Abstract
We describe the construction of a high-resolution radiation hybrid (RH) map of the domestic cat genome, which includes 2,662 markers, translating to an estimated average intermarker distance of 939 kilobases (Kb). Targeted marker selection utilized the recent feline 1.9x genome assembly, concentrating on regions of low marker density on feline autosomes and the X chromosome, in addition to regions flanking interspecies chromosomal breakpoints. Average gap (breakpoint) size between cat-human ordered conserved segments is less than 900 Kb. The map was used for a fine-scale comparison of conserved syntenic blocks with the human and canine genomes. Corroborative fluorescence in situ hybridization (FISH) data were generated using 129 domestic cat BAC-clones as probes, providing independent confirmation of the long-range correctness of the map. Cross-species hybridization of BAC probes on divergent felids from the genera Profelis (serval) and Panthera (snow leopard) provides further evidence for karyotypic conservation within felids, and demonstrates the utility of such probes for future studies of chromosome evolution within the cat family and in related carnivores. The integrated map constitutes a comprehensive framework for identifying genes controlling feline phenotypes of interest, and to aid in assembly of a higher coverage feline genome sequence.
Keywords: domestic cat, radiation hybrid map, comparative mapping, FISH mapping, synteny, chromosome rearrangement, Felidae
Introduction
During the past decade, increasingly detailed genetic and physical maps of the domestic cat genome have provided tools for mapping and discerning the hereditary basis of morphological variation and genetic diseases in cats, which model phenotypes or pathologies observed in other mammals [1; 2; 3; 4]. These include mutations in functional candidate genes that control coat morphology [5; 6; 7], as well as those that are causative for monogenic diseases such as polycystic kidney disease [8], and hypertrophic cardiomyopathy [9]. The maps were also used in genome scans to identify disease genes not previously implicated in human studies at the time of the scan [10; 11], suggesting the value of multiple animal models in understanding the pathogenesis of human disease. The ability to narrow candidate regions and identify causative mutations in the feline model is currently limited by appropriate animal cohorts and by the quality of feline-human comparative gene maps.
Therefore, we have continued to improve the quality and density of domestic cat radiation hybrid maps to define the evolutionary rearrangements that distinguish the cat from other sequenced mammalian genomes, and facilitate positional reasoning in gene and mutation hunting. These maps can be a tool for both the long-range precision and quality control of genome assemblies [12], as well as studies of the dynamics of mammalian chromosome evolution [13]. For example, the feline 1.9x sequence [14] was assembled based on conserved ordered segments defined by the RH-based feline-human and feline-dog comparative maps [3]. However, a large fraction of the genome remains unassembled due primarily to the low coverage of the sequence traces [14] and secondarily to the incomplete (85%) coverage of the previous cat-human comparative maps [3]. The new map reported here provides enhanced coverage of the feline genome (96%), and has been independently validated using FISH data, and by comparison to a new genetic linkage map. This tool will aid the chromosomal assignment and ordering of scaffolds for the draft genome assembly, facilitate identification of genes controlling feline phenotypes, and provide insight into the details of chromosomal evolution that have occurred since the divergence of the cat and dog genomes from the ancestral carnivore karyotype.
Results & Discussion
A Fine-scale Cat Radiation Hybrid Gene Map
We analyzed a final set of 2,674 markers that were evaluated by formal linkage analysis. Twelve of these markers were later dropped while computing the final map in which linkage groups were computed using a two-point LOD score threshold of 9.0. At this threshold, all but five chromosomes comprised complete linkage groups. Chromosomes A1, C2, and D2 were divided into two linkage groups with gaps at the centromeres; these chromosome arms were merged in a single map. Chromosome E1 formed three RH linkage groups with two gaps: the first caused by the nuclear organizer region (NOR), and the second because of the severe changes in retention frequency associated with the RH selectable marker (TK1) that is found on this chromosome. The three RH groups on chromosome E1 were oriented using evidence from the feline linkage maps and FISH data. On chromosome X, the pseudoautosomal region (PAR) comprises a separate linkage group due to increased retention resulting from co-amplification of Y chromosome-bearing fragments in the PAR [3].
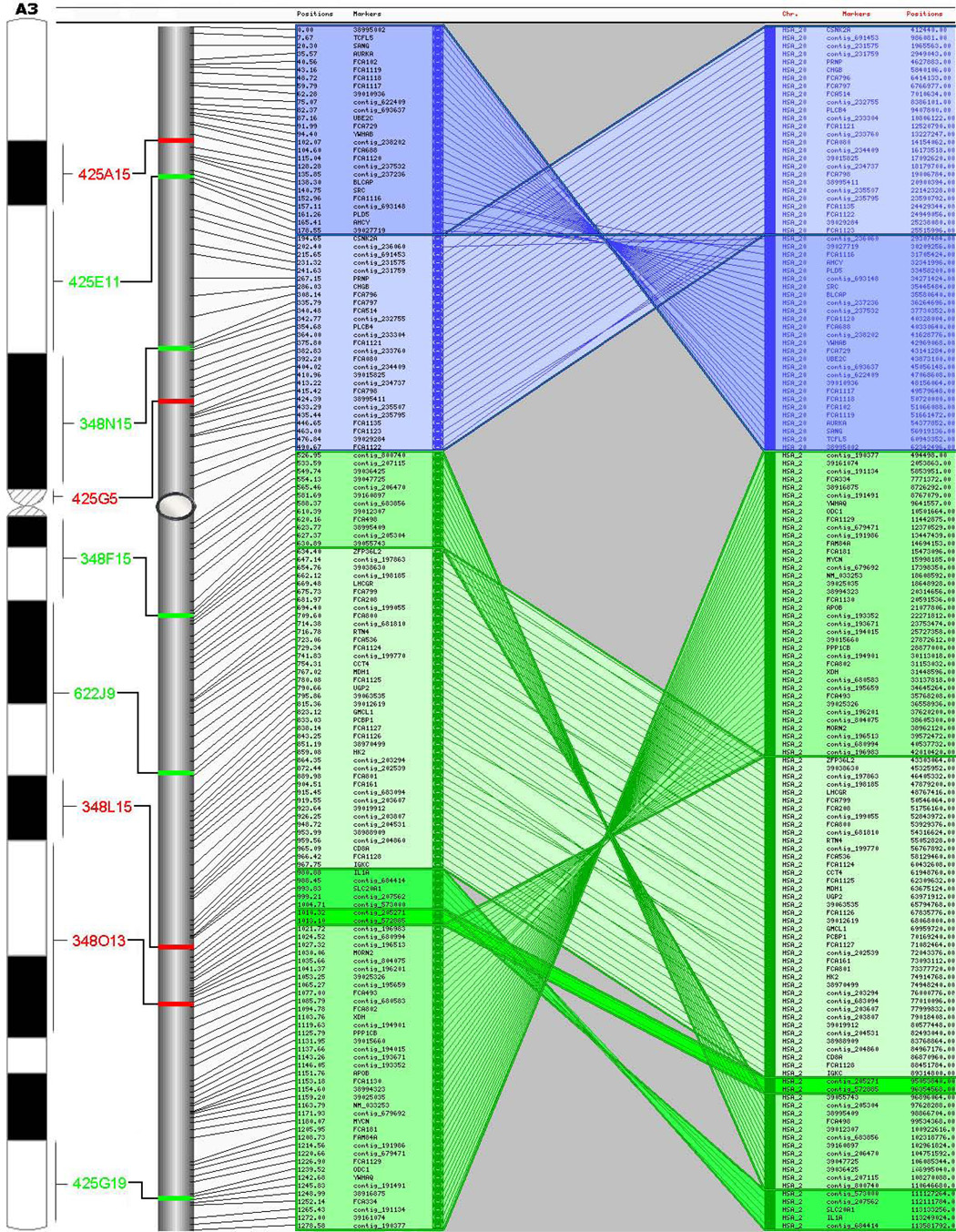
The computed RH map contains 2,662 markers that cover all 18 feline autosomes and the X chromosome, with marker density being fairly uniform across chromosomes (Table 1). The uniform marker density was achieved in part because we computed an intermediate map after 2,550 markers had been genotyped (map not shown here) and the final batch of markers to be developed were targeted to gaps in coverage of that intermediate map. We re-used some marker input data from earlier maps, but we did not assume that markers were in the same order as in previous maps. Of the 2,662 markers, 733 are associated with feline annotated genes or ESTs, 780 are microsatellites, and the remainder are derived from published STS markers or the domestic cat 1.9x genome sequence. The final map has 1,602 markers on the maximum-likelihood (MLE)-consensus framework map, 779 markers placed at centiRay (cR) positions relative to the framework map, and 281 markers assigned to larger bins with respect to the framework map. The 1,602 markers on the framework map are assigned to 1,445 distinct positions; among the 2,381 markers assigned to cR positions, there are 2,184 distinct positions. Therefore, assuming a feline euchromatic genome size of 2.5 gigabases (Gb) [14], all markers comprising the current map result in one marker every 939 kb, 82% of which reside in unique positions. The binned markers are not ordered or assigned a cR position, thus one marker is assigned a cR position every 1050kb. Figure 1 depicts the integrated RH, FISH and human-dog comparative maps for feline chromosome A3. The complete RH comparative maps and cytogenetic maps are available in supplemental material (Supplemental Table 1, Supplemental Figure 1 & Supplemental Figure 2).
Table 1.
Summary Statistics for the Domestic Cat Radiation Hybrid Map
| Domestic Cat Chromosome | Total markers on map | No. of MLE-consensus framework markers | Avg. Marker Density (Mb) | RH Length (cR5000) | Approx. Physical Lengtha (Mb) | cR/Mb | Human CSOs | Dog CSOs |
|---|---|---|---|---|---|---|---|---|
| A1 | 246 | 129 | 1.0 | 1964.4 | 246 | 8.0 | 12 | 13 |
| A2 | 205 | 127 | 0.9 | 1785.2 | 180 | 9.9 | 15 | 10 |
| A3 | 155 | 97 | 0.9 | 1278.6 | 143 | 9.0 | 7 | 4 |
| B1 | 190 | 104 | 1.0 | 1466.2 | 198 | 7.4 | 13 | 19 |
| B2 | 140 | 90 | 1.1 | 1235.3 | 148 | 8.4 | 2 | 6 |
| B3 | 144 | 96 | 1.0 | 950.1 | 143 | 6.7 | 4 | 4 |
| B4 | 148 | 88 | 0.9 | 1040.2 | 138 | 7.6 | 4 | 8 |
| C1 | 197 | 125 | 1.1 | 1626.7 | 220 | 7.4 | 3 | 12 |
| C2 | 158 | 98 | 0.9 | 1302.0 | 148 | 8.8 | 10 | 8 |
| D1 | 133 | 83 | 0.9 | 1089.7 | 123 | 8.9 | 5 | 6 |
| D2 | 105 | 58 | 1.0 | 813.0 | 103 | 7.9 | 8 | 3 |
| D3 | 114 | 80 | 0.9 | 1091.0 | 103 | 10.6 | 10 | 5 |
| D4 | 109 | 69 | 0.9 | 992.4 | 95 | 10.4 | 6 | 3 |
| E1 | 109 | 62 | 0.9 | 1200.9 | 95 | 12.6 | 6 | 4 |
| E2 | 82 | 52 | 0.9 | 552.4 | 78 | 7.1 | 2 | 4 |
| E3 | 69 | 50 | 0.9 | 637.3 | 60 | 10.6 | 7 | 2 |
| F1 | 102 | 56 | 0.7 | 556.5 | 75 | 7.4 | 10 | 5 |
| F2 | 84 | 47 | 0.9 | 608.8 | 75 | 8.1 | 1 | 3 |
| X | 172 | 91 | 0.7 | 960.3 | 128 | 7.5 | 1 | 1 |
| TOTAL | 2662 | 1602 | 0.9 | 21151.0 | 2493 | 8.5 | 126 | 121 |
Assumes a 2.5 Mbp euchromatic genome and the total cytogenetic fraction estimated for each chromosome in the domestic cat genome [Pontius et al. 2007], excluding the Y chromosome.
Figure 1.
Integrated RH, FISH, and comparative maps for feline chromosome A3. CSOs for human chromosome 20 and 2 are shown to the right of each cat chromosome map (only the map scale is shown). Inferred centromere positions are shown by gray ovals. Comparative maps represent the output from AutoGRAPH-based analyses.
Map Order Confirmation using FISH and Linkage Map Comparisons
The accuracy of the long-range marker order of the RH map was verified using two independent approaches. The first approach used FISH mapping of domestic cat bacterial artificial clones (BAC) isolated from the RPCI-86 10x library. BAC end sequencing of 480 random clones was used to identify, by computational comparison to the dog, human and cat 1.9x assemblies, a set of probes distributed relatively evenly across all autosomes and the X chromosome. The positions of 129 feline BAC clones were obtained by FISH (Figure 1, Supplemental Figure 2, Supplemental Table 2). When combined with previously published cytogenetic mapping data (see Supplemental Figure 2), the average spacing of FISH markers is approximately one marker every 18 Mb. The FISH based marker order was identical to the RH-based comparative marker order, confirming the long-range assembly of the RH maps.
A second analysis compared the RH map order with marker order derived from a domestic cat linkage map [15]. A comparison of markers mapped on both maps showed the two maps to be 94% consistent, with only 18 of 319 marker order discrepancies between the two maps (Supplemental Table 3). The vast majority of marker order differences involved flips of closely spaced, adjacent markers (Avg. distance=6.8 cM). Only 3 of 17 discrepantly ordered marker pairs were spaced greater than 10 cM apart, in addition to a single marker, FCA1028, assigned to different chromosomes that could not be resolved (Supplemental Table 3). Overall, the three combined mapping approaches validate the long-range marker order across cat chromosomes, and provide anchor points for each RH map to its respective feline chromosome.
Comparative Synteny Analysis
We identified orthologous positions for 99% of the total 2662 mapped markers in the latest dog and human genome assemblies (Supplemental Table 1). AutoGRAPH [16] was used to visualize chromosome rearrangements between cat, dog and human genomes (Figure 1, Supplemental Figure 1). Using conservative criteria to define blocks of conserved marker order (see Methods), we identified 126 conserved segments ordered (CSOs, or homologous synteny blocks--HSBs [13]) between the cat and human genomes, and 121 between the cat and dog genomes (Supplemental Table 1, Supplemental Figure 1). Though these figures include singletons, we only considered a singleton to represent a CSO if it was present in one comparison species (i.e. human or dog) and part of a multimarker stretch of conserved gene order in the other species. We interpret these singletons to represent evidence of lineage-specific rearrangements.
Comparative coverage of cat CSOs on the human genome assembly was estimated as previously described [17]. Specifically, comparative coverage is defined as the sum of the physical span of human CSOs in cat, divided by the size of the human genome after excluding centromere, telomere, and heterochromatic regions, or regions lacking any cross-species homology in multi-species alignments. Estimated comparative coverage with the human genome is 96%, an 11% increase over the previous cat-human comparative map [3]. The mean gap size is 867 kb (range=0.12–2.3 Mb), with 68% of the gaps being less than 1 Mb, and 97% less than 2 Mb.
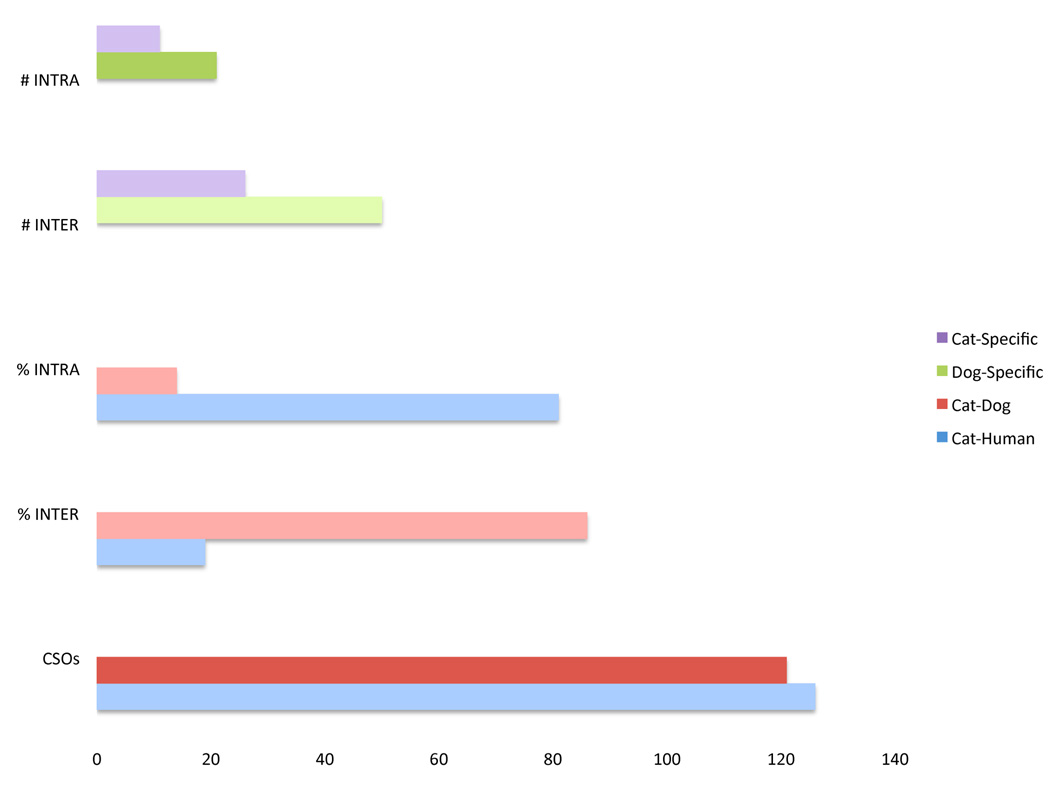
An examination of chromosome rearrangement rates between cat-human and cat-dog, as well as the lineage specific chromosomal breakpoints, is presented in Figure 2. A similar number of CSOs distinguish cat and human and cat and dog genomes (126 versus 121 respectively), despite the fact that dog and cat share a much more recent (55 million years ago [Mya] versus 95 Mya) common ancestry (Figure 2). This is accounted for by the well-documented accelerated rate of canine chromosome evolution [18; 19; 20].
Figure 2.
Lineage specific rearrangement totals for the domestic dog and domestic cat and rearrangement frequencies between the domestic cat, domestic dog, and human genomes. The lineage specific totals are #INTRA=the number of intrachromosomal and #INTER=the number of interchromosomal breakpoints. %INTRA=the percentage of rearrangements between two genomes that are the result of intrachromosomal breakpoints. %INTER=the percentage of rearrangements between two genomes that are the result of interchromosomal breakpoints.
In comparison to human and cat, the dog genome has been punctuated by a very high proportion of interchromosomal rearrangements (Figure 2), while very few blocks of cat-dog conserved synteny are further disrupted by inversions (Figure 1 & Supplemental Figure 1). Specifically, 86% of the rearrangements between cat and dog genomes involve interchromosomal breakpoints, compared to only 19% of human-cat rearrangements, which are predominately inversions (Figure 2). Further, while the relative proportion of interchromosomal to intrachromosomal rearrangements is similar in the two carnivore lineages (roughly twice as many interchromosomal rearrangements), the overall rate of breakage is approximately twice as high in the canid lineage versus the feline lineage. The refined definition of cat-dog-human chromosome breakpoints in this study will allow for a detailed comparative analysis of their sequence properties once high-coverage cat genome sequence becomes available.
Utility of FISH probes in other species of Felidae
We examined the applicability of the domestic cat FISH probes to hybridize to chromosomes of other divergent members of the cat family: serval (Profelis serval), and snow leopard (Panthera uncia). These two species are representatives of felid lineages that last shared a common ancestor with the domestic cat approximately 8 and 10 Mya, respectively, and span the earliest nodes of the felid phylogeny [21]. We tested all probes from two domestic cat chromosomes, B4 and F1, that had differences in G-banding patterns documented between at least two of the three species [22]. It has been suggested that the B4 chromosome of species of the genus Panthera (and several additional felid species) is distinguished from members of the domestic cat lineage and the serval by a pericentric inversion. In addition, domestic cat chromosome F1 is orthologous to chromosomes F3 of serval and snow leopard, but differs in its G-banding pattern, suggesting possible gene order differences between the two chromosomes.
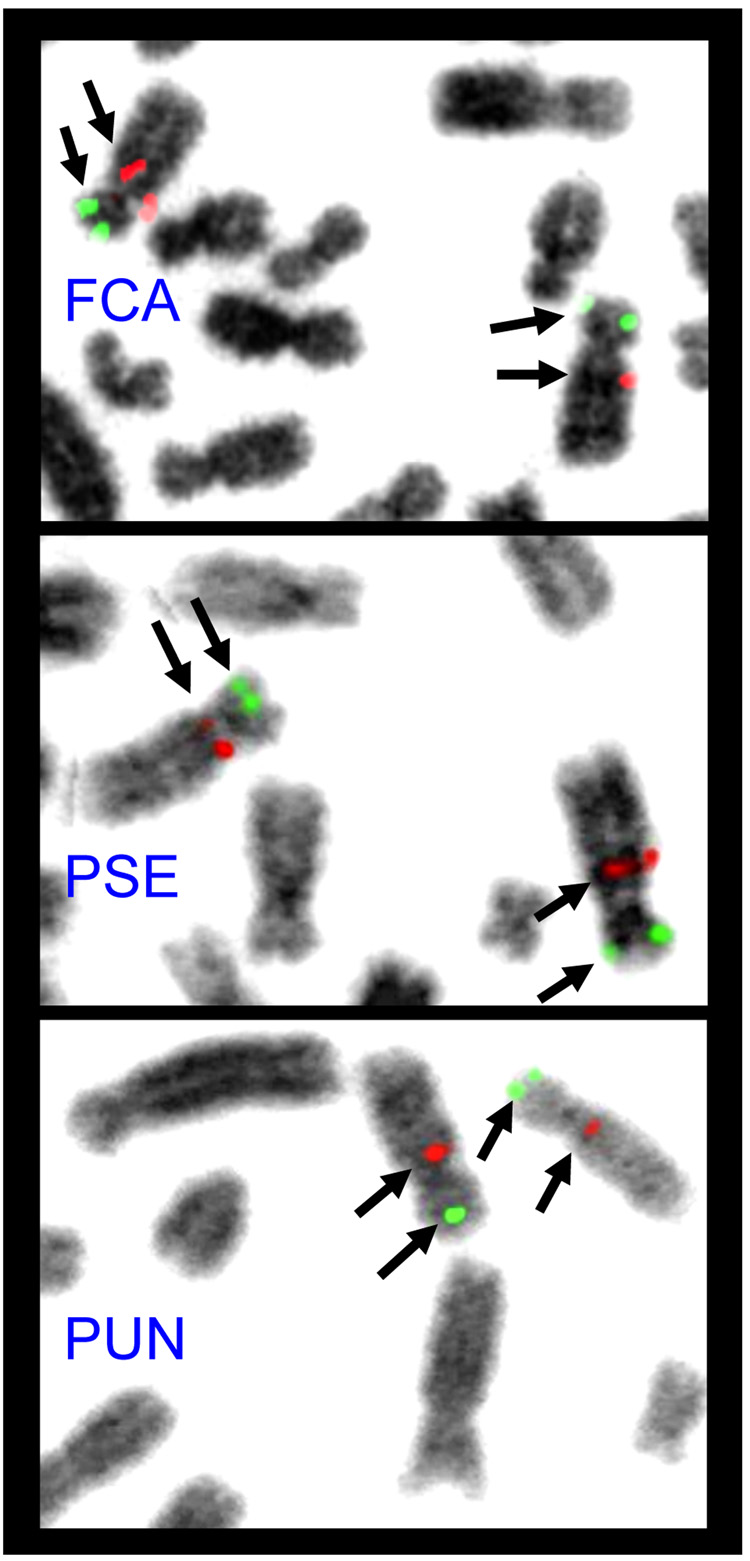
100% of domestic cat BAC probes tested (n=22 independent hybridizations) on serval and snow leopard chromosome preparations produced successful hybridization results (Figure 3). Furthermore, hybridization results suggested the marker order (as shown in Supplemental Figure 2) is conserved across all three felid species on these two chromosomes (Figure 3). Using our probes, we found no evidence of a pericentric inversion distinguishing snow leopard chromosome B4 from domestic cat or serval. Specifically, a BAC probe that hybridizes just below the centromere on domestic cat B4 localizes to the same location of the long arm, rather than the short arm (in the case of a pericentric inversion), of snow leopard chromosome B4 (Figure 3). Furthermore, the marker order was identical across all three cat species for chromosome F1 and its homologue F3. Therefore the observed G-banding differences across the felids studied may not be due to large scale structural changes (inversions), but either to smaller paracentric inversions inside of the markers we applied here, or due to compositional sequence changes between species that do not affect gene order.
Figure 3.
FISH results for two feline BAC clone probes (green: 348A7; red: 348J11) hybridized to B4 orthologous on domestic cat (Fca), serval (Pse) and snow leopard (Pun) metaphase chromosomes. Note the same relative positioning of each probe in all three species. A summary of all FISH results relative to domestic cat chromosomes can be found in Supplementary Figure 2.
Conclusions and Future Directions
We report a 939-kilobase resolution, RH-based, physical map of the feline genome. The most important result of this study is the notable improvement in the comparative genome alignments between the cat-human and cat-dog. The comparative coverage of the human genome is 96%, closing more than two-thirds of the coverage deficiency of the most recent map [3], with evolutionary breakpoints (gaps) resolved to less than 900 kilobase resolution on average. The enhanced map resolution allowed for the identification of several novel small conserved segments generated by intrachromosomal rearrangements not observed in previous maps. The high marker density of this map makes it vital to the construction of the forthcoming assembly and annotation of a higher coverage (than the currently available 1.9x) feline genome sequence. These resources will stimulate and facilitate the identification of feline genes of interest using genetic linkage and selective sweep mapping approaches in domestic and natural populations of felid species.
The high success rate of the domestic cat BAC probes on other felid species suggests they will be a reliable resource for cross-species synteny analysis in all felids. Because results were unable to detect any rearrangement that would explain the observed cytological differences between the orthologous domestic cat F1 and Serval and Panthera F3 chromosomes, these data suggest karyotypic conservatism may be more extreme across Felidae than previously appreciated, and that further application of these, and additional, probes may help resolve other documented karyotypic differences with the cat family. Human BAC probes are routinely applied in other primate lineages that span 40–50 million year divergence times [23]. Our results suggest that a large proportion of domestic cat BAC probes will successfully hybridize to chromosomes of other feliform carnivores (i.e. linsangs, hyenids, herpestids, viverrids), a group that radiated in a similar evolutionary timeframe [24].
Materials and Methods
Marker Selection
Pairs of primers were designed from sequence contigs from the 1.9x feline genome assembly [14] that had reciprocal best hits to one or both orthologous chromosomes of dog (CanFam2) and human (Hs36) genome assemblies. Repetitive sequences were masked using Repeatmasker, and primers were designed with Primer3 [25]. Each primer pair was tested by PCR in cat DNA, hamster DNA, and a 10:1 hamster:cat mixture of DNA. Only those markers showing robust amplification in cat and the 10:1 DNAs were chosen for further genotyping in the RH panel.
RH Genotyping
RH genotyping was performed on the 5,000-rad feline whole genome radiation hybrid panel using previously described methods [4]. Markers were dropped before map computation for one of the following reasons: weak amplification, high hamster background amplification, or excessively high retention frequency (>70% and not predicted to reside on the selectable locus chromosome or near a centromere) or excessively low retention frequency compared to other markers on the same chromosome. These new genotypes were merged with vectors from previous maps for map computation.
Map Construction
Two-point linkage groups were computed at a LOD score of 9.0. Markers within each chromosome or linkage group were ordered using a reduction from the problem of RH mapping to the traveling salesman problem (TSP), as implemented in the software rh_tsp_map [26; 27]. The computations to construct the map were done using programs from the software package rh_tsp_map (ftp://ftp.ncbi.nih.gov/pub/agarwala/rhmapping/rh_tsp_map.tar.gz) and using the package CONCORDE (http://www.isye.gatech.edu/~wcook/rh; [28] linked with QSopt (http://www.isye.gatech.edu/~wcook/qsopt)) to solve the TSP instances to guaranteed optimality. Details of the mapping procedures are described in the tutorial accompanying rh_tsp_map. The three linkage groups on chromosome E1, and the PAR and X-specific region on the X chromosome maps were oriented and ordered using data from linkage maps and FISH.
Before computing the final map shown here, we used similar procedures to compute an intermediate map when approximately 2,550 markers had been developed, amplified, and scored on the 5,000 rad panel. The intermediate map was used for two different purposes that both led to a much-improved final map. First, for any clone and triplet of (apparently) consecutive markers in which the retention patterns showed two obligate breaks, the problematic marker/clone pairs were reevaluated using the strategy described in [17]. This substantial effort in local quality control explains why over 99% of the markers at the start of final map computation could be included in the final map, despite rigorous flips tests of marker order implemented in rh_tsp_map.
Second, the intermediate map was used to identify holes in cat-human and cat-dog map comparisons, so that the last batch of markers to be developed could be targeted to many of these holes. This substantial effort in global quality control explains why the coverage of the new map (96%) is so much higher than the coverage of the previous cat RH map (85%; [3]).
FISH Mapping
480 clones from three randomly selected plates of the domestic cat RPCI-86 BAC library were end-sequenced (SP6 end only) using described protocols [29]. These sequences were repeatmasked and queried against the feline 1.9x genome assembly and human and dog genome assemblies using BLAT to identify best hits and positions in each assembly (Supplemental Table 2). A collection of BAC clones spaced across all cat autosomes and the X chromosome were selected, grown, and DNA extracted using the Qiagen large construct procedure. BAC clone DNA was labeled with biotin and/or digoxigenin (Bio-Nick and Dig-Nick kits; Roche Molecular Biochemicals) and hybridized to domestic cat metaphase chromosomes. Images for a minimum of 30 metaphase spreads were captured and analyzed with a Zeiss Axioplan2 fluorescent microscope equipped with Cytovision/Genus V. 2.7 (Applied Imaging). Identification of cat chromosomes and assignment of markers to specific chromosome bands followed the nomenclature of O'Brien & Nash [30]. A subset of these probes (n=11) were also hybridized to metaphase spreads from individuals representing two divergent felid species: Profelis serval (serval) and Panthera uncia (snow leopard).
Comparative Analysis
For each domestic cat locus, physical positions for orthologous genes were obtained by using either BLAT [31] or discontiguous MegaBLAST (http://www.ncbi.nlm.nih.gov/blast/; [32]) searches to the feline, human and canine reference genome assemblies (felCat3, hg18 and canFam2, respectively). In a small number of cases, BLAT hits to one species (either human or dog) genome were not covered in the top alignment set in the second genome. In these cases, we identified the nearest corresponding stretch of orthology using the dog and human alignment nets of the UCSC Genome Browser. Conserved segments ordered (homologous synteny blocks) were defined by searching for runs of two or more uninterrupted markers on the same chromosome between two species. Inverted segments were defined by runs of three or more markers, each separated by 1 Mbp. Some out of place markers were expected due to mapping/genotyping errors or limitations of RH mapping resolution. These were assigned to the closest conserved segment if the intervening markers did not span more than a few Mb. Markers that were binned or placed with a LOD score <0.5 were not used in determining marker order, though they could be used to determine the extent of comparative coverage. The program AutoGRAPH [16] was used to visualize ordered and unordered conserved segments between cat-human and cat-dog comparisons.
Supplementary Material
Acknowledgments
We thank Keith Durkin, Julie Fronczek, Jan Janecka, William Nash, and Ashley Seabury for helpful advice and technical support. We thank Cynthia King for the serval blood sample. Jim Mullikin provided a list of candidate contigs for primer design. Marilyn Menotti-Raymond shared the marker positions on a new genetic map computed by AAS. This work was supported by funds from the Morris Animal Foundation (Grant D06FE-063) to WJM and the Winn Feline Foundation (Grant 06-026) to WJM, TR, and BPC. This research was supported in part by the intramural research program of the NIH, NLM (RA, AAS).
References
- 1.Menotti-Raymond M, David VA, Agarwala R, Schäffer AA, Stephens R, O'Brien SJ, Murphy WJ. Radiation hybrid mapping of 304 novel microsatellites in the domestic cat genome. Cytogenet Genome Res. 2003;102:272–276. doi: 10.1159/000075762. [DOI] [PubMed] [Google Scholar]
- 2.Menotti-Raymond M, David VA, Roelke ME, Chen ZQ, Menotti KA, Sun S, Schäffer AA, Tomlin JF, Agarwala R, O'Brien SJ, Murphy WJ. Second-generation integrated genetic linkage/radiation hybrid maps of the domestic cat (Felis catus) J Hered. 2003;94:95–106. doi: 10.1093/jhered/esg008. [DOI] [PubMed] [Google Scholar]
- 3.Murphy WJ, Davis B, David VA, Agarwala R, Schäffer AA, Pearks Wilkerson AJ, Neelam B, O'Brien SJ, Menotti-Raymond M. A 1.5-Mb-resolution radiation hybrid map of the cat genome and comparative analysis with the canine and human genomes. Genomics. 2007;89:189–196. doi: 10.1016/j.ygeno.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy WJ, Sun S, Chen Z, Yuhki N, Hirschmann D, Menotti-Raymond M, O'Brien SJ. A radiation hybrid map of the cat genome: implications for comparative mapping. Genome Res. 2000;10:691–702. doi: 10.1101/gr.10.5.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kehler JS, David VA, Schäffer AA, Bajema K, Eizirik E, Ryugo DK, Hannah SS, O'Brien SJ, Menotti-Raymond M. Four independent mutations in the feline fibroblast growth factor 5 gene determine the long-haired phenotype in domestic cats. J Hered. 2007;98:555–566. doi: 10.1093/jhered/esm072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyons LA, Imes DL, Rah HC, Grahn RA. Tyrosinase mutations associated with Siamese and Burmese patterns in the domestic cat (Felis catus) Anim Genet. 2005;36:119–126. doi: 10.1111/j.1365-2052.2005.01253.x. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt-Küntzel A, Eizirik E, O'Brien SJ, Menotti-Raymond M. Tyrosinase and tyrosinase related protein 1 alleles specify domestic cat coat color phenotypes of the albino and brown loci. J Hered. 2005;96:289–301. doi: 10.1093/jhered/esi066. [DOI] [PubMed] [Google Scholar]
- 8.Lyons LA, Biller DS, Erdman CA, Lipinski MJ, Young AE, Roe BA, Qin B, Grahn RA. Feline polycystic kidney disease mutation identified in PKD1. J Am Soc Nephrol. 2004;15:2548–2555. doi: 10.1097/01.ASN.0000141776.38527.BB. [DOI] [PubMed] [Google Scholar]
- 9.Meurs KM, Sanchez X, David RM, Bowles NE, Towbin JA, Reiser PJ, Kittleson JA, Munro MJ, Dryburgh K, Macdonald KA, Kittleson MD. A cardiac myosin binding protein C mutation in the Maine Coon cat with familial hypertrophic cardiomyopathy. Hum Mol Genet. 2005;14:3587–3593. doi: 10.1093/hmg/ddi386. [DOI] [PubMed] [Google Scholar]
- 10.Fyfe JC, Menotti-Raymond M, David VA, Brichta L, Schäffer AA, Agarwala R, Murphy WJ, Wedemeyer WJ, Gregory BL, Buzzell BG, Drummond MC, Wirth B, O'Brien SJ. An ~140-kb deletion associated with feline spinal muscular atrophy implies an essential LIX1 function for motor neuron survival. Genome Res. 2006;16:1084–1090. doi: 10.1101/gr.5268806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menotti-Raymond M, David VA, Schäffer AA, Stephens R, Wells D, Kumar-Singh R, O'Brien SJ, Narfström K. Mutation in CEP290 discovered for cat model of human retinal degeneration. J Hered. 2007;98:211–220. doi: 10.1093/jhered/esm019. [DOI] [PubMed] [Google Scholar]
- 12.Hitte C, Madeoy J, Kirkness EF, Priat C, Lorentzen TD, Senger F, Thomas D, Derrien T, Ramirez C, Scott C, Evanno G, Pullar B, Cadieu E, Oza V, Lourgant K, Jaffe DB, Tacher S, Dreano S, Berkova N, André C, Deloukas P, Fraser C, Lindblad-Toh K, Ostrander EA, Galibert F. Facilitating genome navigation: survey sequencing and dense radiation-hybrid gene mapping. Nat Rev Genet. 2005;6:643–648. doi: 10.1038/nrg1658. [DOI] [PubMed] [Google Scholar]
- 13.Murphy WJ, Larkin DM, Everts-van der Wind A, Bourque G, Tesler G, Auvil L, Beever JE, Chowdhary BP, Galibert F, Gatzke L, Hitte C, Meyers SN, Milan D, Ostrander EA, Pape G, Parker HG, Raudsepp T, Rogatcheva MB, Schook LB, Skow LC, Welge M, Womack JE, O'Brien S J, Pevzner PA, Lewin HA. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- 14.Pontius JU, Mullikin JC, Smith DR, Lindblad-Toh K, Gnerre S, Clamp M, Chang J, Stephens R, Neelam B, Volfovsky N, Schäffer AA, Agarwala R, Narfstrom K, Murphy WJ, Giger U, Roca AL, Antunes A, Menotti-Raymond M, Yuhki N, Pecon-Slattery J, Johnson WE, Bourque G, Tesler G, O'Brien SJ. Initial sequence and comparative analysis of the cat genome. Genome Res. 2007;17:1675–1689. doi: 10.1101/gr.6380007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menotti-Raymond M, David VA, Schäffer AA, Tomlin JF, Eizirik E, Philip C, Wells D, Pontius JU, Hannah SS, O'Brien SJ. A third-generation autosomal genetic linkage map of the domestic cat, Felis silvestris catus. Genomics. doi: 10.1016/j.ygeno.2008.11.004. (Submitted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derrien T, André C, Galibert F, Hitte C. AutoGRAPH: an interactive web server for automating and visualizing comparative genome maps. Bioinformatics. 2007;23:498–499. doi: 10.1093/bioinformatics/btl618. [DOI] [PubMed] [Google Scholar]
- 17.Everts-van der Wind A, Larkin DM, Green CA, Elliott JS, Olmstead CA, Chiu R, Schein JE, Marra MA, Womack JE, Lewin HA. A high-resolution whole-genome cattle-human comparative map reveals details of mammalian chromosome evolution. Proc Natl Acad Sci U S A. 2005;102:18526–18531. doi: 10.1073/pnas.0509285102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nash WG, Menninger JC, Wienberg J, Padilla-Nash HM, O'Brien SJ. The pattern of phylogenomic evolution of the Canidae. Cytogenet Cell Genet. 2001;95:210–224. doi: 10.1159/000059348. [DOI] [PubMed] [Google Scholar]
- 19.Yang F, Graphodatsky AS, O'Brien PC, Colabella A, Solanky N, Squire M, Sargan DR, Ferguson-Smith MA. Reciprocal chromosome painting illuminates the history of genome evolution of the domestic cat, dog and human. Chromosome Res. 2000;8:393–404. doi: 10.1023/a:1009210803123. [DOI] [PubMed] [Google Scholar]
- 20.Graphodatsky AS, Perelman PL, Sokolovskaya NV, Beklemisheva VR, Serdukova NA, Dobigny G, O'Brien SJ, Ferguson-Smith MA, Yang F. Phylogenomics of the dog and fox family (Canidae, Carnivora) revealed by chromosome painting. Chromosome Res. 2008;16:129–143. doi: 10.1007/s10577-007-1203-5. [DOI] [PubMed] [Google Scholar]
- 21.Johnson WE, Eizirik E, Pecon-Slattery J, Murphy WJ, Antunes A, Teeling E, O'Brien SJ. The late Miocene radiation of modern Felidae: a genetic assessment. Science. 2006;311:73–77. doi: 10.1126/science.1122277. [DOI] [PubMed] [Google Scholar]
- 22.Wurster-Hill DH, Centerwall WR. The interrelationships of chromosome banding patterns in canids, mustelids, hyena, and felids. Cytogenet Cell Genet. 1982;34:178–192. doi: 10.1159/000131806. [DOI] [PubMed] [Google Scholar]
- 23.Stanyon R, Rocchi M, Capozzi O, Roberto R, Misceo D, Ventura M, Cardone MF, Bigoni F, Archidiacono N. Primate chromosome evolution: ancestral karyotypes, marker order and neocentromeres. Chromosome Res. 2008;16:17–39. doi: 10.1007/s10577-007-1209-z. [DOI] [PubMed] [Google Scholar]
- 24.Eizirik E, Murphy WJ. Carnivora. In: Hedges SB, Kumar S S, editors. Assembling the Timetree of Life. Oxford: Oxford University Press; in press. [Google Scholar]
- 25.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 26.Agarwala R, Applegate DL, Maglott D, Schuler GD, Schäffer AA. A fast and scalable radiation hybrid map construction and integration strategy. Genome Res. 2000;10:350–364. doi: 10.1101/gr.10.3.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schäffer AA, Rice ES, Cook W, Agarwala R. rh_tsp_map 3.0: end-to-end radiation hybrid mapping with improved speed and quality control. Bioinformatics. 2007;23:1156–1158. doi: 10.1093/bioinformatics/btm077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Applegate DL. The traveling salesman problem : a computational study. Princeton: Princeton University Press; 2006. [Google Scholar]
- 29.Larkin DM, Everts-van der Wind A, Rebeiz M, Schweitzer PA, Bachman S, Green C, Wright CL, Campos EJ, Benson LD, Edwards J, Liu L, Osoegawa K, Womack JE, de Jong PJ, Lewin HA. A cattle-human comparative map built with cattle BAC-ends and human genome sequence. Genome Res. 2003;13:1966–1972. doi: 10.1101/gr.1560203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Brien SJ, Nash WG. Genetic mapping in mammals: chromosome map of domestic cat. Science. 1982;216:257–265. doi: 10.1126/science.7063884. [DOI] [PubMed] [Google Scholar]
- 31.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.