Abstract
A selection of 43 Candida albicans isolates, chosen to represent the four major strain clades of the species and also intraclade diversity, was screened for their virulence in the murine intravenous challenge model of C. albicans infection, for a range of properties measurable in vitro that might relate to virulence, and for the numbers of midrepeat sequences in genes of the ALS and HYR families. Heterozygosity at the mating type locus and low whole-cell acid phosphatase activity and growth rate at 40°C were found to be significantly positively associated with the most virulent isolates. Acid phosphatase activity and growth in 2 M NaCl were statistically significant variables between clades by univariate analysis. Isolates in different clades also differed significantly in midrepeat sequence alleles of ALS2, ALS4, ALS6, ALS7, ALS9, HYR1, and HYR2. There was no association between the midrepeat alleles of any ALS or HYR gene and the virulence of isolates to mice. Genome-wide transcript profiles of 20 isolates (5 per clade) grown under two conditions showed considerable variation between individual isolates, but only a small number of genes showed statistically significant differential gene expression between clades. Analysis of the expression profiles by overall strain virulence revealed 18 open reading frames differing significantly between isolates of high, intermediate, and low virulence. Four of these genes encoded functions related to phosphate uptake and metabolism. This finding and the significant association between whole-cell acid phosphatase activity and virulence led us to disrupt PHO100, which encodes a predicted periplasmic acid phosphatase. The pho100Δ mutant was mildly but significantly attenuated in terms of survival curves in the mouse model. The study has extended the range of properties known to differ between C. albicans clades and suggests a possible but minor role of phosphate metabolism in the virulence of the species.
Candida albicans is the species most commonly associated with superficial and disseminated Candida infections in humans. Isolates of C. albicans analyzed by multilocus sequence typing (MLST) (38, 51) or DNA fingerprinting with the moderately repetitive oligonucleotide Ca3 (5, 43, 48) can be assigned to subsets of closely related strain types, referred to, for convenience, as clades (40, 48). The main clades to which individual strains are assigned are essentially the same by both typing approaches (38, 51), and approximately 70% of the large numbers of C. albicans isolates typed so far belong to one of the four largest clades, numbered 1 to 4 (38). There is, therefore, concordance between the structures and sizes of the DNA repeat sequences recognized by Ca3 and the single-nucleotide polymorphisms determined by sequencing 7 among approximately 6,000 open reading frames (ORFs) in C. albicans.
The most self-evident associations between the four major C. albicans clades and other properties are seen for geographical origins and for the presence of an intron in ribosomal DNA—the so-called ABC type (33, 34). While there is no absolute geographical association between any clade and a definable area where a clade is endemic, a high proportion of isolates within each clade usually originates from a broadly definable region (5, 38, 43, 48, 51). Moreover, there are statistically definable subsets of strains within the broader clade groupings that show more specific geographical origins (50). ABC typing of isolates shows a near-total predominance of type A isolates in clades 1 and 2 and a majority of type B isolates in clade 3 and of type C isolates in clade 4 (38).
Clade-specific associations extend to properties of potential relevance to the role of C. albicans as human commensal and pathogen. Isolates of C. albicans resistant to flucytosine are found predominantly within clade 1 (44, 51). These clade 1 isolates also show a common mechanism of flucytosine resistance, based on an amino acid change (R101C) in the FUR1 gene product, a ribosyltransferase enzyme (13, 23, 51). This mutation is not found in flucytosine-resistant isolates from other clades (13, 51). Amphotericin B resistance shows a less well pronounced but statistically significant association with C. albicans clades (4). Statistically significant differences between clades have been found for the numbers of tandem repeat sequences in alleles of the genes ALS3, ALS5, and ALS6, members of a gene family that encode C. albicans surface proteins that play a role in adhesion to host surfaces (41, 63).
The evidence clearly indicates that C. albicans major clades, delineated by MLST or Ca3 typing, comprise strains that differ in a number of properties not directly related to the particular gene sequences or tandem repeat sequences that are used to define their clade assignation. Each clade therefore comprises strains that have evolved in a manner different from strains in other clades. In this study we have prospectively tested a set of C. albicans isolates representing the four major clades for their virulence in the murine intravenous (i.v.) challenge model of C. albicans infection, for a range of phenotypic attributes, for tandem repeat alleles in several genes encoding glycosylphosphatidylinositol-anchored proteins, and for their gene expression profiles under two growth conditions.
MATERIALS AND METHODS
Fungi and growth conditions.
A panel of 43 C. albicans isolates was selected from a panel typed by MLST (38) to provide representatives of the strains in clades 1 to 4. The isolates (Table 1) were chosen to include, as far as possible for each clade, examples from superficial anatomical sources and bloodstream isolates, isolates from diverse geographical sources, examples of isolates homozygous (a/a or α/α) at the MAT locus as well as the more common a/α types (38), and a mix of types A, B, and C, where this was possible. The MLST data for these 43 isolates were reanalyzed in an P-distance dendrogram made by the unweighted-pair group method using arithmetic averages with phylogeny tested by 1,000 bootstraps in MEGA 3.1 software (25). For the isolates shown in Table 1, the 14 isolates in clade 1 and the 8 isolates in clade 3 coclustered in separate groups with 100% bootstrap values (Fig. 1). For the 10 isolates in clade 2 and the 11 isolates in clade 4, the bootstrap values for the cluster nodes were 90% and 86%, respectively (Fig. 1).
TABLE 1.
Details of the 43 C. albicans isolates included in this study
| Isolate | MLST data
|
Source countryb | Sample or anatomical source | ABC type | MAT type | Inoculation details
|
Survival time (days)
|
Kidney log CFU/g (mean ± SD) | % wt change from day 0 to 3 (mean ± SD) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clade | DSTa | 104 CFU/g (mean ± SD) | No. of repetitionsc | Median | Mean ± SD | |||||||
| YsU751d | 1 | 69 | Malaysia | Urine | A | a/α | 2.5 ± 0.9 | 3 | 8.0 | 7.3 ± 4.1 | 7.5 ± 0.3 | −10.3 ± 9.6 |
| HUN92 | 1 | 37 | UK | Blood | A | a/α | 2.1 ± 0.7 | 3 | 28.0 | 23.0 ± 7.8 | 4.1 ± 1.9 | −2.0 ± 4.9 |
| FJ9 | 1 | 102 | Fiji | Oropharynx | A | a/α | 2.4 ± 1.3 | 3 | 15.5 | 16.5 ± 5.5 | 7.1 ± 1.3 | −6.0 ± 4.8 |
| RIHO9d | 1 | 100 | USA | Blood | A | a/a | 2.3 ± 1.3 | 3 | 28.0 | 25.5 ± 4.5 | 5.3 ± 1.2 | 2.6 ± 3.1 |
| 78/028 | 1 | 73 | UK | Oropharynx | A | a/α | 1.9 ± 0.3 | 3 | 28.0 | 28.0 ± 0.0 | 3.3 ± 1.4 | 3.2 ± 3.4 |
| AM2002/0087d | 1 | 135 | UK | Blood | A | a/α | 2.4 ± 0.9 | 3 | 12.0 | 13.2 ± 8.4 | 7.3 ± 0.5 | −11.7 ± 6.9 |
| HUN96 | 1 | 116 | UK | Blood | A | a/α | 2.0 ± 0.3 | 3 | 28.0 | 28.0 ± 0.0 | 4.0 ± 2.2 | 1.7 ± 3.6 |
| AM2003-013 | 1 | 120 | UK | Oropharynx | A | a/α | 2.3 ± 0.9 | 3 | 11.5 | 13.7 ± 8.0 | 6.4 ± 1.4 | −5.2 ± 6.4 |
| AM2003-016 | 1 | 123 | UK | Oropharynx | A | a/α | 1.8 ± 0.5 | 3 | 28.0 | 24.3 ± 9.0 | 5.4 ± 1.0 | −2.8 ± 2.5 |
| J981301 | 1 | 60 | USA | Vagina | A | a/α | 2.0 ± 0.7 | 3 | 28.0 | 27.2 ± 2.0 | 6.7 ± 1.5 | −0.7 ± 4.4 |
| 81/064d | 1 | 129 | UK | Vagina | A | α/α | 1.3 ± 0.2 | 3 | 28.0 | 24.8 ± 6.0 | 4.2 ± 2.4 | 2.1 ± 4.5 |
| J981318 | 1 | 86 | USA | Vagina | A | a/α | 1.9 ± 0.4 | 3 | 6.5 | 6.7 ± 1.6 | 6.6 ± 0.3 | −13.2 ± 4.6 |
| J951361 | 1 | 44 | UK | Blood | A | a/α | 2.1 ± 0.2 | 2 | 5.0 | 5.3 ± 1.6 | 6.1 ± 1.6 | −15.1 ± 4.6 |
| SC5314d | 1 | 52 | USA | Blood | A | a/α | 1.8 ± 0.3 | 3 | 7.5 | 6.8 ± 1.6 | 6.0 ± 1.0 | −15.7 ± 2.2 |
| AM2003/0089 | 2 | 155 | UK | Blood | A | a/α | 1.5 ± 0.3 | 2 | 11.0 | 16.0 ± 9.4 | 4.9 ± 2.7 | −1.4 ± 4.6 |
| IHEM16972 | 2 | 155 | Rwanda | Oropharynx | A | a/α | 2.7 ± 0.8 | 2 | 9.5 | 12.2 ± 8.0 | 6.3 ± 0.9 | −5.3 ± 4.6 |
| FC22 | 2 | 155 | USA | Vagina | A | a/α | 2.3 ± 0.2 | 2 | 12.5 | 13.8 ± 5.6 | 7.0 ± 0.8 | −2.4 ± 3.1 |
| IHEM16614d | 2 | 206 | Rwanda | Oropharynx | A | a/α | 2.6 ± 0.6 | 2 | 7.0 | 7.3 ± 1.4 | 6.4 ± 0.6 | −9.9 ± 3.3 |
| AM2003/0100d | 2 | 206 | UK | Blood | A | a/α | 1.5 ± 0.7 | 2 | 28.0 | 20.7 ± 11.4 | 5.9 ± 1.1 | −2.6 ± 5.0 |
| IHEM16945d | 2 | 356 | Rwanda | Oropharynx | A | α/α | 1.8 ± 0.5 | 2 | 13.0 | 14.8 ± 6.8 | 6.1 ± 0.9 | −2.1 ± 4.3 |
| AM2004/0028 | 2 | 326 | UK | Oropharynx | A | a/α | 2.7 ± 0.6 | 2 | 22.0 | 20.3 ± 8.1 | 6.1 ± 1.7 | −0.4 ± 2.9 |
| AM2003/0182 | 2 | 225 | UK | Blood | A | a/α | 2.3 ± 0.8 | 2 | 22.0 | 22.0 ± 5.4 | 6.6 ± 0.9 | −0.3 ± 3.5 |
| AM2003/0191d | 2 | 232 | UK | Blood | A | a/α | 2.5 ± 0.1 | 2 | 8.5 | 10.7 ± 5.9 | 6.5 ± 1.2 | −9.1 ± 5.7 |
| AM2003/0069d | 2 | 194 | UK | Vagina | A | a/α | 2.5 ± 0.8 | 2 | 26.0 | 24.5 ± 4.2 | 4.9 ± 1.7 | 0.7−3.2 |
| FC28d | 3 | 159 | USA | Vagina | B | α/α | 2.4 ± 0.8 | 3 | 28.0 | 26.3 ± 4.1 | 4.1 ± 1.8 | 1.1 ± 4.3 |
| 73/025 | 3 | 132 | UK | Vagina | B | a/α | 2.4 ± 0.0 | 2 | 13.0 | 14.0 ± 6.5 | 6.9 ± 0.7 | −3.2 ± 3.3 |
| J981315 | 3 | 56 | USA | Vagina | B | a/α | 2.0 ± 0.1 | 2 | 10.0 | 13.7 ± 8.0 | 5.7 ± 1.7 | −3.4 ± 5.5 |
| b30708/5d | 3 | 186 | UK | Blood | B | a/α | 3.2 ± 0.9 | 2 | 8.0 | 12.8 ± 8.8 | 5.6 ± 0.9 | 3.3 ± 3.3 |
| T101d | 3 | 236 | Canada | Oropharynx | B | a/a | 2.7 ± 0.8 | 2 | 13.5 | 13.7 ± 3.3 | 6.3 ± 1.0 | −2.6 ± 2.6 |
| J990102d | 3 | 45 | Belgium | Vagina | B | a/α | 2.5 ± 0.1 | 2 | 7.0 | 6.5 ± 0.8 | 7.1 ± 0.3 | −10.3 ± 4.3 |
| AM2003/0074 | 3 | 198 | UK | Blood | B | a/α | 2.1 ± 0.0 | 2 | 28.0 | 27.3 ± 1.6 | 6.0 ± 1.2 | 1.1 ± 5.0 |
| AM2003/0165d | 3 | 332 | UK | Blood | B | α/α | 1.5 ± 0.4 | 2 | 26.5 | 21.7 ± 9.0 | 4.2 ± 2.2 | 0.2 ± 6.0 |
| s20152.082 | 4 | 168 | USA | Blood | C | a/α | 2.6 ± 0.9 | 3 | 14.5 | 14.7 ± 5.5 | 7.2 ± 0.4 | −8.0 ± 9.8 |
| s20152.013 | 4 | 167 | USA | Blood | A | a/α | 2.3 ± 0.3 | 3 | 20.0 | 19.3 ± 9.5 | 6.1 ± 1.6 | −10.7 ± 7.7 |
| AM2005/0377d | 4 | 635 | South Africa | Blood | B | a/a | 2.0 ± 0.0 | 2 | 28.0 | 28.0 ± 0.0 | 5.7 ± 0.7 | 2.4 ± 3.5 |
| s20176.079 | 4 | 164 | USA | Blood | A | a/α | 1.8 ± 0.3 | 3 | 21.0 | 20.7 ± 7.5 | 6.0 ± 1.5 | −0.8 ± 8.5 |
| RV4688d | 4 | 87 | Zaire | Blood | C | a/α | 3.3 ± 0.2 | 3 | 28.0 | 28.0 ± 0.0 | 5.3 ± 2.1 | −0.3 ± 3.6 |
| HUN68 | 4 | 95 | UK | Oropharynx | B | a/α | 1.8 ± 0.6 | 3 | 28.0 | 23.2 ± 7.5 | 6.1 ± 1.4 | 0.8 ± 3.6 |
| s20122.073 | 4 | 95 | Switzerland | Blood | B | a/α | 1.8 ± 0.4 | 3 | 5.0 | 12.5 ± 12.0 | 6.0 ± 1.8 | −8.6 ± 11.1 |
| L1086 | 4 | 144 | Saudi Arabia | Wound | B | a/α | 2.9 ± 0.6 | 3 | 10.0 | 10.7 ± 3.1 | 7.3 ± 0.4 | −7.3 ± 5.5 |
| IHEM3742d | 4 | 124 | Zaire | Blood | C | a/α | 1.9 ± 0.4 | 3 | 28.0 | 28.0 ± 0.0 | 5.1 ± 2.2 | 1.8 ± 3.6 |
| s20175.016d | 4 | 124 | Israel | Blood | C | a/α | 2.3 ± 0.1 | 3 | 28.0 | 26.2 ± 3.0 | 6.9 ± 1.2 | −7.0 ± 5.9 |
| AM2003-020d | 4 | 124 | UK | Oropharynx | C | a/α | 2.8 ± 0.6 | 3 | 12.0 | 13.2 ± 8.9 | 7.1 ± 0.8 | −9.9 ± 4.4 |
DST, diploid sequence type.
UK, United Kingdom; USA, United States of America.
The isolate was tested in two groups of three mice (shown as 2) or tested in three groups of two mice (shown as 3).
These isolates were used for expression profiling experiments.
FIG. 1.
Dendrogram showing the similarity clusters (referred to as clades [shown to the right of illustration]) for the 43 C. albicans clinical isolates used in this study. The dendrogram was constructed by the unweighted-pair group method using arithmetic averages with MEGA 3.0 software as previously described (50). The numbers at nodal points indicate bootstrap values (as percentages) for 1,000 replications. The scale bar at the bottom of the figure shows nucleotide substitutions.
All isolates were maintained frozen in 50% glycerol at −80°C. Working subcultures on Sabouraud agar (Oxoid, Basingstoke, United Kingdom) were maintained at 4°C for a maximum of 14 days. For preparation of inocula for all experiments except where indicated otherwise, C. albicans isolates were grown as yeast cells in 5-ml volumes of NGY medium (0.1% Neopeptone, 0.4% glucose, 0.1% yeast extract) at 30°C with constant rotation at 20 rpm (32).
Deletion of PHO100 from C. albicans SC5314 and construction of reintegrant strains.
The CaPHO100 gene was disrupted by the standard “ura-blaster” protocol (ura, uracil) (15). The disruption cassette was produced by amplifying two regions of the gene by PCR; a 5′ region, containing the start codon, was amplified with primer pair DMPHO100_1 and DMPHO100_2 (Table 2), and a 3′ region, containing the stop codon, was amplified with primer pair DMPHO100_3 and DMPHO100_4 (Table 2). Restriction digests of the 5′ region by SacI and BglII and the 3′ region with SalI and SphI allowed the DNA fragments to be cloned into digested pMB7 (15). Digestion of the resulting plasmid with SacI and SphI released the disruption cassette. The disruption cassette consisted of the ura-blaster cassette flanked by complementary sequences of 340 bp at the 5′ end and 460 bp at the 3′ end of PHO100. PHO100 was sequentially disrupted by transformation of strain CAI-4 (15), with the Ura3 marker recycled by selection on SD medium (0.67% [wt/vol] yeast nitrogen base with ammonium sulfate without amino acids but with 2% glucose) containing 5-fluoroorotic acid (1 mg/ml) and uridine (50 μg/ml). To overcome any problems with URA3 expression from the PHO100 locus (6), the Ura-minus null mutant (DMY001; pho100Δ::hisG/pho100Δ::hisG) was transformed with StuI-digested CIp10 plasmid (35), which leads to URA3 expression at the RPS1 locus (DMY002; as DMY001 but RPS1/rps1Δ::CIp10). Correct disruption of the PHO100 alleles was analyzed by PCR (primer pair DMPHO100_1 and DMPHO100_5) and confirmed by Southern analysis. A reintegrant strain for use as a control was created by PCR amplifying the entire PHO100 ORF plus 885-bp promoter and 454-bp terminator sequences (primer pair DMORF_1 and DMORF_2), and cloning it into the pGEM-T vector (Promega Ltd., Southampton, United Kingdom). The insert was released by NotI digestion and cloned into the NotI site in CIp10. The resulting plasmid was digested with StuI and used to transform the Ura-minus pho100Δ null strain (DMY003; as DMY001 but RPS1/rps1Δ::CIp10-PHO100). The control strain for these experiments was NGY152 (6), which is strain CAI-4 transformed with StuI-digested CIp10.
TABLE 2.
Oligonucleotide primers used for gene disruption and screening of C. albicans PHO100
| Primer | Target | Oligonucleotide sequence (5′→3′)a |
|---|---|---|
| DMORF_1 | Reintegrant cassette amplification | ATTAAGGAATCGGCTGCC |
| DMORF_2 | Reintegrant cassette amplification | CATGTCGTCTATCAGTCATCC |
| DMPHO100_1 | Amplification of 5′ region (contains a SacI site) | GACTGAGAGCTCGCAATGAGATTTGTTTAC |
| DMPHO100_2 | Amplification of 5′ region | GAGGCTGGAGTACCGTTG |
| DMPHO100_3 | Amplification of 3′ region (contains a SalI site) | AGCCTTGTCGACGGTCTCGGTGTTGGTTTG |
| DMPHO100_4 | Amplification of 3′ region (contains a SphI site) | CTGAAGGCATGCGTGACGATAAAGGAAGCG |
| DMPHO100_5 | Screening for gene disruption | TGACCAGTCAATCTAGTG |
Restriction sites in primer sequences are underlined.
Virulence testing.
The pathological effects of the 43 C. albicans isolates were determined by murine i.v. challenge in groups of six animals. All animal experimentation was done in accordance with United Kingdom Home Office regulations and was approved by both the Home Office and an institutional ethical review committee. Female BALB/c mice (Harlan, United Kingdom) (weight ranging from 18 to 22 g) were maintained in groups of up to 12 animals per cage; each animal was individually marked by staining fur with picric acid solution, and the mice were weighed daily. The mice were supplied with food and water ad libitum. For preparation of challenge inocula, cells grown in NGY medium were harvested by centrifugation, washed, and resuspended in sterile physiological saline. Yeast counts were adjusted with a hemocytometer to a density such that animals received an i.v. challenge dose of 2 × 104 CFU per g of body weight. The actual numbers injected were determined the following day by counting the viable cells of the inoculum suspensions. The actual range of challenge doses for the 43 isolates was 1.5 × 104 to 3.3 × 104 CFU/g; the median and modal doses were both 2.3 × 104 CFU/g. To keep the numbers of mice infected as low as possible, i.e., groups of six mice, while acknowledging the possibility of batch-to-batch variation in outcomes of challenge, the fungal isolates were each injected either in two mice on three separate occasions or in three mice on two separate occasions (detailed in Table 1).
The mice were observed for up to 28 days postchallenge and were humanely terminated when their body weight fell more than 20% below their initial weight or when the animals showed signs of serious illness. The dates of termination of animals were used to determine survival after challenge. At the time of death, both kidneys were dissected with aseptic precautions and homogenized together in 0.5-ml volumes of sterile water, and tissue burdens were measured by counting the viable cells on Sabouraud agar. The percent change in body weight of each mouse between the day of challenge and 3 days postchallenge was calculated relative to the weight of the mouse on the day of challenge.
The same model was used to test virulence of the pho100Δ mutant, but a single experiment was done with groups of six mice challenged i.v. with control and mutant strains.
Measurement of phenotypic properties.
Several phenotypic properties were characterized for all 43 C. albicans isolates. For all phenotypic tests, cultures of each isolate were grown in 5 ml of NGY medium overnight at 30°C with constant rotation. Cells were harvested and washed with sterile water prior to use in phenotypic tests. Where required, cell numbers were determined by counting cells with a hemocytometer.
Biofilm formation was measured by resuspending cells in RPMI 1640 medium containing 10% (vol/vol) serum (RPMI 1640-10% serum) (1× RPMI 1640 medium, 2 mM l-glutamine, 1.65 mM morpholinepropanesulfonic acid [MOPS], 10% fetal calf serum) to a density of 2 × 106 cells/ml in triplicate wells of a 24-well tissue culture plate. Plates were incubated overnight at 37°C in 5% CO2 without agitation. Biofilm formation was gauged by reading the optical density at 600 nm (OD600) of the supernatant after agitation.
Germ tube formation and mean morphology index were determined for cells incubated for 3 h in fetal calf serum. Washed cells (3 × 106) were resuspended in 1 ml of 100% fetal calf serum (prewarmed to 37°C) and incubated in a 37°C water bath for 3 h. The cultures were fixed with formalin, and the morphology of cells was determined microscopically. For each strain, the morphology index was determined on three separate occasions.
For measurement of growth under proteinase-inducing conditions, overnight cultures in NGY medium were washed twice in sterile water and reinoculated into 5-ml volumes of 1.17% yeast carbon base (Difco)—1% glucose and 0.5% bovine serum albumin (Sigma) at an initial concentration of 104 yeasts/ml. Cultures were incubated for 72 h at 30°C with constant rotation. Turbidity ODs were measured versus a medium blank at 600 nm.
Acid phosphatase activity was determined in intact C. albicans cells. These cells were grown in phosphate-free NGY medium for 16 h, washed once in water, and resuspended in water to one-fifth of the original culture volume. Enzyme activity was measured by the following modification of the method of Lindhardt and Walter (29). The final reaction mixture contained 50 μl of 5.5 mM p-nitrophenyl phosphate (ICN Biomedicals Inc., United Kingdom) in 0.11 M citrate buffer (pH 5.0) and 5 μl cell suspension. The reaction in control wells was immediately stopped by the addition of 245 μl of 0.1 M NaOH. The plate was incubated for 15 min at 37°C, and then the reaction was stopped with the addition of NaOH to the remaining wells. The absorbance of the yellow p-nitrophenyl released was measured at 405 nm, and the amount of p-nitrophenol released was calculated from a standard curve. Experimental measurements were corrected by subtraction of control measurements. Specific activity was defined as nanomoles of p-nitrophenol/15 min/OD600 in the cell suspension.
Alcian blue binding to surface charged groups was measured for yeast cells grown in YPD (1% yeast extract, 2% mycological peptone, 2% glucose) at 30°C for 18 h with constant rotation according to the method of Hobson et al. (21). For each assay, 50 μl of cell suspension (∼1.5 × 107 cells) was harvested, washed in water, and resuspended in 1 ml of 30 μg/ml alcian blue in 20 mM HCl. The cell suspensions were incubated for 10 min at room temperature, and then the cells spun down. A620 values were determined for the supernatants, allowing the amount of alcian blue bound by the cells in the assay to be determined from a standard curve. Specific alcian blue binding was expressed as micrograms of alcian blue bound per OD600 unit of cells.
The growth rates of cells in RPMI 1640 medium were determined by adding 10 μl of washed C. albicans cells to 5 ml RPMI 1640 with low MOPS (1× RPMI 1640, 2 mM l-glutamine, 1.65 mM MOPS [pH 6.5]). For each strain, 200 μl of cell suspension was dispensed into triplicate wells of 96-well flat-bottomed tissue culture plates. Growth curves were plotted at 40°C over 16 h by means of a Fluostar Optima plate reader (BMG Labtech Ltd., Aylesbury, United Kingdom). Growth rates were calculated from the exponential region of the growth curve and were recorded as means from the growth rates determined on three separate occasions.
To measure NaCl tolerance, 10 μl of washed C. albicans cells was added to 5 ml 2× RPMI 1640 with low MOPS (1× RPMI 1640, 2 mM l-glutamine, 1.65 mM MOPS [pH 6.5]), and 100-μl lots were dispensed into wells containing 100 μl 4 M NaCl, such that tolerance to 2 M NaCl was assayed. For each C. albicans isolate, triplicate wells were assayed on at least two occasions.
Adherence of fungi to BECs and catheter material.
Buccal epithelial cells (BECs) were collected by scraping the gums and cheeks of healthy donors, washed, and resuspended (3 × 105 cells/ml) in physiological saline. Cells were monitored microscopically for prior colonization by yeast cells. Washed C. albicans cell suspensions were adjusted to 5 × 106 cells/ml in saline. Adherence assay solutions (600 μl) contained 6 × 104 BECs mixed with 1 × 106 C. albicans cells. The solutions were incubated at 30°C with constant rotation for 1 h. The number of C. albicans cells adherent to each BEC was determined microscopically, with at least 100 BECs examined for each assay. Assays were performed for each strain on at least three separate occasions.
Adherence to catheter material was measured by the method of Seidler et al. (47). Arrow-Howes Hands-Off multilumen central venous catheters (radiopaque polyurethane) were a kind gift from Nigel Webster, University of Aberdeen. Briefly, catheters were cut into 5-mm sections and weighed. Catheter pieces were preconditioned in 100% fetal calf serum at 37°C with agitation overnight. Catheter material was washed twice in phosphate-buffered saline and then immersed in 1 ml C. albicans cell suspension (2 × 106 cells/ml in RPMI 1640-10% serum), and the cell suspension containing catheter material was incubated for 2.5 h at 37°C with shaking to allow adherence. Sections were washed three times with phosphate-buffered saline and then added to 1 ml RPMI 1640-10% serum, and the catheter culture was incubated for 24 h at 37°C under 5% CO2. At this time, cell growth in catheter culture was estimated using Alamar Blue (AbD Serotec, Oxford, United Kingdom) by measuring the percent reduction Alamar Blue per gram catheter plastic.
Expression profiling.
From the full panel of 43 C. albicans isolates, a subset of 20 was chosen for inclusion in expression profiling experiments. The subset comprised five isolates from each of the four clades, selected to represent as much diversity as possible with respect to mouse virulence, MAT homozygosity, and ABC type (Table 1).
For expression profiling, cells were grown either in NGY medium at 30°C or in RPMI 1640-10% serum at 37°C. For NGY-grown cells, an NGY preculture was used to inoculate 100 ml NGY at 30°C to an initial OD600 of approximately 0.15. Cells were grown with constant shaking at 200 rpm until they reached an OD600 of approximately 0.8. For cells grown in RPMI 1640-10% serum, 1 ml of an NGY preculture was harvested and the cells were washed and resuspended in 1 ml distilled H2O. This was used to inoculate 120 ml RPMI 1640-10% serum at 37°C in a tissue culture flask. Cultures were incubated at 37°C under 5% CO2 for 16 h without agitation. Cells were scraped off the flask surface and resuspended in medium. For both growth conditions, cells were harvested by centrifugation, resuspended in residual liquid, and flash-frozen in liquid nitrogen. RNA was extracted by the method of Hauser et al. (20). The quality of the RNA was analyzed by agarose gel electrophoresis, and RNA concentration was measured by spectrophotometry. For each isolate under each growth condition, three independent cultures were grown for RNA extraction.
For cells grown in both NGY medium and RPMI 1640-10% serum, SC5314 RNA samples were used to prepare a pooled control, which consisted of equal quantities of RNA from each of the three samples processed. The pooled control was used for comparison with each isolate, including SC5314. Labeled cDNAs were prepared (11), with 25 μg isolate RNA labeled with Cy3 and 25 μg pooled control labeled with Cy5. For each isolate and growth condition, three microarrays were carried out comparing the three independent RNAs extracted to the relevant pooled control. Microarray hybridizations, washes, and scanning and acquisition of data were performed as described previously (11).
The results of the expression profiling exercise were two sets of triplicate data for 20 C. albicans isolates, each grown in NGY and RPMI 1640 media and profiled with reference to pooled RNA from C. albicans SC5314 triplicate cultures grown under the same conditions. The data were normalized on a per spot and per chip intensity-dependent (Lowess) basis and on a per chip basis to the 50th percentile, and then the expression ratios were converted to base 2 logarithms. The Statistical Analysis of Microarrays (SAM) (54) software package (http://www-stat.stanford.edu/∼tibs/SAM/) was used to determine significantly differentiated genes between groups of isolates. The analyses were done with expression profile data grouped in three strata according to overall virulence and in four strata by clade; multiclass SAM comparisons were done between strata. For each analysis, two approaches were used. In the first, the triplicate data were analyzed directly. In the second, a proprietary computer script was used to exclude the most extreme outlier among each set of triplicates, and SAM analysis was then applied to the most concordant pair from the three data for each gene and isolate. Data sets used in the SAM analysis are provided as tables in the supplemental material. Genes were considered to contribute significantly to differentiation between clades and virulence groups when their q value by SAM was <0.05. Only genes that emerged as significant multiclass differentiators from both the triplicate and the “best two of three” data sets were recorded as likely to represent differences in constitutive gene expression between the strata analyzed. Transcript profiling data can be accessed via ArrayExpress (http://www.ebi.ac.uk/microarray): identification numbers E-MEXP-1933 (RPMI 1640 data) and E-MEXP-1931 (NGY data).
ALS and HYR tandem repeat determinations.
For approximation of the number of tandem repeats for ALS and HYR genes, genomic DNA was extracted from each of the 43 C. albicans isolates (22). PCR mixtures (50 μl) containing 100 ng genomic DNA, 0.4 μM forward and reverse primers, and 1× PCR Mastermix (Thermoscientific, Epsom, Surrey, United Kingdom) were set up for each isolate. Oligonucleotide primer sequences are listed in Table 3. PCR was performed as follows: one step of 1 min at 94°C, followed by 30 cycles (1 cycle consisting of 1 min at 94°C, 1 min at 50°C, and 3 min at 72°C), with a final step of 10 min at 72°C. PCR products were analyzed by agarose gel electrophoresis against appropriate molecular weight DNA markers (Promega UK, Southampton, United Kingdom).
TABLE 3.
Oligonucleotide primers for analysis of cell wall gene repeat regions
| Primera | Target | Oligonucleotide sequence (5′→3′) |
|---|---|---|
| ALS1RFw | ALS1 tandem repeat region | GATTCAATTGACACAGTG |
| ALS1RRv | ALS1 tandem repeat region | TCTTTGAACTTGACATGC |
| ALS2RFw | ALS2 tandem repeat region | GTCATTGTTGCTACAACCCG |
| ALS2RRv | ALS2 tandem repeat region | ATGACACTATTGGTGCCACC |
| ALS3RFw | ALS3 tandem repeat region | CTGCTACAGATGTTAATTCG |
| ALS3RRv | ALS3 tandem repeat region | GTTGGTGTAATGAGGACGAG |
| ALS4RFw | ALS4 tandem repeat region | CCAGTGAATGGACAGGAACA |
| ALS4RRv | ALS4 tandem repeat region | ATTGCCACGCTTGTTTTACC |
| ALS5RFw | ALS5 tandem repeat region | TCCACTTCCAAATCCAAC |
| ALS5RRv | ALS5 tandem repeat region | TATACTTGATGACTGCTC |
| ALS6RFw | ALS6 tandem repeat region | GTTGTACAAGTTCCACTG |
| ALS6RRv | ALS6 tandem repeat region | TATACTTGAAGTCTCGTC |
| ALS7RFw | ALS7 tandem repeat region | CAGCGACAACCACTTACTTCG |
| ALS7RRv | ALS7 tandem repeat region | CCCAATTGAGCTTGATGGAA |
| ALS9RFw | ALS9 tandem repeat region | TGACGGGGATGTGATCGTAG |
| ALS9RRv | ALS9 tandem repeat region | ATCCAGTTCCGAGTGCAGGT |
| HYR1RFw | HYR1 tandem repeat region | GGAGGTAATGAAAGTGGTTC |
| HYR1RRv | HYR1 tandem repeat region | CCTTCATTAGAACCAGAGTG |
| HYR3R1Fw | HYR3 tandem repeat region 1 | TCAGTTAGTGCTTCAGCAGC |
| HYR3R1Rv | HYR3 tandem repeat region 1 | CTCTTACATACAACAGGAGG |
| HYR3R2Fw | HYR3 tandem repeat region 2 | AGTCTTGAAACACCTGTCCC |
| HYR3R2Rv | HYR3 tandem repeat region 2 | TGATGGAACAACCGAGAGTG |
Forward primers and reverse primers are indicated by Fw and Rv suffixes, respectively, at the end of the primer name.
Statistical analysis of data.
SPSS for Windows version 16.0 was used for all statistical analyses. Associations between virulence parameters, such as mean survival times or terminal kidney burdens, and other properties measured for the 43 C. albicans isolates were determined by two-step multiple regression analyses with the SPSS general linear model. The virulence parameter was set as the dependent variable and other properties were input as fixed factors or covariates, according to their type (nominal or continuous variables). The model was set to show the significance of the contribution of each property to main between-subject effects. The four properties with the lowest P values in this first analysis were then reevaluated together to determine which properties still showed a significant association with the virulence parameter being tested. Associations between virulence designations (a nominal variable from 1 to 3 corresponding to high, intermediate, and low overall virulence, respectively), clade assignations, and other properties were determined by the Kruskal-Wallis test for K independent variables. Correlations between data sets were determined by Spearman's parametric r coefficient and by Kendall's nonparametric tau. Because a large number of statistical tests was performed, outcomes were judged as statistically significant only when the probability P that the result occurred by chance was <0.01 in order to avoid type 1 statistical errors. Instances of associations with P < 0.05 but > 0.01 were noted as possibly significant.
For analysis of associations between tandem repeat data and clades, two approaches were used. In the first, results for each isolate were represented as the average number of repeats between the diploid alleles for each isolate; in the second, each allele was analyzed individually, so each isolate was represented in duplicate. The Kruskal-Wallis test was applied in both cases.
Differences in mouse survival were tested by Kaplan-Meier/log rank analyses, and differences in body weight changes and mouse kidney burdens were tested by the Mann-Whitney U test.
RESULTS
Virulence of 43 C. albicans isolates in mice.
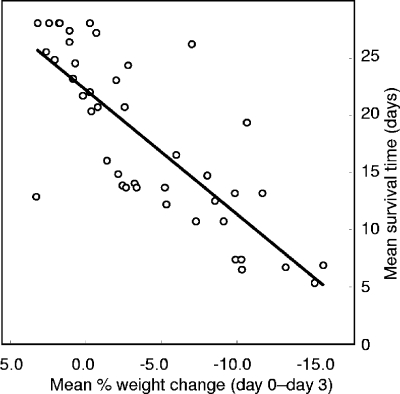
The three parameters related to virulence of each isolate, namely, kidney burden, mean survival time (MST), and weight loss over the first 3 days of infection, are summarized in Table 1. Across the 43 isolates, statistically significant correlations (all P < 0.001 by both Spearman's r and Kendall tau) were found between the three parameters of virulence shown in Table 1. For mean terminal log CFU/g kidney (kidney burden) versus MST, the statistics were r = −0.59 and Τ = −0.43. For mean percentage body weight change from day 0 to day 3 (weight change) and kidney burden, the statistics were r = −0.62 and Τ = −0.50, and for weight change and MST, the statistics were r = 0.79 and Τ = 0.65. The strongest correlation was between weight change and MST (Table 1; Fig. 2), suggesting that the loss of body weight over the first 72 h postinfection was a suitable marker for MST in the BALB/c mouse i.v. challenge model as described in this study. Weight changes over periods shorter than 3 days correlated less effectively with survival times, and from day 4 onwards, mice in some groups began to succumb to infection, resulting in missing data for purposes of analysis. Our use of a 20% fall in body weight as a criterion to determine need for euthanasia of the mice might be thought to have contributed artifactually to the weight change-survival correlation. However, it is clear from Fig. 2 that none of the mean weight changes for the 43 isolates tested reached the −20% level, and therefore, few of the individual weight changes were of this magnitude. Correlations between challenge inoculum and MST, kidney burden, or weight change were all statistically insignificant.
FIG. 2.
Scatterplot of mean change in body weight over 3 days postchallenge versus mean survival time for mice infected with 43 C. albicans isolates. The regression line is shown. The Pearson correlation coefficient r for the data was 0.79; Kendall tau (nonparametric correlation coefficient) was 0.65. For both measures of correlation, P was <0.001.
The C. albicans isolates could be ranked for murine virulence on the basis of the three parameters shown in Table 1, alone or in combination. The ranking approach showed that 14 isolates could be scored as the most consistently virulent in mice by all parameters; 14 could be scored as the least virulent for mice, and the remaining 15 isolates were designated as having “intermediate” mouse virulence (Table 4). There was no obvious association between strain clades and murine virulence.
TABLE 4.
Virulence of 43 C. albicans isolates in mice and other measured characteristics of the isolates tested in vitro
| Isolatea | Virulence rank by:
|
Sum of virulence ranks | Strain characters
|
Growth rate [μmax (h−1)] in RPMI 1640 at:
|
Physiological tests in vitro
|
Adherence tests in vitrof
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MST | Log10 CFU/g kidney | Wt change (day 0 to day 3) | Clade | ABC type | MAT type | 30°C | 37°C | 40°C | Biofilm formation (supernatant OD600) | Growth (OD600) in YCB-BSAe at 30°C | Acid phosphatase sp act (nmol/15 min/OD600) | Alcian blue stain (μg bound per cell OD600) | Growth in 2 M NaCl | BECs with Cal (%) | Cal/BEC | Catheter plasticg | ||
| J990102b | 2 | 5 | 6 | 13 | 3 | B | a/α | 0.18 | 0.23 | 0.18 | 0.07 | 0.09 | 388 | 87 | − | 34 | 2.3 | 1.2 |
| YsU751b | 6 | 1 | 7 | 14 | 1 | A | a/α | 0.17 | 0.23 | 0.11 | 0.02 | 0.07 | 672 | 112 | + | 27 | 1.9 | 1.4 |
| AM2002/0087b | 12 | 2 | 4 | 18 | 1 | A | a/α | 0.17 | 0.23 | 0.13 | 0.03 | 1.41 | 210 | 92 | + | 38 | 1.6 | 1.0 |
| J981318b | 3 | 13 | 3 | 19 | 1 | A | a/α | 0.11 | 0.24 | 0.16 | 0.02 | 0.11 | 319 | 79 | + | 31 | 2.3 | 1.4 |
| J951361b | 1 | 20 | 2 | 23 | 1 | A | a/α | 0.17 | 0.20 | 0.14 | 0.01 | 0.42 | 232 | 72 | − | 30 | 1.7 | 1.4 |
| L1086b | 8 | 3 | 13 | 24 | 4 | B | a/α | 0.16 | 0.23 | 0.18 | 0.04 | 1.45 | 512 | 114 | − | 26 | 1.3 | 1.3 |
| AM2003-020b | 13 | 6 | 9 | 28 | 4 | C | a/α | 0.16 | 0.29 | 0.14 | 0.03 | 0.16 | 296 | 108 | + | 27 | 1.6 | 1.7 |
| IHEM16614b | 5 | 15 | 8 | 28 | 2 | A | a/α | 0.15 | 0.22 | 0.17 | 0.03 | 0.20 | 149 | 131 | − | 21 | 1.8 | 1.6 |
| SC5314b | 4 | 24 | 1 | 29 | 1 | A | a/α | 0.15 | 0.27 | 0.18 | 0.00 | 0.11 | 530 | 76 | + | 27 | 2.0 | 1.3 |
| AM2003/0191b | 7 | 14 | 10 | 31 | 2 | A | a/α | 0.16 | 0.21 | 0.16 | 0.05 | 0.61 | 270 | 122 | + | 35 | 2.5 | 1.1 |
| s20152.082b | 19 | 4 | 12 | 35 | 4 | C | a/α | 0.17 | 0.23 | 0.18 | 0.05 | 0.12 | 436 | 123 | − | 25 | 2.1 | 1.7 |
| IHEM16972b | 9 | 18 | 16 | 43 | 2 | A | a/α | 0.17 | 0.21 | 0.16 | 0.02 | 0.28 | 192 | 70 | − | 22 | 1.8 | 1.0 |
| FJ9b | 22 | 7 | 15 | 44 | 1 | A | a/α | 0.17 | 0.16 | 0.12 | 0.03 | 0.36 | 610 | 72 | + | 35 | 2.1 | 1.1 |
| s20152.013b | 23 | 16 | 5 | 44 | 4 | A | a/α | 0.16 | 0.27 | 0.16 | 0.02 | 0.12 | 320 | 84 | − | 27 | 2.1 | 1.0 |
| 73/025c | 18 | 9 | 19 | 46 | 3 | B | a/α | 0.17 | 0.19 | 0.13 | 0.05 | 0.07 | 509 | 78 | − | 39 | 2.1 | 1.2 |
| AM2003-013c | 14 | 17 | 17 | 48 | 1 | A | a/α | 0.18 | 0.25 | 0.11 | 0.02 | 0.09 | 832 | 101 | + | 28 | 2.5 | 1.1 |
| FC22c | 17 | 8 | 23 | 48 | 2 | A | a/α | 0.18 | 0.23 | 0.16 | 0.06 | 0.64 | 432 | 144 | − | 24 | 1.6 | 1.3 |
| s20122.073c | 10 | 27 | 11 | 48 | 4 | B | a/α | 0.06 | 0.13 | 0.07 | 0.12 | 0.06 | 624 | 111 | − | 36 | 2.7 | 1.1 |
| T101c | 16 | 19 | 21 | 56 | 3 | B | a/a | 0.11 | 0.15 | 0.10 | 0.02 | 1.37 | 680 | 82 | − | 39 | 2.5 | 1.1 |
| s20175.016c | 35 | 10 | 14 | 59 | 4 | C | a/α | 0.13 | 0.23 | 0.11 | 0.25 | 0.11 | 957 | 107 | − | 16 | 1.8 | 0.8 |
| J981315c | 15 | 29 | 18 | 62 | 3 | B | a/α | 0.18 | 0.28 | 0.15 | 0.05 | 0.10 | 455 | 88 | − | 34 | 2.0 | 1.5 |
| IHEM16945c | 20 | 21 | 24 | 65 | 2 | A | α/α | 0.15 | 0.18 | 0.15 | 0.00 | 0.53 | 231 | 134 | − | 26 | 2.0 | 1.3 |
| AM2003/0182c | 28 | 12 | 30 | 70 | 2 | A | a/α | 0.15 | 0.21 | 0.16 | 0.02 | 0.10 | 442 | 110 | + | 36 | 2.0 | 1.2 |
| AM2003/0100c | 25 | 28 | 22 | 75 | 2 | A | a/α | 0.12 | 0.13 | 0.12 | 0.03 | 0.43 | 144 | 61 | − | 33 | 2.5 | 1.5 |
| AM2004/0028c | 24 | 22 | 29 | 75 | 2 | A | a/α | 0.15 | 0.19 | 0.12 | 0.02 | 0.08 | 365 | 97 | − | 29 | 1.9 | 0.7 |
| J981301c | 37 | 11 | 28 | 76 | 1 | A | a/α | 0.18 | 0.26 | 0.11 | 0.01 | 0.08 | 387 | 80 | + | 30 | 2.3 | 0.6 |
| s20176.079c | 26 | 26 | 27 | 79 | 4 | A | a/α | 0.16 | 0.20 | 0.11 | 0.03 | 0.06 | 273 | 108 | + | 20 | 1.5 | 1.4 |
| AM2003-016c | 31 | 32 | 20 | 83 | 1 | A | a/α | 0.16 | 0.25 | 0.11 | 0.01 | 0.09 | 596 | 78 | + | 30 | 2.3 | 1.5 |
| AM2003/0089c | 21 | 37 | 26 | 84 | 2 | A | a/α | 0.16 | 0.20 | 0.14 | 0.01 | 0.32 | 124 | 79 | + | 21 | 1.7 | 1.2 |
| b30708/5d | 11 | 31 | 43 | 85 | 3 | B | a/α | 0.18 | 0.24 | 0.13 | 0.07 | 0.10 | 539 | 92 | − | 34 | 1.5 | 1.5 |
| HUN68d | 30 | 23 | 34 | 87 | 4 | B | a/α | 0.16 | 0.24 | 0.15 | 0.02 | 0.32 | 582 | 109 | − | 23 | 1.6 | 1.8 |
| HUN92d | 29 | 40 | 25 | 94 | 1 | A | a/α | 0.12 | 0.30 | 0.07 | 0.06 | 0.07 | 701 | 112 | + | 21 | 2.1 | 1.4 |
| AM2003/0165d | 27 | 38 | 32 | 97 | 3 | B | α/α | 0.12 | 0.13 | 0.16 | 0.00 | 0.11 | 1049 | 73 | + | 35 | 3.1 | 1.3 |
| AM2003/0074d | 38 | 25 | 35 | 98 | 3 | B | a/α | 0.15 | 0.28 | 0.10 | 0.02 | 0.10 | 703 | 85 | − | 20 | 1.4 | 0.9 |
| AM2003/0069d | 32 | 36 | 33 | 101 | 2 | A | a/α | 0.17 | 0.22 | 0.14 | 0.02 | 0.40 | 143 | 128 | + | 26 | 2.1 | 1.0 |
| RV4688d | 43 | 33 | 31 | 107 | 4 | C | a/α | 0.14 | 0.27 | 0.14 | 0.73 | 0.38 | 1261 | 77 | − | 25 | 1.6 | 1.4 |
| RIHO9d | 34 | 34 | 41 | 109 | 1 | A | a/a | 0.11 | 0.23 | 0.13 | 0.01 | 0.12 | 2060 | 86 | + | 33 | 2.0 | 1.0 |
| AM2005/0377d | 40 | 30 | 40 | 110 | 4 | B | a/a | 0.13 | 0.13 | 0.05 | 0.00 | 0.85 | 542 | 27 | − | 31 | 2.3 | 1.2 |
| 81/064d | 33 | 39 | 39 | 111 | 1 | A | α/α | 0.16 | 0.21 | 0.14 | 0.01 | 0.02 | 388 | 145 | + | 26 | 1.9 | 1.3 |
| FC28d | 36 | 41 | 36 | 113 | 3 | B | α/α | 0.17 | 0.24 | 0.11 | 0.02 | 0.18 | 504 | 81 | − | 42 | 2.0 | 1.2 |
| IHEM3742d | 42 | 35 | 38 | 115 | 4 | C | a/α | 0.13 | 0.22 | 0.13 | 0.83 | 0.43 | 1238 | 74 | − | 28 | 1.8 | 0.3 |
| HUN96d | 41 | 42 | 37 | 120 | 1 | A | a/α | 0.12 | 0.20 | 0.16 | 0.01 | 0.07 | 2522 | 95 | + | 24 | 2.1 | 1.2 |
| 78/028d | 39 | 43 | 42 | 124 | 1 | A | a/α | 0.13 | 0.20 | 0.09 | 0.00 | 1.32 | 225 | 111 | + | 20 | 1.5 | 0.9 |
Isolates are listed in decreasing order of virulence, as determined by the sum of virulence ranks (fifth column from the right).
Isolates ranked as highly virulent.
Isolates ranked as intermediate in virulence.
Isolates ranked as low virulence.
YCB-BSA, yeast carbon base with bovine serum albumin.
Cal, C. albicans.
% reduction Alamar blue per g catheter.
Associations between virulence in mice and other properties of C. albicans isolates.
A range of tests in vitro was applied to the 43-isolate panel to determine whether any property was associated with the virulence of each isolate (Table 4). By the Kruskal-Wallis K-sample test, the data in Table 4 showed a significant association between overall virulence rank and three other variables: MAT heterozygosity versus homozygosity (P = 0.038), acid phosphatase activity (P = 0.019), and growth rate at 40°C (P = 0.004). There was no example of a MAT-homozygous isolate among the 18 most virulent isolates (Table 4). The most virulent isolates tended toward lower acid phosphatase activity: the phosphatase activities (nanomoles/15 min/OD600) (mean ± standard error of the mean [SEM]) were 367 ± 45, 470 ± 64, and 890 ± 190 for the sets of isolates ranked as having high, intermediate, and low virulence, respectively. Finally, the most virulent set of isolates had the highest growth rates in RPMI 1640 medium (mean ± SEM) at 40°C (μ = 0.16 ± 0.01 h−1 for the 14 most virulent isolates versus 0.12 ± 0.01 for the other two groups).
The three variables found to show statistically significant associations with the overall virulence ranking also emerged as significant factors in multiple regression analysis of individual virulence parameters. For MST as the dependent variable, acid phosphatase activity (P = 0.007) and growth at 40°C (P = 0.005) were the only two significantly associated variables in a two-step regression analysis. With weight change as the dependent variable, acid phosphatase activity (P = 0.003) and growth at 40°C (P = 0.003) again emerged as the most significant among the four originally most significant variables, although in this analysis, the clade (P = 0.025) also showed a possibly significant association. Finally, the analysis with kidney burden as the dependent variable showed only MAT heterozygosity (P = 0.002) as a significantly associated factor.
Associations between the clade and other properties of C. albicans isolates.
Kruskal-Wallis analysis of the data (Table 4) for tests in vitro with the C. albicans clade as the grouping variable showed significant associations between the clade and acid phosphatase activity (P = 0.002), ABC type (P < 0.001), and growth in 2 M NaCl (P < 0.001). The percentage of BECs with C. albicans attached (but not the average numbers of C. albicans attached to BECs) showed a possibly significant association with clade (P = 0.033). Only 4 of the 43 isolates made poor or no biofilms on polystyrene under the conditions of the experiment: all were from clade 4, but this association was not significant in the Kruskal-Wallis analysis (P = 0.056). Table 5 summarizes the results of the tests done in vitro, stratified by clade. Data on MAT heterozygosity were not included, since the proportion of homozygous and heterozygous isolates depended on the choice of isolates. The data on ABC type were included as a positive control for validation of the statistical analysis. ABC types A, A, and B are already known to correlate strongly with clades 1, 2, and 3, respectively (38). The association of acid phosphatase activity with clades appeared largely as a generally lower level of activity among clade 2 isolates (Table 5). For clade 2 isolates, mean acid phosphatase specific activity was 249 ± 41 units compared with 735 ± 192, 603 ± 79, and 640 ± 113 units for isolates in clades 1, 3, and 4, respectively. The association of growth in 2 M NaCl with clades was largely the consequence of a much higher proportion of clade 1 isolates that grew at this salt concentration compared with the other clades (Table 5).
TABLE 5.
Summary of results for strain properties and tests in vitro for isolates in each of the four main C. albicans clades
| Strain property or test result | Value for isolates in:
|
|||
|---|---|---|---|---|
| Clade 1 (n = 14) | Clade 2 (n = 10) | Clade 3 (n = 8) | Clade 4 (n = 11) | |
| Type A,a no. of isolates (% of clade) | 14 (100) | 10 (100) | 0 (0) | 2 (18) |
| Type B,a no. of isolates (% of clade) | 0 (0) | 0 (0) | 8 (100) | 4 (36) |
| Type C,a no. of isolates (% of clade) | 0 (0) | 0 (0) | 0 (0) | 5 (45) |
| μmax (mean ± SEM) (h−1) in RPMI 1640 at the following temp: | ||||
| 30°C | 0.15 ± 0.01 | 0.16 ± 0.01 | 0.16 ± 0.01 | 0.14 ± 0.01 |
| 37°C | 0.23 ± 0.01 | 0.20 ± 0.01 | 0.22 ± 0.02 | 0.22 ± 0.02 |
| 40°C | 0.13 ± 0.01 | 0.15 ± 0.01 | 0.13 ± 0.01 | 0.13 ± 0.01 |
| Biofilm formation (mean ± SEM) (supernatant OD600) | 0.017 ± 0.004 | 0.027 ± 0.006 | 0.037 ± 0.030 | 0.191 ± 0.095 |
| Growth (mean ± SEM) (OD600) in YCB-BSAb at 30°C | 0.13 ± 0.13 | 0.36 ± 0.07 | 0.27 ± 0.17 | 0.37 ± 0.14 |
| Acid phosphatase sp acta (mean ± SEM) (nmol/15 min/OD600) | 740 ± 190 | 250 ± 40 | 600 ± 80 | 640 ± 110 |
| Alcian blue (mean ± SEM) (μg bound per cell OD600) | 94 ± 6 | 110 ± 10 | 80 ± 2 | 90 ± 9 |
| Growth in 2 M NaCl,a no. of isolates (% of clade) | 13 (93) | 4 (40) | 1 (13) | 2 (18) |
| No. of BECs positive for C. albicans (mean ± SEM) | 28 ± 1.4 | 27 ± 1.9 | 35 ± 2.5 | 26 ± 1.6 |
| No. of C. albicans per BEC (mean ± SEM) | 2.0 ± 0.1 | 2.0 ± 0.1 | 2.1 ± 0.2 | 1.8 ± 0.1 |
| Adherence to catheter plastic (mean ± SEM) (% reduction Alamar blue per g catheter) | 1.2 ± 0.1 | 1.2 ± 0.1 | 1.3 ± 0.1 | 1.2 ± 0.1 |
These properties were significantly different between clades (Kruskall-Wallis test, P < 0.01).
YCB-BSA, yeast carbon base with bovine serum albumin.
ALS and HYR repeat sequences and their variation between clades and with virulence.
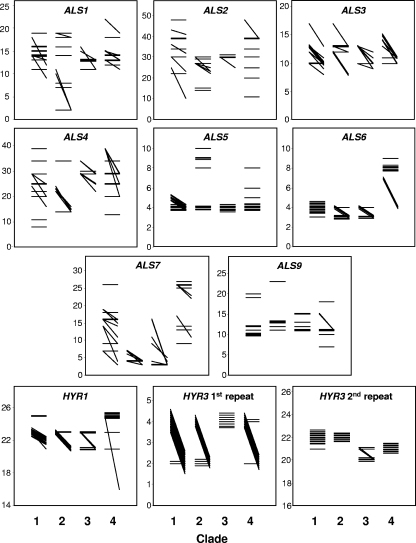
The 43 isolates showed differences in the numbers of many of the ALS and HYR midrepeat sequences for the four C. albicans clades (Fig. 3). Self-evident examples of interclade differences in repeat lengths included ALS6, where isolates in clade 4 all contained at least one allele with approximately twice as many repeats as all alleles in the other three clades, ALS7, where isolates in clade 2 all contained alleles with fewer repeats than for most isolates in the other three clades, the first repeat of HYR3, where clade 3 isolates had uniformly homozygous alleles while almost all the isolates in other clades were heterozygous for this repeat, and the second repeat of HYR3, where there were fewer repeats in the alleles of most isolates in clades 3 and 4 compared to isolates from clades 1 and 2 (Fig. 3). The overall statistics for midrepeat allele data in each clade are shown in Table 6. The data were analyzed for statistically significant differences between clades by applying the Kruskal-Wallis test to alleles considered independently and considered as means of the two alleles per isolate. On the latter, highly conservative basis, the probability that the differences in allele repeats between clades were significant was <0.05 for ALS3 and both HYR3 repeats (Table 6). With the alleles analyzed as independent data by clade, only results for ALS1, ALS3, and ALS4 failed to reach the significance level of P < 0.01.
FIG. 3.
Alleles of midrepeat sequences for genes in the ALS and HYR families. For each of 43 C. albicans isolates, the number of sequence repeats (shown on the y axis) is indicated by a line joining the repeat numbers for the two alleles of each gene. A horizontal line thus indicates homozygous repeats of the numbers indicated in the y axis. A diagonal line indicates an isolate with heterozygous repeats, with the larger of the two allele repeats indicated by the left-hand position of the line. For isolates with identical allele pairs, the results have been jittered vertically to make each isolate line visible. The isolate data are grouped in columns representing results for isolates in clades 1, 2, 3, and 4. Note that some y axes have been compressed to emphasize interclade differences.
TABLE 6.
Summary statistics of ALS and HYR tandem repeat length estimates for 43 C. albicans isolates
| Gene |
P value by cladea
|
Repeat no. for isolates in:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clade 1
|
Clade 2
|
Clade 3
|
Clade 4
|
||||||||||||
| Single alleles | Mean allele | Range | Median | Mode | Range | Median | Mode | Range | Median | Mode | Range | Median | Mode | ||
| ALS1 | 0.062 | 0.95 | 9-19 | 14 | 14 | 2-19 | 12.5 | 2 | 11-16 | 13 | 13 | 11-22 | 14 | 13 | |
| ALS2 | <0.001 | 0.22 | 10-48 | 39 | 39 | 14-30 | 27 | 27 | 25-30 | 30 | 30 | 11-48 | 34 | 39 | |
| ALS3 | 0.078 | 0.016 | 8-17 | 10 | 10 | 8-17 | 13 | 13 | 9-13 | 10.5 | 13 | 10-15 | 11 | 11 | |
| ALS4 | 0.01 | 0.90 | 8-39 | 25 | 25 | 14-34 | 19 | 14 | 22-34 | 29 | 29 | 13-39 | 29 | 29 | |
| ALS5 | 0.002 | 0.071 | 4-5 | 4 | 4 | 4-10 | 6 | 4 | 4-4 | 4 | 4 | 4-8 | 4 | 4 | |
| ALS6 | <0.001 | 0.101 | 3-4 | 4 | 4 | 3-4 | 3 | 3 | 3-4 | 3 | 3 | 4-9 | 8 | 8 | |
| ALS7 | <0.001 | 0.27 | 3-26 | 16 | 16 | 3-7 | 4 | 4 | 3-16 | 3 | 3 | 9-27 | 25 | 26 | |
| ALS9 | 0.007 | 0.44 | 10-20 | 10 | 10 | 11-23 | 13 | 13 | 11-15 | 11.5 | 11 | 7-18 | 11 | 11 | |
| HYR1 | <0.001 | 0.22 | 21-25 | 23 | 22 | 21-23 | 23 | 23 | 21-23 | 22 | 23 | 16-25 | 25 | 25 | |
| HYR3 | |||||||||||||||
| First repeat | <0.001 | 0.018 | 2-4 | 2 | 2 | 2-4 | 2 | 2 | 4-4 | 4 | 4 | 2-4 | 4 | 4 | |
| Second repeat | <0.001 | 0.031 | 21-22 | 22 | 22 | 22-22 | 22 | 22 | 20-21 | 20 | 20 | 21-21 | 21 | 21 | |
Outcome of Kruskall-Wallis test for association between midrepeat numbers and clade, applied to all alleles from all isolates represented separately (single alleles column) or to the average allele size for each (diploid) isolate.
Reanalysis of the midrepeat data grouped according to overall virulence class (Table 4) revealed no statistically significant associations between midrepeat numbers and virulence group (data not shown).
Expression profiling of 20 C. albicans isolates from clades 1 to 4.
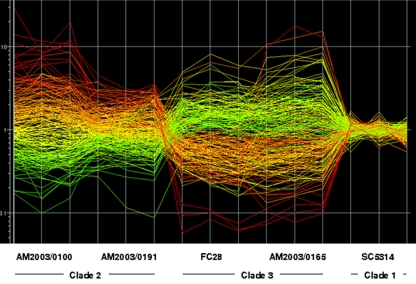
Five isolates selected from each of the four main C. albicans clades were grown to late-exponential phase in NGY and RPMI 1640 media. NGY is the medium used routinely in this laboratory for growth of challenge inocula in the mouse model of disseminated C. albicans infection; it supports the growth of C. albicans purely in the yeast form. It was chosen since the profile observed in this medium is that of cells immediately before they infect experimental animals. RPMI 1640 is a tissue culture medium that mimics the composition of human body fluids and is utilized in applications such as yeast susceptibility testing (37). C. albicans cells grow as mixtures of pseudohyphal and hyphal forms in this medium (7). For both media, expression profiles for the 20 isolates were normalized against the profile for pooled RNA from strain SC5314 grown under the same conditions. The quality of the expression profile data is illustrated in Fig. 4, which shows interpretation graphs for two randomly chosen isolates each in clades 2 and 3 relative to the reference isolate SC5314. Here the relative expression levels of all genes are depicted for 4 of the 20 profiled isolates and the reference isolate SC5314. Figure 4 shows that the replicate quality of the triplicate data was good, and the triplicate SC5314 profiles showed only minor expression variations from the SC5314 RNA pool used to normalize all the data. Figure 4 also illustrates the considerable interisolate diversity found in constitutive gene expression in the present experiments. The expression levels depicted in Fig. 4 suggested extensive interclade gene expression differences, but this level of differential expression was not sustained by analysis of expression profiles for all 20 isolates.
FIG. 4.
Interisolate differences in constitutive gene expression profiles. The figure depicts the GeneSpring interpretation graph for triplicate expression profiles of two C. albicans isolates each from clades 2 and 3, plus reference isolate SC5314, grown in NGY medium. Each line in the graph indicates the relative expression level for a single gene. The expression level of the gene in strain AM2003/0100 compared to SC5314 sets the color of the line, which is kept through all the other strains.
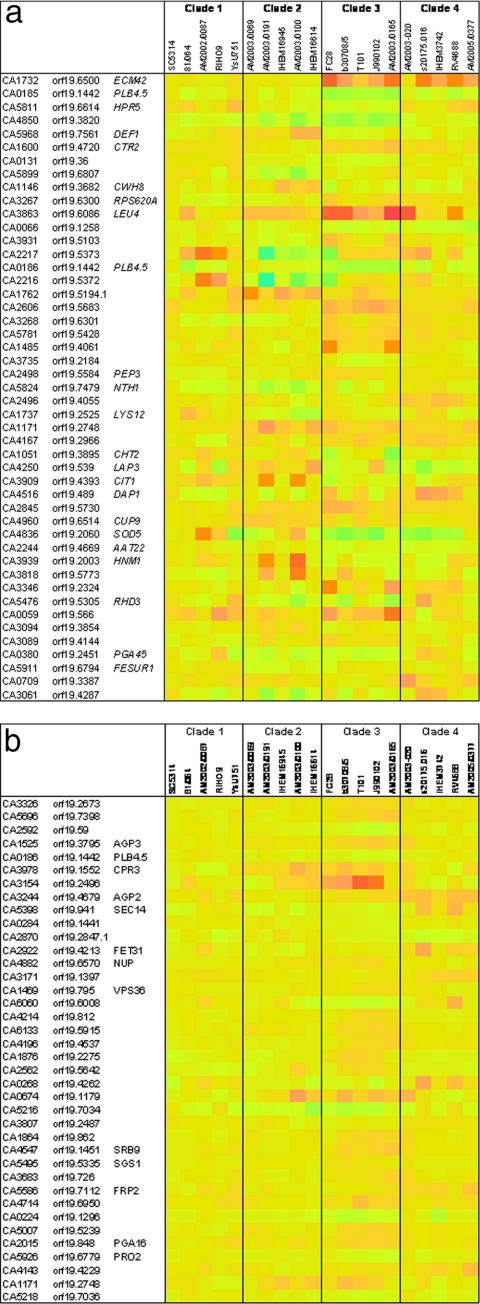
SAM analysis of the 20 expression profile sets stratified by clade identified 134 genes providing multiclass interclade differentiation with a q value of <0.05 for the 20 isolates grown in NGY medium and 43 genes with a q value of <0.05 for the same isolates grown in RPMI 1640 medium. The average differences in expression between significant genes in the multiclass analyses were generally not large; hence, details for only the 50 genes that gave the highest d scores by SAM in the NGY experiment were scrutinized in detail (Fig. 5). Variation in mean expression levels between isolates relative to the control was greater for cells grown in NGY medium than for cells grown in RPMI 1640 medium (compare Fig. 5a and b). Expression of many of the genes illustrated in Fig. 5a appeared to relate particularly to clade 3 isolates, relative to isolates in other clades (Fig. 5a). However, there was also substantial intraclade variation in the levels of expression of many of the genes that emerged from the multiclass SAM analysis for NGY-grown cells.
FIG. 5.
Heat maps indicating the average differences in RNA levels between genes found to be significant interclade differentiators (q < 0.05 by SAM analysis) for 20 C. albicans isolates grown in NGY medium (a) and in RPMI 1640 medium (b). The heat map colors are based on pale yellow indicating no upregulation relative to the pooled SC5314 RNA control. Colors ranging from purple (maximum of 20-fold) through red and orange indicate RNA levels greater than the level for the control, while green through pale blue (20-fold downregulation) colors indicate RNA levels below the level for the control. In both figures, the heat map for SC5314 (mean of triplicate data versus SC5314 RNA pool) is shown in the leftmost column.
The same was not true for cells grown in RPMI 1640 medium (Fig. 5b), where gene expression differences within clades were mostly minor. The five clade 1 isolates, in particular, showed high similarity of expression levels for all of the genes found to be significant contributors to interclade differentiation by SAM analysis (Fig. 5b). Once again, expression levels in clade 3 appeared to offer the most distinctive interclade expression differences.
SAM analysis of the expression profiling data stratified by mouse virulence phenotype (Fig. 6) determined significant multiclass differentiation for 21 ORFs (22 microarray spots) in NGY-grown cells. On the list of differentially expressed genes between the three groups (based on overall virulence ranking; Table 4), genes with reference numbers CA0636 (FDH4.3F), CA0639 (FDH13.3F), and CA3258 (IFC2) are no longer accepted in the current assembly (assembly 21) of the C. albicans genome database (CGD) (http://www.candidagenome.org/). Our finding of differential expression of these ORFs suggest that they may in fact be valid for inclusion in the database. Figure 6 lists the 18 significant and currently accepted genes (one significant gene was represented twice on the arrays, so that the list contains 19 entries) with multiclass differential expression based on the overall virulence of the 20 C. albicans isolates. For RPMI 1640-grown cells, only two genes (ORFs 19.6847 and 19.3075; Fig. 6) met the criteria for inclusion as differentially expressed between isolates in virulence groups 1, 2, and 3.
FIG. 6.
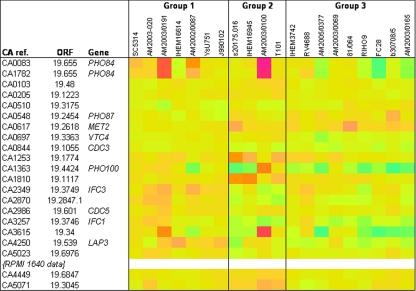
Heat map indicating the average differences in RNA levels between genes found to be significant multiclass differentiators between C. albicans isolates of high (group 1), intermediate (group 2) and low (group 3) virulence in the mouse model (see Table 4). The heat map was set up as explained in the legend to Fig. 5. CA ref., CA reference.
The data in Fig. 6 showed that four genes involved in phosphate uptake, metabolism, and storage were among those with expression significantly differentially associated with virulence. These genes were PHO84 (encoding a protein with similarity to high-affinity phosphate transporters; 26), PHO87 (encoding a protein similar to phosphate permeases; 26), PHO100 (encoding an inducible acid phosphatase; 18), and VTC4, (encoding a polyphosphate synthase; GO annotation, CGD). Other genes with expression significantly different between the three virulence groups were cell cycle genes CDC3 (a yeast neck septin; 12, 55) and CDC5 (involved in filamentous growth; 2), MET2 (encoding a homoserine O-acetyltransferase; 16, 52), LAP3 (encoding an aminopeptidase; 27), and oligopeptide transporter-encoding genes IFC1 and IFC3 (31, 45). It should be noted (see above) that CA3258, another significant gene in our expression array, was formerly known as IFC2.
Characterization of a pho100Δ mutant in vitro and in vivo.
Because the expression profiles possibly associated with virulence contained three upregulated genes associated with phosphate uptake or metabolism, we elected to delete PHO100, which encodes a predicted periplasmic acid phosphatase enzyme (http://www.candidagenome.org). We reasoned that, in this surface location, the phosphatase may be directly involved in host-C. albicans interactions. Yeast cells of the pho100Δ mutant showed elevated acid phosphatase activity in vitro (specific activities of the mutant and pho100::rps1Δ::CIp10-PHO100 reintegrant strains were, respectively, 219% [±29%] and 114% [±37%]) relative to the control strain CAI-4 carrying plasmid CIp10.
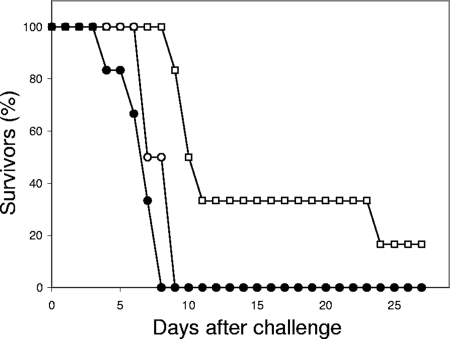
In the mouse intravenous challenge model, the pho100Δ mutant showed modest but statistically significant attenuation of virulence in terms of time to euthanasia (Fig. 7) relative both to the CAI-4 control strain with CIp10 (P < 0.01) and to the pho100::PHO100 reintegrant (P < 0.05). There were no statistically significant differences in percent weight changes between day 0 and day 3 (means ± standard deviations [SDs] were −8.5% ± 4.8%, −13.9% ± 3.1%, and −10.1% ± 3.1%, respectively, for the mutant, CAI-4 control strain with CIp10, and reintegrant strain). Similarly, no significant differences were found between the three strains in terms of kidney fungal burdens at the time of demise. (The log10 CFU/g tissue data were as follows: 5.5 ± 1.6 for the null mutant, 6.0 ± 1.0 for the CAI-4 control strain with CIp10, and 5.8 ± 0.4 for the reintegrant.)
FIG. 7.
Time to euthanasia curves for groups of six mice infected i.v. with the control strain (NGY152; solid circles), pho100::pho100 mutant strain (DMY002; squares), and pho100::PHO100 reintegrant strain (DMY003; open circles).
DISCUSSION
Our study aimed to define possible strain differences between clinical isolates of C. albicans that might relate to their virulence in mice and to their assignment to one of the four major C. albicans clades of related strain types. Because C. albicans is an opportunistic pathogen, depending more on lowered host resistance to infection than on its own inherent virulence attributes to invade and disseminate, we did not expect to find self-evident strain-related properties that would unequivocally associate a particular C. albicans clade or MLST strain type with differential virulence. We anticipated statistical associations between strain attributes and virulence or clade assignment; our data confirmed this expectation.
The basis for clade-level differentiation of C. albicans strains by MLST is partly arbitrary (40) but reflects at least well-established geographical associations suggestive of an independent evolutionary history for isolates in clades 1 to 4 (40, 48, 51). Isolates from the four major clades should therefore exhibit differences detectable by fine-scale measurement but are unlikely to show gross phenotypic differences. In common with previous authors (41, 63), we found statistically significant differences in the numbers of midrepeat sequences for ALS3, ALS5, and ALS6, and we extended the list of genes with clade-associated variable midrepeat sequences to all the other members of the ALS gene family except ALS1 (Table 6) and to three midrepeat sequences in the genes HYR1 and HYR3 (Table 6). Since most of the products encoded by these gene families are surface proteins with known contributions to adherence of C. albicans to various surfaces (58-62) or to yeast-hypha morphogenesis (3, 8), the variations in midrepeat allele lengths between clades might be assumed to contribute to overall virulence of isolates. However, no associations were found between the various midrepeat alleles and strain virulence for mice. Since adherence and morphogenesis are phenomena mediated and regulated by many gene products (1, 9, 14, 19, 28, 42), it is likely that C. albicans possesses considerable functional redundancy with respect to these properties, so while differences may arise between adhesin structures within different strain clades, these are unlikely to translate to detectable effects on strain virulence.
Our expression profiling data generated lists of genes with statistically significant associations with strain clades. Microarray profiling of a complement of more than 6,000 genes with RNA extracted in triplicate from 20 isolates of a fungus might easily result in a set of apparently differential genes arising by chance. Even with appropriate statistical analysis, the sheer number of data points involved offers considerable opportunity for artifactual significant differences in gene expression; hence, we used a highly rigorous approach to data analysis. We compared the SAM outputs from analyses of all triplicate data for each isolate with the outputs for analyses done with the closest pair of results for each isolate-gene combination and accepted from the outputs only genes common to both analyses as potentially significant differentiators at the level of clade or virulence group. One hypothetical expectation for genes with expression differing between clades of strains is that at least some of such genes would encode enzymes in a common biochemical pathway. Our data provide no suggestion of pathway links between any of the genes emerging as differentially expressed between different clades (Fig. 5); the significant multiclass genes listed are all effectively singletons. We have reanalyzed our expression data with the software package GALGO (53), which combines Bayesian and multiple regression approaches to detect sets of genes among expression data that are differentially associated with grouping strata. The software generated a list of genes common with those in Fig. 5 but did not detect any gene combinations that related to strain clades (unpublished data). It is clear from Fig. 5 that expression profiling of none of the genes listed as significant in these experiments could be reliably used to differentiate new isolates at the clade level. However, it is unquestionable that individual isolates of C. albicans can differ markedly in their gene expression profiles when grown under identical conditions (Fig. 4). We cannot exclude the possibility that genes apparently expressed at lower levels in some strains than others may have DNA sequence differences from those of the oligonucleotides in the microarray. Resequencing of a well-selected set of isolates could address this possibility. Overall, we interpret our expression profiles in Fig. 5 as further indication of clade-associated phenomena that represent statistical rather than absolute interclade differences. Profiling for isolates grown under different conditions may, of course, reveal other gene sets that differ between clades.
Among our set of phenotypic properties, levels of acid phosphatase activity and ability to grow in 2 M NaCl were the only parameters significantly associated with strain clades. Average levels of acid phosphatase activity tended to be lower among clade 2 isolates than among isolates from other clades, and all but one of the clade 1 isolates grew in 2 M NaCl compared with 40% in clade 2 and fewer than 20% in clades 3 and 4. Acid phosphatase activity also emerged as a property associated with overall strain virulence (discussed below), but clades themselves were not significantly associated with overall virulence, so we conclude that acid phosphatase should be regarded as a weakly differential interclade property. The finding of generally higher salt tolerance among our clade 1 isolates is similarly difficult to interpret. Clade 1 isolates represent the most commonly encountered C. albicans strain types on a world-wide scale. There have been previous suggestions that clade 1 isolates may be slightly better adapted than others to colonize or superficially infect human hosts (17, 38, 40, 46), but we are not able to suggest how slightly enhanced salt tolerance might contribute to this situation.
Considered overall, the results of this study relating a diverse range of properties to C. albicans clades indicate that direct measurements of genetic diversity, such as determinations of midrepeat sequence lengths, are more successful than transcriptome data and phenotypic testing at revealing interclade differences. Clade-level phenotypic properties are apparent, notably the previously described association of clade 1 isolates with flucytosine resistance and the present finding of much greater salt tolerance among clade 1 isolates. However, none of these represents a characteristic unique to any clade, nor have there been any demonstrable inherent differences in the overall murine virulence of strains from different clades, even in this, the largest examination of interstrain differences in virulence in mice.
Our data relating properties of individual isolates with virulence in mice show more robust associations than the analyses of clade associations. Lower virulence in mice among MAT-homozygous isolates compared with their MAT-heterozygous counterparts has been demonstrated previously. In these tests small numbers of isolates were compared with laboratory-generated MAT-homozygous mutants alone and in experiments with competitive challenge designs. Survival was used as the virulence endpoint (24, 30). Other authors tested a larger panel of clinical isolates and found a lower virulence among MAT-homozygous isolates (57). Our data, based on virulence testing with 43 clinical isolates of C. albicans, fully confirm these previous findings, since no MAT-homozygous isolates were among the 18 most virulent we tested (Table 4). In common with Wu et al. (57), we were unable to confirm a prior suggestion that MAT α/α strains might be even less virulent than a/a types (24). Our results show a mixed distribution of α/α and a/a isolates among the ranks of isolates with intermediate and low virulence in mice (Table 4). The lower virulence of MAT-homozygous isolates is probably a consequence of homozygosity in other genes adjacent to the MAT locus rather than a specific MAT-related event (57).
Phosphate uptake and metabolism emerge from our data as properties associated with mouse virulence in C. albicans. This conclusion is based on two lines of evidence from this study. First, a statistically significant association was found between acid phosphatase activity in vitro and gross virulence of isolates (Table 4). None of the 14 most virulent isolates had acid phosphatase specific activities greater than 700 nmol/15 min/OD600, compared with 7/14 of the least virulent isolates. Second, expression of four genes encoding phosphate-specific activities was shown as significantly related to mouse virulence (Fig. 6): PHO84, PHO87, PHO100, and VTC4, which encode an enzyme involved in phosphate transport, an enzyme involved in phosphate uptake, a phosphomonoesterase, and a polyphosphate synthase, respectively. Acid phosphatase activity detectable in intact C. albicans cells was proposed many years ago as one of several hydrolytic enzymes with theoretical potential as a virulence factor (10). An inducible phosphomonoesterase, which can be released from intact cells by treatment with sulfhydryl reagents (18), was one of the first biological examples of a highly mannosylated protein with enzyme activity (39). In assembly 21 of the C. albicans genome database (http://www.candidagenome.org), five acid phosphatase genes are listed: PHO100, PHO111, PHO112, PHO113, and PHO114. Of these, only PHO100 is inducible and predicted to be located on the surface: we presume this encodes the inducible surface enzyme that has been previously purified and characterized (39) and listed as upregulated in expression profiles of biofilm-forming C. albicans (36). From the C. albicans genome database and our own literature searches, PHO100 has not emerged as a significantly differentially regulated gene in any other studies concerned with C. albicans and virulence. Deletion of the PHO100 gene in the SC5314 background generated a mutant with paradoxically elevated acid phosphatase activity. The mutant showed mild but significant attenuation in virulence in mice as measured by time to euthanasia, and animals infected with the mutant had smaller changes in body weight over the first 3 days postchallenge and the lowest mean kidney fungal burdens compared to the parental reference and PHO100 reintegrant strains, but these differences were not statistically significantly different. The increase in acid phosphatase activity in the pho100 mutant is compatible with the finding of higher acid phosphatase levels in the clinical isolates with the least virulence in mice (Table 4). We think that more-detailed investigation of phosphatase activity is warranted in the context of pathogenesis of C. albicans infections.
The finding of a significant association between gross virulence in mice and an isolate's growth rate in a tissue culture medium at 40°C but not at lower temperatures (Table 4) is compatible with the clinical observation of fever in patients with disseminated C. albicans infections. However, fever is not a characteristic of the mouse i.v. challenge model; body temperature tends to fall with progressive disease (49, 56). We are therefore unable to explain the significance of the high temperature growth rate association with virulence. Monitoring of body temperature to predict the onset of C. albicans terminal disease has been recommended in the mouse model (56). Our own data suggest that mouse body weight change over 3 days postchallenge is also a useful predictor of survival, since the two parameters showed a highly significant correlation. We are currently developing a virulence score based on day 3 weight changes and kidney burdens that will eliminate the need for prolonged survival experiments to determine the virulence of C. albicans strains.
ADDENDUM IN PROOF
A recent update of the Candida genome database lists CA0636 and CA0639 as synonyms of CA1253/orf19.1774 and CA3258 as CA3257/orf19.3746. Since both of these ORFs are already shown in Fig. 6, this new information strengthens the robustness of the data in Fig. 6.
Supplementary Material
Acknowledgments
This work was supported by grants 076954/Z/05/Z and 080088/Z/06/Z from the Wellcome Trust.
Footnotes
Published ahead of print on 16 January 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Argimón, S., J. A. Wishart, R. Leng, S. Macaskill, A. Mavor, T. Alexandris, S. Nicholls, A. W. Knight, B. Enjalbert, R. Walmsley, F. C. Odds, N. A. R. Gow, and A. J. P. Brown. 2007. Developmental regulation of an adhesin gene during cellular morphogenesis in the fungal pathogen Candida albicans. Eukaryot. Cell 6682-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachewich, C., D. Y. Thomas, and M. Whiteway. 2003. Depletion of a polo-like kinase in Candida albicans activates cyclase-dependent hyphal-like growth. Mol. Biol. Cell 142163-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey, D. A., P. J. F. Feldmann, M. Bovey, N. A. R. Gow, and A. J. P. Brown. 1996. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J. Bacteriol. 1785353-5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blignaut, E., J. Molepoa, C. Pujol, D. R. Soll, and M. A. Pfaller. 2005. Clade-related amphotericin B resistance among South African Candida albicans isolates. Diagn. Microbiol. Infect. Dis. 5329-31. [DOI] [PubMed] [Google Scholar]
- 5.Blignaut, E., C. Pujol, S. Lockhart, S. Joly, and D. R. Soll. 2002. Ca3 fingerprinting of Candida albicans isolates from human immunodeficiency virus-positive and healthy individuals reveals a new clade in South Africa. J. Clin. Microbiol. 40826-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brand, A., D. M. MacCallum, A. J. P. Brown, N. A. R. Gow, and F. C. Odds. 2004. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Mol. Microbiol. 3900-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brand, A., J. D. Barnes, K. S. Mackenzie, F. C. Odds, and N. A. R. Gow. 2008. Cell wall glycans and soluble factors determine the interactions between the hyphae of Candida albicans and Pseudomonas aeruginosa. FEMS Microbiol. Lett. 28748-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun, B. R., and A. D. Johnson. 2000. TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics 15557-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, A. J. P., S. Argimón, and N. A. R. Gow. 2007. Signal transduction and morphogenesis in Candida albicans, p. 167-194. In R. J. Howard and N. A. R. Gow (ed.), The Mycota, 2nd ed., vol. VIII. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 10.Chattaway, F. W., F. C. Odds, and A. J. E. Barlow. 1971. An examination of the production of hydrolytic enzymes and toxins by pathogenic strains of Candida albicans. J. Gen. Microbiol. 67255-263. [DOI] [PubMed] [Google Scholar]
- 11.Copping, V. M. S., C. J. Barelle, B. Hube, N. A. R. Gow, A. J. P. Brown, and F. C. Odds. 2005. Exposure of Candida albicans to antifungal agents affects expression of SAP2 and SAP9 secreted proteinase genes. J. Antimicrob. Chemother. 55645-654. [DOI] [PubMed] [Google Scholar]
- 12.DiDomenico, B. J., N. H. Brown, J. Lupisella, J. R. Greene, M. Yanko, and Y. Koltin. 1994. Homologs of the yeast neck filament associated genes: isolation and sequence analysis of Candida albicans CDC3 and CDC10. Mol. Gen. Genet. 242689-698. [DOI] [PubMed] [Google Scholar]
- 13.Dodgson, A. R., K. J. Dodgson, C. Pujol, M. A. Pfaller, and D. R. Soll. 2004. Clade-specific flucytosine resistance is due to a single nucleotide change in the FUR1 gene of Candida albicans. Antimicrob. Agents Chemother. 482223-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ernst, J. F. 2000. Transcription factors in Candida albicans—environmental control of morphogenesis. Microbiology 1461763-1774. [DOI] [PubMed] [Google Scholar]
- 15.Fonzi, W., and M. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.García-Sánchez, S., S. Aubert, I. Iraqui, G. Janbon, J. M. Ghigo, and C. d'Enfert. 2004. Candida albicans biofilms: a developmental state associated with specific and stable gene expression patterns. Eukaryot. Cell 3536-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giblin, L., A. Edelmann, N. X. Zhang, N. B. von Maltzahn, S. B. Cleland, P. A. Sullivan, and J. Schmid. 2001. A DNA polymorphism specific to Candida albicans strains exceptionally successful as human pathogens. Gene 272157-164. [DOI] [PubMed] [Google Scholar]
- 18.Granger, B. L., M. L. Flenniken, D. A. Davis, A. P. Mitchell, and J. E. Cutler. 2005. Yeast wall protein 1 of Candida albicans. Microbiology 1511631-1644. [DOI] [PubMed] [Google Scholar]
- 19.Green, C. B., S. M. Marretta, G. Cheng, F. F. Faddoul, E. J. Ehrhart, and L. L. Hoyer. 2006. RT-PCR analysis of Candida albicans ALS gene expression in a hyposalivatory rat model of oral candidiasis and in HIV-positive human patients. Med. Mycol. 44103-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hauser, N. C., M. Vingron, M. Scheideler, B. Krems, K. Hellmuth, K.-D. Entian, and J. D. Hoheisel. 1998. Transcriptional profiling on all open reading frames of Saccharomyces cerevisiae. Yeast 141209-1221. [DOI] [PubMed] [Google Scholar]
- 21.Hobson, R. P., C. A. Munro, S. Bates, D. M. MacCallum, J. E. Cutler, S. E. M. Heinsbroek, G. D. Brown, F. C. Odds, and N. A. R. Gow. 2004. Loss of cell wall mannosylphosphate in Candida albicans does not influence macrophage recognition. J. Biol. Chem. 27939628-39635. [DOI] [PubMed] [Google Scholar]
- 22.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57267-272. [DOI] [PubMed] [Google Scholar]
- 23.Hope, W. W., L. Tabernero, D. W. Denning, and M. J. Anderson. 2004. Molecular mechanisms of primary resistance to flucytosine in Candida albicans. Antimicrob. Agents Chemother. 484377-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibrahim, A. S., B. B. Magee, D. C. Sheppard, M. Yang, S. Kauffman, J. Becker, J. E. Edwards, and P. T. Magee. 2005. Effects of ploidy and mating type on virulence of Candida albicans. Infect. Immun. 737366-7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 26.Lan, C. Y., G. Newport, L. A. Murillo, T. Jones, S. Scherer, R. W. Davis, and N. Agabian. 2002. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc. Natl. Acad. Sci. USA 9914907-14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lan, C. Y., G. Rodarte, L. A. Murillo, T. Jones, R. W. Davis, J. Dungan, G. Newport, and N. Agabian. 2004. Regulatory networks affected by iron availability in Candida albicans. Mol. Microbiol. 531451-1469. [DOI] [PubMed] [Google Scholar]
- 28.Li, F., M. J. Svarovsky, A. J. Karlsson, J. P. Wagner, K. Marchillo, P. Oshel, D. Andes, and S. P. Palecek. 2007. Eap1p, an adhesin that mediates Candida albicans biofilm formation in vitro and in vivo. Eukaryot. Cell 6931-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindhardt, L., and K. Walter. 1963. Phosphatases, p. 783-785. In H.-U. Bergmeyer (ed.), Methods in enzymatic analysis. Academic Press, New York, NY.
- 30.Lockhart, S. R., W. Wu, J. B. Radke, R. Zhao, and D. R. Soll. 2005. Increased virulence and competitive advantage of a/alpha over a/a or alpha/alpha offspring conserves the mating system of Candida albicans. Genetics 1691883-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorenz, M. C., J. A. Bender, and G. R. Fink. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 31076-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacCallum, D. M., H. Findon, C. C. Kenny, G. Butler, K. Haynes, and F. C. Odds. 2006. Different consequences of ACE2 and SW15 gene disruptions for virulence of pathogenic and nonpathogenic yeasts. Infect. Immun. 745244-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCullough, M., K. V. Clemons, and D. A. Stevens. 1999. Molecular epidemiology of the global and temporal diversity of Candida albicans. Clin. Infect. Dis. 291220-1225. [DOI] [PubMed] [Google Scholar]
- 34.McCullough, M. J., K. V. Clemons, and D. A. Stevens. 1999. Molecular and phenotypic characterization of genotypic Candida albicans subgroups and comparison with Candida dubliniensis and Candida stellatoidea. J. Clin. Microbiol. 37417-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murad, A. M. A., P. R. Lee, I. D. Broadbent, C. J. Barelle, and A. J. P. Brown. 2000. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16325-327. [DOI] [PubMed] [Google Scholar]
- 36.Murillo, L. A., G. Newport, C. Y. Lan, S. Habelitz, J. Dungan, and N. M. Agabian. 2005. Genome-wide transcription profiling of the early phase of biofilm formation by Candida albicans. Eukaryot. Cell 41562-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.NCCLS. 2002. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard—2nd ed., vol. 17 (9). NCCLS, Wayne, PA.
- 38.Odds, F. C., M.-E. Bougnoux, D. J. Shaw, J. M. Bain, A. D. Davidson, D. Diogo, M. D. Jacobsen, M. Lecomte, S.-Y. Li, A. Tavanti, M. C. J. Maiden, N. A. R. Gow, and C. d'Enfert. 2007. Molecular phylogenetics of Candida albicans. Eukaryot. Cell 61041-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Odds, F. C., and J. C. Hierholzer. 1973. Purification and properties of a glycoprotein acid phosphatase from Candida albicans. J. Bacteriol. 114257-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Odds, F. C., and M. D. Jacobsen. 2008. Multilocus sequence typing of pathogenic Candida species. Eukaryot. Cell 71075-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oh, S. H., G. Cheng, J. A. Nuessen, R. Jajko, K. M. Yeater, X. M. Zhao, C. Pujol, D. R. Soll, and L. L. Hoyer. 2005. Functional specificity of Candida albicans Als3p proteins and clade specificity of ALS3 alleles discriminated by the number of copies of the tandem repeat sequence in the central domain. Microbiology 151673-681. [DOI] [PubMed] [Google Scholar]
- 42.Phan, Q. T., C. L. Myers, Y. Fu, D. C. Sheppard, M. R. Yeaman, W. H. Welch, A. S. Ibrahim, J. E. Edwards, and S. G. Filler. 2007. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 5543-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pujol, C., M. Pfaller, and D. R. Soll. 2002. Ca3 fingerprinting of Candida albicans bloodstream isolates from the United States, Canada, South America, and Europe reveals a European clade. J. Clin. Microbiol. 402729-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pujol, C., M. A. Pfaller, and D. R. Soll. 2004. Flucytosine resistance is restricted to a single genetic clade of Candida albicans. Antimicrob. Agents Chemother. 48262-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reuss, O., and J. Morschhauser. 2006. A family of oligopeptide transporters is required for growth of Candida albicans on proteins. Mol. Microbiol. 60795-812. [DOI] [PubMed] [Google Scholar]
- 46.Schmid, J., S. Herd, P. R. Hunter, R. D. Cannon, M. S. M. Yasin, S. Samad, M. Carr, D. Parr, W. McKinney, M. Schousboe, B. Harris, R. Ikram, M. Harris, A. Restrepo, G. Hoyos, and K. P. Singh. 1999. Evidence for a general-purpose genotype in Candida albicans, highly prevalent in multiple geographical regions, patient types and types of infection. Microbiology 1452405-2413. [DOI] [PubMed] [Google Scholar]
- 47.Seidler, M., S. Salvenmoser, and F. M. C. Muller. 2006. In vitro effects of micafungin against Candida biofilms on polystyrene and central venous catheter sections. Int. J. Antimicrob. Agents 28568-573. [DOI] [PubMed] [Google Scholar]
- 48.Soll, D. R., and C. Pujol. 2003. Candida albicans clades. FEMS Immunol. Med. Microbiol. 391-7. [DOI] [PubMed] [Google Scholar]
- 49.Spellberg, B., A. S. Ibrahim, J. E. Edwards, and S. G. Filler. 2005. Mice with disseminated candidiasis die of progressive sepsis. J. Infect. Dis. 192336-343. [DOI] [PubMed] [Google Scholar]
- 50.Takakura, S., S. Ichiyama, J. M. Bain, A. D. Davidson, M. D. Jacobsen, D. J. Shaw, N. A. R. Gow, and F. C. Odds. 2008. Comparison of Candida albicans strain types among isolates from three countries. Int. J. Med. Microbiol. 298663-668. [DOI] [PubMed] [Google Scholar]
- 51.Tavanti, A., A. D. Davidson, M. J. Fordyce, N. A. R. Gow, M. C. J. Maiden, and F. C. Odds. 2005. Population structure and properties of Candida albicans, as determined by multilocus sequence typing. J. Clin. Microbiol. 435601-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tournu, H., G. Tripathi, G. Bertram, S. Macaskill, A. Mavor, L. Walker, F. C. Odds, N. A. R. Gow, and A. J. P. Brown. 2005. Global role of the protein kinase Gcn2 in the human pathogen Candida albicans. Eukaryot. Cell 41687-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trevino, V., and F. Falciani. 2006. GALGO: an R package for multivariate variable selection using genetic algorithms. Bioinformatics 221154-1156. [DOI] [PubMed] [Google Scholar]
- 54.Tusher, V., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to transcriptional responses to ionizing radiation. Proc. Natl. Acad. Sci. USA 985116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Warenda, A. J., and J. B. Konopka. 2002. Septin function in Candida albicans morphogenesis. Mol. Biol. Cell 132732-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Warn, P. A., M. W. Brampton, A. Sharp, G. Morrissey, N. Steel, D. W. Denning, and T. Priest. 2003. Infrared body temperature measurement of mice as an early predictor of death in experimental fungal infections. Laboratory Animals 37126-131. [DOI] [PubMed] [Google Scholar]
- 57.Wu, W., S. R. Lockhart, C. Pujol, T. Srikantha, and D. R. Soll. 2007. Heterozygosity of genes on the sex chromosome regulates Candida albicans virulence. Mol. Microbiol. 641587-1604. [DOI] [PubMed] [Google Scholar]
- 58.Zhao, X., K. J. Daniels, S. H. Oh, C. B. Green, K. M. Yeater, D. R. Soll, and L. L. Hoyer. 2006. Candida albicans Als3p is required for wild-type biofilm formation on silicone elastomer surfaces. Microbiology 1522287-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao, X., S. H. Oh, and L. L. Hoyer. 2007. Deletion of ALS5, ALS6 or ALS7 increases adhesion of Candida albicans to human vascular endothelial and buccal epithelial cells. Med. Mycol. 45429-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao, X., S. H. Oh, K. M. Yeater, and L. L. Hoyer. 2005. Analysis of the Candida albicans Als2p and Als4p adhesins suggests the potential for compensatory function within the Als family. Microbiology 1511619-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao, X. M., S. H. Oh, G. Cheng, C. B. Green, J. A. Nuessen, K. Yeater, R. P. Leng, A. J. P. Brown, and L. L. Hoyer. 2004. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin: functional comparisons between Als3p and Als1p. Microbiology 1502415-2428. [DOI] [PubMed] [Google Scholar]
- 62.Zhao, X. M., S. H. Oh, and L. L. Hoyer. 2007. Unequal contribution of ALS9 alleles to adhesion between Candida albicans and human vascular endothelial cells. Microbiology 1532342-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao, X. M., S. H. Oh, R. Jajko, D. J. Diekerna, M. A. Pfaller, C. Pujol, D. R. Soll, and L. L. Hoyer. 2007. Analysis of ALS5 and ALS6 allelic variability in a geographically diverse collection of Candida albicans isolates. Fungal Genet. Biol. 441298-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.