3σ) after omission of base pairs Gua 1/Cyt6′ and Cyt-1/Gua7′, where intercalation by the side chain of Q138 occurs in the wild type HincII/DNA complex. Residues 138-141 are shown with 2Fo-Fc electron density (light blue, contoured at 1σ). D. Ribbon diagram of N141A HincII (DNA not shown) after coloring by RMSD with wild type HincII using a superposition of the entire dimer onto wild type HincII. Coloring from lowest RMSD in blue (0.08 Å RMSD) to red (7.8 Å RMSD).
Single turnover DNA cleavage measurements
Single turnover DNA cleavage kinetic experiments using three different 16 base pair deoxyoligonucleotides, each containing one of the three HincII recognition sequences, show kinetics consistent with single turnover kinetics, and fit to a single exponential (Fig. 2B and Fig. S1B). Such kinetics occur with a single round of DNA cleavage produced when the enzyme is greater in concentration than the DNA, and binding goes to completion. Two DNA bands are visible on the denaturing polyacrylamide gels, due to the asymmetric position of the HincII cleavage site within the duplex DNA. Cleavage at this site produces the 5′ end labeled products 7 and 9 nucleotides in length. Using this assay, the N141A HincII enzyme does not show any preference among the three HincII cognate sequences, with rate constants of 8.8±1.3×10-5 s-1 (3.2±0.5×10-1 hr-1), 9.2±3.9×10-5 s-1 (3.3±1.4×10-1 hr-1), and 11.0±4.1×10-5 s-1 (4.0±0.2×10-1 hr-1) for DNA containing the HincII sequences GTTAAC, GTCGAC, and the nonpalindromic GTTGAC/GTCAAC, respectively. N141A HincII DNA cleavage activity is approximately 1000 fold reduced relative to that of wild type HincII (Joshi et al., 2006). No cleavage was found in assays with the Noncognate GC DNA after 24 hours incubation (data not shown).
Crystal structure
The three dimensional X-ray crystal structure of N141A HincII bound to DNA was solved using molecular replacement (Table 1). The 2.55 Å structure was refined to an Rfree of 27.9% and an Rcryst of 19.1%. The asymmetric unit contained a single enzyme dimer bound to one duplex of DNA. Only a single Mn2+ ion could be located (Fig. S3A), which does not appear to bind with full occupancy, as Fo-Fc omit maps show only a 4 σ peak (heights over 8 σ are typical for Mn2+). In addition, the refined temperature factor is higher (99 Å2) than surrounding atoms (23-33 Å2), and two ligands of the octahedral ligation shell are missing. However, when modeled as a water molecule, additional difference density occurs nearby and is too close (1.7 Å) to model as hydrogen bonded water molecules. Residues 21-33 and 134-136 of subunit A, and residues 22-32 and 134-135 of subunit B were found to be disordered and excluded from the final model. Simulated annealing omit electron density is shown for the region around residue Gln138 of subunit B (Fig. 2C).
Table I. Crystallographic Data Statistics.
| Space Group | P21 |
| Cell | 46.55 Å, 91.57 Å, 70.59 Å, b=107.47° |
| Resolution | 2.55Å |
| Total Observations | 61,125 |
| Unique Observations | 17,810 |
| % Complete | 96.1% (75.8%) |
| I/sigma | 11 (1.9) |
| Multiplicity | 3.4 |
| Rmerge1 | 9.3% (31.3%) |
| Rcryst2 | 19.1% |
| Rfree3 | 27.9% |
| Overall B factor (Å2, Wilson plot) | 28.7 |
| RMSD-bonds | 0.009 Å |
| RMSD-angles | 1.38° |
| Number of waters | 371 |
| Number of Mn2+ | 1 |
| Protein residues | 485 |
| Nucleotides | 28 |
Rmerge=Σhkl(|<Ihkl>-Ihkl|)/(Σhkl Ihkl) where <Ihkl> is the average intensity over symmetry related and equivalent reflections and Ihkl is the observed intensity for reflection hkl.
Rcyst=Σhkl(||Fobs|-|Fcalc||)/( Σhkl|Fobs|) where |Fobs| and |Fcalc| are the observed and calculated structure factor amplitude for reflection hkl. The sum is carried out over the 95% of the observed reflections which are used in refinement.
Rfree refers to the R factor for the test reflection set (5% of the total observed) which was excluded from refinement.
Structural comparisons with wild type HincII
The overall RMSD of the entire N141A HincII dimer with that of wild type HincII is 2.6 Å. Figures S1C and 2D show ribbon diagrams of N141A HincII (DNA not shown) colored by RMSD with the wild type DNA bound structure, using either superposition of each monomer independently (Fig. S1C), or using the whole dimer in the superposition (Fig. 2D). Using each monomer independently and comparing structural deviations (Fig. S1C), the segments deviating most with wild type occur at the C termini, the loops found at the DNA binding site, and the segments opposite of the C-terminus at the ‘bottom’ of the DNA binding site.
Using the entire dimer for the comparison (Fig. 2D), the location of the largest structural deviations between N141A and wild type HincII are largest at the ‘bottom’ of the DNA binding site. The gradation of RMSD from the ‘top’ of the beta sheet to the ‘bottom’ of the molecule is due to a rigid body rotation of the core of the subunit. The C-terminal helices (which form the dimeric interface at the ‘top’ of the dimer) and the R loops, which bind the DNA in the major groove, and the regions of the protein immediately contacting these segments, appear similar in the two structures and have lower RMSD. The higher values in the dimeric comparison compared to that of the monomeric indicate that the difference in structure is in the positioning of one monomer relative to the other. Figure S1D shows a superposition (wild type in blue/cyan, N141A in red/pink) of the entire dimer to emphasize that the R loops and DNA they contact align, but the more distal DNA and the rest of the protein subunits deviate in the two structures.
The structural comparisons of N141A with wild type HincII indicate that a large part of the difference in structure is from the positioning of one subunit core relative to the other. Rigid body displacement was characterized by superimposing the A subunits of wild type and N141A HincII and calculating the rotation and translation required to align the cores of the B subunits. The difference between DNA bound wild type and N141A HincII enzymes is an 11° rotation about an axis 158° from the vector connecting the centers of mass of the two subunits (COM vector) (Figures 3A and S2A). The rigid body displacement of the core is not however a pure rotation, as it involves also a 6.6 Å translation. Figure 3A shows the rotation axis positioned between the two centers of mass of the B subunits of wild type (blue) and N141A (red) HincII after superposition using the A subunits of both structures (grey).
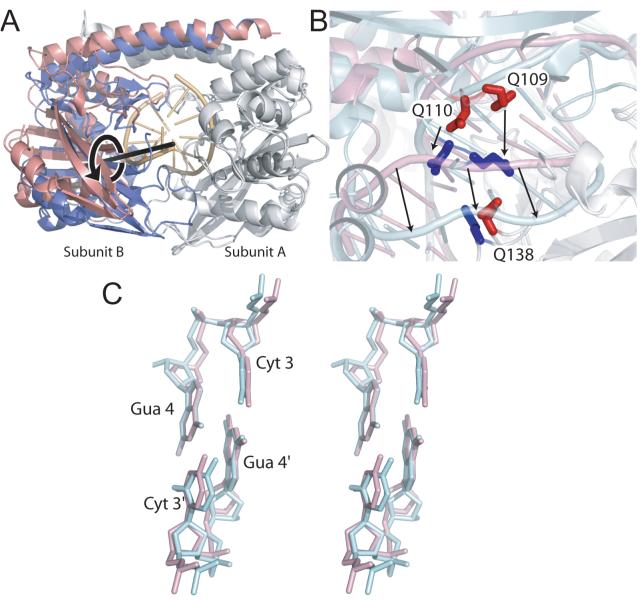
Figure 3. Domain displacement in N141A HincII.
A. Superposition of wild type (blue and grey) HincII and N141A (pink and grey) HincII using all residues of the A subunits (grey). DNA bound to the wild type structure shown in gold. The rotational axis required to align the B subunits (blue and pink) in this superposition is shown as a black rod and has been placed between the two B subunit centers of mass. The direction of rotation about this axis required to align the N141A B subunit with that of wild type HincII is shown in black. B. Superposition described for 3A showing selected residues contacting the DNA. Arrows indicate the difference in the position, between N141A and wild type HincII structures, of the bound DNA and residues 109 and 110 of the B subunits (red, N141A HincII, blue, wild type HincII). Residue Q138 of subunit A also shown (red, N141A HincII, blue, wild type HincII). C. Stereo view of the cross-strand purine stack in DNA bound to N141A HincII (pink), bound to wild type HincII (light blue). Superposition performed with atoms of the base of Gua 4′.
The domain displacement is related to the loss of key protein-DNA interactions in the N141A HincII/DNA complex. Figure 3B shows the same superposition as Fig. 3A using the A subunits of both structures (grey) with a close-up of the DNA and nearby segments. The black arrows show how the backbone of the DNA bound to wild type HincII (light blue, Fig. 3B) is shifted toward the segment containing the intercalating residue Q138 (dark blue, Fig. 3B). This residue is not found to intercalate the DNA bound to N141A HincII (pink, Fig. 3B), although the segment containing residue 138 (red, Fig. 3B) is not badly mispositioned. The lack of intercalation by Q138 appears instead to be related to the rotational/translational displacements of the core subdomains relative to each other. Residues 109-110 of the opposing subunit interact with the DNA on the face opposite from Q138 in the minor groove, while Q138 interacts in the major groove (Fig. S2B). Rotation of subunit B in wild type HincII brings these residues closer into the minor groove, and compresses the DNA onto the intercalating residue Q138.
The structures of the center two base pairs of the DNA bound to N141A HincII (pink) and wild type HincII (light blue), after superposition using the atoms of one of the two Gua 4 bases are shown in Fig. 3C. The roll angle between adjacent bases of the center step is greater in the DNA bound to wild type HincII, however, the purine bases clearly form the CSPS in the DNA bound to N141A HincII.
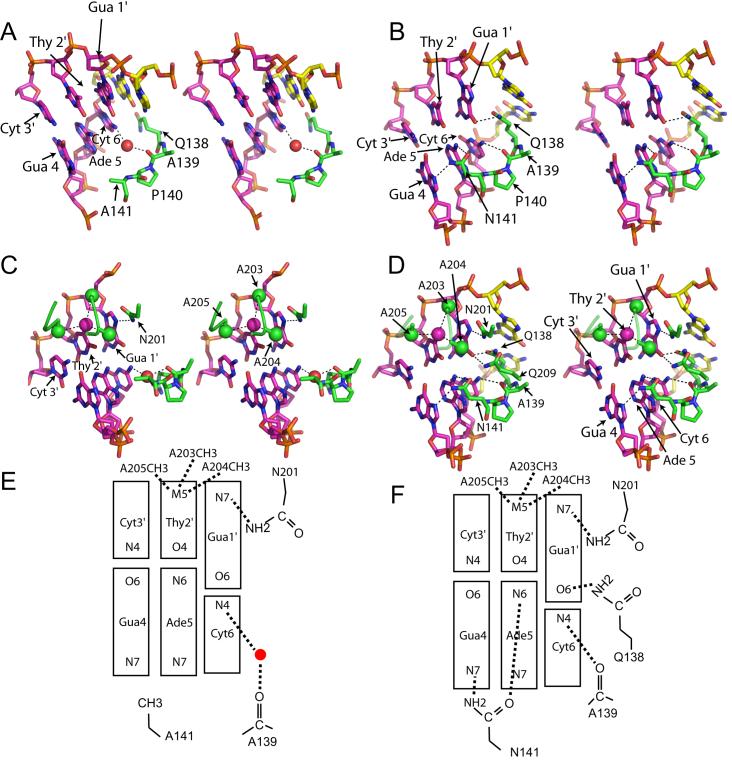
The interactions between N141A or wild type HincII and the DNA bases in the major groove of half of the recognition site are shown in Fig. 4A, C, E (N141A) and 4B, D, F (wild type). (The structures of wild type and N141A HincII dimers bound to the palindromic DNA sequence are symmetric, and the contacts to both halves of the recognition sequence are identical within coordinate error in each structure.) Fig. 4A highlights the interactions with the segment 138-141, where the direct contacts to Gua 1′ (where prime designates the opposing strand in the duplex DNA), Cyt 6, Ade 5, and Gua 4 normally occur. Wild type HincII makes direct contacts between the side chain amide nitrogen of Q138 and the O6 of Gua 1′, the backbone carbonyl of A139 and the N4 of Cyt 6, and the side chain of N141 and the N6 of Ade 5 as well as the N7 of Gua 4 (Fig. 4B,F). The contacts between the side chain of residue 141 and Gua 4 and Ade 5 are lost by the mutation N141A and the direct contacts between A139, Q138 and the DNA are unexpectedly lost as well (Fig. 4A,E). The displacement of the DNA away from this segment of the enzyme appears to increase the separation and allows a water mediated contact to be made between the backbone carbonyl of A139 and the N4 of Cyt 6. The interaction of Q138 with the DNA is especially different, since in wild type HincII this residue intercalates into the DNA just outside of the recognition site, bringing it within hydrogen bonding distance to the O6 of Gua 1′ (Fig. 4B,F). In N141A HincII, the side chain of Q138 is turned away from the Gua 1′ base and toward the protein backbone instead (Fig. 4A). Figures 4C,E show the two direct contacts to the major groove preserved in N141A HincII, namely to the 5 methyl of Thy 2′ by the side chain methyl groups of A203, A204 and A205, and to the N7 of Gua 1′ by the side chain of N201. Similar contacts are seen in the structure of wild type HincII bound to GTCGAC (Fig. 4D,F).
Figure 4. Protein DNA contacts in wild type and N141A HincII/DNA structures.
A. Stereo view of the contacts to the DNA by residues 138-141 of N141A HincII. Colored by atom type: N-blue, O-red, P-orange, C-green in protein, pink in DNA of the recognition site and yellow in flanking base pairs. A water molecule is shown as a red sphere. Dashes indicate hydrogen bonds. B. Stereo view of the contacts to the DNA by residues 138-141 of wild type HincII. Colored as in A. Dashes indicate hydrogen bonds. C. Stereo view of the contacts to the DNA at Thy 2′ and Gua 1′ of N141A HincII. Colored as in A. Methyl groups shown as green or pink spheres, water molecule shown as a red sphere. Dashes indicate hydrogen bonds or van der Waals interactions. D. Stereo view of the contacts to the DNA at Thy 2′ and Gua 1′ of wild type HincII. Colored as in A. Methyl groups shown as green or pink spheres. Dashes indicate hydrogen bonds or van der Waals interactions. E. Cartoon of protein-DNA contacts to bases in the major groove in the N141A HincII/DNA structure. Water molecule shown as a red circle. F. Cartoon of protein-DNA contacts to bases in the major groove in the wild type HincII/DNA structure. Dashes indicate hydrogen bonds or van der Waals interactions.
Discussion
Activity: inactive with Mg2+ but active and sequence specific with Mn2+
Removal of the side chain of N141 was done in order to measure the relative importance of direct versus indirect readout to sequence specific recognition. In the wild type crystal structure, the asparagine side chain of residue 141 hydrogen bonds to the N7 of the center step purine, Gua 4, as well as the N6 of Ade 5. (The numbering for the HincII cognate sequence is shown in Fig. 1C. The numbering of nucleotides in the crystal structure for the same base pairs is 5-10, which has been used in previous reports (Babic, 2008; Horton et al., 2002; Joshi et al., 2006)). Removal of the N141 side chain by mutation to alanine removes the direct readout of these two bases, leaving any residual specificity to be performed solely by indirect readout. Specificity at the Ade 5 is expected to be reduced as a loss of the direct contact, however, the hydrophobic contacts from residues 203-205 to the Ade 5 base pairing partner Thy 2 should result in retention of at least part of the wild type specificity at this base pair. The most significant result expected from the N141A mutation is the loss of some or perhaps all of the specificity for the center base pairs, since the only direct contact to these positions is from the side chain of residue 141. Instead, full specificity is retained, and the N141A HincII enzyme cleaves only the HincII recognition sequences GTYRAC. In addition, although the DNA binding affinity appears greatly weakened, gel shift experiments suggest greater affinity of N141A HincII to DNA containing the cognate sequence CG at the center YR step of the recognition sequence over that with the noncognate GC (Supplementary Material). Therefore, the mutant enzyme uses some indirect means to recognize the DNA sequence at the center step, and it is likely to also be true of the parent wild type enzyme.
The metal ion dependence of DNA cleavage by HincII is altered by the N141A mutation. Plasmid DNA cleavage by N141A HincII occurrs only weakly with 20 mM MgCl2, but goes to near completion with 2 mM MnCl2 under the same conditions. Wild type HincII shows the opposite pattern, with complete cutting in 20 mM MgCl2, but only limited cleavage with MnCl2. The effects of Mn2+ substitution for Mg2+ on Mg2+ dependent endonucleases has been previously noted (Baldwin et al., 1999; Parry et al., 2003; Sam and Perona, 1999; Vermote et al., 1992). For example, maximal turnover of DNA cleavage by EcoRV will occur with Mg2+, despite the fact that the chemical step of DNA cleavage is more rapid with Mn2+ (Sam and Perona, 1999). The reason for the slower turnover in the presence of Mn2+ is greatly slowed product release, presumably due to enhanced affinity for DNA with Mn2+ (Sam and Perona, 1999). The rescue of the DNA cleavage activity of N141A HincII by Mn2+ may occur by a similar effect, as the affinity of N141A HincII for DNA appears greatly reduced as a result of the mutation (Supporting Material). In addition, specificity can often be compromised with Mn2+ substituted for Mg2+(Allingham and Haniford, 2002; Cirino et al., 1995; Hsu and Berg, 1978; Kunkel and Loeb, 1979; Parry et al., 2003; Tabor and Richardson, 1989; Taylor and Halford, 1989; Vaisman et al., 2005; Vermote et al., 1992), suggesting another possible mechanism for the rescue of DNA cleavage activity in severely impaired mutant enzymes (Jeltsch et al., 1993; Parry et al., 2003; Selent et al., 1992; Vermote et al., 1992; Vipond et al., 1996; Xu and Schildkraut, 1991). While Mn2+ substitution is required for full activity, no loss of specificity as a result of the N141A mutation or metal substitution was found, and therefore direct readout of the center YR step is not necessary for sequence discrimination by HincII.
Lack of DNA cleavage in N141A HincII/DNA crystal
The structure appears to be trapped in an inactive state, as the DNA is not cleaved and Mn2+ appears to be only weakly bound, and in only one of the two subunits (Fig. S3). Yet the activity assays show that N141A HincII is capable of cleavage of this sequence in the presence of Mn2+ at 37°C. The trapping of this intermediate may be a result of the temperature of crystallization (17°C). Experiments show that the cleavage reaction is greatly slowed by lowering the temperature 20°C, and only nicking of plasmid DNA occurs using the crystallization conditions as reaction conditions and after overnight incubation at 17°C (data not shown). The higher temperatures may be required for the DNA to be intercalated by HincII, since the absence of the interactions with the DNA as a result of the missing side chain of 141 likely weakens the protein-DNA interface locally.
The Role of Indirect Readout in “Pre-Screening”
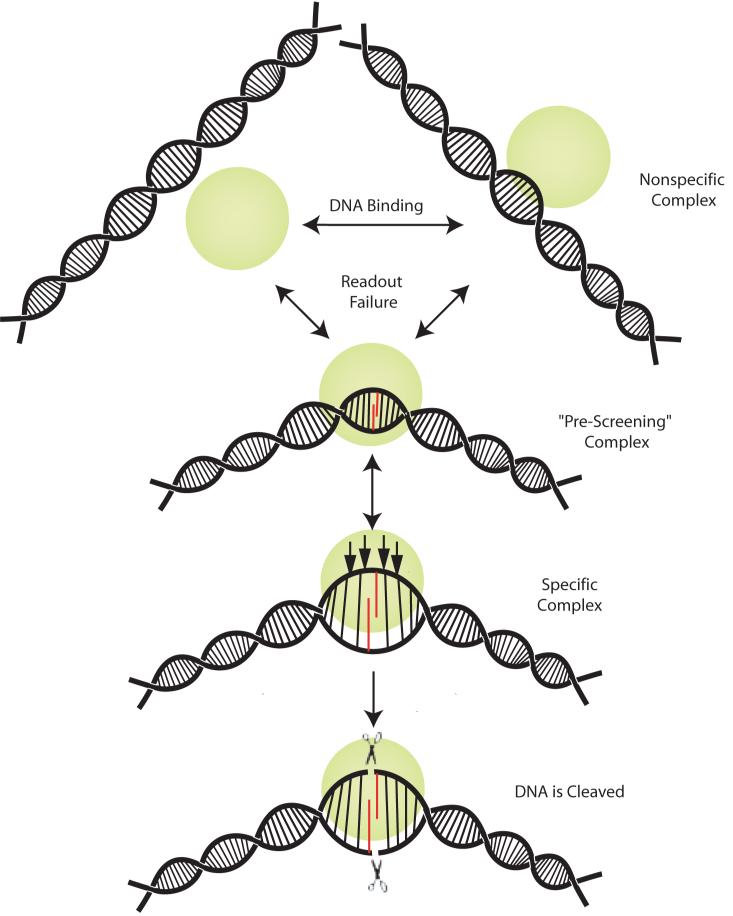
The preserved specificity for YR in the absence of direct readout, and the distortion at the YR step, support the use of the DNA distortion (specifically the CSPS) in the indirect readout of DNA sequence by HincII. Since the conformation of the N141A HincII bound DNA is distorted from B form, but is less distorted from that found in the wild type HincII/DNA complex, the N141A HincII/DNA structure could represent that of an intermediate that normally occurs transiently in the DNA interrogation pathway. Of course, it is not possible to say for certain that the conformation found in the N141A HincII/DNA crystal structure is “on-pathway” to the final active conformation of this mutant or the wild type enzyme, rather, the possibility remains that the conformation found in this structure is an artifact of mutagenesis or crystal packing. However, such an intermediate could function to expedite target site location by eliminating most sites from full interrogation (Fig. 5), and only sites passing this pre-screening would then be fully interrogated. This “pre-screening” intermediate would be distinguished from that proposed using the structure of BstYI bound to hemi-specific DNA (Townson et al., 2007), as the putative HincII “pre-screening” intermediate is on pathway to the final specific complex.
Figure 5. Proposed model of target site location using indirect readout to pre-screen sites for interrogation by direct readout.
HincII (green sphere) binds DNA and may diffuse from the initial binding site to other sites on the DNA (Nonspecific Complex). The DNA sequence may then be interrogated using indirect readout, which includes the formation of the cross-strand purine stack (red), and some limited direct readout contacts (“Pre-screening” Complex). If the sequence fails the early interrogation, HincII may move to a different site, or dissociate from the DNA. If the sequence passes the early interrogation by HincII, it may be tested for sequence by direct readout indicated by the arrows in the Specific Complex. If the sequence passes the direct readout, it will be cleaved by HincII indicated by scissors.
The role of DNA distortion in DNA recognition can now be considered important for at least four functions. First, particular distortions can be used to discriminate in favor of certain sequences by a process termed indirect readout. Secondly, this indirect readout can also be used during target site location by an intermediate in the pathway having limited DNA contacts and distortions. Thirdly, enzymes recognizing similar yet distinct sequences may distort DNA differently to prevent specificity conversion by one or two amino acid substitutions (Townson et al., 2005). Finally, new specificities cannot evolve by merely substituting one amino acid for another at the site of the direct contacts, since the side chains are generally not isosteric. Therefore, changes in either the protein structure or the DNA conformation, or both, may be necessary to re-align any new direct contacts. This phenomenon may explain why members of the type II restriction endonuclease family, having specificities for over 200 different DNA sequences, contain a common core fold but have a high degree of sequence divergence with varied local folding (Niv et al., 2007). Similarly, this may also explain why modular proteins like zinc fingers are limited in the DNA sequences they can recognize (Wolfe et al., 2001).
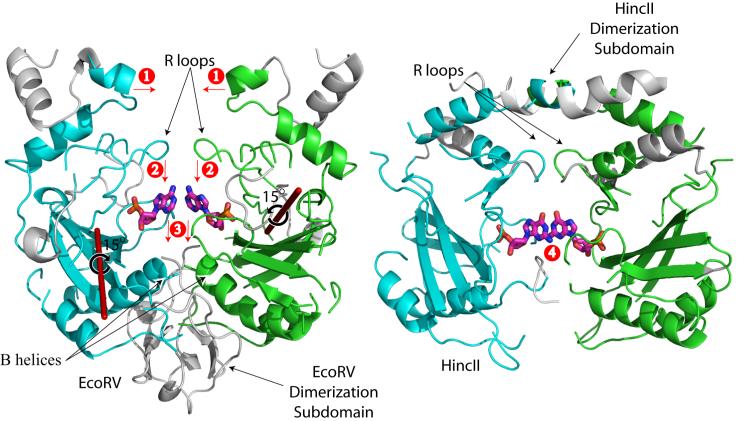
The origin of the HincII induced DNA distortion can be found through comparison of the HincII structure to that of the well characterized restriction endonuclease EcoRV, which is structurally related to HincII (Winkler et al., 1993). Segments of the two enzymes which align using a difference distance approach in the program DALI are colored green (subunit A) and cyan (subunit B) in Fig. 6. After alignment of these segments from both subunits in the EcoRV and HincII dimers, the rotation of each subunit of EcoRV necessary to align with HincII was calculated. The rotation axis thus derived, along with the direction and the degree of rotation (calculated to be 15°) is shown in Fig. 6. The rotation brings the ‘tops’ of the EcoRV subunit closer in HincII, and the ‘bottoms’ further (labeled 1, Fig. 6). Both enzymes use loop structures to contact the DNA in the major groove (R loops). EcoRV also uses indirect readout in DNA recognition at the center step of its target sequence GATATC (Martin et al., 1999). However, the DNA distortion seen in the EcoRV/DNA structure is different from that in the HincII/DNA structure. Comparison of the two structures shows that the orientation of the HincII subunits brings the R loops deeper into the major groove of the HincII bound DNA than the EcoRV bound DNA (labeled 2, Fig. 6), resulting in the shifting of the DNA bases into the minor groove (labeled 3, Fig. 6), which has been shown to be responsible for the formation of the CSPS (Horton et al., 2002)(labeled 4, Fig. 6). In contrast, the orientation of the subunits in EcoRV brings the lower part of the DNA binding site into closer proximity to the DNA, interacting deeper in the minor groove on the opposite side of the DNA (Fig. 6, left). The position of the enzyme dimerization domain correlates with the orientation of the subunits. The dimerization domains occur at the “top” of HincII, where the subunits are rotated to be closer in space, but at the “bottom” in EcoRV, where the subunit rotation brings the pair of the subunits closer together. Therefore the shifting of the base planes to generate the CSPS by HincII may originate from the dimerization domain, leading to a different subunit orientation, and a deeper major groove penetration by the R loops. Analysis of the DNA distortion by EcoRV suggests that the positioning of the B helices, the α-helices at the base of the DNA binding site, are related to the DNA bending and base unstacking proposed to be responsible for indirect readout of the TA step in the EcoRV recognition sequence GATATC (Horton and Perona, 2000). In HincII, this helix is shorter and does not interact directly with the DNA. Hence the type of distortion used to indirectly recognize the center step DNA sequence (YR by HincII, TA by EcoRV) appears to be accomplished by the two proteins differently, despite their common core fold, aided by their differing dimerization domains.
Figure 6. Comparison of domain orientation in EcoRV and HincII.
EcoRV is shown on the left, HincII on the right. Segments which are aligned by the DALI server are colored in green (subunit A) or cyan (subunit B). The nucleotides of the fourth nucleotide of the recognition sequence (Gua 4 in HincII, Ade 4 in EcoRV) are shown as colored sticks (C-pink, N-blue, O-red, P-orange). The axes of rotation required to align the individual subunits of EcoRV onto those of HincII (following superposition of both subunits) are shown as red rods. The degree and direction of rotation is also shown. Rotation of the subunits of EcoRV about these axes, to align with those of HincII, brings the ‘tops’ of the subunits closer (1), the R loops deeper into the major groove of the bound DNA (2), shifting the base planes of the bound DNA into the minor groove to a greater extent (3). The shifting into the minor groove has been related to the CSPS seen in the DNA bound to HincII (4).
Recognition of DNA sequences by proteins involves more than simply direct contacts between the protein and the DNA bases. Proteins can make use of DNA conformational energetics to recognize their target sites in ways that are concurrent with their biological function, often requiring speed in addition to specificity. This conundrum of fast scanning for the correct site and the requirement for interrogation of many possible sites implies that there could be an intermediate in the recognition pathway, what we termed a “pre-screening” complex. Recent studies with single molecules of a human DNA repair enzyme suggest such “negative” selection of sites for interrogation, however through an unknown process (Blainey et al., 2006). We have determined a structure with the properties expected of a “pre-screening” complex that uses both limited direct, as well as indirect readout, to specifically recognize the target site. The potential existence of this intermediate suggests that in addition to direct readout, other mechanisms should be considered when investigating rapid target site location and recognition, and that a better understanding of the less well defined mechanisms of scanning and indirect readout is essential.
Experimental Procedures
Site directed mutagenesis
The N141A mutation was introduced into the HincII gene using the mutagenic primers: JBHincIIN141Afor (ATTTCAGCATATAAGCTTGCTCAGACGTG) and JBHincIIN141Arev (AATAGCGGGTGCTTGAGCTGATTTAC) while in a TA-TOPO PCR2.1 vector (Invitrogen). The primers are designed to also introduce an unique HindIII cut site through silent changes in the coding sequence, and to leave blunt ends after amplification of the entire vector, that when ligated introduce no additional base pairs. The amplification was performed with Pfu Turbo polymerase (Stratagene, Inc.), and the PCR product gel purified, phosphorylated by T4 polynucleotide kinase (New England Biolabs), heated to 65°C for 15 minutes to inactivate the kinase, and then ligated with T4 ligase at room temperature overnight. The ligase reaction mixture was then used to transform TOP10 cells (Invitrogen) and plated onto Luria-Bertani agarose plates supplemented with 50 ug/ml ampicillin. Bacteria colonies that grew overnight were inoculated into 2 mls of Luria-Bertani medium supplemented with 50 ug/ml ampicillin and incubated overnight. DNA was purified using the alkaline lysis method and the presence of the mutation was confirmed by both restriction digest with HindIII and DNA sequencing (Genomic Analysis and Technology Core, University of Arizona).
Expression
The N141A HincII gene was placed into a pETBlue2 vector (Invitrogen, Inc.) which includes the coding sequence for a carboxy terminal hexa-histidine tag. Expression was performed in E. coli TOP10 cells (Invitrogen, Inc.) using infection with CE6 lambda phages (Novagen, Inc.) to introduce T7 RNA polymerase. The following modifications were made to the Novagen protocol: phages were adsorbed at room temp and the host/phage mixture was grown for 8 hours, lysis was never observed, dimethyl sulfoxide was not added to the phage stock, but chloroform was removed from the stocks. A culture of 4 L was grown in Luria-Bertani medium with 0.2% maltose and 50 ug/ml ampicillin to an optical density (at 600 nm) of 0.6, upon which 10 mM MgSO4, and 6.1×1011 CE6 phages were added. Cells were grown for 3 hours, chilled to 15°C, pelleted, and flash frozen.
N141A HincII purification
N141A HincII was successfully purified using Ni2+-chelating sepharose and used in the initial plasmid activity assay, as well as the crystallization experiments. The protein was further purified before the single turnover DNA cleavage assays using SP Sepharose, Q Sepharose, and Heparin resins, resulting in the purified protein shown in Figure S1A. Enzyme purity using this Coomassie stained SDS polyacrylamide gel is estimated at 97%, as the two faster migrating species can be estimated at 0.4 μg each in the 25 μg sample (Fig. S1A). Purified enzyme was dialyzed exhaustively into 50 mM bis-trispropane, pH 7.5 at room temperature, 140 mM NaCl, 1 mM DTT, 1 mM EDTA, and 50% in glycerol, aliquoted, flash frozen in liquid nitrogen, and stored at -80°C until just prior to use where it was dialyzed into 50 mM bis-trispropane, pH 7.5 at room temperature, 140 mM NaCl, 1 mM DTT, 1 mM EDTA, concentrated using Centricon-10 Centrifugal Filter Units (Millipore). The concentration (in terms of monomers) was measured spectrophotometrically using the extinction coefficient ε=36,840 cm-1M-1 at 280 nm and adjusted to the molarity of enzyme dimers.
Preparation of DNA substrates
Plasmid DNA was prepared from E. coli culture using Qiagen midiprep kits (Qiagen, Inc.). Oligonucleotides were obtained from commercial synthetic sources and purified using C18 reverse phase HPLC (Aggarwal, 1990). The concentration of each single strand was then measured spectrophotometrically, with extinction coefficients calculated from standard values for the nucleotides (Fasman, 1975). Equimolar amounts of each strand (typically 100-400 mM each) were mixed and annealed by heating to 90°C for 10 minutes, followed by slow-cooling to 4°C over 4-5 hours in an Eppendorf thermocycler. Radiolabeled oligonucleotides were prepared using γ-32P-ATP (MP Biomedicals, Irvine,CA) and T4 polynucleotide kinase (Fermentas, Hercules, CA). Excess 32P-ATP was removed using Micro Bio-spin 6 Columns P-polyacrylamide P-6 gel (Biorad, Hercules, CA).
DNA substrates used in the binding and single turnover assays are shown below, with the recognition sequence in bold, center step underlined, and cleavage site marked by | :
| Cognate CG | 5′-GGG CCG GTC|GAC CAA C-3′ 3′-CCC GGC CAG|CTG GTT G-5′ |
| Cognate TA | 5′-GGG CCG GTT|AAC CAA C-3′ 3′-CCC GGC CAA|TTG GTT G-5′ |
| Cognate TG/CA | 5′-GGG CCG GTT|GAC CAA C-3′ 3′-CCC GGC CAA|CTG GTT G-5′ |
A DNA substrate containing a non-YR center step was also used in the binding and single turnover assays:
| Noncognate GC | 5′-GGG CCG GTG CAC CAA C-3′ 3′-CCC GGC CAC GTG GTT G-5′ |
Plasmid digestion assay
The plasmid digestion assay consisted of 7 μl of pFastBac-C plasmid DNA (Invitrogen, Inc.) in water mixed with 1 μl of 0.3 mg/ml (or 5 μM) N141A HincII or 1.6 μM wild type HincII (New England Biolabs, Inc.) in buffer containing 10 mM HEPES pH 7.5, 150 mM NaCl, 1 mM DTT, and 50% glycerol, 1 μl of 20 mM MnCl2 or 200 mM MgCl2, and incubated at 37°C overnight. The DNA was electrophoresed on 1 % agarose in 1xTAE (40 mM Tris base, 20 mM acetate, 1 mM EDTA) and visualized with 0.4 mg/ml ethidium bromide.
Single turnover reactions
5′ 32P end-labeled 1.0×10-7 M DNA was incubated with 5.0×10-6 M N141A HincII in reaction buffer (50 mM HEPES (pH 7.5 at 25°C), 1 mM DTT, 25 mM NaCl, 2 mM MnCl2). The reaction was started by mixing preheated (at 37°C) enzyme and DNA, followed by incubation of the reaction at 37°C. At various indicated times after the reaction was started, 10 μl of the reaction were removed and the reaction quenched by the addition of 10 μl of 4 M urea, 50 mM EDTA, 80% formamide. The quenched reaction aliquots were then separated on a 20% acrylamide denaturing polyacrylamide gel (containing 8 M urea) in 1X TBE (89 mM Tris, 89 mM boric acid, 10 mM EDTA, pH 8.3). After autoradiography of the gels, both the cleaved and the uncleaved substrate bands were integrated using ImageQuant (Molecular Dynamics, Sunnyvale, CA), then plotted against the reaction time. Rate constants were determined from fits to a single exponential equation (uncleaved bands: Intensity=C1 +C2*e-kt, cleaved bands: Intensity=C1 +C2*(1-e-kt) where t is time and the constants C1 and C2 and rate constant k are determined by the fit) using the program Kaleidagraph (Synergy Software, Inc.). Each reported rate constant is the average of at least three independent measurements.
Crystallization, Diffraction Data Collection, and Structure Refinement
Crystals were prepared with N141A HincII and DNA containing the cognate sequence 5′-G CCC GTC GAC GGG C-3′, in the presence of 2 mM MnCl2 by mixing a molar ratio of 2:1 DNA:enzyme, with 6 mg/ml of N141A HincII and 1.9 mg/ml DNA. The crystallization conditions included 1.0 μl of the protein:DNA mixture in 10 mM HEPES pH 7.5, 0.15 M NaCl, 1 mM DTT, and 4 mM MnCl2 with 1.0 μl of the precipitating solution: 22 % PEG 4K, 0.1 M imidazole buffer (pH 6.5), and 0.2 M NaCl per drop and placed over 1 ml of the precipitating solution. Crystals grow overnight to 1 week at 17°C. The crystals were then exchanged into a cryoprotection buffer (25% [w/v] polyethylene glycol 4000, 0.1M imidazole buffer (pH 6.5), 0.3 M NaCl, and 30% [v/v] glycerol) and flash-frozen in liquid nitrogen. X-ray diffraction intensities were measured using synchrotron radiation at the Advanced Light Source Beamline 5.0.3 (Berkeley, CA). Data collection was performed while maintaining the crystal at 100K. Image processing and data reduction were performed with HKL2000 (HKL Research, Inc.)(Otwinowski, 1997).
The crystals of N141A bound to cognate DNA are isomorphous with crystals of Q138F bound to cognate DNA (Joshi et al., 2006) and initial electron density maps utilized the structure of Q138F HincII bound to cognate DNA. Model adjustments were made using XtalView (McRee, 1999) and refined with the program CNS (Brünger et al., 1998), PHENIX (Adams et al., 2002; Murshudov et al., 1997) and REFMAC (Murshudov et al., 1997). Simulated annealing omit electron density maps were prepared by deletion of indicated atoms from the model prior to simulated annealing using CNS and cartesian molecular dynamics. A temperature of 2000K was used with a cooling rate of 10K per cycle. Structure figures made with PYMOL (DeLano, 2002).
Acknowledgements
We thank J. Martinez for graphic support. This work was supported by the grant (to N.C.H.) NIH 5R01GM066805. Portions of this work were carried out at the Advanced Light Source. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Aggarwal AK. Crystallization of DNA binding proteins with oligodeoxynucleotides. Methods: A Companion to Methods in Enzymology. 1990;1:83–90. doi: 10.1016/S1046-2023(05)80150-1. [DOI] [PubMed] [Google Scholar]
- Allingham JS, Haniford DB. Mechanisms of metal ion action in Tn10 transposition. J Mol Biol. 2002;319:53–65. doi: 10.1016/S0022-2836(02)00297-8. [DOI] [PubMed] [Google Scholar]
- Babic AC, Little EJ, Manohar VM, Bitinaite J, Horton NC. DNA Distortion and Specificity in a Sequence Specific Endonuclease. J Mol Biol. 2008 doi: 10.1016/j.jmb.2008.08.032. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin GS, Sessions RB, Erskine SG, Halford SE. DNA cleavage by the EcoRV restriction endonuclease: roles of divalent metal ions in specificity and catalysis. J Mol Biol. 1999;288:87–103. doi: 10.1006/jmbi.1999.2672. [DOI] [PubMed] [Google Scholar]
- Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc Natl Acad Sci U S A. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallographica Section D: Biological Crystallography. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cirino NM, Cameron CE, Smith JS, Rausch JW, Roth MJ, Benkovic SJ, Le Grice SF. Divalent cation modulation of the ribonuclease functions of human immunodeficiency virus reverse transcriptase. Biochemistry. 1995;34:9936–9943. doi: 10.1021/bi00031a016. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL User’s Manual. DeLano Scientific; Palo Alto, CA: 2002. [Google Scholar]
- Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erie DA, Yang G, Schultz HC, Bustamante C. DNA bending by Cro protein in specific and nonspecific complexes: implications for protein site recognition and specificity. Science. 1994;266:1562–1566. doi: 10.1126/science.7985026. [DOI] [PubMed] [Google Scholar]
- Fasman GD. CRC Handbook of Biochemistry and Molecular Biology. 3rd edn CRC; Cleveland, OH: 1975. [Google Scholar]
- Garvie CW, Wolberger C. Recognition of specific DNA sequences. Mol Cell. 2001;8:937–946. doi: 10.1016/s1097-2765(01)00392-6. [DOI] [PubMed] [Google Scholar]
- Heinemann U, Lauble H, Frank R, Blocker H. Crystal structure analysis of an A-DNA fragment at 1.8 A resolution: d(GCCCGGGC) Nucleic Acids Res. 1987;15:9531–9550. doi: 10.1093/nar/15.22.9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton NC, Dorner LF, Perona JJ. Sequence selectivity and degeneracy of a restriction endonuclease mediated by DNA intercalation. Nat Struct Biol. 2002;9:42–47. doi: 10.1038/nsb741. [DOI] [PubMed] [Google Scholar]
- Horton NC, Perona JJ. Crystallographic snapshots along a protein-induced DNA-bending pathway. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5729–5734. doi: 10.1073/pnas.090370797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M, Berg P. Altering the specificity of restriction endonuclease: effect of replacing Mg2+ with Mn2+ Biochemistry. 1978;17:131–138. doi: 10.1021/bi00594a019. [DOI] [PubMed] [Google Scholar]
- Jeltsch A, Alves J, Oelgeschlager T, Wolfes H, Maass G, Pingoud A. Mutational analysis of the function of Gln115 in the EcoRI restriction endonuclease, a critical amino acid for recognition of the inner thymidine residue in the sequence - GAATTC- and for coupling specific DNA binding to catalysis. J Mol Biol. 1993;229:221–234. doi: 10.1006/jmbi.1993.1019. [DOI] [PubMed] [Google Scholar]
- Jeltsch A, Wenz C, Stahl F, Pingoud A. Linear diffusion of the restriction endonuclease EcoRV on DNA is essential for the in vivo function of the enzyme. Embo J. 1996;15:5104–5111. [PMC free article] [PubMed] [Google Scholar]
- Joshi HK, Etzkorn C, Chatwell L, Bitinaite J, Horton NC. Alteration of sequence specificity of the type II restriction endonuclease HincII through an indirect readout mechanism. J Biol Chem. 2006;281:23852–23869. doi: 10.1074/jbc.M512339200. [DOI] [PubMed] [Google Scholar]
- Kalodimos CG, Biris N, Bonvin AM, Levandoski MM, Guennuegues M, Boelens R, Kaptein R. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Jr., Smith HO. A restriction enzyme from Hemophilus influenzae. II. Journal of Molecular Biology. 1970;51:393–409. doi: 10.1016/0022-2836(70)90150-6. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Loeb LA. On the fidelity of DNA replication. Effect of divalent metal ion activators and deoxyrionucleoside triphosphate pools on in vitro mutagenesis. J Biol Chem. 1979;254:5718–5725. [PubMed] [Google Scholar]
- Martin AM, Sam MD, Reich NO, Perona JJ. Structural and energetic origins of indirect readout in site-specific DNA cleavage by a restriction endonuclease. Nature Structural Biology. 1999;6:269–277. doi: 10.1038/6707. [DOI] [PubMed] [Google Scholar]
- McRee DE. XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density. Journal of Structural Biology. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Niv MY, Ripoll DR, Vila JA, Liwo A, Vanamee ES, Aggarwal AK, Weinstein H, Scheraga HA. Topology of Type II REases revisited; structural classes and the common conserved core. Nucleic Acids Res. 2007;35:2227–2237. doi: 10.1093/nar/gkm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Schevitz RW, Zhang RG, Lawson CL, Joachimiak A, Marmorstein RQ, Luisi BF, Sigler PB. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988 Oct 27;335(6193):837. doi: 10.1038/335321a0. published erratum appears in. 1988. [DOI] [PubMed] [Google Scholar]; Nature. 335:321–329. [Google Scholar]
- Otwinowski Z.a.M. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW Jr., Sweet RM, editors. Methods in Enzymology. Academic Press; New York: 1997. pp. 307–326. W. [DOI] [PubMed] [Google Scholar]
- Parry D, Moon SA, Liu HH, Heslop P, Connolly BA. DNA recognition by the EcoRV restriction endonuclease probed using base analogues. J Mol Biol. 2003;331:1005–1016. doi: 10.1016/s0022-2836(03)00861-1. [DOI] [PubMed] [Google Scholar]
- Pingoud A, Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter PH, Eigen M. Diffusion controlled reaction rates in spheroidal geometry. Application to repressor--operator association and membrane bound enzymes. Biophysical chemistry. 1974;2:255–263. doi: 10.1016/0301-4622(74)80050-5. [DOI] [PubMed] [Google Scholar]
- Riggs AD, Bourgeois S, Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J Mol Biol. 1970;53:401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- Sam MD, Perona JJ. Mn2+-dependent catalysis by restriction enzymes: pre-steady-state analysis of EcoRV endonuclease reveals burst kinetics and the origins of reduced ativity. J Am Chem Soc. 1999;121:1444–1447. [Google Scholar]
- Selent U, Ruter T, Kohler E, Liedtke M, Thielking V, Alves J, Oelgeschlager T, Wolfes H, Peters F, Pingoud A. A site-directed mutagenesis study to identify amino acid residues involved in the catalytic function of the restriction endonuclease EcoRV. Biochemistry. 1992;31:4808–4815. doi: 10.1021/bi00135a010. [DOI] [PubMed] [Google Scholar]
- Tabor S, Richardson CC. Effect of manganese ions on the incorporation of dideoxynucleotides by bacteriophage T7 DNA polymerase and Escherichia coli DNA polymerase I. Proc Natl Acad Sci U S A. 1989;86:4076–4080. doi: 10.1073/pnas.86.11.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JD, Halford SE. Discrimination between DNA sequences by the EcoRV restriction endonuclease. Biochemistry. 1989;28:6198–6207. doi: 10.1021/bi00441a011. [DOI] [PubMed] [Google Scholar]
- Townson SA, Samuelson JC, Bao Y, Xu SY, Aggarwal AK. BstYI bound to noncognate DNA reveals a “hemispecific” complex: implications for DNA scanning. Structure. 2007;15:449–459. doi: 10.1016/j.str.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Townson SA, Samuelson JC, Xu SY, Aggarwal AK. Implications for switching restriction enzyme specificities from the structure of BstYI bound to a BglII DNA sequence. Structure. 2005;13:791–801. doi: 10.1016/j.str.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Vaisman A, Ling H, Woodgate R, Yang W. Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. Embo J. 2005;24:2957–2967. doi: 10.1038/sj.emboj.7600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermote CL, Vipond IB, Halford SE. EcoRV restriction endonuclease: communication between DNA recognition and catalysis. Biochemistry. 1992;31:6089–6097. doi: 10.1021/bi00141a019. [DOI] [PubMed] [Google Scholar]
- Viadiu H, Aggarwal AK. Structure of BamHI bound to nonspecific DNA: A model for DNA sliding. Molecular cell. 2000;5:889–895. doi: 10.1016/s1097-2765(00)80329-9. [DOI] [PubMed] [Google Scholar]
- Vipond IB, Moon BJ, Halford SE. An isoleucine to leucine mutation that switches the cofactor requirement of the EcoRV restriction endonuclease from magnesium to manganese. Biochemistry. 1996;35:1712–1721. doi: 10.1021/bi9523926. [DOI] [PubMed] [Google Scholar]
- von Hippel PH. From “simple” DNA-protein interactions to the macromolecular machines of gene expression. Annual review of biophysics and biomolecular structure. 2007;36:79–105. doi: 10.1146/annurev.biophys.34.040204.144521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel PH, Berg OG. On the specificity of DNA-protein interactions. Proc Natl Acad Sci U S A. 1986;83:1608–1612. doi: 10.1073/pnas.83.6.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel PH, Berg OG. Facilitated target location in biological systems. J Biol Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- Winkler FK, Banner DW, Oefner C, Tsernoglou D, Brown RS, Heathman SP, Bryan RK, Martin PD, Petratos K, Wilson KS. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. Embo Journal. 1993;12:1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe SA, Grant RA, Elrod-Erickson M, Pabo CO. Beyond the “recognition code”: structures of two Cys2His2 zinc finger/TATA box complexes. Structure (Camb) 2001;9:717–723. doi: 10.1016/s0969-2126(01)00632-3. [DOI] [PubMed] [Google Scholar]
- Xu SY, Schildkraut I. Cofactor requirements of BamHI mutant endonuclease E77K and its suppressor mutants. Journal of bacteriology. 1991;173:5030–5035. doi: 10.1128/jb.173.16.5030-5035.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]