Abstract
Posttranslational modifications (PTMs) of proteins play essential roles in regulating signaling, protein-protein modifications and subcellular localization. In this review, we focus on posttranslational modification of histones and RNA polymerase II (RNAPII) and their roles in gene transcription. A survey of the basic features of PTMs is provided followed by a more detailed account of how PTMs on histones and RNAPII regulate transcription in the model organism Saccharomyces cerevisiae. We emphasize the interconnections between histone and RNAPII PTMs and speculate upon the larger role PTMs have in regulating protein function in the cell.
Keywords: posttranslational modifications, histones, transcription, RNA polymerase II, chromatin
1. Introduction
1.1 Protein modifications in biology
With the completed sequencing of the human genome nearly a decade ago, it is now well established that a relatively small number of genes (<30,000) give rise to a substantially larger number of protein products (∼3×106–3×107) [1, 2]. This great protein diversity is manifested at the level of mRNA by alternative splicing or utilization of different transcription and translation start and stop sites. Once the mRNA is translated into a protein product, further protein diversification can be achieved by covalent modification of amino acid side chains, hence the name “posttranslational modification” (PTM).
Protein modifications are very diverse in nature (totaling greater than 200) and can be found on 15 of the 20 proteinogenic amino acids [3, 4]. Some modifications are familiar (e.g. phosphorylation) while others are found on only a few substrates (e.g. diphthamide) [5]. (For a detailed examination of posttranslational modifications and associated mechanisms see [4] and references therein). Regardless, many PTMs, and their associated biochemical pathways, are evolutionarily conserved between prokaryotes and eukaryotes, suggesting they play indispensable roles in biology.
What are the functions of PTMs? Many modifications have evolved as a way of incorporating more diverse functional groups into proteins such as phosphate or sulfate [4]. Others serve varied biological roles, from controlling protein conformational stability [6] to controlling localization of proteins within the cell [7]. Perhaps the most widely studied role of modifications is their ability to mediate protein-protein interactions. A number of protein domains have evolved to bind PTMs in specific protein contexts, such as SH3 and WW domains that bind to proline-rich domains [8] or SH2 domains that recognize phosphotyrosine [9]. PTMs also appear to play key roles in epigenetic processes as some of these modifications, such as methylation on histones, can be maintained through cell divisions [10].
1.2. Scope of this review
Studies of PTMs are rooted in some of the earliest biochemical research [3, 11-13]. While the types of modifications and associated enzyme mechanisms are well established, in many cases little is known about how these modifications function. This review will highlight some of the modifications for which biological roles have been described and will try to put into a greater context how diverse protein modifications modulate functions within the cell.
We will focus on two biologically connected processes, notably posttranslational modification of histones and transcription by RNA polymerase II (RNAPII). We chose these processes not only for their wonderfully diverse modifications but also because recent research has clearly shown that the two processes are inextricably connected [14-17]. While there is an abundance of literature on these topics, it is also evident that we have only scratched the surface of understanding the full role of modifications in both these processes. Therefore, where appropriate, we will attempt to speculate on what the future might hold for the role of PTMs in these processes.
Since there have been several comprehensive reviews on histone modifications in higher organisms in the recent literature [16, 18, 19], we will focus this review on histone PTMs in the model organism Saccharomyces cerevisiae, as many of the modifications have been extensively studied in this organism. Furthermore, yeast is an attractive model for studies of PTMs as the number of proteins is comparatively small and several whole-proteome studies of PTMs and protein-protein interactions in yeast have already been completed [20, 21]. Much of what we have learned in yeast has proven to be similar in higher organisms; therefore, many of the processes we describe have parallels in mammalian cells as well.
We will review the historical links between histone modification and transcription and briefly describe the common modifications found on RNAPII, focusing primarily on the modification-rich C-terminal domain (CTD), and the core histones (comprehensive reviews of histone modifications can be found elsewhere [18, 22]). We will then put these modifications into greater biological context highlighting several recent findings that demonstrate how modifications of histones dictate changes in chromatin structure leading to changes in gene expression. Additionally we will show how modification of RNAPII dictates changes to histones modifications during transcription.
The goals of this review are twofold. One is to cover many of the major recent findings linking histone modifications with transcription. The second is to convey the enormous role of non-histone PTMs in modulating transcription and chromatin remodeling as well as their likely role in many other biological processes.
1.3. History of chromatin modifications and transcription
Histones have been known to associate with nucleic acids since their discovery in the 1880s by Albrecht Kossel [23]. Their biochemical properties were heavily studied over the next half-century and in 1951, Stedman and Stedman first hypothesized that histones could inhibit nucleic acid-based processes including replication and RNA synthesis [24]. In much of the early histone literature it was noted that they purified in multiple, closely related forms. These forms were found to differ primarily by modifications on the histone side-chains including phosphorylation, acetylation, and methylation. A biological role for histone modifications was first proposed by Allfrey and Mirsky in 1964 when they observed that histone acetylation correlated with increased rates of RNA synthesis [25, 26].
Over the next thirty years, the functions of histone modifications would take a backseat to the volumes of historic work detailing how genetic information is stored and copied. During this time, the enzymes responsible for replication and transcription were extensively studied. Further, the functional association between DNA and histones, the nucleosome, was reported [27, 28]. Understanding this genomic architecture would turn out to be essential to our understanding of the role histone modifications play in regulating not only transcription but other DNA-based processes.
In 1996, the connection between histone modifications and transcription again came to the forefront. Allis and coworkers showed that a well-studied transcriptional activator, Gcn5, was in fact an acetyltransferase enzyme that modified histones [29]. This provided biological context to the observations of Allfrey and Mirsky of the 1960s. In similar work Schreiber and coworkers linked transcriptional repression to deacetylase activity [30]. Thus the idea that histone modifications could modulate gene expression was born. This idea was made manifest a few years later with the “histone code” hypothesis [31-33].
The basic premise of the histone code hypothesis is that modifications on histone tails are recognized by additional protein factors. In turn, these factors alter chromatin structure and consequently, transcriptional output giving rise to complex patterns of gene expression. Some modifications are stable between generations and presumably act as epigenetic marks as well.
The past ten years of research has resulted in the discovery of numerous modifications on histones and the enzymes responsible for their deposition, many of which we will discuss here. Further, many proteins and protein domains that recognize posttranslational modifications have been described. Whether a true “histone code” exists is often a matter of debate, but the data are clear – histones are modified to have complicated patterns of posttranslational marks along their tails and these tails work both independently and in conjunction with one another to recruit other factors to DNA. In the case of transcription, the situation is further complicated as modifications on RNAPII can dictate which chromatin-modifying enzymes are able to gain access to histones in a transcription-dependent manner.
2. Histone modifications associated with transcription
(Note: A new nomenclature for many of the histone-modifying enzymes has been recently adopted. We will refer to proteins according to the new nomenclature [34] and have the more established names in parenthesis to facilitate searching of the published literature).
2.1. The architecture of the nucleosome
The nucleosome is the basic unit of chromatin and is comprised of a protein core made up of histone proteins (2 each of H2A, H2B, H3, and H4) around which approximately 146 base pairs of double stranded DNA is wound. The histone proteins have a well-folded core domain and unstructured tail domains on both the N- and C- termini [35]. While modifications occur within the core domains, the tail regions are rich with protein modifications as depicted in Figure 1A. Modifications on the histone tails are thought to play important roles in controlling chromatin architecture by modulating the stability of nucleosomal particles as well as regulating what proteins are recruited to chromatin.
Figure 1.
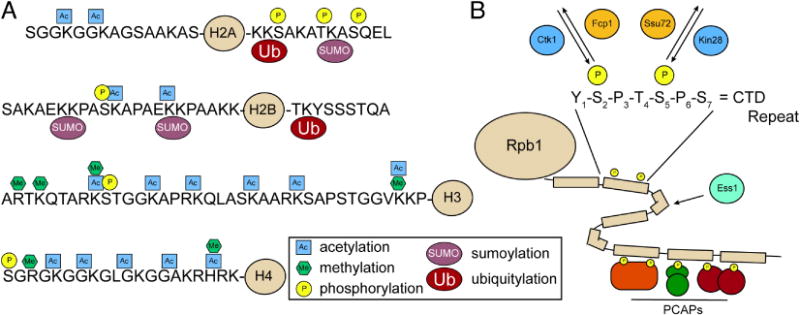
Posttranslational modifications (PTMs) found on histones and RNA polymerase II (RNAPII). (A) Most common PTMs found on the N- and C-terminal tails of the four canonical histones in yeast. (B) Sites of phosphorylation within the 26-repeat C-terminal domain (CTD) of Rpb1, the largest subunit of RNAPII. Phosphorylation of Ser5 is primarily catalyzed by Kin28 and removed by Ssu72. Phosphorylation of Ser2 is primarily catalyzed by Ctk1 and removed by Fcp1. Ess1 is thought to facilitate prolyl isomerization within the CTD. Differential phosphorylation of the CTD leads to recruitment of distinct phosphor-CTD associating proteins (PCAPs) in a spatially and temporally controlled manner to regulate specific transcription-associated processes (e.g. chromatin remodeling and mRNA processing).
2.2. Acetylation and Deacetylation of Histones
Histone acetylation, the most prevalent modification on histones, is catalyzed by a number of enzymes that transfer the acetyl moiety from Acetyl-CoA to the ε-amino group of lysine residues. In yeast, there are 9 reported histone acetyltransferases (HATs) that act on the four core histones. These HAT complexes have different substrate specificities, some recognizing only a few sites while others acting on many lysine residues on multiple tails. HATs are generally found as members of larger protein complexes, thus substrate specificity is most likely regulated through associations with other complex subunits [36]. The intricacies of the individual marks are beyond the scope of this discussion and are described in detail elsewhere [37, 38].
Histone acetylation is generally associated with transcription activation. Acetylation of lysine alters the chemical properties of the amino acid converting the basic residue lysine into the neutral charged ε-N-acetyllysine. In the context of chromatin, studies have shown that acetylation can not only reduce the interaction of histones with DNA but also associations between nucleosomes [39, 40]. Consequently, acetylated nucleosomes are destabilized, promoting both nucleosomal rearrangement by ATP-dependent chromatin remodeling complexes and binding of a diverse set of DNA-binding factors involved in transcription, DNA repair, and numerous other processes [41, 42]. Often these remodeling factors are actually recruited to chromatin through direct binding to the acetyl mark by bromodomains [43-46].
While histone acetylation generally promotes transcription, there is a competing process of histone deacetylation that is generally thought to act in a repressive manner [47, 48]. In yeast, there are nine reported histone deacetylase enzymes (HDACs) belonging to one of two classes [49]. Intuitively, HDAC function is generally associated with nucleosome stabilization and repression of remodeling activities. It is, however, now clear that while HDACs act in opposition to HATs, HDACs also function as activators of gene expression [50-52].
2.3. Histone methylation
Methylation is a considerably more complex modification than acetylation. Several amino acids can be methylated including lysine and arginine on histones [4]. Up to three methyl groups can be added to one lysine residue while arginine can accept up to two methyl groups, with dimethylarginine being either symmetric or asymmetric [53]. Unlike acetylation, the charge of the amino acid sidechain of lysine or arginine is unchanged by methylation. Similar to acetylation, methylated residues serve as binding partners for a number of protein domains including chromodomains [54] and PHD domains [55-57] which bind methyllysine, and Tudor domains which can recognize both methyllysine and methylarginine [58-61].
Lysine methylation of histones in budding yeast has only been identified at a few locations: histone H3 Lys4 (H3K4) [62], Lys36 (H3K36) [63] and Lys79 (H3K79) [64-66]. Fission yeast and higher eukaryotes have several additional sites of methylation, including histone H3 Lys9 (H3K9) [67, 68] and histone H4 Lys20 (H4K20) [69-71]. Lysine methylation differs from acetylation in that it appears that methylation of different lysine residues is catalyzed by different enzymes rather than one enzyme modifying multiple lysine targets. For example, while Kmt2(Set1), Kmt3(Set2) and Kmt4(Dot1) modify histone H3 lysine residues at K4, K36 and K79 respectively, Gcn5(Kat2) is known to have a broad lysine substrate recognition on histone H3 that includes lysine residues K9, K14, K18, K27 and K36 [72, 73]. One reason some histone acetylases likely have such a broad substrate preference is that, in addition to recruiting bromodomain-containing factors, acetylation changes the overall charge of the histone tails, which in turn can regulate nucleosome-nucleosome and histone tail-DNA interactions that drive chromatin decompaction. As lysine methylation has no inherent effect on histone charge, the predominant role of lysine methylation is most likely in recruiting additional factors.
Histone methylation has many diverse roles in transcription. Methylation at H3K4, H3K36, and H3K79, for example, is found associated with active transcription [74, 75]. In contrast, H3K9me in fission yeast, as well as H3K27me in higher eukaryotes, is associated with gene silencing and heterochromatin formation [67, 76, 77]. Even though several sites of methylation on H3 are all associated with active transcription, they have very different roles in regulating transcriptional events. For example H3K4 methylation is known to promote transcription elongation in higher eukaryotes and regulate RNA processing events [78, 79]. H3K36 methylation, however, is associated with recruiting repressive deacetylase activites during transcription elongation [80-82]. We will describe the roles of H3K36me in transcription in greater detail later in this review. To date, the role of H3K79me in transcription is not well understood although the mark can be found associated with promoters and coding regions of most actively transcribed genes and is connected to the process of DNA repair [59, 66, 75, 83, 84].
Arginine methylation is a modification most studied in RNA-binding proteins but also has been reported on histone H4 at Arg3 (H4R3) and on histone H3 at Arg2 (H3R2) in yeast [85, 86]. H4R3me is established by the predominant arginine methyltransferase in yeast, Hmt1. Silver and colleagues have reported a role for H4R3 methylation in gene silencing [86]. Interestingly, H3R2 methylation has also been associated with gene silencing, but through a mechanism that involves antagonizing the establishment of H3K4 methylation at gene promoters [85].
Methylation was believed to be an irreversible mark for many years. Only recently have demethylase enzymes been described that can act on both methyllysine and methylarginine [87, 88]. In yeast, it appears that like the methyltransferases, these enzymes have high specificity and act on particular target residues and even on specific methylation states. For example, the demethylase Kdm5(Jhd2) removes di- and trimethylation from H3K4 [89, 90], while H3K36me is removed by both Kdm2(Jhd1) and Kdm4(Rph1) [91, 92]. While these enzymes clearly act on histones, their roles in chromatin remodeling and transcription are largely unexplored in yeast although much more is known in higher eukaryotes [88].
2.4. Phosphorylation
Phosphorylation of proteins is probably the most recognized protein modification; however, phosphorylation is a relatively rare event on histones. Like in other processes, phosphorylation of histones appears to correspond to changes in the extracellular environment. Phosphorylation of Ser10 on H3 (H3S10) promotes transcription by influencing acetylation at H3K14 in response to changes in carbon source [93]. Phosphorylation of H2B Ser10 (H2BS10) by Ste20 is a signal for apoptosis in response to oxidative stress [94-96]. A significant body of work has been established that shows phosphorylation of the C-terminus of histone H2A is linked to DNA damage repair [97]. Similarly, phosphorylation of histone H4 at Ser1 (H4S1) also accompanies DNA damage [98]. While it is clear that H2A phosphorylation leads to downstream changes in transcription, definitive links between this phosphorylation and the transcription machinery have not been reported. H4S1 phosphorylation, however, has been clearly shown to recruit the NuA4 HAT complex as well as the Swi/Snf chromatin remodeling complex to genes resulting in histone acetylation and recruitment of RNAPII [99, 100].
2.5. Ubiquitination
Ubiquitin is a 76 amino acid protein that is attached to protein lysines through a complex system of protein ligases [101]. These modifications have a variety of functions, including acting as signaling marks, as mediators of protein-protein interaction, and in regulating protein stability. In yeast, ubiquitin has to date only been found on one histone, histone H2B at Lys123 [102]. While the study of H2B ubiquitylation has been restricted to monoubiquitylation (H2Bub1), very recently, polyubiquitylation of H2B has been reported [103]. In higher eukaryotes, H2A is also ubiquitylated on Lys119 [104].
The role of ubiquitin in transcription is very complex. H2Bub1 is catalyzed by the Rad6/Bre1 ubiquitin-ligase complex and functions to promote transcriptional initiation and elongation [102, 105-107]. For example, H2Bub1 facilitates H3K4 methylation and works with the FACT chromatin remodeling complex to promote transcription elongation (Figure 2) [108, 109]. We will further discuss the interplay between H2Bub1, H3K4 and RNAPII modifications later in this review. Many roles of ubiquitination in transcription have recently been reviewed elsewhere [110, 111].
Figure 2.
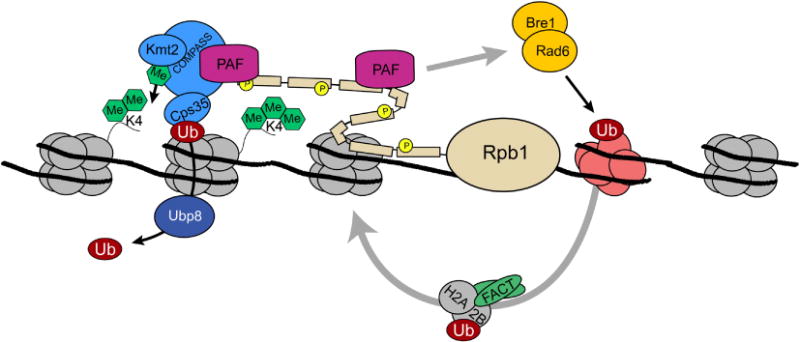
H3K4 methylation and transcriptional activation. The COMPASS methyltransferase is recruited to the promoter regions of genes through interactions with the PAF complex and serine 5 phosphorylation, as well as by making contacts with H2Bub1 through Cps35 (See text). Recruitment of COMPASS leads to the restricted establishment of H3K4me3 near the transcriptional start site This modification may serve to stabilize PIC formation and recruit chromatin remodeling and histone modifying activities that facilitate the transcription process (see text for details). Also associated with COMPASS and the elongating RNAPII complex is the histone chaperone complex, FACT, which disassembles and reassembles nucleosomes to promote transcription. FACT function and COMPASS activity are both functionally regulated, at least in part, through H2Bub1 [110].
2.6. Sumoylation
In addition to ubiquitin, there are several other ubiquitin-like proteins that can be conjugated to proteins in higher eukaryotes [112]. Yeast have two ubiquitin-like proteins, Smt3 and Rub1. Smt3, SUMO (Small Ubiquitin-like Modifer) in higher eukaryotes, has been associated with transcription. In fact, it has been reported that all four histones can be modified by SUMO [113]. While the authors were unable to precisely define the locations of modification on H3, sumoylation of H4 was found on at least 5 locations in the N-terminus H2B sumoylation is thought to occur at Lys6/7 and Lys16/17. On H2A, SUMO is attached to Lys126.
The role of sumoylation in transcription is largely unknown. Berger and coworkers have shown that sumoylation is in general a repressive mark, and in fact antagonizes histone acetylation [113]. However, the exact mechanisms through which SUMO represses transcription are still poorly understood.
2.7. Prolyl Isomerization
Proline isomerization is the only noncovalent posttranslational modification found to occur on histones to date. The ring structure of the proline amino acid allows it to interconvert between two conformations in proteins, either cis or trans. A change in proline conformation can propagate a significant change in protein structure, some of which may serve functions within the cell [114]. For example, the N-terminal tail of histone H3 has several prolines within its sequence. Kouzarides and coworkers have shown that mutation of Pro38 affects the ability of the Kmt3(Set2) enzyme to methylate the nearby H3K36 [115]. They also described the corresponding enzyme, the prolyl isomerase Fpr4, which antagonizes H3K36 methylation by Kmt3(Set2).
2.8. PMTs on Histone Variants
In addition to the canonical histones (H2A, H2B, H3, and H4), there also exist variants of these histones that act in place of one of the four main histones and perform alternate functions. In yeast, there are two, a variant of H2A called H2A.z (Htz1) and an H3 variant called Cse4 (also known as CENP-A in metazoans). Cse4 is localized to centromeres in yeast [116, 117] and while it has been tied to chromatin remodeling complexes, no role for Cse4 in transcription has been described [118, 119]. Htz1 is found to replace H2A in the promoter regions of genes. Htz1-containing nucleosomes are thought to be less stable than H2A-containing nucleosomes and thus probably play some role in marking gene promoters. Htz1 is modified by the NuA4 acetylase complex [120, 121] and has also been reported to be a phospho-accepting substrate of several kinases in vitro [20]. While Htz1 acetylation has been correlated with active transcription, it is not known if Htz1 phosphorylation occurs in vivo and/or functions to regulate nucleosome dynamics in transcription.
2.9. Other modifications
It is important to note that most of the histone modifications described above have been identified in the last decade. It is likely that there are other modifications that have yet to be identified. For example, the modifications described occur almost exclusively on the histone tails. Modifications in the core regions are likely to occur and in fact several new sites of acetylation in the core domains of H3 have been identified recently [122-124].
3. Modifications on Pol II associated with transcription
3.1. The C-terminal domain (CTD) of RNA Polymerase II
The largest subunit of the RNA Polymerase II (RNAPII) holoenzyme comprises not only the catalytic core of RNAPII but also a very long, unstructured C-terminal region that is essential for function. This C-terminal domain (CTD) is made up of multiple repeats (26 in yeast and 52 in humans) of a seven-amino acid consensus sequence YSTPSPS [125] (see Figure 1B). The CTD acts as a scaffold making significant protein-protein interactions that link transcription with PhosphoCTD-associated proteins (PCAPs) that are involved in many biological processes including chromatin remodeling, cell cycle regulation, and mRNA processing (see Figure 1B) [126]. These protein-protein interactions are dictated by a complex set of post-translational modifications that occur on the CTD [125, 127]. Here we will try to summarize CTD modifications and their importance in transcription.
3.2. Ser2 and Ser5 Phosphorylation
Early analysis of the largest subunit of RNAPII (Rpb1) showed that the protein took on many forms as judged by SDS-PAGE electrophoresis [128]. It was later determined that these different gel mobilities signified varying levels of protein phosphorylation on the CTD, specifically at Ser2 and Ser5 [129, 130]. In yeast, Ser5 phosphorylation of RNAPII is catalyzed by the cyclin-dependent kinase Kin28, which is part of the transcription complex TFIIH [131-134]. There also exists supporting data that a factor of the Mediator complex, Srb10, also phosphorylates Ser5, but only when RNAPII is not associated with a gene [135]. Thus phosphorylation by Srb10 is though to prohibit pre-initiation complex (PIC) formation and thus is a negative regulator of transcription.
Ser2 phosphorylation is catalyzed in vivo by the cyclin-dependent kinase Ctk1 [136]. Phosphorylation of Ser2 serves as a mark that denotes transcription elongation and requires many factors, including Ser5 phosphorylation [125], removal of H2Bub1 [137], and association of the PAF complex [138].
The phosphorylation state of RNAPII changes as the transcription machinery moves along a gene. RNAPII bound at promoter regions is largely unphosphorylated [139]. Initiation of transcription coincides with Ser5 phosphorylation by Kin28 [140]. During the elongation phase of transcription, Ctk1 phosphorylates Ser2 to give a RNAPII that is modified at both Ser2 and Ser5 [132]. As RNAPII travels along a gene, phosphorylation on the CTD is removed. The phosphatase Ssu72 has been shown to remove Ser5 phosphorylation [141] while the phosphatase Fcp1 is responsible for removing Ser2 phosphorylation [142]. It is thought that dephosphorylation of the CTD is required for recycling of RNAPII on genes and may play a role in gene looping [143-145].
3.3. Prolyl isomerization by Ess1
As described earlier, prolyl isomerases catalyze the interconversion of the two conformations of proline residues. As is evident from the protein sequence of the RNAPII CTD, there are two prolines in every repeat. Hanes and coworkers found that the prolyl isomerase Ess1 is essential for growth in yeast and binds to the RNAPII CTD [146]. While its activity has never been observed in vivo, modification of the catalytic residues of Ess1 results in a temperature-sensitive phenotype suggesting that at least one function of Ess1 may be to act on the CTD.
3.4. Ubiquitination
As described for H2B, ubiquitylation is a common modification of lysine residues. Monoubiquitylation often serves as a protein-protein interaction mark while polyubiquitylation usually signals for degradation. The largest subunit of RNAPII, Rpb1, associates with members of the ubiquitin-ligase machinery as well as the proteasome (responsible for degrading polyubiquitylated substrates) [147, 148]. WW domains within Rsp5 mediate binding to the elongating RNAPII CTD and Rsp5 then ubiquitylates distant sites on Rpb1 in conjunction with an E2 protein Ubc5, targeting Rpb1 for degradation in response to DNA damage [149]. This process is further regulated by other RNAPII subunits including the nonessential subunit Rpb9 [150]. A recent result from Svejstrup and colleagues shows that deubiquitylation of Rpb1 is also a crucial process, as loss of the ubiquitin protease Ubp3 facilitates removal of stalled RNAPII complexes in response to DNA damage [151].
3.5. Other modifications
The richness of modifications on the CTD leaves one asking whether there are still undiscovered modifications. In fact, late last year, two groups reported that Ser7 of the CTD could be phosphorylated in mammalian cells [152, 153]. Also, there are reports of glycosylation of both Ser2 and Ser5 in higher eukaryotes as well [154, 155]. Tyrosine residues can also be modified in several ways including phosphorylation, sulfation and nitrosylation [4]. The existence of these modifications in yeast has not been reported but clearly these modifications are possible in this organism. Thus, the future will reveal whether these additional modifications are conserved and play important roles in regulating transcription-associated processes or whether we have found evolutionary differences in CTD modifications between yeast and metazoans.
We have chosen to restrict our discussion to modifications on the CTD of the largest subunit of RNA polymerase. However, there are eleven other subunits associated with the RNA polymerase holoenzyme in addition to Rpb1. Several of the other subunits have been reported to be substrates of casein kinase II (CKII) indicating that multiple members of the RNAPII holoenzyme are likely to be modified [156]. Future work will likely be aimed at understanding what role PTMs of the RNAPII holoenzyme play in regulating transcription. While the answers are still unknown, it is likely that these modifications are important signals coordinating transcription with other cellular processes.
4. The Interplay between histone modifications and RNA polymerase
Hundreds of proteins are needed to properly carry out the process of transcription. Hundreds more are required to properly integrate transcription with other processes such as cell cycle progression, cellular metabolism, and RNA processing [157]. As we have already described, transcription is also tightly coupled to changes in chromatin architecture. While researchers have determined most of the chromatin and RNAPII modifications associated with transcription, the mechanisms through which these modifications modulate gene expression have only begun to be elucidated. In this section, we will highlight a few stories for which we have considerable information about how protein modifications and histone modifying enzymes interplay with RNAPII to dictate patterns of gene expression. These examples will serve to emphasize one essential role that protein modifications play in cell physiology.
4.1. H3K4 methylation, H2B ubiquitination and CTD phosphorylation
H3K4 methylation in yeast is catalyzed by the enzyme Kmt2(Set1), which is part of a larger protein complex called COMPASS [62, 138, 158]. It is well established that the presence of H3K4 methylation correlates with nucleosomes residing within genes that are being actively transcribed. As mentioned earlier, lysines can have one (me1), two (me2) or three (me3) methyl groups attached to them. H3K4me3 is most prevalent near promoter regions and then H3K4me2 and H3K4me1 become more abundant in the coding regions of genes [75]. Several questions arise from these findings. For example, do these different methylation states have different functions, and what factors are required for establishing different methyl states?
As we described earlier, it appears that many protein domains are able to recognize methyllysine. Though not confirmed, it is also likely that these binding domains can also distinguish between different methyl states (i.e. me1, me2, me3). Several different proteins have been described that bind to H3K4me both in yeast and humans [57, 159-162]. These proteins have diverse functions including controlling the extent of nucleosome remodeling, demethylation, acetylation, and PIC formation. Therefore, it appears that the different methyl states on H3K4 dictate what factors interact with chromatin. In fact, H3K4 seems to be so important for regulating interactions of binding proteins with nucleosomes that a motif has been recently reported that recognizes unmodified H3K4 in mammalian cells [163].
There are numerous signals that control the methylation of H3K4. H3K4me requires careful coordination of protein modifications not only on histones (i.e. H2Bub) but also RNAPII, as well as recruitment of other transcription factors. COMPASS recruitment to the CTD requires phopshorylation at Ser5, catalyzed by the TFIIH member, Kin28. Kmt2(Set1) does not appear to directly interact with the CTD tail, rather COMPASS appears to be recruited to the CTD through interactions with the PAF transcriptional complex, which is known to directly interact with RNAPII [138]. In addition to CTD phopshorylation, Kmt2(Set1) also requires ubiquitylation of H2B (H2Bub1) for proper methylation of substrates [164-166]. This finding was some of the earliest support for the idea of the “histone code” because it showed that protein modifications on one histone tail directly influenced enzymatic modification of a different tail [167]. This concept has become known as a “trans-tail pathway” and since several other similar mechanisms have been described [168].
Recently, Shilatifard and coworkers uncovered the basis for H2Bub1 regulation of H3K4me [169]. They demonstrated that in the absence of H2Bub1, the Cps35 subunit of COMPASS does not associate with the rest of the complex and thus the complex is not catalytically competent. This outlines a biological role for Cps35, the only essential subunit of COMPASS. According to their model, H2B ubiquitylation directs H3K4 methylation by stabilizing the interaction between Cps35 and the rest of COMPASS, thus making the complex catalytically competent. This finding is significant in that it biochemically defines the factors that “translate” the “trans-tail” pathway. However, these results stimulate many additional questions regarding the role of Cps35 in regulating these processes. For example, as mentioned earlier, Cps35 is the only essential member of COMPASS. It also happens to be a member of the cleavage and polyadenylation factor complex (CPF). Is the essential role for Cps35 in coordinating the activity of COMPASS, in mRNA processing, or perhaps in linking these two processes together? There is likely to be an exciting future in unraveling the many functions of Cps35.
This story demonstrates how numerous modifications on several proteins (including histones and RNAPII) regulate a histone mark that has an important role in gene expression. Even with the considerable knowledge we have concerning the function of H3K4me, there are many questions still largely unanswered. For example, while H3K4me correlates with the promoter and early coding regions of many genes, it is not required for viability. What then is the actual role of H3K4me in transcription? Reinberg and coworkers have uncovered a role for H3K4me in RNA processing [79]. Does something analogous to this occur in yeast?
4.2. Kmt3(Set2) and CTD phosphorylation
The methyltransferase Kmt3(Set2) was described a few years ago as an enzyme responsible for methylating Lys36 of histone H3 (H3K36) [63, 170]. Investigators determined that Kmt3(Set2) methylation of H3K36 was occurring cotranscriptionally and that Kmt3(Set2) associated directly with RNAPII. In a number of structural and biochemical studies, a unique domain of Kmt3(Set2) was described that bound specifically to the CTD of Rpb1 [171]. In collaboration with the Greenleaf laboratory, we showed that Kmt3(Set2) associates preferentially with the CTD phosphorylated at both Ser2 and Ser5 [171]. Deletion of the Ctk1 kinase responsible for Ser2 phosphorylation resulted in a loss of H3K36 methylation, suggesting that binding of Kmt3(Set2) to the CTD is essential for its activity in vivo [172, 173]. Subsequent work determined that H3K36me is most prominently associated with nucleosomes positioned in the mid- to late coding region of actively transcribed genes, consistent with Kmt3(Set2) association to the actively elongating form of RNAPII [75, 172-174].
Like H3K4me, H3K36me is recognized by methyllysine-binding proteins. The Buratowski, Struhl and Workman labs showed that the chromodomain of Eaf3 bound specifically to H3K36me [80-82]. Eaf3 is a component of both the NuA4 HAT complex and the small Rpd3 deacetylase complex known as Rpd3S [81, 82]. It was determined that Rpd3S was the Eaf3-containing complex that interacts with H3K36me, which also depends on an additional histone interactions mediated by a PHD finger in the only Rpd3S-specific factor RcoI [175]. It was also clearly shown that methylation of H3K36 was capable of recruiting Rpd3 to chromatin, which in turn, resulted in deacetylation of nucleosomes [81, 82]. More recently, the Strahl and Mellor labs have reported that H3K36me2 is sufficient to recruit deacetylase activity to genes [176]. Thus methylation of H3K36me2 plays a repressive role in transcriptional processes by recruiting enzymes to remove acetylation from nucleosomes. Whether H3K36me3, which is preferentially associated with actively transcribing genes, plays a distinct role in gene transcription through the recruitment of a second H3K36me-binding protein is unknown but remains an intriguing possibility.
The biological relevance of maintaining the coding regions of RNAPII-regulated genes in a deactylated state was demonstrated by showing that when Kmt3(Set2) or several members of the Rpd3S complex were deleted, nucleosomes in actively transcribed regions were hyperacetylated and cryptic start-sites within genes were inappropriately utilized [82]. These results demonstrate how precise modifications on histone tails can create changes in chromatin that function to direct transcriptional initiation events to the appropriate places within the genome.
This work, highlighting Kmt3(Set2) and H3K36 methylation, shows how modifications on the CTD recruit chromatin-modifying activities, which in turn dictate the expression patterns of certain genes. Thus there exists a complicated regulatory mechanism where at least four different modifications (phosphorylation, methylation, ubiquitylation and acetylation/deacetylation) determine how a given gene is expressed. This mechanism will only become more complex as we learn more about the upstream signals that dictate CTD phosphorylation patterns. For example, our lab has recently found that binding to RNAPII regulates the stability of Kmt3(Set2) protein in a proteasome-specific manner (Fuchs, S.M. and Strahl, B.D., unpublished data). Thus there appear to be additional mechanisms to tightly control Kmt3(Set2) activity in response to changes in CTD phosphorylation.
4.3. H3K14, SAGA and RSC - Chromatin remodeling and gene expression
The first two stories highlighted focus on the factors that regulate methylation of different sites on histone H3. Complex posttranslational regulation extends far beyond histone methylation. As an example, we will highlight some work focusing on how other PTMs, specifically acetylation and phosphorylation coordinate regulation of gene expression.
Kat2(Gcn5), the catalytic subunit of the SAGA histone acetyltranfserase complex, is responsible for acetylation of a number of lysine residues on both histones H3 and H4 [37]. One of these marks, acetylation at lys14 of histone H3 (H3K14ac) has received considerable interest due to its role in recruiting the RSC chromatin remodeling complex [177]. In addition to modifying histones, Kat2(Gcn5) is also known to acetylate non-histone proteins. In the case of RSC, in particular, non-histone acetylation turns out to play a crucial role in gene expression as well. In this section we will describe some of the modifications that contribute to chromatin remodeling by RSC.
As previously stated, histone acetylation is an important factor in loosening chromatin structure that acts to promote transcription. H3K14 is only one of a number of residues that is acetylated by SAGA. Interestingly, SAGA can be specifically recruited to H3K14 by signals elsewhere on the H3 tail. Specifically, phosphorylation of Ser10 on histone H3 (H3S10P) has been shown to increase H3K14ac on certain genes leading to changes in gene expression [178]. H3S10 is phosphorylated by the kinase Snf1 in yeast in response to carbon source to upregulate expression of genes involved in metabolism [93].
How does acetylation at H3K14 cause changes in transcription? At least one answer is by recruiting the additional chromatin remodeling activities of the RSC complex [179]. RSC is a very abundant ATP-dependent chromatin remodeling complex in yeast that plays important roles in regulating gene expression during a number of processes [118, 180]. RSC functions to stimulate transcription elongation by remodeling nucleosomes and recruiting HAT activities [181]. RSC contains at least 15 protein subunits several of which contain domains (bromodomains and BAH domains) that interact with acetylated lysine residues and other unmodified residues within histones. In particular, Cairns and coworkers have shown that tandem bromodomains of Rsc4, an essential member of the RSC complex, specifically interact with H3K14ac [177]. RSC is also known to travel with RNAPII, but unlike Kmt3(Set2), it does not appear to associate with the CTD. Instead, the Rsc4 subunit of RSC interacts with the Rbp5 subunit of RNAPII holoenzyme and that this interaction is critical to RSC function [182]. Thus RSC appears to play an important role in remodeling nucleosomes at sites of H3K14ac.
Regulation of RSC function by Kat2(Gcn5) goes beyond its ability to acetylate H3K14. As a consequence of solving the crystal structure of Rsc4, Cairns and coworkers noted the presence of an acetylated lysine (Lys25) within Rsc4 itself [183]. Acetylation of Rsc4 by Kat2(Gcn5) was found to inhibit Rsc4 binding to H3K14ac. Thus it is appears as if Kat2(Gcn5) acts to both stimulate and inhibit remodeling by RSC. This also appears to be similar to the role of H3S10P as phosphorylation recruits Kat2(Gcn5) yet seems to impede Rsc4 binding. It is therefore likely that H3S10P is a transient mark and an unknown phosphatase removes this mark following H3K14 acetylation to allow Rsc4 recruitment (see Figure 4).
Figure 4.
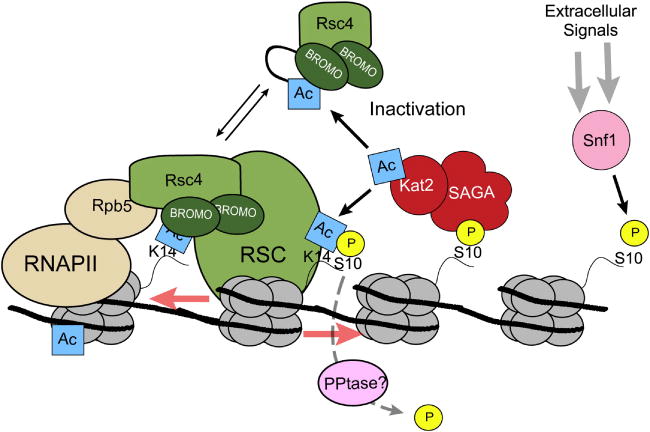
Acetylation both stimulates and inhibits RSC chromatin remodeling activities. Kat2(Gcn5) is recruited to acetylate H3K14 by Snf1-mediated phosphorylation of H3S10. During transcription, RSC chromatin remodeling activities is recruited via Rsc4 which binds to H3K14ac via its tandem bromodomains and with RNAPII through interactions with Rpb5. Rsc4 can also be directly acetylated by Kat2(Gcn5) leading to inactivation of the complex.
The finding that Rsc4 is acetylated and that acetylation regulates its own function raises many further questions about how protein modifications regulate transcription. Are other transcription factors, or regulators of transcription, modified by what are traditionally thought of as “histone-modifying enzymes”? If so, how do these modifications regulate their function? For example, in higher eukaryotes it is now well established that steroid hormone receptors are highly regulated by phosphorylation [184]. The oncogene p53 is known to be phosphorylated as well as methylated, acetylated, sumoylated and ubiquitinated [185]. Therefore, it is clear that the role PTMs extends far beyond modification of just histones and RNAPII.
5. Concluding Remarks
We began by noting that proteomics predictions estimate that there are considerably more proteins in a cell than what are encoded by the DNA. In this review we touched on a number of modifications known to be important for regulating chromatin structure and transcription. However, we limited our discussion to just a handful of modifications on only a few proteins. With the rapid evolution of high-throughput proteomics techniques, our understanding of the extent of protein modification will undoubtedly increase in the next few years. Deciphering the roles of newfound modifications will become more complex as the number of modified proteins increases. Perhaps, understanding the complex networks of protein modifications is the key to understanding how transcription integrates with other cell processes such as mRNA processing, cell-cycle regulation, and signaling events.
Histone tails seem to be the richest sources of protein modifications in cells. This is likely due to their essential roles in maintaining chromatin structure. Are there other modification-rich proteins that have been largely overlooked? Preliminary evidence suggests there are. RNA-binding proteins as well as ribosomal proteins are known to be heavily methylated [186]. What are the roles of methylation in mRNA processing or translation? Structural proteins such as myelin basic protein are decorated with a diverse set of modifications that seem to play essential roles in neurodegenerative disorders [187]. How do modifications of these proteins regulate protein function? Perhaps the most complex modifications are manifest outside the cell. Researchers have spent decades studying how glycosylation patterns on proteins dictate how a cell interacts with its environment [188]. Deciphering glycosylation is key to understanding a number of today's biomedical problems including cancer metastasis and cell differentiation [189].
Protein modifications will likely continue to play a starring role in future stories of chromatin structure and gene regulation. As more modifications are identified, it is likely that these PTMs will be key players in the regulation of gene expression as well as other biological processes in the cell. In that respect, previous and current work on histone modifications will serve as a roadmap for deciphering the function of protein modifications in biology.
Figure 3.
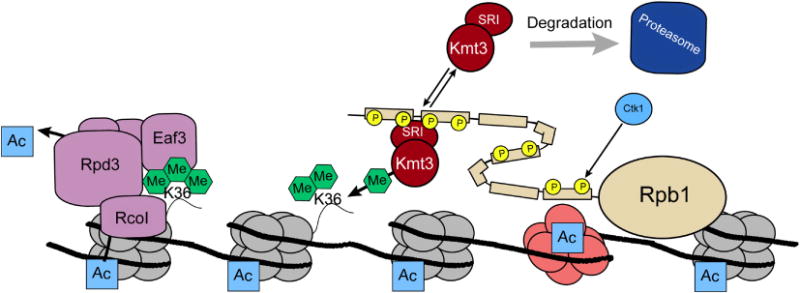
Role of H3K36 methylation in transcriptional dampening. Following CTD phosphorylation by Ctk1, the Kmt3(Set2) methyltransferase is recruited to the elongating form of RNAPII via its SRI domain [171]. Kmt3(Set2) methylates H3K36, leading to Rpd3(S) deacetylase recruitment and deacetylation of ORF specific nucleosomes. This event regulates the removal of elongation-linked acetylation and thereby prevents inappropriate initiation events from occurring in gene bodies. Thus, H3K36me serves to maintain proper transcription initiation at the promoter regions of genes.
Acknowledgments
Work in the Strahl lab is funded by grants from the NIH (to B.D.S. and S.M.F.) and the Pew charitable trusts. B.D.S. is a Pew Scholar in the Biomedical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 2.Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 3.Uy R, Wold F. Posttranslational covalent modification of proteins. Science. 1977;198:890–896. doi: 10.1126/science.337487. [DOI] [PubMed] [Google Scholar]
- 4.Walsh CT. Posttranslational modifications of proteins: expanding nature's inventory. Roberts and Co.; Englewood, CO: 2006. [Google Scholar]
- 5.Dunlop PC, Bodley JW. Biosynthetic labeling of diphthamide in Saccharomyces cerevisiae. J Biol Chem. 1983;258:4754–4758. [PubMed] [Google Scholar]
- 6.Jenkins CL, Raines RT. Insights on the conformational stability of collagen. Nat Prod Rep. 2002;19:49–59. doi: 10.1039/a903001h. [DOI] [PubMed] [Google Scholar]
- 7.McBride AE, Silver PA. State of the arg: protein methylation at arginine comes of age. Cell. 2001;106:5–8. doi: 10.1016/s0092-8674(01)00423-8. [DOI] [PubMed] [Google Scholar]
- 8.Li SS. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem J. 2005;390:641–653. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Machida K, Mayer BJ. The SH2 domain: versatile signaling module and pharmaceutical target. Biochim Biophys Acta. 2005;1747:1–25. doi: 10.1016/j.bbapap.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Bonisch C, Nieratschker SM, Orfanos NK, Hake SB. Chromatin proteomics and epigenetic regulatory circuits. Expert Rev Proteomics. 2008;5:105–119. doi: 10.1586/14789450.5.1.105. [DOI] [PubMed] [Google Scholar]
- 11.Paik WK, Kim S. Protein methylation: chemical, enzymological, and biological significance. Adv Enzymol Relat Areas Mol Biol. 1975;42:227–286. doi: 10.1002/9780470122877.ch5. [DOI] [PubMed] [Google Scholar]
- 12.Saxholm HJ, Pestana A, O'Connor L, Sattler CA, Pitot HC. Protein acetylation. Mol Cell Biochem. 1982;46:129–153. doi: 10.1007/BF00239663. [DOI] [PubMed] [Google Scholar]
- 13.Krebs EG. Historical perspectives on protein phosphorylation and a classification system for protein kinases. Philos Trans R Soc Lond B Biol Sci. 1983;302:3–11. doi: 10.1098/rstb.1983.0033. [DOI] [PubMed] [Google Scholar]
- 14.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 15.Khan AU, Krishnamurthy S. Histone modifications as key regulators of transcription. Front Biosci. 2005;10:866–872. doi: 10.2741/1580. [DOI] [PubMed] [Google Scholar]
- 16.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Sims RJ, 3rd, Mandal SS, Reinberg D. Recent highlights of RNA-polymerase-II-mediated transcription. Curr Opin Cell Biol. 2004;16:263–271. doi: 10.1016/j.ceb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 19.Hartzog GA, Tamkun JW. A new role for histone tail modifications in transcription elongation. Genes Dev. 2007;21:3209–3213. doi: 10.1101/gad.1628707. [DOI] [PubMed] [Google Scholar]
- 20.Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, McCartney RR, Schmidt MC, Rachidi N, Lee SJ, Mah AS, Meng L, Stark MJ, Stern DF, De Virgilio C, Tyers M, Andrews B, Gerstein M, Schweitzer B, Predki PF, Snyder M. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 21.Schwikowski B, Uetz P, Fields S. A network of protein-protein interactions in yeast. Nat Biotechnol. 2000;18:1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- 22.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 23.Kossel A. The chemical composition of the cell nucleus. Elsevier Publishing Company; Amsterdam: 1967. [Google Scholar]
- 24.Stedman E, Stedman E. The Basic Proteins of Cell Nuclei. Phil Trans R Soc Lond B Biol Sci. 1951;235:565–595. [Google Scholar]
- 25.Allfrey VG, Mirsky AE. Structural Modifications of Histones and their Possible Role in the Regulation of RNA Synthesis. Science. 1964;144:559. doi: 10.1126/science.144.3618.559. [DOI] [PubMed] [Google Scholar]
- 26.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and Methylation of Histones and Their Possible Role in the Regulation of Rna Synthesis. Proc Natl Acad Sci USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 28.Kornberg RD, Thomas JO. Chromatin structure; oligomers of the histones. Science. 1974;184:865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- 29.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 30.Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 31.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 32.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 33.Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 34.Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 35.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 36.Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn't fit all. Nat Rev Mol Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 37.Millar CB, Grunstein M. Genome-wide patterns of histone modifications in yeast. Nat Rev Mol Cell Biol. 2006;7:657–666. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- 38.Krebs JE. Moving marks: dynamic histone modifications in yeast. Mol Biosyst. 2007;3:590–597. doi: 10.1039/b703923a. [DOI] [PubMed] [Google Scholar]
- 39.Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 40.Wang X, Hayes JJ. Acetylation mimics within individual core histone tail domains indicate distinct roles in regulating the stability of higher-order chromatin structure. Mol Cell Biol. 2008;28:227–236. doi: 10.1128/MCB.01245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagy Z, Tora L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26:5341–5357. doi: 10.1038/sj.onc.1210604. [DOI] [PubMed] [Google Scholar]
- 42.Carrozza MJ, Utley RT, Workman JL, Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 43.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 44.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 45.Mujtaba S, Zeng L, Zhou MM. Structure and acetyl-lysine recognition of the bromodomain. Oncogene. 2007;26:5521–5527. doi: 10.1038/sj.onc.1210618. [DOI] [PubMed] [Google Scholar]
- 46.Yang XJ. Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays. 2004;26:1076–1087. doi: 10.1002/bies.20104. [DOI] [PubMed] [Google Scholar]
- 47.Thiagalingam S, Cheng KH, Lee HJ, Mineva N, Thiagalingam A, Ponte JF. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann N Y Acad Sci. 2003;983:84–100. doi: 10.1111/j.1749-6632.2003.tb05964.x. [DOI] [PubMed] [Google Scholar]
- 48.Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 49.Kurdistani SK, Grunstein M. Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol. 2003;4:276–284. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- 50.De Nadal E, Zapater M, Alepuz PM, Sumoy L, Mas G, Posas F. The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature. 2004;427:370–374. doi: 10.1038/nature02258. [DOI] [PubMed] [Google Scholar]
- 51.Wang A, Kurdistani SK, Grunstein M. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science. 2002;298:1412–1414. doi: 10.1126/science.1077790. [DOI] [PubMed] [Google Scholar]
- 52.Yoshimoto H, Ohmae M, Yamashita I. The Saccharomyces cerevisiae GAM2/SIN3 protein plays a role in both activation and repression of transcription. Mol Gen Genet. 1992;233:327–330. doi: 10.1007/BF00587597. [DOI] [PubMed] [Google Scholar]
- 53.Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 54.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 55.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, Patel DJ, Aitchison JD, Tackett AJ, Allis CD. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 58.Cote J, Richard S. Tudor domains bind symmetrical dimethylated arginines. J Biol Chem. 2005;280:28476–28483. doi: 10.1074/jbc.M414328200. [DOI] [PubMed] [Google Scholar]
- 59.Grenon M, Costelloe T, Jimeno S, O'Shaughnessy A, Fitzgerald J, Zgheib O, Degerth L, Lowndes NF. Docking onto chromatin via the Saccharomyces cerevisiae Rad9 Tudor domain. Yeast. 2007;24:105–119. doi: 10.1002/yea.1441. [DOI] [PubMed] [Google Scholar]
- 60.Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 61.Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, Allis CD. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 65.Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 67.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 68.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 69.Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 70.Fang J, Feng Q, Ketel CS, Wang H, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Simon JA, Zhang Y. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr Biol. 2002;12:1086–1099. doi: 10.1016/s0960-9822(02)00924-7. [DOI] [PubMed] [Google Scholar]
- 71.Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, Lis JT, Allis CD, Reinberg D. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell. 2002;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- 72.Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- 73.Morris SA, Rao B, Garcia BA, Hake SB, Diaz RL, Shabanowitz J, Hunt DF, Allis CD, Lieb JD, Strahl BD. Identification of histone H3 lysine 36 acetylation as a highly conserved histone modification. J Biol Chem. 2007;282:7632–7640. doi: 10.1074/jbc.M607909200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rando OJ. Global patterns of histone modifications. Curr Opin Genet Dev. 2007;17:94–99. doi: 10.1016/j.gde.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 75.Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 79.Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 81.Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, Boone C, Emili A, Weissman JS, Hughes TR, Strahl BD, Grunstein M, Greenblatt JF, Buratowski S, Krogan NJ. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 82.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 83.Bostelman LJ, Keller AM, Albrecht AM, Arat A, Thompson JS. Methylation of histone H3 lysine-79 by Dot1p plays multiple roles in the response to UV damage in Saccharomyces cerevisiae. DNA Repair (Amst) 2007;6:383–395. doi: 10.1016/j.dnarep.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 84.Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J Biol Chem. 2005;280:9879–9886. doi: 10.1074/jbc.M414453200. [DOI] [PubMed] [Google Scholar]
- 85.Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bahler J, Green RD, Kouzarides T. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu MC, Lamming DW, Eskin JA, Sinclair DA, Silver PA. The role of protein arginine methylation in the formation of silent chromatin. Genes Dev. 2006;20:3249–3254. doi: 10.1101/gad.1495206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- 88.Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- 89.Liang G, Klose RJ, Gardner KE, Zhang Y. Yeast Jhd2p is a histone H3 Lys4 trimethyl demethylase. Nat Struct Mol Biol. 2007;14:243–245. doi: 10.1038/nsmb1204. [DOI] [PubMed] [Google Scholar]
- 90.Seward DJ, Cubberley G, Kim S, Schonewald M, Zhang L, Tripet B, Bentley DL. Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins. Nat Struct Mol Biol. 2007;14:240–242. doi: 10.1038/nsmb1200. [DOI] [PubMed] [Google Scholar]
- 91.Fang J, Hogan GJ, Liang G, Lieb JD, Zhang Y. The Saccharomyces cerevisiae histone demethylase Jhd1 fine-tunes the distribution of H3K36me2. Mol Cell Biol. 2007;27:5055–5065. doi: 10.1128/MCB.00127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim T, Buratowski S. Two Saccharomyces cerevisiae JmjC domain proteins demethylate histone H3 Lys36 in transcribed regions to promote elongation. J Biol Chem. 2007;282:20827–20835. doi: 10.1074/jbc.M703034200. [DOI] [PubMed] [Google Scholar]
- 93.Lo WS, Duggan L, Emre NC, Belotserkovskya R, Lane WS, Shiekhattar R, Berger SL. Snf1--a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science. 2001;293:1142–1146. doi: 10.1126/science.1062322. [DOI] [PubMed] [Google Scholar]
- 94.Ahn SH, Henderson KA, Keeney S, Allis CD. H2B (Ser10) phosphorylation is induced during apoptosis and meiosis in S. cerevisiae. Cell Cycle. 2005;4:780–783. doi: 10.4161/cc.4.6.1745. [DOI] [PubMed] [Google Scholar]
- 95.Ahn SH, Cheung WL, Hsu JY, Diaz RL, Smith MM, Allis CD. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell. 2005;120:25–36. doi: 10.1016/j.cell.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 96.Ahn SH, Diaz RL, Grunstein M, Allis CD. Histone H2B deacetylation at lysine 11 is required for yeast apoptosis induced by phosphorylation of H2B at serine 10. Mol Cell. 2006;24:211–220. doi: 10.1016/j.molcel.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 97.Downs JA, Lowndes NF, Jackson SP. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000;408:1001–1004. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- 98.Cheung WL, Turner FB, Krishnamoorthy T, Wolner B, Ahn SH, Foley M, Dorsey JA, Peterson CL, Berger SL, Allis CD. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr Biol. 2005;15:656–660. doi: 10.1016/j.cub.2005.02.049. [DOI] [PubMed] [Google Scholar]
- 99.Schwabish MA, Struhl K. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol Cell Biol. 2007;27:6987–6995. doi: 10.1128/MCB.00717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Utley RT, Lacoste N, Jobin-Robitaille O, Allard S, Cote J. Regulation of NuA4 histone acetyltransferase activity in transcription and DNA repair by phosphorylation of histone H4. Mol Cell Biol. 2005;25:8179–8190. doi: 10.1128/MCB.25.18.8179-8190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 102.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 103.Geng F, Tansey WP. Polyubiquitylation of Histone H2B. Mol Biol Cell. 2008 doi: 10.1091/mbc.E08-01-0050. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fang J, Chen T, Chadwick B, Li E, Zhang Y. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J Biol Chem. 2004;279:52812–52815. doi: 10.1074/jbc.C400493200. [DOI] [PubMed] [Google Scholar]
- 105.Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, Osley MA. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 2004;18:184–195. doi: 10.1101/gad.1149604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 109.Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 110.Laribee RN, Fuchs SM, Strahl BD. H2B ubiquitylation in transcriptional control: a FACT-finding mission. Genes Dev. 2007;21:737–743. doi: 10.1101/gad.1541507. [DOI] [PubMed] [Google Scholar]
- 111.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 112.Wilson VG, Heaton PR. Ubiquitin proteolytic system: focus on SUMO. Expert Rev Proteomics. 2008;5:121–135. doi: 10.1586/14789450.5.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nathan D, Ingvarsdottir K, Sterner DE, Bylebyl GR, Dokmanovic M, Dorsey JA, Whelan KA, Krsmanovic M, Lane WS, Meluh PB, Johnson ES, Berger SL. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 2006;20:966–976. doi: 10.1101/gad.1404206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schiene C, Fischer G. Enzymes that catalyse the restructuring of proteins. Curr Opin Struct Biol. 2000;10:40–45. doi: 10.1016/s0959-440x(99)00046-9. [DOI] [PubMed] [Google Scholar]
- 115.Nelson CJ, Santos-Rosa H, Kouzarides T. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell. 2006;126:905–916. doi: 10.1016/j.cell.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 116.Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 117.Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 118.Hsu JM, Huang J, Meluh PB, Laurent BC. The yeast RSC chromatin-remodeling complex is required for kinetochore function in chromosome segregation. Mol Cell Biol. 2003;23:3202–3215. doi: 10.1128/MCB.23.9.3202-3215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Baetz KK, Krogan NJ, Emili A, Greenblatt J, Hieter P. The ctf13-30/CTF13 genomic haploinsufficiency modifier screen identifies the yeast chromatin remodeling complex RSC, which is required for the establishment of sister chromatid cohesion. Mol Cell Biol. 2004;24:1232–1244. doi: 10.1128/MCB.24.3.1232-1244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Keogh MC, Mennella TA, Sawa C, Berthelet S, Krogan NJ, Wolek A, Podolny V, Carpenter LR, Greenblatt JF, Baetz K, Buratowski S. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Millar CB, Xu F, Zhang K, Grunstein M. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 2006;20:711–722. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xu F, Zhang K, Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 2005;121:375–385. doi: 10.1016/j.cell.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 123.Hyland EM, Cosgrove MS, Molina H, Wang D, Pandey A, Cottee RJ, Boeke JD. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:10060–10070. doi: 10.1128/MCB.25.22.10060-10070.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ye J, Ai X, Eugeni EE, Zhang L, Carpenter LR, Jelinek MA, Freitas MA, Parthun MR. Histone H4 lysine 91 acetylation a core domain modification associated with chromatin assembly. Mol Cell. 2005;18:123–130. doi: 10.1016/j.molcel.2005.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 126.Phatnani HP, Greenleaf AL. Identifying phosphoCTD-associating proteins. Methods Mol Biol. 2004;257:17–28. doi: 10.1385/1-59259-750-5:017. [DOI] [PubMed] [Google Scholar]
- 127.Phatnani HP, Jones JC, Greenleaf AL. Expanding the functional repertoire of CTD kinase I and RNA polymerase II: novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry. 2004;43:15702–15719. doi: 10.1021/bi048364h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schwartz LB, Roeder RG. Purification and subunit structure of deoxyribonucleic acid-dependent ribonucleic acid polymerase II from the mouse plasmacytoma, MOPC 315. J Biol Chem. 1975;250:3221–3228. [PubMed] [Google Scholar]
- 129.Zhang J, Corden JL. Phosphorylation causes a conformational change in the carboxyl-terminal domain of the mouse RNA polymerase II largest subunit. J Biol Chem. 1991;266:2297–2302. [PubMed] [Google Scholar]
- 130.Cadena DL, Dahmus ME. Messenger RNA synthesis in mammalian cells is catalyzed by the phosphorylated form of RNA polymerase II. J Biol Chem. 1987;262:12468–12474. [PubMed] [Google Scholar]
- 131.Lu H, Zawel L, Fisher L, Egly JM, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 132.O'Brien T, Hardin S, Greenleaf A, Lis JT. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 133.Feaver WJ, Gileadi O, Li Y, Kornberg RD. CTD kinase associated with yeast RNA polymerase II initiation factor b. Cell. 1991;67:1223–1230. doi: 10.1016/0092-8674(91)90298-d. [DOI] [PubMed] [Google Scholar]
- 134.Valay JG, Simon M, Dubois MF, Bensaude O, Facca C, Faye G. The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J Mol Biol. 1995;249:535–544. doi: 10.1006/jmbi.1995.0316. [DOI] [PubMed] [Google Scholar]
- 135.Hengartner CJ, Myer VE, Liao SM, Wilson CJ, Koh SS, Young RA. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 136.Lee JM, Greenleaf AL. A protein kinase that phosphorylates the C-terminal repeat domain of the largest subunit of RNA polymerase II. Proc Natl Acad Sci USA. 1989;86:3624–3628. doi: 10.1073/pnas.86.10.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol Cell. 2007;27:275–288. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 138.Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, Greenblatt JF, Shilatifard A. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 139.Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 140.Lu H, Flores O, Weinmann R, Reinberg D. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc Natl Acad Sci USA. 1991;88:10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M. Ssu72 Is an RNA polymerase II CTD phosphatase. Mol Cell. 2004;14:387–394. doi: 10.1016/s1097-2765(04)00235-7. [DOI] [PubMed] [Google Scholar]
- 142.Kobor MS, Archambault J, Lester W, Holstege FC, Gileadi O, Jansma DB, Jennings EG, Kouyoumdjian F, Davidson AR, Young RA, Greenblatt J. An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol Cell. 1999;4:55–62. doi: 10.1016/s1097-2765(00)80187-2. [DOI] [PubMed] [Google Scholar]
- 143.Ansari A, Hampsey M. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 2005;19:2969–2978. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.O'Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ. Gene loops juxtapose promoters and terminators in yeast. Nat Genet. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 145.Singh BN, Hampsey M. A transcription-independent role for TFIIB in gene looping. Mol Cell. 2007;27:806–816. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 146.Wu X, Rossettini A, Hanes SD. The ESS1 prolyl isomerase and its suppressor BYE1 interact with RNA pol II to inhibit transcription elongation in Saccharomyces cerevisiae. Genetics. 2003;165:1687–1702. doi: 10.1093/genetics/165.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Huibregtse JM, Yang JC, Beaudenon SL. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1997;94:3656–3661. doi: 10.1073/pnas.94.8.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Collins GA, Tansey WP. The proteasome: a utility tool for transcription? Curr Opin Genet Dev. 2006;16:197–202. doi: 10.1016/j.gde.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 149.Somesh BP, Sigurdsson S, Saeki H, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Communication between distant sites in RNA polymerase II through ubiquitylation factors and the polymerase CTD. Cell. 2007;129:57–68. doi: 10.1016/j.cell.2007.01.046. [DOI] [PubMed] [Google Scholar]
- 150.Chen X, Ruggiero C, Li S. Yeast Rpb9 plays an important role in ubiquitylation and degradation of Rpb1 in response to UV-induced DNA damage. Mol Cell Biol. 2007;27:4617–4625. doi: 10.1128/MCB.00404-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kvint K, Uhler JP, Taschner MJ, Sigurdsson S, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Reversal of RNA polymerase II ubiquitylation by the ubiquitin protease Ubp3. Mol Cell. 2008;30:498–506. doi: 10.1016/j.molcel.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 152.Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318:1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- 153.Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kelly WG, Dahmus ME, Hart GW. RNA polymerase II is a glycoprotein. Modification of the COOH-terminal domain by O-GlcNAc. J Biol Chem. 1993;268:10416–10424. [PubMed] [Google Scholar]
- 155.Comer FI, Hart GW. Reciprocity between O-GlcNAc and O-phosphate on the carboxyl terminal domain of RNA polymerase II. Biochemistry. 2001;40:7845–7852. doi: 10.1021/bi0027480. [DOI] [PubMed] [Google Scholar]
- 156.Cabrejos ME, Allende CC, Maldonado E. Effects of phosphorylation by protein kinase CK2 on the human basal components of the RNA polymerase II transcription machinery. J Cell Biochem. 2004;93:2–10. doi: 10.1002/jcb.20209. [DOI] [PubMed] [Google Scholar]
- 157.Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 158.Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci USA. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Martin DG, Baetz K, Shi X, Walter KL, MacDonald VE, Wlodarski MJ, Gozani O, Hieter P, Howe L. The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol Cell Biol. 2006;26:7871–7879. doi: 10.1128/MCB.00573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Shi X, Kachirskaia I, Walter KL, Kuo JH, Lake A, Davrazou F, Chan SM, Martin DG, Fingerman IM, Briggs SD, Howe L, Utz PJ, Kutateladze TG, Lugovskoy AA, Bedford MT, Gozani O. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem. 2007;282:2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 163.Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, Gozani O, Cheng X, Shi Y. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD. Gene silencing: trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- 165.Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- 166.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 167.Henry KW, Berger SL. Trans-tail histone modifications: wedge or bridge? Nat Struct Biol. 2002;9:565–566. doi: 10.1038/nsb0802-565. [DOI] [PubMed] [Google Scholar]
- 168.Sims JK, Houston SI, Magazinnik T, Rice JC. A trans-tail histone code defined by monomethylated H4 Lys-20 and H3 Lys-9 demarcates distinct regions of silent chromatin. J Biol Chem. 2006;281:12760–12766. doi: 10.1074/jbc.M513462200. [DOI] [PubMed] [Google Scholar]
- 169.Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 170.Schaft D, Roguev A, Kotovic KM, Shevchenko A, Sarov M, Shevchenko A, Neugebauer KM, Stewart AF. The histone 3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation. Nucleic Acids Res. 2003;31:2475–2482. doi: 10.1093/nar/gkg372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Shilatifard A, Buratowski S, Greenblatt J. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Rao B, Shibata Y, Strahl BD, Lieb JD. Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol Cell Biol. 2005;25:9447–9459. doi: 10.1128/MCB.25.21.9447-9459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- 176.Youdell ML, Kizer KO, Kisseleva-Romanova E, Fuchs SM, Duro E, Strahl BD, Mellor J. Roles for Ctk1 and Spt6 in regulating the different methylation states of Histone H3 lysine. Mol Cell Biol. 2008;36 doi: 10.1128/MCB.00001-08. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kasten M, Szerlong H, Erdjument-Bromage H, Tempst P, Werner M, Cairns BR. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Prigent C, Dimitrov S. Phosphorylation of serine 10 in histone H3, what for? J Cell Sci. 2003;116:3677–3685. doi: 10.1242/jcs.00735. [DOI] [PubMed] [Google Scholar]
- 179.Ferreira H, Flaus A, Owen-Hughes T. Histone modifications influence the action of Snf2 family remodelling enzymes by different mechanisms. J Mol Biol. 2007;374:563–579. doi: 10.1016/j.jmb.2007.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Koyama H, Itoh M, Miyahara K, Tsuchiya E. Abundance of the RSC nucleosome-remodeling complex is important for the cells to tolerate DNA damage in Saccharomyces cerevisiae. FEBS Lett. 2002;531:215–221. doi: 10.1016/s0014-5793(02)03504-4. [DOI] [PubMed] [Google Scholar]
- 181.Carey M, Li B, Workman JL. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol Cell. 2006;24:481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Soutourina J, Bordas-Le Floch V, Gendrel G, Flores A, Ducrot C, Dumay-Odelot H, Soularue P, Navarro F, Cairns BR, Lefebvre O, Werner M. Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol Cell Biol. 2006;26:4920–4933. doi: 10.1128/MCB.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.VanDemark AP, Kasten MM, Ferris E, Heroux A, Hill CP, Cairns BR. Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation. Mol Cell. 2007;27:817–828. doi: 10.1016/j.molcel.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Weigel NL, Moore NL. Kinases and protein phosphorylation as regulators of steroid hormone action. Nucl Recept Signal. 2007;5:e005. doi: 10.1621/nrs.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Huang J, Berger SL. The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev. 2008 doi: 10.1016/j.gde.2008.01.012. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 186.Xu C, Henry PA, Setya A, Henry MF. In vivo analysis of nucleolar proteins modified by the yeast arginine methyltransferase Hmt1/Rmt1p. RNA. 2003;9:746–759. doi: 10.1261/rna.5020803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Harauz G, Musse AA. A tale of two citrullines--structural and functional aspects of myelin basic protein deimination in health and disease. Neurochem Res. 2007;32:137–158. doi: 10.1007/s11064-006-9108-9. [DOI] [PubMed] [Google Scholar]
- 188.Gabius HJ, Siebert HC, Andre S, Jimenez-Barbero J, Rudiger H. Chemical biology of the sugar code. Chembiochem. 2004;5:740–764. doi: 10.1002/cbic.200300753. [DOI] [PubMed] [Google Scholar]
- 189.Mahal LK. Glycomics: towards bioinformatic approaches to understanding glycosylation. Anticancer Agents Med Chem. 2008;8:37–51. doi: 10.2174/187152008783330806. [DOI] [PubMed] [Google Scholar]


