Abstract
Constitutive androstane receptor (CAR) and peroxisome proliferator activated receptor (PPAR)α are transcription factors known to be primary mediators of liver effects, including carcinogenesis, by phenobarbital-like compounds and peroxisome proliferators, respectively, in rodents. Many similarities exist in the phenotypes elicited by these two classes of agents in rodent liver, and we hypothesized that the initial transcriptional responses to the xenobiotic activators of CAR and PPARα will exhibit distinct patterns, but at later time-points these biological pathways will converge. In order to capture the global transcriptional changes that result from activation of these nuclear receptors over a time course in the mouse liver, microarray technology was used. First, differences in basal expression of liver genes between C57Bl/6J wild-type and Car-null mice were examined and 14 significantly differentially expressed genes were identified. Next, mice were treated with phenobarbital (100 mg/kg by gavage for 24 hrs, or 0.085% w/w diet for 7 or 28 days), and liver gene expression changes with regards to both time and treatment were identified. While several pathways related to cellular proliferation and metabolism were affected by phenobarbital in wild-type mice, no significant changes in gene expression were found over time in the Car-nulls. Next, we determined commonalities and differences in the temporal response to phenobarbital and WY-14,643, a prototypical activator of PPARα. Gene expression signatures from livers of wild-type mice C57Bl6/J mice treated with PB or WY-14,643 were compared. Similar pathways were affected by both compounds; however, considerable time-related differences were present. This study establishes common gene expression fingerprints of exposure to activators of CAR and PPARα in rodent liver and demonstrates that despite similar phenotypic changes, molecular pathways differ between classes of chemical carcinogens.
Keywords: Kupffer cells, PPARα, toxicogenomics, microarrays
INTRODUCTION
Nuclear receptors have been a topic of interest in pharmacology and toxicology due to their important role in cellular signalling and homeostasis. They are the largest known family of transcription factors that function as modulators of tissue gene expression (Urquhart et al., 2007). Numerous xenobiotics may serve as activators of these transcription factors and thus cause dramatic changes in physiological processes (Woods et al., 2007a). Two nuclear receptors, constitutive androstane receptor (CAR) and peroxisome proliferator-activated receptor (PPAR)α, have been implicated as key mediators responsible for non-genotoxic hepatocarcinogenesis in rodents. In addition, activation of these nuclear receptors by various xenobiotics and resultant induction of metabolizing and other immediate response genes yields similar toxicity phenotypes in rodent liver including secondary oxidative stress, cell proliferation, and ultimate development of liver tumors.
Phenobarbital is a prototypical activator of rodent CAR, although it does not exhibit direct binding to the receptor (Kakizaki et al., 2003). Phenobarbital and like compounds had long been observed to induce microsomal enzyme systems, but it was not until the discovery of the CAR gene that the mode of action was uncovered (Honkakoski et al., 1998). Short term administration of phenobarbital to rodents leads to hepatocellular hypertrophy, hyperplasia, and overall hepatomegaly. Chronic exposure to high doses causes hepatocellular adenomas in both mice and rats and hepatocellular carcinomas in some strains of mice (Thorpe and Walker, 1973; Rossi et al., 1977); however, long-term therapy with phenobarbital has not been found to cause human tumors (Whysner et al., 1996). Inter-individual and species differences in the levels of CAR have also been reported, and it was suggested that this may play a role in variability of CAR-dependent liver induction responses (Nuclear Receptors Nomenclature Committee, 1999). Still, since phenobarbital is able to induce xenobiotic metabolizing enzymes in both human and rodent hepatocytes, the molecular basis for species differences in carcinogenic response has yet to be elucidated.
Peroxisome proliferators are a class of diverse molecules which include a wide range of industrial, pharmaceutical and endogenous compounds. Peroxisome proliferators have been extensively studied because of their carcinogenicity in rodents (Lalwani et al., 1981) and uncertain risk to humans (Rusyn et al., 2006). While PPARα is largely responsible for lipid metabolism in liver and other tissues, it has been postulated that activation of this nuclear receptor is a key event in the mode of action of these agents (Peters et al., 2005). There are substantial differences among species in expression, structure and function of PPARα (Palmer et al., 1998), and it is widely believed that these differences may be responsible for susceptibility of rats and mice to liver cancer due to peroxisome proliferators (Gonzalez and Shah, 2008).
Microarray technology has become a useful tool for simultaneously measuring the expression of thousands of genes and has been widely used to establish global transcriptional signatures in response to toxic insult. Indeed, many studies conducted with phenobarbital and like compounds confirm the key role of CAR in activating various xenobiotic metabolism genes (Wei et al., 2000; Ueda et al., 2002; Maglich et al., 2002). In this study we compared global transcriptional changes in response to activation of CAR and PPARα (Woods et al., 2007b) pathways in mouse liver over a time-course. We hypothesized that the initial transcriptional responses to the prototypical xenobiotic activators phenobarbital and WY-14,643 will exhibit distinct patterns of expression changes reflective of activation of CAR and PPARα, respectively; but at later time-points, these biological pathways will converge due to similar phenotypic responses observed in the mouse liver after sub-chronic treatment with the two chemicals. Both a gene-centric and a pathway-centric approach were used to compare biological changes. This study draws parallels and identifies differences in the dynamic changes over a time-course in response to two distinct classes of chemical rodent liver carcinogens. Importantly, while we found that there are similarities between phenobarbital and WY-14,643 in cell proliferation pathways with some temporal differences in their activation, the divergence in metabolism and immune response genes, is profound.
MATERIALS AND METHODS
Chemicals, Animals, Dosing, and Tissue Collection
Phenobarbital sodium and 4-chloro-6-(2,3-xylidino)-pyrimidynylthioacetic acid (WY-14,643) were obtained from Aldrich (Milwaukee, WI). Male C57BL/6J mice 6–8weeks old were purchased from Jackson Laboratories (Bar Harbor, ME) and male Car-null mice (>5 back-crosses to the C57BL/6J strain) were bred in-house. Genotypes were confirmed by PCR amplification of purified DNA extracted from tail and then run on a denaturing gel. Animals were housed up to four per cage in ventilated cages with standard bedding. The animal room was kept on a 12-hr light/dark cycle at a temperature of 22 ± 2°C and relative humidity 50 ± 5%. The facility was maintained by the UNC Division of Laboratory Animal Medicine, and all care was administered in accordance with regulations set forth by the Institutional Animal Care and Use Committee. Animals had free access to purified water throughout the study and the health status of the animals was monitored every other day. Prior to experiments, animals were allowed to acclimate to the animal facility for at least 24 hours and maintained on standard lab chow diet and purified water ad libitum.
Mice were given a single acute dose of 0 (control) or 100 mg/kg phenobarbital sodium in distilled water by oral gavage and sacrificed 24 hrs post-dosing. For sub-chronic treatments, mice were given phenobarbital-containing (0.085% w/w, 850 ppm, equivalent to 70–140 mg/kg daily dose for a 30 g mouse) NIH-07 powdered diet or control NIH-07 powdered diet ad libitum. Animals (n=3–4) were sacrificed after 7 or 28 days of dietary treatment. At sacrifice, mice were anesthetized with pentobarbital (100 mg/kg) and following exsanguination, livers were removed and weighed. Sections from the left lateral lobe, median lobe, and ventral lobe were fixed in 10% formalin. A portion of duodenum was also included as a positive control for proliferation markers used in immunohistochemistry. A small section of the left lateral lobe was also placed in a vinyl specimen mold, embedded in Tissue-Tek OCT compound (Sakuar Finetek, Torrance, CA) and frozen to −80°C. Frozen sections were later used for lipid staining. The remaining tissue was sectioned, placed in eppendorf tubes, and snap frozen in liquid nitrogen. These samples were stored at −80°C until assayed.
Studies with WY-14,643 were described in a previously published report (Woods et al., 2007b) where C57BL/6J mice were administered an acute dose by a single oral gavage of 0 (control) or 50 mg/kg of WY-14,643 in olive oil and sacrificed at 1 day post-dosing. Sub-chronic doses of WY-14,643 were administered in the diet ad libitum. NIH-07 was used as the base for the powdered diet containing either 0% (control) or 0.05% w/w (500 ppm) of WY-14,643. Mice (n=3) were sacrificed after either 7 or 28 days of dietary treatment.
Clinical Chemistry
Blood was collected at sacrifice from the inferior vena cava, placed in collection tubes and spun at 16,000g for 20 minutes at room temperature. Serum was collected over ice and stored at −20°C before analysis. Activity of alanine aminotransferase (ALT) in serum was determined using a standard kinetic assay as described by the manufacturer (Thermo Scientific, Waltham, MA).
Proliferating Cell Nuclear Antigen (PCNA) Immunohistochemistry and Quantification
Formalin-fixed, paraffin-embedded liver sections (5 µm) were mounted onto glass slides. Sections were deparaffinized in xylene, rehydrated in a series of graded alcohol concentrations, and placed in PBS with 1% Tween 20. Immunostaining was performed using DAKO EnVision System HRP (Dako Cytomation, Carpinteria, CA) with primary monoclonal anti-PCNA antibody (Dako, M0879) diluted in PBS containing 1% bovine serum albumin and incubated overnight at 4°C. Slides were counterstained with hematoxylin. Quantitative analysis of immunostained liver sections was performed using BIOQUANT software (BIOQUANT Image Analysis, Nashville, TN) by dividing the number of positively stained nuclei to total nuclei within 10 random fields at 200x magnification.
Oil Red O Staining and Quantification
Frozen liver sections (5–7 mm thick) were mounted onto slides in distilled water and placed into a bath of absolute propylene glycol for 2 minutes. Slides were then immersed in 0.5% Oil Red O solution in 100 mL propylene glycol for one hour, rinsed in 85% propylene glycol for 1 minute, washed with distilled water, and stained with hematoxylin. After rinsing with distilled water, a 1% lithium carbonate-aqueous solution was applied and followed by three rinses with distilled water. Slides were then dehydrated with serial dilutions of ethanol up to xylene and cover slipped with Permount mounting media. Quantitative analysis of stained liver sections was performed using BIOQUANT software (BIOQUANT Image Analysis, Nashville, TN). The percent area stained was determined by dividing the area of positively stained red droplets to total area of the tissue. This was accomplished by randomly selecting 10 fields on each slide at 200x magnification.
RNA Isolation, cDNA Preparation and Microarray Hybridization
Total RNA was isolated from the right lobe of the liver using Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA) according to manufacturer’s protocol. Briefly, approximately 30 mg of frozen tissue was immersed in 600 µl of RLT lysis buffer containing 1% β-mercaptoethanol and homogenized using the Ultra-Turrax® T8 homogenizer (IKA, Wilmington, NC). The homogenate was centrifuged at 16000g for three minutes. Total RNA was extracted from the resulting lysate (supernatant) by ethanol precipitation then bound to a column membrane, washed, and eluted in RNase-free water. RNA purity and integrity were assessed using RNA 6000 Nano Assay LabChips® (Agilent Technologies, Santa Clara, CA) and analyzed on a 2100 Bioanalyzer (Agilent Technologies) according to manufacturer’s protocol.
Preparation of cDNA, labeling and hybridizations were performed using reagents from the low RNA input fluorescent linear amplification kit (Agilent Technologies) based on the manufacturer’s protocol. A pooled mouse RNA sample (Cogenics, RTP, NC) derived from equal amounts of RNA from kidney, spleen, lung, brain, and liver was used as a reference and prepared in parallel to the samples of interest. Samples were analyzed using an Agilent Mouse Oligo Microarray (~21,000 features, catalog# G4121). The hybridized microarrays were washed and scanned using an Agilent G2565BA scanner. Data were extracted from the scanned image using Agilent Feature Extraction software version 9.1. Raw data is available from the UNC Microarray database (genome.unc.edu).
Microarray Data Analysis
Array quality was assessed by Agilent Feature Extraction software and genes with fewer than 70% present data across all arrays were excluded from further analysis. LOWESS normalization was performed to eliminate dye bias. For a given gene, the Cy5/Cy3 ratios were divided by the average Cy5/Cy3 ratio for their time-matched controls. Missing data points were calculated using K-nearest neighbor imputation method. Average-linkage, hierarchical clustering was performed using Cluster software on median centered (by genes) data and visualized by Java Treeview. Batch effects were removed using Partek Genomics Suite in which an ANOVA model was fitted and removed residuals due to batch effects. For analysis comparing gene-by-gene differences in PB versus WY samples, Distance Weighted Discrimination (DWD) was used to combine the two data sets. This method uses a linear discrimination method to classify samples and is beneficial because it avoids data piling (Benito et al., 2004).
Differentially expressed genes were identified using either Significance Analysis of Microarrays (SAM) or Extraction of Differential Gene Expression (EDGE) software (Tusher et al., 2001; Leek et al., 2006). SAM was performed in cases where statistical significance across only one variable (strain, time, or treatment) was being assessed. EDGE was used for identifying differentially expressed genes across two or more variables (i.e., time and treatment). Q-values, which represent the false discover rate (FDR) of less than 0.05 for SAM and EDGE were selected as thresholds for differential expression. Once the list of significant genes was generated by EDGE, a t-statistic was calculated for each gene at each treatment/time combination to determine statistical difference between treatment and control expression. For each treatment/time combination, a list of differentially expressed genes (p<0.05) was used for functional analysis and generation of gene networks.
Functional Analysis of Significant Gene Sets
High-Throughput GOMiner was used to determine biological function of differentially expressed genes, in the context of Gene Onotology (GO) (Zeeberg et al., 2005) and was used for pathway analysis of significant genes lists generated from EDGE time-course analyses. A Q-value, representative of the FDR<0.05 and p<0.05 were the basis for statistical significance from this analysis. Finally, gene networks were generated by Ingenuity Pathways Analysis (Ingenuity Systems, Redwood City, CA). WebGestalt Gene Set Analysis Toolkit, a web-based software developed at Vanderbilt University, was used to visualize chromosomal distribution of significant genes (Zhang et al., 2005). Significantly differentially expressed genes by time-point and treatment were uploaded into Ingenuity Pathways Analysis and identified as focus genes. The gene network algorithm identifies “top networks” limited to 35 genes (“network nodes”) and creates pathways to maximize connections between the focus genes.
RESULTS AND DISCUSSION
Phenotypic changes in response to phenobarbital in liver of wild type and Car-null mice
Phenobarbital elicits numerous characteristic pleiotropic effects on the mouse liver including hepatocellular hypertrophy, hyperplasia, and hyper-proliferation of the smooth endoplasmic reticulum, effects dependent on the presence of a functional Car gene (Wei et al., 2000; Yamamoto et al., 2004). As expected, wild-type animals exhibited a significant increase in liver weight, relative to body weight, at 7 and 28 days of phenobarbital feeding in comparison to their time-matched controls, whereas no such effect was evident in the Car-null (Table 1). Examination of hematoxylin and eosin-stained sections of livers from wild-type phenobarbital-treated mice showed enlarged hepatocytes at 7 and 28 days (data not shown). Immunohistochemical staining with Proliferating Nuclear Cell Antigen (PCNA) antibody was used to quantify the number of nuclei in early G1 phase and S phase of the cell cycle. The phenobarbital-induced hepatocellular proliferation in wild-type C57Bl/6J mice followed a bell-shaped curve with lower-levels at 1 day followed by a proliferative burst at 7 days which then subsided after 28 days exposure. This closely follows the trend of proliferative action observed in C3H/HeJ mice treated with a similar dose and time-course (Peffer et al., 2007). Clinical chemistry markers for liver injury such as alanine aminotransferase (ALT) did not significantly change with PB-treatment in either the wild-type or Car-null mice Supplemental Table 1).
Table 1.
Phenotypic endpoints in livers of C57Bl/6J wild-type and Car-null mice in response to phenobarbital treatment.
| Strain | Treatment | Time | Liver Weight (% of Body Weight) |
Alanine Aminotransferase (U/L) |
Proliferating Cell Nuclear Antigen (% Nuclei Stained) |
Oil Red O (% Area Stained) |
|---|---|---|---|---|---|---|
| 1d | 5.2 ± 0.4 | 50.7 ± 11.4 | 6.2 ± 3.5 | 0.4 ± 0.3 | ||
| Control | 7d | 5.8 ± 0.4 | 33.4 ± 30.8 | 13.5 ± 3.1 | 0.2 ± 0.2 | |
| 850ppm) | 28d | 5.8 ± 0.5 | 16.3 ± 5.4 | 10.5 ± 4.4 | 1.3 ± 1.0 | |
| C57Bl/6J | ||||||
| Phenobarbital | 1d | 5.5 ± 0.1 | 28.5 ± 11.7 | 6.4 ± 4.1 | 3.3 ± 2.9 | |
| (100mg/kg or | 7d | 6.6 ± 0.3* | 24.9 ± 7.1 | 23.6 ± 7.3 | 0.2 ± 0.2 | |
| 28d | 6.8 ± 0.2* | 36.6 ± 13.6 | 16.1 ± 5.3 | 1.7 ± 1.5 | ||
| 1d | 5.5 ± 0.8 | 26.1 ± 13.4 | 10.1 ± 3.4 | 0.3 ± 0.3 | ||
| Control | 7d | 5.4 ± 0.2 | 16.4 ± 4.4 | 18.6 ± 3.6 | 1.5 ± 1.0 | |
| 28d | 5.6 ± 0.1 | 21.1 ± 5.7 | 10.2 ± 3.9 | 3.4 ± 1.6 | ||
| Car-null | ||||||
| Phenobarbital | 1d | 6.0 ± 0.4 | 69.8 ± 23.4 | 12.1 ± 14.1 | 0.7 ± 0.5 | |
| (100mg/kg or | 7d | 5.3 ± 0.5 | 39.8 ± 18.3 | 9.3 ± 5.0 | 0.9 ± 1.0 | |
| 850ppm) | 28d | 5.3 ± 0.2 | 39.8 ± 18.3 | 16.5 ± 6.9 | 1.7 ± 1.6 | |
Values are expressed as mean ± standard deviation
Indicates value significantly different from time-matched control by Student’s t-test, p<0.05.
Differences in basal liver gene expression between wild-type and Car-null mice
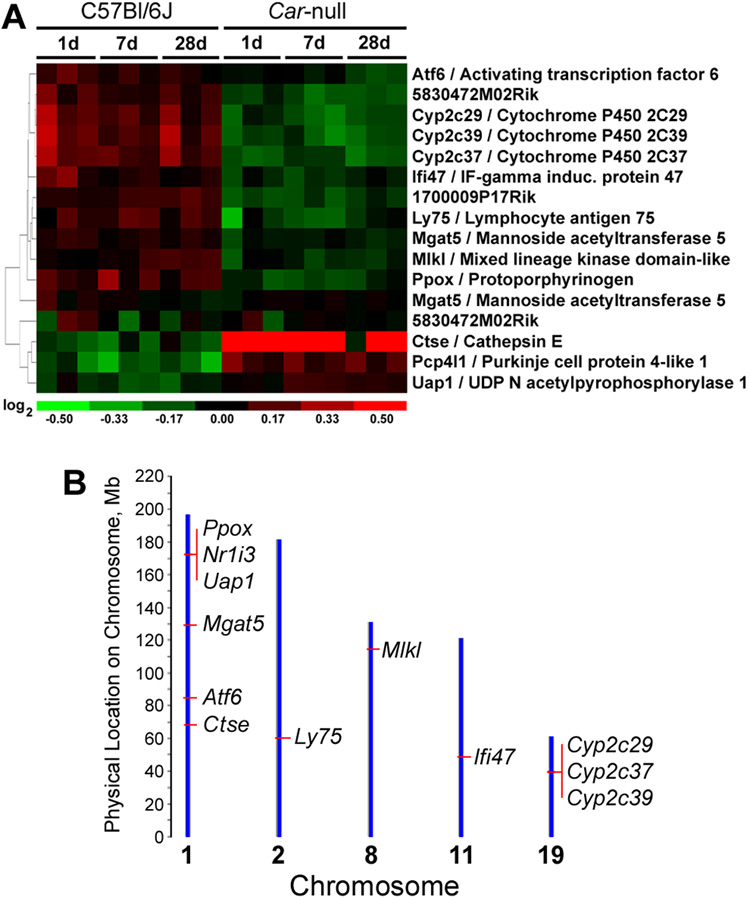
While several studies noted that no overt phenotype results from the deletion of Car in the mouse (Wei et al., 2000), to our knowledge the identification of genes that are dysregulated in mouse liver when Car is lacking has not been reported. Here, we compared gene expression in livers of control wild-type C57BL/6J animals to Car-nulls on C57BL/6J background by using a two-class SAM analysis across all 3 time-points. We found that 14 genes are consistently differentially expressed between wild-type and null animals (Figure 1A). Interestingly, several of these are located on the same chromosome as the Car gene, Nr1i3 (chromosome 1, Figure 1B). The Car-null mice used in this study were originally developed on an Sv129 background (Ueda et al., 2002) and back-crossed to the C57Bl/6J strain for 5 times to achieve accepted levels of congenicity. In constructing a knockout, there is some carryover between the gene target when back-crossing to the desired genetic background, and this does not usually have an effect on major innate differences in the wild-type versus knockout (Bolivar et al., 2001). However, since the congenic interval surrounding Car carries DNA from the 129 strain, which would differ from the Car-wildtype (i.e., C57Bl/6J) DNA, expression differences detected here (Figure 1A) may be, in part, influenced by the genetic regulation of liver gene expression networks between two strains (Gatti et al., 2007). To address this possibility we mapped these genes to the mouse genome (Figure 1B), as well as compared their liver gene expression between control C57Bl/6J and 129X1/SvJ mice. Indeed, UDP N-acetylglucosamine pyrophosphorylase 1 (Uap1), protoporphyrinogen oxidase (Ppox) and mannoside acetyl glucosaminyl transferase 5 (Mgat5) are located in close proximity to the Car (Nr1i3) locus (Figure 1B, Supplemental Figure 1). In addition, the differences in liver expression of these two genes between wild type and Car-null mice (Figure 1A) are also observed between naive C57Bl/6J and 129X1/SvJ mice (i.e., expression levels in the 129X1/SvJ mice is similar to the Car-nulls; data not shown). Thus, we conclude that they are differentially expressed due to the congenic interval, not Car.
Figure 1. Expression of 14 genes differs consistently in liver of C57Bl/6J and Car-null mice.
An unpaired, two-class comparison using Significance Analysis of Microarrays (SAM) was conducted to determine differences in gene expression between control C57Bl/6J (wild-type) and Car-null mice across all time points. (A) Hierarchical clustering (green-colored genes are down-regulated and red-colored genes are up-regulated as compared to time-matched control samples) and (B) chromosomal location (mouse genome build 36) of the differentially expressed genes are shown.
Other genes that were consistently changed in association with Car deletion include a number of cytochrome P450s (Cyp2c29, Cyp2c39, Cyp2c37), interferon-gamma inducible protein 47 (Ifi47), lymphocyte antigen 75 (Ly75), activating transcription factor 6 (Atf6), cathepsin E (Ctse), Purkinje cell protein 4-like (Pcp4l1), and mixed lineage kinase domain-like (Mlkl). These genes are located either distantly to Car locus on chromosome 1 or on other chromosomes, and their expression does not show a distinct pattern of difference between C57Bl/6J or 129X1/SvJ strains (data not shown). Both Cyp2c37 and Cyp2c29 have been shown to be directly regulated by Car upon exposure to phenytoin, a phenobarbital-like compound (Jackson et al., 2004; Jackson et al., 2006). Cyp2c39 is a liver-specific cytochrome P450 that is involved in the metabolism of retinoic acid (Andreola et al., 2004), and thus also is likely to be responsive to changes in Car levels in liver. Other genes have been scarcely studied and their expression in liver may be affected by Car-deficiency in a yet unknown way. More research is needed to identify the significance of the linkages between expression of these genes and Car.
Time-course of Car-specific transcriptional response to phenobarbital in mouse liver
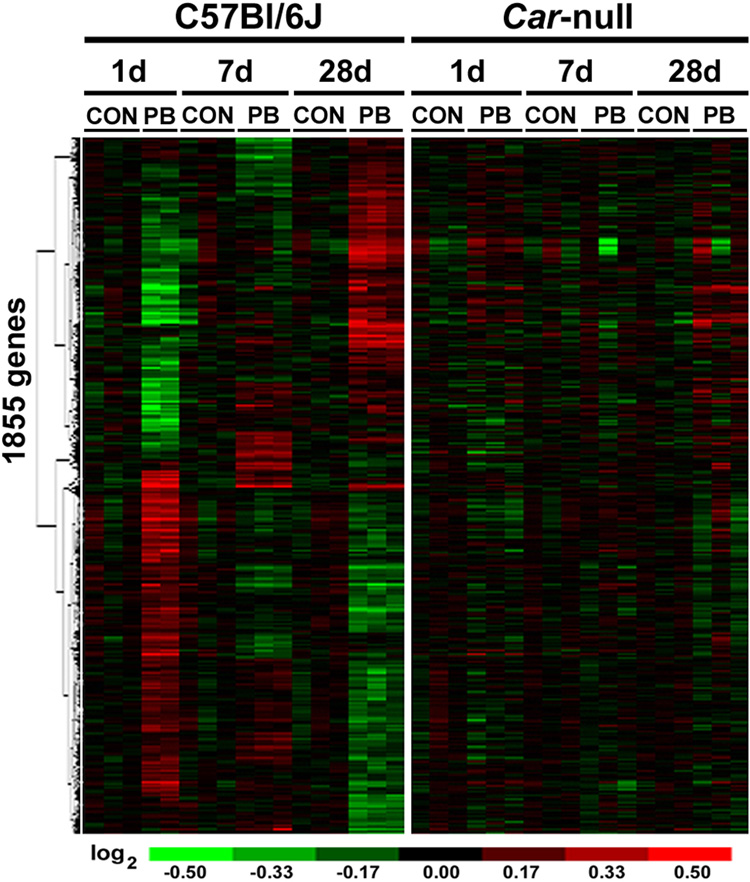
Even though several previous studies have reported differential gene expression in response to phenobarbital in wild type and Car-null mice at a single time-point, we sought to categorize liver genes changing over a time-course at early, intermediate, and subchronic time-points. Extraction of Differential Gene Expression (EDGE) algorithm was used to identify genes that varied in response to both treatment and time. There are 1855 genes (Supplemental Table 2) which are differentially expressed in response to phenobarbital treatment in liver of C57Bl/6J mice over 1, 7, and 28 days. Hierarchical clustering of these genes revealed clear patterns of expression changes across time points (Figure 2, left panel). When these genes were visualized in the Car-null data set and ordered in the same hierarchy, no distinct patterns were evident (Figure 2, right panel). Separate EDGE analysis on the Car-null data set alone produced 16 differentially expressed genes in response to phenobarbital treatment; however, no biological pathways were identified from this gene set (data not shown).
Figure 2. Time course- and Car-dependent gene expression changes in mouse liver caused by phenobarbital.
Gene expression data from Car-null and C57Bl/6J wild type mice treated with phenobarbital (PB) for 1 day (100 mg/kg, i.g.), or 7 and 28 days (both 850 ppm, diet) was used to identify transcripts that were differentially expressed in a time-dependent manner as detailed in Materials and Methods. The analysis produced a list of 1855 genes which are shown as the heat map following hierarchical clustering.
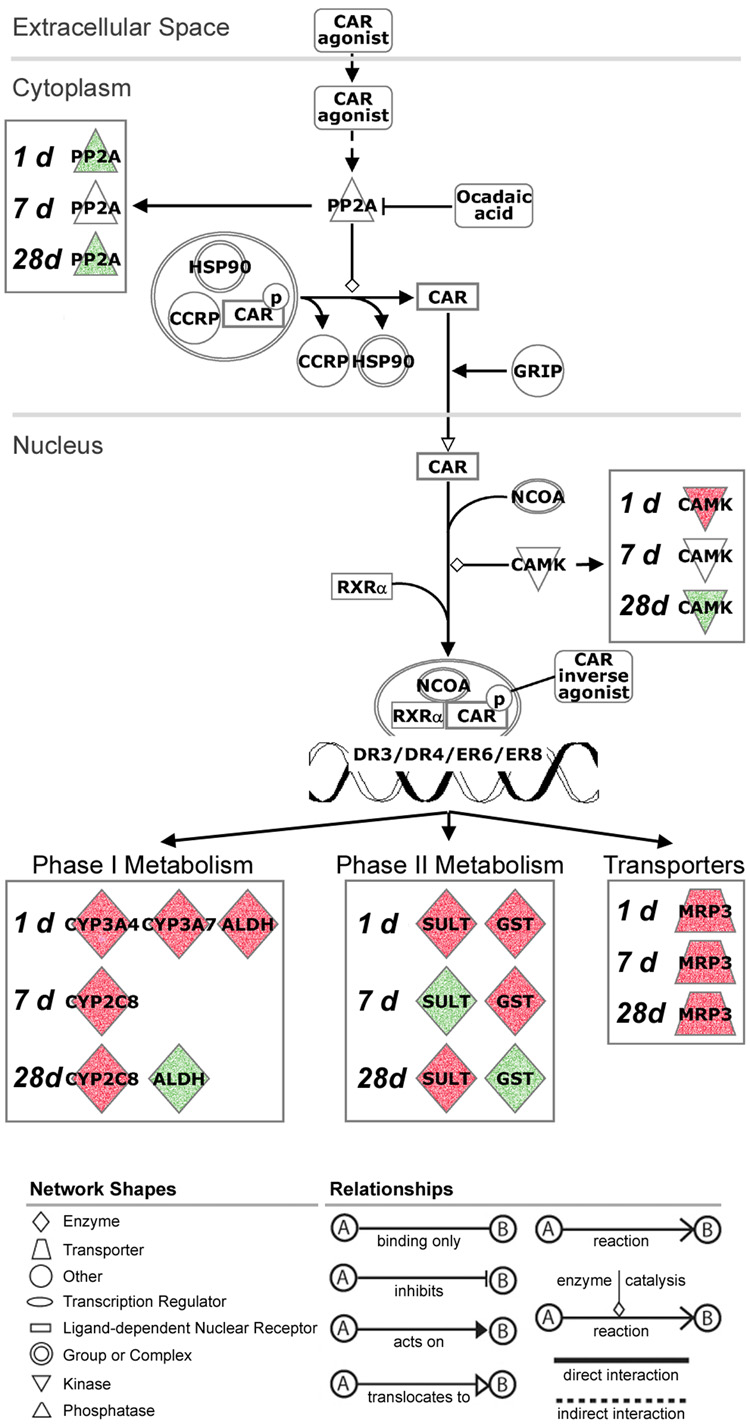
To visualize phenobarbital-related temporal changes in gene expression, a canonical pathway map for the activation of CAR (Ingenuity Pathways Analysis) was used (Figure 3). Various isoforms of CYPs were induced at early versus later time-points with phenobarbital treatment. Cyp3a4 and 7 were upregulated at 1 day, but at 7 and 28 days there was an upregulation of Cyp2c8. Glutathione-s-transferase was upregulated at 1 and 7 days, but downregulated at 28 days, possibly returning to steady state. The transporter multi-drug resistance associated protein 3 (Mrp3, Acc3) was upregulated continually over the time-course. While many studies have suggested that phenobarbital-like compounds may elicit both Car- and Pxr-dependent events in mouse liver (Wei et al., 2002; Maglich et al., 2002), especially early after treatment, our observations suggest Car-specific signature in response to phenobarbital in mouse liver predominates over the sub-chronic time-course.
Figure 3. Time course-dependent changes in Car-agonist-induced xenobiotic metabolism pathway in response to phenobarbital.
Significant genes from EDGE analysis (see Figure 2) were used to visualize phenobarbital-induced temporal changes in the pathway (Ingenuity Pathways Analysis) for Car-mediated xenobiotic metabolism in mouse liver. Biological function and relationships among gene products are identified at the bottom of the figure. Nodes shaded green are down-regulated and red nodes are up-regulated genes as compared to time-matched control samples. Abbreviations: PP2A, serine/threonine protein phosphatase 2A; HSP90, heat shock protein 90; CCRP, DnaJ (Hsp40) homolog, subfamily C, member 7; GRIP, glutamate receptor interacting protein 1; NCOA, nuclear receptor coactivator; CAMK, calcium/calmodulin-dependent protein kinase; RXR-alpha, retinoid x receptor-alpha; CYP3A4/CYP3A7/CYP2C8, cytochrome P450 isoforms; ALDH, aldehyde dehydrogenase; SULT, sulfotransferase; GST, glutathione-s-transferase; MRP3, ATP-binding cassette, sub-family C (CFTR/MRP), member 3.
Comparison of liver gene expression time-course in response to phenobarbital and WY-14,643
It is well established that a number of non-genotoxic rodent liver carcinogens, including phenobarbital-like compounds and peroxisome proliferators, modulate a series of comparable molecular events in liver (Klaunig et al., 1998; Isenberg et al., 2001). The mode of action of these agents is thought to include activation of nuclear receptors with induction of metabolizing and other immediate response genes, secondary oxidative stress, cell proliferation, etc. Studies that discriminate gene expression responses that are nuclear receptor-mediated from those that are not by using mice that lack corresponding receptors have significantly advanced our understanding of the mechanisms of action of many xenobiotics (Cherkaoui-Malki et al., 2001; Wei et al., 2002). In addition, comparative analysis of gene expression data between classes of chemicals has been a valuable and active area of research in toxicology (Hamadeh et al., 2002; Fielden et al., 2007). However, there is limited data comparing time-course sensitive changes in liver gene expression between classes of chemicals which may be important to understand the timing of key events in the mode of action. We have recently reported on the time-course investigation of PPARα- and Kupffer cell-dependent effects of a model peroxisome proliferator compound WY-14,643 in mouse liver (Woods et al., 2007b). Here, we sought to compare responses to phenobarbital and WY-14,643 by closely matching the strain of mice, timing, and doses of exposure.
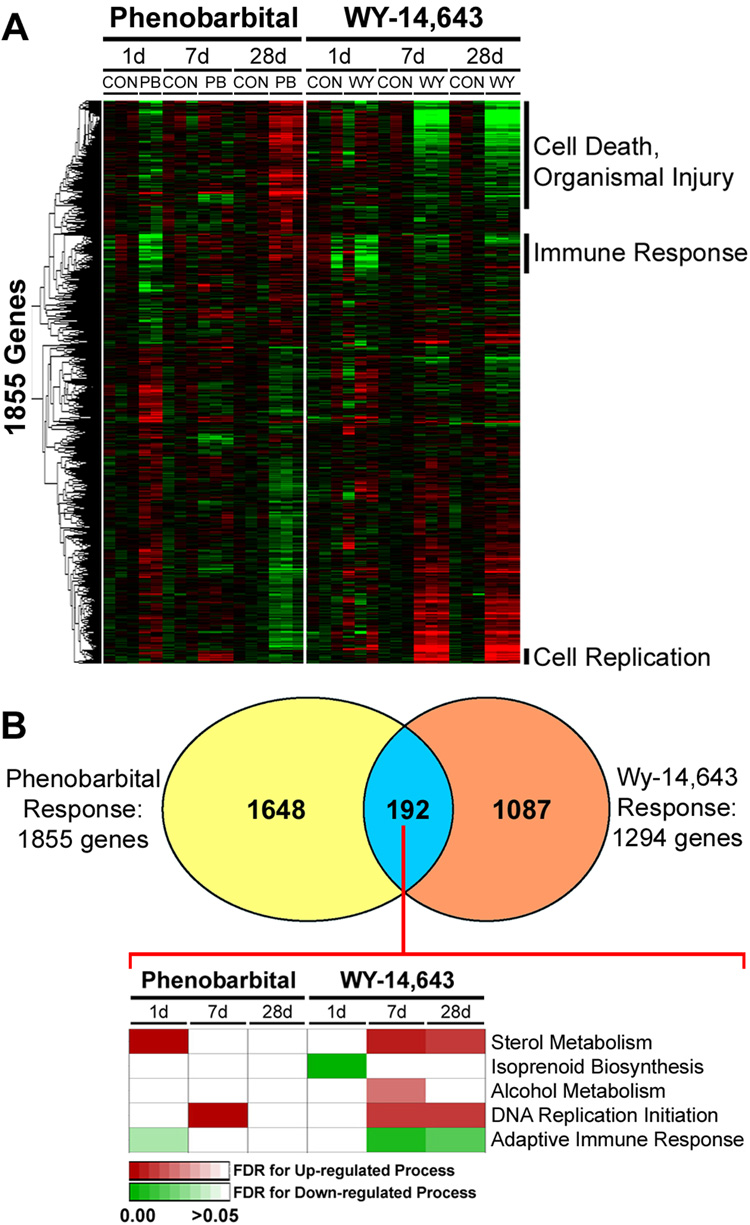
Since the two sets of expression profiling experiments were performed at different times, Distance Weighted Discrimination (DWD) batch correction procedure (Benito et al., 2004) was used to normalize the microarray data sets. The phenobarbital response based on time-course analysis of the significant genes in liver of C57Bl/6J mice includes 1855 genes (Figure 2). These genes were identified in the WY-14,643 data set (Woods et al., 2007b) and ordered in the same hierarchy (Figure 4A). Several clusters of genes exhibited time-dependent changes in expression for both compounds. Interestingly, genes in the cell death and organismal injury, and immune response pathways were time-responsive with both phenobarbital (28 days) and WY-14,643 (7 and 28 days), albeit changing in opposite directions. Gene expression in cell replication pathway shifted in a similar manner for both compounds, especially at day 7 of treatment.
Figure 4. Comparison of liver gene expression time-course in response to phenobarbital or WY-14,643.
Gene expression data from C57Bl/6J mice treated with phenobarbital (see Figure 2) or WY-14,643 (1 day, 50 mg/kg, i.g.; or 7 and 28 days, both 500 ppm, diet) was normalized and used for comparison across treatments. (A) Significant genes identified by EDGE analysis of the phenobarbital-induced temporal changes (see Figure 2) were visualized in both data sets. Gene clusters with similar temporal patterns in expression were selected to identify enriched (based on false discovery rate statistics, FDR<0.05) biological processes. (B) Common genes which exhibited statistically significant expression changes in by both the phenobarbital (this study) and WY-14,643 (Woods et al., 2007b) time-course study (192 genes) were used to determine concordance of changes in enriched biological processes. Processes that were down- (green) and up- (red) regulated are displayed in a heat map where shading reflects the significance (FDR) of a given pathway.
We also sought to identify a common gene expression signature between phenobarbital and WY-14,643 in two ways: on the level of the individual genes and on the level of biological pathways. When time-sensitive response to phenobarbital (1855 genes) and WY-14,643 (1294 genes) are compared for overlap, 192 genes (Supplemental Table 3) are shared (Figure 4B). Surprisingly, there is little concordance in the temporal profile of these common genes and the biological processes in which they are involved (Figure 4B), reflected by the fact that phenobarbital causes a much earlier activation of these pathways than WY-14,643. This can be said for almost all of the genes whose expression is time-dependently altered by phenobarbital (Figure 4A), where significantly more changes in expression were observed at 1 day, compared to WY-14,643. Genes associated with DNA replication show some overlap in the temporal expression profile (Figure 4A) at 7 days, with induction as early as 1 day with phenobarbital, but not sustained beyond 7 days. WY-14,643 shows a much later induction of DNA replication which is sustained with sub-chronic treatment (Woods et al., 2007b).
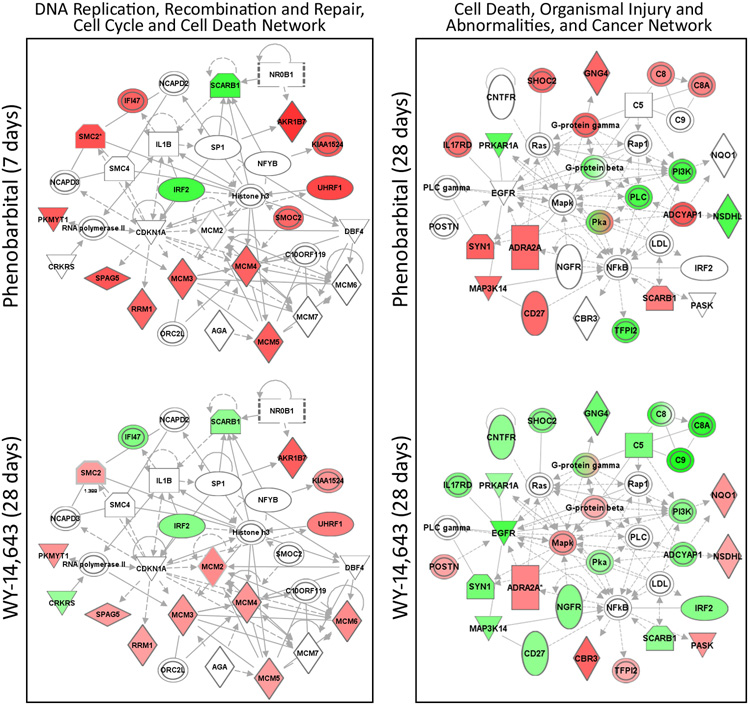
Next we visualized transcriptional changes in response to phenobarbital and WY-14,643 on the level of the biological processes (Figure 5). A number of cell proliferation-related processes were affected similarly by both phenobarbital and WY-14,643 with greatest concordance in responses at 7 days of treatment with phenobarbital, and 7 and 28 days of treatment with WY-14,643. Indeed, the most significant “network” from phenobarbital-response genes produced with Ingenuity Pathways Analysis was the DNA Replication, Recombination, and Repair, Cell Cycle, Cell Death network (Figure 6, left panel; Supplemental Figure 2). Genes in this network showed a nearly identical pattern of changes in response to phenobarbital at 7 days and WY-14,643 at 28 days. Specifically, minichromosome maintenance genes (MCMs) were upregulated in both sets, and these genes are important in DNA replication and cell cycle. Ubiquitin-like, containing PHD and RING finger domains, 1 (Uhrf1) is also upregulated in both sets and is a cell cycle checkpoint gene necessary for liver growth (Sadler et al., 2007). These gene expression data are consistent with phenotypic changes in liver cell proliferation which shows an elevated response at 7 days for phenobarbital (Table 1) and both 7 and 28 days for WY-14,643 (Woods et al., 2007b).
Figure 5. Time- and chemical-dependent changes in biological pathways affected by phenobarbital and WY-14,643 in mouse liver.
Significant genes from EDGE analysis (see Figure 2 for phenobarbital and (Woods et al., 2007b) for WY-14,643) were used to identify enriched biological processes that were enriched in response to each treatment. Supervised hierarchical clustering was performed to organize treatments and chemicals (columns) and biological processes (rows). Processes that were down- (green) and up- (red) regulated are visualized with a heat map where shading reflects the significance (FDR) of enrichment for a given pathway. The pathways are ordered within each category according to Gene Ontology hierarchy.
Figure 6. Comparison of molecular regulatory networks affected in mouse liver by phenobarbital and WY-14,643.
Expression of genes contributing to the DNA Replication, Recombination, and Repair, Cell Cycle, Cell Death Network (left) was visualized using Ingenuity Pathways Analysis software at 7 days of phenobarbital and 28 days of WY-14,643 treatment. Expression of genes contributing to the Cell Death, Organismal Injury and Abnormalities, and Cancer Network (right) is shown at 28 days of phenobarbital or WY treatment. Nodes shaded green are down-regulated and red nodes are up-regulated genes (shades are representative of FDR) as compared to time-matched controls, and white nodes are molecular mediators that were either not measured on the array or did not contribute to this analysis. See Figure 3 for the identification of biological function and relationships among gene products. Abbreviations: NR0B1, nuclear receptor subfamily 0, group B, member 1 (DAX-1); SCARB1, scavenger receptor class B, member 1; NCAPD2, non-SMC condensin I complex, subunit D2; IFI47, interferon gamma inducible protein 47SMC2; IL1B, interleukin 1, beta ; SP1, Sp1 transcription factor ; AKR1B7, aldo-keto reductase family 1, member B7; SMC4, structural maintenance of chromosomes 4; IRF2, interferon regulatory factor 2; NFYB, nuclear transcription factor Y, beta; NCAPD3, non-SMC condensin II complex, subunit D3; UHRF1, ubiquitin-like, containing PHD and RING finger domains, 1; PKMYT1, protein kinase, membrane associated tyrosine/threonine 1; CDKN1A, cyclin-dependent kinase inhibitor 1A (p21, Cip1); MCM2, minichromosome maintenance complex component 2; SMOC2, SPARC related modular calcium binding 2; C10ORF119, chromosome 10 open reading frame 119; MCM minichromosome maintenance complex component; SPAG5, sperm associated antigen 5; CRKRS, Cdc2-related kinase, arginine/serine-rich; RRM1, ribonucleotide reductase M1; ORC2L, origin recognition complex, subunit 2-like (yeast); AGA, aspartylglucosaminidase; MCM, minichromosome maintenance complex component; CNFTR, ciliary neurotrophic factor receptor; SHOC2, soc-2 suppressor of clear homolog; GNG4, guanine nucleotide binding protein, gamma 4; C, complement component; PRKAR1A, protein kinase, cAMP-dependent, regulatory, type I, alpha; IL17RD, interleukin 17 receptor D; PLC, phospholipase C; EGFR, epidermal growth factor receptor; Mapk, mitogen activated protein kinase; PI3K, pi3-kinase; NQO1, NAD(P)H dehydrogenase, quinone 1; NSDHL, NAD(P) dependent steroid dehydrogenase-like; TFPI2, tissue factor pathway inhibitor 2; LDL, low density lipoprotein complex.
However although it was expected that there would be similar pathway induction at later time-points for both phenobarbital and WY-14,643, the timing of changes in a number of pathways, including metabolism, immune response and other biological processes differed greatly between treatments (Figure 5). For example, a biphasic temporal response to sterol, lipid, and cholesterol biosynthetic processes was observed in PB treated groups where there was an upregulation at 1 day followed by down-regulation at 28 days. In contrast, lipid metabolic processes and fatty acid metabolism were only upregulated following WY treatment at 7 and 28 days. Suppression of PPARα pathway, which is thought to be a common effect of CAR activation, could contribute to the opposing lipid metabolism gene expression profiles exhibited by phenobarbital and WY-14,643 (Woods et al., 2007a).
Immune response pathways were similarly downregulated in both treatments, but the time-course was different. Complement activation was down-regulated at 1day of PB treatment whereas down-regulation of this pathway in response to WY was not observed until 7 days and beyond. Similarly, one of the top networks that showed an opposing pattern between treatments at 28 days was the Cell Death, Organismal Injury and Abnormalities, and Cancer network (Figure 6, right panel; Supplemental Figure 3). Many of the genes in this network (e.g., CD27, complement, and interleukin genes) were up-regulated in response to phenobarbital whereas they were mostly down-regulated in response to WY-14,643. This difference may be indicative of a stronger suppression of the inflammatory responses in the WY-14,643-treated animals.
Conclusions
The goal of this study was to identify biological pathways affected in mouse liver by the prototypical nuclear receptor-activators: phenobarbital and WY-14,643, over a time-course by means of gene expression profiling. We hypothesized that initial transcriptional responses to these agents would be different because two different nuclear receptors are activated (CAR versus PPARα), but at later time-points the pathways would converge because the overall modes of action of the two chemicals are similar. While we found that there were parallels in the cellular proliferation pathways that were activated, the temporal profile of activation was somewhat different. Most strikingly, there was great divergence in several metabolism pathways and immune response, suggesting that unique molecular signatures persist despite similarities in the modes of action of these non-genotoxic rodent liver carcinogens.
The pathway mapping is important to examine general changes in gene expression data, as opposed to changes in individual transcripts. Indeed, the approach of analyzing the vast amount of toxicogenomics data using molecular pathway and networks analysis tools in combination with analysis of reference data can increase the understanding of the molecular mechanisms that lead to chemical-induced toxicity and greatly facilitate the application of this knowledge to risk assessment (Ganter et al., 2008). In addition, we argue for the significance of a time-course consideration in toxicogenomics-assisted risk assessment since responses to xenobiotics are highly dynamic and often include transient processes. Thus, assaying gene expression at a single time-point, the approach frequently utilized in toxicogenomic studies, may not be an accurate portrayal of the molecular events. Furthermore, while many studies have reported on gene expression “fingerprints” as a way to classify chemicals which have the ability to cause cancer through various mechanisms (Fielden et al., 2007), our studies indicate that “binning” chemicals based on short-term treatment profiling may not be uniformly predictive because a convergence of molecular events even with sub-chronic treatment may be lacking.
In addition, assessment of the similarities and differences in the dynamic changes over a time-course in response to distinct classes of toxicants provides a valuable resource for (i) determining the effect of a given chemical and its point(s) of interaction with the cell’s gene regulatory and signaling network, (ii) extrapolation of this information across species to determine the relevance of results in different systems, and (iii) analysis of changes in regulatory network structure under different toxicant stimuli. However, development and application of data-driven methods for the inference and high-level modeling of regulatory networks responsive to chemical perturbation is hampered by scarcity of fine-resolution (time and dose) data from toxicological experiments. Thus, to begin the construction of regulatory networks relevant to chemical exposure, integration of gene expression data with a variety of other resources (e.g., transcription factor binding motifs, physical protein-protein/protein-DNA interactions, and other relevant data types) is needed in order to generate as comprehensive a picture as possible of the molecular networks involved in the mechanisms of action of toxicants.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Fred Wright, David Threadgill and Chang Fan, and Ms. Alison Hege with the University of North Carolina-Chapel Hill for valuable assistance with data analysis and interpretation. We wish to thank Mr. Jeffrey Davis at ILS, Research Triangle Park, NC for preparing the diet used in this study. Financial support for these studies was provided, in part, by grants from NIH (ES12686, ES10126 and ES11391) and EPA (RD832720 and RD833825), and by the Intramural Research program of the National Institute of Environmental Health Sciences. The content of the manuscript does not reflect the views and policies of the EPA.
REFERENCES
- Andreola F, Hayhurst GP, Luo G, Ferguson SS, Gonzalez FJ, Goldstein JA, De Luca LM. Mouse liver CYP2C39 is a novel retinoic acid 4-hydroxylase. Its down-regulation offers a molecular basis for liver retinoid accumulation and fibrosis in aryl hydrocarbon receptor-null mice. J Biol Chem. 2004;279:3434–3438. doi: 10.1074/jbc.M305832200. [DOI] [PubMed] [Google Scholar]
- Benito M, Parker J, Du Q, Wu J, Xiang D, Perou CM, Marron JS. Adjustment of systematic microarray data biases. Bioinformatics. 2004;20:105–114. doi: 10.1093/bioinformatics/btg385. [DOI] [PubMed] [Google Scholar]
- Bolivar VJ, Cook MN, Flaherty L. Mapping of quantitative trait loci with knockout/congenic strains. Genome Res. 2001;11:1549–1552. doi: 10.1101/gr.194001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkaoui-Malki M, Meyer K, Cao WQ, Latruffe N, Yeldandi AV, Rao MS, Bradfield CA, Reddy JK. Identification of novel peroxisome proliferator-activated receptor alpha (PPARalpha) target genes in mouse liver using cDNA microarray analysis. Gene Expr. 2001;9:291–304. doi: 10.3727/000000001783992533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielden MR, Brennan R, Gollub J. A gene expression biomarker provides early prediction and mechanistic assessment of hepatic tumor induction by nongenotoxic chemicals. Toxicol Sci. 2007;99:90–100. doi: 10.1093/toxsci/kfm156. [DOI] [PubMed] [Google Scholar]
- Ganter B, Zidek N, Hewitt PR, Muller D, Vladimirova A. Pathway analysis tools and toxicogenomics reference databases for risk assessment. Pharmacogenomics. 2008;9:35–54. doi: 10.2217/14622416.9.1.35. [DOI] [PubMed] [Google Scholar]
- Gatti D, Maki A, Chesler EJ, Kirova R, Kosyk O, Lu L, Manly KF, Williams RW, Perkins A, Langston MA, Threadgill DW, Rusyn I. Genome-level analysis of genetic regulation of liver gene expression networks. Hepatology. 2007;46:548–557. doi: 10.1002/hep.21682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez FJ, Shah YM. PPARalpha: Mechanism of species differences and hepatocarcinogenesis of peroxisome proliferators. Toxicology. 2008;246:2–8. doi: 10.1016/j.tox.2007.09.030. [DOI] [PubMed] [Google Scholar]
- Hamadeh HK, Bushel PR, Jayadev S, Martin K, DiSorbo O, Sieber S, Bennett L, Tennant R, Stoll R, Barrett JC, Blanchard K, Paules RS, Afshari CA. Gene expression analysis reveals chemical-specific profiles. Toxicol Sci. 2002;67:219–231. doi: 10.1093/toxsci/67.2.219. [DOI] [PubMed] [Google Scholar]
- Honkakoski P, Zelko I, Sueyoshi T, Negishi M. The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Mol. Cell Biol. 1998;18:5652–5658. doi: 10.1128/mcb.18.10.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenberg JS, Kamendulis LM, Ackley DC, Smith JH, Pugh G, Jr., Lington AW, McKee RH, Klaunig JE. Reversibility and persistence of di-2-ethylhexyl phthalate (DEHP)- and phenobarbital-induced hepatocellular changes in rodents. Toxicol. Sci. 2001;64:192–199. doi: 10.1093/toxsci/64.2.192. [DOI] [PubMed] [Google Scholar]
- Jackson JP, Ferguson SS, Moore R, Negishi M, Goldstein JA. The constitutive active/androstane receptor regulates phenytoin induction of Cyp2c29. Mol Pharmacol. 2004;65:1397–1404. doi: 10.1124/mol.65.6.1397. [DOI] [PubMed] [Google Scholar]
- Jackson JP, Ferguson SS, Negishi M, Goldstein JA. Phenytoin induction of the cyp2c37 gene is mediated by the constitutive androstane receptor. Drug Metab Dispos. 2006;34:2003–2010. doi: 10.1124/dmd.106.012005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizaki S, Yamamoto Y, Ueda A, Moore R, Sueyoshi T, Negishi M. Phenobarbital induction of drug/steroid-metabolizing enzymes and nuclear receptor CAR. Biochim. Biophys. Acta. 2003;1619:239–242. doi: 10.1016/s0304-4165(02)00482-8. [DOI] [PubMed] [Google Scholar]
- Klaunig JE, Xu Y, Isenberg JS, Bachowski S, Kolaja KL, Jiang J, Stevenson DE, Walborg EF., Jr. The role of oxidative stress in chemical carcinogenesis. Environ. Health Perspect. 1998;106 Suppl 1:289–295. doi: 10.1289/ehp.98106s1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalwani ND, Reddy MK, Qureshi SA, Reddy JK. Development of hepatocellular carcinomas and increased peroxisomal fatty acid beta-oxidation in rats fed [4-chloro-6-(2,3- xylidino)-2-pyrimidinylthio] acetic acid (Wy-14,643) in the semipurified diet. Carcinogenesis. 1981;2:645–650. doi: 10.1093/carcin/2.7.645. [DOI] [PubMed] [Google Scholar]
- Leek JT, Monsen E, Dabney AR, Storey JD. EDGE: extraction and analysis of differential gene expression. Bioinformatics. 2006;22:507–508. doi: 10.1093/bioinformatics/btk005. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, Kliewer SA. Nuclear pregnane x receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol. Pharmacol. 2002;62:638–646. doi: 10.1124/mol.62.3.638. [DOI] [PubMed] [Google Scholar]
- Nuclear Receptors Nomenclature Committee. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- Palmer CN, Hsu MH, Griffin KJ, Raucy JL, Johnson EF. Peroxisome proliferators activated receptor-alpha expression in human liver. Mol. Pharmacol. 1998;53:14–22. [PubMed] [Google Scholar]
- Peffer RC, Moggs JG, Pastoor T, Currie RA, Wright J, Milburn G, Waechter F, Rusyn I. Mouse liver effects of cyproconazole, a triazole fungicide: role of the constitutive androstane receptor. Toxicol Sci. 2007;99:315–325. doi: 10.1093/toxsci/kfm154. [DOI] [PubMed] [Google Scholar]
- Peters JM, Cheung C, Gonzalez FJ. Peroxisome proliferator-activated receptor-alpha and liver cancer: where do we stand? J Mol. Med. 2005;83:774–785. doi: 10.1007/s00109-005-0678-9. [DOI] [PubMed] [Google Scholar]
- Rossi L, Ravera M, Repetti G, Santi L. Long-term administration of DDT or phenobarbital-Na in Wistar rats. Int J Cancer. 1977;19:179–185. doi: 10.1002/ijc.2910190207. [DOI] [PubMed] [Google Scholar]
- Rusyn I, Peters JM, Cunningham ML. Modes of action and species-specific effects of di-(2-ethylhexyl)phthalate in the liver. Crit Rev. Toxicol. 2006;36:459–479. doi: 10.1080/10408440600779065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler KC, Krahn KN, Gaur NA, Ukomadu C. Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, uhrf1. Proc. Natl. Acad Sci U. S. A. 2007;104:1570–1575. doi: 10.1073/pnas.0610774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe E, Walker AI. The toxicology of dieldrin (HEOD). II. Comparative long-term oral toxicity studies in mice with dieldrin, DDT, phenobarbitone, -BHC and -BHC. Food Cosmet. Toxicol. 1973;11:433–442. doi: 10.1016/0015-6264(73)90008-4. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A, Hamadeh HK, Webb HK, Yamamoto Y, Sueyoshi T, Afshari CA, Lehmann JM, Negishi M. Diverse roles of the nuclear orphan receptor CAR in regulating hepatic genes in response to phenobarbital. Mol. Pharmacol. 2002;61:1–6. doi: 10.1124/mol.61.1.1. [DOI] [PubMed] [Google Scholar]
- Urquhart BL, Tirona RG, Kim RB. Nuclear receptors and the regulation of drug-metabolizing enzymes and drug transporters: implications for interindividual variability in response to drugs. J Clin Pharmacol. 2007;47:566–578. doi: 10.1177/0091270007299930. [DOI] [PubMed] [Google Scholar]
- Wei P, Zhang J, Dowhan DH, Han Y, Moore DD. Specific and overlapping functions of the nuclear hormone receptors CAR and PXR in xenobiotic response. Pharmacogenomics. J. 2002;2:117–126. doi: 10.1038/sj.tpj.6500087. [DOI] [PubMed] [Google Scholar]
- Wei P, Zhang J, Egan-Hafley M, Liang S, Moore DD. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature. 2000;407:920–923. doi: 10.1038/35038112. [DOI] [PubMed] [Google Scholar]
- Whysner J, Ross PM, Williams GM. Phenobarbital mechanistic data and risk assessment: enzyme induction, enhanced cell proliferation, and tumor promotion. Pharmacol. Ther. 1996;71:153–191. doi: 10.1016/0163-7258(96)00067-8. [DOI] [PubMed] [Google Scholar]
- Woods CG, Heuvel JP, Rusyn I. Genomic profiling in nuclear receptor-mediated toxicity. Toxicol Pathol. 2007a;35:474–494. doi: 10.1080/01926230701311351. [DOI] [PubMed] [Google Scholar]
- Woods CG, Kosyk O, Bradford BU, Ross PK, Burns AM, Cunningham ML, Qu P, Ibrahim JG, Rusyn I. Time course investigation of PPARalpha- and Kupffer cell-dependent effects of WY-14,643 in mouse liver using microarray gene expression. Toxicol Appl Pharmacol. 2007b;225:267–277. doi: 10.1016/j.taap.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Moore R, Goldsworthy TL, Negishi M, Maronpot RR. The orphan nuclear receptor constitutive active/androstane receptor is essential for liver tumor promotion by phenobarbital in mice. Cancer Res. 2004;64:7197–7200. doi: 10.1158/0008-5472.CAN-04-1459. [DOI] [PubMed] [Google Scholar]
- Zeeberg BR, Qin H, Narasimhan S, Sunshine M, Cao H, Kane DW, Reimers M, Stephens RM, Bryant D, Burt SK, Elnekave E, Hari DM, Wynn TA, Cunningham-Rundles C, Stewart DM, Nelson D, Weinstein JN. High-Throughput GoMiner, an 'industrial-strength' integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID) BMC. Bioinformatics. 2005;6:168. doi: 10.1186/1471-2105-6-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.