Abstract
Tsetse flies (Diptera: Glossinidae) are an ancient taxon of one genus, Glossina, and limited species diversity. All are exclusively haematophagous and confined to sub-Saharan Africa. The Glossina are the principal vectors of African trypanosomes Trypanosoma sp (Kinetoplastida: Trypanosomatidae) and as such, are of great medical and economic importance. Clearly tsetse flies and trypanosomes are coadapted and evolutionary interactions between them are manifest. Numerous clonally reproducing strains of Trypanosoma sp exist and their genetic diversities and spatial distributions are inadequately known. Here I review the breeding structures of the principle trypanosome vectors, G. morsitans s.l., G. pallidipes, G. palpalis s.l. and G. fuscipes fuscipes. All show highly structured populations among which there is surprisingly little detectable gene flow. Rather less is known of the breeding structure of T. brucei sensu lato vis à vis their vector tsetse flies but many genetically differentiated strains exist in nature. Genetic recombination in Trypanosoma via meiosis has recently been demonstrated in the laboratory thereby furnishing a mechanism of strain differentiation in addition to that of simple mutation. Spatially and genetically representative sampling of both trypanosome species and strains and their Glossina vectors is a major barrier to a comprehensive understanding of their mutual relationships.
Keywords: Glossina, Trypanosoma, tsetse, gene flow, African trypanosomiasis
INTRODUCTION
Drosophila melanogaster Mieigen (Diptera: Drosophilidae) is probably the best-known insect, even though its agricultural significance is small. The reasons for its eminence are clear: easy culturing, high reproductive rate, small size, rapid generation time, easily demonstrable polytene chromosomes, and much genetic variation. Drosophila melanogaster is the favored lab animal of many thousands of scientists all over the world. In contrast, tsetse flies (Diptera: Glossinidae) are difficult to culture, have long generation times and low reproductive rates. Only a handful of cultures exist and the numbers of scientists that now work directly with tsetse flies are probably no more than a hundred. Only one laboratory in North America now works with tsetse. Nevertheless, knowledge of tsetse biology is not trivial or insignificant. Tsetse systematics, ecology, physiology, and vector biology have been intensively studied for a hundred and fifty years because of their medical and economic significance.
Tsetse flies are most unusual insects because of their exclusively haematophagous feeding habits and reproductive biology. There are six major texts in English devoted principally to tsetse flies (Buxton, 1955; Glasgow, 1963; Nash, 1969; Ford, 1971; Jordan, 1986; Leak, 1998). These texts review a particularly rich literature on ecology, trypanosomiasis epidemiology and epizootiology, pest management, and history. Investigation of tsetse genetics and physiology, on the other hand, has been greatly inhibited by the difficulty and cost of culturing tsetse flies, their long generation times, low reproductive rates, and sampling of natural populations that has been geographically limited and mostly opportunistic. Earlier reviews of tsetse genetics include Gooding (1984) and Gooding and Krafsur (2004, 2005). Knowledge of trypanosome biology and genetics, on the other hand, is rather well developed because, apart from being medically and intrinsically interesting, trypanosome cultures can be propagated in the laboratory independently of tsetse flies. A thorough understanding of disease epidemiology and management, however, requires information that relates parasite distribution to that of its vectors (Tibayrenc and Ayala, 2002; De Meeus et al., 2007).
Interest in tsetse population genetics has recently developed, fueled by the notion that genetic methods can promote effective area-wide population management by (1) better defining taxonomic units (Gooding and Krafsur, 2005) and estimating degrees of isolation of populations of interest (e.g., Feldmann, 2004; Vreysen, 2006) or (2) modulating vector competence by genetic means (e.g., Aksoy et al., 2001). In these ways it is hoped that the well-known propensity of tsetse dispersal can be overcome and long-term tsetse population management can be achieved, thereby greatly reducing, or eliminating trypanosomiasis altogether.
In this article, I review tsetse fly evolution, trypanosome interactions, and genetics with an emphasis on population and ecological genetics as found in the more recent literature since Gooding and Krafsur’s review (2005).
Systematics and evolution of tsetse flies
TSETSE FLIES are classified into a single genus Glossina Weidemann 1830. There are three extant subgenera, Austenina Townsend, Nemorhina Robineau-Desvoidy, and Glossina Wiedemann that correspond to the Fusca, Palpalis, and Morsitans species groups respectively. A new subgenus, Machadomia Dias 1987, was established to incorporate the anomalous tsetse, G. austeni Newstead. There is an extinct sister group to extant Glossina known from the Florissant shale of Colorado, dating from 35 mya (Grimaldi, 1992) and Glossina-like fossils have recently been recovered from Oligocene strata in Germany (Grimaldi and Engel, 2005). It seems, therefore, that tsetse flies (to include the putative, unnamed, sister taxon) had nearly a worldwide distribution some 30 to 40 million years ago. Their origins, therefore, must predate continental separation in the Cretacious, more than 100 MYA. The higher classification of Glossina is, following McAlpine (1989) and Grimaldi and Engel (2005): suborder Brachycera – infraorder Cyclorrhapha – Section Schizophora – Subsection Calyptrata – Superfamily Pupiparia (= Hippoboscoidea) – Family Glossinidae. An extant sister taxon has not been agreed upon. The other families in the Pupiparia (Hippoboscidae, Streblidae, Nycteribiidae) are more highly specialized morphologically than Glossina and are true ectoparasites. All Pupiparia reproduce by adenotrophic viviparity, such that a zygote develops and hatches in the female’s reproductive tract and the larva feeds on “milk” produced by the female’s reproductive accessory glands until it completes its development. Hippoboscoidea phylogeny and evolution was recently addressed by Peterson et al. (2007).
Thirty-three extant taxa have been described of 22 species that include five species complexes (Gooding and Krafsur, 2004 for review). Leak (1999) lists 31 taxa. Thirty taxa evaluated by Rogers and Robinson (2004) have well-defined habitats in terms of satellite imagery derived variables. The species complexes include G. morsitans morsitans Westwood 1850, G. m. centralis Machado 1970, G. m. submorsitans Newstead 1910 and G. m. submorsitans ugandensis Vanderplank 1949; G. palpalis palpalis Robineau-Desvoidy 1830 and G. p. gambiense Vanderplank 1949. The foregoing taxa are particularly important medically and economically. Other subspecies include G. pallicera pallicera Bigot 1891 and G. p. newsteadi Austen 1929; G. nigrofusca nigrofusca Newstead 1910 and G. n. hopkinsi Van Emden 1944; G. fuscipes fuscipes Newstead 1910, G. f. quanzensis Pires 1948, and G. f. martini Zumpt 1933. Taxa within each species complex are mostly allopatric but morphologically indistinguishable from their sisters, or very nearly so. Results of recent studies suggested incipient speciation in G. p. palpalis although alternative explanations also were offered (Gooding et al., 2004; Ravel et al., 2007). With the likely exception of G. p. palpalis in Bonon, Cote d’Ivoire, sibling species are allopatric, hybrid males are sterile, and the hybrid females vary in their fertilities. The G. morsitans subspecies share little microsatellite variation among them, are reproductively isolated intrinsically and extrinsically, and it was suggested they should be raised to specific rank (Krafsur and Endsley, 2006).
Extant tsetse flies are confined to sub-Saharan Africa, although G. fuscipes fuscipes and G. morsitans submorsitans have been recorded in the southwestern corner of the Arabian Peninsula (Elsen et al., 1990). They are relicts from earlier times when more equable climates prevailed.
Glossinidae, indeed, all Pupiparia, are exclusively haematophagous. Reproduction in tsetse is by adenotrophic viviparity: a larva develops to maturity in its mother over a temperature dependent 7 - 12 day interval. When mature, the mother deposits her larva in an appropriate microhabitat and the animal pupariates in the soil. Adult development requires about 27 days at 25°C. Females become inseminated by the end of their first week of adulthood. The youngest larvapositing female is at least 16 days old, so the minimum time necessary to produce two offspring is about 25 days. Generation time is c. 43 days at 25°C. Compensating for their slow reproduction rates are their adult survival rates, which typically exceed 97% per day (Rogers and Randolph, 1984a, b; 1985).
Medical and economic significance
The impact of tsetse flies is severe in much of sub-Saharan Africa because of their blood feeding habits and their role as vectors of pathogenic trypanosomes. Chief among these is Trypanosoma (Trypanozoon) brucei brucei, T. b. rhodesiense, and T. b. gambiense, two of which are highly pathogenic to man, the agents of ‘sleeping sickness’ or human African trypanosomiasis (HAT). It seems that G. brucei s.l. are not well-defined taxonomic units nor clinical entities, however (Hide and Tait, 2004; Gibson, 2005). T. b. gambiense infection causes a chronic, slow wasting disease that ultimately causes death if untreated. This form of HAT occurs in West and central Africa and is unknown east of the Rift Valley (Welburn et al., 2001). Its chief vectors are Palpalis group flies. Humans are the principal reservoir of T. b. gambiense. The parasite has been isolated from pigs, dogs, and sheep but the epidemiological significance of this remains opaque. T. b. rhodesiense is zoonotic and causes a much more acute disease in humans that is rapidly fatal if not properly treated. T. b. rhodesiense occurs east of the Rift valley. Its vectors are Morsitans group flies. Uganda is the only country where both forms of HAT are known to occur. They do so in separate foci and there is a concern that the two agents soon will overlap because transport of infected cattle can introduce T. b. rhodesiense to western Uganda (Picozzi et al., 2005). T. b. brucei occurs throughout sub-Saharan Africa wherever its vectors may be found. It is not infective to humans because it is rapidly lysed by human serum. The principal vectors of HAT include G. palpalis s.l., G. morsitans s.l., and G. tachinoides Westwood 1850. G. fuscipes fuscipes is the chief vector in Uganda and western Kenya. The principle foci of HAT seem to be static and ancient. The epidemiology of HAT has been comprehensively treated by Welburn et al. (2004) and an epidemiological update is available from the WHO (2006). Trypanosome molecular epidemiology was reviewed by Hide and Tait (2004).
Trypanosomiasis in domestic animals is termed ‘nagana’, a chronic wasting disease that precludes most animal agriculture in much of sub-Saharan Africa but poultry are unaffected by trypanosomiasis. In addition to T. b. brucei and T. b. rhodesiense, tsetse-transmitted agents include T. vivax, T. congolense, T. simiae, and T. godfreyi (Stevens and Brisse, 2004). T. evansi is pathogenic but not transmitted by tsetse flies. Nagana is of much economic significance because where it is prevalent; meat, milk, dung, and draft power production are greatly reduced or lost altogether. The long-term consequence of greatly reducing Nagana is controversial, however, because it has been argued that its elimination will inexorably lead to habitat degradation: deforestation, overgrazing, and erosion (ERGO, 1999).
Tsetse population control methods include aerial application of ultra-low volume insecticide applications, the use of insecticide-laced targets and traps, and the sterile insect technique (Torr et al., 2006). Research efforts are well underway to find ways to employ transgenesis to render vector tsetse flies insusceptible to trypanosome infection and replace natural vector populations with refractory ones (Aksoy et al., 2001). Earlier control methods, no longer extensively applied, included ground spraying of persistent insecticides such as DDT, the clearing of brush, and extermination of native mammals that provide tsetse fly blood meals and act as trypanosome reservoirs.
Vector-parasite-mammalian co-adaptations
Trypanosoma brucei s.l. salivary gland infection rates in tsetse flies are low, typically < 1% in (Maudlin et al., 1998). Much higher infection rates may be expected given the great longevity of the flies (adult daily survival rates typically exceed 97% and half lives exceeding ~ 28 d) and prevalence of trypanosome-infected animals. A likely reason that tsetse infection rates are small is they have a robust innate immune system in which most ingested trypanosomes die in the fly midgut. Once established in the midgut, procyclic trypomastigotes must transform to epimastigotes and proceed via the proventriculus and foregut to the host salivary glands where they develop to infective metacyclic trypanosomes. Only a small fraction of gut infections succeed in developing into infective metacyclic forms lodged in the vector salivary glands. In laboratory studies, injection of E. coli into tsetse flies induced immune stimulation thereby reducing trypanosome-infective feeds and the anti-microbial peptides attacin, defensin, and diptericin were demonstrated in tsetse tissues (Hao et al., 2001). Attacin was shown to provide an important immune response that regulates trypanosome infection in some, but not all, Morsitans group flies (Hu and Aksoy, 2006; Nayduch and Aksoy, 2007; Roditi and Lehane, 2008). The prophenoloxidase cascade (Nigam et al. 1997) and phagocytosis of trypanosomes by haemocytes have also been suggested to play a role in vector competence. Another mechanism that may suppress trypanosome development in tsetse is the production of certain midgut lectins (reviews in Welburn and Maudlin, 1999; Rodite and Lehane, 2008). Lectins are proteins that bind specifically with carbohydrates to form glycoproteins. Lectin titers increase in post-teneral flies and help to explain the insusceptibility of older flies to new trypanosome infections. The foregoing picture, however, has been modified by recent experimental work that demonstrated the addition of antioxidants (e.g. ascorbic acid, uric acid, glutathione, N-acetyl-cysteine) to blood meals increased T. b. brucei midgut infection rates in G. m. morsitans – remarkably to 100% in some experiments. The mechanism of trypanosome suppression by lectins, therefore, is related to their properties as pro-oxidant free radicals induced during digestion of blood meals (MacLeod et al., 2007a). Maturation and migration of midgut forms to the salivary glands, however, also seems to require reactive oxygen species nitric oxide and/or L-cysteine (MacLeod et al., 2007b). Clearly, the pathway from ingestion of blood forms by tsetse flies to the production of infective metacyclic trypomastigotes in the tsetse salivary glands is long and complex, with many critical steps that developing trypanosomes must overcome.
Susceptibility to midgut infection was reported to be under genetic control and inherited matrilineally in G. m. morsitans (Maudlin, 1982) and refractory and susceptible lines have been established experimentally (Welburn and Maudlin, 1999). The mode of matrilineal inheritance can be attributed to the commensal inter- and intracellular bacterium in tsetse flies, Sodalis glossinidius (Dale and Maudlin, 1999). Recent work of MacLeod et al. (2007a), however, raises the question, do the Sodalis flora of natural tsetse fly populations vary in their production of anti-oxidants and is this the underlying mechanism of susceptibility to trypanosome infection in tsetse flies? Could engineered Sodalis eventually be used in a scheme to replace vectorally competent natural tsetse populations with transgenic, insusceptible populations?
Salivary gland infected tsetse flies in the laboratory have a reduced expectation of life and males show greater infection rates than females (Maudlin et al., 1998; Welburn and Maudlin, 1999). Laboratory studies using T. b. brucei and G. m. morsitans have demonstrated that reproductive success by female tsetse can depress the proportion of midgut infections that mature, but midgut infections do not detectably depress tsetse reproductive success (MacLeod et al., 2007b). Co-infection of tsetse flies by more than one trypanosome species and strain is commonly observed in field samples (MacLeod et al., 1999; Lehane et al., 2000). Experimental co-infection and superinfection of G. m. morsitans by two fluorescently marked T. b. brucei strains demonstrated that salivary gland infection by one strain of trypanosomes did not inhibit the salivary gland establishment of another strain; the chief determinant was the insect’s susceptibility to a mature infection (Peacock et al., 2007). The finding is important because genetic recombination between trypanosome strains takes place in the tsetse salivary glands (Gibson and Stevens, 1999) as discussed below.
Are there locally co-adapted trypanosome-tsetse demes distributed throughout a tsetse belt? Genotyping of T. brucei isolates indicate both spatial and host differentiation (e.g., MacLeod et al., 2001). Probably more T. brucei s.l. strains will be uncovered with the development of additional genetic markers and improved techniques. The source of such differentiation is mutation and possibly genetic recombination, first demonstrated in laboratory experiments (e.g., Jenni et al., 1986). Indeed, genetic recombination has been shown to occur among the three G. brucei ‘subspecies’ but the extent of genetic recombination between trypanosome populations in the field remains controversial and largely unknown. It is believed that trypanosome mating and meiosis take place in the tsetse fly salivary glands, probably among epimastigotes (Tait et al., 2007; Gibson et al. 2008). Meiosis in trypanosomes provides an essential adaptive mechanism and may explain the numerous strains that have been described (Gibson, 2001)
Is trypanosome genetic differentiation adaptive, i.e., is it maintained by natural selection? In particular, is it maintained by adaptation to the locally prevalent tsetse vectors? Gene flow among natural T. b. brucei and T. b. rhodesiense populations is said to be absent or greatly limited; indeed, the two are said to be genetically isolated from each other with T. b. brucei showing epidemic population structure and T. b. rhodesiense showing clonal population structure (MacLeod et al., 2000). Estimating the strength and degree of substructuring among T. brucei spp in nature, however, seems a herculean task, and I know of no attempts to evaluate trypanosome population structure over large areas or to evaluate it with respect to its tsetse vectors. Indeed, most trypanosome research is conducted without reference to its vectors. T. brucei s.l. loses its ability to infect tsetse flies after multiple passages in laboratory mice thereby calling into question the epidemiological significance of some findings. Biased sampling of natural T. brucei s.l. populations is a formidable problem such that their population structures are poorly known; the bias arises from a requirement of obtaining large numbers of blood forms from infected hosts and amplifying their numbers in mice before genotyping them (Garcia et al., 2006).
To complete the triangle of co-adaptation, trypanosomes vary in their infectiousness to mammals. For example, T. brucei brucei, ubiquitous in mammals throughout sub-Saharan Africa, is rapidly lysed in human serum but T. b. gambiense and T. b. rhodesiense are not, and therefore are pathogenic to humans as well as many other mammals. A single gene, the serum resistance associated (SRA) gene, is sufficient to confer the trait of human infectivity on T. brucei brucei (Xong et al., 1998); all known strains of T. b. rhodesiense have this gene but T. b. gambiense does not, and therefore must have another mechanism of human serum resistance (Gibson, 2005).
Susceptibility of tsetse species to trypanosome infection in lab trials may not be strongly predictive of their vectorial capacity in nature, for a number of reasons. First, tests of susceptibility/refractoriness typically are based on a single genotype of trypanosome and a well-adapted laboratory strain of Glossina. As we have seen, susceptible and insusceptible lines of a common lab strain of G. m. morsitans have been developed, and it will be shown that tsetse field populations are typically genetically differentiated - often, highly so. A single Glossina sp lab strain is most unlikely to be representative of the species over much of its geographic range and multiple, independently derived lab strains from diverse natural populations are unavailable. Second, even though HAT epidemics are typically of one trypanosome genotype, i.e., clonal (but see Agbo et al., 2003 for contrary evidence), natural populations of T. brucei s.l. are genetically differentiated in ways that remain largely unknown. Indeed, an objective and scientifically adequate appreciation of vector-parasite compatibilities in the field is quite beyond reach at this time because the sampling problems seem insurmountable.
Genetics and cytogenetics of tsetse flies
All tsetse examined have two pairs of metacentric autosomes and a sex bivalent: 2N = 4 + XY. G. m. morsitans, G. m. submorsitans, and G. m. centralis also have small, telocentric, heterochromatic supernumerary chromosomes that vary in number from zero to eight (Warnes and Maudlin, 1992). A lab culture of G. austeni Newstead 1912 had rather more supernumerary chromosomes that varied from 8 to 12 in number (Southern, 1980). Supernumararies vary from 12 to 22 in two taxa of Fusca group flies (review in Gooding and Krafsur, 2005). Sex chromosome karyotypes XXYY, XO, XXY, XYY were found in Nigerian populations of G. palpalis palpalis (Maudlin, 1979). The high frequency of these polymorphisms suggested that they are maintained by some kind of balancing selection instead of high frequencies of spontaneous non-disjunction.
Well-banded polytene chromosomes can be obtained from pupal trichogen cells found in apical scutellar bristles (Southern and Pell, 1974). Gariou-Papalexiou et al. (2007) compared banding patterns among G. pallidipes, G.m. morsitans, G. m. submorsitans, and G. austeni. The karyotypes of these taxa differ by numerous paracentric inversions in the X chromosome and chromosome L1. G. austeni differs from the others by bearing a pericentric inversion on chromosome L2. G. m. morsitans and G. m. submorsitans are homosequential and differ only by a paracentric inversion on the X and another on the L2 chromosomes.
Evidence to date shows abundant chromosome diversity among Morsitans and Palpalis group flies. Chromosome diversity, however, has been examined inadequately among and within species; few tsetse cultures exist and sampling wild populations cytogenetically required rather more interest and support than available. Clearly there is much to be learned by further study, and it is to be hoped that surveys of natural populations can be initiated.
Formal genetics and genetic markers
Five classical morphological markers have been described and these were assigned to two linkage groups in G. p. palpalis, G. m. morsitans, and G. m. submorsitans. Each of thirteen biochemical markers was assigned to one of three linkage groups (Gooding and Rolseth, 1995; Gooding and Challoner, 1999). Composite maps are reviewed and shown in Gooding and Krafsur (2005). The X chromosome map was recently updated with two microsatellite loci in G. palpalis (Gooding et al., 2004). There are three linkage groups in each species examined, but they have not been associated with their physical chromosomes
Population genetics
The forces of evolutionary change include mutation, selection, drift, and migration. When these forces do not operate there is random mating and no change in gene frequencies from one generation to the next. This is the Hardy-Weinberg rule that provides the basic null hypotheses with which to evaluate gene frequencies within and among demes. Natural populations, however, are of finite size and more or less discontinuously distributed so that chance variations in gene frequencies occur from place to place and time to time. Mutations arise and contribute to gene diversity. Matings are more likely within subdivided populations than among them. Selection may act locally and differentially on genotypes thereby contributing to spatial diversity. Migration of reproductives among demes tends to neutralize the forces of drift and local selection. A primer of population genetics applied to molecular epidemiology can be found in an earlier volume of this journal (De Meeus et al., 2007).
Ecological background
Clearly the outcome of Hardy-Weinberg criteria applied to tsetse fly demes will depend on their distribution and abundance. Morsitans group flies are largely savanna and woodland inhabitants, although G. pallidipes may also be found in forests. Morsitans group taxa are adapted to drier habitats than the other two subgenera (Rogers and Robinson, 2004). Palpalis group flies tend to occur in riverine and lacustrine habitats. Fusca group flies largely inhabit moist forests of West Africa although G. brevipalpis occurs discontinuously in East Africa, Zaire, and Mozambique (Jordan, 1963; Leak, 1998). Indeed, maps show that many Glossina taxa are discontinuously distributed according to environmental criteria that include host blood meal sources and abiotic factors. Abiotic factors have been extensively evaluated in terms of satellite imagery (Robinson et al., 1997a, b; Rogers, 2000; Rogers and Robinson, 2004). The chief predictor variables include NDVI, the normalized density vegetation index, elevation, maximum and minimum temperature indices, rainfall, and saturation deficits. Different combinations of variables are necessary for each tsetse species and subspecies. These approaches have allowed a detailed mapping of suitable tsetse habitats in sub-Saharan Africa (Rogers and Robinson, 2004 and references therein). The distribution of tsetse flies spans many ecological zones, among which, different discriminant models and predictor variables best describe the distribution (Robinson et al., 1997a). More accurate predictions of tsetse distributions are possible if an area is first subdivided into ecological zones, and the distribution within each zone treated separately. This suggests the existence of population clusters (i.e., subpopulations or demes) in which the flies have adapted to their differing environments. It seems likely, therefore, that the flies’ adaptations may differ from one cluster to another.
Life history and dispersal
Male G. m. centralis are active only a mean 32 minutes per day (Bursell and Taylor, 1980). Presumably, males and females of the other species show similar activity patterns. Tsetse dispersal is a critical component of models constructed to evaluate efficacies of area-wide population management procedures and to evaluate historical biogeographical events (Hargrove, 1981; Williams et al., 1992; Hargrove, 2000; Vale and Torr, 2005). Dispersion rates are said to be high, compromising otherwise effective control programmes (Williams et al., 1992; Brightwell et al., 1997; Hargrove, 2003). Rogers (1977) modeled dispersion as a random diffusion process in which the distance d moved d ≈ s√t, where s is the step length (the mean daily displacement) and t is time in days. Rogers calculated daily displacement rates for 13 field studies, estimated over a scale of days to 17 years. The overall mean s = 252 m for savannah species. This implies a frontal advance of 1.4 km a month and 4.7 km a year. Riverine taxa are said to disperse at higher rates. A more recent review offered by Hargrove (2000) supports Rogers’ analysis.
The foregoing citations predict significant rates of gene flow. There is a caveat, however. Dispersion is delayed because teneral and young flies are less likely than older flies to disperse because their flight muscles require, at tropical temperatures, 2 – 3 bloodmeals and 6 - 8 days to mature (Hargrove, 2003, 2005). Female flies become receptive at 3 days of age and virtually all are inseminated by 6 - 8 days. Under these conditions, most matings are likely to be between members of a deme or nearby demes. Exchange of flies among demes, therefore, critically depends on post-teneral dispersal rates and the reproductive success of emigrants in their ‘new’ environments.
Genetic variation in tsetse flies
Theory indicates a relationship between population size and genetic diversity (He): He = 4Neν/(4Neν + 1) where Ne is effective population size (typically a large number) and ν is the mutation rate (a very small number, ~10-4 to 10-9). Tsetse populations seem small when compared with Diptera such as Drosophila, Muscoid flies, mosquitoes, etc. Indeed, early studies suggested a paucity of genetic variation in tsetse flies. Morphological mutants were few and allozyme diversity low (Gooding, 1992), features that could be attributed to historically small population sizes. More recent, comprehensive surveys of allozyme variation confirmed the low diversity but disclosed sufficient variation to allow studies of gene flow in natural populations (Krafsur et al., 1997). Comparative genetic statistics for allozymes (Table 1) generally indicate lesser diversities (heterozygosities) than occur in other medically and economically important Diptera (Krafsur and Griffiths, 1997). The four taxa investigated in Morsitans group flies, moreover, showed closely similar heterozygosities although the proportion of polymorphic loci was less in G. swynnertoni Austen 1923. The homogeneous heterozygosities are notable because G. swynnertoni has a much smaller geographic range than the others and its populations have contracted in recent times; indeed, its effective population size is quite small, of tens to hundreds of flies (Marquez et al., 2006). Mitochondrial variation in G. swynnertoni, on the other hand, was less than in other Morsitans group taxa. A similar picture was obtained in Zimbabwe and Zambian populations of G. pallidipes, among which allozyme diversity was greater than that in Kenya populations, but mitochondrial and microsatellite variation was much reduced (Krafsur, 2002a). The conservation of allozyme variation in two tsetse species seems to be an example of balanced polymorphism in which selective advantage accrues to heterozygotes at structural gene loci. A similar phenomenon was observed in Drosophila (Hale and Singh, 1991).
Table 1.
Allozyme variation in tsetse flies compared with other Diptera
| Taxa | No. loci | Percentage polymorphic | Mean alleles per locus | Mean diversity, He | Diversity at polymorphic loci |
|---|---|---|---|---|---|
| G. m. morsitans | 45 | 20.0 | 1.4 | 6.6 | 29.9 |
| G. m. centralis | 31 | 32.3 | 1.4 | 6.0 | 18.7 |
| G. swynnertoni | 34 | 17.6 | 1.2 | 7.2 | 40.5 |
| G. pallidipes | 38 | 26.3 | 1.5 | 6.8 | 25.4 |
| Musca domesticaa | 68 | 53.4 | 2.5 | 18.3 | 36.7 |
| Musca autumnalisb | 50 | 62.0 | 2.4 | 18.6 | 30.5 |
| Stomoxys calcitransc | 38 | 52.6 | 1.8 | 9.6 | 18.2 |
Krafsur, E., Helm, J., Black, W. 1992. Biochem. Genet. 30, 317
Krafsur, E., Black. W. 1992. Biochem. Genet. 30, 625
Krafsur, E. 1993. Biochem. Genet. 31, 231.
Microsatellite variation
Simple sequence repeats (SSRs) occur throughout the nuclear genome. They are codominant and highly polymorphic. SSRs have been developed for the Morsitans group (Baker and Krafsur, 2001; Ouma et al., 2003, 2006). Some primers for Morsitans group tsetse also amplify orthologous loci in the Fusca and Palpalis groups. SSRs have also been developed specifically for G. palpalis gambiensis (Luna et al., 2001) and G. f. fuscipes (Abila et al., 2008).
Diversities in microsatellite loci vary among tsetse taxa and compare with those in house flies, which have very much larger effective populations sizes (Table 2). Rather more loci would be helpful in furthering genetic understanding of tsetse flies. SSRs are expensive and time consuming to identify and evaluate as useful markers. They are typically isolated from untranscribed regions of the genome such that well conserved flanking regions that serve as good priming sites are not abundant, leading to high frequencies of null alleles and poor amplifications in related taxa. Expressed sequence tag libraries are available for tsetse (Lehane et al., 2003) that could be used to find and develop new SSR loci thereby avoiding the problems often associated with loci in unexpressed genomic regions (Kim et al., 2008).
Table 2.
Microsatellite diversities and tests for random matings F in Glossina spp and the house fly
| No. Populations | No. loci | Alleles per locus | Diversity He | Within demes FIS | Among demes FST | |
|---|---|---|---|---|---|---|
| G. m. morsitansa | 6 | 6 | 11.0 ± 5.6 | 0.73 ± 0.06 | 0.03 ± 0.03 | 0.19 ± 0.05 |
| G. m. morsitansb | 9 | 7 | 28.6 ± 4.5 | 0.74 ± 0.05 | 0.17 ± 0.07 | 0.13 ± 0.01 |
| G. m. centralisc | 7 | 7 | 8.8 ± 3.7 | 0.70 ± 0.09 | -0.12 ± 0.04 | 0.19 ± 0.04 |
| G. m. submorsitansa | 7 | 7 | 12.7 ± 6.2 | 0.81 ± 0.04 | 0.03 ± 0.03 | 0.17 ± 0.07 |
| G. pallidipesd | 21 | 8 | 26.8 ± 8.7 | 0.80 ± 0.03 | 0.07 ± 0.03 | 0.18 ± 0.02 |
| G. f. fuscipese | 8 | 5 | 8.2 ± 3.6 | 0.43 ± 0.07 | 0.11 ± 0.05 | 0.22 ± 0.07 |
| Musca domesticaf | 14 | 7 | 7.9 ± 1.1 | 0.86 ± 0.02 | 0.08 ± 0.02 | 0.13 ± 0.02 |
Krafsur and Endsley, 2002. Genotypes scored on acrylamide gels
Ouma et al., 2007. Genotypes scored on ABI 3100 using GeneScan 3.1 and Genotyper 2.5
Krafsur et al., 2001. Genotypes scored on acrylamide gels
Ouma et al., 2005. Genotypes scored on ABI 3100 using GeneScan 3.1 and Genotyper 2.5
Population structure: genetic differentiation and gene flow among demes
Estimators of genetic differentiation include FST (Wright, 1978), GST (Nei, 1987), and theta (θ) (Weir & Cockerham, 1984). FST and it analogues describe departures from random matings among demes at hierarchical population levels. These estimators are based on different assumptions, but each attempts to measure the same thing. FST can be considered as the ratio between within deme diversity HS and the diversity in the total population HT thus (FST = 1 − HS/HT), and as correlations between uniting alleles. In theory, FST can take a value from −1 to 1, but that holds only for a single locus with two alleles. In practice, estimates cannot exceed the mean level of homozygosity in subpopulations, 1 − HS (Hedrick, 1999). Departures from random mating within demes (= subpopulations) are estimated by FIS (individuals in subpopulations).
The geographic scale of sampling tsetse flies varied from hundreds of meters to thousands of kilometers. Often it was found that a nested design was appropriate, for example samples nested in regions. High indices of genetic differentiation can indicate the occurrence of sibling species and must be considered given the substantial number of sibling species (morphologically identical species) that have been detected in other Diptera. Evaluating the question in the Glossinidae is not a simple task, however, because sampling and establishing geographically diverse cultures for genetic analysis is extraordinarily time consuming and expensive. Existing lab cultures are few and longstanding. For example, the G. fuscipes fuscipes culture dates from 1986 or earlier – documentation about its origins are now lost although it is thought the strain originated in the Central African Republic (see Krafsur et al., 2008). G. morsitans s.l. cultures date from the 1960s. The final test of intrinsic reproductive isolation is to cross reciprocally different strains and evaluate the fertilities of the matings and the fertilities of any progeny in backcrosses. Nevertheless, a number of investigations have demonstrated sibling species. A study in chromosome mapping has disclosed the putative existence of two mating types in G. palpalis palpalis, one derived from Nigeria and the other from Zaire (Gooding et al., 2004). Much earlier, G. m. morsitans, G. m. submorsitans, and G. m. centralis were separated initially on the basis of hybrid sterility and shown to be allopatric (Vanderplank, 1949a). More recent studies on the phyletic relationships among G. morsitans s.l. and the closely related G. swynnertoni have been reviewed (Gooding and Krafsur, 2004, 2005).
Population structure in subgenus Morsitans – the Morsitans group
Morsitans group flies occur chiefly in the wooded savannahs of East and southern Africa where they are medically and economically highly significant (Fig. 1). Distribution maps (Rogers and Robinson, 2004) show that only G. longipalpis Weidemann 1830 is exclusively West African; G. pallidipes Austen 1903, found in East and southern Africa, is morphologically and ecologically similar (Vanderplank, 1949a; Leak, 1999) but no crosses between them seem to have been reported. It would be most interesting to investigate their phylogenetic relationships.
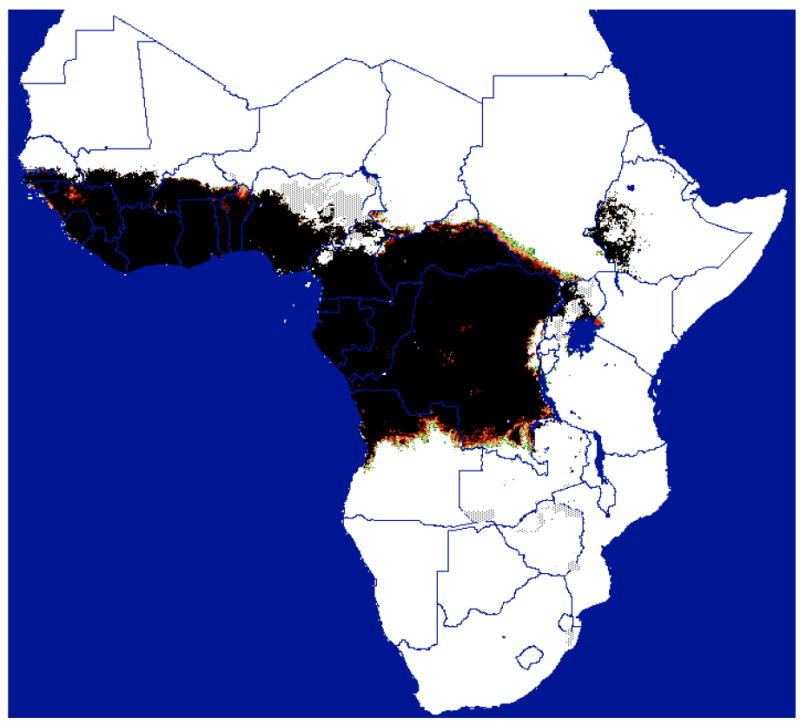
Figure 1.
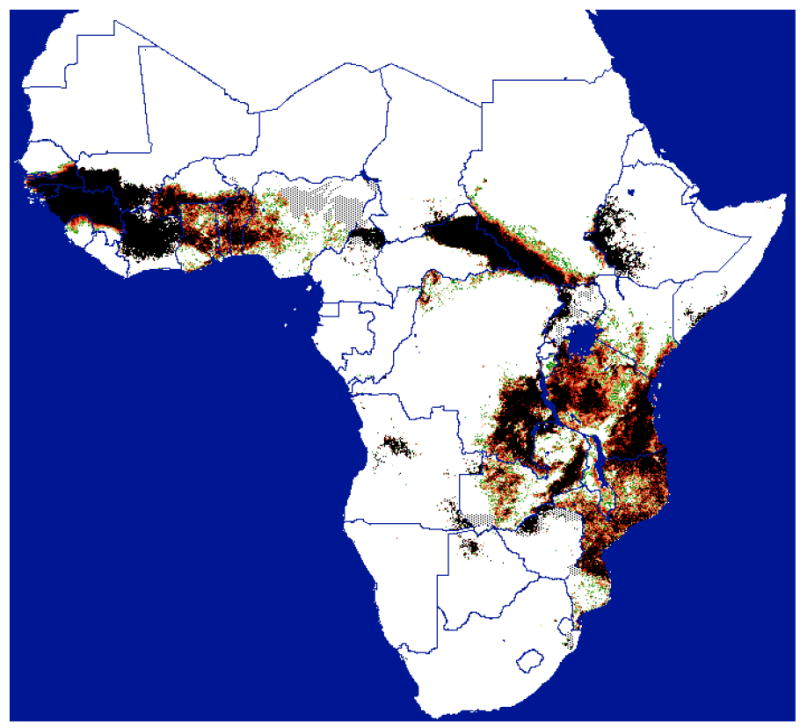
Distribution of Morsitans group tsetse flies predicted from satellite imagery (courtesy of ERGO Ltd and TALA, Oxford, with permission. Available from <http://ergodd.zoo.ox.ac.uk/tseweb/distributions.htm>.
As we have seen, allozyme, microsatellite, and mitochondrial variation have been characterized in G. morsitans s.l. and G. pallidipes. Summary statistics for the foregoing taxa are offered in Tables 2 and 3. In making within-taxon comparisons, it should first be observed that improvements in methods have a strong bearing on summary statistics. In G. m. morsitans, for example, the use of automated nucleotide-sequencing technologies and proprietary allele scoring software doubled the number of alleles detected when compared with silver-stained 4.5% acrylamide gels in the author’s lab. Estimates of diversity He were not affected, but F statistics differed. Use of automated sequencing methods for mitochondrial PCR amplicons provided greater numbers of haplotypes, hence greater diversities, than the SSCP technique. In particular, the number of singletons detected was greater, a demographically important index because a high frequency can indicate recent population expansion or the operation of natural selection.
Table 3.
Mitochondrial diversities and genetic differentiation in wild Glossina species.
| Method | No. populations | No. flies | No. haplotypes | Haplotype diversity, HS | FST/GST | |
|---|---|---|---|---|---|---|
| G. m. morsitansa | SSCP | 5 | 111 | 25 | 0.81 ± 0.04 | 0.09 ± 0.02 |
| G. m. morsitansb | ABI 3730 | 7 | 96 | 33 | 0.81 | 0.40 ± 0.08 |
| G. m. centralisc | SSCP | 6 | 265 | 7 | 0.54 ± 0.16 | 0.81 ± 0.07 |
| G. m. submorsitansd | SSCP | 7 | 282 | 26 | 0.51 ± 0.12 | 0.35 |
| G. pallidipese | SSCP | 21 | 624 | 39 | 0.42 ± 0.02 | 0.52 ± 0.001 |
| G. pallidipesf | ABI 3730 | 23 | 873 | 181 | 0.73 ± 0.09 | 0.47± 0.07 |
| G. p. gambiensisg | SSCP | 13 | 372 | 9 | 0.18 | 0.68 |
| G. swynnertonih | ABI 3730 | 8 | 149 | 18 | 0.59 ± 0.10 | 0.04 |
| G. f. fuscipesi | ABI 3730 | 3 | 79 | 21 | 0.84 ± 0.05 | 0.28 |
| G. f. fuscipesj | ABI 3730 | 9 | 202 | 37 | 0.93 | 0.60 ± 0.07 |
Krafsur and Endsley, 2002
Marquez and Krafsur, unpublished data
G. morsitans sensu lato consist of subspecies principally because they are morphologically difficult to distinguish and intermate freely in cages (Vanderplank,1949a). G. swynnertoni was considered to be only a variant of G. morsitans s.l. for the same reason. Machado (1970) regarded G. morsitans s.l. to be only ‘major geographical races’ even though Vanderplank had demonstrated hybrid sterility, i.e., reproductive isolation. Numerous studies have shown that hybrid males are sterile and the females sterile or semisterile (reviews in Gooding and Krafsur, 2004, 2005). Genetic studies, moreover, provide no tangible evidence of gene flow among natural populations of the three taxa. The subspecies are entirely allopatric. Clearly, these taxa have a common ancestor and share large regions of their genomes, but each has evolved different adaptations. For example, G.m. centralis Machado 1970 occurs in cooler, wetter conditions, compared with G.m. morsitans Westwood 1850, which occurs in hotter and drier areas (Robinson et al. 1997a,b). Krafsur and Endsley (2006) argued that G. morsitans subspecies should be elevated to species rank because genetic evidence indicated complete premating and postmating reproductive isolation among them.
G. m. morsitans is distributed in four broad belts. Two belts occur in Mozambique, one of which extends well into southern and coastal Tanzania. Another large belt occurs from northern Zimbabwe through eastern Zambia (Rogers and Robinson, 2004). This G.m. morsitans belt lies immediately east of two extensive G.m. centralis belts (Robinson et al., 1997a, b). Samples for genetic analysis were obtained from western Mozambique, Zimbabwe, Zambia, and Tanzania at distances of 12 to 917 km (Wohlford et al., 1999; Krafsur and Endsley, 2001; Ouma et al., 2007). Genotypes were obtained for seven microsatellite loci and mitochondrial locus Cox I. Microsatellite allelic diversities (22.6 alleles per locus) and heterozygosities (He = 0.77) were greater in four Tanzanian populations than in five southern African populations (18.7 and 0.71, respectively). Thirty-three Cox I haplotypes were recorded in 96 flies from seven populations (only three southern Africa populations could be amplified by PCR). Only one haplotype was shared among six populations and it occurred in 43 (45%) G.m. morsitans. Seventy haplotypes (73%) were singletons. Tanzania flies had the most private and singular haplotypes; diversity was 0.72 in Tanzania and 0.50 in southern Africa. An hypothesis of isolation by distance (IBD) was not rejected when based on microsatellite loci, but the slope was very shallow. IBD was rejected, however, when based on mitochondrial variation.
G. m. centralis populations are geographically discontinuous. This tsetse occupies an area in central Angola small relative to its chief habitats in western Tanzania, Zambia, and southeastern Democratic Republic of Congo (Zaire). A separate population occurs also in Botswana, the Caprivi Strip of Namibia, and southeastern Angola. Genetic sampling of six populations was carried out in Zambia, Botswana, and Namibia (Krafsur and Griffiths, 1997). No samples were obtained, however, from the huge belt in Tanzania and northern Zambia. Mitochondrial and microsatellite diversities were very low in G. m. centralis, compared with diversities in other Morsitans group tsetses. For example, only seven mitochondrial haplotypes were detected over two loci. Allozyme diversity indices of average alleles per locus and mean heterozygosity, on the other hand, were homogeneous with other G. morsitans s.l. when measured over 31–45 putative loci. Diversity (heterozygosity) at only polymorphic allozyme loci, however, was less in G. m. centralis: 18.7% vs 31.9%. Genetic differentiation was pronounced among the six populations sampled; FST for mitochondrial variation was 0.87 and 0.19 for microsatellites. The difference in estimates can be attributed to high mutation rates, the much greater sensitivity of the mitochondrial genome to demographic change, and homoplasy at microsatellite loci. It is notable that 171 of 174 flies from Botswana and the Caprivi Strip, taken from more than 10 sampling locations, possessed the same haplotype, suggesting the sampled populations originated from an earlier bottlenecked population. Periodic insecticidal treatments were applied aerially to Botswana from 1973 to 1991 with institutional claims of success, but the genetic data are consistent with the ecological analysis of Hargrove (2003), viz, survival of the resident population well within the treated region.
Clearly, G. m. centralis populations are strongly subdivided through southern Africa, a consequence of geographic isolation and genetic drift. It will be interesting to see if genetic diversities among Tanzanian populations are similar to those recorded in Zambian and Botswana populations. G. m. centralis disappeared from large areas of southern Africa (Ford, 1971) because of the rinderpest, and only slowly recovered from very low, undetectable, population numbers. Thus, the paucity of genetic variation was caused by genetic bottlenecks. The species has now been eliminated by aerial applications of the pyrethroid insecticide deltamethrin over 16,000 km2 of the Okavango delta in Botswana in 2001-2002 (Kgori et al., 2006). No tsetse flies have been detected by continuous and intensive sampling since (P. Kgori, Veterinary Services, Botswana, personal communication, 16/5/08). It should be noted that conspecific populations nearby in Namibia and southern Zambia were not treated nor have they yet furnished detectable surviving immigrants to Botswana.
G. m. submorsitans Newstead 1910 occurs in Guinea Bissau east to Uganda and southwestern Ethiopia but its distribution is highly discontinuous. The largest discontinuities occur in Nigeria and southern Sudan (Rogers and Robertson, 2004). Examination of male genitalia led Vanderplank (1949b) to describe four morphologically distinct races, later supported by Machado (1970). The Ugandan forms were designated G. m. submorsitans var ugandensis. Crossing experiments have not adequately tested phylogenetic relationships among the four nominal races, a difficult task because representative laboratory cultures do not exist. Some work has been accomplished on population genetics, however.
Microsatellite and mitochondrial variation was used to investigate differentiation and gene flow in G. m. submorsitans sampled at the eastern and western extremes of its distribution, approximately 5450 km apart (Krafsur et al., 2000; Krafsur and Endsley, 2002). Mitochondrial diversities in five samples from The Gambia (he = 0.27) were only a third of those in two Ethiopian samples (he = 0.84). Here the SSCP method was used to estimate diversity. Genetic differentiation within countries was significant in The Gambia (FST = 0.04) but not in Ethiopia (FST = 0.003) where the two demes were only c. 10 km apart. FST between countries, however, was 0.34. Nei’s (1987) unbiased genetic identity I varies from zero when no variant is shared, to one, when all are shared in equal measure, was I = 0.65 between countries, I = 0.96 among Gambian subpopulations, and about I = 0.985 between the Ethiopian subpopulations. Microsatellite diversities were he = 0.68 in The Gambia and he = 0.72 in Ethiopia. Subpopulation allele frequencies differed significantly within each country. Indices of genetic differentiation for microsatellite loci were FST = 0.34 between The Gambia and Ethiopia and FST = 0.35 among all subpopulations. The low mitochondrial diversities among G. m. submorsitans in The Gambia are indicative of recent contractions in population numbers. Human population increase, loss of fly habitat, and progressive decrease in annual rainfall are likely to have caused the decline in tsetse populations (Krafsur et al., 2000). Genetic diversities among G. m. submorsitans in Ethiopia, on the other hand, seemed typical of a large, longstanding population. Hutchinson (1971) and colleagues found it to be abundant and widespread. Ugandan and Ethiopian G. m. submorsitans are allopatric (see Rogers and Robinson, 2004), so the morphological pattern of Ethiopian G. m. submorsitans could be quite different from the Ugandan morphotype although it has been assumed by some that the Ethiopian race is variety ugandensis. Morphological differentiation has genetic and strong environmental components. Covariances and genotypic by environment interactions further complicates unambiguous genetic inferences of morphological variation. Interpretation of quantal morphological traits therefore requires a formal experimental design and thorough sampling before tentative genetic conclusions may be drawn.
G. swynnertoni occupies only a small region relative to the other Morsitans group tsetse taxa. Two principle belts in north central Tanzania are figured in Rogers and Robinson’s (2004) map. Its preferred habitat is open woodland savannah; its range has contracted because of earlier control programmes and loss of habitat (Marquez et al., 2006). It is now confined largely to national parks and reserves where it remains a serious trypanosomiasis vector. An early report suggested that G. swynnertoni might have experienced a genetic bottleneck (Gooding et al., 1993). The magnitude of allozyme variation in G. swynnertoni, however, was similar to that in other Morsitans group taxa and provided no evidence of a stringent, earlier bottleneck (Table 1). As we shall see, however, allozyme variation is subject to balancing selection that can maintain heterozygosities at high levels even in bottlenecked populations. Mitochondrial variation is more sensitive to demographic phenomena and has been systematically assessed in G. swynnertoni (Marquez et al., 2006). Only 18 haplotypes were detected by sequencing two loci (r16S2 and Cox I) totaling 668 bp among 8 populations totaling 149 flies. Only two haplotypes accounted for 80% of the total number of flies sequenced. Ten of the 18 haplotypes were singletons (56%), indicative of mutation-drift disequilibrium and consistent with the notion that the G. swynnertoni populations examined had experienced an earlier bottleneck and subsequent expansion. The estimate of genetic differentiation among the locations was GST = 0.04, the lowest estimate of any Morsitans group species. Temporal differentiation, however, was four times the magnitude of spatial variation. Drift is inversely proportional to population numbers, so it seems that the sampled populations were quite small. Better resolution of population structure may be afforded by survey of diploid loci. Nine putative microsatellite loci were identified (Baker and Krafsur, 2003; Ouma et al., 2006), but no survey of diversity has yet been carried out.
The temporal flux in G. swynnertoni haplotype frequencies allowed estimates of effective population size, Ne (Table 4). The most reliable estimate was derived from villages in Tarangiri, where successive samplings were made at an interval of approximately 54 generations. Here, the estimate of Ne = 66 reproducing females per generation. The other estimates varied from two to 41 reproducing females. These are not large numbers so it is well to bear in mind that Ne estimates represent harmonic means and so are weighted by the smallest numbers in the sampling intervals.
Table 4.
Effective population numbers (Ne) in Glossina spp
| Location | Years | Interval in generations | Ne | 95% C. L. | |
|---|---|---|---|---|---|
| G. swynnertonia | Tarangiri | 1996 - 99 | 22 | 18 | 14 - 31 |
| Tarangiri | 1999 - 03 | 35 | 19 | 17 - 37 | |
| Tarangiri | 1996 - 04 | 54 | 66 | 54 - 107 | |
| G. pallidipesb | Nguruman | 2000 - 01 | 12 | 333 | 192 - 597 |
| Nguruman | 2000 - 03 | 26 | 679 | 393 - 1190 | |
| Nguruman | 2001 - 03 | 17 | 250 | 157 - 382 | |
| Nguruman | 2000- 03 | 29 | 1026 | 572 - 1871 | |
| Lambwe | 2000 - 02 | 20 | 202 | 122 - 328 | |
| Lambwe | 2000 – 03 | 28 | 522 | 302 - 931 | |
| G. f. fuscipesc | Kenya | - | - | 403 | - |
| Uganda | - | - | 2258 | - |
Marquez et al., 2006. Ne estimated from temporal variances in gene frequencies
Ouma et al., 2006. Ne estimated from temporal variances in gene frequencies
Krafsur et al., 2008. Ne estimated from pairwise differences among mitochondrial haplotypes
G. pallidipes geographic distribution is extremely patchy according to Rogers and Robinson’s (2004) map (Fig. 2). Sizable but discontinuous belts are figured in Ethiopia, Somalia, Kenya, Uganda, Tanzania, Zaire, Mozambique, Zambia, and Zimbabwe. Probabilities of occurrence are small in vast areas of Tanzania and Zambia even though many of these same regions can support G. morsitans. Genetic sampling has been carried out in many of the principal belts. Allozyme variation at eight loci was used to estimate genetic differentiation and gene flow among 11 G. pallidipes populations from Kenya, Zambia, Zimbabwe, and Mozambique (Table 1). These data pointed to a high degree of population structure in which the average level of gene flow Nm was much less than anticipated by a reading of the ecological literature (FST = 0.24, Nm ~ 0.8). Mating was random within the sampled populations thereby falsifying an hypothesis of two or more sibling species within populations. The indices of mean allozyme diversity, HE, and of genetic differentiation, FST, varied between the East African and southern African populations. HE was greater in the southern populations but FST was less. Further investigation compared allozyme, microsatellite, and mitochondrial variation in the same 11 populations (Krafsur, 2002a). Microsatellite diversity and mitochondrial diversity were less in southern Africa than in East Africa and both were strongly correlated with each other, but neither was correlated with allozyme diversity. In contrast to the allozyme diversities, mitochondrial and microsatellite variation was consistent with a severe and prolonged reduction in population sizes in southern Africa. Allozyme variation was conserved, however, and differentiation at allozyme loci among populations was much less than at microsatellite and mitochondrial loci. This is illustrated in Fig. 3, where pairwise estimates of mitochondrial, microsatellite, and allozyme genetic distances are regressed separately on geographic distances. Conservation of allozyme variation in southern African G. pallidipes that had experienced an earlier, severe bottleneck (Ford, 1971) is an example of balancing selection. Allozymes, it seems, are not selectively neutral in tsetse.
Figure 2.
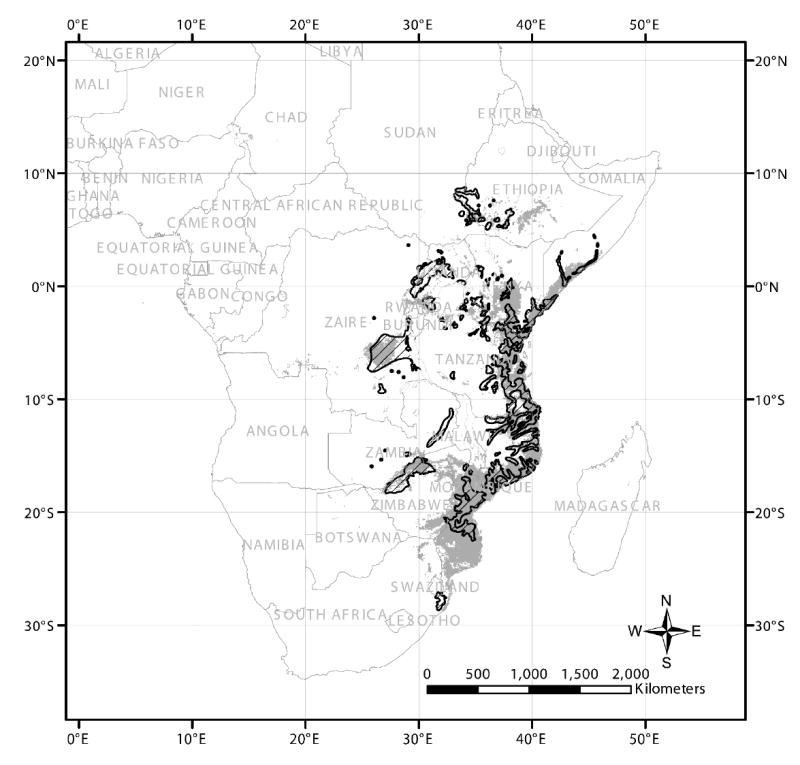
Predicted (grey) and observed (hatched) distribution of G. pallidipes, from Rogers and Robinson (2004) with permission.
Figure 3.
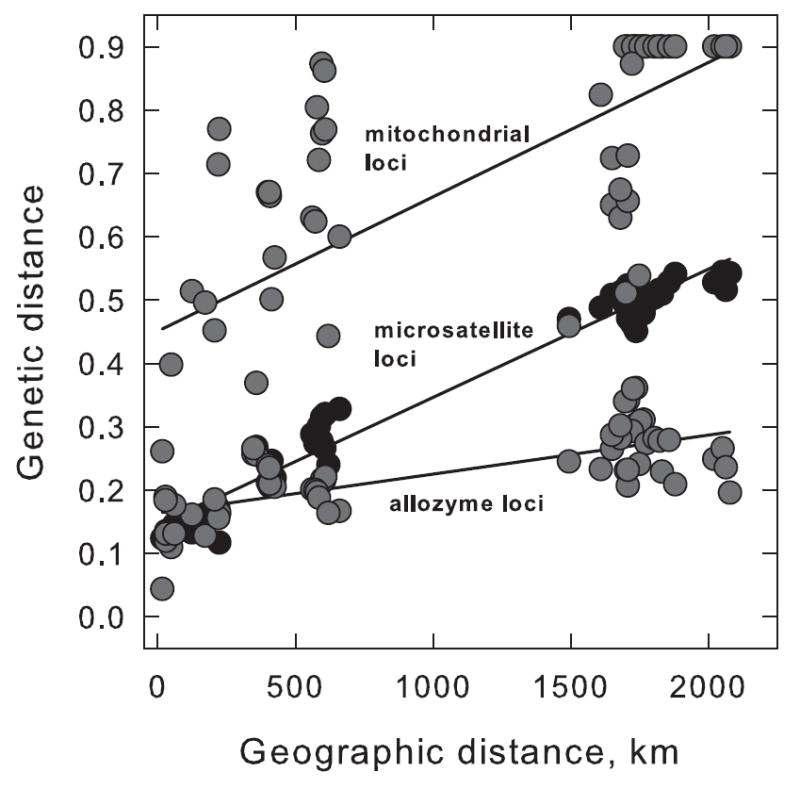
Pair-wise Cavalli-Sforza and Edwards chord genetic distances among G. pallidipes populations regressed on corresponding geographic distances. Note the slope for allozyme loci is much shallower than for mitochondrial and microsatellite loci (from Krafsur, 2002).
Because of its wide, but discontinuous distribution, high degree of subdivision, and economic importance, it was decided to investigate further the population structure of G. pallidipes. Microsatellite and mitochondrial variation was examined in detail at macrogeographic (Krafsur and Wohlford, 1999; Ouma et al., 2005) and microgeographic scales (Ouma et al., 2006). Single strand conformational polymorphisms (SSCPs) were used to characterize mitochondrial variation (later, direct sequencing was adopted to obtain better resolution). The picture afforded by these further studies was consistent with earlier inferences based upon allozyme variation. At the macrogeographic scale, 21 samples representing populations from Ethiopia south to Zimbabwe were placed into five geographically and ecologically homogeneous groups (Table 5). Microsatellite diversities, in terms of mean alleles per locus and heterozygosity, varied significantly among the grouped populations. The eastern populations in Kenya and Tanzania had twice the allelic diversity as the other groups and greater mean heterozygosities. The bottlenecked southern African populations showed similar microsatellite diversity indices as three other groups.
Table 5.
Microsatellite variation in regionally grouped G. pallidipes populations. Means are over 8 loci
| Grouping | No. populations | No. flies | Avg. no.alleles | Diversity | FST |
|---|---|---|---|---|---|
| Ethiopia | 2 | 48 | 10.0 ± 4.8 | 0.69 ± 0.05 | 0.072 |
| Western Kenya | 4 | 144 | 11.5 ± 5.4 | 0.64 ± 0.09 | 0.203 |
| Southwest Kenya | 7 | 176 | 9.9 ± 4.4 | 0.63 ± 0.01 | 0.081 |
| East Kenya, Tanzania | 5 | 240 | 22.4 ± 8.2 | 0.82 ± 0.04 | 0.110 |
| Southern Africa | 3 | 96 | 12.8 ± 5.4 | 0.73 ± 0.06 | 0.096 |
| Totals and means | 21 | 720 | 26.8 ± 8.7 | 0.80 ± 0.03 | 0.213 |
Kruskal-Wallis test of mean alleles per locus among groups χ24 = 66.1, P < 0.001
Kruskal-Wallis test of homogeneity of diversity among groups χ24 = 19.97, P ~ 0.005
Mitochondrial variation is more sensitive to demographic flux than diploid loci because of its uniparental inheritance and single copy number. Thus, of 39 SSCP haplotypes recorded, only four (10%) were found in southern Africa, compared with 23 (59%) in coastal Kenya and Tanzania. Mean diversity was 0.42 and FST = 0.51 over the 21 populations (Table 6). Here, diversity is the probability that two randomly chosen flies have different haplotypes and the estimate of mean gene flow among populations is approximately one reproductive female every two generations.
Table 6.
Mitochondrial variation estimated by single strand conformational polymorphisms in regionally grouped G. pallidipes populations.
| Grouping | No. populations | No. flies | No. haplotypes | Diversity, HS | FST |
|---|---|---|---|---|---|
| Ethiopia | 2 | 48 | 7 | 0.44 ± 0.10 | 0.07 |
| West Kenya | 4 | 160 | 16 | 0.60 ± 0.06 | 0.27 |
| Southwest Kenya | 7 | 152 | 11 | 0.35 ± 0.09 | 0.41 |
| East Kenya, Tanzania | 5 | 168 | 23 | 0.59 ± 0.08 | 0.23 |
| Southern Africa | 3 | 96 | 4 | 0.04 ± 0.03 | 0.48 |
| Totals and means | 21 | 624 | 39 | 0.42 ± 0.02 | 0.51 |
Kruskal-Wallis test of homogeneity of diversities among groups χ24 = 10.97, P = 0.03.
Sequencing r16S2 and Cox I (a sum of 670 bp) yielded 181 haplotypes among the 873 flies sequenced, only 12 (7%) of which occurred in southern Africa and 144 (80%) in Kenya and Tanzania (Marquez and Krafsur, unpublished data). Mean haplotype diversity was much greater at he = 0.73 than estimated by SSCP but FST was about the same at 0.47. There was a high frequency of singleton haplotypes − 64%. Singletons can be indicative of rapidly expanding populations (Slatkin and Hudson, 1991; Rogers and Harpingdon, 1992) or more complex demographic phenomena including selection against mitochondrial mutants. Detection of demographic changes by genetic means, however, is uncertain when expanding populations are subject to pronounced environmental and temporal heterogeneities (Wegmann et al., 2006). Nevertheless, the high frequencies of singular mitochondrial haplotypes is consistent with rapid population expansions and well supported by the historical record relating the African rinderpest epizootic to the disappearance of tsetse fly populations in southern Africa. This is illustrated in regional lineage through time plots (Fig. 4) that show abrupt increases in lineage numbers about 300 to 400 generations ago, corresponding to 41 – 55 years before sampling, in the years 1940 – 1954. The oldest lineages varied from c. 671 generations in Tanzania and southern African G. pallidipes (~ 92 y), 1864 generations in Kenya (~ 255 y), to c. 9200 generations in Ethiopia (~1260 y).
Figure 4.
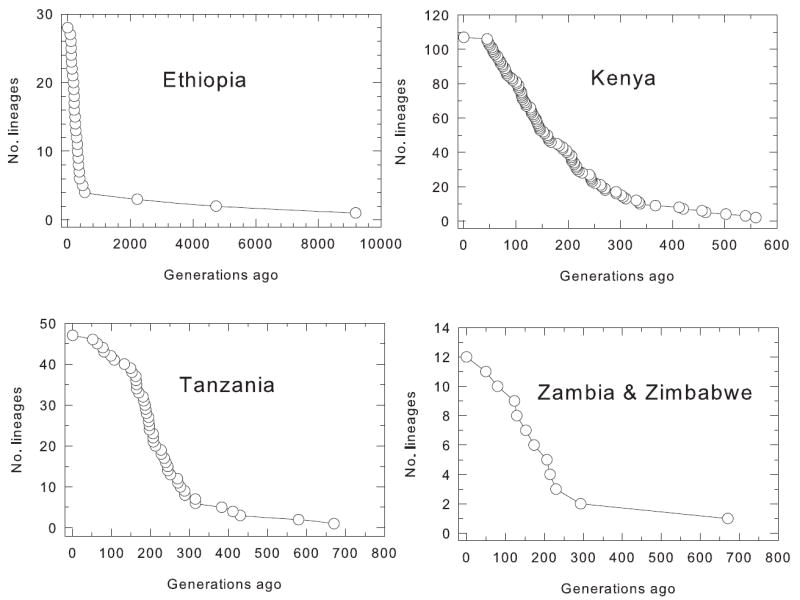
Lineage (haplotype) through time plots showing temporal increases in mitochondrial diversities. Note the small number of lineages (12) in Zimbabwe and Zambia and the rapid increases in diversities 300 – 400 generations ago in each region.
At the microgeographic level, G. pallidipes subpopulations were sampled at scales of 200 m to 14 km (Ouma et al., 2006). The chief locations were Lambwe in the Ruma National Park in western Kenya, and Nguruman, in southwestern Kenya, adjacent to Tanzania. Lambwe and Nguruman are 240 km apart and much of the area between is at least 2000 m elevation and, therefore, inhospitable to tsetse fies. The tsetse populations in Lambwe and Nguruman have been intensively studied (e.g., Turner and Brightwell, 1986; Williams et al., 1992; Brightwell et al., 1992, 1997). Trap catches in Nguruman showed a high degree of spatial aggregation (Odulaja et al., 2001) raising the question, does this aggregation represent a component of genetic differentiation among local demes? G. pallidipes populations closest to Nguruman and Lambwe also were sampled. FST over samples in Lambwe was 0.05; in Nguruman, FST = 0.04, both significantly different from zero (P < 0.0001). Differentiation of allele frequencies among the three blocks in Lambwe was highly significant (χ2(14) = 1067, P < 0.0001). A similar result obtained among the seven blocks in Nguruman (χ2(42) = 1160, P < 0.0001). Even at distances of a few hundred meters, it seems that tsetse flies form demes among which genetic drift is stronger than gene flow. Genetic differentiation of Lambwe G. pallidipes from its nearest conspecific neighbour at Kodera was FST = 0.15. Similarly, the contrast between Nguruman and its neighbour Serengeti yields FST = 0.16. The foregoing estimates are nearly 40% of the maximum values they can take because levels of homozygosity at microsatellite loci were ~ 0.39. Sequencing Cox I and r16S2 loci supported the view of little shared variation (Marquez and Krafsur, unpublished data). Only one of 187 Nguruman G. pallidipes shared its mitochondrial haplotype with one of 31 Serengeti flies.
Estimates of effective population numbers in Nguruman were approximately double those in Lambwe: the modal values were approximately 700 and 350 respectively (Table 4). Taken together with indices of genetic differentiation, the Ne estimates suggest strongly that longstanding, substantial populations existed even in the face of sustained control efforts.
Subgenus Machadomyia
As discussed earlier, G. austeni cytogenetics have been examined (Southern et al., 1972; Gariou-Papalexiou et al., 2007) and chromosome interchanges induced and manipulated experimentally (Curtis et al., 1972). Two subspecies are allopatric and have been described based on differences in body size, color, habitats, and responses to traps (Dias, 1987). The chief systematic question concerning G. austeni is its phylogenetic relationships to the other Glossina (Gooding and Krafsur, 2004, 2005 for reviews). The distribution of G. austeni is largely coastal and highly discontinuous from Natal Province in South Africa north to the River Jubba in Somalia. Representatively sampling this species is difficult because it does not respond well to traps (Vreysen et al., 1998). Its population genetics are uninvestigated but RAPDs were used as markers to detect genetic differences between cultured flies and wild flies on the continent and Zanzibar; unquantified differences in polytene chromosome banding patterns were found among flies from the lab culture and field samples from Zanzibar, Tanga (Tanzania), and Zululand (Gariou-Papalexiou et al., 2000). Sequencing of the r16S2 and Cox I mitochondrial genes in 32 specimens from Pretoria, South Africa and 15 specimens from a lab culture in Tanga, Tanzania yielded only eight haplotypes (Table 7). The Pretoria culture was monomorphic and its haplotype was unshared with the Tanga culture.
Table 7.
Sequence variation at mitochondrial r16S2 (197 bp) and CoxI (321 bp) among Glossina species. The sequences were aligned over species and concatenated.
| Species | No. samples | No. flies | No. haplotypes | Diversity Hs |
|---|---|---|---|---|
| G. swynnertoni | 4 | 150 | 13 | 0.271 |
| G. pallidipes | 23 | 873 | 149 | 0.666 |
| G. m. morsitans | 3 | 88 | 21 | 0.685 |
| G. m. centralis | 1 | 47 | 15 | 0.684 |
| G. m. submorsitans | 3 | 87 | 30 | 0.777 |
| G. austeni (lab culture) | 2 | 47 | 8 | 0.362 |
| G. fuscipes | 4 | 127 | 41 | 0.863 |
| G. p. gambiensis | 2 | 60 | 23 | 0.822 |
| G. longipennis | 1 | 31 | 7 | 0.559 |
| G. brevipalpis | 2 | 72 | 13 | 0.652 |
Kruskal-Wallis test of homogeneity in haplotype numbers: χ29 = 9.49, P = 0.39
Kruskal-Wallis test of homogeneity in diversities: χ29 = 14.4, P = 0.12
Population structure in subgenus Nemorhina – the Palpalis group (Fig. 5)
Figure 5.
Distribution of Palpalis group tsetse flies predicted from satellite imagery (courtesy of ERGO Ltd and TALA, Oxford, with permission. Available from <http://ergodd.zoo.ox.ac.uk/tseweb/distributions.htm>.
A more complex picture obtains for palpalis group than morsitans group tsetses. G. p. gambiensis is riparian and also occurs in humid savannah. Its range includes Senegal, Guinea, Sierra Leone, southwestern Mali, and Burkina Faso. It disperses along watercourses (Cuisance et al., 1985). G. p. palpalis inhabits forest and occurs in Cote d’Ivoire, Benin, Nigeria, Cameroon, Equatorial Guinea and south to Angola (Robinson and Rogers, 2004). Empirical evidence suggested that dispersion and emigration is density-dependent in G. p. palpalis in Cote d’Ivoire (Rogers and Randolph, 1985). Such dispersion would promote gene flow and diminish the effects of genetic drift. No mitochondrial variation was detected by the SSCP technique in G. p. gambiensis from Senegal but nine haplotypes were detected among flies sampled from three river drainages in Mali (Marquez et al., 2004). The Senegal haplotype was detected in only two of 303 flies sampled in Mali. It had been claimed earlier that the Senegal population had been eradicated, but the genetic evidence supports the notion of an expanded, relict population at the time of genetic sampling. The paucity of detectable mitochondrial variation among Senegal tsetse may be indicative of earlier population bottlenecks or of inadequate genetic resolution afforded by SSCP. Sequencing r16S2 and Cox1 mitochondrial genes in 32 Senegalese flies, however, demonstrated 17 haplotypes, and 10 haplotypes among 28 flies from Bado, Mali (Marquez and Krafsur, unpublished data).
Examination of SSCPs indicate that only 0.06% of the genetic variance among Mali G. p. gambiensis was attributed to differences among three river drainages, but GST = 0.17, indicative of significant genetic differentiation. Microsatellite variation at two X-linked loci in G. p. gambiensis showed strong differentiation (FST = 0.18) between a Burkina Faso population and a Senegal population, some 2,000 km distant (Solano et al., 1999). Especially notable was significant differentiation between two sampled populations on the River Koba in Burkina Faso only 15 km apart (FST = 0.12) and an unusually large deficit of heterozygotes (FIS = 0.33) in just one of the two sampled areas. A later report utilizing three microsatellite loci (Solano et al., 2000) confirmed genetic differentiation across sampled demes on the R. Koba (FST = 0.07) and the deficiency of heterozygotes (FIS = 0.20) in the same location as before. Further sampling of G. p. gambiensis in Burkina Faso along 216 km of the River Mouhoun provided a much lesser estimate of FST = 0.012. Of the four sampled demes, two showed significant departures from random matings (FIS = 0.15 and 0.16; Bouyer et al., 2007). Here the deficiencies of heterozygotes were attributed to null alleles. Consistently large heterozygote deficiencies were recorded at each of five loci in G. p. palpalis in Bonon, Cote d’Ivoire (Ravel et al., 2007). Estimates of FIS varied from 0.12 to 0.44 and a mean over loci and samples of 0.34. Ravel et al. (2007) ruled out linkage disequilibrium and null alleles as the primary causes of the extraordinary departures from random mating within all sampled populations (i.e., FIS). Neither gene frequencies nor estimates of genetic differentiation were provided but application of a Bayesian clustering method showed graphically that samples could be reduced to four highly branched clusters. Ravel et al. (2007) concluded that the Wahlund effect explained the extraordinary FIS. Further analysis using Bayesian methods seemed to confirm that individual samples of G. palpalis s.l. included representatives of two or more demes - up to nine, in fact (de Meeus et al., 2007). Thus, it was suggested that even individual fly traps had sampled multiple demes of only one species. As suggested by the authors, heterozygote deficits recorded in G. p. gambiensis and G. p. palpalis could have been examples of the Wahlund effect (the mixing of conspecific demes with differing allele frequencies).
I find the foregoing explanation unlikely, however, and suggest the data indicate sympatric sibling species: Annual cycling of seasonal range expansions and contractions could, in principle, result in the establishment of demes differentiated by founder effects and genetic drift during the favourable wet season, followed by concentration in moist refugia during the dry season. Initial dry season populations could then show the Wahlund effect but subsequent matings, if random, would lead to a population at Hardy-Weinberg equilibrium and FIS would approach zero. Tsetse generation times are of the order of 50 days, so the timing of sampling is important. A full understanding the relative magnitudes of FST and FIS under the foregoing scenario, however, requires sophisticated simulation studies and estimates of deme sizes. Why do only Palpalis group flies in West Africa show extraordinary FIS estimates? Wahlund effects have not been detected in G. f. fuscipes (see below) and in five taxa of morsitans group tsetse.
An alternative hypothesis to mixing of genetically differentiated, conspecific demes in G. palpalis s.l. at some sites but not others is the presence of morphologically identical sibling species, an explanation supported by reciprocal crosses between strains of G. p. palpalis (Gooding et al., 2004). Significant microgeographic genetic differentiation in G. p. gambiensis (i.e., FST ≫ 0) was manifest particularly on the R. Koba, but not the R. Mouhoun (Solano et al., 2000). The uneven spatial distribution in samples of two (or more) unrecognized sibling species would have the effect of inflating both FIS and FST. Colonization of geographic strains of the foregoing taxa followed by cross-matings and backcrosses of surviving progenies need to be carried out to help clarify the picture. Cytological examination of polytene chromosomes would also be helpful in resolving what seems likely, in my opinion, to be two instances of sibling species.
Population structure in G. fuscipes fuscipes
Krafsur et al. (2008) examined the spatial patterns of mitochondrial cytochrome oxidase I (Cox I) and r16S2 variants in two Kenya, one Uganda, and a laboratory population derived from the Central African Republic sometime before 1986. Like many other Nemorhina, G. f. fuscipes is riverine and lacustrine. The East African sampling sites were within 55 km of each other but they shared only four of 21 variants and were genetically differentiated; pairwise FSTs varied from 0 to 0.28. None of its 21 mitochondrial variants was shared between the lab culture and East Africa samples; pairwise FST varied from 0.46 to 0.58. In a geographically more extensive study, Abila et al. (2008) examined microsatellite and mitochondrial variation at cytochrome oxidase II and cytochrome B in nine Uganda populations and in a sample from Sudan. Pairwise FST based on five microsatellite loci varied from 0.02 to 0.39 (mean 0.22) among eight sampled populations. FST based on mitochondrial variation in ten samples varied from 0.01 to 0.91 (mean 0.59). No correlation was observed between tsetse allele frequencies and the distribution of T. b. rhodesiense and T. b. gambiense. Thus, no evidence was obtained to support an hypothesis of local co-adaptation between vector and parasite strains that would reduce gene flow among tsetse nor help to explain the disjunct distribution of T. brucei subspecies.
Population genetics in subgenus Austenina - the Fusca group (Fig. 6)
Figure 6.
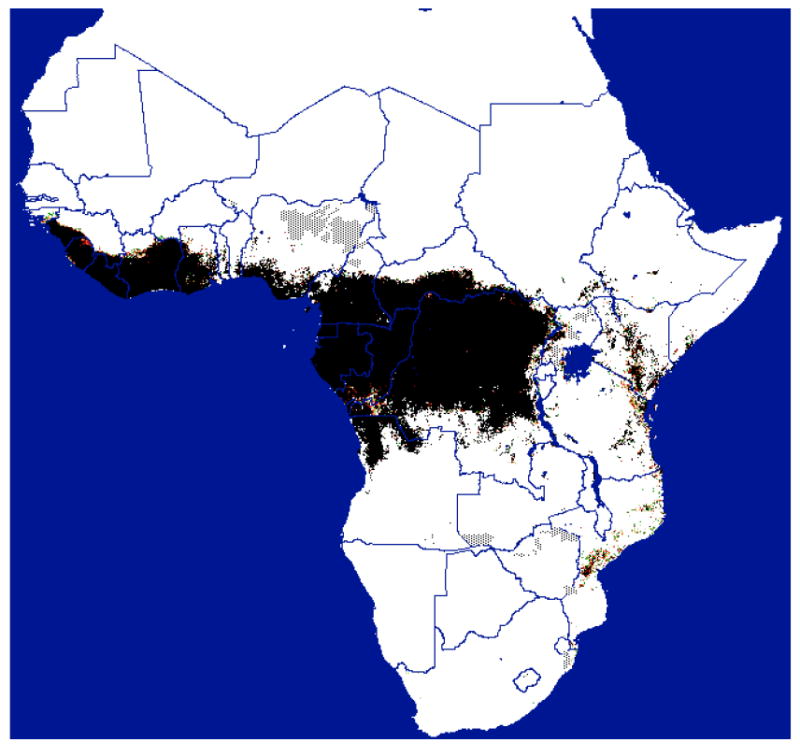
Distribution of Fusca group tsetse flies predicted from satellite imagery (courtesy of ERGO Ltd and TALA, Oxford, with permission. Available from <http://ergodd.zoo.ox.ac.uk/tseweb/distributions.htm>.
It seems that little fieldwork has been reported for any member of the group. Allozyme variation was evaluated in lab stocks of G. brevipalpis Newstead 1910 and G. longipennis Corti 1895 (reviewed in Gooding, 1996). G. brevipalpis is a trypanosome vector and widely but discontinuously distributed from eastern South Africa north to the river deltas in Somalia (Rogers and Robinson, 2004). The largest patches occur in Mozambique, Zaire, northern Tanzania and southeast Kenya. Its population structure would seem to be of interest in anticipation of area-wide control measures. Mitochondrial nucleotide sequences indicated 14 haplotypes and mean diversity Hs = 0.65 in G. brevipalpis; 7 haplotypes and Hs = 0.55 were recorded in G. longipennis (Table 7).
General comments
Estimates of genetic differentiation (FST, GST) were less when estimated at nuclear loci than mitochondrial loci. The mitochondrial genome is single copy and inherited maternally. Moreover, there is no recombination and mutation rates are generally greater than in the nuclear genome. Mitochondrial variation is more sensitive to demographic phenomena. Genetic data based on microsatellite loci tend to underestimate genetic differentiation because FST, GST, and their analogues cannot take values greater than the level of homozygosity (Hedrick, 1999). Microsatellites are also subject to homoplasy (Estoup et al., 1995; Balloux et al., 2000), where two or more alleles become identical in state by convergence from different ancestors. Such alleles cannot be distinguished on gels because they have the same molecular weights thus leading to reductions in estimated diversity. In addition, high frequencies of private and singular alleles bias downward indices of genetic differentiation. A high degree of genetic differentiation was detected at allozyme loci (Krafsur et al., 1997). Balancing selection may occur at some allozyme loci and it tends to minimize estimates of FST (Schierup et al., 2000). In sum, microsatellites and allozymes underestimate genetic differentiation thereby overestimating magnitudes of gene flow (Nagylaki, 1998; Balloux et al., 2000).
Tests for isolation by distance, in which pairwise estimates of FST/(1 − FST) are regressed on log pairwise geographic distances, have generally shown shallow slopes indicating weak relationships between genetic and geographic distances. The results are in keeping with the principal assumptions of the island model of population structure, in which selectively neutral genetic variation is distributed among a very large number of subpopulations of equal size that demonstrate equal rates of gene flow among them. Of course, few, if any, natural populations fit such simplifying assumptions, but the picture in the savannah species is largely consistent with a large number of inhabited patches that exchange reproductive flies at low rates of gene flow.
Because G. pallidipes is considered the most dispersive of all tsetse species (e.g., Vale et al., 1984; Williams et al., 1992; Jordan, 1993; Hargrove, 2003), the high degree of genetic differentiation among its populations is remarkable and calls for further investigation. A full appraisal of its population genetics must include sampling and genetic analysis of heretofore unsampled populations in the DRC (Zaire), Mozambique, southern Somalia, Uganda, northwestern Zambia, and Tanzania. Because many fly belts and geographically small patches throughout its range remain unsampled, it is a valid question to ask if G. pallidipes constitutes a species complex. Perhaps some patches support different mating types of a species complex. It should be noted, however, that no sampled population showed a significant deficiency of heterozygotes, thereby suggesting only one taxonomic unit in each. The suggestion that the West African G. longipalpis may be conspecific with G. pallidipes should be followed up by comparing their polytene chromosome karyotypes, mitochondrial loci, and patterns of microsatellite variation. A compelling test would be to attempt reciprocal crosses and testing the fertilities of any progeny. Earlier attempts to obtain cage matings of G. longipalpis, however, were unsuccessful (Roubaud, 1935).
Effective population sizes (numbers) in Glossina
Effective population size Ne is the harmonic mean number of reproducing individuals in an ideal population that have the same allele frequencies as an actual population under study. Ne determines the rate of genetic drift – the less the Ne, the greater the magnitude of drift. An ideal population has the same rate of inbreeding (i.e., loss of heterozygosity) as the study population and is one in which all Hardy-Weinberg assumptions apply. Clearly, population effective number is bound to be less than the corresponding census number because reproductive success varies enormously among individuals and because of deviations from Hardy-Weinberg assumptions, particularly that of infinite population size.
Estimation of census size is problematic. Accurately estimating Ne is also problematic but for different reasons (Wang, 2005). Moreover, there are a number of estimation methods each of which usually provides numerically quite different values. Short term estimates are based on temporal variance in allele frequencies. The principal assumptions include selective neutrality and a negligible mutation rate in a population closed to immigration. Long-term estimates are based on genetic diversity estimates with the chief assumption of mutation-drift equilibrium and a good estimation of the prevailing mutation rate.
Estimates of mean effective population sizes in G. pallidipes in Lambwe varied from c. 200 to 520, depending principally on the interval between successive samplings (Table 4). At the geographically larger Nguruman site, estimates varied from c. 250 to 1000 (Ouma et al., 2006). Estimates of Ne varied from 18 to 66 in G. swynnertoni (Marquez et al., 2006). A much different approach was used to estimate Ne in G. f. fuscipes (Krafsur et al. 2008). G. f. fuscipes in Kenya and Uganda were estimated to be 403 and 2258 respectively compared with Ne = 1,060 in the International Atomic Energy Agency’s (IAEA) lab culture (Krafsur et al., 2008). It is well to recall that Ne is a harmonic mean value, and is strongly weighted by the least values over numerous generations. The estimates were based on mitochondrial variation, reference to an estimated number of breeders in IAEA’s large lab culture, and estimates of a mutation rate.
The foregoing estimates indicate that tsetse demes are quite small. Especially small effective population numbers seem to characterize G. swynnertoni although observed population densities can be quite high locally (Ndegwa and Mihok, 1999). Estimates of Ne for the genetically isolated G. pallidipes in the Lambwe valley and Nguruman in Kenya suggest longstanding, substantial populations even though intensive control measures were carried out there.
In contrast to the estimates for tsetse flies, consider the effective population size of Leopards (Pantheria pardus) in Tanzania of 38- 48 thousand (Spong et al. 2000). Five island populations of Anopheles gambiae among which gene flow was severely restricted varied from 7 to 25 thousand when estimated by the stepwise mutation model and 3.3 to 8.5 thousand when estimated on the basis of the infinite allele model (Pinto et al., 2002). Ne was estimated in two widely separated Anopheles gambiae populations in Kenya: a population near Lake Victoria was 6.3 or 22.6 thousand, depending on the method of estimation. Another population on the coast, 700 km away, was 4.3 or 7.9 thousand (Lehmann et al., 1998). Thus, the estimation model used can give widely divergent estimates. A further example of Ne is provided by Michel et al. (2006) for Anopheles funestus. Based on mitochondrial variation, effective population sizes for this mosquito in Burkina Faso were estimated at 93.7 to 98.8 thousand and 340 to 311 thousand, depending on the precise long-term model employed. Microsatellite-based estimates in Burkina Faso varied from 174 to 263 thousand. What is clear is that African Anopheles has large effective population numbers even when isolated on islands and surviving annual dry season declines.
Even though Ne varies greatly with the methods used in its calculation, it is plain that tsetse estimates are very much less than other African Diptera and even leopards. Two reasons can be offered. First, tsetse gene frequencies varied greatly from one generation to another. Because genetic variance is inversely proportional to population size N (binomial variance = p(1 − p)/2N), a large estimate of variance is indicative of small N. Second, high FST estimates indicate that gene flow among demes is restricted. Ne is directly proportional to the variance in density and spatial dispersion of a population of interest. As we have seen, in tsetse flies spatial dispersion distances and population densities are small. Forces of genetic drift in Morsitans group (and probably Palpalis group) tsetse clearly overwhelm the forces of dispersion.
Range expansions after the rinderpest epizootic
Range expansions of tsetse flies during the colonial era are discussed in Buxton (1955), Ford (1971), Jordan (1986), and Rogers (1995), based on published and colonial administration records of small refugia after the rinderpest epizootics. Southern African populations of Morsitans group flies showed lesser genetic diversities than conspecific populations from East Africa, consistent with much earlier bottlenecks in population numbers. Do the geographically small but widely distributed populations of G. pallidipes correspond to relict survivors from more extensive, pre-rinderpest populations, founders from earlier surviving populations, or merely the inhabitants of environmentally suitable islands in the midst of unwelcome habitats? No genetic evidence was obtained to support earlier bottlenecks or founder effects in East Africa and Ethiopia because the sampled populations showed abundant variation. Hypotheses of local physiological adaptation via natural selection to prevailing climate offer a mechanism that could explain low rates of gene flow. The question of adaptation was investigated in G. pallidipes.
Physiological adaptation to locally prevailing climates
In principle, adaptation to locally prevailing climates could help to explain the lack of gene flow indicated by spatial patterns of genetic variation, i.e., there may be spatial heterogeneity in natural selection (Lenormand, 2002). Thus, reproductive success of immigrant flies may be poor even though dispersion rates are high. Tests of relevant hypotheses were performed in G. pallidipes in five ecologically diverse sites in Kenya and Zambia where the prevailing climates substantially varied. Hypotheses tested included homogeneity among populations’ thermal tolerances, desiccation tolerances, body water amounts, lipid amounts, and activity levels. Critical thermal limits among the sampled populations were narrow for high temperatures but much less so for low temperatures. Seasonal variation was detected in G. pallidipes in Lambwe and Nguruman and this was attributed to acclimation. Indeed, most among-population variation was explained by phenotypic acclimation rather than genetic differences. Little potential for evolutionary change was detected. Thus, phenotypic plasticity, not genetically determined, heritable, variation explained differences in physiological responses (Terblanche and Chown, 2006; Terblanche et al., 2006).
Cuticular hydrocarbons and lipids are important in water conservation and sex recognition; they were measured quantitatively and qualitatively in the four disparate G. pallidipes populations (Jurenka et al., 2007). Cuticular lipid quantities in flies varied among the sampled populations but did not correlate with environmental measures of normalized density vegetation index or prevailing temperatures and humidities. Desiccation resistance is thought to depend on cuticular lipids. Desiccation rates varied significantly among populations but were uncorrelated with cuticular lipid amounts. Another mechanism that might rationalize high dispersion rates with low rates of gene flow is mate recognition, which is conferred by contact pheromones (Carlson et al., 1984). The chief sex pheromone in G. pallidipes is 12, 23-dimethylpentatricontane and this did not differ among flies from Ethiopia, Zimbabwe, and five independently derived lab cultures (Carlson et al., 2000) nor in four diverse Kenya populations (Jurenka et al., 2007).
To recapitulate, no evidence was obtained that would support the notion that the genetic differentiation of tsetse fly populations is maintained by sex recognition mechanisms or genetically based physiological adaptation to locally prevailing temperatures, saturation deficits, and other measured environmental variables. Instead, phenotypic plasticity and acclimation accounts for tsetse fly responses to short-term environmental variations. Bioclimatic models, however, imply that very long-term responses to environmental heterogeneity include speciation such as that seen among the Morsitans group tsetse.
Summing up
The epidemiological significance of tsetse fly population structures vis a vis population structures of trypanosomes is virtually unknown and requires both laboratory and field research. These endeavours will not be easy, however.
Clearly, demes in two (of nine) Palpalis group taxa, like those in the Morsitans group, are localized, with little detectable gene flow among demes. The extent of genetic sampling in Palpalis group species, however, is limited taxonomically and geographically. Clearly, gene flow in G. p. gambiensis should be investigated further by using markers that afford higher resolution than our use of SSCPs. A number of G. palpalis and G. fusca subspecies, G. tachinoides and G. pallicera s.l., remain to be studied. Our knowledge of Fusca group genetics is virtually nil.
Sampling of Morsitans group flies, while genetically and geographically much greater than Palpalis and Fusca taxa, has many deficiencies. In particular, G. longipalpis and G. austeni require investigation. For Morsitans group taxa, cytogenetical work at a population level should prove highly interesting given the polymorphisms already detected in G. morsitans s.l. and G. pallidipes.
In terms of population biology, G. pallidipes is probably the most thoroughly investigated tsetse fly. The most recent distribution map of G. pallidipes shows highly fragmented populations (Rogers and Robinson, 2004) and compelling genetic evidence indicates little gene flow among them. Yet gene flow among G. pallidipes demes within environmentally suitable patches also seems restricted, as genetic sampling in Ethiopia, Zambia, Tanzania, and Zimbabwe has shown. Distribution maps of the other genetically sampled morsitans group and palpalis group show much less fragmentation than that for G. pallidipes. Indeed, G. morsitans s.l., G. palpalis s.l. and G. fuscipes s.l. distributions are more or less continuous throughout their ranges (Rogers and Robinson, 2004) yet the patterns of genetic differentiation in these taxa do not seem greatly different than G. pallidipes. Tsetse fly demes seem structured much like island populations. To be sure, there must be gene flow, at least historically, but its magnitude seems small. Even populations of species that recovered from earlier, severe bottlenecks are highly structured – for example, G. morsitans s.l. and G. pallidipes in southern Africa.
What may account for this structure? And how well does the hypothesis of little gene flow agree with ecological knowledge of this important group of vector insects? As we have seen earlier in this essay, tsetse dispersal has been measured and thoroughly analyzed (e.g., Rogers, 1977; Williams et al., 1992; Hargrove, 1981, 2000). Based on earlier direct estimates of movement, tsetse dispersal has been simulated (Hargrove, 2000; Vale and Torr, 2004). All the foregoing evidence strongly suggests there should be abundant gene flow among tsetse demes. Are conclusions drawn from the ecological research false? Or do the simplifying assumptions of population genetic theory render false conclusions when applied to tsetse populations?
Of course not. Ecologists and geneticists have measured different things at different scales (Krafsur, 2002b). Ecological estimates are based on scales of one to tens of kilometers measured over short time intervals. The genetic scales are tens to thousands of kilometers and represent demographic phenomena integrated over intervals of many thousands of generations. Let’s go back to the underlying model against which geneticists test hypotheses about gene flow. The model is an ideal population established by Hardy-Weinberg assumptions, viz., populations of infinite size, discrete generations, of randomly mating individuals, and uninfluenced by natural selection. Few taxa, if any, conform to such a model. Genetic inferences about tsetse population structure include small effective population sizes (Ne) of continuously overlapping generations and geographically fragmented populations. Genetic drift is inversely proportional to Ne so is an especially potent force in tsetse population structure, accounting for most, if not all, of the measured differentiation. Also contributing to the genetic differentiation must be numerous and widespread inhospitable patches, in which the precise biotic and abiotic criteria vary greatly (review in Rogers and Randolph, 1985). There is a strong historical component to the picture, that of the rinderpest epizootic beginning in 1887 that killed out 90% of more of the mammalian fauna upon which tsetse feed. Although the epizootic occurred throughout much of sub-Saharan Africa, its effects on tsetse flies seemed to have been particularly severe in southern Africa, as documented by stockmen, entomologists, and veterinarians during the colonial era (Buxton, 1955; Ford, 1971; Jordan, 1986). Morsitans group flies were diminished to only a few, relict refugia. It is important to note that resurgence of G. morsitans s.l. and G. pallidipes there required many years to return to economically significant levels. A more recent example is afforded by results of the Okavango project in which G. m. centralis has failed to reappear after the six years since control operations ceased. Their slow recovery should be an important consideration in examining cost-benefit analysis of thorough, area-wide tsetse control schemes. As pointed out by Rogers and Randolph (2002), tsetse populations are extremely resilient. Adult female average daily survival probabilities are of the order of 98% or more. The half-life of an adult female cohort born simultaneously on day zero is approximately 35 days. Tsetse populations are able to recover from as few as 16 inseminated females, according to Hargrove’s (2005) estimates. Whether that is actually 10 or 100 inseminated females is moot because such small tsetse populations are not likely to be detected even with intensive sampling. Genetic studies support the existence of highly resilient G. pallidipes populations in two ecologically well-studied sites, Lambwe and Nguruman in Kenya. It was shown that populations there shared little genetic variation with the populations nearby. Hypotheses of massive immigration following control measures or seasonal declines were invoked to explain the reappearance of G. pallidipes in Lambwe and Nguruman (Turner and Brightwell, 1986; Williams et al., 1992). Based on genetic evidence, however, it seems those hypotheses are likely to be false.
Genetic models of population structure are gross oversimplifications of natural demographic phenomena, for example, FST and Ne (Whitlock and McCauley, 1999). Simulation modeling has shown that large variances in deme size, caused by environmental or temporal heterogeneity, can produce high values of FST even when dispersion rates are large (Wegmann et al., 2006). It is clear that estimates of Nem are no substitute for estimates based on mark, release, and recapture trials. Based on the oversimplified genetic models, all we can really say about tsetse dispersion is that the subpopulations sampled contain numerically very few individuals that originated in other subpopulations and genetic drift is the dominant force in population structure. A scenario of massive exchange of individuals among any of the populations sampled to date, however, seems to be untenable when based on the genetic evidence.
Acknowledgments
I am indebted to Reg Allsopp and Nigel Griffiths for introducing me to tsetse fly biology. Thanks to William Wint and David Rogers of TALA and ERGO Ltd., Zoology, Oxford, for allowing use of their Glossina distribution maps. Wendy Gibson, Michael Miles, Bruce Christiansen, Mike Lehane, and Ian Maudlin offered helpful comments on trypanosome-tsetse fly interactions. Thanks to my pupils Johnson Ouma, Gerardo Marquez, and Matthew Cummings, and my laboratory assistants David Wohlford and Mark Endsley for their solid contributions and enthusiastic participation in research. Author’s research was supported by NIH grants AI-40048 and AI-52456 from the United States Public Health Service.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE CITED
- Abila P, Slotman M, Parmakelis A, Dion K, Robinson A, Muwanika V, Enyaru J, Lokedi L, Aksoy S, Caccone A. High levels of genetic differentiation between Ugandan Glossina fuscipes fuscipes populations separated by Lake Kyoga. PLoS. 2008 doi: 10.1371/journal.pntd.0000242. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agbo EC, Clausen PH, Buscher P, Majiwa PA, Claassen E, te Pas MF. Population genetic structure and cladistic analysis of Trypanosoma brucei isolates. Infect Genet Evol. 2003;3:165–174. doi: 10.1016/s1567-1348(03)00071-6. [DOI] [PubMed] [Google Scholar]
- Aksoy S, Maudlin I, Dale C, Robinson AS, O’Neil SL. Prospects for control of African trypanosomiasis by tsetse vector manipulation. Trends Parasitol. 2001;17:29–35. doi: 10.1016/s1471-4922(00)01850-x. [DOI] [PubMed] [Google Scholar]
- Baker MD, Krafsur ES. Identification and properties of microsatellite markers in tsetse flies Glossina morsitans sensu lato (Diptera: Glossinidae) Mol Ecol Notes. 2001;1:234–36. doi: 10.1046/j.1471-8278.2001.00087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balloux F, Brunner H, Lugon-Moulin N, Hausser J, Gudet J. Microsatellites can be misleading: an empirical and simulation study. Evolution. 2000;54:1414–1422. doi: 10.1111/j.0014-3820.2000.tb00573.x. [DOI] [PubMed] [Google Scholar]
- Bouyer J, Ravel S, Dujardin J-P, de Meeus T, Vial L, Thevenon S, Guerrini L, Sidibe I, Solano P. Population structuring of Glossina palpalis gambiensis (Diptera: Glossinidae) according to landscape fragmentation in the Mahoun river, Burkina Faso. Med Vet Entomol. 2007;44:788–795. doi: 10.1603/0022-2585(2007)44[788:psogpg]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Brightwell R, Dransfield RD, Williams BG. Factors affecting seasonal dispersal of the tsetse flies, Glossina pallidipes and G. longipennis (Diptera: Glossinidae) at Nguruman, southwest Kenya. Bull Entomol Res. 1992;82:167–182. [Google Scholar]
- Brightwell R, Dransfield RD, Stevenson P, Williams BG. Changes over twelve years in populations of Glossina pallidipes and Glossina longipennis (Diptera: Glossinidae) subject to varying trapping pressure at Nguruman, south-west Kenya. Bull Entomol Res. 1997;87:349–370. [Google Scholar]
- Bursell E, Taylor P. An energy budget for Glossina (Diptera: Glossinidae) Bull Entomol Res. 1980;70:187–196. [Google Scholar]
- Buxton PA. The natural history of tsetse flies. Memoir of the London School of Hygiene and Tropical Medicine no. 10. H.K. Lewis; London: 1955. [Google Scholar]
- Carlson D, Nelson D, Langley P, et al. Contact sex pheromone in the tsetse fly Glossina pallidipes: identification and synthesis. J Chem Ecol. 1984;10:429–450. doi: 10.1007/BF00988090. [DOI] [PubMed] [Google Scholar]
- Carlson D, Sutton B, Bernier U. Cuticular hydrocarbons in Glossina austeni and G. pallidipes: similarities between populations. Insect Sci Applics. 2000;20:281–294. [Google Scholar]
- Cuisance D, Fevrier J, Dejardin J, Filledier J. Dispersion linéaire de Glossina palpalis gambiensis et de Glossina tachinoides dans une galerie forestière en zone soudano-guinéenne (Burkina-Faso) Rev d’Élevage et de Méd Vét des pays tropic. 1985;38:153–172. [Google Scholar]
- Curtis CF, Southern DI, Pell PE, Craig-Cameron TA. Chromosome translocations in Glossina austeni. Genet Res Camb. 1972;20:101–113. doi: 10.1017/s0016672300013616. [DOI] [PubMed] [Google Scholar]
- Dale C, Maudlin I. Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. Int J Systematic Bacteriol. 1999;49:267–275. doi: 10.1099/00207713-49-1-267. [DOI] [PubMed] [Google Scholar]
- De Meeus T, McCoy KD, Prugnolle F, Chevillon C, Durant P, Hurtrez-Bousses SH, Renaud F. Population genetics and molecular epidemiology or how to “debusquer la bête”. Infect Genet Evol. 2007;7:308–322. doi: 10.1016/j.meegid.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Dias JA Travassos Santos. Contribuição para o estudo da sistemãtica do género Glossina Wiedemann, 1830 (Insecta, Brachycera, Cyclorrhapha, Glossinidae) Proposta para a criação deum novo subgénero. Garcia Orta Sér Zool Lisboa. 1987;14:67–78. [Google Scholar]
- Elsen P, Amoudi MA, Leclercq M. First record of Glossina fuscipes Newstead 1910 and Glossina morsitans submorsitans Newstead 1910 in south-western Saudi Arabia. Ann Soc Belge de Med Trop. 1990;70:281–287. [PubMed] [Google Scholar]
- Bourn D, Blench R, editors. ERGO (Environmental Research Group Oxford Ltd. Can livestock and wildlife co-exist? An interdisciplinary review. 1999. [Google Scholar]
- Estoup A, Tailliez C, Cornuet JM, Solignac M. Size homoplasy and mutational processes of interrupted microsatellites in two bee species, Apis mellifera and Bombus terrestris (Apidae) Molec Biol Evol. 1995;12:1074–1084. doi: 10.1093/oxfordjournals.molbev.a040282. [DOI] [PubMed] [Google Scholar]
- Feldmann U. The sterile insect technique as a component of area-wide integerated pest management of tsetse. In: Maudlin I, Holmes P, Miles M, editors. The Trypanosomiases. CAB International; Oxford: 2004. pp. 565–582. [Google Scholar]
- Ford J. The Role of Trypanosomiases in African Ecology. Clarendon Press; Oxford: 1971. [Google Scholar]
- Garcia A, Courtin D, Solano P, Koffi M, Jamonneau V. Human African trypanosomiasis: connecting parasite and host genetics. Trends Parasitol. 2006;22:405–409. doi: 10.1016/j.pt.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Gariou-Papalexiou A, Yannopoulos G, Robinson AS, Zacharopoulou A. Polytene chromosome maps in four species of tsetse flies Glossina austeni, G. pallidipes, G. morsitans morsitans and G. m. submorsitans (Diptera: Glossinidae): a comparative analysis. Genetica. 2007;129:243–251. doi: 10.1007/s10709-006-0004-7. [DOI] [PubMed] [Google Scholar]
- Gariou-Papalexiou A, Yannopoulos G, Robinson AS, Zacharopoulou A. Polytene chromosome maps and RAPD polymorphisms in Glossina austeni. In: Tan KH, editor. Area-wide control of fruit flies and other insect pests. Penerbit universiti Sains Malaysia; Penang: 2000. pp. 239–250. [Google Scholar]
- Gibson W. Sex and evolution in trypanosomes. Int J Parasit. 2001;31:643–647. doi: 10.1016/s0020-7519(01)00138-2. [DOI] [PubMed] [Google Scholar]
- Gibson W. The SRA gene: the key to understanding the nature of Trypanosoma brucei rhodesiense. Parasitology. 2005;131:143–150. doi: 10.1017/s0031182005007560. [DOI] [PubMed] [Google Scholar]
- Gibson W, Stevens J. Genetic exchange in the Trypanosomatidae. Adv Parasitol. 1999;43:1–46. doi: 10.1016/s0065-308x(08)60240-7. [DOI] [PubMed] [Google Scholar]
- Gibson W, Peacock L, Ferris V, Williams K, Bailey M. The use of yellow fluorescent hybrids to indicate mating in Trypanosoma brucei. 2008 doi: 10.1186/1756-3305-1-4. http://www.Parasitesandectors.com/content/1/1/4. [DOI] [PMC free article] [PubMed]
- Glasgow JP. The distribution and abundance of tsetse. Oxford: Pergamon; 1963. p. 252. [Google Scholar]
- Gooding RH. Tsetse genetics: a review. Quaest Entomol. 1984;20:89–128. [Google Scholar]
- Gooding RH. Genetic variation in tsetse flies and its implications for trypanosomiasis. Parasit Today. 1992;8:92–95. doi: 10.1016/0169-4758(92)90246-x. [DOI] [PubMed] [Google Scholar]
- Gooding RH. Genetic analysis of sterility in hybrids from crosses of Glossina morsitans submorsitans and Glossina morsitans centralis (Diptera: Glossinidae) Can J Zool. 1993;71:1963–72. [Google Scholar]
- Gooding RH. Genetic variation in arthropod vectors of disease-causing organisms: obstacles and opportunities. Clin Microb Rev. 1996 July; doi: 10.1128/cmr.9.3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooding RH, Rolseth BM. Genetics of Glossina palpalis palpalis: designation of linkage groups and the mapping of eight biochemical and visible marker genes. Genome. 1995;38:833–37. doi: 10.1139/g95-109. [DOI] [PubMed] [Google Scholar]
- Gooding RH, Challoner CM. Genetics of the tsetse fly, Glossina morsitans submorsitans Newstead (Diptera: Glossinidae): further mapping of linkage groups I, II and III. Can J Zool. 1999;77:1309–13. [Google Scholar]
- Gooding RH, Solano P, Ravel S. X-chromosome mapping experiments suggest occurrence of cryptic species in the tsetse fly Glossina palpalis palpalis. Can J Zool. 2004;82:1902–1909. [Google Scholar]
- Gooding RH, Krafsur ES. In: Tsetse genetics: applications to biology and systematics. Maudlin I, Homes PH, Miles MA, editors. CAB International; 2004. pp. 95–111. [Google Scholar]
- Gooding RH, Krafsur ES. Tsetse genetics: contributions to biology, systematics, and control of tsetse flies. Annu Rev Entomol. 2005;50:101–123. doi: 10.1146/annurev.ento.50.071803.130443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi DA. Vicariance biogeography, geographic extinctions, and the North American Oligocene tsetse flies. In: Novacek MJ, Wheeler QD, editors. Extinction and Phylogeny. NY: Columbia Univ. Press; 1992. pp. 178–204. [Google Scholar]
- Grimaldi D, Engel MS. Evolution of the insects. Cambridge University Press; 2005. p. 755. [Google Scholar]
- Hale LR, Singh RS. A comprehensive study of genic variation in natural populations of Drosophila melanogaster IV. Mitochondrial DNA variation and the role of history vs selection in the genetic structure of geographic populations. Genetics. 1991;129:103–117. doi: 10.1093/genetics/129.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Z, Kasumba I, Lehane MJ, Gibson WC, Kwon J, Aksoy S. Tsetse immune responses and trypanosome transmission: implications for the development of tsetse-based strategies to reduce trypanosomiasis. Proc Natl Acad Sci. 2001;98:12648–12653. doi: 10.1073/pnas.221363798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove JW. Tsetse dispersal reconsidered. J Anim Ecol. 1981;50:351–373. [Google Scholar]
- Hargrove JW. A theoretical study of the invasion of cleared areas by tsetse flies (Diptera: Glossinidae) Bull Entomol Res. 2000;90:201–209. doi: 10.1017/s0007485300000328. [DOI] [PubMed] [Google Scholar]
- Hargrove JW. Research report, DFID Animal Health Programme, Centre for Tropical Veterinary Medicine. University of Edinburgh; UK: 2003. Tsetse eradication: sufficiency, necessity and desirability. [Google Scholar]
- Hargrove J. Tsetse population dynamics. In: Maudlin I, Holmes P, Miles M, editors. The Trypanosomiases. CAB International; Oxford: 2004. pp. 113–137. [Google Scholar]
- Hargrove J. Extinction probabilities and times to extinction for populations of tsetse flies Glossina spp (Diptera: Glossinidae) subjected to various control measures. Bull Entomol Res. 2005;95:1–9. doi: 10.1079/ber2004335. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. Highly variable loci and their interpretation in evolution and conservation. Evolution. 1999;53:313–318. doi: 10.1111/j.1558-5646.1999.tb03767.x. [DOI] [PubMed] [Google Scholar]
- Hide G, Tait A. In: Genetics and molecular epidemiology of trypanosomes. Maudlin I, Homes PH, Miles MA, editors. CAB International; 2004. pp. 77–93. [Google Scholar]
- Hu C, Aksoy S. Innate immune responses regulate trypanosome parasite infection of the tsetse fly Glossina morsitans morsitans. Mol Microbiol. 2006;60:1194–1204. doi: 10.1111/j.1365-2958.2006.05180.x. [DOI] [PubMed] [Google Scholar]
- Hutchinson MP. Trypanosomiasis in south-west Ethiopia (March 1969 – March 1970) Eth Med J. 1971;9:3–69. [PubMed] [Google Scholar]
- Jenni L, Marti S, Schweizer J, Betschart B, Le Page RWF, Wells JM, Tait A, Paindavoines P, Pays E, Steinert M. Hybrid formation betweenAfrican trypanosomed during cyclical transmission. Nature. 1986;322:173–175. doi: 10.1038/322173a0. [DOI] [PubMed] [Google Scholar]
- Jordan AM. The distribution of the Fusca group of tsetse flies in Nigeria and Wests cameroun. Bull Entomol Res. 1963;54:307–323. [Google Scholar]
- Jordan AM. Trypanosomiasis control and African rural development. Longman; London: 1986. p. 357. [Google Scholar]
- Jurenka R, Terblanche JS, Klok CJ, Chown SL, Krafsur ES. Cuticular lipid mass and desiccation rates in Glossina pallidipes: inter-population variation. Physiol Entomol. 2007;32:287–293. doi: 10.1111/j.1365-3032.2007.00571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kgori PM, Modo S, Torr SJ. The use of aerial spraying to eliminate tsetse from the Okavango Delta of Botswana. Acta Tropica. 2006;99:184–199. doi: 10.1016/j.actatropica.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Kim SK, Ratcliffe ST, Frenchy BW, Liu L, Sappington TW. Utility of EST-derived SSRs as population genetics markers in a beetle. J Hered. 2008;99:112–124. doi: 10.1093/jhered/esm104. [DOI] [PubMed] [Google Scholar]
- Krafsur ES. Population structure of the tsetse fly Glossina pallidipes estimated by allozyme, microsatellite, and mitochondrial diversities. Insect Molec Biol. 2002a;11:37–45. doi: 10.1046/j.0962-1075.2001.00307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafsur ES. Genetic diversity and gene flow in morsitans group tsetse flies. Tsetse Tryp Info Quart. 2002b;25:141–146. [PMC free article] [PubMed] [Google Scholar]
- Krafsur ES. Tsetse fly population genetics: an indirect approach to dispersal. Trends Parasitol. 2003;19:162–166. doi: 10.1016/s1471-4922(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Krafsur ES, Cummings MA, Endsley MA, Marquez JG, Nason JD. Geographic differentiation in the house fly estimated by microsatellite and mitochondrial variation. J Heredit. 2005;96:502–512. doi: 10.1093/jhered/esi093. [DOI] [PubMed] [Google Scholar]
- Krafsur ES, Griffiths N. Genetic variation at structural loci in the Glossina morsitans species group. Biochem Genet. 1997;35:1–11. doi: 10.1023/a:1022252311715. [DOI] [PubMed] [Google Scholar]
- Krafsur ES, Wohlford DL. Breeding Structure of Glossina pallidipes Populations Evaluated by Mitochondrial Variation. J Hered. 1999;90:635–642. doi: 10.1093/jhered/90.6.635. [DOI] [PubMed] [Google Scholar]
- Krafsur ES, Madsen M, Wohlford DL, Mihok S, Griffiths NT. Genetic differentiation in G. morsitans submorsitans Newstead. Bull Entomol Res. 2000;90:329–335. doi: 10.1017/s0007485300000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafsur ES, Endsley MA, Wohlford DL, Griffiths NT, Allsopp R. Genetic differentiation of Glossina morsitans centralis populations. Insect Molec Biol. 2001;10:387–398. doi: 10.1046/j.0962-1075.2001.00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafsur ES, Endsley MA. Shared microsatellite loci in Glossina morsitans sensu lato (Diptera: Glossinidae) J Med Entomol. 2006;43:640–642. doi: 10.1603/0022-2585(2006)43[640:smligm]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafsur ES, Marquez JG, Ouma JO. Structure of some East African Glossina fuscipes fuscipes populations. Med Vet Entomol. 2008;22:222–227. doi: 10.1111/j.1365-2915.2008.00739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leak SGA. Tsetse biology and ecology: their role in the epidemiology and control of trypanosomiasis. CABI Publ.; NY: 1998. [Google Scholar]
- Lehane MJ, Msangi AR, Whitaker CJ, Lehane SM. Grouping of trypansome species in mixed infections in Glossina pallidipes. Parasitology. 2000;120:583–592. doi: 10.1017/s0031182099005983. [DOI] [PubMed] [Google Scholar]
- Lehane MJ, Aksoy S, Gibson W, Kerhornou A, Berriman M, Hamilton J, Soares MB, Bonaldo MF, Lehane S, Hall N. Adult midgut expressed sequence tags from the tsetse fly Glossina morsitans morsitans and expression analysis of putative immune response genes. Genome Biol. 2003;4:R63. doi: 10.1186/gb-2003-4-10-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann T, Hawley WA, Grebert H, Collins FH. The effective population size of Anopheles gambiae in Kenya: implication for population structure. Mol Biol Evol. 1998;15:264–276. doi: 10.1093/oxfordjournals.molbev.a025923. [DOI] [PubMed] [Google Scholar]
- Lenormand T. Gene flow and the limits to natural selection. Trends Ecol Evol. 2002;17:183–189. [Google Scholar]
- Luna C, Bonizzoni M, Cheng Q, Robinson AS, Aksoy S, Zheng L. Microsatellite polymorphism in tsetse flies (Diptera: Glossinidae) J Med Entomol. 2001;38:376–81. doi: 10.1603/0022-2585-38.3.376. [DOI] [PubMed] [Google Scholar]
- MacLeod A, Turner CMR, Tait A. A high level of mixed Trypanosoma brucei infections in tsetse flies detected by here hypervariable minisatellites. Molec Biochem Parasitol. 1999;102:237–248. doi: 10.1016/s0166-6851(99)00101-2. [DOI] [PubMed] [Google Scholar]
- MacLeod A, Tweedie A, Welburn SC, Maudlin I, Turner CMR, Tait A. Minisatellite marker analysis of Trypanosoma brucei: reconciliation of clonal, panmictic, and epidemic population genetic structures. Proc Natl Acad Sci (USA) 2000;97:13442–13447. doi: 10.1073/pnas.230434097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod A, Turner CMR, Tait A. The detection of geographical substrucuring of Trypanosoma brucei populations by the analysis of minisatellite polymorphisms. Parasitology. 2001;123:475–482. doi: 10.1017/s0031182001008666. [DOI] [PubMed] [Google Scholar]
- MacLeod ET, Maudlin I, Darby AC, Welburn SC. Antioxidants promote establishment of trypanosome infections in tsetse. Parasitology. 2007a;19:1–5. doi: 10.1017/S0031182007002247. [DOI] [PubMed] [Google Scholar]
- MacLeod ET, Darby AC, Maudlin I, Welburn SC. Factors affecting trypanosome maturation in tsetse flies. PloS One. 2007b Feb;(2):e239. doi: 10.1371/journal.pone.0000239. www.plosone.org. [DOI] [PMC free article] [PubMed]
- Marquez JG, Vreysen MJB, Robinson AS, Bado S, Krafsur ES. Mitochondrial diversity analysis of Glossina palpalis gambiensis from Mali and Senegal. Med Vet Entomol. 2004;18:1–8. doi: 10.1111/j.0269-283X.2004.00508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez JM, Malele II, Ouma JO, Krafsur ES. Glossina swynnertoni Austen: effective population size and gene flow estimated by mitochondrial diversity. Bull Entomol Res. 2006;96:353–360. [PMC free article] [PubMed] [Google Scholar]
- Maudlin I. Chromosome polymorphism and sex determination in a wild population of tsetse. Nature. 1979;277:300–1. doi: 10.1038/277300a0. [DOI] [PubMed] [Google Scholar]
- Maudlin I. Inheritance of susceptibility to Trypanosoma congolense infection in Glossina morsitans. Ann Trop Med Parasitol. 1982;76:225–227. doi: 10.1080/00034983.1982.11687531. [DOI] [PubMed] [Google Scholar]
- Maudlin I, Welburn SC, Milligan PJM. Trypanosome infections and survival in tsetse. Parasitology. 1998;116:S23–S28. doi: 10.1017/s0031182000084912. [DOI] [PubMed] [Google Scholar]
- McAlpine JF. Phylogeny and classification of the Muscomorpha. In: McAlpine JF, editor. Manual of Nearctic Diptera. Vol. 3. Ottawa: Res. Branch Agric. Can; 1989. pp. 1397–518. [Google Scholar]
- Michel AP, Grushko O, guelbeogo WM, Sanon N, Costantini C, Besansky NJ. Effective population size of Anopheles funestus chromosome forms in Burkina Faso. Malaria J. 2006;5:115–120. doi: 10.1186/1475-2875-5-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagylaki T. Fixation indices in subdivided populations. Genetics. 1998;148:1325–1332. doi: 10.1093/genetics/148.3.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash TAM. Africa’s bane: the tsetse fly. Collins; London: 1969. [Google Scholar]
- Nayduch D, Aksoy S. Refractoriness in tsetse flies (Diptera: Glossinidae) may be a matter of timing. J Med Entomol. 2007;44:660–665. doi: 10.1603/0022-2585(2007)44[660:ritfdg]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Ndegwa PN, Mihok S. Development of odour-baited traps for Glossina swynnertoni (Diptera: Glossinidae) Bull Entomol Res. 1999;89:255–261. [Google Scholar]
- Nei M. Molecular population genetics and evolution. New York: Columbia University Press; 1987. [Google Scholar]
- Nigam Y, Maudlin I, Welburn S, Ratcliffe NA. Detection of phenoloxidase activity in the hemolymph of tsetse flies, refractory and susceptible infection with Trypanosoma brucei rhodesiense. J Inverteb Pathol. 1997;69:279–281. doi: 10.1006/jipa.1996.4652. [DOI] [PubMed] [Google Scholar]
- Odulaja A, Baumgartner J, Mihok S, Abu-zinid IM. Spatial and temporal distribution of tsetse fly trap catches at Nguruman, southwest Kenya. Bull Entomol Res. 2001;91:213–220. [PubMed] [Google Scholar]
- Ouma JO, Cummings MA, Jones KC, Krafsur ES. Characterization of microsatellite markers in the tsetse fly, Glossina pallidipes (Diptera: Glossinidae) Molec Ecol Notes. 2003;3:450–453. doi: 10.1046/j.1471-8286.2003.00480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouma JO, Marquez JG, Krafsur ES. Macrogeographic structure of the tsetse fly, Glossina pallidipes (Diptera: Glossinidae) Bull Entomol Res. 2005;95:437–447. doi: 10.1079/BER2005376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouma JO, Marquez JG, Krafsur ES. Microgeographic breeding structure of the tsetse fly, Glossina pallidipes (Diptera: Glossinidae) in south-western Kenya. Med Vet Entomol. 2006;20:138–149. doi: 10.1111/j.1365-2915.2006.00609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouma JO, Marquez GM, Krafsur ES. Patterns of genetic diversity and differentiation in the tsetse fly Glossina morsitans morsitans Westwood populations in East and southern Africa. Genetica. 2007;130:139–151. doi: 10.1007/s10709-006-9001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock L, Ferris V, Bailey M, Gibson W. Dynamics of infection and competition between two strains of Trypanosoma brucei brucei in the tsetse fly observed using fluorescent markers. Kinetoplastid Biol Dis. 2007 doi: 10.1186/1475-9292-6-4. www.kinetoplastids.com/content/6/1/4. [DOI] [PMC free article] [PubMed]
- Petersen FT, Meier R, Kutty SN, Wiegmann BM. The phylogeny and evolution of host choice in the Hippoboscoidea (Diptera) as reconstructed using four molecular markers. Mol Phylog Evol. 2007;45:111–122. doi: 10.1016/j.ympev.2007.04.023. [DOI] [PubMed] [Google Scholar]
- Picozzi K, Fevre EM, Odit M, Carrington M, Eisler MC, Maudlin I, Welburn SC. Sleeping sickness in Uganda: a thin line between two fatal diseases. Brit Med J. 2005;331:1238–1242. doi: 10.1136/bmj.331.7527.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto J, Donnelly MJ, Sousa A, Gil V, Ferreira C, Elissa N, Do Rosario VE, Charlwood JD. Genetic structure of Anopheles gambiae (Diptera: Culicidae) in Sao tome and Principe (West Africa): implications for malaria control. Molec Entomol. 2002;11:2183–2187. doi: 10.1046/j.1365-294x.2002.01587.x. [DOI] [PubMed] [Google Scholar]
- Randolph S, Rogers DJ. Mortality rates and population density of tsetse flies correlated with satellite imagery. Nature. 1981;351:739–741. doi: 10.1038/351739a0. [DOI] [PubMed] [Google Scholar]
- Ravel S, de Meeus T, Dujardin JP, Zeze DG, Gooding RH, Dusfour I, Sane B, Cuny G, Solano P. The tsetse fly Glossina palpalis palpalis is composed of several genetically differentiated small populations in the sleeping sickness focus of Bonon, Cote d’Ivoire. Infect Genet Evol. 2007;7:116–125. doi: 10.1016/j.meegid.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Robinson TP, Rogers DJ, Williams B. Univariate analysis of tsetse habitat in the common fly belt of Southern Africa using climate and remotely sensed vegetation data. Med Vet Entomol. 1997a;11:223–234. doi: 10.1111/j.1365-2915.1997.tb00400.x. [DOI] [PubMed] [Google Scholar]
- Robinson TP, Rogers DJ, Williams B. Mapping tsetse habitat suitability in the common fly belt of Southern Africa using multivariate analysis of climate and remotely sensed vegation data. Med Vet Entomol. 1997b;11:235–245. doi: 10.1111/j.1365-2915.1997.tb00401.x. [DOI] [PubMed] [Google Scholar]
- Roditi i, Lehane MJ. Interactions between trypanosomes and tsetse flies. Curr Opinion Microbiol. 2008;11:1–7. doi: 10.1016/j.mib.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise differences. Molec Biol Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- Rogers DJ. Study of a natural population of tsetse flies and a model for fly movement. J Anim Ecol. 1977;46:309–30. [Google Scholar]
- Rogers DJ. Remote sensing and the changing distribution of tsetse flies in Africa. In: Harington R, Stock NE, editors. Insects in a changing environment. Academic Press; 1995. [Google Scholar]
- Rogers DJ. Satellites, space, time and the African trypanosomes. Adv Parasitol. 2000;47:130–171. doi: 10.1016/s0065-308x(00)47008-9. [DOI] [PubMed] [Google Scholar]
- Rogers DJ, Randolph SE. A review of density-dependent processes in tsetse populations. Insect Sci Appl. 1984;5:397–402. [Google Scholar]
- Rogers DJ, Randolph S. Population ecology of tsetse. Annu Rev Entomol. 1985;30:197–216. doi: 10.1146/annurev.en.30.010185.001213. [DOI] [PubMed] [Google Scholar]
- Rogers DJ, Randolph S. Mortality rates and population density of tsetse flies correlated with satellite imagery. Nature. 1991;351:739–741. doi: 10.1038/351739a0. [DOI] [PubMed] [Google Scholar]
- Rogers DJ, Randolph S. A response to the aim of eradicating tsetse from Africa. Trends Parasitol. 2002;18:534–536. doi: 10.1016/s1471-4922(02)02422-4. [DOI] [PubMed] [Google Scholar]
- Rogers DJ, Robinson TP. Tsetse distribution. In: Maudlin I, Holmes P, Miles M, editors. The Trypanosomiases. CAB International; Oxford: 2004. pp. 139–179. [Google Scholar]
- Roubaud E. Titres et travaux scientifiques. Laval (France), Imprimerie Barnéoud. 1935:212. [Google Scholar]
- Schierup MH, Vekemans X, Charlesworh D. The effect of subdivision on variation at multi-allelic loci under balancing selection. Genet Res. 2000;76:51–62. doi: 10.1017/s0016672300004535. [DOI] [PubMed] [Google Scholar]
- Slatkin M, Hudson RR. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics. 1991;129:555–562. doi: 10.1093/genetics/129.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano P, de La Rocque S, Cuisance D, Geoffroy B, de Meeus T, et al. Intraspecific variability in natural populations of Glossina palpalis gambiensis fromWest Africa, revealed by genetic and morphometric analyses. Med Vet Entomol. 1999;13:401–7. doi: 10.1046/j.1365-2915.1999.00189.x. [DOI] [PubMed] [Google Scholar]
- Solano P, de La Rocque S, de Meeus T, Cuny G, Duvallet G, Cuisance D. Microsatellite DNA markers reveal genetic differentiation among populations of Glossina palpalis gambiensis collected in the agro-pastoral zone of Sideradougou, Burkina Faso. Insect Mol Biol. 2000;9:433–39. doi: 10.1046/j.1365-2583.2000.00205.x. [DOI] [PubMed] [Google Scholar]
- Southern DI. Chromosome diversity in tsetse flies. Insect Cytogenetics 1980 [Google Scholar]; Blackman RL, Hewitt GM, Ashburner M, editors. Symp R Entomol Soc London. Vol. 10. Oxford: Blackwell Sci; pp. 225–43. [Google Scholar]
- Southern DI, Pell PE. Comparative analysis of the polytene chromosomes of Glossina austeni and Glossina morsitans morsitans. Chromosoma. 1974;47:213–26. doi: 10.1007/BF00331808. [DOI] [PubMed] [Google Scholar]
- Spong G, Johansson M, Bjorklund M. High genetic variation in leopards indicates large and long-term stable effective population size. Molec Ecol. 2000;9:1773–1782. doi: 10.1046/j.1365-294x.2000.01067.x. [DOI] [PubMed] [Google Scholar]
- Stevens JR, Brisse S. Systematics of trypanosomes of medical and veterinary importance. In: Maudlin I, Holmes P, Miles M, editors. The Trypanosomiases. CAB International; Oxford: 2004. pp. 1–23. [Google Scholar]
- Tait A, MacLeod A, Tweedie A, Masiga D, Turner CMR. Genetic exchange in trypanosoma brucei: evidence for mating prior to metacyclic stage development. Mol Biochem arasitol. 2007;151:133–136. doi: 10.1016/j.molbiopara.2006.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terblanche JS, Chown SL. The relative contributions of developmental plasticity and adult acclimation to physiological variation in the tsetse fly, Glossina pallidipes (Diptera, Glossinidae) J Exper Biol. 2006;209:1064–1073. doi: 10.1242/jeb.02129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terblanche JS, Klok CJ, Krafsur ES, Chown SL. Phenotypic plasticity and geographic variation in thermal tolerance and water loss of the tsetse fly Glossina pallidipes (Diptera: Glossinidae): implications for distribution modeling. Am J Trop Med Hyg. 2006;74:786–794. [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M, Ayala F. The clonal theory of parasitic protozoa: 12 years on. Trends Parasitol. 2002;18:405–410. doi: 10.1016/s1471-4922(02)02357-7. [DOI] [PubMed] [Google Scholar]
- Torr SJ, Hargrove JW, Vale GA. Towards a rational policy for dealing with tsetse. Trends in Parasitology. 2006;21:537–541. doi: 10.1016/j.pt.2005.08.021. [DOI] [PubMed] [Google Scholar]
- Turner DA, Brightwell R. An evaluation of a sequential aerial spraying operation against Glossina pallidipes Austen (Diptera: Glossinidae) in the Lambwe Valley of Kenya: Aspects of the post-spray recovery and evidence of natural population regulation. Bull Entomol Res. 1986;76:331–349. [Google Scholar]
- Vale GA, Harshey BS, Hargrove JW. The use of small plots to study populations of tsetse (Diptera: Glossinidae) Insect Sci Applic. 1984;5:403–410. [Google Scholar]
- Vale GA, Torr SJ. User-friendly models of the costs and efficacy of tsetse control: application to sterilizing and insecticidal techniques. Med Vet Entomol. 2005;19:293–305. doi: 10.1111/j.1365-2915.2005.00573.x. [DOI] [PubMed] [Google Scholar]
- Vanderplank FL. The classification of Glossina morsitans Westwood Diptera, Muscidae, including a description of a new subspecies, varieties and hybrids. Proc R Entomol Soc Lond B. 1949a;18:56–64. [Google Scholar]
- Vanderplank FL. Variation in the male genitalia of the tsetse fly Glossina pallidipes Austen and a note on G austeni Newstead. Proc R Entomol Soc Lond (B) 1949b;18:65–8. [Google Scholar]
- Vreysen MJB. Prospects for area-wide integrated control of tsetse flies (Diptera: Glossinidae) and trypanosomosis in sub-Saharan Africa. Rev Soc Entomol Argent. 2006;65:1–21. [Google Scholar]
- Vreysen MJB, Zhu Z-R, Saley KM. Field responses of Glossina austeni to sticky panels on Unguja Island, Zanzibar. Med Vet Entomol. 1998;12:407–416. doi: 10.1046/j.1365-2915.1998.00129.x. [DOI] [PubMed] [Google Scholar]
- Wang J. Estimation of effective population sizes from data on genetic markers. Phil Trans Roy Soc B. 2005;360:1395–1409. doi: 10.1098/rstb.2005.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes ML, Maudlin I. An analysis of supernumerary or B-chromosomes of wild and laboratory strains of Glossina morsitans morsitans. Med Vet Entomol. 1992;6:175–176. doi: 10.1111/j.1365-2915.1992.tb00600.x. [DOI] [PubMed] [Google Scholar]
- Wegmann D, Currat M, Excoffier L. Molecular diversity after a range expansion In heterogeneous environments. Genetics. 2006;174:2009–2020. doi: 10.1534/genetics.106.062851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlford DL, Krafsur ES, Griffiths NT, Marquez JG, Baker MD. Genetic differentiation of some Glossina morsitans morsitans populations. Med Vet Entomol. 1999;13:377–385. doi: 10.1046/j.1365-2915.1999.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–70. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Welburn SC, Maudlin I. Tsetse-trypanosome interactions: rites of passage. Parasitol Today. 1999;15:399–403. doi: 10.1016/s0169-4758(99)01512-4. [DOI] [PubMed] [Google Scholar]
- Welburn SC, Fevre EM, Coleman PG, Odiit M, Maudlin I. Sleeping sickness: a tale of two diseases. Trends Parasitol. 2001;17:19–24. doi: 10.1016/s1471-4922(00)01839-0. [DOI] [PubMed] [Google Scholar]
- Welburn SC, Fevre EM, Coleman PG, Maudlin I. In: The Trypanosomiases. Maudlin I, Holmes P, Miles M, editors. CAB International; Oxford: 2004. pp. 219–232. [Google Scholar]
- Whitlock MC, McCauley DE. Indirect measures of gene flow and migration: FST not equal to 1/(4Nm + 1) Heredity. 1999;82:117–125. doi: 10.1038/sj.hdy.6884960. [DOI] [PubMed] [Google Scholar]
- Williams B, Dransfield R, Brightwell R. The control of tsetse flies in relation to fly movement and trapping efficiency. J Appl Ecol. 1992;29:163–179. [Google Scholar]
- WHO. Human African trypanosomiasis (sleeping sickness): epidemiological update. Weekly Epidemiological Record. 2006;81:69–80. [PubMed] [Google Scholar]
- Wright S. Evolution and the genetics of populations, Vol. 4. Variability within and among natural populations. Chicago: Univ Chicago Press; 1978. [Google Scholar]


