In B-cell chronic lymphoid leukemia, patients with 13q14 deletion generally have a favorable outcome. The findings of this study suggest that the number of malignant cells with 13q14 deletion may influence the outcome of patients with this cytogenetic abnormality as a single chromosomal aberration. A high number of malignant cells carrying the 13q14 deletion, as assessed by FISH, appears to be associated with short overall survival and time to progression.
Keywords: B chronic lymphoid leukemia, 13q14 deletion, outcome, proliferation, apoptosis
Abstract
Background
Among patients with B-cell chronic lymphoid leukemia, those with 13q14 deletion have a favorable outcome. However, whether the percentage of cells with 13q- influences the prognosis or the biological characteristics of this disease is unknown. We analyzed the clinico-biological characteristics and outcome of patients with B-cell chronic lymphoid leukemia with loss of 13q as the sole cytogenetic aberration.
Design and Methods
Three hundred and fifty patients with B-cell chronic lymphoid leukemia were studied. Clinical data were collected and fluorescence in situ hybridization and molecular studies were carried out. In addition, a gene expression profile was obtained by microarray-based analysis.
Results
In 109 out of the 350 cases (31.1%) loss of 13q was the sole cytogenetic aberration at diagnosis. In the subgroup of patients with 80% or more of cells with loss of 13q (18 cases), the overall survival was 56 months compared with not reached in the 91 cases in whom less than 80% of cells had loss of 13q (p< 0.0001). The variables included in the multivariate analysis for overall survival were the percentage of losses of 13q14 (p=0.001) and B symptoms (p=0.007). The time to first therapy in the group with 80% or more vs. less than 80% of losses was 38 months vs. 87 months, respectively (p=0.05). In the multivariate analysis the variables selected were unmutated status of IgVH (p=0.001) and a high level of β2microglobulin (p=0.003). Interestingly, these differences regarding overall survival and time to first therapy were also present when other cut-offs were considered. The gene expression profile of patients with a high number of losses in 13q14 showed a high proliferation rate, downregulation of apoptosis-related genes, and dysregulation of genes related to mitochondrial functions.
Conclusions
Patients with B-cell chronic lymphoid leukemia with a high number of losses in 13q14 as the sole cytogenetic aberration at diagnosis display different clinical and biological features: short overall survival and time to first therapy as well as more proliferation and less apoptosis. A quantification of the number of cells showing a genetic abnormality should, therefore, be included in the study of the prognostic factors of B-cell chronic lymphoid leukemia.
Introduction
B-cell chronic lymphoid leukemia (B-CLL) is a heterogeneous disorder both from genetic and prognostic points of view, with some patients displaying an indolent course while others have an aggressive disease with short survival.1,2 In addition to classical prognostic factors,3 new parameters such as immunophenotypic markers (ZAP70, CD38 and CD49d antigens),4–6 molecular markers (mutational status of IGVH genes),7,11 and cytogenetics12–14 have been related to the prognosis in B-CLL.15
B-CLL patients show several cytogenetic aberrations, mainly in the regions of chromosomes 13q, 12, 11q, 17p, 14q and 6q. Some of these abnormalities can be better assessed by means of fluorescence in situ hybridization (FISH), which has shown that 62–80% of patients with B-CLL have cytogenetic abnormalities.10,12 These cytogenetic changes are strongly correlated with the prognosis in terms of overall survival and time to progression (defined as the time to first therapy).12,16–19 Patients with a deletion in 13q14 have a better outcome while patients with deletions in 11q23 or 17p13 have a shorter survival and shorter time to progression.12 Classically, patients with B-CLL and a normal karyotype or trisomy 12 have been considered to have an intermediate prognosis.12 It should, however, be noted that, in some series with a long-term follow-up, patients with B-CLL and a normal karyotype showed a better survival from 12 years on, as compared to patients with 13q-.9 In addition, several studies have demonstrated that the percentage of cells displaying a particular cytogenetic abnormality (e.g. loss of P53)20 or antigenic markers (e.g. CD38 or ZAP-70)7 can be related to prognosis.
For these reasons, we decided to perform an analysis of patients diagnosed with B-CLL and deletion in 13q14, as the sole cytogenetic abnormality. The clinical features, including outcome, and the biological features of the patients displaying different degrees of infiltration by 13q- cells were assessed. Moreover, to gain further insights into the molecular mechanisms involved in 13q14 deletion B-CLL, a gene expression profile study using a microarray-based analysis was also carried out.
Design and Methods
Patients
The study population comprised 350 non-selected patients, from nine Spanish institutions, diagnosed with B-CLL. The diagnosis of B-cell was made according to the World Health Organization (WHO) classification21 and Working Group of National Cancer Institute (NCI) criteria.22 Evidence of persistent lymphocytosis and a compatible immunophenotype were required for the diagnosis. In all cases an immunophenotypic analysis was performed by flow cytometry, including at least the following monoclonal antibodies: CD19, CD5, CD22, CD23, CD38, CD25, CD103, CD11c, FMC7, BCL2, CD10, CD20, and surface immunoglobulins κ and λ23. In addition, FISH studies, including specific probes for the regions 11q21, 12q13, 13q14, 14q32, and 17p13, were carried out. The study protocol was approved by local ethical committees and written informed consent was obtained from the patients.
Clinical data
Clinical data were recorded by reviewing the clinical histories of patients included in the study. In most cases (283 patients; 81%) the FISH study was performed at the time of diagnosis. In more than 95% (61 patients) of the remaining cases, (patients with a long follow-up), the FISH study was normal (25 patients) or showed alterations in 13q14 as the sole cytogenetic aberration (36 patients). Only six patients showed other cytogenetic alterations: two patients had 11q deletions, one had t(14q32) and three patients had two cytogenetic alterations (13q14 deletion plus t(14q32) in two cases and 13q14 deletion plus 11q deletion in the other one). Progression was defined according to previously reported criteria:24 the presence of disease-related symptoms, massive or progressive organomegaly, bone marrow failure or recurrent infections.
Fluorescence in situ hybridization
Interphase FISH was performed on bone marrow samples using commercially available probes for the following regions: 13q14, 12q13, 11q22/ATM, 17p13/P53, and 14q32/IGH (Vysis/Abbott Co, Downers Grove IL, USA). The methods used for the FISH analysis have been described elsewhere.25 14q32 translocations, trisomy 12 and deletions were considered to be present when ≤5%, ≤3% and ≤8% interphase cells showed a split signal, three signals and one signal, respectively. Dual-color FISH using differently-labeled control probes and test probes was performed and signal screening was carried out on at least 200 cells with well-delineated signals. Hybridization was repeated on those slides with less than 80% cells showing two control-probe signals.
Mutation status of IGVH genes
Amplification and sequencing of IGVH genes was performed according to the ERIC recommendations on IGHV gene mutational status analysis in B-CLL.26 Cases were classified as IGVH unmutated if there was at least 98% concordance between the tumor DNA and the respective family sequence, and IGVH mutated if there was less than 98% concordance.
Statistical analysis
Statistical tests were performed with SPSS 13.0 (SPSS, Chicago, IL, USA). The χ2 test was used to assess associations between categorical variables, while continuous variables were analyzed with the Kruskal-Wallis test. The variables with statistical significance related to overall survival and time to first therapy were calculated by the Kaplan-Meier method (log-rank). Results were considered statistically significant for p values ≤0.05. Multivariate analysis of survival and time to first therapy was performed using the Cox regression method.
Gene expression profile analysis
Patients and samples
Bone marrow samples were obtained from 37 patients with B-CLL and deletion of 13q14 as the sole cytogenetic aberration at diagnosis. Fifteen had more than 80% of 13q- cells, while the remaining 22 cases had less than 80% of 13q- cells in the bone marrow. Mononuclear cells from all samples were isolated using Ficoll gradient, snap frozen and stored at −80ºC. Both groups of patients showed more than 80% of clonal B-cell lymphocytes. RNA isolation, labeling and microarray hybridization were performed as previously reported.27
Normalization, signal calculation, significant differential expression, and sample/gene profile clustering
A robust microarray analysis algorithm was used for background correction, intra- and inter-microarray normalization, and expression signal calculation.28–30 Once the absolute expression signal for each gene (i.e., the signal value for each probe set) had been calculated for each microarray, a method, called significance analysis of microarray,31 was applied to calculate significant differential expression and find the gene probe sets that characterized the samples of each compared state. This method uses permutations to provide robust statistical inference of the most significant genes and provides p values adjusted to multiple testing using the false discovery rate (FDR).32 A FDR of less than 0.05 was used for all the differential expression calculations. Finally, the resulting lists of candidate genes associated to a high degree with 13q14 band deletion were tested using another algorithm, the so-called global test,33 which reveals the group of genes that has a global expression pattern most significantly related to the clinical feature studied. We applied all these methods using R and Bioconductor.
The function of the genes included in the expression signature of CLL with a high degree of 13q14 was assigned by applying the GeneCodis program,34 which finds concurrent annotations in GO and KEGG, and thereby derives several groups of genes with functional significance. The functional analysis to identify the most relevant biological mechanisms, pathways and functional categories in the data sets of genes selected by statistical analysis was generated through the use of Ingenuity Pathways Analysis (Ingenuity Systems, Mountain View, CA, USA).
Results
Fluorescence in situ hybridization
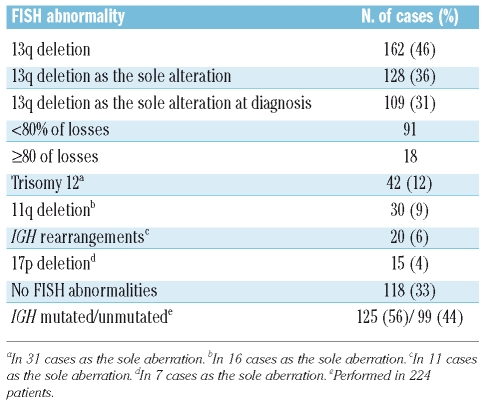
Among the 350 B-CLL patients studied, 162 (46.3%) had 13q deletion. In 128 (36.6%), this aberration was the only cytogenetic abnormality. For the analysis of prognostic factors we restricted the study to the 109 out of these 128 patients in whom the 13q- was analyzed at diagnosis. Biallelic (homozygous) 13q deletion was present in 21.4% of patients, while the remaining patients had a monoallelic (heterozygous) deletion. Table 1 shows the salient cytogenetic features of the whole series of 350 patients.
Table 1.
Incidence of genomic aberrations assessed by FISH in the global series of 350 patients with B-CLL.
Survival and time to progression
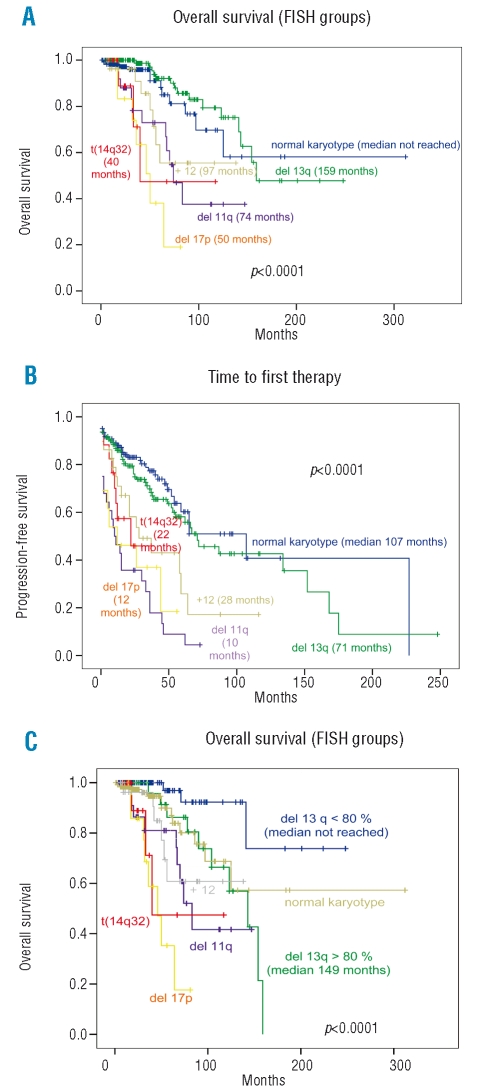
Overall survival and time to first therapy curves, according to cytogenetic abnormalities, in the total series of 350 patients are shown in Figure 1. There was a significant association between overall survival and the cytogenetic groups (p<0.0001). Thus, patients with loss of 13q as the sole anomaly and patients without abnormalities by FISH survived longer (median overall survival 159 months and not reached, respectively; median of total series, 154 months) (Figure 1A). The cytogenetic aberrations also influenced the time to first therapy, such that patients with 13q- as the sole abnormality and those with a normal karyotype had longer time to first therapy (p<0.0001) (Figure 1B).
Figure 1.
(A) Overall survival and (B) time to first therapy in the 350 patients with B-CLL. (C) Overall survival of patients with ≥80% or <80% cells showing the 13q14 deletion against that of patients in the other cytogenetic groups. The median survival and median time to first therapy is shown in months for each cytogenetic group.
Clinical and biological characteristics of patients showing 13q-
At the time of diagnosis 109 patients showed 13q deletion as the sole FISH abnormality, and the study was focused on this group of patients. The recruitment period started in October 1997 and finished in December 2006. All but three patients (with a score of 3) had a CLL immunophenotypic score of either 4 (49 cases) or 5 (58 cases).35 The median age of this group was 65 years (range, 38–90 years) and there was a predominance of males (68%). The majority of patients had asymptomatic disease with clinical and biological characteristics of good prognosis. Thus, 81.7% of patients were in A stage according to the Binet staging system and only seven (6.4%) were in stage C.
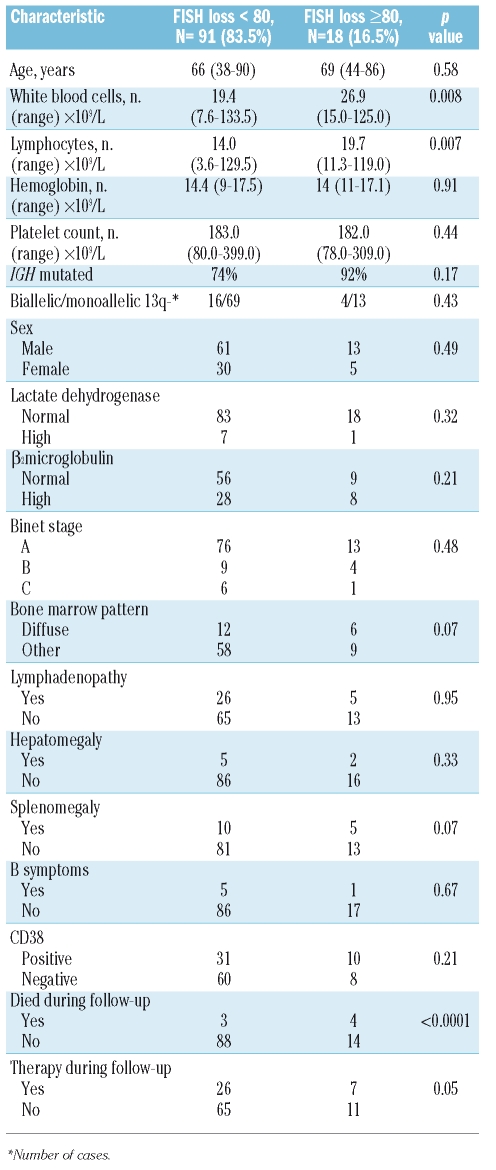
Regarding the percentage of cells with 13q-, the majority of cases (83.5%) had this abnormality in less than 80% of the analyzed cells. No relation was found between monoallelic and biallelic losses (22% in patients with <80% of 13q losses vs. 18% in the ≥80% group) in the 13q- patients. No major differences regarding clinical, biological, immunophenotypic or mutational status features were found in the cases with low (<80%) vs. high infiltration (≥80%) of 13q- cells, except for a high lymphocyte count (median of 14 vs. 19.7×109/L, respectively) (p=0.007) and a trend for an association with a diffuse pattern of bone marrow infiltration (17% vs. 40%; p=0.07) and splenomegaly (11% vs. 28%; p=0.07) in the group with high 13q- (Table 2). Of 109 cases considered, all 33 treated received, fludarabine-based therapies; in three cases allogeneic transplantation with reduced intensity conditioning was also performed.
Table 2.
Characteristics of 109 patients with 13q14 deletion as the sole cytogenetic aberration at diagnosis, divided according to the percentage of losses detected by FISH: < 80% (n=91) or ≥80% (n=18).
Survial and time to progression according to degree of 13q-
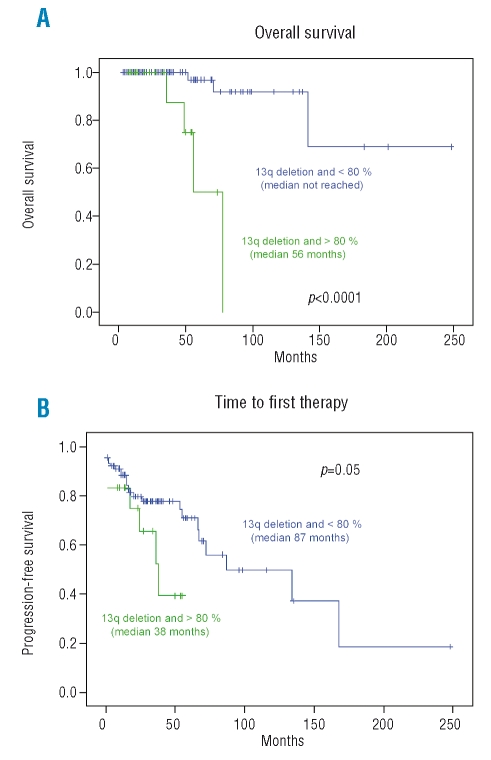
A significantly longer survival was observed in the cohort of patients with losses in 13q in less than 80% of cells. Thus, in the subgroup of patients with 80% or more of cells with loss of 13q the overall survival was 56 months (95% CI: 39–73 months), while in the group of patients in whom less than 80% of cells showed losses in 13q, the overall survival had not been reached (95% CI: 163–254 months) (p<0.0001) (Figure 2A). The proportion of deaths in the two groups was 22.2% and 3.5%, respectively. Univariate analysis showed that six variables were associated with short overall survival (p<0.0001): high percentage of losses in 13q14; high level of serum lactate dehydrogenase; high level of β2microglobulin; diffuse infiltration of the bone marrow; splenomegaly, and presence of B symptoms. In the multivariate analysis, the variables selected as independently related to overall survival were the percentage of losses of 13q14 (p=0.001) and the presence of B symptoms (p=0.007).
Figure 2.
(A) Overall survival and (B) time to first therapy of patients with B-CLL and 13q14 deletion as the sole aberration and <80% or ≥80% FISH losses.
In addition, a significantly shorter time to first thepary was observed in the cohort of patients with 80% or more of cells showing losses in 13q (median of 38 months; 95% CI: 21–55 months) as compared to those cases with less than 80% of 13q- (median of 87 months; 95% CI: 21–153 months) (p=0.05) (Figure 2B). Thus, 38.8% of patients in the group with high infiltration required treatment vs. 28.9% of patients in the group with less than 80% of cells showing 13q- losses. Univariate analysis showed that the variables associated with a short time to first therapy were: a high number of cells with deletion of 13q14 (p=0.05); presence of biallelic losses in 13q14 (p=0.05); non-mutated pattern of IGVH genes; a high level of serum lactate dehydrogenase; a high level of serum β2microglobulin; a positive Coomb’s test; presence of splenomegaly; and a diffuse pattern of bone marrow infiltration (all with a p value <0.0001). In the multivariate analysis, the variables selected as independently related to time to first therapy were the mutational status of IGVH p ( =0.001) and a high β2microglobulin level (p=0.003). Interestingly, the differences were also observed when other cut-offs were analysed (i.e. ≤40%, 41–69%, ≥70%, data not shown, and ≤50%, 51–79% and ≥80%) (Online Supplementary Figure S1).
Gene expression profiles of the subsets of B-CLL patients displaying different degrees of 13q loss
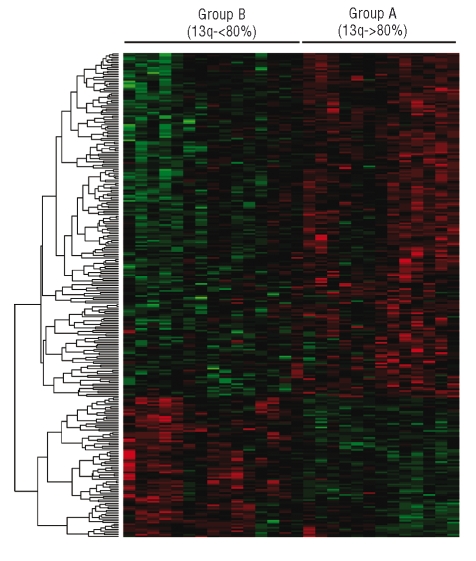
For the gene expression profile analysis two groups of patients with 13q- were compared: those in whom 80% or more of cells showed 13q- (group A) and those in whom less than 80% of cells showed 13q losses (group B). The comparative analysis of the gene expression profile of both groups identified a set of 1755 differentially expressed genes: 1073 genes were upregulated in group A, and 682 were downregulated (Figure 3). The gene function analysis revealed that most of these genes are involved in apoptosis, cellular growth and proliferation, mitochondrial, endoplasmic reticulum and calcium mediated activity (Table 3). Thus, the patients with more than 80% of 13q- cells had overexpression of genes related to cell proliferation and MAP kinase activity, such as ATF2, FGF3, FGF10, FGFR2, RRAS2, GRB2, JUN, MAPT and MAP3K1. By contrast, this group of patients had downregulation of genes related to apoptosis (PDCD10, EGLN3, CASP6, CL, and DAPK1) and cell cycle arrest, such as CDKN2C, GAS2L1, UHMK1, GAS1, GAS2L3, ZAK, and GAS7. Most of the genes involved in proteasome function were downregulated in the group with 13q losses in more than 80% of cells. The proteasome (PSM) genes dysregulated in our study codify proteasomal subunits, both catalytic (PSMA 1–7) and regulatory (PSMD1, PSMD7) ones. The ubiquitin process was also altered. Thus, genes for several enzymes that participate in different steps of the ubiquitin cycle were downregulated in the group with more losses in 13q: peptidases (USP27X), ubiquitin-conjugating enzymes (UBE2), thiolesterases (UCHL3) and ligas-es (HECTD1) (Table 3).
Figure 3.
Unsupervised analysis of patients displaying 13q- in more than 80% of the cells (group A) and the cases with less than 80% of 13q-. Overall, 1755 genes were deregulated, most of them (n=1073) were upregulated in group A.
Table 3.
Most relevant differentially expressed genes in patients with 80% or more cells showing 13q- (Up: overexpressed. Down: underexpressed).
Discussion
In recent years several studies have demonstrated a relationship between genetic changes and outcome in B-CLL patients.9,12,20 As regards cytogenetic abnormalities patients displaying a loss of ATM or TP53 genes have a short survival. By contrast, both loss on 13q and the absence of cytogenetic aberrations assessed by FISH are related to long survival in B-CLL.12 In most of the studies, patients displaying a loss of 13q were observed to have a better prognosis, but in some series with a long follow-up the outcome of patients with normal cytogenetics or 13q14 deletion was similar9 or even better for patients with normal karyotype. These results were reproduced in our series, with a median survival of 159 months for patients showing 13q- vs. a median not reached for those with a normal karyotype In order to better understand the clinical outcome of B-CLL patients with loss of 13q, we carried out a clinical and biological study.
Focusing on patients with loss of 13q as the sole cytogenetic aberration at diagnosis, we observed that the number of malignant cells carrying this genetic lesion influences the disease outcome. Thus, according to the percentage of cells with 13q- two prognostic groups could be established: the patients with a high proportion (≥80%) of 13q- cells had both a shorter overall survival than that of patients with <80% 13q- cells (56 months vs. not reached, p<0.0001) and a shorter time to first therapy (38 months vs. 87 months, p=0.05). These differences persisted when other cut-offs were considered (Online Supplementary Figure S1). The clinical relevance of the percentage of cells displaying a specific genetic abnormality has been recently demonstrated in CLL patients. Thus, the presence of more than 20% of cells with loss of TP53 has been associated with an adverse prognosis, while patients with less than 20% of cells with loss of TP53 had a prognosis similar to that of the global series.20,36 In the present study, a high number of 13q- cells together with the presence of B-symptoms were the only independent adverse factors for a short survival in the multivariate analysis in the group of B-CLL patients with 13q-.
Our data support the concept that patients with 13q-do not constitute a homogeneous group. Thus, patients with a high proportion of 13q losses had a high lymphocyte count and a trend to have more frequently a diffuse pattern of bone marrow infiltration and splenomegaly. In addition, overall survival and time to first therapy in this group were shorter than in patients with a normal karyotype or trisomy 12, being similar to that in patients with 11q deletion, although better than in the rest of cytogenetic subtypes. By contrast, CLL patients with a low number of malignant cells carrying the 13q deletion had a better prognosis than patients with a normal karyotype. It could, therefore, be useful to include quantification of the number of cells showing a genetic abnormality as part of the study of the prognostic factors in this disease.
To test our hypothesis that differences in several cellular functions such as more proliferation and less apoptosis could justify the clinical and prognostic differences found in the subsets of patients with 13q- divided according to the percentage of cells with this loss, we carried out a gene expression profiling analysis. This methodology has been applied in B-CLL showing that patients with specific genomic aberrations, gene expression phenotype or IGVH mutation status have distinct gene expression profiles.37–39
In the present study, we identified important functions differentially deregulated in the two groups of patients displaying 13q-. The main differential pathways involved are related to apoptosis, proliferation, mitochondrial and endopolasmic reticulum function, as well as ubiquitin metabolism. Patients with more 13q- cells had overexpression of genes involved in proliferation. Thus, the MAPK signaling pathway was affected since GRBS, RRAS, JUN or SOS1 were upregulated in the group of B-CLL patients with a high number of losses in 13q. In addition, some genes related to cell cycle arrest, such as CDKN2C, GAS2L1, GAS1, GAS7, ZAK, and GAS2L3, were downregulated in the group of patients with more 13q. Both the upregulation of MAPK and the underexpression of genes related to cell cycle arrest would lead to greater cell proliferation.40 By contrast, genes related to apoptosis (CASP6, CLU, DAPK1, and E2F1) were found to be downregulated in the group of cases with a high proportion of 13q- cells, leading to a decrease in apoptotic activity. The accumulation of mature B cells that have escaped programmed cell death and undergone cell-cycle arrest in the G0/G1 phase is the hallmark of CLL.41 Moreover, in our study we found alterations in other apoptosis-related pathways such as those mediated by mitochondria, endoplasmic reticulum and calcium metabolism. Thus, endoplasmic reticulum and protein-vesicular transport are inhibited because of an upregulation of genes involved in calcium-mediated activities, such as calmodulin binding and calcium ion binding.42,43 Furthermore, mitochondrial oxi-doreductase activity was inhibited in the group of B-CLL patients with a high number of cells with 13q-. The ATP dependency of apoptosis is well-known and both the respiratory chain and ATP synthase itself have been attributed central roles in the apoptotic process.44,45 Therefore, patients with a high number of cells with 13q- not only had more proliferation, but also less apoptosis, which could be related to the more aggressive disease that we observed in the clinical study.
In recent years several hematologic malignancies have been treated with proteasome inhibitors.46 Our study supports the idea that proteasome function is inhibited in the group of B-CLL patients with a high proportion of 13q-.47 It is, therefore, logical to expect that such patients would not benefit from the use of proteasome inhibitors.
The PI3K/NFκB pathway seems to play a pivotal role in B-CLL cell survival and growth.48 In our study, we found deregulation in PI3K β and γ catalytic subunits (PI3KCB and PI3KCG) as well as in other genes involved in this pathway (RAS, c-JUN, PKC and several integrins). This could be essential for mitogenic and antiapoptotic functions. In addition Ras-MAPK and Ras-PI3K signaling pathways are related and both of them lead cells to greater proliferation.
In summary, our findings suggest that the number of malignant cells with 13q- can influence the outcome of B-CLL patients in whom 13q- is the single genetic abnormality. A high number of cells with 13q deletion, as quantified by FISH, is associated with short overall survival and time to progression. To the best of our knowledge, this is the first time that the percentage of losses in 13q14 has been related to survival and progression. In addition, several cellular functions, such as more proliferation and less apoptosis, were found to be altered in the subsets of 13q- patients.
Our results need to be confirmed by additional studies, preferably in the context of large, randomized clinical trials.
Acknowledgments
we thank all the physicians from the Spanish hospitals who contributed clinical data; we are also grateful to Eva Lumbreras, María Pozo, Teresa Prieto, María Ángeles Hernández, Ana Simón, Ana Díez, and Almudena Martín, from “Centro de Investigación del Cáncer, Salamanca” for their technical assistance.
Footnotes
Funding: partially supported by grants from “Proyectos de Investigación Biomédica del SACYL” 106/A/06; “Ayuda predoctoral FIS de formación en investigación” (AR), FI06/00126 and by the “Acción Transversal del Cáncer” project, through an agreement between the Instituto de Salud Carlos III (ISCIII), the Spanish Ministry of Science and Innovation, and the Cancer Research Foundation of Salamanca University”and Redes de Investigación RTIIC (FIS).
Authorship and Disclosures
J-AH designed and performed the research and statistical analyses, recorded and analyzed data and wrote the paper; A-ER: designed and performed the microarray experiments and wrote the paper; CF, JLG, N-CG and JdR performed and interpreted the gene expression profiling studies; MG and J-MH designed and performed research and wrote the paper; VR, MR, GM-N, AGC, RF, JG, IR and FO provided patients’ data and wrote the paper; J-FSM corrected and approved the final version of the manuscript.
The authors reported no potential conflicts of interest.
References
- 1.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352:804–15. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 2.Dighiero G. Unsolved issues in CLL biology and management. Leukemia. 2003;17:2385–91. doi: 10.1038/sj.leu.2403154. [DOI] [PubMed] [Google Scholar]
- 3.Montserrat E. New prognostic markers in CLL. Hematology Am Soc Hematol Educ Program. 2006:279–84. doi: 10.1182/asheducation-2006.1.279. [DOI] [PubMed] [Google Scholar]
- 4.Crespo M, Bosch F, Villamor N, Bellosillo B, Colomer D, Rozman M, et al. ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia. N Engl J Med. 2003;348:1764–75. doi: 10.1056/NEJMoa023143. [DOI] [PubMed] [Google Scholar]
- 5.Durig J, Naschar M, Schmucker U, Renzing-Kohler K, Holter T, Huttmann A, et al. CD38 expression is an important prognostic marker in chronic lymphocytic leukaemia. Leukemia. 2002;16:30–5. doi: 10.1038/sj.leu.2402339. [DOI] [PubMed] [Google Scholar]
- 6.Gattei V, Bulian P, Del Principe MI, Zucchetto A, Maurillo L, Buccisano F, et al. Relevance of CD49d protein expression as overall survival and progressive disease prognosticator in chronic lymphocytic leukemia. Blood. 2008;111:865–73. doi: 10.1182/blood-2007-05-092486. [DOI] [PubMed] [Google Scholar]
- 7.Krober A, Seiler T, Benner A, Bullinger L, Bruckle E, Lichter P, et al. V(H) mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood. 2002;100:1410–6. [PubMed] [Google Scholar]
- 8.Lin K, Sherrington PD, Dennis M, Matrai Z, Cawley JC, Pettitt AR. Relationship between p53 dysfunction, CD38 expression, and IgV(H) mutation in chronic lymphocytic leukemia. Blood. 2002;100:1404–9. doi: 10.1182/blood-2001-11-0066. [DOI] [PubMed] [Google Scholar]
- 9.Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–7. [PubMed] [Google Scholar]
- 10.Oscier DG, Gardiner AC, Mould SJ, Glide S, Davis ZA, Ibbotson RE, et al. Multivariate analysis of prognostic factors in CLL: clinical stage, IGVH gene mutational status, and loss or mutation of the p53 gene are independent prognostic factors. Blood. 2002;100:1177–84. [PubMed] [Google Scholar]
- 11.Wiestner A, Rosenwald A, Barry TS, Wright G, Davis RE, Henrickson SE, et al. ZAP-70 expression identifies a chronic lymphocytic leukemia subtype with unmutated immunoglobulin genes, inferior clinical outcome, and distinct gene expression profile. Blood. 2003;101:4944–51. doi: 10.1182/blood-2002-10-3306. [DOI] [PubMed] [Google Scholar]
- 12.Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–6. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 13.Hernandez JM, Mecucci C, Criel A, Meeus P, Michaux I, Van HA, et al. Cytogenetic analysis of B cell chronic lymphoid leukemias classified according to morphologic and mmunophenotypic (FAB) criteria. Leukemia. 1995;9:2140–6. [PubMed] [Google Scholar]
- 14.Krober A, Bloehdorn J, Hafner S, Buhler A, Seiler T, Kienle D, et al. Additional genetic high-risk features such as 11q deletion, 17p deletion, and V3–21 usage characterize discordance of ZAP-70 and VH mutation status in chronic lymphocytic leukemia. J Clin Oncol. 2006;24:969–75. doi: 10.1200/JCO.2005.03.7184. [DOI] [PubMed] [Google Scholar]
- 15.Hauswirth AW, Jager U. Impact of cytogenetic and molecular prognostic markers on the clinical management of chronic lymphocytic leukemia. Haematologica. 2008;93:14–9. doi: 10.3324/haematol.12319. [DOI] [PubMed] [Google Scholar]
- 16.Cuneo A, Rigolin GM, Bigoni R, De AC, Veronese A, Cavazzini F, et al. Chronic lymphocytic leukemia with 6q- shows distinct hematological features and intermediate prognosis. Leukemia. 2004;18:476–83. doi: 10.1038/sj.leu.2403242. [DOI] [PubMed] [Google Scholar]
- 17.Dohner H, Stilgenbauer S, James MR, Benner A, Weilguni T, Bentz M, et al. 11q deletions identify a new subset of B-cell chronic lymphocytic leukemia characterized by extensive nodal involvement and inferior prognosis. Blood. 1997;89:2516–22. [PubMed] [Google Scholar]
- 18.Cavazzini F, Hernandez JA, Gozzetti A, Russo RA, De AC, Tiseo R, et al. Chromosome 14q32 translocations involving the immunoglobulin heavy chain locus in chronic lymphocytic leukaemia identify a disease subset with poor prognosis. Br J Haematol. 2008;142:529–37. doi: 10.1111/j.1365-2141.2008.07227.x. [DOI] [PubMed] [Google Scholar]
- 19.Cordone I, Masi S, Mauro FR, Soddu S, Morsilli O, Valentini T, et al. p53 expression in B-cell chronic lymphocytic leukemia: a marker of disease progression and poor prognosis. Blood. 1998;91:4342–9. [PubMed] [Google Scholar]
- 20.Catovsky D, Richards S, Matutes E, Oscier D, Dyer MJ, Bezares RF, et al. Assessment of fludarabine plus cyclophosphamide for patients with chronic lymphocytic leukaemia (the LRF CLL4 trial): a randomised controlled trial. Lancet. 2007;370:230–9. doi: 10.1016/S0140-6736(07)61125-8. [DOI] [PubMed] [Google Scholar]
- 21.Harris NL, Jaffe ES, Diebold J, Flandrin G, Muller-Hermelink HK, Vardiman J, et al. World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues: report of the Clinical Advisory Committee meeting-Airlie House, Virginia, November 1997. J Clin Oncol. 1999;17:3835–49. doi: 10.1200/JCO.1999.17.12.3835. [DOI] [PubMed] [Google Scholar]
- 22.Binet JL, Caligaris-Cappio F, Catovsky D, Cheson B, Davis T, Dighiero G, et al. Perspectives on the use of new diagnostic tools in the treatment of chronic lymphocytic leukemia. Blood. 2006;107:859–61. doi: 10.1182/blood-2005-04-1677. [DOI] [PubMed] [Google Scholar]
- 23.Sanchez ML, Almeida J, Gonzalez D, Gonzalez M, Garcia-Marcos MA, Balanzategui A, et al. Incidence and clinicobiologic characteristics of leukemic B-cell chronic lymphoproliferative disorders with more than one B-cell clone. Blood. 2003;102:2994–3002. doi: 10.1182/blood-2003-01-0045. [DOI] [PubMed] [Google Scholar]
- 24.Cheson BD, Bennett JM, Rai KR, Grever MR, Kay NE, Schiffer CA, et al. Guidelines for clinical protocols for chronic lymphocytic leukemia: recommendations of the National Cancer Institute-sponsored working group. Am J Hematol. 1988;29:152–63. doi: 10.1002/ajh.2830290307. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez MB, Hernandez JM, Garcia JL, Lumbreras E, Castellanos M, Hernandez JM, et al. The value of fluorescence in situ hybridization for the detection of 11q in multiple myeloma. Haematologica. 2004;89:1213–8. [PubMed] [Google Scholar]
- 26.Ghia P, Stamatopoulos K, Belessi C, Moreno C, Stilgenbauer S, Stevenson F, et al. ERIC recommendations on IGHV gene mutational status analysis in chronic lymphocytic leukemia. Leukemia. 2007;21:1–3. doi: 10.1038/sj.leu.2404457. [DOI] [PubMed] [Google Scholar]
- 27.Gutierrez NC, Lopez-Perez R, Hernandez JM, Isidro I, Gonzalez B, Delgado M, et al. Gene expression profile reveals deregulation of genes with relevant functions in the different subclasses of acute myeloid leukemia. Leukemia. 2005;19:402–9. doi: 10.1038/sj.leu.2403625. [DOI] [PubMed] [Google Scholar]
- 28.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 29.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 31.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–84. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 33.Goeman JJ, van de Geer SA, de KF, van Houwelingen HC. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93–9. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 34.Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007;8:R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matutes E, Owusu-Ankomah K, Morilla R, Garcia MJ, Houlihan A, Que TH, et al. The immunological profile of B-cell disorders and proposal of a scoring system for the diagnosis of CLL. Leukemia. 1994;8:1640–5. [PubMed] [Google Scholar]
- 36.Thornton PD, Gruszka-Westwood AM, Hamoudi RA, Atkinson S, Kaczmarek P, Morilla RM, et al. Characterisation of TP53 abnormalities in chronic lymphocytic leukaemia. Hematol J. 2004;5:47–54. doi: 10.1038/sj.thj.6200325. [DOI] [PubMed] [Google Scholar]
- 37.Rosenwald A, Alizadeh AA, Widhopf G, Simon R, Davis RE, Yu X, et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med. 2001;194:1639–47. doi: 10.1084/jem.194.11.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klein U, Tu Y, Stolovitzky GA, Mattioli M, Cattoretti G, Husson H, et al. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J Exp Med. 2001;194:1625–38. doi: 10.1084/jem.194.11.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aalto Y, El-Rifa W, Vilpo L, Ollila J, Nagy B, Vihinen M, et al. Distinct gene expression profiling in chronic lymphocytic leukemia with 11q23 deletion. Leukemia. 2001;15:1721–8. doi: 10.1038/sj.leu.2402282. [DOI] [PubMed] [Google Scholar]
- 40.Smal C, Lisart S, Maerevoet M, Ferrant A, Bontemps F, Van Den NE. Pharmacological inhibition of the MAPK/ERK pathway increases sensitivity to 2-chloro-2'-deoxyadeno-sine (CdA) in the B-cell leukemia cell line EHEB. Biochem Pharmacol. 2007;73:351–8. doi: 10.1016/j.bcp.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 41.Caligaris-Cappio F, Hamblin TJ. B-cell chronic lymphocytic leukemia: a bird of a different feather. J Clin Oncol. 1999;17:399–408. doi: 10.1200/JCO.1999.17.1.399. [DOI] [PubMed] [Google Scholar]
- 42.Lao Y, Chang DC. Study of the functional role of Bcl-2 family proteins in regulating Ca(2+) signals in apoptotic cells. Biochem Soc Trans. 2007;35:1038–9. doi: 10.1042/BST0351038. [DOI] [PubMed] [Google Scholar]
- 43.Bernardi P, Rasola A. Calcium and cell death: the mitochondrial connection. Subcell Biochem. 2007;45:481–506. doi: 10.1007/978-1-4020-6191-2_18. [DOI] [PubMed] [Google Scholar]
- 44.McClintock DS, Santore MT, Lee VY, Brunelle J, Budinger GR, Zong WX, et al. Bcl-2 family members and functional electron transport chain regulate oxygen deprivation-induced cell death. Mol Cell Biol. 2002;22:94–104. doi: 10.1128/MCB.22.1.94-104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shchepina LA, Pletjushkina OY, Avetisyan AV, Bakeeva LE, Fetisova EK, Izyumov DS, et al. Oligomycin, inhibitor of the F0 part of H+-ATP-synthase, suppresses the TNF-induced apoptosis. Oncogene. 2002;21:8149–57. doi: 10.1038/sj.onc.1206053. [DOI] [PubMed] [Google Scholar]
- 46.Richardson PG, Sonneveld P, Schuster MW, Irwin D, Stadtmauer EA, Facon T, et al. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N Engl J Med. 2005;352:2487–98. doi: 10.1056/NEJMoa043445. [DOI] [PubMed] [Google Scholar]
- 47.Faderl S, Rai K, Gribben J, Byrd JC, Flinn IW, O’Brien S, et al. Phase II study of single-agent bortezomib for the treatment of patients with fludarabine-refractory B-cell chronic lymphocytic leukemia. Cancer. 2006;107:916–24. doi: 10.1002/cncr.22097. [DOI] [PubMed] [Google Scholar]
- 48.Cuni S, Perez-Aciego P, Perez-Chacon G, Vargas JA, Sanchez A, Martin-Saavedra FM, et al. A sustained activation of PI3K/NF-kappaB pathway is critical for the survival of chronic lymphocytic leukemia B cells. Leukemia. 2004;18:1391–400. doi: 10.1038/sj.leu.2403398. [DOI] [PubMed] [Google Scholar]