Abstract
Guanine-rich nucleic acids can form G-quadruplexes that are important in gene regulation, biosensor design and nano-structure construction. In this article, we report on the development of a nanopore encapsulating single-molecule method for exploring how cations regulate the folding and unfolding of the G-quadruplex formed by the thrombin-binding aptamer (TBA, GGTTGGTGTGGTTGG). The signature blocks in the nanopore revealed that the G-quadruplex formation is cation-selective. The selectivity sequence is K+ > NH4+ ∼ Ba2+ > Cs+ ∼ Na+ > Li+, and G-quadruplex was not detected in Mg2+ and Ca2+. Ba2+ can form a long-lived G-quadruplex with TBA. However, the capability is affected by the cation–DNA interaction. The cation-selective formation of the G-quadruplex is correlated with the G-quadruplex volume, which varies with cation species. The high formation capability of the K+-induced G-quadruplex is contributed largely by the slow unfolding reaction. Although the Na+- and Li+-quadruplexes feature similar equilibrium properties, they undergo radically different pathways. The Na+-quadruplex folds and unfolds most rapidly, while the Li+-quadruplex performs both reactions at the slowest rates. Understanding these ion-regulated properties of oligonucleotides is beneficial for constructing fine-tuned biosensors and nano-structures. The methodology in this work can be used for studying other quadruplexes and protein–aptamer interactions.
INTRODUCTION
Guanine-rich single-stranded nucleic acids can form G-quadruplexes (1–5). These four-stranded complexes repeatedly occur in the human genome, playing an important role in gene regulation (5–9) and serving as targets of drugs for cancer treatment (10–12). Quadruplexes created in vitro are building blocks for nano-structures (13) and nanomachines (14,15). Their high affinity for target proteins make them ideal as powerful biosensors (16) and potent pharmaceuticals (17,18).
Crystallographic and nuclear magnetic resonance imaging show the core of a quadruplex to be the G-tetrad, a planar assembly of four guanine bases networked via hydrogen bonds (1,3,4,19–22). G-tetrads stack one on another, with a cation located between adjacent tetrads in coordinating eight carbonyls of guanine bases for stabilization (Figure 1a). Previous studies using biophysical approaches, such as circular dichroism, UV spectroscopy and differential scanning calorimetry, have outlined the thermodynamic profiles of the folding and unfolding of quadruplexes (2,21,23–25). Fluorescence resonance energy transfer (FRET) (26,27) has been used to study the structural dynamics of quadruplexes at the single-molecule level (28). More recently, the folding/unfolding kinetics of the telomere quadruplex have been determined with a surface plasmon resonance (SPR) biosensor (29). However, the DNA labeling process required in both FRET and SPR detections may affect the measurements (29).
Figure 1.
Molecular structure of TBA and scheme of nanopore detection method. (a) Left, the sequence and structure of TBA G-quadruplex; right, the two G-tetrad planes in the TBA G-quadruplex (DOI 10.2210/pdb1c38/pdb, RCSB Protein Data Bank) (34), with the top tetrad formed by guanines at the position 1, 6, 10 and 15, and the bottom one by guanine 2, 5, 11 and 14. A cation in between is coordinated with eight carbonyls. The cation-carbonyl distance, d, is the one half the mean of inter-carbonyl distances between G1–G11, G2–G10, G5–G15 and G6–G14. d = 2.86 ± 0.7 Å. (b) Diagram of the current trace showing characteristic signature blocks. (c) Long-lived block for capturing a single G-quadruplex in the nanocavity enclosed by the α-hemolysin (αHL) pore; (d) The long block terminal spike produced by translocation of the unfolded G-quadruplex in the nanocavity. (e) Short-lived block formed by translocation of linear form TBA.
The thrombin-binding aptamer (TBA, GGTTGGTGTGGTTGG) is a well-known G-quadruplex. It serves as a highly potent inhibitor to thrombin clotting activity (16) and a sophisticated biosensor for protein detection (30–32). In the presence of cations, this 15-base single-stranded DNA can fold into a two-tetrad, one-cation quadruplex (Figure 1a) (19,23,33,34). The quadruplex formation varies with the cation species. For example, a study using circular dichroism (CD) spectra and melting profiles revealed that cations with ionic radii between 1.3 and 1.5 Å (such as K+, Rb+, NH4+, Sr2+ and Ba2+) fit within the two G-tetrads of the quadruplex better than other ions (such as Li+, Na+, Cs+, Mg2+ and Ca2+). Therefore they can form more stable intramolecular quadruplexes (23). Similar results were also reported with other detection methods, such as capillary electrophoresis (32). Although the formation properties have been well studied, the cation-dependent kinetics of folding and unfolding of the TBA G-quadruplex is still not well understood. Gaining an understanding of the kinetics is important because a properly folded quadruplex is necessary for the molecular recognition involved in many quadruplex functions and is beneficial for designing quadruplex applications.
The αHL nanopore is a receptive single-molecule detector (35) with broad applications, from biosensing (36–38), nucleic acids detection (39–45) and regulation of membrane transportation (46–48), to the study of single-molecule chemistry (49), single-molecule force measurements (50,51) and the construction of biochips(52). The nanopore is also a single-molecule tool for unzipping double-stranded DNA with a transmembrane voltage, capable of detecting DNA unzipping kinetics (41,42,53). Recently, we demonstrated that the nanocavity enclosed by the αHL pore can encapsulate a single TBA G-quadruplex (54). By recognizing the current signatures in the nanopore (Figure 1b), we can discriminate a single DNA molecule, either in the G-quadruplex form (Figure 1c) or the linear form (Figure 1e). In particular, the G-quadruplex trapped in the nanocavity can spontaneously unfold and leave the pore as a linear DNA (Figure 1d). We hypothesized that these single-molecule processes can reveal the G-quadruplex's folding and unfolding properties. In this study, we developed an analytical method in which the lifetime of G-quadruplex and the distributions of the folded and unfolded molecules in the solution can be extracted from the current signatures. Ultimately, we can determine both the equilibrium and folding/unfolding kinetic properties of the G-quadruplex in various cations. This single-molecule detection is non-covalent without DNA labeling.
MATERIALS AND METHODS
We employed an electrophysiology setup and followed a protocol described in a previous study (54) to record pico Ampere currents through a single αHL pore. Briefly, the recording apparatus was composed of two chambers (cis and trans), partitioned with a Teflon film. The planar lipid bilayer of 1,2-diphytanoyl-sn-glycerophosphatidylcholine (Avanti Polar Lipids) was formed spanning a 100–150 μm hole in the center of the partition. Both cis and trans chambers were filled with symmetrical 1 M salt solutions buffered with 10 mM Tris and titrated to pH 7.2. Salts used were LiCl, NaCl, KCl, NH4Cl, Cs Cl, MgCl2, CaCl2 and BaCl2. Protein pores were inserted into the bilayer from the cis side. Oligonucleotides including the thrombin-binding aptamer (TBA, GGTTGGTGTGGTTGG) and control (Ctrl-2, GATTAGTGTGATTAG) were synthesized and electrophoresis-purified by Integrated DNA Technologies, IA. Before testing, DNA solutions were heated at 90°C for 15 min, then cooled to room temperature. Total 2.5 μM DNA was added to the cis solution. The cis chamber was grounded so that a positive voltage drove the translocation of a negatively charged DNA through the pore from cis to trans.
Single-channel currents were recorded with an Axopatch 200A and 200B patch-clamp amplifier (Molecular Device Inc.), filtered with a built-in 4-pole low-pass Bessel Filter at 5 kHz, and acquired with Clampex 9.0 software (Molecular Device Inc.) through a Digidata 1332 A/D converter (Molecular Device Inc.) at a sampling rate of 20 ks−1. The data were analyzed using Clampfit 9.0 (Molecular Device Inc.), Excel (MicroSoft) and SigmaPlot (SPSS) software. The single-channel currents were determined from amplitude histograms by fitting the peaks to Gaussian functions. The duration of short-lived blocks for DNA translocation was obtained by fitting the dwell-time histogram to an exponential distribution. The occurrence of short blocks was given by the reciprocal of the average block interval, which was determined after long blocks were excluded from the trace. Since the numbers of long-lived blocks and their terminal spikes were low (50–100), their duration and occurrence were not obtained from histograms. Instead, the values of the two parameters were represented by the arithmetic means. The terminal spikes were determined by setting the threshold at the level of 50% from the long-block amplitude. Data were given as the mean ± SD, based on at least three separate experiments. The electrophysiology experiments were conducted at 22°C ± 2°C.
RESULTS AND DISCUSSION
Discrimination between the G-quadruplex and linear forms of TBA in various cations
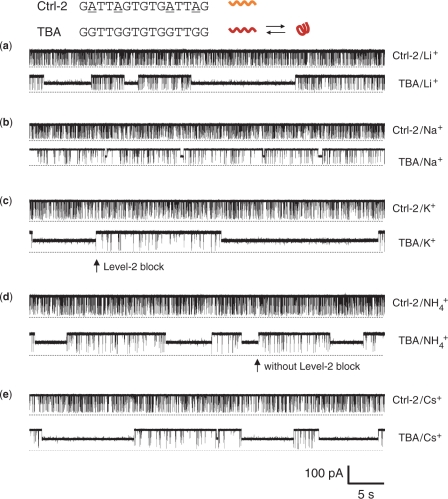
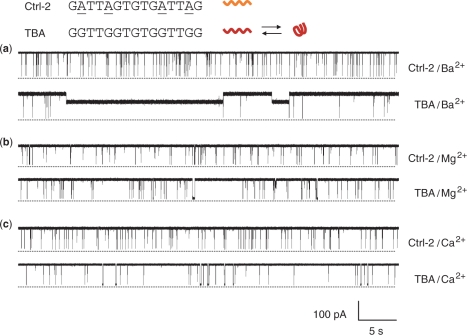
The current traces of TBA and Ctrl-2 were recorded in 1 M LiCl, NaCl, KCl, NH4Cl and CsCl (Figure 2), and MgCl2, CaCl2 and BaCl2 (Figure 3). Ctrl-2 (GATTAGTGTGATTAG) has the identical length (15 nucleotides) and a sequence similar to TBA (GGTTGGTGTGGTTGG), but is unable to fold into the G-quadruplex due to the substitution of guanines at positions 2, 5, 11 and 14 with adenines (55). Ctrl-2 simply traversed the αHL pore by producing full blocks with a conductance (g) of 42–145 pS (Table 1). The translocation duration (τ) in all the monovalent cations was 349–450 μs; and that in divalent cations was prolonged to 1280–5700 μs (Table 1). These durations, particularly in divalent cations, were considerably longer than the previously reported temporal dispersion for linear DNA translocation (56,57). We also found that the translocation of the linear form TBA described below yielded blocks with comparable durations (Table 1).
Figure 2.
Current traces from a single αHL pore showing blocks with Ctrl-2 and TBA in the presence of various monovalent cations. The concentration of both DNAs was 2.5 μM. All traces were recorded at +100 mV in a 1 M salt solution buffered with 10 mM Tris (pH 7.2). The top trace of each panel was recorded for Ctrl-2 and the bottom one for TBA in solutions of (a) LiCl, (b) NaCl, (c) KCl, (d) NH4Cl and (e) CsCl.
Figure 3.
Current traces from a single αHL pore, showing blocks with Ctrl-2 and TBA in the presence of various divalent cations. The recording conditions were the same as Figure 2, and the traces were recorded in solutions of (a) BaCl2, (b) MgCl2 and (c) CaCl2.
Table 1.
Conductance, duration and occurrence of blocks of the αHL pore produced by Ctrl-2 and TBA in various ion solutions (+100 mV)
| DNA | Ion | Short block |
Long Block |
|||
|---|---|---|---|---|---|---|
| g (pS) | τ (μs) | f (s−1) | G (pS) | τ (s) | ||
| Ctrl-2 | Li+ | 53 ± 4 | 450 ± 206 | 29 | ||
| TBA | Li+ | 47 ± 6 | 629 ± 80 | 12 | 355 ± 35 | 15 ± 4 |
| Ctrl-2 | Na+ | 100 ± 12 | 404 ± 95 | 23 | ||
| TBA | Na+ | 89 ± 8 | 597 ± 100 | 7.0 | 438 ± 31 | 0.35 ± 0.14 |
| Ctrl-2 | K+ | 110 ± 27 | 361 ± 103 | 18 | ||
| TBA | K+ | 84 ± 14 | 322 ± 61 | 2.3 | 599 ± 18 | 15 ± 3 |
| Ctrl-2 | NH4+ | 145 ± 11 | 429 ± 91 | 25 | ||
| TBA | NH4+ | 107 ± 16 | 457 ± 65 | 4.5 | 627 ± 18 | 4.0 ± 0.8 |
| Ctrl-2 | Cs+ | 88 ± 30 | 349 ± 111 | 22 | ||
| TBA | Cs+ | 43 ± 9 | 279 ± 51 | 5.7 | 545 ± 25 | 12 ± 1.3 |
| Ctrl-2 | Ba2+ | 50 ± 9 | 1280 ± 220 | 3.3 | ||
| TBA | Ba2+ | 17 ± 7 | 1490 ± 330 | 0.64 | 807 ± 23 | 17 ± 4.4 |
| Ctrl-2 | Mg2+ | 42 ± 18 | 5700 ± 1500 | 1.3 | ||
| TBA | Mg2+ | 13 ± 8 | 4100 ± 390 | 0.9 | ||
| Ctrl-2 | Ca2+ | 55 ± 21 | 4200 ± 500 | 2.5 | ||
| TBA | Ca2+ | 30 ± 16 | 3400 ± 450 | 1.2 | ||
Earlier reports suggested that the longer translocation time is due to certain DNA strands [such as poly(dA)] that require energy to break up their stacked structures before entering the narrow nanopore (56). This interpretation could explain our Ctrl-2 and linear TBA, as the two guanine- and thymine-rich DNA strands may adopt sequence-dependent secondary structures that are different from other DNA sequences containing fewer guanines or thymines. Break up of their secondary structures in the nanopore might cause larger temporal dispersion for translocation. Another possible explanation for the long translocation time in divalent cations is the strong cation–DNA interaction. It is known that the affinities of divalent cations for DNA are 102–103 times greater than for monovalent cations (2,58). For example, pK values for Mg2+- and Ba2+-ATP are 4.6 and 3.3, whereas pK values for Na+-and K+-ATP are 1.1 and 1.3 (58). The bound cations may reduce the negative charges on DNA, resulting in a weakened electrical driving force in the nanopore. Consequently, the velocity of DNA translocation is reduced.
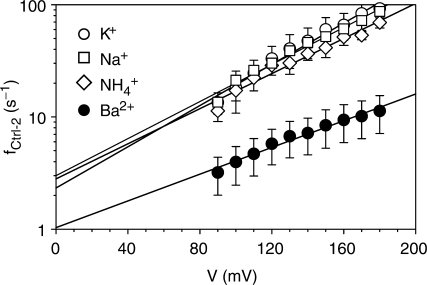
The strong binding of divalent cations may also lessen the occurrence of DNA translocation (f). In monovalent cations, f for the Ctrl-2 blocks was 18–29 s−1 (2.5 μM DNA), while in divalent cations, f was only 1.3–3.3 s−1 (Table 1). The reduced translocation occurrence in divalent cations was also seen over a broad range of voltages (Figure 4). By fitting the data with the Woodhull's equation, ln f(V) = ln f(0)−δzFV/RT, we obtained that the net charge of DNA, z, in Ba2+ (0.34e) is smaller than in K+ (0.5e), Na+ (0.48e) and NH4+ (0.48e) (assuming δ is 1), confirming the fact that DNA in Ba2+ carries a lesser negative charge than in monovalent cations. The occurrence of translocation at 0 mV without electrical drive is represented as f(0). We found f(0) in Ba2+ (1.0 s−1) is only one half or one third of that in K+ (2.3 s−1), Na+ (3.0 s−1) and NH4+ (2.8 s−1). The difference in f(0) might be explained as the formation of higher-order DNA structures in Ba2+, which either enter the pore at a reduced rate or decrease the linear TBA concentration to lower its translocation occurrence. This expectation is supported by the fact that Mg2+ can promote the formation of DNA complexes such as DNA multiplexes (59).
Figure 4.
Voltage-dependent short block occurrences for Ctrl-2 in monovalent cations K+, Na+, NH4+ and divalent cation Ba2+. The concentration of DNA was 2.5 μM and all the occurrences were measured from +90 mV to +180 mV in 1 M salt solution buffered with 10 mM Tris (pH 7.2).
TBA distinguished itself from Ctrl-2 by producing long-lived blocks. The long blocks were observed in all the monovalent ions (Figure 2) and the divalent ion Ba2+ (Figure 3a), but not in other cations Mg2+ and Ca2+ (Figure 3b and c). The long-lived blocks partially reduced the pore conductance to 355–807 pS, with a highly cation-dependent duration that varied almost 50 folds, from 0.35 s in Na+, 4 s in NH4+ 12 s in Cs+, to 15–17 s in Li+, K+ and Ba2+ (Table 1). The long block was produced by arresting a single TBA G-quadruplex in the nanocavity of the αHL pore (54). αHL encloses a nanocavity in the cap domain on the top the transmembrane β-barrel (Figure 1c). The cavity is 4.6 nm wide, with a 2.3 nm opening to the cis solution and a 1.4 opening to the β-barrel in the middle of the pore (60). By comparison, the diagonal distance of TBA G-quadruplex is 2.1 nm (34). Thus a single G-quadruplex can lodge in the cavity from the cis opening and partially block the ion pathway, with the duration of the long blocks representing the lodging time (Figure 1c). Therefore the long-lived block indicated the formation of the TBA G-quadruplex in Li+, Na+, K+, NH4+ and Ba2+, but not in Mg2+ and Ca2+. The fact that no G-quadruplex was detected in Mg2+ or Ca2+, is consistent with the earlier finding that higher concentration of divalent cations (>2 mM) destabilize quadruplexes (2,61). As noted above, the high affinities of divalent cations for DNA may weaken their capability for forming the G-quadruplex with TBA.
Most long-lived blocks were terminated with a short-lived terminal spike (Level-2 block, arrow marked in Figure 2c), that has a duration of 270 ± 114 μs, comparable to that of independent short-lived blocks. The terminal spike has been recognized as the unfolding of the trapped G-quadruplex into a linear DNA, followed by rapid translocation to the trans solution (Figure 1d) (54). The captured G-quadruplex can also escape back to the cis solution without unfolding, as evidenced by long events without a terminal spike (Figure 2d, arrow). The percentage of Level-2 block-terminated long blocks (as part of the total long blocks) was high 81% in Li+, 82% in Na+, 85% in K+, 87% in Ba2+, 75% NH4+ and 83% in Cs+ (+100 mV), suggesting that most of the trapped G-quadruplexes unfold in the cavity rather than escape back to the cis solution.
In addition to long blocks by trapped G-quadruplex, TBA also produced independent short-lived blocks whose conductance and durations were at the same levels as Ctrl-2 blocks: the reduced conductance (g) was 13–107 pS; the block duration (τ) in monovalent ions was 279–629 μs, and that in divalent ions was 1490–4100 μs (Table 1). However, the occurrence of the TBA short blocks in each cation was lower than that of Ctrl-2 blocks. For example, the TBA short blocks in K+ were 2.3 s−1, almost eight times lower than the 18 s−1 for the Ctrl-2 short blocks in the same solution. The occurrence ratio was highly cation-dependent: from low to high, 12% in K+, 18% in NH4+, 19% in Ba2+, 26% in Cs+, 30% in Na+ and 41% in Li+. The TBA-produced short blocks is caused by the translocation of linear form TBA in the nanopore (54) (Figure 1e). Since a portion of the TBA has folded into the G-quadruplex, the concentration of the remaining linear TBA in the solution should be lower than that of Ctrl-2 (both concentrations of TBA and Ctrl-2 were 2.5 μM), resulting in a less translocation occurrence.
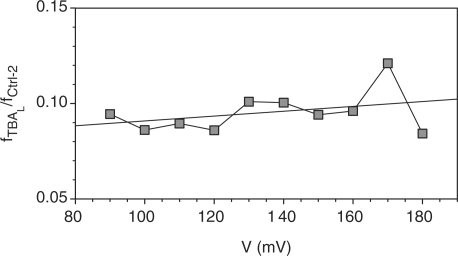
The cation-dependence of translocation occurrence can be utilized to calculate the concentration of linear TBA ([TBAL]) in each cation solution. [TBAL] is proportional to the short block occurrence, i.e. fTBAL = α TBAL •[TBAL]. The coefficient αTBAL is the translocation rate constant. This parameter cannot be determined directly from the current signature. We wished to know if α TBAL can be substituted by α Ctrl - 2, the translocation rate constant for Ctrl-2, which can be determined. Ctrl-2 and TBA not only have the identical length and similar sequences, but their blocks have similar conductance and duration. For example, the conductance and duration for the Ctrl-2 blocks in 1 M KCl were 110 pS and 361 μs, similar to the 84 pS and 322 μs for the TBA short blocks (Table 1). Further study of the voltage dependence showed that the ratio of block occurrences for the two DNAs is independent of the voltage (Figure 5), suggesting the two DNAs exhibit comparable voltage-dependent translocation occurrences. Because of the similar structures and translocation properties between Ctrl-2 and linear TBA, it is reasonable to assume their translocation rate constants are similar. In an approximation, we can use αCtrl- 2 to substitute αTBAL, and determine [TBAL] as
| 1 |
Figure 5.
Voltage-independence of the ratio of occurrences between linear TBA and Ctrl-2 short blocks in K+. The measurement conditions were the same as in Figure 4.
Cation's capacity in G-quadruplex formation and correlation with the G-quadruplex volume
According to the definition, the equilibrium formation constant for the G-quadruplex is Kf = [TBAG]/[TBAL] = ([TBA] - [TBAL])/[TBAL]. In this expression, [TBA] = [Ctrl-2], and [TBAL] can be calculated from Equation (1). Thus Kf will be determined by
| 2 |
The calculated Kf for the G-quadruplex was 7.0 in K+, 4.6 in NH4+, 4.2 in Ba2+, 2.8 in Cs+, 2.3 in Na+ and 1.5 in Li+ (Table 2). Several Kf values are comparable to many of those detected using other techniques. For example, Kf values in K+ and Na+ are similar to those for the telomere G-quadruplex (TTAGGG)4 measured by surface plasma resonance (SPR), 9.0 in K+ and 1.5 in Na+ (25°C) (29). Although the equilibrium constant for Li+•TBA (Kf = 1.5) was much larger than that for Li+•(TTAGGG)4 (Kf = 4.5 × 10−4), the sequence of Kf values for both G-quadruplexes are the same, K+ > Na+ > Li+. The standard free energy ΔG0 for K+•TBA from the single-molecule measurement was −1.1 kcal mol−1. The energy level is also in agreement with that found by ensemble methods. For example, ΔG0 for TBA obtained from the melting property (1 M KCl, 25°C) is −1.7 kcal mol−1 (Figure S1); that by van't Hoff analysis of UV melting curves (25 mM KCl, 37°C) is −1.19 kcal mol−1 (25), and that determined by thermodynamic measurements (100 mM KCl, 20°C) is −2.0 kcal mol−1 (23,24). According to an earlier study, the energy for the formation of the (G3T4G3)2 G-quadruplex with K+ is −4.7 kcal mol−1 and that with Na+ is −2.2 kcal mol−1 (62). Given that this G-quadruplex contains three ion binding sites, these energies are equivalent to −1.6 kcal mol−1 and −0.71 kcal mol−1 per ion, in agreement with our result for TBA, −1.1 kcal mol−1 in K+ and −0.48 kcal mol−1 in Na+.
Table 2.
Equilibrium, folding and unfolding rate constants, and free energy for the TBA G-quadruplex in different cations
| M•TBA | r (Å)a | Kf (no unit) | kf (s−1) | ku (s−1) | ΔG° (kcal mol−1)b |
|---|---|---|---|---|---|
| Li+•TBA | 0.60 | 1.5 | 0.095 | 0.065 | −0.23 |
| Na+•TBA | 0.95 | 2.3 | 6.5 | 2.9 | −0.48 |
| K+•TBA | 1.33 | 7.0 | 0.46 | 0.066 | −1.1 |
| NH4+•TBA | 1.45 | 4.6 | 1.1 | 0.25 | −0.89 |
| Cs+•TBA | 1.69 | 2.8 | 0.23 | 0.082 | −0.61 |
| Ba2+•TBA | 1.35 | 4.2 | 0.25 | 0.061 | −0.84 |
| Mg2+•TBA | 0.65 | ||||
| Ca2+•TBA | 0.99 |
aRef. (72).
bΔG0 was calculated by ΔG0 = RT ln Kf/4.18.
The equilibrium constants suggests the sequence of cation's capability for the G-quadruplex formation is K+ > NH4+ ∼ Ba2+ > Cs+ ∼ Na+ > Li+, and K+ is the most capable cation. This cation selectivity compares with that reached by thermodynamic and optical spectroscopic measurements (2,23). For example, it has been suggested from CD spectroscopy and UV melting profiles that K+, Rb+, NH4+, Sr2+ and Ba2+ are able to form stable intramolecular G-quadruplexes at temperatures above 25°C; the cations Li+, Na+, Cs+, Mg2+ and Ca2+ form weaker complexes at very low temperatures (23). As shown in Figure 6a, Kf is correlated with the ionic radii, consistent with a widely accepted ‘goodness of fit’ model: The cation preference is determined by an optimal fit of cations between the two G-tetrads in coordination with eight guanine carbonyls (23). K+, Ba2+ and NH4+ with similar ionic radii (1.3–1.5 Å) fit well within the two G-tetrads in a G-quadruplex, whereas smaller cations, such as Na+ and Li+ or larger cations such as Cs+, would not fit in (23). On the basis of NMR studies of the G-quadruplex formed by 5′-guanidine mono-phosphate (5′-GMP), the cation selectivity was also interpreted in terms of the ion hydration energy (63,64). For example, fully dehydrated Na+ is supposed to be preferred over K+ in binding with the G-quadruplex. However, Na+ in solution costs much higher energy than K+ to be dehydrated. Thus, the overall energy for Na+ binding with the G-quadruplex is higher than K+, making K+ a preferred cation for G-quadruplex formation. This proposal well explained the selectivity for monovalent cations, but is limited to explain why the divalent cation Ba2+, which has a much larger hydration energy than K+, is still as capable as K+ in the G-quadruplex formation. The intrinsic carbonyl tetrad could be one of the structural determinants to the cation selectivity. This note is supported by similar structures between the G-quadruplex and the potassium-selective ion channels, a class of tetrameric protein pores that selectively transport K+ ions across the cell membrane (63,64). The preferential transport of K+ ions in the K+ channel is governed by a selectivity filter that is assembled by backbone carbonyls from four identical conservative sequences, Thr-Val-Gly-Tyr (65). Similar to the G-quadruplex, the K+ ion in the K+ channel also sits between two carbonyl tetrads, coordinated with eight carbonyls. The K–O distance in the selectivity filter is 2.85 Å, nearly identical to the 2.86 Å for the cation-carbonyl distance in the G-quadruplex (Figure 1a). The cation selectivity in the K+ channel is K+ > NH4+ > Cs+ >> Na+ ∼ Li+ (66), almost identical to the order of ion preference in the G-quadruplex we identified. Like the G-quadruplex, the K+ channel is also highly sensitive to Ba2+. Although Ba2+ is not the transporting ion, it is an important channel inhibitor that blocks the selectivity filter with high affinity. These structural and functional similarities support the idea that the common carbonyl tetrad in both molecules gives rise to their similar ion selectivity. The similarity between the G-quadruplex and the K+ channel are significant for applications in molecular engineering. For example, Davis and co-workers have successfully designed an artificial G-quadruplex species that functions as a transmembrane ion channel (67,68).
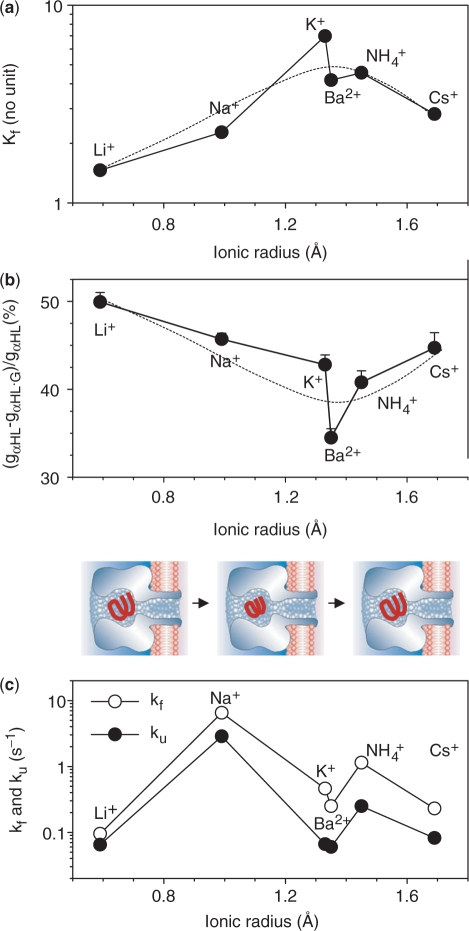
Figure 6.
Equilibrium, conductance blocking and kinetic properties of the TBA G-quadruplex in various cations. The data were from Table 2 and presented in the sequence of ionic radii. (a) The equilibrium formation constant Kf. (b) Blocking percentages, ratios of conductance of the αHL pore with and without G-quadruplexes. The model showed the volume of G-quadruplex that varies with cation species. (c) Folding (kf) and unfolding (ku) rate constants.
We would like to understand if the cation's capacity for G-quadruplex formation is correlated with the G-quadruplex volume. According to the nanopore conductance with (gαHL•G) and without (gαHL) G-quadruplex, we calculated the blocking percentage [(gαHL-gαHL•G)/gαHL]. As shown in Figure 6b, the blocking percentage varies with the cation species: from the lowest 34% in Ba2+ to the highest 50% in Li+, and the sequence is Ba2+ < NH4+ ∼ K+ < Cs+ ∼ Na+ < Li+. The cation-dependent block level may be explained by two mechanisms. First, the volume of G-quadruplex is cation-dependent. Once trapped in the pore, these complexes block the pore with the same size at different conductance levels. Second, the conductivity in the nanopore is altered when it is blocked with the G-quadruplex. The variation of the conductivity gives rise to different block level. To discriminate between the two mechanisms, we examined whether the block of the pore by a molecule with fixed dimension is cation-dependent. β-cyclodextrin (βCD) is qualified for the test because it is a rigid ring-shaped molecule with a fixed diameter of ∼1.5 nm. When lodged in the lumen of αHL, it acted as a molecular adapter for stochastic sensing of pharmaceutical compounds (37,47,52). In contrast to the G-quadruplex, βCD exhibited very similar block levels in Li+ and Ba2+ over a wide range of voltage (Figure S2). For example, the βCD reduced the conductance by 62% in Li+ and 64% in Ba2+ at +40 mV, and 59% in Li+ and 62% in Ba2+ at −40 mV. These levels were also similar to the previous 60% with βCD in 1 M NaCl (−40 mV) (37). The result from testing with βCD suggests that the block percentage by a molecule with a fixed size is independent of the cation species, ruling out the possibility of cation-dependent changes in the pore conductivity. Thus, the block level could reflect different volume of the G-quadruplex, which follow this order, Ba2+•TBA < K+•TBA ∼ NH4+•TBA < Cs+•TBA ∼ Na+•TBA < Li+•TBA. Comparison demonstrated that the blocking percentage (Figure 6b) and Kf (Figure 6a) obey the similar cation sequences, suggesting a correlation between the two properties: the smaller the G-quadruplex volume, the stronger the cation capacity to form a G-quadruplex with TBA. The volume variation could be determined by the guanine−guanine hydrogen bonds and/or the cation-carbonyl distance. For the cation-carbonyl distance (d), the sequence of d would be dBa–O < dK–O ∼ dNH4O < dCs–O ∼ dNa–O < dLi–O. Therefore, it is possible that the greater capability for G-quadruplex formation in Ba2+, K+ and NH4+ than Na+, Li+ and Cs+, is due to the stronger attraction between these cations and the carbonyl, giving rise to their smaller cation-carbonyl distance. At present, there is no published structure to determine every cation-carbonyl distance in the G-quadruplex. However, we can evaluate the range of d based on a simplified model in which the G-quadruplex acts as a ball trapped in a spherical cavity of 4.5 nm with a 2.5 nm cis opening and a 1.5 nm trans opening (Supplementary Data). The results demonstrated that the block percentage increases by 15% from 18% to 33% as the ball radius expands from 2.5 nm to 2.8 nm. Because the average cation-carbonyl distance d is 2.86 Å (Figure 1a), this result is equivalent to a d that varies within 2.86 ± 0.19 Å.
Cation-dependent folding and unfolding of the G-quadruplex
The unfolding rate constant is ku = 1/τG, where τG is the lifetime of the G-quadruplex in aqueous phase. τG cannot be directly measured from the current recording. However, we can measure the duration of long-lived blocks, τ. The long block is produced by trapping a single G-quadruplex in the αHL nanocavity and terminated by releasing unfolded TBA to the solution. Therefore, τ is the lifetime of the G-quadruplex in the pore before unfolding. In the Supplementary Data, we reported on the Monte Carlo simulation to show that the G-quadruplex in the solution is as long-lived as in the nanopore, i.e. τG = τ. This relationship was further interpreted using probability analysis (Supplementary Data). This conclusion is similar to experimental results for DNA duplex formation and deformation in the αHL pore (69): the associate and dissociate rate constants in the nanopore are comparable with that in solution for duplex formation of the same oligonucleotide pair. Thus ku can be determined by Eqn
| 3 |
It should be noted that the two unfolding processes may not be identical because their environments are different. Unfolding of the G-quadruplex in the pore might be affected by the steric constraint in nano-confinement. For example, an earlier report has shown that the activation entropy for the single DNA duplex dissociation in the nanopore is 40% lower than that for a similar dissociation process in the solution (69).
For the folding rate constant, it is barely possible to capture the event for the linear TBA folding into a G-quadruplex during its translocation in 102–103 µs (Table 1). However, because the equilibrium constant [Equation (2)] and unfolding rate constant [Equqtion (3)] have been determined, the folding rate constant, kf, can be obtained from the definition
| 4 |
To compare with Kf (Figure 6a), the calculated kinetic constants kf and ku (Table 2) were plotted against the ionic radii (Figure 6c). Clearly, kf and ku no longer obey the ‘ionic radii’ law as Kf does. Particularly Li+ and Cs+ join with K+ and Ba2+ to possess the lowest ku values. ku was 0.061 s−1 in Ba2+, 0.066 s−1 in K+, 0.065 s−1 in Li+ and 0.082 in Cs+. Because the unfolding rate constant is an indication of the G-quadruplex stability, the G-quadruplexes formed by Ba2+, K+, Li+ and Cs+ should be the most stable. The G-quadruplex in NH4+ is less stable, with a ku value of 0.25 s−1, four times faster than in K+. The most unstable complex is the G-quadruplex in Na+, because it unfolds at ku = 2.9 s−1, forty times faster than in K+, and the fastest of all the G-quadruplexes. Overall, the cation sequence in the order of the quadruplex stability is Ba2+ ∼ K+ ∼ Li+ ∼ Cs+ > NH4+ > Na+.
For the G-quadruplex formation process, TBA did not fold the fastest in K+ or Ba2+, but in Na+ at kf = 6.5 s−1. The folding reaction was the slowest in Li+ at kf = 0.095 s−1. The difference between the two folding speeds is more than 60-fold. In between, the folding rate is 1.1 s−1 in NH4+, 0.46 s−1 in K+ and 0.25 s−1 in Ba2+ and 0.23 s−1 in Cs+. Therefore, the order of folding preference is Na+ > NH4+ > K+ > Ba2+ ∼ Cs+ > Li+.
Because there have been few reports on TBA folding and unfolding rate constants, we compared our data with published kinetic parameters for the telomere quadruplex (TTAGGG)4. kf and ku for TBA in K+, Na+ and Li+ are higher than for the telomere quadruplex determined with SPR (29) and FRET (27,28) techniques. For example, the differences of kf and ku between the two quadruplexes in K+ are one order of magnitude (27–29). This variation could be due to the different quadruplex species investigated. The TBA quadruplex is a double G-tetrad complex, in contrast with the telomere quadruplex that comprises triple G-tetrads. The earlier study using time-dependent spectroscopy showed that TBA folds faster than an HIV-targeted triple G-tetrads aptamer (21), supporting the idea that the quadruplex with fewer tetrads (TBA) can be assembled and disassembled more quickly than one with more tetrads (such as telomere quadruplex).
The salt concentration may also affect the kinetics. Unlike the SPR (29) and FRET (27,28) measurements performed in 100–150 mM salt solutions, the nanopore was measured in a 1 M salt concentration. A higher salt concentration might accelerate the folding reaction, given the finding in the previous study that TBA in 10 mM KCl folds faster than in 1 mM KCl (21). Different kinetic results may also be because of the fact that the encapsulation of a single G-quadruplex in the nanopore nanocavity is a non-covalent process without DNA labeling, different from techniques that require labeled DNAs that may alter the kinetics (27–29).
Comparison of kf and ku suggested that both the folding and unfolding reactions demonstrate similar trends in the cation-dependence (Figure 6c). Although K+•TBA is one of the most stable quadruplexes, as marked by one of the lowest ku values, its folding rate is not the highest. kf in K+ is lower than in Na+ and NH4+. Therefore, the highest equilibrium constant Kf for K+•TBA is achieved mainly through slow unfolding. This result is somewhat different from in the K+-induced telomere quadruplex, because the high Kf value for the telomere quadruplex were shown to be contributed by both slow unfolding and fast folding reactions (29). Furthermore, although the equilibrium constants Kf for Na+•TBA and Li+•TBA are similar, the kf and ku values for the two quadruplexes are remarkably different. Both the kf and ku values for Na+•TBA are the highest of all the quadruplexes, whereas both constants for Li+•TBA are the lowest, revealing that TBA performs the most rapid folding and unfolding reactions in Na+ and the slowest folding and unfolding reactions in Li+. The counteraction between the folding and unfolding reactions results in similar equilibrium constants for the two quadruplexes. Overall, comparison of folding and unfolding processes suggests that Na+ and Li+ play different roles in the kinetic pathway.
CONCLUSIONS AND PERSPECTIVES
Through this nanopore single-molecule study, we determined the cation's capacity for G-quadruplex formation and the cation's regulation of the folding and unfolding of the G-quadruplex. K+, Ba2+ and NH4+ are favorite cations over Cs+, Na+ and Li+ for forming G-quadruplexes with TBA, whereas Mg2+ and Ca2+ did not induce the formation of the G-quadruplex. The cation selectivity in G-quadruplex formation is correlated with the volume of the G-quadruplex, which varies with the cation species. The high formation capability of the K+-induced G-quadruplex may be largely due to the slow unfolding reaction. Although the Na+- and Li+-quadruplexes feature similar equilibrium properties, they undergo radically different pathways. The Na+-quadruplex folds and unfolds most rapidly, while the Li+-quadruplex performs both reactions at the slowest rates. Through this study, the nanopore is proven to be a useful single-molecule tool for probing molecular processes that enrich our understanding of the ion-regulated properties and processes of oligonucleotides. Meanwhile, it is also noted that the nanopore single-molecule method also has an applicability range. The calculation of Kf by counting the linear DNA passing through the nanopore could be more applicable to a ratio of the folded and unfolded TBA that is not too small and not too large, because of the smaller relative deviation of Kf in this range.
This information may prove useful for molecular recognition and design in the aptamer-target complex. The method used in this study may be expanded for the kinetic study of other quadruplexes and their variants. Potential targets include various biologically relevant intramolecular quadruplexes (24), such as the i-motif (quadruplexes formed by cytidine-rich sequences) (70) and chemically modified quadruplexes with unique functionalities (71). The contribution of each guanine to the quadruplex's folding capability may be detected by combining our guest-nanocavity approach with site-directed nucleotide substitution (25). Since the protein-DNA interaction has been probed using a nanopore-based molecular force detector (50,51), analog methods could be introduced for detecting target-quadruplex aptamer interactions. This work has already begun: the influence of thrombin on the encapsulation of the TBA G-quadruplex in the nanocavity has been observed (54). This research may also be helpful in constructing new molecular species with tunable properties for nano-constructions and the manufacture of biosensors (71).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This investigation was supported by NSF (0546165) and National Institutes of Health (GM079613), and was conducted in a facility constructed with support from Research Facilities Improvement Program Grant C06-RR-016489-01 from the National Center for Research Resources, National Institutes of Health. Funding for open access charges: National Institutes of Health.
Conflict of interest statement
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Kent Gates’ laboratory for the great help with the measurement of melting curve of TBA, and Dr Luis Marky's laboratory for the great help with the analysis of DNA melting curve and result interpretation.
REFERENCES
- 1.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hardin CC, Perry AG, White K. Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids. Biopolymers. 2000;56:147–194. doi: 10.1002/1097-0282(2000/2001)56:3<147::AID-BIP10011>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 3.Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 4.Phan AT, Kuryavyi V, Ma JB, Faure A, Andreola ML, Patel DJ. An interlocked dimeric parallel-stranded DNA quadruplex: a potent inhibitor of HIV-1 integrase. Proc. Natl Acad. Sci. USA. 2005;102:634–639. doi: 10.1073/pnas.0406278102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 6.Arthanari H, Bolton PH. Functional and dysfunctional roles of quadruplex DNA in cells. Chem. Biol. 2001;8:221–230. doi: 10.1016/s1074-5521(01)00007-2. [DOI] [PubMed] [Google Scholar]
- 7.Fletcher TM, Sun DK, Salazar M, Hurley LH. Effect of DNA secondary structure on human telomerase activity. Biochemistry. 1998;37:5536–5541. doi: 10.1021/bi972681p. [DOI] [PubMed] [Google Scholar]
- 8.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of Telomerase by G-Quartet Dna Structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 9.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hurley LH. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 11.Neidle S, Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nat. Rev. Drug Dis. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- 12.Kerwin SM. G-quadruplex DNA as a target for drug design. Current Pharma. Des. 2000;6:441–471. doi: 10.2174/1381612003400849. [DOI] [PubMed] [Google Scholar]
- 13.Davis JT, Spada GP. Supramolecular architectures generated by self-assembly of guanosine derivatives. Chem. Soc. Rev. 2007;36:296–313. doi: 10.1039/b600282j. [DOI] [PubMed] [Google Scholar]
- 14.Alberti P, Mergny JL. DNA duplex-quadruplex exchange as the basis for a nanomolecular machine. Proc. Natl Acad. Sci. USA. 2003;100:1569–1573. doi: 10.1073/pnas.0335459100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li JWJ, Tan WH. A single DNA molecule nanomotor. Nano Lett. 2002;2:315–318. doi: 10.1021/nl9011694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded-DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 17.Jing NJ, Zhu QQ, Weerasinghe P, Li YD. Targeting Stat3 with G-quartet oligonucleitides: a potentail novel therapy for human cancers. Clin. Cancer Res. 2005;11:9080S. [Google Scholar]
- 18.Tang ZW, Shangguan D, Wang KM, Shi H, Sefah K, Mallikratchy P, Chen HW, Li Y, Tan WH. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal. Chem. 2007;79:4900–4907. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- 19.Wang KY, Krawczyk SH, Bischofberger N, Swaminathan S, Bolton PH. The tertiary structure of a dna aptamer which binds to and inhibits thrombin determines activity. Biochemistry. 1993;32:11285–11292. doi: 10.1021/bi00093a004. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Patel DJ. Solution structure of a parallel-stranded g-quadruplex DNA. J. Mol. Biol. 1993;234:1171–1183. doi: 10.1006/jmbi.1993.1668. [DOI] [PubMed] [Google Scholar]
- 21.Jing NJ, Hogan ME. Structure-activity of tetrad-forming oligonucleotides as a potent anti-HIV therapeutic drug. J. Biol. Chem. 1998;273:34992–34999. doi: 10.1074/jbc.273.52.34992. [DOI] [PubMed] [Google Scholar]
- 22.Mao XA, Marky LA, Gmeiner WH. NMR structure of the thrombin-binding DNA aptamer stabilized by Sr2+ J. Biomol. Struct. Dynam. 2004;22:25–33. doi: 10.1080/07391102.2004.10506977. [DOI] [PubMed] [Google Scholar]
- 23.Kankia BI, Marky LA. Folding of the thrombin aptamer into a G-quadruplex with Sr2+: stability, heat, and hydration. J. Am. Chem. Soc. 2001;123:10799–10804. doi: 10.1021/ja010008o. [DOI] [PubMed] [Google Scholar]
- 24.Olsen CM, Gmeiner WH, Marky LA. Unfolding of G-quadruplexes: energetic, and ion and water contributions of G-quartet stacking. J. Phys. Chem. B. 2006;110:6962–6969. doi: 10.1021/jp0574697. [DOI] [PubMed] [Google Scholar]
- 25.Smirnov I, Shafer RH. Effect of loop sequence and size on DNA aptamer stability. Biochemistry. 2000;39:1462–1468. doi: 10.1021/bi9919044. [DOI] [PubMed] [Google Scholar]
- 26.Simonsson T, Sjoback R. DNA tetraplex formation studied with fluorescence resonance energy transfer. J. Biol. Chem. 1999;274:17379–17383. doi: 10.1074/jbc.274.24.17379. [DOI] [PubMed] [Google Scholar]
- 27.Green JJ, Ladame S, Ying LM, Klenerman D, Balasubramanian S. Investigating a quadruplex-ligand interaction by unfolding kinetics. J. Am. Chem. Soc. 2006;128:9809–9812. doi: 10.1021/ja0615425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ying LM, Green JJ, Li HT, Klenerman D, Balasubramanian S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA. 2003;100:14629–14634. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Y, Kan ZY, Zeng ZX, Hao YH, Chen H, Tan Z. Determining the folding and unfolding rate constants of nucleic acids by biosensor. Application to telomere G-quadruplex. J. Am. Chem. Soc. 2004;126:13255–13264. doi: 10.1021/ja048398c. [DOI] [PubMed] [Google Scholar]
- 30.Heyduk T, Heyduk E. Molecular beacons for detecting DNA binding proteins. Nat. Biotechnol. 2002;20:171–176. doi: 10.1038/nbt0202-171. [DOI] [PubMed] [Google Scholar]
- 31.Ho HA, Leclerc M. Optical sensors based on hybrid aptamer/conjugated polymer complexes. J. Am. Chem. Soc. 2004;126:1384–1387. doi: 10.1021/ja037289f. [DOI] [PubMed] [Google Scholar]
- 32.Huang CC, Cao ZH, Chang HT, Tan WH. Protein-protein interaction studies based on molecular aptamers by affinity capillary electrophoresis. Anal. Chem. 2004;76:6973–6981. doi: 10.1021/ac049158i. [DOI] [PubMed] [Google Scholar]
- 33.Padmanabhan K, Padmanabhan KP, Ferrara JD, Sadler JE, Tulinsky A. The structure of alpha-thrombin inhibited by a 15-mer single-stranded-DNA aptamer. J. Biol. Chem. 1993;268:17651–17654. doi: 10.2210/pdb1hut/pdb. [DOI] [PubMed] [Google Scholar]
- 34.Marathias VM, Bolton PH. Structures of the potassium-saturated, 2 : 1, and intermediate, 1 : 1,forms of a quadruplex DNA. Nucleic Acids Res. 2000;28:1969–1977. doi: 10.1093/nar/28.9.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bayley H, Cremer PS. Stochastic sensors inspired by biology. Nature. 2001;413:226–230. doi: 10.1038/35093038. [DOI] [PubMed] [Google Scholar]
- 36.Cheley S, Gu LQ, Bayley H. Stochastic sensing of nanomolar inositol 1,4,5-trisphosphate with an engineered pore. Chem. Biol. 2002;9:829–838. doi: 10.1016/s1074-5521(02)00172-2. [DOI] [PubMed] [Google Scholar]
- 37.Gu LQ, Braha O, Conlan S, Cheley S, Bayley H. Stochastic sensing of organic analytes by a pore-forming protein containing a molecular adapter. Nature. 1999;398:686–690. doi: 10.1038/19491. [DOI] [PubMed] [Google Scholar]
- 38.Movileanu L, Howorka S, Braha O, Bayley H. Detecting protein analytes that modulate transmembrane movement of a polymer chain within a single protein pore. Nat. Biotechnol. 2000;18:1091–1095. doi: 10.1038/80295. [DOI] [PubMed] [Google Scholar]
- 39.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl Acad. Sci. USA. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howorka S, Cheley S, Bayley H. Sequence-specific detection of individual DNA strands using engineered nanopores. Nat. Biotechnol. 2001;19:636–639. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- 41.Vercoutere W, Winters-Hilt S, Olsen H, Deamer D, Haussler D, Akeson M. Rapid discrimination among individual DNA hairpin molecules at single-nucleotide resolution using an ion channel. Nat. Biotechnol. 2001;19:248–252. doi: 10.1038/85696. [DOI] [PubMed] [Google Scholar]
- 42.Mathe J, Visram H, Viasnoff V, Rabin Y, Meller A. Nanopore unzipping of individual DNA hairpin molecules. Biophys. J. 2004;87:3205–3212. doi: 10.1529/biophysj.104.047274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakane J, Wiggin M, Marziali A. A nanosensor for transmembrane capture and identification of single nucleic acid molecules. Biophys. J. 2004;87:615–621. doi: 10.1529/biophysj.104.040212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashkenasy N, Sanchez-Quesada J, Bayley H, Ghadiri MR. Recognizing a single base in an individual DNA strand: a step toward DNA sequencing in nanopores. Angew. Chem.-Int. Ed. 2005;44:1401–1404. doi: 10.1002/anie.200462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Astier Y, Braha O, Bayley H. Toward single molecule DNA sequencing: direct identification of ribonucleoside and deoxyribonucleoside 5′-monophosphates by using an engineered protein nanopore equipped with a molecular adapter. J. Am. Chem. Soc. 2006;128:1705–1710. doi: 10.1021/ja057123+. [DOI] [PubMed] [Google Scholar]
- 46.Bezrukov SM, Vodyanoy I, Parsegian VA. Counting polymers moving through a single ion channel. Nature. 1994;370:279–281. doi: 10.1038/370279a0. [DOI] [PubMed] [Google Scholar]
- 47.Gu LQ, Dalla Serra M, Vincent JB, Vigh G, Cheley S, Braha O, Bayley H. Reversal of charge selectivity in transmembrane protein pores by using noncovalent molecular adapters. Proc. Natl Acad. Sci. USA. 2000;97:3959–3964. doi: 10.1073/pnas.97.8.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gu LQ, Cheley S, Bayley H. Electroosmotic enhancement of the binding of a neutral molecule to a transmembrane pore. Proc. Natl Acad. Sci. USA. 2003;100:15498–15503. doi: 10.1073/pnas.2531778100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luchian T, Shin SH, Bayley H. Single-molecule covalent chemistry with spatially separated reactants. Angew. Chem.-Int. Ed. 2003;42:3766–3771. doi: 10.1002/anie.200351313. [DOI] [PubMed] [Google Scholar]
- 50.Hornblower B, Coombs A, Whitaker RD, Kolomeisky A, Picone SJ, Meller A, Akeson M. Single-molecule analysis of DNA-protein complexes using nanopores. Nat. Methods. 2007;4:315–317. doi: 10.1038/nmeth1021. [DOI] [PubMed] [Google Scholar]
- 51.Tropini C, Marziali A. Multi-nanopore force spectroscopy for DNA analysis. Biophys. J. 2007;92:1632–1637. doi: 10.1529/biophysj.106.094060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shim JW, Gu LQ. Stochastic sensing on a modular chip containing a single-ion channel. Anal. Chem. 2007;79:2207–2213. doi: 10.1021/ac0614285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sauer-Budge AF, Nyamwanda JA, Lubensky DK, Branton D. Unzipping kinetics of double-stranded DNA in a nanopore. Phys. Rev. Lett. 2003;90:238101. doi: 10.1103/PhysRevLett.90.238101. [DOI] [PubMed] [Google Scholar]
- 54.Shim JW, Gu LQ. Encapsulating a single G-quadruplex aptamer in a protein nanocavity. J. Phys. Chem. B. 2008;112:8354–8360. doi: 10.1021/jp0775911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vairamani M, Gross ML. G-quadruplex formation of thrombin-binding aptamer detected by electrospray ionization mass spectrometry. J. Am. Chem. Soc. 2003;125:42–43. doi: 10.1021/ja0284299. [DOI] [PubMed] [Google Scholar]
- 56.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Rapid nanopore discrimination between single polynucleotide molecules. Proc. Natl Acad. Sci. USA. 2000;97:1079–1084. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meller A, Nivon L, Branton D. Voltage-driven DNA translocations through a nanopore. Phys. Rev. Lett. 2001;86:3435–3438. doi: 10.1103/PhysRevLett.86.3435. [DOI] [PubMed] [Google Scholar]
- 58.Martell AE. NIST Critically Selected Stability Constants of Metal Complexes Version 8.00. 2004. [Google Scholar]
- 59.Qiu XY, Andresen K, Kwok L, Lamb JS, Park HY, Pollack L. Inter-DNA attraction mediated by divalent counterions. Phys. Rev. Lett. 2007;99:038104. doi: 10.1103/PhysRevLett.99.038104. [DOI] [PubMed] [Google Scholar]
- 60.Song LZ, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 61.Lee JS. The stability of polypurine tetraplexes in the presence of monovalent and divalent-cations. Nucleic Acids Res. 1990;18:6057–6060. doi: 10.1093/nar/18.20.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raghuraman MK, Cech TR. Effect of monovalent cation-induced telomeric DNA-structure on the binding of oxytricha telomeric protein. Nucleic Acids Res. 1990;18:4543–4551. doi: 10.1093/nar/18.15.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hud NV, Smith FW, Anet FAL, Feigon J. The selectivity for K+ versus Na+ in DNA quadruplexes is dominated by relative free energies of hydration: a thermodynamic analysis by H-1 NMR. Biochemistry. 1996;35:15383–15390. doi: 10.1021/bi9620565. [DOI] [PubMed] [Google Scholar]
- 64.Wong A, Wu G. Selective binding of monovalent cations to the stacking G-quartet structure formed by guanosine 5′-monophosphate: a solid-state NMR study. J. Am. Chem. Soc. 2003;125:13895–13905. doi: 10.1021/ja0302174. [DOI] [PubMed] [Google Scholar]
- 65.Doyle DA, Cabral JM, Pfuetzner RA, Kuo AL, Gulbis JM, Cohen SL, Chait BT, Mackinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 66.Hille B. Ion Channels of Excitable Membranes. Sunderland, MA: Sinauer Associates; 2001. [Google Scholar]
- 67.Ma L, Melegari M, Colombini M, Davis JT. Large and stable transmembrane pores from guanosine-bile acid conjugates. J. Am. Chem. Soc. 2008;130:2938–2939. doi: 10.1021/ja7110702. [DOI] [PubMed] [Google Scholar]
- 68.Sidorov V, Kotch FW, El Khouedi M, Davis JT. Toward artificial ion channels: self-assembled nanotubes from calix[4]arene-guanosine conjugates. Chem. Commun. 2000:2369–2370. [Google Scholar]
- 69.Howorka S, Movileanu L, Braha O, Bayley H. Kinetics of duplex formation for individual DNA strands within a single protein nanopore. Proc. Natl Acad. Sci. USA. 2001;98:12996–13001. doi: 10.1073/pnas.231434698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gueron M, Leroy JL. The i-motif in nucleic acids. Curr. Opin. Struct. Biol. 2000;10:326–331. doi: 10.1016/s0959-440x(00)00091-9. [DOI] [PubMed] [Google Scholar]
- 71.Davis JT, Spada GP. Supramolecular architectures generated by self-assembly of guanosine derivatives. Chem. Soc. Rev. 2007;36:296–313. doi: 10.1039/b600282j. [DOI] [PubMed] [Google Scholar]
- 72.Lide DR. CRC Handbook of Chemistry and Physics. Boca Raton, FL: CRC Press; 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.