Abstract
Ribonuclease P (RNase P) is a ribonucleoprotein enzyme that contains a universally conserved, catalytically active RNA component. RNase P RNA requires divalent metal ions for folding, substrate binding, and catalysis. Despite recent advances in understanding the structure of RNase P RNA, no comprehensive analysis of metal-binding sites has been reported, in part due to the poor crystallization properties of this large RNA. We have developed an abbreviated yet still catalytic construct, Bst P7Δ RNA, which contains the catalytic domain of the bacterial RNase P RNA and has improved crystallization properties. We use this mutant RNA as well as the native RNA to map metal-binding sites in the catalytic core of the bacterial RNase P RNA, by anomalous scattering in diffraction analysis. The results provide insight into the interplay between RNA structure and focalization of metal ions, and a structural basis for some previous biochemical observations with RNase P. We use electrostatic calculations to extract the potential functional significance of these metal-binding sites with respect to binding Mg2+. The results suggest that with at least one important exception of specific binding, these sites mainly map areas of diffuse association of magnesium ions.
Keywords: RNA crystallography, RNA electrostatics, RNA–cation complex
INTRODUCTION
Ribonuclease P (RNase P) is a ubiquitous enzyme required for maturation of tRNA. RNase P catalyzes the endonucleolytic hydrolysis of a phosphodiester bond in the substrate RNA to generate the mature, phosphorylated 5′-end, with release of the 5′-leader sequence (for recent reviews, see Evans et al. 2006; Kazantsev and Pace 2006; Torres-Larios et al. 2006; Walker and Engelke 2006). RNase P is a ribonucleoprotein in vivo, but the substrate-binding and catalytic activities are conducted by the RNase P RNA (Guerrier-Takada et al. 1983; Pannucci et al. 1999; Kikovska et al. 2007). All RNase P RNAs are homologs of common ancestry, and they share a highly conserved structure that contains the active site of the ribozyme (Siegel et al. 1996).
RNase P RNA is a metalloenzyme, dependent for its function on divalent metal ions, naturally Mg2+. Mechanistic studies indicate that RNase P RNA catalyzes an SN2-like trans-esterification reaction, with two or three divalent metal ions bound to the transition state (Smith and Pace 1993; Beebe and Fierke 1994; Persson et al. 2003), a mechanism common to all large ribozymes as well as many protein-based phosphoryl transfer enzymes (Steitz and Steitz 1993; Brautigam and Steitz 1998; Nowotny et al. 2005; Stahley and Strobel 2005; Stoddard 2005). Like other functional RNAs, RNase P RNA adopts a complex fold in order to achieve its biological functions of substrate recognition and catalysis. RNA is highly negatively charged, with one negative charge per inter-nucleotide phosphate. Consequently, electrostatic repulsion has considerable impact on RNA structure and function. Electrostatic stabilization of RNA structure is achieved by binding of cations, and both folding and substrate recognition by RNase P have been linked to the binding of metal ions (Pan 1995; Beebe et al. 1996; Fang et al. 1999; Kent et al. 2000). Thus, sites of metal associations are important elements of understanding the structure and function of this ribozyme.
Prior to the development of crystal structures of RNase P RNA, characterization of potential metal-binding sites in the RNase P RNA was limited to indirect methods such as metal ion-induced hydrolysis (Kazakov and Altman 1991; Zito et al. 1993; Ciesiolka et al. 1994; Kufel and Kirsebom 1996b; Brannvall et al. 2001; Kaye et al. 2002), nucleotide analog interference mapping (Harris and Pace 1995; Kazantsev and Pace 1998; Crary et al. 2002), or NMR experiments involving small RNA fragments (Schmitz and Tinoco 2000; Getz et al. 2006). Recent crystal structures (Krasilnikov et al. 2003, 2004) revealed a number of metal-binding sites, identified by anomalous scattering of Ba2+, Pb2+ ions, in the S-domain of RNase P RNA, a highly conserved, independently folding domain of the ribozyme that contributes to substrate recognition (Loria and Pan 1996). Some Os(III) hexamine binding sites also have been observed in the catalytic domain of the RNA, in the low-resolution structures of full-size RNase P RNAs (Kazantsev et al. 2005; Torres-Larios et al. 2005). However, no more comprehensive analysis of metal-binding sites in the catalytic core of RNase P RNA has been reported.
Full-size RNase P RNA has proven to be a poor target for crystallographic analysis of metal-binding sites because of the slow rate of crystal growth and, generally, a low success rate in crystallization experiments. To circumvent this problem, we developed a new crystallization target, Bst P7Δ RNA, which is comprised of the independently folding catalytic domain of the Bacillus stearothermophilus RNase P RNA. This truncated RNA readily crystallizes under low ionic strength conditions suitable for soaking experiments with several metals. We identify, by anomalous scattering, many novel metal-binding sites in the catalytic core of RNase P RNA and evaluate their potential functional significance in the context of the RNase P RNA structure and association with Mg2+, the physiological divalent cation.
RESULTS
Design and crystallographic studies of an abbreviated RNase P RNA
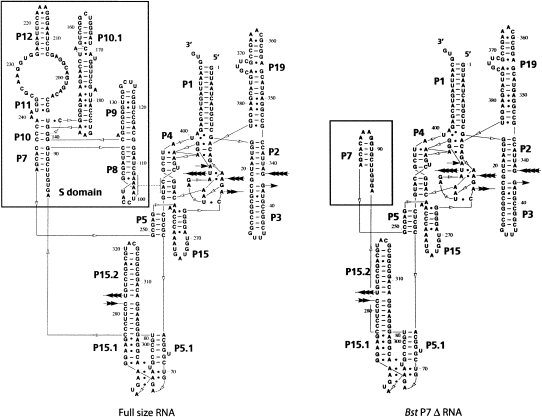
To design an RNA molecule with improved crystallization properties, we deleted, by means of molecular cloning, the regions of the RNA that are disordered in the crystal structure of the full-length B. stearothermophilus RNase P RNA. Several variants of such mutant RNAs were tested for their crystallization properties (Supplemental Fig. 1). One construct, Bst P7Δ, shown in Figure 1, is comprised of only the catalytic domain; all of the S-domain was deleted, and helix P7 was capped with a stabilizing GAAA tetraloop (see Fig. 1 legend for nomenclature of RNA structure). This construct is catalytically active (k cat/K m = 2 ×104 min−1/M−1 at optimal ionic strength 3.0 M potassium acetate, 200 mM magnesium chloride, 0.15 mM spermine tetrachloride, 10 mM Tris, 20 mM HEPES at pH 7.4, 37°C; k cat/K m of native RNAs is, typically, 6 × 107 min−1/M−1 at 1 M monovalent and 25 mM divalent cation at 50°C) and responds to activation by RNase P protein at low ionic strength. The Bst P7Δ RNA readily crystallized from a variety of low and high ionic strength conditions, and crystals diffracted to 3.6 Å resolution.
FIGURE 1.
Secondary structure of the Bst P7 Δ construct compared with the structure of the full-size RNA. Paired regions are labeled P1, P2, etc., in order of their appearance from the 5′-end in the full-size RNA. Inter-helical joining regions are named according to the names of the paired regions that they connect; e.g., J3/4 stands for nucleotides joining P3 and P4. Loops are labeled according to the names of the helices that they cap; e.g., L8 caps helix P8.
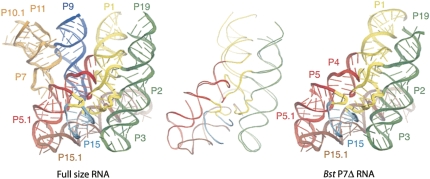
The structure of the Bst P7Δ construct was solved by standard crystallographic techniques from an Os(III) hexamine derivative as described in Materials and Methods and summarized in Supplemental Table 1. As shown in Figure 2, the refined structure of Bst P7Δ RNA is nearly identical to that of the catalytic domain of the full-size RNase P RNA (RMSD for all superimposable atoms is 2.7 Å; for conserved core atoms, 0.75 Å), with the main differences attributed to poorly ordered regions of peripheral helices P1, P3, and P19. An example of the model fit to the electron density map is shown in Supplemental Figure 2.
FIGURE 2.
Crystal structure of the Bst P7Δ construct (PDB ID 3DHS, right) compared with the structure of the full-size RNA (PDB ID 2a64, left). The elements of the secondary structure are labeled and colored accordingly between two different structures. The panel in the middle shows backbone traces of the superimposed structures of the Bst P7Δ RNA and corresponding portion of the full-size RNA.
Mg2+, the physiological divalent cation with RNase P, cannot be seen at the relatively low diffraction limit of the available crystals of RNase P RNA and its derivatives. Consequently, we scanned a variety of di- and trivalent metal ions for their ability to generate anomalously scattering derivatives that could be used to identify metal-binding sites in the Bst P7Δ structure. These included Mn2+, Co2+, Ni2+, Cu2+, Au3+, Pb2+, and the lanthanides (La3+, Pr3+, Sm3+, Gd3+, Dy3+, Yb3+, Lu3+), in addition to Os(III) hexamine used in structure determination. These ions, different in size, charge, and polarizability, are not specific proxies for Mg2+. They can bind to RNA differently than does Mg2+, for instance, through formation of different coordination complexes with RNAs that are formed by Mg2+. Nonetheless, identification of binding sites for these ions can serve to indicate sites in the RNA that attract metals, including potentially Mg2+.
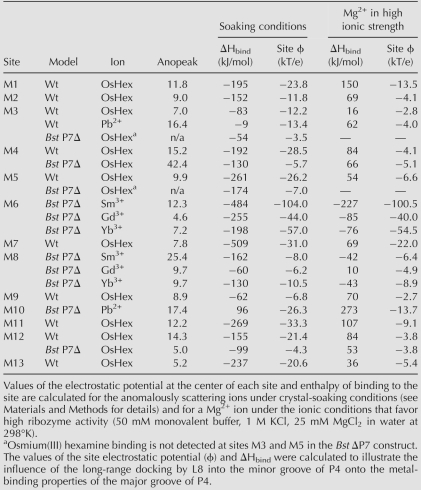
With some of the ions tested, we failed to find cryoprotecting and soaking conditions that would result in a useful derivative. Typical outcomes included loss of diffraction properties as a result of a soaking experiment, or absence of anomalous signal. Overall, five metals, Pb2+, Sm3+, Gd3+, Yb3+, and Os(III) hexamine, produced anomalously diffracting derivatives (Table 1). Metal-binding sites were identified as peaks in anomalous difference maps. We omit from discussion sites involved in crystalline lattice contacts or sites not well defined by anomalous signals.
TABLE 1.
Metal-binding sites in the catalytic core of the bacterial RNase P RNA: Strength of anomalous signal and electrostatic calculations
Overall distribution of metal-binding sites
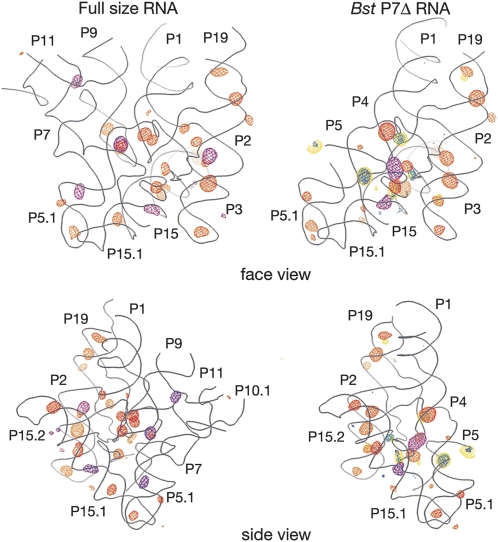
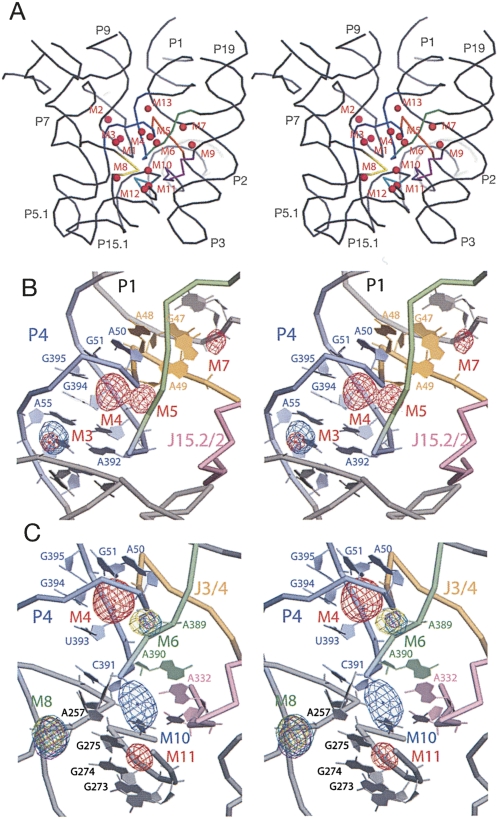
As illustrated in Figure 3, anomalous scattering by cations is localized primarily at the major grooves of helices and at sites where the RNA backbone adopts unusually tightly packed conformations, as occur particularly in the core of the catalytic domain. Despite differences in crystallization and soaking conditions, there is good correlation in the structural distribution of scattering by Os(III) hexamine between the full-size and Bst P7Δ RNA: 12 out of the 15 strongest peaks found in the Bst P7Δ derivative overlap with 12 out of 26 peaks found in the derivative of the full-size RNA. Scattering by lanthanides, analyzed only with the Bst P7Δ construct, does not overlap with the scattering by Os(III) hexamine, although all three ions (Sm3+, Gd3+, and Yb3+) tend to occupy the same sites. This might reflect differences in the coordination chemistry between the lanthanides and Os(III) hexamine, for instance the inability of the Os(III) hexamine to engage in inner-sphere coordination with RNA ligands. Overall, based on the anomalous scattering analysis with the two RNAs, we identify 13 well-resolved metal-binding sites in the catalytic core of the RNase P RNA, shown in Figure 4A (structural details are available in Supplemental Table 2). In the following sections, we describe some of these metal-binding sites that have particular relevance to RNase P RNA structure and function, and biochemical correlations.
FIGURE 3.
Overall distribution of the anomalous scattering. Backbone traces for the models of the full-size RNA and the Bst P7Δ construct are shown as gray wire; two orientations for each construct are shown. (Orange mesh) Anomalous scattering by Os(III) hexamine, (purple mesh) scattering by Pb2+, (yellow mesh) scattering by Sm3+, (blue mesh) scattering by Gd3+, (green mesh) scattering by Yb3+. The RNA-phased anomalous scattering difference maps are contoured at 4.5σ for both derivatives of the full-size RNA and at 4σ for the derivatives of Bst P7Δ, except for the Pb2+-derivative of the Bst P7Δ RNA that is contoured at 5σ.
FIGURE 4.
Metal-binding sites in the catalytic core of the RNase P RNA. (A) Stereo view of the distribution of the core metal-binding sites in the context of the RNase P RNA structure. Highly conserved elements of the structure are highlighted: P4 (blue), J3/4 (orange), J5/15 (yellow), J15/15.1 (teal), CR IV (purple), and J19/4 (green). (B) Stereo view of cation binding at the major groove of helix P4 in the full-size RNase P RNA structure. Anomalous scattering by Pb2+ (contoured at 7σ, blue mesh) and Os(III) hexamine (5σ, red mesh) are shown. Highly conserved elements of the structure: (light blue) P4, (orange) J3/4, (light purple) CR IV, (light green) J19/4. (C) Stereo view of cation binding at the major groove of helix P4 in the Bst P7Δ structure. Anomalous scattering by Pb2+ (contoured at 8 σ, blue mesh) and Os(III) hexamine (7σ, red mesh), Sm3+ (8σ, teal), Gd3+ (4σ, purple), and Yb3+ (5σ, yellow) are shown. Conserved RNA structure elements are colored as above.
Conserved structural center of RNase P RNA (M3–M7, M9, M13)
Helix P4 is formed by long-range pairing of two of the most highly conserved sequences in RNase P RNA and serves as a structural organizing center for the catalytic domain of the RNA. Because of its unusually high conservation among RNase P RNAs, helix P4 and its flanking regions (J3/4 and J19/4) have long been considered a part of the catalytic center of the ribozyme. In the crystal structures, the major groove of P4 is oriented toward the substrate-binding face of the ribozyme, and the flanking sequences are involved in a set of complicated interactions that structurally organize the catalytic center (Kazantsev et al. 2005). Helix P4 is essentially a canonical A-form helix with a bulged U residue (U52) that opens the major groove of the helix to host several metal-binding sites. Metal population of this part of the RNA is seen with osmium hexamine and Pb2+ in the native RNA, and osmium hexamine and the lanthanides in the abbreviated RNA.
Sites in common between the native and mutant RNAs have the same metal coordination features, but the full-length RNA apparently binds metals more avidly than does the mutant RNA. This is seen, for example, with metal-binding sites M3 and M4, which are located adjacent to one another in the major groove of helix P4 (Fig. 4B,C). Site M3, formed primarily by the Hoogsteen faces of bases A55, G57, and G254, is populated by Os(III) hexamine and Pb2+-ions in crystals of the full-size RNA. In contrast, anomalous signal from either ion falls below the threshold of noise in the crystals of Bst P7Δ RNA; the cations bind less tightly, if at all, to that site in the mutant RNA. On the other hand, the adjacent metal-binding site M4 is populated by Os(III) hexamine in both RNAs. Another example of preferential metal binding by the full-length RNA in this locale of the tertiary structure is site M5, located just outside of the major groove of P4, sandwiched by phosphates of U393 and G394 in helix P4 and by phosphates of G47 and A49, adjacent to helix P4 (J3/4, Fig. 4B). This site is occupied by Os(III) hexamine only in crystals of full-sized RNA. The structural reason for these apparently different metal-binding capacities of the native and abbreviated RNAs may lie in the tertiary structure (below).
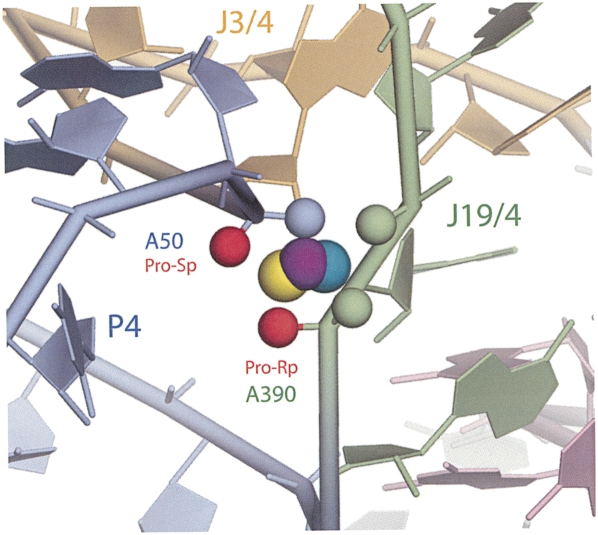
Particularly close backbone packing occurs at the interface between P4 and J19/4, where phosphate atoms of three universally conserved nucleotides (A50, A389, and A390) are brought together within 4.7–4.0 Å of each other. This creates a strongly electronegative center, and we identify a corresponding, compact metal-binding site, M6. As illustrated in Figure 4C, this site is occupied by lanthanide ions (Sm3+, Gd3+, Yb3+) in crystals of the Bst P7Δ RNA. The site is formed primarily by the pro-Rp and pro-Sp nonbridging phosphate oxygens of A50 and A390, as well as by the pro-Sp oxygen of A389 (Supplemental Table 2). This finding provides a structural context and corroboration for previous phosphorothioate modification-interference results that showed the importance of A50 and A390 phosphates in RNase P function (Harris and Pace 1995; Christian et al. 2002; Crary et al. 2002). Those earlier results were interpreted to indicate that these phosphates might participate in catalysis. However, the M6 metal-binding site is some distance (12 Å) from the expected center of chemical catalysis, as collectively painted by numerous photoaffinity cross-linking studies (Burgin and Pace 1990; Kufel and Kirsebom 1996a; Christian et al. 1998; Pomeranz Krummel and Altman 1999; Zahler et al. 2003; Kazantsev et al. 2005), and it is hard to imagine its direct involvement in the chemistry of the reaction without invoking a rather major conformational rearrangement of the enzyme. Nonetheless, site M6 is positioned on the surface of the substrate-binding face of the ribozyme, and is one of the most highly conserved and structurally conspicuous features in the RNase P RNA (see Discussion).
The details of other metal-binding sites in this central portion of the RNase P RNA, some associated with highly conserved structural elements and thereby potentially important sites for the function of the ribozyme, are provided in Supplemental Table 2.
Metal binding promoted by long-range tertiary structure
The loop of Helix P8 (L8) docks into the minor groove of Helix P4 and serves to correctly position the S-domain of RNase P RNA with respect to the catalytic domain (Loria and Pan 1996). This structure is conserved with some variation in all RNase P RNAs (Massire et al. 1998). The fold of loop L8 in the B. stearothermophilus structure, which docks into helix P4, is dominated by two cross-strand purine stacks that position four adenine nucleotides in the loop (A99, A100, A106, A107) for interaction with the minor groove of helix P4. Four consecutive A-minor interactions (Nissen et al. 2001) engage the interacting backbones in consecutive ribose zippers (Tamura and Holbrook 2002). The resulting conformation of loop L8 creates a very narrow major groove, with distances between phosphorous atoms across the groove reduced to 8.6–10 Å, from ∼18.2 Å typical of the canonical A-form helix. This proximity of the phosphates is expected to create a strongly electronegative center; consistent with this expectation, we identify metal-binding sites M1 and M2 in this region, both occupied by Os(III) hexamine ions and only in the full-length RNA (Supplemental Fig. 3). The abbreviated Bst P7Δ RNA lacks helix P8 and so does not form the tertiary structure that creates these metal-binding sites. Site M1 is formed primarily by the nonbridging phosphates of nucleotides C97 and U105, and by the Hoogsteen face of nucleotide G98. Site M2 is more compact, with the closest ion-binding partners being the nonbridging phosphates of nucleotides A99, A100 (pro-Rp oxygen), and A104; the Hoogsteen face of A104; and the ribose of C102 (see Supplemental Table 2 for a detailed list of all potential contacts for each of the metal-binding sites).
The docking of loop L8 into helix P4 contributes not only to the structural organization of the RNA, but also is expected to deliver electronegative potential to that portion of the RNA, enhancing the metal-binding capacity. This electronegative potential is expected to propagate into the major groove of helix P4 and possibly explains the apparently greater affinity of the P4 major groove of the full-length RNA for metals than is seen with the abbreviated mutant RNA. These suggested differences in the electrostatic properties of the full-length and mutant RNAs that may be manifest in metal-binding capacity also are consistent with electrostatic calculations (below).
The catalytic center of RNase P (M10–M12)
The loop of Helix P15 binds the 3′-terminal CCA of tRNA substrates and helps position the substrate phosphate, in the opposite strand of the substrate helix, at the catalytic site. Helix P15 is connected to the catalytic core of the ribozyme by two highly conserved trinucleotide linkers, J5/15 and J15/15.1, which interact with other conserved elements to organize the proposed catalytic center of the ribozyme. Sharp kinks in these linkers and other bonds, and the close packing in this portion of the molecule, bring several phosphate groups into close apposition to result in metal-binding sites. For instance, a sharp backbone kink between nucleotides A257 and C258 in the J15/15.1 linker accommodates metal-binding site M8, occupied by lanthanide ions (Sm3+, Gd3+, and Yb3+) in crystals of the Bst P7Δ RNA (Fig. 4B). This site is formed by nonbridging phosphate oxygens of these adjacent nucleotides, packed within 2.6 Å of each other.
Metal-binding site M10 is of particular interest because it is located at the center of the area identified as the chemically active site of the ribozyme by a variety of biochemical methods (Burgin and Pace 1990; Kufel and Kirsebom 1996a; Christian et al. 1998; Pomeranz Krummel and Altman 1999; Zahler et al. 2003). Site M10 is formed where a backbone kink between nucleotides A255 and A256 in the J5/15 linker brings the phosphate group of A256 within 6.1 Å from the phosphate of C391. This is a structural junction of some of the most conserved elements in RNase P RNAs, including CR IV, J19/4, J5/15, and J15/15.2 (Fig. 4C). Site M10 exhibits anomalous scattering by Pb2+ in crystals of the Bst P7Δ RNA. The Pb2+-binding site is formed by pro-Rp nonbridging phosphate oxygens of nucleotides A255 and A256, along with exocyclic O6 of highly conserved G275, the Hoogsteen face of A332, and both exocyclic carbonyls of U333 of CR IV (Supplemental Table 2). Close by, and partially overlapping with site M10, are sites M11, identified in the native RNA by occupancy with osmium hexamine, and M12, seen in both the native and mutant RNAs with osmium hexamine. This locale of the RNA evidently has a high affinity for metals, even in the absence of a docked substrate molecule. Occlusion by a substrate would be expected to increase substantially the local electronegative potential and consequently the affinity for metals.
Electrostatic calculations
We do not take these metals that we can see by anomalous scattering as specific proxies for Mg2+, the natural divalent cation for RNase P. Indeed, most Mg2+ associates with RNA diffusely and usually is not seen crystallographically. Instead, we interpret these binding sites to indicate localities in the RNA with a particular capacity to attract metals in general, including potentially Mg2+.
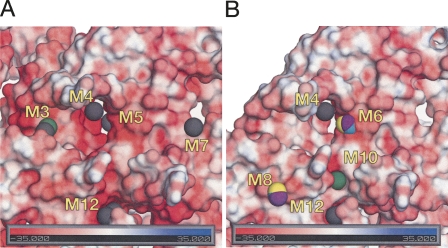
Metal ion affinity of RNA structure is expected to distribute with the electrostatic properties of the solvent-accessible surface. Consequently, to assess if the observed metal-binding sites indeed map areas that are electrostatically favorable for cation binding, we performed electrostatic calculations by numerically solving a nonlinear Poisson–Boltzmann equation for each of the RNA molecules (see Materials and Methods). We initially correlated the distribution of electrostatic potential on the molecular surface and the occurrence of the peaks of anomalous scattering. As illustrated in Figure 5, the metal-binding sites indeed are found in crevices in the RNA surface, where the electrostatic potential is calculated to assume unusually high negative values. As summarized in Supplemental Table 2, peak values of the calculated electrostatic potential on the surface of the metal-binding sites M1–M13 range from −29 to −62 kT/e under the solvent conditions used in the soaking experiments, with most values distributed between −30 and −50 kT/e. This correlates with the negative values of the electrostatic potentials at the center of the metal-binding site (compiled in Table 1).
FIGURE 5.
Cation binding follows the distribution of electrostatic potential on the molecular surface of RNase P RNA. (A) Cation binding to the catalytic core of the full-size RNase P RNA (substrate-binding face). (Gray) Os(III) hexamine peaks, (green) Pb2+ peak. (B) Cation binding to the core of Bst P7Δ RNA. Same orientation as above. (Teal) Sm3+ peaks, (purple) Gd3+ peaks, (yellow) Yb3+ peaks.
We next estimated the electrostatic contribution to the free energy of cation binding under the soaking conditions by calculating, for each bound cation, the difference between the total electrostatic energy of the RNA–cation complex and the unbound states of RNA and the ion. Such calculations are limited by the inability to accurately account for potential competition of different cationic species for the same binding site and for energetically unfavorable entropy changes upon ion binding (Misra et al. 2003) that may be significant. These results are summarized in Table 1. With the single exception of site M10, binding of the anomalously scattering ions to their respective sites is found to be highly electrostatically favorable. This result validates to some extent our use of anomalous scattering ions to map the electrostatic surface for potential binding sites for metals in general. The correspondence between observed binding sites and calculated electrostatic fields also encourages confidence in the application of the electrostatic theory to such a complex system as a large folded RNA in a multicomponent solution.
We next asked whether any of these metal-binding sites are favorable for binding of Mg2+ ions, which differ in charge, size, desolvation energy, and polarizability from the anomalously scattering ions used in this study. Most of the metal-binding sites identified in the catalytic face of RNase P RNA appear to be energetically unfavorable (positive ΔHbind) for specific binding of magnesium under ionic conditions that favor high catalytic activity of the ribozyme (Table 1). Any association with Mg2+ is predicted to be diffuse. The exceptions are the two metal-binding sites M6 and M8, identified by the anomalous scattering of lanthanide ions. Site M6, implicated in RNase P RNA function by phosphorothioate interference studies (above), shows significant enthalpy gain upon binding of magnesium ion (Table 1, up to −227 kJ/mol), consistent with the potential for specific binding of Mg2+ as indicated by modification-interference studies (see Discussion). Site M8, also in the substrate-binding face of the RNA, is only marginally favorable for specific binding (Table 1, up to −43 kJ/mol).
DISCUSSION
Rational design of RNA constructs for crystallization usually involves identification of independently folding domains (Cate et al. 1996; Correll et al. 1998; Kieft et al. 2002) or introduction of crystallization modules at the periphery of RNA to promote lattice contacts (Ferré-D'Amaré et al. 1998). In the case of RNase P RNA, biochemical characterization of the domain structure contributed to crystallization and structure determination of two phylogenetically distinct variants of the S-domain (Krasilnikov et al. 2003, 2004). Here, we sought to derive an RNase P RNA with an intact catalytic domain, but with improved crystallization properties compared with the native RNA. This was approached by excising the native RNA substructures that are poorly ordered in crystals of the full-size RNA.
The winning construct, Bst P7Δ RNA, indeed has improved crystallization properties compared with the full-size RNA, including a relative ease of nucleation, much faster growth rate (a few weeks compared with several months for the full-size RNA), and the ability to crystallize from low ionic strength conditions, while retaining essentially the same structure as the catalytic domain of the full-size RNA. The diffraction limit of the crystals of the best of the designed constructs, Bst P7Δ, did not exceed that of the original crystallization target, full-size RNase P RNA from B. stearothermophilus. However, the faster growth rates of the Bst P7Δ crystals and the higher success rate of the crystallization trials made it a good target for study of metal binding to the catalytic domain of RNase P RNA. Almost all crystallization drops yield diffraction-quality crystals of the Bst P7Δ RNA under optimal conditions, while only ∼5% of drops yielded crystals of the full-size RNA. Diffraction analysis of the anomalously scattering lanthanide derivatives of Bst P7Δ RNA, unavailable in the case of the full-size RNA, identified many metal-binding sites in the catalytic core of the RNA that had not previously been observed.
Overall, we identified 13 unambiguous metal-binding sites in the catalytic core of B. stearothermophilus RNase P RNA. Eleven of these sites are formed by phylogenetically highly conserved structural elements of the RNase P RNA (summarized in Supplemental Table 2), and therefore are expected to be populated by cations to some extent in any bacterial RNase P RNA. This prediction is supported by crystallographic studies of the T. maritima RNase P RNA (Torres-Larios et al. 2005), where the structural homolog of metal-binding site M3, in the major groove of helix P4 (the only one that could be modeled due to low resolution of data), was found to host an osmium hexamine ion. Of course, we acknowledge that magnesium may not occupy the sites of anomalous scattering, and we cannot observe magnesium at the current resolution. Consequently, we asked which of the 13 metal-binding sites that we observe in the core are thermodynamically favorable for binding of the Mg2+ ions that are known to be required for the catalytic activity of RNase P.
A thermodynamic framework for cation interaction with RNA has been well developed by Draper and colleagues (Draper 2004; Draper et al. 2005). Binding of metal ions with closed-shell electron structure, such as Na+, K+, and Mg2+, is primarily influenced by electrostatic interactions with RNA, water, and other ions in the solution. While providing a structurally oversimplified description, two binding modes suffice in adequately modeling the thermodynamics of RNA interaction with these ions: diffuse and specific. In this simple model, diffusely associated ions retain their hydration shell and are loosely associated with the nucleic acid, forming an ionic sheath around the biopolymer. The distribution of diffusely associated ions follows the value of the electrostatic potential around the RNA, with local concentrations of the cation increasing in areas of more negative surface potential.
Specific site binding, in turn, is accompanied by (partial) desolvation of the cation, which allows for compact packing of opposite charges upon binding to RNA, and thus can be electrostatically more favorable than diffuse association. In the case of Mg2+, however, the desolvation cost is very high, due to its small size and consequent high surface charge density. The most common mode of Mg2+ interaction with RNA is diffuse, and site binding can occur only in foci where the electrostatic potential reaches unusually high negative values (∼−60 kT/e or more) (Misra and Draper 2000, 2001; Soto et al. 2007). Despite its loose nature, however, diffuse association of cations with RNA is thermodynamically significant: thermodynamic stabilization of tRNA structure by Mg2+ ions can be adequately modeled by invoking only diffuse association (Misra and Draper 2000).
Despite limitations arising from the complexity of the structures, our calculations suggest that binding of the Os(III) hexamine, Pb2+, and lanthanides in the catalytic core of the bacterial RNase P RNA is mainly electrostatically consistent with true site binding. The only exception to this is the Pb2+ found at site M10. However, Mg2+ association at most of the identified sites is estimated to be diffuse, since the values of the electronegative potential at these sites is insufficient to overcome the large energetic cost of Mg2+ desolvation and release of diffusely bound ions. Thus, these metal-binding sites, although formed by highly conserved structural elements, are expected to represent areas of locally increased concentration of Mg2+ that are structurally important for electrostatic stabilization of the folded catalytic core of the RNA.
Metal-binding site M6 (Fig. 6) stands out in our analysis. This is the only site where specific Mg2+-binding is calculated to be clearly favorable. The site is formed by absolutely conserved elements of the RNase P RNA structure (helix P4 and the inter-helical joining region J19/4) and is accessible from the substrate-binding face of the ribozyme. A number of independent biochemical studies (Harris and Pace 1995; Christian et al. 2002; Crary et al. 2002) have implicated major constituents of this site, pro-Sp oxygen of A50 and pro-Rp oxygen of A390, in direct, inner-sphere coordination of a Mg2+ ion important for the ribozyme activity. Although direct involvement of this metal ion in the chemical step was suggested, structural interpretation of photoaffinity cross-linking experiments (Burgin and Pace 1990; Kufel and Kirsebom 1996a; Christian et al. 1998; Pomeranz Krummel and Altman 1999; Zahler et al. 2003; Kazantsev et al. 2005) suggests that the chemically active site in RNase P may be located ∼12 Å distant from site M6. Aside from hypothetical involvement in the chemistry of the reaction, the specific role of Mg2+ binding at metal-binding site M6 appears to be purely structural, to relieve electrostatic stress associated with the tight packing of the phosphate groups of highly conserved nucleotides A50, A389, and A390, which otherwise may interfere with the tight folding of the catalytic core.
FIGURE 6.
Metal-binding site M6. Lanthanide ions are shown as spheres: Sm3+ (teal), Gd3+ (purple), and Yb3+ (yellow). Nonbridging phosphate oxygen atoms involved in coordination of the lanthanide ions are also shown as spheres colored according to the encompassing structural elements; (highlighted red) atoms identified in phosphorothioate modification-interference experiments as directly coordinating Mg2+ ions.
Where in the RNase P structure are the binding sites for the catalytic magnesium ions? The best candidates at this time are in the vicinity of sites M10 and M11, located at the center of the proposed active site of the ribozyme according to the results of cross-linking experiments. Although in this study these sites are calculated to associate with Mg2+ only diffusely, this is in the absence of the substrate. Packing of the negatively charged substrate into the active site is likely to alter significantly the local electrostatic structure and potentially promote specific coordination of Mg2+ ions required for the catalytic step.
MATERIALS AND METHODS
RNA preparation and annealing
The transcription templates were derived from pBstHH2 (Kazantsev et al. 2005) by standard molecular cloning techniques, including PCR and enzymatic ligations. All resulting constructs bear cis-cleaving hammerhead ribozyme sequences at the 5′ and 3′ termini in order to reduce size heterogeneity of the target RNA upon transcription. All DNA constructs were verified by sequencing. RNA was transcribed in vitro as described elsewhere (Ke and Doudna 2004), purified by electrophoresis in a 4% denaturing polyacrylamide gel, and further purified by gradient ion exchange chromatography on HiTrap Q Sepharose (Amersham-GE). Eluted RNA was concentrated and desalted into 0.2 mM Na-EDTA with Centricon 50 (Millipore), filtered through a 0.2-μm Ultrafree-MC microcentrifuge filter (Millipore), and stored frozen at −20°C. RNA concentration was determined spectrophotometrically. Prior to crystallization, RNA species were annealed at 3–6 mg/mL (28–65 μM) in 10 mM MgCl2 for 2 min at 65°C followed by slow (35–40 min) cooling to 30°C. Crystallization leads were identified by a sparse matrix screen (Scott et al. 1995) and further improved by systematic variation of the crystallization conditions.
Optimized crystallization of Bst P7Δ RNA and heavy-atom soaks
The Bst P7Δ RNA was annealed at 2.6 mg/mL (28 μM) in 10 mM MgCl2 for 2 min at 65°C followed by slow (35–40 min) cooling to 30°C. To the annealed RNA sample, a 1:50 volume of melted 0.5% agarose in 0.2 mM EDTA, pH 8.0, (pre-equilibrated at 37°C) was added, and the sample was immediately mixed by vigorous vortexing. The annealed RNA sample (20 μL) was mixed with 20 μL of the reservoir (50 mM Na-cacodylate at pH 6.5, 100 mM KCl, 0.7–1.0 mM spermine tetrachloride, 21%–23% 1,6-hexanediol) and equilibrated by vapor diffusion against 1 mL of reservoir in the sealed Cryschem Plates (Hampton Research) at 23°C. Diffraction-quality RNA crystals grew in these conditions within a few weeks. Crystals were harvested by transfer to 4 mL of 50 mM Na-cacodylate at pH 6.5, 100 mM KCl, 20 mM MgCl2, 1 mM spermine tetrachloride, 23% 1,6-hexanediol and were cryoprotected by gradually increasing the concentration of 1,6-hexanediol to 31%. Lanthanide derivatives were prepared by soaking the cryoprotected crystals of Bst P7Δ RNA in the cryoprotectant solution containing 0.25 mM SmCl3, GdCl3, or YbCl3 for 6–8 h at 23°C. Pb2+-derivative was prepared as above, except the concentration of the heavy atom compound [Pb(II) acetate] was 10 mM and the concentration of MgCl2 was reduced to 10 mM. The Os(III) hexamine derivative was prepared by transferring the cryoprotected crystals of Bst P7Δ RNA into a solution containing 50 mM Na-N-(2-acetamido)iminodiacetate at pH 6.5, 100 mM KCl, 20 mM MgCl2, 0.30 mM Os(III) hexamine triflate, 1 mM spermine tetrachloride, and 31% 1,6-hexanediol and soaking for 3 d at 23°C. Derivatized crystals were flash-cooled by immersion in liquid nitrogen and stored in liquid nitrogen until data collection.
Data collection, processing, and model refinement
Data collection statistics for the full-size RNA crystals have been reported elsewhere (Kazantsev et al. 2005). Diffraction data from the Bst P7Δ RNA crystals were collected at 100 K at ALS beamlines 8.2.1 and 8.2.2, and SSRL beamline 9.2. Inverse beam strategy was employed to collect anomalous data for all derivatives. All data sets were processed with d*TREK 9 (Pflugrath 1999). Data processing statistics are presented in Supplemental Table 1.
Initial crystallographic phases were solved to 3.6 Å by molecular replacement with CNS (Brunger et al. 1998) using the data collected from the Os(III) hexamine derivative. The search model was generated from the model of the full-size RNA (PDB ID 2A64) by removing excess nucleotides. To lower model bias, an alternative phase solution was generated by a SAD experiment on the same data set. The two sets of phases were combined and the result was further improved by density modification by solvent flipping. Model analysis and rebuilding was performed with the program O (Jones et al. 1991). A typical round of model refinement included restrained simulated annealing by torsion dynamics, crystallographic conjugate gradient minimization, and grouped B-factor refinement. In the last rounds of refinement, the model was scrutinized against a composite annealed omit map (Hodel et al. 1992). After a final round of refinement, the model was deposited with PDB ID 3DHS. The final model statistics are outlined in Supplemental Table 1.
Electrostatic calculations
Electrostatic calculations were performed by solving a nonlinear Poisson–Boltzmann equation with APBS (Baker et al. 2001). Calculations employed a fixed multilevel grid procedure with grid spacing of ≤0.365 Å. The grid dimensions were 109.036 Å × 106.876 Å × 128.479 Å for both RNA models. This placed each RNA molecule inside a box that exceeded the RNA dimensions by at least 20 Å (estimated Debye length under conditions used for modeling did not exceed 8 Å). RNA was treated as a low dielectric medium (ε = 2; Misra and Draper 2001) within a volume enclosed by its water-accessible surface (probe radius 1.4 Å). Hydrogen atoms were added to crystallographic models, and the atomic radii and charges were assigned with pdb2pqr functionality of APBS (Dolinsky et al. 2004) according to AMBER force field parameters. To reduce the sensitivity of computations to the grid setup, cubic B-spline discretization was applied in mapping the RNA charges onto the grid points, and nine-point harmonic averaging was applied to the surface-based dielectric and ion-accessibility coefficients as implemented in APBS. Control calculations with a grid origin shifted by one-third grid spacing in any direction resulted in differences in free energy values by no more than 6%. The surrounding solvent was treated as a high dielectric continuum (ε = 78.5 for water-based solutions and ε = 65 for the solutions containing 30%–31% organic cryoprotectant). Diffusely associated electrolytes were distributed according to a Boltzmann weighted average of mean electrostatic potential, with the following ion-exclusion radii: acetate ion, 3.6 Å; Mg2+ ion, 2.0 Å; K+ ion, 2.2 Å; Cl− ion, 2.0 Å. Dirichlet boundary conditions were set using multiple Debye–Huckel functionality of APBS. The site-bound ions were treated using a Born model with the following radii: Mg2+, 1.45 Å (Misra and Draper 2001); Pb2+, 2.17 Å; Os(III) hexamine, 3.40 Å; Sm3+, 1.93 Å; Gd3+, 1.90 Å; Yb3+, 1.84 Å. The Born radii for lanthanide ions were estimated by adding an empirical value of 0.85 Å to the respective ionic radii (Latimer et al. 1939; Rashin and Honig 1985). The binding enthalpy was calculated as a difference in total electrostatic energy between a complex (RNA plus ion) and free RNA and free ion states, placed at the same respective locations on the grid.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Professor David Draper for fruitful and stimulating comments on the paper. This work has been supported by a grant from the National Institutes of Health.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1331809.
REFERENCES
- Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe J.A., Fierke C.A. A kinetic mechanism for cleavage of precursor tRNAAsp catalyzed by the RNA component of Bacillus subtilis ribonuclease P. Biochemistry. 1994;33:10294–10304. doi: 10.1021/bi00200a009. [DOI] [PubMed] [Google Scholar]
- Beebe J.A., Kurz J.C., Fierke C.A. Magnesium ions are required by Bacillus subtilis ribonuclease P RNA for both binding and cleaving precursor tRNAAsp . Biochemistry. 1996;35:10493–10505. doi: 10.1021/bi960870m. [DOI] [PubMed] [Google Scholar]
- Brannvall M., Mikkelsen N.E., Kirsebom L.A. Monitoring the structure of Escherichia coli RNase P RNA in the presence of various divalent metal ions. Nucleic Acids Res. 2001;29:1426–1432. doi: 10.1093/nar/29.7.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigam C.A., Steitz T.A. Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr. Opin. Struct. Biol. 1998;8:54–63. doi: 10.1016/s0959-440x(98)80010-9. [DOI] [PubMed] [Google Scholar]
- Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Burgin A.B., Pace N.R. Mapping the active site of ribonuclease P RNA using a substrate containing a photoaffinity agent. EMBO J. 1990;9:4111–4118. doi: 10.1002/j.1460-2075.1990.tb07633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate J.H., Gooding A.R., Podell E., Zhou K., Golden B.L., Kundrot C.E., Cech T.R., Doudna J.A. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- Christian E.L., McPheeters D.S., Harris M.E. Identification of individual nucleotides in the bacterial ribonuclease P ribozyme adjacent to the pre-tRNA cleavage site by short-range photo-cross-linking. Biochemistry. 1998;37:17618–17628. doi: 10.1021/bi982050a. [DOI] [PubMed] [Google Scholar]
- Christian E.L., Kaye N.M., Harris M.E. Evidence for a polynuclear metal ion binding site in the catalytic domain of ribonuclease P RNA. EMBO J. 2002;21:2253–2262. doi: 10.1093/emboj/21.9.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesiolka J., Hardt W.D., Schlegl J., Erdmann V.A., Hartmann R.K. Lead-ion-induced cleavage of RNase P RNA. Eur. J. Biochem. 1994;219:49–56. doi: 10.1111/j.1432-1033.1994.tb19913.x. [DOI] [PubMed] [Google Scholar]
- Correll C.C., Munishkin A., Chan Y.L., Ren Z., Wool I.G., Steitz T.A. Crystal structure of the ribosomal RNA domain essential for binding elongation factors. Proc. Natl. Acad. Sci. 1998;95:13436–13441. doi: 10.1073/pnas.95.23.13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crary S.M., Kurz J.C., Fierke C.A. Specific phosphorothioate substitutions probe the active site of Bacillus subtilis ribonuclease P. RNA. 2002;8:933–947. doi: 10.1017/s1355838202025025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky T.J., Nielsen J.E., McCammon J.A., Baker N.A. PDB2PQR: An automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper D.E. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper D.E., Grilley D., Soto A.M. Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- Evans D., Marquez S.M., Pace N.R. RNase P: Interface of the RNA and protein worlds. Trends Biochem. Sci. 2006;31:333–341. doi: 10.1016/j.tibs.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Fang X.W., Pan T., Sosnick T.R. Mg2+-dependent folding of a large ribozyme without kinetic traps. Nat. Struct. Biol. 1999;6:1091–1095. doi: 10.1038/70016. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré A.R., Zhou K., Doudna J.A. A general module for RNA crystallization. J. Mol. Biol. 1998;279:621–631. doi: 10.1006/jmbi.1998.1789. [DOI] [PubMed] [Google Scholar]
- Getz M.M., Andrews A.J., Fierke C.A., Al-Hashimi H.M. Structural plasticity and Mg2+ binding properties of RNase P P4 from combined analysis of NMR residual dipolar couplings and motionally decoupled spin relaxation. RNA. 2006;13:251–266. doi: 10.1261/rna.264207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Harris M.E., Pace N.R. Identification of phosphates involved in catalysis by the ribozyme RNase P RNA. RNA. 1995;1:210–218. [PMC free article] [PubMed] [Google Scholar]
- Hodel A., Kim S.-H., Brunger A.T. Model bias in macromolecular crystal structures. Acta Crystallogr. A. 1992;48:851–858. [Google Scholar]
- Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kaye N.M., Zahler N.H., Christian E.L., Harris M.E. Conservation of helical structure contributes to functional metal ion interactions in the catalytic domain of ribonuclease P RNA. J. Mol. Biol. 2002;324:429–442. doi: 10.1016/s0022-2836(02)01094-x. [DOI] [PubMed] [Google Scholar]
- Kazakov S., Altman S. Site-specific cleavage by metal ion cofactors and inhibitors of M1 RNA, the catalytic subunit of RNase P from Escherichia coli . Proc. Natl. Acad. Sci. 1991;88:9193–9197. doi: 10.1073/pnas.88.20.9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev A.V., Pace N.R. Identification by modification-interference of purine N-7 and ribose 2′-OH groups critical for catalysis by bacterial ribonuclease P. RNA. 1998;4:937–947. doi: 10.1017/s1355838298980384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev A.V., Pace N.R. Bacterial RNase P: A new view of an ancient enzyme. Nat. Rev. Microbiol. 2006;4:729–740. doi: 10.1038/nrmicro1491. [DOI] [PubMed] [Google Scholar]
- Kazantsev A.V., Krivenko A.A., Harrington D.J., Holbrook S.R., Adams P.D., Pace N.R. Crystal structure of a bacterial ribonuclease P RNA. Proc. Natl. Acad. Sci. 2005;102:13392–13397. doi: 10.1073/pnas.0506662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke A., Doudna J.A. Crystallization of RNA and RNA–protein complexes. Methods. 2004;34:408–414. doi: 10.1016/j.ymeth.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Kent O., Chaulk S.G., MacMillan A.M. Kinetic analysis of the M1 RNA folding pathway. J. Mol. Biol. 2000;304:699–705. doi: 10.1006/jmbi.2000.4263. [DOI] [PubMed] [Google Scholar]
- Kieft J.S., Zhou K., Grech A., Jubin R., Doudna J.A. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat. Struct. Biol. 2002;9:370–374. doi: 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- Kikovska E., Svard S.G., Kirsebom L.A. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc. Natl. Acad. Sci. 2007;104:2062–2067. doi: 10.1073/pnas.0607326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasilnikov A.S., Yang X., Pan T., Mondragon A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421:760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- Krasilnikov A.S., Xiao Y., Pan T., Mondragon A. Basis for structural diversity in homologous RNAs. Science. 2004;306:104–107. doi: 10.1126/science.1101489. [DOI] [PubMed] [Google Scholar]
- Kufel J., Kirsebom L.A. Different cleavage sites are aligned differently in the active site of M1 RNA, the catalytic subunit of Escherichia coli RNase P. Proc. Natl. Acad. Sci. 1996a;93:6085–6090. doi: 10.1073/pnas.93.12.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel J., Kirsebom L.A. Residues in Escherichia coli RNase P RNA important for cleavage site selection and divalent metal ion binding. J. Mol. Biol. 1996b;263:685–698. doi: 10.1006/jmbi.1996.0608. [DOI] [PubMed] [Google Scholar]
- Latimer W.M., Pitzer K.S., Slansky C.M. The free energy of hydration of gaseous ions, and the absolute potential of the normal calomel electrode. J. Chem. Phys. 1939;7:108–111. [Google Scholar]
- Loria A., Pan T. Domain structure of the ribozyme from eubacterial ribonuclease P. RNA. 1996;2:551–563. [PMC free article] [PubMed] [Google Scholar]
- Massire C., Jaeger L., Westhof E. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol. 1998;279:773–793. doi: 10.1006/jmbi.1998.1797. [DOI] [PubMed] [Google Scholar]
- Misra V.K., Draper D.E. Mg2+ binding to tRNA revisited: The nonlinear Poisson–Boltzmann model. J. Mol. Biol. 2000;299:813–825. doi: 10.1006/jmbi.2000.3769. [DOI] [PubMed] [Google Scholar]
- Misra V.K., Draper D.E. A thermodynamic framework for Mg2+ binding to RNA. Proc. Natl. Acad. Sci. 2001;98:12456–12461. doi: 10.1073/pnas.221234598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra V.K., Shiman R., Draper D.E. A thermodynamic framework for the magnesium-dependent folding of RNA. Biopolymers. 2003;69:118–136. doi: 10.1002/bip.10353. [DOI] [PubMed] [Google Scholar]
- Nissen P., Ippolito J.A., Ban N., Moore P.B., Steitz T.A. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny M., Gaidamakov S.A., Crouch R.J., Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: Substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Pan T. Higher-order folding and domain analysis of the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry. 1995;34:902–909. doi: 10.1021/bi00003a024. [DOI] [PubMed] [Google Scholar]
- Pannucci J.A., Haas E.S., Hall T.A., Harris J.K., Brown J.W. RNase P RNAs from some Archaea are catalytically active. Proc. Natl. Acad. Sci. 1999;96:7803–7808. doi: 10.1073/pnas.96.14.7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson T., Cuzic S., Hartmann R.K. Catalysis by RNase P RNA: Unique features and unprecedented active site plasticity. J. Biol. Chem. 2003;278:43394–43401. doi: 10.1074/jbc.M305939200. [DOI] [PubMed] [Google Scholar]
- Pflugrath J.W. The finer things in X-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- Pomeranz Krummel D.A., Altman S. Multiple binding modes of substrate to the catalytic RNA subunit of RNase P from Escherichia coli . RNA. 1999;5:1021–1033. doi: 10.1017/s1355838299990416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashin A.A., Honig B. Reevaluation of the Born model of ion hydration. J. Phys. Chem. 1985;89:5588–5593. [Google Scholar]
- Schmitz M., Tinoco I., Jr Solution structure and metal-ion binding of the P4 element from bacterial RNase P RNA. RNA. 2000;6:1212–1225. doi: 10.1017/s1355838200000881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott W.G., Finch J.T., Grenfell R., Fogg J., Smith T., Gait M.J., Klug A. Rapid crystallization of chemically synthesized hammerhead RNAs using a double screening procedure. J. Mol. Biol. 1995;250:327–332. doi: 10.1006/jmbi.1995.0380. [DOI] [PubMed] [Google Scholar]
- Siegel R.W., Banta A.B., Haas E.S., Brown J.W., Pace N.R. Mycoplasma fermentans simplifies our view of the catalytic core of ribonuclease P RNA. RNA. 1996;2:452–462. [PMC free article] [PubMed] [Google Scholar]
- Smith D., Pace N.R. Multiple magnesium ions in the ribonuclease P reaction mechanism. Biochemistry. 1993;32:5273–5281. doi: 10.1021/bi00071a001. [DOI] [PubMed] [Google Scholar]
- Soto A.M., Misra V., Draper D.E. Tertiary structure of an RNA pseudoknot is stabilized by “diffuse” Mg2+ ions. Biochemistry. 2007;46:2973–2983. doi: 10.1021/bi0616753. [DOI] [PubMed] [Google Scholar]
- Stahley M.R., Strobel S.A. Structural evidence for a two-metal-ion mechanism of group I intron splicing. Science. 2005;309:1587–1590. doi: 10.1126/science.1114994. [DOI] [PubMed] [Google Scholar]
- Steitz T.A., Steitz J.A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard B.L. Homing endonuclease structure and function. Q. Rev. Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- Tamura M., Holbrook S.R. Sequence and structural conservation in RNA ribose zippers. J. Mol. Biol. 2002;320:455–474. doi: 10.1016/s0022-2836(02)00515-6. [DOI] [PubMed] [Google Scholar]
- Torres-Larios A., Swinger K.K., Krasilnikov A.S., Pan T., Mondragon A. Crystal structure of the RNA component of bacterial ribonuclease P. Nature. 2005;437:584–587. doi: 10.1038/nature04074. [DOI] [PubMed] [Google Scholar]
- Torres-Larios A., Swinger K.K., Pan T., Mondragon A. Structure of ribonuclease P—A universal ribozyme. Curr. Opin. Struct. Biol. 2006;16:327–335. doi: 10.1016/j.sbi.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Walker S.C., Engelke D.R. Ribonuclease P: The evolution of an ancient RNA enzyme. Crit. Rev. Biochem. Mol. Biol. 2006;41:77–102. doi: 10.1080/10409230600602634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler N.H., Christian E.L., Harris M.E. Recognition of the 5′ leader of pre-tRNA substrates by the active site of ribonuclease P. RNA. 2003;9:734–745. doi: 10.1261/rna.5220703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito K., Huttenhofer A., Pace N.R. Lead-catalyzed cleavage of ribonuclease P RNA as a probe for integrity of tertiary structure. Nucleic Acids Res. 1993;21:5916–5920. doi: 10.1093/nar/21.25.5916. [DOI] [PMC free article] [PubMed] [Google Scholar]